- 1National Key Laboratory of Wheat Improvement, College of Agronomy, Shandong Agricultural University, Tai’an, China
- 2Tai’an Academy of Agricultural Science, Tai’an, China
- 3Rizhao Academy of Agricultural Science, Rizhao, China
- 4Zibo Academy of Agricultural Science, Zibo, China
- 5Xinjiang Academy of Agricultural Sciences, Urumqi, China
Reducing plant height (PH) is one of the core contents of the “Green Revolution”, which began in the 1960s in wheat. A number of 27 reduced-height (Rht) genes have been identified and a great number of quantitative trait loci (QTLs) for PH have been mapped on all 21 chromosomes. Nonetheless, only several genes regulated PH have been cloned. In this study, we found the interval of QTL QPh-1B included an EST-SSR marker swes1079. According to the sequence of swes1079, we cloned the TaOSCA1.4 gene. We developed a CAPS marker to analyze the variation across a natural population. The result showed that the PH was significantly different between the two haplotypes of TaOSCA1.4–1B under most of the 12 environments and the average values of irrigation and rainfed conditions. This result further demonstrated that TaOSCA1.4 was associated with PH. Then, we validated the TaOSCA1.4 via RNAi technology. The average PHs of the wild-type (WT), RNAi lines 1 (Ri-1) and 2 (Ri-2) were 94.6, 83.6 and 79.2 cm, respectively, with significant differences between the WT and Ri-1 and Ri-2. This result indicated that the TaOSCA1.4 gene controls PH. TaOSCA1.4 is a constitutively expressed gene and its protein localizes to the cell membrane. TaOSCA1.4 gene is a member of the OSCA gene family, which regulates intracellular Ca2+ concentration. We hypothesized that knock down mutants of TaOSCA1.4 gene reduced regulatory ability of Ca2+, thus reducing the PH. Furthermore, the cell lengths of the knock down mutants are not significantly different than that of WT. We speculate that TaOSCA1.4 gene is not directly associated with gibberellin (GA), which should be a novel mechanism for a wheat Rht gene.
1 Introduction
Wheat (Triticum aestivum L.) is one of the most important food crops in the world. By the year 2050, the world population is expected to reach 9.3 billion, and the demand for bread wheat is consequently increasing (Dong et al., 2019). Moreover, wheat production faces many difficulties associated with climate, environment and cultivation. Plant height (PH) has a significant effect on plant morphogenesis and yield in the field. The application of dwarf genes in wheat breeding has enhanced the lodging resistance of wheat and greatly increased its yield (Hedden, 2003; Berry et al., 2007; Guedira et al., 2010).
Reducing PH is one of the core contents of the “Green Revolution”, which began in the 1960s in wheat (Peng et al., 1999; Velde et al., 2021). Many efforts have been made to identify genetic loci affecting PH, and 27 wheat reduced-height (Rht) genes have been discovered (Lu et al., 2015; Mo et al., 2018; Song et al., 2023a). The PH of wheat is a quantitatively inherited complex trait that is strongly influenced by interacting genetic and environmental factors. A great number of quantitative trait loci (QTLs) for PH have been mapped on all 21 chromosomes (Börner et al., 2002; Huang et al., 2003; Sourdille et al., 2003; Liu et al., 2011; Griffiffiffiths et al., 2012; Guan et al., 2018; Saini et al., 2022; Singh et al., 2022). Of these, some QTLs may be associated with Rht genes. Using ‘Jingdong 8/Aikang 58’ recombinant inbred lines (RILs), QPh.caas-4D was confirmed to be the RhtD1b locus, and QPh.caas-6A was found to be the Rht24 locus (Li et al., 2015a; Tian et al., 2017). Using ‘Nongda3338/Jingdong6’ doubled haploid (DH) population, QPh.cau-4B.2, QPh.cau-4D.1 and QPh.cau-2D.3 were found to be associated with the dwarfing genes Rht1, Rht2, and Rht8, respectively (Guan et al., 2018).
To date, only several genes for PH have been cloned, which the functions were verified using transgenic technologies. The green revolution genes Rht-B1 and Rht-D1 were the first cloned genes. The dwarfing alleles reduced stem growth by stabilizing the encoded DELLA proteins against degradation through interactions with the growth hormone gibberellin (GA) and its receptor (Peng et al., 1999). Rht-B1 and Rht-D1 have a variety of alleles, including Rht-B1c (Rht3), Rht-D1c (Rht10), Rht-B1e (Rht11), and Rht-B1p (Rht17) (Cao et al., 2009; Pearce et al., 2011; Li et al., 2012a, b, 2013; Bazhenov et al., 2015). The Rht8 gene on chromosome 2DS is considered more influential than the green revolution genes (Botwright et al., 2005; Tian et al., 2017). It was cloned as a ribonuclease h-like domain gene using map-based cloning. It controls the PH and spike length via GA biosynthesis (Chai et al., 2022; Xiong et al., 2022). The TaGA2ox-A14 gene was reported to be a candidate gene of Rht12 based on fine mapping, expression profiling and GA content analysis (Sun et al., 2019). GA2oxA13 (previously named GA2oxA14) was identified as the causal gene of Rht12 through the MutChromSeq approach (Buss et al., 2020). Fine mapping revealed that Rht13 was located on the long arm of chromosome 7B (Ellis et al., 2005). Rht-B13 encodes a nucleotide-binding site/leucine-rich repeat (NB-LRR) gene. Autoactivation of Rht13 leads to the upregulation of pathogenesis-related (PR) genes including class III peroxidases, which may catalyze the crosslinking of cell wall compounds to limit cell elongation and hence reduce height (Borrill et al., 2022). Rht18 gene was identified by mutagenizing the semidwarf durum cultivar Icaro and generating mutants with a range of tall phenotypes. The increased expression of GA2oxA9 in Rht18 results in a reduction of both bioactive GA content and plant height (Ford et al., 2018). Rht24 was isolated by map-based cloning and found to encode the GA2-oxidase TaGA2ox-A9 (Tian et al., 2017). Rht24b conferred higher expression of TaGA2ox-A9 in stems, leading to a reduction of bioactive GA in stems but an elevation in leaves at the jointing stage (Tian et al., 2022). Our group identified a candidate gene ATP-dependent DNA helicase (TaDHL-7B) for PH via QTL mapping and genome-wide association study (GWAS) methods. The knock out mutants of TaDHL-7B significantly reduced the PH without a yield penalty (Guo et al., 2022).
In this study, we mapped a QTL QPh-1B that included an EST-SSR marker swes1079. The full length of cDNA and gDNA were cloned according to the sequence of swes1079, which is the orthologous gene of nonselective hyperosmolality-gate calcium-permeable channel 1.4 (OSCA1.4). Then, we demonstrated that TaOSCA1.4 was associated with PH using a natural population. Finally, we validated that the TaOSCA1.4 gene controls PH via RNAi technology.
2 Materials and methods
2.1 Plant materials and trial design
QTL analysis was performed using a set of RILs derived from a cross of ‘Chuan35050 × Shannong 483’ (C35050 × SN483, C/SN-RILs, 131 lines) that constructed began in 1993 (Li et al., 2007). C35050 was a variety from the Southwestern Winter Wheat Region in China. SN483 was a variety of the Huang-huai Winter Wheat Region, China. SN483 was derived from ‘Ai-Meng-Niu’, one of the most well-known germplasms and backbone parents in Chinese wheat breeding programs. ‘Ai-Meng-Niu’ was developed by Shandong Agricultural University in 1980. More than 16 well-known cultivated varieties, which planted on more than 30 million hectares, have been developed from ‘Ai-Meng-Niu’. The C/SN-RILs and their parents were planted at four environments, Tai’an in 1999 (E1), 2000 (E2), 2001 (E3), and Yantai in 2001 (E4), with two replications (Li et al., 2007). The PH for each line and parent were determined by the sample of 10 plants in each replication.
A natural population of 134 current wheat varieties from the Huang-huai Winter Wheat Region of China was used to test the association between the marker and the PH. The field trials were located at the Experimental Station of Shandong Agricultural University (Tai’an, China), the Zibo Academy of Agricultural Sciences (Zibo, China), and the Xinjiang Academy of Agricultural Sciences (Urumqi, China) in the 2012–2013 and 2013–2014 growing seasons under irrigation and rainfed conditions. These environments were noted as Tai’an irrigation (TAI), Zibo irrigation (ZBI), Urumqi irrigation (URI), Tai’an rainfed (TAR), Zibo rainfed (ZBR) and Urumqi rainfed (URR) in 2013 (13) and 2014 (14), respectively. Seeds were sown on October 5–10, and plants were harvested on June 10–15 of the next year. Each plot consisted of three rows, which were 1.5 m long and spaced 25 cm apart, with two repetitions. Fifty seeds were planted in each row.
2.2 QTL analysis of the C/SN-RILs
A genetic map (Wang et al., 2011) was used to carry out the QTL analysis. The map consisted of 719 markers on 21 chromosomes, with a total length of 4,008.4 cM. The Windows QTL Cartographer 2.5 software (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm) was used to perform the QTL mapping. The parameters were as follows: composite interval mapping (CIM), “model 6 standard analysis”, a walk speed of 1 cM, “forward and backward” regression, up to five control markers, and a blocked window size of 10 cM. The threshold for declaring the presence of a significant QTL was defined by 1,000 permutations at p ≤ 0.05 (Churchill and Doerge, 1994). The LOD threshold for declaring a significant QTL was an LOD≥3.0 (He et al., 2020; Li et al., 2021; Xu et al., 2021).
2.3 DNA/RNA extraction
For the parents of the C/SN-RIL population and the variety ‘Fielder’ (wild-type, WT), total DNA was extracted using a DNA extraction kit (Tiangen, Beijing, China). The quality and concentration of DNA were determined using a NanoDrop 2000c spectrophotometer (Thermo, Wilmington, DE, USA). Total RNA was extracted using the RNAprep Pure Plant Kit (TIANGEN, Beijing, China). The RNA purity was checked using a nanophotometer spectrophotometer (IMPLEN, CA, USA). The RNA integrity was assessed using an RNA Nano 6000 Assay Kit for the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Reverse transcription into cDNA was performed according to the manufacturer’s protocol.
2.4 Amplification and chromosomal localization of candidate genes
The sequence of expressed sequence tag - simple sequence repeat (EST-SSR) marker swes1079 (GenBank ID: CJ623047.1) was used as a query sequence to screen the wheat EST database on GenBank (http://www.ncbi.nlm.nih.gov). We selected the highly overlapping EST sequence with those about swes1079 and used DNAMAN software (http://www.lynnon.com/) to get the splicing sequence. Using the splicing sequence as the starting sequence to continue search the wheat EST datebase and further extension. Then, a putative gene cDNA sequence was assembled. After that, we identified the Open Read Frame (ORF) using the ORF Finder program (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Primer pairs was designed using the Primer Premier 5.0 software to produce the cDNA and gDNA sequence based the ORF. The primers were as follows:
TaOSCA1.4-F: ATGGCGACGCTGCAGGA
TaOSCA1.4-R: TCAATGATCGACTCCAGGGT
Genome-specific primers were designed according to the difference sites of the three gDNA sequences of TaOSCA1.4, and the chromosomal localization was carried out using Chinese Spring nullisomic-tetrasomic lines. The primers were as follows:
TaOSCA1.4–1AF: CGCGTCTACTTCCCCAA
TaOSCA1.4–1AR: ATCTTTTCTCAGGTTCAGTACG
TaOSCA1.4–1BF: GAAGATGGGGTGCAGGTGA
TaOSCA1.4–1BR: GAATACCAAGGACCATTGGAG
TaOSCA1.4–1DF: TTCCGGTCAATGTCTCTGAT
TaOSCA1.4–1DR: ATCAAGTTCGCTAATTCTAGCC
2.5 Knockdown of the TaOSCA1.4 gene by RNAi
The following 94 bp nucleotide sequence, located in the seventh exon, was used as the interference fragment:
TTATGCCAGTTTCTTTTGTGACATTTGACTCAAGATGGGGTGCTGCTGTATGTGCACAGACACAACAGTCAAAGAATCCCACACAATGGCTGAC
The DNA was recovered using an agar-gel DNA recovery kit (Tiangen, Beijing, China). The BP reaction system consisted of 150 ng attB-PCR product, 150 ng pDONR Zeo vector, and 2 μl BP Clonase II enzyme mix in a 10 μl system was supplemented with RNase-free water. The mixture was incubated at 25°C for 2 h. The reaction was terminated by adding 1 μL of proteinase K and incubating at 37°C for 10 min. The reaction mixture was subsequently transformed into E. coli. Pick the clone and verify the sequence. The LR reaction mixture consisted of 150 ng of the entry-target fragment, 150 ng of the pANDA vector, 2 μl of the LR Clonase II enzyme mixture, and 10 μl of system supplemented with RNase-free water. The mixture was incubated at 25°C for 2 h. The reaction was terminated by adding 1 μL of proteinase K and incubating at 37°C for 10 min. The reaction mixture was subsequently transformed into E. coli. Pick the clone and verify the sequence. The plasmid was subsequently transformed into an Agrobacterium strain, after which the wheat genetic transformation experiment and wheat transgenic platform were established by the Crop Research Institute, Shandong Academy of Agricultural Sciences, China. The genome of transgenic wheat was extracted, gene expansion was determined by primers, and the bar gene and target gene were detected via agarose gel electrophoresis. Positive transgenic plants were detected via PCR and 1% agarose gel electrophoresis.
2.6 qRT−PCR, sequence characteristics, and bioinformatics analysis
The transcript levels of TaOSCA1.4 in Fielder organs were analyzed via qRT−PCR. The primers used were as follows:
TaOSCA1.4–1A-qRTF: CAAAGAGTACAGTAATGTGGCC,
TaOSCA1.4–1A-qRTR: AGATGTCGAATGGCTTGAC,
TaOSCA1.4–1B-qRTF: GAGATTACATTTCCTGGCTTCC,
TaOSCA1.4–1B-qRTF: CGGAAGAACTCATCAACTGC,
TaOSCA1.4–1D-qRTF: ACCACTATCTTGGTCAGCAGT
TaOSCA1.4–1D-qRTF: CAGGATGCCTTTCAAACTTC.
The first-strand cDNAs were synthesized from 2 µg of RNA per sample using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, Beijing, China). The qRT−PCR aplifications were performed as described by Zhao et al. (2020). Amplification of TaActin was used as an internal control for data normalization. The experiments were independently replicated three times under identical conditions. The complete alignment of multiple coding sequences and translations of nucleotides into amino acid sequences were performed using the DNAMAN program (version 5.2.2; Lynnon Biosoft, Canada). The prediction of the transmembrane structure was performed using DeepTMHMM (https://dtu.biolib.com/DeepTMHMM). Collinearity analysis was performed using the Triticeae-Gene Tribe (TGT, http://wheat.cau.edu.cn/TGT/).
2.7 Cytological analysis
For the Fielder and RNAi plants, the top of the first internode from the top of the main stem was taken, and 3 biological replicates were performed. Semithin sections were prepared by fixation, dehydration, paraffin impregnation and embedding, dewaxing, plant saffron stain, and then decolorization with an alcohol gradient (50%, 70% and 80%). After the plants were dyed with plant solid green dye solution and dehydrated with anhydrous ethanol, the samples were cleared in xylene and sealed with neutral gum. A Nikon Eclipse Ci-L photographic microscope was used for 400x imaging. After the imaging was completed, Image-Pro Plus 6.0 analysis software was used to count the number of cells, measure the area of cell tissue in the visual field, and calculate the number of cells per unit area = the number of cells/the area of cell tissue in the visual field.
2.8 Subcellular localization
According to the sequence structure characteristics of pBI121-GFP vector, a seamless cloning joint was designed in front of upstream and downstream primers, and a seamless cloning primer was designed:
YXB-3-F: gagagaacacgggggactctagaATGGCGACGCTGCAGG
YXB-3-R: cataagggacrgaccacccggGTGCTCGACTCCAT.
Synthetized Fielder cDNA was used as a template to amplify the cDNA sequences. The upstream primers anneal beginning at the start codon (ATG), and the downstream primers anneal adjacent to the stop codon (not including the stop codon). The reaction system consisted of 5 μl of buffer, 2 μl of cDNA, 0.4 μl of YXB-3-F, 0.4 μl of YXB-3-R, 1 μl of ApexHF HS DNA Polymerase CL, and 1.2 μl of RNase-free water. The PCR procedure was as follows: predenaturation at 94°C for 2 min, followed by 33 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 3 min, with a final extension at 72°C for 5 min.
The pBI121-GFP carrier plasmid was double digested with XmaI and XbaI. The enzyme digestion reactivity was as follows: 5 μl CutSmart Buffer, 33 μl ddH2O, 1 μl XmaI, 1 μl XbaI, and 10 μl pBI121-GFP. After overnight digestion at 37°C, carrier attachment was performed according to the instructions of the LightNingTM DNA Assembly Mix Plus Kit. The GFP fusion vector was subsequently transformed into Trans1-T1 competent E. coli cells, and positive cloning detection primers were designed based on the pBI121-GFP vector sequence:
GFP-F: GTTCCAACCACGTCTTCAAAGC;
GFP-R: CTCGCCGGACACGCTGAACT.
The positive clone bacteria were screened and sent to Sangon Biotech (Shanghai) Co., Ltd for sequencing to verify the correctness of the sequence.
The GFP fusion vector and blank plasmid pBI121-GFP were subsequently transformed into Super EHA105 receptive cells. The specific procedures used referenced the manufacturer’s instructions for the Super EHA105 receptive cells. GFP-F/R was used to detect positive clones, and the strains were preserved.
3 Results
3.1 Acquisition of a candidate gene TaOSCA1.4 for PH from a QTL
3.1.1 QTL location of QPh-1B
QTL analysis was conducted using C/SN-RIL populations, and a stable QTL for PH, QPh-1B, was located on chromosome 1B under two environments (E1 and E4) and average value (AV). This QTL included three molecular markers gdm28, gwm264 and swes1079 (Figure 1A). The markers gdm28 and gwm264 was located in noncoding regions. The swes1079 is a functional EST-SSR marker developed by our research group (Chen et al., 2005).
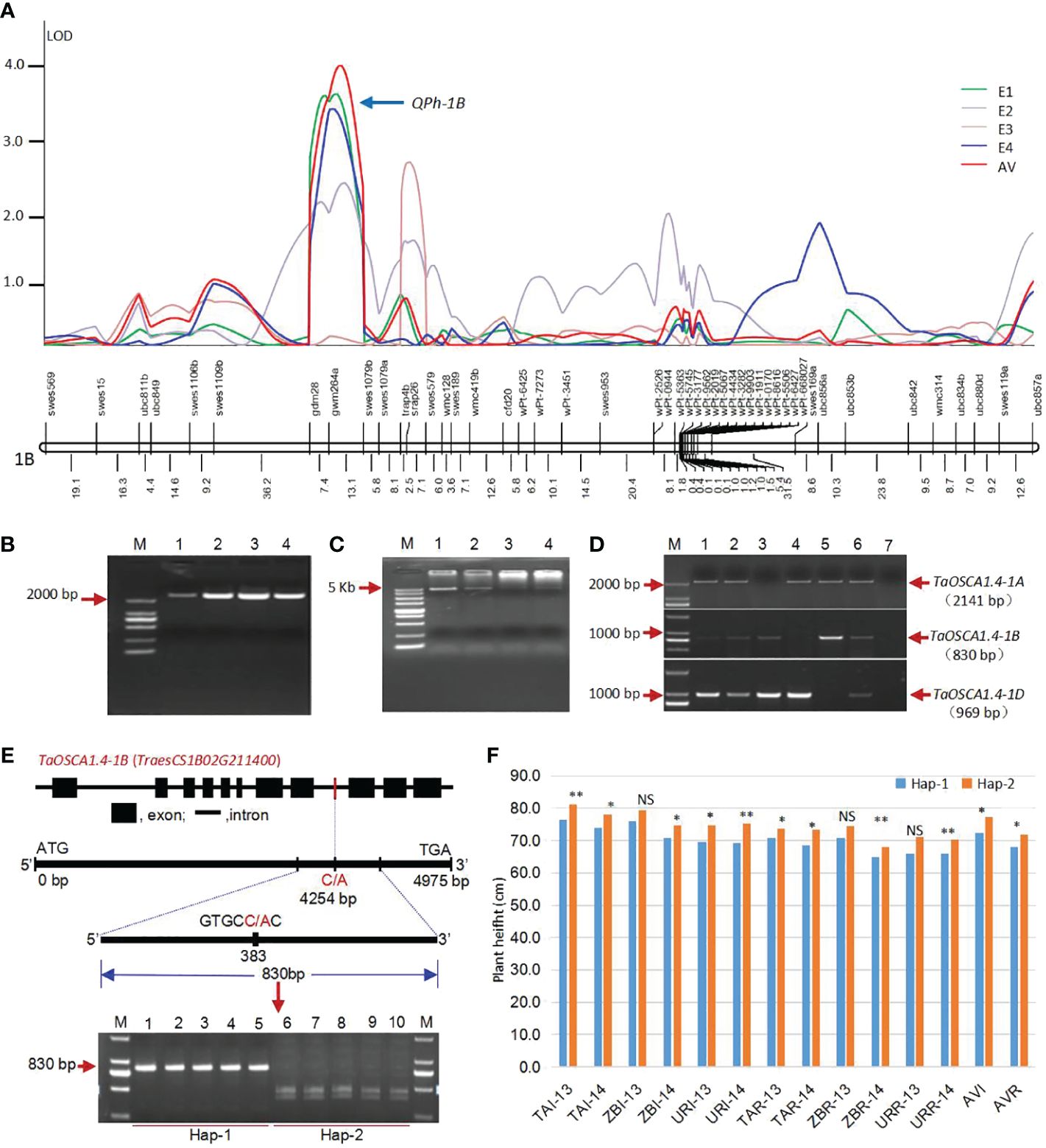
Figure 1 Acquisition of the candidate gene TaOSCA1.4 for PH. (A) QTL location for PH on chromosome 1B under four environments using a genetic map of DNA markers (Wang et al., 2011). (B) Full-length of cDNA sequences for TaOSCA1.4. M, Marker; 1, Wheat variety Tainong18; 2, Wheat variety Linmai6; 3, Wheat variety Chuan35050; 4, Wheat variety Shannong483. (C) Full-length of DNA sequences for TaOSCA1.4. M, Marker; 1, Wheat variety Tainong18; 2, Wheat variety Linmai6; 3, Wheat variety Chuan35050; 4, Wheat variety Shannong483. (D) Chromosome location of TaOSCA1.4 homologous genes using Chinese Spring nullisomic-tetrasomic lines. M, Marker; 1, Wheat variety Tainong18; 2, Wheat variety Linmai6; 3, N1AT1D (Nullisomic 1A-tetrasomic 1D); 4, N1BT1D; 5, N1DT1B; 6, Chinese Spring; 7, ddH2O. (E) Structure of the candidate gene TaOSCA1.4–1B (TraesCS1B02G211400) and the development of a cleaved amplified polymorphism sequence (CAPS) marker. (F) Differences of the PH in the natural population which scanned using the CAPS marker. *, P ≤ 0.05; **, P ≤ 0.01; NS, not significant.
3.1.2 cDNA and gDNA cloning and chromosomal location of TaOSCA1.4
According to the results of searching NCBI database, six wheat ESTs (GenBank ID: CJ623047.1, BJ248974.1, CJ730580, BQ238836.1, CJ726913.1, GH731252.1) were obtained and assembled into a putative cDNA sequence. The cDNA and corresponding gDNA sequences were amplified (Figures 1B, C). It is the previously cloned orthologous gene of nonselective hyperosmolality-gate calcium-permeable channel 1.4 (OSCA1.4) in rice, which was specifically downregulated by drought stress and upregulated by ABA treatment by expression profiles (Li et al., 2015b) (Supplementary Figure 1). Therefore, we named this gene as TaOSCA1.4. Based on the three genome-specific primer pairs, the TaOSCA1.4 genes were located on chromosomes 1A, 1B and 1D using the Chinese Spring nullisomic-tetrasomic lines (Figure 1D). For TaOSCA1.4–1A, the full length of gDNA is 5142 bp and cDNA is 2418 bp, encoding 805 amino acids (AAs). For TaOSCA1.4–1B, the full length of gDNA is 4975 bp and cDNA is 2406 bp, encoding 801 AAs. For TaOSCA1.4–1D, the full length of gDNA is 4961 bp and cDNA is 2409 bp, encoding 802 AAs. After the RefSeq v1.1 genome released in 2018 (IWGSC, 2018), we found that the TaOSCA1.4–1B gene is TraesCS1B02G211400. Its homologous genes on chromosomes 1A and 1D were TraesCS1A02G196800 and TraesCS1D02G200300, respectively. The DNA sequence similarities of TaOSCA1.4 in between A and B, A and D, and B and D were 84.29%, 77.18%, 79.56%, respectively. The AA sequence similarities between A and B, A and D, and B and D were 97.76%, 97.88%, 98.63%, respectively.
3.1.3 Associated analysis between TaOSCA1.4 and PH using a natural population
To further show the relationship between TaOSCA1.4 and PH, we developed a cleaved amplified polymorphism sequence (CAPS) marker from TaOSCA1.4–1B to analyze the variation across a natural population. The CAPS marker divided the natural population into two haplotypes, Hap-1 (830 bp) and Hap-2 (447 and 383 bp) (Figure 1E). PH was significantly different between the two haplotypes under most of the 12 environments and the average values of irrigation (AVI) and rainfed (AVR) conditions (Figure 1F). This result further demonstrated that TaOSCA1.4 was associated with PH.
3.2 Functional confirmation of the TaOSCA1.4 gene using RNAi technology
To obtain the conclusive evidence of the relationship between TaOSCA1.4 and PH, we performed RNA interference analysis of the TaOSCA1.4–1A, TaOSCA1.4–1B and TaOSCA1.4–1D genes using the RNAi system. We planted the WT and RNAi mutant plants of the T4 generation in the field. The qRT−PCR results showed that the expression levels of three homologous genes of TaOSCA1.4 in RNAi lines 1 (Ri-1) and 2 (Ri-2) were significantly decreased (Figure 2A; Supplementary Table 1). According to the results of field trials, the average PHs of the WT, Ri-1 and Ri-2 were 94.6, 83.6 and 79.2 cm, respectively, with significant differences between the WT and Ri-1 and Ri-2 (Figures 2B, C). Compared with those of the WT plants, the heights of Ri-1 and Ri-2 were reduced by 11.0 and 15.4 cm, respectively (Supplementary Table 2). These results indicated that the TaOSCA1.4 gene controls PH. The spike length (SL) was not significantly different between the WT and Ri-1 and Ri-2. The first, second, and third internode length from the top (ILT-1. ILT-2 and ILT-3) decreased by 6.8, 2.1, and 1.3 cm, respectively, in Ri-1; and decreased by 8.0, 3.8, and 1.8 cm, respectively, in Ri-2. These three internode lengths were significantly different from those in the WT (Figures 2D, E; Supplementary Table 2).
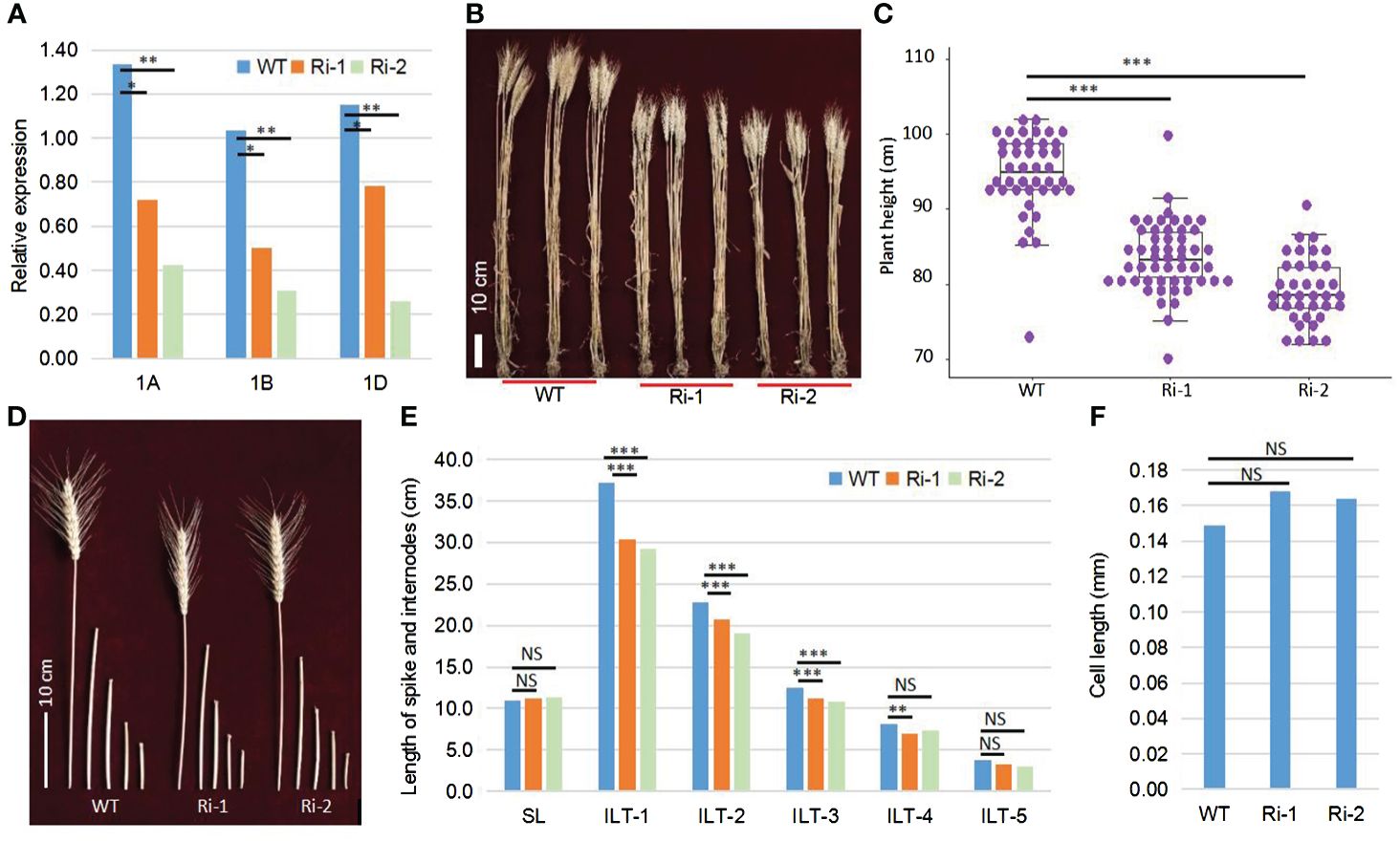
Figure 2 Function confirmation of the TaOSCA1.4 gene using the RNAi system. (A), Expression levels of three homologous genes TaOSCA1.4–1A (1A), TaOSCA1.4–1B (1B) and TaOSCA1.4–1D (1D) in WT (Fielder), Ri-1 and Ri-2 by qRT-PCR. (B) Picture of the plant for WT, Ri-1 and Ri-2. (C) Boxplot of the plant height for WT, Ri-1 and Ri-2. (D) Picture of spikes and internodes for WT, Ri-1 and Ri-2. (E) Difference for the length of spikes and internodes between WT and Ri-1 and Ri-2. (F), Cell length of the first internode from top for WT and Ri-1 and Ri-2. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; NS, not significant.
We observed longitudinal sections of WT and the RNAi lines using the uppermost internodes at the vegetative stage. The cell lengths of the WT, Ri-1 and Ri-2 were 0.149, 0.168 and 0.164 mm, respectively. The cell lengths between WT and Ri-1 and Ri-2 were not significantly different (Figure 2F; Supplementary Table 3).
3.3 Structure, expression, evolution and subcellular localization of TaOSCA1.4
For DNA sequences of TaOSCA1.4–1B between the parents (C35050 and SN483) of the RILs, there are two SNPs (at positions 78 and 4870 bp) and one InDel (6 bp deletion at position 4768 in SN483) in the exons; and are six additional SNPs (at positions 618, 783, 1029, 2817, 3859 and 4255 bp) and four InDels (deletion of 3 bp at position 1110, 8 bp at position 2459 and 1 bp at position 3297 SN 483; and deletion of 1 bp at position 2866 in C35050) in the introns (Supplementary Figure 2). For the AA sequences, two adjacent Q residues were deleted at positions 739 and 740 in SN 483, and an R substitution occurred in C35050 at position 759 (this residue is Q in SN 483) (Supplementary Figure 3). For TaOSCA1.4–1A and TaOSCA1.4–1D, the DNA sequences are not differences between the two parents.
The expression levels of TaOSCA1.4 in different tissues and at different periods determined by qRT-PCR showed that TaOSCA1.4 is a constitutively expressed gene. For TaOSCA1.4–1A, the highest expression is in the spike at the joining stage. For TaOSCA1.4–1B, the highest expression is in the stem at the joining stage. For TaOSCA1.4–1D, the highest expression is in the stem at milk grain stage (Figure 3A).
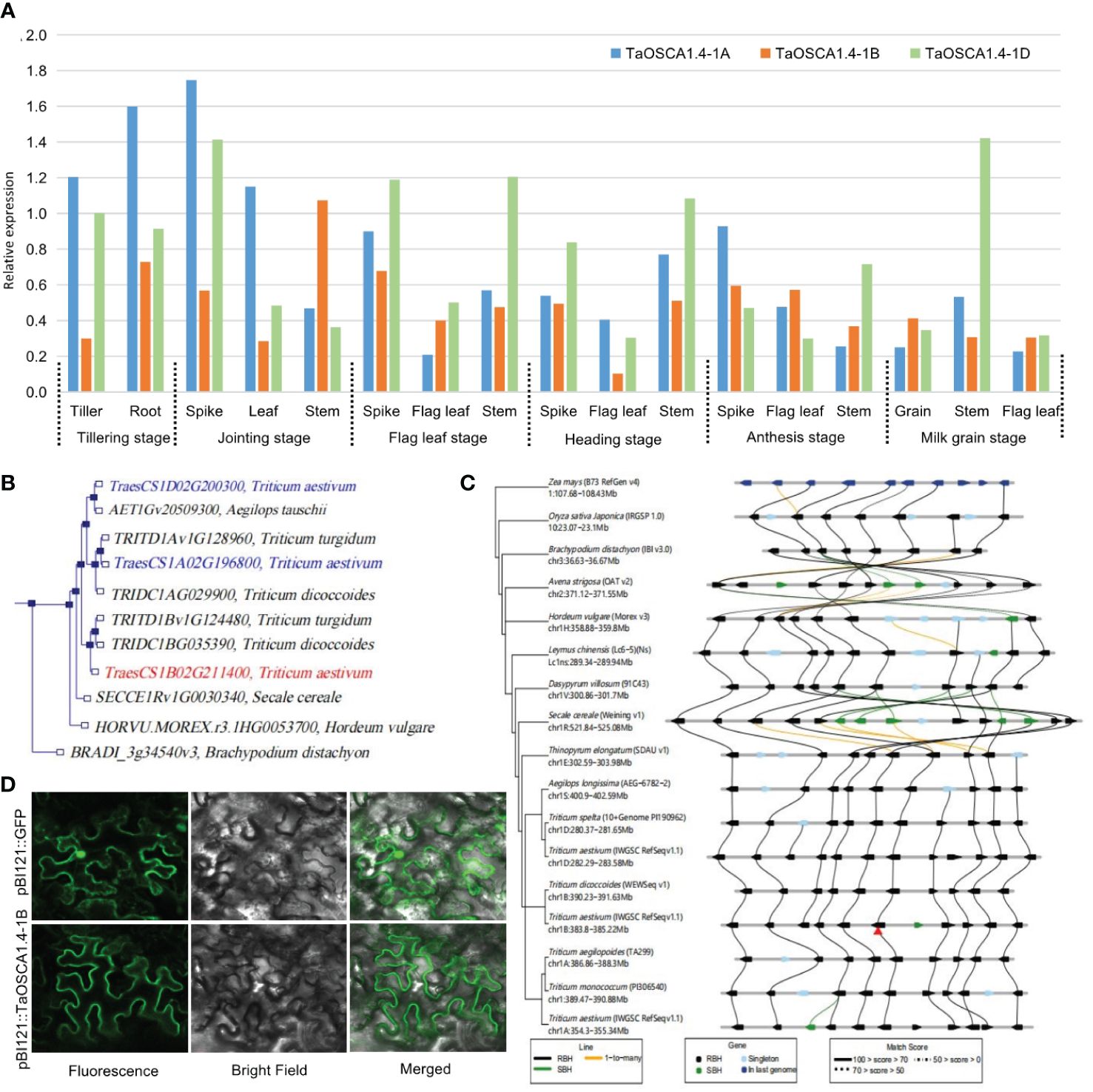
Figure 3 Characteristics of the TaOSCA1.4. (A) Expression levels in different tissues and periods for the three homologous genes TaOSCA1.4–1A, TaOSCA1.4–1B and TaOSCA1.4–1D in WT by qRT-PCR. (B) Phylogenetic tree for three homologous genes TaOSCA1.4–1A, TaOSCA1.4–1B and TaOSCA1.4–1D. (C) Collinearity analysis of TaOSCA1.4. (D) Result of subcellular localization for TaOSCA1.4–1B.
A phylogenetic tree generated using Ensemble (http://plants.ensembl.org/Triticum_aestivum/) revealed that TaOSCA1.4–1B (TraesCS1B02G211400), TaOSCA1.4–1A (TraesCS1A02G196800) and TaOSCA1.4–1D (TraesCS1D02G200300) were on different branches (Figure 3B). TaOSCA1.4–1B is closely related to TRITD1Bv1G124480 of Triticum turgidum and TRIDC1BG035390 of Triticum dicoccoides. TaOSCA1.4–1A is closely related to TRITD1Av1G128960 of Triticum turgidum and TRIDC1AG029900 of Triticum dicoccoides. TaOSCA1.4–1D is closely related to AET1Gv20509300 of Aegilops tauschii. A collinearity analysis using Triticeae-GeneTribe (http://wheat.cau.edu.cn/TGT/) revealed that the TaOSCA1.4–1B region was conserved between genomes A, B, D, and E, but an inversion occurred in Secale cereale (genome R) (Figure 3C).
Subcellular localization was predicted with a transmembrane domain hidden Markov model, which suggested that the TaOSCA1.4–1B protein is a typical transmembrane protein with eight transmembrane domains. According to the subcellular localization results, the green fluorescence presented by the protein encoded by the target gene TaOSCA1.4–1B had a strong signal only on the cell membrane, and no signal was detected in the rest of the cell. This result suggested that the TaOSCA1.4 protein localizes to the cell membrane (Figure 3D).
4 Discussion
Wheat is an allohexaploid species with a large and highly complex genome that severely restricts the isolation of genes via the classical method of map-based cloning. Only a few genes for PH, including Rht8 (Chai et al., 2022; Xiong et al., 2022), Rht13 (Borrill et al., 2022), Rht23 (Zhao et al., 2018) and Rht24 (Tian et al., 2022), have been cloned via map-based cloning. On the other hand, a great number of QTLs for PH have been identified (Saini et al., 2022; Singh et al., 2022). However, the intervals of these QTLs detected using a high-density genetic map of DNA markers are usually include dozens of genes or even more (Choulet et al., 2014). Due to these DNA markers being mainly located in noncoding regions, determining how to use QTLs to clone genes is a challenge. We previously developed EST-SSR markers (Chen et al., 2005), which represent a partial sequence of a gene. We mapped these markers to construct a genetic map (Wang et al., 2011). In this study, we found that the QTL QPh-1B included the EST-SSR marker swes1079, which provided the possibility for identifying the related gene TaOSCA1.4. We then proved this possibility by identifying candidate genes in populations with natural variation. Finally, we validated that TaOSCA1.4 regulate PH via RNAi technology. By the above process, we successfully produced a cloned gene from a QTL. This provides a method for gene cloning. Based on this idea, we constructed a map of unigenes using an RIL population and cloned seven yield component trait genes, TaIFABPL, TaDdRp, TaRLK, TaTD, TaTFC3, TaKMT and TaSPL17 (Zhang et al., 2024), a plant height gene TaDHL (Guo et al., 2022), and a quality gene XIP (Sun et al., 2022) from QTLs. This approach could be an effective method for isolating genes originally from wheat.
The studies of OSCA gene family were focused the expression profiling under osmotic stress in Arabidopsis, rice, wheat and barley, etc. (Li et al., 2015b; Zhai et al., 2020; Tong et al., 2021; Kaur et al., 2022; She et al., 2022). In wheat, Tong et al. (2021) identified 42 OSCA members. The expression profiles showed that 15 TaOSCA members responded to PEG treatment, while TaOSCA12/-39 responded simultaneously to PEG and ABA. Kaur et al. (2022) also identified 42 OSCA genes. They found that a total of 31 TaOSCA genes displayed differential expression in response to the drought, heat, and combined heat–drought stress conditions, and 29 TaOSCA genes showed differential expression in response to salt stress. No report was found that OSCA genes regulating the plant height. Using RNAi technology, we obtained the conclusive evidence that TaOSCA1.4 regulate PH. So, we regard that the TaOSCA1.4 is novel reduced-height (Rht) gene in wheat.
OSCA genes regulate intracellular Ca2+ concentration and play a key role in sensing exogenous and endogenous osmotic changes and regulating plant growth and development. In Arabidopsis, the osca1 mutant exhibited a decreased calcium (Ca2+) concentration under sorbitol or mannitol treatment, inhibited stomatal closure and root growth, indicating that OSCA1 is a plant osmoreceptor (Yuan et al., 2014). In rice, OsOSCA1.1 mediates OICIcyt and SICIcyt in roots, which are critical for stomatal closure, plant survival, and gene expression in shoots, in response to hyperosmolality and the salt stress treatment of roots (Han et al., 2022). When plants are subjected to external stress, Ca2+ flows from high concentrations of extracellular and intracellular calcium reservoirs into the cytoplasm, resulting in an instantaneous increase in the concentration of Ca2+ in the cytoplasm and the generation of calcium signals. The calcium signal is then recognized by three major Ca2+ receptor proteins: calmodulin (CaM) (Dong and Zhang, 2001), calcineurin B-like protein (CBLS) (Desikan et al., 2006; Shabala et al., 2018), and calcium-dependent protein kinases (CDPKs) (Cheng et al., 2002; Chen et al., 2013) and triggers a series of physiological activities. We hypothesized that knock down mutants of TaOSCA1.4 gene reduced regulatory ability of Ca2+, thus reducing the plant height.
Among the cloned Rht genes, Rht-B1 and Rht-D1 (Peng et al., 1999), Rht8 (Chai et al., 2022; Xiong et al., 2022), Rht12 (Buss et al., 2020), Rht18 (Ford et al., 2018), Rht24 (Tian et al., 2022) are involved in GA signaling or metabolism pathway. Rht13, an NB-LRR gene, is not directly involved in GA pathway, and the same mutation in the tomato protein I-2, which impeded ATP hydrolysis and promoted an ATP-bound active form of the protein (Borrill et al., 2022). ZnF-B acts as a brassinosteroid (BR) signaling activator to facilitate proteasomal destruction of the BR signaling repressor TaBKI1, and loss of ZnF stabilizes TaBKI1 to block BR signaling transduction. The deletion of ZnF-B induced the semi-dwarf trait in the absence of the Rht-B1b and Rht-D1b alleles through BR perception (Song et al., 2023b). We previous cloned an Rht gene TaDHL is not directly hormone pathway (Guo et al., 2022). In this study, TaOSCA1.4 gene regulates intracellular Ca2+ concentration. Furthermore, the cell lengths of the knock down mutants is not significantly different than that of WT. We speculate that TaOSCA1.4 gene is not directly associated with GA, which should be a novel mechanism for a wheat Rht gene.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
SL: Writing – original draft, Writing – review & editing, Supervision. GL: Writing – original draft, Data curation, Investigation, Methodology. XJ: Writing – original draft, Data curation, Investigation, Methodology. HW: Investigation, Methodology, Writing – original draft. YW: Data curation, Writing – original draft. QW: Data curation, Software, Writing – original draft. HMW: Investigation, Writing – original draft, Data curation. FJ: Investigation, Writing – original draft. YM: Investigation, Writing – original draft. YA: Methodology, Writing – original draft. MZ: Data curation, Software, Writing – original draft. YG: Writing – original draft, Investigation, Methodology.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2017YFD0100600) and the Mount Taishan Leading Talent Project of Shandong Province, China.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1381243/full#supplementary-material.
References
Bazhenov, M. S., Divashuk, M. G., Amagai, Y., Watanabe, N., Karlov, G. I. (2015). Isolation of the dwarfifing Rht-B1p (Rht17) gene from wheat and the development of an allele-specifific PCR marker. Mol. Breed. 35, 213. doi: 10.1007/s11032–015-0407–1
Berry, P. M., Sylvester-Bradley, R., Berry, S. (2007). Ideotype design for lodging-resistant wheat. Euphytica 154, 165–179. doi: 10.1007/s10681–006-9284–3
Börner, A., Schumann, A. E., Furste, A., Coster, H., Leithold, B., Roder, M. S., et al. (2002). Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.). Theor. Appl. Genet. 105, 921–936. doi: 10.1007/s00122–002-0994–1
Borrill, P., Mago, R., Xu, T. Y., Ford, B., Williams, S. J., Derkx, A., et al. (2022). An autoactive NB-LRR gene causes Rht13 dwarfism in wheat. P. Natl. Acad. Sci. U.S.A. 119, e2209875119. doi: 10.1073/pnas.2209875119
Botwright, T. L., Rebetzke, G. J., Condon, A. G., Richard, R. A. (2005). Inflfluence of the gibberellin-sensitive Rht8 dwarfifing gene on leaf epidermal cell dimensions and early vigour in wheat (Triticum aestivum L.). Ann. Bot. 95, 631–639. doi: 10.1093/aob/mci069
Buss, W., Ford, B. A., Foo, E., Schnippenkoetter, W., Borrill, P., Brooks, B., et al. (2020). Overgrowth mutants determine the causal role of GA2oxidaseA13 in Rht12 dwarfism of wheat. J. Exp. Bot. 47, 7171–7178. doi: 10.1093/jxb/eraa443
Cao, W. G., Somers, D. J., Fedak, G. (2009). A molecular marker closely linked to the region of Rht-Dlc and Ms2 genes in common wheat (Triticum aestivum). Genome 52, 95–99. doi: 10.1139/G08–097
Chai, L. L., Xin, M. M., Dong, C. Q., Chen, Z. Y., Zhai, H. J., Zhuang, J. H., et al. (2022). A natural variation in Ribonuclease H-like gene underlies Rht8 to confer “Green Revolution” trait in wheat. Mol. Plant 15, 377–380. doi: 10.1016/j.molp.2022.01.013
Chen, H. M., Li, L. Z., Wei, X. Y., Li, S. S., Lei, T. D., Hu, H. Z. (2005). Development, chromosome location and genetic mapping of EST-SSR markers in wheat. Chin. Sci. Bull. 50, 2328–2336. doi: 10.1007/BF03183744
Chen, J., Xue, B., Xia, X., Yin, W. L. (2013). A novel calcium-dependent protein kinase gene from Populus euphratica, confers both drought and cold stress tolerance. Biochem. Bioph. Res. Co. 441, 630–636. doi: 10.1016/j.bbrc.2013.10.103
Cheng, S. H., Willmann, M. R., Chen, H. C., Sheen, J. (2002). Calcium signaling through protein kinases. The Arabidopsis calcium dependent protein kinase gene family. Plant Physiol. 129, 469–485. doi: 10.1104/pp.005645
Choulet, F., Alberti, A., Theil, S., Glover, N., Barbe, V., Daron, J., et al. (2014). Structural and functional partitioning of bread wheat chromosome 3B. Science 345, 1249721. doi: 10.1126/science.1249721
Churchill, G. A., Doerge, R. W. (1994). Empirical threshold values for quantitative trait mapping. Genetics 138, 963–971. doi: 10.1093/genetics/138.3.963
Desikan, R., Last, K., Harrett-Williams, R., Tagliavia, C., Harter, K., Hooley, R., et al. (2006). Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J. 47, 907–916. doi: 10.1111/j.1365–313X.2006.02842.x
Dong, H. X., Yan, S. L., Liu, J., Liu, P., Sun, J. Q. (2019). TaCOLD1 defines a new regulator of plant height in bread wheat. Plant Biotechnol. J. 17, 687–699. doi: 10.1111/pbi.13008
Dong, X. J., Zhang, X. S. (2001). Some observations of the adaptations of sandy shrubs to the arid environment in the Mu Us Sandland: leaf water relations and anatomic features. J. Arid Environ. 48, 41–48. doi: 10.1006/jare.2000.0700
Ellis, M. H., Rebetzke, G. J., Azanza, F., Richards, R. A., Spielmeyer, W. (2005). Molecular mapping of gibberellin-responsive dwarfing genes in bread wheat. Theor. Appl. Genet. 111, 423–430. doi: 10.1007/s00122–005-2008–6
Ford, B. A., Foo, E., Sharwood, R., Karafiatova, M., Vrána, J., MacMillan, C., et al. (2018). Rht18 semidwarfism in wheat is due to increased GA 2-oxidaseA9 expression and reduced GA content. Plant Physiol. 177, 168–180. doi: 10.1104/pp.18.00023
Griffiffiffiths, S., Simmonds, J., Leverington, M., Wang, Y. K., Fish, L., Sayers, L., et al. (2012). Meta-QTL analysis of the genetic control of crop height in elite European winter wheat germplasm. Mole. Breeding. 29, 159–171. doi: 10.1007/s11032–010-9534-x
Guan, P., Lu, L., Jia, L. J., Kabir, M. R., Zhang, J. B., Lan, T. Y., et al. (2018). Global QTL analysis identifies genomic regions on chromosomes 4A and 4B harboring stable loci for yield-related traits across different environments in wheat (Triticum aestivum L.). Front. Plant Sci. 9, 529. doi: 10.1007/s11032–010-9534-x
Guedira, M., Brown-Guedira, G., Van, S. D., Sneller, C., Souza, E., Marshall, D. (2010). Distribution of Rht genes in modern and historic winter wheat cultivars from the eastern and central USA. Crop Sci. 50, 1811–1822. doi: 10.2135/cropsci2009.10.0626
Guo, B. J., Jin, X. M., Chen, J. C., Xu, H. Y., Zhang, M. X., Lu, X., et al. (2022). ATP-dependent DNA helicase (TaDHL), a novel reduced-height (Rht) gene in wheat. Genes. 13, 979. doi: 10.3390/genes13060979
Han, Y., Wang, Y. X., Zhai, Y. J., Wen, Z. H., Liu, J., Xi, C., et al. (2022). OsOSCA1.1 mediates hyperosmolality and salt stress sensing in Oryza sativa. Biol. 11, 678. doi: 10.3390/biology11050678
He, X. Y., Kabir, M. R., Roy, K. K., Anwar, M. B., Xu, K. J., Marza, F., et al. (2020). QTL mapping for field resistance to wheat blast in the Caninde#1/Alondra population. Theor. Appl. Genet. 133, 2673–2683. doi: 10.1007/s00122–020-03624-x
Hedden, P. (2003). The genes of the green revolution. Trends Genet. 19, 5–9. doi: 10.1016/S0168–9525(02)00009–4
Huang, X. Q., Cöster, M. H., Ganal, M. W., Roder, M. S. (2003). Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat (Triticum aestivum L.). Theor. Appl. Genet. 106, 1379–1389. doi: 10.1007/s00122–002-1179–7
IWGSC (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191
Kaur, A., Sharma, A., Madhu, Dixit, S., Singh, K., Upadhyay, S. K. (2022). OSCA genes in bread wheat: molecular characterization, expression profiling, and interaction analyses indicated their diverse roles during development and stress response. Int. J. Mol. Sci. 23, 14867. doi: 10.3390/ijms232314867
Li, T., Deng, G. B., Su, Y., Yang, Z., Tang, Y., Wang, J. H., et al. (2021). Identification and validation of two major QTLs for spike compactness and length in bread wheat (Triticum aestivum L.) showing pleiotropic effects on yield-related traits. Theor. Appl. Genet. 134, 3625–3641. doi: 10.1007/s00122–021-03918–8
Li, S. S., Jia, J. Z., Wei, X. Y., Zhang, X. C., Li, L. Z., Chen, H. M., et al. (2007). A intervarietal genetic map and QTL analysis for yield traits in wheat. Mol. Breed. 20, 167–178. doi: 10.1007/s11032–007-9080–3
Li, X. M., Xia, X. C., Xiao, Y. G., He, Z. H., Wang, D. S., Trethowan, R., et al. (2015a). QTL mapping for plant height and yield components in common wheat under water-limited and full irrigation environments. Crop Pasture Sci. 66, 660–670. doi: 10.1071/CP14236
Li, Y. Y., Xiao, J. H., Wu, J. J., Duan, J. L., Liu, Y., Ye, X. G., et al. (2012b). A tandem segmental duplication (TSD) in green revolution gene Rht-D1b region underlies plant height variation. New Phytol. 196, 282–291. doi: 10.1111/j.1469–8137.2012.04243.x
Li, A. X., Yang, W. L., Guo, X. L., Liu, D. C., Sun, J. Z., Zhang, A. M. (2012a). Isolation of a gibberellin-insensitive dwarfing gene, Rht-B1e, and development of an allele-specific PCR marker. Mol. Breeding. 30, 1443–1451. doi: 10.1007/s11032–012-9730-y
Li, A. X., Yang, W. L., Lou, X. Y., Liu, D. C., Sun, J. Z., Guo, X. L., et al. (2013). Novel natural allelic variations at the Rht-1 loci in wheat. J. Integr. Plant Biol. 55, 1026–1037. doi: 10.1111/jipb.12103
Li, Y. S., Yuan, F., Wen, Z. H., Li, Y. H., Wang, F., Zhu, T., et al. (2015b). Genome-wide survey and expression analysis of the OSCA gene family in rice. BMC Plant Boil. 15, 1–13. doi: 10.1186/s12870–015-0653–8
Liu, G., Xu, S. B., Ni, Z. F., Xie, C. J., Qin, D. D., Li, J., et al. (2011). Molecular dissection of plant height QTLs using recombinant inbred lines from hybrids between common wheat (Triticum aestivum L.) and spelt wheat (Triticum spelta L.). Chin. Sci. Bull. 56, 1897. doi: 10.1007/s11434–011-4506-z
Lu, Y., Xing, L. P., Xing, S. J., Hu, P., Cui, C. F., Zhang, M. Y., et al. (2015). Characterization of a putative new semi-dominant reduced height gene, Rht_NM9, in wheat (Triticum aestivum L.). J. Genet. Genomics 42, 685–698. doi: 10.1016/j.jgg.2015.08.007
Mo, Y., Vanzetti, L. S., Hale, L., Spagnolo, E. J., Guidobaldi, F., Ai-Oboudo, J., et al. (2018). Identification and characterization of Rht25, a locus on chromosome arm 6AS affecting wheat plant height, heading time, and spike development. Theor. Appl. Genet. 131, 2021–2035. doi: 10.1007/s00122–018-3130–6
Pearce, S., Saville, R., Vaughan, S. P., Chandler, P. M., Wilhelm, E. P., Sparks, C. A., et al. (2011). Molecular characterization of Rht1 dwarfifing genes in hexaploid wheat. Plant Physiol. 157, 1820–1831. doi: 10.1104/pp.111.183657
Peng, J., Richards, D. E., Hartley, N. M., Murphy, G., Devos, K. M., Flintham, J., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature. 400, 256–261. doi: 10.1038/22307
Saini, D. K., Srivastava, P., Pal, N., Gupta, P. K. (2022). Meta-QTLs, ortho-meta-QTLs and candidate genes for grain yield and associated traits in wheat (Triticum aestivum L.). Theor. Appl. Genet. 135, 1049–1081. doi: 10.1007/s00122–021-04018–3
Shabala, S. (2018). Linking ploidy level with salinity tolerance: NADPH-dependent ‘ROS-Ca2+ hub’ in the spotlight. J. Exp. Bot. 70, 1063–1067. doi: 10.1093/jxb/erz042
She, K., Pan, W. Q., Yan, Y., Shi, T. R., Chu, Y. Q., Cheng, Y., et al. (2022). Genome-wide identification, evolution and expressional analysis of OSCA gene family in barley (Hordeum vulgare L.). Int. J. Mol. Sci. 23, 13027. doi: 10.3390/ijms232113027
Singh, K., Saini, D. K., Saripalli, G., Batra, R., Gautam, T., Singh, R., et al. (2022). WheatQTLdb V2. 0: A supplement to the database for wheat QTL. Mol. Breed. 42, 56. doi: 10.1007/s11032–022-01329–1
Song, J., Li, L., Liu, B. Y., Dong, Y. C., Dong, Y., Li, F. J., et al. (2023a). Fine mapping of reduced height locus RHT26 in common wheat. Theor. Appl. Genet. 136, 62. doi: 10.1007/s00122–023-04331-z
Song, L., Liu, J., Cao, B. L., Liu, B., Zhang, X. P., Chen, Z. Y., et al. (2023b). Reducing brassinosteroid signalling enhances grain yield in semi-dwarf wheat. Nature 617, 118–124. doi: 10.1038/s41586–023-06023–6
Sourdille, P., Cadalen, T., Guyomarc′h, H., Snape, J. W., Perretant, M. R., Charmet, G., et al. (2003). An update of the Courtot/Chinese Spring intervarietal molecular marker linkage map for the QTL detection of agronomic traits in wheat. Theor. Appl. Genet. 106, 530–538. doi: 10.1007/s00122–002-1044–8
Sun, L. H., Yang, W. L., Li, Y. F., Shan, Q. Q., Ye, X. B., Wang, D. Z., et al. (2019). A wheat dominant dwarfing line with Rht12, which reduces stem cell length and affects gibberellic acid synthesis, is a 5AL terminal deletion line. Plant J. 97, 887–900. doi: 10.1111/tpj.14168
Sun, Z. J., Zhang, M. X., An, Y. R., Han, X., Guo, B. J., Lv, G. D., et al. (2022). CRISPR/Cas9-mediated disruption of xylanase inhibitor protein (XIP) gene improved the dough quality of common wheat. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.811668
Tian, X. L., Wen, W. E., Xie, L., Fu, L. P., Xu, D. A., Fu, C., et al. (2017). Molecular mapping of reduced plant height gene Rht24 in bread wheat. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01379
Tian, X. L., Xia, X. C., Xu, D. A., Liu, Y. Q., Xie, L., Hassan, M. A., et al. (2022). Rht24b, an ancient variation of TaGA2ox-A9, reduces plant height without yield penalty in wheat. New Phytol. 233, 783–750. doi: 10.1111/nph.17808
Tong, K., Wu, X. Y., He, L., Qiu, S. Y., Liu, S., Cai, L. N., et al. (2021). Genome-wide identification and expression profile of OSCA gene family members in Triticum aestivum L. Int. J. Mol. Sci. 23, 469. doi: 10.3390/ijms23010469
Velde, K. V. D., Thomas, S. G., Heyse, F., Kaspar, R., Straeten, D. V. D., Rohde, A. (2021). N-terminal truncated RHT-1 proteins generated by translational reinitiation cause semi-dwarfing of wheat green revolution alleles. Mol. Plant 14, 1–9. doi: 10.1016/j.molp.2021.01.002
Wang, Y. Y., Sun, X. Y., Zhao, Y., Kong, F. M., Guo, Y., Zhang, G. Z., et al. (2011). Enrichment of a common wheat genetic map and QTL mapping of genes for fatty acid content in grain. Plant Sci. 181, 65–75. doi: 10.1016/j.plantsci.2011.03.020
Xiong, H. C., Zhou., C. Y., Fu, M. Y., Guo, H. J., Xie, Y. D., Zhao, L. S., et al. (2022). Cloning and functional characterization of Rht8, a ‘‘Green Revolution’’ replacement gene in wheat. Mol. Plant 15, 373–376. doi: 10.1016/j.molp.2022.01.014
Xu, Y. F., La, G. X., Fatima, N., Liu, Z. H., Zhang, L. R., Zhao, L. F., et al. (2021). Precise mapping of QTL for Hessian fly resistance in the hard winter wheat cultivar’Overland’. Theor. Appl. Genet. 134, 3951–3962. doi: 10.1007/s00122–021-03940-w
Yuan, F., Yang, H. M., Xue, Y., Kong, D. D., Ye, R., Li, C. J., et al. (2014). OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature. 514, 44367–44371. doi: 10.1038/nature13593
Zhai, Y., Wen, Z. H., Han, Y., Zhuo, W. Q., Wang, F., Xi., C., et al. (2020). Heterogeneous expression of plasma-membrane-localised OsOSCA1. 4 complements osmotic sensing based on hyperosmolality and salt stress in Arabidopsis osca1 mutant. Cell calcium 91, 102261. doi: 10.1016/j.ceca.2020.102261
Zhang, M. X., Han, X., Wang, H., Sun, J. S., Guo, B. J., Gao, M. G., et al. (2024). Efficient cloning of genes for wheat yield component traits from QTLs via sequencing of the RIL population. bioRxiv. 02. doi: 10.1101/2024.02.22.574000
Zhao, B., Wu, T. T., Ma, S. S., Jiang, D. J., Bie, X. M., Sui, N., et al. (2020). TaD27-B gene controls the tiller number in hexaploid wheat. Plant Biotechnol. J, 18, 513–525. doi: 10.1111/pbi.13220
Keywords: wheat, plant height (PH), reduced-height (Rht) gene, gene cloning, RNA interference (RNAi), quantitative trait locus (QTL), nonselective hyperosmolality-gate calcium-permeable channel 1.4 (TaOSCA1.4)
Citation: Lv G, Jin X, Wang H, Wang Y, Wu Q, Wu H, Jiang F, Ma Y, An Y, Zhang M, Guo Y and Li S (2024) Cloning a novel reduced-height (Rht) gene TaOSCA1.4 from a QTL in wheat. Front. Plant Sci. 15:1381243. doi: 10.3389/fpls.2024.1381243
Received: 03 February 2024; Accepted: 01 May 2024;
Published: 16 May 2024.
Edited by:
Fangpu Han, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Dinesh Kumar Saini, Texas Tech University, United StatesJindong Liu, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Lv, Jin, Wang, Wang, Wu, Wu, Jiang, Ma, An, Zhang, Guo and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sishen Li, c3NsaUBzZGF1LmVkdS5jbg==; Ying Guo, Z3VveWluZzcyOUAxMjYuY29t; Mingxia Zhang, emhhbmdtaW5neGlhMDUwNkAxNjMuY29t
†These authors have contributed equally to this work
 Guangde Lv
Guangde Lv Xuemei Jin1,3†
Xuemei Jin1,3† Hui Wang
Hui Wang Qun Wu
Qun Wu Yanrong An
Yanrong An Ying Guo
Ying Guo Sishen Li
Sishen Li