- 1School of Life Science and Engineering, Southwest University of Science and Technology, Mianyang, China
- 2Maize Research Institute, Southwest University, Chongqing, China
- 3Engineering Research Center of South Upland Agriculture, Ministry of Education, Chongqing, China
- 4Chongqing Key Laboratory of Plant Resource Conservation and Germplasm Innovation, Integrative Science Center of Germplasm Creation in Western China (Chongqing) Science City, School of Life Sciences, Southwest University, Chongqing, China
- 5Key Laboratory of Eco-environments of Three Gorges Reservoir Region, Ministry of Education, School of Life Sciences, Southwest University, Chongqing, China
Isoflavonoids, the major secondary metabolites within the flavonoid biosynthetic pathway, play important roles in plant defense and exhibit free radical scavenging properties in mammals. Recent advancements in understanding the synthesis, transport, and regulation of isoflavonoids have identified their biosynthetic pathways as promising targets for metabolic engineering, offering potential benefits such as enhanced plant resistance, improved biomass, and restoration of soil fertility. This review provides an overview of recent breakthroughs in isoflavonoid biosynthesis, encompassing key enzymes in the biosynthetic pathway, transporters influencing their subcellular localization, molecular mechanisms regulating the metabolic pathway (including transcriptional and post-transcriptional regulation, as well as epigenetic modifications). Metabolic engineering strategies aimed at boosting isoflavonoid content in both leguminous and non-leguminous plants. Additionally, we discuss emerging technologies and resources for precise isoflavonoid regulation. This comprehensive review primarily focuses on model plants and crops, offering insights for more effective and sustainable metabolic engineering approaches to enhance nutritional quality and stress tolerance.
Introduction
Isoflavonoids, a major subclass of flavonoids, play pivotal roles in plant growth, development, and stress defense, with recognized implications for human health (Dixon, 1999; Veitch, 2013; Wang et al., 2022). In plant-microbe interactions, isoflavonoids function as signal molecules perceived by microorganisms (Biala-Leonhard et al., 2021). They act as phytoalexins, inhibiting the growth and reproduction of bacteria and fungi, fortifying plant defense against pathogens. Simultaneously, isoflavonoids attract rhizobia to legume root nodules, establishing symbiotic relationships that enhance plant growth, reduce nitrogen fertilizer use, and improve soil fertility (Abd-Alla et al., 2023). Furthermore, the synthesis and accumulation of isoflavonoids in plants are induced by various biotic and abiotic stresses, enhancing overall adaptability (Dixon and Paiva, 1995). Due to their structural and functional resemblance to endogenous estrogen, isoflavonoids and their derivatives are recognized as phytoestrogens, finding applications in functional foods, nutraceuticals, and medicine for disease prevention and treatment (Dixon, 2004; Griffiths et al., 2014; Yu et al., 2021).
With a profound understanding of the significance of isoflavonoid compounds, current research has increasingly emphasized the biosynthesis, transport, and regulation processes in plants. This emphasis has significantly propelled advancements in metabolic engineering and de novo synthesis studies. While isoflavonoids are predominantly found in leguminous plants, their scarcity in other plants has led researchers to explore metabolic engineering and synthetic biology as promising strategies. These approaches address the challenges posed by the low abundance and difficulty in obtaining large quantities through conventional crop-based manufacturing or chemical synthesis. Prior reviews have provided detailed reviews of the synthesis, regulation, and metabolic engineering of isoflavones (Yu and McGonigle, 2005; Sohn et al., 2021). However, as time progresses, the functions of an increasing number of genes are being reported, and the application of newer technologies are spurring additional breakthroughs in the study of isoflavonoid synthesis and metabolic engineering. This review will focus on a wider variety of isoflavonoid compounds.
The chemical structures and functions of various isoflavonoids
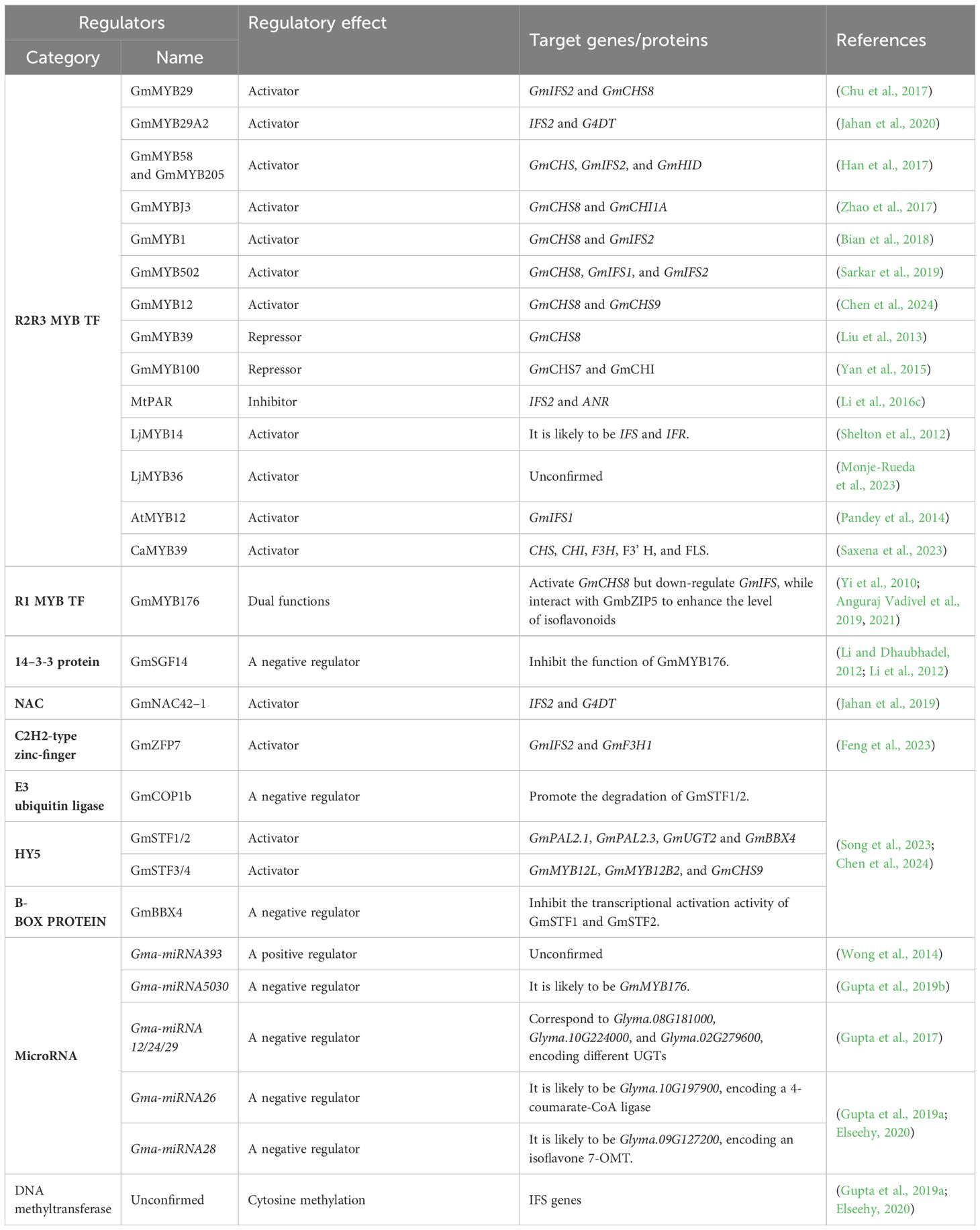
Isoflavonoids, along with flavonoids, lignins, coumarins, and stilbenes, all belong to the class of phenylpropanoid compounds, which are crucial for plant adaptation to terrestrial environments, enabling plants to withstand gravity and offering protection against UV radiation, desiccation, pathogens, and herbivores (Muro-Villanueva et al., 2019; Yadav et al., 2020; Dong and Lin, 2021). Isoflavonoids, also known as 3-phenylchromanes, feature a C6-C3-C6 backbone with the phenyl B-ring attached to position 3 of the heterocyclic pyran ring (the C ring). Based on structural characteristics, they are classified into isoflavones, isoflavans, pterocarpans, rotenoids, coumestans, and derivatives formed through methylation, glycosylation, and acylation (Figure 1).
Figure 1 Structures and classification of isoflavonoids. This basic C6-C3-C6 structure consists of a benzene ring (A-ring), a pyran ring (C-ring), and a phenyl group (B-ring).
Isoflavones, as shown in Figure 2, such as genistein, daidzein, biochanin A, formononetin, daidzin, ononin, etc., are found in various plants, with particularly high amounts in legumes like soybeans, chickpeas (Cicer arietinum), fava beans (Vicia faba L.), alfalfa (Medicago sativa), and medicinal plants like red clover (Trifolium pratense), Glycyrrhiza uralensis, and Astragalus membranaceus (Paiva et al., 1991; Kaufman et al., 1997; Tsao et al., 2006; Gao et al., 2015; Zhang et al., 2018). Almost all isoflavones exhibit antibacterial effects, aiding plants in resisting pathogens. Among them, genistein and daidzein serve as broad-spectrum antimicrobial agents, inhibiting the growth and reproduction of bacteria and fungi, thereby enhancing plant defense against pathogens (Dixon and Ferreira, 2002; Araya-Cloutier et al., 2017).
Figure 2 Biosynthetic pathways of isoflavonoids. PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; IFS, isoflavone synthase; HI4′OMT, 2-hydroxyisoflavanone 4′-O-methyltransferase; HID/IFD, 2-hydroxyisoflavanone dehydratase; IOMT, isoflavonoid O-methyltransferase; UGT, glycosyltransferases; IMaT, isoflavone glucoside malonyltransferase; I2′H, isoflavone 2′-hydroxylase; IFR, isoflavone reductase; VR, vestitone reductase; PTS, pterocarpan synthase; PTR, Pterocarpan reductase; I3S, isoflav-3-ene synthase ; P6aH, pterocarpan 6a-hydroxylase; G2DT, glycinol 2-dimethylallyl transferase; G4DT, glycinol 4-dimethylallyl transferase; GS, glyceollin synthase.
Pterocarpans, as shown in Figure 2, alongside isoflavones, are prevalent isoflavonoids found in various plants, including bitucarpin (A, B) and erybraedin C from Bituminaria morisiana and Bituminaria. bituminosa, medicarpin from Medicago truncatula and alfalfa, glycinol, and glyceollin from soybean, among others (Higgins, 1972; Paiva et al., 1991; Dixon, 1999; Pistelli et al., 2003; Naoumkina et al., 2007; Yoneyama et al., 2016; Sukumaran et al., 2018). In M. truncatula, medicarpin acts as an inducible “phytoalexin,” accumulating during defense responses (Naoumkina et al., 2007; Farag et al., 2008). In soybean, glycinol is formed through the cyclization of daidzein and serves as a precursor to glyceollin. Glyceollin is synthesized via the prenylation of glycinol in response to pathogen infection, enhancing resistance against soybean pathogens like Phytophthora sojae, Diaporthe phaseolorum var. meridionales, Macrophomina phaseolina, and Sclerotinia sclerotiorum (Akashi et al., 2009; Lygin et al., 2010; Yoneyama et al., 2016; Sukumaran et al., 2018). In B. bituminosa, the accumulation of pterocarpans (bitucarpin A and erybraedin C) significantly increases with treatment using arbuscular mycorrhizal fungi, offering a viable method for medicinal pterocarpan production (Pistelli et al., 2003, 2017).
Coumestrol, the principal coumestan compound, is derived from the conversion of unstable precursors isoflav-3-enes (Figure 2). It functions as both a phytoalexin and phytoestrogen, contributing to the well-being of both plants and humans (Martin and Dewick, 1980; Boue et al., 2000; Yuk et al., 2011; Ha et al., 2019; Uchida et al., 2020; Mun et al., 2021). For instance, the accumulation of coumestrol and the expression of genes involved in soybean coumestrol biosynthesis increase during leaf development, senescence, pathogen infection, nodulation, various stress treatments, and hormone treatments such as salicylic acid (SA), methyl jasmonate (MeJA), and ethylene (ET). This suggests that coumestans, including coumestrol, play crucial roles in plant development and stress defense (Boue et al., 2000; Lee et al., 2012; Tripathi et al., 2016; Ha et al., 2019; Mun et al., 2021; Ohta et al., 2021; Lee et al., 2022). Other coumestan compounds, like wedelolactone and psoralidin, exhibit similar functions. However, the key enzymes and regulators involved in their biosynthesis still require further exploration (Perez Rojo et al., 2023; Wang et al., 2023b).
Isoflavans, a specific subclass of isoflavonoids, are 5-deoxyisoflavonoids first discovered in legumes (Figure 2). This category includes glabridin from licorice (Glycyrrhiza glabra), vestitol, and sativan from Lotus japonicus, alfalfa, and others (Dewick and Martin, 1979; Hayashi et al., 1996; Shimada et al., 2000; Akashi et al., 2006; Veitch, 2009, 2013). The synthesis of vestitol in Lotus sp. is notably induced by ultraviolet (UV) radiation and the attachment of the root parasitic plant Striga hermonthica. Pterocarpan reductase (PTR) catalyzes this process, utilizing medicarpin as a substrate (Akashi et al., 2006; Ueda and Sugimoto, 2010; Kaducova et al., 2019; Kaducová et al., 2022). Subsequently, vestitol undergoes methylation by O-methyltransferase to produce sativan, another phytoalexin that accumulates significantly upon fungal pathogen infection in alfalfa and Lotus sp (Ingham and Harborne, 1976; Dewick and Martin, 1979; Saunders and O’neill, 2004; Trush et al., 2023). Glabridin, a prenylated isoflavan found in licorice plants, exhibits notable fungicidal activity against various phytopathogenic fungi such as Fusarium graminearum, Sclerotinia sclerotiorum, and Corynospora cassiicola (Simmler et al., 2013; Yang et al., 2021; Li et al., 2021a).
Rotenone and its derivative deguelin, the most common rotenoid compounds in Derris sp., are extensively employed as insecticides due to their capability to inhibit electron transport in the respiratory chain (Anzeveno, 1979; Hail and Lotan, 2004; Preston et al., 2017; Russell et al., 2020);. Additionally, rotenone exhibits anticancer activity in vitro but its application is limited due to neurotoxicity associated with its ability to cross the blood-brain barrier (Cannon et al., 2009; Deng et al., 2010). However, hydroxylated rotenoids are more hydrophilic and less likely to readily cross the blood-brain barrier, potentially acting as cell-selective killers to inhibit the proliferation of cancer cells (Naguib et al., 2018; Zhang et al., 2022a).
Indeed, the diversity and complexity of isoflavonoid compounds are largely due to chemical modifications such as methylation, glycosylation, and acylation. Differences in modification sites, types, and numbers of modifying groups can lead to changes in the physicochemical properties and biological activities of these derivatives. Therefore, studying the structure-function relationships of isoflavonoid derivatives and the related enzymes will contribute significantly to the efficient targeted synthesis of complex isoflavonoids in synthetic biology.
Biosynthesis of isoflavonoids in plants
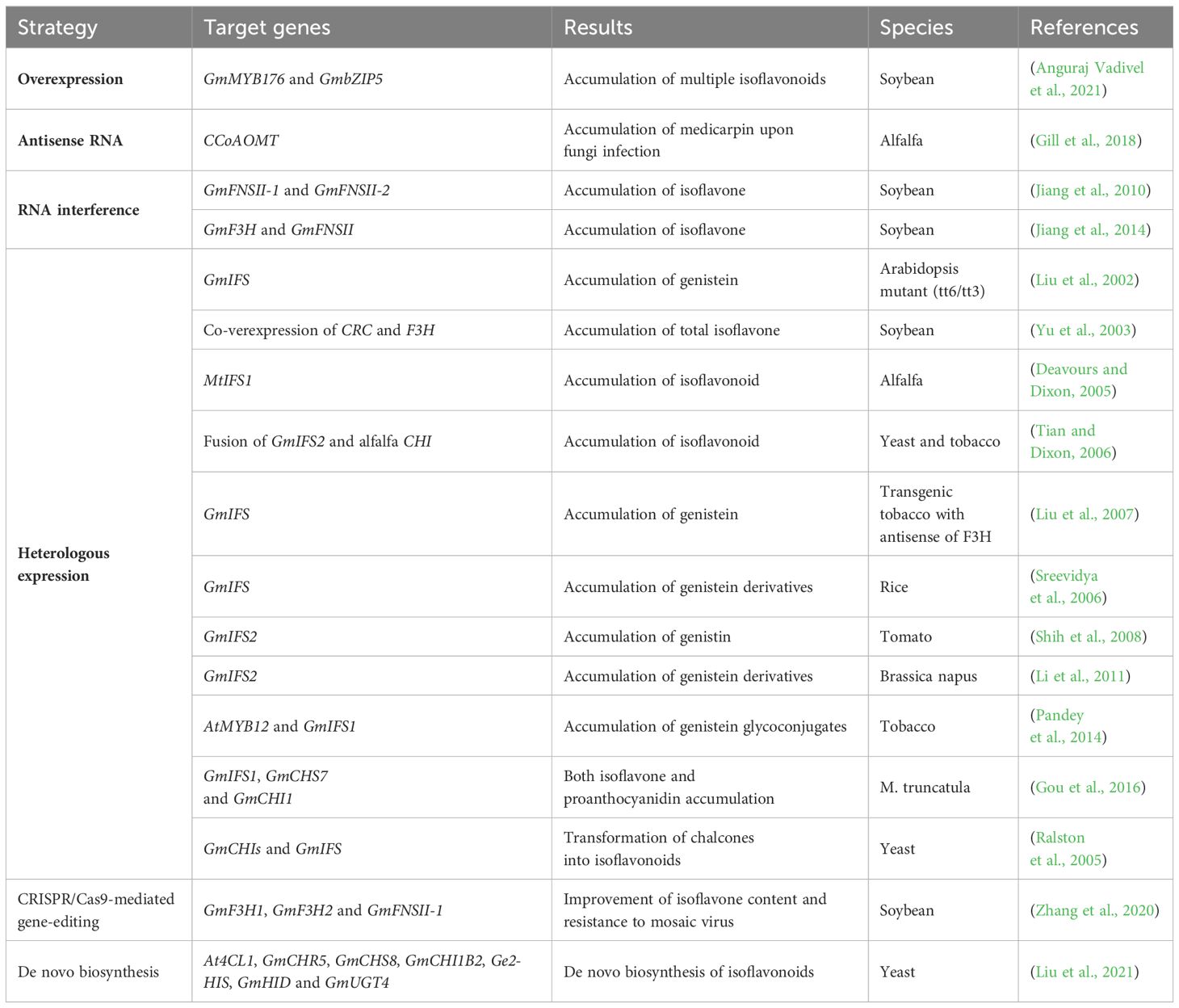
The biosynthesis of isoflavonoids begins with L‐phenylalanine and forms compounds with a classical C6-C3 core, which is called phenylpropanoid because of containing a benzene ring (phenyl) and a propionic acid side chain. The chemical intermediate p-coumaroyl-CoA catalyzed by 4-coumarate: CoA ligase (4CL) is a highly activated molecule, which acts as a key hub to determine the metabolic flow in response to developmental or environmental signals (Figure 2). One is to form molecules with a C6-C3 core structures, such as lignin monomers (including paracoumaryl alcohol, coniferyl alcohol, and sinapyl alcohol), and the other is to combine with Malonyl-CoA to form molecules with a C6-C3-C6 core, such as flavonoids (2-phenylchromen-4-one and derivatives) and isoflavonoids (3-phenylchromen-4-one and derivatives) (Figure 2).
Isoflavonoids can be divided into “phytoanticipins,” which are pre-existing compounds, including genistein, daidzein, formononetin, lupin, etc., or inducible “phytoalexins,” which are produced upon infection by pathogens or insects, such as medicarpin, pisatin, sativan, vestitol, glyceollin, and coumestrol (Dixon and Ferreira, 2002). While predominantly found in leguminous plants, isoflavonoids have been identified in various plant species beyond the legume family, including iridaceous, compositous, moraceous plants, barley (Hordeum vulgare), and others (Reynaud et al., 2005; Mackova et al., 2006; Darbour et al., 2007; Abderamane et al., 2011; Picmanov et al., 2012; Polturak et al., 2023). A recent study in wheat unveiled a pathogen-induced biosynthetic gene cluster with seven enzymes, including chalcone synthase (CHS1), non-induced chalcone isomerase (CHI), two cytochrome P450s, and three O-methyltransferases (OMTs), resulting in the production of a wheat-specific isoflavonoid named triticein (Polturak et al., 2023). This breakthrough provides new opportunities for isoflavonoid synthesis in non-legume crop plants.
As shown in Figure 2, the isoflavonoid biosynthetic pathway initiates with isoflavone synthase (IFS), catalyzing the migration of the B-ring from the 2- to the 3-position of the C ring, yielding 2-hydroxyisoflavanone (Jung et al., 2000; Subramanian et al., 2006). Subsequent dehydration by 2‐hydroxyisoflavanone dehydratase (HID/IFD) results in isoflavones like genistein and daidzein (Hakamatsuka et al., 1998; Akashi et al., 2005; Shimamura et al., 2007). In soybeans (Glycine max), another HID/IFD converts 4′-methoxylated 2-hydroxyisoflavanones into formononetin or biochanin A (Akashi et al., 2005). Diverse isoflavonoid compounds result from multiple O-methyltransferases (OMTs) methylating at the C3′-,4′, 3-, or 7-hydroxyl group. Various derivatives, such as 3’-methoxy-puerarin, 4′-O-methylated isoflavonoid formononetin, and 7-O-methylated isoflavone isoformononetin, are formed (He and Dixon, 2000; Liu and Dixon, 2001; Zubieta et al., 2001; Akashi et al., 2003; Liu et al., 2006; Li et al., 2016a). Subsequently, methylated formononetin transforms into medicarpin through isoflavone reductase (IFR), vestitone reductase (VR), and pterocarpan synthase (PTS) (Paiva et al., 1991; Oommen et al., 1994; Dixon et al., 1995; Guo and Paiva, 1995; Nakamura et al., 1999; Uchida et al., 2017). Glycosyltransferases (UGTs) further contribute to derivatives synthesis, e.g., 7-O-glucosyltransferase converting daidzein to daidzin and 8-C-glucosyltransferase forming puerarin from daidzein (He and Dixon, 2000; Noguchi et al., 2007; Li et al., 2014; Funaki et al., 2015; Wang et al., 2017). Acyltransferases add acyl groups (prenyl or acetyl) to produce prenylated or acetylated isoflavonoids, enhancing antimicrobial activities (Dixon, 1999; Mukne et al., 2011; Shen et al., 2012; Ahmad et al., 2017; Araya-Cloutier et al., 2017; Mouffouk et al., 2017; Araya-Cloutier et al., 2018; Kalli et al., 2021).
A large number of isoflavonoid compounds are produced in plants during specific developmental stages or in response to environmental signals, which are accompanied by re-distribution of substrates and induced phytoalexin synthesis (Piślewska et al., 2002; Gutierrez-Gonzalez et al., 2009, 2010). In many times, the encoding genes of these enzymes only function after being induced by speific factors (Shimada et al., 2007; Shelton et al., 2012). Therefore, key enzymes in the isoflavonoid biosynthetic pathway, especially those involved in the modification of isoflavonoids such as glycosyltransferases and methyltransferases for the modification of -OH groups at C-7, C-5, or C-4’, glycosidases for hydrolysis glycosylated isoflavonoids, and acyltransferase for malonylation or acetylation of isoflavonoids are still not fully characterized, and their functions require further elucidation (Figures 2, 3). To comprehensively explore these biosynthetic pathways and regulatory mechanisms, germplasm resources of legumes can be collected or established, and integrated approaches such as genome resequencing techniques, transcriptomics, and metabolomics can be applied to identify the genes responsible for synthesizing key enzymes.
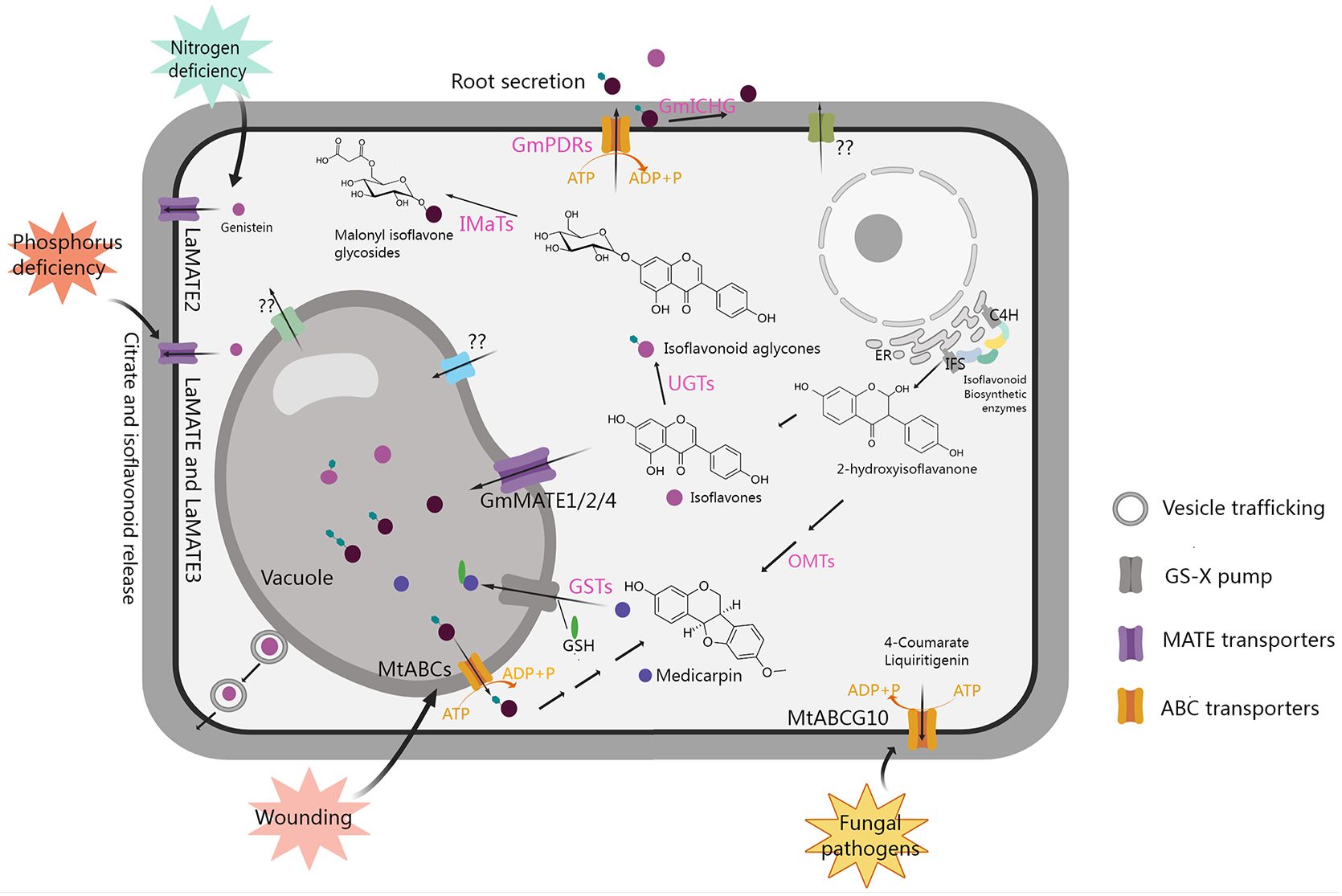
Figure 3 Transport and accumulation of isoflavonoids in legumes. GS-X pump, glutathione conjugates pump; PDRs, pleiotropic drug resistance transporters belonging to the ABCG subfamily; GSTs, glutathione S-transferases; ER, endoplasmic reticulum. The figure was created with Medpeer (https://image.medpeer.cn/).
Transport of isoflavonoids in legumes
Extensive studies have indicated that glycosylation and acylation modifications of isoflavonoids not only enhance transporter affinity and efficiency, but also improve water solubility and stability, facilitating storage in vacuoles or extracellular secretion (Dixon, 1999; Zhao et al., 2011; Le Roy et al., 2016; Ahmad et al., 2017; Ku et al., 2020; Ahmad et al., 2021). Additionally, isoflavonoids can act as signaling molecules by translocating to the nucleus and influencing the expression of downstream genes (Naoumkina et al., 2007; Naoumkina and Dixon, 2008; Yu et al., 2008; Zhao and Dixon, 2010). As shown in Figure 3, various transport mechanisms, including vesicle trafficking, ATP-binding cassette (ABC) transporters, multidrug and toxic compound extrusion (MATE) transporters, and glutathione S-transferase (GST), located in the vacuolar membrane or plasma membrane, have been reported to play roles in the transport and distribution of isoflavonoids (Li et al., 1997; Naoumkina et al., 2007; Naoumkina and Dixon, 2008; Zhao and Dixon, 2010; Zhao et al., 2011; Banasiak et al., 2013; Zhao, 2015; Biala-Leonhard et al., 2021).
In leguminous plants, isoflavonoids released by the roots serve as signaling molecules, influencing rhizobia to promote symbiosis. Identified transporters play crucial roles in the release of isoflavonoids from roots. For instance, in Lupinus albus, phosphorus deficiency induces the expression of LaMATE, LaMATE2, and LaMATE3, as well as the release of genistein from roots. The individual silencing of these genes reduces genistein release. Among them, LaMATE2 has been confirmed to transport genistein using yeast microsomal membrane vesicles, while the isoflavone transport functions of LaMATE and LaMATE3 require further exploration (Figure 3). Additionally, LaMATE2 is induced by nitrogen deficiency, and its silencing also reduces the number of nodules, indicating that LaMATE2 facilitates the export of isoflavonoids into the rhizosphere, fostering nodule formation (Biala-Leonhard et al., 2021; Zhou et al., 2021). In M. truncatula, a plasma membrane-localized ABC transporter MtABCG10, which belongs to the G subfamily, is involved in medicarpin precursor transport, modulates the distribution of 4-coumarate and liquiritigenin during nodule formation and plant defense against fungal pathogens (Jasinski et al., 2009; Banasiak et al., 2013; Biala et al., 2017). LjABCG1, which is homologous to the MtABCG10 in L. japonicus, is associated with pathogenesis rather than symbiosis, but the substrate remains unclear and requires further investigation (Sugiyama et al., 2015). In soybean, 13 pleiotropic drug resistance genes (PDRs) encoding ABCG transporters were found to be expressed in roots. One of them was confirmed to participate in membrane transport of genistein, daidzein, and other isoflavonoid aglycones, initiating legume-rhizobium symbiosis formation (Sugiyama et al., 2007, 2008). Additionally, the β-glucosidase GmICHG in soybean cell walls hydrolyzes genistin, contributing to isoflavone secretion in roots (Suzuki et al., 2006).
Transporters also play a crucial role in directing isoflavonoids into the vacuole for storage. In soybean, GmMATE1, GmMATE2, and GmMATE4, localized in the vacuolar membrane, are implicated in transporting isoflavones for accumulation through yeast uptake assay. In addition, it has also been confirmed that the total isoflavone content in seeds is significantly increased due to the overexpression of GmMATE1 in transgenic soybean, while it is significantly decreased due to GmMATE1 mutation (Ng et al., 2021; Ku et al., 2022). The utilization of maize GST-mediated conjugation with GSH (medicarpin-GS) significantly enhances the uptake of isoflavonoids by vacuoles (Li et al., 1997). When plants are exposed to stress-induced signals, they utilize store isoflavonoids to synthesize phytoalexin to enhance their resistance. For example, wound signal MeJA induces a decrease in the content of isoflavone glycosides, while it can also induce the accumulation of medicarpin in alfalfa, which is accompanied by the up-regulation the expression of multiple ABC transporter genes and β-glucosidase genes, suggesting that ABC transporters may be involved in transport of isoflavone glycosides from the vacuole to the cytoplasm for the synthesis of medicarpin, thereby increasing plant resistance to wounding (Naoumkina et al., 2007). However, the specific ABC transporters involved in this process remain unclear and require further exploration.
As shown in Figure 3, the transporters that have been reported so far are mainly involved in the transport of isoflavones such as genistein, while there is scarce research on proteins that transport other types of isoflavonoids. Among them, although MtABCs may be involved in the transport of medicarpin, it is still unclear which ABC protein is responsible. There are also some transporter proteins whose encoding gene expression levels can affect the change in isoflavone content, but whether they have a direct transport function needs further confirmation. Overall, a more in-depth understanding of the transporters involved in their conveyance, accumulation, and extracellular secretion is essential for unraveling their physiological and pathological actions in plants.
Regulation of isoflavonoid biosynthesis
The biosynthesis of isoflavonoids in plants is intricately regulated by diverse environmental factors (such as UV radiation, fungal infection, nitrogen, and phosphorus deficiencies), through transcriptional regulation, post-translational modifications, and epigenetic changes (Dastmalchi et al., 2017; Su et al., 2021; Wang et al., 2023a).
Numerous studies have demonstrated the accumulation of isoflavonoid in plants is also induced by hormonal signals (Ng et al., 2015; Boivin et al., 2016; Yuk et al., 2016; Jeong et al., 2018; Kurepa et al., 2023). The dynamic presence or absence of isoflavonoids significantly influences plant resistance to fungi and environmental stresses, shaping plant growth and development through intricate modulation of auxin transport in vivo (Mathesius, 2001; Wasson et al., 2006; Gao et al., 2021; Zhang et al., 2022b). For instance, in alfalfa, exposure to a fungal elicitor promptly triggers the accumulation of the phytoalexin medicarpin, subsequently undergoing glycosylation and malonylation to form isoflavonoid conjugates (Kessmann et al., 1990). In M. truncatula, fungal infection triggers de novo medicarpin biosynthesis, while wound signals induce the downstream genes converting formononetin and isoflavone glycosides into medicarpin (Naoumkina et al., 2007; Farag et al., 2008). RNA interference-induced silencing of chalcone synthase gene (CHS) in M. truncatula amplifies auxin transport, leading to an impaired ability to form nodules and a deficiency in (iso)flavonoids, particularly formononetin, daidzein and medicarpin (Wasson et al., 2006). When using these compounds and their glycoside forms to treat the wild-type roots, only the free formononetin significantly inhibited auxin transport. However, compared to the wild type, CHS silencing increased auxin transport in roots, indicating that isoflavonoid acts as an auxin transport inhibitor in M. truncatula (Laffont et al., 2010). Additionally, transcriptome analysis of M. truncatula root hairs during rhizobial infection reveals the induction of genes associated with auxin signaling, strigolactone (SL), gibberellic acid (GA), brassinosteroid (BR), and medicarpin biosynthesis, accompanied by the repression of genes involved in lignin biosynthesis. This emphasizes the pivotal roles of (iso) flavonoids and plant hormones, particularly auxin, in the context of rhizobial infection (Breakspear et al., 2014).
An optimal auxin gradient is required for the formation and development of legume nodule primordia (Benkova et al., 2003; Kohlen et al., 2018). In soybean, the involvement of the PIN-FORMED auxin transporter GmPIN1 in nodulation has been elucidated. This process is mediated by two nodulation regulators, (iso)flavonoids (genistein and 7,4′-dihydroxyflavone), which expand GmPIN1 distribution, and cytokinin, which rearranges the cellular polarity of GmPIN1. This orchestration establishes an appropriate auxin gradient, fostering soybean nodulation (Gao et al., 2021). Furthermore, GmPIN1 is involved in the polar transport of auxin from the leaf to the petiole base, resulting in an asymmetric distribution of auxin in the upper and lower petiole cells. This asymmetry significantly impacts cell expansion and the leaf petiole angle. Light-induced (iso)flavonoids accumulate more in the upper petiole cells, inhibiting GmPIN1 expression and disrupting its distribution, leading to reduced auxin in the upper petiole cells. Conversely, lower petiole cells, with lower (iso)flavonoid levels, accumulate more auxin, promoting cell expansion (Zhang et al., 2022b). In addition, cytokinin signaling induces the expression of (iso)flavonoid synthesis genes, influencing the accumulation of (iso)flavonoids and auxin transport, thereby impacting nodule formation (Goyal and Ramawat, 2008; Ng et al., 2015).
As previously discussed, (iso)flavonoid accumulation is induced in response to light or UV radiation, a process mediated by proteins engaged in light signal transduction pathways. As shown in Figure 4, these include UV-A and blue-light photoreceptors cryptochromes (CRYs) and phototropins (PHOTs), along with CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1, an E3 ubiquitin ligase), ELONGATED HYPOCOTYL 5 (HY5, a basic leucine-zipper transcription factor), and B-BOX CONTAINING PROTEINs (BBXs) (Xu, 2020; Liu et al., 2023). A recent study confirmed that the photoreceptors–COP1–HY5-BBX4 regulatory module could regulate the isoflavonoid biosynthesis in soybean (Song et al., 2023). GmSTF1 and GmSTF2 (HY5 orthologs) serve as positive regulators of isoflavonoid synthesis, activating the expression of GmPAL2.1, GmPAL2.3, and GmUGT2 while repressing GmBBX4 expression. GmBBX4, in turn, inhibits the transcriptional activation activity of GmSTF1/2 through direct interaction. Photoreceptors CRYs and PHOTs play positive regulatory roles in light-signal-mediated isoflavonoid biosynthesis. Conversely, COP1, acting as their genetically downstream component, negatively regulates isoflavonoid synthesis by promoting the degradation of GmSTF1/2. Furthermore, GmSTF3/4 are involved in UV-mediated isoflavonoid synthesis, responding to UV-B light through the UV-B photoreceptor (UVR8) in shoots. This signal is then transmitted to the roots, activating the expression of GmMYB12B2 and GmCHS9, subsequently increasing the isoflavone content (Chen et al., 2024). Investigating the impact of light on isoflavone accumulation provides both a theoretical basis and technical support for intercropping. For instance, in maize-soybean intercropping, shading effects from maize growth, limiting photosynthesis, were found to decrease mildew incidence on soybean pods, attributed to the accumulation of isoflavones in soybean under shading conditions, particularly during the vegetative stage (Li et al., 2021b).
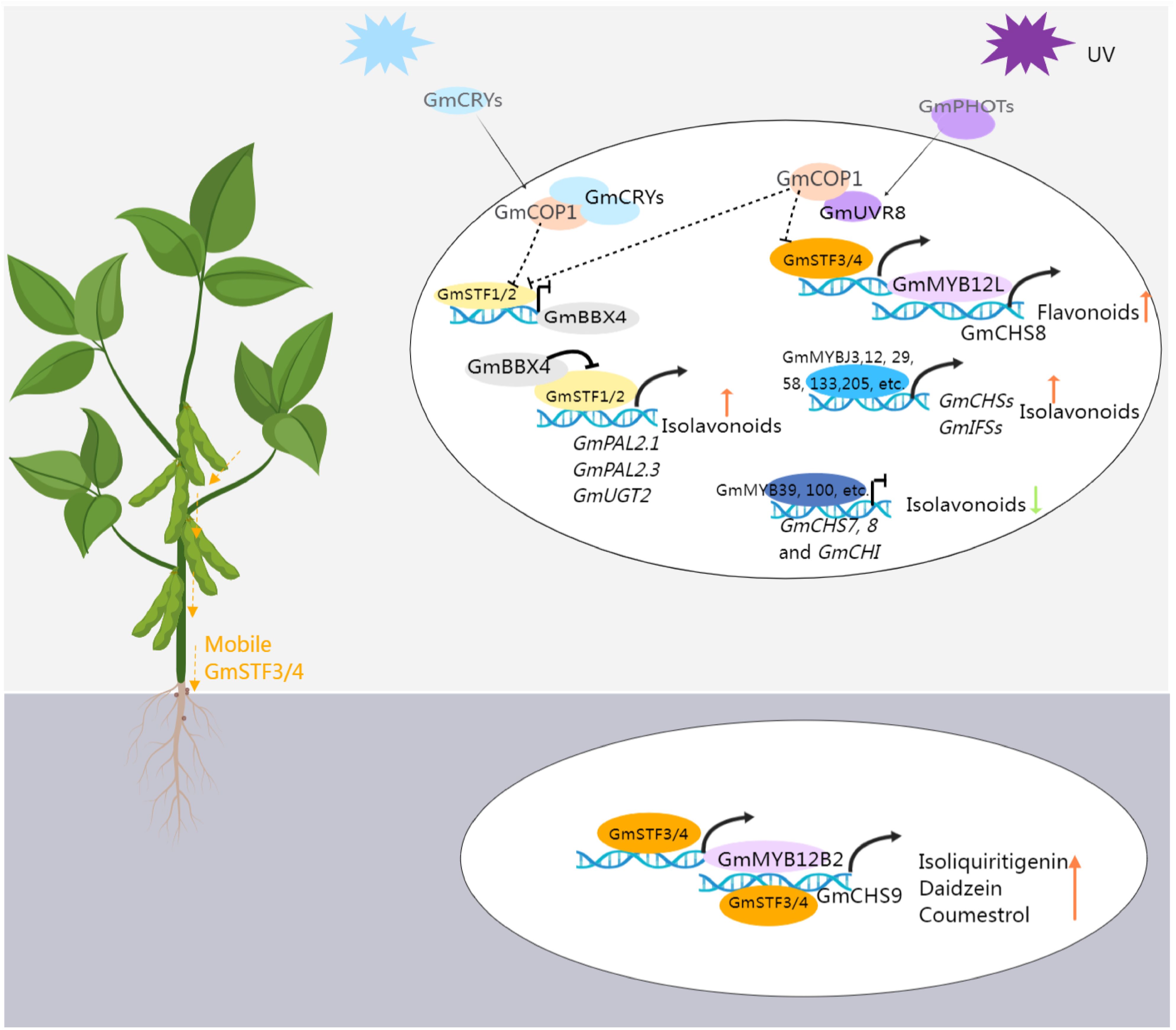
Figure 4 The transcriptional regulation pattern of isoflavonoid biosynthesis in soybean. Blue light activates the GmCRVs receptors, which interacts with GmCOP1, releasing GmSTF1/2 from the STF-COP1 complex. This activation leads to the expression of GmPAL2 and GmUGT2 by GmSTF1/2, promoting the accumulation of isoflavonoids. Additionally, at the transcriptional level, GmSTF1/2 can inhibit the expression of GmBBX4. At the protein level, GmBBX4 interacts with GmSTFs and suppresses their ability to activate target genes related to isoflavonoid synthesis, creating a negative feedback loop. UV activation of the URV receptor allows it to interact with COP1, releasing STF3/4 from the STF-COP1 complex. STF3/4 then activates GmMYB12L, which in turn activates the expression of GmCHS8, promoting the synthesis of flavonoids. Moreover, STF3/4 can move from the shoot to the soybean root, activating the expression of GmMYB12B2 and GmCHS9, promoting the synthesis of (iso)flavonoid. The figure was created with Medpeer (https://image.medpeer.cn/).
Key enzymes in isoflavonoid biosynthesis, such as CHS, CHI, and IFS, are regulated by specific transcription factors (TFs). Notably, MYB TFs have emerged as direct regulators of isoflavonoid biosynthesis genes. As shown in Table 1, GmMYB29, GmMYB58, GmMYB205, GmMYBJ3, GmMYB12, GmMYB133, and GmMYB502 act as activators to increase the expression of CHS and IFS genes, thereby promoting the accumulation of isoflavonoids in soybean, while GmMYB39 and GmMYB100 are repressors. It is worth mentioning that GmMYB176, an R1 MYB TF, plays a dual role in isoflavonoid biosynthesis. And it decreases the isoflavonoids accumulation by down-regulating the expression of GmIFS, and increases the flavanone liquiritigenin content by activating the expression of GmCHS8, respectively. In the presence of GmbZIP5, however, GmMYB176 acts as a positive regulator to enhance the accumulation of some other isoflavonoids, such as glyceollin, isowighteone and a unique O-methylhydroxy isoflavone (Anguraj Vadivel et al., 2019, 2021). Besides, the intracellular localization of GmMYB176 or GmMYB133 can be modulated through interactions with 14–3-3 proteins, such as GmSGF14. This interaction subsequently hinders their regulatory role in isoflavonoid biosynthesis (Li and Dhaubhadel, 2012; Li et al., 2012; Bian et al., 2018). In other plant species, various MYB TFs have been documented, including positive regulators LjMYB14 and LjMYB36 in L. japonicus, CaMYB39 in chickpea, and a negative regulator MtPAR in M. truncatula (Table 1). MtPAR serves as a switch for proanthocyanidin synthesis, capable of directly inhibiting the expression of IFS2. Conversely, it activates the expression of anthocyanidin reductase (ANR) encoding gene in the presence of MtTT8 and MtWD40–1, leading to a reduction in isoflavone and anthocyanidin levels, thereby channeling metabolic flux towards proanthocyanidin biosynthesis (Verdier et al., 2012; Li et al., 2016b, c).
Additional regulators, including NAC and C2H2-type zinc-finger TFs, are also implicated in isoflavonoid biosynthesis. For instance, the expression of GmNAC42–1 responds to both abiotic and biotic elicitors, stimulating the synthesis of pterocarpan glyceollin by activating IFS2 and G4DT (encoding glycinol 4-dimethylallyl transferase) in soybean (Jahan et al., 2019). Notably, GmNAC42–1 is under positive regulation by GmMYB29A2, which itself acts as a positive regulator in the glyceollin biosynthetic pathway (Jahan et al., 2020). Another player, GmZFP7, a C2H2 zinc-finger TF, has been reported to modulate isoflavone accumulation by activating GmIFS2 and Flavanone 3 β-hydroxylase 1 (GmF3H1) in soybean (Feng et al., 2023).
Recent studies also reveal the involvement of microRNAs (miRNAs) in post-transcriptional regulation of isoflavonoid biosynthesis. In soybean, P. sojae infection induced the expression of Gma-miRNA393 in roots. Knockdown of Gma-miRNA393 reduced isoflavonoid content and downregulated the gene expression of GmHID1 and GmIFS1, while increasing susceptibility to P. sojae. This suggests that Gma-miRNA393 acts as a positive regulator of isoflavonoid biosynthesis; however, its downstream target genes remain unidentified (Wong et al., 2014). Table 1 delineates additional miRNAs, including Gma-miRNA12, Gma-miRNA24, Gma-miRNA29, Gma-miRNA26, and Gma-miRNA28, acting as negative regulators by interfering with the expression of their target genes. These target genes encode key enzymes or transcription factors crucial in isoflavonoid biosynthesis (Gupta et al., 2017; Gupta et al., 2019b).
Moreover, epigenetic regulation, including DNA methylation and histone modifications, has been implicated in the control of isoflavonoid accumulation (Baulcombe and Dean, 2014; Chang et al., 2020). A comprehensive comparative analysis across various soybean genotypes exhibiting distinct isoflavone contents revealed a positive correlation between the expression of the IFS gene and the cytosine methylation level within its coding region (Gupta et al., 2019a). This finding underscores the potential positive regulatory impact of epigenetic modifications on the intricate process of isoflavonoid biosynthesis. In the first generation (T1) of transgenic wheat (Triticum aestivum) overexpressing IFS, methylation levels in the exogenous promoter region exhibited a negative correlation with IFS expression (Elseehy, 2020), implying that T1 plants can reconstitute gene expression by altering the methylation status of the exogenous promoter.
Collectively, as shown in Figure 4 and Table 1, although many genes involved in the regulation of isoflavonoid synthesis and their target genes have been reported, some regulatory mechanisms are still unclear. These regulatory genes often affect the synthesis of multiple isoflavonoid compounds simultaneously. Therefore, in future, more specific factors need to be explored, such as those that uniquely control the synthesis of the subclass of isoflavonoids. In addition, as mentioned earlier, environmental factors and hormone signals play a significant role in isoflavonoid synthesis, regulation, and transport, which is also an important direction for future exploration.
Metabolic engineering of isoflavonoids biosynthesis
Isoflavonoids, recognized for their benefits in plants, livestock, and human health, have spurred research in metabolic engineering. As shown in Table 2, strategies like antisense RNA, RNA interference, CRISPR/Cas9-mediated gene editing, co-expression, and heterologous expression have been employed for isoflavonoid engineering in legumes, non-legume plants, and microorganisms (Dixon and Steele, 1999; Zhang et al., 2020; Liu et al., 2021).
The main strategies for forage legumes breeding to increase the nutritional value and digestibility of forage include reducing anti-nutritional factors, such as lectins, saponins, oxalic acid, and condensed tannins, increasing crude protein concentrations, enhancing stress tolerance, and changing cell wall structure and composition to improve the degradability of cell wall polysaccharides (Kumar, 2011; Kulkarni et al., 2018; Katoch, 2022). Interestingly, most of these breeding goals can be achieved through metabolic engineering of phenylpropanoid biosynthesis (Dixon and Steele, 1999; Du et al., 2010). In alfalfa, overexpression of the encoding gene of isoflavone O-methyltransferase (IOMT) results in heightened levels of formononetin and medicarpin, enhancing disease resistance to Phoma medicaginis in transgenic plants (He and Dixon, 2000). Heterologous expression of MtIFS1 can lead to the enhanced accumulation of medicarpin in transgenic alfalfa plants upon P. medicaginis infection, indicating that this modification is beneficial for plant response to stress (Deavours and Dixon, 2005). Moreover, Gou et al. disrupted the limitation of precursors by simultaneously overexpressing of GmIFS1, GmCHS7 and GmCHI1 in M. truncatula, promoted both isoflavone and proanthocyanidin accumulation, which are beneficial for ruminant animals (Gou et al., 2016). As shown in Table 2, the key enzyme genes involved in isoflavonoid biosynthesis are important target genes for metabolic engineering, Caffeoyl-CoA O-methyltransferase (CCoAOMT) encoding a key enzyme of lignin pathway also has an important impact on isoflavonoid synthesis. Down-regulated of CCoAOMT via antisense RNA technology, leading to a decrease in the content of guaiacyl (G) lignin and an increase in syringyl to guaiacyl ratio (S/G) (Guo et al., 2001). When wild-type and CCoAOMT downregulated plants are infected with fungi, the expression of medicarpin biosynthesis genes is upregulated in both, but more significantly in the CCoAOMT downregulated plants. This leads the lignin modified alfalfa to redirect metabolic flux towards the medicarpin pathway upon fungal infection, thereby improving the availability of cell wall polysaccharides and resistance against fungal disease (Guo et al., 2001; Gill et al., 2018).
In soybean, the overexpression of F3H alone does not significantly affect the total isoflavone content. However, the overexpression of a fusion gene of the maize C1 and R (CRC) increases the total isoflavone content in transgenic soybean seeds by approximately 2-fold. Co-verexpression of CRC and F3H can enhance the total isoflavone content by about 4-fold (Yu et al., 2003). RNA interference was used to generate silence FNSs (encoding flavone synthases) in soybean, which reduced the synthesis of apigenins and anthocyanins from naringenin, thus promoting the accumulation of isoflavones (Jiang et al., 2010). Notably, silencing alone of FNSII or F3H results in a ~1.3- or ~1.9-fold increase in isoflavone content, while double silencing of FNSII and F3H can result in a ~2.2-fold increase in isoflavone production compared to transgenic soybean hairy roots containing empty vectors (Jiang et al., 2014). Co-overexpression of GmMYB176 and GmbZIP5 results in an approximate 1.4-fold increase in the total isoflavonoid content in hairy roots (Anguraj Vadivel et al., 2021).
Non-legume plants and microorganisms can also synthesize large amounts of isoflavonoids through metabolic engineering, which involves utilizing the existing flavonoid biosynthesis pathway to provide precursors and introducing the key enzyme genes for isoflavonoid biosynthesis. For example, heterologous expression GmIFS in Arabidopsis thaliana could accumulate a small amount of genistein glycosides while introducing the GmIFS gene into tt6/tt3 double mutant, where expression of F3H and dihydroflavonol reductase (DFR) was abolished, resulted in a large accumulation of genistein, which provides an important idea for the isoflavonoid accumulation via metabolic engineering (Liu et al., 2002). Heterologous expression of the IFS/CHI fusion gene in tobacco results in higher levels of genistein and its glycoside compounds compared to expressing IFS alone (Tian and Dixon, 2006). When AtMYB12 and GmIFS1 are co-overexpressed in tobacco, the expression of key enzyme genes in the flavonoid pathway is significantly upregulated, leading to a substantial increase in the content of flavonoid compounds and synthesizing approximately 0.05 mg/g of genistein in the fresh tissues (Pandey et al., 2014). In addition, isoflavonoids could be synthesized and accumulated in non-legume plants through the heterologous expression of GmIFSs in rice, tomato and Brassica napus (Sreevidya et al., 2006; Tian and Dixon, 2006; Liu et al., 2007; Shih et al., 2008; Li et al., 2011; Pandey et al., 2014). These results indicate that heterologous expression of key enzyme genes can achieve the synthesis of isoflavones in non-leguminous plants. However, to achieve high content, a strategy of co-expressing multiple structural genes or a combination of transcription factors with structural genes can be employed. Additionally, the activity of key isoflavone biosynthetic enzymes may vary in different non-leguminous plants, which may potentially affect the yields. For example, CHIs are divided into two groups (type I and type II). Type I CHIs, which are found in both legumes and non-legumes, function to isomerize only 6′-hydroxychalcone to 5-hydroxyflavanone (naringenin). Whereas, type II CHIs belong to a legume-specific group that are active on both 6′-deoxychalcone and 6′-hydroxychalcone, yielding 5-deoxyflavanone (liquiritigenin) and 5-hydroxyflavanone, respectively (Shimada et al., 2003). The experiments in yeast or E. coli strains successfully demonstrate that they have significant differences in enzymatic activity (Shimada et al., 2003; Tian and Dixon, 2006).
Recently, microorganisms such as Saccharomyces cerevisiae and Escherichia coli have been adapted and engineered for heterologous isoflavonoid synthesis, overcoming the complexity associated with biosynthesis and accumulation in non-endogenous plants through advancements in synthetic biology. The de novo synthesis of parent isoflavonoids, such as genistein and quercetin, has been achieved in engineered yeast strains by overexpressing at least seven enzymes (PAL/TAL, 4CL, CHS, CHI, CHR, IFS, and IFD) (Trantas et al., 2009; Rodriguez et al., 2017). Co-cultivation of an IFS-expressing S. cerevisiae strain with a naringenin-producing E. coli strain resulted in the accumulation of genistein (6 mg/L) (Katsuyama et al., 2007). Additionally, de novo biosynthesis of bioactive isoflavonoids and the hops bioactive flavonoid xanthohumol has been achieved in yeast (Liu et al., 2021; Yang et al., 2024). These studies demonstrate that the optimal combination of key enzyme genes from various plants, the copy number of these genes, the physical distance between adjacent key enzymes, and the accommodation of membrane proteins by the endoplasmic reticulum are factors influencing the efficient synthesis of isoflavonoids. These are all important considerations for future de novo synthesis of (iso)flavonoidand other complex natural products.
New technologies and resources
The CRISPR/Cas systems, known for their high efficiency and versatility, have found extensive applications in various plant genome editing and metabolic engineering endeavors (Wada et al., 2022). In Fagopyrum tataricum, the CRISPR/Cas9-mediated knockout of FtMYB45 resulted in a reduction of flavonoids (Wen et al., 2022). In soybean, precise editing of key enzymes involved in isoflavonoid biosynthesis, including GmF3H1, GmF3H2, GmFNS-1, and Gm-IFS, was achieved through CRISPR/Cas9-directed mutagenesis (Zhang et al., 2020). This targeted mutagenesis led to a 2-fold increase in isoflavone content in soybean leaves and enhanced resistance to soybean mosaic virus. The study emphasized the roles of genes in isoflavonoid biosynthesis and phytohormones influencing growth effects (Mipeshwaree Devi et al., 2023).
Machine learning and multiomics approaches have also been incorporated into isoflavonoid research. Nearly thirty flavor molecule databases and various models have been identified (Kou et al., 2023). Over 1200 natural flavonoid compounds have been cataloged in the customized Flavonoid Astringency Prediction Database (FAPD, Guo et al., 2023). The establishment of this database facilitates an understanding of the relationship between the molecular structure of flavonoid compounds and their astringency in foods. Key genes in crops that influence astringency can be screened by integrating transcriptomic and metabolomic analyses. Subsequently, transgenic or gene editing approaches are utilized to verify the functions of these genes, facilitating the breeding of superior crop varieties that are both healthy and flavorful (Qin et al., 2022; Qiu et al., 2023). Given that the distribution of isoflavonoids and the genes involved in biosynthesis, regulation and transport are strongly induced by environmental factors, and exhibit tissue specificity and developmental stage specificity in leguminous plants. The integration of single-cell sequencing and spatial transcriptomics is poised to provide robust support for further elucidation and metabolic engineering of the isoflavonoid biosynthetic pathway. For instance, the iflavonoids in the roots of leguminous plants are closely related to the formation of root nodules. Through single-cell sequencing technology, researchers can analyze the gene expression patterns of specific cell types during the root nodule formation process at the single-cell level. Combined with transcriptomic sequencing, it is possible to further explore the gene expression patterns related to isoflavonoid synthesis, regulation, transport, and secretion.
Conclusion and future prospects
Isoflavonoids play a crucial role in plant adaptation to complex environmental stimuli, with leguminous plant roots utilizing them to regulate nodule formation and influence overall growth. Consequently, exploring isoflavonoid metabolic engineering holds promise for genetic improvements in both legume and non-legume crops. However, it is important to note that changes in the composition and content of flavonoids, isoflavonoids, and lignin, which are interconnected through the phenylpropanoid pathway, could potentially have adverse effects on plants, such as reduced biomass, an imbalance between disease resistance and stress tolerance, and altered flavor. Therefore, it is necessary to consider the entire growth and developmental state of the plant, rather than focusing solely on changes in isoflavonoid content. Recent innovative approaches, exemplified by Sulis et al.’s work using a multiscale model of lignin biosynthesis, showcase effective multiplex CRISPR-editing strategies (Sulis et al., 2023). This enables the reduction of lignin levels in poplar without compromising growth, enhancing cell wall degradability, promoting cellulose utilization for papermaking and bioenergy production, and minimizing environmental impact. These advances provide valuable insights for precise and efficient crop genetic breeding.
What is the relationship between the molecular structure of isoflavonoid compounds and their bioactivity and flavor? Which genes determine the production of specific isoflavonoids? Which genes influence the transformation between free isoflavonoids and their modifications? What is the connection between the accumulation or secretion of plant isoflavonoids and the environment? Can the synthesis of isoflavonoids in non-leguminous plants achieve symbiosis with rhizobia to enhance nitrogen fixation? These are all subjects that require further research and exploration. Further exploration of diverse isoflavonoids and derivatives through metabolic engineering or synthetic biology requires the identification of key enzyme genes and regulators. Integration of machine learning and database predictions may expedite the discovery of additional enzymes and compounds. The future lies in the collaborative efforts of synthetic biology and metabolic engineering for efficient and sustainable isoflavonoid production. Advancements in multi-omics technologies are anticipated to unravel key insights into isoflavonoid biosynthesis, transport, and accumulation.
Author contributions
LW: Funding acquisition, Resources, Writing – original draft, Data curation, Writing – review & editing. CL: Funding acquisition, Writing – original draft, Resources, Writing – review & editing. KL: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Natural Science Foundation of China (31901333, 32372040), the Natural Science Foundation of Southwest University of Science and Technology (19zx7120), the Fundamental Research Funds for the Central Universities of Southwest University (SWU-KQ22074).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abd-Alla, M. H., Al-Amri, S. M., El-Enany, A.-W. E. (2023). Enhancing rhizobium–legume symbiosis and reducing nitrogen fertilizer use are potential options for mitigating climate change. Agriculture 13, 2092. doi: 10.3390/agriculture13112092
Abderamane, B., Tih, A. E., Ghogomu, R. T., Blond, A., Bodo, B. (2011). Isoflavonoid derivatives from Lophira alata stem heartwood. Z Naturforsch. C J. Biosci. 66, 87–92. doi: 10.1515/znc-2011-3-401
Ahmad, M. Z., Li, P., Wang, J., Rehman, N. U., Zhao, J. (2017). Isoflavone malonyltransferases GmIMaT1 and GmIMaT3 differently modify isoflavone glucosides in soybean (Glycine max) under various stresses. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00735
Ahmad, M. Z., Zhang, Y., Zeng, X., Li, P., Wang, X., Benedito, V. A., et al. (2021). Isoflavone malonyl-CoA acyltransferase GmMaT2 is involved in nodulation of soybean by modifying synthesis and secretion of isoflavones. J. Exp. Bot. 72, 1349–1369. doi: 10.1093/jxb/eraa511
Akashi, T., Aoki, T., Ayabe, S. (2005). Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis. Plant Physiol. 137, 882–891. doi: 10.1104/pp.104.056747
Akashi, T., Koshimizu, S., Aoki, T., Ayabe, S. (2006). Identification of cDNAs encoding pterocarpan reductase involved in isoflavan phytoalexin biosynthesis in Lotus japonicus by EST mining. FEBS Lett. 580, 5666–5670. doi: 10.1016/j.febslet.2006.09.016
Akashi, T., Sasaki, K., Aoki, T., Ayabe, S., Yazaki, K. (2009). Molecular cloning and characterization of a cDNA for pterocarpan 4-dimethylallyltransferase catalyzing the key prenylation step in the biosynthesis of glyceollin, a soybean phytoalexin. Plant Physiol. 149, 683–693. doi: 10.1104/pp.108.123679
Akashi, T., Sawada, Y., Shimada, N., Sakurai, N., Aoki, T., Ayabe, S. (2003). cDNA cloning and biochemical characterization of S-adenosyl-L-methionine: 2,7,4’-trihydroxyisoflavanone 4’-O-methyltransferase, a critical enzyme of the legume isoflavonoid phytoalexin pathway. Plant Cell Physiol. 44, 103–112. doi: 10.1093/pcp/pcg034
Anguraj Vadivel, A. K., McDowell, T., Renaud, J. B., Dhaubhadel, S. (2021). A combinatorial action of GmMYB176 and GmbZIP5 controls isoflavonoid biosynthesis in soybean (Glycine max). Commun. Biol. 4, 356. doi: 10.1038/s42003–021-01889–6
Anguraj Vadivel, A. K., Renaud, J., Kagale, S., Dhaubhadel, S. (2019). GmMYB176 regulates multiple steps in isoflavonoid biosynthesis in soybean. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00562
Anzeveno, P. B. (1979). Rotenoid interconversion. Synthesis of deguelin from rotenone. J. Organic Chem. 44, 2578–2580. doi: 10.1021/jo01328a056
Araya-Cloutier, C., den Besten, H. M., Aisyah, S., Gruppen, H., Vincken, J. P. (2017). The position of prenylation of isoflavonoids and stilbenoids from legumes (Fabaceae) modulates the antimicrobial activity against Gram positive pathogens. Food Chem. 226, 193–201. doi: 10.1016/j.foodchem.2017.01.026
Araya-Cloutier, C., Vincken, J. P., van de Schans, M. G. M., Hageman, J., Schaftenaar, G., den Besten, H. M. W., et al. (2018). QSAR-based molecular signatures of prenylated (iso)flavonoids underlying antimicrobial potency against and membrane-disruption in Gram positive and Gram negative bacteria. Sci. Rep. 8, 9267. doi: 10.1038/s41598–018-27545–4
Banasiak, J., Biala, W., Staszkow, A., Swarcewicz, B., Kepczynska, E., Figlerowicz, M., et al. (2013). A Medicago truncatula ABC transporter belonging to subfamily G modulates the level of isoflavonoids. J. Exp. Bot. 64, 1005–1015. doi: 10.1093/jxb/ers380
Baulcombe, D. C., Dean, C. (2014). Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 6, a019471. doi: 10.1101/cshperspect.a019471
Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. doi: 10.1016/S0092-8674(03)00924-3
Biala, W., Banasiak, J., Jarzyniak, K., Pawela, A., Jasinski, M. (2017). Medicago truncatula ABCG10 is a transporter of 4-coumarate and liquiritigenin in the medicarpin biosynthetic pathway. J. Exp. Bot. 68, 3231–3241. doi: 10.1093/jxb/erx059
Biala-Leonhard, W., Zanin, L., Gottardi, S., de Brito Francisco, R., Venuti, S., Valentinuzzi, F., et al. (2021). Identification of an isoflavonoid transporter required for the nodule establishment of the rhizobium-fabaceae symbiotic interaction. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.758213
Bian, S., Li, R., Xia, S., Liu, Y., Jin, D., Xie, X., et al. (2018). Soybean CCA1-like MYB transcription factor GmMYB133 modulates isoflavonoid biosynthesis. Biochem. Biophys. Res. Commun. 507, 324–329. doi: 10.1016/j.bbrc.2018.11.033
Boivin, S., Fonouni-Farde, C., Frugier, F. (2016). How auxin and cytokinin phytohormones modulate root microbe interactions. Front. Plant Sci. 7 1240. doi: 10.3389/fpls.2016.01240
Boue, S. M., Carter, C. H., Ehrlich, K. C., Cleveland, T. E. (2000). Induction of the soybean phytoalexins coumestrol and glyceollin by Aspergillus. J. Agric. Food Chem. 48, 2167–2172. doi: 10.1021/jf9912809
Breakspear, A., Liu, C., Roy, S., Stacey, N., Rogers, C., Trick, M., et al. (2014). The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell 26, 4680–4701. doi: 10.1105/tpc.114.133496
Cannon, J. R., Tapias, V., Na, H. M., Honick, A. S., Drolet, R. E., Greenamyre, J. T. (2009). A highly reproducible rotenone model of Parkinson’s disease. Neurobiol. Dis. 34, 279–290. doi: 10.1016/j.nbd.2009.01.016
Chang, Y. N., Zhu, C., Jiang, J., Zhang, H., Zhu, J. K., Duan, C. G. (2020). Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 62, 563–580. doi: 10.1111/jipb.12901
Chen, J., Xu, H., Liu, Q., Ke, M., Zhang, Z., Wang, X., et al. (2024). Shoot-to-root communication via GmUVR8-GmSTF3 photosignaling and flavonoid biosynthesis fine-tunes soybean nodulation under UV-B light. New Phytol. 241, 209–226. doi: 10.1111/nph.19353
Chu, S., Wang, J., Zhu, Y., Liu, S., Zhou, X., Zhang, H., et al. (2017). An R2R3-type MYB transcription factor, GmMYB29, regulates isoflavone biosynthesis in soybean. PloS Genet. 13, e1006770. doi: 10.1371/journal.pgen.1006770
Darbour, N., Bayet, C., Rodin-Bercion, S., Elkhomsi, Z., Lurel, F., Chaboud, A., et al. (2007). Isoflavones from ficus nymphaefolia. Nat. Prod Res. 21, 461–464. doi: 10.1080/14786410601086871
Dastmalchi, M., Chapman, P., Yu, J., Austin, R. S., Dhaubhadel, S. (2017). Transcriptomic evidence for the control of soybean root isoflavonoid content by regulation of overlapping phenylpropanoid pathways. BMC Genomics 18, 70. doi: 10.1186/s12864-016-3463-y
Deavours, B. E., Dixon, R. A. (2005). Metabolic engineering of isoflavonoid biosynthesis in alfalfa. Plant Physiol. 138, 2245–2259. doi: 10.1104/pp.105.062539
Deng, Y. T., Huang, H. C., Lin, J. K. (2010). Rotenone induces apoptosis in MCF-7 human breast cancer cell-mediated ROS through JNK and p38 signaling. Mol. Carcinogenesis: Published cooperation Univ. Texas MD Anderson Cancer Center 49, 141–151. doi: 10.1002/mc.20583
Dewick, P. M., Martin, M. (1979). Biosynthesis of pterocarpan and isoflavan phytoalexins in Medicago sativa: the biochemical interconversion of pterocarpans and 2′-hydroxyisoflavans. Phytochemistry 18, 591–596. doi: 10.1016/S0031-9422(00)84266-1
Dixon, R. A. (1999). Isoflavonoids: biochemistry, molecular biology and biological functions. Compr. Natural products Chem. 1, 773–823. doi: 10.1016/B978-0-08-091283-7.00030-8
Dixon, R. A. (2004). Phytoestrogens. Annu. Rev. Plant Biol. 55, 225–261. doi: 10.1146/annurev.arplant.55.031903.141729
Dixon, R. A., Ferreira, D. (2002). Genistein. Phytochemistry 60, 205–211. doi: 10.1016/S0031-9422(02)00116-4
Dixon, R. A., Harrison, M. J., Paiva, N. L. (1995). The isoflavonoid phytoalexin pathway: from enzymes to genes to transcription factors. Physiologia Plantarum 93, 385–392. doi: 10.1111/j.1399-3054.1995.tb02243.x
Dixon, R. A., Paiva, N. L. (1995). Stress-induced phenylpropanoid metabolism. Plant Cell 7, 1085–1097. doi: 10.2307/3870059
Dixon, R. A., Steele, C. L. (1999). Flavonoids and isoflavonoids - a gold mine for metabolic engineering. Trends Plant Sci. 4, 394–400. doi: 10.1016/S1360-1385(99)01471-5
Dong, N. Q., Lin, H. X. (2021). Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 63, 180–209. doi: 10.1111/jipb.13054
Du, H., Huang, Y., Tang, Y. (2010). Genetic and metabolic engineering of isoflavonoid biosynthesis. Appl. Microbiol. Biotechnol. 86, 1293–1312. doi: 10.1007/s00253-010-2512-8
Elseehy, M. M. (2020). Differential transgeneration methylation of exogenous promoters in T1 transgenic wheat (Triticum aestivum). Cytol. Genet. 54, 493–504. doi: 10.3103/S0095452720050151
Farag, M. A., Huhman, D. V., Dixon, R. A., Sumner, L. W. (2008). Metabolomics reveals novel pathways and differential mechanistic and elicitor-specific responses in phenylpropanoid and isoflavonoid biosynthesis in Medicago truncatula cell cultures. Plant Physiol. 146, 387–402. doi: 10.1104/pp.107.108431
Feng, Y., Zhang, S., Li, J., Pei, R., Tian, L., Qi, J., et al. (2023). Dual-function C2H2-type zinc-finger transcription factor GmZFP7 contributes to isoflavone accumulation in soybean. New Phytol. 237, 1794–1809. doi: 10.1111/nph.18610
Funaki, A., Waki, T., Noguchi, A., Kawai, Y., Yamashita, S., Takahashi, S., et al. (2015). Identification of a highly specific isoflavone 7-O-glucosyltransferase in the soybean (Glycine max (L.) Merr.). Plant Cell Physiol. 56, 1512–1520. doi: 10.1093/pcp/pcv072
Gao, Z., Chen, Z., Cui, Y., Ke, M., Xu, H., Xu, Q., et al. (2021). GmPIN-dependent polar auxin transport is involved in soybean nodule development. Plant Cell 33, 2981–3003. doi: 10.1093/plcell/koab183
Gao, Y., Yao, Y., Zhu, Y., Ren, G. (2015). Isoflavone content and composition in chickpea (Cicer arietinum L.) sprouts germinated under different conditions. J. Agric. Food Chem. 63, 2701–2707. doi: 10.1021/jf5057524
Gill, U. S., Uppalapati, S. R., Gallego-Giraldo, L., Ishiga, Y., Dixon, R. A., Mysore, K. S. (2018). Metabolic flux towards the (iso)flavonoid pathway in lignin modified alfalfa lines induces resistance against Fusarium oxysporum f. sp. medicaginis. Plant Cell Environ. 41, 1997–2007. doi: 10.1111/pce.13093
Gou, L., Zhang, R., Ma, L., Zhu, F., Dong, J., Wang, T. (2016). Multigene synergism increases the isoflavone and proanthocyanidin contents of Medicago truncatula. Plant Biotechnol. J. 14, 915–925. doi: 10.1111/pbi.12445
Goyal, S., Ramawat, K. (2008). Synergistic effect of morphactin on cytokinin-induced production of isoflavonoids in cell cultures of Pueraria tuberosa (Roxb. ex. Willd.) DC. Plant Growth Regul. 55, 175–181. doi: 10.1007/s10725-008-9271-x
Griffiths, K., Wilson, D. W., Singh, R. B., De Meester, F. (2014). Effect of dietary phytoestrogens on human growth regulation: imprinting in health & disease. Indian J. Med. Res. 140 Suppl, S82–S90.
Guo, D., Chen, F., Inoue, K., Blount, J. W., Dixon, R. A. (2001). Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa. Impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13, 73–88. doi: 10.1105/tpc.13.1.73
Guo, L., Paiva, N. L. (1995). Molecular cloning and expression of alfalfa (Medicago sativa L.) vestitone reductase, the penultimate enzyme in medicarpin biosynthesis. Arch. Biochem. biophys. 320, 353–360. doi: 10.1016/0003-9861(95)90019-5
Guo, T., Pan, F., Cui, Z., Yang, Z., Chen, Q., Zhao, L., et al. (2023). FAPD: an astringency threshold and astringency type prediction database for flavonoid compounds based on machine learning. J. Agric. Food Chem. 71, 4172–4183. doi: 10.1021/acs.jafc.2c08822
Gupta, O. P., Dahuja, A., Sachdev, A., Jain, P. K., Kumari, S., Vinutha, T., et al. (2019a). Cytosine methylation of isoflavone synthase gene in the genic region positively regulates its expression and isoflavone biosynthesis in soybean seeds. DNA Cell Biol. 38, 510–520. doi: 10.1089/dna.2018.4584
Gupta, O. P., Dahuja, A., Sachdev, A., Kumari, S., Jain, P. K., Vinutha, T., et al. (2019b). Conserved miRNAs modulate the expression of potential transcription factors of isoflavonoid biosynthetic pathway in soybean seeds. Mol. Biol. Rep. 46, 3713–3730. doi: 10.1007/s11033-019-04814-7
Gupta, O. P., Nigam, D., Dahuja, A., Kumar, S., Vinutha, T., Sachdev, A., et al. (2017). Regulation of isoflavone biosynthesis by miRNAs in two contrasting soybean genotypes at different seed developmental stages. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00567
Gutierrez-Gonzalez, J. J., Guttikonda, S. K., Tran, L.-S. P., Aldrich, D. L., Zhong, R., Yu, O., et al. (2010). Differential expression of isoflavone biosynthetic genes in soybean during water deficits. Plant Cell Physiol. 51, 936–948. doi: 10.1093/pcp/pcq065
Gutierrez-Gonzalez, J. J., Wu, X., Zhang, J., Lee, J.-D., Ellersieck, M., Shannon, J. G., et al. (2009). Genetic control of soybean seed isoflavone content: importance of statistical model and epistasis in complex traits. Theor. Appl. Genet. 119, 1069–1083. doi: 10.1007/s00122-009-1109-z
Ha, J., Kang, Y. G., Lee, T., Kim, M., Yoon, M. Y., Lee, E., et al. (2019). Comprehensive RNA sequencing and co-expression network analysis to complete the biosynthetic pathway of coumestrol, a phytoestrogen. Sci. Rep. 9, 1934. doi: 10.1038/s41598–018-38219–6
Hail, N., Jr., Lotan, R. (2004). Apoptosis induction by the natural product cancer chemopreventive agent deguelin is mediated through the inhibition of mitochondrial bioenergetics. Apoptosis 9, 437–447. doi: 10.1023/B:APPT.0000031449.57551.e1
Hakamatsuka, T., Mori, K., Ishida, S., Ebizuka, Y., Sankawa, U. (1998). Purification of 2-hydroxyisoflavanone dehydratase from the cell cultures of Pueraria lobata in honour of Professor GH Neil Towers 75th birthday. Phytochemistry 49, 497–505. doi: 10.1016/S0031-9422(98)00266-0
Han, X., Yin, Q., Liu, J., Jiang, W., Di, S., Pang, Y. (2017). GmMYB58 and GmMYB205 are seed-specific activators for isoflavonoid biosynthesis in Glycine max. Plant Cell Rep. 36, 1889–1902. doi: 10.1007/s00299-017-2203-3
Hayashi, H., Hiraoka, N., Ikeshiro, Y., Yamamoto, H. (1996). Organ specific localization of flavonoids in Glycyrrhiza glabra L. Plant Sci. 116, 233–238. doi: 10.1016/0168-9452(96)04387-7
He, X. Z., Dixon, R. A. (2000). Genetic manipulation of isoflavone 7-O-methyltransferase enhances biosynthesis of 4’-O-methylated isoflavonoid phytoalexins and disease resistance in alfalfa. Plant Cell 12, 1689–1702. doi: 10.1105/tpc.12.9.1689
Higgins, V. J. (1972). Role of the phytoalexin medicarpin in three leaf spot diseases of alfalfa. Physiol. Plant Pathol. 2, 289–300. doi: 10.1016/0048-4059(72)90012-4
Ingham, J. L., Harborne, J. B. (1976). Phytoalexin induction as a new dynamic approach to the study of systematic relationships among higher plants. Nature 260, 241–243. doi: 10.1038/260241a0
Jahan, M. A., Harris, B., Lowery, M., Coburn, K., Infante, A. M., Percifield, R. J., et al. (2019). The NAC family transcription factor GmNAC42–1 regulates biosynthesis of the anticancer and neuroprotective glyceollins in soybean. BMC Genomics 20, 149. doi: 10.1186/s12864-019-5524-5
Jahan, M. A., Harris, B., Lowery, M., Infante, A. M., Percifield, R. J., Kovinich, N. (2020). Glyceollin transcription factor GmMYB29A2 regulates soybean resistance to Phytophthora sojae. Plant Physiol. 183, 530–546. doi: 10.1104/pp.19.01293
Jasinski, M., Banasiak, J., Radom, M., Kalitkiewicz, A., Figlerowicz, M. (2009). Full-size ABC transporters from the ABCG subfamily in Medicago truncatula. Mol. Plant Microbe Interact. 22, 921–931. doi: 10.1094/MPMI-22-8-0921
Jeong, Y. J., An, C. H., Park, S. C., Pyun, J. W., Lee, J., Kim, S. W., et al. (2018). Methyl jasmonate increases isoflavone production in soybean cell cultures by activating structural genes involved in isoflavonoid biosynthesis. J. Agric. Food Chem. 66, 4099–4105. doi: 10.1021/acs.jafc.8b00350
Jiang, Y., Hu, Y., Wang, B., Wu, T. (2014). Bivalent RNA interference to increase isoflavone biosynthesis in soybean (Glycine max). Braz. Arch. Biol. Technol. 57, 163–170. doi: 10.1590/S1516-89132013005000018
Jiang, Y. N., Wang, B., Li, H., Yao, L. M., Wu, T. L. (2010). Flavonoid production is effectively regulated by RNAi interference of two flavone synthase genes from Glycine max. J. Plant Biol. 53, 425–432. doi: 10.1007/s12374-010-9132-9
Jung, W., Yu, O., Lau, S. M., O’Keefe, D. P., Odell, J., Fader, G., et al. (2000). Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat. Biotechnol. 18, 208–212. doi: 10.1038/72671
Kaducová, M., Eliašová, A., Trush, K., Bačovčinová, M., Sklenková, K., Pal’ove-Balang, P. (2022). Accumulation of isoflavonoids in Lotus corniculatus after UV-B irradiation. Theor. Exp. Plant Physiol. 34, 53–62. doi: 10.1007/s40626-021-00228-8
Kaducova, M., Monje-Rueda, M. D., Garcia-Calderon, M., Perez-Delgado, C. M., Eliasova, A., Gajdosova, S., et al. (2019). Induction of isoflavonoid biosynthesis in Lotus japonicus after UV-B irradiation. J. Plant Physiol. 236, 88–95. doi: 10.1016/j.jplph.2019.03.003
Kalli, S., Araya-Cloutier, C., Hageman, J., Vincken, J. P. (2021). Insights into the molecular properties underlying antibacterial activity of prenylated (iso)flavonoids against MRSA. Sci. Rep. 11, 14180. doi: 10.1038/s41598–021-92964–9
Katoch, R. (2022). “Approaches for Nutritional Quality Improvement in Forages,” in Nutritional Quality Management of Forages in the Himalayan Region (Singapore: Springer Singapore), 167–192. doi: 10.1007/978-981-16-5437-4
Katsuyama, Y., Miyahisa, I., Funa, N., Horinouchi, S. (2007). One-pot synthesis of genistein from tyrosine by coincubation of genetically engineered Escherichia coli and Saccharomyces cerevisiae cells. Appl. Microbiol. Biotechnol. 73, 1143–1149. doi: 10.1007/s00253-006-0568-2
Kaufman, P. B., Duke, J. A., Brielmann, H., Boik, J., Hoyt, J. E. (1997). A comparative survey of leguminous plants as sources of the isoflavones, genistein and daidzein: implications for human nutrition and health. J. Altern. Complement Med. 3, 7–12. doi: 10.1089/acm.1997.3.7
Kessmann, H., Edwards, R., Geno, P. W., Dixon, R. A. (1990). Stress responses in alfalfa (Medicago sativa L.): V. Constitutive and elicitor-induced accumulation of isoflavonoid conjugates in cell suspension cultures. Plant Physiol. 94, 227–232. doi: 10.1104/pp.94.1.227
Kohlen, W., Ng, J. L. P., Deinum, E. E., Mathesius, U. (2018). Auxin transport, metabolism, and signalling during nodule initiation: indeterminate and determinate nodules. J. Exp. Bot. 69, 229–244. doi: 10.1093/jxb/erx308
Kou, X., Shi, P., Gao, C., Ma, P., Xing, H., Ke, Q., et al. (2023). Data-driven elucidation of flavor chemistry. J. Agric. Food Chem. 71, 6789–6802. doi: 10.1021/acs.jafc.3c00909
Ku, Y. S., Cheng, S. S., Cheung, M. Y., Niu, Y., Liu, A., Chung, G., et al. (2022). The poly-glutamate motif of GmMATE4 regulates its isoflavone transport activity. Membranes (Basel) 12, 206. doi: 10.3390/membranes12020206
Ku, Y. S., Ng, M. S., Cheng, S. S., Lo, A. W., Xiao, Z., Shin, T. S., et al. (2020). Understanding the composition, biosynthesis, accumulation and transport of flavonoids in crops for the promotion of crops as healthy sources of flavonoids for human consumption. Nutrients 12, 1717. doi: 10.3390/nu12061717
Kulkarni, K. P., Tayade, R., Asekova, S., Song, J. T., Shannon, J. G., Lee, J. D. (2018). Harnessing the potential of forage legumes, alfalfa, soybean, and cowpea for sustainable agriculture and global food security. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01314
Kumar, S. (2011). Biotechnological advancements in alfalfa improvement. J. Appl. Genet. 52, 111–124. doi: 10.1007/s13353-011-0028-2
Kurepa, J., Shull, T. E., Smalle, J. A. (2023). Friends in arms: Flavonoids and the auxin/cytokinin balance in terrestrialization. Plants (Basel) 12, 517. doi: 10.3390/plants12030517
Laffont, C., Blanchet, S., Lapierre, C., Brocard, L., Ratet, P., Crespi, M., et al. (2010). The compact root architecture1 gene regulates lignification, flavonoid production, and polar auxin transport in medicago truncatula. Plant Physiol. 153, 1597–1607. doi: 10.1104/pp.110.156620
Lee, H. I., Lee, J. H., Park, K. H., Sangurdekar, D., Chang, W. S. (2012). Effect of soybean coumestrol on Bradyrhizobium japonicum nodulation ability, biofilm formation, and transcriptional profile. Appl. Environ. Microbiol. 78, 2896–2903. doi: 10.1128/AEM.07336-11
Lee, E. J., Song, M. C., Rha, C. S. (2022). Mass biosynthesis of coumestrol derivatives and their isomers via soybean adventitious root cultivation in bioreactors. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.923163
Le Roy, J., Huss, B., Creach, A., Hawkins, S., Neutelings, G. (2016). Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00735
Li, Z. S., Alfenito, M., Rea, P. A., Walbot, V., Dixon, R. A. (1997). Vacuolar uptake of the phytoalexin medicarpin by the glutathione conjugate pump. Phytochemistry 45, 689–693. doi: 10.1016/S0031-9422(97)00031-9
Li, X., Chen, L., Dhaubhadel, S. (2012). 14–3-3 proteins regulate the intracellular localization of the transcriptional activator GmMYB176 and affect isoflavonoid synthesis in soybean. Plant J. 71, 239–250. doi: 10.1111/j.1365-313X.2012.04986.x
Li, P., Chen, B., Zhang, G., Chen, L., Dong, Q., Wen, J., et al. (2016b). Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 210, 905–921. doi: 10.1111/nph.13816
Li, X., Dhaubhadel, S. (2012). 14–3-3 proteins act as scaffolds for GmMYB62 and GmMYB176 and regulate their intracellular localization in soybean. Plant Signal Behav. 7, 965–968. doi: 10.4161/psb.20940
Li, P., Dong, Q., Ge, S., He, X., Verdier, J., Li, D., et al. (2016c). Metabolic engineering of proanthocyanidin production by repressing the isoflavone pathways and redirecting anthocyanidin precursor flux in legume. Plant Biotechnol. J. 14, 1604–1618. doi: 10.1111/pbi.12524
Li, J., Li, C., Gou, J., Zhang, Y. (2016a). Molecular cloning and functional characterization of a novel isoflavone 3’-O-methyltransferase from Pueraria lobata. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00793
Li, J., Li, Z., Li, C., Gou, J., Zhang, Y. (2014). Molecular cloning and characterization of an isoflavone 7-O-glucosyltransferase from Pueraria lobata. Plant Cell Rep. 33, 1173–1185. doi: 10.1007/s00299-014-1606-7
Li, X., Qin, J. C., Wang, Q. Y., Wu, X., Lang, C. Y., Pan, H. Y., et al. (2011). Metabolic engineering of isoflavone genistein in Brassica napus with soybean isoflavone synthase. Plant Cell Rep. 30, 1435–1442. doi: 10.1007/s00299-011-1052-8
Li, X., Yang, C., Chen, J., He, Y., Deng, J., Xie, C., et al. (2021b). Changing light promotes isoflavone biosynthesis in soybean pods and enhances their resistance to mildew infection. Plant Cell Environ. 44, 2536–2550. doi: 10.1111/pce.14128
Li, A., Zhao, Z., Zhang, S., Zhang, Z., Shi, Y. (2021a). Fungicidal activity and mechanism of action of glabridin from Glycyrrhiza glabra L. Int. J. Mol. Sci. 22, 10966. doi: 10.3390/ijms222010966
Liu, C. J., Blount, J. W., Steele, C. L., Dixon, R. A. (2002). Bottlenecks for metabolic engineering of isoflavone glycoconjugates in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99, 14578–14583. doi: 10.1073/pnas.212522099
Liu, C. J., Deavours, B. E., Richard, S. B., Ferrer, J. L., Blount, J. W., Huhman, D., et al. (2006). Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses. Plant Cell 18, 3656–3669. doi: 10.1105/tpc.106.041376
Liu, C. J., Dixon, R. A. (2001). Elicitor-induced association of isoflavone O-methyltransferase with endomembranes prevents the formation and 7-O-methylation of daidzein during isoflavonoid phytoalexin biosynthesis. Plant Cell 13, 2643–2658. doi: 10.1105/tpc.010382
Liu, R., Hu, Y., Li, J., Lin, Z. (2007). Production of soybean isoflavone genistein in non-legume plants via genetically modified secondary metabolism pathway. Metab. Eng. 9, 1–7. doi: 10.1016/j.ymben.2006.08.003
Liu, Q., Liu, Y., Li, G., Savolainen, O., Chen, Y., Nielsen, J. (2021). De novo biosynthesis of bioactive isoflavonoids by engineered yeast cell factories. Nat. Commun. 12, 6085. doi: 10.1038/s41467-021-26361-1
Liu, Y., Singh, S. K., Pattanaik, S., Wang, H., Yuan, L. (2023). Light regulation of the biosynthesis of phenolics, terpenoids, and alkaloids in plants. Commun. Biol. 6, 1055. doi: 10.1038/s42003-023-05435-4
Liu, X., Yuan, L., Xu, L., Xu, Z., Huang, Y., He, X., et al. (2013). Over-expression of GmMYB39 leads to an inhibition of the isoflavonoid biosynthesis in soybean (Glycine max. L). Plant Biotechnol. Rep. 7, 445–455. doi: 10.1007/s11816-013-0283-2
Lygin, A. V., Hill, C. B., Zernova, O. V., Crull, L., Widholm, J. M., Hartman, G. L., et al. (2010). Response of soybean pathogens to glyceollin. Phytopathology 100, 897–903. doi: 10.1094/PHYTO-100-9-0897
Mackova, Z., Koblovska, R., Lapcik, O. (2006). Distribution of isoflavonoids in non-leguminous taxa - an update. Phytochemistry 67, 849–855. doi: 10.1016/j.phytochem.2006.01.020
Martin, M., Dewick, P. M. (1980). Biosynthesis of pterocarpan, isoflavan and coumestan metabolites of Medicago sativa: The role of an isoflav-3-ene. Phytochemistry 19, 2341–2346. doi: 10.1016/S0031-9422(00)91023-9
Mathesius, U. (2001). Flavonoids induced in cells undergoing nodule organogenesis in white clover are regulators of auxin breakdown by peroxidase. J. Exp. Bot. 52, 419–426. doi: 10.1093/jexbot/52.suppl_1.419
Mipeshwaree Devi, A., Khedashwori Devi, K., Premi Devi, P., Lakshmipriyari Devi, M., Das, S. (2023). Metabolic engineering of plant secondary metabolites: prospects and its technological challenges. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1171154
Monje-Rueda, M. D., Pal’ove-Balang, P., Trush, K., Marquez, A. J., Betti, M., Garcia-Calderon, M. (2023). Mutation of MYB36 affects isoflavonoid metabolism, growth, and stress responses in Lotus japonicus. Physiol. Plant 175, e14084. doi: 10.1111/ppl.14084
Mouffouk, S., Marcourt, L., Benkhaled, M., Boudiaf, K., Wolfender, J.-L., Haba, H. (2017). Two new prenylated isoflavonoids from Erinacea anthyllis with antioxidant and antibacterial activities. Natural Product Commun. 12, 1934578X1701200716. doi: 10.1177/1934578X1701200716
Mukne, A. P., Viswanathan, V., Phadatare, A. G. (2011). Structure pre-requisites for isoflavones as effective antibacterial agents. Pharmacogn Rev. 5, 13–18. doi: 10.4103/0973-7847.79095
Mun, B. G., Kim, H. H., Yuk, H. J., Hussain, A., Loake, G. J., Yun, B. W. (2021). A potential role of coumestrol in soybean leaf senescence and its interaction with phytohormones. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.756308
Muro-Villanueva, F., Mao, X., Chapple, C. (2019). Linking phenylpropanoid metabolism, lignin deposition, and plant growth inhibition. Curr. Opin. Biotechnol. 56, 202–208. doi: 10.1016/j.copbio.2018.12.008
Naguib, A., Mathew, G., Reczek, C. R., Watrud, K., Ambrico, A., Herzka, T., et al. (2018). Mitochondrial complex I inhibitors expose a vulnerability for selective killing of PTEN-null cells. Cell Rep. 23, 58–67. doi: 10.1016/j.celrep.2018.03.032
Nakamura, K., Akashi, T., Aoki, T., Kawaguchi, K., Ayabe, S. (1999). Induction of isoflavonoid and retrochalcone branches of the flavonoid pathway in cultured Glycyrrhiza eChinata cells treated with yeast extract. Biosci. Biotechnol. Biochem. 63, 1618–1620. doi: 10.1271/bbb.63.1618
Naoumkina, M., Dixon, R. A. (2008). Subcellular localization of flavonoid natural products: a signaling function? Plant Signaling Behav. 3, 573–575. doi: 10.4161/psb.3.8.5731
Naoumkina, M., Farag, M. A., Sumner, L. W., Tang, Y., Liu, C. J., Dixon, R. A. (2007). Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 104, 17909–17915. doi: 10.1073/pnas.0708697104
Ng, J. L., Hassan, S., Truong, T. T., Hocart, C. H., Laffont, C., Frugier, F., et al. (2015). Flavonoids and auxin transport inhibitors rescue symbiotic nodulation in the Medicago truncatula cytokinin perception mutant cre1. Plant Cell 27, 2210–2226. doi: 10.1105/tpc.15.00231
Ng, M. S., Ku, Y. S., Yung, W. S., Cheng, S. S., Man, C. K., Yang, L., et al. (2021). MATE-type proteins are responsible for isoflavone transportation and accumulation in soybean seeds. Int. J. Mol. Sci. 22, 12017. doi: 10.3390/ijms222112017
Noguchi, A., Saito, A., Homma, Y., Nakao, M., Sasaki, N., Nishino, T., et al. (2007). A UDP-glucose:isoflavone 7-O-glucosyltransferase from the roots of soybean (Glycine max) seedlings. Purification, gene cloning, phylogenetics, and an implication for an alternative strategy of enzyme catalysis. J. Biol. Chem. 282, 23581–23590. doi: 10.1074/jbc.M702651200
Ohta, T., Uto, T., Tanaka, H. (2021). Effective methods for increasing coumestrol in soybean sprouts. PloS One 16, e0260147. doi: 10.1371/journal.pone.0260147
Oommen, A., Dixon, R. A., Paiva, N. L. (1994). The elicitor-inducible alfalfa isoflavone reductase promoter confers different patterns of developmental expression in homologous and heterologous transgenic plants. Plant Cell 6, 1789–1803. doi: 10.1105/tpc.6.12.1789
Paiva, N. L., Edwards, R., Sun, Y. J., Hrazdina, G., Dixon, R. A. (1991). Stress responses in alfalfa (Medicago sativa L.) 11. Molecular cloning and expression of alfalfa isoflavone reductase, a key enzyme of isoflavonoid phytoalexin biosynthesis. Plant Mol. Biol. 17, 653–667. doi: 10.1007/BF00037051
Pandey, A., Misra, P., Khan, M. P., Swarnkar, G., Tewari, M. C., Bhambhani, S., et al. (2014). Co-expression of Arabidopsis transcription factor, AtMYB12, and soybean isoflavone synthase, GmIFS1, genes in tobacco leads to enhanced biosynthesis of isoflavones and flavonols resulting in osteoprotective activity. Plant Biotechnol. J. 12, 69–80. doi: 10.1111/pbi.12118
Perez Rojo, F., Pillow, J. J., Kaur, P. (2023). Bioprospecting microbes and enzymes for the production of pterocarpans and coumestans. Front. Bioeng. Biotechnol. 11. doi: 10.3389/fbioe.2023.1154779
Piślewska, M., Bednarek, P., Stobiecki, M., Zieliśska, M., Wojtaszek, P. (2002). Cell wall-associated isoflavonoids and β-glucosidase activity in Lupinus albus plants responding to environmental stimuli. Plant Cell Environ. 25, 20–40. doi: 10.1046/j.0016-8025.2001.00808.x
Picmanov, M., Honys, D., Koblovsk, R., Lapcik, O. (2012). “Isoflavone synthase genes in legumes and non-leguminous plants: isoflavone synthase,” in 2012 International Conference on Biomedical Engineering and Biotechnology. (Macau, Macao: IEEE). 344–347. doi: 10.1109/iCBEB.2012.257
Pistelli, L., Noccioli, C., Appendino, G., Bianchi, F., Sterner, O., Ballero, M. (2003). Pterocarpans from Bituminaria morisiana and Bituminaria bituminosa. Phytochemistry 64, 595–598. doi: 10.1016/S0031-9422(03)00190-0
Pistelli, L., Ulivieri, V., Giovanelli, S., Avio, L., Giovannetti, M., Pistelli, L. (2017). Arbuscular mycorrhizal fungi alter the content and composition of secondary metabolites in Bituminaria bituminosa L. Plant Biol. (Stuttg) 19, 926–933. doi: 10.1111/plb.12608
Polturak, G., Misra, R. C., El-Demerdash, A., Owen, C., Steed, A., McDonald, H. P., et al. (2023). Discovery of isoflavone phytoalexins in wheat reveals an alternative route to isoflavonoid biosynthesis. Nat. Commun. 14, 6977. doi: 10.1038/s41467-023-42464-3
Preston, S., Korhonen, P. K., Mouchiroud, L., Cornaglia, M., McGee, S. L., Young, N. D., et al. (2017). Deguelin exerts potent nematocidal activity via the mitochondrial respiratory chain. FASEB J. 31, 4515–4532. doi: 10.1096/fj.201700288R
Qin, W., Yang, Y., Wang, Y., Zhang, X., Liu, X. (2022). Transcriptomic and metabolomic analysis reveals the difference between large and small flower taxa of Herba Epimedii during flavonoid accumulation. Sci. Rep. 12, 2762. doi: 10.1038/s41598–022-06761-z
Qiu, S., Cai, Y., Yao, H., Lin, C., Xie, Y., Tang, S., et al. (2023). Small molecule metabolites: discovery of biomarkers and therapeutic targets. Signal Transduct Target Ther. 8, 132. doi: 10.1038/s41392-023-01399-3
Ralston, L., Subramanian, S., Matsuno, M., Yu, O. (2005). Partial reconstruction of flavonoid and isoflavonoid biosynthesis in yeast using soybean type I and type II chalcone isomerases. Plant Physiol. 137, 1375–1388. doi: 10.1104/pp.104.054502
Reynaud, J., Guilet, D., Terreux, R., Lussignol, M., Walchshofer, N. (2005). Isoflavonoids in non-leguminous families: an update. Nat. Prod Rep. 22, 504–515. doi: 10.1039/b416248j
Rodriguez, A., Strucko, T., Stahlhut, S. G., Kristensen, M., Svenssen, D. K., Forster, J., et al. (2017). Metabolic engineering of yeast for fermentative production of flavonoids. Bioresour Technol. 245, 1645–1654. doi: 10.1016/j.biortech.2017.06.043
Russell, D. A., Bridges, H. R., Serreli, R., Kidd, S. L., Mateu, N., Osberger, T. J., et al. (2020). Hydroxylated rotenoids selectively inhibit the proliferation of prostate cancer cells. J. Nat. Prod 83, 1829–1845. doi: 10.1021/acs.jnatprod.9b01224
Sarkar, M. A. R., Watanabe, S., Suzuki, A., Hashimoto, F., Anai, T. (2019). Identification of novel MYB transcription factors involved in the isoflavone biosynthetic pathway by using the combination screening system with agroinfiltration and hairy root transformation. Plant Biotechnol. (Tokyo) 36, 241–251. doi: 10.5511/plantbiotechnology.19.1025a
Saunders, J., O’neill, N. (2004). The characterization of defense responses to fungal infection in alfalfa. BioControl 49, 715–728. doi: 10.1007/s10526-004-5281-4
Saxena, S., Pal, L., Naik, J., Singh, Y., Verma, P. K., Chattopadhyay, D., et al. (2023). The R2R3-MYB-SG7 transcription factor CaMYB39 orchestrates surface phenylpropanoid metabolism and pathogen resistance in chickpea. New Phytol. 238, 798–816. doi: 10.1111/nph.18758
Shelton, D., Stranne, M., Mikkelsen, L., Pakseresht, N., Welham, T., Hiraka, H., et al. (2012). Transcription factors of Lotus: regulation of isoflavonoid biosynthesis requires coordinated changes in transcription factor activity. Plant Physiol. 159, 531–547. doi: 10.1104/pp.112.194753
Shen, G., Huhman, D., Lei, Z., Snyder, J., Sumner, L. W., Dixon, R. A. (2012). Characterization of an isoflavonoid-specific prenyltransferase from Lupinus albus. Plant Physiol. 159, 70–80. doi: 10.1104/pp.112.195271
Shih, C. H., Chen, Y., Wang, M., Chu, I. K., Lo, C. (2008). Accumulation of isoflavone genistin in transgenic tomato plants overexpressing a soybean isoflavone synthase gene. J. Agric. Food Chem. 56, 5655–5661. doi: 10.1021/jf800423u
Shimada, N., Akashi, T., Aoki, T., Ayabe, S. (2000). Induction of isoflavonoid pathway in the model legume Lotus japonicus: molecular characterization of enzymes involved in phytoalexin biosynthesis. Plant Sci. 160, 37–47. doi: 10.1016/S0168-9452(00)00355-1
Shimada, N., Aoki, T., Sato, S., Nakamura, Y., Tabata, S., Ayabe, S. (2003). A cluster of genes encodes the two types of chalcone isomerase involved in the biosynthesis of general flavonoids and legume-specific 5-deoxy(iso)flavonoids in Lotus japonicus. Plant Physiol. 131, 941–951. doi: 10.1104/pp.004820
Shimada, N., Sato, S., Akashi, T., Nakamura, Y., Tabata, S., Ayabe, S.-i., et al. (2007). Genome-wide analyses of the structural gene families involved in the legume-specific 5-deoxyisoflavonoid biosynthesis of Lotus japonicus. DNA Res. 14, 25–36. doi: 10.1093/dnares/dsm004
Shimamura, M., Akashi, T., Sakurai, N., Suzuki, H., Saito, K., Shibata, D., et al. (2007). 2-Hydroxyisoflavanone dehydratase is a critical determinant of isoflavone productivity in hairy root cultures of Lotus japonicus. Plant Cell Physiol. 48, 1652–1657. doi: 10.1093/pcp/pcm125
Simmler, C., Pauli, G. F., Chen, S.-N. (2013). Phytochemistry and biological properties of glabridin. Fitoterapia 90, 160–184. doi: 10.1016/j.fitote.2013.07.003
Sohn, S. I., Pandian, S., Oh, Y. J., Kang, H. J., Cho, W. S., Cho, Y. S. (2021). Metabolic engineering of isoflavones: an updated overview. Front. Plant Sci. 12, 670103. doi: 10.3389/fpls.2021.670103
Song, Z., Zhao, F., Chu, L., Lin, H., Xiao, Y., Fang, Z., et al. (2023). The GmSTF1/2-GmBBX4 negative feedback loop acts downstream of blue-light photoreceptors to regulate isoflavonoid biosynthesis in soybean. Plant Commun. 5, 100730. doi: 10.1016/j.xplc.2023.100730
Sreevidya, V. S., Srinivasa Rao, C., Sullia, S. B., Ladha, J. K., Reddy, P. M. (2006). Metabolic engineering of rice with soybean isoflavone synthase for promoting nodulation gene expression in rhizobia. J. Exp. Bot. 57, 1957–1969. doi: 10.1093/jxb/erj143
Su, L., Liu, S., Liu, X., Zhang, B., Li, M., Zeng, L., et al. (2021). Transcriptome profiling reveals histone deacetylase 1 gene overexpression improves flavonoid, isoflavonoid, and phenylpropanoid metabolism in Arachis hypogaea hairy roots. PeerJ 9, e10976. doi: 10.7717/peerj.10976
Subramanian, S., Stacey, G., Yu, O. (2006). Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant J. 48, 261–273. doi: 10.1111/j.1365-313X.2006.02874.x
Sugiyama, A., Fukuda, S., Takanashi, K., Yoshioka, M., Yoshioka, H., Narusaka, Y., et al. (2015). Molecular characterization of LjABCG1, an ATP-binding cassette protein in Lotus japonicus. PloS One 10, e0139127. doi: 10.1371/journal.pone.0139127
Sugiyama, A., Shitan, N., Yazaki, K. (2007). Involvement of a soybean ATP-binding cassette-type transporter in the secretion of genistein, a signal flavonoid in legume-rhizobium symbiosis. Plant Physiol. 144, 2000–2008. doi: 10.1104/pp.107.096727
Sugiyama, A., Shitan, N., Yazaki, K. (2008). Signaling from soybean roots to Rhizobium: An ATP-binding cassette-type transporter mediates genistein secretion. Plant Signaling Behav. 3, 38–40. doi: 10.4161/psb.3.1.4819
Sukumaran, A., McDowell, T., Chen, L., Renaud, J., Dhaubhadel, S. (2018). Isoflavonoid-specific prenyltransferase gene family in soybean: GmPT01, a pterocarpan 2-dimethylallyltransferase involved in glyceollin biosynthesis. Plant J. 96, 966–981. doi: 10.1111/tpj.14083
Sulis, D. B., Jiang, X., Yang, C., Marques, B. M., Matthews, M. L., Miller, Z., et al. (2023). Multiplex CRISPR editing of wood for sustainable fiber production. Science 381, 216–221. doi: 10.1126/science.add4514
Suzuki, H., Takahashi, S., Watanabe, R., Fukushima, Y., Fujita, N., Noguchi, A., et al. (2006). An isoflavone conjugate-hydrolyzing beta-glucosidase from the roots of soybean (Glycine max) seedlings: purification, gene cloning, phylogenetics, and cellular localization. J. Biol. Chem. 281, 30251–30259. doi: 10.1074/jbc.M605726200
Tian, L., Dixon, R. A. (2006). Engineering isoflavone metabolism with an artificial bifunctional enzyme. Planta 224, 496–507. doi: 10.1007/s00425-006-0233-0
Trantas, E., Panopoulos, N., Ververidis, F. (2009). Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metab. Eng. 11, 355–366. doi: 10.1016/j.ymben.2009.07.004
Tripathi, P., Rabara, R. C., Reese, R. N., Miller, M. A., Rohila, J. S., Subramanian, S., et al. (2016). A toolbox of genes, proteins, metabolites and promoters for improving drought tolerance in soybean includes the metabolite coumestrol and stomatal development genes. BMC Genomics 17, 102. doi: 10.1186/s12864-016-2420-0
Trush, K., Eliašová, A., Monje-Rueda, M., Kolarčík, V., Betti, M., Paľove-Balang, P. (2023). Chitosan is involved in elicitation of vestitol production in Lotus japonicus. Biol. plantarum 67, 75–86. doi: 10.32615/bp.2023.007
Tsao, R., Papadopoulos, Y., Yang, R., Young, J. C., McRae, K. (2006). Isoflavone profiles of red clovers and their distribution in different parts harvested at different growing stages. J. Agric. Food Chem. 54, 5797–5805. doi: 10.1021/jf0614589
Uchida, K., Akashi, T., Aoki, T. (2017). The missing link in leguminous pterocarpan biosynthesis is a dirigent domain-containing protein with isoflavanol dehydratase activity. Plant Cell Physiol. 58, 398–408. doi: 10.1093/pcp/pcw213
Uchida, K., Aoki, T., Suzuki, H., Akashi, T. (2020). Molecular cloning and biochemical characterization of isoflav-3-ene synthase, a key enzyme of the biosyntheses of (+)-pisatin and coumestrol. Plant Biotechnol. (Tokyo) 37, 301–310. doi: 10.5511/plantbiotechnology.20.0421a
Ueda, H., Sugimoto, Y. (2010). Vestitol as a chemical barrier against intrusion of parasitic plant Striga hermonthica into Lotus japonicus roots. Biosci. biotechnol. Biochem. 74, 1662–1667. doi: 10.1271/bbb.100285
Veitch, N. C. (2009). Isoflavonoids of the leguminosae. Natural Product Rep. 26, 776–802. doi: 10.1039/b616809b
Veitch, N. C. (2013). Isoflavonoids of the leguminosae. Nat. Prod Rep. 30, 988–1027. doi: 10.1039/c3np70024k
Verdier, J., Zhao, J., Torres-Jerez, I., Ge, S., Liu, C., He, X., et al. (2012). MtPAR MYB transcription factor acts as an on switch for proanthocyanidin biosynthesis in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 109, 1766–1771. doi: 10.1073/pnas.1120916109
Wada, N., Osakabe, K., Osakabe, Y. (2022). Expanding the plant genome editing toolbox with recently developed CRISPR-Cas systems. Plant Physiol. 188, 1825–1837. doi: 10.1093/plphys/kiac027
Wang, J., Li, L., Wang, Z., Feng, A., Li, H., Qaseem, M. F., et al. (2023a). Integrative analysis of the metabolome and transcriptome reveals the molecular regulatory mechanism of isoflavonoid biosynthesis in Ormosia henryi Prain. Int. J. Biol. Macromol. 246, 125601. doi: 10.1016/j.ijbiomac.2023.125601
Wang, X., Li, C., Zhou, C., Li, J., Zhang, Y. (2017). Molecular characterization of the C-glucosylation for puerarin biosynthesis in Pueraria lobata. Plant J. 90, 535–546. doi: 10.1111/tpj.13510
Wang, Y., Luo, Y., Zhang, Y., Xu, C., Yao, Y., Gu, J., et al. (2023b). A novel flavonoid prenyltransferase gene PcPT11 with broad substrate promiscuity in Psoralea corylifolia L. Ind. Crops Products 199, 116746. doi: 10.1016/j.indcrop.2023.116746
Wang, X., Pan, H., Sagurthi, S., Paris, V., Zhuo, C., Dixon, R. A. (2022). The protein conformational basis of isoflavone biosynthesis. Commun. Biol. 5, 1249. doi: 10.1038/s42003-022-04222-x
Wasson, A. P., Pellerone, F. I., Mathesius, U. (2006). Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18, 1617–1629. doi: 10.1105/tpc.105.038232
Wen, D., Wu, L., Wang, M., Yang, W., Wang, X., Ma, W., et al. (2022). CRISPR/Cas9-mediated targeted mutagenesis of FtMYB45 promotes flavonoid biosynthesis in tartary buckwheat (Fagopyrum tataricum). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.879390
Wong, J., Gao, L., Yang, Y., Zhai, J., Arikit, S., Yu, Y., et al. (2014). Roles of small RNAs in soybean defense against Phytophthora sojae infection. Plant J. 79, 928–940. doi: 10.1111/tpj.12590
Xu, D. (2020). COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol. 228, 1748–1753. doi: 10.1111/nph.16296
Yadav, V., Wang, Z., Wei, C., Amo, A., Ahmed, B., Yang, X., et al. (2020). Phenylpropanoid pathway engineering: an emerging approach towards plant defense. Pathogens 9, 312. doi: 10.3390/pathogens9040312
Yan, J., Wang, B., Zhong, Y., Yao, L., Cheng, L., Wu, T. (2015). The soybean R2R3 MYB transcription factor GmMYB100 negatively regulates plant flavonoid biosynthesis. Plant Mol. Biol. 89, 35–48. doi: 10.1007/s11103-015-0349-3
Yang, S., Chen, R., Cao, X., Wang, G., Zhou, Y. J. (2024). De novo biosynthesis of the hops bioactive flavonoid xanthohumol in yeast. Nat. Commun. 15, 253. doi: 10.1038/s41467-023-44654-5
Yang, C., Xie, L., Ma, Y., Cai, X., Yue, G., Qin, G., et al. (2021). Study on the fungicidal mechanism of glabridin against Fusarium graminearum. Pesticide Biochem. Physiol. 179, 104963. doi: 10.1016/j.pestbp.2021.104963
Yi, J., Derynck, M. R., Li, X., Telmer, P., Marsolais, F., Dhaubhadel, S. (2010). A single-repeat MYB transcription factor, GmMYB176, regulates CHS8 gene expression and affects isoflavonoid biosynthesis in soybean. Plant J. 62, 1019–1034. doi: 10.1111/j.1365-313X.2010.04214.x
Yoneyama, K., Akashi, T., Aoki, T. (2016). Molecular characterization of soybean pterocarpan 2-dimethylallyltransferase in glyceollin biosynthesis: Local gene and whole-genome duplications of prenyltransferase genes led to the structural diversity of soybean prenylated isoflavonoids. Plant Cell Physiol. 57, 2497–2509. doi: 10.1093/pcp/pcw178
Yu, X. H., Chen, M. H., Liu, C. J. (2008). Nucleocytoplasmic-localized acyltransferases catalyze the malonylation of 7-O-glycosidic (iso)flavones in Medicago truncatula. Plant J. 55, 382–396. doi: 10.1111/j.1365-313X.2008.03509.x
Yu, O., McGonigle, B. (2005). Metabolic engineering of isoflavone biosynthesis. Adv. Agron. 86, 147–190. doi: 10.1016/S0065-2113(05)86003-1
Yu, L., Rios, E., Castro, L., Liu, J., Yan, Y., Dixon, D. (2021). Genistein: dual role in women’s health. Nutrients 13, 3048. doi: 10.3390/nu13093048
Yu, O., Shi, J., Hession, A. O., Maxwell, C. A., McGonigle, B., Odell, J. T. (2003). Metabolic engineering to increase isoflavone biosynthesis in soybean seed. Phytochemistry 63, 753–763. doi: 10.1016/S0031-9422(03)00345-5
Yuk, H. J., Lee, J. H., Curtis-Long, M. J., Lee, J. W., Kim, Y. S., Ryu, H. W., et al. (2011). The most abundant polyphenol of soy leaves, coumestrol, displays potent α-glucosidase inhibitory activity. Food Chem. 126, 1057–1063. doi: 10.1016/j.foodchem.2010.11.125
Yuk, H. J., Song, Y. H., Curtis-Long, M. J., Kim, D. W., Woo, S. G., Lee, Y. B., et al. (2016). Ethylene induced a high accumulation of dietary isoflavones and expression of isoflavonoid biosynthetic genes in soybean (Glycine max) leaves. J. Agric. Food Chem. 64, 7315–7324. doi: 10.1021/acs.jafc.6b02543
Zhang, P., Du, H., Wang, J., Pu, Y., Yang, C., Yan, R., et al. (2020). Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soyabean mosaic virus. Plant Biotechnol. J. 18, 1384–1395. doi: 10.1111/pbi.13302
Zhang, Z., Gao, L., Ke, M., Gao, Z., Tu, T., Huang, L., et al. (2022b). GmPIN1-mediated auxin asymmetry regulates leaf petiole angle and plant architecture in soybean. J. Integr. Plant Biol. 64, 1325–1338. doi: 10.1111/jipb.13269
Zhang, J., Liu, L., Wang, J., Ren, B., Zhang, L., Li, W. (2018). Formononetin, an isoflavone from Astragalus membranaceus inhibits proliferation and metastasis of ovarian cancer cells. J. Ethnopharmacol. 221, 91–99. doi: 10.1016/j.jep.2018.04.014
Zhang, P., Zhang, M., Mellich, T. A., Pearson, B. J., Chen, J., Zhang, Z. (2022a). Variation in rotenone and deguelin contents among strains across four tephrosia species and their activities against aphids and whiteflies. Toxins (Basel) 14, 339. doi: 10.3390/toxins14050339
Zhao, J. (2015). Flavonoid transport mechanisms: how to go, and with whom. Trends Plant Sci. 20, 576–585. doi: 10.1016/j.tplants.2015.06.007
Zhao, J., Dixon, R. A. (2010). The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci. 15, 72–80. doi: 10.1016/j.tplants.2009.11.006
Zhao, J., Huhman, D., Shadle, G., He, X. Z., Sumner, L. W., Tang, Y., et al. (2011). MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. Plant Cell 23, 1536–1555. doi: 10.1105/tpc.110.080804
Zhao, M., Wang, T., Wu, P., Guo, W., Su, L., Wang, Y., et al. (2017). Isolation and characterization of GmMYBJ3, an R2R3-MYB transcription factor that affects isoflavonoids biosynthesis in soybean. PloS One 12, e0179990. doi: 10.1371/journal.pone.0179990
Zhou, Y., Olt, P., Neuhäuser, B., Moradtalab, N., Bautista, W., Uhde-Stone, C., et al. (2021). Loss of LaMATE impairs isoflavonoid release from cluster roots of phosphorus-deficient white lupin. Physiol. Plant 173, 1207–1220. doi: 10.1111/ppl.13515
Keywords: isoflavonoids, biosynthesis, metabolic engineering, bio-function, regulation
Citation: Wang L, Li C and Luo K (2024) Biosynthesis and metabolic engineering of isoflavonoids in model plants and crops: a review. Front. Plant Sci. 15:1384091. doi: 10.3389/fpls.2024.1384091
Received: 08 February 2024; Accepted: 10 June 2024;
Published: 25 June 2024.
Edited by:
Deyu Xie, North Carolina State University, United StatesReviewed by:
Juan Guo, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaHiroshi A. Maeda, University of Wisconsin-Madison, United States
Copyright © 2024 Wang, Li and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chaofeng Li, Y2ZsaTE5ODhjYXNAc3d1LmVkdS5jbg==; Keming Luo, a2VtaW5nbEBzd3UuZWR1LmNu
 Lijun Wang
Lijun Wang Chaofeng Li
Chaofeng Li Keming Luo
Keming Luo