- 1Key Laboratory of Agricultural Water Resources, Hebei Key Laboratory of Soil Ecology, Center for Agricultural Resources Research, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Shijiazhuang, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3State Key Laboratory of Nutrient Use and Management, College of Resources and Environmental Sciences, National Academy of Agriculture Green Development, China Agricultural University, Beijing, China
- 4Key Laboratory of Plant-Soil Interactions, Ministry of Education, China Agricultural University, Beijing, China
- 5National Observation and Research Station of Agriculture Green Development, Quzhou, China
Soil salinization poses a critical problem, adversely affecting plant development and sustainable agriculture. Plants can produce soil legacy effects through interactions with the soil environments. Salt tolerance of plants in saline soils is not only determined by their own stress tolerance but is also closely related to soil legacy effects. Creating positive soil legacy effects for crops, thereby alleviating crop salt stress, presents a new perspective for improving soil conditions and increasing productivity in saline farmlands. Firstly, the formation and role of soil legacy effects in natural ecosystems are summarized. Then, the processes by which plants and soil microbial assistance respond to salt stress are outlined, as well as the potential soil legacy effects they may produce. Using this as a foundation, proposed the application of salt tolerance mechanisms related to soil legacy effects in natural ecosystems to saline farmlands production. One aspect involves leveraging the soil legacy effects created by plants to cope with salt stress, including the direct use of halophytes and salt-tolerant crops and the design of cropping patterns with the specific crop functional groups. Another aspect focuses on the utilization of soil legacy effects created synergistically by soil microorganisms. This includes the inoculation of specific strains, functional microbiota, entire soil which legacy with beneficial microorganisms and tolerant substances, as well as the application of novel technologies such as direct use of rhizosphere secretions or microbial transmission mechanisms. These approaches capitalize on the characteristics of beneficial microorganisms to help crops against salinity. Consequently, we concluded that by the screening suitable salt-tolerant crops, the development rational cropping patterns, and the inoculation of safe functional soils, positive soil legacy effects could be created to enhance crop salt tolerance. It could also improve the practical significance of soil legacy effects in the application of saline farmlands.
1 Introduction
In recent years, land degradation caused by climate change has posed a huge challenge to agricultural production. In the absence of major technological breakthroughs in agriculture, existing arable land resources are hardly sufficient to support global food security (German et al., 2017; Hartmann and Six, 2023). Saline farmland is an important reserve resource of arable land with great potential for ensuring food security and sustainable agricultural development (Negacz et al., 2022). Therefore, finding solutions to increase the productivity of saline farmland and improve crop tolerance to saline stress has become an important research topic currently (Munns and Tester, 2008).
Soil salinization is a global environmental problem, with more than 833 million hectares of soil and more than 10% of farmland affected by salinization (FAO, 2021), causing at least 25% of crops to suffer from varying degrees of yield loss due to persistent salt stress, with a serious impact on food security (Farooq et al., 2017; Kumar et al., 2022). Soil salinization leads to reduced crop yield because the significant negative impacts on seed germination by disrupting the membrane permeability of the seed embryo and increasing the osmotic stress on seeds (Deng et al., 2014). For salt-sensitive crops, seed germination rate, germination time, and the length of the plumule are all affected by salt stress (Abbas et al., 2012). Persistent salt stress during the crop growth phase leads to crop water loss and ion toxicity due to increased cellular osmotic pressure and disruption of cell membranes (Läuchli and Grattan, 2007). Salt stress also reduces nutrient uptake by inhibiting crop root growth (Burssens et al., 2000; West et al., 2004), inhibits photosynthesis by decreasing the crop’s leaf area (Hu et al., 2022), and ultimately affects crop yield and quality.
Moreover, the survival of microorganisms is directly associated with plant and soil environments (Pulleman et al., 2012). Salt stress can reduce the abundance and activity of soil microbial communities (Rietz and Haynes, 2003), affecting the composition of functional soil microbes (Zhang et al., 2019), and disrupting the stability of microbial networks (Li et al., 2023a). This disruption affects nutrient cycling (Bai et al., 2012) and material utilization (Elmajdoub and Marschner, 2013) ultimately affecting the ecological functions of soil microbial communities (Zhang et al., 2023). Weakened ecological functions of microbial community, in turn, affect plant-microbe interactions (Etesami and Beattie, 2017), as manifested by reduced microbial colonization (Li et al., 2023a) and impaired plant growth (Jansson et al., 2023).
Both plants and soil microorganisms have developed specific abilities and mutualistic associations to cope with various stresses (Zhao et al., 2020; Liu et al., 2022). Halophytes and salt-tolerant plants, as the dominant vegetation in saline environments, are better adapted to saline stresses and have formed unique strategies improving their adaptability through such pathways as salt gland excretion (Yuan F. et al., 2016), ionic and osmotic regulation (Zhu, 2016), antioxidant defenses (Apse and Blumwald, 2002) and root structural modifications (Yu et al., 2022). Soil microorganisms also have various salt-tolerance strategies, such as salt accumulation and synthesis of organic osmotic material to adapt to high-salt environments (Gunde-Cimerman et al., 2018). Meanwhile, beneficial microorganisms can influence performance of their host plants under harsh conditions (Wang and Song, 2022). For example, arbuscular mycorrhizal fungi can help host plants to cope with abiotic stresses like drought, salt, etc., by improving plant water utilization, regulating photosynthesis and maintaining osmotic balance (Borde et al., 2017).
In addition, soil legacy effects are microbiological and functional substance traits retained in the soil by the plants, which influence the growth of succeeding plants (Van der Putten et al., 2013). The formation of soil legacy effects is the process of plant-microbe interactions in which plants respond to stressful stimuli and mobilize the required metabolites and functional microorganisms, thus promoting the growth of their own and succeeding plants as well as increasing their tolerance (Bakker et al., 2018). So, the application of soil legacy effects may also help to refine the way we cultivate and manage crops for agricultural production (Mariotte et al., 2018; Carrión et al., 2019; Cordovez et al., 2019). Therefore, based on the theoretical foundation of soil legacy effects in natural ecosystems, it is important to further explore the mechanism of crop-soil-microbe interactions in saline farmlands, which has profound implication for mitigating crop salt stress, increasing crop productivity and improving the environment of saline farmlands (Vukicevich et al., 2016).
2 Formation and role of soil legacy effects in natural ecosystems
In natural ecosystems, plants and soil organisms have various effects to soil legacy (Wardle et al., 2004; Faucon et al., 2017). Plant species with different root structures, growth habits and ways of interacting with soil organisms have important impacts on soil legacy effects (Oliver et al., 2021), while plant species composition and diversity also significantly modify such effects at the community level (Kowalchuk et al., 2002; Lange et al., 2015). Soil organisms, playing important roles in soil ecosystems, influence soil legacy effects by affecting soil organic matter decomposition, nutrient cycling and soil structure (Bardgett and Wardle, 2010).
Therefore, natural ecosystems have become a ‘database’ for exploring the mechanisms of soil legacy effects in the context of a highly diversified plants, microorganisms and soil environmental factors. An increasing number of studies have been carried out on the growth characteristics, resource utilization and survival strategies of plants and microorganisms that contribute to a better understanding about the soil legacy effects (Bezemer et al., 2006; Cortois et al., 2016; Bezemer et al., 2018; Heinen et al., 2020). The diversity of plant species, plant functional traits and soil microorganisms in natural ecosystems contributes to extensive research on species interactions and stress adaptations. The intricate interactions between plants and soil microorganisms play a crucial role in promoting the stabilization of soil ecosystems (Grayston et al., 1998; Berg and Smalla, 2009; Berendsen et al., 2012). Above- and below-ground interactions of plants have long-term legacy effects on biotic stresses in natural ecosystems and can improve plant performance and resistance by manipulating soil microbial communities (Wurst and Ohgushi, 2015; Pineda et al., 2017). For abiotic stresses, soil microorganisms are able to implement a variety of mechanisms to fight against them and keep soil fertility as well as plant development in good condition (Abdul Rahman et al., 2021). For example, drought stress-induced dominance of fungal communities can influence succeeding plant drought adaptation by maintaining higher rates of litter decomposition and soil respiration (Mariotte et al., 2015). Inoculation of drought-conditioned phyllosphere and soil microbial communities can make plants capable of coping with repeated drought stress (Li et al., 2022).
Plant functional group is a common concept in the study of soil legacy effects, which refers to a group of plants that respond similarly to ecological processes and environmental changes, such as the grasses, forbs and legumes that are frequently mentioned in the literature (Kulmatiski et al., 2008; Cortois et al., 2016). Different plant functional groups can create positive or negative soil legacy effects by accumulating soil pathogens, recruiting beneficial microorganisms and regulating interactions with insects, etc (Petermann et al., 2008; Latz et al., 2012; Heinen et al., 2019). Such soil legacy effects, mediated by aboveground plant functional groups and soil microorganisms, play a role for succeeding plant growth in terms of soil physical properties, soil nutrient availability, soil microbial community structure, stress tolerance and competitive coexistence relationships (Byun et al., 2013; Strecker et al., 2015; Fischer et al., 2018; Mackie et al., 2018; Adomako and Yu, 2023).
Different plant functional groups play distinct roles in shaping soil legacy effects. For instance, grasses may improve soil physical structure and water retention through dense root systems (Hanamant et al., 2022), while legumes retain soil nutrients through nitrogen fixation (Spehn et al., 2002). The soil legacy effects resulting from these changes in the soil environment create more favorable conditions for succeeding plant growth. Simultaneously, the interaction between various plant functional groups and soil microorganisms yields diverse soil legacy effects. Grasses and forbs secrete different carbon compounds into the soil, recruiting different soil microorganisms (Philippot et al., 2013). For example, the presence of the grasses Lolium perenne not only increased the density of active bacteria in the soil but also elevated the expression of biocontrol genes associated with these bacteria, thereby contributing to the productivity of succeeding plant communities (Latz et al., 2015). Moreover, grasses positively influence other plant functional groups by altering soil microbial communities and soil nutrients (Cortois et al., 2016). Forbs, however, with more decomposers and higher concentrations of chemicals in their litter, may negatively impact succeeding plants (Bonanomi et al., 2006).
To foster positive soil legacy effects, it is essential to manage specific plant functional groups, regulate appropriate levels of beneficial microorganisms, decomposers and pathogenic microorganisms, and develop diverse plant-microbe community interactions (Carrión et al., 2019; De la Fuente Cantó et al., 2020; Xiong et al., 2020; Song et al., 2021). However, there is a current lack of studies exploring the application of the principle of soil legacy effects in understanding plant salt tolerance. Most studies have focused on the mechanism of plant’s intrinsic salt tolerance and the utilization of specific microorganisms to enhance salt tolerance in laboratory and simulation experiments (Li et al., 2020a; Li et al., 2020b; Li et al., 2021a; Schmitz et al., 2022). Therefore, it is important to address how the rules of soil legacy effects can be developed and applied in saline farmlands.
3 Processes of plant response to salt stress
Plants have various strategies to cope with salt stress, involving refinement in their cellular physiology, phenotypic structures, osmoregulation, antioxidant production, and the regulation of signaling pathways (Van Zelm et al., 2020; Zhao et al., 2020). For instance, plants eliminate excess salt through a salt excretion mechanism to minimize salt-damage (Dassanayake and Larkin, 2017). Plants can also modify their root structure, such as developing deeper root systems to increase water uptake and mitigate the impact of salinity (Galvan-Ampudia and Testerink, 2011). In addition, plants respond to salt stress-induced damage by producing antioxidants, osmotic substances and protective enzymes (Hasegawa et al., 2000). ABA-dependent protein kinases are activated in response to salt stress, affecting cellulose distribution, controlling root tip cells, thus promoting salt avoidance in plant (Yu et al., 2022). Plant roots also secrete peptides that are transferred to the leaves to induce ABA accumulation, thereby driving stomatal closure to prevent leaf (Takahashi et al., 2018; Yu et al., 2020). Therefore, the combined application of these strategies enables plants to better adapt and survive in high-salt environments.
Besides plant innate responses, the complex microbial communities in rhizosphere soil play a critical role in host performance and tolerance to stresses (Durán et al., 2018; Carrión et al., 2019). These microbial communities help plants adapt to harsh conditions by forming mutualistic relationships, participating in nutrient uptake, producing beneficial compounds, and inducing immune responses that support plants against stress (Hou et al., 2021).
In terms of salinity tolerance, microorganisms establish mutually beneficial symbiotic relationships with plants through various mechanisms, assisting them in adapting to high salt environments. Rhizosphere microorganisms can secrete specific compounds, such as bacterial exopolysaccharides (EPS), which improve plant ion balance, promote soil aggregation, and thus maintain plant growth in high-salt (Morcillo and Manzanera, 2021). Arbuscular mycorrhizal fungi (AMF) enhance host plant salt tolerance by manipulating the osmotic balance through mycelium, improving access to water and nutrients (Hammer et al., 2011; Ruiz-Lozano et al., 2012). Moreover, rhizosphere microorganisms also play a role in physiological regulation and defense processes (Mishra et al., 2021). Plant growth promoting rhizobacteria (PGPR) can stimulate root development and enhance nutrient utilization under salt stress. For instance, the IAA-overproducing strain Sinorhizobium meliloti has been found to enhancive salt tolerance of alfalfa in saline soils by stimulating root proliferation (Bianco and Defez, 2009). Under salt stress conditions, the increase in the number and weight of root nodules in Acacia gerrardii inoculated with Bacillus subtilis contributed to the enhancement of nitrogen fixation by the roots, as well as uptake and systemic translocation of phosphorus by the plant (Hashem et al., 2016a, Hashem et al., 2016b). AMF can activate an antioxidant protection system, maintaining cell membrane stability by decreasing permeability and malondialdehyde (MDA) content in plants (Yang et al., 2014).
These complex processes converting salinity tolerance cannot be separated from the dynamic interactions between plants and microorganisms (Liu et al., 2022). In the context of climate change-induced stress, introducing new microbial taxa had been shown to improve plant survival in stressful environments, and plant tolerance can be predicted by the climatic history of the microbial community (Allsup et al., 2023). Building on this, plant-soil-microbe interactions in salt-stressed environments may result in a history of stress response for soil microbes and the soil environment, generating soil legacy effects that aid succeeding plants in overcoming salt stress (Figure 1; Li et al., 2021a; Jing et al., 2022).
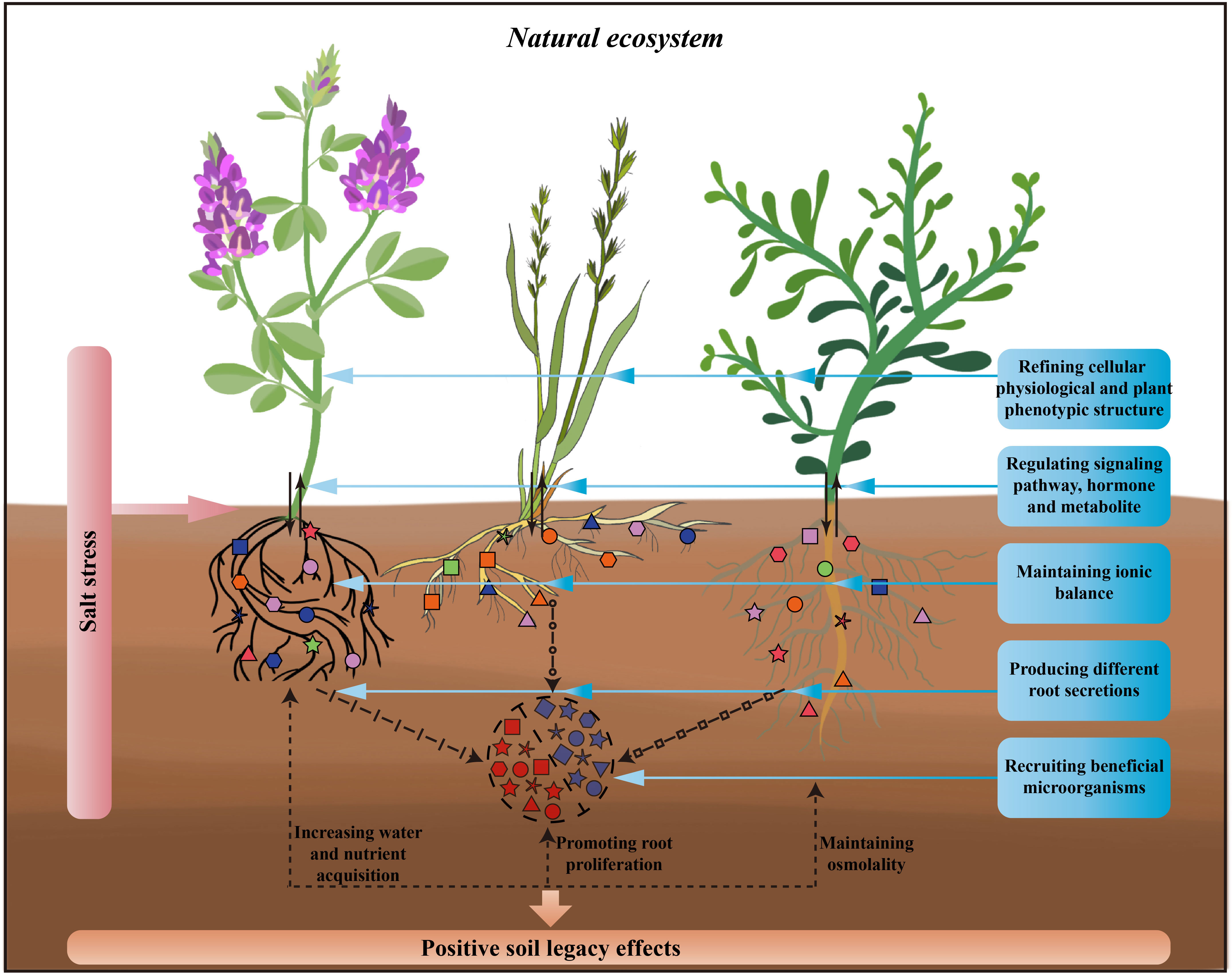
Figure 1 Processes of plant response to salt stress in natural ecosystems and possible soil legacy effects by plants. This figure shows, from left to right, three different plant functional groups, legume, grass, and forb, which respond simultaneously through above-ground and below-ground parts to salt stress. For above-ground parts of the plant, by refining cellular physiological and plant phenotypic structure, regulating signaling pathway, hormone and metabolite and thus responding to salt stress. For below-ground parts of the plant, by maintaining ionic balance, producing different root secretions, recruiting beneficial microorganisms and thus responding to salt stress. The response of above- and below-ground parts to salt stress simultaneously with increasing the plant’s own acquisition of soil water and nutrients, promoting plant root proliferation, and maintaining the osmolality of the plant as well as the rhizosphere, thus creating the positive soil legacy effects through this favourable response processes.
4 Creating soil legacy effects to improve crop salt tolerance
Farmlands vulnerable to saline stress often experience extreme environmental conditions and undergo specific agricultural management practices. These practices include high surface evapotranspiration, low precipitation, elevated ambient temperatures, and the application of chemicals, along with heavy irrigation during production (Arora et al., 2018; Enebe and Babalola, 2018). In contrast to natural ecosystems, the production function of farmland directly determines its monoculture structure, resulting in low plant diversity and nutrient use efficiency, imbalanced dynamics between above-ground crops and below-ground soil food webs, and altered crop defense mechanisms (Savary et al., 2019). Crops cultivated in farmlands tend to prioritize growth over defense compared to their wild counterparts of the same species. This preference, combined with the monoculture structure, increases the likelihood of negative soil legacy effects between previous and succeeding crops (Mariotte et al., 2018). The multiple stresses of saline farmlands challenge the growth of crops and soil microbes, and there is a need to rethink how to create soil legacy environments that are conducive to crop growth, while optimizing agricultural practices and fostering sustainable methods to enhance soil health and crop (Li et al., 2014).
4.1 The use of plants to create soil legacy effects
The productivity constraints of saline farmlands primarily result from the highly stressful environment directly impacting the growth of aboveground crops. Most staple crops in agricultural production, such as maize, wheat, and rice, show high sensitivity to salinity stress. This sensitivity manifests itself in increased crop water loss, plant and fruit wilting, reduced crop photosynthesis, lowered carbon fixation, inhibited crop nutrient uptake, and slowed growth (Atta et al., 2023). To overcome the production bottlenecks in saline farmlands, it is necessary to harness biological resources with inherent salt tolerance found in natural environments. Additionally, establishing positive soil legacy effects through the introduction of specific plant species and plant functional groups is crucial (Figure 2).
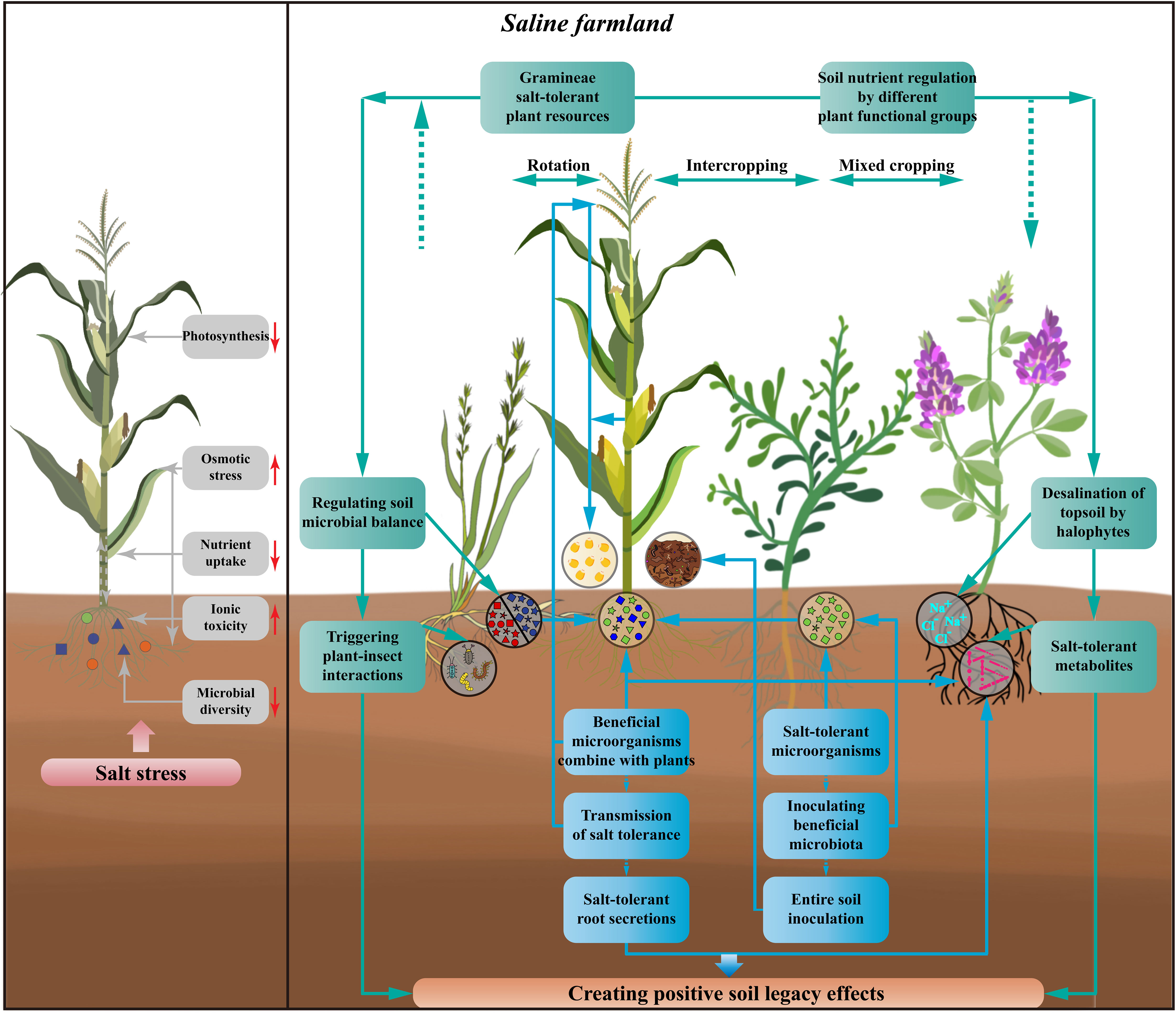
Figure 2 Effects of salt stress on crops and how to create soil legacy effects as well as improve crop salt tolerance in saline farmlands. The harmful effects of salt stress on crops include weakening crop photosynthesis, increasing osmotic stress, reducing crop nutrient uptake, adding ionic toxicity, and declining rhizosphere microbial diversity. By using halophytes, salt-tolerant plants, and plants of different functional groups, and developing the cropping patterns of rotating, intercropping, and mixed cropping with crops, the interactions between above- and below-ground parts of the plants can achieve the regulation of soil nutrients in saline farmlands, the desalination of surface soils, the secretion of salt-tolerant metabolite, and thus regulating the balance of soil microorganisms, as well as triggering the interactions between plants and insects. The improvement of salt tolerance in crops can also be achieved by screening for salt-tolerant microorganisms, inoculation with beneficial microbiota or entire soil inoculation. At the same time, new cultivation techniques could be used to combine the beneficial microorganisms directly with the plants and to transmit the tolerance. Crops with improved tolerance continue to produce salt-tolerant root secretions and to recruit beneficial microorganisms, thus creating an effective recycle of crop salt tolerance. All of these processes can create positive soil legacy effects through beneficial interactions between the above-ground and below-ground parts of the crop and influence succeeding crop.
4.1.1 The use of salt-tolerant biological resources
One feasible approach is to utilize the ability of halophytes and salt-tolerant plants in the natural environment. Firstly, some halophytes can absorb salt ions, and they are effective in reducing surface soil salinity while fighting against the increase in ion levels in tissue cells through leaf succulence (Song and Wang, 2015). Halophytes also dissolve calcium in the soil through root respiration, where calcium ions replace sodium ions in the cation-exchange complex, and ultimately improve soil physical properties in the plant’s root zone (Qadir et al., 2005). Desalinated soils resulting from these processes contribute favorably to the subsequent growth of plants. Secondly, both halophytes and salt-tolerant plants boast robust root systems with strong penetration and water-holding capacity, thus enhancing soil structure (Silva et al., 2016). This improvement increases soil permeability and water retention post-planting, with the positive effects on soil structure persisting over an extended period (Liang and Shi, 2021). Finally, certain salt-tolerant plants, such as the forage crop sweet sorghum, can develop salt tolerance through hormonal signaling and secondary metabolites (Chen et al., 2022). Notably, stress-induced plant secondary metabolites have demonstrated legacy effects on succeeding plant growth by manipulating the composition of soil microbiome (Hu et al., 2018). Consequently, the utilization of halophytes and salt-tolerant plants presents opportunities to desalinate saline farmlands, improve soil conditions, or directly leverage the soil legacy effects created by the metabolites they produce to enhance crop resilience to salinity.
4.1.2 Introducing plant functional groups into crop rotation systems
The soil legacy effects observed in natural ecosystems, facilitated by specific functional groups of plants, can significantly impact succeeding plants (Bezemer et al., 2006). This insight has inspired the development of effective cropping patterns for saline farmlands, especially considering that traditional monoculture patterns have contributed to soil resource depletion and decreased farmland productivity (Guo and Zhou, 2022). Grasses, have a solid research base in the field of ecology, known for carbon sequestration, nutrient cycling and improved soil stability (Franzluebbers, 2012; Hanamant et al., 2022). As the understanding of grassland ecosystem functioning continues to improve, forbs, representing a large proportion of species and functional richness, have also been recognized for their stress tolerance, indication of overgrazing, and maintenance of insect diversity (Siebert et al., 2021). Legumes, aside from being high-quality food and forage resources, are consistently recognized for sequestering nutrients and increasing diversity in cropping systems (Stagnari et al., 2017).
The crop rotation system of grasses and crops increased soil organic matter and earthworm numbers, resulting in improved soil structure compared to conventional crop rotations (Van Eekeren et al., 2008). This legacy effect of the grasses’ influence on soil properties, then, increased the yield and seed nitrogen content of succeeding crops (Christensen et al., 2009). Legumes are even more beneficial to agricultural production by providing diverse services. One aspect is that the nitrogen-fixing capacity of legumes can continually increase the nitrogen yield of succeeding crops (Fox et al., 2020). Moreover, the growth process of legumes releases organic acids and other compounds, directly activate nutrients and indirectly promote the activity of soil microorganisms, thus increasing crop yields and soil fertility (Latati et al., 2016). Studies have shown that the deposition of rhizosphere nitrogen in legumes accounts for 70% of the total plant nitrogen (Fustec et al., 2010). These deposited nitrogens have mechanisms for transfer to other crops, affecting agricultural production potential. Although there are fewer practices on the involvement of forbs in crop rotation, studies have shown that forbs are rather less affected by changes in nutrient conditions than grasses due to their ability to store nutrients in their roots (Herz et al., 2017). Forbs are also important for maintaining the diversity of arthropods in the environment and some forb communities are more resistant to herbivores (Potts et al., 2010; Van Coller et al., 2018). Therefore, introducing these plant functional groups, such as grasses, forbs, and legumes, during crop rotation can strategically change soil nutrient levels or indirectly regulate the biotic and abiotic environment of saline farmlands.
Moreover, grasses and forbs exhibit different abiotic stress tolerance mechanisms and growth strategies. Due to obvious differences in growth, development and physiological structure between grasses and forbs, applying knowledge of forbs to improve salt tolerance in major cereal crops becomes challenging (Tester and Bacic, 2005). Meanwhile, the ability of grasses to accumulate salt ions in shoots and leaves may be weaker than that of forbs due to fewer salt glands (Semenova et al., 2010). So, although the planting of forbs like Suaeda salsa can effectively reduce soil salinity, it is difficult to apply the mechanism of salt ion accumulation and succulence in shoots of forbs to crops of grasses.
However, Poaceae, particularly within the functional group of grasses, has a unique history of salt tolerance, including major halophytic taxa identified as sources of halophytes (Flowers et al., 1986). Compared to the forbs, grasses usually maintain ion levels in aboveground tissues by limiting sodium uptake, having high potassium/sodium selectivity, and efficient potassium utilization, essential for survival under saline conditions (Flowers and Colmer, 2008). Many wild-type grasses are naturally tolerate to salt stress (Landi et al., 2017). For example, the study found that its close wild relatives Tripsacum dactyloides and Zea perennis both showed strong salt tolerance compared to maize (Li et al., 2023b). The leaf surface of wild rice, Porteresia coarctata, can excrete salts, maintaining intercellular ion concentrations and lower sodium to potassium ratios (Sengupta and Majumder, 2010). Grasses have been reported to produce positive soil legacy effects by altering soil microbial communities, influencing nutrient transfer, and even triggering interactions between above-ground plants and insects (Kos et al., 2015; Cortois et al., 2016; Schmid et al., 2021). Also, the ionic changes that occur in grasses during salt tolerance are closely related to their rhizosphere microorganisms (Hamdia et al., 2004; Paul and Lade, 2014). Thus, by introducing plant functional groups into the crop rotation system and combining their different ecological functions and salt-tolerate characteristics, positive soil legacy effects can be generated. This provides broader thinking for the improving the soil environment in saline farmland and enhancing of crop salt tolerance.
4.1.3 Introducing plant functional groups into crop intercropping system
The combination of plant functional groups within the same time and space can exert a significant influence on succeeding crops. One notable example is the legume and grass forage matching system, a typical forage mixing approach where the growth of grasses synergistically enhances both the symbiotic nitrogen fixation of legumes and the competitive nitrogen uptake of grasses (De Deyn et al., 2012; Suter et al., 2015). Beyond improving soil nutrient use efficiency, the extended growing period of mixed legumes and grasses also helps suppress topsoil salt accumulation, thereby enhancing soil quality (Li et al., 2021b).
While there are fewer studies on crop tillage systems and salt tolerance, similar to forage mixes, crop intercropping can weaken the negative impacts of saline farmland and may have legacy effects on succeeding crops. Firstly, intercropping systems increase the biodiversity of farmland ecosystems by direct introducing companion plants, such as differential crops or salt-tolerant plants, which provide services for saline farmland and the main crop (Yang et al., 2021). The introduction of different plants diversifies the rhizosphere environment, and the recruited microbial community can promote nutrient cycling, salt transformation, and degradation in the soil, thereby alleviating the damage of the saline environment to the crops. For example, the introduction of legumes can improve intercropping system resilience and resource use efficiency by enhancing crop growth and tolerance to abiotic stresses through root distribution, vegetative cover, and nutrient activation (Chamkhi et al., 2022). Furthermore, the intercropping of the halophyte Suaeda salsa with maize significantly transferred more sodium ions to the rhizosphere of Suaeda salsa, thereby reducing the salt content of the maize rhizosphere (Wang S. et al., 2021). Regarding the rhizosphere enrichment by intercropping systems, it was shown that legume-grass crop intercropping (maize/faba bean) increased the abundance of rhizobia and reduced pathogens in the soil. The soil legacy effects it produced could be one of the reasons for the observed yield advantage in intercropping systems (Wang et al., 2020). Particularly under salt stress, the beneficial microorganisms recruited by the intercropping system (sorghum/peanut) achieved increased crop tolerance by altering the composition and content of metabolites (Shi et al., 2023). Therefore, the potential positive soil legacy effects of salt-tolerant forage mixtures and salt-tolerant crops of different functional groups can help to develop efficient intercropping systems for saline farmlands.
4.2 The use of soil microorganisms to synergistically create soil legacy effects
The presence of soil microorganisms in natural ecosystems depends on the soil environment, chemical signals provided by plants and nutrient resources (Bai et al., 2022). In response to the direct release of stress-responsive signals and compounds in plants, the associated soil microorganisms undergo specific changes (Hartman and Tringe, 2019). These changes are closely related to plants, especially alterations in rhizosphere microorganisms, and are critical to support the growth and recovery potential of plants under stress (Park et al., 2023). Salt-tolerant microorganisms capable of thriving and multiplying in high-salt environments, directly aiding plants in tolerating salt stress through their salt-tolerance mechanisms (Sharma et al., 2015; Wang R. et al., 2021). Plants in traditional environments, when confronted with salt stress, also respond by directly recruiting beneficial microorganisms through root secretions (Kumar et al., 2023). Furthermore, the mechanism by which soil microorganisms regulate plant salt tolerance also involves osmotic regulators, nutrients and soluble salts they provide to plants. These pathways can indirectly influence plant hormones and metabolism, stimulate plant growth and help plants overcome salt stress (Glick, 2012; Shrivastava and Kumar, 2015). These actions not only alleviate the negative effects of salinity but also establish soil legacy effects that confer tolerance to succeeding plants (see Figure 2; Zhalnina et al., 2018; Otlewska et al., 2020). Considering this, the question arises: How can we apply the direct and indirect effects of soil microorganisms on plant salt tolerance to saline farmland? What measures can be taken to sustain these positive effects in the farmland?
4.2.1 Direct utilization of soil microorganisms
Soil microorganisms play a crucial role in defending against saline stress, and saline soils serve as a significant source of salt-tolerant microorganisms (Zhang et al., 2023). Current research has successfully isolated several culturable salt-tolerant strains. For instance, 70% of the culturable strains of the root endophyte from the coastal perennial grass Festuca rubra exhibit salt tolerance (Pereira et al., 2019). The core microorganisms of the rhizosphere of Suaeda salsa have been found to harbor genes encoding salt stress adaptation and nutrient solubilization processes (Yuan Z. et al., 2016). Microbial inoculation is a direct method of utilizing these specialized salt-tolerant microbial resources, which can be applied to enhance plant salt stress adaptation and promote growth. Studies have demonstrated that inoculation with the salt-tolerant endophyte Sphingomonas prati significantly increases the salt tolerance of Suaeda salsa by improving the antioxidant enzyme system (Guo et al., 2021). Curvularia sp., isolated from Suaeda salsa, can establish a beneficial symbiotic relationship with poplar and promote its growth (Pan et al., 2018). Moreover, the inoculation of salt-tolerant microorganisms has been gradually extended to major crops, including soybean, maize, wheat, and peanut. Its positive effect in mitigating salt stress has been consistently verified in numerous indoor simulation experiments (Ramadoss et al., 2013; Goswami et al., 2014; Zerrouk et al., 2016; Khan et al., 2019; Shabaan et al., 2022).
In addition to the salt-tolerant microbial resources associated with saline soils and halophytes, salt stress is alleviated by the recruitment of beneficial microorganisms to the rhizosphere of plants when they face with salt stress in normal environments (Ilangumaran and Smith, 2017; Santoyo, 2021). For example, it has been shown that 1-aminocyclopropane-1-carboxylate (ACC), a stress-related amino acid in plants, can reshape the soil microbiome, enhancing plant tolerance to salinity stress (Liu et al., 2019). In addition, rice influences rhizosphere microorganisms by producing metabolites such as salicin and arbutin, enabling rhizosphere microorganisms associated salt stress tolerance (Lian et al., 2020). Moreover, beneficial rhizosphere microorganisms in plants can not only enhance salt-tolerant properties but also synergistically improve plant responses to salt stress by altering physiological growth processes, including seed germination, morphological structure, and biomass accumulation and partitioning (Pan et al., 2020). Regarding the inoculation of beneficial microbial strains to help crops tolerant salinity, studies have demonstrated that inoculation with Pseudomonas flavescens D5 strain effectively increased the biomass and antioxidant enzyme activities of barley, while reducing the adverse effects of salt stress on barley (Ignatova et al., 2022). Inoculation of candidate strains of Azotobacter has also been found to increase the potassium-sodium ratio, polyphenol and chlorophyll content, and decrease proline concentration in maize, thereby alleviating salt stress in maize by integrating multiple mechanisms (Rojas-Tapias et al., 2012).
Indeed, successful microbial inoculation often requires a combination of strains rather than a single strain to enhance the sustainability of its impact on (Verbruggen et al., 2012; Finkel et al., 2017). Notably, double inoculation with Rhizobium and Pseudomonas has been observed to elicit positive adaptive responses in alfalfa under salt stress (Younesi et al., 2013). Similarly, dual inoculation of plant growth-promoting bacteria with Bradyrhizobium strains has proven more effective in enhancing salt tolerance in soybean, reducing salt-induced ethylene production, and improving nutrient uptake (Win et al., 2023). Further studies have found that inoculation with species-specific microbiomes or whole-soil inoculation can assist plants in coping with various biotic and abiotic stresses (De Vries et al., 2020; Ma et al., 2020; Trivedi et al., 2020). The introduction of microbiomes or the whole-soil achieves more complex ecological functions by coordinating microbial interactions (Pineda et al., 2019; Trivedi et al., 2021), and it avoids the potential issue of single strains struggling to survive inoculation into foreign soil (Mallon et al., 2018). However, it is crucial to acknowledge the possibility that introducing exotic microbial communities may reshape functions within the native microbial community (Amor et al., 2020). Recent evidence suggests that the beneficial effects of microbial inoculation on plant growth are best explained as changes in native microorganisms rather than direct effects on plants (Hu et al., 2021). This underscores the importance of understanding the intricate interactions occurring within the microbial community and their influence on plant health and resilience.
While practical examples of microorganism inoculation for saline farmland improvement are limited, the concept of soil legacy effects suggests that enhancing saline farmland and crops can be achieved through microbial-mediated processes. By inoculating salt-tolerant microbial strains and communities of beneficial microorganisms, and even inoculating the entire soil including most microorganisms, it becomes possible to modulate crop responses to salt stress and enhance salt tolerance. Concurrently, synergistic changes with the inoculated microorganisms involve stress response-related metabolites and alterations in the crop rhizosphere environments. These changes encompass crop rhizosphere secretions, microbial metabolites, and native microbial communities. Their persistent influence on succeeding crop growth in the form of soil legacy effects contributes to ongoing salt stress mitigation in saline farmland. Thus, the application of microbial interventions holds promise for sustainable improvements in saline farmland and crop resilience (Cuddington, 2011; Trivedi et al., 2020).
4.2.2 Indirect utilization of soil microorganisms
Alongside traditional plant- and microorganism-based methods for restoring saline farmlands, advanced modern agricultural techniques with their high efficiency and precision have also found application agricultural production (Varshney et al., 2011; Ahanger et al., 2017). Research has focused on integrating and applying the active components of rhizosphere exudates to soil microbial systems, revealing improvements in soil physicochemical environments and microbial communities associated with rhizosphere exudates. These improvements are speculated to have an impact on plant growth (Shi et al., 2011). Similar findings were observed in maize system, where a significant increase in bacterial density and altered metabolic potential in the maize rhizosphere after application of maize rhizosphere exudates (Baudoin et al., 2003). In terms of enhancing crop tolerance, research has shown that introducing the ability of releasing volatile organic compounds (VOCs) into maize varieties that do not release specific VOCs can reduce the threat of pests (Degenhardt et al., 2009). This suggests that the introduction of tolerant metabolites is not limited to rhizosphere exudates, and the application of below-ground volatiles, as well as other tolerant signals, offers additional possibilities for improving salt tolerance in crops on saline farmlands. The advances in agricultural technology have also inspired the exploration of beneficial root traits in wild relatives of crops, the introduction of which may solve the problems faced by saline farmlands (Preece and Peñuelas, 2020).
In the past decade, cultivation techniques have gradually emerged, pointing to the unique microbiome existing in plant seeds and how it spreads from generation to generation, aiding plants in adapting to their environment and increasing tolerance (Gopal and Gupta, 2016; Abdelfattah et al., 2023). In this context, delivering endophytes to the next generation of crops and ensuring the persistence of their tolerance has been achieved by combining relevant beneficial microorganisms with plants (Wei and Jousset, 2017). For example, a suspension of Paraburkholderia phytofirmans PsJN was sprayed in plots at the flowering stage of wheat in field experiment, and thus the maturation of its progeny plants was accelerated by the introduction of this endophytic bacteria into the flowers of the wheat parents (Mitter et al., 2017). The advantage of this approach lies in the ability of seed endophytes to avoid competition with native soil microorganisms, establishing closer interactions with the plant early on. While there is currently limited research related to this approach concerning salt tolerance in progeny plants, seed endophytes have long been shown to provide plants with tolerance against a wide range of stresses, participate in plant adaptation mechanisms, and enhance plant competitiveness (Samreen et al., 2021). Therefore, the use of these new bioculture techniques and the genetic mechanisms of plant microbes offer innovative avenues for improving saline farmland. These approaches are closely related to plant-microbe interactions and are centered around the concept of creating positive soil legacy effects.
Inspired by the mentioned approaches, microorganisms can be used indirectly, such as through the recruitment of microorganisms by plant rhizosphere exudates and intergenerational dissemination of beneficial microorganisms, to create positive soil legacy effects in saline farmland. However, it is acknowledged that microbial-related methods of creating soil legacy effects are imperfect, and their processes may introduce soil pathogens or other responsive substances, necessitating further in-depth research to explore safer methods of creating soil legacy effects (Jing et al., 2022).
5 Conclusion and future prospects
This paper provides a summary of the ways in which plants, in collaboration with soil microorganisms in natural ecosystems, jointly respond to salt stress. It suggests enhancing the salt tolerance of crops in saline farmlands through the perspective of soil legacy effects. The focus is on meeting the salt tolerance needs of crops by creating well-considered soil legacy effects. The paper explores both the direct use of plants and the synergistic use of soil microorganisms to establish positive soil legacy effects, offering innovative insights to boost production potential and improve the ecological environment of saline farmland. The emphasis lies on creating positive soil legacy effects through the selection of suitable salt-tolerant crops, the development of planting patterns with a rational match of crop functional groups, the inoculation of functional microorganisms, the inoculation of safe and efficacious soils, and the application of advanced agricultural technologies and bio-cultivation methods. This approach underscores the practical utility of crop-soil microorganism interactions in agricultural production. In addition to plants and associated soil microorganisms, the role of soil animals in constructing soil food webs is acknowledged. These soil animals, through direct or indirect interactions with microorganisms and plants, contribute to the cycling of soil nutrient resources, influencing soil ecosystem function (Du et al., 2018). Multi-trophic interactions between mycorrhizal fungi, fungus-eating protozoa, and nematodes in the soil can enhance crop nutrient uptake, crop yield, and tolerance (Jiang et al., 2020). This suggests that future studies can more precisely and directly leverage soil legacy effects to trigger positive tolerant responses by regulating specific species or soil fauna in the soil food web of saline farmlands, or even by controlling certain trophic levels.
Author contributions
YM: Writing – original draft, Writing – review & editing. CZ: Funding acquisition, Writing – review & editing. YB: Funding acquisition, Writing – review & editing. CS: Funding acquisition, Writing – review & editing. FZ: Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Young Scientists Project of National Key Research and Development Program of China (2021YFD1900200), the National Key Research and Development Program of China (2022YFF1302800, 2022YFD1900300), the Strategic Priority Research Program of Chinese Academy of Sciences (XDA26040103), the Special Key Grant Project of Technology Research and Development of Zhangjiakou City (“Take-and-lead” Program) (No.2022J001) and the “100 Talents Project” of Chinese Academy of Sciences.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbas, M. K., Ali, A. S., Hasan, H. H., Ghal, R. H. (2012). Salt tolerance study of six cultivars of rice (Oryza sativa L.) during germination and early seedling growth. J. Agric. Sci. 5, 250. doi: 10.5539/jas.v5n1p250
Abdelfattah, A., Tack, A. J., Lobato, C. B., Wassermann, B., Berg, G. (2023). From seed to seed: the role of microbial inheritance in the assembly of the plant microbiome. Trends Microbiol. 31, 346–355. doi: 10.1016/j.tim.2022.10.009
Abdul Rahman, N. S., Abdul Hamid, N. W., Nadarajah, K. K. (2021). Effects of abiotic stress on soil microbiome. Int. J. Mol. Sci. 22, 9036. doi: 10.3390/ijms22169036
Adomako, M. O., Yu, F. (2023). Functional group dominance-mediated N:P effects on community productivity depend on soil nutrient levels. Rhizosphere 26, 100692. doi: 10.1016/j.rhisph.2023.100692
Ahanger, M. A., Akram, N. A., Ashraf, M., AlYemeni, M. N., Wijaya, L., Ahmad, P. (2017). Plant responses to environmental stresses-from gene to biotechnology. AoB Plants 9, plx025. doi: 10.1093/aobpla/plx025
Allsup, C. M., George, I. F., Lankau, R. A. (2023). Shifting microbial communities can enhance tree tolerance to changing climates. Science 380, 835–840. doi: 10.1126/science.adf2027
Amor, D. R., Ratzke, C., Gore, J. (2020). Transient invaders can induce shifts between alternative stable states of microbial communities. Sci. Adv. 6, eaay8676. doi: 10.1126/sciadv.aay8676
Apse, M. P., Blumwald, E. (2002). Engineering salt tolerance in plants. Curr. Opin. Biotechnol. 13, 146–150. doi: 10.1016/s0958-1669(02)00298-7
Arora, N. K., Fatima, T., Mishra, I., Verma, M. K., Mishra, J., Mishra, V. (2018). Environmental sustainability: challenges and viable solutions. Environ. Sustainability 1, 309–340. doi: 10.1007/s42398-018-00038-w
Atta, K., Mondal, S., Gorai, S., Singh, A. P., Kumari, A., Ghosh, T., et al. (2023). Impacts of salinity stress on crop plants: improving salt tolerance through genetic and molecular dissection. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1241736
Bai, J., Gao, H., Xiao, R., Wang, J., Huang, C. (2012). A review of soil nitrogen mineralization as affected by water and salt in coastal wetlands: issues and methods. Clean-Soil Air Water 40, 1099–1105. doi: 10.1002/clen.201200055
Bai, B., Liu, W., Qiu, X., Zhang, J., Zhang, J., Bai, Y. (2022). The root microbiome: Community assembly and its contributions to plant fitness. J. Integr. Plant Biol. 64, 230–243. doi: 10.1111/jipb.13226
Bakker, P. A., Pieterse, C. M., Jonge, R. D., Berendsen, R. L. (2018). The soil-borne legacy. Cell 172, 1178–1180. doi: 10.1016/j.cell.2018.02.024
Bardgett, R. D., Wardle, D. A. (2010). Aboveground-belowground linkages: Biotic interactions, ecosystem processes, and global change (New York: Oxford University Press).
Baudoin, E., Benizri, E., Guckert, A. (2003). Impact of artificial root exudates on the bacterial community structure in bulk soil and maize rhizosphere. Soil Biol. Biochem. 35, 1183–1192. doi: 10.1016/s0038-0717(03)00179-2
Berendsen, R. L., Pieterse, C. M., Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berg, G., Smalla, K. (2009). Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 68, 1–13. doi: 10.1111/j.1574-6941.2009.00654.x
Bezemer, T. M., Jing, J., Bakx-Schotman, J. M., Bijleveld, E. (2018). Plant competition alters the temporal dynamics of plant-soil feedbacks. J. Ecol. 106, 2287–2300. doi: 10.1111/1365-2745.12999
Bezemer, T. M., Lawson, C. S., Hedlund, K., Edwards, A. R., Brook, A. J., Igual, J. M., et al. (2006). Plant species and functional group effects on abiotic and microbial soil properties and plant-soil feedback responses in two grasslands. J. Ecol. 94, 893–904. doi: 10.1111/j.1365-2745.2006.01158.x
Bianco, C., Defez, R. (2009). Medicago truncatula improves salt tolerance when nodulated by an indole-3-acetic acid-overproducing Sinorhizobium meliloti strain. J. Exp. Bot. 60, 3097–3107. doi: 10.1093/jxb/erp140
Bonanomi, G., Sicurezza, M. G., Caporaso, S., Esposito, A., Mazzoleni, S. (2006). Phytotoxicity dynamics of decaying plant materials. New Phytol. 169, 571–578. doi: 10.1111/j.1469-8137.2005.01611.x
Borde, M., Dudhane, M., Kulkarni, M. V. (2017). “Role of arbuscular mycorrhizal fungi (AMF) in salinity tolerance and growth response in plants under salt stress conditions,” in Mycorrhiza-Eco-Physiology, Secondary Metabolites, Nanomaterials. Eds. Varma, A., Prasad, R., Tuteja, N. (Springer, Cham), 71–86.
Burssens, S., Himanen, K., van de Cotte, B., Beeckman, T., Van Montagu, M., Inzé, D., et al. (2000). Expression of cell cycle regulatory genes and morphological alterations in response to salt stress in Arabidopsis thaliana. Planta 211, 632–640. doi: 10.1007/s004250000334
Byun, C., Blois, S. D., Brisson, J. (2013). Plant functional group identity and diversity determine biotic resistance to invasion by an exotic grass. J. Ecol. 101, 128–139. doi: 10.2307/23354673
Carrión, V. J., Perez-Jaramillo, J., Cordovez, V., Tracanna, V., de Hollander, M., Ruiz-Buck, D., et al. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 366, 606–612. doi: 10.1126/science.aaw9285
Chamkhi, I., Cheto, S., Geistlinger, J., Youssef, Z. (2022). Legume-based intercropping systems promote beneficial rhizobacterial community and crop yield under stressing conditions. Ind. Crops Products 183, 114958. doi: 10.1016/j.indcrop.2022.114958
Chen, C., Shang, X., Sun, M., Tang, S., Khan, A., Zhang, D., et al. (2022). Comparative transcriptome analysis of two sweet sorghum genotypes with different salt tolerance abilities to reveal the mechanism of salt tolerance. Int. J. Mol. Sci. 23, 2272. doi: 10.3390/ijms23042272
Christensen, B. T., Rasmussen, J., Eriksen, J., Hansen, E. M. (2009). Soil carbon storage and yields of spring barley following grass leys of different age. Eur. J. Agron. 31, 29–35. doi: 10.1016/j.eja.2009.02.004
Cordovez, V., Dini-Andreote, F., Carrión, V. J., Raaijmakers, J. M. (2019). Ecology and evolution of plant microbiomes. Annu. Rev. Microbiol. 73, 69–88. doi: 10.1146/annurev-micro-090817-062524
Cortois, R., Schröder-Georgi, T., Weigelt, A., Putten, W. H., De Deyn, G. B. (2016). Plant-soil feedbacks: role of plant functional group and plant traits. J. Ecol. 104, 1608–1617. doi: 10.1111/1365-2745.12643
Cuddington, K. (2011). Legacy effects: the persistent impact of ecological interactions. Biol. Theory 6, 203–210. doi: 10.1007/s13752-012-0027-5
Dassanayake, M., Larkin, J. C. (2017). Making plants break a sweat: the structure, function, and evolution of plant salt glands. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00406
De Deyn, G. B., Quirk, H., Oakley, S., Ostle, N., Bardgett, R. D. (2012). Increased plant carbon translocation linked to overyielding in grassland species mixtures. PloS One 7, e45926. doi: 10.1371/journal.pone.0045926
Degenhardt, J., Hiltpold, I., Köllner, T. G., Frey, M., Gierl, A., Gershenzon, J., et al. (2009). Restoring a maize root signal that attracts insect-killing nematodes to control a major pest. Proc. Natl. Acad. Sci. United States America 106, 13213–13218. doi: 10.1073/pnas.0906365106
De la Fuente Cantó, C., Simonin, M., King, E., Moulin, L., Bennett, M. J., Castrillo, G., et al. (2020). An extended root phenotype: the rhizosphere, its formation and impacts on plant fitness. Plant J. 103, 951–964. doi: 10.1111/tpj.14781
Deng, Y., Yuan, F., Feng, Z., Ding, T. L., Song, J., Wang, B. (2014). Comparative study on seed germination characteristics of two species of Australia saltbush under salt stress. Acta Ecologica Sin. 34, 337–341. doi: 10.1016/J.CHNAES.2013.07.011
De Vries, F. T., Griffiths, R. I., Knight, C. G., Nicolitch, O., Williams, A. (2020). Harnessing rhizosphere microbiomes for drought-resilient crop production. Science 368, 270–274. doi: 10.1126/science.aaz5192
Du, X., Li, Y., Liu, F., Su, X. L., Li, Q. (2018). Structure and ecological functions of soil micro-food web. J. Appl. Ecol. 29, 403–411. doi: 10.13287/j.1001-9332.201802.033
Durán, P., Thiergart, T., Garrido-Oter, R., Agler, M. T., Kemen, E., Schulze-Lefert, P., et al. (2018). Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175, 973–983. doi: 10.1016/j.cell.2018.10.020
Elmajdoub, B., Marschner, P. (2013). Salinity reduces the ability of soil microbes to utilise cellulose. Biol. Fertil. Soils 49, 379–386. doi: 10.1007/s00374-012-0734-x
Enebe, M. C., Babalola, O. (2018). The influence of plant growth-promoting rhizobacteria in plant tolerance to abiotic stress: a survival strategy. Appl. Microbiol. Biotechnol. 102, 7821–7835. doi: 10.1007/s00253-018-9214-z
Etesami, H., Beattie, G. A. (2017). “Plant-microbe interactions in adaptation of agricultural crops to abiotic stress conditions,” in Probiotics and Plant Health. Eds. Kumar, V., Kumar, M., Sharma, S., Prasad, R. (Springer, Singapore). doi: 10.1007/978-981-10-3473-2_7
Farooq, M., Gogoi, N., Hussain, M., Barthakur, S., Paul, S., Bharadwaj, N., et al. (2017). Effects, tolerance mechanisms and management of salt stress in grain legumes. Plant Physiol. Biochem. 118, 199–217. doi: 10.1016/j.plaphy.2017.06.020
Faucon, M., Houben, D., Lambers, H. (2017). Plant functional traits: soil and ecosystem services. Trends Plant Sci. 22, 385–394. doi: 10.1016/j.tplants.2017.01.005
Finkel, O. M., Castrillo, G., Herrera Paredes, S., Salas González, I., Dangl, J. L. (2017). Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 38, 155–163. doi: 10.1016/j.pbi.2017.04.018
Fischer, C., Leimer, S., Roscher, C., Ravenek, J. M., De Kroon, H., Kreutziger, Y., et al. (2018). Plant species richness and functional groups have different effects on soil water content in a decade-long grassland experiment. J. Ecol. 107, 127–141. doi: 10.1111/1365-2745.13046
Flowers, T. J., Colmer, T. D. (2008). Salinity tolerance in halophytes. New Phytol. 179, 945–963. doi: 10.1111/j.1469-8137.2008.02531.x
Flowers, T. J., Hajibagheri, M. A., Clipson, N. J. W. (1986). Halophytes. Q. Rev. Biol. 61, 313–337. doi: 10.1086/415032
Food and Agriculture Organization of the United Nations (2021) Global Map of Salt-affected Soils. Available online at: https://www.fao.org/global-soil-partnership/gsasmap/en.
Fox, A. F., Suter, M., Widmer, F., Lüscher, A. (2020). Positive legacy effect of previous legume proportion in a ley on the performance of a following crop of Lolium multiflorum. Plant Soil 447, 497–506. doi: 10.1007/s11104-019-04403-4
Franzluebbers, A. J. (2012). Grass roots of soil carbon sequestration. Carbon Manage. 3, 9–11. doi: 10.4155/cmt.11.73
Fustec, J., Lesuffleur, F., Mahieu, S., Cliquet, J. (2010). Nitrogen rhizodeposition of legumes. A review. Agron. Sustain. Dev. 30, 57–66. doi: 10.1051/agro/2009003
Galvan-Ampudia, C. S., Testerink, C. (2011). Salt stress signals shape the plant root. Curr. Opin. Plant Biol. 14, 296–302. doi: 10.1016/j.pbi.2011.03.019
German, R. N., Thompson, C., Benton, T. G. (2017). Relationships among multiple aspects of agriculture’s environmental impact and productivity: a meta-analysis to guide sustainable agriculture. Biol. Rev. Cambridge Philos. Soc. 92, 716–738. doi: 10.1111/brv.12251
Glick, B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012, 963401. doi: 10.6064/2012/963401
Gopal, M., Gupta, A. (2016). Microbiome selection could spur next-generation plant breeding strategies. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01971
Goswami, D., Dhandhukia, P., Patel, P. B., Thakker, J. N. (2014). Screening of PGPR from saline desert of Kutch: growth promotion in Arachis hypogea by Bacillus licheniformis A2. Microbiological Res. 169, 66–75. doi: 10.1016/j.micres.2013.07.004
Grayston, S. J., Wang, S. S., Campbell, C. D., Edwards, A. C. (1998). Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 30, 369–378. doi: 10.1016/S0038-0717(97)00124-7
Gunde-Cimerman, N., Plemenitaš, A., Oren, A. (2018). Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 42, 353–375. doi: 10.1093/femsre/fuy009
Guo, J., Chen, Y., Lu, P., Liu, M., Sun, P., Zhang, Z. (2021). Roles of endophytic bacteria in Suaeda salsa grown in coastal wetlands: Plant growth characteristics and salt tolerance mechanisms. Environ. pollut. 287, 117641. doi: 10.1016/j.envpol.2021.117641
Guo, Z., Zhou, H. (2022). A review of saline-alkali land resources and improvement measures. Sci. J. Humanities Soc. Sci. 4, 258–262. Available at: http://www.sjohss.org/download/SJOHSS-4-2-258-262.pdf
Hamdia, M. A., Shaddad, M. A., Doaa, M. M. (2004). Mechanisms of salt tolerance and interactive effects of Azospirillum brasilense inoculation on maize cultivars grown under salt stress conditions. Plant Growth Regul. 44, 165–174. doi: 10.1023/B:GROW.0000049414.03099.9b
Hammer, E. C., Nasr, H., Pallon, J., Olsson, P. A., Wallander, H. (2011). Elemental composition of arbuscular mycorrhizal fungi at high salinity. Mycorrhiza 21, 117–129. doi: 10.1007/s00572-010-0316-4
Hanamant, M. H., Govindasamy, P., Chaudhary, M., Srinivasan, R., Prasad, M., Wasnik, V. K., et al. (2022). Range grasses to improve soil properties, carbon sustainability, and fodder security in degraded lands of semi-arid regions. Sci. Total Environ. 851, 158211. doi: 10.1016/j.scitotenv.2022.158211
Hartman, K. J., Tringe, S. G. (2019). Interactions between plants and soil shaping the root microbiome under abiotic stress. Biochem. J. 476, 2705–2724. doi: 10.1042/BCJ20180615
Hartmann, M., Six, J. (2023). Soil structure and microbiome functions in agroecosystems. Nat. Rev. Earth Environ. 4, 1–15. doi: 10.1038/s43017-022-00366-w
Hasegawa, P. M., Bressan, R. A., Zhu, J., Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499. doi: 10.1146/annurev.arplant.51.1.463
Hashem, A., Abd_Allah, E. F., Alqarawi, A. A., Al-Huqail, A. A., Wirth, S., Egamberdieva, D. (2016b). The interaction between arbuscular mycorrhizal fungi and endophytic bacteria enhances plant growth of Acacia gerrardii under salt stress. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01089
Hashem, A., Abd Allah, E. F., Alqarawi, A. A., Al-Huqail, A. A., Shah, M. A. (2016a). Induction of osmoregulation and modulation of salt stress in Acacia gerrardii Benth by arbuscular mycorrhizal fungi and Bacillus subtilis (BERA 71). BioMed. Res. Int. 2016, 1–11. doi: 10.1155/2016/6294098
Heinen, R., Biere, A., Bezemer, T. M. (2019). Plant traits shape soil legacy effects on individual plant-insect interactions. Oikos 129, 261–273. doi: 10.1111/oik.06812
Heinen, R., Hannula, S. E., De Long, J. R., Huberty, M. R., Jongen, R., Kielak, A. M., et al. (2020). Plant community composition steers grassland vegetation via soil legacy effects. Ecol. Lett. 23, 973–982. doi: 10.1111/ele.13497
Herz, K., Dietz, S., Haider, S., Jandt, U., Scheel, D., Bruelheide, H. (2017). Drivers of intraspecific trait variation of grass and forb species in German meadows and pastures. J. Vegetation Sci. 28, 705–716. doi: 10.1111/jvs.12534
Hou, S., Wolinska, K. W., Hacquard, S. (2021). Microbiota-root-shoot-environment axis and stress tolerance in plants. Curr. Opin. Plant Biol. 62, 102028. doi: 10.1016/j.pbi.2021.102028
Hu, D., Li, R., Dong, S., Zhang, J., Zhao, B., Ren, B., et al. (2022). Maize (Zea mays L.) responses to salt stress in terms of root anatomy, respiration and antioxidative enzyme activity. BMC Plant Biol. 22, 602. doi: 10.1186/s12870-022-03972-4
Hu, L., Robert, C. A., Cadot, S., Zhang, X., Ye, M., Li, B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9, 2738. doi: 10.1038/s41467-018-05122-7
Hu, J., Yang, T., Friman, V., Kowalchuk, G. A., Hautier, Y., Li, M., et al. (2021). Introduction of probiotic bacterial consortia promotes plant growth via impacts on the resident rhizosphere microbiome. Proc. R. Soc. B 288, 20211396. doi: 10.1098/rspb.2021.1396
Ignatova, L. V., Usmanova, A. M., Brazhnikova, Y., Omirbekova, A., Egamberdieva, D., Mukasheva, T. D., et al. (2022). Plant probiotic endophytic Pseudomonas flavescens D5 strain for protection of barley plants from salt stress. Sustainability 4, 15881. doi: 10.3390/su142315881
Ilangumaran, G., Smith, D. L. (2017). Plant growth promoting rhizobacteria in amelioration of salinity stress: a systems biology perspective. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01768
Jansson, J. K., McClure, R., Egbert, R. G. (2023). Soil microbiome engineering for sustainability in a changing environment. Nat. Biotechnol. 41, 1716–1728. doi: 10.1038/s41587-023-01932-3
Jiang, Y., Luan, L., Hu, K., Liu, M., Chen, Z., Geisen, S., et al. (2020). Trophic interactions as determinants of the arbuscular mycorrhizal fungal community with cascading plant-promoting consequences. Microbiome 8, 142. doi: 10.1186/s40168-020-00918-6
Jing, J., Cong, W., Bezemer, T. M. (2022). Legacies at work: plant-soil-microbiome interactions underpinning agricultural sustainability. Trends Plant Sci. 27, 781–792. doi: 10.1016/j.tplants.2022.05.007
Khan, M. A., Asaf, S., Khan, A., Adhikari, A., Jan, R., Ali, S., et al. (2019). Halotolerant rhizobacterial strains mitigate the adverse effects of NaCl stress in soybean seedlings. BioMed. Res. Int. 2019, 1–15. doi: 10.1155/2019/9530963
Kos, M., Tuijl, M. A., Roo, J. D., Mulder, P. P., Bezemer, T. M. (2015). Species-specific plant-soil feedback effects on above-ground plant-insect interactions. J. Ecol. 103, 904–914. doi: 10.1111/1365-2745.12402
Kowalchuk, G. A., Buma, D. S., de Boer, W., Klinkhamer, P. G., van Veen, J. A. (2002). Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie Van Leeuwenhoek 81, 509–520. doi: 10.1023/A:1020565523615
Kulmatiski, A., Beard, K. H., Stevens, J. R., Cobbold, S. M. (2008). Plant-soil feedbacks: a meta-analytical review. Ecol. Lett. 11, 980–992. doi: 10.1111/j.1461-0248.2008.01209.x
Kumar, P., Choudhary, M. K., Halder, T., Prakash, N. R., Singh, V., V., V. T., et al. (2022). Salinity stress tolerance and omics approaches: revisiting the progress and achievements in major cereal crops. Heredity 128, 497–518. doi: 10.1038/s41437-022-00516-2
Kumar, N., Haldarb, S., Saikia, R. (2023). Root exudation as a strategy for plants to deal with salt stress: an updated review. Environ. Exp. Bot. 216, 105518. doi: 10.1016/j.envexpbot.2023.105518
Landi, S., Hausman, J., Guerriero, G., Esposito, S. (2017). Poaceae vs. abiotic stress: focus on drought and salt stress, recent insights and perspectives. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01214
Lange, M., Eisenhauer, N., Sierra, C. A., Bessler, H., Engels, C., Griffiths, R. I., et al. (2015). Plant diversity increases soil microbial activity and soil carbon storage. Nat. Commun. 6, 6707. doi: 10.1038/ncomms7707
Latati, M., Bargaz, A., Belarbi, B., Lazali, M., Benlahrech, S., Tellah, S., et al. (2016). The intercropping common bean with maize improves the rhizobial efficiency, resource use and grain yield under low phosphorus availability. Eur. J. Agron. 72, 80–90. doi: 10.1016/j.eja.2015.09.015
Latz, E., Eisenhauer, N., Rall, B. C., Allan, E., Roscher, C., Scheu, S., et al. (2012). Plant diversity improves protection against soil-borne pathogens by fostering antagonistic bacterial communities. J. Ecol. 100, 597–604. doi: 10.1111/j.1365-2745.2011.01940.x
Latz, E., Eisenhauer, N., Scheu, S., Jousset, A. (2015). Plant identity drives the expression of biocontrol factors in a rhizosphere bacterium across a plant diversity gradient. Funct. Ecol. 29, 1225–1234. doi: 10.1111/1365-2435.12417
Läuchli, A., Grattan, S. R. (2007). “Plant growth and development under salinity stress,” in Advances in Molecular Breeding Toward Drought and Salt Tolerant Crops. Eds. Jenks, M. A., Hasegawa, P. M., Jain, S. M. (Springer, Dordrecht). doi: 10.1007/978-1-4020-5578-2_1
Li, X., Jimoh, S. O., Li, Y., Duan, J., Cui, Y., Jin, K., et al. (2022). Stress memory and phyllosphere/soil legacy underlie tolerance and plasticity of Leymus chinensis to periodic drought risk. Agric. For. Meteorol. 312, 108717. doi: 10.1016/j.agrformet.2021.108717
Li, C., Jin, L., Zhang, C., Li, S., Zhou, T., Hua, Z., et al. (2023a). Destabilized microbial networks with distinct performances of abundant and rare biospheres in maintaining networks under increasing salinity stress. iMeta 2, e79. doi: 10.1002/imt2.79
Li, H., La, S., Zhang, X., Gao, L., Tian, Y. (2021a). Salt-induced recruitment of specific root-associated bacterial consortium capable of enhancing plant adaptability to salt stress. ISME J. 15, 2865–2882. doi: 10.1038/s41396-021-00974-2
Li, P., Lu, Y., Chen, H., Day, B. (2020a). The lifecycle of the plant immune system. Crit. Rev. Plant Sci. 39, 72–100. doi: 10.1080/07352689.2020.1757829
Li, J., Pu, L., Han, M., Zhu, M., Zhang, R., Xiang, Y. (2014). Soil salinization research in China: advances and prospects. J. Geographical Sci. 24, 943–960. doi: 10.1007/s11442-014-1130-2
Li, X., Wang, X., Ma, Q., Zhong, Y., Zhang, Y., Zhang, P., et al. (2023b). Integrated single-molecule real-time sequencing and RNA sequencing reveal the molecular mechanisms of salt tolerance in a novel synthesized polyploid genetic bridge between maize and its wild relatives. BMC Genomics 24, 55. doi: 10.1186/s12864-023-09148-0
Li, A., Wu, Y. Z., Tai, X. S., LI, J., LI, C. M., GE, C. M., et al. (2021b). Effects of planting legume-grass mixtures on soil salinity and nutrients in irrigated areas along the Yellow River in Gansu province. Acta Agrestia Sin. 29, 664–670. doi: 10.11733/j.issn.1007-0435.2021.04.004
Li, J., Zhou, H., Zhang, Y., Li, Z., Yang, Y., Guo, Y. (2020b). The GSK3-like kinase BIN2 is a molecular switch between the salt stress response and growth recovery in Arabidopsis thaliana. Dev. Cell 55, 367–380. doi: 10.1016/j.devcel.2020.08.005
Lian, T., Huang, Y., Xie, X., Huo, X., Shahid, M. Q., Tian, L., et al. (2020). Rice SST variation shapes the rhizosphere bacterial community, conferring tolerance to salt stress through regulating soil metabolites. mSystems 5, e00721-20. doi: 10.1128/mSystems.00721-20
Liang, J., Shi, W. (2021). Cotton/halophytes intercropping decreases salt accumulation and improves soil physicochemical properties and crop productivity in saline-alkali soils under mulched drip irrigation: A three-year field experiment. Field Crops Res. 262, 108027. doi: 10.1016/j.fcr.2020.108027
Liu, H., Khan, M. Y., Carvalhais, L. C., Delgado-Baquerizo, M., Yan, L., Crawford, M. H., et al. (2019). Soil amendments with ethylene precursor alleviate negative impacts of salinity on soil microbial properties and productivity. Sci. Rep. 9, 6892. doi: 10.1038/s41598-019-43305-4
Liu, Y., Xun, W., Chen, L., Xu, Z., Zhang, N., Feng, H., et al. (2022). Rhizosphere microbes enhance plant salt tolerance: Toward crop production in saline soil. Comput. Struct. Biotechnol. J. 20, 6543–6551. doi: 10.1016/j.csbj.2022.11.046
Ma, H., Pineda, A., Hannula, S. E., Kielak, A. M., Setyarini, S., Bezemer, T. M. (2020). Steering root microbiomes of a commercial horticultural crop with plant-soil feedbacks. Appl. Soil Ecol. 150, 103468. doi: 10.1016/j.apsoil.2019.103468
Mackie, K. A., Zeiter, M., Bloor, J. M., Stampfli, A. (2018). Plant functional groups mediate drought resistance and recovery in a multisite grassland experiment. J. Ecol. 107, 937–949. doi: 10.1111/1365-2745.13102
Mallon, C. A., Roux, X. L., Doorn, G. S., Dini-Andreote, F., Poly, F., Salles, J. F. (2018). The impact of failure: unsuccessful bacterial invasions steer the soil microbial community away from the invader’s niche. ISME J. 12, 728–741. doi: 10.1038/s41396-017-0003-y
Mariotte, P., Mariotte, P., Mehrabi, Z., Bezemer, T. M., Deyn, G. B., Kulmatiski, A., et al. (2018). Plant-soil feedback: bridging natural and agricultural sciences. Trends Ecol. Evol. 33, 129–142. doi: 10.1016/j.tree.2017.11.005C
Mariotte, P., Robroek, B. J., Jassey, V. E., Buttler, A. (2015). Subordinate plants mitigate drought effects on soil ecosystem processes by stimulating fungi. Funct. Ecol. 29, 1578–1586. doi: 10.1111/1365-2435.12467
Mishra, P., Mishra, J., Arora, N. K. (2021). Plant growth promoting bacteria for combating salinity stress in plants-Recent developments and prospects: A review. Microbiological Res. 252, 126861. doi: 10.1016/j.micres.2021.126861
Mitter, B., Pfaffenbichler, N., Flavell, R. B., Compant, S., Antonielli, L., Petric, A., et al. (2017). A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00011
Morcillo, R. J. L., Manzanera, M. (2021). The effects of plant-associated bacterial exopolysaccharides on plant abiotic stress tolerance. Metabolites 11, 337. doi: 10.3390/metabo11060337
Munns, R., Tester, M. A. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Negacz, K., Malek, Ž., de Vos, A., Vellinga, P. (2022). Saline soils worldwide: identifying the most promising areas for saline agriculture. J. Arid Environ. 203, 104775. doi: 10.1016/j.jaridenv.2022.104775
Oliver, I. C., Knox, O. G., Flavel, R. J., Wilson, B. R. (2021). “Rhizosphere legacy: plant root interactions with the soil and its biome,” in Rhizosphere biology: interactions between microbes and plants, Rhizosphere Biology. Eds. Sharma, A. K., Gupta, V. (Springer, Singapore). doi: 10.1007/978-981-15-6125-2_6
Otlewska, A., Migliore, M., Dybka-Stępień, K., Manfredini, A., Struszczyk-Świta, K., Napoli, R., et al. (2020). When salt meddles between plant, soil, and microorganisms. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.553087
Pan, J., Huang, C. H., Peng, F., You, Q. G., Liu, F. Y., Xue, X. (2020). Mechanisms of salt tolerance and growth promotion in plant induced by plant growth-promoting rhizobacteria. Biotechnol. Bull. 36, 75–87. doi: 10.13560/j.cnki.biotech.bull.1985.2020-0511
Pan, X., Qin, Y., Yuan, Z. (2018). Potential of a halophyte-associated endophytic fungus for sustaining Chinese white poplar growth under salinity. Symbiosis 76, 109–116. doi: 10.1007/s13199-018-0541-8
Park, I., Seo, Y., Mannaa, M. (2023). Recruitment of the rhizo-microbiome army: assembly determinants and engineering of the rhizosphere microbiome as a key to unlocking plant potential. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1163832
Paul, D., Lade, H. (2014). Plant-growth-promoting rhizobacteria to improve crop growth in saline soils: a review. Agron. Sustain. Dev. 34, 737–752. doi: 10.1007/s13593-014-0233-6
Pereira, E. C., Vázquez de Aldana, B. R., San Emeterio, L., Zabalgogeazcoa, I. (2019). A survey of culturable fungal endophytes from Festuca rubra subsp. pruinosa, a grass from marine Cliffs, reveals a core microbiome. Front. Microbiol. 9, 3321. doi: 10.3389/fmicb.2018.03321
Petermann, J. S., Fergus, A., Turnbull, L. A., Schmid, B. (2008). Janzen-connell effects are widespread and strong enough to maintain diversity in grasslands. Ecology 89, 2399–2406. doi: 10.1890/07-2056.1
Philippot, L., Raaijmakers, J. M., Lemanceau, P., Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Pineda, A., Kaplan, I., Bezemer, T. M. (2017). Steering soil microbiomes to suppress aboveground insect pests. Trends Plant Sci. 22, 770–778. doi: 10.1016/j.tplants.2017.07.002
Pineda, A., Kaplan, I., Hannula, E. S., Ghanem, W., Bezemer, M. T. (2019). Conditioning the soil microbiome through plant-soil feedbacks suppresses an aboveground insect pest. New Phytol. 226, 595–608. doi: 10.1111/nph.16385
Potts, S. G., Biesmeijer, J. C., Kremen, C., Neumann, P., Schweiger, O., Kunin, W. E. (2010). Global pollinator declines: trends, impacts and drivers. Trends Ecol. Evol. 25, 345–353. doi: 10.1016/j.tree.2010.01.007
Preece, C., Peñuelas, J. (2020). A return to the wild: root exudates and food security. Trends Plant Sci. 25, 14–21. doi: 10.1016/j.tplants.2019.09.010
Pulleman, M. M., Creamer, R. E., Hamer, U., Helder, J., Pelosi, C., Pérès, G., et al. (2012). Soil biodiversity, biological indicators and soil ecosystem services-An overview of European approaches. Curr. Opin. Environ. Sustainability 4, 529–538. doi: 10.1016/J.COSUST.2012.10.009
Qadir, M., Noble, A. D., Oster, J. D., Schubert, S., Ghafoor, A. (2005). Driving forces for sodium removal during phytoremediation of calcareous sodic and saline-sodic soils: a review. Soil Use Manage. 21, 173–180. doi: 10.1111/j.1475-2743.2005.tb00122.x
Ramadoss, D., Lakkineni, V., Bose, P., Ali, S., Annapurna, K. (2013). Mitigation of salt stress in wheat seedlings by halotolerant bacteria isolated from saline habitats. Springer Plus 2, 6. doi: 10.1186/2193-1801-2-6
Rietz, D., Haynes, R. (2003). Effects of irrigation-induced salinity and sodicity on soil microbial activity. Soil Biol. Biochem. 35, 845–854. doi: 10.1016/S0038-0717(03)00125-1
Rojas-Tapias, D. F., Moreno-Galván, A., Pardo-Díaz, S., Obando, M., Rivera, D., Bonilla, R. (2012). Effect of inoculation with plant growth-promoting bacteria (PGPB) on amelioration of saline stress in maize (Zea mays L.). Appl. Soil Ecol. 61, 264–272. doi: 10.1016/j.apsoil.2012.01.006
Ruiz-Lozano, J. M., Porcel, R., Azcón, C., Aroca, R. (2012). Regulation by arbuscular mycorrhizae of the integrated physiological response to salinity in plants: new challenges in physiological and molecular studies. J. Exp. Bot. 63, 4033–4044. doi: 10.1093/jxb/ers126
Samreen, T., Naveed, M., Nazir, M. Z., Asghar, H. N., Khan, M. I., Zahir, Z. A., et al. (2021). Seed associated bacterial and fungal endophytes: diversity, life cycle, transmission, and application potential. Appl. Soil Ecol. 168, 104191. doi: 10.1016/j.apsoil.2021.104191
Santoyo, G. (2021). How plants recruit their microbiome? New insights into beneficial interactions. J. Advanced Res. 40, 45–58. doi: 10.1016/j.jare.2021.11.020
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P. D., McRoberts, N., Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Schmid, M. W., van Moorsel, S. J., Hahl, T., de Luca, E., De Deyn, G. B., Wagg, C., et al. (2021). Effects of plant community history, soil legacy and plant diversity on soil microbial communities. J. Ecol. 109, 3007–3023. doi: 10.1111/1365-2745.13714
Schmitz, L., Yan, Z., Schneijderberg, M. A., de Roij, M., Pijnenburg, R., Zheng, Q., et al. (2022). Synthetic bacterial community derived from a desert rhizosphere confers salt stress resilience to tomato in the presence of a soil microbiome. ISME J. 16, 1907–1920. doi: 10.1038/s41396-022-01238-3
Semenova, G. A., Fomina, I. R., Biel, K. Y. (2010). Structural features of the salt glands of the leaf of Distichlis spicata ‘Yensen 4a’ (Poaceae). Protoplasma 240, 75–82. doi: 10.1007/s00709-009-0092-1
Sengupta, S., Majumder, A. L. (2010). Porteresia coarctata (Roxb.) Tateoka, a wild rice: a potential model for studying salt-stress biology in rice. Plant Cell Environ. 33, 526–542. doi: 10.1111/j.1365-3040.2009.02054.x
Shabaan, M., Asghar, H. N., Zahir, Z. A., Zhang, X., Sardar, M. F., Li, H. (2022). Salt-tolerant PGPR confer salt tolerance to maize through enhanced soil biological health, enzymatic activities, nutrient uptake and antioxidant defense. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.901865
Sharma, A., Singh, P., Kumar, S., Kashyap, P. L., Srivastava, A. K., Chakdar, H., et al. (2015). Deciphering diversity of salt-tolerant Bacilli from saline soils of eastern indo-gangetic plains of India. Geomicrobiol. J. 32, 170–180. doi: 10.1080/01490451.2014.938205
Shi, S., Richardson, A. E., O’Callaghan, M., DeAngelis, K. M., Jones, E. E., Stewart, A., et al. (2011). Effects of selected root exudate components on soil bacterial communities. FEMS Microbiol. Ecol. 77, 600–610. doi: 10.1111/j.1574-6941.2011.01150.x
Shi, X., Zhou, Y., Zhao, X., Guo, P., Ren, J., Zhang, H., et al. (2023). Soil metagenome and metabolome of peanut intercropped with sorghum reveal a prominent role of carbohydrate metabolism in salt-stress response. Environ. Exp. Bot. 209, 105274. doi: 10.1016/j.envexpbot.2023.105274
Shrivastava, P. V., Kumar, R. (2015). Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 22, 123–131. doi: 10.1016/j.sjbs.2014.12.001
Siebert, F., Morris, C., Chamane, S., Ntuli, N., Siebert, S. J. (2021). “The functional importance of forbs in grassland ecosystems,” in The Joint XXIV International Grassland and XI Rangeland 2021 Congress, Proceedings XXIV, vol. 1-2. (Kenya: UKnowledge), 35. Theme.
Silva, Y. J., Silva, Y. J., Freire, M. B., Lopes, E. A., Santos, M. A. (2016). Atriplex nummularia Lindl. as alternative for improving salt-affected soils conditions in semiarid environments: a field experiment. Chilean J. Agric. Res. 76, 343–348. doi: 10.4067/S0718-58392016000300012
Song, C., Jin, K., Raaijmakers, J. M. (2021). Designing a home for beneficial plant microbiomes. Curr. Opin. Plant Biol. 62, 102025. doi: 10.1016/j.pbi.2021.102025
Song, J., Wang, B. (2015). Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann. Bot. 115, 541–553. doi: 10.1093/aob/mcu194
Spehn, E. M., Scherer-Lorenzen, M., Schmid, B., Hector, A., Caldeira, M. D., Dimitrakopoulos, P. G., et al. (2002). The role of legumes as a component of biodiversity in a cross-European study of grassland biomass nitrogen. Oikos 98, 205–218. doi: 10.1034/J.1600-0706.2002.980203.X
Stagnari, F., Maggio, A., Galieni, A., Pisante, M. (2017). Multiple benefits of legumes for agriculture sustainability: an overview. Chem. Biol. Technol. Agric. 4, 1–13. doi: 10.1186/s40538-016-0085-1
Strecker, T., Barnard, R. L., Niklaus, P. A., Scherer-Lorenzen, M., Weigelt, A., Scheu, S., et al. (2015). Effects of plant diversity, functional group composition, and fertilization on soil microbial properties in experimental grassland. PloS One 10, e0125678. doi: 10.1371/journal.pone.0125678
Suter, M., Connolly, J., Finn, J. A., Loges, R., Kirwan, L., Sebastià, M. T., et al. (2015). Nitrogen yield advantage from grass-legume mixtures is robust over a wide range of legume proportions and environmental conditions. Global Change Biol. 21, 2424–2438. doi: 10.1111/gcb.12880
Takahashi, F., Suzuki, T., Osakabe, Y., Betsuyaku, S., Kondo, Y., Dohmae, N., et al. (2018). A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556, 235–238. doi: 10.1038/s41586-018-0009-2
Tester, M. A., Bacic, A. (2005). Abiotic stress tolerance in grasses. From model plants to crop plants. Plant Physiol. 137, 791–793. doi: 10.1104/pp.104.900138
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., Singh, B. K. (2020). Plant-microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 1–15. doi: 10.1038/s41579-020-0412-1
Trivedi, P., Mattupalli, C., Eversole, K. A., Leach, J. E. (2021). Enabling sustainable agriculture through understanding and enhancement of microbiomes. New Phytol. 230, 2129–2147. doi: 10.1111/nph.17319
Van Coller, H., Siebert, F., Scogings, P. F. (2018). Herbaceous responses to herbivory, fire and rainfall variability differ between grasses and forbs. South Afr. J. Bot. 119, 94–103. doi: 10.1016/j.sajb.2018.08.024
Van der Putten, W. H., Bardgett, R. D., Bever, J. D., Bezemer, T. M., Casper, B. B., Fukami, T., et al. (2013). Plant-soil feedback: the past, the present and future challenges. J. Ecol. 101, 265–276. doi: 10.1111/1365-2745.12054
Van Eekeren, N., Bommelé, L., Bloem, J., Schouten, T., Rutgers, M., De Goede, R. G., et al. (2008). Soil biological quality after 36 years of ley-arable cropping, permanent grassland and permanent arable cropping. Appl. Soil Ecol. 40, 432–446. doi: 10.1016/j.apsoil.2008.06.010
Van Zelm, E., Zhang, Y., Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005
Varshney, R. K., Bansal, K. C., Aggarwal, P. K., Datta, S. K., Craufurd, P. (2011). Agricultural biotechnology for crop improvement in a variable climate: hope or hype? Trends Plant Sci. 16, 363–371. doi: 10.1016/j.tplants.2011.03.004
Verbruggen, E., van der Heijden, M. G., Weedon, J. T., Kowalchuk, G. A. (2012). Community assembly, species richness and nestedness of arbuscular mycorrhizal fungi in agricultural soils. Mol. Ecol. 21, 2341–2353. doi: 10.1111/j.1365-294X.2012.05534.x
Vukicevich, E., Lowery, T., Bowen, P. A., Úrbez-Torres, J. R., Hart, M. M. (2016). Cover crops to increase soil microbial diversity and mitigate decline in perennial agriculture. A review. Agron. Sustain. Dev. 36, 48. doi: 10.1007/s13593-016-0385-7
Wang, G., Bei, S., Li, J., Bao, X., Zhang, J., Schultz, P. A., et al. (2020). Soil microbial legacy drives crop diversity advantage: Linking ecological plant-soil feedback with agricultural intercropping. J. Appl. Ecol. 58, 496–506. doi: 10.1111/1365-2664.13802
Wang, Z., Song, Y. (2022). Toward understanding the genetic bases underlying plant-mediated “cry for help” to the microbiota. iMeta 1, e8. doi: 10.1002/imt2.8
Wang, R., Wang, C., Feng, Q., Liou, R. M., Lin, Y. (2021). Biological inoculant of salt-tolerant bacteria for plant growth stimulation under different saline soil conditions. J. Microbiol. Biotechnol. 31, 398–407. doi: 10.4014/jmb.2009.09032
Wang, S., Zhao, Z. Z., Ge, S., Zhang, K., Tian, C., Mai, W. (2021). Suaeda salsa/Zea mays L. intercropping in saline soil on plant growth and rhizospheric physiological processes. Agriculture 12, 107. doi: 10.21203/rs.3.rs-393380/v1
Wardle, D. A., Bardgett, R. D., Klironomos, J., Setälä, H., Putten, W. H., Wall, D. H. (2004). Ecological linkages between aboveground and belowground biota. Science 304, 1629–1633. doi: 10.1126/science.1094875
Wei, Z., Jousset, A. (2017). Plant breeding goes microbial. Trends Plant Sci. 22, 555–558. doi: 10.1016/j.tplants.2017.05.009
West, G., Inzé, D., Beemster, G. T. (2004). Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol. 135, 1050–1058. doi: 10.1104/pp.104.040022
Win, K. T., Wasai-Hara, S., Tanaka, F., Oo, A. Z., Minamisawa, K., Shimoda, Y., et al. (2023). Synergistic N2-fixation and salt stress mitigation in soybean through dual inoculation of ACC deaminase-producing Pseudomonas and Bradyrhizobium. Sci. Rep. 13, 17050. doi: 10.1038/s41598-023-43891-4
Wurst, S., Ohgushi, T. (2015). Do plant- and soil-mediated legacy effects impact future biotic interactions? Funct. Ecol. 29, 1373–1382. doi: 10.1111/1365-2435.12456
Xiong, W., Song, Y., Yang, K., Gu, Y., Wei, Z., Kowalchuk, G. A., et al. (2020). Rhizosphere protists are key determinants of plant health. Microbiome 8, 27. doi: 10.1186/s40168-020-00799-9
Yang, H. X., Liu, R. J., Guo, S. X. (2014). Effects of arbuscular mycorrhizal fungus Glomus mosseae on the growth characteristics of Festuca arundinacea under salt stress conditions. Acta Prataculturae Sin. 23, 195–203. doi: 10.11686/cyxb20140424
Yang, H., Zhang, W. P., Li, L. (2021). Intercropping: feed more people and build more sustainable agroecosystems. Front. Agric. Sci. Eng. 8, 373–386. doi: 10.15302/j-fase-2021398
Younesi, O., Chaichi, M. R., Postini, K. (2013). Salt tolerance in alfalfa following inoculation with Pseudomonas. Middle East J. Sci. Res. 16, 101–107. doi: 10.5829/idosi.mejsr.2013.16.01.11626
Yu, Z., Duan, X., Luo, L., Dai, S., Ding, Z., Xia, G. (2020). How plant hormones mediate salt stress responses. Trends Plant Sci. 25, 1117–1130. doi: 10.1016/j.tplants.2020.06.008
Yu, B., Zheng, W., Xing, L., Zhu, J., Persson, S., Zhao, Y. (2022). Root twisting drives halotropism via stress-induced microtubule reorientation. Dev. Cell 57, 2412–2425. doi: 10.1101/2022.06.05.494861
Yuan, Z., Druzhinina, I. S., Labbé, J. L., Redman, R. S., Qin, Y., Rodriguez, R., et al. (2016). Specialized microbiome of a halophyte and its role in helping non-host plants to withstand salinity. Sci. Rep. 6, 32467. doi: 10.1038/srep32467
Yuan, F., Leng, B., Wang, B. (2016). Progress in studying salt secretion from the salt glands in recretohalophytes: how do plants secrete salt? Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00977
Zerrouk, I. Z., Benchabane, M., Khelifi, L., Yokawa, K., Ludwig-Müller, J., Baluška, F. (2016). A Pseudomonas strain isolated from date-palm rhizospheres improves root growth and promotes root formation in maize exposed to salt and aluminum stress. J. Plant Physiol. 191, 111–119. doi: 10.1016/j.jplph.2015.12.009
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhang, G., Bai, J., Zhai, Y., Jia, J., Zhao, Q., Wang, W., et al. (2023). Microbial diversity and functions in saline soils: a review from a biogeochemical perspective. J. Advanced Res. doi: 10.1016/j.jare.2023.06.015
Zhang, W., Wang, C., Xue, R., Wang, L. (2019). Effects of salinity on the soil microbial community and soil fertility. J. Integr. Agric. 18, 1360–1368. doi: 10.1016/S2095-3119(18)62077-5
Zhao, C., Zhang, H., Song, C., Zhu, J., Shabala, S. (2020). Mechanisms of plant responses and adaptation to soil salinity. Innovation 1, 100017. doi: 10.1016/j.xinn.2020.100017
Keywords: saline soil, halophytes, salt-tolerant crops, beneficial microorganism, salt tolerance
Citation: Ma Y, Zheng C, Bo Y, Song C and Zhu F (2024) Improving crop salt tolerance through soil legacy effects. Front. Plant Sci. 15:1396754. doi: 10.3389/fpls.2024.1396754
Received: 06 March 2024; Accepted: 22 April 2024;
Published: 10 May 2024.
Edited by:
Dinesh Yadav, Deen Dayal Upadhyay Gorakhpur University, IndiaReviewed by:
Yuri Shavrukov, Flinders University, AustraliaAbhishek Joshi, Mohanlal Sukhadia University, India
Copyright © 2024 Ma, Zheng, Bo, Song and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Zhu, emh1ZmVuZ0BzanppYW0uYWMuY24=
 Yue Ma1,2
Yue Ma1,2 Chunyan Zheng
Chunyan Zheng Chunxu Song
Chunxu Song Feng Zhu
Feng Zhu