- Division of Plant Science and Technology, Bond Life Sciences Center, and Interdisciplinary Plant Group, University of Missouri, Columbia, MO, United States
Phytopathogenic microbes use secreted effector proteins to increase their virulence in planta. If these effectors or the results of their activity are detected by the plant cell, the plant will mount an immune response which applies evolutionary pressure by reducing growth and success of the pathogen. Bacterial effector proteins in the AvrRps4 family (AvrRps4, HopK1, and XopO) have commonly been used as tools to investigate plant immune components. At the same time, the in planta functions of this family of effectors have yet to be fully characterized. In this minireview we summarize current knowledge about the AvrRps4 effector family with emphasis on properties of the proteins themselves. We hypothesize that the HopK1 C-terminus and the AvrRps4 C-terminus, though unrelated in sequence and structure, are broadly related in functions that counteract plant defense responses.
Introduction
Plants constantly interact with new challenges presented by their surroundings, and perhaps one of the most relevant interactions for humanity is that between a plant and a pest or pathogen attempting to benefit from the unique position of plants as primary nutrient sources. In the case of microbial biotrophic pathogens, the association between host and co-evolved pathogen is accompanied by intricate manipulation of the plant for the pathogen’s benefit (Jones and Dangl, 2006; Ebert and Fields, 2020). Usually manipulation involves secretion by the attacking pathogen of proteins called effectors into the plant cell which, broadly, attempt to improve the attacker’s virulence and ultimate ecological success by blocking plant recognition and resistance and/or by increasing availability of resources the attacker needs (McCann and Guttman, 2008; Feng and Zhou, 2012; Kazan and Lyons, 2014; Wang et al., 2022). Successful pathogens cause diseased crops, reduced yield and, in extreme cases, total crop loss. Identifying the in planta virulence targets, or the functions, of these effectors is the first step along a pathway that leads to developing crops with greater resistance to disease and eventually to greater food security. In this review, we take this first step by examining the well-known AvrRps4 effector family and reviewing published information that may provide insight into the in planta functions of each effector domain.
As the first isolated bacterial effector known to trigger resistance through a Toll/Interleukin-1 Receptor - Nucleotide Binding - Leucine Rich Repeat (TNL) resistance protein, AvrRps4 was used widely among plant-microbe researchers as a tool for the discovery and investigation of plant resistance components (Gassmann et al., 1999). Though AvrRps4 has been invaluable to the field of molecular plant-pathogen interactions, many facets of the effector’s virulence and avirulence targets in planta remain unknown.
The three members of the AvrRps4 effector family (AvrRps4, HopK1, and XopO) are characterized by high N-terminal homology to each other followed by a GGGKRVY motif. After secretion into a plant cell via the type III secretion system (T3SS), all members of the family are processed by an unknown protease(s) that cleaves the N-terminus from the C-terminus between GG and GKRVY. This occurs between G133 and G134 (G123 and 124 in XopO) and for unknown reasons requires an arginine at position 112 (R112) (R111 in XopO) (Figure 1) (Roden et al., 2004; Sohn et al., 2009; Li et al., 2014; Halane et al., 2018; Su et al., 2021; Nguyen et al., 2022). AvrRps4 and XopO possess homologous AvrRps4-type N- and C-termini (AvrRps4N and AvrRps4C), but HopK1 only shares N-terminal homology with AvrRps4. HopK1 C-terminus (HopK1C) instead shares homology with XopAK (Figure 1) and RipBQ, a Ralstonia effector not discussed in this minireview due to limited space and literature (Sabbagh et al., 2019). Although XopAK is not a proper member of the AvrRps4 effector family due to lack of a GGGKRVY motif or any homology to AvrRps4N, it is present in multiple important bacterial crop pathogens, including those that cause banana Xanthomonas wilt, maize bacterial leaf streak, bacterial leaf streak of rice, and all currently known citrus canker strains (Moreira et al., 2010; Studholme et al., 2010; Jalan et al., 2011, 2013; Bart et al., 2012; Darrasse et al., 2013; Wichmann et al., 2013; Gagnevin et al., 2014; Aritua et al., 2015). We believe its inclusion will provide useful insights into the in planta function(s) of HopK1C.
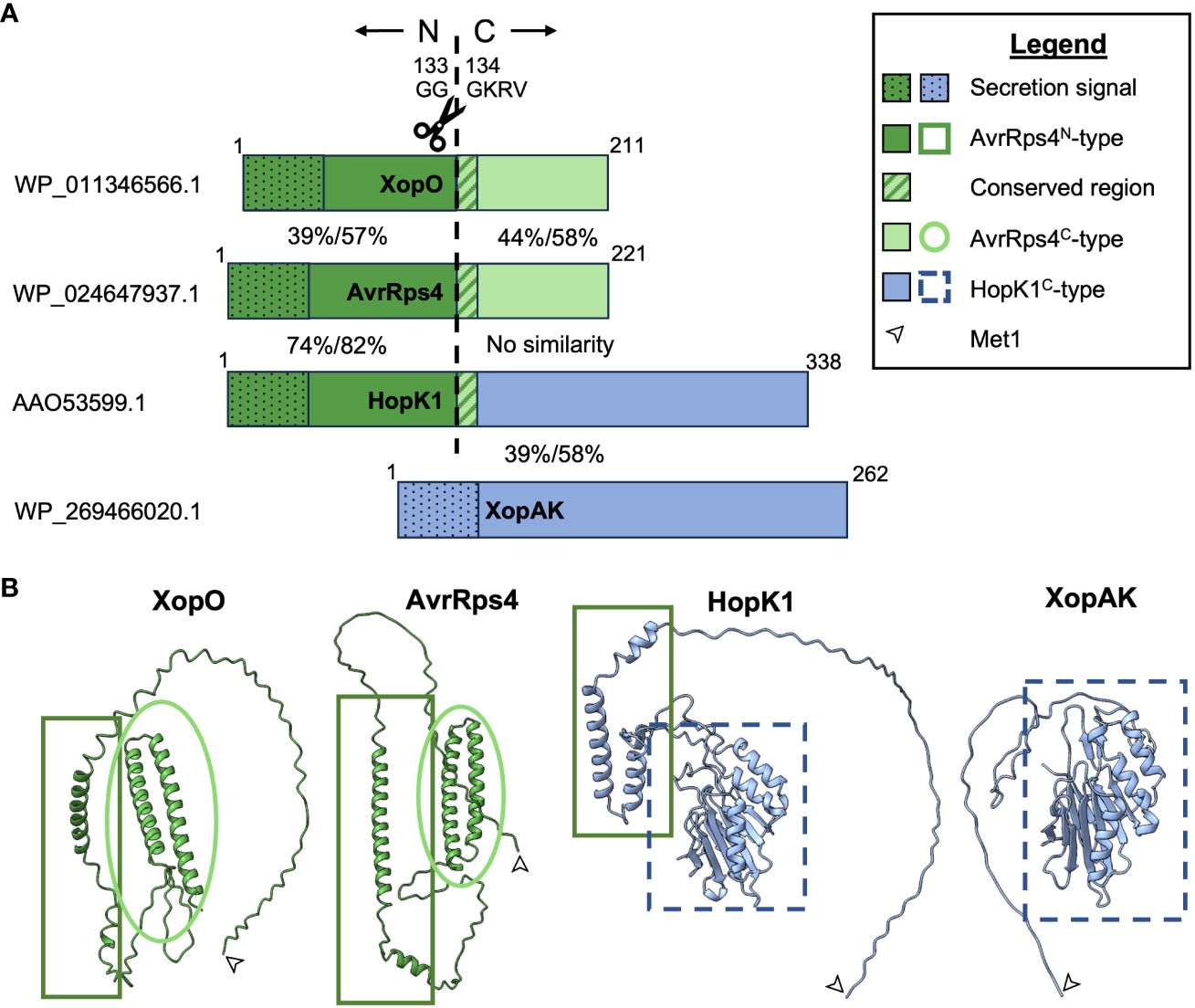
Figure 1 Visual representations of AvrRps4 effector family homology. (A) Proportional amino acid sequence homology indicated by color and pattern. AvrRps4N-type sequences are indicated in dark green, AvrRps4C-type sequences are indicated in light green, and HopK1C-type sequences are indicated in light blue. NCBI accession numbers for proteins used in this comparison are listed on the left. Percentages between the termini indicate % identity/% positives for each pair as predicted by NCBI’s blastp (Altschul et al., 1990). The ~50AA secretion signals (represented by dots) are included in this comparison but the 12AA central homologous regions (represented by stripes) are not. (B) Protein structural homology with Met1 indicated by an arrow. Structural predictions were made with ColabFold and visualized using ChimeraX (Pettersen et al., 2021; Mirdita et al., 2022).
This summary is intended to be used as a collection of knowledge on the topic and to be viewed as a description of possibilities to be explored further. We strive to consider the preponderance of evidence available rather than focusing on any singular result when we discuss these effectors below.
AvrRps4
The founding member of the family, the avrRps4 avirulence gene from Pseudomonas syringae pv. pisi (Psp) str. 151, was described in 1996 and was one of the first avirulence genes shown to be present in all tested strains of a pathovar (Hinsch and Staskawicz, 1996). In both Psp 151 and P. syringae pv. phaseolicola str. 1448A, avrRps4 is located on a plasmid (Kim et al., 1998; Joardar et al., 2005).
After secretion and subsequent processing inside the plant cell, both AvrRps4 termini are largely nucleo-cytoplasmically localized (Bhattacharjee et al., 2011; Heidrich et al., 2011; Sohn et al., 2012; Li et al., 2014; Park et al., 2017; Halane et al., 2018). Interestingly, Li et al., 2014 also reported N-terminus-dependent localization of transgenically expressed AvrRps4 and HopK1 to the chloroplast. Independently, in vitro import assays showed localization of HopK1 in chloroplasts (De Torres Zabala et al., 2015). More recent work has shown that deliberate AvrRps4 mislocalization to the plasma membrane by replacing the N-terminus does not prevent processing, accumulation of the C-terminus in the nucleus, or contribution to resistance (Halane et al., 2018). AvrRps4N (aa 1-136) in particular has been used with varied success in the past as part of the effector detector vector system (pEDV) for identification of candidate effectors from other bacteria (Kim et al., 2023), oomycetes (Sohn et al., 2007; Fabro et al., 2011; Kemen et al., 2011; Badel et al., 2013; Upadhyaya et al., 2014), fungi (Sharma et al., 2013; Zhang et al., 2020), insects (Navarro-Escalante et al., 2020; Peng et al., 2023), and nematodes (Shi et al., 2018b, 2018a). This, alongside the apparent similarities between N-terminal secretion signals of type III secreted effector proteins and chloroplast/mitochondrial transit peptides (Guttman et al., 2002) indicate that while a portion of the cellular pool of AvrRps4 may be targeted to the chloroplast for its virulence function, presence of AvrRps4N is likely not the only factor leading to the observed chloroplast localization.
Further, the AvrRps4 N-terminus appears to have a greater role in virulence than exclusively as a signal peptide. Transgenic expression of AvrRps4N alone in Arabidopsis thaliana (Arabidopsis) Col-0 increased virulence of P. syringae pv. tomato (Pto) strain DC3000 hopk1- (Halane et al., 2018). Furthermore, AvrRps4N was necessary and sufficient to trigger a strong hypersensitive response (HR) in resistant lettuce cultivars (Halane et al., 2018; Su et al., 2021). When co-expressed in resistant lettuce with AvrRps4C, this AvrRps4N-triggered HR phenotype was reduced. In contrast, full avirulence in Arabidopsis Col-0 requires the presence of both AvrRps4N and AvrRps4C (Halane et al., 2018). These dual roles of AvrRps4N are the hallmarks of an effector: benefiting the pathogen while being detrimental enough to some plants that they have evolved to detect it. As for a virulence function, reports directly examining the N-terminus suggest AvrRps4N interacts with EDS1 (Bhattacharjee et al., 2011; Halane et al., 2018), an important immune regulator for TNL proteins and basal resistance (Dongus and Parker, 2021). Interaction with EDS1 has also been reported for the unrelated P. syringae pv. glycinea effector AvrA1 when transiently expressed in N. benthamiana (Wang et al., 2014). For both AvrRps4N and AvrA1 the immediate functional effects of their interactions with EDS1 in terms of virulence or avirulence have not been determined. Taken together, these works highlight a need for greater understanding of AvrRps4N functions.
Contrastingly, AvrRps4C functions are well understood. An electronegative patch (Sohn et al., 2012) interacts with defense-related WRKY transcription factors (Kim et al., 2024) and with the C-terminal integrated WRKY decoy domain of resistance protein RRS1 (Sarris et al., 2015; Mukhi et al., 2021; Kim et al., 2024), providing a mechanism for earlier observations of AvrRps4-dependent suppression of HR in tobacco (Fujikawa et al., 2006; Cvetkovska and Vanlerberghe, 2012), pattern-triggered immunity (PTI) in Arabidopsis, and enhancement of pathogen growth within N. benthamiana (Sohn et al., 2009). Processing of AvrRps4 is not necessary for AvrRps4C-specific triggering of HR in cultivars of Brassica rapa (turnip) nor for resistance to be triggered in Arabidopsis, but is necessary for its virulence function (Sohn et al., 2009). Other work has shown that AvrRps4C directly interacts with BRUTUS (BTS), an iron-sensing protein in the nucleus, causing interference with the degradation of bHLH115 and ILR3 and, when not recognized, leading to increased levels of apoplastic iron (Xing et al., 2021).
XopO
XopO was first described in 2004 from Xanthomonas euvesicatoria pv. vesicatoria (Xev) str. 85-10 and is only known to exist in this strain (formerly: Xanthomonas axonopodis pv. vesicatoria and Xanthomonas campestris pv. vesicatoria) and Xanthomonas oryzae pv. oryzicola (Xoc), neither of which appear to be greatly affected by its deletion (Roden et al., 2004; Hajri et al., 2009, 2012; Liao et al., 2020). Indeed, in multiple strains XopO was found to be mutationally inactivated or to possess insertions or deletions within the first 100 amino acids of the protein, suggesting an evolutionary advantage in removing the effector from the secretion repertoire (Wang et al., 2011, p. 20; Barak et al., 2016).
It is not known whether XopOC interacts with WRKY proteins in the same manner as AvrRps4C, but we do know that XopOC does not trigger HR in turnip as AvrRps4C does and likewise does not appear to weaken the recognition of XopON in lettuce to the same degree that AvrRps4C does for AvrRps4N (Sohn et al., 2009; Nguyen et al., 2022). These amino acid polymorphisms and the differential recognition between AvrRps4C and XopOC may be related to functionality within specific plant hosts, and future functional analyses of these effectors should take this into consideration.
HopK1
HopK1 (previously HopPtoK) was identified in 2002 from Pto DC3000 (Petnicki-Ocwieja et al., 2002). HopK1 in its full length is found almost exclusively within pathovars of P. syringae, although not every pathovar or strain carries it. After AvrRps4, HopK1 is the most well-researched bacterial effector protein in the AvrRps4 family.
In Pto DC3000, the hopK1 gene is located on the chromosome and is encoded in a 24 kb Tn6022-like element, but in other strains hopK1 may occur on a plasmid (Li et al., 2014; Peters et al., 2014). In the case of Pto DAPP-PG 215, hopK1 is borne on p107, a small plasmid which possesses two prophage regions and some precursors for coronatine production in addition to the effector (Orfei et al., 2023).
Pto DC3000 hopk1- strains are significantly reduced in their ability to grow and cause disease within plants. The full wild-type growth of the mutant in Arabidopsis Col-0 was not restored by simultaneous transgenic expression of HopK1C but was restored by expression in the same manner of the full-length protein. The N-terminus was also necessary for reduction in reactive oxygen species (ROS) and callose deposition, both indicators of PTI, indicating that HopK1N is indispensable for effector virulence activities (Li et al., 2014). Presence of HopK1 in an otherwise effectorless Pto DC3000 strain did not contribute to nor inhibit growth compared to Pto DC3000 with no effectors at all, nor did it reduce production of ROS in N. benthamiana. Nevertheless, authors reported a moderate reduction in “non-avirulence-related” cell death caused by other effectors when HopK1 was present (Wei et al., 2018). Finally, expression of HopK1 did not suppress flg22-mediated activation of FRK1-LUC, another PTI indicator, in Arabidopsis protoplasts (He et al., 2006). Together, these findings may indicate that HopK1N works synergistically with an unknown Pto DC3000 effector(s) in order to counteract early PTI events. This synergistic interaction resulting in reduced ROS and callose may also be required for maximum effectiveness of HopK1, as HopK1C has been shown to travel intercellularly up to one cell layer away by using plasmodesmata as long as movement is not restricted by excess callose deposition (Li et al., 2021; Iswanto et al., 2022).
When infiltrated into Arabidopsis Col-0 alongside AvrRps4C, HopK1N fully complements the resistance response seen when AvrRps4N and AvrRps4C are infiltrated together. The central conserved portion of the N-terminus containing the positively charged R112 is required for recognition in lettuce (Halane et al., 2018; Su et al., 2021; Nguyen et al., 2022). However, processing is not required for full pathogenicity (Li et al., 2014).
As mentioned above with AvrRps4, HopK1 was bioinformatically predicted to localize to the chloroplast and was shown to localize to the chloroplast stroma when inducibly expressed by transgenic Arabidopsis, yet nucleo-cytoplasmically when expressed by A. tumefaciens in a constitutive transient manner in N. benthamiana (Li et al., 2014; De Torres Zabala et al., 2015).
XopAK
Perhaps representative of its presence in many Xanthomonas species with different life histories, xopAK is variable in length and number of accumulated mutations. For example, the gene showed very few polymorphic sites among strains of X. axonopodis manihotis (Bart et al., 2012; Trujillo et al., 2014) but is believed to be inactivated in X. phaseoli pv. dieffenbachiae due to a frameshift (Constantin et al., 2017) and is in some cases missing a significant coding region that nearly halves the length of the protein (Fan et al., 2022). Machine learning predicted the presence of a deaminase catalytic domain at the C-terminus of XopAK and at least one potential myristoylation or palmitoylation motif which could target the effector to the plasma membrane (Teper et al., 2015; Barak et al., 2016). This putative deaminase domain is well within the region of homology with HopK1C and may therefore contribute to a more thorough picture of the AvrRps4 family’s in planta functions.
Like HopK1, XopAK had no significant effect on flg22-triggered activation of FRK1 in Arabidopsis protoplasts (He et al., 2006; Popov et al., 2016). Unlike hopK1, deletion of xopAK from Xoc caused no visible changes in disease symptoms on its host (rice) (Liao et al., 2020).
Discussion
The AvrRps4-type N-terminus is a metaphorical black box for researchers of this family. Its proposed functional region is very small, only about 37 aa long (Su et al., 2021), yet it provokes a strong HR from lettuce (Halane et al., 2018; Su et al., 2021), interacts with a major TNL resistance hub (EDS1) (Bhattacharjee et al., 2011; Halane et al., 2018), and appears more important for the growth of Pto DC3000 than HopK1C alone (Li et al., 2014; Halane et al., 2018), but we still know almost nothing about its function. Intriguingly, using AvrRps4N similarly to pEDV for delivery of the full-length unrelated effector XopQ into N. benthamiana cells resulted in a weaker response by the plant than when XopQ was delivered by the AvrRpt2 signal peptide (Gantner et al., 2018). Authors did not test further but hypothesized that this weakness could be the result of differences in stability or translocated amount of protein. While this may be the case, it is also possible that AvrRps4N interaction with EDS1 interferes with the EDS1-mediated recognition of XopQ (Adlung et al., 2016), but more study is needed.
We also do not yet know how detrimental simply possessing an endogenous avrRps4N might be for bacteria themselves. xopO and xopAK both naturally occur within the genome of Xev 85-10, but the same is not true for avrRps4 and hopK1, which to date have never been found to naturally co-occur. There are no examples of a strain possessing multiple members of the AvrRps4 effector family (sensu stricto), multiple copies of a single member, the avrRps4-type C-terminus without an attached N-terminus, or even a single copy of the avrRps4-type N-terminus without an attached C-terminus of any type. It is tempting to conclude from this that AvrRps4N may be detrimental to the bacterium over evolutionary time, and we look forward to any future work examining AvrRps4N more thoroughly.
Neither HopK1C nor XopAK cause any significant effect on flg22-triggered FRK1 activation (He et al., 2006; Popov et al., 2016), an early PTI response, yet HopK1 has still been shown to block or reduce HR and PTI (Jamir et al., 2004; Li et al., 2014; Gimenez-Ibanez et al., 2018; Wei et al., 2018). This reduction appears to be related to the action of HopK1N and to be essential for the full virulence of HopK1C, yet its homologue XopAK occurs in many more species of Xanthomonas without the N-terminus-bearing XopO than it does with it. In fact, their co-occurrence is limited to the only two known pathovars possessing XopO: Xev and Xoc. This raises the question whether XopAK benefits from another effector with redundant function when XopO is not present, or if polymorphisms between the homologues render them dissimilar enough that a ‘helper’ effector is not necessary for XopAK. In the latter case, addition of XopO may have presented an opportunity for range expansion by the effector and the pathogen alike.
As summarized in this review, the presence of xopO and xopAK within the same strains of Xanthomonas, hopK1 association with transposable elements (Peters et al., 2014; Orfei et al., 2023), and avrRps4 and hopK1’s occasional location on plasmids (Kim et al., 1998; Chang et al., 2005; Orfei et al., 2023) seem to indicate that our modern chimeric effectors may have been formed in a Xanthomonas melting pot and then shared with bacteria inhabiting a similar niche, such as Pseudomonas, via horizontal gene transfer (HGT). HGT has been implicated in the evolution of Pseudomonas (Kim et al., 1998; Rohmer et al., 2004; Dillon et al., 2019) and Xanthomonas (Merda et al., 2017; Ruh et al., 2017; Chen et al., 2018) pathogenicity in the past, though not specifically for the AvrRps4 effector family. The rapidity and therefore the agricultural implications of these transfers could be uncovered with examination of the evolutionary history of the family.
As the 30th anniversary of the discovery and cloning of avrRps4 approaches, we hope investigators will revisit the last 3 decades of AvrRps4 research and open questions with renewed interest, paving the way for (at least)! 3 more decades of insightful molecular plant-pathogen interactions research.
Author contributions
KH: Conceptualization, Data curation, Writing – original draft, Writing – review & editing. WG: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by a University of Missouri Life Sciences Graduate Fellowship (KNH) and grants from NSF (IOS-1456181) and USDA-NIFA (PBI 2022-11933) (WG).
Acknowledgments
We apologize to colleagues whose work we were unable to include. We thank members of the Gassmann lab and Michael Pisias for discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adlung, N., Prochaska, H., Thieme, S., Banik, A., Blüher, D., John, P., et al. (2016). Non-host resistance induced by the Xanthomonas effector XopQ is widespread within the genus Nicotiana and functionally depends on EDS1. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01796
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Aritua, V., Harrison, J., Sapp, M., Buruchara, R., Smith, J., Studholme, D. J. (2015). Genome sequencing reveals a new lineage associated with lablab bean and genetic exchange between Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01080
Badel, J. L., Piquerez, S. J. M., Greenshields, D., Rallapalli, G., Fabro, G., Ishaque, N., et al. (2013). In planta effector competition assays detect Hyaloperonospora arabidopsidis effectors that contribute to virulence and localize to different plant subcellular compartments. Mol. Plant Microbe Interact. 26, 745–757. doi: 10.1094/MPMI-06-12-0154-R
Barak, J. D., Vancheva, T., Lefeuvre, P., Jones, J. B., Timilsina, S., Minsavage, G. V., et al. (2016). Whole-genome sequences of Xanthomonas euvesicatoria strains clarify taxonomy and reveal a stepwise erosion of type 3 effectors. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01805
Bart, R., Cohn, M., Kassen, A., McCallum, E. J., Shybut, M., Petriello, A., et al. (2012). High-throughput genomic sequencing of cassava bacterial blight strains identifies conserved effectors to target for durable resistance. Proc. Natl. Acad. Sci. 109, E1972–E1979. doi: 10.1073/pnas.1208003109
Bhattacharjee, S., Halane, M. K., Kim, S. H., Gassmann, W. (2011). Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334, 1405–1408. doi: 10.1126/science.1211592
Chang, J. H., Urbach, J. M., Law, T. F., Arnold, L. W., Hu, A., Gombar, S., et al. (2005). A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc. Natl. Acad. Sci. 102, 2549–2554. doi: 10.1073/pnas.0409660102
Chen, N. W. G., Serres-Giardi, L., Ruh, M., Briand, M., Bonneau, S., Darrasse, A., et al. (2018). Horizontal gene transfer plays a major role in the pathological convergence of Xanthomonas lineages on common bean. BMC Genomics 19, 606. doi: 10.1186/s12864-018-4975-4
Constantin, E. C., Haegeman, A., Van Vaerenbergh, J., Baeyen, S., Van Malderghem, C., Maes, M., et al. (2017). Pathogenicity and virulence gene content of Xanthomonas strains infecting Araceae, formerly known as Xanthomonas axonopodis pv. dieffenbachiae. Plant Pathol. 66, 1539–1554. doi: 10.1111/ppa.12694
Cvetkovska, M., Vanlerberghe, G. C. (2012). Coordination of a mitochondrial superoxide burst during the hypersensitive response to bacterial pathogen in Nicotiana tabacum. Plant Cell Environ. 35, 1121–1136. doi: 10.1111/j.1365-3040.2011.02477.x
Darrasse, A., Carrère, S., Barbe, V., Boureau, T., Arrieta-Ortiz, M. L., Bonneau, S., et al. (2013). Genome sequence of Xanthomonas fuscans subsp. fuscans strain 4834-R reveals that flagellar motility is not a general feature of xanthomonads. BMC Genomics 14, 761. doi: 10.1186/1471-2164-14-761
De Torres Zabala, M., Littlejohn, G., Jayaraman, S., Studholme, D., Bailey, T., Lawson, T., et al. (2015). Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat. Plants 1, 15074. doi: 10.1038/nplants.2015.74
Dillon, M. M., Almeida, R. N. D., Laflamme, B., Martel, A., Weir, B. S., Desveaux, D., et al. (2019). Molecular evolution of Pseudomonas syringae type III secreted effector proteins. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00418
Dongus, J. A., Parker, J. E. (2021). EDS1 signalling: At the nexus of intracellular and surface receptor immunity. Curr. Opin. Plant Biol. 62, 102039. doi: 10.1016/j.pbi.2021.102039
Ebert, D., Fields, P. D. (2020). Host–parasite co-evolution and its genomic signature. Nat. Rev. Genet. 21, 754–768. doi: 10.1038/s41576-020-0269-1
Fabro, G., Steinbrenner, J., Coates, M., Ishaque, N., Baxter, L., Studholme, D. J., et al. (2011). Multiple candidate effectors from the Oomycete pathogen Hyaloperonospora arabidopsidis suppress host plant immunity. PloS Pathog. 7, e1002348. doi: 10.1371/journal.ppat.1002348
Fan, Q., Bibi, S., Vallad, G. E., Goss, E. M., Hurlbert, J. C., Paret, M. L., et al. (2022). Identification of genes in Xanthomonas euvesicatoria pv. rosa that are host limiting in tomato. Plants 11, 796. doi: 10.3390/plants11060796
Feng, F., Zhou, J.-M. (2012). Plant–bacterial pathogen interactions mediated by type III effectors. Biot. Interact. 15, 469–476. doi: 10.1016/j.pbi.2012.03.004
Fujikawa, T., Yamashita, T., Tsuyumu, S. (2006). Hypersensitive response suppression by type III effectors of plant pathogenic bacteria. J. Gen. Plant Pathol. 72, 176–179. doi: 10.1007/s10327-005-0268-2
Gagnevin, L., Bolot, S., Gordon, J. L., Pruvost, O., Vernière, C., Robène, I., et al. (2014). Draft genome sequence of Xanthomonas axonopodis pv. allii strain CFBP 6369. Genome Announc 2, e00727-14. doi: 10.1128/genomea.00727-14
Gantner, J., Ordon, J., Ilse, T., Kretschmer, C., Gruetzner, R., Löfke, C., et al. (2018). Peripheral infrastructure vectors and an extended set of plant parts for the Modular Cloning system. PloS One 13, e0197185. doi: 10.1371/journal.pone.0197185
Gassmann, W., Hinsch, M. E., Staskawicz, B. J. (1999). The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 20, 265–277. doi: 10.1046/j.1365-313X.1999.t01-1-00600.x
Gimenez-Ibanez, S., Hann, D. R., Chang, J. H., Segonzac, C., Boller, T., Rathjen, J. P. (2018). Differential suppression of Nicotiana benthamiana innate immune responses by transiently expressed Pseudomonas syringae type III effectors. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00688
Guttman, D. S., Vinatzer, B. A., Sarkar, S. F., Ranall, M. V., Kettler, G., Greenberg, J. T. (2002). A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295, 1722–1726. doi: 10.1126/science.295.5560.1722
Hajri, A., Brin, C., Hunault, G., Lardeux, F., Lemaire, C., Manceau, C., et al. (2009). A «repertoire for repertoire» hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas. PloS One 4, e6632. doi: 10.1371/annotation/92d243d0-22b2-44da-9618-83b4aa252724
Hajri, A., Brin, C., Zhao, S., David, P., Feng, J.-X., Koebnik, R., et al. (2012). Multilocus sequence analysis and type III effector repertoire mining provide new insights into the evolutionary history and virulence of Xanthomonas oryzae. Mol. Plant Pathol. 13, 288–302. doi: 10.1111/j.1364-3703.2011.00745.x
Halane, M. K., Kim, S. H., Spears, B. J., Garner, C. M., Rogan, C. J., Okafor, E. C., et al. (2018). The bacterial type III-secreted protein AvrRps4 is a bipartite effector. PloS Pathog. 14, e1006984. doi: 10.1371/journal.ppat.1006984
He, P., Shan, L., Lin, N.-C., Martin, G. B., Kemmerling, B., Nürnberger, T., et al. (2006). Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125, 563–575. doi: 10.1016/j.cell.2006.02.047
Heidrich, K., Wirthmueller, L., Tasset, C., Pouzet, C., Deslandes, L., Parker, J. E. (2011). Arabidopsis EDS1 connects pathogen effector recognition to cell compartment–specific immune responses. Science 334, 1401–1404. doi: 10.1126/science.1211641
Hinsch, M., Staskawicz, B. (1996). Identification of a new Arabidopsis disease resistance locus, RPs4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi. Mol. Plant Microbe Interact. 9, 55–61. doi: 10.1094/MPMI-9-0055
Iswanto, A. B. B., Vu, M. H., Pike, S., Lee, J., Kang, H., Son, G. H., et al. (2022). Pathogen effectors: what do they do at plasmodesmata? Mol. Plant Pathol. 23, 795–804. doi: 10.1111/mpp.13142
Jalan, N., Aritua, V., Kumar, D., Yu, F., Jones, J. B., Graham, J. H., et al. (2011). Comparative genomic analysis of Xanthomonas axonopodis pv. citrumelo F1, which causes citrus bacterial spot disease, and related strains provides insights into virulence and host specificity. J. Bacteriol 193, 6342–6357. doi: 10.1128/JB.05777-11
Jalan, N., Kumar, D., Andrade, M. O., Yu, F., Jones, J. B., Graham, J. H., et al. (2013). Comparative genomic and transcriptome analyses of pathotypes of Xanthomonas citri subsp. citri provide insights into mechanisms of bacterial virulence and host range. BMC Genomics 14, 551. doi: 10.1186/1471-2164-14-551
Jamir, Y., Guo, M., Oh, H.-S., Petnicki-Ocwieja, T., Chen, S., Tang, X., et al. (2004). Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J. 37, 554–565. doi: 10.1046/j.1365-313X.2003.01982.x
Joardar, V., Lindeberg, M., Jackson, R. W., Selengut, J., Dodson, R., Brinkac, L. M., et al. (2005). Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol 187, 6488–6498. doi: 10.1128/JB.187.18.6488-6498.2005
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kazan, K., Lyons, R. (2014). Intervention of phytohormone pathways by pathogen effectors. Plant Cell 26, 2285–2309. doi: 10.1105/tpc.114.125419
Kemen, E., Gardiner, A., Schultz-Larsen, T., Kemen, A. C., Balmuth, A. L., Robert-Seilaniantz, A., et al. (2011). Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PloS Biol. 9, e1001094. doi: 10.1371/journal.pbio.1001094
Kim, B., Yu, W., Kim, H., Dong, Q., Choi, S., Prokchorchick, M., et al. (2023). A plasma membrane nucleotide-binding leucine-rich repeat receptor mediates the recognition of the Ralstonia pseudosolanacearum effector RipY in Nicotiana benthamiana. Plant Commun. 4, 100640. doi: 10.1016/j.xplc.2023.100640
Kim, H., Kim, J., Choi, D. S., Kim, M.-S., Deslandes, L., Jayaraman, J., et al. (2024). Molecular basis for the interference of the Arabidopsis WRKY54-mediated immune response by two sequence-unrelated bacterial effectors. Plant J. 118, 839-855. doi: 10.1111/tpj.16639
Kim, J. F., Charkowski, A. O., Alfano, J. R., Collmer, A., Beer, S. V. (1998). Sequences related to transposable elements and bacteriophages flank avirulence genes of Pseudomonas syringae. Mol. Plant Microbe Interact. 11, 1247–1252. doi: 10.1094/MPMI.1998.11.12.1247
Li, G., Froehlich, J. E., Elowsky, C., Msanne, J., Ostosh, A. C., Zhang, C., et al. (2014). Distinct Pseudomonas type-III effectors use a cleavable transit peptide to target chloroplasts. Plant J. 77, 310–321. doi: 10.1111/tpj.12396
Li, Z., Variz, H., Chen, Y., Liu, S.-L., Aung, K. (2021). Plasmodesmata-dependent intercellular movement of bacterial effectors. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.640277
Liao, Z.-X., Li, J.-Y., Mo, X.-Y., Ni, Z., Jiang, W., He, Y.-Q., et al. (2020). Type III effectors xopN and avrBS2 contribute to the virulence of Xanthomonas oryzae pv. oryzicola strain GX01. Res. Microbiol. 171, 102–106. doi: 10.1016/j.resmic.2019.10.002
McCann, H. C., Guttman, D. S. (2008). Evolution of the type III secretion system and its effectors in plant–microbe interactions. New Phytol. 177, 33–47. doi: 10.1111/j.1469-8137.2007.02293.x
Merda, D., Briand, M., Bosis, E., Rousseau, C., Portier, P., Barret, M., et al. (2017). Ancestral acquisitions, gene flow and multiple evolutionary trajectories of the type three secretion system and effectors in Xanthomonas plant pathogens. Mol. Ecol. 26, 5939–5952. doi: 10.1111/mec.14343
Mirdita, M., Schütze, K., Moriwaki, Y., Heo, L., Ovchinnikov, S., Steinegger, M. (2022). ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682. doi: 10.1038/s41592-022-01488-1
Moreira, L. M., Almeida, N. F., Potnis, N., Digiampietri, L. A., Adi, S. S., Bortolossi, J. C., et al. (2010). Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii. BMC Genomics 11, 238. doi: 10.1186/1471-2164-11-238
Mukhi, N., Brown, H., Gorenkin, D., Ding, P., Bentham, A. R., Stevenson, C. E. M., et al. (2021). Perception of structurally distinct effectors by the integrated WRKY domain of a plant immune receptor. Proc. Natl. Acad. Sci. U. S. A 118, e2113996118. doi: 10.1073/pnas.2113996118
Navarro-Escalante, L., Zhao, C., Shukle, R., Stuart, J. (2020). BSA-seq discovery and functional analysis of candidate Hessian Fly (Mayetiola destructor) avirulence genes. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00956
Nguyen, Q.-M., Iswanto, A. B. B., Son, G. H., Vuong, U. T., Lee, J., Kang, J.-H., et al. (2022). AvrRps4 effector family processing and recognition in lettuce. Mol. Plant Pathol. 23, 1390–1398. doi: 10.1111/mpp.13233
Orfei, B., Pothier, J. F., Fenske, L., Blom, J., Moretti, C., Buonaurio, R., et al. (2023). Race-specific genotypes of Pseudomonas syringae pv. tomato are defined by the presence of mobile DNA elements within the genome. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1197706
Park, E., Lee, H.-Y., Woo, J., Choi, D., Dinesh-Kumar, S. P. (2017). Spatiotemporal monitoring of Pseudomonas syringae effectors via type III secretion using split fluorescent protein fragments. Plant Cell 29, 1571–1584. doi: 10.1105/tpc.17.00047
Peng, Z., Su, Q., Ren, J., Tian, L., Zeng, Y., Yang, Y., et al. (2023). A novel salivary effector, BtE3, is essential for whitefly performance on host plants. J. Exp. Bot. 74, 2146–2159. doi: 10.1093/jxb/erad024
Peters, J. E., Fricker, A. D., Kapili, B. J., Petassi, M. T. (2014). Heteromeric transposase elements: generators of genomic islands across diverse bacteria. Mol. Microbiol. 93, 1084–1092. doi: 10.1111/mmi.12740
Petnicki-Ocwieja, T., Schneider, D. J., Tam, V. C., Chancey, S. T., Shan, L., Jamir, Y., et al. (2002). Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. 99, 7652–7657. doi: 10.1073/pnas.112183899
Pettersen, E. F., Goddard, T. D., Huang, C. C., Meng, E. C., Couch, G. S., Croll, T. I., et al. (2021). UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. Publ. Protein Soc. 30, 70–82. doi: 10.1002/pro.3943
Popov, G., Fraiture, M., Brunner, F., Sessa, G. (2016). Multiple Xanthomonas euvesicatoria type III effectors inhibit flg22-triggered immunity. Mol. Plant Microbe Interact. 29, 651–660. doi: 10.1094/MPMI-07-16-0137-R
Roden, J. A., Belt, B., Ross, J. B., Tachibana, T., Vargas, J., Mudgett, MB, et al. (2004). A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proc. Natl. Acad. Sci. 101, 16624–16629. doi: 10.1073/pnas.0407383101
Rohmer, L., Guttman, D. S., Dangl, J. L. (2004). Diverse evolutionary mechanisms shape the type III effector virulence factor repertoire in the plant pathogen Pseudomonas syringae. Genetics 167, 1341–1360. doi: 10.1534/genetics.103.019638
Ruh, M., Briand, M., Bonneau, S., Jacques, M.-A., Chen, N. W. G. (2017). Xanthomonas adaptation to common bean is associated with horizontal transfers of genes encoding TAL effectors. BMC Genomics 18, 670. doi: 10.1186/s12864-017-4087-6
Sabbagh, C. R. R., Carrere, S., Lonjon, F., Vailleau, F., Macho, A. P., Genin, S., et al. (2019). Pangenomic type III effector database of the plant pathogenic Ralstonia spp. PeerJ 7, e7346. doi: 10.7717/peerj.7346
Sarris, P. F., Duxbury, Z., Huh, S. U., Ma, Y., Segonzac, C., Sklenar, J., et al. (2015). A Plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161, 1089–1100. doi: 10.1016/j.cell.2015.04.024
Sharma, S., Sharma, S., Hirabuchi, A., Yoshida, K., Fujisaki, K., Ito, A., et al. (2013). Deployment of the Burkholderia glumae type III secretion system as an efficient tool for translocating pathogen effectors to monocot cells. Plant J. 74, 701–712. doi: 10.1111/tpj.12148
Shi, Q., Mao, Z., Zhang, X., Ling, J., Lin, R., Zhang, X., et al. (2018a). The novel secreted Meloidogyne incognita effector MiISE6 targets the host nucleus and facilitates parasitism in Arabidopsis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00252
Shi, Q., Mao, Z., Zhang, X., Zhang, X., Wang, Y., Ling, J., et al. (2018b). A Meloidogyne incognita effector MiISE5 suppresses programmed cell death to promote parasitism in host plant. Sci. Rep. 8, 7256. doi: 10.1038/s41598-018-24999-4
Sohn, K. H., Hughes, R. K., Piquerez, S. J., Jones, J. D. G., Banfield, M. J. (2012). Distinct regions of the Pseudomonas syringae coiled-coil effector AvrRps4 are required for activation of immunity. Proc. Natl. Acad. Sci. 109, 16371–16376. doi: 10.1073/pnas.1212332109
Sohn, K. H., Lei, R., Nemri, A., Jones, J. D. G. (2007). The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell 19, 4077–4090. doi: 10.1105/tpc.107.054262
Sohn, K. H., Zhang, Y., Jones, J. D. G. (2009). The Pseudomonas syringae effector protein, AvrRPS4, requires in planta processing and the KRVY domain to function. Plant J. 57, 1079–1091. doi: 10.1111/j.1365-313X.2008.03751.x
Studholme, D. J., Kemen, E., MacLean, D., Schornack, S., Aritua, V., Thwaites, R., et al. (2010). Genome-wide sequencing data reveals virulence factors implicated in banana Xanthomonas wilt. FEMS Microbiol. Lett. 310, 182–192. doi: 10.1111/j.1574-6968.2010.02065.x
Su, J., Nguyen, Q.-M., Kimble, A., Pike, S. M., Kim, S. H., Gassmann, W. (2021). The conserved arginine required for AvrRps4 processing is also required for recognition of its N-terminal fragment in lettuce. Mol. Plant Microbe Interact. 34, 270–278. doi: 10.1094/MPMI-10-20-0285-R
Teper, D., Burstein, D., Salomon, D., Gershovitz, M., Pupko, T., Sessa, G. (2015). Identification of novel Xanthomonas euvesicatoria type III effector proteins by a machine-learning approach. Mol. Plant Pathol. 17, 398–411. doi: 10.1111/mpp.12288
Trujillo, C. A., Ochoa, J. C., Mideros, M. F., Restrepo, S., López, C., Bernal, A. (2014). A complex population structure of the cassava pathogen Xanthomonas axonopodis pv. manihotis in recent years in the Caribbean region of Colombia. Microb. Ecol. 68, 155–167. doi: 10.1007/s00248-014-0411-8
Upadhyaya, N. M., Mago, R., Staskawicz, B. J., Ayliffe, M. A., Ellis, J. G., Dodds, P. N. (2014). A bacterial type III secretion assay for delivery of fungal effector proteins into wheat. Mol. Plant Microbe Interact. 27, 255–264. doi: 10.1094/MPMI-07-13-0187-FI
Wang, Y., Pruitt, R. N., Nürnberger, T., Wang, Y. (2022). Evasion of plant immunity by microbial pathogens. Nat. Rev. Microbiol. 20, 449–464. doi: 10.1038/s41579-022-00710-3
Wang, J., Shine, M. B., Gao, Q.-M., Navarre, D., Jiang, W., Liu, C., et al. (2014). Enhanced Disease Susceptibility1 mediates pathogen resistance and virulence function of a bacterial effector in soybean. Plant Physiol. 165, 1269–1284. doi: 10.1104/pp.114.242495
Wang, Y., Zhang, Q., Sun, M., Guo, D. (2011). High-accuracy prediction of bacterial type III secreted effectors based on position-specific amino acid composition profiles. Bioinformatics 27, 777–784. doi: 10.1093/bioinformatics/btr021
Wei, H.-L., Zhang, W., Collmer, A. (2018). Modular study of the type III effector repertoire in Pseudomonas syringae pv. tomato DC3000 reveals a matrix of effector interplay in pathogenesis. Cell Rep. 23, 1630–1638. doi: 10.1016/j.celrep.2018.04.037
Wichmann, F., Vorhölter, F.-J., Hersemann, L., Widmer, F., Blom, J., Niehaus, K., et al. (2013). The noncanonical type III secretion system of Xanthomonas translucens pv. graminis is essential for forage grass infection. Mol. Plant Pathol. 14, 576–588. doi: 10.1111/mpp.12030
Xing, Y., Xu, N., Bhandari, D. D., Lapin, D., Sun, X., Luo, X., et al. (2021). Bacterial effector targeting of a plant iron sensor facilitates iron acquisition and pathogen colonization. Plant Cell 33, 2015–2031. doi: 10.1093/plcell/koab075
Keywords: effector, pathogen, Pseudomonas, Xanthomonas, virulence, HopK1, XopO, XopAK
Citation: Horton KN and Gassmann W (2024) Greater than the sum of their parts: an overview of the AvrRps4 effector family. Front. Plant Sci. 15:1400659. doi: 10.3389/fpls.2024.1400659
Received: 14 March 2024; Accepted: 26 April 2024;
Published: 10 May 2024.
Edited by:
Choong-Min Ryu, Korea Research Institute of Bioscience and Biotechnology (KRIBB), Republic of KoreaReviewed by:
Hee-Kyung Ahn, The Sainsbury Laboratory, United KingdomTakaki Maekawa, University of Cologne, Germany
Jennifer D. Lewis, Agricultural Research Service (USDA), United States
Copyright © 2024 Horton and Gassmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Walter Gassmann, Z2Fzc21hbm53QG1pc3NvdXJpLmVkdQ==
 Katie N. Horton
Katie N. Horton Walter Gassmann
Walter Gassmann