- 1Jilin Provincial Key Laboratory of Tree and Grass Genetics and Breeding, College of Forestry and Grassland Science, Jilin Agricultural University, Changchun, China
- 2State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University, Harbin, China
- 3The Forest Tree Genetics and Breeding Laboratory, Tongliao Forestry and Grassland Science Research Institute, Tongliao, China
Background: Populus simonii, a notable native tree species in northern China, demonstrates impressive resistance to stress, broad adaptability, and exceptional hybridization potential. DOF family is a class of specific transcription factors that only exist in plants, widely participating in plant growth and development, and also playing an important role in abiotic stress response. To date, there have been no reported studies on the DOF gene family in P. simonii, and the expression levels of this gene family in different tissues of poplar, as well as its expression patterns under cold, heat, and other stress conditions, remain unclear.
Methods: In this study, DOF gene family were identified from the P. simonii genome, and various bioinformatics data on the DOF gene family, gene structure, gene distribution, promoters and regulatory networks were analyzed. Quantitative real time PCR technology was used to verify the expression patterns of the DOF gene family in different poplar tissues.
Results: This research initially pinpointed 41 PSDOF genes in P. simonii genome. The chromosomal localization results revealed that the PSDOF genes is unevenly distributed among 19 chromosomes, with the highest number of genes located on chromosomes 4, 5, and 11. A phylogenetic tree was constructed based on the homology between Arabidopsis thaliana and P. simonii, dividing the 41 PSDOF genes into seven subgroups. The expression patterns of PSDOF genes indicated that specific genes are consistently upregulated in various tissues and under different stress conditions, suggesting their pivotal involvement in both plant development and response to stress. Notably, PSDOF35 and PSDOF28 serve as pivotal hubs in the interaction network, playing a unique role in coordinating with other genes within the family.
Conclusion: The analysis enhances our comprehension of the functions of the DOF gene family in tissue development and stress responses within P. simonii. These findings provide a foundation for future exploration into the biological roles of DOF gene family.
1 Introduction
The DNA binding with one finger (DOF) transcription factors are a group of specific transcription factors exclusively present in plants, encompassing gymnosperms, angiosperms, and certain lower algae. DOF transcription factors belong to a subfamily within the zinc finger protein family, characterized by the presence of oligomerization sites, nuclear localization signals, and two principal domains encompassing a total of 200 – 400 amino acids (Zou and Sun, 2023). One of the domains is called the N-terminal DNA-binding domain, also known as the DOF domain, which exhibits a highly conserved C2-C2 single zinc finger structure composed of the CX2CX21CX2C motifs. This single zinc finger structure endows it with unique DNA recognition abilities, especially the ability to specifically recognize the AAAG/CTTT functional element in plant promoter sequences. Moreover, there are four absolutely conserved Cys residues and a Zn+ covalently bound in this single zinc finger structure. Zn+ and Cys residues are necessary for the DOF protein to maintain its activity (Khan et al., 2023). The other domain is the transcriptional regulatory domain located at the C-terminal of the DOF protein. Its amino acid sequence is relatively variable and lacks conservatism, which leads to the functional diversity of the DOF protein in plants (Noguero et al., 2013; Gupta et al., 2015). The distinctive structures and functionalities of the DOF gene family allow it to bind to DNA and interact with regulatory elements, participate in various physiological and biochemical processes in plants, and play a crucial role in regulating gene expression at all stages of plant growth and development, as well as in resisting stress responses (Zou et al., 2019; Ruta et al., 2020).
Based on the findings of genome-wide analysis, DOF genes are abundantly present across the genomes of diverse plants. For instance, in model plants, we observed the presence of 36 DOF genes in Arabidopsis thaliana, whereas 30 members of the DOF family were identified in Oryza sativa (Lijavetzky et al., 2003). DOF family members have also been discovered in ornamental plants, including Chrysanthemum morifolium, where a total of 20 DOF genes were identified (Song et al., 2016). Furthermore, the DOF family is ubiquitous in numerous crop species. Specifically, 33 DOF family members have been identified in Capsicum annuum (Wu et al., 2016), while 20 members were detected in Spinacia oleracea (Yu et al., 2021). And, 74 family members were discovered in Musa acuminata (Dong et al., 2016), and 36 family members in Cucumis sativus (Wen et al., 2016). Moreover, the DOF family’s presence has been confirmed in woody plants as well. Specifically, 25 DOF genes have been detected in Prunus persica (Chen et al., 2017), while 24 members of the DOF family have been identified in Prunus sibirica (Li et al., 2022a). There are also related studies in Salicaceae plants. For example, Wang et al. identified the 41 DOF genes in Populus trichocarpa and studied the expression level of it under osmotic stress and abscisic acid stress (Wang et al., 2017). In addition, Wang et al. also identified the 44 DOF genes in Populus simonii × Populus nigra and elucidated the role of DOF genes in promoting nitrogen assimilation and utilization (Wang et al., 2022). These extensive studies indicate that DOF family genes are universally distributed throughout the plant kingdom, laying a solid foundation for further investigations into their diverse functions and roles. Nonetheless, there have been no reported studies on the DOF gene family in P. simonii, and the expression levels of this gene family in different tissues of poplar, as well as its expression patterns under cold, heat, and other stress conditions, remain unclear.
Numerous investigations have underscored the pivotal role of DOF genes across a spectrum of biological processes, spanning from plant growth and development to stress responses and gene regulation. Currently, the involvement of DOF family members in the developmental processes and stress tolerance of diverse plants, such as grape, tea, petunia, poplar, and A. thaliana, has been firmly established. In grape, a comprehensive analysis identified 25 DOF genes, among which VaDOF17d was found to notably elevate raffinose levels in callus under cold stress conditions (Wang et al., 2021). After experiencing varying degrees of drought stress, the expression levels of specific DOF family members in tea underwent significant changes. Notably, the expression of CsDOF10 was significantly upregulated under moderate drought conditions, indicating its crucial role in coping with drought stress (Yu et al., 2020). In petunias, DOF family members participate in the reproductive development of plants. Specifically, the expression of PiDOF14 effectively promotes the degradation of the tapetal layer, thereby inhibiting meiosis of microspore cells and ultimately resulting in a male sterility phenotype (Yue et al., 2021). In A. thaliana, the overexpression of AtDOF4.1 leads to significant delays in flowering and developmental defects in reproductive organs. Simultaneously, the overexpression line of this gene exhibits reduced organs, including leaves, flowers, and stems, indicating that AtDOF4.1 functions as a transcription suppressor (Ahmad et al., 2013). Furthermore, the overexpression of the gene PnDOF30, originating from P. simonii × P. nigra, in A. thaliana notably enhances the growth of transgenic A. thaliana plants when subjected to low nitrogen conditions (Wang et al., 2022).
P. simonii, a notable native tree species in northern China, demonstrates impressive resistance to stress, broad adaptability, and exceptional hybridization potential (Chen et al., 2013). Considering the substantial impact of DOF genes on growth and stress responses in various plant species, coupled with the scarcity of research on this gene family in P. simonii, it becomes imperative to further explore the functions and contributions of DOF genes within this particular species. To comprehend the unique traits and functions of these genes within P. simonii, we explored multiple aspects, including identifying family members, phylogenetic relationships, chromosome localization, gene structure, specific expression profiles, protein-protein interactions, and conducting qRT-PCR analysis. This study provides valuable insights for further research on the potential roles of the DOF gene family in the growth, development, and stress resistance of P. simonii.
2 Materials and methods
2.1 Identification of DOF genes in P. simonii
In order to identify the DOF family members within the P. simonii genome, we obtained the full DOF protein sequence of A. thaliana from the Tair database (https://www.arabidopsis.org/). Moreover, we constructed a high-quality reference genome of P. simonii, featuring a contig N50 of 24 Mb (Cai et al., 2023). Initially, we used the BlastP program (e-value, 1 × e−5) in Tbtools software (v2.080) to identify PSDOF protein sequences, employing A. thaliana DOF proteins as query sequences (Chen et al., 2020). From the search results, we eliminated duplicate sequences to obtain the presumptive member sequences. Subsequently, to further validate the candidate protein sequences, we utilized the NCBI “batch Web CD-Search Tool” (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi). This tool can be used to detect and confirm whether each candidate protein is a bona fide DOF protein based on the presence of conserved domains. Finally, we employed the online tools SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi) and Pfam (http://pfam-legacy.xfam.org/) to perform rigorous verification of the conserved domains within our search results. Through this extensive verification process, we were able to identify the potential PSDOF genes. Additionally, using Tbtools software, we performed comprehensive biochemical characterization of each PSDOF protein, analyzing important parameters including amino acid composition, molecular weight, and instability index.
2.2 Phylogenetic analyses of DOF genes in P. simonii
To explore the evolutionary relationships among PSDOF proteins, we conducted an extensive analysis utilizing the candidate protein sequences. Initially, we conducted multiple sequence alignment using the Muscle program within MEGA software (v11.0), following its default parameters (Tamura et al., 2021). Next, phylogenetic trees were constructed in MEGA using the neighbor-joining (NJ) method. To improve the tree topologies’ dependability and consistency, a bootstrapping process involving 1000 iterations was executed. For better visual understanding and analysis of phylogenetic relationships, the ultimate phylogenetic tree of the PSDOF genes underwent refinement and annotation via the Interactive Tree of Life (iTOL) web tool (https://itol.embl.de/).
2.3 Chromosomal localization and synteny analysis of PSDOF genes
The Tbtools software was utilized to identify the chromosomal locations of DOF genes on 19 chromosomes in P. simonii, employing General Feature Format (GFF) data for visual representation. Following this, utilizing the locational data of genes on chromosomes, the PSDOF genes were methodically rebranded as PSDOF1 to PSDOF41. For an in-depth examination of PSDOF genes’ synteny within P. simonii, pairs of syntenic genes were pinpointed using McscanX analysis. Subsequently, the synteny ircus plot produced was meticulously developed using Tbtools software’s “Advanced Circos” program. Furthermore, Tbtools software’s “One-step MCScanX” program enabled the syntenic examination of DOF genes in P. simonii, P. trichocarpa, and A. thaliana. The results of this study were visually depicted through the “Multiple Synteny Plot” program of Tbtools software.
2.4 Gene structure, conserved motif and promoter cis-elements analysis
The “Gene Structure View” program of Tbtools software was employed to visually depict the exon/intron arrangement of PSDOF genes, utilizing genomic structural data along with gene identification information. Additionally, a web-based application called “Multiple Em for Motif Elicitation” (MEME v5.5.3), available at http://memesuite.org/tools/meme, was utilized to examine the preserved motif patterns in proteins produced by PSDOFs.
Furthermore, the promoter region sequences of PSDOF genes, covering a 2000 base pair area before the translational start site (ATG), were isolated from the P. simonii genome. Foreseeing cis-elements in these promoters was made easier using the PlantCARE tool, available at https://bioinformatics.psb.ugent.be/webtools/plantcare/html/. Following this, Tbtools software was utilized to graphically depict these cis-elements.
2.5 Expression analysis and interaction network construction of PSDOF genes
RNA sequencing was conducted on various tissues, including terminal buds (NT), axillary buds (NB), leaves (NL), stems (NS), phloem (NP), and roots (NR) of P. simonii, to assess the expression level of PSDOF genes, more details about the plant materials and sample collection can be found in Cai et al., 2023. Subsequently, the expression levels were graphically depicted using heat maps generated with Tbtools software. In order to delve deeper into how PSDOF genes are expressed under various stress scenarios, the data gathered included information from heat, cold, and salt stress treatments. Subsequently, the data underwent normalization and was employed to create an expression heatmap, effectively depicting the transcriptional reactions of all PSDOF genes linked to stress scenarios.
Furthermore, to forecast the interactions between 41 PSDOF protein sequences, protein-protein interaction (PPI) networks were developed via the STRING website (https://www.string-db.org/). Subsequently, the derived network files were rendered visible through Cytoscape software (v0.9.2) (Kohl et al., 2011).
2.6 RNA extraction and qRT-PCR analysis
The samples’ total RNA was extracted utilizing the plant total RNA extraction kit provided by Takara (Beijing, China). In order to validate our earlier expression profile results, we randomly selected 15 genes and conducted reverse transcription-quantitative PCR (RT-qPCR) analysis using specifically designed primers. The Tbtools software was employed to create customized primers for each gene, using Actin as the benchmark internal gene (Regier and Frey, 2010) (Supplementary Table S1). The ABI 7500 RT PCR system was utilized in our qRT-PCR analysis. The comparative expression levels of the chosen genes were determined through the 2-ΔΔCt method (Pfaffl, 2001). For the purpose of maintaining consistency and dependability, three biological duplicates were employed in the analysis of gene expression.
3 Results
3.1 Identification of PSDOF genes and its physicochemical properties
Using the protein sequences of A. thaliana as a reference, we successfully identified 41 members of the DOF family in P. simonii through alignment and screening processes. Subsequently, each of the 41 PSDOF genes was named in sequential order based on their homologous chromosome positions, ranging from PSDOF1 to PSDOF41. Delving deeper into PSDOF genes’ characteristics, along with other basic information, were summarized in Supplementary Table S2. The analysis revealed variations among the PSDOF genes. Specifically, the number of amino acids ranged from 159 to 504, with PSDOF40 showed the largest count with 504 amino acids, and PSDOF10 was the smallest with 159. The lipid amino acid index exhibited a wide range, varying from 43.58 to 65.29, whereas the predicted isoelectric point (pI) ranged from 4.76 to 9.48. Additionally, the instability index displayed significant variation, spanning from 38.85 to 72.38. Lastly, the molecular weights exhibited a range from 17.70 to 55.21 kDa, with PSDOF1 exhibiting the lowest molecular weight of 17.70 kDa and PSDOF41 displaying the highest at 55.21 kDa.
3.2 Phylogenetic analysis of PSDOF genes
Based on the homology of DOF proteins within the phylogenetic tree, the 41 PSDOF genes were classified into seven distinct subfamilies, specifically designated as Group I, Group II-A, Group II-B, Group III, Group IV, Group V-A, and Group V-B (Figure 1). The subfamily containing the largest gene count encompassed eight members of the DOF family. On the other hand, the subfamily with the least number of members were Group II-A and Group V-A, both having four members from the DOF family.
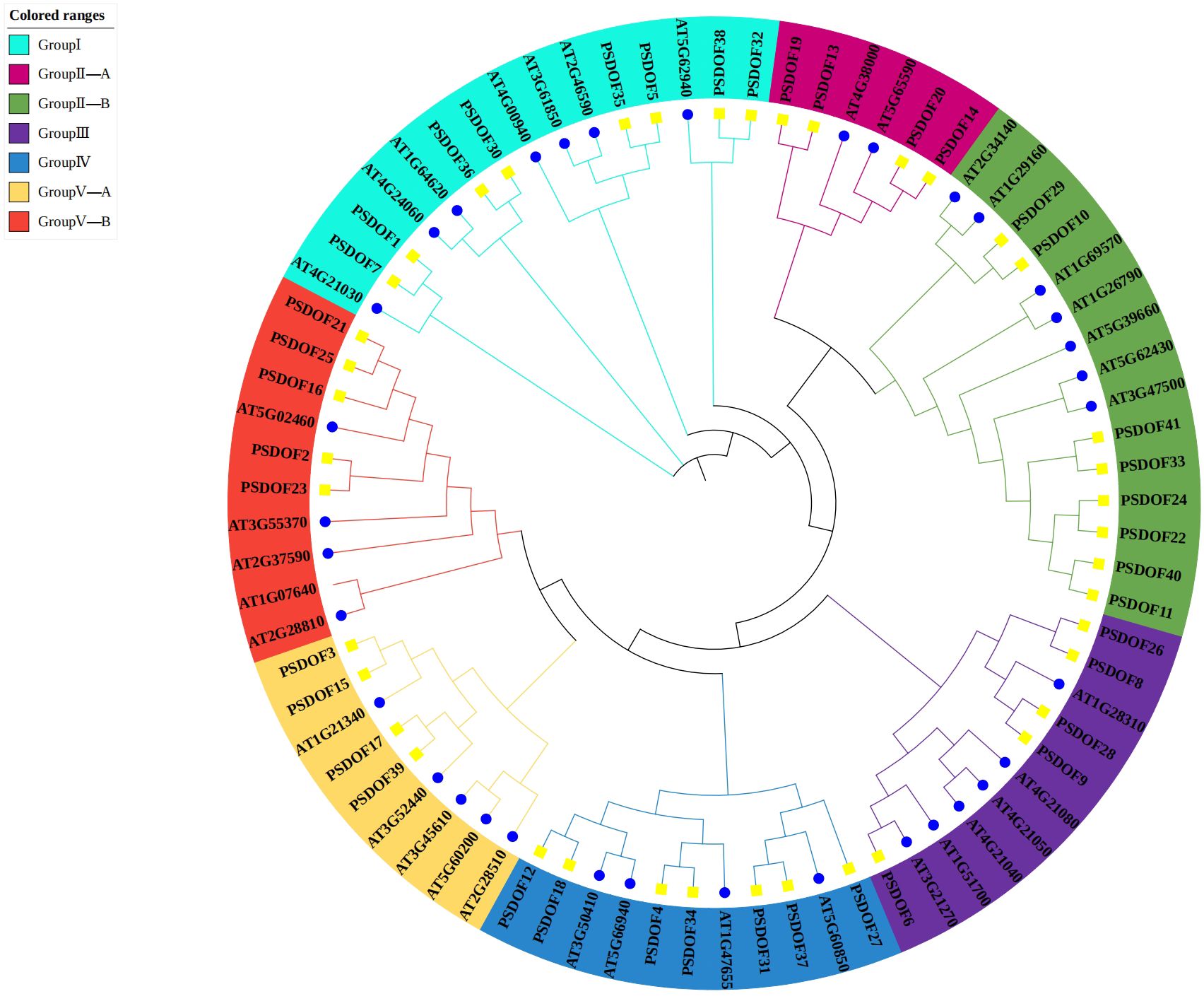
Figure 1 Phylogenetic relationship among PSDOF genes. Seven distinct subfamilies, symbolized by cyan, pink, green, purple, blue, yellow, and red arcs.
3.3 Motif composition of PSDOF genes
To further explore the functional domain of PSDOF proteins, we undertook a thorough analysis of the amino acid motifs (Figure 2A). The majority of PSDOF genes are grouped together on a solitary clade, exhibiting a uniform motif composition at corresponding positions, hinting at potential similarities in their biological functionalities. However, there are significant differences in motif composition among different groups. For example, the most abundant and complex motif arrangement is observed in Group II-B, while Group II-A, Group IV, and Group V-A display the fewest number of motifs and the simplest arrangement. Moreover, particular motifs demonstrated specificity towards specific groups. For instance, motif 8 is present only in all genes of Group I, while motif 7 is detected exclusively in every gene of the Group V-B branch, thus distinguishing it from other branches.
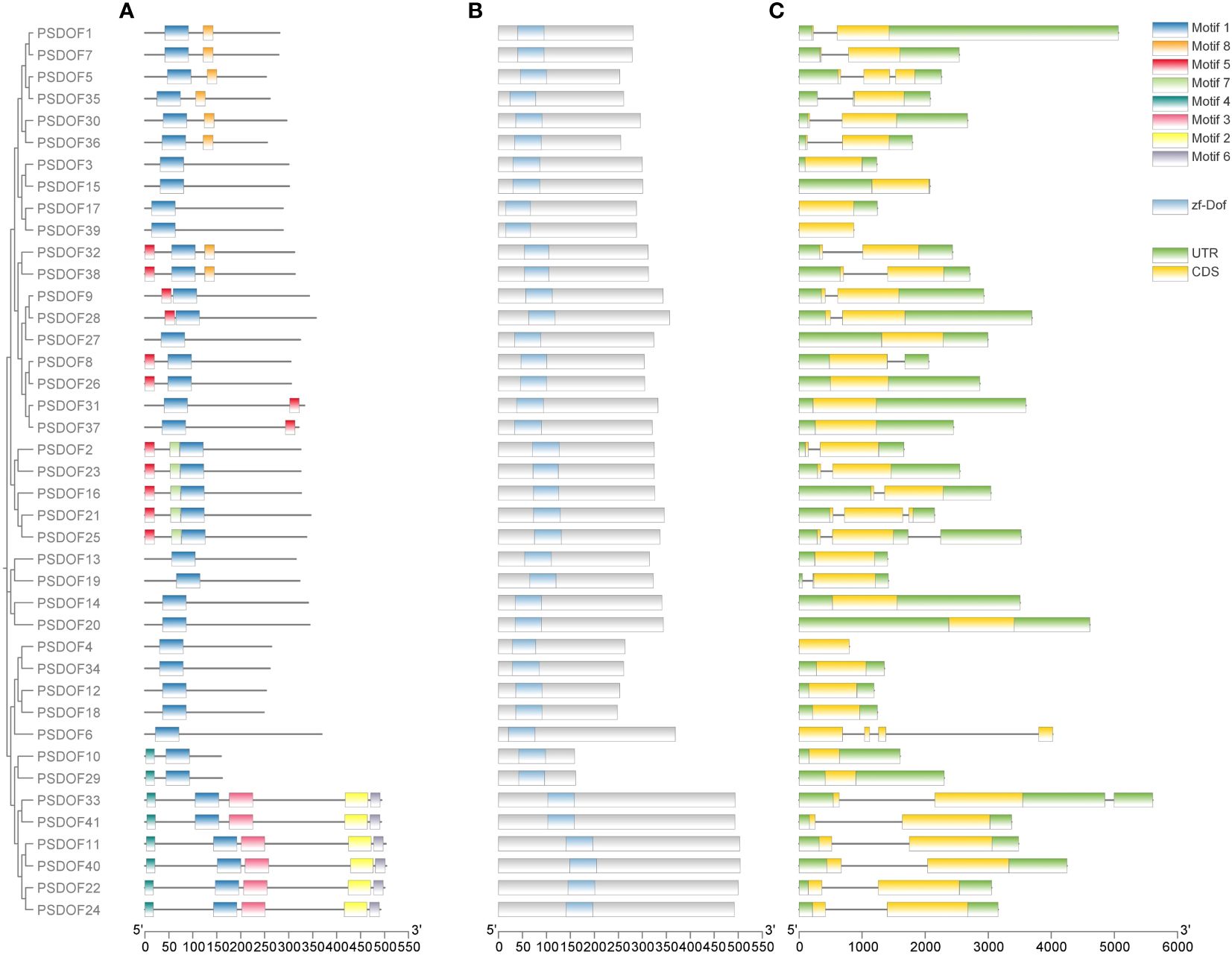
Figure 2 Motif, conserved domain, and gene structure analysis of PSDOFs family. (A) The arrangement of motifs in the PSDOF protein, motif 1-6 is depicted through boxes in various colors. (B) Conserved domain of PSDOF genes. (C) Structure of exons and introns in PSDOF genes. The areas of UTR, exons, and introns are depicted using green boxes, yellow boxes, and black lines, in that order.
3.4 Structure composition of PSDOF genes
In order to gain deeper insights into the local structural characteristics of the DOF domain, we conducted a comparative analysis of PSDOF protein sequences. The results observed that each gene harbors a common conserved domain zf-DOF, indicating a high degree of conservation within the PSDOF family domain (Figure 2B).
Furthermore, an exon-intron analysis was performed on PSDOF genes to elucidate their structural diversity (Figure 2C). The research identified introns in 24 genes, with most genes having only one intron, as seen in PSDOF1 and PSDOF7, while a minority contained three introns, such as PSDOF6. Additionally, each gene exhibited varying numbers of exons, ranging from 1 to 4. For instance, PSDOF6 possessed the highest number of exons with 4 exons, whereas others like PSDOF5 and PSDOF35 contained only one exon. The majority of family members typically contained two or three exons. In the same group, a considerable proportion of PSDOF genes exhibit similar gene structures in terms of exon count. For example, all family members in Group II-B possessed two exons except PSDOF10 and PSDOF29. Notably, PSDOF6 possesses exons and introns but lacks untranslated regions (UTRs), suggesting it may have undergone significant evolution.
3.5 Analysis of cis-elements in the promoter regions of PSDOF genes
For exploring the biological roles of PSDOFs, the PlantCARE database was employed to examine cis-elements in the preceding 2 kb sequences of each gene. In the promoter region upstream, seven unique cis-elements were discovered, mainly consisting of elements responsive to growth, hormones, and abiotic stress (Figure 3). The element influencing plant growth encompassed responsiveness to light. Hormone response elements included responsiveness to auxin, MeJA, gibberellin, abscisic acid, and salicylic acid. Abiotic stress response elements mainly consisted of defense and stress responsiveness. The varied nature of cis-elements indicates the vital involvement of PSDOF genes in numerous biological functions, especially in a range of hormonal reactions and other vital biological routes.
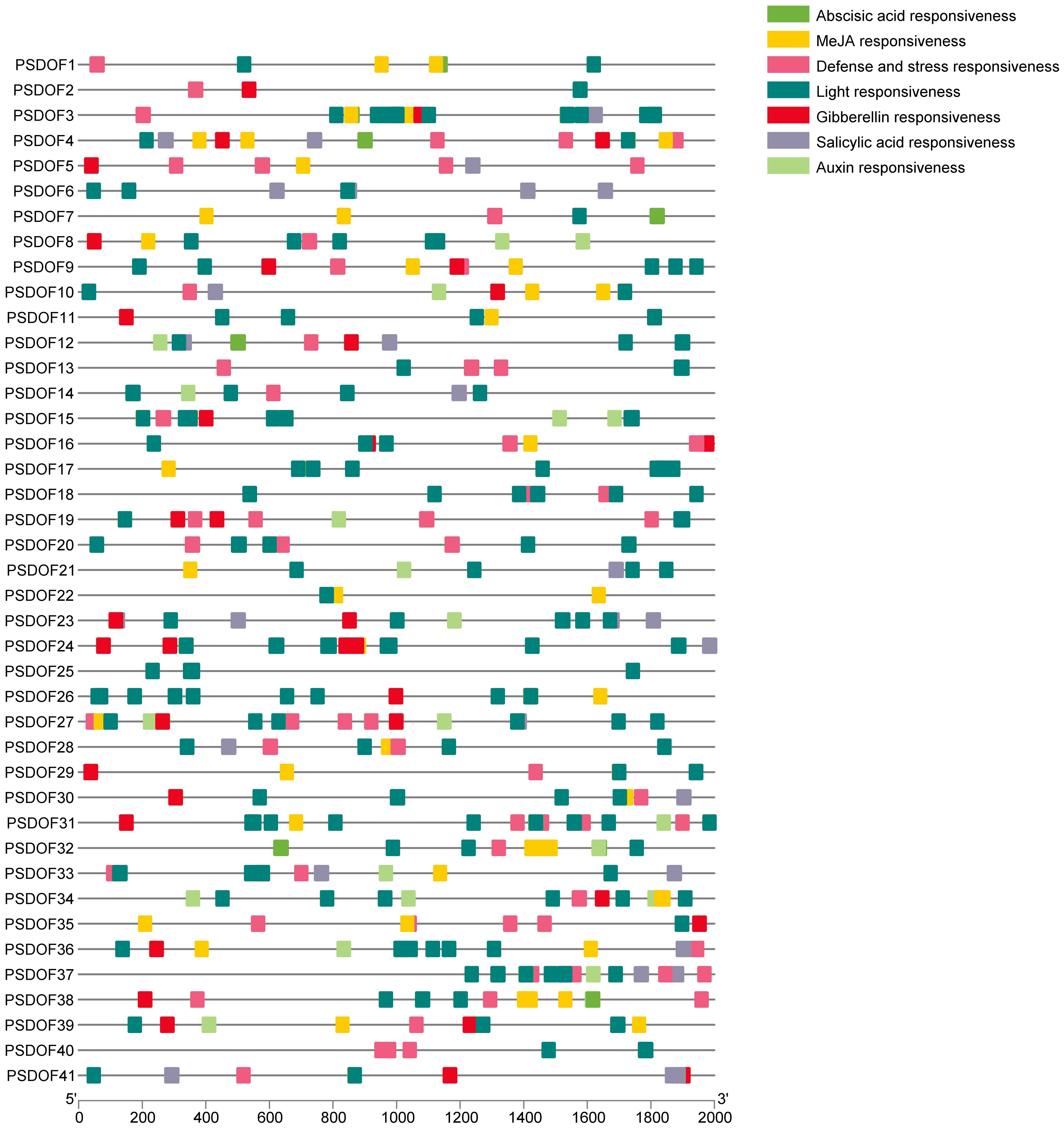
Figure 3 Analysis of cis-acting elements of PSDOFs promoter. The brightly hued squares to the right represent cis-elements, each serving distinct roles.
3.6 Chromosome localization and synteny relationship of PSDOF genes
By analyzing the genomic data of P. simonii, we unveiled the chromosomal distribution of PSDOF genes, as illustrated in Figure 4. Among the 19 chromosomes of P. simonii, except for chromosome 18 where the PSDOF genes was not found, the number of PSDOF genes distributed on the remaining 18 chromosomes varies. Notably, PSDOF genes were most densely distributed on chromosomes 4, 5, and 11, each containing four genes. However, certain chromosomes harbored only one PSDOF gene, such as chromosome 9 and 16. Furthermore, the largest chromosome, chromosome 1, contained only two genes, PSDOF1 and PSDOF2.
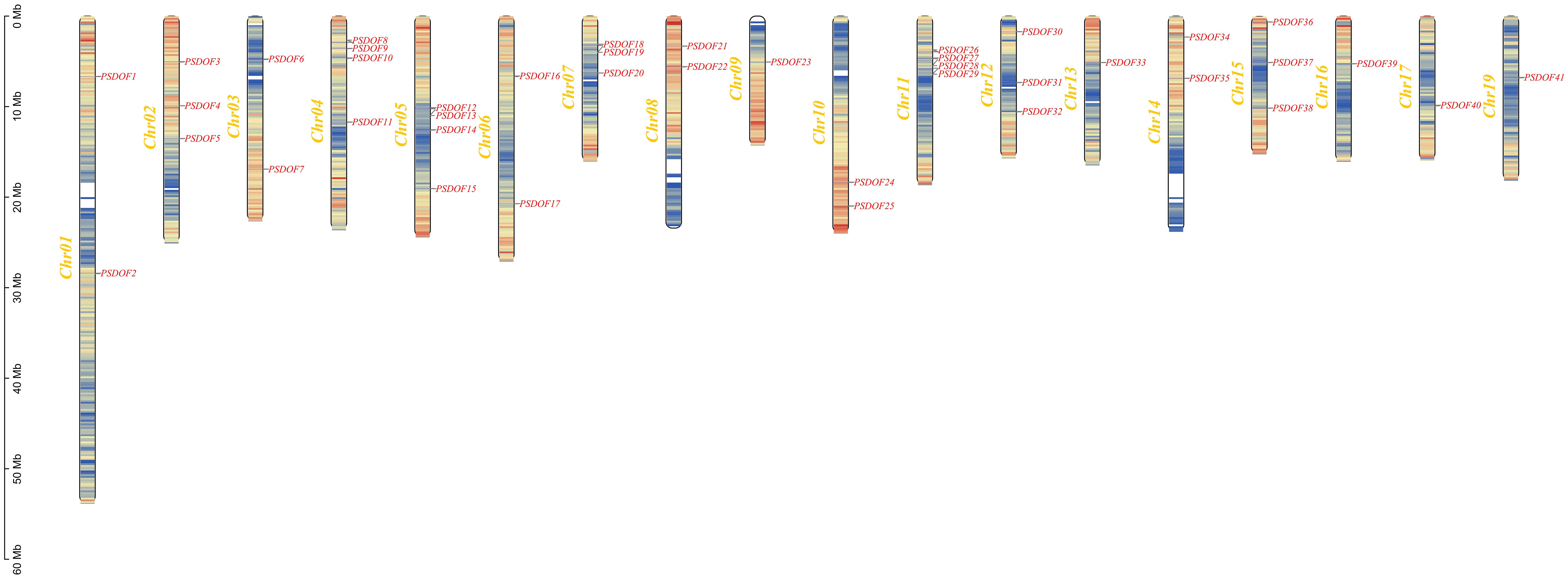
Figure 4 Chromosome distribution of PSDOFs family. Each gene’s genomic position is accurately charted on the chromosome, determined by its distinct physical coordinates. The left-side displays the chromosome count (Chr01-Chr19), while the right-side lists all the genes.
For an in-depth exploration of the evolutionary connections within the PSDOF gene family, synteny analysis was performed (Figure 5). The study uncovered 37 pairs of homologous genes within the 41 PSDOF genes, predominantly on chromosomes 2 and 14, with a higher prevalence of homologous gene pairs, succeeded by chromosome 8. In contrast, chromosome 13 had a relatively small count of homologous gene pairs. It’s notable that certain PSDOF genes show associations with multiple genes situated on different chromosomes.
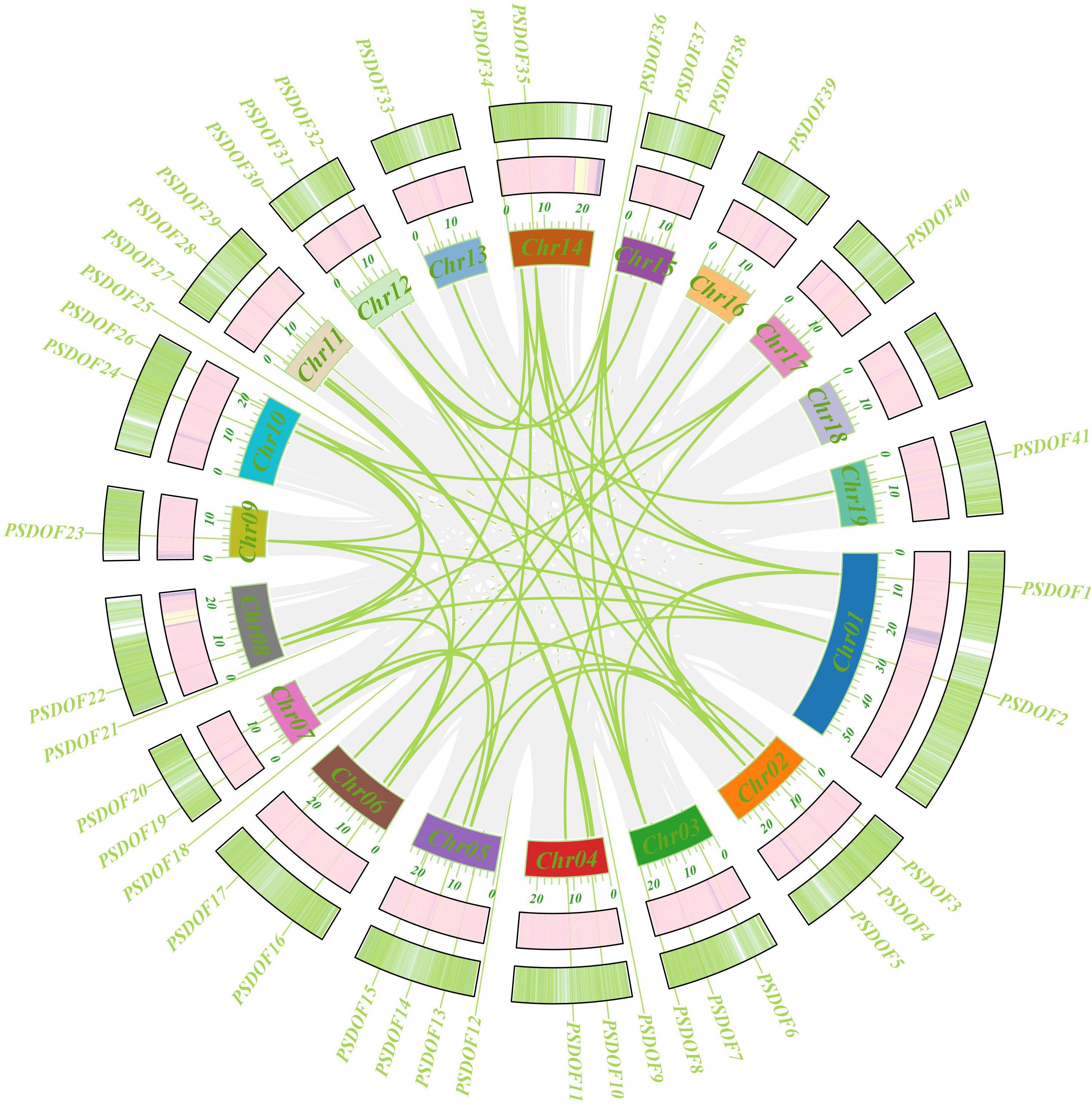
Figure 5 Circso diagram of PSDOF genes. The external to internal heat map illustrates how gene density and GC ratio are distributed along the chromosome. Syntenic segments in the P. simonii genome are denoted by gray lines, while green lines represent pairs of PSDOF genes that are collinear.
3.7 Synteny relationships of DOF genes in P. simonii and different species
Aiming to delve more profoundly into the development of the PSDOF gene family, we created a comparative chart that includes P. simonii and two notable species, P. trichocarpa and A. thaliana (Figure 6). Of the 41 PSDOF genes, 62 gene pairs were identified as homologous to A. thaliana, while 119 gene pairs were found to be homologous to P. trichocarpa. These homologous gene pairs with P. simonii displayed varying distributions on each chromosome. Notably, homologous genes in A. thaliana and P. trichocarpa were predominantly concentrated on chromosome 8 of P. simonii, with 12 and 16 pairs, respectively. Conversely, homologous gene pairs between A. thaliana and P. simonii were dispersed on chromosome 7, 15, and 17 of P. simonii, with only two pairs of homologous genes. Notably, there was no distribution of homologous gene pairs on chromosome 19 of P. simonii.
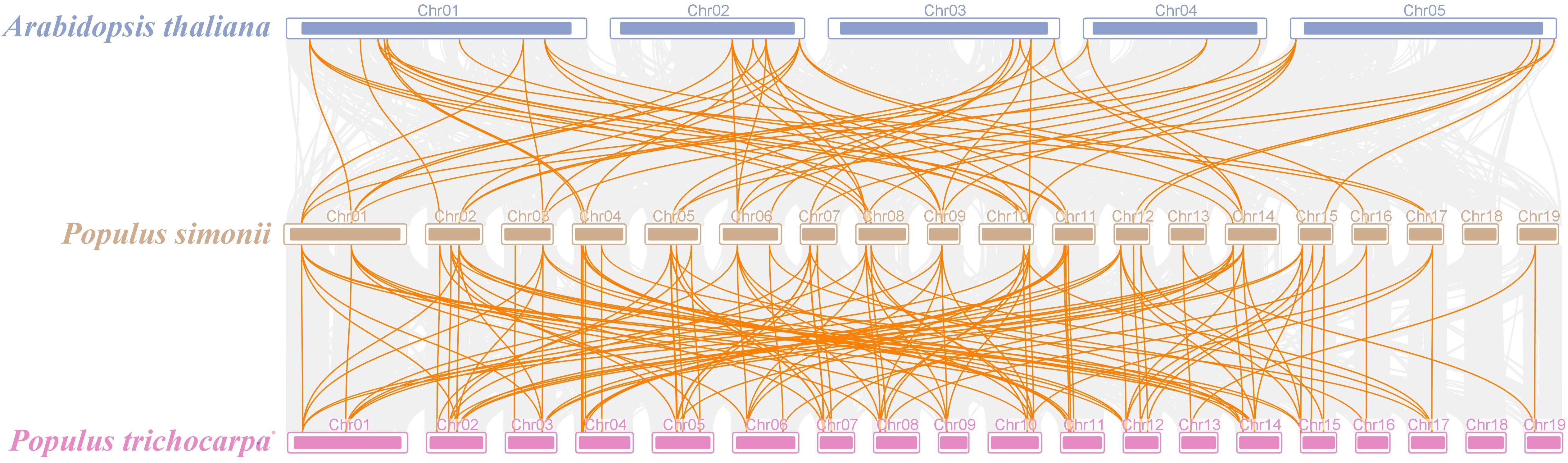
Figure 6 Synteny relationship of PSDOF genes among P. simonii, A. thaliana and P. trichocarpa. Orange lines indicate pairs of DOF genes aligned in a straight line, while gray lines show segments of P. simonii that are similar in structure to the genomes of both plants.
Analogously, the distribution of homologous genes between P. trichocarpa and P. simonii exhibited variation, as there were a higher number of homologous genes located on chromosomes 1-14 and a lower number on chromosomes 16, 17, and 19. Intriguingly, the homology in DOF genes between P. simonii and P. trichocarpa was markedly greater compared to those with A. thaliana. The disparity could be ascribed to their more intimate familial ties and evolutionary background.
3.8 Analysis of PSDOF genes regulatory network
Based on the homology of PSDOF genes with A. thaliana proteins, we constructed a protein interaction network encompassing all PSDOF genes, aiming to gain deeper insights into the potential connections between these genes. The results, depicted in Figure 7, revealed that 16 out of 41 PSDOF genes were implicated in the construction of the gene regulatory network. Among these genes, PSDOF25, PSDOF28, PSDOF35, and PSDOF38 exhibited stronger interactions with others, playing pivotal roles in the protein interaction network. As an example, PSDOF25 functioned as a central gene, engaging with eight additional genes, such as PSDOF29, PSDOF35, and PSDOF36. Furthermore, PSDOF20 also played a pivotal role in the network by engaging in interactions with seven other genes. Conversely, PSDOF37 and PSDOF40 exhibited limited interactions with other genes, with PSDOF37 interacting solely with PSDOF39 and PSDOF40 interacting exclusively with PSDOF41.
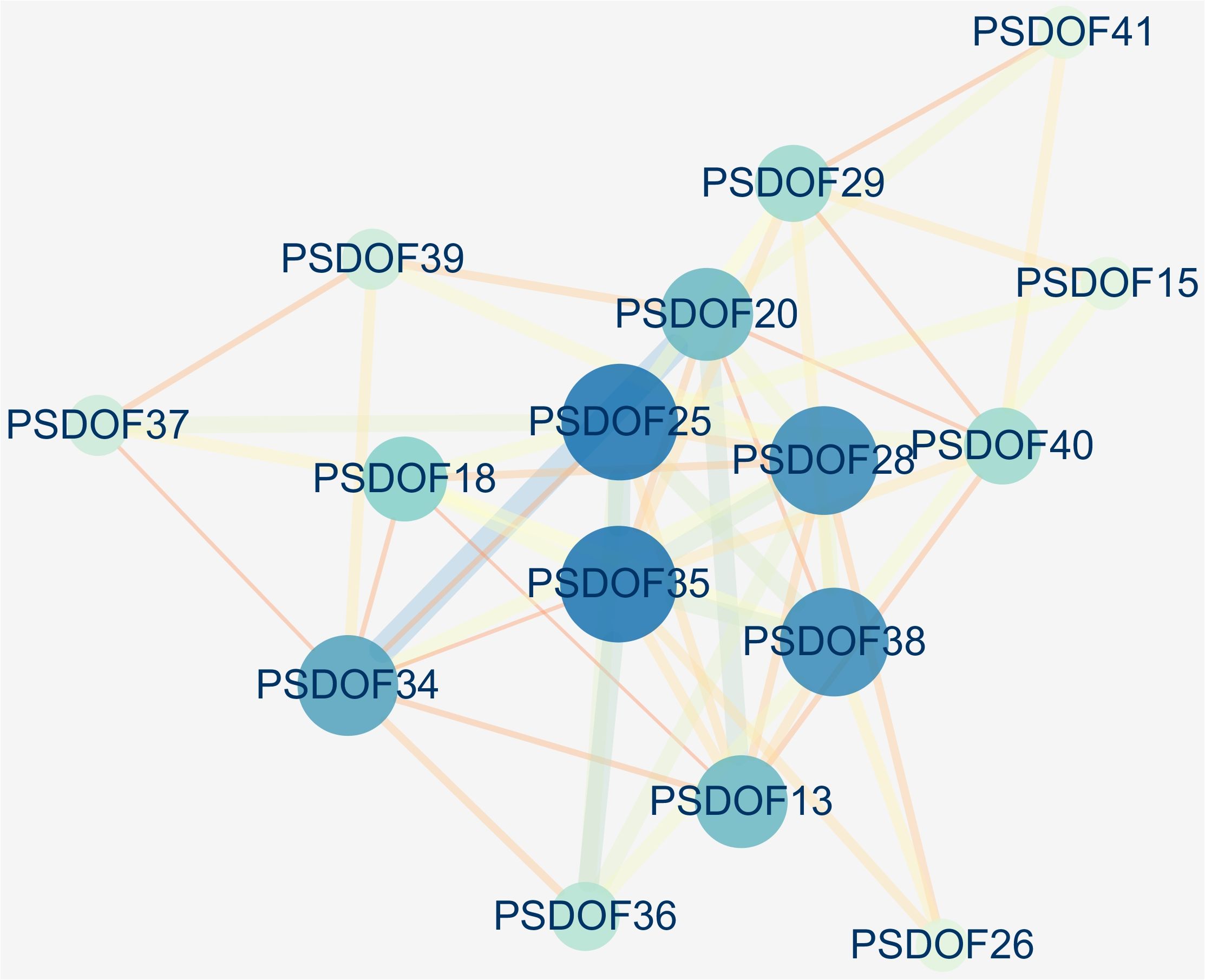
Figure 7 Interaction network of PSDOF genes. Network nodes represent proteins, while edges represent interactions between proteins in the network.
3.9 Analysis of PSDOF genes expression profiles in different tissues
A thorough investigation into the functions of DOF genes in plant development and stress response is imperative for enhancing our comprehension of the biological roles played by these genes. For an in-depth analysis of the distinct roles of DOF genes in P. simonii, transcriptome sequencing was employed to thoroughly investigate these genes’ expression profiles in different tissues of P. simonii.
The results demonstrated in Figure 8A showed that PSDOF genes were expressed in all tissues of P. simonii, but their expressions were significantly different. For example, the genes PSDOF20, PSDOF14, and PSDOF11, which have higher expression in the NL, have significantly lower expression in other tissues such as NS, and NP. On the other hand, genes such as PSDOF7, PSDOF13, and PSDOF34 were highly expressed in two tissues, namely NS and NR. It was further observed that most of the genes, such as PSDOF27, PSDOF23, and PSDOF33, had relatively high expression in the NR and NP, low expression in the NT and NB. It is noteworthy that compared to other genes, the hub gene PSDOF28 exhibits relatively high expression levels across various tissues, indicating its potential predominant role in the growth and development of P. simonii. Meanwhile, the other three hub genes such as PSDOF25 show relatively high expression levels specifically in roots, suggesting their potentially pronounced roles in root function.
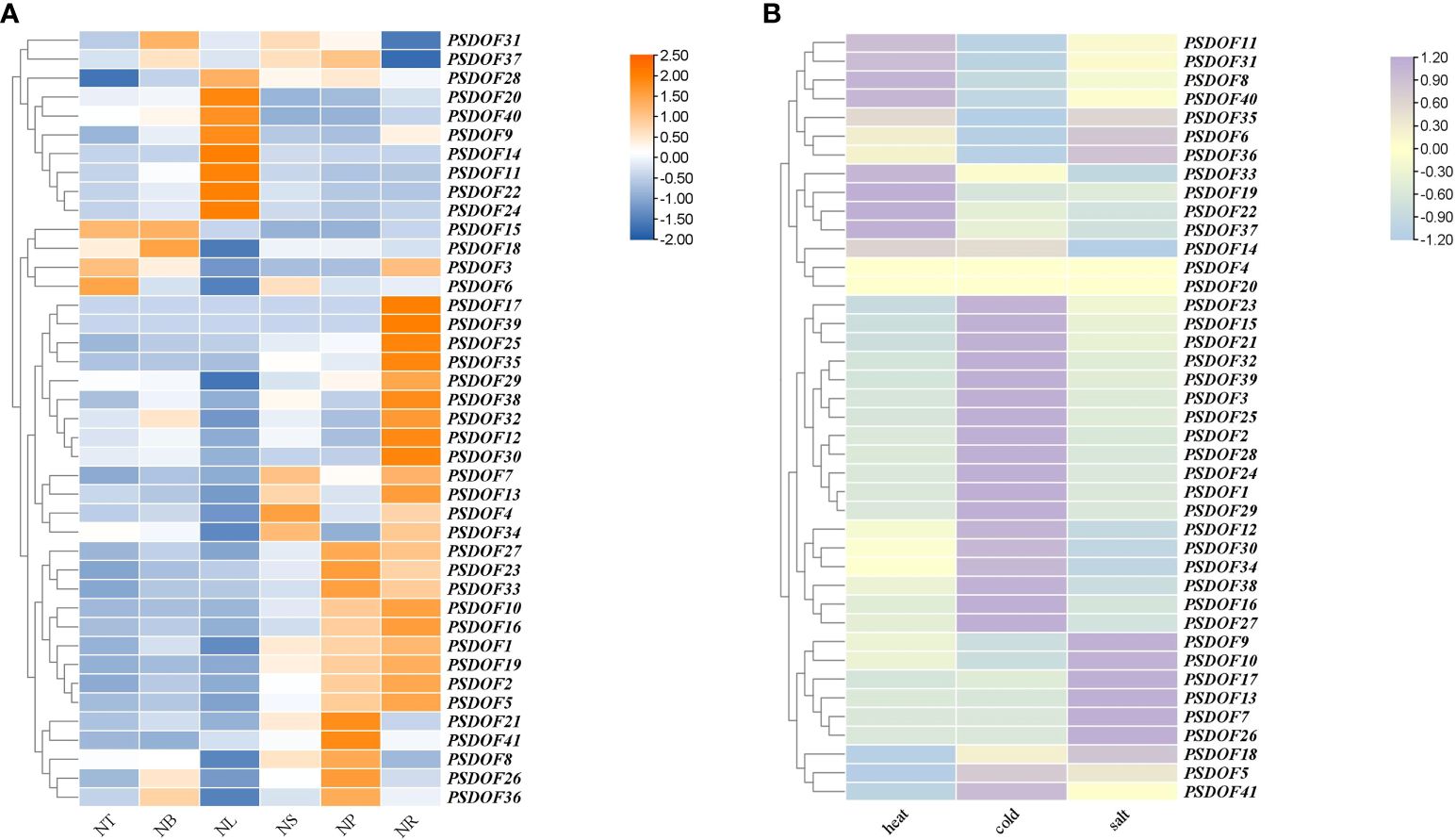
Figure 8 Expression patterns of PSDOF genes. (A) A thermal chart depicting PSDOF genes across various tissues. The shift in the color spectrum from blue to orange signifies varying degrees of expression, ranging from low to high. (B) Patterns of PSDOF genes expression under varying stress conditions. The shift in color from blue to purple symbolizes the evolution of expression levels within the spectrum, ranging from low to high.
3.10 Analysis of PSDOF genes expression profiles in different stress conditions
Through the examination of P. simonii transcriptome data in conditions of heat, cold, and salt stress, the expression patterns of PSDOF genes under different stressors were extensively studied (Figure 8B). The results indicate that under heat stress conditions, the expression levels of eight genes including PSDOF11, PSDOF31, and PSDOF8 are elevated, while the expression levels of the remaining members of the PSDOF family are relatively low. Twenty PSDOF genes, including PSDOF23, PSDOF15, and PSDOF21, among others, exhibited elevated expression levels under cold stress conditions. Additionally, the expression of genes such as PSDOF9, and PSDOF17 was upregulated under salt stress conditions. Additionally, we observed that the hub gene PSDOF35 exhibits elevated expression levels under three types of stress, suggesting its involvement in stress response. It can be found that the genes distributed on the same branch from phylogenetic tree have a certain similarity in their expression to the same stress condition. For example, the genes PSDOF11, PSDOF31, and PSDOF8 belonged to the same branch in the evolutionary tree, and their expression under heat stress conditions was significantly higher than that under cold and salt stress conditions.
3.11 Quantitative reverse transcription polymerase chain reaction analysis
Fifteen PSDOF genes were chosen at random for qRT-PCR analysis with tailored primers, enhancing our comprehension of PSDOF gene expression patterns in various tissues. The way these PSDOF genes are expressed across various tissues aligns well with the gene expression patterns identified earlier via transcriptome sequencing, confirming the utility of RNA-seq data in evaluating these genes’ expression levels (Figure 9).
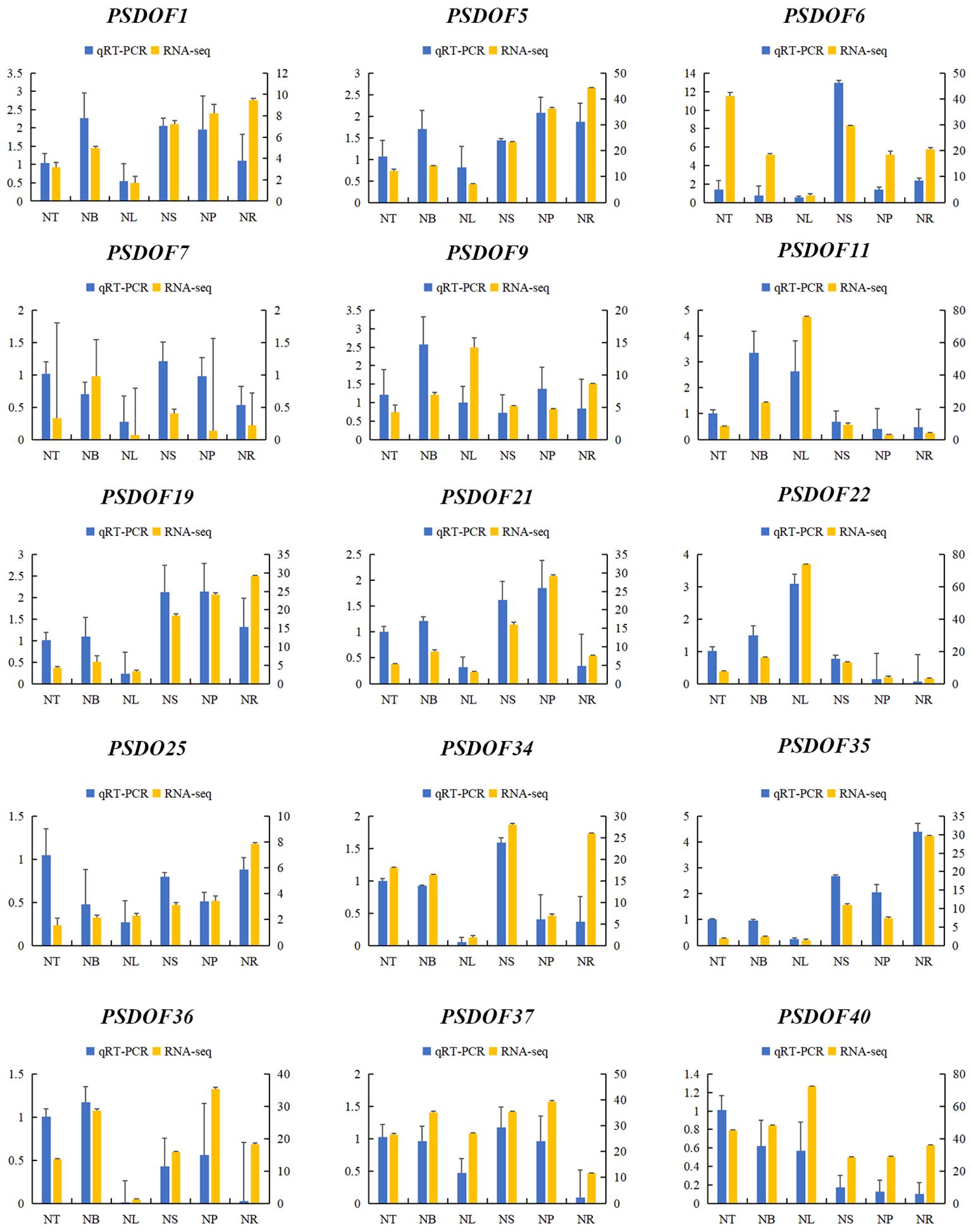
Figure 9 Gene expression profiles derived from RNA-seq and qRT-PCR. On the X-axis, six distinct tissues are illustrated. The expression data from qRT-PCR is displayed on the left y-axis, whereas the right y-axis illustrates the comparative expression levels of genes confirmed via RNA-seq. The error bars represent the standard error.
4 Discussion
The DOF gene family is a unique gene family in plants, playing crucial roles in plant growth, development, and adaptation to adversity. Although some members of the DOF family have been identified in other plant species, comprehensive genomic analysis of DOF genes in P. simonii has not been conducted, and their regulatory functions remain unclear. This study conducted a comprehensive investigation and expression analysis of the DOF family using genome data, providing a foundational understanding for further exploration of the development and stress responses of P. simonii.
Based on earlier research, the structure and number of gene families are not only related to the genome of a species, but also to the evolutionary process of a plant (Lohani et al., 2021; Waschburger et al., 2024). The research revealed 41 DOF genes in P. simonii, but the count of DOF genes varied across different species. When compared with reported numbers in other species, the number of DOF family members in P. simonii exceeds those reported in tea (16) (Yu et al., 2020), chickpea (37) (Nasim et al., 2016), and watermelon (36) (Zhou et al., 2020). Conversely, the count of DOF family members is lower than that reported in Brassica napus (117) (Lohani et al., 2021), cassava (45) (Zou et al., 2019), and P. simonii × P. nigra (44) (Wang et al., 2022). Additionally, the number of DOF genes in P. trichocarpa (41) (Wang et al., 2017) was similar to that in P simonii in this study, indicating that DOF genes were relatively conserved in P simonii.
Exploring the protein motifs within PSDOFs will deepen our comprehension of their unique functions in developmental and stress adaptation. In studies of motif composition, it was found that all members of the PSDOFs possess motif 1, indicating its potential conservatism and its determination of the functional role of DOF genes. Similar studies have also been conducted in sweet potato (Zhang et al., 2023) and sunflower (Song et al., 2024), where individual motifs were found to be present in all family members. This finding parallels our research results, suggesting that the motif may be a key motif within the DOF family, playing a crucial role in the functionality of DOF genes. Furthermore, the conserved motifs within the same subfamily of DOF proteins is largely consistent, suggesting that members of the DOF protein family may have similar functions in plant growth, development, and stress responses. The presence of specific motifs found exclusively among members of a particular subgroup, such as motif 8 being present in all genes of Group I, indicates the unique evolutionary significance of these motifs within that subgroup. This phenomenon has also been observed in Brassica napus, consistent with our research findings (Lohani et al., 2021).
The analysis of the gene structure of PSDOFs will help us to further explore its specific function in the process of evolution (Li et al., 2021). The analysis of the amino acid sequence revealed that the PSDOF gene possesses a highly conserved zf-DOF domain, suggesting a relatively conserved evolutionary trajectory for the DOF family in plants. The majority of PSDOF genes belonging to the same subgroup exhibit comparable intron-exon structures, yet a few genes demonstrate distinct and specific structures. Additionally, the number and length of introns and exons vary among genes in different branches, potentially due to extended evolutionary processes. In this study, the number of introns in all genes except PSDOF6 was between 0 and 2, which was similar to the number of introns in maize (Chen and Cao, 2015), rice (Khan et al., 2021), pepper (Kang et al., 2016), and Cerasus humilis (Liu et al., 2023). This similarity suggests a relatively stable gene structure for the PSDOF family. Additionally, studies have reported that the presence of introns is advantageous for organismal functionality. For instance, due to alternative splicing, organisms can increase protein diversity, resulting in enhanced functional capabilities of genes (Lohani et al., 2021).
The role of cis-acting elements in gene expression is crucial, and analyzing gene promoters is key to comprehending gene expression in plants. During this study, we identified active elements, including light responsive, hormone responsive, and abiotic stress responsive elements, within PSDOFs. These elements play crucial roles in plant growth and development, closely resembling the cis-acting elements analyzed in wheat (Liu et al., 2020), lotus (Cao et al., 2022), and blueberry (Li et al., 2022b). These studies also indicate that the composition of DOF gene promoter elements is relatively conserved, and the associated DOF genes play roles in growth, development, and stress responses.
By examining the expression profiles of PSDOF genes across various tissues of P. simonii, we gain valuable insights into the dynamic gene expression patterns that underlie its growth and development. The findings revealed that PSDOF genes exhibit elevated expression levels in specific tissues of P. simonii, thus highlighting their crucial role in the plant’s growth and developmental processes. At the same time, a small number of PSDOF family members show different tissue-specific expression patterns, which means that this small number of genes may have a specific function. Similar findings have been reported in many studies as well. In A. thaliana, DOF2.1 pro, DOF5.3 pro, DOF4.6 pro gene expression in leaves an early stage plays an important role in vascular development (Gardiner et al., 2010). The expression of OeuDOF4 gene was significantly upregulated in olive, which indicated that OEUDOF 4 gene was involved in the development of olive leaves (Mariyam et al., 2021). Moreover, prior studies have shown the pivotal involvement of DOF genes across multiple stages of grape fruit development and ripening (Da Silva et al., 2016). The DOF genes plays a role in the production of starch in maize. When ZmDOF36 is overexpressed, it elevates starch levels while diminishing reduced sugar and soluble sugar, aiding in the development of methods to control starch production in maize endosperm (Wu et al., 2019). Additionally, in Brassica napus, BnCDF1 plays a transcriptional regulatory role in flowering time and frost resistance. Overexpression of BnCDF1 in A. thaliana delays flowering time by modulating the expression patterns of CO and FT flowering time control genes, leading to significantly enhanced frost tolerance (Xu and Dai, 2016). These results indicate that DOF genes play irreplaceable roles in the growth and development of various plants.
DOF genes are crucial in controlling the non-living stress response of plants (Ma et al., 2015). In this study, we found that the expression levels of most PSDOF genes are significantly upregulated under stress conditions, indicating their involvement in stress adaptation. Similarly, PSDOF genes have been found to be induced by stress in other plants. Under salt stress, except for PgDOF8, the expression of other PgDOFs genes in pearl millet leaves was up-regulated, and the up-regulation of PgDOF5 was the most significant (Anup et al., 2017). Under high temperature stress in passion fruit, the majority of PeDOF members were inducible, and the expression of PeDOF11 was upregulated most significantly (Chen et al., 2023). In walnut, research has revealed that JrDOF3 contributes to enhancing the heat stress response of JrGRAS2. Arabidopsis plants overexpressing JrGRAS2 exhibited increased tolerance to heat stress (Yang et al., 2018). In Gossypium hirsutum, research has identified changes in the gene expression levels of GhDOFD9.6 in response to salt stress. Additionally, genes such as GhDOFA5.7, GhDOFA7.4, and GhDOFD11.3 were found to significantly respond to low-temperature stress (Li et al., 2018). Moreover, it has been hypothesized that certain TaDOFs in wheat serve as dynamic modulators of ROS clearance pathways, as indicated by their responsive behavior towards heavy metal stress, as reported by Liu et al. (2020). These expression analyses further confirm the roles played by DOFs in stress regulation. In addition, RT-qPCR analysis was conducted to confirm the RNA-seq findings, with an emphasis on the expression patterns of PSDOF genes.
5 Conclusion
This research led to the identification and categorization of 41 DOF genes in P. simonii, examining them across multiple dimensions including phylogenetic evolution, gene architecture, and evolutionary connections. The findings indicate that certain PSDOF family members are crucial in the growth of plant tissues and their reaction to diverse stress scenarios. Notably, PSDOF35 and PSDOF28 serve as pivotal hubs in the interaction network, playing a unique role in coordinating with other genes within the family. The findings of our research offer valuable insights for delving deeper into the biological roles of DOF genes in P. simonii.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
KC: Writing – original draft, Writing – review & editing. XX: Data curation, Writing – review & editing. LH: Data curation, Software, Writing – review & editing. JC: Data curation, Investigation, Writing – review & editing. JZ: Formal Analysis, Methodology, Writing – review & editing. HY: Data curation, Formal Analysis, Writing – review & editing. JS: Validation, Writing – review & editing. YR: Software, Visualization, Writing – review & editing. XZ: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Technological Innovation 2030-Major Project for Agricultural Biotechnology Breeding (2022ZD0401504).
Acknowledgments
Thanks to the members of the State Key Laboratory of Tree Genetics and Breeding for their assistance during laboratory works and for fruitful discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1412175/full#supplementary-material
References
Ahmad, M., Rim, Y., Chen, H., Kim, J. (2013). Functional characterization of Arabidopsis Dof transcription factor AtDof4. 1. Russian J. Plant Physiol. 60, 116–123. doi: 10.1134/S1021443712060027
Anup, P. C., Melvin, P., Kini, K. R. (2017). Reference gene selection and validation for gene expression studies in downy mildew infected pearl millet by quantitative real-time PCR. Australas. Plant Pathol. 46, 441–452. doi: 10.1007/s13313-017-0506-z
Cai, K., Zhao, Q., Zhang, J., Yuan, H., Li, H., Han, L., et al. (2023). Unraveling the guardians of growth: A comprehensive analysis of the aux/IAA and ARF gene families in populus simonii. Plants 12, 3566. doi: 10.3390/plants12203566
Cao, X., Wan, W., Mao, H., Yin, D., Deng, X., Yan, H., et al. (2022). Genome-wide identification and expression analysis of dof transcription factors in lotus (Nelumbo nucifera gaertn.). Plants 11, 2057. doi: 10.3390/plants11152057
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, G., Xu, Y., Gui, J., Huang, Y., Ma, F., Wu, W., et al. (2023). Characterization of Dof transcription factors and the heat-tolerant function of peDof-11 in passion fruit (Passiflora edulis). Int. J. Mol. Sci. 24, 12091. doi: 10.3390/ijms241512091
Chen, J., Song, Y., Zhang, H., Zhang, D. (2013). Genome-wide analysis of gene expression in response to drought stress in Populus simonii. Plant Mol. Biol. Rep. 31, 946–962. doi: 10.1007/s11105-013-0563-6
Chen, M., Liu, X., Huan, L., Sun, M., Liu, L., Chen, X., et al. (2017). Genome-wide analysis of Dof family genes and their expression during bud dormancy in peach (Prunus persica). Scientia Hortic. 214, 18–26. doi: 10.1016/j.scienta.2016.11.014
Chen, Y., Cao, J. (2015). Comparative analysis of Dof transcription factor family in maize. Plant Mol. Biol. Rep. 33, 1245–1258. doi: 10.1007/s11105-014-0835-9
Da Silva, D. C., da Silveira Falavigna, V., Fasoli, M., Buffon, V., Porto, D. D., Pappas, G. J., et al. (2016). Transcriptome analyses of the Dof-like gene family in grapevine reveal its involvement in berry, flower and seed development. Horticulture Res. 3, 16042. doi: 10.1038/hortres.2016.42
Dong, C., Hu, H., Xie, J. (2016). Genome-wide analysis of the DNA-binding with one zinc finger (Dof) transcription factor family in bananas. Genome 59, 1085–1100. doi: 10.1139/gen-2016-0081
Gardiner, J., Sherr, I., Scarpella, E. (2010). Expression of DOF genes identifies early stages of vascular development in Arabidopsis leaves. Int. J. Dev. Biol. 54, 1389–1396. doi: 10.1387/ijdb.093006jg
Gupta, S., Malviya, N., Kushwaha, H., Nasim, J., Bisht, N. C., Singh, V., et al. (2015). Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 241, 549–562. doi: 10.1007/s00425-014-2239-3
Kang, W.-H., Kim, S., Lee, H.-A., Choi, D., Yeom, S.-I. (2016). Genome-wide analysis of Dof transcription factors reveals functional characteristics during development and response to biotic stresses in pepper. Sci. Rep. 6, 33332. doi: 10.1038/srep33332
Khan, A., Bibi, F., Jan, S. A., Shinwari, Z. K. (2023). Role of Dof transcription factors under abiotic stresses. Proc. Pakistan Acad. Sciences: B. Life Environ. Sci. 60, 367–376. doi: 10.53560/PPASB(60-3)905
Khan, I., Khan, S., Zhang, Y., Zhou, J. (2021). Genome-wide analysis and functional characterization of the Dof transcription factor family in rice (Oryza sativa L.). Planta 253, 1–14. doi: 10.1007/s00425-021-03627-y
Kohl, M., Wiese, S., Warscheid, B. (2011). Cytoscape: Software for Visualization and Analysis of Biological Networks. Data Min. Proteomics: Standards to Appl. 696, 291–303. doi: 10.1007/978-1-60761-987-1_18
Li, H., Dou, L., Li, W., Wang, P., Zhao, Q., Xi, R., et al. (2018). Genome-wide identification and expression analysis of the Dof transcription factor gene family in Gossypium hirsutum L. Agronomy 8, 186. doi: 10.3390/agronomy8090186
Li, H., Gou, N., Chen, C., Liu, K., Wang, L., Wuyun, T. (2022a). Genome-wide identification and characterization of the DOF gene family in prunus sibirica. Pak. J. Bot. 54, 1375–1383. doi: 10.30848/PAK.J.BOT
Li, T., Wang, X., Elango, D., Zhang, W., Li, M., Zhang, F., et al. (2022b). Genome-wide identification, phylogenetic and expression pattern analysis of Dof transcription factors in blueberry (Vaccinium corymbosum L.). PeerJ 10, e14087. doi: 10.7717/peerj.14087
Li, X., Cai, K., Pei, X., Li, Y., Hu, Y., Meng, F., et al. (2021). Genome-wide identification of NAC transcription factor family in Juglans mandshurica and their expression analysis during the fruit development and ripening. Int. J. Mol. Sci. 22, 12414. doi: 10.3390/ijms222212414
Lijavetzky, D., Carbonero, P., Vicente-Carbajosa, J. (2003). Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC evolutionary Biol. 3, 1–11. doi: 10.1186/1471-2148-3-17
Liu, W., Ren, W., Liu, X., He, L., Qin, C., Wang, P., et al. (2023). Identification and characterization of Dof genes in Cerasus humilis. Front. Plant Sci. 14, 1152685. doi: 10.3389/fpls.2023.1152685
Liu, Y., Liu, N., Deng, X., Liu, D., Li, M., Cui, D., et al. (2020). Genome-wide analysis of wheat DNA-binding with one finger (Dof) transcription factor genes: evolutionary characteristics and diverse abiotic stress responses. BMC Genomics 21, 1–18. doi: 10.1186/s12864-020-6691-0
Lohani, N., Babaei, S., Singh, M. B., Bhalla, P. L. (2021). Genome-wide in silico identification and comparative analysis of Dof gene family in Brassica napus. Plants 10, 709. doi: 10.3390/plants10040709
Ma, J., Li, M.-Y., Wang, F., Tang, J., Xiong, A.-S. (2015). Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics 16, 1–15. doi: 10.1186/s12864-015-1242-9
Mariyam, S., Haseeb, M., Atif, R. M., Naqvi, S.A.A.A.A., Ali, N., Javed, M. A., et al. (2021). Genome-wide identification and characterization of a plant-specific Dof transcription factor gene family in olive (Olea europaea) and its comparison with Arabidopsis. Horticulture Environment Biotechnol. 62, 949–968. doi: 10.1007/s13580-021-00366-7
Nasim, J., Malviya, N., Kumar, R., Yadav, D. (2016). Genome-wide bioinformatics analysis of Dof transcription factor gene family of chickpea and its comparative phylogenetic assessment with Arabidopsis and rice. Plant Systematics Evol. 302, 1009–1026. doi: 10.1007/s00606-016-1314-6
Noguero, M., Atif, R. M., Ochatt, S., Thompson, R. D. (2013). The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 209, 32–45. doi: 10.1016/j.plantsci.2013.03.016
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45–e45. doi: 10.1093/nar/29.9.e45
Regier, N., Frey, B. (2010). Experimental comparison of relative RT-qPCR quantification approaches for gene expression studies in poplar. BMC Mol. Biol. 11, 1–8. doi: 10.1186/1471-2199-11-57
Ruta, V., Longo, C., Lepri, A., De Angelis, V., Occhigrossi, S., Costantino, P., et al. (2020). The DOF transcription factors in seed and seedling development. Plants 9, 218. doi: 10.3390/plants9020218
Song, A., Gao, T., Li, P., Chen, S., Guan, Z., Wu, D., et al. (2016). Transcriptome-wide identification and expression profiling of the DOF transcription factor gene family in Chrysanthemum morifolium. Front. Plant Sci. 7, 177455. doi: 10.3389/fpls.2016.00199
Song, H., Ji, X., Wang, M., Li, J., Wang, X., Meng, L., et al. (2024). Genome-wide identification and expression analysis of the Dof gene family reveals their involvement in hormone response and abiotic stresses in sunflower (Helianthus annuus L.). Gene 910, 148336. doi: 10.1016/j.gene.2024.148336
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Wang, H., Zhao, S., Gao, Y., Yang, J. (2017). Characterization of Dof transcription factors and their responses to osmotic stress in poplar (Populus trichocarpa). PloS One 12, e0170210. doi: 10.1371/journal.pone.0170210
Wang, S., Wang, R., Yang, C. (2022). Selection and functional identification of Dof genes expressed in response to nitrogen in Populus simonii× Populus nigra. Open Life Sci. 17, 756–780. doi: 10.1515/biol-2022-0084
Wang, Z., Wang, Y., Tong, Q., Xu, G., Xu, M., Li, H., et al. (2021). Transcriptomic analysis of grapevine Dof transcription factor gene family in response to cold stress and functional analyses of the VaDof17d gene. Planta 253, 1–14. doi: 10.1007/s00425-021-03574-8
Waschburger, E. L., Filgueiras, J. P. C., Turchetto-Zolet, A. C. (2024). DOF gene family expansion and diversification. Genet. Mol. Biol. 46, e20230109. doi: 10.1590/1678-4685-gmb-2023-0109
Wen, C.-l., Cheng, Q., Zhao, L., Mao, A., Yang, J., Yu, S., et al. (2016). Identification and characterisation of Dof transcription factors in the cucumber genome. Sci. Rep. 6, 23072. doi: 10.1038/srep23072
Wu, J., Chen, L., Chen, M., Zhou, W., Dong, Q., Jiang, H., et al. (2019). The DOF-domain transcription factor ZmDOF36 positively regulates starch synthesis in transgenic maize. Front. Plant Sci. 10, 442398. doi: 10.3389/fpls.2019.00465
Wu, Z., Cheng, J., Qin, C. (2016). Genome-wide identification and expression profile of Dof transcription factor gene family in pepper (Capsicum annuum L.). Front. Plant Sci. 7, 185464. doi: 10.3389/fpls.2016.00574
Xu, J., Dai, H. (2016). Brassica napus Cycling Dof Factor1 (BnCDF1) is involved in flowering time and freezing tolerance. Plant Growth Regul. 80, 315–322. doi: 10.1007/s10725-016-0168-9
Yang, G., Gao, X., Ma, K., Li, D., Jia, C., Zhai, M., et al. (2018). The walnut transcription factor JrGRAS2 contributes to high temperature stress tolerance involving in Dof transcriptional regulation and HSP protein expression. BMC Plant Biol. 18, 1–14. doi: 10.1186/s12870-018-1568-y
Yu, H., Ma, Y., Lu, Y., Yue, J., Ming, R. (2021). Expression profiling of the Dof gene family under abiotic stresses in spinach. Sci. Rep. 11, 14429. doi: 10.1038/s41598-021-93383-6
Yu, Q., Li, C., Zhang, J., Tian, Y., Wang, H., Zhang, Y., et al. (2020). Genome-wide identification and expression analysis of the Dof gene family under drought stress in tea (Camellia sinensis). PeerJ 8, e9269. doi: 10.7717/peerj.9269
Yue, Y., Du, J., Li, Y., Thomas, H. R., Frank, M. H., Wang, L., et al. (2021). Insight into the petunia Dof transcription factor family reveals a new regulator of male-sterility. Ind. Crops Products 161, 113196. doi: 10.1016/j.indcrop.2020.113196
Zhang, C., Dong, T., Yu, J., Hong, H., Liu, S., Guo, F., et al. (2023). Genome-wide survey and expression analysis of Dof transcription factor family in sweetpotato shed light on their promising functions in stress tolerance. Front. Plant Sci. 14, 1140727. doi: 10.3389/fpls.2023.1140727
Zhou, Y., Cheng, Y., Wan, C., Li, J., Yang, Y., Chen, J. (2020). Genome-wide characterization and expression analysis of the Dof gene family related to abiotic stress in watermelon. PeerJ 8, e8358. doi: 10.7717/peerj.8358
Zou, X., Sun, H. (2023). DOF transcription factors: Specific regulators of plant biological processes. Front. Plant Sci. 14, 1044918. doi: 10.3389/fpls.2023.1044918
Keywords: Populus simonii, DOF family, phylogenetic analysis, chromosome localization, expression pattern
Citation: Cai K, Xie X, Han L, Chen J, Zhang J, Yuan H, Shen J, Ren Y and Zhao X (2024) Identification and functional analysis of the DOF gene family in Populus simonii: implications for development and stress response. Front. Plant Sci. 15:1412175. doi: 10.3389/fpls.2024.1412175
Received: 04 April 2024; Accepted: 24 April 2024;
Published: 08 May 2024.
Edited by:
Qingzhang Du, Beijing Forestry University, ChinaReviewed by:
Shuxia Jia, Beihua University, ChinaShengji Wang, Shanxi Agricultural University, China
Wenjing Yao, Nanjing Forestry University, China
Jinhui Chen, Hainan University, China
Copyright © 2024 Cai, Xie, Han, Chen, Zhang, Yuan, Shen, Ren and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiyang Zhao, emhhb3h5cGhkQDE2My5jb20=
†These authors have contributed equally to this work
 Kewei Cai
Kewei Cai Xiaoyu Xie1†
Xiaoyu Xie1† Xiyang Zhao
Xiyang Zhao