- 1Syngenta Ltd, Jealott’s Hill International Research Centre, Bracknell, Berkshire, United Kingdom
- 2Syngenta España S.A., Carmona-Lora de Rio, Sevilla, Spain
- 3Syngenta Agro SLR, Bucharest, Romania
- 4Syngenta Crop Protection AG, Werk Rosental, Basel, Switzerland
The development and commercialisation of sunflower varieties tolerant to acetolactate synthase (ALS)-inhibiting herbicides some 20 years ago provided farmers with an alternative method for the cost-effective control of Orobanche cumana. In 2020, however, two independent sunflower broomrape populations from Drama (GR-DRA) and Orestiada (GR-ORE), Greece, were reported to be heavily infested with O. cumana after application of the ALS-inhibiting herbicide imazamox. Here we have investigated the race of GR-DRA and GR-ORE and determined the basis of resistance to imazamox in the two Greek O. cumana samples. Using a set of five diagnostic sunflower varieties characterised by different resistant genes with respect to O. cumana infestation, we have clearly established that the GR-ORE and GR-DRA populations belong to the invasive broomrape races G and G+, respectively. Live underground tubercles and emerged shoots were identified at the recommended field rate of imazamox for GR-DRA and GR-ORE but not for two other standard sensitive populations in a whole plant dose response test using two different herbicide-tolerant sunflower hybrids as hosts. Sequencing of the ALS gene identified an alanine 205 to aspartate mutation in all GR-ORE samples. Most GR-DRA tubercles were characterised by a second serine 653 to asparagine ALS mutation whilst a few GR-DRA individuals contained the A205D mutation. Mutations at ALS codons 205 and 653 are known to impact on the binding and efficacy of imazamox and other imidazolinone herbicides. The knowledge generated here will be important for tracking and managing broomrape resistance to ALS-inhibiting herbicides in sunflower growing regions.
Introduction
Sunflower broomrape, Orobanche cumana Wallr., is the most important biotic constraint to sunflower (Helianthus annuus L.) seed production outside North and South America (Molinero-Ruiz et al., 2015). It is an obligate holoparasite that infects the roots of sunflower early in the growing season and hinders the growth and development of the species by depleting the host of essential nutrients (Joel, 2009). At physiological maturity, one single broomrape plant can produce several tens of thousands of seeds which can remain viable for up to 20 years (Fernández-Martínez et al., 2015; Molinero-Ruiz et al., 2015). Sunflower seed losses caused by broomrape infestation can exceed 50% in susceptible H. annuus hybrids (Domínguez, 1996).
O. cumana is generally managed through breeding of genetic resistance into the host (Kaya, 2014). New sunflower varieties are regularly developed and released to manage O. cumana populations that have overcome current resistant traits (Molinero-Ruiz et al., 2015). Genetic resistance to O. cumana is in most cases qualitative (vertical) and controlled by major dominant genes, allowing classification of sunflower broomrape populations, so far, into eight races with increasing virulence from A to H (Vranceanu et al., 1981; Pérez-Vich et al., 2013; Martín-Sanz et al., 2016; Duca et al., 2022). The second way to control sunflower broomrape is via the use of herbicides on sunflower varieties that contain a resistant acetolactate synthase (ALS) target enzyme and some level of tolerance due to non-target-site based mechanisms (Kaya, 2015; Breccia et al., 2017). The first ALS-resistant sunflower was created via conventional breeding of a resistant trait from wild Helianthus annuus into elite cultivated sunflower varieties (Tan et al., 2005). The ALS-resistant wild sunflower population was discovered in Kansas in 1996 in a soybean field that was treated with imazethapyr for seven consecutive years (Al-Khatib et al., 1998). The wild Kansas H. annuus population was subsequently found to contain the alanine to valine mutation at ALS codon position 205 (Kolkman et al., 2004). The A205V target-site mutant sunflower, released as IMISUN or Clearfield variety, is resistant to imidazolinone subclass of ALS-inhibitors allowing in-crop use of herbicides such as imazamox for controlling broomrape and other important sunflower broadleaf and grass weeds (Mitkov et al., 2016). A second generation of herbicide-resistant sunflower (Clearfield Plus or CLHA-PLUS) was developed following EMS mutagenesis and selection with imazapyr (Sala et al., 2008a). CLHA-PLUS is characterized by the A122T mutation and shows increased tolerance to imidazolinones (IMI) herbicides compared to IMISUN and Clearfield sunflowers (Sala et al., 2008b; Pfenning et al., 2012). A third (SURES) and fourth (ExpressSun) class of sunflowers containing the P197L ALS resistant mutation was developed through introgression of the resistant allele from a wild H. annuus population and EMS mutagenesis, respectively (Miroslava, 2022). The SURES and ExpressSun varieties are tolerant to some sulfonylurea herbicides such as tribenuron-methyl but are sensitive to IMI herbicides. The use of IMISUN and CLHA-PLUS tolerant hybrids and IMI herbicides has proved very successful to control sunflower broomrape since their launch in 2003 and 2013, respectively (Molinero-Ruiz et al., 2015). As the technology also allows the simultaneous control of key non-parasitic weeds, their utilisation has exceeded 70% in some Mediterranean sunflower growing areas (Kaya, 2014).
During the last years highly aggressive broomrape populations that have overcome the existing resistance genes have been identified thus rendering the recalcitrant O. cumana populations prone to evolve resistance to IMI herbicides as well (Molinero-Ruiz et al., 2015; Martín-Sanz et al., 2016; Duca et al., 2022). Recently two Greek fields cultivated with the IMISUN and CLHA-PLUS sunflower hybrids were identified with severe attacks of O. cumana affecting more than 75% of the plants despite the use of imazamox. The objectives of this research therefore were to (a) assess the virulence of these two Greek sunflower broomrape populations using a set of diagnostic sunflower genotypes, (b) confirm imazamox resistance in the said populations under controlled conditions, and (c) identify any potential target-site resistance mechanism(s) to the imidazolinone herbicide.
Materials and methods
Orobanche cumana seed populations
Sunflower broomrape seeds were collected in 2020 from two Greek populations that have escaped a recommended application of imazamox (Pulsar®40, BASF). The populations were located at Drama (41°03’41.0”N 24°10’41.5”E) and Orestiada (41°39’44.0”N 26°13’19.2”E). The Drama and Orestiada broomrape seed populations are respectively referred to as GR-DRA and GR-ORE throughout the paper. An IMISUN hybrid was planted at Drama and several IMISUN and CLHA-PLUS hybrids were grown at Orestiada. A broomrape population from Seville province, Spain (SP-SE1) and another from Tulcea province Romania (RO-TU1) that are sensitive to imazamox were used as standards in herbicide resistance evaluations. Additionally, SP-SE1 and RO-TU1 which are both characterised as race G+ were used alongside three other broomrape populations from Seville province, Spain (SP-SE2), Haskovo province, Bulgaria (BU-HAS), and Tulcea province, Romania (RO-TU2) in race classification. SP-SE2, BU-HAS and RO-TU2 belong to races F, G and G+, respectively.
Sunflower lines and hybrids
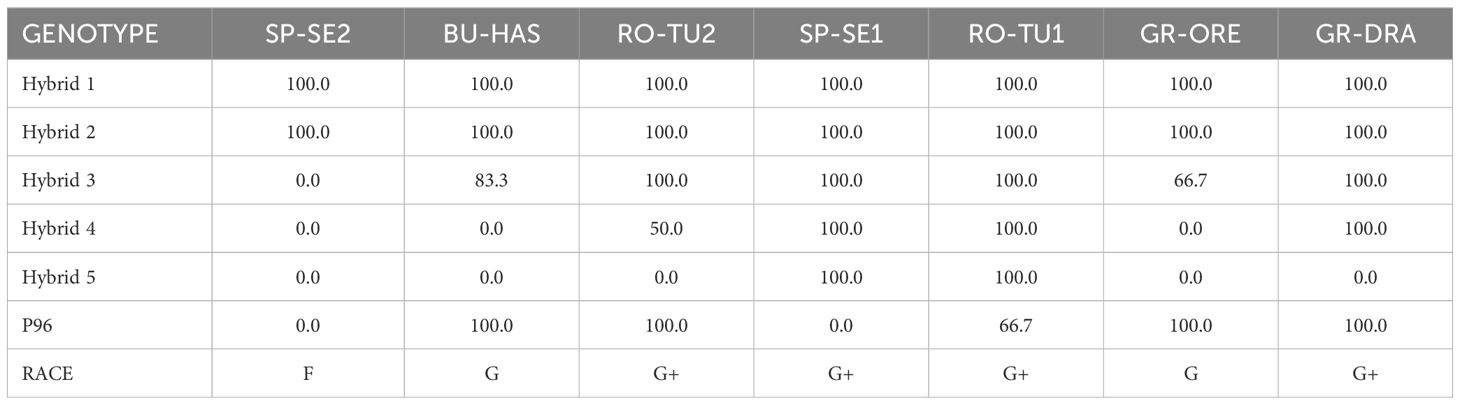
Five Syngenta commercial hybrids and one inbred line (P96) were used to assess the virulence of the two Greek O. cumana populations. The hybrids consisted of Hybrid 1, an old cultivar without any known resistance gene and susceptible to all evaluated broomrape populations according to Syngenta’s internal data; Hybrid 2, resistant to races A-E (Or5 resistance gene); Hybrid 3, resistant to races A-F (Or7); Hybrid 4, resistant to races A-G (Or5 and Or7); and Hybrid 5 (Or5, Or7 and other unknown resistance genes), resistant to broomrape populations that are more aggressive than race G. The inbred line P96 is a genotype commonly used as a race F differential (Martín-Sanz et al., 2016).
The Sunflower Hybrid A (IMISUN) and Hybrid B (CLHA-PLUS) were used for the herbicide phenotypic evaluations. Both hybrids harbour the Or5 resistance gene and are resistant to broomrape races A to E.
The different hybrids were selected based on extensive field trial data from several countries in addition to further tests performed in control glasshouse conditions. The hybrids were also evaluated against broomrape populations classified as race FGV, GGV and GTK in other studies (Martín-Sanz et al., 2016, Martín-Sanz et al., 2020; Fernández-Melero et al., 2023).
Racial characterisation
The virulence of GR-DRA and GR-ORE was determined using known broomrape standards RO-TU1 (race G+), SP-SE1 (race G+), SP-SE2 (race F), BU-HAS (race G) and RO-TU2 (race G+), and five differential sunflower varieties (Hybrid 1, Hybrid 2, Hybrid 3, Hybrid 4 and Hybrid 5) and P96 line. Twelve plants of each sunflower line or hybrid were evaluated for each broomrape population. The experiments were conducted in 1 L pots containing a mixture of sand and peat (1:1 by volume). The soil was artificially contaminated with seeds from each broomrape population at a concentration of 20 ppm (20 mg seed kg-1 soil) and fertilised at a concentration of 6 gL-1 using Nutricote (Projar). The contaminated soil for each broomrape population was mixed to homogeneity in a cement mixer for 30 minutes. Sunflower seeds were germinated on moistened filter paper in Petri-dishes. Single emerged sunflower seedlings were transplanted in pre-prepared 1 L pots and placed in a growth chamber at 25/20°C (day/night) with a 16h photoperiod, photon flux density at 300 µmol m-2s-1 and irrigated as required. Phenotypic evaluation of virulence was determined 65 days after planting by calculating the incidence (percentage of sunflower plants infected by broomrape) and severity (average number of broomrape tubercles in the sunflower infected plants). Only broomrape tubercles with a differentiated stem were considered for estimation of incidence and severity.
Evaluation of imazamox-resistance
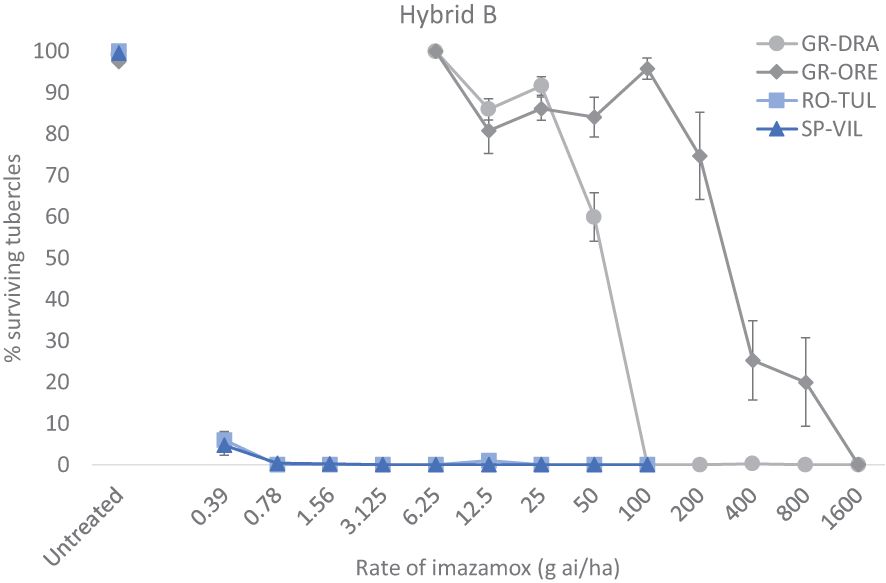
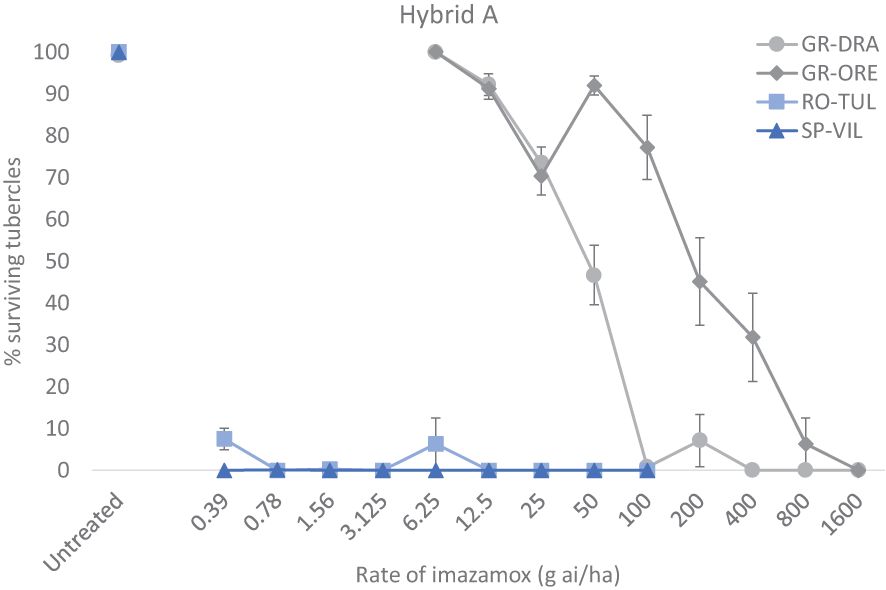
Broomrape populations GR-DRA and GR-ORE were assessed for imazamox resistance alongside RO-TU1 and SP-SE1 which are known sensitive standards for the ALS-inhibiting herbicide. The four different populations were produced in the Hybrid B (CLHA-PLUS) as described above except for the pot volume which was 50 cm3. When the sunflower plants were at the four true leaf-stage (around 25 days after planting) they were sprayed with imazamox (Pulsar®40, BASF) using a Euro-Pulvé backpack sprayer, (model 68130) at a flow rate of 300 L ha-1. Sensitive standards SP-SE1 and RO-TU1 were treated with imazamox at 0, 0.39, 0.78, 1.56, 3.12, 6.25, 12.5, 25, 50 100 g ai ha-1 whilst the two suspected resistant populations GR-DRA and GR-ORE were tested at 0, 6.25, 12.5, 25, 50, 100, 200, 400, 800, 1600 g ai ha-1. Sixteen replicate pots were employed for each herbicide treatment (broomrape population X sunflower hybrid X herbicide dose) and 32 replicate pots were used for untreated controls. Thirty days after imazamox treatment the percentage of alive broomrape tubercles was recorded for each pot. This whole experiment for confirming broomrape resistance in the two Greek samples was replicated using the Hybrid A (IMISUN) as the sunflower host.
Mechanism of imazamox resistance
The acetolactate synthase gene was sequenced to determine whether resistance to imazamox in the two Greek broomrape populations could be due to a target-site mutation. Thirty-two broomrape tubercles were analysed in all including five each from untreated RO-TU1 and SP-SE1 pots and 11 each from GR-DRA and GR-ORE collected from different imazamox treatments as shown in Table 1. Each individual O. cumana tubercle was chilled in liquid nitrogen and was pulverised into a fine, homogeneous powder using mortar and pestle. Total genomic DNA was extracted using Qiagen’s DNeasy Plant Mini Kit (Cat. No. 69104, Qiagen, Germany) following the manufacturer’s instructions. DNA integrity was checked on a gel and quantified with Nanodrop. The quasi-complete ALS gene was amplified by PCR using the forward 5’ CACCGTCGACAATGGCGG 3’ and reverse 5’ ATCACATCCTTAAACGCACC 3’ primer pairs derived from Orobanche minor, the closest species for which the ALS gene was available (GenBank reference: IADW01087971.1). NEB Q5 Polymerase (Cat. No. M0491S, NEB, MA, USA) was employed to amplify the high GC content of the Orobanche minor ALS gene. PCRs were performed on an Eppendorf Master Cycle Gradient Thermocycler Model 96 programmed for an initial denaturation step of 98°C for 1 min, followed by 30 cycles of 30 s at 98°C, 10 s at 64°C and 1 min at 72°C. A final extension step for 2 min at 72°C was also included. PCR product was checked on a gel and column-purified with Invitrogen’s PureLink® PCR Purification Kit (Cat. No K310001, Invitrogen, MA, USA) and sent for Sanger sequencing at Genwiz. The PCR fragment was fully sequenced using four forward and two reverse internal sequencing primers.
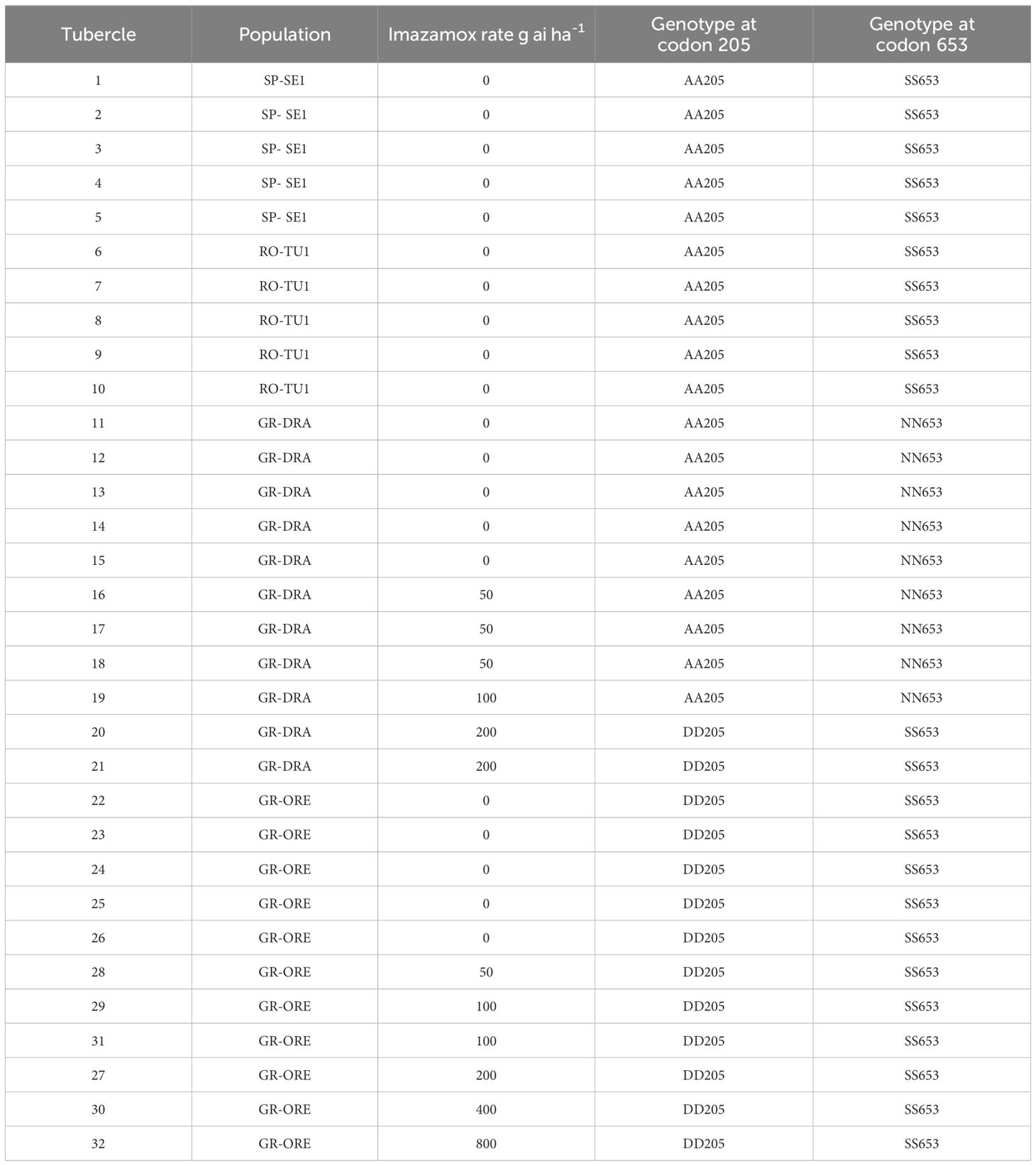
Table 1 Origin and genotypes at ALS codons 205 and 653 of 32 tubercles collected at different imazamox rates.
Results
Virulence of the two Greek broomrape populations
The five reference broomrape populations employed in the racial characterisation study behaved as expected. The reference populations, resistant to race F or above, were able to attach to the sunflower roots and develop a differentiated stem in every susceptible Hybrid 1 and race A-E resistant Hybrid 2 plant (Table 2). The average number of broomrape tubercles observed in Hybrid 1 ranged from 4.3 for RO-TU2 to as many as 21.3 for SP-SE2 (Table 3). A respectively low and high number of tubercles were also detected for RO-TU2 (5.8) and SP-SE2 (20.8) in Hybrid 2. Consistent with its race F status, SP-SE2 could not infest Hybrid 3, Hybrid 4, Hybrid 5 and P96 which are resistant to races F or above. BU-HAS, a race G broomrape population, was able to colonise Hybrid 3 and P96. RO-TU2 (race G+) parasitised all sunflower varieties except Hybrid 5 which is resistant to broomrapes that are more aggressive than race G. RO-TU1 parasitised all sunflower genotypes consistent with its race G+ status. SP-SE1, in agreement with its distinct G+ profile, could infest all sunflower genotypes except P96. The Greek population GR-ORE being investigated parasitised Hybrid 1, Hybrid 2, the race F-resistant Hybrid 3 and P96 sunflowers but not Hybrid 4 and Hybrid 5 hybrids which are resistant to race G broomrapes. The average number of GR-ORE tubercles was comparatively low at 2.7 in Hybrid 3 but as high as 21.2 in P96 (Table 3). GR-ORE was thus classified as a race G broomrape. With its ability to parasitise all but the sunflower Hybrid 5, the Greek population GR-DRA was assigned to race G+. An average of 5.3 GR-DRA tubercles was recorded in Hybrid 4 (resistant to race G) and 26 in P96 (resistant to race F).
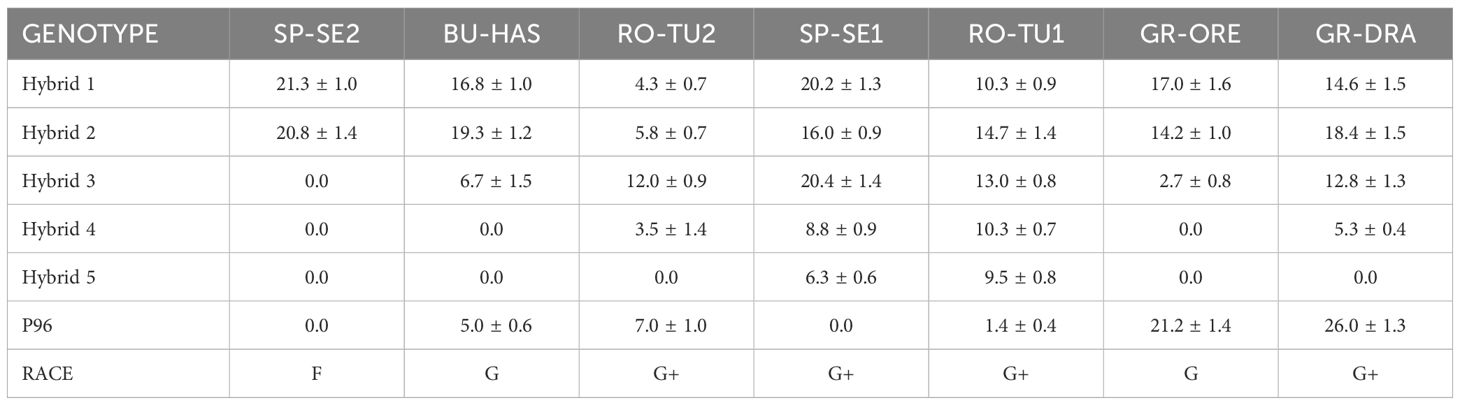
Table 3 Average number of O. cumana attachments with a differentiated stem in six sunflower hybrids and lines (severity).
Investigation of imazamox resistance in the two Greek Orobanche populations
The two standard sensitive populations SP-SE1 (race G+) and RO-TU1 (race G+) and two GR-DRA (race G+) and GR-ORE (race G) populations being investigated adequately parasitised the Hybrid A and Hybrid B hosts allowing determination of imazamox resistance in the two Greek samples. However, there was a relatively large variation in the total number of tubercles among replicate pots for all four populations and herbicide rates tested. On average, the total number of broomrape tubercles was around 50-65 for GR-ORE, GR-DRA and SP-SE1 and half as much for RO-TU1 in untreated sunflower pots. The percentage of live tubercles decreased rapidly as the rate of imazamox increased, especially for the two sensitive broomrape populations in both sunflower hybrids (Figures 1, 2). In Hybrid B for example, only 4.7% of SP-SE1 and 6.0% of RO-TU1 were recorded at the lowest rate of 0.39 g ai ha-1 of imazamox tested (Figure 1). From 0.78 g ai/ha imazamox onwards no live RO-TU1 tubercles were observed whilst only 2.4% of live SP-SE1 tubercles were detected at the second rate of the IMI herbicide. A few live SP-SE1 individuals were still observed at 1.56 and 3.3 g imazamox rates but these only amounted to 0.3 and 0.2% of the total number of tubercles. In contrast, all GR-DRA and GR-ORE tubercles were alive at 6.5 g of imazamox. At the recommended field rate of 50 g ai ha-1 imazamox, 59.9% of GR-ORE and 84% of GR-DRA tubercles survived the herbicide treatment. No GR-DRA survivors were found beyond 100 g ai ha-1 imazamox whilst as much as 20% of GR-ORE live tubercles was recorded at 800 g ai ha-1 of the herbicide. The broomrape resistance profiles in Hybrid A were comparable with those in the Hybrid B with the two sensitive standard populations being killed at relative low rates of imazamox (Figure 2). In Hybrid A, GR-DRA survivors were identified with up to 50 g ai ha-1 imazamox but not beyond and live GR-ORE tubercles were present up to 800 g ai ha-1 of the IMI herbicide. The large difference in herbicide response between GR-ORE and GR-DRA on the one side, and the two known susceptible SP-SE1 and RO-TU1 populations on the other side, confirmed imazamox resistance in the two Greek populations. The relatively higher level of imazamox resistance in GR-ORE as compared to GR-DRA suggests that two different mechanisms may be at play.
Mechanism of imazamox resistance
Thirty-two individual broomrape tubercles collected from untreated and treated GR-DRA, GR-ORE, SP-SE1 and RO-TU1 were sequenced to determine whether a target-site mutation was responsible for imazamox resistance in the two Greek populations (Table 1). Polymerase chain reactions generated a 1945 base pair fragment that covered 98% of the acetolactate synthase gene. The amplified fragment encompassed 11 bases of the 5’ UTR and the first base of codon 645 of ALS. Analysis of the ALS sequences from O. cumana showed on average 67% and 75% homologies at the nucleotide and protein levels respectively, with publicly available ALS from O. minor. The ALS gene was very conserved among all 32 O. cumana samples, except for a cytosine to an adenine transversion at the second bases of codons 205 and 653 (Arabidopsis equivalent), resulting in an alanine 205 to aspartate mutation and a serine 653 to asparagine mutation, respectively. The A205D mutation was present at the homozygous state in all untreated and treated GR-ORE samples and in two GR-DRA tubercules that have survived a high rate (200 g ai ha-1) of imazamox treatment (Table 1). The eight GR-DRA tubercles collected from untreated pots or at a relatively lower rate of imazamox were homozygous for the S653N mutation. No A205D and S653N double mutants were identified among the GR-DRA or GR-ORE samples. The nucleotide sequences of all 10 tubercules from the standard sensitive SP-SE1 and RO-TU1 populations were identical and characterised by homozygous wild type AA205 and SS653 alleles.
Discussion
Racial characterisation of the two Greek O. cumana populations
In Southeast Europe, O. cumana has become an increasingly troublesome species affecting the yield of sunflower (Molinero-Ruiz et al., 2015; Rubiales, 2020). Investigation of broomrape virulence from sunflower growing countries is important to guide the breeding and selection of tolerant varieties for effectively managing O. cumana (Perez-de-Luque et al., 2009; Shi and Zhao, 2020). Over the years, several surveys have been conducted from broomrape samples collected from Spain, Turkey, Romania, Bulgaria, Serbia and Russia (Malek et al., 2017; Škorić et al., 2021). Here, the racial evaluation of the two Greek populations to a set of differential sunflower host genotypes showed that the population from Drama belonged to race G+ while the other population from Orestiada belonged to race G. The identification of highly virulent broomrape strains from Greece is in line with a recent investigation which showed that the two races were prevalent in most South-eastern European countries except for Serbia whereby the strains were found to belong to race E or were less virulent than E (Duca et al., 2022). It is noteworthy that the race G strain identified in the Drama population differed from some Spanish race G populations which could not infest P96, a key race F differential. The distinct Spanish race G strain was baptised race GGV (Martín-Sanz et al., 2016). The virulence identified in the Greek populations in part explicate why they could infest the race A-E resistant sunflower hybrids planted at Drama and Orestiada. The appearance of increasingly virulent populations in recent years could be explained by the intensity of sunflower breeding for resistance to new strains of broomrape, thereby exerting immense selection pressure on the parasite to evolve more virulent races (Molinero-Ruiz et al., 2015; Miraille et al., 2022).
Evolution of imazamox resistance in O. cumana
Our detailed study here confirmed high levels of imazamox resistance in the two Greek populations. This is the first instance of herbicide resistance identified in sunflower broomrape and the second case of resistance evolution in a holoparasite. The first occurrence of herbicide failure in a holoparasite was in an Israeli field dodder, Cuscuta campestris, population that was also found to be resistant to ALS-inhibiting herbicides (Rubin, 1995).
Resistance to ALS herbicides is by far the most common among all other modes of action with as many as 172 different species, including O. cumana, being affected (Powles and Yu, 2010; Heap, 2023). The rapid and widespread evolution of resistance to ALS herbicides is attributed to the large number of ALS-inhibiting herbicides developed and commercialised over the last 40 years and their intensive use in all major crops and growing regions worldwide (Zhou et al., 2007; Mendes et al., 2022). ALS herbicides are potent inhibitors, often with long residual activity, providing effective kill of susceptible individuals, thus exerting high selection pressure on weed populations to evolve resistance (Whitcomb, 1999; Zhou et al., 2007). The intrinsic prevalence of ALS resistant genes in untreated weed populations is much higher than with other modes of action and several mutations that endow target-site resistance have little to no impact on plant fitness due to the non-competitive inhibition of the ALS enzyme (Powles and Yu, 2010; Gaines et al., 2020). In highly outcrossing and heterogeneous species, such as Alopecurus myosuroides and Lolium rigidum resistance was identified within a few years of ALS herbicide application (Heap, 2023). Compared to most other species, resistance evolution to ALS herbicides in O. cumana has been relatively slow mainly because chemical control is generally combined with sunflower resistance genes, including OrSII and OrDEB2 that impact on the attachment and development of the holoparasite (Martín-Sanz et al., 2020; Fernández-Aparicio et al., 2022). This double system of protection explains why the vast majority of O. cumana populations is still effectively controlled with ALS herbicides even after 20 years of use. Other contributing factors for the slow evolution of resistance include the relatively lower selection pressure exerted on O. cumana by ALS herbicides. Sunflower broomrape is a non-photosynthetic root parasite plant that relies entirely on the host for its nutrients and water (Fernández-Martínez et al., 2015). Chemical control of O. cumana is complicated because it produces a large number of seeds that can survive in the soil for several years and sprout when stimulants from the host roots are detected (Plaza et al., 2004; Wu et al., 2022). For effective control of O. cumana, the herbicide must be sprayed at the right growth stage of the sunflower plant so it can be optimally translocated to the host roots and suppress the developing broomrape (Joel et al., 2007). The level of O. cumana control is also very much dependent on the number and depth of the holoparasite attachment (Eizenberg et al., 2009). Altogether, the level of kill and selection pressure from the application of ALS herbicides on O. cumana is not as high as with other non-parasitic weeds due to escapes of susceptible individuals that ultimately contribute to the O. cumana seed bank. Additionally, sunflower is very rarely grown in the same field year after year in Southeast Europe. Instead, the oilseed crop is often rotated with corn, cotton, winter oilseed rape and small grain cereal crops. The relatively long crop rotation and deep cultivation often practiced may explain the slow evolution of resistance to imazamox in sunflower broomrape, and also why only a few cases of ALS resistance have been recorded in other key sunflower weeds such as Ambrosia artemisiifolia and Amaranthus retroflexus in Southeast Europe (Heap, 2023).
Mechanism of imazamox resistance in the Greek broomrape populations
An alanine 205 to aspartate amino acid change was found to be present in all samples from the imazamox resistant population from Orestiada. A few individuals of the population from Drama also contained the A205D mutation whilst most samples were characterised by the S653N mutation. The role of the S653N mutation in conferring resistance to IMI herbicides has clearly been established in several published studies (Powles and Yu, 2010). The heterologous expression of the Arabidopsis ALS gene bearing the wild serine and mutant asparagine alleles in E. coli and inhibition assays of the corresponding proteins showed that the S653N endowed high levels of resistance to IMIs whilst being mostly susceptible to sulfonylurea herbicides (Duggleby et al., 2008). The S653N mutation has also been documented in several weed species that have evolved resistance to ALS herbicides (Heap, 2023). In Amaranthus tuberculatus for example, whole plant dose response tests showed that the S653N mutation conferred resistance to imazethapyr but was susceptible to thifensulfuron (Patzoldt and Tranel, 2007). The S653N ALS mutation is the basis of resistance in some IMI-tolerant crops (Tan et al., 2005). ALS-tolerant Clearfield wheat bearing the S653N mutation, for instance, is resistant to imazamox but sensitive to tribenuron-methyl. In contrast, the importance of the A205D mutation in conferring resistance to ALS-inhibiting herbicides has not been fully investigated so far. The only occurrence of this mutation was in a single plant of an A. palmeri population from Spain (Barco-Antoñanzas et al., 2022). The A205D mutation was present as a double mutant alongside the W574L mutation that is known to confer broad resistance to all ALS-inhibiting herbicides. Therefore, the exact role of the A205D mutation in conferring resistance to ALS herbicides could not be assessed adequately. However, wild type alanine 205 is known to be a key determinant in the binding of some ALS inhibiting herbicides since two mutations at this codon position were found to affect the efficacy of some ALS-inhibitors (Heap, 2023). A Conyza canadensis population carrying the A205V mutation was found to be highly resistant to pyrithiobac-sodium (pyrimidinyl-thiobenzoate) and imazapyr (imidazolinone) and moderately resistant to sulfometuron-methyl (sulfonylurea) (Matzrafi et al., 2015). The A205V mutation is the basis of tolerance to imazamox in IMISUN sunflower indicating that the A205D mutation may also be conferring resistance to imazamox to O. cumana (Sala et al., 2012). Similarly, the Ala-205-Phe substitution identified for the first time in a Poa annua population, expressed in E. coli and assayed in vitro with a range of ALS herbicides was found to be resistant to all five ALS subclasses of herbicides except for florasulam (Brosnan et al., 2016). When present in the same A. palmeri population and genetic background, the S653N change endowed higher levels of resistance to the IMI herbicide imazethapyr compared to the A205V mutation (Palmieri et al., 2022). Here, it appears that the A205D mutation confers higher levels of resistance than the S653N mutation as a larger shift in imazamox efficacy was identified in the A205D mutant Orestiada population versus the Drama population which is essentially S653N mutant with some individuals containing the A205D mutation. This is not surprising given that resistance to ALS-inhibiting herbicides tends to be compound and mutation specific (Powles and Yu, 2010; Gaines et al., 2020).
Detection of herbicide resistance in broomrape
Given the importance of ALS herbicides for the control of O. cumana, it is imperative to determine whether resistance is also present in other sunflower regions and countries where the holoparasite is prevalent. Here, resistance to imazamox was confirmed using the time consuming, space-inefficient and laborious classical whole plant pot assay (Burgos et al., 2013). Furthermore, many pots were required for resistance confirmation as the number of tubercles among replications was appreciably variable even for untreated samples. The need for a sensitive sunflower host that can be effectively parasitised by O. cumana adds another level of difficulty in herbicide resistance investigations. As with some other weed species, sunflower broomrape tested under controlled glasshouse conditions was found to be very sensitive to imazamox with the susceptible populations being killed in some instances with as low as 0.39 g ai ha-1. Thus, using a single recommended rate of 50 g ai ha-1 could cause over-kill and lead to false negatives of resistance especially for populations characterised by target-site mutations or other mechanisms that provide lower levels of resistance than the A205D or S653N amino acid changes. The classical whole plant pot assay with a wide range of herbicide rates and replicate pots cannot be sustainably employed if large numbers of broomrape populations need to be tested. Instead, a Petri-dish seed-based assay could be envisaged using proven sunflower broomrape germination stimulants such as GR24 and a wide range of pre-determined imazamox rates (Lachia et al., 2014; Chen et al., 2021). If successful, minimal efforts and space would be required, and test results would be available in less than half the time required for a whole plant pot test. The seed assay will also facilitate the determination of cross-resistance endowed by different ALS resistance mechanisms as a sunflower host with specific herbicide tolerances would no longer be required. Alternatively, if resistance is found to be mainly due to known target-site mutations, high throughput DNA-based molecular assays could be developed for the quick and efficient determination of resistance in large number of populations (Kaundun et al., 2019). This will be relatively straightforward and highly applicable across populations given the extreme conservation of the ALS gene among O. cumana populations and individuals.
Sustainable use of ALS herbicides for controlling O. cumana
Chemical control of O. cumana in sunflower is currently limited to a few imidazolinone herbicides (Kolkman et al., 2004). Combined with sunflower resistant genes which were regularly discovered and bred into elite sunflower varieties, herbicide use has considerably improved management of O. cumana in Southeastern Europe for the best part of the last two decades (Fernández-Martínez et al., 2012; Molinero-Ruiz et al., 2015). The appearance of progressively more virulent populations has significantly increased the risk of resistance evolution to ALS herbicides in O. cumana. Therefore, perpetual breeding and selection efforts are required to keep pace with the development and spread of new sunflower broomrape races. Recent progress in sunflower genomics provides additional tools for plant breeders to find durable solutions against broomrape spread and virulence in sunflower (Cvejić et al., 2020). The discovery and breeding of multiple resistant genes via a pyramiding approach will drastically decrease the risk of evolution of virulent broomrape races (Velasco et al., 2016; Miraille et al., 2022). For the long-term management of the holoparasite, it is imperative to adopt an integrated approach combining chemical and genetic control with sound agronomic practices, including rotation with trap crops, use of clean seeds and measures to limit seed spread via agricultural machinery and tillage tools (Rubiales et al., 2009; Habimana et al., 2014). The construction of a computer-based model of O. cumana that accounts for the full life cycle of the species and diverse control methods will help design useful strategies for the sustainable management of sunflower broomrape.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, accession number ID is 2803848.
Author contributions
SK: Conceptualization, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing. AM: Conceptualization, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft. MR: Investigation, Methodology, Writing – review & editing. TS: Investigation, Resources, Writing – review & editing. JM: Investigation, Methodology, Writing – review & editing. EM: Formal analysis, Investigation, Writing – review & editing. GL: Conceptualization, Methodology, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors would like to thank Antonio Sayago for technical assistance, Leonardo Velasco for providing some of the O. cumana strains used in this study and several Syngenta colleagues for reviewing manuscript.
Conflict of interest
All authors are employed by companies of Syngenta Group.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Al-Khatib, K., Baumgartner, J. R., Peterson, D. E., Currie, R. S. (1998). Imazethapyr resistance in common sunflower (Helianthus annuus). Weed Sci. 46, 403–407. doi: 10.1017/S0043174500090809
Barco-Antoñanzas, M., Gil-Monreal, M., Eceiza, M. V., Royuela, M., Zabalza, A. (2022). Primary metabolism in an Amaranthus palmeri population with multiple resistance to glyphosate and pyrithiobac herbicides. Plant Sci. 318, 111212. doi: 10.1016/j.plantsci.2022.111212
Breccia, G., Gil, M., Vega, T., Altieri, E., Bulos, M., Picardi, L., et al. (2017). Contribution of non-target-site resistance in imidazolinone-resistant Imisun sunflower. Bragantia 76, 536–542. doi: 10.1590/1678-4499.2016.336
Brosnan, J. T., Vargas, J. J., Breeden, G. K., Grier, L., Aponte, R. A., Tresch, S., et al. (2016). A new amino acid substitution (Ala-205-Phe) in acetolactate synthase (ALS) confers broad spectrum resistance to ALS-inhibiting herbicides. Planta 243, 149–159. doi: 10.1007/s00425-015-2399-9
Burgos, N. R., Tranel, P. J., Streibig, J. C., Davis, V. M., Shaner, D., Norsworthy, J. K., et al. (2013). Review: confirmation of resistance to herbicides and evaluation of resistance levels. Weed Sci. 61, 4–20. doi: 10.1614/WS-D-12-00032.1
Chen, Y., Kuang, Y., Shi, L., Wang, X., Fu, H., Yang, S., et al. (2021). Synthesis and evaluation of new halogenated GR24 analogs as germination promotors for Orobanche cumana. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.725949
Cvejić, S., Radanović, A., Dedić, B., Jocković, M., Jocić, S., Miladinović, D. (2020). Genetic and genomic tools in sunflower breeding for broomrape resistance. Genes 11, 152. doi: 10.3390/genes11020152
Domínguez, J. (1996). “Estimating effects on yield and other agronomic parameters in sunflower hybrids infested with the new races of sunflower broomrape,” in Symposium on Disease Tolerance in Sunflower, Beijing, China (Paris, France: International Sunflower Association). 118–123.
Duca, M., Clapco, S., Joita-Pacureanu, M. (2022). Racial status of Orobanche cumana Wallr. in some countries other the world. Helia 45, 1–22. doi: 10.1515/helia-2022-0002
Duggleby, R. G., McCourt, J. A., Guddat, L. W. (2008). Structure and mechanism of inhibition of plant acetohydroxyacid synthase. Plant Physiol. Biochem. 46, 309–324. doi: 10.1016/j.plaphy.2007.12.004
Eizenberg, H., Hershenhorn, J., Ephrath, J. (2009). Factors affecting the efficacy of Orobanche cumana chemical control in sunflower. Weed Res. 49, 308–315. doi: 10.1111/j.1365-3180.2009.00701.x
Fernández-Aparicio, M., del Moral, L., Muños, S., Velasco, L., Pérez-Vich, B. (2022). Genetic and physiological characterization of sunflower resistance provided by the wild-derived OrDeb2 gene against highly virulent races of Orobanche cumana Wallr. Theor. Appl. Genet. 135, 501–525. doi: 10.1007/s00122-021-03979-9
Fernández-Martínez, J. M., Pérez-Vich, B., Velasco, L. (2015). “Sunflower broomrape (Orobanche cumana Wallr.),” Sunflower Oilseed. Chemistry, Production, Processing and Utilization. Eds. Martínez-Force, E., Dunford, N. T., Salas, J. J. (Champaign, IL: AOCS Press), 129–156.
Fernández-Martínez, J. M., Velasco, L., Pérez-Vich, B. (2012). Progress in research on breeding for resistance to sunflower broomrape. Helia 35, 47–56. doi: 10.2298/HEL1257047F
Fernández-Melero, B., Martín-Sanz, A., Del Moral, L., Pérez-Vich, B., Velasco, L. (2023). A novel sunflower broomrape race with unusual virulence potentially caused by a mutation. Front. Plant Sci. 14, 1236511. doi: 10.3389/fpls.2023.1236511
Gaines, T., Duke, S. O., Morran, S., Rigon, C. A. G., Tranel, P. J., Küpper, A., et al. (2020). Mechanisms of evolved herbicide resistance. J. Biol. Chem. 295, 10307–10330. doi: 10.1074/jbc.REV120.013572
Habimana, S., Nduwumuremyi, A., Chinama R, J. (2014). Management of orobanche in field crops: A review. J. Soil Sci. Plant Nutr. 14, 43–62. doi: 10.4067/S0718-95162014005000004
Heap, I. M. (2023) International survey of Herbicide-Resistant Weeds. Available at: http://www.weedscience.com.
Joel, D. (2009). New nomenclature of orobanche and phelipanche. Weed Res. 49, 6–7. doi: 10.1111/j.1365-3180.2009.00748.x
Joel, D., Hershenhorn, J., Eizenberg, H., Aly, R., Ejeta, G., Rich, P., et al. (2007). Biology and management of weedy root parasites. Hortic. Rev. 33, 267–349. doi: 10.1002/9780470168011.ch4
Kaundun, S. S., Marchegiani, E., Hutchings, S.-J., Baker, K. (2019). Derived Polymorphic Amplified Cleaved Sequence (dPACS): a novel PCR-RFLP procedure for detecting known single nucleotide and deletion-insertion polymorphisms. Int. J. Mol. Sci. 20, 3193. doi: 10.3390/ijms20133193
Kaya, Y. (2014). “Current situation of sunflower broomrape around the world,” in Proceedings of the 3rd International Symposium on Broomrape (Orobanche spp.) in Sunflower, Paris, France (Paris, France: International Sunflower Association). 9–18.
Kaya, Y. (2015). Herbicide resistance breeding in sunflower, current situation and future directions. Buletinul Academiei Ştiinţe Moldovei. Ştiinţele vieţii 326, 101–106.
Kolkman, J. M., Slabaugh, M. B., Bruniard, J. M., Berry, S., Bushman, B. S., Olungu, C., et al. (2004). Acetohydroxyacid synthase mutations conferring resistance to imidazolinone or sulfonylurea herbicides in sunflower. Theor. Appl. Genet. 109, 1147–1159. doi: 10.1007/s00122-004-1716-7
Lachia, M., Wolf, H. C., De Mesmaeker, A. (2014). Synthesis of strigolactones analogues by intramolecular [2+ 2] cycloaddition of ketene-iminium salts to olefins and their activity on Orobanche cumana seeds. Bioorg Med. Chem. 24, 2123–2128. doi: 10.1016/j.bmcl.2014.03.044
Malek, J., Del Moral, L., Fernandez-Escobar, J., Perez-Vich, B., Velasco, L. (2017). Racial characterization and genetic diversity of sunflower broomrape populations from Northern Spain. Phytopathol. Mediterr 56, 70–76. Available at: https://www.jstor.org/stable/44809278.
Martín-Sanz, A., Malek, J., Fernández-Martínez, J. M., Pérez-Vich, B., Velasco, L. (2016). Increased virulence in sunflower broomrape (Orobanche cumana Wallr.) populations from Southern Spain is associated with greater genetic diversity. Front. Plant Sci. 7, 589. doi: 10.3389/fpls.2016.00589
Martín-Sanz, A., Pérez-Vich, B., Rueda, S., Fernández-Martínez, J. M., Velasco, L. (2020). Characterization of post-haustorial resistance to sunflower broomrape. Crop Sci. 60, 1188–1198. doi: 10.1002/csc2.20002
Matzrafi, M., Lazar, T. W., Sibony, M., Rubin, B. (2015). Conyza species: distribution and evolution of multiple target-site herbicide resistances. Planta 242, 259–267. doi: 10.1007/s00425-015-2306-4
Mendes, K. F., Mielke, K. C., La Cruz, R. A. D., Alberto da Silva, A., Ferreira, E. A., Vargas, L. (2022). Applied Weed and Herbicide Science. Eds. Mendes, K. F., Alberto da Silva, A. (Cham: Springer International Publishing), 207–253. doi: 10.1007/978-3-031-01938-8
Miraille, C., Auriac, M.-C., Boniface, M.-C., Delgrange, S., Folletti, T., Jardinaud, M.-F., et al. (2022). Wild Helianthus species: A reservoir of resistance genes for sustainable pyramidal resistance to broomrape in sunflower. Front. Plant Sci. 13, 21. doi: 10.3389/fpls.2022.1038684
Miroslava, H.-C. (2022). Pesticides. Eds. Marcelo, L. L., Sonia, S. (Rijeka: IntechOpen). doi: 10.5772/intechopen.104478
Mitkov, A., Yanev, M., Tonev, T., Tityanov, M. (2016). Weed control in sunflower fields by Clearfield technology. Agric. Sci VIII 19, 167–173.
Molinero-Ruiz, L., Delavault, P., Pérez-Vich, B., Pacureanu-Joita, M., Bulos, M., Altieri, E., et al. (2015). History of the race structure of Orobanche cumana and the breeding of sunflower for resistance to this parasitic weed: A review. Span. J. Agric. Res. 13, e10R01. doi: 10.5424/sjar/2015134-8080
Palmieri, V. E., Alvarez, C. E., Permingeat, H. R., Perotti, V. E. (2022). A122S, A205V, D376E, W574L and S653N substitutions in acetolactate synthase (ALS) from Amaranthus palmeri show different functional impacts on herbicide resistance. Pest Manag Sci. 78, 749–757. doi: 10.1002/ps.6688
Patzoldt, W. L., Tranel, P. J. (2007). Multiple ALS mutations confer herbicide resistance in waterhemp (Amaranthus tuberculatus). Weed Sci. 55, 421–428. doi: 10.1614/WS-06-213.1
Perez-de-Luque, A., Fondevilla, S., Perez-Vich, B., Aly, R., Thoiron, S., Simier, P., et al. (2009). Understanding Orobanche and Phelipanche–host plant interactions and developing resistance. Weed Res. 49, 8–22. doi: 10.1111/j.1365-3180.2009.00738.x
Pérez-Vich, B., Velasco, L., Rich, P. J., Ejeta, G. (2013). Parasitic Orobanchaceae: parasitic mechanisms and control strategies. Eds. Joel, D. M., Gressel, J., Musselman, L. J. (Berlin, Heidelberg: Springer Berlin Heidelberg), 369–391.
Pfenning, M., Tan, S., Perez-Brea, J. (2012). Weed control in Clearfield-Plus sunflowers with superior herbicide solutions (Mar del Plata-Balcarce, Argentina: Proc XVIII Sunflower Conf.), 535–538.
Plaza, L., Fernández, I., Juan, R., Pastor, J., Pujadas, A. (2004). Micromorphological studies on seeds of Orobanche species from the Iberian Peninsula and the Balearic Islands, and their systematic significance. Ann. Bot. 94, 167–178. doi: 10.1093/aob/mch124
Powles, S., Yu, Q. (2010). Evolution in action: plants reistant to herbicides. Annu. Rev. Plant Biol. 61, 317–347. doi: 10.1146/annurev-arplant-042809-112119
Rubiales, D. (2020). Broomrape threat to agriculture. Outlooks Pest Manag 31, 141–145. doi: 10.1564/v31_jun_12
Rubiales, D., Fernandez-Aparicio, M., Wegmann, K., Joel, D. M. (2009). Revisiting strategies for reducing the seedbank of Orobanche and Phelipanche spp. Weed Res. 49, 23–33. doi: 10.1111/j.1365-3180.2009.00742.x
Rubin, B. (1995). “Herbicide resistance outside north America and Europe : Causes and Significance,” in De Prado, R., Jorrín, J., Garcia-Torres, L., Marshall, G., eds. Proceedings international symposium on weed and crop resistance to herbicides (Cordoba, Spain: University of Córdoba).
Sala, C. A., Bulos, M., Altieri, E., Ramos, M. L. (2012). Genetics and breeding of herbicide tolerance in sunflower. Helia 35, 57–70. doi: 10.2298/HEL1257057S
Sala, C., Bulos, M., Echarte, M., Whitt, S., Ascenzi, R. (2008b). Molecular and biochemical characterization of an induced mutation conferring imidazolinone resistance in sunflower. Theor. Appl. Genet. 118, 105–112. doi: 10.1007/s00122-008-0880-6
Sala, C., Bulos, M., Echarte, A., Whitt, S., Budziszewski, G., Howie, W., et al. (2008a). “Development of CLHA-Plus: a novel herbicide tolerance trait in sunflower conferring superior imidazolinone tolerance and ease of breeding,” in Proceedings of XVII International Sunflower Conference, Cordoba, Espana (Paris, France: International Sunflower Association). 489–494.
Shi, B., Zhao, J. (2020). Recent progress on sunflower broomrape research in China. OCL 27, 30. doi: 10.1051/ocl/2020023
Škorić, D., Joiţa-Păcureanu, M., Gorbachenko, F., Gorbachenko, O., Maširević, S. (2021). Dynamics of change in broomrape populations (Orobanche cumana Wallr.) in Romania and Russia (Black Sea area). Helia 44, 1–14. doi: 10.1515/helia-2020-0025
Tan, S., Evans, R. R., Dahmer, M. L., Singh, B. K., Shaner, D. L. (2005). Imidazolinone-tolerant crops: history, current status and future. Pest Manag Sci. 61, 246–257. doi: 10.1002/ps.993
Velasco, L., Pérez-Vich, B., Fernández Martínez, J. M. (2016). Research on resistance to sunflower broomrape: an integrated vision. OCL - Oilseeds Fats Crops Lipids 23, D203. doi: 10.1051/ocl/2016002
Vranceanu, A., Tudor, V., Stoenescu, F., Pirvu, N. (1981). “Virulence groups of Orobanche cumana Wallr.[root parasite], differential hosts and resistance sources and genes in sunflower”. In proceedings of the 9th International Conference of Sunflower Torremolinos, 8-13 Jun 1980, Instituto Nacional de Investigaciones Agrarias, Madrid (Spain).
Whitcomb, C. E. (1999). An introduction to ALS-inhibiting herbicides. Toxicol. Ind. Health 15, 231–239. doi: 10.1191/074823399678846592
Wu, W., Huang, H., Su, J., Yun, X., Zhang, Y., Wei, S., et al. (2022). Dynamics of germination stimulants dehydrocostus lactone and costunolide in the root exudates and extracts of sunflower. Plant Signal Behav. 17, 2025669. doi: 10.1080/15592324.2022.2025669
Keywords: sunflower broomrape, Orobanche cumana, virulence, imazamox, herbicide resistance
Citation: Kaundun SS, Martin-Sanz A, Rodríguez M, Serbanoiu T, Moreno J, Mcindoe E and le Goupil G (2024) First case of evolved herbicide resistance in the holoparasite sunflower broomrape, Orobanche cumana Wallr.. Front. Plant Sci. 15:1420009. doi: 10.3389/fpls.2024.1420009
Received: 19 April 2024; Accepted: 21 May 2024;
Published: 04 June 2024.
Edited by:
Prem Lal Kashyap, Indian Institute of Wheat and Barley Research (ICAR), IndiaReviewed by:
Atsushi Okazawa, Osaka Metropolitan University, JapanDobrinka Anastasova Balabanova, University of Hasselt, Belgium
Copyright © 2024 Kaundun, Martin-Sanz, Rodríguez, Serbanoiu, Moreno, Mcindoe and le Goupil. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiv Shankhar Kaundun, ZGVlcGFrLmthdW5kdW5Ac3luZ2VudGEuY29t
†These authors have contributed equally to this work and share first authorship
 Shiv Shankhar Kaundun
Shiv Shankhar Kaundun Alberto Martin-Sanz
Alberto Martin-Sanz Maribel Rodríguez2
Maribel Rodríguez2 Jose Moreno
Jose Moreno