- 1School of Biological Science and Technology, Liupanshui Normal University, Liupanshui, China
- 2School of Public Health, Jining Medical University, Jining, China
Kiwifruit growth and development are severely affected by various biotic and abiotic stresses, especially cold stress and the bacterial disease caused by Pseudomonas syringae pv. actinidiae (Psa). Metallothioneins (MTs) are a group of cysteine-rich proteins that play crucial roles in stress response, metal detoxification, and homeostasis in plants. However, the protective role of these MTs in kiwifruit remains to be elucidated. In the present study, four AcMT genes were identified in the Hongyang kiwifruit genome, namely, two Type 2 isoforms (AcMT2 and AcMT2a) and two Type 3 isoforms (AcMT3a and AcMT3b) located separately on four different chromosomes. The hormones and stress response cis-elements within the promoter regions of these AcMTs were characterized. It was revealed that the four AcMT genes exhibited different expression patterns in different tissues: AcMT2 and AcMT2a were expressed at much higher levels in the fruit, male flower, female flower, root, and bark, while AcMT3a was expressed mainly in the fruit and AcMT3b was expressed highly in the bark. The expression patterns of these AcMT genes after exposure to Psa infection and different phytohormones, including gibberellic acid A3(GA3), ethylene (ET), and abscisic acid (ABA), were evaluated. It was revealed that in response to Psa infection, the main AcMTs in each tissue (those with expression levels higher compared to the other MTs in that tissue) were downregulated during the early stage in kiwifruits, followed by a recovery phase. In addition, most AcMTs were downregulated after exposure to ET and GA3, while type 2 AcMTs (AcMT2 and AcMT2a) were upregulated after treatment with ABA. The overexpression of AcMTs in Escherichia coli presented a higher tolerance to H2O2, heavy metals, low temperature, and high temperature. Collectively, these findings demonstrated the protective roles of AcMTs in terms of stress resistance conferred through plant hormone-related signal pathways.
1 Introduction
Metallothioneins (MTs) are a group of small, cysteine-rich proteins with a high binding affinity for metal ions, particularly the ones toxic to cells, such as cadmium (Cd), lead (Pb), copper (Cd), and mercury (Hg) (Hassinen et al., 2011). MT proteins were first identified in horse kidneys, following which studies revealed that MT genes are conserved across various organisms, including plants, animals, bacteria, and fungi (Cobbett and Goldsbrough, 2002; Zhou et al., 2006; Leszczyszyn et al., 2013; Barbosa et al., 2017). In plants, MTs are classified into four types based on the distribution of Cys residues in these proteins, and these different types of MTs reportedly exhibit distinct tissue distribution with potentially diverse functions (Usha et al., 2009; Freisinger, 2011). In Arabidopsis, seven MT genes were characterized and categorized into four types: AtMT1a and AtMT1c in Type 1 MT, AtMT2a and AtMT2b in Type 2 MT, AtMT3 in Type 3 MT, and AtMT4a and AtMT4b in Type 4 MT. These four types of MTs were reportedly expressed predominantly in the roots, leaves, fruits, and seeds, respectively (Ledger and Gardner, 1994; Zhou and Goldsbrough, 1994; Hsieh et al., 1995; Zhou and Goldsbrough, 1995; Hsieh et al., 1996). The roles of MTs in plant’s responses to various environmental signals and developmental processes have been studied extensively in recent studies, which have revealed that the expressions of MTs were precisely regulated in response to different stresses (Leszczyszyn et al., 2013; Benatti et al., 2014; Barbosa et al., 2017). MTs play essential roles in plant survival, development, and adaptation to changing environmental conditions. The presence of different motifs in the promoters of MT genes implies that these genes are important for the regulation and execution of a normal developmental process and stress responses in plants (Cheng et al., 2021; Gao et al., 2022).
The roles of MT genes, specifically in plant stress tolerance, are multifaceted. MTs were initially studied because of their abundant Cys residues, which could reversibly bind toxic and essential metal ions, enabling MTs to play crucial roles in the homeostasis and detoxification of metal ions in cells (Hamer, 1986; Hassinen et al., 2009). This functional ability of MTs to sequester toxic metals and thereby prevent toxicity in various processes occurring within the cells is crucial for plant stress tolerance, especially in environments with high metal concentrations. MT overexpression in different model systems, including Arabidopsis, tobacco, yeast, and Escherichia coli, has been demonstrated to contribute to the metal tolerance and homeostasis mechanisms. Specifically, the overexpression of HbMT2 in E. coli reportedly enhanced the tolerance of these bacteria to heavy metals (Zn2+ and Cu2+) (Huang et al., 2018). The overexpression of CrMT in Canavalia rosea enhanced the tolerance of this yeast to heavy metals owing to the metal-chelating ability of the CrMT that was overexpressed in the yeast cells (Zou et al., 2022). The overexpression of the tobacco MT gene NtMT2F in E. coli and Arabidopsis thaliana reportedly enhanced tolerance to Cd stress (Li et al., 2023). In addition to metal-binding properties, scavenging of reactive oxygen species (ROS) is conferred to the MTs due to the abundance of Cys residues (Hassinen et al., 2011; Lv et al., 2013). This enables the MTs to play a protective role against oxidative stress. ROS accumulation occurs during multiple stress conditions, such as drought and exposure to UV radiation and various pollutants. MTs scavenge the ROS and thereby assist in maintaining the redox balance in cells, contributing to cellular resilience. An increasing number of researchers have begun investigating the roles of genes encoding those MTs that serve as ROS scavengers in various physiological processes to understand the contribution of these genes to plant defense responses against different biotic and abiotic stresses (Huang et al., 2018; Zou et al., 2022); cell growth, proliferation, and regulation (Grennan, 2011); root development (Yuan et al., 2008); fruit ripening (Moyle et al., 2005); and senescence (Guo et al., 2003). Transgenic plants with OsMT2b overexpression reportedly exhibited increased vulnerability to bacterial blight and blast fungus due to a sharp decline in the generated ROS (Wong et al., 2004). SbMT-2 overexpression in tobacco significantly improved the tolerance to salt, osmotic, and metal stresses in transgenic tobacco plants (Ahn et al., 2012). In addition, salinity stress reportedly induced the transcriptional activation of OsMT1e-P, the overexpression of which enhanced the tolerance of tobacco to salinity, drought, cold, and heat stresses (Kumar et al., 2012). Collectively, the above findings related to different MT genes from various plants suggest that MTs improve stress tolerance in plants through their ROS-scavenging function.
Kiwifruits have a high nutritional and commercial value, and are, therefore, cultivated widely across different parts of the world, including New Zealand, Italy, Chile, Iran, and China (Ferguson, 2014). “Hongyang” kiwifruit, derived from Actinidia chinensis var. chinensis (Li et al., 2017), is becoming a recent favorite among people owing to its delicious taste, unique flavor, and high content of beneficial components such as vitamin C, anthocyanins, amino acids, and minerals. However, in their natural habitat, “Hongyang” kiwifruits frequently encounter various biotic and abiotic stresses, especially those related to bacterial diseases (infection with Psa, Pseudomonas syringae pv. actinidiae), drought, heavy metal exposure, and extreme temperatures, which significantly impacts their growth and development. Previous studies reported that an MT-like protein gene exhibited higher expression during kiwifruit ripening (Ledger and Gardner, 1994) and was also expressed differentially in Psa-infected kiwifruits (Wang et al., 2018). Therefore, considering the above findings and the crucial role of MTs against multiple stresses, the present study was conducted to investigate the roles of kiwifruit MTs in response to Psa infection and other stresses. Four AcMT genes were screened out from the “Hongyang” kiwifruit genome and then analyzed for their expression patterns in different tissues and in response to Psa infection and exposure to various phytohormones. Furthermore, the identified AcMTs were overexpressed in E. coli to investigate their roles in the plant’s response against stresses due to heavy metals, temperature, and ROS, thereby obtaining further insights into the functions of MTs in kiwifruit.
2 Materials and methods
2.1 Plant materials and treatments
The 8-year-old A. chinensis cv. Hongyang selected for the present study was planted (26.28 N and 104.48 E) at the kiwifruit plantation in Shaomi town, Liupanshui City, Guizhou Province, China. In order to clone the MT genes and analyze their tissue distribution, the female flowers, male flowers, leaves, roots, and bark of the plants were harvested at the flowering stage; the fruits were harvested at 118 days after flowering; and the seeds were harvested after the fruit matured and softened. In order to study the effects of hormone treatments on the expressions of the AcMTs in the fruits, the fruits harvested at 18 days after flowering were steeped for 3 s with 20 mg/L CPPU, 50 mg/L ethephon (ET, an ethylene-releasing compound), 50 mg/L GA3, and 10 mg/L ABA at 0, 3, 6, and 9 h prior to sampling. Fruits from the treated plants were collected at each time point and subjected to RNA extraction, and untreated plants were used as controls. The experiment was repeated three times with three technical replicates (five trees per replicate).
2.2 RNA extraction and cDNA synthesis
Total RNA was extracted from kiwifruit tissues as described in a previous study (Liu et al., 2020). First-strand cDNA was then synthesized from the extracted RNA using the RevertAid First Strand cDNA Synthesis Kit (ThermoFisher, Shanghai, China), according to the manual.
2.3 Isolation of the MT genes in kiwifruit
The sequences of the MT genes identified in other plant species were downloaded from the National Center for Biotechnology Information (NCBI) database and used as query sequences for the basic local alignment search tool (BLAST) analysis of the collective transcriptome database established by our research group and the Kiwifruit Genome Database (http://kiwifruitgenome.org/) (Yue et al., 2020). The full-length cDNA sequences of four AcMTs were thus obtained. Gene identification was confirmed through an RT-PCR conducted using the primers provided in Supplementary Table S1, followed by DNA sequencing.
2.4 Multiple sequence alignment and bioinformatics analysis
The obtained cDNA sequences were submitted to the NCBI database for BLAST searches. The amino acid sequences of the four AcMTs and the MTs of other plant species were aligned using ClustalW (v2.0) software (Larkin et al., 2007). A phylogenetic tree was constructed using the neighbor-joining method in MEGA 7.0 with 1,000 bootstrap replicates. The molecular weights (MWs) and the theoretical isoelectric point values of the AcMTs were calculated using the ProtParam online tool (http://www.expasy.ch/tools/protparam.html). The AcMT promoters were characterized by obtaining 2,000-bp genomic regions upstream of the initiation codon (ATG) of each of the four AcMT genes from the Kiwifruit Genome Database (http://kiwifruitgenome.org/) (Yue et al., 2020). The cis-elements were predicted using Plantcare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). TBtools (https://github.com/CJ-Chen/TBtools) was employed to predict gene structures and conserved domains (Chen et al., 2020). The conserved motifs were analyzed using MEME (http://meme-suite.org/) with the following parameters: maximum number, 5; site distribution, any number of repetitions; minimum width, 6; maximum width, 50 (Brown et al., 2013). TBtools was then employed to draw the location figure, and the chromosomal positions of the AcMT genes were obtained from the GFF file (Chen et al., 2020). MCScanX, Circos, and Dual Synteny Plotter programs in TBtools were employed to calculate and draw the collinear genes in the kiwifruit genome and among the different species (including A. thaliana, Rosa chinensis, Zea mays, Musa acuminata, and Populus trichocarpa) (Ma et al., 2023). The KaKs_Calculator2.0 was employed to calculate the Ka/Ks value of each gene pair (Wang et al., 2010).
2.5 Expressions of the AcMTs in response to Psa infection
The effects of bacterial infection on the expressions of the AcMTs were evaluated, for which the RNA-seq data (Wang et al., 2017; Michelotti et al., 2018; Song et al., 2019) in the Kiwifruit Genome Database (http://kiwifruitgenome.org/) were downloaded (Yue et al., 2020). The data were from the shoot, leaf, and seedling samples of kiwifruit infected with Psa (datasets PRJNA514180, PRJNA328414, and PRJNA436459, respectively). The raw RNA-seq reads were processed using Trimmomatic to remove adapters and low-quality bases, and the trimmed reads shorter than 80% of the original length were discarded. Gene transcript levels were expressed as the expected number of reads per kilobase of the transcript sequence per million base pairs sequenced (RPKM), and the corresponding heat map was generated using R (v3.4.0) with log2(FC). The p-values were adjusted using Benjamini and Hochberg’s approach to constrain the false discovery rate (FDR). The genes with an adjusted p-value (FDR) of <0.05, as determined using DESeq, were designated as the differentially expressed genes.
2.6 The qRT-PCR analysis
The expressions of the AcMT genes were determined using the qRT-PCR analysis, performed as described in a previous study (Liu et al., 2017). The primers used in the qRT-PCR are listed in Supplementary Table S1. The relative expression levels of the AcMT genes were calculated using the 2−ΔΔCT method (Trick et al., 2021).
2.7 Expression of the AcMTs in E. coli
The coding region of each of the AcMTs was amplified through PCR using primer pairs listed in Supplementary Table S2. The PCR products were then inserted into the expression vector pET-28a(+) as described by Huang et al. (2018). The recombination proteins were expressed in E. coli BL21 under induction with 0.4 mM isopropyl β-D-thiogalactopyranoside (IPTG) at 37°C for 4 h, followed by SDS-PAGE for protein identification (Laemmli, 1970). The tolerance of the transformed E. coli BL21 cells to abiotic stress was investigated by comparing the bacterial growth status of the strains containing pET-28a-AcMTs with that of the strains harboring the blank vector alone, as described in a previous report (Huang et al., 2018). The strains were subjected to abiotic stresses as described by Huang et al. (2018). Bacterial growth was monitored and quantified by measuring the OD600 after incubation at 20°C or 45°C for 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, and 3.5 h.
2.8 Statistical analysis
The resulting data were expressed as means ± standard errors (SEs) of three independent biological replicates. Statistical analysis was performed using one-way ANOVA in GraphPad Prism 8.0, and the values indicated using different letters represented significant differences (p < 0.05).
3 Results
3.1 Identification and sequence analysis of AcMTs
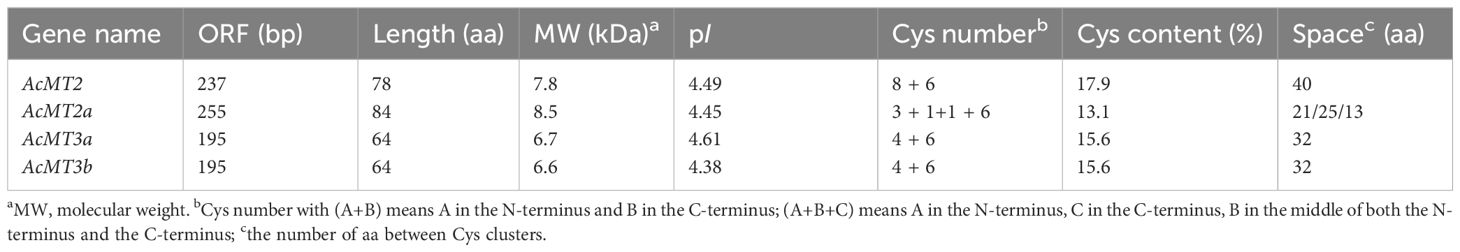
Four AcMTs were identified from the A. chinensis cv. Hongyang genome: AcMT2, AcMT2a, AcMT3a, and AcMT3b. The identification was based on the characteristics of encoded proteins. These four AcMTs were classified according to the sequences of their corresponding Arabidopsis homologs into Type 2 (AcMT2 and AcMT2a) and Type 3 (AcMT3a and AcMT3b) MT proteins in plants (Figure 1A). The length of the open reading frame (ORF) of the four AcMTs ranged from 195 to 255 base pairs (bps). These ORFs encoded proteins containing 64–84 amino acids (aa) with MWs ranging from 6.6 to 8.5 kDa. The theoretical isoelectric point (pI) values of these proteins ranged between 4.38 and 4.61 (Table 1). The Cys residue content in the four AcMTs ranged from 13.1% to 17.9% (Table 1), which is comparable to the range reported for MTs from other plants. AcMT2 contained 14 Cys residues, 8 of which were located in the N-terminus, and 6 were located in the C-terminus, which is characteristic of a typical Type 2 MT protein. AcMT2a, on the other hand, had 11 Cys residues, which were split into four Cys-rich domains by three Cys-free spacers with 25, 21, and 13 aa. Three of the Cys residues were located in the N-terminus, and six were located in the C-terminus. Two Cys-rich domains were located in the middle of the N-terminus. The C-terminus contained one Cys residue. AcMT3a and AcMT3b had 10 Cys residues each, and among the 10 residues in each protein, 4 were located in the N-terminus, and 6 were located in the C-terminus of the protein, which is characteristic of a typical Type 3 MT protein (Figure 1B).
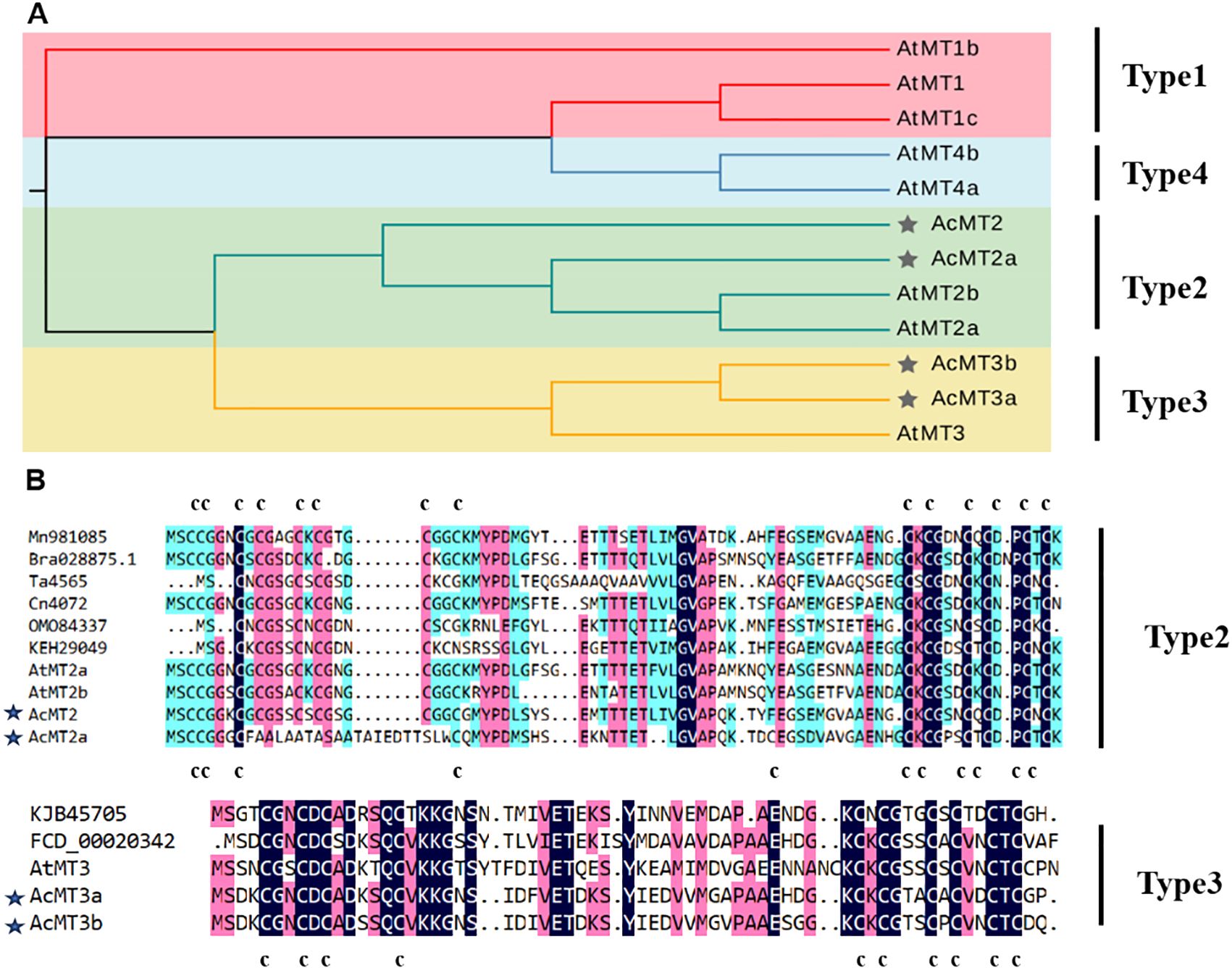
Figure 1. Multiple sequence alignment results, the phylogenetic tree, and the gene structure of kiwifruit MTs. (A) Comparison of the deduced amino acids of kiwifruit MTs, along with their homologs from other plant species. The conserved cysteine residues are marked by the letter C. Identical or conserved amino acids are shaded in black, pink, and green. (B) The phylogenetic tree for the proteins in kiwifruit and Arabidopsis MTs, constructed using MEGA version 6.0. The AcMTs are depicted using a black pentagram.
The structures of the AcMT genes were analyzed (Figure 2B). It was observed that all AcMT genes had a similar exon–intron arrangement. The MEME program was then applied to screen the conserved protein motifs among the AcMTs. The results revealed that each of the AcMTs contained five conserved motifs, among which motif 1 and motif 3 were conserved in all four AcMT proteins (Figure 2A). Motifs 1, 3, and 4 were present in Type 3 MTs, while Type 2 MTs contained Motifs 1, 2, 3, and 5. AcMTs of the same type were revealed to share the conserved gene structure and protein domains. The chromosome distribution of the four AcMTs is presented in Figure 2C. The four AcMTs were mapped independently on four different chromosomes. AcMT2, AcMT2a, AcMT3a, and AcMT3b were distributed on chromosomes 22, 8, 12, and 17, respectively, each located in the forward direction.
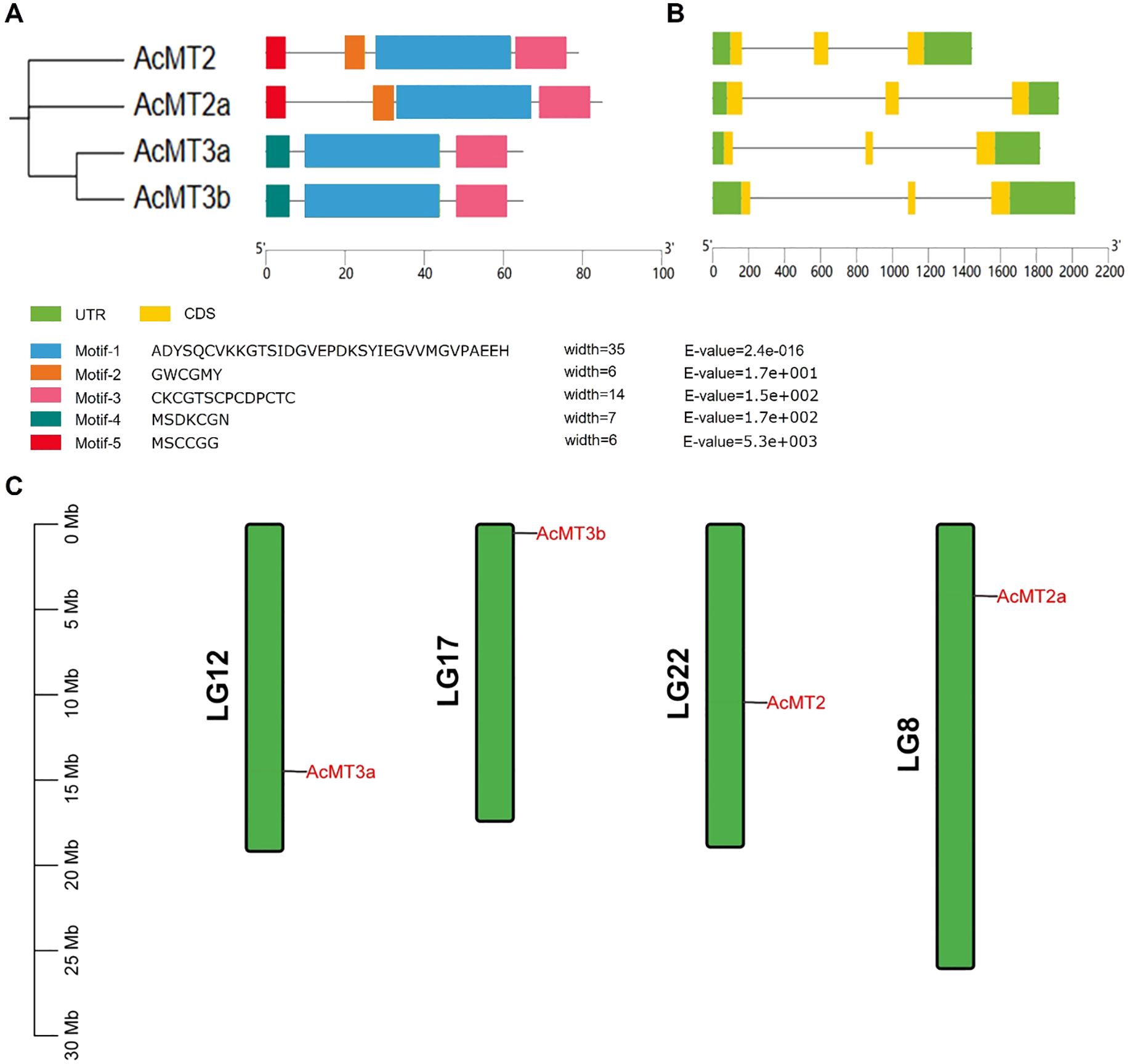
Figure 2. The conserved protein motifs, the gene structure, and the chromosomal distribution of the four AcMTs in kiwifruit. (A) The conserved protein motifs of the four AcMTs. Five patterns of the conserved protein motifs are depicted within different colored boxes. (B) The gene structure of the four AcMTs. (C) The chromosome distribution of the AcMT genes, with each of the four AcMT genes mapped onto four different chromosomes in kiwifruit.
3.2 Phylogenetic analysis of the four AcMTs
The phylogenetic relationships of the four AcMTs were studied through the multiple sequence alignments of 34-aa MT domains (Supplementary Table S3) from 16 species. As depicted in Figure 3, the 34 MT genes could be classified into four groups (Type 1, Type 2, Type 3, and Type 4). However, the analysis revealed that the AcMTs were grouped in only Type 2 and Type 3, with no Type 1 and Type 4 noted. The phylogenetic relationships of different species were determined by constructing a phylogenetic tree. According to the phylogenetic tree, during evolution, the AcMT proteins diverged into two subgroups, Type 2 and Type 3, which remained conserved in kiwifruit.
Figure 3. Phylogenetic analysis of 36 MT gene-encoding proteins. The blank circles represented the AcMTs. The different color circles represent different species. AcMT2, AcMT2a, AcMT3a, and AcMT3b were from A. chinensis; AtMT2a, AtMT2b, AtMT3, AtMT4a, and AtMT4b were from A. thaliana; CaMT1 and CaMT2 were from Capsicum annuum; ClMT was from Citrullus lanatus; CnMT was from Codonopsis lanceolata; FcMT1, FcMT2, and FcMT3 were from Ficus carica; GmMT was from Glycine max; GsMT1, GsMT2, GsMT3, and GsMT4 were from Glycine soja; HbMT1 and HbMT2 were from Hevea brasiliensis; HvMT was from Hordeum vulgare subsp. vulgare; NaMT1 and NaMT2 were from Nicotiana attenuata; OsMT1 was from the Oryza sativa Japonica Group; OsMT2 was from the Oryza sativa Indica Group; RcMT1 and RcMT2 were from R. chinensis; StMT was from Solanum tuberosum; TcMT1, TcMT2, TcMT3, and TcMT4 were from Theobroma cacao; ZmMT was from Solanum Z. mays.
3.3 Duplications and synteny analysis of the AcMTs
The collinear relationship analysis provides important insights into the functions and evolution of the AcMT families. As depicted in Figure 4A, two pairs (AcMT2/AcMT2a and AcMT3a/AcMT3b) of collinear relationships existed between the AcMT families. These two pairs of collinear relationships belonged to the segmental duplication of different chromosomes, suggesting the existence of segmental duplication between these AcMTs. The non-synonymous (Ka) and synonymous (Ks) values were utilized to evaluate the positive selection pressure of the duplication events. The Ka/Ks value of each duplicated AcMT pair was calculated (Supplementary Table S4), and the following were the indications: Ka/Ks = 1, neutral selection; Ka/Ks < 1, purifying selection; Ka/Ks > 1, positive selection (Gao et al., 2022). The Ka/Ks values of AcMT2/AcMT2a and AcMT3a/AcMT3b were 0.5093 and 1.2202, respectively. These values suggested that the evaluated AcMTs were subjected to purifying or positive selection pressure during evolution. Collectively, the above findings suggested that duplication events were the main reason for the amplification of AcMTs. In order to further explore the syntenic relationships of the AcMT families with other plant species, syntenic maps were constructed to explore the homology among A. chinensis, A. thaliana, R. chinensis, Z. mays, M. acuminata, and P. trichocarpa (Figure 4B). The results indicated that the four AcMTs had five pairs of MT collinearity with P. trichocarpa, three pairs of collinearity with M. acuminata, and one pair of collinearity with A. thaliana, R. chinensis, and Z. mays (Supplementary Table S5). The highest number of AcMT homologs was observed for P. trichocarpa, while A. thaliana, R. chinensis, and Z. mays presented the lowest number of homologs. These results demonstrated that the MT gene family members of different species might have originated from the same ancestor and performed a similar function.
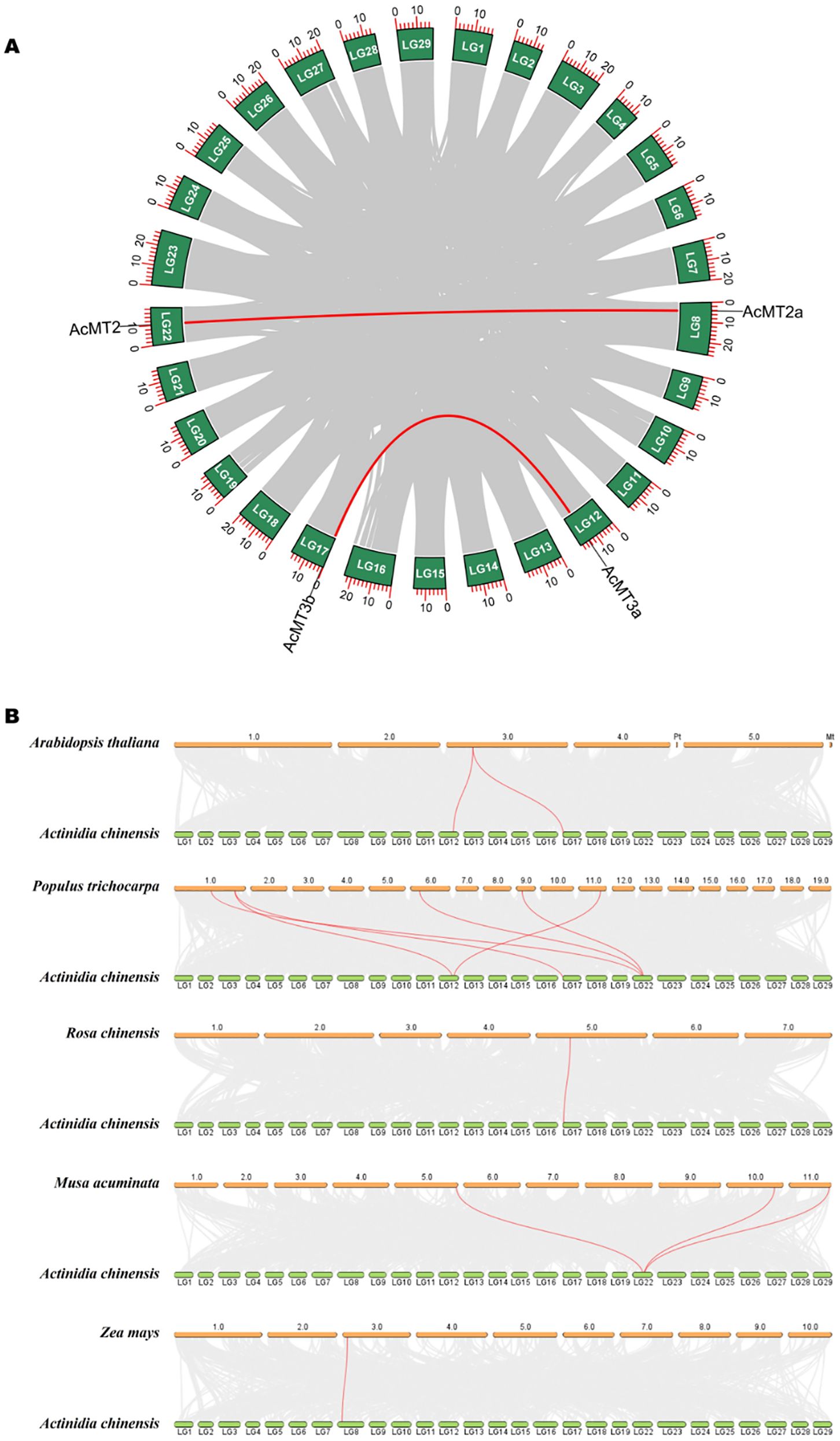
Figure 4. Duplication analysis and synteny analysis of the AcMT genes. (A) Duplication analysis of MTs in kiwifruit. Duplicate blocks are depicted using gray lines, while the duplicate AcMT gene pairs are depicted using red lines. (B) Syntenic maps between kiwifruit and other species. The syntenic MT gene pairs are depicted using red lines.
3.4 Cis-elements predicted in the AcMTs promoters
The cis-elements within the promoter region of the AcMTs related to biotic and abiotic stresses were identified by obtaining and analyzing the upstream 2-kb genomic sequence prior to the start codon of each AcMT gene. The predicted cis-elements are depicted in Figure 5A and were observed to be associated with response to hormone and stress exposure (Supplementary Table S6). Five types of cis-elements were related to the plant response to hormone exposure: ABA response element, auxin response element, MeJA response element, GA3 response element, and SA response element. The remaining cis-elements were related to drought stress, anaerobic induction, light response, meristem expression, zein metabolism regulation, regulation of flavonoid biosynthetic genes, endosperm expression, and seed-specific regulation. These cis-elements were configured with different combinations within the AcMT promoter regions (Figure 5A).
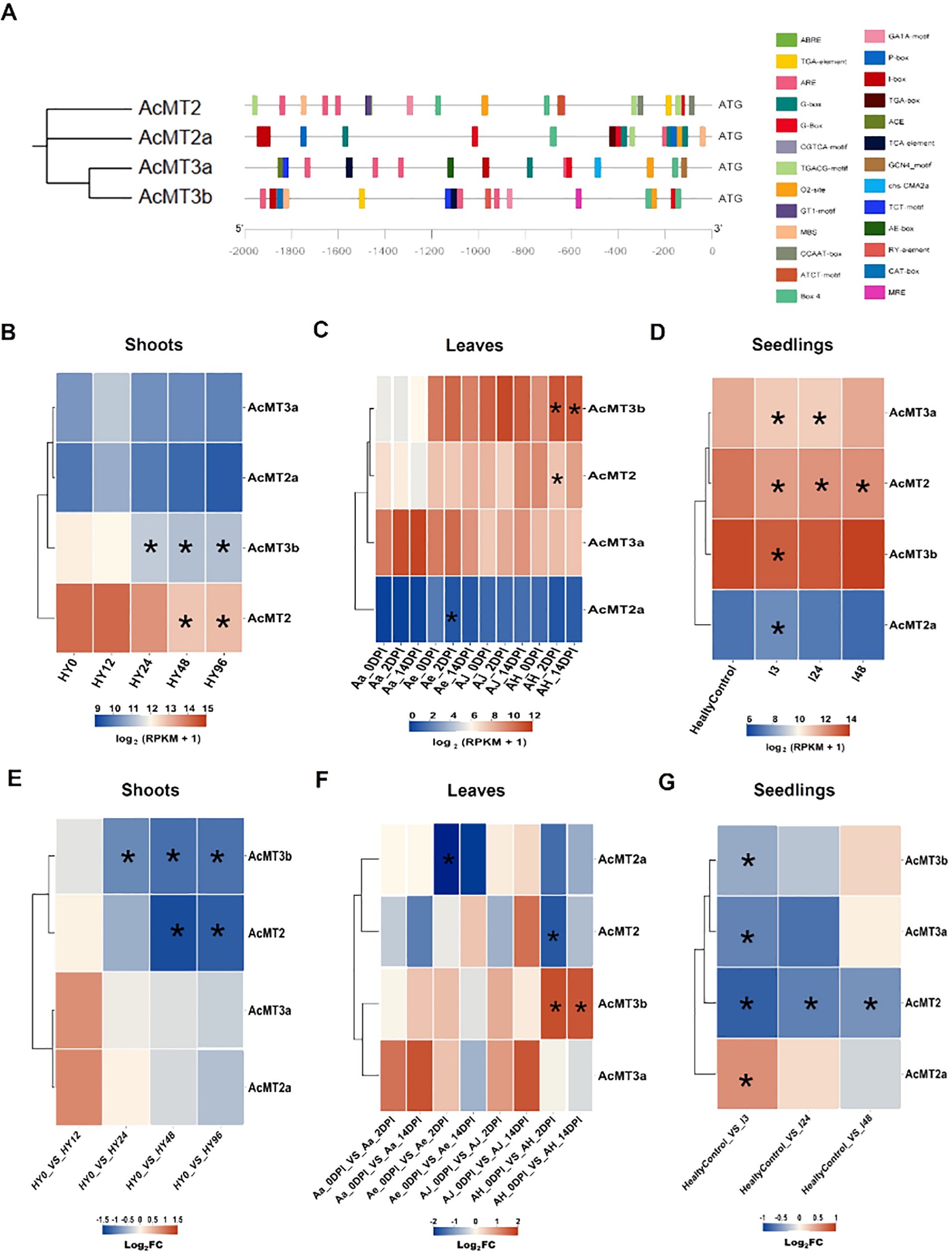
Figure 5. The analysis of the regulatory cis-elements and the expressions of AcMTs in response to Psa infection in four Actinidia species. (A) The predicted regulatory cis-elements in the promoters of the AcMTs in response to exposure to different plant hormones and stresses. (B, E) Heat maps generated at different time points for the levels of AcMTs in response to Psa infection in “Hongyang” shoot samples at levels of RPKM and log2(FC), respectively. HY0, HY12, HY24, HY48, and HY96 represent 0, 12, 24, 48, and 96 h post-Psa inoculation in HY. (C, F) Heat maps generated at different time points for the levels of AcMTs in response to Psa infection in the leaves of four Actinidia species/cultivars at levels of RPKM and log2(FC), respectively. AH, samples from the cultivar “Hongyang” of A. chinensis; AJ, samples from the cultivar “Jinyan” of A. chinensis; Ae, Actinidia eriantha; Aa, Actinidia arguta. 0DPI, 2DPI, and 14DPI were inoculated leaves for 0, 2, and 14 days, respectively. (D, G) Heat maps generated at different time points for the levels of AcMTs in response to Psa infection in the seedlings of A. chinensis var. chinensis at levels of RPKM and log2(FC), respectively. HealtyControl, healthy control plants; I3, I24, and I48 were inoculated plants for 3, 24 and 48 h post-inoculation, respectively. The gene expression levels were determined using the RPKM (expected number of reads per kilobase of the transcript sequence per million base pairs sequenced) method. The transcript levels are depicted using different colors on the scale, with blue and red indicating low and high expression levels, respectively. FDR cutoff value was set at 0.05, and asterisks in the middle of the heat maps meant points below the selected cutoff as significantly different expression. Asterisks in the middle of the heat maps meant points below the selected cutoff as significantly different expression.
3.5 Expression patterns of AcMTs in response to Psa infection
In order to determine the expression patterns of AcMTs in response to Psa infection, the RNA-seq data available in the Sequence Read Archive (SRA) database were analyzed for the expression of AcMTs in response to Psa infection in the shoots of “Hongyang” kiwifruit; the leaves of “Hongyang” kiwifruit, “Jinyan” kiwifruit, “Maohua” kiwifruit, and “Ruanzao” kiwifruit; and the seedlings of A. chinensis var. chinensis (Supplementary Table S7). The analysis of the RNA-seq dataset for the kiwifruit shoots with Psa inoculation (PRJNA514180) revealed (Figures 5B–E) that the expressions of AcMT3b and AcMT2 were higher than those of the other two MTs, with a decreasing trend noted for the former two at the early stage (2 to 48 h) after Psa infection, followed by a recovery phase that began at 96 h. The transcriptional data (PRJNA328414) from the leaves of four kiwifruit varieties with Psa inoculation were analyzed. The results (Figures 5C, F) revealed that the regular expression of Type 2 MTs (AcMT2 and AcMT2a) in the leaves of four kiwifruit varieties did not differ significantly after the Psa infection, while the Type 3 MTs (AcMT3a or AcMT3b) were upregulated in the leaves at 2 and 14 days after the Psa infection. In contrast, a sharp decline was noted in the expression of AcMT3a in the “Ruanzao” and “Maohua” kiwifruit and the expression of AcMT3b in “Maohua”, “Hongyang”, and “Jinyan” kiwifruit varieties. The analysis of the RNA-seq dataset for the Psa-inoculated kiwifruit seedlings (PRJNA436459) revealed (Figures 5D, G) that AcMT2, AcMT3a, and AcMT3b were downregulated during the early stage after the Psa infection, following which an increase was noted in the expressions of these MTs to the levels that were higher compared to the respective normal levels. The only exception to this was AcMT2a, the expression of which remained at a low level after the Psa infection.
3.6 Expression patterns of AcMTs in different tissues
The expression patterns of the four AcMTs in seven selected kiwifruit tissues, namely, bark, leaf, fruit, male flower, female flower, root, and seed, were determined. As depicted in Figure 6, the four AcMTs exhibited distinct expression patterns across the selected tissues. AcMT2 was expressed at the highest level in the male flower, followed by root, female flower, bark, and fruit. The expressions of AcMT2a and AcMT3a were much higher in the fruit compared to the other tissues, while AcMT3b exhibited the highest expression level in the bark (Figure 6D).
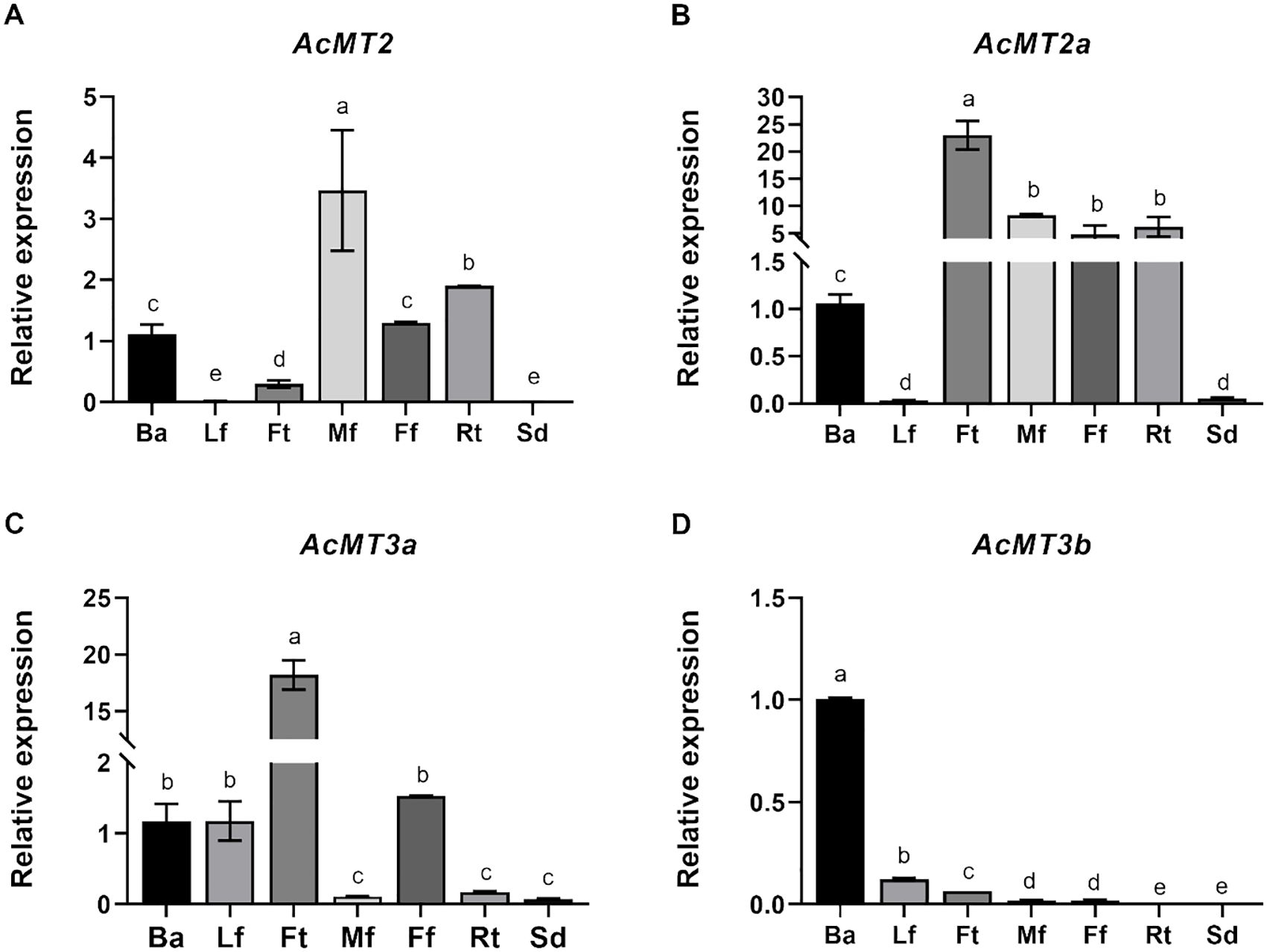
Figure 6. Expression patterns of the four AcMT genes in different tissues of kiwifruit. (A) AcMT2, (B) AcMT2A, (C) AcMT3a, and (D) AcMT3b. Ba, bark; Lf, leaf; Ff, Ft, fruit; Mf, male flower; Ff, female flower; Rt, root; Sd, seed. The values presented are means ± SEs of three independent experiments. Different letters indicate significant differences (p < 0.05) revealed in one-way analysis of variance.
3.7 Expressions of the four AcMTs in response to treatment with different plant hormones
The effects of treatment with different plant hormones on the expressions of the four AcMT genes were determined through qRT-PCR. The fruits of kiwifruit were treated with three plant hormones (ET, ethylene; GA3, gibberellic acid A3; ABA, abscisic acid) in this analysis (Figure 7). It was observed that ET treatment led to a decrease in the expressions of all AcMTs during the early stage after hormone exposure. The decrease in the expression of AcMT2 was noted only at 9 h after treatment with ET. The expressions of the Type 3 MT genes (AcMT3a and AcMT3b) recovered to the baseline levels at 12 h (Figure 7A). Furthermore, as presented in Figure 7B, AcMT2, AcMT2a, and AcMT3a significantly downregulated after treatment with GA3. Notably, the expression of AcMT3b increased significantly and peaked at 6 h after treatment, followed by a decline to the baseline level. After treatment with ABA, the Type 2 MT genes (AcMT2 and AcMT2a) exhibited significant upregulation, with the peak expression for each gene noted at 6 h, which was followed by a decline. At 12 h after treatment with ABA, the expression of AcMT2 declined to a level lower than that noted at 0 h. In contrast, the Type 3 MT genes (AcMT3a and AcMT3b) were downregulated at 3 h. The expression of AcMT3a recovered to the baseline level at 12 h, while that of AcMT3b remained at a lower level compared to the baseline level until 12 h (Figure 7C).
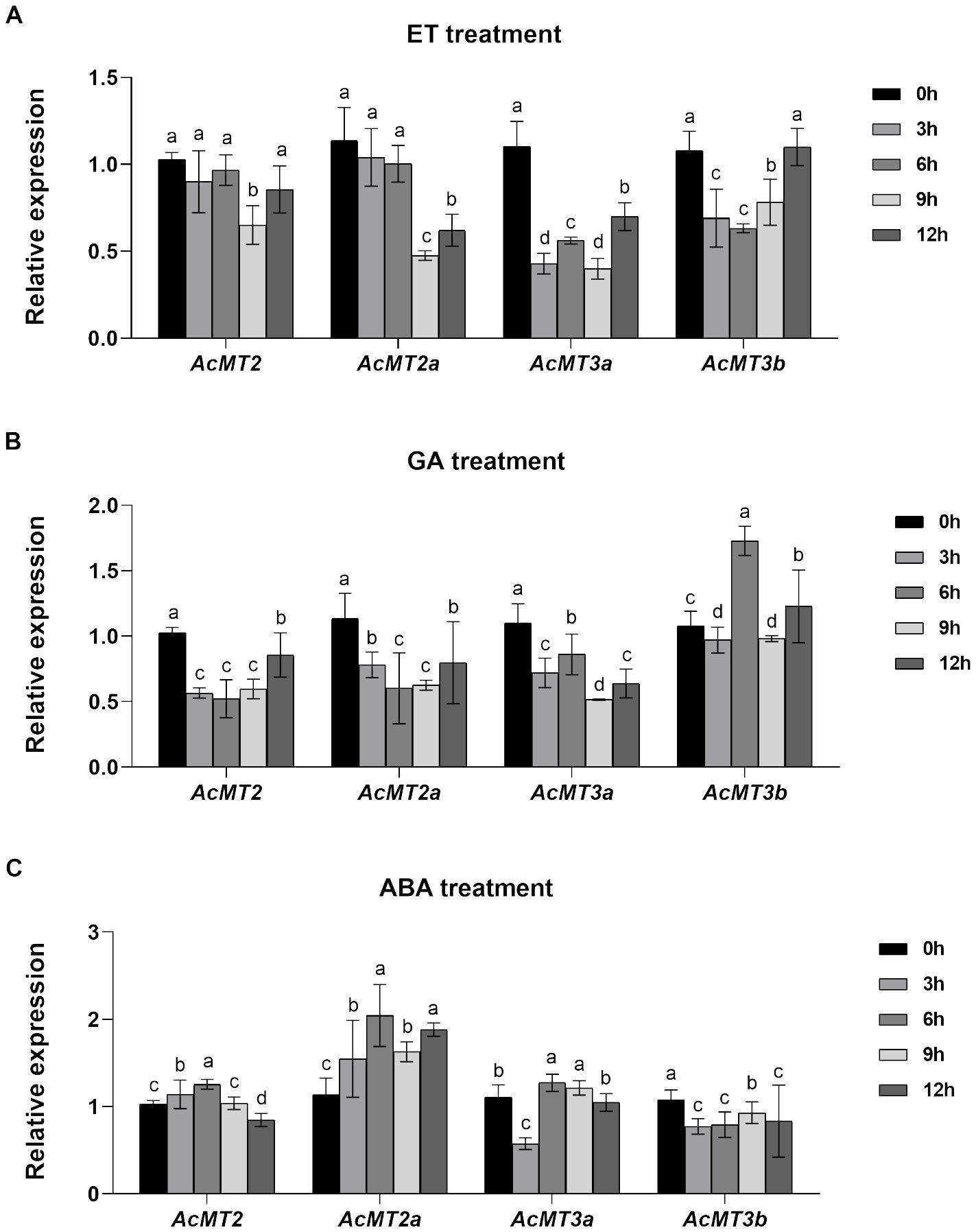
Figure 7. The effects of treatments with ET(A), GA3 (B), and ABA (C) on the expressions of the four AcMTs in the fruit determined through qPCR in kiwifruit. The values presented are means ± SEs of three independent biological replicates. Different letters indicate significant differences (p < 0.05) revealed in one-way analysis of variance.
3.8 Overexpression of AcMTs in E. coli
In order to further explore the effects of the four AcMTs on the tolerance to heavy metals, temperature, and ROS, the recombinant pET28a-AcMTs fusion plasmid was overexpressed in E. coli (Figure 8A). It was observed that under normal growth conditions (LB broth, 37°C), the E. coli strain with AcMT3b overexpression exhibited the highest growth among all strains evaluated (Figure 8B). Moreover, under heavy metal stress (LB broth with 500 μM copper or zinc, 37°C) and high-temperature stress (LB broth, 45°C) conditions, the E. coli strains with overexpressed AcMTs exhibited better growth compared to the control with the overexpression of tag peptides (Figures 8C, F, H). The E. coli strains with AcMT2 overexpression exhibited a higher growth ratio compared to the other strains upon treatment with 500 μM copper and zinc (Figures 8C, F). Furthermore, upon treatment with 1 mM H2O2 and 500 mM NaCl, the growth of all strains was greatly repressed, although the E. coli strain with AcMT3b overexpression exhibited better growth compared to the other strains (Figures 8D, E). It was also observed that none of the recombinant HbMT proteins (except for AcMT3b) were advantageous to E. coli growth under low-temperature (20°C) conditions (Figure 8G).
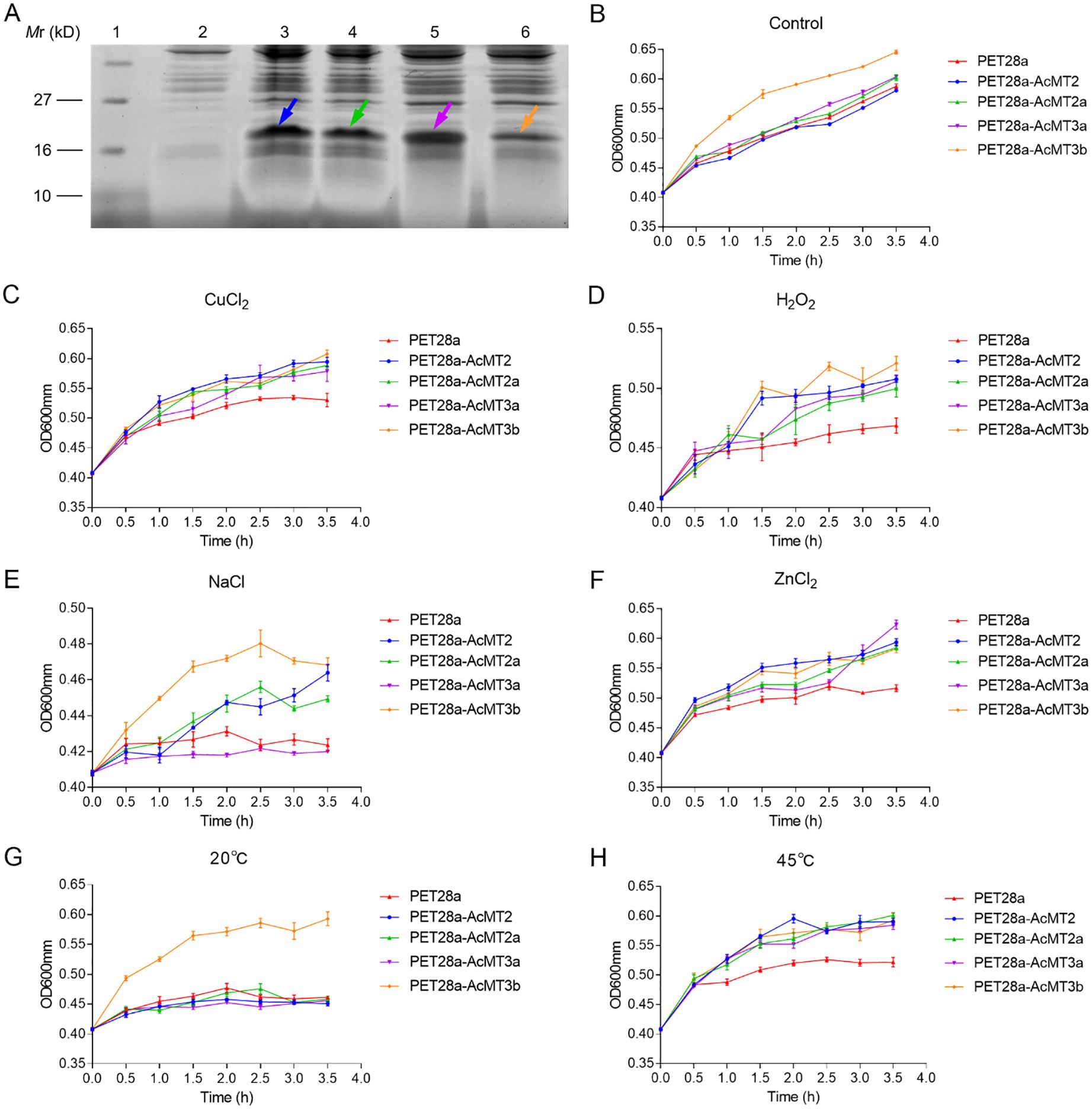
Figure 8. Effects of the overexpression of AcMTs on the growth of E. coli under heavy metal, H2O2, salt, and temperature stresses. (A) SDS-PAGE analysis of recombinant AcMT over-production in the E. coli cells harboring the blank vector pET-28a (Lane 2) or recombinant vectors pET-28a-AcMTs after IPTG induction (Lanes 3–6). Lane 1: Protein molecular weight marker; Lane 2: pET-28a- AcMT2; Lane 3: pET-28a-AcMT2; Lane 4: pET-28a-AcMT2a; Lane 5: pET-28a-AcMT3a; Lane 6: pET-28a-AcMT3b. Arrows indicate the respective tag peptide (His.Tag-thrombin recognition site-T7.Tag)–AcMT fusion proteins. (B) The relative populations of E. coli cells overexpressing different tag peptide–AcMT fusion proteins under culture in an LB medium without supplements. (C) The relative populations of E. coli cells overexpressing different tag peptide–AcMT fusion proteins under culture in the LB medium supplemented with 500 μM CuCl2 or 1 mM H2O2 (D), 500 mM NaCl (E), or 500 μM ZnCl2 (F). (G) The relative populations of the E. coli cells overexpressing different tag peptide–AcMT fusion proteins under culture in the LB medium at 20°C or 45°C (H). The results presented are the means ± SEs from three independent experiments.
4 Discussion
The levels of MT genes in a given plant species are dependent on the genomic ploidy of the species, regardless of whether it is a dicot or monocot (Huang et al., 2018; Gao et al., 2022). In the present study, four MT members were identified in “Hongyang” kiwifruit, including two Type 2 MTs (AcMT2 and AcMT2a) and two Type 3 MTs (AcMT3a and AcMT3b), while no Type 1 and Type 4 MTs were detected (Figure 1B) (Cobbett and Goldsbrough, 2002). Similarly, previous studies have identified certain plant species lacking one or two types of MTs, such as rubber tree (without Type 1 and Type 4), Phoenix dactylifera (without Type 1 and Type 3), and Medicago truncatula (without Type 4) (Huang et al., 2018). The lack of Type 1 and Type 4 MTs in “Hongyang” kiwifruit could be attributed to the functional complementarity and redundancy among different types of MTs during the evolution of kiwifruit.
The number and distribution of cysteine residues in the N-terminus and C-terminus regions of MTs are known to be conserved and related to the function of these MTs. In the present study, AcMT3a and AcMT3b presented cysteine residue characteristics similar to those of the other Type 3 MTs, with four Cys located at the N-terminal region and six Cys located at the C-terminal region. AcMT2 and AcMT2a had the Cys characteristics of other Type2 MTs, with three Cys-X-Cys motifs at the C-terminus of their sequences. Meanwhile, AcMT2a presented a unique structure with eight Cys residues (CCXXXCXCXXXCXCXXXCXXCXXX) at the N-terminus of its structure. AcMT2a was, therefore, regarded as an atypical MT. In previous studies, the Cys residues in MTs were demonstrated to affect their heavy metal-chelating function (De Oliveira et al., 2020). In addition, mutations in the Cys residues of NtMT2F were reported to significantly reduce the tolerance of A. thaliana to heavy metals (Li et al., 2023). Cysteine substitution with tyrosine (C3Y) in PtMT2b reportedly enhanced the Cd tolerance in the transformed yeast (De Oliveira et al., 2020). The missing Cys residues at the C-terminus of ScMT10 could enhance the freezing tolerance in the heterogeneously expressed tobacco plants (Feng et al., 2022). Similarly, in the present study, the bacterial strain overexpressing AcMT2 exhibited faster growth compared to the bacterial strains overexpressing AcMT2a under Cu and Zn stresses. These results indicated that the substitution of Cys residues in AcMT2a significantly reduced its ability to chelate heavy metals. Further studies should, therefore, be conducted to elucidate the role of Cys residues in MTs in influencing tolerance to heavy metals in plants.
An increasing number of studies have recently reported that the same types of plant MT genes exhibit varied expression patterns in different plant tissues. HbMT2a, for instance, was reported to exhibit a flower-specific expression (Huang et al., 2018), while NtMT2F is primarily expressed in roots and old leaves, according to reports (Yu et al., 2021). CsMT2, on the other hand, exhibits a relatively universal expression in different tissues (Zhou et al., 2019). In the present study, AcMT2 and AcMT2a exhibited the highest expression levels in the fruit, followed by male flower, female flower, root, and bark, although both MT genes were expressed in low levels in the leaf and seed. Meanwhile, AcMT3a, similar to the other members of Type 3 MT genes, exhibited the highest expression in the fruit, while AcMT3b exhibited the highest accumulation of its transcripts in the bark. It is noteworthy that the highest expression of AcMT2 was noted in the male flower, the highest expressions of AcMT2a and AcMT3a were noted in the fruit, and the highest expression of AcMT3b was noted in the bark, indicating that AcMTs have discrete and overlapping roles in different tissues. The high expression of AcMT2 in the roots might be a contributor to the enhanced resistance of plants to heavy metal exposure, while the high expression of AcMT3b in the bark could serve as a barrier against cold stress and pathogen invasion, although further studies are warranted to confirm these hypotheses.
The bacterial canker caused by Psa has been detected in the main kiwifruit-producing nations since 2007 (Vanneste et al., 2011; Chapman et al., 2012). The rapid transmission and strong pathogenicity of Psa have led to serious production and economic losses in several nations, causing Psa to become a major limiting factor in the development of the kiwifruit industry (Mazzaglia et al., 2012; Vanneste, 2012). Certain genes involved in the plant’s response to Psa infection have been characterized in previous studies, including the genes encoding pentatricopeptide repeat (PPR) proteins. These PPR genes are thought to regulate plant–pathogen interactions by controlling the degree of RNA editing, particularly the composition of the NDH complex (Zhang et al., 2023). According to previous studies, the MTs in plants play protective roles against pathogen attacks (Reymond et al., 2000; Wong et al., 2004). In the present study, the expression levels of the main AcMTs (with a higher expression level compared to the other MTs) in each tissue of the kiwifruit were revealed to be downregulated during the early stage after Psa infection, followed by a recovery phase, indicating that different combinations of AcMTs serve as the main barriers against Psa infection in different tissues of the kiwifruits. These findings were similar to those reported previously for the leucine-rich repeat receptor-like proteins (LRR-RLPs) in kiwifruit (Cao et al., 2024). Previous studies have also demonstrated that while ROS accumulation causes severe cellular damage through protein oxidation, lipid peroxidation, nucleic acid damage, and enzyme inhibition (Sharma et al., 2012), it plays a central role in plant defense against pathogens by limiting the spread of pathogenic bacteria (Lamb and Dixon, 1997). The regulation of ROS levels is considered important for plant disease resistance. All AcMTs evaluated in the present study exhibited ROS-scavenging abilities, as evidenced by the significant tolerance of the E. coli strain with overexpression of each AcMT to the ROS stress. The downregulation of the main AcMTs in each tissue during the early stage after Psa infection might be beneficial to the ROS accumulation in the plant for acting against the pathogen invasion. Once the ROS levels are too high to cause oxidative damage, the expression levels of the main AcMTs in each tissue begin to return to the baseline levels, resulting in the observed recovery phase.
MTs are involved in numerous physiological processes, such as cell growth proliferation and regulation, plant growth and defense against pathogens, homeostasis, and heavy metal toxicity protection (Khalid et al., 2020). These functions of MTs are similar to those of the other functional genes involved in plant growth and stress resistance in kiwifruit, which are reportedly regulated precisely through plant hormone-related signaling pathways. CsMT4 (Zhou et al., 2019) and BrMT1 (Ahn et al., 2012), for example, are reported to exhibit increased levels upon ABA treatment, conferring the plants with the ability to withstand osmotic pressure and drought resistance. HbMT2b levels (Huang et al., 2018) were markedly depressed upon ethephon treatment in a previous study, which increased the risk of oxidative stress in tapped rubber trees. In the present study, ET and GA3 significantly downregulated most of the AcMT genes, indicating that during fruit ripening or stages of rapid plant growth, kiwifruits were more susceptible to damage due to external stress when the levels of ET and GA3 were higher. In contrast, when the synthesis of ABA increased, causing the plant growth to be inhibited and the plant to enter into the dormancy stage, the synthesis of Type 2 MTs (AcMT2 and AcMT2a) was probably increased to enhance the ability of kiwifruit to withstand the external stresses.
5 Conclusions
In the present study, four AcMTs were identified in kiwifruit, and their sequence characteristics were determined. The expression profiles of these four AcMTs in response to Psa infection and treatment with plant hormones were determined, and these profiles implied the roles of these AcMTs in plant defense against pathogens. Furthermore, the E. coli strains overexpressing the AcMTs exhibited better growth under H2O2, heavy metal, and extreme temperature stresses, indicating that these AcMTs could enhance the tolerance of the bacteria to these stresses. These findings provide insights into the potential functions of the MT genes in kiwifruits, and the information would also be useful for elucidating the mechanism underlying stress tolerance in kiwifruits.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
LL: Formal analysis, Funding acquisition, Writing – original draft. SS: Data curation, Formal analysis, Investigation, Writing – original draft. NY: Data curation, Formal analysis, Methodology, Writing – original draft. BH: Data curation, Writing – original draft. LX: Writing – review & editing, Conceptualization. YH: Conceptualization, Funding acquisition, Investigation, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Scientific Research and Cultivation Project of Liupanshui Normal University (LPSSY2023KJZDPY07), the National Natural Science Foundation of China (Grant No. 32360740), the Science and Technology Program of Guizhou Province (QianKeHeJiChu[2020]1Y115), the Natural Science Research Project of the Education Department of Guizhou Province (Qianjiaoji[2022]054), the High-Level Talents Research Start-up Fund Project of Liupanshui Normal University (LPSSYKYJJ202205), and the Science and Technology Program of Liupanshui (52020-2022-PT-03 and 52020-2023-0-2-15).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1425072/full#supplementary-material
References
Ahn, Y. O., Kim, S. H., Lee, J., Kim, H., Lee, H. S., Kwak, S. S. (2012). Three Brassica rapa metallothionein genes are differentially regulated under various stress conditions. Mol. Biol. Rep. 39, 2059–2067. doi: 10.1007/s11033-011-0953-5
Barbosa, B. C. F., Silva, S. C., De Oliveira, R. R., Chalfun, A. (2017). Zinc supply impacts on the relative expression of a metallothionein-like gene in Coffea arabica plants. Plant Soil 411, 179–191. doi: 10.1007/s11104-016-2983-1
Benatti, M. R., Yookongkaew, N., Meetam, M., Guo, W. J., Punyasuk, N., Abuqamar, S., et al. (2014). Metallothionein deficiency impacts copper accumulation and redistribution in leaves and seeds of Arabidopsis. New Phytol. 202, 940–951. doi: 10.1111/nph.12718
Brown, P., Baxter, L., Hickman, R., Beynon, J., Moore, J. D., Ott, S. (2013). MEME-LaB: motif analysis in clusters. Bioinformatics 29, 1696–1697. doi: 10.1093/bioinformatics/btt248
Cao, Y., Zhang, C., Liu, F., Li, D., Zhang, A., Li, L., et al. (2024). Genome-wide identification and expression analysis of kiwifruit leucine-rich repeat receptor-like proteins reveal their roles in biotic and abiotic stress responses. Int. J. Mol. Sci. 25, 4497. doi: 10.3390/ijms25084497
Chapman, J., Taylor, R., Weir, B., Romberg, M., Vanneste, J., Luck, J., et al. (2012). Phylogenetic relationships among global populations of Pseudomonas syringae pv. actinidiae. Phytopathology 102, 1034–1044. doi: 10.1094/PHYTO-03-12-0064-R
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, M., Yuan, H., Wang, R., Zou, J., Liang, T., Yang, F., et al. (2021). Genome-wide identification and analysis of the metallothionein genes in Oryza genus. Int. J. Mol. Sci. 22, 9651. doi: 10.3390/ijms22179651
Cobbett, C., Goldsbrough, P. (2002). Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 53, 159–182. doi: 10.1146/annurev.arplant.53.100301.135154
De Oliveira, V. H., Ullah, I., Dunwell, J. M., Tibbett, M. (2020). Bioremediation potential of Cd by transgenic yeast expressing a metallothionein gene from Populus trichocarpa. Ecotoxicol. Environ. Saf. 202, 110917. doi: 10.1016/j.ecoenv.2020.110917
Feng, M., Yu, Q., Chen, Y., Fu, Z., Xu, L., Guo, J. (2022). ScMT10, a metallothionein-like gene from sugarcane, enhances freezing tolerance in Nicotiana tabacum transgenic plants. Environ. Exp. Bot. 194, 104750. doi: 10.1016/j.envexpbot.2021.104750
Ferguson, A. (2014). “Kiwifruit in the world-2014,” in VIII International Symposium on Kiwifruit 1096), Belgium: International Society for Horticultural Science. 33–46.
Freisinger, E. (2011). Structural features specific to plant metallothioneins. J. Biol. Inorganic. Chem. 16, 1035–1045. doi: 10.1007/s00775-011-0801-z
Gao, C., Gao, K., Yang, H., Ju, T., Zhu, J., Tang, Z., et al. (2022). Genome-wide analysis of metallothionein gene family in maize to reveal its role in development and stress resistance to heavy metal. Biol. Res. 55, 1–13. doi: 10.1186/s40659-021-00368-w
Grennan, A. K. (2011). Metallothioneins, a diverse protein family. Plant Physiol. 155, 1750–1751. doi: 10.1104/pp.111.900407
Guo, W. J., Bundithya, W., Goldsbrough, P. B. (2003). Characterization of the Arabidopsis metallothionein gene family: tissue-specific expression and induction during senescence and in response to copper. New Phytol. 159, 369–381. doi: 10.1046/j.1469-8137.2003.00813.x
Hamer, D. H. (1986). Metallothionein. Annu. Rev. Biochem. 55, 913–951. doi: 10.1146/annurev.bi.55.070186.004405
Hassinen, V., Tervahauta, A., Schat, H., Kärenlampi, S. (2011). Plant metallothioneins–metal chelators with ROS scavenging activity? Plant Biol. 13, 225–232. doi: 10.1111/j.1438-8677.2010.00398.x
Hassinen, V., Tuomainen, M., Peräniemi, S., Schat, H., Kärenlampi, S., Tervahauta, A. (2009). Metallothioneins 2 and 3 contribute to the metal-adapted phenotype but are not directly linked to Zn accumulation in the metal hyperaccumulator, Thlaspi caerulescens. J. Exp. Bot. 60, 187–196. doi: 10.1093/jxb/ern287
Hsieh, H.-M., Liu, W. K., Chang, A., Huang, P. (1996). RNA expression patterns of a type 2 metallothionein-like gene from rice. Plant Mol. Biol. 32, 525–529. doi: 10.1007/BF00019104
Hsieh, H.-M., Liu, W. K., Huang, P. (1995). A novel stress-inducible metallothionein-like gene from rice. Plant Mol. Biol. 28, 381–389. doi: 10.1007/BF00020388
Huang, Y., Fang, Y., Long, X., Liu, L., Wang, J., Zhu, J., et al. (2018). Characterization of the rubber tree metallothionein family reveals a role in mitigating the effects of reactive oxygen species associated with physiological stress. Tree Physiol. 38, 911–924. doi: 10.1093/treephys/tpy003
Khalid, M., Nan, H., Kayani, S. I., Kexuan, T. (2020). Diversity and versatile functions of metallothioneins produced by plants: A review. Pedosphere 30, 577–588. doi: 10.1016/S1002-0160(20)60022-4
Kumar, G., Kushwaha, H. R., Panjabi-Sabharwal, V., Kumari, S., Joshi, R., Karan, R., et al. (2012). Clustered metallothionein genes are co-regulated in rice and ectopic expression of OsMT1e-P confers multiple abiotic stress tolerance in tobacco via ROS scavenging. BMC Plant Biol. 12, 1–16. doi: 10.1186/1471-2229-12-107
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Lamb, C., Dixon, R. A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Biol. 48, 251–275. doi: 10.1146/annurev.arplant.48.1.251
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Ledger, S. E., Gardner, R. C. (1994). Cloning and characterization of five cDNAs for genes differentially expressed during fruit development of kiwifruit (Actinidia deliciosa var. deliciosa). Plant Mol. Biol. 25, 877–886. doi: 10.1007/BF00028882
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Leszczyszyn, O. I., Imam, H. T., Blindauer, C. A. (2013). Diversity and distribution of plant metallothioneins: a review of structure, properties and functions. Metallomics 5, 1146–1169. doi: 10.1039/c3mt00072a
Li, W., Ding, Z., Ruan, M., Yu, X., Peng, M., Liu, Y. (2017). Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis. Sci. Rep. 7, 16861. doi: 10.1038/s41598-017-16905-1
Li, R., Yang, Y., Cao, H., Peng, X., Yu, Q., He, L., et al. (2023). Heterologous expression of the tobacco metallothionein gene NtMT2F confers enhanced tolerance to Cd stress in Escherichia coli and Arabidopsis thaliana. Plant Physiol. Biochem. 195, 247–255. doi: 10.1016/j.plaphy.2023.01.027
Liu, L., Huang, Y., Huang, X., Yang, J., Wu, W., Xu, Y., et al. (2017). Characterization of the dioscorin gene family in Dioscorea alata reveals a role in tuber development and environmental response. Int. J. Mol. Sci. 18, 1579. doi: 10.3390/ijms18071579
Liu, L., Yang, N., Dai, Y., Luo, C., Cheng, T., Huang, Y. (2020). An efficient method for isolating high-quality total RNA from different tissues of kiwifruit (Actinidia chinensis). Acta Agricult. Jiangxi. 32, 30–34. doi: 10.19386/j.cnki.jxnyxb.2020.09.06
Lv, Y., Deng, X., Quan, L., Xia, Y., Shen, Z. (2013). Metallothioneins BcMT1 and BcMT2 from Brassica campestris enhance tolerance to cadmium and copper and decrease production of reactive oxygen species in Arabidopsis thaliana. Plant Soil 367, 507–519. doi: 10.1007/s11104-012-1486-y
Ma, Y., Xue, M., Zhang, X., Chen, S. (2023). Genome-wide analysis of the metallothionein gene family in cassava reveals its role in response to physiological stress through the regulation of reactive oxygen species. BMC Plant Biol. 23, 227. doi: 10.1186/s12870-023-04174-2
Mazzaglia, A., Studholme, D. J., Taratufolo, M. C., Cai, R., Almeida, N. F., Goodman, T., et al. (2012). Pseudomonas syringae pv. actinidiae (Psa) isolates from recent bacterial canker of kiwifruit outbreaks belong to the same genetic lineage. PloS One 7, e36518. doi: 10.1371/journal.pone.0036518
Michelotti, V., Lamontanara, A., Buriani, G., Orrù, L., Cellini, A., Donati, I., et al. (2018). Comparative transcriptome analysis of the interaction between Actinidia chinensis var. chinensis and Pseudomonas syringae pv. actinidiae in absence and presence of acibenzolar-S-methyl. BMC Genomics 19, 1–22. doi: 10.1186/s12864-018-4967-4
Moyle, R., Fairbairn, D. J., Ripi, J., Crowe, M., Botella, J. R. (2005). Developing pineapple fruit has a small transcriptome dominated by metallothionein. J. Exp. Bot. 56, 101–112. doi: 10.1093/jxb/eri015
Reymond, P., Weber, H., Damond, M., Farmer, E. E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–719. doi: 10.1105/tpc.12.5.707
Sharma, P., Jha, A. B., Dubey, R. S., Pessarakli, M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 1, 217037. doi: 10.1155/2012/217037
Song, Y., Sun, L., Lin, M., Chen, J., Qi, X., Hu, C., et al. (2019). Comparative transcriptome analysis of resistant and susceptible kiwifruits in response to Pseudomonas syringae pv. actinidiae during early infection. PloS One 14, e0211913. doi: 10.1371/journal.pone.0211913
Trick, A. Y., Chen, F. E., Schares, J. A., Freml, B. E., Lor, P., Yun, Y., et al. (2021). High resolution estimates of relative gene abundance with quantitative ratiometric regression PCR (qRR-PCR). Analyst 146, 6463–6469. doi: 10.1039/D1AN01397A
Usha, B., Venkataraman, G., Parida, A. (2009). Heavy metal and abiotic stress inducible metallothionein isoforms from Prosopis juliflora (SW) DC show differences in binding to heavy metals in vitro. Mol. Genet. Genomics 281, 99–108. doi: 10.1007/s00438-008-0398-2
Vanneste, J. (2012). Pseudomonas syringae pv. actinidiae (Psa): a threat to the New Zealand and global kiwifruit industry. New Z. J. Crop Hortic. Sci. 40, 265–267. doi: 10.1080/01140671.2012.736084
Vanneste, J., Poliakoff, F., Audusseau, C., Cornish, D., Paillard, S., Rivoal, C., et al. (2011). First report of Pseudomonas syringae pv. actinidiae, the causal agent of bacterial canker of kiwifruit in France. Plant Dis. 95, 1311–1311. doi: 10.1094/PDIS-03-11-0195
Wang, Z., Liu, Y., Li, D., Li, L., Zhang, Q., Wang, S., et al. (2017). Identification of circular RNAs in kiwifruit and their species-specific response to bacterial canker pathogen invasion. Front. Plant Sci. 8, 413. doi: 10.3389/fpls.2017.00413
Wang, T., Wang, G., Jia, Z. H., Pan, D. L., Zhang, J. Y., Guo, Z. R. (2018). Transcriptome analysis of kiwifruit in response to Pseudomonas syringae pv. actinidiae infection. Int. J. Mol. Sci. 19, 373. doi: 10.3390/ijms19020373
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteomics Bioinf. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wong, H. L., Sakamoto, T., Kawasaki, T., Umemura, K., Shimamoto, K. (2004). Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol. 135, 1447–1456. doi: 10.1104/pp.103.036384
Yu, Q., He, L., Huo, C., Jiang, X., Chen, H., Wang, R., et al. (2021). Genome-wide identification and expression analysis of heavy metal stress–responsive metallothionein family genes in Nicotiana tabacum. Plant Mol. Biol. Rep. 39, 443–454. doi: 10.1007/s11105-020-01262-7
Yuan, J., Chen, D., Ren, Y., Zhang, X., Zhao, J. (2008). Characteristic and expression analysis of a metallothionein gene, OsMT2b, down-regulated by cytokinin suggests functions in root development and seed embryo germination of rice. Plant Physiol. 146, 1637–1650. doi: 10.1104/pp.107.110304
Yue, J., Liu, J., Tang, W., Wu, Y. Q., Tang, X., Li, W., et al. (2020). Kiwifruit Genome Database (KGD): a comprehensive resource for kiwifruit genomics. Horticult. Res. 7, 117. doi: 10.1038/s41438-020-0338-9
Zhang, A., Xiong, Y., Liu, F., Zhang, X. (2023). A genome-wide analysis of the Pentatricopeptide Repeat Protein gene family in two kiwifruit species with an emphasis on the role of RNA editing in pathogen stress. Int. J. Mol. Sci. 24, 13700. doi: 10.3390/ijms241813700
Zhou, J., Goldsbrough, P. B. (1994). Functional homologs of fungal metallothionein genes from Arabidopsis. Plant Cell 6, 875–884. doi: 10.1105/tpc.6.6.875
Zhou, J., Goldsbrough, P. B. (1995). Structure, organization and expression of the metallothionein gene family in Arabidopsis. Mol. Gen. Genet. 248, 318–328. doi: 10.1007/BF02191599
Zhou, Y., Liu, J., Liu, S., Jiang, L., Hu, L. (2019). Identification of the metallothionein gene family from cucumber and functional characterization of CsMT4 in Escherichia coli under salinity and osmotic stress. 3. Biotech. 9, 1–11. doi: 10.1007/s13205-019-1929-8
Zhou, G., Xu, Y., Li, J., Yang, L., Liu, J. Y. (2006). Molecular analyses of the metallothionein gene family in rice (Oryza sativa L.). BMB. Rep. 39, 595–606. doi: 10.5483/BMBRep.2006.39.5.595
Keywords: kiwifruit, regulation, AcMT genes, ROS, stress response
Citation: Liu L, Song S, Yang N, He B, Xin L and Huang Y (2024) Metallothionein family genes in kiwifruit: characterization and determining their roles in plant’s response to different stresses. Front. Plant Sci. 15:1425072. doi: 10.3389/fpls.2024.1425072
Received: 29 April 2024; Accepted: 30 September 2024;
Published: 18 October 2024.
Edited by:
Gianfranco Romanazzi, Marche Polytechnic University, ItalyReviewed by:
Xiujun Zhang, Chinese Academy of Sciences (CAS), ChinaJulian Mario Peña-Castro, Universidad del Papaloapan, Mexico
Copyright © 2024 Liu, Song, Yang, He, Xin and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lusheng Xin, eGlubHVzaGVuZ0AxMjYuY29t; Yacheng Huang, eWFjaGVuZ2h1YW5nMTMxNEAxMjYuY29t
†These authors have contributed equally to this work
 Linya Liu
Linya Liu Shuyi Song
Shuyi Song Na Yang
Na Yang Bin He
Bin He Lusheng Xin
Lusheng Xin Yacheng Huang
Yacheng Huang