- Department of Systematics, Biodiversity and Evolution of Plants (with Herbarium), University of Goettingen, Göttingen, Germany
Introduction: The eleven plastid ndh genes encode for subunits of the ndh (NAD(P)H dehydrogenase-like) complex, which mediates electron flow in photosystem I. The loss of ndh genes in plants was observed in many different lineages of Viridiplantae. In lineages of Orchidaceae, the loss of ndh genes was often associated with myco-heterotrophy. However, in previous studies on this topic only a few slipper orchids were included. Our study aimed to analyze the loss of ndh genes within Cypripedioideae, a subfamily that is assumed to be fully autotroph.
Methods: Based on a comprehensive sampling of 100 published plastomes representing 60% of Cypripedioideae species, the phylogenetic relationships were revealed on three levels. For family and subfamily levels, 57 and 66 plastid genes, respectively, were extracted and concatenated in Geneious, while for the genus-level phylogeny, complete plastomes were used to calculate a maximum likelihood tree. Additionally, divergence time estimates were performed to illuminate the evolutionary timeframe of the gene loss. The prevalence, pseudogenization and loss of ndh genes were assessed and visualized along the phylogenetic trees.
Results: The results confirmed the four analyzed genera of Cypripedioideae to be monophyletic and could increase the resolution within the genera compared to previous studies. The diversification of the subfamily started at about 30 Ma with genus Paphiopedilum displaying the most recent diversification starting at about 11 Ma and showing most speciation events around 4 Ma. The rapid loss of plastid ndh genes within the subfamily Cypripedioideae, particularly in the genera Mexipedium, Phragmipedium and Paphiopedilum could be illustrated. Furthermore, the results illustrated that Cypripedioideae are in an early stage of plastid degradation.
Discussion and conclusions: Recent studies showed that partial myco-heterotrophy (mixotrophy) is far more common in plant lineages than originally assumed. Based on our findings, we suggest that the possibility of a mixotrophic lifestyle within (sub-)tropical slipper orchids should be reevaluated. Further research regarding the reasons behind plastid gene loss in slipper orchids could provide a better understanding of the ecological evolution of Cypripedioideae.
1 Introduction
The plastome of land plants encodes for approximately 100 protein coding genes and 30-50 RNA genes, and is assumed to be highly conserved in gene content and order (Rivas et al., 2002). However, recent studies showed that plastomes might vary a lot regarding gene presence and gene order (Ruhlman and Jansen, 2021). Hu et al. (2022) indicated that the evolution of the plastid genome may be an adaptive response to abiotic stresses, i.e. temperature or water stress. The plastome contains highly conserved sequences and is utilized in a large number of phylogenetic studies, e.g., for further understanding of the molecular and adaptive evolution of Orchidaceae Juss (Guo et al., 2021). The eleven ndh genes encode for subunits of the NDH (NAD(P)H dehydrogenase-like) complex, which is important for the photosynthetic electron transport chain, and are present in all lineages of higher plants. Furthermore, ndh genes enable a better protection of the plant against environmental, i.e. oxidative, stress. Plastid ndh genes appear to have homologs in cyanobacteria, which is in accord with the endosymbiotic theory of the evolution of the chloroplast (Burrows, 1998). The NDH complex is one of the many adaptations of land plants to the challenges of populating terrestrial habitats, i.e. ultraviolet radiation, water limitation and drastic fluctuations in light intensity, but is now of low biological significance in extant plants (Ruhlman et al., 2015). Under favorable growth conditions, plants can grow without the NDH complex (Burrows, 1998; Ifuku et al., 2011). Furthermore, Martín et al. (2015) suggested that ndh genes could also be dispensable at low atmospheric CO2 concentrations. While the loss of plastid ndh genes was observed in various lineages, i.e. Geraniaceae Juss., Orchidaceae, Orobanchaceae Vent., Petrosaviaceae Hutch., Simmondsiaceae Tiegh. and Triuridaceae Gardner (Blazier et al., 2011; Lin et al., 2017; Kharabian-Masouleh et al., 2023), the majority of plants have all eleven plastid ndh genes. This may hint towards a selective advantage of a functioning NDH complex. The absence of ndh genes in many independent lineages shows the different possible reasons and functions, e.g., in Cactaceae ndh loss was observed and it was suggested to be related to CAM photosynthesis as an adaptation to dry habitats (Köhler et al., 2020). In other lineages, plants with missing or pseudogenized ndh genes tend to have a heterotrophic lifestyle in form of myco-heterotrophs or parasitism (Wicke et al., 2016; Lin et al., 2017). Such a correlation was also observed in Orchidaceae (Kim et al., 2015; Lin et al., 2017).
The family Orchidaceae comprises five subfamilies, namely Apostasioideae, Vanilloideae, Cypripedioideae, Orchidoideae and Epidendroideae. With over 25,000 species and approximately 800 genera, Orchidaceae are the second largest family of flowering plants and only exceeded by Asteraceae Bercht. & J.Presl (Cozzolino and Widmer, 2005; Chase et al., 2015). Since new orchid genera are being described each year, the phylogeny of orchids is far from being fully resolved (Chase et al., 2015; Pérez-Escobar et al., 2024). Orchids are not only distributed on five different continents, but their habitats and lifeforms vary greatly: there are orchid species which are terrestrial, lithophytic or, predominantly found in tropical areas, epiphytic; furthermore, myco-heterotrophic lifestyles can be found in nearly every subfamily (Kim et al., 2020). The loss of ndh genes is usually associated with a myco-heterotrophic lifestyle in Orchidaceae. Since many lineages of orchids contain pseudogenized or lost ndh genes, the individual lineages experienced independent gene losses (Wicke et al., 2016; Kim et al., 2020). Additionally, in orchid species that lacked all ndh genes, the chlororespiration function was lost (Kim et al., 2015). So far, the prevalence of ndh genes has been primarily investigated in Orchidoideae and Epidendroideae, which are the two largest subfamilies of Orchidaceae, while the other subfamilies like Cypripedioideae were not subject to such comprehensive studies or were represented by a very small and restricted sampling (Kim et al., 2020).
The slipper orchids (Cypripedioideae) contain five monophyletic genera (Cypripedium L., Mexipedium V.A.Albert & M.W.Chase, Paphiopedilum Pfitzer, Phragmipedium Rolfe and Selenipedium Rchb.f.) and 169 species, of which 136 occur in the Old World and 33 in the New World (Chase et al., 2015; Pérez-Escobar et al., 2024). The name slipper orchid is derived from one of their distinct morphological synapomorphies, the pouch-like labellum, which resembles a slipper (Figure 1). Further morphological synapomorphies regarding the flower are a shield-like staminode, two fertile stamens, and united lateral sepals or a synsepa (Cox et al., 1997). The temperate species of slipper orchids are terrestrial, while most (sub-)tropical species grow as epiphytes or lithophytes. Cypripedioideae occupy diverse habitats and are successfully adapted to different environments. Based on a recent study using target sequencing data, Cypripedioideae diversified about 30 Ma and where once wide spread in the boreotropics, while they show today an ancient relictual range (Pérez-Escobar et al., 2024). The genus Paphiopedilum is regarded as the most recent diversifying genus within slipper orchids (5-7 Ma; Tsai et al., 2020). The ancestor of the slipper orchids likely had a continuous distribution in the boreotropics followed by migration southwards to both sides of the Pacific Ocean due to the climatic cooling in the late Cenozoic (Guo et al., 2012; Pérez-Escobar et al., 2024). The current distribution of Cypripedium differs highly from the other genera of slipper orchids (Figure 1). Cypripedium is distributed in temperate and subtropical habitats of the northern hemisphere, with a main distribution in alpine areas, where it is exposed to high light intensity and rarely suffers from water stress (Brieger and Schlechter, 1992; Guo et al., 2012; Hu et al., 2022). Phragmipedium and the monotypical genus Mexipedium can be found in tropical Central and South American habitats (Guo et al., 2012).
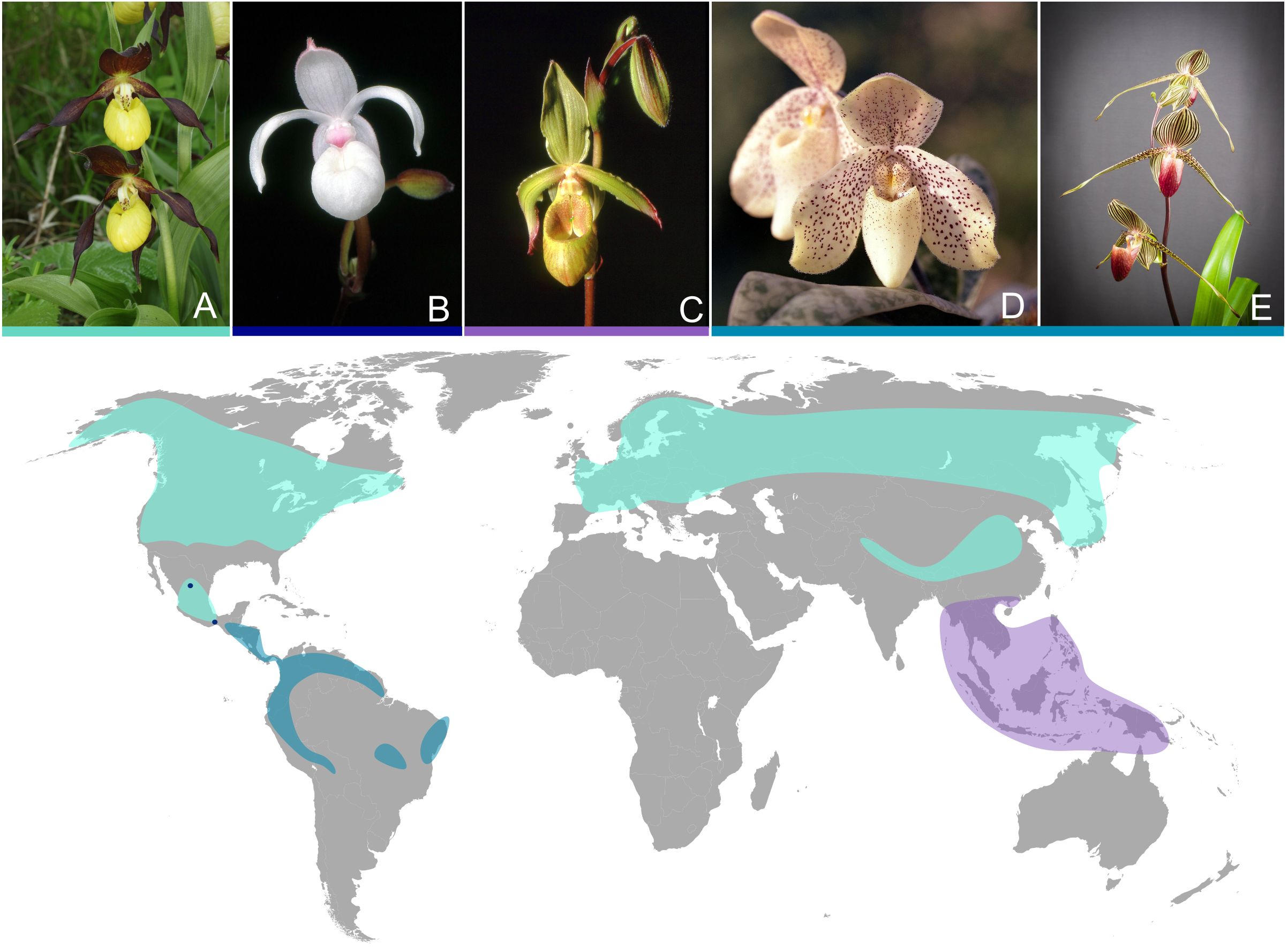
Figure 1. Five species of the subfamily Cypripedioideae. (A) Cypripedium calceolus, (B) Mexipedium xerophyticum, (C) Phragmipedium lindleyanum, (D) Paphiopedilum concolor and (E) Paphiopedilum rothschildianum. The map shows the distribution ranges of genera Cypripedium (light blue), Mexipedium (dark blue dots), Phragmipedium (petrol) and Paphiopedilum (purple). For the distribution ranges of the genera of Cypripedioideae we utilized the GBIF database. Orchid pictures (A-E) were taken from Wikimedia and fall under figure licenses (Krisp, 2013; Orchi, 2005; Orchi, 2006a; Orchi, 2006b; Takebayashi, 2018).
Paphiopedilum is distributed in tropical areas of South East Asia from India eastward to the Philippean and Suda Isles and southward to New Guinea and the Solomon Isles (Brieger and Schlechter, 1992). Paphiopedilum species grow terrestrial, as lithophytes or as epiphytes and were because of its ornamental value subject to several recent studies (Tsai et al., 2020; Hu et al., 2022; Liu et al., 2022). The strongly differing habitats of slipper orchids and a distribution around multiple contintents makes them an ideal group to study the adaptive evolution of plastomes, specifically the loss of ndh genes.
The loss of ndh genes within Orchidaceae has been the topic of numerous scientific studies, but so far Cypripedioideae were only sparsely sampled (Kim et al., 2020). This study aims to gain further insight into the loss of plastid ndh genes within the subfamily, which will help to understand general mechanisms of plastid gene loss, to develop ideas on compensatory strategies to survive without an NDH complex and to increase our knowledge on evolution and adaptation of slipper orchids to different habitats. We want to analyze whether the gene loss happened gradually or if one or multiple sudden gene losses occurred during the evolutionary history of the subfamily. Futhermore, if the loss of ndh genes happened multiple times independently and lastly, we want to assess the gene loss in a temporal time frame.
2 Material and methods
2.1 Sampling
The 163 plastomes used in this study were derived from NCBI Genbank. In detail, three plastomes of Vanilloideae, 101 of Cypripedioideae, 15 of Orchidoideae and 43 of Epidendroideae. The plastomes were downloaded in the GenBank (full) format. Additionally, one sample of Allium cepa and one sample of Iris sanguinea were included as outgroups. Only one sample per species was selected, since an interspecific variation of ndh genes seemed very unlikely. Additionally, the large majority of Cypripedioideae species in the NCBI database were represented by only one sample and using consistently one sample per species avoided sampling bias. For Vanilloideae, at least one sample for each of the two tribes was selected. The samples represented approximately 60% of Cypripedioideae species. Three of the four Orchidaceae tribes and twelve of 16 Epidendroideae tribes were represented. A full list of samples and their reference names can be found in Supplementary Material (Supplementary Table S1).
2.2 Gene extraction
Since the plastome showed high variation between the different species of Orchidaceae and Cypripedioideae regarding plastome length as well as order, length and prevalence of plastid genes, a subset of genes was extracted for the phylogenetic analysis. The respective genes were chosen because of their presence in at least 90% of samples of either Orchidaceae or Cypripedioideae. This way we received well aligned genes and avoided too many gaps. The resulting subset for Orchidaceae consisted of 57 plastome genes (Supplementary Table S2.1) from 85 samples (three Vanilloideae, 22 Cypripedioideae, 16 Orchidoideae and 42 Epidendroideae plus two outgroup samples). For the phylogenetic analysis of subfamily Cypripedioideae, 66 genes were extracted from the plastome samples (Supplementary Table S2.2). These were in detail three Vanilloideae as outgroup taxa and 101 Cypripedioideae samples representing eleven Cypripedium, one Mexipedium, six Phragmipedium and 83 Paphiopedilum species. Individual plastid genes were extracted from the full plastomes with Geneious 2023.2 (Kearse et al., 2012). The genes were aligned separately with MAFFT v7 (Katoh and Standley, 2013), implemented in Geneious 2023.2, using the FFT-NS-2 algorithm. For a better alignment process, poorly aligned sections were removed manually within Geneious 2023.2. Then the genes were aligned a second time with MAFFT v7, this time using the L-INS-i algorithm. Afterwards, the gene alignments were concatenated to one supermatrix with Geneious 2023.2.
On family and subfamily level the plastome show many structural differences and huge inversions or rearrangements, therefore the alignment of full plastomes was impossible. However, on genus level within subfamily Cypripedioideae the plastome structure was conserved and thus it was possible to use complete plastomes for genus Paphiopedilum to increase the phylogenetic signal for this closely related group. The 83 samples were completed with three Phragmipedium samples to root the tree, that is Ph. longifolium Rolfe, Ph. lindenii Dressler & N.H.Williams and Ph. kovachii J.T.Atwood, Dalström & Ric.Fernández. All 86 plastomes were aligned with MAFFT v7. The calculation was done on the High Performance Cluster (HPC) of the GWDG (Gesellschaft fuer wissenschaftliche Datenverarbeitung Göttingen), Göttingen.
2.3 Phylogenetic analysis and ultrametric tree calculation
To analyze the relationships of species and the evolutionary history of the ndh genes, we performed maximum likelihood analyses for all plastid gene alignments (Orchidaceae, Cypripedioideae) and the full plastome alignment of Paphiopedilum. The maximum likelihood (ML) trees were calculated with RAxML 8.2.12 (Stamatakis, 2014) with the maximum likelihood model GRT-GAMMA. 100 rapid bootstrap replicates were calculated, followed by a ML search. The resulting trees were visualized with FigTree 1.4.4 (Rambaut, 2014).
The dated phylogeny was based on the dataset for Cypripedioideae. We roughly followed the settings of Pérez-Escobar et al. (2021). BEAUTI v.2.6 (Bouckaert et al., 2014) was used to set a GTR nucleotide substitution model with Γ distribution rate with four categories. We used an uncorrelated log-normal strict molecular clock in combination with a prior clock rate interval of 0.0001–0.001 substitutions/site/Ma, modelled by a uniform distribution. A birth–death tree process, modelled by a uniform distribution for the birth and relative death rates was used. For the secondary calibration we set the most recent common ancestor (MRCA) prior to 30Ma for the crown of Cypripedioideae following previously published dated phylogenies on Orchidaceae (Zhang et al., 2023; Pérez-Escobar et al., 2024), modelled by normal distributions (σ = 1). We run 10 million generations, sampling every 1 000 generation. Finally, we used TREEANNOTATOR v.2.6 (https://www.beast2.org/treeannotator/) with a burn-in threshold of 10% to compute a maximum credibility consensus tree from the Markov chain Monte Carlo (MCMC) posterior trees.
2.4 ndh gene assessment
The plastomes downloaded from Genbank were fully annotated. The prevalence of ndh genes was assessed with Geneious 2023.2 based on the existing gene annotations from Genbank entries. Since the length of genes varied a lot within Orchidaceae, we defined an internal reference gene length for the ndh genes based on the Cypripedioideae samples. The gene annotations were used to determine what gene length should be set as reference gene length for comparison. A gene length was set as reference gene length if the exact number of bp was present in at least 50% of Cypripedioideae samples, which was the case for ndhB, ndhC, ndhE, ndhG, ndhH, ndhI and ndhJ. For the ndh genes with strongly varying lengths, namely ndhA, ndhD, ndhF and ndhK, the following procedure was used to determine the standard gene length: The average gene length was calculated two times. First, the average gene length was calculated based on all Cypripedioideae samples that had the gene annotated. The second calculation excluded samples that differed in their gene length by at least 30% from the first calculated average gene length. Finally, the second calculated average gene lengths were selected as reference gene length. Thus, the reference gene lengths were determined as following: 2,263 bp for ndhA, 2,242 bp for ndhB, 363 bp for ndhC, 1,513 bp for ndhD, 306 bp for ndhE, 2,118 bp for ndhF, 531 bp for ndhG, 1,182 bp for ndhH, 495 bp for ndhI, 477 bp for ndhJ and 764 bp for ndhK. Afterwards, the actual presence of ndh genes in Cypripedioideae was determined manually by comparing the samples with up to three Cypripedium samples, namely Cypripedium lichiangense S.C.Chen & P.J.Cribb, Cypripedium formosanum Hayata and Cypripedium calceolus L. This was done to discover annotation errors in the Cypripedioideae samples. For the Orchidaceae and outgroup samples, the prevalence of ndh genes was based on the existing annotations of the samples and a correct annotation was assumed The ndh genes with a gene length below 80% or above 120% reference bp length were defined as pseudogenized. This included indels, sequence duplications and changes in reading frames. For Cypripedioideae, it was determined in more detail, if the pseudogenization resulted from truncation, i.e., that stop codons occurred within the annotated sequence, caused by mutations or frame shifts, resulting in (very) short functional reading frames. Genes with a length below 20% of the reference gene length or not found at all were defined as lost. The results of the gene assessment were visualized alongside the phylogenetic trees.
3 Results
3.1 Alignment statistics
The length of the alignment for the Orchidaceae family tree based on 85 samples and 57 plastid genes that passed our thresholds was 60,203 bp long including 24,995 SNPs (41.5%) and 13.2% missing data (incl. outgroup). For the Cypripedioideae the alignment of 66 genes resulted in 74,028 bp length, containing 12,067 SNPs (16.3%) and 11.24% missing data. Finally, the full plastome alignment for genus Paphiopedilum showed a length of 265,241 bp (150,136-163,948 bp), 35,289 SNPs (13.3%) and 40.34% missing data (incl. outgroups).
3.2 Phylogenetic trees and dated phylogeny
The generated maximum likelihood tree represented four of the five subfamilies of Orchidaceae, i.e., Vanilloideae, Cypripedioideae, Orchidoideae and Epidendroideae (Figure 2, Supplementary Figure S2 with full branch length). Three of the four subfamilies were highly supported as monophyletic in the ML tree, with a bootstrap support (BS) of 99-100%. Vanilloideae were the sister clade to the remaining three subfamilies. Cypripedioideae were shown to be in sister relationship with the clade containing Orchidoideae and Epidendroideae, however on an unsupported short branch. Within Cypripedioideae all four genera appeared monophyletic (BS=100%). The branches of the Gastroideae tribe (Didymoplexis pallens Griff. and Gastrodia elata Blume) were much longer than of all other Orchidaceae samples and therefore shortened in Figure 2 (see Supplementary Figure S2 with full branch length). The clades within Epidendroideae were poorly to well supported and the backbone of this subfamily showed very short branch lengths (Figure 2). Within Orchidoideae, clades were well supported (BS=100%). The majority of included tribes appeared to be monophyletic, except for Collabieae (Calanthe sylvatica (Thouars) Lindl., Chrysoglossum ornatum Blume, Phaius takeoi (Hayata) H.J.Su. and Tainia cordifolia Hook.f.), which was revealed as being polyphyletic.
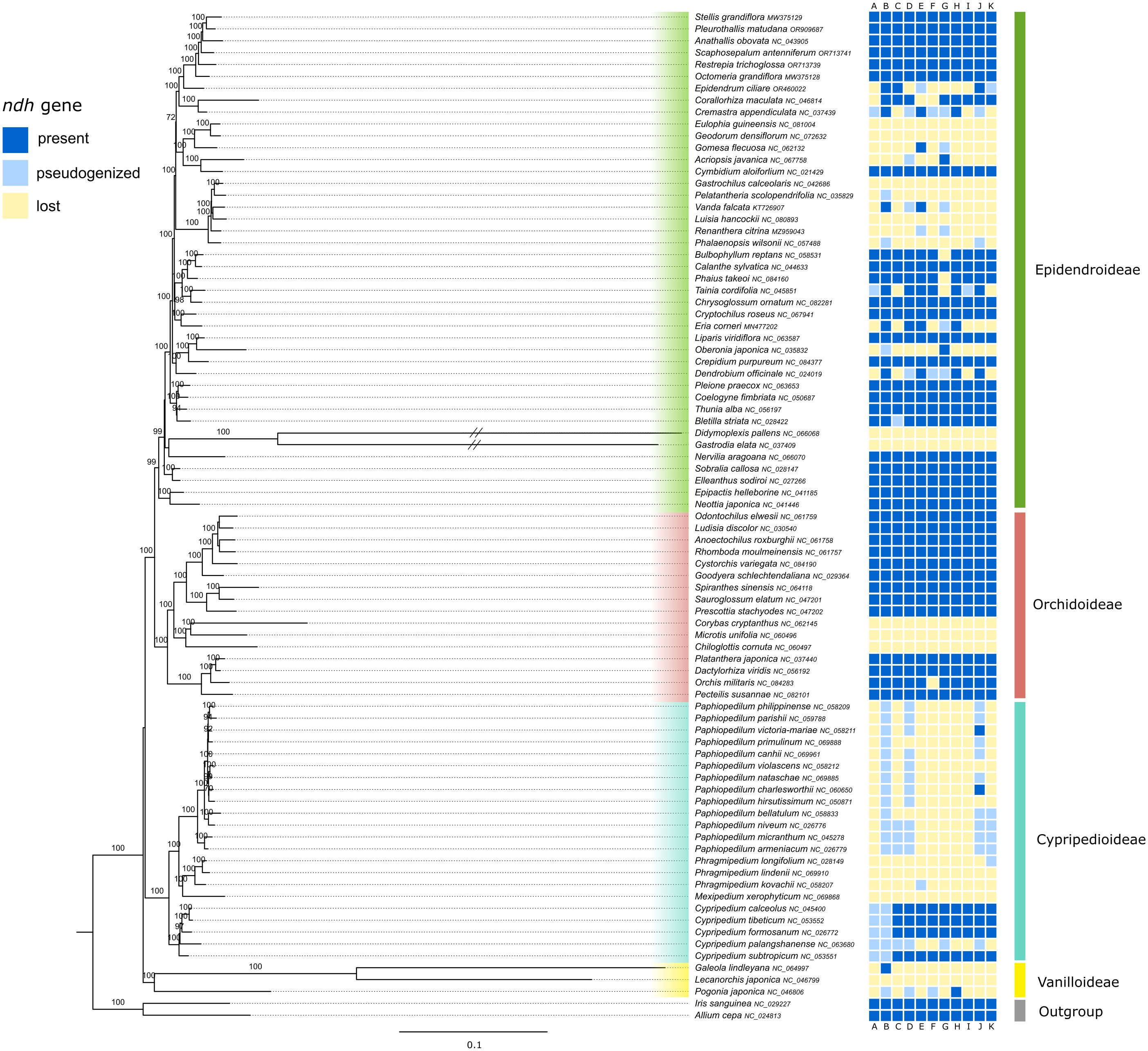
Figure 2. Phylogenetic relationships inferred from a RAxML analysis based on 57 plastid genes and 85 taxa of Orchidaceae. The tree was rooted with Allium and Iris used as outgroup. The four subfamilies were color-coded. The prevalence/loss of ndh genes is shown in the colored boxes next to taxon names following the legend in the upper left corner. Bootstrap values from 100 replicates are shown above branches. The branch length is based on the number of substitutions per site and then normalized (see RAxML manual).
The generated ML tree of Cypripedioideae represents four of the five genera of the subfamily (Cypripedium, Mexipedium, Phragmipedium and Paphiopedilum, Figure 3, Supplementary Figure S3). The three sampled Vanilloideae species, Pogonia japonica Rchb.f., Galeola lindleyana Rchb.f. and Lecanorchis japonica Blume, were used as outgroup. The long branches were shortened in Figure 3, see Supplementary Figure S3 for full branch length. The monophyly of the four slipper orchid genera was supported by very high bootstrap support values (BS=100%). Cypripedium was sister to the remaining genera, followed by a clade containing Mexipedium and Phragmipedium (BS=100%), to which Paphiopedilum was in sister position. Bootstrap values within Cypripedium were overall very high (BS=100%), with two exceptions of slightly lower bootstrap support (BS=89%). Cypripedium debile Rchb.f. was sister to these two clades. The genus Paphiopedilum was monophyletic, however, branch lengths within this genus in the ML tree of Cypripedioideae were very short (Figure 3). Therefore, we performed a phylogenetic tree based on full plastomes for this genus.
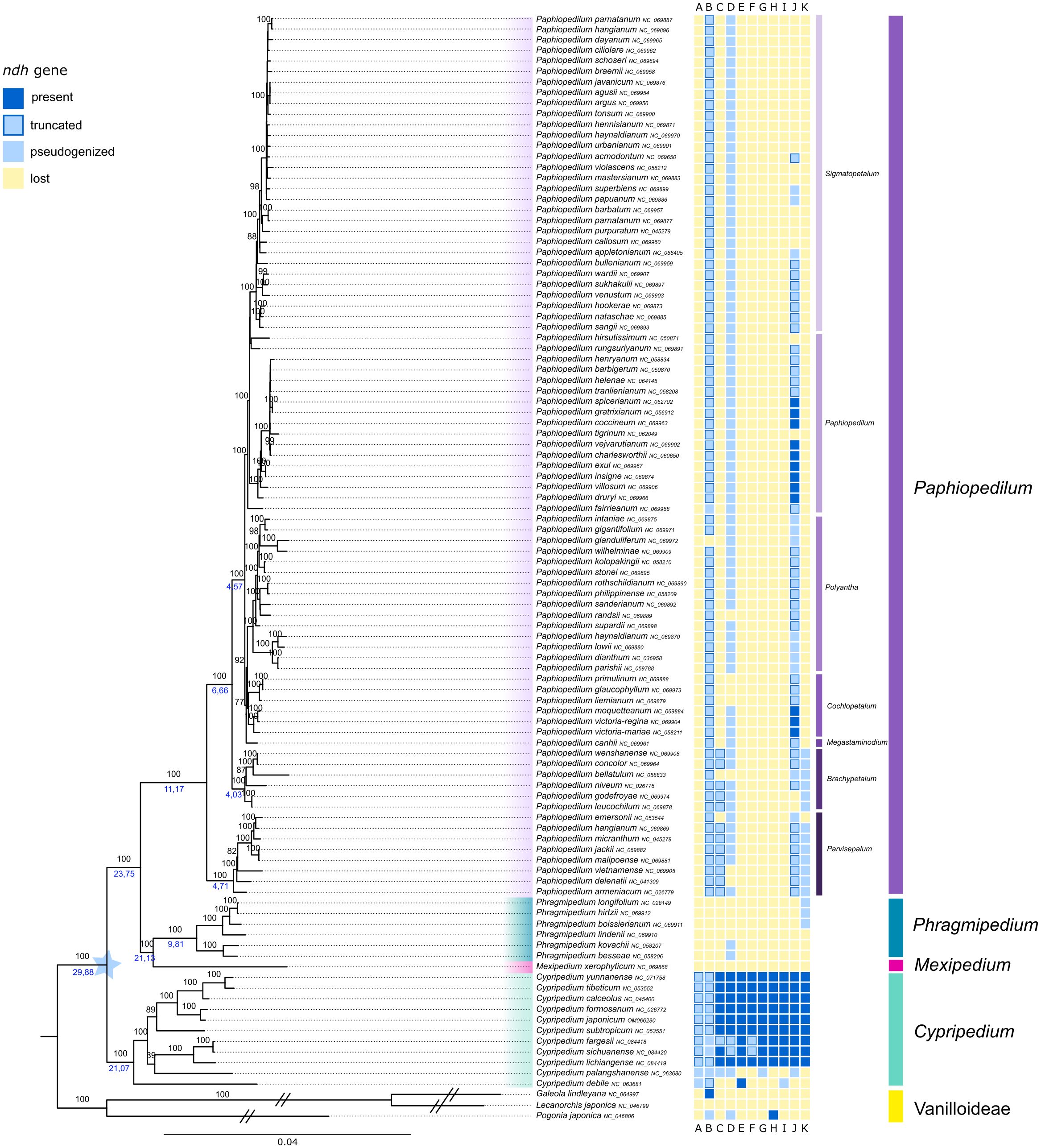
Figure 3. Phylogenetic relationships inferred from a RAxML analysis based on 66 plastid genes and 101 taxa of Cypripedioideae. The tree was rooted with three samples of Vanillioideae used as outgroup (branches shortened for illustration). The four genera were color-coded. The prevalence/loss of ndh genes is shown in the colored boxes next to taxon names following the legend in the upper left corner. Bootstrap values are shown above branches, node ages below selected branches (taken from BEAST analysis, Supplementary Figure S1). The branch length is based on the number of substitutions.
The Paphiopedilum phylogeny based on full plastomes contained 83 species plus three outgroup taxa and showed well resolved clades (Figure 4). The seven subgenera of Paphiopedilum all appeared as monophyletic with a very high bootstrap support (BS=100%), the exceptions being subg.
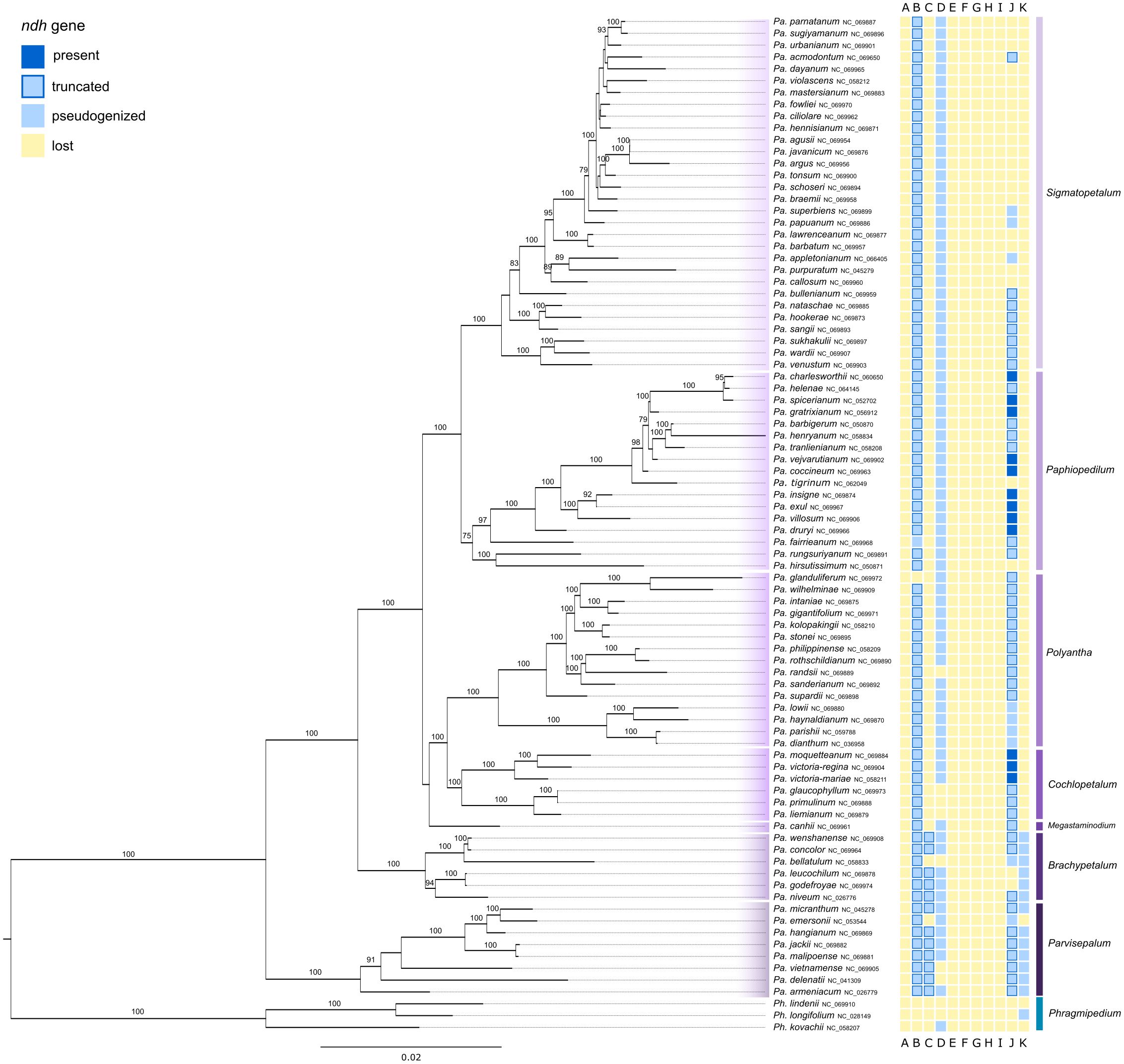
Figure 4. Phylogenetic relationships revealed from a RAxML analysis based on full plastomes of 85 taxa of Paphiopedilum. The tree was rooted with three samples of Phragmipedium used as outgroup. The subgenera were color-coded. The prevalence/loss of ndh genes is shown in the colored boxes next to taxon names following the legend in the upper left corner. Bootstrap values are shown above branches. The branch length is based on the number of substitutions.
Paphiopedilum with a slightly lower bootstrap support (BS=75%) and subg. Megastaminodium with low bootstrap support. Within subg. Parvisepalum, bootstrap values were very high (BS=91-100%) with only one clade being of lower support. Similar results can be observed in subg. Brachypetalum, where only one clade was of slightly lower but still good bootstrap support (BS=94%). Subg. Megastaminodium, subg. Cochlopetalum and subg. Polyantha formed one clade and each of the three subgenera was monophyletic with a varying bootstrap support. Within subg. Cochlopetalum all clades were highly supported (BS=100%). Subg. Polyantha was of very high bootstrap support (BS=100%). The clade containing subg. Paphiopedilum and subg. Sigmatopetalum had a very high bootstrap support as well (BS=100%). Subg. Paphiopedilum was well supported (BS=75-100%). Subg. Sigmatopetalum showed variable bootstrap values, ranging between low support and 100% BS.
The full results of the dated phylogeny of the Cypripedioideae using BEAST are shown in Supplementary Figure S1 and the node ages for the main clade were also illustrated in Figure 3. The subfamily started to diverge about 30 Ma. The split of Mexipedium, Phragmipedium and Paphiopedilum happened about 24 Ma. The genus Phragmipedium showed a crown age of about 10 Ma, while genus Paphiopedilum diversified 11 Ma. Within the latter genus, the clade containing the five subgenera Megastaminodium, Cochlopetalum, Polyantha, Paphiopedilum and Sigmatopetalum started to diverge at about 4.5 Ma.
3.3 ndh gene assessment
Within the family Orchidaceae, the loss of ndh genes can be observed in all four represented subfamilies (Figure 2). While the two outgroup samples contain all eleven plastid ndh genes, the loss of ndh genes was observed in all three samples of Vanilloideae. Within Orchidoideae, the three samples of Diurideae have lost all eleven plastid ndh genes, while many of the other sampled species still contain all genes. Within the subfamily Epidendroideae, only the tribes Neottieae, Sobralieae and Nervilieae showed the presence of all ndh genes. However, several species lost part or all ndh genes, especially in tribes Cymbidieae, Gastroideae and Vandeae. Overall, there was a strong variation in the degradation of plastid ndh genes, as can be seen in Figure 2.
Within the slipper orchids, the majority of ndh genes were lost at least twice. Cypripedium was the only genus that retained most of the eleven ndh genes in the majority of sampled species. Both Cypripedium debile Rchb.f. and Cypripedium palangshanense Tang & F.T.Wang lost numerous ndh genes, with the remaining ones, except for ndhE in C. debile, being pseudogenized. The genes ndhA and ndhB were pseudogenized in every Cypripedium sample.
Within the clade containing Mexipedium and Phragmipedium, all ndh genes were lost in the majority of samples. The gene ndhD was found pseudogenized in two Phragmipedium samples and lost in the remaining samples. Three other samples of Phragmipedium contained functional fractions of the ndhK gene (Figure 3). All Paphiopedilum samples have lost the ndhA, ndhE, ndhF, ndhG, ndhH and ndhI genes. The gene ndhB was truncated in the majority of samples within this genus. NdhC was truncated in subg. Parvisepalum and subg. Brachypetalum, and lost in the other subgenera. The gene ndhD was pseudogenized in the majority of Paphiopedilum samples. Seven samples did not have a functional ndhD. NdhJ was functional in twelve samples of this genus (within subg. Paphiopedilum and subg. Cochlopetalum), truncated in 39 samples and pseudogenized or lost in the remaining samples (Figure 3). Only the samples within subg. Parvisepalum and subg. Brachypetalum contained a pseudogenized ndhK, the remaining samples have lost this gene. The better resolved phylogeny based on full plastomes revealed the same pattern (Figure 4).
4 Discussion
In our study we aimed to analyze the ndh gene loss in members of the subfamily Cypripedioideae and put it in an evolutionary frame work. One sample per species was selected for the calculations of the phylogenetic trees. Previous studies showed slight variation in the retention of ndh genes between different samples of one species within Orchidaceae, but not within Cypripedioideae (Kim et al., 2020; Hu et al., 2022).
4.1 Phylogenetic relationships and ndh gene loss in Orchidaceae
The phylogenetic analysis of Orchidaceae was based on 91 samples representing four of the five subfamilies and was based on 64 plastid genes (Supplementary Table S2). The phylogenetic relationships of the subfamilies and tribes of Orchidaceae were congruent with the consensus phylogeny of Orchidaceae presented in Chase et al. (2015) and a recent phylogeny based on more than 600 nuclear genes (Zhang et al., 2023). The long branches of Didymoplexis pallens and Gastrodia elata, both Epidendroideae tribe Gastrodieae, resulted from the drastically reduced plastome size (35.304 bp & 51.241 bp) and thus missing data, probably due to their myco-heterotrophic lifestyle (Leake, 1994).
The presence of ndh genes within Orchidaceae was similar to Kim et al. (2020), except that their study included more samples of myco-heterotrophic orchids and thus showed more variation in ndh gene presence within Cranichideae and Neottieae. In contrast to Kim et al. (2020), our study conducted a more representative tree of Orchidaceae by including samples of multiple genera belonging to a tribe (Figure 2), while Kim et al. (2020) represented multiple tribes by only one genus, namely Diurideae, Nervillieae, Gastrodieae and Collabieae. Given the diversity of species within a genus, the inclusion of several samples is necessary to draw a more complete picture of ndh gene loss in Orchidaceae. Lin et al. (2017) suggested that the loss of ndh genes is related or possibly even necessary for the heterotrophic phase orchids have to go through at their early life stages. This does seem rather unlikely, as the ndh genes have been shown to be present in many independent orchid lineages (Figure 2; Kim et al., 2020).
4.2 Phylogenetic relationships and ndh gene loss in slipper orchids
The phylogenetic tree of Cypripedioideae was based on 66 plastid genes and 101 samples of slipper orchids, representing four out of the five genera (Figure 3). The phylogenetic relationships have strong similarities with previous phylogenetic studies of Cypripedioideae (Cox et al., 1997; Tsai et al., 2020; Guo et al., 2021; Szlachetko et al., 2021; Hu et al., 2022). The results of the BEAST analysis used a crown age of 30 Ma for Cypripedioideae (Zhang et al., 2023; Pérez-Escobar et al., 2024). The split of the clade containing Phragmipedium and Mexipedium, and Paphiopedilum happened 24 Ma. The latter genus started diversifying 11 Ma, which is younger than with former studies based on plastid data (Guo et al., 2012), however, similar to more recent studies on Orchidaceae evolution (Pérez-Escobar et al., 2024). We are aware that the similar diversification times might be the result of the calibration and clock model we applied following Pérez-Escobar et al. (2024). Like in the Paphiopedilum phylogenies based on plastid protein coding genes (Tsai et al., 2020; Hu et al., 2022), many clades of subg. Sigmatopetalum were of low support. The branches of the genus Paphiopedilum were relatively short, which suggests a recent radiation of this genus (Tsai et al., 2020), which is in accordance with the recent divergence of this subgenus in our study (c. 3 Ma). It may even be a hint that the Paphiopedilum genus is in its preliminary stage of speciation (Guo et al., 2021). Over the decades, the phylogeny of Paphiopedilum has repeatedly been proven to be difficult to resolve (Cox et al., 1997). Additionally, phylogenetic studies based on plastids do not provide enough genetic variation to study closely related species (Provan et al., 2001). Furthermore, reticulate evolution and clonal propagation, which are not uncommon within this rather young genus, can have yet unknown effects on the plastome genome evolution (Guo et al., 2021). However, a recent study on Paphiopedilum based on 41 full plastomes resolved the relationships of this genus (Liu et al., 2022). We followed their example and to increase resolution within the genus, we complemented our study with a genus-level tree for Paphiopedilum based on full plastomes (Figure 4). The phylogenetic relationships of the Paphiopedilum subgenera were in accord with previous phylogenetic studies of this genus (Cox et al., 1997; Guo et al., 2012; Tsai et al., 2020; Hu et al., 2022; Liu et al., 2022). However, here we included a comprehensive sampling of 83 species and the resolution and bootstrap support of Paphiopedilum subgenera were much higher than in previous studies with similar sample sizes regarding the phylogeny of Paphiopedilum or Cypripedioideae (Cox et al., 1997; Hu et al., 2022; Liu et al., 2022).
Our results regarding the loss of ndh genes within Cypripedioideae (Figure 3) showed that most ndh genes were found functional in Cypripedium, but were either lost or pseudogenized in the genera Mexipedium, Phragmipedium and Paphiopedilum, which confirms previous findings (Kim et al., 2020; Guo et al., 2021; Hu et al., 2022). The loss of ndh genes was not gradually as expected, but presumably happened rapidly and at least twice. First, in the early-branching lineages of Cypripedium, namely in C. palangshanense and C. debile, and second, in the common ancestor of Mexipedium, Phragmipedium and Paphiopedilum about 24 Ma. The pattern of gene loss in Paphiopedilum in comparison to Phragmipedium and Mexipedium differs in the presence of the genes ndh B, C and J (Figure 3), indicating that more than two events of ndh losses occurred more recently during the evolution of slipper orchids, i.e., 21 and 11 Ma, respectively. In a recent study on the evolution of epiphytism in Orchidaceae, Zhang et al. (2023) stated a shift of terrestrial to litophytic and epiphytic lifestyle within Cypripedioideae. While Cypripedium is exclusively terrestrial, Mexipedium is regarded as lithophytic and the genera Phragmipedium and Paphiopedilum contain also epiphytic species. The genera Phragmipedium and Mexipedium occur in the New World and diversified 21 Ma, however, the crown age of Phragmipedium is much younger (9.8Ma). The loss of all ndh genes might be the result of the shift to the subtropical environment. The transition in lifestyle could also be an explanation for the rapid loss of ndh genes within the subfamily. In this case we would assume to find a pattern of lifestyle and ndh gene loss in our study, i.e., that epiphytic species lost more genes than non-epiphytic ones. However, within Paphiopedilum and Phragmipedium, we do not see such a lifestyle related pattern of ndh loss. Therefore, the lifestyle is no exclusive explanation for the rapid loss of many ndh genes in both genera. Liu et al. (2022) stated that the ndh gene variations were found in IR (inverted repeat) boundaries in Paphiopedilum species and thus were considered to be attributed to the recombination of IRs. Next, they speculated that the degradation of ndh genes in Paphiopedilum was the result of adaptive evolution to a low light environment. Species of Paphiopedilum living beneath dense canopies with insufficient light and exhibited the characteristics of shade plants (Liu et al., 2022).
A large majority of Cypripedioideae retained an intact copy of ndhB, ndhD and ndhJ. This observation hints toward a possible higher importance of these specific genes and should be material for further study, which could bring new insights regarding the functions of these genes. Horváth et al. (2000) showed that tobacco plants with a defective ndhB gene were inhibited in their photosynthesis when growing under low air humidity, leading to a possible explanation for why the selection toward an intact ndhB gene was observed. However, Paphiopedilum is only distributed in tropical habitats, in which low air humidity is not a defining characteristic. Still, environmental factors and the adaptation to them are probably an important driving force in the evolution of the plastome within slipper orchids (Hu et al., 2022). This theory is further supported by the major difference in ndh degradation between Cypripedium and the other three genera analyzed in this study (Figure 3). The habitats and distribution ranges between these two clades are distinctly different from each other (Figure 1). The loss of ndh genes does infer with a plants` adaptability to changing environmental factors like sudden changes in light intensity or strong fluctuations of temperature (Yamori et al., 2011; Martín et al., 2015). The habitats of tropical species of slipper orchids provide much less fluctuations in temperature and light intensity, whereas the temperate montane habitats of Cypripedium usually provide stronger fluctuations of environmental factors. This may be a reason why the ndh genes have been retained in the majority of the analyzed Cypripedium samples. Additionally, a correlation between the loss of ndh genes and a natural distribution in shady habitats has also been observed in other genera, i.e. Allium L (Scobeyeva et al., 2021). Thus, adaptations to different light and temperature conditions might play a role in the degradation or retention of ndh genes.
Kim et al. (2015) theorized that the ndh genes may have been transferred to the nuclear genome. However, it is known that orchids that lost their plastid ndh genes have lost their nuclear ndh genes in most cases as well (Lin et al., 2017). Although a study conducted by Lin et al. (2015) found that fragments of chloroplast genes have been transferred to the mitochondrial genome, these genes do not get translated into functional proteins. However, no samples of Cypripedioideae were used in the study conducted by Lin et al. (2017), and the orchid species represented in their study were not fully autotroph. In a study on Paphiopedilum, the authors found fragments of ndh genes transferred to the nucleus and assumed that gene transfer in Paphiopedilum might be explained by the decreased demands on photosynthesis and plastid translational capacities due to the adaptation to the unique habitat of the species – without giving any further explanation (Liu et al., 2022). Further studies are needed to analyze the possible transfer of plastid ndh genes into the nuclear genome within the autotrophic Cypripedioideae, but so far it is assumed the NDH complex loses its function without functioning plastid ndh genes. Our study also showed that the majority of ndh genes were either lost or pseudogenized in the genus Paphiopedilum, which makes it unlikely that the species of this genus have a functioning NDH complex. Physiological characteristics suggested a non-functioning NDH complex; when compared to Cypripedium leaves, Paphiopedilum leaves had a significantly lower photosynthetic capacity (Guan et al., 2011; Yang et al., 2018). These results may correlate with the loss of ndh genes, as has been shown in Lin et al. (2017). Thus, the lower photosynthetic capacity in Paphiopedilum can be seen as further proof of the loss of a functioning NDH complex. Since the NDH complex is not only important for photosynthesis but also in the protection of the plant against environmental stress, it is of interest to observe whether Paphiopedilum shows inferred abilities regarding these functions as well, or if there are any yet unknown factors compensating for it. According to Wicke et al. (2016), the loss of plastid ndh genes is the first step of gene loss associated with photosynthetic genes and congruent with the transition to a heterotrophic lifestyle. The loss of ndh genes within Cypripedioideae observed in this study could hence be a sign of a heterotrophic lifestyle within this subfamily. However, current research states no signs of a heterotrophic lifestyle within any Cypripedioideae species (Guo et al., 2021). The cultivation of myco-heterotrophic orchids has repeatedly been proven to be challenging (Brieger and Schlechter, 1992), but Cypripedioideae species and cultivars are regularly sold as outdoor and indoor plants. If Cypripedioideae had an obligate myco-heterotrophic lifestyle, they would not be such a common ornamental plant due to the difficulty of cultivation. This does not necessarily mean that all Cypripedioideae are solely autotrophic plants. Wicke et al. (2016) observed that some photosynthetic parasites (hemiparasites) can grow without a host plant and thus are considered to be facultative parasites. This may also be the case for the slipper orchid genera Mexipedium, Phragmipedium and Paphiopedilum. Some myco-heterotrophic plants obtain the majority of carbon from photosynthesis and only a small portion from mycorrhizal fungi, often to compensate for low photosynthetic rates resulting from shady habitats; this is also called mixotrophy (Selosse and Roy, 2009). The previously named genera of slipper orchids are known to grow in shady habitats, which make a myco-heterotrophic lifestyle within these genera more likely. Merckx and Freudenstein (2010) also suggested that myco-heterotrophy and partial myco-heterotrophy may have been an adaptation to shady grounds of forests as an alternative way to collect carbon. Furthermore, a recent study conducted by Tian et al. (2023) regarding the soil fungal community structures of Paphiopedilum subg. Brachypetalum found major differences between the habitats of each species. This indicates a high specificity regarding mycorrhizal partners in Paphiopedilum, which is often observed in myco-heterotrophic plants. However, the prospect of mixotrophic or hemiparasitic plants is not much studied, although recent studies showed that mixotrophy may be far more widespread among plants than originally assumed, i.e. in Ranunculaceae Juss (Giesemann et al., 2020).
Based on previous findings and the results from this study, we suggest reevaluating the possibility of a mixotrophic or facultative myco-heterotrophic lifestyle for Cypripedioideae, particularly in Mexipedium, Phragmipedium and Paphiopedilum. This theory should be tested in future studies to gain more understanding of the ecology of slipper orchids, along with research regarding their habitats and ecological adaptation. By understanding how and with which mechanisms slipper orchids adapted to their habitats and what genetic changes lead to vulnerabilities regarding environmental stressors, the conservation of this orchid subfamily will become feasible. Further, such studies will help to increase our understanding regarding the plants reaction to a changing climate.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
VR: Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. NW: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received forthe research and/or publication of this article. The authors acknowledge support by the Open Access Publication Funds of the Göttingen University.
Acknowledgments
We thank Katharina Nargar for the invitation to contribute to this special research topic. We thank Pia Marincek for advice regarding the dating of the phylogenetic tree. We thank reviewers for their useful comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1507415/full#supplementary-material
References
Blazier, C. J., Guisinger, M. M., Jansen, R. K. (2011). Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Mol. Biol. 76, 263–272. doi: 10.1007/s11103-011-9753-5
Bouckaert, R., Vaughan, T., Wu, C., Xie, D., Suchard, M. A., Rambaut, A., et al. (2014). BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Computational Biology. 10 (4), 1–6. doi: 10.1371/journal.pcbi.1003537
Brieger, F. G., Schlechter, R. (1992). Botanische Grundlagen der Orchideenforschung. Taxonomischer Teil. 3rd Edn (Berlin and Hamburg: Verlag Paul Parey).
Burrows, P. A. (1998). Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 17, 868–876. doi: 10.1093/emboj/17.4.868
Chase, M. W., Cameron, K. M., Freudenstein, J. V., Pridgeon, A. M., Salazar, G., van den Berg, C., et al. (2015). An updated classification of Orchidaceae. Botanical J. Linn. Soc. 177, 151–174. doi: 10.1111/boj.12234
Cox, A. V., Pridgeon, A. M., Albert, V. A., Chase, M. W. (1997). Phylogenetics of the slipper orchids (Cypripedioideae, Orchidaceae): Nuclear rDNA ITS sequences. Plant Systematics Evol. 208, 197–223. doi: 10.1007/BF00985442
Cozzolino, S., Widmer, A. (2005). Orchid diversity: an evolutionary consequence of deception? Trends Ecol. Evol. 20, 487–494. doi: 10.1016/j.tree.2005.06.004
Giesemann, P., Rasmussen, H. N., Liebel, H. T., Gebauer, G. (2020). Discreet heterotrophs: green plants that receive fungal carbon through Paris -type arbuscular mycorrhiza. New Phytol. 226, 960–966. doi: 10.1111/nph.16367
Guan, Z.-J., Zhang, S.-B., Guan, K.-Y., Li, S.-Y., Hu, H. (2011). Leaf anatomical structures of Paphiopedilum and Cypripedium and their adaptive significance. J. Plant Res. 124, 289–298. doi: 10.1007/s10265-010-0372-z
Guo, Y.-Y., Luo, Y.-B., Liu, Z.-J., Wang, X.-Q. (2012). Evolution and Biogeography of the Slipper Orchids: Eocene vicariance of the conduplicate genera in the Old and New World tropics. PloS One 7, e38788. doi: 10.1371/journal.pone.0038788
Guo, Y.-Y., Yang, J.-X., Bai, M.-Z., Zhang, G.-Q., Liu, Z.-J. (2021). The chloroplast genome evolution of Venus slipper (Paphiopedilum): IR expansion, SSC contraction, and highly rearranged SSC regions. BMC Plant Biol. 21, 248. doi: 10.1186/s12870-021-03053-y
Horváth, E. M., Peter, S. O., Joët, T., Rumeau, D., Cournac, L., Horváth, G. V., et al. (2000). Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol. 123, 1337–1350. doi: 10.1104/pp.123.4.1337
Hu, C., Jiao, Z., Deng, X., Tu, X., Lu, A., Xie, C., et al. (2022). The ecological adaptation of the unparalleled plastome character evolution in slipper orchids. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1075098
Ifuku, K., Endo, T., Shikanai, T., Aro, E.-M. (2011). Structure of the chloroplast NADH dehydrogenase-like complex: nomenclature for nuclear-encoded subunits. Plant Cell Physiol. 52, 1560–1568. doi: 10.1093/pcp/pcr098
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kharabian-Masouleh, A., Furtado, A., Alsubaie, B., Al-Dossary, O., Wu, A., Al-Mssalem, I., et al. (2023). Loss of plastid ndh genes in an autotrophic desert plant. Comput. Struct. Biotechnol. J. 21, 5016–5027. doi: 10.1016/j.csbj.2023.10.023
Kim, H. T., Kim, J. S., Moore, M. J., Neubig, K. M., Williams, N. H., Whitten, W. M., et al. (2015). Seven new complete plastome sequences reveal rampant independent loss of the ndh gene family across Orchids and associated instability of the inverted repeat/small single-copy region boundaries. PloS One 10, e0142215. doi: 10.1371/journal.pone.0142215
Kim, Y.-K., Jo, S., Cheon, S.-H., Joo, M.-J., Hong, J.-R., Kwak, M., et al. (2020). Plastome evolution and phylogeny of Orchidaceae, with 24 new sequences. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00022
Köhler, M., Reginato, M., Souza-Chies, T. T., Majure, L. C. (2020). Insights into chloroplast genome evolution across Opuntioideae (Cactaceae) reveals robust yet sometimes conflicting phylogenetic topologies. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00729
Krisp, H. (2013). Frauenschuh Cypripedium calceolus [Figure 1]. Wikimedia Commons. Available online at: https://commons.wikimedia.org/wiki/File:Frauenschuh_Cypripedium_calceolus.jpg (Accessed September 23, 2024).
Leake, J. R. (1994). The biology of myco-heterotrophic (‘saprophytic’) plants. New Phytol. 127, 171–216. doi: 10.1111/j.1469-8137.1994.tb04272.x
Lin, C., Chen, J. J. W., Chiu, C., Hsiao, H. C. W., Yang, C., Jin, X., et al. (2017). Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J. 90, 994–1006. doi: 10.1111/tpj.13525
Lin, C.-S., Chen, J. J. W., Huang, Y.-T., Chan, M.-T., Daniell, H., Chang, W.-J., et al. (2015). The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 5, 9040. doi: 10.1038/srep09040
Liu, H., Ye, H., Zhang, N., Ma, J., Wang, J., Hu, G., et al. (2022). Comparative analyses of chloroplast genomes provide comprehensive insights into the adaptive evolution of paphiopedilum (Orchidaceae). Horticulturae 8, 391. doi: 10.3390/horticulturae8050391
Martín, M., Noarbe, D. M., Serrot, P. H., Sabater, B. (2015). The rise of the photosynthetic rate when light intensity increases is delayed in ndh gene-defective tobacco at high but not at low CO2 concentrations. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00034
Merckx, V., Freudenstein, J. V. (2010). Evolution of mycoheterotrophy in plants: a phylogenetic perspective. New Phytol. 185, 605–609. doi: 10.1111/j.1469-8137.2009.03155.x
Orchi (2005). Paphiopedilum concolor Orchi 01 [Figure 1]. Wikimedia Commons. Available online at: https://commons.wikimedia.org/wiki/File:Paphiopedilum_concolor_Orchi_01.jpg (Accessed September 23, 2024).
Orchi (2006a). Mexipedium xerophyticum orchi 01 [Figure 1]. Wikimedia commons, Creative Commons Attribution-Share Alike 3.0 Unported.
Orchi (2006b). Phragmipedium lindleyanum Orchi 02 [Figure 1]. Wikimedia Commons. Available online at: https://commons.wikimedia.org/wiki/File:Phragmipedium_lindleyanum_Orchi_02.jpg (Accessed September 23, 2024).
Pérez-Escobar, O. A., Bellot, S., Przelomska, N. A. S., Flowers, J. M., Nesbitt, M., Ryan, P., et al. (2021). Molecular clocks and archaeogenomics of a Late Period Egyptian date palm leaf reveal introgression from wild relatives and add timestamps on the domestication. Mol. Biol. Evol. 38 (10), 4475–4492. doi: 10.1093/molbev/msab188
Pérez-Escobar, O. A., Bogarín, D., Przelomska, N. A. S., Ackerman, J. D., Balbuena, J. A., Bellot, S., et al. (2024). The origin and speciation of orchids. New Phytol. 242, 700–716. doi: 10.1111/nph.19580
Provan, J., Powell, W., Hollingsworth, P. M. (2001). Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol. Evol. 16, 142–147. doi: 10.1016/S0169-5347(00)02097-8
Rambaut, A. (2014). Figtree, a graphical viewer of phylogenetic trees. Available online at: http://tree.bio.ed.ac.uk/software/figtree (Accessed September 23, 2024).
Rivas, J.D.L., Lozano, J. J., Ortiz, A. R. (2002). Comparative analysis of chloroplast genomes: Functional annotation, genome-based phylogeny, and deduced evolutionary patterns. Genome Res. 12, 567–583. doi: 10.1101/gr.209402
Ruhlman, T. A., Chang, W.-J., Chen, J. J., Huang, Y.-T., Chan, M.-T., Zhang, J., et al. (2015). NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biol. 15, 100. doi: 10.1186/s12870-015-0484-7
Ruhlman, T. A., Jansen, R. K. (2021). “Plastid genomes of flowering plants: essential principles,” in Chloroplast Biotechnology. Methods in Molecular Biology, vol. 2317. Ed. Maliga, P.. (Humana, New York, NY). doi: 10.1007/978-1-0716-1472-3_1
Scobeyeva, V. A., Artyushin, I. V., Krinitsina, A. A., Nikitin, P. A., Antipin, M. I., Kuptsov, S. V., et al. (2021). Gene loss, pseudogenization in plastomes of genus Allium (Amaryllidaceae), and putative selection for adaptation to environmental conditions. Front. Genet. 12. doi: 10.3389/fgene.2021.674783
Selosse, M.-A., Roy, M. (2009). Green plants that feed on fungi: facts and questions about mixotrophy. Trends Plant Sci. 14, 64–70. doi: 10.1016/j.tplants.2008.11.004
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Szlachetko, D. L., Górniak, M., Kowalkowska, A. K., Kolanowska, M., Jurczak-Kurek, A., Archila Morales, F. (2021). The natural history of the genus Cypripedium (Orchidaceae). Plant Biosyst. - Int. J. Dealing all Aspects Plant Biol. 155, 772–796. doi: 10.1080/11263504.2020.1785963
Takebayashi, N. (2018). Paphiopedilum rothschildianum, (38895499920) [Figure 1]. Wikimedia Commons. Available online at: https://commons.wikimedia.org/wiki/File:Paphiopedilum_rothschildianum_(38895499920).jpg (Accessed September 23, 2024).
Tian, L., An, M., Wu, M., Liu, F., Zhang, Y. (2023). Habitat ecological characteristics and soil fungal community structure of Paphiopedilum subgenus Brachypetalum Hallier (Orchidaceae) plants in Southwest China. Plant Signal Behav. 18 (1), 2227365. doi: 10.1080/15592324.2023.2227365
Tsai, C.-C., Liao, P.-C., Ko, Y.-Z., Chen, C.-H., Chiang, Y.-C. (2020). Phylogeny and historical biogeography of Paphiopedilum Pfitzer (Orchidaceae) based on nuclear and plastid DNA. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00126
Wicke, S., Müller, K. F., dePamphilis, C. W., Quandt, D., Bellot, S., Schneeweiss, G. M. (2016). Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc. Natl. Acad. Sci. 113, 9045–9050. doi: 10.1073/pnas.1607576113
Yamori, W., Sakata, N., Suzuki, Y., Shikanai, T., Makino, A. (2011). Cyclic electron flow around photosystem I via chloroplast NAD(P)H dehydrogenase (NDH) complex performs a significant physiological role during photosynthesis and plant growth at low temperature in rice. Plant J. 68, 966–976. doi: 10.1111/j.1365-313X.2011.04747.x
Yang, Z.-H., Huang, W., Yang, Q.-Y., Chang, W., Zhang, S.-B. (2018). Anatomical and diffusional determinants inside leaves explain the difference in photosynthetic capacity between Cypripedium and Paphiopedilum, Orchidaceae. Photosynth Res. 136, 315–328. doi: 10.1007/s11120-017-0466-8
Keywords: gene loss, ultrametric tree, plastome, phylogeny, Paphiopedilum, myco-heterotrophic lifestyle
Citation: Ringelmann VE and Wagner ND (2025) Rapid loss of plastid ndh genes in slipper orchids (Cypripedioideae, Orchidaceae). Front. Plant Sci. 16:1507415. doi: 10.3389/fpls.2025.1507415
Received: 07 October 2024; Accepted: 31 March 2025;
Published: 22 April 2025.
Edited by:
Stefan Kusch, RWTH Aachen University, GermanyReviewed by:
Cássio Van Den Berg, State University of Feira de Santana, BrazilMatias Köhler, Federal University of Rio Grande do Sul, Brazil
Copyright © 2025 Ringelmann and Wagner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natascha D. Wagner, bmF0YXNjaGEud2FnbmVyQHVuaS1nb2V0dGluZ2VuLmRl
†ORCID: Victoria E. Ringelmann, orcid.org/0009-0005-4380-2462
Natascha D. Wagner, orcid.org/0000-0001-6623-7623
 Victoria E. Ringelmann
Victoria E. Ringelmann Natascha D. Wagner
Natascha D. Wagner