- Weed Research Laboratory, College of Life Sciences, Nanjing Agricultural University, Nanjing, China
Introduction: Bidirectional gene flow via pollen between transgenic rice and weedy rice could occur in natural fields. Gene flow from transgenic rice to weedy rice has been confirmed in many studies, and thus results showed that F1 hybrids could persist in natural agroecosystems due to their unimpaired reproductive ability. However, the reverse gene flow from weedy rice to transgenic rice is rarely reported.
Method: We quantified reverse gene flow from three weedy rice accessions to transgenic rice line T1c-19 with cry1C*/bar. In field trials with alternating layout of cultivating transgenic rice and weedy rice accessions and adjacent layout cultivating them in a close vicinity, the reverse gene flow was detected. And the fitness of reverse F1 (RF1) hybrids obtained by manual pollination using T1c-19 as maternal plants and weedy rice as paternal plants was evaluated in field.
Result: No gene flow from WRTZ was observed, while gene flows from WRMM were observed at 0.0508% and 0.0808%, respectively, and those from WRYY were 0.0692% and 0.1008%, respectively. RF1 plants exhibited significantly higher composite fitness compared to their weedy rice counterparts, due to enhanced fecundity-related traits observed under both insect pressure and no-insect pressure conditions. However, the impact of reverse gene flow may be limited because RF1 hybrid seeds presented lower seed shattering, and therefore most of it would be harvested by combine harvester.
Discussion: Our study revealed that gene flow from three weedy rice accessions to T1c-19 could produce RF1 hybrids with greater composite fitness. Any loss of seeds into agroecosystem may result in a greater risk of RF1 hybrids due to their morphological similarity and high fitness.
1 Introduction
Rice (Oryza sativa L.) is a worldwide food crop that plays a pivotal role in China’s food production. A large number herbicide resistant (HR), insect resistant (IR), even stacked traits rice lines have been bred by transgenic or mutation technique and the HR rice by mutation technique have been planted in China (Tang et al., 2006; Chen et al., 2021). While the commercial release of HR rice will bring economic and social benefits, it also poses certain ecological risks (Nicolia et al., 2014; Merotto et al., 2016; Clark and Maselko, 2020). One of the major concerns in relation to the ecological risks is HR gene flow via pollen to its weedy relative weedy rice (Oryza sativa L.) (Messeguer, 2003; Sun et al., 2015; Nam et al., 2019). Once the HR gene flows into weedy rice, hybrids carrying the HR gene could survive under herbicide selection, making the control of weedy rice more challenging. Meanwhile, HR gene may persist and spread within the weedy rice population by both gene flow via pollen and seed dispersal. This could result in an aggravated weedy rice problem that, combined with the technical difficulties of control, could jeopardize the HR rice technology itself. This has been the case with non-transgenic imidazolinone resistant rice (IMR-rice, Clearfield®) (Rajguru et al., 2005; Roso et al., 2010; Busconi et al., 2012; Merotto et al., 2016; Avila et al., 2021; Unan et al., 2024). Thus, HR weedy rice may become a significant barrier to continued HR cultivated rice (Messeguer et al., 2004; Busconi et al., 2014; Merotto et al., 2016; Dauer et al., 2018).
Weedy rice is present in most rice-growing areas worldwide (Arrieta-Espinoza et al., 2005; Olsen et al., 2007; Shivrain et al., 2009; Karn et al., 2020). It has widely evolved from cultivated rice via de-domestication (Sun et al., 2019; Qiu et al., 2017, 2020; Wu et al., 2021). Wild rice (Oryza rufipogon) hybridization with weedy rice has contributed substantially to the evolution of Southeast Asian weedy rice, with some strains acquiring weed-adaptive traits through introgression from the wild progenitor (Li et al., 2024). Weedy rice has strong feral characteristics including robust seed shattering and dormancy, seedling vigor, early maturity, and greater competitive advantage, which are critical for its ability to survive and persist in rice fields (Thurber et al., 2011; Dai et al., 2014, 2017; Zhao et al., 2018, 2020, 2021). It has become the primary weed-related factor constraining increased rice yields in China, including Jiangsu, Heilongjiang, Ningxia and Guangdong to the whole East China, Northeast China, Northwest China and South China, causing increasing losses to rice production (Ma et al., 2005; Wang et al., 2023; Cai et al., 2023).
Whether the HR gene can successfully flow to weedy relatives and pose a threat to agricultural production and the environment depends on the rate at which the transgenic crop can hybridize with weedy relatives and, particularly, on the fitness of the hybrids (Song et al., 2011; Zuo et al., 2011; Liu et al., 2016; Dauer et al., 2018; Huang et al., 2019). Fitness is the ability of an individual to survive and reproduce under specific environmental conditions. It determines whether hybrids can survive and establish populations in nature (Jenczewski et al., 2003; Lu and Yang, 2009; Xia et al., 2011). Numerous studies have determined that the fitness of hybrids carrying a HR gene is closely related to several factors, including parental genetic background, and environmental conditions (with or without various selection pressures, competition) (Cao et al., 2009; Vila-Aiub et al., 2009; Song et al., 2011; Yang et al., 2011; Darmency et al., 2015; Vila-Aiub, 2019).
The HR gene flow via pollen from cultivated rice to weedy rice has been confirmed in many studies, with an outcrossing rate usually less than 1% (Zhang et al., 2003; Messeguer, 2003; Chen et al., 2004; Zhang et al., 2006; Shivrain et al., 2007, 2008; Zuo et al., 2011; Sun et al., 2015; Nam et al., 2019). F1 hybrids frequently exhibit heterosis for vegetative traits but have lower reproductive capacity than the corresponding weedy rice parent because of their lower seed set (Zhang et al., 2008; Sanchez-Olguin et al., 2009). However, some F1 hybrids display similar seed set as well as composite agronomic performance (Song et al., 2011; Liu et al., 2016) or produced significantly more and heavier grains (Nam et al., 2019). For insect-resistant transgenic rice and weedy rice, hybrids under low insect pressure exhibited fitness cost, and fitness advantage under high insect pressure (Cao et al., 2009; Yang et al., 2011). These researchers suggested that the HR or IR F1 hybrids could persist under natural agroecosystem because these F1 hybrids presented certain reproductive ability even unimpaired one.
Gene flow between HR cultivated rice and weedy rice is bidirectional; direct gene flow occurs from cultivated to weedy rice, while indirect or reverse gene flow occurs in the opposite direction. Reverse gene flow under field conditions could result in the transfer of dominant weedy traits to HR cultivars, potentially leading to the emergence of HR weedy rice (Shivrain et al., 2009; Serrat et al., 2013; Zhang et al., 2018).
This reverse gene flow might significantly impact the evolutionary dynamics of weedy rice populations. The segregating progeny of HR transgenic hybrid rice pollinated by weedy rice rapidly develop into weedy forms carrying the HR transgene under natural conditions (Zhang et al., 2018). Although bidirectional gene flow between HR cultivated rice and weedy rice can occur and produce HR weedy rice, reverse gene flow has received less attention compared to direct gene flow from HR cultivated rice to weedy rice. One of the main reasons is the more complex methods required to verify reverse F1 hybrids (RF1) produced by reverse gene flow, compared to F1 hybrids resulting from direct gene flow, which can be tested by herbicide application or PCR analysis for the HR gene.
Reverse gene flow from imidazolinone-resistant rice (IMI-R; Clearfield® rice) to red rice has been documented in two cases (Shivrain et al., 2009; Serrat et al., 2013). The initially selected putative reverse flow (PRF) seedlings were identified based on their most important phenotypic characteristic—vigorous growth, characterized by faster and taller growth. PRF plants were then analyzed using the Oryza simple sequence repeat (SSR) primers RM234 and RM253 or by determining their molecular fingerprint patterns through AFLP analysis. These studies supported the possibility that HR weedy rice could arise through reverse gene flow from weedy rice to HR rice.
Red pericarp is a typical and convergent characteristic of weedy rice (Sun et al., 2019; Qiu et al., 2020). The Rc gene, located on rice chromosome 7, contains eight exons and confers a red pericarp. The Rc allele rc, found in most white pericarp rice genotypes, is characterized by the absence of a 14 bp fragment in the seventh exon (Sweeney et al., 2007; Li et al., 2014). If gene flow occurred from weedy rice to HR rice, both fragments of 118 bp and 104 bp should be present in RF1 hybrids (Huang et al., 2019). Therefore, both alleles can serve as markers for detecting reverse gene flow.
The insect-resistant and glufosinate-resistant stacked transgenic rice line T1c-19 was successfully developed by Huazhong Agricultural University (Tang et al., 2006). The weediness of T1c-19 under no selection pressure, gene flow from T1c-19 to weedy rice, and the fitness of its F1 hybrids have been reported (Huang et al., 2015; 2016; 2019). T1c-19 did not exhibit weedy characteristics; it had weak overwintering ability, low seed shattering, and failed to establish volunteer plants. The gene flow from T1c-19 rice to weedy rice ranged from 0.230% to 0.106%, depending on the genotype of the weedy rice and field planting design. Compared to their weedy rice counterparts, the F1 hybrids displayed either higher performance or non-significant changes in the presence or absence of insect pressure in mixed planting with weedy rice. The F1 hybrids also showed non-significant changes regardless of glufosinate pressure, insect pressure, in monoculture planting. The potential risk of gene flow from T1c-19 to weedy rice should be considered due to the greater fitness advantage of F1 hybrids in most cases.
However, reverse gene flow from weedy rice to T1c-19 and the fitness of reverse F1 hybrids (RF1) using weedy rice as the pollen donor have not been reported. In this study, the method for verifying the RF1 hybrids was established based on the Rc gene. A field experiment was conducted to assess the reverse gene flow from three weedy rice accessions to T1c-19 and to compare the fitness of the RF1 hybrids, under no insect and natural insect selection pressures, with their paternal weedy rice counterparts. The results from this study should be useful for regulatory authorities to consider reverse gene flow and develop relevant policies for assessing the risks associated with bidirectional gene flow. These findings are also helpful for regulatory authorities in making decisions regarding the authorization of T1c-19. Furthermore, the data provide an experimental basis for developing risk assessment standards for stacked-trait rice.
2 Materials and methods
2.1 Plant materials
The insect-resistant and glufosinate-resistant stacked transgenic rice line T1c-19 contains two tightly linked genes: the cry1C* insect resistance gene and the bar glufosinate resistant gene. This line was generated through Agrobacterium-mediated co-transformation of the indica rice (O. sativa ssp. indica) cultivar MH63, an elite cytoplasmic male-sterile (CMS) restorer line (Tang et al., 2006). T1c-19 was provided by the State Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, China.
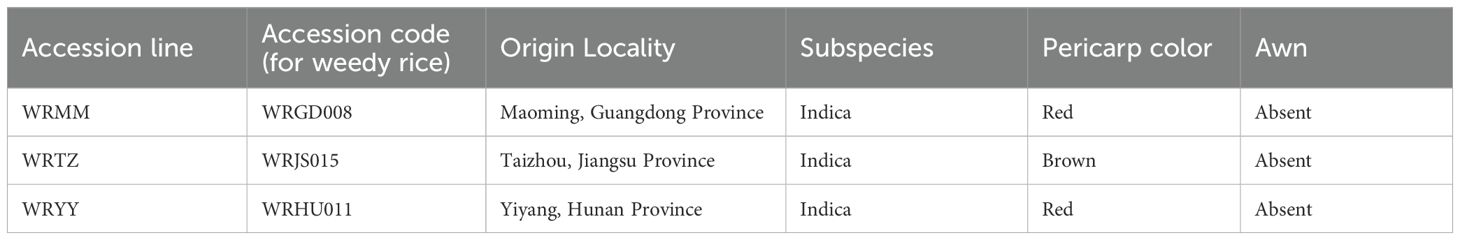
Three weedy rice accessions, WRMM, WRTZ, and WRYY, were selected from the germplasm collection of the Weed Research Laboratory at Nanjing Agricultural University (NJAU), China (Table 1).
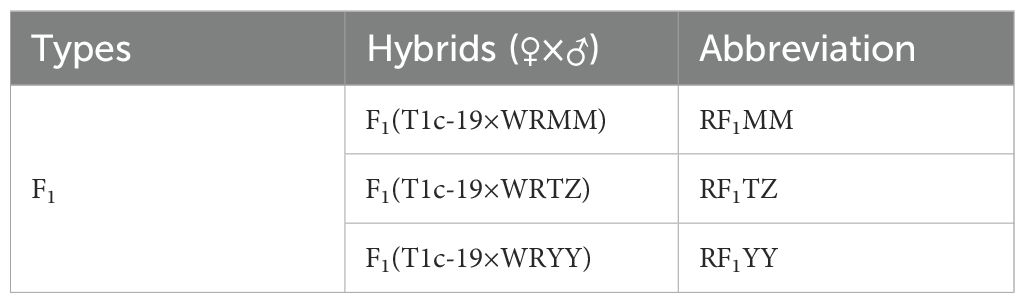
The F1 hybrids obtained by manual pollination between weedy rice (as the paternal parent) and T1c-19 (as the maternal parent) were designated as RF1 to distinguish from the F1 hybrids obtained by manual pollination between weedy rice (as maternal parent) and T1c-19 (as paternal parent) (Table 2). All RF1 plants were transgene positive. Since three weedy rice accessions were used as paternal parents, three different RF1 hybrids were obtained by hand pollination in 2014 and further identified by the site of origin of the male parent (Table 2). Mature seeds were harvested, dried at 25 °C, and stored at 4°C until use.
2.2 Methods
2.2.1 Gene flow from weedy rice to T1c-19
2.2.1.1 Arrangement of field experiment
To detect the maximum possible gene flow frequency from weedy rice to T1c-19, two different experimental designs, alternating and adjacent cultivation, were conducted at the Jiangpu Experimental Station (N32.011569 E118.624535), part of the Weed Research Laboratory of Nanjing Agricultural University, China. The experimental fields, where the studies were conducted from May to October in 2016, were authorized by Ministry of Agriculture and Rural Affairs of the People’s Republic of China (MOA). The experimental area was surrounded by a 100 m wide corn crop, providing a buffer zone without rice crop. All rice cultivation practices, including irrigation, fertilization, and pest management followed those common in rice production in China. Weed control was performed manually as needed.
T1c-19 was sown on 18th June, three days earlier than both weedy rice sown on 21st June for the alternating cultivation design trial and the first batch weedy rice in adjacent cultivation design trial. This adjustment was necessary because T1c-19 flowers three days earlier than three weedy rice accessions, according to our preliminary experiments. The second batch weedy rice seeds in the adjacent cultivation design trial were sown on 26th June (five days after the first batch) to ensure flowering synchronization with the pollen recipient T1c-19. One-month-old seedlings were transplanted according to the following experimental designs. Each experiment involving a weedy rice accession was conducted in field using a complete randomized block design with four replications.
2.2.1.2 Alternating cultivation design trial
The seedlings of each weedy rice accession and T1c-19 were transplanted into individual 5×6 m plots in alternating rows, planted at a density of one seedling per hole. The inter-row and interplant distances were 30 cm and 20 cm, respectively (Figure 1). The rows were aligned with the prevailing wind direction (from South to North). Each plot contained 20 rows, with 26 seedlings per row. Plots were separated by a distance of 200 cm and each plot was replicated four times resulting in a total of 12 experimental plots. This design mitigated potential biases in gene flow due to the equal distance between each transgenic and weedy rice plant.
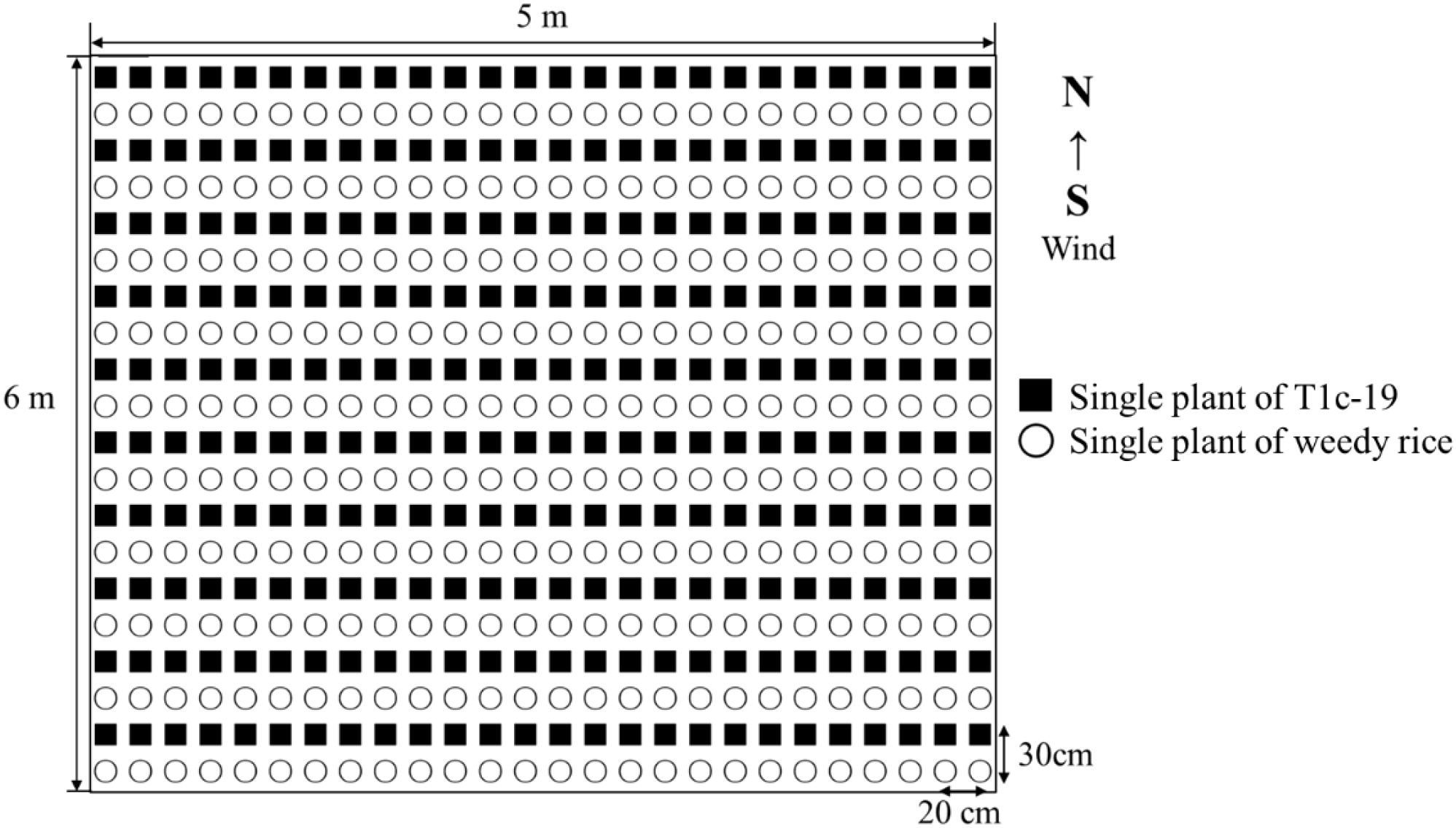
Figure 1. Layout of an individual plot in the alternating cultivation for gene flow from weedy rice (○) to T1c-19 (■).
2.2.1.3 Adjacent cultivation design trial
The seedlings of each weedy rice accession and T1c-19 were transplanted into individual 5.7×3.0 m plots, including four blocks (Figure 2). The seedlings were manually transplanted at a spacing of 30 cm between rows and 20 cm between plants. Each plot contained 84 seedlings of T1c-19, with 96 and 140 seedlings in the first and second batches of weedy rice, respectively. Plots were separated from each other by 60 cm. This experiment was conducted using a complete randomized block design with four replications, for a total of 12 experimental plots.
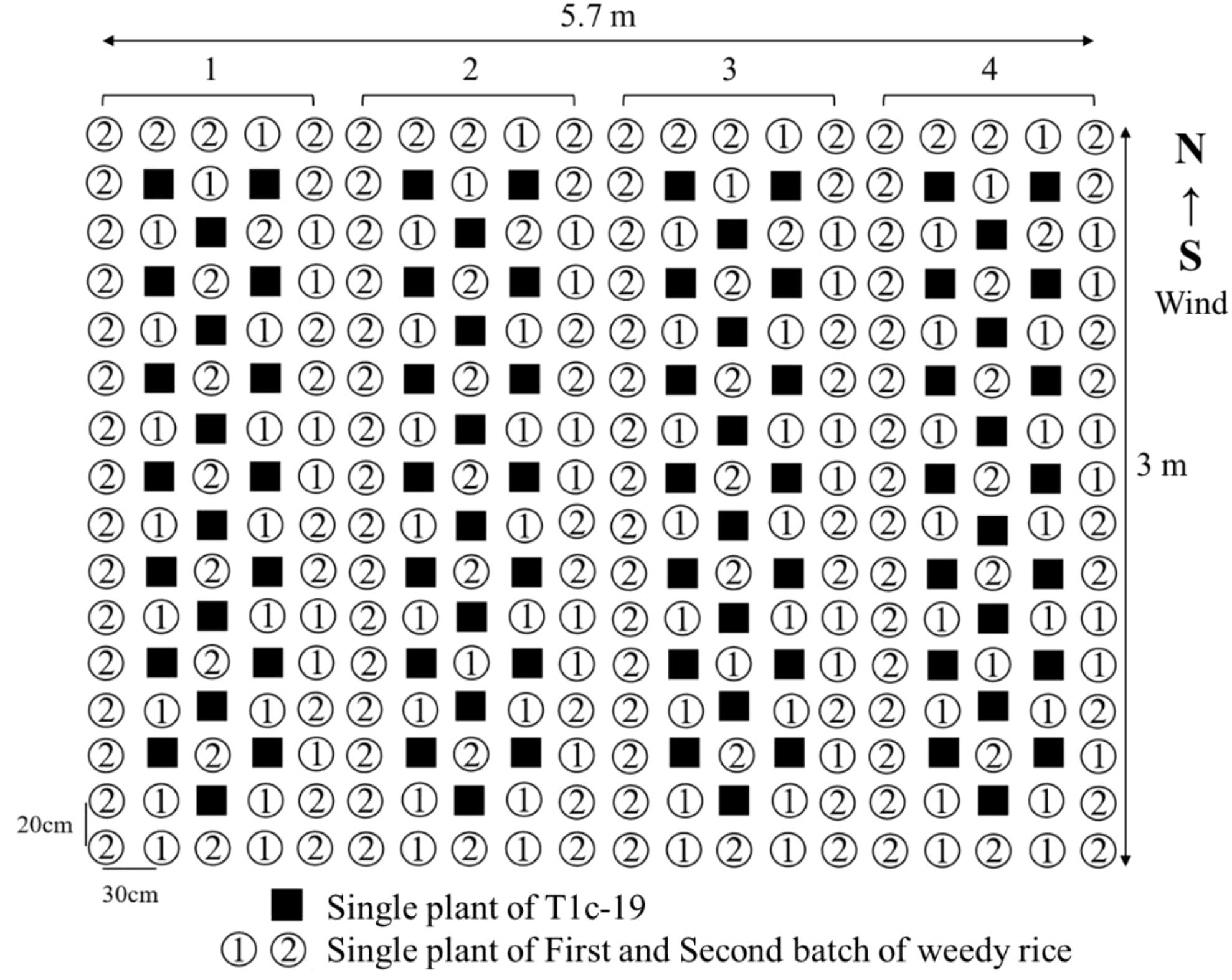
Figure 2. Layout of an individual plot in the adjacent cultivation for gene flow from two batches weedy rice (① and ②) to T1c-19 (■).
2.2.1.4 Field data and seed collection
Plant height of both T1c-19 and weedy rice individuals was measured at the end of the flowering period. The dates of first, peak, and last flowering were recorded to estimate the days of flowering overlap between the pollen donor and recipient plants. The first (approximately 5% of total flowers per panicle opening in one day), peak (approximately 30–80% of total flowers per panicle opening in one day) and the final flowering time (approximately 95% of total flowers per panicle opening in one day) was observed in the adjacent cultivation experiment according to Sun et al. (2015).
Seeds from the pollen recipient T1c-19 in the two experiments were harvested in October and thoroughly mixed for each replication, in order to ensure reliability for testing gene flow frequency. To calculate gene flow frequency, approximately 30,000 seeds were randomly selected from each replication, which is the number of seeds required for testing gene flow frequency if it exceeds 0.05% (Wang et al., 2020).
2.2.1.5 Procedures to verify reverse gene flow
The three weedy rice accessions possess the Rc gene that confers a red pericarp, while T1c-19 has the known 14-bp deletion in the Rc gene that confers the white pericarp characteristic of cultivated rice. To enhance the efficiency of testing the Rc gene in seeds collected from pollen recipients T1c-19, an optimized method for detecting hybrids produced via pollen from weedy rice to T1c-19 was established (Supplementary File). Sixty seeds, randomly selected from those collected from T1c-19 within the same plot, were grouped into a total of 500 groups, amounting to 30,000 seeds. These seeds were first dehulled, and each group seeds were ground completely to pass through a 60-mesh sieve. Subsequently, genomic DNA was extracted from ground tissue using the DP305 DNA Miniprep Kit produced by Tiangen Company (Shanghai, China). The method for the PCR analysis of the Rc gene followed that reported by Huang et al. (2019).
2.2.2 Fitness of RF1 hybrids
All RF1 hybrids were obtained by hand pollination in 2014 using T1c-19 as the maternal parent and three weedy rice accessions as the paternal parent. The fitness of RF1 hybrids obtained in 2014 was evaluated in the same manner as the fitness of F1 in that year (Huang et al., 2019). Field trials were conducted at the Jiangpu Experimental Station (32.011569°N, 118.624535°E) of the Weed Research Laboratory of NJAU. The experimental studies were conducted from May to November 2014 and were authorized by the MOA. The experimental area was isolated by a 100-m-wide corn crop. Pregerminated seeds of hybrids were sown in seedling trays (54 cm×29 cm×5 cm) filled with field soil at a density of approximately 1400 individuals per m2.
Both expected amplification fragments of the Rc gene (118 bp, and 104 bp) should be present in true F1 hybrids, with T1c-19 as maternal plants and the three weedy rice accessions as paternal parents. At the four to five leaf stage, all RF1 hybrids were tested for the Rc gene. T1c-19 and WRMM accessions were used as positive and negative controls, respectively. The methods for testing the Rc gene in RF1 were the same as those described by Huang et al. (2019).
2.2.2.1 Variables measured in the field
At the five–six leaf stage, selected uniform RF1 plants were transplanted into the experimental field. The plot experiments included two forms of selection pressures: with and without natural insect pressure. Under monoculture planting, plots contained 36 individuals in a 6×6 grid with a 20-cm spacing.
In plots with natural insect pressure, plants were not treated with insecticides or any other insect control measures. Absence of natural insect pressure was achieved by covering the experimental plots with an anti-insect net and supplementing this with insecticide application as described as Huang et al. (2019).
Insect damage was assessed by counting the blasted tillers and folded leaves at the heading stage within the weedy rice plants. An insect index was calculated as follows: Insect index (%)=Leaf folded (%)+Tillers blasted (%) (Xia et al., 2011).
For each plant type, 10 individuals per plot were randomly selected at maturity for measuring fitness variables, excluding for those in the border rows to avoid edge effects. All fitness variables were measured according to the methods described by Song et al. (2011). These variables included plant height, effective panicle number per plant, filled grain number per panicle, yield per plant, 1000-grain weight and seed set.
2.2.2.2 Statistical analysis
The mean plant height of T1c-19 and weedy rice was calculated in the gene flow experiment. One-way ANOVA (Dunnett’s Multiple Range Test, DMRT) was used to examine significant differences in plant height between T1c-19 and two batches of weedy rice in the adjacent cultivation. The t-test was used to examine significant differences in plant height between T1c-19 and weedy rice in the adjacent cultivation experiment.
Composite fitness across the six fitness-related traits measured from vegetative to mature stages was calculated using the methods outlined for F1 by Huang et al. (2019).
One-way ANOVA was used to examine differences in the insect index among the hybrids and the respective weedy rice accessions under natural insect pressure. The means of each trait and composite fitness for the hybrids and their respective weedy rice accessions, along with the values for composite fitness of hybrids, were compared using the t-test for independent samples in SPSS version 17 (SPSS Inc., 2008).
Given that the fitness of RF1 and F1 was assessed in the same year under identical experimental conditions, the relative fitness of RF1/F1 was compared using the same methodology as that used RF1 and their weedy rice counterparts.
3 Results
3.1 Gene flow from weedy rice to T1c-19
3.1.1 Flowering period
Due to being sown and transplanted on the same day in both the alternating the adjacent experiments, T1c-19 began flowering on 22 September, with a flowering duration of 20 days in both experiments. The flowering overlap between T1c-19 and WRMM, WRYY and WRTZ in the alternating experiment was 5, –6 and –8 days, respectively (Table 3). In adjacent experiment, the overlap between T1c-19 and WRMM, WRYY and WRTZ in the first and second batches was 5, 6, and 8 days, and 6, 8 and 11 days, respectively (Table 4).
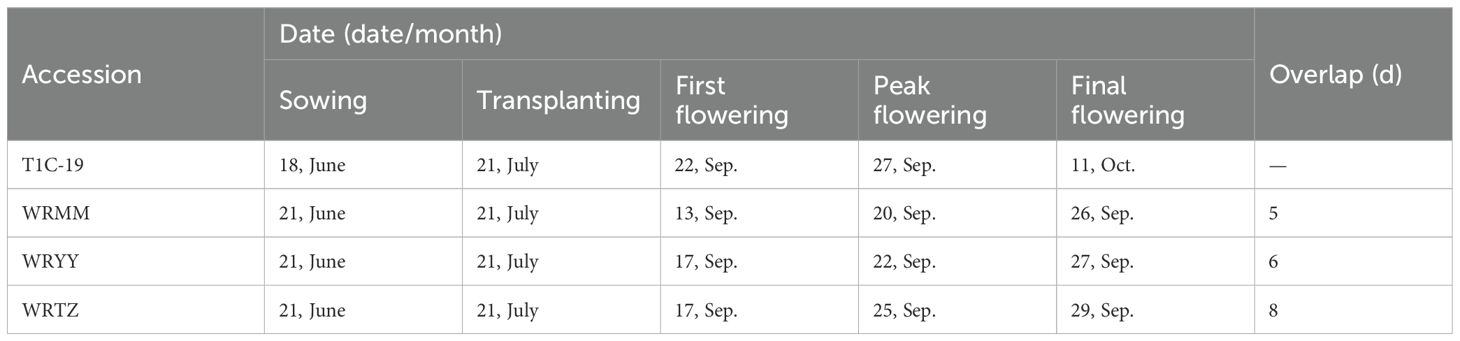
Table 3. Flowering and overlaps periods of weedy rice with T1c-19 in alternation cultivation experiment for gene flow from weedy rice to T1c-19.
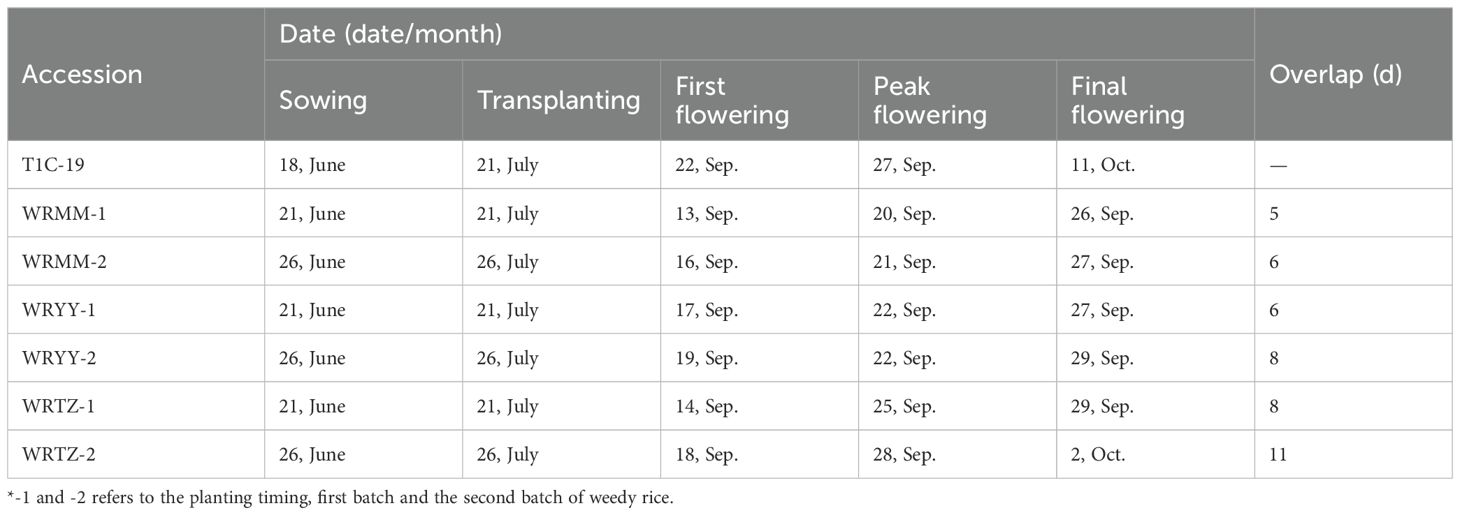
Table 4. Flowering periods and overlaps of weedy rice accessions with T1c-19 in adjacent cultivation experiment for gene flow from weedy rice to T1c-19.
3.1.2 Flowering time
In both the alternating and adjacent cultivation, T1c-19 and the three weedy rice accessions had similar flowering patterns (Figure 3). T1c-19 began flowering at 09:30, reached peak flowering at 10:30, and ceased flowering by 11:30. The daily flowering rhythms of WRMM and WRYY were consistent with that of T1c-19. WRTZ began and ceased flowering synchronously with T1c-19 but peaked at 10:00, which was 30 min earlier than T1c-19. The flowering time period of T1c-19 overlapped with the three weedy rice by approximately two hours.
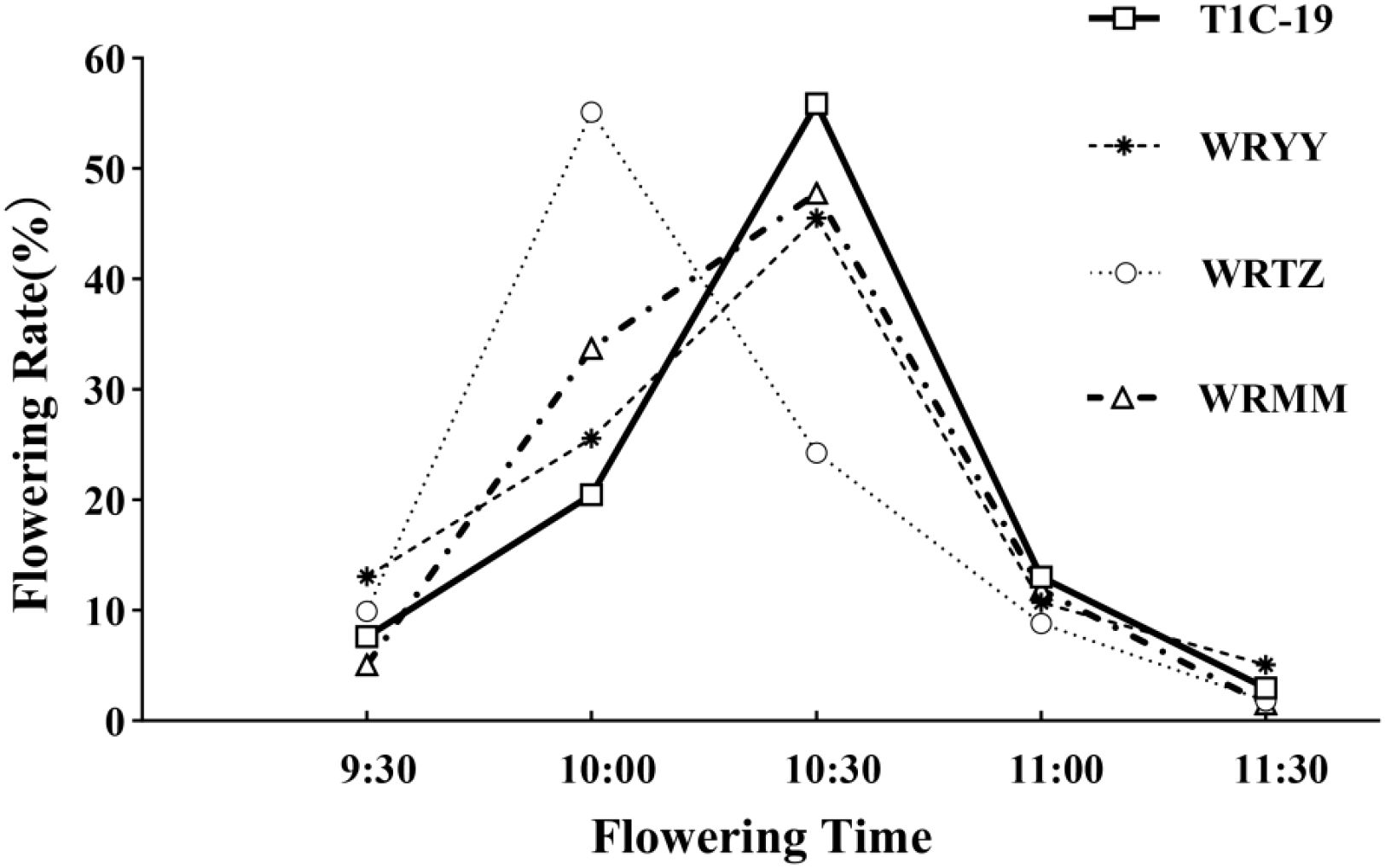
Figure 3. Daily flowering rhythm of weedy rice in relation to T1c-19 in alternation and adjacent cultivation experiments for gene flow from weedy rice to T1c-19.
3.1.3 Plant height
In the alternating experiment, the plant heights of T1c-19 with WRMM, WRTZ and WRYY were 110.4 cm、97.9 cm、108 cm, respectively. The height of T1c-19 was significantly lower by 45.20 cm compared to WRMM, 17.90 cm higher than WRTZ, and similar to WRYY (Figure 4).
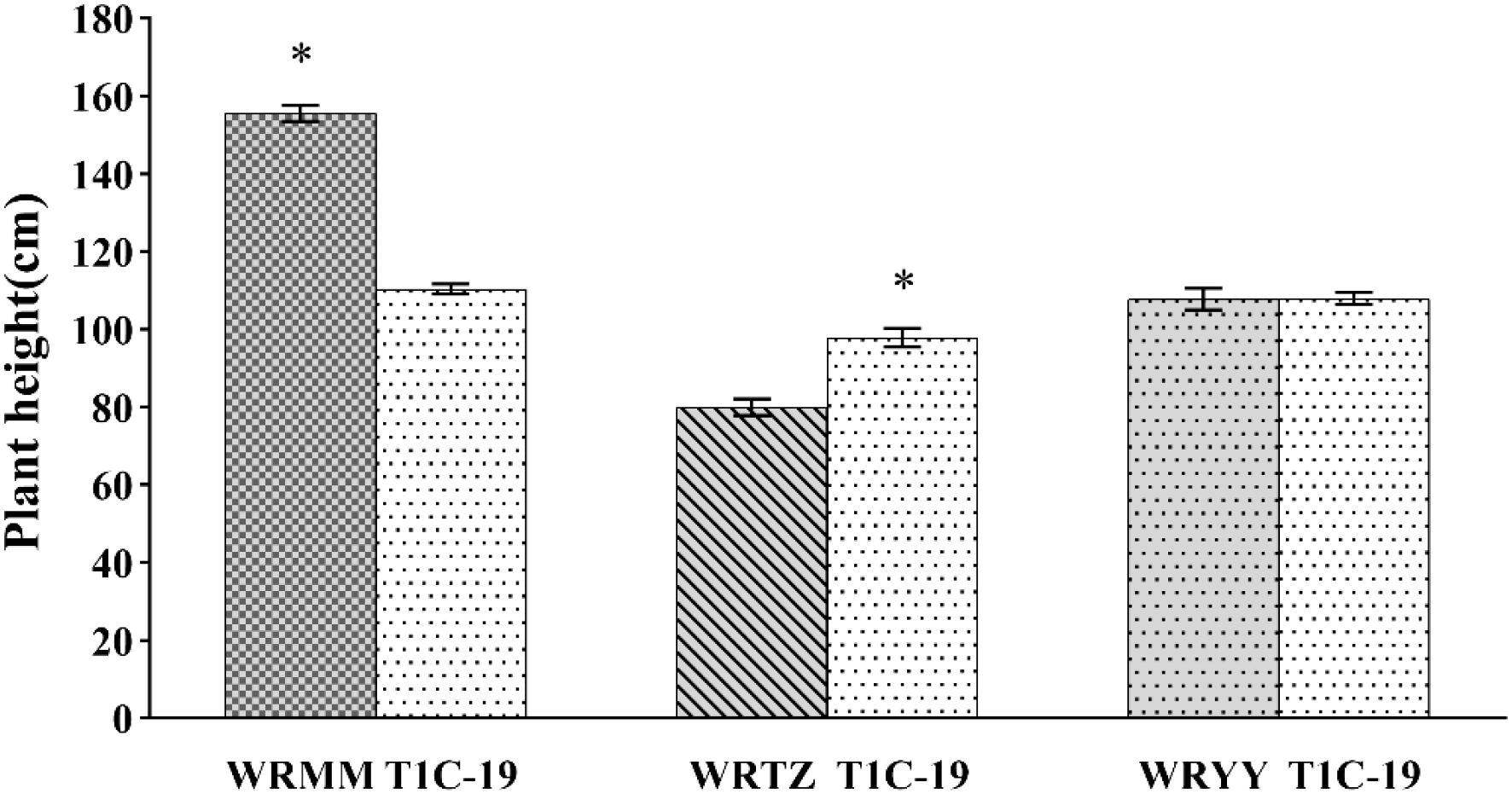
Figure 4. Plant height (mean ± SE) of weedy rice with T1c-19 in the alternating cultivation experiment for gene flow from weedy rice to T1c-19. * means significant difference at 0.05 between weedy rice and T1c-19 by using t-test.
In the adjacent experiment, T1c-19 was significantly shorter by 62 cm than WRMM-1 and WRMM-2, 24.8 and 24 cm taller than WRTZ-1 and WRTZ-2, and similar in height to WRYY (Figure 5).
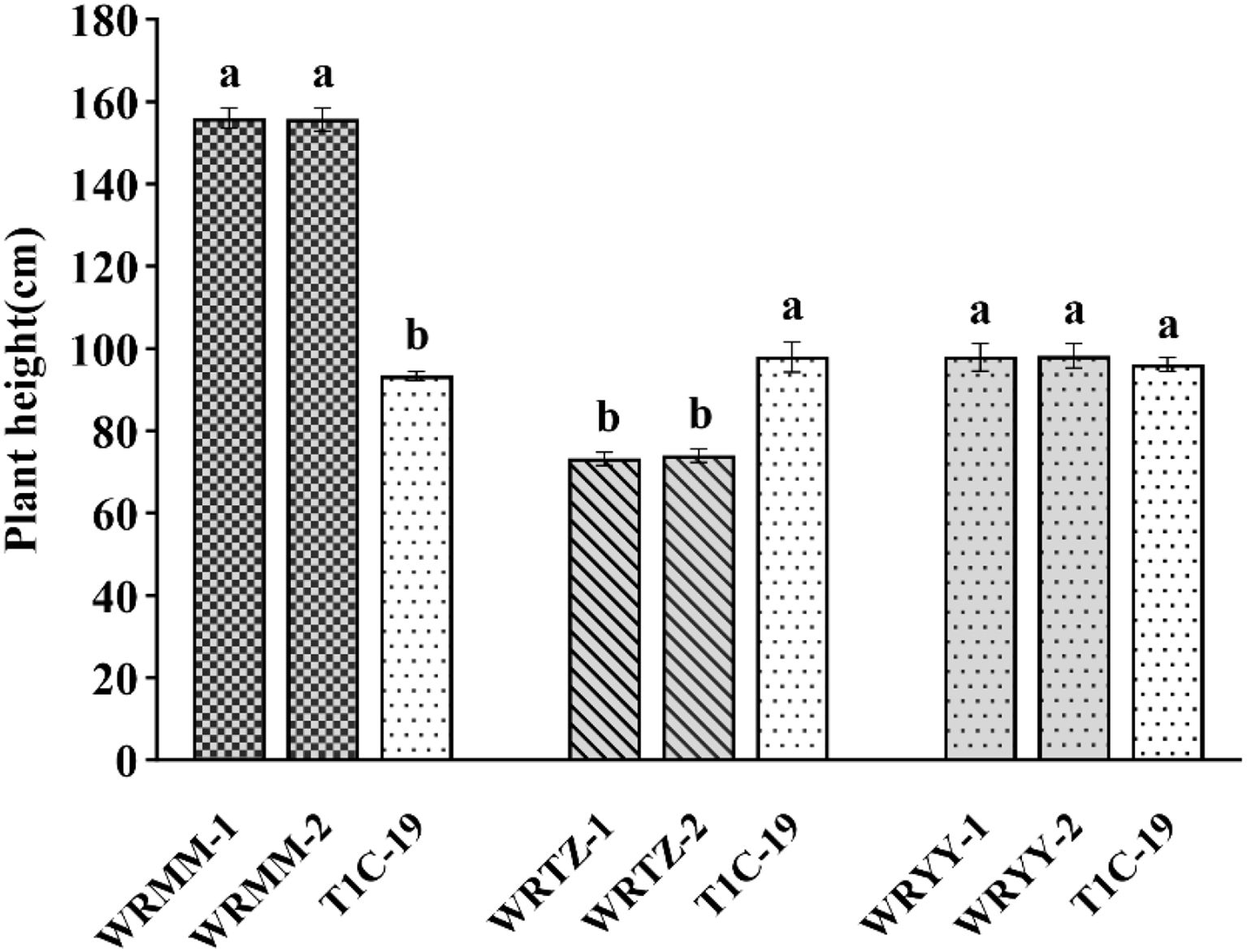
Figure 5. Plant height (mean ± SE) of weedy rice with T1c-19 in the alternating adjacent cultivation experiment for gene flow from weedy rice to T1c-19. The same letters indicate no significant difference at 0.05% by DMRT.
3.1.4 Verification of gene flow
The 118 bp Rc gene fragment, present in the weedy rice, was not detected in any of the 30,000 T1c-19 seeds collected from each plot in both the alternating and adjacent experiments with WRTZ. This indicates that gene flow from WRTZ to T1c-19 did not occur. For WRMM, out of 500 tests per plot, the 104 bp and 118 bp fragments were detected 13, 15, 16, and 17 times in the alternating experiment and 26, 22, 25, and 24 times in the adjacent experiment. The average gene flow frequency was 0.0508% in the alternating experiment and 0.0808% in the adjacent experiment. For WRYY, the fragments were detected 23, 21, 20, and 19 times in the alternating experiment and 29, 32, 33, and 27 times in the adjacent experiment, resulting in average gene flow frequency of 0.0692% and 0.1008%, respectively (Figure 6).
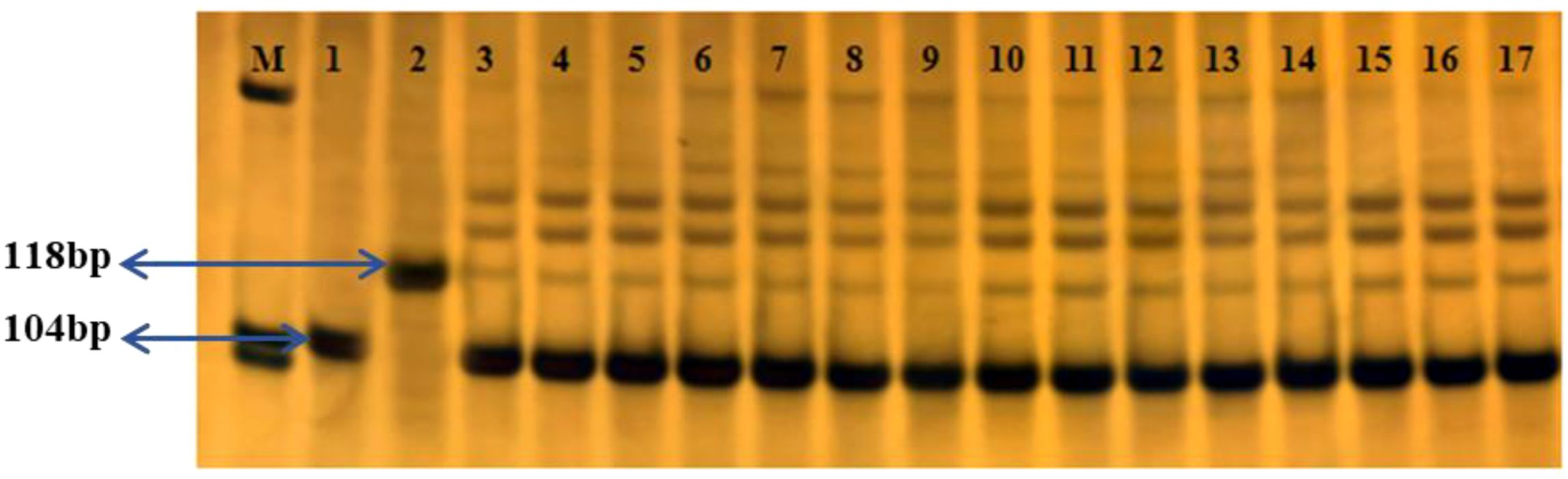
Figure 6. The gene Rc Polymerase chain reaction (PCR) for the detection of the Rc gene in seeds of T1c-19 in the alternating and adjacent cultivation experiments for gene flow from weedy rice to T1c-19. M: Marker; 1: transgenic rice T1c-19; 2: WRYY; 3-6: T1c-19 and WRMM in the alternating cultivation; 7-10: T1c-19 and WRYY in the alternating cultivation; 11-14: T1c-19 and WRMM in the adjacent cultivation; 15-17: T1c-19 and WRYY in the adjacent cultivation.
3.2 Fitness of RF1
3.2.1 Verification of RF1 hybrids
All tested RF1 hybrids using T1c-19 as the maternal plant, carried the expected 118 bp and 104 bp fragments (Figure 7). Therefore, all putative RF1 used in the experiment were confirmed as true RF1 hybrids.
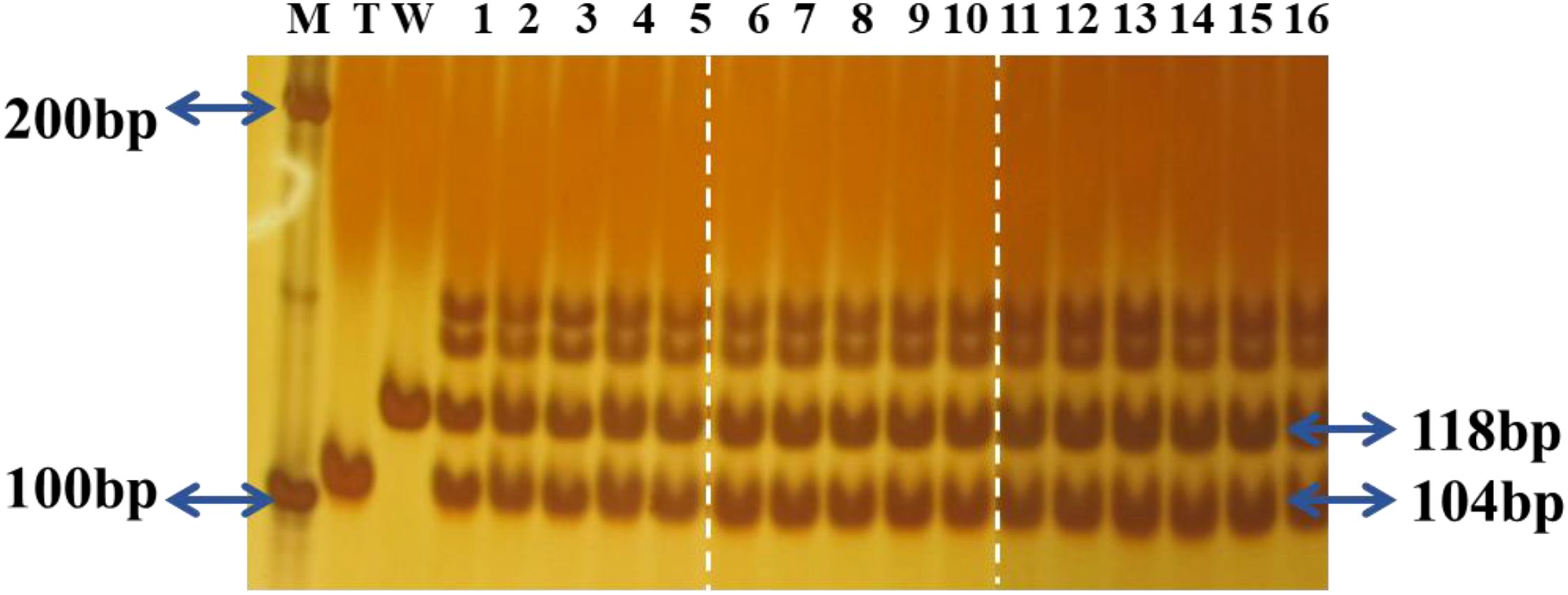
Figure 7. Polymerase chain reaction (PCR) detection of the Rc gene in F1 hybrids using T1c-19 as maternal plants in the experiment. M: Marker; T:T1c-19; W: WRMM; 1-5: RF1MM; 6-10: RF1TZ; 11-16: RF1YY.The reassembled figure presents different regions from the same gel, with white dotted lines indicating the cropping boundaries.
3.2.2 Target insect pressure
As expected, no target insect pressure was observed in the plots protected where insects were physically excluded. Under natural insect pressure, the target insect pressure on transgene-positive RF1 plants (average 1.75%) was significantly lower than on weedy rice (average 19.57%) (Figure 8).
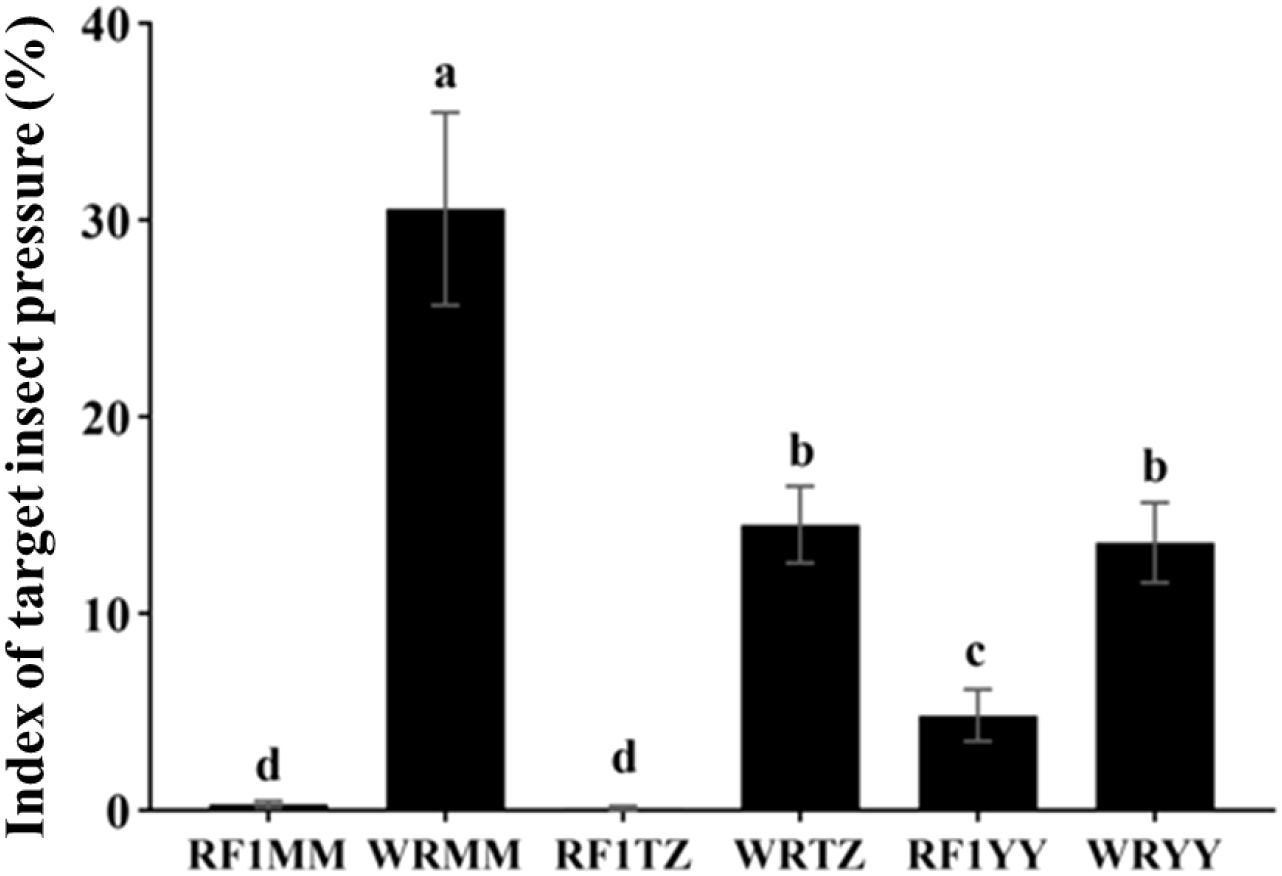
Figure 8. Index of target insect pressure under natural insect pressure in the monoculture planting. WRMM, WRTZ, and WRYY are weedy rice collected from Maoming, Taizhou and Yiyang, respectively. RF1MM, RF1TZ and RF1YY are RF1 hybrids of T1c-19 (as maternal plant) and weedy rice accessions (as paternal plants). Data are presented as mean ± SE (n=4). Different letters indicate significant differences among RF1 and weedy rice at P<0.05 by one-way ANOVA.
3.2.3 Fitness of RF1 and weedy rice
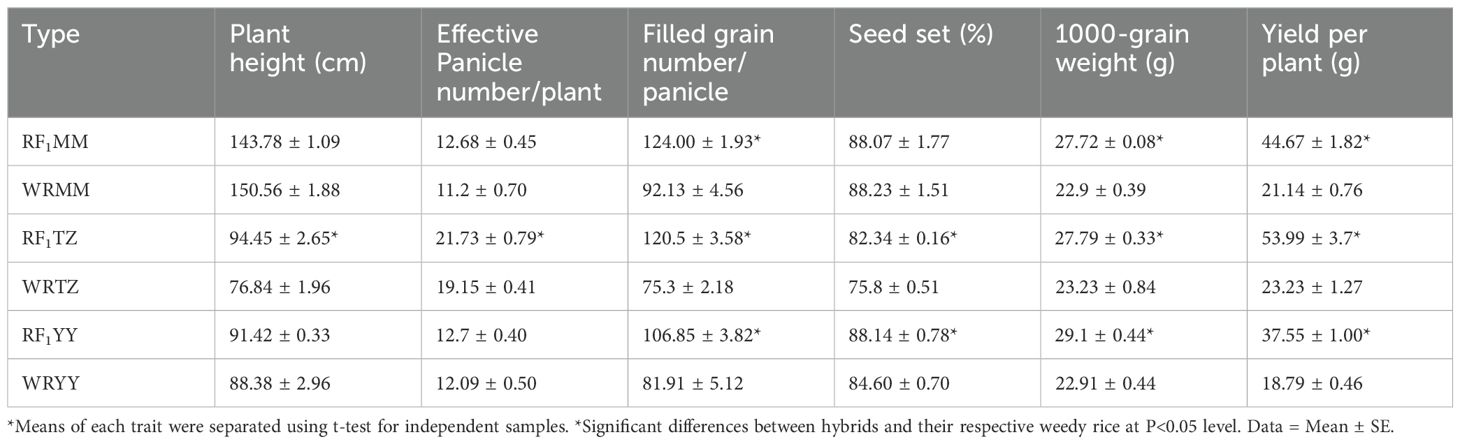
In the absence of insect pressure, compared to their respective weedy rice parents, RF1MM had approximately 21% greater 1000-grain weight, produced 34.6% more filled grains per panicle, and had 1.11 times more yield per plant compared to WRMM. RF1TZ exhibited superiority across all tested variables compared to WRTZ. RF1YY produced 30.4% more filled grains per panicle, 4.2% higher seed set, 27% greater 1000-grain weight, and nearly 1 time more yield per plant compared to WRYY (Table 5).
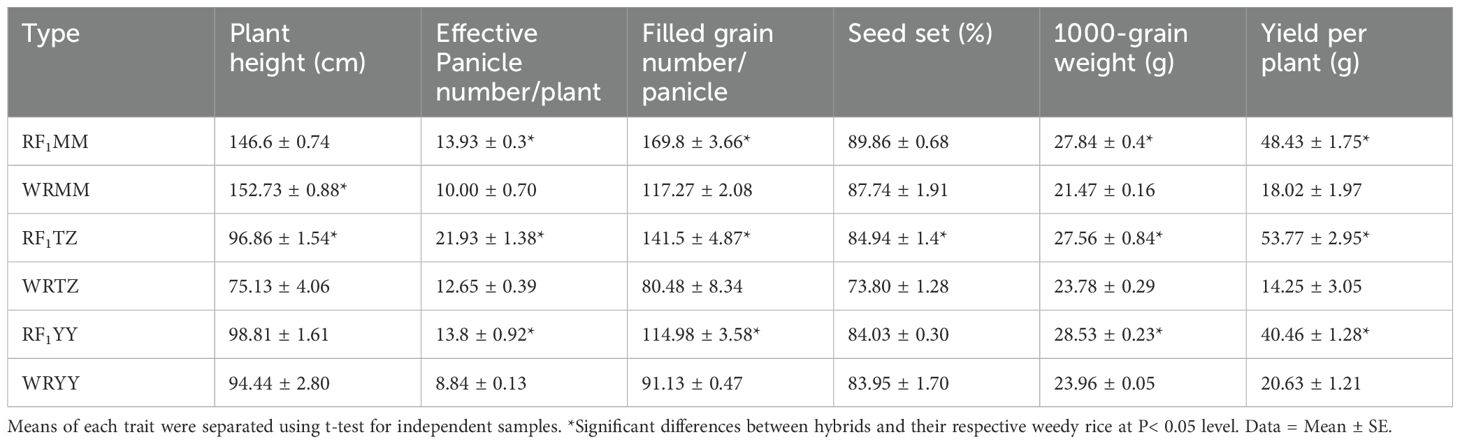
Table 6. Fitness-related traits of RF1 and weedy rice under natural insect pressure in pure planting.
Under natural insect pressure, RF1 plants produced approximately 39.3-73.4% more panicles per plant and 15.9-29.7% greater 1000-grain weight compared to their respective weedy rice parents. Additionally, RF1MM, RF1TZ, and RF1YY produced 44.8%, 75.8%, and 26.2% more filled grains per panicle and 168.8%, 277.3% and 96.1% yield per plant, respectively, than their weedy rice counterparts (Table 6).
In general, RF1 plants had significantly greater composite fitness than their weedy rice counterparts, attributable to higher fecundity-related traits under both insect pressure or no insect pressure (Figure 9).
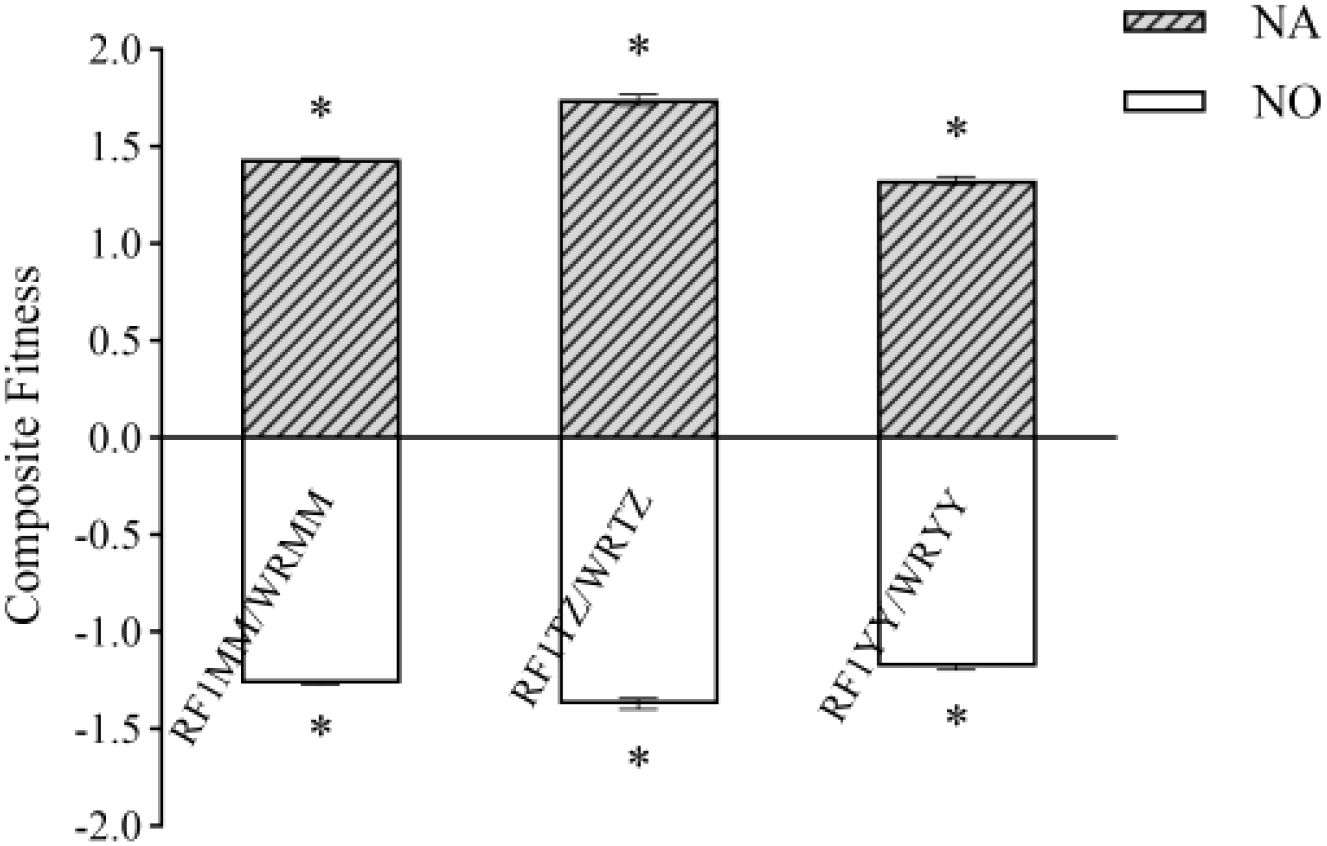
Figure 9. Fitness related traits of RF1 and its weedy rice paternal parent under natural or no insect pressure in monoculture planting. NA indicates under natural insect pressure; NO indicates under no insect pressure. (P < 0.05, error bar: SE, n=4). Means of composite fitness was separated using t-test for independent samples. * Significant differences between hybrids and their respective weedy rice at P < 0.05 level.
3.2.4 Fitness of RF1 and F1
When comparing the composite fitness of RF1 and F1, RF1 plants were significantly more fit than F1 plants under both under insect or no insect pressure (Figure 10).
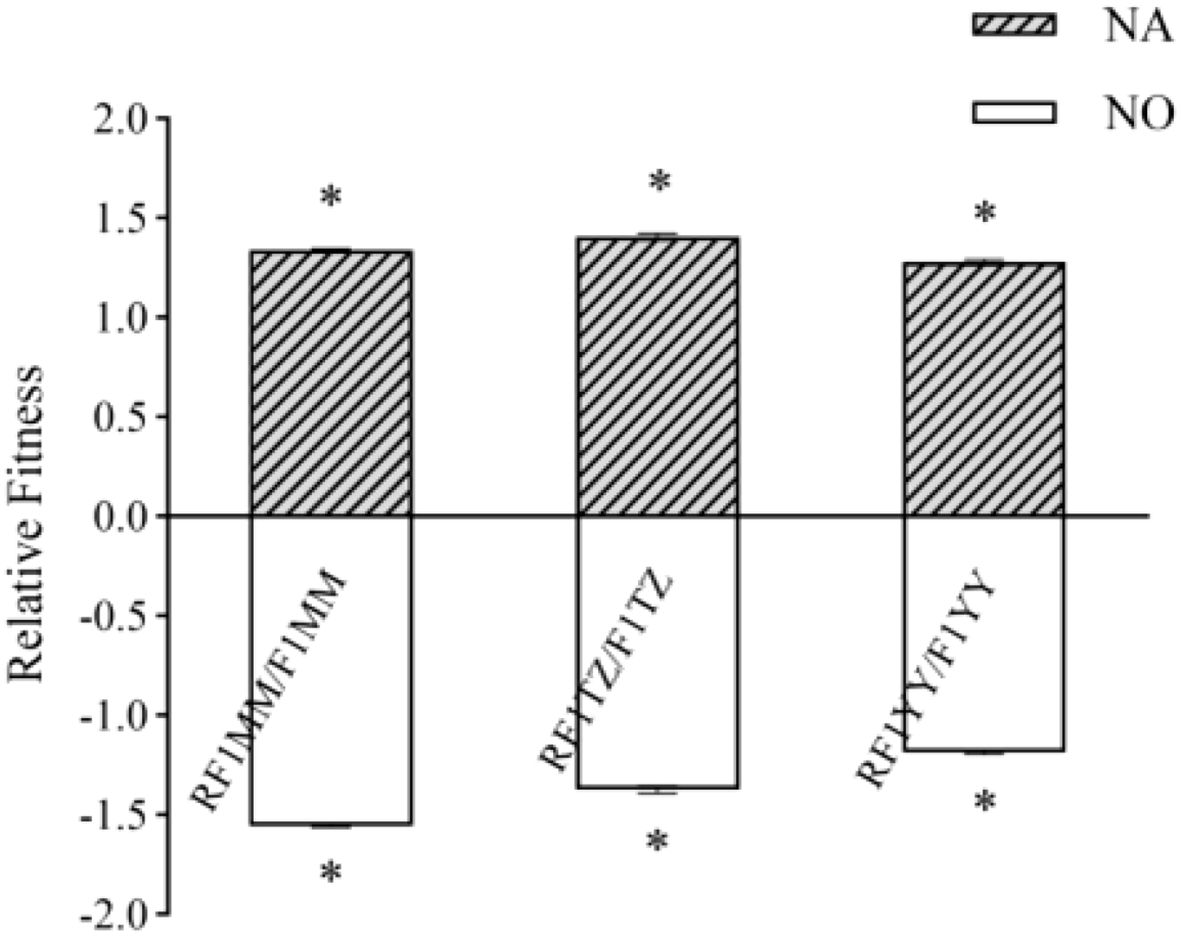
Figure 10. Fitness related traits of RF1 (T1c-19 as maternal parent) and F1 (T1c-19 as paternal parent) under natural or no insect pressure in monoculture planting. NA indicates under natural insect pressure; NO indicates under no insect pressure (P < 0.05, error bar: SE, n=4). Means of composite fitness was separated using t-test for independent samples. * Significant differences between RF1 hybrids and F1 hybrids at P <0.05 level.
3.3 Seed shattering of RF1 and weedy rice plants
Among the three weedy rice accessions, WRTZ had the highest seed shattering (19.80%), while WRMM and WRYY had similar seed shattering of 12.93% and 12.00%, respectively. Compared to their weedy rice counterparts, the seed shattering of the three RF1 plants was significantly lower by 5.78-11.12%. There was no significant difference in seed shattering among the RF1 plants (Figure 11).
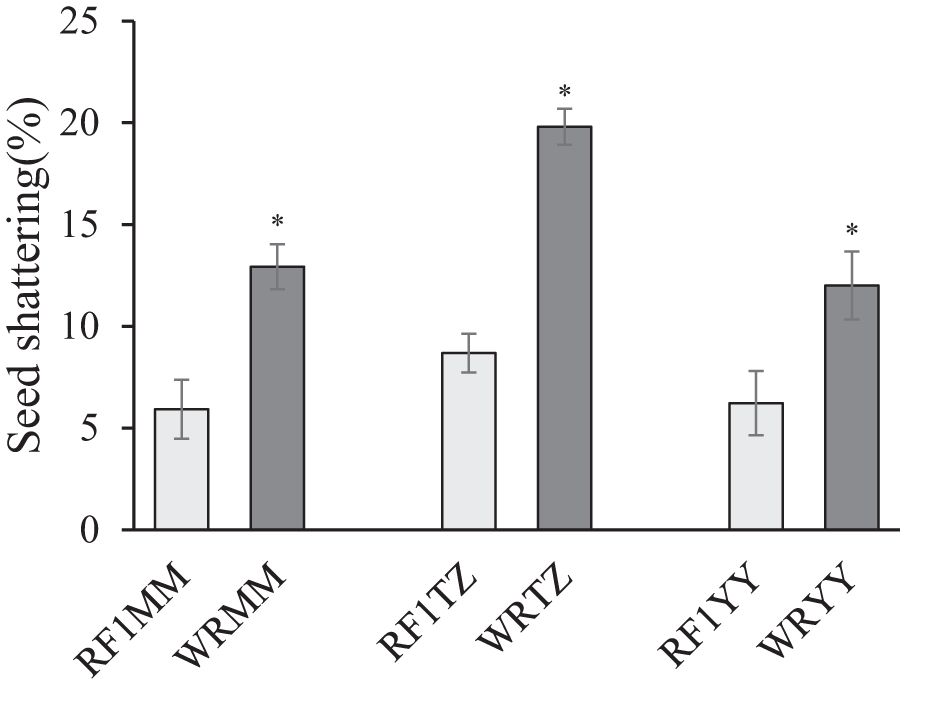
Figure 11. Seed shattering of weedy rice accessions and RF1 hybrids with T1c-19 (as the maternal plant) and weedy rice accessions (as paternal plants). WRMM, WRTZ and WRYY are weedy rice collected from Maoming, Taizhou and Yiyang, respectively. Data are mean ± SE (n=4). Means was separated using t-test for independent samples. * Significant differences between hybrids and their respective weedy rice at P< 0.05 level.
4 Discussion
4.1 Gene flow from weedy rice to cultivated rice
With respect to HR crops, gene flow does not differ whether the HR trait is introduced via genetic engineering or via conventional breeding techniques (Mallory-Smith and Olguin, 2011). Once the HR hybrids are produced through pollen-mediated gene flow from weedy rice to HR cultivated rice, the HR gene could further disperse to other weedy rice populations increasing the frequency of HR weedy rice.
In our preliminary research, we established a method for verifying F1 hybrids produced by reverse gene flow from weedy rice with red pericarp to cultivated rice (with white pericarp). This method used the 118 bp Rc gene of weedy rice as a marker. Sixty seeds collected from pollen recipient HR rice were grouped, ground to pass through a 60-mesh sieve, and tested for the Rc gene. This method proved effective, high-resolution, and cost-efficient for rapidly screening closely related cultivated rice hybrids produced by reverse gene flow.
In current research, reverse gene flow from three weedy rice accessions to T1c-19 differed: it was absent from WRTZ, while the frequencies were 0.0508% and 0.0808% from WRMM, and 0.0692% and 0.1008% from WRYY in alternating and adjacent cultivation designs, respectively. Thus, T1c-19 had the highest potential to evolve into HR weedy rice by accepting the pollen from WRYY, followed by WRMM. The probability of T1c-19 hybridizing with WRTZ was the lowest among the three weedy rice accessions.
Gene flow frequency varies depending on several factors, including reproductive compatibility between the rice cultivar and weedy rice biotype, sympatry, flowering synchrony, and length of overlapping flowering periods, plant height, experimental design, pollen source hectarage, and weather conditions (Zhang et al., 2003; Messeguer, 2003; Chen et al., 2004; Zhang et al., 2006; Shivrain et al., 2007, 2008; Zuo et al., 2011; Sun et al., 2015; Nam et al., 2019). Sexual reproduction of angiosperm involves a series of stages from pollen germination on the stigma to the development of the embryo and endosperm. Species that produce more seed under controlled pollination have a higher reproductive compatibility (Song et al., 2009). The differences of reverse gene flow among three weedy rice accessions likely relate to their sexual compatibility. In our previous experiment, seed setting was 39.8%, 10% and 6.3% when T1c-19 was manually crossed with WRYY, WRMM and WRTZ, respectively (unpublished data). The extent of sexual compatibility followed the same trend as the reverse gene flow frequency. Therefore, to avoid reverse gene flow, HR rice with lower sexual compatibility with local weedy rice should be cultivated.
Besides sexual compatibility, gene flow frequency is also influenced by pollen load (the number of pollen grains reaching the spikes of pollen recipients), which is related to overlapping flowering periods, coinciding daily flowering rhythms, and relative plant height of pollen donor and recipient (Shivrain et al., 2009; Zuo et al., 2011; Sun et al., 2015; Nam et al., 2019).
In this study, compared to T1c-19, WRMM was taller, and WRYY was similar, which likely facilitated the pollen of WRYY and WRMM in reaching the spikes of the shorter T1c-19, thereby increasing the chance for outcrossing. Conversely, WRTZ was shorter than T1c-19, likely decreasing the chance for outcrossing. This was the main reason for the absence of gene flow from T1c-19 to WRTZ.
In our previous research, in alternating and adjacent cultivations, the gene flow frequencies from T1c-19 to WRYY was 0.164 and 0.230%, and to WRTZ, 0.106 and 0.211%, respectively. However, no gene flow was detected with WRMM. The differences between direct and reverse gene flow results may be attributed to the shorter overlapping flowering periods between WRMM and T1c-19, and the much taller stature of WRMM compared to the pollen donor T1c-19. Additionally, the sexually compatibility of T1c-19 as maternal plant may differ with that as paternal plant, and variations during the two experimental periods, despite similar experimental designs, may have contributed to the results (Huang et al., 2015).
In conclusion, bidirectional gene flow between T1c-19 and three weedy rice accessions could produce HR weedy rice, complicating the evolution of HR weedy rice.
4.2 Effects of stacked transgenes on the fitness of F1 hybrids
The ecological risk of gene flow from cultivated crops to compatible weedy relatives is determined by the fitness of the F1 hybrids and subsequent generations (Song et al., 2009; 2011; Xia et al., 2011; Shukla et al., 2020). If the genes provide a selective advantage to crop-weed hybrids, greater fitness could lead to increased weediness (Chapman and Burke, 2006). The fitness of hybrids between insect or herbicide resistant crop and weedy relatives is affected by herbivore damage or herbicide pressure (Dechaine et al., 2009; Londo et al., 2010; Yang et al., 2011; 2012; 2015). Therefore, evaluating hybrids fitness under different selection pressures is essential for assessing gene flow risk for insect or herbicide resistant crops.
Compared to weedy rice, RF1 had significantly higher 1000-grain weight, yield per plant, and filled grain number per panicle, both with or without insect pressure. Thus, each plant of RF1 could produce 2–3 times of filled grains than weedy rice. It implied that RF1 hybrids could have a greater potential to produce more offspring, and the herbicide- and insect-resistant weedy rice populations could be more likely to establish population in field. Therefore, the higher composite fitness of RF1 means RF1 could cause potential ecological risk (Liu et al., 2016). Shivrain et al. (2009) reported that most hybrids produced by gene flow from weedy to cultivated rice yielded similar amounts of seed to that of the weedy parents. The differences in results between studies may be due to differences in parental genotype.
When comparing RF1 and F1, RF1MM and RF1TZ hybrids had 18.8-91.0% more panicles per plant, 26.4-31.3% more filled grains per panicle and 44.5-125.3% higher yield per plant than their F1 counterparts, regardless of insect pressure. RF1YY had 19.5% more panicles per plant under insect pressure and 12.3%-16.30% more filled grains per panicle, with a 30.7-35.0% higher yield per plant compared to RF1YY with or without insect pressure (Huang et al., 2019). In general, RF1 hybrids have a greater potential to produce more offspring, which could favor the establishment of herbicide- and insect-resistant weedy rice populations in field. Therefore, reverse gene flow may pose a significant ecological risk. This may be attributed to maternal effects in reciprocal crosses. Hybrids using cultivated crops as female parents result in weedy progenies with predominantly cultivated crop genes (De Wet and Harlan, 1975). Therefore, reverse gene flow from HR rice to weedy rice should receive careful attention. It is recommended that the reverse gene flow of HR rice, regardless of the method for cultivating it, should be evaluated before commercial release.
High seed shattering is a key trait for weedy rice, ensuring successful and efficient offspring dispersal (Wu et al., 2021). If weedy rice/cultivated rice hybrids have equal or higher seed shattering, they are more likely to escape harvesting and disperse seeds (Song et al., 2011). However, in this study, seed shattering characteristics of RF1 hybrids was partially suppressed compared to their weedy rice counterparts. The reduction in seed shattering could be unfavorable for RF1 hybrids, reducing their ability to avoid removal during grain harvest and persist in paddy fields. These findings are consistent with previous research on the seed shattering of F1 hybrids of weedy rice × T1c-19 (direct gene flow) (Huang et al., 2019), as well as studies on weedy rice × cultivated IMI-R rice (Shivrain et al., 2009; Serrat et al., 2013). The reason may be that F1 hybrids inherit the domestication trait of reduced seed shattering from cultivated rice. The major genes regulating seed shattering are Sh4 and qSH10 (Konishi et al., 2006; Li et al., 2006), Additionally, qSH1, OsCPL1, OsXTH8 and OsCel9 are important gene for seed shattering (Nunes et al., 2013). Other genes, including SH5, SHAT1, SSH1, SH1, OSH15, GRF4, and NPC1, are also involved in the loss of seed shattering (Wu et al., 2021). Further exploration of changes in these genes in hybrids compared to their parents is needed.
The lower seed shattering of RF1 hybrids implies that their seed would remain in the spike and thus be more likely removed during harvesting. This underscores the importance of using certificated cultivated rice seed to prevent the infestation of HR weedy rice mixed with cultivated rice seed (Serrat et al., 2013).
Seed dormancy, another key adaptive trait, regulates the timing of germination across seasons, enabling weedy rice to persist in agroecosystems (Gu et al., 2004; Pipatpongpinyo et al., 2020). Seed dormancy in rice interrelates to the weedy red pericarp color (Gu et al., 2005), and the pleiotropic locus most likely controls the dormancy and pigment traits by regulating ABA and flavonoid biosynthetic pathways, respectively (Gu et al., 2011). Furthermore, the natural genes controlling seed dormancy in weedy rice are involved in regulation of soil seedbank longevity (Pipatpongpinyo et al., 2020). Although the RF1 hybrids may display lower seed dormancy compared to its weedy rice counterparts, which could be unfavorable for their persistence in field, the accumulation of seed dormancy genes from weedy rice in HR transgenic cultivated rice could result in persistence of RF1 in rice tillage systems. Seed dormancy in RF1 was not investigated in this study and should be further researched.
In conclusion, gene flow via pollen from weedy rice to transgenic HR and IR rice can occur in natural field conditions, and HR hybrids produced by this reverse gene flow could survive in rice fields. Thus, measures, such as cultivating transgenic crop varieties which was cleistogamy, male sterility, chloroplast targeting and tandemly coupled mitigation genes, transgene excision, deleting transgene, or displayed lower genetic compatibility, or asynchronous flowering period with the associated weedy relatives, should be implemented (Madsen et al., 2002; Gressel and Valverde, 2009; Moon et al., 2010; Kwit et al., 2011; Clark and Maselko, 2020; Yang et al., 2021; Huang et al., 2023; Duan et al., 2023). Meanwhile, effective integrated weed management needs to be considered, such as using uncontaminated, certified seeds, applying the herbicide pre-emergence followed by post-emergence, clean machinery and crop rotation, must be taken to mitigate both reverse and direct gene flow (Roso et al., 2010; Sudianto et al., 2013; Avila et al., 2021; Unan et al., 2024).
Besides gene flow, herbicide selection drives the evolution of HR weedy rice (Qiu et al., 2020). Weedy rice can become acquired herbicide resistance through target-site resistance (TSR) or non–target site resistance (NTSR) when the Clearfield® (acetolactate synthase, ALS-inhibiting herbicide imidazolinone) or Provisia™ (acetyl-coenzyme A carboxylase, ACCase-inhibiting herbicide) rice is planted (Shivrain et al., 2010; Kaloumenos et al., 2013; Yean et al., 2021; Ruzmi et al., 2020, 2021; Gonzalez-Torralva and Norsworthy, 2023; Unan et al., 2024). HR weedy rice plants will be reduced effectiveness of the HR rice technology, and increased production costs and reduced yield (Avila et al., 2021; Ruzmi et al., 2020, 2021; Gonzalez-Torralva and Norsworthy, 2023; Unan et al., 2024).
HR weedy rice may evolve through bidirectional gene flow between rice and weedy rice, accompanied by repeated herbicide selection. Intensive adoption of HR rice technology, whether transgenic or non-transgenic, necessitates full attention for the management of HR weedy rice populations.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author/s.
Author contributions
G-LX: Formal analysis, Data curation, Writing – original draft, Writing – review & editing. J-QS: Data curation, Writing – original draft, Writing – review & editing. MW: Investigation, Data curation, Formal analysis, Writing – original draft. J-KL: Investigation, Data curation, Formal analysis, Writing – original draft. YH: Investigation, Data curation, Writing – original draft. SQ: Supervision, Project administration, Writing – review & editing, Funding acquisition. X-LS: Supervision, Validation, Methodology, Funding acquisition, Project administration, Writing – original draft, Writing – review & editing. W-MD: Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Jiangsu Provincial Key Research and Development Program (BE2022388: Creation and Application of Remote Quantitative Monitoring System for Green Weed Control Technology in Rice/Wheat Rotation Fields) and the National Natural Science Foundation of China (32071656).
Acknowledgments
The authors would like to express their deep appreciation to Bernal E. Valverde, the guest professor of Nanjing Agricultural University, China and Professor Do-Soon Kim, Seoul National University for the insightful comments and English editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1513367/full#supplementary-material.
References
Arrieta-Espinoza, G., Sánchez, E., Vargas, S., Lobo, J., Quesada, T., and Espinoza, A. M. (2005). The weedy rice complex in Costa Rica. I. Morphological study of relationships between commercial rice varieties, wild Oryza relatives and weedy types. Genet. Resour. Crop Evol. 52, 575–587. doi: 10.1007/s10722-004-6109-x
Avila, L., Marchesan, E., Camargo, E., Merotto, A., Ulguim, A., Noddin, J., et al. (2021). Eighteen years of Clearfield rice in Brazil: what have we learned? Weed Sci. 69, 585–597. doi: 10.1017/wsc.2021.49
Busconi, M., Baldi, G., Lorenzoni, C., Fogher, C., and Mendel, R. (2014). Gene flow from transgenic rice to red rice (Oryza sativa L.) in the field. Plant Biol. 16, 22–27. doi: 10.1111/plb.12021
Busconi, M., Rossi, D., Lorenzoni, C., Baldi, G., and Fogher, C. (2012). Spread of herbicide-resistant weedy rice (red rice, Oryza sativa L.) after 5 years of Clearfield rice cultivation in Italy. Plant Biol. 14, 751–759. doi: 10.1111/j.1438-8677.2012.00570.x
Cai, X. X., Wang, Z., Yuan, Y., Pang, L. H., Wang, Y., and Lu, B. R. (2023). Crop–Weed introgression plays critical roles in genetic differentiation and diversity of weedy rice: A case study of human-influenced weed evolution. Biology 12, 744. doi: 10.3390/biology12050744
Cao, Q. J., Xia, H., Yang, X., and Lu, B. R. (2009). Performance of hybrids between weedy rice and insect-resistant transgenic rice under field experiments: implication for environmental biosafety assessment. J. Integr. Plant Biol. 51, 1138–1148. doi: 10.1111/j.1744-7909.2009.00877.x
Chapman, M. A. and Burke, J. M. (2006). Letting the gene out of the bottle: the population genetics of genetically modified crops. New Phytol. 170, 429–443. doi: 10.1111/j.1469-8137.2006.01710.x
Chen, L., Gu, G., Wang, C. X., Chen, Z. F., Yan, W., Jin, M., et al. (2021). Trp548Met mutation of acetolactate synthase in rice confers resistance to a broad spectrum of ALS-inhibiting herbicides. Crop J. 9, 750–758. doi: 10.1016/j.cj.2020.11.003
Chen, L. J., Lee, D. S., Song, Z. P., Suh, H. S., and Lu, B. R. (2004). Gene flow from cultivated rice (Oryza sativa) to its weedy and wild relatives. Ann. Bot. 93, 67–73. doi: 10.1093/aob/mch006
Clark, M. and Maselko, M. (2020). Transgene biocontainment strategies for molecular farming. Front. Plant Sci. 11, 210. doi: 10.3389/fpls.2020.00210
Dai, L., Dai, W., Song, X., Lu, B., and Qiang, S. (2014). A comparative study of competitiveness between different genotypes of weedy rice (Oryza sativa L.) and cultivated rice. Pest Manage. Sci. 70, 113–122. doi: 10.1002/ps.3534
Dai, L., Song, X., He, B., et al. (2017). Enhanced photosynthesis endows seedling growth vigour contributing to the competitive dominance of weedy rice over cultivated rice. Pest Manage. Sci. 73, 1410–1420. doi: 10.1002/ps.4471
Darmency, H., Menchari, Y., Corre, V. L., and Delye, C. (2015). Fitness cost due to herbicide resistance may trigger genetic background evolution. Evolution 69-1, 271–278. doi: 10.1111/evo.12531
Dauer, J., Hulting, A., Carlson, D., Mankin, L., Hardencand, J., and Mallory-Smith, C. (2018). Gene flow from single and stacked herbicide-resistant rice (Oryza sativa): modeling occurrence of multiple herbicide-resistant weedy rice. Pest Manage. Sci. 74, 348–355. doi: 10.1002/ps.4711
Dechaine, J. M., Burger, J. C., Chapman, M. A., Seiler, G. J., Brunick, R., Knapp, S. J., et al. (2009). Fitness effects and genetic architecture of plant-herbivore interactions in sunflower crop-wild hybrids. New Phytol. 184, 828–841. doi: 10.1111/j.1469-8137.2009.02964.x
De Wet, J. M. J. and Harlan, J. R. (1975). Weeds and domesticates: evolution in the man-made habitat. Economic Bot. 29, 99–108. doi: 10.1007/BF02863309
Duan, Z. Z., He, M. Y., Akbar, S., Zhao, D. G., Zhang, M. Q., Li, Y., et al. (2023). Confirmation of ‘Pollen- and Seed-Specific Gene Deletor’ system efficiency for transgene excision from transgenic Nicotiana tabacum under field conditions. Int. J. Mol. Sci. 24, 1160. doi: 10.3390/ijms24021160
Gonzalez-Torralva, F. and Norsworthy, J. K. (2023). Quizalofop resistance in weedy rice (Oryza sativa L.) is mainly conferred by an Ile1781Leu mutation. Plant Sci. 336, 111838. doi: 10.1016/j.plantsci.2023.111838
Gressel, J. and Valverde, B. R. (2009). A strategy to provide long-term control of weedy rice while mitigating herbicide resistance transgene flow, and its potential use for other crops with related weeds. Pest Manage. Sci. 65, 723–731. doi: 10.1002/ps.1754
Gu, X. Y., Foley, M. E., Horvath, D. P., Anderson, J. V., Feng, J., Zhang, L., et al. (2011). Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates abscisic acid and flavonoid synthesis in weedy red rice. Genetics 189, 1515–1524. doi: 10.1534/genetics.111.131169
Gu, X. Y., Kianian, S. F., and Foley, M. E. (2004). Multiple loci and epistases control genetic variation for seed dormancy in weedy rice. Genetics 166, 1503–1516. doi: 10.1534/genetics.166.3.1503
Gu, X. Y., Kianian, S. F., and Foley, M. E. (2005). Seed dormancy imposed by covering tissues interrelates to shattering and seed morphological characters in weedy rice. Crop Sci. 45, 948–955. doi: 10.2135/cropsci2004.0339
Huang, D. B., Gao, L. W., McAdams, J., Zhao, F. Z., Lu, H. Y., Wu, Y. H., et al. (2023). Engineered cleistogamy in Camelina sativa for bioconfinement. Horticulture Res. 10, uhac280. doi: 10.1093/hr/uhac280
Huang, Y., Li, J. K., Qiang, S., Dai, W. M., and Song, X. L. (2016). Transgenic restorer rice line T1c-19 with stacked cry1C*/bar genes has low weediness potential without selection pressure. J. Integr. Agric. 15, 1046–1058. doi: 10.1016/S2095-3119(15)61219-9
Huang, Y., Li, J. K., Qiang, S., Luo, T. P., and Song, X. L. (2015). Gene flow from transgenic rice T1c-19 with stacked ow from transgenic rice T1c-19 with stacked cry1C*/bar ry1C*/bar genes to weedy genes to weedy and cultivated rice species and cultivated rice species. Chin. J. Appl. Environ. Biol. 21, 1112–1119. doi: 10.3724/SP.J.1145.2015.05030
Huang, Y., Wang, Y. Y., Qiang, S., Song, X. L., and Dai, W. M. (2019). Fitness of F1 hybrids between stacked transgenic rice T1c-19 with cry1C*/bar genes and weedy rice. J. Integr. Agric. 18, 2793–2805. doi: 10.1016/S2095-3119(19)62662-6
Jenczewski, E., Ronfort, J., and Chèvre, A. M. (2003). Crop-to-wild gene flow, introgression and possible fitness effects of transgenes. Environ. Biosafety Res. 2, 9–24. doi: 10.1051/ebr:2003001
Kaloumenos, S. N., Capote, N., Aguado, A., and Eleftherohorinos, G. I. (2013). Red rice (Oryza sativa) cross-resistance to imidazolinone herbicides used in resistant rice cultivars grown in northern Greece. Pesticide Biochem. Physiol. 105, 177–183. doi: 10.1016/j.pestbp.2013.01.008
Karn, E., De Leon, T., Espino, L., Al-Khatib, K., and Brim-DeForest, W. (2020). Phenotypic diversity of weedy rice (Oryza sativa f. spontanea) biotypes found in California and implications for management. Weed Sci. 68, 485–495. doi: 10.1017/wsc.2020.43
Konishi, S., Izawa, T., Lin, S. Y., Ebana, K., Fukuta, Y., Sasaki, T., et al. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312, 1392–1396. doi: 10.1126/science.1126410
Kwit, C., Moon, H. S., Warwick, S. I., and Stewart, C. N. (2011). Transgene introgression in crop relatives: molecular evidence and mitigation strategies. Trends Biotechnol. 29, 284–293. doi: 10.1016/j.tibtech.2011.02.003
Li, L. F., Pusadee, T., Wedger, M. J., Li, Y. L., Li, M. R., Lau, Y. L., et al. (2024). Porous borders at the wild-crop interface promote weed adaptation in Southeast Asia. Nat. Commun. 15, 1182. doi: 10.1038/s41467-024-45447-0
Li, X. Y., Qiang, S., Song, X. L., Cai, K., Sun, Y. N., Shi, Z. H., et al. (2014). Allele types of Rc gene of weedy rice from Jiangsu Province, China. Rice Sci. 21, 252–261. doi: 10.1016/S1672-6308(13)60183-3
Li, C. B., Zhou, A. L., and Sang, T. (2006). Rice domestication by reducing shattering. Science 311, 1936–1939. doi: 10.1126/science.1123604
Liu, S. N., Song, X. L., Hu, Y. H., Dai, W. M., and Qiang, S. (2016). Fitness of hybrids between two types of transgenic rice and six japonica and indica weed rice accessions. Crop Sci. 56, 2571–2765. doi: 10.2135/cropsci2015.11.0719
Londo, J. P., Bautista, N. S., Sagers, C. L., Lee, E. H., and Watrud, L. S. (2010). Glyphosate drift promotes changes in fitness and transgene gene flow in canola (Brassica napus) and hybrids. Ann. Bot. 106, 957–965. doi: 10.1093/aob/mcq190
Lu, B. R. and Yang, C. (2009). Gene flow from transgenic rice to its wild relatives: Assessing potential ecological consequences. Biotechnol. Adv. 27, 1083–1091. doi: 10.1016/j.biotechadv.2009.05.018
Ma, D. R., Chen, W. F., Xu, Z. J., and Zhang, W. Z. (2005). Occurrence and control measures of weedy rice in Liaoning Province. China Agric. Bull. 21, 358–360.
Madsen, K. H., Valverde, B. E., and Jensen, J. E. (2002). Risk assessment of herbicide-resistant crops: a Latin American perspective using rice (Oryza sativa) as a model. Weed Technol. 16, 215–223. doi: 10.1614/0890-037X(2002)016[0215:RAOHRC]2.0.CO;2
Mallory-Smith, C. A. and Olguin, E. S. (2011). Gene flow from herbicide-resistant crops: it’s not just for transgenes. J. Agric. Food Chem. 59, 5813–5818. doi: 10.1021/jf103389v
Merotto, A., Goulart, I. C. G. R., Nunes, A. L., Kalsing, A., Markus, C., Menezes, V. G., et al. (2016). Evolutionary and social consequences of introgression of nontransgenic herbicide resistance from rice to weedy rice in Brazil. Evolutionary Appl. 9, 837–846. doi: 10.1111/eva.12387
Messeguer, J. (2003). Gene flow assessment in transgenic plants. Plant Cell Tissue Organ Culture 73, 201–212. doi: 10.1023/A:1023007606621
Messeguer, J., Marfà, V., Català, M. M., Guiderdoni, E., and Melé, E. (2004). Field study of pollen-mediated gene flow from Mediterranean transgenic rice to conventional rice and the red rice weed. Mol. Breed. 13, 103–112. doi: 10.1023/B:MOLB.0000012285.39859.9d
Moon, H. S., Li, Y., and Stewart, C. N., Jr. (2010). Keeping the genie in the bottle: Transgene biocontainment by excision in pollen. Trends Biotechnol. 28, 3–8. doi: 10.1016/j.tibtech.2009.09.008
Nam, K. H., Kim, D. Y., Moon, Y. S., Pack, I. S., Jeong, S. C., Park, K. W., et al. (2019). Gene flow from transgenic PPO-inhibiting herbicide-resistant rice to weedy rice, and agronomic performance by their hybrids. J. Plant Biol. 62, 286–296. doi: 10.1007/s12374-019-0013-6
Nicolia, A., Manzo, A., Veronesi, F., and Rosellin, D. (2014). An overview of the last 10 years of genetically engineered crop safety research. Crit. Rev. Biotechnol. 34, 77–88. doi: 10.3109/07388551.2013.823595
Nunes, A. L., Delatorre, C. A., and Merotto, A. (2013). Gene expression related to seed shattering and the cell wall in cultivated and weedy rice. Plant Biol. 16, 888–896. doi: 10.1111/plb.12133
Olsen, K. M., Caicedo, A. L., and Jia, Y. (2007). Evolutionary Genomics of weedy rice in the USA. J. Integr. Plant Biol. 49, 811–816. doi: 10.1111/j.1744-7909.2007.00506.x
Pipatpongpinyo, W., Korkmaz, U., Wu, H., Kena, A., Ye, H., Feng, J., et al. (2020). Assembling seed dormancy genes into a system identified their effects on seedbank longevity in weedy rice. Heredity 124, 135–145. doi: 10.1038/s41437-019-0253-8
Qiu, J., Jia, L., Wu, D., Weng, X., Chen, L. J., Sun, J., et al. (2020). Diverse genetic mechanisms underlie worldwide convergent rice feralization. Genome Biol. 21, 70. doi: 10.1186/s13059-020-01980-x
Qiu, J., Zhou, Y. J., Mao, L. F., Ye, C. Y., Wang, W. D., Zhang, J. P., et al. (2017). Genomic variation associated with local adaptation of weedy rice during de-domestication. Nat. Communication 24, 8:15323. doi: 10.1038/ncomms15323
Rajguru, S. N., Burgos, N. R., Shivrain, V. K., and Stewart, J.M. (2005). Mutations in the red rice ALS gene associated with resistance to imazethapyr. Weed Sci. 53, 567–577. doi: 10.1614/WS-04-111R1.1
Roso, A. C., Meroto, A., Delatorre, C. A., and Menezes, V. G. (2010). Regional scale distribution of imidazolinone herbicide-resistant alleles in red rice (Oryza sativa L.) determined through SNP markers. Field Crops Res. 119, 175–182. doi: 10.1016/j.fcr.2010.07.006
Ruzmi, R., Ahmad-Hamdani, M. S., Abidin, M. Z. Z., and Burgos, N. R. (2021). Evolution of imidazolinone-resistant weedy rice in Malaysia: The current status. Weed Sci. 69 (5), 598–608. doi: 10.1017/wsc.2021.33
Ruzmi, R., Ahmad-Hamdani, M. S., and Mazlan, N. (2020). Ser-653-Asn substitution in the acetohydroxyacid synthase gene confers resistance in weedy rice to imidazolinone herbicides in Malaysia. PloS One 15, e0227397. doi: 10.1371/journal.pone.0227397
Sanchez-Olguin, E. R., Arrieta-Espinoza, G., Lobo, J. A., and Espinoza-Esquivel, A. M. (2009). Assessment of gene flow from a herbicide-resistant indica rice (Oryza sativa L.) to the Costa Rican weedy rice (Oryza sativa) in Tropical America: factors affecting hybridization rates and characterization of F1 hybrids. Transgenic Res. 18, 633–647. doi: 10.1007/s11248-009-9255-2
Serrat, X., Esteban, R., Messeguer, J., Peas, G., Catala, M. M., Melé, E., et al. (2013). Direct and reverse pollen-mediated gene flow between GM rice and red rice weed. AoB Plants 5, 1–12. doi: 10.1093/aobpla/plt050
Shivrain, V. K., Burgos, N. R., Anders, M. M., Rajguru, S. N., Moore, J., and Sales, M. A. (2007). Gene flow between Clearfield rice and red rice. Crop Prot. 26, 349–356. doi: 10.1016/j.cropro.2005.09.019
Shivrain, V. K., Burgos, N. R., Gealy, D. R., Moldenhauer, K. A. K., and Baquireza, C. J. (2008). Maximum outcrossing rate and compatibility between red rice (Oryza sativa) biotypes and Clearfield rice. Weed Sci. 56, 807–813. doi: 10.1614/WS-08-026.1
Shivrain, V. K., Burgos, N. R., Gealy, D. R., Sales, M. A., and Smith, K. L. (2009). Gene flow from weedy red rice (Oryza sativa L.) to cultivated rice and fitness of hybrids. Pest Manage. Sci. 65, 1124–1129. doi: 10.1002/ps.1802
Shivrain, V. K., Burgos, N. R., Sales, M. A., and Kuk, Y. I. (2010). Polymorphisms in the ALS gene of weedy rice (Oryza sativa L.) accessions with differential tolerance to imazethapyr. Crop Prot. 29, 336–341. doi: 10.1016/j.cropro.2009.10.002
Shukla, K., Sbrizzi, S., Laursen, A. E., Benavides, J., and Campbell, L. G. (2020). Hybridization slows rate of evolution in crop-wild compared to wild populations of weedy Raphanus across a moisture gradient. Front. Agron. 2, 600346. doi: 10.3389/fagro.2020.600346
Song, X. L., Liu, L. L., Wang, Z., and Qiang, S. (2009). Potential gene flow from transgenic rice (Oryza sativa L.) to different weedy rice (Oryza sativa f. spontanea) accessions based on reproductive compatibility. Pnsect Manage. Sci. 65, 862–869. doi: 10.1002/ps.1766
Song, X. L., Wang, Z., and Qiang, S. (2011). Agronomic performance of F1, F2 and F3 hybrids between weedy rice and transgenic glufosinate-resistant rice. Pest Manage. Sci. 67, 921–931. doi: 10.1002/ps.2132
Sudianto, E., Beng-Kah, S., Ting-Xiang, N., Saldain, E. N., Scott, C. R., and Burgos, R. N. (2013). Clearfield® rice: Its development, success, and key challenges on a global perspective. Crop Prot. 49, 40–51. doi: 10.1016/j.cropro.2013.02.013
Sun, G. H., Dai, W. M., Cui, R. R., Qiang, S., and Song, X. L. (2015). Gene flow from glufosinate-resistant transgenic hybrid rice Xiang 125S/Bar68–1 to weedy rice and cultivated rice under different experimental designs. Euphytica 204, 211–227. doi: 10.1007/s10681-015-1370-y
Sun, J., Ma, D., Tang, L., Zhao, M., Zhang, G., Wang, W., et al. (2019). Population genomic analysis and De novo assembly reveal the origin of weedy rice as an evolutionary game. Mol. Plant 12, 632–647. doi: 10.1016/j.molp.2019.01.019
Sweeney, M. T., Thomson, M. J., Cho, Y. G., Park, Y. J., Williamson, S. H., Bustamante, C. D., et al. (2007). Global dissemination of a single mutation conferring white pericarp in rice. PloS Genet. 3, 121–133. doi: 10.1371/journal.pgen.0030133
Tang, W., Chen, H., Xu, C. G., Li, X. H., Lin, Y. J., and Zhang, Q. F. (2006). Development of insect-resistant transgenic indica rice with a synthetic cry1C* gene. Mol. Breed. 18, 1–10. doi: 10.1007/s11032-006-9002-9
Thurber, C. S., Hepler, P. K., and Caicedo, A. L. (2011). Timing is everything: early degradation of abscission layer is associated with increased seed shattering in U.S. weedy rice. BMC Plant Biol. 11, 14. doi: 10.1186/1471-2229-11-14
Unan, R., Azapoglu, O., Deligoz, I., Mennan, H., and Al-Khatib, K. (2024). Gene flow and spontaneous mutations are responsible for imidazolinone herbicide-resistant weedy rice (Oryza sativa L.). Pesticide Biochem. Physiol. 198, 105746. doi: 10.1016/j.pestbp.2023.105746
Vila-Aiub, M. M. (2019). Fitness of herbicide-resistant weeds: current knowledge and implications for management. Plants 8, 469. doi: 10.3390/plants8110469
Vila-Aiub, M. M., Neve, P., and Powles, S. B. (2009). Fitness costs associated with evolved herbicide resistance alleles in plants. New Phytol. 184, 751–767. doi: 10.1111/j.1469-8137.2009.03055.x
Wang, M., DAI, W. M., Qiang, S., and Song, X. L. (2020). Research on sampling technique for verifying gene flow frequency of transgenic rice. J. Nanjing Agric. Univ. 43, 260–266. doi: 10.7685/jnau.201904024
Wang, H. Q., Dai, W. M., Zhang, Z. X., Li, M. S., Meng, L. C., Zhang, Z., et al. (2023). Occurrence pattern and morphological polymorphism of Chinese weedy rice. J. Integr. Agric. 22, 149–169. doi: 10.1016/j.jia.2022.08.001
Wu, D., Lao, S., and Fan, L. (2021). De-domestication: an extension of crop evolution. Trends Plant Sci. 26, 560–574. doi: 10.1016/j.tplants.2021.02.003
Xia, H., Lu, B. R., Xu, K., Wang, W., Yang, X., Yang, C., et al. (2011). Enhanced yield performance of Bt rice under target-insect attacks: implications for field insect management. Transgenic Res. 20, 655–664. doi: 10.1007/s11248-010-9449-7
Yang, C., Ge, J., Fu, X. K., Luo, K. M., and Xu, C. Z. (2021). Dual reproductive Cell-Specific Promoter-Mediated Split-Cre/LoxP System suitable for exogenous gene deletion in hybrid progeny of transgenic Arabidopsis. Int. J. Mol. Sci. 22, 5080. doi: 10.3390/ijms22105080
Yang, X., Li, L., Cai, X. X., Wang, F., Su, J., and Lu, B. R. (2015). Efficacy of insect-resistance Bt/CpTI transgenes in F5-F7 generations of rice crop-weed hybrid progeny: implications for assessing ecological impact of transgene flow. Sci. Bull. 60, 1563. doi: 10.1007/s11434-015-0885-x
Yang, X., Wang, F., Su, J., and Lu, B. R. (2012). Limited fitness advantages of crop-weed hybrids containing insect-resistant transgenes (Bt/CpTI) in transgenic rice field. PloS One 7, 398–398. doi: 10.1371/journal.pone.0041220
Yang, X., Xia, H., Wang, W., Wang, F., Su, J., Snow, A. A., et al. (2011). Transgenes for insect resistance reduce herbivory and enhance fecundity in advanced generations of crop-weed hybrids of rice. Evolutionary Appl. 4, 672–684. doi: 10.1111/j.1752-4571.2011.00190.x
Yean, R., Dilipkumar, M., Rahman, S., and Song, B. K. (2021). A two-in-one strategy: target and nontarget site mechanisms both play important role in IMI-resistant weedy rice. Int. J. Mol. Sci. 22, 982. doi: 10.3390/ijms22030982
Zhang, J. X., Kang, Y., Valverde, E. B., Dai, W. M., Song, X. L., and Qiang, S. (2018). Feral rice from introgression of weedy rice genes into transgenic herbicide-resistant hybrid-rice progeny. J. Exp. Bot. 69, 3855–3865. doi: 10.1093/jxb/ery210
Zhang, N. Y., Linscombe, S., and Oard, J. (2003). Out-crossing frequency and genetic analysis of hybrids between transgenic glufosinate herbicide-resistant rice and the weed, red rice. Euphytica 130, 35–45. doi: 10.1023/A:1022371104679
Zhang, W. Q., Linscombe, S. D., and Oard, J. H. (2008). Genetic and agronomic analyses of red rice-Clearfield hybrids and their progeny produced from natural and controlled crosses. Euphytica 164, 659–668. doi: 10.1007/s10681-008-9661-1
Zhang, W. Q., Linscombe, S. D., Webster, E., Tan, S. Y., and Oard, J. (2006). Risk assessment of the transfer of imazethapyr herbicide tolerance from Clearfield rice to red rice (Oryza sativa). Euphytica 152, 75–86. doi: 10.1007/s10681-006-9180-x
Zhao, C., Xu, W. R., Meng, L. C., Qiang, S., Dai, W. M., Zhang, Z., et al. (2020). Rapid endosperm development promotes early maturity in weedy rice (Oryza sativa f. spontanea). Weed Sci. 68, 168–178. doi: 10.1017/wsc.2020.5
Zhao, C., Xu, W. R., Song, X. L., Dai, W. M., Dai, L., Zhang, Z., et al. (2018). Early flowering and rapid grain filling determine early maturity and escape from harvesting in weedy rice. Pest Manage. Sci. 74, 465–476. doi: 10.1002/ps.4730
Zhao, C., Xu, W., Zhang, Z., Meng, L. C., Dai, W. M., Qiang, S., et al. (2021). The rapid cytological process of grain determines early maturity in weedy rice. Front. Plant Sci. 12, 711321. doi: 10.3389/fpls.2021.711321
Keywords: transgenes, herbicide-resistant rice, T1c-19, weedy rice, reverse gene flow, reverse F1 hybrids, fitness
Citation: Xie G-L, Shen J-Q, Wang M, Li J-K, Huang Y, Qiang S, Song X-L and Dai W-M (2025) Gene flow from weedy rice to T1c-19 transgenic rice stacked with cry1C*/bar genes and fitness of F1 hybrids. Front. Plant Sci. 16:1513367. doi: 10.3389/fpls.2025.1513367
Received: 07 November 2024; Accepted: 12 June 2025;
Published: 22 July 2025.
Edited by:
Dayun Tao, Yunnan Academy of Agricultural Sciences, ChinaReviewed by:
Tian Qing Zheng, Chinese Academy of Agricultural Sciences, ChinaShamseldeen Shehabeldin Eltaher, University of Sadat City, Egypt
Copyright © 2025 Xie, Shen, Wang, Li, Huang, Qiang, Song and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Ling Song, c3hsQG5qYXUuZWR1LmNu; Wei-Min Dai, ZGFpd2VpbWluNEBuamF1LmVkdS5jbg==
 Guang-Le Xie
Guang-Le Xie Jia-Qi Shen
Jia-Qi Shen Xiao-Ling Song
Xiao-Ling Song