- Department of Biology, KU Leuven, Leuven, Belgium
Vernalisation is a prolonged cold exposure that synchronises flowering with favourable seasonal conditions, protecting reproductive development from winter stress and optimising crop yields. By ‘recording’ winter, this biological process displays memory and becomes progressively more difficult to reverse. In the well-studied model Arabidopsis, vernalisation epigenetically silences the gene locus of the floral repressor FLC. Temperate cereals, including crops such as wheat and barley, respond to a similar vernalisation cue with a memory property and recent functional studies also support an epigenetic mechanism. Current evidence points to the flowering promoter VRN1 as the primary site for storing this memory. Because vernalisation in cereals appears to rely on epigenetic activation of VRN1, rather than repression as in Arabidopsis, the specific histone marks responsible for storing this epigenetic memory are possibly different. This highlights the need for further research to identify the specific genes and histone modifications involved, and to fully elucidate the mechanisms underlying vernalisation memory in cereals. The goal of this review is to synthesise recent advances in our understanding of the epigenetic regulation of vernalisation in temperate cereals. Therefore, this review focuses on the roles of key genes such as VRN1, VRN3, and ODDSOC2, and examines the dynamic chromatin landscape associated with vernalisation-induced flowering. In particular, we investigate the possible interplay of chromatin marks involved in the epigenetic activation of VRN1. By synthesising current knowledge and highlighting unresolved questions, this review aims to provide a framework for future research in the field of cereal vernalisation memory.
Introduction
Plants, as immobile organisms, have evolved sophisticated mechanisms to sense and integrate environmental cues, ensuring that key developmental transitions occur under favourable conditions. A prime example of such a sophisticated mechanisms is vernalisation, the requirement for a prolonged period of cold to induce flowering (Chouard, 1960; Sung and Amasino, 2004). By gradually sensing prolonged exposure to cold temperatures, plants initiate physiological changes that accelerate the transition from the vegetative to the reproductive stage, ensuring that flowering occurs only in spring. This adaptive strategy helps to protect developing floral structures from harsh winter conditions and prevents premature seed set (Sung and Amasino, 2004; Trevaskis et al., 2007). Vernalisation is particularly critical for winter varieties of temperate cereal crops, such as wheat, barley, and oats, that are cultivated in regions with distinct seasonal changes and temperate climates. These winter varieties are preferred in these climates because they demonstrate superior yield, explained by the prolonged vegetative growth they undergo before transitioning to reproduction. The ability to synchronise flowering with favourable seasonal conditions not only underscores the fundamental biological importance of vernalisation but also highlights its critical role in agriculture, where flowering time directly influences crop yield and stability (Wang et al., 2023; Wu et al., 2017). As climate patterns grow increasingly unpredictable, understanding the mechanisms of vernalisation acquires further importance in developing resilient crop varieties.
Through the mechanism behind vernalisation, plants acquire the ability to “remember” winter. Because epigenetic mechanisms change at a slower rate than e.g. transcriptional changes, the memory property of vernalisation is probably encoded at the epigenetic level in the different species that developed a vernalisation response. Vernalisation research in Arabidopsis thaliana (Arabidopsis) has been pivotal in uncovering the molecular basis of this epigenetic memory. In this species, the floral repressor FLOWERING LOCUS C (FLC) is epigenetically silenced during extended cold, thereby lifting repression and permitting flowering when temperatures rise (Song et al., 2012; Sung and Amasino, 2004). The degree of histone modification at the FLC locus quantitatively reflects the duration of cold exposure, acting as a molecular record that ensures flowering only after the vernalisation requirement is met. This model system has also illuminated the roles of specific histone modifiers, non-coding RNAs, and feedback mechanisms that stabilise FLC repression and maintain the vernalised state (Whittaker and Dean, 2017).
In contrast, the mechanisms underlying vernalisation memory in temperate cereals differ in two fundamental ways. First, while Arabidopsis stores vernalisation memory through the stable deposition of repressive histone marks at a single locus, it remains unclear whether vernalisation memory in cereals is confined to VRN1 or involves coordinated regulation across multiple genes and genomic loci. Second, rather than repressing a floral inhibitor, temperate cereals appear to rely on the epigenetic activation of a floral promoter gene, VERNALISATION1 (VRN1), which promotes the transition to flowering (Dixon et al., 2019; Shitsukawa et al., 2007; Trevaskis et al., 2003; Yan et al., 2003). Furthermore, the specific histone modifications responsible for maintaining this irreversible active state in cereals have yet to be identified. These fundamental differences underscore the complexity and diversity of vernalisation mechanisms among plant species and highlights significant gaps in our current understanding of cereal vernalisation memory.
In this review, we synthesise recent advances in the field, with a particular emphasis on the roles of key genes such as VRN1, VRN3, and ODDSOC2, and the dynamic chromatin landscape that underpins vernalisation-induced flowering. We discuss the interplay of chromatin marks involved in the epigenetic activation of VRN1, highlight unresolved questions, and propose a framework for future research to advance our understanding of vernalisation memory in cereals.
Genetic basis of vernalisation
The transition to flowering in temperate cereals is a tightly regulated developmental process that ensures reproductive development takes place only when environmental conditions are optimal. This precise timing is orchestrated by a complex genetic network centred around the VERNALISATION (VRN) genes: VRN1, VRN2, and VRN3 (Dennis and Peacock, 2009; Greenup et al., 2009). Notably, these genes are distinct from the vernalisation genes of Arabidopsis, even though their names are identical. The fact that different genes control the same process suggests that the vernalisation pathways in temperate cereals and dicots have evolved independently (Andrés and Coupland, 2012; Dennis and Peacock, 2009).
VRN1 encodes a protein from the MADS-box transcription factor family, homologous to the Arabidopsis genes AP1 and FUL (Trevaskis et al., 2003; Yan et al., 2003). VRN1 possesses a characteristic MIKC-type domain structure, which includes four conserved domains: the MADS (M), Intervening (I), Keratin-like (K), and C-terminal (C) domains (Figure 1) (Egea-Cortines et al., 1999; Melzer et al., 2009; Riechmann and Meyerowitz, 1997; Shore and Sharrocks, 1995). The MADS domain is a highly conserved DNA-binding motif crucial for recognising and regulating the expression of target genes involved in floral development (West et al., 1997). The I domain provides specificity for protein dimerisation, helping VRN1 select its interaction partners. The K domain is essential for protein-protein interactions, facilitating the formation of higher-order complexes with other MADS-box proteins, which is vital for the regulation of flowering. The C-terminal domain contributes to transcriptional activation and further protein-protein interactions. Importantly, MADS-box transcription factors such as VRN1 often function not as single proteins but as part of multimeric complexes, typically dimers or higher-order assemblies, which enable combinatorial control of gene expression and allow for precise regulation of developmental processes (Immink et al., 2009; Smaczniak et al., 2012; Theißen and Saedler, 2001). In cereals, VRN1 directly binds to promoters of key flowering genes, including VRN3, and flowering repressors such as VRN2 and ODDSOC2, thereby promoting flowering after vernalisation (Figure 2) (Chen and Dubcovsky, 2012; Deng et al., 2015; Oliver et al., 2009; Woods et al., 2016). This modular MIKC structure enables VRN1 to function as a transcriptional regulator, integrating vernalisation signals by altering gene expression profiles that trigger the transition from vegetative to reproductive growth (Distelfeld and Dubcovsky, 2009; Trevaskis et al., 2007).
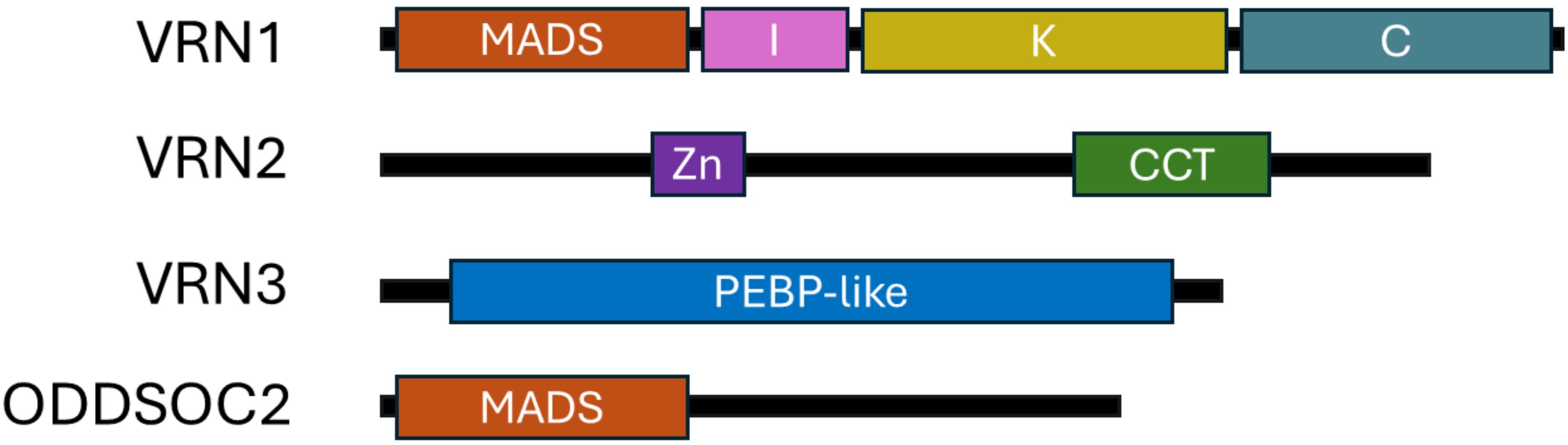
Figure 1. Structural domain composition VRN1, VRN2, VRN3, and ODDSOC2. Schematic representation of the structural domains present in VRN1, VRN2, VRN3, and ODDSOC2, showing the arrangement of MADS, intervening (I), keratin-like (K), C-terminal, and PEBP domains to highlight differences in domain composition among these key flowering regulators from Brachypodium distachyon. Created in BioRender. Daneels, T. (2025) https://BioRender.com/lo4b3yy.
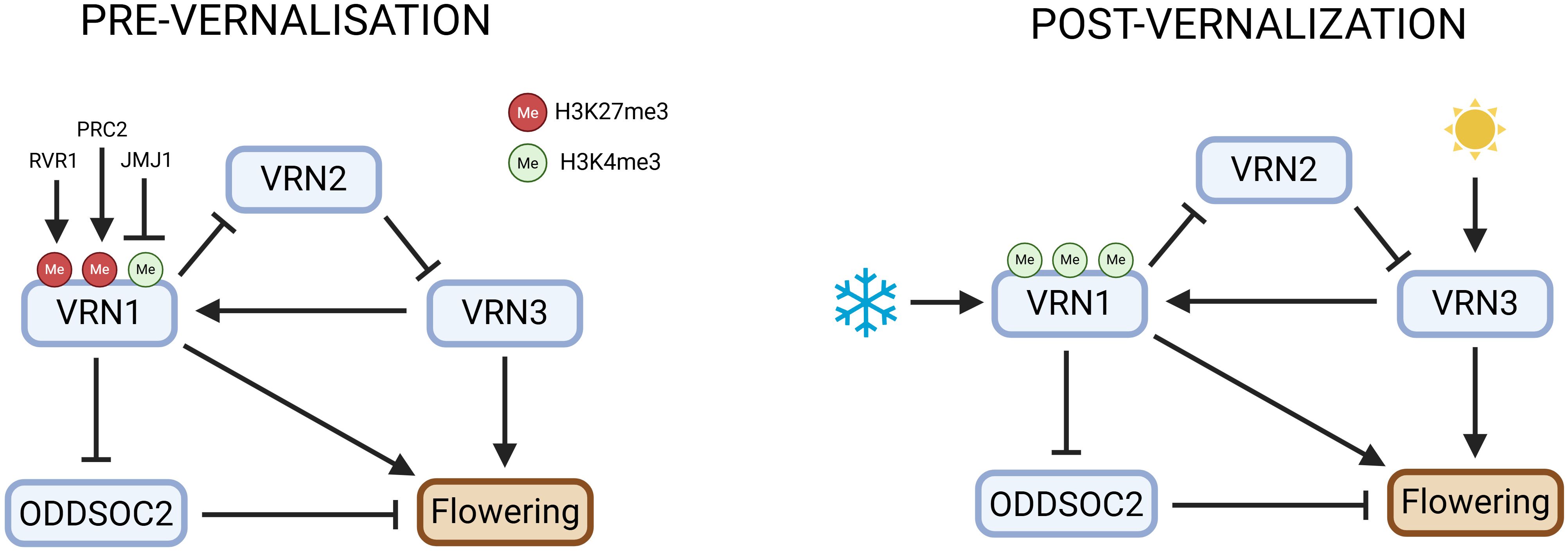
Figure 2. Genetic and epigenetic network controlling vernalisation and flowering in temperate cereals. This figure illustrates the genetic and epigenetic regulation of flowering in temperate cereals in response to cold and light: before vernalisation, VRN1 is repressed by high H3K27me3 (via PRC2/EZL1), as well as by the pre-vernalisation repressors RVR1 and JMJ1, with JMJ1 demethylating H3K4 to further silence VRN1; this maintains active VRN2 and ODDSOC2, which keep VRN3 inhibited. During and after cold exposure, VRN1 is activated through loss of H3K27me3 and gain of H3K4me3, leading to stable expression and winter memory; VRN1 then represses VRN2 and ODDSOC2, releasing VRN3 from inhibition. VRN3 activation is marked by increased H3K4me3, especially under long days. ODDSOC2 repression involves increased H3K27me3 in Brachypodium, while in wheat and barley, its downregulation is associated with reduced H3K4me3 and is mediated by VRN1. These coordinated genetic and chromatin changes, regulated by EZL1, RVR1, and JMJ1, integrate cold and photoperiod signals to precisely control flowering time. Created in BioRender. Daneels, T. (2025) https://BioRender.com/0z70j0s.
VRN2, in contrast, encodes a CCT domain protein, containing both a zinc finger and a CCT (CONSTANS, CONSTANS-like, and TIMING OF CAB1) domain (Figure 1) (Fu et al., 2005; Yan et al., 2006). The CCT domain is essential for nuclear localisation and protein-protein interactions, particularly with NF-Y transcription factors, and is a hallmark of proteins involved in photoperiod and circadian regulation (Griffiths et al., 2003; Tiwari et al., 2010). The repressor activity of VRN2 depends on the integrity of the CCT domain, and mutations in this region can abolish the vernalisation requirement (Chen and Dubcovsky, 2012; Fu et al., 2005). The zinc finger domain contributes to DNA binding, further supporting VRN2’s function as a floral repressor.
VRN3 encodes a member of the phosphatidylethanolamine-binding protein (PEBP) family, homologous to Arabidopsis FLOWERING LOCUS T (Figure 1) (Yan et al., 2006). The PEBP domain enables VRN3 to act as a mobile florigen, interacting with 14-3–3 proteins and the bZIP transcription factor FD to form a complex that migrates from leaves to the shoot apical meristem, where it activates downstream flowering genes, including VRN1 (Distelfeld and Dubcovsky, 2009; Yan et al., 2006).
ODDSOC2, along with ODDSOC1 and MADS37, represents a group of FLC-like MADS-box transcription factors in cereals. While ODDSOC2 is a truncated MADS-box protein, it retains the core DNA-binding domain necessary for its function as a potent floral repressor (Figure 1) (Greenup et al., 2010).
The structural features of these proteins directly inform their genetic interactions, creating a tightly regulated feedback loop that integrates environmental cues to control flowering time (Figure 2). In autumn, VRN2 is highly expressed and acts as a floral repressor by downregulating VRN3, thereby preventing premature flowering (Dubcovsky, 2005; Yan et al., 2004). In winter varieties of wheat and barley, VRN1 is expressed at low levels prior to winter but is strongly induced by prolonged cold exposure, a response that is central to the vernalisation process (Dixon et al., 2019; Shitsukawa et al., 2007; Yan et al., 2003). This cold-induced upregulation of VRN1 is essential for initiating the transition from vegetative to reproductive growth, and its high expression is maintained after vernalisation, ensuring that flowering is irreversibly initiated, after the plant has experienced sufficient winter cold. In contrast, spring varieties often carry deletions or mutations in the VRN1 promoter or first intron, which allows VRN1 to be expressed even in the absence of cold exposure, thereby reducing or abolishing the requirement for vernalisation (Fu et al., 2005; Muterko et al., 2016; Xu and Chong, 2018). The epigenetic regulation of VRN1 is reminiscent of the FLC pathway in Arabidopsis: VRN1 upregulation is associated with a decrease in repressive H3K27me3 (trimethylation of histone H3 at lysine 27) chromatin marks and an increase in active H3K4me3 (trimethylation of histone H3 at lysine 4) marks, supporting the importance of chromatin state changes in vernalisation-induced gene expression (Huan et al., 2018; Liu et al., 2023; Oliver et al., 2009). High levels of VRN1 suppress VRN2 expression, allowing VRN3 activation and flowering. Notably, this VRN1-mediated suppression of VRN2 is unique to wheat and barley, as in other pooid grasses such as Brachypodium, VRN2 is not repressed by VRN1 during or after vernalisation (Greenup et al., 2009; Woods et al., 2016).
With VRN2 repressed, VRN3 is upregulated, especially under long-day conditions in spring, a process further promoted by photoperiod genes such as PPD1 and CO (Chen and Dubcovsky, 2012; Dubcovsky et al., 2006). VRN3, acting as a mobile florigen, migrates to the shoot apex and directly promotes VRN1 transcription, reinforcing its own activation through a positive feedback loop (Trevaskis et al., 2007; Turner et al., 2005; Yan et al., 2006). This feedback ensures a robust and irreversible commitment to flowering once vernalisation and favourable photoperiod conditions are met (A. Chen and Dubcovsky, 2012; Distelfeld and Dubcovsky, 2009).
ODDSOC2, as a floral repressor, is also tightly regulated by this network. Following vernalisation, VRN1 is required to maintain the repression of ODDSOC2, ensuring that this repressor remains downregulated and thus permitting rapid flowering (Deng et al., 2015; Dixon et al., 2019; Greenup et al., 2010). In vrn1 mutants or in cases of incomplete vernalisation, ODDSOC2 expression can rise again, leading to delayed flowering and reduced plant growth. Mechanistically, ODDSOC2 represses FPF1-like genes, which are positive regulators of floral development and cell elongation, providing a direct link between ODDSOC2 activity and the inhibition of flowering (Greenup et al., 2010).
Vernalisation memory
A classic example of vernalisation memory is found in Arabidopsis, where prolonged cold leads to the stable silencing of the floral repressor FLC (Bastow et al., 2004; Michaels and Amasino, 1999; Sheldon et al., 2000). During vernalisation, specific histone modifications accumulate at the FLC locus. Most notably, the repressive mark H3K27me3 spreads across the gene, locking FLC into a silent state that persists even after the plant returns to warmer temperatures (Angel et al., 2011; Bastow et al., 2004; De Lucia et al., 2008).
Histone methylation, such as H3K27me3, is a particularly stable modification. It can persist through cell divisions, providing the molecular basis for long-term epigenetic memory and ensuring continued repression of FLC after vernalisation (Cheung and Lau, 2005; Whittaker and Dean, 2017). In contrast, histone acetylation is highly dynamic and reversible, allowing for rapid but short-lived changes in gene activity (Chen and Tian, 2007). In addition, activating marks like H3K4me3 are reduced at FLC during vernalisation (De Lucia et al., 2008). Together, these histone modifications influence how tightly DNA is packaged around histone proteins, ultimately determining whether genes are accessible for transcription or kept in a silenced state.
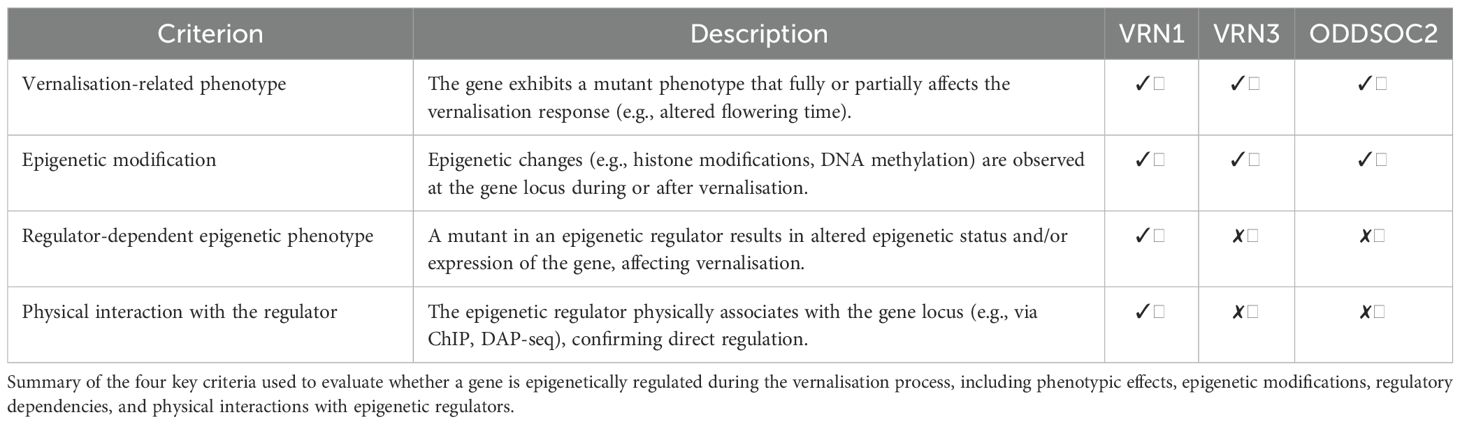
Turning to temperate cereals, recent genome-wide studies have shown that vernalisation leads to widespread changes in histone modifications and gene expression at several important loci, including VRN1, VRN3, and ODDSOC2 (Oliver et al., 2009; Sharma et al., 2017). While VRN1 remains a central focus, these findings raise the possibility that the epigenetic memory of vernalisation in cereals could involve a broader network of genes, rather than being limited to a single locus. This highlights the need for further investigation in this area. Given this complexity, it is important to set out clear criteria to help identify which genes may be involved in storing vernalisation memory. In the next section, we outline such criteria, providing a framework for pinpointing candidate memory genes and guiding future research into the molecular basis of vernalisation memory in temperate cereals.
Criteria for establishing epigenetic regulation
First, there should be phenotypic evidence from knockout lines that demonstrates the gene’s importance in the vernalisation process. In Arabidopsis, for instance, FLC knockout plants lose their vernalisation requirement and flower early, highlighting the gene’s pivotal role (Michaels and Amasino, 1999). Similar phenotypic evidence should be sought in cereals, where loss-of-function mutants in candidate genes would be expected to eliminate the vernalisation response. Second, there should be direct evidence of epigenetic changes at this gene locus during vernalisation. At FLC, vernalisation leads to the accumulation of repressive histone marks at the gene locus, which correlates with stable gene silencing (Bastow et al., 2004; Shindo et al., 2006; Sung and Amasino, 2005). Detecting comparable chromatin changes at candidate loci in cereals is crucial. Third, phenotypes in mutants of epigenetic regulators, such as histone modification writers and erasers, should be considered. In Arabidopsis, mutations in chromatin-modifying proteins that affect FLC silencing also disrupt vernalisation memory (Mylne et al., 2004). The identification of similar regulator mutants in cereals strengthens the case for the role of the corresponding epigenetic modifications in establishing memory. Fourth, there should ideally be evidence of physical interaction between the regulators and the gene locus. For FLC, direct binding of chromatin-modifying proteins to the gene during vernalisation has been demonstrated (Angel et al., 2011; Franco-Echevarría et al., 2023; Schulten et al., 2024). Evidence of such interactions at cereal loci would further support their involvement. These criteria, illustrated by the case of FLC in Arabidopsis, provide a robust framework for dissecting the molecular basis of vernalisation memory in cereals.
Targets of epigenetic regulation
Having outlined the criteria for identifying genes that may store the epigenetic memory of vernalisation, we now evaluate the main candidate genes in temperate cereals. Genome-wide studies have shown that vernalisation triggers widespread changes in histone modifications and gene expression (Oliver et al., 2009). However, while many genes display such changes during cold treatment, only a few have been directly linked to a vernalisation phenotype. Applying our criteria helps distinguish which genes are already known to be central to the epigenetic regulation of flowering by vernalisation. Currently, the strongest candidates for storing vernalisation memory in temperate cereals are VRN1, VRN3, and ODDSOC2 (Table 1).
Knockout lines of VRN1 show a pronounced delay in flowering, confirming its essential role in the vernalisation response (Trevaskis et al., 2003; Yan et al., 2003). Epigenetic regulation of VRN1 during vernalisation is characterised by dynamic changes in histone marks, which are orchestrated by specific chromatin modifiers. Before vernalisation, the VRN1 locus in Brachypodium, barley, and wheat is marked by high levels of the repressive histone modification H3K27me3, which keeps the gene silent (see Table 2 for a detailed overview of epigenetic changes at the VRN1 locus) (Lomax et al., 2018; Oliver et al., 2009). During prolonged cold exposure, these repressive marks are progressively reduced, while activating marks such as H3K4me3, H3K36me3 (trimethylation of histone H3 at lysine 36), and H3K27Ac (acetylation of histone H3 at lysine 27) become increasingly enriched at the VRN1 promoter and first exon. The extent of H3K27me3 loss and H3K4me3 gain is closely linked to the duration of cold, correlating with a cumulative and possibly stable activation of VRN1.
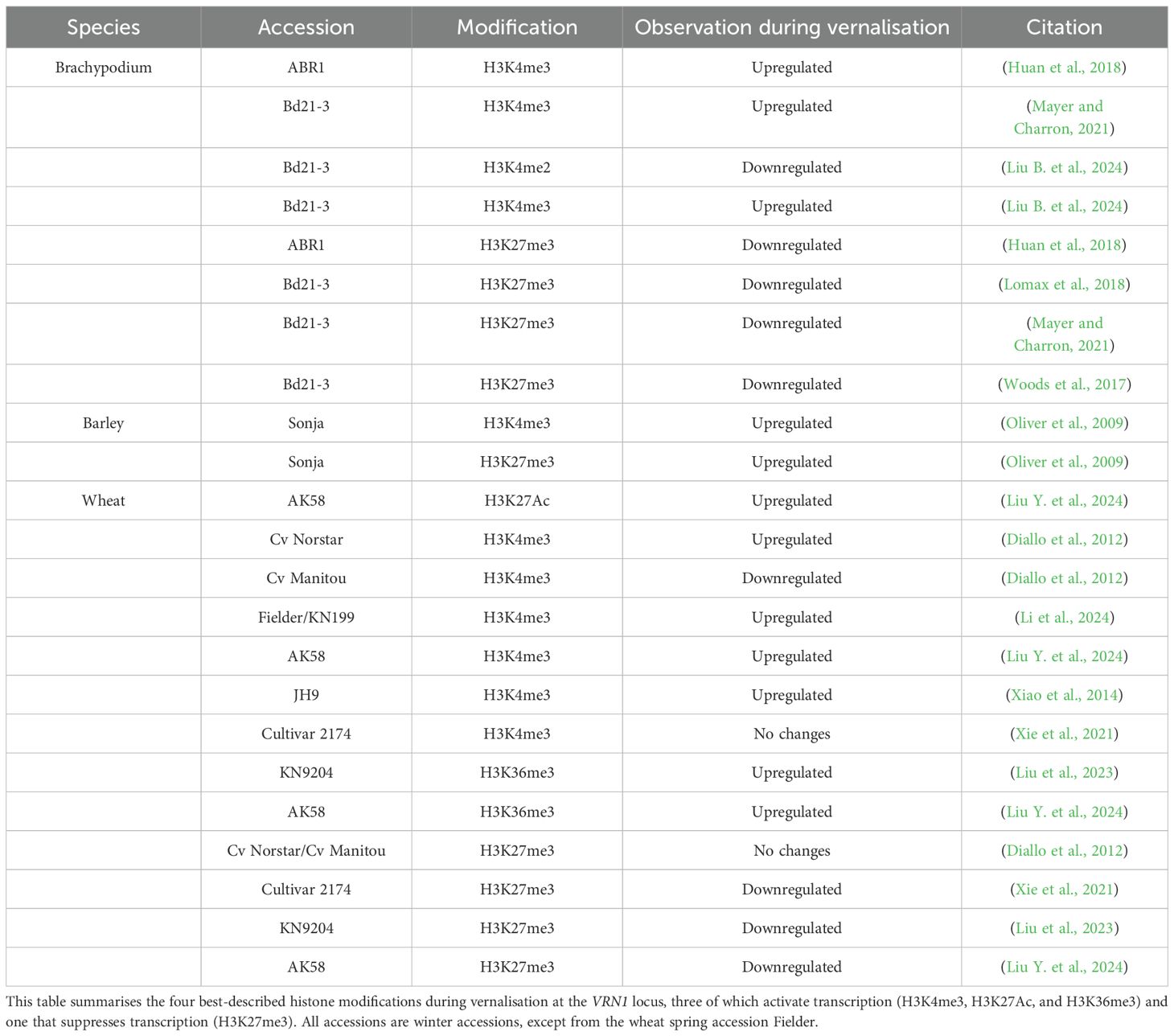
Table 2. Epigenetic modifications at the VRN1 locus during vernalisation in Brachypodium, barley, and wheat.
The establishment and maintenance of histone marks is mediated by several key epigenetic regulators. ENHANCER OF ZESTE-LIKE 1 (EZL1), a homolog of CURLY LEAF in Arabidopsis, acts as the catalytic subunit of the Polycomb Repressive Complex 2 (PRC2), which deposits H3K27me3 at the VRN1 locus (Lomax et al., 2018). In Brachypodium, mutations in EZL1 result in reduced H3K27me3 levels and early flowering even without vernalisation, highlighting its role in repressing VRN1 prior to cold exposure (Lomax et al., 2018). However, since PRC2 does not deposit activating marks, EZL1 is mainly involved in establishing the vernalisation requirement rather than maintaining post-vernalisation memory.
REPRESSOR OF VERNALISATION1 (RVR1) is another key regulator that helps maintain H3K27me3 levels at the VRN1 locus before vernalisation in Brachypodium (Woods et al., 2017). Loss-of-function mutations in RVR1 lead to an overall reduction in H3K27me3, which mimics the vernalised state and results in early flowering even without cold exposure (Woods et al., 2017). This demonstrates that RVR1 acts as a pre-vernalisation repressor, ensuring that VRN1 is only activated after sufficient cold exposure. Its inactivation during vernalisation permits the epigenetic activation of VRN1, thereby linking environmental cues to reproductive timing (Woods et al., 2017).
JUMONJI1 (JMJ1) is a Jumonji C (JmjC)-domain-containing histone demethylase that plays a crucial role in regulating flowering time in Brachypodium (Liu B. et al., 2024). This enzyme specifically removes methyl groups from the histone marks H3K4me2 and H3K4me3 at target genes, including the key flowering regulator VRN1 (Liu B. et al., 2024). JMJ1 helps to repress VRN1 before vernalisation by maintaining low levels of activating histone marks at the VRN1 locus. When JMJ1 is mutated or absent, H3K4me2 and H3K4me3 accumulate at VRN1, leading to increased VRN1 expression and earlier flowering (Liu B. et al., 2024). Chromatin immunoprecipitation experiments have shown that JMJ1 directly binds to the chromatin of VRN1 and another flowering gene, INDETERMINATE1 (ID1), confirming its direct regulatory role (Liu B. et al., 2024). Genome-wide analyses indicate that H3K4me3 is generally associated with higher gene expression, while H3K4me2 is negatively correlated with transcript levels in Brachypodium (Liu B. et al., 2024). Thus, JMJ1-mediated demethylation fine-tunes the chromatin environment at VRN1, ensuring that flowering occurs at the appropriate time in response to seasonal cues (Liu B. et al., 2024).
VRN1 fulfills the criteria for an epigenetic memory locus in vernalisation. Numerous studies have shown that VRN1 remains stably upregulated following vernalisation, supporting its role in the long-term memory of winter conditions (Lomax et al., 2018; Woods et al., 2017). In some cases, VRN1 expression may decrease slightly when plants return to warm temperatures, but this typically occurs only after insufficient cold exposure, resulting in incomplete vernalisation and potential loss of memory (Woods et al., 2017).
The regulation of VRN1 expression involves a tight interplay between repressive and activating histone marks, coordinated by the epigenetic modifiers EZL1, RVR1, and JMJ1. EZL1 and RVR1 maintain repression of VRN1 through H3K27me3 prior to vernalisation, while JMJ1 reinforces this silent state by removing activating histone marks (Lomax et al., 2018; Woods et al., 2017). During cold exposure, the coordinated reduction of repressive marks and accumulation of activating marks at VRN1 enable stable gene activation (Lomax et al., 2018; Woods et al., 2017). Mutations in these histone-modifying regulators alter flowering time, and direct interactions between these complexes and the VRN1 locus have been demonstrated. However, although these mutants clearly disrupt histone modification patterns and flowering time, they do not yet fully explain how the stable epigenetic memory of vernalisation is established and maintained at the VRN1 locus. The precise molecular mechanisms underlying the long-term maintenance of this memory remain unresolved and require further investigation.
VRN3 is another important gene associated with the vernalisation response and is considered a potential target for epigenetic regulation. While knockout lines for VRN3 have not yet been generated, Yan et al. (2006) demonstrated that overexpression of VRN3 in wheat results in early flowering. However, the evidence for epigenetic regulation of VRN3 varies across species. While a single study in barley did not observe changes in epigenetic marks (Oliver et al., 2009), evidence for epigenetic regulation of VRN3 has been reported in other species, such as wheat and Brachypodium (Huan et al., 2018; Liu Y. et al., 2024). Notably, in Brachypodium, these epigenetic changes were observed only when vernalisation was combined with inductive long-day conditions, making it unclear whether vernalisation alone is sufficient to induce such regulation (Huan et al., 2018). As a result, while VRN3 can be epigenetically regulated in some temperate cereals, it does not fully meet all four proposed criteria.
ODDSOC2 acts as a flowering repressor that is downregulated during vernalisation. In Brachypodium, its stable repression is associated with increased H3K27me3 at the ODDSOC2 locus (Sharma et al., 2017). In contrast, in barley and wheat, ODDSOC2 downregulation during vernalisation does not involve direct H3K27me3-mediated silencing. Instead, in barley, repression is primarily achieved through VRN1-mediated transcriptional regulation, with additional evidence for epigenetic involvement as vernalisation reduces H3K4me3 levels at the ODDSOC2 locus, correlating with decreased expression (Deng et al., 2015; Greenup et al., 2010). In wheat, the initial downregulation of ODDSOC2 is triggered by cold exposure independently of VRN1, but long-term repression after vernalisation depends on VRN1 activity, again without evidence for H3K27me3 involvement (Chen and Dubcovsky, 2012; Greenup et al., 2010; Liu et al., 2023).
To summarise the evaluation of these candidates, VRN1 demonstrates the strongest evidence for correlative epigenetic modifications alongside stable changes in expression during vernalisation. It is the only gene that fully meets all four criteria and thus serves as the primary candidate for storing vernalisation memory in temperate cereals. This is reminiscent of the conclusive experiments in Arabidopsis (Berry et al., 2015), which demonstrated that the epigenetic memory of winter is stored at the FLC locus through localised and heritable changes in H3K27me3. Such an experiment in cereals would be able to directly test whether vernalisation memory is confined to a single locus like VRN1, or distributed among multiple sites. However, it cannot be excluded at this stage that other genes such as VRN3 and ODDSOC2 may also contribute to vernalisation memory, potentially indicating a broader network of epigenetically regulated genes. Further research will be required to determine whether memory is restricted to a single locus or distributed among multiple sites.
Epigenetic gene activation of VRN1
In temperate cereals, the epigenetic regulation of vernalisation memory centres on the activation of the flowering promoter VRN1, in contrast to the silencing-based mechanism observed in Arabidopsis. Upon exposure to prolonged cold, H3K27me3 levels at the VRN1 locus gradually decrease, relieving repression and allowing the gene to be activated (Figure 3) (Huan et al., 2018; Oliver et al., 2009). This loss of the repressive mark is an early and necessary step in the transition from a silent to an active chromatin state. As H3K27me3 is removed, activating histone modifications, most notably H3K4me3, accumulate at the VRN1 promoter and 5′ region (Liu Y. et al., 2024; Oliver et al., 2009). The presence of both H3K27me3 and H3K4me3 at the VRN1 locus may represent a poised or bivalent chromatin state, keeping the gene responsive to environmental cues and primed for full activation (Azuara et al., 2006; Bernstein et al., 2006).
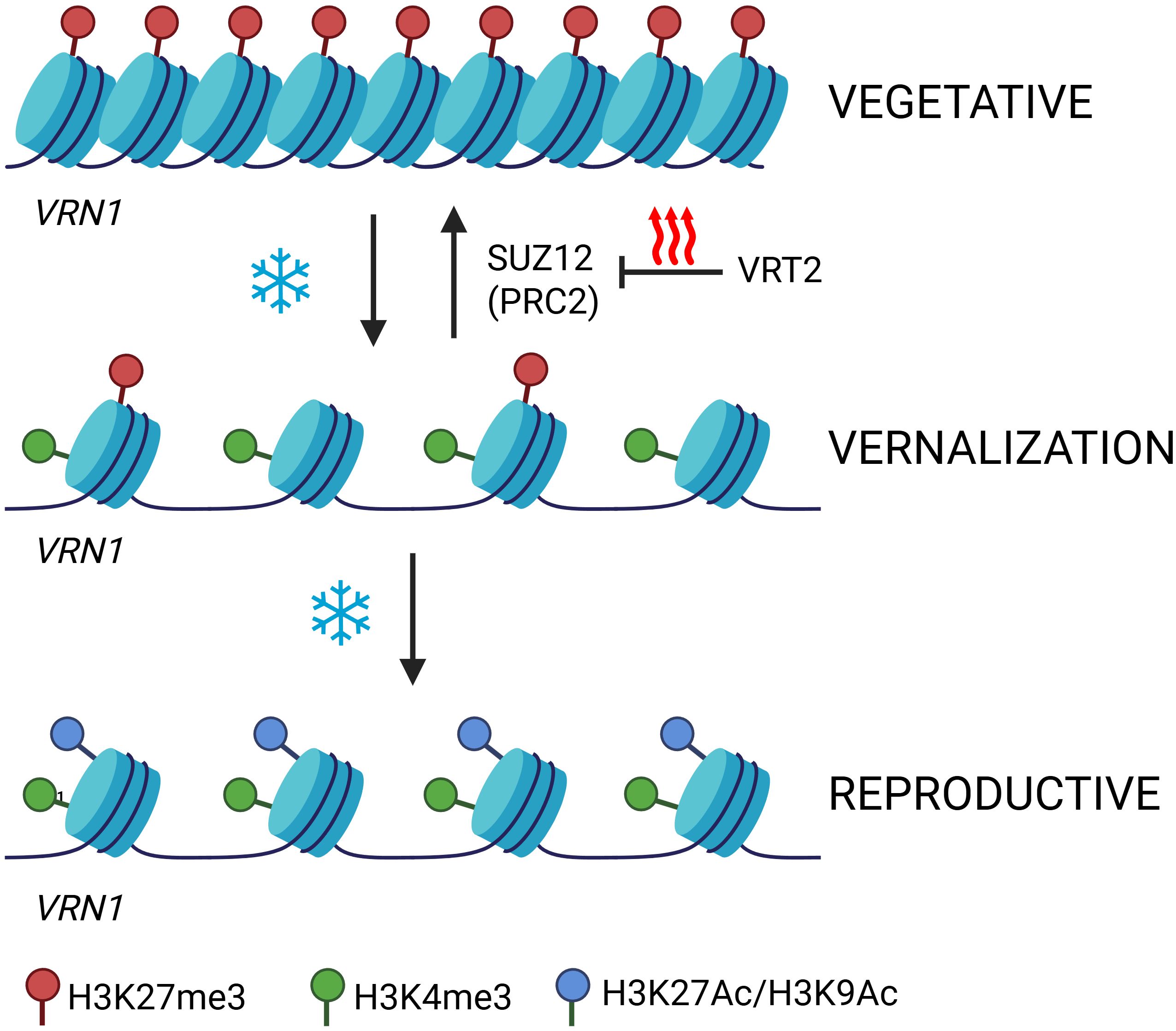
Figure 3. Epigenetic activation VRN1 locus by vernalisation. The figure depicts the epigenetic regulation of the VRN1 locus during vernalisation in temperate cereals as a continuous process. Before cold exposure, VRN1 chromatin is compact and enriched in the repressive mark H3K27me3 (red), keeping the gene silent. During prolonged cold, H3K27me3 levels gradually decrease while activating marks such as H3K4me3 (green) and H3K27Ac or H3K9Ac (blue) accumulate, leading to chromatin opening and VRN1 activation. After vernalisation, in warm conditions, VRN1 remains actively transcribed with sustained H3K4me3, H3K27Ac, H3K9Ac, and H3K36me3 marks, which stabilise expression and prevent re-silencing. The transcription factor VRT2 regulates the PRC2 component SUZ12 to block re-deposition of H3K27me3, ensuring the vernalised, active state is maintained. Colours: red, H3K27me3; green, H3K4me3; blue, acetylation. Created in BioRender. Daneels, T. (2025) https://BioRender.com/v14g358.
As vernalisation progresses, additional activating marks such as H3K27ac and H3K9ac (acetylation of histone H3 at lysine 9) are enriched at the VRN1 promoter, further loosening chromatin structure and facilitating transcriptional activation (Liu Y. et al., 2024; Wang et al., 2009). After transcription is initiated, H3K36me3 is deposited across the gene body, supporting transcriptional fidelity and preventing the re-establishment of repressive marks (Finogenova et al., 2020; Yuan et al., 2011). This sequence, initial loss of H3K27me3, accumulation of H3K4me3 and histone acetylation, followed by H3K36me3 deposition, suggests a stepwise transition from a repressed to an active and stably expressed state. However, the precise chromatin mark or combination of marks responsible for storing the epigenetic memory of vernalisation in cereals remains to be fully determined.
However, the crosstalk and feedback between these histone marks add a layer of complexity, as the presence or removal of one modification can influence the recruitment or exclusion of enzymes responsible for other marks (Janssen and Lorincz, 2022). For example, the removal of H3K27me3 may facilitate the recruitment of H3K4 methyltransferases (Schmitges et al., 2011), while the establishment of H3K36me3 can inhibit the re-deposition of H3K27me3 (Yuan et al., 2011), reinforcing the active state. These dynamic and interdependent relationships underscore that epigenetic regulation is rarely dictated by a single mark, but rather by coordinated modifications that collectively determine chromatin state and gene expression. Recent work in Arabidopsis further illustrates that not only the interplay but also the timing of these histone modifications is critical; dynamic changes in chromatin marks can serve as molecular timers during developmental transitions, ensuring that gene activation is precisely integrated with developmental cues (Huang et al., 2021).
A defining feature of vernalisation memory is its persistence: once established, the vernalised state is generally maintained even if plants are subsequently exposed to high temperatures. Recent research in Brachypodium has shed light on the molecular mechanisms that safeguard this memory from reversal. Kennedy et al. (2024) demonstrated that the MADS-box transcription factor VRT2 plays a crucial role in this process by transcriptionally regulating the PRC2 component SUZ12. By modulating SUZ12 expression, VRT2 may influence the deposition of H3K27me3 at key flowering loci, thereby preventing the re-establishment of a repressive chromatin state and ensuring the irreversibility of vernalisation memory even under fluctuating temperatures. This finding highlights VRT2 as a potential guardian of vernalisation memory, acting to stabilise the epigenetic state acquired during cold exposure (Kennedy et al., 2024). Future studies will be important to further elucidate the precise molecular mechanisms underlying this regulation and to explore how these pathways can be manipulated to improve crop adaptation and flowering time control.
Conclusion and future perspectives
It is still unclear whether vernalisation memory in temperate cereals is stored at a single gene locus or spread across multiple genes. However, the strongest evidence points to a central role for VRN1 and the dynamic changes in its chromatin state. The balance between repressive and activating histone marks at VRN1 appears to be critical, although the exact order and mechanisms remain to be fully defined. Future research should focus on understanding how these epigenetic modifications are established and maintained, and whether other genes also contribute to vernalisation memory. Such discoveries will be vital for deepening our knowledge of plant adaptation to seasonal cues and for improving crop resilience in a changing climate.
Author contributions
TD: Project administration, Writing – original draft, Writing – review & editing. GM-B: Writing – original draft, Writing – review & editing. CB: Writing – review & editing. KG: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. TD and The Geuten Lab are supported by KU Leuven grant C24E/21/004. GM-B is supported by Conahcyt, reference number 2021-000013-01EXTF-00183.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Andrés, F. and Coupland, G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. doi: 10.1038/nrg3291
Angel, A., Song, J., Dean, C., and Howard, M. (2011). A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476, 105–109. doi: 10.1038/nature10241
Azuara, V., Perry, P., Sauer, S., Spivakov, M., Jørgensen, H. F., John, R. M., et al. (2006). Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8, 532–538. doi: 10.1038/ncb1403
Bastow, R., Mylne, J. S., Lister, C., Lippman, Z., Martienssen, R. A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. doi: 10.1038/nature02269
Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. doi: 10.1016/j.cell.2006.02.041
Berry, S., Hartley, M., Olsson, S. G., Dean, C., and Howard, M. (2015). Local chromatin environment of a Polycomb target gene instructs its own epigenetic inheritance. Elife 83, 143–148. doi: 10.7554/eLife.07205.001
Chen, A. and Dubcovsky, J. (2012). Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PloS Genet. 8. doi: 10.1371/journal.pgen.1003134
Chen, Z. J. and Tian, L. (2007). Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim. Biophys. Acta. 1769, 295–307 doi: 10.1016/j.bbaexp.2007.04.007
Cheung, P. and Lau, P. (2005). Epigenetic regulation by histone methylation and histone variants. In Mol. Endocrinol. 19, 563–573. doi: 10.1210/me.2004-0496
Chouard, P. (1960). Vernalization and its relations to dormancy. Plant Physiol. 11, 191–238. Available online at: www.annualreviews.org.
De Lucia, F., Crevillen, P., Jones, A. M. E., Greb, T., and Dean, C. (2008). A PHD-Polycomb Repressive Complex 2 triggers the epigenetic silencing of FLC during vernalization. PNAS 105, 16831–16836. Available online at: www.pnas.org/cgi/content/full/.
Deng, W., Casao, M. C., Wang, P., Sato, K., Hayes, P. M., Finnegan, E. J., et al. (2015). Direct links between the vernalization response and other key traits of cereal crops. Nat. Commun. 6. doi: 10.1038/ncomms6882
Dennis, E. S. and Peacock, W. J. (2009). Vernalization in cereals. J. Biol. 8. doi: 10.1186/jbiol156
Diallo, A. O., Ali-Benali, M. A., Badawi, M., Houde, M., and Sarhan, F. (2012). Expression of vernalization responsive genes in wheat is associated with histone H3 trimethylation. Molecular Genetics Genomics, 287 (7), 575–590. doi: 10.1007/s00438-012-0701-0
Distelfeld, A. and Dubcovsky, J. (2009). Characterization of the maintained vegetative phase deletions from diploid wheat and their effect on VRN2 and FT transcript levels. Mol. Genet. Genomics 283, 223–232. doi: 10.1007/s00438-009-0510-2
Dixon, L. E., Karsai, I., Kiss, T., Adamski, N. M., Liu, Z., Ding, Y., et al. (2019). VERNALIZATION1 controls developmental responses of winter wheat under high ambient temperatures. Dev. (Cambridge). 146. doi: 10.1242/dev.172684
Dubcovsky (2005). Regulation of flowering time in wheat and barley. Comp. Biochem. And. Physiol. A-Mol. Integr. Physiol. 141. doi: 10.1007/s00438-004-1095-4
Dubcovsky, J., Loukoianov, A., Fu, D., Valarik, M., Sanchez, A., and Yan, L. (2006). Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Mol. Biol. 60, 469–480. doi: 10.1007/s11103-005-4814-2
Egea-Cortines, M., Saedler, H., and Sommer, H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18, 5370–5379. doi: 10.1093/emboj/18.19.5370
Finogenova, K., Bonnet, J., Poepsel, S., Schäfer, I. B., Finkl, K., Schmid, K., et al. (2020). Structural basis for prc2 decoding of active histone methylation marks h3k36me2/3. ELife 9, 1–30. doi: 10.7554/eLife.61964
Franco-Echevarría, E., Nielsen, M., Schulten, A., Cheema, J., Morgan, T. E., Bienz, M., et al. (2023). Distinct accessory roles of Arabidopsis VEL proteins in Polycomb silencing. Genes Dev. 37, 801–817. doi: 10.1101/gad.350814.123
Fu, D., Szűcs, P., Yan, L., Helguera, M., Skinner, J. S., Von Zitzewitz, J., et al. (2005). Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol. Genet. Genomics 273, 54–65. doi: 10.1007/s00438-004-1095-4
Greenup, A., Peacock, W. J., Dennis, E. S., and Trevaskis, B. (2009). The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann. Bot. 103, 1165–1172. doi: 10.1093/aob/mcp063
Greenup, A. G., Sasani, S., Oliver, S. N., Talbot, M. J., Dennis, E. S., Hemming, M. N., et al. (2010). ODDSOC2 is a MADS box floral repressor that is down-regulated by vernalization in temperate cereals. Plant Physiol. 153, 1062–1073. doi: 10.1104/pp.109.152488
Griffiths, S., Dunford, R. P., Coupland, G., and Laurie, D. A. (2003). The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 131, 1855–1867. doi: 10.1104/pp.102.016188
Huan, Q., Mao, Z., Chong, K., and Zhang, J. (2018). Global analysis of H3K4me3/H3K27me3 in Brachypodium distachyon reveals VRN3 as critical epigenetic regulation point in vernalization and provides insights into epigenetic memory. New Phytol. 219, 1373–1387. doi: 10.1111/nph.15288
Huang, R., Huang, T., and Irish, V. F. (2021). Do epigenetic timers control petal development? Front. Plant Sci. 12. doi: 10.3389/fpls.2021.709360
Immink, R. G. H., Tonaco, I. A. N., de Folter, S., Shchennikova, A., van Dijk, A. D. J., Busscher-Lange, J., et al. (2009). SEPALLATA3: The “glue” for MADS box transcription factor complex formation. Genome Biol. 10. doi: 10.1186/gb-2009-10-2-r24
Janssen, S. M. and Lorincz, M. C. (2022). Interplay between chromatin marks in development and disease. Nat. Rev. Genet. 23, 137–153. doi: 10.1038/s41576-021-00416-x
Kennedy, A., Li, M., Vandeperre, A., Usama Hameed, M., Van Dyck, M., Preston, J. C., et al. (2024). Transcription factor VRT2 reinitiates vernalization when interrupted by warm temperatures in a temperate grass model. Plant Physiology, 196 (4). doi: 10.1093/plphys/kiae498/7774039
Li, Y., Jin, L., Liu, X., He, C., Bi, S., Saeed, S., et al. (2024). Epigenetic control on transcription of vernalization genes and whole-genome gene expression profile induced by vernalization in common wheat. Plant Diversity, 46 (3), 386–394. doi: 10.1016/j.pld.2024.02.005
Liu, B., Li, C., Li, X., Wang, J., Xie, W., Woods, D. P., et al. (2024). The H3K4 demethylase JMJ1 is required for proper timing of flowering in Brachypodium distachyon. Plant Cell 36, 2729–2745. doi: 10.1093/plcell/koae124
Liu, X., Lin, X., Deng, M., Shi, B., Chen, J., Li, H., et al. (2023). Distinct roles of H3K27me3 and H3K36me3 in vernalization response, maintenance and resetting in winter wheat. Science China Life Sciences, 67, 2251–2266. doi: 10.1101/2023.12.19.572364
Liu, Y., Liu, P., Gao, L., Li, Y., Ren, X., Jia, J., et al. (2024). Epigenomic identification of vernalization cis-regulatory elements in winter wheat. Genome Biol. 25. doi: 10.1186/s13059-024-03342-3
Lomax, A., Woods, D. P., Dong, Y., Bouché, F., Rong, Y., Mayer, K. S., et al. (2018). An ortholog of CURLY LEAF/ENHANCER OF ZESTE like-1 is required for proper flowering in Brachypodium distachyon. Plant J. 93, 871–882. doi: 10.1111/tpj.13815
Mayer, B. F. and Charron, J. B. (2021). Transcriptional memories mediate the plasticity of cold stress responses to enable morphological acclimation in Brachypodium distachyon. New Phytol. 229, 1615–1634. doi: 10.1111/nph.16945
Melzer, R., Verelst, W., and Theißen, G. (2009). The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in ‘floral quartet’-like complexes in vitro. Nucleic Acids Res. 37, 144–157. doi: 10.1093/nar/gkn900
Michaels, S. D. and Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. Available online at: www.plantcell.org.
Muterko, A., Kalendar, R., and Salina, E. (2016). Novel alleles of the VERNALIZATION1 genes in wheat are associated with modulation of DNA curvature and flexibility in the promoter region. BMC Plant Biol. 16, 66–81. doi: 10.1186/s12870-015-0691-2
Mylne, J., Greb, T., Lister, C., and Dean, C. (2004). Epigenetic regulaton in the control of flowering. 69, 457–64. Available online at: www.annualreviews.org.
Oliver, S. N., Finnegan, E. J., Dennis, E. S., Peacock, W. J., and Trevaskis, B. (2009). Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. PNAS. 106, 8386–8391. doi: 10.1073/pnas.0903566106
Riechmann, J. L. and Meyerowitz, E. M. (1997). MADS domain proteins in plant development. J. Biol. Chem. 378, 1079–101. doi: 10.1091/mbc.8.7.1243
Schmitges, F. W., Prusty, A. B., Faty, M., Stützer, A., Lingaraju, G. M., Aiwazian, J., et al. (2011). Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42, 330–341. doi: 10.1016/j.molcel.2011.03.025
Schulten, A., Jang, G.-J., Payne-Dwyer, A., Fiedler, M., Nielsen, M. L., Bienz, M., et al. (2024). Functional specialization of Arabidopsis VEL polymerization domains in the switch to Polycomb silencing. doi: 10.1101/2024.02.15.580496
Sharma, N., Ruelens, P., D’Hauw, M., Maggen, T., Dochy, N., Torfs, S., et al. (2017). A flowering locus C homolog is a vernalization-regulated repressor in Brachypodium and is cold regulated in Wheat. Plant Physiol. 173, 1301–1315. doi: 10.1104/pp.16.01161
Sheldon, C. C., Rouse, D. T., Finnegan, E. J., Peacock, W. J., and Dennis, E. S. (2000). The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). PNAS 97, 3753–3758. doi: 10.1073/pnas.97.7.3753
Shindo, C., Lister, C., Crevillen, P., Nordborg, M., and Dean, C. (2006). Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev. 20, 3079–3083. doi: 10.1101/gad.405306
Shitsukawa, N., Ikari, C., Shimada, S., Kitagawa, S., Sakamoto, K., Saito, H., et al. (2007). The einkorn wheat (Triticum monococcum) mutant, maintained vegetative phase, is caused by a deletion in the VRN1 gene. Genes Genet. Syst. 82, 167–170. doi: 10.1266/ggs.82.167
Shore, P. and Sharrocks, A. D. (1995). The MADS-box family of transcription factors. Eur. J. Biochem. 229, 1–13. doi: 10.1111/j.1432-1033.1995.0001l.x
Smaczniak, C., Immink, R. G. H., Muiño, J. M., Blanvillain, R., Busscher, M., Busscher-Lange, J., et al. (2012). Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. United. States America 109, 1560–1565. doi: 10.1073/pnas.1112871109
Song, J., Angel, A., Howard, M., and Dean, C. (2012). Vernalization - a cold-induced epigenetic switch. J. Cell Sci. 125, 3723–3731. doi: 10.1242/jcs.084764
Sung, S. and Amasino, R. M. (2004). Vernalization and epigenetics: How plants remember winter. Curr. Opin. Plant Biol. 7, 4–10. doi: 10.1016/j.pbi.2003.11.010
Sung, S. and Amasino, R. M. (2005). Remembering winter: Toward a molecular understanding of vernalization. Annu. Rev. Plant Biol. 56, 491–508. doi: 10.1146/annurev.arplant.56.032604.144307
Tiwari, S. B., Shen, Y., Chang, H. C., Hou, Y., Harris, A., Ma, S. F., et al. (2010). The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187, 57–66. doi: 10.1111/j.1469-8137.2010.03251.x
Trevaskis, B., Bagnall, D. J., Ellis, M. H., Peacock, W. J., and Dennis, E. S. (2003). MADS box genes control vernalization-induced flowering in cereals. 100, 13099–13104. Available online at: www.pnas.org.
Trevaskis, B., Hemming, M. N., Dennis, E. S., and Peacock, W. J. (2007). The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 12, 352–357. doi: 10.1016/j.tplants.2007.06.010
Turner, A., Beales, J., Faure, S., Dunford, R. P., and Laurie, D. A. (2005). The pseudo-response regulator ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1029–1031. doi: 10.1126/science.1117682
Wang, F., Li, S., Kong, F., Lin, X., and Lu, S. (2023). Altered regulation of flowering expands growth ranges and maximizes yields in major crops. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1094411
Wang, Z., Zang, C., Cui, K., Schones, D. E., Barski, A., Peng, W., et al. (2009). Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031. doi: 10.1016/j.cell.2009.06.049
West, A. G., Shore, P., and Sharrocks, A. D. (1997). DNA binding by MADS-box transcription factors: a molecular mechanism for differential DNA bending. Mol. And. Cell. Biol. 17, 2876–2887. doi: 10.1128/MCB.17.5.2876
Whittaker, C. and Dean, C. (2017). The FLC locus: A platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 12, 555–575. doi: 10.1146/annurev-cellbio-100616
Woods, D. P., McKeown, M. A., Dong, Y., Preston, J. C., and Amasino, R. M. (2016). Evolution of VRN2/Ghd7-like genes in vernalization-mediated repression of grass flowering. Plant Physiol. 170, 2124–2135. doi: 10.1104/pp.15.01279
Woods, D. P., Ream, T. S., Bouché, F., Lee, J., Thrower, N., Wilkerson, C., et al. (2017). Establishment of a vernalization requirement in Brachypodium distachyon requires REPRESSOR OF VERNALIZATION1. Proc. Natl. Acad. Sci. United. States America 114, 6623–6628. doi: 10.1073/pnas.1700536114
Wu, X., Liu, H., Li, X., Tian, Y., and Mahecha, M. D. (2017). Responses of winter wheat yields to warming-mediated vernalization variations across temperate europe. Front. Ecol. Evol. 5. doi: 10.3389/fevo.2017.00126
Xiao, J., Xu, S., Li, C., Xu, Y., Xing, L., Niu, Y., et al. (2014). O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nature Communications, 5. doi: 10.1038/ncomms5572
Xie, L., Zhang, Y., Wang, K., Luo, X., Xu, D., Tian, X., et al. (2021). TaVrt2, an SVP-like gene, cooperates with TaVrn1 to regulate vernalization-induced flowering in wheat. New Phytologist. 231 (2), 834–848. doi: 10.1111/nph.16339
Xu, S. and Chong, K. (2018). Remembering winter through vernalisation. Nat. Plants 4, 997–1009. doi: 10.1038/s41477-018-0301-z
Yan, L., Fu, D., Li, C., Blechl, A., Tranquilli, G., Bonafede, M., et al. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. 103, 19581–19586. Available online at: www.pnas.org/cgi/content/full/.
Yan, L., Loukoianov, A., Blechl, A., Tranquilli, G., Ramakrishna, W., Sanmiguel, P., et al. (2004). The wheat VRN2 gene is a flowering repressor downregulated by vernalization. Science 303, 1677–1686. Available online at: www.sciencemag.org.
Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T., and Dubcovsky, J. (2003). Positional cloning of the wheat vernalization gene VRN1. PNAS 100, 6263–6268. doi: 10.1073/pnas.0937399100
Keywords: vernalisation, temperate cereals, histone modification, VRN1, devernalisation
Citation: Daneels T, Martinez-Barrales G, Bosmans C and Geuten K (2025) Targets and mechanisms of epigenetic regulation in the temperate cereal vernalisation process. Front. Plant Sci. 16:1520593. doi: 10.3389/fpls.2025.1520593
Received: 31 October 2024; Accepted: 17 July 2025;
Published: 05 August 2025.
Edited by:
Liangsheng Zhang, Zhejiang University, ChinaReviewed by:
Ruirui Huang, University of San Francisco, United StatesChris Helliwell, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Australia
Copyright © 2025 Daneels, Martinez-Barrales, Bosmans and Geuten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Koen Geuten, a29lbi5nZXV0ZW5Aa3VsZXV2ZW4uYmU=
†These authors have contributed equally to this work and share first authorship
 Tomas Daneels
Tomas Daneels Gustavo Martinez-Barrales
Gustavo Martinez-Barrales Cederic Bosmans
Cederic Bosmans Koen Geuten
Koen Geuten