- 1College of Forestry, Gansu Agricultural University, Lanzhou, China
- 2Wolfberry Harmless Cultivation Engineering Research Center of Gansu Province, Lanzhou, China
Wolfberry (Lycium barbarum), a member of the Solanaceae family, is recognized as a pioneering tree species for afforestation in saline-alkali soils and holds significant economic value as a forest species. Its fruit is abundant in bioactive compounds that contribute in both ecological health and human well-being. The WRKY gene family has been extensively studied across various species, with its members’ functions increasingly elucidated. However, limited research has focused on the role of the WRKY genes of L. barbarum, particularly in resistance to root rot. This study identified the bioinformatics of 104 WRKY genes in wolfberry, encompassing phylogenetics, conserved motifs, gene structures, synteny, and collinearity. Based on structural and phylogenetic, the 104 LbWRKYs are divided into three main groups, Group I, II and III, with 26, 62 and 15 members, respectively. Synteny analysis revealed high homology between LbWRKY and tomato SlWRKY, with a total of 117 pairs of homologous genes identified. Cis-acting elements analysis demonstrated that subgroup II LbWRKY genes contained a higher number of plant hormone-related regulatory elements. Furthermore, 28 LbWRKY genes were found to respond to the infection of Fusarium solani. Protein-protein interaction prediction and correlation analyses revealed that associations between LbWRKY genes and flavonoid and phenylpropanoid synthesis-related genes, and the results showed that LbWRKY8/100/63/84/102/42/45 was involved in the mechanism of root rot resistance. Expression analysis following Fusarium solani inoculation confirmed that these genes participate in root rot resistance in L. barbarum. This study provides valuable insights into the functional roles of LbWRKY genes, and establishing a foundation for future research on their involvement in secondary metabolite synthesis and their role in enhancing the disease resistance of L. barbarum.
Introduction
Wolfberry Lycium barbarum L., a perennial shrub of the Solanaceae family, is valuable medicinal and edible plant (Hu et al., 2014). It is primarily distributed in arid to semi-arid regions of the temperate zones, and its fruit is widely utilized in the pharmaceutical and health care industries (Levin and Miller, 2005; Ma R et al., 2023). Due to its saline-alkaline and drought tolerance, along with its high economic value, wolfberry has become a widely cultivated fruit crop in Northwest China (Yao et al., 2017; Cao et al., 2021; Gong et al., 2022). However, the increasing acreage is accompanied by frequent diseases affecting wolfberry. The root rot, caused by Fusarium species, is a common soil-borne fungal disease that poses a serious threat to wolfberry production, leading to reduced yields and compromised crop health (Cichy et al., 2007; Eliane, 2017). Breeding resistant varieties is considered the most effective approach to controlling plant diseases (Agbowuro et al., 2020). Therefore, identifying key disease resistance genes is critical for developing wolfberry varieties resistant to root rot.
Transcription factors (TFs) are regulatory proteins that bind to specific cis-elements in their target genes and modulate the expression level of genes under particular stress conditions. These “cis-trans” interactions play a crucial role in enhancing plant survival under adverse environmental conditions (Shrestha et al., 2018). Several TF families have been identified as key regulators of plant adaptation to stress, including WRKY, bHLH, AP2/ERF, ARF, NAC, MYB, bZIP, among others (Xiao et al., 2023; Ahmad et al., 2024; Bian et al., 2024; Li et al., 2024; Ou et al., 2024; Wang et al., 2024b; Ye et al., 2024). The WRKY family, recognized as one of the most critical transcription factor families, is extensively involved in plant responses to biotic, abiotic and hormonal stresses (Ahuja et al., 2010; Chen et al., 2017; Jiang et al., 2017). For instance, 103 CpWRKYs in Cucurbita pepo L. are implicated in responses to cold and drought stresses (Liu et al., 2024). Members of the wheat WRKY gene family play a crucial role in the tolerance mechanisms to drought and osmotic stress (Gahlaut et al., 2016; Wani et al., 2018). Similarly, 102 MeWRKYs identified in Manihot esculenta confer resistance to Xanthomonas axonopodis pv. Manihotis (Zhu et al., 2022b). Moreover, 63 AtWRKYs in Akebia trifoliata have been reported to play a significant role in plant disease resistance (Zhu et al., 2022a).
The N-terminus of WRKY DNA-binding domain (DBD) contains a highly conserved sequence, with WRKYGQK as the core motif. Mutants with amino acid substitutions and the featuring either a C-terminus C2H2 or C2HC zinc-binging motif have also been reported (Rushton et al., 2010; Song et al., 2018). Based on the number of WRKY domains and the type of zinc finger motif, WRKY TFs are categorized into three groups, with Group II are further subdivided into five subgroups (IIa–IIe) (Kazuhiko et al., 2005). WRKY TF are involved in phytohormone-mediated signaling pathways, the regulation of pathogenesis-related proteins, and biosynthesis of secondary metabolites by acting on downstream target genes in response to biotic stresses (Li et al., 2020; Nisha and Paramjit, 2021; Luan et al., 2023). In plant, the phenylpropanoid pathway participates in the biosynthesis of secondary metabolites related to disease defense (Zhang et al., 2021). Through the combined analysis of transcriptomic and metabolomic data, LbPAL, LbCHS and LbUGT involved in the biosynthesis of flavonoids and polysaccharides, and their expression was significantly upregulated in response to pathogen infection (Zhang et al., 2021).
Lycium barbarum is an important traditional medicinal and food supplement in China, and its genome has recently been released (Cao et al., 2021). Based on this genomic resource, several gene families in L. barbarum have been identified, and their biological functions partially elucidated. For instance, 28 LbaBBXs and 137 LbaR2R3MYBs were identified, with specific members such as LbaBBX2 and LbaBBX4, (Lba11g0183 and Lba02g01219) shown to play regulatory roles in the carotenoids biosynthesis (Yin et al., 2022a, b). Additionally, 12 LbCCOs (carotenoid cleavage oxygenase genes) were identified as being involved in regulation of phytohormones, pigments, and aromatic substances (Li et al., 2023). He et al. revealed that 38 LbAQPs (aquaporins) are involved in developmental processes and responses to abiotic stress (He et al., 2022). In Solanaceae, the overexpression or silencing of WRKY genes significantly impacts plant defense. For instance, SlWRKY12 contributes to tomato resistance against Botrytis cinerea by positively regulating related defense genes (Liu et al., 2014). Recent studies have demonstrated that the regulation of CaWRKY40 in pepper and its associated resistance to Ralstonia solanacearum depend on the transcriptional activation of CaWRKY06 (Cai et al., 2015). However, comprehensive studies on WRKY genes in wolfberry have not yet been reported.
In the current study, we conducted a comprehensive genome-wide identification and analysis of the WRKY gene family in L. barbarum. We systematically examined their chromosomal location, classifications, conserved protein domains, motif composition, gene structures, phylogenetic relationships, and duplication events. Additionally, we investigated the expression profile of WRKY genes in response to inoculation with F. solani and explored their potential roles in flavonoid and phenylpropanoid synthesis. These findings identify potential candidate genes for further functional studies and enhance our understanding of the molecular mechanisms underlying lignin synthesis in plants.
Materials and methods
Identification and characterization of LbWRKY genes in the L. barbarum genome
Genome and gene annotation files of L. barbarum were downloaded from the NCBI datebase https://www.ncbi.nlm.nih.gov/, the HMMs (PF03106) of the WRKY TFs downloaded from the pfam database http://pfam.xfam.org/ were utilized as a blast search query for WRKY using a threshold E-value of 1e−5. The candidate genes were then further validated using the SMART http://smart.embl-heidelberg.de/ and NCBI-CDD https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (Lu et al., 2020). Arabidopsis WRKY protein sequences were downloaded from TAIR https://www.arabidopsis.org/, Solanum lycopersicum (Sl) ITAG5.0 and Capsicum annuum (Ca) were downloaded from Phytozome13 https://phytozome-next.jgi.doe.gov/ and NCBI datebase https://www.ncbi.nlm.nih.gov/. The protein sequences of LbWRKY members were analyzed with Multiple Protein Profiler 1.0 (MPP) https://mproteinprofiler.microbiologyandimmunology.dal.ca/ Predict the physicochemical properties of the proteins and visualize (Sganzerla et al., 2024). The subcellular localization of these proteins was predicted and analyzed using the Plant-mPLoc 2.0 http://www.csbio.sjtu.edu.cn/bioinf/plant/ (Chou and Shen, 2008). TBtools was used to visualize the position of the candidate WRKY genes on the chromosome (Chen et al., 2020b).
Multiple sequence alignment, phylogenetic analysis, gene structure and conserved motif analyze of the WRKY genes in L. bararum
The LbWRKYs protein sequences were aligned using ClustalW in MEGA7.0, and analyzed the conserved WRKY core domain (60 amino acids) using DNAMAN. The phylogenetic tree of wolfberry and Arabidopsis thaliana (At) was constructed using the Maximum Composite Likelihood (MCL) model of neighbor-joining (NJ) algorithm in MEGA 7.0 software, and 1,000 iterations of bootstrapping were performed (Chen et al., 2020b). The Evolview 3.0 https://www.evolgenius.info/evolview/#/treeview was used to further modify and enrich the evolutionary tree (Subramanian et al., 2019).
The WRKY protein sequences and Genome file of wolfberry were sent to the Visualize Gene Structure in TBtools software to investigate exon-intron structures (Chen et al., 2020b). The conserved motifs of the LbWRKY protein sequences were analyzed using MEME https://meme-suite.org/meme/tools/ meme online website with a number of motifs of 10 and optimum width of 6 to 50 bp (Bailey et al., 2009).
Gene duplication, collinearity analyses and cis-acting elements in the promoters of LbWRKY family
The collinearity of WRKYs within L. barbarum was analyzed by Circos and TBtools software, and the synteny maps of WRKYs between L. barbarum, A. thaliana, S. lycopersium and C. annuum were depicted through MCscanX in TBtools (Krzywinski et al., 2009; Chen et al., 2020a). The Adobe Illustrator 2021 software was used to further modify and enrich.
The 2000 bp gene sequence upstream of the initiation codon (ATG) of LbWRKYs was identified as the gene promoter sequence. Cis-acting regulatory elements of LbWRKYs regions were searched for within 2000bp upstream using PlantCARE tool and then visualized with TBtools (Lescot et al., 2002; Chen et al., 2020b).
Expression profile of LbWRKY family in different developmental stages of Lycium barbarum and prediction of miRNA target and SSR
The expression profiles of the LbWRKY gene family in different developmental stages of L. barbarum were analyzed using the Genevestigator platform (Hruz et al., 2008). Transcriptome data were obtained from the RefSeq: NCBI reference Sequence Database, and further analyzed using TBtools software. The 3D structure models of key LbWRKY family members are predicted by AlphaFold Protein Structure Database https://alphafold.ebi.ac.uk/ (Jumper et al., 2021). MiRanda software and Trf software were used to predict miRNA and SSR respectively (Enright et al., 2003; Saliminejad et al., 2019). Tomato miRNA was downloaded from plant microRNA database http://bioinformatics.cau.edu.cn/PMRD/ (Zhang et al., 2009).
RNA-seq analysis
The plant materials used in this study were annual seedlings of Ningqi 7 and provided by Wolfberry producting areas in Baiyin City, Gansu Province. The pathogenic was Fusarium solani, which is preserved by the Wolfberry Harmless Cultivation Engineering Research Center of Gansu Province, Lanzhou, China. The leaves of L. bararum were collected at 30 d post inoculation (dpi), and stored at -80°C after freezing in liquid nitrogen. The RNA-seq in this study was commissioned by Beijing Biomarker Technologies Co., LTD to complete the library construction, and the cDNA library was sequenced using the Illumina NovaSeq high-throughput sequencing platform.
Correlation analysis of DEGs of disease resistance and regulatory network of LbWRKYs
The differential genes DEGs (|Log2FC| >1.5, FDR < 0.05) in response to the infection of F. solani were analyzed in the transcriptome database, and Pearson correlation analysis in Origin 2022 was used to detect the relationship between resistance-related genes. Solanum lycopersicum was used as the reference sequence for string http://string-db.org/ prediction network interaction maps (Szklarczyk et al., 2023). Network diagram was visualized using Cytoscape v3.9.1 (Shannon et al., 2003).
RNA extraction and quantitative RT-PCR
RNA was extracted from leaves of L. barbarum using a Plant RNA Kit (Omega Bio-Tek, Guangzhou, China). PrimeScriptTM RT reagent Kit and TB Green® Premix Ex TaqTM II were used for gDNA erasure and quantitative RT-PCR (Shanghai BioScience Co., Ltd). The primers were designed using NCBI online website Supplementary Table S9 https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi. For each reaction, three independent biological and technical replicates were used. The relative expression of each gene for RT-qPCR analysis was calculated by 2−ΔΔCt method, Origin 2022 was used for statistical analysis.
Results
Identification and characteristics of LbWRKY genes
The WRKY domain HMM profile PF03106 and BLSATP analysis using 71 WRKY Protein sequences from Arabidopsis thaliana were employed to identify WRKY genes in L. barbarum genome. A total of 104 putative LbWRKY genes were identified (Supplementary Table S1), and named LbWRKY1 to LbWRKY104 based on their chromosomal locations. These genes were unevenly distributed across twelve chromosomes (Figure 1), with most localized at the ends of chromosomes. Chromosome 11 contained the highest number of LbWRKYs genes (13.5%), followed by chromosomes 4 and 5, which each harbored 12 genes, (11.5%). Chromosomes 8 and 12 housed 10 LbWRKYs each, while chromosome 10 had the fewest, with only 4 genes (3.8%). The remaining chromosomes 1, 3, 7, 2 and 9 contained separately 9, 9, 6, 5 and 5 LbWRKY genes, respectively. Additionally, 47 LbWRKY genes, such as LbWRKY4/5, LbWRKY25/26, LbWRKY27/28/29, etc. were clustered into 22 tandam duplication regions on chromosome 1 and 4-12, suggesting potential duplication events.
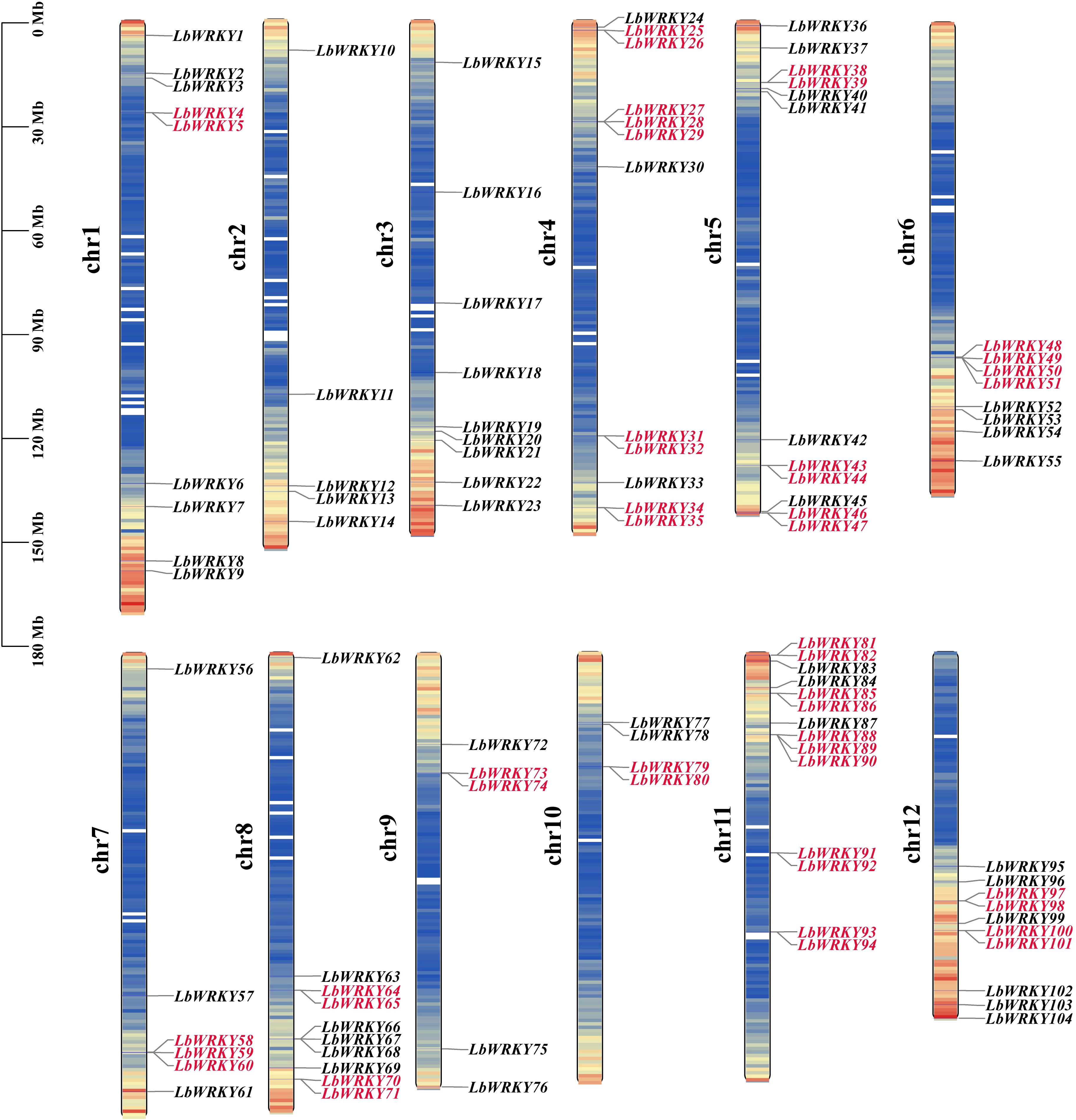
Figure 1. Chromosomal location of LbWRKY genes in Wolfberry. At the left of the chromosome is the number of that chromosome. On the right side of every chromosome are the names of the genes. Different colors on the chromosome indicated varying gene densities, with red highlighting the tandem repeat genes.
We analyzed and visualized various physiochemical properties of 104 LbWRKY proteins with MPP profiler (Supplementary Table S1 and Supplementary Figure S1). The amino acid lengths of LbWRKY proteins ranged from 165 (LbWRKY30) to 729 (LbWRKY34), with predicted molecular weights ranging from 18.96 to 79.73 kDa. Most LbWRKY proteins were acidic, with a pI ratio of approximately 2:1. The GRAVY (grand average of hydropathicity) values for all LbWRKY proteins were negative, indicating there hydrophilic nature. Instability index analysis showed that most of these proteins were unstable. Subcellular localization results revealed that all LbWRKY proteins were located in the nucleus.
Protein domains and phylogenetic analysis of LbWRKY proteins
To further understand the evolutionary relationship of WRKY gene in L. barbarum, a total of 175 WRKY genes, including 104 from wolfberry, 71 from in Arabidopsis thaliana, were used to construct an unrooted phylogenetic tree (Figure 2). The tree organized WRKY into three main groups. Group I included 39 members (26 LbWRKY) and (13 AtWRKY genes), and was characterized by two conserved WRKY domains (Supplementary Figure S2). Group II consisted 108 members, which were further subdivided into five subgroups: IIa (n = 21), IIb (n = 20), IIc (n = 28), IId (n = 16), and IIe (n = 23). Group III was the smallest containing 28 members: 15 WRKY genes from wolfberry and 13 from Arabidopsis. These results suggested that L. barbarum has a significantly greater number of WRKY genes compared to A. thaliana. This difference is likely due to the larger genome size of wolfberry, which has facilitated the expansion of WRKY genes during genome evolution.
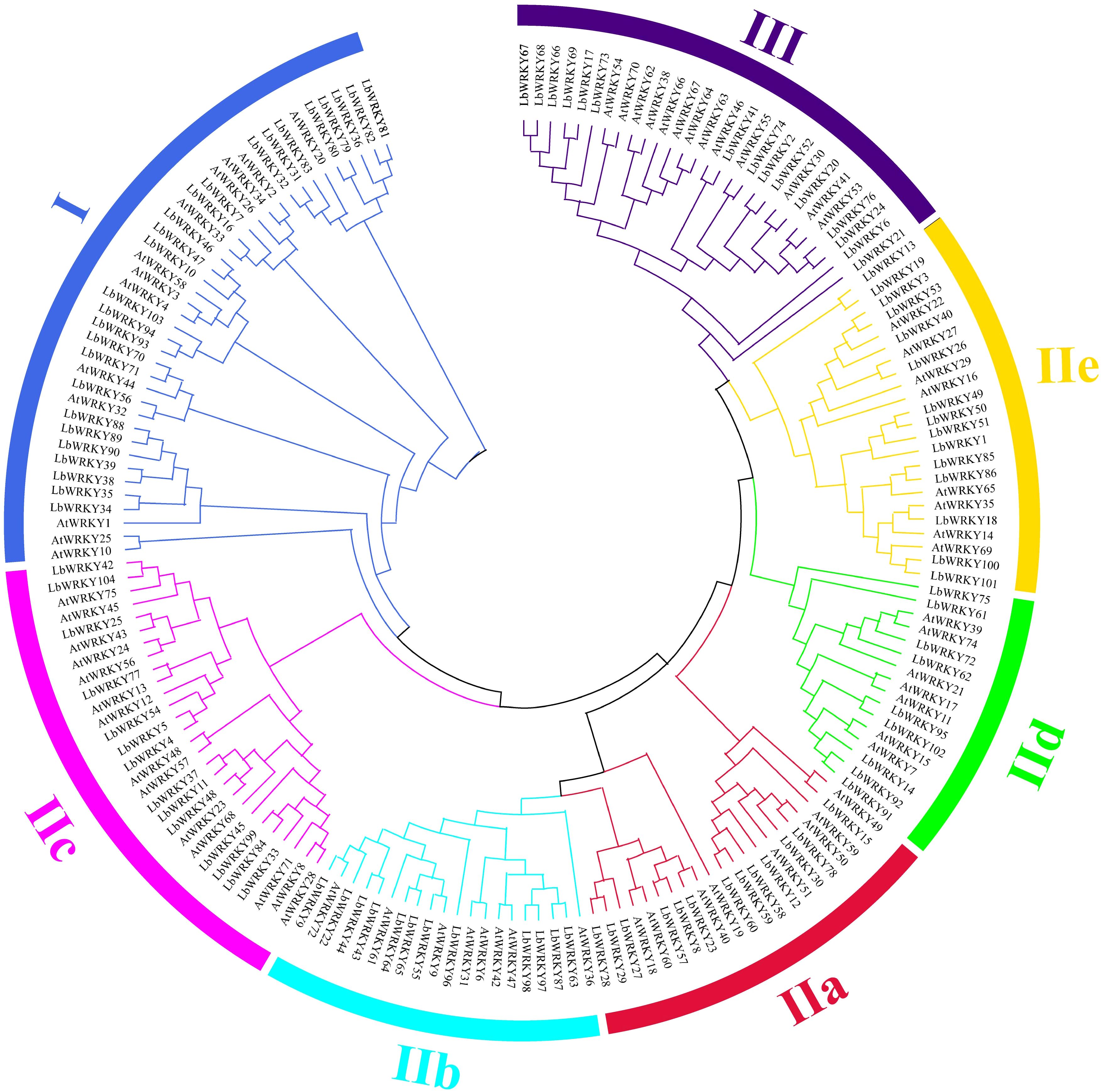
Figure 2. The phylogenetic tree of WRKY proteins from wolfberry and Arabidopsis thaliana. These proteins were clustered in three main groups represented by different colors, with each subgroup marked outside the circle as I, II and III. Lb, Lycium barbarum; At, Arabidopsis thaliana.
Gene structure and protein motif of WRKYs in L. barbarum
To investigate of structural diversity LbWRKY genes, the analyses of exon-intron structures and conserved motifs were performed, followed by a phylogenetic analysis (Figure 3). The results indicated that the classification of 104 LbWRKY protein domains based on the multiple sequence alignment (Figure 3A) was consistent with the grouping in Figure 1. Members within the same subgroup exhibited similar gene structures and motif modules. Gene structure analysis showed that each LbWRKY consisted of a variable number of exons and introns (Figure 3C). The number of introns LbWRKY genes ranged from 0 to 5, with varying intron sizes. Group I featured 4–6 exons, LbWRKY32 and LbWRKY83 specifically contained 6 exons. Group II subgroups (IIa-e) contained 1–6 exons, with subgroup IId having the highest exon count: 4 members contained 6 exons, and 5 members had 5 exons. In Group III, only LbWRKY6 and LbWRKY74 contained 4 exons, while all other members had 3 exons. Apart from IId, most of the other groups contain 3 exons.
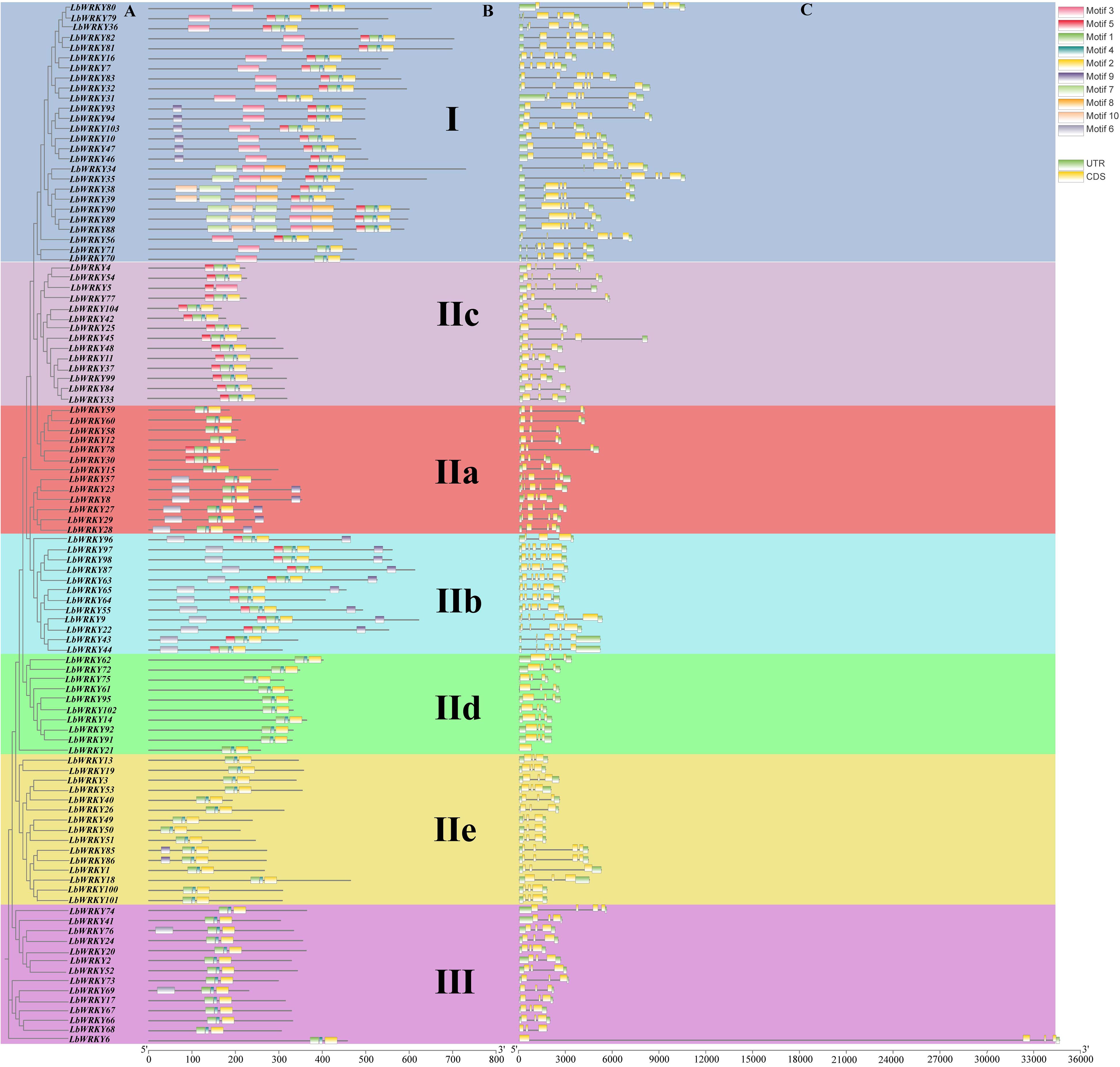
Figure 3. Phylogenetic clustering, conserved protein motifs and gene structure of LbWRKY genes. (A) A phylogenetic tree was constructed of LbWRKYs. Different groups and subgroups are displayed in different colors. (B) Use different color boxes to represent the motif. (C) Exon-intron structure of the LbWRKYs. Untranslated 5 ′ -and 3 ′ - regions, exons and introns Subs are represented by green box, yellow box and black line respectively.
Using the MEME online tool, 10 conserved motifs in LbWRKY genes were predicted and named as motif 1 to motif 10 (Supplementary Figure S3). The results revealed that all members of the LbWRKY gene family contained the motif 1, 2 and 4. However, these motifs varied across the phylogenetic groups (Figure 3B). Motif 1and 3 contained the WRKYGQK sequence, proteins in Group I predominantly contained the motifs 1-5, while Group IId, IIe and III mainly contained motifs 1, 2 and 4, and Group IIc consisted of motif 1, 2, 4 and 5, except for LbWRKY5, which contained only two motifs, motifs 3 and 5. The similarity in gene structure, motif content and distribution patterns within each phylogenetic group indicates a shared evolutionary origin. Conversely, the uniqueness of motifs within phylogenetic groups reflects the functional differences among LbWRKY members.
Collinearity and synteny analyses
Segmented, scattered or tandem gene replication events in the genome are regarded as a major driving of evolution (Panchy et al., 2016). Segmental duplication analysis of the 104 LbWRKY genes by TBtools and Circos identified 30 homologous sites and 22 pairs of segmental duplication events (Figure 4A; Supplementary Table S3). However, 19 LbWRKY genes lacated on chromosomes 8, 9 and 10 were derived from dispersed and tandem chromosomal distributions. The results of the Ka/Ks ratio analysis for homologous genes showed that 18 ratios were less than 1.0, showing that the LbWRKY gene family may have undergone strong purifing during evolution (Supplementary Table S2).
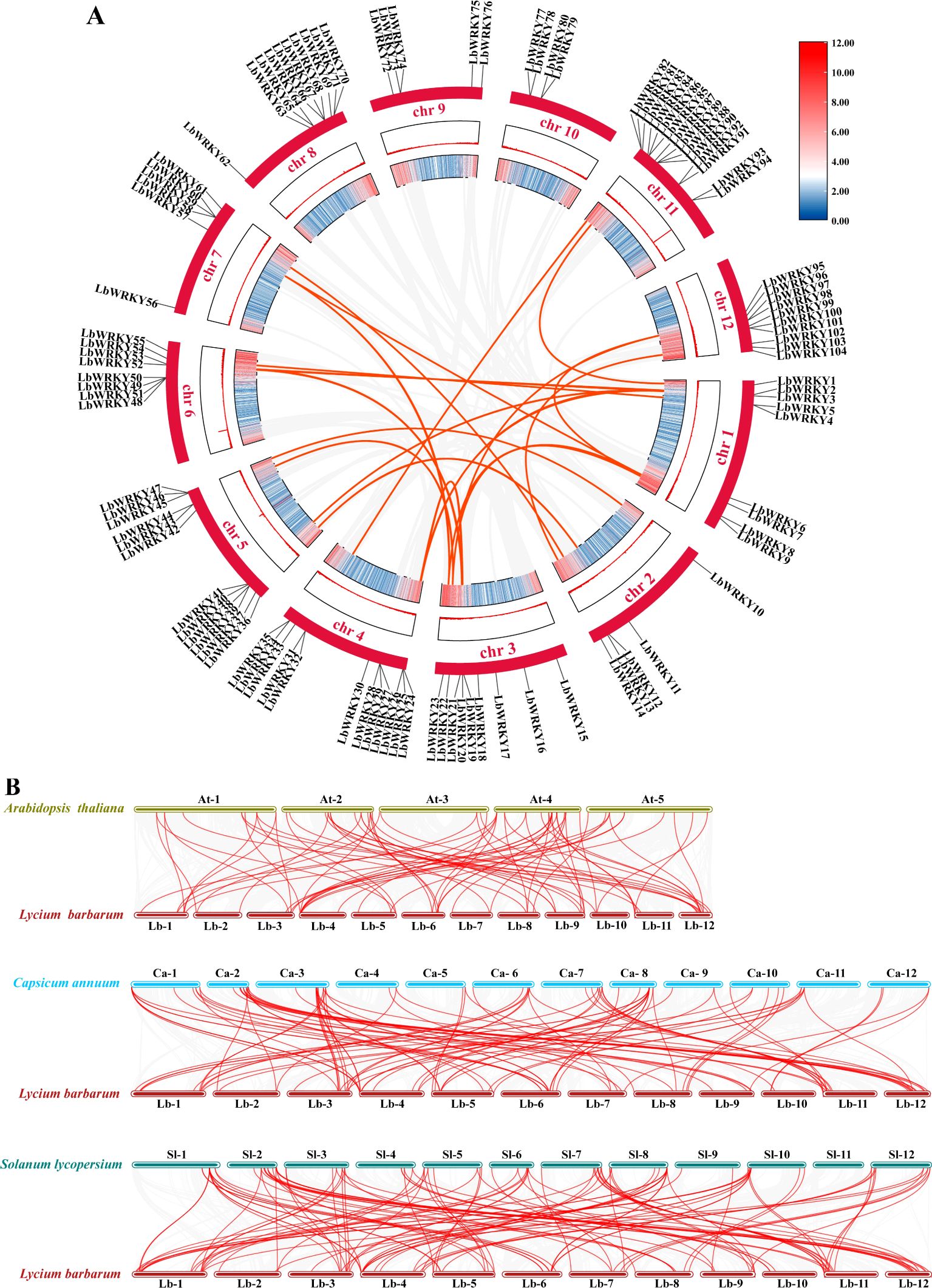
Figure 4. (A) The outermost circle represented the identified members of the WRKY gene family in Lycium barbarum, the red bars represent different chromosomes, and the inner colored bars represented the gene density on the chromosomes. (B) Colinearity analysis of wolfberry with Arabidopsis, pepper and tomato, with colored lines above and below each set collinearity represented chromosomes of different species.
The collinearity of L. barbarum WRKYs with those of A. thaliana, C. annuum and S. lycopersium was investigated (Figure 4B), and there are 71, 98 and 117 pairs of colinear genes, respectively (Supplementary Table S4). A significant number of genes exhibited collinearity across three selected species, suggesting that these genes may play a critical role in the evolution of LbWRKY gene family.
Cis-acting elements analysis of LbWRKY genes promotors
Gene expression can be regulated by the binding of transcription factors (TF)to cis-acting elements (Wang et al., 2014), which is closely related to their adaptability to developmental and stress responses. The cis-acting elements of 104 LbWRKY genes were analyzed using PlantCARE. As shown in Figure 5, a total of 992 cis-acting elements were identified and classified into three basic categories: including plant growth and development, phytohormone responsive and abiotic and biotic stresses (Supplementary Figure S4, Supplementary Table S5). Among these, cis-acting elements related to phytohormone responsiveness accounted for 41.18% in second subgroup. LbWRKY38 and LbWRKY39 contained the biggest number of cis-elements (21), while LbWRKY26, LbWRKY28, LbWRKY30 and LbWRKY41 contained the lowest number (3). Phytohormone response elements such as ABRE 21.67% and TGACG-motif 11.59%, antioxidant response elements ARE 15.12% were widely distributed in the promoter regions of LbWRKY genes. However, only LbWRKY10 and LbWRKY80 contained cis-acting elements involved in plant growth and development, such as the HD-ZIP 3 element, which is associated with meristem formation, lateral organ morphogenesis, and vascular development.
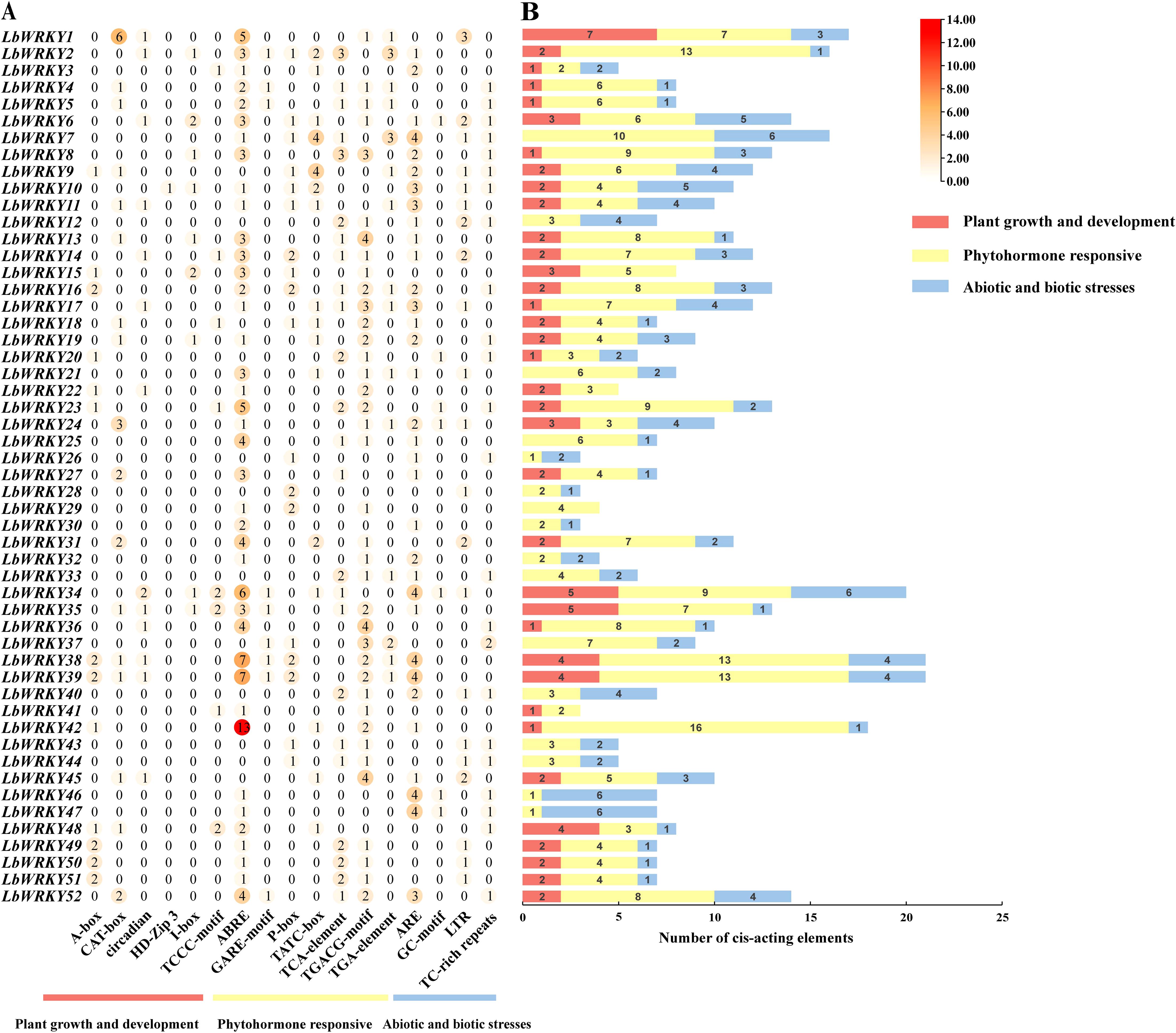
Figure 5. Analysis of cis-acting elements in promoter regions of LbWRKY1-52. (A) Three categories of cis-acting elements in the LbWRKYs. Different number and color of the circles representing the number of different cis elements. (B) The accumulation map of cis-acting elements in each LbWRKY gene. Different colors representing different categories of cis-acting elements.
Expression profile of LbWRKY gene family in different developmental stages of Lycium barbarum and prediction of miRNA target and SSR
To elucidate the expression profiles of LbWRKY members in response to Fusarium solani infection during the development of L. barbarum, the expression patterns of 28 LbWRKY genes were analyzed using the Genevestigator platform (Supplementary Figure S7). The results indicated that the LbWRKY family was widely expressed across different tissues and developmental stages of L. barbarum. Notably, LbWRKY7, LbWRKY8 and LbWRKY14 exhibited significantly higher expression levels, suggesting their crucial roles in the growth and development. Then, the 3D structural models of LbWRKY7, LbWRKY8 and LbWRKY14 are predicted by AlphaFold tool. The results showed that the protein tertiary structure contain alpha helix and random coil (Supplementary Figure S8).
MicroRNAs (miRNAs) plays a significant role in plant gene regulation (Millar, 2020). In this study, tomato miRNA was used to predict miRNA targets of LbWRKY genes in Lycium barbarum. A total of 32 different miRNA (Supplementary Table S6) were predicted, of which LbWRKY72, LbWRKY100 and LbWRKY101 contained the largest number of miRNA targets (n=9). Using Trf software to analyze the SSR of LbWRKY family, there are 5 kinds of SSR sequences, which are CAA, AAACAA, CCA, GCA and TAA, among which CCA sequence has the most replication times, with a total of 21 times. In addition, the sequence similarity between the repeats of LbWRKY62 and LbWRKY18 was the highest, which was 93%.
Analysis expression pattern of LbWRKY genes after inoculation with F. solani
Previous research has demonstrated that WRKY transcription are widely involved in plant disease resistance pathways (Rushton et al., 2010). We analyzed the transcriptome of L. barbarum inoculated with F. solani, a total of 4271 genes were differentially expressed, of which 2144 were up-regulated and 2127 down-regulated (Figure 6A). These differential genes were clustered into seven modules according to their similar expression patterns (Figure 6B). These results indicated that pathogen invasion triggers significant changes in gene expression in L. barbarum.
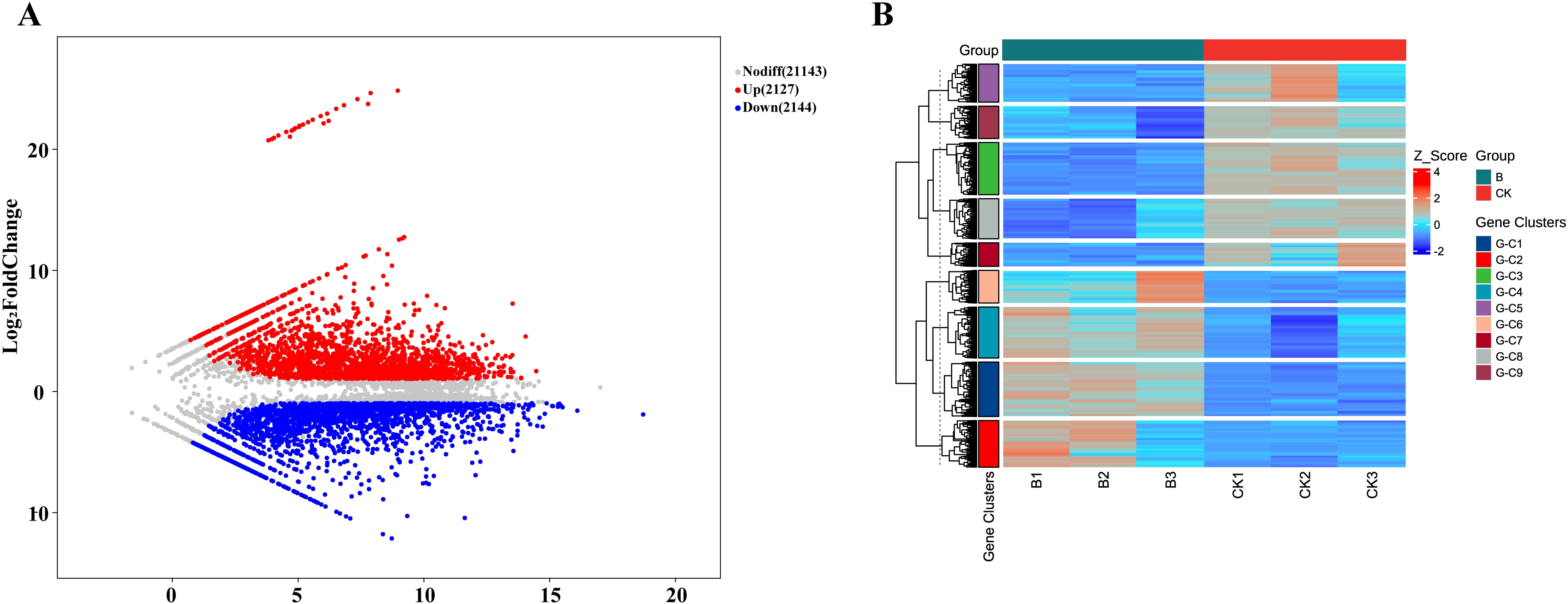
Figure 6. DEGs expression pattern of Wolfberry under root rot pathogen stress. (A) MA plot (M-versus-A plot) of DEGs. Color indicates whether the gene is up-regulated, down-regulated or non-significantly differentially expressed. (B) Cluster analysis heat map of DEGs. Gene clusters, Gene clustering classification tags, genes with similar expression patterns are clustered into a cluster (G-C1, C2 etc).
KEGG pathway analysis of the transcriptome data highlighted several enriched pathways, including plant hormone signal transduction, phenylpropanoid biosynthesis and flavonoid biosynthesis, suggesting that the synthesis of stimulant metabolites in wolfberry plays an important role in the resistance to F. solani (Supplementary Figure S5). In addition, statistical analysis of TF families showed significant changes in 38 TFs, among which most members of the ERF, MYB, bHLH families involved in the resistance response (Supplementary Figure S6). Notably, the expression of 28 WRKY members was up-regulated, making them the second most up-regulated TF family after to ERF.
Correlation analysis of DEGs between LbWRKYs and involved in flavonoid biosynthesis and phenylpropanoid biosynthesis
As a key components of plant defense, secondary metabolites play a crucial role in plant disease resistance (Hou et al., 2024). It has been demonstrated that WRKY TF regulate the synthesis of secondary metabolites (Lu et al., 2023; Zhao et al., 2024). To elucidate the role of WRKY family members in regulating flavonoids and phenylpropane synthesis in L. barbarum for enhanced resistance to root rot, we analyzed the correlation between WRKYs and synthesis-related genes in response to F. solani (Supplementary Table S7, Supplementary Table S8). The expression of 28 LbWRKY genes changed significantly, with some showing strong correlations with the key genes of flavonoid and phenylpropanoid synthesis (Figure 7). LbWRKY63 displayed a significant positive correlation with PER21 and GT5, while LbWRKY84 and LbWRKY22 were positively correlated with 4CL2 and AHT1. A highly significant positive correlation was observed between LbWRKY97 and CAD6. conversely, most relationships between AT1, C4H2 and HHT1 genes and these LbWRKYs were negatively correlated. These results showed that LbWRKY transcription factors regulate the synthesis of flavonoids and phenylpropanoids in L. barbarum, thereby contributing to resistance against F. solani.
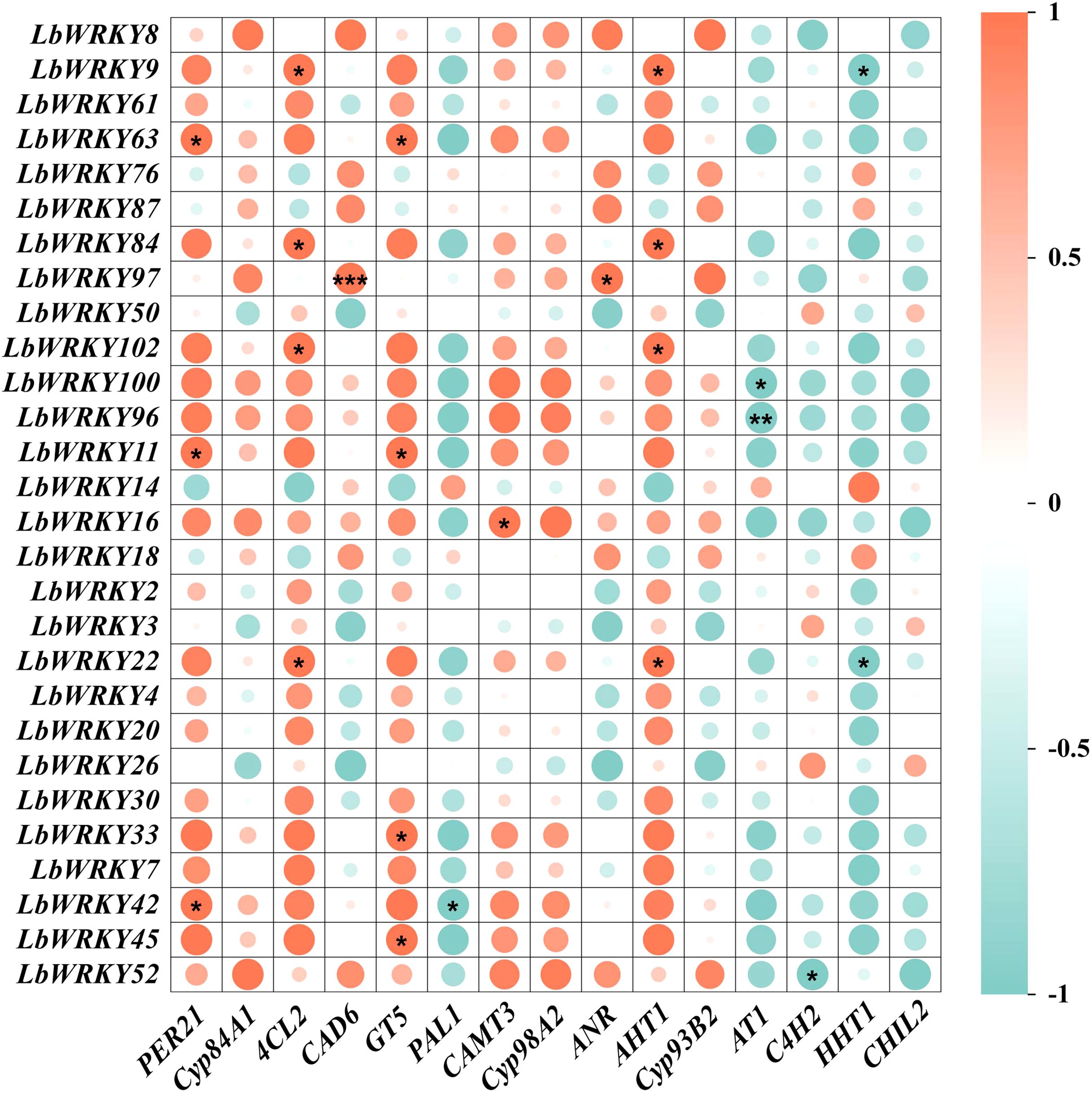
Figure 7. The correlation analysis of DEGs between LbWRKYs and involved in flavonoid biosynthesis and phenylpropanoid biosynthesis. The depth of color represents the strength of correlation. *P < 0.05, **P < 0.01, ***P < 0.001.
Prediction regulatory network of LbWRKYs and synthesis-related genes of flavonoids and phenylpropane
Utilizing the STRING database, the regulatory network of LbWRKY genes and synthesis-related genes in response to F. solani was predicted, with Solanum lycopersicum serving as a reference species (Figure 8). This network analysis enabled us to infer the potential functions of LbWRKY genes in the synthesis of secondary metabolites. LbWRKY8, LbWRKY100, LbWRKY16, LbWRKY3, LbWRKY33 and LbWRKY63 exhibited a strong correlation to synthesis of secondary metabolites, particularly phenylpropanoid biosynthesis, as deduced from their roles in S. lcopersicum. These findings suggested that LbWRKY genes may be involved in regulating plant growth and development and stress resistance process.
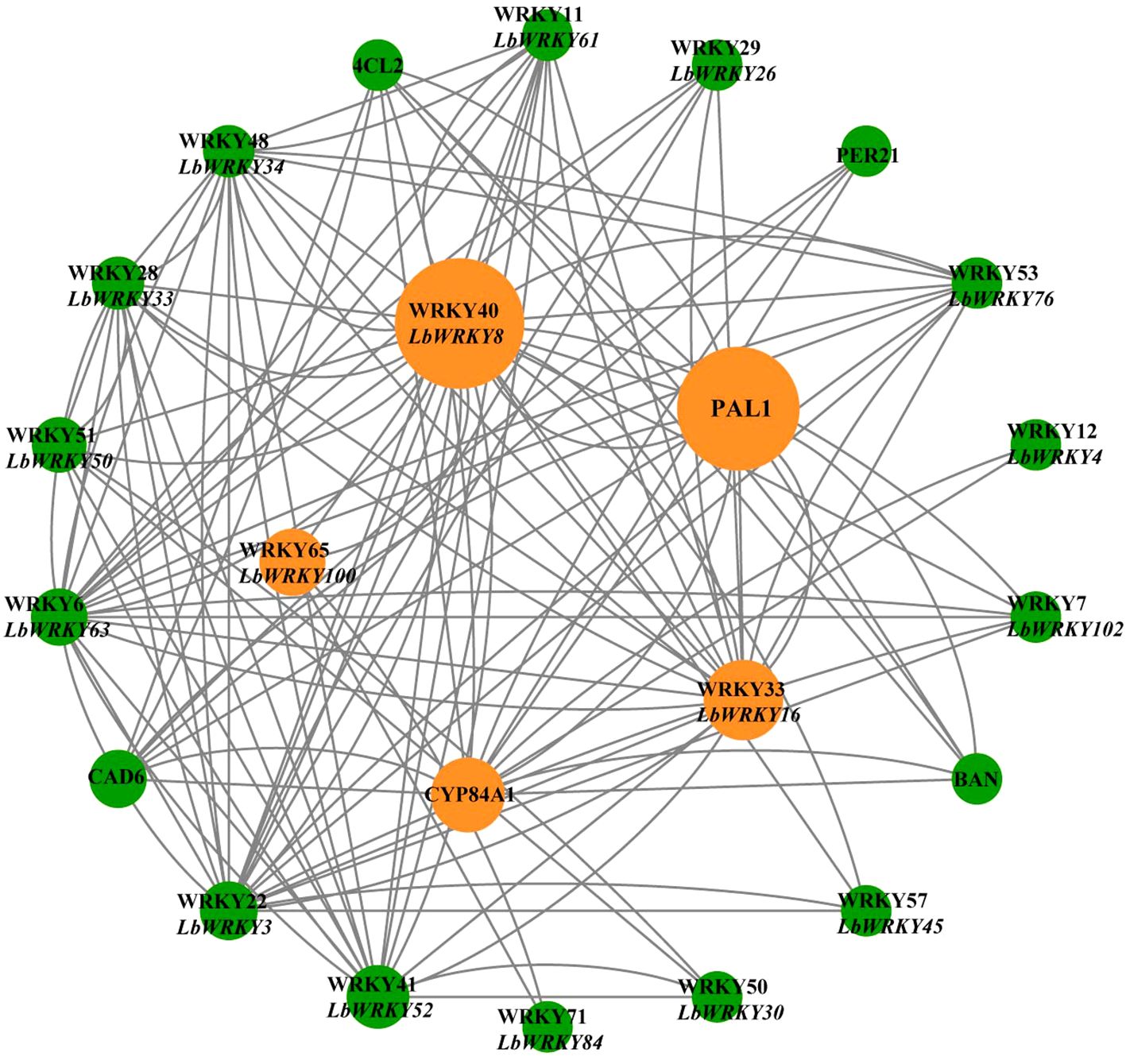
Figure 8. Protein regulatory network of LbWRKYs and synthesis-related genes of flavonoids and phenylpropane. Different color circles represented WRKYs members and flavonoid phenylpropanoid synthesis-related genes in response to F. solani, and the grey lines represent possible regulatory relationships. The inner circle represents the genes with more regulatory relationships. The size of the circle represents the degree of connection. If the circle was larger, the degree of connection was higher.
Expression profile of LbWRKY genes in response to Fusarium solani
In order to clarify the expression characteristics of LbWRKY genes in L. barbarum during the incubation and disease development stages, we analyzed relative gene expression levels at 7 dpi and 28 dpi following inoculation with F. solani. The results showed that the expression of LbWRKY genes increased significantly after inoculation with F. solani, with some genes, including LbWRKY22, LbWRKY33, LbWRKY42, LbWRKY16 and LbWRKY84, showing continuous upregulation as the infection progressed (Figure 9A). It speculated that LbWRKYs play crucial roles in the disease resistance. Additionally, a standard curve was established to compare the expression levels of 12 related genes obtained through RNA-seq and RT-qPCR, yielding a correlation coefficient of 0.9493. The high correlation indicated strong consistency between the RNA-seq and RT-qPCR results (Figure 9B).
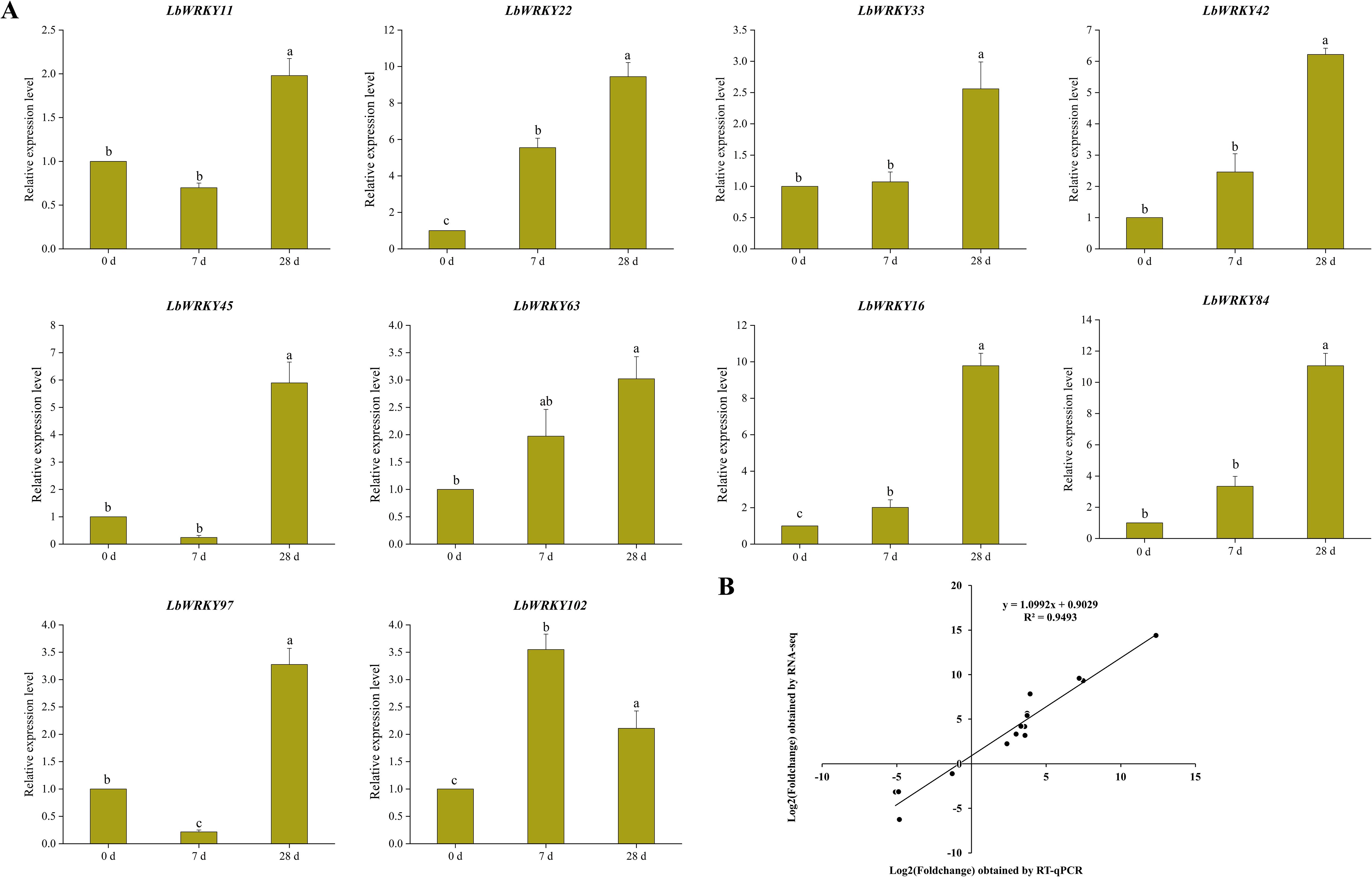
Figure 9. Response of relative expression of LbWRKY genes to F. solani and verification of RNA-seq data. (A) The expression levels of LbWRKY genes at 0 dpi, 7 dpi and 28 dpi, Lowercase letters indicate significant differences among different inoculation time (p-value<0.05). (B) Standard curve of log2Foldchange between RNA-Seq and qRT-PCR values. Lowercase letters indicate significant differences (P<0.05).
Discussion
WRKY TFs are one of the most prominent families in higher plants, playing crucial roles in plant growth and responses to both biotic and abiotic stresses (Ahmed et al., 2022). With the advancements in genomics and transgenic technology, the functions of WRKY gene family members in plant adaptation to environmental stress have become perspicuity. There were 76, 73, 81 and 116 WRKY gene family members in Cucurbita pepo (Chen et al., 2024), Capsicum chinense (Zhang et al., 2023), solanum lycopersicum (Khan et al., 2023) and Gossypium (Dou et al., 2014), they have been shown to play an active role in the biological process. In the present study, a total of 104 LbWRKY genes were identified in the L. bararum genome, 28 of which were responsive to the infection of F. solani.
The WRKY gene family is categorized into three groups based on highly conserved zinc finger motifs and WRKY domains (Eulgem et al., 2000). The LbWRKY gene family was divided into group I, II (IIa, IIb, IIc, IId, IIe), and III, with the proportion of WRKY members in each group being 25%, 60.6%, and14.4%, respectively. Notably, members of group II constitute more than half of the total. However, in Arabidopsis, group II members account for only 40% (Wang et al., 2011). It is speculated that members of group II underwent more gene duplication processes in wolfberry. Three variations of heptapeptide sequence, WRKYGKK, WRKYGMK and WRKYGQN were identified in LbWRKY genes, which may be associated with the amplification of WRKY gene in L. barbarum. In pepper (Capsicum annuum), the replacement of Q with M in the conserved WRKYGQK in CaWRKY27b prevents its binding to W-boxes. Instead, CaWRKY27b interacts with CaWRKY40 in the nucleus, positively regulates the tolerance of pepper to high temperature and the resistance to Ralstonia solanacearum (Yang et al., 2022). These variations are hypothesized to influence the expression of stress-responsive genes targeted by LbWRKY transcription factors. Further investigation is needed to elucidate the interaction mechanism of LbWRKY proteins in L. barbarum.
Conserved motif analysis provides essential information for understanding the evolution of gene family (Hu et al., 2015). The analysis of protein motifs showed that nearly all members of the group II contained motifs 1,2 and 4, but motif 5 was specific to subgroups IIc and IIb. In addition, there were motifs 6 and 9 were present in certain members of IIa and all members of IIb. The similarity in the distribution of conserved motifs among members of the same group indicated functional similarities. The promoter regions of WRKY gene contains stress-related cis-acting elements (Hayashi et al., 2013). We identified 992 cis-acting elements related to plant growth and development, plant hormone response, and abiotic and biotic stress. Notably, plant hormone-related elements accounted for 41.18%, indicating that the LbWRKY genes are involved in various plant hormones regulation pathways.
Plant-specific WRKY TFs play diverse roles in various plant processes, including growth, development, and stress signaling through both autonomic and cross-regulation of numerous genes (Bakshi and Oelmüller, 2014). Previous researches have demonstrated that WRKY TFs participate in the response to abiotic stress. For instance, sugarcane transcription factor ScWRKY4 has been shown to negatively regulate resistance to pathogen infection (Wang et al., 2024a). In Panax notoginseng, PnWRKY9 positively regulates the resistance to root rot caused by F. solani (Zheng et al., 2022). In S. lycopersicum, SlWRKY30 and SlWRKY81 work synergistically against modulate immunity to Ralstonia solanacearum (Dang et al., 2023). In this study, 28 LbWRKY members responded to F. solani, including LbWRKY2, LbWRKY8, LbWRKY33, LbWRKY30 and LbWRKY50, among others. This finding is consistent with previous studies, suggesting a close association between the LbWRKY gene family and the resistance of wolfberry to pathogens. In addition, transcriptome analysis of L. barbarum after inoculation with F. solani revealed differential expression of genes related to flavonoid and phenylpropanoid synthesis pathways. Notably, there were strong correlations between WRKY members and genes related to flavonoid and phenylpropanoid synthesis in response to F. solani. These results provide further evidence that WRKY members regulate the synthesis of flavonoids and phenylpropanoids thereby enhancing disease resistance (Wen et al., 2021; Ma F et al., 2023).
Protein-protein interactions (PPI) are essential for the regulation of plant growth, and the stimulation of transcription factors to proteins activates various stress pathways in plants (Braun et al., 2013; Francois et al., 2020). The PPIs of LbWRKY genes and synthesis-related genes were analyzed using String database, revealing strong correlations. For example, WRKY40 had high degree value of 211.02. Among these, the homologue of LbWRKY8 in S. lycopersicum was identified as WRKY40 (Solyc03g116890), which acts as positive regulator in effector-triggered immunity against the bacterial pathogen Pseudomonas syringae DC3000 (Moritz et al., 2013). In other Solanaceae plants, WRKY40 plays a positive role in disease resistance. For instance, CaWRKY40 mediates autoregulation during the response to R. solanacearum in pepper (Liu et al., 2020). In addition, CaWRKY40 binds to the W4-box element of the ChiIV3 promoter region, activating transcription and enhancing resistance to R. solanacearum (Liu et al., 2019). In summary, it speculated that LbWRKY gene family may play an active role in resistance to root rot. Interestingly, a significant correlation was observed between LbWRKY genes and genes involved in flavonoid and phenylpropanoid biosynthesis, including PAL, 4CL2, CAD6 and PER21. The heterologous expression of CcWRKY25 from Capsicum chinense in Arabidopsis promoted the expression of PAL, 4CL1, 4CL2, 4CL3, CCR and CCoAOMT, leading to the accumulation of lignin and flavonoids (Zhang et al., 2023). Similarly, in Ocimum sanctum, overexpression of OscWRKY1 positively regulated genes in the phenylpropanoid pathway and enhances resistance to pathogens (Joshi et al., 2022). These researches further illustrate that the LbWRKY gene family in L. barbarum regulates resistance to F. solani by modulating flavonoid and phenylpropanoid synthesis pathways.
Conclusion
In the current study, the WRKY gene family in the whole genome of Lycium barbarum was identified, comprising a total of 104 members. The bioinformatics identification and characteristics of the LbWRKY genes were conducted, and collinearity analysis with the WRKY gene families of other Solanaceae plants showed their conserved evolutionary nature. Transcriptome data revealed 28 LbWRKY genes responding to Fusarium solani, and played a key role in regulating flavonoid and phenylpropanoid synthesis pathways in Lycium barbarum. These results provide valuable insights into the functional roles of LbWRKY genes in root rot resistance in L. bararum and lay a foundation for the breeding disease-resistant cultivars of wolfberry.
Data availability statement
The original contribution presented in this study is included in the article/Supplementary Material, and the raw RNA-seq data are freely available in the NCBI database under accession no. PRJNA1260582.
Author contributions
XW: Writing – original draft. NL: Formal analysis, Visualization, Writing – review & editing. BW: Writing – review & editing. WC: Visualization, Writing – review & editing. CZ: Writing – original draft, Data curation, Formal analysis. YS: Visualization, Writing – original draft. JH: Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32060341), Gansu Provincial Department of Science and Technology Major Project (23ZDNA002), Gansu Province Education Science and Technology Innovation Project in 2025 (2025CXZX-833)and Gansu Provincial Science and Technology Department Rural Revitalization Project (Grant No. 24CXNA018).
Acknowledgments
The author would like to thank the facilities, support and encouragement provided by the Engineering Research Center of Harmless Cultivation of L. barbarum in Gansu Province.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1543373/full#supplementary-material
References
Agbowuro, G. O., Afolabi, M. S., Olamiriki, E. F., and Awoyemi, S. O. (2020). Rice blast disease (Magnaporthe oryzae): A menace to rice production and humanity. Int. J. Pathogen Res. 4, 32–39. doi: 10.9734/ijpr/2020/v4i330114
Ahmad, Y., Haider, S., Iqbal, J., Naseer, S., Attia, K. A., Mohammed, A. A., et al. (2024). In-silico analysis and transformation of OsMYB48 transcription factor driven by CaMV35S promoter in model plant - Nicotiana tabacum L. conferring abiotic stress tolerance. GM Crops Food 15, 130–149. doi: 10.1080/21645698.2024.2334476
Ahmed, K. M., Amjad, H., Nurhasanah, R. F., Qurban, A., Malook, C. M., M., A. R., et al. (2022). WRKY transcription factors (TFs): molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1039329
Ahuja, I., Vos, R.C.H.d., Bones, A. M., and Hall, R. D. (2010). Plant molecular stress responses face climate change. Trends Plant Sci. 15, 664–674. doi: 10.1016/j.tplants.2010.08.002
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bakshi, M. and Oelmüller, R. (2014). WRKY transcription factors: Jack of many trades in plants. Plant Signal Behav. 9, e27700. doi: 10.4161/psb.27700
Bian, S. L., Zhao, M. L., Zhang, H. J., and Ren, Y. J. (2024). Differentially expressed genes identification of Kohlrabi Seedlings (Brassica oleracea var. caulorapa L.) under polyethylene glycol osmotic stress and AP2/ERF transcription factor family analysis. Plants (Basel) 13, 1167. doi: 10.3390/plants13081167
Braun, P., Aubourg, S., Van, L. J., De, J. G., and Lurin, C. (2013). Plant protein interactomes. Annu. Rev. Plant Biol. 64, 161–187. doi: 10.1146/annurev-arplant-050312-120140
Cai, H., Yang, S., Yan, Y., Xiao, Z., Cheng, J., Wu, J., et al. (2015). CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 66, 3163–3174. doi: 10.1093/jxb/erv125
Cao, Y. L., Li, Y. l., Fan, Y. F., Li, Z., Yoshida, K., Wang, J. Y., et al. (2021). Wolfberry genomes and the evolution of Lycium (Solanaceae). Commun. Biol. 4, 671. doi: 10.1038/s42003-021-02152-8
Chen, C. H., Chen, X. Q., Han, J., Lu, W. L., and Ren, Z. H. (2020a). Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 20, 443. doi: 10.1186/s12870-020-02625-8
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020b). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, F., Hu, Y., Vannozzi, A., Wu, K. C., Cai, H. Y., Qin, Y., et al. (2017). The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 36, 311–335. doi: 10.1080/07352689.2018.1441103
Chen, J. H., Tao, F., Xue, Y. Y., Xu, B. L., and Li, X. W. (2024). Genome-wide identification of the WRKY gene family and functional characterization of CpWRKY5 in Cucurbita pepo. Int. J. Mol. Sci. 25, 4177. doi: 10.3390/ijms25084177
Chou, K. C. and Shen, H. B. (2008). Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 3, 153–162. doi: 10.1038/nprot.2007.494
Cichy, K. A., Snapp, S. S., and Kirk, W. W. (2007). Fusarium root rot incidence and root system architecture in grafted common bean lines. Plant Soil 300, 233–244. doi: 10.1007/s11104-007-9408-0
Dang, F., Lin, J., Li, Y., Jiang, R., Fang, Y., Ding, F., et al. (2023). SlWRKY30 and SlWRKY81 synergistically modulate tomato immunity to Ralstonia solanacearum by directly regulating SlPR-STH2. Hortice Res. 10, uhad050. doi: 10.1093/hr/uhad050
Dou, L. L., Zhang, X. H., Pang, C. Y., Song, M. Z., Wei, H. L., Fan, S. L., et al. (2014). Genome-wide analysis of the WRKY gene family in cotton. Mol. Genet. Genomics 289, 1103–1121. doi: 10.1007/s00438-014-0872-y
Eliane, T. B. (2017). Root rot diseases in plants: A review of common causal agents and management strategies. Agric. Res. Technol. 5, 1–8. doi: 10.19080/ARTOAJ.2017.05.555661
Enright, A. J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D. S. (2003). MicroRNA targets in drosophila. Genome Biol. 5, R1. doi: 10.1186/gb-2003-5-1-r1
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/s1360-1385(00)01600-9
Francois, M., Donovan, P., and Fontaine, F. (2020). Modulating transcription factor activity: Interfering with protein-protein interaction networks. Semin. Cell Dev. Biol. 99, 12–19. doi: 10.1016/j.semcdb.2018.07.019
Gahlaut, V., Jaiswal, V., Kumar, A., and Gupta, P. K. (2016). Transcription factors involved in drought tolerance and their possible role in developing drought tolerant cultivars with emphasis on wheat (Triticum aestivum L.). Theor. Appl. Genet. 129, 2019–2042. doi: 10.1007/s00122-016-2794-z
Gong, H. G., Haiguang, G., Fazal, R., Yun, M., Biao, A., Shaohua, Z., et al. (2022). Germplasm resources and strategy for genetic breeding of Lycium species: A review. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.802936
Hayashi, S., Takahashi, H., Wakasa, Y., Kawakatsu, T., and Takaiwa, F. (2013). Identification of a cis-element that mediates multiple pathways of the endoplasmic reticulum stress response in rice. Plant J. 74, 248–257. doi: 10.1111/tpj.12117
He, W., Liu, M. Y., Qin, X. Y., Liang, A. H., Chen, Y., Yin, Y., et al. (2022). Genome-wide identification and expression analysis of the aquaporin gene family in Lycium barbarum during fruit ripening and seedling response to heat stress. Curr. Issues Mol. Biol. 44. doi: 10.3389/fpls.2023.1305299
Hou, J., Ai, M. T., Li, J. B., Cui, X. M., Liu, Y., and Yang, Q. (2024). Exogenous salicylic acid treatment enhances the disease resistance of Panax Vietnamensis by regulating secondary metabolite production. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1428272
Hruz, T., Laule, O., Szabo, G., Wessendorp, F., Bleuler, S., Oertle, L., et al. (2008). Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinf. 2008, 420747. doi: 10.1155/2008/420747
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, N., Zheng, J., Li, W. C., and Suo, Y. R. (2014). Isolation, stability, and antioxidant activity of anthocyanins from Lycium ruthenicum murray and Nitraria Tangutorum bobr of Qinghai-Tibetan plateau. Sep Sci. Technol. 49, 2897–2906. doi: 10.1080/01496395.2014.943770
Jiang, J. J., Ma, S. H., Ye, N. H., Jiang, M., Cao, J. S., and Zhang, J. H. (2017). WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59, 86–101. doi: 10.1111/jipb.12513
Joshi, A., Jeena, G. S., Shaikha, Kumar, R. S., Pandey, A., and Shukla, R. (2022). Ocimum sanctum, OscWRKY1, regulates phenylpropanoid pathway genes and promotes resistance to pathogen infection in Arabidopsis. Plant Mol. Biol. 110, 235–251. doi: 10.1007/s11103-022-01297-2
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature. 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kazuhiko, Y., Takanori, K., Makoto, I., Masaru, T., Tomoko, Y., Takashi, Y., et al. (2005). Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell 17, 944–956. doi: 10.1105/tpc.104.026435
Khan, I. N., Asaf, S., Jan, R., Bilal, S., Lubna, Khan, A. L., et al. (2023). Genome-wide annotation and expression analysis of WRKY and bHLH transcriptional factor families reveal their involvement under cadmium stress in tomato (Solanum lycopersicum L.). Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1100895
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Levin, R. A. and Miller, J. S. (2005). Relationships within tribe Lycieae (Solanaceae): paraphyly of Lycium and multiple origins of gender dimorphism. Am. J. Bot. 92, 2044–2053. doi: 10.3732/ajb.92.12.2044
Li, W. N., Che, J. H., Lian, Q. L., Wang, C. P., Dai, G. L., and Chen, J. H. (2023). Identification and expression pattern of the carotenoid cleavage oxygenase gene family in Lycium suggest CCOs respond to abiotic stress and promote carotenoids degradation. Forests 14, 983. doi: 10.3390/f14050983
Li, S. L., Khoso, M. A., Wu, J. B., Yu, B. G., Wagan, S., and Liu, L. J. (2024). Exploring the mechanisms of WRKY transcription factors and regulated pathways in response to abiotic stress. Plant Stress 12, 100429. doi: 10.1016/j.stress.2024.100429
Li, H. P., Wu, J. J., Shang, X. F., Geng, M. M., Gao, J., Zhao, S. Q., et al. (2020). WRKY transcription factors shared by BTH-induced resistance and NPR1 -mediated acquired resistance improve broad-spectrum disease resistance in Wheat. Mol. Plant Microbe Interact. 33, 433–443. doi: 10.1094/MPMI-09-19-0257-R
Liu, B., Hong, Y. B., Zhang, Y. F., Li, X. H., Huang, L., Zhang, H. J., et al. (2014). Tomato WRKY transcriptional factor SlDRW1 is required for disease resistance against Botrytis cinerea and tolerance to oxidative stress. Plant Sci. 227, 145–156. doi: 10.1016/j.plantsci.2014.08.001
Liu, Z. Q., Shi, L. P., Weng, Y. H., Zou, H. S., Li, X., Yang, S., et al. (2019). ChiIV3 acts as a novel target of WRKY40 to mediate pepper immunity against Ralstonia solanacearum infection. Mol. Plant Microbe Interact. 32, 1121–1133. doi: 10.1094/MPMI-11-18-0313-R
Liu, Z. Q., Shi, L. P., Yang, S., Qiu, S. S., Ma, X. L., Cai, J. S., et al. (2020). A conserved double-W box in the promoter of CaWRKY40 mediates autoregulation during response to pathogen attack and heat stress in pepper. Mol. Plant Pathol. 22, 3–18. doi: 10.1111/mpp.13004
Liu, J. T., Wang, Y. Q., Ye, X. R., Zhang, Q. R., Li, Y. P., Chen, M. D., et al. (2024). Genome-wide identification and expression analysis of the WRKY gene family in response to low-temperature and drought stresses in Cucurbita pepo L. Sci. Hortic. 330, 113048. doi: 10.1016/j.scienta.2024.113048
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–d268. doi: 10.1093/nar/gkz991
Lu, Z. G., Wang, X. W., Mostafa, S., Noor, I., Lin, X. Y., Ren, S. X., et al. (2023). WRKY Transcription Factors in Jasminum sambac: an insight into the regulation of aroma synthesis. Biomolecules 13, 1679. doi: 10.3390/biom13121679
Luan, Y. T., Chen, Z. J., Meng, J. S., Tao, J., and Zhao, D. Q. (2023). PoWRKY17 promotes drought tolerance in Paeonia ostii by modulating lignin accumulation. Industrial Crops Products 204, 117228. doi: 10.1016/j.indcrop.2023.117228
Ma, R. H., Zhang, X. X., Ni, Z. J., Thakur, K., Wang, W., Yan, Y. M., et al. (2023). Lycium barbarum (Goji) as functional food: a review of its nutrition, phytochemical structure, biological features, and food industry prospects. Crit. Rev. Food Sci. Nutr. 63, 10621–10635. doi: 10.1080/10408398.2022.2078788
Ma, F. N., Zhou, H. W., Yang, H. T., Huang, D. M., Xing, W. T., Wu, B., et al. (2023). WRKY transcription factors in passion fruit analysis reveals key PeWRKYs involved in abiotic stress and flavonoid biosynthesis. Int. J. Biol. Macromol 256, 128063–128063. doi: 10.1016/j.ijbiomac.2023.128063
Moritz, S., Armin, T., Celia, D., Charlotte, R., Lore, W., Marcel, W., et al. (2013). Analyses of wrky18 wrky40 plants reveal critical roles of SA/EDS1 signaling and indole-glucosinolate biosynthesis for Golovinomyces orontii resistance and a loss-of resistance towards Pseudomonas syringae pv. tomato AvrRPS4. Mol. Plant Microbe Interact. 26, 758–767. doi: 10.1094/MPMI-11-12-0265-R
Nisha, N. and Paramjit, K. (2021). A salicylic acid inducible mulberry WRKY transcription factor, MiWRKY53 is involved in plant defence response. Plant Cell Rep. 40, 1–21. doi: 10.1007/s00299-021-02710-8
Ou, X. G., Sun, L. X. Y., Chen, Y., Zhao, Z. W., and Jian, W. (2024). Characteristics of NAC transcription factors in Solanaceae crops and their roles in responding to abiotic and biotic stresses. Bio Bioph Res. Co 709, 149840. doi: 10.1016/j.bbrc.2024.149840
Panchy, N., Lehti-Shiu, M., and Shiu, S. H. (2016). Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316. doi: 10.1104/pp.16.00523
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Saliminejad, K., Khorram Khorshid, H. R., Soleymani, F. S., and Ghaffari, S. H. (2019). An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell Physiol. 234, 5451–5465. doi: 10.1002/jcp.27486
Sganzerla, M. G., Dutt, M., Kumar, A., and Kelvin, D. J. (2024). Multiple Protein Profiler 1.0 (MPP): A Webserver for predicting and visualizing physiochemical properties of proteins at the proteome Level. Protein J. 43, 711–717. doi: 10.1007/s10930-024-10214-z
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shrestha, A., Khan, A., and Dey, N. (2018). Cis–trans engineering: Advances and perspectives on customized transcriptional regulation in plants. Mol. Plant 11, 886–898. doi: 10.1016/j.molp.2018.05.008
Song, H., Sun, W. H., Yang, G. F., and Sun, J. (2018). WRKY transcription factors in legumes. BMC Plant Biol. 18, 243. doi: 10.1186/s12870-018-1467-2
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., and Chen, W. H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275. doi: 10.1093/nar/gkz357
Szklarczyk, D., Kirsch, R., Koutrouli, M., Nastou, K., Mehryary, F., Hachilif, R., et al. (2023). The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646. doi: 10.1093/nar/gkac1000
Wang, T. T., Liu, Z. H., Yan, J. W., Chen, J. Y., Wang, L., Yu, W. W., et al. (2024b). A Torreya grandis ARF transcription factor TgARF15 enhances drought stress tolerance. Environ. Exp. Bot. 222, 105761. doi: 10.1016/j.envexpbot.2024.105761
Wang, D. J., Wang, W., Zang, S. J., Qin, L. Q., Liang, Y. L., Lin, P. X., et al. (2024a). Sugarcane transcription factor ScWRKY4 negatively regulates resistance to pathogen infection through the JA signaling pathway. Crop J. 12, 164–176. doi: 10.1016/j.cj.2023.12.002
Wang, Q. S., Wang, M. H., Zhang, X. Z., Hao, B. J., Kaushik, S. K., and Pan, Y. Y. (2011). WRKY gene family evolution in Arabidopsis thaliana. Genetica 139, 973–983. doi: 10.1007/s10709-011-9599-4
Wang, W., Wu, Y., and Messing, J. (2014). RNA-Seq transcriptome analysis of Spirodela dormancy without reproduction. BMC Genomics 15, 60. doi: 10.1186/1471-2164-15-60
Wani, S. H., Tripathi, P., Zaid, A., Challa, G. S., Kumar, A., Kumar, V., et al. (2018). Transcriptional regulation of osmotic stress tolerance in wheat (Triticum aestivum L.). Plant Mol. Biol. 97, 469–487. doi: 10.1007/s11103-018-0761-6
Wen, W. W., Wang, R. Y., Su, L. T., Lv, A. M., Zhou, P., and An, Y. (2021). MsWRKY11, activated by MsWRKY22, functions in drought tolerance and modulates lignin biosynthesis in alfalfa (Medicago sativa L.). Environ. Exp. Bot. 184, 104373. doi: 10.1016/j.envexpbot.2021.104373
Xiao, R. Y., Sun, Y., Yang, S., Yang, Y. X., Wang, D. H., Wang, Z. Z., et al. (2023). Systematic identification and functional analysis of the Hypericum perforatum L. bZIP gene family indicating that overexpressed HpbZIP69 enhances drought resistance. Int. J. Mol. Sci. 24, 14238. doi: 10.3390/ijms241814238
Yang, S., Cai, W., Shen, L., Cao, J., Liu, C., Hu, J., et al. (2022). A CaCDPK29-CaWRKY27b module promotes CaWRKY40-mediated thermotolerance and immunity to Ralstonia solanacearum in pepper. New Phytol. 233, 1843–1863. doi: 10.1111/nph.17891
Yao, R., Heinrich, M., and Weckerle, C. S. (2017). The genus Lycium as food and medicine: A botanical, ethnobotanical and historical review. J. Ethnopharmacol 212, 50–66. doi: 10.1016/j.jep.2017.10.010
Ye, D. L., Liu, J. N., Tian, X. B., Wen, X. L., Zhang, Y. Y., Zhang, X. L., et al. (2024). Genome-wide identification of bHLH gene family and screening of candidate gene in response to salt stress in kiwifruit. Environ. Exp. Bot. 222, 105774. doi: 10.1016/j.envexpbot.2024.105774
Yin, Y., Guo, C., Shi, H. Y., Zhao, J. H., Ma, F., An, W., et al. (2022a). Genome-wide comparative analysis of the R2R3-MYB gene family in five Solanaceae species and identification of members regulating carotenoid biosynthesis in Wolfberry. Int. J. Mol. Sci. 23, 2259. doi: 10.3390/ijms23042259
Yin, Y., Shi, H. Y., Mi, J., Qin, X. Y., Zhao, J. H., Zhang, D. K., et al. (2022b). Genome-Wide identification and analysis of the BBX gene family and its role in carotenoid biosynthesis in Wolfberry (Lycium barbarum L.). Int. J. Mol. Sci. 23, 8440. doi: 10.3390/ijms23158440
Zhang, H. Y., Liu, F. R., Wang, J. J., Yang, Q. R., Wang, P., Zhao, H. J., et al. (2021). Salicylic acid inhibits the postharvest decay of goji berry (Lycium barbarum L.) by modulating the antioxidant system and phenylpropanoid metabolites. Postharvest Biol. Technol. 2021, 178(1). doi: 10.1016/j.postharvbio.2021.111558
Zhang, L. P., Wu, D., Zhang, W., Shu, H. Y., Sun, P. X., Huang, C., et al. (2023). Genome-wide identification of WRKY gene family and functional characterization of CcWRKY25 in Capsicum chinense. Int. J. Mol. Sci. 24, 11389. doi: 10.3390/ijms241411389
Zhang, Z. H., Yu, J. Y., Li, D. F., Zhang, Z. Y., Liu, F. X., Zhou, X., et al. (2009). PMRD: plant microRNA database. Nucleic Acids Res. 38, D806–D813. doi: 10.1093/nar/gkp818
Zhao, P. P., Zhao, M., Gao, X. Y., Shan, Y., Li, F. J., Tian, X. L., et al. (2024). GhWRKY1bD improves drought tolerance by co-regulation of ABA, ROS, and proline homeostasis in cotton (Gossypium hirsutum). Ind Crops Prod 220, 119179. doi: 10.1016/j.indcrop.2024.119179
Zheng, L. L., Qiu, B. L., Su, L. L., Wang, H. L., Cui, X. M., Ge, F., et al. (2022). Panax notoginseng WRKY transcription factor 9 is a positive regulator in responding to root rot pathogen Fusarium solani. Front Plant Sci. 13, 930644. doi: 10.3389/fpls.2022.930644
Zhu, S. S., Fan, R. C., Xiong, X., Li, J. J., Xiang, L., Hong, Y. H., et al. (2022b). MeWRKY IIas, subfamily genes of WRKY transcription factors from Cassava, play an important role in disease resistance. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.890555
Keywords: Lycium barbarum, WRKY gene family, Fusarium solani, flavonoid and phenylpropanoid synthesis, expression analysis
Citation: Wu X, Li N, Wang B, Chen W, Zhang C, Sun Y and He J (2025) Genome-wide identification of WRKY gene family and expression analysis of key WRKY genes in response to Fusarium solani infection in Lycium barbarum. Front. Plant Sci. 16:1543373. doi: 10.3389/fpls.2025.1543373
Received: 11 December 2024; Accepted: 16 April 2025;
Published: 19 May 2025.
Edited by:
Yasser Nehela, Tanta University, EgyptReviewed by:
Anuj Kumar, Dalhousie University, CanadaJun Tang, Jiangsu Academy of Agricultural Sciences, China
Pradeep Kumar Singh, Eternal University, India
Copyright © 2025 Wu, Li, Wang, Chen, Zhang, Sun and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing He, aGVqaW5nMjY4QGFsaXl1bi5jb20=
 Xia Wu
Xia Wu Nan Li1
Nan Li1 Jing He
Jing He