- 1Department of Genetics and Plant Breeding, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
- 2Department of Plant Biotechnology, Centre for Plant Molecular Biology and Biotechnology, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
- 3Department of Rice, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
- 4Department of Plant Pathology, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
- 5Department of Crop Physiology, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
Rice (Oryza sativa. L) is a staple crop globally, but blast disease caused by fungal pathogens Magnaporthe oryzae is one of the most devastating and results in severe economic losses in rice production worldwide. Recent technological advancements have opened new possibilities for developing blast resistance. The dynamic and highly adaptable nature of M. oryzae allows it to overcome plant defense mechanisms rapidly, posing a major threat to global food security and agricultural sustainability. While foundational to early resistance development, traditional breeding approaches have been limited by their time-consuming nature and reliance on phenotypic selection. These methods often require several generations to establish stable resistance traits. However, with the emergence of molecular breeding technologies, resistance breeding has experienced significant acceleration and precision. Tools such as marker-assisted selection (MAS), marker-assisted backcross breeding (MABB), and quantitative trait locus (QTL) mapping allow for the identification and introgression of resistance genes (R genes) more efficiently and accurately. Recent advances in genome engineering techniques, particularly CRISPR-Cas 9, have transformed the capability to manipulate resistance genes directly, enabling targeted editing and stacking of multiple genes (gene pyramiding) for durable resistance. Moreover, omics technologies—including genomics, transcriptomics, proteomics, and metabolomics—offer a comprehensive understanding of the molecular interactions between host and pathogen, facilitating the discovery of novel resistance mechanisms and regulatory pathways. The integration of allele mining with advanced biotechnological tools has further promoted the development of cisgenic and intragenic plants, where resistance genes from related cultivars or wild species are introduced without foreign DNA, thus addressing public concerns over transgenic crops. These strategies enhance resistance and help retain the desirable agronomic traits of elite rice varieties. Despite these advancements, the high mutation rate and genetic plasticity of M. oryzae enable it to evolve and overcome resistance provided by single R genes. Therefore, understanding host–pathogen interactions at the molecular and cellular levels remains essential. Emerging technologies such as nanotechnology show promise in developing targeted fungicide delivery systems and innovative diagnostic tools. Synthetic biology opens avenues for constructing synthetic resistance pathways or deploying plant biosensors. Additionally, machine learning and artificial intelligence (AI) algorithms are increasingly used to predict disease outbreaks, model gene interactions, and optimize breeding strategies based on large datasets. Thus, managing rice blast disease necessitates a holistic approach combining conventional breeding wisdom with modern molecular tools and emerging technologies. The synergy among these approaches holds promise to enhance resistance durability and protect global rice production against evolving fungal threats. This review emphasizes recent advancements in managing rice blast disease, offering valuable insights to sustain resilient breeding programs against this pathogen.
Introduction
Rice (Oryza sativa L.) is considered the most crucial staple food crop, sustaining over half of the global population (Maurya et al., 2024). Biotic and abiotic stresses significantly impact yield losses in food production, thus necessitating the improvement of stress tolerance in crops. Among these stresses, disease stands out as a primary limiting stress factor in rice crop production (Zeng et al., 2023). Rice is susceptible to more than 70 diseases caused by various fungi, nematodes, viruses, and bacteria (Rijal and Devkota, 2020). Rice blast, caused by Magnaporthe oryzae (Anamorph: Pyricularia oryzae), is a particularly widespread and notorious fungal pathogen that can reduce grain yield and quality by 70%–80% (Simkhada and Thapa, 2022). This disease affects all parts of the rice plants and can lead to complete yield loss under favorable conditions worldwide. Leaf blast and panicle blast are two economically significant forms of disease (Ding et al., 1999). Leaf blast impairs photosynthesis and reduces carbohydrate production (Bastiaans, 1993), typically resulting in 1% to 10% yield losses (Savary et al., 2000); severe epidemics can cause leaf death or kill entire plants in early stages (Ou, 1985). Panicle blast, which is generally more economically impactful, can diminish yield by hindering grain filling, especially near the panicle base, potentially leading to the loss of the entire panicle (Zeigler et al., 1994). Approximately 30% of the yield loss is attributed to collar and neck blasts caused by spores produced later in the growth season (Srivastava et al., 2017). Climate change may alter pathogen distribution and growth rates and affect host plant resistance, growth, and metabolism. Lower temperatures in humid tropics and warm, humid subtropical regions increase the risk of blast epidemics (Ninomiya et al., 2020).
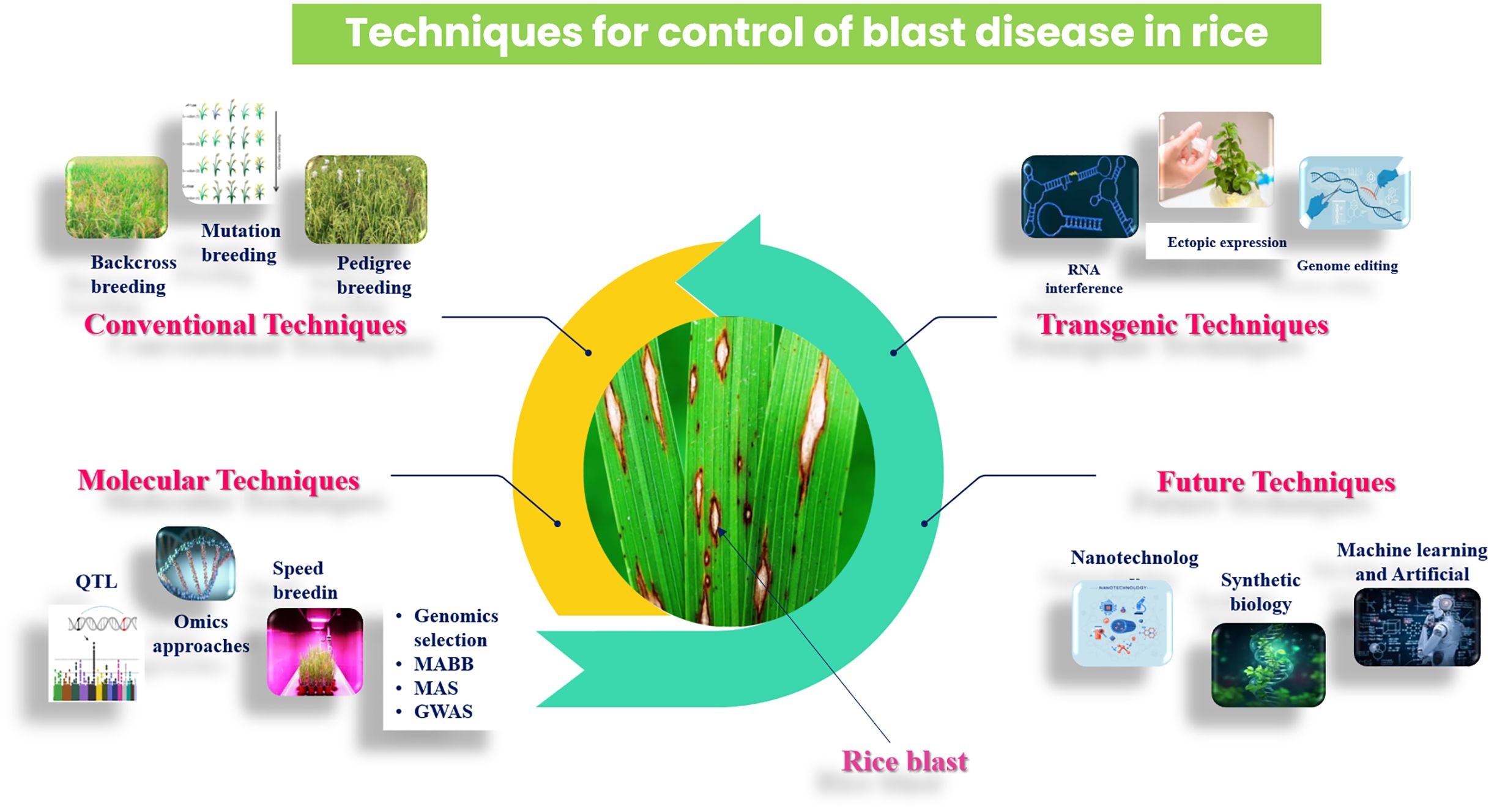
The management of rice blast disease primarily relies on traditional breeding techniques, such as pedigree, backcross, mutation, and recurrent selection, which often face challenges with linkage drag, where undesirable traits are inadvertently transferred along with resistance genes. These conventional methods have limitations in effectively controlling blast disease and are associated with high labor costs and a time-consuming process. Consequently, researchers have employed molecular markers to identify blast resistance using various techniques, including QTL mapping, genetic transformation, and marker-aided selection (MAS). Integrating marker-assisted selection (MAS) with traditional breeding methods has enabled the accumulation of R genes in elite rice cultivars, enhancing their resistance to blast and improving durability. These approaches involve enhanced selection processes that focus on improved quality and desirable traits, leading to the development of blast-resistant rice varieties (Ashkani et al., 2015). Thus, understanding the molecular interactions between the pathogen and the host plant is essential to develop effective strategies to manage and control rice blast disease. Additionally, recent studies have identified specific blast resistance genes, offering new opportunities for breeding programs (Ramkumar et al., 2015; Amoghavarsha et al., 2022). This review briefly examines conventional methods and molecular approaches such as transgenic techniques, CRISPR/Cas9 technology, nanotechnology, allele mining, and omics strategies that contribute to the management of rice blast disease, offering insights into its control.
Blast in rice
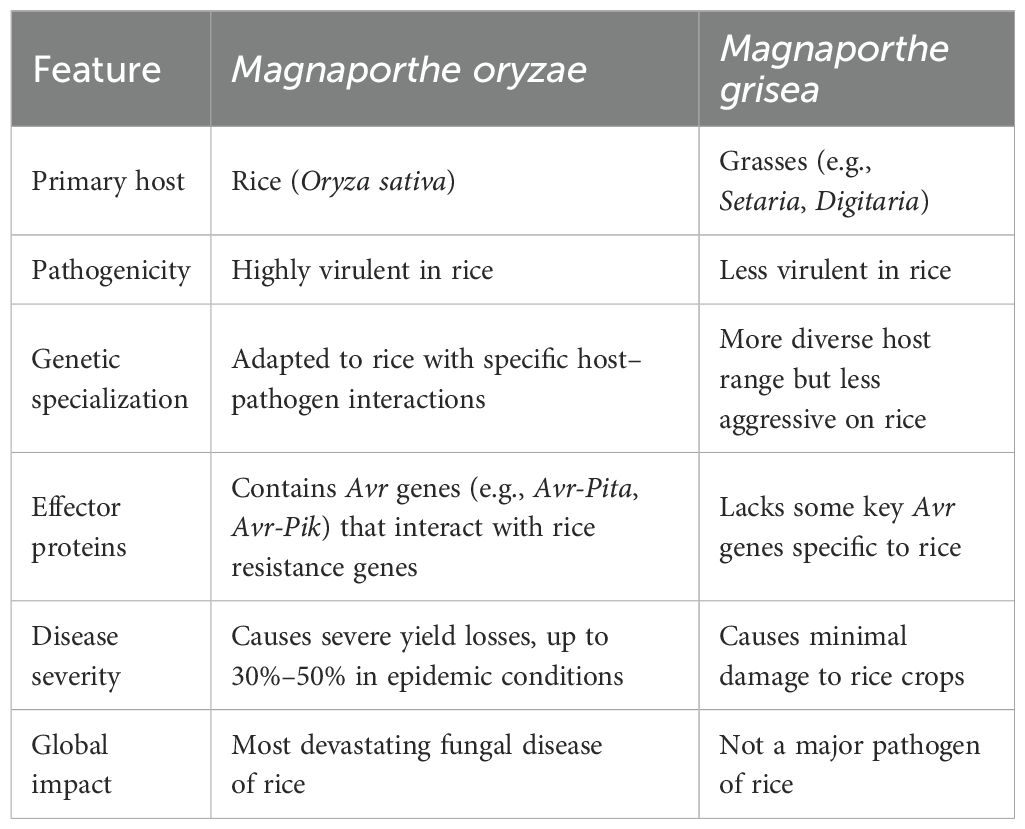
Magnaporthe oryzae and Magnaporthe grisea are closely related fungal pathogens known to cause rice blast disease. However, M. oryzae is considered the primary and more aggressive pathogen responsible for rice blast, whereas M. grisea is generally associated with other grasses and is less effective in infecting rice (Choi et al., 2013). Their pathogenicity and host specificity differ significantly.
Host specificity and pathogenicity
Magnaporthe oryzae: Primarily responsible for rice blast disease, M. oryzae infects a broad range of grasses, including economically important crops such as wheat, barley, millet, and maize. Its adaptability to various hosts makes it a significant threat to global cereal production (Chung et al., 2020).
Magnaporthe grisea: Initially isolated from crabgrass (Digitaria sanguinalis), M. grisea exhibits a more restricted host range, predominantly infecting species within the Digitaria genus (Zhang et al., 2015).
Comparative effects on rice blast disease
In summary, Magnaporthe oryzae is more adept at causing rice blast disease than Magnaporthe grisea. This conclusion is supported by recent studies highlighting the broader host range and higher pathogenicity of M. oryzae in rice and other cereal crops.
Pathogen description and diversity
The presence of multiple races within a blast pathogen intensifies its interaction with the host, overpowering the host’s defense system. A total of 25 cultivars, viz., recombinant inbred lines, commercial cultivars, and donors, are generally used to monitor the virulence of blast pathogens, according to the All India Coordinated Rice Improvement Programme (Hasan et al., 2024). This pathogen is highly variable and rapidly evolves into novel pathotypes. In the 1970s, the identification of a novel race group took place—IJ, which emerged from Indian isolates of M. oryzae, with the IC3 and ID1 races standing out prominently (Padmanabhan et al., 1970). Notably, ID-17 was prevalent within Indian paddy ecosystems among five pathogenic race groups—ID-1, ID-2, IB-4, IC-17, and IC-25. Evaluation of the genetic heterogeneity of Magnaporthe species in both rice and finger millet ecosystems in southern India involved the utilization of simple sequence repeats (SSRs) and repetitive DNA-based markers targeting pathogenicity genes (Palanna et al., 2024). To establish a durable system to protect against blast disease since 2006, the Japan International Research Center for Agricultural Sciences (JIRCAS) has been conducting a collaborative study, “Blast Research Network for Stable Rice Production,” targeting Southeast and East Asia. According to the gene for gene hypothesis, the complex interaction occurs between host resistance and fungus virulence in rice blast pathogens; every resistance gene in the host corresponds to an avirulence gene in the pathogen. Based on this theory, differential varieties (DVs), which can be used to distinguish pathotypes (races) by their reaction patterns to each pathogen strain, have been developed to identify the blast pathogen population structure and predict the emergence of new blast races. Using several sets of DVs, pathogenicity studies of blast isolates have been performed in China and Southeast Asia. Using 12 Japanese differential varieties (DVs) for Pia, Pik-s, Pii, Pik, Pik-m, Piz, Pita, Pita-2, Piz-t, Pik-p, Pib, and Pit (Kiyosawa, 1984; Yamada et al., 1976; Tsunematsu et al., 2000), 12 kinds of blast race have been identified among 129 isolates collected from all over the Mekong River Delta area of Vietnam. A total of 25 monogenic lines harboring 23 resistance genes, namely, Pish, Pib, Pit, Pia, Pii, Pi3, Pi5(t), Pik-s, Pik-m, Pi1, Pik-h, Pik, Pik-p, Pi7(t), Pi9(t), Piz, Piz-5, Piz-t, Pita-2, Pita, Pi12(t), Pi19(t), and Pi20(t), were developed as a new set of international DVs by several backcrosses using the Chinese susceptible rice variety LTH and a set of LTH NILs carrying 11 resistance genes (Pib, Piz-5, Pi9(t), Pi3, Pia, Pik-s, Pik, Pik-h, Pi7(t), Pita, and Pita-2) developed by Telebanco-Yanoria et al. (2010). However, there has been no research into blast races in Cambodia, nor has any information on blast disease or genotypes of rice varieties been collected. The pathogenicity of blast isolates was based on inoculation test using differential varieties in near-isogenic lines in Japan (Kawasaki-Tanaka et al., 2016), Laos (Xangsayasane et al., 2020), Vietnam (Fukuta et al., 2020; Nguyet et al., 2020), Indonesia, Bangladesh (Khan et al., 2016), West Africa (Odjo et al., 2014), and Kenya (Fukuta et al., 2019). These DVs had the common genetic background of LTH (Lijiangxintuanheigu), meaning that the influence of genetic background on the appearance of blast symptoms was minimized. The targets of the DV sets and other useful materials released are major blast resistance genes, which have been used internationally as part of JIRCAS’s research. These monogenic lines and LTH NILs are being used to develop differential systems in each country through (1) pathogenicity analysis of blast isolates, (2) elucidation of blast race distribution, and (3) selection of standard differential blast isolates. In the context of Magnaporthe oryzae interactions with rice differentials in the CO39 background, several virulence and avirulence (AVR) genes that play crucial roles in determining the pathogen’s specificity and the host’s resistance response have been identified. Near-isogenic lines (NILs) were developed to study the interaction between rice and Magnaporthe oryzae by introgressing individual resistance (R) genes from diverse donor parents while maintaining a uniform genetic background. CO39, an Indica rice variety, was selected as the recurrent parent due to its high susceptibility to blast disease, allowing a clear evaluation of the effects of individual R genes. Each NIL carries a single R gene introduced from different donor parents through successive backcrossing and marker-assisted selection. NILs are composed of 14 R genes: Pish, Pib, Piz-5, Piz-t, Pi5(t), Pik-s, Pik, Pik-h, Pik-m, Pik-p, Pi1, Pi7(t), Pita, and Pita-2, each derived from different donor varieties such as Toride 1, Fukunishiki, Kanto 51, K60, Tetep, and Tsuyuake. Telebanco-Yanoria et al. (2011) developed blast-resistant rice varieties through multiple backcrossing cycles and selecting specific M. oryzae isolates to confirm the presence of the targeted R genes. Further phenotypic evaluations showed that these NILs displayed reaction patterns akin to monogenic lines containing identical R genes when tested against standard Philippine isolates. This resemblance highlights their significance as differential varieties in studying blast resistance, especially in areas where Japonica-type varieties such as LTH are less impactful (Yang et al., 2022). Within the framework of CO39 rice cultivar, AVR1-CO39 has been studied as a significant locus that regulates the extensive avirulence of M. oryzae strain 2539 on cultivated rice. This gene aligns with the resistance gene Pi-CO39(t) in rice, where it has been genetically mapped to a specific locus on the short arm of chromosome 11 and has shown effectiveness against various isolates, highlighting its evolutionary importance (Zheng et al., 2011). Consequently, it can be inferred that AVR1-CO39 is specific to the species rather than the cultivar, functioning as a host-specific AVR locus for M. oryzae in rice (Tosa et al., 2005). Notably, both AVR1-CO39 and AVR-Pia are recognized by the rice resistance proteins RGA4 and RGA5, which collaboratively initiate immune responses (Petit-Houdenot and Fudal, 2017). Additionally, the AVR-Pik locus exhibits high haplotype diversity, with novel variants evolved through stepwise base substitutions, enabling the pathogen to overcome Pik-mediated resistance (Li J et al., 2019). Unraveling the intricacies of AVR gene functions is crucial to develop effective strategies to combat blast disease resistance in rice.
Geographic distribution
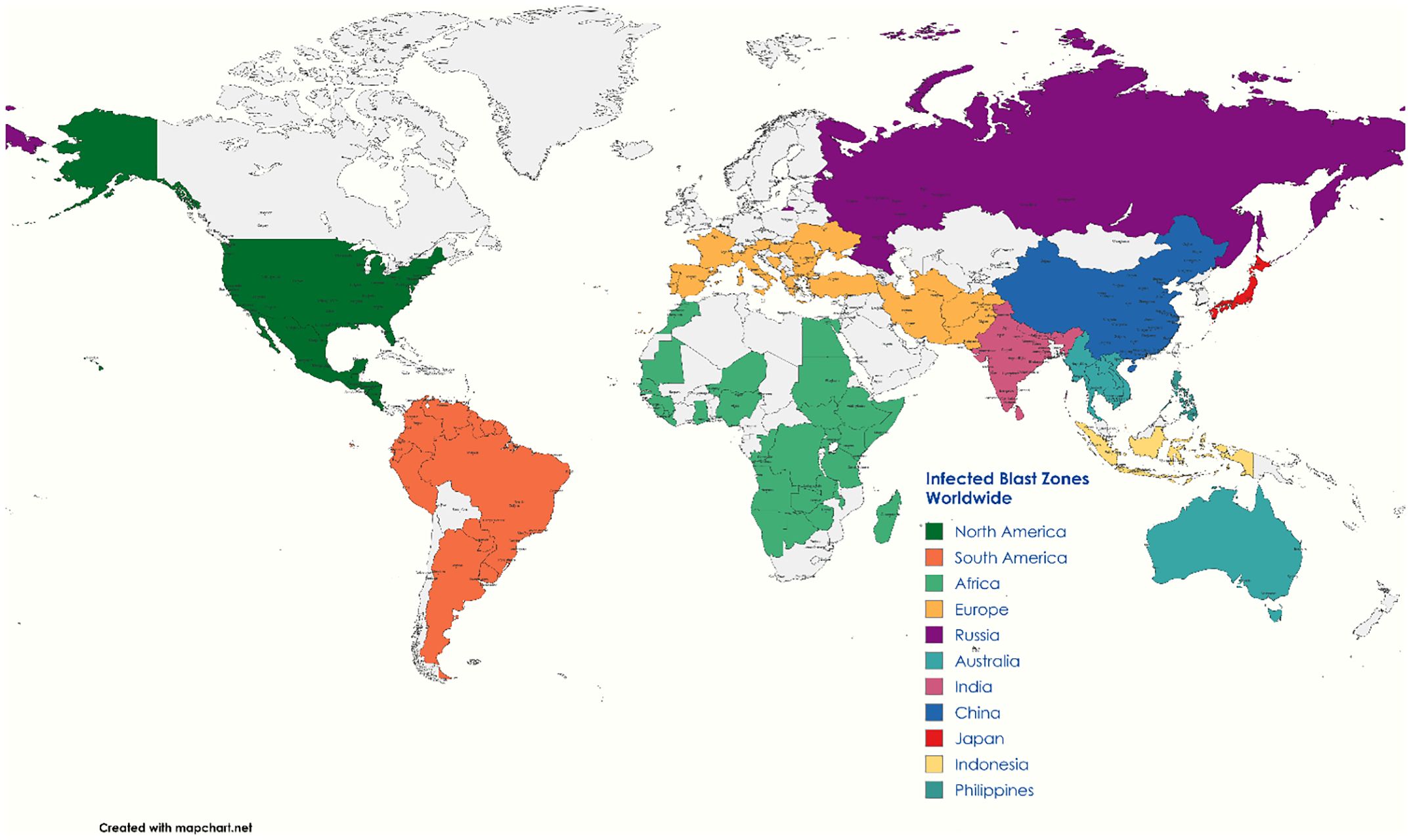
Rice production in West Africa, the world’s largest rice producer, is severely affected by blast disease, causing yield losses of 3%–77% (Shahriar et al., 2020). This disease, also referred to as rice seedling blight (Zeigler et al., 1994) and rice rotten neck (Talbot, 2003), was first documented as “rice fever” in China in 1637 and later as Imochi-byo in Japan in 1704. It gained global recognition with Italy experiencing its first epidemic in 1828 and the Tanjore delta in India identifying it in 1919. Currently, the disease impacts approximately 85 countries worldwide, particularly in South Asia and Africa (Wang et al., 2017), with annual yield losses ranging from 10% to 80% (Simkhada and Thapa, 2022). These losses are influenced by various factors such as varietal susceptibility, infection severity, fungicide application timing, high humidity, drought, heavy dew, elevated mean temperatures, high plant density, and excessive nitrogen fertilizer use. In India, rice blast epidemics can result in yield losses of up to 50%. During natural epidemics in the wet season, disease incidence ranges from 14% to 27% (exceeding the economic threshold), leading to yield losses of about 27%–35%. Severe epidemics occurred between 1980 and 1987 in several Indian states, such as Himachal Pradesh, Andhra Pradesh, Tamil Nadu, and Haryana, causing substantial financial losses. It is estimated that the annual yield reduction due to rice blast disease could feed approximately 60 million people each year. In the United States, Arkansas, Louisiana, and Mississippi are the most affected states, with yield losses ranging from 6% to 50% and an average annual loss of USD 69.34 million due to blast. The disease is also a significant concern in European countries like Italy, Spain, Portugal, Greece, and France, where it has been observed to reduce the milling yield by 20% to 50% (Devanna et al., 2022) (Figure 1).
Interaction between host plant and blast pathogen in rice
Once the rice is infected by M. oryzae, pattern recognition receptors (PRRs) on the cell surface can specifically recognize pathogen-associated molecule patterns (PAMPs) and activate defense response by cell wall modification, callose deposition, and via the expression of defense-related proteins in host cells, which is termed PAMP-triggered immunity (PTI) (Figure 2). However, PTI is a weak and non-specific resistance mechanism (Bernoux et al., 2011). In many cases, M. oryzae can secrete certain effectors to inhibit PAMP-induced PTI and break resistance responses (Jones and Dangl, 2006; Mentlak et al., 2012). At the same time, rice has acquired more specific resistance proteins that directly or indirectly recognize pathogen-effector proteins. This recognition mechanism activates a second layer of the defense response in rice, known as effector-triggered immunity (ETI), which results in the production of ion (Ca2+, K+, and H+) currents, superoxide, nitric oxide, and programmed cell death at the site of invasion (Nurnberger et al., 2004). ETI is a highly specialized disease resistance mechanism in the host (Boller and He, 2009), which is activated in the gene-for-gene model upon recognition by an R (resistance) protein of the corresponding effector protein of M. oryzae. Effector proteins are often encoded by avirulence genes in M. oryzae. The R genes in rice correspond to the avirulence (AVR) genes in M. oryzae in a gene-for-gene manner (Flor, 1971), which ensures that the interaction between a specific R protein in rice and the corresponding AVR effector in the pathogen renders resistance. The R protein encoded by R genes interacts directly or indirectly with the effector protein, thus sensing pathogen invasion and inducing disease resistance. Despite the deployment of resistant varieties, blast epidemics can still occur due to a lapse in host resistance and the emergence of new virulent pathotypes (Chuma et al., 2011). Thus, the effectiveness of R genes depends on the respective AVR gene.
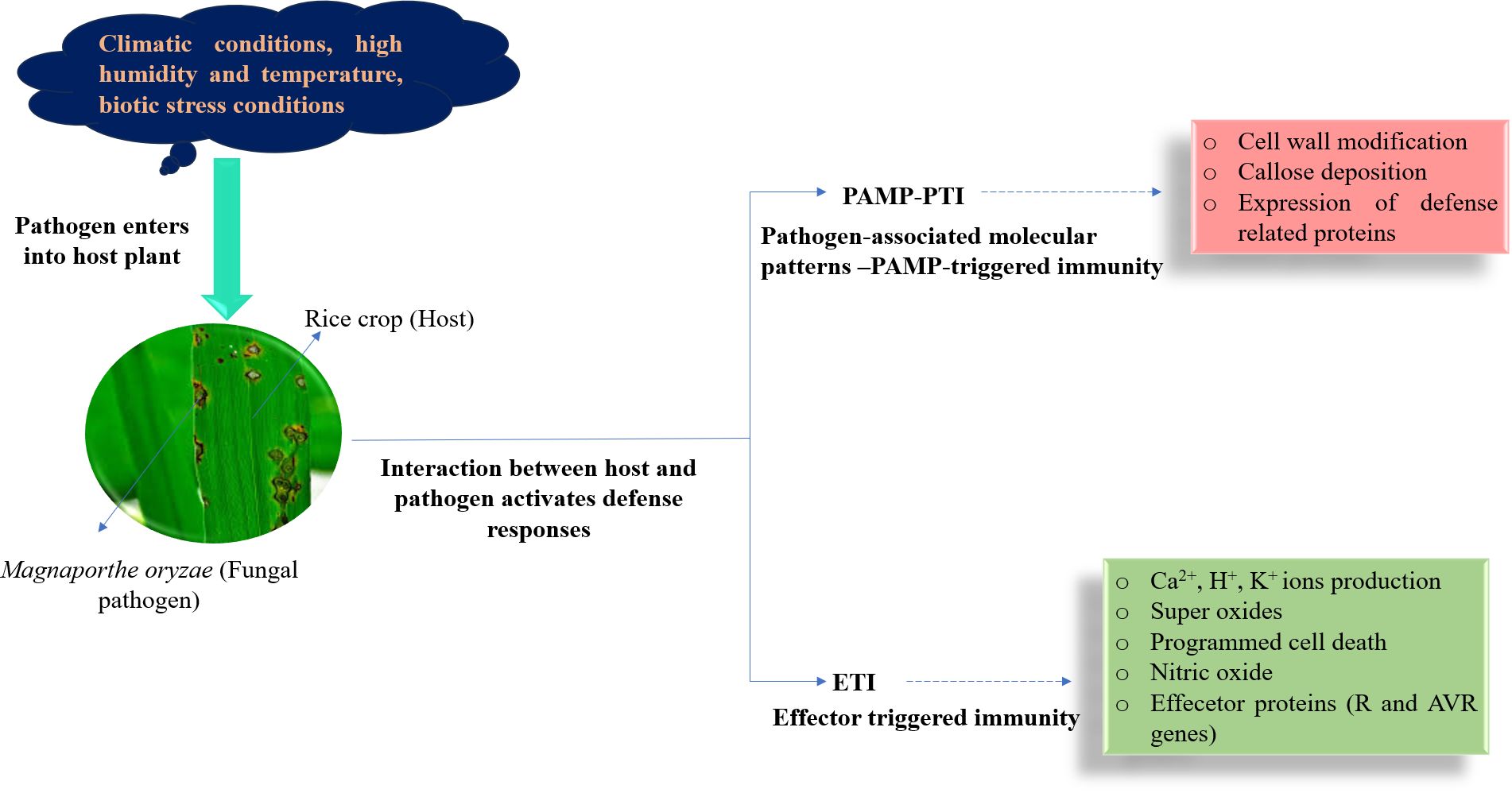
Figure 2. Different defense mechanisms occur when the interaction between Magnaporthe oryzae and rice.
Various mechanisms underlying Magnaporthe oryzae and rice plant
Rice plants have developed complex defense mechanisms to recognize and combat the rice blast fungus, Magnaporthe oryzae. Understanding these mechanisms is crucial to develop effective breeding methods aimed at producing disease-resistant rice varieties (Saklani et al., 2023). In their interaction with M. oryzae, rice plants utilize pattern recognition receptors (PRRs) to detect pathogen-associated molecular patterns (PAMPs). This detection triggers PAMP-triggered immunity (PTI), which starts early defensive responses—for instance, when rice PRRs identify chitin fragments from the fungal cell wall, it activates signaling pathways that promote the generation of reactive oxygen species (ROS) and the expression of defense-related genes (Zhang L et al., 2024).
Pattern-triggered immunity
Plants possess sophisticated immune systems that can identify and respond to pathogenic threats through two connected mechanisms: pattern-triggered immunity (PTI) and Effector-Triggered Immunity (ETI). PTI activates when pattern recognition receptors (PRRs) on the surface of plant cells detect conserved pathogen-associated molecular patterns (PAMPs), such as bacterial flagellin or fungal chitin. This detection triggers a series of defensive actions, including the production of reactive oxygen species, reinforcement of the cell wall, and the activation of defense-related genes, collectively impeding pathogen invasion. To bypass PTI, pathogens excrete effector proteins that can interfere with these defenses. In response, plants have developed intracellular nucleotide-binding leucine-rich repeat (NB-LRR) proteins that can detect these effectors, which leads to ETI. ETI generally manifests as a hypersensitive response, characterized by localized cell death at the infection site, limiting the pathogens’ spread. The dynamic interaction between PTI and ETI forms a robust defense network that enables plants to recognize various pathogens and initiate appropriate immune responses (Dodds et al., 2024). In rice, the initial defense mechanism relies on pattern recognition receptors (PRRs) on the plasma membrane to identify pathogen-associated molecular patterns (PAMPs). A notable PRR in rice is the chitin elicitor binding protein (CEBiP), which specifically binds to chitin fragments from fungal cell walls. Upon recognizing chitin, CEBiP associates with another receptor-like kinase, CERK1, which activates a signaling cascade that leads to PAMP-triggered immunity (PTI). This immune response includes the generation of reactive oxygen species (ROS), activation of mitogen-activated protein kinases (MAPKs), and the expression of defense-related genes, all contributing to the prevention of pathogen invasion (Mentlak et al., 2012).
Effector-triggered immunity
To inhibit PTI, Magnaporthe oryzae secretes effector proteins into the host cells. Rice plants have evolved resistance (R) genes that encode nucleotide-binding leucine-rich repeat (NLR) proteins that specifically detect these effectors. This recognition activates effector-triggered immunity (ETI), a strong defense mechanism that often leads to localized cell death, referred to as the hypersensitive response (HR), to thwart the pathogen (Tian et al., 2018). Effector-triggered immunity (ETI) plays a crucial role in plant defense, enabling the identification of specific pathogen effectors, particularly in the interaction between rice (Oryza sativa) and Magnaporthe oryzae. In this gene-for-gene model, rice plants contain resistance (R) genes coding for intracellular NLR proteins that recognize corresponding avirulence (AVR) effectors released by M. oryzae. This interaction triggers a vigorous immune response, often resulting in localized cell death to manage the pathogen, known as the hypersensitive response. A notable instance is the relationship between the rice R gene Piz-t and the M. oryzae AVR effector AvrPiz-t (Liu X et al., 2024). The AvrPiz-t gene encodes a secreted protein that when identified by the Piz-t protein in rice, initiates ETI, effectively stopping disease development. However, M. oryzae can escape this defense by modifying, deleting, or inserting transposons into its AVR genes, leading to altered effectors that the plant’s R proteins cannot recognize. This ongoing evolutionary struggle necessitates continuous monitoring of AVR gene changes and the creation of rice cultivars with diverse durable R genes to manage blast disease effectively (Zhang and Xu, 2014). Reactive oxygen species (ROS) play a critical role in PTI and ETI, serving as signaling molecules and direct agents against pathogens. In PTI, ROS are generated in a swift burst that strengthens cell walls and activates defense genes. In ETI, ROS accumulation is prolonged, contributing to the hypersensitive response, reinforcing cell walls, and triggering programmed cell death to control the pathogen.
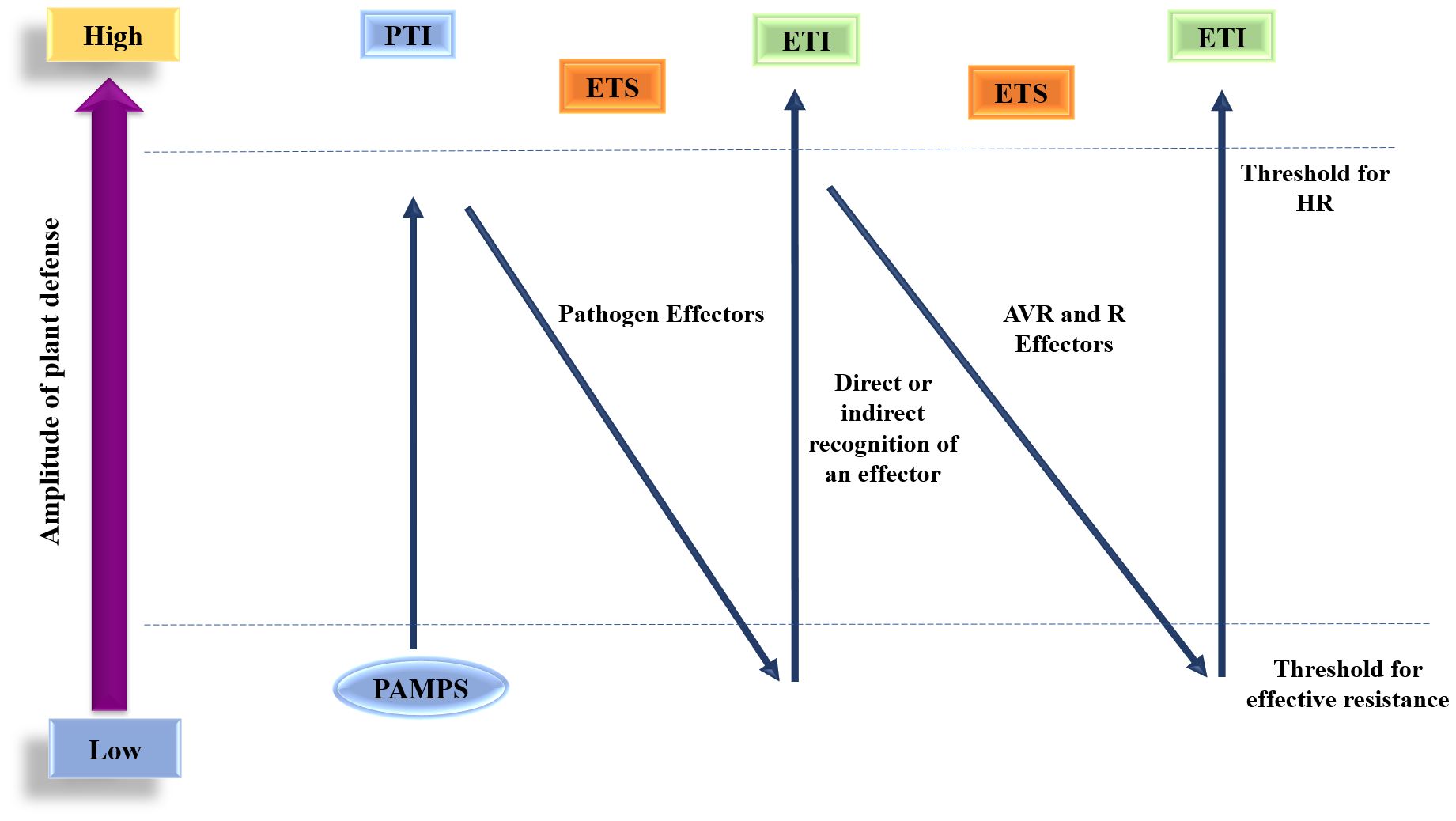
The zig-zag model, initially proposed by Jones and Dangl in 2006, describes how the recognition of PAMPs initiates primary defense mechanisms based on PTI, which help to reduce pathogen growth, though they do not completely eliminate it. Pathogens that flourish have developed effector or virulence factors that promote their growth by suppressing PTI, resulting in effector-triggered susceptibility (ETS). In response to certain pathogen effectors, plants have developed ETI, primarily by recognizing altered self-products from ETS via NB-LRR. This continuous evolutionary battle between host and pathogen manifests in repeating cycles of ETS and subsequent ETI. The outcome of the plant-pathogen interaction is influenced by the sum of ([PTI − ETS] + ETI) (Figure 3).
Understanding these molecular interactions provides essential insights for breeding strategies. By identifying and integrating R genes that can detect a broad spectrum of M. oryzae effectors, breeders can develop rice varieties with enduring resistance. Additionally, enhancing the expression of PRRs like CEBiP or other components involved in ROS production may strengthen basic immunity, creating a multi-tiered defense against the pathogen. Breeding approaches that incorporate multiple resistance genes, coupled with understanding the dynamic interplay between rice and M. oryzae, can enhance durable resistance. Future research should focus on integrating these molecular insights into advanced breeding techniques such as CRISPR-Cas9 genome editing to achieve broad-spectrum and long-lasting resistance in rice.
Interaction of host R genes with AVR genes
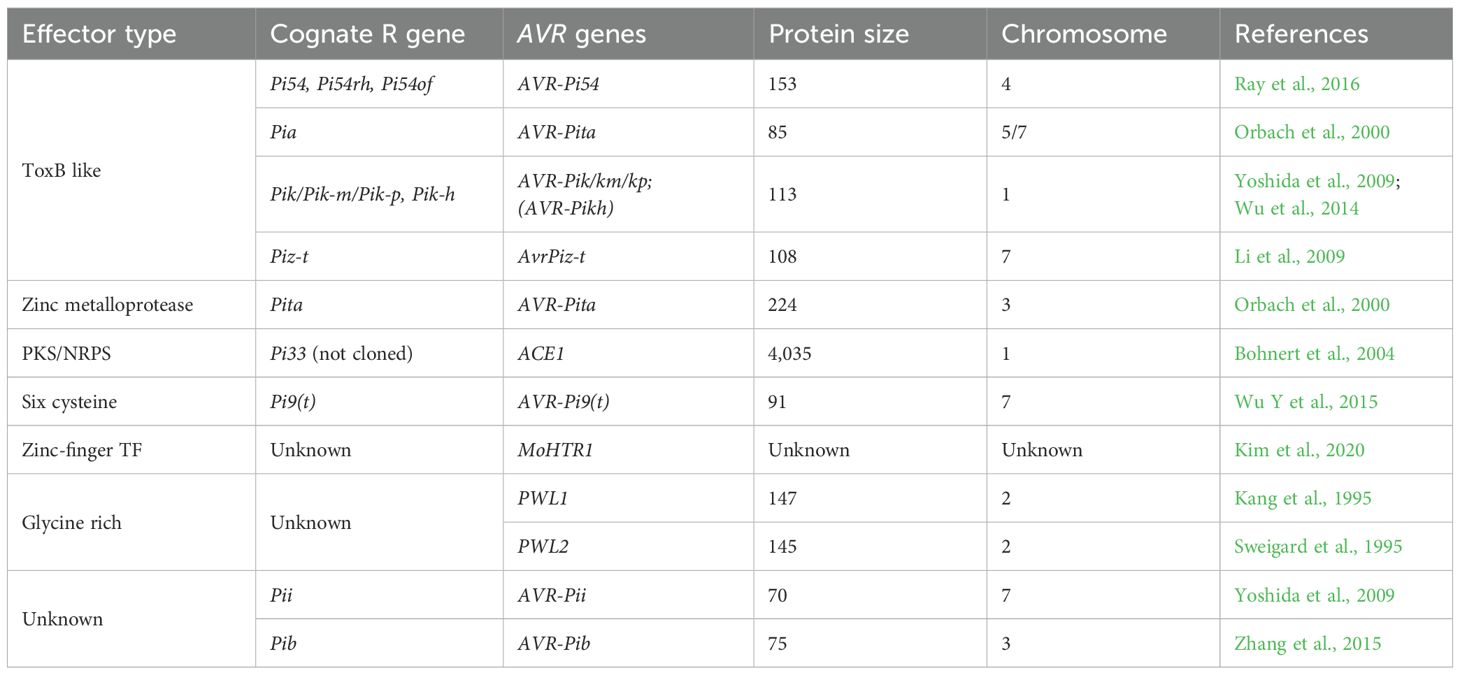
To date, 26 Avr/effector genes have been mapped in M. oryzae, and 14 of them, including two unmapped Avrs, MoHTR1 and MoHTR2, have been cloned and characterized (Table 1). The first discovered Pwl effectors (Pwl1–Pwl4) belong to a small, glycine-rich, rapidly evolving effector family that provides avirulence on weeping lovegrass and finger millet but does not affect rice. Except for cell death induction/suppression or interaction with resistance protein features, identifying candidate effector proteins is a difficult task due to their unique sequence features. Among the 26 reported Avr-genes, 15 were mapped near the chromosome ends, and five of the cloned Avr genes were flanked by transposons. These transposons are active companions of the Avr genes and play a role in the loss and gain of these genes. The molecular interaction studies of the reported seven R-Avr pairs showed that five of them, namely, Pi-ta/AVR-Pita, Pik/AVR-Pik, Pia/AVR-Pia, Pi-CO39/AVR1-CO39, and Pi54/AVR Pi54 interact directly, whereas Piz-t/AvrPiz-t and Pii/AVR-Pii have indirect interaction.
Common R–AVR pairs and their role against rice blast resistance
1. Pi-ta and AVR-Pita
AVR-Pita and Pi-ta from the fungal pathogen M. oryzae are among the first R–Avr interactions to be thoroughly explored (Wang et al., 2019). This interaction has played a pivotal role in establishing a fundamental understanding of the complex dynamics between plants and pathogens, specifically about disease initiation and developing resistance mechanisms. The gene AVR-Pita, which is located near to telomeres and is responsible for encoding a protein, secretes and possesses a unique domain known as Zn metalloprotease (Khang et al., 2008). The Avr-Pita protein attains its mature state as a protease, consisting of a sequence of 176 amino acids located at the C-terminus (Razzaq et al., 2019). Avr-Pita is a member of a unique subclass within the AVR-Pita gene family, comprising three different genes: AVR-Pita1, AVR-Pita2, and AVR-Pita3. The first two genes mentioned possess functional properties that initiate Pi-ta-mediated resistance, whereas the third gene is a pseudogene lacking Avr functionality (Ribaut, 2006). The Pi-ta R gene counterpart is a conventional NLR (928 amino acids) receptor situated in the cytoplasm and generally exhibits constitutive expression (Wu J et al., 2015). The direct interaction between the leucine-rich domain (LRD) of the Pi-ta protein and the AVR Pita176 protein leads to the activation of downstream signaling cascades. The utilization of site-directed mutagenesis has facilitated the functional validation of AVR-Pita, leading to the identification of two critical amino acid substitutions, AVR-pita176E177D and AVR pita176M178W, which result in the loss of its virulence function. In a similar vein, the presence of a mutated form of the Pi-ta R gene, characterized by a single amino acid substitution (LRDA918S), has been observed to reduce the physical interaction between the AVR-Pita176 and Pi-ta LRD proteins. This finding underscores the significance of the interplay between R–Avr pairs in the establishment of immunity against M. oryzae (Hossain et al., 2018).
2. Pia and AVR-Pia
This is the second class of interaction in which two NLRs, RGA4 and RGA5, interact with a single Avr protein (Mutiga et al., 2021; Wang et al., 2019). The encoded secretory protein of AVR-Pia contains an N-terminal SP (Martin et al., 2003). Different isolates of M. oryzae that are resistant to Pia genes in rice have different numbers of copies, ranging from one to three; this depends on the isolate. For instance, the avirulent strain Ina168 possesses three copies of AVR-Pia genes (Ribot et al., 2013). The NMR (nuclear magnetic resonance)- determined structure of AVR-Pia reveals a MAX effector β-sandwich-like structure, while Pia is composed of RGA4 and RGA5 protein genes, oriented face-to-face in opposite directions (Mutiga et al., 2021). Furthermore, two isoforms of RGA5 called RGA5-A and RGA5-B are the consequences of RGA5 alternative splicing, in which only RGA5-A mediates Pia resistance. In in vitro experiments, it has been observed that the continuous production of RGA4 leads to the initiation of cell death. However, in the absence of infection, this cell death is suppressed by RGA5 in planta. It is important to note that the NB (nucleotide-binding) domain of RGA4 is essential for the induction of cell death (Fukoka et al., 2009; Hayashi K et al., 2010). Physical contact between AVR-Pia and the non-LRR C-terminal domain of RGA5 facilitates the inhibition and promotion of RGA4-mediated cell damage.
3. Pii and AVR-Pii
This is the third type of interaction in which the R–Avr pair (Pii and AVR-Pii) mediates the immune response through an indirect interaction (Martin et al., 2003). The secreted protein encoded by AVR-Pii belongs to a protein family known as pex33. The protein structure comprises four homologs with two conserved motifs (Yoshida et al., 2009). Conversely, the protein encoded by Pii is a common NLR consisting of 1025 amino acids (Mutiga et al., 2021). Two forms (I and II) of AVR-Pii exist in different isolates. Form I is a hybrid of rice proteins (OsExo70-F2 and OsExo70-F3) and AVR-Pii. Though both rice proteins are required for the immune response, the latter (OsExo70-F3) instead of the former rice protein, induces a Pii-mediated immune response. This result implies that OsExo70 serves as a helper protein in the interaction of Pii/AVR-Pii (Hossain et al., 2018).
4. Piz-t and AVR-Piz-t
One such form of R–AVR contact pertains to the indirect interaction that occurs between Piz-t and AVR Piz-t, which is a classic example of a plant–pathogen interaction where a single, broad-spectrum R gene recognizes and interacts with multiple variants of an AVR gene (Liu et al., 2014). The AVR Piz-t protein has secretory characteristics akin to those of other well-known AVR genes. The structure of AVR Piz-t and similar ToXB genes was determined via NMR. AVR Piz-t comprises a β-sheet consisting of six disulfide chains from Cys62 to Cys75. Single point mutations on any cysteine residue reduce the toxicity of AVR Piz-t (Sone et al., 2013). Piz-t functions as a broad-spectrum NLR gene. The LRR domain of Piz-t exhibits 18 amino acid alterations, which determine the activation of resistance and differentiate Piz-t from Pi-2 (Zhou et al., 2006). Being a broad-spectrum R gene, twelve different interacting proteins of AVR Piz-t (APIPs) interact with AVR Piz-t in various lines of rice. The nature of resistance or immunological response is contingent upon the specific AVR Piz-t protein and the genetic composition of the rice host harboring the Piz-t gene. In the context of Piz-t-lacking Nippon bare rice, the suppression of PTI is observed as a result of the interaction between AVR Piz-t and PTI. Conversely, in the presence of Piz-t, PTI is stabilized when the rice plant is infected by M. oryzae (Fujisaki et al., 2015).
Cloning of blast resistance genes in rice
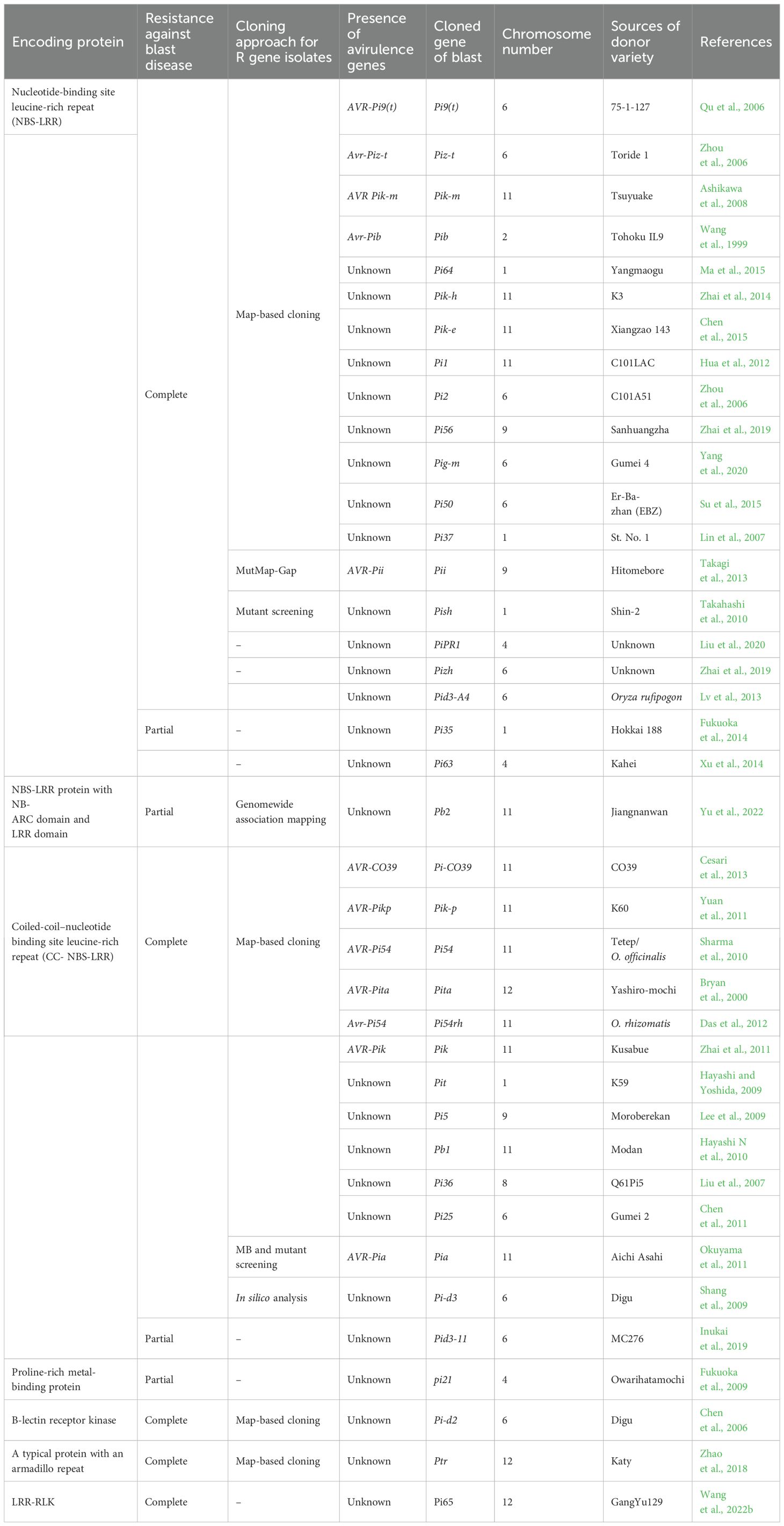
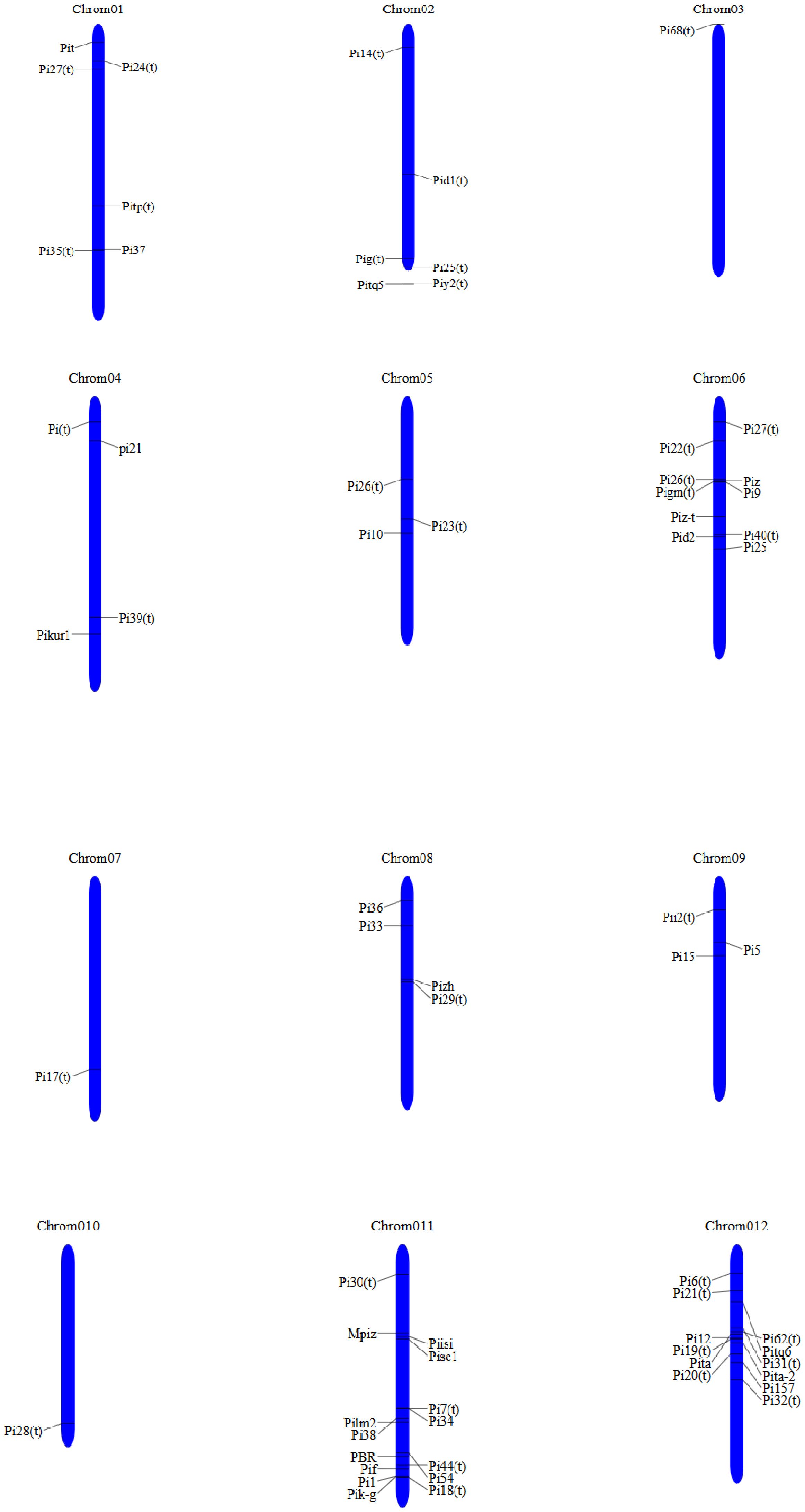
Analysis of rice germplasm with different races reveals complete resistance conferred by significant blast resistance (R) gene. However, the resistance may be broken down due to its single R gene locus having race-specific characteristics. Because of the progress in molecular marker development and functional genomics, blast resistance genetics in rice have been strengthened (Qian et al., 2023). R genes serve as the cornerstone of disease resistance. To date, 146 resistance genes of the blast have been identified; among them, 41 of these genes (Pib, Pit, Pish, Pita, Pi54, Pi-d2, Ptr, Pi9(t), Pia, Pi- C039, Pi65, Pi2, Piz-t, Piz-h, Pig-m, Pi50, Pi36, Pi37, Pik-h, Pik-m, Pi1, Pik-e, Pi56, Pi-d3, Pi25, pi21, Pb1, Pi5(t), Pii, Pik, Pid3A4, Pi35, PiPR1, Pi64, Pik-p, Pi63, Pid3-11, Pb2, Ptr, and Pi54-rh) have been cloned and functionally authenticated (Sahu et al., 2022; Younas et al., 2023). Among the array of identified resistance genes against blast disease, Pi54 and Pi2 are highly effective and provide broad-spectrum resistance (Aleena et al., 2023). The genetic analysis, mapping, and cloning of rice blast resistance have been extensively explored since the discovery of three independently inherited R genes, namely, Pia, Pii, and Pik, during the 1960s. R genes are distributed across 11 chromosomes of the rice genome, with a notable clustering on chromosomes 6, 11, and 12, representing 18%, 25%, and 21%, respectively (Ashkani et al., 2016). Since the cloning of the first R gene, Pib, 41 R genes have been successfully identified (Table 2). Except for pi21, which acts as a recessive R gene, the remaining 37 R genes exhibit dominance. All of the genes, excluding pi21, Pi35, Pi63, Pb1, and Pid3-I1, confer complete resistance. Pi-d2 encodes a B-lectin kinase domain protein (Chen et al., 2006), while pi21 encodes a proline-rich protein with a heavy metal domain (Fukuoka et al., 2012), and Ptr encodes an atypical protein with an armadillo repeat (Zhao et al., 2018); the remaining genes encode nucleotide-binding site leucine-rich repeat (NBS-LRR) domain proteins. Several R genes, including Pik, Pikm, Pik-p, Pi1, Pike, Pi5(t), Pia, and Pi-CO39, contain two NBS-LRR protein structural genes for blast resistance (Cesari et al., 2013). The Pi5-1, Pb1, pi21, and Pi63 genes are induced by pathogen infection, whereas the remaining genes are constitutively expressed. Most cloned R genes confer resistance against leaf blast at the seedling stage, while only a few, such as Pb1, Pi25, and Pi64, provide resistance to panicle blast (Cao et al., 2019).
Donors for resistance against rice blast
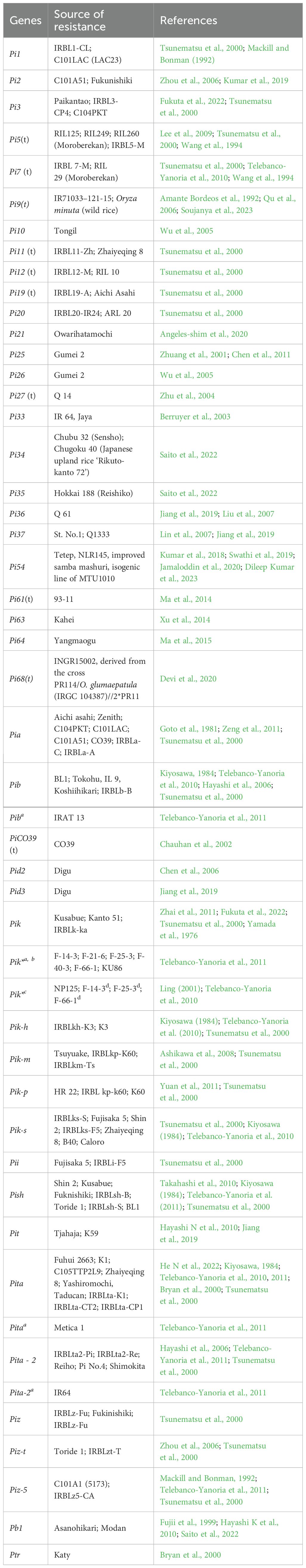
The development of resistant rice varieties through genetic improvement is a sustainable option for managing plant diseases. Since there are no genotypes with absolute resistance, the identification of reliable resistance sources must be confined to moderate to high levels of tolerance in the germplasm. There are several such genotypes reported (Table 3) that are being used in breeding blast-resistant cultivars. Among the cultivated species, the indica cultivars are reported to show better resistance than the Japonica type (Takahashi et al., 2010; Patroti et al., 2019). Additionally, some accessions of wild species, such as O. rufipogon, O. minuta and O. glumaepatula have been reported to be resistant to blast disease.
Breeding approaches
Rice breeders employ various strategies to combat blast disease, focusing on the development of resistant cultivars through both conventional and molecular methods. These approaches are designed to improve the longevity of resistance and respond to the changing virulence of Magnaporthe oryzae. The following essential breeding techniques are discussed below.
Traditional breeding
Conventional breeding is one of the oldest methods vital for developing new genetic variants, including blast-resistant rice varieties. This approach depends on interactions between genotype and the environment and helps in phenotypic selection among cultivars. In this scenario, breeders considered factors such as the pathogen’s race, the plant genotype, and resistance (qualitative or quantitative) to disease (Nizolli et al., 2021). Conventional methods are crucial for ensuring genetic diversity, conserving wild germplasm, hybridization, and induce mutations. Over the past 30 years, traditional breeding has produced elite cultivars of IRRIs with a wide array of disease-resistance genes. Various techniques, such as the pedigree method, backcrossing, recurrent selection, and mutation breeding have been frequently employed in conventional breeding programs in recent decades (Table 4).
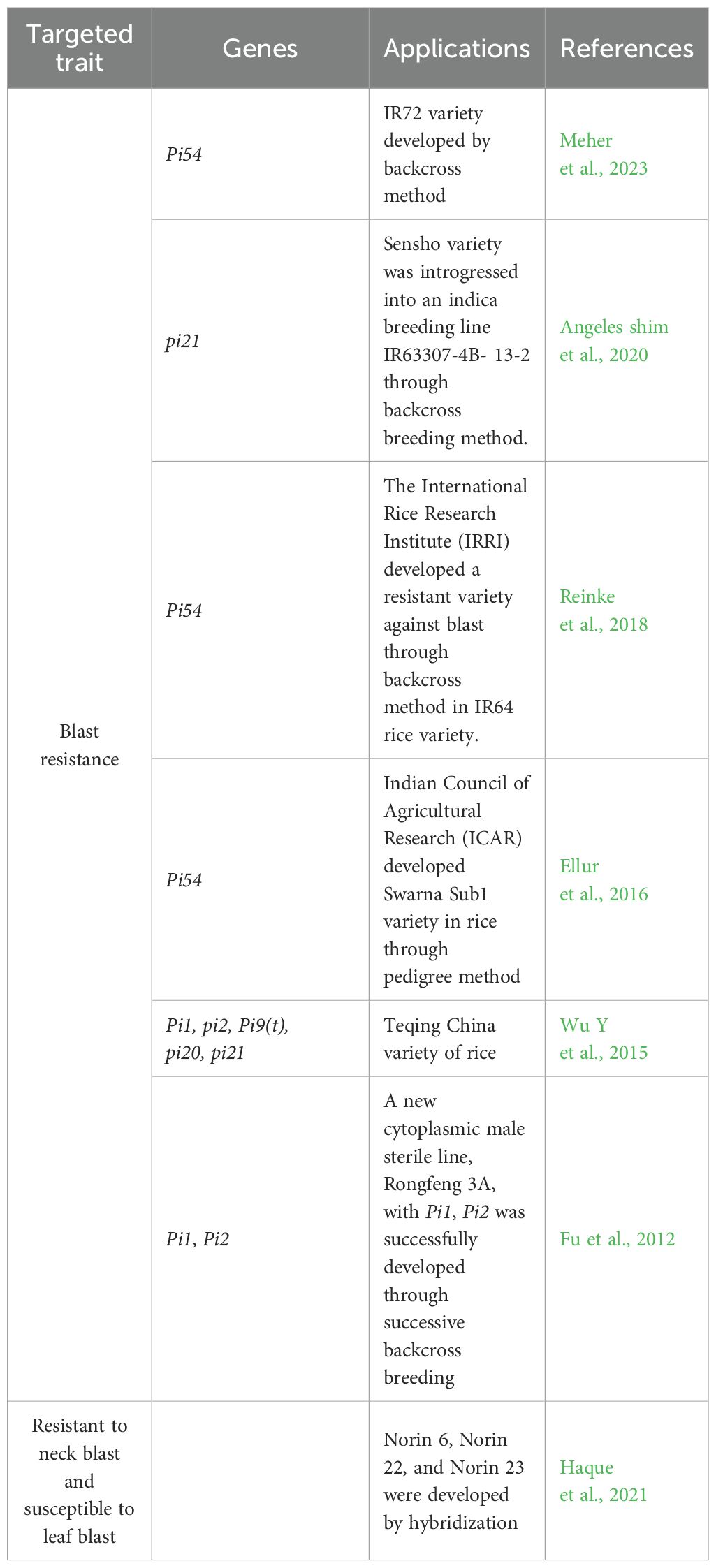
Table 4. Successful examples of rice varieties developed through conventional breeding methods against blast disease.
The pedigree method is well-suited for determining traits controlled by major genes. It is extensively used in rice improvement to develop resistance to insects and diseases (Srivastava et al., 2017). However, a major drawback is the time-consuming process of evaluating lines throughout the growing season and maintaining records for selection at maturity. This method requires an interaction between genotype and environment on trait expression. This method may not be the most efficient for traits governed by multiple genes. Backcrossing is a widely employed technique in rice breeding for the introduction or substitution of a target gene from a donor parent to a recipient. This method aims to reduce the donor genome content in progeny, offers a precise way to enhance varieties across multiple traits. Backcross breeding has been widely adopted in southern and southeast Asia as a breeding strategy to improve the resistance of elite varieties such as KDML105, Basmati, and Manawthukha to blast. In addition to backcross breeding, recurrent selection is another method in rice breeding for disease control. This approach facilitates shorter breeding cycles and more precise genetic gains, while promoting a broad range of genetic diversity in breeding lines. Several blast-resistant cultivars, such as the upland cultivar CG-91, have been developed through recurrent selection. Numerous major genes, including Pib, Pita, Pia, Pi1, Pikh, Pi2(t), and Pi4(t), have been successfully identified and introgressed into rice varieties for blast resistance via conventional breeding methods (Miah et al., 2017). Mutation breeding in rice complements conventional breeding by effectively improving major traits such as agronomic characteristics, resistance to pests and diseases, and grain quality parameters. This method is particularly valuable for generating new alleles to develop new varieties. The incorporation of a blast resistance gene into the high-yielding variety Ratna (IR8/TKm 6) was achieved through chemo-mutagenesis (Kaur et al., 1976) using 0.1% and 0.2% ethyl-methane sulfonate (EMS). In China, the mutant rice variety Zhefu, characterized by high resistance to rice blast, was developed through gamma-ray irradiation of the variety Simei 2 (Ahloowalia et al., 2004). Major blast resistance genes including Pib, Pita, Pia, Pi1, Pikh, Pi2, and Pi4 have been successfully introduced into rice varieties via conventional breeding programs. The durability of multiline varieties is influenced by the rate at which blast races develop, the proportion of lines present in a mixture and the size of the planted area. Attempts have been made to develop multiline varieties using blast resistant isogenic lines for “Nipponbare” (Higashi et al., 1981; Horisue et al., 1984), “Toyonishiki” (Nakajima, 1994) and “Sasanishiki” (Matsunaga, 1996). Studies have confirmed that blast control is achieved through the use of multiple line varieties (Nakajima et al., 1996). Specifically, the “Sasanishiki” multiple-line variety has been commercially grown on a market scale since 1995. In addition, new isogenic lines have been developed and a detailed examination of the races of the blast pathogen has been carried out, which is crucial for stable use (Ashizawa et al., 2007). A cross combination of Koshihikari blast-resistant isogenic lines (BLs) was developed (Ishizaki et al., 2005). The BLs were developed by crossing with Sasanishiki (Pia), Todorokiwase (Pii), Pi4 (Pita-2), Niigatawase (Piz), Koshiminori (Pik), Tsuyuake (Pik-m), Toride 1 (Piz-t) and BL1 (Pib) as the donor parent respectively, and then repeated back-crossings with “Koshihikari” as the pollen parent were performed.
Gene strategy implementation involves utilizing different blast resistance mechanisms in various rice varieties and arranging them in specific temporal or spatial patterns. Rice cultivation practices incorporate seasonal and regional preferences for location-specific varieties. This approach allows for the development of distinct varieties using diverse blast resistance sources. Even within varieties used for a particular season, those with different maturity periods should incorporate unique blast resistance sources. This method slows the evolution of new virulent races and enhances the durability of blast resistance in current varieties. Among various strategies, deploying distinct genes in different maturity groups may improve the longevity of blast resistance in newly developed rice varieties. However, traditional resistance breeding has notable limitations, including extended breeding cycles, inefficient selection processes, and challenges in distant crossing. These drawbacks result in a lag between the creation of new resistant cultivars and the emergence of virulent pathotypes of the causative pathogen.
Molecular approaches
QTL mapping and GWAS
Rice germplasm harbors both qualitative and quantitative types of blast resistance genes. Blast resistance can be categorized into complete resistance, governed by major genes (R genes) and exhibiting race specificity; and partial resistance, governed by numerous genes known as quantitative trait loci (QTLs). Partial disease resistance offers durable protection against a wide range of pathogens, promising avenue for sustainable rice production in future. QTL detection serves as a valuable tool for mapping major or minor genes responsible for disease resistance (Mora et al., 2016). The mapping and tagging of QTLs linked to blast resistance can facilitate the cloning of major disease resistance genes and aid in marker-assisted breeding programs for the development of resistant cultivars. Many major blast resistance genes are qualitative in nature and have been identified and mapped within the rice genome (Figure 4) (Ashkani et al., 2016).
Genome-Wide Association Studies (GWAS) are a powerful tool used to identify genetic loci associated with blast resistance in rice by analyzing genetic variations across diverse rice populations. This approach has revolutionized resistance breeding by uncovering novel resistance (R) genes and quantitative trait loci (QTLs) associated with blast resistance. GWAS evaluates the statistical association between genetic markers (e.g., SNPs) and phenotypic traits (e.g., resistance to blast disease). QTLs are identified through GWAS and can be targeted for breeding. The discovery of new loci in wild rice species, such as Oryza rufipogon and Oryza nivara, offers novel sources of resistance. In the United States, a set of 151 accessions were used with 156 SSR markers for association with seed weight, plant height, and heading against blast resistance (R) genes, Pi-ta marker in rice (Wang et al., 2015). GWAS along with RNA sequencing analysis was performed to identify novel marker–trait associations against blast resistance in rice, where Pi5 and Pi56(t) was on chromosome 9 and 127 associations and 283 upregulated genes were revealed. The expression level increased after fungal inoculations; 401 downregulated genes significantly decreased against blast disease (Lu et al., 2019). In Japonica rice varieties from European countries, 311 accessions were screened and 14 marker–trait associations for blast resistance were identified using both field and growth chamber screenings (Volante et al., 2020). A nitrogen-induced susceptibility (NIS1) locus was analyzed in 139 Japonica rice strains by using the GWAS technique and conferred blast resistance by the identification of novel loci (NIS2, NIS3, and RRobN1 on chromosomes 5, 10, and 6) to be involved in the rice against blast fungus under different nitrogen regimes (Frontini et al., 2021). The significant associations were identified as the candidate loci in 48 accessions of rice for the blast resistance in rice that will serve as an important genetic resistance source to be introduced into an elite rice line in future breeding programs for deciphering blast resistance in rice. This GWAS helped to uncover significant gene regions which encode proteins to resist blast infection in rice plants (Barua et al., 2024). GWAS in rice identified 43 QTLs significantly associated with resistance to panicle blast genes for OsAKT1, OsRACK1A, Bsr-k1, and Pi25/Pid3 (Jinlong et al., 2024). Pangenome-wide association study (panGWAS) was carried out on nine blast resistance-related phenotypes using 414 international diverse rice accessions from an international rice panel, and 74 QTLs associated with rice blast resistance were identified. The significant potential QTL (qPBR1–6 candidate genes) confers resistance to both panicle and leaf blast throughout their growth period, and 3,311 differentially expressed genes are involved against blast resistance (Wang J et al., 2024). From these references, it can be inferred that GWAS loci are combined with known Pi-genes to develop varieties with durable, broad-spectrum blast resistance in rice.
Marker-assisted selection
Marker-assisted selection (MAS) is a tool and often controlled by a single or few genes. The MAS leverages the specific interaction between resistance (R) genes and avirulence (AVR) genes in host–pathogen interactions, thereby enhancing blast control (Petit-Houdenot and Fudal, 2017). By identifying molecular markers linked to desired traits, MAS improves the efficiency of conventional breeding methods—for instance, a set of SSR markers (RM168, RM8225, RM1233, RM6836, RM5961, and RM413) associated with blast resistance has been identified and could be utilized in MAS programs. These molecular markers, combined with MAS strategies, play a critical role in the development of durable blast-resistant plant varieties. The three rice varieties, namely, BRRI dhan48, BRRI dhan58 (recurrent parent), and IRBL9-W (Donor parent), were crossed for blast resistance, and introgression of genes was confirmed by marker-assisted selection (Nadim et al., 2024). The drought variety of rice Huhan 1516 had the blast Pi2 gene introgressed by using this technique, where Huhan 1509 was the donor parent (Li A et al., 2024). The water-saving and drought-resistant rice core parents were Hanhui 3, BL675-1-127, and B5. BL5 carries the Pi1 and Pi2 genes, BL675-1-127 carries the Pi9(t) gene, and B5 carries the Bph14 and Bph15 genes; these were introgressed through MAS (Liu G et al., 2024). Fengshun et al. (2024) had genes introduced into the Japonica rice cultivar Anhui for blast resistance genes Pita, Pi5(t), Pi1, Pia, Pik, Pi54, and Pb1. To introduce the broad-spectrum blast resistance gene R6 into the early indica rice thermosensitive genic male sterile (TGMS) line HD9802S was used by employing MAS (Chen et al., 2023). The Mushk budji cultivar was used as recipient parent where the blast resistance genes Pi9(t) and Pi54 were introgressed (Shafi et al., 2023). A set of 119 rice main varieties (94 Japonica and 25 indica) were used to introduce the 14 major blast resistance genes Pit, Pish, Pib, Pi1, Pia, Pi54, Pita, Pi9(t), Pi2, Pikm, Pigm, Pi5, Pb1, and Piz-t through marker-assisted selection, which were screened by SSR markers (Qi et al., 2023). The Pi9(t), Pi5, and Pi54 genes were introduced into Huhan 1s and Huhan 74S (Liu et al., 2021). According to Wu et al. (2016), the Yangdao 6 rice variety was where broad-spectrum durable resistance genes Piz-t, Pi2, Pigm, Pi40, Pi9(t), and Piz were introgressed and validated with SSR markers. From these inferences, we can conclude that blast resistance genes like Pib, Pi1, Pia, Pi54, Pita, Pi40, Pi9(t), Pi2, Pikm, Pigm, Pi5(t), Pb1, and Piz-t, etc., were introgressed into different suitable recipient cultivars, and these resistance varieties can further help in the development of new varieties that are useful in crop breeding programs.
Marker-assisted backcross breeding
Marker-assisted backcrossing (MABB) is a simplified breeding method in which molecular markers are used to precisely target specific genetic loci, which reduces the length of donor segments containing these loci and allows us to efficiently recover desired traits from the recurrent parent genome. The aim of this approach is to transfer targeted genes while reducing donor segment size and retaining recurrent parental characteristics (Hasan et al., 2015). MABB offers precision, efficiency, and time savings over traditional backcrossing methods by employing tightly linked molecular markers for key traits. This approach has been widely adopted to transfer resistance genes into popular rice varieties worldwide (Table 5). Numerous studies have demonstrated the efficacy of marker-assisted selection in developing new rice varieties. The blast and bacterial blight resistant lines, two blast resistance genes (Pi9(t) and Pb1) and three bacterial blight resistance genes (xa5, xa13, and Xa21), were pyramided in the background of premium quality rice variety BRRI dhan63 through marker-assisted backcross breeding. Pi9(t)-US2 and Pb1-US2 were used as donor parents for Pi9(t) and Pb1, respectively, and for xa5, xa13, and Xa21, IRBB60 was used as the donor parent (Nihad et al., 2024). Sowmiya et al. (2024) examined that the resistant varieties through MABB were blast resistance gene Pi54 introgressed into ADT43 from RP-Bio-Patho-2. The mega rice variety with high-yielding Swarna lines was introgressed with blast and blight genes Pi54 and Xa21 into the near-isogenic line of improved samba mature through this MABB technique (Kousik et al., 2024). Dileep Kumar et al. (2023) reported on the introgression of blast resistance gene Pi54 (isogenic line of MTU 1010 as donor parents) into Jaya rice variety as the recurrent parent.
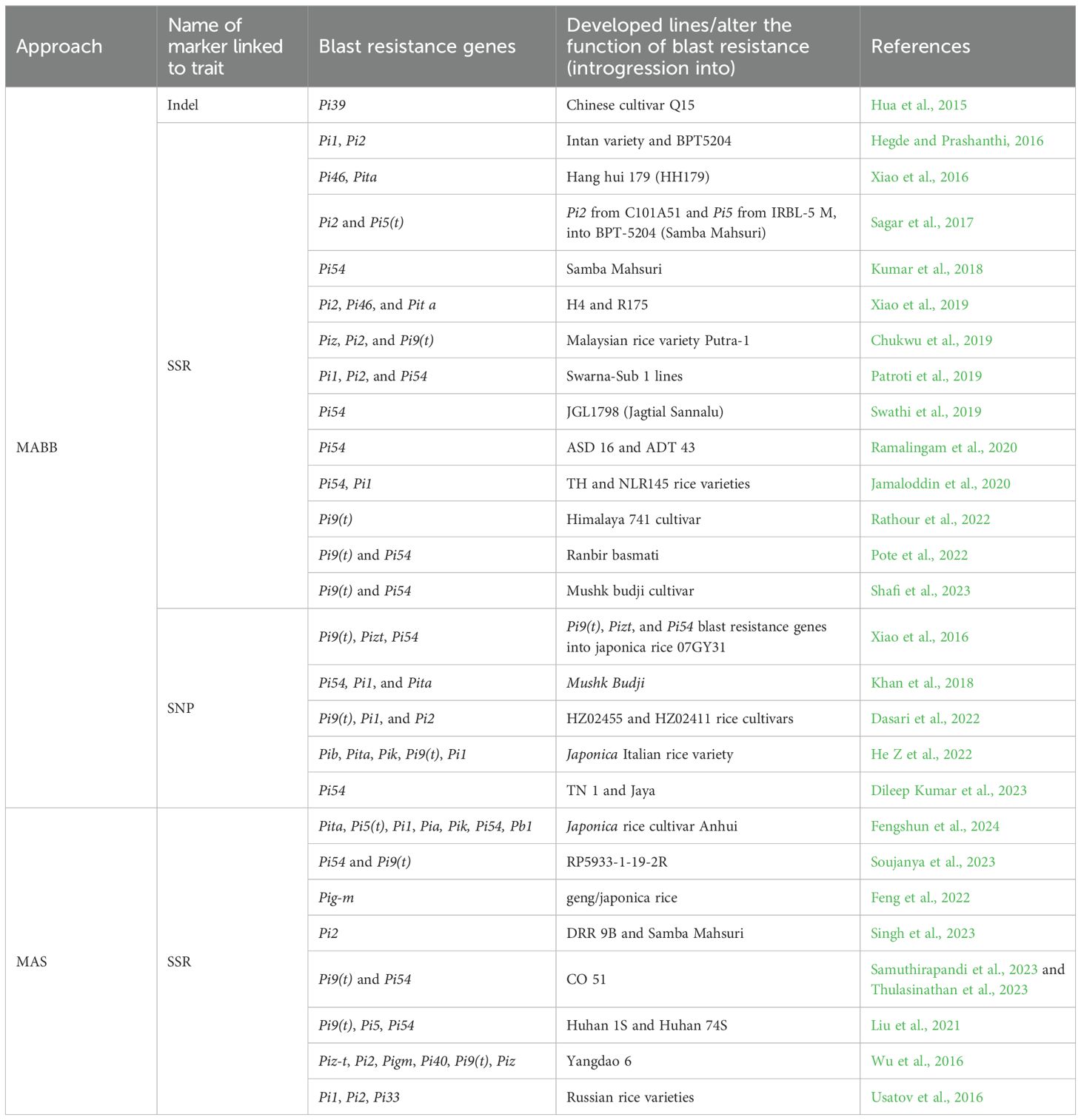
Table 5. Successful examples for introgression of blast-resistant genes in rice through marker- assisted selection (MAS) and marker-assisted backcross breeding (MABB).
The sd1 gene for semi-dwarfism and the Pi9(t) and Pi54 genes for blast resistance were incorporated into a traditional basmati rice variety, Ranbir Basmati (Pote et al., 2022). Dasari et al. (2022) revealed that blast resistance was developed by introducing the Pi9(t), Pi1, and Pi2 genes in HZ02455 and HZ02411 rice cultivars, as confirmed using SNP markers. The Japonica rice cultivar was also introgressed with blast resistance genes Pib, Pita, Pik, Pi9(t), and Pi1, as validated by SNP markers (He Z et al., 2022). The broad-spectrum resistance locus Pi9(t) was transferred from a Basmati donor, PB1637, into the cold-tolerant variety Himalayan 741 (Rathour et al., 2022). The Pi1 and Pikh blast resistance genes were incorporated into the Jyothi and Kanchana rice varieties from Parambuvattan through marker-assisted backcross breeding (MABB) (Anusha, 2022). Tellahamsa, a cold-tolerant variety, served as the recurrent parent, while Improved Samba Mahsuri (Xa21 and xa13) and NLR 145 (Pi54 and Pi1) were selected as donor parents for introgression using MABB (Jamaloddin et al., 2020). In ASD 16 and ADT43, the Pi54 blast genes were introduced and confirmed by SSR markers (Ramalingam et al., 2020). Sagar et al. (2020) revealed that the introgression of two genes, each governing resistance to major rice diseases, bacterial blight (BB) (xa13 and Xa21) and blast (Pi2 and Pi54), was achieved in the popular basmati cultivar Pusa Basmati 1509 through marker-assisted backcross breeding (MABB). The Malaysian rice variety Putra-1 harbors blast resistance genes Piz, Pi2, and Pi9(t) (Chukwu et al., 2019). Xiao et al. (2016) reported the introgression of the Pi9(t), Pizt, and Pi54 blast resistance genes into japonica rice 07GY31. Hence, these selected plants can be forwarded for further generations to develop high-yielding blast-resistant rice lines. Hence, addressing blast resistance in genes is typically focused on the successful integration of resistant genes Piz, Pi2, Pi54, Pi1, and Pi9(t) into high-yielding or any locally adapted cultivars through MABB which is helpful to enhance the ability to withstand the pathogen M. oryzae. Consequently, this strategy serves as an effective tool in modern rice breeding programs, significantly enhancing blast resistance while ensuring high productivity and climate adaptability.
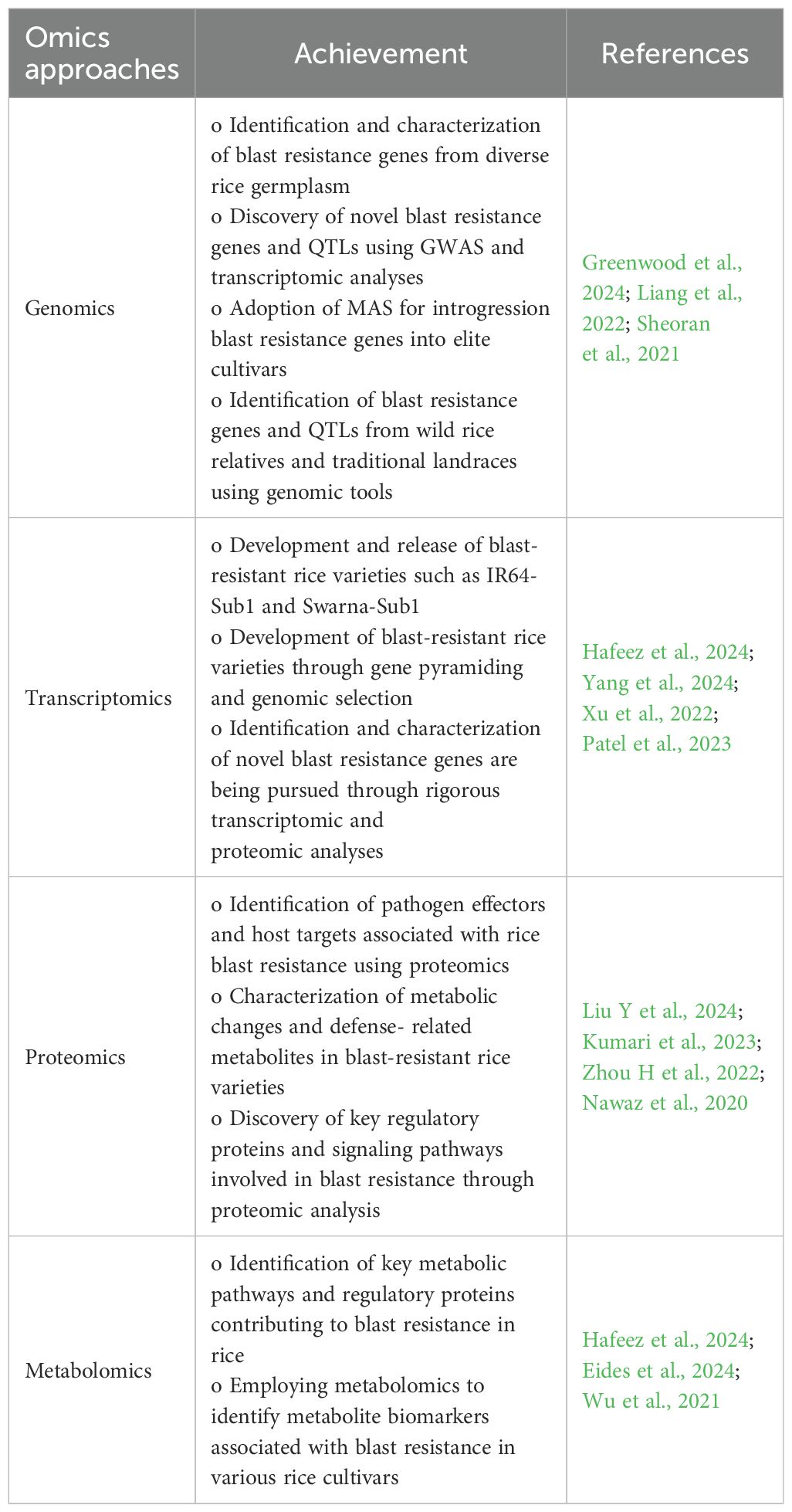
Omics approaches
Generally, omics approaches, including genomics, transcriptomics, proteomics, and metabolomics, have drastically revolutionized the study of blast resistance in rice by providing comprehensive insights into the genetic and molecular mechanisms underlying host–pathogen interactions (Greenwood et al., 2024) (Table 6). Genomic studies have identified key genes and genomic regions associated with blast resistance in rice. Genome-wide association studies (GWASs), quantitative trait loci (QTLs) mapping, and comparative genomics are often used to identify candidate genes and alleles conferring resistance (Wei et al., 2021). High-throughput genotyping techniques such as SNP arrays and next-generation sequencing facilitate the identification of genetic markers associated with resistance traits. Genome-wide association studies (GWASs) have been utilized to identify natural allelic variations associated with blast resistance across diverse rice germplasms, offering valuable insights into the genetic framework governing resistance (Abhijith et al., 2022). Genomic selection (GS) leverages genome-wide marker information to predict the breeding value for blast resistance in rice breeding programs. It enables the early selection of superior genotypes based on genomic estimated breeding values (GEBVs) for crop improvement (Tabassum et al., 2021). Transcriptomic studies have provided dynamic gene expression changes against rice blast infection. RNA sequencing (RNA-seq) and microarray analysis have been used to profile gene expression patterns during different stages of infection and in response to various pathogens (Zhu et al., 2024) and are also helpful in identifying differentially expressed genes involved in defense signaling, pathogen recognition, and secondary metabolite biosynthesis, providing insights into the molecular basis of rice defense mechanisms against blast disease (Chandrakanth et al., 2024). Transcriptomics facilitates identifying and characterizing transcription factors (TFs) and regulatory elements involved in blast resistance. TFs such as WRKY, NAC, and bZIP families are key regulators of defense gene expression and play crucial roles in coordinating immune responses (Thapa et al., 2024).
Proteomic studies are helpful in identifying the complex network of proteins involved in rice defense responses against blast. High-throughput proteomic techniques, including quantitative proteomics and phospho-proteomics, are being used to identify key signaling components and defense proteins (Zhang et al., 2022). Recent proteomic analyses have revealed post-translational modifications and protein–protein interactions that regulate the activation of defense pathways in rice upon blast infection (Wang et al., 2022a) and also the identification of defense-related proteins, such as chitinases, peroxidases, and pathogenesis-related (PR) proteins that are upregulated in response to blast infection. Techniques such as two-dimensional gel electrophoresis (2-DE) and liquid chromatography–mass spectrometry (LC–MS/MS) facilitate the comprehensive profiling of proteins, leading to the discovery of key defense proteins and signaling pathways against rice blast disease (Zhao et al., 2021). Metabolomics is the analysis of small molecules in biological systems, elucidating the metabolic changes underlying blast resistance in rice. Advances in metabolite profiling techniques are enabling the identification of defense-related metabolites and metabolic pathways, phytohormones, and redox-active compounds helpful in disease response against blast in rice (Shi et al., 2024). High-throughput analytical techniques such as liquid chromatography–mass spectrometry (LC–MS) and gas chromatography–mass spectrometry (GC-MS) allow for the comprehensive profiling of metabolites, facilitating the discovery of biomarkers and key metabolic pathways. Metabolomic studies have shown changes in the levels of defense-related metabolites, such as phytoalexins, phenolics, volatile organic compounds (VOCs), and flavonoids, in response to blast infection. Numerous studies have been done by using these multi-omic approaches. In rice, overexpressing phytochrome-interacting factor-like 1 (OsPIL1) rice lines were evaluated in terms of their impact on growth, grain development, and resistance to Magnaporthe oryzae. Multi-omics analysis (RNA-seq, metabolomics, and CUT&Tag) and RT-qPCR validated the OsPIL1 target genes and key metabolites (Zhao et al., 2024). Multi-omics approach, especially proteomics, was used to comprehensively analyze MoKin1 function, and the results revealed that MoKin1 affected the cellular response to endoplasmic reticulum stress (ER stress), of which the downregulated proteins in ΔMokin1 mutant were enriched mainly in response to ER stress triggered by the unfolded protein. Therefore, the phosphorylation of various proteins regulating the transcription of ER stress-related genes and mRNA translation was significantly downregulated (Zhang L et al., 2024). Integration of transcriptomic, proteomic, and phosphor-proteomic analysis of Mowanggu was performed after inoculation with M. oryzae, revealing that differentially expressed genes and proteins were upregulated and significantly enriched in protein phosphorylation, peroxisome, plant–pathogen interactions, phenylpropanoid metabolism, phenylalanine biosynthesis pathways, reactive oxygen species (ROS), glycolysis, MAPK signaling pathways, and amino acid biosynthesis against rice blast resistance (Peng et al., 2023). Hence, from these, it was concluded that omics approaches viz., genomics, transcriptomics, proteomics, and metabolomics, have revolutionized research on blast resistance in rice required for GWAS (identification of resistance, susceptibility genes, QTLs) linked to blast, paving the way for marker-assisted breeding and genome editing, differential gene expression, and identification of proteins (pathogenesis-related proteins, ROS pathways), secondary metabolites, and signaling molecules that contribute to the development of resistant lines against blast disease.
Allele mining
Allele mining is a widely employed molecular technique for identifying allelic variations or novel alleles within a targeted gene. This approach involves thoroughly characterizing a large set of germplasm collections used for allele mining. Tilling and eco-tilling techniques are used to identify induced point mutations in the targeted gene by heteroduplexes of alleles during DNA replication process for identifying allelic variations. Sharma et al. (2012) reported the allelic variation for genes responsible for rice blast resistance, such as Pi-ta, Pi-kh, and Pi-z(t), among Indian land races using allele mining technique. Their findings revealed a substantial variation in Pi-kh and Pi-z(t) alleles compared to Pi-ta alleles. Effective allele mining uses genetically diverse materials with prior knowledge of gene sequence information. This approach aids in the detection of superior alleles from both wild and cultivated rice species available thus far for blast resistance genes. A large-scale screening of new blast resistance alleles was conducted across 2,000 rice accessions from major rice-producing areas in China. Sequence-based allele mining was used to identify the allelic variants of major rice blast resistance genes at the Pi5 locus of chromosome 9. Six novel alleles were identified from 64 accessions, and 153 accessions showed moderate resistance against blast (Zhou Y et al., 2024). The alleles from seven varieties showing high resistance were selected for transformation into the susceptible variety J23B to construct near-isogenic lines (NILs). There is a large-scale screen of rice blast resistance in about 2,000 rice accessions; among them, 247 accessions showed at least medium resistance, and seven novel Pik alleles were identified as blast resistant. The rate of Pik-R0/ME/7017 donors was greater than 80% (Ying et al., 2022).
These NILs showed resistance in a field test in Enshi and Yichang, indicating that the seven novel rice-blast-resistance tandem-repeat regions at the Pi2/Pi9(t) locus of chromosome 6 could potentially serve as a genetic resource for molecular breeding of resistance to rice blast (Zhou et al., 2020). Allele mining for blast-resistant gene Pi9(t) was performed in 338 rice landraces, among them 136 polymorphic sites comprising of transitions, transversions, and insertion and deletions (InDels) were identified in the 2.9-kb sequence of Pi9(t) alleles (Imam et al., 2016). The AC134922 locus is nucleotide-binding-site leucine-rich-repeat (NBS-LRR) gene family in rice genome where six rice blast resistance (R) genes have been cloned from this locus and two resistance candidate genes, Pi34 and Pi47, are also mapped. A total of 22 genes from 12 cultivars based on allele-mining strategy was cloned at this locus, and six rice blast R genes were identified, with four of them recognizing more than one isolate (Wang et al., 2014). PCR-based allele mining for blast resistance gene Pi54 from six cultivated rice lines and eight wild rice species was carried out to understand its structural variation and its impact on the phenotypes in Tetep (Pi54 genes) in which the sequence analysis showed more interspecies variation of cultivated and wild species. The structural analysis of alleles showed the presence of a variable number of open reading frames (0–2) principally having point mutations in the leucine-rich repeats (LRR) regions, and these resistance alleles can be used in the effective management of rice blast disease through gene pyramiding (Kumari et al., 2013). The identification of novel alleles of rice blast resistance genes Pikh and Pita genes with linked markers RM206, TRS26, TRS33, YL153, YL154, YL155, and YL87 for Pita was also used to screen materials based on marker profiles to downsize the number of genotypes for allele mining (Ramkumar et al., 2010). Therefore, it concluded that allele mining serves as a cornerstone helpful in identifying and characterizing genetic variations in blast resistance genes that contribute to combat the fungal pathogen M. oryzae.
Genomic selection
Genomic selection (GS) is an advanced breeding approach that utilizes genome-wide genetic information to predict the performance of rice cultivars. This technique has revolutionized rice breeding, enabling the rapid development of blast-resistant varieties by predicting and selecting resistance traits without the need for extensive phenotypic evaluation to improve breeding efficiency by predicting the individuals’ genetic potential based on their genome-wide marker data (Meuwissen et al., 2001). The main principle of this technique is using a training population with known phenotypic and genotypic data to develop prediction models. These models are then applied to selection candidates to predict their resistance to blast disease. GS allows the simultaneous selection of multiple quantitative trait loci (QTLs) associated with blast resistance, speeding up the breeding process and enabling the accumulation of favorable alleles (Escola et al., 2023). Genomic selection integrates genomic information from resistance genes, such as Pi genes (Pi9(t), Pi54, and Pi33), which are associated with blast resistance. High-throughput genotyping methods like SNP arrays and next-generation sequencing are used to identify markers linked to blast resistance traits. Models are trained on datasets combining phenotypic resistance data (from artificial or natural blast infections) with genome-wide marker profiles (Younas et al., 2024).
Advanced statistical techniques, such as ridge regression best linear unbiased prediction (RR-BLUP), Bayesian models, and machine learning algorithms, improve the accuracy of predictions (Xu et al., 2021). GS complements traditional marker-assisted selection (MAS) by allowing breeders to select complex traits controlled by multiple genes, such as partial resistance to blast, which is often governed by polygenes (Hickey et al., 2017). The main components of GS are (i) utilizing genome-wide markers simultaneously to develop a genotype–phenotype relationship model in one population (called training population) accounting for genome-wide linkage disequilibrium (LD) among markers and (ii) predicting the genomic estimated breeding values (GEBV) based on the model in future candidates of other related populations (called breeding population) (Heffner et al., 2009). The success of GS depends on the accuracy of prediction and predictability of models adapted to different crops (Crossa et al., 2017). A set of 162 rice lines from USDA and 237 African lines was evaluated for blast resistance as determined by genomic estimated breeding values (GEBVs) by RR-BLUP model and confirms that the accuracy of genomic selection for blast resistance in rice varies from germplasm to germplasm which ranges from 0.29 to 0.59 (Balimponya, 2015). Genomic selection studies by using GBLUP statistical method in 161 African rice accessions found resistance against blast disease in rice (Huang et al., 2019). Hence, it confirms that different statistical models are helpful for a selection of traits controlled by multiple genes, QTLs against blast resistance in rice, which leads to the development of broad-spectrum varieties by genomic selection.
The International Rice Research Institute (IRRI) has implemented genomic selection to pyramid blast resistance genes (Pi9(t) and Pi54) into high-yielding rice varieties. This approach has successfully developed improved varieties like NSIC Rc222 with enhanced resistance and better yield potential (www.irri.org). Chinese researchers also used this technique to identify and incorporate QTLs associated with broad-spectrum blast resistance into elite varieties. Hybrid rice breeding programs in India and Southeast Asia use genomic selection to predict blast resistance in parental lines (Xiao et al., 2021). This has enabled the development of hybrids with improved blast resistance and high yields. Wild relatives of rice (Oryza rufipogon and Oryza nivara) have been incorporated into GS programs to introduce novel blast resistance genes (Balimponya, 2015).
Speed breeding
Speed breeding is an innovative technique designed to accelerate the traditional breeding cycle, enabling the rapid development of rice varieties with enhanced resistance to diseases like blast (Magnaporthe oryzae) (Ahmar et al., 2020). The approach leverages controlled environments, such as growth chambers or greenhouses, to shorten generation time by optimizing environmental factors, which is helpful for the deployment of blast-resistant varieties. Speed breeding aims to increase the rate of genetic improvement by shortening the time, particularly useful for traits like disease resistance, which needs continuous improvement due to evolving pathogens (Sharma et al., 2022). By optimizing photoperiod, light intensity, temperature, and humidity, this technique allows rice plants to flower and mature more quickly than under traditional field conditions. Growth chambers or controlled greenhouses that allow for extended day-length conditions (up to 22 h of light per day) and higher temperatures promote faster plant development (Jahne et al., 2020). Speed breeding can be integrated with MAS to identify plants carrying specific Pi-genes or other resistance loci (Chimmili et al., 2022). The University of Queensland has developed a speed breeding platform that allows researchers to rapidly generate rice lines with enhanced resistance to diseases like blast. The International Rice Research Institute (IRRI) and the University of Sydney use speed breeding methods to shorten breeding cycles and rapidly incorporate blast resistance genes into high-yielding varieties. Speed breeding allows breeders to introduce blast resistance genes into elite varieties much faster than conventional methods. The breeding cycle is shortened from 5 to 6 years to as little as 8–12 months, allowing the quicker deployment of resistant varieties to combat blast outbreaks. Faster breeding cycles allow for the pyramiding of multiple resistance genes (Pi-genes and others) into a single variety, leading to more durable and broad-spectrum resistance (Zainuddin et al., 2024). Hence, the combination of speed breeding with genomics-assisted breeding (MAS and GWAS) can further accelerate the development of blast-resistant varieties.
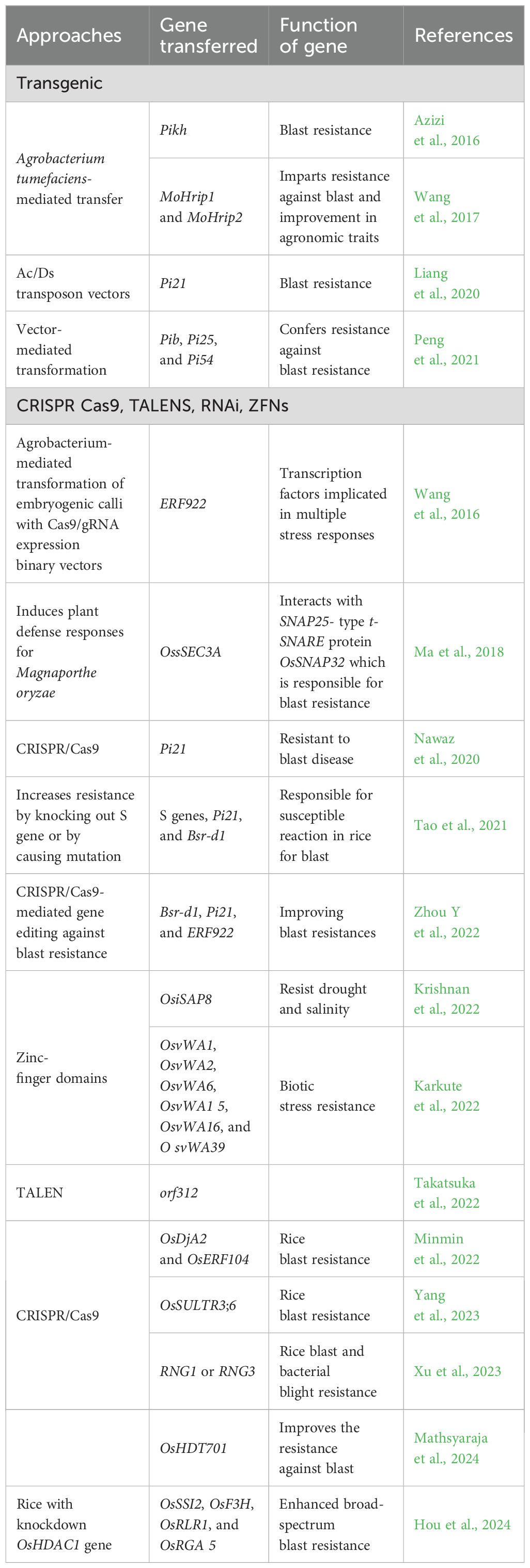
Transgenic approaches
These transgenic approaches are vital in mitigating blast disease and yield loss by enhancing resistance and improving crop resilience. Hence, to address this issue, improving high yield and climate resilience traits can be approached through efficient strategies, i.e., RNA interference, transgenic, and genome editing.
RNAi interference
RNA interference (RNAi) has emerged as a potent and efficient tool for combating various challenges caused by small microbial organisms such as viruses, bacteria, and fungi. In RNAi, short interfering RNA (siRNA) molecules are used to silence targeted gene expression. In rice blast management, siRNA molecules target essential genes in the fungus, disrupting its growth and virulence (Jain et al., 2017). Tissue-specific gene silencing is induced by employing gene-specific promoters to simultaneously silence several genes. Successful reports have proven the efficiency of RNAi in achieving integrated biotic resistance against major diseases and pests in plants. The dsRNA (PyDCL2–863 bp) was synthesized for the silencing of DCL2 transcript of P. oryzae through RNA interference and showed potential for PyDCL2-dsRNA to be developed as a new fungicide for the sustainable disease management of rice blast (Pushpanjie et al., 2024). HIGS (host-induced gene silencing) is used for six genes (CRZ1, PMC1, MAGB, LHS1, CYP51A, and CYP51B) that play important roles in the pathogenicity and development of M. oryzae. HIGS vectors were transformed into rice calli through Agrobacterium-mediated transformation, and T0, T1, and T2 generations of transgenic rice plants were generated. Following infection with M. oryzae of HIGS transgenic plants, the expression levels of target genes were reduced as demonstrated by quantitative RT-PCR. In addition, treating M. oryzae with small RNA derived from the target genes inhibited fungal growth. These findings suggest that RNA silencing signals can be transferred from the host to an invasive fungus and that HIGS has the potential to generate resistant rice against M. oryzae (Wang and Dean, 2022). A transient dsRNA supplementation system for the targeted knockdown of MoDES1, a host-defense suppressor pathogenicity gene from M. oryzae, was carried out by spray-induced silencing (Sarkar and Roy-Barman, 2021). Thus, these findings confirm that RNAi-based gene expression approaches are a powerful tool for blast disease control in rice. Spray-induced gene silencing (SIGS) utilizing double-stranded RNA (dsRNA) or small interfering RNA (siRNA) is gaining interest because of its low cost and straightforward preparation in transgenic plants. Once dsRNA is applied to the leaf surface, it can either directly target pathogen cells or be absorbed by plant cells and transferred to the pathogens. This technique is further useful for silencing blast resistance genes and is helpful for the development of broad-spectrum durable resistance varieties in rice.
Genome editing
Genome editing techniques like CRISPR/Cas9, TALEN, and ZFNs have been used to develop various rice varieties for blast resistance (Table 7) (Viana et al., 2019). Among these, CRISPR/Cas9 techniques are more widely used in rice than TALEN and ZFNs. Rapid and adaptable genome modification is a potent approach for gaining comprehensive insights into molecular mechanisms in biological studies. Recently, genome editing facilitated by CRISPR/Cas9 has emerged as a dependable method for genetic manipulation across various biological research fields, including investigations of filamentous fungi. The CRISPR/Cas9 system consists of a Cas9 protein and a single-guide RNA (sgRNA), with the Cas9/sgRNA complex inducing a DNA double-strand break at the intended genomic site. This protocol outlines a fundamental CRISPR/Cas9 methodology, encompassing target sequence design, CRISPR/Cas9 expression vector construction, and transformation for genome editing in Pyricularia (Magnaporthe) oryzae. This approach enables efficient targeted gene disruption, base editing, and reporter gene knock-in without necessitating additional modifications to host components. The protocol also applies to implementing other CRISPR/Cas technologies and diverse functional genomics studies in P. oryzae. An 84-bp arginyl (Arg)-tRNA promoter-driven CRISPR/Cas9 system enables efficient and cost-effective gene editing in P. oryzae. By using the Mo_tRNAArg24-gRNA-Cas9 cassette, the Ppg1 gene disruption rate was increased up to 75.9% (Wang R et al., 2024). Developing Bsr-d1 knockout mutants via CRISPR/Cas9 enhances broad-spectrum resistance to rice blast in Northeast China (Zhang Y et al., 2024). CRISPR/Cas9-induced mutations in the MIR827 gene altogether abolish miR827 production and confer resistance to M. oryzae infection. This resistance is accompanied by the reduction of leaf Pi content compared to wild-type plants, whereas Pi levels increase in the leaves of the blast-susceptible miR827 overexpressed or plants. In wild-type plants, miR827 accumulation in leaves decreases during the biotrophic phase of the infection process (Bundo et al., 2024). 58B was edited by CRISPR/Cas9, targeting a Pi21 gene and effector-binding element (EBE) of the OsSULTR3;6 gene, and the mutants 58b were obtained by Agrobacterium-mediated method, but the expression of defense-responsive genes was significantly upregulated after infection with rice blast (Yang et al., 2023). A simple single-guide RNA (sgRNA) was designed to create oss5h1oss5h2oss5h3 triple mutants through CRISPR/Cas9-mediated gene mutagenesis. oss5h1oss5h2oss5h3 exhibited stronger blast resistance in rice to Xoo than single oss5h mutants and also significantly upregulated OsWRKY45 and pathogenesis-related (PR) genes in OsS5H gene editing (Liu et al., 2023). CRISPR/Cas9-mediated gene editing is employed to rapidly install mutations in three known broad-spectrum blast-resistant genes, Bsr-d1, Pi21, and ERF922, in an indica thermosensitive genic male sterile (TGMS) rice line Longke638S (LK638S) (Zhou et al., 2022). Hence, it infers that CRISPR-Cas9 technology emerges as a transformative tool for editing and the targeted modifications of blast resistance genes (Pi genes) and is also useful in gene knockout of susceptibility genes (S genes), stacking of resistant genes, which accelerates the development of blast-resistant rice varieties.
Ectopic expression
The most notable advancement in varietal development for disease resistance is the use of genetic engineering to develop transgenic rice with enhanced disease resistance. This method is advantageous for introducing disease resistance into elite rice cultivars, as transgenic plants can acquire a single desired trait without altering the original genetic background. Several studies have been performed to confer the disease resistance in rice against M. oryzae (Peng et al., 2021; Li et al., 2021; Wang et al., 2017). Transgenic technology enables the precise manipulation of genes encoding the desired traits of interest by inserting foreign genes from unrelated species or silencing specific gene expression. Agrobacterium transformation and biolistic methods are the most commonly employed techniques for transferring a gene of interest into selected plant cells. Agrobacterium-mediated transformation ensures the stable integration of new genes into the targeted genome—for instance, enhanced resistance to blast fungus was achieved in rice by expressing genes such as rice chitinase [rice class-I chitinase gene, Cht-2 or Cht-3]. Coca et al. (2006) developed blast-resistant transgenic rice by transferring the ER-CecA gene from the giant silk moth Hyalophora cecropia. This gene was optimized to produce Cecropin A peptides in paddy, which is an are a member of antimicrobial protein families and which is a good indicator of the direct effect of a gene on the pathogen. Moreover, Wang et al. (2017) transferred MoHrip1 and MoHrip2 genes into rice through an Agrobacterium tumefaciens-based method used against blast resistance to produce the transgenic paddy plants and constrain the growth of fungal hyphae and also had a high water-retention capacity. Furthermore, marker-free transgenic rice was generated using maize’s Ac/Ds transposon vectors carrying fluorescent protein (GFP) and red fluorescent protein (mCherry) genetic markers to generate marker-free transgenic plants. Pi21 gene was expressed in these transgenic plants to generate resistance against rice blasts. The transformed lines had good resistance against M. oryzae (Li et al., 2021).
Notably, three Pi genes, viz., Pib, Pi25, and Pi54, were transferred together into two rice varieties, the indica variety Kasalath and the japonica variety Zhenghan 10. The transformed varieties exhibited a good level of resistance against blast pathogens, but this gene pyramiding came with its baggage of linkage drag and pleiotropic effects of these genes. The transgenic plants were impairing many gene transcriptions, which ultimately interrupted the normal development of the plants (Peng et al., 2021). Glucan plays an important role in the growth and development of fungi, whereas glucanase can inhibit the growth of fungi by breaking glycosidic bonds and may be a promising target for developing rice varieties with broad-spectrum disease resistance. Researchers engineered a codon-optimized β-1,6-glucanase gene (GluM) derived from myxobacteria and introduced it into the japonica rice variety Zhonghua11 (ZH11). They generated numerous individual transgenic lines overexpressing GluM. From these, three single-copy, homozygous lines were selected at the T₃ generation for disease resistance analysis. The key outcomes compared to the non-transgenic ZH11 control are blast lesion area was reduced by ~82.71%, indicating a substantial enhancement against this major fungal disease in rice (Shen et al., 2023). Huang et al. (2023) revealed that transgenic plants overexpressing the ATP2 gene were generated via genetic transformation in the Zhonghua11 (ZH11) genetic background, and the blast resistance and immune response of ATP2- overexpressing lines and wild-type plants were compared. When infected by the rice blast fungus, the transgenic rice plants exhibited stronger antioxidant enzyme activity and a greater ratio of chlorophyll a to chlorophyll b. The first cloning of a synthetic maize chitinase 1 gene and its insertion in rice cv. (Basmati 385) is via Agrobacterium-mediated transformation to confer resistance to the rice blast pathogen, Pyricularia oryzae. Transgenic lines were analyzed using molecular and functional techniques and confirmed by polymerase chain reaction with primer sets specific to chitinase and hpt genes. Furthermore, real-time PCR analysis of transformants indicated a strong association between transgene expression and elevated levels of resistance to rice blast in transgenic Basmati 385 plants (Anwaar et al., 2024). The potential future for transgenic crops can be used to generate new varieties or to create strong resistance barriers against blast disease.
The adoption of transgenic approaches for controlling rice blast disease, though promising, faces significant regulatory, ethical, and social challenges across countries due to their biosafety laws, public perception, and agricultural policies. Regulatory hurdles include stringent approval processes imposed by authorities such as America’s USDA (United States Department of Agriculture) and APHIS (Animal and Plant Health Inspection Service), Europe’s EFSA (European Food Safety Authority), and India’s GEAC (Genetic Engineering Appraisal Committee), which evaluate GM rice for potential environmental risks, allergenicity, and unintended gene flow to wild relatives (Singh et al., 2022). Ethical concerns arise over intellectual property rights, as transgenic rice varieties are often patented, raising fears of corporate monopolization and reduced access for smallholder farmers, particularly in Asia and Africa where rice is a staple food (Shukla et al., 2018). Additionally, public opposition to GM crops persists in regions like Europe and Japan, where consumers demand “GM-free” labels due to perceived health risks and environmental concerns despite scientific evidence supporting the safety of GM crops (Sabat and Tripathy, 2024). In contrast, China and the Philippines have begun approving GM rice, driven by food security needs and government-backed research (ISAAA, 2024). Conversely, India has maintained a strict stance against GM food crops, with only GM cotton approved. Simultaneously, public protests and concerns over corporate control hinder the approval of GM rice (Dutta et al., 2016). Developing rice varieties resistant to pests and diseases will help protect farmers from the adverse impacts of chemical insecticides and fungicides. Additionally, challenges posed by abiotic stressors like drought, extreme temperatures, and salinity, factors that hinder rice cultivation, can be mitigated by creating GM rice featuring genes that enhance tolerance to these stresses. Nonetheless, the commercialization of GM crops remains a divisive topic, as global acceptance continues to evolve (Dutta et al., 2016). Overall, the outlook for GM rice is encouraging; however, social acceptance remains a bottleneck, with misinformation and a lack of farmer awareness slowing the adoption rates (Sandhu et al., 2024). Thus, while transgenic rice holds immense potential for durable blast resistance, overcoming these regulatory, ethical, and social barriers will require transparent policies, farmer education, and region-specific regulatory frameworks that balance innovation with public trust.
Hence, it concluded that all approaches for blast-resistant rice varieties through genetic engineering and marker-assisted breeding have significantly progressed the field of rice cultivation. Nevertheless, the long-term stability of these resistance traits faces challenges due to the adaptive evolution of Magnaporthe oryzae, the organism responsible for rice blast disease. The pathogen’s considerable genetic variability allows it to overcome resistance provided by singular major resistance (R) genes, leading to the gradual breakdown of resistance in rice cultivars over time. For instance, the Pi-ta gene, which has demonstrated effectiveness against specific strains of M. oryzae, has been compromised due to mutations in the corresponding avirulence gene present in the pathogen, thus underscoring the coevolutionary dynamics between host resistance and pathogen virulence (Jia et al., 2016). To enhance the durability of blast resistance, strategies such as pyramiding multiple R genes and integrating quantitative trait loci (QTLs) associated with partial resistance have been adopted. This methodology aims to establish a broader and more sustainable defense against varied pathogen populations. Innovations in genomic tools, including CRISPR/Cas9-mediated gene editing, have expedited the precise stacking of resistance genes, thereby presenting a promising pathway for the development of rice varieties with enhanced and enduring blast resistance (Zhou et al., 2022). Ongoing surveillance of M. oryzae populations is essential to identify emerging virulent strains promptly. The integration of these strategies ensures the long-term stability of blast resistance traits in rice, consequently safeguarding global rice production against this widespread disease (Pedrozo et al., 2025).
Future emerging diagnostic tools
Nanotechnology
Nanotechnology has emerged as a promising avenue for the effective management of rice blast that poses a substantial threat to global rice production. Various nanosized materials, which include quantum dots, metallic nanoparticles, silica nanospheres, magnetic nanoparticles, silicon nanowires, carbon nanotubes, nanopores, graphene, nanostructured surfaces, and metal films, have exhibited immense potential in revolutionizing the development of diverse in vitro diagnostic assays for rice blast pathogens. These materials offer unique properties, such as a high surface area-to-volume ratio, tunable surface chemistry, enhanced sensitivity, and specific targeting capabilities, which are essential for the accurate detection and characterization of rice blast pathogens. By harnessing nanotechnology, diagnostic procedures that empower the effective management and mitigation of rice blast infection in global rice production are streamlined (Kashyap et al., 2017b). Utilizing nanotechnology principles, Yang et al. (2014) developed an electrochemical device for the early detection of blast fungal infection in rice, offering targeted delivery and reduced environmental impact. This nanodevice employed palladium nanoparticles (PdNPs) as catalysts within a 3,3′,5,5′-tetramethylbenzidine sulfate/H2O2 system, alongside immobilized mannose-binding jacalin-related lectin (Osmbl) from rice. Rhamnolipid (RL)-modified silica nanoparticles (SiO2NPs) based on excellent antimicrobial effectiveness against various phytopathogens, helping to reduce plant diseases (Li S. et al., 2024). We investigated the functions and mechanisms of RL@SiO2NPs in alleviating rice blast disease, resulting in a remarkable 10.80% decrease in disease incidence, 97.05% reduction in fungal growth, and 13.33% increase in shoot dry biomass, collectively lowering the infection pressure from the blast fungus. This treatment also enhanced the plants’ nutritional condition and bolstered their disease resistance by restoring nutrient balance and maintaining ion homeostasis. This is supported by a 23.84% increase in potassium levels in leaves, a 60.34% rise in silica concentration in roots, and reductions of 11.89% in magnesium and of 11.89% in iron levels in rice leaves. In summary, our results indicate that RL0.1@SiO2NPs enhance rice plant resistance to blast by boosting the in vivo antifungal activity, activating the antioxidant defense system, and improving nutrient absorption in rice seedlings. Shahid et al. (2025) reported that silver nanoparticles (AgNPs) act as a preventive measure to reduce rice blast in Kocuria species. In vitro antifungal activity showed that AgNPs significantly reduced the mycelial growth and conidial germination of M. oryzae. Their antifungal effects were more pronounced than those of propiconazole, a commonly used demethylation inhibitor fungicide for rice diseases. Similarly, zinc oxide nanoparticles (ZnO NPs) have been recognized as effective agents against M. oryzae, the causal agent of rice blast. They exhibit direct antifungal properties by inhibiting fungal conidiation and appressorium formation. Additionally, they enhance the basal resistance of rice plants by inducing the accumulation of reactive oxygen species and upregulating defense-related genes and also reduce abscisic acid levels, leading to increased stress tolerance in rice seedlings (Qiu et al., 2023). The synthesis of nickel–chitosan nanoparticles (Ni–Ch NPs) was carried out using nickel chloride, and its effectiveness in promoting plant growth and inhibiting Pyricularia oryzae (the blast pathogen) was evaluated. A significant increase in germination and growth characteristics, including shoot and root lengths as well as the number of lateral roots, was noted in paddy seeds treated with Ni–Ch NPs compared to the control group, highlighting an improved photosynthetic rate. Additionally, Asian rice exhibited reduced blast symptoms on leaves treated with NPs under glasshouse conditions, demonstrating 64% mycelial inhibition in Petri plates. These findings suggest that nickel–chitosan nanoparticles could act as an effective plant growth promoter in managing rice blast disease (Parthasarathy et al., 2023). Nano-particles derived from Chaetomium sp. extracts exhibited significant antifungal activity against M. oryzae (Song et al., 2018). These findings emphasize the potential of nanotechnology to create sustainable and efficient methods for managing rice blast, providing alternatives to traditional chemical treatments. Therefore, machine learning and artificial intelligence techniques are crucial for identifying and developing networks designed to detect blast disease in rice.
Synthetic biology
Synthetic biology is a growing discipline which includes biology, genetic engineering, and computer science to design and construct new biological processes or perform specific functions (Benner and Sismour, 2005). This technique helps introgression or modify resistance (R) genes to improve the resistance against Magnaporthe oryzae—for instance, the Pi54 gene has been identified and cloned from the Indica rice line Tetep, conferring resistance to blast disease. Researchers have developed DNA markers and isolated new alleles, such as Pi54rh and Pi54 of, from wild rice species, contributing to the development of resistant rice varieties. The rice callus platform provides a unique opportunity to test strategies for the metabolic engineering of synthetic carotenoid pathways, leading to novel carotenoid-biofortified crops (Zhu et al., 2022). In transgenic rice plants, flowering time can be controlled by specific agrochemicals and yield-related traits like grain number, plant height, and heading date, which may lead to the production of crops suitable for growth in different climates and facilitate breeding for various agronomical traits by using synthetic biology (Okada et al., 2017). Hence, this perspective reveals the synthetic biology approach mostly used for the development of genetic engineering against traits and to explore the potential of protein construction to address the invasion and proliferation of M. oryzae, with the goal of identifying new drug targets and designing small-molecule compounds to manage this disease (Yan et al., 2024). This emerging technique is still not used for the development of resistance varieties against blast broad-spectrum disease.
Machine learning and artificial intelligence
Machine learning and artificial intelligence have emerged as a powerful tool for managing blast disease caused by Magnaporthe oryzae (Rajpoot et al., 2023). Advances in automated and high-throughput imaging technologies have resulted in a deluge of high-resolution images and sensor data of plants. However, extracting patterns and features from this large corpus of data requires machine learning (ML) tools to enable data assimilation and feature identification for stress phenotyping (Gill et al., 2022). The four stages of the decision cycle in plant stress phenotyping and plant breeding activities where different ML approaches can be deployed are (i) identification, (ii) classification, (iii) quantification, and (iv) prediction (ICQP). We provide a comprehensive overview and user-friendly taxonomy of ML tools to enable the plant community to correctly and quickly apply the appropriate ML tools and best-practice guidelines for various biotic and abiotic stress traits (Singh et al., 2016). These technologies enable accurate disease detection, prediction, and decision-making by analyzing large datasets, including phenotypic, environmental, and genotypic information. Recently, the convolutional neural networks (CNNs) model has been used to differentiate healthy and infected leaves more efficiently and accurately. These AI models analyze multi-omics data to identify candidate genes and pathways involved in blast disease and utilize drones for mapping disease severity and are helpful for the amount of fungicide application against blast resistance in rice. Faster R-CNN, Cascade R-CNN, and YOLOv3 were used as the primary detection frameworks of deep learning methods used for spore identification against blast fungal disease in rice (Zhou H et al., 2024). The prediction of the severity of blast disease to classes 0, 1, 2, and 3, respectively, in rice was performed by using a linear SVM (support vector machine) machine (Varsha et al., 2024). Integration of a support vector machine classifier and convolutional neural networks is used to recognize and classify specific varieties of paddy plant diseases (Haridasan et al., 2023; Maheswaran et al., 2022). The visual patterns on the rice plants are processed using machine learning classifiers such as support vector machine (SVM), logistic regression, decision tree, naïve Bayes, random forest, linear discriminant analysis (LDA), and principal component analysis (PCA), and based on the classification results, plants are recognized as healthy or unhealthy for blast disease (Kumar et al., 2021). Deep learning and hyperspectral imaging methods, including the UeAMNet (Unsupervised Extraction Attention-based Mixed CNN) model, assess the efficacy of identifying infected and healthy cases of rice blast disease (Yin et al., 2025). Integrating UAV remote sensing with deep learning allows for efficient high-throughput field phenotyping in breeding for rice blast resistance (Zhang et al., 2025). Kaur and Sivia (2024) found that four deep learning convolutional neural networks (CNNs)—AlexNet, Inception, Xception, and Visual Geometry Group (VGG)—paired with machine learning models like decision tree (DT), random forest (RF), and support vector machine (SVM), are based on supervised learning in the automatic detection of leaf blast disease in rice. Automatic leaf identification of blast diseases is essential for several reasons, including minimizing yield loss, monitoring and forecasting infections, recognizing host resistance, and investigating fundamental host–pathogen interactions. Artificial neural networks (ANN) performance is enhanced by selecting parameters using the adaptive sunflower optimization (ASFO) algorithm. Subsequently, the infected area is delineated using a level set segmentation algorithm. Our evaluation of this method, based on accuracy, sensitivity, and specificity, indicates that the proposed technique achieves a maximum accuracy of 97.94% in predicting rice blast disease (Zheng et al., 2011). Lee et al. (2022) reported that machine learning methods utilize three artificial neural network (ANN)-based models to predict rice blast. These models incorporate two types of ANN—the feed-forward neural network (FFNN) and long short-term memory (LSTM)—using various input datasets for performance comparison. The Blast Weather FFNN model attained the highest recall score of 66.3% in rice blast prediction. The effectiveness of ANN-based disease prediction models improved by applying appropriate machine learning techniques and hyperparameter tuning to optimize input data. A machine learning algorithm facilitates the early detection of rice blast disease. For this system, images of both healthy rice leaves and those affected by the disease are collected. Features are extracted from the healthy parts and diseased areas of the rice leaves. The simulation results indicate an accuracy of 99% for images showing blast infection and 100% for healthy images in the training phase. In the testing phase, accuracy is recorded at 90% for infected images and 86% for healthy ones, respectively (Ramesh and Vydeki, 2018).
Challenges
Blast disease in rice presents significant challenges due to its complex nature, geographical distribution, and the ever-changing characteristics of its pathogen. The fungus Magnaporthe oryzae is highly adaptable, showing rapid genetic variability that undermines the effectiveness of resistance in rice varieties. Differences in environmental conditions, such as humidity, temperature, and cropping systems across various regions, further complicate disease prediction and management strategies. Traditional management practices, including the use of fungicides, face issues related to environmental sustainability, the development of resistance in the pathogen, and high costs, particularly in resource-limited areas. Although biotechnological interventions show promise, they are hindered by the need for extensive research to identify durable resistance genes, the high costs associated with genetic engineering technologies, and regulatory challenges surrounding genetically modified organisms (GMOs) (Figure 5). Furthermore, breeding approaches require significant time investments, and integrating resistance genes often leads to trade-offs with yield or other agronomic traits. Many rice-growing regions also face limited access to high-throughput phenotyping, advanced diagnostic tools, and precise disease forecasting systems, which further hampers effective interventions. Socioeconomic factors, such as a lack of awareness among farmers, limited infrastructure, and difficulties in adopting advanced technologies, increase the challenges of achieving sustainable disease management on a global scale.
Conclusions and future perspectives
Traditional control strategies prove ineffective at the commercial scale. Employing resistant varieties is the most efficient approach to mitigate disease occurrence and avoid pesticide-related risks. Through conventional plant breeding methods, numerous blast-resistant cultivars have been developed. However, molecular-level advancements in rice have opened new avenues for enhancing rice production systems. Molecular breeding techniques, including MAS, MABB, transgenic approaches, and genome editing, have effectively managed resistance. Furthermore, cloning R and Avr genes and studies of their products will deepen our understanding of host–pathogen interactions. Rice varieties carrying a single R gene for a specific pathogen race often lose their resistance over time due to the appearance of new virulent strains.
Understanding the genetic identity of the new M. oryzae race is important for the accurate employment of rice cultivars with different R genes. Stacking of multiple R genes will provide long-lasting resistance. In rice breeding programs against blast disease, various combinations of resistance R genes in a single host plant should be considered. Incorporating multiple race-specific R genes into elite rice cultivars is widely recognized as the most effective strategy for developing broad-spectrum and durable resistance to blast disease. Nevertheless, this approach may inadvertently foster the evolution of new pathogen races, potentially including super races that could overcome multiple major R genes and trigger severe epidemics. As a result, it is imperative to strategically employ race-specific R genes in breeding programs to preserve rice cultivars’ blast resistance, which is still poorly understood. To achieve broad-spectrum and long-lasting resistance, a strategic blend of significant R genes and minor QTLs is essential, leveraging conventional integrated breeding techniques, state-of-the-art genomic approaches, and gene editing tools.
Marker-assisted foreground and background selection can accelerate the development of near-isogenic lines (NILs). More expertise could be involved in performing recently developed transgenic and genome editing technologies that could enhance their application in creating blast-resistant rice through precise genetic modifications. Some recent techniques, such as allele mining, association mapping, and genome editing, will also play a vital role in controlling blast disease. Emerging technologies such as nanotechnology, machine learning, and artificial intelligence offer promising advancements that can help reduce blast disease in rice. Nanotechnology approaches encompass nanoformulated pesticides, nanosensors for early disease detection, and enhanced resistance mechanisms. These methods provide precise and eco-friendly strategies for managing diseases and improving plant immunity through nano-based formulations. Machine learning enables early prediction and precise disease identification by assessing environmental and genetic factors. Artificial intelligence improves disease management by refining control strategies, advancing resistance breeding, and supporting decision-making through predictive modeling. By integrating these cutting-edge technologies, rice farmers can manage diseases proactively, minimize chemical use, and boost crop yields. Future studies should focus on merging nanomaterials with AI-powered smart sensors to develop real-time, field-ready diagnostic tools. Moreover, deep learning models utilizing extensive field data can enhance disease prediction and refine disease management strategies.
However, due to the variable nature of pathogens, the need for regular research on the advancement of sustainable resistant cultivars will always be a never-ending process due to the co-evolution of pathogens. Future investigations should identify such S genes in rice to harness them through genome modification techniques for the development of blast-resistant varieties. These advanced diagnostic techniques are useful for the early detection of blast disease, which is helpful for prevention and effective strategies against the development of broad-spectrum durable blast resistance in rice.
Author contributions
SR: Writing – original draft, Writing – review & editing. AL: Conceptualization, Supervision, Writing – review & editing. MS: Writing – review & editing, Supervision. GC: Conceptualization, Supervision, Writing – review & editing, Validation. RV: Writing – review & editing, Supervision. RJ: Writing – review & editing, Conceptualization, Supervision, Validation.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. Funding support from the Department of Biotechnology (grant number BT/PR47823/NER/95/1965/2022), Government of India, Ministry of Science and Technology, New Delhi.
Acknowledgments
We are thankful to all authors for the assistance, guidance, and support in writing this review article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abhijith, K. P., Gopala Krishnan, S., Ravikiran, K. T., Dhawan, G., Kumar, P., Vinod, K. K., et al. (2022). Genome-wide association study reveals novel genomic regions governing agronomic and grain quality traits and superior allelic combinations for basmati rice improvement. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.994447
Ahloowalia, B. S., Maluszynski, M., and Nichterlein, K. (2004). Global impact of mutation-derived varieties. Euphytica 135, 187–204. doi: 10.1023/B:EUPH.0000014914.85465.4f
Ahmar, S., Gill, R. A., Jung, K. H., Faheem, A., Qasim, M. U., Mubeen, M., et al. (2020). Conventional and molecular techniques from simple breeding to speed breeding in crop plants: recent advances and future outlook. Int. J. Mol. Sci. 21, 2590. doi: 10.3390/ijms21072590
Aleena, D., Padma, V., Rekha, G., Prasad, M. S., Madhav, M. S., Punniakoti, E., et al. (2023). Identification and validation of blast resistance Pi1 gene in the derived mapping population of rice variety, NLR34449 through molecular markers. Indian Phytopathol. 76, 1–7. doi: 10.1007/s42360-023-00688-9
Amante-Bordeos, A., Sitch, L. A., Nelson, R., Dalmacio, R. D., Oliva, N. P., Aswidinnoor, H., et al. (1992). Transfer of bacterial blight and blast resistance from the tetraploid wild rice Oryza minuta to cultivated rice, Oryza sativa. Theor. Appl. Genet. 84, 345–354. doi: 10.1007/BF00229493
Amoghavarsha, C., Pramesh, D., Sridhara, S., Patil, B., Shil, S., Naik, G. R., et al. (2022). Spatial distribution and identification of potential risk regions to rice blast disease in different rice ecosystems of Karnataka. Sci. Rep. 12, 7403. doi: 10.1038/s41598-022-11453-9
Angeles-Shim, R. B., Reyes, V. P., Del Valle, M. M., Lapis, R. S., Shim, J., Sunohara, H., et al. (2020). Marker-assisted introgression of quantitative resistance gene pi21 confers broad spectrum resistance to rice blast. Rice Sci. 27, 113–123. doi: 10.1016/j.rsci.2020.01.002
Anusha, T. (2022). Incorporation of blast resistance into Jyothi and Kanchana rice (Oryza sativa L.) varieties through marker assisted breeding. Department of Plant Breeding and Genetics, College of Agriculture, Vellanikkara.
Anwaar, S., Jabeen, N., Ahmad, K. S., Shafique, S., Irum, S., Ismail, H., et al. (2024). Cloning of maize chitinase 1 gene and its expression in genetically transformed rice to confer resistance against rice blast caused by Pyricularia oryzae. PloS One 19, e0291939. doi: 10.1371/journal.pone.0291939
Ashikawa, I., Hayashi, N., Yamane, H., Kanamori, H., Wu, J., Matsumoto, T., et al. (2008). Two adjacent nucleotide-binding site–leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 180, 2267–2276. doi: 10.1534/genetics.108.095034
Ashizawa, T., Sasahara, M., Ohba, A., Hori, T., Ishikawa, K., Sasaki, Y., et al. (2007). Lesion-based analysis of leaf blast suppression in mixture of rice cultivar and a resistant near-isogenic line. J. Gen. Plant Pathol. 73, 15–21. doi: 10.1007/s10327-006-0321-9
Ashkani, S., Rafii, M. Y., Shabanimofrad, M., Ghasemzadeh, A., Ravanfar, S. A., and Latif, M. A. (2016). Molecular progress on the mapping and cloning of functional genes for blast disease in rice (Oryza sativa L.): current status and future considerations. Crit. Rev. Biotechnol. 36, 353–367. doi: 10.3109/07388551.2014.961403
Ashkani, S., Rafii, M. Y., Shabanimofrad, M., Miah, G., Sahebi, M., Azizi, P., et al. (2015). Molecular breeding strategy and challenges towards improvement of blast disease resistance in rice crop. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00886
Azizi, P., Rafii, M. Y., Abdullah, S. N. A., Hanafi, M. M., Maziah, M., Sahebi, M., et al. (2016). Over-expression of the Pikh gene with a CaMV 35S promoter leads to improved blast disease (Magnaporthe oryzae) tolerance in rice. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00773
Balimponya, E. G. (2015). Application of genomic selection and association mapping to breeding for resistance to rice blast and bacterial blight of rice (Oryza sativa L.). The Ohio State University.
Barua, P., Phukon, M., Munda, S., Ranga, V., Sruthi, R., Borah, J. L., et al. (2024). Identification of significant SNPs and candidate loci for blast disease resistance via GWAS and population structure analysis in ARC panel of Oryza sativa. Physiol. Mol. Biol. Plants 30, 1673–1689. doi: 10.1007/s12298-024-01518-6
Bastiaans, L. (1993). Effects of leaf blast on growth and production of a rice crop: 1. Determining the mechanism of yield reduction. Netherlands J. Plant Pathol. 99, 323–334. doi: 10.1007/BF01974313
Benner, S. A. and Sismour, A. M. (2005). Synthetic biology. Nat. Rev. Genet. 6, 533–543. doi: 10.1038/nrg1637
Bernoux, M., Ellis, J. G., and Dodds, P. N. (2011). New insights in plant immunity signaling activation. Curr. Opin. Plant Biol. 14, 512 ± 518. doi: 10.1016/j.pbi.2011.05.005
Berruyer, R., Adreit, H., Milazzo, J., Gaillard, S., Berger, A., Dioh, W., et al. (2003). Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor. Appl. Genet. 107, 1139–1147. doi: 10.1007/s00122-003-1349-2
Boöhnert, H. U., Fudal, I., Dioh, W., Tharreau, D., Notteghem, J. L., and Lebrun, M. H. (2004). A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16, 2499–2513. doi: 10.1105/tpc.104.022715
Boller, T. and He, S. Y. (2009). Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science, 324: 742 ± 744. distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12, 2033–2045. doi: 10.1105/tpc.12.11.2033
Bryan, G. T., Wu, K. S., Farrall, L., Jia, Y., Hershey, H. P., McAdams, S. A., et al. (2000). A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12, 2033–2045. doi: 10.1105/tpc.12.11.2033
Bundó, M., Val-Torregrosa, B., Martín-Cardoso, H., Ribaya, M., Campos-Soriano, L., Bach-Pages, M., et al. (2024). Silencing Osa-miR827 via CRISPR/Cas9 protects rice against the blast fungus Magnaporthe oryzae. Plant Mol. Biol. 114, 105. doi: 10.1007/s11103-024-01496-z
Cao, W. L., Yu, Y., Li, M. Y., Luo, J., Wang, R. S., Tang, H. J., et al. (2019). OsSYP121 accumulates at fungal penetration sites and mediates host resistance to rice blast. Plant Physiol. 179, 1330–1342. doi: 10.1104/pp.18.01013
Cesari, S., Thilliez, G., Ribot, C., Chalvon, V., Michel, C., Jauneau, A., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463 ± 1481. doi: 10.1105/tpc.112.107201
Chandrakanth, R., Sunil, L., and Devaki, N. S. (2024). Transcriptomic profiling of resistant and susceptible rice cultivar blast resistance genes during Magnaporthe oryzae infection. Plant Mol. Biol. Rep. 42, 111–121. doi: 10.1007/s11105-023-01404-7
Chauhan, R., Farman, M. A., Zhang, H. B., and Leong, S. (2002). Genetic and physical mapping of a rice blast resistance locus, Pi-CO39 (t), that corresponds to the avirulence gene AVR1-CO39 of Magnaporthe grisea. Mol. Genet. Genomics 267, 603–612. doi: 10.1007/s00438-002-0691-4
Chen, P., Gao, G., Lou, G., Hu, J., Wang, Y., Liu, R., et al. (2023). Improvement of rice blast resistance in TGMS line HD9802S through optimized anther culture and molecular marker-assisted selection. Int. J. Mol. Sci. 24, 14446. doi: 10.3390/ijms241914446
Chen, J., Peng, P., Tian, J. S., He, Y. G., Zhang, L. P., Liu, Z. X., et al. (2015). Pike, a rice blast resistance allele consisting of two adjacent NBS-LRR genes, was identified as a novel allele at the Pik locus. Mol. Breed. 35, 117. doi: 10.1007/s11032-015-0305-6
Chen, X., Shang, J., Chen, D., Lei, C., Zou, Y., Zhai, W., et al. (2006). AB-lectin receptor kinase gene conferring rice blast resistance. Plant J. 46, 794–804. doi: 10.1111/j.1365-313X.2006.02739.x
Chen, J., Shi, Y. F., Liu, W. Z., Chai, R. Y., Fu, Y. P., Zhuang, J. Y., et al. (2011). A Pid3 allele from rice cultivar Gumei 2 confers resistance to Magnaporthe oryzae. J. Genet. Genomics 38, 209–216. doi: 10.1016/j.jgg.2011.03.010
Chimmili, S. R., Kanneboina, S., Hanjagi, P. S., Basavaraj, P. S., Sakhare, A. S., Senguttuvel, P., et al. (2022). “Integrating advanced molecular, genomic, and speed breeding methods for genetic improvement of stress tolerance in rice,” in Next-generation plant breeding approaches for stress resilience in cereal crops (Springer Nature Singapore, Singapore), 263–283. doi: 10.1007/978-981-19-1445-4_8
Choi, J., Park, S. Y., Kim, B. R., Roh, J. H., Oh, I. S., Han, S. S., et al. (2013). Comparative analysis of pathogenicity and phylogenetic relationship in Magnaporthe grisea species complex. PloS One 8, e57196. doi: 10.1371/journal.pone.0057196
Chukwu, S. C., Rafii, M. Y., Ramlee, S. I., Ismail, S. I., Oladosu, Y., Kolapo, K., et al. (2019). Marker-assisted introgression of multiple resistance genes confers broad spectrum resistance against bacterial leaf blight and blast diseases in Putra-1 rice variety. Agronomy 10, 42. doi: 10.3390/agronomy10010042
Chuma, I., Isobe, C., Hotta, Y., Ibaragi, K., Futamata, N., Kusaba, M., et al. (2011). Multiple translocation of the AVR-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PloS Pathogen 7, e1002147. doi: 10.1371/journal.ppat.1002147
Chung, H., Goh, J., Han, S. S., Roh, J. H., Kim, Y., Heu, S., et al. (2020). Comparative pathogenicity and host ranges of Magnaporthe oryzae and related species. Plant Pathol. J. 36, 305. doi: 10.5423/PPJ.FT.04.2020.0068
Coca, M., Peñas, G., Gómez, J., Campo, S., Bortolotti, C., Messeguer, J., et al. (2006). Enhanced resistance to the rice blast fungus Magnaporthe grisea conferred by expression of a cecropin A gene in transgenic rice. Planta 223, 392–406. doi: 10.1007/s00425-005-0069-z
Crossa, J., Pérez-Rodríguez, P., Cuevas, J., Montesinos-López, O., Jarquín, D., De Los Campos, G., et al. (2017). Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 22, 961–975. doi: 10.1016/j.tplants.2017.08.011
Das, A., Soubam, D., Singh, P., Thakur, S., Singh, N., and Sharma, T. (2012). A novel blast resistance gene, Pi54rh cloned from wild species of rice, Oryza rhizomatis confers broad spectrum resistance to Magnaporthe oryzae. Funct. Integr. Genomics 12, 215–228. doi: 10.1007/s10142-012-0284-1
Dasari, A., Vemulapalli, P., Gonuguntla, R., Thota, D. K., Elumalai, P., Muppavarapu, K., et al. (2022). Improvement of bacterial blight resistance of the popular variety, Nellore Mahsuri (NLR34449) through marker-assisted breeding. J. Genet. 101, 1–11. doi: 10.1007/s12041-021-01340-z
Devanna, B. N., Jain, P., Solanke, A. U., Das, A., Thakur, S., Singh, P. K., et al. (2022). Understanding the dynamics of blast resistance in rice-Magnaporthe oryzae interactions. J. Fungi 8, 584. doi: 10.3390/jof8060584
Devi, S. R., Singh, K., Umakanth, B., Vishalakshi, B., Rao, K. V., Suneel, B., et al. (2020). Identification and characterization of a large effect QTL from Oryza glumaepatula revealed Pi68(t) as putative candidate gene for rice blast resistance. Rice 13, 1–3. doi: 10.1186/s12284-020-00378-4
Dileep Kumar, G. D., Kasarla, C., Viswanatha, K. P., Darmagaru, S., Patwari, P., Bupalli, S., et al. (2023). Introgression of genes associated with yield enhancement and resistance against bacterial leaf blight and blast diseases into an elite rice variety ‘Jaya’ through marker assisted backcrossing. Res. square, 1–20. doi: 10.21203/rs.3.rs-3648037/v1
Ding KeJian, D. K., Tan GenJia, T. G., and Wu Jian, W. J. (1999). A study on yield loss caused by rice blast. Acta Phytophylacica Sinica 26, 60–64.
Dodds, P. N., Chen, J., and Outram, M. A. (2024). Pathogen perception and signaling in plant immunity. Plant Cell 36, 1465–1481. doi: 10.1093/plcell/koae020
Dutta, S. S., Pale, G., Iangrai, B., Aochen, C., Rai, M., and Pattanayak, A. (2016). Current status and future prospects of research on genetically modified rice: A review. Agric. Rev. 37, 10–18. doi: 10.0.73.117/ar.v37i1.9259
Eides, J. R., Ikehara, B. R., Almeida, N. R., Macedo, W. R., and Pinto, F. G. (2024). Metabolomics as a tool for analysis of wheat leaves from different cultivars infected with Pyricularia oryzae. ACS Agric. Sci. Technol. 4, 535–543. doi: 10.1021/acsagscitech.3c00580
Ellur, R. K., Khanna, A., Yadav, A., Pathania, S., Rajashekara, H., Singh, V. K., et al. (2016). Improvement of Basmati rice varieties for resistance to blast and bacterial blight diseases using marker assisted backcross breeding. Plant Sci. 242, 330–341. doi: 10.1016/j.plantsci.2015.08.020
Escolà, G., González-Miguel, V. M., Campo, S., Catala-Forner, M., Domingo, C., Marqués, L., et al. (2023). Development and genome-wide analysis of a blast-resistant japonica rice variety. Plants 12, 3536. doi: 10.3390/plants12203536
Feng, Z., Li, M., Xu, Z., Gao, P., Wu, Y., Wu, K., et al. (2022). Development of rice variety with durable and broad-spectrum resistance to blast disease through marker-assisted introduction of pigm gene. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.937767
Fengshun, S., Ran, Z., Hassan, M. A., Quan, G., Cuixiang, L., Bin, T., et al. (2024). Analysis of rice blast resistance dominant genes and their combinations in Japonica rice. Physiologia Plantarum 176, e14156. doi: 10.1111/ppl.14156
Flor, H. H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. doi: 10.1146/annurev.py.09.090171.001423
Frontini, M., Boisnard, A., Frouin, J., Ouikene, M., Morel, J. B., and Ballini, E. (2021). Genome-wide association of rice response to blast fungus identifies loci for robust resistance under high nitrogen. BMC Plant Biol. 21, 1–12. doi: 10.1186/s12870-021-02864-3
Fu, C., Wu, T., Liu, W., Wang, F., Li, J., Zhu, X., et al. (2012). Genetic improvement of resistance to blast and bacterial blight of the elite maintainer line Rongfeng B in hybrid rice (Oryza sativa L.) by using marker-assisted selection. Afr. J. Biotechnol. 11, 13104–13124. doi: 10.5897/AJB12.1465
Fujii, K., Hayano Saito, Y., Sugiura, N., Hayashi, N., Saka, N., Tooyama, T., et al. (1999). Gene analysis of panicle blast resistance in rice (Oryza sativa) cultivars with rice stripe resistance. Breed. Res. (Japan) 1, 203–210.
Fujisaki, K., Abe, Y., Ito, A., Saitoh, H., Yoshida, K., Kanzaki, H., et al. (2015). Rice Exo70 interacts with a fungal effector, AVR-Pii, and is required for AVR-Pii-triggered immunity. Plant J. 83, 875–887. doi: 10.1111/tpj.12934
Fukuoka, S., Mizobuchi, R., Saka, N., Ivan, S., Matsumoto, T., Okuno, K., et al. (2012). A multiple gene complex on rice chromosome 4 is involved in durable resistance to rice blast. Theor. Appl. Genet. 125, 551–559. doi: 10.1007/s00122-012-1852-4
Fukuoka, S., Saka, N., Koga, H., Ono, K., Shimizu, T., Ebana, K., et al. (2009). Loss of function of a proline- containing protein confers durable disease resistance in rice. Science 325, 998–1001. doi: 10.1126/science.1175550
Fukuoka, S., Yamamoto, S. I., Mizobuchi, R., Yamanouchi, U., Ono, K., Kitazawa, N., et al. (2014). Multiple functional polymorphisms in a single disease resistance gene in rice enhance durable resistance to blast. Sci. Rep. 4, 1 ± 7. doi: 10.1038/srep04550
Fukuta, Y., Nguyen, T. L., Pham, T. T. H., Trinh, T. L., du Pham, V., Hayashi, N., et al. (2020). Pathogenicity of rice blast (Pyricularia oryzae Cavara) isolates from Mekong River Delta, Vietnam. Japan Agric. Res. Q. 54, 239–252. doi: 10.6090/jarq.54.239
Fukuta, Y., Telebanco-Yanoria, M. J., Hayashi, N., Yanagihara, S., Machungo, C. W., and Makihara, D. (2019). Pathogenicities of rice blast (Pyricularia oryzae Cavara) isolates from Kenya. Plant Dis. 103, 3181–3188. doi: 10.1094/PHP-05-20-0041-RS
Fukuta, Y., Telebanco-Yanoria, M. J., Koide, Y., Saito, H., Kobayashi, N., Obara, M., et al. (2022). Near-isogenic lines for resistance to blast disease, in the genetic background of the Indica Group rice (Oryza sativa L.) cultivar IR64. Field Crops Res. 282, 108506. doi: 10.1016/j.fcr.2022.108506
Gill, T., Gill, S. K., Saini, D. K., Chopra, Y., de Koff, J. P., and Sandhu, K. S. (2022). A comprehensive review of high throughput phenotyping and machine learning for plant stress phenotyping. Phenomics 2, 156–183. doi: 10.1007/s43657-022-00048-z
Goto, I., Jaw, Y. L., and Baluch, A. A. (1981). Genetic studies on resistance of rice plant to blast fungus IV. Linkage analysis of four genes, Pia, Pi-k, Pi-z and Pi-i. Annuals Phytopathol. Soc. 47, 252. doi: 10.3186/jjphytopath.47.252
Greenwood, J. R., Lacorte-Apostol, V., Kroj, T., Padilla, J., Telebanco-Yanoria, M. J., Glaus, A. N., et al. (2024). Genome-wide association analysis uncovers rice blast resistance alleles of Ptr and Pia. Commun. Biol. 7, 607. doi: 10.1038/s42003-024-06244-z
Hafeez, R., Guo, J., Ahmed, T., Ibrahim, E., Ali, M. A., Rizwan, M., et al. (2024). Integrative transcriptomic and metabolomic analyses reveals the toxicity and mechanistic insights of bioformulated chitosan nanoparticles against Magnaporthe oryzae. Chemosphere 356, 141904. doi: 10.1016/j.chemosphere.2024.141904
Haque, M. A., Rafii, M. Y., Yusoff, M. M., Ali, N. S., Yusuff, O., Datta, D. R., et al. (2021). Recent advances in rice varietal development for durable resistance to biotic and abiotic stresses through marker-assisted gene pyramiding. Sustainability 13, 10806. doi: 10.3390/su131910806
Haridasan, A., Thomas, J., and Raj, E. D. (2023). Deep learning system for paddy plant disease detection and classification. Environ. Monit. Assess. 195, 120. doi: 10.1007/s10661-022-10656-x
Hasan, M. M., Akter, M., Saha, S., Malek, M. A., Islam, N., and Alam, M. A. (2024). Morpho-molecular characterization revealed distinct diversity among rice blast (Pyricularia oryzae) isolates. Indian Phytopathol. 9, 1–9. doi: 10.1007/s42360-024-00749-7
Hasan, M. M., Rafii, M. Y., Ismail, M. R., Mahmood, M., Rahim, H. A., Alam, M. A., et al. (2015). Marker-assisted backcrossing: A useful method for rice improvement. Biotechnol. Biotechnol. Equipument 29, 237–254. doi: 10.1080/13102818.2014.995920
Hayashi, N., Inoue, H., Kato, T., Funao, T., Shirota, M., Shimizu, T., et al. (2010). Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 64, 498–510. doi: 10.1111/j.1365-313X.2010.04348.x
Hayashi, K., Yasuda, N., Fujita, Y., Koizumi, S., and Yoshida, H. (2010). Identification of the blast resistance gene Pit in rice cultivars using functional markers. Theor. Appl. Genet. 121, 1357–1367. doi: 10.1007/s00122-010-1393-7
Hayashi, K. and Yoshida, H. (2009). Re functionalization of the ancient rice blast disease resistance gene Pit by the recruitment of a retrotransposon as a promoter. Plant J. 57, 413 ± 425. doi: 10.1111/j.1365-313X.2008.03694.x
Hayashi, K., Yoshida, H., and Ashikawa, I. (2006). Development of PCR-based allele-specific and InDel marker sets for nine rice blast resistance genes. Theor. Appl. Genet. 113, 251–260. doi: 10.1007/s00122-006-0290-6
He, N., Huang, F., Yu, M., Zhu, Y., Li, Q. Q., and Yang, D. (2022). Analysis of a rice blast resistance gene Pita-Fuhui2663 and development of selection marker. Sci. Rep. 12, 14917. doi: 10.1038/s41598-022-19004-y
He, Z., Xin, Y., Wang, C., Yang, H., Xu, Z., and Cheng, J. (2022). Genomics-Assisted improvement of super high-yield hybrid rice variety “super 1000” for resistance to bacterial blight and blast diseases. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.881244
Heffner, E. L., Sorrells, M. E., and Jannink, J. L. (2009). Genomic selection for crop improvement. Crop Sci. 49, 1–12. doi: 10.2135/cropsci2008.08.0512
Hegde, S. S. and Prashanthi, S. K. (2016). Identification of polymorphic markers and introgression of Pi1 and Pi2 genes for blast resistance in rice. J. Farm Sci. 29, 327–331. doi: 10.5555/20173060456
Hickey, J. M., Chiurugwi, T., Mackay, I., and Powell, W. (2017). Genomic prediction unifies animal and plant breeding programs to form platforms for biological discovery. Nat. Genet. 49, 1297–1303. doi: 10.1038/ng.3920
Higashi, T., Sato, H., Horisue, N., and Fujimaki, H. (1981). Breeding of isogenic lines for blast resistance in rice. 1. Comparison of characters between B4F2 lines and their recurrent,”Nipponbare. Breed. Sci. 31, 46.
Horisue, N., Higashi, T., Sato, H., and Koizumi, S. J. B. S. (1984). Breeding of isogenic lines for blast resistance in rice. 2. Agronomical characteristics of kanto-IL1-14. Breed. Sci. 34, 316.
Hossain, M. A., Li, Z. G., Hoque, T. S., Burritt, D. J., Fujita, M., and Munné-Bosch, S. (2018). Heat or cold priming-induced cross-tolerance to abiotic stresses in plants: key regulators and possible mechanisms. Protoplasma 255, 399–412. doi: 10.1007/s00709-017-1150-8
Hou, J., Xiao, H., Yao, P., Ma, X., Shi, Q., Yang, J., et al. (2024). Unveiling the mechanism of broad-spectrum blast resistance in rice: The collaborative role of transcription factor OsGRAS30 and histone deacetylase OsHDAC1. Plant Biotechnol. J. 22, 1740–1756. doi: 10.1111/pbi.14299
Hua, L. X., Liang, L. Q., He, X. Y., Wang, L., Zhang, W. S., Liu, W., et al. (2015). Development of a marker specific for the rice blast resistance gene Pi39 in the Chinese cultivar Q15 and its use in genetic improvement. Biotechnol. Biotechnol. Equip. 29, 448–456. doi: 10.1080/13102818.2015.1011894
Hua, L. X., Wu, J. Z., Chen, C. X., Wu, W. H., He, X. Y., Lin, F., et al. (2012). The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor. Appl. Genet. 125, 1047–1055. doi: 10.1007/s00122-012-1894-7
Huang, M., Balimponya, E. G., Mgonja, E. M., McHale, L. K., Luzi-Kihupi, A., Wang, G. L., et al. (2019). Use of genomic selection in breeding rice (Oryza sativa L.) for resistance to rice blast (Magnaporthe oryzae). Mol. Breed. 39, 1–16. doi: 10.1007/s11032-019-1023-2
Huang, Q., Chen, C., Wu, X., Qin, Y., Tan, X., Zhang, D., et al. (2023). Overexpression of ATP synthase subunit beta (Atp2) confers enhanced blast disease resistance in transgenic rice. J. Fungi 10, 5. doi: 10.3390/jof10010005
Imam, J., Mandal, N. P., Variar, M., and Shukla, P. (2016). Allele mining and selective patterns of Pi9 gene in a set of rice landraces from India. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01846
Inukai, T., Nagashima, S., and Kato, M. (2019). Pid3-I1 is a race-specific partial-resistance allele at the Pid3 blast resistance locus in rice. Theor. Appl. Genet. 132, 395 ± 404. doi: 10.1007/s00122-018-3227-y
Ishizaki, K., Hoshi, T., Abe, S., Sasaki, Y., Kobayashi, K., Kasaneyama, H., et al. (2005). Breeding of blast resistant isogenic lines in rice variety “Koshihikara” and evaluation of their characters. Breed. Sci. 55, 371–377. doi: 10.1270/jsbbs.55.371
Jähne, F., Hahn, V., Würschum, T., and Leiser, W. L. (2020). Speed breeding short-day crops by LED-controlled light schemes. Theor. Appl. Genet. 133, 2335–2342. doi: 10.1007/s00122-020-03601-4
Jain, P., Sharma, V., Dubey, H., Singh, P. K., Kapoor, R., Kumari, M., et al. (2017). Identification of long non-coding RNA in rice lines resistant to rice blast pathogen Maganaporthe oryzae. Bioinformation 13, 249. doi: 10.6026/97320630013249
Jamaloddin, M., Durga Rani, C. V., Swathi, G., Anuradha, C., Vanisri, S., Rajan, C. P. D., et al. (2020). Marker Assisted Gene Pyramiding (MAGP) for bacterial blight and blast resistance into mega rice variety “Tellahamsa. PloS One 15, e0234088. doi: 10.1371/journal.pone.0234088
Jia, Y., Zhou, E., Lee, S., and Bianco, T. (2016). Coevolutionary dynamics of rice blast resistance gene Pi-ta and Magnaporthe oryzae avirulence gene AVR-Pita 1. Phytopathology 106, 676–683. doi: 10.1094/PHYTO-02-16-0057-RVW
Jiang, H., Li, Z., Liu, J., Shen, Z., Gao, G., Zhang, Q., et al. (2019). Development and evaluation of improved lines with broad-spectrum resistance to rice blast using nine resistance genes. Rice 12, 1–1. doi: 10.1186/s12284-019-0292-z
Jinlong, H., Yu, Z., Ruizhi, W., Xiaoyu, W., Zhiming, F., Qiangqiang, X., et al. (2024). A genome-wide association study of panicle blast resistance to Magnaporthe oryzae in rice. Mol. Breed. 44, 49. doi: 10.1007/s11032-024-01486-5
Jones, J. D. and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kang, S., Sweigard, J. A., and Valent, B. (1995). The PWL host specificity gene family in the blast fungus Magnaporthe grisea. MPMI-Molecular. Plant Microbe Interact. 8, 939–948. doi: 10.1016/j.rsci.2021.11.007
Karkute, S. G., Kumar, V., Tasleem, M., Mishra, D. C., Chaturvedi, K. K., Rai, A., et al. (2022). Genome-wide analysis of von willebrand factor A gene family in rice for its role in imparting biotic stress resistance with emphasis on rice blast disease. Rice Sci. 29, 375–384. doi: 10.1016/j.rsci.2021.11.007
Kashyap, P. L., Kumar, S., and Srivastava, A. K. (2017b). Nano diagnostics for plant pathogens. Environ. Chem. Letter 15, 7–13. doi: 10.1007/s10311-016-0580-4
Kaur, S., Padmanabhan, S. Y., and Rao, M. (1976). Research co-ordination meeting on induced mutations for disease resistance in crop plants. Ames, Iowa, USA.
Kaur, G. and Sivia, J. S. (2024). Development of deep and machine learning convolutional networks of variable spatial resolution for automatic detection of leaf blast disease of rice. Comput. Electron. Agric. 224, 109210. doi: 10.1016/j.compag.2024.109210
Kawasaki-Tanaka, A., Hayashi, N., Yanagihara, S., and Fukuta, Y. (2016). Diversity and distribution of rice blast (Pyricularia oryzae Cavara) races in Japan. Plant Dis. 100, 816–823. doi: 10.1094/PDIS-04-15-0442-RE
Khan, M. A., Ali, M. A., Monsur, M. A., Kawasaki-Tanaka, A., Hayashi, N., Yanagihara, S., et al. (2016). Diversity and distribution of rice blast (Pyricularia oryzae Cavara) races in Bangladesh. Plant Dis. 100, 2025–2033. doi: 10.1094/PDIS-12-15-1486-RE
Khan, G. H., Shikari, A. B., Vaishnavi, R., Najeeb, S., Padder, B. A., Bhat, Z. A., et al. (2018). Marker-assisted introgression of three dominant blast resistance genes into an aromatic rice cultivar Mushk Budji. Sci. Rep. 8, 4091. doi: 10.1038/s41598-018-22246-4
Khang, C. H., Park, S. Y., Lee, Y. H., Valent, B., and Kang, S. (2008). Genome organization and evolution of the AVR-Pita avirulence gene family in the Magnaporthe grisea species complex. Mol. Plant-Microbe Interact. 21, 658–670. doi: 10.1094/MPMI-21-5-0658
Kim, S., Kim, C. Y., Park, S. Y., Kim, K. T., Jeon, J., Chung, H., et al. (2020). Two nuclear effectors of the rice blast fungus modulate host immunity via transcriptional reprogramming. Nat. Commun. 11, 5845. doi: 10.1038/s41467-020-19624-w
Kiyosawa, S. (1984). Establishment of differential varieties for pathogenicity test of rice blast fungus. Rice Genet. Newslett. 1, 95–96.
Kousik, M., Punniakotti, E., Rekha, G., Chaitra, K., Kumar, T. D., Hajira, S. K., et al. (2024). Screening of high-yielding Swarna lines developed through MABB for resistance against bacterial blight and blast diseases. Int. J. Advanced Biochem. Res. 8, 566–570. doi: 10.33545/26174693.2024.v8.i2g.630
Krishnan, S. R., Muthuramalingam, P., Priya, A. M., Prasanth, M. I., Gopinath, K., Mohan, C., et al. (2022). Expressing OsiSAP8, a zinc-finger associated protein gene, mitigates stress dynamics in existing elite rice varieties of the ‘green revolution’. Sustainability 14, 10174. doi: 10.3390/su141610174
Kumar, R., Baloch, G., Buriro, A. B., and Bhatti, J. (2021). Fungal blast disease detection in rice seed using machine learning. Int. J. Advanced Comput. Sci. Appl. 12, 248–258. doi: 10.14569/IJACSA.2021.0120232
Kumar, S. V., Prasad, M. S., Rambabu, R., Bhaskar, B., Sundaram, R. M., Prakasam, V., et al. (2019). Marker assisted introgression of broad-spectrum blast resistance gene Pi2 into an elite rice cultivar, Samba Mahsuri. Indian J. Plant Prot. 47, 154–163.
Kumar, S. V., Rambabu, R., Bhaskar, B., Madhavi, K. R., Srikanth, S., Prakasam, V., et al. (2018). Introgression of durable blast resistance gene Pi54 into indica rice cv. samba mahsuri, through Marker Assisted Backcross Breeding. Electronic J. Plant Breed. 9, 705–715. doi: 10.5958/0975-928X.2018.00084.4
Kumari, A., Das, A., Devanna, B. N., Thakur, S., Singh, P. K., Singh, N. K., et al. (2013). Mining of rice blast resistance gene Pi54 shows effect of single nucleotide polymorphisms on phenotypic expression of the alleles. Eur. J. Plant Pathol. 137, 55–65. doi: 10.1007/s10658-013-0216-5
Kumari, M., Kapoor, R., Devanna, B. N., Varshney, S., Kamboj, R., Rai, A. K., et al. (2023). iTRAQ based proteomic analysis of rice lines having single or stacked blast resistance genes: Pi54/Pi54rh during incompatible interaction with Magnaporthe oryzae. Physiol. Mol. Biol. Plants 29, 871–887. doi: 10.1007/s12298-023-01327-3
Lee, K. T., Han, J., and Kim, K. H. (2022). Optimizing artificial neural network-based models to predict rice blast epidemics in Korea. Plant Pathol. J. 38, 395. doi: 10.5423/PPJ.NT.04.2022.0062
Lee, S. K., Song, M. Y., Seo, Y. S., Kim, H. K., Ko, S., Cao, P. J., et al. (2009). Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil-nucleotide-binding leucine-rich repeat genes. Genetics 181, 1627 ± 1638. doi: 10.1534/genetics.108.099226
Li, S., Gu, X., Wang, S., Wang, L., Lin, Y., Liang, X., et al. (2024). Rhamnolipid modified silica nanoparticles control rice blast disease by enhancing antifungal activity in vivo and antioxidant defense system of rice (Oryza sativa L.). ACS Appl. Materials Interfaces 17, 1792–1802. doi: 10.1021/acsami.4c11833
Li, X., Pan, L., Bi, D., Tian, X., Li, L., Xu, Z., et al. (2021). Generation of marker-free transgenic rice resistant to rice blast disease using Ac/Ds transposon-mediated transgene reintegration system. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.644437
Li, J., Wang, Q., Li, C., Bi, Y., Fu, X., and Wang, R. (2019). Novel haplotypes and networks of AVR-Pik alleles in Magnaporthe oryzae. BMC Plant Biol. 19, 1–12. doi: 10.1186/s12870-019-1817-8
Li, W., Wang, B., Wu, J., Lu, G., Hu, Y., Zhang, X., et al. (2009). The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol. Plant- Microbe Interact. 22, 411–420. doi: 10.1094/MPMI-22-4-0411
Li, A., Zhu, P., Kong, D., Wang, L., Zhang, A., Liu, Y., et al. (2024). Using marker-assisted selection to develop a drought-tolerant rice line with enhanced resistance to blast and brown planthopper. Agronomy 14, 2566. doi: 10.3390/agronomy14112566
Liang, T., Chi, W., Huang, L., Qu, M., Zhang, S., Chen, Z. Q., et al. (2020). Bulked segregant analysis coupled with whole-genome sequencing (BSA-Seq) mapping identifies a novel pi21 haplotype conferring basal resistance to rice blast disease. Int. J. Mol. Sci. 21, 2162. doi: 10.3390/ijms21062162
Liang, M., Zhang, H., Chen, J., Dai, D., Du, C., Wang, H., et al. (2022). Developing fragrant early Indica TGMS line with blast resistance by using CRISPR/Cas9 technology. Chin. J. Rice Sci. 36, 248–258. doi: 10.16819/j.1001-7216.2022.211007
Lin, F., Chen, S., Que, Z., Wang, L., Liu, X., and Pan, Q. (2007). The blast resistance gene Pi37 encodes a nucleotide binding site–leucine-rich repeat protein and is a member of a resistance gene cluster on rice chromosome 1. Genetics 177, 1871–1880. doi: 10.1534/genetics.107.080648
Ling, Z. Z. (2001). Development of Chinese near isogenic lines of rice and their differentiating ability of pathogenic races of blast fungus. Chin. Agric. Sci. 2001, 50–56.
Liu, M. H., Kang, H., Xu, Y., Peng, Y., Wang, D., Gao, L., et al. (2020). Genome-wide association study identifies an NLR gene that confers partial resistance to Magnaporthe oryzae in rice. Plant Biotechnol. J. 18, 1376–1383. doi: 10.1111/pbi.13300
Liu, X., Lin, F., Wang, L., and Pan, Q. (2007). The in-silico map-based cloning of Pi36, a rice coiled-coil–nucleotide-binding site–leucine-rich repeat gene that confers race- specific resistance to the blast fungus. Genetics 176, 2541–2549. doi: 10.1534/genetics.107.075465
Liu, W., Liu, J., Triplett, L., Leach, J. E., and Wang, G. L. (2014). Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 52, 213–241. doi: 10.1146/annurev-phyto-102313-045926
Liu, Y., Sun, T., Li, Y., Huang, J., Wang, X., Bai, H., et al. (2024). Proteomic analysis revealed the function of PoElp3 in development, pathogenicity, and autophagy through the tRNA-mediated translation efficiency in the rice blast fungus1. J. Integr. Agric. 24 (4), 1–29. doi: 10.1016/j.jia.2024.01.027
Liu, X., Yu, Y., Yao, W., Yin, Z., Wang, Y., Huang, Z., et al. (2023). CRISPR/Cas9-mediated simultaneous mutation of three salicylic acid 5-hydroxylase (OsS5H) genes confers broad-spectrum disease resistance in rice. Plant Biotechnol. J. 21, 1873–1886. doi: 10.1111/pbi.14099
Liu, Y., Zhang, F., and Luo, X. (2021). Molecular Breeding of a novel PTGMS line of WDR for broad-spectrum resistance to blast using Pi9, Pi5, and Pi54 genes. Rice 14, 96. doi: 10.1186/s12284-021-00537-1
Liu, G., Zhu, P., Liu, Y., Kong, D., Wang, J., Luo, L., et al. (2024). Molecular marker-assisted selection of a new water-saving and drought-resistant rice (WDR) restoration line, hanhui 8200, for enhanced resistance to rice blast. Agronomy 14, 1504. doi: 10.3390/agronomy14071504
Lu, Q., Wang, C., Niu, X., Zhang, M., Xu, Q., Feng, Y., et al. (2019). Detecting novel loci underlying rice blast resistance by integrating a genome-wide association study and RNA sequencing. Mol. Breed. 39, 1–10. doi: 10.1007/s11032-019-0989-0
Lv, Q., Xu, X., Shang, J., Jiang, G., Pang, Z., Zhou, Z., et al. (2013). Functional analysis of Pid3-A4, an ortholog of rice blast resistance gene Pid3 revealed by allele mining in common wild rice. Phytopathology 103, 594–599. doi: 10.1094/PHYTO-10-12-0260-R
Ma, J., Jia, M. H., and Jia, Y. (2014). Characterization of rice blast resistance gene Pi61 (t) in rice germplasm. Plant Disease 98 (9), 1200–1204. doi: 10.1094/PDIS-09-13-1014-RE
Ma, J., Chen, J., Wang, M., Ren, Y., Wang, S., Lei, C., et al. (2018). Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. J. Exp. Bot. 69, 1051–1064. doi: 10.1093/jxb/erx458
Ma, J., Lei, C., Xu, X., Hao, K., Wang, J., Cheng, Z., et al. (2015). Pi64, encoding a novel CC-NBS-LRR protein, confers resistance to leaf and neck blast in rice. Mol. Plant-Microbe Interact. 28, 558–568. doi: 10.1094/MPMI-11-14-0367-R
Mackill, D. J. and Bonman, J. M. (1992). Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 82, 746–749. doi: 10.1094/Phyto-82-746
Maheswaran, S., Sathesh, S., Rithika, P., Shafiq, I. M., Nandita, S., and Gomathi, R. D. (2022). “Detection and classification of paddy leaf diseases using deep learning (cnn),” in International Conference on Computer, Communication, and Signal Processing, Cham. 60–74 (Chennai, India: Springer International Publishing). doi: 10.1007/978-3-031-11633-9_6
Martin, G. B., Bogdanove, A. J., and Sessa, G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54, 23–61. doi: 10.1146/annurev.arplant.54.031902.135035
Mathsyaraja, S., Lavudi, S., and Vutukuri, P. R. (2024). Enhancing resistance to blast disease through CRISPR/Cas9 gene editing technology in OsHDT701 gene in RPBio-226 rice cv. (Oryza sativa L.). J. Appl. Biol. Biotechnol. 12, 289–294. doi: 10.7324/JABB.2024.154893
Matsunaga, K. (1996). Breeding of a multiline rice cultivar “Sasanishiki BL” and its use for control of blast disease in Miyagi prefecture. J. Agric. Sci. 51, 173–176.
Maurya, R., Singh, M., and Srivastava, D. (2024). A Review of conventional and molecular approaches for the management of rice blast disease. Fungal Dis. Rice Their Manage., 91–102.
Meher, J., Sarkar, A., and Sarma, B. K. (2023). Binding of stress-responsive OsWRKY proteins through WRKYGQK heptapeptide residue with the promoter region of two rice blast disease resistance genes Pi2 and Pi54 is important for development of blast resistance. 3 Biotechnol. 13, 294. doi: 10.1007/s13205-023-03711-y
Mentlak, T. A., Kombrink, A., Shinya, T., Ryder, L. S., Otomo, I., Saitoh, H., et al. (2012). Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 24, 322–335. doi: 10.1105/tpc.111.092957
Meuwissen, T. H., Hayes, B. J., and Goddard, M. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829. doi: 10.1093/genetics/157.4.1819
Miah, G., Rafii, M. Y., Ismail, M. R., Sahebi, M., Hashemi, F. S. G., Yusuff, O., et al. (2017). Blast disease intimidation towards rice cultivation: a review of pathogen and strategies to control. J. Anim. Plant Sci. 27, 1058–1066.
Mora, F., Quitral, Y. A., Matus, I., Russell, J., Waugh, R., and Del Pozo, A. (2016). SNP-based QTL mapping of 15 complex traits in barley under rain-fed and well-watered conditions by a mixed modeling approach. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00909
Mutiga, S. K., Rotich, F., Were, V. M., Kimani, J. M., Mwongera, D. T., Mgonja, E., et al. (2021). Integrated strategies for durable rice blast resistance in sub-Saharan Africa. Plant Dis. 105, 2749–2770. doi: 10.1094/PDIS-03-21-0593-FE
Nadim, M. K. A., Islam, M. M., Hoque, M. I., Hasan, M. J., and Uddin, M. I. (2024). Development of blast-resistant rice varieties through marker-assisted selection: Development of blast-resistant rice varieties. Bangladesh J. Agric. 49, 41–51. doi: 10.3329/bjagri.v49i1.74041
Nakajima, T. (1994). Mechanism of rice blast disease control by multilines. Jornal Agric. Sci. 49, 390–395.
Nakajima, T., Sonoda, R., Yaegashi, H., and Saito, H. (1996). Factors related to suppression of leaf blast disease with a multiline of rice cultivar Sasanishiki and its isogenic lines. Japanese J. Phytopathol. 62, 360–364. doi: 10.3186/jjphytopath.62.360
Nawaz, G., Usman, B., Peng, H., Zhao, N., Yuan, R., Liu, Y., et al. (2020). Knockout of Pi21 by CRISPR/Cas9 and iTRAQ-based proteomic analysis of mutants revealed new insights into M. oryzae resistance in elite rice line. Genes 11, 735. doi: 10.3390/genes11070735
Nguyet, N. T., Long, H. H., Ngoc, N. B., Nhai, N. T., Thuy, N. T., Hayashi, N., et al. (2020). Diversity and distribution of rice blast (Pyricularia oryzae Cavara) races in Vietnam. Plant Dis. 104, 381–387. doi: 10.1094/PDIS-05-19-1008-RE
Nihad, S. A. I., Hasan, M. A. I., Anik, T. R., Rashid, M. M., Khan, M. A. I., Islam, M. R., et al. (2024). Pyramiding of blast and bacterial blight resistance genes in premium quality rice variety, BRRI dhan63 through marker-assisted breeding approach. Euphytica 220, 13. doi: 10.1007/s10681-023-03255-5
Ninomiya, A., Urayama, S. I., Suo, R., Itoi, S., Fuji, S. I., Moriyam, H., et al. (2020). Mycovirus-induced tenuazonic acid production in a rice blast fungus Magnaporthe oryzae. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01641
Nizolli, V. O., Pegoraro, C., and Oliveira, A. C. D. (2021). Rice blast: Strategies and challenges for improving genetic resistance. Crop Breed. Appl. Biotechnol. 21. doi: 10.1590/1984-70332021v21Sa22
Nurnberger, T., Brunner, F., Kemmerling, B., and Piater, L. (2004). Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 198, 249 ± 266. doi: 10.1111/j.0105-2896.2004.0119.x
Odjo, T., Kawasaki-Tanaka, A., Noda, T., Ahohuendo, B. C., Sere, Y., Kumashiro, T., et al. (2014). Pathogenicity analysis of blast (Pyricularia oryzae Cavara) isolates from West Africa. Japan Agric. Res. Q. 48, 403–412. doi: 10.6090/jarq.48.403
Okada, R., Nemoto, Y., Endo-Higashi, N., and Izawa, T. (2017). Synthetic control of flowering in rice independent of the cultivation environment. Nat. Plants 3, 1–7. doi: 10.1038/nplants.2017.39
Okuyama, Y., Kanzaki, H., Abe, A., Yoshida, K., Tamiru, M., Saitoh, H., et al. (2011). A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 66, 467 ± 479. doi: 10.1111/j.1365-313X.2011.04502.x
Orbach, M. J., Farrall, L., Sweigard, J. A., Chumley, F. G., and Valent, B. (2000). A telomeric avirulence gene determines efficacy for the rice blast resistance gene. Pi-ta Plant Cell 12, 2019–2032. doi: 10.1105/tpc.12.11.2019
Padmanabhan, S. Y., Chakrabarti, N. K., Mathur, S. C., and Veeraraghavan, J. (1970). Identification of pathogenic races of Pyricularia oryzae. India Phytopathol. 60, 1574–1577. doi: 10.1094/Phyto-60-1574
Palanna, K. B., Vinaykumar, H. D., Koti, P. S., Jeevan, B., Rajashekara, H., Raveendra, H. R., et al. (2024). Geographic distribution, host preference and phylogenetic relationships among Pyricularia species inciting millet and rice blast disease in India. Plant Pathol. 73, 692–705. doi: 10.1111/ppa.13830
Parthasarathy, R., Jayabaskaran, C., Manikandan, A., and Anusuya, S. (2023). Synthesis of nickel-chitosan nanoparticles for controlling blast diseases in Asian rice. Appl. Biochem. Biotechnol. 195, 2134–2148. doi: 10.1007/s12010-022-04198-8
Patel, A., Sahu, K. P., Mehta, S., Javed, M., Balamurugan, A., Ashajyothi, M., et al. (2023). New insights on endophytic microbacterium-assisted blast disease suppression and growth promotion in rice: revelation by polyphasic functional characterization and transcriptomics. Microorganisms 11, 362. doi: 10.3390/microorganisms11020362
Patroti, P., Vishalakshi, B., Umakanth, B., Suresh, J., Senguttuvel, P., and Madhav, M. S. (2019). Marker-assisted pyramiding of major blast resistance genes in Swarna-Sub1, an elite rice variety (Oryza sativa L.). Euphytica 215, 1–11. doi: 10.1007/s10681-019-2487-1
Pedrozo, R., Osakina, A., Huang, Y., Nicolli, C. P., Wang, L., and Jia, Y. (2025). Status on genetic resistance to rice blast disease in the post-genomic era. Plants 14, 807. doi: 10.3390/plants14050807
Peng, M., Lin, X., Xiang, X., Ren, H., Fan, X., and Chen, K. (2021). Characterization and Evaluation of transgenic rice pyramided with the Pi Genes Pib, Pi25 and Pi54. Rice 14, 78. doi: 10.1186/s12284-021-00512-w
Peng, W., Wang, Y., Zeng, X., Li, W., Song, N., Liu, J., et al. (2023). Integrative transcriptomic, proteomic, and phosphoproteomic analysis on the defense response to Magnaporthe oryzae reveals different expression patterns at the molecular level of durably resistant rice cultivar Mowanggu. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1212510
Petit-Houdenot, Y. and Fudal, I. (2017). Complex interactions between fungal avirulence genes and their corresponding plant resistance genes and consequences for disease resistance management. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01072
Pote, T. D., Kaachra, A., Thakur, K., Salgotra, R. K., Krishnan, S. G., and Rathour, R. (2022). Genetic improvement of traditional Basmati rice Ranbir Basmati for semi-dwarfism and blast resistance through molecular breeding. Plant Gene 32, 100386. doi: 10.1016/j.plgene.2022.100386
Pushpanjie, K., Hong, L. W., Saad, N. S., Jin, H., and Wong, M. Y. (2024). Spray-Induced Gene Silencing against Rice Blast Disease Targeting Dicer-like Protein 2 (DCL2) of Pyricularia oryzae. Res. Square. doi: 10.21203/rs.3.rs-4757955/v1
Qi, Z., Du, Y., Yu, J., Zhang, R., Yu, M., Cao, H., et al. (2023). Molecular detection and analysis of blast resistance genes in rice main varieties in Jiangsu province, China. Agronomy 13, 157. doi: 10.3390/agronomy13010157
Qian, H., Sun, L., Wu, M., Zhao, W., Liu, M., Liang, S., et al. (2023). The COPII subunit MoSec24B is involved in development, pathogenicity and autophagy in the rice blast fungus. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1074107
Qiu, J., Chen, Y., Liu, Z., Wen, H., Jiang, N., Shi, H., et al. (2023). The application of zinc oxide nanoparticles: An effective strategy to protect rice from rice blast and abiotic stresses. Environmental Pollution 331, 121925. doi: 10.1016/j.envpol.2023.121925
Qu, S. H., Liu, G. F., Zhou, B., Bellizzi, M., Zeng, L. R., Dai, L. Y., et al. (2006). The broad-spectrum blast resistance gene Pi9encodes a nucleotide-binding site–leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172, 1901–1914. doi: 10.1534/genetics.105.044891
Rajpoot, V., Tiwari, A., and Jalal, A. S. (2023). Automatic early detection of rice leaf diseases using hybrid deep learning and machine learning methods. Multimedia Tools Appl. 82, 36091–36117. doi: 10.1007/s11042-023-14969-y
Ramalingam, J., Raveendra, C., Savitha, P., Vidya, V., Chaithra, T. L., Velprabakaran, S., et al. (2020). Gene pyramiding for achieving enhanced resistance to bacterial blight, blast, and sheath blight diseases in rice. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.591457
Ramesh, S. and Vydeki, D. (2018). “Rice blast disease detection and classification using machine learning algorithm,” in 2018 2nd International Conference on Micro-Electronics and Telecommunication Engineering (ICMETE). 255–259 (Ghaziabad, India: IEEE). doi: 10.1109/ICMETE.2018.00063
Ramkumar, G., Biswal, A. K., Mohan, K. M., Sakthivel, K., Sivaranjani, A., Neeraja, C. N., et al. (2010). Identifying novel alleles of rice blast resistance genes Pikh and Pita through allele mining. Int. Rice Res. Notes 117, 4185.
Ramkumar, G., Prahalada, G. D., Hechanova, S. L., Vinarao, R., and Jena, K. K. (2015). Development and validation of SNP-based functional codominant markers for two major disease resistance genes in rice (O. sativa L.). Mol. Breed. 35, 1–11. doi: 10.1007/s11032-015-0323-4
Rathour, R., Kumar, R., Thakur, K., and Pote, T. D. (2022). Genetic improvement for blast resistance in high-yielding cold-tolerant rice (Oryza sativa L.) cultivar Himalaya 741 by marker-assisted backcross breeding. 3 Biotechnol. 12, 165. doi: 10.1007/s13205-022-03244-w
Ray, S., Singh, P. K., Gupta, D. K., Mahato, A. K., Sarkar, C., Rathour, R., et al. (2016). Analysis of Magnaporthe oryzae genome reveals a fungal effector, which is able to induce resistance response in transgenic rice line containing resistance gene, Pi54. Front. Plant Sci. 8. doi: 10.3389/fpls.2016.01140
Razzaq, A., Saleem, F., Kanwal, M., Mustafa, G., Yousaf, S., Imran Arshad, H. M., et al. (2019). Modern trends in plant genome editing: an inclusive review of the CRISPR/Cas9 toolbox. Int. J. Mol. Sci. 20, 4045. doi: 10.3390/ijms20164045
Reinke, R., Kim, S. M., and Kim, B. K. (2018). Developing japonica rice introgression lines with multiple resistance genes for brown planthopper, bacterial blight, rice blast, and rice stripe virus using molecular breeding. Mol. Genet. Genomics 293, 1565–1575. doi: 10.1007/s00438-018-1470-1
Ribot, C., Césari, S., Abidi, I., Chalvon, V., Bournaud, C., Vallet, J., et al. (2013). The Magnaporthe oryzae effector AVR 1–CO 39 is translocated into rice cells independently of a fungal-derived machinery. Plant J. 74, 1–12. doi: 10.1111/tpj.12099
Rijal, S. and Devkota, Y. (2020). A review on various management method of rice blast disease. Malaysian J. Sustain. Agric. 4, 14–18. doi: 10.26480/mjsa.01.2020.29.33
Sabat, M. and Tripathy, A. (2024). Genetically modified and gene-edited food crops: recent status and future prospects. Food Production Diversity Saf. Under Climate Change 34, 211–222. doi: 10.1007/978-3-031-51647-4_18
Sagar, V., Dhawan, G., Gopala Krishnan, S., Vinod, K. K., Ellur, R. K., Mondal, K. K., et al. (2020). Marker assisted introgression of genes governing resistance to bacterial blight and blast diseases into an elite Basmati rice variety, ‘Pusa Basmati 1509’. Euphytica 216, 1–18. doi: 10.1007/s10681-019-2549-4
Sagar Krishna Murthy, P., Deshmukh, D. B., Yashvanth Kumar, K. J., Patil, S., Jakkeral, S., Nemappa, G. H., et al. (2017). Introgression of Pi2 and Pi5 genes for blast (Magnaporthe oryzae) resistance in rice and field evaluation of introgression lines for resistance and yield traits. J. Phytopathol. 165, 397–405. doi: 10.1111/jph.12573
Sahu, P. K., Sao, R., Choudhary, D. K., Thada, A., Kumar, V., Mondal, S., et al. (2022). Advancement in the breeding, biotechnological and genomic tools towards development of durable genetic resistance against the rice blast disease. Plants 11, 2386. doi: 10.3390/plants11182386
Saito, H., Tomita, A., Yoshida, T., Nakamura, M., Suzuki, T., Ikeda, A., et al. (2022). Characterization of six partial resistance genes and one quantitative trait locus to blast disease using near isogenic lines with a susceptible genetic background of indica group rice (Oryza sativa). PhytoFrontiers™ 2, 230–241. doi: 10.1094/PHYTOFR-06-21-0042-R
Saklani, B. K., Ray, S., Arora, K., Asthana, R. K., and Sharma, T. R. (2023). Magnaporthe oryzae encoded effector protein AvrPi54 interacts in vivo with rice encoded cognate resistance protein Pi54 at the host plasma membrane. J. Plant Biochem. Biotechnol. 32, 274–283. doi: 10.1007/s13562-022-00803-3
Samuthirapandi, S., Thiyagarajan, T., Viswabharathy, S., Bharathi, A., Rohit, K., Ranjani, R. V., et al. (2023). Improving rice blast resistance by stacking of two broad spectrum resistance gene Pi9and Pi54 in cultivar CO 51 through marker assisted selection. Electronic J. Plant Breed. 14, 695–706. doi: 10.37992/2023.1402.081
Sandhu, R., Chaudhary, N., Shams, R., and Dash, K. K. (2024). Genetically modified crops and sustainable development: navigating challenges and opportunities. Food Sci. Biotechnol. 34, 1–17. doi: 10.1007/s10068-024-01669-y
Sarkar, A. and Roy-Barman, S. (2021). Spray-induced silencing of pathogenicity gene MoDES1 via exogenous double-stranded RNA can confer partial resistance against fungal blast in rice. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.733129
Savary, S., Willocquet, L., Elazegui, F. A., Castilla, N. P., and Teng, P. S. (2000). Rice pest constraints in tropical Asia: quantification of yield losses due to rice pests in a range of production situations. Plant Dis. 84, 357–369. doi: 10.1094/pdis.2000.84.3.357
Shafi, A., Khan, R. S., Mir, S., Khan, G. H., Masoodi, K. Z., Sofi, N. R., et al. (2023). Gene expression of near-isogenic lines (NILs) carrying blast resistance genes Pi9and Pi54 in the background of rice cultivar Mushk Budji. Mol. Biol. Rep. 50, 1–15. doi: 10.1007/s11033-023-08475-5
Shahid, M., Goetze, P. K., Shafqat, U., and Jo, Y. K. (2025). Biogenic silver nanoparticles to control rice blast caused by Magnaporthe oryzae. Physiol. Mol. Plant Pathol. 138, 102636. doi: 10.1016/j.pmpp.2025.102636
Shahriar, S. A., Imtiaz, A. A., Hossain, M. B., Husna, A., Eaty, M., and Khatun, N. (2020). Rice blast disease. Annu. Res. Rev. Biol. 35, 50–64. doi: 10.9734/arrb/2020/v35i130180
Shang, J. J., Tao, Y., Chen, X. W., Zou, Y., Lei, C. L., Wang, J., et al. (2009). Identification of a new rice blast resistance gene, Pid3, by genome wide comparison of paired nucleotide-binding site- leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes. Genetics 182, 1303 ± 1311. doi: 10.1534/genetics.109.102871
Sharma, S., Kumar, A., Dhakte, P., Raturi, G., Vishwakarma, G., Barbadikar, K. M., et al. (2022). Speed breeding opportunities and challenges for crop improvement. J. Plant Growth Regul. 42, 46–59. doi: 10.1007/s00344-021-10551-8
Sharma, T. R., Rai, A. K., Gupta, S. K., and Singh, N. K. (2010). Broad spectrum blast resistance gene Pi-kh cloned from rice line Tetep designated as Pi54. J. Plant Biochem. Biotechnol. 19, 87 ± 89. doi: 10.1007/BF03323441
Sharma, T. R., Rai, A. K., Gupta, S. K., Vijayan, J., Devanna, B. N., and Ray, S. (2012). Rice blast management through host-plant resistance: retrospect and prospects. Agric. Res. 1, 37–52. doi: 10.1007/s40003-011-0003-5
Shen, E., Wang, X., Lu, Z., Zhou, F., Ma, W., Cui, Z., et al. (2023). Overexpression of a beta-1, 6-glucanase gene GluM in transgenic rice confers high resistance to rice blast, sheath blight and false smut. Pest Manage. Sci. 79, 2152–2162. doi: 10.1002/ps.7394
Sheoran, N., Ganesan, P., Mughal, N. M., Yadav, I. S., and Kumar, A. (2021). Genome assisted molecular typing and pathotyping of rice blast pathogen, Magnaporthe oryzae, reveals a genetically homogenous population with high virulence diversity. Fungal Biol. 125, 733–747. doi: 10.1016/j.funbio.2021.04.007
Shi, Q., Wang, R., Lu, W., Zhu, J., Zhang, H., Xiong, Q., et al. (2024). Metabolomics analysis of variation in grain quality of high-quality Japonica rice. Agronomy 14, 430. doi: 10.3390/agronomy14030430
Shukla, M., Al-Busaidi, K. T., Trivedi, M., and Tiwari, R. K. (2018). Status of research, regulations and challenges for genetically modified crops in India. GM Crops Food 9, 173–188. doi: 10.1080/21645698.2018.1529518
Simkhada, K. and Thapa, R. (2022). Rice blast, a major threat to the rice production and its various management techniques. Turkish J. Agriculture-Food Sci. Technol. 10, 147–157. doi: 10.24925/turjaf.v10i2.147-157.4548
Singh, A., Ganapathysubramanian, B., Singh, A. K., and Sarkar, S. (2016). Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 21, 110–124. doi: 10.1016/j.tplants.2015.10.015
Singh, R., Kaur, N., Praba, U. P., Kaur, G., Tanin, M. J., Kumar, P., et al. (2022). A prospective review on selectable marker-free genome engineered rice: past, present and future scientific realm. Front. Genet. 13. doi: 10.3389/fgene.2022.882836
Singh, A. K., Ponnuswamy, R., Srinivas Prasad, M., Sundaram, R. M., Hari Prasad, A. S., Senguttuvel, P., et al. (2023). Improving blast resistance of maintainer line DRR 9B by transferring broad spectrum resistance gene Pi2 by marker assisted selection in rice. Physiology and Molecular Biology of Plants 29 (2), 253–262. doi: 10.1007/s12298-023-01291-y
Sone, T., Takeuchi, S., Miki, S., Satoh, Y., Ohtsuka, K., Abe, A., et al. (2013). Homologous recombination causes the spontaneous deletion of AVR-Pia in Magnaporthe oryzae. FEMS Microbiol. Lett. 339, 102–109. doi: 10.1111/1574-6968.12058
Song, J., Kanokmedhakul, S., Kanokmedhkul, K., Poeaim, S., and Soytong, K. (2018). Nano-particles constructed from Chaetomium cochliodes CTh05 against Magnaporthe oryzae causing rice blast. bioRxiv, 339283. doi: 10.1101/339283
Soujanya, T., Madamsetty, S. P., Hemalatha, V., Yamini, K. N., Bandela, E., Hari Prasad, A. S., et al. (2023). Improving hybrid rice parental lines for blast resistance by introgression of broad-spectrum resistance genes Pi54 and Pi9(t) by marker-assisted selection. Plant Breed. 142, 300–311. doi: 10.1111/pbr.13093
Sowmiya, C. A., Ramalingam, J., Pushpam, R., Shoba, D., Kumar, K. K., and Arumugam Pillai, M. (2024). Introgression of blast and bacterial blight disease resistance genes in a rice genotype ADT43 through marker assisted back cross breeding. Physiol. Mol. Biol. Plants 30, 1–17. doi: 10.1007/s12298-024-01461-6
Srivastava, D., Shamim, M. D., Kumar, M., Mishra, A., Pandey, P., Kumar, D., et al. (2017). Current status of conventional and molecular interventions for blast resistance in rice. Rice Sci. 24, 299–321. doi: 10.1016/j.rsci.2017.08.001
Su, J., Wang, W. J., Han, J. L., Chen, S., Wang, C. Y., Zeng, L. X., et al. (2015). Functional divergence of duplicated genes results in a novel blast resistance gene Pi50 at the Pi2/9 locus. Theor. Appl. Genet. 128, 2213 ± 2225. doi: 10.1007/s00122-015-2579-9
Swathi, G., Durga Rani, C. V., Md, J., Madhav, M. S., Vanisree, S., Anuradha, C., et al. (2019). Marker-assisted introgression of the major bacterial blight resistance genes, Xa21 and xa13, and blast resistance gene, Pi54, into the popular rice variety, JGL1798. Mol. Breed. 39, 1–12. doi: 10.1007/s11032-019-0950-2
Sweigard, J. A., Carroll, A. M., Kang, S., Farrall, L., Chumley, F. G., and Valent, B. (1995). Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell 7, 1221–1233. doi: 10.1105/tpc.7.8.1221
Tabassum, J., Ahmad, S., Hussain, B., Mawia, A. M., Zeb, A., and Ju, L. (2021). Applications and potential of genome-editing systems in rice improvement:current and future perspectives. Agronomy 11, 1359. doi: 10.3390/agronomy11071359
Takagi, H., Uemura, A., Yaegashi, H., Tamiru, M., Abe, A., Mitsuoka, C., et al. (2013). MutMapGap: Whole-genome resequencing of mutant F2 progeny bulk combined with de novo assembly of gap regions identifies the rice blast resistance gene Pii. New Phytol. 200, 276 ± 283. doi: 10.1111/nph.12369
Takahashi, A., Hayashi, N., Miyao, A., and Hirochika, H. (2010). Unique features of the rice blast resistance Pish locus revealed by large scale retrotransposon-tagging. BMC plantbiol. 10, 1–14. doi: 10.1186/1471-2229-10-175
Takatsuka, A., Kazama, T., Arimura, S. I., and Toriyama, K. (2022). TALEN-mediated depletion of the mitochondrial gene f312 proves that it is a Tadukan-type cytoplasmic male sterility-causative gene in rice. Plant J. 110, 994–1004. doi: 10.1111/tpj.15715
Talbot, N. J. (2003). On the trail of a cereal killer: exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 57, 177–202. doi: 10.1146/annurev.micro.57.030502.090957
Tao, H., Shi, X., He, F., Wang, D., Xiao, N., Fang, H., et al. (2021). Engineering Broad-spectrum disease-resistant rice by editing multiple susceptibility genes. J. Integr. Plant Biol. 63, 1639–1648. doi: 10.1111/jipb.13145
Telebanco-Yanoria, M. J., Koide, Y., Fukuta, Y., Imbe, T., Kato, H., Tsunematsu, H., et al. (2010). Development of near-isogenic lines of Japonica-type rice variety Lijiangxintuanheigu as differentials for blast resistance. Breed. Sci. 60, 629–638. doi: 10.1270/jsbbs.60.629
Telebanco-Yanoria, M. J., Koide, Y., Fukuta, Y., Imbe, T., Tsunematsu, H., Kato, H., et al. (2011). A set of near-isogenic lines of Indica-type rice variety CO 39 as differential varieties for blast resistance. Mol. Breed. 27, 357–373. doi: 10.1007/s11032-010-9437-x
Thapa, R., Tabien, R. E., Thomson, M. J., and Septiningsih, E. M. (2024). Genetic factors underlying anaerobic germination in rice: Genome-wide association study and transcriptomic analysis. Plant Genome 17, e20261. doi: 10.1002/tpg2.20261
Thulasinathan, T., Ayyenar, B., Kambale, R., Manickam, S., Chellappan, G., Shanmugavel, P., et al. (2023). Marker assisted introgression of resistance genes and phenotypic evaluation enabled identification of durable and broad-spectrum blast resistance in elite rice cultivar, CO51. Genes 14, 719. doi: 10.3390/genes14030719
Tian, D., Yang, L., Chen, Z., Chen, Z., Wang, F., Zhou, Y., et al. (2018). Proteomic analysis of the defense response to Magnaporthe oryzae in rice harboring the blast resistance gene Piz-t. Rice 11, 1–13. doi: 10.1186/s12284-018-0240-3
Tosa, Y., Osue, J., Eto, Y., Oh, H. S., Nakayashiki, H., Mayama, S., et al. (2005). Evolution of an avirulence gene, AVR1-CO39, concomitant with the evolution and differentiation of Magnaporthe oryzae. Mol. Plant-Microbe Interact. 18, 1148–1160. doi: 10.1094/MPMI-18-1148
Tsunematsu, H., Yanoria, M. J. T., Ebron, L. A., Hayashi, N., Ando, I., Kato, H., et al. (2000). Development of monogenic lines of rice for blast resistance. Breed. Sci. 50, 229–234. doi: 10.1270/jsbbs.50.229
Usatov, A. V., Kostylev, P. V., Azarin, K. V., Markin, K. V., Markareno, M. S., Khachumov, V. A., et al. (2016). Introgression of rice blast resistance gene Pi1, Pi2 and Pi33 into Russian rice varieties by marker assisted selection. Indian J. Genet. Plant Breed. 76, 18–23. doi: 10.5958/0975-6906.2016.00003.1
Varsha, M., Basavaraju, P., and MP, P. K. (2024). “Advanced Machine Learning Techniques for Disease Management in Paddy Crop for Sustainable Rice Production: Prediction of Rice Blast Disease Severity,” in Natural Resource Management Issues in Human-Influenced Landscapes (IGI Global), 64–87. doi: 10.4018/978-1-6684-7051-0.ch004
Viana, V. E., Pegoraro, C., Busanello, C., and Costa de Oliveira, A. (2019). Mutagenesis in rice: the basis for breeding a new super plant. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01326
Volante, A., Tondelli, A., Desiderio, F., Abbruscato, P., Menin, B., Biselli, C., et al. (2020). Genome wide association studies for japonica rice resistance to blast in field and controlled conditions. Rice 13, 1–17. doi: 10.1186/s12284-020-00431-2
Wang, M. and Dean, R. A. (2022). Host induced gene silencing of Magnaporthe oryzae by targeting pathogenicity and development genes to control rice blast disease. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.959641
Wang, D., Guo, C., Huang, J., Yang, S., Tian, D., and Zhang, X. (2014). Allele-mining of rice blast resistance genes at AC134922 locus. Biochem. Biophys. Res. Commun. 446, 1085–1090. doi: 10.1016/j.bbrc.2014.03.056
Wang, Z., Han, Q., Zi, Q., Lv, S., Qiu, D., and Zeng, H. (2017). Enhanced disease resistance and drought tolerance in transgenic rice plants overexpressing protein elicitors from Magnaporthe oryzae. PloS One 12, e0175734. doi: 10.1371/journal.pone.0175734
Wang, J., Hu, H., Jiang, X., Zhang, S., Yang, W., Dong, J., et al. (2024). Pangenome-wide association study and transcriptome analysis reveal a novel QTL and candidate genes controlling both panicle and leaf blast resistance in rice. Rice 17, 27. doi: 10.1186/s12284-024-00707-x
Wang, X., Jia, M. H., Ghai, P., Lee, F. N., and Jia, Y. (2015). Genome-wide association of rice blast disease resistance and yield-related components of rice. Mol. Plant-Microbe Interact. 28, 1383–1392. doi: 10.1094/MPMI-06-15-0131-R
Wang, L., Li, J., Zhang, H., Wang, J., Wang, D., and Guo, Z. (2022a). Phosphoproteomic analysis reveals protein phosphorylation changes associated with rice blast resistance. Phytopathol. Res. 4, 1–12. doi: 10.1186/s42483-021-00106-w
Wang, L., Ma, Z., Kang, H., Gu, S., Mukhina, Z., Wang, C., et al. (2022b). Cloning and functional analysis of the novel rice blast resistance gene Pi65 in Japonica rice. Theor. Appl. Genet. 135, 173–183. doi: 10.1007/s00122-021-03957-1
Wang, G. L., Mackill, D. J., Bonman, J. M., McCouch, S. R., Champoux, M. C., and Nelson, R. J. (1994). RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar. Genetics 136, 1421–1434. doi: 10.1093/genetics/136.4.1421
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., et al. (2016). Enhanced rice blast resistance by CRISPR/Cas9-Targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PloS One 11, e0154027. doi: 10.1371/journal.pone.0154027
Wang, Z. X., Yano, M., Yamanouchi, U., Iwamoto, M., Monna, L., Hayasaka, H., et al. (1999). The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J. 19, 55–64. doi: 10.1046/j.1365-313X.1999.00498.x
Wang, R. J., Zhao, J., Bhadauria, V., and Peng, Y. L. (2024). Efficient gene editing with an Arg-tRNA promoter-driven CRISPR/Cas9 in the rice blast fungus Pyricularia oryzae. Phytopathol. Res. 6, 56. doi: 10.1186/s42483-024-00271-8
Wang, L., Zhao, L., Zhang, X., Zhang, Q., Jia, Y., Wang, G., et al. (2019). Large-scale identification and functional analysis of NLR genes in blast resistance in the Tetep rice genome sequence. Proc. Natl. Acad. Sci. 116, 18479–18487. doi: 10.1073/pnas.1910229116
Wei, X., Qiu, J., Yong, K., Fan, J., Zhang, Q., Hua, H., et al. (2021). A quantitative genomics map of rice provides genetic insights and guides breeding. Nat. Genet. 53, 243–253. doi: 10.1038/s41588-020-00769-9
Wu, J. L., Fan, Y. Y., Li, D. B., Zheng, K. L., Leung, H., and Zhuang, J. Y. (2005). Genetic control of rice blast resistance in the durably resistant cultivar Gumei 2 against multiple isolates. Theor. Appl. Genet. 111, 50–56. doi: 10.1007/s00122-005-1971-2
Wu, J., Kou, Y., Bao, J., Li, Y., Tang, M., Zhu, X., et al. (2015). Comparative genomics identifies the Magnaporthe oryzae avirulence effector AvrPi9(t) that triggers Pi9-mediated blast resistance in rice. New Phytol. 206, 1463–1475. doi: 10.1111/nph.13310
Wu, Z., Wang, G., Zhang, B., Dai, T., Gu, A., Li, X., et al. (2021). Metabolic mechanism of plant defense against rice blast induced by probenazole. Metabolites 11, 246. doi: 10.3390/metabo11040246
Wu, W., Wang, L., Zhang, S., Li, Z., Zhang, Y., Lin, F., et al. (2014). Stepwise arms race between AvrPik and Pik alleles in the rice blast pathosystem. Mol. Plant-Microbe Interact. 27, 759–769. doi: 10.1094/MPMI-02-14-0046-R
Wu, Y., Xiao, N., Yu, L., Pan, C., Li, Y., Zhang, X., et al. (2015). Combination patterns of major R genes determine the level of resistance to the M. oryzae in rice (Oryza sativa L.). PloS One 10, e0126130. doi: 10.1371/journal.pone.0126130
Wu, Y. Y., Yu, L., Pan, C. H., Dai, Z. Y., Li, Y. H., Xiao, N., et al. (2016). Development of near isogenic lines with different alleles of Piz locus and analysis of their breeding effect under Yangdao 6 background. Mol. Breed. 36, 1–12. doi: 10.1007/s11032-016-0433-7
Xangsayasane, P., Boualaphanh, C., Bounphanousay, C., Bounphanousay, V., Manivong, P., Voradeth, S., et al. (2020). Genetic variation of rice blast (Pyricularia oryzae) isolates in Laos. Plant Health Prog. 21, 248–255. doi: 10.1016/S0885-5765(05)80007-4
Xiao, W. M., Luo, L. X., Wang, H., Guo, T., Liu, Y. Z., Zhou, J. Y., et al. (2016). Pyramiding of Pi46 and Pita to improve blast resistance and to evaluate the resistance effect of the two R genes. J. Integr. Agric. 15, 2290–2298. doi: 10.5555/20173004564
Xiao, N., Pan, C., Li, Y., Wu, Y., Cai, Y., Lu, Y., et al. (2021). Genomic insight into balancing high yield, good quality, and blast resistance of japonica rice. Genome Biol. 22, 1–22. doi: 10.1186/s13059-021-02488-8
Xiao, W., Yang, Q., Huang, M., Guo, T., Liu, Y., Wang, J., et al. (2019). Improvement of rice blast resistance by developing monogenic lines, two-gene pyramids and three-gene pyramid through MAS. Rice 12, 1–11. doi: 10.1186/s12284-019-0336-4
Xu, Y., Bai, L., Liu, M., Liu, Y., Peng, S., Hu, P., et al. (2023). Identification of two novel rice S genes through combination of association and transcription analyses with gene-editing technology. Plant Biotechnol. J. 21, 1628–1641. doi: 10.1111/pbi.14064
Xu, X., Hayashi, N., Wang, C. T., Fukuoka, S., Kawasaki, S., Takatsuji, H., et al. (2014). Rice blast resistance gene Pikahei-1 (t), a member of a resistance gene cluster on chromosome 4, encodes a nucleotide-binding site and leucine-rich repeat protein. Mol. Breed. 34, 691–700. doi: 10.1007/s11032-014-0067-6
Xu, Y., Ma, K., Zhao, Y., Wang, X., Zhou, K., Yu, G., et al. (2021). Genomic selection: A breakthrough technology in rice breeding. Crop J. 9, 669–677. doi: 10.1016/j.cj.2021.03.008
Xu, Y., Miao, Y., Tian, X., Wang, Q., Hu, Y., and Luo, Q. (2022). Transcriptomic and epigenomic assessment reveals epigenetic regulation of WRKY genes in response to Magnaporthe oryzae infection in rice. Curr. Genomics 23, 182–194. doi: 10.2174/1389202923666220510195910
Yamada, M., Kiyosawa, S., Yamaguchi, T., Hirano, T., Kobayashi, T., Kushibuchi, K., et al. (1976). Proposal of a new method of differentiating races of Pyricularia oryzae Cavara in Japan. Annuals Phytopathol. Soc. Japan 42, 216–219. doi: 10.3186/jjphytopath.42.216
Yan, J., Li, L., Bao, J., Wang, J., Liu, X., Lin, F., et al. (2024). A glance at structural biology in advancing rice blast fungus research. Virulence 15, 2403566. doi: 10.1080/21505594.2024.2403566
Yang, W., Yang, Z., Yang, L., Li, Z., Zhang, Z., Wei, T., et al. (2024). Genomic and transcriptomic analyses of the elite rice variety Huizhan provide insight into disease resistance and heat tolerance. Genomics. 116 (5), 110915. doi: 10.1016/j.ygeno.2024.110915
Yang, J., Fang, Y., Wu, H., Zhao, N., Guo, X., Mackon, E., et al. (2023). Improvement of resistance to rice blast and bacterial leaf streak by CRISPR/Cas9-mediated mutagenesis of Pi21 and OsSULTR3 in rice (Oryza sativa L.). Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1209384
Yang, D., Li, S., Lu, L., Fang, J., Wang, W., Cui, H., et al. (2020). Identification and application of the Pigm-1 gene in rice disease resistance breeding. Plant Biol. 22, 1022–1029. doi: 10.1111/plb.13170
Yang, W., Zhang, H., Li, M., Wang, Z., Zhou, J., Wang, S., et al. (2014). Early diagnosis of blast fungus, Magnaporthe oryzae in rice plant by using an ultrasensitive electrically magnetic-controllable electrochemical biosensor. Analytica Chimica Acta 850, 85–91. doi: 10.1016/j.aca.2014.08.040
Yang, L., Zhao, M., Sha, G., Sun, Q., Gong, Q., Yang, Q., et al. (2022). The genome of the rice variety LTH provides insight into its universal susceptibility mechanism to worldwide rice blast fungal strains. Comput. Struct. Biotechnol. J. 20, 1012–1026. doi: 10.1016/j.csbj.2022.01.030
Yin, Y., Wang, R., Jiang, Y., Suo, Y., Li, Y., Wang, Z., et al. (2025). A method for the rapid identification of rice seed blast using deep learning and hyperspectral imagery. Agronomy 15, 290. doi: 10.3390/agronomy15020290
Ying, Z., Tao, W., Bin, Y., Fang, L., Meijuan, C., Qiong, W., et al. (2022). Improving rice blast resistance by mining broad-spectrum resistance genes at pik locus. Rice Sci. 29, 133–142. doi: 10.1016/j.rsci.2022.01.002
Yoshida, K., Saitoh, H., Fujisawa, S., Kanzaki, H., Matsumura, H., Yoshida, K., et al. (2009). Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 21, 1573–1591. doi: 10.1105/tpc.109.066324
Younas, M. U., Wang, G., Du, H., Zhang, Y., Ahmad, I., Rajput, N., et al. (2023). Approaches to reduce rice blast disease using knowledge from host resistance and pathogen pathogenicity. Int. J. Mol. Sci. 24, 4985. doi: 10.3390/ijms24054985
Younas, M. U., Ahmad, I., Qasim, M., Ijaz, Z., Rajput, N., Parveen Memon, S., et al. (2024). Progress in the management of rice blast disease: The role of avirulence and resistance genes through gene-for-gene interactions. Agronomy 14 (1), 163. doi: 10.3390/agronomy14010163
Yu, Y., Ma, L., Wang, X., Zhao, Z., Wang, W., Fan, Y., et al. (2022). Genome-wide association study identifies a rice panicle blast resistance gene, Pb2, encoding NLR protein. Int. J. Mol. Sci. 23, 5668. doi: 10.3390/ijms23105668
Yuan, B., Zhai, C., Wang, W., Zeng, X., Xu, X., Hu, H., et al. (2011). The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes. Theor. Appl. Genet. 122, 1017–1028. doi: 10.1007/s00122-010-1506-3
Zainuddin, F. A., Ismail, M. R., Hatta, M. A. M., and Ramlee, S. I. (2024). Advancement in modern breeding and genomic approaches to accelerate rice improvement: speed breeding focus. Euphytica 220, 109. doi: 10.1007/s10681-024-03353-y
Zeigler, R. S., Leong, S. A., and Teng, P. S. (1994). Rice blast disease (International Rice Research Institute).
Zeng, N., Gong, G., Zhou, G., and Hu, C. (2023). An accurate classification of rice diseases based on ICAI-V4. Plants 12, 2225. doi: 10.3390/plants12112225
Zeng, X., Yang, X., Zhao, Z., Lin, F., Wang, L., and Pan, Q. (2011). Characterization and fine mapping of the rice blast resistance gene Pia. Sci. China Life Sci. 54, 372–378. doi: 10.1007/s11427-011-4154-1
Zhai, K., Deng, Y., Liang, D., Tang, J., Liu, J., Yan, B., et al. (2019). RRM transcription factors interact with NLRs and regulate broad-spectrum blast resistance in rice. Mol. Cell 74, 996–1009. doi: 10.1016/j.molcel.2019.03.013
Zhai, C., Lin, F., Dong, Z., He, X., Yuan, B., Zeng, X., et al. (2011). The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol. 189, 321–334. doi: 10.1111/j.1469-8137.2010.03462.x
Zhai, C., Zhang, Y., Yao, N., Lin, F., Liu, Z., Dong, Z., et al. (2014). Function and interaction of the coupled genes responsible for Pik-h encoded rice blast resistance. PloS One 9, e98067. doi: 10.1371/journal.pone.0098067
Zhang, J., Du, Q., Wu, Y., Shen, M., Gao, F., Wang, Z., et al. (2025). Ubiquitin ligase gene osPUB57 negatively regulates rice blast resistance. Plants 14, 758. doi: 10.3390/plants14050758
Zhang, X., He, R., Li, Y., Ren, S., Xiang, S., Zheng, J., et al. (2025). tRNA thiolation optimizes appressorium-mediated infection by enhancing codon-specific translation in Magnaporthe oryzae. Nucleic Acids Res. 53, gkae1302. doi: 10.1093/nar/gkae1302
Zhang, Y., Lin, X. F., Li, L., Piao, R. H., Wu, S., Song, A., et al. (2024). CRISPR/Cas9-mediated knockout of Bsr-d1 enhances the blast resistance of rice in Northeast China. Plant Cell Rep. 43, 100. doi: 10.1007/s00299-024-03192-0
Zhang, S., Wang, L., Wu, W., He, L., Yang, X., and Pan, Q. (2015). Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Sci. Rep. 5, 11642. doi: 10.1038/srep11642
Zhang, S. and Xu, J. R. (2014). Effectors and effector delivery in Magnaporthe oryzae. PloS Pathog. 10, e1003826. doi: 10.1371/journal.ppat.1003826
Zhang, X., Xue, C., Wang, R., Shen, R., and Lan, P. (2022). Physiological and proteomic dissection of the rice roots in response to iron deficiency and excess. J. Proteomics 267, 104689. doi: 10.1016/j.jprot.2022.104689
Zhang, L., Zhang, Y., Liu, Y., Miao, W., Ai, J., Li, J., et al. (2024). Multi-omics analysis revealed that the protein kinase MoKin1 affected the cellular response to endoplasmic reticulum stress in the rice blast fungus, Magnaporthe oryzae. BMC Genomics 25, 449. doi: 10.1186/s12864-024-10337-8
Zhang, P., Zhou, Z., Huang, H., Yang, Y., Hu, X., Zhuang, J., et al. (2025). Hybrid integrated feature fusion of handcrafted and deep features for rice blast resistance identification using uav imagery. IEEE J. Selected Topics Appl. Earth Observations Remote Sens. 18, 7304–7317. doi: 10.1109/JSTARS.2025.3543190
Zhao, Q., Lin, J., Yousaf, L., Xue, Y., and Shen, Q. (2021). Protein structural properties and proteomic analysis of rice during storage at different temperatures. Food Chem. 361, 130028. doi: 10.1016/j.foodchem.2021.130028
Zhao, T., Tang, P., Liu, C., Zuo, R., Su, S., Zhong, Y., et al. (2024). Multi-omics approach reveals osPIL1 as a regulator promotes rice growth, grain development, and blast resistance. J. Agric. Food Chem. 72, 1822–1843. doi: 10.1021/acs.jafc.3c07330
Zhao, H. J., Wang, X. Y., Jia, Y. L., Minkenberg, B., Wheatley, M., Fan, J. B., et al. (2018). The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat. Communication 9, 2039. doi: 10.1038/s41467-018-04369-4
Zheng, Y., Zheng, W., Lin, F., Zhang, Y., Yi, Y., Wang, B., et al. (2011). AVR1-CO39 is a predominant locus governing the broad avirulence of Magnaporthe oryzae 2539 on cultivated rice (Oryza sativa L.). Mol. Plant-Microbe Interact. 24, 13–17. doi: 10.1094/MPMI-10-09-0240
Zhou, H., Hwarari, D., Zhang, Y., Mo, X., Luo, Y., and Ma, H. (2022). Proteomic analysis reveals salicylic acid as a pivotal signal molecule in rice response to blast disease infection. Plants 11, 1702. doi: 10.3390/plants11131702
Zhou, H., Lai, Q., Huang, Q., Cai, D., Huang, D., and Wu, B. (2024). Automatic detection of rice blast fungus spores by deep learning-based object detection: models, benchmarks and quantitative analysis. Agriculture 14, 290. doi: 10.3390/agriculture14020290
Zhou, Y., Lei, F., Wang, Q., He, W., Yuan, B., and Yuan, W. (2020). Identification of novel alleles of the rice blast-resistance gene Pi9 through sequence-based allele mining. Rice 13, 1–15. doi: 10.1186/s12284-020-00442-z
Zhou, B., Qu, S., Liu, G., Dolan, M., Sakai, H., Lu, G., et al. (2006). The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol. Plant-Microbe Interact. 19, 1216–1228. doi: 10.1094/MPMI-19-1216
Zhou, Y., Xu, S., Jiang, N., Zhao, X., Bai, Z., Liu, J., et al. (2022). Engineering of Rice varieties with enhanced resistances to both blast and bacterial blight diseases via CRISPR/Cas9. Plant Biotechnol. J. 20, 876–885. doi: 10.1111/pbi.13766
Zhou, Y., Xu, Y., Wang, X., Kou, S., Huang, P., Qiu, W., et al. (2024). Allele mining for blast-resistance gene at Pi5 locus in rice. Plant Stress 12, 100465. doi: 10.1016/j.stress.2024.100465
Zhu, C., Bai, C., Gomez-Gomez, L., Sandmann, G., Baysal, C., Capell, T., et al. (2022). Rice callus as a high-throughput platform for synthetic biology and metabolic engineering of carotenoids. Methods enzymol. 671, 511–526. doi: 10.1016/bs.mie.2021.09.016
Zhu, Q., Li, Y., Hassan, M. A., Fang, W., and Wang, S. (2024). Comparative physiological and transcriptomic characterization of rice (Oryza sativa L.) root seedlings under phosphorus deficiency. Plant Stress 12, 100508. doi: 10.1016/j.stress.2024.100508
Zhu, M., Wang, L., and Pan, Q. (2004). Identification and characterization of a new blast resistance gene located on rice chromosome 1 through linkage and differential analyses. Phytopathology 94, 515–519. doi: 10.1094/PHYTO.2004.94.5.515
Keywords: rice, blast disease, MAS, MABB, CRISPR/Cas9, omics approaches, nanotechnology, artificial intelligence
Citation: Ragulakollu S, Loganathan A, Swaminatham M, Chellappan G, Veeraswamy R and Jegadeesan R (2025) Molecular breeding approaches for sustainable rice blast management: recent advances and challenges. Front. Plant Sci. 16:1551018. doi: 10.3389/fpls.2025.1551018
Received: 24 December 2024; Accepted: 31 July 2025;
Published: 23 September 2025.
Edited by:
Brigitte Mauch-Mani, Retired, Fribourg, SwitzerlandReviewed by:
Joseph Bigirimana, International Rice Research Institute (IRRI), PhilippinesPankaj Kumar Singh, Indian Institute of Technology (BHU), India
Copyright © 2025 Ragulakollu, Loganathan, Swaminatham, Chellappan, Veeraswamy and Jegadeesan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ramalingam Jegadeesan, cmFtYWxpbmdhbS5qQHRuYXUuYWMuaW4=
†ORCID: Ramalingam Jegadeesan, orcid.org/0000-0002-0887-8286
 Sravanthi Ragulakollu
Sravanthi Ragulakollu Arul Loganathan
Arul Loganathan Manonmani Swaminatham
Manonmani Swaminatham Gopalakrishnan Chellappan
Gopalakrishnan Chellappan Ravichandran Veeraswamy
Ravichandran Veeraswamy Ramalingam Jegadeesan
Ramalingam Jegadeesan