- 1Plant Breeding and Genetics, Punjab, Agricultural University, Ludhiana, Punjab, India
- 2Indian Council of Agricultural Research (ICAR)-Indian Institute of Maize Research, Ludhiana, Punjab, India
- 3Department of Agricultural Botany, Mahatma Phule Krishi Vidyapeeth, Rahuri, Maharashtra, India
- 4Indian Council of Agricultural Research (ICAR)-Indian Institute of Agricultural Biotechnology, Ranchi, Jharkhand, India
Lysine and tryptophan, two essential amino acids, are generally deficient in normal maize but enriched in opaque2 (o2) mutants. However, these o2 mutants are linked to undesirable effects like soft endosperm and yield loss. To circumvent this, researchers introgressed o2 modifiers (Mo2s) into mutant maize and developed Quality Protein Maize (QPM). This study identifies genomic regions linked to Mo2 governing kernel hardness, opacity, and tryptophan content. Two QPM lines (DQL 2104–1 and DQL 2034), contrasting for these traits, were crossed to develop a 138 F2 and 109 F2:3 mapping population. Genotyping with 141 informative SSR markers resulted in 2417.01 cM genetic map with an average marker distance of 20.66 cM between markers. Inclusive composite interval mapping (ICIM) detected 11 QTLs across six different chromosomes: seven QTLs for kernel opacity (chromosomes 1, 2, 4, 7), three for hardness (chromosomes 7, 8, 9), and one for tryptophan (chromosome 9). These QTLs co-localized with candidate genes (opaque1, opaque11, floury1, floury2, floury4, mucronate1, and waxy1). The identified QTLs provide foundational targets for marker-assisted breeding. Few QTLs like qHRD9.1 (PVE = 14.18%) and qTRP9.1 (PVE = 10.69%) are prime candidates for improving hardness and tryptophan. These loci can be pyramided into elite lines using SSR markers; genomic selection could be used to optimize trait stacking. Future fine-mapping and functional studies will refine these regions, accelerating the development of high-yielding QPM with vitreous kernels and enhanced nutritional quality.
1 Introduction
Maize (Zea mays L.) is one of the world’s largest crops, serves as a staple crop for a billion people in Sub-Sahara Africa, Latin America, and Asian countries. Furthermore, besides being a staple food, it acts as a major vehicle for providing nutrients to humans and livestock (Shiferaw et al., 2011). Maize proteins generally lack sufficient levels of two important amino acids: lysine and tryptophan. The lysine level in maize kernel is typically 0.16-0.26%, while the tryptophan content is between 0.02-0.06%, which is less than half of the recommended dose specified for human nutrition (Mertz et al., 1964; Bjarnason and Vasal, 1992; Hossain et al., 2008; Vivek, 2008). The primary cause of the lack of lysine and tryptophan is mostly attributed to zein (prolamin fraction), a storage protein that makes up to 70% of all storage proteins (Gibbon and Larkins, 2005). This nature of maize kernel changed with the discovery of the natural opaque (o2) mutant, which led to the subsequent development of genotypes with increased lysine and tryptophan in maize endosperm (Mertz et al., 1964; Vasal, 2000; Gupta et al., 2009). The o2 mutant has more accumulation of non-zein protein fractions, achieving the desired amino acid composition (Wang and Larkins, 2001). However, these mutant maize lines also have undesirable pleiotropic effects, such as soft and chalky kernels, and susceptibility to pests and diseases, which ultimately reduces yields (Gupta et al., 2013). To combat these effects, plant breeders introgressed quantitative trait loci (QTLs) known as o2 modifiers (Mo2s) in o2 mutant maize, developing what is now known as quality protein maize (QPM). This modifier-genetic system in the genetic background of the o2 gene can transform the soft endosperm into a hard and vitreous endosperm without altering the increased protein content (Paez et al., 1969; Villegas et al., 1992; Or et al., 1993; Vivek, 2008; Babu et al., 2005; Krivanek et al., 2007; Pandey et al., 2016).
The inheritance of these o2 modifiers has been proven to be highly complex but effective in improving the negative pleiotropic effects of o2 genes (Vasal et al., 1980; Gibbon and Larkins, 2005). The modifier genes have been associated with elevated γ-zein, which renders the vitreousness in maize endosperm (Babu and Prasanna, 2014). Number of studies have aimed to comprehend the genetics behind the endosperm modifiers and their influence on grain quality in QPM with different genetic backgrounds. Lopes et al., 1995, using bulked segregant analysis (BSA), showed that chromosome 7 contains regions for endosperm modification. The first locus was found to encode the 27-kDa γ-zein and was located close to the centromere. The second locus was identified by using a limited set of restriction fragment length polymorphism (RFLP) markers, positioned near the telomeric ends of the long arm of chromosome 7 (Lopes et al., 1995). A number of studies have suggested that the γ-zeins play a major role in the regulation of endosperm modification (Geetha et al., 1991; Burnett and Larkins, 1999). Dannenhoffer et al. (1995) observed 27 kDa gamma-zeins to be involved in the development of the protein bodies responsible for the formation of the vitreous endosperm. Holding et al. (2008) examined individual F2:3 kernels for hardness and seed vitreousness and mapped seven opaque2 modifier (Opm) QTLs on chromosomes 1, 5, 7, 9, and 10 using bulked sequencing analysis (BSA). Babu et al. (2015) utilized gene-based SSR markers of lysine and tryptophan and genomic SSRs on the F2:3 mapping population to identify five QTLs on chromosomes 5, 7, and 9. Later, it was shown that silencing or deletion of 27-kDa gamma zein expression abolished the formation of vitreous endosperm in QPM (Wu et al., 2010; Yuan et al., 2014). Zein proteins are stored in rough endoplasmic reticulum-retained protein bodies (PBs) in the maize endosperm (Wolf et al., 1967; Larkins and Dalby, 1975; Burr and Burr, 1976; Wu and Messing, 2010). The 27-kDa γ-zeins, along with 16-kDa γ-zein and 15-kDa β-zein, play an important role in the initiation and stabilization of protein bodies (PB) (Lending et al., 1992; Coleman et al., 1997). A significant QTL, namely qy27, was discovered to be a duplication of the 27-kDa γ-zein gene (Li et al., 2018). It was observed that this 15.26 kb duplicated segment regulated the elevated expression of the 27-kDa γ-zein gene in QPM. Furthermore, the correlation between the degree of modification and the level of 27-kDa γ-zein expression in different populations created by a cross between QPM and a starchy o2 mutant suggested that the qy27 controlling enhanced expression of 27-kDa gamma zein potentially houses an o2 modifier (Li et al., 2018). However, still the total number of modifier genes affecting the endosperm modification is unknown.
To strengthen our knowledge of the genetic mechanism of the modifiers, we analyzed two different QPM lines, DQL 2104–1 and DQL 2034, with significantly varying kernel opacity, kernel hardness, and tryptophan content. Crosses were made to develop the F2:3 population for phenotypic and genotypic analysis. This study is distinct from earlier reports as it focuses on the features of kernel opacity, hardness, and amino acids in a population derived from the opaque2 inbred lines. The objective of the present study is to identify genomic regions linked with endosperm modifiers affecting kernel opacity, tryptophan, and hardness that could be further utilized for the identification of key genes responsible for increased amino acid content and vitreous kernels and the development of better-performing QPM lines.
2 Materials and methods
2.1 Plant material
Two inbred lines, DQL 2104–1 and DQL 2034, significantly differing for kernel opacity, kernel hardness, and tryptophan content were crossed during Kharif 2021 ta Ladhowal Fields, ICAR-Indian Institute of Maize Research, Ludhiana, to generate F1 seeds (Anonymous, 2021). Approximately 350 F1 seeds were sown at the Agricultural Research Station (ARS), Peddapuram, Andhra Pradesh, during Rabi 2021. However, a reduced germination rate by ~40.59% was observed, yielding 138 F2 plants. These F2 individuals were self-pollinated at Ladhowal Farm in the Spring 2022 to produce F2:3 seeds. Due to heavy rainfall and waterlogging at the flowering stage, the seed set was impacted, resulting in only 109 F2:3 ears with >= 30 seeds per cob. The harvested F2:3 ears were used to examine kernel opacity, hardness, and tryptophan content. All the seeds were thoroughly dried and stored at 4°C.
2.2 Measurement of kernel opacity
The kernel opacity of F2:3 seeds was recorded using the lightbox (Bjarnason and Vasal, 1992) in three replications. The light box is used to differentiate between opaque2 and normal grains based on their endosperm textures. The distorted three-dimensional structure of the kernel endosperm shows opaque seeds, whereas normal seeds are vitreous under the lightbox screening. For the screening, 10 randomly selected seeds per replication were examined. A total of thirty seeds were uniformly spread on the lightbox screen in such a way that the germ (embryo) part was facing down. The degree of opaqueness of seeds was calculated with the formula:
Where, N100, N75, N50, N25, and N0 are the numbers of seeds with 100%, 75%, 50%, 25%, and 0% opacity, respectively (Hossain et al., 2008; Sarika et al., 2018).
2.3 Measurement of kernel hardness
Three seeds per replication were randomly selected and evaluated for kernel hardness by using a digital portable kernel hardness tester. The hardness was measured at a grain moisture content of approximately 14%. The kernels were placed in the chamber with the germ side up on a plate, and the crushing strength of individual seeds. To minimize the error, care was taken by placing all the seeds in the same direction. A test load of 5 to 500 N was applied, and breaking force was measured using the device’s frequency. The first peak force (N, Newton) in the force deformation curve was defined as kernel hardness (Zunjare et al., 2015).
2.4 Tryptophan estimation
The tryptophan content was estimated according to the colorimetric method (Hernandez and Bates, 1969). Ten seeds per genotype were randomly selected and were kept in distilled water overnight. The pericarp layer and germ layer were removed the following morning, and the seeds were left to dry in a hot air oven. The dried seeds were grounded with the help of a maize seed grinder, followed by defatting for 3 days (72 hours). The defatted samples (100 mg) were subjected to protein digestion (adding 4 mL of papain solution) and kept in an incubator at 65°C overnight. The following day, samples were processed using Hernandez and Bates, 1969. The standard curve for the tryptophan was taken using a standard tryptophan solution of known concentration against a blank at 545 nm. Thereafter, the factor was calculated using the standard curve to calculate percent tryptophan.
2.5 DNA extraction
Leaf samples were collected from all 138 F2 plants in the same season (Rabi 2021) - 30 days after germination (plant height: 15–20 cm) and were immediately stored at -80°C. Genomic DNA was extracted in two replicates per sample using the CTAB method (Murray and Thompson, 1980). DNA pellet was suspended in TE buffer (pH 8.0), and treated with RNase (10μg/mL) at 37°C for 35–40 minutes to remove RNA contamination. Purified DNA samples (including replication) were stored at -20°C in a 1.5 mL microcentrifuge tube. DNA quality was assessed by the intensity of DNA bands on a 0.8% agarose gel, while quantity was measured using a nanodrop spectrophotometer (ND 2000).
2.6 SSR marker selection and genotyping
A total of 496 Simple Sequence Repeats (SSR) markers, uniformly spread across all chromosomes, were used for parental polymorphism. Of these, 141 markers (28.43%) showed polymorphism and were selected for genotyping. From the original 138 F2 plants, 109 individuals were chosen for further analysis based on the F2:3 progeny available (genotypes having at least 30 seeds per cob).
2.7 PCR amplification and electrophoresis
Polymerase chain reaction (PCR) was carried out in a 10 μL reaction volume containing 1.5 μL template DNA, 1 μL forward and reverse primer each, 5 μL TaKaRa™ master mix (2X), and 1.5 μL nuclease-free water (NFW) in an Eppendorf MasterCycler™. The amplification conditions of the PCR reaction are as follows: first, initial denaturation at 95°C for 5 minutes, then 35 cycles of denaturation at 94°C for 40 seconds, annealing (temperature as per primer) for 40 seconds, extension at 72°C for 40 seconds and a final extension at 72°C for 8 minutes. The amplified products were resolved on 4% agarose gel stained with ethidium bromide and visualized under UV light.
2.8 Statistical analysis
The chi-square (χ2) test was used to determine deviations of observed genotypic ratios from the expected Mendelian segregation (1:2:1 for the F2 population) (Arora et al., 2021). Markers showing significant distortion (p<0.05) were eliminated from the further analysis.
2.9 Linkage map and QTL analysis
From the initial 141 polymorphic SSR markers (www.maizegdb.org), 25 markers showing significant segregation distortion (χ2) were removed, resulting in 116 informative markers. These markers were used to construct a genetic linkage map for the F2 population derived from the cross between DQL 2104–1 and DQL 2034. Linkage analysis was conducted using QTL IciMapping software version 4.2 (Wang et al., 2016) with a logarithm of odds (LOD) threshold of 3.0 for grouping and ordering markers. The Kosambi function was used to measure the map distance and was expressed in centimorgans (cM) (Kosambi, 2016).
3 Results
3.1 Phenotypic evaluation
Phenotypic analysis of F2:3 progeny shows significant variation in three key endosperm traits: kernel opacity, hardness, and tryptophan content were estimated in F2:3 seeds (Table 1). The parent lines showed contrasting traits: DQL 2104–1 was fully opaque (100%) with lower hardness (Hardness: 95.83 N), while parent DQL 203 showed reduced kernel opacity (25%) and higher hardness of 163.17 N. The lightbox testing of parents and population is shown in Figure 1. The F2:3 population exhibited a broad range for kernel opacity (16.67 -100%), hardness (62.50-235.13 N), and tryptophan content (0.01-0.11%), demonstrating transgressive segregation (Figure 2). A negative relationship between opacity and hardness was observed, with seeds having lower opacity observed to have hard endosperm. The average tryptophan content was 0.081% in DQL 2104-1, 0.054% in DQL 2034, and 0.044% in the F2:3 population. The data about the kernel opacity, hardness, and tryptophan content of F2:3 population seeds is given in Supplementary Table S1.
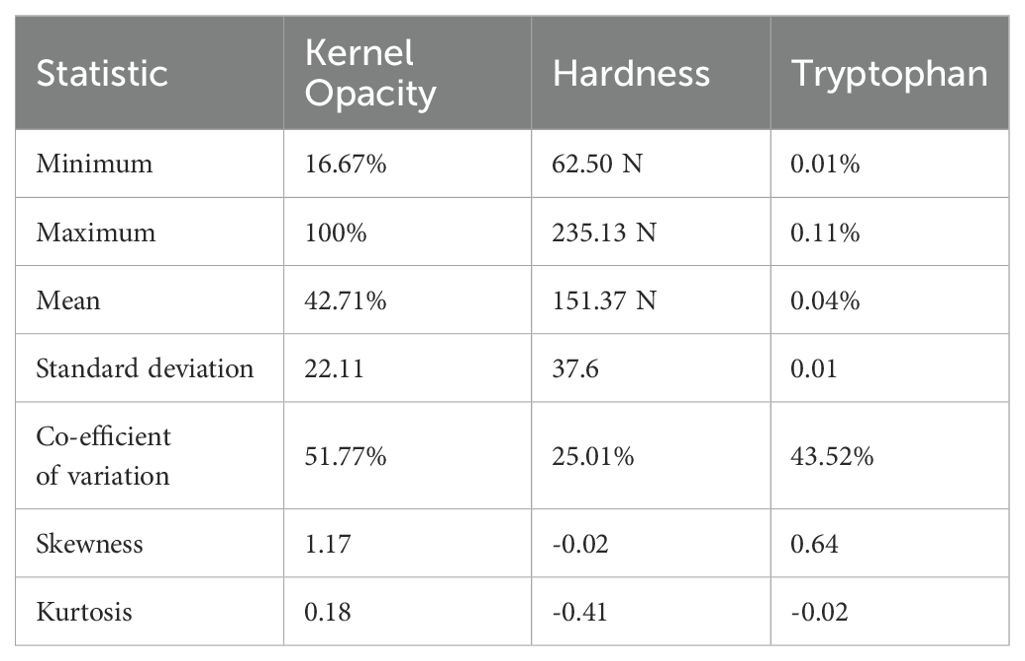
Table 1. Descriptive statistics of the kernel opacity, hardness, and tryptophan content of the quality protein maize F2:3 mapping population.
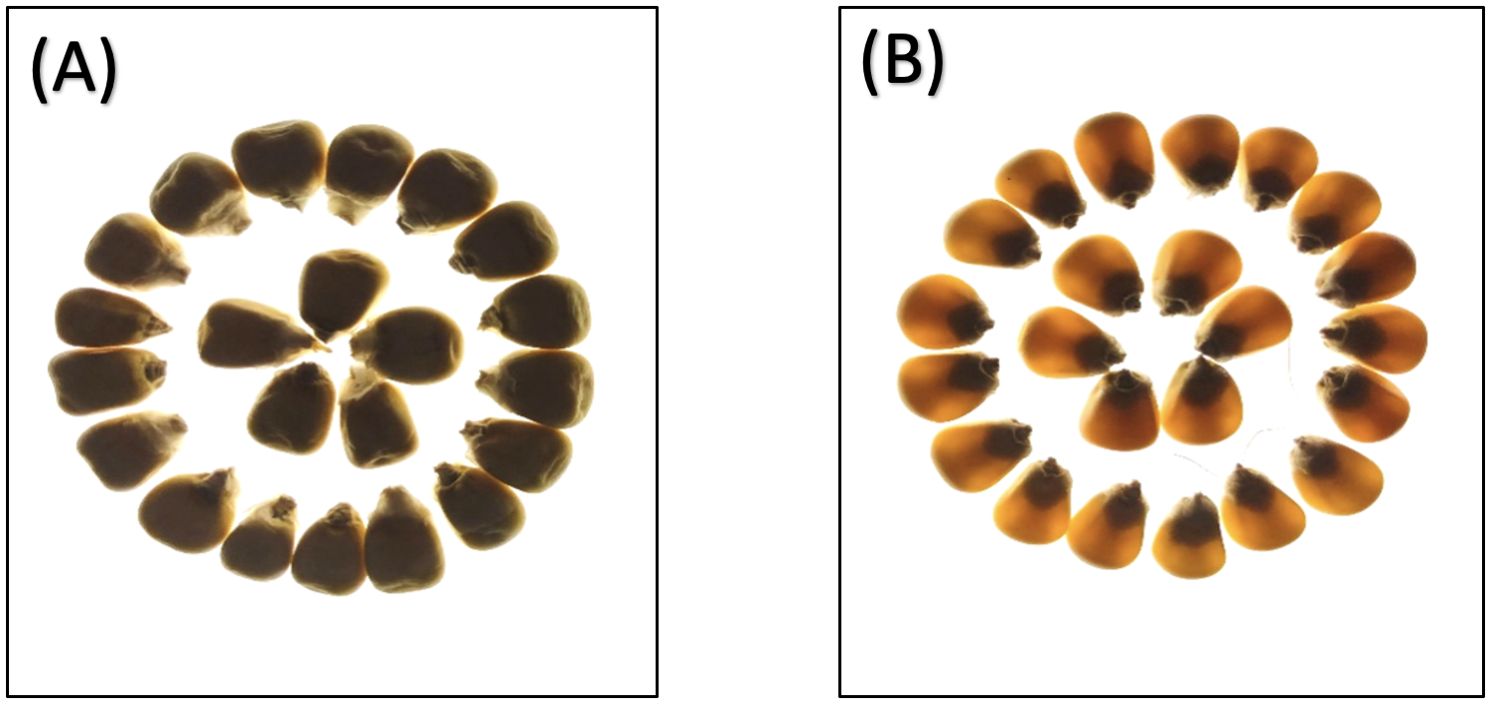
Figure 1. Light box testing of parents seeds: (A) Parent 1 – DQL2104-1 - 100% kernel opacity, (B) Parent 2 – DQL2034 - 25% kernel opacity.
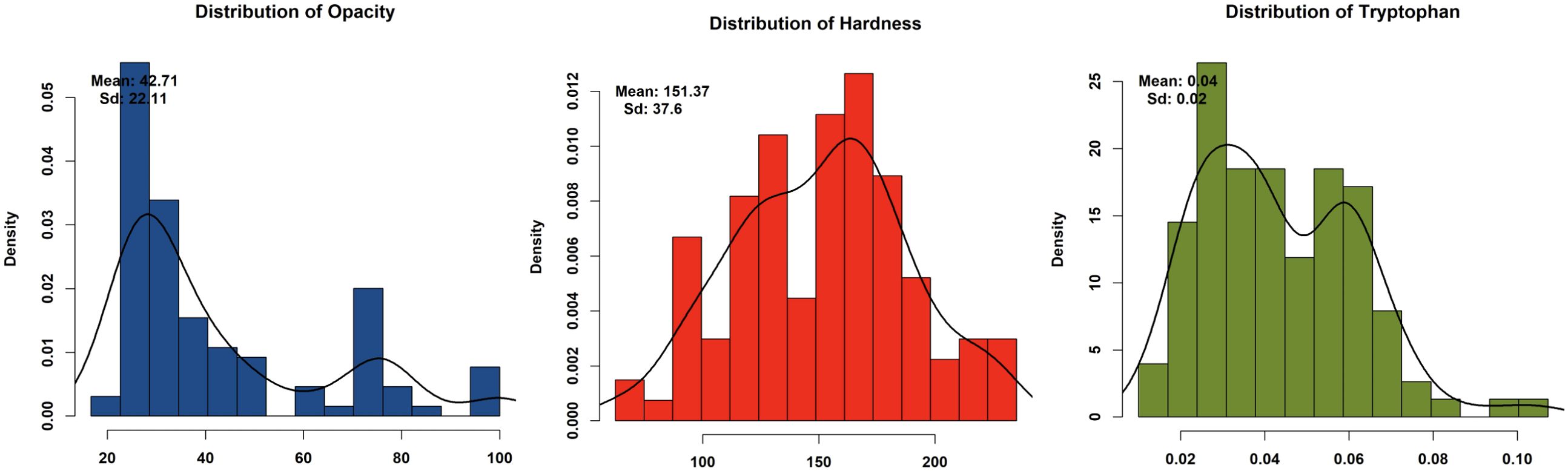
Figure 2. Graphical representation of density histogram of F2:3 population for kernel opacity, kernel hardness, and tryptophan content.
3.2 Correlation among the traits
The correlation analysis showed a significant correlation among traits (Figure 3). A weak but positive correlation (r = 0.29) was observed between tryptophan and opacity, indicating that an increase in kernel opacity is associated with an increase in tryptophan content. However, 25 genotypes (22.9%) do not follow the trend, exhibiting below-average opacity (<42.71%) coupled with higher tryptophan content (>0.045%), which might imply the role of genetic modifiers. Strong negative correlations were observed between kernel opacity and kernel hardness (r = -0.74) and kernel hardness and tryptophan content (r = -0.27), highlighting the inverse relationship between endosperm density and nutritional quality.
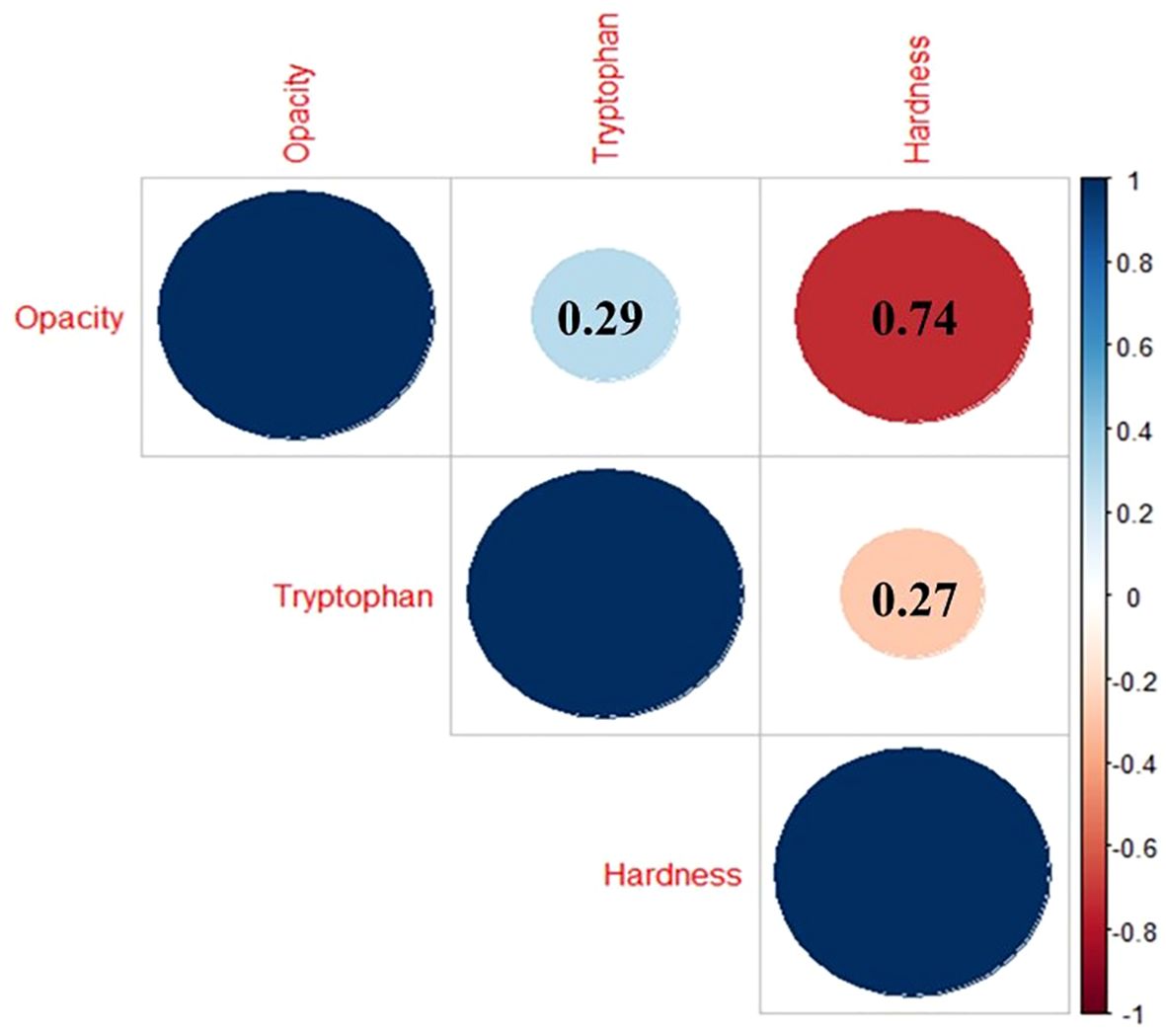
Figure 3. Correlation matrix between kernel opacity, kernel hardness, and tryptophan content (The color represents the direction of the correlation: the blue gradient indicates positive correlations, while the red gradient signifies negative correlations. The size of the circle indicated magnitude of the correlation within the matrix.).
3.3 Genotyping and linkage map analysis
3.3.1 opaque2 gene confirmation in parental lines
First, to validate the genetic basis of phenotypic divergence, both parents were screened with opaque2-specific co-dominate SSR markers, i.e., umc1066 (bin 7.01) and phi057 (bin 7.01) (Magulama and Sales, 2009). Monomorphic banding pattern for both markers confirmed the presence of the o2 allele in both DQL 2104–1 and DQL 2034 (Figure 4). This result confirms that differences in kernel opacity, hardness, and tryptophan content between the parents are independent of o2 presence, implicating additional genetic factor like modifiers, might be responsible for modulating these traits.
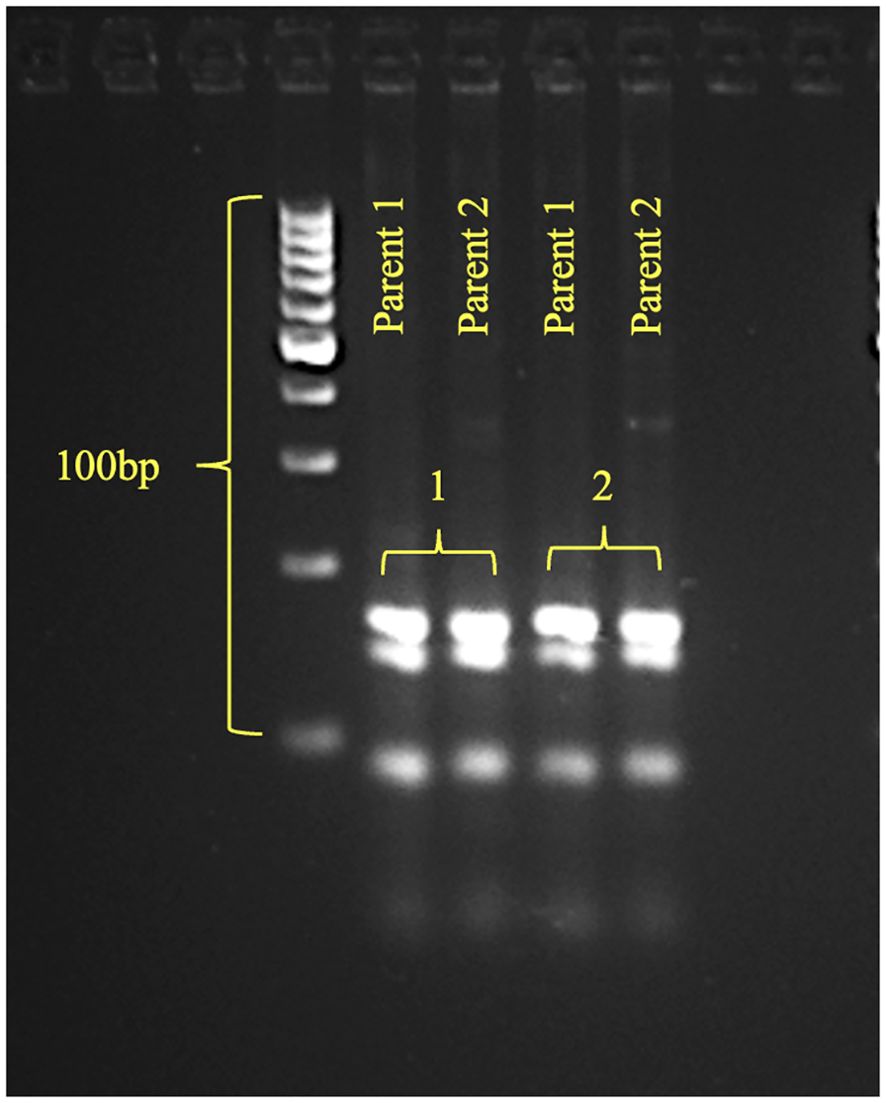
Figure 4. The gel pattern of parents (Parent 1 - DQL2104-1; Parent 2 - DQL2034) using opaque2 SSR markers (1-phi057, 2-umc1066).
3.3.2 Identification of polymorphic markers
To identify genomic regions linked to endosperm modifiers and parents, DQL 2104–1 and DQL 2034 were screened using a total of 496 SSR loci, covering all ten maize chromosomes. Of these, 141 markers (28.43%) exhibited polymorphism and were selected for genotyping 109 F2 individuals (Figure 5). Chromosome 7 showed the highest polymorphism, with fourteen out of thirty-one (45.16%) markers and chromosome 4 showed the lowest (20.41%) with ten out of forty-nine markers.
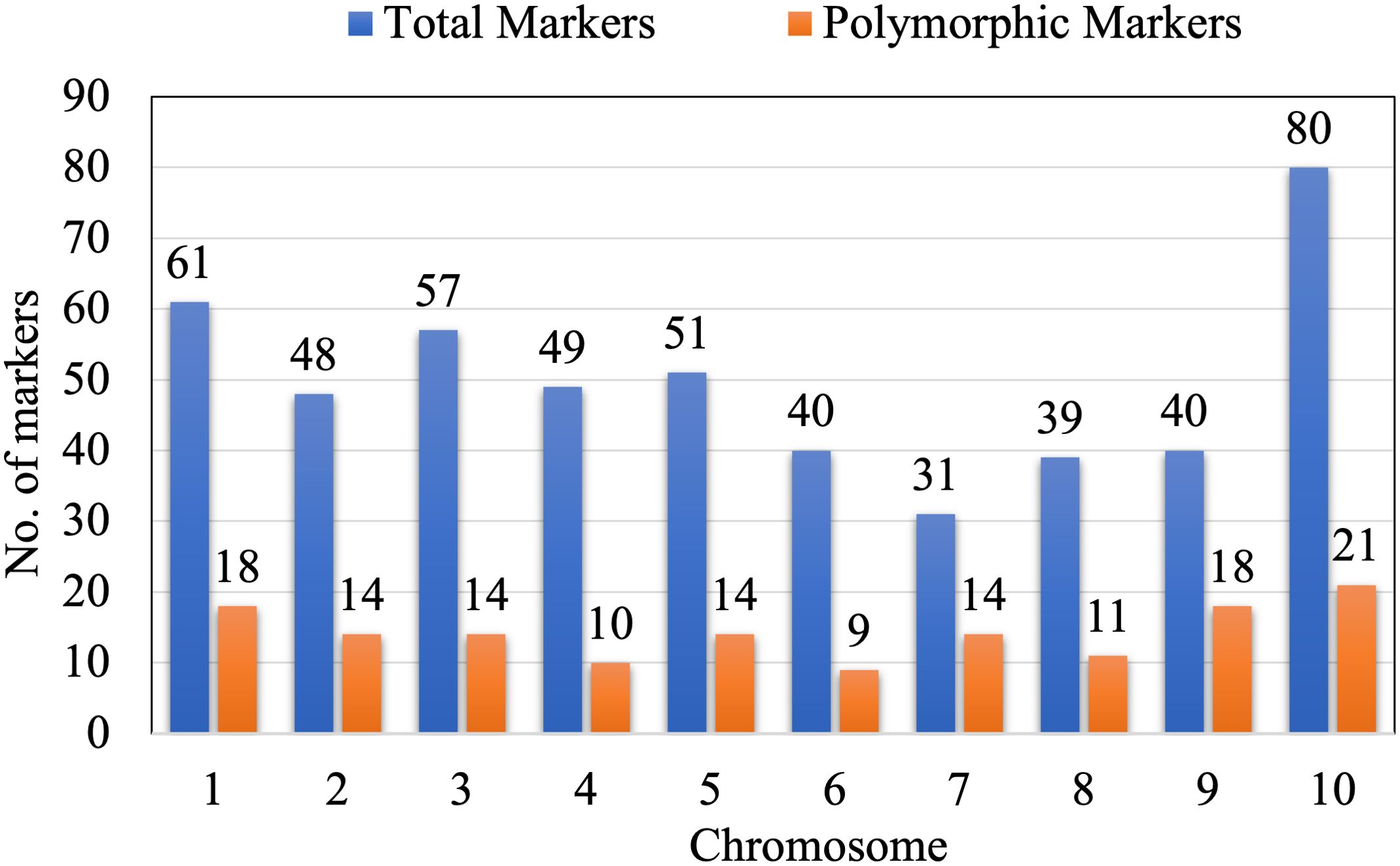
Figure 5. Graphical representation of total markers and polymorphic markers per chromosome of maize.
3.3.3 Chi-square (χ2) test for segregation distortion
To determine the segregation distortion of the marker loci, chi-square (χ2) was performed. Out of the total 141 SSR markers, twenty-five markers (17.73%) were removed from the analysis due to distorted chi-square values, resulting in 116 high-confidence polymorphic SSR marker loci spread across all the ten chromosomes (Figure 5). These markers were used to construct a linkage map with a total length of 2417.01 cM and an average distance of 20.66 cM between markers using IciMapping Version 4.2 software (Wang et al., 2016). Chromosome one was the longest linkage group (475.52 cM), while chromosome 6 was the shortest (155.33 cM). The information on the polymorphic primers, including their bin location, sequence, and annealing temperature, is summarized in Supplementary Table S2.
3.4 QTL analysis
The QTL mapping analysis was done using IciMapping V4.2 software (Wang et al., 2016). Inclusive composite interval mapping (ICIM) based on kernel opacity, hardness, and tryptophan content of 109 F2:3 individuals and genotypic data of 116 SSR markers was performed. Based on the analysis, we identified eleven QTLs on chromosomes 1, 2, 4, 7, 8, and 9 for all the traits (Figure 6). Seven minor QTLs were detected for kernel opacity on chromosomes 1, 2, 4, and 7, explaining a total phenotypic variance (PVE) of 20.76% (Table 2). Out of all seven, three QTLs on chromosome 1 for kernel opacity were detected between loci umc1630 and umc1374 with a LOD of 3.25, umc2237 and bnlg1598 with a LOD of 3.42, and bnlg1178 and bnlg2057 with a LOD 6.21 comprising total PVE of 9.81%. One QTL each on chromosomes 2 and 4 was identified for kernel opacity between marker loci umc2246 and umc2214 with an LOD value of 5.12 and between umc1109 and umc1008 with an LOD value of 3.97, respectively. The remaining two QTLs for kernel opacity were detected on chromosome 7 between marker loci umc1412 and umc1407 with an LOD value of 4.06 and a PVE value of 1.16% and between umc2190 and phi116 with an LOD value of 5.89 and a PVE value of 3.31%. A major QTL for tryptophan was mapped to chromosome 9, with a PVE of 10.69% and an LOD of 2.53, which is the second highest among all the QTLs identified. Three QTLs for kernel hardness were observed on chromosomes 7, 8, and 9. The QTL between the loci bnlg1525 and dup029 on chromosome 9 had a LOD value of 5 and a PVE value of 14.18%, which is the highest among all QTLs reported here and showed an additive effect contributed by DQL 2034. The QTL for kernel opacity with the LOD of 4.06 on chromosome 7 between the loci umc1412 and umc1407 coincides with the QTL for kernel hardness, indicating its pleiotropic nature. Another QTL for kernel hardness was detected on chromosome 8 between marker loci bnlg1607 and umc1149, with an LOD of 2.99 and a PVE of 8.43% (Table 2). All the QTLs having higher LOD value (threshold value = 3) shows robust effect.
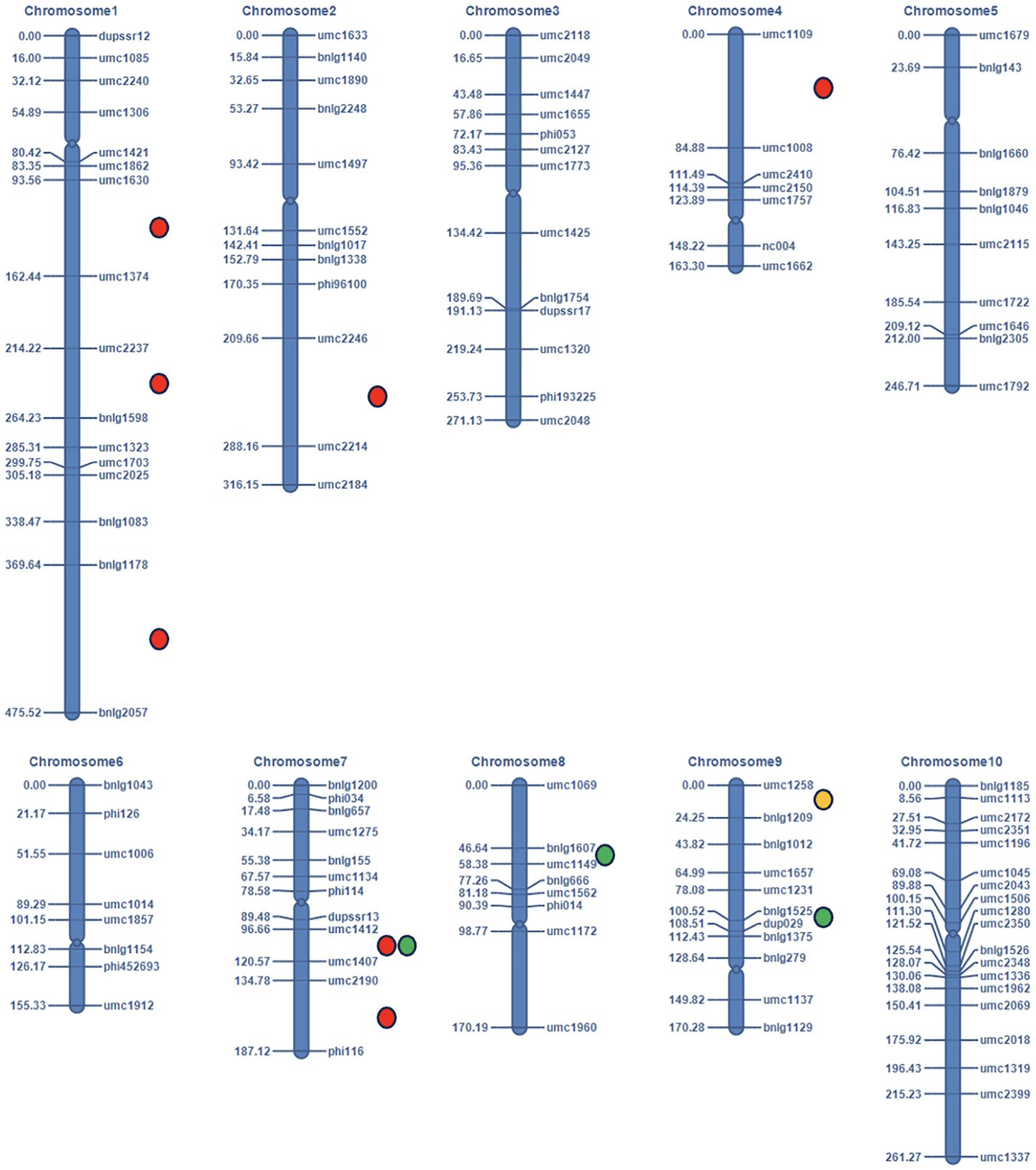
Figure 6. The linkage map (in cM) and location of the QTLs identified in the F2:3 mapping population of QPM maize using 116 polymorphic SSR markers (Colored circles are used to represent the QTLs for different traits: Red dots represent for kernel opacity, green for hardness, and yellow for tryptophan content).
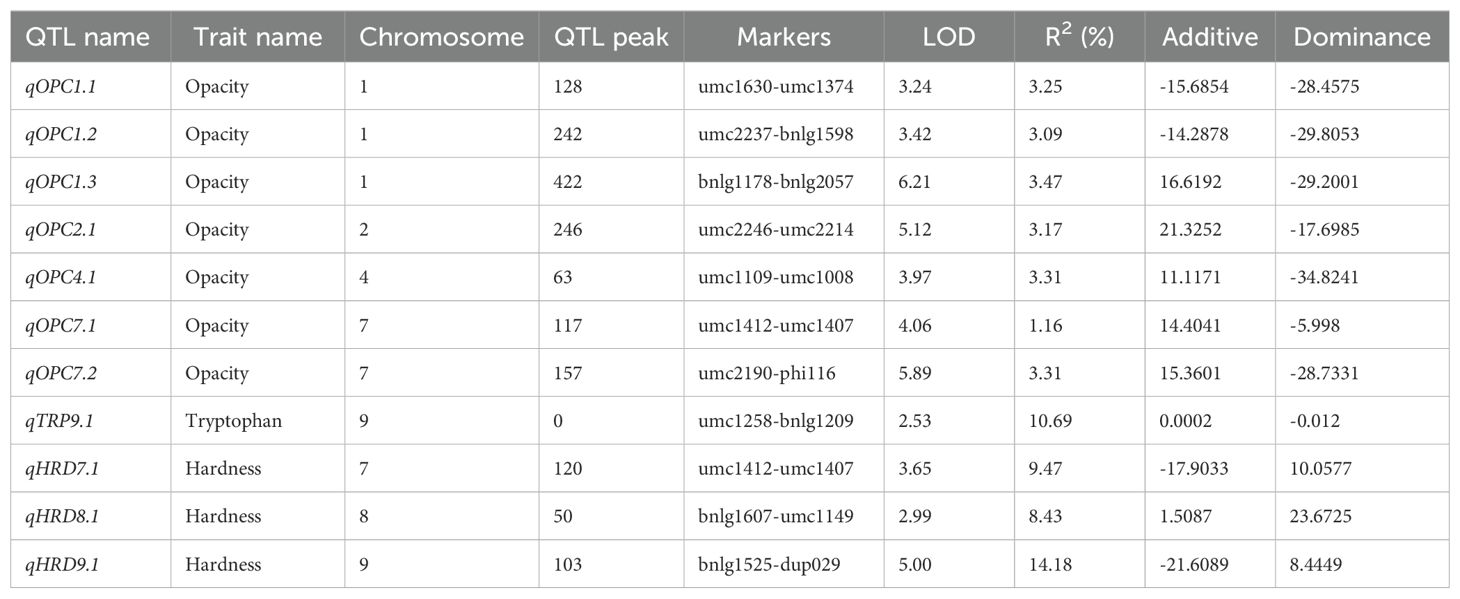
Table 2. The details of the identified QTLs for all three kernel traits with their LOD score, phenotypic variance (R2), additive effect, and dominance effect.
4 Discussion
The nature and position of endosperm modifiers are very complex in most of the QPM germplasm, and it has been observed that opaque2 modifiers often have an impact on several characteristics, including amino acids, kernel opacity, density, hardness, and vitreousness (Vasal et al., 1980; Gutierrez-Rojas et al., 2008; Holding et al., 2011). While opaque2 (o2) mutations elevate lysine and tryptophan by suppressing α- zein synthesis, modifier genes restore kernel hardness without compromising nutritional quality. These endosperm modifiers are present along the ten chromosomes of maize, with dosage effects and cytoplasmic effects, adding to its complexity (Babu et al., 2015). The main objective of this study was to identify the genomic regions influencing the o2 modifiers for three major traits, viz., kernel opacity, kernel hardness, and tryptophan content in an F2:3 mapping population derived from QPM inbreds DQL 2104-1 (high opacity, soft kernels) and DQL 2034 (low opacity, hard kernels. Kernel opacity was examined with the help of a lightbox, which is a reliable method used to differentiate individual kernels based on different degrees of modification. The seeds having 100% opacity resulted in a completely black, opaque region, whereas normal seeds did not show any opaqueness. The seeds were characterized depending on the degree of opaqueness at 100%, 75%, 50%, 25%, and 0%. The second trait, i.e., kernel hardness, was analyzed by the digital portable tablet hardness tester for 109 genotypes of the F2:3 population. About 52.29% of F2:3 genotypes were observed to have more hardness than the average. The third trait, tryptophan content, was estimated using a biochemical method, and it was observed that the 109 F2:3 population differed significantly.
A positive correlation (r = 0.29) between kernel opacity and tryptophan content was observed. The major storage proteins in maize are known as zeins, which are structurally divided into three types – alpha (α), beta (β), and gamma (γ) zeins. It has been studied that the o2 mutation is responsible for the downregulation of α-zein proteins, which results in opaque endosperm (Hunter et al., 2002). Other studies reported that mutations in the transcriptional activator of the o2 gene cause the concentrations of lysine and tryptophan to double (Henry et al., 2005; Maqbool et al., 2021). Conversely, the negative correlation between opacity and hardness was observed with r = - 0.74, underscoring the trade-off between nutritional quality and kernel density. This has been supported by numerous studies that have demonstrated a relationship between the phenotype of the kernel and the deposition of protein bodies during endosperm development (Tsai et al., 1978; Woo et al., 2001). Our result in this study is in agreement with previous studies suggesting a positive correlation between amino acid content and degree of opaqueness (Scott et al., 2004; Gutiérrez-Rojas et al., 2010; Babu et al., 2015) and a negative correlation between hardness and opacity (Holding et al., 2008; Kaur et al., 2020). However, a small number of genotypes were found to deviate from this pattern, which could be accounted for by the impact of endosperm modifiers in the population. The o2 mutation usually decreases the α-zein protein in the kernel, which results in a soft and opaque kernel with a high amino acid content. However recent research has revealed that a 15.26 kb duplicated section (qy27) at the 27 kDa locus of γ-zein is a key modifier of QTL that induces endosperm hardness and increases protein expression (Liu et al., 2016). Due to the modifier genes, the soft and dull kernels get converted into the hard and vitreous type without affecting the amino acid content, hence explaining the few deviations in our study pattern.
4.1 QTL analysis for kernel opacity
The genetic complexity of kernel opacity in maize is emphasized by the identification of seven QTLs acting as modifiers through their influence on starch metabolism, storage protein synthesis, and endosperm architecture. These are distributed across chromosomes 1, 2, 4, and 7 and are overlapped with functionally validated genes for kernel texture to nutritional quality. The QTL qOPC1.1 on chromosome 1 is correlated with the OHP1 gene (opaque2 heterodimerizing protein 1), a bZIP-type transcriptional factor (TF). OHP1 was first identified as a paralog of the O2-box motif in the promoters of zein genes, playing a critical role in regulating the 27-kDa gamma zein genes (Pysh et al., 1993; Pysh and Schmidt, 1996; Yang et al., 2016; Li and Song, 2020). Studies have shown that OHPs directly impact tryptophan content by synergistically coactivating the expression of zein genes, and its silencing reduces 27-kDa gamma-zein proteins (Zhang et al., 2015; Yang et al., 2016; Zhan et al., 2018). Therefore, OHP is crucial for 27 kDa-zein transcription, which promotes the concentration of amino acids in QPM lines and facilitates the synthesis of protein bodies and the efficient accumulation of zein storage protein (Li et al., 2018). This locus plays a dual role in maintaining vitreous endosperm while enhancing amino acid profiles. The QTL on chromosome 2, qOPC1.2 aligns with earlier studies reporting loci in bin 1.06 (umc1335) that influence lysine and kernel opacity/vitreousness (Gutierrez-Rojas et al., 2010; Salazar-Salas et al., 2014). The QTLs having lower PVE value, despite a robust LOD score may not have major affect but may have synergetic effect with other loci.
The identification of qOPC2.1 on chromosome 2 is a novel finding, as no QTL for kernel opacity has previously been reported on chromosome 2. This discovery highlights the complex nature of genetic modifiers and suggests allelic diversity in current maize germplasm unmasked this locus. This locus overlaps with three functionally characterized genes - namely, floury1 (fl1), opaque11 (o11), and mucronate 1 (Mc1) were found within the genomic interval of qOPC2.1. Each gene plays a distinct role in endosperm development and composition. The opaque11 (o11) gene encodes for the basic helix-loop-helix (bHLH) transcription factor unique to cereal endosperm. Mutation in o11 results in smaller, opaque endosperm that contains less starch and protein. Further, this transcription factor coordinates and regulates cell development and storage metabolism and has a positive role in starch accumulation (Feng et al., 2018). The mucronate (Mc) gene, another candidate within this interval, is a dominant mutation that reduces the amount of zein protein synthesis (a major class of maize storage protein) (Zhang and Boston, 1992; Kim et al., 2006). This reduction in zein protein disturbs the formation of protein bodies (PBs). The antagonistic relationship between these genes exemplifies how modifier loci balance starch and protein deposition. Previous studies have also demonstrated that the interaction of PBs, cytoplasm, and starch granules is important for the development of opaque endosperm (Wolf et al., 1967; Holding, 2014; Zhang et al., 2018).
A single QTL on chromosome 4, qOPC4.1, coincides with three important genes - floury2 (fl2), floury4 (fl4), and Opaque1 (O1). Semi-dominate mutation involving the fl2 and fl4 genes results in the formation of soft and starchy maize endosperm due to the accumulation of novel 24-kDa α-zein proteins, which disrupts the PB formation and starch-protein interactions (Lopes et al., 1994; Coleman et al., 1995). The O1, encoding a Myosin XI Motor Protein, regulates the endoplasmic reticulum (ER), influencing PB formation and endosperm texture. Mutation in O1 disrupts the ER and leads to the formation of PB formation and opaque kernels (Wang et al., 2012). The association of these loci with qOPC4.1 emphasizes its role as a key modifier of kernel opacity.
On chromosome 7, which was previously reported as a hub for kernel modifiers (Lopes et al., 1995; Holding et al., 2008; Babu et al., 2015), two additional loci, namely, qOPC7.1 and qOPC7.2, were mapped. It has been discovered that this QTL is pleiotropic, influencing both kernel opacity and kernel hardness. Also, this locus coincided with genomic regions reported in earlier studies (Holding et al., 2008), where markers like umc2190 were linked to kernel vitreousness. The second locus qOPC7.2, showed additive and dominant effects which aligned with previous studies (Holding et al., 2008).
These findings highlight the genetic complexity of kernel opacity but also suggest the direct impact of these loci on amino acid enrichment and starch-protein balance. While the study successfully identified QTLs associated with kernel opacity, the broad confidence interval (> 50 cM) observed for several QTLs reflects the limitation of low marker density and recombination events in the F2 population. To resolve this, further studies of fine mapping using advanced populations and high-density markers are required to narrow down intervals, enabling precise identification of candidate genes. These loci can be incorporated into predictive models to exploit additive effects for kernel opacity.
4.2 QTL analysis for kernel hardness
Three QTLs, viz., qHRD7.1, qHRD8.1, and qHRD9.1, were identified for kernel hardness on chromosomes 7, 8, and 9, respectively. The qHRD7.1 co-localized with the kernel opacity QTL on chromosome 7, suggesting that this locus could have a pleiotropic effect, a shared genetic mechanism influencing both hardness and opacity. Therefore, it is a strategic target for breeders aiming to simultaneously improve kernel texture and nutritional traits. On chromosome 8, a novel QTL (qHRD8.1) coincides with OHP3 (O2-heterodimerizing protein 3), another transcriptional regulator for zein genes. While previous QTL on chromosome 8 reported QTLs for texture, opacity, and vitreousness (Gutierrez-Rojas et al., 2010), qHRD8.1 also has a pleiotropic effect for both hardness and opacity. The most impactful locus, qHRD9.1, on chromosome 9 has the highest PVE among all identified QTLs and exhibits a strong additive effect contributed by parent DQL2034.
4.3 QTL analysis for tryptophan content
One highly effective QTL was identified for tryptophan content on chromosome 9, explaining a phenotypic variance of 10.69%, which is the second highest among the QTLs. This locus coincides with the QTL identified in a previous study (Babu et al., 2015). The locus co-localized with the waxy 1 (Wx1) gene, encoding for granule-bound starch synthase 1 (GBSS1), a key enzyme influencing both amino acid metabolism and starch composition (Babu et al., 2015). While Wx1 is traditionally linked with amylose synthesis and vitreous endosperm formation (Gibbon et al., 2003), our findings suggest a dual role in influencing tryptophan levels. This aligns with the prior studies (Babu et al., 2015), where Wx1 was linked to opaque2 modifiers, further validating its importance in maize improvement.
The major QTLs in our investigation are identified on chromosomes 1, 7, and 9, in accordance to past studies (Holding et al., 2008; Babu et al., 2015), established that these chromosomes are the major hub of the endosperm modifiers. These loci, including qHRD9.1 (chromosome 9, PVE = 14.18%) and qTRP9.1 (chromosome 9, PVE = 10.69%), which align with earlier findings that highlight the importance of γ-zein genes and their regulators in protein body formation and starch-protein interactions (Geetha et al., 1991; Lopes and Larkins, 1995; Burnett and Larkins, 1999; Hunter et al., 2002; Adunola et al., 2017). Protein 27-kDa γ-zein has been shown to be crucial for the development of protein bodies and the formation of the protein network that encircles starch grains in the endosperm (Dannenhoffer et al., 1995; Gutiérrez-Rojas et al., 2010; Pandey et al., 2016). Furthermore, it has been noted that variations in the polypeptide levels of the enzymes responsible for starch production and the branching pattern of starch have been explained by variations in phenotypes (Gibbon et al., 2003).
While the major genes could be utilized in Marker-Assisted Selection (MAS) and gene pyramiding, the minor genes (with low PVE value) may have epistasis effects to enhance traits as these QTLs could synergistically enhance both kernel hardness and lysine/tryptophan levels. Despite small individual effects, these QTLs have potential in breeding program. Notably, three loci qOPC7.1, qHRD7.1, qHRD8.1) may have pleiotropic effects, emphasizing there shared role in genetic mechanisms between kernel texture and nutritional traits. These loci could be used in pyramiding in elite lines. Genomic Selection (GS) can be used to integrate these loci into predictive models, allowing breeders an ability to exploit additive genetic variance across the genome.
In this study, low recombination event and low marker density contributed to broad confidence interval in QTL regions associated with endosperm modifications and amino acids. The current findings advance our understanding of the regions governing the endosperm modifiers in quality protein maize. To enhance the efficiency of QTL mapping, further strategies like fine mapping, haplotype analysis, genome-wide association studies (GWAS) can be done to get more detail information of genetics of endosperm modifiers. Additionally functional validation of candidate genes gives deeper insight into genetic regulation of endosperm modifications. This will accelerates the development of improved QPM varieties with improved nutritional quality an agronomical performance compared to traditional breeding methods.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author contributions
DJ: Conceptualization, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. SK: Investigation, Writing – review & editing. YK: Investigation, Writing – review & editing. AD: Conceptualization, Methodology, Project administration, Supervision, Writing – review & editing. AS: Methodology, Supervision, Writing – review & editing. DP: Methodology, Supervision, Writing – review & editing. SS: Investigation, Writing – review & editing. PS: Methodology, Supervision, Writing – review & editing. SR: Methodology, Supervision, Writing – review & editing. RK: Conceptualization, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The lab facilities and material provided by the director of the ICAR-Indian institute of Maize Research, PAU Campus, Ludhiana, are gratefully acknowledged.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1553512/full#supplementary-material
References
Adunola, P. M., Akinyele, B. O., Odiyi, A. C., Fayeun, L. S., and Akinwale, M. G. (2017). Introgression of opaque-2 gene into the genetic background of popcorn using marker assisted selection. Ondo State, Nigeria: Federal University of Technology, Akure.
Anonymous (2021). Package of practices for kharif crops (Ludhiana: Punjab Agricultural University), 26–31.
Arora, S., Kaur, S., Dhillon, G. S., Singh, R., Kaur, J., Sharma, A., et al. (2021). Introgression and genetic mapping of leaf rust and stripe rust resistance in Aegilops triuncialis. J. Genet. 100, 1–11. doi: 10.1007/s12041-020-01253-3
Babu, B. K., Agrawal, P. K., Saha, S., and Gupta, H. S. (2015). Mapping QTLs for opaque2 modifiers influencing the tryptophan content in quality protein maize using genomic and candidate gene-based SSRs of lysine and tryptophan metabolic pathway. Plant Cell Rep. 34, 37–45. doi: 10.1007/s00299-014-1685-5
Babu, R., Nair, S. K., Kumar, A., Venkatesh, S., Sekhar, J. C., Singh, N. N., et al. (2005). Two-generation marker-aided backcrossing for rapid conversion of normal maize lines to quality protein maize (QPM). Theor. Appl. Genet. 111, 888–897. doi: 10.1007/s00122-005-0011-6
Babu, R. and Prasanna, B. (2014). Molecular Breeding for Quality Protein Maize (QPM). In: Tuberosa, R., Graner, A., Frison, E. (eds) Genomics of Plant Genetic Resources. (Dordrecht: Springer), 489–505. doi: 10.1007/978-94-007-7575-6_21
Bjarnason, M. and Vasal, S. K. (1992). “Breeding of quality protein maize (QPM),” in Plant breeding reviews, vol. 9 . Ed. Janick (Wiley, London), 181–216.
Burnett, R. J. and Larkins, B. A. (1999). Opaque2 modifiers alter transcription of the 27-kDa γ-zein genes in maize. Mol. Genomics Genet. 261, 908–916. doi: 10.1007/s004380051038
Burr, B. and Burr, F. A. (1976). Zein synthesis in maize endosperm by polyribosomes attached to protein bodies. Proc. Natl. Acad. Sci. U.S.A. 73, 515–519. doi: 10.1073/pnas.73.2.515
Coleman, C. E., Clore, A. M., Ranch, J. P., Higgins, R., Lopes, M. A., and Larkins, B. A. (1997). Expression of a mutant α-zein creates the floury 2 phenotype in transgenic maize. Proc. Natl. Acad. Sci. U.S.A. 94, 7094–7097. doi: 10.1073/pnas.94.13.7094
Coleman, C. E., Lopes, M. A., Gillikin, J. W., Boston, R. S., and Larkins, B. A. (1995). A defective signal peptide in the maize high-lysine mutant floury 2. Proc. Natl. Acad. Sci. U.S.A. 92, 6828–6831. doi: 10.1073/pnas.92.15.6828
Dannenhoffer, J. M., Bostwick, D. E., Or, E., and Larkins, B. A. (1995). opaque-15, a maize mutation with properties of a defective opaque-2 modifier. Proc. Natl. Acad. Sci. U.S.A. 92, 1931–1935. doi: 10.1073/pnas.92.6.1931
Feng, F., Qi, W., Lv, Y., Yan, S., Xu, L., Yang, W., et al. (2018). OPAQUE11 is a central hub of the regulatory network for maize endosperm development and nutrient metabolism. Plant Cell 30, 375–396. doi: 10.1105/tpc.17.00616
Geetha, K. B., Lending, C. R., Lopes, M. A., Wallace, J. C., and Larkins, B. A. (1991). opaque-2 modifiers increase gamma-zein synthesis and alter its spatial distribution in maize endosperm. Plant Cell 3, 1207–1219. doi: 10.1105/tpc.3.11.1207
Gibbon, B. C. and Larkins, B. A. (2005). Molecular genetic approaches to developing quality protein maize. Trends Genet. 4, 227–233. doi: 10.1016/j.tig.2005.02.009
Gibbon, B. C., Wang, X., and Larkins, B. A. (2003). Altered starch structure is associated with endosperm modification in quality protein maize. Proc. Natl. Acad. Sci. U.S.A. 100, 15329–15334. doi: 10.1073/pnas.2136854100
Gupta, H. S., Agrawal, P. K., Mahajan, V., Bisht, G. S., Kumar, A., Verma, P., et al. (2009). Quality protein maize for nutritional security: rapid development of short duration hybrids through molecular marker assisted breeding. Curr. Sci., 230–237. Available at: chrome-extension://efaidnbmnnnibpcajpcglclefindmkaj/https://d1wqtxts1xzle7.cloudfront.net/54383107/Quality_protein_Maize_for_Nutritional_Se20170908-6308-9ky4ea-libre.pdf?1504931225=&response-content-disposition=inline%3B+filename%3DQuality_protein_maize_for_nutritional_se.pdf&Expires=1746628552&Signature=aFM6q2o8KdApfyvRzpsLQPNCbsplLGPhflgkuS7fsjTJwbiu2VpZe2AgteNfag3J0rN0O1tjVsO~B1nHjjd0WNBRGpjfCNg6z771fMaBIadSklHy5RyxnDSV8aAfomFkYRQEXA9LDnx71WW8z1ShIG9-Elh8vxh2mz5cqmPWJMPK73b4W-JTAwQLl2gYCoTT1TPm0qHjijEAcx~rWQfhBElyQ17gxr51dKiIP2te29FndLGbmqv8rzwwvTaBSR4UdfjvJpDFEMAQjHb0h-bgvTy9UX1Udxi8WmbeqfSyC3~ZzLdQW9diA9oBn0~dRsoJBxpLbA6P6Bdr5Xf6cB23BQ__&Key-Pair-Id=APKAJLOHF5GGSLRBV4ZA.
Gupta, H. S., Raman, B., Agrawal, P. K., Mahajan, V., Hossain, F., and Thirunavukkarasu, N. (2013). Accelerated development of quality protein maize hybrid through marker-assisted introgression of opaque-2 allele. Plant Breed. 132, 77–82. doi: 10.1111/pbr.12009
Gutiérrez-Rojas, A., Betrán, J., Scott, M. P., Atta, H., and Menz, M. (2010). Quantitative trait loci for endosperm modification and amino acid contents in quality protein maize. Crop Sci. 50, 870–879. doi: 10.2135/cropsci2008.10.0634
Gutierrez-Rojas, A., Scott, M. P., Leyva, O. R., Menz, M., and Betrán, J. (2008). Phenotypic characterization of quality protein maize endosperm modification and amino acid contents in a segregating recombinant inbred population. Crop Sci. 48, 1714–1722. doi: 10.2135/cropsci2007.08.0429
Henry, A. M., Manicacci, D., Falque, M., and Damerval, C. (2005). Molecular evolution of the opaque-2 gene in Zea mays L. J. Mol. Evol. 61, 551–558. doi: 10.1007/s00239-005-0003-9
Hernandez, H. H. and Bates, L. S. (1969). A modified method for a rapid tryptophan analysis in maize Vol. 13 (Mexico City, Mexico: CIMMYT Research Bulletin).
Holding, D. R. (2014). Recent advances in the study of prolamine storage protein organization and function. Front. Plant Sci. 5, 276. doi: 10.3389/fpls.2014.00276
Holding, D. R., Hunter, B. G., Chung, T., Gibbon, B. C., Ford, C. F., Bharti, A. K., et al. (2008). Genetic analysis of opaque2 modifier loci in quality protein maize. Theor. Appl. Genet. 117, 157–170. doi: 10.1007/s00122-008-0762-y
Holding, D. R., Hunter, B. G., Klingler, J. P., Wu, S., Guo, X., Gibbon, B. C., et al. (2011). Characterization of opaque2 modifier QTLs and candidate genes in recombinant inbred lines derived from the K0326Y quality protein maize inbred. Theor. Appl. Genet. 122, 783–794. doi: 10.1007/s00122-010-1486-3
Hossain, F., Prasanna, B. M., Kumar, R., and Singh, B. B. (2008). Genetic analysis of kernel modification in quality protein maize (QPM) genotypes. Indian J. Genet. Plant Breed. 68, 1–9. Available online at: https://www.isgpb.org/journal/index.php/IJGPB/article/view/1252.
Hunter, B. G., Beatty, M. K., Singletary, G. W., Hamaker, B. R., Dilkes, B. P., Larkins, B. A., et al. (2002). Maize opaque endosperm mutations create extensive changes in patterns of gene expression. Plant Cell 14, 2591–2612. doi: 10.1105/tpc.003905
Kaur, R., Kaur, G., Vikal, Y., Gill, G. K., Sharma, S., Singh, J., et al. (2020). Genetic enhancement of essential amino acids for nutritional enrichment of maize protein quality through marker assisted selection. Physiol. Mol. Biol. Plants 26, 2243–2254. doi: 10.1007/s12298-020-00897-w
Kim, C. S., Gibbon, B. C., Gillikin, J. W., Larkins, B. A., Boston, R. S., and Jung, R. (2006). The maize Mucronate mutation is a deletion in the 16-kDa γ-zein gene that induces the unfolded protein response 1. Plant J. 48, 440–451. doi: 10.1111/j.1365-313X.2006.02884.x
Kosambi, D. D. (2016). “The estimation of map distances from recombination values,” in DD Kosambi. Ed. Ramaswamy, R. (Springer, , New Delhi), 125–130. doi: 10.1007/978-81-322-3676-4_16
Krivanek, A. F., DeGroote, H., Gunaratna, N. S., Diallo, A. O., and Friesen, D. (2007). Breeding and disseminating quality protein maize (QPM) for Africa. J. Biotechnol. 6, 312–324. Available online at: https://www.cabidigitallibrary.org/doi/full/10.5555/20073208051.
Larkins, B. A. and Dalby, A. (1975). In vitro synthesis of zein-like protein by maize polyribosomes. Biochem. Biophys. Res. Commun. 66, 1048–1054. doi: 10.1016/0006-291X(75)90746-9
Lending, C. R., Wallace, J. C., and Larkins, B. A. (1992). Synthesis of zeins and their potential for amino acid modification. Plant Protein Eng. 1, 209. Available online at: https://books.google.com/books?hl=en&lr=&id=1jLTUizO1HwC&oi=fnd&pg=PA209&dq=Lending,+C.+R.,+Wallace,+J.+C.,+and+Larkins,+B.+A.+(1992).+Synthesis+of+zeins+and+their+potential+for+amino+acid+modification.+Plant+Protein+Eng.+1,+209.&ots=xJo8d5xjJS&sig=tGTldqfJzHbP5clno-_wjqwCSZE#v=onepage&q&f=false.
Li, C. and Song, R. (2020). The regulation of zein biosynthesis in maize endosperm. Theor. Appl. Genet. 133, 1443–1453. doi: 10.1007/s00122-019-03520-z
Li, C., Yue, Y., Chen, H., Qi, W., and Song, R. (2018). The ZmbZIP22 transcription factor regulates 27-kD γ-zein gene transcription during maize endosperm development. Plant Cell 30, 2402–2424. doi: 10.1007/s00122-019-03520-z
Liu, H., Shi, J., Sun, C., Gong, H., Fan, X., Qiu, F., et al. (2016). Gene duplication confers enhanced expression of 27-kDa γ-zein for endosperm modification in quality protein maize. Proc. Natl. Acad. Sci. U.S.A. 113 (18), 4964–9.
Lopes, M. A., Coleman, C. E., Kodrzycki, R., Lending, C. R., and Larkins, B. A. (1994). Synthesis of an unusual α-zein protein is correlated with the phenotypic effects of the floury2 mutation in maize. Mol. Gen. Genet. 245, 537–547. doi: 10.1007/BF00282216
Lopes, M. A. and Larkins, B. A. (1995). Genetic analysis of opaque2 modifier gene activity in maize endosperm. Theor. Appl. Genet. 91, 274–281. doi: 10.1007/BF00220889
Lopes, M. A., Takasaki, K., Bostwick, D. E., Helentjaris, T., and Larkins, B. A. (1995). Identification of two opaque2 modifier loci in quality protein maize. Mol. Gen. Genet. 247, 603–613. doi: 10.1007/BF00290352
Magulama, E. E. and Sales, E. K. (2009). Marker-assisted introgression of opaque2 gene into elite maize inbred lines. Res. Dev. 17, 131–135. Available online at: chrome-extension://efaidnbmnnnibpcajpcglclefindmkaj/https://d1wqtxts1xzle7.cloudfront.net/55275290/Marker-assisted_introgression_of_opaque_2_gene_into_elite_maize_inbred_lines-libre.pdf?1513129933=&response-content-disposition=inline%3B+filename%3DMarker_assisted_introgression_of_opaque.pdf&Expires=1746628009&Signature=U41kwl1M6oVuK1o5uwrl948bbj224NhdIaSkknerKDIoK6QS1yStOZJ9Nediu1-YhT7QMe3twfTqDPi-MSnE-4hAJ~sI1yY0QSktSigRKRgN-KU0DWA8zg29ml9tzpDFnYddrEEx-jS7OZhzgdIdmeGsi0KZ1rXWCFgB8Pme4f4v5a8W9NX4uXnQhvSNd-Epn5z5L2O4rR9ep3p50PMcNoJhxSOnDCTYItHbr69-pGnsp7ukKmAF9PnnNQxh1g6GQGZV9caFe8aKgWAgxu~mztXvqL~RSApAy0C8FAoJdKRxhosM~rr7n5bMrLdQpHfqvph5EKeOxEi5MzKHf7Eexg__&Key-Pair-Id=APKAJLOHF5GGSLRBV4ZA.
Maqbool, M. A., Beshir Issa, A., and Khokhar, E. S. (2021). Quality protein maize (QPM): importance, genetics, timeline of different events, breeding strategies and varietal adoption. Plant Breed. 140, 375–399. doi: 10.1111/pbr.v140.3
Mertz, E. T., Bates, L. S., and Nelson, O. E. (1964). Mutant gene that changes protein composition and increases lysine content of maize endosperm. Science 145, 279–280. doi: 10.1126/science.145.3629.27
Murray, M. G. and Thompson, W. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Or, E., Boyer, S. K., and Larkins, B. A. (1993). opaque2 modifiers act post-transcriptionally and in a polar manner on gamma-zein gene expression in maize endosperm. Plant Cell 5, 1599–1609. doi: 10.1105/tpc.5.11.1599
Paez, A. V., Helm, J. L., and Zuber, M. S. (1969). Lysine content of opaque-2 maize kernels having different phenotypes. Crop Sci. 9, 251–252. doi: 10.2135/cropsci1969.0011183X000900020045x
Pandey, N., Hossain, F., Kumar, K., Vishwakarma, A. K., Muthusamy, V., Saha, S., et al. (2016). Molecular characterization of endosperm and amino acids modifications among quality protein maize inbreds. Plant Breed. 135, 47–54. doi: 10.1111/pbr.12328
Pysh, L. D., Aukerman, M. J., and Schmidt, R. J. (1993). OHP1: a maize basic domain/leucine zipper protein that interacts with opaque2. Plant Cell 5, 227–236. doi: 10.1105/tpc.5.2.227
Pysh, L. D. and Schmidt, R. J. (1996). Characterization of the maize OHP1 gene: evidence of gene copy variability among inbreds. Gene 177, 203–208. doi: 10.1016/0378-1119(96)00302-2
Salazar-Salas, N. Y., Pineda-Hidalgo, K. V., Chavez-Ontiveros, J., Gutierrez-Dorado, R., ReyesMoreno, C., Bello-Pérez, L. A., et al. (2014). Biochemical characterization of QTLs associated with endosperm modification in quality protein maize. J. Cereal Sci. 60, 255–263. doi: 10.1016/j.jcs.2014.04.004
Sarika, K., Hossain, F., Muthusamy, V., Zunjare, R. U., Baveja, A., Goswami, R., et al. (2018). Opaque16, a high lysine and tryptophan mutant, does not influence the key physico-biochemical characteristics in maize kernel. PloS One 13, e0190945. doi: 10.1371/journal.pone.0190945
Scott, P., Bhatnager, S., Betrán, J. O., Scott, M. P., and Betran, J. (2004). Tryptophan and methionine levels in quality protein maize breeding germplasm. Maydica 49, 303–311. Available online at: chrome-extension://efaidnbmnnnibpcajpcglclefindmkaj/https://oaktrust.library.tamu.edu/server/api/core/bitstreams/925ce8cb-26ec-4980-8fe2-4c7ae9dcf453/content.
Shiferaw, B., Prasanna, B. M., Hellin, J., and Bänziger, M. (2011). Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food Secur. 3, 307–327. doi: 10.1007/s12571-013-0263-y
Tsai, C. Y., Huber, D. M., and Warren, H. L. (1978). Relationship of the kernel sink for N to maize productivity. Crop Sci. 18, 399–404. doi: 10.2135/cropsci1978.0011183X001800030011x
Vasal, S. K. (2000). The quality protein maize story. Food Nutr. Bull. 21, 445–450. doi: 10.1177/1564826500021004
Vasal, S. K., Villegas, E., Bjarnason, M., Gelaw, B., and Goertz, P. (1980). “Genetic modifiers and breeding strategies in developing hard endosperm opaque-2 materials,” in Improvement of quality traits of maize for grain and silage use. Eds. Pollmer, W. G. and Phipps, R. H. (Springer, Dordrecht, Netherlands), 37–73.
Villegas, E., Vasal, S. K., Bjarnason, M., and Mertz, E. T. (1992). Quality protein maize—what is it and how was it developed. Qual. Protein Maize, 27–48. Available online at: https://www.cabidigitallibrary.org/doi/full/10.5555/19931645130.
Vivek, B. S. (2008). Breeding quality protein maize (QPM): protocols for developing QPM cultivars (Mexico: CIMMYT).
Wang, X. L. and Larkins, B. A. (2001). Genetic analysis of amino acid accumulation in opaque-2 maize endosperm. Plant Physiol. 125, 1766–1777. doi: 10.1104/pp.125.4.1766
Wang, J., Li, H., Zhang, L., and Meng, L. (2016). User’s manual of QTL Ici-Mapping ver (Beijing/Mexico City: Quantitative Genetics Group, Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS)/Genetic Resources Program, International Maize and Wheat Improvement Center (CIMMYT).
Wang, G., Wang, F., Wang, G., Wang, F., Zhang, X., Zhong, M., et al. (2012). Opaque1 encodes a myosin XI motor protein that is required for endoplasmic reticulum motility and protein body formation in maize endosperm. Plant Cell 24, 3447–3462. doi: 10.1105/tpc.112.101360
Wolf, M. J., Khoo, U., and Seckinger, H. L. (1967). Subcellular structure of endosperm protein in high-lysine and normal corn. Science 157, 556–557. doi: 10.1126/science.157.3788.556
Woo, Y. M., Hu, D. W. N., Larkins, B. A., and Jung, R. (2001). Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. The Plant Cell 13 (10), 2297–2317.
Wu, Y., Holding, D. R., and Messing, J. (2010). γ-Zeins are essential for endosperm modification in quality protein maize. Proc. Natl. Acad. Sci. U.S.A. 107, 12810–12815. doi: 10.1073/pnas.1004721107
Wu, Y. and Messing, J. (2010). RNA interference-mediated change in protein body morphology and seed opacity through loss of different zein proteins. Plant Physiol. 153, 337–347. doi: 10.1104/pp.110.154690
Yang, J., Ji, C., and Wu, C. (2016). Divergent transactivation of maize storage protein zein genes by the transcription factors Opaque2 and OHPs. Genetics 204, 581–591. doi: 10.1534/genetics.116.192385
Yuan, L., Dou, Y., Kianian, S. F., Zhang, C., and Holding, D. R. (2014). Deletion mutagenesis identifies a haploinsufficient role for γ-zein in opaque2 endosperm modification. Plant Physiol. 164, 119–130. doi: 10.1104/pp.113.230961
Zhan, J., Li, G., Ryu, C. H., Ma, C., Zhang, S., Lloyd, A., et al. (2018). Opaque-2 regulates a complex gene network associated with cell differentiation and storage functions of maize endosperm. Plant Cell 30, 2425–2446. doi: 10.1105/tpc.18.00392
Zhang, F. and Boston, R. S. (1992). Increases in binding protein (BiP) accompany changes in protein body morphology in three high-lysine mutants of maize. Protoplasma 171, 142–152. doi: 10.1007/BF01403729
Zhang, Z., Yang, J., and Wu, Y. (2015). Transcriptional regulation of zein gene expression in maize through the additive and synergistic action of opaque2, prolamine-box binding factor, and O2 heterodimerizing proteins. Plant Cell 27, 1162–1172. doi: 10.1105/tpc.15.00035
Zhang, S. S., Zhan, J. P., and Yadegari, R. (2018). Maize opaque mutants are no longer opaque. Plant Reprod. 31, 319–326. doi: 10.1007/s00497-018-0344-3
Keywords: QTL mapping, quality protein maize (QPM), o2 endosperm modifiers, kernel opacity, kernel hardness, tryptophan content
Citation: Jasrotia D, Kumar S, Kaur Y, Das AK, Singh A, Paul D, Shukla S, Sharma P, Rakshit S and Kumar R (2025) Identification of QTLs associated with opaque2 modifiers influencing kernel opacity, kernel hardness, and tryptophan content in quality protein maize. Front. Plant Sci. 16:1553512. doi: 10.3389/fpls.2025.1553512
Received: 30 December 2024; Accepted: 21 April 2025;
Published: 21 May 2025.
Edited by:
Xuehai Zhang, Henan Agricultural University, ChinaReviewed by:
Jindong Liu, Chinese Academy of Agricultural Sciences, ChinaXiangjin Wei, China National Rice Research Institute (CAAS), China
Copyright © 2025 Jasrotia, Kumar, Kaur, Das, Singh, Paul, Shukla, Sharma, Rakshit and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ramesh Kumar, cmsucGhhZ25hQGdtYWlsLmNvbQ==
†Present address: Diksha Jasrotia, Plant and Environmental Sciences, Clemson University, SC, United States
‡ORCID: Ramesh Kumar, orcid.org/0000-0002-0350-3573
 Diksha Jasrotia1†
Diksha Jasrotia1† Abhijit Kumar Das
Abhijit Kumar Das Alla Singh
Alla Singh Dharam Paul
Dharam Paul Shanu Shukla
Shanu Shukla Priti Sharma
Priti Sharma Sujay Rakshit
Sujay Rakshit Ramesh Kumar
Ramesh Kumar