- 1Sanya Institute of Breeding and Multiplication, School of Tropical Agriculture and Forestry, Hainan University, Haikou, China
- 2Hainan Seed Industry Laboratory, Sanya, China
NODULE-INCEPTION-Like Protein (NLP) is a plant-specific transcription factor that plays a crucial role in nitrate uptake and stress responses. However, studies of NLP gene family in coconut are lacking. In this study, 7 NLP genes in the Aromatic Dwarf and 6 NLP genes in Hainan Tall coconut varities were identified. Furthermore, protein interaction network analysis suggested that CnD-NLP3/4/5 and CnT-NLP3/4/5 potentially interact with NRG2, HRS1, and LBD37, which may significantly influence nitrogen utilization in coconut, analysis of the promoters of CnD-NLPs and CnT-NLPs revealed many cis-acting elements associated with abiotic stress responses. Additionally, expression profiling showed that CnD-NLPs and CnT-NLPs were mainly expressed in mature leaves. Moreover, qRT-PCR analysis demonstrated that all identified CnD-NLPs responded to nitrogen deficiency, drought, and salt stress. Specifically, CnD-NLP5 may be a potentially critical gene in nitrogen deficiency responses, CnD-NLP4/5/6 and CnD-NLP1 may play a pivotal role in drought and salt stress response, respectively. And screening of homologous genes showed that CnD-NLP5 and AtNLP8, CnD-NLP4 and AtNLP7, CnD-NLP1 and OsNLP2 are homologous genes, with similar functions, further supporting their involvement in nitrogen deficiency, drought, and salt stress responses. These results lay the foundation for future research on the NLPs function and potential applications in coconut.
1 Introduction
The coconut (Cocos nucifera L., 2n=32), a member of the Palmaceae family, is an important perennial tropical fruit and oil crop, often revered as the “tree of life” due to its extensive economic value (Beveridge et al., 2022). Thriving in tropical coastal regions, coconut trees are primarily categorized into two types-dwarf and tall-based on their morphological and growth characteristics (Beveridge et al., 2022; Mat et al., 2022).
Tall coconuts, which can grow up to 20-30 meters, are slower to mature, typically bearing fruit after 6-10 years (Harries, 1978; DebMandal and Mandal, 2011). In contrast, dwarf coconuts, known for their rapid growth, reach heights of 10-15 meters and start bearing numerous small nuts within just 4-5 years (Swaminathan and Nambiar, 1961). This early productivity makes them a preferred choice for breeding new varieties (Xiao et al., 2017). However, as global climate change, coconut palms are facing increasing challenges from abiotic stresses such as nitrogen deficiency, drought, and salt stress. These stresses severely impact their growth and productivity (Sousa Santos et al., 2020). For example, nitrogen deficiency in coconut plants leads to yellowing of mature leaves and a reduction in the number of female flowers, which ultimately hinders their normal growth and productivity (Broschat, 2009; Mohandas, 2012; Ramírez-Silva et al., 2021). Concurrently, coconut palms, predominantly distributed in tropical coastal regions, are susceptible to the challenges posed by drought and saline stress. Drought and salt stress initially leads to the death of leaf margins and tips, followed by wilting and yellowing (Wang et al., 2020; Samarasinghe et al., 2021; Yang et al., 2021). Interestingly, tall coconut varieties tend to exhibit greater drought tolerance compared to dwarf varieties (Sousa Santos et al., 2020), as well as the Hainan tall coconut, a local variety from Hainan Island, exhibits better salt tolerance than certain dwarf coconut varieties like aromatic coconut, yellow dwarf coconut and red dwarf coconut (Sun et al., 2024).
The NODULE-INCEPTION-Like Protein (NLP), a transcription factor unique to plants, is crucial for the transduction of nitrate signals and their assimilation, as well as for coping with abiotic stress and modulating plant growth and development (Konishi and Yanagisawa, 2014; Jia et al., 2023). In various plant species, including Arabidopsis, rice, apple, and tea, 9, 6, 6, and 33 NLP gene have been identified, respectively (Schauser et al., 2005; Wang et al., 2019; Jagadhesan et al., 2020; Li et al., 2024). The NLP gene mainly contains two conserved domains: the RWP-RK domain, which is responsible for DNA binding and specifically interacts with nitrate response cis-acting elements in the promoter region of the target genes (Chardin et al., 2014), and the PB1 domain, which is involved in nitrate-induced protein-protein interaction (Konishi and Yanagisawa, 2019). Additionally, a highly conserved GAF domain has been found at the N-terminal of some NLP proteins, which linked to signal transduction (Ho et al., 2000). The tissue-specific expression patterns of NLP genes in tomato, watermelon, and Brassica napus indicated that the expression patterns of NLP genes vary among different plant species. Specifically, NLP genes in tomato and watermelon are predominantly expressed in roots and leaves (Liu et al., 2021; Yuan et al., 2022), whereas in Brassica napus, NLPs are mainly expressed in leaves (Liu et al., 2018).
Functional studies have confirmed the key role of NLP genes in nitrogen response and assimilation processes. For instance, in Arabidopsis, AtNLP regulates primary nitrogen responses by binding to the nitrogen-responsive cis-acting elements (NRES) in the promoter region of target genes (Konishi and Yanagisawa, 2013). AtNLP2 plays a critical role in nitrogen assimilation processes based on the availability of nitrate in the soil (Durand et al., 2023). In addition, AtNLP7 interacts with key genes in the nitrogen metabolism pathway, such as ANR1, NRT1.1, NRT2, and LBD37/38, to regulate the expression of downstream genes (Marchive et al., 2013; Yu et al., 2016). AtNLP8 has been demonstrated to directly bind to the promoter of abscisic acid catabolic enzyme genes, thereby reducing abscisic acid levels in a nitrate-dependent manner, and consequently regulating nitrate-promoted seed germination (Yan et al., 2016). Under conditions of persistent nitrate availabiliy and nitrogen starvation, AtNLP6 and AtNLP7 interact with another key transcriptional regulator TCP20 and play an important role in regulating the expression of nitrate response genes (Guan et al., 2017; Kim et al., 2023). In rice, overexpression of OsNLP1 has been associated with enhanced plant growth, increased grain yield, and improved nitrogen use efficiency, while the knockout of the OsNLP1 gene under nitrogen restriction leads to a reduction in both grain yield and nitrogen utilization efficiency (Alfatih et al., 2020). Similarly, under conditions of nitrogen deficiency, the expression of SlNLP1/2/4/6 genes in tomato was up-regulated, underscoring their important role in plant’s response to nitrogen limitation (Liu et al., 2021).
Notably, NLPs gene are involved in plant response to abiotic stress. For example, AtNLP7 mutants are more tolerant to drought and salt than wild-type plants (Castaings et al., 2009; Le et al., 2022). Elephant grass and alfalfa studies have demonstrated that NLP genes respond to abiotic stresses (Jin et al., 2023; Yu et al., 2023). Specifically, CpNLP5.2 in elephant grass exhibits rapid induction under drought and salt stress conditions (Jin et al., 2023). Similarly, in alfalfa, the expression of MsNLP24 gene increases over time in response to drought and salt stress. In contrast, MsNLP48 gene expression initially decreases, subsequently increases, but remains lower than the initial level, with the lowest expression observed at 6 h post-treatment (Yu et al., 2023). This suggests that salt and drought stress significantly induced or inhibited the expression of MsNLPs genes.
Despite the advances in understanding the role of NLP transcription factors in various biochemical and physiological processes in plants, there remains a significant gap regarding the function of NLP transcription factors in coconut. Comprehensive identification and systematic analysis of NLP gene family in coconut have yet to be conducted. Therefore, this study aims to systematically investigate the evolutionary characteristics, tissue-specific expression patterns, and responses to nitrogen deficiency, drought, and salt stress of NLP gene family members at the genome-wide level using the genomes of Aromatic Dwarf and Hainan Tall coconut. Additionally, we will explore the evolutionary relationship between dwarf coconut and tall coconut to identify the key genes involved in stress responses. Our goal is to provide insights into the evolutionary history and potential functional diversity of NLP gene in coconut.
2 Materials and methods
2.1 Identification and analysis of CnNLP family
Genome-wide data and gene structure annotation files of Aromatic Dwarf and Hainan Tall coconut varieties were downloaded from the China National Center for Bioinformation (https://ngdc.cncb.ac.cn/gwh/; accessed on 19 September 2023) under accession numbers GWHBEBT00000000 and GWHBEBU00000000, respectively (Wang et al., 2021). Protein sequences of NLP gene family members in Arabidopsis thaliana were sourced from The Arabidopsis Information Resource Database (https://www.arabidopsis.org/; accessed on 19 September 2023), and the candidate NLP proteins in coconut was identified using BLAST search. Furthermore, Hidden Markov Models (HMMs) for the RWP-RK (PF02042) and PB1 (PF00564) conserved domain of NLP protein were downloaded from the Pfam Database (https://www.ebi.ac.uk/interpro/entry/pfam/; accessed on 19 September 2023). The HMM search was performed with an E value threshold set at 1×10-5 to identify candidate NLP proteins in the coconut genome. To ensure accuracy, all candidate protein sequences obtained by the two methods were further verified using the NCBI Batch CD-search (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi; accessed on 19 September 2023).
The physicochemical properties of NLP gene family members in coconut, including the length of amino acids, molecular weight, isoelectric point, instability index, and grand average of hydropathicity (GRAVY), were analyzed using the ExPASy online tool (http://web.expasy.org/protparam/; accessed on 19 September 2023). The subcellular localization of these genes was predicted with the Plant-mPLoc online tool (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/; accessed on 19 September 2023). Furthermore, the chromosome gene density and the chromosome location of NLP genes in coconut were visualized using TBtools-II v2.086 (Chen et al., 2023).
2.2 Prediction of CnNLP protein structure
The secondary structure of NLP protein was predicted using the SOPMA online tool (https://npsa.lyon.inserm.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html; accessed on 12 October 2023). Additionally, the tertiary structure of NLP protein was predicted by SWISS-MODEL online tool (https://swissmodel.expasy.org/interactive; accessed on 12 October 2023).
2.3 Prediction of interaction network of CnNLP protein
The interactions among NLP family member proteins were predicted using the functional protein association network online database STRING (https://cn.string-db.org/; accessed on 12 October 2023), with the model species selected as Arabidopsis.
2.4 Phylogenetic analysis of CnNLP
Download the protein sequences of NLP gene family in rice from the Rice Genome Annotation Project Database (http://rice.uga.edu/; accessed on 4 October 2023). Additionally, protein sequences of the validated NLP gene family from Elaeis guineensis, Phoenix dactylifera, Spirodela polyrhiza, Ananas comosus, Musa acuminate, Vitis vinifera, Pyunus persica, Theobroma cacao, Sorghum bicolor, Glycine max, and Populus trichocarpa were obtained from the Plant Transcription Factor Database (https://planttfdb.gao-lab.org/; accessed on 4 October 2023). The evolutionary tree was constructed using Neighbour-Joining method with 1000 bootstrap replicates, employing MEGA11 software (Tamura et al., 2021). The FigTree v1.4.4 software (Andrew Rambaut, IEB, UoE, UK) was utilized to visualize and analyze the evolutionary relationships.
2.5 Structural characteristics and conservative motifs of CnNLP
The evolutionary tree was constructed by Neighbour-Joining method (1000 bootstrap replicates) in MEGA11 software. The conserved motifs of NLP gene family proteins in coconut were analyzed using the online MEME tool (https://meme-suite.org/meme/tools/meme; accessed on 5 October 2023) with the number of conserved motifs set to 10 and the other parameters left at their default values. The exon/intron information of NLP gene, including mRNA, coding sequence (CDS), and untranslated region (UTR), was extracted from the coconut genome structure file.
2.6 Collinearity analysis of CnNLP
The KaKs_Calculator 2.0 (Wang et al., 2010) was utilized to evaluate the nucleotide substitution parameters of duplication genes: non-synonymous substitution rate (Ka) and synonymous substitution rate (Ks), and their ratio (Ka/Ks) was calculated. Using the method of Yao et al (Yao et al., 2021), the gene duplication events within the NLP family in coconut and the collinearity analysis among Arabidopsis thaliana, Oryza sativa, Ananas comosus, Elaeis guineensis, and Cocos nucifera were evaluated respectively. After that, we analyzed the origin within the CnNLP gene family using BLAST comparison. The genome-wide data and gene structure annotation file of Ananas comosus were obtained from the Phytozome Database (https://phytozome-next.jgi.doe.gov/; accessed on 5 October 2023), and those for Elaeis guineensis were downloaded from the National Center for Biotechnology Information Database (https://www.ncbi.nlm.nih.gov/; accessed on 5 October 2023).
2.7 Analysis of CnNLP gene promoter
The 2000 bp sequence upstream of the gene transcription start site was extracted as a putative promoter region. Subsequently, the extracted sequence was submitted to PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/; accessed on 4 October 2023) for cis-acting element prediction (Wu et al., 2021; Rao et al., 2021).
2.8 Plant growth and treatments
The experiment was conducted in December 2023 at Sanya Breeding Institute of Hainan University (18° 30’ N, 109° 60’ E). Coconut seedlings cultivated for one year (Cocos nucifera L. cv. Aromatic Dwarf and Cocos nucifera L. cv. Hainan Tall) were selected as the experimental materials. Fresh samples of roots, petioles, young, and mature leaves from both Aromatic Dwarf and Hainan Tall seedlings were collected, with three replicates for each treatment, and then stored at -80°C. In addition, Aromatic Dwarf seedlings were incubated in clean water for 3 days before being transferred to an abiotic stress environment for further cultivation. The treatments included nitrogen deficiency stress using nitrogen-deficient Hoagland nutrient solution, salt stress with a normal Hoagland nutrient solution containing 300 mmol/L NaCl, and drought stress using a normal Hoagland nutrient solution with 25% PEG6000. Roots, petioles, young, and mature leaves were collected at 0, 3, 6, 12, 24, and 48 h post-treatment, respectively, and stored in the refrigerator at -80 °C after being preserved liquid nitrogen.
2.9 qRT-PCR analysis and homologous gene screening
Total RNA was extracted using an RNA extraction kit according to the manufacturer’s protocol (Tiangen, Beijing, China, DP437). RNA was reverse-transcribed into cDNA using HiScripv II Q RT SuperMix for quantitative real-time PCR (qRT-PCR) (Vazyme, Nanjing, China, R223). The SYBR Green I chimeric fluorescence method was employed for quantitative qRT-PCR reaction on qTOWER3G Real-Time PCR Thermal Cycler (Analytik Jena, Germany). The β-actin gene of coconut was selected as the internal control. The relative expression was calculated by 2−ΔΔCTmethod. Three biological replicates were set up for each sample. The primers used in this experiment are listed in Supplementary Table S1. Homologous genes of NLP in coconut were screened using the Quick Find Best Homology functions in TBtools (Chen et al., 2023). Building an evolutionary tree using MEGA11 software (Tamura et al., 2021).
2.10 Statistical analysis
All data were statistically analyzed using Microsoft Excel 2019 (Microsoft, Redmond, WA, USA). The analysis software Statistix 9 (Analytical Software, Tallahassee, FL, USA) was used to conduct one-way ANOVA. Graphs in this study were visualized using GraphPad Prism 8.0.2.263 (GraphPad Software Inc., San Diego, CA, USA), Origin 2024 (OriginLab Corporation, Northampton, MA, USA), and TBtools-II v2.086 (Chen et al., 2023).
3 Results
3.1 Identification and basic characteristics of CnNLP transcription factor family
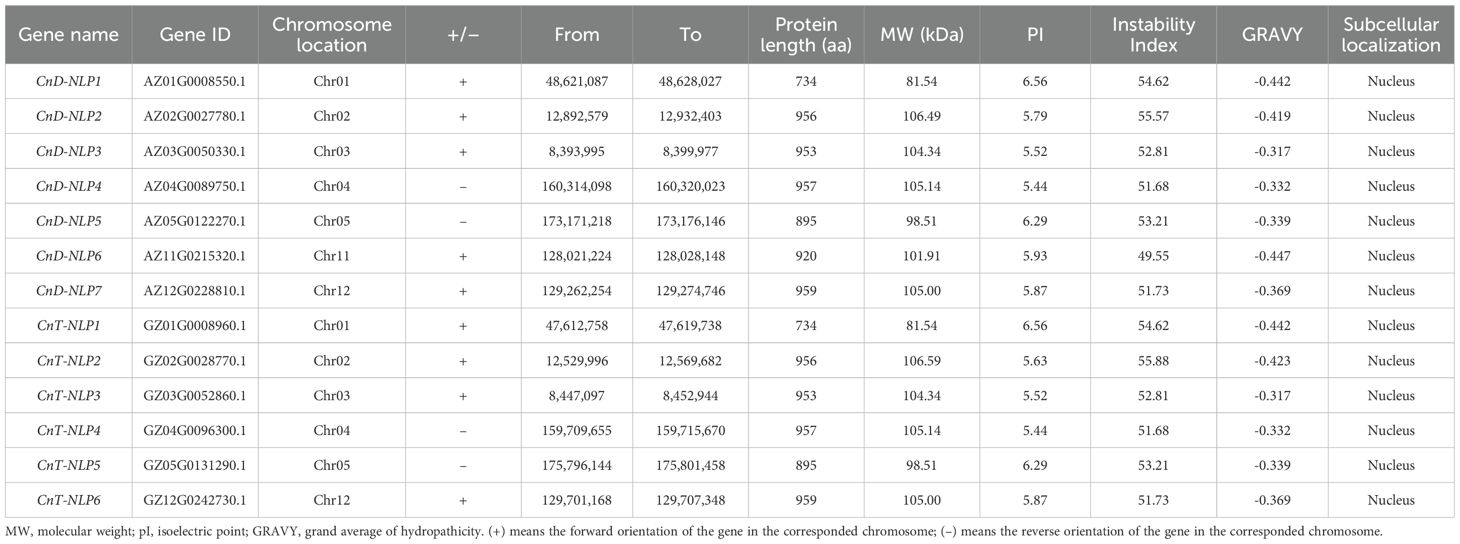
After performing BLAST sequence alignment, HMM search, redundancy removal, and protein domain screening, 7 NLP genes were identified in Aromatic Dwarf and 6 NLP genes in Hainan Tall. Based on their chromosomal positions (Figure 1), the 7 NLP genes in Aromatic Dwarf were designated as CnD-NLP1 to CnD-NLP7, while the 6 NLP genes in Hainan Tall were designated as CnT-NLP1 to CnT-NLP6. These genes were distributed across different chromosomes, with most of them located near the chromosome ends (Figure 1). Protein characterization analysis revealed that the CnD-NLP gene family members have amino acid lengths ranging from 734 (CnD-NLP1) to 959 (CnD-NLP7) and relative molecular weights between 81.54 kDa (CnD-NLP1) and 106.49 kDa (CnD-NLP2). The isoelectric points of these proteins range from 5.44 (CnD-NLP4) to 6.56 (CnD-NLP1), and their instability indices range from 49.55 (CnD-NLP6) to 55.57 (CnD-NLP2). The grand average of hydropathicity (GRAVY) values for these proteins range from -0.447 (CnD-NLP6) to -0.317 (CnD-NLP3). Similarly, the CnT-NLP gene family encodes proteins with amino acid lengths from 734 (CnT-NLP1) to 959 (CnT-NLP6) and relative molecular weights between 81.54 kDa (CnT-NLP1) and 106.59 kDa (CnT-NLP2). Their isoelectric points range from 5.44 (CnT-NLP4) to 6.56 (CnT-NLP1), and their instability indices range from 51.68 (CnT-NLP4) to 55.88 (CnT-NLP2), with GRAVY values ranging from -0.442 (CnT-NLP1) to -0.317 (CnT-NLP3). Both CnD-NLP and CnT-NLP gene family members are characterized as acidic proteins with isoelectric points below 7, unstable proteins with instability indices above 40, and hydrophilic proteins with negative GRAVY values. According to Plant-mPLoc prediction, all CnD-NLPs and CnT-NLPs proteins are localized in the nucleus (Table 1).
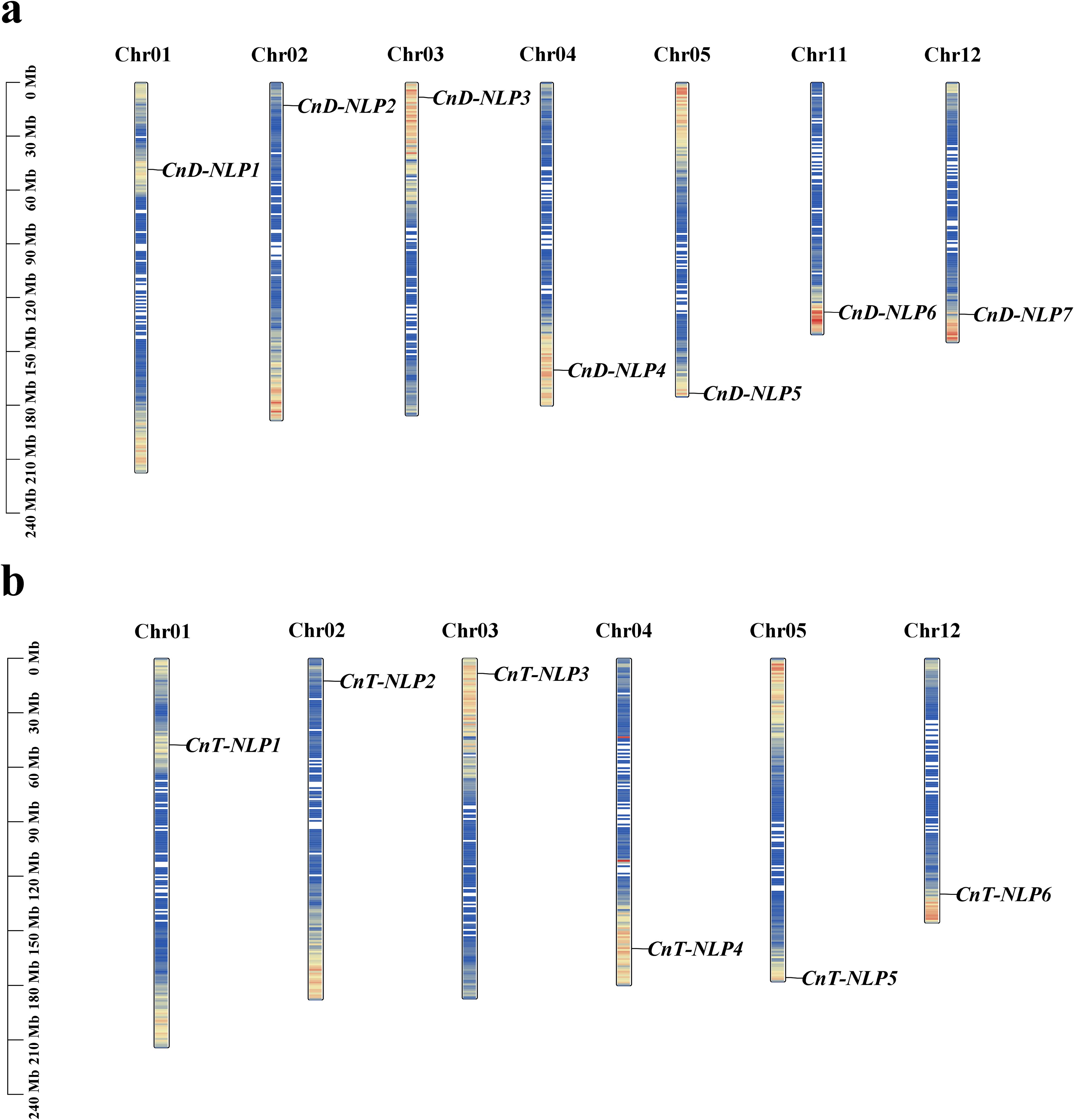
Figure 1. Chromosome location and distribution of NLP gene in Aromatic Dwarf (a) and Hainan Tall (b). These bars represent chromosomes. The chromosome number is displayed at the top and the gene name is displayed on the right. The relative chromosomal location of each NLP gene is located on the black line on the right. Blue to red on the chromosome indicates gene density.
3.2 Structure and interaction network analysis of CnNLP protein
The secondary structure prediction of CnNLP proteins revealed that the 13 NLP proteins were mainly consisted of random coil and α-helix, with random coils being the most prevalent, ranging between 62.49% to 66.63%. In addition, the secondary structure included extended strands and β-turn, with β-turn representing the smallest proportion among all NLP members, ranging from 0.84% to 1.77%. Notabaly, the proportion of these secondary structures were largely consistent across the proteins (Figures 2a, b; Supplementary Table S2). Using SWISS-MODEL, we predicted the tertiary structures of these proteins. The GMQE values for the 13 models ranged from 0.59 to 0.62, with sequence similarities exceeding 70% (Supplementary Table S3), indicating that the predicted tertiary structures are reliable. The structural analysis showed that there are certain differences in the tertiary structure among proteins within the same variety, suggesting potential diversity among the members of the NLP gene family. Furthermore, we found that CnD-NLP1 and CnT-NLP1, CnD-NLP2 and CnT-NLP2, CnD-NLP3 and CnT-NLP3, CnD-NLP4 and CnT-NLP4, CnD-NLP5 and CnT-NLP5, CnD-NLP7 and CnT-NLP6 are homologous genes with highly similar sequences and the same predicted tertiary structure (Figure 2c), indicating that these may have similar functions.
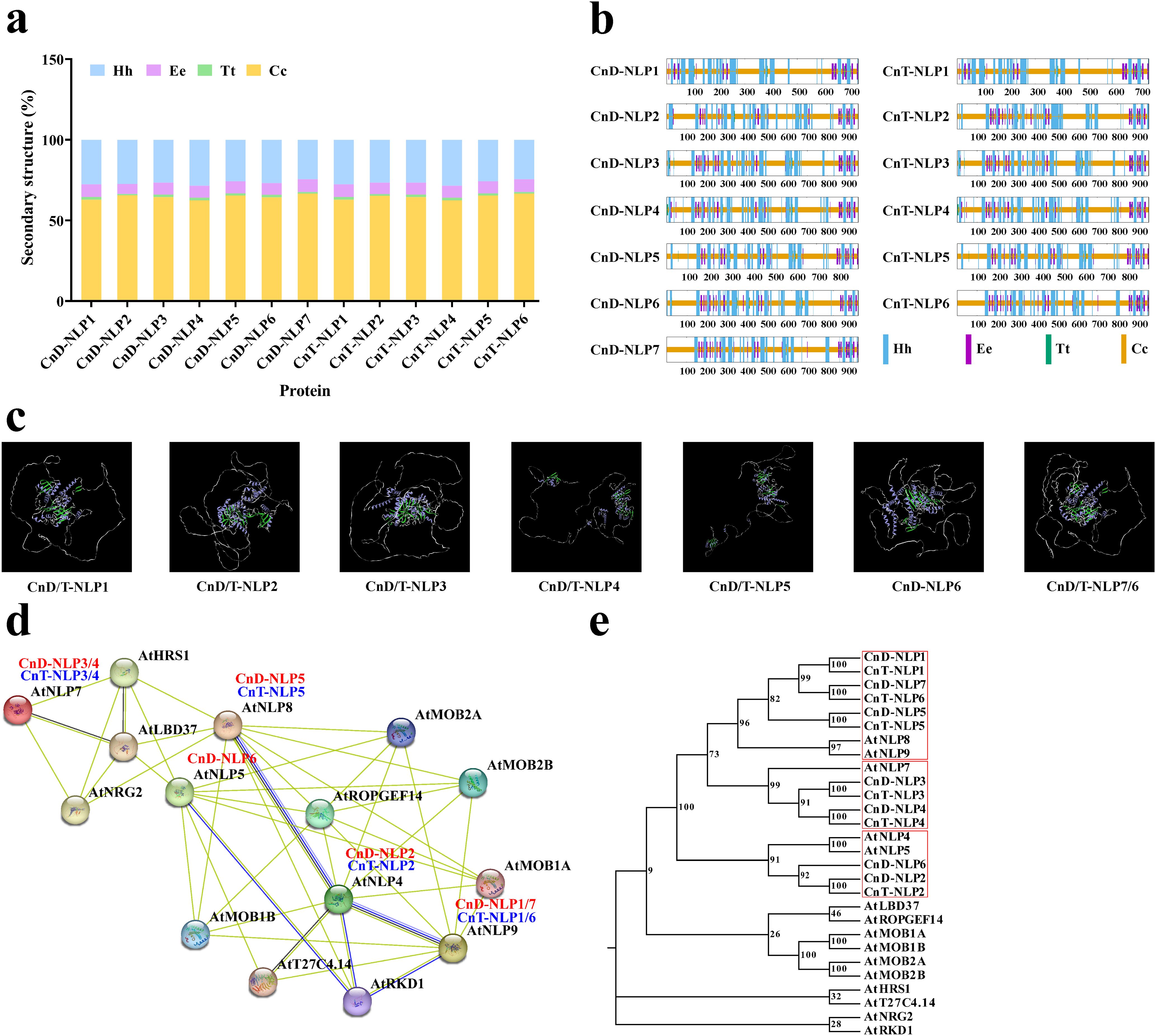
Figure 2. Protein structure and protein interaction network of NLP in coconut. (a, b) Proportion and distribution of secondary structure element. Hh: Alpha helix; Ee: Extended strand; Tt: Beta turn; Cc: Random coil. (c) The predicted tertiary structure of NLP protein in Aromatic Dwarf and Hainan Tall. Purple represents the Alpha helix. Green represents the β-sheet. Gray represents the Random coil. (d) Protein interaction network of NLP in Aromatic Dwarf and Hainan Tall constructed by referring to AtNLPs. The different data sources of predicted interactions are indicated by lines with diverse colors. Dark blue represents gene co-occurrence. Chartreuse represents text mining. Black represents co-expression. Light blue represents protein homolog. (e) Phylogenetic analysis of NLP proteins from coconut, Arabidopsis, and their interacting proteins.
To predict the potential function of NLP gene in coconut, we constructed an interaction network of coconut NLP proteins using the homologous sequences of the model species Arabidopsis thaliana, employing the STRING database (Figure 2d). Subsequently, we constructed a phylogenetic tree that includes the NLP proteins of Arabidopsis and their interacting proteins, as well as the NLP protein sequences from coconut (Figure 2e). In this network, CnD-NLP3/4 and CnT-NLP3/4 corresponds to AtNLP7, CnD-NLP5 and CnT-NLP5 corresponds to the AtNLP8 (Figures 2d, e; Supplementary Table S4). The network diagram illustrates that the AtNLP7/8 protein can interact with the nitrate-regulating gene AtNRG2, as well as transcription factors AtHRS1 and AtLBD37, among others. Based on these interactions, we predict that CnD-NLP3/4/5 and CnT-NLP3/4/5 play key roles in nitrogen utilization in coconut.
3.3 Phylogenetic and structural analysis of NLP gene
To evaluate the evolutionary relationship among plant NLP genes, we constructed a phylogenetic tree using 111 NLP amino acid sequences from 15 angiosperms species encompassing both monocots and dicots (Figure 3a; Supplementary Table S5). Following the classification from Arabidopsis thaliana (Schauser et al., 2005), the NLP family was divided into three groups: Group I (in green) includes CnD-NLP2/6 and CnT-NLP2; Group II (in purple) includes CnD-NLP3/4 and CnT-NLP3/4; and Group III (in orange) includes CnD-NLP1/5/7 and CnT-NLP1/5/6. Furthermore, the dendrogram also revealed closely related orthologs among Cocos nucifera L. cv. Aromatic Dwarf, Cocos nucifera L. cv. Hainan Tall, Elaeis guineensis, and Phoenix dactylifera (for instance, CnD-NLP1/CnT-NLP1/EgNLP3/PdNLP2), suggesting that these genes may share similar functions. To further understand the evolution of NLP genes, we counted the number of NLP genes in 15 herbaceous and woody plants (Figure 3b). The number of NLP genes varied among the plants, with Populus trichocarpa having the highest number (14), while Spirodela polyrhiza (4), Vitis vinifera (4), Pyunus persica (4), and Theobroma cacao (4) each having the lowest numbers.
Figure 3. Phylogenetic and structural analysis of NLP genes. (a) Phylogenetic analysis of the NLP proteins from different species. The phylogenetic tree was divided into three groups, each with a different colour. The values in the branch represent bootstrap values. (b) Statistics of NLP gene numbers across 15 plant species. (c) Phylogenetic tree, gene structure and conserved motif of NLP gene family in coconut. UTR, untranslated region; CDS, coding sequence.
The exon-intron distribution of the CnNLP genes was analyzed, revealing minor variations in the number of exons and introns among different genes The CnNLP genes contained four to five exons and three to six introns. Notably, Group II and Group III of CnNLP genes each contained five exons, whereas Group I of CnNLP gene contained four exons. Additionally, the number of CnNLP exons within the same group was consistent across the genes (Figure 3c). Furthermore, the sequence motifs in CnNLP were analyzed using the MEME website, and 10 motifs were identified, named as Motif1-Motif10 (Figure 3c; Supplementary Figure S1). Most genes contained all 10 conserved motifs, except for CnD-NLP4 and CnT-NLP4 (which lacked Motif10 and thus had only 9 conserved motifs), CnD-NLP1 and CnT-NLP1 (which lacked Motif7 and also had only 9 conserved motifs).
3.4 Collinear analysis of CnNLP
Collinearity analysis found that seven members of the CnD-NLP gene family have four repetitive gene pairs (Figure 4a), and six members of the CnT-NLP gene family have four repetitive gene pairs (Figure 4b). To further explore the evolutionary origins of the CnNLP gene family, we selected chromosomal fragments containing the CnNLP gene to identify regions of collinearity. Our analysis revealed that the fragment containing CnD-NLP1 on chromosome 01 exhibited high collinearity with the fragment containing CnD-NLP7 on chromosome 12. Similarly, the fragment containing CnD-NLP2 on chromosome 02 was found to be highly collinear with the fragment containing CnD-NLP6 on chromosome 11, and the fragment containing CnD-NLP3 on chromosome 03 was highly collinear with the fragment containing CnD-NLP4 on chromosome 04. Notably, the fragment containing CnD-NLP5 on chromosome 05 was not identified (Supplementary Figure S2a). Subsequently, we compared the chromosomal fragment containing the CnNLP gene with that containing the AcNLP gene. This comparison demonstrated that the fragment containing CnD-NLP1 on chromosome 01 was highly collinear with the fragment containing AcNLP1 on chromosome 17. The fragment containing CnD-NLP2 on chromosome 02 was highly collinear with the fragment containing AcNLP6 on chromosome 03, and the fragment containing CnD-NLP3 on chromosome 03 was highly collinear with the fragment containing AcNLP3 on chromosome 11. Additionally, the fragment containing CnD-NLP5 on chromosome 05 was found to be highly collinear with the fragment containing AcNLP4 on chromosome 01 (Supplementary Figure S2b). Therefore, we conclude that CnD-NLP5 likely originated prior to species differentiation, while other members of the CnD-NLP gene family may have originated from whole-genome duplication (WGD) events. Similarly, the CnT-NLP gene family also showed similar patterns of collinearity, leading us to inferr that CnT-NLP2 and CnT-NLP5 may have originated before species differentiation, while other members of CnT-NLP gene family potentially originated from WGD events (Supplementary Figures S2c, d). Further analysis of the selection types for these repetitive gene pairs whthin the CnD-NLP and CnT-NLP gene families revealed that the Ka/Ks values of all repetitive gene pairs were less than 1, indicating that the expansion of these gene families was mainly affected by purifying selection (Figures 4a, b; Supplementary Table S6).
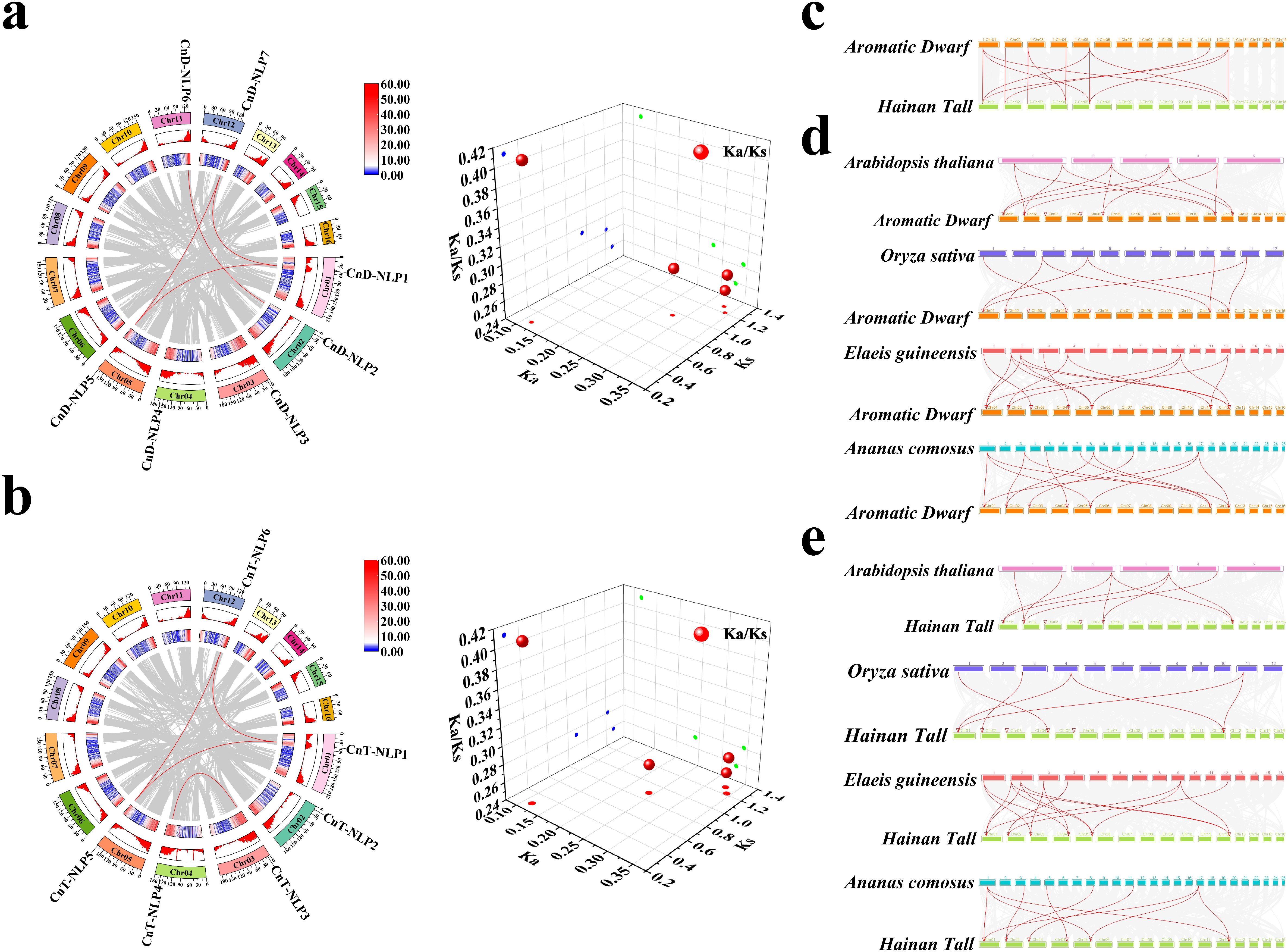
Figure 4. Gene collinearity analysis. (a, b) Collinear analysis and Ka/Ks analysis of 3D scatter plots of NLP gene in Aromatic Dwarf and Hainan Tall. The gray lines in the background represent collinear blocks in the coconut genome. The red lines indicate that the repeated NLP gene pairs of coconut are collinear. The number of chromosomes is shown inside each chromosome. The bars and heat maps of the outer circle represent the gene density on the chromosome. (c–e) Homology analysis of NLP gene between coconut and other representative plants. The gray lines in the background represent collinear blocks in the genomes of coconuts and other plants. The red lines represent the regions where the CnNLP gene is significantly collinear between plant genomes. The red triangle represents the position of the CnNLP gene on the coconut chromosome.
To further investigate the evolutionary relationship of NLP genes across different plant species, including Arabidopsis thaliana, Oryza sativa, Elaeis guineensis, Ananas comosus, and Cocos nucifera, a collinear map was constructed. The results showed that 7 CnD-NLP genes and 6 CnT-NLP genes formed homologous pairs with 14 collinear relationships (Figure 4c). Furthermore, 5 CnD-NLP genes and 5 AtNLP genes formed homologous pairs with 12 collinear relationships, 5 CnD-NLP genes and 5 OsNLP genes formed homologous pairs with 8 collinear relationships, 7 CnD-NLP genes and 7 EgNLP genes formed homologous pairs with 15 collinear relationships, 7 CnD-NLP genes and 7 AcNLP genes formed homologous pairs with 13 collinear relationships (Figure 4d). Similarly, 4 CnT-NLP genes and 5 AtNLP genes formed homologous pairs with 9 collinear relationships, 4 CnT-NLP genes and 4 OsNLP genes formed homologous pairs with 6 collinear relationships, 6 CnT-NLP genes and 7 EgNLP genes formed homologous pairs with 15 collinear relationships, and 6 CnT-NLP genes and 6 AcNLP genes formed homologous pairs with 10 collinear relationships (Figure 4e). These findings suggest that the NLP gene family in coconut is closely related to the NLP gene family in oil palm, indicating that these genes may share similar functions.
3.5 Analysis of CnNLP gene cis-regulatory elements
To further explore the potential function of the CnNLP genes, the upstream 2000 bp sequence was extracted as the promoter region and analyzed using PlantCare to predict cis-acting elements. Various cis-acting elements were identified, including light responsive, hormone responsive, stress responsive, and growth and development response elements. In the CnNLP gene family, CnD-NLP6/7 and CnT-NLP6 contained defense-related stress elements (TC-rich repeats), while CnD-NLP1/4/7 and CnT-NLP1/4/6 contained multiple drought response elements (MBS), suggesting that these genes may be involved in the coconut’s stress response (Supplementary Figure S3).
3.6 Effects of abiotic stress on coconut seedlings growth
To understand the impact of abiotic stress on the growth of different coconut varieties, Aromatic Dwarf and Hainan Tall were subjected to nitrogen deficiency, drought stress, and salt stress. Compared to the control, the plant height, petiole length, leaf fresh weight, and petiole fresh weight of both varieties of decreased significantly (Figure 5b). Under nitrogen deficiency stress, the lower leaves of both varieties withered. Under drought stress, the leaves of Aromatic Dwarf wilted extensively, while only the tips of the upper and lower leaves of the Hainan Tall wilted. Under salt stress, some leaves of both coconut varieties turned yellow and dry (Figure 5a). Together, Hainan Tall showed strong tolerance to different abiotic stresses, leading to the selection of Aromatic Dwarf as the further research object.
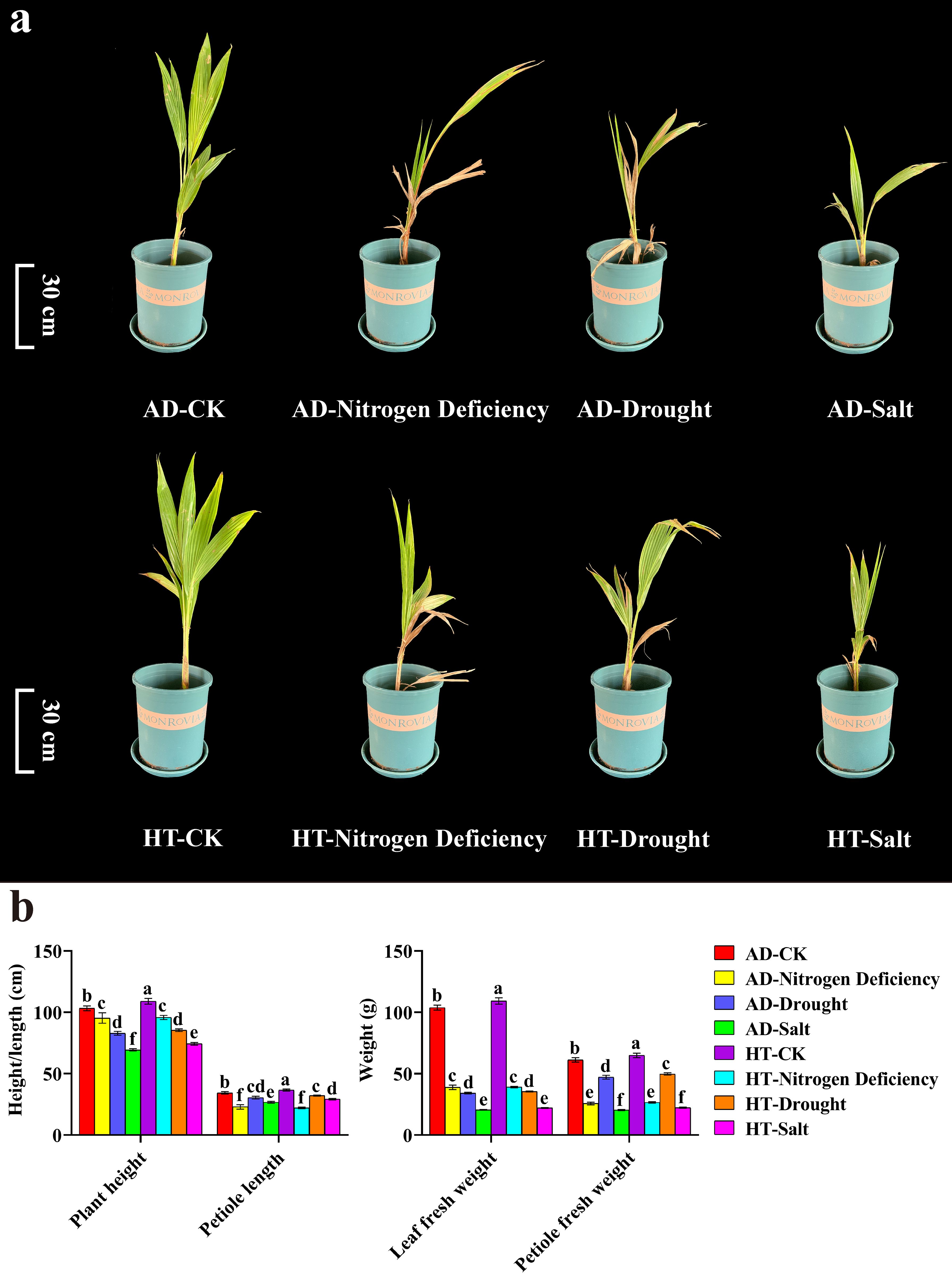
Figure 5. Growth and development of coconut under different varieties and treatments. AD stands for Aromatic Dwarf. HT stands for Hainan Tall. (a) Phenotypic changes of Coconut under nitrogen deficiency, drought, and salt stress. (b) The plant height, petiole length, leaf fresh weight, and petiole fresh weight under CK, nitrogen deficiency, drought, and salt treatments. In the one-way ANOVA, the values represented by different lowercase letters were significantly different at p < 0.05.
3.7 Gene expression profiles of CnNLP transcription factor family and key gene screening
Through bioinformatics analysis, it was found that CnD-NLP1 and CnT-NLP1, CnD-NLP2 and CnT-NLP2, CnD-NLP3 and CnT-NLP3, CnD-NLP4 and CnT-NLP4, CnD-NLP5 and CnT-NLP5, CnD-NLP7 and CnT-NLP6 were homologous genes. The expression patterns of NLP genes in Aromatic Dwarf and Hainan Tall were studied using qRT-PCR across tissues, including roots, petioles, young leaves, and mature leaves (Figures 6a, b). These genes exhibited tissue-specific transcriptional accumulation patterns. Notably, the expression profiles of CnD-NLP2 and CnT-NLP2/5 were similar across different tissues. The expression levels of other genes were higher in leaves (including young leaves and mature leaves) compared to in roots and petioles. CnD-NLP3 and CnT-NLP1 were highly expressed in young leaves, whereas CnD-NLP1/4/5/6/7 and CnT-NLP3/4/6 were highly expressed in mature leaves. The specific expression of CnNLP genes in leaves suggested their involvement in leaf growth and development.
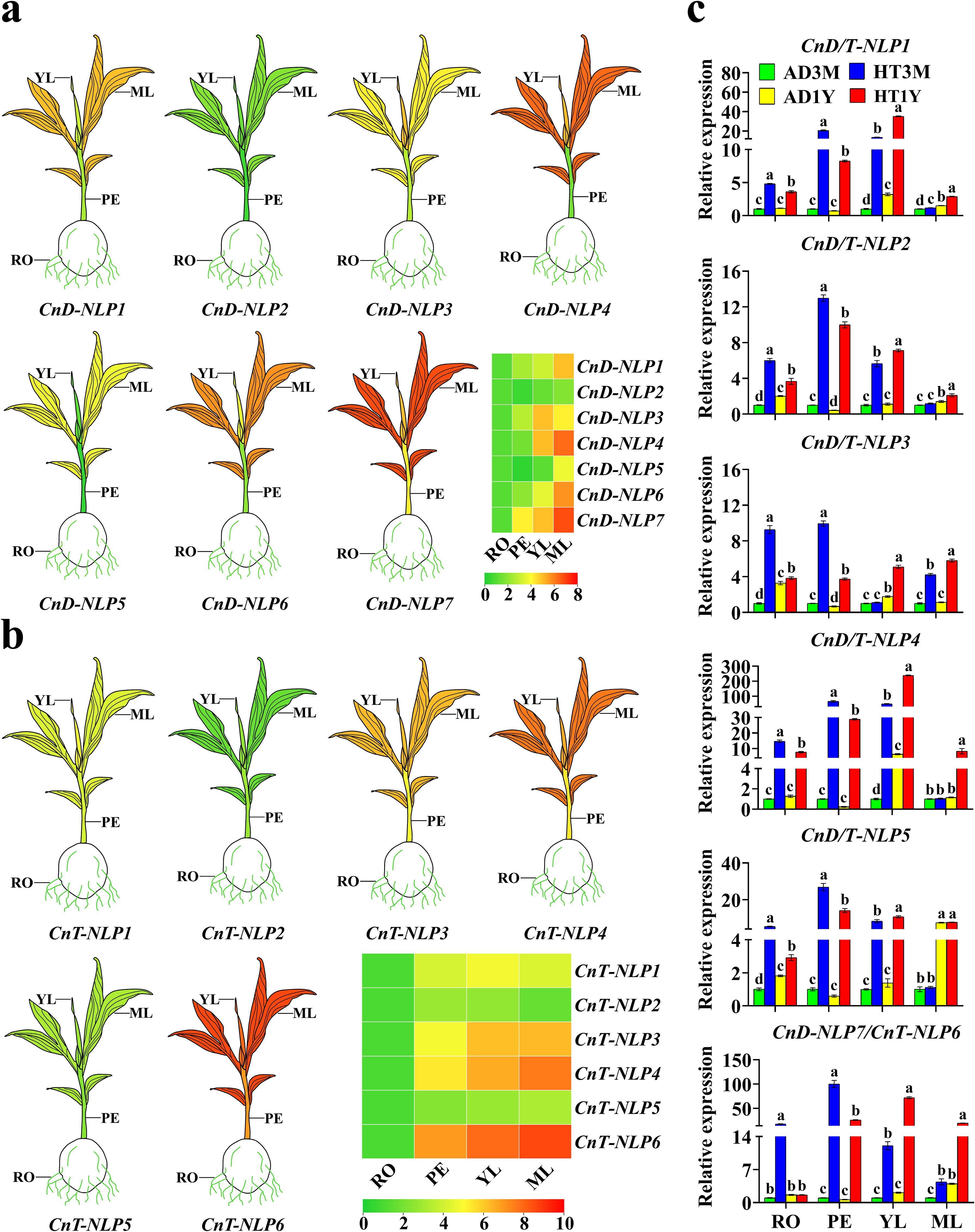
Figure 6. Gene expression analysis of NLP gene in coconut. AD stands for Aromatic Dwarf, HT stands for Hainan Tall; 3M represents coconut cultivated for three months, 1Y represents coconut cultivated for one year; RO, roots; PE, petioles; YL, young leaves; MF, mature leaves. (a, b) Differential expression of representative CnNLP genes in different tissues. The mean expression value was calculated from three independent biological replicates and is expressed in relate to that in the roots. Data is converted by Log to generate the heatmap. The red represents high expression levels, and the green represents low expression levels. (c) Analysis of NLP gene expression in different growth stages of coconut. The mean expression values were calculated from three independent biological replicates and are relative to those of the AD3M controls. In the one-way ANOVA, the values represented by different lowercase letters were significantly different at p < 0.05.
To further understand the role of NLP genes in coconut development, the qRT-PCR technique was employed to analyze the expression of CnD-NLPs and CnT-NLPs at two growth stages. The results indicated that, at the same growth stage, NLP gene expression was higher in Hainan Tall’s tissues than in Aromatic Dwarf’s, indicating that Hainan Tall possessed stronger stress resistance. In Aromatic Dwarf seedlings cultured for 3 months and 1 year, NLP genes expression in AD3M petioles was higher than in AD1Y, while the expression in AD3M roots, young leaves, and mature leaves was lower than in AD1Y. Consequently, Aromatic Dwarf seedlings cultured for 1 year were selected for abiotic stress study (Figure 6c). The results showed that the expression of 7 CnD-NLP genes varied across tissues with the passage of time under abiotic stress (Figure 7a). Notably, under nitrogen deficiency, drought, and salt stress, the expression of CnD-NLP5 gene in young leaves increased continuously under nitrogen deficiency stress, while the expression of other genes in different tissues increased at first and then decreased, and reached the peak at a specific time point. This shows that the response of NLP to nitrogen deficiency, drought, and salt stress is mainly within 24 hours, and the response is the most intense in 3-6 hours. Under nitrogen deficiency stress, the expression of CnD-NLP5 in petioles, young leaves, and mature leaves changed significantly. After 24 hours of stress, CnD-NLP5 expression in petioles and mature leaves reached the maximum level (16.2 and 14.7 times of the expression level at 0 h, respectively), and after 48 hours, the expression of CnD-NLP5 in young leaves reached the maximum level (17.1 times of the expression level at 0 h). It is speculated that CnD-NLP5 may be the key gene in its response to nitrogen deficiency stress. Under drought stress, the expression of CnD-NLP4/5/6 genes in roots changed significantly. After 3 hours, CnD-NLP6 reached its maximum level in roots (10.3 times of the expression level at 0 h), and after 6 hours, the expression of CnD-NLP4/5 in roots reached the maximum level (9.6 and 8.0 times of the expression level at 0 h, respectively). Therefore, these three genes may play a key role in response to drought stress. Under salt stress, CnD-NLP1 gene expression in petioles, young leaves, and mature leaves showed significant changes. After 3 hours, the expression of CnD-NLP1 reached its peak in young leaves and mature leaves, which were 10.8 times and 6.4 times higher than that at 0 h, respectively. In petioles, the expression level of CnD-NLP1 reached its highest level 12 h after stress, which was 7.4 times higher than that at 0 h. This suggests that CnD-NLP1 may be a key gene in response to salt stress. In addition, we found that the expression of all genes in young leaves peaked after 3 hours of salt stress and then gradually decreased, indicating that salt stress can quickly induce the expression of NLP genes in coconut young leaves.
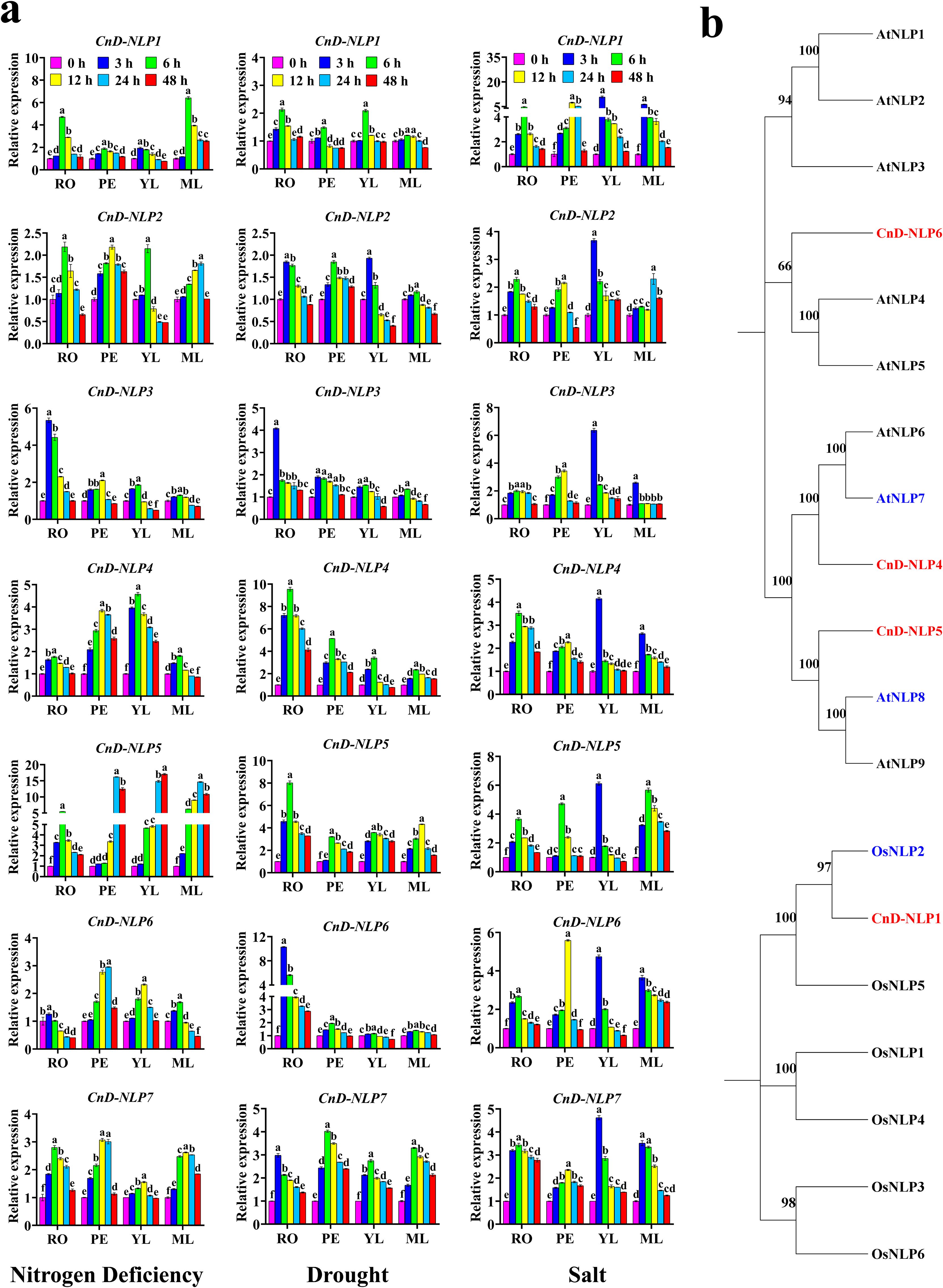
Figure 7. Key gene analysis. (a) Expression profiles of CnD-NLP genes in response to different stress treatments. RO, roots; PE, petioles; YL, young leaves; MF, mature leaves. The mean expression values were calculated from three independent biological replicates and are relative to those of the 0 h controls. In the one-way ANOVA, the values represented by different lowercase letters were significantly different at p < 0.05. (b) Screening of homologous genes of key genes. The red mark represents key genes in the CnNLP gene family that may respond to nitrogen deficiency, drought and salt stress, and the blue mark represents Arabidopsis and rice NLP genes with similar functions. AtNLP8 is a nitrogen-related gene, AtNLP7 is a drought-resistant gene, OsNLP2 responds to salt stress.
Homology screening of the key genes revealed that CnD-NLP5 shared homology with AT2G43500.5 (AtNLP8) (Figure 7b). Previous studies have shown that AtNLP8 reduces abscisic acid levels in a nitrate-dependent manner to regulate nitrate-promoted seed germination (Yan et al., 2016), indicating that CnD-NLP5 may serve a similar function. Additionally, CnD-NLP4 has a high degree of homology with AT4G24020.1 (AtNLP7) after strict screening (Figure 7b). It is known that AtNLP7 mutants exhibit greater drought tolerance compared to wild-type plants (Castaings et al., 2009), indicating that CnD-NLP4 may play a key role in responding to drought stress. Furthermore, the homologous gene LOC_Os04g41850.1 (OsNLP2) of CnD-NLP1 was identified in the rice genome (Figure 7b). Previous studies found that salt stress induced OsNLP2, which upregulated the expression of OsNR, thereby promoting salt tolerance during seed germination (Yi et al., 2022). This indicates that OsNLP2 responds to salt stress, suggesting that CnD-NLP1 may a similar function. These results further demonstrate the role of CnD-NLP5, CnD-NLP4, and CnD-NLP1 in coconut’s responses to nitrogen deficiency, drought stress, and salt stress.
4 Discussion
In this study, 7 CnD-NLP genes and 6 CnT-NLP genes were identified in Aromatic Dwarf and Hainan Tall varieties, respectively (Table 1). Previous studies have revealed that there are 9 NLP genes in Arabidopsis and maize (Schauser et al., 2005; Wang et al., 2018), 6 NLP genes in rice and apple (Wang et al., 2019; Jagadhesan et al., 2020), which is comparable to the members of NLP genes in coconut. However, NLP gene numbers vary significantly among species, including 53 in alfalfa, 33 in tea, and 15 in elephant grass (Jin et al., 2023; Yu et al., 2023; Li et al., 2024), indicating that species-specific variation in NLP genes numbers.
Interestingly, NLP family genes were found to encode longer proteins and have lower isoelectric points in many species (Schauser et al., 2005; Chardin et al., 2014; Cheng et al., 2014), and similar results were observed in NLP genes of coconut (Table 1). This suggests that this transcription factor may be more active under acidic conditions. Subcellular localization prediction showed that all CnD-NLPs and CnT-NLPs are located in the nucleus (Table 1), consistent with the characteristics of transcription factors. Conserved motif analysis of NLP members in two coconuts varieties revealed similar distributions of conserved motifs within the same group (Figure 3c), indicating that NLP members in the same group may have similar biological functions. Gene structure analysis revealed significant differences in the lengths of coconut NLP genes, with most varying from CnD-NLP5 (4929 bp) to CnD-NLP7 (12493 bp), and a few, such as CnD-NLP2 (39825 bp) and CnT-NLP2 (39687 bp), having longer sequences (Table 1; Figure 3c). Previous studies have shown that the acquisition or loss of exons or introns contributes to structural differences and functional diversity of genes (Xu et al., 2012). A comprehensive analysis of the sequence length and exon-intron structure of NLP genes in coconut revealed that introns, particularly those in CnD-NLP2 and CnT-NLP2, significantly affect sequence length, indicating their importance in exon splicing regulation. Phylogenetic analysis showed that coconut NLPs were divided into three groups. Coconut, oil palm, and date palm NLPs were clustered in close branches in each group, reflecting their close relationship as members of Palmaceae (Figure 3a). Gene duplication events play a crucial role in the development of the genome, allowing new functions and evolutionary pathways to be acquired (Qiao et al., 2018, 2019). This study suggests that CnD-NLP5, CnT-NLP2 and CnT-NLP5 likely originated prior to species differentiation, while other members of the CnD-NLP and CnT-NLP gene family may have originated from the WGD event. We speculate that the amplification of the CnNLP gene family may have been driven by WGD (Supplementary Figure S2).
Tissue-specific expression of NLP genes has been studied in variety plants. In Arabidopsis thaliana, AtNLP8 and AtNLP9 are highly expressed in senescent leaves and seeds, but lower in other organs. In rice, OsNLP1 and OsNLP3 are preferentially expressed in the source organs (Schauser et al., 2005). Similarly, in this study, CnD-NLP2 and CnT-NLP2/5 showed similar expression patterns across tissues, with most other genes exhibiting higher expression in leaves than in roots and petioles. Specifically, CnD-NLP3 and CnT-NLP1 were highly expressed in young leaves, while CnD-NLP1/4/5/6/7 and CnT-NLP3/4/6 were highly expressed in mature leaves (Figures 6a, b), indicating that these genes may be involved in leaves growth and development. In addition, lower NLP expression in coconut roots compared to leaf tissues suggests a primary role in nitrate transport rather than absorption (Kumar et al., 2018). In addition, in the study of the MeNLP gene of tropical crop cassava, it was found that the number of members of the MeNLP gene family was similar to that of CnNLP, but MeNLP was mainly expressed in root, which may be caused by species differences (Lin et al., 2023).
In the NLP protein interaction network map of coconut, AtNLP7/8 interacted with the nitrate regulatory protein AtNRG2 and transcription factors AtHRS1 and AtLBD37 (Figure 2d). Our results indicated that CnD-NLP3/4/5 and CnT-NLP3/4/5, which are homologous to AtNLP7/8 in coconut, may also participate in nitrate response. In addition, we analyzed the promoters of the NLP gene family members in coconut and identified several abiotic stress responses cis-acting elements, suggesting that NLP gene may be regulated by these cis-acting elements in their promoter under abiotic stress conditions (Supplementary Figure S3). Studies have shown that OsNLP1, a key gene for nitrogen utilization in rice, is rapidly induced under nitrogen starvation (Alfatih et al., 2020). Moreover, previous research indicated that Arabidopsis thaliana lacking AtNLP7 exhibited greater drought and salt tolerance than the wild type (Castaings et al., 2009; Le et al., 2022), suggesting that the expression of NLP genes affects drought and salt tolerance in plants. In this study, it was observed that under nitrogen deficiency, drought, and salt stress, the expression of CnD-NLP5 gene in young leaves continuously increased under nitrogen deficiency stress, while the expression of other genes in different tissues initially increased and then decreased, peaking at a specific time points. Under nitrogen deficiency stress, CnD-NLP5 expression in petioles and leaves changed significantly, indicating that its potential as a key gene in nitrogen deficiency response. The expression of CnD-NLP4/5/6 gene in roots changed significantly under drought stress, suggesting that these may be key genes in the drought stress response. Similarly, CnD-NLP1 expression in petioles and leaves changed significantly under salt stress, indicating that it may be the key gene in response to salt stress (Figure 7a). These results indicate that the NLP gene family plays a key role in tissue development and stress response, which lays a foundation for further study of the function of NLPs in coconut.
5 Conclusions
Genome-wide analysis identified 7 NLP members in Aromatic Dwarf and 6 NLP members in Hainan Tall. Bioinformatics analysis revealed the physicochemical properties, protein structure, protein interaction, conserved motifs, gene structure, and evolutionary relationship of these NLP members. Functional predictions indicated that these NLP genes are closely related to responses to abiotic stress. Tissue expression profile analysis showed that CnD-NLPs were mainly expressed in leaves. Stress response analysis showed that NLP transcription factors regulate responses to nitrogen deficiency, drought, and salt stress. Specifically, CnD-NLP5 may be a key gene in response to nitrogen deficiency, CnD-NLP4/5/6 may be a key gene in response to drought stress, and CnD-NLP1 may be a key gene in response to salt stress. These findings lay a foundation for further study of the function of NLP gene family in coconut.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
YBL: Writing – original draft. YJL: Writing – original draft. XH: Investigation, Writing – review & editing. CB: Investigation, Writing – review & editing. JZ: Investigation, Writing – review & editing. SW: Writing – review & editing. XZ: Writing – review & editing. PL: Writing – review & editing. CS: Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Project of Sanya Yazhou Bay Science and Technology City, grant number SCKJ-JYRC-2023-24; the National Natural Science Foundation of China, grant number 32060716; the Hainan Province Science and Technology Special Fund, grant number ZDYF2022XDNY264.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1565559/full#supplementary-material
References
Alfatih, A., Wu, J., Zhang, Z.-S., Xia, J.-Q., Jan, S. U., Yu, L.-H., et al. (2020). Rice NIN-LIKE PROTEIN 1 rapidly responds to nitrogen deficiency and improves yield and nitrogen use efficiency. J. Exp. Bot. 71, 6032–6042. doi: 10.1093/jxb/eraa292
Beveridge, F. C., Kalaipandian, S., Yang, C., Adkins, S. W. (2022). Fruit biology of coconut (Cocos nucifera L.). Plants 11 (23), 3293. doi: 10.3390/plants11233293
Broschat, T. K. (2009). Palm nutrition and fertilization. HortTechnology 19, 690–694. doi: 10.21273/HORTTECH.19.4.690
Castaings, L., Camargo, A., Pocholle, D., Gaudon, V., Texier, Y., Boutet-Mercey, S., et al. (2009). The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 57, 426–435. doi: 10.1111/j.1365-313X.2008.03695.x
Chardin, C., Girin, T., Roudier, F., Meyer, C., Krapp, A. (2014). The plant RWP-RK transcription factors: key regulators of nitrogen responses and of gametophyte development. J. Exp. Bot. 65, 5577–5587. doi: 10.1093/jxb/eru261
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Cheng, F., Wu, J., Wang, X. (2014). Genome triplication drove the diversification of Brassica plants. Horticult. Res. 1, 14024. doi: 10.1038/hortres.2014.24
DebMandal, M., Mandal, S. (2011). Coconut (Cocos nucifera L.: Arecaceae): In health promotion and disease prevention. Asian Pac. J. Trop. Med. 4, 241–247. doi: 10.1016/S1995-7645(11)60078-3
Durand, M., Brehaut, V., Clement, G., Kelemen, Z., Macé, J., Feil, R., et al. (2023). The Arabidopsis transcription factor NLP2 regulates early nitrate responses and integrates nitrate assimilation with energy and carbon skeleton supply. Plant Cell 35, 1429–1454. doi: 10.1093/plcell/koad025
Guan, P., Ripoll, J.-J., Wang, R., Vuong, L., Bailey-Steinitz, L. J., Ye, D., et al. (2017). Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. U.S.A. 114, 2419–2424. doi: 10.1073/pnas.1615676114
Harries, H. (1978). The evolution, dissemination and classification of Cocos nucifera L. Bot. Rev. 44, 265–319. doi: 10.1007/BF02957852
Ho, Y. S., Burden, L. M., Hurley, J. H. (2000). Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 19, 5288–5299. doi: 10.1093/emboj/19.20.5288
Jagadhesan, B., Sathee, L., Meena, H. S., Jha, S. K., Chinnusamy, V., Kumar, A., et al. (2020). Genome wide analysis of NLP transcription factors reveals their role in nitrogen stress tolerance of rice. Sci. Rep. 10, 9368. doi: 10.1038/s41598-020-66338-6
Jia, Y., Qin, D., Zheng, Y., Wang, Y. (2023). Finding balance in adversity: nitrate signaling as the key to plant growth, resilience, and stress response. Int. J. Mol. Sci. 24 (19), 14406. doi: 10.3390/ijms241914406
Jin, Y., Luo, J., Yang, Y., Jia, J., Sun, M., Wang, X., et al. (2023). The evolution and expansion of RWP-RK gene family improve the heat adaptability of elephant grass (Pennisetum purpureum Schum.). BMC Genomics 24, 510. doi: 10.1186/s12864-023-09550-8
Kim, Y., Wang, J., Ma, C., Jong, C., Jin, M., Cha, J., et al. (2023). GmTCP and gmNLP underlying nodulation character in soybean depending on nitrogen. Int. J. Mol. Sci. 24 (9), 7750. doi: 10.3390/ijms24097750
Konishi, M., Yanagisawa, S. (2013). Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 4, 1617. doi: 10.1038/ncomms2621
Konishi, M., Yanagisawa, S. (2014). Emergence of a new step towards understanding the molecular mechanisms underlying nitrate-regulated gene expression. J. Exp. Bot. 65, 5589–5600. doi: 10.1093/jxb/eru267
Konishi, M., Yanagisawa, S. (2019). The role of protein-protein interactions mediated by the PB1 domain of NLP transcription factors in nitrate-inducible gene expression. BMC Plant Biol. 19, 90. doi: 10.1186/s12870-019-1692-3
Kumar, A., Batra, R., Gahlaut, V., Gautam, T., Kumar, S., Sharma, M., et al. (2018). Genome-wide identification and characterization of gene family for RWP-RK transcription factors in wheat (Triticum aestivum L.). PloS One 13, e0208409. doi: 10.1371/journal.pone.0208409
Le, Q. T., Lee, W. J., Choi, J. H., Nguyen, D. T., Truong, H. A., Lee, S.-A., et al. (2022). The loss of function of the NODULE INCEPTION-like PROTEIN 7 enhances salt stress tolerance in arabidopsis seedlings. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.743832
Li, D., Jin, Y., Lu, Q.-H., Ren, N., Wang, Y.-Q., Li, Q.-S. (2024). Genome-wide identification and expression analysis of NIN-like protein (NLP) genes: Exploring their potential roles in nitrate response in tea plant (Camellia sinensis). Plant Physiol. Biochem. 207, 108340. doi: 10.1016/j.plaphy.2024.108340
Lin, C., Guo, X., Yu, X., Li, S., Li, W., Yu, X., et al. (2023). Genome-wide survey of the RWP-RK gene family in cassava (Manihot esculenta crantz) and functional analysis. Int. J. Mol. Sci. 24, 12925. doi: 10.3390/ijms241612925
Liu, M., Chang, W., Fan, Y., Sun, W., Qu, C., Zhang, K., et al. (2018). Genome-wide identification and characterization of NODULE-INCEPTION-like protein (NLP) family genes in brassica napus. Int. J. Mol. Sci. 19 (8), 2270. doi: 10.3390/ijms19082270
Liu, M., Zhi, X., Wang, Y., Wang, Y. (2021). Genome-wide survey and expression analysis of NIN-like Protein (NLP) genes reveals its potential roles in the response to nitrate signaling in tomato. BMC Plant Biol. 21, 347. doi: 10.1186/s12870-021-03116-0
Marchive, C., Roudier, F., Castaings, L., Bréhaut, V., Blondet, E., Colot, V., et al. (2013). Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 4, 1713. doi: 10.1038/ncomms2650
Mat, K., Kari, Z. A., Rusli, N. D., Harun, H. C., Wei, L. S., Rahman, M. M., et al. (2022). Coconut palm: food, feed, and nutraceutical properties. Animals 12 (16), 2107. doi: 10.3390/ani12162107
Mohandas, S. (2012). Effect of NPK fertilizer levels on mineral nutrition and yield of hybrid (Tall x Dwarf) coconut. Madras Agric. J. 99, 87–91. doi: 10.29321/MAJ.10.100021
Qiao, X., Li, Q., Yin, H., Qi, K., Li, L., Wang, R., et al. (2019). Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 20, 38. doi: 10.1186/s13059-019-1650-2
Qiao, X., Yin, H., Li, L., Wang, R., Wu, J., Wu, J., et al. (2018). Different modes of gene duplication show divergent evolutionary patterns and contribute differently to the expansion of gene families involved in important fruit traits in pear (Pyrus bretschneideri). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00161
Ramírez-Silva, J. H., Cortazar-Ríos, M., Ramírez-Jaramillo, G., Oropeza-Salín, C. M., Rondón-Rivera, D. D. (2021). Soil organic matter and nitrogen content as related to coconut nutrition in Guerrero, Mexico. Open Access Library J. 8, 1–16. doi: 10.4236/oalib.1107727
Rao, S., Wu, X., Zheng, H., Lu, Y., Peng, J., Wu, G., et al. (2021). Genome-wide identification and analysis of Catharanthus roseus RLK1-like kinases in Nicotiana benthamiana. BMC Plant Biol. 21, 425. doi: 10.1186/s12870-021-03208-x
Samarasinghe, C. R. K., Meegahakumbura, M. K., Kumarathunge, D. P., Dissanayaka, H.D.M.A.C., Weerasinghe, P. R., Perera, L. (2021). Genotypic selection approach made successful advancement in developing drought tolerance in perennial tree crop coconut. Sci. Hortic. 287, 110220. doi: 10.1016/j.scienta.2021.110220
Schauser, L., Wieloch, W., Stougaard, J. (2005). Evolution of NIN-like proteins in arabidopsis, rice, and lotus japonicus. J. Mol. Evol. 60, 229–237. doi: 10.1007/s00239-004-0144-2
Sousa Santos, M. M., Lacerda, C. F., Rocha Neves, A. L., Carvalho de Sousa, C. H., Ribeiro, A., Bezerra, M. A., et al. (2020). Ecophysiology of the tall coconut growing under different coastal areas of northeastern Brazil. Agric. Water Manage. 232, 106047. doi: 10.1016/j.agwat.2020.106047
Sun, X., Kaleri, G. A., Mu, Z., Feng, Y., Yang, Z., Zhong, Y., et al. (2024). Comparative transcriptome analysis provides insights into the effect of epicuticular wax accumulation on salt stress in coconuts. Plants 13 (1), 141. doi: 10.3390/plants13010141
Swaminathan, M. S., Nambiar, M. C. (1961). Cytology and origin of the dwarf coconut palm. Nature 192, 85–86. doi: 10.1038/192085a0
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11 molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Wang, X., Chen, X.-x., Li, H.-l., Zhang, F.-j., Zhao, X.-y., Han, Y.-p., et al. (2019). Genome-wide identification and expression pattern analysis of NLP (Nin-like protein) transcription factor gene family in apple. Sci. Agric. Sin. 52, 4333–4349. doi: 10.3864/j.issn.0578-1752.2019.23.014
Wang, L., Lee, M., Ye, B., Yue, G. H. (2020). Genes, pathways and networks responding to drought stress in oil palm roots. Sci. Rep. 10 (1), 21303. doi: 10.1038/s41598-020-78297-z
Wang, S., Xiao, Y., Zhou, Z.-W., Yuan, J., Guo, H., Yang, Z., et al. (2021). High-quality reference genome sequences of two coconut cultivars provide insights into evolution of monocot chromosomes and differentiation of fiber content and plant height. Genome Biol. 22, 304. doi: 10.1186/s13059-021-02522-9
Wang, Z., Zhang, L., Sun, C., Gu, R., Mi, G., Yuan, L. (2018). Phylogenetic, expression and functional characterizations of the maize NLP transcription factor family reveal a role in nitrate assimilation and signaling. Physiol. Plant 163, 269–281. doi: 10.1111/ppl.12696
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., Yu, J. (2010). KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteomics Bioinf. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wu, X., Lai, Y., Rao, S., Lv, L., Ji, M., Han, K., et al. (2021). Genome-wide identification reveals that nicotiana benthamiana hypersensitive response (HR)-like lesion inducing protein 4 (NbHRLI4) mediates cell death and salicylic acid-dependent defense responses to turnip mosaic virus. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.627315
Xiao, Y., Xu, P., Fan, H., Baudouin, L., Xia, W., Bocs, S., et al. (2017). The genome draft of coconut (Cocos nucifera). GigaScience 6 (11), 1–11. doi: 10.1093/gigascience/gix095
Xu, G., Guo, C., Shan, H., Kong, H. (2012). Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. U.S.A. 109, 1187–1192. doi: 10.1073/pnas.1109047109
Yan, D., Easwaran, V., Chau, V., Okamoto, M., Ierullo, M., Kimura, M., et al. (2016). NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 7, 13179. doi: 10.1038/ncomms13179
Yang, Y., Bocs, S., Fan, H., Armero, A., Baudouin, L., Xu, P., et al. (2021). Coconut genome assembly enables evolutionary analysis of palms and highlights signaling pathways involved in salt tolerance. Commun. Biol. 4 (1), 105. doi: 10.1038/s42003-020-01593-x
Yao, Y., Sun, L., Wu, W., Wang, S., Xiao, X., Hu, M., et al. (2021). Genome-wide investigation of major enzyme-encoding genes in the flavonoid metabolic pathway in tartary buckwheat (Fagopyrum tataricum). J. Mol. Evol. 89, 269–286. doi: 10.1007/s00239-021-10004-6
Yi, Y., Peng, Y., Song, T., Lu, S., Teng, Z., Zheng, Q., et al. (2022). NLP2-NR module associated NO is involved in regulating seed germination in rice under salt stress. Plants 11 (6), 795. doi: 10.3390/plants11060795
Yu, L.-H., Wu, J., Tang, H., Yuan, Y., Wang, S.-M., Wang, Y.-P., et al. (2016). Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and -sufficient conditions by enhancing nitrogen and carbon assimilation. Sci. Rep. 6, 27795. doi: 10.1038/srep27795
Yu, J., Yuan, Y., Dong, L., Cui, G. (2023). Genome-wide investigation of NLP gene family members in alfalfa (Medicago sativa L.): evolution and expression profiles during development and stress. BMC Genomics 24 (1), 320. doi: 10.1186/s12864-023-09418-x
Keywords: NLP transcription factors, coconut, phylogenetic analysis, abiotic stress, gene expression profiles
Citation: Li Y, Li Y, Hai X, Bao C, Zhao J, Wang S, Zhou X, Liu P and Si C (2025) Based on two coconut (Cocos nucifera L.) genome-wide investigation of NODULE-INCEPTION-like protein family: evolution and expression profiles during development and stress. Front. Plant Sci. 16:1565559. doi: 10.3389/fpls.2025.1565559
Received: 23 January 2025; Accepted: 31 March 2025;
Published: 16 April 2025.
Edited by:
Xinyang Wu, China Jiliang University, ChinaReviewed by:
Su Yang, China Jiliang University, ChinaQiaomei Ma, Zhejiang Academy of Agricultural Sciences, China
Copyright © 2025 Li, Li, Hai, Bao, Zhao, Wang, Zhou, Liu and Si. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengcheng Si, Y2NzaUBoYWluYW51LmVkdS5jbg==; Penghui Liu, cGVuZ2h1aS5saXVAaGFpbmFudS5lZHUuY24=
†These authors contributed equally to this work
 Yubin Li
Yubin Li Yujie Li2†
Yujie Li2† Shouchuang Wang
Shouchuang Wang Penghui Liu
Penghui Liu Chengcheng Si
Chengcheng Si