- 1Australian Research Council Research Hub for Medicinal Agriculture, Department of Ecological Plant and Animal Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC, Australia
- 2La Trobe Institute of Sustainable Agriculture and Food, Department of Ecological Plant and Animal Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC, Australia
- 3Australian Research Council Research Hub for Protected Cropping, Department of Ecological Plant and Animal Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC, Australia
- 4Australian Research Council Research Hub for Sustainable Crop Protection, Department of Ecological Plant and Animal Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC, Australia
Phytocannabinoids are medically important specialized defense compounds that are sparsely distributed among plants, yet Cannabis sativa can synthesize unprecedented amounts of these compounds within highly specialized surface cell factories known as glandular trichomes. The control mechanisms that allow for this high level of productivity are poorly understood at the molecular level, although increasing evidence supports the role of oxylipin metabolism in phytocannabinoid production. Oxylipins are a large class of lipid-based oxygenated biological signaling molecules. Although some oxylipins are known to participate in plant defense, roles for the majority of the ca. 600 plant oxylipins are largely unknown. In this review, we examine oxylipin gene expression within glandular trichomes and identify key oxylipin genes that determine the fate of common lipid precursors. Mechanisms by which oxylipins may be interacting with phytocannabinoid metabolism, as well as specialized plant metabolism more broadly, are discussed and a model summarizing these contributions proposed.
1 Introduction
Cannabis sativa L. (Cannabis) is a chemically complex plant that has a repertoire of more than 500 diverse molecules that span several classes of specialized metabolites, including a large class of medically important isoprenylated resorcinol aliphatic polyketides known as phytocannabinoids (PCs). PCs serve as the active pharmaceutical ingredients in medicines used as adjunct therapies for drug-resistant epilepsies (Epidiolex) and multiple sclerosis (Sativex), as well as nausea, pain, and loss of appetite (Dronabinol) (Lichtman et al., 2018; Thiele et al., 2018). In plants, PCs play a critical role in defense responses, including protection from UV radiation, herbivorous insects, and microbial pathogens, but the exact role of PCs in Cannabis is poorly understood (Tanney et al., 2021). While many of the enzymes responsible for producing these metabolites have been elucidated, the control mechanisms governing their synthesis remain underexplored (Stout et al., 2012; Welling et al., 2023). Increasing evidence suggests that PC production may be mediated by oxylipins (Bailey, 2019; Booth et al., 2020; Burgel et al., 2020; Apicella et al., 2022; Garrido et al., 2022; Welling et al., 2023; Table 1).
Oxylipins are structurally and functionally diverse, lipid-based biological signaling molecules that participate in plant immunity (Blée, 2002; Christensen et al., 2016; Ameye et al., 2018; Wasternack and Feussner, 2018; Toporkova et al., 2024; Figure 1). Oxylipins play vital roles in regulating biological functions in plants including growth and development, innate and induced defense mechanisms as response to diverse environmental elicitors and as secondary messengers activating defense compounds (McGarry et al., 2024; Wang et al., 2024; Yokoyama et al., 2025). They are produced by the activity of lipoxygenases (LOXs) and various enzymes of the cytochrome P450 CYP74 family, such as allene oxide synthase (AOS) and hydroperoxide lyase (HPL) (Song and Brash, 1991; Howe et al., 2000; Wasternack and Feussner, 2018). Jasmonates (JAs), including jasmonate-isoleucine (JA-Ile), jasmonic acid, and its methylated form methyl jasmonate (MeJA) are oxylipin-derived phytohormones formed from the LOX-AOS cascade (Wasternack and Hause, 2013; Wasternack and Feussner, 2018; Ghorbel et al., 2021).
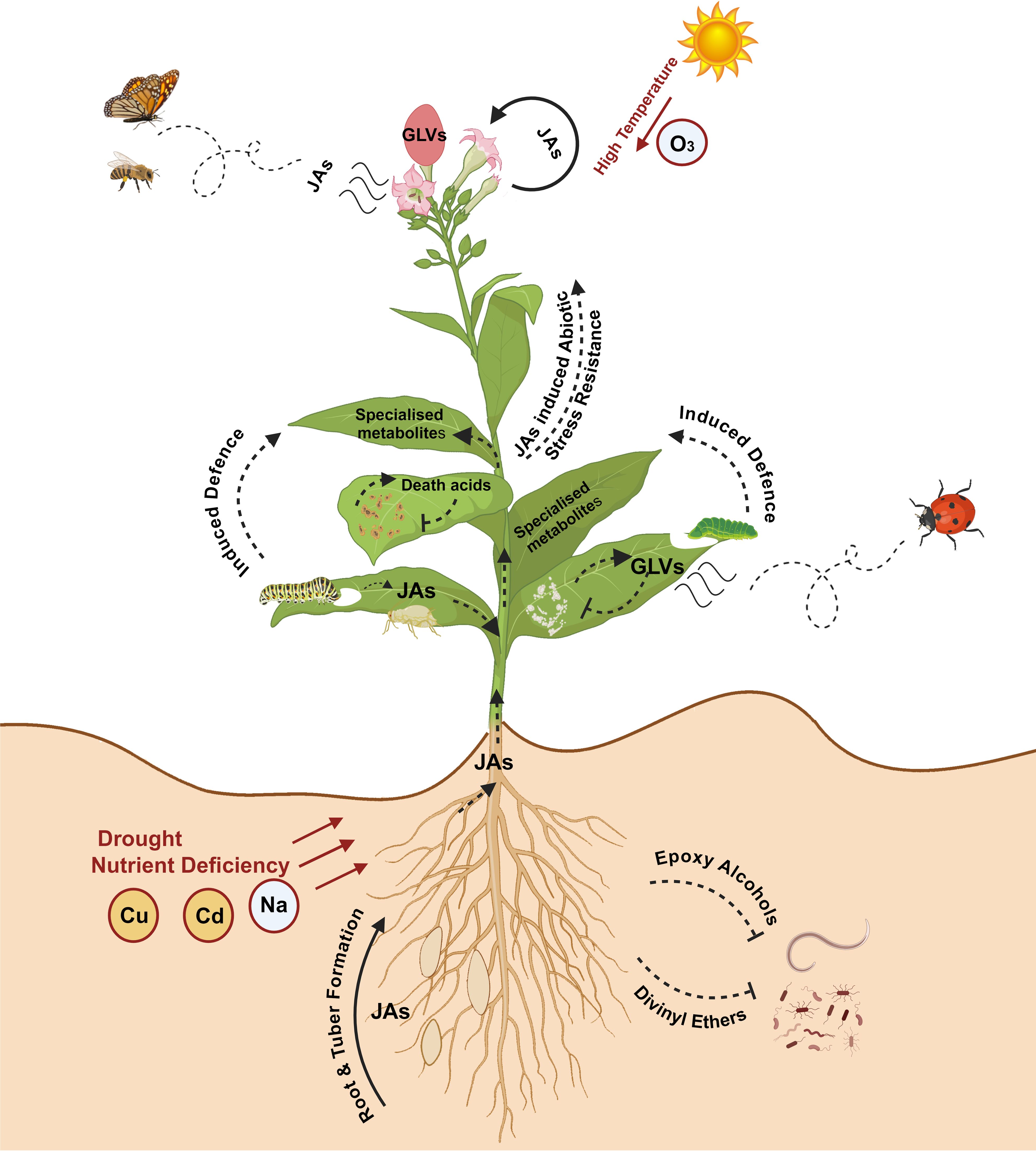
Figure 1. Role of oxylipins in plant growth, development, biotic and abiotic stress tolerance. Oxylipins play multifaceted roles in regulating plant growth, development, and responses to biotic and abiotic stresses. Jasmonates (JAs) mediate developmental processes, including lateral root formation, tuber formation, flowering, and trichome development. Several JAs are also known to attract pollinators and influence plant reproductive strategies. JAs activate defense genes in plants to protect against biotic and abiotic stress (nutrient deficiency, heavy metals [copper (Cu), cadmium (Cd)] and ozone (O3)). Green leaf volatiles (GLVs) contribute to fruit ripening and act as direct defense molecules against pathogenic microbes. GLVs also induce other defense responses, such as attracting natural predators and promoting the production of specialized metabolites. Oxylipins like epoxy alcohols and divinyl ethers serve as direct defense molecules against various pests and pathogens. Image created in BioRender.com.
JAs induce specialized metabolism in diverse plant lineages, including Solanum lycopersicum (tomato) and Lavandula spp. (lavender), while also regulating the development of glandular trichomes and flowers where PCs are concentrated (Boughton et al., 2005; Chen et al., 2018; Dong et al., 2022a; Kianersi et al., 2022; Yan et al., 2022). More recently, JAs have been shown to enhance the levels of PCs, although the intricate mechanism(s) driving this increase in PC production is not fully understood (Table 1). There is also an emerging hypothesis that green leaf volatiles (GLVs) formed via the LOX-HPL branch of oxylipin metabolism may provide substrate for the synthesis of hexanoic acid, the polyketide starter unit for PC production, although only limited evidence has been reported supporting this interaction (Stout et al., 2012; Welling et al., 2023). There has been considerable gene expansion in the LOX family in Cannabis compared with other species, and expression of some LOXs have been localized to the glandular trichomes where PCs are synthesized, suggesting that these may have acquired specialized roles in trichome biology and PC metabolism (Livingston et al., 2020; Borrego et al., 2023; Fayaz et al., 2023; Welling et al., 2023). Collectively, these data serve as evidence for the distinct yet incompletely understood role of the oxylipin pathway in PC production. In this review, we use a combination of phylogenetic and transcriptomic analyses to investigate the mechanisms by which oxylipins may influence PC production in Cannabis and propose a model highlighting these potential interactions.
2 Cannabis: a versatile and underutilized crop
2.1 Origin, classification and uses
Cannabis is a predominantly dioecious, herbaceous annual plant in the Cannabaceae family that originated in central Asia and now has a broad distribution (Kovalchuk et al., 2020). This genus comprises only one species but exhibits high polymorphism, with subspecies: C. sativa ssp. sativa, C. sativa ssp. indica, and C. sativa ssp. rudelaris characterized by varying chemical and morphological features (Small, 2015; Hazekamp et al., 2016). Although the plant is known for its intoxicating properties, it has been used over millennia for medicine, fiber, and oil extraction (Omare et al., 2021; Shakil et al., 2021). Current applications also extend to phytoremediation, functional foods, and as ornamental plants (Frassinetti et al., 2018; Wu et al., 2021; Hesami et al., 2022b). Classification of Cannabis has often been based on its end use, with recreational drug types typically referred to as ‘marijuana’ and fiber or seed types ‘industrial hemp’. Plants can also be classified into chemical phenotypes (chemotypes) based on their proportions of Δ9-tetrahydrocannabinol (Δ9-THC) or cannabidiol (CBD) PCs, or on the level of Δ9-THC, the principal intoxicant found in Cannabis (Jin et al., 2021; Hesami et al., 2022a; Salamone et al., 2022).
Despite the multiple end uses of Cannabis, research on this plant has been constrained over several decades due to its narcotic status, which has limited germplasm exchange and the application of modern genetic improvement strategies (Welling et al., 2016). Recent reforms to the cultivation and use of Cannabis by many legislators have enabled strong growth in licit Cannabis markets and these are projected to increase rapidly in the coming decades (de Brito Siqueira et al., 2023). Similarly, legislative reform has allowed for an increase in Cannabis research, with recent efforts focusing on establishing genomic resources for Cannabis and improving understanding on factors driving PC yield, such as flowering behavior as well as trichome initiation and hormonal regulation (Livingston et al., 2020; Hurgobin et al., 2021; Livingston et al., 2021; Apicella et al., 2022; Toth et al., 2022; Sands et al., 2023; Steel et al., 2023; Dowling et al., 2024; Huang et al., 2024).
2.2 PCs: important medicinal compounds
Extensive research has been conducted on the pharmacology of the two major PCs Δ9-THC and CBD, both of which have modulatory effects on the human endocannabinoid system (ECS). Δ9-THC is a partial agonist of type 1 G protein-coupled cannabinoid receptors (Tagen and Klumpers, 2022). These receptors have important roles in regulating memory, mood, sleep, appetite, inflammation, and pain sensation, and their activation has been shown to have analgesic, neuroprotective, and anti-nausea effects (Mechoulam and Parker, 2013). In contrast, CBD is also thought to modulate the ECS via a less understood mechanism involving other ECS targets and has been reported to counteract the intoxicating properties of Δ9-THC (Devinsky et al., 2014). Medicines containing Δ9-THC and CBD are currently used in the treatment of pediatric epilepsies (e.g., Dravet syndrome and Lennox-Gastaut syndrome) and to alleviate side-effects of chemotherapy, with many other PCs, such as the varinoid PCs cannabidivarin (CBDV; GWP42006) and tetrahydrocannabivarin (THCV; GWP42004), at various levels of drug development (Chanda et al., 2019; Urits et al., 2019; Britch et al., 2021). Other non-intoxicating and low abundant or minor PCs, including cannabichromene (CBC), cannabigerol (CBG) and their carboxylated analogues, cannabichromenic acid (CBCA) and cannabigerolic acid (CBGA), are also considered promising therapeutics (Borrelli et al., 2013; Anderson et al., 2021; Cabrera et al., 2021).
2.3 Glandular trichomes are multicellular structures that synthesize, secrete, and store enormous amounts of PCs
Cannabis produces glandular as well as non-glandular trichomes on the surface of leaves and other parts of the plant body (Livingston et al., 2020; Xie et al., 2023). However, it is the capitate stalked glandular trichomes on modified leaves (perigonal bracts) surrounding female flowers that are the primary site for PC biosynthesis (Figure 2A). These trichomes are multicellular organs, consisting of a stalk, a disc of secretory cells, and a large globular head containing an extracellular cavity formed by cell wall delamination that stores the resinous compounds (Hammond and Mahlberg, 1973; Livingston et al., 2020; Figures 2B, C). These specific trichome morphotypes can produce substantial amounts of PCs, with these compounds contributing up to 40% of their dry weight (w/w) (Livingston et al., 2020). Capitate stalked glandular trichome initiation and development are coordinated with the onset of flowering and likely influenced by several phytohormones, including gibberellins, cytokinins, and JAs, as have been established in numerous plant species (Pattanaik et al., 2014; Li et al., 2021; Huang et al., 2024).
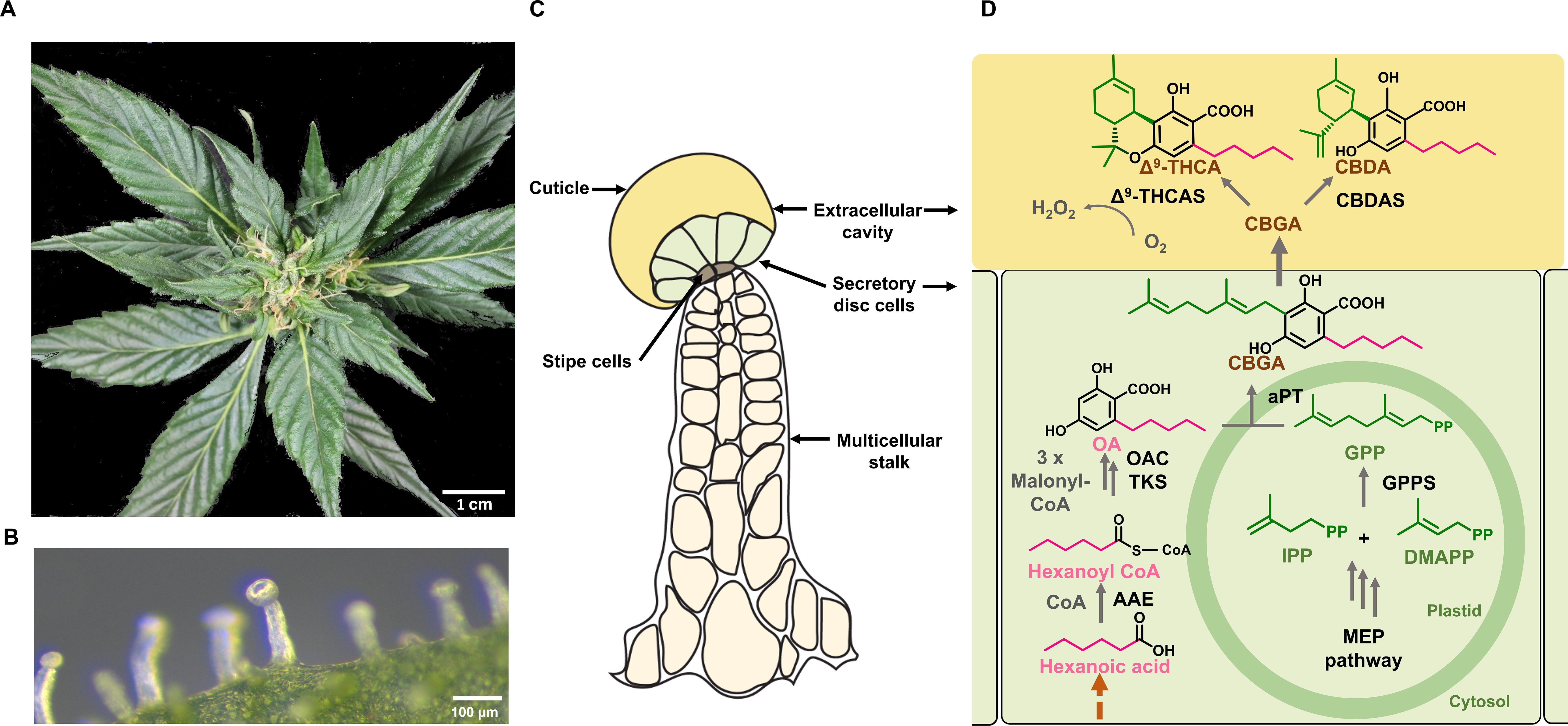
Figure 2. Spatial localization of the phytocannabinoid (PC) pathway in Cannabis. (A) Image of Cannabis female inflorescence. (B) Microscopic view of a capitate stalked glandular trichome on a perigonal bract. (C) Schematic representation of a capitate stalked glandular trichome, the major site of PC biosynthesis and storage. This consists of multicellular stalk (cream), stipe cells at the base of the trichome head (grey), secretory disc cells (light green) and large extracellular storage cavity (yellow). (D) The PC biosynthesis pathway in Cannabis. The methylerythritol phosphate (MEP) pathway takes place in non-photosynthetic chloroplasts of glandular trichome disc cells, while the polyketide pathway is localized in the cytosol. Geranyl pyrophosphate (GPP) is formed from the MEP pathway. The polyketide pathway requires an activated fatty acid starter unit (e.g., hexanoyl-CoA) and three molecules of malonyl-CoA and gives rise to olivetolic acid (OA). Prenylation with GPP by an aromatic prenyltransferase forms CBGA. This is exported to the extracellular cavity where cannabinoid synthases perform oxidative cyclization of the isoprenoid moiety of CBGA, forming the major acid PCs; CBDA and Δ9-THCA. In the final reaction, H2O2 is produced as a biproduct when CBGA is converted to PCs in the presence of O2. Solid dark grey arrows represent known steps in pathways while the brown dashed arrows represent steps of the pathway that have not been resolved. OA, olevetolic acid; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; GPP, geranyl pyrophosphate; CBGA, cannabigerolic acid; CBDA, cannabidiolic acid; Δ9-THCA, Δ9-tetrahydrocannabinolic acid; AAE, acyl activating enzyme; TKS, tetraketide synthase; OA, olivetolic acid cyclase; GPPS, geranyl pyrophosphate synthase; aPT, aromatic prenyltransferase; CBGAS, cannabigerolic acid synthase, CBDAS, cannabidiolic acid synthase; D9-THCAS, D9-tetrahydrocannabinolic acid synthase.
The biosynthesis of PCs traverses across several organelles within the secretory disc cells of glandular trichomes (Figure 2D). The precursors of PCs, olivetolic acid (OA), and geranyl pyrophosphate (GPP) are synthesized by the polyketide pathway in the cytosol and the methylerythritol 4-phosphate (MEP) pathway in plastids, while the final step occurs in the extracellular cavity (Livingston et al., 2022; Figures 2C, D). OA is formed by a type III polyketide synthase (tetraketide synthase; TKS), which produces a tetraketide intermediate, while the accessory protein, OA cyclase (OAC) catalyzes a C2-C7 aldol condensation (Gagne et al., 2012). Together, these enzymes form OA using three molecules of malonyl-CoA and an activated fatty acid, hexanoyl-CoA (Stout et al., 2012). Prenylation of OA with GPP produce the first PC, CBGA, a step catalyzed by an aromatic prenyltransferase, CBGA synthase (CBGAS) (Luo et al., 2019; Sands et al., 2023). CBGA is then transported to the extracellular cavity and converted to Δ9- THCA and CBDA by Δ9- THCA and CBDA synthases, respectively (Taura et al., 1996; Sirikantaramas et al., 2005). This compartmentalization is thought to be attributed to the cytotoxicity of PCs and their bi-product H2O2 which is formed at a molar ratio of 1:1 during the oxidative cyclisation of the isoprenoid residue of CBGA (Sirikantaramas et al., 2004, 2005; Figure 2D). The acidic PCs are then non-enzymatically decarboxylated over time or when subjected to heat, forming the bioactive neutral PCs, such as Δ9-THC and CBD (Sirikantaramas and Taura, 2017).
3 The origin of PC fatty acid precursors is unresolved
A key unanswered question in PC biosynthesis is the origin of the fatty acid hexanoic acid, which, once activated, serves as the C6 carbon starter unit for OA (Welling et al., 2023; Figure 2D). Several metabolic pathways could synthesize hexanoic acid within the secretory disc cells of capitate stalked trichomes. In the trichome glands of Petunia, C6-C8 straight-chain acyl acids are formed from branched-chain amino acid catabolism and α-ketoacid elongation (Kroumova et al., 1994). This pathway requires the activity of four proteins, yet transcripts for these genes have not been reported in Cannabis trichomes (Kroumova et al., 1994; Marks et al., 2009). A second pathway involves de novo fatty acid synthesis. This is supported by trichome-specific expressed sequence tags for a 3-oxoacyl- [acyl-carrier-protein (ACP)] reductase (fabG), as well as the association of this gene with changes in PC composition in a genome-wide association study (Marks et al., 2009; Welling et al., 2020). While C6-specific thioesterases required to terminate fatty acid elongation have not been recovered from trichome cells, this enzyme could reduce the β-keto group following the first two condensation reactions of acetyl CoA, forming hexanoic acid, and, instead of leaving the condensing enzyme, participate in additional reactions with malonyl CoA (Horper and Marner, 1996; Stout et al., 2012). Contrary to this hypothesis is the functional validation of a cytosolic-localized hexanoic acid-specific AAE (hexanoyl-CoA synthetase) which is highly expressed in Cannabis trichome secretory disc cells (Stout et al., 2012).
A third hypothetical pathway could involve an oxylipin-based origin (Stout et al., 2012; Welling et al., 2023). This would involve the degradation of polyunsaturated fatty acids (PUFAs), such as linoleic or linolenic acid, and the activity of LOX to form hydroperoxy PUFAs that can be cleaved into C6 aldehydes and progenitors of hexanoic acid (Nakashima et al., 2013; Welling et al., 2023; Figure 3). Genes encoding desaturases, plastid-localized LOXs, and HPL that participate in these reactions are highly expressed in Cannabis trichomes (Stout et al., 2012; Balcke et al., 2017), while the secondary cleavage product of LOX/HPL, α-oxo acid 12-oxo-(10E)-dodecenoic acid ((10E)-traumatin) (Figure 3), has recently been correlated with PC content in Cannabis inflorescences following treatment with the oxylipin phytohormone MeJA (Welling et al., 2023; Table 1). C18 PUFAs and their oxylipin derivatives are also highly abundant in type VI stalked glandular trichomes of Solanum spp. and genes encoding acyl-hydrolyzing GDSL-type lipases that can release free C18 fatty acid substrates from galactolipids have been identified in Cannabis quantitative trait loci (QTL) for PC content (Cropano et al., 2022). The following sections provide a detailed discussion of oxylipins in Cannabis and highlight key areas of research that could be used to understand the interaction between oxylipins and PC production.
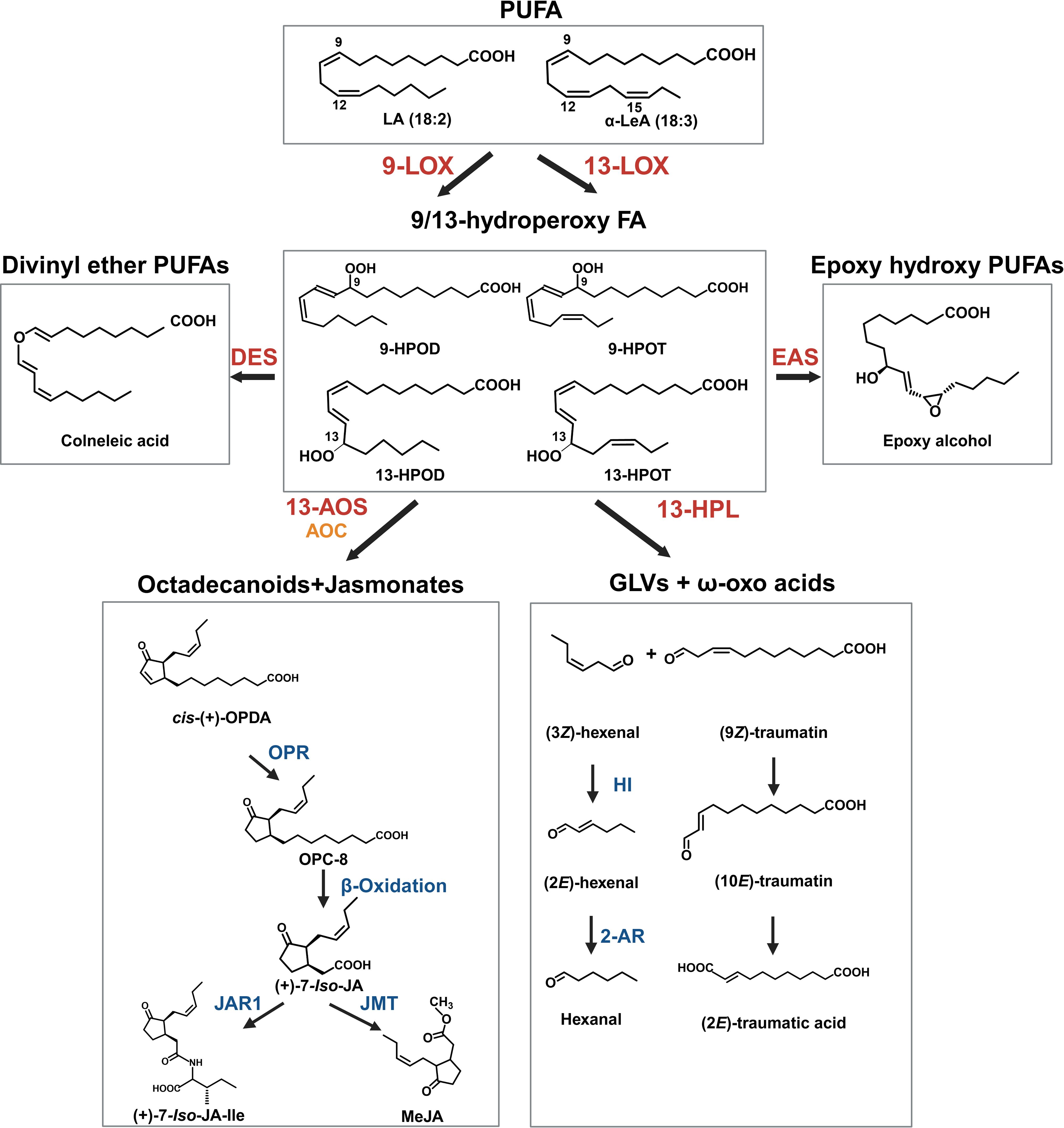
Figure 3. Key oxylipins in the oxylipin pathway. The oxylipin pathway starts with the hydroperoxidation of polyunsaturated fatty acids (PUFA) by lipoxygenase (LOX) enzymes. The position of oxygenation is either at the 9th or 13th carbon from the carbonyl end of the fatty acid. The resulting polyunsaturated hydroperoxides include 9-/13-hydroperoxy linoleic acid (9/13-HPOD) and 9-/13-hydroperoxy linolenic acid (9/13-HPOT). This reaction represents a key branchpoint for oxylipin biosynthesis and provides substrate for several enzymatic pathways, including allene oxide synthase (AOS), hydroperoxide lyase (HPL), divinyl ether synthase (DES), epoxy alcohol synthase (EAS) and peroxygenase (POX) pathways. The AOS and allene oxide cyclase (AOC) cascade leads to the formation of jasmonates (JAs), while the HPL branch leads to the production of green leaf volatiles (GLVs) and 12-oxo acids/traumatins. The other three pathways produce several groups of molecules that act in plant defense and signaling, including epoxy alcohols and divinyl ethers (e.g., colneleic acid). Abbreviations for compounds: α-LeA, α-linolenic acid; LA, Linoleic acid, 9-HPOD, (9S)-hydroperoxy-(10E,12Z)-octadecadienoic acid; 9-HPOT, (9S)-hydroperoxy-(10E,12Z,15Z)-octadecatrienoic acid; 13-HPOD, (13S)-hydroperoxy-(9Z,11E)-octadecadienoic acid; 13-HPOT, (13S)-hydroperoxy-(9Z,11E,15Z)- octadecatrienoic acid; epoxy alcohol, 13(S)-epoxy-9(S)-hydroxy-10(E)-octadecenoic acid; cis-(+)-OPDA, cis-(+)-12-oxo-phytodienoic acid; OPC-8, 3-oxo-2-(2-pentenyl)-cyclopentane-1-octanoic acid; (+)-7-iso-JA, jasmonic acid; (+)-7-iso-JA-Ile, jasmonic acid isoleucine conjugate;. (9Z)-traumatin, 12-oxo-(9Z)-dodecenoic acid; (10E)-traumatin, 12-oxo-(10E)-dodecenoic acid, (2E)-traumatic acid; (2E)-dodecanedioic acid; JAR1, jasmonoyl amino acid conjugate synthase; OPR, 12-oxo-phytodienoic acid reductase; AOC, allene oxide cyclase, HI, hexenal isomerase. 2-AR, 2-alkyl reductase.
4 Oxylipin pathway and metabolism
4.1 Key metabolic branch points determine the fate of oxylipin molecules and their biological functions
In plants, oxylipins are generated through the oxidation of PUFAs. PUFA oxidation can occur enzymatically by dioxygenases or nonenzymatic by the activity of free radicals (e.g., singlet oxygen (1O2)) (Brash, 1999; Zoeller et al., 2012; Figure 3). The resulting hydroperoxides are highly reactive and act as key substrates in oxylipin metabolism, with the fate of oxylipin molecules decided by several enzymatic branch points including (1) the previously described HPL branch that forms C6 aldehydes, alcohols and acyl esters (collectively known as GLVs) concomitant with 12-oxo-(9Z)-dodecenoic acids ((9Z)-traumatin), (2) the JAs-forming AOS pathway, (3) the divinyl ether synthase (DES) pathway that forms divinyl ethers, and (4) the epoxy alcohol synthase/peroxygenase pathways that produce epoxy hydroxy PUFAs (Wasternack and Hause, 2013; Christensen et al., 2015, 2016; Figure 3). Oxylipins within each branch can have distinct roles (Schlotzhauer et al., 1996; Figure 3). For example, divinyl ethers and epoxy alcohols act as defense compounds in roots and leaves (Blée, 1998; Hamberg, 1999; Weber et al., 1999; Hamberg, 2004; Sanadhya et al., 2021; Gorina et al., 2022; Toporkova et al., 2024; Figure 1). The HPL and AOS branches are among the most extensively researched, with AOS-derived JAs having broad roles as signaling molecules, in reproduction, plant growth, and development (Scala et al., 2013; Ghasemi Pirbalouti et al., 2014; Ameye et al., 2018; Raza et al., 2021; Figures 1, 3).
4.2 LOX serves as the critical juncture dictating oxylipin fate
LOXs, such as linoleate 13S-lipoxygenase (EC 1.13.11.12), serve as primary enzymes that mediate the fate of lipid precursors (Figure 3). These are monomeric non-heme iron (Fe)-containing dioxygenases widely distributed in plants that convert PUFAs into hydroperoxides in a two-step reaction, involving the reduction of Fe3+ to Fe2+ by proton-coupled electron transfer, followed by oxygen insertion (Egmond et al., 1973; Goldsmith et al., 2002). LOXs are categorized into two main functional classes based on the carbon position they catalyze. For the 9-LOX and 13-LOX this is at the 9th and 13th position from the carbonyl carbon in the fatty acid, respectively, and can also be distinguished as type I and type II based on subcellular localization (Figure 3). In addition, different LOX paralogs with the same regiospecificity can control different branches of the oxylipin pathway but are spatially separated. For example, in Zea mays, the JA-producing 13-LOX ZmLOX8 is localized to the chloroplast, while a GLV-producing 13-LOX ZmLOX10 is localized to non-photosynthetic plastids (Christensen et al., 2013). While often associated with defense process, impaired LOX function has resulted in changes in plant reproduction, fruit ripening and root development (Kolomiets et al., 2001; Fortes et al., 2004; Liu and Han, 2010; Caldelari et al., 2011; Vogt et al., 2013; Liu et al., 2020).
5 Phylogenetic insights and structural features of Cannabis LOX proteins
5.1 Phylogenetic analysis of the Cannabis LOX protein family reveals distinct clades that may have specific metabolic functions
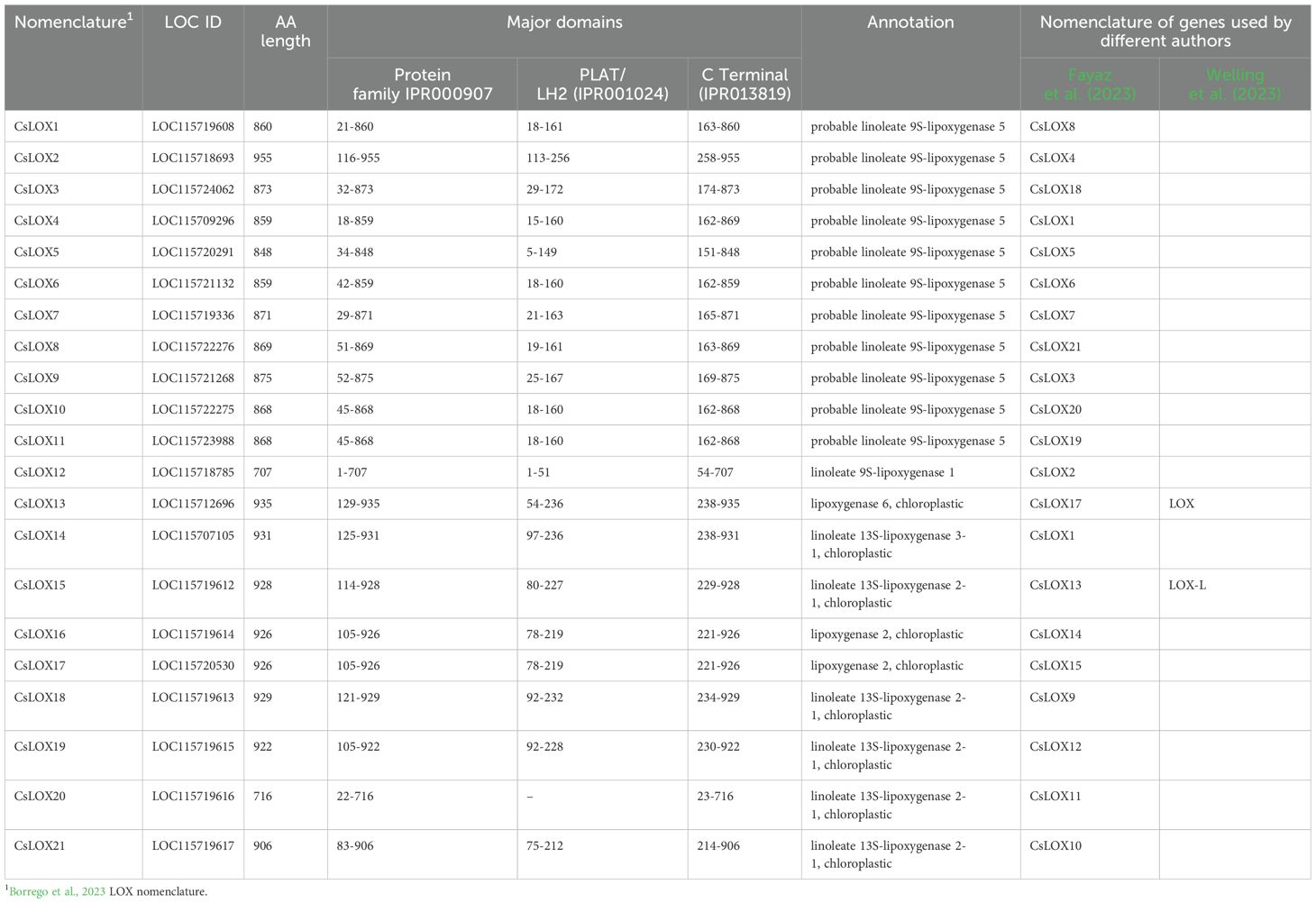
To explore the diversity of LOX gene family in Cannabis, a phylogenetic tree was constructed using protein sequences predicted from the Cannabis var. CBDRx genome [NCBI GenBank Accession: GCA_900626175.2] (Supplementary Table S1). Of the twenty-one Cannabis LOXs, all had the PLAT/LH2 domain (IPR001024) at the N terminus except CsLOX20 (Table 2, Supplementary Table S1). They also contained the C-terminal domain IPR013819 and were classified as LOX super family (IPR000907) members, which is consistent with authentic LOX proteins (Chen et al., 2015; Song et al., 2016). Twelve were clustered with the type-I cytosolic-localized 9-LOXs, while nine were clustered with the type-II plastidial-localized 13-LOX sub-class (Figure 4A). Within the 13-LOX sub-group, CsLOX13 and CsLOX14 form a clade with LOX orthologs associated with JA synthesis, while CsLOX15-CsLOX17 form a distinct clade that shows homology with AtLOX2, a dual functioning ortholog involved in both JA and GLV synthesis (Chen et al., 2004; Allmann et al., 2010; Chauvin et al., 2013; Christensen et al., 2013; Shen et al., 2014; Mochizuki et al., 2016; He et al., 2020; Figure 4A).
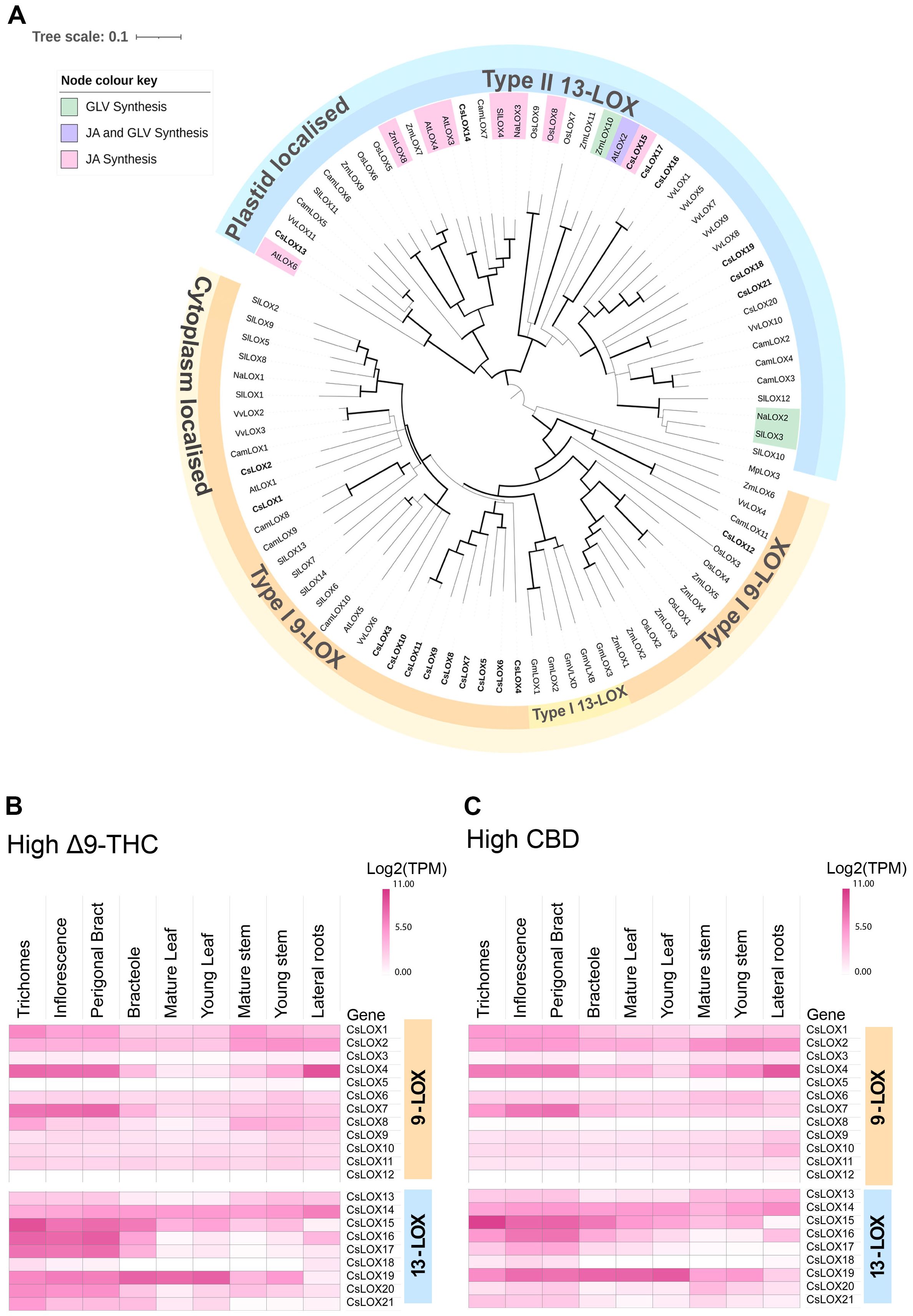
Figure 4. Phylogenetic relationship of LOX proteins and tissue series gene expression in Cannabis. (A) Phylogenetic relationship of lipoxygenase (LOX) proteins in Cannabis to several plant species, including Arabidopsis thaliana (AtLOX), Camelia sinensis (CamLOX), Nicotiana attenuata (NaLOX), Oryza sativa (OsLOX) Solanum lycopersicum (SlLOX), Vitis vinifera (VvLOX) and Zea mays (ZmLOX). Branches with Bootstrap values above 80 shown by greater line thickness (black). Cannabis LOX proteins are shown in bold. Functionally validated genes for corresponding LOXs are shaded as indicated by the key. Absolute expression (Log2 (mean TPM) of LOX genes in different tissue types of (B) high Δ9-THC and (C) high CBD chemotypes. RNA-Seq data reported by Jost et al. (2025). Protein sequences of Arabidopsis thaliana (At), Cannabis sativa (Cs), Camelia sinensis (Cam), Nicotiana attenuata (Na), Glycine max wild type (Gm) Oryza sativa (Os), Solanum lycopersicum (Sl) Zea mays (Zm) and Vitis vinifera (Vv) were used for phylogenetic analysis and PLAT/LH2 and lipoxygenase (LOX) domains identified using InterPro1 (v.98.0). Non-Cannabis sequences without both domains were removed from the analysis. Ninety-one higher plant LOX proteins were used for phylogenetic analysis. The Marchantia polymorpha (common liverwort) LOX3 protein served as an outgroup. The alignment was trimmed of spurious sequences and only residues of the Interpro lipoxygenase super family (IPR000907) domain was compared. The multiple sequence alignment was generated using clustal omega (ver. 1.2.2) in Geneious prime (ver. 2023.0.4), and the phylogenetic tree was generated using the neighbor-joining method (Sievers and Higgins, 2021) and visualized using Interactive Tree Of Life (iTOL; v. 6.8.1) (Letunic and Bork, 2007).
5.2 Enhanced expression of Cannabis LOX in trichomes and floral tissues
We hypothesized that LOXs which have high expression in PC producing tissues, such as trichomes, perigonal bracts and inflorescences may have important roles in PC production. For example, some 9-LOX isoforms are exclusively expressed in roots where they participate in the synthesis of defense compounds (e.g., etherolenic acid, colnelenic acid) against root pathogens (Grechkin et al., 1997; Sanadhya et al., 2021). CsLOX7, CsLOX15, CsLOX16 and CsLOX17 consistently showed high levels of expression in the PC-producing tissues across two medicinal cannabis chemotypes, while CsLOX15 was predominantly expressed in the trichomes (Figures 4B, C), indicating its potential involvement in trichome biochemistry. The recent heterologous expression of CsLOX15 in Nicotiana attenuata and N. benthamiana plants indicate that this paralog encodes a functional enzyme, although activity was assessed using a single substrate, linoleic acid (Fayaz et al., 2023). Elucidating the molecular context of CsLOX15 in its native environment will be critical to disentangle its precise role within Cannabis glandular trichomes and subsequent impact on PC production.
5.3 CsLOX15 structure - conserved residues in catalytic region
To further understand the structure and function of CsLOX15 and other Cannabis LOXs, we compared the predicted structures of these Cannabis proteins with the crystal structure from the Glycine max (soybean) GmLOX1. As expected, many of the functional residues were conserved across the CsLOXs, with minor exceptions (Meyer et al., 2008; Offenbacher et al., 2017; Ruddat et al., 2004; Li et al., 2018) (Supplementary Table S2, Supplementary Figure S1). The predicted 3D structure of CsLOX15 shows the β-sheet rich N-terminal domain as well as the α-helix rich long carboxylic domain within the catalytic site, which facilitates electron transfer and the insertion of oxygen (Figures 5A, B, Supplementary Figure S1). The predicted 3D structure of CsLOX15, however, indicates that the side chain of W585 near the Fe ligand interacts with residue H584 (GmLOX1: H499) (Figures 5C, D). This may reduce space around Fe ligand by distorting the octahedral geometry, which could impact electron transfer (Steczko et al., 1992; Minor et al., 1996; Tomchick et al., 2001; Liavonchanka and Feussner, 2006; Figure 5A). We also observed that the N780 residue (GmLOX1: N694 involved in Fe binding) was positioned further away from the Fe ligand which could further weaken contact and increase flexibility (Figures 5C, D), Modification of N694H in GmLOX1 reduced flexibility due to stronger H bonding in the coordination sphere and resulted in reduced catalytic activity (Holman et al., 1998; Schenk et al., 2003; Segraves et al., 2006). The application of electron paramagnetic resonance (EPR) and magnetic circular dichroism (MCD) hydrogen deuterium exchange mass spectrometry will further aid in resolving the 3D structure and catalytic center of LOXs in non-model species such as Cannabis (Holman et al., 1998; Hu et al., 2019; Gaffney, 2020).
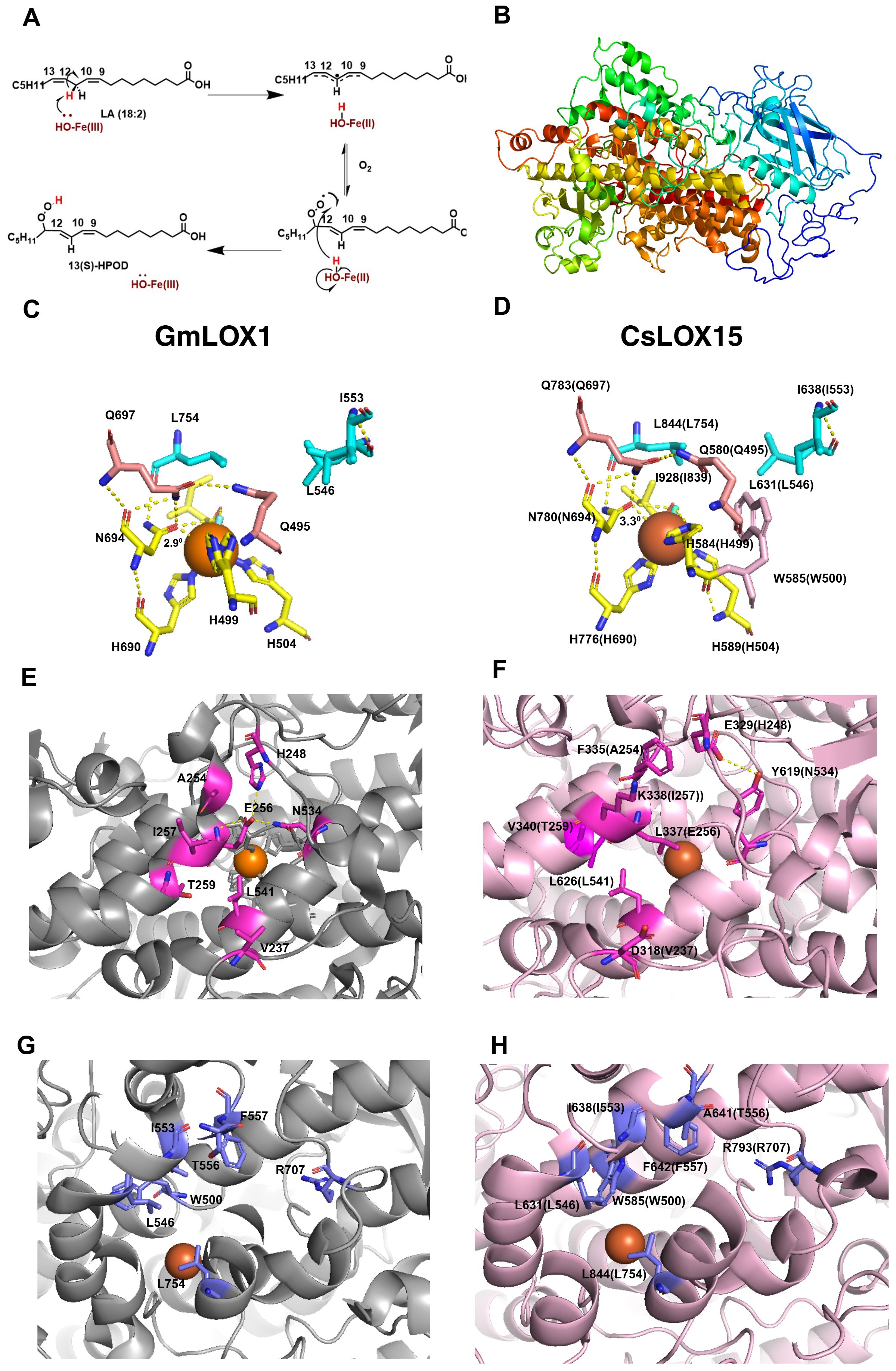
Figure 5. Structural features of GmLOX1 and CsLOX15. (A) Lipoxygenase (LOX) reaction mechanism, 13-LOX convert polyunsaturated fatty acids i.e. linoleic acid (LA) into (13S)-hydroperoxy-(9Z,11E)-octadecadienoic acid ((13S)-HPOD) by a two-step reaction involving the reduction of Fe3+ to Fe2+ by proton coupled electron transfer and then insertion of oxygen. (B) In silico 3D structure of CsLOX15 showing β- sheet rich N-terminal domain and α- helix rich C-terminal domain. (C) Coordination geometry of the catalytic site and Fe ligand of WT GmLOX1, showing important residues involved in iron coordination (H499, H504, H690, N694, I839, OH/H2O, Q495, Q697, N694) and substrate binding (I553, L546, L754). (D) In silico coordination geometry of CsLOX15, around the Fe ligand (superimposed to (5 A0)) showing iron coordination, substrate binding residues, and an additional side chain residue W585. (E) Amino acid residues in substrate entry of GmLOX1 facilitating H binding network. (F) Probable amino acid residues in CsLOX15 substrate entry. (G) Key amino acid residues within GmLOX1 substrate cavity. (H) Key amino acid residues within CsLOX15 substrate cavity. Orange sphere, LOX Fe ligand; blue rod, water molecule; yellow dashed lines, polar contacts. The secondary structure was predicted using Phyre2 (V 2.0) (normal mode) with 100% confidence (Kelley et al., 2015) and the validation of the model was carried out using PROCHECK program in SAVES v6.12 (Supplementary Figure 2). The 3D-structure was modelled using PYMOL (Ver. 3.3) by superimposing with GmLOX1 (PDB entry 1F8N).
Analysis of the in silico predicted 3D structure of CsLOX15 also indicates changes in the substrate entry pocket which could potentially impact substrate specificity. Strong H bonds from residues H248-E256-N534 at the entrance of the substrate binding site of GmLOX1 have been partially lost in CsLOX15 (Figures 5E, F) (Skrzypczak-Jankun et al., 2001). Replacement of these with hydrophobic residues leucine and threonine as seen in CsLOX15 may alter the H bond network and expand the PUFA entry point (Youn et al., 2006). Replacement of V340 with the bulkier T259 residue (GmLOX1) also appears to have expanded the entry pocket (Figures 5E, F). Substrate orientation may also be affected in CsLOX15. For example, there was less distance between A641 (GmLOX1: T556) and R797 (GmLOX1: R707) within the substrate cavity which could potentially affect pocket volume, substrate entry and orientation (Hornung et al., 1999; Hughes et al., 2001; Hershelman et al., 2019; Figures 5G, H). Understanding the impact of these changes is complicated by the fact that there are no crystal structures of plant LOXs bound with linoleic acid or α-linolenic acid (Knapp and Klinman, 2003; Newie et al., 2016). Using in-silico tools such as Autodock to model the interaction between fatty acid and LOX proteins would provide deeper insights into substrate-enzyme interactions (Naimuzzaman et al., 2025).
Variations in the predicted tertiary structure of CsLOX15 compared to GmLOX1 suggest potential for differences in catalytic activity that might allow CsLOX15 to catalyze reactions utilizing numerous fatty acid substrates. As Cannabis glandular trichome disc cells are proposed to have non-photosynthetic chloroplasts, the role of CsLOX15 may facilitate the mobilization of storage lipids to provide carbon for producing defense compounds (Livingston et al., 2022). To meet the carbon demand, trichomes reliant on the supply of photosynthates from source organs, may be partially dependent on locally stored non-structural carbohydrates to maintain carbon supply (Huang et al., 2020). Hence, trichome-specific LOXs could be providing an alternate carbon source through hydrolysis of storage lipids.
6 The cytochrome P450 enzymatic step may represent an important branch point to study the effects of oxylipins on PC production
Both HPL and AOS are homologous cytochrome P450 enzymes that do not require molecular oxygen or the reducing power of NADPH reductase (Li et al., 2008). Structural analysis of the A. thaliana AOS protein shows that one point mutation in the catalytic site converts AOS to HPL (Li et al., 2008). While both enzymes compete for the same substrate (e.g. hydroperoxy fatty acids), competition can be controlled through temporal regulation, with changes in the time of expression observed in HPL- or AOS-specific LOXs in N. attenuata, or through spatial discrimination, as described previously in Z. mays and tomato (Froehlich et al., 2001; Allmann et al., 2010). Silencing HPL in soybean, rice, and potato impaired GLV production while increasing JAs synthesis, suggesting crosstalk between these pathways (Vancanneyt et al., 2001; Tong et al., 2012; Wang et al., 2020). As Cannabis is predicted to contain only a single copy of HPL (CYP74B) and AOS (CYP74A) (Borrego et al., 2023), silencing CsHPL or CsAOS2 would be beneficial to determine compensatory mechanisms from loss of function of either gene and subsequent impact on PC production. To elucidate sequential steps up and downstream of these CYP74 enzymes, protein-protein interactions could also be explored by chemical cross-linking coupled with mass spectrometry (MS)-cleavable tag analysis (Piersimoni et al., 2021).
7 JAs and other oxylipins in PC production
7.1 The precise mechanism(s) by which JAs mediate PC production remains unresolved
There are several mechanisms by which JAs such as MeJA may increase PC content (Table 1). For example, through glandular trichome induction, increasing inflorescence compactness by reducing internode length, and/or the spacing of repeating phytomer units that host these specialized structures (Spitzer-Rimon et al., 2019; Welling et al., 2023; Huang et al., 2024). The complexity of Cannabis floral architecture makes measuring JA responses challenging. Consequently, there have only been limited attempts to quantify trichome density on floral leaves (e.g., perigonal bracts/calyxes), and there are mixed reports of the effectiveness of MeJA to induce trichomes on the foliar fan leaves (Welling et al., 2023; Hahm et al., 2024; Huang et al., 2024). Promisingly, a nearly two-fold increase in trichome density of calyxes was observed in 100 µM MeJA treated plants (Hahm et al., 2024). However, trichome imaging was limited to five calyxes per plant, which may not be representative, and density measurements were calculated based on a 6 x 4 mm leaf surface area (Hahm et al., 2024). The application of deep learning models and high spatial resolution imaging technologies could be used to accurately measure changes in floral architecture following exogenous JA exposure (Huang et al., 2024; Matsumoto et al., 2024).
JAs may also directly increase PC content by inducing PC gene expression and synthesis. To date, no significant changes in the expression of PC-related genes have been reported following a MeJA treatment (Table 1). However, spatiotemporal analysis of PC and precursor pathway genes within target tissues, such as the trichome secretory cells, following JA exposure are lacking (Garrido et al., 2022; Welling et al., 2023). JA related molecular responses are often associated with JA pathway genes (LOX, AOS, AOC, JAZ (JASMONATE-ZIM-DOMAIN), COI1 (CORONATINE INSENSITIVE 1) interacting with transcription factors like MYC2 and basic helix–loop–helix (bHLH)148, and a similar model is predicted for Cannabis (Schuurink and Tissier, 2020; Wang et al., 2020; Han et al., 2022; Song et al., 2022; Xie et al., 2023; Huang et al., 2024). The regulatory functions of JA signaling are typically initiated by the synthesis of JA-Ile, which promotes formation of the SCFCOI1–JAZ co-receptor complex (Hickman et al., 2017). Degradation of JAZ repressor proteins by SCFCOI1 results in the release of JAZ-mediated transcription factors, including MYC2, which activates the expression of JA-responsive genes by targeting their promoters (Hickman et al., 2017).
In Cannabis, the application of exogenous MeJA has increased the expression of JA-responsive genes like JAZ and COI1 and the key regulator of these genes, CsMYC4 (Huang et al., 2024). This suggests that CsMYC4 may have a similar function to MYC orthologs in tomato and L. angustifolia, both of which influence glandular trichome size and density (Xu et al., 2018; Dong et al., 2022b; Huang et al., 2024). Members of the WRKY gene family, which have roles in glandular trichome formation in Artemisia annua, have also been shown to be responsive to MeJA in Cannabis (Xie et al., 2021; Rashid et al., 2023), indicating that a much larger network of transcription factors may be contributing to JA-mediated trichome formation. To fully understand these complexes, it will be necessary to comprehensively reconstruct the gene regulatory networks that form in response to JA exposure in Cannabis.
7.2 Mechanisms by which oxylipins may be interacting with PC production
Despite significant progress towards understanding the influence of oxylipin metabolism on PC production (Table 1), there are multiple avenues by which these lipid molecules may be driving metabolite production (Figure 6). The following interactions are proposed: (i) JAs could induce PC production through transcription regulatory mechanism, either indirectly through JA-induced responses, or directly by inducing the biosynthesis pathways. Similarly, HPL derived oxo acids and GLVs could interact with JAs and induce JA mediated responses or be directly involved as signal molecules inducing PCs, (ii) oxylipin-induced cleavage of fatty acids may form aldehydes that act as a direct carbon source for polyketide and PC synthesis, and (iii) oxylipins could be involved in the detoxification of reactive oxygen species (ROS) in the microenvironment of glandular trichomes (Balcke et al., 2017), as LOXs consume H2O2, the biproduct of PC synthesis (Taura et al., 1996; Sirikantaramas et al., 2005). ROS could also be generated in response to JAs and in return JAs can modulate ROS homeostasis (Denness et al., 2011; Corpas et al., 2015; Demiwal et al., 2024; Zhao et al., 2025). The interplay between ROS and JAs may lead to complex signaling pathways that could activate specialized metabolism in Cannabis. Additionally, JA signaling is closely aligned with the regulation of other phytohormones, including ethylene and salicylic acid, all of which can affect LOX expression and PC production (Yamamoto et al., 2020; Li et al., 2021; Jeyasri et al., 2023; Rodrigues Magalhães et al., 2023).
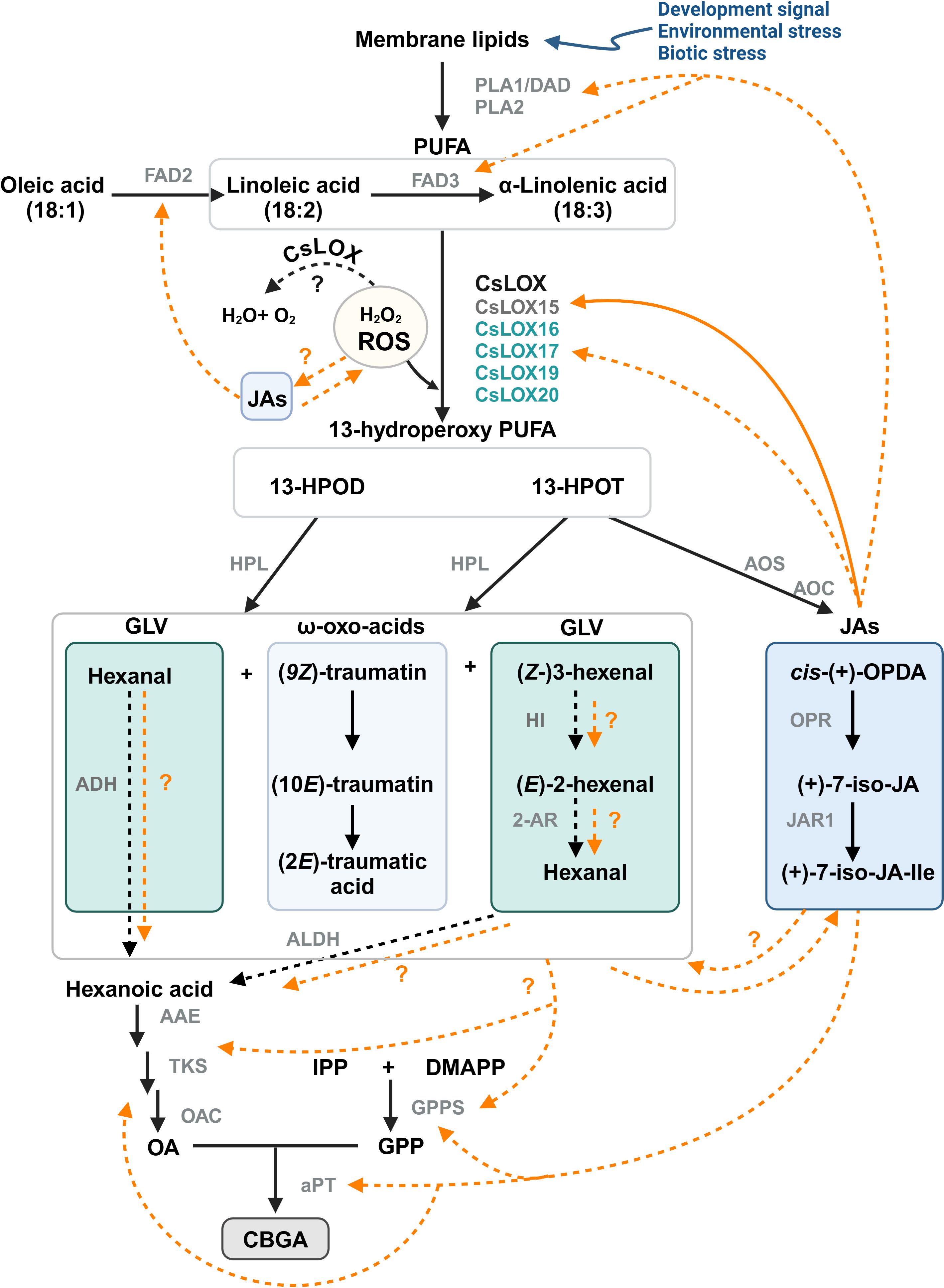
Figure 6. Proposed model for the interactions between oxylipin and phytocannabinoid metabolism in Cannabis. LOX-AOS and LOX-HPL branches of the oxylipin pathway may be interacting with phytocannabinoid biosynthesis. Potential mechanisms could include JA-mediated transcriptional regulation of phytocannabinoid biosynthesis genes or by providing a carbon source via the breakdown of PUFA. Solid orange lines represent known interactions; orange dashed lines indicate other possible interactions. Trichome-specific Cannabis LOX candidates for GLV and JA synthesis are shown (Table 1); Black dashed lines represent enzymatic reaction not yet fully explored in Cannabis. CsLOX15 (dark grey) has been functionally characterized, while other CsLOXs (turquoise) remain uncharacterized. Image created in BioRender.com PUFA, poly unsaturated fatty acids; 13-HPOD, (13S)-hydroperoxy-(9Z,11E)-octadecadienoic acid; 13-HPOT, (13S)-hydroperoxy-(9Z,11E,15Z)- octadecatrienoic acid; cis-(+)-OPDA, cis-(+)-12-oxo-phytodienoic acid; (+)-7-iso-JA, jasmonic acid; (+)-7-iso-JA-Ile, jasmonic acid isoleucine conjugate; (9Z)-traumatin, 12-oxo-(9Z)-dodecenoic acid; (10E)-traumatin, 12-oxo-(10E)-dodecenoic acid, (2E)-traumatic acid; (2E)-dodecanedioic acid; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; OA, olivetolic acid; GPP, geranyl pyrophosphate; DAD, defective in anther dehiscence protein, a phospholipase (PLA1); FAD2/3, fatty acid desaturase 2/3; CsLOX, Cannabis lipoxygenase; HPL, hydroperoxide lyase; AOS, allene oxide synthase; AOC, allene oxide cyclase; OPR, 12-oxo-phytodienoic acid reductase; JAR1, jasmonoyl amino acid conjugate synthase; HI, hexenal isomerase; 2-AR, 2-alkynal reductase; ALDH, aldehyde dehydrogenase; AAE, acyl activating enzyme; TKS, tetraketide synthase; OAC, olivetolic acid cyclase; GPPS, geranyl pyrophosphate synthase; aPT, aromatic prenyltransferase.
8 Conclusion
Despite being a multi-use crop with huge potential as a source for food, fiber and medicine, research on Cannabis has been constrained for more than 50 years. There exists a substantial knowledge gap on molecular drivers responsible for the diverse array of chemicals synthesized by Cannabis glandular trichomes. Cannabis has an extensive number of LOXs, and several are highly expressed in trichomes and are responsive to JAs (Fayaz et al., 2023). We identified the trichome-specific CsLOX15, which together with CsLOX16 and CsLOX17, forms a distinct phylogenetic clade. The use of chemical inhibitors and gene silencing approaches, such as virus-induced gene-silencing (Schachtsiek et al., 2019; Alter et al., 2022), targeting these LOXs and other key branch points of oxylipin biosynthesis will be essential in understanding the contribution of oxylipins to PC production. Structure-function analysis of candidate LOXs by site-directed mutagenesis may also prove useful in understanding sequence variation among trichome-specific LOXs (Farmer et al., 1994; Li et al., 2020).
Determining the influence of oxylipin products among different trichomes types or tissues would also be interesting avenues for future research. However, due to high reactivity and volatile nature of these compounds, challenges exist in examining these in their native form. The use of spatial omics analyses of glandular trichomes and adjacent tissues may also help to resolve the contextual molecular components of these highly specialized cell types and aid in understanding of the molecular interactions between oxylipin and PC pathways. To trace the fate of oxylipin products, it will be necessary to use isotopic tracers to label PUFA precursors. Disentangling these interactions could provide important knowledge on the partitioning of carbon resources between primary and specialized metabolism and offer new opportunities for the biotechnological enhancement of Cannabis. These innovations could ultimately facilitate the development of elite chemical phenotypes for both industrial and medicinal applications.
Author contributions
GS: Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. AG: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. KJ: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. MW: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Australian Research Council (ARC) Research Hub for Medicinal Agriculture (IH180100006). Cann Group Limited are an Industry Partner Organization of IH180100006. GIS is supported by a La Trobe University ARC Research Hub for Medicinal Agriculture Graduate Research Scholarship and a La Trobe University Full Fee Research Scholarship. Work in ARGs laboratory is also supported by the Australian Research Council Research Hub for Sustainable Crop Production (IH19100022).
Acknowledgments
Authors like to acknowledge Prof. Matthew Lewsey, Dr. Oliver Berkowitz, Dr. Ricarda Jost, and Dr. Muluneh Oli for providing RNA sequence data for the LOX gene tissue expression analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1568548/full#supplementary-material
Abbreviations
AOS, Allene oxide synthase; CBC, Cannabichromene; CBCA, Cannabichromenic acid; CBD, Cannabidiol; CBDA, Cannabidiolic acid; CBDAS, Cannabidiolic acid synthase; CBGA, Cannabigerolic acid; CBGAS, Cannabigerolic acid synthase; ECS, Endocannabinoid system; GLV, Green leaf volatiles; GPP, Geranyl pyrophosphate; GPPS, Geranyl pyrophosphate synthase; HPL, Hydroperoxide lyase; JA/JAs, Jasmonate/Jasmonates; JA-Ile, Jasmonic acid isoleucine; LOX, Lipoxygenase; MeJA, Methyl jasmonate; MEP, Methylerythritol 4-phosphate; OA, Olivetolic acid; OAC, Olivetolic acid cyclase; cis-(+)-OPDA, cis-(+)-12-oxo-phytodienoic acid; PC, Phytocannabinoid; PUFA, Polyunsaturated fatty acid; ROS, Reactive oxygen species; Δ9-THC, Δ9-Tetrahydrocannabinol; Δ9-THCA, Δ9-Tetrahydrocannabinolic acid; TKS, Tetraketide synthase.
Footnotes
References
Allmann, S., Halitschke, R., Schuurink, R. C., Baldwin, I. T. (2010). Oxylipin channelling in Nicotiana attenuata: lipoxygenase 2 supplies substrates for green leaf volatile production. Plant Cell Environ. 33, 2028–2040. doi: 10.1111/j.1365-3040.2010.02203.x
Alter, H., Peer, R., Dombrovsky, A., Flaishman, M., Spitzer-Rimon, B. (2022). Tobacco rattle vrus as a tool for rapid reverse-genetics screens and analysis of gene function in Cannabis sativa L. Plants 11, 327. doi: 10.3390/plants11030327
Ameye, M., Allmann, S., Verwaeren, J., Smagghe, G., Haesaert, G., Schuurink, R. C., et al. (2018). Green leaf volatile production by plants: a meta-analysis. New Phytol. 220, 666–683. doi: 10.1111/nph.14671
Anderson, L. L., Ametovski, A., Luo, J. L., Everett-Morgan, D., McGregor, I. S., Banister, S. D., et al. (2021). Cannabichromene, related phytocannabinoids, and 5-fluoro-cannabichromene have anticonvulsant properties in a mouse model of Dravet syndrome. ACS Chem. Neurosci. 12, 330–339. doi: 10.1021/acschemneuro.0c00677
Apicella, P. V., Sands, L. B., Ma, Y., Berkowitz, G. A. (2022). Delineating genetic regulation of cannabinoid biosynthesis during female flower development in. Cannabis sativa. Plant Direct 6, e412. doi: 10.1002/pld3.412
Bailey, R. (2019). The effect of elicitor stimulation on cannabinoid production by industrial hemp (Cannabis sativa) varieties in a hydroponic system (University Honors College, Middle Tennessee State University). Available online at: https://jewlscholar.mtsu.edu/server/api/core/bitstreams/932cdea6-01bd-4bea-ab88-85c5d1ce4ca9/content (Accessed 25 Dec. 2024).
Balcke, G. U., Bennewitz, S., Bergau, N., Athmer, B., Henning, A., Majovsky, P., et al. (2017). Multi-omics of tomato glandular trichomes reveals distinct features of central carbon metabolism supporting high productivity of specialized metabolites. Plant Cell 29, 960–983. doi: 10.1105/tpc.17.00060
Behr, M., Lutts, S., Hausman, J. F., Guerriero, G. (2018). Jasmonic acid to boost secondary growth in hemp hypocotyl. Planta 248, 1029–1036. doi: 10.1007/s00425-018-2951-5
Blée, E. (1998). Biosynthesis of phytooxylipins: the peroxygenase pathway. Lipid/Fett 100, 121–127. doi: 10.1002/(SICI)1521-4133(19985)100:4/5%3C121::AID-LIPI121%3E3.0.CO;2-4
Blée, E. (2002). Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 7, 315–322. doi: 10.1016/S1360-1385(02)02290-2
Booth, J. K., Yuen, M. M. S., Jancsik, S., Madilao, L. L., Page, J. E., Bohlmann, J. (2020). Terpene synthases and terpene variation in Cannabis sativa. Plant Physiol. 184, 130–147. doi: 10.1104/pp.20.00593
Borrego, E. J., Robertson, M., Taylor, J., Schultzhaus, Z., Espinoza, E. M. (2023). Oxylipin biosynthetic gene families of Cannabis sativa. PLoS One 18, e0272893. doi: 10.1371/journal.pone.0272893
Borrelli, F., Fasolino, I., Romano, B., Capasso, R., Maiello, F., Coppola, D., et al. (2013). Beneficial effect of the non-psychotropic plant cannabinoid cannabigerol on experimental inflammatory bowel disease. Biochem. Pharmacol. 85, 1306–1316. doi: 10.1016/j.bcp.2013.01.017
Boughton, A. J., Hoover, K., Felton, G. W. (2005). Methyl jasmonate application induces increased densities of glandular trichomes on tomato, Lycopersicon esculentum. J. Chem. Ecol. 31, 2211–2216. doi: 10.1007/s10886-005-6228-7
Brash, A. R. (1999). Lipoxygenases: occurrence, functions, catalysis, and acquisition of substrate. J. Biol. Chem. 274, 23679–23682. doi: 10.1074/jbc.274.34.23679
Britch, S. C., Babalonis, S., Walsh, S. L. (2021). Cannabidiol: pharmacology and therapeutic targets. Psychopharmacol. 238, 9–28. doi: 10.1007/s00213-020-05712-8
Burgel, L., Hartung, J., Schibano, D., Graeff-Hönninger, S. (2020). Impact of different phytohormones on morphology, yield and cannabinoid content of Cannabis sativa L. Plants 9, 725. doi: 10.3390/plants9060725
Cabrera, C. L. R., Keir-Rudman, S., Horniman, N., Clarkson, N., Page, C. (2021). The anti-inflammatory effects of cannabidiol and cannabigerol alone, and in combination. Pulm. Pharmacol. Ther. 69, 102047. doi: 10.1016/j.pupt.2021.102047
Caldelari, D., Wang, G., Farmer, E. E., Dong, X. (2011). Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 75, 25–33. doi: 10.1007/s11103-010-9701-9
Chanda, D., Neumann, D., Glatz, J. F. (2019). The endocannabinoid system: overview of an emerging multi-faceted therapeutic target. Prostaglandins Leukot. Essent. Fatty Acids 140, 51–56. doi: 10.1016/j.plefa.2018.11.016
Chauvin, A., Caldelari, D., Wolfender, J. L., Farmer, E. E. (2013). Four 13-lipoxygenases contribute to rapid jasmonate synthesis in wounded Arabidopsis thaliana leaves: a role for lipoxygenase 6 in responses to long-distance wound signals. New Phytol. 197, 566–575. doi: 10.1111/nph.12029
Chen, G., Hackett, R., Walker, D., Taylor, A., Lin, Z., Grierson, D. (2004). Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol. 136, 2641–2651. doi: 10.1104/pp.104.041608
Chen, G., Klinkhamer, P. G. L., Escobar-Bravo, R., Leiss, K. A. (2018). Type VI glandular trichome density and their derived volatiles are differently induced by jasmonic acid in developing and fully developed tomato leaves: implications for thrips resistance. Plant Sci. 276, 87–98. doi: 10.1016/j.plantsci.2018.08.007
Chen, Z., Chen, X., Yan, H., Li, W., Li, Y., Cai, R., et al. (2015). The lipoxygenase gene family in poplar: identification, classification, and expression in response to MeJA treatment. PLoS One 10, e0125526. doi: 10.1371/journal.pone.0125526
Christensen, S. A., Huffaker, A., Hunter, C. T., Alborn, H. T., Schmelz, E. A. (2016). A maize death acid, 10-oxo-11-phytoenoic acid, is the predominant cyclopentenone signal present during multiple stress and developmental conditions. Plant Signal. Behav. 11, e1120395. doi: 10.1080/15592324.2015.1120395
Christensen, S. A., Huffaker, A., Kaplan, F., Sims, J., Ziemann, S., Doehlemann, G., et al. (2015). Maize death acids, 9-lipoxygenase-derived cyclopente(a)nones, display activity as cytotoxic phytoalexins and transcriptional mediators. Proc. Natl. Acad. Sci. U.S.A. 112, 11407–11412. doi: 10.1073/pnas.1511131112
Christensen, S. A., Nemchenko, A., Borrego, E., Murray, I., Sobhy, I. S., Bosak, L., et al. (2013). The maize lipoxygenase, ZmLOX10, mediates green leaf volatile, jasmonate and herbivore-induced plant volatile production for defense against insect attack. Plant J. 74, 59–73. doi: 10.1111/tpj.12101
Corpas, F. J., Gupta, D. K., Palma, J. M. (2015). “Production sites of reactive oxygen species (ROS) in organelles from plant cells,” in Reactive Oxygen Species and Oxidative Damage in Plants Under Stress. Eds. Gupta, D. K., Palma, J. M., Corpas, F. J. (Springer, Cham), 1–22. doi: 10.1007/978-3-319-20421-5_1
Cropano, C., Carrera, D.Á., George, G. M., Katsir, L., Vogt, M. M., Ruckle, M. E., et al. (2022). Quantitative trait loci (QTLs) associated with a high-varin trait in Cannabis. WO2022180532A1. Available online at: https://patents.google.com/patent/WO2022180532A1/en?assignee=puregene&oq=puregene (Accessed November 10, 2024).
de Brito Siqueira, A. L., Cremasco, P. V., Bahú, J. O., da Silva, A. P., de Andrade, L. R. M., González, P. G., et al. (2023). Phytocannabinoids: Pharmacological effects, biomedical applications, and worldwide prospection. J. Tradit. Complement. Med. 13, 575–587. doi: 10.1016/j.jtcme.2023.08.006
Demiwal, P., Nabi, S. U., Mir, J. I., Verma, M. K., Yadav, S. R., Roy, P., et al. (2024). Methyl jasmonate improves resistance in scab-susceptible Red Delicious apple by altering ROS homeostasis and enhancing phenylpropanoid biosynthesis. Plant Physiol. Biochem. 207, 108371. doi: 10.1016/j.plaphy.2024.108371
Denness, L., McKenna, J. F., Segonzac, C., Wormit, A., Madhou, P., Bennett, M., et al. (2011). Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species-and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 156, 1364–1374. doi: 10.1104/pp.111.175737
Devinsky, O., Cilio, M. R., Cross, H., Fernandez-Ruiz, J., French, J., Hill, C., et al. (2014). Cannabidiol: pharmacology and potential therapeutic role in epilepsy and other neuropsychiatric disorders. Epilepsia 55, 791–802. doi: 10.1111/epi.12631
Dong, Y., Li, J., Zhang, W., Bai, H., Li, H., Shi, L. (2022a). Exogenous application of methyl jasmonate affects the emissions of volatile compounds in lavender (Lavandula angustifolia). Plant Physiol. Biochem. 185, 25–34. doi: 10.1016/j.plaphy.2022.05.022
Dong, Y., Zhang, W., Li, J., Wang, D., Bai, H., Li, H., et al. (2022b). The transcription factor LaMYC4 from lavender regulates volatile Terpenoid biosynthesis. BMC Plant Biol. 22, 289. doi: 10.1186/s12870-022-03660-3
Dowling, C. A., Shi, J., Toth, J. A., Quade, M. A., Smart, L. B., McCabe, P. F., et al. (2024). A FLOWERING LOCUS T ortholog is associated with photoperiod-insensitive flowering in hemp (Cannabis sativa L.). Plant J. 119, 383–403. doi: 10.1111/tpj.16769
Egmond, M., Veldink, G. A., Vliegenthart, J. F. G., Boldingh, J. (1973). C-11 H-abstraction from linoieic acid, the rate-limiting step in lipoxygenase catalysis. Biophys. Res. Commun. 54, 1178–1184. doi: 10.1016/0006-291X(73)90816-4
Fayaz, M., Kundan, M., Gani, U., Sharma, P., Wajid, M. A., Katoch, K., et al. (2023). Identification of Lipoxygenase gene repertoire of Cannabis sativa and functional characterization of CsLOX13 gene. Plant Sci. 334, 111780. doi: 10.1016/j.plantsci.2023.111780
Farmer, E. E., Caldelari, D., Pearce, G., Walker-Simmons, M., Ryan, C. A. (1994). Diethyldithiocarbamic acid inhibits the octadecanoid signaling pathway for the wound induction of proteinase inhibitors in tomato leaves. Plant Physiol. 106 (1), 337–342. doi: 10.1104/pp.106.1.337
Flores-Sanchez, I. J., Pec, J., Fei, J., Choi, Y. H., Dusek, J., Verpoorte, R., et al (2009). Elicitation studies in cell suspension cultures of Cannabis sativa L. J. Biotechnol. 143 (2), 157–168. doi: 10.1016/j.jbiotec.2009.05.006
Fortes, A. M., Coronado, M. J., Testillano, P. S., Risueño, M. D. C., Pais, M. S. (2004). Expression of lipoxygenase during organogenic nodule formation from hop internodes. J. Histochem. Cytochem. 52, 227–241. doi: 10.1177/002215540405200211
Frassinetti, S., Moccia, E., Caltavuturo, L., Gabriele, M., Longo, V., Bellani, L., et al. (2018). Nutraceutical potential of hemp (Cannabis sativa L.) seeds and sprouts. Food Chem. 262, 56–66. doi: 10.1016/j.foodchem.2018.04.078
Froehlich, J. E., Itoh, A., Howe, G. A. (2001). Tomato allene oxide synthase and fatty acid hydroperoxide lyase, two cytochrome P450s involved in oxylipin metabolism, are targeted to different membranes of chloroplast envelope. Plant Physiol. 125, 306–317. doi: 10.1104/pp.125.1.306
Gaffney, B. J. (2020). EPR spectroscopic studies of lipoxygenases. Chem. Asian J. 15, 42–50. doi: 10.1002/asia.201901461
Gagne, S. J., Stout, J. M., Liu, E., Boubakir, Z., Clark, S. M., Page, J. E. (2012). Identification of olivetolic acid cyclase from Cannabis sativa reveals a unique catalytic route to plant polyketides. Proc. Natl. Acad. Sci. U.S.A. 109, 12811–12816. doi: 10.1073/pnas.1200330109
Garrido, J., Rico, S., Corral, C., Sanchez, C., Vidal, N., Martinez-Quesada, J. J., et al. (2022). Exogenous application of stress-related signaling molecules affect growth and cannabinoid accumulation in medical cannabis (Cannabis sativa L.). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1082554
Ghasemi Pirbalouti, A., Sajjadi, S. E., Parang, K. (2014). A review (research and patents) on jasmonic acid and its derivatives. Arch. Pharm. (Weinheim). 347, 229–239. doi: 10.1002/ardp.201300287
Ghorbel, M., Brini, F., Sharma, A., Landi, M. (2021). Role of jasmonic acid in plants: the molecular point of view. Plant Cell Rep. 40, 1471–1494. doi: 10.1007/s00299-021-02687-4
Goldsmith, C. R., Jonas, R. T., Stack, T. D. P. (2002). C– H bond activation by a ferric methoxide complex: modeling the rate-determining step in the mechanism of lipoxygenase. J. Am. Chem. Soc 124, 83–96. doi: 10.1021/ja016451g
Gorina, S. S., Mukhtarova, L. S., Iljina, T. M., Toporkova, Y. Y., Grechkin, A. N. (2022). Detection of divinyl ether synthase CYP74H2 biosynthesizing (11Z)-etheroleic and (1′Z)-colnelenic acids in asparagus (Asparagus officinalis L.). Phytochemistry 200, 113212. doi: 10.1016/j.phytochem.2022.113212
Grechkin, A. N., Ilyasov, A. V., Hamberg, M. (1997). On the mechanism of biosynthesis of divinyl ether oxylipins by enzyme from garlic bulbs. Eur. J. Biochem. 245, 137–142. doi: 10.1111/j.1432-1033.1997.00137.x
Hahm, S., Lee, Y., Lee, K., Park, J. (2024). Optimization of cannabinoid production in hemp through methyl jasmonate application in a vertical farming system. Horticulturae 10, 1165. doi: 10.3390/horticulturae10111165
Hamberg, M. (1999). An epoxy alcohol synthase pathway in higher plants: biosynthesis of antifungal trihydroxy oxylipins in leaves of potato. Lipids 34, 1131–1142. doi: 10.1007/s11745-999-0464-7
Hamberg, M. (2004). Isolation and structures of two divinyl ether fatty acids from Clematis vitalba. Lipids 39, 565–569. doi: 10.1007/s11745-004-1264-9
Hammond, C. T., Mahlberg, P. G. (1973). Morphology of glandular hairs of Cannabis sativa from scanning electron microscopy. Am. J. Bot. 60, 524–528. doi: 10.1002/j.1537-2197.1973.tb05953.x
Han, G., Li, Y., Yang, Z., Wang, C., Zhang, Y., Wang, B. (2022). Molecular mechanisms of plant trichome development. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.910228
Hazekamp, A., Tejkalová, K., Papadimitriou, S. (2016). Cannabis: from cultivar to chemovar II—a metabolomics approach to cannabis classification. Cannabis Cannabinoid Res. 1, 202–215. doi: 10.1089/can.2016.0017
He, Y., Borrego, E. J., Gorman, Z., Huang, P. C., Kolomiets, M. V. (2020). Relative contribution of LOX10, green leaf volatiles and JA to wound-induced local and systemic oxylipin and hormone signature in Zea mays (maize). Phytochemistry 174, 112334. doi: 10.1016/j.phytochem.2020.112334
Hershelman, D., Kahler, K. M., Price, M. J., Lu, I., Fu, Y., Plumeri, P. A., et al. (2019). Oxygenation reactions catalyzed by the F557V mutant of soybean lipoxygenase-1: evidence for two orientations of substrate binding. Arch. Biochem. Biophys. 674, 108082. doi: 10.1016/j.abb.2019.108082
Hesami, M., Pepe, M., Baiton, A., Jones, A. M. P. (2022a). Current status and future prospects in cannabinoid production through in vitro culture and synthetic biology. Biotechnol. Adv. 108074. doi: 10.1016/j.bioteChadv.2022.108074
Hesami, M., Pepe, M., Baiton, A., Salami, S. A., Jones, A. M. P. (2022b). New insight into ornamental applications of cannabis: Perspectives and challenges. Plants 11 2383. doi: 10.3390/plants11182383
Hickman, R., Van Verk, M. C., Van Dijken, A. J., Mendes, M. P., Vroegop-Vos, I. A., Caarls, L., et al. (2017). Architecture and dynamics of the jasmonic acid gene regulatory network. Plant Cell. 29, 2086–2105. doi: 10.1105/tpc.16.00958
Holman, T. R., Zhou, J., Solomon, E. I. (1998). Spectroscopic and functional characterization of a ligand coordination mutant of soybean lipoxygenase-1: first coordination sphere analogue of human 15-lipoxygenase. J. Am. Chem. Soc. 120, 12564–12572. doi: 10.1021/ja982844c
Hornung, E., Walther, M., Kühn, H., Feussner, I. (1999). Conversion of cucumber linoleate 13-lipoxygenase to a 9-lipoxygenating species by site-directed mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 96, 4192–4197. doi: 10.1073/pnas.96.7.4192
Horper, W., Marner, F. J. (1996). Biosynthesis of primin and miconidin and its derivatives. Phytochemistry 41, 451–456. doi: 10.1016/0031-9422(95)00590-0
Howe, G. A., Lee, G. I., Itoh, A., Li, L., DeRocher, A. E. (2000). Cytochrome P450-dependent metabolism of oxylipins in tomato. Cloning and expression of allene oxide synthase and fatty acid hydroperoxide lyase. Plant Physiol. 123, 711–724. doi: 10.1104/pp.123.2.711
Hu, S., Offenbacher, A. R., Thompson, E. M., Gee, C. L., Wilcoxen, J., Carr, C. A., et al. (2019). Biophysical characterization of a disabled double mutant of soybean lipoxygenase: the “undoing” of precise substrate positioning relative to metal cofactor and an identified dynamical network. J. Am. Chem. Soc. 141, 1555–1567. doi: 10.1021/jacs.8b10992
Huang, X., Chen, W., Zhao, Y., Chen, J., Ouyang, Y., Li, M., et al. (2024). Deep learning-based quantification and transcriptomic profiling reveal a methyl jasmonate-mediated glandular trichome formation pathway in Cannabis sativa. Plant J. 118, 1155–1173. doi: 10.1111/tpj.16663
Huang, J., Rücker, A., Schmidt, A., Gleixner, G., Gershenzon, J., Trumbore, S., et al. (2020). Production of constitutive and induced secondary metabolites is coordinated with growth and storage in Norway spruce saplings. Tree Physiol. 40, 928–942. doi: 10.1093/treephys/tpaa040
Hughes, R., Lawson, D. M., Hornostaj, A. R., Fairhurst, S. A., Casey, R. (2001). Mutagenesis and modelling of linoleate-binding to pea seed lipoxygenase. Eur. J. Biochem. 268, 1030–1040. doi: 10.1046/j.1432-1327.2001.01964.x
Hurgobin, B., Tamiru-Oli, M., Welling, M. T., Doblin, M. S., Bacic, A., Whelan, J., et al. (2021). Recent advances in Cannabis sativa genomics research. New Phytol. 230, 73–89. doi: 10.1111/nph.17140
Jeyasri, R., Muthuramalingam, P., Karthick, K., Shin, H., Choi, S. H., Ramesh, M. (2023). Methyl jasmonate and salicylic acid as powerful elicitors for enhancing the production of secondary metabolites in medicinal plants: an updated review. Plant Cell Tissue Organ Cult. 153, 447–458. doi: 10.1007/s11240-023-02485-8
Jin, D., Henry, P., Shan, J., Chen, J. (2021). Identification of chemotypic markers in three chemotype categories of cannabis using secondary metabolites profiled in inflorescences, leaves, stem bark, and roots. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.699530
Jost, R., Berkowitz, O., Pegg, A., Hurgobin, B., Tamiru-Oli, M., Welling, M. T., et al. (2025). Sink strength, nutrient allocation, cannabinoid yield, and associated transcript profiles vary in two drug-type Cannabis chemovars. J. Exp. Bot. 76, 152–174. doi: 10.1093/jxb/erae367
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kianersi, F., Azarm, D. A., Pour-Aboughadareh, A., Poczai, P. (2022). Change in secondary metabolites and expression pattern of key rosmarinic acid related genes in Iranian lemon balm (Melissa officinalis L.) ecotypes using methyl jasmonate treatments. Molecules 27, 1715. doi: 10.3390/molecules27051715
Knapp, M. J., Klinman, J. P. (2003). Kinetic studies of oxygen reactivity in soybean lipoxygenase-1. Biochemistry 42, 11466–11475. doi: 10.1021/bi0300884
Kolomiets, M. V., Hannapel, D. J., Chen, H., Tymeson, M., Gladon, R. J. (2001). Lipoxygenase is involved in the control of potato tuber development. Plant Cell 13, 613–626. doi: 10.1105/tpc.13.3.613
Kovalchuk, I., Pellino, M., Rigault, P., Van Velzen, R., Ebersbach, J., Ashnest, J. R., et al. (2020). The genomics of Cannabis and its close relatives. Annu. Rev. Plant Biol. 71, 713–739. doi: 10.1146/annurev-arplant-081519-040203
Kroumova, A. B., Xie, Z., Wagner, G. J. (1994). A pathway for the biosynthesis of straight and branched, odd-and even-length, medium-chain fatty acids in plants. Proc. Natl. Acad. Sci. U.S.A. 91, 11437–11441. doi: 10.1073/pnas.91.24.11437
Letunic, I., Bork, P. (2007). Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127,128. doi: 10.1093/bioinformatics/btl529
Li, L., Chang, Z., Pan, Z., Fu, Z. Q., Wang, X. (2008). Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase. Proc. Natl. Acad. Sci. U.S.A. 105, 13883–13888. doi: 10.1073/pnas.0804099105
Li, P., Soudackov, A. V., Hammes-Schiffer, S. (2018). Impact of mutations on the binding pocket of soybean lipoxygenase: implications for proton-coupled electron transfer. J. Phys. Chem. Lett. 9, 6444–6449. doi: 10.1021/acs.jpclett.8b02945
Li, J., Wang, X., Jiang, R., Dong, B., Fang, S., Li, Q., et al. (2021). Phytohormone-based regulation of trichome development. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.734776
Li, Y., Qiu, L., Zhang, Q., Zhuansun, X., Li, H., Chen, X., et al. (2020). Exogenous sodium diethyldithiocarbamate, a Jasmonic acid biosynthesis inhibitor, induced resistance to powdery mildew in wheat. Plant Direct. 4 (4), e00212. doi: 10.1002/pld3.212
Liavonchanka, A., Feussner, I. (2006). Lipoxygenases: occurrence, functions and catalysis. J. Plant Physiol. 163, 348–357. doi: 10.1016/j.jplph.2005.11.006
Lichtman, A. H., Lux, E. A., McQuade, R., Rossetti, S., Sanchez, R., Sun, W., et al. (2018). Results of a double-blind, randomized, placebo-controlled study of nabiximols oromucosal spray as an adjunctive therapy in advanced cancer patients with chronic uncontrolled pain. J. Pain Symptom Manage. 55, 179–188.e171. doi: 10.1016/j.jpainsymman.2017.09.001
Liu, S., Han, B. (2010). Differential expression pattern of an acidic 9/13-lipoxygenase in flower opening and senescence and in leaf response to phloem feeders in the tea plant. BMC Plant Biol. 10, 1–15. doi: 10.1186/1471-2229-10-228
Liu, J., Zhou, Y., Li, J., Wang, F., Yang, Y. (2020). Comprehensive genomic characterization and expression analysis of the lipoxygenase gene family in watermelon under hormonal treatments. Agriculture 10, 429. doi: 10.3390/agriculture10100429
Livingston, S. J., Bae, E. J., Unda, F., Hahn, M. G., Mansfield, S. D., Page, J. E., et al. (2021). Cannabis glandular trichome cell walls undergo remodeling to store specialized metabolites. Plant Cell Physiol. 62, 1944–1962. doi: 10.1093/pcp/pcab127
Livingston, S. J., Quilichini, T. D., Booth, J. K., Wong, D. C., Rensing, K. H., Laflamme-Yonkman, J., et al. (2020). Cannabis glandular trichomes alter morphology and metabolite content during flower maturation. Plant J. 101, 37–56. doi: 10.1111/tpj.14516
Livingston, S. J., Rensing, K. H., Page, J. E., Samuels, A. L. (2022). A polarized supercell produces specialized metabolites in cannabis trichomes. Curr. Biol. 32, 4040–4047.e4044. doi: 10.1016/j.cub.2022.07.014
Luo, X., Reiter, M. A., d’Espaux, L., Wong, J., Denby, C. M., Lechner, A., et al. (2019). Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature 567, 123–126. doi: 10.1038/s41586-019-0978-9
Marks, M. D., Tian, L., Wenger, J. P., Omburo, S. N., Soto-Fuentes, W., He, J., et al. (2009). Identification of candidate genes affecting Δ9-tetrahydrocannabinol biosynthesis in Cannabis sativa. J. Exp. Bot. 60, 3715–3726. doi: 10.1093/jxb/erp210
Matsumoto, S., Utsumi, Y., Kozuka, T., Iwamura, M., Nakai, T., Yamauchi, D., et al. (2024). CT image-based 3D inflorescence estimation of Chrysanthemum seticuspe. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1374937
McGarry, R. C., Lin, Y.-T., Kaur, H., Higgs, H., Arias-Gaguancela, O., Ayre, B. G. (2024). Disrupted oxylipin biosynthesis mitigates pathogen infections and pest infestations in cotton (Gossypium hirsutum). J. Exp. Bot. 75, 7365–7380. doi: 10.1093/jxb/erae394
Mechoulam, R., Parker, L. A. (2013). The endocannabinoid system and the brain. Annu. Rev. Psychol. 64, 21–47. doi: 10.1146/annurev-psych-113011-143739
Meyer, M. P., Tomchick, D. R., Klinman, J. P. (2008). Enzyme structure and dynamics affect hydrogen tunneling: The impact of a remote side chain (I553) in soybean lipoxygenase-1. Proc. Natl. Acad. Sci. U.S.A. 105, 1146–1151. doi: 10.1073/pnas.0710643105
Minor, W., Steczko, J., Stec, B., Otwinowski, Z., Bolin, J. T., Walter, R., et al. (1996). Crystal structure of soybean lipoxygenase L-1 at 1.4 Å resolution. Biochemistry 35, 10687–10701. doi: 10.1021/bi960576u
Mochizuki, S., Sugimoto, K., Koeduka, T., Matsui, K. (2016). Arabidopsis lipoxygenase 2 is essential for formation of green leaf volatiles and five-carbon volatiles. FEBS Lett. 590, 1017–1027. doi: 10.1002/1873-3468.12133
Naimuzzaman, M., Hasan, M. M., Kumer, A., Hossin, A. Y., Harun-Ur-Rashid, M., Roy, S. K., et al. (2025). Computational and In silico study of novel fungicides against combating root rot, gray mold, fusarium wilt, and cereal rust. PloS One 20, e0316606. doi: 10.1371/journal.pone.0316606
Nakashima, A., von Reuss, S. H., Tasaka, H., Nomura, M., Mochizuki, S., Iijima, Y., et al. (2013). Traumatin-and dinortraumatin-containing galactolipids in Arabidopsis: their formation in tissue-disrupted leaves as counterparts of green leaf volatiles. J. Biol. Chem. 288, 26078–26088. doi: 10.1074/jbc.M113.487959
Newie, J., Andreou, A., Neumann, P., Einsle, O., Feussner, I., Ficner, R. (2016). Crystal structure of a lipoxygenase from Cyanothece sp. may reveal novel features for substrate acquisition. J. Lipid Res. 57, 276–287. Available online at: https://www.jlr.org/article/S0022-2275(20)35463-8/fulltext.
Offenbacher, A. R., Hu, S., Poss, E. M., Carr, C. A., Scouras, A. D., Prigozhin, D. M., et al. (2017). Hydrogen–deuterium exchange of lipoxygenase uncovers a relationship between distal, solvent exposed protein motions and the thermal activation barrier for catalytic proton-coupled electron tunneling. ACS Cent. Sci. 3, 570–579. doi: 10.1021/acscentsci.7b00142
Omare, M. O., Kibet, J. K., Cherutoi, J. K., Kengara, F. O. (2021). Current trends in the use of Cannabis sativa: beyond recreational and medicinal applications. Open Access Libr. J. 8, 1–15. doi: 10.4236/oalib.1107132
Oultram, J. M. J., Pegler, J. L., Eamens, A. L., Gordon, R., Korbie, D. J., Grof, C. P. L. (2024). Exogenously applied gibberellic acid alters cannabinoid profile in Cannabis sativa L. Agronomy 14, 2417. doi: 10.3390/agronomy14102417
Pattanaik, S., Patra, B., Singh, S. K., Yuan, L. (2014). An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00259
Piersimoni, L., Kastritis, P. L., Arlt, C., Sinz, A. (2021). Cross-linking mass spectrometry for investigating protein conformations and protein–protein interactions─ a method for all seasons. Chem. Rev. 122, 7500–7531. doi: 10.1021/acs.chemrev.1c00786
Rashid, A., Khajuria, M., Ali, V., Faiz, S., Jamwal, S., Vyas, D. (2023). Genome-wide identification and expression profiling of WRKY family suggest their potential role in cannabinoid regulation in Cannabis sativa L. Ind. Crops Prod. 206, 117706. doi: 10.1016/j.indcrop.2023.117706
Raza, A., Charagh, S., Zahid, Z., Mubarik, M. S., Javed, R., Siddiqui, M. H., et al. (2021). Jasmonic acid: a key frontier in conferring abiotic stress tolerance in plants. Plant Cell Rep. 40, 1513–1541. doi: 10.1007/s00299-020-02614-z
Rodrigues Magalhães, H. C., Alves Filho, E. G., Rivero Meza, S. L., Oliveira, A., Garruti, D. S., Purgatto, E. (2023). Effect of methyl jasmonate on the biosynthesis of volatile compounds associated with the ripening of grape tomato fruits. J. Agric. Food Chem. 71, 4696–4705. doi: 10.1021/acs.jafc.2c06215
Ruddat, V. C., Mogul, R., Chorny, I., Chen, C., Perrin, N., Whitman, S., et al. (2004). Tryptophan 500 and arginine 707 define product and substrate active site binding in soybean lipoxygenase-1. Biochemistry 43, 13063–13071. doi: 10.1021/bi0489098
Salamone, S., Waltl, L., Pompignan, A., Grassi, G., Chianese, G., Koeberle, A., et al. (2022). Phytochemical characterization of Cannabis sativa L. chemotype V reveals three new dihydrophenanthrenoids that favorably reprogram lipid mediator biosynthesis in macrophages. Plants 11, 2130. doi: 10.3390/plants11162130
Sanadhya, P., Kumar, A., Bucki, P., Fitoussi, N., Carmeli-Weissberg, M., Borenstein, M., et al. (2021). Tomato divinyl ether-biosynthesis pathway is implicated in modulating of root-knot nematode Meloidogyne javanica’s parasitic ability. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.670772
Sands, L. B., Haiden, S. R., Ma, Y., Berkowitz, G. A. (2023). Hormonal control of promoter activities of Cannabis sativa prenyltransferase 1 and 4 and salicylic acid mediated regulation of cannabinoid biosynthesis. Sci. Rep. 13, 8620. doi: 10.1038/s41598-023-35303-4
Scala, A., Allmann, S., Mirabella, R., Haring, M. A., Schuurink, R. C. (2013). Green leaf volatiles: a plant’s multifunctional weapon against herbivores and pathogens. Int. J. Mol. Sci. 14, 17781–17811. doi: 10.3390/ijms140917781
Schachtsiek, J., Hussain, T., Azzouhri, K., Kayser, O., Stehle, F. (2019). Virus-induced gene silencing (VIGS) in Cannabis sativa L. Plant Methods 15, 157. doi: 10.1186/s13007-019-0542-5
Schenk, G., Neidig, M. L., Zhou, J., Holman, T. R., Solomon, E. I. (2003). Spectroscopic characterization of soybean lipoxygenase-1 mutants: the role of second coordination sphere residues in the regulation of enzyme activity. Biochemistry 42, 7294–7302. doi: 10.1021/bi027380g
Schlotzhauer, W. S., Pair, S. D., Horvat, R. J. (1996). Volatile constituents from the flowers of Japanese honeysuckle (Lonicera japonica). J. Agric. Food Chem. 44, 206–209. doi: 10.1021/jf950275b
Schuurink, R., Tissier, A. (2020). Glandular trichomes: micro-organs with model status? New Phytol. 225, 2251–2266. doi: 10.1111/nph.16283
Segraves, E. N., Chruszcz, M., Neidig, M. L., Ruddat, V., Zhou, J., Wecksler, A. T., et al. (2006). Kinetic, spectroscopic, and structural investigations of the soybean lipoxygenase-1 first-coordination sphere mutant, Asn694Gly. Biochemistry 45, 10233–10242. doi: 10.1021/bi060577e
Shakil, S. S. M., Gowan, M., Hughes, K., Azam, M. N. K., Ahmed, M. N. (2021). A narrative review of the ethnomedicinal usage of Cannabis sativa Linnaeus as traditional phytomedicine by folk medicine practitioners of Bangladesh. J. Cannabis Res. 3, 1–12. doi: 10.1186/s42238-021-00063-3
Shen, J., Tieman, D., Jones, J. B., Taylor, M. G., Schmelz, E., Huffaker, A., et al. (2014). A 13-lipoxygenase, TomloxC, is essential for synthesis of C5 flavour volatiles in tomato. J. Exp. Bot. 65, 419–428. doi: 10.1093/jxb/ert382
Sievers, F., Higgins, D. G. (2021). “The Clustal Omega Multiple Alignment Package.” in Multiple Sequence Alignment, ed. Katoh, K (New York, NY: Humana). Methods Mol. Biol. 2231. doi: 10.1007/978-1-0716-1036-7_1
Sirikantaramas, S., Morimoto, S., Shoyama, Y., Ishikawa, Y., Wada, Y., Shoyama, Y., et al. (2004). The gene controlling marijuana psychoactivity: molecular cloning and heterologous expression of Δ1-tetrahydrocannabinolic acid synthase from Cannabis sativa L. J. Biol. Chem. 279, 39767–39774. doi: 10.1074/jbc.M403693200
Sirikantaramas, S., Taura, F. (2017). “Cannabinoids: biosynthesis and biotechnological applications,” in Cannabis sativa L. - Botany and Biotechnology. Eds. Chandra, S., Lata, H., ElSohly, M. A. (Springer Nature, Cham), 183–206. doi: 10.1007/978-3-319-54564-6_8
Sirikantaramas, S., Taura, F., Tanaka, Y., Ishikawa, Y., Morimoto, S., Shoyama, Y. (2005). Tetrahydrocannabinolic acid synthase, the enzyme controlling marijuana psychoactivity, is secreted into the storage cavity of the glandular trichomes. Plant Cell Physiol. 46, 1578–1582. doi: 10.1093/pcp/pci166
Skrzypczak-Jankun, E., Bross, R. A., Carroll, R. T., Dunham, W. R., Funk, M. O. (2001). Three-dimensional structure of a purple lipoxygenase. J. Am. Chem. Soc 123, 10814–10820. doi: 10.1021/ja011759t
Small, E. (2015). Evolution and classification of Cannabis sativa (marijuana, hemp) in relation to human utilization. Bot. Rev. 81, 189–294. doi: 10.1007/s12229-015-9157-3
Song, W. C., Brash, A. R. (1991). Purification of an allene oxide synthase and identification of the enzyme as a cytochrome P-450. Science 253, 781–784. doi: 10.1126/science.1876834
Song, C., Cao, Y., Dai, J., Li, G., Manzoor, M. A., Chen, C., et al. (2022). The multifaceted roles of MYC2 in plants: toward transcriptional reprogramming and stress tolerance by jasmonate signaling. Front.Plant Sci. 13. doi: 10.3389/fpls.2022.868874
Song, H., Wang, P., Li, C., Han, S., Lopez-Baltazar, J., Zhang, X., et al. (2016). Identification of lipoxygenase (LOX) genes from legumes and their responses in wild type and cultivated peanut upon Aspergillus flavus infection. Sci. Rep. 6, 35245. doi: 10.1038/srep35245
Spitzer-Rimon, B., Duchin, S., Bernstein, N., Kamenetsky, R. (2019). Architecture and florogenesis in female Cannabis sativa plants. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00350
Steczko, J., Donoho, G. P., Clemens, J. C., Dixon, J. E., Axelrod, B. (1992). Conserved histidine residues in soybean lipoxygenase: functional consequences of their replacement. Biochemistry 31, 4053–4057. doi: 10.1021/bi00131a022
Steel, L., Welling, M., Ristevski, N., Johnson, K., Gendall, A. (2023). Comparative genomics of flowering behavior in Cannabis sativa. Front. Plant Sci. 14, 1227898. doi: 10.3389/fpls.2023.1227898
Stout, J. M., Boubakir, Z., Ambrose, S. J., Purves, R. W., Page, J. E. (2012). The hexanoyl-CoA precursor for cannabinoid biosynthesis is formed by an acyl-activating enzyme in Cannabis sativa trichomes. Plant J. 71, 353–365. doi: 10.1111/j.1365-313X.2012.04949.x
Tagen, M., Klumpers, L. E. (2022). Review of delta-8-tetrahydrocannabinol (Δ8-THC): Comparative pharmacology with Δ9-THC. Br. J. Pharmacol. 179, 3915–3933. doi: 10.1111/bph.15865
Tanney, C. A., Backer, R., Geitmann, A., Smith, D. L. (2021). Cannabis glandular trichomes: A cellular metabolite factory. Front. Plant Sci. 12, 1923. doi: 10.3389/fpls.2021.721986
Taura, F., Morimoto, S., Shoyama, Y. (1996). Purification and characterization of cannabidiolic-acid synthase from Cannabis sativa L.: Biochemical analysis of a novel enzyme that catalyzes the oxidocyclization of cannabigerolic acid to cannabidiolic acid. J. Biol. Chem. 271, 17411–17416. doi: 10.1074/jbc.271.29.17411
Thiele, E. A., Marsh, E. D., French, J. A., Mazurkiewicz-Beldzinska, M., Benbadis, S. R., Joshi, C., et al. (2018). Cannabidiol in patients with seizures associated with Lennox-Gastaut syndrome (GWPCARE4): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet 391, 1085–1096. doi: 10.1016/S0140-6736(18)30136-3
Tomchick, D. R., Phan, P., Cymborowski, M., Minor, W., Holman, T. R. (2001). Structural and functional characterization of second-coordination sphere mutants of soybean lipoxygenase-1. Biochemistry 40, 7509–7517. doi: 10.1021/bi002893d
Tong, X., Qi, J., Zhu, X., Mao, B., Zeng, L., Wang, B., et al. (2012). The rice hydroperoxide lyase OsHPL3 functions in defense responses by modulating the oxylipin pathway. Plant J. 71, 763–775. doi: 10.1111/j.1365-313x.2012.05027.x
Toporkova, Y. Y., Smirnova, E. O., Gorina, S. S. (2024). Epoxyalcohol synthase branch of lipoxygenase cascade. Curr. Issues Mol. Biol. 46, 821–841. doi: 10.3390/cimb46010053
Toth, J. A., Stack, G. M., Carlson, C. H., Smart, L. B. (2022). Identification and mapping of major-effect flowering time loci Autoflower1 and Early1 in Cannabis sativa L. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.991680
Urits, I., Borchart, M., Hasegawa, M., Kochanski, J., Orhurhu, V., Viswanath, O. (2019). An update of current cannabis-based pharmaceuticals in pain medicine. Pain Ther. 8, 41–51. doi: 10.1007/s40122-019-0114-4
Vancanneyt, G., Sanz, C., Farmaki, T., Paneque, M., Ortego, F., Castañera, P., et al. (2001). Hydroperoxide lyase depletion in transgenic potato plants leads to an increase in aphid performance. Proc. Natl. Acad. Sci. U.S.A. 98, 8139–8144. doi: 10.1073/pnas.141079498
Vogt, J., Schiller, D., Ulrich, D., Schwab, W., Dunemann, F. (2013). Identification of lipoxygenase (LOX) genes putatively involved in fruit flavour formation in apple (Malus domestica). Tree Genet. Genomes 9, 1493–1511. doi: 10.1007/s11295-013-0653-5
Wang, Y., Liu, M., Ge, D., Akhter-Bhat, J., Li, Y., Kong, J., et al. (2020). Hydroperoxide lyase modulates defense response and confers lesion-mimic leaf phenotype in soybean (Glycine max (L.) Merr.). Plant J. 104, 1315–1333. doi: 10.1111/tpj.15002
Wang, J., Song, L., Gong, X., Xu, J., Li, M. (2020). Functions of jasmonic acid in plant regulation and response to abiotic stress. Int. J. Mol. Sci. 21, 1446. doi: 10.3390/ijms21041446
Wang, Y., Wang, D., Li, S., Zhai, Y., Zhao, Y., Zhong, F., et al. (2024). Methyl jasmonate and salicylic acid enhance the total flavonoid, phenolics, and cannabidiol contents of Cannabis sativa L. adventitious roots. Plant Cell Tissue Organ Cult. 159, 1–14. doi: 10.1007/s11240-024-02909-z
Wang, X., Zeng, Y., Li, L., Fan, R., Yan, Y., Guo, L., et al. (2024). Unraveling jasmonate-mediated plant immunity against green peach aphid (Myzus persicae) in peach: an integrated transcriptomic and RNA sequencing approach. Plant Growth Regul. 104, 1245–1259. doi: 10.1007/s10725-024-01198-y
Wasternack, C., Feussner, I. (2018). The oxylipin pathways: biochemistry and function. Annu. Rev. Plant Biol. 69, 363–386. doi: 10.1146/annurev-arplant-042817-040440
Wasternack, C., Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Weber, H., Chételat, A., Caldelari, D., Farmer, E. E. (1999). Divinyl ether fatty acid synthesis in late blight–diseased potato leaves. Plant Cell 11, 485–493. doi: 10.1105/tpc.11.3.485
Welling, M. T., Deseo, M. A., O’Brien, M., Clifton, J., Bacic, A., Doblin, M. S. (2023). Metabolomic analysis of methyl jasmonate treatment on phytocannabinoid production in Cannabis sativa. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1110144
Welling, M. T., Liu, L., Kretzschmar, T., Mauleon, R., Ansari, O., King, G. J. (2020). An extreme-phenotype genome-wide association study identifies candidate cannabinoid pathway genes in Cannabis. Sci. Rep. 10, 18643. doi: 10.1038/s41598-020-75271-7
Welling, M. T., Shapter, T., Rose, T. J., Liu, L., Stanger, R., King, G. J. (2016). A belated green revolution for Cannabis: virtual genetic resources to fast-track cultivar development. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01113
Wu, Y., Trejo, H. X., Chen, G., Li, S. (2021). Phytoremediation of contaminants of emerging concern from soil with industrial hemp (Cannabis sativa L.): a review. Environ. Dev. Sustain. 23, 114405–14435. doi: 10.1007/s10668-021-01289-0
Xie, Z., Mi, Y., Kong, L., Gao, M., Chen, S., Chen, W., et al. (2023). Cannabis sativa: origin and history, glandular trichome development, and cannabinoid biosynthesis. Horticult. Res. 10, uhad150. doi: 10.1093/hr/uhad150
Xie, L., Yan, T., Li, L., Chen, M., Ma, Y., Hao, X., et al. (2021). The WRKY transcription factor AaGSW2 promotes glandular trichome initiation in Artemisia annua. J. Exp. Bot. 72, 1691–1701. doi: 10.1093/jxb/eraa523
Xu, J., van Herwijnen, Z. O., Dräger, D. B., Sui, C., Haring, M. A., Schuurink, R. C. (2018). SlMYC1 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells. Plant Cell 30, 2988–3005. doi: 10.1105/tpc.18.00571
Yamamoto, F., Iwanaga, F., Al-Busaidi, A., Yamanaka, N. (2020). Roles of ethylene, jasmonic acid, and salicylic acid and their interactions in frankincense resin production in Boswellia sacra Flueck. trees. Sci. Rep. 10, 16760. doi: 10.1038/s41598-020-73993-2
Yan, X., Cui, L., Liu, X., Cui, Y., Wang, Z., Zhang, H., et al. (2022). NbJAZ3 is required for jasmonate-meditated glandular trichome development in Nicotiana benthamiana. Physiol. Plant. 174, e13666. doi: 10.1111/ppl.13666
Yokoyama, M., Kurusu, T., Ohno, H., Ifuku, O., Harada, R., Tada, Y. (2025). Oxylipin KODA enhances the early growth of rice (Oryza sativa L.) under low-temperature stress at night to simulate a natural temperature condition. Plant Biotechnol. 42, 73–77. doi: 10.5511/plantbiotechnology.24.1218a
Youn, B., Sellhorn, G. E., Mirchel, R. J., Gaffney, B. J., Grimes, H. D., Kang, C. (2006). Crystal structures of vegetative soybean lipoxygenase VLX-B and VLX-D, and comparisons with seed isoforms LOX-1 and LOX-3. Proteins 65, 1008–1020. doi: 10.1002/prot.21182
Zhao, Y., Wu, Y., Zhang, X., Zhu, X., Hou, Y., Chen, J., et al. (2025). Methyl jasmonate attenuates chilling injury of prune fruit by maintaining ROS homeostasis and regulating GABA metabolism and energy status. Postharvest Biol. Technol. 220, 113303. doi: 10.1016/j.postharvbio.2024.113303
Zoeller, M., Stingl, N., Krischke, M., Fekete, A., Waller, F., Berger, S., et al. (2012). Lipid profiling of the Arabidopsis hypersensitive response reveals specific lipid peroxidation and fragmentation processes: biogenesis of pimelic and azelaic acid. Plant Physiol. 160, 365–378. doi: 10.1104/pp.112.202846
Keywords: glandular trichomes, green leaf volatiles, jasmonates, lipoxygenase, oxylipins, specialized metabolites
Citation: Senevirathne GI, Gendall AR, Johnson KL and Welling MT (2025) Understanding the role of oxylipins in Cannabis to enhance cannabinoid production. Front. Plant Sci. 16:1568548. doi: 10.3389/fpls.2025.1568548
Received: 30 January 2025; Accepted: 28 March 2025;
Published: 24 April 2025.
Edited by:
Xin Fang, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Dr. Pawan Kumar, Agricultural Research Organization (ARO), IsraelTianyue An, Binzhou Medical University, China
Copyright © 2025 Senevirathne, Gendall, Johnson and Welling. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kim L. Johnson, ay5qb2huc29uQGxhdHJvYmUuZWR1LmF1; Matthew T. Welling, bS53ZWxsaW5nQGxhdHJvYmUuZWR1LmF1
 Gayathree I. Senevirathne
Gayathree I. Senevirathne Anthony R. Gendall
Anthony R. Gendall Kim L. Johnson
Kim L. Johnson Matthew T. Welling
Matthew T. Welling