- 1Institute of Crop Science, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
- 2Zhejiang Key Laboratory of Crop Germplasm Innovation and Utilization, Hangzhou, China
- 3Zhejiang University Zhongyuan Research Institute, Zhengzhou, China
The important roles of JDP members in regulating abiotic and biotic stress tolerance have been demonstrated in many plants. However, fewer studies have explored the JDP gene family and its role in the salt stress response in barley, a crop known for its superior salt tolerance compared to other major cereals. Here, we identified a total of 109 putative JDP genes (nine HvDJAs, eight HvDJBs, 92 HvDJCs) in barley. Promoter analysis of HvDJs suggested that HvDJs might be involved in the processes of hormone regulation and stress response. Tandem and segmental duplications appear to be the driving forces behind JDP gene family expansion. RNA-seq analysis showed that the expression of 37 HvDJs was salt-induced, and HvDJB06, HvDJC58, and HvDJC59 were the most differentially expressed under salt stress. Protein–protein interaction analysis indicated that HvDJA09 and HvDJA05 play core roles in the complex regulatory network. Taken together, the current study provides valuable information for a deeper understanding of the function of HvDJs in regulating salt stress tolerance in barley.
1 Introduction
Soil salinity is a major abiotic stress, severely restricting crop growth and production. Salt stress can cause internal dehydration and disrupt metabolic processes (Munns, 2002). Correspondingly, plants have evolved multiple defense strategies to cope with salt stress, especially through diverse signal transduction pathways. It is well documented that transcription factors, protein kinases, and protein phosphatases are involved in a complex regulatory network in response to salt stress (Liang et al., 2024; Pieterse et al., 2012; Zhou et al., 2024). However, research exploring the impact of heat-shock proteins (Hsps) on salt tolerance remains quite limited.
J-domain proteins, also known as Hsp40s (Heat Shock Protein 40s), are characterized by the presence of an evolutionary conserved J-domain located near the N-terminus and composed of approximately 70 amino acid residues (Cyr et al., 1992). The invariant tripeptide (HPD) is the hallmark of the J-domain. The J-domain can interact with HSP70 and stimulate its ATPase activity to facilitate protein folding, unfolding, translocation, and degradation (Qiu et al., 2006; Wang et al., 2004). J-domain proteins consist of three domains: the highly conserved J-domain at the N-terminus, the CxxCxGxG zinc-finger domain (C, cysteine; G, glycine; X, other amino acid residues), and the C-terminal domain (Rosenzweig et al., 2019). JDP family proteins are usually classified into four categories based on their conserved domains: DJ-A, DJ-B, DJ-C, and DJ-D. DJ-A proteins are characterized by a J-domain, a CxxCxGxG zinc-finger domain, and a C-terminal domain. DJ-B proteins contain a J-domain plus either a zinc-finger domain or a C-terminal domain, whereas DJ-C proteins contain only a J-domain (Walsh et al., 2004). Additionally, DJ-D proteins contain a J-like domain that lacks the critical HPD tripeptide (Kampinga and Craig, 2010).
JDPs have been widely reported to be involved in resistance to biotic and abiotic stresses, such as pests, pathogens, drought, salt, low temperature, and heat. In Arabidopsis, DJA5 and DJA6 proteins are essential for chloroplast iron-sulfur cluster biogenesis (Zhang et al., 2021). In tomato, a chloroplast-targeted J-domain protein, LeCDJ1, can enhance heat tolerance and maintain the stability of photosystem II under chilling stress (Kong et al., 2014a, b). Overexpression of tobacco MsDJLP enhances chilling and heat tolerance (Lee et al., 2018). A putative J-domain protein in Nicotiana tabacum can facilitate drought tolerance by regulating the expression of drought-responsive genes (Xia et al., 2014). AtJ3 can maintain pH homeostasis by directly interacting with PKS5, thereby enhancing salt and alkaline stress tolerance (Yang et al., 2010). Additionally, ERdjB has been reported to play a role in maintaining normal anther development in Arabidopsis under high temperatures (Yamamoto et al., 2020). In rice, OsDnaJ15 can facilitate the formation of the OsSUVH7–OsBAG4–OsMYB106 transcriptional complex to activate OsHKT1;5 and enhance salt tolerance (Liu et al., 2023). These findings highlight the crucial roles of J-domain proteins in regulating tolerance to biotic and abiotic stresses.
Barley (Hordeum vulgare) is the fourth-largest cereal crop worldwide and is extensively used for human food, animal feed, and brewing material (Cai et al., 2020). Compared with other cereal crops (i.e., rice, wheat, maize), barley can withstand salt concentrations exceeding 200 mM, making it an ideal model crop for deciphering salt tolerant mechanisms (Fu et al., 2018; Munns and Tester, 2008). Previous studies have identified 129 JDP homologs in Arabidopsis (Zhang et al., 2018), 115 in rice (Luo et al., 2019), 76 in pepper (Fan et al., 2020), 236 in wheat (Liu et al., 2022), 86 in citrus (Tian et al., 2024), and 91 in maize (Li et al., 2024). However, limited studies has been conducted on the amount and functions of JDP genes in barley, despite its significantly higher salt tolerance compared to other cereal crops, including rice and wheat. Thus, it is imperative to determine the possible roles of HvDJ genes in response to salt stress.
In this study, we conducted a genome-wide analysis of the JDP gene family in barley and identified a total of 109 J-domain proteins. Phylogenetic relationships, gene structures, protein motifs, cis-regulatory elements, and chromosomal locations of these HvDJs were analyzed. We also found that tandem and segmental duplications extensively promoted the expansion of HvDJs. In addition, the expression profiles of HvDJs in response to salt stress were analyzed, identifying 37 salt-responsive HvDJ genes, including downregulated HvDJC58 and HvDJC59, and upregulated HvDJC46. The protein structures of these three HvDJs were predicted using AlphaFold3. Finally, protein–protein interacting network identified hub HvDJ genes (HvDJA09 and HvDJA05) within complex regulatory networks. These results highlight the biological functions of HvDJ in response to salt stress in barley.
2 Materials and methods
2.1 Genome-wide identification of HvDJ gene family in barley
The genomic sequences of barley were obtained from EnsemblPlants (http://plants.ensembl.org/index.html). The Hidden Markov Model (HMM) profile of the J-domain (PF00226), downloaded from the Pfam protein family database, was used as a query sequence to search for putative barley J-protein genes with an e-value < 1 × 10−5. The putative HvDJs were then verified using the NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi), SMART (http://smart.embl-heidelberg.de/), and Pfam (https://pfam.xfam.org/). Finally, 109 genes were identified as members of the JDP gene family in barley. Furthermore, the amino acid lengths (aa), molecular weights (MW), and isoelectric points (pI) of the identified JDP proteins were analyzed using the ExPasy website (http://web.expasy.org/protparam/).
2.2 Phylogenetic analysis of HvDJ proteins
Multiple sequence alignment of the HvDJ protein sequences was conducted using MEGA7 software with the ClustalW algorithm (Kumar et al., 2016; Thompson et al., 1994). The aligned sequences were then subjected to phylogenetic analysis using the neighbor-joining (NJ) method through MEGA7 software with 1,000 bootstrap replicates.
2.3 Gene structure and conserved motif analysis of HvDJ genes
Gene structure features of the 109 HvDJs were extracted using TBtools software based on the barley gene feature format (GFF) files. In addition, conserved motifs were predicted and analyzed using the MEME online tool (http://meme-suite.org/tools/meme). The number of motifs was set to 10. TBtools was used to visualize the gene structure and MEME results (Chen et al., 2023).
2.4 Cis-elements analysis on the promoter region of HvDJs
Upstream 2-kb sequences of the 109 HvDJ genes were extracted from the barley genome database. The PlantCARE program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to analyze the sequences and identify putative cis-regulatory elements. TBtools was used to visualize the results (Chen et al., 2023).
2.5 Chromosomal distribution and gene duplication of HvDJ genes
HvDJ genes were mapped to barley chromosomes using TBtools based on barley genomic data (Chen et al., 2023). Tandem and segmental duplication events of the HvDJ genes, as well as genome collinearity between barley and other species (rice, maize, sorghum, wheat, and Arabidopsis), were analyzed using the Multiple Collinearity Scan ToolKit-X (MCScanX) with default parameters (Wang et al., 2012) and visualized using Circos and the Dual Synteny Plot in TBtools (Chen et al., 2023; Krzywinski et al., 2009). Nonsynonymous (Ka) and synonymous (Ks) substitution rates were calculated using the simple Ka/Ks calculator in TBtools (Chen et al., 2023).
2.6 Expression pattern analysis of HvDJ genes
Transcriptome data from salt-treated barley were obtained from published sources (Zhang et al., 2020). FastQC was used for quality control, and HISAT2 (v2.2.1) was then used to map clean reads to the reference barley genome (Morex) (Pertea et al., 2016). Transcriptome assembly was conducted using StringTie (v2.2.1). DESeq2 (v1.30.0) was used to identify differentially expressed genes (DEGs) based on a criteria two-fold change and an adjusted p-value <0.05. Heatmaps were generated using TBtools software (Chen et al., 2023).
2.7 qRT-PCR analysis
Four JDP genes identified as salt-induced in the RNA-seq data were selected for qRT-PCR validation. A barley cultivar “Golden Promise” was used as a plant material and treated with 100 mM NaCl at 0 h, 6 h, and 48 h from 14-day-old seedlings, following Shen et al. (2020). Total RNA waS extracted from barley roots using the Easy-Do Plant Total RNA Rapid Extraction Kit, and reverse transciption was performed using reverse transcriptase and universal oligo(dT) primers (9769 and RR037A, Takara). qRT-PCR reactions were prepared following the SYBR Green Supermix (RR820, Takara) protocol and run on a Roche LightCycler 480 II system. Primer sequences are listed in Supplementary Table S6, with the α-tubulin gene used as an internal reference. Relative expression levels were calculated using the 2−ΔΔCT method.
2.8 Protein structure prediction and interaction network analysis of HvDJs
Protein structure prediction was performed using AlphaFold3 (Abramson et al., 2024). Protein–protein interactions (PPIs) of the HvDJ protein family were analyzed using STRING v12.0 (https://string-db.org). Cytoscape v3.10.0 (Shannon et al., 2003), was used to visualize the putative interaction network.
3 Results
3.1 Identification and characterization of HvDJs
In total, 109 JDP genes were identified from the reference genome of the barley cultivar Morex (Mascher et al., 2017) (Table 1; Supplementary Table S1). Based on the J-domain and their chromosomal positions, they were classified into three types (DJA, DJB, and DJC), with each type harboring nine, eight, and 92 members, respectively (Table 1; Supplementary Table S1). Their protein lengths vary from 99 (HvDJC17) to 2,577 (HvDJC05) amino acids (aa), with molecular weights (MWs) ranging from 8.82 (HvDJC89) to 281.4 (HvDJC05) kDa (Table 1). In addition, the isoelectric points (pIs) of these HvDJ proteins ranged from 4.22 (HvDJC61) to 11.18 (HvDJC66) (Table 1). All HvDJs contained a J-domain consisting of an average of 61 aa, with HvDJC47 having the shortest J-domain (32 aa) and HvDJB07 the longest (105 aa) (Supplementary Table S1). The GRAVY values of all 109 J-domain proteins were below zero (except HvDJC60), indicating that these proteins are hydrophilic.
3.2 Structure and motif analysis of HvDJs
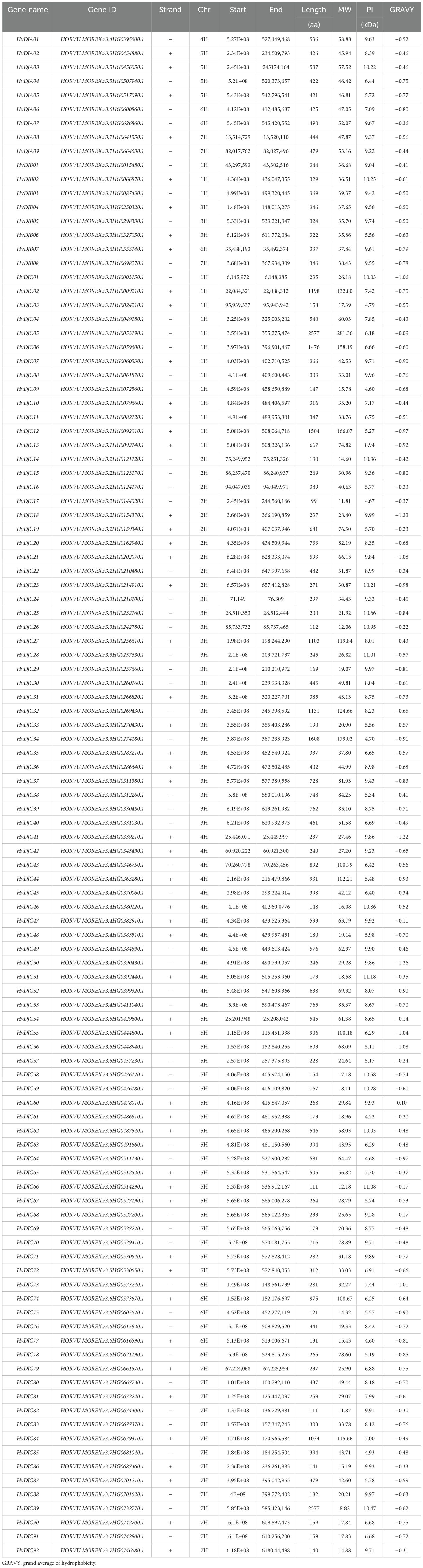
We further analyzed the conserved motifs and gene structures of HvDJ protein sequences, followed by a phylogenetic tree analysis for HvDJs (Figure 1A). Gene structure analysis showed that HvDJs harbored 1–22 exons and 0–21 introns (Figure 1B). In details, among 109 HvDJ genes, 17 had no introns, and the others contained two to 22 exons (13 with two exons, 16 with three, five with four, 12 with five, six with six, six with seven, eight with eight, seven with nine, five with 10, seven with 11, two with 12, one each with 13, 17, 18, 19, and 22 exons). Ten conserved motifs were identified in 109 HvDJ protein sequences (Figure 1B; Supplementary Table S2). Among these motifs, motif1 and motif2 were the most frequently present, appearing 82 and 87 times, respectively, indicating that these two motifs were highly conserved in the core JDP genes (Figure 1B).

Figure 1. The phylogenetic relationship, gene structure, and motif compositions of HvDJs. (A) The phylogenetic tree was constructed using the full-length sequences of HvDJ proteins. (B) Purple rectangles, orange rectangles, and black lines indicate UTRs (untranslated region), CDSs (coding sequence or exons), and introns, respectively (C) Ten amino acid motifs in HvDJ proteins are shown in different colored boxes, and black lines indicate amino acid length.
3.3 Cis-elements analysis of HvDJs
To understand the transcriptional regulation of HvDJ genes, we analyzed the cis-elements in the promoter regions of the HvDJs. A total of 19 types of cis-regulatory elements were identified in the upstream 2,000-bp sequences of 109 HvDJs (Figure 2; Supplementary Table S3). These elements are involved in hormone (auxin, abscisic acid, methyl jasmonate, gibberellin, and salicylic acid), stress (anaerobic, anoxic, defense, drought, and low temperature), tissue (endosperm, palisade mesophyll cells, and seed), circadian rhythm, cell cycle, light, zein, as well as transcription factor binding sites (MYB, MYC) (Figure 2). Notably, most HvDJs harbored MYB- and light-responsive cis-elements, indicating that HvDJs may be regulated by MYB transcription factors and light signals.
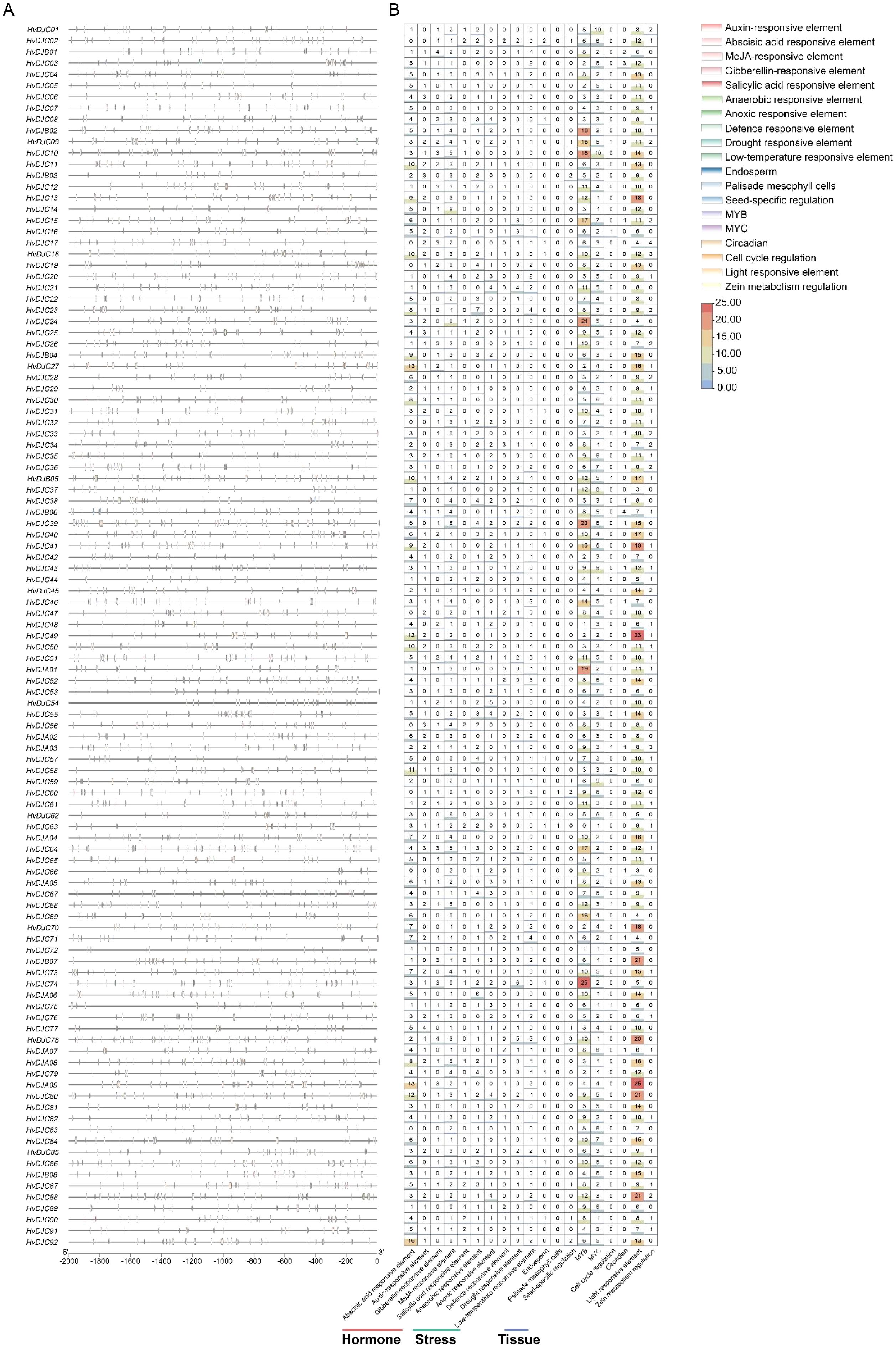
Figure 2. The cis-regulatory elements predicted in the promoter regions of HvDJs. (A) Distribution of predicted cis-regulatory elements in the HvDJ gene family. (B) The number of each cis-regulatory element in the HvDJ gene family.
3.4 Chromosomal distribution and gene duplication analysis of HvDJs
The 109 HvDJ genes were unevenly distributed on the seven chromosomes (Figure 3A; Table 1), with 1H, 2H, 3H, 4H, 5H, 6H, and 7H containing 16, 10, 20, 14, 23, 9, and 17 HvDJ genes, respectively. Additionally, we identified three pairs of tandemly duplicated HvDJ genes—HvDJC28 and HvDJC29, HvDJC67 and HvDJC68, and HvDJC68 and HvDJC69 (Figure 3A). These genes were closely distributed on the chromosomes and formed clusters on the phylogenetic tree. Segmental duplication analysis of the 109 HvDJ genes identified 21 pairs of segmental duplication events (Figure 3B). The ratios of non-synonymous (Ka) to synonymous (Ks) substitutions (Ka/Ks) in these two tandem duplication and 10 segmental duplication gene pairs were less than 1 (Supplementary Table S4), indicating that purifying selection is likely stronger than positive selection in the evolution of the HvDJ genes.
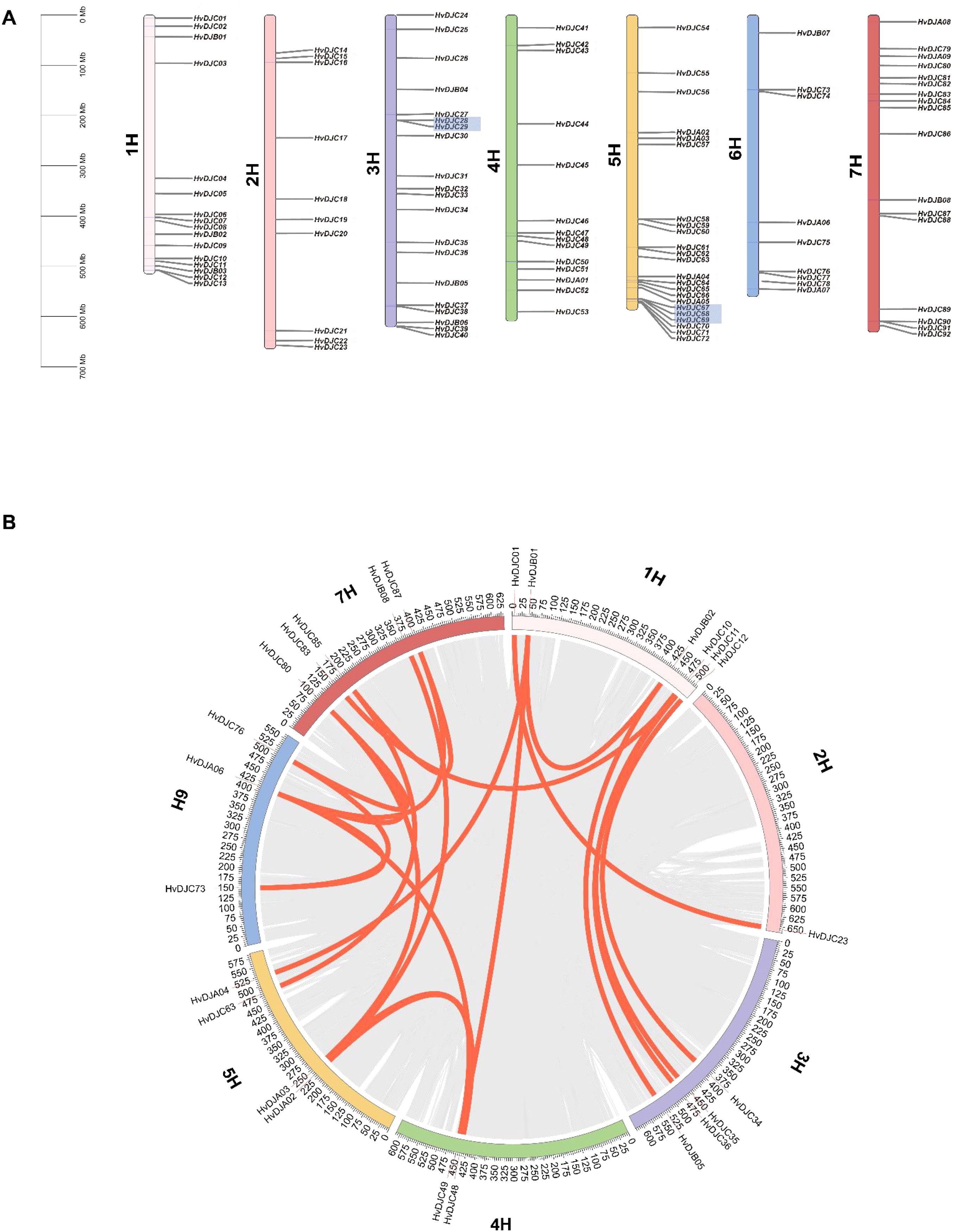
Figure 3. (A) The distribution of HvDJ genes across the chromosomes. 1–7H represent the seven barley chromosomes; shaded areas under gene names represent tandem duplications, and the 0 Mb–700 Mb scale indicates chromosome length. (B) Synteny analysis of HvDJ genes in the barley genome. Gray lines represent all synteny blocks in the barley genome. Orange lines represent duplicated HvDJ gene pairs.
3.5 Synteny analysis of JDP genes
To determine the evolutionary trajectory of the JDP family in barley and other plant species, we performed an evolutionary relationship analysis of JDP genes. In detail, we compared four monocotyledonous species (rice, maize, sorghum, and wheat) and one dicotyledonous plant (Arabidopsis) (Figure 4). The results showed that barley shared 88, 101, 90, 280, and eight collinear genes with rice, maize, sorghum, wheat, and Arabidopsis, respectively, indicating that JDPs in barley are more closely related to these in wheat in terms of evolution relationship.
Figure 4. Synteny analysis of JDP genes between barley and six other plants species (Oryza sativa, Zea mays, Sorghum bicolor, Triticum aestivum, and Arabidopsis thaliana). Gray lines between barley and the other species represent collinear blocks across broad genomic regions, while colored lines indicate the synteny of JDP genes.
3.6 Expression profiles of HvDJs in response to salt stress
To explore the response of HvDJ genes to salt stress, we investigated their expression using public data (Zhang et al., 2020). Barley seedlings were sampled at four time points (0 h, 1 h, 6 h, and 24 h) of salt exposure for RNA-seq analysis. Differentially expressed genes (DEGs) were identified by comparing salt-stressed samples to the control. In total, we identified 37 salt-responsive HvDJs (Figure 5; Supplementary Table S5). Among them, HvDJC09, HvDJB03, HvDJC33, HvDJB06, HvDJC46, HvDJC58, and HvDJC59 were differentially expressed at 1 h, 6 h, and 24 h after salt stress. Four genes—HvDJC09, HvDJB03, HvDJC33, and HvDJC46—were significantly upregulated in response to salt stress, with HvDJC46 being the most upregulated, while HvDJB06, HvDJC58, and HvDJC59 were significantly downregulated, with HvDJB06 exhibiting the greatest downregulation. To further validate their response patterns under short-term salt stress, four upregulated HvDJ genes (HvDJC09, HvDJB03, HvDJC33, and HvDJC46) were selected for detailed qRT-PCR analysis (Figure 6). The four HvDJs exhibited upregulated expression patterns under salt stress, with the highest expression at 48 h (Figure 6).
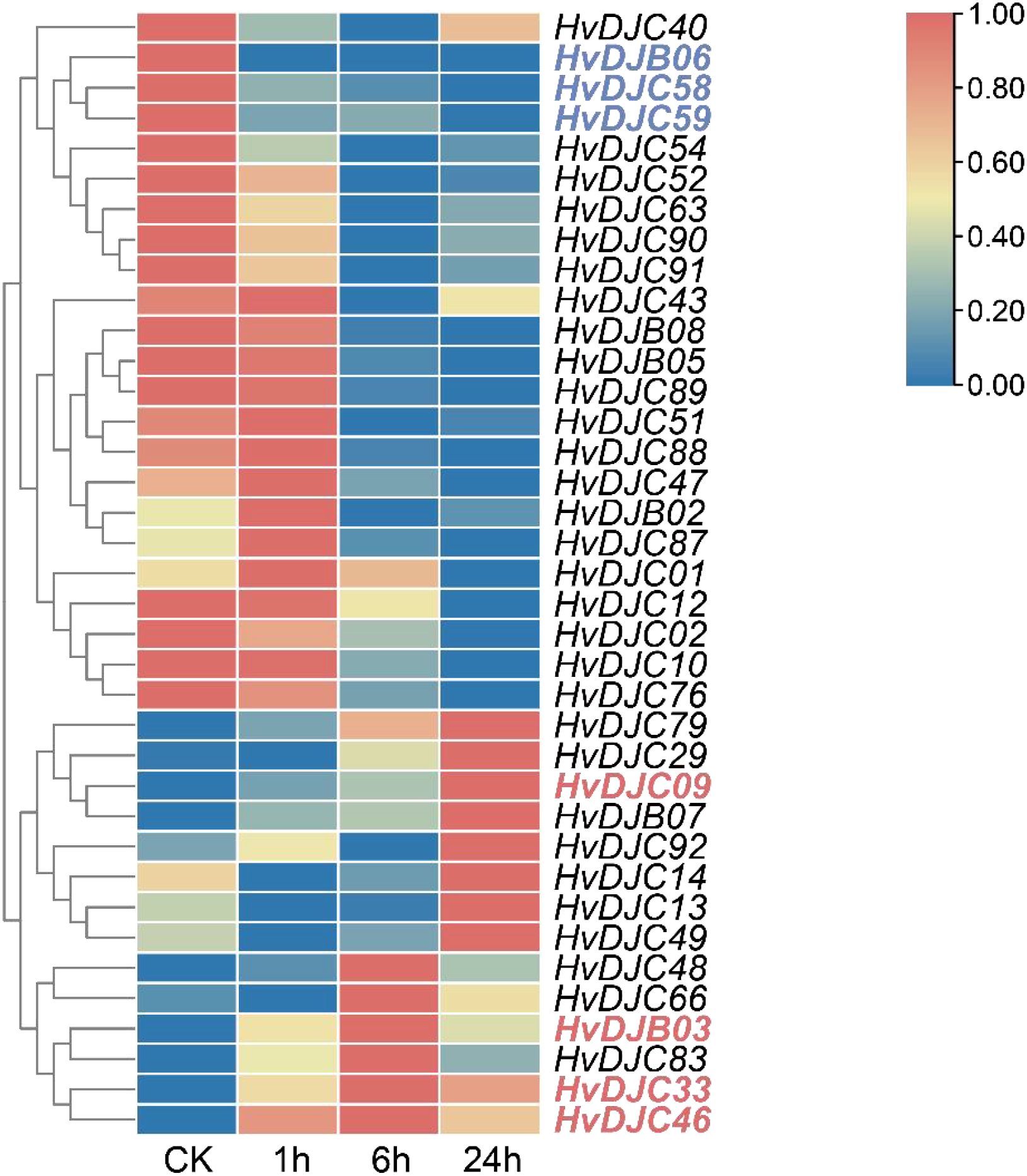
Figure 5. Expression profiles of HvDJ genes in response to salt stress. TPM values of HvDJs genes are scaled individually from 0 to 1. Blue and red represent lower and higher expression levels, respectively.
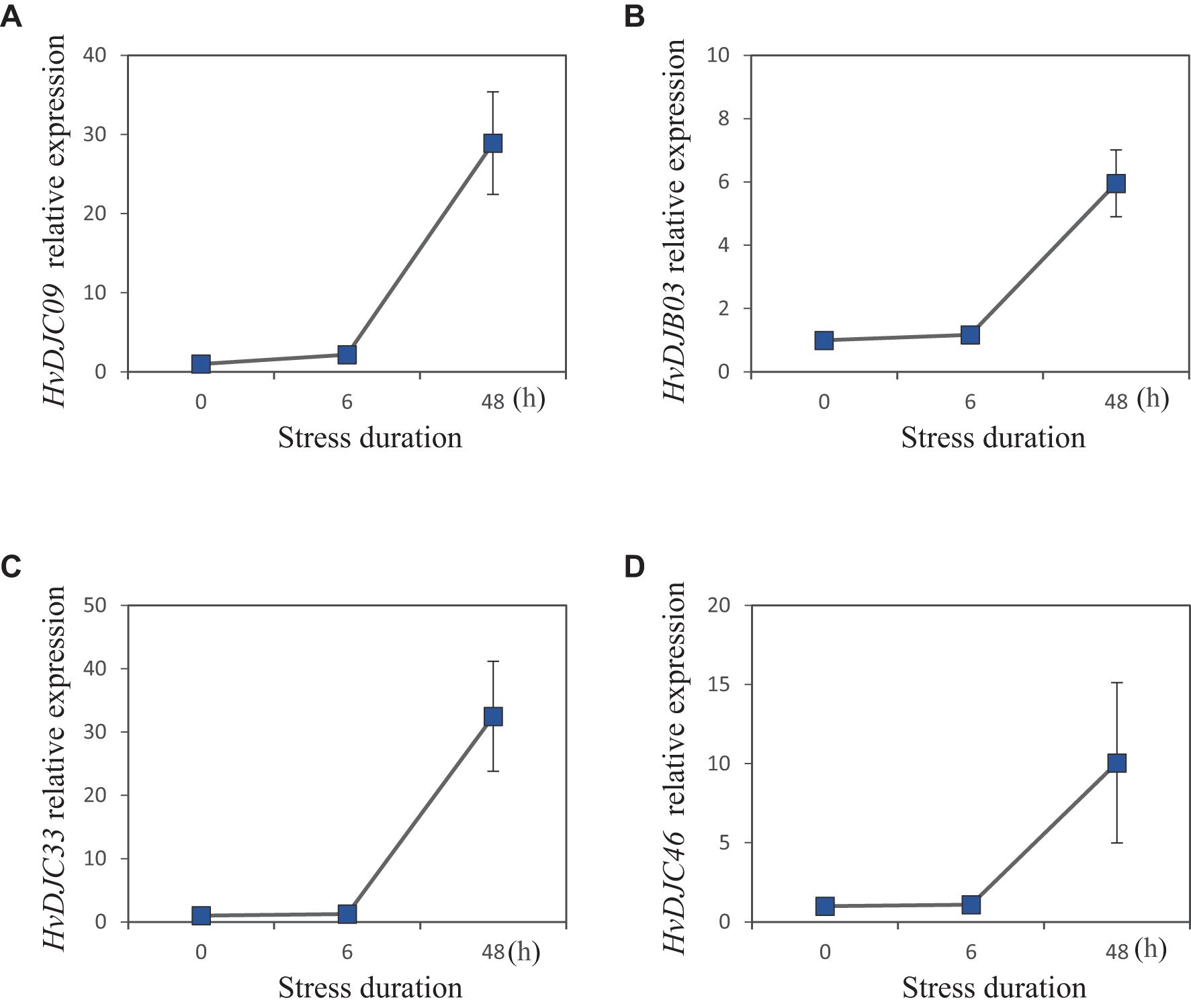
Figure 6. Expression patterns of four selected JDP genes in barley under 200 mM salt stress at 0h, 6h, and 48h. (A) HvDJC09, (B) HvDJB03, (C) HvDJC33, and (D) HvDJC46 (n =4,±SE).
To structurally analyze these salt-responsive JDPs, we attempted to identify key similarities and differences in their three-dimensional conformations, aiming to provide a structural basis for their functional characterization. We analyzed the protein structure of HvDJC09, HvDJB03, HvDJC33, HvDJC46, HvDJB06, HvDJC58, and HvDJC59 using AlphaFold3 (Figure 7). Interestingly, we observed highly similar protein structures among the five proteins (HvDJC09, HvDJC33, HvDJC46, HvDJC58, and HvDJC59), all harboring at least four α-helices (Figure 7). Meanwhile, we found that the a-helices of HvDJC46, HvDJC58, and HvDJC59 were unevenly distributed at the C- terminal (Figures 7D, F, G), whereas those of HvDJC09 and HvDJC33 were localized at the N-terminus (Figures 7A, C). HvDJB03 and HvDJB06 were predicted to harbor similar protein structures, with both α-helices and β-sheets unevenly distributed (Figures 7B, E).
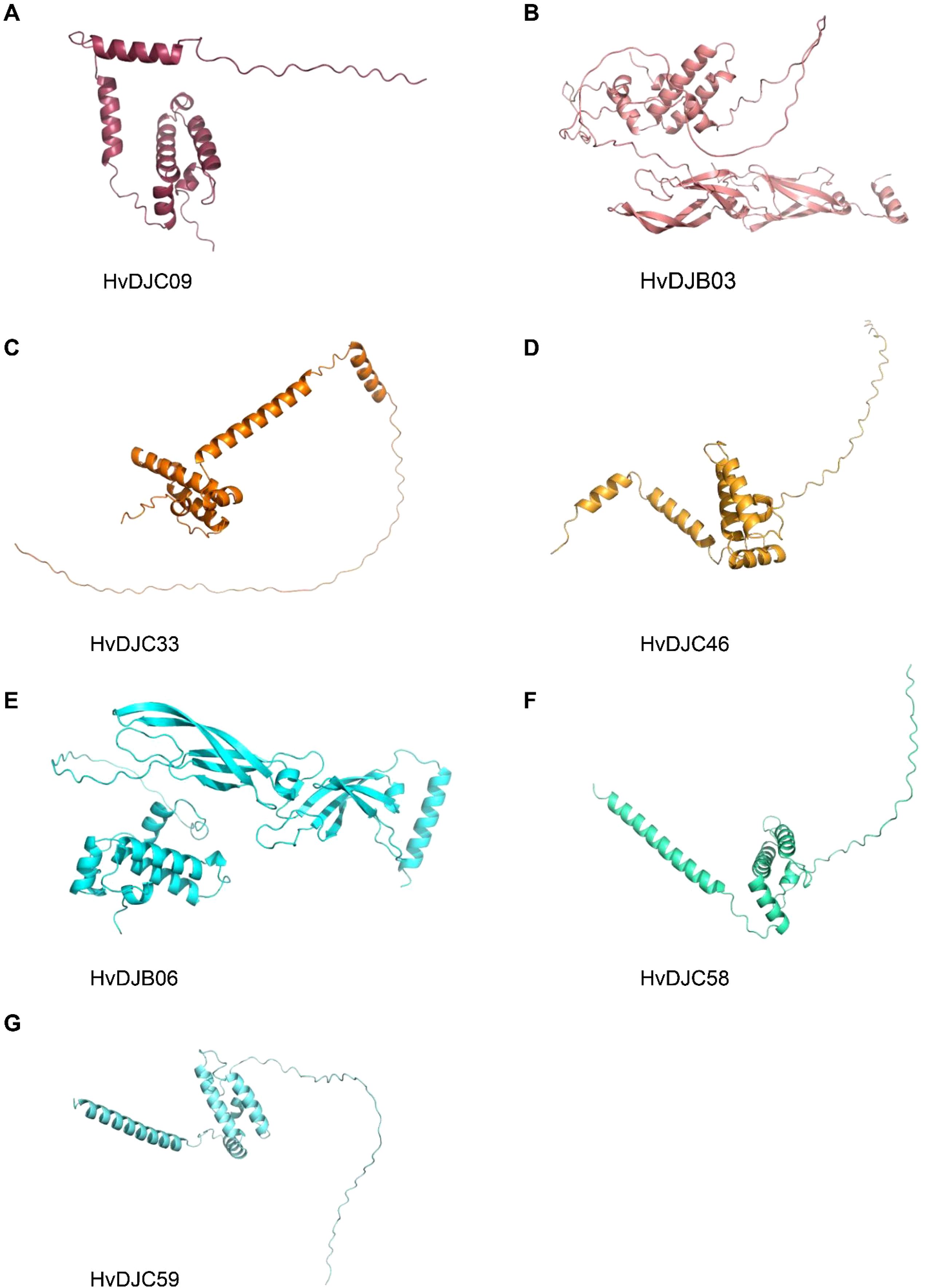
Figure 7. Schematic structures of HvDJC09, HvDJB03, HvDJC33, HvDJC46, HvDJB06, HvDJC58, and HvDJC59 predicted by AlphaFold3.
3.7 Regulatory network analysis of HvDJ genes
STRING integrates both experimental and computational evidence, including high-throughput experimental data (e.g., yeast two-hybrid, affinity purification-mass spectrometry), computational predictions (e.g., gene co-expression, conserved genomic context, phylogenetic profiling), text-mining of published literature, and database-curated interactions from known pathways. To decipher the molecular regulatory networks of HvDJs, we used the STRING database to predict potential interactions among the HvDJ proteins (Figure 8). There are 41 nodes in the HvDJ protein interaction network, each capable of interacting with the others. We also found several HvDJs exhibiting direct interactions, including HvDJA09 with HvDJC07, HvDJC22, HvDJC51 and HvDJA05, HvDJC25 with HvDJC30, HvDJA05 with HvDJC67, HvDJC74, HvDJC75, HvDJC77, HvDJC78, and HvDJB08, HvDJC73 with HvDJC77. Core genes function as central hubs that play pivotal roles in network modules. Among them, HvDJA09 and HvDJA05 played core and pivotal roles in the complex regulatory network. Additionally, other proteins such as HSP70-7, HSP70-15, HSP70-17, and DJC82 were also identified as targets in the core network of J-domain proteins.
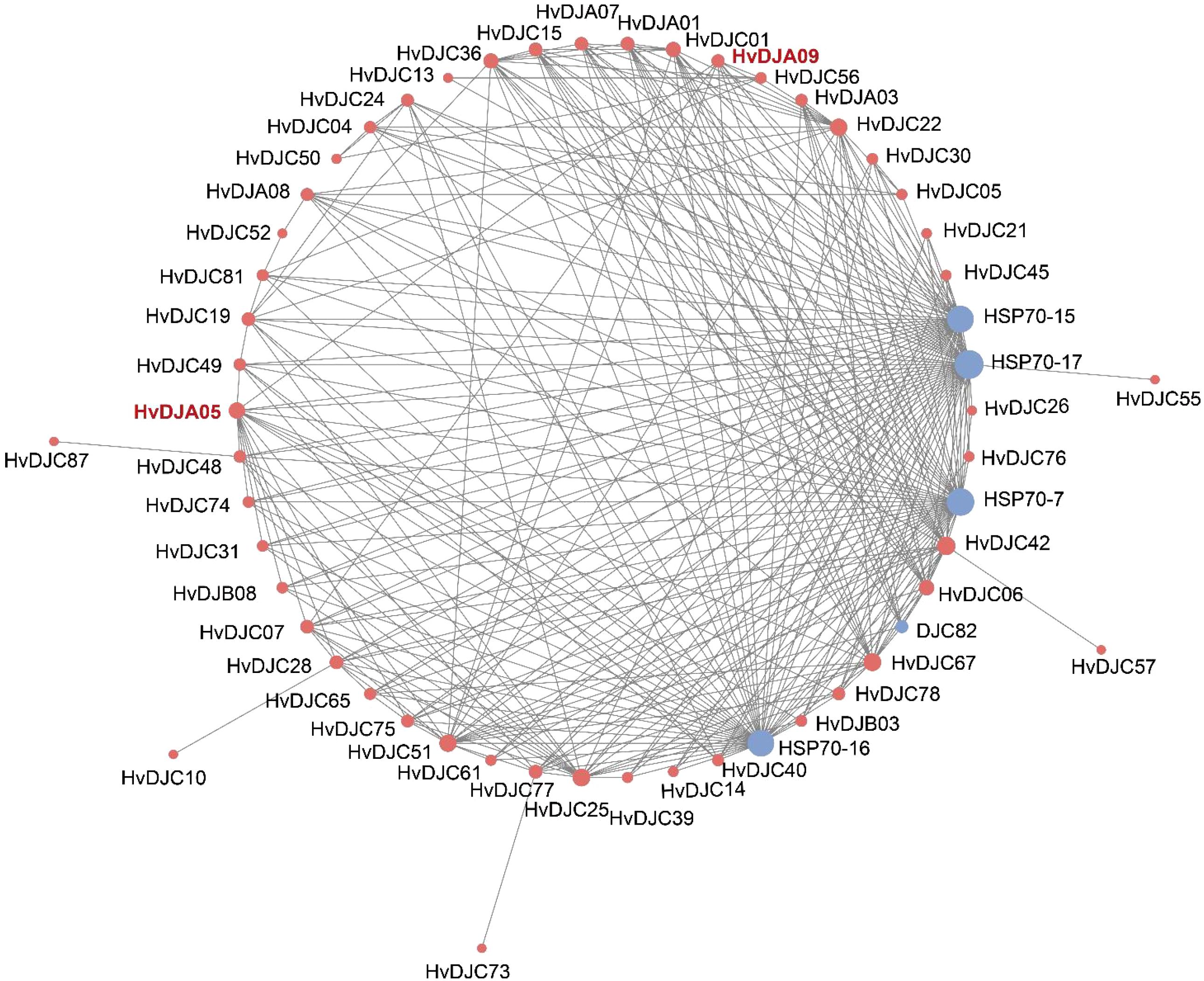
Figure 8. Protein–protein interaction networks of HvDJ proteins. Red indicates HvDJs and blue indicates other interacting proteins.
4 Discussion
With the rapid development of gene sequencing technology, an increasing number of plant genomes have been published, facilitating the identification of the variable gene families. To date, 129 JDP homologs in Arabidopsis (Zhang et al., 2018), 115 in rice (Luo et al., 2019), 76 in pepper (Fan et al., 2017), 236 in wheat (Liu et al., 2022), 86 in citrus (Tian et al., 2024), and 91 in maize (Li et al., 2024) have been identified and characterized. In this study, 109 HvDJ genes were identified in the barley genome (Table 1), providing valuable genetic information for a deeper understanding of their functions. Barley JDPs exhibit a wide range of sequence lengths and significant differences in exon numbers (Figure 1). Moreover, there are dramatic differences in motif distribution and number among HvDJs (Figure 1). With respect to isoelectric point (pI), HvDJ proteins range from 4.22 (HvDJC61) to 11.18 (HvDJC66) (Table 1), with 65% exhibiting a pI >7, similar to those of TaDnaJs and CbuDnaJs (Liu et al., 2022; Yang et al., 2023).
Features in gene and protein structure can elucidate the characteristics of gene families and guide functional research (Rogozin et al., 2005). Here, gene structural analysis of HvDJs revealed considerable variation in the number and distribution of introns and exons (Figure 1B), suggesting functional divergence in their response to salt stress. It is well documented that cis-elements in gene promoters regulate gene expression during plant growth and development, as well as adaption to environmental stimuli (Guo et al., 2024; Li et al., 2018). In this study, cis-elements within the 2-kb upstream regions of HvDJ genes were analyzed using the PlantCARE program (Lescot et al., 2002). Four major groups of cis-elements were identified: hormone-responsive, stress-related, tissue-specific, and transcription factor-binding elements (Figure 2; Supplementary Table S3). These findings suggest that HvDJs may be involved in the responses to abiotic stress, hormone regulation, and transcriptional regulation.
Gene duplication serves not only as a primary source of evolutionary innovation but also as a major driving force for gene family expansion (Schmutz et al., 2010). In barley, 109 JDP genes were unevenly distributed across the seven chromosomes (Figure 3A). Thirty HvDJ genes have undergone gene duplication, including both tandem duplication and segmental duplication events (Figure 3). Three tandem and 21 segmental duplication events were observed. These results suggest that both tandem and segmental duplications have played vital roles in the expansion of the HvDJ gene family in barley. Collinearity analysis revealed that HvDJ genes are more closely related to monocotyledonous plants, particularly wheat (Figure 4). These findings highlight the evolutionary origins and genetic relationships of JDPs between barley and other plant species.
JDPS have been reported to be involved in responses to various biotic and abiotic stresses. Silencing of NtMPIP1, a DnaJ-like protein in tobacco, significantly inhibited infection by tobacco mosaic virus (TMV) (Shimizu et al., 2009). Overexpression of soybean HSP40 induced hypersensitive response (HR)-like cell death in tobacco leaves (Liu and Whitham, 2013). Cotton GhDNAJ1 positively regulates resistance to V. dahlia (Feng et al., 2021). In addition, JDPs have been reported to play important roles in regulating abiotic stress tolerance, including responses to heat, drought, chilling, and salt stress (Kong et al., 2014a; Lee et al., 2018; Liu et al., 2023; Wang et al., 2015; Yamamoto et al., 2020). To examine the expression profiles of HvDJ genes in response to salt stress, RNA-seq data were obtained from barley seedlings sampled at four time points of salt exposure (0 h, 1 h, 6 h, and 24 h) (Zhang et al., 2020). A total of 37 HvDJ genes were identified as salt-responsive, exhibiting distinct expression patterns across the time points. Eight genes (HvDJC09, HvDJC11, HvDJB03, HvDJC33, HvDJB06, HvDJC46, HvDJC58, and HvDJC59) exhibited differential expression at 1 h, 6 h, and 24 h after salt stress exposure. Among them, HvDJC09, HvDJB03, HvDJC33, and HvDJC46 were upregulated, while HvDJB06, HvDJC58, and HvDJC59 were downregulated (Figure 5). qRT-PCR further confirmed that HvDJC09, HvDJB03, HvDJC33, and HvDJC46 were upregulated by salt stress, with peak expression observed after 48 h of exposure (Figure 6). Additionally, –12 genes (HvDJB02, HvDJC29, HvDJB05, HvDJC40, HvDJC47, HvDJC66, HvDJC79, HvDJB08, HvDJC88, HvDJC89, HvDJC90, and HvDJC91) were differentially expressed at 6 h and 24 h, with only HvDJC40 showing downregulation at both 1 h and 6 h. Seventeen genes were differentially expressed at only one time point following salt stress (Figure 5). These findings suggest that HvDJ genes vary in their expression patterns and function in response to salt stress. Furthermore, we predicted the protein structures of seven differentially expressed genes (HvDJC09, HvDJB03, HvDJC33, HvDJB06, HvDJC46, HvDJC58, and HvDJC59) at three time points (1 h, 6 h. and 24 h) to explore their potential roles in salt stress response (Figure 7). Notably, several potential genes for salinity tolerance were identified on chromosome 4H, including HvDJC53 (Fan et al., 2016). In addition, QTLs for grain yield relative to control conditions were found near the QTL for salinity tolerance score on chromosome 3H, where HvDJC10, HvDJB04, and HvDJC28 are located (Liu et al., 2017). Additionally, 41 HvDJs were predicted to interact with one another, with the interaction of HvDJA09 and HvDJA05 serving as the central node in the complex regulatory network (Figure 8). Overall, the results of protein structure and interaction analysis provided new insight into the biological functions of HvDJs.
5 Conclusion
In this study, 109 JDP genes in barley were identified and characterized. Our results showed that tandem and segmental duplications are the driving forces behind JDP gene family expansion. A total of 37 HvDJs showed differential expression under salt stress, with HvDJB06 and HvDJC46 showing the highest expression levels. In total, 41 nodes were identified in the HvDJ protein interaction network, with HvDJA09 and HvDJA05 playing central roles.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
YX: Conceptualization, Formal Analysis, Software, Writing – original draft. HS: Investigation, Writing – original draft. LS: Investigation, Writing – original draft. BW: Investigation, Writing – original draft. LL: Investigation, Writing – original draft. GZ: Conceptualization, Writing – review & editing. QS: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32171929); the Key Research Foundation of the Science and Technology Department of Zhejiang Province of China (2021C02064-3); the Fundamental Research Funds for the Central Universities; and the Jiangsu Collaborative Innovation Center for Modern Crop Production (JCIC-MCP).
Acknowledgments
We thank Dr. Enhui Shen (Zhejiang University) for support with bioinformatics and big data technology.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1574097/full#supplementary-material
References
Abramson, J., Adler, J., Dunger, J., Evans, R., Green, T., Pritzel, A., et al. (2024). Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500. doi: 10.1038/s41586-024-07487-w
Cai, K., Chen, X., Han, Z., Wu, X., Zhang, S., Li, Q., et al. (2020). Screening of worldwide barley collection for drought tolerance: the assessment of various physiological measures as the selection criteria. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01159
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one “bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Cyr, D., Lu, X., and Douglas, M. (1992). Regulation of HSP70 function by a Eukaryotic DnaJ homolog. J. Biol. Chem. 267, 20927–20931. doi: 10.1016/S0021-9258(19)36777-8
Fan, F. F., Yang, X., Cheng, Y., Kang, Y. Y., and Chai, X. R. (2017). The DnaJ Gene Family in Pepper (Capsicum annuum L.): Comprehensive Identification, Characterization and Expression Profiles. Front. Plant Sci. 8, 689. doi: 10.3389/fpls.2017.00689
Fan, F., Liu, F., Yang, X., Wan, H., and Kang, Y. (2020). Global analysis of expression profile of members of DnaJ gene families involved in capsaicinoids synthesis in pepper (Capsicum annuum L). BMC Plant Biol. 20, 326. doi: 10.1186/s12870-020-02476-3
Fan, Y., Zhou, G. F., Shabala, S., Chen, Z. H., Cai, S. G., Li, C. D., et al. (2016). Genome-wide association study reveals a new QTL for salinity tolerance in Barley (Hordeum vulgare L.) Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00946
Feng, H. J., Li, C., Zhou, J. L., Yuan, Y., Feng, Z. L., Shi, Y. Q., et al. (2021). A cotton WAKL protein interacted with a DnaJ protein and was involved in defense against Verticillium dahlia. Int. J. Biol. Macromol. 167, 633–643. doi: 10.1016/j.ijbiomac.2020.11.191
Fu, L., Shen, Q., Kuang, L., Yu, J., Wu, D., and Zhang, G. (2018). Metabolite profiling and gene expression of Na/K transporter analyses reveal mechanisms of the difference in salt tolerance between barley and rice. Plant Physiol. Biochem. 130, 248–257. doi: 10.1016/j.plaphy.2018.07.013
Guo, M., Yang, F., Zhu, L., Wang, L., Li, Z., Qi, Z., et al. (2024). Loss of cold tolerance is conferred by absence of the WRKY34 promoter fragment during tomato evolution. Nat. Commun. 15, 6667. doi: 10.1038/s41467-024-51036-y
Kampinga, H. and Craig, E. (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592. doi: 10.1038/nrm2941
Kong, F., Deng, Y., Wang, G., Wang, J., Liang, X., and Meng, Q. (2014a). LeCDJ1, a chloroplast DnaJ protein, facilitates heat tolerance in transgenic tomatoes. J. Integr. Plant Biol. 56, 63–74. doi: 10.1111/jipb.12119
Kong, F., Deng, Y., Yin, B., Lv, W., and Meng, Q. (2014b). Suppression of a tomato chloroplast DnaJ protein gene reduced heat tolerance in transgenic tomatoes. Plant Physiol. J. 50, 68–76.
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., and Tamure, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, K., Rahman, M., Kim, K., Choi, G., Cha, J., Cheong, M., et al. (2018). Overexpression of the alfalfa DnaJ-like protein (MsDJLP) gene enhances tolerance to chilling and heat stresses in transgenic tobacco plants. Turk J. Biol. 42, 12–22. doi: 10.3906/biy-1705-30
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, G., Chen, Z., Guo, X., Tian, D., Li, C., Lin, M., et al. (2024). Genome-wide identification and analysis of maize DnaJ family genes in response to salt, heat, and cold at the seedling stage. Plants-Basel 13, 2488. doi: 10.3390/plants13172488
Li, N., Wei, S., Chen, J., Yang, F., Kong, L., Chen, C., et al. (2018). OsASR2 regulates the expression of a defence-related gene, Os2H16, by targeting the GT-1 cis-element. Plant Biotechnol. J. 16, 771–783. doi: 10.1111/pbi.12827
Liang, X., Li, J., Yang, Y., Jiang, C., and Guo, Y. (2024). Designing salt stress-resilient crops: current progress and future challenges. J. Integr. Plant Biol. 66, 303–329. doi: 10.1111/jipb.13599
Liu, X. H., Fan, Y., Mak, M., Babla, M., Holford, P., Wang, F. F., et al. (2017). QTLs for stomatal and photosynthetic traits related to salinity tolerance in barley. BMC Genomics 18, 9. doi: 10.1186/s12864-016-3380-0
Liu, Y., Li, M., Yu, J., Ma, A., Wang, J., Yun, D., et al. (2023). Plasma membrane-localized Hsp40/DNAJ chaperone protein facilitates OsSUVH7-OsBAG4-OsMYB106 transcriptional complex formation for OsHKT1;5 activation. J. Integr. Plant Biol. 65, 265–279. doi: 10.1111/jipb.13403
Liu, J. Z. and Whitham, S. A. (2013). Overexpression of a soybean nuclear localized type-III DnaJ domain-containing HSP40 reveals its roles in cell death and disease resistance. Plant J. 74, 110–121. doi: 10.1111/tpj.12108
Liu, T., Xu, M., Gao, S., Zhang, Y., Hu, Y., Jin, P., et al. (2022). Genome-wide identification and analysis of the regulation wheat DnaJ family genes following wheat yellow mosaic virus infection. J. Integr. Agric. 21, 153–169. doi: 10.1016/S2095-3119(21)63619-5
Luo, Y., Fang, B., Wang, W., Yang, Y., Rao, L., and Zhang, C. (2019). Genome-wide analysis of the rice J-protein family: Identification, genomic organization, and expression profiles under multiple stresses. 3 Biotech. 9, 358. doi: 10.1007/s13205-019-1880-8
Mascher, M., Gundlach, H., Himmelbach, A., Beier, S., Twardziok, S., Wicker, T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433. doi: 10.1038/nature22043
Munns, R. (2002). Comparative physiology of salt and water stress. Plant Cell Environ. 25, 239–250. doi: 10.1046/j.0016-8025.2001.00808.x
Munns, R. and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Pertea, M., Kim, D., Pertea, G., Leek, J., and Salzberg, S. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Pieterse, C., van der Does, D., Zamioudis, C., Leon-Reyes, A., and Van Wees, S. (2012). Hormonal modulation of plant immunity, Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Qiu, X., Shao, Y., Miao, S., and Wang, L. (2006). The diversity of the Dnaj/HSP40 family, the crucial partners for HSP70 chaperones. Cell. Mol. Life Sci. 63, 2560–2570. doi: 10.1007/s00018-006-6192-6
Rogozin, I. B., Sverdlov, A. V., Babenko, V. N., and Koonin, E. V. (2005). Analysis of evolution of exon-intron structure of eukaryotic genes. Brief. Bioinform. 6, 118–134. doi: 10.1093/bib/6.2.118
Rosenzweig, R., Nillegoda, N., Mayer, M., and Bukau, B. (2019). The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 20, 665–680. doi: 10.1038/s41580-019-0133-3
Schmutz, J., Cannon, S., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Shannon, P., Markiel, A., Ozier, O., Baliga, N., Wang, J., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shen, Q. F., Fu, L. B., Su, T. T., Ye, L. Z., Huang, L., Kuang, L. H., et al. (2020). Calmodulin HvCaM1 negatively regulates salt tolerance via modulation of HvHKT1s and HvCAMTA4. Plant Physiol. 183, 1650–1662. doi: 10.1104/pp.20.00196
Shimizu, T., Yoshii, A., Sakurai, K., Hamada, K., Yamaji, Y., Suzuki, M., et al. (2009). Identification of a novel tobacco DnaJ-like protein that interacts with the movement protein of tobacco mosaic virus. Arch. Virol. 154, 959–967. doi: 10.1007/s00705-009-0397-6
Thompson, J., Higgins, D., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tian, Y., Wang, X., Huang, H., Deng, X., Zhang, B., Meng, Y., et al. (2024). Genome-Wide identification of the DnaJ gene family in citrus and functional characterization of ClDJC24 in response to citrus Huanglongbing. Int. J. Mol. Sci. 25, 11967. doi: 10.3390/ijms252211967
Walsh, P., Bursac, D., Law, Y., Cyr, D., and Lithgow, T. (2004). The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5, 567–571. doi: 10.1038/sj.embor.7400172
Wang, G., Kong, F., Zhang, S., Meng, X., Wang, Y., and Meng, Q. (2015). A tomato chloroplast-targeted DnaJ protein protects Rubisco activity under heat stress. J. Exp. Bot. 66, 3027–3040. doi: 10.1093/jxb/erv102
Wang, Y., Tang, H., Debarry, J., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, X., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Xia, Z., Zhang, X., Li, J., Su, X., and Liu, J. (2014). Overexpression of a tobacco J-domain protein enhances drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 83, 100–106. doi: 10.1016/j.plaphy.2014.07.023
Yamamoto, M., Uji, S., Sugiyama, T., Sakamoto, T., Kimura, S., Endo, T., et al. (2020). ERdj3B-mediated quality control maintains anther development at high temperatures. Plant Physiol. 182, 1979–1990. doi: 10.1104/pp.19.01356
Yang, Y., Qin, Y., Xie, C., Zhao, F., Zhao, J. F., Liu, D., et al. (2010). The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell 22, 1313–1332. doi: 10.1105/tpc.109.069609
Yang, Y., Zhao, L., Wang, J., Lu, N., Ma, W., Ma, J., et al. (2023). Genome-wide identification of DnaJ gene family in Catalpa bungei and functional analysis of CbuDnaJ49 in leaf color formation. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1116063
Zhang, J., Bai, Z. C., Ouyang, M., Xu, X., Xiong, H., Wang, Q., et al. (2021). The DnaJ proteins DJA6 and DJA5 are essential for chloroplast iron-sulfur cluster biogenesis. EMBO J. 40, e106742. doi: 10.15252/embj.2020106742
Zhang, H. W., Feng, H., Zhang, J. W., Ge, R. C., Zhang, L. Y., Wang, Y. X., et al. (2020). Emerging crosstalk between two signaling pathways coordinates K+ and Na+ homeostasis in the halophyte Hordeum brevisubulatum. J. Integr. Plant Biol. 71, 4345–4358. doi: 10.1093/jxb/eraa191
Zhang, B., Qiu, H. L., Qu, D. H., Ruan, Y., and Chen, D. (2018). Phylogeny-dominant classification of J-proteins in Arabidopsis thaliana and Brassica oleracea. Genome 61, 405–415. doi: 10.1139/gen-2017-0206
Keywords: HvDJ, barley, cis-regulatory elements, phylogenetic analysis, salt response
Citation: Xu Y, Sun H, Shen L, Wan B, Liu L, Zhang G and Shen Q (2025) The potential functions of HvDJ genes in regulating salt tolerance in barley. Front. Plant Sci. 16:1574097. doi: 10.3389/fpls.2025.1574097
Received: 10 February 2025; Accepted: 11 June 2025;
Published: 10 July 2025.
Edited by:
Meng Jiang, Zhejiang University, ChinaReviewed by:
Chandan Sahi, Indian Institute of Science Education and Research, Bhopal, IndiaBen Zhang, Shanxi University, China
Meixue Zhou, University of Tasmania, Australia
Dawei Xue, Hangzhou Normal University, China
Feng Xue, Qingdao Agricultural University, China
Xiaoyan He, Qingdao Agricultural University, China
Copyright © 2025 Xu, Sun, Shen, Wan, Liu, Zhang and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiufang Shen, c2hlbnFmQHpqdS5lZHUuY24=
 Yunfeng Xu
Yunfeng Xu Haoran Sun
Haoran Sun Ling Shen
Ling Shen Boyan Wan
Boyan Wan Lijun Liu
Lijun Liu Guoping Zhang
Guoping Zhang Qiufang Shen
Qiufang Shen