- 1Wuliangye Yibin Co., Ltd, Yibin, China
- 2College of Biological Engineering, Sichuan University of Science & Engineering, Yibin, China
- 3State Key Laboratory for Crop Stress Resistance and High-Efficiency Production, College of Plant Protection, Northwest A&F University, Xianyang, Shaanxi, China
Phytosulfokine (PSK) is an important plant growth factor regulating plant stress response, PSKR gene family plays an important role in the PSK signaling pathway, but there are few reports in wheat (Triticum aestivum L.). In this study, 149 TaPSKR genes were identified by using hidden Markov models (HMMs) and sequence homology, and their evolutionary relationship, structural characteristics and stress regulation mechanism were systematically analyzed. Our results indicated that genes were unevenly distributed across 21 chromosomes of the wheat genome. Phylogenetic tree and conserved motif analysis revealed that the gene motifs, domains, and structures were relatively conserved, and 75.2% of TaPSKR genes clustered in one clade with monocotyledons. Chromosome distribution showed that genes were enriched in chromosomes 6A (20.13%), 6B (16.11%) and 6D (18.79%). A total of 6,463 cis-acting elements were discovered in the promoter regions of PSKR genes, indicating that these genes are regulated by various developmental, environmental, and hormonal factors, as well as light responses. Expression profile results demonstrated that these genes were influenced by various abiotic stressors, such as PEG6000, salt, cold, heat, and different hormones, including gibberellic acid, jasmonic acid, abscisic acid, 6-benzylaminopurine, and salicylic acid. Additionally, quantitative real-time PCR was performed to further confirm the transcriptomic data. TaPSKR genes may participated in abiotic stress response and phytohormone regulation pathway, which provided key gene resources and theoretical basis for the analysis of PSKR signaling pathway and molecular breeding for stress resistance in wheat.
1 Introduction
The escalating threats of rising temperatures and water scarcity are significantly impacting crop productivity worldwide. Due to their inability to escape like animals, plants have evolved numerous mechanisms to adapt to the dynamic environment, thus guaranteeing survival under unfavorable conditions. Plants respond to biotic and abiotic stressors through signal perception, physical adaption, and molecular and cellular changes (Ahuja et al., 2010; Takahashi and Shinozaki, 2019). An increasing number of studies have investigated peptide hormones, such as phytosulfokine (PSK), as a central component of cell to cell communication that plays an important role in regulating plant growth and reproduction (Sauter, 2015; Hohmann et al., 2017; Godel-Je˛drychowska et al., 2019).
PSK, which is a disulfated pentapeptide, is considered a crucial plant growth factor. It was first identified in Asparagus officinalis mesophyll cells grown in a culture medium (Matsubayashi and Sakagami, 1996). PSK, which is encoded by the PSK gene family, is composed of five residues, including two sulfated tyrosines (Tyr (SO3H)-IleTyr (SO3H)-Thr-Gln) (Yang et al., 2001; Matsubayashi et al., 2006; Kutschmar et al., 2009). PSK is processed from an ~80-amino acid precursor protein and matured through proteolytic cleavage and post-translational sulfonation of its tyrosine residues (Srivastava et al., 2008). Tyrosine sulfation occurs in the trans-Golgi network, is catalyzed by a tyrosylprotein sulfotransferase, and is further cleaved by cytoplasm-localized subtilisin-like serine proteases (Kutschmar et al., 2009). Subsequently, mature PSK peptides are perceived by membrane-bound receptors (PSKRs), which were first isolated from Daucus carrota, and they are evolutionarily conserved in different species (Hanai et al., 2000; Kaufmann and Sauter, 2019). PSKRs are localized at the plasma membrane and belong to the canonical leucine-rich repeat receptor kinases (LRR-RK) family, which contains extracellular LRRs and a PSK-binding domain, a transmembrane domain, an intercellular kinase domain, and an island domain (Kwezi et al., 2011). A total of 2, 2, and 15 potential PSK receptors have been predicted in Arabidopsis, tomato, and rice, respectively (Matsubayashi et al., 2006; Amano et al., 2007; Zhang et al., 2018; Nagar et al., 2020).
In plants, PSK participates in growth and development regulation, including callus formation, cell proliferation, sexual reproduction, lateral root development, and biotic and abiotic stress responses (Matsubayashi and Sakagami, 1996; Matsubayashi et al., 1999; Stührwohldt et al., 2011; Sauter, 2015). In Arabidopsis, PSK is induced by wounding, and the PSKR1 mutant inhibits callus formation, demonstrating that signaling through PSKR1 is essential for cell proliferation (Matsubayashi et al., 2006; Loivama et al., 2010). Additionally, PSK-α positively regulates root growth, and the knockout of any PSKR results in short roots due to reduced root cell length (Kutschmar et al., 2009). Recent publications have shown that PSK signaling pathways fine-tune plant growth by acting together with other phytohormones (Rodiuc et al., 2016). For example, the PSKR1 mutant is sensitive to the hemibiotrophic bacterial pathogen Pseudomonas syringae but insensitive to the necrotrophic fungal pathogen Alternaria brassicicola, suggesting that PSK has an antagonistic effect on salicylic acid (SA)-mediated defense responses (Mosher et al., 2013). In addition, PSK suppresses ethylene (ET) synthesis, regulates copper homeostasis, and stimulates non-embryogenic proliferation in cooperation with auxin (Eun et al., 2003). In Arabidopsis, ectopic expression of OsPSKR15 from rice positively regulated drought tolerance by controlling stomatal movements in reactive oxygen species (ROS)-dependent abscisic acid (ABA) signaling (Nagar et al., 2022). OsPSKR has mutual antagonism with OsPEPR1 signaling, thereby modulating the transition of defense signals to growth signals during wounding (Chitthavalli et al., 2024). In tomato, the PSK-PSKR signaling pathway triggers auxin-dependent immune responses against Botrytis cinerea by inducing intracellular Ca2+ release (Zhang et al., 2018). PSK is induced by drought and regulates flower drop (Reichardt et al., 2020).
Thus far, studies on the PSK-PSKR signaling pathway have mainly focused on Arabidopsis and rice. Wheat, one of the most important food crops, has a higher salt tolerance than rice. However, in wheat, there are few reports of the PSKR gene family, which plays an important role in the PSK signaling pathway and regulates plant growth and development and biotic and abiotic stress responses. In this study, the PSKR gene family was identified in wheat based on the latest genome sequence. We performed genome-wide characterization of PSKRs in wheat; determined the phylogenetic relationships between PSKRs in wheat, Arabidopsis, and rice; and identified the conserved motifs in wheat PSKRs and the cis-acting elements in the 2000-bp promoter of wheat PSKR genes. In addition, we determined the PSKR gene expression profile under NaCl, heat, cold and ABA treatment. Our results are expected to provide a basis for further revealing the function and molecular mechanism of abiotic stress tolerance of TaPSKR genes.
2 Materials and method
2.1 Plant material growth and abiotic stress conditions
Wheat (T. aestivum) cultivar ‘Fielder’ seedlings were grown in a growth chamber (white fluorescent tubes, 200 - 300 μmol m−2 s−1) at maintaining 14°C/12°C day/night temperatures (16/8 h photoperiod) at 60% humidity or in the field (natural long-day conditions). Seedlings at the 3-leaf stage were treated with 200 mM NaCl, 4°C, 24°C, and 100 µM ABA, respectively. All 3 leaves from at least 5 plants were separately collected 0, 1, 3, 6, 12, 24, and 48 h after treatment. All leaf samples were flash frozen in liquid nitrogen and ground in a mortar for RNA isolation.
2.2 Identification and characterization of TaPSKR genes in wheat
To identify the potential PSKR genes in T. aestivum, hidden Markov models (HMMs) and sequence homology were used. Two AtPSKR genes (At2g02220 and At5g53890) and fifteen OsPSKR genes were retrieved from TAIR (https://www.arabidopsis.org/) and the Rice Genome Annotation Project (https://rice.uga.edu/index.shtml), respectively, and used to search for PSKR members in T. aestivum (IWGSC.57) using HMMER (hmmer.org). The protein sequences were downloaded from Ensembl plants (release 59). TBtools-II was used to filter genes dissimilar to other species with PSKR proteins (Chen et al., 2023).
2.3 Phylogenetic analysis and chromosomal localization of TaPSKR genes
A phylogenetic tree of PSKR genes in wheat, Arabidopsis, and rice was constructed using the maximum likelihood (ML) method with 1000 bootstrap replicates and combined with the protein domain structure and gene structure using iTOL (https://itol.embl.de/). The TaPSKR genes of wheat were mapped to 21 chromosomes using genome data from TBtools-II (Chen et al., 2023).
2.4 Gene structure, conserved motifs, and cis-element analysis of PSKR genes
Gene structure annotation was performed using the GFF3 file of the wheat genome and visualized using TBtools v2.025 (Chen et al., 2023). MEME (https://meme-suite.org/meme/) was used to identify the conserved motifs of TaPSKR proteins. The potential cis-elements predicted in the 2-kb region upstream of the start codon of TaPSKR genes in the wheat genome were used in promoter sequence analysis with the PlantCARE database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
2.5 Expression profile analysis of TaPSKR genes under different abiotic stress and phytohormone treatments
The transcriptome data of TaPSKR genes in wheat under different abiotic stress and phytohormone treatments were downloaded from the WheatOmics 1.0 database (http://202.194.139.32/). A heat map was constructed using TBtools-II (Chen et al., 2023).
2.6 Quantitative real-time PCR analysis
RNA was extracted from the leaves of both the control and treatment groups. Quantitative real-time PCR (qRT-PCR) was performed to analyze the expression of 13 randomly selected TaPSKR genes in wheat under 200 mM NaCl, 100 µM ABA, cold (4°C), and heat (24°C) treatments. The total qRT-PCR volume was 20 µL: 10 µL of SYBR Premix Ex Taq (RR820 Takara, China), 0.5 µL of each primer (primers are listed in Supplementary Table S1), 1 µL of cDNA template, and 8 µL of ddH2O. PCR was performed as follows: initial pre-denaturation at 95°C for 2 min; 40 cycles of 95°C for 10 s, annealing at 56°C for 30 s, and extension at 72°C for 15 s. The experimental results were normalized using the 2−▵▵Ct method.
3 Results
3.1 Identification of PSKR genes in wheat
In this paper, we used the HMM and BLASTp approaches. A total of 149 PSKR genes, named TaPSKR1 to TaPSKR149, were identified in the wheat genome based on 2 Arabidopsis thaliana and 15 Oryza sativa gene sequence searches (Supplementary Table S2). We then analyzed detailed information on the 149 TaPSKR genes. Consistent with previous studies, transmembrane domain prediction showed that all proteins had a single membrane spanning domain (https://services.healthtech.dtu.dk/services/DeepTMHMM-1.0/; Matsubayashi et al., 2006; Hallgren et al., 2022). The number of amino acids in these proteins ranged from 155 (TraesCS2A02G550500/TaPSKR9) to 1205 (TraesCS6A02G129000/TaPSKR88), with an average of 789 amino acids. The isoelectric point (pI) ranged from 4.88 (TraesCS6B02G154700/TaPSKR102) to 9.66 (TraesCS1B02G193000/TaPSKR3), with an average of 6.52. The molecular weight (MW) ranged from 16.76 (TraesCS6A02G128600/TaPSKR79) to 130.18 KD (TraesCS6A02G129000/TaPSKR80), with an average of 81.2 KD. In addition, we exported the cDNA, coding, and protein sequences.
3.2 Phylogenetic, motif distribution, conserved motif, and exon-intron structure analysis of wheat PSKR proteins
To investigate the relationships between these PSKR proteins, we constructed a phylogenetic tree using the protein sequence of 149 TaPSKR, 2 AtPSKR, and 15 OsPSKR genes (Figure 1). These TaPSKR genes were divided into 3 subfamilies using tree-based distances in TreeCluster (https://github.com/niemasd/TreeCluster; Balaban et al., 2019). Significant variations were observed in the number of genes within each branch. The most members were in branch I, which contained 112 TaPSKR genes (accounting for 75.2% of the total). Notably, all AtPSKRs and OsPSKRs were distributed in branch I, highlighting the evolutionary conservation of PSKR genes across species in branch I. In branch I, most TaPSKR proteins formed a cluster with OsPSKR proteins, separate from the orthologs in Arabidopsis, suggesting that TaPSKR genes have more sequence similarity with monocotyledons than dicotyledons.
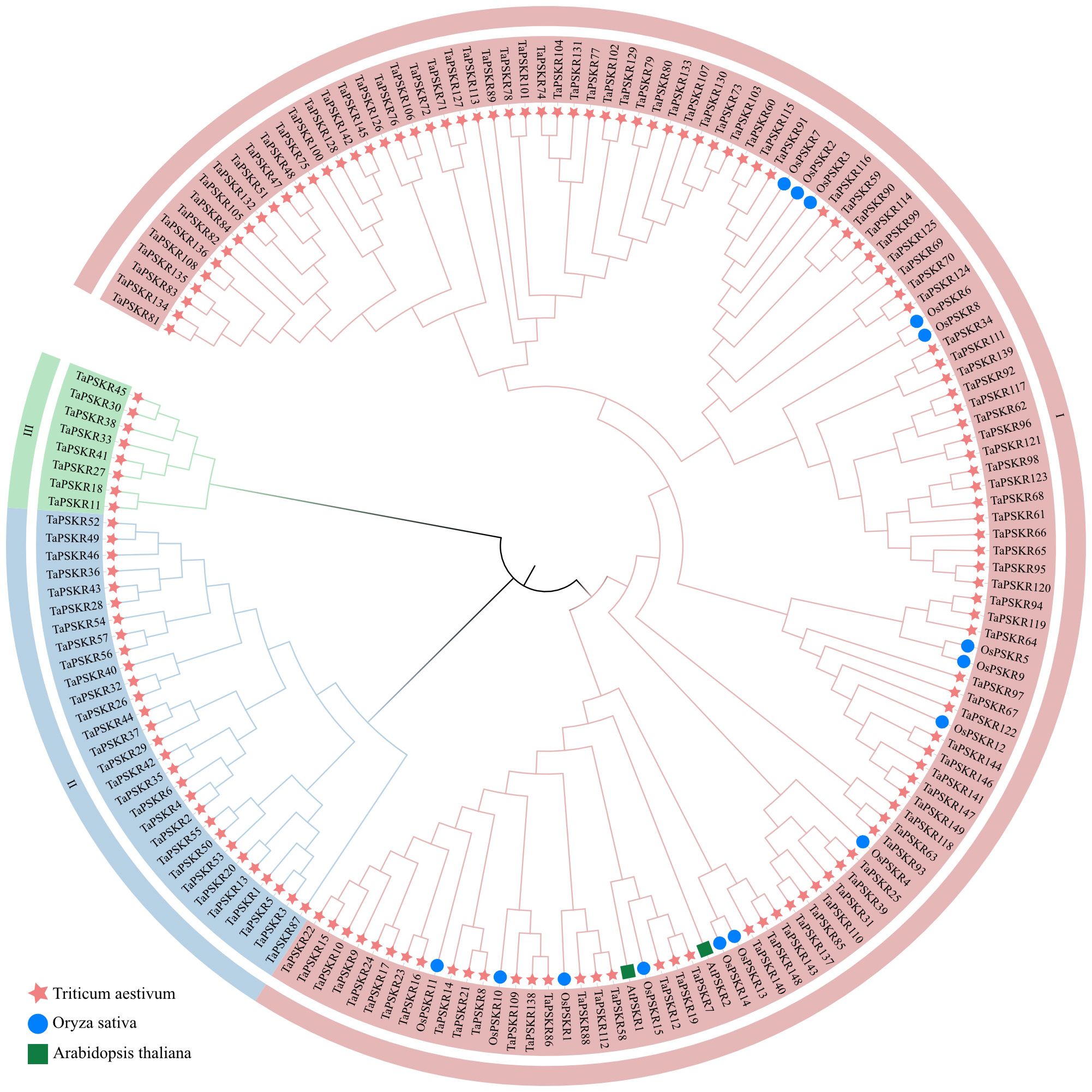
Figure 1. Phylogenetic analysis of PSKR proteins in wheat, Arabidopsis, and rice. The phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstrap replications. In the tree, the TaPSKR proteins were divided into 3 subgroups, which are represented by different colored clusters within the tree.
3.3 Gene structure and conserved motifs in TaPSKR genes
To understand the structural composition of the TaPSKR gene family, a phylogenetic tree was constructed to elucidate the relationships among all TaPSKR genes (Figure 2A). Based on tree topology, we further analyzed the conserved motifs, domains, and exon-intron structure features of TaPSKR genes. A total of 20 conserved motifs (named motifs 1-20) were generated for all TaPSKR genes using the MEME tool (Figure 2B; Supplementary Table S3). The conserved motifs ranged from 1 (TraesCS2A02G550500/TaPSKR9) to 20, and 42 TaPSKR proteins contained less than 10 motifs. A total of 58 TaPSKR proteins contained 10–19 motifs, and 49 TaPSKR proteins contained 20 conserved motifs. Motifs 2, 5, 6, 12, 14, 15, and 16 were located within the C-terminal, and motifs 10, 13, 17, 18, 19, and 20 were located within the N-terminal. Consistent with the gene structure distribution, TaPSKR proteins containing a similar conserved motif were clustered together in the phylogenetic tree (Figure 2B). Consistent with other LRR-RLKs, all TaPSKR proteins contained a single transmembrane domain (TMHilex) and diverse kinase domain types, such as the Ser-Thr kinase superfamily, PknB_PASTA kinase superfamily (Matsubayashi et al., 2006; Burastero et al., 2022), SPS1 superfamily, and WAK-associated superfamily. In addition, other domains, such as the LRR superfamily, LapA domain superfamily response to heat shock, Gnk2-like superfamily response to abiotic stress (Serrazina et al., 2022), GUB_WAK_bind superfamily response to salt stress (Tang et al., 2022), and the PLN0034 and PLN00113 superfamily, were detected in these proteins (Figure 2C; Supplementary Table S4). The gene structure displayed significant diversity among the TaPSKR genes; the exon number ranged from 1 to 18, with the majority containing only 1 exon (98, 65.77%), 6 containing 2 exons, TaPSKR85 and TaPSKR87 containing 3 exons, TaPSKR38 and TaPSKR45 containing 18 exons, and the remainder containing between 3 and 17 exons (Figure 2D; Supplementary Table S5). Diversity variations likely reflect the different functional roles and evolutionary trajectories of these genes, suggesting diverse regulatory mechanisms within the TaPSKR gene family related to plant growth and development. Generally, the same subgroup in the phylogenetic tree shared a similar exon-intron structure.

Figure 2. Gene structure and conserved domain organization of PSKR genes in wheat. (A) Their phylogenetic relationships based on the neighbor-joining method. (B) Motif distribution of the TaPSKR proteins. Different motifs (1-20) are indicated by different colors. (C) Conserved domain organization in TaPSKR proteins. (D) Exon–intron structures of TaPSKR genes.
3.4 Chromosomal location analysis of TaPSKR genes
To analyze the relative position of each TaPSKR gene copy on the chromosome, we labeled their physical placements on wheat A, B, and D chromosomes. As shown in Figure 3, the TaPSKR genes were mapped on the 21 chromosomes with a significantly uneven distribution. Only 1 gene (5.26%) was located on chromosomes 5B and 5D, and most of the TaPSKR genes were mapped on chromosome 6A (30/20.13%), followed by chromosome 6B (24/16.11%) and chromosome 6D (28/18.79%).
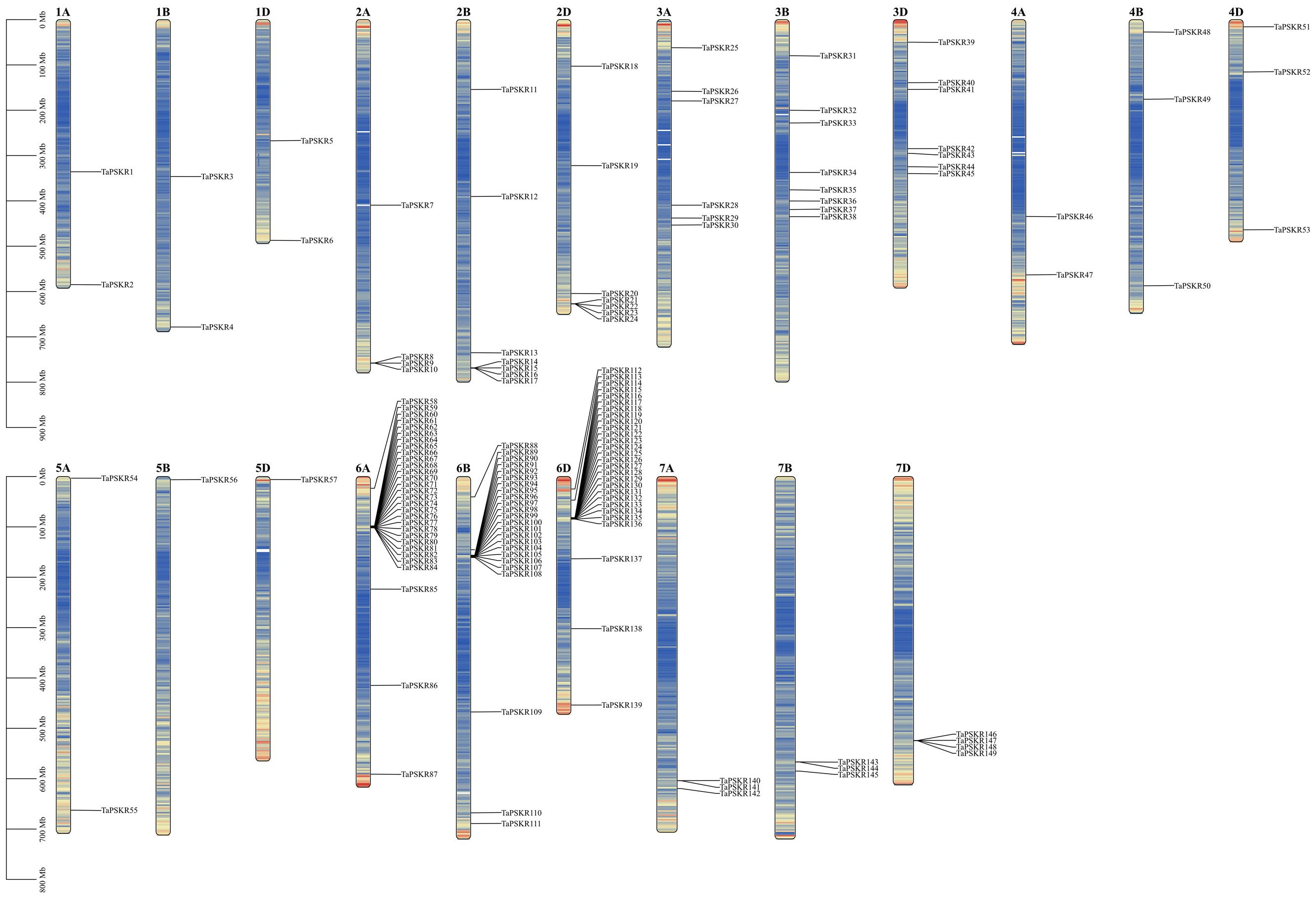
Figure 3. Chromosomal distribution of TaPSKR gene copies. Only chromosomes containing TaPSKR genes are represented.
3.5 Prediction of cis-regulatory elements of TaPSKR genes
Cis-acting elements in the promoter are crucial to regulating gene expression and its function (Lescot et al., 2002). To further predict the bio-function of TaPSKR genes, we analyzed the cis-acting elements in the 2-kb promoter region of all TaPSKR genes using the online database PlantCARE. As shown in Figure 4, a total of 6463 cis-acting were identified across 149 TaPSKR genes (Supplementary Table S6). These cis-acting elements were classified into 54 types and further categorized into five main functional groups: RNA polymerase-binding site, related to development, environmental stress response, hormone response, and light response (Figure 4). Among them, a large number of core elements (4277, 66.18%) were associated with RNA polymerase-binding sites, such as AT-rich elements, CAAT-box, and TAAT-box, indicating that it could enable genes to complete the normal transcription process. Of the cis-elements, 58 (0.9%) responded to development regulation, consistent with a previous study. AACA and GCN4 motifs control the endosperm starch structure in maize (Wu et al., 2015; Liu et al., 2018). Of the cis-elements, 402 (6.22%) were involved in the environmental stress response, of which the MBS and TC-rich repeat motifs in the AsGRAS gene family (GRAS gene family in A. sativa) regulate the salt stress response. A total of 729 (11.28%) cis-elements, such as ABA-responsive elements (ABREs), gibberellin-responsive elements (TCTC-box, P-box, and GARE-motif), auxin-responsive elements (TGA-element), and SA-responsive elements (TCA-element), responded to hormones (Zhang et al., 2024), and 997 (15.43%) cis-elements responded to light. These results suggest that TaPSKR genes are involved in plant growth and abiotic stress responses.
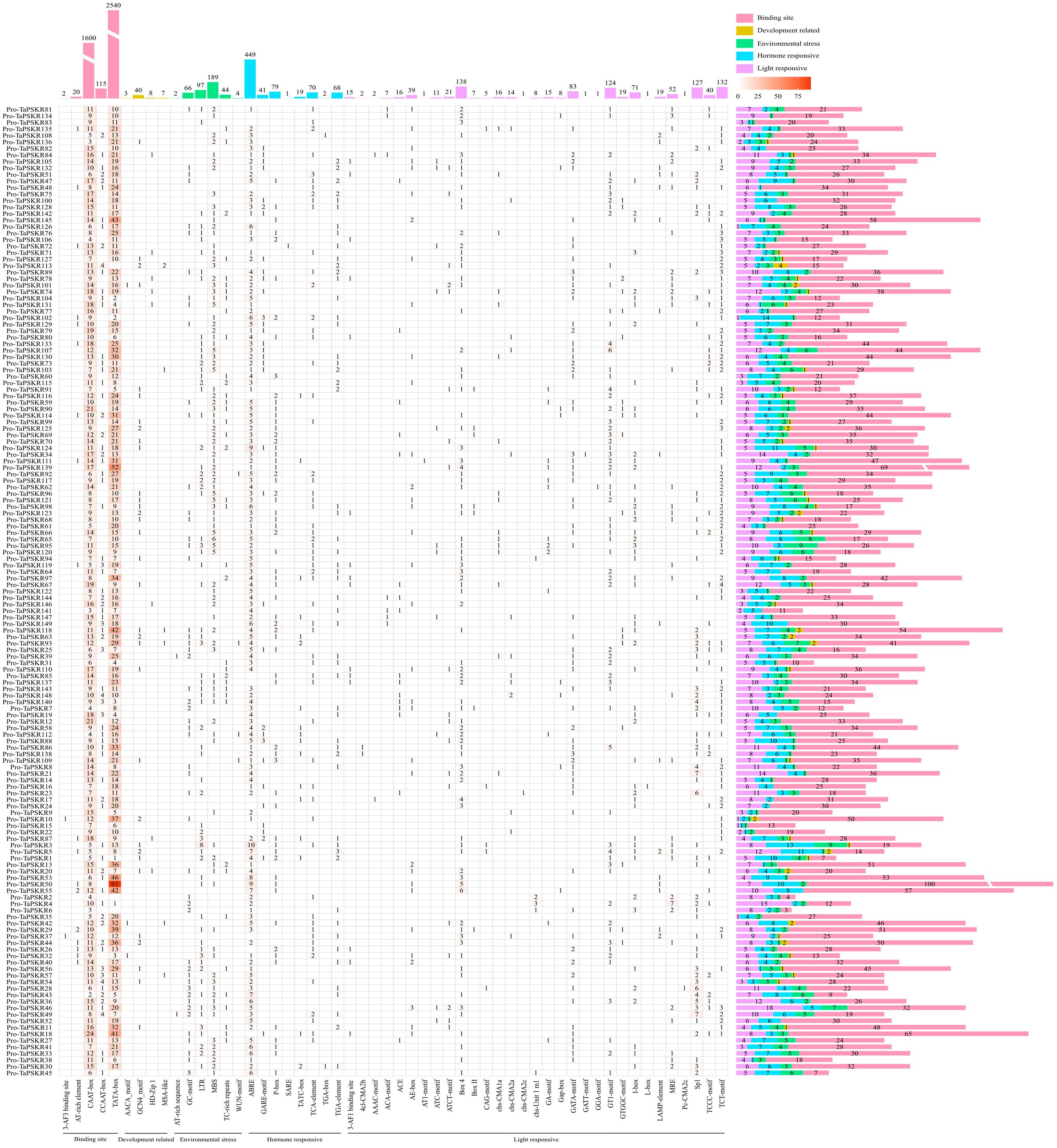
Figure 4. Number and composition of cis-acting regulatory elements in the promotor regions of TaPSKR genes. The 2000-bp promoter region of each gene copy is displayed.
3.6 Expression profile of TaPSKR genes under abiotic stress
To investigate the abiotic stress response of TaPSKR genes, we obtained transcriptome data for 149 TaPSKR genes from wheat under abiotic stress (such as PEG 6000, salt stress, heat stress, and cold stress) and phytohormone treatments (such as gibberellic acid (GA), jasmonic acid (JA), 6-benzylaminopurine (6BA), ABA, and SA) from WheatOmics. In the tolerant cultivar “Giza 168”, PEG6000 treatment at 2 h and 12 h up-regulated 42 and 32 genes and down-regulated 87 and 102 genes, respectively, whereas in the sensitive cultivar “Gemmiza 10”, 135 TaPSKR genes responded to PEG6000, with 61 up-regulated and 74 down-regulated at 2 h, and 87 up-regulated and 48 down-regulated at 12 h. Heat treatment up-regulated 40 and 42 genes and down-regulated 59 and 71 genes in the thermotolerant “HD2985” and thermosusceptible “HD2329” varieties, respectively. Under salt stress, the salt-sensitive ‘Chinese Spring’ showed up-regulation of 96, 90, and 53 genes and down-regulation of 38, 43, and 80 genes at 6, 12, and 24 h, respectively, while the salt-tolerant “Qingmai 6” exhibited up-regulation of 83, 80, 59, and 50 genes and down-regulation of 51, 57, 80, and 83 genes at 6, 12, 24, and 48 h, respectively. Phytohormones significantly influenced TaPSKR expression: GA and JA treatment at 1 h up-regulated 17 and 45 genes and down-regulated 102 and 74 genes, respectively, while ABA and SA treatment at 3 h up-regulated 24 and 76 genes and down-regulated 76 and 32 genes, respectively. These results suggest that TaPSKR genes are involved in abiotic stress responses and phytohormone regulatory pathways.
3.7 Validation of abiotic-responsive TaPSKRs through qRT-PCR analysis
To investigate the abiotic stress response of TaPSKR genes, we randomly selected 13 TaPSKR genes, namely TaPSKR8, TaPSKR9, TaPSKR10, TaPSKR11, TaPSKR15, TaPSKR19, TaPSKR41, TaPSKR64, TaPSKR75, TaPSKR83, TaPSKR87, TaPSKR140, and TaPSKR143, and verified their expression using qRT-PCR under different abiotic stress conditions, such as cold stress, heat stress, salt stress, and ABA treatment (Figure 5).
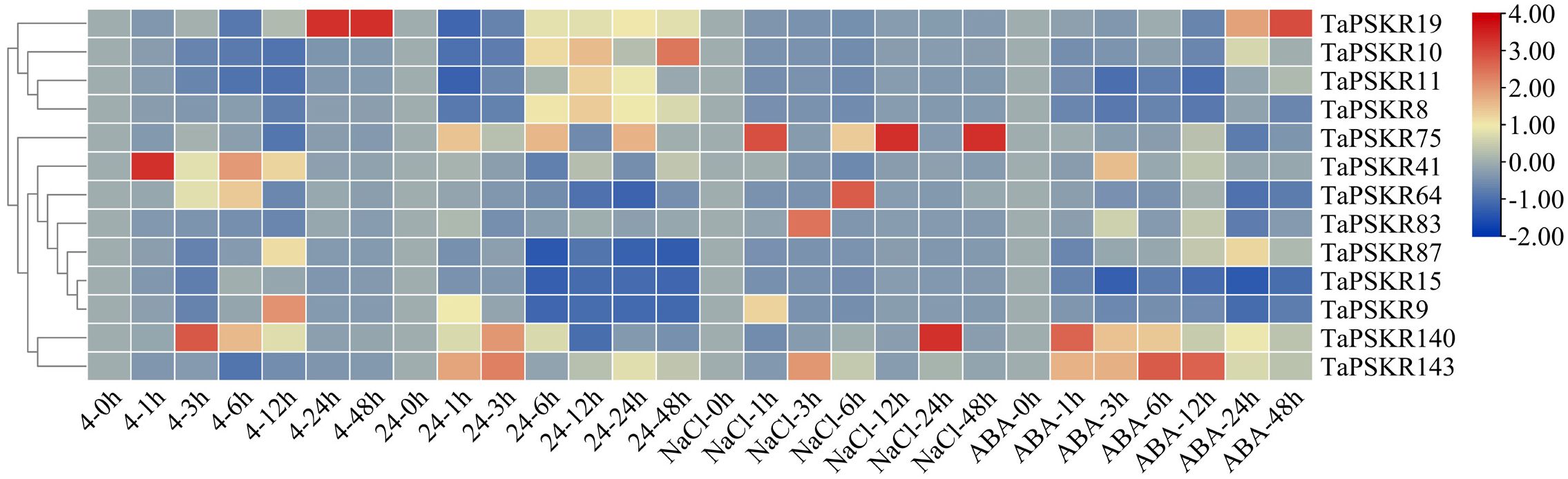
Figure 5. Relative expression levels of 13 randomly selected TaPSKR genes under abiotic stress (4°C, 24°C, 200 mM NaCl) and 100 µM ABA conditions using qRT-PCR analysis.
Cold stress disrupts root water uptake and leads to inadequate water supply to the stem, resulting in drought stress (Aroca et al., 2012). Each year, spring frost takes place in March and April at the early booting stage, affecting approximately 85% of the wheat-sown area in the world (Yue et al., 2016). Among these 13 TaPSKRs, 7 were less affected by cold stress, the rest of 6 TaPSKR genes induced by cold stress (Figure 5). Among them, TaPSKR41 reached the highest expression at 1 h. TaPSKR140, TaPSKR64, and TaPSKR9 showed the highest expression at 3, 6, and 12 h, respectively. TaPSKR19 expression was induced by cold stress at 24 h.
Heat stress increases ROS and lipid peroxidation products in the coleoptile and developing organs and inhibits root and first leaf development at the seedling stage (Savicka and Skute, 2010). Additionally, heat stress alters wheat grain quality, such as grain weight, nutrients, fiber, protein content, and starch granule composition (Akter et al., 2017). In this study, TaPSKR10 induced by heat at 6 h, and reached the highest expression at 48 h, TaPSKR143 rapidly response to heat at 1 h, TaPSKR87, TaPSKR15, and TaPSKR9 were inhibited by heat stress.
Soil salinity affects approximately 7% of the land mass worldwide and has become a serious environmental stressor increasingly affecting crop production (Yang and Guo, 2018; Hickey et al., 2019). Among the TaPSKR genes, TaPSKR19 TaPSKR10, TaPSKR11, TaPSKR8, TaPSKR41, TaPSKR87, and TaPSKR15 were less affected by salt stress, while the other 6 TaPSKR genes were significantly affected by salt stress. TaPSKR9 displayed the highest expression at 1 h, and TaPSKR143 and TaPSKR83 showed the highest expression at 3 h. The expression of TaPSKR64, TaPSKR75, and TaPSKR140 was highest at 6, 12, and 24 h, respectively. The diverse expression patterns of these TaPSKRs imply their different roles in the salt stress response.
ABA is a major phytohormone affecting the response to environmental stress and regulating cellular, physiological, and developmental processes (Stone, 2019). Our results indicated that TaPSKR11, TaPSKR8, TaPSKR15, and TaPSKR9 were down-regulated by ABA. TaPSKR140, TaPSKR41, TaPSKR143, and TaPSKR19 reached the highest expression under 100 µM ABA condition at 1, 3, 6, and 24 h, respectively.
4 Discussion
4.1 Systematic identification and comprehensive analysis of TaPSKR genes in wheat
Signal transduction is an important way for plants to experience sensory environmental stimuli, which is essential for the regulation of plant growth and development processes and environmental stress responses. PSKR proteins were first isolated in microsomal fractions from carrot cell cultures (Daucus carota) (Matsubayashi et al., 2002). A total of 2 and 15 ortholog genes have been identified in Arabidopsis and rice, respectively (Matsubayashi et al., 2006; Nagar et al., 2020). Previous studies have shown that PSKR genes participate in disease resistance and the drought response, regulating the balance between growth and defense in rice (Yang et al., 2019; Nagar et al., 2022; Harshith et al., 2024), modulating the defense response, and initiating auxin-dependent immunity in tomato (Zhang et al., 2018; Hu et al., 2023). AtPSKR1 and AtPSKR2 promote plant growth and callus formation, control pollen tube growth and funicular pollen tube guidance, and regulate immune responses against pathogens (Mosher et al., 2013; Stuhrwohldt et al., 2015; Rodiuc et al., 2016).
In this study, 149 TaPSKR genes were identified in the wheat genome (Supplementary Table S1). The phylogenetic tree constructed with 149 TaPSKR, 2 AtPSKR, and 15 OsPSKR genes showed that 112 (75.2%) TaPSKR genes clustered with Arabidopsis and rice genes, forming a distinct branch (Figure 1). Clustering suggested that TaPSKR genes in wheat had more evolutionary diversification, thereby indicating their functional diversity. Conserved motif analysis of 149 TaPSKR genes revealed that the genes grouped within the same branch of the phylogenetic tree had similar conserved motifs (Figures 2A, B). This suggests that members with similar conserved motifs may have functional redundancy. Additionally, all TaPSKR genes have a single membrane spanning domain (Figure 2C), consistent with the structural characteristics of the LRR-RLK family (Sauter, 2015). Among the 149 TaPSKR genes, 98 (65.77%) and 51 (34.23%) contained only 1 exon and more than 2 exons, respectively (Figure 2D), suggesting that the structural evolution and gene expansion of the TaPSKR gene family are relatively conserved (Li et al., 2024).
4.2 Expression profile analysis of TaPSKR genes in wheat under abiotic stress
Plant growth and development are finely regulated by external environment factors and internal gene expression levels (Li et al., 2024). To further clarify the bio-functions of TaPSKR genes in wheat, we predicted the cis-elements of 149 TaPSKR genes. The results revealed that TaPSKR genes may be involved in development regulation, environment stress response, hormone response, and light response (Figure 4), suggesting that TaPSKR genes have diverse functions and play an important role in environment factor response. Additionally, three elements associated with stress responses were identified: MBS, ABRE, and TC-rich repeats. Among these elements, 189 MBS elements and 44 TC-rich repeat elements were identified in 149 TaPSKR genes and are involved in multiple environmental stress responses in plants (Figure 3). A total of 449 ABREs that participated in the ABA response pathway and abiotic stress responses were identified in the promoters of 149 TaPSKR genes. Other response elements, including 41 GARE-motif, 70 P-Box, and 97 LTR endogenous retroviral elements, were also identified. The expression profile results showed that the expression of more than 100 TaPSKR genes was influenced by abiotic stress and phytohormone treatments, consistent with the results of our qPCR analysis (Figures 6 and 5), indicating that TaPSKR genes are involved in a variety of biological processes throughout the life cycle of wheat.
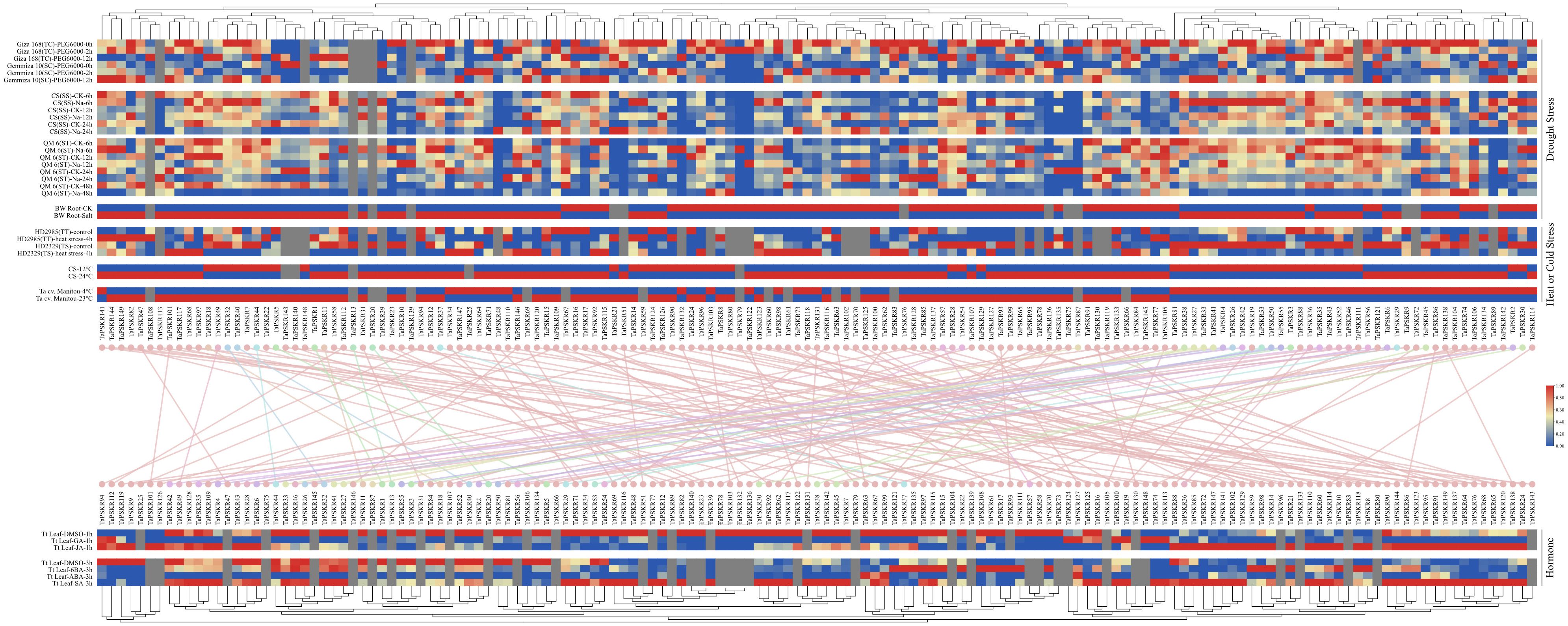
Figure 6. Expression patterns of TaPSKR genes under abiotic stress and phytohormone treatment conditions. TC, tolerant cultivar; SC, sensitive cultivar; TT, thermotolerant; TS, thermosusceptible; SS, salt sensitive; ST, salt tolerant; CS, Chinese Spring; QM6, Qing Mai 6; BW, bread wheat; Ta cv., Triticum aestivum cv; and Tt, Triticum turgidum.
5 Conclusion
In this study, 149 TaPSKR genes were identified in the wheat genome; among them, 112 TaPSKR genes clustered in one group with AtPSKR and OsPSKR genes. Conserved motif analysis revealed that TaPSKR genes within the same branch shared similar conserved motifs, further validating the robustness of the phylogenetic tree. Additionally, each TaPSKR gene contained a single transmembrane domain, consistent with the structural characteristics of the LRR-RLK family. Chromosome localization analysis revealed an uneven distribution of the 149 TaPSKR genes across 21 chromosomes, with the lowest number on chromosomes 5B and 5D and the highest on chromosome 6A. Cis-element prediction and expression profiling demonstrated that TaPSKR genes exhibited complex regulatory mechanisms under various abiotic stress and phytohormone treatments. Our qPCR analysis of 13 randomly selected TaPSKR genes supported this result. In summary, our results provide a comprehensive analysis of TaPSKR genes in wheat and contribute to molecular breeding for improving abiotic stress response.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/material.
Author contributions
ZM: Conceptualization, Data curation, Formal Analysis, Methodology, Visualization, Writing – original draft. JW: Data curation, Investigation, Writing – original draft. JZ: Methodology, Resources, Writing – original draft. JS: Formal Analysis, Visualization, Writing – original draft. DZ: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded by Wuliangye Yibin Co., Ltd.
Conflict of interest
Authors ZM, JZ, JS, and DZ were employed by Wuliangye Yibin Co., Ltd.
The remaining author declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1582433/full#supplementary-material
References
Ahuja, I., Vos Bones, A. M., and Hall, R. D. (2010). Plant molecular stress responses face climate change. Trends Plant Sci. 15, 664–674. doi: 10.1016/j.tplants.2010.08.002
Akter, N., Rafiqul Islam, M., and Islam, R. (2017). Heat stress efects and management in wheat. A review. Agron. Sustain. Dev. 37, 37. doi: 10.1007/s13593-017-0443-9
Amano, Y., Tsubouchi, H., Shinohara, H., Ogawa, M., and Matsubayashi, Y. (2007). Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 18333–18338. doi: 10.1073/pnas.0706403104
Aroca, R., Porcel, R., and Ruiz-Lozano, J. M. (2012). Regulation of root water uptake under abiotic stress conditions. J. Exp. Bot. 63, 43–57. doi: 10.1093/jxb/err266
Balaban, M., Moshiri, N., Mai, U., Jia, X. F., and Mirarab, S. (2019). Tree Cluster: Clustering biological sequences using phylogenetic trees. PloS One 14, e0221068. doi: 10.1371/journal.pone.0221068
Burastero, O., Cabrera, M., Lopez, E. D., Defelipe, L. A., Arcon, J. P., Durán, R., et al. (2022). Specificity and reactivity of mycobacterium tuberculosis serine/threonine kinases pknG and pknB. J. Chem. Inf. modeling 62, 1723–1733. doi: 10.1021/acs.jcim.1c01358
Chen, C. J., Wu, Y., Li, J. W., Wang, X., Zeng, Z. H., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Chitthavalli, Y., Harshith, A. P., Monoswi, C., Ashwin, N., Steffi, R., and Padubidri, V. S. (2024). Wound-induced small-peptide-mediated signaling cascade, regulated by OsPSKR, dictates balance between growth and defense in rice. Cell Rep. 43, 114515. doi: 10.1016/j.celrep.2024.114515
Eun, C. H., Ko, S. M., Matsubayashi, Y., Sakagami, Y., and Kamada, H. (2003). Phytosulfokine-α requires auxin to stimulate carrot non-embryogenic cell proliferation. Plant Physiol. Biochem. 41, 447–452. doi: 10.1016/S0981-9428(03)00052-4
Godel-Je˛drychowska, K., Mackowska, K., Kurczynska, E., and Grzebelus, E. (2019). Composition of the reconstituted cell wall in protoplastderived cells of daucus is affected by phytosulfokine (PSK). Int. J. Mol. Sci. 20, 5490. doi: 10.3390/ijms20215490
Hallgren, J., Konstantinos, D., Tsirigos, Mads, D. P., Armenteros, J. J. A., Marcatili, P., et al. (2022). DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv. doi: 10.1101/2022.04.08.487609
Hanai, H., Matsuno, T., Yamamoto, M., Matsubayashi, Y., Kobayashi, T., Kamada, H., et al. (2000). A secreted peptide growth factor, phytosulfokine, acting as a stimulatory factor of carrot somatic embryo formation. Plant Cell Physiol. 41, 27–32. doi: 10.1093/pcp/41.1.27
Harshith, C. Y., Pal, A., Chakraborty, M., Nair, A., Raju, S., and Shuvaprasad, P. V. (2024). Wound induced small-peptide mediated signalling cascade regulated by OsPSKR, dictates balance between growth and defense in rice. Cell Rep. 43, 114515. doi: 10.1016/j.celrep.2024.114515
Hickey, L. T., Hafeez, N., Robinson, H., Jackson, S. A., Leal-Bertioli, S. C. M., Tester, M., et al. (2019). Breeding crops to feed 10 billion. Nat. Biotechnol. 37, 744–754. doi: 10.1038/s41587-019-0152-9
Hohmann, U., Lau, K., and Hothorn, M. (2017). The structural basis of ligand perception and signal activation by receptor kinases. Annu. Rev. Plant Biol. 68, 109–137. doi: 10.1146/annurev-arplant-042916-040957
Hu, Z. J., Fang, H. M., Zhu, C. G., Gu, S. H., Ding, S. T., Yu, J. Q., et al. (2023). Ubiquitylation of PHYTOSULFOKINE RECEPTOR 1 modulates the defense response in tomato. Plant Physiol. 192, 2507–2522. doi: 10.1093/plphys/kiad188
Kaufmann, C. and Sauter, M. (2019). Sulfated plant peptide hormones. J. Exp. Bot. 70, 4267–4277. doi: 10.1093/jxb/erz292
Kutschmar, A., Rzewuski, G., Stuhrwohldt, N., Beemster, G. T., Inze, D., and Sauter, M. (2009). PSK-alpha promotes root growth in Arabidopsis. New Phytol. 181, 820–831. doi: 10.1111/j.1469-8137.2008.02710.x
Kwezi, L., Ruzvidzo, O., Wheeler, J. I., Govender, K., Iacuone, S., Thompson, P. E., et al. (2011). The phytosulfokine (PSKα) receptor is capable of guanylate cyclase activity and enabling cyclic GMP-dependent signaling in plants. J. Biol. Chem. 286, 22580–22588. doi: 10.1074/jbc.M110.168823
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van Der Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, Y., Fu, Q., Li, X., Zhang, Q., Zhao, Q., Ding, Y., et al. (2024). Exploring the guardian of abiotic stress: Genome-wide identification of the basic helix-loop-helix transcription factor family in Juglans mandshurica. Sci. Hortic. 331, 113154. doi: 10.1016/j.scienta.2024.113154
Liu, X., Li, S., Yang, W., Mu, B., Jiao, Y., Zhou, X. J., et al. (2018). Synthesis of seed-specific bidirectional promoters for metabolic engineering of anthocyanin-rich maize. Plant Cell Physiol. 59, 1942–1955. doi: 10.1093/pcp/pcy110
Loivamaki, M., Stu€hrwohldt, N., Deeken, R., Steffens, B., Roitsch, T., Hedrich, R., et al. (2010). A role for PSK signaling in wounding and microbial interactions in Arabidopsis. Physiol. Plant 139, 348–357. doi: 10.1111/j.1399-3054.2010.01371.x
Matsubayashi, Y., Ogawa, M., Kihara, H., Niwa, M., and Sakagami, Y. (2006). Disruption and overexpression of arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 142, 45–53. doi: 10.1104/pp.106.081109
Matsubayashi, Y., Ogawa, M., Morita, A., and Sakagami, Y. (2002). An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296, 1470–1472. doi: 10.1126/science.1069607
Matsubayashi, Y. and Sakagami, Y. (1996). Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc. Natl. Acad. Sci. U.S.A. 93, 7623–7627. doi: 10.1073/pnas.93.15.7623
Matsubayashi, Y., Takagi, L., Omura, N., Morita, A., and Sakagami, Y. (1999). The endogenous sulfated pentapeptide phytosulfokine-a stimulates tracheary element differentiation of isolated mesophyll cells of zinnia. Plant Physiol. 120, 1043–1048. doi: 10.1104/pp.120.4.1043
Mosher, S., Seybold, H., Rodriguez, P., Stahl, M., Davies, K. A., Dayaratne, S., et al. (2013). The tyrosine-sulfated peptide receptors PSKR1 and PSY1R modify the immunity of Arabidopsis to biotrophic and necrotrophic pathogens in an antagonistic manner. Plant J. 73, 469–482. doi: 10.1111/tpj.12050
Nagar, P., Kumar, A., Jain, M., Kumari, S., and Mustafiz, A. (2020). Genome-wide analysis and transcript profiling of PSKR gene family members in Oryza sativa. PloS One 15, e0236349. doi: 10.1371/journal.pone.0236349
Nagar, P., Sharma, N., Jain, M., Sharma, G., Prasad, M., and Mustafiz, A. (2022). OsPSKR15, a phytosulfokine receptor from rice enhances abscisic acid response and drought stress tolerance. Physiol. Plantarum 174, e13569. doi: 10.1111/ppl.13569
Reichardt, S., Piepho, H.-P., Stintzi, A., and Schaller, A. (2020). Peptide signaling for drought-induced tomato flower drop. Science 367, 1482–1485. doi: 10.1126/science.aaz5641
Rodiuc, N., Barlet, X., Hok, S., Perfus-Barbeoch, L., Allasia, V., Engler, G., et al. (2016). Evolutionarily distant pathogens require the Arabidopsis phytosulfokine signalling pathway to establish disease. Plant Cell Environ. 39, 1396–1407. doi: 10.1111/pce.12627
Sauter, M. (2015). Phytosulfokine peptide signalling. J. Exp. Bot. 66, 5161–5169. doi: 10.1093/jxb/erv071
Savicka, M. and Skute, N. (2010). Efects of high temperature on malondialdehyde content, superoxide production and growth changes in wheat seedlings (Triticum aestivum L.). Ekologija. 56, 26–33. doi: 10.2478/v10055-010-0004-x
Serrazina, S., Martinez, M. T., Cano, V., Malhó, R., Costa, R. L., and Corredoira, E. (2022). Genetic transformation of quercus ilex somatic embryos with a gnk2-like protein that reveals a putative anti-oomycete action. Plants 11, 304. doi: 10.3390/plants11030304
Srivastava, R., Liu, J. X., and Howell, S. H. (2008). Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. Plant J. 56, 219–227. doi: 10.1111/j.1365-313X.2008.03598.x
Stone, S. L. (2019). Role of the ubiquitin proteasome system in plant response to abiotic stress. Int. Rev. Cell Mol. Biol. 343, 65–110. doi: 10.1016/bs.ircmb.2018.05.012
Stuhrwohldt, N., Dahlke, R. I., Kutschmar, A., Peng, X., Sun, M. X., and Sauter, M. (2015). Phytosulfokine peptide signaling controls pollen tube growth and funicular pollen tube guidance in Arabidopsis thaliana. Physiol. Plant 153, 643–653. doi: 10.1111/ppl.12270
Stührwohldt, N., Dahlke, R. I., Steffens, B., Johnson, A., and Sauter, M. (2011). Phytosulfokine-a controls hypocotyl length and cell expansion in Arabidopsis thaliana through phytosulfokine receptor 1. PloS One 6, e21054. doi: 10.1371/journal.pone.0021054
Takahashi, F. and Shinozaki, K. (2019). Long-distance signaling in plant stress response. Curr. Opin. Plant Biol. 47, 106–111. doi: 10.1016/j.pbi.2018.10.006
Tang, Y., Chen, H., Deng, T., Chang, Y., Sun, K. T., Ditta, A., et al. (2022). Genome-wide identification and analysis of the GUB_WAK_bind gene family in Gossypium hirsutum. Mol. Biol. Rep. 49, 6405–6413. doi: 10.1007/s11033-022-07449-3
Wu, J. D., Jiang, C. P., Zhu, H. S., Jiang, H. Y., Cheng, B. J., and Zhu, S. W. (2015). Cloning and functional analysis of the promoter of a maize starch synthase III gene (ZmDULL1). Genet. Mol. Res. 14, 5468–5479. doi: 10.4238/2015
Yang, Y. Q. and Guo, Y. (2018). Elucidating the molecular mechanism smediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Yang, H., Matsubayashi, Y., Nakamura, K., and Sakagami, Y. (2001). Diversity of Arabidopsis genes encoding precursors for phytosulfokine, a peptide growth factor. Plant Physiol. 127, 842–851. doi: 10.1104/pp.010452
Yang, W., Zhang, B. G., Qi, G. H., Shang, L. Y., Liu, H. F., Ding, X., et al. (2019). Identification of the phytosulfokine receptor 1 (OsPSKR1) confers resistance to bacterial leaf streak in rice. Planta 250, 1603–1612. doi: 10.1007/s00425-019-03238-8
Yue, Y., Zhou, Y., Wang, J., and Ye, X. (2016). Assessing wheat frost risk with the support of gis: an approach coupling a growing season meteorological index and a hybrid fuzzy neural network model. Sustainability 8, 1308. doi: 10.3390/su8121308
Zhang, H., Hu, Z., Lei, C., Zheng, C. F., Wang, J., Shao, S. J., et al. (2018). A plant phytosulfokine peptide initiates auxin-dependent immunity through cytosolic ca2+ Signaling in tomato. Plant Cell 30, 652–667. doi: 10.1105/tpc.17.00537
Keywords: wheat, TaPSKR genes, abiotic stress, expression profile, qRT-PCR
Citation: Ma Z, Wei J, Zheng J, Su J and Zhao D (2025) Genome-wide identification of PSKR genes in wheat and differential expression under abiotic stress conditions. Front. Plant Sci. 16:1582433. doi: 10.3389/fpls.2025.1582433
Received: 24 February 2025; Accepted: 26 August 2025;
Published: 12 September 2025.
Edited by:
Zefeng Yang, Yangzhou University, ChinaReviewed by:
Cheng Liu, Shandong Academy of Agricultural Sciences, ChinaHouping Wang, Yunnan University, China
Copyright © 2025 Ma, Wei, Zheng, Su and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenbing Ma, bWF6aGVuYmluZzFAMTYzLmNvbQ==; Dong Zhao, MzMzMjE1MjNAcXEuY29t
†These authors share first authorship
 Zhenbing Ma
Zhenbing Ma Jingren Wei
Jingren Wei Jia Zheng
Jia Zheng Jian Su1
Jian Su1