- 1Department of Plant Sciences, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
- 2National Institute of Genomics and Advance Biotechnology, National Agriculture Research Centre, Islamabad, Pakistan
- 3Biology Department, College of Science, United Arab Emirates University, Al Ain, United Arab Emirates
- 4Chemistry Department, College of Science, United Arab Emirates University, Al Ain, United Arab Emirates
The dual challenges of climate change and population growth have intensified both biotic and abiotic stresses on crops resulting in disruptions of water dissipation patterns, lessen growth, yield, productivity and food security. Therefore, smart and sustainable agriculture practices for climate resilient and high yielding crops is the need of time. For this purpose, Innovation in biotechnological strategies is essential for sustainable agricultural development. Traditional breeding techniques have evolved through molecular approaches like marker-assisted selection (MAS) and quantitative trait loci (QTL) mapping, which accelerate the identification of trait-specific improvements. Mutational breeding, although effective in generating genetic diversity but lacks the precision, accuracy and effectiveness. Transgenic breeding allows for the transfer of beneficial genes across species, but recent advancements have shifted focus toward more refined approaches, such as RNA interference (RNAi) and genome editing tools like CRISPR-Cas9. These technologies enable precise, controlled genetic modifications to enhance traits like stress tolerance, disease resistance, and nutritional content. The integration of cutting-edge multi-omics platforms, including transcriptomics, proteomics, metabolomics combined with robust artificial intelligence (AI) based methods has revolutionizing crop genome elucidation. AI-driven analysis of large-scale biological data has revealed intricate genetic networks and regulatory pathways that underpin stress responses, growth, yield and genetics circuit patterns. These innovations in biotechnology from conventional breeding to advanced data-trait elucidation integrated methods are pushing the boundaries of climate resilient and next generation crop development. This review focused on the future of resilient and sustainable agriculture that lies in the convergence of conventional and molecular breeding, biotechnology approaches and AI’s driven strategies that enabling scientists to understand the genomics circuits of crops. These next generationally evolved crops bridging gaps from laboratory to field application with reduced reliance on chemical fertilizers, lessen yield gaps, climate resilience and promising nutritional enrichment. Such crops thrive under harsh environment paving the way for resilient and sustainable crop system development in constantly populating and warming ecosystem.
1 Introduction
Agriculture is at a crossroads, confronting unprecedented challenges posed by a rapidly growing global population and increasingly erratic climate patterns. Biotic and abiotic stressors including extreme temperature, drought, soil salinity and flooding now severely devastate crop yield, quality, and food security. These environmental disruptions compromise ecosystem stability and jeopardize life on Earth (Davis et al., 2021; Lesk et al., 2022). Addressing these multifaceted challenges necessitate transformative approach of advanced biotechnologies by integrating traditional practices with cutting-edge innovations in crop improvement and their real-world application can be proven by significantly field validation (Figure 1). While traditional breeding has historically underpinned agricultural advancement and are now getting apparent in era of climatic instability and escalating global food security. Climatic Implications and limitation are insisting the researchers for developing advanced genetics tool aimed at enhancing crop resilience, productivity, nutritional value and sustainable agriculture (Yu and Li, 2022). Classical breeding including marker-assisted selection (MAS) and quantitative trait loci (QTL) mapping enabling the precise identification and propagation of desirable traits (Figure 1). There are several crops with agronomic superiority for tolerance to different stress conditions based on QTLs. Surprisingly, Identified QTLs were failed in actual field research of versatile climatic pressure including barley for yield related characteristics (Genievskaya et al., 2025). This indicates the importance of counter check for laboratory-based breakthroughs and complementation with field trials. Emergence of mutagenesis breeding opened new possibilities to generate genetic diversity for improving stress tolerance in crops. The unexpected phenomenon of wheat yield reductions was observed from 10-28% while performing height and drought related function accordingly after treatment to gamma radiations. Mutagenesis accomplishments for trait characterization required repeatedly seasonal diversity, multiple geological location testing and critical legislatory framework along with molecular correlation (Ahumada-Flores et al., 2021). Transgenic approaches, particularly RNA interference (RNAi), have been instrumental in silencing deleterious genetic circuits but have shown compromised response in variable field trials. Survey of 5 years for RNAi edited rice grown in Asian temperate environment shown effective yield but failed in tropical region with 30-40% efficiency (Davidson and Mccray, 2011; Tardin-Coelho et al., 2025). Genome-editing technologies including zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and CRISPR-Cas9 systems have redefined the scope of crop genetic modification. Experimental to commercial application of CRISPR derived crops revealed challenges and promises including drought tolerance and sensitivity issue in various conditions conclusively highlighting epigenetic role (Wang et al., 2017). There are success stories for CRISPRCas9 and its variant based editing in Rice (DST, SPL10, NAC041 for salt tolerance and LCT1, HAK1, PRX2, NRAMP5, ARM1 for abiotic stress tolerance) (Rahman et al., 2022; Kumar et al., 2023), Maize (ARGOS8 for drought tolerance and PAP1 for flavone content improvement) (Rasheed et al., 2023; Mackon et al., 2023), Wheat (DEP1 and LOX2 for Nitrogen use efficiency) (Li et al., 2022a; Bharat et al., 2020) and tomatoes (AGL and CBF1 temperature and drought tolerance) (Wang et al., 2019; Zhao et al., 2021). CRISPR’s advancements i.e., base and prime editing paving way to unparalleled precision in correcting genetic sequences (Ahmar et al., 2024; Pacesa et al., 2024). Prime editing, a groundbreaking innovation and establishing a new frontiers in crop genetic engineering by targeted modifications without inducing double-stranded DNA breaks (Li et al., 2024). These tools have significantly accelerated the development of stress-resilient crops with enhanced yield potential and entered to legislation period in EU and US. The convergence of multi-omics technologies, synthetic biology and artificial intelligence (AI) heralds a new era in crop science. Integrating genomic, transcriptomic, proteomic and metabolomic enabling researchers to elucidate and enhance stress resilience at the molecular to field level (Lee et al., 2012). Biosynthetic engineering enabling reconstruct and optimize novel biosynthetic pathways to with improved traits, boosting both yield and nutritional content. Nitrogen fixation and use efficiency of cereal crops including maize can be enhance by genetic circuit optimization with soil microbiome (Wen et al., 2021). AI-driven predictive models are further refining these efforts and offering unprecedented insights into plant-environment interactions (Gao, 2021; Sun et al., 2022). Landmark investigation coupling artificial intelligence and machine learning models with phenomics data from various crops including rice, wheat and maize predicted accurate yield in variable climatic condition over the globe (Xu et al., 2022). In this review, we try to address the remarkable evolution of agriculture biotechnology with emphasis on conventional crop improvement to modern laboratory generated molecular analysis, integration of artificial intelligent multi-omics platforms and essential role of field validation in transforming data driven breakthroughs into sustainable development and solutions. The review predominantly highlights critical barriers for laboratory to farmer and field research results validation to better understand biotechnological potential of crops. The current era of potential development required to boost agriculture and resilient crop biology by artificial intelligence based cutting edge technology integration. Such integration would bring robust laboratory trials, field testing and creating continuous feedback mechanism between stakeholder and policy makers to implement agriculture governance. Rarely such advanced approaches are hope to make us withstand against extreme climate and develop true resilient crops to meet nutritional demand and food security of a growing 21st century.
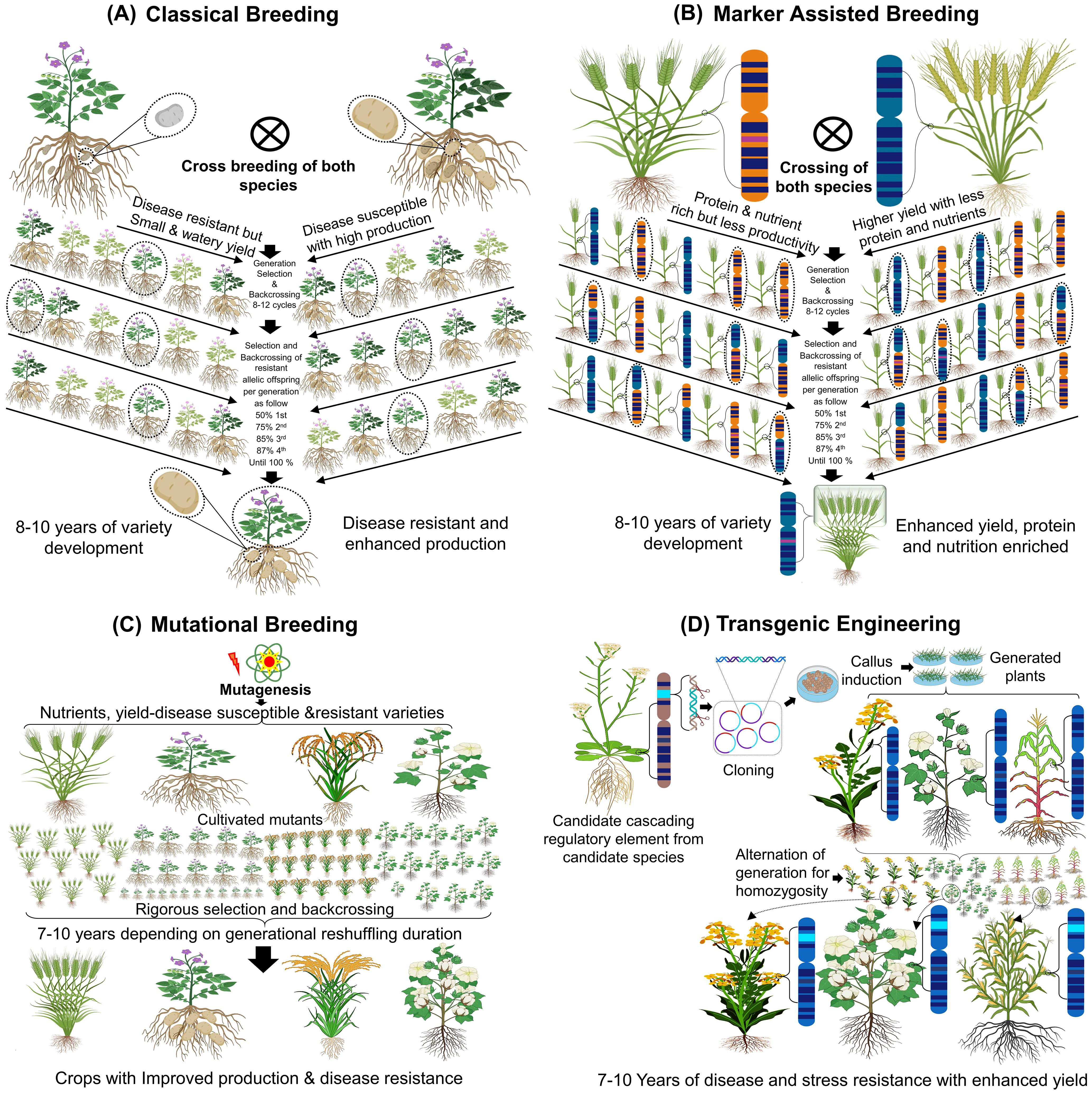
Figure 1. Stress and disease resilient high yielding crop development. (A) Selecting and cross-breeding of potato to improve fruit and disease traits through natural variation. (B) Molecular marker breeding to select plants with yield, nutrient and protein rich genetic traits. (C) Inducing genetic mutations by radioactive material for yield, nutrient and disease resistant trait. (D) Inserting foreign genes or manipulation of genes within the genome to develop nutrient rich, disease tolerant, high yielding and resilience crops.
2 Environmental cues limiting crop productivity
The ecosystem governed by competitive selection pressures, the challenges of survival, adaptation, and resource availability are naturally amplified. In the current era of overpopulation and erratic climate fluctuations, stress influencers such as extreme temperatures, drought, barren soils, and floods have drastically reduced both the yield and quality of vital food supplies with denatured soil structure. While traditional crop breeding methods have been instrumental in agricultural development, they are time-intensive and often insufficient to address the escalating demands for productivity, disease resistance, and biomass generation (Zhang et al., 2018). This highlights the pressing need to harness modern and safe biotechnological tools for developing stress-resistant and high-yielding crops. Despite significant advancements, achieving robust resilience to biotic and abiotic stresses remains a formidable challenge. The molecular and physiological responses of crops to stressors are intricate and multifaceted, often involving overlapping and interdependent pathways (Zhang et al., 2017, 2020). The simultaneous activation of different stress-response pathways can exacerbate declines in yield, quality, and crop texture, further complicating breeding efforts. Decoding these complex stress tolerance mechanisms requires a collaborative effort from plant breeders, genetic engineers, and molecular researchers. Emerging technologies, such as artificial intelligence (AI), machine learning, deep learning, and systems biology, are revolutionizing the analysis of vast datasets to uncover the genetic and molecular pathways underpinning stress resilience (Seleiman et al., 2021). These tools enable researchers to dissect plant physiological, molecular, and metabolic responses to stress, providing critical insights for sustaining and enhancing resilience traits in dynamic environments. Recent progress in molecular and evolutionary biology has yielded cultivars with enhanced tolerance to diverse biotic and abiotic stressors (Zhang et al., 2023). Innovative genome-editing technologies, such as CRISPR-Cas9 systems, have emerged as powerful tools for creating crops with superior resilience and improved productivity. These advancements have paved the way for engineering plants capable of withstanding multiple stressors, ensuring stable food production amid growing global challenges. Understanding the intricate molecular mechanisms of stress tolerance and leveraging biotechnological innovations hold immense potential for the future of agriculture. By integrating cutting-edge genomic tools with insights from systems biology, it is possible to cultivate resilient, high-quality crops that thrive under adverse environmental conditions (Lohani et al., 2022). Such innovations are crucial for safeguarding food security and meeting the nutritional demands of a rapidly expanding population.
3 Plant genetics and breeding approaches for productivity and environmental resilience
Classical breeding, also referred to as conventional or traditional breeding, involves the development of new cultivars by introducing desired traits, such as stress tolerance and high yield, through crossbreeding or hybridization (Figure 1) (Wuest et al., 2021). The domestication of plants via conventional breeding began in the early 20th century, spearheaded by Gregor Mendel, the father of classical genetics, who developed the first high-yielding and nutritious crop cultivars (Van Dijk et al., 2018). In the 1960s, N. Borlaug advanced this field by producing high-yielding wheat and rice varieties (Swaminathan, 2009). Classical breeding focuses on selecting specific traits or phenotypes, such as biotic and abiotic stress resilience or high yield, and linking these traits to genetic changes resulting from extensive crossing between closely related species over successive generations. Stabilizing these traits often requires 5–6 cycles of selfing, with the process of developing stable, high-performing cultivars taking 12–15 years (Gao, 2021; Husaini, 2022). While classical breeding has successfully produced disease-resistant and high-yielding crops such as potatoes (Sli gene elucidation (Eggers et al., 2021)), salt-tolerant wheat, and drought-tolerant barley (As explained in Figure 1). This limitation has prompted the development of advanced technologies to accelerate crop improvement.
3.1 Marker-assisted selection and quantitative trait loci for crop improvement
Marker-assisted selection (MAS) utilizes molecular markers linked to specific traits, such as disease resistance or stress tolerance, to enhance breeding efficiency. MAS integrates classical genetics with molecular biology, relying on phenotypic, biochemical, or DNA markers to select for specific traits (Su et al., 2019; Xu et al., 2022). It has been instrumental in improving complex traits such as stress tolerance and disease resistance, especially with the advent of high-throughput genotyping and association mapping. Quantitative trait loci (QTL) are genomic regions associated with phenotypic traits. High-throughput phenotyping technologies link genetic information to specific traits, aiding in the identification of QTLs governing stress tolerance, yield, or disease resistance (Tardieu and Tuberosa, 2010). Techniques such as linkage disequilibrium (LD) mapping and genome-wide association studies (GWAS) enable precise identification of single nucleotide polymorphisms (SNPs) associated with traits like heat stress tolerance (Younessi-Hamzekhanlu and Gailing, 2022). Combining MAS, QTL, and GWAS accelerates the development of resilient crop varieties (Scott et al., 2020; Luo et al., 2022).
3.2 Mutational breeding for better traits
Mutational breeding introduces genetic variations through chemical, physical, or biological mutagenesis. Techniques such as gamma or X-ray irradiation, chemical mutagens like ethyl methane sulfonate (EMS), and site-directed mutagenesis by Agrobacterium T-DNA transformation have been used to modify traits related to yield, disease resistance, and stress tolerance (Jankowicz-Cieslak et al., 2017). One prominent reverse genetics approach, Targeted Induced Local Lesions in Genomes (TILLING), identifies mutations in specific genes. Coupling TILLING with next-generation sequencing (NGS) has facilitated the discovery of allelic variations crucial for stress resilience (Toppino et al., 2022). Mutational breeding (Figure 1), has successfully produced crops like rice, tomato, cotton, and wheat with improved traits (Liu and Zhang, 2022).
3.3 Transgenic manipulation and engineering for quality traits
Transgenic engineering integrates molecular biology techniques to create crops with desired traits by introducing specific genes or genomic elements into the target plant. Advances in recombinant DNA technology, such as GATEWAY cloning, Gibson assembly, and seamless ligation-independent cloning, have enabled the precise manipulation of plant genomes (Furmanek-Blaszk et al., 2009). Agrobacterium-mediated transformation and biolistic methods are the most widely used techniques for crop genetic engineering. Agrobacterium tumefaciens delivers T-DNA constructs into the host genome, allowing for the stable expression of transgenes for traits like stress tolerance and higher yield (Hoekema et al., 1983). Biolistic transformation, or particle bombardment, directly introduces DNA into plant cells, bypassing genotypic barriers, and has been used to develop crops like wheat, maize, and rice with enhanced traits (Garvin et al., 2008; Zhi et al., 2022). Both methods have contributed to developing transgenic crops capable of thriving under adverse environmental conditions, ensuring higher productivity and resilience. The flexibility of these techniques allows the use of diverse regulatory elements, such as promoters and enhancers, to achieve trait-specific expression, further advancing crop improvement efforts.
4 RNA interference for controlling negative traits and crop improvement
RNA interference (RNAi) is a breakthrough mechanism for downregulating, silencing, upregulating, or controlling gene expression in a specific manner (Figure 2). This phenomenon was first identified by R. Jorgensen in 1990 while attempting to enhance the color of petunia flowers by introducing multiple copies of the chalcone synthase (Chls A) gene. Instead of producing dark purple flowers, the experiment yielded white and patchy flowers, a result of gene silencing at homologous, endogenous, and exogenous loci (Napoli et al., 1990). The pivotal discovery of RNAi as a molecular mechanism was made by Fire and Mello in 1998, who observed that double-stranded RNA (dsRNA) silenced homologous genes in C. elegans (Fire et al., 1998; Tabara et al., 1999). Their work earned them the Nobel Prize in Physiology or Medicine in 2006. They demonstrated that dsRNA triggered gene silencing via a sequence-specific mechanism, leading to translational inhibition of target mRNA (Devanapally et al., 2021). RNAi operates through sequence complementarity between dsRNA and the target mRNA. This specificity inhibits gene expression by cleaving mRNA into fragments, serving as templates for RNAi activity (Gu et al., 2012). The molecular mechanism comprised of Dicer-like Proteins (DCL), in which RNAse III enzymes process dsRNA into small interfering RNAs (siRNAs), typically 18–24 base pairs in length. RNA-Induced Silencing Complex (RISC) in which siRNAs guide this protein complex, containing Argonaute (AGO) proteins, to bind complementary mRNA sequences. The activated RISC-miRNA complex cleaves or represses the target mRNA, halting translation. Then, RNA-Dependent RNA Polymerase (RdRP) enzyme amplifies the silencing signal, ensuring sustained gene suppression. RNAi results in either post-transcriptional gene silencing (mRNA degradation) or transcriptional repression via chromatin rearrangements (Sontheimer, 2005; Wei et al., 2023). RNAi has revolutionized genetic engineering for developing stress-tolerant, high-yield, and disease-resistant crops. RNAi enables precise regulation of gene expression by introducing dsRNA complementary to specific target genes. The wide accepted benefits of RNAi for crop improvement includes, the development of heat, drought, salt resistant crops and Improved resistance to pathogens and pests. Metabolic pathway redirection by enhancing production of desired metabolites and improving crop quality. There are several methods for RNAi induction in Plants mainly include virus induced gene silencing (VIGS) that utilize viral vectors to introduce dsRNA for gene silencing. Agrobacterium-Mediated Transformation that deliver RNAi constructs into plant genomes via Agrobacterium tumefaciens. Direct Spray/Biolistic Bombardment involved direct application of RNAi molecules or constructs to plant tissues. These methods have enabled significant advancements in understanding gene function and enhancing crop resilience to environmental challenges (Chung et al., 2021; Lopez-Gomollon and Baulcombe, 2022).
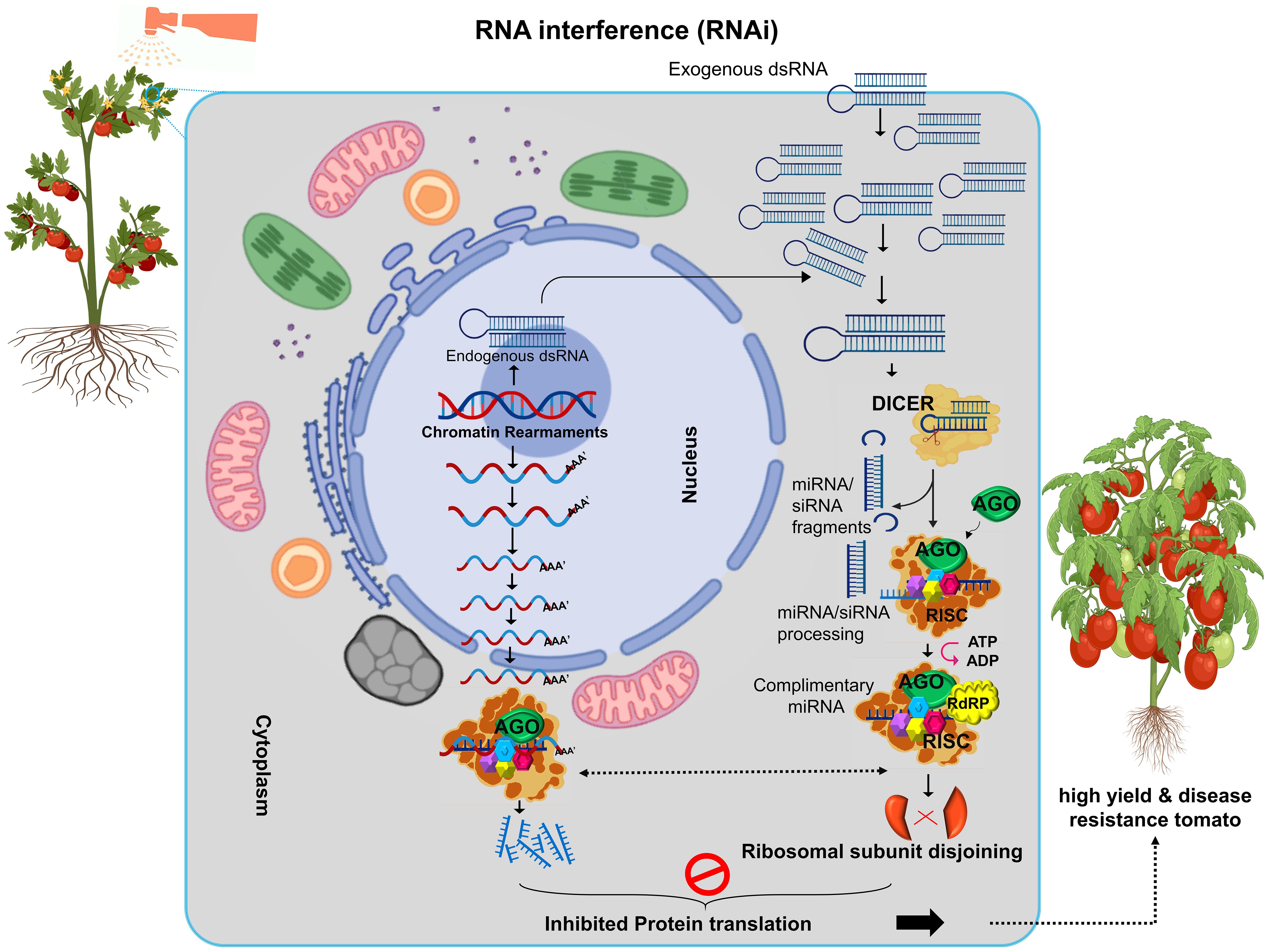
Figure 2. RNA interference (RNAi) technology used in crops to silence specific genes, enhancing resilience to both internal (endogenous) and external (exogenous) stresses. RNAi helps plants withstand environmental challenges like drought, heat and pests, making it a valuable tool for breeding stress-tolerant crops.
5 Genome editing revolutionizing crop resilience via (ZFNs), (TALENs) and (CRISPR)
Genome editing technologies (Figure 3), have gained wide spread acceptance for enhancing crop traits, disease resistance, food production, and environmental adaptability. These techniques, which date back to the 1980s, have evolved significantly over the years, with advancements continually improving precision and efficiency in molecular biology and crop development. Various genome manipulation techniques enabled gene insertion, deletion and overexpression to enhance crop traits for resilience and superior performance. There are several examples of CRISPR-Cas9 and its advanced variant-based editing of traits in rice, maize, wheat and other domestic to model crops (Table 1). These genetically modified organisms (GMOs) can exhibit improved traits but also raise concerns, such as potential allergenicity or microbial resistance due to genomic reshuffling (Rozas et al., 2022). The prominent genome editing technologies are zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeat (CRISPR) with various examples of varieties and gene trait testing are documented (Table 1). CRISPR stands out for its ability to induce desired traits without introducing foreign genetic material, making it more widely accepted. However, each technique has its advantages and limitations, contributing uniquely to developing customized crops with higher yields, stability, and tolerance to environmental and physiological stresses (Lee et al., 2016; Ricroch, 2019). ZFNs were introduced in the 1990s by Sangamo Biosciences which hold intellectual property rights (Scott, 2005). This technique relies on restriction enzymes composed of zinc finger DNA-binding domains and nonspecific cleavage motifs from FokI endonuclease. A single zinc finger unit recognizes 4–6 base pairs, with a pair recognizing up to 24 base pairs, creating double-stranded breaks (DSBs) through FokI dimerization. Crops like Arabidopsis thaliana and Zea mays have been edited using ZFNs, resulting in herbicide tolerance, enhanced yield, and resistance to biotic and abiotic stresses (Ahmar et al., 2020). However, ZFNs face limitations, including high costs, time-consuming development, and off-target mutations, reducing their accuracy and efficiency. These shortcomings prompted the development of newer genome editing technologies (Urnov et al., 2010). TALENs, discovered by D.F. Voytas, offer more accurate genome editing than ZFNs. These enzymes use transcription activator-like effector (TALE) proteins fused with FokI nucleases for DNA cleavage. Unlike ZFNs, TALENs can edit longer DNA sequences with greater specificity and reduced off-target effects (Li et al., 2022b). TALE proteins consist of a DNA-binding domain with tandem repeats of 34 amino acids, specifying target recognition. Nuclear localization signals. Activation domains for transcriptional activity. TALENs have been successfully used to edit rice (Oryza sativa), achieving biallelic modifications in a single generation. This method is preferred for its cost-effectiveness, adaptability, and ability to target DNA regions without protospacer adjacent motif (PAM) site restrictions (Yee, 2016; Iqbal et al., 2020).
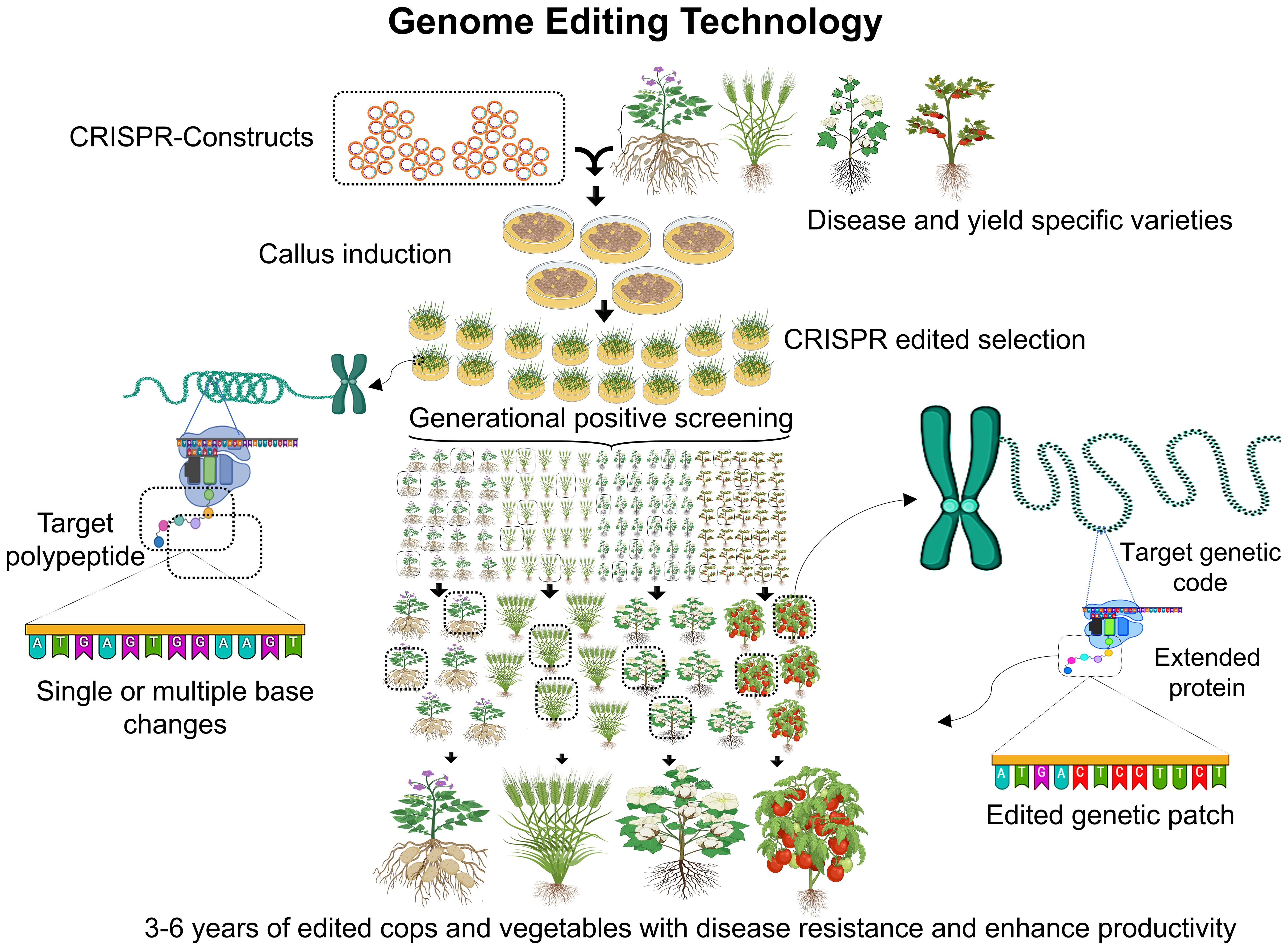
Figure 3. An illustration of CRISPR-based genome editing for resilient and targeted crop traits. The process begins with the creation of CRISPR constructs, followed by callus induction and the selection of CRISPR-edited plants. Generational positive screening ensures disease-resistant and high-yield crop varieties through precise genetic modifications, including single or multiple base changes and targeted genetic patching. The timeline for producing enhanced crops and vegetables with improved productivity and resistance ranges from 3 to 6 years.
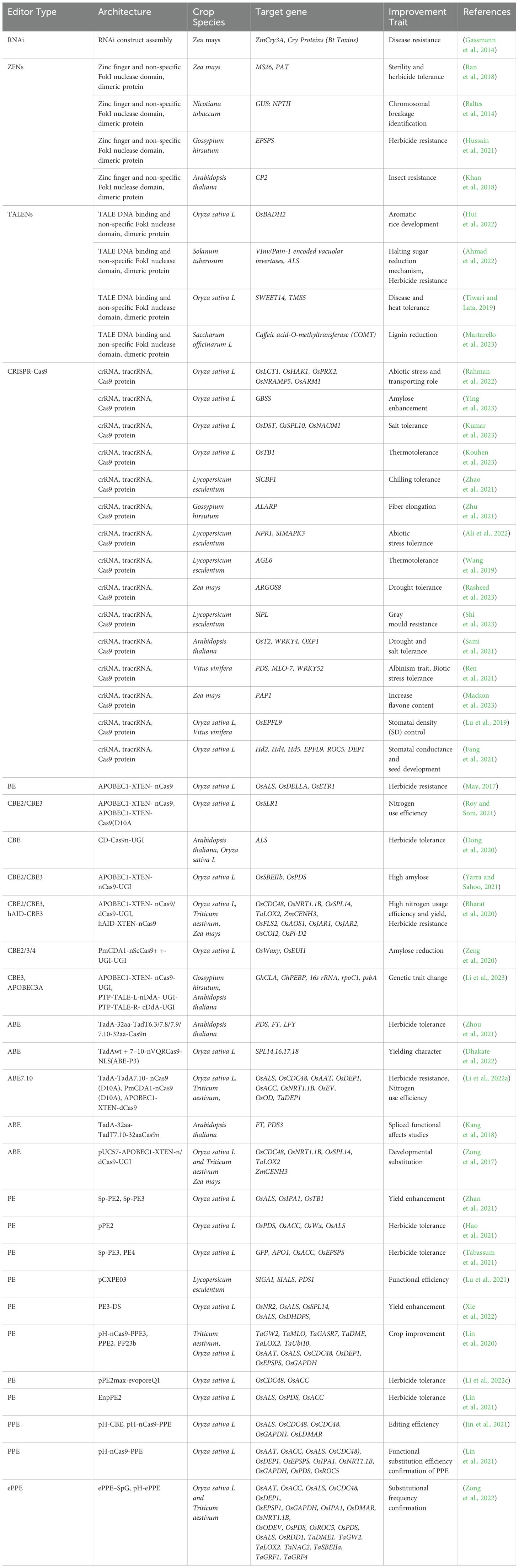
Table 1. Trait based improvement of different model and domestic crops by conventional and advanced genome editing methods.
CRISPR/Cas9, the latest and most widely accepted genome editing technique, revolutionized crop improvement due to its simplicity, precision, and affordability (Al-Attar et al., 2011). Unlike earlier methods, CRISPR does not rely on foreign DNA, using the organism’s native machinery to induce or modify gene expression (Ishii and Ishii, 2022). The CRISPR mechanism involves the guide RNA (gRNA), A sequence-specific RNA that directs the Cas9 protein to the target DNA. Cas9 Protein endonuclease introduces DSBs near the PAM sequence (NGG) (Pickar-Oliver and Gersbach, 2019). CRISPR is classified into two main classes in class I involves multiple effector proteins and II, relies on a single effector protein like Cas9, making it more efficient and widely used (Shmakov et al., 2017). Upon introducing DSBs, the cellular machinery repairs the breaks through three primary mechanisms, 1): non-homologous end joining (NHEJ), which is quick repair but prone to indels, causing frameshift mutations (Peterka et al., 2022), 2):Microhomology-mediated end joining (MMEJ) create deletions and chromosomal rearrangements, often independent of Ku proteins or DNA ligase 4 (Stinson et al., 2020). The third is homology directed repair (HDR), which is precise mechanism using homologous templates for error-free repair, primarily active during the S and G2 phases of the cell cycle (Chen et al., 2022). CRISPR has enabled breakthroughs in crop development, allowing researchers to create resistant, high-yield, and stress-tolerant crops. Despite its successes, some sub-mechanisms, like MMEJ, remain under investigation to further enhance its utility (Molla et al., 2021). Genome editing technologies continue to transform agricultural research and crop development. Each method including ZFNs, TALENs, and CRISPR, offers unique contributions to creating resilient and superior crops, addressing global food security challenges in an environmentally sustainable manner.
5.1 Base editing to avoid drastic genomic complexity
The improvement of agronomic traits in crops has been revolutionized by genome editing technologies, particularly through single nucleotide polymorphism (SNP) determination and confirmation (Gao, 2021). Substituting bases offers immense potential for introducing new crop varieties with enhanced traits. Base editing is a transformative molecular tool, enabling precise, programmed modifications to plant genomes (Zhu et al., 2020). Its rapid adoption and global acceptance stem from its precision, accuracy, and functional durability. Base editing achieves permanent and targeted single DNA or RNA base conversions without requiring double-strand breaks (DSBs) or repair mechanisms. This process involves DNA and RNA base editors, which utilize inactive CRISPR-Cas9 modules (dead Cas9, Cas9 nickase, or Cas9 variants) fused with cytosine and adenosine deaminases (Li et al., 2018; Pacesa et al., 2024). In developing new crop traits, base editors are categorized into cytosine base editors (CBEs) (Figure 4) and adenosine base editors (ABEs) (Figure 4). Cytosine base editors (CBEs) are formed by fusing cytidine deaminase with the inactive domain of CRISPR-Cas9. CBEs induce deamination, converting cytosine (C) to uracil (U), which is recognized as thymine (T) during DNA replication, thereby enabling C.G to T.A substitutions (Neugebauer et al., 2023). Adenosine base editors (ABEs) consist of Cas9 nickase, sgRNA, and transfer RNA (tRNA) adenosine deaminase (TadA), responsible for deaminating adenosine (A) to inosine (I). Inosine is interpreted as guanine (G) during DNA replication, allowing precise A.T to G.C conversions (Rees and Liu, 2018). There are number of examples for editing genes within the crops are reported in by various researchers (Table 1).
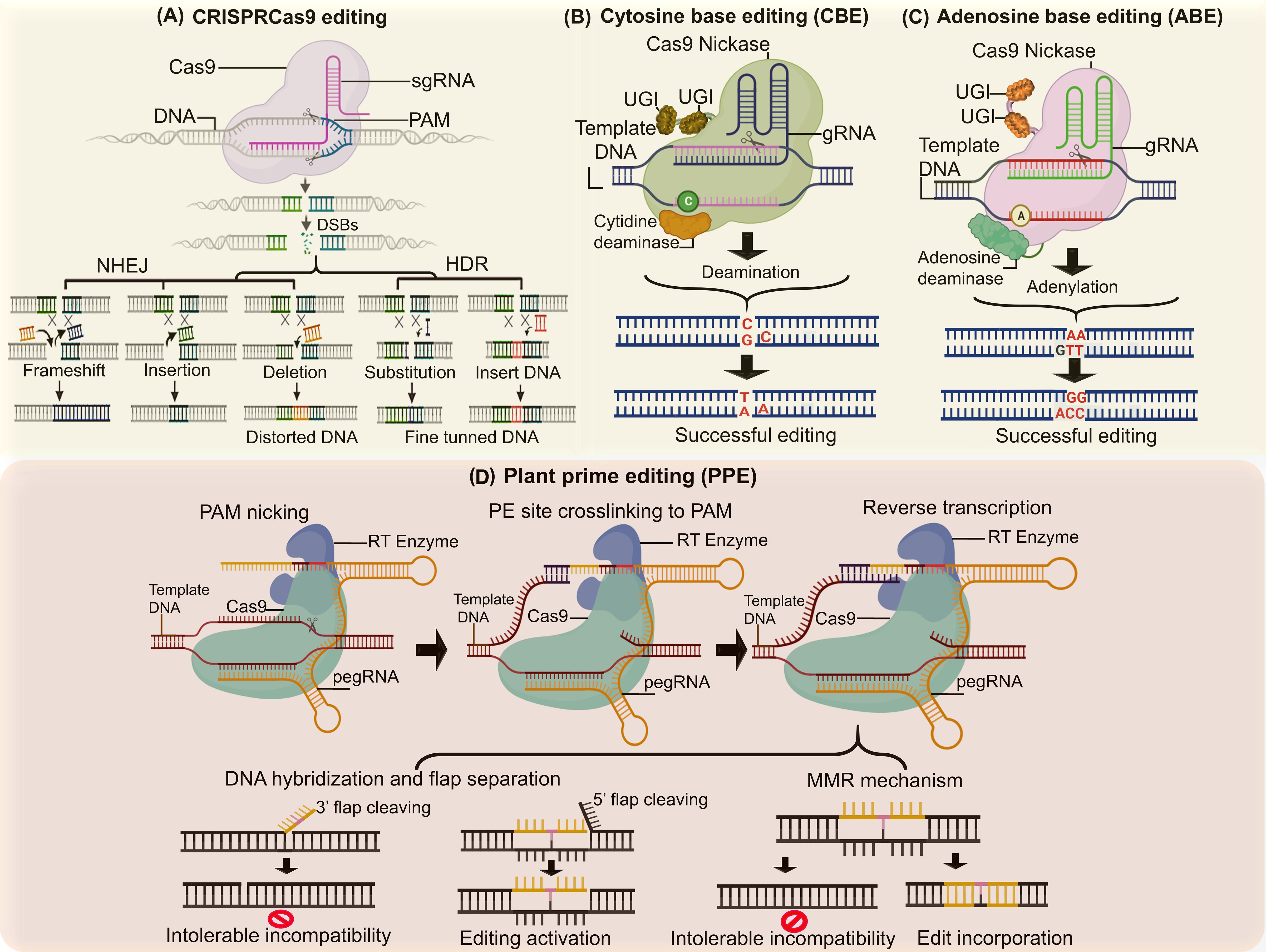
Figure 4. Base editing approaches spanning single base edits to multiple patches or ORF for targeted trait change. (A) CRISPR editing enables precise DNA cuts to add, remove or modify genes, facilitating targeted improvements in traits like disease resistance and yield. (B) Cytosine base editors convert cytosine (C) to thymine (T) in DNA without cuts, allowing precise alterations of genes linked to development and stress tolerance. (C) Adenosine base editors change adenine (A) to guanine (G) without cutting DNA, enabling specific modifications in traits like flowering time and nutrient efficiency. (D) Prime editors perform precise edits by inserting, deleting, or altering bases without double-strand breaks and facilitating complex trait enhancements in crops.
5.2 RNA editing to elucidate developmental perturbation in plants
RNA editing involves post-transcriptional modifications by altering nucleotides in mRNA to produce like plastids and mitochondria. RNA editing in coding sequences of mRNA is evolutionarily stable and functionally significant, often restoring gene functions lost due to mutations (Gommans et al., 2009). This natural repair mechanism, combined with the precision and reduced off-target effects of base editing, makes it a promising tool for developing stress-resilient crops under changing environmental conditions.
5.3 Prime editing and editors’ evolution as an additional strategy for smart crops
Prime editing (PE) is an advanced CRISPR-based genome editing tool (Figure 4), with higher accuracy and efficiency, coupled with minimal off-target effects that was firstly introduced by David R. Liu’s research group (Chen and Liu, 2023; Huang and Liu, 2023). PE employs a “search and replace” mechanism to modify target sites (Anzalone et al., 2019). It enables four base transitions (e.g., A→G, C→T) and eight transversions (e.g., G→C, A→T) and supports insertions and deletions of 80–40 base pairs (Nelson et al., 2022a). The major components of PE system comprise prime editing guide RNA (pegRNA) that identifies and guides nucleotide replacement. The fusion protein which consists of a Cas9 H840A nickase fused with murine leukemia virus (M-MLV) reverse transcriptase. Single guide RNA (sgRNA) directs Cas9 H840A nickase to nick the target DNA strand. Molecular mechanism of PE governs by Cas9 H840A nickase with an H840A substitution, inactivating the NHH domain to induce single-stranded nicks via the RuvC domain (Shao et al., 2018). In very next, M-MLV reverse transcriptase transcribes RNA templates into DNA. prime editing guide RNA (pegRNA) provides a primer-binding site (PBS) and a reverse transcriptase template, ensuring precise insertion, deletion, or base alteration without requiring donor templates or DSBs (Oh et al., 2022). In this way, mismatch repair mechanism (MMR) incorporates the new sequence into the genome (Zhu et al., 2020; Yan et al., 2024). Prime editors have garnered significant attention for their potential to repair genetic diseases, develop therapeutic compounds, and redirect plant development and signaling cascades, ultimately improving genome performance. However, prime editing faces limitations due to DNA mismatch repair (MMR), which can lead to insertions or deletions (indels), negatively impacting editing accuracy and efficiency (Ahmad et al., 2023). Despite these challenges, the editing efficiency of prime editors has driven their evolution toward customizable systems that enhance expression of genetic traits under various conditions (Tingting et al., 2023).
Several innovative prime editing systems, (Table 1) have been developed to improve functionality and minimize off-target effects. These systems, collectively referred to as prime plant editors (PPE) (Figure 4), include variants such as PE1-5, NPE, epegRNA, dual-pegRNA, TWIN-PE, enpPE2, and ePPE (Zeng et al., 2024). The first prime editor (PE1) linked wild-type moloney murine leukemia virus (M-MLV-RT) reverse transcriptase to the Cas9 H840A nickase at the C-terminal domain, achieving notable editing efficiency (Grünewald et al., 2023). PE2 system introduced five amino acid mutations in the M-MLV-RT enzyme, enhancing PE1 functionality and producing the Cas9-H840A-M-MLV-RT complex with improved editing efficiency (Zhao et al., 2023). The challenges of MMR were halted by PE3 which introduce an additional single guide RNA (sgRNA) to direct the Cas9 nickase to nick the original DNA strand near the editing site. Although this approach enhanced repair, it also increased off-target effects (Chen and Liu, 2023). The system PE4 was variant of PE2, that incorporated an additional plasmid encoding a dominant-negative MLH1, which suppressed endogenous MMR by knocking out MLH1, thus enhancing editing efficiency and reducing off-target effects (Truong et al., 2024). PE5 was then introduced relying on PE3 and PE4, that further minimized MMR-related issues and off-target effects while achieving higher editing efficiency (Park et al., 2024). NPE (Nuclease Prime Editor) relies on a Cas9 nuclease requiring only one pegRNA, enabling double-stranded DNA nicking with high stability and efficiency, unlike PE3, which uses a double-nick approach (Rahimi et al., 2024). Engineered pegRNA (epegRNA) method introduces structural modifications at the 3′ ends of pegRNA to reduce degradation and enhance editing precision. Dual-pegRNA: This approach employs NGG-pegRNA and CCN-pegRNA to simultaneously edit forward and reverse DNA strands, improving accuracy (Nelson et al., 2022). Twin Prime Editing (TWIN-PE) utilizes a single editing protein and two pegRNAs to directly replace double-stranded DNA, bypassing MMR mechanisms (Anzalone et al., 2022). Enhanced Plant Prime Editor 2 (enpPE2) uses composite promoters for pegRNA expression and adjustable editing architecture to improve efficiency in plants. Engineered system (ePPE) degrades the ribonuclease H (RNase H) domain and incorporates viral NC proteins, preventing pegRNA degradation and enhancing editing accuracy (Zong et al., 2022). Prime editing systems enable precise genomic modifications, including base substitutions, insertions, deletions, and inversions, with minimal off-target effects. These features make them ideal tools for trait development in horticultural crops and other plants. Zhong et al, demonstrated the stability, accuracy, and high efficiency of the ePPE strategy for prime editing in plants. However, prime editing is not without limitations. Challenges such as window size, target specificity, molecular influences from other proteins, selection pressure, and the sustainability of edited traits remain. Despite these hurdles, the potential of prime editing to develop agronomic crops with enhanced traits and value-added nutrients holds great promise for addressing global food insecurity (Zhao et al., 2023).
6 Multi omics, artificial intelligence and synthetic biology for resilient crops
The advent of synthetic biology has revolutionized genomic research and genetic engineering, offering unprecedented opportunities to manipulate biosynthetic pathways for producing essential compounds and regulating intricate biological processes. The integration of molecular components of networks and pathways, synthetic biology enables the reprogramming of cellular systems to address critical challenges in the development of agronomically relevant and stress-resilient crops (Figure 5; Goold et al., 2018). This interdisciplinary field bridges the gap between engineering and biological sciences, facilitating the modular design and rationalization of genetic devices and molecular frameworks. Such innovations aim to develop high-yielding, nutritionally enhanced crops that withstand biotic and abiotic stressors (Lu et al., 2009; Khalil and Collins, 2010). Synthetic biology provides robust tools to decode plant genomes and integrate traits for disease resistance and environmental resilience through multi-gene assembly within plant genomes. This technological advancement has enabled the engineering of complex gene circuits to introduce novel traits in horticultural crops, promoting enhanced productivity and stress tolerance (Yasmeen et al., 2023). Breakthroughs in cost-efficient DNA synthesis, coupled with the development of advanced genetic regulators, such as metabolites, transcription factors, and promoters, have catalyzed the production of crops with superior nutritional profiles, extended shelf life, and improved storage stability under climatic stress (Kaufmann et al., 2010; Li et al., 2024).
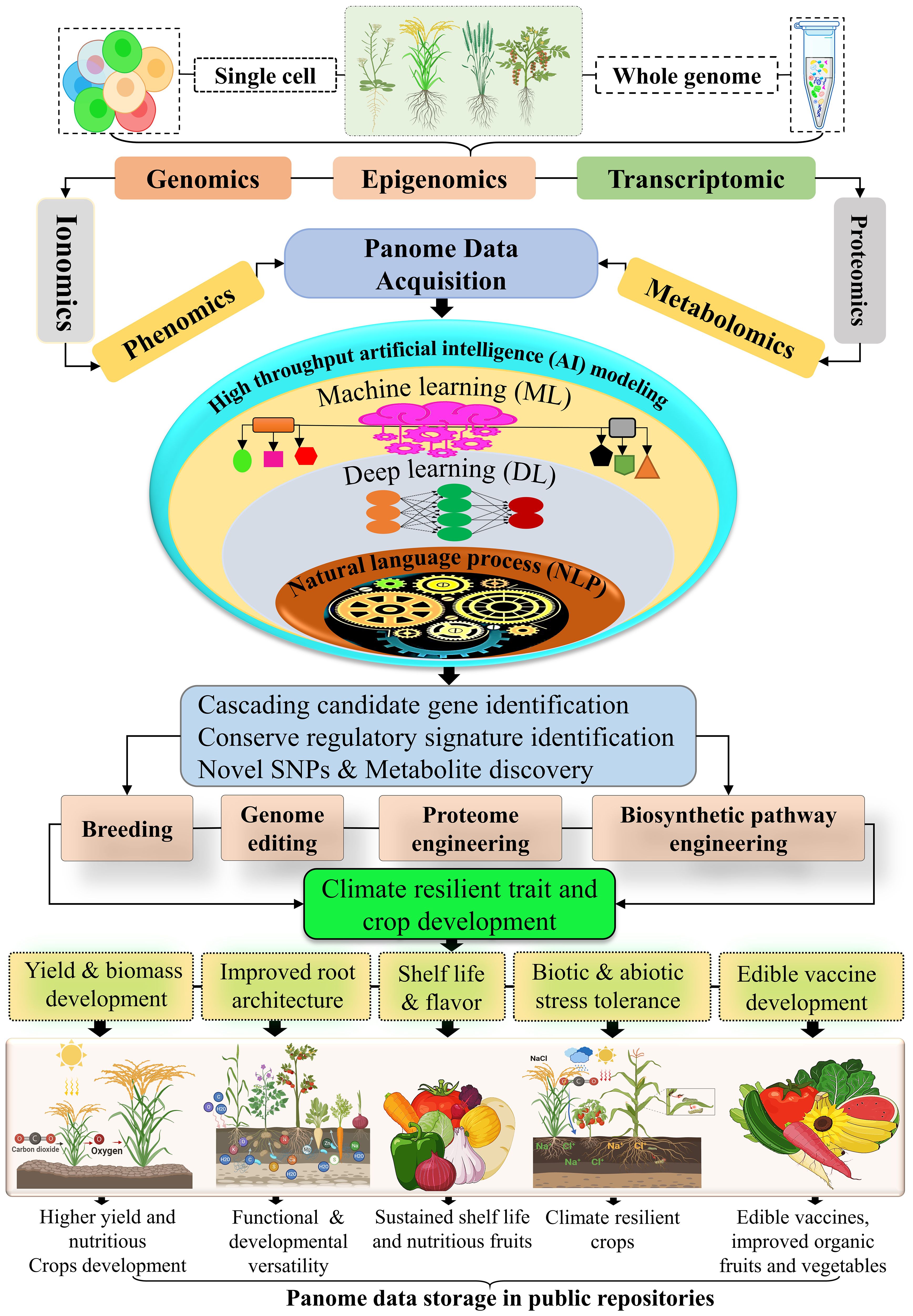
Figure 5. Multiomics integrating genomic, transcriptomic, proteomic, and metabolomic data driven AI and machine learning approaches are transforming crop resilience mechanisms. By analyzing whole-genome and single-cell data, researchers gain comprehensive insights into plant biology. These datasets are processed using advanced omics platforms to identify regulatory elements, cascading genes, SNPs, and metabolites linked to key traits. This information aids in developing climate- and stress-resilient crops with improved yields, taste, texture, shelf life, root efficiency and disease tolerance. High-quality data from these methods are shared in public repositories, supporting global food security and sustainable agriculture.
The integration of synthetic biology with machine learning and single-cell transcriptomics (scRNA) (Figure 5), has further optimized the utilization of metabolic pathways from stress-tolerant organisms like bacteria and algae, enhancing the ability of crops to adapt to environmental challenges and food insecurity. These advancements enable precision design of biological pathways and gene networks, unlocking new avenues for stress resilience, yield improvement, and sustainable agriculture (Long et al., 2022; Sha et al., 2024). High-throughput sequencing technologies, combined with machine learning algorithms, are shaping the future of precision agriculture, fostering global crop resilience (Vazquez-Vilar et al., 2016). Understanding plant stress tolerance mechanisms requires a multi-dimensional exploration at the genome level. High-throughput next-generation sequencing (NGS) approaches, encompassing genomics, epigenomics, transcriptomics, proteomics, metabolomics, and phenomics, have advanced the identification of stress-responsive genomic variants, including substitutions, insertions, deletions, and copy number variations (Kwoji et al., 2023). These multi-omics and AI-driven methodologies elucidate the intricate hierarchies and functional networks required for developing climate-resilient crops (Raza, 2020). Genomics involves deciphering genome-scale regulatory pathways, revealing the influence of intergenic and “dark matter” regions on stress responses (Purugganan and Jackson, 2021). Epigenomics highlights reversible modifications, such as DNA methylation and histone modification, that regulate gene expression without altering the DNA sequence, maintaining critical physiological processes like DNA repair under stress (Luo et al., 2020).
Transcriptomics focuses on RNA-seq analysis to monitor dynamic transcriptome changes under stress, unraveling the cascade of signals that drive defense mechanisms and physiological adaptations. Advanced RNA sequencing technologies facilitate genome-wide profiling of coding and non-coding RNAs, regulatory regions, and enhancers, contributing to a deeper understanding of molecular pathways involved in plant development (Zhou et al., 2022). Proteomics provides insight into cellular states during stress, detailing post-translational modifications, protein-protein interactions, and regulatory networks essential for stress adaptation and developmental processes (Derbyshire et al., 2022). Metabolomics investigates the diversity of primary and secondary metabolites, revealing their roles in stress resilience, including pathways like shikimate, acetate-malonate, and the TCA cycle, which synthesize compounds such as alkaloids, terpenoids, and phenolics. These metabolites, along with exogenous applications of compounds like proline, tryptophan, and glycine betaine, improve crop productivity and stress tolerance (Nowicka et al., 2018; Mohammadzadeh et al., 2022). Phenomics, driven by AI, captures high-throughput phenotypic data to link stress-induced molecular changes with observable traits. Advanced AI models enable precise phenotypic predictions, facilitating the breeding of resilient crops (Harfouche et al., 2023; Zavafer et al., 2023). Integration of scMulti-omics allows simultaneous quantification of genomic, transcriptomic, and proteomic data, enabling the dissection of complex molecular mechanisms underlying environmental resilience (Cao and Gao, 2022).
Lastly, ionomics explores the role of nutrient and ion homeostasis in stress adaptation. High-throughput AI-driven approaches identify biomarkers and cascading pathways involved in ion transport, metal homeostasis, and rhizosphere interactions, paving the way for genetic innovations in nutrient-efficient and stress-tolerant crop development (Xu et al., 2022). These synergistic advancements in synthetic biology, omics technologies, and artificial intelligence have the potential to revolutionize agricultural productivity, ensuring food security while addressing the challenges posed by climate change and environmental stresses. By enhancing the precision and scalability of crop improvement strategies, these technologies hold promise for developing resilient, high-yielding crops capable of thriving in the face of global agricultural challenges.
7 Conclusions and future perspective
Biotechnology is revolutionizing agriculture, offering transformative solutions to pressing global challenges like food security and climate resilience. Advances in gene editing, such as CRISPR, enable precise trait modifications, while multi-omics approaches and artificial intelligence (AI) are unlocking key regulatory networks and stress-tolerant traits. These innovations promise to deliver nutrient-rich, environmentally resilient and high-yield crops capable of thriving under extreme environmental conditions. However, significant challenges remain. Public skepticism, regulatory hurdles, and ecological uncertainties still hinder the widespread acceptance of gene-edited crops. Furthermore, the underrepresentation of diverse crop species and the limited exploration of biotechnological interactions with complex environments constrain global impact. Addressing these gaps requires a balanced approach that combines cutting-edge technologies with traditional agricultural practices and localized strategies. The future lies in integrating AI-driven analytics with multi-omics data to identify pivotal molecules and pathways essential for crop resilience. This synergy will accelerate crop development tailored to regional needs while fostering global food security. Equally important is engaging the public to build trust in biotechnology and ensuring innovations are accessible and sustainable. By addressing current limitations and fostering interdisciplinary collaboration, biotechnology has the potential to reshape agriculture, creating resilient, productive, and climate-adapted crops to meet the demands of a growing population. The innovative and environmentally conscious methods ensuring agricultural advancements to benefit both humanity and the planet are critical need of future.
Author contributions
MR: Conceptualization, Data curation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. EY: Data curation, Methodology, Resources, Visualization, Writing – original draft, Writing – review & editing. BS: Data curation, Investigation, Validation, Visualization, Writing – review & editing. MH: Formal analysis, Methodology, Resources, Validation, Writing – review & editing. MS: Formal analysis, Methodology, Writing – review & editing. RS: Data curation, Methodology, Software, Writing – review & editing. GA: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. JK: Investigation, Resources, Validation, Visualization, Writing – review & editing. MG: Conceptualization, Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The current research was financially supported by United Arab Emirate University (UAE) from SURE PLUS Research Program 2024, grant No.G00004708.
Acknowledgments
The current research was financially supported by United Arab Emirate University (UAE) from SURE PLUS Research Program 2024, grant No.G00004708.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. Generative AI was utilized solely for language refinement and proofreading of this manuscript, ensuring clarity and coherence while preserving the originality and integrity of the content. All scientific ideas, analyses and conclusions remain entirely by the author.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmad, D., Zhang, Z., Rasheed, H., Xu, X., and Bao, J. (2022). Recent advances in molecular improvement for potato tuber traits. Int. J. Mol. Sci. 23, 9982. doi: 10.3390/ijms23179982
Ahmad, N., Awan, M. J. A., and Mansoor, S. (2023). Improving editing efficiency of prime editor in plants. Trends Plant Sci. 28, 1–3. doi: 10.1016/j.tplants.2022.09.001
Ahmar, S., Saeed, S., Khan, M. H. U., Ullah Khan, S., Mora-Poblete, F., Kamran, M., et al. (2020). A revolution toward gene-editing technology and its application to crop improvement. Int. J. Mol. Sci. 21, 5665. doi: 10.3390/ijms21165665
Ahmar, S., Usman, B., Hensel, G., Jung, K.-H., and Gruszka, D. (2024). CRISPR enables sustainable cereal production for a greener future. Trends Plant Sci. 29, 179–195. doi: 10.1016/j.tplants.2023.10.016
Ahumada-Flores, S., Gómez Pando, L. R., Parra Cota, F. I., de la Cruz Torres, E., Sarsu, F., and De Los Santos Villalobos, S. (2021). Technical note: Gamma irradiation induces changes of phenotypic and agronomic traits in wheat (Triticum turgidum ssp. durum). Appl. Radiat. Isotopes 167, 109490. doi: 10.1016/j.apradiso.2020.109490
Al-Attar, S., Westra, E. R., van der Oost, J., and Brouns, S. J. (2011). Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. J. Biol. Chem. 392, 277–289. doi: 10.1515/bc.2011.042
Ali, Z., Rai, S., Jan, S., and Raina, K. (2022). An assessment on CRISPR Cas as a novel asset in mitigating drought stress. Genet. Resour. Crop Evol. 69, 2011–2027. doi: 10.1007/s10722-022-01364-z
Anzalone, A. V., Gao, X. D., Podracky, C. J., Nelson, A. T., Koblan, L. W., Raguram, A., et al. (2022). Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat. Biotechnol. 40, 731–740. doi: 10.1038/s41587-021-01133-w
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. doi: 10.1038/s41586-019-1711-4
Baltes, N. J., Gil-Humanes, J., Cermak, T., Atkins, P. A., and Voytas, D. F. (2014). DNA replicons for plant genome engineering. Plant Cell 26, 151–163. doi: 10.1105/tpc.113.119792
Bharat, S. S., Li, S., Li, J., Yan, L., and Xia, L. (2020). Base editing in plants: Current status and challenges. Crop J. 8, 384–395. doi: 10.1016/j.cj.2019.10.002
Cao, Z. J. and Gao, G. (2022). Multi-omics single-cell data integration and regulatory inference with graph-linked embedding. Nat. Biotechnol. 40, 1458–1466. doi: 10.1038/s41587-022-01284-4
Chen, J., Li, S., He, Y., Li, J., and Xia, L. (2022). An update on precision genome editing by homology-directed repair in plants. Plant Physiol. 188, 1780–1794. doi: 10.1093/plphys/kiac037
Chen, P. J. and Liu, D. R. (2023). Prime editing for precise and highly versatile genome manipulation. Nat. Rev. Genet. 24, 161–177. doi: 10.1038/s41576-022-00541-1
Chung, S. H., Feng, H., and Jander, G. (2021). Engineering pest tolerance through plant-mediated RNA interference. Curr. Opin. Plant Biol. 60, 102029. doi: 10.1016/j.pbi.2021.102029
Davidson, B. L. and Mccray, P. B. (2011). Current prospects for RNA interference-based therapies. Nat. Rev. Genet. 12, 329–340. doi: 10.1038/nrg2968
Davis, K. F., Downs, S., and Gephart, J. A. (2021). Towards food supply chain resilience to environmental shocks. Nat. Food 2, 54–65. doi: 10.1038/s43016-020-00196-3
Derbyshire, M. C., Batley, J., and Edwards, D. (2022). Use of multiple ‘omics techniques to accelerate the breeding of abiotic stress tolerant crops. Curr. Plant Biol. 32 (44), 100262. doi: 10.1016/j.cpb.2022.100262
Devanapally, S., Raman, P., Chey, M., Allgood, S., Ettefa, F., Diop, M., et al. (2021). Mating can initiate stable RNA silencing that overcomes epigenetic recovery. Nat. Commun. 12, 4239. doi: 10.1038/s41467-021-24053-4
Dhakate, P., Sehgal, D., Vaishnavi, S., Chandra, A., Singh, A., Raina, S. N., et al. (2022). Comprehending the evolution of gene editing platforms for crop trait improvement. Front. Genet. 13, 876987. doi: 10.3389/fgene.2022.876987
Dong, H., Wang, D., Bai, Z., Yuan, Y., Yang, W., Zhang, Y., et al. (2020). Generation of imidazolinone herbicide resistant trait in Arabidopsis. PloS One 15, e0233503. doi: 10.1371/journal.pone.0233503
Eggers, E.-J., van der Burgt, A., Van Heusden, S., De Vries, M. E., Visser, R. G. F., Bachem, C. W. B., et al. (2021). Neofunctionalisation of the Sli gene leads to self-compatibility and facilitates precision breeding in potato. Nat. Commun. 12, 4141. doi: 10.1038/s41467-021-24267-6
Fang, J., Guo, T., Xie, Z., Chun, Y., Zhao, J., Peng, L., et al. (2021). The URL1–ROC5–TPL2 transcriptional repressor complex represses the ACL1 gene to modulate leaf rolling in rice. Plant Physiol. 185, 1722–1744. doi: 10.1093/plphys/kiaa121
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi: 10.1038/35888
Furmanek-Blaszk, B., Boratynski, R., Zolcinska, N., and Sektas, M. (2009). M1. MboII and M2. MboII type IIS methyltransferases: different specificities, the same target. Microbiology 155, 1111–1121. doi: 10.1099/mic.0.025023-0
Gao, C. (2021). Genome engineering for crop improvement and future agriculture. Cell 184, 1621–1635. doi: 10.1016/j.cell.2021.01.005
Garvin, D. F., Gu, Y. Q., Hasterok, R., Hazen, S. P., Jenkins, G., Mockler, T. C., et al. (2008). Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Crop Sci. 48, S–69-S-84. doi: 10.2135/cropsci2007.06.0332tpg
Gassmann, A. J., Petzold-Maxwell, J. L., Clifton, E. H., Dunbar, M. W., Hoffmann, A. M., Ingber, D. A., et al. (2014). Field-evolved resistance by western corn rootworm to multiple Bacillus thuringiensis toxins in transgenic maize. Proc. Natl. Acad. Sci. U.S.A. 111, 5141–5146. doi: 10.1073/pnas.1317179111
Genievskaya, Y., Abugalieva, S., and Turuspekov, Y. (2025). Identification of QTLs associated with grain yield-related traits of spring barley. BMC Plant Biol. 25, 554. doi: 10.1186/s12870-025-06588-6
Gommans, W. M., Mullen, S. P., and Maas, S. (2009). RNA editing: a driving force for adaptive evolution? Bioessays 31, 1137–1145. doi: 10.1002/bies.200900045
Goold, H., Wright, P., and Hailstones, D. (2018). Emerging opportunities for synthetic biology in agriculture. Genes 9, 341. doi: 10.3390/genes9070341
Grünewald, J., Miller, B. R., Szalay, R. N., Cabeceiras, P. K., Woodilla, C. J., Holtz, E. J. B., et al. (2023). Engineered CRISPR prime editors with compact, untethered reverse transcriptases. Nat. Biotechnol. 41, 337–343. doi: 10.1038/s41587-022-01473-1
Gu, S. G., Pak, J., Guang, S., Maniar, J. M., Kennedy, S., and Fire, A. (2012). Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat. Genet. 44, 157–164. doi: 10.1038/ng.1039
Hao, L., Pu, X., and Song, J. (2021). Introduction of mutations in plants with prime editing. Methods 194, 83–93. doi: 10.1016/j.ymeth.2021.03.014
Harfouche, A. L., Nakhle, F., Harfouche, A. H., Sardella, O. G., Dart, E., and Jacobson, D. (2023). A primer on artificial intelligence in plant digital phenomics: embarking on the data to insights journey. Trends Plant Sci. 28, 154–184. doi: 10.1016/j.tplants.2022.08.021
Hoekema, A., Hirsch, P. R., Hooykaas, P. J., and Schilperoort, R. A. (1983). A binary plant vector strategy based on separation of vir-and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303, 179–180. doi: 10.1038/303179a0
Huang, Z. and Liu, G. (2023). Current advancement in the application of prime editing. Front. Bioeng. Biotechnol. 11, 1039315. doi: 10.3389/fbioe.2023.1039315
Hui, S., Li, H., Mawia, A. M., Zhou, L., Cai, J., Ahmad, S., et al. (2022). Production of aromatic three-line hybrid rice using novel alleles of BADH2. Plant Biotechnol. J. 20, 59–74. doi: 10.1111/pbi.13695
Husaini, A. M. (2022). High-value pleiotropic genes for developing multiple stress-tolerant biofortified crops for 21st-century challenges. Heredity 128, 460–472. doi: 10.1038/s41437-022-00500-w
Hussain, A., Ding, X., Alariqi, M., Manghwar, H., Hui, F., Li, Y., et al. (2021). Herbicide resistance: another hot agronomic trait for plant genome editing. Plants 10, 621. doi: 10.3390/plants10040621
Iqbal, Z., Iqbal, M. S., Ahmad, A., Memon, A. G., and Ansari, M. I. (2020). New prospects on the horizon: genome editing to engineer plants for desirable traits. Curr. Plant Biol. 24, 100171. doi: 10.1016/j.cpb.2020.100171
Ishii, M. and Ishii, T. (2022). Proving that a genome-edited organism is not GMO. Trends Biotechnol. 40, 525–528. doi: 10.1016/j.tibtech.2021.11.001
Jankowicz-Cieslak, J., Mba, C., and Till, B. J. (2017). “Mutagenesis for crop breeding and functional genomics,” in Biotechnologies for plant mutation breeding (Springer, Cham), 3–18.
Jin, S., Lin, Q., Luo, Y., Zhu, Z., Liu, G., Li, Y., et al. (2021). Genome-wide specificity of prime editors in plants. Nat. Biotechnol. 39, 1292–1299. doi: 10.1038/s41587-021-00891-x
Kang, B.-C., Yun, J.-Y., Kim, S.-T., Shin, Y., Ryu, J., Choi, M., et al. (2018). Precision genome engineering through adenine base editing in plants. Nat. Plants 4, 427–431. doi: 10.1038/s41477-018-0178-x
Kaufmann, K., Pajoro, A., and Angenent, G. C. (2010). Regulation of transcription in plants: mechanisms controlling developmental switches. Nat. Rev. Genet. 11, 830–842. doi: 10.1038/nrg2885
Khalil, A. S. and Collins, J. J. (2010). Synthetic biology: applications come of age. Nat. Rev. Genet. 11, 367. doi: 10.1038/nrg2775
Khan, Z., Khan, S. H., Mubarik, M. S., and Ahmad, A. (2018). Targeted genome editing for cotton improvement. Past Present Future Trends Cotton Breed. 11. doi: 10.5772/intechopen.73600
Kouhen, M., García-Caparrós, P., Twyman, R. M., Abdelly, C., Mahmoudi, H., Schillberg, S., et al. (2023). Improving environmental stress resilience in crops by genome editing: insights from extremophile plants. Crit. Rev. Biotechnol. 43, 559–574. doi: 10.1080/07388551.2022.2042481
Kumar, M., Prusty, M. R., Pandey, M. K., Singh, P. K., Bohra, A., Guo, B., et al. (2023). Application of CRISPR/Cas9-mediated gene editing for abiotic stress management in crop plants. Front. Plant Sci. 14, 1157678. doi: 10.3389/fpls.2023.1157678
Kwoji, I. D., Aiyegoro, O. A., Okpeku, M., and Adeleke, M. A. (2023). ‘Multi-omics’ data integration: applications in probiotics studies. NPJ Sci. Food 7, 25. doi: 10.1038/s41538-023-00199-x
Lee, J., Chung, J. H., Kim, H. M., Kim, D. W., and Kim, H. (2016). Designed nucleases for targeted genome editing. Plant Biotechnol. J. 14, 448–462. doi: 10.1111/pbi.2016.14.issue-2
Lee, J. W., Na, D., Park, J. M., Lee, J., Choi, S., and Lee, S. Y. (2012). Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat. Chem. Biol. 8, 536–546. doi: 10.1038/nchembio.970
Lesk, C., Anderson, W., Rigden, A., Coast, O., Jägermeyr, J., Mcdermid, S., et al. (2022). Compound heat and moisture extreme impacts on global crop yields under climate change. Nat. Rev. Earth Environ. 3, 872–889. doi: 10.1038/s43017-022-00368-8
Li, J., Chen, L., Liang, J., Xu, R., Jiang, Y., Li, Y., et al. (2022c). Development of a highly efficient prime editor 2 system in plants. Genome Biol. 23, 161. doi: 10.1186/s13059-022-02730-x
Li, Q., Duncan, S., Li, Y., Huang, S., and Luo, M. (2024). Decoding plant specialized metabolism: new mechanistic insights. Trends Plant Sci. 29, 535–545. doi: 10.1016/j.tplants.2023.11.015
Li, A., Hao, C., Wang, Z., Geng, S., Jia, M., Wang, F., et al. (2022a). Wheat breeding history reveals synergistic selection of pleiotropic genomic sites for plant architecture and grain yield. Mol. Plant 15, 504–519. doi: 10.1016/j.molp.2022.01.004
Li, J., Zhang, C., He, Y., Li, S., Yan, L., Li, Y., et al. (2023). Plant base editing and prime editing: The current status and future perspectives. J. Integr. Plant Biol. 65, 444–467. doi: 10.1111/jipb.13425
Li, H., Zhu, Z., Li, S., Li, J., Yan, L., Zhang, C., et al. (2022b). Multiplex precision gene editing by a surrogate prime editor in rice. Mol. Plant 15, 1077–1080. doi: 10.1016/j.molp.2022.05.009
Li, C., Zong, Y., Wang, Y., Jin, S., Zhang, D., Song, Q., et al. (2018). Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 19, 59. doi: 10.1186/s13059-018-1443-z
Lin, Q., Jin, S., Zong, Y., Yu, H., Zhu, Z., Liu, G., et al. (2021). High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 39, 923–927. doi: 10.1038/s41587-021-00868-w
Lin, Q., Zong, Y., Xue, C., Wang, S., Jin, S., Zhu, Z., et al. (2020). Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585. doi: 10.1038/s41587-020-0455-x
Liu, H. and Zhang, J. (2022). Is the mutation rate lower in genomic regions of stronger selective constraints? Mol. Biol. Evol. 39, msac169. doi: 10.1093/molbev/msac169
Lohani, N., Singh, M. B., and Bhalla, P. L. (2022). Biological parts for engineering abiotic stress tolerance in plants. BioDesign Res. 2022, 1–41. doi: 10.34133/2022/9819314
Long, B., Fischer, B., Zeng, Y., Amerigian, Z., Li, Q., Bryant, H., et al. (2022). Machine learning-informed and synthetic biology-enabled semi-continuous algal cultivation to unleash renewable fuel productivity. Nat. Commun. 13, 541. doi: 10.1038/s41467-021-27665-y
Lopez-Gomollon, S. and Baulcombe, D. C. (2022). Roles of RNA silencing in viral and non-viral plant immunity and in the crosstalk between disease resistance systems. Nat. Rev. Mol. Cell Biol. 23, 645–662. doi: 10.1038/s41580-022-00496-5
Lu, J., He, J., Zhou, X., Zhong, J., Li, J., and Liang, Y.-K. (2019). Homologous genes of epidermal patterning factor regulate stomatal development in rice. J. Plant Physiol. 234, 18–27. doi: 10.1016/j.jplph.2019.01.010
Lu, T. K., Khalil, A. S., and Collins, J. J. (2009). Next-generation synthetic gene networks. Nat. Biotechnol. 27, 1139. doi: 10.1038/nbt.1591
Lu, Y., Tian, Y., Shen, R., Yao, Q., Zhong, D., Zhang, X., et al. (2021). Precise genome modification in tomato using an improved prime editing system. Plant Biotechnol. J. 19, 415–417. doi: 10.1111/pbi.13497
Luo, C., Fernie, A. R., and Yan, J. (2020). Single-cell genomics and epigenomics: technologies and applications in plants. Trends Plant Sci. 25, 1030–1040. doi: 10.1016/j.tplants.2020.04.016
Luo, H., Hu, L., Brito, L. F., Dou, J., Sammad, A., Chang, Y., et al. (2022). Weighted single-step GWAS and RNA sequencing reveals key candidate genes associated with physiological indicators of heat stress in Holstein cattle. J. Anim. Sci. Biotechnol. 13, 1–13. doi: 10.1186/s40104-022-00748-6
Mackon, E., Mackon, G.C.J.D.E., Guo, Y., Ma, Y., Yao, Y., and Liu, P. (2023). Development and application of CRISPR/Cas9 to improve anthocyanin pigmentation in plants: opportunities and perspectives. Plant Sci. 333, 111746. doi: 10.1016/j.plantsci.2023.111746
Martarello, D. C., Almeida, A. M., Sinzker, R. C., Oliveira, D. M., Marchiosi, R., Dos Santos, W. D., et al. (2023). The known unknowns in lignin biosynthesis and its engineering to improve lignocellulosic saccharification efficiency. Biomass Convers. Biorefin. 13, 2497–2515. doi: 10.1007/s13399-021-01291-6
Mohammadzadeh, V., Barani, M., Amiri, M. S., Yazdi, M. E. T., Hassanisaadi, M., Rahdar, A., et al. (2022). Applications of plant-based nanoparticles in nanomedicine: A review. Sustain. Chem. Pharm. 25, 100606. doi: 10.1016/j.scp.2022.100606
Molla, K. A., Sretenovic, S., Bansal, K. C., and Qi, Y. (2021). Precise plant genome editing using base editors and prime editors. Nat. Plants 7, 1166–1187. doi: 10.1038/s41477-021-00991-1
Napoli, C., Lemieux, C., and Jorgensen, R. (1990). Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell 2, 279–289. doi: 10.2307/3869076
Nelson, J. W., Randolph, P. B., Shen, S. P., Everette, K. A., Chen, P. J., Anzalone, A. V., et al. (2022). Engineered pegRNAs improve prime editing efficiency. Nat. Biotechnol. 40, 402–410. doi: 10.1038/s41587-021-01039-7
Neugebauer, M. E., Hsu, A., Arbab, M., Krasnow, N. A., Mcelroy, A. N., Pandey, S., et al. (2023). Evolution of an adenine base editor into a small, efficient cytosine base editor with low off-target activity. Nat. Biotechnol. 41, 673–685. doi: 10.1038/s41587-022-01533-6
Nowicka, B., Ciura, J., Szymańska, R., and Kruk, J. (2018). Improving photosynthesis, plant productivity and abiotic stress tolerance–current trends and future perspectives. J. Plant Physiol. 231, 415–433. doi: 10.1016/j.jplph.2018.10.022
Oh, Y., Lee, W.-J., Hur, J. K., Song, W. J., Lee, Y., Kim, H., et al. (2022). Expansion of the prime editing modality with Cas9 from Francisella novicida. Genome Biol. 23, 92. doi: 10.1186/s13059-022-02644-8
Pacesa, M., Pelea, O., and Jinek, M. (2024). Past, present, and future of CRISPR genome editing technologies. Cell 187, 1076–1100. doi: 10.1016/j.cell.2024.01.042
Park, J.-C., Kim, Y.-J., Hwang, G.-H., Kang, C. Y., Bae, S., and Cha, H.-J. (2024). Enhancing genome editing in hPSCs through dual inhibition of DNA damage response and repair pathways. Nat. Commun. 15, 4002. doi: 10.1038/s41467-024-48111-9
Peterka, M., Akrap, N., Li, S., Wimberger, S., Hsieh, P.-P., Degtev, D., et al. (2022). Harnessing DSB repair to promote efficient homology-dependent and-independent prime editing. Nat. Commun. 13, 1–9. doi: 10.1038/s41467-022-28771-1
Pickar-Oliver, A. and Gersbach, C. A. (2019). The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 20, 490–507. doi: 10.1038/s41580-019-0131-5
Purugganan, M. D. and Jackson, S. A. (2021). Advancing crop genomics from lab to field. Nat. Genet. 53, 595–601. doi: 10.1038/s41588-021-00866-3
Rahimi, S., Balusamy, S. R., Perumalsamy, H., Ståhlberg, A., and Mijakovic, I. (2024). CRISPR-Cas target recognition for sensing viral and cancer biomarkers. Nucleic Acids Res. 52, 10040–10067. doi: 10.1093/nar/gkae736
Rahman, M.-U., Zulfiqar, S., Raza, M. A., Ahmad, N., and Zhang, B. (2022). Engineering abiotic stress tolerance in crop plants through CRISPR genome editing. Cells 11, 3590. doi: 10.3390/cells11223590
Ran, Y., Patron, N., Kay, P., Wong, D., Buchanan, M., Cao, Y. Y., et al. (2018). Zinc finger nuclease-mediated precision genome editing of an endogenous gene in hexaploid bread wheat (Triticum aestivum) using a DNA repair template. Plant Biotechnol. J. 16, 2088–2101. doi: 10.1111/pbi.2018.16.issue-12
Rasheed, A., Jie, H., Ali, B., He, P., Zhao, L., Ma, Y., et al. (2023). Breeding drought-tolerant maize (Zea mays) using molecular breeding tools: recent advancements and future prospective. Agronomy 13, 1459. doi: 10.3390/agronomy13061459
Raza, A. (2020). Metabolomics: a systems biology approach for enhancing heat stress tolerance in plants. Plant Cell Rep. 41, 1–23. doi: 10.1007/s00299-020-02635-8
Rees, H. A. and Liu, D. R. (2018). Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788. doi: 10.1038/s41576-018-0059-1
Ren, C., Liu, Y., Guo, Y., Duan, W., Fan, P., Li, S., et al (2021). Optimizing the CRISPR/Cas9 system for genome editing in grape by using grape promoters. Horticulture Research. 8 (1), 52. doi: 10.1038/s41438-021-00489-z
Ricroch, A. (2019). Global developments of genome editing in agriculture. Transgenic Research. 28 (Suppl 2), 45–52. doi: 10.1007/s11248-019-00133-6
Roy, S. and Soni, P. (2021). Genome editing for biofortification of rice: current implications and future aspects. Genome Eng. Crop Improv., 297–313. doi: 10.1002/9781119672425.ch17
Rozas, P., Kessi-Pérez, E. I., and Martínez, C. (2022). Genetically modified organisms: adapting regulatory frameworks for evolving genome editing technologies. Biol. Res. 55, 31. doi: 10.1186/s40659-022-00399-x
Sami, A., Xue, Z., Tazein, S., Arshad, A., He Zhu, Z., Ping Chen, Y., et al. (2021). CRISPR–Cas9-based genetic engineering for crop improvement under drought stress. Bioengineered 12, 5814–5829. doi: 10.1080/21655979.2021.1969831
Scott, C. T. (2005). The zinc finger nuclease monopoly. Nat. Biotechnol. 23, 915–919. doi: 10.1038/nbt0805-915
Scott, M. F., Ladejobi, O., Amer, S., Bentley, A. R., Biernaskie, J., Boden, S. A., et al. (2020). Multi-parent populations in crops: a toolbox integrating genomics and genetic mapping with breeding. Heredity 125, 396–416. doi: 10.1038/s41437-020-0336-6
Seleiman, M. F., Al-Suhaibani, N., Ali, N., Akmal, M., Alotaibi, M., Refay, Y., et al. (2021). Drought stress impacts on plants and different approaches to alleviate its adverse effects. Plants 10, 259. doi: 10.3390/plants10020259
Sha, Y., Qiu, Y., Zhou, P., and Nie, Q. (2024). Reconstructing growth and dynamic trajectories from single-cell transcriptomics data. Nat. Mach. Intell. 6, 25–39. doi: 10.1038/s42256-023-00763-w
Shao, Y., Wang, L., Guo, N., Wang, S., Yang, L., Li, Y., et al. (2018). Cas9-nickase-mediated genome editing corrects hereditary tyrosinemia in rats. J. Biol. Chem. 293, 6883–6892. doi: 10.1074/jbc.RA117.000347
Shi, Y., Li, B.-J., Grierson, D., and Chen, K.-S. (2023). Insights into cell wall changes during fruit softening from transgenic and naturally occurring mutants. Plant Physiol., kiad128. doi: 10.1093/plphys/kiad128
Shmakov, S., Smargon, A., Scott, D., Cox, D., Pyzocha, N., Yan, W., et al. (2017). Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol. 15, 169–182. doi: 10.1038/nrmicro.2016.184
Sontheimer, E. J. (2005). Assembly and function of RNA silencing complexes. Nat. Rev. Mol. Cell Biol. 6, 127–138. doi: 10.1038/nrm1568
Stinson, B. M., Moreno, A. T., Walter, J. C., and Loparo, J. J. (2020). A mechanism to minimize errors during non-homologous end joining. Mol. Cell 77, 1080–1091. doi: 10.1016/j.molcel.2019.11.018
Su, J., Jiang, J., Zhang, F., Liu, Y., Ding, L., Chen, S., et al. (2019). Current achievements and future prospects in the genetic breeding of chrysanthemum: a review. Hortic. Res. 6, 109. doi: 10.1038/s41438-019-0193-8
Sun, D., Xu, Y., and Cen, H. (2022). Optical sensors: deciphering plant phenomics in breeding factories. Trends Plant Sci. 27, 209–210. doi: 10.1016/j.tplants.2021.06.012
Swaminathan, M. S. (2009). Norman E. Borlaug, (1914–2009). Nature 461, 894–894. doi: 10.1038/461894a
Tabara, H., Sarkissian, M., Kelly, W. G., Fleenor, J., Grishok, A., Timmons, L., et al. (1999). The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132. doi: 10.1016/S0092-8674(00)81644-X
Tabassum, J., Ahmad, S., Hussain, B., Mawia, A. M., Zeb, A., and Ju, L. (2021). Applications and potential of genome-editing systems in rice improvement: current and future perspectives. Agronomy 11, 1359. doi: 10.3390/agronomy11071359
Tardieu, F. and Tuberosa, R. (2010). Dissection and modelling of abiotic stress tolerance in plants. Curr. Opin. Plant Biol. 13, 206–212. doi: 10.1016/j.pbi.2009.12.012
Tardin-Coelho, R., Fletcher, S., Manzie, N., Gunasekara, S. N., Fidelman, P., Mitter, N., et al. (2025). A systematic review on public perceptions of RNAi-based biopesticides: Developing Social Licence to Operate. NPJ Sustain. Agric. 3, 15. doi: 10.1038/s44264-025-00057-1
Tingting, L., Jinpeng, Z., Xi, Y., Kejian, W., Yuchun, R., and Chun, W. (2023). Development and application of prime editing in plants. Rice Sci. 30, 509–522. doi: 10.1016/j.rsci.2023.07.005
Tiwari, S. and Lata, C. (2019). Genome engineering in rice: applications, advancements and future perspectives. Mol. Approaches Plant Biol. Environ. Challenges, 323–337. doi: 10.1007/978-981-15-0690-1_15
Toppino, L., Barchi, L., and Rotino, G. L. (2022). “Next generation breeding for abiotic stress resistance in eggplant,” in Genomic Designing for Abiotic Stress Resistant Vegetable Crops (Cham, Switzerland: Springer), 115–151.
Truong, D.-J. J., Geilenkeuser, J., Wendel, S. V., Wilming, J. C. H., Armbrust, N., Binder, E. M. H., et al. (2024). Exonuclease-enhanced prime editors. Nat. Methods 21, 455–464. doi: 10.1038/s41592-023-02162-w
Urnov, F. D., Rebar, E. J., Holmes, M. C., Zhang, H. S., and Gregory, P. D. (2010). Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646. doi: 10.1038/nrg2842
Van Dijk, P. J., Weissing, F. J., and Ellis, T. H. N. (2018). How Mendel’s interest in inheritance grew out of plant improvement. Genetics 210, 347–355. doi: 10.1534/genetics.118.300916
Vazquez-Vilar, M., Bernabé-Orts, J. M., Fernandez-Del-Carmen, A., Ziarsolo, P., Blanca, J., Granell, A., et al. (2016). A modular toolbox for gRNA–Cas9 genome engineering in plants based on the GoldenBraid standard. Plant Methods 12, 1–12. doi: 10.1186/s13007-016-0101-2
Wang, L., Chen, L., Li, R., Zhao, R., Yang, M., Sheng, J., et al. (2017). Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 65, 8674–8682. doi: 10.1021/acs.jafc.7b02745
Wang, T., Zhang, H., and Zhu, H. (2019). CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 6, 77. doi: 10.1038/s41438-019-0159-x
Wei, X., Li, J., and Li, Y. (2023). RNA interfering machinery: new insight into the Dicer-2 complexes in the dsRNA-processing cycle. Signal Transduct. Target. Ther. 8, 2. doi: 10.1038/s41392-022-01272-9
Wen, A., Havens, K. L., Bloch, S. E., Shah, N., Higgins, D. A., Davis-Richardson, A. G., et al. (2021). Enabling biological nitrogen fixation for cereal crops in fertilized fields. ACS Synthetic Biol. 10, 3264–3277. doi: 10.1021/acssynbio.1c00049
Wuest, S. E., Peter, R., and Niklaus, P. A. (2021). Ecological and evolutionary approaches to improving crop variety mixtures. Nat. Ecol. Evol. 5, 1068–1077. doi: 10.1038/s41559-021-01497-x
Xie, Y., Haq, S. I. U., Jiang, X., Zheng, D., Feng, N., Wang, W., et al. (2022). Plant genome editing: CRISPR, base editing, prime editing, and beyond. Grassland Res. 1, 234–243. doi: 10.1002/glr2.12034
Xu, Y., Zhang, X., Li, H., Zheng, H., Zhang, J., Olsen, M. S., et al. (2022). Smart breeding driven by big data, artificial intelligence, and integrated genomic-enviromic prediction. Mol. Plant 15, 1664–1695. doi: 10.1016/j.molp.2022.09.001
Yan, J., Oyler-Castrillo, P., Ravisankar, P., Ward, C. C., Levesque, S., Jing, Y., et al. (2024). Improving prime editing with an endogenous small RNA-binding protein. Nature 628, 639–647. doi: 10.1038/s41586-024-07259-6
Yarra, R. and Sahoo, L. (2021). Base editing in rice: current progress, advances, limitations, and future perspectives. Plant Cell Rep. 40, 595–604. doi: 10.1007/s00299-020-02656-3
Yasmeen, E., Wang, J., Riaz, M., Zhang, L., and Zuo, K. (2023). Designing artificial synthetic promoters for accurate, smart, and versatile gene expression in plants. Plant Commun. 4, 100558. doi: 10.1016/j.xplc.2023.100558
Yee, J. K. (2016). Off-target effects of engineered nucleases. FEBS J. 283, 3239–3248. doi: 10.1111/febs.2016.283.issue-17
Ying, Y., Hu, Y., Zhang, Y., Tappiban, P., Zhang, Z., Dai, G., et al. (2023). Identification of a new allele of soluble starch synthase IIIa involved in the elongation of amylopectin long chains in a chalky rice mutant. Plant Sci. 328, 111567. doi: 10.1016/j.plantsci.2022.111567
Younessi-Hamzekhanlu, M. and Gailing, O. (2022). Genome-wide SNP markers accelerate perennial forest tree breeding rate for disease resistance through marker-assisted and genome-wide selection. Int. J. Mol. Sci. 23, 12315. doi: 10.3390/ijms232012315
Yu, H. and Li, J. (2022). Breeding future crops to feed the world through de novo domestication. Nat. Commun. 13, 1171. doi: 10.1038/s41467-022-28732-8
Zavafer, A., Bates, H., Mancilla, C., and Ralph, P. J. (2023). Phenomics: conceptualization and importance for plant physiology. Trends Plant Sci. 28, 1004–1013. doi: 10.1016/j.tplants.2023.03.023
Zeng, H., Daniel, T. C., Lingineni, A., Chee, K., Talloo, K., and Gao, X. (2024). Recent advances in prime editing technologies and their promises for therapeutic applications. Curr. Opin. Biotechnol. 86, 103071. doi: 10.1016/j.copbio.2024.103071
Zeng, D., Li, X., Huang, J., Li, Y., Cai, S., Yu, W., et al. (2020). Engineered Cas9 variant tools expand targeting scope of genome and base editing in rice. Plant Biotechnol. J. 18, 1348. doi: 10.1111/pbi.13293
Zhan, X., Lu, Y., Zhu, J. K., and Botella, J. R. (2021). Genome editing for plant research and crop improvement. J. Integr. Plant Biol. 63, 3–33. doi: 10.1111/jipb.13063
Zhang, H., Li, Y., and Zhu, J.-K. (2018). Developing naturally stress-resistant crops for a sustainable agriculture. Nat. Plants 4, 989–996. doi: 10.1038/s41477-018-0309-4
Zhang, K., Raboanatahiry, N., Zhu, B., and Li, M. (2017). Progress in genome editing technology and its application in plants. Front. Plant Sci. 8, 177. doi: 10.3389/fpls.2017.00177
Zhang, H., Zhao, Y., and Zhu, J.-K. (2020). Thriving under stress: how plants balance growth and the stress response. Dev. Cell 55, 529–543. doi: 10.1016/j.devcel.2020.10.012
Zhang, C., Zhong, X., Li, S., Yan, L., Li, J., He, Y., et al. (2023). Artificial evolution of OsEPSPS through an improved dual cytosine and adenine base editor generated a novel allele conferring rice glyphosate tolerance. J. Integr. Plant Biol. 65, 2194–2203. doi: 10.1111/jipb.13543
Zhao, X., Jayarathna, S., Turesson, H., Fält, A.-S., Nestor, G., González, M. N., et al. (2021). Amylose starch with no detectable branching developed through DNA-free CRISPR-Cas9 mediated mutagenesis of two starch branching enzymes in potato. Sci. Rep. 11, 4311. doi: 10.1038/s41598-021-83462-z
Zhao, Z., Shang, P., Mohanraju, P., and Geijsen, N. (2023). Prime editing: advances and therapeutic applications. Trends Biotechnol. 41, 1000–1012. doi: 10.1016/j.tibtech.2023.03.004
Zhi, H., Zhou, S., Pan, W., Shang, Y., Zeng, Z., and Zhang, H. (2022). The promising nanovectors for gene delivery in plant genome engineering. Int. J. Mol. Sci. 23, 8501. doi: 10.3390/ijms23158501
Zhou, F.-Y., Han, Y.-J., Wang, Y.-H., Yao, C.-C., and Zhang, Y. (2021). Overexpression of Polypogon fugax Type I–Like MADS-Box Gene PfAGL28 affects flowering time and pod formation in transgenic Arabidopsis. Plant Mol. Biol. Rep., 1–9. doi: 10.1007/s11105-021-01312-8
Zhou, R., Jiang, F., Niu, L., Song, X., Yu, L., Yang, Y., et al. (2022). Increase crop resilience to heat stress using omic strategies. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.891861
Zhu, H., Li, C., and Gao, C. (2020). Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 21, 661–677. doi: 10.1038/s41580-020-00288-9
Zhu, S., Li, Y., Zhang, X., Liu, F., Xue, F., Zhang, Y., et al. (2021). GhAlaRP, a cotton alanine rich protein gene, involves in fiber elongation process. Crop J. 9, 313–324. doi: 10.1016/j.cj.2020.08.007
Zong, Y., Liu, Y., Xue, C., Li, B., Li, X., Wang, Y., et al. (2022). An engineered prime editor with enhanced editing efficiency in plants. Nat. Biotechnol. 40, 1394–1402. doi: 10.1038/s41587-022-01254-w
Keywords: molecular breeding, food security, genome editing, climate resilience, sustainable agriculture, artificial intelligence
Citation: Riaz M, Yasmeen E, Saleem B, Hameed MK, Saeed Almheiri MT, Saeed Al Mir RO, Alameri G, Khamis Alghafri JS and Gururani MA (2025) Evolution of agricultural biotechnology is the paradigm shift in crop resilience and development: a review. Front. Plant Sci. 16:1585826. doi: 10.3389/fpls.2025.1585826
Received: 03 March 2025; Accepted: 15 May 2025;
Published: 19 June 2025.
Edited by:
Steven Maina Runo, Kenyatta University, KenyaReviewed by:
Dang Fengfeng, South China Agricultural University, ChinaSuresh L. M., The International Maize and Wheat Improvement Center (CIMMYT), Kenya
Copyright © 2025 Riaz, Yasmeen, Saleem, Hameed, Saeed Almheiri, Saeed Al Mir, Alameri, Khamis Alghafri and Gururani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mayank Anand Gururani, Z3VydXJhbmlAdWFldS5hYy5hZQ==
†These authors have contributed equally to this work
‡ORCID: Mayank Anand Gururani, orcid.org/0000-0003-4254-6450
 Muhammad Riaz
Muhammad Riaz Erum Yasmeen1†
Erum Yasmeen1† Bilal Saleem
Bilal Saleem Mayank Anand Gururani
Mayank Anand Gururani