- 1Tobacco Research Institute, Chinese Academy of Agricultural Sciences, Qingdao, China
- 2Leaf Tobacco Technology Extension Department, Deyang Company of Sichuan Provincial Tobacco Corporation, Deyang, China
- 3Institute of Tobacco Science, Fujian Provincial Tobacco Company of China National Tobacco Corporation, Fuzhou, China
Pentatricopeptide repeat (PPR) gene family is one of the largest gene families in higher plants. The Restoration of fertility like (RFL) clade of the family plays a crucial role in restoring fertility of cytoplasmic male sterility (CMS) lines in plants. Common tobacco (Nicotiana tabacum L.) is an important economic crop of which the CMS hybrids have been widely used in commercial cultivation. However, the restorer line of tobacco and the regulatory mechanism of fertility restoration remain elusive. In addition, PPR and RFL genes have not been illustrated in common tobacco. In this study, a total of 1002 NtPPR genes were identified, of which 27 NtRFLs belonging to P subfamily were demonstrated. The collinearity analysis showed that a total of 15 pairs of NtRFL genes had collinear relationship and unevenly distributed in 9 linkage groups. Cis-element analysis revealed that a large number of environmental stress and phytohormone response elements were located in the promoter of NtRFLs. By combining the RNA-seq and qPCR analysis, NtRFL3 was further selected as the candidate gene due to its significantly higher expression at early anther development in the fertile line MF1. NtRFL3 was predicted to be localized in mitochondria and shared high sequence similarity with the known fertility-restorer PPR592 in petunia. Our results provided new gene targets for molecular breeding of tobacco restorer lines and for illustration of molecular mechanism on fertility restoration of plant CMS lines.
Introduction
Pentatricopeptide repeat (PPR) proteins represent an extensive plant-specific protein family predominantly found in terrestrial plants (Small and Peeters, 2000). These sequence-specific RNA-binding proteins localize to semi-autonomous organelles and are crucial for plant growth and developmental processes (Liu et al., 2021). Following their initial discovery in Arabidopsis, subsequent studies have expanded our understanding of PPR proteins across various plant species. Structurally, PPR proteins comprise three distinct regions: an N-terminal targeting signal, a central tandem repeat domain containing 2-30 conserved motifs (each spanning 30-40 amino acids), and a C-terminal functional domain (Lurin et al., 2004). PPR gene family can be divided into P and PLS subfamily based on their motif structure (Han et al., 2020). The P subfamily contains P motifs common to all eukaryotes (Cheng et al., 2016), and PLS subfamily can be divided into four subgroups: PLS, E, E+, and DYW based on the different non-PPR domains at the C-terminus. The variable N-terminal region determines subcellular localization to either mitochondria or chloroplasts (Saha et al., 2007). Contemporary research has elucidated PPR proteins multifaceted roles in organellar gene expression regulation. These proteins mediate post-transcriptional processes including RNA editing, splicing, stabilization, and translation, while also influencing embryogenesis and chloroplast biogenesis (Hao et al., 2019; Hayes et al., 2015; Ichinose et al., 2012; Tavares-Carreón et al., 2008).
Cytoplasmic male sterility (CMS) is a maternal genetic trait in which mitochondrial gene abnormalities lead to stamen degeneration, pollen abortion or functional infertility in plants while pistils function normally (Baranwal et al., 2012). Some studies have found that CMS lines usually accumulate reactive oxygen species (ROS) in anthers and other tissues to form oxidative stress (Jiang et al., 2007), which is a direct cause of pollen abortion. Male sterility can be restored by the expression of restorer-of-fertility (Rf) genes, so that plants produce normal pollen (Dahan and Mireau, 2013). Numerous genes have been identified as Rf genes to play pivotal roles in mediating fertility restoration mechanisms in plants, such as the Zea mays RF2 (Cui et al., 1996) and Beta vulgaris RF1 (Kitazaki et al., 2015) belonging to the Aldehyde dehydrogenase and Peptidase-like family, respectively. The Restoration of fertility like (RFL) genes are also characterized as Rfs, derived from P subfamily of PPRs. Some RFLs can specifically regulate the transcription of male sterility genes in mitochondria and restore plant fertility (Fujii et al., 2011). Plant genomes generally encode 10-30 RFL proteins (Dahan and Mireau, 2013), most of which belong to P subfamily of PPRs. Arabidopsis thaliana has 26 AtRFLs, While there are 53 BnRFL and 38 RFLs in Brassica napus (Ning et al., 2020) and Solanum tuberosum (Ding, 2014), respectively. However, several RFL proteins, including Rf1 (Klein et al., 2005) in Sorghum bicolor and Rfm1 (Ui et al., 2015) in Hordeum vulgare have been functionally characterized as members of the PLS subfamily.
Current studies have identified the primary mechanisms by which Rfs restore plant fertility: Firstly, it involves expression suppression of sterility genes by Rfs binding and processing their RNA. The resulted metabolic reprogramming could compensate the cellular energy deficits caused by the expression of CMS-associated transcripts. In the radish Ogura CMS system (one CMS type in cruciferous crops), the Rf gene orf687 encodes a protein that directly binds to the transcript of the sterility gene orf138, suppressing its expression at the post-transcriptional level (Desloire et al., 2003; Yamagishi et al., 2021). For rice CMS line (BT-type) caused by the mitochondrial gene orf79, RF1a mediates endonucleolytic cleavage of atp6-orf79 mRNA, while RF1b promotes their rapid degradation, collectively restoring fertility of rice CMS lines (Akagi et al., 2004; Wang et al., 2006; Kazama et al., 2008).
Heterosis has been extensively exploited in Nicotiana tabacum, where F1 hybrids exhibit significantly enhanced growth vigor and stress resistance compared to the mid-parental values (Hancock and Lewis, 2017). The reliable production of these hybrids depends on CMS systems, which ensure seed purity by preventing paternal pollen contamination. The sterility genes originate from diverse sources, including wild species cytoplasm, natural mutation of sterile plants, and impaired coordination between nuclear and cytoplasm. For example, the sterility of tobacco CMS lines (designated as sua-CMS) is due to the wild species Nicotiana suaveolens-derived cytoplasm. Previous studies have identified mitochondrial respiratory chain-related genes as sterility genes which presented aberrant open reading frames (ORFs) leading to programmed cell death (PCD)-related pollen abortion and insufficient ATP synthesis in tobacco (Liu, 2022). However, the nuclear factors coordinating mitochondrial-nuclear interactions for fertility restoration remain poorly characterized.
With the deepening of the whole genome sequencing of tobacco, it provides a good basis for tobacco bioinformatic analysis. Ding (2014) systematically identified the PPR family members from Nicotiana tomentosiformis and cloned six NtomRfs (Ding, 2014). Through genome-wide analysis, we characterized PPR family members, especially RFL genes in cultivated tobacco, with their expression patterns validated by both transcriptomics and qPCR at different anther developmental stages. The findings would provide new gene targets for molecular breeding of tobacco restorer lines and theoretical foundation for illustration of regulatory network on fertility restoration of plant CMS lines.
Materials and methods
Identification of PPR family members in Nicotiana tabacum L.
The Hidden Markov Model (HMM) of PPR domain (PF01535) (Che et al., 2022) was downloaded from Pfam (http://pfam.xfam.org/). The HMM profile was used to identify all the potential PPR protein sequences through the Simple HMM Search in TBtools (Chen et al., 2020), using the genome data of Nicotiana tabacum cultivar ZY300 (Zan et al., 2025). The conserved domain of all identified NtPPRs were analyzed by NCBI Batch-CDD tools (http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) using the CDD-62466 position-specific scoring matrix (PSSMs) database. Key parameters were set as follows: E-value threshold set to 0.01, maximum matching sequences limited to 500, with remaining parameters maintained at default ones. The screened protein domains were further confirmed by SMART (http://smart.embl.de/smart/set_mode.cgi?GENOMIC=1) and Expasy (https://web.expasy.org/protparam) tools.
Chromosomal localization and gene structure analysis
The chromosomal localization and gene structure information of NtPPRs was acquired in the annotation file of ZY300. The results were visualized by Map Gene 2 Chrom v2.1 (Chao et al., 2015) and Gene Structure Viewer in TBtools, respectively.
Subcellular localization prediction
The Cell-Ploc (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/), TargetP (http://www.cbs.dtu.dk/services/TargetP/), Predotar (https://urgi.versailles.inra.fr/predotar/predotar.html) were used to predict protein subcellular localization.
Phylogenetic and conserved motif analysis
The PPR protein sequences of Solanum lycopersicum were downloaded from Sol Genomics Network (https://solgenomics.sgn.cornell.edu/). The sequences of five Rf-PPRs, RPF1 (Arabidopsis thaliana), Rf_PPR592 (Petunia hybrida), Rf1a (Oryza sativa), Rfo (Raphanus sativus), and PPR1 (Capsicum annuum), were downloaded from NCBI database (https://www.ncbi.nlm.nih.gov/). The protein sequences were aligned by MUSCLE using MEGA 11, and then subjected to genetic distance analysis using compute pairwise distance. Bootstrap analysis was selected with a default setting of 1000 replicates, and the p-distance method was chosen for the calculation. The phylogenetic analysis was performed by Neighbor-Joining (NJ) method with 500 bootstrap iterations. The ITOL (https://itol.embl.de/) was used to modify the output phylogenetic tree.
The MEME suite (http://meme-suite.org/tools/meme) was used to predict the conserved motifs of NtPPRs (Bailey et al., 2009). The seven conserved motifs of Arabidopsis were used as primary sequences to discover NtPPR motifs.
Identification and physicochemical property analysis of NtRFLs
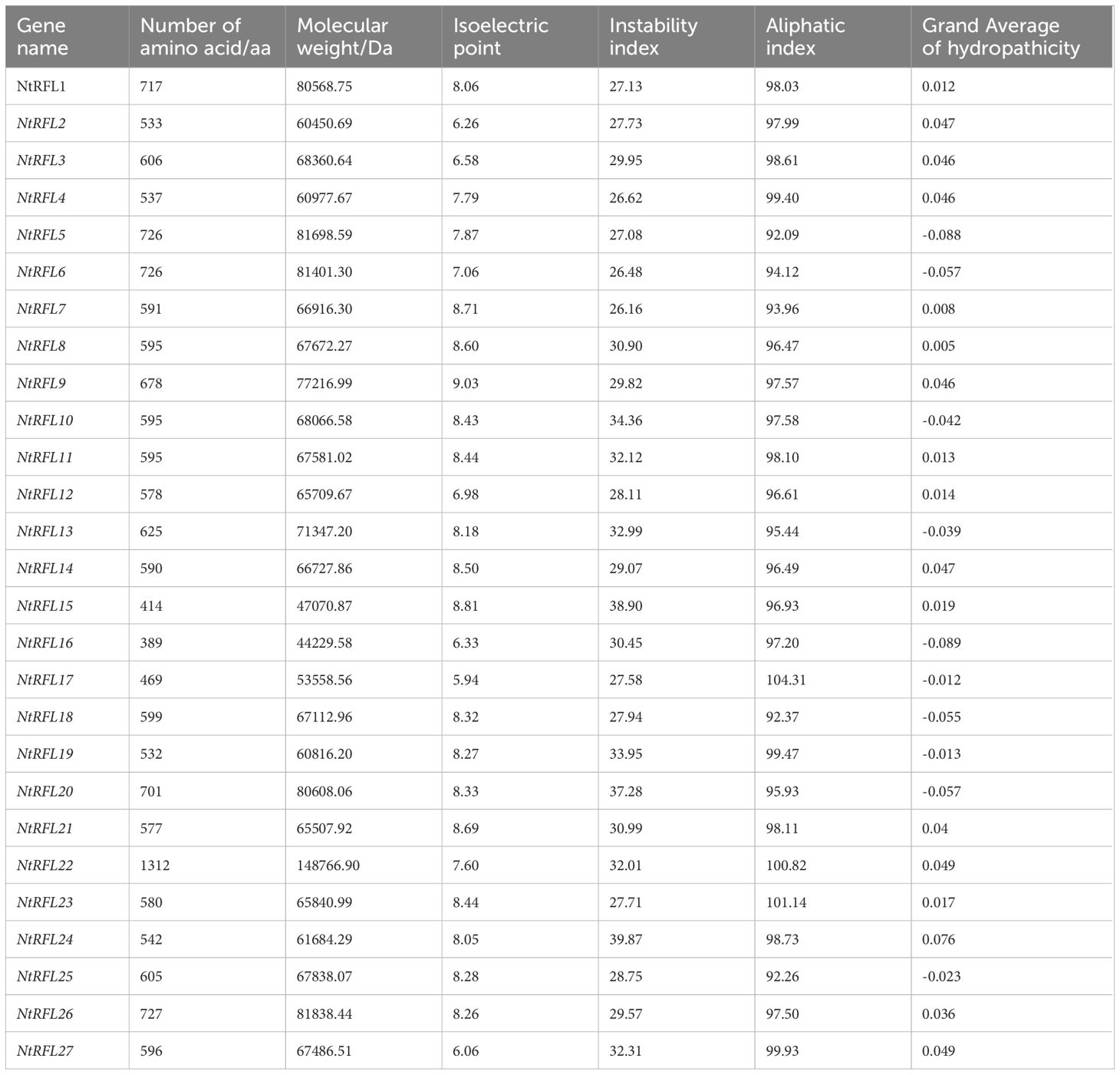
The protein sequences of AtRFL1-AtRFL26 and Rf-PPR592 were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/), and were used for BLAST searches against the protein sequences of ZY300 in TBtools with E-value<e-100. The isoelectric point, molecular weight, amino acid number, instability index, aliphatic index and grand average of hydropathicity of NtRFLs were analyzed by ProtParam (https://web.expasy.org/computepi/).
Collinearity analysis of NtRFLs
Genomic data of Solanum tuberosum and Arabidopsis thaliana were downloaded from Sol Genomics Network and TAIR (https://www.arabidopsis.org), respectively. The One Step MCScanX in TBtools was used to analyze gene duplication events. The syntenic relationship of RFL genes between tobacco and the other two species was determined using the Dual Synteny Plotter tool in TBtools. The results were visualized by the Advanced circos in TBtools.
Prediction of cis-acting elements in NtRFLs
The promoter sequence of NtRFLs (2000 bp upstream sequences of CDS) were extracted by TBtools according to the annotation file of ZY300. The PlantCARE online tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) was used to predict the cis-acting elements of promoter, and Rstudio was used for statistical and visual analysis of each element.
Cytological observation of flower buds in tobacco cultivars with different fertility
The 2-3 mm, 3-5 mm, 5-7 mm of flower buds, corresponding to sporogenous cell-microsporocyte period, meiosis-tetrad period, and the mid uninucleate period, were collected and subsequently embedded into paraffin according to Yuan (Yuan et al., 2018). The toluidine blue stained sections was observed using the inverted microscope.
Total RNA extraction, cDNA synthesis and qPCR analysis
Flower buds (2-3 mm, 3-5 mm, 5-7 mm in size) were sampled from three tobacco varieties: the fertility line F609 and two sterile line MS609 (which possesses stamens but lacks pollen grains), and MSG28 (stamenless). Three biological replicates were performed for each line. Samples were stored at -80°C after quick freezing in liquid nitrogen.
Total RNA with three biological replicates were extracted using TRNpure reagent (Nobelab, China) according to the manufacturer’s instructions. 1 μg RNA was used for cDNA synthesis using Evo M-MLV Mix Kit with gDNA Clean for qPCR (AccurateBiology, AG11728, China). All reactions were performed with two technical replicates using SYBR Green Premix Pro Taq HS qPCR Kit (AccurateBiology, AG11701, China), with primers indicated in Supplementary Table S1. qPCR was conducted on the LightCycler 96 Instrument. Relative gene expression was normalized to the expression level of Actin.
Transcriptome analysis
RNA samples extracted from 2-3 mm flower buds were sequenced by the High-Throughput Sequencing Instruments using the Illumina NovaSeq 6000 (illumina, USA) platform, obtained an average yield of 5.9 Gb data per sample. Sequencing adapter contamination, reads with N bases, and low-quality reads were removed using fastp (version 0.19.7). Clean reads were aligned against ZY300 reference genome using the Hisat2 (V2.0.5) software (Mortazavi et al., 2008). Gene expression levels were log-transformed and normalized using DESeq2 (1.20.0) software. The FPKM value of NtRFL genes were used to characterize their expression patterns.
Results
Identification of PPR family members in Nicotiana tabacum L.
Through a comprehensive analysis combining hidden Markov model (HMM) profiling and conserved domain verification, we systematically identified 1002 pentatricopeptide repeat (PPR) genes in the tobacco genome. These NtPPR genes were chromosomally mapped and numerically designated from NtPPR1 to NtPPR1002 according to their genomic positions (Figure 1a). Structural characterization revealed distinct intron-exon patterns: 58.5% of NtPPR genes existed as single-exon structures, 19.0% contained one intron, while the remaining 22.5% possessed two or more introns (maximum 18 introns per gene) (Figure 1b). Prediction of subcellular localization showed that 69.36% of NtPPRs were localized either in chloroplasts or in mitochondria (Figure 1c).
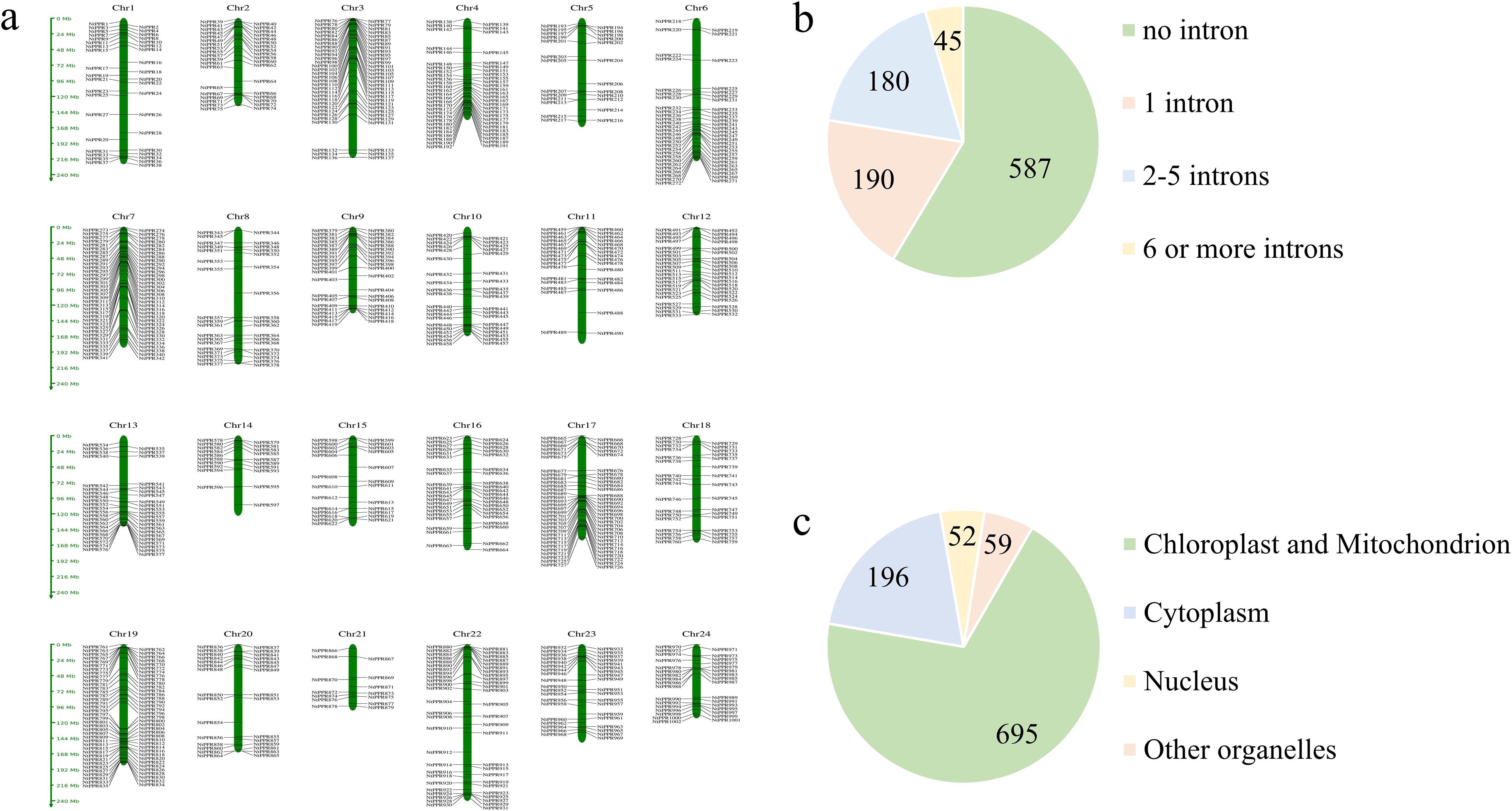
Figure 1. Chromosomal distribution, gene structure and subcellular localization of PPR genes in tobacco. (a) Chromosomal localization of NtPPR genes. (b) Number of NtPPR genes with different intron numbers. (c) Number of NtPPRs with predicted subcellular localization.
Phylogenetic analysis and classification of NtPPRs
To clarify the subfamilies of NtPPRs, phylogenetic tree was constructed using the identified 1002 NtPPRs and 471 known Solanum lycopersicum PPR proteins. The analysis revealed clear separation into two major subfamilies: the P subfamily (530 members) and PLS subfamily (472 members) (Figure 2a). Through protein conservation analysis, we identified six conserved motifs that enabled further subdivision of the PLS subfamily (Figure 2c‐h). The PLS subfamily was subsequently divided into four distinct subgroups: PLS (39 members), E (220), E+ (43), and DYW (170), as shown in (Figure 2b).
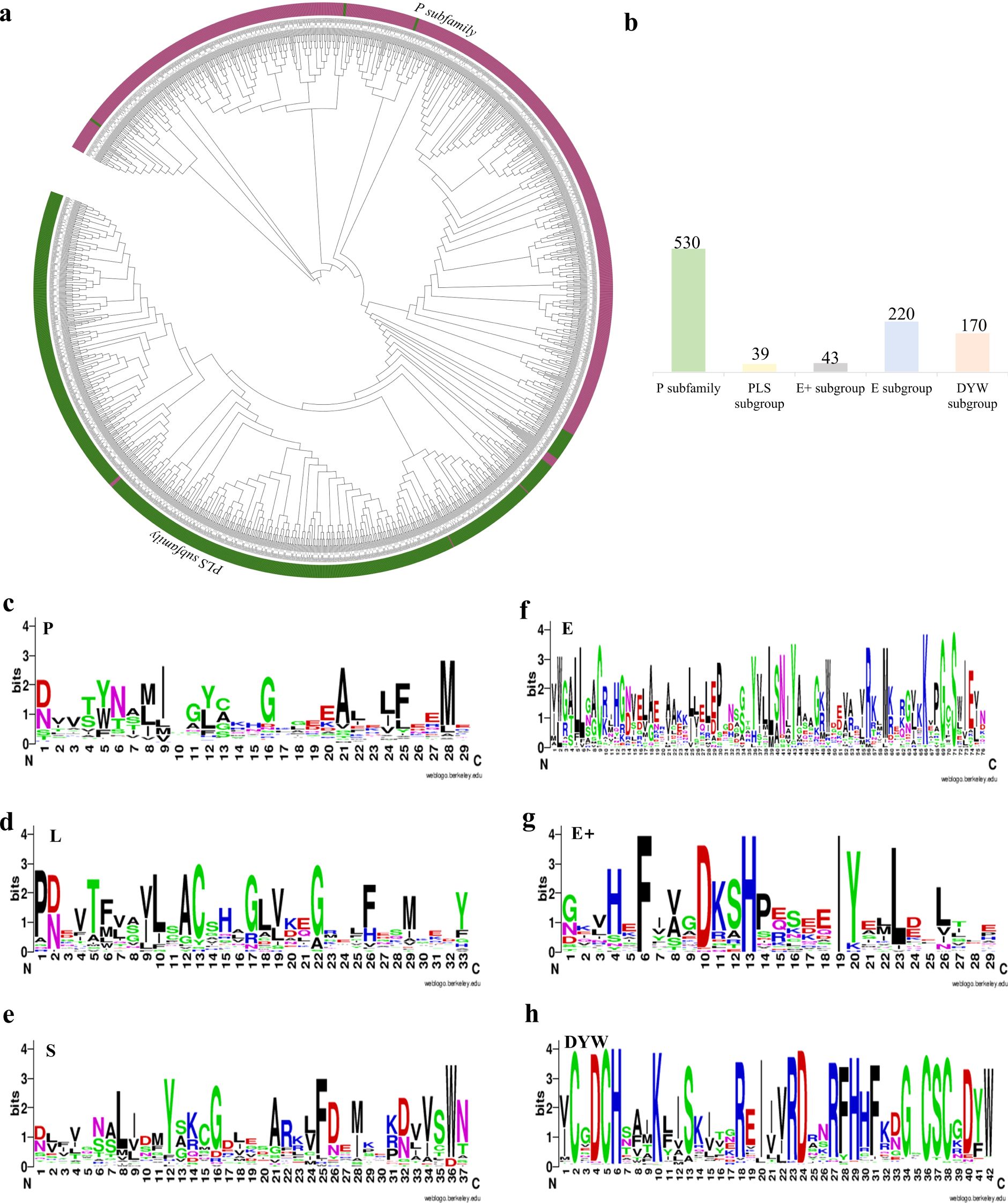
Figure 2. Phylogenetic relationships and conserved motif analysis among the NtPPR family genes. (a) PPR phylogenetic tree. 1002 NtPPRs and 471 SlPPRs were aligned. P and PLS subfamilies were highlighted in purple and green, respectively. (b) Number of NtPPR proteins belonging to the P subfamily and PLS subgroups. (c-h) Conserved motif analysis among the NtPPR family genes. The height of characters correlates to the conservation of the amino acid, the higher the character height, the more conservative of the amino acid.
Identification and physicochemical property analysis of tobacco RFL-PPR members
We performed the NtRFLs identification based on their homology with Arabidopsis RFLs and petunia Rf-PPR592. Among the 26 AtRFLs, 9 had no homologs in tobacco, including AtRFL1, AtRFL8, AtRFL10, AtRFL19, AtRFL20, AtRFL22, AtRFL23, AtRFL24, and AtRFL26. By using Rf-PPR592 as a reference, 20 NtRFLs were identified. Combining both blast results, a total of 27 NtRFLs were identified in tobacco all belonging to P subfamily. These genes were designated as NtRFL1 to NtRFL27. The molecular weight of NtRFLs ranged from 442 to 148.8 KDa, with NtRFL16 and NtRFL22 as smallest and largest proteins, respectively. The theoretical isoelectric points of NtRFLs varied from 5.94 to 9.03, with 6 NtRFLs as acidic proteins having pI values below 7, while the rest were alkaline. The instability index of NtRFL proteins were all below 40, suggesting their high stability. The coefficients of lipid solubility differed from 92.09 to 104.31, and the negative GRAVY values of 10 RFL proteins demonstrated their hydrophilic nature (Table 1).
The collinearity analysis of tobacco RFLs
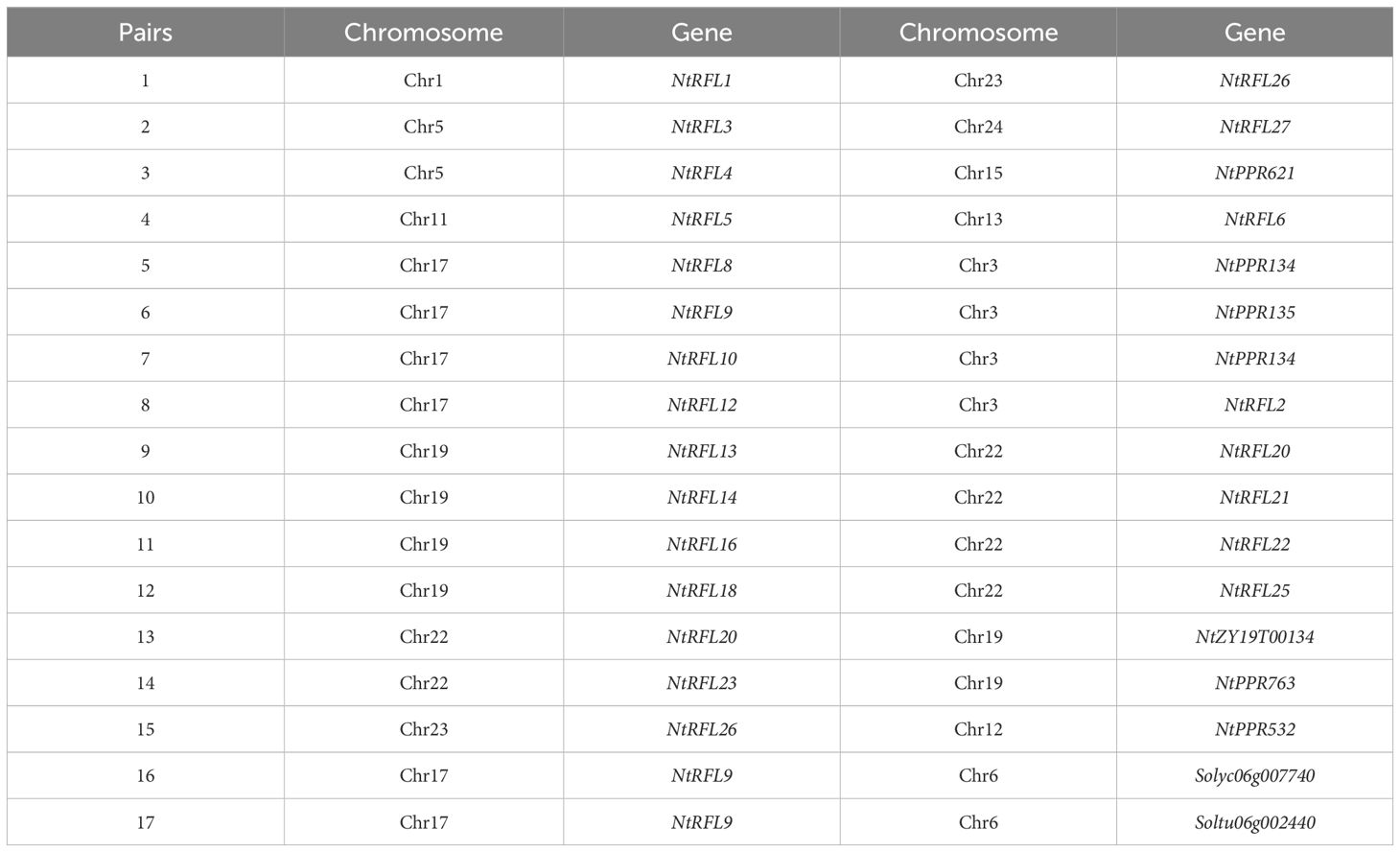
To investigate the evolutionary patterns of tobacco RFLs, we performed collinearity analysis of gene duplication events. The results (Figure 3a) revealed 15 collinear gene pairs unevenly distributed across 9 linkage groups. There were 6 NtRFLs distributed on Chr19, 4 NtRFLs on Chr17, and only 1 NtRFL on Chr1, Chr11, Chr12, Chr15 and Chr24 respectively. NtRFL26 had collinearity with both NtRFL1 and NtPPR532 indicating that they may evolve distinct roles due to gene replication events. For the interspecific collinearity of RFLs, it showed that no collinearity was found in RFLs between tobacco and Arabidopsis. While NtRFL9 forms homologous gene pairs with both Solyc06g007740 in tomato and Soltu06g002440 in potato suggesting potential conserved functions of these genes between different species (Table 2).
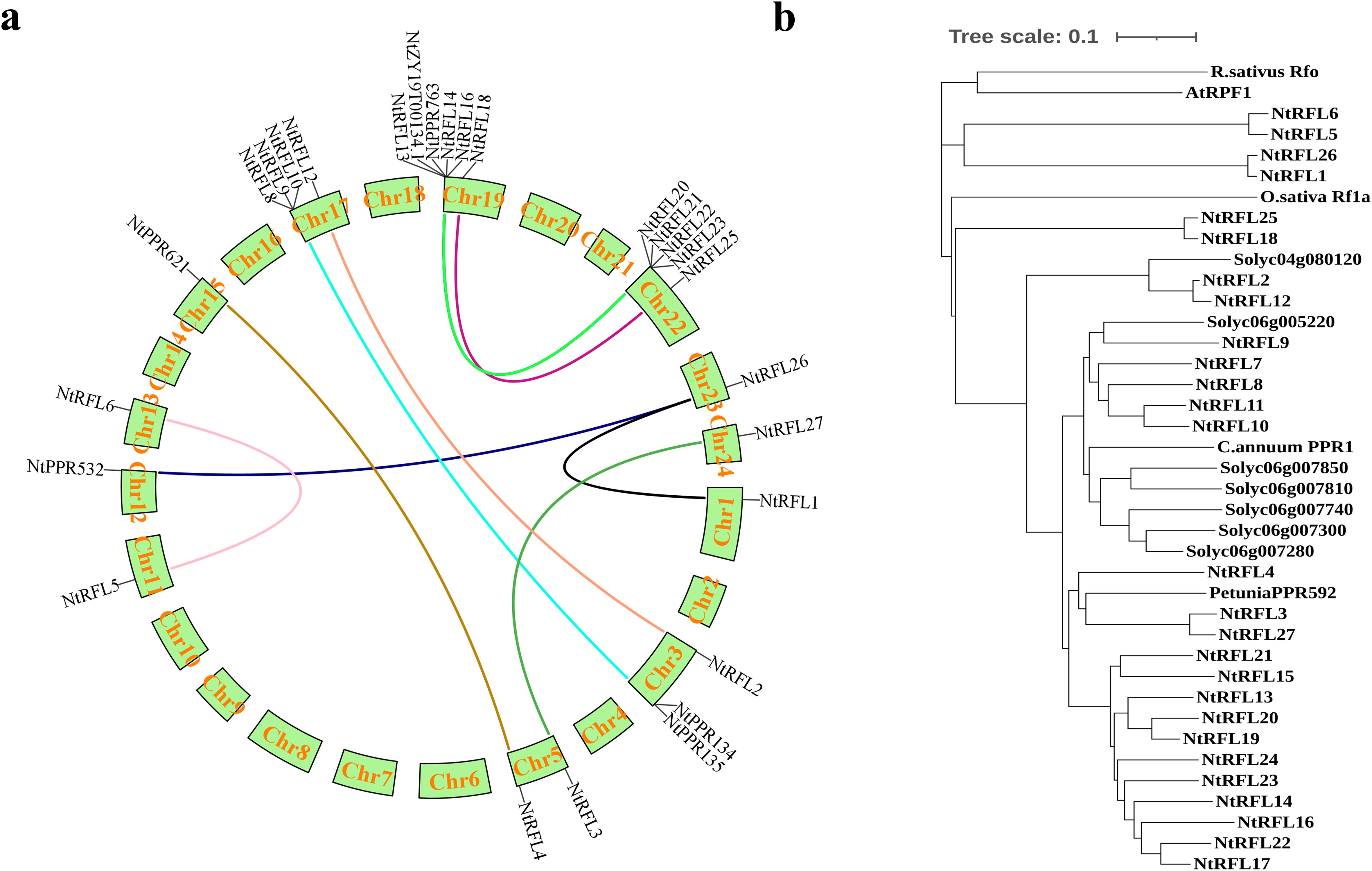
Figure 3. Evolutionary relationship of RFL genes in tobacco. (a) Distribution of collinear RFL gene pairs on tobacco chromosomes. The different colored lines indicate collinear RFL gene pairs. (b) Phylogenetic tree of Rf proteins in multiple species. Rs, Raphanus sativus; Os, Oryza sativa; At, Arabidopsis thaliana; Ca, Capsicum annuum; Ph, Petunia hybrida; Solyc, Solanum lycopersicum.
We also examined the homology of NtRFLs with various known Rfs in multiple species (Figure 3b). The results demonstrated that the RFL genes of tobacco predominantly clustered with those of tomato, pepper, and petunia, all belong to Solanacea, while being distant from those of rice and Arabidopsis. Among them, 14 NtRFLs were clustered with petunia PPR592 which is encoded by a tandem array of 14 PPR motifs and is able to restore fertility to CMS plants by decreasing the accumulation of petunia mitochondrial fused gene (PCF) (Bentolila et al., 2002). This indicates that they may be the orthologs of PPR592 and play similar roles in regulating plant fertility.
Analysis of cis-acting elements of NtRFLs
To further explore the regulatory mechanism of RFLs gene expression in tobacco, we first performed cis-acting element analysis of their promoters (Figure 4). After removing non-functional elements, 13 types of cis-acting elements were identified in this study. These elements were mainly related to environmental stress and phytohormone responses, which perhaps collectively regulate the expression of the NtRFLs in tobacco.
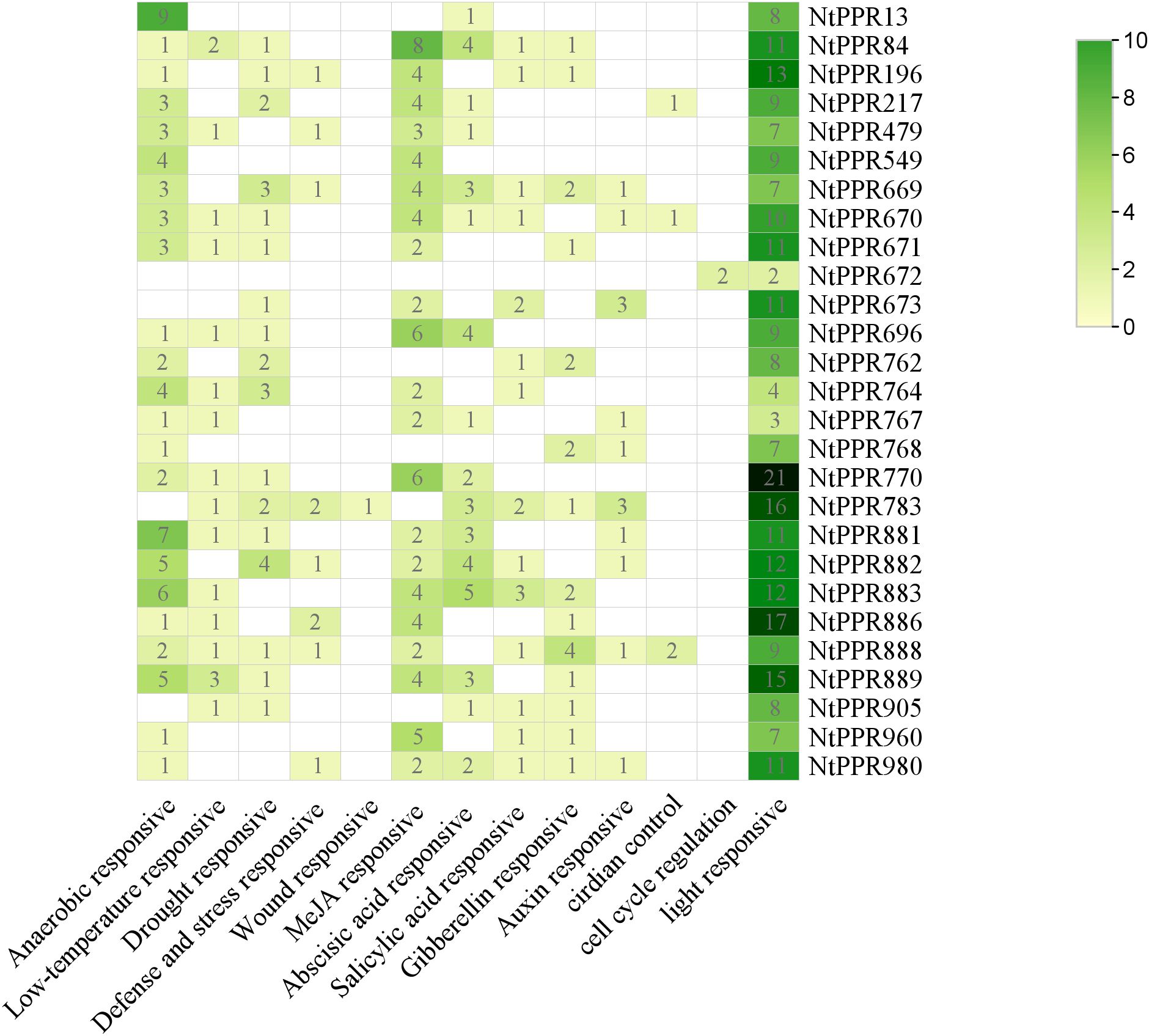
Figure 4. Prediction results of cis-acting elements of NtRFL genes. The number represents the element numbers in each category contained in NtRFLs.
Several thermo-sensitive male-sterile genes have been cloned and reported. However, how the environmental signals contribute to the fertility restoration remains unclear. We found that tobacco RFL genes, especially NtRFL2 and NtRFL24, contained both low-temperature and light responsive elements, suggesting their potential role in pollen fertility recovery in a thermo-mediated way. Additionally, there are many cis-elements in response to phytohormones such as jasmonic acid, abscisic acid, salicylic acid, gibberellin and auxin. Zang et al. (2023) demonstrated that excessive activation of auxin signaling may inhibit pollen development, while inhibiting auxin signaling partially promoted pollen development in CMS-D2 cotton (Zang et al., 2023). Moreover, the fertility of Rice could be restored by applying exogenous methyl jasmonate (Pak et al., 2021). Therefore, the genes possessing hormone-responsive cis-elements are potential targets for studying the regulation of pollen fertility in response to various phytohormones in tobacco.
Cytological observation of flower buds in tobaccos with different fertilily
To explore the expression profile of NtRFLs during anther development, three tobacco lines displaying diverse fertility as well as collected at different stages were used. The anther of fertile line MF1 developed normally with distinct cytological structures in each stage. Firstly, a transition stage including both sporogenous cell and microsporocyte periods was presented in the small buds (2-3 mm) of MF1. This was followed by meiosis and tetrad periods when the tapetum was degradated gradually in the buds of 3-5 mm. At the mid uninucleate stage exhibited in flower buds of 5-7 mm, the tapetum was almost disappeared, and the anther became mature with large number of pollen grains inside (Figures 5a, d–f). In contrast with MF1, the semi-sterile line CMS1 showed an irregular structure of tapetum resulting into disappeared pollen sacs (Figures 5g–h). At the mid unicucleate stage, a lamellar structure without pollen grains was formed as it’s shown in Figures 5b, i. Instead of showing three distinct stages described above, sterile line CMS2 with small buds was still at an earlier time point when the stamen primordium haven’t differentiated into sporogenous cells (Figure 5j). The cells were degenerated later and no stamens formed completely (Figures 5c, k–l).
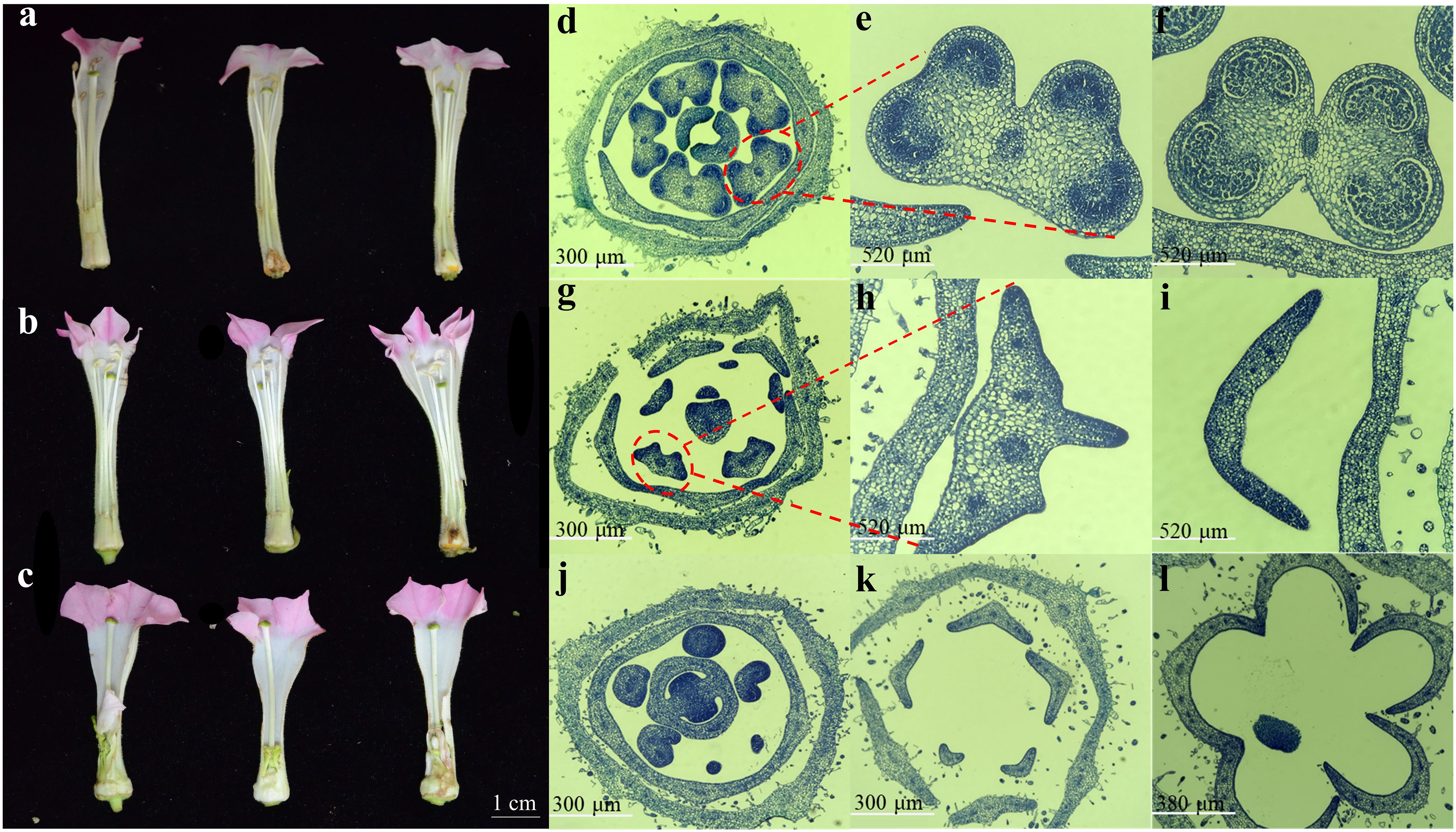
Figure 5. Morphological and cytological observation of anther development in three different tobacco lines. Floral phenotypes of (a) MF1 (fertile line), (b) CMS1 (semi-sterile line which possesses stamens but lacks pollen grains), and (c) CMS2 (the sterile line which is stamenless). (d-l) The cross sections of flower buds at the sporogenous cell-microsporocyte period, meiosis-tetrad period, and the mid uninucleate stage in MF1 (d-f), CMS1 (g-i) and CMS2 (j-l), respectively.
Expression analysis of NtRFLs in tobaccos with different fertilily
Previous studies have found that cotton restoration gene n-PPR-1, n-PPR-2 is specifically expressed in early anther development (Gao et al., 2022). It is speculated that fertility related genes are expressed at sporogenous cell-microsporocyte period or even earlier for determining the fate of embryonic cells. We therefore analyzed the transcriptome data in flower buds of 2-3 mm from the above lines. It was shown that 21 out of 27 NtRFLs were highly expressed in fertile line MF1, while expressed in lower level in CMS sterile lines (Figure 6).
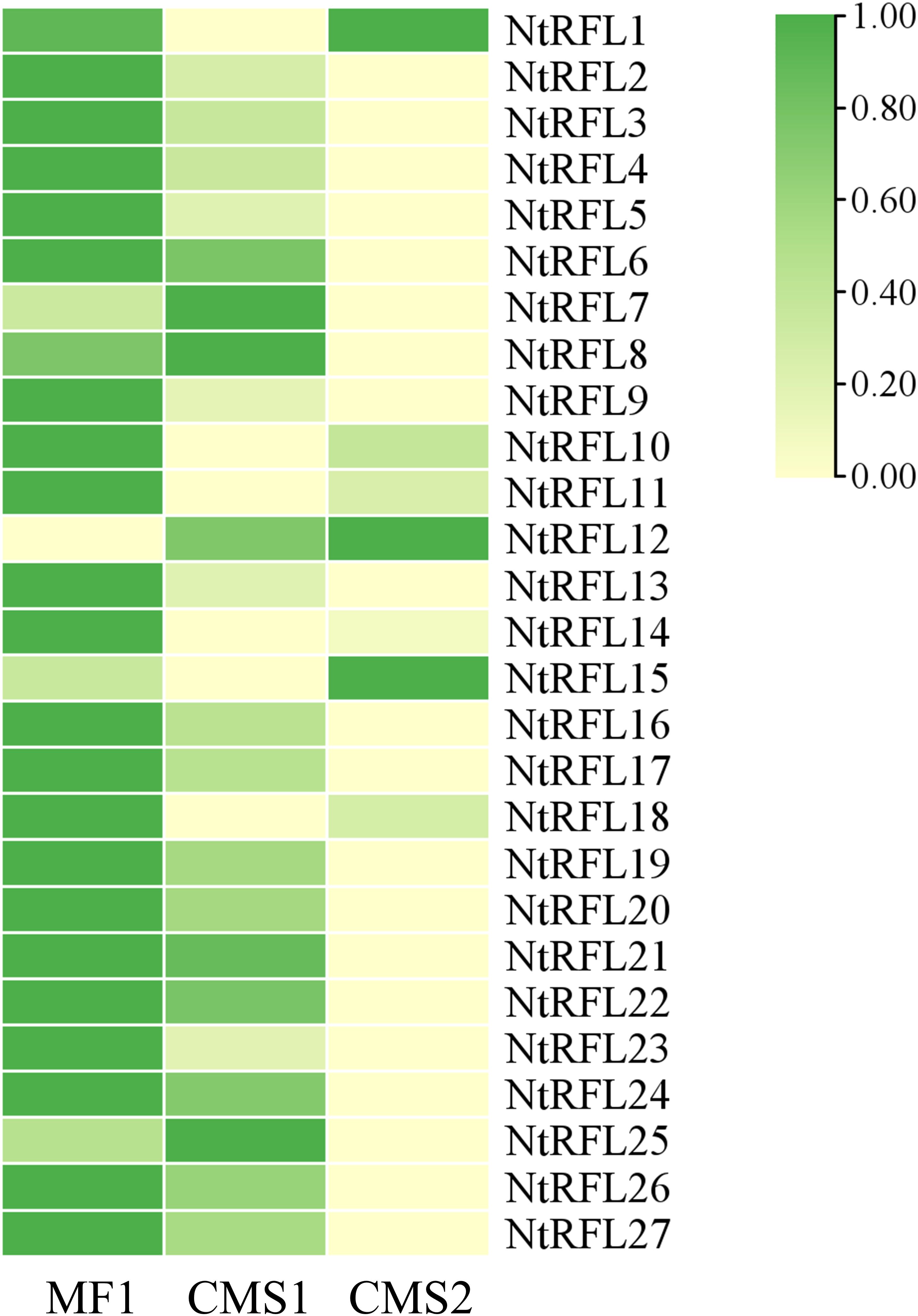
Figure 6. RNA-seq analysis of NtRFL genes in tobacco lines with different fertility. The up- and down-regulation were presented as green and yellow, respectively. MF1, the fertile line; CMS1, the semi-sterile line which possesses stamens but lacks pollen grains; CMS2, the sterile line which is stamenless.
To validate the RNA-seq results, we carried on qPCR analysis of all NtRFLs and found that 10 genes had higher expression in MF1 compared to CMS lines (Figures 7a–j). Whether their expression were specific to early time point was tested in samples including all three stages mentioned above. The results demonstrated that NtRFL1 and NtRFL3 were highly active at sporogenous cell-microsporocyte period of MF1, while there was no or less expression shown in CMS lines (Figures 7k–l).
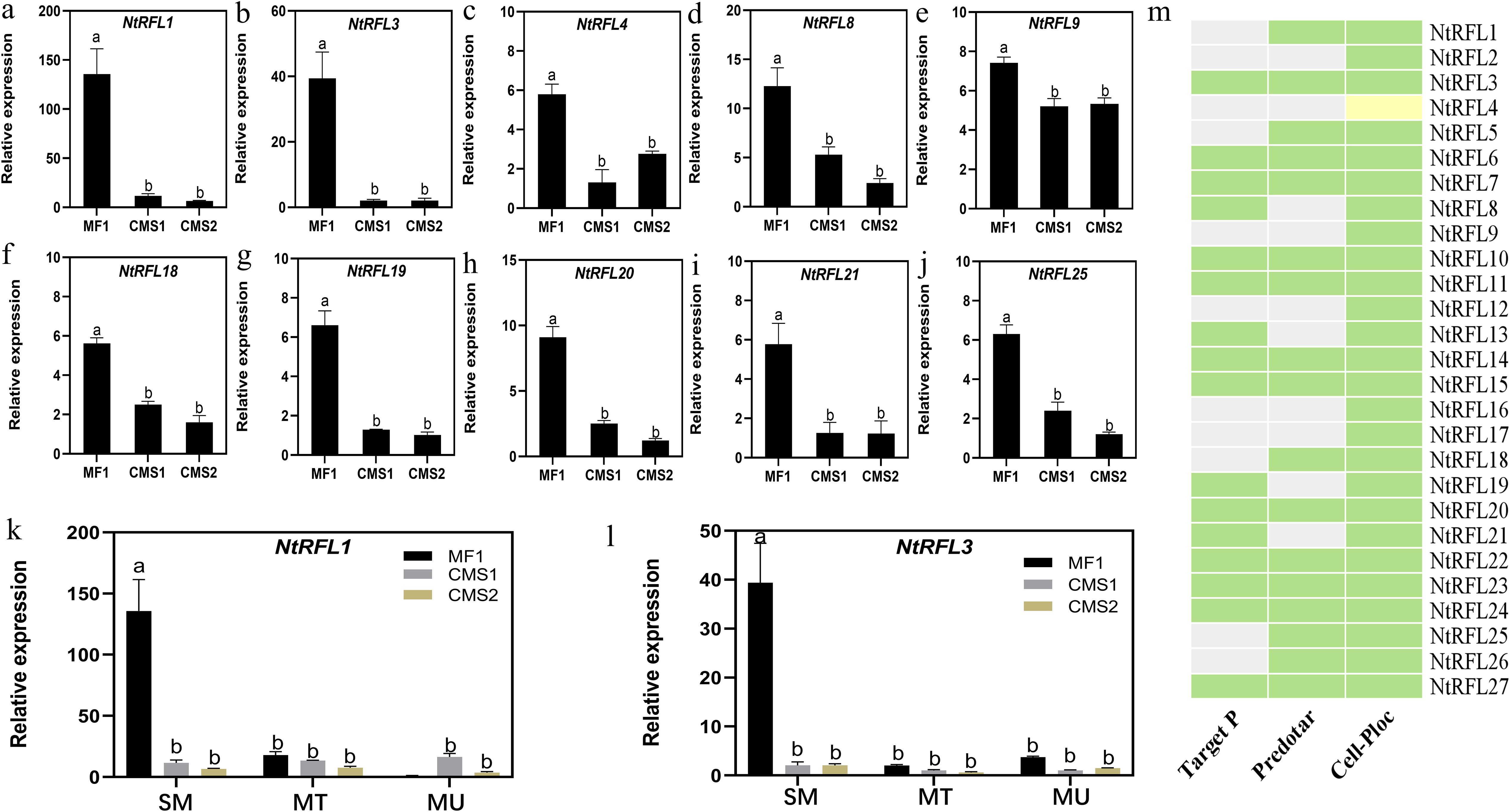
Figure 7. qPCR analysis and subcellular localization prediction of NtRFLs. (a-l) The expression of NtRFLs at sporogenous cell-microsporocyte stage (a-j) or at all three developmental stages (k-l) in MF1 (fertile line), CMS1 (semi-sterile line which possesses stamens but lacks pollen grain) and CMS2 (the sterile line which is stamenless). SM, sporogenous cell-microsporocyte stage; MT, meiosis-tetrad stage; MU, mid uninucleate stage. (m) Subcellular localization of NtRFLs. The green denoted mitochondria; the yellow are chloroplasts; the grey meant unsure.
RFLs can specifically regulate the transcription of male sterility genes in mitochondria and restore plant fertility (Fujii et al., 2011). The subcellular localization prediction of TargetP and Predotar software were proved to be highly consistent with that of fluorescence protein localization experiments (Lurin et al., 2004). Here, three different web tools were used for NtRFL1 and NtRFL3 subcellular localization prediction. It was demonstrated that they may localize in mitochondria (Figure 7m). As NtRFL3 had consistent expression patterns in both transcriptome data and qPCR analysis, and shared high similarity (79%) with petunia PPR592 which has been functionally characterized as a fertility restorer gene (Figure 3b), it was further selected as the candidate gene and speculated as a key regulator in early anther development and determination of plant fertility.
Discussion
Taking the advantage of next-generation sequencing technology, it is possible to identify and study gene families at the whole genome-wide level. At present, 441 PPR genes have been identified in Arabidopsis (Lurin et al., 2004), 491 PPRs in rice (O'Toole et al., 2008), 626 PPRs in poplar (Xing et al., 2018), 1079 PPRs in Brassica napus (Ning et al., 2020), 181 PPRs in grape (Che et al., 2022) and 105 in moss (Sugita et al., 2013). Common tobacco is derived from the natural genome doubling after hybridization between Nicotiana tomentosiformis and Nicotiana sylvestris (Clarkson et al., 2005). In this study, we identify 1002 PPR genes in allotetraploid common tobacco which is more than twice the number of diploid tomato (471 PPR) and Nicotiana tomentosiformis (487 PPR) (Ding, 2014), both belonging to Solanacea. Chromosome localization analysis showed that PPR genes distributed more widely on chromosomes and did not appear in clusters, so it was speculated that the functions of tobacco PPR gene family members may be more complex and diverse.
The Rf-PPR proteins constitute evolutionarily unique protein subgroups in the PPR family of angiosperms (Dahan and Mireau, 2013). In this study, 27 NtRFLs were identified in N.tabacum cultivar ZY300 through bioinformatics tools, all of which belonged to the P subfamily. However, the number of NtRFLs was much less than 1/10 of the members of the P subfamily which was indicated in previous studies. This may be related to the fact that Solanaceae species including tobaccos experienced a whole genome triploid event (Gebhardt, 2016), resulting in the loss of some RFL genes.
Rfs is a high linkage on chromosomes (Lee et al., 2014). Studies presented that multiple genetically linked Rf genes restoring identical CMS localized within the same restorer locus. For instance, two identified Rf genes found to colocate in Mimulus guttatus. The genetically linked Rf1 and Rf2 mapped in Mimulus guttatus were found to reside in chromosomal loci containing 12 and 6 Rf-like genes, respectively (Barr and Fishman, 2010). Similarly, the sorghum Rf5 locus was mapped to a 584-kb DNA fragment where researchers identified a cluster of 6 PPR exhibiting strong homology with the rice Rf1a (Jordan et al., 2011). Here, the synteny analysis revealed clustered distribution patterns of NtRFLs along chromosomes, with 15 NtRFL gene pairs unevenly distributed across 9 linkage groups.
Anther morphogenesis exhibits direct correlation with plant fertility. Developmental defects in anthers at any growth stage may lead to male sterility. CMS plants demonstrate diverse phenotypes in their impaired male reproductive organs, such as abnormal anther morphogenesis or defects in functional pollen development. In rice CMS systems, immature pollen grains show CMS-type-specific developmental arrest, with cytological differences in starch accumulation levels. PCD is crucial for normal anther formation, especially in tapetum and pollen sac wall cells (Ma, 2005). Premature or delayed tapetal PCD has been documented to induce male sterility through disrupted sporopollenin deposition and pollen wall patterning (Ji et al., 2013). In this study, we observed that tobacco CMS2 line remained undifferentiated with visible stamen primordium in 2-3 mm flower buds. The following developmental process ceased and the stamens were found to be completely degenerated when its flower became mature. Conversely, intermediate type CMS1 exhibited an undeveloped anther with lighter color and not filled with pollen grains, which was already noticed at the meiosis-tetrad period. These findings corroborate previous research indicating that stamens in male-sterile lines exhibit developmental anomalies at the floral bud stage, manifesting as tissue fusion with the pistil base and failure to undergo further differentiation (Cui et al., 2023).
Research evidence indicates that early anther developmental stages are critical for microspore fertility. Cytological studies have shown that pollen abortion predominantly initiates during the microspore mother cell differentiation phase and tetrad formation stage, highlighting the importance of early anther development in determining the fate of plant fertility (Li, 2015). The expression pattern analysis in this study showed that NtRFL1 and NtRFL3 were highly active at sporogenous cell-microsporocyte period of MF1, but near half of NtRFLs also had high expression in tobacco CMS lines. This has been seen in a previous study where the expression of NtomRf in MS K326 is higher than that in the maintainer line (Ding, 2014). It is hypothesized that the Rf gene remains expressed in male-sterile lines, but fails to be translated into functional proteins or form effective complexes.
Notably, gametophyte development involves exceptional energy demands. Tapetal cells exhibit mitochondrial densities up to 40-fold higher than somatic cells (Duroc et al., 2005). Such extraordinary mitochondrial proliferation likely provides essential energy support for pollen development. However, CMS associated gene products disrupt mitochondrial biogenesis, leading to metabolic dysfunction. This interference ultimately triggers premature PCD in the tapetum, resulting in pollen abortion. Studies demonstrates that all characterized restorer PPR proteins localize to mitochondria and function by specifically reducing the accumulation of CMS-associated mitochondrial RNA or protein. Furthermore, the protein-protein interaction network of Arabidopsis RFL proteins suggests their potential recruitment of chaperone partners (Hölzle et al., 2011; Fujii et al., 2016). RFL proteins lack intrinsic endonuclease activity, indicating their functional dependence on forming multimeric complexes with auxiliary protein factors. At present, the cloning and regulatory mechanism of CMS restoration genes in Nicotiana tabacum have not yet been realized. The molecular mechanism of how the NtRFL regulates the expression of CMS genes in mitochondria and affects plant fertility is still unknown, which requires further investigation.
Data availability statement
All relevant data is contained within the article: The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
MW: Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. YJ: Conceptualization, Investigation, Writing – original draft, Writing – review & editing. CZ: Investigation, Validation, Writing – review & editing. SD: Investigation, Writing – review & editing. RG: Investigation, Writing – review & editing. JW: Resources, Writing – review & editing. JL: Investigation, Writing – review & editing. QZ: Resources, Writing – review & editing. YL: Investigation, Writing – review & editing. AY: Validation, Writing – review & editing. YC: Project administration, Funding acquisition, Writing – review & editing. XZ: Supervision, Funding acquisition, Writing – review & editing. GL: Conceptualization, Supervision, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Agricultural Science and Technology Innovation Program (ASTIP-TRIC01), Natural Science Foundation of Shandong Province (ZR2022MC145), Science and Technology Project of China National Tobacco Corporation Sichuan Corporation (SCYC202302, SCYC202421, SCYC202508, YCQTSC202402), Science and Technology Project of China National Tobacco Corporation Fujian Corporation (2023350000200076), The Agricultural Science and Technology Innovation Program (ASTIP-TRIC-QH-2023B05) and International Foundation of Tobacco Research Institute of Chinese Academy of Agricultural Sciences (IFT202403).
Conflict of interest
Authors CZ, JW, QZ were employed by Deyang Company of Sichuan Provincial Tobacco Corporation. Author ZC was employed by Fujian Provincial Tobacco Company of China National Tobacco Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1591130/full#supplementary-material
References
Akagi, H., Nakamura, A., Yokozeki-Misono, Y., Inagaki, A., Takahashi, H., Mori, K., et al. (2004). Positional cloning of the rice rf-1 gene, a restorer of bt-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor. Appl. Genet. 108, 1449–1457. doi: 10.1007/s00122-004-1591-2
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME suite: tools for motif discovery and searching. Nucleic. Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Baranwal, V. K., Mikkilineni, V., Zehr, U. B., Tyagi, A. K., and Kapoor, S. (2012). Heterosis: emerging ideas about hybrid vigour. J. Exp. Bot. 63, 6309–6314. doi: 10.1093/jxb/ers291
Barr, C. M. and Fishman, L. (2010). The nuclear component of a cytonuclear hybrid incompatibility in mimulus maps to a cluster of Pentatricopeptide Repeat genes. Genetics 184, 455–465. doi: 10.1534/genetics.109.108175
Bentolila, S., Alfonso, A. A., and Hanson, M. R. (2002). A Pentatricopeptide Repeat-Containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. U. S. A. 99, 10887–10892. doi: 10.1073/pnas.102301599
Chao, J. T., Kong, Y. Z., Wang, Q., Sun, Y. H., Gong, D. P., Lv, J., et al. (2015). Mapgene2chrom, a tool to draw gene physical map based on perl and svg languages. Yi Chuan 37, 91–97. doi: 10.16288/j.yczz.2015.01.013
Che, L., Lu, S., Liang, G., Gou, H., Li, M., Chen, B., et al. (2022). Identification and expression analysis of the grape Pentatricopeptide Repeat (ppr) gene family in abiotic stress. Physiol. Mol. Biol. Plants 28, 1849–1874. doi: 10.1007/s12298-022-01252-x
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). Tbtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, S., Gutmann, B., Zhong, X., Ye, Y., Fisher, M. F., Bai, F., et al. (2016). Redefining the structural motifs that determine RNA binding and RNA editing by Pentatricopeptide Repeat proteins in land plants. Plant J. 85, 532–547. doi: 10.1111/tpj.13121
Clarkson, J. J., Lim, K. Y., Kovarik, A., Chase, M. W., Knapp, S., and Leitch, A. R. (2005). Long-term genome diploidization in allopolyploid Nicotiana section repandae (Solanaceae). New Phytol. 168, 241–252. doi: 10.1111/j.1469-8137.2005.01480.x
Cui, F. F., Meng, L. F., Liu, M. M., Zhang, J. Q., Wang, J. G., and Liu, Q. Y. (2023). Characteristics of MADS-box and SUPERMAN genes in tobacco cytoplasmic male sterile line K326. Crop J. 49, 3204–3214.
Cui, X., Wise, R. P., and Schnable, P. S. (1996). The Rf2 nuclear restorer gene of male-sterile t-cytoplasm maize. Science 272, 1334–1336. doi: 10.1126/science.272.5266.1334
Dahan, J. and Mireau, H. (2013). The Rf and Rf-like PPR in higher plants, a fast-evolving subclass of PPR genes. RNA Biol. 10, 1469–1476. doi: 10.4161/rna.25568
Desloire, S., Gherbi, H., Laloui, W., Marhadour, S., Clouet, V., Cattolico, L., et al. (2003). Identification of the fertility restoration locus, Rfo, in radish, as a member of the Pentatricopeptide-Repeat protein family. EMBO Rep. 4, 588–594. doi: 10.1038/sj.embor.embor848
Ding, A. M. (2014). Identification of PPR gene family in N.tomentosiformis and tomato and function analysis of Rf-related genes in tobacco. Chin. Acad. Agric. Sci. 2014, 33, 48.
Duroc, Y., Gaillard, C., Hiard, S., DeFrance, M. C., Pelletier, G., and Budar, F. (2005). Biochemical and functional characterization of orf138, a mitochondrial protein responsible for Ogura cytoplasmic male sterility in Brassiceae. Biochimie 87, 1089–1100. doi: 10.1016/j.biochi.2005.05.009
Fujii, S., Bond, C. S., and Small, I. D. (2011). Selection patterns on Restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc. Natl. Acad. Sci. U. S. A. 108, 1723–1728. doi: 10.1073/pnas.1007667108
Fujii, S., Suzuki, T., Giegé, P., Higashiyama, T., Koizuka, N., and Shikanai, T. (2016). The Restorer-of-fertility-like2 Pentatricopeptide Repeat protein and RNase P are required for the processing of mitochondrial orf291 RNA in Arabidopsis. Plant J. 86, 504–513. doi: 10.1111/tpj.13185
Gao, B., Ren, G., Wen, T., Li, H., Zhang, X., and Lin, Z. (2022). A super PPR cluster for restoring fertility revealed by genetic mapping, homocap-seq and de novo assembly in cotton. Theor. Appl. Genet. 135, 637–652. doi: 10.1007/s00122-021-03990-0
Gebhardt, C. (2016). The historical role of species from the solanaceae plant family in genetic research. Theor. Appl. Genet. 129, 2281–2294. doi: 10.1007/s00122-016-2804-1
Han, Z., Qin, Y., Li, X., Yu, J., Li, R., Xing, C., et al. (2020). A genome-wide analysis of Pentatricopeptide Repeat (PPR) protein-encoding genes in four Gossypium species with an emphasis on their expression in floral buds, ovules, and fibers in upland cotton. Mol. Genet. Genomics 295, 55–66. doi: 10.1007/s00438-019-01604-5
Hancock, W. G. and Lewis, R. S. (2017). Heterosis, transmission genetics, and selection for increased growth rate in a N.tabacum × synthetic tobacco cross. Mol. Breed. 37, 53. doi: 10.1007/s11032-017-0654-4
Hao, Y., Wang, Y., Wu, M., Zhu, X., Teng, X., Sun, Y., et al. (2019). The nuclear-localized PPR protein OsNPPR1 is important for mitochondrial function and endosperm development in rice. J. Exp. Bot. 70, 4705–4720. doi: 10.1093/jxb/erz226
Hayes, M. L., Dang, K. N., Diaz, M. F., and Mulligan, R. M. (2015). A conserved glutamate residue in the C-terminal deaminase domain of Pentatricopeptide Repeat proteins is required for RNA editing activity. J. Biol. Chem. 290, 10136–10142. doi: 10.1074/jbc.M114.631630
Hölzle, A., Jonietz, C., Törjek, O., Altmann, T., Binder, S., and Forner, J. (2011). A Restorer of fertility-like PPR gene is required for 5'-end processing of the Nad4 mRNA in mitochondria of Arabidopsis thaliana. Plant J. 65, 737–744. doi: 10.1111/j.1365-313X.2010.04460.x
Ichinose, M., Tasaki, E., Sugita, C., and Sugita, M. (2012). A PPR-DYW protein is required for splicing of a group ii intron of COX1 pre-mRNA in physcomitrella patens. Plant J. 70, 271–278. doi: 10.1111/j.1365-313X.2011.04869.x
Ji, C., Li, H., Chen, L., Xie, M., Wang, F., Chen, Y., et al. (2013). A novel rice BHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development. Mol. Plant 6, 1715–1718. doi: 10.1093/mp/sst046
Jiang, P., Zhang, X., Zhu, Y., Zhu, W., Xie, H., and Wang, X. (2007). Metabolism of reactive oxygen species in cotton cytoplasmic male sterility and its restoration. Plant Cell Rep. 26, 1627–1634. doi: 10.1007/s00299-007-0351-6
Jordan, D. R., Klein, R. R., Sakrewski, K. G., Henzell, R. G., Klein, P. E., and Mace, E.S. (2011). Mapping and characterization of Rf5: a new gene conditioning pollen fertility restoration in A1 and A2 cytoplasm in sorghum (Sorghum bicolor (l.) Moench). Theor. Appl. Genet. 123, 383–396. doi: 10.1007/s00122-011-1591-y
Kazama, T., Nakamura, T., Watanabe, M., Sugita, M., and Toriyama, K. (2008). Suppression mechanism of mitochondrial orf79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. Plant J. 55, 619–628. doi: 10.1111/j.1365-313X.2008.03529.x
Kitazaki, K., Arakawa, T., Matsunaga, M., Yui-Kurino, R., Matsuhira, H., Mikami, T., et al. (2015). Post-translational mechanisms are associated with fertility restoration of cytoplasmic male sterility in sugar beet (Beta vulgaris). Plant J. 83, 290–299. doi: 10.1111/tpj.12888
Klein, R. R., Klein, P. E., Mullet, J. E., Minx, P., Rooney, W. L., and Schertz, K.F. (2005). Fertility restorer locus Rf1 [corrected] of sorghum (Sorghum bicolor l.) Encodes a Pentatricopeptide Repeat protein not present in the colinear region of rice chromosome12. Theor. Appl. Genet. 111, 994–1012. doi: 10.1007/s00122-005-2011-y
Lee, Y. P., Cho, Y., and Kim, S. (2014). A high-resolution linkage map of the Rfd1, a restorer-of-fertility locus for cytoplasmic male sterility in radish (Raphanus sativus l.) produced by a combination of bulked segregant analysis and RNA-seq. Theor. Appl. Genet. 127, 2243–2252. doi: 10.1007/s00122-014-2376-x
Liu, D. D., Wang, J. Y., Tang, R. J., Chen, L., and Ma, C. L. (2021). Genome-wide identification of PPR gene family and expression analysis of albino related genes in tea plants. J. Tea Sci. 41 (02), 159–172. doi: 10.13305/j.cnki.jts.2021.02.002
Li, F. X. (2015). Important genes of tobacco: 9. Male sterility and fertility restoration genes of tobacco. Chin. Tobacco Sci. 36, 106–108.
Liu, Y. F. (2022). Tobacco sua-CMS infertility specification specific gene function analysis and inflammatory cytoplasm evaluation. Chin. Acad. Agric. Sci. doi: 10.27630/d.cnki.gznky.2022.000176
Lurin, C., Andrés, C., Aubourg, S., Bellaoui, M., Bitton, F., Bruyère, C., et al. (2004). Genome-wide analysis of arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 16, 2089–2103. doi: 10.1105/tpc.104.022236
Ma, H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56, 393–434. doi: 10.1146/annurev.arplant.55.031903.141717
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Ning, L., Wang, H., Li, D., Li, Y., Chen, K., Chao, H., et al. (2020). Genome-wide identification of the Restorer-of-Fertility-Like (RFL) gene family in Brassica napus and expression analysis in Shaan2a cytoplasmic male sterility. BMC Genomics 21, 765. doi: 10.1186/s12864-020-07163-z
O'Toole, N., Hattori, M., Andres, C., Iida, K., Lurin, C., Schmitz-Linneweber, C., et al. (2008). On the expansion of the Pentatricopeptide Repeat gene family in plants. Mol. Biol. Evolutionmolecular Biol. Evol. 25, 1120–1128. doi: 10.1093/molbev/msn057
Pak, H., Wang, H., Kim, Y., Song, U., Tu, M., Wu, D., et al. (2021). Creation of male-sterile lines that can be restored to fertility by exogenous methyl jasmonate for the establishment of a two-line system for the hybrid production of rice (Oryza sativa l.). Plant Biotechnol. J. 19, 365–374. doi: 10.1111/pbi.13471
Saha, D., Prasad, A. M., and Srinivasan, R. (2007). Pentatricopeptide Repeat proteins and their emerging roles in plants. Plant Physiol. Biochem. 45, 521–534. doi: 10.1016/j.plaphy.2007.03.026
Small, I. D. and Peeters, N. (2000). The PPR motif-a TPR-related motif prevalent in plant organellar proteins. Trends Biochem.Sci. 25, 46–47. doi: 10.1016/s0968-0004(99)01520-0
Sugita, M., Ichinose, M., Ide, M., and Sugita, C. (2013). Architecture of the PPR gene family in the moss Physcomitrella patens. RNA Biol. 10, 1439–1445. doi: 10.4161/rna.24772
Tavares-Carreón, F., Camacho-Villasana, Y., Zamudio-Ochoa, A., Shingú-Vázquez, M., Torres-Larios, A., Pérez-Martínez, X., et al. (2008). The Pentatricopeptide Repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. J. Biol. Chem. 283, 1472–1479. doi: 10.1074/jbc.M708437200
Ui, H., Sameri, M., Pourkheirandish, M., Chang, M. C., Shimada, H., Stein, N., et al. (2015). High-resolution genetic mapping and physical map construction for the fertility restorer Rfm1 locus in barley. Theor. Appl. Genet. 128, 283–290. doi: 10.1007/s00122-014-2428-2
Wang, Z., Zou, Y., Li, X., Zhang, Q., Chen, L., Wu, H., et al. (2006). Cytoplasmic male sterility of rice with boroii cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell. 18, 676–687. doi: 10.1105/tpc.105.038240
Xing, H., Fu, X., Yang, C., Tang, X., Guo, L., Li, C., et al. (2018). Genome-wide investigation of Pentatricopeptide Repeat gene family in poplar and their expression analysis in response to biotic and abiotic stresses. Sci. Rep. 8, 2817. doi: 10.1038/s41598-018-21269-1
Yamagishi, H., Jikuya, M., Okushiro, K., Hashimoto, A., Fukunaga, A., Takenaka, M., et al. (2021). A single nucleotide substitution in the coding region of ogura male sterile gene, orf138, determines effectiveness of a fertility restorer gene, Rfo, in radish. Mol. Genet. Genomics 296, 705–717. doi: 10.1007/s00438-021-01777-y
Yuan, Y. R., Xu, H. J., Hou, X., Liu, K., Liu, C. L., Zhao, H. Y., et al. (2018). Detection of tissue structure of tobacco leaf by microwave rapid paraffin sectioning method. Acta Tabacaria Sin. 24, 139–140. doi: 10.16472/j.Chinatobacco.2017.394
Zan, Y., Chen, S., Ren, M., Liu, G., Liu, Y., Han, Y., et al. (2025). The genome and genebank genomics of allotetraploid Nicotiana tabacum provide insights into genome evolution and complex trait regulation. Nat. Genet. 57, 986–996. doi: 10.1038/s41588-025-02126-0
Keywords: pentatricopeptide repeat (PPR) gene family, restoration of fertility like (RFL) gene, cytoplasmic male sterility (CMS), anther development, tobacco
Citation: Wu M, Ji Y, Zhang C, Du S, Gong R, Wang J, Li J, Zhong Q, Li Y, Yang A, Cheng Y, Zhang X and Liu G (2025) Genome-wide analysis of Rf-PPR-like genes in Nicotiana tabacum and their potential roles in anther development. Front. Plant Sci. 16:1591130. doi: 10.3389/fpls.2025.1591130
Received: 10 March 2025; Accepted: 13 May 2025;
Published: 11 September 2025.
Edited by:
Yong Jia, Murdoch University, AustraliaReviewed by:
Ira Vashisht, Jawaharlal Nehru University, IndiaZhao Xutao, Yunnan Academy of Agricultural Sciences, China
Copyright © 2025 Wu, Ji, Zhang, Du, Gong, Wang, Li, Zhong, Li, Yang, Cheng, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoxiang Liu, bGl1Z3VveGlhbmdAY2Fhcy5jbg==; Xingwei Zhang, emhhbmd4aW5nd2VpQGNhYXMuY24=; Yazhi Cheng, Zmp5Y2N5ekAxNjMuY29t
†These authors have contributed equally to this work
 Mengting Wu
Mengting Wu Yan Ji
Yan Ji Chengbei Zhang2†
Chengbei Zhang2† Jun Wang
Jun Wang Aiguo Yang
Aiguo Yang