- 1State Key Laboratory of Tree Genetics and Breeding, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, China
- 2Jiangsu Key Laboratory for the Research and Utilization of Plant Resources, Institute of Botany, Jiangsu Province and Chinese Academy of Sciences (Nanjing Botanical Garden Mem. Sun Yat-Sen), Nanjing, China
- 3Jiangxi Provincial Key Laboratory of Improved Variety Breeding and Efficient Utilization of Native Tree Species, Institute of Biological Resources, Jiangxi Academy of Sciences, Nanchang, China
Cinnamomum camphora (Lauraceae), an evergreen arborescent species endemic to East Asian ecosystems, is ecologically and economically prized for three cardinal attributes: decay-resistant xylem, aesthetic canopy architecture, and pharmacologically active terpenoid emissions. The plant-specific Lateral Organ Boundaries Domain (LBD) transcription factors mediate phylogenetically conserved developmental pathways governing lateral organogenesis and secondary metabolism across embryophytes. Despite multiple published C. camphora genome assemblies, functional characterization of LBD transcription factors in this species remains limited. We systematically identified 40 LBD genes through whole-genome analysis and characterized their structural features, evolutionary relationships, and expression patterns. Five are intron-free, while seven genes harbor two or more introns each. Detailed annotation of CcLBD promoter regions identified 33 cis-regulatory elements linked to hormone signaling and stress adaptation. Transcriptional dynamics of the 40 CcLBD genes were profiled across seven tissues of the camphor tree using short-read RNA-Seq, revealing that 22 genes were highly expressed in flowers and 12 were predominantly expressed in roots, suggesting potential roles in reproductive organ development and root formation in C. camphora. Phylogenetic analysis classified all CcLBD proteins into two clades, each harboring a conserved lateral organ boundaries (LOB) domain. Integrative omics analyses (small RNA-seq and degradome data) further implicated miR408 and miR2950c in post-transcriptional regulation of CcLBD5 via mRNA cleavage. These results establish a framework for the functional dissection of LBD-mediated developmental and stress-response pathways in C. camphora.
1 Introduction
Lateral organ boundaries domain (LBD) proteins, alternatively termed ASYMMETRIC LEAVES2-LIKE (ASL) proteins, are defined by a conserved lateral organ boundaries (LOB) domain (Iwakawa et al., 2002). This structural module contains three conserved elements: a zinc finger-like CX2CX6CX3C motif essential for DNA binding, a Gly-Ala-Ser (GAS) block, and a leucine zipper motif LX6LX3LX6L critical for protein dimerization (Majer and Hochholdinger, 2011). Phylogenetic analyses classify the LBD family into Class I (retaining functional leucine zippers) and Class II (lacking this motif), with Class I subdivided into five evolutionarily distinct subclasses (Ia-Ie) that constitute the majority of LBD members (Landschulz et al., 1988; Iwakawa et al., 2002; Shuai et al., 2002; Chanderbali et al., 2015; Liu et al., 2019).
As plant-exclusive transcription factors, these proteins orchestrate lateral organ formation in shoots and roots. They also regulate biological processes, including tissue regeneration and pollen maturation (Oh et al., 2010; Majer and Hochholdinger, 2011; Fan et al., 2012; Kim et al., 2015; Xu et al., 2016). AtLBD18 interacts with auxin response factors AtARF7 and AtARF19, enhancing their transcriptional activity to promote lateral root development (Pandey et al., 2018). Ectopic expression of AtLBD30 and AtLBD18 drives nonvascular cell reprogramming into tracheary elements, the core structural components of xylem vessels (Soyano et al., 2008). Recently, a phylogenetically related LBD member, similar to Arabidopsis AtLBD17 and AtLBD29, has been identified as crucial for lateral root and adventitious root development in tomato (Solanum lycopersicum) (Omary et al., 2022). Furthermore, another member of the Class IB LBD transcription factors, MtLBD16 in Medicago truncatula and ASL18/LjLBD16a in Lotus japonicus, plays critical roles in lateral root development and nodule formation (Schiessl et al., 2019; Soyano et al., 2019). Additionally, LBD and NAC proteins exhibit a crucial positive feedback regulatory mechanism essential for the growth modulation of Arabidopsis thaliana by regulating xylem cell differentiation (Soyano et al., 2008; Yamaguchi et al., 2010). In woody plants, LBD transcription factors are pivotal for secondary vascular development. Overexpression of PtaLBD1 in Populus tremula × P. alba promotes secondary phloem and xylem growth (Yordanov et al., 2010). In P. trichocarpa, PtrLBD39 and PtrLBD22 contribute to tension wood formation by mediating transcriptional responses to mechanical stress (Yu et al., 2022), while in Eucalyptus grandis, EgLBD22, EgLBD29, and EgLBD37 regulate phloem and xylem differentiation, influencing wood formation (Lu et al., 2018). Despite these advances, the functional characterization of LBD transcription factors in perennial woody species remains limited. Given the unique developmental traits of trees, including sustained secondary growth, vascular complexity, and long-term environmental adaptation, further research is needed to elucidate the evolutionary and functional diversification of LBD transcription factors in woody lineages.
MicroRNAs (miRNAs) are evolutionarily conserved small non-coding RNAs (18–24 nucleotides) that fine-tune gene expression post-transcriptionally (O’Brien et al., 2018). By binding complementary mRNA sequences, they induce transcript cleavage or translational repression, thereby modulating diverse biological processes (Rong et al., 2024). In Medicago truncatula, the miR390/TAS3 module regulates MtLBD17/29a via ARF2, coordinating lateral root and nodule development (Kirolinko et al., 2024). Similarly, in poplar, miR408 suppresses LBD15 expression, influencing lignification and biomass accumulation (Guo et al., 2023). While several miRNAs indirectly regulate LBD genes by targeting upstream transcription regulators (e.g., NAC1 or ARF family members), no direct miRNA–LBD interactions have been experimentally validated.
The Lauraceae family includes ecologically and economically vital species that dominate tropical and subtropical forests, serving as key resources in timber, medicinal compounds, spices, and essential oils. Cinnamomum camphora (camphor tree) is a particularly valuable species due to its aromatic properties and ecological significance (Meng et al., 2021). Widely cultivated in East Asia, it is a cornerstone of subtropical evergreen broadleaved forests and a major global producer of essential oils (Wang et al., 2022).
Despite the availability of multiple high-quality C. camphora genome assemblies (Wei-Hong et al., 2021; Jiang et al., 2022; Wang et al., 2022; Li et al., 2023), the functional roles of its LBD transcription factors remain largely unexplored. In this study, we present the first genome-wide identification and characterization of 40 LBD genes in C. camphora. Our comprehensive analysis revealed conserved LOB domain architectures, tissue-specific expression divergence, and cis-regulatory element landscapes within promoter regions. Evolutionary assessments resolved phylogenetic relationships among LBD subfamilies, while integrated sRNA-degradome data uncovered post-transcriptional regulation of CcLBD5 mediated by miR408/miR2950c through cleavage events. These findings provide a foundational framework for investigating LBD gene function in woody plants and shed new light on multilayered regulatory networks to govern organ development and stress responses in perennial species.
2 Materials and methods
2.1 Plant materials
The tissues and organs utilized in this study were sourced from there-year-old seedlings propagated from cultivar ‘Gantong1’ cuttings cultivated at the Jiangxi Academy of Sciences Nursery Base. To ensure reliability and statistical significance, three biological replicates were performed.
2.2 Sequence alignment and homolog identification
The genome and protein sequences of C. camphora cultivar ‘Gantong1’ were sourced from our prior study (https://ngdc.cncb.ac.cn/gwh/Assembly/23429/show) (Shen et al., 2022), which were then employed to construct a local BLAST database. Homology screening employed A. thaliana LBD proteins from TAIR as BLASTp queries, with a stringent e-value threshold of 1e-20 (Lamesch et al., 2011). Candidate LBD proteins in ‘Gantong1’ were screened using the LOB domain-specific hidden Markov model (PF03195) (Mistry et al., 2020). Domain validation was performed via SMART and HMMER tools under default settings (Finn et al., 2011; Letunic et al., 2011).
For functional characterization, the Expasy Protparam server predicted coding sequences (CDS), isoelectric points (pI), molecular weights (MW), and hydropathy indices (GRAVY) of all CcLBDs (Kyte and Doolittle, 1982; Wilkins et al., 1999). Subcellular localization predictions were further conducted using Cell-PLoc 2.0 (Chou and Shen, 2008).
2.3 Phylogenetic and conserved domains analysis
Protein sequences of A. thaliana and C. camphora were aligned with ClustalX 2.0 (Larkin et al., 2007). A Neighbor-Joining phylogenetic tree was constructed in MEGA-X, with node support evaluated by 1000 bootstrap replicates (Saitou and Nei, 1987; Kumar et al., 2018). Conserved motifs were identified through MEME suite analysis, configured to detect motifs of 6–50 amino acid widths with a six-motif limit per sequence (Bailey et al., 2009). Sequence logos representing motif conservation patterns were generated via WebLogo (Crooks et al., 2004).
2.4 Gene structure and promoter Cis-acting elements
The chromosomal distribution of CcLBD genes was analyzed using TBtools (v1.1047) (Chen et al., 2020). Gene structures (exon/intron organization) were annotated with GSDS 2.0, followed by visualization in TBtools (Hu et al., 2014; Chen et al., 2020). Promoter sequences (2000 bp upstream) were extracted from the C. camphora genome via Tbtools (Chen et al., 2020). Cis-regulatory elements were predicted using PlantCare, with statistically filtered results presented as a heatmap (Lescot et al., 2002).
2.5 Gene expression analysis
Transcriptome sequencing data from multiple C. camphora tissues (flowers, leaves, fruits, roots, young stems, developing xylem, and trunk phloem) were obtained from our previous study (Shen et al., 2022). Expression levels were normalized and converted to log10(TPM+1). Candidate CcLBD expression profiles were analyzed using RNA-Seq short-read data and visualized through pheatmap (v1.0.12).
For experimental validation, total RNA was isolated from high-polyphenol/polysaccharide-enriched tissues with the RNAprep Pure Plant Plus Kit (Tiangen), followed by first-strand cDNA synthesis (TaKaRa 5X PrimeScript™ RT Master Mix). Gene-specific primers designed via Beacon Designer 8 enabled qRT-PCR amplification (ViiA 7 Real-Time PCR System, Applied Biosystems) using PowerUp™ SYBR™ Green Master Mix. Relative expression levels were normalized to reference genes via the 2−ΔΔCt method.
2.6 Prediction and identification of miRNA-binding sites
Degradome and small RNA sequencing data (Accession: SRP127892) (Chen et al., 2018) were analyzed to identify miRNA-CcLBD interactions. Potential miRNA binding sites were predicted using psRNATarget (https://www.zhaolab.org/psRNATarget/). High-confidence targets were defined as alignments with CleaveLand4 (v4.5) (Addo-Quaye et al., 2008) category ≤2 and p-value ≤0.05.
3 Results
3.1 Identification of LBD genes in C. camphora
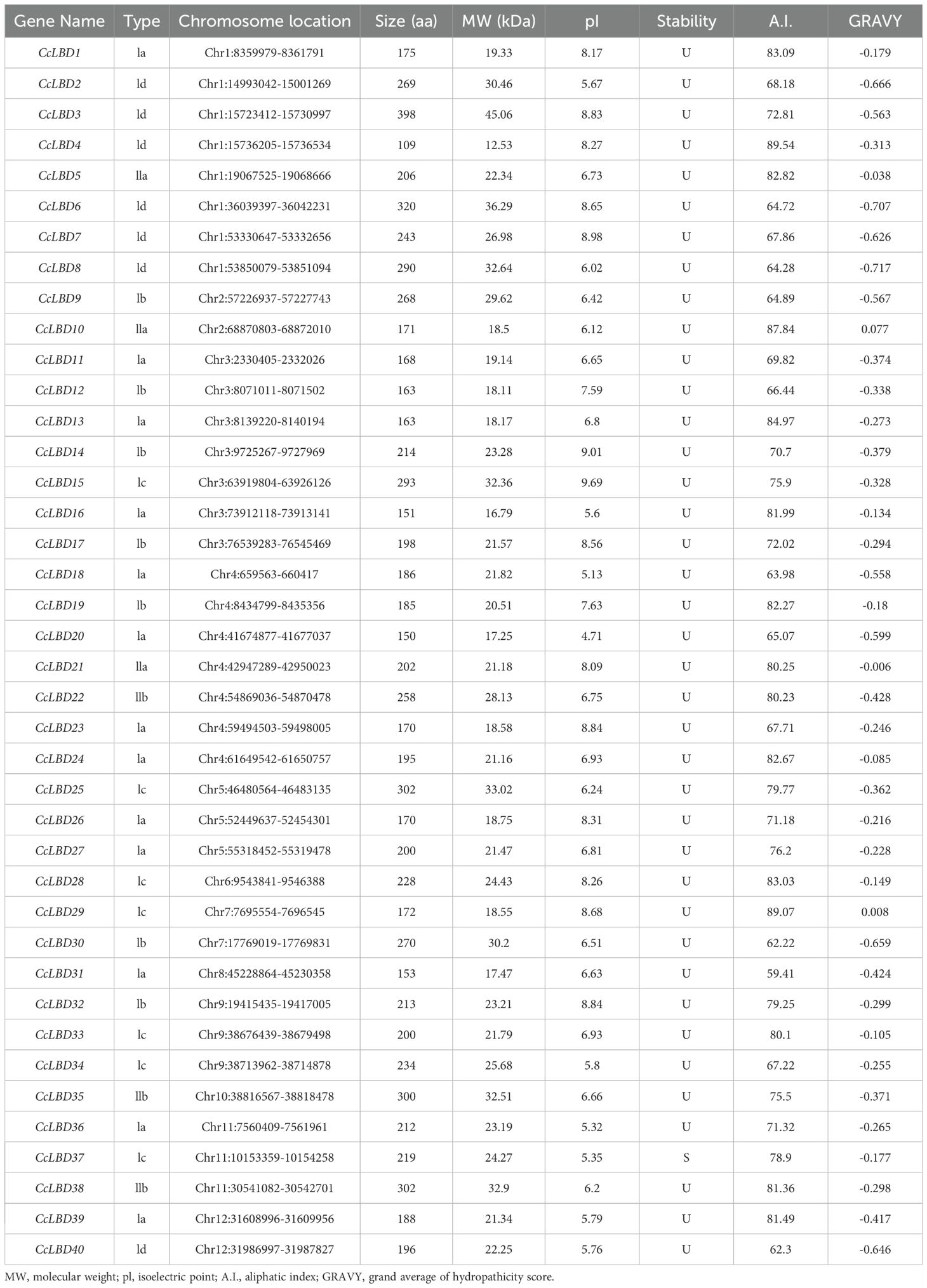
Through a systematic genome-wide investigation of C. camphora, we identified 40 unique LBD genes, designated CcLBD1 to CcLBD40 based on their chromosomal distribution. The encoded proteins exhibited substantial size diversity, with molecular weights spanning 12.53 kDa (CcLBD4, 109 amino acid residues) to 45.06 kDa (CcLBD3, 398 amino acid residues). Theoretical pI values spanned from 4.71 (CcLBD20) to 9.69 (CcLBD15), highlighting substantial electrostatic divergence within the LBD family. A striking 95% of family members exhibited negative GRAVY indices (-0.89 to -0.12), consistent with hydrophilic character. Subcellular localization predicted by Cell-PLoc uniformly placed all LBD members within the nuclear compartment (Table 1).
3.2 Phylogenetic relationships and conserved motif analysis
To delineate the phylogenetic relationships of LBD proteins, Neighbor-Joining analysis was performed using 43 A. thaliana and 40 C. camphora LBD homologs (Figure 1) (Shuai et al., 2002). The CcLBD family was phylogenetically stratified into Class I (subdivided into Ia:13, Ib:7, Ic:7, Id:7) and Class II (IIa:3, IIb:3), revealing lineage-specific expansion patterns. The absence of subclass Ie in C. camphora indicates evolutionary divergence from A. thaliana.
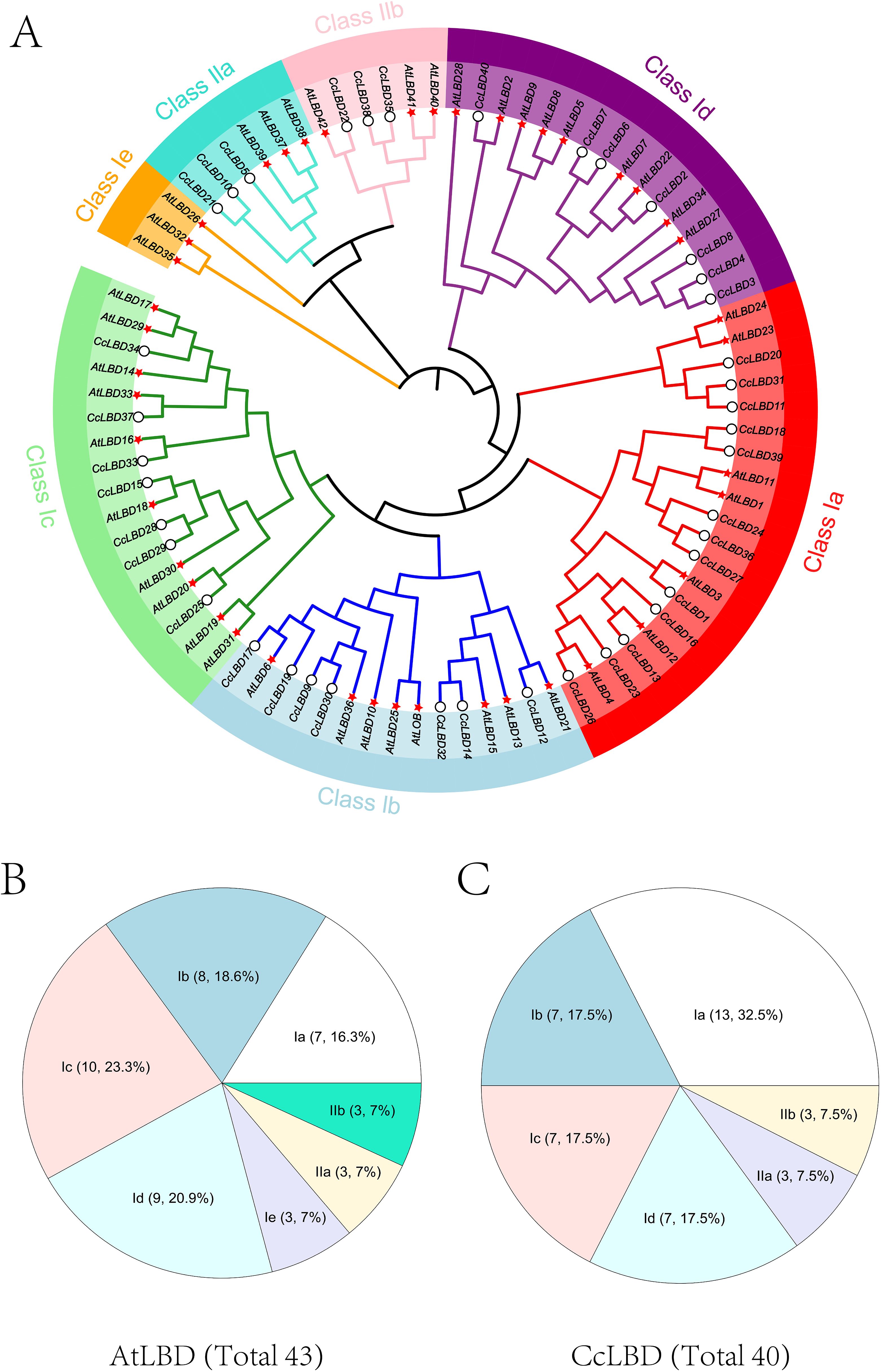
Figure 1. Phylogenetic relationships and subclass distribution of LBD proteins in A. thaliana (At) and C. camphora (Cc). (A) The circular cladogram delineates seven conserved clades (Class Ia, Ib, Ic, Id, Ie, IIa, IIb), with lineage-specific expansions highlighted by distinct colors. (B) Companion pie chart quantifies clade proportions: A. thaliana LBDs exhibit dominant Ic (23.3%) and Ib (18.6%) subclasses. (C) Companion pie chart quantifies clade proportions: C. camphora LBDs are enriched in Ia (32.5%) and IIb (17.5%) subclasses. Representative protein IDs (e.g., AtLBD39, CcLBD42) are annotated on terminal branches.
Conserved domain analysis delineated two signature architectures across CcLBD proteins: a 100-aa N-terminal LOB domain (Figure 2A) and a zinc finger-like motif (Figure 2B). Evolutionary conservation was evident as Class I members retained the angiosperm-typical leucine zipper domain, while complementary MEME analysis resolved six conserved motifs (Figure 3A). Notably, motifs 1–3 (LOB-associated) showed 97.5% conservation, whereas CcLBD3 uniquely harbored motifs 1–5, contrasting with the Class II-specific motif 6 (Supplementary Figure S1). Subclass specialization was observed, exemplified by motif six exclusivity to subclass II.
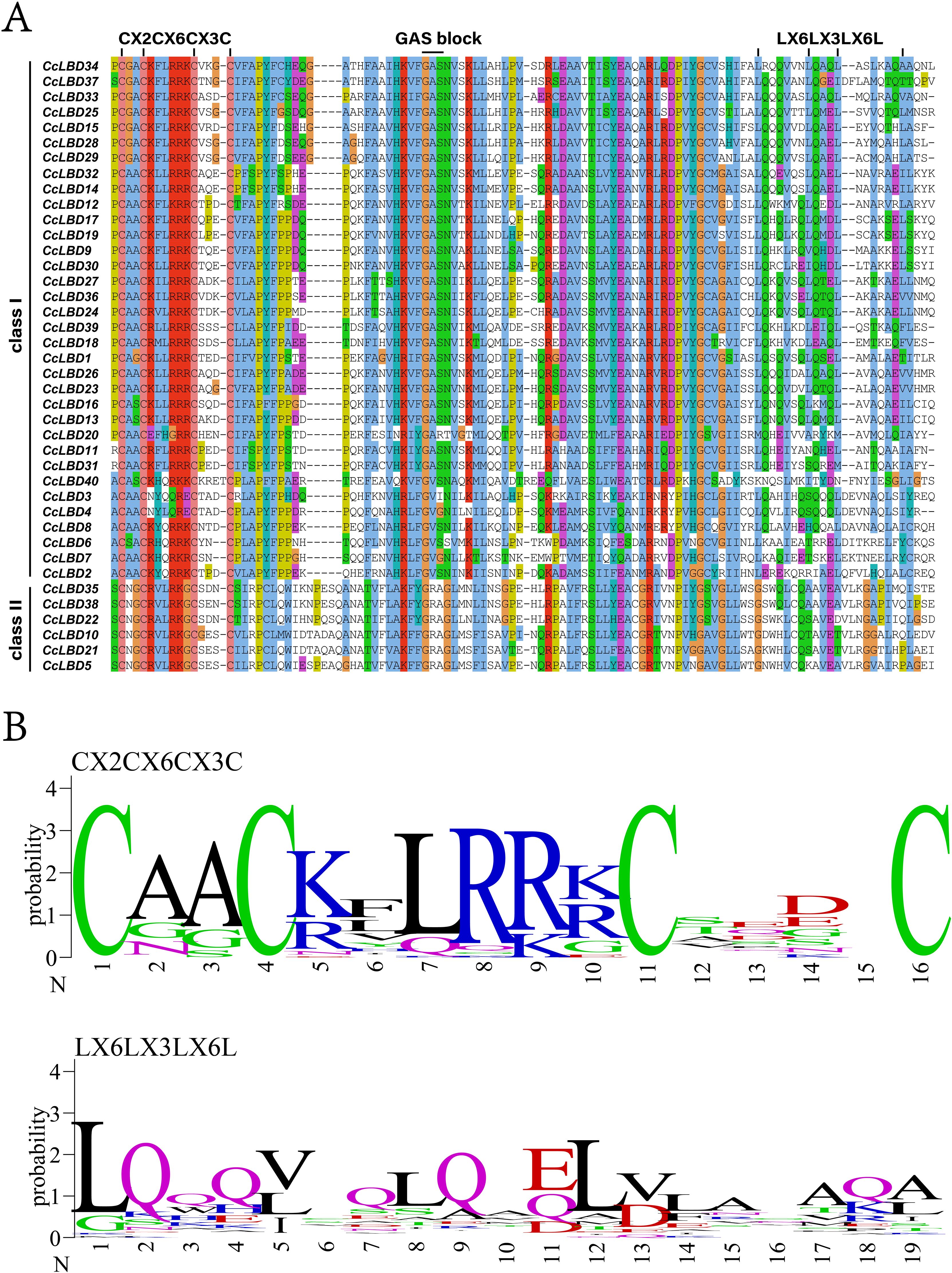
Figure 2. Conserved domain architecture of C. camphora LBD proteins. (A) Ubiquitous zinc finger-like CX2CX6CX3C domain across all 40 CcLBDs (color-coded), contrasting with Class I-specific leucine zipper-like LX6LX3LX6L motifs. (B) Domain sequence alignment (ClustalX 2.0) and corresponding motif logos (WebLogo-generated).
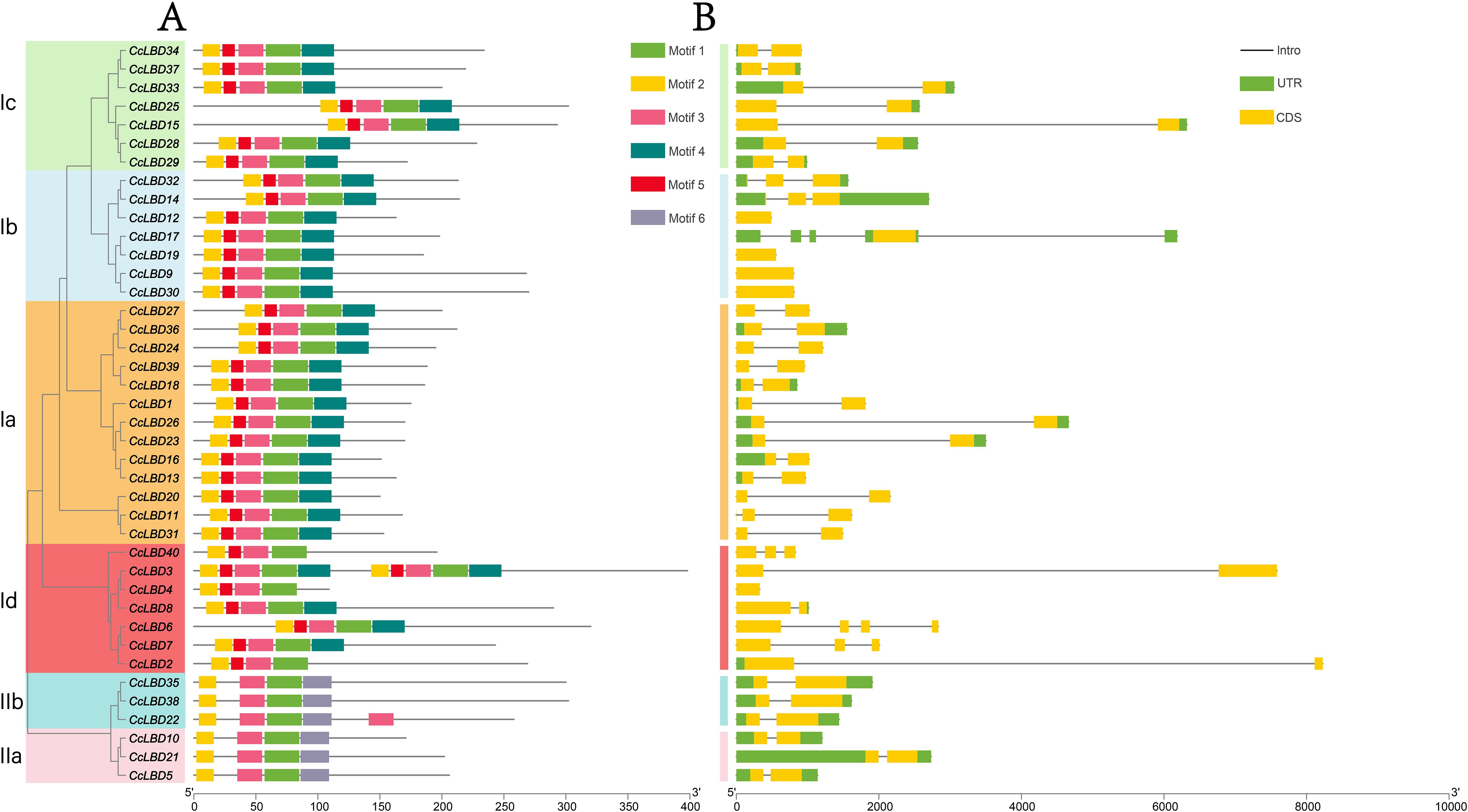
Figure 3. Conserved motifs and gene structures of CcLBDs. (A) Distribution of conserved motifs. The zinc finger-like CX2CX6CX3C domain is ubiquitous across all 40 CcLBDs, whereas the leucine zipper-like LX6LX3LX6L motif is restricted to Class I members. (B) Gene structure organization. Exon-intron architectures were analyzed via ClustalX2-based alignment and WebLogo-generated motif logos. Scale bars indicate gene length (bp) and protein sequence length (aa).
3.3 Gene structure and promoter elements
Gene structure investigation offers essential perspectives on the evolutionary trajectories of plant gene families (Li et al., 2017). Systematic alignment of CcLBD coding sequences with genomic DNA delineated the architectural organization of UTRs, exons, and introns (Figure 3B). Exon number variation was observed across the 40 CcLBDs, spanning 1–5 exons: 72.5% (29 genes) exhibited a two-exon configuration, contrasting with minority populations of single-exon (12.5%, five genes), three-exon (10%, four genes), and rare four-/five-exon (2.5%, one gene each) architectures. Subfamily-specific conservation of exon-intron structures was evident, displaying homologous exon length distributions and intron retention patterns. Mechanistically, intronic sequences mediated alternative splicing events and transcriptional fine-tuning, thereby expanding proteomic diversity through isoform generation (Greenwood and Kelsoe, 2003).
Promoter cis-element profiling of 2000-bp upstream regions from all CcLBD genes revealed 33 functional elements through PlantCare database annotation (Figure 4). Hormone-related elements encompassed ABRE (abscisic acid), CGTCA (MeJA), GARE (gibberellic acid), TCA (salicylic acid), and TGA (auxin) motifs. Stress-responsive elements included those associated with defense mechanisms, salt tolerance, and abiotic stress adaptation. These results indicate that CcLBD gene expression is modulated by cis-regulatory networks coordinating developmental processes and stress responses.
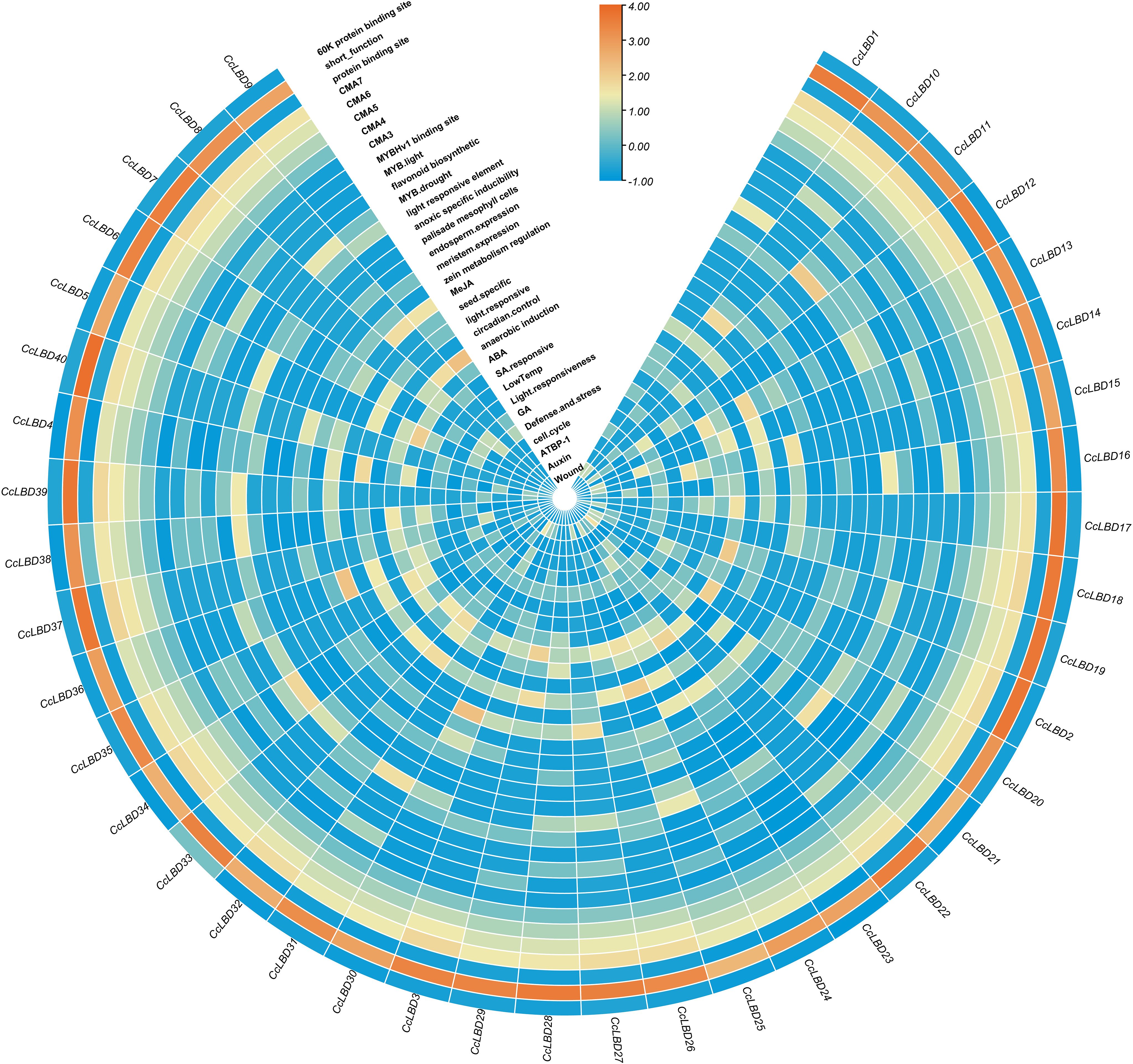
Figure 4. Cis-regulatory element profiling in C. camphora LBD promoters. The heatmap illustrates 33 functionally annotated cis-regulatory elements identified within 2,000-bp upstream promoter regions of 40 CcLBD genes. These elements include hormone-responsive motifs (e.g., ABRE, CGTCA, GARE, TCA, TGA) and stress-related elements involved in defense signaling, salt tolerance, and abiotic stress responses. Values represent log2-transformed counts of each element per gene, followed by row-wise Z-score normalization.
3.4 Genomic location and gene duplication events
The 40 CcLBD genes in the camphor tree are unevenly distributed across 12 chromosomes (Figure 5). Specifically, chromosomes 6, 8, and 10 each contain one CcLBD gene; chromosomes 2, 7, and 12 each harbor two genes; chromosomes 5, 9, and 11 each have three genes; chromosomes 3 and 4 each possess seven genes; and chromosome 1 harbors the highest number, with eight genes.
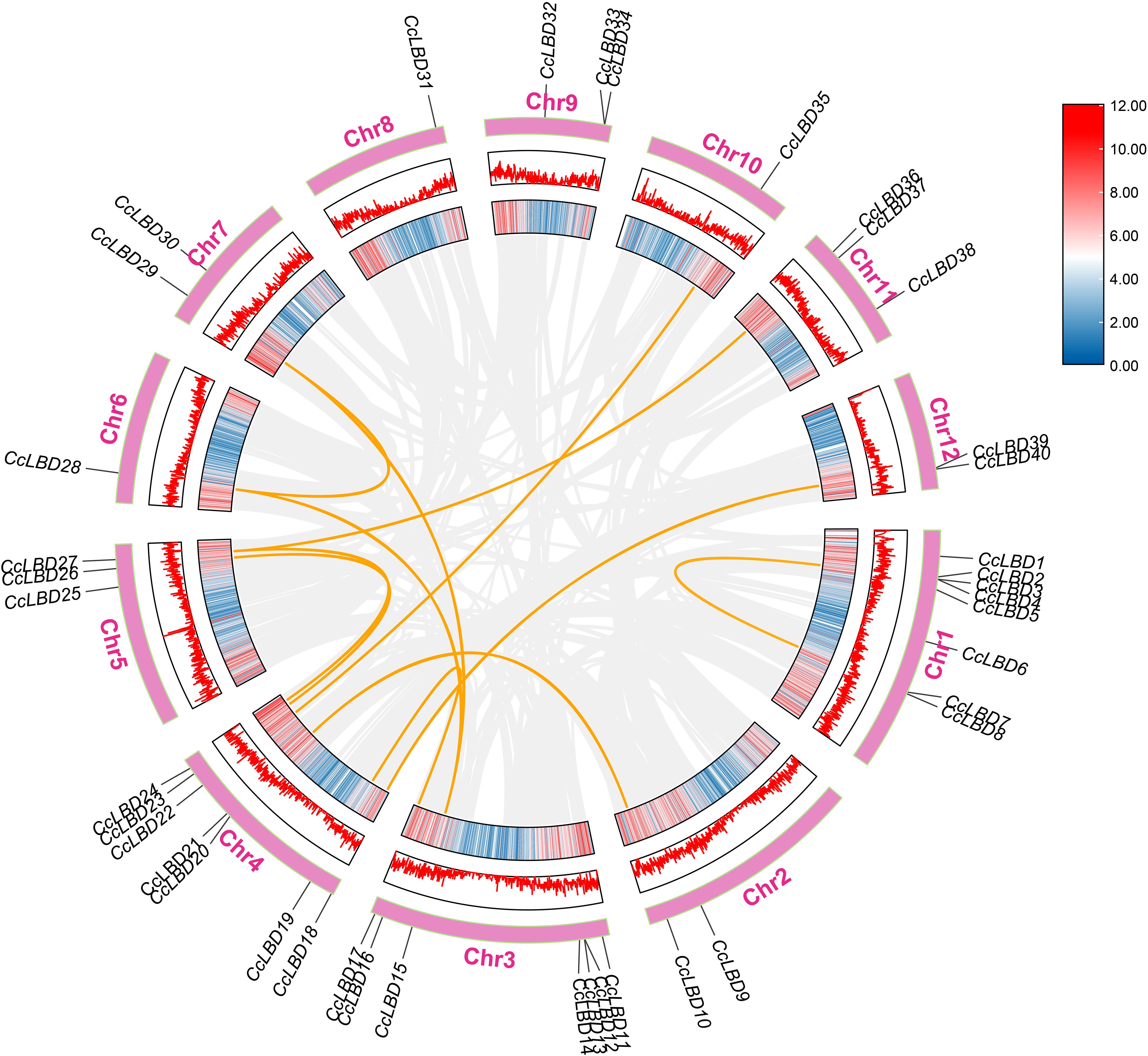
Figure 5. Chromosomal distribution and intraspecific collinearity of C. camphora LBD genes. CcLBD loci are annotated on chromosomes, with orange connectors marking tandem duplication events. The grey background lines represent syntenic relationships among C. camphora chromosomes, highlighting conserved genomic regions and gene collinearity across the genome.
Evolutionary analysis revealed duplication events among CcLBD genes (Figure 5). Among the 40 CcLBD genes, 27.5% (11 genes) originated from duplication events. Chromosomal distribution analysis revealed that chromosomes 8 and 9 lacked duplicated CcLBDs, whereas chromosome 4 harbored the highest duplication frequency (six gene pairs).
Evolutionary analysis of CcLBD duplicates employed Ka/Ks substitution rate calculations to estimate divergence timing. The Ka/Ks ratio (nonsynonymous/synonymous substitution rates) for duplicated CcLBD gene pairs spanned 0.0139–0.3828, with all ratios < 1 (mean = 0.157), demonstrating strong purifying selection (Supplementary Table S1) (Zhang et al., 2006).
3.5 Differential expression profiles
Transcriptome profiling is critical for elucidating gene functions in plant growth and development (Tong et al., 2013). Here, RNA-Seq was employed to assess transcriptional dynamics of 40 CcLBD genes across seven organs: flowers, leaves, fruits, roots, young stems, developing xylem, and trunk phloem. Expression levels were normalized to Transcripts Per Million (TPM) and visualized via heatmap (Figure 6). Distinct tissue-specific patterns emerged: 22 CcLBDs showed elevated expression in flowers, whereas 12 genes were root-enriched, suggesting organ-specific regulatory roles in C. camphora development. Intriguingly, genes within the same phylogenetic clade exhibited divergent expression patterns across tissues, implying functional diversification of CcLBD paralogs. Transcriptome profiling revealed pronounced tissue-specific expression heterogeneity among CcLBDs, with 22 genes preferentially expressed in floral tissues and 12 genes showing root-specific activation (Figure 6), suggesting specialized roles in organ development.
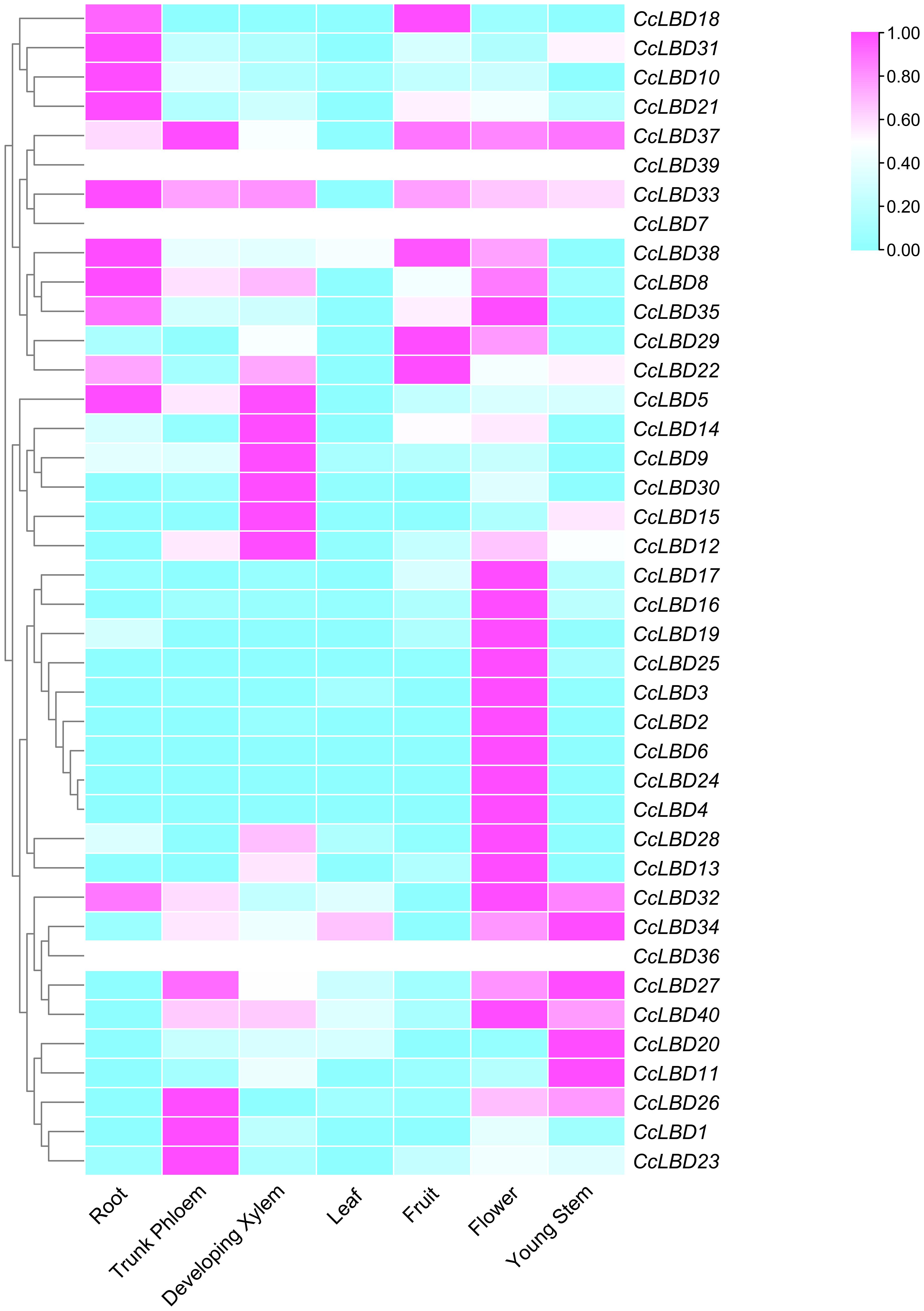
Figure 6. Transcriptional landscape of CcLBD genes in C. camphora. The heatmap displays log10(TPM+1) normalized expression values for 40 CcLBDs across seven tissues (root, trunk phloem, developing xylem, leaf, fruit, flower, young stem). Expression gradients are color-coded: purple (high) to blue (low).
To validate the transcriptomic findings, we conducted qRT-PCR analysis on seven representative CcLBD genes across multiple tissues (Figure 7; Supplementary Table S2). The results corroborated the RNA-Seq data, revealing pronounced tissue-specific expression patterns and suggesting potential subfunctionalization within the CcLBD family. CcLBD8 was exclusively expressed in flowers, indicating a specialized role in floral organ development. CcLBD21 displayed broad expression, with the highest transcript abundance in roots, followed by fruits and trunk phloem, suggesting its involvement in root development and secondary tissue formation. CcLBD22 and CcLBD38 were both highly expressed in fruits, implying a possible function in fruit maturation. CcLBD23 and CcLBD27 exhibited predominant expression in trunk phloem, with CcLBD27 also showing moderate expression in young stems, implying a coordinated role in phloem differentiation and early secondary vascular development. CcLBD33 demonstrated root-specific expression, pointing to a potential function in root patterning. These findings not only validate the RNA-Seq results but also underscore the functional divergence among CcLBD members, with distinct expression profiles reflecting specialized roles in organ development and physiological regulation in C. camphora.
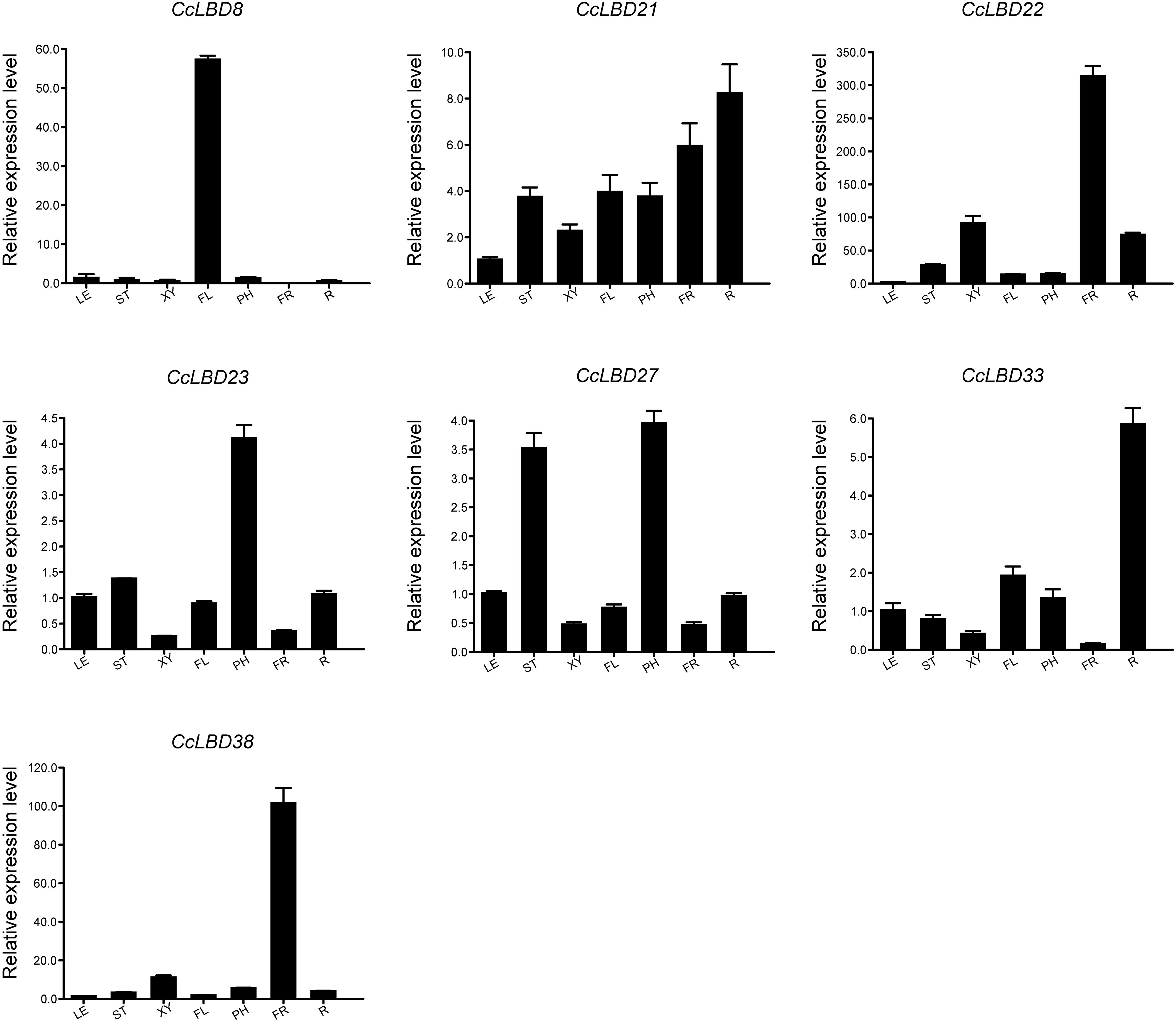
Figure 7. Tissue-specific expression profiles of seven CcLBD genes in C. camphora. Tissues analyzed: root, trunk phloem, developing xylem, leaf, fruit, flower, and young stem. Data represent mean ± SD from three biological replicates.
3.6 Regulation of CcLBD5 by microRNAs in the camphor tree
In this study, CleaveLand4 (version 4.5), a computational tool, was employed to categorize miRNA-mRNA interactions into three confidence levels (0, 1, and 2), facilitating a more accurate identification of potential regulatory miRNA-target pairs. Through degradome sequencing and subsequent computational analysis, two specific miRNA-CcLBD gene interactions were identified and characterized (Figure 8). Degradome analysis identified miR408 and miR2950c as post-transcriptional regulators of CcLBD5, with cleavage sites at positions 296 and 309, respectively. These findings demonstrate miRNAs’ regulatory impact on CcLBD gene networks, likely mediating C. camphora’s developmental plasticity and stress adaptation.
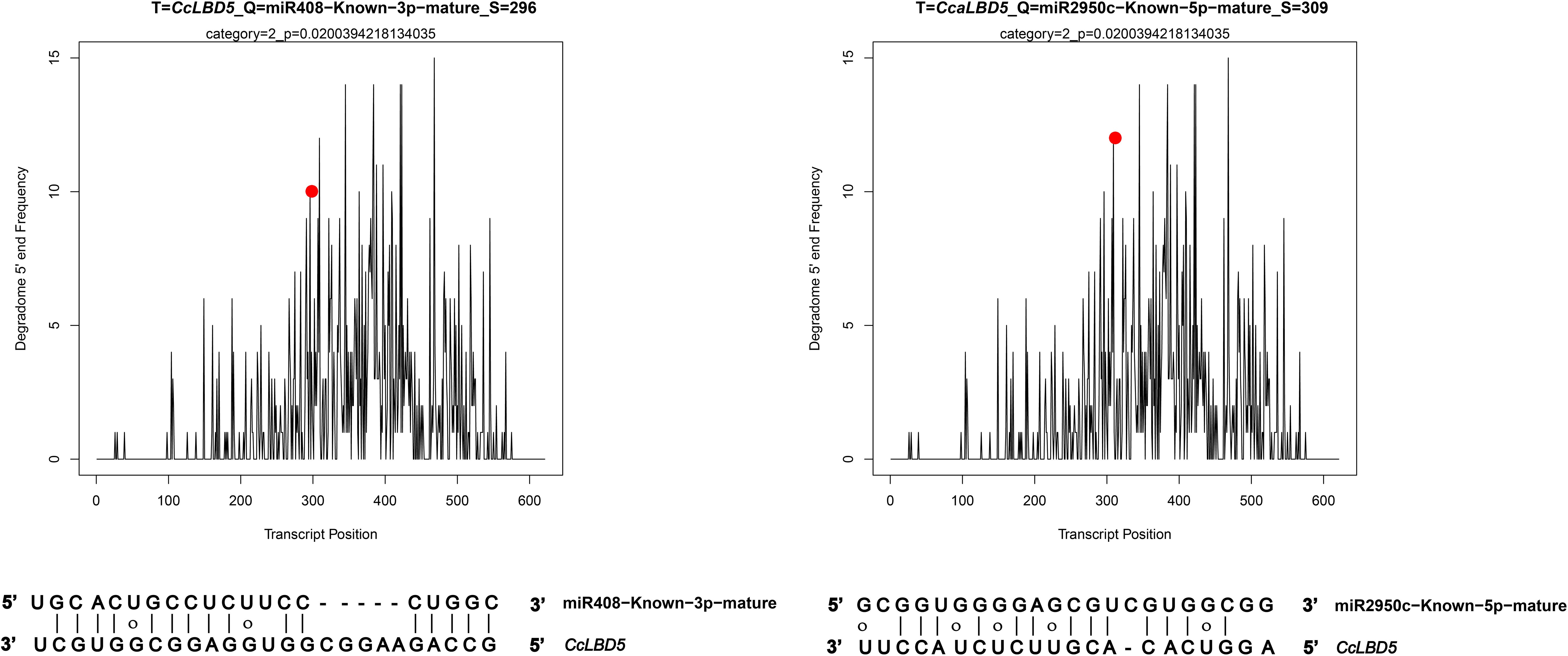
Figure 8. Degradome sequencing t-plots validate miR408 and miR2950c targeting CcLBD5 at positions 296 and 309, respectively.
4 Discussion
The LBD transcription factor family regulates plant-specific developmental pathways and demonstrates evolutionary conservation across angiosperms, as evidenced in A. thaliana and Oryza sativa (Majer and Hochholdinger, 2011; Xu et al., 2016). Utilizing whole-genome annotation approaches, we systematically identified 40 CcLBD genes in the C. camphora genome, revealing conserved domain architectures characteristic of this plant-specific TF family. These proteins were categorized by a conserved C-terminal leucine zipper-like domain (LX6LX3LX6L), with Class I containing 85% of members and Class II comprising the remaining 15%. This distribution reveals strong evolutionary selection for Class I expansion, mirroring patterns observed in model systems like A. thaliana and O. sativa (Shuai et al., 2002; Yang et al., 2006).
While the core domains of LBD proteins remain evolutionarily conserved, their biological functions have undergone significant diversification across plant lineages. In annual species like A. thaliana, key LBD proteins (e.g., AtLBD18, AtLBD29, and AtLBD30) primarily regulate lateral root formation and tracheary element differentiation through auxin-mediated signaling pathways (Soyano et al., 2008; Pandey et al., 2018). Similarly, in species such as S. lycopersicum and M. truncatula, LBD homologs are involved in adventitious root formation and nodule development (Schiessl et al., 2019; Soyano et al., 2019; Omary et al., 2022). In contrast, in perennial woody species such as Populus and Eucalyptus, LBD proteins are predominantly associated with secondary vascular development and wood formation, particularly through the regulation of xylem and phloem differentiation (Yordanov et al., 2010; Lu et al., 2018; Yu et al., 2022). Our qRT-PCR analysis revealed that several CcLBD genes (CcLBD21, CcLBD23, and CcLBD27) exhibit high expression in trunk phloem and developing xylem, suggesting their involvement in secondary growth regulation. These findings are consistent with the established roles of LBD proteins in other woody species and support the hypothesis that LBD proteins have evolved specialized functions to govern long-term growth processes in perennial plants.
Structural analysis of the CcLBD genes revealed a relatively conserved gene organization, with members of each subgroup exhibiting comparable exon/intron configurations. This structural uniformity among the CcLBD genes is in line with previous findings in A. thaliana, O. sativa, and Malus domestica (Shuai et al., 2002; Yang et al., 2006; Wang et al., 2013), suggesting a conserved structural basis for the LBD family across different angiosperms. Conserved motif analysis revealed that all CcLBD subclasses share motifs 1-3, which collectively constitute the conserved LOB domain architecture. Subclass-specific motifs were observed, with motif six exclusively localized to Class II members, reflecting functional divergence within the LBD family. A highly conserved CX2CX6CX3C zinc finger-like motif, critical for DNA binding, further underscores the transcriptional regulatory capacity of these proteins.
The promoter regions of CcLBD genes exhibit significant enrichment of signal-responsive cis-elements, suggesting coordinated transcriptional regulation during environmental adaptation. In Arabidopsis, LBD15 enhances drought tolerance via ABA-mediated transcriptional activation of ABSCISIC ACID INSENSITIVE4 (ABI4) (Guo et al., 2020). Similarly, ZmLBD5 adjusts gibberellin and abscisic acid homeostasis to mediate drought adaptation in Zea mays (Feng et al., 2022). In root development, AtLBD16/28/29/33 orchestrate lateral root formation via auxin signaling, a process dependent on ARF7 and ARF19-mediated auxin response elements (Okushima et al., 2007; Lee et al., 2009). Additionally, methyl jasmonate (MeJA) regulates LBD genes in Gossypium species, underscoring the intricate crosstalk between phytohormones and transcriptional networks (Li et al., 2020). Genome-wide screening identified 41 cis-regulatory elements associated with phytohormone signaling pathways (ABA, MeJA, IAA) in CcLBD promoter regions. Notably, 15 ABA-responsive elements (ABREs) and 11 stress-related motifs were systematically annotated, with ABREs ubiquitously present across all CcLBD promoters. This comprehensive cis-element landscape suggests that CcLBD genes orchestrate C. camphora’s transcriptional reprogramming during abiotic stress adaptation.
MiRNAs are well-established as crucial post-transcriptional regulators of plant development, primarily through their targeting of transcription factors that, in turn, regulate downstream gene expression networks (Rong et al., 2024). Previous studies have demonstrated that miRNAs such as miR390 and miR408 are indirect modulators of LBD-mediated developmental pathways, acting via upstream regulators including members of the ARF and NAC transcription factor families (Guo et al., 2023; Kirolinko et al., 2024). In this study, we report the first evidence of potential direct miRNA–LBD interactions in C. camphora. Through integrated degradome sequencing and computational analyses, we identified miR408 and miR2950c as putative direct regulators of CcLBD5, suggesting a lineage-specific post-transcriptional regulatory mechanism in this woody species. While these predictions are supported by bioinformatic evidence and partial degradome data, further experimental validation is required to confirm these interactions. Future research should employ 5′ RACE (Rapid Amplification of cDNA Ends) to definitively characterize miRNA-guided cleavage events. Moreover, comprehensive screening of miRNA-binding sites across the entire CcLBD gene family could reveal the broader regulatory network mediated by miRNAs in perennial woody plants, providing crucial insights into their unique developmental regulation.
5 Conclusion
This study establishes the first genome-wide inventory of 40 CcLBD genes in C. camphora, phylogenetically classified into six distinct subclasses (Ia, Ib, Ic, Id, IIa, IIb). Comprehensive characterization integrated chromosomal distributions, exon-intron organizational patterns, conserved protein domain architectures, promoter cis-regulatory landscapes, and tissue-preferential expression dynamics. Expanding beyond transcriptional regulation, miRNA interaction networks were mapped to reveal post-transcriptional control nodes targeting CcLBDs, notably identifying miR408 and miR2950c as key post-transcriptional regulators. These collective insights advance the functional annotation of LBD genes in camphor trees and provide a mechanistic scaffold for probing their roles in lineage-specific developmental adaptations.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
JZ: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. LS: Investigation, Software, Validation, Visualization, Writing – review & editing. HQ: Data curation, Investigation, Methodology, Validation, Writing – review & editing. CC: Conceptualization, Funding acquisition, Resources, Software, Validation, Writing – review & editing. YZ: Conceptualization, Funding acquisition, Resources, Validation, Writing – review & editing. MX: Conceptualization, Formal Analysis, Investigation, Project administration, Resources, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the National Natural Science Foundation of China (Grant Nos. 32160397), the Scientific Research Funding of Provincial Talent Training of Jiangxi Academy of Sciences (2023YRCS004), and the National Government Guidance Fund for Regional Science and Technology Development (20192ZDD01004).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1591736/full#supplementary-material
References
Addo-Quaye, C., Miller, W., and Axtell, M. J. (2008). CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25, 130–131. doi: 10.1093/bioinformatics/btn604
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Chanderbali, A. S., He, F., Soltis, P. S., and Soltis, D. E. (2015). Out of the water: origin and diversification of the LBD gene family. Mol. Biol. Evol. 32, 1996–2000. doi: 10.1093/molbev/msv080
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C., Zheng, Y., Zhong, Y., Wu, Y., Li, Z., Xu, L.-A., et al. (2018). Transcriptome analysis and identification of genes related to terpenoid biosynthesis in Cinnamomum camphora. BMC Genomics 19, 550. doi: 10.1186/s12864-018-4941-1
Chou, K.-C. and Shen, H.-B. (2008). Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 3, 153–162. doi: 10.1038/nprot.2007.494
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Fan, M., Xu, C., Xu, K., and Hu, Y. (2012). LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 22, 1169–1180. doi: 10.1038/cr.2012.63
Feng, X., Xiong, J., Zhang, W., Guan, H., Zheng, D., Xiong, H., et al. (2022). ZmLBD5, a class-II LBD gene, negatively regulates drought tolerance by impairing abscisic acid synthesis. Plant J. 112, 1364–1376. doi: 10.1111/tpj.v112.6
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Greenwood, T. A. and Kelsoe, J. R. (2003). Promoter and intronic variants affect the transcriptional regulation of the human dopamine transporter gene. Genomics 82, 511–520. doi: 10.1016/S0888-7543(03)00142-3
Guo, Y., Wang, S., Yu, K., Wang, H.-L., Xu, H., Song, C., et al. (2023). Manipulating microRNA miR408 enhances both biomass yield and saccharification efficiency in poplar. Nat. Commun. 14, 4285. doi: 10.1038/s41467-023-39930-3
Guo, Z., Xu, H., Lei, Q., Du, J., Li, C., Wang, C., et al. (2020). The Arabidopsis transcription factor LBD15 mediates ABA signaling and tolerance of water-deficit stress by regulating ABI4 expression. Plant J. 104, 510–521. doi: 10.1111/tpj.v104.2
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2014). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., et al. (2002). The ASYMMETRIC LEAVES2 gene of arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43, 467–478. doi: 10.1093/pcp/pcf077
Jiang, R., Chen, X., Liao, X., Peng, D., Han, X., Zhu, C., et al. (2022). A chromosome-level genome of the camphor tree and the underlying genetic and climatic factors for its top-geoherbalism. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.827890
Kim, M.-J., Kim, M., Lee, M. R., Park, S. K., and Kim, J. (2015). LATERAL ORGAN BOUNDARIES DOMAIN (LBD)10 interacts with SIDECAR POLLEN/LBD27 to control pollen development in Arabidopsis. Plant J. 81, 794–809. doi: 10.1111/tpj.2015.81.issue-5
Kirolinko, C., Hobecker, K., Cueva, M., Botto, F., Christ, A., Niebel, A., et al. (2024). A lateral organ boundaries domain transcription factor acts downstream of the auxin response factor 2 to control nodulation and root architecture in Medicago truncatula. New Phytol. 242, 2746–2762. doi: 10.1111/nph.v242.6
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kyte, J. and Doolittle, R. F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. doi: 10.1016/0022-2836(82)90515-0
Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2011). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210. doi: 10.1093/nar/gkr1090
Landschulz, W. H., Johnson, P. F., and McKnight, S. L. (1988). The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins Science 240, 1759–1764. doi: 10.1126/science.3289117
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee, H. W., Kim, N. Y., Lee, D. J., and Kim, J. (2009). LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 151, 1377–1389. doi: 10.1104/pp.109.143685
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Doerks, T., and Bork, P. (2011). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. doi: 10.1093/nar/gkr931
Li, P., Jiang, Z., Wang, Y., Deng, Y., Van Nostrand, J. D., Yuan, T., et al. (2017). Analysis of the functional gene structure and metabolic potential of microbial community in high arsenic groundwater. Water Res. 123, 268–276. doi: 10.1016/j.watres.2017.06.053
Li, H., Li, J., Chen, J., Yan, L., and Xia, L. (2020). Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol. Plant 13, 671–674. doi: 10.1016/j.molp.2020.03.011
Li, D., Lin, H.-Y., Wang, X., Bi, B., Gao, Y., Shao, L., et al. (2023). Genome and whole-genome resequencing of Cinnamomum camphora elucidate its dominance in subtropical urban landscapes. BMC Biol. 21, 192. doi: 10.1186/s12915-023-01692-1
Liu, H., Cao, M., Chen, X., Ye, M., Zhao, P., Nan, Y., et al. (2019). Genome-wide analysis of the lateral organ boundaries domain (LBD) gene family in solanum tuberosum. Int. J. Mol. Sci. 20, 5360. doi: 10.3390/ijms20215360
Lu, Q., Shao, F., Macmillan, C., Wilson, I. W., van der Merwe, K., Hussey, S. G., et al. (2018). Genomewide analysis of the lateral organ boundaries domain gene family in Eucalyptus grandis reveals members that differentially impact secondary growth. Plant Biotechnol. J. 16, 124–136. doi: 10.1111/pbi.2018.16.issue-1
Majer, C. and Hochholdinger, F. (2011). Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 16, 47–52. doi: 10.1016/j.tplants.2010.09.009
Meng, J., Li, M., Guo, J., Zhao, D., and Tao, J. (2021). Predicting suitable environments and potential occurrences for cinnamomum camphora (Linn.) presl. Forests 12, 1126. doi: 10.3390/f12081126
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar Gustavo, A., Sonnhammer, E. L. L., et al. (2020). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
O’Brien, J., Hayder, H., Zayed, Y., and Peng, C. (2018). Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 9, 402. doi: 10.3389/fendo.2018.00402
Oh, S. A., Park, K. S., Twell, D., and Park, S. K. (2010). The SIDECAR POLLEN gene encodes a microspore-specific LOB/AS2 domain protein required for the correct timing and orientation of asymmetric cell division. Plant J. 64, 839–850. doi: 10.1111/j.1365-313X.2010.04374.x
Okushima, Y., Fukaki, H., Onoda, M., Theologis, A., and Tasaka, M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19, 118–130. doi: 10.1105/tpc.106.047761
Omary, M., Gil-Yarom, N., Yahav, C., Steiner, E., Hendelman, A., and Efroni, I. (2022). A conserved superlocus regulates above- and belowground root initiation. Science 375, eabf4368. doi: 10.1126/science.abf4368
Pandey, S. K., Lee, H. W., Kim, M.-J., Cho, C., Oh, E., and Kim, J. (2018). LBD18 uses a dual mode of a positive feedback loop to regulate ARF expression and transcriptional activity in Arabidopsis. Plant J. 95, 233–251. doi: 10.1111/tpj.2018.95.issue-2
Rong, F., Lv, Y., Deng, P., Wu, X., Zhang, Y., Yue, E., et al. (2024). Switching action modes of miR408-5p mediates auxin signaling in rice. Nat. Commun. 15, 2525. doi: 10.1038/s41467-024-46765-z
Saitou, N. and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Schiessl, K., Lilley, J. L. S., Lee, T., Tamvakis, I., Kohlen, W., Bailey, P. C., et al. (2019). NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in medicago truncatula. Curr. Biol. 29, 3657–3668.e3655. doi: 10.1016/j.cub.2019.09.005
Shen, T, Qi, H, Luan, X, Xu, W, Yu, F, Zhong, Y, and Xu, M. (2022). The chromosome-level genome sequence of the camphor tree provides insights into Lauraceae evolution and terpene biosynthesis. Plant Biotechnol. J 20(2), 244–246. doi: 10.1111/pbi.13749
Shuai, B., Reynaga-Peña, C. G., and Springer, P. S. (2002). The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 129, 747–761. doi: 10.1104/pp.010926
Soyano, T., Shimoda, Y., Kawaguchi, M., and Hayashi, M. (2019). A shared gene drives lateral root development and root nodule symbiosis pathways in Lotus. Science 366, 1021–1023. doi: 10.1126/science.aax2153
Soyano, T., Thitamadee, S., Machida, Y., and Chua, N.-H. (2008). ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 20, 3359–3373. doi: 10.1105/tpc.108.061796
Tong, C., Wang, X., Yu, J., Wu, J., Li, W., Huang, J., et al. (2013). Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genomics 14, 689. doi: 10.1186/1471-2164-14-689
Wang, X.-D., Xu, C.-Y., Zheng, Y.-J., Wu, Y.-F., Zhang, Y.-T., Zhang, T., et al. (2022). Chromosome-level genome assembly and resequencing of camphor tree (Cinnamomum camphora) provides insight into phylogeny and diversification of terpenoid and triglyceride biosynthesis of Cinnamomum. Horticult Res. 9, uhac216. doi: 10.1093/hr/uhac216
Wang, X., Zhang, S., Su, L., Liu, X., and Hao, Y. (2013). A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES domain) gene family in malus domestica with a functional characterization of mdLBD11. PloS One 8, e57044. doi: 10.1371/journal.pone.0057044
Wei-Hong, S., Xiang, S., Zhang, Q.-G., Xiao, L., Zhang, D., Zhang, P., et al. (2021). The camphor tree genome enhances the understanding of magnoliid evolution. J. Genet. Genomics. 49, 249–253. doi: 10.1016/j.jgg.2021.11.001
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J.-C., Williams, K. L., Appel, R. D., et al. (1999). “Protein identification and analysis tools in the exPASy server,” in 2-D Proteome Analysis Protocols. Eds. Link, A. J. and Totowa, N. J. (New Jersey, USA: Humana Press), 531–552.
Xu, C., Luo, F., and Hochholdinger, F. (2016). LOB domain proteins: beyond lateral organ boundaries. Trends Plant Sci. 21, 159–167. doi: 10.1016/j.tplants.2015.10.010
Yamaguchi, M., Ohtani, M., Mitsuda, N., Kubo, M., Ohme-Takagi, M., Fukuda, H., et al. (2010). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in arabidopsis. Plant Cell 22, 1249–1263. doi: 10.1105/tpc.108.064048
Yang, Y., Yu, X., and Wu, P. (2006). Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenet. Evol. 39, 248–262. doi: 10.1016/j.ympev.2005.09.016
Yordanov, Y. S., Regan, S., and Busov, V. (2010). Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in Populus. Plant Cell 22, 3662–3677. doi: 10.1105/tpc.110.078634
Yu, J., Zhou, C., Li, D., Li, S., Jimmy Lin, Y.-C., Wang, J. P., et al. (2022). A PtrLBD39-mediated transcriptional network regulates tension wood formation in Populus trichocarpa. Plant Commun. 3, 100250. doi: 10.1016/j.xplc.2021.100250
Keywords: camphor tree, Lateral Organ Boundaries (LOB), microRNA (miRNA), transcription factors, miRNA targeting
Citation: Zhang J, Shan L, Qi H, Chen C, Zhong Y and Xu M (2025) Genome-wide identification, transcriptional profiling, and miRNA-binding site analysis of the LBD gene family in the camphor tree. Front. Plant Sci. 16:1591736. doi: 10.3389/fpls.2025.1591736
Received: 11 March 2025; Accepted: 02 June 2025;
Published: 19 June 2025.
Edited by:
Changlin Liu, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Mingyang Quan, Beijing Forestry University, ChinaShichao Liu, Chinese Academy of Tropical Agricultural Sciences, China
Copyright © 2025 Zhang, Shan, Qi, Chen, Zhong and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meng Xu, eHVtQG5qZnUuZWR1LmNu; Yongda Zhong, emhvbmd5b25nZGEwNTA0QDE2My5jb20=
 Jiaqi Zhang1
Jiaqi Zhang1 Meng Xu
Meng Xu