- 1Institute of Forest Biotechnology, Forestry College, Hebei Agricultural University, Baoding, China
- 2Hebei Key Laboratory for Tree Genetic Resources and Forest Protection, Baoding, China
- 3Flower Research Institute, Langfang Academy of Agriculture and Forestry Sciences, Langfang, China
- 4State-owned Assets Supervision and Administration, Hebei Agricultural University, Baoding, China
The plant-specific NAC transcription factor family has the largest number of members, they are involved in regulating the entire process of plant growth and development, and the responses to biotic and abiotic stressors. This article reviews the progress regarding the elucidation of the structure and function of the NAC transcription factor family and the regulatory mechanisms through which NAC transcription factors affect various processes in woody plants, including the formation of the secondary cell wall, seed development, flowering, and fruit ripening. Additionally, this article encompasses the current research status of NAC transcription factors in response to abiotic stressors, such as salt and drought, as well as biotic stressors, such as pathogens and pests in woody plants. Most of the research is still at the stages of gene cloning, structural identification, and functional analysis; the specific downstream target genes and their molecular mechanisms remain unclear. Future research should focus on exploring unknown functions and action mechanisms, to promote an understanding of the regulatory network of NAC transcription factors in woody plants, thereby providing a theoretical basis and gene resources for research on NAC transcription factors and the creation of new forest germplasm.
Transcription factors (TFs), also called trans-acting factors, are DNA-binding protein molecules with sequence specificity that regulate gene expression. NAC TFs are among the largest TF family in plants (Duval et al., 2002). The name is derived from the initials of the NAM (no apical meristem) gene in Petunia hybrida (Souer et al., 1996), the ATAF1/2 (Arabidopsis transcription activation factor) genes in Arabidopsis thaliana (Jensen et al., 2013), and the CUC2 (cup-shaped cotyledon) gene in A. thaliana (Aida et al., 1997). The N-terminus of NAC TFs contains a highly conserved region of approximately 150 amino acid residues called the NAC domain, while the C-terminus is a diverse transcriptional regulatory region. NAC TFs have been found in dozens of plants, and their structure, expression characteristics, and functions reveal remarkable results (Zhao et al., 2020a). The NAC TF family genes are promising for plant breeding. Although many studies have reported on the functions of NAC TF genes, the functions of most members remain unclear, particularly in woody plants. Therefore, this review elaborates on the discovery and structural characteristics of NAC TFs, further discusses the gene classification and identification of NAC TFs in woody plants, and summarizes their regulatory roles in various growth and development processes, including seed germination, organ senescence, secondary-wall formation, flowering, fruit ripening, regulation of plant height, root development, and hormonal regulation in woody plants. This article also elaborates on the key roles of NAC TFs in the responses of woody plants to biotic and abiotic stressors.
1 Discovery and structural features of NAC transcription factors
In 1996, researchers first isolated (Souer et al., 1996) the NAC TF NAM from Petunia hybrida and reported an association with embryonic development (Zhong et al., 2023). Subsequently, researchers successively identified a variety of NAC-type TFs from woody plants, such as Eucommia ulmoides (Zhang et al., 2023b) Camellia nitidissima (Liu et al., 2025), and Salix matsudana (Qian et al., 2024). Many NAC TF genes have been discovered and cloned. The plant TF database has 19,997 NAC TFs documented from 150 species (Liu, 2023).
The common feature of these TFs is the presence of the NAC domain at the N-terminus, which consists of 150 highly conserved amino acid residues. NAC TFs possess unique structural characteristics. A typical NAC TF consists of a conserved N-terminal protein domain and a variable C-terminal transcriptional regulatory region. The N terminus is involved in the specific binding of cis-acting elements, while the C terminus is responsible for regulating transcriptional activation (Ooka et al., 2003) as shown in Figure 1A (a). In contrast, as depicted in Figure 1A (b–f). An atypical NAC TF may contain a single NAC domain, two tandemly repeated NAC domains, or have the NAC domain at the C-terminus and a highly variable transcription regulation (TR) region at the N-terminus (Zhao et al., 2020a).The NAC domain is relatively complex, consisting of five subdomains (A, B, C, D, and E), while the NAM domain is composed of subdomains A, B, C, and D, among which subdomains A, C, and D are highly conserved in different species, and the C and D subdomains contain nuclear localization signals. However, subdomains B and E are less conserved (Li et al., 2018). ANAC019 from Arabidopsis thaliana is the first NAC protein to have its crystal structure determined. X-ray analysis revealed that the NAC domain is not a classical helix-turn-helix structure but a reverse parallel β-fold structure surrounded by several helices (Ernst et al., 2004) (Figure 1B), which form a conservative functional protein dimer through hydrogen bonding or a salt bridge between arginine (Arg) and glutamic (Glu) acid. In addition, many NAC proteins have dimer binding sites similar to ANAC019, and most NAC proteins bind to DNA in the form of homodimers, thus playing a regulatory role (Zhao et al., 2020a) (Figure 1C). The C-terminus of NAC TFs is a highly diverse transcriptional regulatory region, but some simple amino acids are frequently repeated. This region is rich in serine, threonine, proline, and glutamate, which leads to the loss of intrinsic disorder and a stable three-dimensional structure (Olsen et al., 2005).
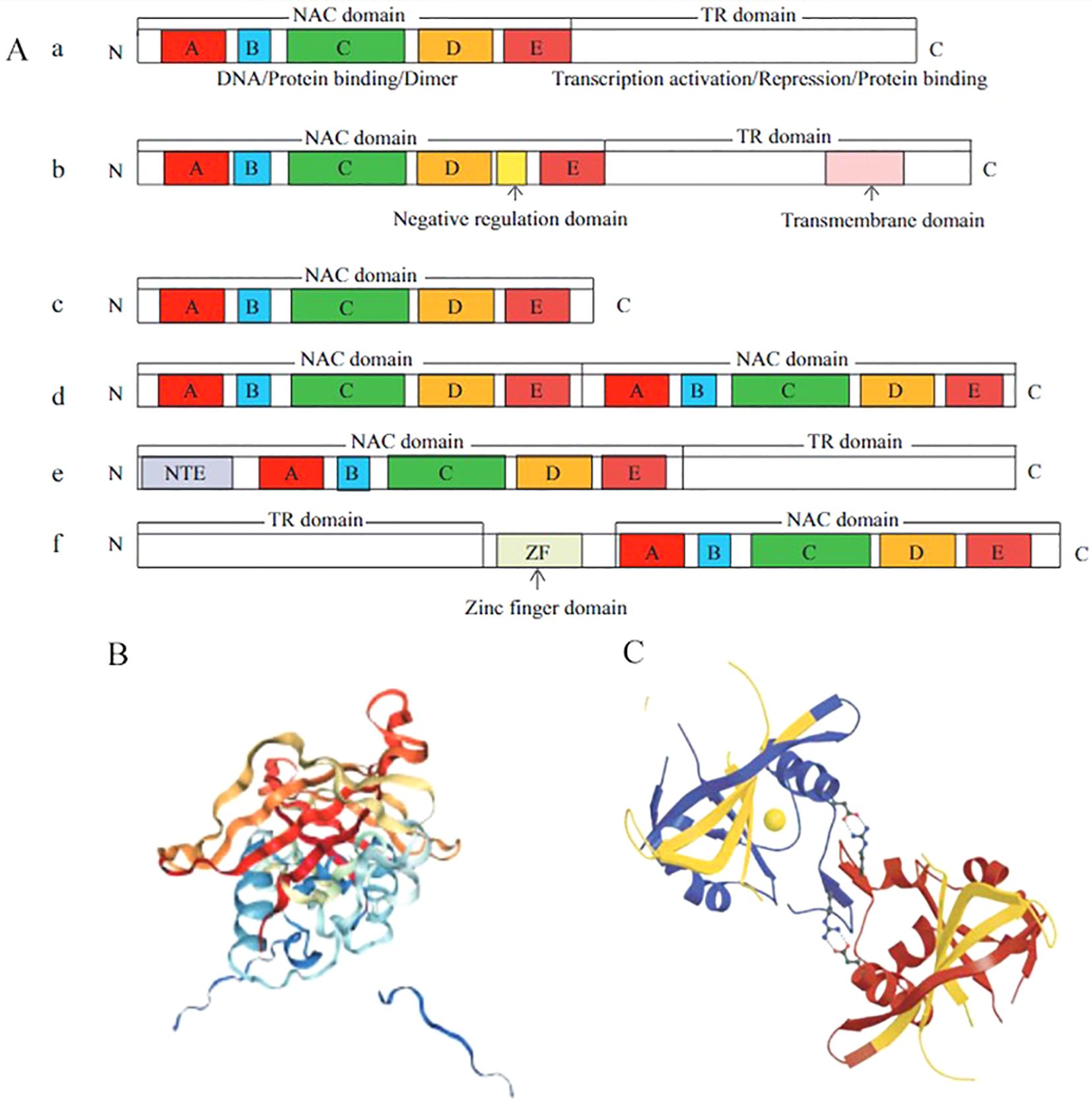
Figure 1. Structural characteristics of NAC transcription factors (Zhao et al., 2020a). Structural characteristics of NAC proteins (A), TR: a highly variable transcription regulation (A); Three-dimensional monomer structure of ANAC019 protein (B); Homodimer structure of ANAC019 protein (C).
Among the many NAC TFs, only a few have clear functions. Research on most NAC TF members is still at the level of gene cloning, structural identification, and expression analysis, and the downstream target genes and upstream regulatory factors of NAC TFs are poorly understood (Xu, 2008).
2 Classification and identification of NAC transcription factors
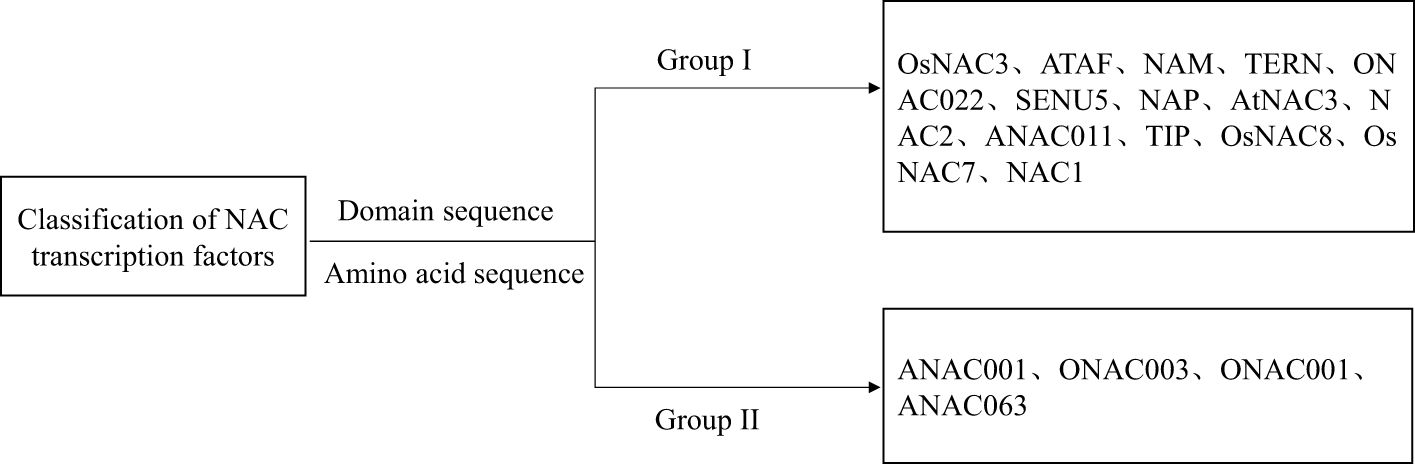
The NAC TF family has been extensively studied in the model plants A. thaliana and rice, and its classification is rather complex (Huang et al., 2017). The most commonly used method is to classify based on the characteristics of the NAC domain and a phylogenetic tree analysis. According to the similarity of the NAC domain and the protein amino acid sequences between Oryza sativa and A. thaliana, Ooka et al. (2003) divided the gene family into 2 categories (group I and group II) and 18 subgroups (group I contains 14 subgroups and group II contains 4 subgroups) (Figure 2). All subsequent classifications of NAC-TF subfamilies are primarily based on this study (Lin et al., 2023). For example, Rushton et al. (2008) analyzed the phylogeny of NAC TF family members in Arabidopsis, tobacco, rice, and other plants, and divided them into seven subfamilies, six of which exist in tobacco and other plants, and the seventh subfamily is unique to Solanaceae. Fang et al. (2008) conducted a phylogenetic tree analysis of NAC TFs in rice based on the coding amino acid sequences and divided them into groups I to V. Group I is related to plant growth and development and is further divided into 5 subgroups: I-1 (OsNAC7), I-2 (NAC1), I-3 (NAM/CUC), I-4 (GRAB2), and I-5 (NAC2). The tree structure of Group II is more complex than that of Group I, and it does not contain the published NAC sequences. Group III contains NAC genes related to stress (such as ANAC019, ANAC055, and ANAC072 in A. thaliana), so it is named stress-related NAC (SNAC). Group IV contains 14 NAC members, and Group V has two NAC members. Most scholars use this method as the classification standard for the NAC gene family in woody plants.
For example, Lu et al. (2018) cloned and identified the SNAC genes PeNAC034, PeNAC045, and PeNAC036 from Populus euphratica. Among them, PeNAC034 and PeNAC045 belong to the ATAF subgroup, while PeNAC036 belongs to the ANAC072 subgroup. Shang (2023) identified 21 LiNAC genes within the Lagerstroemia indica transcriptome data. The phylogenetic tree classified the NAC genes into two groups and 14 subgroups. The LiNACs genes in the subgroups exhibited similar conserved domains, gene structures, and analogous biological functions. Using the A. thaliana NAC genes as a reference, Zhang et al. (2023d) screened 76 NAC members and classified them into 17 subfamilies in Cerasus humilis, mainly including OsNAC7, ANAC001, ONAC003and other subfamilies, with the number of ChNAC family members in each subfamily varying and their functions potentially differing (Zhang et al., 2023d). Qi (2012) conducted a systematic analysis of the NAC family in Populus. A total of 163 NAC gene family members were identified. Phylogenetic analysis showed that these genes were clustered into 18 sub-families.
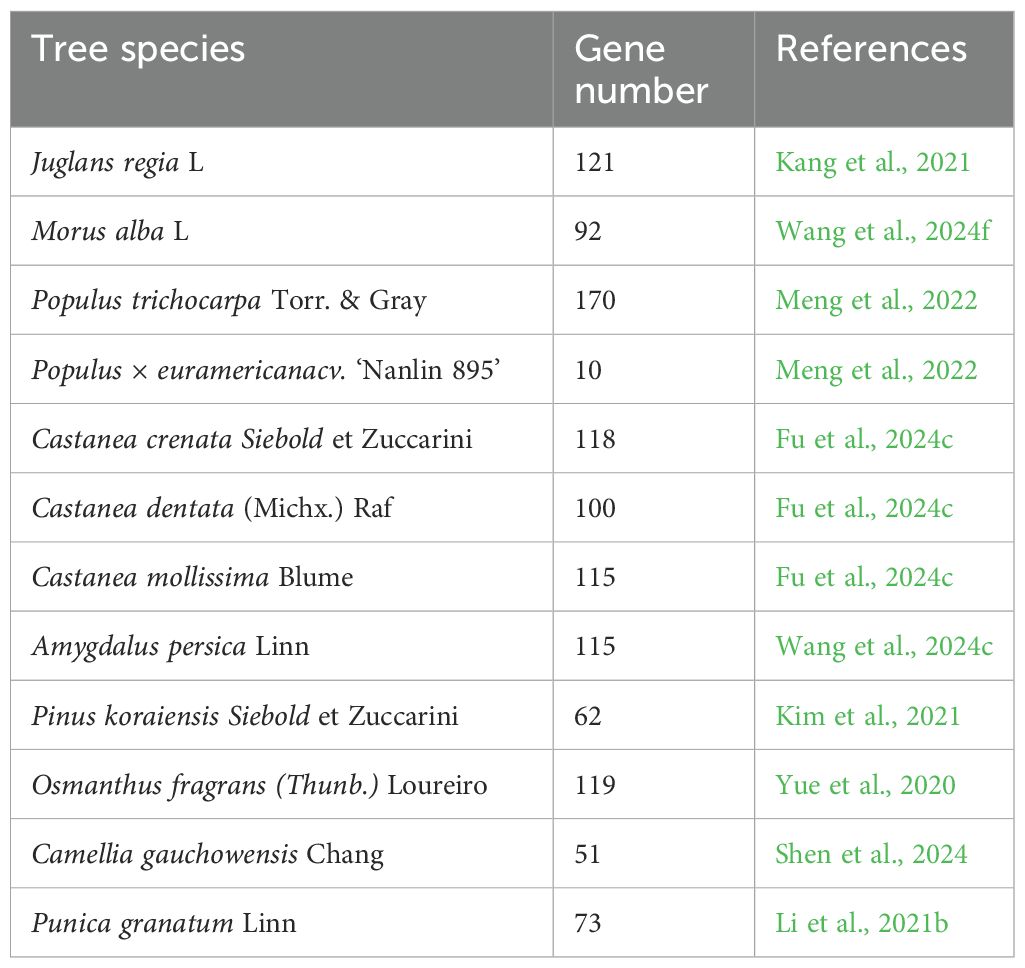
Many NAC TFs have been discovered through research on the woody plant genome. Researchers have identified different numbers of NAC TFs in woody plants (Table 1), and have explored the functions and mechanisms of the NAC TFs in the growth, development, and stress responses of woody plants. For example, Kang et al. (2021) identified 121 members in the NAC family within the entire Juglans regia genome. Among them, genes such as JrNAC2, JrNAC8, JrNAC115, and JrNAC97, may be key genes in the development of Juglans regia floral organs. Wang et al. (2024f) utilized bioinformatics methods to identify 92 members of the NAC gene family from Morus alba. They speculated that genes such as MnNAC75, MnNAC104 and MnNAC7, play roles in resisting drought stress. Meng et al. (2022) identified 170 NAC genes from Populus trichocarpa, cloned 10 PdNAC genes from Populus × euramericana ‘Nanlin 895’, and analyzed their expression patterns under drought stress. Fu et al. (2024c) identified the NAC gene families of three Castanea species. Among them, Castanea crenata Siebold contains 118 NAC genes, Castanea dentata contains 100 NAC genes, and Castanea mollissima contains 115 NAC genes. They analyzed the evolutionary relationships, selection pressures, and codon preferences of the NAC gene family members in Castanea plants. Wang et al. (2024c) conducted a transcriptome analysis of different tissues in Prunus persica and screened out PpNAC genes that are specifically expressed in the fruit from 115 members of the PpNAC gene family, and analyzed the changes in their expression levels at different developmental stages. In addition, researchers have identified 62, 119, 51, and 73 NAC TFs from woody plants, such as Pinus koraiensis Siebold, Osmanthus fragrans, Camellia gauchowensis, and Punica granatum, respectively, and analyzed their roles in growth, development, and the stress responses (Kim et al., 2021; Yue et al., 2020; Shen et al., 2024; Li et al., 2021b).
Most of the research on the functions of NAC TFs is concentrated on model plants, such as A. thaliana, Nicotiana tabacum, and O. sativa, while research on forest trees is lagging. Moreover, there are differences in the numbers, functions, and action mechanisms of NAC TFs among woody plants.
3 NAC transcription factors regulate the growth and development of woody plants
3.1 Participation in the formation of secondary walls
A group of TFs, including PtrNAC150, PtrNAC156, PtrNAC157, and PtrMYB18 have been revealed in poplar, a woody model plant (Figure 3A). These TFs activate the gene promoters of all three secondary wall biosynthetic pathways in the transient transactivation system using Arabidopsis protoplasts (Zhong et al., 2011) and regulate the gene expression of each component through a hierarchical NAC-MYB family transcription factor regulatory network (Hu, 2023). BpNAC012 in Betula platyphylla is crucial for formation of the secondary wall. Once BpNAC012 expression is inhibited, the deposition of the secondary wall in the stem fibers decreases significantly, and overexpression of BpNAC012 activates the expression of downstream genes related to the secondary wall by directly binding to the site of the NAC binding element of the secondary wall, inducing ectopic deposition of the secondary wall in the stem epidermis (Hu et al., 2019). The NAC family member SND2, an indirect target of SND1 and a key regulator of fiber secondary cell wall formation, plays a crucial role in the development of the secondary wall. Overexpression of Arabidopsis SND2 increases the secondary wall thickness of fiber cells in Eucalyptus. This may be because eucalyptus trees have a stronger tolerance to high levels of SND2 and/or the levels of SND2 co-regulatory factors in the xylem, and the transcriptional level of SND2 in eucalyptus trees remains moderate (Hussey et al., 2011). SND2-B2 is a homolog of Arabidopsis SND2 in poplar and promotes the synthesis of the secondary cell wall. When SND2-B2 is overexpressed, the expression of genes related to synthesis of the secondary cell wall, such as the cellulose synthetic genes PtrCesA17 and PtrCesA18, as well as the lignin biosynthetic genes PtrCAld5H1 and PtrCAld5H2, are upregulated. The cell wall of secondary xylem fibers in transgenic plants becomes thicker; however, when the expression of SND2-B2 is suppressed, the expression levels in transgenic plants are downregulated to varying degrees, in which the expression of xylan synthesis gene PtrIRX9 was significantly down-regulated, and the secondary cell wall becomes thinner (Han, 2021). Alternative splicing of PtrWND1B in Populus trichocarpa occurs only in secondary xylem fiber cells. Overexpression of the normal short transcript of PtrWND1B-S in Populus trichocarpa enhances the thickening of the fibrous cell wall while overexpressing the PtrWND1B-1 long transcript inhibits thickening of the fibrous cell wall (Zhao et al., 2014). In Populus tomentosa, genes encoding NAC domain proteins were isolated and named the PtVNS (VND-, NST/SND-, and SMB-related proteins) genes and the PtrWND genes includingVND and NST groups are driven under control of the cauliflower mosaic virus (CaMV) 35S promoter and induce thickening of the ectopic secondary walls in transgenic poplar leaves (Ohtani et al., 2011). The lignin content of the transgenic line overexpressing PtrNAC128 in P. trichocarpa is significantly higher than that of the wild-type, which plays an important regulatory role in the synthesis of secondary cell wall components by activating the expression of key enzyme genes and TFs involved in lignin and cellulose biosynthesis (Li et al., 2020). The wood-related NAC domain protein 3 (PdWND3A) in Populus deltoides is a homologous sequence of VND4 and VND5 in A. thaliana and is involved in regulating synthesis of the secondary cell wall (Yang et al., 2019). The GUS activity driven by the PtNAC068 promoter in transgenic Populus tomentosa Carr is present in the vascular tissues of stems, leaves, petioles, and roots. In contrast, the GUS activity driven by the PtNAC154 promoter only occurs in the secondary xylem of stems and leaf veins, indicating that upregulation of PtNAC068 and PtNAC154 is related to the secondary growth in poplars (Han et al., 2012). The NAC TF in Pteroceltis tatarinowii plays an important regulatory role in secondary growth (Zhou et al., 2025).
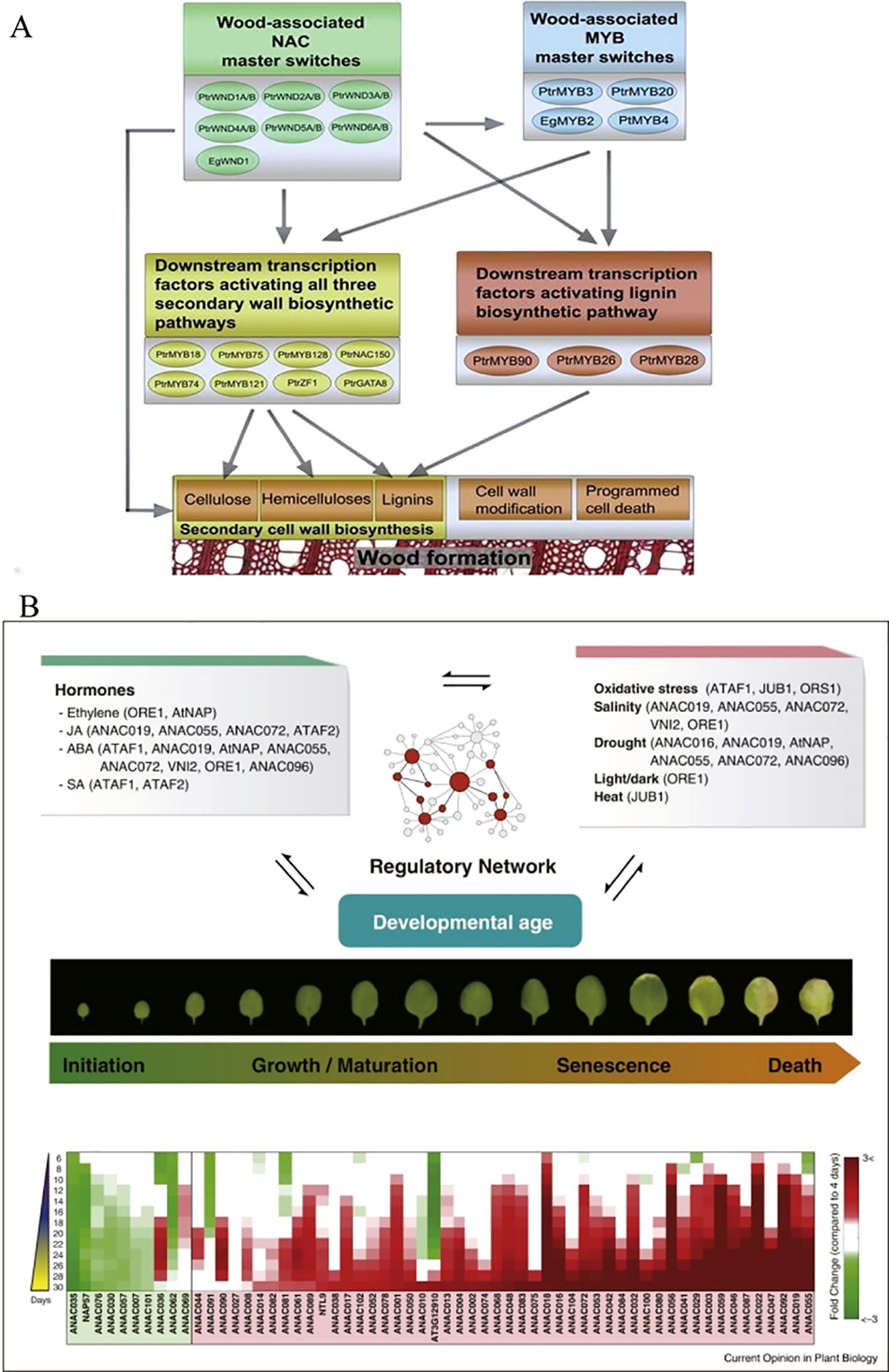
Figure 3. Role model of NAC in regulating secondary wall formation (Zhong et al., 2011) (A) and leaf senescence (Kim et al., 2016) (B).
The secondary cell wall of plants is mainly composed of cellulose, lignin and hemicellulose. NAC TFs mainly participate in secondary cell wall formation by activating downstream TFs, increasing gene expression related to the synthesis of secondary cell wall components, such as cellulose and lignin, and regulating the biosynthesis of each component. Although some progress has been made in the biosynthesis of secondary wall components in model plants such as Arabidopsis and poplar, many pathways are unknown and need to be discussed in the fine regulatory framework of their component synthesis.
3.2 Participation in the regulation of organ aging
NAC TFs are important regulators of leaf senescence (Kim et al., 2016) (Figure 3B). The alternative Populus splicing variant PtRD26IR regulates leaf senescence by interacting with several central Sen-NAC TFs and inhibiting their DNA binding activities (Wang et al., 2021a). The heterologous expression of DRL1 encoding a NAC TF in Vitis vinifera inhibits abscisic acid (ABA) synthesis, regulates the expression of related genes, and the content levels of 9-cis-epoxycarotenoid dioxygenase (NCED1), NCED5, zeaxanthin epoxidase 1 (ZEP1), ABA DEFICIENT2 (ABA2), ABA4, and ABA INSENSITIVE 2 (ABI2) decrease significantly, which reduces the sensitivity of plants to ABA, and delays leaf senescence in tobacco (Zhu et al., 2019). The V. vinifera VviNAC33 gene is negatively regulated by miRNA164, which induces leaf chlorosis, inhibits organ growth, and directly activates the expression of STAY-GREEN protein 1, which is involved in the degradation of chlorophyll and the photosystems, and autophagy-related protein 8f (ATG8f), which participates in plant senescence (D’Incà et al., 2021). CpNAC68 in Chimonanthus praecox, which is most highly expressed in old leaves and flowers, regulates physiological factors that provide stress tolerance during senescence (Lin et al., 2021). Tectona grandis TgNAC01 as a transcriptional activator of the ABA-mediated regulation and induces leaf senescence (Matias et al., 2022). In Malus pumila, MdNAC4 directly binds to the promoter of the senescence-associated gene MdSAG39, upregulating its expression, and positively regulates nitrogen deficiency-induced leaf senescence by enhancing ABA biosynthesis (Wen et al., 2023).
The leaves of woody plants are one of the most sensitive organs to senescence. Hormones, such as cytokinins, ethylene, jasmonic acid (JA), and salicylic acid, play important roles in organ senescence. NAC TFs play important roles in regulating secondary cell wall and organ senescence in woody plants, primarily by regulating the expression of related genes and plant hormone signal transduction pathways (ABA, ethylene, and JA). Most studies have focused on leaf senescence in annual plants, but the molecular mechanism of the regulation of leaf senescence in perennial woody plants remains unclear.
3.3 Participation in the regulation of seed development
Perennial woody plants often have dormant seeds, and there is relatively little research into the specific molecular mechanisms. Seed dormancy and development are regulated by NAC TFs (Figure 4). The PcNAC30 protein in Pyrus calleryana directly binds to the PcHAB1 promoter, regulates expression of the PcHAB1 gene, and controls release from seed dormancy (Bian, 2021). PpNAC5 may participate in the gibberellin (GA) and reactive oxygen species (ROS) synthetic pathways. PpNAC65 regulates the GA synthetic pathway, and PpNAC138 is involved in the GA and ABA synthetic pathways, thereby regulating the process of breaking seed dormancy in P. calleryana (Sun, 2019b). VvNAC26 in Vitis vinifera may regulate seed development by affecting several hormone pathways and interacting with VvMADS9 (Zhang et al., 2021a). In Cerasus humilis, genes such as ChNAC68, ChNAC72, and ChNAC76 contain seed-specific cis-acting elements that are likely to play important roles in endosperm development (Zhang et al., 2023d). Under normal culture, Arabidopsis lines transfected with HaNAC38 from Haloxylon ammodendron exhibit significantly lower germination rates than wild-type lines, suggesting that NACs may delay or inhibit seed germination (Luo et al., 2023).
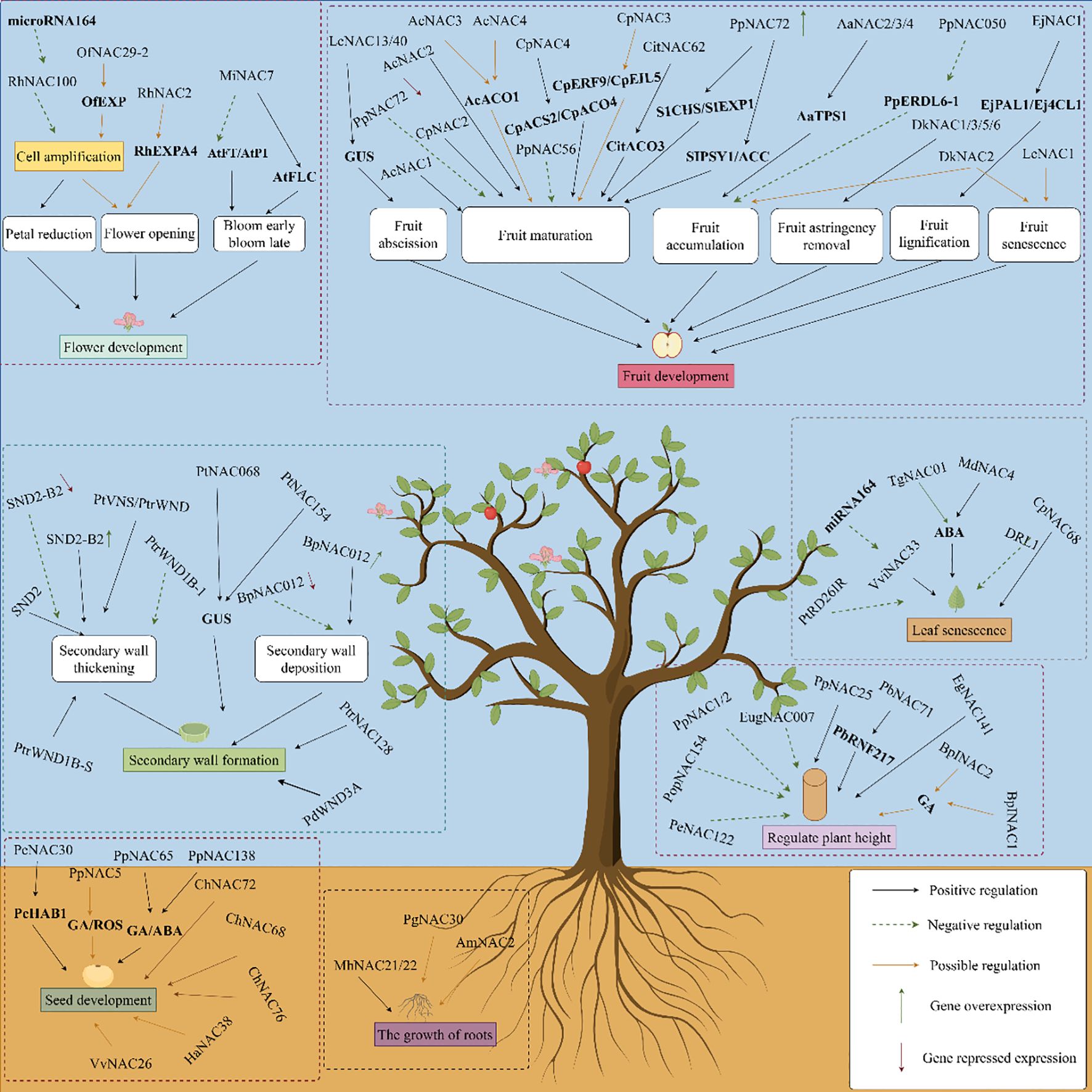
Figure 4. NAC transcription factors participate in the regulation of growth and development in woody plants (by Figdraw).
As plant-specific regulatory factors, NAC TFs play a key role in various processes by regulating the expression of related genes or interacting with other TFs, synthesizing hormones, and participating in hormone signal transduction pathways to induce and maintain seed dormancy and promote seed germination and other developmental processes.
3.4 Participation in the regulation of flowering
NAC TFs are also involved in the flowering of woody plants (Figure 4). The relevant OfNAC genes have been screened from the Osmanthus fragrans transcriptome. Among them, the OfNAC29–2 gene may act like a regulator of OfEXPA2 and OfEXLA1 to control OfEXP and affect the expansion of petal cells. It has been speculated that OfNAC participates in the regulation of flower opening in O. fragrans (Miao et al., 2021). The RhNAC2 gene in Rosarugosa Thunb may be involved in flower opening by regulating the expression of RhEXPA4 (Dai et al., 2012). Ethylene regulates the accumulation of RhNAC100 transcripts in Rosarugosa Thunb through microRNA164. Moreover, overexpression of RhNAC100 in Arabidopsis inhibits the expansion of petal cells and negatively regulates their growth, leading to smaller petals (Pei et al., 2013). In Mangifera indica, MiNAC7 affects the flowering of transgenic plants by regulating the expression of flowering-related genes (AtFT, AtAP1, and AtFLC) (Huang et al., 2023).
NAC TFs modulate the flowering process in woody plants by regulating the expression of related genes and adjusting post-transcriptional levels through miRNA164, thereby regulating hormone synthesis and flowering. However, the specific regulatory network needs further research.
3.5 Participation in the regulation of fruit development
NAC TFs play an important role in fruit growth and development (Figure 4). For example, promoters of the LcNAC13/40 genes in Litchi chinensis drive the downstream GUS gene to be expressed in several organ abscission/shedding sites of A. thaliana, such as the abscission zones of floral organs, the abscission zones of cauline leaves, the seed abscission sites, and the pod dehiscent zones (Wen, 2019). Overexpression of PpNAC72 in Amygdalus persica regulates the expression of a series of genes related to fruit development and maturation. The expression levels of the S1CHS, SlEXP1, SlPSY1, and ACC genes promote the growth and development of fruit, and speed up fruit maturation (Wu, 2021). When the AdNAC3 gene from Actinidia chinensis is overexpressed in tomatoes, this gene promotes tomato fruit ripening, enhances the expression of genes related to ethylene synthesis, and accelerates the degradation of pectin (Yang et al., 2025). In Citrus reticulata, CitNAC62 and CitWRKY1 transactivate the CitAco3 promoter, thus degrading citric acid during the development and ripening of citrus fruits (Li et al., 2017). Instantaneous silencing of the PpNAC56 and PpNAC72 genes in A. persica inhibits fruit ripening by reducing ethylene release and delaying fruit softening (Wang, 2024d). The TFs AcNAC1 and AcNAC2 in Actinidia chinensis participate in the ripening and softening processes by activating endo-transglucosylase/hydrolase genes (Fu et al., 2024a). Ethylene-induced CpNAC4 mediates ethylene production by binding to and activating the promoters of ethylene biosynthetic-related genes CpACS2 and CpACO4, thus participating in ripening of Pseudocydonia sinensis (Fu et al., 2023). A new MADS-box gene in Pseudocydonia sinensis, named CpMADS4, specifically binds and activates the promoters of ethylene signaling genes CpERF9 and CpEIL5, together with CpNAC3, thereby possibly regulating fruit ripening (Fu et al., 2021). CpEIN3a interacts with CpNAC2 to individually or synergistically activate the transcription of gene subgroups related to carotenoid biosynthesis, thereby participating in the regulation of Pseudocydonia sinensis fruit ripening (Fu et al., 2017). Ethylene-induced AcNAC3 and AcNAC4 are transcriptional activators that may participate in ripening of kiwifruit by activating AcACO1 (Fu et al., 2024b). AaNAC2, AaNAC3, and AaNAC4 in Actinidia arguta bind to the terpene synthetase 1 promoter (AaTPS1), giving rise to the accumulation of more monoterpene volatiles in fruit (Nieuwenhuizen et al., 2015). The expression of DkNAC1, DkNAC3, DkNAC5, and DkNAC6 is highly correlated with fruit astringency in persimmon, and DkNAC2 may participate in processes related to fruit ripening or aging, but not in the removal of astringency (Min et al., 2015). PpNAC050 increases fructose accumulation in flesh by inhibiting PpERDL6–1 expression (Liu et al., 2024). EjNAC1 in Eriobotrya japonica regulates the expression of related genes (EjPAL1 and Ej4CL1) and realizes fruit lignification (Xu et al., 2015). The expression of LcNAC1 in L. chinensis fruit increases during the aging of the peel and flesh, which may promote fruit aging (Jiang et al., 2017).
As plant-specific regulatory factors, NAC TFs play key roles in a variety of processes. They regulate developmental processes, such as the induction and maintenance of seed dormancy and the promotion of seed germination, as well as a series of flowering and fruit development processes in woody plants. NAC TFs achieve this by regulating the expression of related genes, interacting with other TFs, and participating in hormone synthesis and hormone signal transduction pathways. However, the molecular mechanisms of how NAC TFs regulate the growth and development of woody plants are poorly understood, so further research is needed.
3.6 Regulation of plant height and root development
NAC TFs participate in the regulation of plant dwarfism traits (Figure 4). The PpNAC1 and PpNAC2 genes have been screened from Prunus persica. When the PpNAC1 gene was transferred into poplar trees using the Agrobacterium-mediated method, stem thickness shrank, plant height decreased, and the leaves curled and wrinkled in the transformed Populus trees. Stem diameter and plant height also decrease in Populus trees transformed with the PpNAC2 gene (Yang, 2022a). PbRNF217 in Pyrus promotes the ubiquitination and degradation of PbNAC71 dependent on the 26S proteasome, regulating the development of xylem and vessels and, thereby, affecting plant height (Cong, 2023). In eucalyptus, EgNAC141 is a positive regulatory factor in wood formation and participates in lignin synthesis (Sun et al., 2021). EugNAC007 in E. urophylla × E. grandis is dominantly expressed in the xylem, while overexpression of this gene in poplar inhibits plant growth (He et al., 2023a). Overexpression of the PpNAC25 gene increases the height of Populus trees, by increasing the number of internodes and promoting growth and development (Geng, 2023). Overexpression of the PopNAC122 gene in Arabidopsis, a homolog of XND1 in Populus, reduces the number of phloem fibers, xylem cell size and number, and vessel number while overexpressing the PopNAC154 gene decreases plant height and increases the relative proportion of bark to xylem (Grant et al., 2010). In Betula platyphylla, BpINAC1 and BpINAC2 are induced by GA3, may participate in the GA signaling pathway, and regulate xylem development (Guo et al., 2015). When PeNAC122 is overexpressed in Populus euphratica, plant height decreases, the xylem thickens, lignin accumulates, and the expression of genes related to secondary cell wall biosynthesis is upregulated (Chen et al., 2022).
Roots are one of the important organs for plant growth. Studies have shown that NAC genes affect the stress response in woody plants by regulating the growth and development of roots. The AmNAC2 protein in Ammopiptanthus mongolicus is highly expressed in the roots of wild plants and it may be involved in the growth and development of roots and leaves and related physiological activities (Zhang et al., 2021c). PgNAC30 in Punica granatum likely plays an important role in the development of the root system (Li et al., 2021b). The NAC TFs in Casuarina equisetifolia may be related to nitrogen fixation in root nodules, thereby participating in growth and development of plant roots (Xiao et al., 2025). Heterologous transformation of A. thaliana with MhNAC21/22 was carried out with Malus hupehensis, and overexpressing MhNAC21/22 promoted the formation of lateral roots (Cheng, 2024).
3.7 Regulation of hormonal balance
NAC TFs participate in a variety of hormone signal transduction processes. They exhibit complex regulatory functions during stress responses. NAC TFs also participate in the GA, ABA, ethylene, auxin, and cytokinin signal transduction pathways. For example, in Carica papaya, CpNAC2 directly binds to the CpEIN3a promoter, activates the transcription of a subgroup of genes related to carotenoid biosynthesis, such as CpPDS2/4, CpZDS, CpLCY-e and CpCHY-b, and regulates the synthesis of carotenoids during fruit ripening (Fu et al., 2017). CpNAC4 is an ethylene-induced transcriptional activator that may mediate the production of ethylene by activating genes related to ethylene biosynthesis (Fu et al., 2023). CpMADS4 (a novel MADS-box gene) and CpNAC3 specifically bind to and activate the promoters of the ethylene signaling genes CpERF9 and CpEIL5, thereby regulating the ethylene signaling transduction pathway during ripening of papaya fruit (Fu et al., 2021). The CpNAC1 gene may promotes the synthesis of carotenoids during ripening in Carica papaya by regulating the expression of the CpPDS2/4 gene (Fu et al., 2016). Ethylene-induced AcNAC3 and AcNAC4 in kiwifruit may participate in ripening and ethylene biosynthesis by activating AcACO1 (Fu et al., 2024b). The MdNAC42 gene promotes the synthesis of anthocyanins and proanthocyanidins in Malus pumila and regulates phenylalanine content in apples by controlling anthocyanidin reductase (Zhang et al., 2020b). MdNAC18.1 indirectly regulates the transcriptional activities of key ethylene synthetic genes MdACO1-like and MdACS1 by activating MdNAC72 and MdMYC2. This forms a regulatory cascade that affects ethylene content (Zhang et al., 2025). The MdNAC52 transcript level increases during fruit coloring. Overexpression of MdNAC52 promotes the accumulation of anthocyanins in the Malus pumila callus (Sun et al., 2019a). VcNAC072 interacts with the AtPAP1 promoter and activates its expression, positively regulating the accumulation of anthocyanins in Vaccinium vitis-idaea fruit (Song et al., 2019). NAC6 in Mangifera indica may inhibit the expression of carotenoid cleavage dioxygenase, thus slowing the degradation rate of carotenoid components (Liang et al., 2024a). The AsNAC22 gene in Aquilaria sinensis plays a role in the formation of secondary metabolites, such as chromones or sesquiterpenes (Yang et al., 2023). Heterologous expression of the VaNAC26 gene from Vitis amurensis in A. thaliana upregulates the expression of JA synthesis genes and genes in the JA signaling pathway, thereby increasing JA content in A. thaliana (Fang et al., 2016).
NAC TFs mediate the production of hormones by activating the expression of relevant synthetic genes. They regulate the balance of hormones through multiple mechanisms and participate in their signal transduction pathways, playing an important role in growth, development, and stress responses.
4 NAC transcription factors are involved in the stress responses of woody plants
4.1 The salt stress response
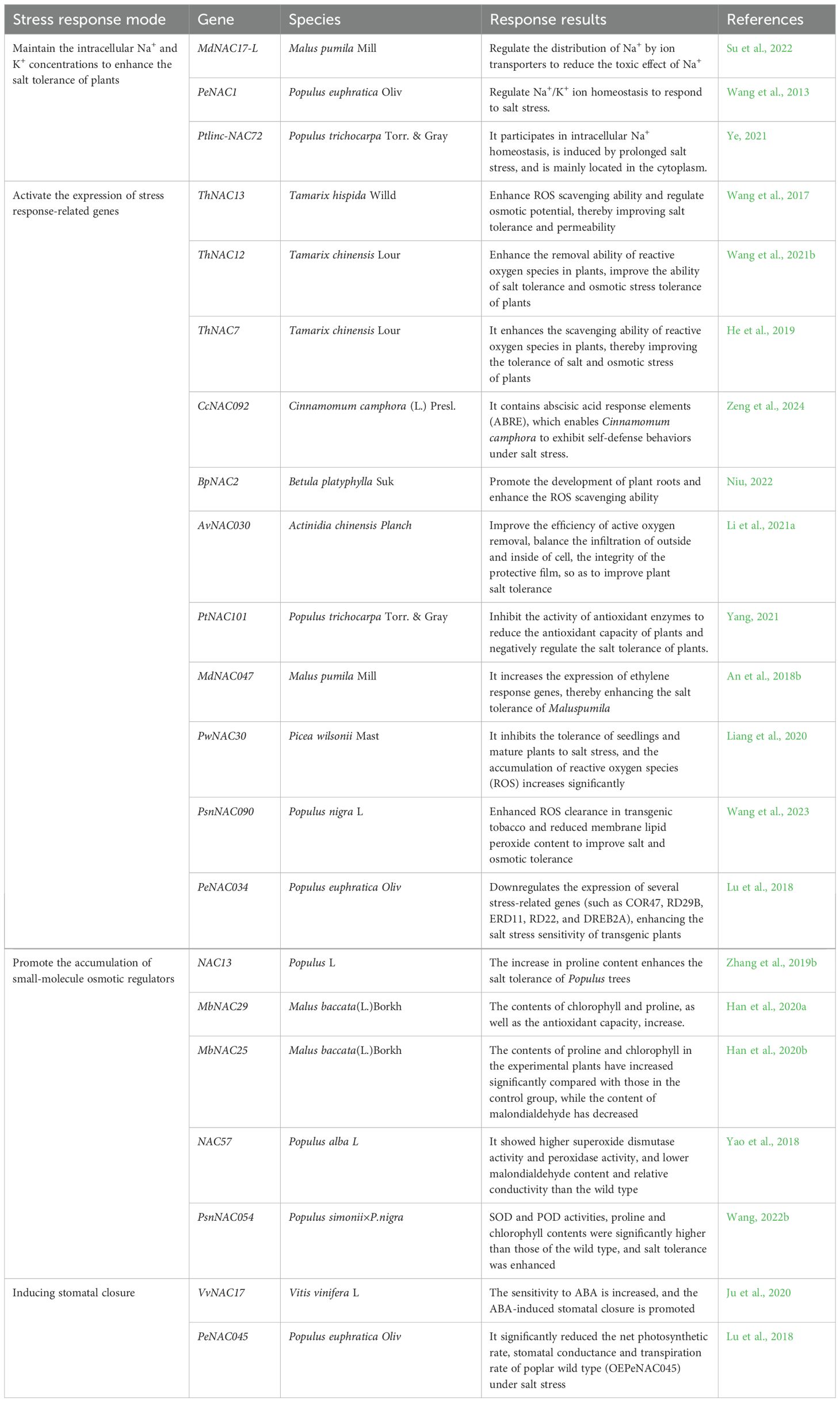
NAC TFs respond to salt stress as shown in Table 2.
First, NAC TFs enhance salt tolerance in plants by maintaining the intracellular concentrations of Na+ and K+ (Figure 5A). When plants take in excessive amounts of Na+, NAC TFs distribute Na+ by regulating ion transporters. The protein complex formed by MdWRKY55 and MdNAC17-L in Malus pumila promotes the expression of the downstream MdNHX1 gene, alleviating the toxic effects of Na+ (Su et al., 2022). The PeNAC1 TF in Populus euphratica responds to salt stress by regulating Na+/K+ ion homeostasis (Wang et al., 2013). Ptlinc-NAC72 in Populus trichocarpa is involved in intracellular Na+ homeostasis. It is induced by long-term salt stress and is mainly located in the cytoplasm (Ye, 2021).
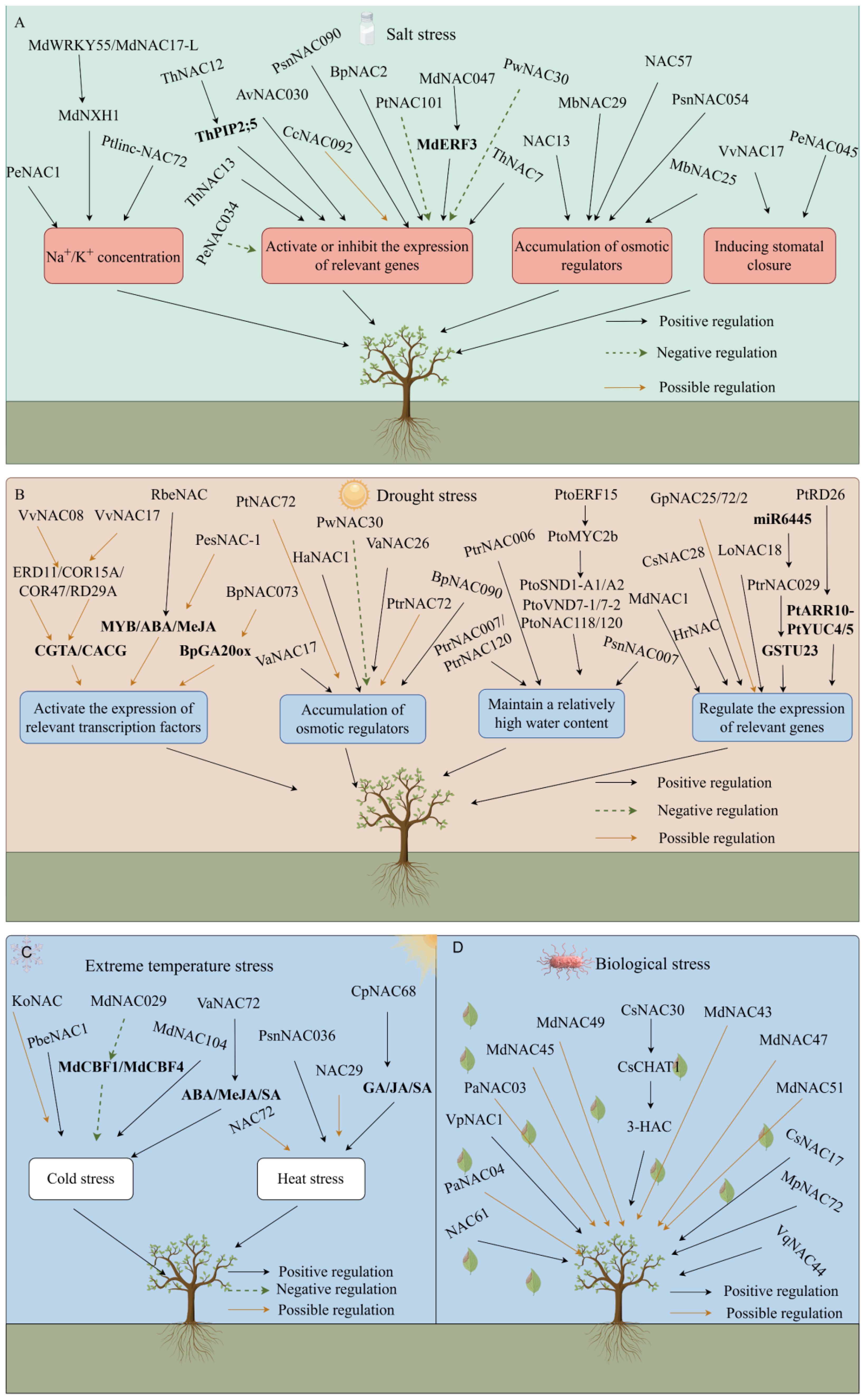
Figure 5. Interaction between NAC genes and various stresses (by Figdraw). Salt stress (A); Drought stress (B); Extreme temperature stress (C); Biological stress (D).
NAC TFs maintain a relatively stable intracellular environment by activating the expression of stress response-related genes, such as SOD, POD, ABA, RD22, and ethylene (Figure 5A). Overexpression of ThNAC13 TF in Tamarix hispida induces the activity of superoxide dismutase (SOD) and peroxidase (POD), improves salt tolerance and permeability by enhancing ROS scavenging ability and regulates osmotic potential (Wang et al., 2017). The ThNAC12 TFs directly regulate the expression of the ThPIP2;5 genes, to scavenge ROS and improve salt tolerance (Wang et al., 2021b). Overexpression of ThNAC7 enhances ROS scavenging, thereby improving tolerance to salt (He et al., 2019). CcNAC092 contains an ABA response element, which prompts Cinnamomum camphora’s self-defense behavior under salt stress (Zeng et al., 2024). Overexpression of the BpNAC2 gene promotes the development of roots and enhances ROS scavenging ability to improve salt tolerance in Betula platyphylla (Niu, 2022). In Actinidia chinensis, AvNAC030 enhances salt tolerance by improving the removal efficiency of ROS, maintaining the osmotic balance inside and outside the cells, and protecting the integrity of the membrane (Li et al., 2021a). In Populus trichocarpa, PtNAC101 inhibits antioxidant enzyme activities, reduces the antioxidant capacity of plants, and negatively regulates salt tolerance (Yang, 2021). MdNAC047 in Malus pumila directly binds to the MdERF3 promoter (ethylene response factor) and activates its transcription, increasing the expression of ethylene-responsive genes, through a new regulatory pathway called MdNAC047-MdERF3-ethylene-salt tolerance, thereby enhancing salt stress tolerance in Malus pumila (An et al., 2018b). Transgenic A. thaliana with overexpressed PwNAC30 from Picea wilsonii significantly inhibits salt stress tolerance in seedlings and mature plants, and ROS accumulation increases significantly (Liang et al., 2020). In Populus nigra, PsnNAC090 improves salt and osmotic tolerance by enhancing ROS scavenging and reducing membrane lipid peroxide content in transgenic tobacco (Wang et al., 2023). In Populus euphratica, PeNAC034 downregulates the expression of several stress-related genes (such as COR47, RD29B, ERD11, RD22, and DREB2A), thereby enhancing the salt stress sensitivity of transgenic plants (Lu et al., 2018).
NAC TFs promote the accumulation of small molecular osmotic regulators (such as betaine, proline, and soluble sugars) to protect the stability of cell structures (Figure 5A). Proline content increases in transgenic Populus trees overexpressing NAC13 under salt stress, which enhances salt tolerance (Zhang et al., 2019b). Overexpressing MbNAC29 from Malus baccata in A. thaliana results in higher chlorophyll and proline contents and antioxidant capacity under salt stress (Han et al., 2020a). When the MbNAC25 gene of M. baccata is overexpressed in A. thaliana under high salt stress, proline and chlorophyll contents increase significantly compared with those of control plants. Additionally, malondialdehyde (MDA) content decreases, ROS species scavenging ability is enhanced, and the tolerance of plants to salt stress is promoted (Han et al., 2020b). Overexpression of Populus alba NAC57 in Arabidopsis exhibits higher SOD and POD activity. The MDA content and relative electrical conductivity were lower than those of the wild-type, indicating that the NAC57 gene plays an important role in the salt stress response (Yao et al., 2018). Under salt stress, plants overexpressing the PsnNAC054 gene in Populus simonii × P. nigra have significantly higher SOD and POD activities, and higher proline and chlorophyll contents than wild-type plants, enhancing their salt tolerance (Wang, 2022b).
NAC TFs regulate stomatal closure to balance the water status within the plant (Figure 5A). The heterologously expressed Vitis vinifera VvNAC17 gene increases the tolerance of A. thaliana to salt, osmotic, and freezing stress, increases ABA sensitivity, and promotes the ABA-induced stomatal closure (Ju et al., 2020). In Populus euphratica, Overexpression of PeNAC045 leads to a significant decrease in the net photosynthetic rate, stomatal conductance, and transpiration rate of wild-type poplar (OEPeNAC045) under salt stress conditions (Lu et al., 2018).
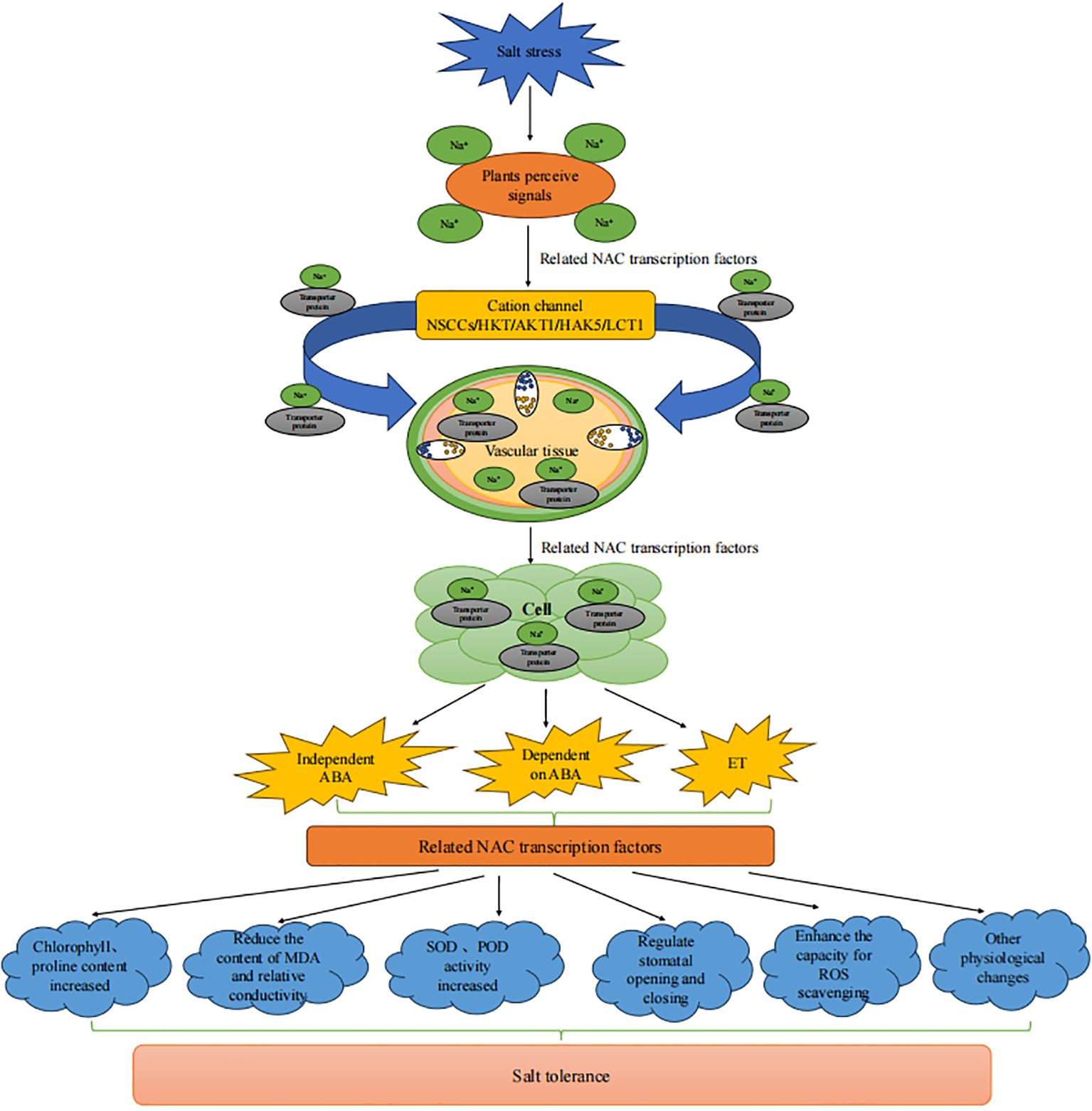
Under salt stress, external Na+ primarily enters the root cells via cation channels and transporters, such as nonselective cation channels (NSCCs), high-affinity K+ transporters (HKT), shaker-type potassium channels (AKT1), high-affinity K+ transporters (HAK5), low-affinity cation transporters (LCT1), etc (Kronzucker and Britto, 2011; Mian et al., 2011; Isayenkov and Maathuis, 2019). It then moves into plants through xylem loading and phloem recycling in stems (Van Zelm et al., 2020; Zhao et al., 2020b; Figure 6). NAC TFs simultaneously regulate multiple downstream stress-responsive genes and activate the salt tolerance response in plants. For example, NAC TFs respond to salt stress damage by regulating the transport of Na+ through cation transporters, activating the expression of stress response-related genes, mediating the ABA signaling pathway, accumulating osmotic regulators, balancing water status, and eliminating ROS through oxidative defense mechanisms to reduce oxidative damage.
4.2 The drought stress response
Plant NAC TFs improve plant drought tolerance through four pathways, as shown in Table 3.
NAC TFs bind to the cis-acting elements of key TFs responding to drought stress. The expression of related TFs is activated to improve plant drought tolerance, such as ABA, MYB, DREB, and WRKY (Figure 5B). The VvNAC08 and VvNAC17 TFs in V. vinifera may regulate the expression of these genes by binding to the core response elements ([CGTA/CACG]) of the promoter sequences of downstream target genes (ERD11, COR15A, COR47, and RD29A), thereby enhancing tolerance to drought stress (Ju, 2019). The RbeNAC promoter in Rosa berberifolia contains numerous MYB binding sites and ABA and methyl jasmonate elements are involved in drought-induced responses, indicating that they play an important role in drought stress (Feng et al., 2023). PeSNAC-1 in Phyllostachys edulis may enhance drought tolerance in transgenic rice by regulating genes in the ABA-dependent and ABA-independent signaling pathways (Hou et al., 2020). In Betula platyphylla, BpNAC073 and BpDNMT1 proteins interact, potentially affecting BpGA20ox1 expression via methylation to enhance drought tolerance (Zhao, 2024).
NAC TFs increase the contents of osmotic regulators, such as proline and betaine, within the plant, preventing protein denaturation and increasing the survival rate under drought stress (Figure 5B). The MDA content of PtNAC72-overexpressing Citrus sinensis plants is significantly higher than that of the wild-type, while the contents of proline, POD, and CAT are lower than those of the wild type, indicating that PtNAC72 may negatively affects the drought stress response in Poncirus trifoliata (Xiong, 2020). Ectopic expression of HaNAC1 in Haloxylon ammodendron leads to enhanced drought tolerance in transgenic overexpressing lines, the expression of stress-induced marker genes is upregulated, and the contents of proline, indole acetic acid, and ABA increase (Gong et al., 2020). PtrNAC72 in trifoliate Poncirus trifoliata is a repressor of putrescine biosynthesis and may negatively regulates the drought stress response by modulating putrescine-related ROS homeostasis (Wu et al., 2016a). BpNAC090 in Betula platyphylla reduces the contents of ROS and hydrogen peroxide (H2O2) and increases the synthesis of proline to mitigate the damage caused by drought-induced osmotic stress in B. platyphylla (Wang, 2022c). The TF VaNAC17 enhances drought tolerance of transgenic A. thaliana in Vitis amurensis by regulating JA synthesis (Su et al., 2020). VaNAC26 upregulates genes involved in JA synthesis and signaling, increases JA content in A. thaliana, and enhances drought tolerance (Fang et al., 2016). Picea wilsonii PwNAC30 weakens the drought tolerance of transgenic A. thaliana by increasing the accumulation of ROS and inhibiting the expression of stress genes (Liang, 2021).
NAC TFs maintain a relatively high water content by inhibiting the outflow of water from cells (Figure 5B). PtrNAC007 and PtrNAC120 in Populus trichocarpa are highly induced by drought. Transgenic plants overexpressing these genes enhance their drought tolerance by altering the number and pore size of vessel cells (Li et al., 2019b). The soluble lignin and total lignin contents of PtrNAC006 overexpressing plants increase significantly. After drought stress, the stomatal conductance of plants decreases, water loss decreases, water use efficiency increases, and the decline in hydraulic conductivity is mitigated, indicating that PtrNAC006 overexpressing plants have stronger anti-embolism ability, thereby enhancing drought tolerance (He, 2023b). Transgenic Populus simonii × P. nigra plants overexpressing PsnNAC007 reduce water loss by decreasing transpiration and stomatal conductance and increasing water use efficiency (Wu et al., 2024). Kong et al. (2023) identified PtoERF15 and its target gene PtoMYC2b in Populus tomentosa. Three homologous gene groups in the NAC family, PtoSND1-A1/A2, PtoVND7-1/7-2, and PtoNAC118/120, are PtoMYC2b targets and regulate vessel morphology. They maintain the stem water potential by controlling development of xylem vessels, which enhances drought tolerance.
NAC TFs reduce the stress damage to plants by regulating the expression of drought stress-related response genes (Figure 5B). Overexpression of the MdNAC1 gene in Malus pumila enhances antioxidant capacity under drought stress and alleviates the damage caused by drought stress (Jia, 2019). Overexpression of CsNAC28 in Camellia sinensis confers hypersensitivity to ABA treatment in Arabidopsis and reduces the accumulation of ROS, thereby improving dehydration tolerance (Zhang et al., 2023c). Drought stress induces HrNAC in Hippophae rhamnoides, which activates the expression of drought-resistant related genes, thereby enhancing drought tolerance in sea buckthorn (Zhang et al., 2023a). The PtRD26 promoter in Populus drives the expression of the isopentenyl transferase gene IPT to produce overexpressing plants. Transgenic plants improve drought tolerance in Populus by inducing the synthesis of cytokinin and PtARR10-PtYUC4/5-mediated ROS scavenging (Wang et al., 2022a). Expression of the LoNAC18 gene in Larix olgensis is upregulated under drought stress conditions, and the contents of SOD, POD, soluble proteins, and soluble sugars increase, which participate in regulating the L. olgensis response to PEG-simulated drought stress (Zhang et al., 2020a). In poplar, PtrNAC029 binds to the glutathione S-transferase GSTU23 promoter and inhibits its expression. miR6445 directly targets PtrNAC029, and its upregulation downregulates PtrNAC029, leading to higher GSTU23 expression. These three factors collectively respond to drought stress in poplar (Niu et al., 2024). Under drought stress, the expression levels of GpNAC25, GpNAC72, and GpNAC2 in Gymnocarpos przewalskii are upregulated, indicating their potential roles in the drought response (Yang, 2024).
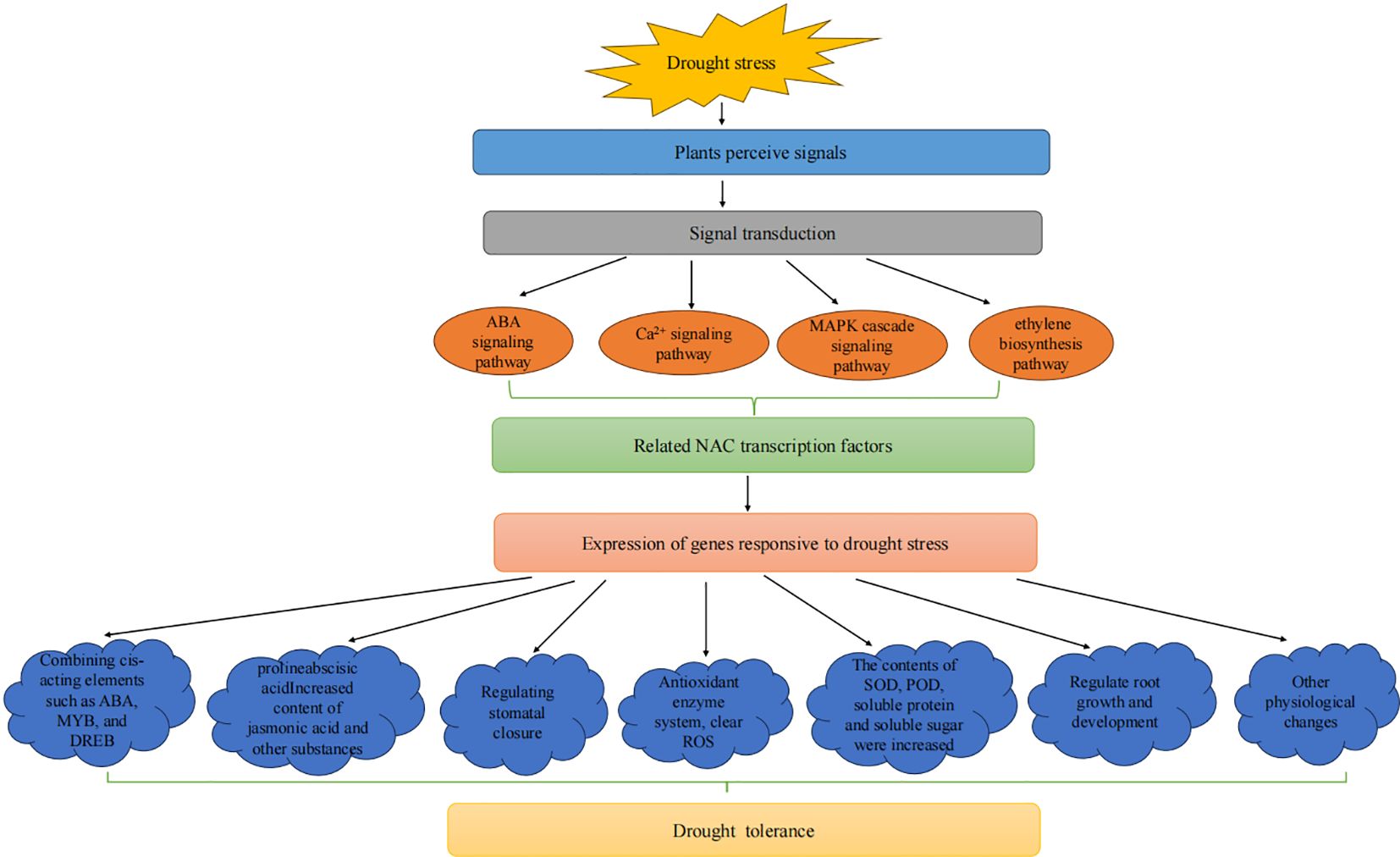
Plants mobilize various signaling pathways to respond to drought stress, including ABA, calcium ion, MAPKs cascade, and ethylene signaling pathways (Zhao, 2024; Liang et al., 2024b). NAC TFs also play essential roles in regulating these complex processes to respond to drought stress under varying environmental conditions (Figure 7). For example, ethylene affects osmotic regulatory substances and antioxidant enzyme systems, regulates ROS content, and participates in regulating stomatal opening and closing and inducing the expression of related genes by influencing ABA synthesis. In addition, NAC genes also affect the ability of plants to defend against stress by regulating the growth and development of roots. The MiNAC1 gene in Mangifera indica improves the tolerance of transgenic plants to drought stress, salt stress, and low-temperature stress by increasing the length and number of lateral roots (Yang et al., 2022b). PwNAC11 has been isolated from Picea wilsonii and affects drought tolerance in transgenic A. thaliana by affecting root development (Zhang, 2019a).
4.3 The extreme temperature stress response
When plants are subjected to low-temperature stress, TFs activate the expression of low-temperature response genes by binding to cis-acting elements on gene promoters, thereby regulating the signal transduction pathways to improve low-temperature tolerance (Figure 5C). Overexpression of PbeNAC1 in Pyrus betulifolia enhances tolerance to cold stress (Jin et al., 2017). Overexpression of MdNAC029 in Malus pumila significantly inhibits the expression of MdCBF1 and MdCBF4 in the CBF pathway and, thus, the cold tolerance of plants is significantly downregulated (An et al., 2018a). Overexpression of MdNAC104 affects the accumulation of osmotic regulatory substances in the stems and leaves of M. pumila under low-temperature freezing conditions, thereby enhancing cold tolerance (Mei, 2023). The KoNAC gene in Kandelia obovata contains a low-temperature response element LTR, which may play a role in the response to low-temperature stress (Sun et al., 2022). Under low-temperature stress, the expression of 10 NAC genes was significantly upregulated in plum blossoms, while the expression of five others was inhibited, indicating that the NAC TFs respond to low-temperature stress (Zhuo et al., 2018). In Vitis amurensis, VaNAC72, a TF regulated by the ABA, MeJA, and SA signaling pathways, positively regulates cold tolerance by acting upstream of VaCP17 under cold stress (Qin et al., 2023).
Plants induce the expression of heat shock proteins under high-temperature conditions to maintain the stability of the cell membranes and remove excess ROS, thereby enhancing heat tolerance (Figure 5C). Overexpression of the PsnNAC036 gene from P. simonii × P. nigra in tobacco promotes the growth of tobacco and enhances salt and heat tolerance (Zhang et al., 2021b). Woody plants also enhance their heat adaptive ability under various stressors through the ABA-dependent pathway. They close their stomata to reduce water transpiration and withstand heat stress. For example, NAC29 and NAC72 are upregulated under heat stress in Rosa chinensis, indicating that ABA signaling is likely to be involved in the heat stress response (Li et al., 2019c). The CpNAC68 gene in Chimonanthus praecox, which activates nuclear protein transcription, positively regulates heat tolerance without affecting growth and development. It is induced by treatments, such as GA, JA, and SA (Lin et al., 2021).
NAC TFs have potential application value in improving the tolerance of woody plants to temperature stress. However, most research has focused on cold tolerance in woody plants. Only a few NAC TFs have been demonstrated to respond to high-temperature stress in woody plants, and the specific mechanisms of signal transduction and transcriptional regulatory networks require further study.
4.4 Participation in other abiotic stress responses
Woody plants are also subjected to other abiotic stressors in the environment. For example, Salix integra is a short-rotation woody plant with a high potential for phytoremediation of lead (Pb). SiNAC has a significant response to Pb, among them, the seven candidate transcripts of ATAF and NAP subfamilies, as well as the SiNAC137 transcript of SEIN5 subfamily, showed significant positive responses to lead stress and were significantly induced within 4 hours to 14 days after lead treatment. In addition, OsNAC7 and other subfamilies containing negative response transcripts may also participate in the lead response of integrated Streptomyces by reducing the activation of target genes (Xin et al., 2023). MdNAC1 in Malus domestica is significantly induced by iron deficiency stress, enhances root acidification and promotes iron absorption by upregulating genes such as AHA2/MDAHA12 and FRO2 (Li et al., 2022). In Dangshan Su pears (Pyrus bretschneideri), the PbrNAC gene may respond to abiotic stressors, such as iron deficiency, by regulating root physiology and affecting chemical secretion and signal molecule transmission (Guo et al., 2023). SmNAC007 in Salix matsudana may positively regulates responses to flooding stress by upregulating downstream genes involved in enzymatic detoxification and cellular protection, and increasing antioxidant proteins to reduce cell damage (Qian et al., 2024). Overexpressing MhNAC21/22 in Arabidopsis thaliana from Malus hupehensis enhances root penetration ability, indicating positive regulation of root compression tolerance (Cheng, 2024). Under low nitrogen stress, the NAC1 gene in Betula luminifera is regulated by miR164, and its expression rapidly increases to promote root development and enhance nitrogen absorption in seedlings (Wu et al., 2016b).
4.5 Participation in the biotic stress response
Plants are immobile, so they suffer adverse effects from the external environment during growth and development. Bacteria and fungi are significant biotic stressors (Figure 5D). For example, overexpression of VpNAC1 in Vitis vinifera enhances the tolerance of transgenic tobacco to powdery mildew fungi and Phytophthora nicotianae (Zhu et al., 2012). The NAC TF NAC61 in grapevine regulates the Botrytis cinerea-susceptibility gene WRKY52 (Foresti et al., 2023). VqNAC44 enhances stilbene synthesis and disease tolerance in Chinese wild grapes by interacting with VqMYB15 (Wang et al., 2024b). PaNAC03 and PaNAC04 may enhance tolerance to white rot infection in Picea abies (Dalman et al., 2017). The NAC genes MdNAC43, MdNAC45, MdNAC47, MdNAC49, and MdNAC51 identified in Malus pumila are differentially expressed during infection with the Alternaria alternata apple pathotype, and their relative expression levels increase significantly, indicating that they may play a regulatory role in the tolerance of M. pumila to fungal diseases (Li et al., 2019a). Overexpressing MpERF105 and MpNAC72 boosts anthocyanin accumulation and enhances apple rust tolerance in leaves (Wang et al., 2024e). After Ectropis obliqua feeding, the tea plant (Camellia sinensis) activates JA signaling, upregulating CsNAC30 and CsTCP11. This positively regulates CsCHAT1 transcription, promotes (Z)-3-hexenyl acetate (3-HAC) synthesis in leaves, and enhances tolerance to E. obliqua (Gu et al., 2025). The interaction between CsNAC17 and CsbHLH62 activates CsRPM1, triggering the hypersensitive response and H2O2 accumulation, enhancing tea plant leaf tolerance to anthracnose (Han et al., 2024).
Under biotic stress, NAC TFs mainly participate in hormone-mediated disease resistance pathways, activating or repressing related gene expression to enhance plant adaptability and resistance. However, the specific mechanisms and pathways remains unknown.
5 Existing problems and prospects
NAC TFs have been discovered in dozens of plants. Remarkable achievements have been made in research on their structures, expression characteristics, and functions. They are a very promising family of genes for plant breeding. Through combined analysis of multiomics, such as genomics and proteomics, the structural features and characteristics of many forest tree NAC TFs have been identified, and it is more accurate to conduct functional verification research on forest tree NAC TFs with the development of gene editing technology. Many studies have reported on the gene functions of forest tree NAC TFs. However, most of the research has focused on whether NAC genes achieve the expected target effects in terms of growth, development, and stress responses after being transferred into plants, and their specific functions are still unclear. In particular, their roles and mechanisms in woody plants have not been fully elucidated. Most studies on the regulatory network and action mechanism of NAC TFs are concentrated at the levels of gene cloning, structural identification, and expression analysis, and there is a lack of in-depth research on specific downstream target genes and upstream regulatory factors of NAC TFs. Although previous studies have shown that NAC TFs play an important role in the growth, development, and stress responses of woody plants, their specific molecular mechanisms remain unclear, particularly in terms of the regulation of hormones and the signal transduction pathways. Therefore, in the future more forest tree NAC TFs will be mined through methods such as reverse genetics, genomics, and proteomics, and the functions of specific NAC TFs will be further explored through gene editing and transgenic technologies. It will be necessary to establish the regulatory network of NAC TFs, identify their downstream target genes and upstream regulatory factors, and combine multiomics data, such as the transcriptome, proteome, and metabolome, to analyze systematically the regulatory mechanisms and action pathways of NAC TFs, to understand more fully their roles in plant growth, development, and the stress responses, gradually improve the functional analysis of the NAC family in woody plants.
Cultivating superior forest tree varieties is currently the focus of forest tree genetic breeding research. The existing single superior traits are not meeting social needs, and comprehensive improvements in multiple target traits are required. Therefore, fully mining the key NAC genes from forest trees by combining existing superior forest tree varieties through genetic engineering methods and breeding forest tree varieties with high yield, strong adaptability, and stress tolerance, and applying them to production are needed. Most of the receptor plants for verifying NAC gene function are model plants, such as tobacco and A. thaliana. There are relatively more poplars among woody plants, while relatively fewer other plants are used in genetic transformation experiments, and there are even fewer reports on molecular breeding research in forest trees. Therefore, cross-species comparative studies are lacking, which prevents a comprehensive understanding of the functional differences in the NAC TFs in different woody plants. In recent years, genetic transformation and regeneration systems in common woody plants, such as poplar and pine, have progressed, but challenges remain. Most genetic transformations use Agrobacterium-mediated methods with antibiotics like kanamycin and hygromycin as markers, but this often results in false positives and chimeras, complicating screening and causing unstable gene expression. Transformation rates are low and cycles are long. While gene-editing technologies, such as CRISPR/Cas9, are widely used, their efficiency and stability need improvement and vary across species. In the future, this could be used as an entry point to mine more forest tree NAC TFs with high ecological and commercial value, conduct comparative studies of NAC TFs in different woody plants, explore their evolutionary processes and functional diversity, and establish an efficient, universal genetic transformation and regeneration system for co-transformation of multiple genes within a species or the same gene across species, improving the efficiency of forest tree traits. Additionally, clarifying the molecular mechanisms by which NAC TFs regulate the growth of forest trees and provide genetic resources, and establishing a theoretical basis for understanding their roles in plant adaptive evolution and forest tree trait improvement.
Most experiments on the regulation of growth, development, and stress responses in woody plants by NAC TFs use potted seedlings of common main cultivated varieties, so the conclusions apply to the seedling stage. However, woody plants are perennial with long growth cycles, and there are clear differences between the physiological functions and stress response mechanisms of seedlings and mature plants (Wu et al., 2015). As the plants grow, the types and content levels of secondary metabolites and hormones, as well as the functions of organs, such as roots, stems, leaves, flowers, and fruits, change accordingly. Similarly, it remains unclear whether the functions expressed by transgenic plants during the seedling stage also change later. Currently, the field trials of genetically modified forest trees mainly focus on the research of improving plant stress resistance. Whether there will be changes in their long-term physiological states and organ functions still requires more long-term field trials for monitoring. Therefore, in the future, the focus of experiments should not be limited to the seedling stage. Instead, transgenic plants should be planted for field trials. This will make it easier to observe whether the trait characteristics exhibited by transgenic plants under laboratory conditions can be maintained under long-term natural stress. Transgenic plants should be field-tested to observe if lab-demonstrated traits persist under long-term natural stress. This will allow for observations that cannot be seen during short-term stress at the seedling stage, such as damage to macromolecules (cell membranes, proteins, DNA) due to stomatal closure, impaired photosynthesis, and ROS accumulation, leading to a decline in physiological functions (Wei et al., 2020).
Hetero and homologous expression genes enable transgenic plants to obtain excellent tolerance and adapt more effectively to the environment. However, while the target genes are being expressed, they may interfere with the normal expression of host genes, leading to the upregulation or downregulation of genes, producing unexpected effects. For example, after some genes are transferred into plants, overexpression (strong constitutive promoters) leads to negative effects in the plants, as slow growth and organ senescence occur, harmful secondary metabolites accumulate, and adaptability of the stress response is weaker than expected (Shao et al., 2015). Some foreign NAC genes promote or inhibit stress tolerance without affecting growth. For example, CpNAC68 in Chimonanthus praecox enhances heat tolerance without impacting growth (Lin et al., 2021). Conversely, some foreign genes affect the growth and development of plants while also enhancing stress tolerance, such as by regulating root growth in transgenic plants (Han et al., 2015; Zhang, 2019a). NAC TFs often regulate plant growth, development, and stress responses. Under natural conditions, most plants face multiple simultaneous stressors, such as drought, high temperature, and salinity (Shao et al., 2015). Under these conditions, enabling transgenic plants to cope with several stressors without hindering growth or even positively regulating it, to achieve expected benefits remains a challenge. The focus of future research should shift toward more in-depth research on the molecular mechanisms that produce unexpected effects, and whether it is possible to counteract some adverse effects by using weaker or specific promoters to drive gene overexpression. Moreover, as plants grow, they have some self-repair ability against negative impacts from the introduction of foreign genes. It remains unclear whether foreign gene expression and early-stage traits change with growth and environmental change. Therefore, we should continuously observe gene function and expression in adult trees to understand more fully the changing effects of foreign genes on recipient plants and obtain high-quality trees with excellent genetic traits.
Currently, research on NAC TFs in woody plants has focused on growth, development, and stress responses, with less attention on ecological adaptability. Introducing a foreign gene into a recipient plant could impact nearby genes, affecting certain plant traits. Within the gene flow range of genetically modified plants, if there are weed species that are genetically compatible for hybridization, field planting of transgenic plants may cause gene flow, leading to mutations in surrounding plants and weed overgrowth. Large-scale cultivation would reduce the number of traditional plants, disrupting ecological balance (Wang et al., 2024a; Huang, 2022). Therefore, long-term ecological monitoring is needed in the later stage to assess its impact on the ecosystem. The adaptability of plants relies on the integration of various stress response pathways under a constantly changing environment (Xu et al., 2024, 2025). Domestic and foreign scholars have relatively few research reports on the extreme temperature stress response of woody plant NAC TFs. Due to limited research on the heat stress response, only a few NAC genes have been confirmed to respond to extreme temperatures in woody plants. Moreover, current research is mostly biased toward the regulatory role of NAC genes under a single high-temperature stress, and further research is required to elucidate their related signal transmission and transcriptional regulatory networks. Relying on gene editing technologies, such as CRISPR/Cas9, will allow us to identify and utilize extreme temperature stress-related NAC TFs, and study the role of woody plant NAC TFs in ecological adaptation, particularly their performance under climate change and other environmental pressures, thus providing a theoretical basis for the cultivation and protection of woody plants.
In conclusion, although great progress has been made in research on NAC TFs since 1996, many mysteries are yet to be solved. Solving these mysteries and conducting relevant research will help to deepen our understanding of NAC TFs in woody plants and provide important theoretical bases and gene resources for plant breeding and ecological protection. With the development of molecular biology techniques, and employing genetic engineering methods such as genetic transformation and utilizing the characteristics of the NAC TFs, novel biotechnological approaches will be developed to improve certain plant traits and obtain superior new varieties, thereby enhancing the stress tolerance and productivity of woody plants, promoting the creation of new forest germplasm and providing broad application prospects in plant breeding.
Author contributions
HZ: Writing – original draft, Resources, Conceptualization, Data curation. JZ: Data curation, Conceptualization, Writing – original draft, Resources. TZ: Writing – original draft, Data curation, Resources, Investigation. GW: Investigation, Writing – review & editing, Data curation, Resources. ZH: Writing – review & editing. YM: Writing – review & editing. JB: Writing – review & editing. YR: Conceptualization, Writing – review & editing, Supervision, Funding acquisition. MY: Supervision, Conceptualization, Writing – review & editing, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Hebei Natural Science Foundation (C2023204136), Key Research and Development Program of Hebei Province (21326301D, 22326318D), the Basic Scientific Research Operating Expenses Research Project of Hebei Provincial Colleges and Universities (KY2023034) and the Talents Special Fund of Hebei Agricultural University (YJ2021028).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell. 9, 841–857. doi: 10.1105/tpc.9.6.841
An, J. P., Li, R., Qu, F. J., You, C. X., Wang, X. F., and Hao, Y. J. (2018a). An apple NAC transcription factor negatively regulates cold tolerance via CBF-dependent pathway. J. Plant Physiol. 221, 74–80. doi: 10.1016/j.jplph.2017.12.009
An, J. P., Yao, J. F., Xu, R. R., You, C. X., Wang, X. F., and Hao, Y. J. (2018b). An apple NAC transcription factor enhances salt stress tolerance by modulating the ethylene response. Physiol. Plant 164, 279–289. doi: 10.1111/ppl.12695
Bian, Y. H. (2021). Mining of differential genes related to dormancy release of Pyrus calleryana seeds and preliminary exploration of the regulatory mechanism of the PcNAC30 gene (Yangzhou: Yangzhou Univ). doi: 10.27441/d.cnki.gyzdu.2021.001897
Chen, Z., Peng, Z., Liu, S., Leng, H., Luo, J., Wang, F., et al. (2022). Overexpression of PeNAC122 gene promotes wood formation and tolerance to osmotic stress in poplars. Physiol. Plant 174, 13751. doi: 10.1111/ppl.13751
Cheng, Y. J. (2024). MiR164a-MhNAC21/22 module regulates the mechanism of root response to mechanical pressure in Pingyi sweet tea (Taian: Shandong Agric. Univ). doi: 10.27277/d.cnki.gsdnu.2024.001138
Cong, L. (2023). Research on the function and mechanism of PbNAC71 in regulating the dwarf traits of Pyrus spp (Xianyang: Northwest A&F Univ). doi: 10.27409/d.cnki.gxbnu.2023.002380
D’Incà, E., Cazzaniga, S., Foresti, C., Vitulo, N., Bertini, E., Galli, M., et al. (2021). VviNAC33 promotes organ de-greening and represses vegetative growth during the vegetative-to-mature phase transition in grapevine. New Phytol. 231, 726–746. doi: 10.1111/nph.17263
Dai, F. W., Zhang, C. Q., Jiang, X. Q., Kang, M., Yin, X., Lu, P., et al. (2012). RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals. Plant Physiol. 160, 2064–2082. doi: 10.1104/pp.112.207720
Dalman, K., Wind, J. J., Nemesio-Gorriz, M., Hammerbacher, A., Lundén, K., Ezcurra, I., et al. (2017). Overexpression of PaNAC03, a stress induced NAC gene family transcription factor in Norway spruce leads to reduced flavonol biosynthesis and aberrant embryo development. BMC Plant Biol. 17, 1–17. doi: 10.1186/s12870-016-0952-8
Duval, M., Hsieh, T. F., Kim, S. Y., and Thomas, T. L. (2002). Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 50, 237–248. doi: 10.1023/A:1016028530943
Ernst, H. A., Olsen, N. A., Skriver, K., and Lo, L. L. (2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5, 297–303. doi: 10.1038/sj.embor.7400093
Fang, L., Su, L., Sun, X., Li, X., Sun, M., Karungo, S. K., et al. (2016). Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis. J. Exp. Bot. 67, 2829–2845. doi: 10.1093/jxb/erw122
Fang, Y., You, J., Xie, K., Xie, W., and Xiong, L. (2008). Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics 280, 547–563. doi: 10.1007/s00438-008-0386-6
Feng, C. T., Jiang, L., Liu, X. Y., Luo, L., Pan, H. T., Zhang, Q. X., et al. (2023). Identification of the NAC gene family in Rosa persica and analysis of its response to drought stress. Biotechnol. Bull. 39, 283–296. doi: 10.13560/j.cnki.biotech.bull.1985.2023-0531
Foresti, C., Orduña, L., Matus, J. T., Vandelle, E., Danzi, D., Bellon, O., et al. (2023). NAC61 regulates late-and post-ripening osmotic, oxidative, and biotic stress responses in grapevine. J. Exp. Bot. 75, 2330–2350. doi: 10.1093/jxb/erad507
Fu, C. C., Chen, H. J., Gao, H. Y., Wang, S. L., Wang, N., Jin, J. C., et al. (2021). Papaya CpMADS4 and CpNAC3 co-operatively regulate ethylene signal genes CpERF9 and CpEIL5 during fruit ripening. Postharvest Biol. Technol. 175, 111485. doi: 10.1016/j.postharvbio.2021.111485
Fu, C. C., Han, Y. C., Fan, Z. Q., Chen, J. Y., Chen, W. X., Lu, W. J., et al. (2016). The Papaya transcription factor CpNAC1 modulates carotenoid biosynthesis through activating phytoene desaturase genes CpPDS2/4 during fruit ripening. J. Agric. Food Chem. 64, 5454–5463. doi: 10.1021/acs.jafc.6b01020
Fu, C. C., Han, Y. C., Kuang, J. F., Chen, J. Y., and Lu, W. J. (2017). Papaya CpEIN3a and CpNAC2 Co-operatively regulate carotenoid biosynthesis-related genes CpPDS2/4, CpLCY-e and CpCHY-b during fruit ripening. Plant Cell Physiol. 58, 2155–2165. doi: 10.1093/pcp/pcx149
Fu, C., Han, C., Wei, Y., Liu, D., and Han, Y. (2024a). Two NAC transcription factors regulated fruit softening through activating xyloglucan endotransglucosylase/hydrolase genes during kiwifruit ripening. Int. J. Biol. Macromol. 263, 130678. doi: 10.1016/j.ijbiomac.2024.130678
Fu, C., Han, C., Yu, Z., Liu, D., Wei, Y., and Han, Y. (2024b). Ethylene induced AcNAC3 and AcNAC4 take part in ethylene synthesis through mediating AcACO1 during kiwifruit (Actinidia chinensis) ripening. J. Sci. Food Agric. 104, 7367–7374. doi: 10.1002/jsfa.13557
Fu, L. Z., Yang, Y., Guo, J. H., Liu, J., Wang, M., Wang, X., et al. (2024c). Identification of the NAC gene family in three species of Castanea plants and analysis of codon bias. SW China J. Agric. Sci. 37, 1677–1689. doi: 10.16213/j.cnki.scjas.2024.8.004
Fu, C., Yu, Z., Han, C., and Han, Y. (2023). Ethylene induced CpNAC4 participates in ethylene synthesis by regulating CpACS2 and CpACO4 during papaya fruit ripening. Postharvest Biol. Technol. 206, 112582. doi: 10.1016/j.postharvbio.2023.112582
Geng, M. X. (2023). Identification of the NAC25 gene in Prunus persica and phenotypic analysis of transgenic Populus (Zhengzhou: Henan Agric Univ). doi: 10.27117/d.cnki.ghenu.2023.000914
Gong, L., Zhang, H., Liu, X., Gan, X., Nie, F., Yang, W., et al. (2020). Ectopic expression of HaNAC1, an ATAF transcription factor from Haloxylon ammodendron, improves growth and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 151, 535–544. doi: 10.1016/j.plaphy.2020.04.008
Grant, E. H., Fujino, T., Beers, E. P., and Brunner, A. M. (2010). Characterization of NAC domain transcription factors implicated in control of vascular cell differentiation in Arabidopsis and Populus. Planta 232, 337–352. doi: 10.1007/s00425-010-1181-2
Gu, H., Li, J., Qiao, D., Li, M., Yao, Y., Xie, H., et al. (2025). A defensive pathway from NAC and TCP transcription factors activates a BAHD acyltransferase for (Z)-3-hexenyl acetate biosynthesis to resist herbivore in tea plant (Camellia sinensis). New Phytol. 245, 1232–1248. doi: 10.1111/nph.20283
Guo, H., Wang, Y., Liu, H., Hu, P., Jia, Y., Zhang, C., et al. (2015). Exogenous GA3 application enhances xylem development and induces the expression of secondary wall biosynthesis related genes in Betula platyphylla. Int. J. Mol. Sci. 16, 22960–22975. doi: 10.3390/ijms160922960
Guo, G. L., Yu, T., Tang, X. M., Liu, L., Ye, Z. F., Jia, B., et al. (2023). Expression analysis of NAC transcription factors during the regreening process of ‘Dangshan Suli’ yellow leaves. J. Northw. Bot. 43, 1994–2004. doi: 10.7606/j.issn.1000-4025.20230458
Han, Z. (2021). Regulation of secondary cell wall synthesis by the Populus SND2-B2 gene (Taian: Shandong Agric. Univ). doi: 10.27277/d.cnki.gsdnu.2021.000365
Han, D., Du, M., Zhou, Z., Wang, S., Li, T., Han, J., et al. (2020a). An NAC transcription factor gene from Malus baccata, MbNAC29, increases cold and high salinity tolerance in Arabidopsis. In Vitro Cell Dev-Pl. 56, 588–599. doi: 10.1007/s11627-020-10105-9
Han, D., Du, M., Zhou, Z., Wang, S., Li, T., Han, J., et al. (2020b). Overexpression of a Malus baccata NAC transcription factor gene MbNAC25 increases cold and salinity tolerance in Arabidopsis. Int. J. Mol. Sci. 21, 1198. doi: 10.3390/ijms21041198
Han, X., Feng, Z., Xing, D., Yang, Q., Wang, R. G., Qi, L. W., et al. (2015). Two NAC transcription factors from Caragana intermedia altered salt tolerance of the transgenic Arabidopsis. BMC Plant Biol. 15, 208. doi: 10.1186/s12870-015-0591-5
Han, X., He, G., Zhao, S., Guo, C., and Lu, M. (2012). Expression analysis of two NAC transcription factors PtNAC068 and PtNAC154 from poplar. Plant Mol. Biol. Rep. 30, 370–378. doi: 10.1007/s11105-011-0350-1
Han, R., Mei, H., Huang, Q., Ma, C., Zhao, Y., Jeyaraj, A., et al. (2024). CsNAC17 enhances resistance to Colletotrichum gloeosporioides by interacting with CsbHLH62 in Camellia sinensis. Hortic. Res. 12, uhae295. doi: 10.1093/hr/uhae295
He, X. (2023b). Establishment of genetic transformation system of Populus × beichengensis and functional study of PtrNAC006 transcription factor in response to drought stress (Harbin: Northeast forest. Univ). doi: 10.27009/d.cnki.gdblu.2023.001035
He, Z., Li, Z., Lu, H., Huo, L., Wang, Z., Wang, Y., et al. (2019). The NAC protein from Tamarix hispida, ThNAC7, confers salt and osmotic stress tolerance by increasing reactive oxygen species scavenging capability. Plants 8, 221. doi: 10.3390/plants8070221
He, S. E., OU, Y. L. N., Yang, J. Q., Zheng, J. Q., and Chen, S. X. (2023a). Cloning and functional analysis of the EugNAC007 gene in Eucalyptus urophylla × Eucalyptus grandis. Eucalypt. Sci. Technol. 40, 1–8. doi: 10.13987/j.cnki.askj.2023.04.001
Hou, D., Zhao, Z., Hu, Q., Li, L., Vasupalli, N., Zhuo, J., et al. (2020). PeSNAC-1 a NAC transcription factor from moso bamboo (Phyllostachys edulis) confers tolerance to salinity and drought stress in transgenic rice. Tree Physiol. 40, 1792–1806. doi: 10.1093/treephys/tpaa099
Hu, Z. B. (2023). Functional study of the PxNAC73 gene of Populus simonii × P. nigra in the wood formation process (Harbin: Northeast Forest. Univ). doi: 10.27009/d.cnki.gdblu.2023.000260
Hu, P., Zhang, K., and Yang, C. (2019). BpNAC012 positively regulates abiotic stress responses and secondary wall biosynthesis. Plant Physiol. 179, 700–717. doi: 10.1104/pp.18.01167
Huang, Y. L. (2022). Growth and ecological safety evaluation of transgenic multi-resistant gene Populus × euramericana ‘107’ (Baoding: Hebei Agric. Univ). doi: 10.27109/d.cnki.ghbnu.2022.000204
Huang, J., Deng, J., Zhu, L. W., and Chen, Q. F. (2017). Research progress of NAC family transcription factors related to plant seed development. Seeds 36, 51–55. doi: 10.16590/j.cnki.1001-4705.2017.11.051
Huang, X., Xu, H. H., Li, K. J., He, X. H., Xia, L. M., Lu, T. T., et al. (2023). Expression pattern of Mangifera indica MiNAC7 gene and functional analysis in flowering and abiotic stress responses. Chin. J. Trop. Crops. 44, 2256–2264. doi: 10.3969/j.issn.1000-2561.2023.11.014
Hussey, S. G., Mizrachi, E., Spokevicius, A. V., Bossinger, G., Berger, D. K., and Myburg, A. A. (2011). SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol. 11, 1–17. doi: 10.1186/1471-2229-11-173
Isayenkov, S. V. and Maathuis, F. J. M. (2019). Plant salinity stress: Many unanswered questions remain. Front. Plant Sci. 10, 80. doi: 10.3389/fpls.2019.00080
Jensen, M. K., Lindemose, S., de Masi, F., Reimer, J. J., Nielsen, M., Perera, V., et al. (2013). ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio. 3, 321–327. doi: 10.1016/j.fob.2013.07.006
Jia, D. F. (2019). Research on the regulation of drought stress response and dwarf traits by NAC transcription factors in apple (Xianyang: Northwest A&F Univ).
Jiang, G. X., Yan, H. L., Wu, F. W., Zhang, D. D., Zeng, W., Qu, H. X., et al. (2017). Litchi fruit LcNAC1 is a target of LcMYC2 and regulator of fruit senescence through its interaction with LcWRKY1. Plant Cell Physiol. 58, 1075–1089. doi: 10.1093/pcp/pcx054
Jin, C., Li, K. Q., Xu, X. Y., Zhang, H. P., Chen, H. X., Chen, Y. H., et al. (2017). A novel NAC transcription factor, PbeNAC1, of pyrus betulifolia confers cold and drought tolerance via interacting with PbeDREBs and activating the expression of stress-responsive genes. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01049
Ju, Y. L. (2019). Functional study of Vitis vinifera VvNAC08 and VvNAC17 transcription factors in response to drought stress (Xianyang: Northwest A&F Univ). doi: 10.27409/d.cnki.gxbnu.2019.000249
Ju, Y. L., Yue, X. F., Min, Z., Wang, X. H., Fang, Y. L., and Zhang, J. X. (2020). VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 146, 98–111. doi: 10.1016/j.plaphy.2019.11.002
Kang, C., Guo, C. H., Zhang, X. M., Liu, J. X., Yuan, X., Quan, S. W., et al. (2021). Genome-wide identification and analysis of the Juglans regia NAC gene family. J. Fruit Sci. 38, 1444–1458. doi: 10.13925/j.cnki.gsxb.20210158
Kim, H. J., Nam, H. G., and Lim, P. O. (2016). Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 33, 48–56. doi: 10.1016/j.pbi.2016.06.002
Kim, M. H., Tran, T. N. A., Cho, J. S., Park, E. J., Lee, H., Kim, D. G., et al. (2021). Wood transcriptome analysis of Pinus densiflora identifies genes critical for secondary cell wall formation and NAC transcription factors involved in tracheid formation. Tree Physiol. 41, 1289–1305. doi: 10.1093/treephys/tpab001
Kong, L., Song, Q., Wei, H., Wang, Y., Lin, M., Sun, K., et al. (2023). The AP2/ERF transcription factor PtoERF15 confers drought tolerance via JA-mediated signaling in Populus. New Phytol. 240, 1848–1867. doi: 10.1111/nph.19251
Kronzucker, H. J. and Britto, D. T. (2011). Sodium transport in plants: A critical review. New Phytol. 189, 54–81. doi: 10.1111/j.1469-8137.2010.03540.x
Li, H. L., Chen, X. X., Ji, X. L., Qiao, Z. W., Liu, R. X., Wang, X. F., et al. (2022). BTB protein MdBT2 negatively regulates iron homeostasis by interacting with MdNAC1 in apple. Environ. Exp. Bot. 195, 104778. doi: 10.1016/j.envexpbot.2021.104778
Li, H. L., Chen, X. X., Ji, X. L., Qiao, Z. W., Liu, R. X., Wang, X. F., et al. (2019a). Cloning and expression analysis of 13 Apple NAC transcription factor genes. J. Plant Genet. Resour. 20, 1041–1051. doi: 10.13430/j.cnki.jpgr.20181205001
Li, Y., Chen, J. H., Jin, Z., Hou, J. Y., Jiang, Y. S., and Xing, H. T. (2020). Function of the Populus trichocarpa NAC128 gene in secondary wall formation. Sci. Silv Sin. 56, 62–72. doi: 10.3969/j.issn.0517-6611.2025.02.022
Li, W., Li, X. X., Chao, J. T., Zhang, Z. L., Wang, W. F., and Guo, Y. F. (2018). NAC family transcription factors in Tobacco and their potential role in regulating leaf senescence. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01900
Li, S., Lin, Y. J., Wang, P., Zhang, B., Li, M., Chen, S., et al. (2019b). The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in Populus trichocarpa. Plant Cell. 31, 663–686. doi: 10.1105/tpc.18.00437
Li, S. L., Wang, C. M., and Li, X. J. (2021b). Bioinformatics analysis of the NAC transcription factor family in Punica granatum. Mol. Plant Breed. 19, 88–99. doi: 10.13271/j.mpb.019.000088
Li, M., Wu, Z., Gu, H., Cheng, D., Guo, X., Li, L., et al. (2021a). AvNAC030, a NAC domain transcription factor, enhances salt stress tolerance in Kiwifruit. Int. J. Mol. Sci. 22, 11897–11897. doi: 10.3390/ijms222111897
Li, Z. Q., Xing, W., Luo, P., Zhang, F. J., Jin, X. L., and Zhang, M. H. (2019c). Comparative transcriptome analysis of Rosa chinensis’ Slaters’ crimson China’ provides insights into the crucial factors and signaling pathways in heat stress response. Plant Physiol. Biochem. 142, 12–331. doi: 10.1016/j.plaphy.2019.07.002
Li, S. J., Yin, X. R., Wang, W. L., Liu, X. F., Zhang, B., and Chen, K. S. (2017). Citrus CitNAC62 cooperates with CitWRKY1 to participate in citric acid degradation via up-regulation of CitAco3. J. Exp. Bot. 68, 3419–3426. doi: 10.1093/jxb/erx187
Liang, K. H. (2021). Functional research and validation of the NAC transcription factors PwNAC30/PwNAC31 in Picea wilsonii (Beijing: Beijing Forest Univ). doi: 10.26949/d.cnki.gblyu.2020.000783
Liang, M. H., Liang, R. J., Yang, Z. F., and Deng, H. L. (2024a). Expression and influence of NAC6 transcription factor in the metabolic process of carotenoids in postharvest mango fruits. Food Sci. 45, 77–87. doi: 10.7506/spkx1002-6630-20230816-098
Liang, X. M., Qin, S. S., Wei, F., Wei, G. L., Lin, Q., and Liang, Y. (2024b). Research progress on the function of transcription factors in plant drought stress. Biol. Resour. 46, 220–230. doi: 10.14188/j.ajsh.2024.03.002
Liang, K. H., Wang, A. B., Yuan, Y. H., Miao, Y. H., and Zhang, L. Y. (2020). Picea wilsonii NAC transcription factor PwNAC30 negatively regulates abiotic stress tolerance in transgenic Arabidopsis. Plant Mol. Biol. Rep. 38, 554–571. doi: 10.1007/s11105-020-01216-z
Lin, H., Luan, X., Chen, C., Gong, X., Li, X., Li, H., et al. (2023). A systematic genome-wide analysis and screening of the NAC family genes related to wood formation in Cinnamomum camphora. Genomics 115, 110631. doi: 10.1016/j.ygeno.2023.110631
Lin, J., Liu, D. F., Wang, X., Ahmed, S. J., Li, M. Y., Kovinich, N., et al. (2021). Transgene CpNAC68 from wintersweet (Chimonanthus praecox) improves Arabidopsis survival of multiple abiotic stresses. Plant (Basel). 10, 1403–1403. doi: 10.3390/plants10071403
Liu, F. G. (2023). Expression and functional analysis of the Oryza sativa NAC transcription factor gene OsNAC071 (Kunming: Yunnan Agric. Univ).
Liu, J. H., Jing, Y. F., Liu, Y. X., Xu, Y. G., Yu, Y., Ge, X. X., et al. (2024). Identification of the NAC gene family in Prunus persica and research on PpNAC050 promoting fructose accumulation in fruits. AHS 51, 1983–1996. doi: 10.16420/j.issn.0513-353x.2023-0085
Liu, H. X., Xu, F. F., Yang, Z. L., Wu, X. M., Xu, S. Y., and Li, B. (2025). Identification and analysis of the NAC gene family in Camellia nitidissima based on full-length transcriptome sequencing. J. Anhui Agric. Sci. 53, 97–103 + 126. doi: 10.3969/j.issn.0517-6611.2025.02.022
Lu, X., Zhang, X., Duan, H., Lian, C., Liu, C., Yin, W., et al. (2018). Three stress-responsive NAC transcription factors from Populus euphratica differentially regulate salt and drought tolerance in transgenic plants. Physiol. Plant 162, 73–97. doi: 10.1111/ppl.12613
Luo, G. K., Mu, R. B., Xue, B., Zhang, H., Ren, Y. P., and Ma, H. (2023). Functional and characteristic analysis of NAC transcription factor HaNAC38 in Haloxylon ammodendron. J. Agric. Sci. Technol. 25, 65–73. doi: 10.13304/j.nykjdb.2022.0266
Matias, F., Novais de Oliveira, P., Gómez-Espinoza, O., Galeano, E., and Carrer, H. (2022). Overexpression of the Tectona grandis TgNAC01 regulates growth, leaf senescence and confer salt stress tolerance in transgenic tobacco plants. Peer J. 10, e13039–ee1039. doi: 10.7717/peerj.13039
Mei, C. (2023). Research on the function and regulatory mechanism of MdNAC104 transcription factor in Malus pumila under low temperature stress (Xianyang: Northwest A&F Univ). doi: 10.27409/d.cnki.gxbnu.2023.000186
Meng, L., Chen, S., Li, D., Huang, M., and Zhu, S. (2022). Genome-wide characterization and evolutionary expansion of poplar NAC transcription factors and their tissue-specific expression profiles under drought. Int. J. Mol. Sci. 24, 253–253. doi: 10.3390/ijms24010253
Mian, A., Oomen, R. J., Isayenkov, S., Sentenac, H., Maathuis, F. J., and Véry, A. A. (2011). Over-expression of an Na+- and K+-permeable HKT transporter in barley improves salt tolerance. Plant J. 68, 468–479. doi: 10.1111/j.1365-313X.2011.04701.x
Miao, Y. F., Zhou, D., Dong, B., and Zhao, H. B. (2021). Identification of OfNAC transcription factors in Osmanthus fragrans and their expression analysis during flower opening stages. J. Zhejiang A&F Univ. 38, 433–444. doi: 10.11833/j.issn.2095-0756.20200474
Min, T., Wang, M. M., Wang, H. X., Liu, X. F., Fang, F., Grierson, D., et al. (2015). Isolation and expression of NAC genes during persimmon fruit postharvest astringency removal. Int. J. Mol. Sci. 16, 1894–1906. doi: 10.3390/ijms16011894
Nieuwenhuizen, N. J., Chen, X. Y., Wang, M. Y., Matich, A. J., Perez, R. L., Allan, A. C., et al. (2015). Natural variation in monoterpene synthesis in kiwifruit: transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-like transcription factors. Plant Physiol. 167, 1243–1258. doi: 10.1104/pp.114.254367
Niu, Y. N. (2022). Regulatory mechanism of BpNAC2 transcription factor on salt tolerance of Betula platyphylla (Harbin: Northeast Forest. Univ). doi: 10.27009/d.cnki.gdblu.2022.000491
Niu, M. X., Feng, C. H., He, F., Zhang, H., Bao, Y., Liu, S. J., et al. (2024). The miR6445-NAC029 module regulates drought tolerance by regulating the expression of glutathione S-transferase U23 and reactive oxygen species scavenging in Populus. New Phytol. 242, 2043–2058. doi: 10.1111/nph.19703
Ohtani, M., Nishikubo, N., Xu, B., Yamaguchi, M., Mitsuda, N., Goue, N., et al. (2011). A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J. 67, 499–512. doi: 10.1111/j.1365-313X.2011.04614.x
Olsen, A. N., Ernst, H. A., Leggio, L. L., and Skriver, K. (2005). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. doi: 10.1016/j.tplants.2004.12.010
Ooka, H., Satoh, K., Doi, K., Nagata, T., Otomo, Y., Murakami, K., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247. doi: 10.1093/dnares/10.6.239
Pei, H. X., Ma, N., Tian, J., Luo, J., Chen, J. W., Li, J., et al. (2013). An NAC transcription factor controls ethylene-regulated cell expansion in flower petals. Plant Physiol. 163, 775–791. doi: 10.1104/pp.113.223388
Qi, G. (2012). Analysis of the Populus NAC gene family and research on the functions of PdCSLD5 and PdCSLD6 genes. Univ. Chin. Acad. Sci. Available online at: https://d.wanfangdata.com.cn/thesis/ChhUaGVzaXNOZXdTMjAyNDA5MjAxNTE3MjUSCFkyMzY4NzgxGghhNWMxNWJzaw==.
Qian, C. N., Xu, S. R., Li, M. R., Deng, M. C., Sun, S., Liu, G. Y., et al. (2024). Genome-wide identification of the NAC gene family in Salix matSudana and its expression analysis under waterlogging and salt stress. J. South. Agric. 55, 3056–3070. doi: 10.3969/j.issn.2095-1191.2024.10.018
Qin, H., Cui, X., Shu, X., and Zhang, J. (2023). The transcription factor VaNAC72-regulated expression of the VaCP17 gene from Chinese wild Vitis amurensis enhances cold tolerance in transgenic grape (V. vinifera). Plant Physiol. Biochem. 200, 107768–107768. doi: 10.1016/j.plaphy.2023.107768
Rushton, P. J., Bokowiec, M. T., Han, S., Zhang, H., Brannock, J. F., Chen, X., et al. (2008). Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae. Plant Physiol. 147, 280–295. doi: 10.1104/pp.107.114041
Shang, L. X. (2023). Identification of the NAC and TCP gene families in Lagerstroemia indica and the study of their expression patterns regulating the weeping trait (Hangzhou: Zhejiang A&F Univ). doi: 10.27756/d.cnki.gzjlx.2023.000213
Shao, H., Wang, H., and Tang, X. (2015). NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00902
Shen, H. J., Deng, J. Q., Liao, B. Y., Zhang, B. P., Li, Y. Q., and Wang, Y. (2024). Identification and bioinformatics analysis of the NAC transcription factor family in Camellia gauchowensis. Hunan Agric. Sci. 07, 7–14. doi: 10.16498/j.cnki.hnnykx.2024.007.002
Song, Y., Liu, H. D., Wang, H. B., Zhang, H. J., and Liu, F. Z. (2019). Cloning of VcNAC072 from Vaccinium myrtillus and functional analysis of its promotion on anthocyanin accumulation. Sci. Agric. Sin. 52, 503–511. doi: 10.3864/j.issn.0578-1752.2019.03.010
Souer, E., van Houwelingen, A., Kloos, D., Mol, J., and Koes, R. (1996). The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170. doi: 10.1016/S0092-8674(00)81093-4
Su, L., Fang, L., Zhu, Z., Zhang, L., Sun, X., Wang, Y., et al. (2020). The transcription factor VaNAC17 from grapevine (Vitis amurensis) enhances drought tolerance by modulating jasmonic acid biosynthesis in transgenic Arabidopsis. Plant Cell Rep. 39, 621–634. doi: 10.1007/s00299-020-02519-x
Su, M., Wang, S., Liu, W., Yang, M., Zhang, Z., Wang, N., et al. (2022). Interaction between MdWRKY55 and MdNAC17-L enhances salt tolerance in apple by activating MdNHX1 expression. Plant Sci. 320, 111282–111282. doi: 10.1016/j.plantsci.2022.111282
Sun, Y. (2019b). Study on the role and application of NACs in the seed development and dormancy release process of Pyrus calleryana (Yangzhou: Yangzhou Univ). doi: 10.27441/d.cnki.gyzdu.2019.001380
Sun, Y., Jiang, C., Jiang, R., Wang, F., Zhang, Z., and Zeng, J. (2021). A novel NAC transcription factor from Eucalyptus, EgNAC141, positively regulates lignin biosynthesis and increases lignin deposition. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.642090
Sun, Q., Jiang, S., Zhang, T., Xu, H., Fang, H., Zhang, J., et al. (2019a). Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 289, 110286. doi: 10.1016/j.plantsci.2019.110286
Sun, M. M., Liu, X., Huang, X. J., Yang, J. J., Qin, P. T., Zhou, H., et al. (2022). Genome-Wide Identification and Expression Analysis of the NAC Gene Family in Kandelia obovata, a Typical Mangrove Plant. Curr Issues Mol Biol. 44, 5622–5637. doi: 10.3390/cimb44110381
Van Zelm, E., Zhang, Y., and Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005
Wang, X. Y. (2022b). Functional analysis of PsnNAC054 gene in salt stress tolerance of Populus simonii × P. nigra (Harbin: Northeast Forest. Univ). doi: 10.27009/d.cnki.gdblu.2022.001434
Wang, Z. B. (2022c). Molecular mechanism of BpNAC090 transcription factor regulating drought tolerance of Betula platyphylla (Harbin: Northeast Forest. Univ). doi: 10.27009/d.cnki.gdblu.2022.001885
Wang, X. F. (2024d). Mechanism analysis of Tandem genes PpNAC56 and PpNAC72 regulating the maturation of Prunus persica fruit (Zhengzhou: Henan Agric. Univ). doi: 10.27117/d.cnki.ghenu.2024.000602
Wang, Y., An, H., Yang, Y., Yi, C., Duan, Y., Wang, Q., et al. (2024e). The MpNAC72/MpERF105-MpMYB10b module regulates anthocyanin biosynthesis in Malus ‘Profusion’ leaves infected with Gymnosporangium yamadae. Plant J. 118, 1569–1588. doi: 10.1111/tpj.16697
Wang, L. F., Chen, Z. J., Xu, Q., Shen, J. R., Zhao, Y. N., and Liu, J. X. (2024a). Study on insect resistance of transgenic Populus × euramericana’107’. For. Ecol. Sci. 39, 161–168. doi: 10.13320/j.cnki.hjfor.2024.0019
Wang, Y. L., Do, H., Deng, P., Zhou, X., and Quan, J. E. (2024f). Identification of the NAC gene family in Morus alba and expression pattern analysis under treatment with rooting powder. J. Fujian Agric. For. Univ. (Nat. Sci. Ed.). 53, 629–640. doi: 10.13323/j.cnki.j.fafu(nat.sci.).202311015
Wang, X. F., Gao, L. Y., Liu, N., Cheng, J., Wang, W., Tan, B., et al. (2024c). Identification of the peach gene PpNAC and its expression analysis at different developmental stages. J. Henan Agric. Univ. 03, 412–423. doi: 10.16445/j.cnki.1000-2340.20240301.002
Wang, L., Li, Z., Lu, M., and Wang, Y. (2017). ThNAC13, a NAC transcription factor from Tamarix hispida, confers salt and osmotic stress tolerance to transgenic Tamarix and. Arabidopsis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00635
Wang, J. Y., Wang, J. P., and He-Yuan (2013). A Populus euphratica NAC protein regulating Na+/K+ homeostasis improves salt tolerance in Arabidopsis thaliana. Gene 521, 265–273. doi: 10.1016/j.gene.2013.03.068
Wang, H. L., Yang, Q., Tan, S., Wang, T., Zhang, Y., Yang, Y., et al. (2022a). Regulation of cytokinin biosynthesis using PtRD26 pro-IPT module improves drought tolerance through PtARR10-PtYUC4/5-mediated reactive oxygen species removal in Populus. J. Integr. Plant Biol. 64, 771–786. doi: 10.1111/jipb.13218
Wang, Y., Zang, W., Li, X., Wang, C., Wang, R., Jiang, T., et al. (2023). Ectopic expression of PsnNAC090 enhances salt and osmotic tolerance in transgenic tobacco. Int. J. Mol. Sci. 24, 8985. doi: 10.3390/ijms24108985
Wang, L., Zhang, M., Li, J., Luo, Q., Yao, Q., Huang, Q., et al. (2024b). VqNAC44 enhances stilbene synthesis and disease resistance in Chinese wild grape by interacting with VqMYB15. Plant Sci. 341, 111994. doi: 10.1016/j.plantsci.2024.111994
Wang, R., Zhang, Y., Wang, C., Wang, Y. C., and Wang, L. Q. (2021b). ThNAC12 from Tamarix hispida directly regulates ThPIP2; 5 to enhance salt tolerance by modulating reactive oxygen species. Plant Physiol. Biochem. 163, 27–35. doi: 10.1016/j.plaphy.2021.03.042
Wang, H. L., Zhang, Y., Wang, T., Yang, Q., Yang, Y. L., Li, Z., et al. (2021a). An alternative splicing variant of PtRD26 delays leaf senescence by regulating multiple NAC transcription factors in Populus. Plant Cell. 33, 1594–1614. doi: 10.1093/plcell/koab046
Wei, Z. L., Han, Y. J., and Bu, Y. Y. (2020). Research progress on stress resistance in woody plants. Mol. Plant Breed. 18, 2382–2387. doi: 10.13271/j.mpb.018.002382
Wen, Z. X. (2019). Screening and functional analysis of NAC transcription factors related to Litchi chinensis fruit abscission. SCAU. doi: 10.27152/d.cnki.ghanu.2019.000866
Wen, B. B., Zhao, X. H., Gong, X. Y., Zhao, W. Z., Sun, M. Y., Chen, X. D., et al. (2023). The NAC transcription factor MdNAC4 positively regulates nitrogen deficiency-induced leaf senescence by enhancing ABA biosynthesis in apple. Mol. Hortic. 3, 5–5. doi: 10.1186/s43897-023-00053-4
Wu, X. Q. (2021). Cloning and functional analysis of PpNAC72 gene in Prunus persica (Tianjin: Tianjin Agric. Univ). doi: 10.27717/d.cnki.gtjnx.2019.000047
Wu, H., Fu, B., Sun, P., Xiao, C., and Liu, J. H. (2016a). A NAC transcription factor represses putrescine biosynthesis and affects drought tolerance. Plant Physiol. 172, 1532–1547. doi: 10.1104/pp.16.01096
Wu, X. Q., He, X., Gao, J. H., and Li, S. (2024). Creation and characterization analysis of Populus simonii × Populus nigra germplasm with high drought tolerance by transforming PsnNAC007. BBR 44, 349–360. doi: 10.7525/j.issn.1673-5102.2024.03.004
Wu, F. Z., Wang, H. X., Xu, G. H., and Zhang, Z. C. (2015). Research progress on the physiological and molecular mechanisms of woody plants under low temperature stress. Sci. Silvae Sin. 51, 116–128. doi: 10.11707/j.1001-7488.20150713
Wu, J., Zhang, J. H., Huang, M. H., Zhu, M. H., and Tong, Z. K. (2016b). Expression analysis of miR164 and its target gene NAC1 in Betula luminifera under low nitrogen stress. Hereditas 38, 155–162. doi: 10.16288/j.yczz.15-231
Xiao, S. W., Wang, H. Y., Li, T. H., Jin, Y. D., Wu, J., Zhang, Z., et al. (2025). Identification and expression of NAC gene family in Casuarina equisetifolia. Mol. Plant Breed., 1–12. Available at: http://kns.cnki.net/kcms/detail/46.1068.S.20241111.1253.011.html.
Xin, Y., Huang, R., Xu, M., and Xu, L. (2023). Transcriptome-Wide identification and response pattern analysis of the salix integra NAC transcription factor in response to pb stress. Int. J. Mol. Sci. 24, 11334. doi: 10.3390/ijms241411334
Xiong, J. (2020). Research on the mechanism of PtAOS1 and PtNAC72 genes of Poncirus trifoliata in response to drought stress (Changsha: Hunan Agric Univ). doi: 10.27136/d.cnki.ghunu.2020.000162
Xu, M. (2008). Research on gene expression regulation of adventitious root development in Populus (Nanjing: Nanjing Forest Univ).
Xu, L., Liu, Y., Feng, S., Liu, C., Zhong, X., Ren, Y., et al. (2024). The relationship between atmospheric particulate matter, leaf surface microstructure, and the phyllosphere microbial diversity of Ulmus L. BMC Plant Biol. 24, 566. doi: 10.1186/s12870-024-05232-z
Xu, Q., Wang, W. Q., Zeng, J. K., Zhang, J., Grierson, D., Li, X., et al. (2015). A NAC transcription factor, EjNAC1, affects lignification of loquat fruit by regulating lignin. Post Biol. Technol. 102, 25–31. doi: 10.1016/j.postharvbio.2015.02.002
Xu, L., Zhong, X., Liu, C., Ren, Y., Huang, Y., Liu, Y., et al. (2025). Iron deficiency and toxicity trigger divergent metabolic responses and adaptive plasticity in Ulmus pumila: Insights from integrated transcriptomic and metabolomic analyses. Plant Physiol. Biochem. 221, 109601. doi: 10.1016/j.plaphy.2025.109601
Yang, J. (2021). Functional analysis of the PtNAC101 gene of Populus trichocarpa under high salt stress (Yantai: Ludong Univ). doi: 10.27216/d.cnki.gysfc.2021.000616
Yang, L. (2022a). Research on the mechanism of PpNAC1 and PpNAC2 regulating the thickening of xylem in Prunus persica branches (Zhengzhou: Henan Agric. Univ). doi: 10.27117/d.cnki.ghenu.2022.000459
Yang, C. X. (2024). Study on NAC transcription factor gene family and forage quality in Gymnocarpos przewalskii (Inner Mongolia: Inner Mong. Norm. Univ). doi: 10.27230/d.cnki.gnmsu.2024.000036
Yang, Z., Cai, C. H., Zeng, J., Dai, H. F., Song, X. Q., Ding, X. P., et al. (2023). Cloning and expression analysis of AsNAC22 gene from Aquilaria sinensis. Mol. Plant Breed., 1–17. Available online at: http://kns.cnki.net/kcms/detail/46.1068.S.20231115.1429.020.html (Accessed March 30, 2025).
Yang, Y., Chen, M., Zhu, Q., Lv, Y., Liu, C., Wei, Y., et al. (2025). The transcription factors AdNAC3 and AdMYB19 regulate kiwifruit ripening through brassinosteroid and ethylene signaling networks. Plant Physiol. 197, kiaf084. doi: 10.1093/plphys/kiaf084
Yang, Y., Yoo, C. G., Rottmann, W., Winkeler, K. A., Collins, C. M., Gunter, L. E., et al. (2019). PdWND3A, a wood-associated NAC domain-containing protein, affects lignin biosynthesis and composition in. Populus. BMC Plant Biol. 219, 486. doi: 10.1186/s12870-019-2111-5
Yang, X. Z., Zhou, S. L., He, X. H., Liu, Y., Yu, H. X., Lu, T. T., et al. (2022b). Functional study of the Mangifera indica MiNAC1 gene. Chin. J. Trop. Crops. 43, 1527–1534. doi: 10.3969/j.issn.1000-2561.2022.08.001
Yao, W. J., Zhao, K., Cheng, Z., Li, X. Y., Zhou, B., and Jiang, T. B. (2018). Transcriptome analysis of poplar under salt stress and over-expression of transcription factor NAC57 gene confers salt tolerance in transgenic Arabidopsis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01121
Ye, X. X. (2021). Research on the salt response rules and molecular mechanisms of lncRNAs in Populus trichocarpa analyzed by big data (Harbin: Northeast Forest. Univ). doi: 10.27009/d.cnki.gdblu.2021.000095
Yue, Y. Z., Li, L., Li, Y. L., Li, H. Y., Ding, W. J., Shi, T. T., et al. (2020). Genome-wide analysis of NAC transcription factors and characterization of the cold stress response in Sweet Osmanthus. Plant Mol. Biol. Rep. 38, 314–330. doi: 10.1007/s11105-020-01195-1
Zeng, W. W., Li, Y. F., Zhu, Y. M., Lin, Y. Q., Chen, S. Y., and Zou, S. Q. (2024). Genome-wide identification of the NAC gene family in Cinnamomum camphora and expression analysis under salt stress. Sw. China J. Agr. Sci. 37, 1393–1408. doi: 10.16213/j.cnki.scjas.2024.7.001
Zhang, M. J. (2019a). Cloning and preliminary functional analysis of transcription factors PwNAC11/PwNAC38 in Picea wilsonii (Beijing: Beijing Forest Univ). doi: 10.26949/d.cnki.gblyu.2019.001283
Zhang, S., Chen, Y., Zhao, L., Li, C., Yu, J., Li, T., et al. (2020b). A novel NAC transcription factor, MdNAC42, regulates anthocyanin accumulation in red-fleshed apple by interacting with MdMYB10. Tree Physiol. 40, 413–423. doi: 10.1093/treephys/tpaa004
Zhang, X., Cheng, Z., Yao, W., Zhao, K., Wang, X., and Jiang, T. (2021b). Functional characterization of PsnNAC036 under salinity and high temperature stresses. Int. J. Mol. Sci. 22, 2656. doi: 10.3390/ijms22052656
Zhang, X., Cheng, Z., Zhao, K., Yao, W., Sun, X., Jiang, T., et al. (2019b). Functional characterization of poplar NAC13 gene in salt tolerance. Plant Sci. 281, 1–8. doi: 10.1016/j.plantsci.2019.01.003
Zhang, S., Dong, R., Wang, Y., Li, X., Ji, M., and Wang, X. (2021a). NAC domain gene VvNAC26 interacts with VvMADS9 and influences seed and fruit development. Plant Physiol. Biochem. 164, 63–72. doi: 10.1016/j.plaphy.2021.04.031
Zhang, Z. X., Guo, X. W., Wang, Z. W., Wang, P. F., Zhang, J. C., Du, J. J., et al. (2023d). Identification and expression analysis of the NAC gene family in Cerasus humilis. J. Fruit Sci. 40, 206–222. doi: 10.13925/j.cnki.gsxb.20220404
Zhang, X., Li, L., Lang, Z., Li, D., He, Y., Zhao, Y., et al. (2023c). Corrigendum: Genome-wide characterization of NAC transcription factors in Camellia sinensis and the involvement of CsNAC28 in drought tolerance. Front. Plant Sci. 14. doi: 10.3389/fpls.2022.1065261
Zhang, D., Ma, H. R., Duan, X. J., Qiao, W. F., Ye, F. F., Ma, Q., et al. (2023a). The spatiotemporal expression pattern of Hippophae rhamnoides NAC genes under drought stress. Acta Agric. Boreali-occident. Sin. 32, 878–886. doi: 10.7606/j.issn.1004-1389.2023.06.006
Zhang, B., Wang, X., Yue, Q., Zhang, W., Liu, H., Zhang, T., et al. (2025). Autosuppression of MdNAC18.1 endowed by a 61-bp promoter fragment duplication delays maturity date in apple. Plant Biotechnol. J. 23, 1216–1229. doi: 10.1111/pbi.14580
Zhang, L., Xiong, H. H., Cao, Q., Zhao, J. L., and Zhang, H. G. (2020a). Research on the drought tolerance of Larix olgensis plants with instantaneous genetic transformation of NAC genes. BBR 40, 394–400. doi: 10.7525/j.issn.1673-5102.2020.03.010
Zhang, S., Xu, T., Ren, Y., Song, L., Liu, Z., Kang, X., et al. (2023b). The NAC transcription factor family in Eucommia ulmoides: Genome-wide identification, characterization, and network analysis in relation to the rubber biosynthetic genes. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1030298
Zhang, Y., Zhou, K. W., Ren, M. Y., Ma, L., Tang, K. G., Guo, H. Q., et al. (2021c). Analysis of transcriptional activation activity of AmNAC2 protein and expression of its encoding gene in Ammopiptanthus mongolicus. Genomics Appl. Biol. 40, 3576–3583. doi: 10.13417/j.gab.040.003576
Zhao, L. F. (2024). Study on the regulation of drought resistance and secondary cell wall formation by NAC073 transcription factor in Betula platyphylla (Harbin: Northeast. For. Univ). doi: 10.27009/d.cnki.gdblu.2024.000019
Zhao, C. M., Huang, X. Q., Yin, F. Y., Li, D. Q., Chen, Y., and Ling, L. (2020a). Research progress of the oryza sativa NAC transcription factor family. J. Plant Sci. 38, 278–287. doi: 10.11913/PSJ.2095-0837.2020.20278
Zhao, Y., Sun, J., Xu, P., Zhang, R., and Li, L. G. (2014). Intron-mediated alternative splicing of WOOD-ASSOCIATED NAC TRANSCRIPTION FACTOR1B regulates cell wall thickening during fiber development in Populus species. Plant Physiol. 164, 765–776. doi: 10.1104/pp.113.231134
Zhao, C., Zhang, H., Song, C., Zhu, J. K., and Shabala, S. (2020b). Mechanisms of plant responses and adaptation to soil salinity. Inno 1, 100017. doi: 10.1016/j.xinn.2020.100017
Zhong, Q. W., Ma, J. K., Jia, T., Cheng, Q. Q., Du, X. J., and Nie, Z. X. (2023). Gene cloning of Toona ciliata var. pubescens TcNAC2 and its expression analysis under drought stress. Acta Agric. Univ. Jiangxiensis. 45, 1051–1060. doi: 10.13836/j.jjau.2023097
Zhong, R. Q., McCarthy, R. L., Lee, C., and Ye, Z. H. (2011). Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol. 157, 1452–1468. doi: 10.1104/pp.111.181354
Zhou, G. F., Li, G. H., Zhao, Q. S., and Guo, W. (2025). Identification of NAC transcription factor genes and their expression in vascular development in Pteroceltis tatarinowii. Mol. Plant Breed 1-9. Available at: http://kns.cnki.net/kcms/detail/46.1068.S.20231219.1351.029.html (Accessed March 30, 2025).
Zhu, Z., Li, G., Yan, C., Liu, L., Zhang, Q., Han, Z., et al. (2019). DRL1, encoding a NAC transcription factor, is involved in leaf senescence in grapevine. Int. J. Mol. Sci. 20, 2678. doi: 10.3390/ijms20112678
Zhu, Z., Shi, J., He, M., Cao, J., and Wang, Y. (2012). Isolation and functional characterization of a transcription factor VpNAC1 from Chinese wild Vitis Pseudoreticulata. Biotechnol. Lett. 34, 1335–1342. doi: 10.1007/s10529-012-0890-y
Keywords: NAC transcription factors, woody plants, growth and development, adversity and coercion, regulatory mechanisms
Citation: Zhang H, Zhao J, Zhang T, Wang G, Han Z, Meng Y, Bi J, Ren Y and Yang M (2025) Research progress of NAC transcription factors in woody plants. Front. Plant Sci. 16:1592898. doi: 10.3389/fpls.2025.1592898
Received: 13 March 2025; Accepted: 05 May 2025;
Published: 04 June 2025.
Edited by:
Yi-Hong Wang, University of Louisiana at Lafayette, United StatesReviewed by:
Qi Jia, Fujian Agriculture and Forestry University, ChinaDeguo Han, Northeast Agricultural University, China
Tao Xue, Huaibei Normal University, China
Copyright © 2025 Zhang, Zhao, Zhang, Wang, Han, Meng, Bi, Ren and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yachao Ren, cnljeXhAMTI2LmNvbQ==; Minsheng Yang, eWFuZ21zMTAwQDEyNi5jb20=
 Huan Zhang
Huan Zhang Jia Zhao1,2
Jia Zhao1,2 Guiying Wang
Guiying Wang Yachao Ren
Yachao Ren Minsheng Yang
Minsheng Yang