- 1Department of Silviculture, Faculty of Forestry and Environment, Institut Pertanian Bogor (IPB University), Bogor, West Java, Indonesia
- 2Molecular Science Research Group, Advanced Research Laboratory, Institut Pertanian Bogor (IPB University), Bogor, West Java, Indonesia
- 3Department of Biochemistry, Faculty of Mathematics and Natural Sciences, Institut Pertanian Bogor (IPB University), Bogor, West Java, Indonesia
- 4Research Centre for Applied Botany, National Research and Innovation Agency/Badan Riset dan Inovasi Nasional (BRIN), Cibinong, Bogor, West Java, Indonesia
1 Introduction
The Neolamarckia cadamba (Roxb.) Bosser, commonly known as white jabon, and Neolamarckia macrophylla (Roxb.) Bosser, referred to as red jabon, are fast-growing tree species native to Indonesia, belonging to the Rubiaceae family. In recent years, these trees have drawn significant attention for their versatile applications in industrial plantations, community forests, and projects focused on forest and land rehabilitation (Kallio et al., 2011; Irawan and Purwanto, 2014; Sarjono et al., 2017). Both N. cadamba and N. macrophylla are used for various purposes, including the production of wood for the pulp industry, plywood manufacturing, and construction and carpentry materials (Soerianegara and Lemmens, 1993; Kartawinata, 1994; Lempang, 2014). Additionally, N. cadamba is recognized for its potential medicinal benefits, including pain relief, anti-inflammatory properties, antipyretic effects (Mondal et al., 2009), as well as antimicrobial (Acharyya et al., 2011), and antibacterial (Mishra and Siddique, 2011) activities.
The cultivation of N. cadamba and N. macrophylla is still hindered by the lack of access to superior seeds from breeding programs. Most seeds used for various planting initiatives are sourced from natural forests or plantation forests that are classified as Identified Seed Stands (TBT), where no tree breeding activities have been conducted (UPT Perbenihan Tanaman Hutan Jawa Timur, 2022). As a result, these stands often exhibit high growth variability, along with low productivity and poor wood quality. To address these issues, prioritizing the development of fast-growing native species should be a focus for tree breeding and silviculture research. This approach can help create superior native forest plant species that could potentially reduce the dominance of exotic species in virtually all industrial plantation forests across Indonesia. However, there is still limited genetic information available for N. cadamba and N. macrophylla from Indonesia, particularly concerning genome sequencing and microsatellite markers, also known as Simple Sequence Repeats (SSRs). This lack of data poses challenges for developing effective cultivation strategies, as these data are needed for genetic research and breeding.
Chloroplast genomes are valuable for phylogenetic analysis because they are predominantly maternally inherited, possess a conserved gene structure and content, and exhibit a low mutation frequency (Palmer et al., 1988). Moreover, chloroplast genomes provide essential data for population genetics, molecular identification, and genetic engineering (Powell et al., 1995; Daniell et al., 2016; Cao et al., 2022). On the other hand, microsatellite markers play crucial role in cultivar identification, assessing genetic diversity, genome mapping, quantitative trait loci (QTL) analysis, paternity analysis, cross-species transferability, segregation analysis, phylogenetic relationships, and identification of wild cross hybrids in plant species (Miah et al., 2013; Ahmad et al., 2018).
To date, several studies have focused on whole-genome sequencing of two species. One study characterized the complete genome of Neolamarckia macrophylla from South Sulawesi, Indonesia, using the Illumina HiSeq Nova platform and examined its phylogenetic relationship with N. cadamba and other species (Shi et al., 2020). Another study characterized the complete chloroplast genome of Neolamarckia cadamba from Guangdong province of China using Illumina pair-end sequencing (Li et al., 2018). However, both studies lack the development of microsatellite markers. This gap highlights the critical need to investigate the genomes and microsatellites of N. cadamba and N. macrophylla from Indonesia, as these populations may harbor unique genetic characteristics. In this study, whole genome sequencing data from N. cadamba and N. macrophylla were used to complete the chloroplast genome and screen microsatellite (or Simple Sequence Repeats, SSRs) markers for future molecular studies.
1.1 Plant material, DNA extraction, and sequencing
The plant material used in this study consisted of silica-gel dried leaf samples collected from the adult tree of Neolamarckia cadamba (white jabon) planted in Kediri Forest Management Unit (KPH), Kediri Regency, East Java Province, Indonesia (-7°55’38,61163” S, 112°11’46,13546” E) and the adult tree of Neolamarckia macrophylla (red jabon) planted in Special Purpose Forest Area (KHDTK) Parung Panjang, Bogor Regency, West Java Province, Indonesia (-6°23’9,54262” S, 106°31’23,97738” E). The Neolamarckia cadamba tree used in this study originated from Nusakambangan Island, Indonesia (Sample code: AI NJ18), while the Neolamarckia macrophylla tree was sourced from Laeya District, South Konawe Regency, Southeast Sulawesi Province, Indonesia (Sample code: S016–1 Prov1 Blok3). The silica-gel dried leaf samples from the two species underwent genomic DNA extraction using the Cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle, 1990). Initial quantification and purity of the genomic DNA were observed using Nanodrop 2000 (Thermo Scientific) and visualized through agarose gel electrophoresis. Qubit dsDNA BR Assay Kits (Thermo Scientific) were used for accurate DNA quantification. The genomic DNA of N. cadamba (concentration of 120 ng/μL and amount of 2,23 μg) and N. macrophylla (concentration of 7,96 ng/μL and amount of 0,398 μg) that passed the quality check were then subjected to library preparation and whole genome sequencing utilizing the Illumina NextSeq 500 System, producing a data output of 6 GB per sample.
1.2 Chloroplast genome assembly and annotation
Sequencing data were uploaded to the Galaxy web platform, specifically the public server at usegalaxy.org version 23.1.2.dev0 (https://usegalaxy.eu/) for analysis (Afgan et al., 2016). The quality of raw reads was assessed using FASTQC version 0.12.1 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) (Andrews, 2010), and clean reads were filtered with Fastp version 0.23.2 (https://github.com/OpenGene/fastp) (Chen et al., 2018) using the default parameters. Clean reads were assembled using SPAdes version 3.15.3 (Bankevich et al., 2012) and NOVOPlasty version 4.3.1 (Dierckxsens et al., 2017), both with default parameters. The assembly results were then annotated using GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) (Tillich et al., 2017). The fully annotated genome was illustrated using OrganellarGenomeDRAW v1.3.1 (Greiner et al., 2019).
1.3 Microsatellite marker
Microsatellite (SSR) markers were extracted using Krait: Microsatellites Investigation and Primer Design version v1.5.1 (Du et al., 2018) from the scaffolds of N. cadamba and N. macrophylla. The minimum repetition rates were set as follows: six for motifs with two bases and five for motifs with three, four, five, and six bases. A minimum gap of 100 bases was maintained between different microsatellite motifs. Sequences containing these microsatellite motifs were selected based on two criteria: (i) the flanking regions must be at least 150 base pairs (bp) in length on both sides and (ii) the microsatellite repeats should have the longest repeat motif. The microsatellite markers were then designed using Krait: Microsatellites Investigation and Primer Design, version 1.5.1 (Du et al., 2018), with default parameters.
2 Results
The chloroplast genome of N. cadamba exhibited a typical quadripartite structure (Figure 1) with a total length of 154,973 bp. This genome comprises small single copy (SSC: 17,845 bp) and large single copy (LSC: 85,861 bp) regions, separated by a pair of inverted repeat regions: inverted repeat A (IRA: 25.633 bp) and inverted repeat B (IRB: 25,634 bp) (Figure 1). The N. cadamba chloroplast genome contained 126 genes in total, including 84 protein-coding genes (78 of which are unique), 36 transfer RNA (tRNA) genes (29 unique), and 8 ribosomal RNA (rRNA) genes, comprising 4 unique rRNA sequences (Supplementary Table 1). The GC content of the N. cadamba sequence is 37.6% (LSC: 35.4%; SSC: 31.6%; IR: 43.2%). These findings are consistent with previous results reported by Li et al. (2018), that the size of the chloroplast genome of N. cadamba was 154,999 bp, harboring an SSC of 17,851 bp, LSC of 85,880 bp, and a pair of inverted repeats (IRs) of 25,634 bp. Li et al. (2018) also reported the presence of 130 genes, where 96 were unique and 17 were duplicated in the IRs. The coding regions comprised 79 protein genes, 30 tRNA genes, and 4 rRNA genes, and the overall GC content of the chloroplast genome was 37.6%.
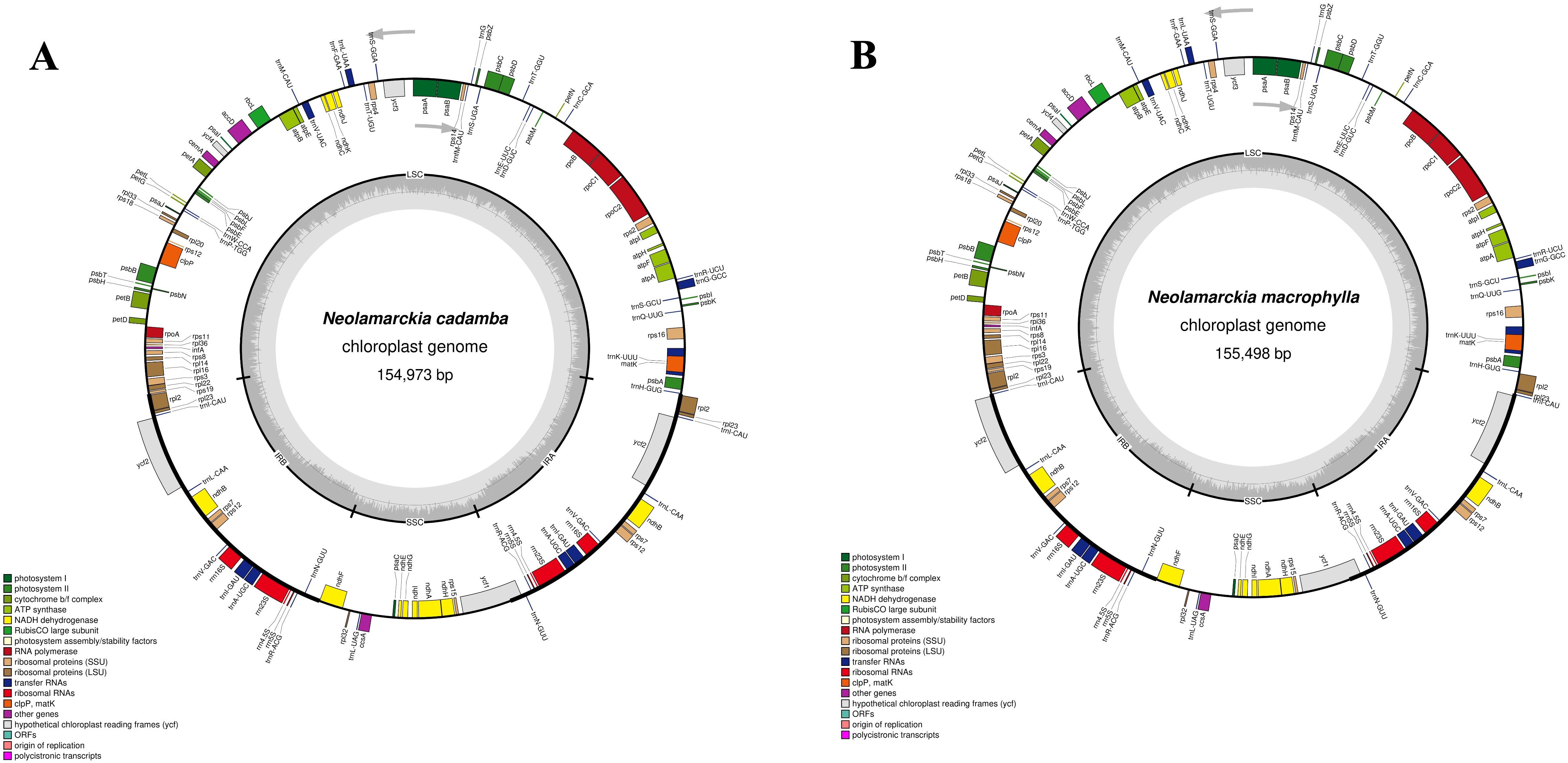
Figure 1. The complete chloroplast genomes of the two Neolamarckia species: (A) Neolamarckia cadamba and (B) Neolamarckia macrophylla.
Similarly, the chloroplast genome of N. macrophylla also displays a quadripartite structure (Figure 1) with a length of 155,498 bp. This genome includes small single copy (SSC: 18,100 bp) and large single copy (LSC: 88,847 bp) regions, similarly separated by inverted repeat regions: IRA (24,275 bp) and IRB (24,276 bp) (Figure 1). The N. macrophylla chloroplast genome consisted of 126 genes, encompassing 84 protein-coding sequences with 29 unique sequences, 36 transfer RNA (tRNA) genes (29 unique), and 8 ribosomal RNA (rRNA) genes comprising 4 unique rRNA sequences (Supplementary Table 1). The GC content of the N. macrophylla sequence is 37.5% (LSC: 35.6%; SSC: 31.6%; IR: 43.4%). These findings align with the study by Shi et al. (2020), which revealed that the complete chloroplast genome of N. macrophylla is 155,406 bp and includes an SSC of 18,063 bp, an LSC of 86,013 bp, and a pair of IR regions of 25,665 bp each. Shi et al. (2020) also identified 128 genes, including 8 rRNA, 36 tRNA, and 84 protein-coding genes, and reported that the overall GC content of the chloroplast genome was 37.56%.
The analysis of microsatellites (SSRs) in N. macrophylla and N. cadamba (Table 1) revealed that N. macrophylla contained a greater number of SSRs, totaling 157,972, compared to 112,439 for N. cadamba. Additionally, the number of sequences containing SSRs was also greater in N. macrophylla, with 50,133 sequences compared to 34,884 in N. cadamba. Furthermore, candidate microsatellite markers were selected based on the richness of T/C content, consisting of 20 markers for N. cadamba (Supplementary Table 2) and 20 markers for N. macrophylla (Supplementary Table 3). The selected microsatellite marker candidates for each species will play a crucial role in future comprehensive analyses aimed at unraveling the genetic diversity of each species. This investigation will enhance the understanding of their unique genetic structures and evolutionary relationships, thereby providing invaluable support for advanced breeding programs.
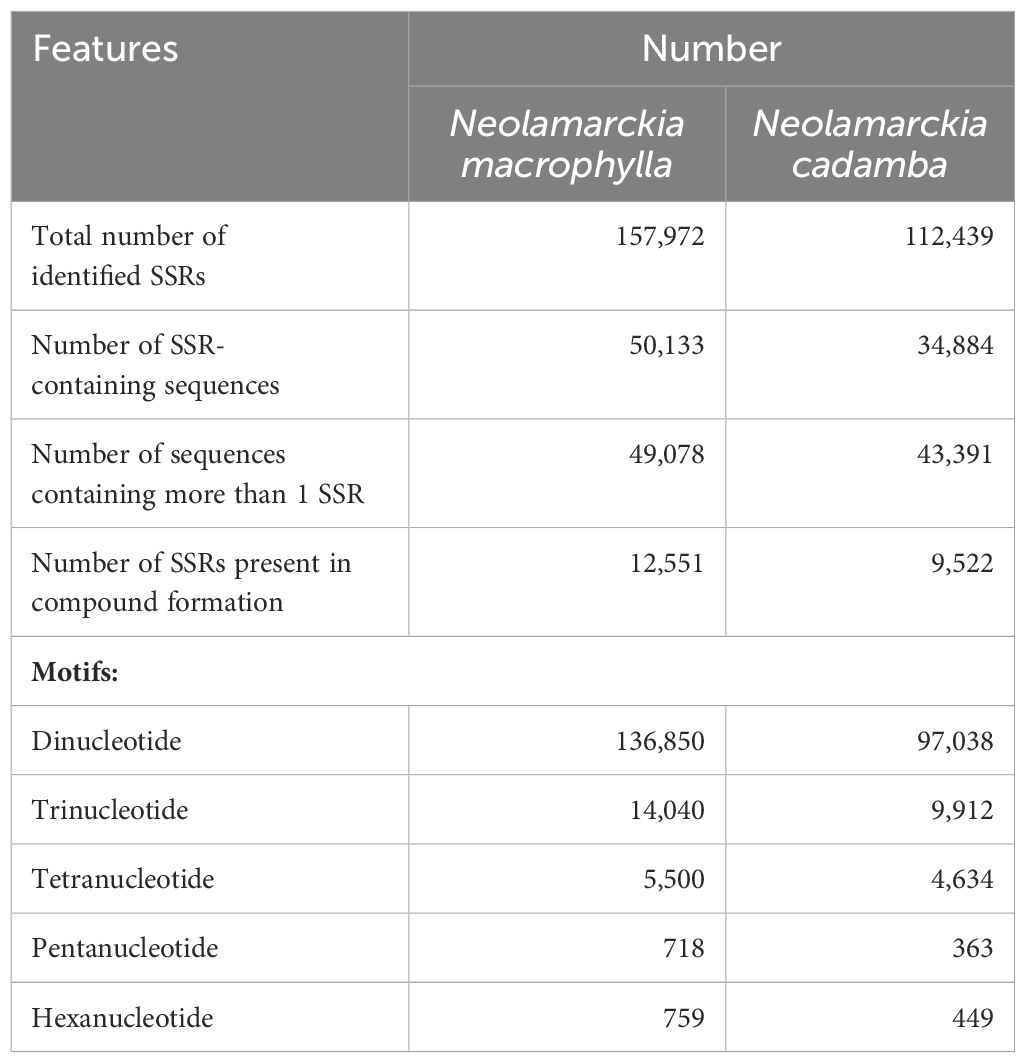
Table 1. Summary of identified microsatellites (SSRs) from the genome assembly of Neolamarckia cadamba and Neolamarckia macrophylla.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the DNA Data Bank of Japan (DDBJ) under the Bioproject number PRJDB20739 (https://ddbj.nig.ac.jp/search/entry/bioproject/PRJDB20739). The Bio-sample number for Neolamarckia cadamba (white jabon) is SAMD00911405 (https://ddbj.nig.ac.jp/search/entry/biosample/SAMD00911405), and the Bio-sample number for Neolamarckia macrophylla (red jabon) is SAMD00911406 (https://ddbj.nig.ac.jp/search/entry/biosample/SAMD00911406).
Author contributions
FGD: Investigation, Conceptualization, Methodology, Resources, Writing – original draft, Data curation. IK: Formal Analysis, Writing – original draft, Visualization, Software. RP: Validation, Software, Writing – review & editing. DS: Data curation, Writing – review & editing. ER: Resources, Project administration, Data curation, Writing – review & editing. RD: Data curation, Writing – review & editing. IZS: Supervision, Writing – review & editing. DJS: Data curation, Writing – review & editing, Resources, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the RIIM (Riset dan Inovasi untuk Indonesia Maju) Research Grant Batch-2 (2022-2025) under Contract Number: B-1738/II.7.5/FR/11/2022 and the Regular Fundamental Research Scheme for 2025 (Skema Penelitian Fundamental Reguler 2025), which was awarded by the Ministry of Higher Education, Science, and Technology of the Republic of Indonesia (Kemendiktisaintek) under Grant No: 006/C3/DT.05.00/PL/2025 (between Kemendiktisaintek and IPB University), as well as Grant No: 23032/IT3.D10/PT.01.03/P/B/2025 (between Directorate of Research and Innovation IPB University and the Principal Investigator, IZS), entitled “Genome and Species Distribution Model Study of Endangered Tropical Trees for Climate Change Mitigation and Species Conservation (Studi Genome dan Species Distribution Model Pohon Tropis Terancam Punah untuk Mitigasi Perubahan Iklim dan Konservasi Spesies)”.
Acknowledgments
The authors express their gratitude to the Indonesia Endowment Fund for Education Agency (LPDP) and the National Research and Innovation Agency of the Republic of Indonesia (BRIN) for their support throughout the research. The authors are also grateful to the Center for the Application of Environmental and Forestry Instrument Standards (BPSILHK) in Bogor-Indonesia and Perhutani Forestry Institute – Excellent Forestry Institute (PeFI Perhutani) for their vital role in providing the necessary research permit. Additionally, the authors sincerely thank Andrea Ajeng Eirenne Kristianti for her valuable assistance with DNA extraction.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1608577/full#supplementary-material
References
Acharyya, S., Rathore, D., Kumar, H. K., and Panda, N. (2011). Screening of Anthocephalus cadamba (Roxb.) Miq. root for antimicrobial and anthelmintic activities. Int. J. Res. Pharm. BioMed. Sci. 2, 297–300.
Afgan, E., Baker, D., van den Beek, M., Blankenberg, D., Bouvier, D., Čech, M., et al. (2016). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44, W3–W10. doi: 10.1093/nar/gkw343
Ahmad, A., Wang, J.-D., Pan, Y.-B., Sharif, R., and Gao, S.-J. (2018). Development and use of simple sequence repeats (SSRS) markers for sugarcane breeding and genetic studies. Agronomy 8, 260. doi: 10.3390/agronomy8110260
Andrews, S. (2010). FASTQC: A quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (Accessed April 21, 2024).
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Cao, D.-L., Zhang, X.-J., Xie, S.-Q., Fan, S.-J., and Qu, X.-J. (2022). Application of chloroplast genome in the identification of traditional Chinese medicine Viola philippica. BMC Genomics 23, 540. doi: 10.1186/s12864-022-08727-x
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Daniell, H., Lin, C.-S., Yu, M., and Chang, W.-J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134. doi: 10.1186/s13059-016-1004-2
Dierckxsens, N., Mardulyn, P., and Smits, G. (2017). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18. doi: 10.1093/nar/gkw955
Doyle, J. J. and Doyle, J. L. (1990). Isolation of plant DNA from fresh tissue. Focus (Madison). 12, 13–15.
Du, L., Zhang, C., Liu, Q., Zhang, X., and Yue, B. (2018). Krait: an ultrafast tool for genome-wide survey of microsatellites and primer design. Bioinformatics 34, 681–683. doi: 10.1093/bioinformatics/btx665
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Irawan, U. S. and Purwanto, E. (2014). White jabon (Anthocephalus cadamba) and Red Jabon (Anthocephalus macrophyllus) for community land rehabilitation: Improving local propagation efforts. Agric. Sci. 2, 36–45. doi: 10.12735/as.v2i3p36
Kallio, M. H., Krisnawati, H., Rohadi, D., and Kanninen, M. (2011). Mahogany and kadam planting farmers in South Kalimantan: The link between silvicultural activity and stand quality. Small-scale For. 10, 115–132. doi: 10.1007/s11842-010-9137-8
Kartawinata, K. (1994). The use of secondary forest species in rehabilitation of degraded forest lands. J. Trop. For Sci. 7, 76–86. Available at: http://www.jstor.org/stable/43581793 (Accessed April 20, 2024).
Lempang, M. (2014). Basic properties and potential uses of jabon merah wood. J. Penelit. Kehutan. Wallacea 3, 163–175. doi: 10.18330/jwallacea.2014.vol3iss2pp163-175
Li, J., Zhang, D., Ouyang, K., and Chen, X. (2018). The complete chloroplast genome of the miracle tree Neolamarckia cadamba and its comparison in Rubiaceae family. Biotechnol. Biotechnol. Equip. 32, 1087–1097. doi: 10.1080/13102818.2018.1496034
Miah, G., Rafii, M. Y., Ismail, M. R., Puteh, A. B., Rahim, H. A., Islam, K., et al. (2013). A review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. Int. J. Mol. Sci. 14, 22499–22528. doi: 10.3390/ijms141122499
Mishra, R. P. and Siddique, L. (2011). Antibacterial properties of Anthocephalus cadamba fruits. Asian J. Plant Sci. Res. 1, 1–7. Available at: https://api.semanticscholar.org/CorpusID:7241153 (Accessed April 19, 2024).
Mondal, S. P., Dash, G. K., and Acharyya, S. (2009). Analgesic, anti-inflammatory and antipyretic studies of Neolamarckia cadamba Barks. J. Pharm. Res. 2, 1133–1136. Available at: https://api.semanticscholar.org/CorpusID:30375842 (Accessed April 19, 2024).
Palmer, J. D., Jansen, R. K., Michaels, H. J., Chase, M. W., and Manhart, J. R. (1988). Chloroplast DNA variation and plant phylogeny. Ann. Missouri Bot. Gard. 75, 1180–1206. doi: 10.2307/2399279
Powell, W., Morgante, M., McDevitt, R., Vendramin, G. G., and Rafalski, J. A. (1995). Polymorphic simple sequence repeat regions in chloroplast genomes: Applications to the population genetics of pines. Proc. Natl. Acad. Sci. U.S.A. 92, 7759–7763. doi: 10.1073/pnas.92.17.7759
Sarjono, A., Sunoto, A., and Devitra, J. (2017). Logs production and land expectation value of jabon (Anthocephalus cadamba) at PT Intraca Hutani Lestari. J. Hutan Trop. 5, 22–30. Available at: https://ppjp.ulm.ac.id/journal/index.php/jht/article/view/4052/3689 (Accessed April 20, 2024).
Shi, S., Larekeng, S. H., Lv, P., Nie, Y., Restu, M., and Yang, H. (2020). The complete chloroplast genome of Neolamarckia macrophylla (Rubiaceae). Mitochondrial DNA Part B Resour. 5, 1611–1612. doi: 10.1080/23802359.2020.1745709
Soerianegara, I. and Lemmens, R. H. M. (1993). Plant resources of South-East Asia 5(1): Timber trees: Major commercial timbers (Wageningen: Pudoc Scientific Publishers). doi: 10.2307/4113897
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11. doi: 10.1093/nar/gkx391
UPT Perbenihan Tanaman Hutan Jawa Timur (2022). Menjadi “Si Paling Paham Benih” Melalui Pelatihan Teknis Perbenihan Tanaman Hutan untuk Penyuluh Kehutanan. Available online at: https://uptpth.dishut.jatimprov.go.id/2022/10/24/menjadi-si-paling-paham-benih-melalui-pelatihan-teknis-perbenihan-tanaman-hutan-untuk-penyuluh-kehutanan/ (Accessed April 19, 2024).
Keywords: chloroplast, genome, illumina, microsatellite, Neolamarckia, plant breeding
Citation: Dwiyanti FG, Kamal I, Pratama R, Syaputra D, Rustam E, Damayanti RU, Siregar IZ and Sudrajat DJ (2025) Whole genome sequencing of Neolamarckia macrophylla (Roxb.) Bosser and Neolamarckia cadamba (Roxb.) Bosser from Indonesia: a vital resource for completing chloroplast genomes and mining microsatellite markers. Front. Plant Sci. 16:1608577. doi: 10.3389/fpls.2025.1608577
Received: 09 April 2025; Accepted: 27 May 2025;
Published: 20 June 2025.
Edited by:
Yuri Shavrukov, Flinders University, AustraliaReviewed by:
Lei Ding, Université Catholique de Louvain, BelgiumHoang Dang Khoa Do, Nguyen Tat Thanh University, Vietnam
Copyright © 2025 Dwiyanti, Kamal, Pratama, Syaputra, Rustam, Damayanti, Siregar and Sudrajat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fifi Gus Dwiyanti, ZmlmaV9kd2l5YW50aUBhcHBzLmlwYi5hYy5pZA==
 Fifi Gus Dwiyanti
Fifi Gus Dwiyanti Irsyad Kamal
Irsyad Kamal Rahadian Pratama
Rahadian Pratama Dhika Syaputra
Dhika Syaputra Evayusvita Rustam
Evayusvita Rustam Ratna Uli Damayanti
Ratna Uli Damayanti Iskandar Z. Siregar
Iskandar Z. Siregar Dede J. Sudrajat
Dede J. Sudrajat