- 1Jilin Collaborative Innovation Center for Antibody Engineering, Jilin Medical University, Jilin, China
- 2College of Laboratory, Jilin Medical University, Jilin, China
Superoxide dismutase (SOD) serves as a critical regulator of plant stress adaptation to salinity, drought, and heavy metal toxicity. Flax (Linum usitatissimum L.), a globally cultivated oilseed and fiber crop, lacks comprehensive genomic characterization of its SOD gene family. Here, we systematically identified 12 LuSOD genes in the flax genome. Phylogenetic reconstruction of SOD homologs across diverse plant species classified these genes into three evolutionarily conserved subgroups: Cu/Zn-SOD (6 LuCSD), Fe-SOD (3 LuFSD), and Mn-SOD (3 LuMSD). Comparative analysis of exon-intron architectures and conserved motifs revealed high structural conservation among LuSOD members within each clade. Promoter cis-element profiling identified predominant associations with phytohormone signaling (abscisic acid, methyl jasmonate) and abiotic stress responses, including hypoxia, drought, and low-temperature adaptation. MicroRNA target prediction identified lus-miR159 as the primary regulatory miRNA interacting with LuSOD genes. Gene ontology (GO) enrichment highlighted LuSOD roles in stress perception, metal ion chelation, and enzymatic reactive oxygen species (ROS) scavenging. Transcriptomic profiling demonstrated ubiquitous high expression of LuSOD genes in leaf tissues. qRT-PCR validation under cold, drought, and salt stresses revealed significant upregulation of nine LuSOD genes, implicating their involvement in antioxidant defense mechanisms. Functional characterization of LuCSD3 in transgenic Arabidopsis confirmed its role in enhancing salt tolerance through ROS homeostasis modulation. This study provides foundational insights into LuSOD-mediated stress resilience, serving as a valuable resource for molecular breeding and functional genomics in flax.
1 Introduction
Abiotic stress induced reactive oxygen species (ROS) overproduction in plants, causing oxidative impairment to biomacromolecules, membrane integrity, and cellular ultrastructure, ultimately triggering programmed cell death (Kar, 2011). Superoxide dismutase (SOD), a pivotal enzyme in ROS homeostasis, constituted the first enzymatic barrier against oxidative damage by catalyzing ROS detoxification, thereby safeguarding cellular components from oxidative injury (Abreu and Cabelli, 2010; Gill et al., 2015). Plant SODs were classified into three phylogenetically distinct isoforms—Cu/Zn-SOD, Fe-SOD, and Mn-SOD—based on their metal cofactor specificity. These isoforms exhibited divergent amino acid sequences, subcellular compartmentalization, tertiary structures, and hydrogen peroxide sensitivity profiles (Jiang et al., 2019).
Recent research underscored the pivotal role of Superoxide Dismutases (SODs) in safeguarding plants against diverse abiotic stressors, such as extreme temperatures, drought, salinity, and hormonal fluctuations (Qin, 2023). Certain SOD isoforms operated within specialized cellular compartments to mitigate oxidative stress (Corpas et al., 2017). Cu/Zn-SOD (CSD), the most ubiquitous isoform, localized to chloroplasts, mitochondria, and the cytosol. Mn-SOD (MSD) predominantly resided in mitochondria and peroxisomes, playing essential roles in drought and salinity tolerance (Asensio et al., 2012). Fe-SOD was primarily chloroplast-localized, while Mn-SOD also occurred in peroxisomes. Cu/Zn-SOD additionally occupied extracellular spaces and peroxisomes (Huseynova et al., 2014; Case, 2017). In rape, genomic studies had identified 31 SOD genes, with eight exhibiting pronounced upregulation under hormonal and non-biomass stress conditions. Similarly, Salvia miltiorrhiza was found to harbor eight SOD genes that displayed distinct responsiveness to cold, salt, drought, heavy metals, and phytohormonal changes (Han et al., 2020). There are 7 members of SOD gene family in barley, among which HvSOD1, HvSOD4 and HvSOD5 expression changed significantly under drought and salt stress (Zhang et al., 2021). In tomatoes, among nine SlSOD genes, SlSOD1 was uniquely upregulated under stress, while SlSOD2, SlSOD5, SlSOD6, and SlSOD8 responded specifically to salt stress (Feng et al., 2016). Almost all HbSOD genes of rubber tree have high expression level under drought stress (Yu et al., 2022). The drought and salt tolerance of overexpressed peanut AhCu/ZnSOD in tobacco was significantly higher than that of wild type tobacco, and it could survive for a long time under water shortage (Negi et al., 2015). In tea plants, flavonoid biosynthesis genes and flavonoid levels correlated strongly with SOD activity, a relationship further evidenced in Arabidopsis, where EkFLS overexpression boosted both flavonoids and SOD expression under drought (Wang et al., 2021). Additionally, trehalose was shown to modulate tomato Cu/ZnSOD expression during cold stress (Liu et al., 2020). These findings indicate that bolstering SOD gene activity and its elevated expression are key in enhancing plant stress resistance.
It has been found that miRNA-mediated ROS transcription regulation plays an important role in improving crop yield and stress resistance (Ravichandran et al., 2019; Cui et al., 2020; Ding et al., 2020). In Arabidopsis, two Cu/ZnSOD genes are targeted by miR398. There are 20 miRNA targeting 14 SOD genes in cotton (Wang et al., 2017). Ghr-miR414c, ghr-miR7267, ghr-miR0081, ghr-miR0166, ghr-miR0362, ghr-miR0362 plays an important role in cotton fiber development (Li et al., 2012). Inhibition of miR398 expression in Arabidopsis induced up-regulation of copper/zinc SOD gene CSD1 and CSD2, and improved plant antioxidant stress ability (Sunkar et al., 2006). These findings indicate that miRNA plays an important role in environmental signaling and plant development by modifying the SOD gene.
Flax (Linum usitatissimum L.), a historically significant crop with global cultivation spanning temperate zones, has been utilized for oilseed, fiber, and dual-purpose applications (Huis et al., 2010). Based on its agronomic applications, flax is functionally categorized into three types: oilseed, fiber, and dual-purpose varieties (Chytilova et al., 2013). Flaxseeds are nutritionally dense, containing bioactive compounds such as lignans, dietary fiber, and alpha-linolenic acid (ALA)—an essential omega-3 fatty acid critical for human metabolic and cardiovascular health (Santos et al., 2020). However, in flax cultivation, abiotic stresses such as cold, salinity, and drought severely limit its yield (Yadav et al., 2022). Prior to this study, the SOD gene family in flax remained genomically uncharacterized. Here, we systematically investigated the SOD gene family in flax using an integrated bioinformatics approach. Analyses encompassed evolutionary relationships, chromosomal distribution, protein physicochemical properties, conserved motifs, promoter cis-elements, protein interaction networks, and tissue-specific expression profiles. Our findings revealed that flax SOD genes exhibit transcriptional responsiveness to low-temperature, salinity, and drought stresses, providing mechanistic insights into their roles in stress adaptation. This study represents the first comprehensive genomic characterization of the SOD gene family in flax, establishing a foundation for future functional studies aimed at enhancing stress resilience in this economically vital crop.
2 Materials and methods
2.1 Plant material and statistical analysis
The experiments were conducted using the flax cultivar ‘Longyan 10’ from the Gansu Academy of Agricultural Sciences (Zhang et al., 2020). Seeds were surface-sterilized by immersion in 75% ethanol for 10 minutes, followed by three rinses with sterile deionized water, and subsequently sown into autoclaved nutrient soil. Growth chamber conditions were maintained at 26°C (day)/18°C (night) with a 16-hour photoperiod. Stress treatments were initiated when seedlings reached 6–7 cm in height. For drought simulation, plants were gently uprooted, washed with distilled water to eliminate soil particles, and transferred to hydroponic systems containing 10% (w/v) polyethylene glycol-6000 (PEG-6000). Salt stress was imposed using an identical protocol with 100 mM sodium chloride (NaCl) solution. Control plants were maintained in distilled water, while low-temperature stress groups were incubated at 4°C. Leaf tissues were harvested at 0, 6, 12, and 24 hours post-treatment using synchronous sampling across all groups to eliminate diurnal rhythm interference, with three biological replicates per time point. All samples were flash-frozen in liquid nitrogen and archived at -80°C for subsequent molecular analyses.
All experimental procedures were conducted with a minimum of three biological replicates. Quantitative data are presented as mean ± standard deviation (SD) derived from triplicate measurements. Statistical analyses and graphical representations were performed using GraphPad Prison 8 software (version 8.4.3; GraphPad Software). Significant differences between groups were determined by two-tailed Student’s t-test, with asterisks denoting the following probability thresholds: *P < 0.05, **P < 0.01, ***P < 0.01.
2.2 Identification of SOD gene in flax
The complete genome assembly of flax (Longya10) was retrieved from the NCBI database under accession number QMEI02000000, while genome annotation files were sourced from the Figshare repository (https://figshare.com/articles/dataset/Annotation_files_for_Longya-10_genome/13614311). Eight Arabidopsis SOD homologs were analyzed via the TAIR database (Garcia-Hernandez et al., 2002). Candidate flax SOD genes were identified through BLASTP homology searches against the flax proteome (E-value cutoff: 1e-5). Hidden Markov Models (HMMs) for Cu/Zn-SOD (PF00080) and Fe/Mn-SOD (PF02777, PF00081) domains were acquired from the Pfam database (http://pfam.xfam.org/) (Mistry et al., 2021). The HMMER3.0 hmmsearch algorithm (Potter et al., 2018) was employed to predict SOD homologs in flax. Predicted sequences were further validated using SMART (http://smart.embl.de/smart/batch.pl) and the Conserved Domain Database (CDD; https://www.ncbi.nlm.nih.gov/cdd/), yielding a final set of 12 non-redundant LuSOD genes. Biophysical parameters of the LuSOD proteins—including coding sequence length, amino acid count, molecular weight (MW), theoretical isoelectric point (pI), and grand average of hydropathy (GRAVY)—were computationally derived using ExPASy ProtParam (https://web.expasy.org/protparam/) (Fink and Scandalios, 2002). Subcellular localization predictions were performed via the BUSCA web server (http://www.busca.cn).
2.3 Phylogeny, chromosome location, conserved domain and conserved motif of LuSOD gene
The full-length genomic sequences of rice and soybean were acquired from the Phytozome platform (https://phytozome-next.jgi.doe.gov/) (Goodstein et al., 2012). SOD gene families in these species were characterized using identical bioinformatics workflows. Multiple sequence alignment of SOD proteins from Arabidopsis, rice, soybean, and flax was performed using ClustalW in MEGA11 with default parameters (Yuan et al., 1999). A maximum likelihood (ML)-based phylogenetic tree was generated in MEGA11 under standard configurations (neighbor-joining algorithm; 1,000 bootstrap replicates) (Tamura et al., 2021). Chromosomal localization of LuSOD genes was mapped by integrating the flax genomic FASTA file with GFF3 annotations. Conserved structural domains were predicted via the NCBI CD-Search tool (https://www.ncbi.nlm.nih.gov/Structure/BWRPSB/BWRPSB.cgi). Conserved protein motifs were identified using MEME Suite (http://alternate.meme-suite.org/tools/meme), and visualization was executed with TBtools v2.069 (Chen et al., 2020).
2.4 Genome-wide replication and collinear analysis of LuSOD gene
The genome and annotation files of Arabidopsis were retrieved from the TAIR database (https://www.arabidopsis.org/). Collinear relationships were predicted using the Multiple Collinearity Scan (MCScanX) algorithm (Wang et al., 2012). Genome-wide duplication (WGD) events involving LuSOD genes were identified through whole-genome synteny analysis. Tandem duplication events were defined as chromosomal regions harboring two or more homologous genes within a 100-kb span, with no intervening non-homologous genes. Segmental duplication events (BLASTN E-value < 1e-5) were detected by analyzing 100-kb genomic regions (50 kb upstream and downstream) flanking coding sequences (CDS) using BLASTN alignments. Repetitive genes were classified based on sequence alignment length ≥200 base pairs (bp) and nucleotide sequence identity exceeding 85% (Khaja et al., 2006).
2.5 MiRNA prediction and cis-acting element analysis
Potential miRNA targets of the LuSOD gene family were predicted by aligning miRNA sequences with the 5’ and 3’ untranslated regions (UTRs) and coding sequences (CDS) using the psRNATarget platform (Plant MicroRNA Target Analysis Server; https://www.zhaolab.org/psRNATarget/analysis?function=3) under default parameters (Griffiths-Jones et al., 2006). The 2,000-bp upstream promoter regions of LuSOD genes were isolated from the flax genome using the TBtools software suite. To elucidate the transcriptional regulatory mechanisms of LuSOD genes under environmental stress, cis-acting elements within the 2.0-kb promoter sequences upstream of the translation start site were annotated via the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). Identified motifs were visualized using TBtools (version 2.069) for comparative analysis.
2.6 Construction of protein interaction network and GO enrichment analysis
To delineate the protein interaction network of the LuSOD gene family, orthologous Arabidopsis SOD genes were employed as reference sequences. Functional protein-protein interaction (PPI) networks were reconstructed using the STRING database (v11.5; https://string-db.org/) under standard configurations (Franceschini et al., 2013). For Gene Ontology (GO) enrichment analysis, the flax proteome was annotated via the eggNOG-mapper platform (http://eggnog-mapper.embl.de/) using the GO-base.ob reference file integrated within TBtools, yielding comprehensive functional annotations (Cantalapiedra et al., 2021). Resultant datasets were visualized using TBtools.
2.7 Expression pattern analysis of the LuSOD gene family and construction of LuCSD3-transgenic Arabidopsis
In this study, five flax transcriptional groups were sequenced: (a) pistil, stamen, fruit and stem tip tissues (PRJNA1002756) (https://www.ncbi.nlm.nih.gov/sra/?term=); (b) floral tissues at 30, 20, 10 and 5 days after anthesis (PRJNA833557); (c) different flax embryo tissues, anther and seed tissues (PRJNA663265); (d) root and leaf tissues after salt stress (PRJNA977728) (e) Stem tissue (PRJNA874329) after heat stress. The data were filtered by fastp, then compared to the Longya10 reference genome (Chen et al., 2018). Transcript abundance was quantified as FPKM (Fragments Per Kilobase Million), and log2-transformed FPKM values were visualized as clustered heatmaps using TBtools.
Transgenic Arabidopsis was obtained via the floral dip method (Clough and Bent, 1998). The Agrobacterium harboring the intact 35S::LuCSD3 construct was inoculated into 150 mL of LB liquid medium and cultured at 200 rpm and 28°C for 16 hours. Cells were collected by centrifugation at 5000 rpm for 10 minutes and resuspended in a 5% sucrose solution adjusted to OD600 = 1.0, supplemented with 0.01% Silwet-77 surfactant. Flower buds at the pre-bolting stage were immersed in the transformation solution for 2 minutes, with three rounds of infection per week. T1 generation plants were obtained and screened. T2 lines that produced 100% hygromycin-resistant plants in the T3 generation were identified as homozygous transgenic lines.
2.8 RNA extraction and fluorescence quantitative PCR analysis
Total RNA was isolated from flax leaf tissues using the Trizol reagent-based protocol. The SPARKscript II RT Plus Kit (With gDNA Eraser) (Shandong Sparkjade Biotechnology Co., Ltd.) was utilised to create cDNA. Gene-specific primers (Supplementary Table S5) were designed using Oligo 7 primer design software, and quantitative real-time PCR amplification was conducted with TB Green® Premix Ex Taq™ II. Relative gene expression levels were calculated via the 2−ΔΔCT method (Livak and Schmittgen, 2001), with GAPDH serving as the internal reference gene. Three technical replicates were analyzed per sample to determine cycle threshold (Ct) values.
2.9 Phenotypic analysis and NBT staining experiment of Arabidopsis with LuCSD3 gene transfer
To assess the salt stress tolerance phenotypes of T3 transgenic Arabidopsis, two-week-old wild-type (Col-0) and overexpression lines (OE) were divided into treatment and control groups. The treatment cohort was subjected to 200 mM NaCl irrigation every three days for 15 days, while controls received equivalent volumes of distilled water. Following the stress regimen, the third fully expanded rosette leaves from both groups were harvested for nitroblue tetrazolium (NBT) staining. The NBT working solution was formulated by dissolving 0.05 g NBT powder in 0.5 mL phosphate-buffered solution (PBS, pH 7.8), followed by dilution to 50 mL with deionized water. Leaf samples were incubated in the NBT solution for 30–60 minutes under dark conditions. Subsequently, stained leaves were destained in 95% ethanol until complete chlorophyll removal for histological observation. All experimental procedures were performed with three biological replicates to ensure statistical robustness.
2.10 Physiological and biochemical indicators detection of transgenic Arabidopsis with LuCSD3 gene
The physiological and biochemical indicators of the control group and OE lines (OE-1 and OE-5) were measured after 15 days of 200 mM NaCl stress. The parameters included malondialdehyde (MDA) content, proline (Pro) content, superoxide dismutase (SOD) activity, peroxidase (POD) activity, and hydrogen peroxide (H2O2) content. All reagent kits were provided by Beijing Boxbio Science & Technology Co.,Ltd. The measurements were conducted following the methods described in previous studies (Lu et al., 2025). Each sample was analyzed in triplicate.
3 Results
3.1 Identification and phylogenetic analysis of SOD gene family in flax
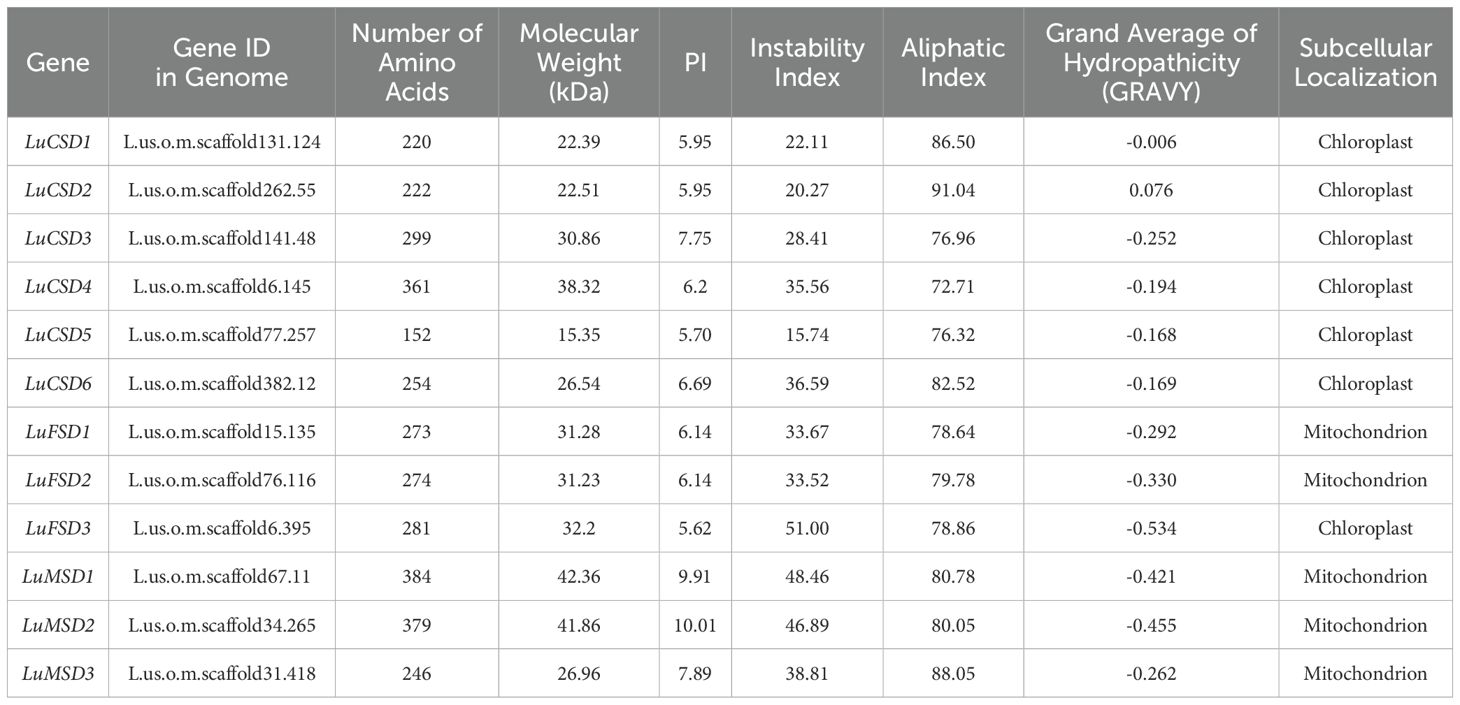
Utilizing Hidden Markov Model (HMM) profiles of Cu/ZnSOD (PF00080) and Fe/MnSOD (PF02777, PF00081), 12 SOD genes were identified in the flax (Longya10) genome. Phylogenetic orthology with Arabidopsis SOD homologs led to their nomenclature as LuCSD1–LuCSD6 (Cu/Zn-SOD), LuFSD1–LuFSD3 (Fe-SOD), and LuMSD1–LuMSD3 (Mn-SOD) (Table 1; Figure 1). Biochemical characterization of the encoded proteins revealed substantial variation: LuMSD1 encoded the longest polypeptide (384 amino acids), while LuCSD5 represented the shortest (152 amino acids). Molecular weights of LuSOD proteins spanned 15.35–42.36 kDa. Isoelectric point (pI) analysis indicated that only four proteins (LuCSD3, LuMSD1, LuMSD2, and LuMSD3) exhibited pI values >7, suggesting a predominance of acidic amino acids in the LuSOD family. Instability indices ranged from 15.74 (stable) to 51.0 (unstable), with all LuSOD proteins except LuFSD3 classified as stable. Aliphatic indices varied between 72.71 and 91.04, reflecting differences in thermostability. Hydropathicity analysis predicted LuCSD2 as the sole hydrophobic protein, whereas others were hydrophilic. Subcellular localization predictions assigned five genes (LuFSD1, LuFSD2, LuMSD1–LuMSD3) to mitochondria, with remaining members localized to chloroplasts.
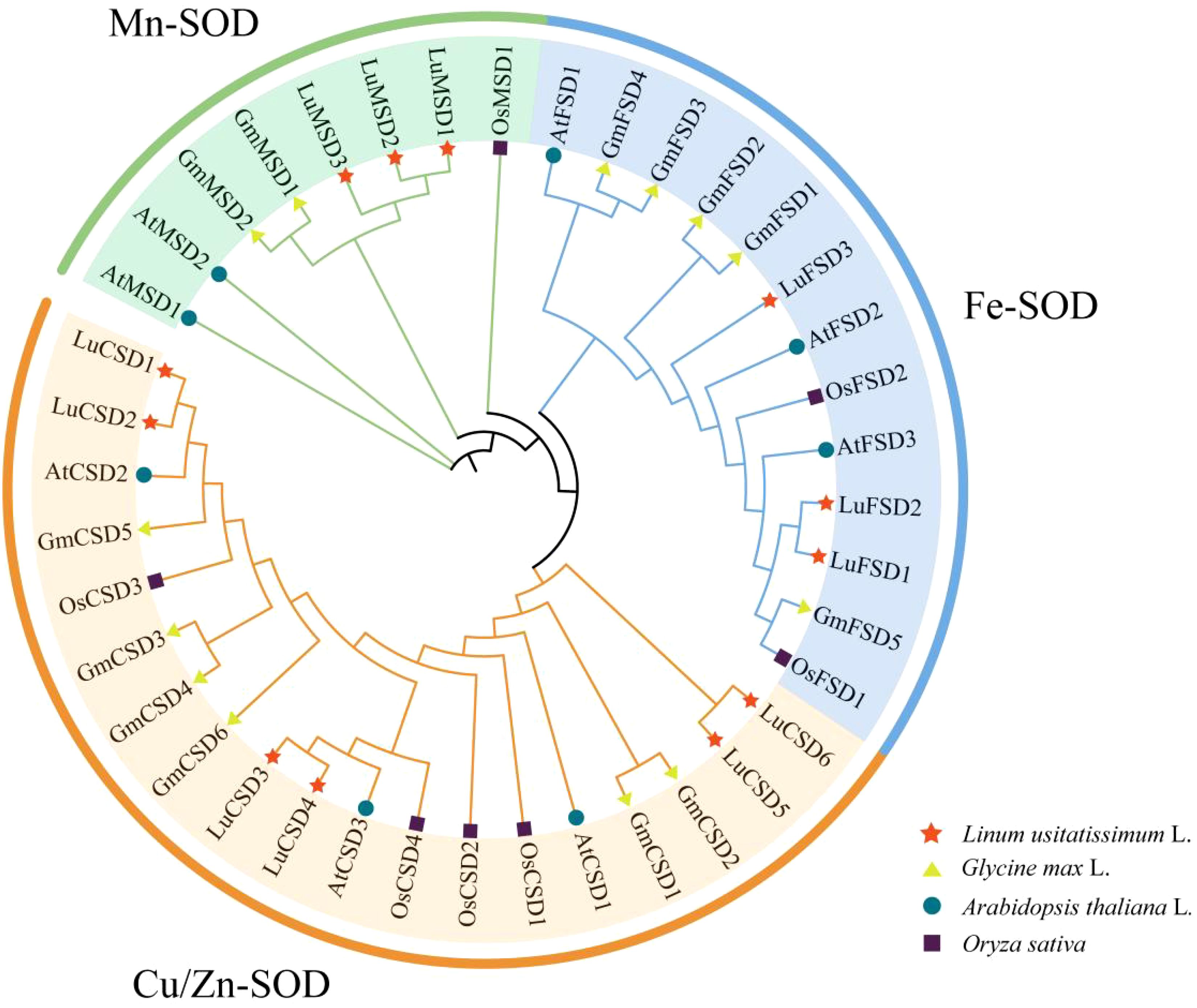
Figure 1. SOD protein phylogenetic tree of four species. The pentagram represents the flax gene, the circle represents the Arabidopsis gene, the triangle represents the soybean, and the square represents the rice gene. All SOD genes can be divided into three subfamilies: Cu/Zn-SOD, Mn-SOD and Fe-SOD, which are represented by different colors.
To resolve evolutionary relationships, a phylogenetic tree was reconstructed from 40 SOD protein sequences across Arabidopsis (8), Glycine max (13), Oryza sativa (7), and Linum usitatissimum (12) (Figure 2). According to Arabidopsis SOD protein subfamily classification (Supplementary Table S1), 12 SOD genes in flax were divided into three subfamilies, named Mn-SOD, Fe-SOD and Cu/Zn-SOD respectively. Twelve LuSOD genes were distributed in each subfamily, and the largest number of genes was in the Cu/Zn-SOD subfamily, including 6 members (LuCSD1-LuCSD6). Further studies showed that there was a close relationship between flax and SOD family of Arabidopsis among the three middle species.
3.2 Analysis of gene structure and conservative motif of LuSOD
To characterize the protein structural features of the flax SOD gene family, the amino acid sequences of 12 LuSOD homologs were subjected to conserved motif analysis using the MEME online platform. Ten evolutionarily conserved motifs (annotated as Motif 1–Motif 10) were systematically identified across the LuSOD protein sequences (Figures 2A, B; Supplementary Table S2). LuCSD gene contains only three domains (motif1, motif3 and motif6), among which motif3 and motif6 are unique domains of LuCSD gene. We also found that both LuMSD and LuFSD genes contain motif2 and motif4, in which motif7 and motif8 are unique domains of LuMSD gene. Among LuFSD1 and LuFSD2 genes, motif9 and motif10 are their unique domains. There are great differences in protein domains among different subfamilies of LuSOD genes, but the same subfamilies have the same domain, which also proves that different subfamilies have different biological functions.
The analysis of the structure of LuSOD gene shows that the number of exons is between 5 and 11, and the number of introns is between 4 and 10 (Figure 2C). Both LuMSD1 and LuMSD2 contain 11 exons and 10 introns, four genes (LuCSD1, LuCSD2, LuCSD3 and LuFSD3) contain 8 exons and 7 introns, two genes (LuCSD6 and LuFSD2) contain 7 exons and 6 introns, LuCSD5 and LuCSD4 both contain 6 exons and 5 introns. The least number of exons and introns in LuMSD3 are 5 and 4 respectively.
3.3 Chromosome mapping and collinearity analysis of LuSOD gene
Chromosomal localization of the LuSOD gene family was mapped using the flax reference genome, revealing an uneven distribution of 12 LuSOD genes across six chromosomes (Figure 2D). Chromosome 7 harbored the highest number of LuSOD genes (3 genes, 25% of the total), followed by chromosomes 1 and 5 (2 genes each, 16.67%), while chromosomes 3, 9, and 11 each contained a single LuSOD locus (8.33% per chromosome). To investigate duplication events, a collinearity analysis of the LuSOD family was performed via Circos visualization (Figure 3A), identifying eight segmental duplication pairs, indicative of substantial gene family expansion. To elucidate evolutionary conservation, syntenic relationships between flax and Arabidopsis SOD homologs were analyzed (Figure 3B). Nine collinear ortholog pairs were identified, with flax chromosomes 1, 3, 5, 7, 9, and 11 exhibiting synteny to Arabidopsis chromosomes 2, 3, and 5. Notably, no collinear SOD gene pairs were detected on Arabidopsis chromosomes 1 and 4. These findings collectively demonstrate strong chromosomal conservation of SOD genes between flax and Arabidopsis, with lineage-specific divergence in genomic organization.
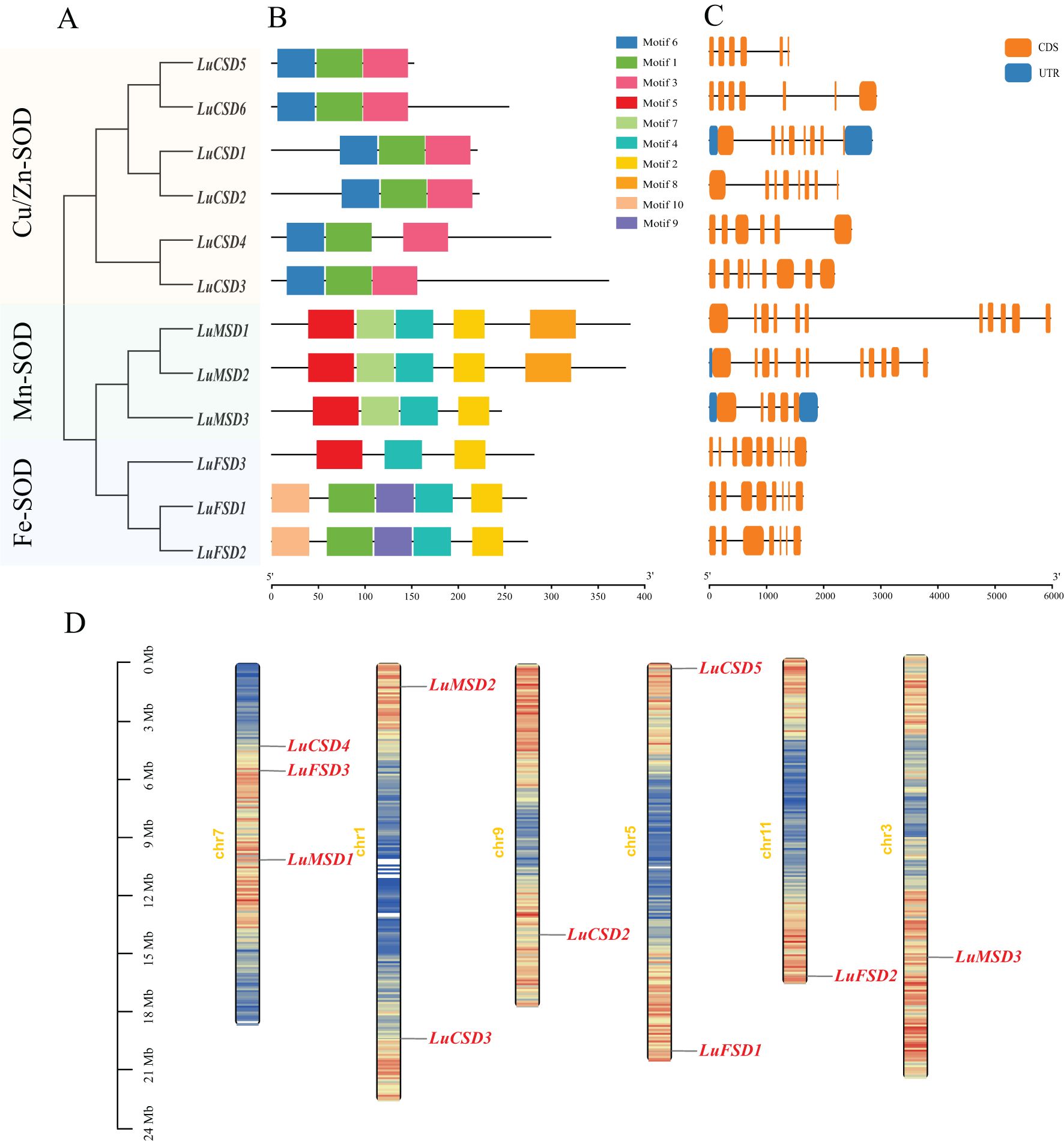
Figure 2. Structural and genomic characterization of the LuSOD gene family. (A) Phylogenetic clustering of LuSOD genes. (B) Conserved motif distribution, with gray lines indicating gene length. (C) Exon-intron architectures, highlighting structural conservation within subfamilies. (D) Chromosomal localization, visualized with a 100-kb sliding window; base density gradients are color-mapped from red (highest) to blue (lowest).
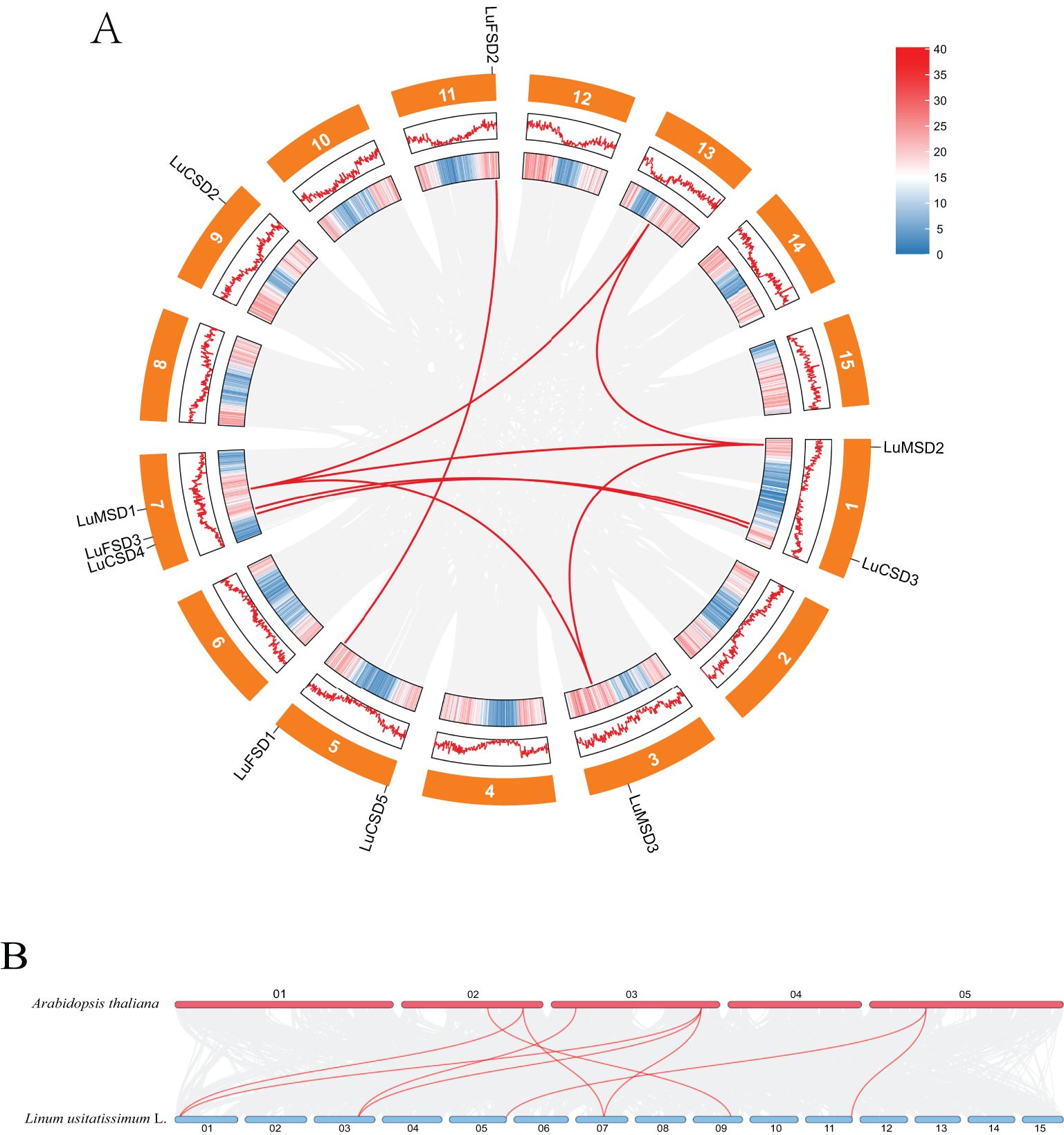
Figure 3. Syntenic relationships of LuSOD genes. (A) Intra-genomic collinearity among LuSOD loci. (B) Cross-species collinearity between flax and Arabidopsis SOD genes. Gray lines denote genome-wide syntenic blocks, while red lines highlight LuSOD-specific orthologous pairs.
3.4 LuSOD regulatory element profiling and functional network analysis
To elucidate the regulatory mechanisms of the LuSOD gene family under abiotic stress, promoter regions spanning 2000 bp upstream of the LuSOD loci were analyzed and visualized (Figure 4; Supplementary Table S3). Due to genomic proximity, the upstream sequences of LuCSD1 and LuCSD6 could not be isolated, likely resulting from overlapping or truncated intergenic regions. Consequently, promoter analyses focused on the remaining 10 LuSOD genes, excluding ubiquitous elements such as CAAT-box and TATA-box. A total of 259 cis-regulatory elements were identified and categorized into four functional groups: developmental regulation, environmental stress adaptation, phytohormone signaling, and light responsiveness (Figure 4). Light-responsive elements constituted the predominant category (91 elements, 35.14%), with conserved motifs including G-box, I-box, and Box 4. The second largest group comprised hormone-related elements (89 elements, 34.36%), dominated by methyl jasmonate (MeJA)-responsive motifs (TGACG and CGTCA), alongside abscisic acid (ABRE), auxin (TGA-element, AuxRR-core), gibberellin (GARE-motif), and salicylic acid (TCA-element) response elements. Notably, MeJA-associated motifs were the most abundant hormonal regulators in LuSOD promoters. Environmental stress-responsive elements (57 elements, 22.01%) included anaerobic induction (ARE), drought-inducible MYB binding sites (MBS), low-temperature response (LTR), MYBHv1 recognition sites (CCAAT-box), and defense/stress-related TC-rich repeats. Developmental elements (22 elements, 8.5%) encompassed zein metabolism regulators (O2-site), meristem-specific motifs (CAT-box), and endosperm activity markers (GCN4_motif).
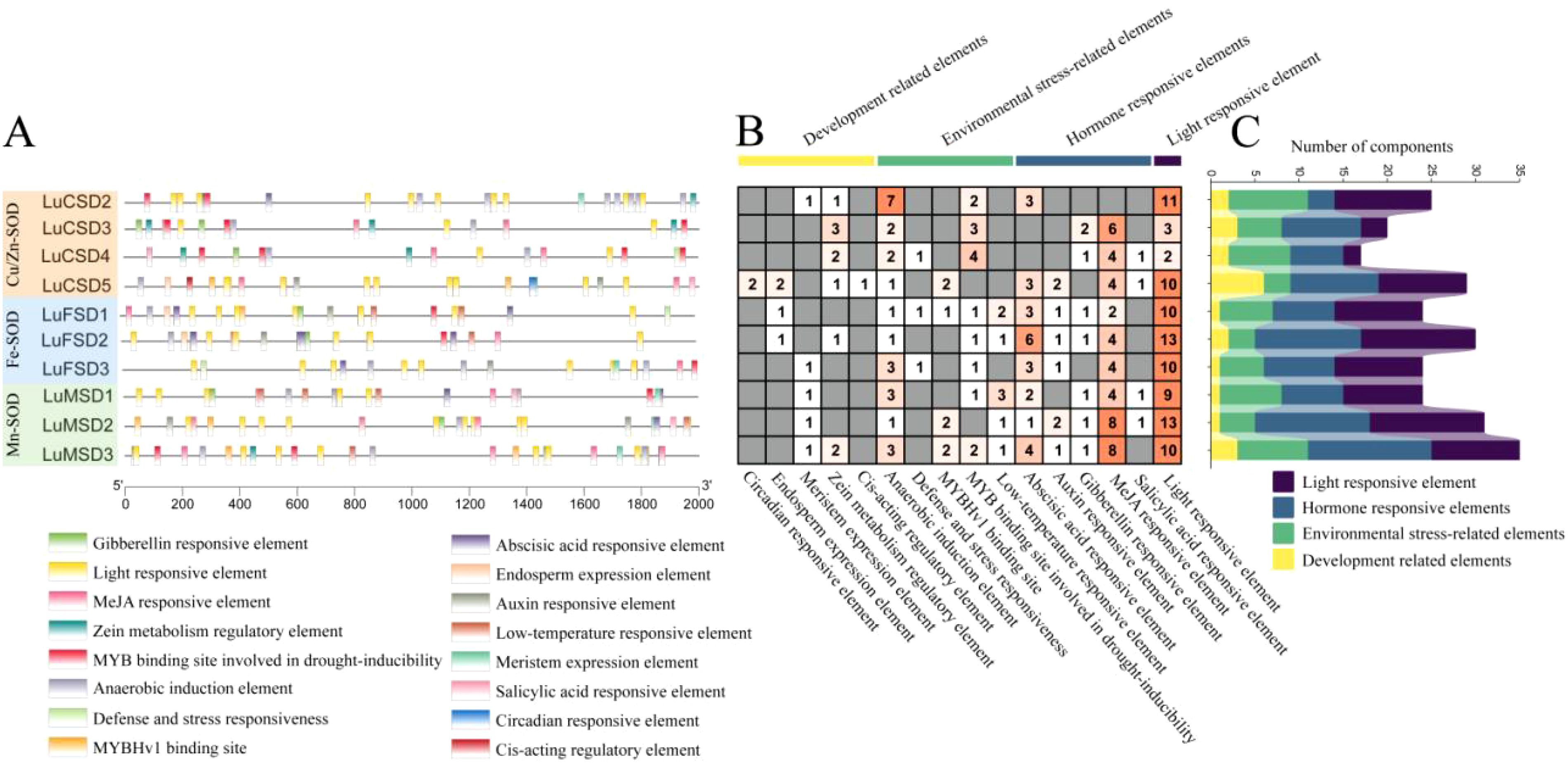
Figure 4. Cis-acting regulatory element profiling of LuSOD promoters. (A) Subfamily-specific distribution of cis-elements. (B, C) Quantitative analysis of motifs associated with developmental regulation (yellow), environmental stress (green), hormonal signaling (dark blue), and light response (purple).
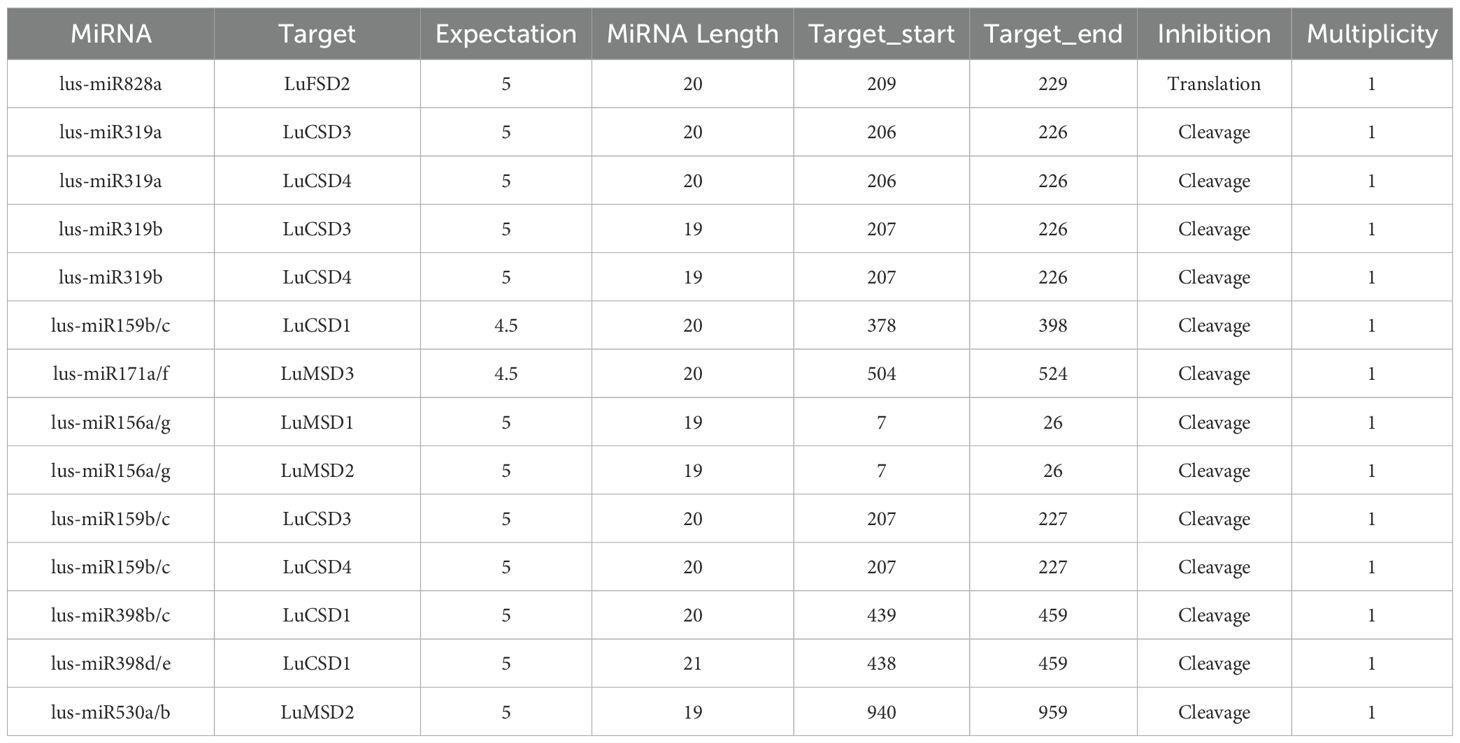
MiRNA prediction results showed that among the 12 LuSOD gene families, only 7 family members (58.33%) predicted 17 miRNA targets (Table 2). Among them, LuCSD1 gene predicted the most miRNA targets, including lus-miR159b/c and lus-miR398b/c/d/e. LuFSD2 gene has the least target and contains only one miRNA target (lus-miR828a). We found that the same LuSOD gene can be targeted by different miRNA. For example, LuCSD1 can be targeted by both lus-miR159 and lus-miR398, and LuMSD2 gene can be targeted by lus-miR156 and lus-miR530 at the same time. We also found that different LuSOD genes can be targeted by the same miRNA. For example, lus-miR319 can target both LuCSD3 and LuCSD4 genes, lus-miR156 can target both LuMSD1 and LuMSD2 genes, and lus-miR159 can target LuCSD1, LuCSD3 and LuCSD4 at the same time. The results show that lus-miR159 is the main target miRNA of LuSOD gene family.light response elements.
To investigate functional linkages between flax SOD proteins and their regulatory roles, a protein-protein interaction (PPI) network was constructed using Arabidopsis homologs as a reference framework (Figure 5A). The analysis revealed pairwise interactions among six LuSOD genes (LuCSD2, LuCSD3, LuCSD6, LuFSD2, LuFSD3, and LuMSD2), whereas the remaining six genes showed no connectivity. The strong evolutionary conservation between LuSOD and AtSOD proteins suggested functional parallels, particularly given the established role of Arabidopsis SODs in mitigating biotic and abiotic stressors. These findings implied that the six interacting LuSOD genes may mediate analogous stress-responsive mechanisms in flax.
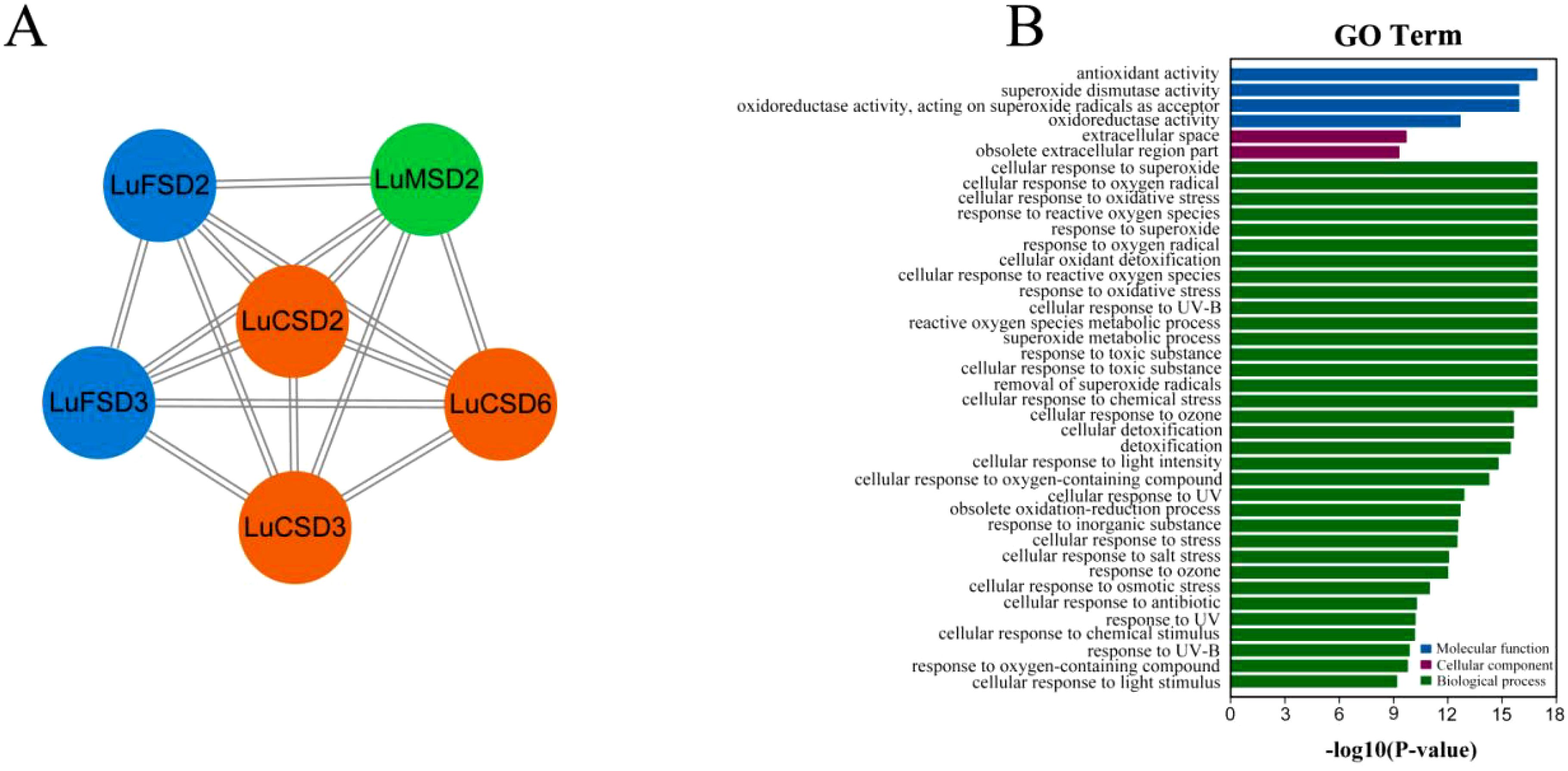
Figure 5. Protein interaction networks and functional annotation of LuSOD genes. (A) Predicted protein-protein interaction (PPI) networks based on Arabidopsis homologs, color-coded by subfamily: LuCSD (orange), LuMSD (green), and LuFSD (blue). (B) GO enrichment, categorized into molecular function (blue), cellular component (purple), and biological process (green).
Gene Ontology (GO) enrichment analysis further delineated the functional spectrum of LuSOD genes across three domains: molecular functions, cellular components, and biological processes (Figure 5B; Supplementary Table S4). Molecular functions were primarily associated with antioxidant activity and superoxide dismutase activity. Cellular component annotations highlighted localization to the extracellular space and archived extracellular regions (obsolete classification). Biological processes predominantly involved cellular responses to superoxide radicals, oxygen radical detoxification, and oxidative stress adaptation, underscoring the pivotal role of LuSOD genes in stress resilience.
3.5 LuSOD expression patterns and abiotic stress responses in flax
Transcriptomic analysis of the LuSOD gene family under abiotic stress revealed distinct expression dynamics: under heat stress, only LuFSD1 exhibited elevated expression in stems, while salt stress significantly suppressed most LuSOD genes in leaves and stems (Figure 6A). However, four genes (LuCSD3, LuCSD4, LuCSD5, and LuCSD6) were upregulated in roots, and three (LuMSD3, LuCSD5, and LuCSD6) showed marked induction in leaves under salt stress, with the LuFSD subfamily displaying pronounced sensitivity to salt-induced repression. Tissue-specific expression profiling further uncovered spatiotemporal regulation: in floral tissues (Figures 6B, C), all LuSOD genes except LuCSD4 and LuFSD2 were highly expressed at 5 days post-anthesis (DPA), followed by seven genes (LuCSD4, LuFSD2, LuCSD2, LuMSD1, LuCSD3, LuFSD1, and LuMSD2) at 10 DPA, three (LuCSD1, LuCSD5, and LuMSD3) at 20 DPA, and universal downregulation by 30 DPA. Vegetative tissues showed dominant LuSOD expression in leaves (notably LuCSD3), contrasting with minimal activity in roots, stems, anthers, stamens, and seeds. Embryo development stages revealed specialized roles: LuMSD3 peaked in mature and cotyledon-stage embryos; LuFSD3 and LuFSD2 in heart-stage embryos; LuFSD2 and LuMSD1 in globular embryos; and the LuFSD subfamily in torpedo-stage embryos. Reproductive organs exhibited coordinated upregulation of LuCSD5 and LuCSD6 in ovaries, pistils, and fruits. Collectively, the robust expression of LuSOD genes, particularly LuCSD3, in leaf tissues underscored their pivotal role in maintaining ROS homeostasis, positioning leaves as central hubs for antioxidant defense in flax.
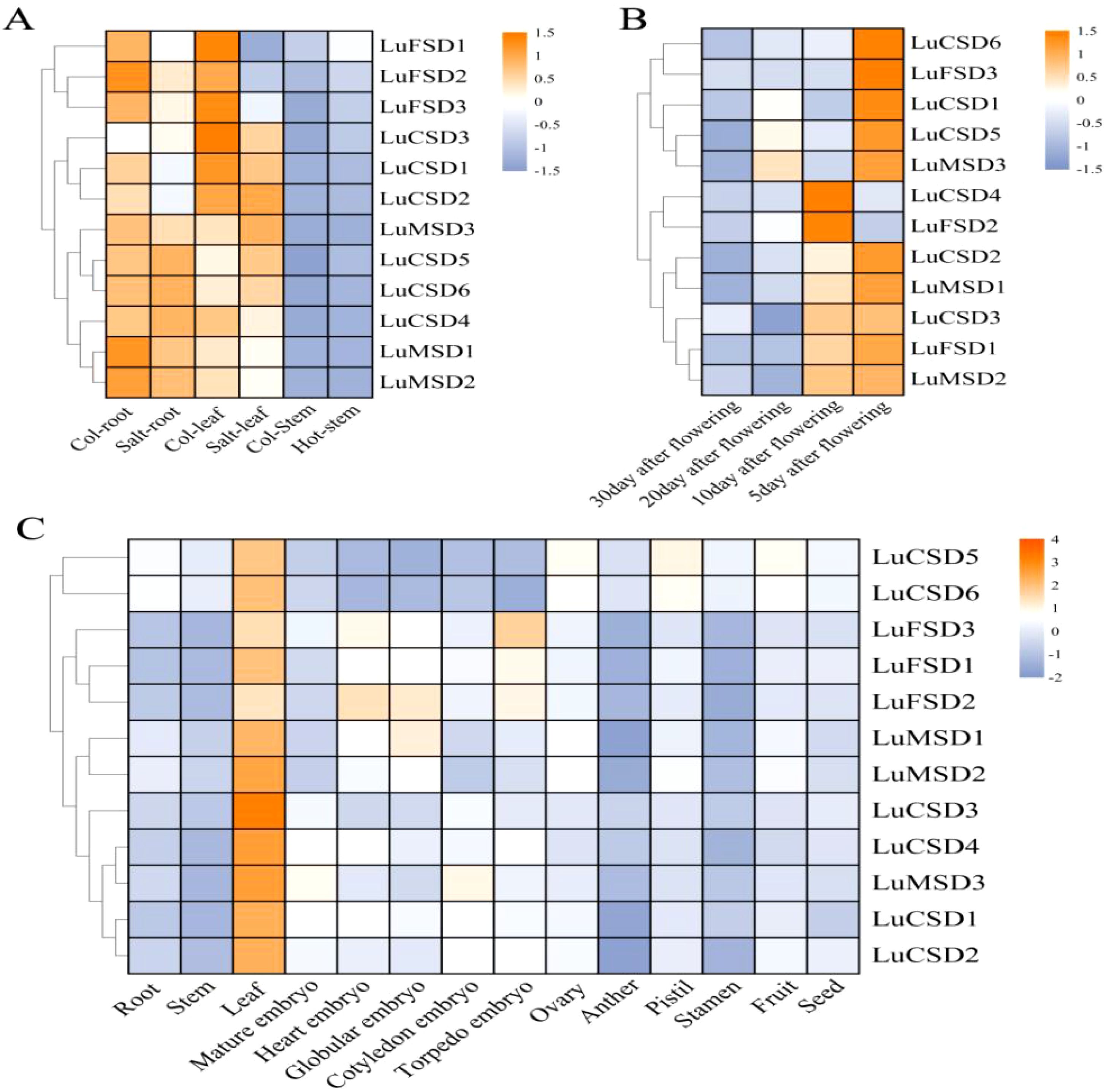
Figure 6. Spatiotemporal expression dynamics of LuSOD genes. (A) Transcriptional responses to salt and heat stress. (B) Expression profiles in post-anthesis floral tissues. (C) Tissue-specific expression patterns across flax organs. Expression levels are normalized as log2-transformed FPKM values, color-scaled from high (orange) to low (blue-purple).
To evaluate the involvement of LuSOD genes in abiotic stress responses (salt, cold, drought), qRT-PCR was employed to quantify their relative expression levels in flax leaf tissues under salt stress. Transcriptional dynamics were assessed at 0, 6, 12, and 24 hours post-stress induction, with expression levels normalized to the 0-hour control (Figures 7–9). Salt stress triggered time-dependent transcriptional reprogramming within the LuSOD family (Figure 7). Six genes (LuCSD4, LuCSD5, LuCSD6, LuFSD2, LuMSD1, and LuMSD3) displayed pronounced upregulation under 12-hour salt stress, peaking at 2.3-, 1.3-, 1.25-, 2.4-, and 3.1-fold increases relative to controls, respectively. Two genes (LuCSD3 and LuFSD1) exhibited maximal induction (2.1- and 2.3-fold increases) after 24 hours of salt exposure. In contrast, LuCSD1 showed no transcriptional response to salt stress in leaves. Conversely, three genes (LuCSD2, LuFSD3, and LuMSD2) demonstrated substantial downregulation, reaching minimal expression levels at 24 hours.
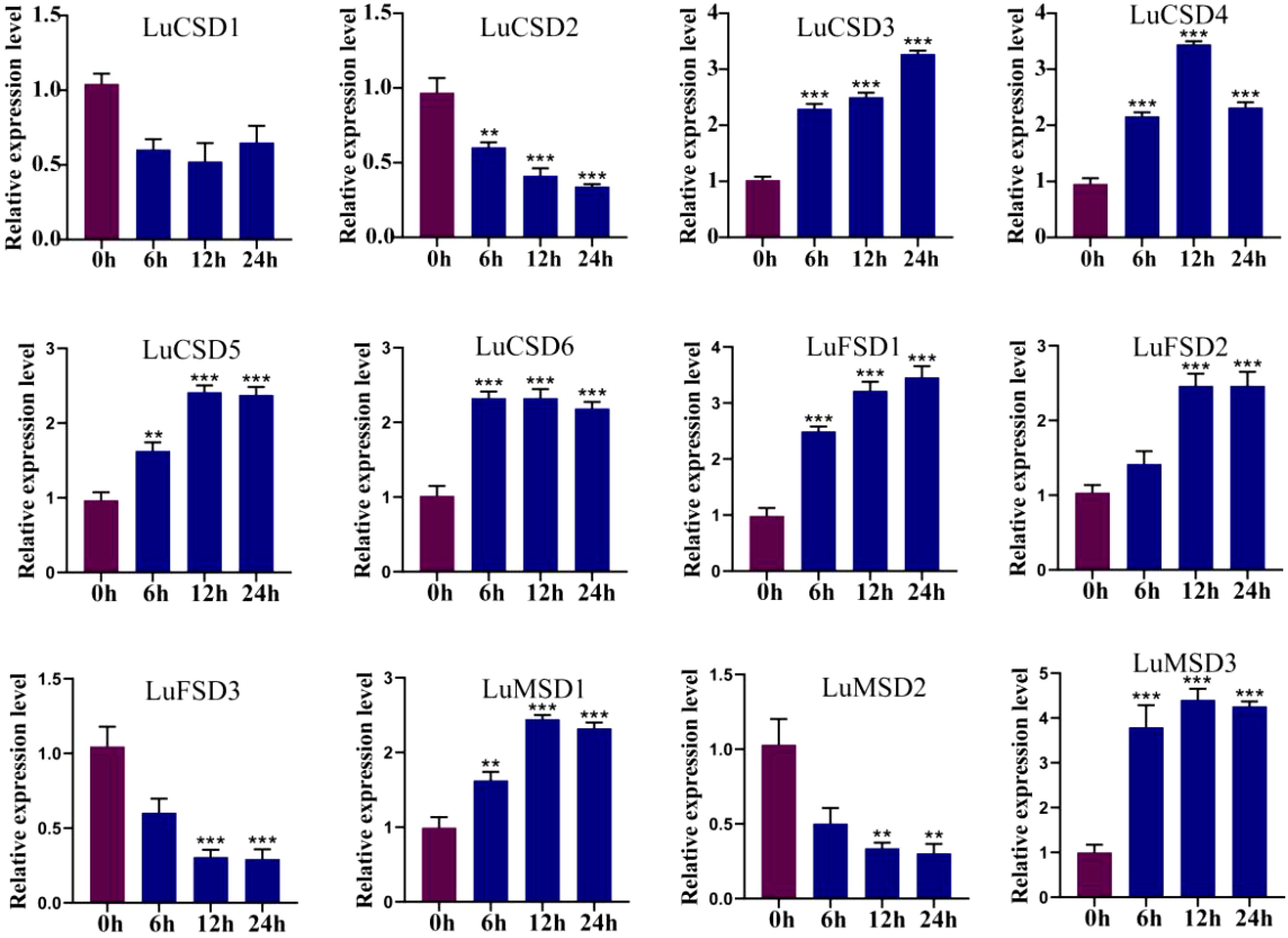
Figure 7. LuSOD gene expression under salt stress. Dark purple and dark blue bars represent untreated controls and salt-stressed samples, respectively. Asterisks denote statistically significant differences (Student’s t-test: **p < 0.01; ***p < 0.001).
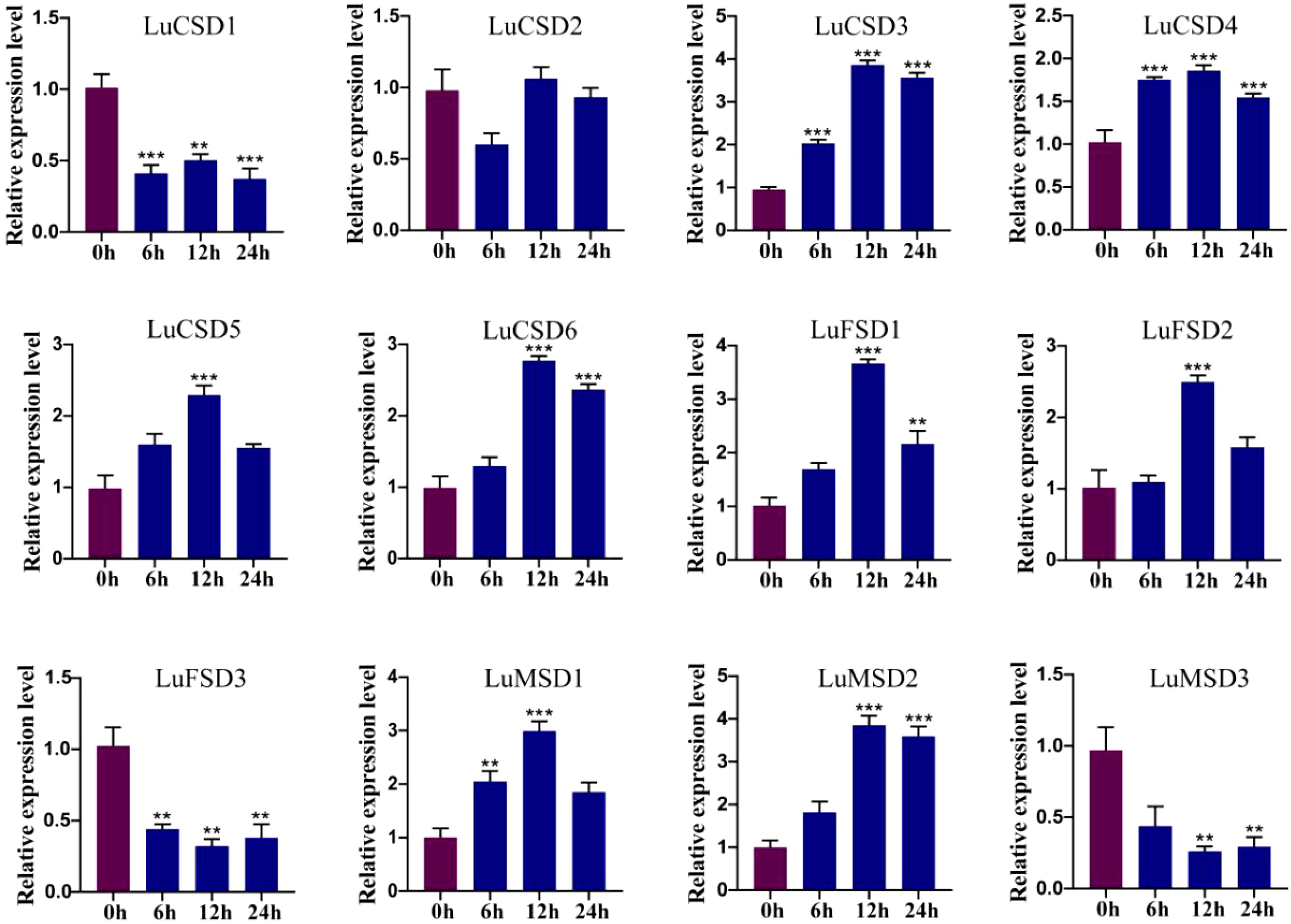
Figure 8. LuSOD gene expression under cold stress. Dark purple and dark blue bars represent untreated controls and salt-stressed samples, respectively. Asterisks denote statistically significant differences (Student’s t-test: **p < 0.01; ***p < 0.001).
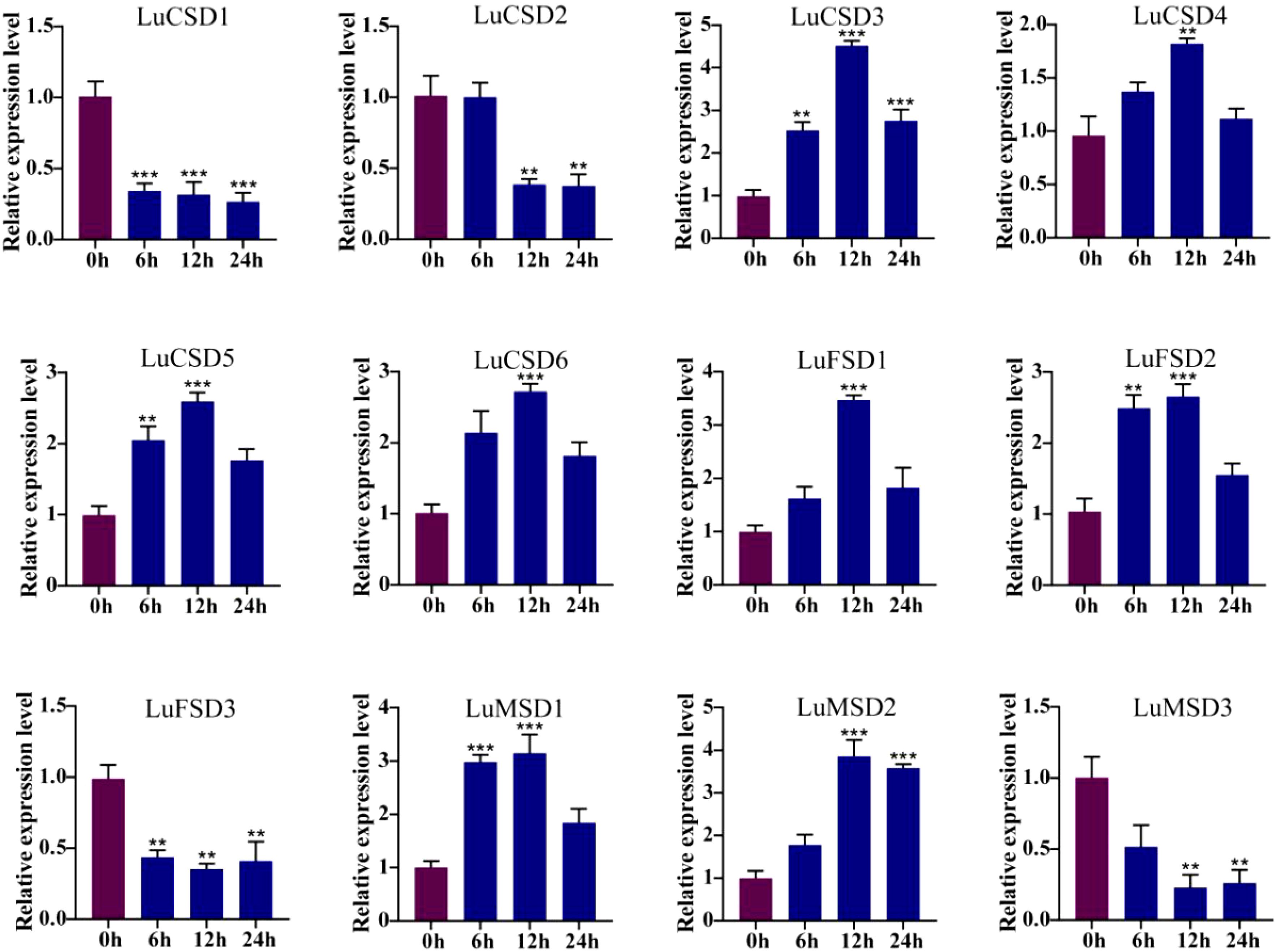
Figure 9. LuSOD gene expression under drought stress. Dark purple and dark blue bars represent untreated controls and salt-stressed samples, respectively. Asterisks denote statistically significant differences (Student’s t-test: **p < 0.01; ***p < 0.001).
Under prolonged abiotic stress conditions, the LuSOD gene family exhibited dynamic transcriptional regulation in flax leaf tissues. During cold stress (Figure 8), eight genes (LuCSD3, LuCSD4, LuCSD5, LuCSD6, LuFSD1, LuFSD2, LuMSD1, and LuMSD2) showed pronounced upregulation at 12 hours, peaking at 2.8-, 0.8-, 1.2-, 1.6-, 2.5-, 1.4-, 1.9-, and 2.8-fold induction, respectively, followed by gradual attenuation, while LuCSD2 remained unresponsive. Three genes (LuCSD1, LuFSD3, and LuMSD3) were significantly downregulated, reaching minimal expression levels at 24 hours, suggesting their potential role in cold adaptation through negative regulatory mechanisms. Similarly, under drought stress (Figure 9), the same eight genes displayed marked upregulation at 12 hours, with peak expression levels of 3.3-, 0.7-, 1.4-, 1.5-, 2.4-, 1.5-, 2.1-, and 2.8-fold increases, respectively, whereas LuCSD1, LuFSD3, and LuMSD3 exhibited sustained downregulation, implicating their involvement in drought-responsive suppression pathways. Notably, LuCSD3 consistently demonstrated robust upregulation under both stress conditions, highlighting its central role in stress adaptation. These results collectively underscore the functional divergence of LuSOD genes in mediating stress-specific transcriptional reprogramming, with select members acting as positive regulators of antioxidant defense and others contributing to stress tolerance through negative feedback mechanisms.
3.6 Constructing transgenic plants with LuCSD3 gene and its role in salt stress response
Our experimental findings revealed that the LuCSD3 gene conferred robust stress tolerance in flax. To functionally characterize LuCSD3, we engineered a binary expression vector (pCAMBIA3301-LuCSD3; Figure 10A) and stably transformed it into wild-type Arabidopsis (Col-0) via Agrobacterium-mediated floral dip. Primary transformants screening identified eight independent T1 lines showing constitutive LuCSD3 overexpression. Molecular characterization of T3-generation progeny confirmed two homozygous lines (OE-1 and OE-5) with maximal transgene expression levels (Figures 10B, C). Salt tolerance tests were conducted on Col and OE lines (OE-1 and OE-5) by exposing plants to 200 mM NaCl stress for 15 days. The results showed that the OE plants exhibited superior growth phenotypes under salt stress, with significantly less wilting compared to the control group (Col) (Figure 10D).
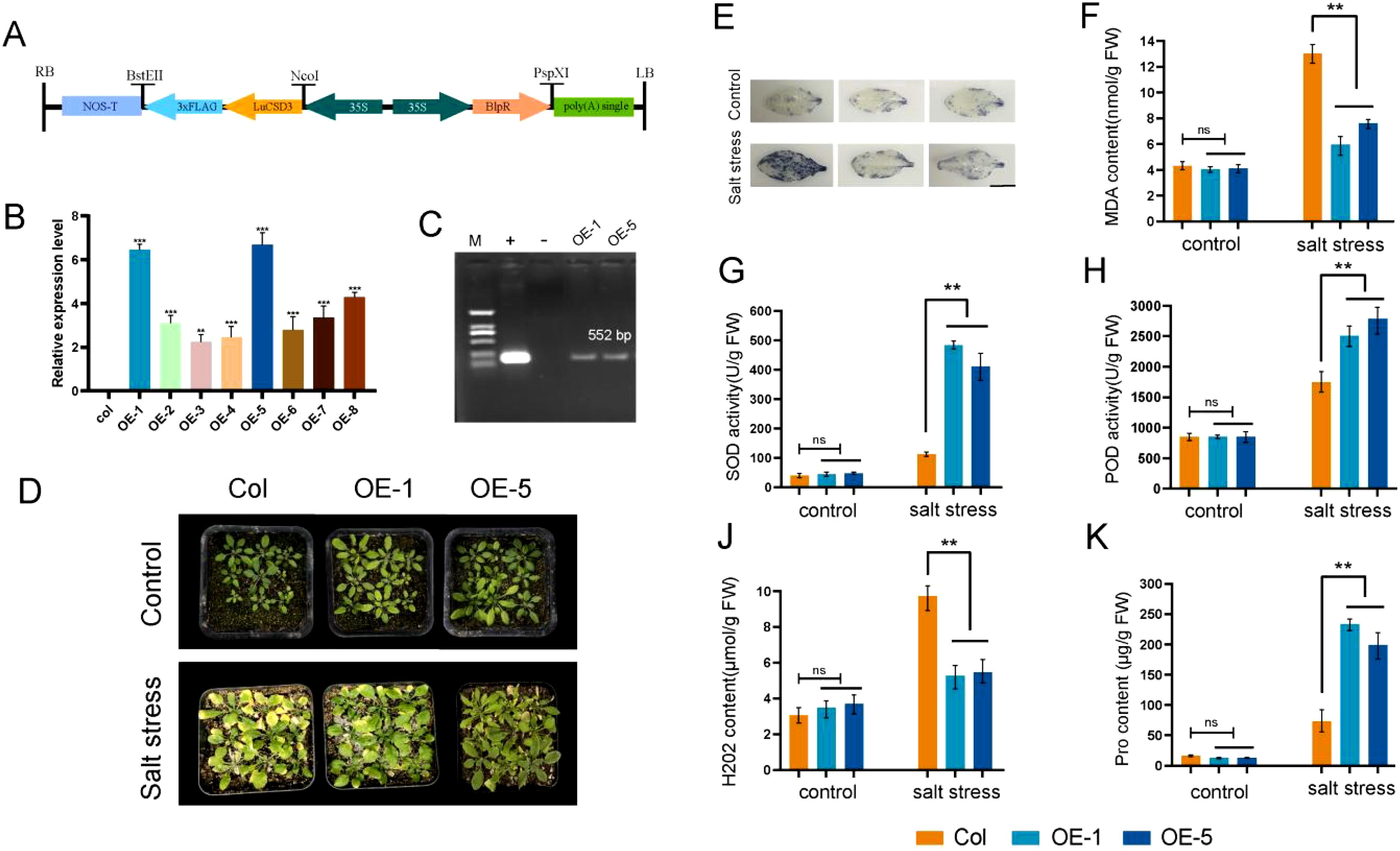
Figure 10. Identification and salt tolerance analysis of LuCSD3-overexpressing Arabidopsis. (A) Schematic representation of the pCAMBIA3301-LuCSD3 vector. Arrows indicate expression elements, and horizontal lines represent vector restriction sites. (B) Gene expression analysis of T3-generation transgenic LuCSD3 Arabidopsis lines. (C) PCR verification of LuCSD3 gene expression in 35S::LuCSD3 overexpression lines (OE-1 and OE-5). M: DNA Marker DL2000; +: recombinant plasmid pCAMBIA3301-LuCSD3; -: empty vector pCAMBIA3301 negative control. (D) Salt tolerance analysis of overexpression (OE) lines and control (Col) plants. Two-week-old Col and OE plants were watered with or without 200 mM NaCl every three days for 15 days. Photographs were taken to monitor stress phenotypes and measure physiological indices. (E) NBT staining of Col and OE leaves under salt stress to detect ROS. Scale bar=1 cm. (F) Malondialdehyde (MDA) content. (G) Analysis of superoxide dismutase (SOD) activity. (H) Peroxidase (POD) activity. (J) Hydrngen peroxide (H2O2) content. (K) Proline (Pro) content. Statistical significance of differences is indicated by asterisks: *p < 0.05; **p < 0.01; ***p < 0.001.
3.7 Overexpression of LuCSD3 gene reduces ROS accumulation
To assess the physiological and biochemical impacts of LuCSD3 overexpression, malondialdehyde (MDA) and proline (Pro) levels were quantified in wild-type (Col) and transgenic lines (OE-1, OE-5) under salt stress (Figures 10F, K). Transgenic plants exhibited markedly reduced MDA concentrations and significantly elevated Pro accumulation compared to Col controls. To evaluate LuCSD3-mediated regulation of reactive oxygen species (ROS), hydrogen peroxide (H2O2) content, superoxide dismutase (SOD), and peroxidase (POD) activities were examined. Under non-stress conditions, no statistically significant variations in SOD, POD, or H2O2 levels were observed between Col and OE lines (Figures 10G, H, J). However, salt-stressed OE lines demonstrated a pronounced increase in SOD (1.8-fold) and POD (2.3-fold) enzymatic activities, coupled with a 40% reduction in H2O2 content relative to controls. These findings were further supported by NBT staining assays, which revealed reduced ROS accumulation in OE leaf tissues under stress (Figure 10E). Collectively, these results indicate that LuCSD3 overexpression enhances salt stress resilience in plants by augmenting antioxidant enzyme activities (SOD, POD) to neutralize ROS and modulating osmoprotectant (Pro) synthesis while minimizing oxidative damage (reduced MDA). This dual mechanism underscores LuCSD3 as a key regulator of ROS homeostasis under abiotic stress.
3.8 Expression pattern analysis of key genes in salt stress pathway in OE-LuCSD3 strain
When plants encounter salt stress, they employ various mechanisms to resist adverse conditions. The expression levels of five selected salt stress-related genes (NHX1, HKT1, SOS1, SOS2, and SOS3) were analyzed in OE-LuCSD3 lines (Figure 11). The results indicated that the expression levels of SOS2 and SOS3 were not correlated with the expression of LuCSD3. However, three key genes (NHX1, HKT1, and SOS1) showed significantly upregulated expression under salt stress. These findings suggest that the LuCSD3 gene enhances salt tolerance by mitigating ROS accumulation in plants.
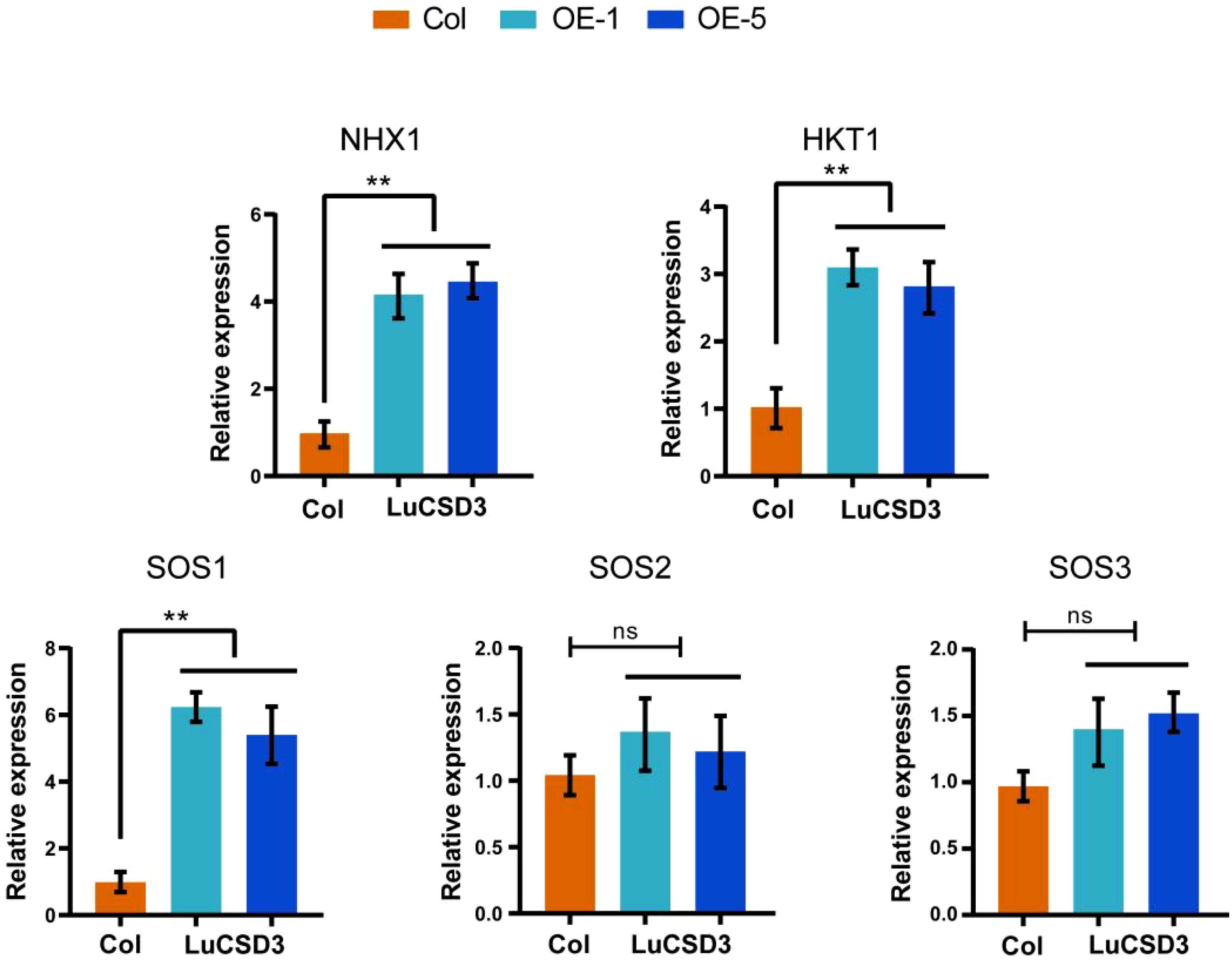
Figure 11. Expression of salt-responsive genes in LuCSD3-overexpressing lines. Relative expression levels of NHX1, HKT1, SOS1, SOS2, and SOS3 in transgenic (OE-LuCSD3) and wild-type (Col) plants under salt stress. Data are means ± SD (n = 3). Asterisks denote significance (*p < 0.05; **p < 0.01; ***p < 0.001).
4 Discussion
Abiotic stress has long been a major constraint on agricultural productivity. Superoxide dismutase (SOD), a critical enzyme in plant stress responses, mitigates oxidative damage caused by salt, drought, and heavy metal toxicity (Gill and Tuteja, 2010). In the past few years, peroxidase family genes in different plants have been identified, such as five peroxidase genes in seaweed (Zang et al., 2020), seven genes in Medicago truncatula (Song et al., 2018), seven SOD genes in Rosa chinensis (Rafique et al., 2023), nine genes in tomato (Feng et al., 2016), 18 genes in cotton (Wang et al., 2017) and 25 genes in banana (Feng et al., 2015), 26 genes were identified in wheat (Jiang et al., 2019), 29 genes were identified in B. juncea (Verma et al., 2019), 31 genes were identified in Brassica napus (Su et al., 2021). The subfamily distribution of the LuSOD genes included six Cu/Zn SOD genes, three Fe SOD genes, and three Mn SOD genes (Table 1) (Figure 1). This classification aligns with the six SOD members reported in Medicago truncatula (Song et al., 2018). This relatively moderate number of genes suggested that flax might have relied more on transcriptional and post-transcriptional regulation, rather than extensive gene duplication, to modulate SOD-mediated stress responses. In addition, a large number of studies have shown that Cu/ZnSOD gene is mainly distributed in mitochondria, cytoplasmic sol, peroxisome and chloroplast, FeSOD gene is mainly distributed in mitochondria and chloroplast, while MnSOD gene is almost distributed in mitochondria (Corpas et al., 2006; Wang et al., 2017). This was consistent with the results of this study. The predicted subcellular localizations in the chloroplast, mitochondrion, and cytoplasm emphasized the compartment-specific detoxification of ROS, a well-established strategy for fine-tuning redox signaling under stress conditions (Noctor et al., 2014). Cu/Zn SODs localized to the chloroplast were particularly important in responding to light-induced oxidative stress, while mitochondrial Mn SODs helped alleviate ROS generated during respiration (Alscher et al., 2002).
Analysis of gene architecture demonstrated that the LuSOD family in flax exhibited variable exon-intron configurations, with exon numbers ranging from 5 to 11 and introns from 4 to 10 (Figure 2). Structurally, the LuSOD genes exhibited conserved exon–intron organization and motif composition within each subfamily, reflecting strong purifying selection during evolution. This conservation supported the hypothesis that SOD isoforms were functionally constrained due to their critical roles in redox buffering (Fink and Scandalios, 2002). Divergence in LuSOD gene organization may reflect evolutionary dynamics driven by exon/intron indels or splicing variations (Xu et al., 2012). These findings align with prior reports on SOD gene structure in tomato (Feng et al., 2016) and cotton (Wang et al., 2017), reinforcing the role of structural conservation in stress-responsive gene families.
Gene duplication events played a pivotal role in the evolutionary diversification of SOD genes, with repetitive gene copies often driving functional innovation to enhance plant adaptive responses to environmental stressors. Gene duplication of SOD genes has also been identified in cotton and rapeseed (Wang et al., 2016; Verma et al., 2019). In this study, eight segmental duplication pairs were detected within the LuSOD gene family (Figure 3A), indicating that segmental duplication served as the primary driver of LuSOD family expansion. Synteny analysis, a robust approach for reconstructing gene evolutionary trajectories, revealed nine collinear ortholog pairs between flax and Arabidopsis. These conserved syntenic relationships suggested shared ancestral origins prior to species divergence (Jiao et al., 2014), further underscoring the high evolutionary conservation of LuSOD genes in flax.
Cis-regulatory elements within promoter regions orchestrate transcriptional regulation, primarily through sequence-specific interactions between transcription factors and their cognate DNA motifs, a process central to stress-responsive gene expression (Punia et al., 2021). Bioinformatic interrogation of LuSOD promoters revealed a predominant enrichment of cis-elements associated with phytohormonal signaling (abscisic acid and methyl jasmonate pathways) and abiotic stress adaptation, including hypoxia response, drought inducibility, and low-temperature tolerance (Figure 4). Functional annotation of stress-related motifs identified three core regulatory modules: LTR (low-temperature responsiveness), ARE (anaerobic induction), and TC-rich repeats (drought and defense signaling) (Zhang et al., 2018; Dhatterwal et al., 2021). Notably, the majority of LuSOD promoters harbored ARE, LTR, and TC-rich repeat elements, underscoring their conserved role in mediating oxidative stress resilience and environmental adaptation (Osakabe et al., 2014).
MicroRNAs have been recognized as pivotal regulators in harmonizing plant developmental processes and environmental interactions (Song et al., 2019). Our analysis shows that lus-miR159 is the main target miRNA of the LuSOD gene family (Table 2). In tobacco, miR159-GAMYB pathway plays a role in biological defense response, which is activated after miR159 inhibition (Zheng et al., 2020). In maize, overexpression of miR159 leads to grain enlargement in transgenic plants, indicating that miR159-ZmMYB module, as the hub of endosperm development, is involved in endosperm cell division and proliferation (Wang et al., 2023). However, a broader perspective suggested that the SOD–miRNA regulatory module was evolutionarily conserved across plant species and was frequently modulated under abiotic stress conditions. For example, in Arabidopsis, miR398 played a central role in oxidative stress signaling by targeting CSD1 and CSD2. Its downregulation under stress led to increased SOD expression and enhanced stress tolerance (Beauclair et al., 2010). Comparable modules were reported in cotton, where miR398 regulated GhCSD1 during salt and drought conditions (Shumayla et al., 2017). These findings highlighted the evolutionary conservation of miRNA-mediated regulation of SOD genes across diverse plant taxa. Therefore, the miRNA–LuSOD interactions in flax might have represented a broader regulatory circuit involved in stress responses, warranting further experimental validation. Protein-protein interactions were essential not only for maintaining functional integrity but also for predicting functional diversification of proteins (Nobeli et al., 2009). In this study, protein interaction network analysis revealed six LuSOD genes with significant pairwise interactions (Figure 5A), suggesting their central roles in combating diverse biotic and abiotic stresses. GO enrichment analysis corroborated these findings (Figure 5B), aligning with conserved SOD functions observed in other crops (Feng et al., 2016; Jiang et al., 2019; Verma et al., 2019). This study underscored the critical role of LuSOD genes in flax adaptation to environmental adversities, reinforcing their universal importance in plant stress physiology.
Transcriptomic profiling revealed tissue-specific expression dynamics of LuSOD genes across flax organs, with predominant transcriptional activity observed in leaves and floral tissues at 5 and 10 days post-anthesis (Figures 6B, C). Salt stress markedly downregulated LuSOD expression in leaf and stem tissues, while four genes exhibited upregulated expression in roots and three in leaves under salinity, consistent with prior findings in other species (Su et al., 2021; Yu et al., 2022; Rafique et al., 2023). The ubiquitous expression of these genes across flax tissues suggested their functional involvement in developmental regulation. Under abiotic stress (salt, cold, drought), qRT-PCR analysis of leaf tissues demonstrated significant upregulation of nine LuSOD genes, except LuCSD1, LuCSD2, and LuFSD3 (Figures 7-9). Notably, LuMSD2 and LuMSD3 exhibited completely opposite transcriptional responses under salt stress compared to cold and drought conditions. LuMSD2 was significantly downregulated under salt stress but showed strong upregulation in response to cold and drought. In contrast, LuMSD3 displayed markedly increased expression under salt stress, while its expression was suppressed during cold and drought exposure (Figures 7-9). This contrasting behavior suggested possible functional divergence despite their shared subcellular localization. Cis-regulatory analysis of their promoter regions revealed distinct features: LuMSD2 contained a higher abundance of low-temperature and drought-responsive elements such as LTR and MBS, whereas LuMSD3 was enriched with salinity-related stress-responsive motifs, including ARE and TC-rich repeats (Dhatterwal et al., 2021). The functional divergence might have reflected subtle differences in their roles within mitochondria under specific stress conditions. Under salt stress, mitochondrial metabolism underwent rapid alterations, which might have required a distinct SOD isoform to more effectively buffer ROS or to help maintain mitochondrial integrity (Suzuki et al., 2012). LuMSD3 might have played a more prominent role in acute ROS scavenging, whereas LuMSD2 might have been involved in slower or regulatory ROS signaling under cold and drought stress conditions, where mitochondrial ROS functioned as secondary messengers (Huang et al., 2019). Comparative analyses across species demonstrated conserved SOD upregulation under stress: cucumber SODs responded to cold, heat, salt, and drought (Zhou et al., 2017); tomato SODs were induced by salt and drought (Feng et al., 2016); wheat SODs showed systemic activation under similar conditions (Jiang et al., 2019). In Brassica napus, all SODs except BnCSD6 and BnFSD1 were stress-inducible, while Rosa chinensis roots exhibited elevated RcCSD1, RcCSD3, and RcFSD3 expression under salinity (Rafique et al., 2023). Furthermore, this study revealed that the overexpression of the LuCSD3 gene enhanced the salt stress response in Arabidopsis, ultimately improving the plant’s salt tolerance (Figure 10). Overexpression of LuCSD3 in Arabidopsis enhanced salinity tolerance by modulating ROS scavenging, analogous to the stress-responsive MeCSOD2 in cassava (Zheng et al., 2023). ROS, critical for cellular homeostasis and signaling, accumulated under stress, inducing oxidative damage (Yang et al., 2015). In this study, transgenic overexpression lines (OE) displayed markedly elevated superoxide dismutase (SOD) and peroxidase (POD) enzymatic activities under salt stress, concurrent with a pronounced reduction in hydrogen peroxide (H2O2) accumulation. These observations were further validated through nitroblue tetrazolium (NBT) staining assays (Figure 10). The LuCSD3 gene enhances abiotic stress tolerance by upregulating peroxidase and antioxidant enzyme activities in leaves, thereby mitigating ROS accumulation. To elucidate salt adaptation mechanisms, the expression profiles of five well-characterized salt-responsive genes (AtSOS1, AtSOS2, AtSOS3, AtNHX1, and AtHKT1) were analyzed in both wild-type (Col) and OE lines (Lu et al., 2023; Ma et al., 2023). Compared to the control group, the expression levels of three key genes (NHX1, HKT1, and SOS1) were significantly upregulated under salt stress conditions (Figure 11). These findings demonstrated conclusively that LuCSD3 plays a pivotal role in mediating flax’s response to abiotic stressors. Collectively, this investigation established a framework for deciphering the molecular mechanisms underlying SOD-mediated stress resistance in flax, providing critical insights for future functional studies of the LuSOD gene family.
5 Conclusions
In this study, the Superoxide Dismutase (SOD) gene family in flax was systematically characterized for the first time. A total of 12 LuSOD genes were annotated in the flax genome. Evolutionary relationships, exon-intron architectures, conserved protein motifs, and syntenic patterns were investigated through comparative bioinformatics analyses. miRNA target screening revealed lus-miR159 as the predominant miRNA regulating the LuSOD family. Protein interaction networks identified functional linkages among six LuSOD members (LuCSD2, LuCSD3, LuCSD6, LuMSD2, LuFSD2, and LuFSD3). GO enrichment highlighted LuSOD involvement in stress response pathways, metal ion binding, and enzymatic antioxidant activity. Promoter cis-element profiling demonstrated enrichment of motifs associated with phytohormone signaling (MeJA and ABA) and abiotic stress adaptation. Transcriptomic and qRT-PCR analyses revealed that most LuSOD genes exhibited tissue-specific expression patterns, particularly in leaves and floral tissues at 5 days post-anthesis, and responded dynamically to environmental stressors (cold, drought, salt). Functional validation in Arabidopsis demonstrated that LuCSD3 overexpression enhanced salt tolerance by modulating ROS scavenging. Collectively, this comprehensive investigation elucidated the structural and functional diversity of the LuSOD family, offering novel insights into SOD-mediated mechanisms underlying flax development and stress resilience.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author contributions
YZ: Writing – original draft, Writing – review & editing. RW: Writing – review & editing, Writing – original draft. HPW: Writing – review & editing, Writing – original draft. HYW: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Science & Technology Development, Jilin Province (No. 20240404022ZP).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1609085/full#supplementary-material
References
Abreu, I. A. and Cabelli, D. E. (2010). Superoxide dismutases-a review of the metal-associated mechanistic variations. Biochim. Biophys. Acta 1804, 263–274. doi: 10.1016/j.bbapap.2009.11.005
Alscher, R. G., Erturk, N., and Heath, L. S. (2002). Role of superoxide dismutases (sods) in controlling oxidative stress in plants. J. Exp. Bot. 53, 1331–1341. doi: 10.1093/jexbot/53.372.1331
Asensio, A. C., Gil-Monreal, M., Pires, L., Gogorcena, Y., Aparicio-Tejo, P. M., and Moran, J. F. (2012). Two fe-superoxide dismutase families respond differently to stress and senescence in legumes. J. Plant Physiol. 169, 1253–1260. doi: 10.1016/j.jplph.2012.04.019
Beauclair, L., Yu, A., and Bouche, N. (2010). Microrna-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mrna in arabidopsis. Plant J. 62, 454–462. doi: 10.1111/j.1365-313X.2010.04162.x
Cantalapiedra, C. P., Hernandez-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). Eggnog-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829. doi: 10.1093/molbev/msab293
Case, A. J. (2017). On the origin of superoxide dismutase: an evolutionary perspective of superoxide-mediated redox signaling. Antioxidants 6(4), 82. doi: 10.3390/antiox6040082
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., and Xia, R. (2020). Tbtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one fastq preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chytilová, M., Mudroňová, D., Nemcová, R., Gancarčíková, S., Buleca, V., Koščová, J., and Tkáčiková, Ľ. (2013). Anti-inflammatory and immunoregulatory effects of flax-seed oil and lactobacillus plantarum - biocenol lp96 in gnotobiotic pigs challenged with enterotoxigenic escherichia coli. Res. Vet. Sci. 95, 103–109. doi: 10.1016/j.rvsc.2013.02.002
Clough, S. J. and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Corpas, F. J., Barroso, J. B., and Palma Rodriguez-Ruiz, J. M.M. (2017). Plant peroxisomes: a nitro-oxidative cocktail. Redox Biol. 11, 535–542. doi: 10.1016/j.redox.2016.12.033
Corpas, F. J., Fernandez-Ocana, A., Carreras, A., Valderrama, R., Luque, F., et al. (2006). The expression of different superoxide dismutase forms is cell-type dependent in olive (olea europaea l.) Leaves. Plant Cell Physiol. 47, 984–994. doi: 10.1093/pcp/pcj071
Cui, C., Wang, J. J., Zhao, J. H., Fang, Y. Y., He, X. F., Guo, H. S., and Duan, C. G. (2020). A brassica mirna regulates plant growth and immunity through distinct modes of action. Mol. Plant 13, 231–245. doi: 10.1016/j.molp.2019.11.010
Dhatterwal, P., Mehrotra, S., Miller, A. J., and Mehrotra, R. (2021). Promoter profiling of arabidopsis amino acid transporters: clues for improving crops. Plant Mol. Biol. 107, 451–475. doi: 10.1007/s11103-021-01193-1
Ding, Y., Ding, L., Xia, Y., Wang, F., and Zhu, C. (2020). Emerging roles of micrornas in plant heavy metal tolerance and homeostasis. J. Agric. Food. Chem. 68, 1958–1965. doi: 10.1021/acs.jafc.9b07468
Feng, K., Yu, J., Cheng, Y., Ruan, M., Wang, R., Ye, Q., Wan, H., et al. (2016). The sod gene family in tomato: identification, phylogenetic relationships, and expression patterns. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01279
Feng, X., Lai, Z., Lin, Y., Lai, G., and Lian, C. (2015). Genome-wide identification and characterization of the superoxide dismutase gene family in musa acuminata cv. Tianbaojiao (aaa group). BMC Genomics 16, 823. doi: 10.1186/s12864-015-2046-7
Fink, R. C. and Scandalios, J. G. (2002). Molecular evolution and structure–function relationships of the superoxide dismutase gene families in angiosperms and their relationship to other eukaryotic and prokaryotic superoxide dismutases. Arch. Biochem. Biophys. 399, 19–36. doi: 10.1006/abbi.2001.2739
Franceschini, A., Szklarczyk, D., Frankild, S., Kuhn, M., Simonovic, M., Roth, A., Jensen, L. J., et al. (2013). String v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic. Acids Res. 41, D808–D815. doi: 10.1093/nar/gks1094
Garcia-Hernandez, M., Berardini, T., Chen, G., Crist, D., Doyle, A., Huala, E., et al. (2002). Tair: a resource for integrated arabidopsis data. Funct. Integr. Genomics 2, 239–253. doi: 10.1007/s10142-002-0077-z
Gill, S. S., Anjum, N. A., Gill, R., Yadav, S., Hasanuzzaman, M., Fujita, M., Tuteja, N., et al. (2015). Superoxide dismutase–mentor of abiotic stress tolerance in crop plants. Environ. Sci. pollut. Res. 22, 10375–10394. doi: 10.1007/s11356-015-4532-5
Gill, S. S. and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., Rokhsar, D. S., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic. Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Griffiths-Jones, S., Grocock, R. J., van Dongen, S., Bateman, A., and Enright, A. J. (2006). Mirbase: microrna sequences, targets and gene nomenclature. Nucleic. Acids Res. 34, D140–D144. doi: 10.1093/nar/gkj112
Han, L. M., Hua, W. P., Cao, X. Y., Yan, J. A., Chen, C., and Wang, Z. Z. (2020). Genome-wide identification and expression analysis of the superoxide dismutase (sod) gene family in salvia miltiorrhiza. Gene 742, 144603. doi: 10.1016/j.gene.2020.144603
Huang, H., Ullah, F., Zhou, D. X., Yi, M., and Zhao, Y. (2019). Mechanisms of ros regulation of plant development and stress responses. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00800
Huis, R., Hawkins, S., and Neutelings, G. (2010). Selection of reference genes for quantitative gene expression normalization in flax (linum usitatissimum l.). BMC Plant Biol. 10, 71. doi: 10.1186/1471-2229-10-71
Huseynova, I. M., Aliyeva, D. R., and Aliyev, J. A. (2014). Subcellular localization and responses of superoxide dismutase isoforms in local wheat varieties subjected to continuous soil drought. Plant Physiol. Biochem. 81, 54–60. doi: 10.1016/j.plaphy.2014.01.018
Jiang, W., Yang, L., He, Y., Zhang, H., Li, W., Chen, H., et al. (2019). Genome-wide identification and transcriptional expression analysis of superoxide dismutase (sod) family in wheat (triticum aestivum). Peerj 7, e8062. doi: 10.7717/peerj.8062
Jiao, Y., Li, J., Tang, H., and Paterson, A. H. (2014). Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell. 26, 2792–2802. doi: 10.1105/tpc.114.127597
Kar, R. K. (2011). Plant responses to water stress: role of reactive oxygen species. Plant Signal. Behav. 6, 1741–1745. doi: 10.4161/psb.6.11.17729
Khaja, R., MacDonald, J. R., Zhang, J., and Scherer, S. W. (2006). Methods for identifying and mapping recent segmental and gene duplications in eukaryotic genomes. Methods Mol. Biol. 338, 9–20. doi: 10.1385/1-59745-097-9:9
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., Rombauts, S., et al. (2002). Plantcare, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic. Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, Q., Jin, X., and Zhu, Y. X. (2012). Identification and analyses of mirna genes in allotetraploid gossypium hirsutum fiber cells based on the sequenced diploid g. Raimondii genome. J. Genet. Genomics 39, 351–360. doi: 10.1016/j.jgg.2012.04.008
Liu, T., Ye, X., Li, M., Li, J., Qi, H., and Hu, X. (2020). H2o2 and no are involved in trehalose-regulated oxidative stress tolerance in cold-stressed tomato plants. Environ. Exp. Bot. 171, 103961. doi: 10.1016/j.envexpbot.2019.103961
Livak, K. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative pcr and the 2–δδct method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, K. K., Song, R. F., Guo, J. X., Zhang, Y., Zuo, J. X., Chen, H. H., Liu, W. C., et al. (2023). Cycc1;1-wrky75 complex-mediated transcriptional regulation of sos1 controls salt stress tolerance in arabidopsis. Plant Cell. 35, 2570–2591. doi: 10.1093/plcell/koad105
Lu, J., Xiaoyang, C., Li, J., Wu, H., Wang, Y., Di, P., et al. (2025). Whole-genome identification of the flax fatty acid desaturase gene family and functional analysis of the lufad2.1 gene under cold stress conditions. Plant Cell Environ. 48, 2221–2239. doi: 10.1111/pce.15284
Ma, L., Han, R., Yang, Y., Liu, X., Li, H., Zhao, X., et al. (2023). Phytochromes enhance sos2-mediated pif1 and pif3 phosphorylation and degradation to promote arabidopsis salt tolerance. Plant Cell. 35, 2997–3020. doi: 10.1093/plcell/koad117
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L., et al. (2021). Pfam: the protein families database in 2021. Nucleic. Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Negi, N. P., Shrivastava, D. C., Sharma, V., and Sarin, N. B. (2015). Overexpression of cuznsod from arachis hypogaea alleviates salinity and drought stress in tobacco. Plant Cell Rep. 34, 1109–1126. doi: 10.1007/s00299-015-1770-4
Nobeli, I., Favia, A. D., and Thornton, J. M. (2009). Protein promiscuity and its implications for biotechnology. Nat. Biotechnol. 27, 157–167. doi: 10.1038/nbt1519
Noctor, G., Mhamdi, A., and Foyer, C. H. (2014). The roles of reactive oxygen metabolism in drought: not so cut and dried. Plant Physiol. 164, 1636–1648. doi: 10.1104/pp.113.233478
Osakabe, Y., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. P. (2014). Aba control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 202, 35–49. doi: 10.1111/nph.12613
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). Hmmer web server: 2018 update. Nucleic. Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Punia, H., Tokas, J., Malik, A., Sangwan, S., Rani, A., Yashveer, S., et al. (2021). Genome-wide transcriptome profiling, characterization, and functional identification of nac transcription factors in sorghum under salt stress. Antioxidants 10(10), 1605. doi: 10.3390/antiox10101605
Qin, Q. (2023). Ros: important factor in plant stem cell fate regulation. J. Plant Physiol. 289, 154082. doi: 10.1016/j.jplph.2023.154082
Rafique, M. U., Nahid, N., Azeem, F., Fiaz, S., Attia, K. A., Zameer, R., et al. (2023). Genome wide analysis for the identification and characterization of superoxide-dismutase gene family in rosa chinensis ascertains the role of salinity-responsive rcmsd1 protein and its interaction with peroxyl radical. Plant Stress 10, 100218. doi: 10.1016/j.stress.2023.100218
Ravichandran, S., Ragupathy, R., Edwards, T., Domaratzki, M., and Cloutier, S. (2019). Microrna-guided regulation of heat stress response in wheat. BMC Genomics 20, 488. doi: 10.1186/s12864-019-5799-6
Santos, H. O., Price, J. C., and Bueno, A. A. (2020). Beyond fish oil supplementation: the effects of alternative plant sources of omega-3 polyunsaturated fatty acids upon lipid indexes and cardiometabolic biomarkers-an overview. Nutrients 12(10), 3159. doi: 10.3390/nu12103159
Shumayla Sharma, S., Taneja, M., Tyagi, S., Singh, K., and Upadhyay, S. K. (2017). Survey of high throughput rna-seq data reveals potential roles for lncrnas during development and stress response in bread wheat. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01019
Song, X., Li, Y., Cao, X., and Qi, Y. (2019). Micrornas and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 70, 489–525. doi: 10.1146/annurev-arplant-050718-100334
Song, J., Zeng, L., Chen, R., Wang, Y., and Zhou, Y. (2018). In silico identification and expression analysis of superoxide dismutase (sod) gene family in medicago truncatula. 3 Biotech. 8, 348. doi: 10.1007/s13205-018-1373-1
Su, W., Raza, A., Gao, A., Jia, Z., Zhang, Y., Hussain, M. A., et al. (2021). Genome-wide analysis and expression profile of superoxide dismutase (sod) gene family in rapeseed (brassica napus l.) Under different hormones and abiotic stress conditions. Antioxidants 10(8), 1182. doi: 10.3390/antiox10081182
Sunkar, R., Kapoor, A., and Zhu, J. K. (2006). Posttranscriptional induction of two cu/zn superoxide dismutase genes in arabidopsis is mediated by downregulation of mir398 and important for oxidative stress tolerance. Plant Cell. 18, 2051–2065. doi: 10.1105/tpc.106.041673
Suzuki, N., Koussevitzky, S., Mittler, R., and Miller, G. (2012). Ros and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270. doi: 10.1111/j.1365-3040.2011.02336.x
Tamura, K., Stecher, G., and Kumar, S. (2021). Mega11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Verma, D., Lakhanpal, N., and Singh, K. (2019). Genome-wide identification and characterization of abiotic-stress responsive sod (superoxide dismutase) gene family in brassica juncea and b. Rapa. BMC Genomics 20, 227. doi: 10.1186/s12864-019-5593-5
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). Mcscanx: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic. Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, Q., Wan, J., Dang, K., Meng, S., Hu, D., Lin, Y., et al. (2023). Zma-mir159 targets zmmyb74 and zmmyb138 transcription factors to regulate grain size and weight in maize. Plant Physiol. 193, 2430–2441. doi: 10.1093/plphys/kiad455
Wang, W., Xia, M., Chen, J., Deng, F., Yuan, R., Zhang, X., and Shen, F. (2016). Data set for phylogenetic tree and rampage ramachandran plot analysis of sods in gossypium raimondii and g. Arboreum. Data Brief 9, 345–348. doi: 10.1016/j.dib.2016.05.025
Wang, W., Zhang, X., Deng, F., Yuan, R., and Shen, F. (2017). Genome-wide characterization and expression analyses of superoxide dismutase (sod) genes in gossypium hirsutum. BMC Genomics 18, 376. doi: 10.1186/s12864-017-3768-5
Wang, M., Zhang, Y., Zhu, C., Yao, X., Zheng, Z., Tian, Z., and Cai, X. (2021). Ekfls overexpression promotes flavonoid accumulation and abiotic stress tolerance in plant. Physiol. Plant 172, 1966–1982. doi: 10.1111/ppl.13407
Xu, G., Guo, C., Shan, H., and Kong, H. (2012). Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. U. S. A. 109, 1187–1192. doi: 10.1073/pnas.1109047109
Yadav, B., Kaur, V., Narayan, O. P., Yadav, S. K., Kumar, A., and Wankhede, D. P. (2022). Integrated omics approaches for flax improvement under abiotic and biotic stress: current status and future prospects. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.931275
Yang, L., Fountain, J. C., Wang, H., Ni, X., Ji, P., Lee, R. D., et al. (2015). Stress sensitivity is associated with differential accumulation of reactive oxygen and nitrogen species in maize genotypes with contrasting levels of drought tolerance. Int. J. Mol. Sci. 16, 24791–24819. doi: 10.3390/ijms161024791
Yu, W., Kong, G., Chao, J., Yin, T., Tian, H., Ya, H., et al. (2022). Genome-wide identification of the rubber tree superoxide dismutase (sod) gene family and analysis of its expression under abiotic stress. Peerj 10, e14251. doi: 10.7717/peerj.14251
Yuan, J., Amend, A., Borkowski, J., DeMarco, R., Bailey, W., Liu, Y., et al. (1999). Multiclustal: a systematic method for surveying clustal w alignment parameters. Bioinformatics 15, 862–863. doi: 10.1093/bioinformatics/15.10.862
Zang, Y., Chen, J., Li, R., Shang, S., and Tang, X. (2020). Genome-wide analysis of the superoxide dismutase (sod) gene family in zostera marina and expression profile analysis under temperature stress. Peerj 8, e9063. doi: 10.7717/peerj.9063
Zhang, J., Qi, Y., Wang, L., Wang, L., Yan, X., Dang, Z., et al. (2020). Genomic comparison and population diversity analysis provide insights into the domestication and improvement of flax. Iscience 23, 100967. doi: 10.1016/j.isci.2020.100967
Zhang, X., Zhang, L., Chen, Y., Wang, S., Fang, Y., Zhang, X., et al. (2021). Genome-wide identification of the sod gene family and expression analysis under drought and salt stress in barley. Plant Growth Regul. 94, 49–60. doi: 10.1007/s10725-021-00695-8
Zhang, H., Zheng, J., Su, H., Xia, K., Jian, S., and Zhang, M. (2018). Molecular cloning and functional characterization of the dehydrin (ipdhn) gene from ipomoea pes-caprae. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01454
Zheng, L., Hamidou, A. A., Zhao, X., Ouyang, Z., Lin, H., Li, J., et al. (2023). Superoxide dismutase gene family in cassava revealed their involvement in environmental stress via genome-wide analysis. Iscience 26, 107801. doi: 10.1016/j.isci.2023.107801
Zheng, Z., Wang, N., Jalajakumari, M., Blackman, L., Shen, E., Verma, S., et al. (2020). Mir159 represses a constitutive pathogen defense response in tobacco. Plant Physiol. 182, 2182–2198. doi: 10.1104/pp.19.00786
Keywords: flax, superoxide dismutase, phylogeny analysis, salt stress, reactive oxygen species
Citation: Zhang Y, Wang R, Wang H and Wang H (2025) LuCSD3 enhances salt stress tolerance in flax: genome-wide profiling and functional validation of the SOD gene family. Front. Plant Sci. 16:1609085. doi: 10.3389/fpls.2025.1609085
Received: 10 April 2025; Accepted: 30 May 2025;
Published: 03 July 2025.
Edited by:
Shalini Tiwari, University of the Ozarks, United StatesReviewed by:
Vijay Sheri, Texas Tech University, United StatesWu Shenjie, Shanxi Agricultural University, China
Copyright © 2025 Zhang, Wang, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiyan Wang, d2h5YW5AamxtdS5lZHUuY24=
 Yuan Zhang
Yuan Zhang Ruinan Wang2
Ruinan Wang2 Huiyan Wang
Huiyan Wang