- 1Crop Diseases, Pests and Genetics Research Unit, San Joaquin Valley Agricultural Sciences Center, U.S. Department of Agriculture-Agricultural Research Service, Parlier, CA, United States
- 2Oak Ridge Institute for Science and Engineering, U.S. Department of Energy, Parlier, CA, United States
- 3Department of Plant Science, California State University, Fresno, CA, United States
- 4Chemistry Research Unit, Center for Medical, Agricultural and Veterinary Entomology, U.S. Department of Agriculture-Agricultural Research Service, Gainesville, FL, United States
Introduction: Grapevine fungal trunk diseases are cosmopolitan and act to reduce vineyard yields over time. Additionally, Pierce’s disease, caused by Xylella fastidiosa, is a fatal disease of grapevines and a major threat wherever it is endemic. These grapevine diseases are generally managed via cultural practices and chemical applications. However, management can be costly due to labor costs or are becoming less effective due to pathogen resistance to pesticides. Thus, there is increasing interest in biological control agents to manage grapevine diseases. Therefore, novel isolates of Trichoderma species were collected from grapevine tissues in California with the intention that these would be likely to survive and thrive in the semi-arid and very hot climate present throughout much of the state.
Methods: Genetic analyses and morphology were utilized to identify Californian vineyard-acquired isolates to species or species complex, which yielded several different species: two isolates of Trichoderma harzianum, two isolates of Trichoderma capillare, and two putative novel Trichoderma species. These were examined for activity against fungal trunk pathogens Diplodia seriata, Eutypa lata, and Neofusicoccum parvum via co-plating and spent media assays. Follow-up greenhouse studies also assessed the ability of isolates to limit fungal pathogen canker development and Xylella fastidiosa success over six months. Lastly, field studies tested the ability to limit or remove fungal trunk pathogen colonization of pruned spurs by the Trichoderma isolates from this study and two isolates from another study, which were an isolate of Trichoderma asperellum and a member of the Trichoderma saturnisporopsis species complex.
Results and Discussion: Results potentially yielded Trichoderma isolates with some ability to limit fungal pathogens in culture, greenhouse plants, and pruned spurs in the field, and with the ability to be re-isolated after a full field season. However, these isolates were not able to consistently limit Xylella fastidiosa titers or Pierce’s disease symptoms. Taken together, these experiments demonstrated the ability of California Trichoderma isolates to be deployed as locally sourced biological control agents to protect Californian vineyards as well as those in similar climates.
1 Introduction
Grapevines encounter a variety of serious trunk diseases, including those that are rarely fatal but result in decreased yields over time, such as those collectively known as fungal trunk diseases, and those that cause fatal blocking of xylem vessels, namely, Pierce’s disease caused by the bacterium Xylella fastidiosa (Sipiora and Cuellar, 2014; Kaplan et al., 2016; Rapicavoli et al., 2018; Baumgartner et al., 2019). The former set of diseases includes Botryosphaeria diebacks (caused by pathogens such as Diplodia seriata and Neofusicoccum parvum), Eutypa dieback (caused by Eutypa lata), and esca disease (caused by fungi such as Phaeomoniella chlamydospora) (Mugnai et al., 1999; Urbez-Torres, 2011; Trotel-Aziz et al., 2019; Martinez-Diaz et al., 2021; Ye et al., 2021). These fungal diseases are spread via conidia during wet weather in the springtime, often due to open pruning wounds within vineyards, as well as through other vineyard cultural practices. Initial introductions generally occur via infected nursery stock. Incidence can quickly build toward 100% in vineyards (approximately by year 7 after planting), and severity can gradually reduce vine productivity until the potential need for replanting the entire vineyard to maintain profitability becomes required (Baumgartner et al., 2019). Currently, there is no cure for fungal trunk diseases, with management generally focused on attempts to reduce spread via delayed or double pruning, better sanitation, occasional applications of pruning wound protectants, and synthetic fungicides (Spagnolo et al., 2017; Travadon et al., 2022). These methods are becoming increasingly costly, as pruning or sanitation practices incur large labor costs, and synthetic pesticides are observed to be becoming less effective due to potential pathogen resistance and are being phased out in many jurisdictions (Urbez-Torres and Gubler, 2009; Brown et al., 2020; Travadon et al., 2022).
As for Pierce’s disease, it is caused by the bacterium X. fastidiosa ssp. fastidiosa and is spread by the vectoring sharpshooter insects, such as the glassy-winged sharpshooter (Homalodisca vitripennis) (Hopkins and Purcell, 2002; Rapicavoli et al., 2018). Pierce’s disease is often fatal, as infections by X. fastidiosa affect water transportation in vines, leading to desiccation and death (Sicard et al., 2018). Current management consists of vector control generally by synthetic insecticides, but these are becoming limited in use due to an increasing regulatory environment and building insect resistance (Andreason et al., 2018).
Ultimately, long-term, sustainable management of these diseases would involve improved grapevine materials with natural resistance. Indeed, new cultivars have been released to express a tolerant phenotype to X. fastidiosa infection and, as a result, do not develop Pierce’s disease, albeit these do not cover all grape marketing classes (Krivanek et al., 2006).
Until the development of improved cultivars, another possible management option could be the use of biological control agents. Perhaps the most studied and utilized biological control agents are those from the fungal genus Trichoderma, which are widely used in agriculture (Woo et al., 2014; Verma et al., 2017). These work generally by direct predation or active antagonization of other fungi, as well as the indirect inhibition of other microbial growth (Harman et al., 2004). The latter involves the production of secreted antibiotic compounds that may move systemically throughout colonized plants as well as via induction of plant defenses (Guzman-Guzman et al., 2019).
For grapevines, several studies have tested Trichoderma, generally isolates of Trichoderma atroviride or Trichoderma harzianum, or isolates later often reclassified as Trichoderma afroharzianum (Chaverri et al., 2015), for control of different fungal trunk diseases, such as black foot, esca, Eutypa dieback, and nursery diseases (Fourie et al., 2001; Di Marco et al., 2004; John et al., 2005; Fourie and Halleen, 2006; Berlanas et al., 2018; Marraschi et al., 2019; Berbegal et al., 2020; Blundell and Eskalen, 2022). However, it should be noted that previous studies have been limited in the number of isolates tested and limited in the climates where said studies were performed. This likely results in a challenge in determining the suitability of various Trichoderma species to serve as effective controls in a particular vineyard. As such, efforts have been undertaken to isolate a greater variety of Trichoderma isolates specifically for tests in vineyards, from different climates, such as in Canada (Pollard-Flamand et al., 2022) and Italy (Urbez-Torres et al., 2020). This could provide better biological control agents, as the use of Trichoderma isolates from the same climate and host should presumably be better suited to limit pathogens (Pollard-Flamand et al., 2022; Chan et al., 2023) and could even discover novel species endemic to the region that are usable for effective biological control (Pollard-Flamand et al., 2022). Furthermore, the use of isolates within the same region collected could greatly simplify the registration process, as exploiting an organism as a biological control agent in its original region should pose no additional threat to non-target organisms, and therefore, such testing will not be necessary.
Therefore, this study was initiated to obtain new Trichoderma isolates for use as biological control agents in vineyards in the semi-arid climate of California. In addition to examining the ability of novel isolates to limit fungal trunk pathogens, their capacity to reduce the development of Pierce’s disease was also examined. Gained knowledge should be useful in determining whether the region-specific isolation of Trichoderma could be viable to generate new biological control agents to manage various grapevine diseases.
2 Materials and methods
2.1 Fungal isolations and culturing conditions
In the summer of 2020, leaf, stem, and root tissues from grapevines growing throughout central California were collected. Locations sampled included vineyards within and near Bakersfield, Delano, King City, Parlier, Reedley, San Luis Obispo, and Tooleville, CA, USA. This involved surface-sterilizing 1–2-cm segments of stems or roots, or small discs (1-cm diameter) of leaves, in 10% bleach for 1 minute, followed by two washes in sterile deionized water. The tissues were then placed onto sterile potato dextrose agar (PDA; Difco, Thermo-Fisher, Waltham, MA, USA). Emergent fungi were hyphal tip-cultured to obtain pure cultures. Cultures that had green sporulation were then separated, as these would most likely be Trichoderma species. Morphological assessments further confirmed the cultures that belonged to the Trichoderma genus following a dichotomous key (Samuels and Hebbar, 2016). This resulted in 30 potential Trichoderma isolates across all vineyards and tissues. Isolates similar in morphology and from the same sites and tissues were considered to be potentially the same isolate, and therefore, only one of these was tested further, with the others kept in glycerol freezer stocks. Trichoderma isolates, as well as pathogen isolates of D. seriata, E. lata, N. parvum, or P. chlamydospora, were maintained on PDA plates grown in the dark at 24 °C. Descriptions of these isolates are provided in Table 1.
2.2 Preliminary culture antagonism assays and further Trichoderma identifications
Initial culture antagonism assays were conducted to screen and reduce the total number of Trichoderma isolates to a more manageable number for further assays. These consisted of plating a Trichoderma isolate approximately 2cm from the edge of a 100-mm PDA plate and then placing D. seriata, E. lata, or N. parvum approximately 2cm from the opposite end of the plate. Negative controls involved plating the pathogens by themselves. Four replicate plates of each Trichoderma isolate and pathogen combination were used. Radiuses in two directions of the pathogen colonies were measured at 3 days of growth. Percent inhibition was calculated as the difference between the mean radius of a pathogen colony co-plated with a Trichoderma isolate and the mean radius of a pathogen colony grown by itself, divided by the radiuses of the colonies on the co-plated experimental plates, and multiplied by 100%.
In addition to co-plating assays, the ability of compounds produced by six different Trichoderma isolates was assessed to reduce the colony growth of D. seriata, E. lata, or N. parvum on plates amended with spent media. The co-plating assays observed that the presence of a fungal pathogen increased the pigmented metabolite production of Trichoderma in the growth media. Thus, to acquire Trichoderma compounds, N. parvum was first grown in 500 mL of potato dextrose broth (PDB; Difco) in 1,000-mL Erlenmeyer flasks on a shaker (100 rpm) at 26 °C for 1 week. These colonies were then autoclaved to kill the N. parvum pathogen colonies, after which one of the six isolates of Trichoderma was added and grown for 1 week under similar conditions, with other media left sterile as a control. Following the week of Trichoderma isolate growth, the PDB cultures were filtered through 1-μm filters (Millipore Sigma, St. Louis, MO, USA) to obtain sterile spent media. Potato dextrose agar with half of the usual water added was autoclaved, and afterward, the sterile spent media were added to make up for the missing water prior to pouring the culture plates (resulting in a medium comprised of 50% of the original concentration of the spent broth). Colonies of D. seriata, E. lata, or N. parvum were then started in the middle of these plates, with growth measured as colony radiuses after 3 days of growth. Each treatment had 10 plates used. Percent inhibition was calculated similarly to the co-plating assays.
The six isolates with the most inhibitory capacity (i.e., DL1-3, KC1-1, KC2-2, PAR3, PAR10, and SLO1-1) were chosen for their ability to reduce infections in greenhouse and field assays. They were also sequenced to identify the isolates to species. DNA was extracted using the Quick-DNA Plant Kit (Zymo Research, Tustin, CA, USA), with DNA quantified by Invitrogen (Waltham, MA, USA) Qubit and quality-assessed using an Agilent (Santa Clara, CA, USA) TapeStation following the manufacturer’s protocols. The high-quality DNA was then sent for short-read sequencing using an Illumina (Torrance, CA, USA) NovaSeq 6000 via a commercial provider, Novogene (Durham, NC, USA), for DL1-3, KC1-1, PAR3, PAR10, and SLO1-1. Sequence reads were assembled using SPAdes ver. 3.14.0 (Bankevich et al., 2012), with quality of the assembly assessed using QUAST (Gurevich et al., 2013). For KC2-2, DNA was sequenced using the Oxford Nanopore (San Francisco, CA, USA) PromethION instrument (with R10.4.1 flow cells and v14 chemistry), with Flye ver. 2.9.1 (Kolmogorov et al., 2019) used for selecting high-quality reads, Medaka ver. 1.8.0 (Oxford Nanopore) for polishing, and Busco ver. 5.7.1 (Simao et al., 2015) for quality assessment. For all sequences, stand-alone BLAST+ (Camacho et al., 2009) was then used to identify the internal transcribed spacer (ITS) region, the RNA polymerase II (rpb2), and the translation elongation factor 1-α (tef1) genes. The iterative process described by Cai and Druzhinina (2021) was then used to identify Trichoderma to species. This involved first determining if the percent identity of the ITS region was greater than 76% between unknown isolates and at least one known Trichoderma species (Cai and Druzhinina, 2021). The ITS sequences were aligned by inserting those from these isolates into the Nexus file provided as Supplementary Data Sheet in Cai and Druzhinina (2021). Next, curated collections of closely related Trichoderma sp. for both the rpb2 and tef1 genes were obtained. These were used to determine if the percent identities would be over 99% for rpb2 and over 97% for tef1 when comparing the sequence from an unidentified isolate with that of known Trichoderma sp., in which case a match that met the criteria would identify the isolate’s species (Cai and Druzhinina, 2021). If not, phylogenetic trees would be examined to determine if an isolate is a putative novel species (Cai and Druzhinina, 2021). Alignment of these sequences is available in Supplementary Data Sheet 1 and 2. To further confirm species, colony morphology (with pictures available in Supplementary Data Sheet 3) and microscopy were performed to verify that the characteristics of colonies were consistent with the sequencing identifications (Samuels and Hebbar, 2016).
2.3 Greenhouse assays to assess the capacity of Trichoderma isolates to reduce pathogen infections
Grapevines used in all greenhouse experiments were Cabernet Sauvignon grafted onto 101–14 MG rootstocks and were kept in climate-controlled conditions (temperatures averaging 20 °C to 30 °C, humidity approximately 25%, and supplemental lighting set to maintain 14 hours of daylight).
Greenhouse experiments were replicated in full twice, once in 2023 and once in 2024. In 2023, only one isolate of each species was examined (i.e., DL1-3, KC1-1, PAR10, and SLO1-1). In 2024, all isolates from this study were used (i.e., DL1-3, KC1-1, KC2-2, PAR3, PAR10, and SLO1-1) as well as two additional promising isolates from California identified in another study: RSI (previously identified as Trichoderma saturnisporopsis) and TLI (previously identified as Trichoderma asperellum) (Antrim et al., 2025).
All treatments were placed in a completely randomized block experimental design, with two spatial blocks used per experiment. Treatments consisted of a first inoculation treatment of either a mock inoculation (on five plants in 2023 or 10 plants in 2024) or inoculation of one of the Trichoderma isolates (on five plants each during both years). Inoculations consisted of creating a 10-mm wound in the bark of the grapevine with a cork borer in the middle of the first internode of the scion after the scion-rootstock graft site. To this wound, a colonized 8-mm PDA plug (or uncolonized sterile plug for mock-inoculated controls) was applied, mycelium-side down, and wrapped in parafilm to allow colonization to occur.
Two weeks after the Trichoderma inoculations, pathogen inoculations occurred. This involved a mock inoculation, inoculations by the fungal pathogens D. seriata or N. parvum (performed similarly with colonized agar plugs that were performed with the Trichoderma inoculations), or inoculation by the bacteria X. fastidiosa ssp. fastidiosa strain SL (Hopkins, 2001). The fungal inoculations or mock controls were applied to the first internode after the scion-rootstock graft on a different branch than the Trichoderma sites or, when only one branch was available, applied to two internodes above the Trichoderma inoculation site.
Prior to inoculation, X. fastidiosa was maintained on periwinkle wilt media for 2 weeks in darkness at 24 °C. X. fastidiosa inoculations were performed via the pin-prick method. In brief, bacteria were harvested from culture plates, and a suspension of approximately 1 × 105 CFUs/mL was made in sterile water. An 18-gauge needle was used to make five wound sites (on locations similar to where the fungal pathogens were applied as described above), and approximately 10 µL of bacterial suspension was applied to those wounds, allowing the bacteria to enter the branch via capillary action.
Six months after the pathogen treatments, disease symptoms were assessed by photographing all grapevines. Disease severity was rated on a 0–5 scale, similar to a commonly used scale for Pierce’s diseases (Wallis and Chen, 2012), with “0” meaning no symptoms or leaf damage, “4” meaning severe damage, and “5” meaning plant death. In addition to foliar damage and discoloration, this scale also incorporated relative decreases in new internode lengths and leaf size, as these could be symptoms of Pierce’s disease or fungal canker disease.
Following symptom assessment, all plants were destructively harvested to quantify infections. At the fungal inoculation sites, development of cankers was assessed using a ruler to measure both external cankers, and developing internal discoloration was reviewed after de-barking the infected internode segment. Approximately 1-cm segments in the apical and basal sides of the developing canker were collected, surface-sterilized in 10% bleach for 1 minute, washed in sterile water twice, and then plated on sterile PDA to confirm Trichoderma or pathogen fungal recovery. Confirmations were made via morphology observations. Isolate-treated vines that were negative for Trichoderma were removed from further analyses, as well as when non-Trichoderma controls were positive for Trichoderma. Likewise, mock-inoculated vines that failed to test positive for the subsequent pathogen infections were also removed from analyses.
For X. fastidiosa-inoculated grapevines, stem segments from the initial inoculation site were pulverized in liquid nitrogen, and DNA was extracted as described above. Real-time quantitative PCR was then performed using the primers and methods as described in Chen et al. (2005) using a Bio-Rad (Hercules, CA, USA) qPCR thermocycler and associated software.
2.4 Field efficacy of Trichoderma to control fungal pathogens in grapevine spurs
The six Trichoderma isolates selected from this study (DL1-3, KC1-1, KC2-2, SLO1-1, PAR3, and PAR10), as well as the two from a previous study (RSI and TLI), were assessed for their ability to prevent or remove naturally occurring fungal canker pathogens in an over 10-year-old Cabernet Sauvignon vineyard. The experiment was performed in both 2023 and 2024.
A total of 24 vines in 2023 and 12 vines in 2024 received one of the following treatments on six (for 2023) or three (for 2024) freshly pruned spurs per vine in May, arranged in a completely randomized block design with three spatial blocks: inoculation with one of the eight Trichoderma isolates (at a concentration of 100,000 spores/mL), mock inoculation with sterile water, or treatment with a fungicide (Topsin M, or thiophanate-methyl, at 1.8 g/L). Spore suspensions, water, or fungicide was applied using a micropipette with enough liquid to completely cover a cut spur, approximately 0.5 to 1 mL of solution per spur.
Six months after treatment, spurs were harvested. For each spur, two horizontal segments (approximately 10mm thick) were collected. The apical segment was surface-sterilized in 10% bleach for 1 minute, washed twice in sterile water, and plated on PDA. Plates were observed at 5 days, with the emergent fungi noted and identified via morphology. Pictures were taken to assist in data collection.
2.5 Statistical analyses
JMP version 17 (SAS Institute, Cary, NC, USA) was used for all statistical analyses, with α = 0.05. For analyses that required normal distributions, normality of the data was assessed using a quantile–quantile (QQ) plot and the Shapiro–Wilk test. Differences between growth inhibition, external canker lengths, internal discoloration, and pathogen recoveries or Trichoderma recoveries between treatments were determined using analysis of variance (ANOVA) tests with follow-up least significant difference (LSD) mean separation tests. Differences in Pierce’s disease symptoms and X. fastidiosa titers were determined by non-parametric Kruskal–Wallis tests with follow-up Wilcoxon pairwise comparisons, as the data were non-continuous or did not meet normality assumptions.
3 Results
3.1 Trichoderma isolate species identification
Every isolate that was sequenced had a percent similarity index greater than 76% with multiple existing Trichoderma species, and therefore, all were concluded to be members of the Trichoderma genus.
DL1–3 had the greatest similarity index value of rpb2 of 97.49% with Trichoderma guizhouense and a similarity index of tef1 intron 4 of 95.95% with Trichoderma rifaii. The findings were precise, accurate, and unambiguous that a putative new species had non-concordant phylogenies of rpb2 and tef1. DL1–3 was therefore considered a putative novel species, Trichoderma sp. DL1-3. Because of the potential as a biological control agent, we propose a provisional name of Trichoderma kernensis, after Kern County, California, USA, where it is from, although a great morphological description is needed.
KC1–1 and PAR3 were 100% similar for the rpb2 and tef1 sequences. Both had the greatest similarity of rpb2 of 99.30% to T. harzianum and similarity of tef1 intron 4 of 100% with T. harzianum. The identification was precise, accurate, and unambiguous that KC1–1 and PAR3 were isolates of T. harzianum.
KC2–2 and SLO1–1 had identical rpb2 and tef1 sequences. They had the greatest similarity of rpb2 of 99.58% with Trichoderma capillare and similarity of tef1 intron 4 of 99.28% with T. capillare. Therefore, the results were precise, accurate, and unambiguous that KC2–2 and SLO1–1 were identified as T. capillare.
PAR10 had the greatest similarity of rpb2 of 96.94% with three species (T. guizhouense, Trichoderma pyramidale, and Trichoderma simmonsii) and similarity of tef1 intron 4 of 90.44% with T. pyramidale. These results were precise, accurate, and unambiguous and suggested that PAR10 is a new species, related to Trichoderma pyramidale, and described as Trichoderma sp. PAR10. Due to the potential importance in biological control, we propose the name of Trichoderma parlierensis, after Parlier, CA, USA, the town from which it was isolated, although a greater morphological description is needed.
3.2 Ability of Trichoderma isolates to inhibit pathogen growth on culture plates
All selected isolates successfully reduced pathogen growth (or had a greater mean inhibition) when co-plated with pathogens D. seriata (F6, 26 = 52.619; p < 0.001) and N. parvum (F6, 26 = 36.829; p < 0.001), but not significantly so for E. lata (F6, 26 = 1.852; p = 0.137) (Figure 1). However, follow-up LSD tests did reveal that when E. lata colonies were co-plated with DL1-3, PAR3, and PAR10, the sizes were significantly smaller according to LSD tests than when E. lata was not co-plated. For D. seriata, colonies co-plated with DL1–3 were also significantly smaller than colonies co-plated with either PAR10 or SLO1-1. For N. parvum, colonies co-plated with either KC1–1 or SLO1–1 were significantly smaller than colonies co-plated with PAR10.
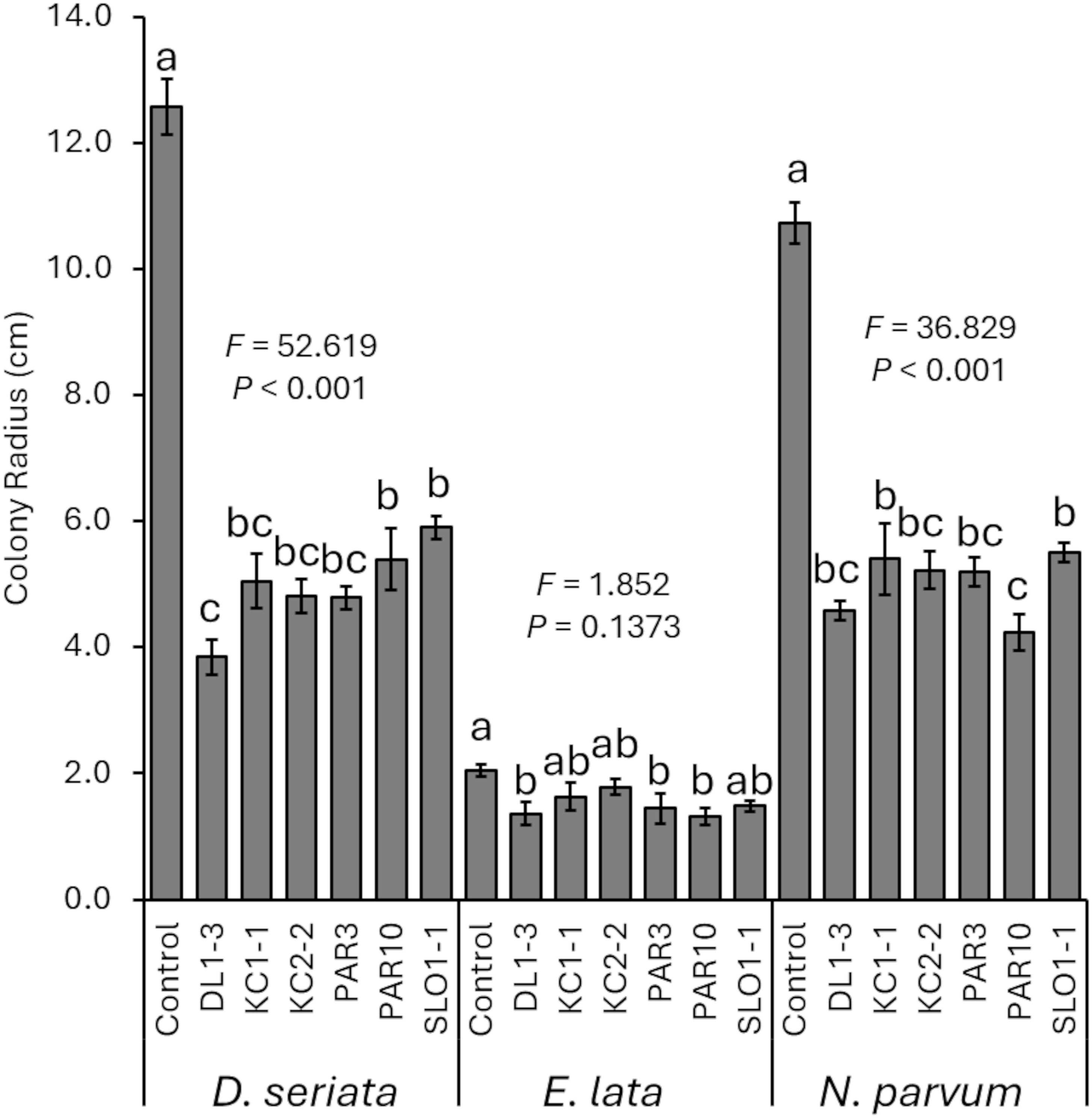
Figure 1. Mean radius of pathogen colonies when co-plated with one of the six Trichoderma isolates. Error bars represent standard errors. ANOVA statistics are provided for each fungal pathogen, and different letters represent significant differences by LSD tests. LSD, least significant difference.
When grown on PDA plates amended with media that previously had the Trichoderma isolates grown (“spent media”), D. seriata (F6, 28 = 10.479; p < 0.001), E. lata (F6, 28 = 4.689; p = 0.002), and N. parvum (F6, 25 = 9.979; p < 0.001) had reduced colony growth (greater inhibition of growth) compared with controls (Figure 2). For D. seriata, LSD tests revealed significant differences in colonies grown on control agar compared with those grown on spent media from all isolates except KC1–1 and KC2-2. In addition, colonies grown on spent media from DL1–3 or PAR3 were smaller than those grown on spent media with the other isolates. For E. lata, LSD tests revealed that colonies grown on control media were significantly larger than those grown on spent media from KC2–2 or SLO1-1. The use of spent media from SLO1–1 also reduced E. lata colonies more than spent media from all others except DL1–3 and KC2-2, and spent media from KC2–2 reduced growth more than spent media from all other isolates except SLO1-1. Lastly, N. parvum colonies grown on control plates were significantly larger than those on colonies grown on spent media plates. N. parvum colonies grown on KC1–1 spent media were also significantly larger than those grown on DL1-3, KC2-2, or PAR3 spent media.
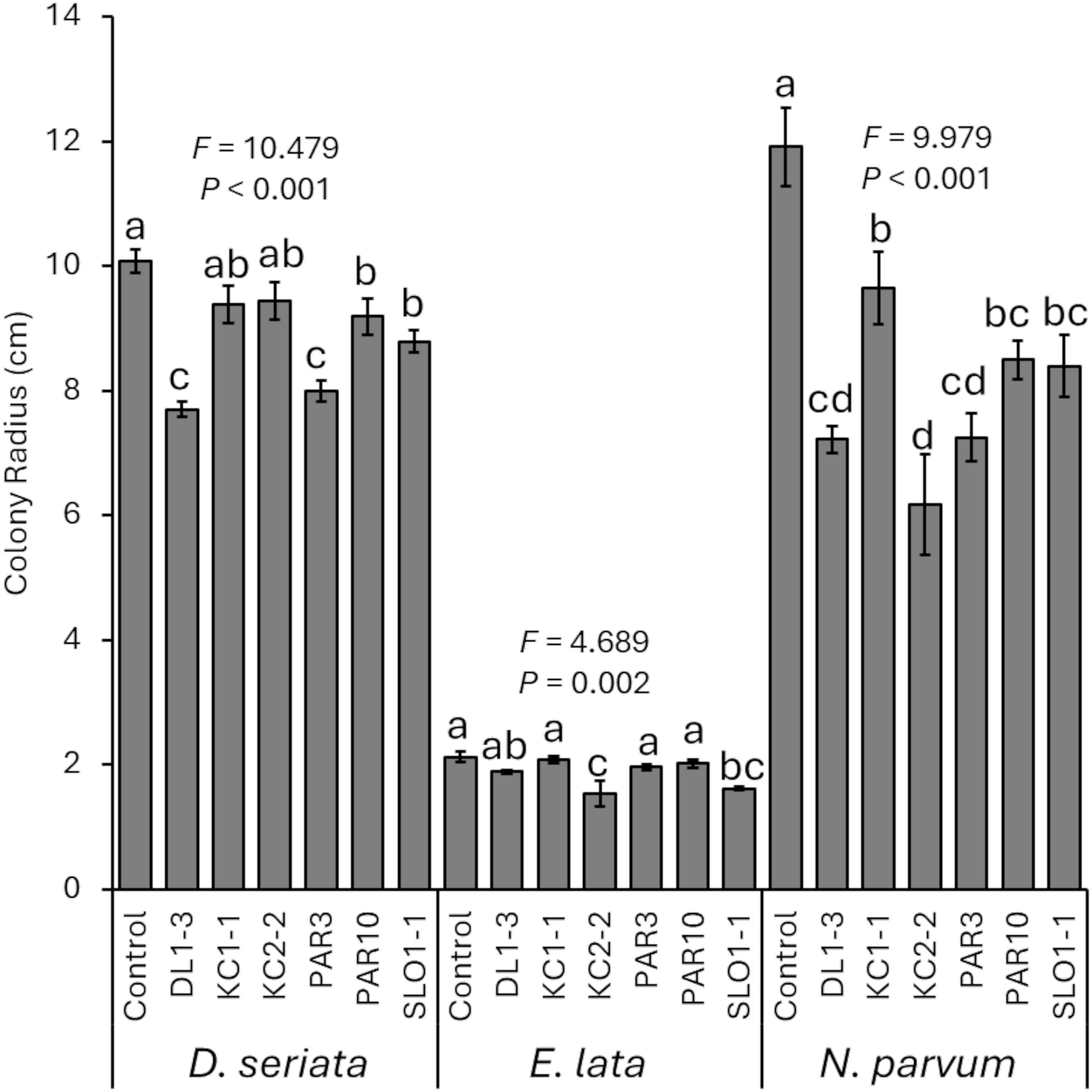
Figure 2. Mean percent reduction of pathogen colonies when grown on spent media from one of the six Trichoderma isolates. Error bars represent standard errors. ANOVA statistics are provided for each fungal pathogen, and different letters represent significant differences by LSD tests. LSD, least significant difference.
3.3 Capacity of Trichoderma stem inoculations to reduce disease
For 2023, the ability of one isolate each of T. harzianum (KC1-1), T. capillare (SLO1-1), and the two putative new species (Trichoderma sp. DL1–3 and Trichoderma sp. PAR10) was assessed to reduce fungal canker pathogen diseases or Pierce’s diseases. For 2024, the additional isolates of T. harzianum (PAR3) and T. capillare (KC2-2), as well as the two isolates from Antrim et al. (2025) (T. asperellum isolate TLI and T. saturnisporopsis isolate RSI), were added to the 2023 tested isolates in the experimental trial.
In 2023, prior inoculation of any of the four isolates (DL1-3, KC1-1, PAR10, or SLO1-1) was able to generally significantly reduce subsequent D. seriata cankers compared to controls, whether measured as external cankers (F4, 24 = 7.977; p = 0.001) (Figure 3A) or interior discoloration (F4, 24 = 8.720; p < 0.001) (Figure 3B). In addition, vines treated initially with DL1–3 had smaller subsequent D. seriata cankers developed than those treated initially with PAR10.
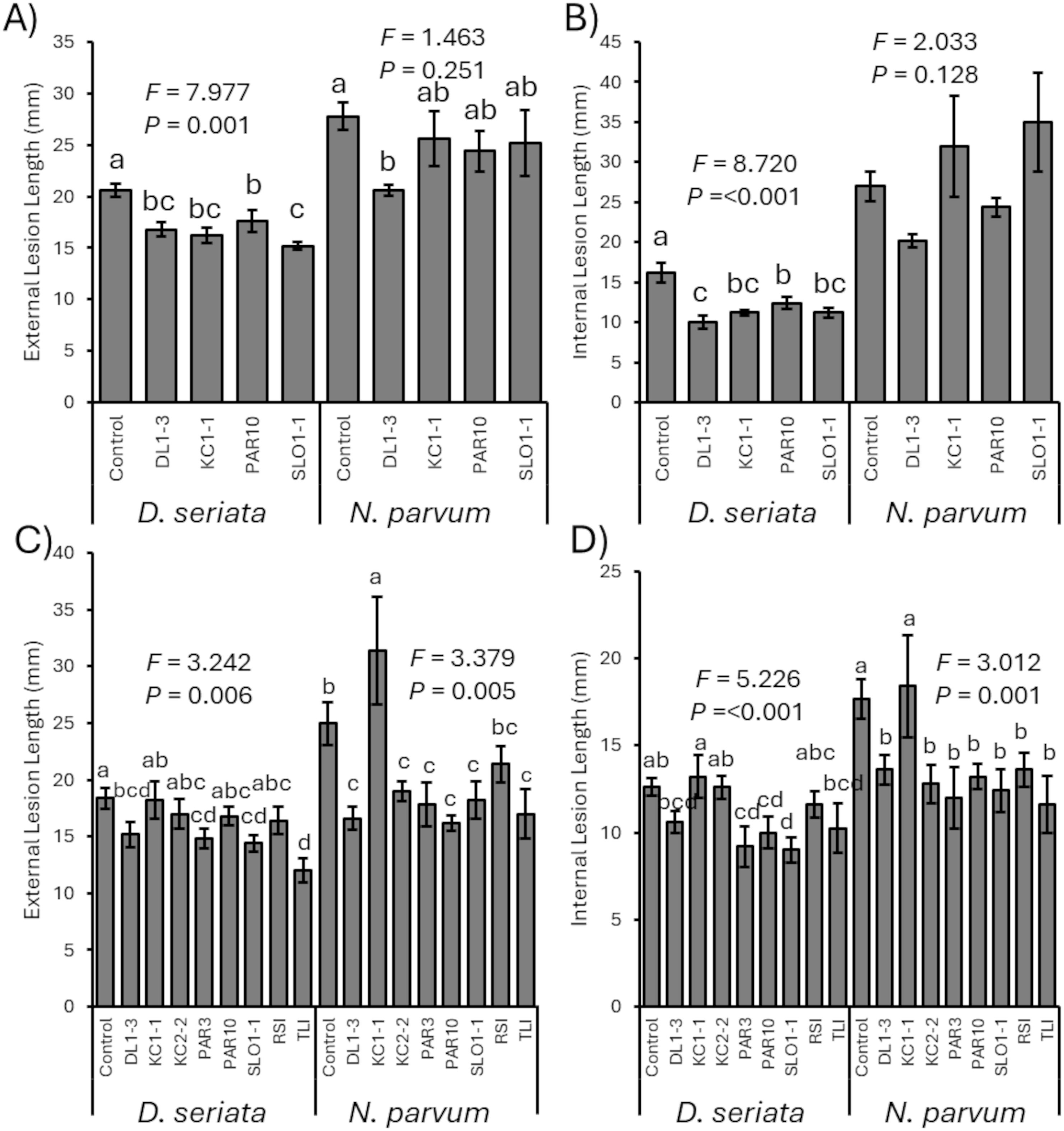
Figure 3. (A) Mean developing external canker length of Diplodia seriata or Neofusicoccum parvum infections in 2023 trials. (B) Mean internal discoloration caused by D. seriata or N. parvum infections in 2023 trials. (C) Mean developing external canker length of D. seriata or N. parvum infections in 2024 trials. (D) Mean internal discoloration caused by D. seriata or N. parvum infections in 2024 trials. Error bars represent standard errors. ANOVA statistics are provided for each fungal pathogen, and different letters represent significant differences by LSD tests. LSD, least significant difference.
Also in 2023, vines inoculated initially with any of the Trichoderma isolates did not have significant differences in canker development of subsequent N. parvum inoculations, measured as either external cankers (F4, 24 = 1.463; p = 0.251) (Figure 3A) or internal discoloration (F4, 24 = 2.033; p = 0.128) (Figure 3B). However, LSD tests did suggest that prior inoculation with isolate DL1–3 had reduced N. parvum external cankers compared with control vines that did not have Trichoderma inoculation.
For the 2024 experimental trials, vines that were not inoculated with a Trichoderma isolate had greater D. seriata external lesions than those inoculated with isolates DL1-3, PAR3, SLO1-1, and TLI (F8, 40 = 3.242; p = 0.006) (Figure 3C), and control vines had greater internal discoloration than vines inoculated with PAR3, PAR10, and SLO1-1 (F8, 40 = 5.226; p < 0.001) (Figure 3D). Vines inoculated with KC1–1 had more D. seriata canker growth and internal discoloration than vines inoculated with other isolates. For N. parvum, both mock-inoculated and KC1-1-inoculated vines had greater lesion lengths (F8, 40 = 3.379; p = 0.005) (Figure 3C) and internal discoloration (F8, 40 = 3.012; p = 0.001) (Figure 3D) than those inoculated with any of the Trichoderma isolates except KC1-1 (albeit mock-inoculated and RSI-treated vines did not have different external lesions).
In both 2023 and 2024, prior treatment with the Trichoderma isolates did not alter the development of Pierce’s disease symptoms that followed X. fastidiosa inoculation when compared to controls (for 2023, χ2 = 2.626, p = 0.6222, N=25; for 2024, χ2 = 5.855, p = 0.663, N=50). Nevertheless, in 2023, measurements of X. fastidiosa titers by qPCR suggested significant differences in Ct values (with lower Ct values representing greater bacterial titers) between vines previously inoculated with isolate DL1–3 and those not previously inoculated or inoculated with isolate SLO1–1 or PAR10 (χ2 = 11.046, p = 0.026, N=24) (Figure 4A). However, no significant differences were observed (χ2 = 9.098, p = 0.334, N=50) in X. fastidiosa titers in the 2024 experimental trial (Figure 4B).
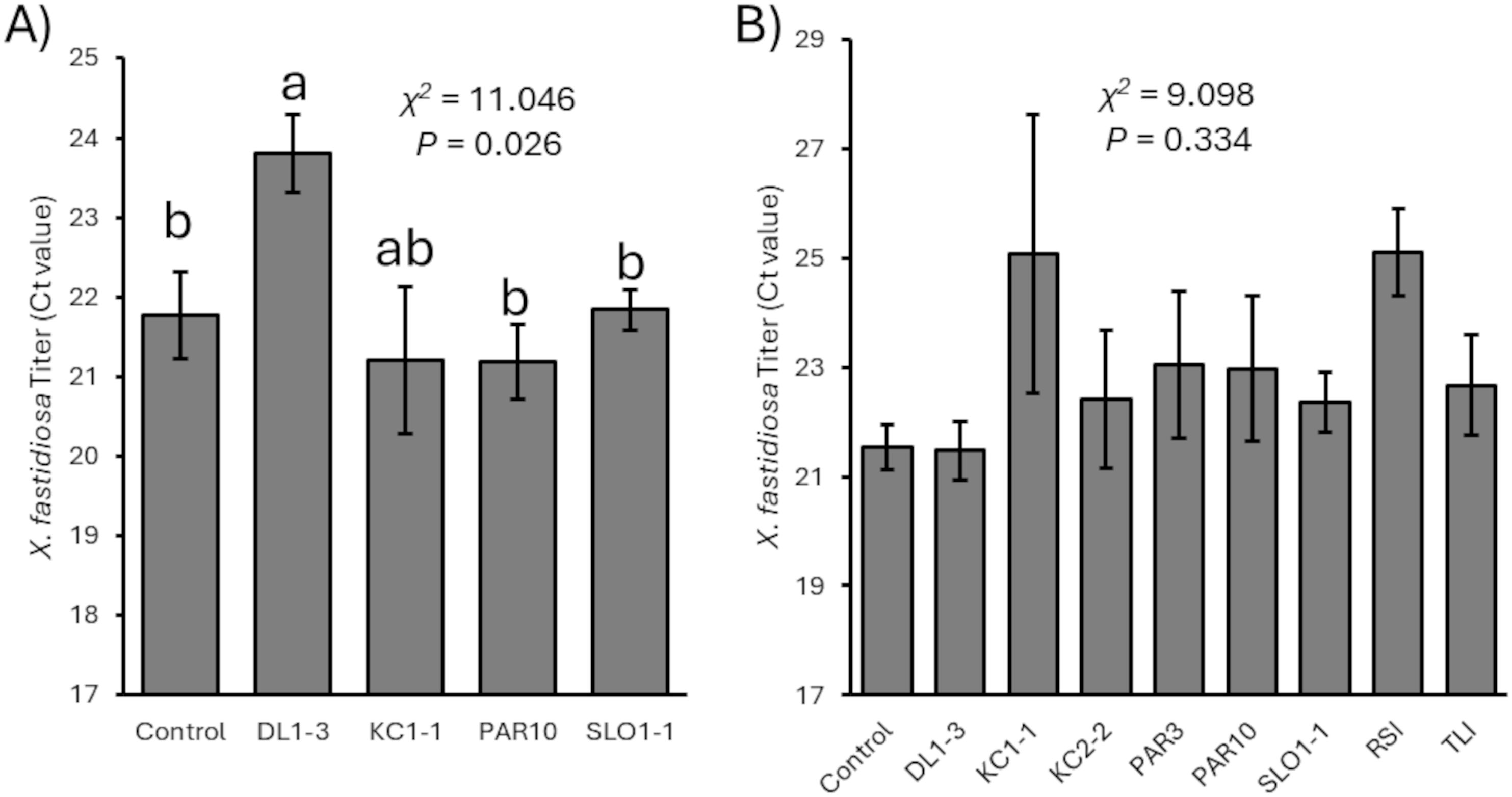
Figure 4. Mean Ct values related to Xylella fastidiosa titers (with greater Ct values representing lower titers) for (A) 2023 and (B) 2024 vines that were mock-inoculated or inoculated with a Trichoderma isolate. Error bars represent standard errors. Mann–Whitney U test statistics are provided, and different letters represent significant differences by Wilcoxon non-parametric pairwise tests. There were no significant differences in 2024 vines.
3.4 Capacity of Trichoderma isolates to prevent colonization of cut spurs
To assess the ability of the six Trichoderma isolates from this study and the two other promising isolates (Antrim et al., 2025), freshly cut spurs were inoculated, treated with fungicide, or mock-inoculated with water in May 2023 or 2024. Over 6 months later, the spurs were collected and assessed for the recovery of fungal pathogens or Trichoderma isolates. Note that negative controls may have already been infected with pre-existing fungal pathogens or Trichoderma.
In 2023, six of the eight Trichoderma isolates and the fungicide significantly reduced the recovery of the pathogen compared to the mock-inoculated control (F9, 1113 = 7.181; p < 0.001) (Figure 5A). Only isolates RSI and SLO1–1 did not result in reduced pathogen recoveries from the spurs compared with the water-only negative controls. Furthermore, all Trichoderma isolates were recovered in greater numbers in inoculated spurs rather than fungicide-treated spurs or controls over 6 months after application (F9, 1113 = 20.607; p < 0.001) (Figure 5B).
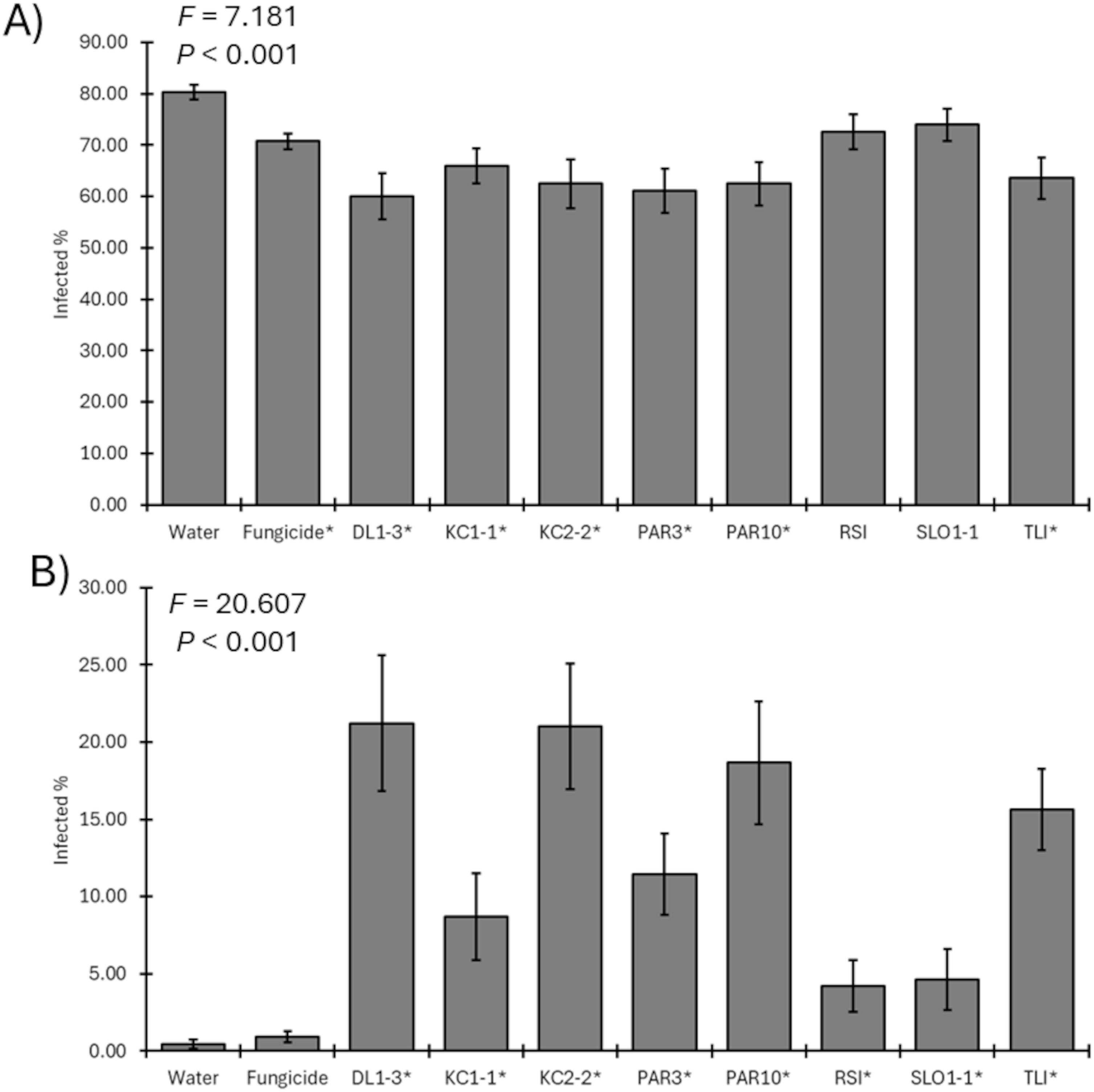
Figure 5. (A) Mean percent recovery of fungal pathogens in plants treated by mock inoculation, fungicide, or inoculation by a Trichoderma isolate for 2023 trial. ANOVA statistics are provided, and an asterisk represents isolates where recovery was lower than that of water-only negative controls. (B) Mean recovery percent of a Trichoderma isolate in plants treated by mock inoculation, fungicide, or isolate inoculation for 2023 trial. ANOVA statistics are provided, and an asterisk represents isolates where recovery was greater than that of water-only negative controls. Error bars represent standard errors.
For the 2024 field trial, five of the eight Trichoderma isolates significantly reduced the recovery of the pathogen compared to the mock-inoculated control (F9, 356 = 4.616; p < 0.001) (Figure 6A). Only isolates PAR3, RSI, and TLI did not result in reduced pathogen recoveries from the spurs compared with the water-only negative controls. Unlike 2023, the percent recovery of Trichoderma from treated spurs was only significantly greater for DL1-3, KC2-2, and PAR10 than Trichoderma recoveries from water-only negative controls (F9, 356 = 2.139; p = 0.026) (Figure 6B).

Figure 6. (A) Mean percent recovery of fungal pathogens in plants treated by mock inoculation, fungicide, or inoculation by a Trichoderma isolate for 2024 trial. ANOVA statistics are provided, and an asterisk represents isolates where recovery was lower than that of water-only negative controls. (B) Mean recovery percent of a Trichoderma isolate in plants treated by mock inoculation, fungicide, or isolate inoculation for 2024 trial. ANOVA statistics are provided, and an asterisk represents isolates where recovery was greater than that of water-only negative controls. Error bars represent standard errors.
4 Discussion
These results observed the promise of a couple to several novel Trichoderma sp. isolates to outcompete or predate fungal pathogens to provide health benefits to grapevine hosts. This was demonstrated by observing direct predation via co-plating assays, the capacity of Trichoderma metabolites to inhibit pathogen growth, the reduction of developing pathogen lesions or titers in greenhouse experiments, or reduced pathogen recovery combined with Trichoderma recovery in treated spurs in vineyard studies. However, some inconsistencies were observed between years, implying the role of fluctuating weather conditions that may have influenced the results. For instance, it could be hypothesized that warmer, drier weather likely reduced the recovery of specific Trichoderma isolates and made them overall less likely to impair the targeted fungal pathogens. Furthermore, confirmation by sequencing of the recovered fungi would have been desirable and preferred in future follow-up studies, but could not be performed due to a large number of analyzed spurs (over 2,000 in 2023 and over 500 in 2024). Regardless, due to the sample size and previous studies that did sequence to confirm recoveries with great accuracy of over 95% (Wallis et al., 2022b; Sinclair et al., 2025), the statistical conclusions reached from this study would likely be similar.
Regardless, the objective of this study was to identify novel isolates of Trichoderma collected from grapevine tissues for re-application to grapevines to limit pathogen development in the hot, dry climate found in central California. To this end, promising isolates were identified and determined to be mostly in the T. harzianum complex, as well as T. capillare, T. asperellum, and T. saturnisporopsis. This was similar to studies by Pollard-Flamand et al. (2022) and Urbez-Torres et al. (2020), albeit these isolates were extracted directly from plant tissues in a different geographic location.
Regarding the differences in the isolates, T. harzianum and, to some degree, T. asperellum appeared to function differently than T. capillare SLO1-1. The former isolates appeared to aggressively predate the pathogens, whether by overrunning the plates in co-plating assays or perhaps overrunning spurs to remove and prevent pathogen establishment at those sites. By contrast, T. capillare SLO1–1 produced a more visibly pigmented compound in the spent media assays, which appeared to dramatically reduce pathogen growth. Likewise, systemic effects when applied to stem wounds appeared quite effective at reducing lesion sizes. However, SLO1–1 was not as readily recoverable in the spur experiments, nor was it able to remove pathogens from spurs as effectively.
Regarding X. fastidiosa and resultant Pierce’s disease, only one isolate (DL1-3) statistically reduced titers, implying a likely systemic effect to reduce the bacterial infections. It was possible that DL1–3 had produced its own compounds to affect X. fastidiosa directly, was able to induce changes in grapevine host physiology to alter the infection process, or some combination of both. Further research is warranted to explore what may be the cause of the observations.
In conclusion, these results identified different Trichoderma isolates that were observed to directly reduce pathogen growth and infection success and also had the capacity to survive in grapevine tissues throughout an entire growing season. Further studies will be needed to develop these further into potential products, and an improved understanding of isolate genomics and metabolomics will allow further conclusions on how they function to be reached.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
CW: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing. RS: Investigation, Methodology, Writing – review & editing. ME: Investigation, Validation, Writing – review & editing. ZG: Investigation, Methodology, Writing – review & editing. NM: Investigation, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was funded partly by funds allocated by the Consolidated Central Valley Table Grape Pest and Disease Control District, the California Department of Food and Agriculture, Pierce’s Disease and Glassy-winged Sharpshooter Research Board (Agreement 22-0554-000-SA), and funds allocated to the USDA-Agricultural Research Service (Project 2034-22000-015-00D).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Correction note
This article has been corrected with minor changes. These changes do not impact the scientific content of the article.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture (USDA). The USDA is an equal opportunity provider and employer.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1609693/full#supplementary-material
Supplementary Data Sheet 1 | File in the fasta format of the used rpb2 sequences to perform alignments and allow the putative determination of collected Trichoderma strains for species identification purposes.
Supplementary Data Sheet 2 | File in the fasta format of the used rpb2 sequences to perform alignments and allow the putative determination of collected Trichoderma strains for species identification purposes.
Supplementary Data Sheet 3 | Representative photographs of culture of each Trichoderma strain grown on potato dextrose agar (PDA) (left) or Spezieller Nahrstoffarmer agar (SNA) (right) after 1 week at 24 °C in darkness.
References
Andreason, S. A., Prabhaker, N., Castle, S. J., Ganjisaffar, F., Haviland, D. R., Stone-Smith, B., et al. (2018). Reduced susceptibility of Homalodisca vitripennis (Hemiptera: Cicadellidae) to commonly applied insecticides. J. Econ. Entomol. 111, 2340–2348. doi: 10.1093/jee/toy192, PMID: 29982564
Antrim, E., Ellis, M. L., and Wallis, C. M. (2025). Evaluating the ability of Californian grapevine-isolated Trichoderma saturnisporopsis strain RSI and Trichoderma asperellum strain TLI to reduce fungal trunk diseases. PhytoFrontiers. doi: 10.1094/PHYTOFR-04-25-0034-SC
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021, PMID: 22506599
Baumgartner, K., Hillis, V., Lubell, M., Norton, M., and Kaplan, J. (2019). Managing grapevine trunk diseases in California’s Southern San Joaquin valley. Am. J. Enol. Vitic. 70, 267–276. doi: 10.5344/ajev.2019.18075
Berbegal, M., Ramon-Albalat, A., Leon, M., and Armengol, J. (2020). Evaluation of long-term protection from nursery to vineyard provided by Trichoderma atroviride SC1 against fungal grapevine trunk pathogens. Pest Manage. Sci. 76, 967–977. doi: 10.1002/ps.5605, PMID: 31472038
Berlanas, C., Andres-Sodupe, M., Lopez-Manzanares, B., Maldonado-Gonzalez, M. M., and Gramaje, D. (2018). Effect of white mustard cover crop residue, soil chemical fumigation and Trichoderma spp. root treatment on black-foot disease control in grapevine. Pest Manage. Sci. 74, 2864–2873. doi: 10.1002/ps.5078, PMID: 29781195
Blundell, R. and Eskalen, A. (2022). Evaluation of biological and chemical pruning wound protectants to control grapevine trunk disease pathogens Eutypa lata and Neofusicoccum parvum. Plant Health Prog. 23, 197–205. doi: 10.1094/PHP-08-21-0113-RS
Brown, A. A., Travadon, R., Lawrence, D., Torres, G., Zhuang, G., and Baumgartner, K. (2020). Pruning-wound protectants for trunk-disease management in California table grapes. Crop Prot. 141, 105490. doi: 10.1016/j.cropro.2020.105490
Cai, F. and Druzhinina, I. S. (2021). In honor of John Bissett: authoritative guidelines on molecular identification of Trichoderma. Fungal Diversity 107, 1–69. doi: 10.1007/s13225-020-00464-4
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10, 421. doi: 10.1186/1471-2105-10-421, PMID: 20003500
Chan, M. E., Tan, J. Y., Lee, Y. Y., Lee, D., Fong, Y. K., Mutwil, M., et al. (2023). Locally isolated Trichoderma harzianum species have broad spectrum biocontrol activities against the wood rot fungal species through both volatile inhibition and mycoparasitism. J. Fungi (Basel) 9, 675. doi: 10.3390/jof9060675, PMID: 37367611
Chaverri, P., Branco-Rocha, F., Jaklitsch, W. M., Degenkolb, T., and Samuels, G. J. (2015). Systematics of the Trichoderma harzianum species complex and the identification of commercial biocontrol strains. Mycologia 107, 558–590. doi: 10.3852/14-147, PMID: 25661720
Chen, J., Groves, R., Civerolo, E., Viveros, M., Freeman, M., and Zheng, Y. (2005). Two Xylella fastidiosa genotypes are associated with almond leaf scorch disease on the same location in California. Phytopathology 95, 708–714. doi: 10.1094/PHYTO-95-0708, PMID: 18943788
Chen, S., Morgan, D. P., Hasey, J. K., Anderson, K., and Michailides, T. J. (2014). Phylogeny, morphology, distribution, and pathogenicity of Botryosphaeriaceae and Diaporthaceae from English walnut in California. Plant Dis. 98, 636–652. doi: 10.1094/PDIS-07-13-0706-RE, PMID: 30708543
Czemmel, S., Galarneau, E. R., Travadon, R., McElrone, A. J., Cramer, G. R., and Baumgartner, K. (2015). Genes expressed in grapevine leaves reveal latent wood infection by the fungal pathogen Neofusicoccum parvum. PloS One 10, 1–21. doi: 10.1371/journal.pone.0121828, PMID: 25798871
Di Marco, S., Osti, F., and Cesari, A. (2004). Experiments on the control of esca by Trichoderma. Phytopathol. Mediterr. 43, 108–115. doi: 10.36253/phyto-5040
Fourie, P. H. and Halleen, F. (2006). Chemical and biological protection of grapevine propagation material from trunk disease pathogens. Eur. J. Plant Pathol. 116, 255–265. doi: 10.1007/s10658-006-9057-9
Fourie, P. H., Halleen, F., van der Vyver, J., and Schreuder, W. (2001). Effect of Trichoderma treatments on the occurrence of decline pathogens in the roots and rootstocks of nursery grapevines. Phytopathol. Mediterr. 40, S473–S478.
Galarneau, E. R., Lawrence, D. P., Travadon, R., and Baumgartner, K. (2019). Drought exacerbates Botryosphaeria dieback symptoms in grapevines and confounds host-based molecular markers of infection by Neofusicoccum parvum. Plant Dis. 103, 1738–1745. doi: 10.1094/PDIS-09-18-1549-RE, PMID: 31082329
Gorman, Z., Chen, J., Perez de Leon, A. A., and Wallis, C. M. (2023). Comparison of assembly platforms for the assembly of the nuclear genome of Trichoderma harzianum strain PAR3. BMC Genomics 24, 454. doi: 10.1186/s12864-023-09544-6, PMID: 37568116
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086, PMID: 23422339
Guzman-Guzman, P., Porras-Troncoso, M. D., Olmedo-Monfil, V., and Herrera-Estrella, A. (2019). Trichoderma species: versatile plant symbionts. Phytopathology 109, 6–16. doi: 10.1094/PHYTO-07-18-0218-RVW, PMID: 30412012
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., and Lorito, M. (2004). Trichoderma species-opportunistic, avirulent plant symbiont. Nat. Rev. Microbiol. 2, 43–56. doi: 10.1038/nrmicro797, PMID: 15035008
Hopkins, D. (2001). “Xylella fastidiosa,” in Laboratory guide for identification of plant pathogenic bacteria, 3rd ed. Eds. Schaad, N. W., Jones, J. B., and Chun, W. (American Phytopathological Society, St. Paul, MN, USA), 201–213.
Hopkins, D. L. and Purcell, A. H. (2002). Xylella fastidiosa: cause of Pierce’s disease of grapevine and other emergent diseases. Plant Dis. 86, 1056–1066. doi: 10.1094/PDIS.2002.86.10.1056, PMID: 30818496
John, S., Wicks, T. J., Hunt, J. S., Lorimer, M. F., Oakey, H., and Scott, E. S. (2005). Protection of grapevine pruning wounds from infection by Eutypa lata using Trichoderma harzianum and Fusarium lateritium. Australas. Plant Pathol. 34, 569–575. doi: 10.1071/AP05075
Kaplan, J., Travadon, R., Cooper, M., Hillis, V., Lubell, M., and Baumgartner, K. (2016). Identifying economic hurdles to early adoption of preventative practices: The case of trunk diseases in California winegrape vineyards. Wine Econ. Policy. 5, 127–141. doi: 10.1016/j.wep.2016.11.001
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8, PMID: 30936562
Krivanek, A. F., Riaz, S., and Walker, M. A. (2006). Identification and molecular mapping of PdR1, a primary resistance gene to Pierce’s disease in Vitis. Theor. App. Genet. 112, 1125–1131. doi: 10.1007/s00122-006-0214-5, PMID: 16435126
Marraschi, R., Ferreira, A. B. M., da Silva Bueno, R. N., Leite, J. A. B. P., Lucon, C. M. M., Harakava, R., et al. (2019). A protocol for selection of Trichoderma spp. to protect grapevine pruning wounds against Lasiodiplodia theobromae. Braz. J. Microbiol. 50, 213–221. doi: 10.1007/s42770-018-0029-y, PMID: 30637650
Martinez-Diaz, M., Diaz-Losada, E., Diaz-Fernandez, A., Bouzas-Cid, Y., and Gramaje, D. (2021). Protection of grapevine pruning wounds against Phaeomoniella chlamydospora and Diplodia seriata by commercial biological and chemical methods. Crop Prot. 143, 105464. doi: 10.1016/j.cropro.2020.105465
Morales-Cruz, A., Amrine, K. C. H., Blanco-Ulate, B., Lawrence, D. P., Travadon, R., Rolshausen, P. E., et al. (2015). Distinctive expansion of gene families associated with plant cell wall degradation, secondary metabolism, and nutrient uptake in the genomes of grapevine trunk pathogens. BMC Genomics 16, 469. doi: 10.1186/s12864-015-1624-z, PMID: 26084502
Mugnai, L., Graniti, A., and Surico, G. (1999). Esca (black measles) and brown wood-streaking: two old and elusive diseases of grapevines. Plant Dis. 83, 404–418. doi: 10.1094/PDIS.1999.83.5.404, PMID: 30845530
Pollard-Flamand, J., Boule, J., Hart, M., and Urbez-Torres, J. R. (2022). Biocontrol activity of Trichoderma species isolated from grapevines in British Columbia against Botryosphaeria dieback fungal pathogens. J. Fungi (Basel) 8, 409. doi: 10.3390/jof8040409, PMID: 35448640
Rapicavoli, J., Ingel, B., Blanco-Ulate, B., Cantu, D., and Roper, M. C. (2018). Xylella fastidiosa: an examination of a reemerging plant pathogen. Mol. Plant Pathol. 19, 786–800. doi: 10.1111/mpp.12585, PMID: 28742234
Samuels, G. and Hebbar, P. (2016). Trichoderma: Identification and agricultural applications (St. Paul, MN, USA: APS Press).
Sicard, A., Zeilinger, A. R., Vanhove, M., Schartel, T. E., Beal, D. J., Daugherty, M. P., et al. (2018). Xylella fastidiosa: insights into an emerging plant pathogen. Ann. Rev. Phytopathol. 56, 181–202. doi: 10.1146/annurev-phyto-080417-045849, PMID: 29889627
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351, PMID: 26059717
Sinclair, G. C., Travadon, R., Eschen, P. J., Wallis, C., Baumgartner, K., Delmas, C. E. L., et al. (2025). Differential physiological responses of resistant and susceptible grape cultivars to Eutypa dieback. J. Exp. Bot. 76, 3172–3185. doi: 10.1093/jxb/eraf103, PMID: 40099478
Spagnolo, A., Mondello, V., Larignon, P., Villaume, S., Rabenoelina, F., Clement, C., et al. (2017). Defense responses in grapevine (cv. Mourvedre) after inoculation with the Botryosphaeria dieback pathogens Neofusicoccum parvum and Diplodia seriata and their relationship with flowering. Int. J. Mol. Sci. 18, 393. doi: 10.3390/ijms18020393, PMID: 28208805
Travadon, R. and Baumgartner, K. (2015). Molecular polymorphism and phenotypic diversity in the Eutypa dieback pathogen Eutypa lata. Phytopathology 105, 255–264. doi: 10.1094/PHYTO-04-14-0117-R, PMID: 25084304
Travadon, R., Lawrence, D. P., Moyer, M. M., Fujiyoshi, P. T., and Baumgartner, K. (2022). Fungal species associated with grapevine trunk diseases in Washington wine grapes and California table grapes, with novelties in the genera Cadophora, Cytospora, and Sporocadus. Front. Fungal Biol. 3, 1018140. doi: 10.3389/ffunb.2022.1018140, PMID: 37746176
Travadon, R., Rolshausen, P. E., Gubler, W. D., Cadle-Davidson, L., and Baumgartner, K. (2013). Susceptibility of cultivated and wild Vitis spp. to wood infection by fungal trunk pathogens. Plant Dis. 97, 1529–1536. doi: 10.1094/PDIS-05-13-0525-RE, PMID: 30716856
Trotel-Aziz, P., Abou-Mansour, E., Courteaux, B., Rabenoelina, F., Clement, C., Fontaine, F., et al. (2019). Bacillus subtilis PTA-271 Counteracts Botryosphaeria dieback in grapevine. Front. Plant Sci. 10, 25. doi: 10.3389/fpls.2019.00025, PMID: 30733727
Urbez-Torres, J. R. (2011). The status of Botryosphaeriaceae species grapevines. Phytopathol. Mediterr. 50, S5–S45.
Úrbez-Torres, J. R., Leavitt, G. M., Voegel, T. M., and Gubler, W. D. (2006). Identification and distribution of Botryosphaeria spp. associated with grapevine cankers in California. Plant Dis. 90, 1490–1503. doi: 10.1094/PD-90-1490, PMID: 30780967
Urbez-Torres, J. R. and Gubler, W. D. (2009). Double pruning, a potential method to control Bot canker disease of grapes, and susceptibility of grapevine pruning wounds to infection by Botryosphaeriaceae. Phytopathol. Mediterr. 48, 185.
Urbez-Torres, J. R., Tomaselli, E., Pollard-Flamand, J., Boule, J., Gerin, D., and Pollastro, S. (2020). Characterization of Trichoderma isolates from southern Italy, and their potential biocontrol activity against grapevine trunk disease fungi. Phytopathol. Mediterr. 59, 425–439.
Verma, M., Brar, S. K., Tyagi, R. D., Surampalli, R. Y., and Valero, J. R. (2017). Antagonistic fungi, Trichoderma spp.: panoply of biological control. Biochem. Eng. J. 37, 1–20. doi: 10.1016/j.bej.2007.05.012
Wallis, C. M. and Chen, J. (2012). Grapevine phenolic compounds in xylem sap and tissues are significantly altered during infection by Xylella fastidiosa. Phytopathology 102, 816–826. doi: 10.1094/PHYTO-04-12-0074-R, PMID: 22671027
Wallis, C. M., Chen, J., and Perez de Leon, A. A. (2022a). Mitochondrial genome resource of a grapevine strain of Trichoderma harzianum, a potential biological control agent for fungal canker diseases. PhytoFrontiers 2, 143–146. doi: 10.1094/PHYTOFR-08-21-0052-A
Wallis, C. M., Gorman, Z., Galarneau, E. R. A., and Baumgartner, K. (2022b). Mixed infections of fungal trunk pathogens and induced systemic phenolic compound production in grapevines. Front. Fungal Biol. 3, 1001143. doi: 10.3389/ffunb.2022.1001143, PMID: 37746162
Woo, S. L., Ruocco, M., Vinale, F., Nigro, M., Marra, R., Lombardi, N., et al. (2014). Trichoderma-based products and their widespread use in agriculture. Open Mycol. J. 8, 71–126. doi: 10.2174/1874437001408010071
Keywords: biological control agents, Bot canker, dieback, fungal trunk disease, Pierce’s disease
Citation: Wallis CM, Shinde R, Ellis ML, Gorman Z and Mekdara N (2025) Assessment of the potential of novel Californian grapevine Trichoderma isolates to reduce colonization of fungal trunk canker pathogens and Xylella fastidiosa. Front. Plant Sci. 16:1609693. doi: 10.3389/fpls.2025.1609693
Received: 10 April 2025; Accepted: 20 August 2025;
Published: 09 September 2025; Corrected: 09 December 2025.
Edited by:
Esther Menendez, University of Salamanca, SpainReviewed by:
Khalid Hameed, Duke University, United StatesShitou Xia, Hunan Agricultural University, China
Copyright © 2025 Wallis, Shinde, Ellis, Gorman and Mekdara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christopher M. Wallis, Y2hyaXN0b3BoZXIud2FsbGlzQHVzZGEuZ292
 Christopher M. Wallis
Christopher M. Wallis Ranjeet Shinde2
Ranjeet Shinde2 Zachary Gorman
Zachary Gorman