- 1Department of Biology, Faculty of Science, Mahasarakham University, Kantharawichai, Maha Sarakham, Thailand
- 2The Northeastern Soil Salinity Research Unit, Faculty of Science, Mahasarakham University, Kantharawichai, Maha Sarakham, Thailand
- 3Faculty of Science, Omics Science and Bioinformatics Center, Chulalongkorn University, Bangkok, Thailand
Eucalyptus is an economic plant of Thailand that can tolerate salt and drought stress. This work aims to report on the study of biodiversity among endophytic actinobacteria isolated from Eucalyptus camaldulensis grown in saline soil, as well as their properties for promoting plant growth and inhibiting fungal pathogens in vitro. Root, twig, and leaf samples of five plants, E. camaldulensis grown in Kalasin Province, Thailand, were collected. The soil samples of each plant were collected, and soil salinity was evaluated by the electrical conductivity of a saturated soil extract (ECe). It was found that the ECe of the soil was between 4.5 and 12 dS/m, and the pH of the soil ranged between 3.9 and 5.2. Based on their morphology and 16S rRNA gene sequences, the majority of the isolates (552, 96.7%) were identified as the genus Streptomyces. The remaining isolates (19, 3.3%), which included ten genera: Cellulosimicrobium (3), Kocuria (3), Brevibacterium (2), Micrococcus (2), Microbacterium (2), Peterkaempfera (2), Tsukamurella (2), Brachybacterium (1), Curtobacterium (1), and Gordonia (1). Two hundred and seventy-three isolates were tested for antifungal activity against two eucalyptus pathogens, Pseudoplagiostroma eucalypti LS6 and Cladosporium sp. LB1. Most isolates showed antifungal activity against P. eucalypti LS6. The plant growth-promoting (PGP) study of 154 selected strains showed that 154 (100%), 18 (11.7%), 14 (9.1%), and 7 (4.5%) isolates could produce indole-3-acetic acid (IAA), 1-aminocyclopropane (ACC) deaminase, solubilize phosphate, and fix nitrogen in vitro, respectively. Identification of non-actinobacteria isolates based on 16S rRNA gene sequence analysis indicated that 10 genera were obtained: Aureimonas, Bacillus, Chryseobacterium, Deinococcus, Massilia, Methylobacterium, Pseudomonas, Serratia, Staphylococcus, and Stenotrophomonas. One selected Streptomyces strain, EWL5.1, was selected for a seed germination test in salinity stress and PGP in planta. The result indicated that this strain could support the seedling length vigor index (SLVI) of eucalyptus seedlings in salinity conditions and significantly increase the fresh weight of eucalyptus seedlings in planta. Four representative Streptomyces strains and one strain of Micrococcus were sequenced for their genomes. The result indicated that these four Streptomyces strains comprise various biosynthesis gene clusters (BGCs) of antibiotic production. Genome data mining also reveals that all strains contain genes encoding PGP properties. These potential strains can be applied to be used as PGPB to support eucalyptus growth in the future.
Introduction
Endophytes are microorganisms living in the intercellular spaces of plant tissues without causing any disease symptoms. Some of these endophytes may benefit their hosts by producing plant growth-promoting (PGP) agents and inhibiting plant pathogens. Endophytes can be found in plants ranging from woody tree species to herbaceous crops. Actinomycetes, also known as Actinobacteria, are Gram-staining positives that comprise the Phylum Actinomycetota. These are spore-forming filamentous bacteria whose spores can survive in dry and stressed environments (Singh and Dubey, 2018). Actinobacteria are a major group that produce known antibiotics (more than 70%) to inhibit bacteria and fungi, including PGP properties. Actinobacteria produce phytohormones like auxin, cytokinin, and gibberellin; ACC deaminase to get rid of ethylene gas; osmoprotectants like proline, glycine-betaine, and polyamine; exopolysaccharides (EPS); and antioxidative enzymes to lower oxidative stress. These properties make it possible for plants to grow in salty conditions (Atouei et al., 2019; Feikema and Baker, 2011). Soil salinity distribution in wide areas of Thailand is a major problem for land use in agricultural areas. The majority area of saline soil is the Northeast, especially Kalasin and Maha Sarakham provinces. The high soil salinity affects the growth of any plants and leads to low productivity yield. Gum tree, or Eucalyptus, is an economic plant of Thailand. Eucalyptus camaldulensis, a widely distributed species in Thailand, produces wood pulp for the paper industry. There are many applications of eucalyptus, including paper pulp, cooking charcoal, crutch, timber, furniture, fly wood, chopped wood, and industrial fuel. E. camaldulensis can grow quickly and has the ability to tolerate drought and salinity (Marcar et al., 2002). Eucalyptus tree is a high-potential plant species with a deep root system for water absorption to prevent the salt rise-up, a large bushy cover to cover the surface area preventing the evaporation rate, and to detoxify the toxic salt ions by organic matter. It was known that planting gum trees at the ridge of the paddy field could protect it from salt erosion. However, plant diseases are the major problems of eucalyptus plantations by reducing plant yield. There were about 30 species of fungi that can cause eucalyptus diseases. Fungi can destroy eucalyptus plants in different stages of growth, ranging from seedling, cutting, and tree. The severity of diseases is different depending on fungal strains, susceptible plant variety, and climate in each area. It was reported that the most serious disease in Eucalyptus trees was leaf and shoot blight caused by Pseudoplagiostroma eucalypti, Cylindrocladium reteaudii, and Teratosphaeria destructans (Old et al., 2003). After the infection of these fungi, plants are weak and may be infected by other fungi such as Kirramyces destructans, Cytospora sp., Lasiodiplodia theobromae, and Phomopsis sp. to destruct stem and bark (Old et al., 2003). These fungal pathogens can cause severe symptoms and reduce plant yield dramatically. The bacterial pathogen, Ralstonia solanacearum, which causes eucalyptus wilt, is a secondary infection that occurs after fungi infect the plant in younger trees. Plant symptoms showed wilted and foliar necrosis or defoliation of the lower portion of the canopy (Santiago et al., 2014).
To the best of our knowledge, there is no report of studying the biodiversity of endophytic actinobacteria isolated from Eucalyptus grown in a salinity area. This is the first study of the biodiversity of endophytic actinobacteria from eucalyptus grown in a salinity area. The project’s goal is to find the beneficial endophytic actinobacteria that will promote growth of Eucalyptus trees and inhibit fungal pathogens in salinity conditions. This study focused on studying species biodiversity of endophytic actinobacteria from surface sterilized E. camaldulensis tissues grown at moderately and highly saline soil at Yang Talat district, Kalasin province, Thailand, studying antimicrobial activity of actinobacteria against important fungal pathogens, Pseudoplagiostroma eucalypti and opportunistic fungi causing leaf blight of eucalyptus, Cladosporium sp. in vitro, and studying plant growth-promoting properties of selected actinobacteria in vitro. We also tested the selected strain for seedling growth under salinity conditions and its ability to promote plant growth in Planta. Additionally, we studied the genomes of five selected strains from each plant sample.
Materials and methods
Plant sample collection
Root, twig, and leaf samples of Eucalyptus camaldulensis were collected from Ponsim Village, Tumbol Hua Nakham, Yang Talat District, Kalasin Province. Plant samples were collected in April 2022. We collected four plant samples from moderately saline soil and one from highly saline soil. Plant samples were collected, kept in paper bags, and processed within 24 hr. The information on salt contents in each area in this village is based on the previous study of Dr. Winya Dungkaew (data not reported). Supplementary Figure S1 displays the latitudes and longitudes of each plant sample, EB, EC, EK, ES, and EW, along with its corresponding pictures.
Collection of soil and salt concentration analysis
We collected soil samples from the eucalyptus growing area by digging 30 cm below the soil surface. Three soil samples were collected per sample site. Soil was air-dried for 2 weeks and then ground by mortar and pestle. Soil was sieved to discard contaminants by using a sieve size of 2 x 2 mm. The evaluation of soil salinity is measured by the electrical conductivity of a saturated soil extract (ECe) following the method of Rhoades et al. (1999). pH of soil samples was measured in Soil-H2O system (1:5) followed the method of FAO (2021) and soil solution was measured pH using pH meter.
Isolation of endophytic actinobacteria
Samples of plants were surface sterilized using the method of Kaewkla and Franco (2013). Samples of roots, leaves, and twigs were properly cleaned using tap water and then in sterile RO water. Barks from twigs and roots were pulled off and dried on paper towels. The samples were surface sterilized by immersing them in 70% ethanol for five minutes, then in a solution of 6% sodium hypochlorite (freshly produced and 6% available chloride) for five minutes. The chemicals were then removed by washing them five times in sterile RO water. To break up plant tissues and slow the growth of endophytic fungi, the samples are soaked in 10% (w/v) NaHCO3 for ten minutes. They are then rinsed twice in sterile RO water. The surface sterilization is verified by aseptically rolling surface-sterilized plant tissues over each of the isolation media and tryptone soy agar. Each sample was chopped into small pieces measuring roughly 0.5 x 0.5 cm using a sterile scalpel. They were then crushed with a sterile mortar and pestle and plated on isolation media.
Each sterilized root, twig, and leaf sample was plated in triplicate onto four isolation media. Humic acid vitamin B agar (HVA) (Hayakawa and Nonomura, 1987), VL70 gellan gum with amino acid mixture (AA, comprising 17 amino acids), VL70 gellan gum with carboxymethyl cellulose, and starch casein nitrate agar (Mohseni et al., 2013) were used as the isolation media. 10 µg/mL of icotranazol was added as an antifungal agent. The VL 70 media composition is derived from previous works (Hudson et al., 1989; Joseph et al., 2003; Schoenborn et al., 2004). The plates were incubated at 30°C in small, airtight plastic boxes lined with wet towel paper to retain moisture for eight weeks. Colonies of actinobacteria were selected from isolation media and streaked onto half-strength potato dextrose agar (HPDA) plates to purify cultures. Pure cultures were maintained at 4°C on HPDA slants. The cells and spores were stored in 50% glycerol and frozen at -80°C for a long time.
Identification of actinobacteria isolates and non-filamentous bacteria based on morphological characterization and molecular techniques
Morphological descriptions of all actinobacteria-like strains on ISP 2, ISP 3, and HPDA are regularly studied following the general guidelines of the International Streptomyces Project (ISP) (Shirling and Gottlieb, 1966). These include pigment or melanin production, the presence or absence of sporulation, and the color and growth characteristics of the mycelia. Non-filamentous bacteria grown on isolation plates were purified on nutrient agar and kept in nutrient agar slant at 4°C and prolonged-kept in 50% glycerol at -80°C. They were tested for Gram staining, and both Gram-staining positive and negative were kept for identification by 16S rRNA gene sequence analysis.
For the 16S rRNA gene sequence analysis, we chose strains that were typical of each morphological group of actinobacteria and non-filamentous bacteria. Genomic DNA was extracted from bacterial cells by using the GF1-DNA extraction kit (Vivantis). The 16S rRNA gene was amplified by PCR procedure using primer pairs 27f and 1492r and sequenced as described by Kaewkla and Franco (2013). The resultant sequences were compared to an online database using the EzbioCloud server (https://www.ezbiocloud.net; Yoon et al., 2017).
Estimated relative abundance in genus level
The relative abundance (RA) at the genus level is the proportional number of each genus in each plant sample. The RA of each genus is expressed as
The variable ni represents the number of individuals within the same genus, while N represents the total number of individuals across all genera.
Phylogenetic tree construction of 16S rRNA gene
The resulting sequences were compared to an online database using the EzTaxon-e service (Yoon et al., 2017). The 16S rRNA gene sequences of isolates from each genus were compared with the corresponding sequences of their closely related type strains available in GenBank/EMBL using CLUSTAL X (Thompson et al., 1997). The 16S rRNA gene phylogenetic trees of isolates belonging to genera Streptomyces and Peterkaempfera were generated using neighbor joining (NJ) (Saitou and Nei, 1987), maximum likelihood (ML) (Tamura and Nei, 1993), and maximum parsimony (MP) (Nei and Kumar, 2000) methods using the MEGA version 11 software tool (Tamura et al., 2021). Embleya scabrisporus DSM 41855T was the outgroup.
The NJ tree computed the evolutionary distances using the Maximum Composite Likelihood method (Tamura et al., 2004), and the rate variation among sites was modeled with a gamma distribution (shape parameter=1). The ML tree was inferred by using the Maximum Likelihood method, and initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Tamura-Nei model. The MP tree was obtained using the Tree-Bisection-Regrafting (TBR) algorithm with search level 1. The tree’s topology was assessed by a bootstrap approach (Felsenstein, 1985) including 1000 replications.
The 16S rRNA gene phylogenetic trees of isolates belonging to non-Streptomyces genera (9 genera) were generated using NJ, ML, and MP methods as described for the Streptomyces group with Nocardia nova JCM 6044T as the outgroup.
Antifungal and antibacterial assay in vitro
The dual culture method described by Kampapongsa and Kaewkla (2016) was used to test all actinobacteria to evaluate antifungal activity against Pseudoplagiostroma eucalypti LS6 and Cladosporium sp. LB1. The percent inhibition was calculated as follows:
Antibacterial activity against leaf wilt disease of eucalyptus, Ralstonia solanacearum TISTR 2069, was tested by the dual culture technique described by Kampapongsa and Kaewkla (2016).
Screening for plant growth promoting traits of selected strains
Actinobacterial strains that showed good and strong inhibition against at least one tested fungal pathogen were selected to test for 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase production, indole acetic acid (IAA) production, nitrogen fixation, phosphate solubilization, and cellulase production in vitro.
ACC deaminase production and indole acetic acid (IAA production)
The capacity of actinobacteria to produce ACC deaminase was evaluated utilizing DF agar according to methods of Penrose and Glick (2003) and Kaewkla et al. (2022).
IAA production was investigated in accordance with the methodology of Kaewkla et al. (2022). One 8 mm actinomycete disc from a 7-day culture on HPDA was inoculated into ISP 2 broth with 0.2% (w/v) L-tryptophan and incubated on a rotary shaker at 150 rpm for 7 days in the dark at 28°C. The cultures were centrifuged at 6000 g for 10 minutes. The one milliliter of supernatant was mixed with two milliliters of Salkowski’s reagent and incubated in the dark for 30 minutes at ambient temperature. The reaction mixture’s absorbance was measured at 530 nm using a spectrophotometer. The concentration of IAA generated per milliliter of culture (µg/ml) was determined utilizing a standard curve of indole acetic acid (20–100 µg/ml).
Phosphate solubilization and Nitrogen fixation assay
The capacity of actinobacteria for phosphate solubilization was evaluated using the methodology of Castagno et al. (2011). NBRIP agar was utilized, and a positive result was determined by measuring the halo zone surrounding the colony of each strain after a 14-day incubation duration. The method described by Baldani et al. (1997) was used to apply the nitrogen fixation in vitro. Nitrogen-free semi-solid (NFb) medium was used to inoculate actinobacteria and incubated it at 30°C for 14 days. The dark blue color of the media and the robust growth of the bacteria indicated a positive result. The culture was sub-cultured to the same media three times if it obtained a positive result.
Cellulase production
The potential of actinobacteria to degrade cellulose was evaluated using ISP 2 agar supplemented with carboxymethyl cellulose 1%. Each bacterial isolate was cultivated for 14 days by spotting at the middle of the Petri dish. The hydrolysis of CMC was evaluated using a flood agar plate containing 0.1% Congo red, incubated for 10 minutes. Congo red was eliminated, and 10 ml of 1 M NaCl was flooded to the agar plate. The positive result demonstrated a halo zone surrounding the colony of actinobacteria.
Growth on different sodium chloride concentration
Thirty-five selected strains of actinobacteria, which showed good PGP traits, were tested for their growth on different concentrations of sodium chloride at 1, 3, 5, 7, 9, and 11% (w/v) on ISP 2 medium pH 7.2. Actinobacterial cells were streaked in three lines on each agar plate and incubated at 30°C for 14 days. Growth was observed and evaluated at scores 0, 1, 2, and 3 for no growth, weak, moderate, and good growth, respectively. The experiment was done in triplicate.
Seed germination test
Streptomyces strain EWL5.16, which showed good antifungal activity and PGP in vitro, was selected for a seed germination test with salinity stress in different sodium chloride concentrations. Also, the genome of this strain comprises many BGCs of antifungal compounds. E. camaldulensis seeds were gained from the forest research and development office, the Royal Forest Department, Thailand. Seeds were separated from seed debris by hand and soaked in RO water overnight and surface sterilized. Briefly, seeds were soaked in 70% ethanol for one min and rinsed three times with sterilized RO water (1 min in each wash). After that, seeds were immersed in 8% sodium hypochlorite for 5 mins and washed with sterilized RO water four times. Then, seeds were soaked in 2% sodium thiosulfate for 3 mins and washed in sterilized RO water. Strain EWL5.16 was cultured on HPDA for seven days, and spore suspension was prepared in sterilized RO water. Spore suspension was filtered and passed through a sterilized 10 mL syringe filled with sterilized cotton to separate the spores from the mycelia, and a hemocytometer was used to count the spores to 108 spores per ml. After that, surface-sterilized Eucalyptus seeds were soaked in spore suspension for 1 hr., and RO water was used as a control.
Different concentrations of NaCl (0, 50, 100, 150, and 200 mM) were used in this study. Fifteen bacterized seeds from the above experiment were placed on sterilized Whatman No. 1 paper lined on a sterilized glass Petri dish (9 cm diameter) and moistened with 5 mL of each concentration of salt. The experiment was carried out in five replicates, and Petri dishes were located in a growth chamber in the dark two days before being exposed to light. After that, plates were exposed with cool daylight (12/12 light/dark cycles) at 28°C for 8 days. Seedlings were measured for seedling length (SL) (shoot and root lengths) at day 8. The experiment was done in a factorial design (2 factors with 5 levels of salt), and plates were arranged in a completely randomized design (CRD). Seeds were counted every day for eight days. The Seeding Length Vigor Index (SLVI) after eight days was calculated following some modifications from Zou et al. (2023).
In the Equations 1–3, M is the total number of test seeds (M=15); m1 is the number of normally germinated seeds (root length ≥ 2 mm) within 3 days; m2 is the number of normally germinated seeds within 8 days; SL is the average seedling length (cm) after 8 days of germinated seeds. Germination potential (GP) in Equation 1 was followed (Zou et al., 2023), while germination rate (GR) in Equation 2 and SLVI in Equation 3 were described by Kerbab et al. (2021).
Plant growth promoting in planta
Strain EWL5.16 was also selected to study the growth of eucalyptus seedlings in soil. The spore suspension of strain EWL5.16 was filtered and counted to 108 spores/ ml according to a method described above. E. camaldulensis seeds were prepared and surface sterilized according to the method of seed germination test. Surface-sterilized eucalyptus seeds were soaked with strain EWL5.16 for 60 minutes and immersed in sterile RO water as a control experiment according to the method described above. Bacterized seeds were germinated in a sterilized plastic box (22 x 33 x 10 cm) lined with sterilized Whatman paper no. 1 and moistened with sterilized RO water pH 7.2 by placing 200 seeds per box for 7 days at 28°C in the dark.
Fine sand was sieved and autoclaved at 121°C for 30 minutes and left overnight and completely autoclaved for two other times. Sterilized sand was packed into 17 x 26 cm trays, 7 x 5 blocks per tray (3 x 3 cm per block). The experiment was conducted in six replicates per treatment (water and strain EWL5.16). The seedlings with even growth grown on filter paper prepared above were planted (2 seedlings per block). The experiment was arranged in a CRD. Plants were kept in the dark for 2 days before being exposed to cool daylight. The planting trays were kept in a growth chamber with cool daylight (12/12 light/dark cycles) at 28°C for fourteen days. Plants were watered with sterilized RO water one time per day. Although seedlings were surface sterilized, some seed-borne fungi destroyed seedlings, and plants started dying with root rots and decoloring leaves on day 7 after sowing. After fourteen days, survival seedlings with normal growth, considering plants with green leaves, were counted. Seedlings were gently taken from sand, and their roots were thoroughly washed to discard sand. Plants were measured for seedling lengths (shoot and root lengths) and plant fresh weight. As seedlings were still small, the dry weight could not be measured after drying the plant.
Statistical analysis
The seed germination test applied a factorial design with two treatments (bacteria and without bacteria) and five levels of salt with a completely randomized design arrangement. IBM SPSS Statistics version 29 (Mahasarakham University License) was applied for statistical analysis. Data were submitted to normality and homogeneity of variances tests before the analysis of multivariate analysis of variance (MANOVA). The statistical analyses were carried out using GLM (General Linear Model) to determine the effectiveness of bacteria (seed priming with bacteria and salt) on seeding length vigor index. The Tukey method compared the means to identify a significant group with a P-value of less than 0.05 (P<0.05). For the PGP test in planta, data were submitted to normality and homogeneity of variances tests before analysis. The Independent-samples T-test was used to analyze data, and a P-value of less than 0.05 (P<0.05) showed significant differences.
Genome sequencing, assembly, and annotation
As a model strain for genome sequencing, Streptomyces strain EKR5.2 was taken from sample EK. This strain isolated from plant sample grew in high-salinity soil and produced ACC deaminase and IAA. We chose strain ESS7.8 as the representative strain of Streptomyces from sample ES because it was good at killing fungal pathogens and was closely related to Streptomyces ardesiacus NRRL B-1773T, which was the most common species in four plant samples. We selected the representative strains of Streptomyces from the EC and EW strains, ECR2.10 and EWL5.16, for genome sequencing. These strains were a unique species of Streptomyces spp., closely related to Streptomyces roietensis WES2T. Also, these two strains showed activities of ACC deaminase and IAA production and good activity against fungal pathogens.
The representative strain of the non-Streptomyces genus from sample EW which contained the highest number of non-Streptomyces genera, Micrococcus strain EWR3.9.1, was selected for genome sequencing. We extracted the genomic DNA from Streptomyces strains EKR5.2, ESS7.8, ECR2.10, EWL5.16, and Micrococcus strain EWR3.9.1, using the previously described method as above. A short insert size library was prepared for genome sequencing of these five strains. Samples were sequenced at the Omics, Chulalongkorn University, using an Illumina NovaSeq sequencer (2 x 150 bp paired-end reads). We applied Unicycler (0.5.1) for de novo assembly of the reads (Wick et al., 2017). Genomes of these five strains were annotated using the Clusters of Orthologous Groups (COGs) on the Eggnog-Mapper database (V.2.1.9) (Cantalapiedra et al., 2021) and were evaluated for functional protein categories.
Genome comparison study
The Genome-to-Genome Distance calculator (GGDC 2.1; BLAST + method) was used to analyze the digital DNA-DNA hybridization (dDDH) value between Streptomyces strains EKR5.2, ESS7.8, ECR2.10, EWL5.16, and Micrococcus strain EWR3.9.1 and its closely related type strains of the genus Streptomyces and Micrococcus. Formula 2 (identities/HSP length) was applied to the analysis (Meier-Kolthoff et al., 2013).
The phylogenetic tree of the genomes of Streptomyces strains EKR5.2, ESS7.8, ECR2.10, and EWL5.16 was constructed by the Type (Strain) Genome Server (TYGS) with other closely related type strains, with Embleya scabrispora DSM 41855T as the outgroup (Lefort et al., 2015; Meier-Kolthoff and Göker, 2019). Also, the phylogenetic tree of the genome of Micrococcus strain EWR3.9.1 and its closely related type strains of the genus Micrococcus with Tersicoccus phoenicis DSM 30849T as the outgroup was constructed by the Type (Strain) Genome Server (TYGS) (Lefort et al., 2015; Meier-Kolthoff and Göker, 2019).
The Average Nucleotide Identity values (ANI) blast (ANIb) and ANI MUMmer (ANIm) with pairwise genome alignment between Streptomyces strains EKR5.2, ESS7.8, ECR2.10, EWL5.16, and Micrococcus strain EWR3.9.1 and its closely related type strain of the genus Streptomyces and Micrococcus were analyzed using the JSpeicesWS web service (Richter and Rosselló-Móra, 2009; Richter et al., 2016).
Biosynthetic Gene cluster analysis and in silico gene prediction
Secondary metabolite analysis Shell (anti-SMASH) version 7.0 (Blin et al., 2023) was used to predict biosynthetic gene clusters (BGCs) of Streptomyces strains EKR5.2, ESS7.8, ECR2.10, and EWL5.16 and Micrococcus strain EWR3.9.1. The genomes of these five strains were then annotated by the Clusters of Orthologous Groups (COGs). These genomes were then used to search for genes encoding for metabolite products that support plant growth, produce antibiotics and bioactive compounds, break down proteins, etc (Cantalapiedra et al., 2021).
Results and discussion
Soil salinity test and pH of soil
The EW, EC, ES, and EB soil samples were all moderately saline soil because their saturation extract conductivity was 4.5, 6.6, 7.1, and 7.4 dS/m, respectively. The pH of these soil samples was 4.6, 5.2, 4.2, and 5.0, respectively, which is strongly acid soil. The EK soil sample, on the other hand, was a strongly saline soil because its ECe value was 12 dS/m. The pH of the EK soil sample was 3.9, which was extremely acidic (Table 1). The pH of soil was classified as extremely acid (pH ≤ 4.0), very strongly or strongly acid soils (4.0 < pH ≤ 5.5), moderately acid, slightly acid and neutral soils (5.5 < pH ≤ 7.3), slightly alkaline and moderately alkaline soils (7.3 < pH ≤ 8.5), and strongly and very strongly alkaline soils (pH > 8.5) (Batjes, 1995; Mosley et al., 2024). It was reported that moderately saline soil (ECe 4–8 dS/m) affects yields of many crops, while strongly saline soil (ECe 8–16 dS/m) affects many plants, and only salt-tolerant crops can grow satisfactorily (Abrol et al., 1998). In this study, the soil sample of the EK plant was extremely acidic, and the soil of the other four plants (pH ≤ 5.5) was strongly acidic soil. Acidic soils primarily contribute to reductions in plant productivity due to deficits in important nutrients, including phosphorus (P), calcium (Ca), and magnesium (Mg). Moreover, metal toxicities, specifically from manganese (Mn) and aluminum (Al), intensify these difficulties. Soil acidification continues to be a significant concern in sustainable agriculture, negatively impacting soil health, crop yield, and environmental stability (Chen and Huang, 2024).
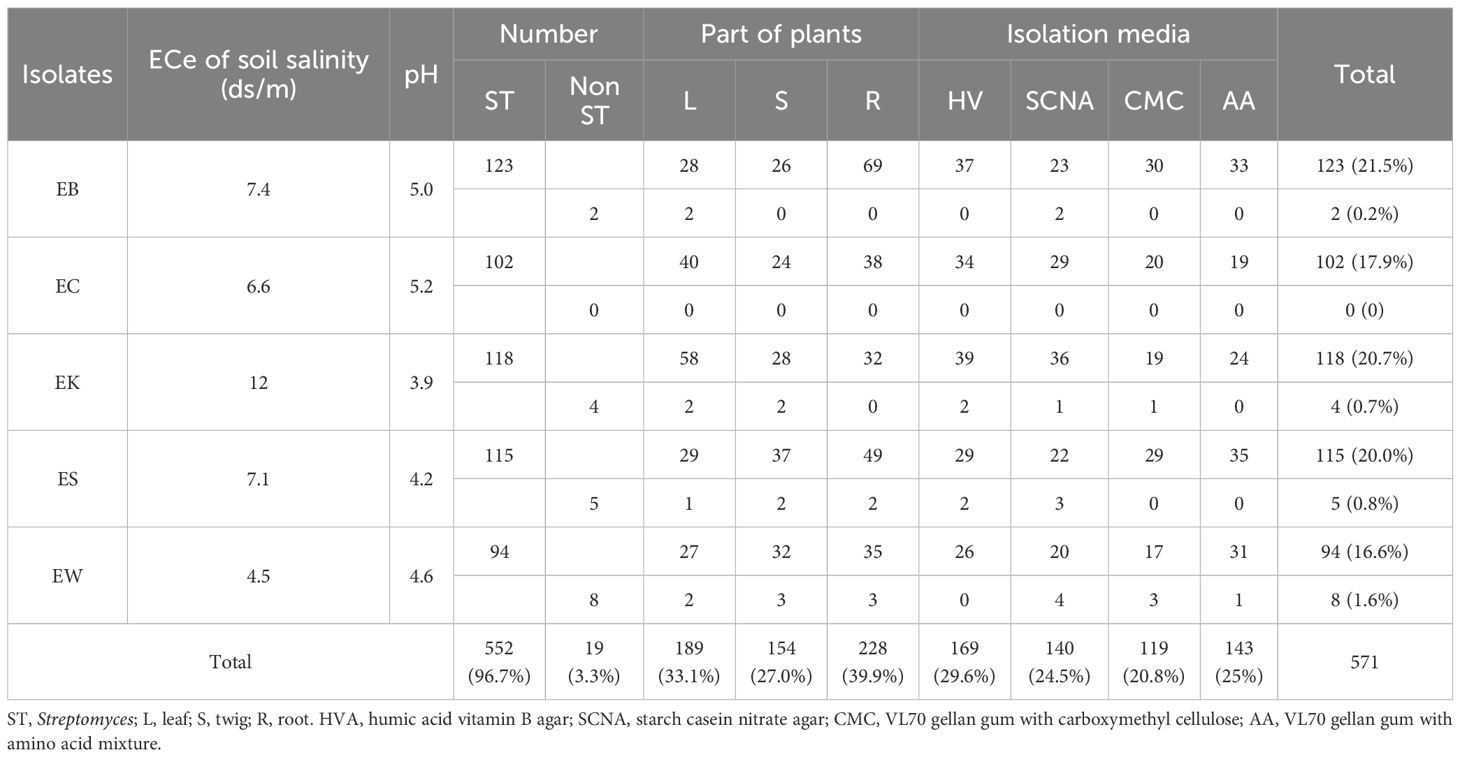
Table 1. The isolation numbers of endophytic actinobacteria isolated from five eucalyptus samples from three different media and plant parts.
Number of endophytic actinobacteria
Five hundred and seventy-one isolates of endophytic actinobacteria were isolated from five eucalyptus plants. Sample EB yielded the highest number of isolates at 125 (21.9%), with the rest of the isolates coming from samples EK (n=122), ES (n=120), EC (n=102), and EW (n=102). Most isolates came from roots (228), while the remaining isolates, 189 and 154, came from leaves and twigs, respectively. Three plant samples, EB, ES, and EW, gave the highest numbers of actinobacteria from roots, while plant samples EC and EK yielded the highest numbers of actinobacteria from leaves (Table 1). There were many studies reporting that most endophytic actinobacteria were obtained from root tissue (Kaewkla and Franco, 2013; Saikia et al., 2022; El-Akshar et al., 2024). The EK sample grows in highly saline soil (12 dS/m) and extremely acidic soil, which might affect actinobacteria to adapt to colonize in leaf tissues for their survival. This correlates with the work of Shan et al. (2018), who isolated most strains of endophytic actinobacteria from leaf and stem tissue. The other report showed that most isolates of endophytic actinobacteria were obtained from the leaf tissues of E. camaldulensis (Kaewkla and Franco, 2013).
Identification of endophytic actinobacteria
Identification of all actinobacteria based on their morphology and 16S rRNA gene sequences showed that most isolates belonged to the genus Streptomyces (552, 96.7%), and the rest of the isolates were non-Streptomyces (19, 3.3%), comprising ten genera: Cellulosimicrobium (3), Kocuria (3), Brevibacterium (2), Micrococcus (2), Microbacterium (2), Peterkaempfera (2), Tsukamurella (2), Brachybacterium (1), Curtobacterium (1), and Gordonia (1) (Table 2; Figure 1). The details of the closest match of each isolate are shown in Table 2. It was found that Streptomyces was the most common genus of endophytic actinobacteria, which was also found in other studies (Himaman et al., 2016; Shan et al., 2018; Hazarika et al., 2022; Saikia et al., 2022).
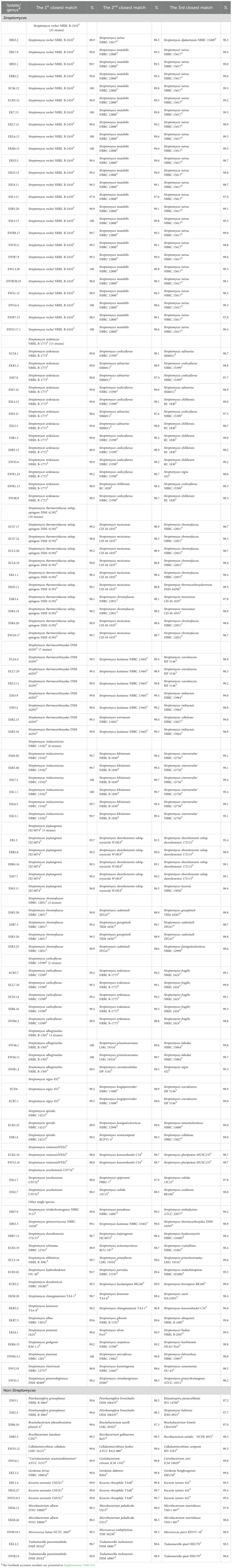
Table 2. The three closest matches of type strains of endophytic actinobacteria isolated from surface-sterilized tissues based on 16S rRNA gene similarity.
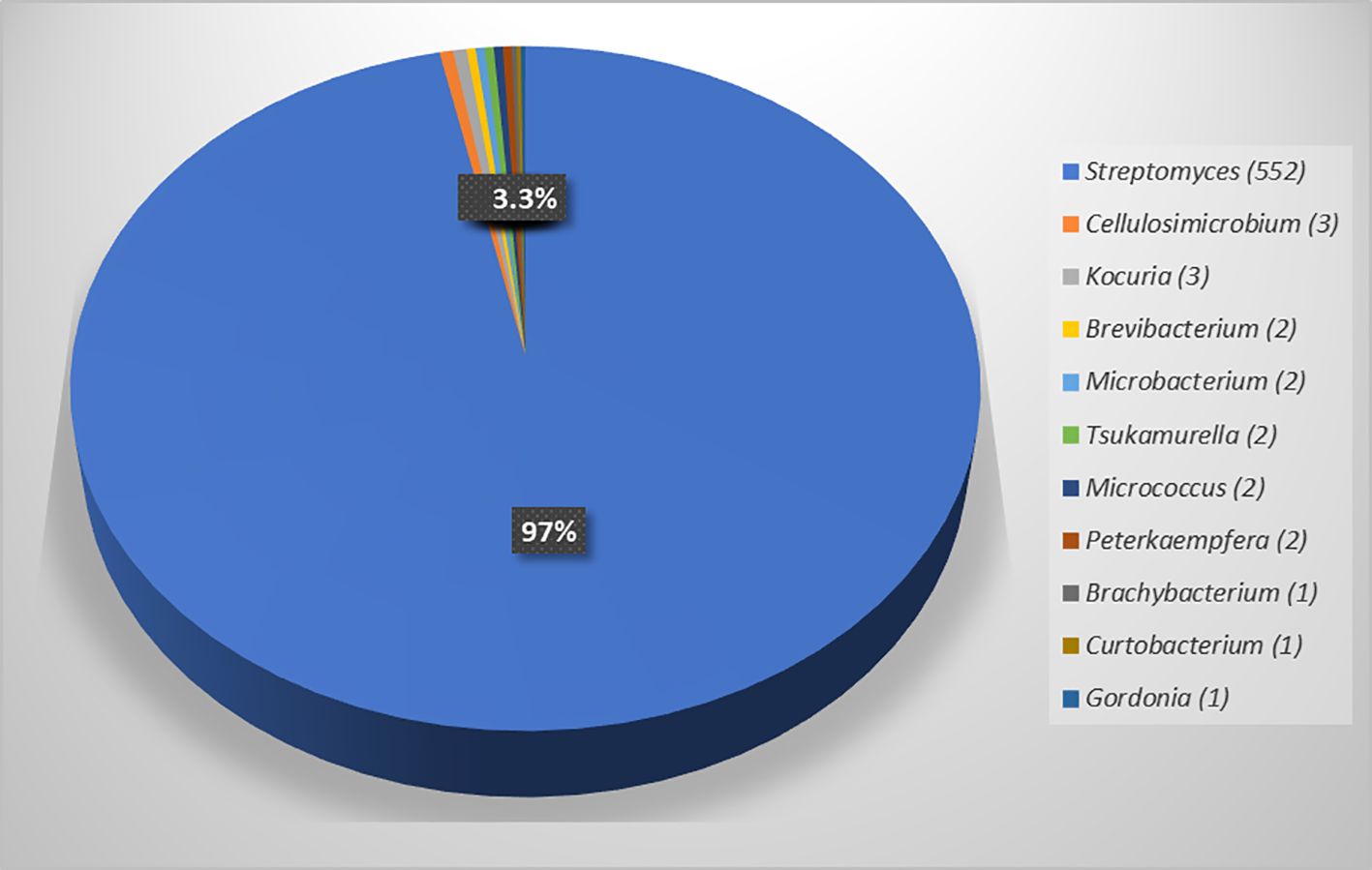
Figure 1. Numbers of endophytic actinobacteria in eleven genera and percentage of genus Streptomyces and non-Streptomyces genera isolated from five plant samples of Eucalyptus camaldulensis..
16S rRNA gene and phylogenetic tree analysis of Streptomyces and Peterkaempfera
One hundred and one isolates of the Streptomyces morphological groups were sequenced, and the 16S rRNA gene sequence analysis showed that these strains shared 16S rRNA gene similarity with the validly published species in the genus Streptomyces between 95.7% and 100% (Table 3). NJ, ML, and MP phylogenetic trees of the 16S rRNA gene constructed of all the strains of Streptomyces and Peterkaempfera along with their most closely related type strains are shown in Figures 2; Supplementary Figures S2, S3. The result showed that most of the Streptomyces strains from eucalyptus tissues position in the same clade or cluster with their closest type strains on three phylogenetic trees.
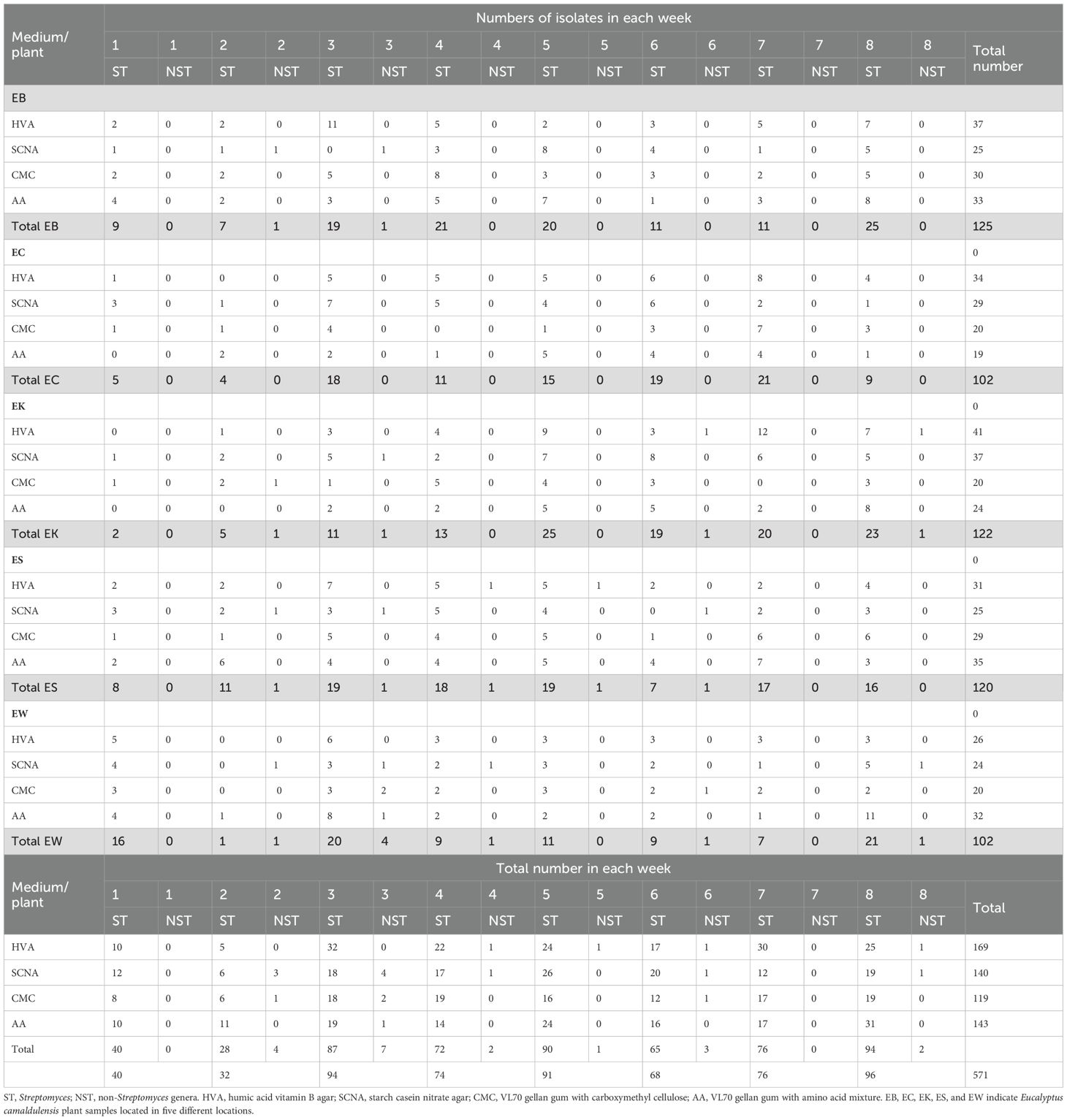
Table 3. Numbers of endophytic actinobacteria isolated from each plant and each medium between week 1 and week 8.
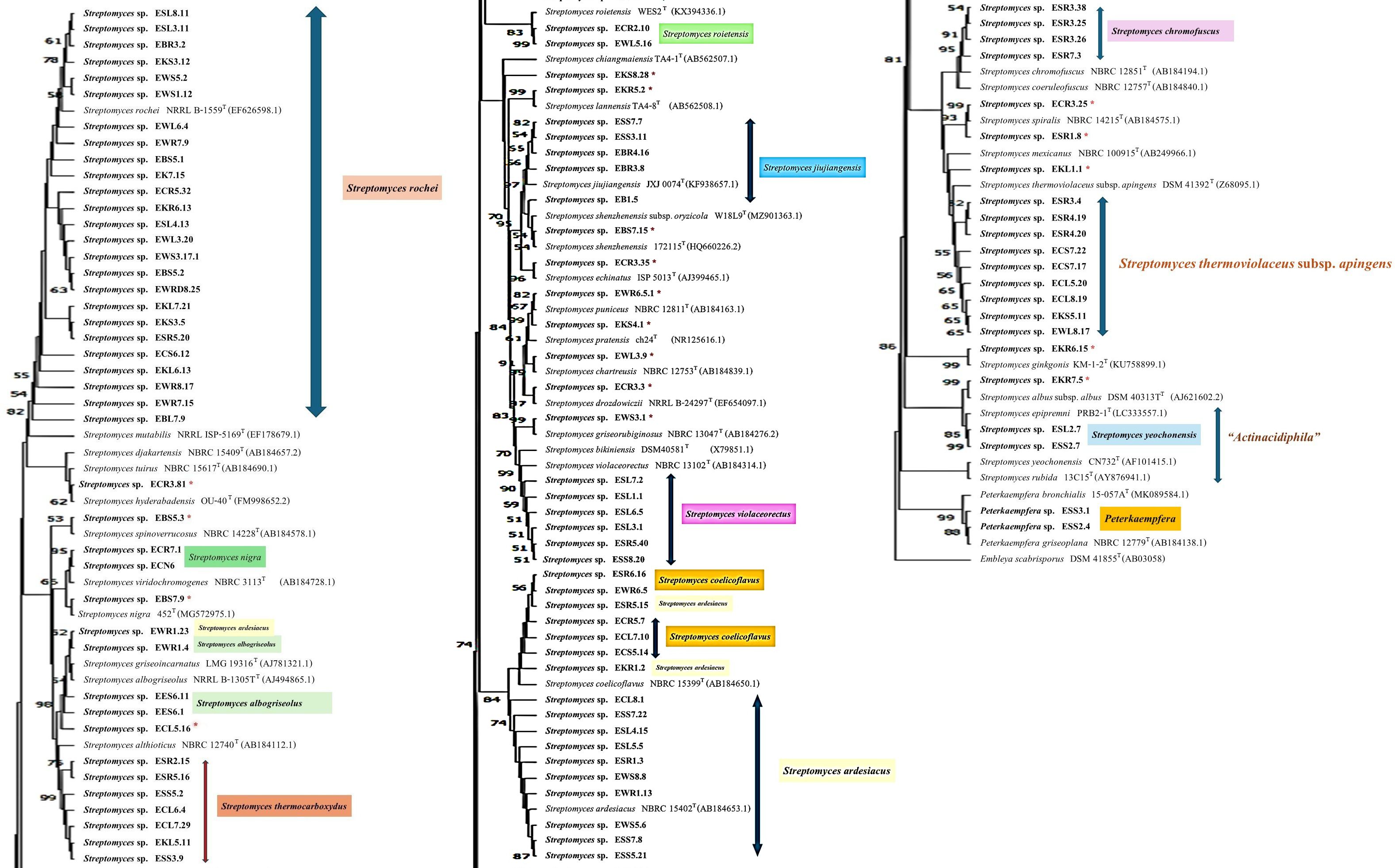
Figure 2. The neighbor-joining phylogenetic tree is based on 16S rRNA gene sequences of 101 strains of Streptomyces and 2 strains of the genus Peterkaemfera and their closely related members in the genera Streptomyces and Peterkaemfera and Embleya scabrisporus DSM 41855T as the out-group. Bootstrap values based on 1000 replicates are shown at the branch nodes. *indicate a single strain of a unique species of Streptomyces..
There were 25 isolates that were closely related to Streptomyces rochei NRRL B-2410T, sharing 16S rRNA gene similarity with this type strain between 97.8% and 100%. All plant samples contained isolates that were closely related to this type strain. These isolates were clustered with the closest type strain on the NJ tree with low bootstrap numbers. However, strains EBS5.1 and EBL7.9 were positioned in a different cluster with the type strain, S. rochei NRRL B-2410T, on the MP tree, while strain EBL7.9 was positioned in a different cluster with the closest type strain on the ML tree with low bootstrap numbers.
The next largest group comprised 13 isolates that shared the highest 16S rRNA gene similarity with Streptomyces ardesiacus NRRL B-1773T, between 98.8% and 99.9%. The members of this group were divergent. Ten isolates were grouped in the same cluster with this closest type strain with low bootstrap numbers. However, two isolates, ESR5.15 and EKR1.2, were positioned in the same cluster with Streptomyces coelicoflavus NBRC 15399T on the NJ tree. Strain ESR5.15 is also positioned in the same cluster of S. coelicoflavus NBRC 15399T on ML and MP trees with low bootstrap numbers (Supplementary Figures S2, S3). Isolate EWR1.23 was positioned in the same cluster of type strain, Streptomyces albogriseolus NRRL B-1305T, on NJ, ML, and MP trees.
Ten isolates shared the highest 16S rRNA gene similarity with the type strain, Streptomyces thermoviolaceus subsp. apingens DSM 41392T, between 98.1% and 99.2%. Nine isolates were grouped together and positioned in the same cluster with the type strain with low bootstrap numbers on NJ, ML, and MP trees. One isolate, EKL1.1, formed a separate clade with these nine isolates, but it was positioned close to the closest type strain on all phylogenetic trees. There were seven isolates that were closely related to Streptomyces thermocarboxydus DSM 44293T, sharing 16S rRNA gene similarity between 99.3% and 99.8%. These isolates were clustered together with low bootstrap numbers on NJ, ML, and MP trees. They were isolated from EC, ES, and EK samples.
There were 6, 5, 5, 4, and 3 isolates that were closely related to Streptomyces violaceorectus NBRC 13102T, Streptomyces jiujiangensis JXJ 0074T, Streptomyces coelicoflavus NBRC 15399T, Streptomyces chromofuscus NBRC 12851T, and Streptomyces albogriseolus NRRL B-1305T, respectively. The six isolates, closely related to S. violaceorectus NBRC 13102T, were positioned in the same cluster as the type strains on NJ trees, which have high bootstrap numbers. However, isolate ESR5.40 was located in a separate phylogenetic tree, distant from the other isolates on both the ML and MP trees. Five isolates, which were closely related to S. jiujiangensis JXJ 0074T, were divergent. Four isolates were positioned in the same cluster and close to the type strain with high bootstrap numbers on the NJ and ML trees but showed low bootstrap numbers on the MP tree. One isolate, EB1.5, was positioned in a different cluster with the type strains, and the other four isolates were on NJ, ML, and MP trees.
Five isolates, which were closely related to S. coelicoflavus NBRC 15399T, were positioned in the same cluster with the type strain on NJ, ML, and MP trees with high bootstrap numbers. They were closely related to members of the S. ardesiacus group. Three isolates, which were closely related to S. albogriseolus NRRL B-1305T, were positioned in the same cluster of this type strain on NJ, ML, and MP trees with low bootstrap numbers.
There were two isolates; each belonged to Streptomyces spiralis NBRC 14215T and Streptomyces roietensis WES2T. The position of these isolates was close to their type strains on NJ, ML, and MP trees with high bootstrap support. Two isolates each were closely related to Streptomyces yeochonensis CN732T and Streptomyces nigra 452T, but they were positioned in the different cluster of these closest type strains on NJ, ML, and MP trees with low bootstrap numbers. S. yeochonensis CN732T was previously reclassified as the genus Actinacidiphila, but it was a heterotypic synonym of Streptomyces (Madhaiyan et al., 2022).
There were fifteen isolates of Streptomyces, which were a unique species isolated from each plant sample. All of them are positioned close to their closest type strain on the NJ tree with high bootstrap numbers. Only strain ECL5.16 was positioned in the different cluster with its closest type strain, Streptomyces althioticus NBRC 12740T, on the ML and MP trees with low bootstrap numbers. Two isolates of the ES sample belonged to the genus Peterkaempfera, which are closely related to the closest type strain, Peterkaempfera griseoplana NBRC 12779T, and formed the same cluster with this type strain with high bootstrap numbers on NJ, ML, and MP trees. Peterkaempfera was previously identified as the genus Streptomyces and later reclassified as the genus Peterkaempfera (Madhaiyan et al., 2022).
16S rRNA gene and phylogenetic analysis of non-Streptomyces strains
We constructed 16S rRNA gene NJ, ML, and MP phylogenetic trees that included all strains from ten non-Streptomyces genera. The result showed that each isolate was in the same cluster with its closest type strain with high bootstrap support on NJ, ML, and MP trees (Figures 3; Supplementary Figures S4, S5). In addition, two isolates of Kocuria from different plant samples (EB and EW) were in the same clades. Similarly, two isolates of Tsukamurella from plants (EK and EW) were in the same clade with high bootstrap support on NJ, ML, and MP trees, but they are in a different clade with the closest type strain on the NJ tree.
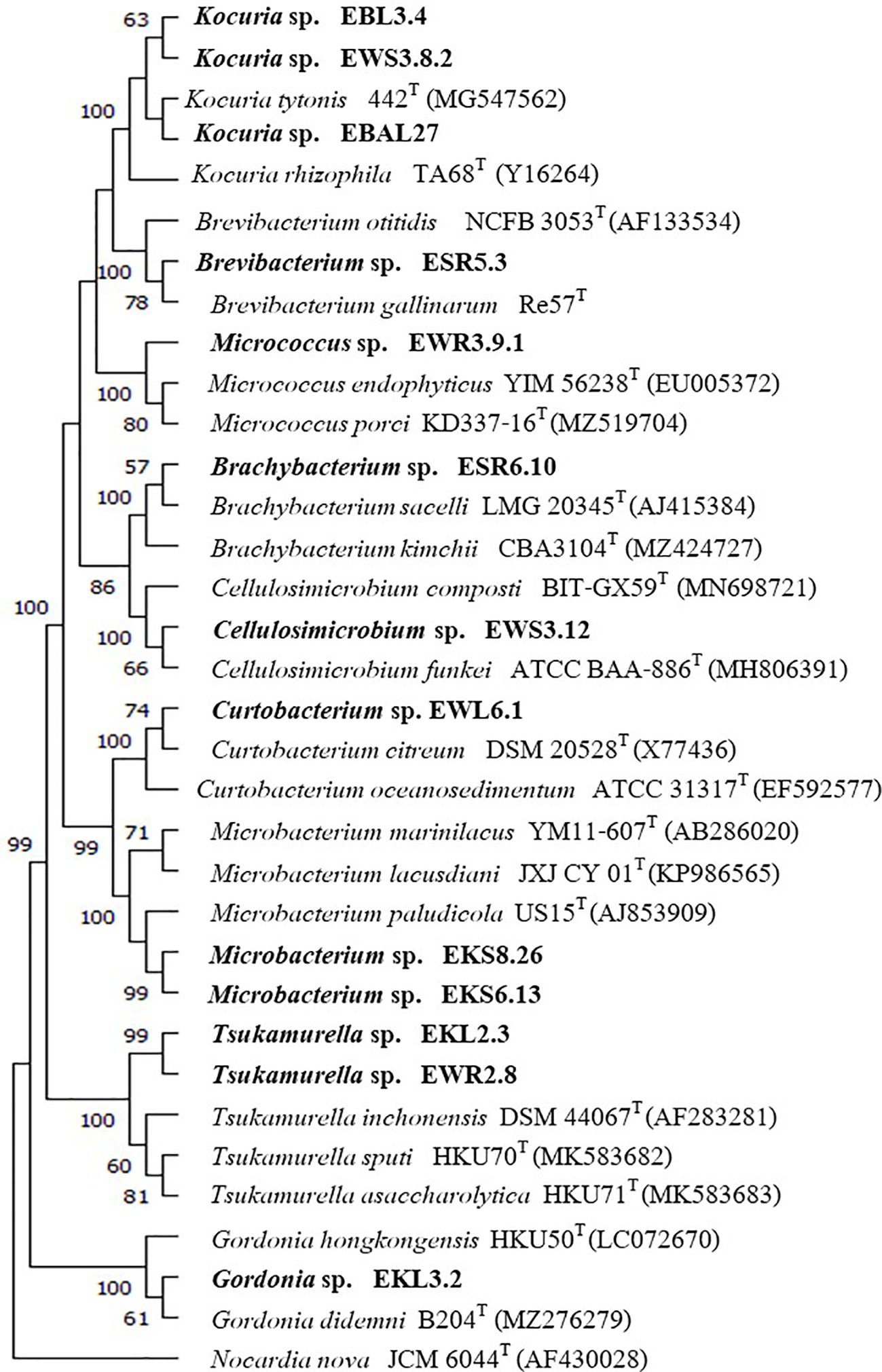
Figure 3. Neighbor-joining phylogenetic tree is based on the 16S rRNA gene sequences of 13 strains from 9 genera and their closely related members, with Nocardia nova JCM6044T serving as the out-group. Bootstrap values based on 1000 replicates are shown at the branch nodes.
Identification of non-actinobacteria isolates
We identified non-actinobacteria isolates based on 16S rRNA gene sequence analysis. There were 10 genera obtained: Aureimonas, Bacillus, Chryseobacterium, Deinococcus, Massilia, Methylobacterium, Pseudomonas, Serratia, Staphylococcus, and Stenotrophomonas. Supplementary Table S1 displays the details of the closest match for each isolate. The EK sample, which was grown in high-salinity soil and extremely acidic soil, comprised the most different genera of non-actinobacteria, at 8 genera: Bacillus, Chryseobacterium, Deinococcus, Methylobacterium, Pseudomonas, Seratia, Staphylococcus, and Stenotrophomonas. The EW sample was the second-best plant, which comprised five different genera, while the ES and EB samples contained four and three different genera, respectively. The non-actinobacteria strain was not isolated from the EC sample. EK and EW samples contained a unique genus of Stenotrophomonas and Aureimonas, respectively.
The study reported isolating the Aureimonas strain C2P003 from the leaf of the Fraxinus excelsior tree. This strain may suppress the colonization of ash dieback caused by the invasive pathogen, Hymenoscyphus fraxineus (Becker et al., 2022). Deinococcus was isolated from EK and EW plants. It was reported that Deinococcus are known for their resistance to extreme stresses, including radiation, oxidative stress, desiccation, and high temperature (Gerber et al., 2015). It was correlated from this study that the EK plant grown in high-salt soil yielded extremophile bacteria.
Biodiversity of endophytic actinobacteria isolated from each medium
HVA was the best medium for yielding endophytic actinobacteria at 170 isolates, while SCNA and AA media yielded similar numbers at 140 and 143 isolates, respectively. CMC medium yielded the lowest numbers of endophytic actinobacteria at 118 isolates (Table 1). The majority of Streptomyces isolates emerged from plant tissues in weeks 3, 5, and 8 at 87, 90, and 94 isolates, respectively. Most of the non-Streptomyces isolates emerged at week 3 at seven isolates.
HVA gave the highest numbers of Streptomyces at weeks 3, 4, and 7. SCNA gave the highest numbers of Streptomyces at weeks 1, 5, and 6, while AA medium had the highest numbers at weeks 2 and 8. SCNA was the best medium to yield the highest numbers of non-Streptomyces at weeks 2 and 3 (Table 3).
Figure 4 shows the total numbers of isolates at weeks 2, 4, 6, and 8. In the first two weeks, AA medium was the best to give the highest numbers of Streptomyces. HVA gave the highest numbers of total Streptomyces isolates at weeks 4, 6, and 8 (N=68, 109, and 164). SCNA was the best medium to give the total numbers of non-Streptomyces isolates at weeks 2, 4, 6, and 8 (N=3, 8, 9, and 10). HVA gave the highest numbers of Streptomyces for the EK plant, which had a soil sample that was strongly saline and extremely acidic. Soil samples from the EB, EC, ES, and EW plants were moderately saline, with the highest numbers of isolates obtained from HVA for EB and EC, and from AA for ES and EW (Table 3).
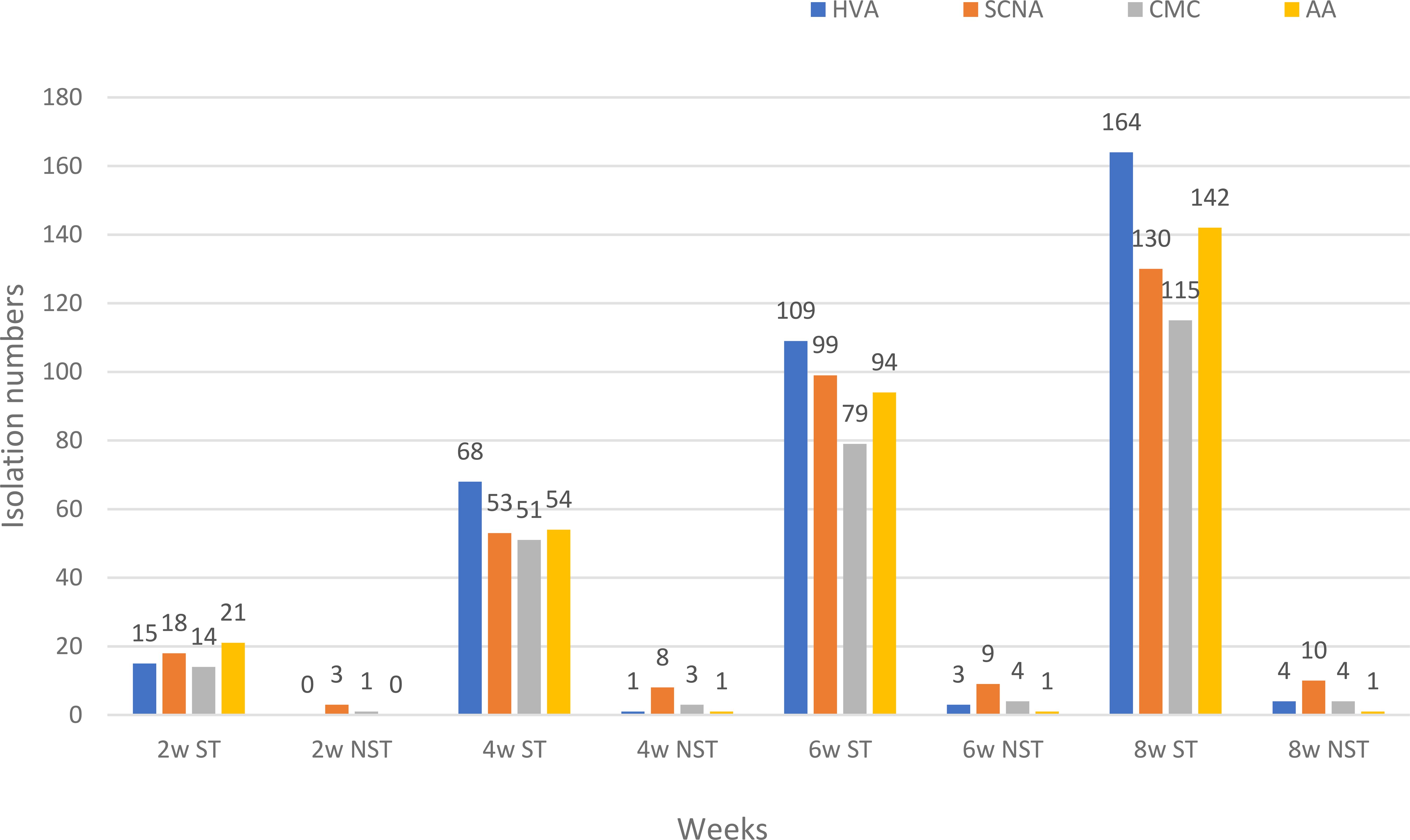
Figure 4. The total numbers of endophytic actinobacteria isolated from each plant sample at weeks 2, 4, 6, and 8 on four different media. HVA, humic acid vitamin B agar; SCNA, starch casein nitrate agar; CMC, VL70 gellan gum with carboxymethyl cellulose; AA, VL70 gellan gum with amino acid mixture; w, week; ST, genus Streptomyces; NST, Non-Streptomyces genera.
The influence of media on the biodiversity of unique species of Streptomyces and non-Streptomyces isolates was studied. The result indicated that SCNA was the best to yield seven unique species of Streptomyces, followed by HVA (n=4), AA (n=3), and CMC (n=1) (Table 4). Furthermore, SCNA was the best medium to gain the highest number of non-Streptomyces at 10, followed by HVA (n=4), CMC (n=4), and AA (n=1) (Table 4). SCNA also gave the highest numbers of different genera of non-Streptomyces at seven, in which Kocuria, Cellulosimicrobium, Tsukamurella, Micrococcus, Peterkaempfera, and Brachybacterium are rare genera. HVA and AA media gave only 2 and 1 genera of non-Streptomyces isolates, respectively. Although CMC gave the lowest number of unique species of Streptomyces, it gave one unique isolate of Curtobacterium, which was a rare genus. Currently, there are only thirteen validly published species of Curtobacterium (Parte et al., 2020) (accessed July 10, 2025).
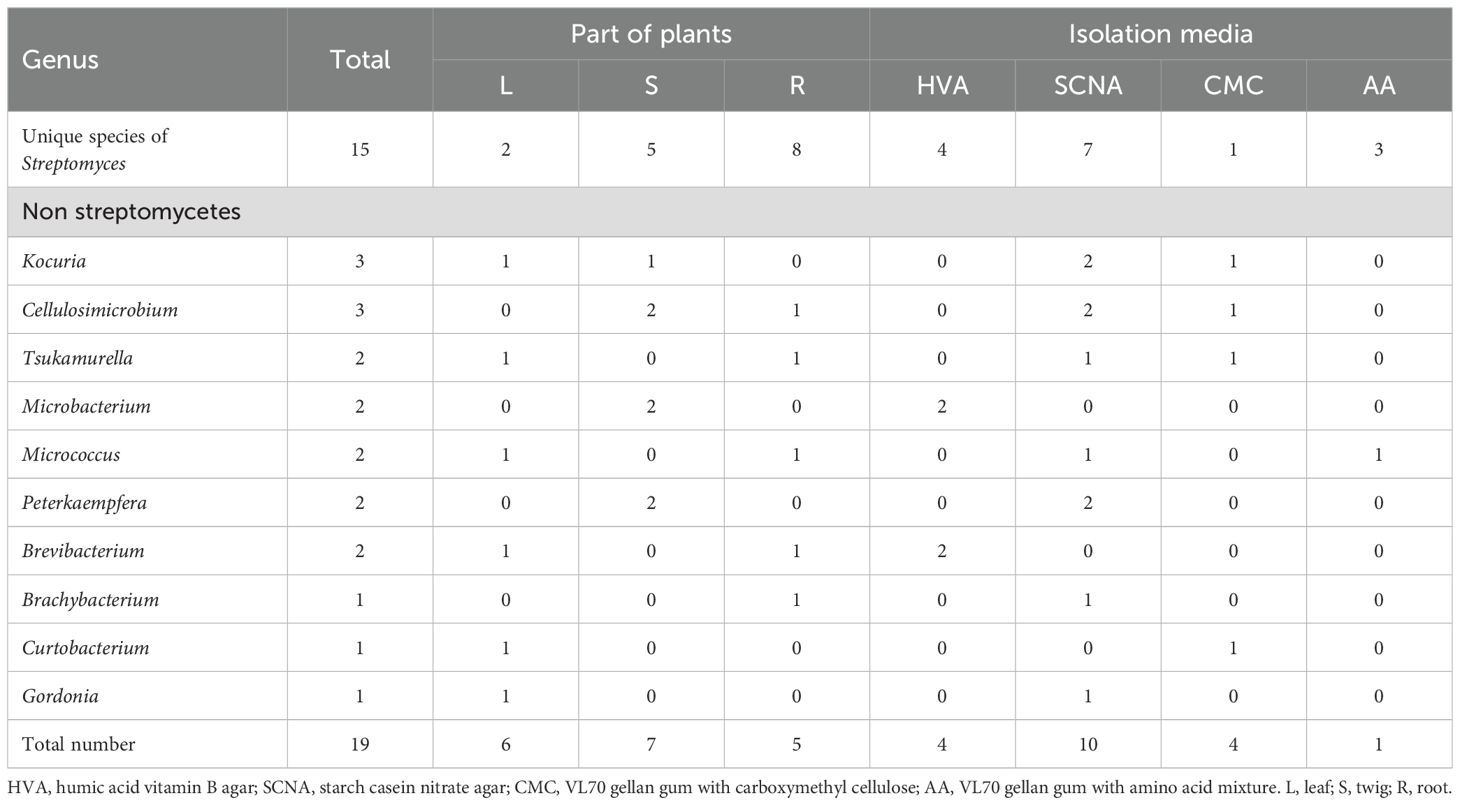
Table 4. Numbers of unique species of Streptomyces and all non-Streptomyces isolates isolated from each part of plants and each isolation medium.
Overall, for this study, HVA and SCNA were suitable media to isolate the highest number of Streptomyces and non-Streptomyces isolates, respectively. AA medium was the second one, which gave the highest number of Streptomyces isolates. Moreover, SCNA was the best media to yield the different genera of actinobacteria, including different species of Streptomyces. In addition, CMC media also gave the unique member of a rare genus. Therefore, the variety of isolation media can influence both numbers of isolates and numbers of different genera, including rare genera. We suggested that HVA and SCNA were the first choices to isolate endophytic actinobacteria from plants grown in saline soil. Also, CMC and AA media should be included as isolation media to enhance the number of rare genera and substantial biodiversity of Streptomyces species.
According to Qin et al. (2011) and Kaewkla and Franco (2013), isolation media have influenced the biodiversity of endophytic actinobacteria, including high numbers of unique species and rare genera. There were many reports showing that HVA was the best medium for isolating actinobacteria (Kaewkla and Franco, 2013; Himaman et al., 2016; Vo et al., 2021). Sardi et al. (1992) used starch casein medium to isolate endophytic actinobacteria from the roots of 28 plant species and obtained a good number of non-Streptomyces strains.
Biodiversity of endophytic actinobacteria isolated from each plant sample
Plant samples in this study have influenced the biodiversity of both Streptomyces and non-Streptomyces isolates. EB gave the highest number of Streptomyces isolates at 123, with the rest from EK, ES, EC, and EW at 118, 115, 102, and 94 isolates, respectively. Although the EB sample comprised the highest numbers of Streptomyces, the biodiversity of Streptomyces species groups was the lowest at 4 groups. The EC sample contained the highest number of Streptomyces species groups at 12 groups, while the ES, EK, and EW samples contained 10, 9, and 9 Streptomyces species groups, respectively. Moreover, EC and EK samples comprised the highest numbers of unique Streptomyces species groups at 5, while the EW sample comprised unique species groups at 4. ES and EB samples comprised 3 and 2 unique Streptomyces species groups, respectively (Table 5).
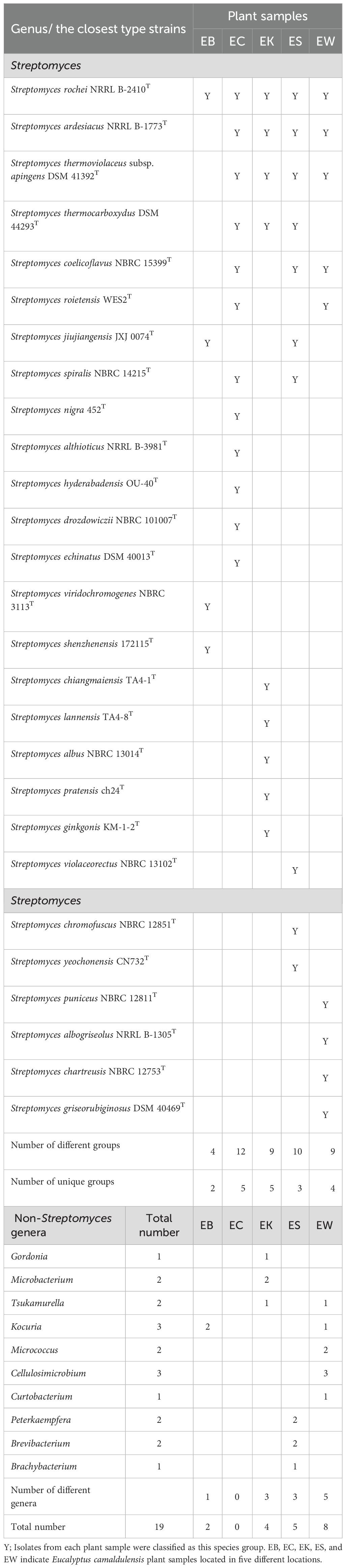
Table 5. Streptomyces species groups based on 16S rRNA gene similarity and numbers of non-Streptomyces from each plant sample.
Endophytic actinobacteria from the EK sample that were grown in highly salty soil at 12 dS/m and low pH at 3.9 may have adapted to survive in stressful conditions. Then, it would contain a greater biodiversity of actinobacteria than other moderately saline areas. A study by Himaman et al. (2016) obtained actinomycetes from the roots (23 samples) and rhizosphere soil (27 samples) of healthy Eucalyptus (E. camaldulensis) trees in different provinces of Thailand. There were 439 isolates that belonged to the genus Streptomyces, while 38 isolates were non-Streptomyces in nine different genera. Kaewkla and Franco (2013) isolated endophytic actinobacteria from surface-sterilized plant tissues of Eucalyptus microcarpa (grey box) and Eucalyptus camaldulensis (red gum) by using low-nutrient agar and long incubation of isolation plates. Although these two plant samples were the same genus, there were different numbers of actinobacterial isolates from gray box and red gum: 191 and 39 isolates, respectively. The grey box comprised Streptomyces (N=122) and non-Streptomyces (N=69) in nine different genera, while the red gum comprised Streptomyces (N=37) and non-Streptomyces (N=2) in two different genera.
There were nineteen non-Streptomyces isolates obtained in ten genera. The EW sample gave the highest number of non-Streptomyces at eight isolates, and the EW sample also gave the highest number of different genera at 5. There were 5, 4, and 2 isolates from ES, EK, and EB samples, respectively. ES, EK, and EB samples gave different numbers of genera at 3, 3, and 1, respectively. The EC sample, which comprised the highest numbers of different group species of Streptomyces, did not contain non-Streptomyces isolates. The ES plant sample was a valuable source because it yielded two rare genera: Peterkaempfera and Brachybacterium. These genera had only two and thirty-one valid published species at the time of this writing (Parte et al., 2020) (accessed July 10, 2025). EW and EK plant samples were also prospective sources to yield the rare genus Tsukamurella, which comprises 22 validly published species. We isolated Cellulosimicrobium, a rare genus with only 10 validly published species, exclusively from the EW plant. This suggests that the EW plant may harbor unique microbial diversity that could be crucial for further taxonomic studies. Additionally, the presence of these rare genera highlights the potential ecological significance of the EW plant’s environment, warranting further investigation into its role in supporting microbial life.
The relative genus abundance of endophytic actinobacteria from five plant samples revealed that Streptomyces was the predominant genus discovered from all plant samples, which ranged from 92.2 to 100% (Figure 5). Moreover, non-Streptomyces genera in the order Micrococcales were a predominant group recovered from four plant samples: EB, EK, ES, and EW. This group includes seven genera: Brevibacterium, Brachybacterium, Cellulosimicrobium, Curtobacterium, Kocuria, Microbacterium, and Micrococcus. However, the relative genus abundance of these genera in each plant sample was very low, between 1 and 2%. Moreover, the members of the order Mycobacteriales, comprising two genera, Gordonia and Tsukamurella, were found in low relative abundance in EK and EW plants (1-2%). The members of the genus Peterkaempfera were discovered only within the tissues of the ES plant sample with low relative abundance at 1%. This genus was a rare one and belonged to the family Streptomycetaceae, which is the same family as the genus Streptomyces.
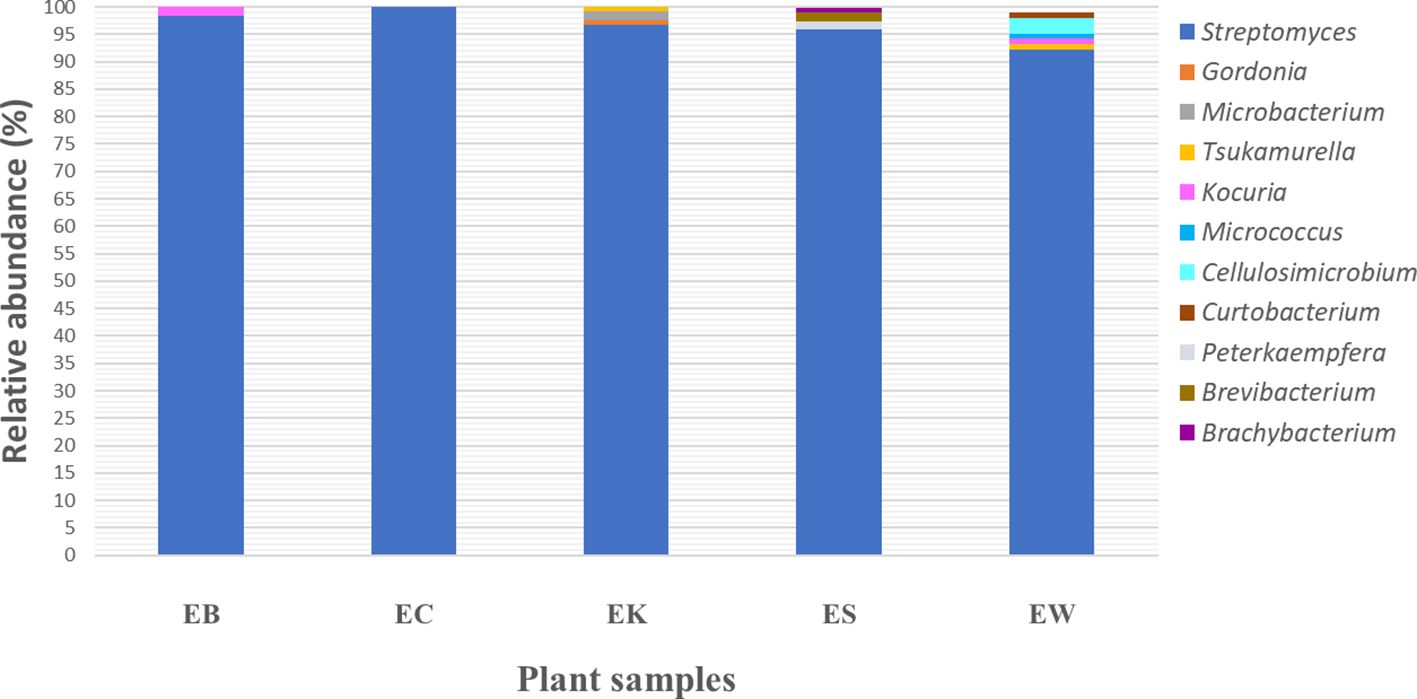
Figure 5. Bacterial community relative abundance analysis at the genus level from each plant sample. Different colors indicate eleven different genera. EB, EC, EK, ES, and EW indicate samples of the Eucalyptus camaldulensis plant from five different locations.
The analysis of plant parts comprising unique species of Streptomyces and non-Streptomyces genera is presented in Table 4. Root tissues gave the highest numbers of unique species of Streptomyces at eight different species. Stem tissues gave the highest numbers of non-Streptomyces at 7, followed by leaf and root tissues at 6 and 5 isolates, respectively.
There were some reports of non-Streptomyces genera that exhibited plant-growth-promoting capabilities. These findings suggest that the diversity of microbial sources for potential plant growth promoters may be broader than previously thought, opening new avenues for an agricultural area. Microbacterium was isolated from Limonium sinense, enhancing salinity tolerance in the plant (Qin et al., 2018). Brachybacterium paraconglomeratum strain SMR20 isolated from Chlorophytum borivilianum improved the salt tolerance and yield of the host plant (Barnawal et al., 2016). Cellulosimicrobium 60I1 was a promising strain for preparing a bioinoculant to promote the growth of a pepper plant (Lobato-Ureche et al., 2023). Curtobacterium albidum strain SRV4 had the ability to fix nitrogen (N2) and produce exopolysaccharide (EPS), hydrogen cyanide (HCN), IAA, and ACC deaminase. Moreover, Curtobacterium strain SRV4 promotes paddy plants by enhancing plant growth parameters, photosynthetic pigment efficiency, membrane stabilization index, and proline content (Vimal et al., 2019). Kocuria strain ST19, which was isolated from halophytes, could show efficiency to mitigate salt stress conditions in tomato plants (Dif et al., 2021). In addition, Brevibacterium sediminis strain IBGE3C, which was isolated from a saline area, promoted the seedling growth of rice (Mahmud-Ur-Rahman et al., 2022). Therefore, non-Streptomyces strains from this study might have the ability to promote eucalyptus growth in salinity conditions, but it requires further study to prove their properties.
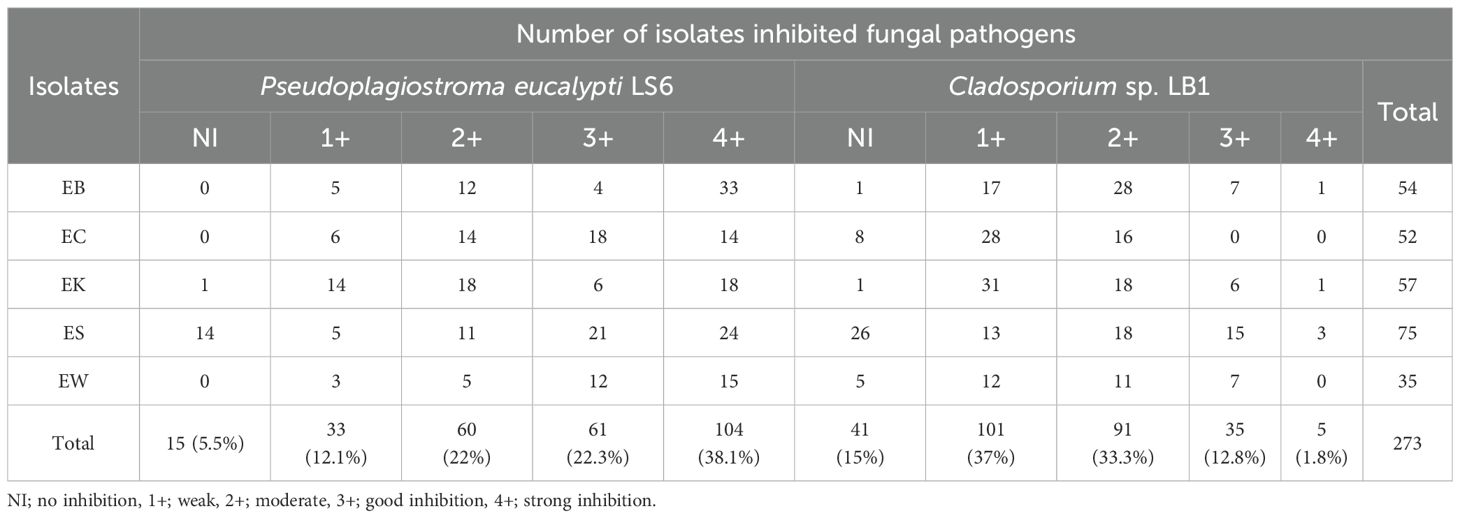
Antifungal activity
Two hundred and seventy-three isolates of endophytic actinobacteria were tested for antifungal activity against two eucalyptus pathogens, P. eucalypti LS6 and Cladosporium sp. LB1. There were more isolates showed good activity against P. eucalypti LS6 than Cladosporium sp. LB1. There were 61 (22.3%) and 104 (38.1%) isolates that showed good and strong inhibition against P. eucalypti LS6, while 35 (12.8%) and 5 (1.8%) isolates demonstrated good and strong inhibition against Cladosporium sp. LB1 (Table 6). There were EB isolates (n=33) that showed strong activity against P. eucalypti LS6 (Supplementary Figure S6), while there were ES isolates (n=3) that showed strong inhibition against Cladosporium sp. LB1 (Supplementary Figure S7). We isolated the fungal strains LS6 and LB1 from eucalyptus leaves with leaf spot and leaf blight symptoms, respectively. We tested both strains for disease symptoms on the surface-sterilized eucalyptus leaves using the detached leaf assay. The result showed that P. eucalypti LS6 and Cladosporium sp. LB1 caused severe and moderate lesions on the leaves, respectively (data not shown). Pseudoplagiostoma eucalypti was first named Cryptosporiopsis eucalypti by Sankaran et al. (1995), but it was later reclassified as Pseudoplagiostoma eucalypti. This fungus causes severe leaf spots on eucalyptus trees and is common in both tropical and temperate countries (Cheewangkoon et al., 2010). Lueangpraplut et al. (2013) reported that P. eucalypti caused leaf spot and shoot blight diseases on eucalyptus in Thailand. Zauza et al. (2023) reported the severe disease and dieback of Eucalyptus spp. in Brazil, caused by P. eucalypti. It was recently reported that this fungus showed the first serious outbreak of eucalyptus disease after more than two decades in northern India (Negi et al., 2024). Cladosporium spp. are endophytic, or dormant, fungi that have been found to spread disease on dead parts of many different host plants (Bensch et al., 2010). Eucalyptus ficifolia was infected by Cladosporium sp. in New Zealand (Farm Forestry New Zealand, 1992). All these actinobacterial isolates, which showed good and strong inhibition against these fungal pathogens, belonged to the genus Streptomyces. This was in line with other studies that found most isolates of the genus Streptomyces could inhibit fungi (Himaman et al., 2016; Pushpalatha et al., 2023; Shahid et al., 2021). It was reported that 273 isolates of endophytic actinobacteria from Eucalyptus microcarpa and Eucalyptus camaldulensis were tested for their antifungal activity against plant pathogens Phytophthora palmivora and Fusarium oxysporum (Kaewkla, 2009). The result showed that most strains belonging to the genus Streptomyces had good activity inhibiting these two fungal pathogens in vitro. Himaman et al. (2016) studied endophytic actinobacteria from eucalyptus by testing them against three types of fungi: P. eucalypti (Cryptosporiopsis eucalypti), Cylindrocladium sp., and Teratosphaeria destructans. As a result, 57%, 24.7%, and 50.5% of the isolates were able to inhibit these fungal pathogens, respectively, and most of these strains belonged to the genus Streptomyces.
Plant growth promoting study and bacterial inhibition
For the PGP study in vitro, 154 isolates were chosen because they showed good or strong activity against at least one tested fungus. They were also studied antibacterial activity against R. solanacearum TISTR 2069, which causes eucalyptus leaves to wilt. There were only two isolates (1.3%) from the EW sample showing good inhibition against this bacterium (Supplementary Figure S8), and they belonged to the genus Streptomyces (Table 7). It was reported that leaf wilt disease occurrence got 40% of the eucalyptus field, and destroyed plants exhibited reddening and wilting of the foliage, leaf drop, and branch dieback with wilting symptoms (Santiago et al., 2014). It was found that strains UFV-56 (Bacillus thuringiensis) and UFV-62 (Bacillus cereus), along with a commercial mixture of several rhizobacteria called Rizolyptus®, inhibit the bacterial wilt in eucalyptus trees (Santiago et al., 2015). Ralstonia wilt of chili pepper caused by R. solanacearum was inhibited by Streptomyces philanthi RL-1-178 (Boukaew et al., 2011).
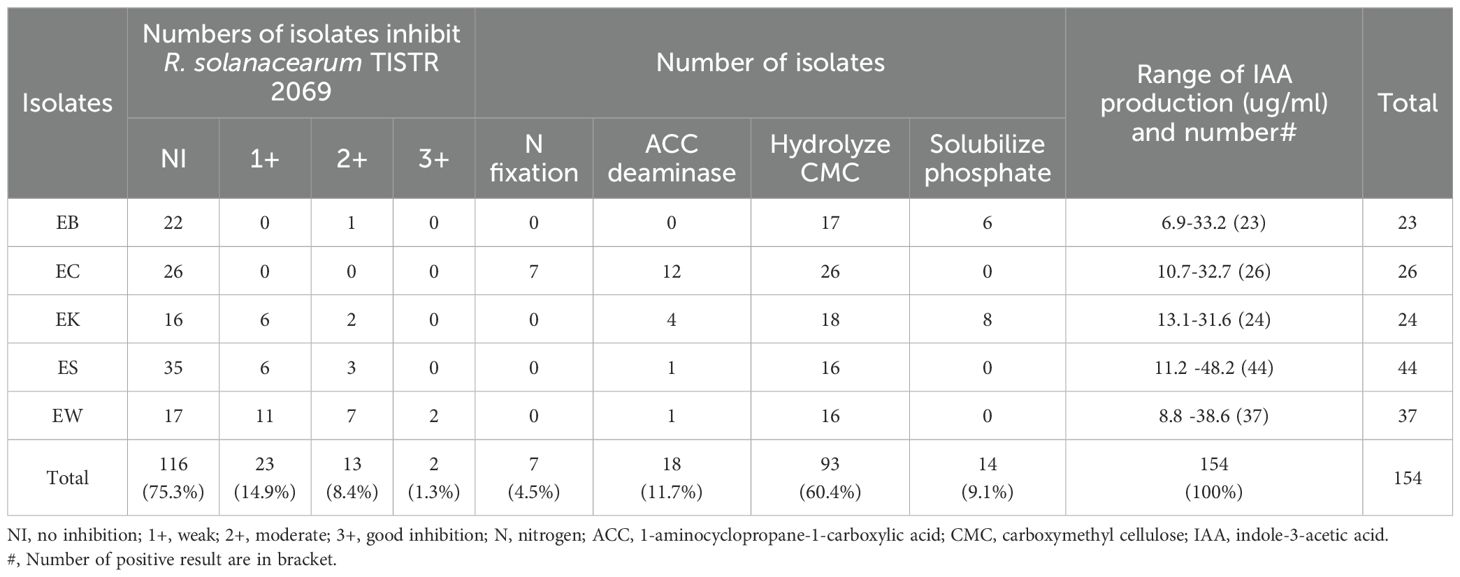
Table 7. Numbers of endophytic actinobacteria inhibited a bacterial pathogen and presented plant growth promoting properties.
The result showed that all these endophytic actinobacteria produced IAA which ranged from 6.9 to 48.2 ug/ml. ES isolate produced the highest IAA at 48.2 ug/ml. There were 93 strains (60.4%) that could hydrolyze CMC, and all isolates from EC samples could hydrolyze CMC. There were 18 isolates that could produce ACC deaminase and 14 isolates that could dissolve phosphate (Table 7; Figure 6). Most isolates from the EC sample produced ACC deaminase (66.7%). Only isolates from EK and EB samples (8 and 6 isolates) could solubilize phosphate. Only isolates from the EC sample could fix nitrogen (n=7). The mechanisms of actinobacteria to support plant growth in salinity stress include phytohormone production, especially auxin. Production of 1-aminocyclopropane-1-carboxylate (ACC) deaminase reduce ethylene gas and accumulation of osmoprotectant compounds such as proline, glycine-betaine, and polyamine. Some strains accumulate exopolysaccharides (EPS) and production of anti-oxidative enzymes to reduce oxidation stress (Atouei et al., 2019; Feikema and Baker, 2011). Thus, selected strains of endophytic actinobacteria producing IAA and ACC deaminase may have the ability to promote plant growth in salinity conditions in planta. Himaman et al. (2016) studied endophytic actinobacteria from eucalyptus tissues and rhizosphere soil for siderophores and IAA production and phosphate solubilization. The result showed that the Streptomyces sp. strain EUSKR2S82 could strongly inhibit all tested fungi and displayed plant growth-promoting traits in vitro and in planta.
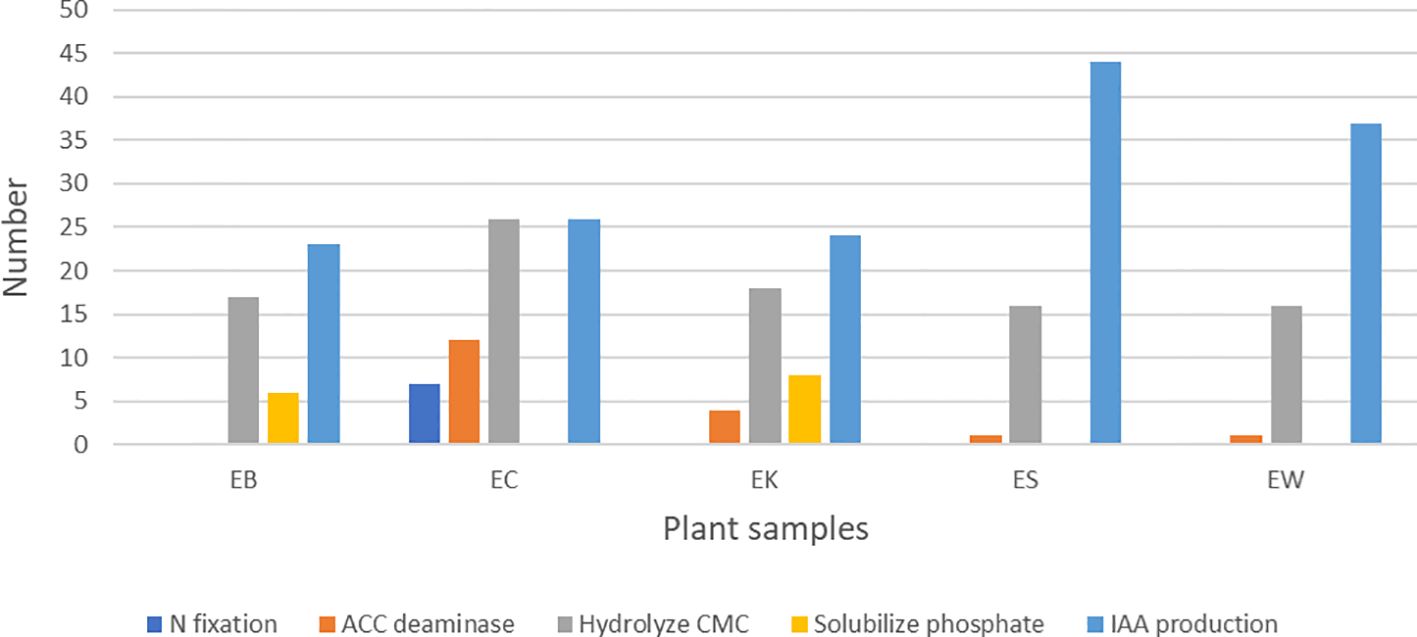
Figure 6. Numbers of endophytic actinobacteria present plant growth promoting traits in vitro from each plant sample. EB, EC, EK, ES, and EW indicate Eucalyptus camaldulensis plant samples located in different locations. N, nitrogen; ACC, 1-aminocyclopropane-1-carboxylic acid; CMC, carboxymethyl cellulose; IAA, indole-3-acetic acid.
Salt tolerance study
Based on the result of plant-growth-promoting properties, thirty-five potential strains of endophytic actinobacteria were selected to test on different sodium chloride concentrations up to 11% (w/v). Most isolates could grow well at 5% NaCl (w/v), while 6, 4, and 1 isolate could grow well at 7%, 9%, and 11%, respectively (Supplementary Table S2). Then, based on the PGP traits and ability of salt tolerance, the promising strains will be selected to test for PGP in planta in the further study.
Seed germination test
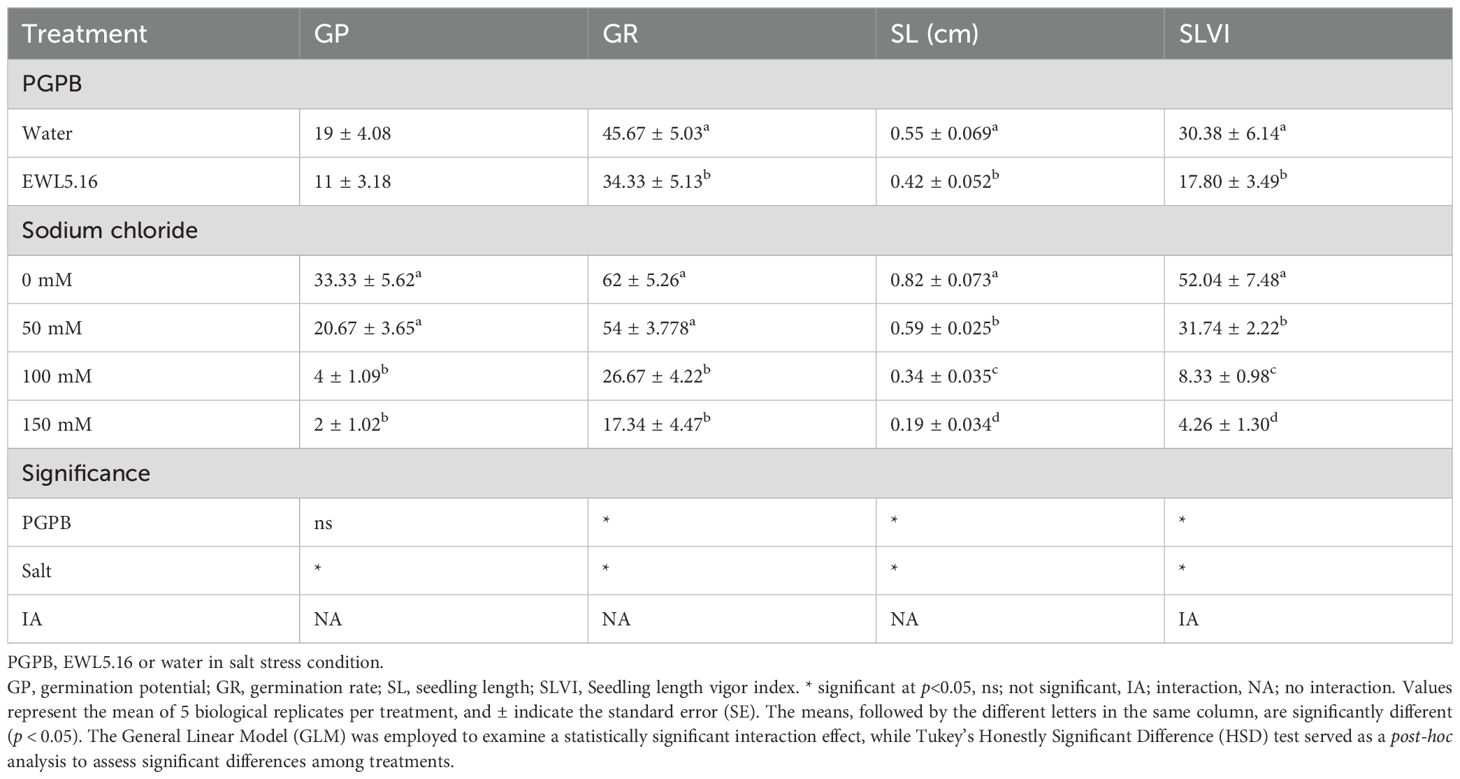
The test for germination potential (GP), germination rate (GR), and seedling length (SL) showed that there was no interaction between actinobacteria and salt (Supplementary Figure S9). The only parameter that had an interaction was the seedling length vigor index (SLVI) (Table 8). The treatment of Streptomyces strain EWL5.16 suppressed seed germination and seedling growth, as GR and SL were significantly lower than the control (water) (P<0.05), but it was not significant for GP. The concentration of salt between 100 mM and 150 mM did not significantly affect (P<0.05) GP, GR, and SLVI, while SL was significantly reduced (P<0.05) by these salt concentrations. However, treatment with strain EWL5.16 supported the SLVI by reducing the effect of salt at 50 mM compared with water (Figure 7). The SLVI of the treatment with strain EWL5.16 at 0 mM and 50 mM NaCl was not significantly different (P<0.05), while the SLVI of the control treatment at 50 mM was significantly lower than the SLVI at 0 mM NaCl (P<0.05).
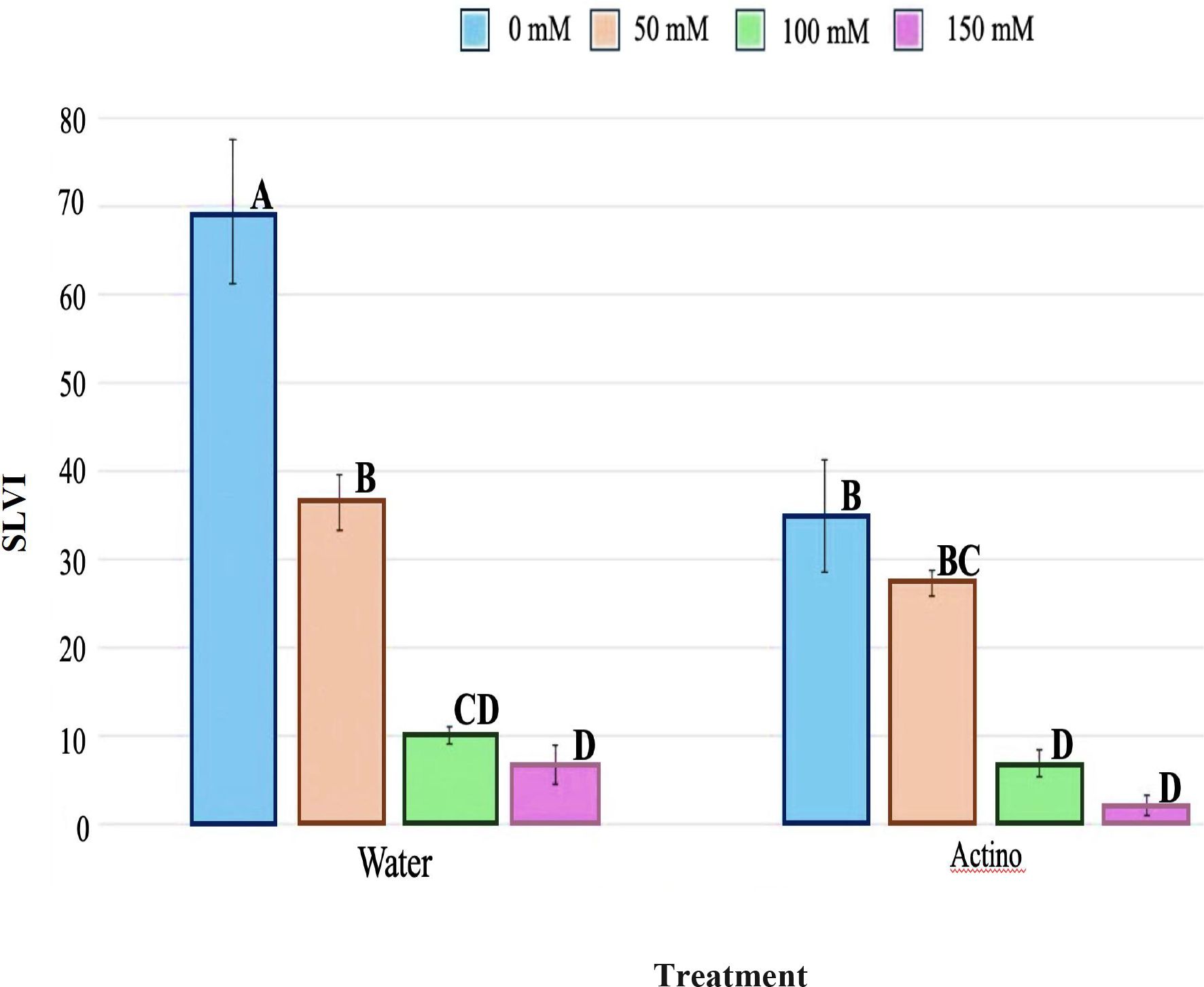
Figure 7. Effect of inoculation on seedling length vigor index (SLVI) on eucalyptus seedlings treated with strain EWL5.16 (Actino) or without strain EWL5.16 (Water) at 0, 50, 100, and 150 mM NaCl. Values represent the mean of five biological replicates per treatment, and bars indicate the standard error (SE). Means followed by the different letters for treatments are significantly different according to the Tukey Honestly Significant Difference (HSD) test (p < 0.05).
Plant growth promoting in planta
The PGP study in planta showed similar results with the seed germination test. Seedlings of the Streptomyces EWL5.16 treatment had lower shoot length, root length, and seedling length than the control, but these growth parameters were not significantly different from the control (Table 9). These findings suggested that while the Streptomyces EWL5.16 treatment may not adversely affect seedling growth in a statistically significant manner, it could still influence the overall development patterns of the plants. Further investigation is needed to explore the underlying mechanisms and potential long-term effects of this treatment on plant growth. However, the fresh weight of seedlings in strain EWL5.16 treatment was significantly higher than the control (P<0.05) (Table 9). Moreover, seed-borne fungi, Fusarium sp., severely infected seedlings, and most plants died at day 7 after planting (Figure 8). The survival rate of seedlings in strain EWL5.16 treatment (36.1%) was higher than in the control treatment (27.4%). In this study, the Streptomyces strain EWL5.16 suppressed seed germination and seedling growth, which might involve bioherbicide or cellulose production to inhibit root cell walls. Moreover, IAA production of strain EWL5.16 may negatively impact plant growth. The further study by varying the lower spore inoculum for coating seeds might reveal if this strain can be a potential inoculum to promote eucalyptus seedlings in the salinity stress. According to Bo et al. (2019), the Streptomyces strain 329 showed the potential bioherbicidal efficacy by testing on grass and broadleaf weeds for phytotoxic activity. At pre-emergence application, the phytotoxic efficacy of strain 329 to Digitaria sanguinalis and Sorghum bicolor on seed germination was 90.4% and 81.3%, respectively. Streptomyces sp. KRA16–334 was grown on M3 medium, and its ability to kill 10 weeds was tested using the culture filtrate. The result showed that the 2-fold dilution applied to leaves resulted in complete control of nine weed species (Kim et al., 2024).
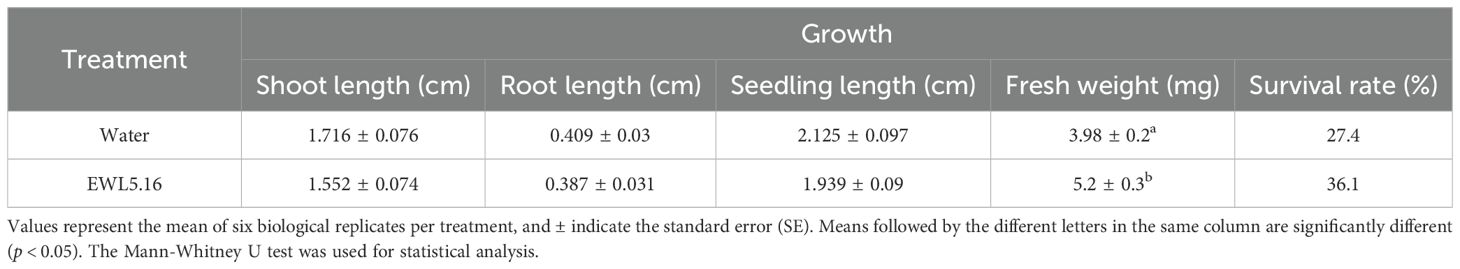
Table 9. Effect of inoculation on plant growth parameters and survival rate (%) of eucalyptus seedlings treated with water or strain EWL5.16.

Figure 8. Plant growth-promoting test of endophytic actinobacteria on eucalyptus seedlings grown on sterilized sand for 14 days. (A) Plant growing tray with seedlings treated with strain EWL5.16. (B) Plant growing tray treated with water (red arrows: seed-borne fungi destroyed seedlings). (C) Eucalyptus seedling treated with water. (D) Eucalyptus seedling treated with strain EWL5.16.
Genome comparison study
The genomes of Streptomyces strains EKR5.2 and ESS7.8 were compared with their closest type strains. Strain EKR5.2 had the highest dDDH, ANIb, and ANIm values at 96.2%, 99.3%, and 99.6% with Streptomyces lannensis JCM16578T. Strain ESS7.8 shared the highest dDDH, ANIb, and ANIm values at 94.2%, 99.0%, and 99.3% with Streptomyces ardesiacus NBRC 15402T. Streptomyces strains ECR2.10 and EWL5.1 were very close and had a 99.9% similarity in their 16S rRNA gene. The dDDH, ANIb, and ANIm values for these two strains were 99.9%, 99.9%, and 99.99%, respectively. The closest type strain, which shared the highest 16S rRNA gene similarity at 99.0% with these two strains, was Streptomyces roietensis WES2T. However, this type strain has not been sequenced in the genome. Therefore, there is no data on the genome comparison study between this type strain and strains ECR2.10 and EWL5.1.
Micrococcus strain EWR3.9.1 shared the highest dDDH, ANIb, and ANIm values with the type strain Micrococcus luteus ATCC 4698T at 76.4%, 96.7%, and 97.4%. The species-level definition should have dDDH and ANI values lower than the threshold of 70% (Meier-Kolthoff et al., 2013) and 95-96% (Richter and Rosselló-Móra, 2009), respectively. Therefore, Streptomyces strains EKR5.2 and ESS7.8 and Micrococcus strain EWR3.9.1 belonged to the known species. Streptomyces strains ECR2.10 and EWL5.1 were the same species. However, to confirm their novelty, a genome comparison with the closest species, S. roietensis WES2T, was necessary.
The TYGS phylogenomic tree of Streptomyces strains EKR5.2, ESS7.8, ECR2.10, and EWL5.1 also supports the dDDH, ANIb, and ANIm values between these strains and their closest type strains. Strains EKR5.2 and ESS7.8 were in the same clade with their closest type strains with high bootstrap support. Also, strains ECR2.10 and EWL5.1 were in the same clade and in the separated cluster with other type strains (Figure 9). The TYGS phylogenomic tree of Micrococcus strain EWR3.9.1 showed that it formed the same clade as the closest type strain, M. luteus ATCC 4698T, with the high bootstrap number (Figure 10).
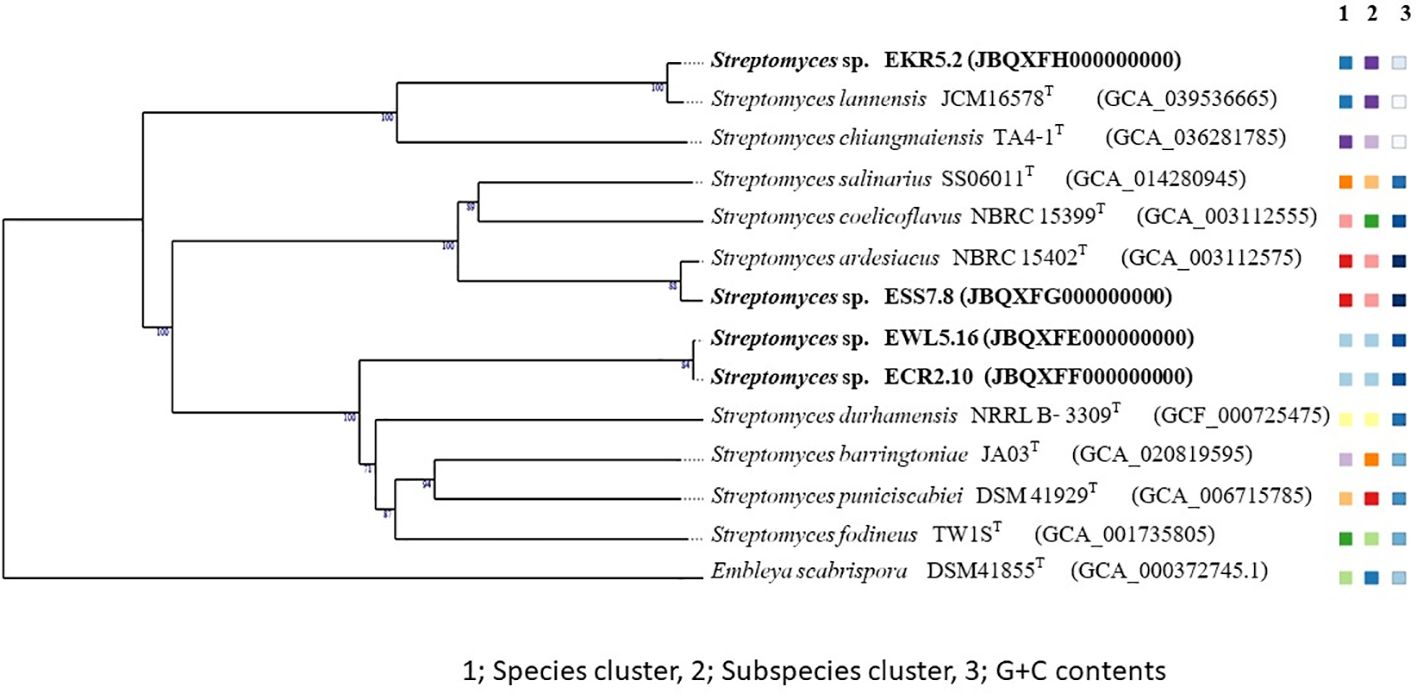
Figure 9. A phylogenomic tree based on the TYGS result shows the relationship between Streptomyces strains EKR5.2, ESS7.8, ECR2.10, and EWL5.16, as well as their closely related type strains. The tree was inferred with FastME 2.1.6.1 (Lefort et al., 2015) from GBDP distances calculated from genome sequences. The numbers above branches are GBDP pseudo-bootstrap support values > 60% from 100 replications, with an average branch support of 92.1%. The tree was rooted at the midpoint (Farris, 1972).
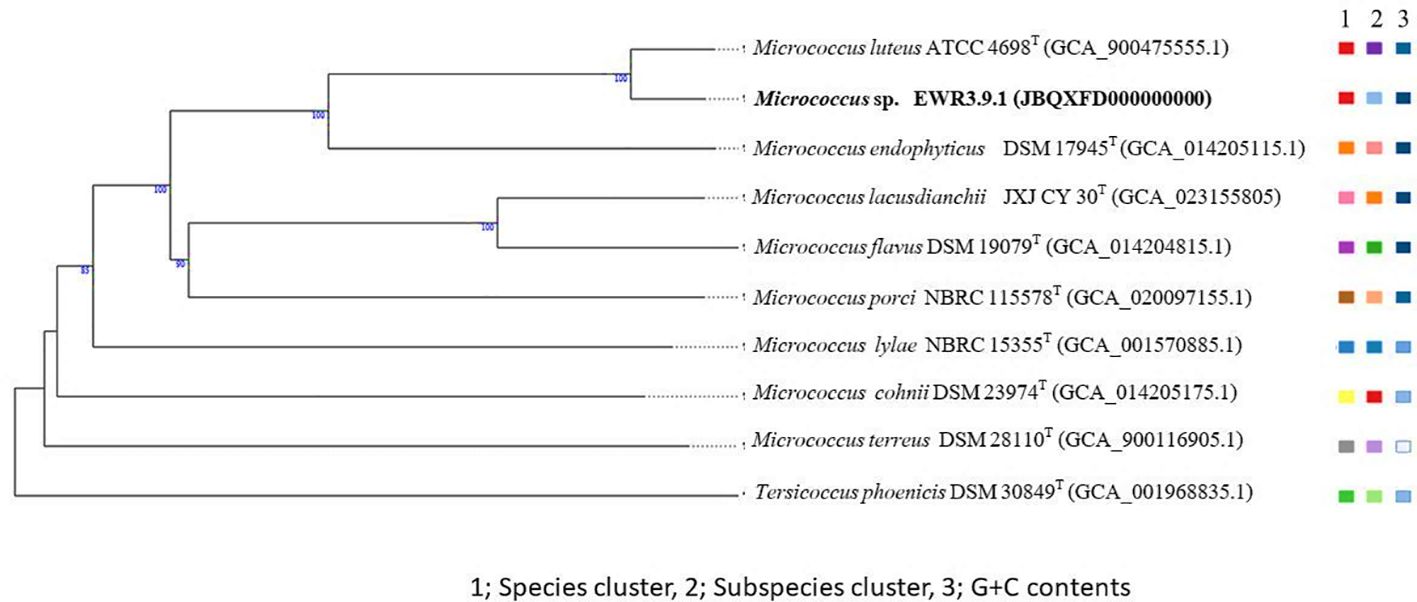
Figure 10. A phylogenomic tree based on the TYGS result shows the relationship between Micrococcus EWR3.9.1 and their closely related type strains. The tree was inferred with FastME 2.1.6.1 (Lefort et al., 2015) from GBDP distances calculated from genome sequences. The numbers above branches are GBDP pseudo-bootstrap support values > 60% from 100 replications, with an average branch support of 89.4%. The tree was rooted at the midpoint (Farris, 1972).
The GenBank accession numbers for the genomes of Streptomyces strains EKR5.2, ESS7.8, ECR2.10, and EWL5.1, as well as Micrococcus strain EWR3.9.1, are JBQXFH000000000, JBQXFG000000000, JBQXFF000000000, JBQXFE000000000, and JBQXFD000000000, respectively.
Biosynthetic Gene Clusters and gene prediction
All Streptomyces strains, EKR5.2, ESS7.8, ECR2.10, and EWL5.1, contain BGCs of hopene (53, 100, 92, 92%) and geosmin (100% for all), which are common compounds found in many strains of Streptomyces (Supplementary Tables S3–S5). Strains EKR5.2 and ESS7.8 comprise BGCs of spore pigment (66, 66%), while only strains EKR5.2 and ECR2.10 have BGCs of melanin compound (42, 71%). Strains EKR5.2, ESS7.8, ECR2.10, and EWL5.1 contain BGCs of informipeptin (42, 42, 100, 100%) and albaflavenone (100% for all), which is a tricyclic sesquiterpene antibiotic with antibacterial activity produced by Streptomyces (Moody et al., 2012). All four strains have BGCs of germicidin (100% for all), which is an auto-regulative suppressor of spore germination in the genus Streptomyces (Petersen et al., 1993). The genome of strain EKR5.2 comprises BGCs of the invaluable compound ϵ-Poly-L-lysine (100%), which is a microbial peptide that is used as an antimicrobial compound to preserve packaged food. It was reported that ϵ-PL is competitive to use widely worldwide because it shows broad antimicrobial activity against Gram-negative and Gram-positive bacteria, yeasts, and molds. However, the production of this compound is limited because the yield of the commercial strain was low (Ye et al., 2013). Therefore, strain EKR5.2 will be a beneficial strain to develop as a commercial strain in the future.
The genome of strain ESS7.8 comprises higher numbers of BGCs than the genomes of the other three strains. Of these BGCs, BGCs of thiazostatin, watasemycins A and B (100%), new antibiotics produced by Streptomyces sp. TP-A059, which showed antibacterial activity against Gram-staining positive and negative bacteria, and yeast are detected in the genome of strain ESS7.8 (Sasaki et al., 2002). The genome of strain ESS7.8 comprises BGCs of coelibactin (100%), a bacterial toxin that is produced by some strains of bacteria living in the human gut and induces prophage to be in the lytic development stage (Silpe et al., 2022). The genome of strain ESS7.8 contains BGCs of butyrolactol A (86%), which showed activity against fungal human pathogens, such as Candida albicans and Trichophyton mentagrophytes (Harunari et al., 2017). Moreover, the genome of strain ESS7.8 comprises a BGC of enterocin (95%). It was reported that the genome of Streptomyces qinglanensis 172205, isolated from mangroves, comprises BGCs of enterocin, a bacteriocin that can inhibit activity against β-amyloid protein (Aβ1-42) fibrillation and moderate cytotoxicity against HeLa and HepG2 (Xu et al., 2015). In addition, the genomes of strains ESS7.8 and ECR2.10 contain BGCs of alkylresorcinols (Ars) (100, 100%), which are polyphenolic compounds with the potential to be used as the regulation of host metabolism. The study showed that the intestinal microbiota could produce several Ars related to olivetol supplementation and microbiota metabolic activity. Moreover, Ars are potential quorum-sensing molecules, which support gut microbiota composition and host metabolism (Zabolotneva et al., 2023). Genomes of strains ESS7.8, ECR2.10, and EWL5.1 contain BGC of 1,3,6,8-tetrahydroxynaphthalene (THN) (100% for all), which is a precursor of melanin production. It was reported that THN derivative IBR33 produced by Nocardia sp. CS682 showed promising UV protection effects (Mishra et al., 2019).
Streptomyces strains ECR2.10 and EWL5.1 were close to each other (99.9% dDDH value). The BGCs of strains ECR2.10 and EWL5.1 were mostly the same, but the BGCs of melanin, methylated alkyl-resorcinol, and aurantimycin A were only found in strain ECR2.10. Many strains of Streptomyces produce Pentalenolactone, a sesquiterpenoid antibiotic that is detected in the genomes of strains ECR2.10 and EWL5.1 (58, 58%). Pentalenolactone inhibited both Gram-positive and Gram-negative bacteria, as well as pathogenic and saprophytic fungi (Takamatsu et al., 2011). The genome of strain ECR2.10 comprises the BGC of Aurantimycin (ATM), produced by Streptomyces aurantiacus JA 4570. ATM showed strong activity against some strains of Gram-positive bacteria and possessed cytotoxic activity against L-929 mouse fibroblast cells. The tandem overexpression of genes artB and artX increases the production of ATM about 2.5-fold in S. aurantiacus JA 4570 (Zhao et al., 2016). Genomes of strains ECR2.10 and EWL5.1 contain interesting BGCs of Cystargolides A and B (90, 90%), which are rarely reported. Cystargolides A and B were initially isolated from Kitasatospora cystarginea NRRL B16505 and showed activity to inhibit the human proteasome and the caseinolytic protease ClpP in the micromolar range (Beller et al., 2024). These findings highlight the diverse biosynthetic capabilities of the analyzed Streptomyces strains, suggesting their potential for producing a variety of bioactive compounds. Further investigation into the specific roles of these BGCs could lead to new discoveries in antibiotic development and natural product chemistry.
These Streptomyces strains; EKR5.2, ESS7.8, ECR2.10, and EWL5.1, all have BGCs that code for compounds that are related to PGP traits, like ectoine (100% for all). The strains ESS7.8, ECR2.10, and EWL5.1 all have BGCs of the siderophore desferrioxamin B and E (100% for all), while the strain EKR5.2 has BGCs of the NI-siderophore FW0622 (62%), which is a new siderophore from the marine species Verrucosispora sp. FIM060022 (Zhao et al., 2019). Interestingly, strain ESS7.8 comprises the BGC of 6-methylsalicyclic acid (6-MeSA). Researchers reported that salicylates and related compounds could induce disease resistance in plants through the Induced Systematic Resistant pathway. The treatment of tobacco leaves with 6-MeSA increased the accumulation of the pathogenesis-related (PR) proteins PR1, β-1,3-glucanase, and chitinase and supported the plant’s resistance to the tobacco mosaic virus (Yalpani et al., 2001).
The genome of Micrococcus strain EWR3.9.1 comprises only one BGC of a terpene known as carotenoid (66%). The study of Kandasamy and Kathirvel (2024) showed that crude yellow pigment from endophytic Micrococcus luteus associated with Avicennia marina showed significant dose-dependent antioxidant and anticancer activity. Strain EWR3.9.1 may produce carotenoid which may comprise the antioxidant properties of host plant. Further research could elucidate the mechanisms behind this relationship and explore the potential applications of these findings in agricultural practices.
Genome data mining
Supplementary Table S6 displays the COG functional category classification of Streptomyces strains EKR5.2, ESS7.8, ECR2.10, and EWL5.1. These four strains comprise the highest number of genes relating to category K: transcription, with the following categories: E: amino acid metabolism and transport, and G: carbohydrate metabolism and transport. Strains ECR2.10 and EWL5.1 comprise more sequences at 402 and 329, respectively, belonging to category Q: secondary structure than strains EKR5.2 and ESS7.8, which contain only 219 and 229 sequences, respectively. The COG functional category classification of Micrococcus strain EWR3.9.1 showed different results when compared to these strains of Streptomyces. The most genes were found in category L, which is for replication and repair. The next most genes were found in categories E, which is for amino acid metabolism and transport, and the last few were found in category J, which is for translation. Additionally, the ratio of genes associated with category P, which pertains to inorganic ion transport and metabolism, is higher than that of the other four Streptomyces strains.
Functional annotation showed that four Streptomyces strains and one strain of Micrococcus contain genes relating to plant growth promotion to reduce stress on plants under drought and saline conditions (Supplementary Tables S7–S11). In the genomes of these five strains, genes that encode the osmo-protectant glycine betaine and proline have been found to help plants under stress. However, there were more genes encoding these proteins detected in Streptomyces strains than in Micrococcus. Genes encoding heat and osmotic pressure proteins were also detected in five strains, including chaperone proteins. However, only four strains of Streptomyces comprised more genes encoding ectoine production, of which only one gene was detected in the genome of Micrococcus. Also, only three strains of Streptomyces; EKR5.2, ECR2.10, and EWL5.1, have an acds gene that encodes 1-aminocyclopropane-1-carboxylate deaminase. This correlated with the phenotypic data of these strains that produce ACC deaminase in vitro. These five strains also contain genes that encode biodegradation enzymes like amylase, cellulase, chitinase, and xylose isomerase, which find application in various industrial sectors.
In all strains of Streptomyces and Micrococcus EWR3.9.1, there are genes that encode antioxidants such as ferredoxin, flavodoxin, glutaredoxin, and thioredoxin. Only Micrococcus strain EWR3.9.1 contains redoxin. Thioredoxin and peroxiredoxins are non-enzymatic antioxidants that can detoxify any excess reactive oxidative scavenger (ROS) (Hopkins and Neumann, 2019). Glutaredoxins (Grxs) are small disulfide reductase enzymes that use glutathione and NADPH as cofactors. A few strains of bacteria produce Grxs as antioxidants to defend against oxidative stress and reveal the physiological roles of GrxD in oxidative stress protection (Saninjuk et al., 2023).
Streptomyces strain EKR5.2, which was isolated from high-salinity soil, had a gene that encodes peroxiredoxin. Peroxiredoxins (Prxs) or thioredoxin peroxidases (TPXs) are a group of thiol-specific antioxidant enzymes that help protect cells from oxidative damage (Kim et al., 2013).
Streptomyces strain ESS7.8 had genes that encoded cupredoxin and rubredoxin. Rubredoxin plays an important role in the reduction of superoxide and protect bacterial cell from oxidative (Coulter and Kurtz, 2001). These unique rubredoxins likely may play a crucial role in the survival and metabolic flexibility of Streptomyces strain ESS7.8, enabling them to thrive in environments where oxygen levels fluctuate.
Three strains of Streptomyces, EKR5.2, ECR2.10, and EWL5.1, including Micrococcus strain EWR3.9.1, comprise genes encoding IAA production. Streptomyces strain ESS7.8 and Micrococcus strain EWR3.9.1 contain genes encoding siderophore production. Only Streptomyces strains ESS7.8, ECR2.10, and EWL5.1 comprise genes encoding phenazine production, which is an antimicrobial compound. Moreover, the genomes of Streptomyces strain ESS7.8 and Micrococcus strain EWR3.9.1 contain genes encoding L-asparaginase II, an enzyme that can treat cancer (Darvishi et al., 2022). The genomes of these two strains comprise genes encoding lycopene production. The genomes of strains EKR5.2, ECR2.10 and EWL5.1 have genes encoding exopolysaccharide production. It was reported that exopolysaccharide can help plants to tolerate drought and salinity conditions (Bhagat et al., 2021). Seedling length vigor index (SLVI) of seedlings treated with Streptomyces strain EWL5.1 showed that this strain helped seedlings grow even when they were under 50 mM of salt stress. It may produce exopolysaccharide to help plants in salt stress conditions. This suggests that strain EWL5.1 may have positive effects due to both its production of IAA and ACC deaminase. Moreover, this strain had ability to produce exopolysaccharides, which may help the plant more resilient in harsh environments. We need to conduct further studies to clarify the mechanisms underlying this interaction and investigate the potential uses of these strains in agricultural practices. Micrococcus strain EWR3.9.1 contained genes related to metal resistance, copper resistance, and cadmium resistance transporters. This strain comprises the Nramp gene, which encodes a natural resistance-associated macrophage protein. In prokaryotes, Nramp plays a complex role in both nutrition and defense mechanisms. It was reported that the Nramps of Salmonella typhimurium and Escherichia coli displayed transport activity for Mn and are significantly stimulated after macrophage invasion (Kehres et al., 2000).
Conclusion
Eucalyptus grown in different soil salinity levels yielded different numbers and biodiversity of both Streptomyces and non-Streptomyces strains. The endophytic actinobacteria isolated from plants grown on high salinity soil contained a substantial biodiversity of Streptomyces species groups and non-Streptomyces, including non-actinobacteria genera. Endophytic actinobacteria from this study were a good source of antifungal activity to inhibit severe leaf spot fungi in Eucalyptus. In vitro studies indicated that selected strains had PGP traits to promote plant growth in salinity conditions, such as ACC deaminase and IAA production. All isolates could produce IAA, which was a phytohormone to help the plant tolerate stressful conditions. One selected Streptomyces strain could support the seedling length vigor index (SLVI) of eucalyptus seedlings in salinity conditions and significantly increase the fresh weight of eucalyptus seedlings in planta. The genome analysis of four representative Streptomyces strains revealed that these strains contain various biosynthetic gene clusters (BGCs) involved in antibiotic production. The result of genome data mining of four Streptomyces strains and a strain of Micrococcus also reveals that these strains contain genes encoding PGP properties such as glycine-betaine, proline, IAA, and ACC deaminase production including antioxidants. In conclusion, endophytic actinobacteria found in Eucalyptus trees could inhibit fungal pathogens and exhibit PGP traits that could be used in the future as plant inoculum in saline soil. This suggests a promising avenue for agricultural practices, particularly in regions facing salinity challenges. By harnessing the beneficial properties of these endophytic actinobacteria, farmers may enhance crop resilience and productivity in adverse environmental conditions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
OK: Resources, Investigation, Writing – original draft, Software, Formal analysis, Funding acquisition, Visualization, Project administration, Data curation, Conceptualization, Validation, Writing – review & editing, Methodology, Supervision. KK: Visualization, Data curation, Validation, Software, Investigation, Formal analysis, Writing – review & editing. SC: Visualization, Writing – review & editing, Investigation. BK: Software, Investigation, Visualization, Formal analysis, Writing – review & editing. PK: Investigation, Visualization, Writing – review & editing, Formal analysis, Methodology, Software. WD: Writing – review & editing, Formal analysis, Methodology, Data curation, Visualization, Investigation.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research project is funded by National Research Council of Thailand (NRCT) and Mahasarakham University, Thailand (Grant number; N42A650338).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1610327/full#supplementary-material
References
Abrol, I. P., Yadav, J. S. P., and Massoud, F. I. (1998). Salt-affected soils and their management (Rome, Italy: FAO).
Atouei, M. T., Pourbabaee, A. A., and Shorafa, M. (2019). Alleviation of salinity stress on some growth parameters of wheat by exopolysaccharide-producing bacteria. Iran. J. Sci. Technol. A. 43, 2725–2733. doi: 10.1007/s40995-019-00753-x
Baldani, J. I., Caruso, L., Baldani, V. L. D., Goi, S. R., and Döbereiner, J. (1997). Recent advances in BNF with non-legume plants. Soil Biol. Biochem. 29, 911–922. doi: 10.1016/S0038-0717(96)00218-0
Barnawal, D., Bharti, N., Tripathi, A., Pandey, S. S., Chanotiya, C. S., and Kalra, A. (2016). ACC-deaminase-producing endophyte Brachybacterium paraconglomeratum strain SMR20 ameliorates chlorophytum stress via altering phytohormone generation. J. Plant Growth Regul. 35, 553–564. doi: 10.1007/s00344-015-9560-3
Batjes, N. H. (1995). A global data set of soil pH properties (Wageningen: International Soil Reference and Information Centre). Technical paper 27.
Becker, R., Ulrich, K., Behrendt, U., Schneck, V., and Ulrich, A. (2022). Genomic characterization of Aureimonas altamirensis C2P003-A specific member of the microbiome of Fraxinus excelsior trees tolerant to ash dieback. Plants 11, 3487. doi: 10.3390/plants11243487
Beller, P., Fink, P., Wolf, F., Männle, D., Helmle, I., Kuttenlochner, W., et al. (2024). Characterization of the cystargolide biosynthetic gene cluster and functional analysis of the methyltransferase CysG. J. Biol. Chem. 300, 105507. doi: 10.1016/j.jbc.2023.105507
Bensch, K., Groenewald, J. Z., Dijksterhuis, J., Starink-Willemse, M., Andersen, B., Summerell, B. A., et al. (2010). Species and ecological diversity within the Cladosporium cladosporioides complex (Davidiellaceae, Capnodiales). Stud. Mycol. 67, 1–94. doi: 10.1016/S0166-0616(14)60026-9
Bhagat, N., Raghav, M., Dubey, S., and Bedi, N. (2021). Bacterial exopolysaccharides: Insight into their role in plant abiotic stress tolerance. J. Microbiol. Biotechnol. 31, 1045–1059. doi: 10.4014/jmb.2105.05009
Blin, K., Shaw, S., Augustijn, H. E., Reitz, Z. L., Biermann, F., Alanjary, M., et al. (2023). AntiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 51, W46–W50. doi: 10.1093/nar/gkad344
Bo, A. B., Kim, J. D., Kim, Y. S., Sin, H. T., Kim, H. J., Khaitov, B., et al. (2019). Isolation, identification and characterization of Streptomyces metabolites as a potential bioherbicide. PloS One 23, e0222933. doi: 10.1371/journal.pone.0222933
Boukaew, S., Chuenchit, S., and Petcharat, V. (2011). Evaluation of Streptomyces spp. for biological control of Sclerotium root and stem rot and Ralstonia wilt of chili pepper. BioControl 56, 365–374. doi: 10.1007/s10526-010-9336-4
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). EggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829. doi: 10.1093/molbev/msab293
Castagno, L. N., Estrella, M. J., Sannazzaro, A. I., Grassano, A. E., and Ruiz, O. A. (2011). Phosphate-solubilization mechanism and in vitro plant growth promotion activity mediated by Pantoea eucalypti isolated from Lotus tenuis rhizosphere in the Salado river basin (Argentina). J. Appl. Microbiol. 110, 1151–1165. doi: 10.1111/j.1365-2672.2011.04968.x
Cheewangkoon, R., Groenewald, J. Z., Verkley, G. J. M., Hyde, K. D., Wingfield, M. J., Gryzenhout, M., et al. (2010). Re-evaluation of Cryptosporiopsis eucalypti and Cryptosporiopsis-like species occurring on Eucalyptus leaves. Fungal Divers. 44, 89–105. doi: 10.1007/s13225-010-0041-5
Chen, Z. C. and Huang, C. F. (2024). Sustainable utilization of acid soils. Plant Soil 504, 1–3. doi: 10.1007/s11104-024-07051-5
Coulter, E. D. and Kurtz, D. E., Jr. (2021). A role for rubredoxin in oxidative stress protection in Desulfovibrio vulgaris: catalytic electron transfer to rubrerythrin and two-iron superoxide reductase. Arch. Biochem. Biophys. 394, 7686. doi: 10.1006/abbi.2001.2531
Darvishi, F., Jahanafrooz, Z., and Mokhtarzadeh, A. (2022). Microbial L-asparaginase as a promising enzyme for treatment of various cancers. Appl. Microbiol. Biotechnol. 106, 5335–5347. doi: 10.1007/s00253-022-12086-8
Dif, G., Belaouni, H. A., Goudjal, Y., Yekkour, A., Djemouai, N., and Zitouni, A. (2021). Potential for plant growth promotion of Kocuria arsenatis strain ST19 on tomato under salt stress conditions. S. Afr. J. Bot. 138, 94–104. doi: 10.1016/j.sajb.2020.12.014
El-Akshar, E. A., El-Meihy, R. M., Tewfike, T. A., Husnain, L. A., Alkahtani, M. D. F., Bouqellah, N. A., et al. (2024). Endophytic chitinase and antifungal metabolites-producing actinobacteria for biological control of cucumber damping off disease. J. Plant Pathol. 107, 469–490. doi: 10.1007/s42161-024-01790-1
FAO (2021). Standard operating procedure for soil pH determination (Rome: Food and Agriculture Organization of the United Nations). Available online at: https://openknowledge.fao.org/server/api/core/bitstreams/6ad6862a-eadc-437c-b359-ef14cb687222/content (Accessed March 10, 2025).
Farm forestry Newzealand (1992). New Leafspots on Eucalyptus, From Forest health News No.10, June 1992. Available online at: https://www.nzffa.org.nz/farm-forestry-model/the-essentials/forest-health-pests-and-diseases/forestry-diseases/cladosporium/new-leafspots-on-eucalyptus/ (Accessed March 10, 2025).
Farris, J. S. (1972). Estimating phylogenetic trees from distance matrices. Am. Nat. 106, 645–668. doi: 10.1086/282802
Feikema, P. M. and Baker, T. G. (2011). Effect of soil salinity on growth of irrigated plantation Eucalyptus in south-eastern Australia. Agric. Water Manage. 98, 1180–1188. doi: 10.1016/j.agwat.2011.03.005
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Gerber, E., Bernard, R., Castang, S., Chabot, N., Coze, F., Dreux-Zigha, A., et al. (2015). Deinococcus as new chassis for industrial biotechnology: biology, physiology and tools. J. Appl. Microbiol. 119, 1–10. doi: 10.1111/jam.12808
Harunari, E., Komaki, H., and Igarashi, Y. (2017). Biosynthetic origin of butyrolactol A, an antifungal polyketide produced by a marine-derived Streptomyces. Beilstein J. Org. Chem. 13, 441–450. doi: 10.3762/bjoc.13.47
Hayakawa, M. and Nonomura, H. (1987). Humic acid vitamin agar, a new method for the selective isolation of soil actinomycetes. J. Ferment. Technol. 65, 501–509. doi: 10.1016/0385-6380(87)90108-7
Hazarika, S. N., Saikia, K., and Thakur, D. (2022). Characterization and selection of endophytic actinobacteria for growth and disease management of tea (Camellia sinensis L.). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.989794
Himaman, W., Thamchaipenet, A., Pathomm-aree, W., and Duangmal, K. (2016). Actinomycetes from Eucalyptus and their biological activities for controlling Eucalyptus leaf and shoot blight. Microbiol. Res. 188-189, 42–52. doi: 10.1016/j.micres.2016.04.011
Hopkins, B. L. and Neumann, C. A. (2019). Redoxins as gatekeepers of the transcriptional oxidative stress response. Redox Biol. 21, 101104. doi: 10.1016/j.redox.2019.101104
Hudson, J. A., Schofield, K. M., Morgan, H. W., and Daniel, R. M. (1989). Thermonema lapsum gen. nov., sp. nov., a thermophilic gliding bacterium. Int. J. Syst. Bacteriol. 39, 485–487. doi: 10.1099/00207713-39-4-485
Joseph, S. J., Hugenholtz, P., Sangwan, P., Osborne, C. A., and Janssen, P. H. (2003). Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environl. Microbiol. 69, 7210–7215. doi: 10.1128/AEM.69.12.7210-7215.2003
Kaewkla, O. (2009). Isolation and identification of endophytic actinobacteria from Australian native plants and their antimicrobial activity. The Flinders University, Adelaide, South Australia 257.
Kaewkla, O. and Franco, C. M. M. (2013). Rational approaches to improving the isolation of endophytic actinobacteria from Australian native trees. Microb. Ecol. 65, 384–393. doi: 10.1007/s00248-012-0113-z
Kaewkla, O., Sukpanoa, S., SuriyaChadkun, C., Chamroensaksi, N., Chumroenphat, T., and Franco, C. M. M. (2022). Streptomyces spinosus sp. nov. and Streptomyces shenzhenensis subsp. oryzicola subsp. nov. endophytic actinobacteria isolated from Jasmine rice and their genome mining for potential as antibiotic producers and plant growth promoters. Antonie Van Leeuwenhoek 115, 871–888. doi: 10.1007/s10482-022-01741-9
Kampapongsa, D. and Kaewkla, O. (2016). Biodiversity of endophytic actinobacteria from Jasmine rice (Oryza sativa L. KDML 105) grown in Roi-et province, Thailand and their antimicrobial activity against rice pathogen. Ann. Microbiol. 66, 587–595. doi: 10.1007/s13213-015-1140-z
Kandasamy, G. D. and Kathirvel, P. (2024). Production, characterization and in vitro biological activities of crude pigment from endophytic Micrococcus luteus associated with Avicennia marina. Arch. Microbiol. 206, 26. doi: 10.1007/s00203-023-03751-1
Kehres, D. G., Zaharik, M. L., Finlay, B. B., and Maguire, M. E. (2000). The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol. Microbiol. 36, 1085–1100. doi: 10.1046/j.1365-2958.2000.01922.x
Kerbab, S., Silini, A., Bouket, A. C., Cherif-Silini, H., Eshelli, M., Rabhi, N. E. H., et al. (2021). Mitigation of NaCl stress in wheat by rhizosphere engineering using salt habitat adapted PGPR halotolerant bacteria. Appl. Sci. 11, 1034. doi: 10.3390/app11031034
Kim, I. S., Kim, Y. S., and Yoon, H. S. (2013). Expression of salt-induced 2-Cys peroxiredoxin from Oryza sativa increases stress tolerance and fermentation capacity in genetically engineered yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 97, 3519–3533. doi: 10.1007/s00253-012-4410-8
Kim, Y. S., Jang, K. S., and Choi, J. S. (2024). Culture optimization of Streptomyces sp. KRA16–334 for increased yield of new herbicide 334-W4. PloS One 19, e0301104. doi: 10.1371/journal.pone.0301104
Lefort, V., Desper, R., and Gascuel, O. (2015). FastME 2.0: A comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 32, 2798–2800. doi: 10.1093/molbev/msv150
Lobato-Ureche, M. A., Pérez-Rodriguez, M. M., Segura, D., Monasterio, R., and Cohen, A. C. (2023). Pseudomonas 42P4 and Cellulosimicrobium 60I1 as a sustainable approach to increase growth, development, and productivity in pepper plants. Front. Sustain. Food Syst. 6. doi: 10.3389/fsufs.2022.1111573
Lueangpraplut, S., Unartngam, A., and Unartngam, J. (2013). Molecular identification of Pseudoplagiostoma eucalypti causing leaf spot and shoot blight diseases on eucalyptus in Thailand based on ITS rDNA sequence. J. Agric. Technol. 9, 165–175.
Madhaiyan, M., Saravanan, V. S., See-Too, W. S., Volpiano, C. G., Sant'Anna, F. H., Mota, F. F., et al. (2022). Genomic and phylogenomic insights into the Family Streptomycetaceae lead to the proposal of six novel genera. Int. J. Syst. Evol. Microbiol. 72. doi: 10.1099/ijsem.0.005570
Mahmud-Ur-Rahman, Naser, I. B., Mahmud, N. U., Sarker, A., Hoque, M. N., and Islam, T. (2022). A highly salt-tolerant bacterium Brevibacterium sediminis promotes the growth of rice (Oryza sativa L.) seedlings. Stresses 2, 275–289. doi: 10.3390/stresses2030020
Marcar, N. E., Zohar, Y., Guo, J., and Crawford, D. F. (2002). Effect of NaCl and high pH on seedling growth of 15 Eucalyptus camaldulensis Dehnh. provenances. New Forests 23, 193–206. doi: 10.1023/A:1020334418883
Meier-Kolthoff, J. P. M., Auch, A. F., Klenk, H. P., and Goker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 14, 60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P. and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10, 2182. doi: 10.1038/s41467-019-10210-3
Mishra, R., Dhakal, D., Han, J. M., Lim, H. N., Jung, H. J., Yamaguchi, T., et al. (2019). Production of a novel tetrahydroxynaphthalene (THN) derivative from Nocardia sp. CS682 by metabolic engineering and its bioactivities. Molecules 24, 244. doi: 10.3390/molecules24020244
Mohseni, M., Norouzi, H., Hamedi, J., and Roohi, A. (2013). Screening of antibacterial producing actinomycetes from sediments of the Caspian sea. Int. J. Mol. Cell Med. 2, 64–71.
Moody, S. C., Zhao, B., Lei, L., Nelson, D. R., Mullins, J. G. L., Waterman, M. R., et al. (2012). Investigating conservation of the albaflavenone biosynthetic pathway and CYP170 bifunctionality in streptomycetes. FEBS. J. 279, 1640–1649. doi: 10.1111/j.1742-4658.2011.08447.x
Mosley, L. M., Rengasamy, P., and Fitzpatrick, R. (2024). Soil pH: Techniques, challenges and insights from a global dataset. Eur. J. Soil Sci. 75, e70021. doi: 10.1111/ejss.70021
Negi, N., Ramkrishna, Meena, R. K., Pandey, A., Bhandari, M. S., and Pandey, S. (2024). Pseudoplagiostoma eucalypti: an emerging pathogen of Eucalyptus in northern India. For. Pathol. 54, e12856. doi: 10.1111/efp.12856
Nei, M. and Kumar, S. (2000). Molecular Evolution and Phylogenetics (New York: Oxford University Press).
Old, K. M., Wingfield, M. J., and Yuan, Z. Q. (2003). A Manual of Diseases of Eucalypts in South-East Asia (Bogor: Center for International Forestry Research, Jarkarta), 98.
Parte, A. C., Carbasse, J. S., Meier-Kolthoff, J. P., Reimer, L. C., and Göker, M. (2020). List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612. doi: 10.1099/ijsem.0.004332
Penrose, D. M. and Glick, B. R. (2003). Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant 118, 10–15. doi: 10.1034/j.1399-3054.2003.00086.x
Petersen, F., Zähner, H., Metzger, J. W., Freund, S., and Hummel, R. P. (1993). Germicidin, an autoregulative germination inhibitor of Streptomyces viridochromogenes NRRL B-1551. J. Antibiot. 46, 1126–1138. doi: 10.7164/antibiotics.46.1126
Pushpalatha, H. G., Naveen, J., Geetha, N., Hithamani, G., and Shetty, H. S. (2023). Plant growth promotion and biological control of Sclerospora graminicola in pearl millet by endophytic Streptomyces spp. Indian Phytopathol. 76, 521–530. doi: 10.1007/s42360-023-00616-x
Qin, S., Feng, W. W., Zhang, Y. J., Wang, T. T., Xiong, Y. W., and Xing, K. (2018). Diversity of bacterial microbiota of coastal halophyte Limonium sinense and amelioration of salinity stress damage by symbiotic plant growth-promoting actinobacterium Glutamicibacter halophytocola KLBMP 5180. Appl. Environ. Microbiol. 84, e01533–e01518. doi: 10.1128/AEM.01533-18
Qin, S., Xing, K., Jiang, J. H., Xu, L. H., and Li, W. J. (2011). Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Appl. Microbiol. Biotechnol. 89, 457–473. doi: 10.1007/s00253-010-2923-6
Rhoades, J. D., Chanduvi, F., and Lesch, S. (1999). Soil salinity assessment: methods and interpretation of electrical conductivity measurements. FAO Irrigation Drainage Paper 57, 150.
Richter, M. and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Richter, M., Rosselló-Móra, R., Oliver, G. F., and Peplies, J., Jr. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinform. 32, 929–931. doi: 10.1093/bioinformatics/btv681
Saikia, J., Mazumdar, R., and Thakur, D. (2022). Phylogenetic affiliation of endophytic actinobacteria associated with selected orchid species and their role in growth promotion and suppression of phytopathogens. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1058867
Saitou, N. and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Saninjuk, K., Romsang, A., Duang-nkern, J., Wongsaroj, L., Leesukon, P., Dubbs, J. M., et al. (2023). Monothiol glutaredoxin is essential for oxidative stress protection and virulence in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 89, e01714-22. doi: 10.1128/aem.01714-22
Sankaran, K. V., Sutton, B. C., and Balasundaran, M. (1995). Cryptosporiopsis eucalypti sp. nov., causing leaf spots of eucalypts in Australia, India and USA. Mycol. Res. 99, 827–830. doi: 10.1016/S0953-7562(09)80735-1
Santiago, T. R., Grabowski, C., and Mizubuti, E. S. G. (2014). First report of bacterial wilt caused by Ralstonia solanacearum on Eucalyptus sp. in Paraguay. New Dis. Rep. 29, 2. doi: 10.5197/j.2044-0588.2014.029.002
Santiago, T. R., Grabowski, C., Rossato, M., Romeiro, R. S., and Mizubuti, E. S. G. (2015). Biological control of eucalyptus bacterial wilt with rhizobacteria. Biol. Control 80, 14–22. doi: 10.1016/j.biocontrol.2014.09.007
Sardi, P., Saracchi, M., Quaroni, S., Petrolini, B., Borgonovi, G. E., and Merli, S. (1992). Isolation of endophytic Streptomyces strains from surface-sterilized roots. Appl. Environ. Microbiol. 58, 2691–2693. doi: 10.1128/aem.58.8.2691-2693.1992
Sasaki, T., Igarashi, Y., Saito, N., and Furumai, T. (2002). Watasemycins A and B, new antibiotics produced by Streptomyces sp. TP-A0597. J. Antibiot. 55, 249–255. doi: 10.7164/antibiotics.55.249
Schoenborn, L., Yates, P. S., Grinton, B. E., Hugenholtz, P., and Janssen, P. H. (2004). Liquid serial dilution is inferior to solid media for isolation of cultures representative of the Phylum-level diversity of soil bacteria. Appl. Environ. Microbiol. 70, 4363–4366. doi: 10.1128/AEM.70.7.4363-4366.2004
Shahid, M., Singh, B. N., Verma, S., Choudhary, P., Das, S., Chakdar, H., et al. (2021). Bioactive antifungal metabolites produced by Streptomyces amritsarensis V31 help to control diverse phytopathogenic fungi. Braz. J. Microbiol. 52, 1687–1699. doi: 10.1007/s42770-021-00625-w
Shan, W., Zhou, Y., Liu, H., and Yu, X. (2018). Endophytic actinomycetes from tea plants (Camellia sinensis): isolation, abundance, antimicrobial, and plant-growth-promoting activities. BioMed. Res. Int. 2018, 1470305. doi: 10.1155/2018/1470305
Shirling, E. B. and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Silpe, J. E., Wong, J. W. H., Owen, S. V., Baym, M., and Balskus, E. P. (2022). The bacterial toxin colibactin triggers prophage induction. Nature 603, 315–320. doi: 10.1038/s41586-022-04444-3
Singh, R. and Dubey, A. K. (2018). Diversity and applications of endophytic actinobacteria of plants in special and other ecological niches. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01767
Takamatsu, S., Xu, L. H., Fushinobu, S., Shoun, H., Komatsu, M., Cane, D. E., et al. (2011). Pentalenic acid is a shunt metabolite in the biosynthesis of the pentalenolactone family of metabolites: hydroxylation of 1-deoxypentalenic acid mediated by CYP105D7 (SAV_7469) of Streptomyces avermitilis. J. Antibiot. 64, 65–71. doi: 10.1038/ja.2010.135
Tamura, K. and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526. doi: 10.1093/oxfordjournals.molbev.a040023
Tamura, K., Nei, M., and Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. PNAS 101, 11030–11035. doi: 10.1073/pnas.0404206101
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Vimal, S. R., Patel, V. K., and Singh, J. S. (2019). Plant growth promoting Curtobacterium albidum strain SRV4: an agriculturally important microbe to alleviate salinity stress in paddy plants. Ecol. Indic. 105, 553–562. doi: 10.1016/j.ecolind.2018.05.014
Vo, Q. A. T., Ballard, R. A., Barnett, S. J., and Franco, C. M. M. (2021). Isolation and characterisation of endophytic actinobacteria and their effect on the growth and nodulation of chickpea (Cicer arietinum). Plant Soil 466, 357–371. doi: 10.1007/s11104-021-05008-6
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Xu, D. B., Ma, M., Deng, Z. X., and Hong, K. (2015). Genotype-driven isolation of enterocin with novel bioactivities from mangrove-derived Streptomyces qinglanensis 172205. Appl. Microbiol. Biotechnol. 99, 5825–5832. doi: 10.1007/s00253-015-6574-5
Yalpani, N., Altier, D. J., Barbour, E., Cigan, A. L., and Scelonge, C. J. (2001). Production of 6-methylsalicylic acid by expression of a fungal polyketide synthase activates disease resistance in tobacco. Plant Cell. 13, 1401–1410. doi: 10.1105/TPC.010015
Ye, R., Xu, H., Wan, C., Peng, S., Wang, L., Xu, H., et al. (2013). Antibacterial activity and mechanism of action of ϵ-poly-l-lysine. Biochem. Biophys. Res. Commun. 439, 148–153. doi: 10.1016/j.bbrc.2013.08.001
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Zabolotneva, A. A., Gaponov, A. M., Roumiantsev, S. A., Vasiliev, I. Y., Grigoryeva, T. V., Kit, O. I., et al. (2023). Alkylresorcinols as new modulators of the metabolic activity of the gut microbiota. Int. J. Mol. Sci. 24, 14206. doi: 10.3390/ijms241814206
Zauza, E. A. V., Guimarães, L. M. S., Sales, N. L. P. S., Santos, S. A. S., Alfenas, R. F., and Alfenas, A. C. (2023). Outbreak of shoot blight and dieback of Eucalyptus spp., caused by Pseudoplagiostoma eucalypti in Brazil. For Pathol. 53, e12806. doi: 10.1111/efp.12806
Zhao, W., Peng, F., Wang, C., Xie, Y., Lin, R., Fang, Z., et al. (2019). FW0622, a new siderophore isolated from marine Verrucosispora sp. by genomic mining. Nat. Prod. Res. 34, 3082–3088. doi: 10.1080/14786419.2019.1608541
Zhao, H., Wang, L., Wan, D., Qi, J., Gong, R., Deng, Z., et al. (2016). Characterization of the aurantimycin biosynthetic gene cluster and enhancing its production by manipulating two pathway-specific activators in Streptomyces aurantiacus JA 4570. Microb. Cell Fact 15, 160. doi: 10.1186/s12934-016-0559-7
Keywords: endophyte, Eucalyptus camaldulensis, saline soil, plant growth promoting, Streptomyces, genome insight
Citation: Kaewkla O, Kiakhunthod K, Chookhampaeng S, Klinjantasorm B, Klankeo P and Dungkaew W (2025) Phylogenetic affiliation of endophytic actinobacteria associated with red gum tree grown in salinity area and their plant growth promoting properties and suppression of phytopathogens, and genome data mining of selected strains. Front. Plant Sci. 16:1610327. doi: 10.3389/fpls.2025.1610327
Received: 11 April 2025; Accepted: 29 September 2025;
Published: 26 November 2025.
Edited by:
Ricardo Aroca, Spanish National Research Council (CSIC), SpainReviewed by:
Ali Budhi Kusuma, Associação do Instituto Superior Técnico de Investigação e Desenvolvimento (IST-ID), PortugalIshu Khangwal, Baba Mastnath University, India
Copyright © 2025 Kaewkla, Kiakhunthod, Chookhampaeng, Klinjantasorm, Klankeo and Dungkaew. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Onuma Kaewkla, b251bWEua0Btc3UuYWMudGg=
 Onuma Kaewkla
Onuma Kaewkla Kawintip Kiakhunthod1,2
Kawintip Kiakhunthod1,2