- 1Center for Agricultural Genetic Resources Research, Shanxi Agricultural University, Taiyuan, Shanxi, China
- 2Germplasm Enhancement on Loess Plateau, Ministry of Agriculture and Rural Affairs, Taiyuan, Shanxi, China
- 3Houji Lab of Shanxi Province, Taiyuan, Shanxi, China
- 4College of Agriculture, Shanxi Agricultural University, Jinzhong, Shanxi, China
Background: The Brassinazole-resistant (BZR) family of transcription factors acts as key regulators in brassinosteroid (BR) signaling, influencing plant growth, development, biotic and abiotic stresses. However, systematic analysis of the BZR genes in oat has not been conducted. Moreover, little is known about their functions in osmotic stress, which is a major abiotic stress affecting oat production.
Methods: In this study, we performed a genome-wide analysis of the BZR gene family in oat. Their chromosome locations, gene structures, phylogenetic relationships, conserved domains, promoter cis-elements, and gene duplication events were analyzed. Furthermore, the expression patterns of BZR genes under osmotic stress were characterized, and the subcellular localization of AsBZR12 was investigated in Nicotiana benthamiana.
Results and discussion: In this study, we mapped 14 members of the BZR gene family across 12 oat chromosomes, and classified them into three groups based on phylogenetic analysis. The BZR proteins displayed group-specific patterns in their exon-intron structures and conserved motifs. Furthermore, cis-acting element analysis revealed that AsBZR genes are primarily involved in phytohormone responses and environmental stress adaptation. Examination of gene duplication revealed that segmental duplications drove the expansion of the BZR gene family in the oat genome, with evidence of strong purifying selection pressure during evolutionary development. The qRT-PCR analysis demonstrated varied expression patterns among AsBZR members. Specifically, AsBZR12 was significantly upregulated in roots, stems, and leaves, with nuclear localization. In summary, our study provides a comprehensive analysis of the AsBZR genes and characterizes their expression patterns under osmotic stress conditions, thereby identifying potential candidate genes for future research. This research provides comprehensive insights into BZR gene structure and evolution, establishing a foundation for understanding their osmotic stress responses in oat.
1 Introduction
Brassinosteroids (BRs) are an indispensable class of plant steroidal hormones that play crucial roles in regulating plant growth and development, as well as in coping with biotic and abiotic stresses (Manoli et al., 2018; Feng et al., 2023). The Brassinazole-resistant (BZR) transcription factor family has been extensively studied for its critical role in modulating BR signaling pathways (Wang et al., 2024). ATP-binding cassette (ABC) transporter ABCB19 mediates the transport of BRs to the apoplast, where these molecules bind to the plasma membrane-localized receptor BRASSINOSTEROID INSENSITIVE1 (BRI1) and its coreceptor BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1), thereby initiating the BR signaling pathway (Li et al., 2002; Nam and Li, 2002; Ying et al., 2024). This interaction activates key transcription factors such as BRASSINAZOLE RESISTANT1 (BZR1) and BRI1-EMS-SUPPRESSOR1 (BES1)/BZR2, which are central to the regulation of gene expression in response to BR signaling (Lee et al., 2024). Despite sharing 88% amino acid sequence similarity, and 97% consistency in the DNA binding domain, BZR1 and BES1/BZR2 exhibit distinct functions (Yin et al., 2002; Li et al., 2024). BZR1 functions as a transcriptional repressor that targets the BR response element (CGTGT/CG sequence). BZR1 regulates BR homeostasis and signaling through a dual-action mechanism: it activates the expression of growth-promoting genes by recruiting the BRAHMA-associated SWI/SNF chromatin-remodeling complex to open chromatin, while repressing BR-responsive genes by interacting with TOPLESS to recruit histone deacetylase 19 and form a transcriptional repression complex (He et al., 2005; Zhu T. et al., 2024). In contrast, BES1/BZR2 binds to E box (CANNTG) sequences and acts as a negative regulator of the BR pathway (Yin et al., 2005).
In the context of modern agriculture, climate change has emerged as a pivotal determinant of crop productivity. Rising global temperatures and erratic precipitation patterns have increased the frequency of drought conditions, making drought one of the most significant abiotic stresses affecting plant growth, development, and yield (Singh and Laxmi, 2015; He Z. H. et al., 2024; Zhan et al., 2024). An increasing number of studies have demonstrated that members of the BZR gene family play significant roles in plants’ responses to drought stress. In Setaria italica, SiBZR1 functions as a negative regulator of drought tolerance by directly targeting SiPLT-L1, whose overexpression enhances drought resistance (Zhao et al., 2021). Similarly, in Zanthoxylum armatum DC, BZR genes are upregulated under drought stress, and overexpression of ZaBZR1 in transgenic Nicotiana benthamiana confers enhanced drought tolerance (Jin et al., 2024). Moreover, TaBZR2 in wheat positively regulates drought tolerance by activating TaPPR13, which enhances drought tolerance through facilitating ABA-mediated stomatal movement (Cui et al., 2019; Hou et al., 2025). Additionally, the response of BZR gene family members to drought stress has been reported in several species, including Glycine max, Cucumis sativus, Populus trichocarpa, and Solanum tuberosum (Li et al., 2018; Luo et al., 2023; Li et al., 2024; Wang et al., 2024). However, the role of the BZR gene family in drought stress response in oat remains unclear.
Oat (Avena sativa L.) is an allohexaploid (AACCDD, 2n = 6x = 42) plant with a large and complex genome. As a highly nutritious cereal crop, it plays a significant role in both human food and animal feed (Peng et al., 2022). The prominence of oats in scientific research and industry has increased owing to their health-promoting attributes, such as the cholesterol-lowering effects of oat beta-glucan and their potential benefits in managing type-2 diabetes (Othman et al., 2011; Guo et al., 2023). Moreover, oats contain unique phytochemicals, including avenanthramides and steroidal saponins, which exhibit antioxidant, anti-inflammatory, and potential anticancer activities (Paudel et al., 2021). Oats demonstrate robust adaptability to diverse soil and climatic conditions, enabling their cultivation in areas with harsh environmental stressors that are unsuitable for other food crops (Chen et al., 2024). Predominantly grown in the arid regions of northern, northwestern, and southwestern China, oat yields are significantly limited by dry climatic conditions (Zhang et al., 2021). The availability of a high-quality oat reference genome has greatly enhanced our understanding of their evolutionary history and facilitated the identification of functional genes (Kamal et al., 2022; Peng et al., 2022).
Recent research has increasingly focused on various oat gene families, such as the glutathione reductase gene family, WRKY transcription factor family, catalase gene family, MYB transcription factor family, and aquaporin gene family (Liu et al., 2022; Sun et al., 2022; Ghorbel et al., 2023; Chen et al., 2024; Zhou et al., 2024). To date, the BZR gene family in oat has not been comprehensively identified or analyzed, and the functional studies of AsBZR genes in osmotic stress response remain uncharacterized. In this study, we conducted a genome-wide characterization of BZR genes in A. sativa, elucidating their chromosome locations, gene structures, phylogenetic relationships, conserved domains, promoter cis-elements, and gene duplication events. Additionally, we examined the expression patterns of AsBZR genes under osmotic stress, revealing that AsBZR12 was significantly upregulated in roots, stems, and leaves. Subsequently, we examined the subcellular localization of AsBZR12 in Nicotiana tabacum. The primary objective of this study was to conduct a comprehensive analysis of the AsBZR genes and to identify potential osmotic stress-responsive candidate genes by examining their expression patterns under PEG-induced osmotic stress conditions. These findings lay a foundation for future functional studies on the AsBZR genes in osmotic stress response.
2 Materials and methods
2.1 Identification and characterization of BZR gene family members in A. sativa
To perform genome-wide analysis in A. sativa, genome and annotation data were retrieved from the GrainGenes database (https://wheat.pw.usda.gov/GG3/content/avena-sang-download) (Kamal et al., 2022). To identify all possible AsBZR genes in A. sativa, both BLASTP and Hidden Markov Model (HMM) searches were performed. Known BZR protein sequences, including 6 from Arabidopsis thaliana (https://www.arabidopsis.org/) (Supplementary Table S1) and 6 from Oryza sativa (https://rice.uga.edu/) (Supplementary Table S1), were used as query sequences for BLASTP analysis with an e-value threshold set to less than or equal to 1e-5 (Ullah et al., 2022). In addition, oat protein sequences were searched against HMM profiles of the BZR domain (PF05687). To validate the presence of BZR domains in the candidate AsBZR genes, we performed manual domain annotation using three independent databases: Pfam (https://pfam-legacy.xfam.org/), the Conserved Domain Database (CDD) of the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), and SMART (http://smart.embl-heidelberg.de/) (Letunic et al., 2021). This analysis confirmed that all candidate genes contained the characteristic BZR conserved domain. The physicochemical properties of the AsBZR protein were calculated using the Protein Parameter Calculator in TBtools (Chen et al., 2023). Furthermore, the subcellular localization of the AsBZR proteins was predicted using Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/) (Chou and Shen, 2008).
2.2 Conserved motifs and gene structure analysis of AsBZR
The conserved motifs in the AsBZR proteins were identified using the MEME online program (https://meme-suite.org/meme/tools/meme) with the maximum number of motifs set to 10. The gene structure of the AsBZR genes was analyzed using the Visualize Gene Structure program in TBtools software based on A. sativa annotation data. The conserved motifs and gene structure information were visualized and mapped using the Gene Structure View program in TBtools.
2.3 Cir-acting and chromosome localization analysis
To identify the cis-acting elements in the promoter region, we screened 2000 bp upstream sequences of the AsBZR genes using PlantCARE tool (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Based on the A. sativa annotation data, the chromosomal locations of the AsBZR genes were determined using the Gene Location Visualization tool in TBtools.
2.4 Phylogenetic tree construction and collinearity analysis
To analyze the evolutionary relationships and origins of BZR transcription factors from A. sativa, Triticum aestivum, O. sativa, and A. thaliana, we aligned their full-length amino acid sequences using the ClustalW program. The phylogenetic tree was then constructed with 1000 bootstrap replicates using MEGA 7.0 software with the neighbor-joining method. The phylogenetic tree was edited and visualized using the Interactive Tree Of Life (iTOL) platform (https://itol.embl.de/). Gene duplication events within the AsBZR gene family and synteny analysis of orthologous BZR genes from A. sativa, A. thaliana, T. aestivum, and O. sativa were performed using the One Step MCScanX program in TBtools, with an e-value threshold of 1e-10. The non-synonymous substitution (Ka) and synonymous substitution (Ks) substitution rates for each duplicated gene pair were calculated using the Simple Ka/Ks Calculator in TBtools.
2.5 Plant materials and treatments
A. sativa cv. Pinyan No.8 (abbreviated as PY8), provided by the Center for Agricultural Genetic Resources Research, Shanxi Agricultural University, was used in this study. Oat seeds were surface-sterilized with 1.0% sodium hypochlorite for 20 min and then washed three times with sterile water. The seeds were germinated on filter paper in petri dishes for 4 days and then transplanted to a germination box containing modified Hoagland nutrient solution. The experiment was conducted in a greenhouse under a 16 h light/8 h darkness cycle with a light intensity of 250 μmol m-2 s-1. At the one-leaf stage, PY8 seedlings were treated with 15% polyethylene glycol 6000 (PEG 6000) solutions. The first leaves and roots of PY8 seedlings were sampled at five time points (6, 12, 24, 48, and 72 hours), with three biological replicates in both treated and untreated groups. Additionally, the stems were collected at the 24-hour time point, with three biological replicates for each group. Each sample was immediately frozen in liquid nitrogen and stored at −80°C for further analysis.
2.6 Quantitative reverse transcription PCR
Total RNA was extracted from each sample using the DP432 kit (Tiangen, China), and cDNA was synthesized using HiScript III 1st-Strand cDNA Synthesis Kit (Vazyme, China). qRT-PCR was performed on the QuantStudion 6 Flex Real-Time PCR System (Thermo Fisher Scientific, USA) using 2X RealStar Fast SYBR qPCR Mix (Genstar, China). The primers used to evaluate the transcript levels are listed in Supplementary Table S2. The AsActin gene was used as the endogenous control. Relative expression levels were determined using the (2−ΔΔCt) ± SEM method. Each sample was analyzed in three biological replicates.
2.7 Subcellular localization analysis
To determine the molecular characteristics of the AsBZR12 protein, the coding sequence of AsBZR12 was cloned and fused into the pCAMBIA1302-GFP vector. The recombinants and pCAMBIA1302-GFP empty controls were subjected to Agrobacterium (GV3101)-mediated transient expression in N. tabacum leaves, together with the nucleus marker plasmid NLS-mCherry. The transformed N. tabacum leaves were observed using confocal laser-scanning microscopy (LSM800, Carl Zeiss, Germany). The subcellular localization of AsBZR12 protein was determined at three times.
3 Results
3.1 Identification of BZR gene family in A. sativa
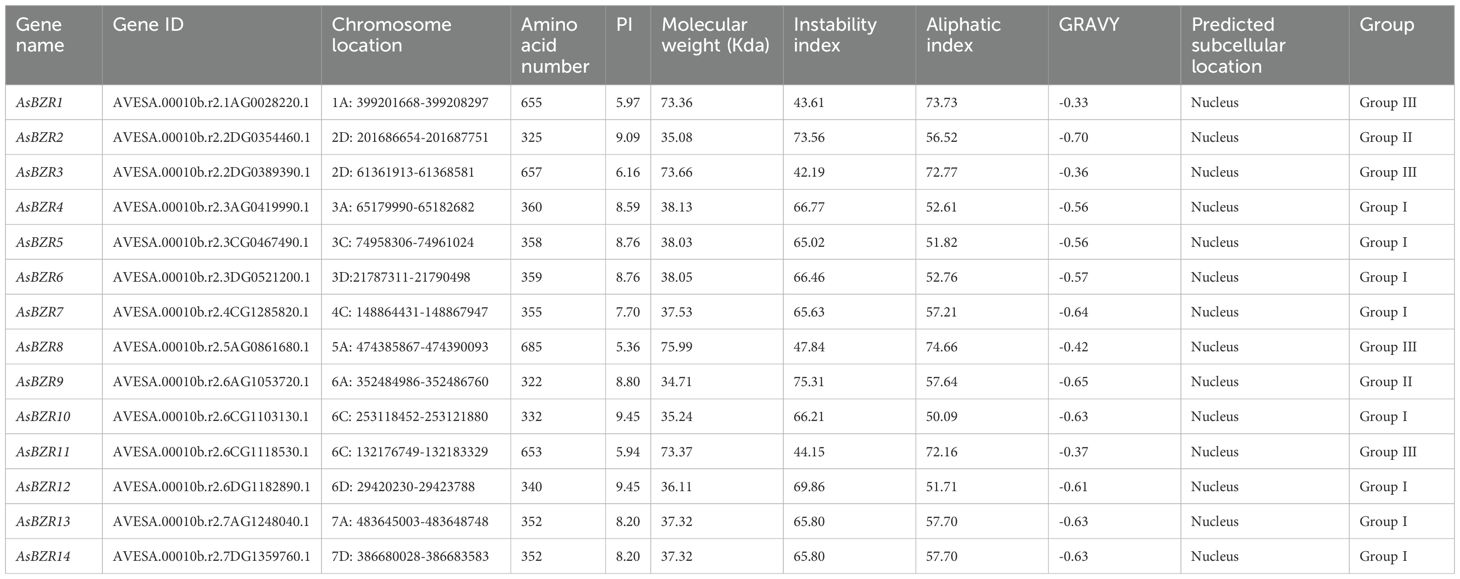
According to BLASTP and HMM searches, followed by examination of conserved domains in Pfam, NCBI-CDD, and SMART, a total of 14 BZR genes were identified in A. sativa. The AsBZR gene family members were systematically assigned and named AsBZR1 to AsBZR14 based on their respective chromosomal locations (Table 1). The amino acid length of AsBZR proteins ranged from 322 to 685 residues, with isoelectric points (PI) varying from 5.36 to 9.45, averaging 7.89. The predicted molecular weight of BZR proteins ranged from 34.71 kDa to 75.99 kDa, with a mean of 47.42 kDa. Proteins with an instability index above 40 are considered unstable and prone to degradation (Ullah et al., 2022). As the instability index of all AsBZR proteins was above 40, these proteins are likely unstable in A. sativa. Additionally, the grand average of hydropathicity (GRAVY) values for all AsBZR proteins were negative, indicating their hydrophilic nature. Subcellular localization prediction showed that all AsBZR genes were located in the nucleus.
3.2 Chromosomal distribution of AsBZR genes
The 14 identified AsBZR genes were mapped to 12 of the 21 oat chromosomes (Table 1), revealing an uneven distribution across the genome. The AsBZR genes were found on chromosomes 1A, 2D, 3A, 3C, 3D, 4C, 5A, 6A, 6C, 6D, 7A, and 7D, with each chromosome containing one to two genes. Specifically, chromosomes 2D and 6C each harbored 2 AsBZR genes, while single genes were present on the remaining chromosomes. Notably, homoeologous group 6 contained the highest number of AsBZR genes, comprising AsBZR9, AsBZR10, AsBZR11, and AsBZR12. The AsBZR genes demonstrated relatively uniform distribution among the A, C, and D sub-genomes, with 5 genes in both A and D sub-genomes and 4 genes in the C sub-genome.
3.3 Phylogenetic analysis of AsBZR genes
To elucidate the evolutionary relationships of BZR genes across species, the phylogenetic tree was constructed incorporating 14 AsBZR, 16 TaBZR, 6 OsBZR, and 6 AtBZR proteins (Ullah et al., 2022; Yin et al., 2005). The phylogenetic analysis revealed three distinct groups among the 14 AsBZR proteins. Among the three groups, Group I was the largest, with eight members: AsBZR4, AsBZR5, AsBZR6, AsBZR7, AsBZR10, AsBZR12, AsBZR13, and AsBZR14. Group II contained two members (AsBZR2 and AsBZR9), and Group III included four members (AsBZR1, AsBZR3, AsBZR8, and AsBZR11) (Figures 1, 2A).
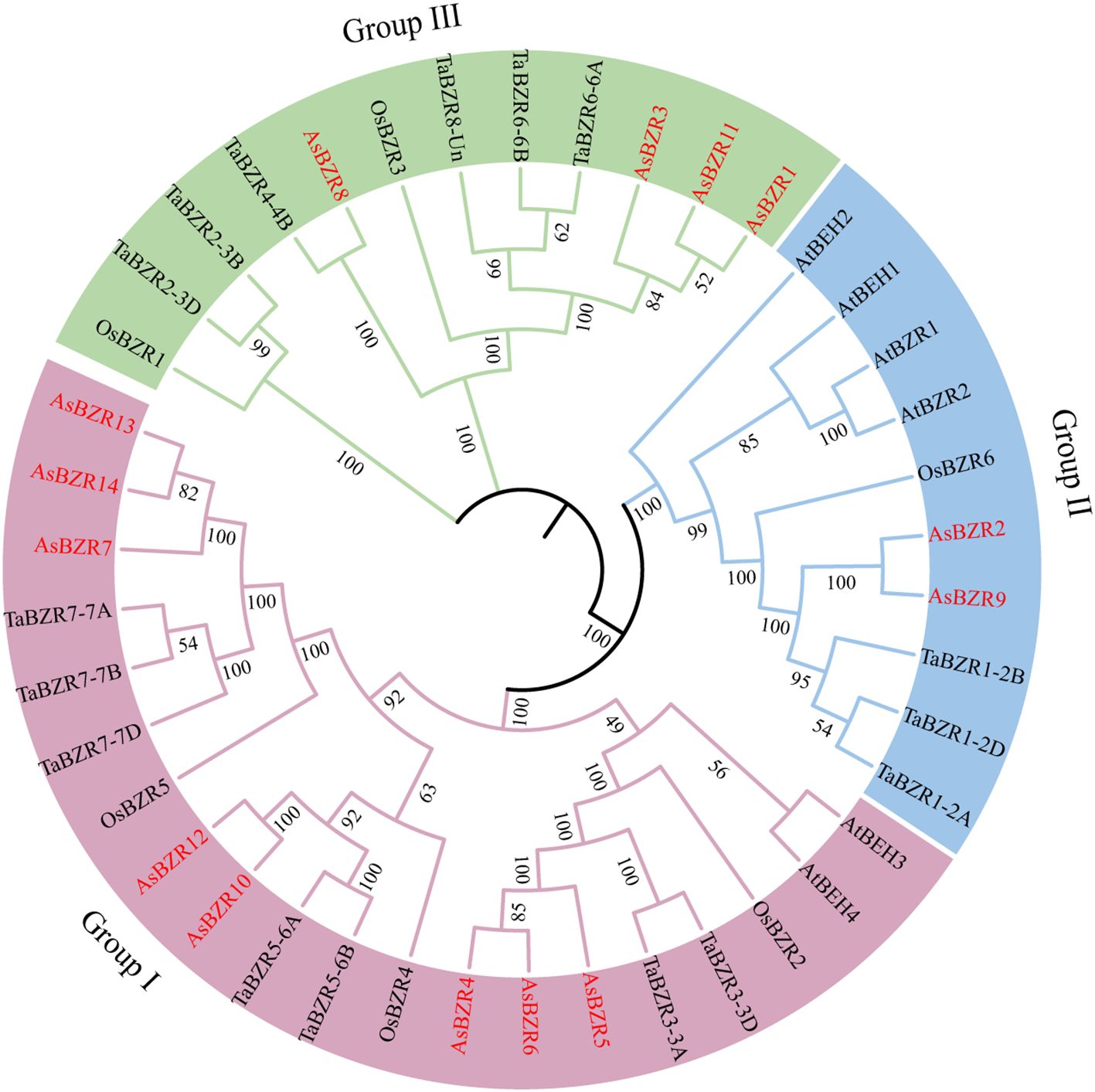
Figure 1. Phylogenetic relationship of BZR proteins in the A. sativa, A. thaliana, O. sativa, and T. aestivum. All AsBZR proteins are highlighted in red, and different colors represent different groups.
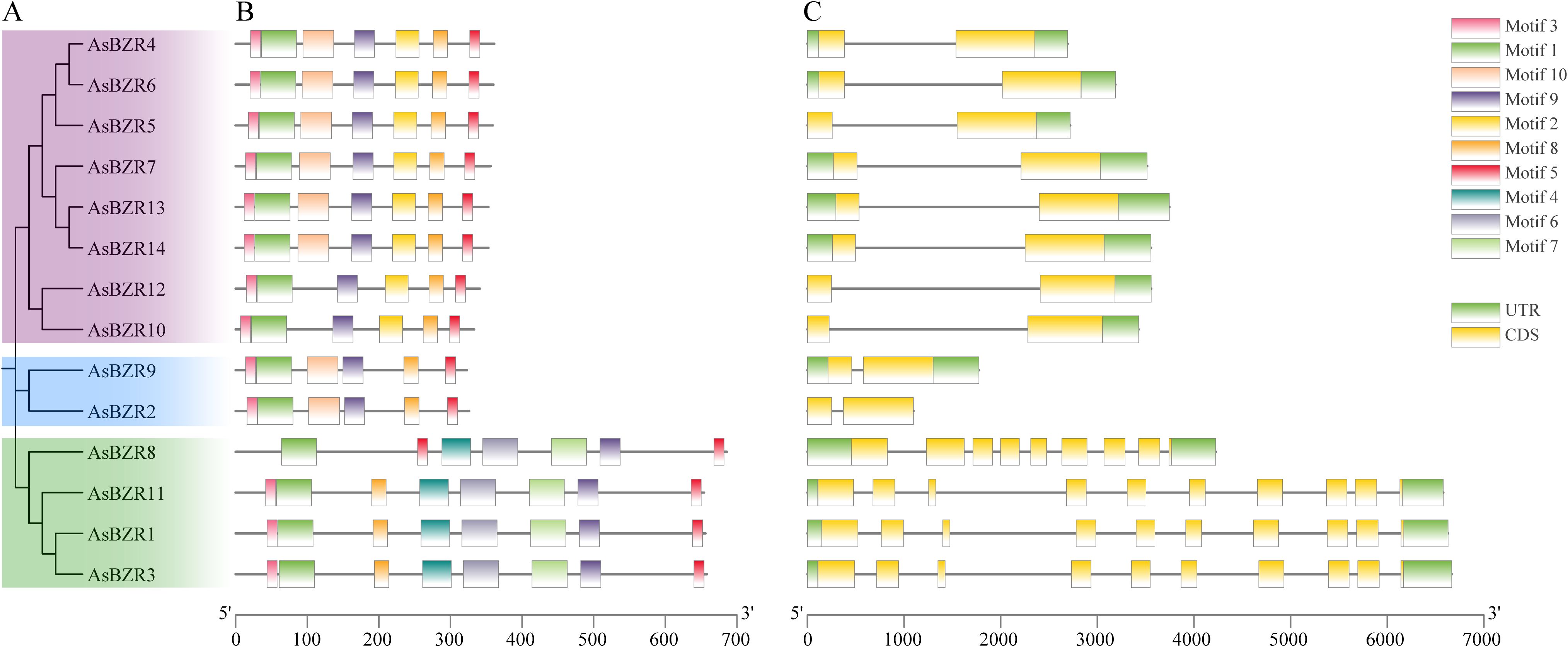
Figure 2. Phylogenetic tree, conserved motifs, and gene structure of AsBZR genes. (A) Phylogenetic tree constructed using the AsBZR protein sequences. (B) Distribution of conserved motifs for AsBZR proteins. The different motifs are shown in different color boxes. (C) Gene structure of AsBZR genes. Green boxes indicate UTR regions, yellow boxes indicate exons, grey lines indicate introns.
3.4 Gene structure and motif analysis of AsBZR genes
Analysis of sequence conservation in AsBZR genes revealed distinct patterns in their conserved motifs (Figure 2B; Supplementary Figure S1). Motifs 1, 5, 8, and 9 were widely distributed across all AsBZR domains, indicating their high conservation among the genes. Members within the same subgroup displayed similar patterns in both type and number of conserved motifs, suggesting functional similarities. The key distinction between Group I and Group II members was the presence of motif 2 exclusively in Group I, while motifs 4, 6, and 7 were characteristic of Group III.
To investigate the evolution of the BZR gene family in oat, we analyzed the exon-intron structures of 14 AsBZR genes. The analysis revealed that all AsBZR genes contained between two to ten exons (Figure 2C). AsBZR genes within the same group exhibited similar structural patterns. While AsBZR genes in both Group I and Group II contained one intron and two exons, Group I displayed longer intron lengths compared to Group II. Group III members contained nine to ten exons (AsBZR8 with nine exons, AsBZR1, AsBZR11, and AsBZR3 with ten exons). The phylogenetic analyses, conserved motif patterns, and similar gene architectures among BZR gene family members within clusters provide substantial evidence supporting the accuracy of the taxonomic classification.
3.5 Cis−acting elements analysis
To better understand the potential regulatory mechanism of the BZR gene family in oat, we examined the regulatory elements located 2000 bp upstream of the promoters for all 14 AsBZR genes (Figure 3; Supplementary Figure S2). The results showed that the promoters of AsBZR genes contain numerous phytohormones-responsive cis-elements, including those responsive to salicylic acid, abscisic acid, zein metabolism, methyl jasmonate (MeJA), gibberellin, and auxin. Additionally, cis-elements associated with environmental stress signal responsiveness were identified, including light, low-temperature, circadian, anoxic, anaerobic, and mixed stress (defense and stress responsiveness) elements. Several AsBZR genes contained tissue-specific expression elements for meristem, seed, root, and palisade mesophyll. Moreover, cis-elements associated with transcription factors, such as MYB, were found to bind to BZR gene promoters. The findings indicate that AsBZR family members likely participate in diverse stress and hormone signaling pathways, suggesting significant roles in stress responses and development.
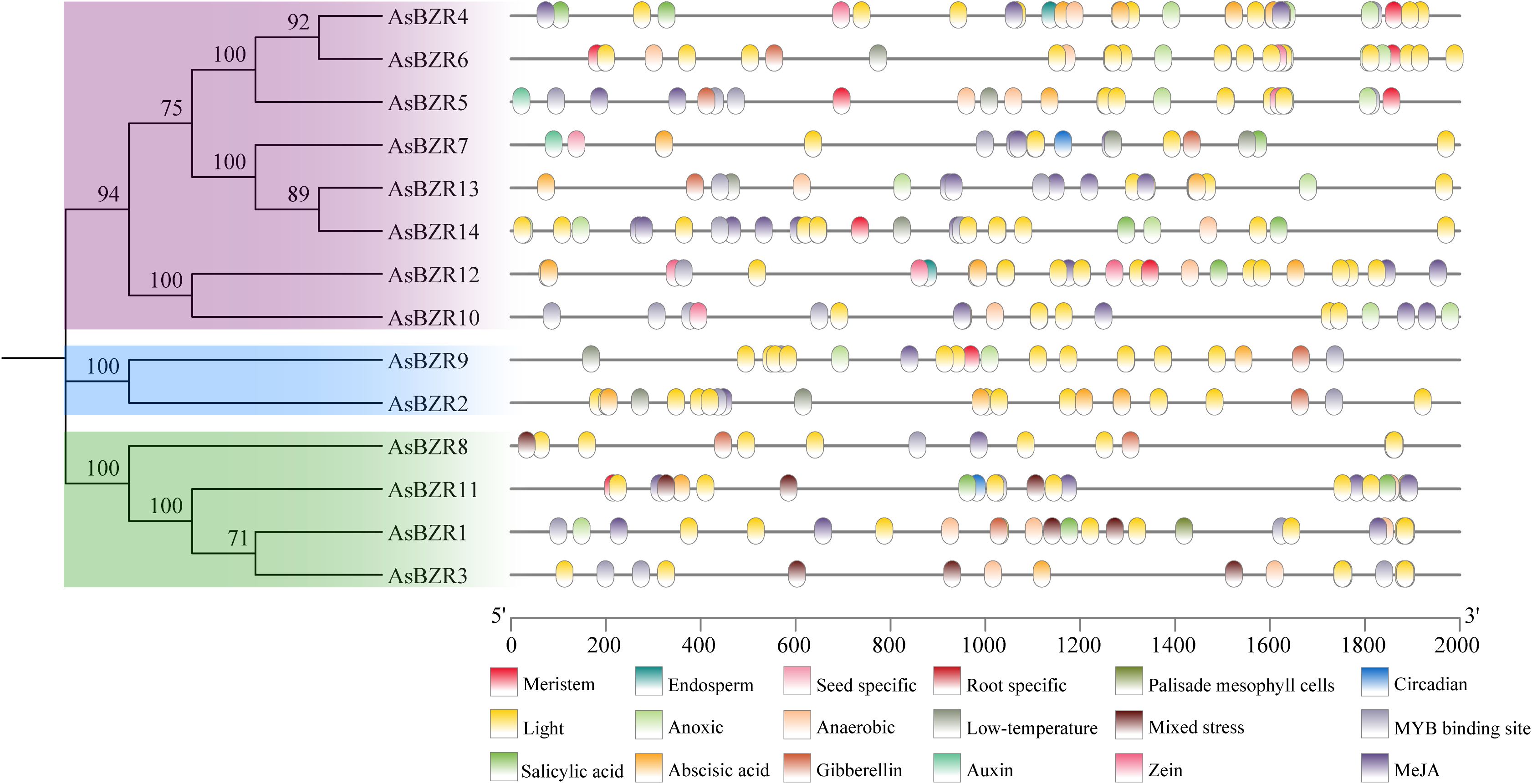
Figure 3. Cis-acting elements in the promoters of AsBZR genes. Different colors represent different elements.
3.6 Gene duplication and collinearity analysis of BZR gene family
To elucidate the relationship between the expansion of the BZR gene family and the polyploidization process in oat, we performed a genome-wide analysis of duplication events involving AsBZR genes (Figure 4A). The analysis revealed 67,910 regions of collinearity in the oat genome, including those on the Un chromosomes. The study identified 19 segmental duplication events among 14 AsBZR genes, with some AsBZR genes having multiple repeats. No tandem duplication pairs were detected (Supplementary Table S3). Among these, 15 gene pairs were comprised of AsBZR genes at both ends, while the remaining pairs contained an AsBZR gene at one end and a non-AsBZR gene at the other. This indicates that the BZR gene family expansion in the oat genome occurred exclusively through segmental duplication, with no evidence of tandem duplication events.
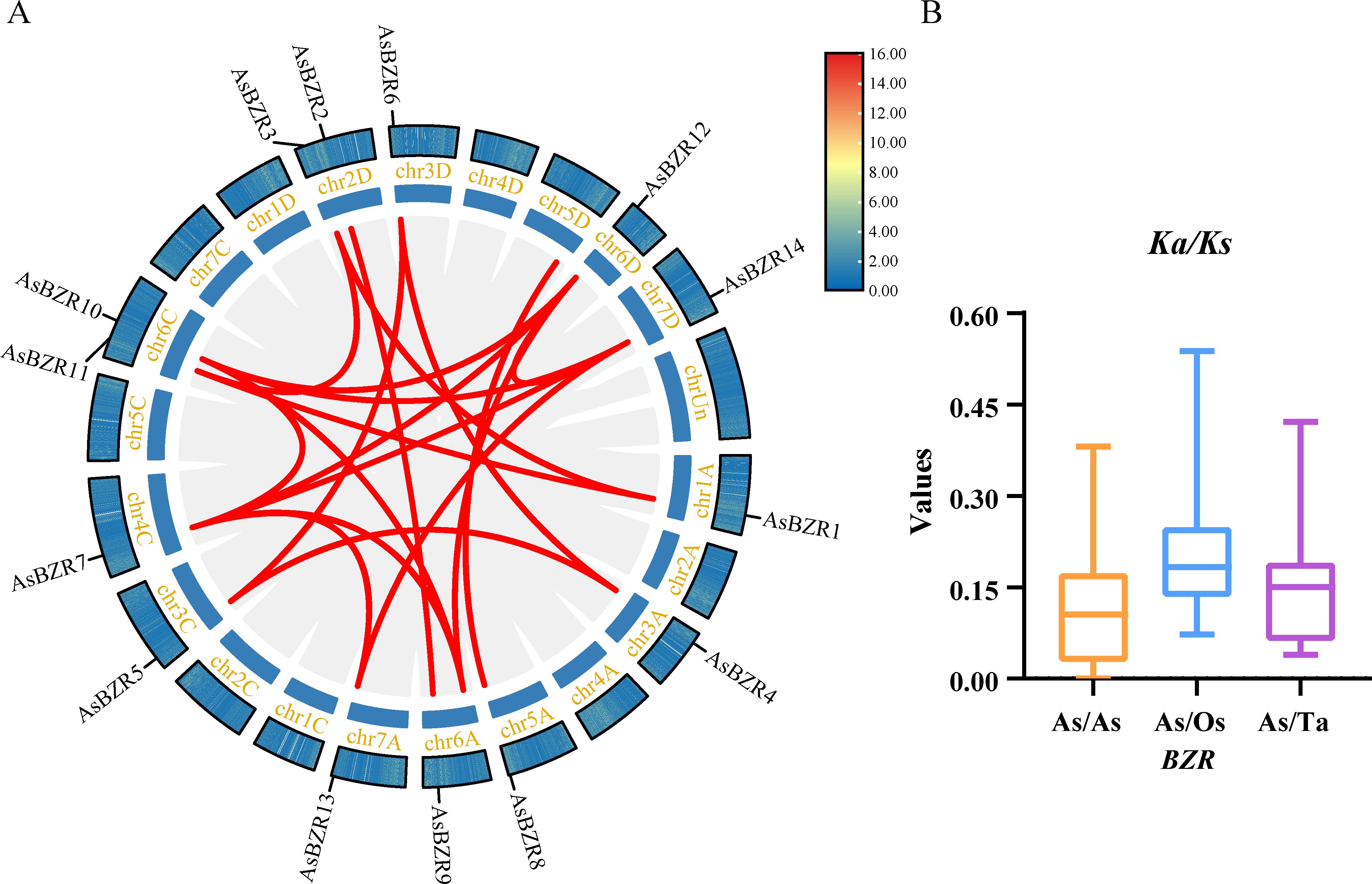
Figure 4. Duplication events and evolutionary rates of the AsBZR genes. (A) Duplication event in A. sativa. The gray lines indicate collinearity and the red lines highlight the duplication events of AsBZR genes. (B) Ka/Ks ratios of AsBZR gene pairs with TaBZR and OsBZR.
To further elucidate the evolutionary dynamics and selective pressures affecting the duplicated AsBZR genes, we calculated the ratios of Ka and Ks substitutions for the 15 duplicated AsBZR gene pairs (Supplementary Table S4). The value of Ka/Ks > 1 indicates positive selection, = 1 indicates neutral selection, and < 1 suggests purifying selection (Liu et al., 2022). The results indicated that Ka/Ks values for the 15 gene pairs ranged from 0.00 and 0.38. Since these Ka/Ks values were all less than 1 (Figure 4B), this suggests that the oat BZR gene family has experienced strong purifying selection pressure during evolution. Additionally, we performed Ka/Ks ratio analyses of the BZR genes in oat compared with their orthologs in O. sativa, and T. aestivum (Figure 4B; Supplementary Table S8). The results showed that the Ka/Ks values were all less than 1, indicating that purifying selection has been a significant factor in the evolution of the BZR genes.
To investigate the evolutionary mechanisms and orthologous relationships of the oat BZR gene family, comparative synteny maps were constructed between oat and A. thaliana, T. aestivum, and O. sativa (Figure 5). The collinearity analysis revealed 7,418, 56,774, and 94,487 collinearity pairs between oat and A. thaliana, O. sativa, and T. aestivum, respectively. Additionally, three AsBZR genes (AsBZR4, AsBZR5, and AsBZR6) demonstrated collinear gene pairs with AtBEH3 in A. thaliana and oat. A total of 13 AsBZR genes exhibited syntenic relationships with those in O. sativa and T. aestivum. The analysis identified 3, 26, and 34 orthologous pairs between oat and A. thaliana (Supplementary Table S5), O. sativa (Supplementary Table S6), and T. aestivum (Supplementary Table S7) respectively. Several AsBZR genes showed associated with at least three syntenic pairs in O. sativa and T. aestivum.
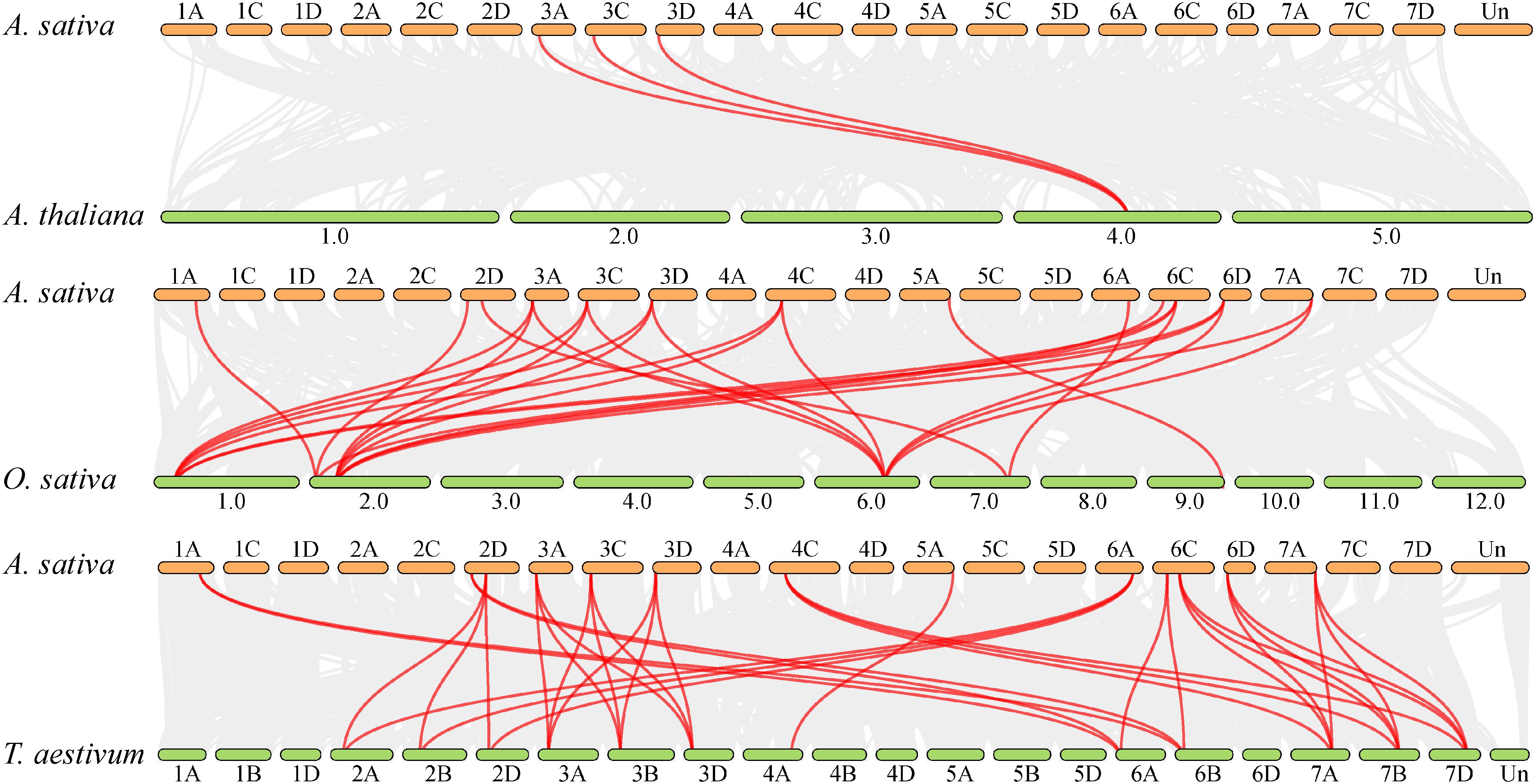
Figure 5. Synteny analysis of BZR genes between A. sativa, A. thaliana, O. sativa, and T. aestivum. The gray lines show colinear blocks within A. sativa with other plant genomes, while the red lines highlight the synteny BZR gene pairs. Chromosome numbers are labelled above or below each chromosome.
3.7 Expression pattern of AsBZR genes under osmotic stress
To elucidate the response of AsBZR genes to osmotic stress, we examined their expression patterns at various time points (6, 12, 24, 48, and 72 h) and in different organs (roots, stems, and leaves) in oat. Relative to the control group, osmotic treatment significantly upregulated AsBZR10, AsBZR12, and AsBZR13 in roots, while AsBZR1 and AsBZR11 were downregulated. The remaining genes exhibited variable upregulation or downregulation at different time points post-treatment (Figure 6). In leaves, AsBZR1, AsBZR3, AsBZR4, AsBZR8, AsBZR11, AsBZR12, AsBZR13, and AsBZR14 were significantly upregulated, while AsBZR2, AsBZR6, and AsBZR9 were significantly downregulated (Figure 7). At 24 h post-treatment, AsBZR1, AsBZR3, AsBZR4, AsBZR5, AsBZR8, AsBZR9, AsBZR10, AsBZR11, AsBZR12, and AsBZR14 were upregulated in stems, with AsBZR2 being downregulated (Figure 8). Notably, AsBZR12 maintained consistent upregulation across all tested organs. The comprehensive expression analysis suggests that AsBZR12 may play a crucial role in osmotic response.
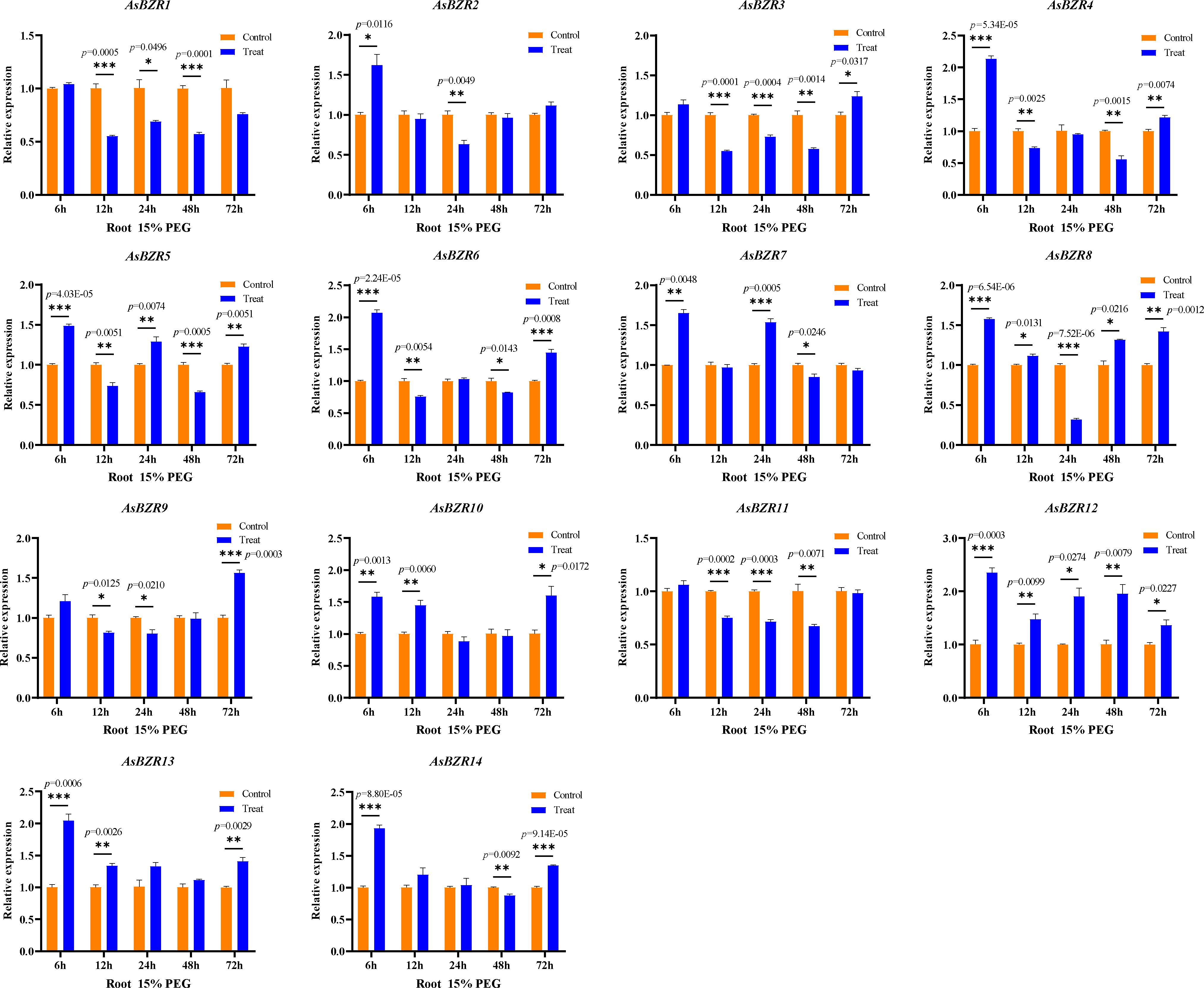
Figure 6. Expression patterns of osmotic stress in roots of A. sativa at different time points. The ‘control’ and ‘treat’ groups represent samples of the untreated and treated with 15% PEG 6000, respectively. The AsActin gene was used as a control. The error bars indicate the means ± SEMs from three independent experiments. Asterisks indicate significant differences between the ‘treat’ group and the ‘control’ group (*P < 0.05, **P < 0.01, ***P < 0.01, Student’s t-test).
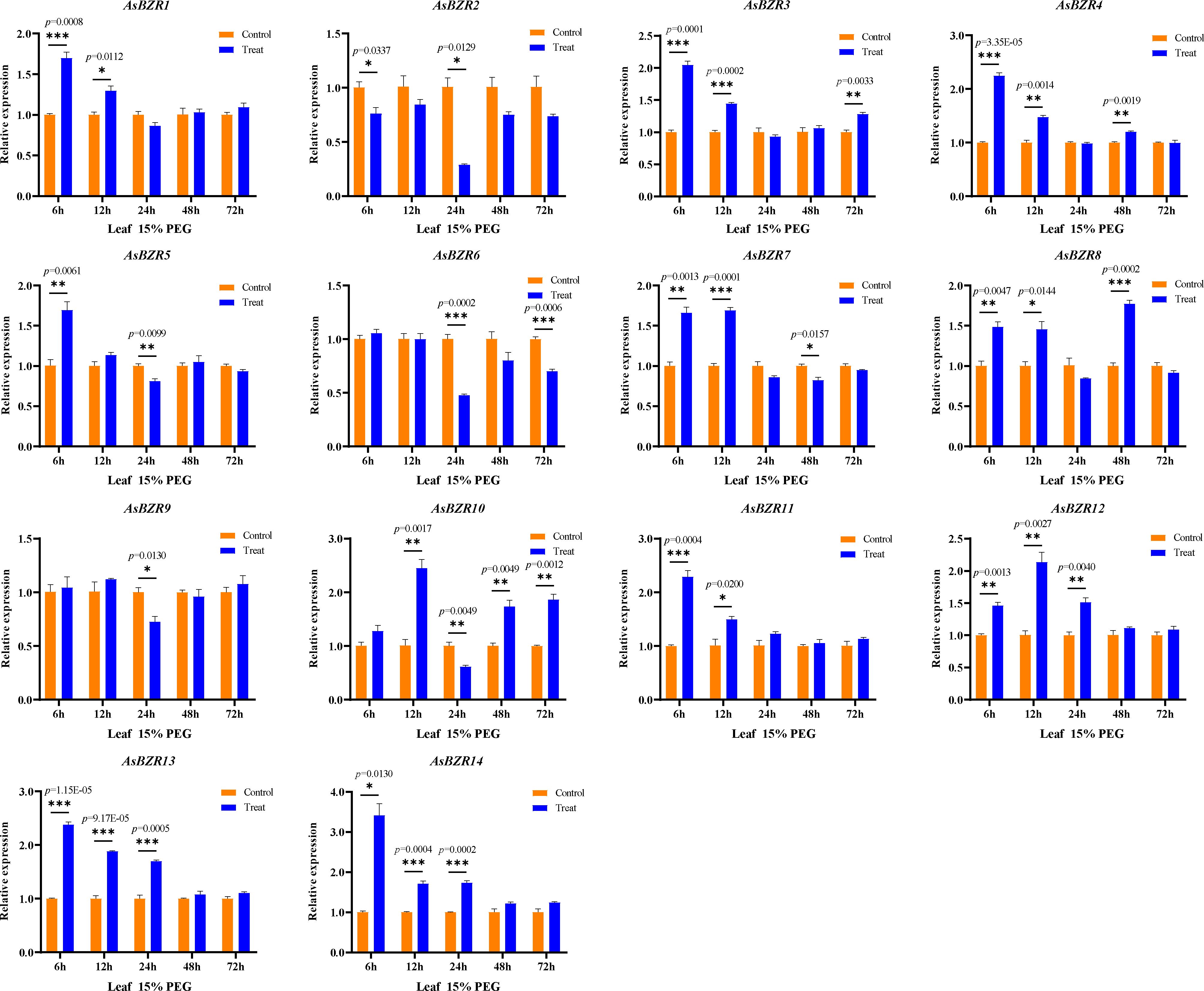
Figure 7. Expression patterns of osmotic stress in leaves of A. sativa at different time points. The ‘control’ and ‘treat’ groups represent samples of the untreated and treated with 15% PEG 6000, respectively. The AsActin gene was used as a control. The error bars indicate the means ± SEMs from three independent experiments. Asterisks indicate significant differences between the ‘treat’ group and the ‘control’ group (*P < 0.05, **P < 0.01, ***P < 0.01, Student’s t-test).

Figure 8. Expression heatmap of osmotic stress in stems of A. sativa. Data collected after 24 h of PEG treatment.
3.8 Subcellular localization of the AsBZR12 protein
The expression patterns of AsBZR genes revealed that AsBZR12 exhibited consistently high expression levels across all tested organs, suggesting that AsBZR12 may have a critical function. To investigate the subcellular localization of AsBZR12, we generated an AsBZR12-GFP fusion construct and transiently expressed it in N. tabacum leaves. The results revealed that the AsBZR12-GFP fusion protein was localized in the nucleus, confirming our initial prediction (Figure 9).
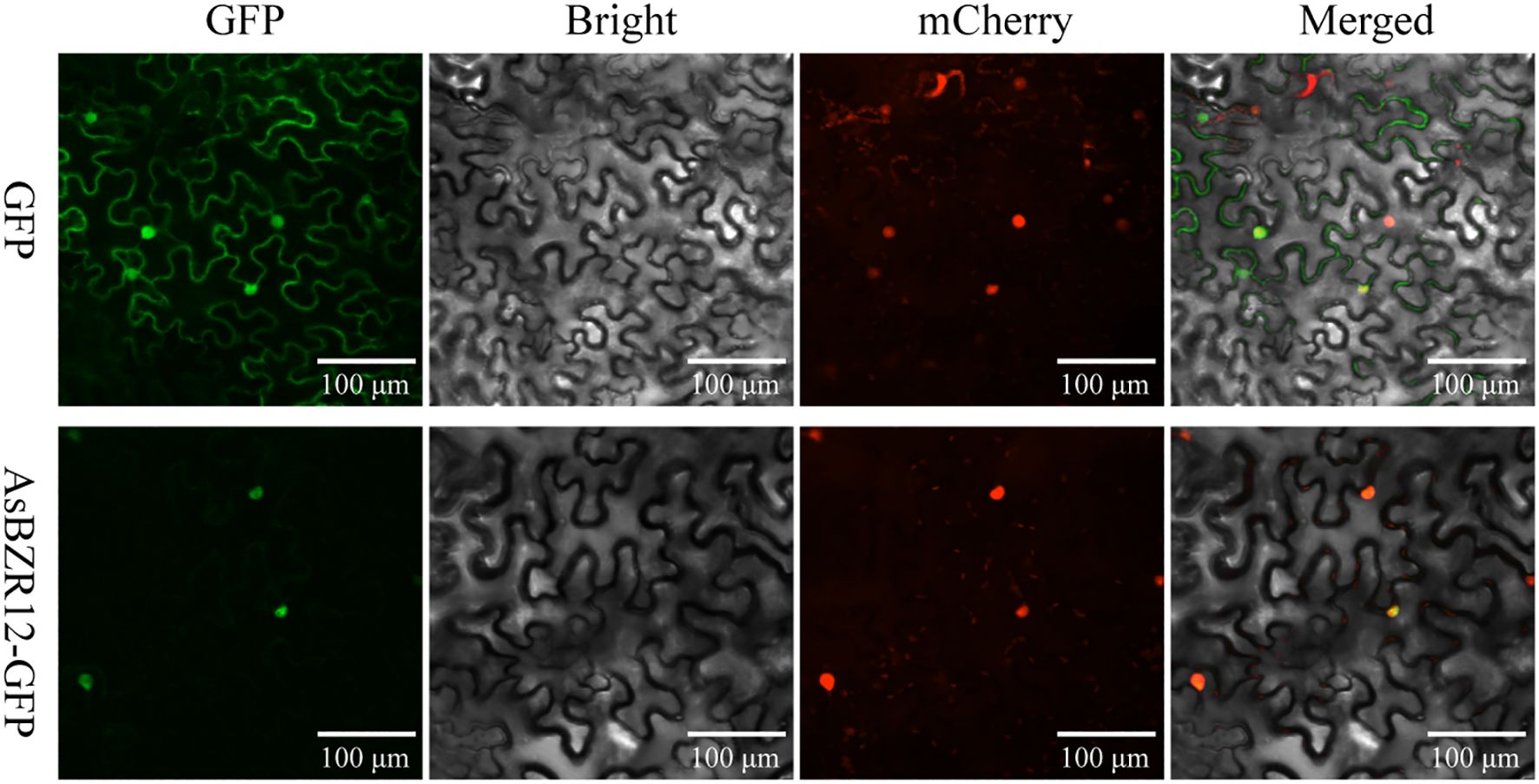
Figure 9. Subcellular localization of AsBZR12. GFP was used as a control. Three independent experiments were performed. Scale bars, 100 μm.
4 Discussion
The BZR gene family is a key component in the BR signaling pathway, playing an indispensable role in plant growth and development as well as in response to stress. A comprehensive whole-genome analysis of the BZR gene family is essential for understanding the functions of BZR proteins in plants. Such analyses have been performed in various species, including A. thaliana (Yin et al., 2005), T. aestivum (Ullah et al., 2022), O. sativa (Ullah et al., 2022), Zea mays (Manoli et al., 2018), S. italica (Liu et al., 2021) and Beta vulgaris (Wang et al., 2019). The number of BZR genes identified varies among species: 6 in A. thaliana, O. sativa and B. vulgaris, 16 in T. aestivum, 11 in Z. mays, and 7 in S. italica. The genome size of oat is approximately 11 Gb (He Q. et al., 2024), which is smaller than that of T. aestivum but larger than those of O. sativa, Z. mays, S. italica, and B. vulgaris. In this study, we identified 14 members of the BZR gene family in A. sativa, distributed across 12 different chromosomes. The number of members in the BZR gene family may be correlated with the size of the plant genome.
To understand the evolutionary relationships and potential functional divergence within the BZR gene family in oat, phylogenetic analysis classified the 14 AsBZR genes into three groups. This tripartite classification aligns with findings in C. sativus (Luo et al., 2023), B. vulgaris (Wang et al., 2019), and four dicot species (Wang et al., 2024): A. thaliana, Carica papaya, Vitis vinifera, and Populus trichocarpa. Phylogenetic analysis of the AsBZR protein and its orthologs in A. thaliana, O. sativa, and T. aestivum revealed that the T. aestivum orthologs within a more closely related branch, indicating a stronger phylogenetic relationship between AsBZR and the T. aestivum orthologs. Furthermore, the gene structure and conserved motif identification corroborate the classification results. The widespread presence of motifs 1, 5, 8, and 9 in all AsBZR domains emphasizes their significance in maintaining core functions of the BZR gene family. Group-specific motifs, such as motif 2 in Group I and motifs 4, 6, and 7 specific in Group III, suggest functional specialization. Additionally, the promoter regions of AsBZR genes contain diverse cis-acting elements, including hormone-responsive elements, stress resistance response elements, and tissue-specific expression elements, indicating their critical roles in regulating hormone signaling, responding to stress, and controlling development.
Oats represent a vital food and forage crop in northern China’s arid and semi-arid regions, where drought stress significantly impacts yield and quality (Xu et al., 2024; Zhu S. S. et al., 2024). Studies have shown that drought stress induces the expression of the BZR transcription factor in multiple plant species. BZR transcription factors play a regulatory role in Arabidopsis responses to drought (Ye et al., 2017). Drought stress upregulates SbBZR1 expression, and its overexpression in A. thaliana enhances drought tolerance through activation of antioxidant genes (Li et al., 2025). ZmBES1/BZR1–4 binds to the downstream target genes ZmMBP1 and ZmPum6 to activate their transcription, while simultaneously recruiting the ZmTLP5 protein to form a regulatory complex that further enhances transcriptional activity, thereby negatively regulating drought response (Feng et al., 2025). To elucidate the role of AsBZR genes in oat osmotic responses, we analyzed their expression levels under osmotic stress conditions. The expression patterns demonstrate a complex regulatory response to osmotic stress. In roots, most genes exhibit an oscillatory pattern of upregulation, downregulation, and upregulation, except for AsBZR1, AsBZR10, AsBZR11, and AsBZR12. In leaves, AsBZR5, AsBZR7, and AsBZR10 display a similar oscillatory pattern. Notably, AsBZR1 and AsBZR11 demonstrate tissue-specific responses, with downregulation in roots but upregulation in leaves. AsBZR12 shows significant upregulation across roots, stems, and leaves, with nuclear localization, suggesting its potential role in coordinating osmotic response through regulation of downstream target genes and enhancement of plant osmotic resilience.
We investigated the relationships between AsBZR12 and its orthologous pairs in wheat and found that AsBZR12 has three orthologs in wheat, namely TraesCS7A02G354800 (TaBZR7-7A), TraesCS7B02G272900 (TaBZR7-7B), and TraesCS7D02G368000 (TaBZR7-7D). Sequence alignment revealed that these orthologs share a highly conserved BES1_N domain at the N-terminus (Supplementary Figure S3). Gene function validation often relies on mutant and transgenic studies. For instance, the bzr1-1D mutant has been used to demonstrate that BZR1 inhibits flowering primarily through the suppression of CO and FT and the activation of FLC (Xu et al., 2025). Similarly, mutant and transgenic analyses have confirmed that SlBZR1.7 promotes tomato fruit elongation by positively regulating SUN gene expression (Yu et al., 2022). In the present study, we conducted expression analysis but did not perform in-depth functional studies. Future work should focus on creating mutants and conducting transgenic analyses of AsBZR12 to validate its function.
Gene duplication is a major driving force for gene family expansion and has played a significant role in the evolutionary process of plants (Chen et al., 2024). In wheat, the expansion of the BZR gene family is likely mediated by whole-genome duplication (WGD) and segmental duplication events (Kesawat et al., 2021). Oat, like wheat, is hexaploid and have experienced WGD during their evolutionary history. The identification of three pairs of orthologous genes between AsBZR2, AsBZR7, AsBZR9, AsBZR10, AsBZR12, AsBZR13 and TaBZR suggests that the BZR syntenic orthologs may have originated from segmental duplication events. Therefore, we hypothesize that the expansion of the BZR gene family in oat is similarly driven by WGD and segmental duplication.
To investigate the genetic relationships and evolutionary dynamics of BZR genes across different species, we constructed a collinearity relationship among A. thaliana, rice, and wheat. Comparative analysis reveals that more orthologous BZR gene pairs are identified between oat and wheat than between oat and either A. thaliana or rice. This finding indicates a closer genetic relationship between AsBZR and TaBZR. To further explore the evolutionary rates of BZR genes, we calculated the Ka/Ks ratios for AsBZR gene pairs and for orthologous BZR gene families between oat and rice or wheat. The Ka/Ks ratios were consistently less than 1, indicating that BZR genes are under purifying selection pressure. Similarly, the Ka/Ks ratios of TaBZR gene pairs were also less than 1 (Kesawat et al., 2021). These results imply that these genes are highly conserved and likely play crucial roles in maintaining their fundamental functions. Additionally, the Ka/Ks ratios between AsBZR and TaBZR genes are relatively lower than those between AsBZR and OsBZR genes, further supporting the closer genetic relationship between oat and wheat. This difference may be attributed to the genomic characteristics and evolutionary history of oat, which share many similarities with wheat.
5 Conclusion
In this study, we performed a genome-wide identification and characterization of the BZR gene family in A. sativa, identifying 14 AsBZR genes divided into three groups. The promoters of all AsBZR genes contain enriched elements response to phytohormone and environmental stress, demonstrating strong purifying selection pressure throughout evolution. Through qRT-PCR analysis, AsBZR12 demonstrated significant involvement in osmotic stress response, with AsBZR12 showing nuclear localization. These findings elucidate the sequence characteristics of the AsBZR gene family and establish a foundation for future investigations into the osmotic stress resistance functions of AsBZR genes in oat.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
SX: Methodology, Writing – review & editing, Writing – original draft, Conceptualization, Project administration. ZW: Investigation, Formal Analysis, Writing – original draft. MM: Writing – original draft, Investigation, Formal Analysis, Visualization. LZ: Formal Analysis, Writing – original draft, Visualization, Investigation. ZL: Visualization, Investigation, Formal Analysis, Writing – original draft. LL: Project administration, Writing – review & editing, Resources, Conceptualization.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This project was supported by the PhD of Shanxi Agricultural University Scientific Research Start-up Project (2024BQ50), Shanxi Province Doctoral Work Award-Scientific Research Project (SXBYKY2024114), the China Agriculture Research System of MOF and MARA (CARS-07-A-2), Key R&D Project in Shanxi Province (2022ZDYF110), and Hou Ji Laboratory in Shanxi Province (202304010930003).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1616026/full#supplementary-material
References
Chen, C. J., Wu, Y., Li, J. W., Wang, X., Zeng, Z. H., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Chen, Y., Li, A. X., Yun, P., Chen, Q., Pan, D. Y., Guo, R., et al. (2024). Genome-wide analysis of MYB transcription factor family and AsMYB1R subfamily contribution to ROS homeostasis regulation in Avena sativa under PEG-induced drought stress. BMC Plant Biol. 24, 632. doi: 10.1186/s12870-024-05251-w
Chou, K. C. and Shen, H. B. (2008). Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 3, 153–162. doi: 10.1038/nprot.2007.494
Cui, X. Y., Gao, Y., Guo, J., Yu, T. F., Zheng, W. J., Liu, Y. W., et al. (2019). BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 180, 605–620. doi: 10.1104/pp.19.00100
Feng, W. Q., Zhang, H. W., Cao, Y., Liu, Y., Zhao, Y. R., Sun, F. A., et al. (2023). Maize ZmBES1/BZR1–1 transcription factor negatively regulates drought tolerance. Plant Physiol. Biochem. 205, 108188. doi: 10.1016/j.plaphy.2023.108188
Feng, W. Q., Zhou, Y. H., Duan, H. M., Zhou, W. X., Zhang, X., Liu, Y., et al. (2025). Maize ZmBES1/BZR1–4 recruits ZmTLP5 to regulate drought tolerance and seed development by regulating ZmPum6 and ZmMBP1. Plant J. 122, e70162. doi: 10.1111/tpj.70162
Ghorbel, M., Zribi, I., Chihaoui, M., Alghamidi, A., Mseddi, K., and Brini, F. (2023). Genome-wide investigation and expression analysis of the catalase gene family in oat plants (Avena sativa L.). Plants 12, 3694. doi: 10.3390/plants12213694
Guo, H. Q., Wu, H. L., Hou, Y. B., Hu, P. L., Du, J. E., Cao, L. J., et al. (2023). Oat β-D-glucan ameliorates type II diabetes through TLR4/PI3K/AKT mediated metabolic axis. Int. J. Biol. Macromol. 249, 126039. doi: 10.1016/j.ijbiomac.2023.126039
He, J. X., Gendron, J. M., Sun, Y., Gampala, S. S. L., Gendron, N., Sun, C. Q., et al. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634–1638. doi: 10.1126/science.1107580
He, Q., Li, W., Miao, Y. Q., Wang, Y., Liu, N. K., Liu, J. N., et al. (2024). The near-complete genome assembly of hexaploid wild oat reveals its genome evolution and divergence with cultivated oats. Nat. Plants. 10, 2062–2078. doi: 10.1038/s41477-024-01866-x
He, Z. H., Zhang, P., Jia, H. T., Zhang, S. L., Nishawy, E., Sun, X. P., et al. (2024). Regulatory mechanisms and breeding strategies for crop drought resistance. New Crops. 1, 100029. doi: 10.1016/j.ncrops.2024.100029
Hou, Z. H., Zheng, W. J., Zheng, L., Wang, J. Y., Zhang, S. X., Wei, J. T., et al. (2025). TaPPR13, a pentatricopeptide repeat protein gene activated by TaBZR2, confers drought stress tolerance by enhancing the antioxidant defense system and promoting retrograde signaling in wheat (Triticum aestivum). Adv. Sci. 29, e02984. doi: 10.1002/advs.202502984
Jin, Z. Y., Zhou, T., Chen, J. J., Lang, C. T., Zhang, Q. Q., Qin, J., et al. (2024). Genome-wide identification and expression analysis of the BZR gene family in Zanthoxylum armatum DC and functional analysis of ZaBZR1 in drought tolerance. Planta 260, 41. doi: 10.1007/s00425-024-04469-0
Kamal, N., Renhuldt, N. T., Bentzer, J., Gundlach, H., Haberer, G., Juhász, A., et al. (2022). The mosaic oat genome gives insights into a uniquely healthy cereal crop. Nature 606, 113–119. doi: 10.1038/s41586-022-04732-y
Kesawat, M. S., Kherawat, B. S., Singh, A., Dey, P., Kabi, M., Debnath, D., et al. (2021). Genome-wide identification and characterization of the Brassinazole-resistant (BZR) gene family and its expression in the various developmental stage and stress conditions in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 22, 8743. doi: 10.3390/ijms22168743
Lee, S. H., Kim, S. H., Park, T. K., Kim, Y. P., Lee, J. W., and Kim, T. W. (2024). Transcription factors BZR1 and PAP1 cooperate to promote anthocyanin biosynthesis in Arabidopsis shoots. Plant Cell. 36, 3654–3673. doi: 10.1093/plcell/koae172
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, X., Chen, C., Cao, B., Feng, S., Zhang, Y. L., Bin, T. T., et al. (2025). Drought-activated BZR1 reprograms flavonoid metabolism via transcriptional cascades to amplify baicalin biosynthesis in Scutellaria baicalensis. Plant Physiol. Biochem. 227, 110196. doi: 10.1016/j.plaphy.2025.110196
Li, Y. Y., He, L. L., Li, J., Chen, J. H., and Liu, C. N. (2018). Genome-wide identification, characterization, and expression profiling of the legume BZR transcription factor gene family. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01332
Li, J., Wen, J. Q., Lease, K. A., Doke, J. T., Tax, F. E., and Walker, J. C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. doi: 10.1016/S0092-8674(02)00812-7
Li, R. N., Zhang, B. L., Li, T., Yao, X. Y., Feng, T. T., Ai, H., et al. (2024). Identification and characterization of the BZR transcription factor genes family in potato (Solanum tuberosum L.) and their expression profiles in response to abiotic stresses. Plants 13, 407. doi: 10.3390/plants13030407
Liu, D., Cui, Y. J., Zhao, Z. L., Li, S. Y., Liang, D., Wang, C. L., et al. (2021). Genome-wide identification and characterization of the BES/BZR gene family in wheat and foxtail millet. BMC Genomics 22, 682. doi: 10.1186/s12864-021-08002-5
Liu, K. Q., Ju, Z. L., Jia, Z. F., Liang, G. L., Ma, X., and Liu, W. H. (2022). Genome-wide identification and characterization of the oat (Avena sativa L.) WRKY transcription factor family. Genes 13, 1918. doi: 10.3390/genes13101918
Luo, S. L., Zhang, G. B., Zhang, Z. Y., Wan, Z. L., Liu, Z. C., Lv, J., et al. (2023). Genome-wide identification and expression analysis of BZR gene family and associated responses to abiotic stresses in cucumber (Cucumis sativus L.). BMC Plant Biol. 23, 214. doi: 10.1186/s12870-023-04216-9
Manoli, A., Trevisan, S., Quaggiotti, S., and Varotto, S. (2018). Identification and characterization of the BZR transcription factor family and its expression in response to abiotic stresses in Zea mays L. Plant Growth Regul. 84, 423–436. doi: 10.1007/s10725-017-0350-8
Nam, K. H. and Li, J. M. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. doi: 10.1016/S0092-8674(02)00814-0
Othman, R. A., Moghadasian, M. H., and Jones, P. J. (2011). Cholesterol-lowering effects of oat β-glucan. Nutr. Rev. 69, 299–309. doi: 10.1111/j.1753-4887.2011.00401.x
Paudel, D., Dhungana, B., Caffe, M., and Krishnan, P. (2021). A review of health-beneficial properties of oats. Foods 10, 2591. doi: 10.3390/foods10112591
Peng, Y. Y., Yan, H. H., Guo, L. C., Deng, C., Wang, C. L., Wang, Y. B., et al. (2022). Reference genome assemblies reveal the origin and evolution of allohexaploid oat. Nat. Genet. 54, 1248–1258. doi: 10.1038/s41588-022-01127-7
Singh, D. and Laxmi, A. (2015). Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00895
Sun, M., Sun, S. J., Jia, Z. C., Ma, W., Mao, C. L., Ou, C. M., et al. (2022). Genome-wide analysis and expression profiling of glutathione reductase gene family in oat (Avena sativa) indicate their responses to abiotic stress during seed imbibition. Int. J. Mol. Sci. 23, 11650. doi: 10.3390/ijms231911650
Ullah, U., Shalmani, A., Ilyas, M., Raza, A., Ahmad, S., Shah, A. Z., et al. (2022). BZR proteins: identification, evolutionary and expression analysis under various exogenous growth regulators in plants. Mol. Biol. Rep. 49, 12039–12053. doi: 10.1007/s11033-022-07814-2
Wang, L. N., Lin, M., Zou, L. N., Zhang, S. R., Lan, Y. G., Yan, H. W., et al. (2024). Comprehensive investigation of BZR gene family in four dicots and the function of PtBZR9 and PtBZR12 under drought stress. Plant Physiol. Biochem. 207, 108360. doi: 10.1016/j.plaphy.2024.108360
Wang, W., Sun, Y. Q., Li, G. L., and Zhang, S. Y. (2019). Genome-wide identification, characterization, and expression patterns of the BZR transcription factor family in sugar beet (Beta vulgaris L.). BMC Plant Biol. 19, 191. doi: 10.1186/s12870-019-1783-1
Xu, W. W., Guo, L. C., Wang, C. L., Wei, L. M., Wang, Q., Ren, Q. Y., et al. (2024). Transcriptome analysis reveals drought-responsive pathways and key genes of two oat (Avena sativa) varieties. Plants 13, 177. doi: 10.3390/plants13020177
Xu, X. W., Jiang, W. B., Chen, Y. B., Tian, H., Yang, Z. J., Liu, S., et al. (2025). BRASSINAZOLE RESISTANT 1 delays photoperiodic flowering by repressing CONSTANS transcription. Plant Physiol. 197, kiaf032. doi: 10.1093/plphys/kiaf032
Ye, H. X., Liu, S. Z., Tang, B. Y., Chen, J. N., Xie, Z. L., Nolan, T. M., et al. (2017). RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun. 8, 14573. doi: 10.1038/ncomms14573
Yin, Y. H., Vafeados, D., Tao, Y., Yoshida, S., Asami, T., and Chory, J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120, 249–259. doi: 10.1016/j.cell.2004.11.044
Yin, Y. H., Wang, Z. Y., Mora-Garcia, S., Li, J. M., Yoshida, S., Asami, T., et al. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191. doi: 10.1016/S0092-8674(02)00721-3
Ying, W., Wang, Y. W., Wei, H., Luo, Y. M., Ma, Q., Zhu, H. Y., et al. (2024). Structure and function of the Arabidopsis ABC transporter ABCB19 in brassinosteroid export. Science 383, eadj4591. doi: 10.1126/science.adj4591
Yu, T., Ai, G., Xie, Q. M., Wang, W. Q., Song, J. W., Wang, J. Y., et al. (2022). Regulation of tomato fruit elongation by transcription factor BZR1.7 through promotion of SUN gene expression. Horticult. Res. 9, uhac121. doi: 10.1093/hr/uhac121
Zhan, C., Liang, C., Zhao, L., Jiang, S. Z., and Zhang, Y. L. (2024). Differential responses of crop yields to multi-timescale drought in mainland China: spatiotemporal patterns and climate drivers. Sci. Total. Environ. 906, 167559. doi: 10.1016/j.scitotenv.2023.167559
Zhang, D. J., Cheng, Y. C., Lu, Z. Y., Wang, J. G., Ye, X. S., Zhang, X. Q., et al. (2021). Global insights to drought stress perturbed genes in oat (Avena sativa L.) seedlings using RNA sequencing. Plant Signaling Behav. 16, 1845934. doi: 10.1080/15592324.2020.1845934
Zhao, Z. Y., Tang, S., Li, W., Yang, X. R., Wang, R. J., Diao, X. M., et al. (2021). Overexpression of a BRASSINAZOLE RESISTANT 1 homolog attenuates drought tolerance by suppressing the expression of PLETHORA-LIKE 1 in Setaria italica. Crop J. 9, 1208–1213. doi: 10.1016/j.cj.2021.02.006
Zhou, X. Y., Yi, D. X., Ma, L., and Wang, X. M. (2024). Genome-wide analysis and expression of the aquaporin gene family in Avena sativa L. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1305299
Zhu, S. S., Mi, J. Z., Zhao, B. P., Wang, Z. M., Yang, Z. X., Wang, M. X., et al. (2024). Integrative transcriptome and metabolome analysis reveals the mechanism of fulvic acid alleviating drought stress in oat. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1439747
Keywords: Avena sativa, BZR transcription factor, genome-wide analysis, osmotic stress, gene expression
Citation: Xu S, Wei Z, Ma M, Zhang L, Liu Z and Liu L (2025) Genome-wide identification and characterization of the Brassinazole-resistant gene family and associated responses to osmotic stress in Avena sativa. Front. Plant Sci. 16:1616026. doi: 10.3389/fpls.2025.1616026
Received: 22 April 2025; Accepted: 19 July 2025;
Published: 14 August 2025.
Edited by:
Yaroslav B. Blume, National Academy of Sciences of Ukraine (NAN Ukraine), UkraineReviewed by:
Min May Wong, Iowa State University, United StatesAkhilesh Kumar Pandey, Sardar Vallabhbhai Patel University of Agriculture and Technology, India
Ping Yun, University of Western Australia, Australia
Copyright © 2025 Xu, Wei, Ma, Zhang, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Longlong Liu, bGxsb25nNzgxMjExQHNpbmEuY29t
†These authors have contributed equally to this work
 Shirui Xu
Shirui Xu Zihao Wei1,4†
Zihao Wei1,4†