- 1School of Biological Sciences, The University of Western Australia, Perth, WA, Australia
- 2The Institute of Agriculture, The University of Western Australia, Perth, WA, Australia
- 3Centre for Applied Bioinformatics, The University of Western Australia, Perth, WA, Australia
Brassica species, which include economically important Brassica crops grown around the globe, are important as popular vegetables, forage, and oilseed crops, supplying food for humans and animals. Despite their importance, these crops face increasing challenges from biotic and abiotic stresses, exacerbated by climate change and the evolving threat of crop pathogens. Enhancing crop resilience against these stresses has become a key priority to ensure stable crop production. Recent advancements in genomic studies on Brassica crops and their pathogens have facilitated the deployment of CRISPR/Cas systems in breeding major Brassica crops. This review highlights recent progress in CRISPR/Cas-based gene editing technologies to improve resistance to pathogens and enhance tolerance to drought, salinity, and extreme temperatures. It also summarises the molecular mechanisms underlying crop responses to these stresses. Furthermore, the review discusses the workflow for employing the CRISPR/Cas system to boost stress tolerance and resistance, outlines the associated challenges, and explores prospects based on gene editing research in Brassica species.
1 Introduction
The Brassica genus in the Brassicaceae family comprises around 159 species, many of which are of economic importance, providing seed oil, condiments, and vegetables (Bhattacharya, 2019; Borges et al., 2023). Brassica vegetables are cultivated and consumed worldwide, involving different cultivars of cabbage, cauliflower, broccoli, brussels sprouts (Brassica oleracea var. gemmifera), and kale (Podsędek, 2007). Meanwhile, B. napus (canola) is one of the most important oilseed crops planted globally and is the leading crop amongst other Brassica oilseed crops such as winter turnip rape (B. rapa subsp. oleifera), swede (B. napus spp. napobrassica), Indian mustard (B. juncea), Ethiopian mustard (B. carinata) and black mustard (B. nigra) cultivated in selected regions of the world (Kirkegaard et al., 2021). The genomic relationships of these Brassica species described by U, N, (1935) comprises three diploid and three allopolyploid species, including B. oleracea (CC, 2n = 18), B. rapa (AA genome, 2n = 20), B. nigra (BB, 2n = 16), B. carinata (BBCC, 2n = 34), B. juncea (AABB, 2n = 36), and B. napus (AACC, 2n = 38).
Both oilseed and vegetable Brassica species play important roles in global food security, with a steady increase in their production since 2000 (FAOSTAT, 2023). Brassica vegetables are good sources of vitamins and antioxidants and protect against certain cancers and inflammation (Francisco et al., 2017). According to FAOSTAT (2023), cauliflower and broccoli are among the world’s primary crops, with a total production of approximately 26 million tonnes (MT) in 2022. Based on the average production of cauliflower and broccoli during the 1994–2022 period, China was the leading producer with around 7.5 MT, followed by India (6.3 MT), the US (1.1 MT), Spain (0.5 MT) and Italy (0.4 MT). China is the leading cabbage producer, with 29.9 MT, followed by India (7.0 MT) and Russia (2.9 MT) (FAOSTAT, 2023). Brassica vegetable production plays an important economic and social role in developing countries, such as African countries. In Kenya, cabbage is cultivated on small to medium-scale farms and is an important crop for tackling nutrition poverty due to its highly nutritious components (Daniel et al., 2023). Brassica oil crops, such as B. napus and B. juncea, provide canola oil for multiple industries, such as food, feed, fertiliser, and biodiesel. Canola oil is the third most consumed oil after soybean and palm oil (Adwiyah et al., 2023; USDA, 2023). Recently, Brassica carinata has been cultivated as a rotated crop for biofuel production (Basili and Rossi, 2018). Demand for vegetable and oilseed Brassica species is forecast to increase in the next decade due to the increasing global population. Besides, recent changes in renewable energy policies in some countries have led to the expansion of the biofuel market (OECD-FAO, 2023) and allowed the use of canola oil in biofuel production, boosting canola oil demand. However, global Brassica crop production faces challenges, such as limitations in arable land and consequences of climate change, such as the emergence of more virulent plant pathogens and more intensive abiotic stresses. Diseases caused by microbial pathogens can cause up to 90% of yield losses in Brassica species (Greer et al., 2021), while heat stress and UV-B radiation cause significant yield reduction (Secchi et al., 2023; Sehgal et al., 2022).
Breeding for disease resistance and abiotic stress tolerance in Brassica crops has been dramatically improved with advancements in genomics (Boivin et al., 2004; Das et al., 2024) and genome editing tools (Bao et al., 2019; Chen et al., 2019; Bhadauria et al., 2024). Nonetheless, the adverse consequences of climate change continue to challenge Brassica crop yield and have been linked to the emergence of novel strains of plant microbial pathogens that overcome current resistance in cultivated crops (Borges et al., 2023; Guerret et al., 2017; Van de Wouw et al., 2014). Fortunately, recent advancements in functional genomics of Brassica crops have revealed complex genetic networks and molecular mechanisms underlying their response to abiotic and biotic stressors, from which key genes and genomic factors controlling the response have been identified, allowing for the intervention of novel gene editing technologies (Wu et al., 2020; Varanda et al., 2021; Tan et al., 2024; Wang et al., 2024). Since 2012, the clustered regularly interspaced short palindromic repeats/CRISPR-associated protein (CRISPR/Cas) systems modified from the bacterial adaptive immune system of bacteria and archaea (Jinek et al., 2012) have been the most widely used genome editing tool worldwide (Talakayala et al., 2022; Wada et al., 2022). Compared to genome editing nucleases, such as zinc-finger nucleases (ZFNs) (Carroll, 2008; Cathomen and Keith, 2008) and transcription activation-like (TAL) effector nucleases (TALENs) (Christian et al., 2012; Li and Yang, 2013), RNA-directed CRISPR-Cas systems are more popular due to their versatility, simplicity, and feasibility (Li et al., 2025; Guo et al., 2023; Nerkar et al., 2022). Briefly, a typical CRISPR/Cas system for genome editing involves two main components a single Cas effector protein with one or two nuclease domain(s) and one or multiple CRISPR RNA (crRNA) sequences, with an additional trans-activating CRISPR RNA (tracrRNA) in case of the Cas9 systems (Charpentier et al., 2015; Jiang and Doudna, 2017). Each crRNA contains a spacer (30–40 bp) at the 5’ end, complementary to a sequence of a foreign DNA source, and a CRISPR repeat (25–35 bp) at the 3’ end (Jiang and Doudna, 2017). For the Cas9 and most Cas12 systems, the base pairing of the repeat sequence of crRNA with tracrRNA forms guide RNA (gRNA). The gRNA directs the Cas protein to a complementary DNA site (~20 nucleotides), or DNA targets flanked by specific protospacer adjacent motifs (PAMs) (Asmamaw and Zawdie, 2021). For the Cas9 and Cas12 systems, Cas protein in complex with gRNA cleaves the double-stranded DNA (dsDNA) target and generates a double-stranded break (DSB). Modifications (e.g. indels, base editing) are introduced at DSBs through natural cellular DNA repair pathways called non-homologous end joining (NHEJ), where the ends of cleaved DNA are re-joined, and homologous recombination (HR), where the DNA gaps are synthesised based on the homologous template (Cathomen and Keith, 2008; Li and Heyer, 2008). Therefore, Cas9 and Cas12 systems are powerful tools for plant functional genomics and improving crop traits (Nerkar et al., 2022; Hussain et al., 2018). In the RNA targeting systems, such as CRISPR/Cas13a-d, Cas13 protein is directed to its target mainly by gRNA and is less dependent on PAMs for target recognition, of which the Cas13c and Cas13d system has no PAM preference (Mahas et al., 2019; Cox et al., 2017). The single-stranded RNA target is cleaved upon being bound by the CRISPR/Cas13 (Cox et al., 2017). Hence, the CRISPR/Cas13 has been quickly adopted for targeting RNA viruses to improve plant resistance to viruses and diagnose viral diseases (Hagit et al., 2024; Zhan et al., 2023; Shihong Gao et al., 2021).
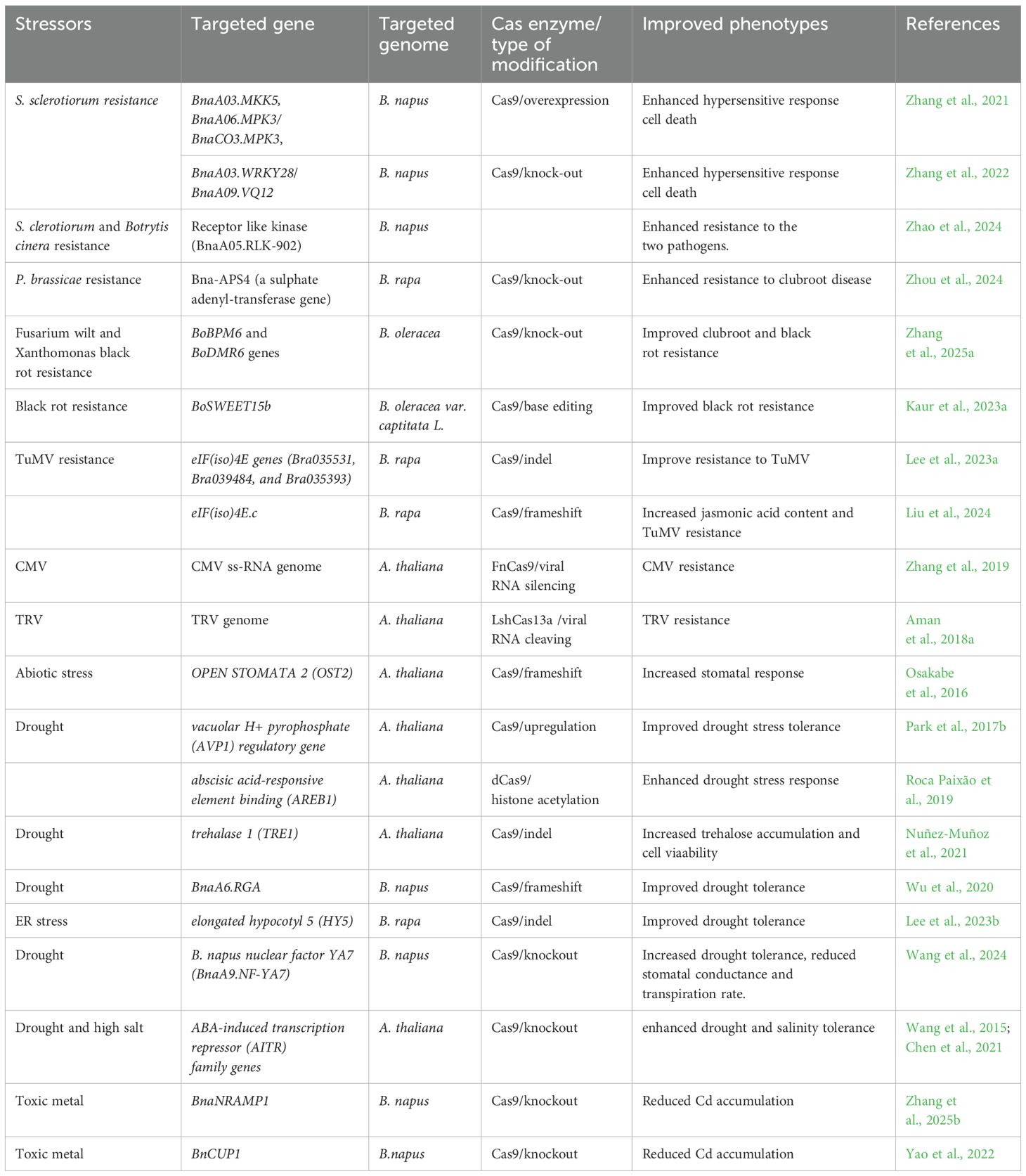
Over the past two decades, CRISPR/Cas-based precise genome modification has rapidly evolved in its application in plant crops, including Brassica species (Li et al., 2024). Benefiting from the Brassica omics findings, the CRISPR/Cas tools further characterise functions of candidate genes and key genetic elements regulating plant resilience traits (Amas et al., 2023; Zhang et al., 2023), enable precise mutation induction in the Brassica genomes that conferring resistance/tolerance to biotic and abiotic stress. This review highlights the recent progress in deploying CRISPR/Cas tools in Brassica crops, focusing on resistance to microbial pathogens and tolerance to major abiotic stresses, such as heat, drought, and salinity which reveal potential genes controlling these processes were summarised in Table 1. The workflow for utilising the endonuclease system is also discussed.
2 Improvement of biotic stress tolerance in Brassica
To cope with pathogen infection, plants have developed two layers of immune systems. The first layer, pathogen-associated molecular patterns (PAMPs) triggered immunity (PTI), is activated by surface-localised pattern recognition receptors (PRRs) upon recognising (PAMPs), microbe/damage-associated molecular patterns (MAMPs and DAMPs) (Iqbal et al., 2021; Ngou et al., 2022). The second layer is effector-triggered immunity (ETI), which is induced by recognition of pathogen-secreted effector proteins by plant resistance proteins. The resistance proteins are usually receptors consisting of nucleotide-binding leucine rich repeat (NLR) domains (Dalio et al., 2021). Most identified resistance genes mentioned below are NLRs.
2.1 Alternatives for enhancing resistance to fungal diseases
The fungal pathogens Alternaria spp., Fusarium oxysporum, Leptosphaeria maculans, and Sclerotinia sclerotiorum are the primary pathogens affecting Brassica crop yield worldwide (Zheng et al., 2020a). To overcome plant host PTI, fungi produce specialised virulence factors called effector proteins that interact with plant host resistance proteins encoded by (R) genes, activating ETI (Tsuda and Katagiri, 2010; Spoel and Dong, 2012);. Resistance (R) genes are the key components of the plant immune response involved in pathogen recognition and the activation of the defence response (Chisholm et al., 2006).
As an efficient system to generate mutants, CRISPR/Cas9 has contributed to recent developments in the knowledge of plant-fungal pathogen interactions, unveiling the intricate dynamics of these processes. This ribonucleoprotein (RNP) complex has been utilised to eludcidate plant response mechanisms to to identify and confirm the functions of major host and fungal factors involved in the interactions. Chitin, a major component of fungal cell walls, functions as a PAMP in plants and animals (Liu et al., 2020). Host plants counteract fungal invasions by degrading these molecules with chitinases (Punja and Zhang, 1993; Zhu et al., 2021). To counteract, fungal pathogens secrete chitin-binding proteins to protect their hyphae from hydrolysis and avoid activating escalated defence responses (Cunnac et al., 2009). Cas9-mediated knockout mutants of a L. maculans gene (LmCBP1) encoding chitin-binding protein resulting in two mutants with reduced pathogenicity in B. napus (Liu et al., 2020). Similarly, the RNP complex has been used to confirm the roles of glucosinolate biosynthetic genes of B. oleracea, like ST5b-Bol026202 and MYB34-Bol017062 in resistance to Mycosphaerella brassicicola (Abuyusuf et al., 2018) and modify effector genes (OBR08294 and OBR06881) in Colletotrichum higginsianum, a hemibiotrophic fungal pathogen responsible for anthracnose disease in Brassicas, to identify the potential effectors involved in the pathogenicity of the disease (Bhadauria et al., 2024).
Towards improving fungal resistance in Brassica crops, plant host factors, such as Calmodulin-binding transcription activators (CAMTAs), BnWRKY transcription factors in B. napus and PRRs also play important roles in various plant biological processes including biotic stress tolerance and disease resistance (Finkler et al., 2007; Jose et al., 2020; Noman et al., 2021; Chen et al., 2021). Several CAMTA genes have been identified in B. napus, from which BnCAMTA3, also found in B. oleracea, may play a significant role in resistance against Sclerotinia stem rot infection (Rahman et al., 2016). This assumption is based on observations on S. sclerotiorum resistance of Arabidopsis plants with the CAMTA3 mutation (Atcamta1-6), where the camta3 plants were more resistant to S. sclerotiorum compared to wild-type and the other camta mutant plants (Rahman et al., 2016).
Based on the understanding of plant defense mechanisms and available Brassica genomic data, many candidate resistance genes have been identified. CRISPR/Cas9 have been used to introduce mutations to these genes to confirm their functions. In some cases, modifying genes that negatively control host resistance result in mutants with improved resistance to fungal pathogens. The WRKY family is one of the largest plant transcription factor gene families, of which several members are involved in regulating the defence response (Chen et al., 2021). In B. napus, BnWRKY28 and BnWRKY33 have antagonistic roles in the response of B. napus to S. sclerotiorum invasion (Zhang et al., 2022), in which BnWRKY28 on chromosome A03 (BnaA03.WRKY28) functions negatively in resistance to S. sclerotiorum in B. napus. The CRISPR/Cas9 system was used to mutate six sites in the target gene, resulting in the BnaA03.WRKY28 knockout mutant lines with enhanced resistance to S. sclerotiorum, the organism responsible for stem canker (Zhang et al., 2022). By similar approach, improved resistance to Sclerotinia was also reported in transgenic B. napus plants containing BnaWRKY70 mutants (Sun et al., 2018).
Plant host factors have different effects on fungal infection. During fungal infection, a lysin motif-containing PRR, chitin elicitor receptor kinase 1 (CERK1), perceives and binds to chito-oligosaccharides (COs) elicitors, which activates the downstream signalling cascade and, hence, mounts immune responses (Liu et al., 2012; Yang et al., 2022).While most RLKs have been reported to have positive roles in empowering plant resistance during fungal infection (Soltabayeva et al., 2022), a few RLKs were reported with adverse effects. A receptor-like protein kinase 902 (RLK-902) in B. napus has been suggested to assist S. sclerotiorum and Botrytis cinerea pathogenicity (Zhao et al., 2024). The function of RLK-902 in disease progression was confirmed in CRISPR/Cas9-based RLK-902 knockout mutants in B. napus through pathogenicity tests with S. sclerotiorum and B. cinerea. The RLK902 knockout lines significantly improved disease resistance without compromising plant growth and development (Zhao et al., 2024; Ye et al., 2024). Overexpression of lysine motif RLK 4 in Sinapis alba, an inactive kinase from B. juncea, significantly induced resistance to Alternaria brassicicola compared with susceptible B. juncea in the same conditions (De et al., 2021).
Incorporating CRISPR-Cas9 technology into Brassica breeding programs offers tremendous possibilities for creating resilient crop varieties. In contrast to traditional breeding techniques, which can be time-consuming and less accurate, CRISPR gene editing enables specific genetic alterations conferring the expected resistance in T0 generationand significantly reduce the time for developing germplasm of resistant cultivars.
2.2 Bacteria/protists
Diseases induced by bacteria and protists are among the many challenges facing the sustainable cultivation of Brassica crops globally. Although there are fewer examples of bacterial and protist diseases of Brassica crops than those caused by fungi, they still require careful management to prevent significant yield losses. Clubroot, caused by the biotrophic obligate parasite protist Plasmodiophora brassicae (Javed et al., 2023), is a widespread disease of canola and vegetable Brassicas, which is particularly devastating to the Canadian canola industry (Zheng et al., 2020b) with yield loss in canola ranging from 60% to 90% (Pageau et al., 2006). It has emerged as one of the biggest threats to canola production, and its management alone has forced significant resource investment (Botero-Ramirez et al., 2022). The bacterial pathogen Xanthomonas campetris pv. campestris (Xcc) causes black rot, a major disease primarily affecting vegetable Brassicas, such as B. oleracea var capitata (Vicente and Holub, 2013; Sun et al., 2021). In addition, several species of bacteria belonging to the genera Erwinia, Pseudomonas and Pectobacterium result in bacterial leaf spot and soft rot in various Brassica species (Ren et al., 2001; Takikawa and Takahashi, 2014; Klair et al., 2021). Coordinating with recent advance in genomics, CRISPR/Cas tools have accelerated the identification, validation and utilization of clubroot and black rot resistance genes.
Deploying genetically resistant cultivars has been one of the primary management strategies to control clubroot (Hwang et al., 2014). This is because there is currently no way to eradicate the pathogen from infested soil, and soil amendments only provide limited disease control (Hasan et al., 2021). Therefore, there have been significant efforts to identify sources of natural resistance and the genes underlying clubroot resistance (see Hasan et al., 2021 for a comprehensive review of clubroot resistance genes). used a CRISPR/Cas9-based vector system to introduce the clubroot R gene Rcr1 into the susceptible B. napus line DH12075. The modified plants in T2 generation were selection marker-free and showed stable resistance to clubroot. Beside Rcr1, two resistant genes from B. rapa, CRa and Crr1, encoding Toll-Interleukin1 receptor/nucleotide-binding site/leucine-rich-repeat (TIR-NBS-LRR; TNL) have been isolated (Yu et al., 2016). By transcriptomic analysis and comparative analysis of cell wall components in clubroot resistant B.napus, and Tu et al. (2024) suggested that Rcr1 and Crr1rutb in canola mediated the induction of lignin accumulations and possibly interact with genes involved in the phenylpropanoid pathway. Over 10 resistance genes and over 20 QTLs were identified within Brassica species, especially in B. rapa (Yu et al., 2016). Hu’s study (Hu et al, 2024) demonstrates an efficient Cas9-based platform to quickly introduce a clubroot resistant line by integrating the resistance gene into a susceptible plant and a Cas9-assisted breeding framework that help avoiding rounds of back crossing, with the resistant line achievable in just two generations.
Another recent study examining clubroot resistance identified a microRNA-target pair that was thought to regulate clubroot resistance in the B. rapa cultivar ECD04 (Zhou et al., 2024). The authors generated a CRISPR/Cas9 construct by cloning their sgRNA into the pYLCRISPR/Cas9 expression vector and used it to disrupt Bna-APS4, a sulphate adenylyl-transferase gene that is targeted by miR395a. Loss-of-function mutants displayed increased resistance toward clubroot, implicating Bna-APS4 as a negative regulator of clubroot resistance (Zhou et al., 2024). These studies demonstrate multiple applications of CRISPR/Cas9 to investigate resistance mechanisms toward clubroot in Brassica species. Their findings deepen the understanding of resistance mechanisms and can support the accelerated identification of novel clubroot receptors or genes that mediate clubroot resistance.
Cas9 from Streptococcus canis (ScCas9) was successfully employed to introduce broad-spectrum resistance against Fusarium wilt, black rot and clubroot in B. oleracea by knocking out the BoBPM6 and BoDMR6 genes (Zhang et al., 2025a). The BPM6 gene, BTB/POZ (Broad complex, Tramtrack, Bric-a-brac/Pox virus and Zinc finger)-MATH 6 (BPM6), was differently expressed and induced by Fusarium wilt and black rot of B. oleracea. Meanwhile, Downy Mildew Resistant 6 (DMR6) has been known as a conserved S gene (Zhang et al. 2025a; Thomazella et al., 2021). Subsequent inoculation experiments with homozygous mutants in T1 generation resulted in decreased disease indexes (DIs), as compared to DIs for wild-type plants, after being challenged with the three pathogens. The bodmr6 mutant presented significant DI changes with black rot and clubroot infection (from 79.3 to 55.1, and from 90.7 to 57.6), while the bobpm6 showed significant DI decrease with Fusarium wilt and clubroot (from 65.4 to 14.5, and from 53.8 to 20.9). These results suggest Cas9-based editing as a powerful in breeding disease resistance in B. oleracea.
Another study by the same authors employed CRISPR/Cas9 to optimise the modification of a potential host susceptibility gene, BoSWEET15b (SWEET15b gene in B. oleracea), in the cabbage cultivar ‘Ohgane’ (Kaur et al., 2023b). In plants, Sugars Will Eventually be Exported Transporter (SWEET) proteins have been known to involve in biological processes (e.g. regulation of pollen development, nectar secretion, seed development, phloem loading, and leaf senescence) and poteintial regulatory roles under biotic and abiotic stress (Song et al., 2022). The Cas9 ribonucleoprotein and sgRNA were transferred via a PEG-mediated delivery system, and a 39% insertion/deletion frequency was achieved in BoSWEET15b (Kaur et al., 2023b). The search for black rot resistance in cabbage is sped up supported by advancement in genomics (Ma et al., 2025). Therefore, the finding by Kaur et al (2023a) laid the foundation for employing CRISPR/Cas9 to both characterise potential black rot resistance genes and breeding black rot resistance in B. oleracea. Although in its infancy, applying CRISPR/Cas technology to enhance resistance to bacterial pathogens in Brassica crops is a promising tool to accelerate future resistance improvements.
2.3 Viruses
Viral diseases can cause up to 84% yield loss in rapeseed (B. napus and B. juncea) (Jones, 2021) and 65% in vegetable Brassicas (Das et al, 2024), which are often caused by turnip mosaic virus (TuMV, family Potyviridae), cucumber mosaic virus (CMV; family Bromoviridae), cauliflower mosaic virus (CaMV, family Caulimoviridae), Turnip yellow virus (TuYV), and Brassica yellow virus (BrYV) (Bruckner et al., 2024; Wu et al., 2024). The last two viruses belong to the genus Polerovirus of the family Luteoviridae (Kidanemariam and Abraham, 2023). In natural conditions, aphids are the most common vector for transmission of the mentioned viruses (Ng and Perry, 2004; Latham et al., 2003), and mixed infections with TuMV, TuYV and CaMV often occur in Brassica crops worldwide (Raybould et al., 1999; Broadbent, 2015; Latham et al., 2003; Z Meena et al., 2021; Bruckner et al., 2024).
While persistently transmitted viruses, such as luteoviruses and poleroviruses, or semi-persistently transmitted viruses, such as TuYV and CaMV, can be managed using chemical treatment (Fereres and Raccah, 2015). Such treatment is ineffective for non-persistent viruses, such as TuMV and CMV (Hooks and Fereres, 2006). Additionally, the widespread of TuMV in Brassica growing areas worldwide, especially in cultivated B. napus and B. rapa, has necessitated the search for TuMV resistance genes and breeding TuMV resistance (Walsh et al., 2023; Palukaitis and Kim, 2021). Virus infection induces layers of plant response, such as PTI, RNA silencing and PAMP-triggered response. Upon TuMV infection, Brassica species response and develop symptom differently depending on viral strains. Most of the naturally identified virus resistance is conferred by R genes. Being transmitted by more than 80 aphid species have made TuMV become a concern for Brassica crops worldwide (Nellist et al., 2022).
As a model for RNA virus study and its economic importance to canola and cabbage crops, many research groups have focus on TuMV genome structure and TuMV resistance. The intensive studies revealed 16 dominant resistant loci or quantitative trait loci (QTLs) associated with TuMV resistance in the A genome of B. rapa, 5 resistance loci in the A or C genome of B. napus, and one in B. juncea (Palukaitis and Kim, 2021). Among the mapped resistance loci, the dominant and recessive resistance genes, ConTR01 and retr01 shared the same loci with the eukaryotic initiation factor 4E (eIF4E) and eIF(Iso)4E, respectively (Rusholme et al., 2007). This effort has been facilitated by advancements in sequencing and gene editing technologies, and Brassica genomic data, which deepens the understanding of molecular interactions between virus and plant host factors during the viral infection. In TuMV, viral genome-linked protein (VPg) functions like 5’-cap binding to eukaryotic translation initiation factors (eIF) and initiates viral protein translation (Okade et al., 2009; Leoonard et al., 2000);. Mutations in the binding sites of viral VPg or plant host eIF affect virus infectivity (Leoonard et al., 2000; Zhang R. et al., 2022; Lee et al., 2023b). Resistance to potyviruses, including TuMV, conferred by natural and artificial mutated eIF4E and eIF(iso)4E in different plant species, including A. thaliana and B. rapa, was thoroughly reviewed by Sanfaçon (2015). Therefore, currently, there are two approaches to utilise CRISPR/Cas tool for introducing virus resistance trait in plant host, either modifying plant genome to interfere virus multiplication and movement or directly cleaving genome of the infecting virus. The Cas9 systems are often selected for the first approach, while Cas13 systems are used for the later approach (Nie et al., 2024; Ton, 2024; Robertson et al., 2022; Mahas et al., 2019).
With climate change happening, general virus control measures become less effective, of which the most reliable ones are host resistance genes and control of insect vectors (Jose et al., 2020). Additionally, RNA viruses are prone to mutations that lead to the rise of novel strains (McDonald and Lindie, 2002). New TuMV strains overcoming known resistances in B. napus was reported in Australia and Korea (Song et al., 2022; Guerret et al., 2017). These facts necessitate the CRISPR/Cas tool deployment in the arms race between viruses and Brassica crops.
Induced sequence-specific point mutations at eIF(iso)4E locus in A. thaliana showed complete resistance to the GFP-tagged infectious TuMV clone pCB-TuMV-GFP with no TuMV detection by viral GFP imaging and quantitative RT-PCR. At the same time, TuMV-GFP systemic infection was visible in wild-type Arabidopsis plants (Pyott et al., 2016). There was no difference in plant growth between the mutants and the wild-type in this study, implying no compensations for growth in the eIF(iso)4E mutant line. Following this approach, Lee et al (2023b) utilised CRISPR/Cas9 vector targeting the eIF(iso)4E genes (Bra035531, Bra039484, and Bra035393) of B. rapa. The eIF(iso)4E-T1 edited plants were successfully generated and exhibited resistance to TuMV under the experimental conditions. Both studies showed no phenotypic differences between the mutant lines and the wild-type plants of Arabidopsis and B. rapa. However, single-gene resistance is not durable and overcome easily by novel virus strains (Palukaitis and Kim, 2021). Fortunately, the advancement of Brassica genomic data has supported the search for new candidate resistance genes. Shopan et al (2020) revealed that new eIF subgroups, eIF2β, eIF2α, eIF2Bβ, eEF1A, and poly(A)-binding proteins (PABPs) could be the targets for antiviral strategies in B. juncea. Meanwhile, defect/mutated eIF2Bβ, eIF4E, eIF(iso)4E, eIF4G, and eIF(iso)4G have been found to confer recessive resistance to plant viral infections (Shopan et al., 2017, Shopan et al., 2020).
In addition to modifying the plant genome, virus resistance can be achieved by using RNA-targeting CRISPR/Cas systems to target and cleave viral genomes in infected plant hosts. A reprogrammed Cas9 protein from Francisella novicida (FnCas9) inhibited CMV in Arabidopsis, conferring virus resistance (Zhang et al., 2018a). The functionality of CRISPR-Cas13 systems in plant cells encouraged the use of this tool in tackling RNA plant viruses. Aman et al. (2018a) investigated the ability of the Cas13a effector from Leptotrichia shahii (LshCas13a) to target TuMV in vitro and planta on tobacco plants (N. benthamiana). This study used a fusion clone of TuMV-GFP to visualise the virus movement in the plants expressing crRNA-LshCas13a. Four crRNAs targeting TuMV-GFP at the sites encoding for HC-pro, CP, GFP1 and GFP2 were cloned as uniplexes or a multiplex of 3 crRNAs (HC-pro, GFP1 and GFP2) into tobacco rattle virus (TRV) vectors. Transient expression assays with Nicotiana benthamiana leaves agro-infiltrated with a mixture of A. tumacien strain GV3010 containing TuMV-GFP, LshCas13a vector, and TRV-crRNAs showed an apparent reduction in GFP intensity at seven days post-inoculation compared with the control treatments with neither LshCas13a expression nor the TRV-crRNAs. In pCas13a-overexpressed tobacco plants, leaf agro-infiltration with TuMV-GFP and uniplex TRV-crRNAs or multiplex TRV-crRNA produced a similar result as the transient assay. Transgenic A. thaliana lines expressing crRNA- LshCas13a targeting HC-Pro also had reduced TuMV accumulation upon being infected with TuMV-GFP (Aman et al., 2018b). The results on both model plants showed higher TuMV inhibition efficiencies with crRNAs binding HC-pro and GFP-T2 targets compared to CP and GFP-T1 targets. The LshCas13a systems have been successful in inhibiting the tobacco mosaic virus (TMV), southern rice black-streaked dwarf virus (SRBSDV), and rice stripe mosaic virus (RSMV) invasion in N. benthamiana and rice (Zhang et al., 2019). Together, these studies have revealed the potential use of Cas13 tools to confer resistance to RNA viruses in monocot and dicot plants, advancing crop breeding strategies for RNA virus resistance.
3 Abiotic stress tolerance in Brassica
Climate change consequences have intensified abiotic stress on Brassica crops and vegetables. Stress conditions such as drought, high temperature, and high salinity adversely affect plan physiological, metabolic and biochemical processes, resulting in significant reduction in yield and productivity (Raza et al., 2021). Plants possess different mechanisms in response to each individual stress. Recent transcriptomic studies on Brassica under abiotic stress and application of CRISPR/Cas9 in elucidating the role of stress responsive genes involved have provided insights into the molecular mechanisms underlying the stress responses. The developing understanding on the key genes controlling these mechanisms will facilitate the application of modern molecular breeding techniques, such as genetically transformation and genome editing (Sato et al., 2024).
3.1 Drought tolerance
Drought is caused by several reasons, including low soil moisture, salinity, high and low temperature (Salehi-Lisar and Bakhshayeshan-Agdam, 2016). Drought stress worsened by climate change significantly challenges these crops, affecting growth, development, and productivity (Wang et al., 2024). Annually, drought causes a minimum 30% reduction in canola yield (Farooq et al., 2009). Therefore, it is crucial to study Brassica crops’ responses to drought stress and strategies to enhance their drought resistance.
Drought inhibits photosynthesis, reducing biomass and yield, and disrupts biochemical pathways (Liu et al., 2004; Wahab et al., 2022; Zahra et al., 2023). However, plants have evolved adaptations to combat drought stress effectively (Yang et al., 2021), primarily through regulating abscisic acid (ABA) biosynthesis and signalling cascades (Wei et al., 2025; Soma et al., 2021) Drought condition induces the expression of many ABA biosynthesis genes along with binding factors and transcription factors (TF), leading to an increase in ABA levels and ABA-orchestrating mechanisms (Muhammad Aslam et al., 2022). Regulating stomatal closure to avoid dehydration is the primary plant response to drought (Sato et al., 2024). However, long-term photosynthesis disruption due to stomatal closure increases oxidative stress, activating stress-responsive genes like superoxide dismutase, catalase, dehydrins, late embryogenesis abundant (LEA) and DELLA proteins (Sun et al., 2021; Wang et al., 2020). Therefore, the activation of reactive oxygen species (ROS) scavenging pathways and increased biosynthesis of the protective genes represent drought tolerance. Genes responsive to drought and abscisic acid (ABA) are vital for plant protection, influencing LEA proteins, chaperones, osmo-protectants, sugar and proline transporters, aquaporins, and ROS-detoxifying enzymes (Muhammad Aslam et al., 2022; Kim et al., 2024).
Tolerance to drought stress is a complex quantitative trait (Rahman et al., 2022). Generating knock-out mutants of key stress responsive genes by CRISPR/Cas9 is a time efficient approach to elucidate functions of genes responsive to drought stress, which have been demonstrated through multiple studies in Arabidopsis. ABA mediated stomatal closure is the popular target for improving drought tolerance in crops (Hyun, 2020). OPEN STOMATA 2 (OST2) gene, which encodes a plasma membrane H+ATPase AHA1, activates many secondary transporters involve in ion and metabolite uptake and prevents ABA-mediated stomatal closure (Merlot et al., 2007). Osakabe et al (2016) successfully generated homozygous OST2 mutated Arabidopsis using CRISPR/Cas9. The homozygous line with 1-bp frameshift OST2 mutation showed a lower rate of transpirational water loss compared with that of the wild-type. Drought tolerance enhancement can be achieved by editing activation of the vacuolar H+-pyrophosphate (AVP1) regulatory gene, abscisic acid-responsive element binding (AREB1) gene, and silencing of the trehalase 1 (TRE1) gene, as well as editing of the STL1 structural gene (Park et al., 2017a; Roca Paixão et al., 2019; Nuñez-Muñoz et al., 2021). Increased drought tolerance phenotypes include plant survival, growth, and development after drought treatment. In case of overexpressing AVP1 gene, the modified plants survived and restored growth after 8 days without watering, while the wild type died after seven days (Park et al., 2017a). Regarding plant growth, the edited plants resulted in 2–5 fold increases in expression, additional four leaves, double size in single-leaf area, and enhanced drought tolerance compared to the wild-type (Park et al., 2017a). These preliminary studies on the model plants provide platforms for employing CRISPR/Cas9-assisted breeding towards drought tolerance in cultivated Brassica species.
The CRISPR-Cas9 systems are powerful tools to characterise functions of members of gene family contributing to stress response in an allotetraploid species, such as B. napus. As an example, CRISPR/Cas9 was deployed to edit BnaA6.RGA. B. napus contains 10 DELLA genes, including four homologs of RGA: BnaA6.RGA, BnaC7.RGA, BnaA9.RGA, and BnaC9.RGA. Previously, CRISPR/Cas9 technology was used to create mutants of these BnaRGAs (Yang et al., 2017). Subsequently, Wu et al. (2020) confirmed gain-of-function mutants of BnaA6.RGA and BnaC7.RGA among the BnaRGA mutants, which positively regulate drought tolerance in B. napus.
In a study, Lee et al (2023b) used CRISPR/Cas9 technology to investigate the role of the ELONGATED HYPOCOTYL 5 (HY5) gene in B. rapa under endoplasmic reticulum (ER) stress conditions which generally result from abiotic stresses (eg., increasing temperature, drought, salinity and pathogen infection). Researchers targeted the HY5 gene with sgRNAs and confirmed mutations. When subjected to ER stress using tunicamycin (TM), wild-type and hy5 mutant plants showed increased growth inhibition with higher TM concentrations, but the hy5 mutants had less severe inhibition. Staining methods revealed that hy5 mutants produced lower reactive oxygen species levels under ER stress. Additionally, these mutants exhibited lower expression levels of genes related to the unfolded protein response and cell death. The study concludes that editing the HY5 gene can reduce stress-related growth inhibition, potentially improving crop quality and yield.
In a recent study (Wang et al., 2024), using CRISPR/Cas9 to introduce a nonsynonymous substitution (M63I) in the target gene, researchers confirmed a transcription factor called BnaA9.NF-YA7 (nuclear factor-Y) in B. napus that negatively affects drought tolerance. They used a genome-wide association study to pinpoint this factor, highlighting two specific SNPs within a CCAAT cis-element that reduced expression of the B. napus nuclear factor YA7 (BnaA9.NF-YA7) under drought conditions. Additionally, they discovered a genetic substitution (M63I) that activates BnaA4.DOR inhibits abscisic acid (ABA)-induced stomatal closure, thereby affecting water regulation in the plant. Furthermore, the study revealed that Bna.ABF3/4s directly control the expression of BnaA9.NF-YA7. Interestingly, BnaA9.NF-YA7 indirectly suppresses Bna.ABF3/4s expression through its regulation of Bna.ASHH4s. These findings underscore the role of BnaA9.NF-YA7 is used to maintain ABA signal balance during drought stress in canola. The study suggests that targeting BnaA9.NF-YA7 could be a promising strategy for breeding drought-tolerant varieties of B. napus.
Although some drought responsive genes have been cloned and functionally characterised in B. napus and B. rapa, the molecular signalling during stress response in Brassica species is largely unexplored (Wang et al., 2024). The dependence on Agrobacterium-mediated transformation of CRISPR/Cas delivery and genotype-dependence of transformant regeneration conditions may hindered the Cas-based breeding process. Moreover, exploring functions and roles of homologous genes controlling a complex trait like drought tolerance has recently initiated in Brassica along with the evolution of sequencing technologies (Zhang et al., 2023; Kayum et al., 2015). To date, CRIPSPR/Cas9 have been mainly utilised to elucidate functions of potential genes involved in the drought response.
3.2 Salinity and metal toxicity tolerance
Salinity and metal toxicity are other persistent problems affecting Brassica crop production. Like other abiotic stresses, they affect key physiological processes, ultimately affecting yield outcomes in these crops (Dahlawi et al., 2023; Pavlović et al., 2019). Losses to salinity stress have been reported to reach up to 50% (Chakraborty et al., 2016), while toxic concentrations of heavy metals significantly reduce plant development, resulting in severe yield and quality reduction (Dahlawi et al., 2023).
Salinity stress is commonly due to high concentration of Na+ and Cl− in the soil, resulting in hyperosmotic and hyperionic conditions (Ismail et al., 2014; Yang et al, 2018). Salt stress tolerance in Brassica spp is a complex trait that varies among species, of which allotetraploid species (B. juncea, B. carinata, and B. napus) are relatively more tolerant to salt stress as compared to their diploid parents, such as B. campestris, B. nigra, and B. oleracea (Shahzad et al., 2022). In B. napus, salt stress induced photosynthesis reduction, leaf gas exchange, and high ROS production (Shahzad et al., 2022; Wani et al., 2013). The comparison between halophyte and glycophytic plants (quinoa and Atriplex versus sugar beet and bean) highlight typical features of salt stress tolerance, such as high K+ retention in leaf mesophyll associated with higher vacuolar Na+ sequestration and less H+ pumping.
The deployment of CRISPR/Cas systems in studying salinity tolerance in Brassica is an emerging field, with pioneering works mainly relying on the knowledge gained from the related model species A. thaliana. For instance, knock-down of transcription factors (TFs) WRKY3, WRKY4, WRKY66 and AITR (ABA-induced transcription repressor) using CRISPR/Cas9 resulted in severe salt stress in A. thaliana, indicating the critical role of regulatory elements in mediating salt tolerance in plants (Chen et al., 2021; Li et al., 2021a; Zhang et al., 2023). WRKY proteins are key regulators in developmental processes, such as trichome initiation, embryp morphogenesis, senescence, and plant hormone-mediated signal transduction processes by GA, ABA, or SA (Kayum et al., 2015). Orthologs of these genes have been found in Brassica species and can be potentially targeted to improve salt tolerance in these crops (Zhang et al., 2023). These findings enhance the understanding of the complicated genetic control of salinity tolerance in crops extending beyond the known mechanisms, such as osmotic regulation and ion sequestration (Shah et al., 2018). Rewiring the regulatory network where these genetic elements participate can potentially create novel salt-tolerant phenotypes valuable for breeding (Hetti-Arachchilage et al., 2022).
CRISPR/Cas has also helped investigate the interaction of salt stress with other trace elements essential for plant growth, including boron, which has been reported to alleviate salt stress in crops (Qu et al., 2024). In B. napus, boron application upregulated the expression of BnaA2.HKT1, a gene involved in sodium (Na) unloading in plant cells (Hua et al., 2024). CRISPR/Cas9-knockout mutants of this gene were severely affected by salt stress despite the presence of elevated boron concentration. This suggests that boron is a positive regulator of Na unloading mechanisms, increasing the plant’s tolerance to salt stress. This finding informs the implementation of an optimal soil fertilisation strategy to enhance salinity tolerance in B. napus, translating CRISPR/Cas research into an actual field management practice.
Similarly, CRISPR/Cas systems have proven useful in identifying genes involved in metal toxicity tolerance in Brassica crops. Loss of function mutation in the gene BnaNRAMP1 using CRISPR/Cas9 resulted in low cadmium (Cd) accumulation in B. napus plants (Zhang et al., 2025b). Further analysis indicates this gene is crucial in detoxifying Cd by reducing its toxic levels within plant cells. Another gene that regulates Cd absorption is BnCUP1 (Yao et al., 2022), which was previously implicated in chelating excess copper in plant cells. The disruption of this gene using CRISPR/Cas9 in B. napus reduced the accumulation of Cd in both roots and shoots without negatively affecting the agronomic characteristics of the genome-edited plants. From the field experiment, in comparison with observations in the wild-type, BnCUP1-edited lines accumulated less Cd with reduction by 52% in roots and by 77% in shoots and increased in the biomass (by 42%) and yield (by 47%). Furthermore, the other key elements, including iron, zinc, and manganese, were maintained in typical concentrations, suggesting that BnCUP1 does not affect the absorption of these elements to maintain normal plant growth. The created BnCUP1-edited lines are important germplasm for breeding Cd safe edible and fodder oilseed rape.
While applying the CRISPR/Cas systems for studying salt and metal toxicity tolerance in Brassica crops is still a developing field, the abovementioned studies provide the groundwork for effectively implementing genome editing strategies to develop cultivars better adapting to these stresses. This will be further supported by the continuous development of high-quality genomic resources (Amas et al., 2023), which enables faster identification of candidate genes for various agronomic traits, including tolerance to various stresses.
3.3 Extreme temperature tolerance
Under current climate change and global warming scenarios, the impact of higher temperatures on crop production, particularly Brassica crops, is a critical concern. Climate model simulations indicate that by 2100, the Earth’s average temperature may rise by 1.1 to 5.4°C, and projections suggest a 50% increase in drought-affected areas (Battisti and Naylor, 2009; Herring, 2012).
The upper threshold warm temperature varies based on plant species. In general, an increase by 5–10 °C exceeding a plant’s optimal growth temperature triggers ROS burst and irreversible cellular oxidative damage, especially in photosynthesis systems I and II (Kan et al., 2023). Numerous studies have shown that heat stress, often coupled with drought, significantly affects crop production by impacting plant metabolism, reproduction, and physiology (Nadeem et al., 2019; Kourani et al., 2022). Studies have reported substantial crop yield reductions (Frenck et al., 2011; Hasanuzzaman et al., 2014). As cool-season crops, Brassicas can be extremely sensitive to high temperatures, significantly impacting their production (Yu et al., 2014; Ahmed et al., 2020). Hence, due to global population growth, expanding crop varieties that can withstand environmental stresses is crucial, using traditional breeding methods and advanced biotechnological tools such as CRISPR/Cas9.
The CRISPR/Cas9 technology has been applied extensively to enhance heat tolerance in various crops, however, its application in Brassica species for this trait remains relatively limited. Heat shock proteins (HSPs) act as molecular chaperones that aid cellular survival by transporting, folding, and degrading proteins under heat stress (Xu et al., 2011). Increased expression of HSP70 genes has been shown to provide more excellent resistance to abiotic stresses, including high temperatures (Wang et al., 2016; Zhao et al., 2019). Under heat stress condition, heat stress transcription factors (HSFs) bind to with HSP gene promoters and interact with heat shock factor binding proteins (HSBP) genes. HSF-mediated stress tolerance is negatively regulated by HSBPs, which affects the DNA-binding capacity activation activity of HSFs (Muthusamy et al., 2023). Although, the loss-of-function mutant of BrHSBP1 in B. rapa was indistinguishable from the BrHSBP1 overexpressed line in heat-stress responsive phenotypes, the Muthusamy et al. (2023) suggested the positive role of BrHSBP1 in drought-stress response through supporting raffinose biosynthesis, through which enhanced yield and stress tolerance can be achieved.
Heat stress significantly reduces seed production in Brassica species by altering the typical structure of floral organs. In a mutation known as sepal carpal modification (scm) observed in B. rapa, four of the five sepals merge to form a ring structure enclosing abnormal stamens and a pistil, ultimately leading to diminished seed yield. This mutation affects homologues of the BrAP2 gene, which are orthologous to the APETALA (AP2) gene in Arabidopsis. Researchers used CRISPR/Cas9 technology to generate knockout plants of four BnAP2 gene homologues in rapeseed, aiming to explore their roles in sepal and petal development (Zhang et al., 2018b). In another study, the expression of BrRH22, a chloroplast-targeted DEAD-box RNA helicase, in cabbage (B. rapa) was markedly increased by heat, drought, salt or cold stress (Nawaz et al., 2018). BrRH22 has been known to contribute positively to seed germination and plant vigor under varied abiotic stress conditions (Nawaz et al., 2018). DEAD-box RNA helicases (RHs) are nucleus-encoded chloroplast-targeted RNA-binding proteins (RBPs) involve in regulating chloroplast gene expression.
In conclusion, efforts to enhance heat tolerance through advanced biotechnological tools like CRISPR/Cas9 show promise. Future research should focus on expanding these technologies to commercial Brassica crops, such as B. napus, to mitigate the adverse effects of heat stress on yield and ensure food security in a changing climate. Integrating traditional breeding methods with cutting-edge biotechnology remains pivotal in developing resilient Brassica varieties capable of withstanding anticipated climatic challenges.
4 CRISPR gene editing in modern plant breeding
CRISPR genome editing can be integrated into plant breeding to improve resistance to biotic and abiotic stress. This involves a thorough literature review of the transcriptomic and genomic data to identify targets involved in regulating plant’s mechanisms to enhance resistance to biotic and abiotic stress (Kumar et al., 2023). Advances in plant molecular biology techniques have helped decipher the principles underlying plant signalling and regulatory pathways involved in abiotic and biotic stress responsiveness (Aroca and García, 2023). The co-occurrence of biotic and abiotic stresses sometimes results in the activation of signalling and regulatory pathways that are either unrelated, additive, or conflicting to the stress (Li et al., 2019; Yang et al., 2019). Transcriptome analysis of the plants exposed to individual and combined stresses identifies genes and transcriptional factors involved in the stress response (Rizhsky et al., 2004). On the other hand, techniques such as RNA-seq, genome-wide association studies (GWAS), and quantitative trait loci QTL mapping help to determine the significantly upregulated and down-regulated genes in plants (Meng et al., 2020). QTL mapping has identified factors involved in resistance to Sclerotinia stem rot on the chromosome locus SRC06 (Wu et al., 2013). CRISPR can be used to validate the function of the candidate genes on the locus in a short time and choose the right gene responsible for providing resistance.
The next step to validate gene function is to create a knock-out in resistant or stable transformants of the gene in susceptible plants. CRISPR/Cas genome editing offers great potential for targeted gene editing in plants (Zhang et al., 2018). The outcome of these studies is then utilized in plant breeding through traditional breeding approaches such as backcrossing and selfing to incorporate the mutated gene (Kumar et al., 2023). The plants with mutated genes are then tested in the fields under real growth conditions and challenged by the abiotic or biotic stress to assess their yield and performance. This is done to ensure that the overall yield and growth of the plant is not affected by the desired mutation and the trait has successfully been passed on to the next generation.
Genome-wide identification and characterisation studies have identified several transcription factors (TFs) involved in biotic and abiotic stress responsiveness in Brassica. Some of these studies have highlighted the function of TFs such as BRASSINAZOLE-RESISTANT (BZR) (Saha et al., 2016), NUCLEAR FACTOR (NF-Y) (Xu et al., 2014), bZIPs, MYB, NAC, WRKYs and EREBPs (Meraj et al., 2020) in biotic and abiotic stresses as well as several regulatory pathways. BZR is a positive regulator of the brassinosteroid (BR) signaling pathway in different plant families (Saha et al., 2016). Members of the auxin-responsive transcription factors (ARF) transcription factors play role in altering the expression of genes involved in auxin, abscisic acid, MeJA, salicylic acid and ethylene (Li et al., 2021). These integrative analyses have facilitated the identification of several key genes involved in both biotic and abiotic stresses. Which genes in mutated forms/altering expression level can enhance plant growth during stress conditions, then, CRISPR/cas9 can be used for gene editing.
The genome of Brassica is polyploid, complex, and redundant, making it a challenge for genome editing. As there are multiple copies of the same genes, they need to be eliminated at the same time for functional studies (Khan et al., 2021). Moreover, the chromosomal rearrangement and epigenetic modification in polyploid plants often produce transcriptional changes such as activation of transposable elements, duplication, and neo-functionalization of genes and variable expression (Wendel et al., 2018), making it difficult to link genotype to phenotype and characterize gene function (May et al., 2023). In polyploid crops such as Brassica, the exact number of homologues and homeologous genes and their function remains unclear due to gene redundancy (Schaart et al., 2021).
The availability of well-curated databases such as Brassica Database (Cheng et al., 2011) for genetics and genomics, BrassicaEDB (Cheng et al., 2011) for gene expression database, and BrassicaTED (Murukarthick et al., 2014) a public database for the utilisation of miniature transposable elements in Brassica species, have helped to narrow down targets for gene editing. EnsemblePlants and NCBI’s GEO (functional genomics dataset) have further improved gene search by providing access to gene models and synteny with related species, such as A. thaliana (Bolser et al., 2016; Clough et al., 2024). These genetics and functional genomic databases not only provide a valuable tool for designing gRNAs with high specificity and minimal off-target effects but also differentiate between homeologous gene copies, which is a common challenge in polyploid Brassica genomes.
CRISPR-based tools have a significant advantage for genome editing in Brassica due to precise, homeolog-specific gene editing, providing researchers the ability to target genes and their copies on the subgenomes, evaluating their response to different stresses (Ahmad et al., 2023a). More advanced gene editing techniques, such as CRISPRa (CRISPR activation) of Resistance genes, and CRISPRi (CRISPR interference) of susceptible genes, can be effectively employed to improve disease resistance in Brassica crops (Zafar et al., 2020) Figure 1. Multiplex gene editing of JAGGED genes in B. napus through CRISPR has helped understand the role of several homeologs of the genes in pod shattering resistance. The results showed that a single mutation of one of the JAG genes on ChrA08 helped to improve pod shattering resistance (Zaman et al., 2019). In most reported studies, by targeting the key genes regulating the stress-responsive pathways, scientists can improve a plant’s ability to withstand adverse environmental conditions. Cas-edited mutants often have loss or gain functions, while some are fine-tune in their functions. The precision and flexibility in genome editing provide a huge potential for developing crops with enhanced yield, improved resistance to biotic and abiotic stress, and better quality.
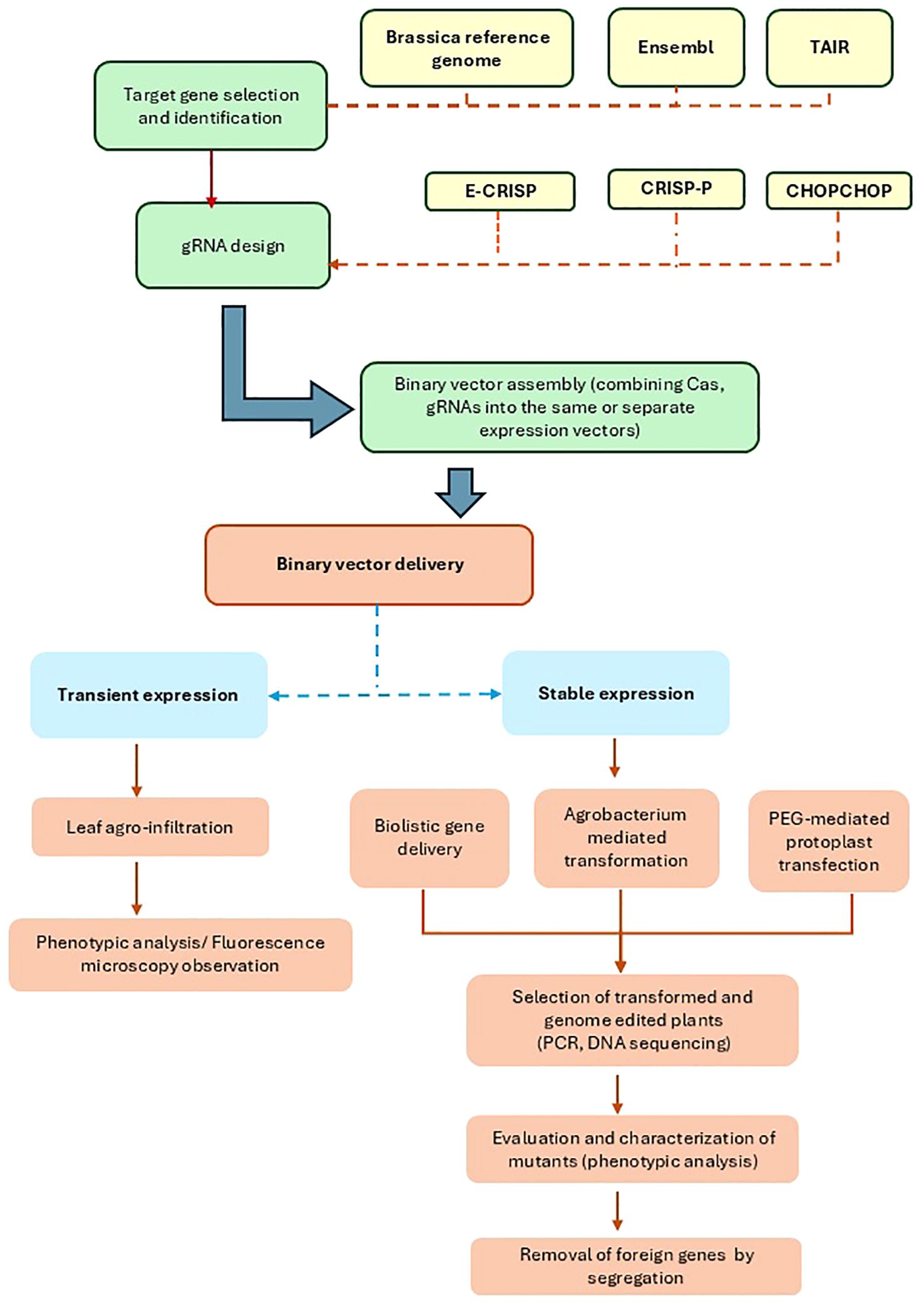
Figure 1. The flowchart shows a typical CRISPR/Cas-based genome editing pipeline in Brassica. Candidate genes are selected through transcriptomic and genomic analysis and used as targets for gRNA design. gRNA prediction tools, such as E-CRISP, CRISP-P and CHOPCHOP are used for designing gRNA. Then, CRISPR/Cas constructs specifically targeting the candidate genes are incorporated into transformation vectors. CRISPR/Cas constructs can be delivered and active through either transient (agro-infiltration) or stable transformation methods (biolistic gene delivery, Agrobacterium-mediated transformation and protoplast transformation). Following the transformation, modified plants are genotyped and selected on based on selection markers (e.g. fluorescence signal) and phenotypes. They are then multiplied and crossed for a few generations to confirm the transfer of the desired trait and removal of foreign DNA.
5 Workflow for CRISPR/Cas application in Brassica
CRISPR/Cas-mediated genome editing in plants, including Brassica species, is often coupled with transformation. Therefore, the workflow for CRISPR/Cas-based modification in Brassica involves three main stages, as depicted in Figure 1: constructing a vector carrying an endonuclease system for targeted modifications, transforming vectors encoding the CRISPR/Cas components into plant cells, transgenic plant generating and evaluating the transformation and genome editing efficiency (Ahmad et al., 2023b).
Firstly, developing a CRISPR/Cas construct may involve the following steps: identifying genomic targets, selecting a CRISPR/Cas tool, and designing and evaluating CRISPR/Cas vectors Hanna and Doench, 2020. The availability of Arabidopsis reference genome TAIR10 and the evolution of Brassica genomic data within recent years, especially an increasing number of published Brassica reference genomes and pangenomes, has benefited choices for the genetic improvement of Brassica crops via genome editing tools (Niu et al., 2024). Typical online Cas9-gRNA predicting tools are E-CRISP v.5.4 (Heigwer et al., 2014), CHOPCHOP (Labun et al., 2019), and Optimized CRISPR Plant Design Tool/CRISPR-P (http://cbi.hzau.edu.cn/cgi-bin/CRISPR; Liu et al., 2017; Lei et al., 2014). These tools employ published genome data as input for PAM scanning and gRNA designing. In 2022, Müller Paul et al., 2022 introduced CROPSR, a gRNA design and validation tool specialised for crop genomes, claiming this open-source tool written in Python 3.7 as the first developed tool for genome-wide generation and validation of sgRNA for crop genome editing and outperforming CHOPCHOP.
Following gRNA design, assembling CRISPR/Cas components is crucial in determining genome editing efficiency. A typical CRISPR/Cas expression vector can range from 9 to 16 kb (Zhang et al., 2024; Sun et al., 2018; Ma et al., 2015), containing Cas, gRNA, selection marker, and/or reporter gene sequences. Promoters control the expression of these genes. Choices of promoters, especially those controlling the expression of Cas enzymes and gRNAs, contribute to efficient genome editing. CaMV 35S, a strong constitutive promoter, is widely used for Cas expression in Brassica crops and vegetables (Villao-Uzho et al., 2023; Zhang D. et al., 2020). Use of other promoter types, such as tissue-specific promoters, inducible promoters, and Arabidopsis ubiquitin 10 promoter (Ubi10), were also reported (Tu et al., 2024; LeBlanc et al., 2018; Kurokawa et al., 2021; Papikian et al., 2019). Due to the small size of gRNA (20–30 nt), A. thaliana RNA polymerase III promoters (U3, U6) are often used to control the expression of gRNA cassettes in Arabidopsis and other Brassica species beside ubiquitin promoters (Li et al., 2022; Ma et al., 2019). In Arabidopsis, the gene editing efficiency of Cas9 driven by DD45 (subgroup of cysteine-rich peptide sequences, CRP) and the ribosomal protein S5A (RPS5A) promoters showed an increase in gene editing efficiency up to 30-fold compared to 35S and ubiquitin promoters (Ordon et al., 2020). Likewise, cell-specific promoters, such as pYAO and pEC, also showed similar effect (Chen et al., 2021, Chen et al., 2021; Wang et al., 2015). Cas and sgRNA(s) constructs of a CRISPR/Cas system can be expressed in the same or separate expression vectors. Other main components the expression vector includes are the localisation signal (NLS) and terminators of Cas and gRNA expression cassettes, which also attribute to efficient genome modification (Ordon et al., 2020).
For Brassica and other plant species, Agrobacterium-mediated transformation is the most commonly used method (Li et al., 2024; Li et al., 2022; Huang et al., 2020) for permanent and transient expression of CRISPR/Cas systems (Zhang K. et al., 2020; Aman et al., 2018a). Other methods such as PEG-mediated transformation and bombardment have been successfully applied for delivering CRISPR/Cas constructs directly into Arabidopsis protoplasts (Zhang Y. et al., 2022), B. oleracea (Romero and Pérez, 2024; Stelmach-Wityk et al., 2024; Stajič and Kunej, 2023), and B. napus (Li et al., 2021b). Yu et al. (2024) optimised a PEG-mediated protoplast transformation system for the transient expression of CRISPR/Cas9 vectors in B. oleracea L. (Chinese cabbage), Chinese kale and B. rapa (turnips) with improved protoplast yield and high transfection efficiencies (50 – 80%).
A non-transformation approach for CRISPR/Cas delivery, such as leaf infiltration and virus-based Cas-gRNA vectors, can be utilised to quickly evaluate the CRISPR/Cas system expression and skip many steps of in vitro plant regeneration. The small size of virus vectors (~ 1 kb) allows more efficient CRISPR/Cas delivery than Agrobacterium vectors (~ 8.9 kb). However, their package capacity is often below 4 kb (Varanda et al., 2021; Kirigia et al., 2014). Therefore, in a typical functional genomic study, gRNA constructs were cloned into virus-derived vectors, while Cas9 were delivered into plant cells via different vectors. Ali et al. (2015) developed the RNA2 genome of the bipartite genome of the tobacco rattle virus (TRV) into a vehicle for sgRNAs delivery in Nicotiana benthamiana. This TRV-mediated system also successfully delivered multiple sgRNAs into Arabidopsis leaves, leading to high mutation frequencies of targeted sequences (Ali et al., 2018). Other plant virus-derived CRISPR/Cas systems, also known as effective plant genome editing tools, such as pea early browning virus (PEBV), tobacco mosaic virus (TMV), and Sonchus yellow net rhabdovirus (SYNV), caused high frequency of target mutations in N. benthamiana (Ma, 2020; Ali et al., 2018). However, these have not been applied in Brassica plants.
To obtain a CRISPR/Cas mutant line, in most cases, transformed cells/tissues need to constitutively express CRISPR/Cas components, grow on selection media, and develop into whole plants (Romero and Pérez, 2024; Bao et al., 2019). The putative transformed plants are then genotyped and checked for the expression of the Cas and gRNA sequences using PCR and qPCR (Yu et al., 2024; Li et al., 2022; Hussain et al., 2018). Genome editing efficiency is calculated based on the number of plants containing targeted mutants and the total number of transgenic plants (Zhang et al., 2019). Mutated lines of the T0 population are then sequenced to confirm the CRISPR/Cas-induced mutations. For commercial, breeding or study purposes, several rounds of crossing and backcrossing of T0 progenies may be needed, as shown in Figure 2, to remove foreign genetic insertions in the plant genome, such as selection marker genes, gRNA and Cas sequences, and to obtain homozygous mutant plants as shown in Figure 2 (Romero and Pérez, 2024). Transgene-free CRISPR/Cas-mutated plants can be directly obtained in T0 generation with protoplast-based genome editing by transient expression of CRISPR/Cas without incorporating the foreign gene construct into plant genome (Romero and Pérez, 2024). However, Brassica regeneration from protoplasts is more challenging than other explants, such as cotyledons and hypocotyl segments, and is a time-consuming process (Li et al., 2021b, Li et al, 2022). Therefore, Agrobacterium-mediated transformation using cotyledon or hypocotyl explants remains the dominant method (Zhang K. et al., 2020). Despite the availability of many regeneration protocols for the main Brassica crops (B. napus, B. oleracea, and B. rapa), their transformation efficiency and in vitro growth are mostly genotype-dependent (Farooq et al., 2019), presenting a major challenge for developing a CRISPR/Cas genome-edited Brassica crop.
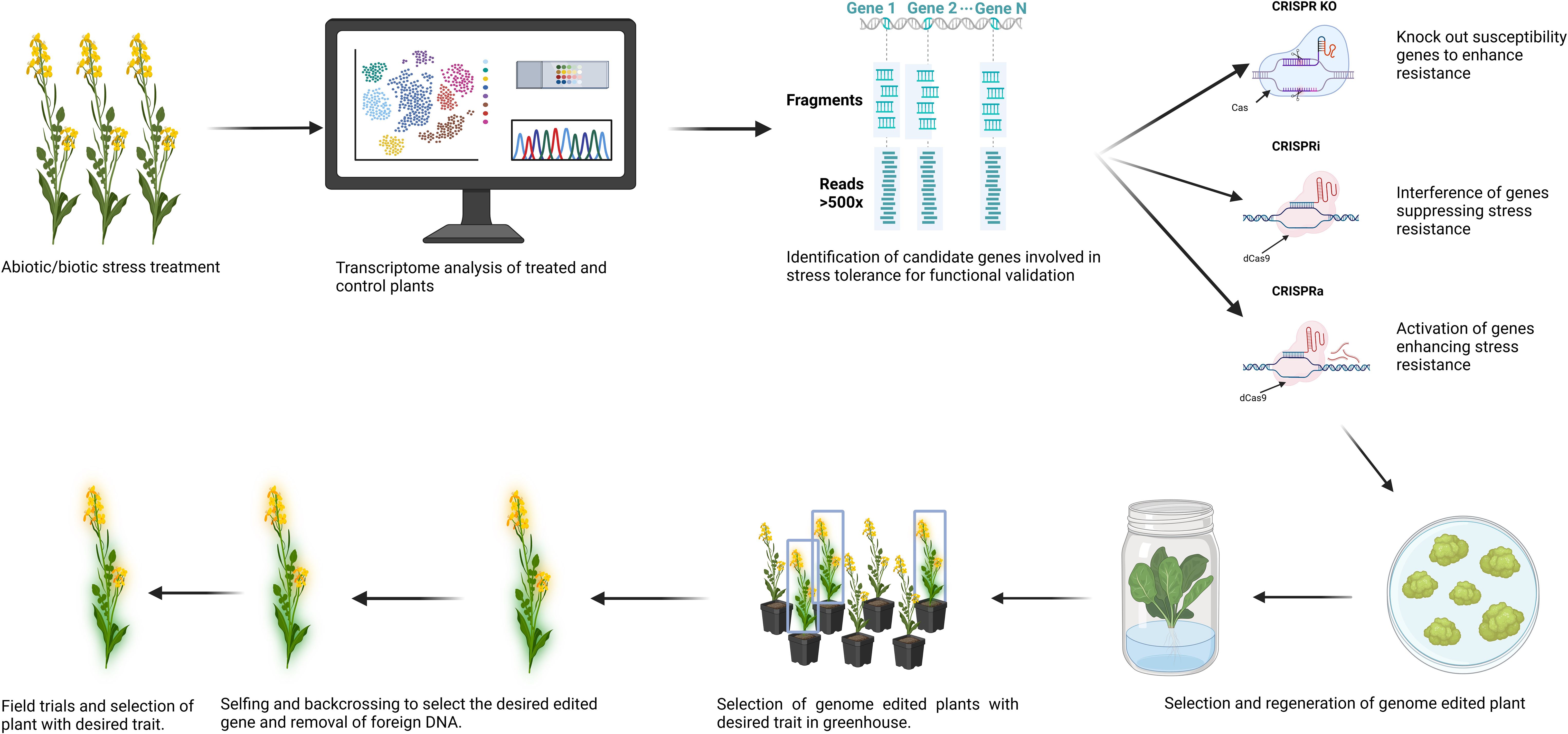
Figure 2. The workflow highlights key steps in employing advanced CRISPR/Cas tools (CIRSPR-KO, CRISPRi, and CRISPRa) in breeding abiotic/biotic stress tolerant Brassica plants, from genetic target identification, CRISPR/Cas vector construction and delivery, transformant selection and confirmation, characterization of genetic modifications, and the generation of “transgene-free” and homologous mutants of edited plants. Created in BioRender. Qayyum, Z. (2025) https://BioRender.com/3jb9miy.
6 Conclusion
CRISPR/Cas systems are efficient tools for exploring functions of Brassica genomes, especially the complex polyploid genomes of B. napus. These tools also help develop the knowledge of interactions between brassica hosts and their pathogens and the genetic mechanisms underlying abiotic stress responses in Brassica crops. This knowledge widens CRISPR/Cas-based approaches for the genetic control of Brassica phenotypes, improving resistance and resilience to biotic and abiotic stress. Although limited Brassica genes with known functions are being exploited for these breeding targets, several Brassica genomic resources are being constructed, and the evolution of Brassica multi-omics will provide more information for CRISPR/Cas genome editing interventions. Regarding tackling plant pathogens, CRISPR/Cas tools can target plant hosts or pathogens to improve plant defence capacity. At the same time, components of the CRISPR/Cas expression constructs are also being optimised for different research purposes and tailored for more efficient and controllable mutation inductions in Brassica. Genotype-dependence in transformation and regeneration of both Brassica explants and protoplasts remains a challenge, prolonging these processes. A transgene-free protoplast-based genome editing approach is still being pursued by many research groups to develop CRISPR/Cas-based edited Brassica crops for commercial release. Platforms for applying CRISPR/Cas genome editing tools in Brassica are urgently needed to accelerate the improvement of these crops under the negative consequences of climate change.
Author contributions
LT: Writing – original draft, Conceptualization, Validation, Investigation. ZQ: Writing – original draft. JA: Writing – original draft. WT: Writing – original draft. DE: Supervision, Writing – review & editing, Conceptualization. JB: Funding acquisition, Writing – review & editing, Supervision, Conceptualization. AD: Supervision, Writing – review & editing, Conceptualization, Writing – original draft, Visualization.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The authors declare that this study received funding from the Australian Research Council projects DP210100296, FL230100030 and Grain Research and Development Corporation projects UWA1905-006RTX, UWA2307-002RTX. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Acknowledgments
L. B. Ton acknowledges the support of the University of Western Australia through the UWA International Fees Scholarship, UWA Postgraduate Awards (UPA) and Underwood PhD Completion Scholarship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abuyusuf, M., Robin, A. H. K., Kim, H.-T., Islam, M. R., Park, J.-I., and Nou, I.-S. (2018). Altered glucosinolate profiles and expression of glucosinolate biosynthesis genes in ringspot-resistant and susceptible cabbage lines. Int. J. Mol. Sci. 19.2833 doi: 10.3390/ijms19092833
Adwiyah, R., Syaukat, Y., Indrawan, D., and Mulyati, H. (2023). Examining sustainable supply chain management (SSCM) performance in the palm oil industry with the triple bottom line approach. Sustainability 15, 13362. doi: 10.3390/su151813362
Ahmad, N., Fatima, S., Mehmood, M. A., Zaman, Q. U., Atif, R. M., Zhou, W., et al. (2023a). Targeted genome editing in polyploids: lessons from Brassica. Front. Plant Sci. 14,1152468. doi: 10.3389/fpls.2023.1152468
Ahmad, N., Fatima, S., Mehmood, M. A., Zaman, Q. U., Atif, R. M., Zhou, W., et al. (2023b). Targeted genome editing in polyploids: lessons from Brassica. Front. Plant Sci. 14, 1152468. doi: 10.3389/fpls.2023.1152468
Ahmed, W., Xia, Y., Li, R., Bai, G., Siddique, K. H. M., and Guo, P. (2020). Non-coding RNAs: functional roles in the regulation of stress response in Brassica crops. Genomics 112, 1419–1424. doi: 10.1016/j.ygeno.2019.08.011
Ali, Z., Abul-Faraj, A., Li, L., Ghosh, N., Piatek, M., Mahjoub, A., et al. (2015). Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol. Plant 8, 1288–1291. doi: 10.1016/j.molp.2015.02.011
Ali, Z., Eid, A., Ali, S., and Mahfouz, M. M. (2018). Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis. Virus Res. 244, 333–337. doi: 10.1016/j.virusres.2017.10.009
Aman, R., Ali, Z., Butt, H., Mahas, A., Aljedaani, F., Khan, M. Z., et al. (2018a). RNA virus interference via CRISPR/Cas13a system in plants. Biol. 19. doi: 10.1186/s13059-017-1381-1
Aman, R., Mahas, A., Butt, H., Aljedaani, F., and Mahfouz, M. (2018b). Engineering RNA virus interference via the CRISPR/Cas13 machinery in Arabidopsis. Viruses 10. doi: 10.3390/v10120732
Amas, J. C., Thomas, W. J. W., Zhang, Y., Edwards, D., and Batley, J. (2023). Key advances in the new era of genomics-assisted disease resistance improvement of Brassica species. Phytopathology 113, 771–785. doi: 10.1094/PHYTO-08-22-0289-FI
Aroca, A. and García, I. (2023). Advances in plant molecular biology: Towards new challenges. J. Exp. Bot. 74, 5949–5954. doi: 10.1093/jxb/erad350
Asmamaw, M. and Zawdie, B. (2021). Mechanism and applications of CRISPR/Cas-9-mediated genome editing. Biologics 15, 353–361. doi: 10.2147/BTT.S3264222
Bao, A., Burritt, D. J., Chen, H., Zhou, X., Cao, D., and Tran, L.-S. P. (2019). The CRISPR/Cas9 system and its applications in crop genome editing. Crit. Rev. Biotechnol. 39, 321–336. doi: 10.1080/07388551.2018.1554621
Basili, M. and Rossi, M. A. (2018). Brassica carinata-derived biodiesel production: economics, sustainability and policies. The Italian case. J. Clean. Prod. 191, 40–47. doi: 10.1016/j.jclepro.2018.03.306
Battisti, D. S. and Naylor, R. L. (2009). Historical warnings of future food insecurity with unprecedented seasonal heat. Science 323, 240–244. doi: 10.1126/science.1164363
Bhadauria, V., Han, T., Li, G., Ma, W., Zhang, M., Yang, J., et al. (2024). A gln-tRNA-based CRISPR/Cas9 knockout system enables the functional characterization of genes in the genetically recalcitrant brassica anthracnose fungus Colletotrichum higginsianum. Int. J. Biol. Macromol. 254, 127953. doi: 10.1016/j.ijbiomac.2023.127953
Bhattacharya, S. (2019). Brassica-aphid interaction: Challenges and prospects of genetic engineering for integrated aphid management. Physiol. Mol. Plant Pathol. 108, 101442. doi: 10.1016/j.pmpp.2019.101442
Boivin, K., Acarkan, A., Mbulu, R. S., Clarenz, O., and Schmidt, R. (2004). The Arabidopsis genome sequence as a tool for genome analysis in Brassicaceae. A comparison of the Arabidopsis and Capsella rubella genomes. Plant Physiol. 135, 735–744. doi: 10.1104/pp.104.040030
Bolser, D., Staines, D. M., Pritchard, E., and Kersey, P. (2016). Ensembl plants: integrating tools for visualizing, mining, and analyzing plant genomics data. Plant bioinformatics: Methods Protoc., 115–140. doi: 10.1007/978-1-4939-3167-5_6
Borges, C. E., Von dos Santos Veloso, R., da Conceição, C. A., Mendes, D. S., Ramirez-Cabral, N. Y. Z., Shabani, F., et al. (2023). Forecasting Brassica napus production under climate change with a mechanistic species distribution model. Sci. Rep. 13, 12656. doi: 10.1038/s41598-023-38910-3
Botero-Ramirez, A., Hwang, S. F., and Strelkov, S. E. (2022). Effect of clubroot (Plasmodiophora brassicae) on yield of canola (Brassica napus). Can. J. Plant Pathol. 44, 372–385. doi: 10.1080/07060661.2021.1989801
Broadbent, L. (2015). Investigation of virus diseases of Brassica crops (No. 14) (Cambridge, UK: Cambridge University Press).
Bruckner, F. P., Barbosa, T. M. C., Eiras, M., Zanardo, L. G., and Awasthi, L. P. (2024). “Chapter 51 - broccoli, cabbage and cauliflower,” in Viral diseases of field and horticultural crops ed. Awasthi, L. P. (London, UK: Academic Press), 427–436. doi: 10.1016/B978-0-323-90899-3.00022-7
Carroll, D. (2008). Progress and prospects: Zinc-finger nucleases as gene therapy agents. Gene Ther. 15, 1463–1468. doi: 10.1038/gt.2008.145
Cathomen, T. and Keith, J. J. (2008). Zinc-finger nucleases: the next generation emerges. Mol. Ther. 16, 1200–1207. doi: 10.1038/mt.2008.114
Chakraborty, K., Sairam, R. K., and Bhaduri, D. (2016). Effects of different levels of soil salinity on yield attributes, accumulation of nitrogen, and micronutrients in Brassica spp. J. Plant Nutr. 39, 1026–1037. doi: 10.1080/01904167.2015.1109105
Charpentier, E., Richter, H., van der Oost, J., and White, M. F. (2015). Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity. FEMS Microbiol. Rev. 39, 428–441. doi: 10.1093/femsre/fuv023
Chen, H., Wang, Y., Liu, J., Zhao, T., Yang, C., Ding, Q., et al. (2021). Identification of WRKY transcription factors responding to abiotic stresses in Brassica napus L. Planta 255, 3. doi: 10.1007/s00425-021-03733-x
Chen, K., Wang, Y., Zhang, R., Zhang, H., and Gao, C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. doi: 10.1146/annurev-arplant-050718-100049
Chen, S., Zhang, N., Zhou, G., Hussain, S., Ahmed, S., Tian, H., et al. (2021). Knockout of the entire family of AITR genes in Arabidopsis leads to enhanced drought and salinity tolerance without fitness costs. BMC Plant Biol. 21, 1–15. doi: 10.1186/S12870-021-02907-9/FIGURES/9
Cheng, F., Liu, S., Wu, J., Fang, L., Sun, S., Liu, B., et al. (2011). BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 11, 1–6. doi: 10.1186/1471-2229-11-136
Chisholm, S. T., Coaker, G., Day, B., and Staskawicz, B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814. doi: 10.1016/j.cell.2006.02.008
Christian, M. L., Demorest, Z. L., Starker, C. G., Osborn, M. J., Nyquist, M. D., Zhang, Y., et al. (2012). Targeting G with TAL effectors: a comparison of activities of TALENs constructed with NN and NK repeat variable di-residues. PloS One 7, e45383. doi: 10.1371/journal.pone.0045383
Clough, E., Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., et al. (2024). NCBI GEO: archive for gene expression and epigenomics data sets: 23-year update. Nucleic Acids Res. 52, D138–D144. doi: 10.1093/nar/gkad965
Cox, D. B. T., Gootenberg, J. S., Abudayyeh, O. O., Franklin, B., Kellner, M. J., Joung, J., et al. (2017). RNA editing with CRISPR-cas13. Science 358, 1019–1027. doi: 10.1126/science.aaq0180
Cunnac, S., Lindeberg, M., and Collmer, A. (2009). Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr. Opin. Microbiol. 12, 53–60. doi: 10.1016/j.mib.2008.12.003
Dahlawi, S., Sadiq, M., Sabir, M., Farooqi, Z. U. R., Saifullah, Q. A.A., and Faraj, T. K. (2023). Differential response of Brassica cultivars to potentially toxic elements and their distribution in different plant parts irrigated with metal-contaminated water. Sustainability 15.1966 doi: 10.3390/SU15031966
Dalio, R. J. D., Paschoal, D., Arena, G. D., Magalhães, D. M., Oliveira, T. S., Merfa, M. V., et al. (2021). Hypersensitive response: From NLR pathogen recognition to cell death response. Ann. Appl. Biol. 178, 268–280. doi: 10.1111/aab.12657
Daniel, K. A. M., Muindi, E. M. D., and Muti, S. M. D. (2023). Cabbage (Brassica oleracea) production in Kenya: a review of its economic importance, ecological requirement and production constraints. Int. J. Plant Soil Sci. 35, 245–254. doi: 10.9734/ijpss/2023/v35i183287
Das, S., Kundu, A., and Podder, S. (2024). Impact of biotic stresses on the Brassicaceae family and opportunities for crop improvement by exploiting genotyping traits. Planta 259, 97. doi: 10.1007/s00425-024-04379-1
De, A., Maity, A., Mazumder, M., Mondal, B., Mukherjee, A., and Ghosh, S. (2021). Overexpression of LYK4, a lysin motif receptor with non-functional kinase domain, enhances tolerance to Alternaria brassicicola and increases trichome density in Brassica juncea. Plant Sci. 309, 110953. doi: 10.1016/j.plantsci.2021.110953
FAOSTAT (2023). Crops and livestock products. Available online at: www.fao.org/faostat/en/data/QCL/visualize (Accessed November 2024).
Farooq, N., Nawaz, M. A., Mukhtar, Z., Ali, I., Hundleby, P., and Ahmad, N. (2019). Investigating the in vitro regeneration potential of commercial cultivars of Brassica. Plants 8, 558. doi: 10.3390/plants8120558
Farooq, M., Wahid, A., Kobayashi, N., Fujita, D., and Basra, S. M. A. (2009). Plant drought stress: effects, mechanisms and management. Agron. Sustain. Dev. 29, 185–212. doi: 10.1051/agro:2008021
Finkler, A., Ashery-Padan, R., and Fromm, H. (2007). CAMTAs: Calmodulin-binding transcription activators from plants to human. FEBS Lett. 581, 3893–3898. doi: 10.1016/j.febslet.2007.07.051
Francisco, M., Tortosa, M., Martínez-Ballesta, M., d., C., Velasco, P., García-Viguera, C., et al. (2017). Nutritional and phytochemical value of Brassica crops from the agri-food perspective. Ann. Appl. Biol. 170, 273–285. doi: 10.1111/aab.12318
Frenck, G., van der Linden, L., Mikkelsen, T. N., Brix, H., and Jørgensen, R. B. (2011). Increased [CO2] does not compensate for negative effects on yield caused by higher temperature and [O3] in Brassica napus L. Eur. J. Agron. 35, 127–134. doi: 10.1016/j.eja.2011.05.004
Greer, S. F., Hackenberg, D., Gegas, V., Mitrousia, G., Edwards, D., Batley, J., et al. (2021). Quantitative trait locus mapping of resistance to turnip yellows virus in Brassica rapa and Brassica oleracea and introgression of these resistances by resynthesis into allotetraploid plants for deployment in Brassica napus [Original Research. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.781385
Guerret, M. G. L., Nyalugwe, E. P., Maina, S., Barbetti, M. J., van Leur, J. A. G., and Jones, R. A. C. (2017). Biological and molecular properties of a Turnip mosaic virus (TuMV) strain that breaks TuMV resistances in Brassica napus. Plant Dis. 101, 674–683. doi: 10.1094/pdis-08-16-1129-re
Guo, Y., Zhao, G., Gao, X., Zhang, L., Zhang, Y., Cai, X., et al. (2023). CRISPR/Cas9 gene editing technology: a precise and efficient tool for crop quality improvement. Planta 258, 36. doi: 10.1007/s00425-023-04187-z
Hagit, H., Steffen, O., Anton, R., Shany Ishgur, G., Gur, P., Julia, K., et al. (2024). Rapid, direct, and sequence-specific identification of RNA viruses in various crop plants using CRISPR/Cas13a. bioRxiv 2024, 2002.2022.581525. doi: 10.1101/2024.02.22.581525
Hanna, R. E. and Doench, J. G. (2020). Design and analysis of CRISPR–Cas experiments. Nat. Biotechnol. 38, 813–823. doi: 10.1038/s41587-020-0490-7
Hasan, J., Megha, S., and Rahman, H. (2021). Clubroot in Brassica: Recent advances in genomics, breeding, and disease management. Genome 64, 735–760. doi: 10.1139/gen-2020-0089
Hasanuzzaman, M., Nahar, K., Alam, M. M., and Fujita, M. (2014). Modulation of antioxidant machinery and the methylglyoxal detoxification system in selenium-supplemented Brassica napus seedlings confers tolerance to high temperature stress. Biol. Trace Element Res. 161, 297–307. doi: 10.1007/s12011-014-0120-7
Heigwer, F. and Kerr, G. (2014). and Boutros, M. E-CRISP: fast CRISPR target site identification. Nat. Methods 11, 122–123. doi: 10.1038/nmeth.2812
Hetti-Arachchilage, M., Challa, G. S., and Marshall-Colón, A. (2022). Rewiring network plasticity to improve crops. Plant Breed. Rev. 45, 143–183. doi: 10.1002/9781119828235.CH3
Herring, D. (2012).Climate change: Global temperature projections. Available online at: www.climate.gov/news-features/understanding-climate/climate-change-global-temperature (Accessed November 15, 2024).
Hooks, C. R. R. and Fereres, A. (2006). Protecting crops from non-persistently aphid-transmitted viruses: A review on the use of barrier plants as a management tool. Virus Res. 120, 1–16. doi: 10.1016/j.virusres.2006.02.006
Hu, H. and Yu, F. (2024). Studies on the temporal, structural, and interacting features of the clubroot resistance gene Rcr1 using CRISPR/Cas9-based systems. Hortic. Plant J. 10, 1035–1048. doi: 10.1016/j.hpj.2024.04.001
Hu, H., Zhang, Y., and Yu, F. (2024). A CRISPR/Cas9-based vector system enables the fast breeding of selection-marker-free canola with Rcr1-rendered clubroot resistance. J. Exp. Bot. 75, 1347–1363. doi: 10.1093/jxb/erad471
Hua, Y., Pei, M., Song, H., Liu, Y., Zhou, T., Chao, H., et al. (2024). Boron confers salt tolerance through facilitating BnaA2.HKT1-mediated root xylem Na+ unloading in rapeseed (Brassica napus L.). Plant J. 120, 1326–1342. doi: 10.1111/TPJ.17052
Huang, H., Cui, T., Zhang, L., Yang, Q., Yang, Y., Xie, K., et al. (2020). Modifications of fatty acid profile through targeted mutation at BnaFAD2 gene with CRISPR/Cas9-mediated gene editing in Brassica napus. Theor. Appl. Genet. 133, 2401–2411. doi: 10.1007/s00122-020-03607-y
Hussain, B., Lucas, S. J., and Budak, H. (2018). CRISPR/Cas9 in plants: at play in the genome and at work for crop improvement. Brief. Funct. Genomics 17, 319–328. doi: 10.1093/bfgp/ely016
Hwang, S. F., Howard, R. J., Strelkov, S. E., Gossen, B. D., and Peng, G. (2014). Management of clubroot (Plasmodiophora brassicae) on canola (Brassica napus) in western Canada. Can. J. Plant Pathol. 36, 49–65. doi: 10.1080/07060661.2013.863806
Hyun, T. K. (2020). CRISPR/Cas-based genome editing to improve abiotic stress tolerance in plants. Bot. Serbica 44, 121–127. doi: 10.2298/BOTSERB2002121H
Iqbal, Z., Iqbal, M. S., Hashem, A., Abd_Allah, E. F., and Ansari, M. I. (2021). Plant defense responses to biotic stress and its interplay with fluctuating dark/light conditions. Front. Plant Sci. 12, 631810. doi: 10.3389/fpls.2021.631810
Ismail, A., Takeda, S., and Nick, P. (2014). Life and death under salt stress: Same players, different timing? J. Exp. Bot. 65, 2963–2979. doi: 10.1093/jxb/eru159
Javed, M. A., Schwelm, A., Zamani-Noor, N., Salih, R., Silvestre Vañó, M., Wu, J., et al. (2023). The clubroot pathogen Plasmodiophora brassicae: A profile update. Mol. Plant Pathol. 24, 89–106. doi: 10.1111/mpp.13283
Jiang, F. and Doudna, J. A. (2017). CRISPR–Cas9 structures and mechanisms. Annu. Rev. Biophys. 46, 505–529. doi: 10.1146/annurev-biophys-062215-010822
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jones, R. A. C. (2021). Global plant virus disease pandemics and epidemics. Plants 10, 233. doi: 10.3390/plants10020233
Jose, J., Ghantasala, S., and Roy Choudhury, S. (2020). Arabidopsis transmembrane receptor-like kinases (RLKs): A bridge between extracellular signal and intracellular regulatory machinery. Int. J. Mol. Sci. 21, 4000. doi: 10.3390/ijms21114000
Kan, Y., Mu, X. R., Gao, J., Lin, H. X., and Lin, Y. (2023). The molecular basis of heat stress responses in plants. Mol. Plant 16, 1612–1634. doi: 10.1016/j.molp.2023.09.013
Kaur, C., Lee, M., Jeon, Y., and Lee, G. J. (2023b). Enhancing black-rot resistance in cabbage using CRISPR/Cas9 system. Hortic. Sci. Technol. 41, 423.
Kaur, C., Lee, M., Yao, J., Nam, D., and Lee, G. J. (2023a). CRISPR/Cas12-mediated detection of Xanthomonascampestris pv. campestris (race 1 and 4) for improved pathogen diagnosis in cabbage. Hortic. Sci. Technol. 41, 338.
Kayum, M. A., Jung, H. J., Park, J. I., Ahmed, N. U., Saha, G., Yang, T. J., et al. (2015). Identification and expression analysis of WRKY family genes under biotic and abiotic stresses in Brassica rapa. Mol. Genet. Genom. 290, 79–95. doi: 10.1007/s00438-014-0898-1
Khan, M. H. U., Hu, L., Zhu, M., Zhai, Y., Khan, S. U., Ahmar, S., et al. (2021). Targeted mutagenesis of EOD3 gene in Brassica napus L. regulates seed production. J. Cell. Physiol. 236, 1996–2007. doi: 10.1002/jcp.29986
Kidanemariam, D. and Abraham, A. (2023). “Chapter 3 luteoviruses,” in Plant RNA viruses, ed. Gaur, R. K., Patil, B. L., and Selvarajan, R.. (London, UK: Selvarajan (Academic Press), 57–77.
Kim, J. S., Kidokoro, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2024). Regulatory networks in plant responses to drought and cold stress. Plant Physiol. 195, 170–189. doi: 10.1093/plphys/kiae105
Kirigia, D., Runo, S., and Alakonya, A. (2014). A virus-induced gene silencing (VIGS) system for functional genomics in the parasitic plant Striga hermonthica. Plant Methods 10, 1–8. doi: 10.1186/1746-4811-10-16
Kirkegaard, J. A., Lilley, J. M., Berry, P. M., and Rondanini, D. P. (2021). “Canola,” in Crop physiology case histories for major crops. Eds. Sadras, V. O. and Calderini, D. F. (London, UK: Academic Press), 518–549.
Klair, D., Boluk, G., Silva, J., Arizala, D., Dobhal, S., and Arif, M. (2021). First report of bacterial soft rot disease on pak choi (Brassica rapa subsp. chinensis) caused by Pectobacterium brasiliense in the United States. Plant Dis. 105. doi: 10.1094/PDIS-08-20-1854-PDN
Kourani, M., Mohareb, F., Rezwan, F. I., Anastasiadi, M., and Hammond, J. P. (2022). Genetic and physiological responses to heat stress in Brassica napus. Front. Plant Sci. 13, 832147. doi: 10.3389/fpls.2022.832147
Kumar, M., Prusty, M. R., Pandey, M. K., Singh, P. K., Bohra, A., Guo, B., et al. (2023). Application of CRISPR/Cas9-mediated gene editing for abiotic stress management in crop plants. Front. Plant Sci. 14, 1157678. doi: 10.3389/fpls.2023.1157678
Kurokawa, S., Rhaman, H., Yamanaka, N., Ishizaki, C., Islam, S., Aiso, T., et al. (2021). A simple heat treatment increases spCas9-mediated mutation efficiency in arabidopsis. Plant Cell Physiol. 62, 1676–1686. doi: 10.1093/pcp/pcab123
Labun, K., Montague, T. G., Krause, M., Torres Cleuren, Y. N., Tjeldnes, H., and Valen, E. (2019). CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171–W174. doi: 10.1093/nar/gkz365
Latham, L. J., Smith, L. J., and Jones, R. A. C. (2003). Incidence of three viruses in veget able brassica plantings and associated wild radish weeds in south-west Australia. Australas. Plant Pathol. 32, 387–391. doi: 10.1071/ap03031
Leoonard, S., Plante, D., Wittmann, S., Daigneault, N., Fortin, M. G., and Laliberte, J. F. (2000). Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 74, 7730–7737. doi: 10.1128/jvi.74.17.7730-7737.2000
LeBlanc, C., Zhang, F., Mendez, J., Lozano, Y., Chatpar, K., Irish, V. F., et al. (2018). Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J. 93, 377–386. doi: 10.1111/tpj.13782
Lee, Y. R., Ko, K. S., Lee, H. E., Lee, E. S., Han, K., Yoo, J. Y., et al. (2023b). CRISPR/Cas9-mediated HY5 gene editing reduces growth inhibition in Chinese cabbage (Brassica rapa) under ER Stress. Int. J. Mol. Sci. 24, 13105. doi: 10.3390/ijms241713105
Lee, Y. R., Siddique, M. I., Kim, D. S., Lee, E. S., Han, K., Kim, S. G., et al. (2023a). CRISPR/Cas9-mediated gene editing to confer turnip mosaic virus (TuMV) resistance in Chinese cabbage (Brassica rapa). Hortic. Res. 10, uhad078. doi: 10.1093/hr/uhad078
Lei, Y., Lu, L., Liu, H. Y., Li, S., Xing, F., and Chen, L. L. (2014). CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7, 1494–1496. doi: 10.1093/mp/ssu044
Li, N., Han, X., Feng, D., Yuan, D., and Huang, L.-J. (2019). Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: do we understand what they are whispering? Int. J. Mol. Sci. 20, 671. doi: 10.3390/ijms20030671
Li, X. and Heyer, W.-D. (2008). Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 18, 99–113. doi: 10.1038/cr.2008.1
Li, W., Huai, X., Li, P., Raza, A., Mubarik, M. S., Habib, M., et al. (2021). Genome-wide characterization of glutathione peroxidase (GPX) gene family in rapeseed (Brassica napus L.) revealed their role in multiple abiotic stress response and hormone signaling. Antioxidants 10, 1481. doi: 10.3390/antiox10091481
Li, P., Li, X., and Jiang, M. (2021a). CRISPR/Cas9-mediated mutagenesis of WRKY3 and WRKY4 function decreases salt and Me-JA stress tolerance in Arabidopsis thaliana. Mol. Biol. Rep. 48, 5821–5832. doi: 10.1007/S11033-021-06541-4/FIGURES/6
Li, X., Sandgrind, S., Moss, O., Guan, R., Ivarson, E., Wang, E. S., et al. (2021b). Efficient protoplast regeneration protocol and CRISPR/Cas9-mediated editing of glucosinolate transporter (GTR) genes in rapeseed (Brassica napus L.). Front. Plant Sci. 12, 680859. doi: 10.3389/fpls.2021.680859
Li, T. and Yang, B. (2013). TAL effector nuclease (TALEN) engineering. Enzyme Engineering: Methods Protoc. 978, 63–72. doi: 10.1007/978-1-62703-293-3_5
Li, J., Yu, X., Zhang, C., Li, N., and Zhao, J. (2022). The application of CRISPR/Cas technologies to Brassica crops: current progress and future perspectives. aBIOTECH 3, 146–161. doi: 10.1007/s42994-022-00076-3
Li, L., Zhang, D., Zhang, Z., and Zhang, B. (2025). CRISPR/Cas: a powerful tool for designing and improving oil crops. Trends Biotechnol. 43, 773–789. doi: 10.1016/j.tibtech.2024.09.007
Liu, H., Ding, Y., Zhou, Y., Jin, W., Xie, K., and Chen, L.-L. (2017). CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant 10, 530–532. doi: 10.1016/j.molp.2017.01.003
Liu, H. S., Li, F. M., and Xu, H. (2004). Deficiency of water can enhance root respiration rate of drought sensitive but not drought-tolerant spring wheat. Agric. Water Manage. 64, 41–48. doi: 10.1016/s0378-3774(03)00143-4
Liu, T., Liu, Z., Song, C., Hu, Y., Han, Z., She, J. I., et al. (2012). Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164. doi: 10.1126/science.1218867
Liu, F., Selin, C., Zou, Z., and Dilantha Fernando, W. G. (2020). LmCBP1, a secreted chitin-binding protein, is required for the pathogenicity of Leptosphaeria maculans on Brassica napus. Fungal Genet. Biol. 136, 103320. doi: 10.1016/j.fgb.2019.103320
Liu, Y., Xin, X., Li, P., Wang, W., Yu, Y., Zhao, X., et al. (2024). Editing of eIF (iso) 4E. c confers resistance against Turnip mosaic virus in Brassica rapa. Hortic. Plant J. 10, 1020–1034. doi: 10.1016/j.hpj.2024.05.001
Ma, X. (2020). Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat. Plants. 6, 773–779. doi: 10.1038/s41477-020-0704-5
Ma, H., Kong, C., Deng, S., Zhao, T., Ji, J., Wang, Y., et al. (2025). Resistance screening of cabbage to black rot and inheritance pattern analysis. Scientia Hortic. 345, 114129. doi: 10.1016/j.scienta.2025.114129
Ma, C., Liu, M., Li, Q., Si, J., Ren, X., and Song, H. (2019). Efficient BoPDS gene editing in cabbage by the CRISPR/Cas9 system. Hortic. Plant J. 5, 164–169. doi: 10.1016/j.hpj.2019.04.001
Ma, X. L., Zhang, Q. Y., Zhu, Q. L., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Mahas, A., Aman, R., and Mahfouz, M. (2019). CRISPR-Cas13d mediates robust RNA virus interference in plants. Genome Biol. 20, 263. doi: 10.1186/s13059-019-1881-2
May, D., Paldi, K., and Altpeter, F. (2023). Targeted mutagenesis with sequence-specific nucleases for accelerated improvement of polyploid crops: Progress, challenges, and prospects. Plant Genome 16, e20298. doi: 10.1002/tpg2.20298
McDonald, B. A. and Linde, C. (2002). Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40, 349–379. doi: 10.1146/annurev.phyto.40.120501.101443
Meena, P. D., Mehta, N., and Saharan, G. S. (2021). Minor pathogens: a worldwide challenge to cultivation of crucifers. Agric. Res. J. 58, 557–580. doi: 10.5958/2395-146x.2021.00081.8
Meng, J.-Y., Liang, G.-W., He, Y.-J., and Qian, W. (2020). QTL mapping of salt and drought tolerance related traits in Brassica napus L. Acta Agron. Sin. (China) 47, 462–471. doi: 10.3724/SP.J.1006.2021.04034
Meraj, T. A., Fu, J., Raza, M. A., Zhu, C., Shen, Q., Xu, D., et al. (2020). Transcriptional factors regulate plant stress responses through mediating secondary metabolism. Genes 11, 346. doi: 10.3390/genes11040346
Merlot, S., Leonhardt, N., Fenzi, F., Valon, C., Costa, M., Piette, L., et al. (2007). Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 26, 3216–3226. doi: 10.1038/sj.emboj.7601750
Muhammad Aslam, M., Waseem, M., Jakada, B. H., Okal, E. J., Lei, Z., Saqib, H. S. A., et al. (2022). Mechanisms of abscisic acid-mediated drought stress responses in plants. Int. J. Mol. Sci. 23, 1084. doi: 10.3390/ijms23031084
Müller Paul, H., Istanto, D. D., Heldenbrand, J., and Hudson, M. E. (2022). CROPSR: an automated platform for complex genome-wide CRISPR gRNA design and validation. BMC Bioinf. 23, 74. doi: 10.1186/s12859-022-04593-2
Murukarthick, J., Sampath, P., Lee, S. C., Choi, B.-S., Senthil, N., Liu, S., et al. (2014). BrassicaTED-a public database for utilization of miniature transposable elements in Brassica species. BMC Res. Notes 7, 1–11. doi: 10.1186/1756-0500-7-379
Muthusamy, M., Son, S., Park, S. R., and Lee, S. I. (2023). Heat shock factor binding protein BrHSBP1 regulates seed and pod development in Brassica rapa. Front. Plant Sci. 14, 1232736. doi: 10.3389/fpls.2023.1232736
Nadeem, M., Li, J., Yahya, M., Wang, M., Ali, A., Cheng, A., et al. (2019). Grain legumes and fear of salt stress: focus on mechanisms and management strategies. Int. J. Mol. Sci. 20, 799. doi: 10.3390/ijms20040799
Nawaz, G., Lee, K., Park, S. J., Kim, Y. O., and Kang, H. (2018). A chloroplast-targeted cabbage DEAD-box RNA helicase BrRH22 confers abiotic stress tolerance to transgenic Arabidopsis plants by affecting translation of chloroplast transcripts. Plant Physiol. Biochem. 127, 336–342. doi: 10.1016/j.plaphy.2018.04.007
Nellist, C. F., Ohshima, K., Ponz, F., and Walsh, J. A. (2022). Turnip mosaic virus, a virus for all seasons. Ann. Appl. Biol. 180, 312–327. doi: 10.1111/aab.12755
Nerkar, G., Devarumath, S., Purankar, M., Kumar, A., Valarmathi, R., Devarumath, R., et al. (2022). Advances in crop breeding through precision genome editing. Front. Genet. 13. doi: 10.3389/fgene.2022.880195
Ng, J. C. K. and Perry, K. L. (2004). Transmission of plant viruses by aphid vectors. Mol. Plant Pathol. 5, 505–511. doi: 10.1111/j.1364-3703.2004.00240.x
Ngou, B. P. M., Jones, J. D. G., and Ding, P. (2022). Plant immune networks. Trends Plant Sci. 27, 255–273. doi: 10.1016/j.tplants.2021.08.012
Nie, X., Wang, D., Pan, Y., Hua, Y., Lü, P., and Yang, Y. (2024). Discovery, classification and application of the CPISPR-Cas13 system. Technol. Health Care 32, 525–544. doi: 10.3233/THC-230258
Noman, M., Aysha, J., Ketehouli, T., Yang, J., Du, L., Wang, F., et al. (2021). Calmodulin binding transcription activators: An interplay between calcium signalling and plant stress tolerance. J. @ Plant Physiol. 256, 153327. doi: 10.1016/j.jplph.2020.153327
Nuñez-Muñoz, L., Vargas-Hernández, B., Hinojosa-Moya, J., Ruiz-Medrano, R., and Xoconostle-Cázares, B. (2021). Plant drought tolerance provided through genome editing of the trehalase gene. Plant Signal. Behav. 16, 1877005. doi: 10.1080/15592324.2021.1877005
OECD-FAO (2023). OECD-FAO agricultural outlook 2023-2032 (Paris: OECD Publishing). doi: 10.1787/08801ab7-en
Okade, H., Fujita, Y., Miyamoto, S., Tomoo, K., Muto, S., Miyoshi, H., et al. (2009). Turnip mosaic virus genome-linked protein VPg binds C-terminal region of cap-bound initiation factor 4E orthologue without exhibiting host cellular specificity. J. Biochem. 145, 299–307. doi: 10.1093/jb/mvn180
Ordon, J., Bressan, M., Kretschmer, C., Dall’Osto, L., Marillonnet, S., Bassi, R., et al. (2020). Optimized Cas9 expression systems for highly efficient Arabidopsis genome editing facilitate isolation of complex alleles in a single generation. Funct. Integr. Genomics 20, 151–162. doi: 10.1007/s10142-019-00665-4
Osakabe, Y., Watanabe, T., Sugano, S. S., Ueta, R., Ishihara, R., Shinozaki, K., et al. (2016). Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep. 6, 26685. doi: 10.1038/srep26685
Pageau, D., Lajeunesse, J., and Lafond, J. (2006). Impact of clubroot [Plasmodiophora brassicae] on the yield and quality of canola. Can. J. Plant Pathol. 28 , 137–143.
Palukaitis, P. and Kim, S. (2021). Resistance to turnip mosaic virus in the Family Brassicaceae. Plant Pathol. J. 37, 1–23. doi: 10.5423/PPJ.RW.09.2020.0178
Papikian, A., Liu, W., Gallego-Bartolomé, J., and Jacobsen, S. E. (2019). Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat. Commun. 10, 729. doi: 10.1038/s41467-019-08736-7
Park, J.-J., Dempewolf, E., Zhang, W., and Wang, Z.-Y. (2017a). RNA-guided transcriptional activation via CRISPR/dCas9 mimics overexpression phenotypes in Arabidopsis. PloS One 12, e0179410. doi: 10.1371/journal.pone.0179410
Park, J. J., Dempewolf, E., Zhang, W., and Wang, Z. Y. (2017b). RNA-guided transcriptional activation via CRISPR/dCas9 mimics overexpression phenotypes in Arabidopsis. PloS One 12, pe0179410. doi: 10.1371/journal.pone.0179410
Pavlović, I., Mlinarić, S., Tarkowská, D., Oklestkova, J., Novák, O., Lepeduš, H., et al. (2019). Early Brassica crops responses to salinity stress: A comparative analysis between chinese cabbage, white cabbage, and kale. Front. Plant Sci. 10. doi: 10.3389/FPLS.2019.00450/BIBTEX
Podsędek, A. (2007). Natural antioxidants and antioxidant capacity of Brassica vegeta bles: A review. LWT-Food Sci. Technol. 40, 1–11. doi: 10.1016/j.lwt.2005.07.023
Punja, Z. K. and Zhang, Y. Y. (1993). Plant chitinases and their roles in resistance to fungal diseases. J. Nematol. 25, 526–540.
Pyott, D. E., Sheehan, E., and Molnar, A. (2016). Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 17, 1276–1288. doi: 10.1111/mpp.12417
Qu, M., Havshøi, N. W., Huang, X., Shabala, L., Yu, M., Fuglsang, A. T., et al. (2024). Understanding the mechanistic basis of ameliorative effects of boron on salinity in barley (Hordeum vulgare). Environ. Exp. Bot. 220, 105690. doi: 10.1016/J.ENVEXPBOT.2024.105690
Rahman, H., Xu, Y.-P., Zhang, X.-R., and Cai, X.-Z. (2016). Brassica napus genome possesses extraordinary high number of CAMTA genes and CAMTA3 contributes to PAMP triggered immunity and resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 7, 581. doi: 10.3389/fpls.2016.00581
Rahman, M. U., Zulfiqar, S., Raza, M. A., Ahmad, N., and Zhang, B. (2022). Engineering abiotic stress tolerance in crop plants through CRISPR genome editing. Cells. 11, 3590. doi: 10.3390/cells11223590
Raybould, A. F., Maskell, L. C., Edwards, M.-L., Cooper, J. I., and Gray, A. J. (1999). The prevalence and spatial distribution of viruses in natural populations of Brassica oleracea. New Phytol. 141, 265–275. doi: 10.1046/j.1469-8137.1999.00339.x
Raza, A., Razzaq, A., Mehmood, S. S., Hussain, M. A., Wei, S., He, H., et al. (2021). Omics: The way forward to enhance abiotic stress tolerance in Brassica napus L. GM Crops Food 12, 251–281.
Ren, J., Petzoldt, R., and Dickson, M. H. (2001). Screening and identification of resistance to bacterial soft rot in Brassica rapa. Euphytica 118, 271–280. doi: 10.1023/A:1017522501229
Rizhsky, L., Liang, H., Shuman, J., Shulaev, V., Davletova, S., and Mittler, R. (2004). When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 134, 1683–1696. doi: 10.1104/pp.103.033431
Robertson, G., Burger, J., and Campa, M. (2022). CRISPR/Cas-based tools for the targeted control of plant viruses. Mol. Plant Pathol. 23, 1701–1718. doi: 10.1111/mpp.13252
Roca Paixão, J. F., Gillet, F.-X., Ribeiro, T. P., Bournaud, C., Lourenço-Tessutti, I. T., Noriega, D. D., et al. (2019). Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a histone acetyltransferase. Sci. Rep. 9, 8080. doi: 10.1038/s41598-019-44571-y
Romero, M. and Pérez, M. (2024). Optimizing Brassica oleracea L. breeding through somatic hybridization using cytoplasmic male sterility (CMS) Lines: from protoplast isolation to plantlet regeneration. Plants 13, 3247. doi: 10.3390/plants13223247
Rusholme, R. L., Higgins, E. E., Walsh, J. A., and Lydiate, D. J. (2007). Genetic control of broad-spectrum resistance to turnip mosaic virus in Brassica rapa (Chinese cabbage). J. Gen. Virol. 88, 3177–3186. doi: 10.1099/vir.0.83194-0
Saha, G., Park, J.-I., Ahmed, N. U., Kayum, M. A., Kang, K.-K., and Nou, I.-S. (2016). Characterization and expression profiling of MYB transcription factors against stresses and during male organ development in Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Physiol. Biochem. 104, 200–215. doi: 10.1016/j.plaphy.2016.03.021
Salehi-Lisar, S. Y. and Bakhshayeshan-Agdam, H. (2016). Drought stress in plants: Causes, consequences, and tolerance. Drought Stress tolerance Plants (Cham) 1, 1–16. doi: 10.1007/978-3-319-28899-4_1
Sanfaçon, H. (2015). Plant translation factors and virus resistance. Viruses 7, 3392–3419. doi: 10.3390/v7072778
Sato, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2024). Complex plant responses to drought and heat stress under climate change. Plant J. 117, 1873–1892. doi: 10.1111/tpj.16612
Schaart, J. G., van de Wiel, C. C. M., and Smulders, M. J. M. (2021). Genome editing of polyploid crops: prospects, achievements and bottlenecks. Transgenic Res. 30, 337–351. doi: 10.1007/s11248-021-00251-0
Secchi, M. A., Fernandez, J. A., Stamm, M. J., Durrett, T., Prasad, P. V. V., Messina, C. D., et al. (2023). Effects of heat and drought on canola (Brassica napus L.) yield, oil, and protein: A meta-analysis. Field Crops Res. 293, 108848. do. doi: 10.1016/j.fcr.2023.108848
Sehgal, A., Reddy, K. R., Walne, C. H., Barickman, T. C., Brazel, S., Chastain, D., et al. (2022). Climate stressors on growth, yield, and functional biochemistry of two Brassica species, kale and mustard. Life (Basel) 12. doi: 10.1038/srep26685
Shah, N., Anwar, S., Xu, J., Hou, Z., Salah, A., Khan, S., et al. (2018). The response of transgenic Brassica species to salt stress: a review. Biotechnol. Lett. 40, 1159–1165. doi: 10.1007/S10529-018-2570-Z/TABLES/1
Shahzad, B., Rehman, A., Tanveer, M., Wang, L., Park, S. K., and Ali, A. (2022). Salt stress in Brassica: Effects, tolerance mechanisms, and management. J. Plant Growth Regul. 41, 781–795. doi: 10.1007/s00344-021-10338-x
Shihong Gao, D., Zhu, X., and Lu, B. (2021). Development and application of sensitive, specific, and rapid CRISPR-Cas13-based diagnosis. J. Med. Virol. 93, 4198–4204. doi: 10.1002/jmv.26889
Shopan, J., Liu, C., Hu, Z., Zhang, M., and Yang, J. (2020). Identification of eukaryotic translation initiation factors and the temperature-dependent nature of Turnip mosaic virus epidemics in allopolyploid Brassica juncea. 3 Biotech. 10, 75. doi: 10.1007/s13205-020-2058-0
Shopan, J., Mou, H., Zhang, L., Zhang, C., Ma, W., Walsh, J. A., et al. (2017). Eukaryotic translation initiation factor 2B-beta (eIF2Bβ), a new class of plant virus resistance gene. Plant J. 90, 929–940. doi: 10.1111/tpj.13519
Soltabayeva, A., Dauletova, N., Serik, S., Sandybek, M., Omondi, J. O., Kurmanbayeva, A., et al. (2022). Receptor-like Kinases (LRR-RLKs) in response of plants to biotic and abiotic stresses. Plants (Basel) 11. doi: 10.3390/plants11192660
Soma, F., Takahashi, F., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2021). Cellular phosphorylation signaling and gene expression in drought stress responses: ABA-dependent and ABA-independent regulatory systems. Plants 10, 756. doi: 10.3390/plants10040756
Song, Z.-X., Chu, S.-J., Seo, E.-Y., Hu, W.-X., Lim, Y. P., Park, T.-S., et al. (2022). Construction of full-length infectious clones of turnip mosaic virus isolates infecting Perilla frutescens and genetic analysis of recently emerged strains in Korea. Arch. Virol. 167, 1089–1098. doi: 10.1007/s00705-021-05356-9
Spoel, S. H. and Dong, X. (2012). How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12, 89–100. doi: 10.1038/nri3141
Stajič, E. and Kunej, U. (2023). Optimization of cabbage (Brassica oleracea var. capitata L.) protoplast transformation for genome editing using CRISPR/Cas9. Front. Plant Sci. 14, 1245433. doi: 10.3389/fpls.2023.1245433
Stelmach-Wityk, K., Szymonik, K., Grzebelus, E., and Kiełkowska, A. (2024). Development of an optimized protocol for protoplast-to-plant regeneration of selected varieties of Brassica oleracea L. BMC Plant Biol. 24, 1279. doi: 10.1186/s12870-024-06005-4
Sun, Z., Li, S., Chen, W., Zhang, J., Zhang, L., Sun, W., et al. (2021). Plant dehydrins: expression, regulatory networks, and protective roles in plants challenged by abiotic stress. Int. J. Mol. Sci. 22, 12619. doi: 10.3390/ijms222312619
Sun, Q., Lin, L., Liu, D., Wu, D., Fang, Y., Wu, J., et al. (2018). CRISPR/Cas9-mediated multiplex genome editing of the BnWRKY11 and BnWRKY70 genes in Brassica napus L. Int. J. Mol. Sci. 19, ), 2716. doi: 10.3390/ijms19092716
Takikawa, Y. and Takahashi, F. (2014). Bacterial leaf spot and blight of crucifer plants (Brassicaceae) caused by Pseudomonas syringae pv. maculicola and P. cannabina pv. alisalensis. J. Gen. Plant Pathol. 80, 466–474. doi: 10.1007/s10327-014-0540-4
Talakayala, A., Ankanagari, S., and Garladinne, M. (2022). CRISPR-Cas genome editing system: a versatile tool for developing disease resistant crops. Plant Stress 3, 100056. doi: 10.1016/j.stress.2022.100056
Tan, Z., Han, X., Dai, C., Lu, S., He, H., Yao, X., et al. (2024). Functional genomics of Brassica napus: progress, challenges, and perspectives. J. Integr. Plant Biol. 66, 484–509. doi: 10.1111/jipb.13635
Thomazella, D. P. D. T., Seong, K., Mackelprang, R., Dahlbeck, D., Geng, Y., Gill, U. S., et al. (2021). Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. U. S. A. 118, e2026152118. doi: 10.1073/pnas.2026152118
Ton, L. (2024). Establishment of a CRISPR/CasRx system to modify Turnip mosaic virus (TuMV) resistance in Brassica napus. [dissertation]. [Perth, Western Australia]: The University of Western Australia.
Tsuda, K. and Katagiri, F. (2010). Comparing signalling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. doi: 10.1016/j.pbi.2010.04.006
Tu, M., Wang, R., Guo, W., Xu, S., Zhu, Y., Dong, J., et al. (2024). A CRISPR/Cas9-induced male-sterile line facilitating easy hybrid production in polyploid rapeseed (Brassica napus). Hortic. Res. 11, uhae139. doi: 10.1093/hr/uhae139
U, N (1935). Genome-analysis in Brassica with special reference to the experimental formation of Brassica napus and peculiar mode of fertilization. J. Jpn. Bot. 7, 389–452.
USDA (2023). European Union: oilseeds and products annual. Available online at: https://fas.usda.gov/data/european-union-oilseeds-and-products-annual-3 (Accessed October 20, 2024).
Van de Wouw, A. P., Marcroft, S. J., Ware, A., Lindbeck, K., Khangura, R., and Howlett, B. J. (2014). Breakdown of resistance to the fungal disease, blackleg, is averted in commercial canola (Brassica napus) crops in Australia. Field Crops Res. 166, 144–151. doi: 10.1016/j.fcr.2014.06.023
Varanda, C. M. R., Félix, M., d., R., Campos, M. D., Patanita, M., and Materatski, P. (2021). Plant Viruses: from targets to tools for CRISPR. Viruses 13, 141. doi: 10.3390/v13010141
Vicente, J. G. and Holub, E. B. (2013). Xanthomonas campestris pv. Campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Mol. Plant Pathol. 14, 2–18. doi: 10.1111/j.1364-3703.2012.00833.x
Villao-Uzho, L., Chávez-Navarrete, T., Pacheco-Coello, R., Sánchez-Timm, E., and Santos-Ordóñez, E. (2023). Plant promoters: their identification, characterization, and role in gene regulation. Genes 14, 1226. doi: 10.3390/genes14061226
Wada, N., Osakabe, K., and Osakabe, Y. (2022). Expanding the plant genome editing toolbox with recently developed CRISPR–Cas systems. Plant Physiol. 188, 1825–1837. doi: 10.1093/plphys/kiac027
Wahab, A., Abdi, G., Saleem, M. H., Ali, B., Ullah, S., Shah, W., et al. (2022). Plants’ physio-biochemical and phyto-hormonal responses to alleviate the adverse effects of drought stress: a comprehensive review. Plants (Basel) 11, 1620. doi: 10.3390/plants11131620
Walsh, H. M., Rönkä, A., and Walsh, J. A. (2023). Identification and genetic inheritance of a new source of broad-spectrum extreme resistance to turnip mosaic virus (TuMV) in Brassica rapa. Eur. J. Plant Pathol. 165, 693–699. doi: 10.1007/s10658-022-02634-3
Wang, Z., Liu, L., Cheng, C., Ren, Z., Xu, S., and Li, X. (2020). GAI functions in the plant response to dehydration stress in Arabidopsis thaliana. Int. J. Mol. Sci. 21,819. doi: 10.3390/ijms21030819
Wang, J., Mao, L., Li, Y., Lu, K., Qu, C., Tang, Z., et al. (2024). Natural variation in BnaA9.NF-YA7 contributes to drought tolerance in Brassica napus L. Nat. Commun. 15, 2082. doi: 10.1038/s41467-024-46271-2
Wang, Z. P., Xing, H. L., Dong, L., Zhang, H. Y., Han, C. Y., Wang, X. C., et al. (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Gen. Biol. 16, 144. doi: 10.1186/s13059-015-0715-0
Wang, X., Yan, B., Shi, M., Zhou, W., Zekria, D., Wang, H., et al. (2016). Overexpression of a Brassica campestris HSP70 in tobacco confers enhanced tolerance to heat stress. Protoplasma 253, 637–645. doi: 10.1007/s00709-015-0867-5
Wani, A. S., Ahmad, A., Hayat, S., and Fariduddin, Q. (2013). Salt-induced modulation in growth, photosynthesis and antioxidant system in two varieties of Brassica juncea. Saudi J. Biol. Sci. 20, 183–193. doi: 10.1016/j.sjbs.2013.01.006
Wei, Y. S., Javed, T., Liu, T. T., Ali, A., and Gao, S. J. (2025). Mechanisms of abscisic acid (ABA)-mediated plant defense responses: An updated review. Plant Stress 15, 100724. doi: 10.1016/j.stress.2024.100724
Wendel, J. F., Lisch, D., Hu, G., and Mason, A. S. (2018). The long and short of doubling down: polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr. Opin. Genet. Dev. 49, 1–7. doi: 10.1016/j.gde.2018.01.004
Wu, J., Cai, G., Tu, J., Li, L., Liu, S., Luo, X., et al. (2013). Identification of QTLs for resistance to Sclerotinia stem rot and BnaC. IGMT5. a as a candidate gene of the major resistant QTL SRC6 in Brassica napus. PloS One 8, e67740. doi: 10.3389/fpls.2020.00577
Wu, G., Fang, X., Yu, T., Chen, J., and Yan, F. (2024). Turnip mosaic virus pathogenesis and host resistance mechanisms in Brassica. Hortic. Plant J. 10, 947–960. doi: 10.1016/j.hpj.2024.03.001
Wu, J., Yan, G., Duan, Z., Wang, Z., Kang, C., Guo, L., et al. (2020). Roles of the Brassica napus DELLA protein BnaA6. RGA, in modulating drought tolerance by interacting with the ABA signaling component BnaA10. ABF2. Front. Plant Sci. 11, 577.
Xu, L., Lin, Z., Tao, Q., Liang, M., Zhao, G., Yin, X., et al. (2014). Multiple NUCLEAR FACTOR Y transcription factors respond to abiotic stress in Brassica napus L. PloS One 9, e111354. doi: 10.1371/journal.pone.0111354
Xu, Y., Zhan, C., and Huang, B. (2011). Heat shock proteins in association with heat tolerance in grasses. Int. J. Proteom. 2011, 529648. doi: 10.1155/2011/529648
Yang, J., Duan, G., Li, C., Liu, L., Han, G., Zhang, Y., et al. (2019). The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 10, 1349. doi: 10.3389/fpls.2019.01349
Yang, X., Lu, M., Wang, Y., Wang, Y., Liu, Z., and Chen, S. (2021). Response mechanism of plants to drought stress. Horticulturae 7, 50. doi: 10.3390/horticulturae7030050
Yang, C., Wang, E., and Liu, J. (2022). CERK1, more than a co-receptor in plant–microbe interactions. New Phytol. 234, 1606–1613. doi: 10.1111/nph.18074
Yang, H., Wu, J.-J., Tang, T., Liu, K.-D., and Dai, C. (2017). CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 7 7489. doi: 10.1038/s41598-017-07871-9
Yang, Y., Zhu, K., Li, H., Han, S., Meng, Q., Khan, S. U., et al. (2018). Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development. Plant Biotechnol. J. 16, 1322–1335. doi: 10.1111/pbi.12872
Yao, J., Bai, J., Liu, S., Fu, J., Zhang, Y., Luo, T., et al. (2022). Editing of a novel Cd uptake-related gene CUP1 contributes to reducing Cd accumulations in Arabidopsis thaliana and Brassica napus. Cells 11, 3888. doi: 10.3390/CELLS11233888/S1
Ye, W., Hossain, R., Pröbsting, M., Ali, A. A. M., Han, L., Miao, Y., et al. (2024). Knock-out of BnHva22c reduces the susceptibility of Brassica napus to infection with the fungal pathogen Verticillium longisporum. Crop J. 12, 503–514. doi: 10.1016/j.cj.2024.02.012
Yu, E., Fan, C., Yang, Q., Li, X., Wan, B., Dong, Y., et al. (2014). Identification of heat responsive genes in Brassica napus siliques at the seed-filling stage through transcriptional profiling. PloS One 9, e101914. doi: 10.1371/journal.pone.0101914
Yu, F., Zhang, X., Huang, Z., Chu, M., Song, T., Falk, K. C., et al. (2016). Identification of genome-wide variants and discovery of variants associated with Brassica rapa clubroot resistance gene Rcr1 through bulked segregant RNA sequencing. PloS One 11, e0153218.
Yu, X., Yu, J., Lu, Y., Li, W., Huo, G., Zhang, J., et al. (2024). An efficient and universal protoplast-based transient gene expression system for genome editing in Brassica crops. Hortic. Plant J. 10, 983–994. doi: 10.1016/j.hpj.2024.06.001
Zafar, S. A., Zaidi, S., Gaba, Y., Singla-Pareek, S. L., Dhankher, O. P., Li, X., et al. (2020). Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J. Exp. Bot. 71, 470–479. doi: 10.1093/jxb/erz476
Zahra, N., Hafeez, M. B., Kausar, A., Al Zeidi, M., Asekova, S., Siddique, K. H., et al. (2023). Plant photosynthetic responses under drought stress: Effects and management. J. Agron. Crop Sci. 209, 651–672. doi: 10.1111/jac.12652
Zaman, Q. U., Chu, W., Hao, M., Shi, Y., Sun, M., Sang, S.-F., et al. (2019). CRISPR/Cas9-mediated multiplex genome editing of JAGGED gene in Brassica napus L. Biomolecules 9, 725. doi: 10.3390/biom9110725
Zhan, X., Liu, W., Nie, B., Zhang, F., and Zhang, J. (2023). Cas13d-mediated multiplex RNA targeting confers a broad-spectrum resistance against RNA viruses in potato. Commun. Biol. 6, 855. doi: 10.1038/s42003-023-05205-2
Zhang, K., He, J., Liu, L., Xie, R., Qiu, L., Li, X., et al. (2020). A convenient, rapid and efficient method for establishing transgenic lines of Brassica napus. Plant Methods 16, 43. doi: 10.1186/s13007-020-00585-6
Zhang, Y., Huang, S., Wang, X., Liu, J., Guo, X., Mu, J., et al. (2018b). Defective APETALA2 genes lead to sepal modification in Brassica crops. Front. Plant Sci. 9, 367. doi: 10.3389/fpls.2018.00367
Zhang, X., Li, J., Cao, Y., Huang, J., and Duan, Q. (2023). Genome-wide identification and expression analysis under abiotic stress of BrAHL genes in Brassica rapa. Int. J. Mol. Sci. 24. doi: 10.3390/ijms241512447
Zhang, Y., Liu, J., Li, Y., Ma, H., Ji, J., Wang, Y., et al. (2025a). Generation of novel bpm6 and dmr6 mutants with broad-spectrum resistance using a modified CRISPR/Cas9 system in Brassica oleracea. J. Integr. Plant Biol. 67, 1214–1216. doi: 10.1111/jipb.13842
Zhang, K., Zhuo, C., Wang, Z., Liu, F., Wen, J., Yi, B., et al. (2021). BnaA03. WRKY28, interacting with BnaA09.VQ12, acts as a brake factor of activated BnWRKY33-mediated resistance outburst against Sclerotinia sclerotiorum in Brassica napus. Plant 11, 609. doi: 10.3390/plants11050609
Zhang, K., Liu, F., Wang, Z., Zhuo, C., Hu, K., Li, X., et al. (2022). Transcription factor WRKY28 curbs WRKY33-mediated resistance to Sclerotinia sclerotiorum in Brassica napus. Plant Physiol. 190, 2757–2774. doi: 10.1093/plphys/kiac439
Zhang, Y., Massel, K., Godwin, I. D., and Gao, C. (2018). Applications and potential of genome editing in crop improvement. Genome Biol. 19, 210. doi: 10.1186/s13059-018-1586-y
Zhang, L., Meng, S., Liu, Y., Han, F., Xu, T., Zhao, Z., et al. (2024). Advances in and perspectives on transgenic technology and CRISPR-Cas9 gene editing in broccoli. Genes 15, 668. doi: 10.3390/genes15060668
Zhang, K., Nie, L., Cheng, Q., Yin, Y., Chen, K., Qi, K, et al. (2019). Effective editing for lysophosphatidic acid acyltransferase 2/5 in allotetraploid rapeseed (Brassica napus L.) using CRISPR-Cas9 system. Biotechnol. Biofuels 12, 225. doi: 10.1186/s13068-019-1567-8
Zhang, Y., Wang, R., Luo, T., Fu, J., Yin, M., Wang, M., et al. (2025b). CRISPR-mediated BnaNRAMP1 homologous copies editing create a low Cd-accumulation oilseed rape germplasm with unaffected yield. J. Integr. Agric. 24, 1704–1717. doi: 10.1016/J.JIA.2024.05.016
Zhang, R., Zhang, C., Lyu, S., Wu, H., Yuan, M., Fang, Z., et al. (2022). BcTFIIIA Negatively regulates turnip mosaic virus infection through interaction with viral CP and VPg proteins in pak choi (Brassica campestris ssp. chinensis). Genes (Basel) 13. doi: 10.3390/genes13071209
Zhang, D., Zhang, Z., Unver, T., and Zhang, B. (2020). CRISPR/Cas: A powerful tool for gene function study and crop improvement. J. Adv. Res. 29, 207–221. doi: 10.1016/j.jare.2020.10.003
Zhang, T., Zheng, Q., Yi, X., An, H., Zhao, Y., Ma, S., et al. (2018a). Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol. J. 16, 1415–1423. doi: 10.1111/pbi.12881
Zhao, D., Xia, X., Su, J., Wei, M., Wu, Y., and Tao, J. (2019). Overexpression of herbaceous peony HSP70 confers high temperature tolerance. BMC Genome 20, 70. doi: 10.1186/s12864-019-5448-0
Zhao, C., Zhang, Y., Gao, L., Xie, M., Zhang, X., Zeng, L., et al. (2024). Genome editing of RECEPTOR-LIKE KINASE 902 confers resistance to necrotrophic fungal pathogens in Brassica napus without growth penalties. Plant Biotechnol. J. 22, 538–540. doi: 10.1111/pbi.14253
Zheng, X., Koopmann, B., Ulber, B., and von Tiedemann, A. (2020a). A global survey on diseases and pests in oilseed rape-current challenges and innovative strategies of control. Front. Agron. 2, 590908. doi: 10.3389/fagro.2020.590908
Zheng, M., Zhang, L., Tang, M., Liu, J., Liu, H., Yang, H., et al. (2020b). Knockout of two Bna MAX 1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotechnol. J. 18, 644–654.
Zhou, X., Zhong, T., Wu, M., Li, Q., Yu, W., Gan, L., et al. (2024). Multiomics analysis of a resistant European turnip ECD04 during clubroot infection reveals key hub genes underlying resistance mechanism. Front. Plant Sci. 15, 1396602. doi: 10.3389/fpls.2024.1396602
Keywords: Brassica, CRiSPR/Cas, genome editing, disease resistance, stress, transformation, resistance genes, plant breeding
Citation: Ton LB, Qayyum Z, Amas J, Thomas WJW, Edwards D, Batley J and Dolatabadian A (2025) Applications of CRISPR/Cas tools in improving stress tolerance in Brassica crops. Front. Plant Sci. 16:1616526. doi: 10.3389/fpls.2025.1616526
Received: 23 April 2025; Accepted: 14 July 2025;
Published: 02 September 2025.
Edited by:
Aytug Tuncel, University of Maryland, College Park, United StatesReviewed by:
Nobuya Koizuka, Tamagawa University, JapanKateřina Mácová, Central European Institute of Technology (CEITEC), Czechia
Copyright © 2025 Ton, Qayyum, Amas, Thomas, Edwards, Batley and Dolatabadian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aria Dolatabadian, YXJpYS5kb2xhdGFiYWRpYW5AdXdhLmVkdS5hdQ==
†ORCID: Linh Bao Ton, orcid.org/0000-0002-8094-927X
 Linh Bao Ton1†
Linh Bao Ton1† Zuhra Qayyum
Zuhra Qayyum Junrey Amas
Junrey Amas William J. W. Thomas
William J. W. Thomas David Edwards
David Edwards Jacqueline Batley
Jacqueline Batley Aria Dolatabadian
Aria Dolatabadian