- Department of Plant Pathology, University of Minnesota – Twin Cities, St. Paul, MN, United States
Sclerotinia sclerotiorum is a devastating fungal pathogen capable of causing substantial yield loss on a wide range of agronomically important crops worldwide. S. sclerotiorum’s impressive virulence across its broad host range is primarily due to the abundance of pathogenic strategies at its disposal. These pathogenic strategies include the use of organic acids, hydrolytic enzymes, and various effector molecules that work in concert during host attack. While plants have evolved sophisticated defense mechanisms, complete resistance to S. sclerotiorum remains elusive among the more than 400 known plant hosts. Among these hosts, soybean, canola, and sunflower are the most important oilseed crops severely affected by S. sclerotiorum infection, which can result in 94% crop loss in extreme cases. Current management strategies rely on chemical fungicides, crop rotations, and partially resistant varieties, albeit with varying levels of success. Despite extensive research on individual host-pathogen interactions, there is a notable gap in comparative studies exploring defense mechanisms across plant families. This review seeks to address this gap by providing an overview of known defense strategies against Sclerotinia stem rot (SSR) in soybean and canola, as well as head rot (SHR), mid-stalk rot (MSR), and basal stalk rot (BSR) in sunflower. By identifying commonalities and differences among distantly related hosts, this comparative analysis aims to deepen our understanding of key plant defense strategies against S. sclerotiorum, thereby highlighting areas requiring future research.
1 Introduction
Sclerotinia sclerotiorum is a hemibiotrophic, filamentous ascomycete that is a formidable plant pathogen worldwide. It is currently estimated to infect 425 plant species comprising 74 families, mainly within dicotyledonous plants, although a few monocotyledonous species are also affected (Derbyshire et al., 2022). S. sclerotiorum was first described by Libert in 1837 (Libert, 1837). In 1945, Whetzel defined the pathogen as the primary species of the genus Sclerotinia (Whetzel, 1945). This soil-borne fungal pathogen is named for its durable, melanized resting structures known as sclerotia, which germinate myceliogenically or carpogenically to form hyphae or fruiting bodies, respectively. Hyphae are hyaline, septate, and multinucleate and develop in a branched pattern (Maxwell et al., 1972; Willetts and Wong, 1980). The fruiting bodies of S. sclerotiorum are known as apothecia, which are cup-shaped and sit atop a stalk that rises from the germinated sclerotia (The Canola Council of Canada, 2020). The diseases caused by S. sclerotiorum are often referred to as white mold due to the production of cottony masses on infected tissues (Figure 1). White mold is most devastating in temperate regions throughout the world, although it has been reported on every continent except Antarctica (Seiler et al., 2017). White mold is a formidable pathogen of many crops and ranks globally among oilseed crops as one of the most significant, economically devastating fungal pathogens (Gulya et al., 2016; Roth et al., 2020; Zheng et al., 2020).
Three of its most economically important oilseed crop hosts are soybean, canola, and sunflower. Yield reductions among these hosts vary depending on environmental conditions and management practices. In severe cases, such as the one seen in Wisconsin in 2009, it was estimated that 10% of the soybeans grown that year were lost to white mold (Crop Protection Network, 2023). The National Sclerotinia Initiative reports that combined soybean, canola, and sunflower losses can be as high as $424 million in the United States in the year 2021. Individually, losses have totaled $300 million in soybean, $24 million in canola, and $100 million in sunflower (U.S. Department of Agriculture-Agricultural Research Service, 2022).
S. sclerotiorum’s devastating nature is the result of numerous compounding factors. The pathogen’s polyphagous diet and widespread distribution are likely due to the multitude of phytotoxic metabolites, hydrolytic enzymes, and effectors that it uses during attack of its host. S. sclerotiorum’s ability to infect such a wide range of hosts is not fully understood. However, current understanding regarding its broad host expansion suggests preadaptation of Sclerotinia spp., in which the species developed virulence factors capable of targeting highly conserved plant defense mechanisms (Derbyshire et al., 2022).
Although this pathogen has adopted many complex and robust virulence mechanisms, plant hosts have also evolved an array of defense strategies in this evolutionary arms race. Among these divergent hosts, many conserved and overlapping defense response pathways exist, including the production of polygalacturonase-inhibiting proteins (PGIPs), antimicrobial phytoalexins, and flavonoids. However, variation and divergence among these orthologous systems are widespread, and the evolution of clade or family-specific defenses is not uncommon. For example, glyceollins comprise a class of phytoalexins specific to the genus Glycine that have demonstrated inhibition of S. sclerotiorum growth in culture (Kim et al., 2010b; Lygin et al., 2010). Glucosinolates, a secondary metabolite produced strictly by crucifers, have also been shown to increase the defense responses of B. napus when challenged with SSR (Bednarek et al., 2009).
Conversely, in sunflower, S. sclerotiorum does not cause one disease. Instead, it can result in one of three different diseases depending on the location of infection. These diseases include basal stalk rot (BSR), mid-stalk rot (MSR), and head rot (SHR), demonstrating the pathogen’s ability to infect more than just stem tissues in this host. Understanding what makes all plant organs of sunflower susceptible while other hosts are able to prevent root infection requires comprehensive comparative studies of the diverse hosts’ physiological and molecular resistance and susceptibility mechanisms against S. sclerotiorum.
This review discusses past and recent progress in oilseed crop management against SSR pertaining to the many virulence mechanisms of S. sclerotiorum and the defense mechanisms employed by these three oilseed crop hosts. By exploring individual pathosystems and identifying the similarities and differences among host responses, we hope to motivate comprehensive comparative studies to better understand how to develop resistance against S. sclerotiorum.
2 Life cycle and pathogenic strategies of Sclerotinia sclerotiorum
S. sclerotiorum is commonly referred to as a necrotrophic pathogen as it spends the majority of its life cycle necrotizing host tissues. However, recent transcriptomics studies reveal a potentially more nuanced disease progression, demonstrating a marked transition in gene expression following infection and before pathogen-induced host cell death, characteristic of a hemibiotrophic lifestyle (Kabbage et al., 2013). S. sclerotiorum overwinters in the soil or within infected debris as either mycelium or sclerotia, which can survive up to 8 years (Adams and Ayers, 1979). The infection cycle for above-ground plant organs is largely similar for soybean and canola, as well as two of the three sunflower diseases, MSR and SHR. Conversely, the disease BSR in sunflower, which begins below the soil surface, exhibits a rather unique disease cycle (Figure 2).
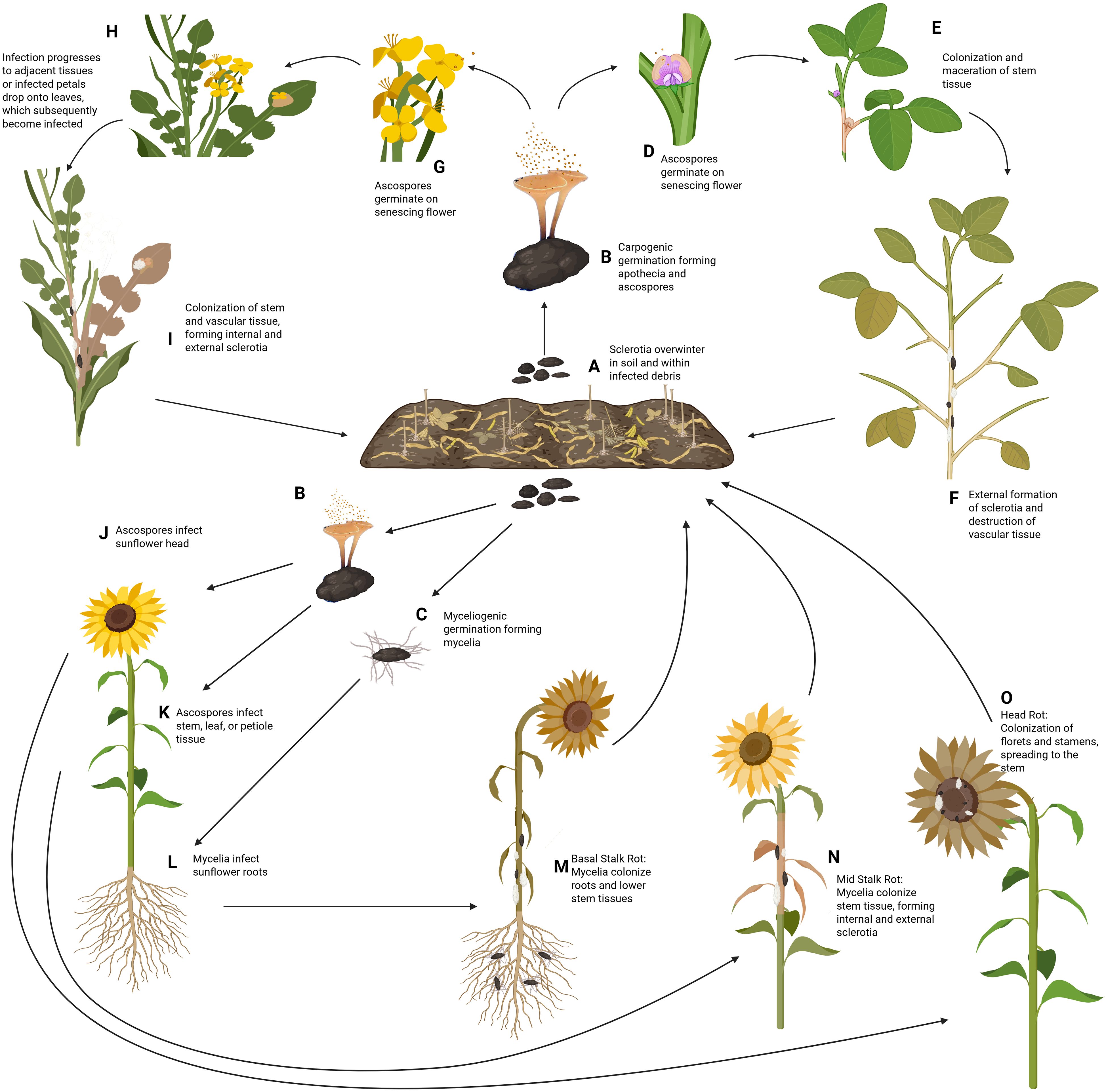
Figure 2. Disease Cycle of S. sclerotiorum on soybean, canola, and sunflower. (A) S. sclerotiorum survives in the soil and within infected debris as sclerotia, remaining viable for up to eight years. (B) Under conducive conditions, these sclerotia germinate carpogenically in the spring, to form a fruiting structure known as apothecia. Each apothecia can produce more than 10 million ascospores which are then wind disseminated to the host plant. (C) Alternatively, sclerotia can germinate myceliogenically, producing hyphae that will grow outward in the soil towards plant roots. In soybean (D), under natural conditions, the ascospores will land on senescing flowers, which release nutrients utilized by the germinating ascospore. Following germination, germ tubes form, and mycelia begin to colonize the plant tissues. (E) Mycelia will continue spreading from the point of infection, colonizing and macerating the stem tissues. (F) The water-soaked lesions will begin to dry out as the pathogen radiates from the point of infection, girdling the stem and replacing the pith with sclerotia, which will be deposited back into the soil for subsequent seasons. In canola (G), like soybean, the ascospores are deposited on senescing flowers during the rosette phase, where the released nutrients are utilized for hyphal formation. (H) Following infection of the flower, the pathogen can spread to the stem directly from the flower. Alternatively, the flower petals may detach before reaching the stem. However, if the infected petal lands on a leaf below, the mycelia will penetrate the leaf and spread to the remainder of the plant. (I) Further colonization of the stem results in a destruction of vascular tissue, death of the plant, and formation of sclerotia internally and externally, which can be deposited into the soil for the following year. In sunflower (J), ascospores can be deposited on the flower head, where they utilize nutrients from saccharose glands to germinate. (K) The ascospores can also be deposited and initiate infection on the stems, leaves, and petioles of the plant. (L) Alternatively, the sclerotia can directly produce hyphae via myceliogenic germination which can penetrate the sunflower roots, starting its colonization of the host from the base. (M) Infection of the roots results in the disease known as basal stalk rot, where the mycelia destroy the root system and work upwards along the stem. (N) Infection of the main stem results in the disease, mid stalk rot, which rapidly colonizes and destroys the vascular system, similar to what is seen in soybean and canola. (O) Finally, infection of the flower results in sclerotinia head rot of sunflower, where the flower tissues are destroyed, prior to colonization of the stem tissue. Sclerotia formed from all three diseases of sunflower will be deposited into the soil during harvest, where the sclerotia can overwinter and begin its cycle again the following year. Created with BioRender.com.
In the above-ground diseases, ascospores serve as the primary source of infection in the spring and early summer. Favorable conditions for the carpogenic germination of sclerotia include approximately 10 days of consistent moisture and temperatures ranging from 15-25°C. Apothecia will sprout from the germinated sclerotia following germination, where each apothecium can produce more than 10 million ascospores, for a period of two to three weeks (Agrios, 2005). The spores are then wind disseminated to the host plant, where their germination relies on readily available nutrient sources and moisture. In soybean and canola, successful infection will rely on the spore’s delivery to senescing plant organs, which release their nutrients during senescence. In HR, the sunflower petals have saccharose-producing glands on exterior cells, which is thought to be the main source of nutrients for the germinating ascospore, although senescing petals and pollen may also serve as food sources (Rodríguez et al., 2004). Finally, sunflower MSR is the only one of the above-ground diseases that does not directly involve a host flower. Here, the ascospores will be deposited on the stem or leaf tissue instead. In this case, the nutrients provided for the germination of the ascospore rely on senescing leaves or any pollen that may have fallen from the flower above (Martínez-Force et al., 2015). In all of these scenarios, the acquisition of nutrients by the ascospore provides it with the energy needed to develop an appressorium, which directly penetrates the host tissue allowing the mycelia to begin colonization of the host. The ascospores can also enter the host via natural openings such as stomata, hydathodes, and wounds, although these are less common (Rodríguez et al., 2004; Agrios, 2005; Wang et al., 2015; The Canola Council of Canada, 2020).
Following entrance into the host, the pathogen grows biotrophically and intercellularly for approximately 12–24 hours before undergoing a metabolic transition to a necrotrophic phase (Kabbage et al., 2013). Symptoms in soybean, canola, and sunflower MSR begin subtly with the formation of water-soaked lesions as the hyphae progresses into the stem, releasing numerous effectors and virulence factors into the host tissues. Water-soaked areas quickly transition to pale brown lesions of necrotic tissue that expand radially from the point of infection, eventually girdling the stem, destroying the vascular tissue, and replacing the pith with sclerotia. In these scenarios, the destruction of the vascular tissue will commonly result in wilting, lodging, and eventual death of the plant. In sunflower SHR, the pathogen will proliferate through the florets or the stamens of the flower and will spread to the stem of the plant. Colonization results in water-soaked lesions of the infected flower parts, eventually leading to necrosis and death of the infected tissues. While SHR doesn’t typically result in severe wilting or lodging, infection of the flower head can cause a significant reduction in yield, and large amounts of sclerotia can form in the receptacle of the flower (Bolton et al., 2006).
Alternative to the above-ground diseases is sunflower BSR, where the primary source of infection is mycelia. Basal stalk rot of sunflower is a rather unique disease for S. sclerotiorum as sunflower is one of the few hosts that can be colonized myceliogenically through the roots in a natural environment (Bolton et al., 2006). Myceliogenic germination of the sclerotia commonly occurs during periods of moderate temperatures and high relative humidity >80% for a minimum of 12 hours (Saharan and Mehta, 2008). Following germination, the mycelia will colonize the host root tissues via direct penetration of the cuticle, where the pathogen spreads from the roots to the base of the stem, macerating the tissue as it progresses. Maceration of the taproot and lateral roots results in eventual wilting and death of the host, and colonization of the stem base compromises the stem’s integrity, leading to lodging (Lumsden, 1979).
In all of these diseases, secondary infections can occur via mycelia when there is direct contact between healthy and infected plants. In above-ground diseases, dense cropping systems facilitate direct contact of healthy stems with mycelia growing externally on a nearby infected plant. Below ground, in BSR, contact between plant roots is the main cause of the pathogen spreading from one plant to another (Tourneau, 1979).
3 Key virulence strategies of S. sclerotiorum
During host invasion, S. sclerotiorum has ample strategies to evade host defenses during the early stages of colonization. This is followed by an onslaught of virulence factors, including hydrolytic enzymes, phytotoxic metabolites, and effector proteins (Figure 3). All of these factors contribute to the pathogen’s devastating nature (Table 1). We will discuss each of these virulence strategies of S. sclerotiorum below.
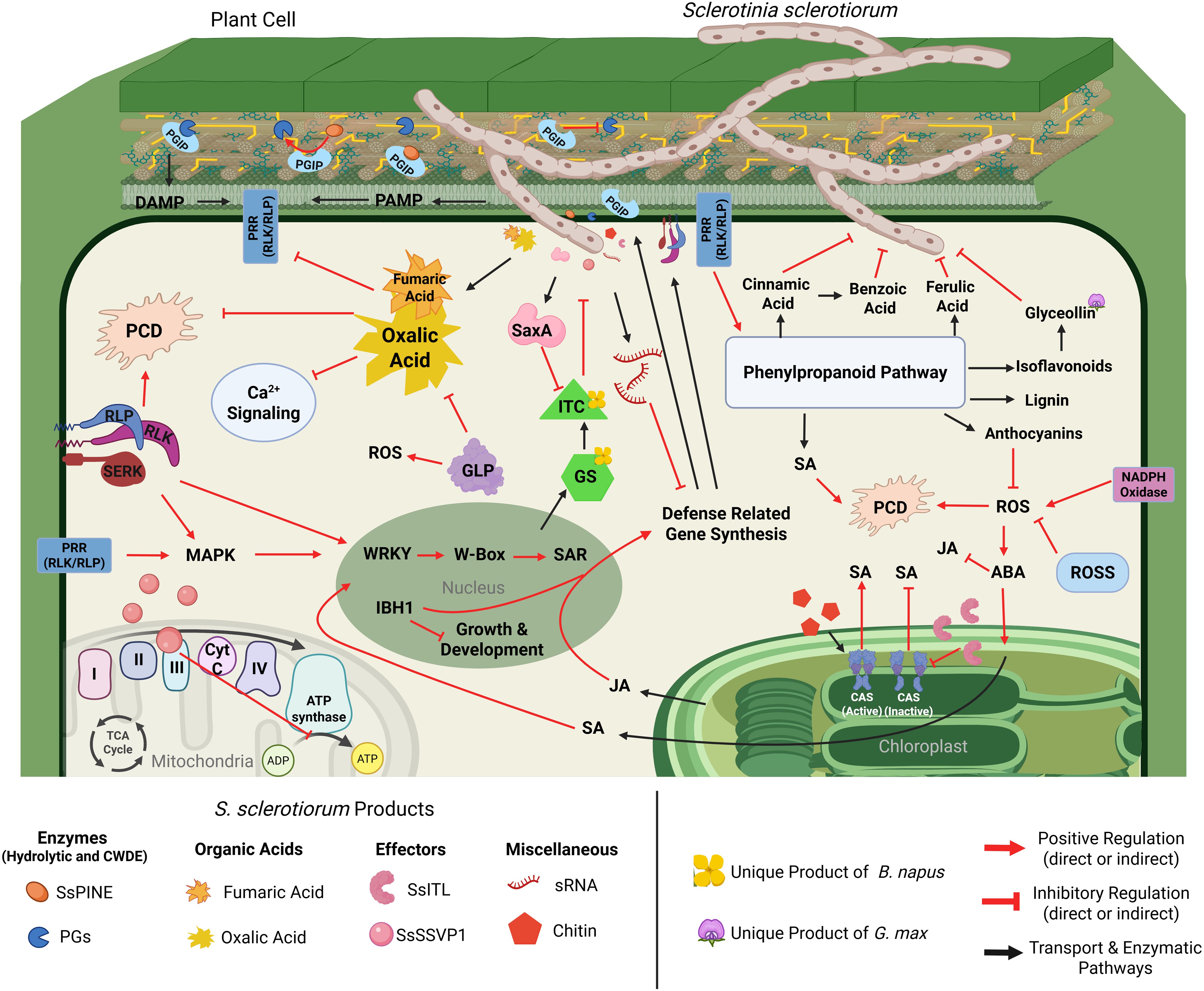
Figure 3. Cellular interaction model detailing S. sclerotiorum virulence mechanisms and corresponding defense responses of soybean, canola, and sunflower. Upon penetration of the host cell, S. sclerotiorum begins production of plant cell wall degrading enzymes (PCWDEs), including polygalacturonases (PGs), which are responsible for the degradation of the structural cell wall component pectin (yellow lines), resulting in decreased cell stability and production of damage-associated molecular patterns (DAMPs). DAMPs are recognized by host pattern recognition receptors (PRRs), which initiate MAPK signaling cascades that activate transcription factors (TFs) such as WRKYs and IBH1. Activation of TFs results in the synthesis of defense-related genes and hormone signaling. Additionally, certain TFs, such as IBH1, will produce downstream signals to halt the synthesis of genes related to growth and development S. sclerotiorum. Defense-related genes such as polygalacturonase inhibiting proteins (PGIPs) are synthesized and shuttled to the cell wall, where they will bind and deactivate PGs. However, S. sclerotiorum produces PGIP-inactivating effectors (SsPINE1), which possess a higher binding affinity to PGIP than PGs, resulting in the displacement of bound PGs, furthering the severity of cell wall degradation. Further interference of defense-related gene synthesis can be seen in the production of small RNAs (sRNA) by S. sclerotiorum. Some sRNAs produced by S. sclerotiorum are complementary to defense-related gene sequences and interfere with their production. Another major product of S. sclerotiorum includes oxalic acid and potentially other acids, such as fumaric acid, which inhibit several host defense mechanisms. Deposition of oxalic acid leads to acidification of the host cell, promoting Ca2+ chelation, resulting in the interference of Ca2+ signaling, dampening host immune responses, and weakening calcium pectate in the middle lamella, assisting in the hydrolysis of pectin via PGs. Furthermore, the suboptimal pH caused by oxalic acid interferes with host protein function and prevents programmed cell death (PCD), further impairing the host’s ability to defend itself. One defense strategy these hosts possess is the production of germin-like proteins (GLPs), which play many roles in the defense against S. sclerotiorum infection, but notably, facilitate the production of reactive oxygen species (ROS) and have predicted oxalate oxidase activities, which could help slow the acidification of the host cell. Host cell receptors also initiate defense responses via activation and reprogramming of the phenylpropanoid pathway. The phenylpropanoid pathway produces several defense-related compounds, including cinnamic acid, benzoic acid, and ferulic acid, which have inhibitory activity on S. sclerotiorum’s growth and development. Lignin produced from the phenylpropanoid pathway provides structural defense within the cell wall, slowing the incursion of the pathogen. Anthocyanins from the phenylpropanoid pathway have antioxidant properties, which degrade reactive oxygen species, slowing the rate of PCD. Glyceollins, a glycine-specific isoflavonoid (denoted by the soybean flower) are products of the phenylpropanoid pathway, which have been shown to play a vital role in defense against S. sclerotiorum, although the exact mechanism is unknown. Finally, the phenylpropanoid pathway is one of the metabolic pathways in plants that produce salicylic acid (SA), which is a critical defense signaling hormone. SA is also produced within the chloroplasts, where changes in calcium concentrations within the thylakoid membrane are detected via host calcium-sensing receptors (CAS). Additionally, CAS activation can occur via the detection of MAMPs such as chitin. CAS activation positively regulates the production of SA in the chloroplasts, improving host defenses. However, S. sclerotiorum produces an integrin-like protein (SsITL), which localizes inside of the chloroplast, binding to the CAS proteins and preventing their activation and subsequent mobilization of defense signaling. Further, hormone signaling control is modulated by abscisic acid (ABA), which is understood to be a modulator of plant hormone response. ABA signaling is induced by the production of ROS, where it shifts hormone signals away from growth and development while promoting the further production of SA and antagonizing JA signaling. Another strategy employed by S. sclerotiorum is the interruption of host cell energy metabolism via the small secreted virulence-like protein (SsSSVP1), which interacts with the QCR8 subunit of the cytochrome III complex, preventing subcellular localization of the complex and interrupting electron transport and transmembrane proton exchange, inhibiting the production of ATP. A defense-related response specific to plants within the order brassicales (denoted by the canola flower) is the production of glucosinolates (GS). Glucosinolates do not directly inhibit S. sclerotiorum; however, the breakdown of GSs results in the formation of isothiocyanates (ITC), which have been shown to inhibit mycelial growth and sclerotial development. In response to the production of ITCs, S. sclerotiorum synthesizes enzymes that hydrolyze ITCs (SsSaxA), detoxifying the antifungal metabolites. Created with BioRender.com.
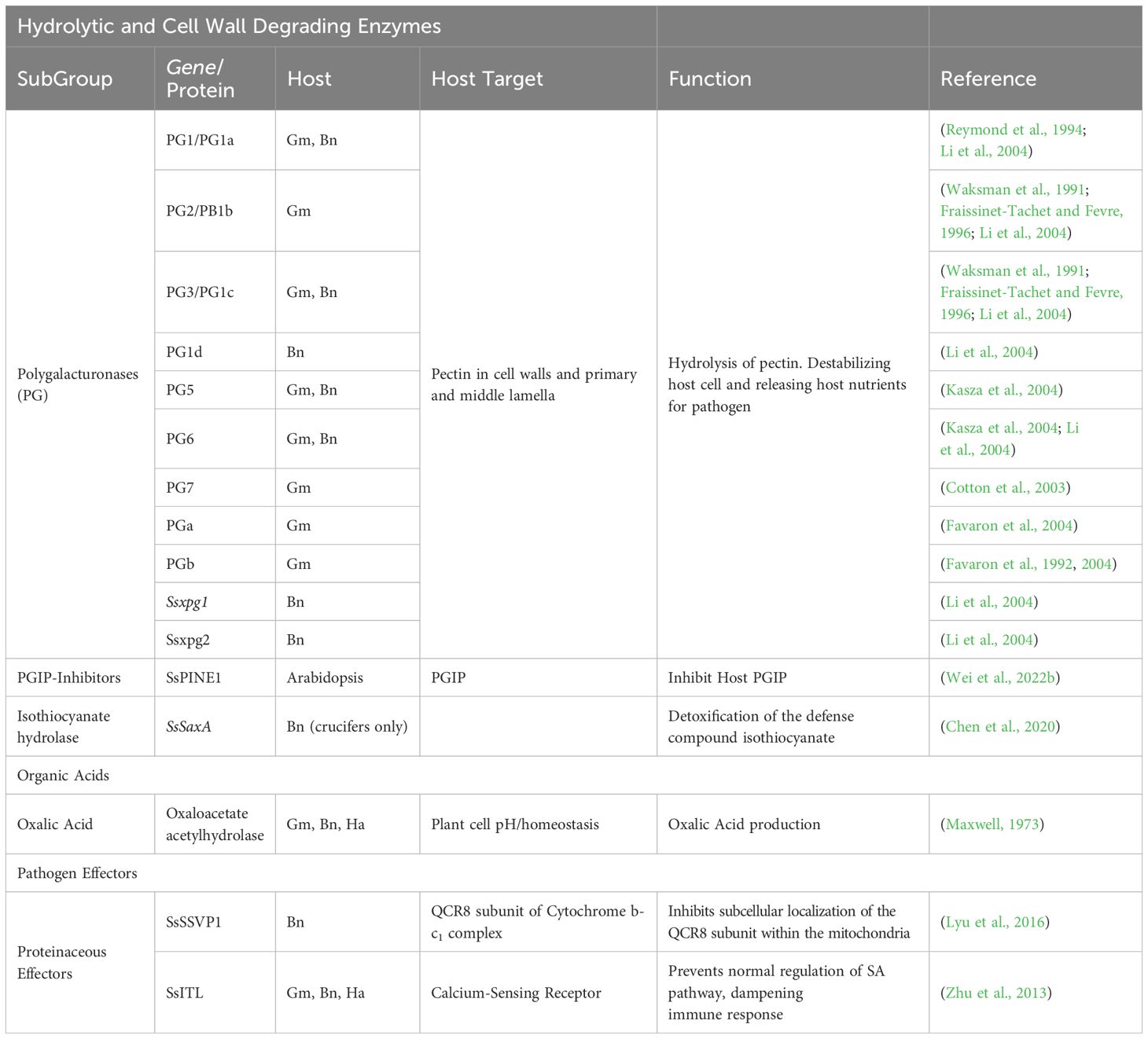
Table 1. Summary of known S. sclerotiorum virulence factors against the plant hosts, Soybean (Gm), Canola (Bn), and Sunflower (Ha).
3.1 Hydrolytic and cell wall degrading enzymes
As a predominantly necrotrophic pathogen, S. sclerotiorum possesses an array of plant cell wall degrading enzymes (PCWDEs). The largest class of these enzymes are the polygalacturonases (PGs), of which S. sclerotiorum has at least 16 (Bolton et al., 2006). PGs are responsible for the degradation of pectin, a critical structural component of plant cell walls, as well as the primary and middle lamella (Kasza et al., 2004). Hydrolysis of structural pectin within the plant cell leads to the decline of cell stability, resulting in the subsequent release of nutrients into the apoplastic environment, which suggests the importance of PGs in cell penetration and fungal proliferation, potentially in the early stages of infection before the necrotrophic phase (Kars and van Kan, 2007). Pectin is a heteropolysaccharide composed of linear chains of D-galacturonic acid residues with various attached side chains (Mohnen, 2008). Wei et al., 2020 identified four S. sclerotiorum D-galacturonic acid catabolizing enzymes, namely Ssgar1, Ssgar2, Sslgd1and Sslga1, and showed that Ssgar2, Sslgd1, and Sslga1 are essential for its virulence on soybean and peas (Wei et al., 2020). In the constant battle between plants and pathogens, plants have evolved a class of cell wall leucine-rich repeat proteins (LRR) known as polygalacturonase inhibiting proteins (PGIPs) (Lorenzo et al., 2001). PGIPs will be discussed in more detail in the plant defense sections of this review. However, PGIPs are effective against PGs in two key ways. First, PGIPs can directly bind to PGs, blocking their ability to degrade cell walls and thereby inhibiting fungal growth. Second, PGs trigger the production of oligosaccharides, which plants can recognize as damage-associated molecular patterns (DAMPs). These DAMPs, in turn, lead to increased production of PGIPs, further elevating the defense response. Incidentally, S. sclerotiorum has evolved an extracellular effector capable of inactivating plant defense systems via inhibition of PGIPs (Wei et al., 2022b). Wei and colleagues discovered an S. sclerotiorum PGIP-inactivating effector 1 (SsPINE1) that the pathogen utilizes to dampen host immune responses. They determined that SsPINE1 acts as a competitive inhibitor that is preferentially bound by PGIP, allowing it to bind and even replace PG from PGIP after the PG-PGIP complex has formed. The dissociation of PG from PGIP further increases the presence of PGs in the host and significantly contributes to virulence by enhancing PG activity (Wei et al., 2022b). Homologs of SsPINE have been identified in other necrotrophic fungal pathogens such as Botrytis cinerea, which suggests ancient evolutionary origins of the gene and sheds light on the timescale of these arms race interactions (Wei et al., 2022b). This impressive adaptation against a highly conserved plant defense mechanism once again demonstrates the sophistication of S. sclerotiorum while also lending another potential explanation for its cosmopolitan nature. Another key hydrolytic enzyme at S. sclerotiorum’s disposal is isothiocyanate hydrolase (ICTase), an enzyme thought to play a crucial role in the pathogen’s ability to protect itself during infection of plants in the order Brassicales. Isothiocyanates (ITC) are a class of defensive toxins unique to the Brassicales and are synthesized upon herbivory and pathogen attack via glucosinolate activation (Rask et al., 2000; Bednarek et al., 2009). There exists a relative wealth of knowledge on how herbivorous insects circumvent these toxins, including conjugation of hydrolyzed glucosinolate products and, in some cases, by employing symbiotic gut bacteria capable of detoxifying ITC as it is consumed, preventing ill effects in the insect (Ratzka et al., 2002; Wittstock et al., 2004, 2016; Jeschke et al., 2017). ITCs have also been shown to have toxic effects on fungal pathogens, although there is currently very little information regarding fungal detoxification of ITCs (Rahmanpour et al., 2014). Chen et al., 2020 explored the role of glucosinolates and ITCs in host defense against S. sclerotiorum as well as the pathogen’s ability to efficiently detoxify ITCs (Chen et al., 2020). They identified and characterized the gene responsible for ITC hydrolysis, naming it SsSaxA (S. sclerotiorum survival in Arabidopsis extracts) after the hydrolase. They determined from their work that SsSaxA can efficiently hydrolyze and detoxify both aliphatic and aromatic forms of ITC, allowing the pathogen to circumvent the antifungal metabolites produced during the plant defense reaction.
Additionally, they observed that ITC produced by the plants began accumulating as soon as 6 hours post inoculation (hpi) and reached a peak at 24hpi, followed by a substantial decrease at 48hpi, which they attributed to SsSaxA activity as the drop in ITC levels coincided with an increase in hydrolyzed ITC products within the leaves. Further work is needed to confirm the peak expression timing of this hydrolytic enzyme. However, this study reveals SsSaxA as a critical player during the later stages of the infection process (Chen et al., 2020).
3.2 Organic acids
The non-specific phytotoxin oxalic acid (OA) is arguably the most well-studied and commonly discussed metabolite of S. sclerotiorum. Oxalic acid’s contribution to S. sclerotiorum’s pathogenicity is a direct result of the acidification of the host cell (Dutton and Evans, 1996; Palmieri et al., 2019). Suboptimal pH within the plant cell promotes the chelation of Ca2+, which interferes with calcium signaling and chelates calcium pectate within the middle lamella, facilitating the hydrolysis of pectate via pathogen polygalacturonases (Magro et al., 1984; Dutton and Evans, 1996; Monazzah et al., 2018a). There exists a debate on whether OA should be considered a virulence factor or simply a modulator of host physiology (Callahan and Rowe, 1991). Regardless of exact classification, it is clear that the disruption of host cell homeostasis imparted by the release of OA results in the loss of a plant’s ability to defend itself effectively, increasing the pathogen’s virulence. The role of oxalic acid in the virulence of S. sclerotiorum has been covered extensively in the past, and detailed reviews have been completed by Xu et al. in the Annual Review of Phytopathology in 2018 and Gokul et al. in the Physiological and Molecular Plant Pathology Journal in 2024 (Xu et al., 2018; Daniel et al., 2024).
Continuing the investigation into host cell acidification and its impact on virulence, Xu et al., in 2015, produced mutants defective in oxaloacetate acetylhydrolase (OAH), the enzyme responsible for OA production in S. sclerotiorum (Xu et al., 2015)). Their study discovered that oah mutants lost their ability to accumulate OA during infection. However, contrary to their hypothesis that OA is S. sclerotiorum’s sole virulence factor, these mutants retained their ability to induce symptoms in Faba bean and pea but almost entirely lost their virulence in soybean. The proposed explanation for this difference was that soybeans, compared to the other hosts in this experiment, have the highest pH and buffering capacity, thus disallowing S. sclerotiorum from properly modulating its environment. This observed maintenance of virulence in specific hosts coincided with an accumulation of fumaric acid. They demonstrated that fumaric acid, a relatively weaker acid, accumulated in infected leaves to such an extent that the tissue pH reached nearly that of leaves infected by WT strains. This finding does not directly disprove the importance of OA; instead, it further demonstrates the pathogen’s aggressiveness and adaptability. While S. sclerotiorum clearly displays a preference for the production of OA in establishing a conducive environment for infection, it seems fully capable of producing fumaric acid as an alternate virulence factor. This adaptability could further explain this pathogen’s lack of host preference.
3.3 Proteinaceous effectors
In nature, pathogens secrete effectors, most often as proteins, to infect and survive in the host plants (Giraldo et al., 2013). These effectors are utilized by pathogens to manipulate host cellular processes to their advantage. The list of effector proteins in S. sclerotiorum’s arsenal is robust and ever-growing. A recent study that produced an updated version of the complete genome sequence of S. sclerotiorum reported a total of 70 putative effectors via EffectorP analysis (Derbyshire et al., 2017). Pathogen effectors have many functions throughout the disease cycle, ranging from evading host detection, dampening immune responses, and hijacking biochemical pathways for their benefit (Rovenich et al., 2014). A large majority of the proteinaceous effectors described for S. sclerotiorum are involved in disrupting immune signaling and induction of host cell death. One such effector protein described by Lyu et al. is the Small Secreted Virulence-like Protein 1 (SsSSVP1). SsSSVP1 was found to be highly expressed as soon as 3 hours post-inoculation (hpi) and reached its highest expression level at 12hpi, inducing rapid cell death following its translocation into the host cell (Lyu et al., 2016). SsSSVP1 interacts with a QCR8 subunit of the cytochrome b-c1 complex, inhibiting subcellular localization within the mitochondria. The authors suggest that small secretory protein has two distinct yet compounding effects during infection. The first, as a toxin, leads directly to rapid cell death. The second, as an impediment to plant energy metabolism, results in the gradual weakening of the cell and, eventually, cell death. Because this protein is expressed early in the infection process, it could potentially serve to exhaust the plant’s defense machinery before S. sclerotiorum releases its other effectors, increasing its virulence and resulting in a more rapid collapse of the plant upon its transition to necrotrophy.
Another major secretory protein employed by S. sclerotiorum is the integrin-like protein SsITL, first described by Zhu et al., 2013. While SsITL contains homology to that of other integrin proteins, it does not contain a transmembrane domain and was determined to function as a secretory protein that localizes within the host cells during infection. Expression analysis indicated that SsITL is highly expressed 90 minutes following infection and displayed peak expression levels at 3hpi. Initial findings demonstrated that SsITL suppressed host immune responses very early in the disease cycle via an interruption of jasmonic acid (JA) and ethylene (Et) signaling, prevention of salicylic acid (SA) accumulation, and interference of crosstalk between the two antagonistic pathways. However, the mechanism for this interference was unknown until 2020, when Tang et al. discovered that SsITL interacts with a calcium-sensing receptor (CAS), which functions as a positive regulator of SA signaling and, therefore, plant immune responses (Tang et al., 2020). The CAS receptor recognizes and responds to the presence of the microbe-associated molecular pattern (MAMP) chitin, rapidly promoting the expression of SA biosynthesis genes (Nomura et al., 2012). Rapid accumulation of SsITL during early infection allows S. sclerotiorum to immediately disarm the plant’s ability to sense and respond to the invasion. These examples show S. sclerotiorum possesses many robust strategies for suppressing host immune responses. Moreover, a common theme emerges, demonstrating that the first organism to react in the host-pathogen interaction will likely prevail.
3.4 Small RNAs
Small RNAs (sRNA) are RNA molecules ranging from 20 to 30 nucleotides in length and are non-coding (Dang et al., 2011). While sRNAs do not directly code for genes, they are known to play a crucial role in endogenous gene expression via RNA interference (RNAi). The binding of complementary sequences by sRNAs results in the silencing or repression of gene expression, which is crucial for growth and development, as well as response to the environment (Baulcombe, 2015). In addition to endogenous interactions, sRNAs secreted by fungal pathogens into host tissues can result in targeted host gene silencing, impeding host defense mechanisms and facilitating infection (Weiberg et al., 2013). In this sense, sRNAs can function similarly to pathogen-effector proteins. Yet more work needs to be done to discover sRNAs of S. sclerotiorum. In 2019, Derbyshire et al. conducted the first known study into the production of sRNA by S. sclerotiorum during plant infection (Derbyshire et al., 2019). This study revealed 374 sRNA sequences that were significantly upregulated during the infection of both Arabidopsis thaliana and Phaseolus vulgaris. Furthermore, the predicted targets of these sRNAs in A. thaliana were involved in host disease resistance. This was tested by silencing two of these predicted targets, SERK2 and SNAK2 in A. thaliana, resulting in increased susceptibility in the mutant lines. This work clearly demonstrates the need for further study into the utilization of sRNA by S. sclerotiorum, especially in other important oil seed hosts, to better understand and defend against the pathogenic mechanisms of S. sclerotiorum.
4 Defense mechanism of soybean to S. sclerotiorum
In soybean, S. sclerotiorum causes stem rot known as Sclerotinia stem rot (SSR). SSR consistently ranks among the top yield-limiting soybean diseases in the continental United States. In 2021, SSR ranked first as the most devastating fungal disease of soybean in the U.S.A., resulting in a loss of more than 25.7 million bushels, totaling nearly $335,000,000, and ranked as the second most important yield-limiting soybean disease (Crop Protection Network, 2023).
For an effective defense response, the most crucial time during soybean infection is within the first 12 hours following germination of the ascospore (Rothmann and McLaren, 2018). Partially resistant soybean varieties show significant and rapid changes in gene expression and metabolite accumulation following infection compared to susceptible varieties (Kabbage et al., 2013; Ranjan et al., 2019; Zhang et al., 2022b). This observation suggests that the timing of the response might be more impactful than the quantity or quality of the response, although that has yet to be directly studied. Furthermore, the transition of S. sclerotiorum’s ephemeral biotrophic phase to a necrotrophic lifestyle places even greater importance on a plant’s ability to recognize the threat, respond quickly, and shift rapidly from a biotrophic defense strategy to one that defends more effectively against necrotrophic pathogens. In studying this transition, Kabbage et al. reported that during the biotrophic phase, Arabidopsis plants initiated a hypersensitive (HR) response, typical of a biotrophic pathogen response, resulting in the initiation of programmed cell death (PCD) meant to quarantine the pathogen (Kabbage et al., 2013). However, upon transition to necrotrophy, the pathogen leverages this response to procure nutrients from these cells, fortifying its necrotizing abilities.
Studies conducted on quantitative trait loci (QTL) are a powerful tool in understanding pathogen-host interactions for uncovering the mechanisms behind disease resistance. Numerous QTL studies have been conducted concerning SSR and soybean interactions. In 2010, Li et al. identified three QTLs in the relatively tolerant soybean line Maple Arrow, associated with soluble pigment content and SSR resistance (Li et al., 2010). Reports prior to this study have mainly focused on QTLs that are related to disease escape mechanisms, including flowering date, canopy architecture, and maturity groups, leading Li et al. to focus on QTLs associated with resistance rather than escape mechanism phenotypes, which can be agronomically useful in various systems but do not directly relate to resistance response (Kim et al., 1999). The utility of identifying QTLs associated with soluble pigment content lies in secondary metabolite and anthocyanin production in response to pathogen attack (Li et al., 2010). A follow-up study exploring the soluble pigment content of soybean stems identified a QTL named Qswm13-1, located on chromosome 13, was found to explain 23.62% of stem pigment variation, which the authors believed to be related to candidate genes beneficial to resistance response (Zhao et al., 2015). Future studies should focus on the characterization of candidate genes associated with this marker for utilization in breeding programs and engineering for improved resistance.
While no completely resistant soybean varieties have been identified, these plants still have several defense strategies at their disposal when facing S. sclerotiorum infection (Table 2). These responses include the production of phytoalexins, intermediary metabolites, pathogenesis-related proteins (PRRs), reactive oxygen species (ROS) scavengers, and countless transcription factors (Figure 3). These plant defense strategies have been discussed below in detail.
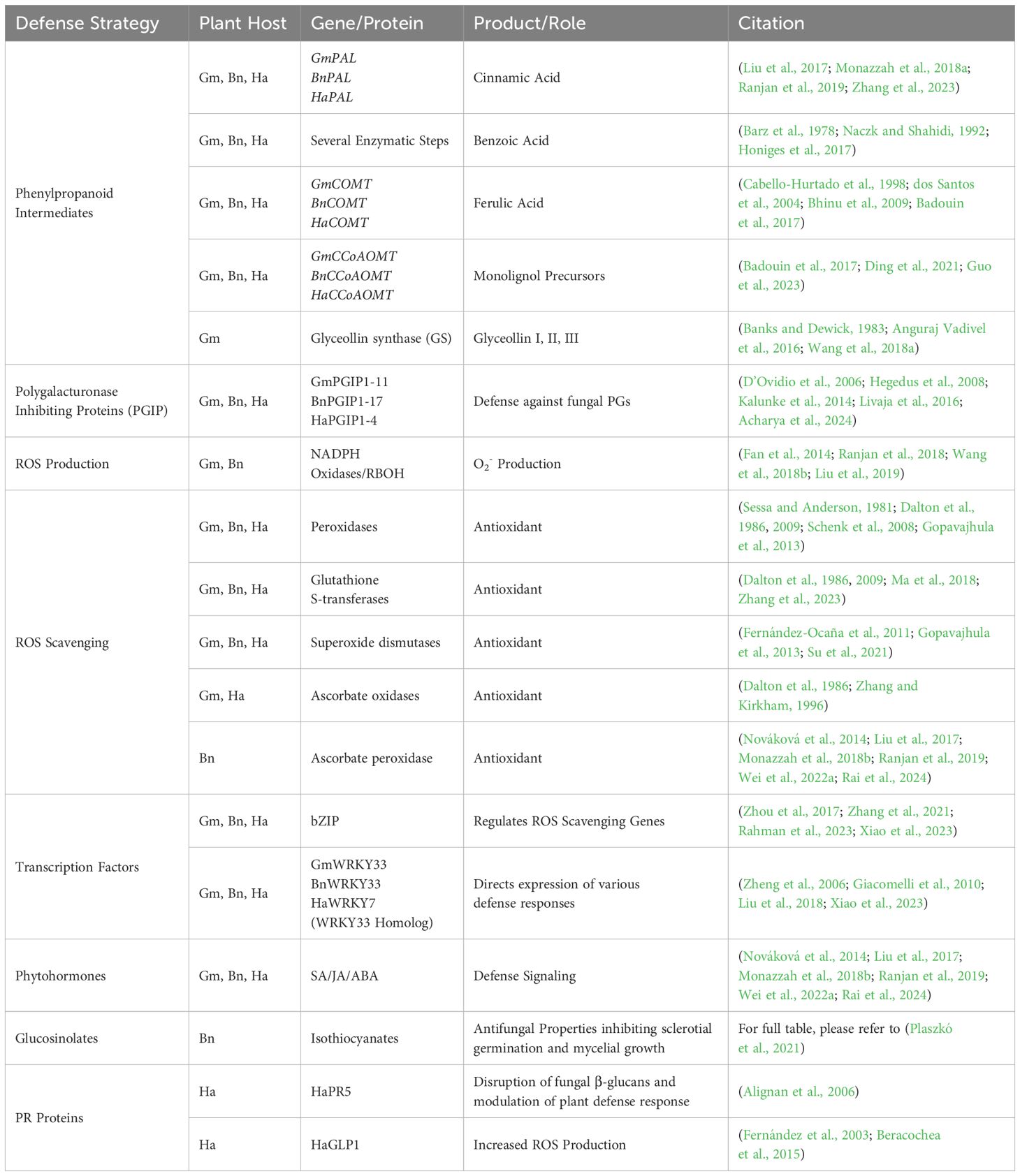
Table 2. Summary of the plant hosts soybean (Gm), canola (Bn), and Sunflower (Ha) defense strategies against Sclerotinia sclerotiorum.
4.1 Secondary metabolites (phenylpropanoid products and lignin intermediates)
Plant-derived secondary metabolites play a crucial role in defending against microbial pathogens. One of the major plant secondary metabolite biosynthesis pathways is the phenylpropanoid biosynthesis pathway. The phenylpropanoid biosynthesis pathway produces several key metabolites, such as flavonoids, phytoalexins, and lignin precursors, which have also shown promise in the search for improved pathogen resistance. In a transcriptomics and metabolomics study comparing resistant and susceptible responses to SSR infection, resistant lines produced increased levels of cinnamic acid, benzoic acid, and ferulic acid. In resistant lines, metabolite production nearly doubled for cinnamic acid as soon as 6hpi. In contrast, ferulic acid had doubled by 72 hpi, and benzoic acid displayed an impressive 6-fold increase at 72 hpi (Ranjan et al., 2019). Furthermore, all three intermediates inhibited S. sclerotiorum growth in amended agarose plates, demonstrating their antifungal properties (Ranjan et al., 2019; Wang et al., 2019). The timing and production of these antimicrobial intermediates are of great interest, as cinnamic acid is immediately upstream of benzoic acid in the phenylpropanoid biosynthesis pathway. However, their peak heights are 66 hours apart, potentially implicating cinnamic acid’s role during early infection against biotrophic pathogens, whereas, at 72 hpi, S. sclerotiorum’s necrotrophic machinery is in complete control (Ranjan et al., 2019).
In addition to these antimicrobial metabolites, soybeans produce many vital enzymes that facilitate defense responses. Of these, S-adenosyl-L-methionine-dependent methyltransferases superfamily proteins (SAM-Mtases), also known as caffeoyl coenzyme A O-methyltransferases (CCoAOMT), and calcium-binding proteins have all shown significant association with S. sclerotiorum disease response, via QTL analysis (Wang et al., 2015; Zhang et al., 2022b). The CCoAOMT gene family is positioned within the phenylpropanoid pathway and catalyzes the conversion of caffeoyl-CoA to feruloyl-CoA by adding an O-methoxyl group to the 3’ position of the aromatic ring (Vanholme et al., 2010). Feruloyl-CoA’s production directs the production of various aldehydes, monolignols, and eventually syringyl and guaiacyl lignin (Kim et al., 2010a); Ranjan et al., 2019). Lignin and aldehyde intermediates have shown great importance in plant structure and defense. However, the GmCCoAOMT gene family has not been fully characterized, and work in this area may shed light on novel strategies for improving SSR resistance in soybean. Phytoalexins are a broad class of toxic antimicrobial compounds produced by plants in response to pathogen infection. In legumes, isoflavonoids comprise the majority of phytoalexin products and are produced as secondary metabolites from the phenylpropanoid pathway. Among these isoflavonoids exists a class of Glycine-specific metabolites known as glyceollins, which are synthesized from the isoflavonoid branch of the phenylpropanoid pathway (Banks and Dewick, 1983; Lygin et al., 2010; Anguraj Vadivel et al., 2016). Glyceollin production was upregulated in response to a range of pathogens, demonstrating its relation to soybean defense response. Moreover, S. sclerotiorum and five other fungal pathogens of soybean all displayed growth inhibition when grown in cultures amended with glyceollin (Darvill and Albersheim, 1984; Sharma and Salunkhe, 1991; Lygin et al., 2010). However, in the same study, Lygin et al. found that S. sclerotiorum could metabolize glyceollin in culture, reporting that less than 2% of the glyceollin added to the plate remained once fully colonized, further exemplifying the devastating nature and impressive adaptations of this fungal pathogen. While the exact mode of inhibition is unknown, it is clear that glyceollins play a vital role in defense response. Therefore, further study of its specific function and mode of action could reveal critical insights into developing durable resistance.
4.2 Polygalacturonase-inhibiting proteins
Another group of key defense molecules produced by soybean is a group of membrane proteins known as polygalacturonase-inhibiting proteins (PGIPs) (Kalunke et al., 2015). Many fungal pathogens, including S. sclerotiorum, produce PGs along with other pectin-degrading enzymes during pathogenesis, contributing to their virulence and facilitating nutrient acquisition. To defend themselves, soybeans produce PGIPs, which are members of the leucine-rich repeat (LRRs) glycoproteins commonly found in the plant cell wall (Lorenzo et al., 2001). To date, 11 PGIP genes have been identified in G. max which are spread across four chromosomes. Four copies are located on chromosomes 5 and 8, two on chromosome 15 and one on chromosome 19 (Acharya et al., 2024). The multiple PGIPs found on chromosomes 5, 8, and 15 are likely due to the two gene duplication events (D’Ovidio et al., 2006; Kalunke et al., 2014; Acharya et al., 2024). Kalunke et al. expanded the list of GmPGIPs from four to six in 2014, where they performed sequencing and transcriptomics experiments to identify and characterize the expression changes of the six PGIPs in response to S. sclerotiorum infection. As a group of resistance-related proteins, it is unsurprising that they observed increased expression levels of the PGIP genes following infection. Moreover, they showed that the different genes showed peak expression increases at various times, demonstrating that they may function independently or at least distinctly against the variety of PGs used by the pathogen. Importantly, they observed that the expression level of GmPGIP5 steadily decreased from 8 hpi to 24 hpi, followed by a 1000-fold increase from 24 hpi to 48 hpi. In GmPGIP7, expression levels remained low yet steadily increased from 8 hpi to 24 hpi, with a 30-fold increase from 24 hpi to 48 hpi. While this gene family is considered to be crucial for resistance against necrotrophic fungal pathogens, specifically containing PGs, the authors report that by the time the expression levels rose, around the 48-hour mark, the plant tissues were already displaying necrosis and maceration by S. sclerotiorum, indicating that the response may have been too late to be fully effective (Kalunke et al., 2014). These results are consistent with the hemibiotrophic model of S. sclerotiorum’s lifestyle as well as the idea that response timing may be a more critical factor than the type of response or possession of proper machinery. While not explored in the aforementioned study, it is not hard to imagine that S. sclerotiorum evolved an array of effectors, which during early biotrophic stages of infection, signal to the plant that it is facing a biotrophic pathogen. This trojan horse-style strategy could explain the decrease in PGIP5 expression in the first 24 hours, followed by the rapid and drastic increase in PGIP5 and PGIP7 expression at 48 hours once the pathogen has transitioned to the necrotrophic stage. Moreover, once the pathogen begins producing and releasing its CWDEs, PGs, and effectors around 24 hpi, the plant clearly responds with a significant increase in defense gene expression; however, these results demonstrate that the response, while bountiful, was too late to confer any meaningful protection.
While this research on PGIP expression and gene duplication is important, no further studies have been conducted on the relationship between soybean defense against S. sclerotiorum and PGIPs, highlighting the lack of current knowledge in an area that is likely to hold a repository of crucial information about the improvement of resistance responses to SSR.
4.3 Reactive oxygen species
Reactive oxygen species (ROS) production is ubiquitous in the plant kingdom and is produced at relatively lower levels during normal cellular metabolism (Scandalios et al., 1997). While plants possess scavenging mechanisms to degrade these basal levels of ROS, they are known to be highly overproduced during a stress response, reaching toxic levels (Kotchoni and Gachomo, 2006; Mittler et al., 2011; Noctor et al., 2014; Xia et al., 2015; Sewelam et al., 2016). The toxic accumulation of ROS results in localized cell death, a typical plant response for quarantining and defending against biotrophic pathogens (Ranjan et al., 2018). However, programmed cell death via ROS bursts may facilitate infection of necrotrophic pathogens, such as S. sclerotiorum. ROS in plants can come from several different sources, but one of the major contributors includes the membrane-bound NADPH oxidases, which catalyze the conversion of O2 to O2- (Sagi and Fluhr, 2001). In 2018, Ranjan et al. identified and characterized four respiratory burst oxidase homologs of soybean (GmRBOH) that are significantly upregulated upon S. sclerotiorum infection, confirming their role in the infection process. Additionally, they found that virus-induced gene silencing (VIGS) of this group of GmRBOH genes resulted in increased resistance to the fungal pathogen (Ranjan et al., 2018). These results suggest that ROS production via NADPH oxidases may aggravate host infection by facilitating the necrotrophic phase of this fungal pathogen.
However, plants possess several mechanisms to prevent the overaccumulation of various oxidative stressors with the use of antioxidant scavenging molecules. The ability to modulate ROS accumulation via scavenging improves the plants’ ability to fine-tune its response to biotic and abiotic stresses that commonly trigger ROS bursts. In soybean, genes related to ROS scavenging include peroxidases, glutathione S-transferases (GST), ascorbate oxidases, and superoxide dismutases, to name a few (Sessa and Anderson, 1981; Dalton et al., 1986, 2009; Gopavajhula et al., 2013). Additionally, anthocyanins have long been recognized for their antioxidant capacity in plants. Ranjan et al. explored the differential expression of these genes in response to S. sclerotiorum infection in resistant and susceptible lines. They found that resistant soybean lines showed significantly different expression of these genes during defense response compared to susceptible plants. They identified five GSTs, five peroxidases, five ascorbate oxidases, and two superoxide dismutases that showed significant upregulation between 24-96hpi, demonstrating the ability of the resistant lines to efficiently respond and detoxify the ROS accumulation related to SSR infection. Regarding anthocyanins, they found five of seven anthocyanin-related genes to be upregulated following expression, importantly, most reaching their peak expression levels at 96hpi. This evidence provides another line of reasoning pointing to the hijacking of ROS accumulation by S. sclerotiorum at later stages of infection while demonstrating the resistant lines’ ability to recognize and respond to these accumulations. Further work should be completed to explore how to increase the antioxidant capacity of soybean and other hosts in defense against S. sclerotiorum attack (Ranjan et al., 2019).
4.4 Transcription factors
Transcription factors (TFs) are class of proteins regulating genes’ transcript levels (Latchman, 1997). Transcription factors related to plant-pathogen defense are activated in response to biotic stimuli, resulting in effective signal transduction and cellular communication within the plant (Schluttenhofer and Yuan, 2015; Zhang et al., 2018). In 2021, Zhang et al. characterized the soybean bZIP transcription factor GmbZIP15 in its role in S. sclerotiorum defense response. They found that upregulation of GmbZIP15 increased the resistance response to S. sclerotiorum in soybean by increasing the transcription of ROS scavenging genes, allowing the plant to better defend against RBOH responses to SSR. Furthermore, they confirmed that introducing phytohormones, such as SA, JA, and ethylene, induced the function of this bZIP TF (Zhang et al., 2021). In a separate study, Xiao et al. explored the role of GmSWEET genes in S. sclerotiorum response. SWEET transporters are known for their role in carbohydrate transport across cell membranes. These transport genes have been extensively studied in other plant-pathogen interactions, namely those between rice and Xanthomonas oryzae pv. oryzae; however, their role in soybean-Sclerotinia interactions had not previously been reported. They observed that gmsweet15 mutants had reduced lesion area and pathogen colonization compared to WT lines, which they explained to be the result of drastic transcriptional reprogramming due to the mutation. Specifically, they found that transcription factors such as WRKY33 and heat stress transcription factors (HSF5, HSF36) were positively correlated with the phenotype observed in gmsweet15 mutants. In contrast, bZIP and MYB transcription factors were negatively correlated with the mutant phenotype (Xiao et al., 2023). As we progress our knowledge of the many defense-related interactions between soybean and Sclerotinia, studies like these add to our list of potential quantitative factors related to disease resistance in this pathosystem while also highlighting the vast number of interactions that remain unexplored.
4.5 Phytohormones
Phytohormones facilitate a plant’s ability to respond to both biotic and abiotic changes to its environment (Bari and Jones, 2009; Denancé et al., 2013). Key phytohormones commonly associated with defense signaling in response to pathogens include, but are not limited to salicylic acid (SA), jasmonic acid (JA), and abscisic acid (ABA) (Jones and Dangl, 2006; Zheng et al., 2006; Bari and Jones, 2009). In a 2022 study, Wie et al. found that upon infection by S. sclerotiorum gene-ontology terms for JA were enriched in both resistant and susceptible genotypes as soon as 8 hours post infection (Wei et al., 2022a). In another study by Ranjan et al. in 2019, it was found that the phytohormones salicylic acid (SA), jasmonic acid (JA), and abscisic acid (ABA) were differentially utilized in defense response when comparing resistant and susceptible lines, further implicating the importance of hormone signaling in pathogen defense. In a metabolite analysis study conducted by Ranjan et al., they report several findings related to the quantity and timing of phytohormone response, looking at hormone production dynamics at 6, 12, 24, 48, and 72hpi. Beginning with SA, they observed that susceptible lines produced nearly a two-fold increase in SA production across all time points when compared to the resistant variety, which, in comparison, showed a steady decrease in SA production. When assessing JA production, the resistant and susceptible lines were indistinguishable until 48 and 72hpi, where the susceptible line shifted from around 200 pmoles/g of tissue to nearly 800 pmole/g, while the resistant line dropped to nearly zero by 72hpi. Finally, ABA showed a significant increase in production in the susceptible line at 12hpi; however, the resistant line began producing more ABA at both 48hpi and 72hpi. As an overall modulator of plant hormone response, it can be understood that the peak in ABA in the resistant line took control at the later time points, antagonizing and reducing the JA signal (Ranjan et al., 2019). Future research should focus on the precise mechanisms controlling the timing and quantity of phytohormone signaling to better improve the defense response reactions in the presence of pathogen attack.
5 Defense mechanisms of canola to S. sclerotiorum
Sclerotinia stem rot (SSR) of canola is often referred to as the most destructive threat to canola production in Canada, where disease prevalence of up to 79% has been reported (The Canola Council of Canada, 2020). B. napus has several defense mechanisms that can be used in defending against SSR, and some relatively tolerant canola varieties have been identified in wild relatives such as B. oleracea (Mei et al., 2015; Taylor et al., 2018). Still, as in other hosts, complete resistance has yet to be found. Of the defense mechanisms that canola can employ, it has a range of inducible phytoalexins, including flavonoids and terpenoids, which possess antimicrobial activity (Figure 3). Additionally, canola can produce key metabolites such as glucosinolates and brassinosteroids, which are unique to the brassica family (Table 2).
Other defense strategies of canola that are seldom discussed in soybean and sunflower include various avoidance strategies. When the ascospore begins colonization of the senescing flower, the flower may be shed before the pathogen reaches the stem. Additionally, if a lateral branch is infected, the plant may abscise the branch, successfully stopping the pathogen from reaching the main stem and avoiding the death of the whole plant. However, these detached flowers and stems may land on a leaf below them or a neighboring plant, which would, again, result in further infection (Wang et al., 2015). Understanding how canola can sense and respond to the oncoming invasion may provide useful insights into developing plants that have improved avoidance strategies.
5.1 Glucosinolates
Glucosinolates are a class of organic compounds derived from glucose and amino acid precursors. Glucosinolates do not possess direct inhibitory properties by themselves but instead are enzymatically altered within the plant to produce bioactive defense compounds (Halkier and Du, 1997; Wittstock et al., 2016). The most well-studied inhibitory product from glucosinolate breakdown is isothiocyanate (ITC), which was reviewed thoroughly by (Plaszkó et al., 2021). However, there are a few key points worth mentioning. These antimicrobial compounds have been shown to inhibit mycelial growth and sclerotial germination of S. sclerotiorum, demonstrating their importance in the defense response to SSR infection (Manici et al., 1997; Kurt et al., 2011; Dhingra et al., 2013; Sotelo et al., 2015). Although, Rahmanpour et al. found that repeated exposures to ITC lead to the pathogen developing significant tolerance to high doses of ITC, exemplifying the difficulties in combating this pathogen (Rahmanpour et al., 2009). However, glucosinolates possess significant variation in their side chain chemistry, resulting in a multitude of degradation products that can be produced in varying quantities (Plaszkó et al., 2021). These studies have focused on one or a few variations of ITCs individually, and further work should focus on investigating the many permutations of ITC combinations and concentrations, as it is unlikely that S. sclerotiorum could recognize and detoxify all of them simultaneously.
5.2 Polygalacturonase-inhibiting proteins
Similar to soybean, canola utilizes PGIPs to defend against pathogen attack by inactivating fungal PGs (Li et al., 2004; Hegedus et al., 2008). At the time of writing, 17 BnPGIPs have been identified, having differential responses to various biotic and abiotic stimuli (Hegedus et al., 2008; Wang et al., 2021). Given the range of stimuli PGIPs can respond to, Hegedus et al. sought to identify which BnPGIPs were induced during S. sclerotiorum infection. Their results indicated that the 16 BnPGIPs clustered into three groups that were named BnPGIP1, BnPGIP2, or intermediate, based on sequence similarity. Among these groups, they found that the BnPGIP2 group, consisting of BnPGIP2, 5, 6, 9, 12, and 16, as well as BnPGIP8 (intermediate), all showed increased expression levels following S. sclerotiorum infection (Hegedus et al., 2008). Given the range of BnPGIPs and SsPGs, Wang et al. assessed the interactions between BnPGIPs and SsPGs. Additionally, they performed functional studies assessing the expression levels and overexpression phenotypes of BnPGIP2, 5, and 10 in relation to S. sclerotiorum infection. First, they determined that BnPGIP2 and 10 were greatly upregulated in leaf tissues following S. sclerotiorum infection, while BnPGIP5 only showed slight increases in expression levels compared to non-infected tissues. Next, in assessing the disease responses of canola lines overexpressing each of the three PGIPs they observed that BnPGIP2-OE lines developed significantly smaller lesions on both stem and leaf tissues, confirming BnPGIP2’s role in an improved defense response. Perhaps unexpectedly, BnPGIP5-OE lines also had significant reductions in lesion size, where BnPGIP10-OE was not different from the wild type at any time point or tissue type. Finally, in determining which BnPGIPs interact with SsPGs they found that BnPGIP2 and BnPGIP5, but not BnPGIP10, directly inhibit the function of SsPG crude extracts, proving their importance in defense against S. sclerotiorum infection (Wang et al., 2021). These studies highlight the importance of performing functional characterization to determine the role and interactions of host and pathogen proteins and further exemplify the nuances that exist within these systems.
5.3 Mitogen-activated protein kinases
Mitogen-activated protein kinases (MAPKs) are well known in the plant kingdom for their role in innate plant immunity responses (Jones and Dangl, 2006). MAPK cascades are mediated by effector-triggered immunity (ETI), and pathogen-associated molecular pattern (PAMP) triggered immunity (PTI), where a cascade of phosphorylation events converts MAPKKK to MAPKK to MAPK, eventually leading to the phosphorylation and subsequent activation of defense-related transcription factors (Bigeard et al., 2015; Cui et al., 2015). While this immune response pathway is generally recognized, its role in B. napus response to S. sclerotiorum was poorly understood until relatively recently. In 2022, Zhang et al. identified and characterized the BnaA03.MKK5 and BnaA06.MPK3/BnaC03.MPK3, determining their roles in positive modulation in response to SSR. It was determined that MAPKK peaks at 12hpi, followed by a MAPK peak at 24hpi, confirming this phosphorylation cascade and its importance in plant-pathogen defense in this pathosystem. Additionally, they found that overexpression of BnaA03.MKK5 delayed lesion expansion compared to WT lines when infected with S. sclerotiorum (Zhang et al., 2022b).
5.4 Transcription factors
WRKY transcription factors are key players in many plant-microbe interactions that regulate the transcription levels of various defense and stress-related genes, allowing plants to respond appropriately to a wide range of elicitors. WRKYs are proteins that contain a DNA binding domain consisting of a conserved WRKYGQK motif at their N-terminus and a zinc-binding motif at their C-terminus. This N-terminus DNA binding domain interacts with W-box cis-element in the promoter region of targeted defense-related genes to activate or inhibit their transcription (Dong et al., 2024). The WRKY family includes several types of WRKY proteins, the largest group being transcriptional regulators, commonly associated with response to biotic and abiotic stimuli. Additionally, WRKYs facilitate the regulation of pathogenesis-related gene expression. Regulation of WRKY genes is controlled by phosphorylation events via MAPK cascades, protein-protein interaction, and protein degradation via the proteasome, which limits the duration of activation or repression of genes by transcription factors (Dong et al., 2024). JA and SA-dependent signal responses to pathogen attack require transcriptional reprogramming, which includes transcriptional factors such as WRKY. In Arabidopsis, WRKY factors act in a complex defense response network as both positive and negative regulators. At present, 79 WRKY genes have been found in Arabidopsis. Notably, AtWRKY33 functions as a positive regulator of resistance toward the necrotrophic fungi Alternaria brassicicola and Botrytis cinerea, and AtWRKY53 and AtWRKY70 both positively modulated SAR (Wang et al., 2006; Zheng et al., 2006). SA biosynthesis and SA-dependent defenses are regulated by WRKY TFs. BnWRKY expression was examined in response to S. sclerotiorum infection, and He et al. found that BnWRKY33 expression was increased 12-fold within one hour following treatment with a rapid alkalinization factor (RALF) associated with S. sclerotiorum, further demonstrating the role of WRKY transcription factors in response to attack by necrotrophic pathogens (He et al., 2016; Liu et al., 2018). However, when studying the interactions of BnWRKYs in response to S. sclerotiorum infection, Zhang et al. found that while BnWRKY33 expression spikes during early infection, BnaA03.WRKY28 responds by binding to the promoter of BnWRKY33, decreasing its expression, resulting in a shift away from defense and back towards growth and development. In this way, they determined that BnaA03.WRKY28 negatively regulates defense to S. sclerotiorum (Zhang et al., 2022a). Further studies need to be conducted to identify the precise regulation of TFs in response to S. sclerotiorum infection. Identification and characterization of AtWRKY53 and AtWRKY70 homologs in B. napus could provide insight into improving resistance against necrotrophic pathogens.
5.5 Phytohormones
Phytohormone signaling in plants drives growth and development as well as response to biotic and abiotic stressors (Bari and Jones, 2009; Denancé et al., 2013). As small signaling molecules, phytohormones direct the expression of countless genes within plants (Xia et al., 2015). There are many classes of plant hormones which are categorized by their chemical structures and role within the plant. Of the many plant hormones, salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) are commonly associated with defense signaling in plants, where SA is traditionally associated with response to biotrophic pathogens and JA/ET signaling are associated with necrotrophic pathogen response (Jones and Dangl, 2006). However, given the transient biotrophic phase of S. sclerotiorum, both pathways are likely crucial for improving resistance. A study conducted in 2014 measured the hormone levels within B. napus following infection by S. sclerotiorum, and they observed that infected plants produced significantly elevated levels of SA at 12, 24, and 48hpi. Further, they observed a slight decrease in JA production at 12hpi, followed by a significant increase at 24hpi (Nováková et al., 2014). These findings demonstrate that both pathways are induced following infection. To further explore the role of SA and JA in defense response, they pretreated plants with SA or JA derivatives to induce these hormone pathways prior to infection. Interestingly, contrary to the traditional roles of SA and JA, they observed that SA induction significantly improved the resistance response in detached leaf assays, where only a slight and non-significant decrease in infection was observed in the JA pretreatment group. The results from this study exemplify two key points: first, this demonstrates the importance of phytohormones in plant disease response, and second, phytohormones are capable of producing different reactions in different pathosystems and oversimplifications stating that SA and JA are only meaningful in biotrophic and necrotrophic responses, respectively, need to be reevaluated for each host-pathogen interaction being assessed.
6 Defense mechanisms of sunflower to S. sclerotiorum
In contrast to soybean and canola interactions with S. sclerotiorum, this pathogen causes three distinct diseases in sunflowers. S. sclerotiorum can infect the flower directly, resulting in head rot (SHR), the stem causing mid-stalk rot (MSR), or it can infect through the roots, resulting in basal stalk rot (BSR) (Gulya et al., 2016; Harveson et al., 2016). While S. sclerotiorum is not the greatest threat to crop loss in sunflower production, it is capable of causing up to 50% yield reductions in severely infected fields (Research: USDA ARS, 2022). Similar to soybean and canola, current strategies for combating these three diseases rely primarily on the use of fungicides, rotations, and avoidance. However, the prevalence of the disease in sunflower fields remains a significant problem as genetic resistance to HR, MSR, and BSR are thought to be physiologically distinct, and developing resistance to one disease is not likely to protect against the others in infected fields (Talukder et al., 2014).
Interestingly, several wild Helianthus relatives have been identified, each displaying variable tolerance levels to the three sunflower disease complexes. Still, none of the varieties display resistance to all three complexes, and differences in Helianthus ploidy levels hinder traditional resistance breeding.
6.1 Metabolites and intermediates
Plant metabolites and intermediates from the citric acid cycle (TCA) serve as the precursors for several key amino acids, cell wall molecules, and phytohormones (Scheideler et al., 2002; Rhoads et al., 2006; Zhu et al., 2008). The differential responses of plant metabolite production in response to pathogen attack can offer essential insights into plant response, as well as comparisons between susceptible and resistant responses. Peluffo et al. found key differences in resistant and susceptible sunflower varieties in their metabolite production in response to S. sclerotiorum infection. Their analysis found that a resistant line RHA801 produced significantly higher levels of trehalose, inositol, glycerate, citrate, isocitrate, and succinate compared to the susceptible line HA89. Interestingly, the susceptible line showed increased production of tyrosine and chlorogenate, which are associated with the production of phenolic compounds, auxins, and SA. In correspondence with these results, they also found a significant decrease (nearly 50%) in phenylalanine ammonia-lyase activity in RHA801 at 1-day post-inoculation compared to mock-inoculated plants, indicating a shift away from the phenylpropanoid biosynthesis pathway (Peluffo et al., 2010).
6.2 Transcription factors and non-coding RNAs
Using RNA-seq analysis, DEGs were identified in susceptible and moderately resistant lines. Some key DEGs identified in the tolerant lines were the transcription factor IBH1, while another was a non-coding RNA (Fass et al., 2020). In Arabidopsis, the IBH1 TF was determined to inhibit a different transcription factor, HBI1, which functions in the promotion of growth and inhibition of immune response (Fan et al., 2014). Therefore, it can be understood that the IBH1 TF facilitates the transition between growth and development and immune response, although more work needs to be done to confirm that these molecular mechanisms are equivalent across the different hosts. The ncRNAs were the most consistent DEG signal found in the study by Fass et al., 2020. Although this research did not test the role these ncRNAs play in disease response. However, it is well established that ncRNAs function as genetic regulators in addition to serving as precursors to sRNAs, microRNAs, and small-interfering RNAs, all of which have known defense response roles in other systems.
Further, they identified a hypoxia-responsive protein. This family of proteins is thought to be associated with group VII Ethylene response factors, which can be understood to be related to necrotrophic pathogen response signaling (Fass et al., 2020).
6.3 Pathogenesis-related proteins
Pathogenesis-related proteins are a well-known group of proteins that play a crucial role in plant defense and were first discovered in tobacco by Van Loon and Van Kammen in 1970 (van Loon, 1985). To date, 16 recognized families of PR proteins have been identified, grouped by their sequence similarity and enzymatic activities (Edreva and Kostoff, 2005). Proteins within the PR-5 group are believed to contribute to biotic defense by disrupting the pathogens’ plasma membrane permeability and are known to localize in various tissues, including roots and flowers (Sudisha et al., 2012; Musa-Khalifani et al., 2021). In a study comparing PR5 gene expression in susceptible and moderately resistant accessions of sunflowers, it was found that PR5 showed a 5-fold increase in expression in the resistant lines 3 hours after infection, implicating the role of PR5 in defense against S. sclerotiorum attack (Table 2) (Musa-Khalifani et al., 2021). While the utility of this PR protein is clear, there are still many more PRs of sunflower that require identification and functional analysis in the search for improved defense strategies against white mold. Another important group of PR proteins includes the germin-like proteins (GLPs). Germin-like proteins comprise a large class of water-soluble plant proteins that are known to play important roles in plant defense response to biotic and abiotic stressors (Zhang et al., 1995; Schweizer et al., 1999). To date, 16 HaGLPs have been suggested, based on EST motifs, although they have not all been confirmed and only two have been characterized for function (Beracochea et al., 2015). A sunflower GLP, HaGLP1 was first described in 2003 by Fernandez et al. in the search for resistance-related genes to Sclerotinia head rot (Fernández et al., 2003). The function of HaGLP1 was functionally studied in 2015 where Beracochea et al. transformed Arabidopsis thaliana with the HaGLP1 gene and challenged them with S. sclerotiorum. While both the transgenic and wild-type plants showed notable lesion expansion, the transgenic lines showed statistically improved disease response compared to the WT plants. It was further determined that the expression of HaGLP1 in A. thaliana resulted in increased production of O2- and H2O2, demonstrating its role in the generation of reactive oxygen species in the presence of S. sclerotiorum (Beracochea et al., 2015). Another GLP, HaGLP3, was reported in an association mapping study conducted by Filippi et al. in 2020. In this study, the marker for this gene was found to be associated with increased resistance response to Sclerotinia head rot, although functional studies were not conducted (Filippi et al., 2020). Future work should focus on the discovery of other GLPs in sunflower, along with functional studies to elucidate the mechanism by which HaGLPs modulate resistance responses.
6.4 Phytohormones of helianthus
Phytohormone signaling in plant defense response has been well characterized in many systems, where it is generally accepted that hormones such as salicylic acid (SA) are utilized against biotrophic pathogens to induce local immune responses and initiate systemic acquired resistance (Bari and Jones, 2009; Denancé et al., 2013). Other hormones such as jasmonic acid (JA) are also used in pathogen defense signaling, commonly suggested in the response to herbivory and necrotrophic pathogens, which result in transcriptional reprogramming and biosynthesis of defense-related secondary metabolites. Another important phytohormone includes abscisic acid (ABA). Abscisic acid is commonly discussed in the context of plant growth regulation, stomatal conductance, and response to abiotic stressors. However, it also plays a crucial role in pathogen defense, as ABA pathways are highly interconnected with SA and JA signaling pathways (Rai et al., 2024). In a study comparing the phytohormone responses of resistant and susceptible lines of sunflower following S. sclerotiorum infection, it was determined that following infection, ABA levels continuously increased in resistant lines, where susceptible lines showed a strong peak at 12 hpi, followed by a drop in ABA below that of the untreated controls. When observing SA production, the resistant line showed significant increases from the control at 24hpi, followed by a return to untreated levels. Interestingly, the SA levels in the susceptible line were decreased at nearly every time point and were significantly lower than the controls, demonstrating a positive correlation between SA and ABA (Liu et al., 2017). This data further demonstrates the importance of SA signaling during the early stages of infection. In assessing the role of JA in defense response, Monazzah et al. assessed the expression levels of the helianthus plant defensin gene (PDF1.2) in resistant and susceptible varieties following infection. PDF1.2 is considered a marker gene for the JA/ET pathway and can be used as a proxy for indirect measurement of JA production. Their results demonstrated that the partially resistant varieties had significantly increased expression levels of PDF1.2 at 48 and 72hpi, where the susceptible variety showed significant downregulation at those same time points. It can be understood that the ability of the resistant lines to properly initiate a JA/ET response confers a stronger defense response in this system (Monazzah et al., 2018b).
7 Conclusion
Taken together, individual plant families have developed both common and specific strategies to combat SSR, potentially holding a repository of undiscovered quantitative resistance genes. However, complete resistance to S. sclerotiorum is unknown in any of its hosts, driving the need for comparative studies of these important interactions. In this review, common defense strategies of these hosts were evaluated, demonstrating overlapping defense strategies such as the use of phenylpropanoid intermediates, PGIPs, and phytohormones. Additionally, host specific defense strategies were explored such as glyceollins and glucosinolates, found in soybean and canola, respectively. In contrast, sunflowers were not found to have any species/family specific defense strategies. In fact, they are unique in their susceptibility compared to the other two hosts, being vulnerable to three diseases on different tissue regions. A study conducted in 2022 aiming to elucidate the root susceptibility of sunflower, found that root infection by S. sclerotiorum is specific to plants of the Asteraceae family, while hosts outside of this family such as canola and dry bean were able to halt mycelial spread to the stem following root infection.
Author contributions
AR: Conceptualization, Resources, Investigation, Funding acquisition, Supervision, Writing – review & editing, Project administration, Writing – original draft. NT: Methodology, Investigation, Conceptualization, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Funding for this study was provided by USDA, Agricultural Research Service (USDA–ARS) through the National Sclerotinia Initiative, Agreement No. 58-3060-1-025. This fund supported Nick Talmo.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acharya, S., Troell, H. A., Billingsley, R. L., Lawrence, K. S., McKirgan, D. S., Alkharouf, N. W., et al. (2024). Glycine max polygalacturonase inhibiting protein 11 (GmPGIP11) functions in the root to suppress Heterodera glycines parasitism. Plant Physiol. Biochem. 213, 108755. doi: 10.1016/j.plaphy.2024.108755
Adams, P. B. and Ayers, W. A. (1979). Ecology of Sclerotinia species. Phytopathology 69, 896–899. Available at: https://www.apsnet.org/publications/phytopathology/backissues/Documents/1979Articles/Phyto69n08_896.pdf (Accessed February 11, 2024).
Agrios, G. N. (2005). Plant Pathology (Burlington MA, USA: Elsevier). Available at: https://play.google.com/store/books/details?id=CnzbgZgby60C (Accessed August 22, 2022).
Alignan, M., Hewezi, T., Petitprez, M., Dechamp-Guillaume, G., and Gentzbittel, L. (2006). A cDNA microarray approach to decipher sunflower (Helianthus annuus) responses to the necrotrophic fungus Phoma macdonaldii. New Phytol. 170, 523–536. doi: 10.1111/j.1469-8137.2006.01696.x
Anguraj Vadivel, A. K., Sukumaran, A., Li, X., and Dhaubhadel, S. (2016). Soybean isoflavonoids: role of GmMYB176 interactome and 14-3–3 proteins. Phytochem. Rev. 15, 391–403. doi: 10.1007/s11101-015-9431-3
Badouin, H., Gouzy, J., Grassa, C. J., Murat, F., Staton, S. E., Cottret, L., et al. (2017). The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546, 148–152. doi: 10.1038/nature22380
Banks, S. W. and Dewick, P. M. (1983). Biosynthesis of glyceollins I, II and III in soybean. Phytochemistry 22, 2729–2733. doi: 10.1016/s0031-9422(00)97682-9
Bari, R. and Jones, J. D. G. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Barz, W., Schlepphorst, R., Wilhelm, P., Kratzl, K., and Tengler, E. (1978). Metabolism of benzoic acids and phenols in cell suspension cultures of soybean and mung bean. Z. Naturforsch. C 33, 363–367. doi: 10.1515/znc-1978-5-610
Bednarek, P., Pislewska-Bednarek, M., Svatos, A., Schneider, B., Doubsky, J., Mansurova, M., et al. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106. doi: 10.1126/science.1163732
Beracochea, V. C., Almasia, N. I., Peluffo, L., Nahirñak, V., Hopp, E. H., Paniego, N., et al. (2015). Sunflower germin-like protein HaGLP1 promotes ROS accumulation and enhances protection against fungal pathogens in transgenic Arabidopsis thaliana. Plant Cell Rep. 34, 1717–1733. doi: 10.1007/s00299-015-1819-4
Bhinu, V.-S., Li, R., Huang, J., Kaminskyj, S., Sharpe, A., and Hannoufa, A. (2009). Perturbation of lignin biosynthesis pathway in Brassica napus (canola) plants using RNAi. Can. J. Plant Sci. 89, 441–453. doi: 10.4141/cjps08164
Bigeard, J., Colcombet, J., and Hirt, H. (2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8, 521–539. doi: 10.1016/j.molp.2014.12.022
Bolton, M. D., Thomma, B. P. H. J., and Nelson, B. D. (2006). Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16. doi: 10.1111/j.1364-3703.2005.00316.x
Cabello-Hurtado, F., Durst, F., Jorrín, J. V., and Werck-Reichhart, D. (1998). Coumarins in helianthus tuberosus: characterization, induced accumulation and biosynthesis. Phytochemistry 49, 1029–1036. doi: 10.1016/s0031-9422(97)01036-4
Callahan, F. E. and Rowe, D. E. (1991). Use of a host-pathogen interaction system to test whether oxalic acid is the sole pathogenic determinant in the exudate of Sclerotinia trifoliorum. Phytopathology 81, 1546–1550. Available at: https://www.apsnet.org/publications/phytopathology/backissues/Documents/1991Articles/Phyto81n12_1546.pdf (Accessed May 15, 2025).
Chen, J., Ullah, C., Reichelt, M., Beran, F., Yang, Z.-L., Gershenzon, J., et al. (2020). The phytopathogenic fungus Sclerotinia sclerotiorum detoxifies plant glucosinolate hydrolysis products via an isothiocyanate hydrolase. Nat. Commun. 11, 3090. doi: 10.1038/s41467-020-16921-2
Cotton, P., Kasza, Z., Bruel, C., Rascle, C., and Fèvre, M. (2003). Ambient pH controls the expression of endopolygalacturonase genes in the necrotrophic fungus Sclerotinia sclerotiorum. FEMS Microbiol. Lett. 227, 163–169. doi: 10.1016/s0378-1097(03)00582-2
Crop Protection Network (2023).Estimates of crop yield losses due to diseases and invertebrate pests: an online tool. Available online at: https://loss.cropprotectionnetwork.org/ (Accessed April 11, 2024).
Cui, H., Tsuda, K., and Parker, J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
D’Ovidio, R., Roberti, S., Di Giovanni, M., Capodicasa, C., Melaragni, M., Sella, L., et al. (2006). The characterization of the soybean polygalacturonase-inhibiting proteins (Pgip) gene family reveals that a single member is responsible for the activity detected in soybean tissues. Planta 224, 633–645. doi: 10.1007/s00425-006-0235-y
Dalton, D. A., Boniface, C., Turner, Z., Lindahl, A., Kim, H. J., Jelinek, L., et al. (2009). Physiological roles of glutathione s-transferases in soybean root nodules. Plant Physiol. 150, 521–530. doi: 10.1104/pp.109.136630
Dalton, D. A., Russell, S. A., Hanus, F. J., Pascoe, G. A., and Evans, H. J. (1986). Enzymatic reactions of ascorbate and glutathione that prevent peroxide damage in soybean root nodules. Proc. Natl. Acad. Sci. U.S.A. 83, 3811–3815. doi: 10.1073/pnas.83.11.3811
Dang, Y., Yang, Q., Xue, Z., and Liu, Y. (2011). RNA interference in fungi: pathways, functions, and applications. Eukaryot. Cell 10, 1148–1155. doi: 10.1128/EC.05109-11
Daniel, A. I., Basson, G., Keyster, M., Klein, A., and Gokul, A. (2024). Molecular mechanism of oxalic acid synthesis as virulence factor of Sclerotinia sclerotiorum. Physiol. Mol. Plant Pathol. 134, 102412. doi: 10.1016/j.pmpp.2024.102412
Darvill, A. G. and Albersheim, P. (1984). Phytoalexins and their elicitors-A defense against microbial infection in plants. Annu. Rev. Plant Physiol. 35, 243–275. doi: 10.1146/annurev.pp.35.060184.001331
Denancé, N., Sánchez-Vallet, A., Goffner, D., and Molina, A. (2013). Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00155
Derbyshire, M., Denton-Giles, M., Hegedus, D., Seifbarghy, S., Rollins, J., van Kan, J., et al. (2017). The complete genome sequence of the phytopathogenic fungus Sclerotinia sclerotiorum reveals insights into the genome architecture of broad host range pathogens. Genome Biol. Evol. 9, 593–618. doi: 10.1093/gbe/evx030
Derbyshire, M., Mbengue, M., Barascud, M., Navaud, O., and Raffaele, S. (2019). Small RNAs from the plant pathogenic fungus Sclerotinia sclerotiorum highlight host candidate genes associated with quantitative disease resistance. Mol. Plant Pathol. 20, 1279–1297. doi: 10.1111/mpp.12841
Derbyshire, M. C., Newman, T. E., Khentry, Y., and Owolabi Taiwo, A. (2022). The evolutionary and molecular features of the broad-host-range plant pathogen Sclerotinia sclerotiorum. Mol. Plant Pathol. 23, 1075–1090. doi: 10.1111/mpp.13221
Dhingra, O. D., Schurt, D. A., Oliveira, R. D. L., and Rodrigues, F. A. (2013). Potential of soil fumigation with mustard essential oil to substitute biofumigation by cruciferous plant species. Trop. Plant Pathol. 38, 337–342. doi: 10.1590/S1982-56762013005000014
Ding, Y., Yu, S., Wang, J., Li, M., Qu, C., Li, J., et al. (2021). Comparative transcriptomic analysis of seed coats with high and low lignin contents reveals lignin and flavonoid biosynthesis in Brassica napus. BMC Plant Biol. 21, 246. doi: 10.1186/s12870-021-03030-5
Dong, X., Yu, L., Zhang, Q., Yang, J., Gong, Z., Niu, X., et al. (2024). Structural basis for the regulation of plant transcription factor WRKY33 by the VQ protein SIB1. Commun. Biol. 7, 561. doi: 10.1038/s42003-024-06258-7
dos Santos, W. D., Ferrarese, M., de, L. L., Finger, A., Teixeira, A. C. N., and Ferrarese-Filho, O. (2004). Lignification and related enzymes in Glycine max root growth-inhibition by ferulic acid. J. Chem. Ecol. 30, 1203–1212. doi: 10.1023/b:joec.0000030272.83794.f0
Dutton, M. V. and Evans, C. S. (1996). Oxalate production by fungi: its role in pathogenicity and ecology in the soil environment. Can. J. Microbiol. 42, 881–895. doi: 10.1139/m96-114
Edreva, A. and Kostoff, D. (2005). Pathogenesis-related proteins: Research progress in the last 15 years. Gen. Appl. Plant Physiol. 31, 105–124. Available at: http://www.bio21.bas.bg/ipp/gapbfiles/v-31/05_1-2_105-124.pdf?origin (Accessed April 23, 2024).
Fan, M., Bai, M.-Y., Kim, J.-G., Wang, T., Oh, E., Chen, L., et al. (2014). The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern–triggered immunity in arabidopsis. Plant Cell 26, 828–841. doi: 10.1105/tpc.113.121111
Fass, M. I., Rivarola, M., Ehrenbolger, G. F., Maringolo, C. A., Montecchia, J. F., Quiroz, F., et al. (2020). Exploring sunflower responses to Sclerotinia head rot at early stages of infection using RNA-seq analysis. Sci. Rep. 10, 13347. doi: 10.1038/s41598-020-70315-4
Favaron, F., Alghisi, P., and Marciano, P. (1992). Characterization of two Sclerotinia sclerotiorum polygalacturonases with different abilities to elicit glyceollin in soybean. Plant Sci. 83, 7–13. doi: 10.1016/0168-9452(92)90056-r
Favaron, F., Sella, L., and D’Ovidio, R. (2004). Relationships among endo-polygalacturonase, oxalate, pH, and plant polygalacturonase-inhibiting protein (PGIP) in the interaction between Sclerotinia sclerotiorum and soybean. Mol. Plant Microbe Interact. 17, 1402–1409. doi: 10.1094/MPMI.2004.17.12.1402
Fernández, P., Paniego, N., Lew, S., Hopp, H. E., and Heinz, R. A. (2003). Differential representation of sunflower ESTs in enriched organ-specific cDNA libraries in a small scale sequencing project. BMC Genomics 4, 40. doi: 10.1186/1471-2164-4-40
Fernández-Ocaña, A., Chaki, M., Luque, F., Gómez-Rodríguez, M. V., Carreras, A., Valderrama, R., et al. (2011). Functional analysis of superoxide dismutases (SODs) in sunflower under biotic and abiotic stress conditions. Identification of two new genes of mitochondrial Mn-SOD. J. Plant Physiol. 168, 1303–1308. doi: 10.1016/j.jplph.2011.01.020
Filippi, C. V., Zubrzycki, J. E., Di Rienzo, J. A., Quiroz, F. J., Puebla, A. F., Alvarez, D., et al. (2020). Unveiling the genetic basis of Sclerotinia head rot resistance in sunflower. BMC Plant Biol. 20, 322. doi: 10.1186/s12870-020-02529-7
Fraissinet-Tachet, L. and Fevre, M. (1996). Regulation by galacturonic acid of pectinolytic enzyme production by Sclerotinia sclerotiorum. Curr. Microbiol. 33, 49–53. doi: 10.1007/s002849900073
Giacomelli, J. I., Ribichich, K. F., Dezar, C. A., and Chan, R. L. (2010). Expression analyses indicate the involvement of sunflower WRKY transcription factors in stress responses, and phylogenetic reconstructions reveal the existence of a novel clade in the Asteraceae. Plant Sci. 178, 398–410. doi: 10.1016/j.plantsci.2010.02.008
Giraldo, M. C., Dagdas, Y. F., Gupta, Y. K., Mentlak, T. A., Yi, M., Martinez-Rocha, A. L., et al. (2013). Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat. Commun. 4, 1996. doi: 10.1038/ncomms2996
Gopavajhula, V. R., Chaitanya, K. V., Akbar Ali Khan, P., Shaik, J. P., Reddy, P. N., and Alanazi, M. (2013). Modeling and analysis of soybean (Glycine max. L) Cu/Zn, Mn and Fe superoxide dismutases. Genet. Mol. Biol. 36, 225–236. doi: 10.1590/S1415-47572013005000023
Gulya, T. J., Mathew, F., Harveson, R., Markell, S., and Block, C. (2016). “Diseases of sunflower,” in Handbook of Florists’ crops diseases, handbook of plant disease management (Springer International Publishing, Cham), 1–20. Available at: https://www.researchgate.net/profile/Robert-Harveson/publication/310902022_Diseases_of_Sunflower/links/63a49b97097c7832ca5913a1/Diseases-of-Sunflower.pdf (Accessed April 11, 2024).
Guo, Z.-W., Si, X.-Y., Jiao, L.-P., and Liu, D.-W. (2023). Cloning and bioinformatics of CCoAOMT relating to resistance of soybean to cyst nematodes. Fujian Journal of Agricultural Sciences. 38 (5), 616–623. doi: 10.5555/20230318537
Halkier, B. A. and Du, L. (1997). The biosynthesis of glucosinolates. Trends Plant Sci. 2, 425–431. doi: 10.1016/s1360-1385(97)90026-1
Harveson, R. M., Markell, S. G., Block, C. C., and Gulya, T. J. (Eds.) (2016). Compendium of sunflower diseases and pests (Minnesota, USA: The American Phytopathological Society). doi: 10.1094/9780890545096
He, Y., Mao, S., Gao, Y., Zhu, L., Wu, D., Cui, Y., et al. (2016). Genome-Wide Identification and Expression Analysis of WRKY Transcription Factors under Multiple Stresses in Brassica napus. PLoS One 11, e0157558. doi: 10.1371/journal.pone.0157558
Hegedus, D. D., Li, R., Buchwaldt, L., Parkin, I., Whitwill, S., Coutu, C., et al. (2008). Brassica napus possesses an expanded set of polygalacturonase inhibitor protein genes that are differentially regulated in response to Sclerotinia sclerotiorum infection, wounding and defense hormone treatment. Planta 228, 241–253. doi: 10.1007/s00425-008-0733-1
Honiges, A., Sirbu, V., Rahota, D., Luca, C. M., and Pallag, A. (2017). Biochemical study of Helianthus annuus L. Exudate, related to orobanche cumana wallr. Biol. control 13, 15. Available at: http://bch.ro/pdfRC/HONIGES%20A%2012%2016.pdf (Accessed May 22, 2025).
Jeschke, V., Kearney, E. E., Schramm, K., Kunert, G., Shekhov, A., Gershenzon, J., et al. (2017). How glucosinolates affect generalist lepidopteran larvae: growth, development and glucosinolate metabolism. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01995
Jones, J. D. G. and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kabbage, M., Williams, B., and Dickman, M. B. (2013). Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PloS Pathog. 9, e1003287. doi: 10.1371/journal.ppat.1003287
Kalunke, R. M., Cenci, A., Volpi, C., O’Sullivan, D. M., Sella, L., Favaron, F., et al. (2014). The pgip family in soybean and three other legume species: evidence for a birth-and-death model of evolution. BMC Plant Biol. 14, 189. doi: 10.1186/s12870-014-0189-3
Kalunke, R. M., Tundo, S., Benedetti, M., Cervone, F., De Lorenzo, G., and D’Ovidio, R. (2015). An update on polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein that protects crop plants against pathogens. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00146
Kars, I. and van Kan, J. A. L. (2007). “Extracellular enzymes and metabolites involved in pathogenesis of botrytis,” in Botrytis: Biology, Pathology and Control. Eds. Elad, Y., Williamson, B., Tudzynski, P., and Delen, N. (Springer Netherlands, Dordrecht), 99–118. doi: 10.1007/978-1-4020-2626-3_7
Kasza, Z., Vagvölgyi, C., Févre, M., and Cotton, P. (2004). Molecular characterization and in planta detection of Sclerotinia sclerotiorum endopolygalacturonase genes. Curr. Microbiol. 48, 208–213. doi: 10.1007/s00284-003-4166-6
Kim, H. S., Sneller, C. H., and Diers, B. W. (1999). Evaluation of soybean cultivars for resistance to Sclerotinia stem rot in field environments. Crop Sci. 39, 64–68. doi: 10.2135/cropsci1999.0011183x003900010010x
Kim, H. J., Suh, H.-J., Lee, C. H., Kim, J. H., Kang, S. C., Park, S., et al. (2010b). Antifungal activity of glyceollins isolated from soybean elicited with Aspergillus sojae. J. Agric. Food Chem. 58, 9483–9487. doi: 10.1021/jf101694t
Kim, B.-G., Sung, S. H., Chong, Y., Lim, Y., and Ahn, J.-H. (2010a). Plant flavonoid O-methyltransferases: substrate specificity and application. J. Plant Biol. 53, 321–329. doi: 10.1007/s12374-010-9126-7&casa_token=68Ut1GJ4NhEAAAAA:_yV3OxNPoUNgAddWA9JxekyTW4L7Gz1_mX0hXQROoAMSgNeTpCAK4piLRNgQRP8lo9oLw7nVOsiHsglsnw
Kotchoni, S. O. and Gachomo, E. W. (2006). The reactive oxygen species network pathways:an essential prerequisite for perception of pathogen attack and the acquired disease resistance in plants. J. Biosci. 31, 389–404. doi: 10.1007/BF02704112
Kurt, S., Güneş, U., and Soylu, E. M. (2011). In vitro and in vivo antifungal activity of synthetic pure isothiocyanates against Sclerotinia sclerotiorum. Pest Manage. Sci. 67, 869–875. doi: 10.1002/ps.2126
Latchman, D. S. (1997). Transcription factors: an overview. Int. J. Biochem. Cell Biol. 29, 1305–1312. doi: 10.1016/s1357-2725(97)00085-x
Li, R., Rimmer, R., Buchwaldt, L., Sharpe, A. G., Séguin-Swartz, G., and Hegedus, D. D. (2004). Interaction of Sclerotinia sclerotiorum with Brassica napus: cloning and characterization of endo- and exo-polygalacturonases expressed during saprophytic and parasitic modes. Fungal Genet. Biol. 41, 754–765. doi: 10.1016/j.fgb.2004.03.002
Li, D., Sun, M., Han, Y., Teng, W., and Li, W. (2010). Identification of QTL underlying soluble pigment content in soybean stems related to resistance to soybean white mold (Sclerotinia sclerotiorum). Euphytica 172, 49–57. doi: 10.1007/s10681-009-0036-z
Liu, F., Li, X., Wang, M., Wen, J., Yi, B., Shen, J., et al. (2018). Interactions of WRKY15 and WRKY33 transcription factors and their roles in the resistance of oilseed rape to Sclerotinia infection. Plant Biotechnol. J. 16, 911–925. doi: 10.1111/pbi.12838
Liu, J., Lu, H., Wan, Q., Qi, W., and Shao, H. (2019). Genome-wide analysis and expression profiling of respiratory burst oxidase homologue gene family in Glycine max. Environ. Exp. Bot. 161, 344–356. doi: 10.1016/j.envexpbot.2018.07.015
Liu, J., Zhang, Y., Meng, Q., Shi, F., Ma, L., and Li, Y. (2017). Physiological and biochemical responses in sunflower leaves infected by Sclerotinia sclerotiorum. Physiol. Mol. Plant Pathol. 100, 41–48. doi: 10.1016/j.pmpp.2017.06.001
Livaja, M., Steinemann, S., and Schön, C.-C. (2016). Sunflower polygalacturonase-inhibiting proteins (HaPGIP) are genetically conserved in cultivated sunflower (Helianthus annuus L.) but diverse in wild species. Mol. Breed. 36, 1–19. doi: 10.1007/s11032-016-0444-4
Lorenzo, G. D., De Lorenzo, G., D’Ovidio, R., and Cervone, F. (2001). The role of polygalacturonase-inhibiting proteins (Pgips) in defense against pathogenic fungi. Annu. Rev. Phytopathol. 39, 313–335. doi: 10.1146/annurev.phyto.39.1.313
Lumsden, R. D. (1979). Histology and physiology of pathogenesis in plant diseases caused by Sclerotinia species. Phytopathology 69, 890–895. doi: 10.1094/Phyto-69-890
Lygin, A. V., Hill, C. B., Zernova, O. V., Crull, L., Widholm, J. M., Hartman, G. L., et al. (2010). Response of soybean pathogens to glyceollin. Phytopathology® 100, 897–903. doi: 10.1094/phyto-100-9-0897
Lyu, X., Shen, C., Fu, Y., Xie, J., Jiang, D., Li, G., et al. (2016). A small secreted virulence-related protein is essential for the necrotrophic interactions of sclerotinia sclerotiorum with its host plants. PLoS Pathog. 12, e1005435. doi: 10.1371/journal.ppat.1005435
Ma, L., Zhang, Y., Meng, Q., Shi, F., Liu, J., and Li, Y. (2018). Molecular cloning, identification of GSTs family in sunflower and their regulatory roles in biotic and abiotic stress. World J. Microbiol. Biotechnol. 34, 109. doi: 10.1007/s11274-018-2481-0
Magro, P., Marciano, P., and Lenna, P. (1984). Oxalic acid production and its role in pathogenesis ofSclerotinia sclerotiorum. FEMS Microbiol. Lett. 24, 9–12. doi: 10.1111/j.1574-6968.1984.tb01234.x
Manici, L. M., Lazzeri, L., and Palmieri, S. (1997). In vitro fungitoxic activity of some glucosinolates and their enzyme-derived products toward plant pathogenic fungi. J. Agric. Food Chem. 45, 2768–2773. doi: 10.1021/jf9608635
Martínez-Force, E., Dunford, N. T., and Salas, J. (2015). Sunflower: chemistry, production, processing, and utilization (Urbana IL, USA: AOCS Press). Available at: https://books.google.com/books?hl=en&lr=&id=k9JcCgAAQBAJ&oi=fnd&pg=PP1&ots=LfuO0mM-Uq&sig=dT6fnVb8AZzSrbXXXYf0Ks9YjEg (Accessed May 20, 2025).
Maxwell, D. (1973). Oxalate formation in Whetzelinia sclerotiorum by oxaloacetate acetylhydrolase. Physiologial Plant Pathol. 3, 279–288. doi: 10.1016/0048-4059(73)90090-8
Maxwell, D. P., Williams, P. H., and Maxwell, M. D. (1972). Studies on the possible relationships of microbodies and multivesicular bodies to oxalate, endopolygalacturonase, and cellulase (Cx) production by Sclerotinia sclerotiorum. Can. J. Bot. 50, 1743–1748. doi: 10.1139/b72-216
Mei, J., Liu, Y., Wei, D., Wittkop, B., Ding, Y., Li, Q., et al. (2015). Transfer of sclerotinia resistance from wild relative of Brassica oleracea into Brassica napus using a hexaploidy step. Züchter Genet. Breed. Res. 128, 639–644. doi: 10.1007/s00122-015-2459-3
Mittler, R., Vanderauwera, S., Suzuki, N., Miller, G., Tognetti, V. B., Vandepoele, K., et al. (2011). ROS signaling: the new wave? Trends Plant Sci. 16, 300–309. doi: 10.1016/j.tplants.2011.03.007
Mohnen, D. (2008). Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11, 266–277. doi: 10.1016/j.pbi.2008.03.006
Monazzah, M., Soleimani, M. J., Tahmasebi Enferadi, S., and Rabiei, Z. (2018a). Effects of oxalic acid and culture filtrate of Sclerotinia sclerotiorum on metabolic changes in sunflower evaluated using FT-IR spectroscopy. J. Gen. Plant Pathol. 84, 2–11. doi: 10.1007/s10327-017-0755-2
Monazzah, M., Tahmasebi Enferadi, S., and Rabiei, Z. (2018b). Enzymatic activities and pathogenesis-related genes expression in sunflower inbred lines affected by Sclerotinia sclerotiorum culture filtrate. J. Appl. Microbiol. 125, 227–242. doi: 10.1111/jam.13766
Musa-Khalifani, K., Darvishzadeh, R., and Abrinbana, M. (2021). Resistance against Sclerotinia basal stem rot pathogens in sunflower. Trop. Plant Pathol. 46, 651–663. doi: 10.1007/s40858-021-00463-z
Naczk, M. and Shahidi, F. (1992). “Phenolic constituents of rapeseed,” in Plant Polyphenols (Springer US, Boston, MA), 895–910. doi: 10.1007/978-1-4615-3476-1_53
Noctor, G., Mhamdi, A., and Foyer, C. H. (2014). The roles of reactive oxygen metabolism in drought: not so cut and dried. Plant Physiol. 164, 1636–1648. doi: 10.1104/pp.113.233478
Nomura, H., Komori, T., Uemura, S., Kanda, Y., Shimotani, K., Nakai, K., et al. (2012). Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat. Commun. 3, 926. doi: 10.1038/ncomms1926
Nováková, M., Sašek, V., Dobrev, P. I., Valentová, O., and Burketová, L. (2014). Plant hormones in defense response of Brassica napus to Sclerotinia sclerotiorum - reassessing the role of salicylic acid in the interaction with a necrotroph. Plant Physiol. Biochem. 80, 308–317. doi: 10.1016/j.plaphy.2014.04.019
Palmieri, F., Estoppey, A., House, G. L., Lohberger, A., Bindschedler, S., Chain, P. S. G., et al. (2019). Oxalic acid, a molecule at the crossroads of bacterial-fungal interactions. Adv. Appl. Microbiol. 106, 49–77. doi: 10.1016/bs.aambs.2018.10.001
Peluffo, L., Lia, V., Troglia, C., Maringolo, C., Norma, P., Escande, A., et al. (2010). Metabolic profiles of sunflower genotypes with contrasting response to Sclerotinia sclerotiorum infection. Phytochemistry 71, 70–80. doi: 10.1016/j.phytochem.2009.09.018
Plaszkó, T., Szűcs, Z., Vasas, G., and Gonda, S. (2021). Effects of glucosinolate-derived isothiocyanates on fungi: A comprehensive review on direct effects, mechanisms, structure-activity relationship data and possible agricultural applications. J. Fungi 7, 539. doi: 10.3390/jof7070539
Rahman, S., Rehman, A., Waqas, M., Mubarik, M. S., Alwutayd, K., AbdElgawad, H., et al. (2023). Genome-wide exploration of bZIP transcription factors and their contribution to alkali stress response in Helianthus annuus. Plant Stress 10, 100204. doi: 10.1016/j.stress.2023.100204
Rahmanpour, S., Backhouse, D., and Nonhebel, H. M. (2009). Induced tolerance ofSclerotinia sclerotiorumto isothiocyanates and toxic volatiles fromBrassicaspecies. Plant Pathol. 58, 479–486. doi: 10.1111/j.1365-3059.2008.02015.x
Rahmanpour, S., Backhouse, D., and Nonhebel, H. M. (2014). Toxicity of hydrolysis volatile products of Brassica plants to Sclerotinia sclerotiorum, in vitro. Arch. Phytopathol. Plant Prot. 47, 1860–1865. doi: 10.1080/03235408.2013.860723
Rai, G. K., Khanday, D. M., Choudhary, S. M., Kumar, P., Kumari, S., Martínez-Andújar, C., et al. (2024). Unlocking nature’s stress buster: Abscisic acid’s crucial role in defending plants against abiotic stress. Plant Stress 11, 100359. doi: 10.1016/j.stress.2024.100359
Ranjan, A., Jayaraman, D., Grau, C., Hill, J. H., Whitham, S. A., Ané, J.-M., et al. (2018). The pathogenic development of Sclerotinia sclerotiorum in soybean requires specific host NADPH oxidases. Mol. Plant Pathol. 19, 700–714. doi: 10.1111/mpp.12555
Ranjan, A., Westrick, N. M., Jain, S., Piotrowski, J. S., Ranjan, M., Kessens, R., et al. (2019). Resistance against Sclerotinia sclerotiorum in soybean involves a reprogramming of the phenylpropanoid pathway and up-regulation of antifungal activity targeting ergosterol biosynthesis. Plant Biotechnol. J. 17, 1567–1581. doi: 10.1111/pbi.13082
Rask, L., Andréasson, E., Ekbom, B., Eriksson, S., Pontoppidan, B., and Meijer, J. (2000). Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 42, 93–113. doi: 10.1023/A:1006380021658
Ratzka, A., Vogel, H., Kliebenstein, D. J., Mitchell-Olds, T., and Kroymann, J. (2002). Disarming the mustard oil bomb. Proc. Natl. Acad. Sci. 99, 11223–11228. doi: 10.1073/pnas.172112899
Research: USDA ARS (2022). Available online at: https://www.ars.usda.gov/plains-area/fargo-nd/etsarc/docs/national-sclerotinia-initiative-folder/research/ (Accessed October 12, 2023).
Reymond, P., Deléage, G., Rascle, C., and Fèvre, M. (1994). Cloning and sequence analysis of a polygalacturonase-encoding gene from the phytopathogenic fungus Sclerotinia sclerotiorum. Gene 146, 233–237. doi: 10.1016/0378-1119(94)90298-4
Rhoads, D. M., Umbach, A. L., Subbaiah, C. C., and Siedow, J. N. (2006). Mitochondrial reactive oxygen species. Contribution to oxidative stress and interorganellar signaling. Plant Physiol. 141, 357–366. doi: 10.1104/pp.106.079129
Rodríguez, M. A., Venedikian, N., Bazzalo, M. E., and Godeas, A. (2004). Histopathology of Sclerotinia sclerotiorum attack on flower parts of Helianthus annuus heads in tolerant and susceptible varieties. Mycopathologia 157, 291–302. doi: 10.1023/b:myco.0000024177.82916.b7
Roth, M. G., Webster, R. W., Mueller, D. S., Chilvers, M. I., Faske, T. R., Mathew, F. M., et al. (2020). Integrated management of important soybean pathogens of the United States in changing climate. J. Integr. Pest Manag 11, 17. doi: 10.1093/jipm/pmaa013
Rothmann, L. A. and McLaren, N. W. (2018). Sclerotinia sclerotiorum disease prediction: A review and potential applications in South Africa. S. Afr. J. Sci. 114, 9. doi: 10.17159/sajs.2018/20170155
Rovenich, H., Boshoven, J. C., and Thomma, B. P. H. J. (2014). Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Curr. Opin. Plant Biol. 20, 96–103. doi: 10.1016/j.pbi.2014.05.001
Sagi, M. and Fluhr, R. (2001). Superoxide production by plant homologues of the gp91phox NADPH oxidase. Modulation of activity by calcium and by tobacco mosaic virus infection. Plant Physiol. 126, 1281–1290. doi: 10.1104/pp.126.3.1281
Saharan, G. S. and Mehta, N. (2008). Sclerotinia Diseases of Crop Plants: Biology, Ecology and Disease Management (Netherlands: Springer Science & Business Media). Available at: https://play.google.com/store/books/details?id=uvaQCSybm_gC (Accessed August 24, 2022).
Scandalios, J. G., Guan, L., and Polidoros, A. (1997). Catalases in plants: Gene structure, properties, regulation, and expression. Cold Spring Harbor Monograph Arch. 34, 343–406. doi: 10.1101/087969502.34.343
Scheideler, M., Schlaich, N. L., Fellenberg, K., Beissbarth, T., Hauser, N. C., Vingron, M., et al. (2002). Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J. Biol. Chem. 277, 10555–10561. doi: 10.1074/jbc.M104863200
Schenk, P. M., Thomas-Hall, S. R., Nguyen, A. V., Manners, J. M., Kazan, K., and Spangenberg, G. (2008). Identification of plant defence genes in canola using Arabidopsis cDNA microarrays. Plant Biol. (Stuttg.) 10, 539–547. doi: 10.1111/j.1438-8677.2008.00056.x
Schluttenhofer, C. and Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167, 295–306. doi: 10.1104/pp.114.251769
Schweizer, P., Christoffel, A., and Dudler, R. (1999). Transient expression of members of the germin-like gene family in epidermal cells of wheat confers disease resistance. Plant J. 20, 541–552. doi: 10.1046/j.1365-313x.1999.00624.x
Seiler, G. J., Qi, L. L., and Marek, L. F. (2017). Utilization of sunflower crop wild relatives for cultivated sunflower improvement. Crop Sci. 57, 1083–1101. doi: 10.2135/cropsci2016.10.0856
Sessa, D. J. and Anderson, R. L. (1981). Soybean peroxidases: purification and some properties. J. Agric. Food Chem. 29, 960–965. doi: 10.1021/jf00107a019
Sewelam, N., Kazan, K., and Schenk, P. M. (2016). Global plant stress signaling: reactive oxygen species at the cross-road. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00187
Sharma, R. P. and Salunkhe, D. K. (1991). Mycotoxins & Phytoalexins (USA: CRC-Press). Available at: https://play.google.com/store/books/details?id=56_wAAAAMAAJ (Accessed August 22, 2022).
Sotelo, T., Lema, M., Soengas, P., Cartea, M. E., and Velasco, P. (2015). In vitro activity of glucosinolates and their degradation products against brassica-pathogenic bacteria and fungi. Appl. Environ. Microbiol. 81, 432–440. doi: 10.1128/AEM.03142-14
Su, W., Raza, A., Gao, A., Jia, Z., Zhang, Y., Hussain, M. A., et al. (2021). Genome-wide analysis and expression profile of superoxide dismutase (SOD) gene family in rapeseed (Brassica napus L.) under different hormones and abiotic stress conditions. Antioxidants (Basel) 10, 1182. Available at: https://www.mdpi.com/2076-3921/10/8/1182 (Accessed May 23, 2025).
Sudisha, J., Sharathchandra, R. G., Amruthesh, K. N., Kumar, A., and Shetty, H. S. (2012). “Pathogenesis related proteins in plant defense response,” in Plant Defence: Biological Control. Eds. Mérillon, J. M. and Ramawat, K. G. (Springer Netherlands, Dordrecht), 379–403. doi: 10.1007/978-94-007-1933-0_17
Talukder, Z. I., Hulke, B. S., Marek, L. F., and Gulya, T. J. (2014). Sources of resistance to sunflower diseasesin a global collection of domesticated USDA plant introductions. Crop Sci. 54, 694–705. doi: 10.2135/cropsci2013.07.0506
Tang, L., Yang, G., Ma, M., Liu, X., Li, B., Xie, J., et al. (2020). An effector of a necrotrophic fungal pathogen targets the calcium-sensing receptor in chloroplasts to inhibit host resistance. Mol. Plant Pathol. 21, 686–701. doi: 10.1111/mpp.12922
Taylor, A., Rana, K., Handy, C., and Clarkson, J. P. (2018). Resistance to Sclerotinia sclerotiorum in wild Brassica species and the importance of Sclerotinia subarctica as a Brassica pathogen. Plant Pathol. 67, 433–444. doi: 10.1111/ppa.12745
The Canola Council of Canada (2020). The Canola Council of Canada. Available online at: https://www.canolacouncil.org/canola-encyclopedia/diseases/sclerotinia-stem-rot/ (Accessed August 10, 2022).
Tourneau, L. (1979). Morphology, cytology, and physiology of Sclerotinia species in culture. Phytopathology 69, 887–890. doi: 10.1094/Phyto-69-887
U.S. Department of Agriculture-Agricultural Research Service (2022). 2022 National Sclerotinia Initiative Annual Meeting Bookle. Available online at: https://www.ars.usda.gov/ARSUSERFILES/30600500/SCLEROTINIA/2022%20NSI%20ANNUAL%20MEETING/2022%20NSI%20ANNUAL%20MEETING%20BOOKLET%20-%20FINAL.PDF (Accessed April 11, 2024).
Vanholme, R., Demedts, B., Morreel, K., Ralph, J., and Boerjan, W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153, 895–905. doi: 10.1104/pp.110.155119
van Loon, L. C. (1985). Pathogenesis-related proteins. Plant Mol. Biol. 4, 111–116. doi: 10.1007/BF02418757
Waksman, G., Keon, J. P., and Turner, G. (1991). Purification and characterization of two endopolygalacturonases from Sclerotinia sclerotiorum. Biochim. Biophys. Acta 1073, 43–48. doi: 10.1016/0304-4165(91)90180-o
Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2, e123. doi: 10.1371/journal.ppat.0020123
Wang, Liu, Ding, Wang, and Wan (2015). Advance in Sclerotinia stem rot of rapeseed. J. Northwest Anthr. 43, 85–93. Available at: https://www.cabdirect.org/cabdirect/abstract/20163017260 (Accessed August 10, 2022).
Wang, K., Peng, Q., Qiao, Y., Li, Y., Suo, D., and Shi, B. (2018a). Different glyceollin synthesis-related metabolic content and gene expressions in soybean callus suspension cultures and cotyledon tissues induced by alginate oligosaccharides. Process Biochem. 73, 188–196. doi: 10.1016/j.procbio.2018.08.013
Wang, Y., Sun, Y., Wang, J., Zhou, M., Wang, M., and Feng, J. (2019). Antifungal activity and action mechanism of the natural product cinnamic acid against Sclerotinia sclerotiorum. Plant Dis. 103, 944–950. doi: 10.1094/PDIS-08-18-1355-RE
Wang, Z., Wan, L., Zhang, X., Xin, Q., Song, Y., Hong, D., et al. (2021). Interaction between Brassica napus polygalacturonase inhibition proteins and Sclerotinia sclerotiorum polygalacturonase: implications for rapeseed resistance to fungal infection. Planta 253, 34. doi: 10.1007/s00425-020-03556-2
Wang, W., Zhang, H., Wei, X., Yang, L., Yang, B., Zhang, L., et al. (2018b). Functional characterization of calcium-dependent protein kinase (CPK) 2 gene from oilseed rape (Brassica napus L.) in regulating reactive oxygen species signaling and cell death control. Gene 651, 49–56. doi: 10.1016/j.gene.2018.02.006
Wei, W., Pierre-Pierre, N., Peng, H., Ellur, V., Vandemark, G. J., and Chen, W. (2020). The D-galacturonic acid catabolic pathway genes differentially regulate virulence and salinity response in Sclerotinia sclerotiorum. Fungal Genet. Biol. 145, 103482. doi: 10.1016/j.fgb.2020.103482
Wei, W., Wu, X., Blahut-Beatty, L., Simmonds, D. H., and Clough, S. J. (2022a). Transcriptome profiling reveals molecular players in early soybean-Sclerotinia sclerotiorum interaction. Phytopathology 112, 1739–1752. doi: 10.1094/PHYTO-08-21-0329-R
Wei, W., Xu, L., Peng, H., Zhu, W., Tanaka, K., Cheng, J., et al. (2022b). A fungal extracellular effector inactivates plant polygalacturonase-inhibiting protein. Nat. Commun. 13, 2213. doi: 10.1038/s41467-022-29788-2
Weiberg, A., Wang, M., Lin, F.-M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Whetzel, H. H. (1945). A synopsis of the genera and species of the sclerotiniaceae, A family of stromatic inoperculate discomycetes. Mycologia 37, 648–714. doi: 10.1080/00275514.1945.12024025
Willetts, H. J. and Wong, J. A. L. (1980). The biology ofSclerotinia sclerotiorum,S. trifoliorum, andS. minor with emphasis on specific nomenclature. Bot. Rev. 46, 101–165. doi: 10.1007/bf02860868
Wittstock, U., Agerbirk, N., Stauber, E. J., Olsen, C. E., Hippler, M., Mitchell-Olds, T., et al. (2004). Successful herbivore attack due to metabolic diversion of a plant chemical defense. Proc. Natl. Acad. Sci. U.S.A. 101, 4859–4864. doi: 10.1073/pnas.0308007101
Wittstock, U., Kurzbach, E., Herfurth, A.-M., and Stauber, E. J. (2016). Glucosinolate breakdown. Adv. Botanical Res. 80, 125–169. doi: 10.1016/bs.abr.2016.06.006
Xia, X.-J., Zhou, Y.-H., Shi, K., Zhou, J., Foyer, C. H., and Yu, J.-Q. (2015). Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 66, 2839–2856. doi: 10.1093/jxb/erv089
Xiao, K., Qiao, K., Cui, W., Xu, X., Pan, H., Wang, F., et al. (2023). Comparative transcriptome profiling reveals the importance of GmSWEET15 in soybean susceptibility to Sclerotinia sclerotiorum. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1119016
Xu, L., Li, G., Jiang, D., and Chen, W. (2018). Sclerotinia sclerotiorum: an evaluation of virulence theories. Annu. Rev. Phytopathol. 56, 311–338. doi: 10.1146/annurev-phyto-080417-050052
Xu, L., Xiang, M., White, D., and Chen, W. (2015). pH dependency of sclerotial development and pathogenicity revealed by using genetically defined oxalate-minus mutants of Sclerotinia sclerotiorum. Environ. Microbiol. 17, 2896–2909. doi: 10.1111/1462-2920.12818
Zhang, Z., Collinge, D. B., and Thordal-Christensen, H. (1995). Germin-like oxalate oxidase, a H2O2-producing enzyme, accumulates in barley attacked by the powdery mildew fungus. Plant J. 8, 139–145. doi: 10.1046/j.1365-313X.1995.08010139.x
Zhang, J.-X. and Kirkham, M. (1996). Enzymatic responses of the ascorbate-glutathione cycle to drought in sorghum and sunflower plants. Plant Sci. 113, 139–147. doi: 10.1016/0168-9452(95)04295-4
Zhang, M., Liu, Y., Li, Z., She, Z., Chai, M., Aslam, M., et al. (2021). The bZIP transcription factor GmbZIP15 facilitates resistance against Sclerotinia sclerotiorum and Phytophthora sojae infection in soybean. iScience 24, 102642. doi: 10.1016/j.isci.2021.102642
Zhang, M., Liu, Y., Shi, H., Guo, M., Chai, M., He, Q., et al. (2018). Evolutionary and expression analyses of soybean basic Leucine zipper transcription factor family. BMC Genomics 19, 159. doi: 10.1186/s12864-018-4511-6
Zhang, K., Liu, F., Wang, Z., Zhuo, C., Hu, K., Li, X., et al. (2022a). Transcription factor WRKY28 curbs WRKY33-mediated resistance to Sclerotinia sclerotiorum in Brassica napus. Plant Physiol. 190, 2757–2774. doi: 10.1093/plphys/kiac439
Zhang, Y., Wang, Y., Zhou, W., Zheng, S., and Ye, R. (2022b). Detection of candidate gene networks involved in resistance to Sclerotinia sclerotiorum in soybean. J. Appl. Genet. 63, 1–14. doi: 10.1007/s13353-021-00654-z
Zhang, H., Zhang, X., Zhao, H., Hu, J., Wang, Z., Yang, G., et al. (2023). Genome-wide identification and expression analysis of phenylalanine ammonia-lyase (PAL) family in rapeseed (Brassica napus L.). BMC Plant Biol. 23, 481. doi: 10.1186/s12870-023-04472-9
Zhao, X., Han, Y., Li, Y., Liu, D., Sun, M., Zhao, Y., et al. (2015). Loci and candidate gene identification for resistance to Sclerotinia sclerotiorum in soybean (Glycine max L. Merr.) via association and linkage maps. Plant J. 82, 245–255. doi: 10.1111/tpj.12810
Zheng, X., Koopmann, B., Ulber, B., and von Tiedemann, A. (2020). A global survey on diseases and pests in oilseed rape—Current challenges and innovative strategies of control. Front. Agron. 2. doi: 10.3389/fagro.2020.590908
Zheng, Z., Qamar, S. A., Chen, Z., and Mengiste, T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605. doi: 10.1111/j.1365-313X.2006.02901.x
Zhou, Y., Xu, D., Jia, L., Huang, X., Ma, G., Wang, S., et al. (2017). Genome-wide identification and structural analysis of bZIP transcription factor genes in Brassica napus. Genes (Basel) 8, 288. doi: 10.3390/genes8100288
Zhu, L., Liu, X., Liu, X., Jeannotte, R., Reese, J. C., Harris, M., et al. (2008). Hessian fly (Mayetiola destructor) attack causes a dramatic shift in carbon and nitrogen metabolism in wheat. Mol. Plant Microbe Interact. 21, 70–78. doi: 10.1094/MPMI-21-1-0070
Keywords: Sclerotinia sclerotiorum, resistance mechanisms, effectors, organic acids, host pathogen interaction, soybean, canola, sunflower
Citation: Talmo N and Ranjan A (2025) Comparative insights into soybean and other oilseed crops’ defense mechanisms against Sclerotinia sclerotiorum. Front. Plant Sci. 16:1616824. doi: 10.3389/fpls.2025.1616824
Received: 23 April 2025; Accepted: 02 June 2025;
Published: 24 June 2025.
Edited by:
Katarzyna Otulak-Kozieł, Warsaw University of Life Sciences, PolandReviewed by:
Maria Victoria Aguilar Pontes, University of Cordoba, SpainNavin Chandra Gupta, Indian Council of Agricultural Research, India
Guohong Cai, Agricultural Research Service (USDA), United States
Copyright © 2025 Talmo and Ranjan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashish Ranjan, YXJhbmphbkB1bW4uZWR1
 Nick Talmo
Nick Talmo Ashish Ranjan
Ashish Ranjan