- 1Key Laboratory of Tropical Fruit Biology, Ministry of Agriculture and Rural Affairs, South Subtropical Crops Research Institute, Chinese Academy of Tropical Agricultural Sciences, Zhanjiang, Guangdong, China
- 2Key Laboratory of Hainan Province for Postharvest Physiology and Technology of Tropical Horticultural Products, South Subtropical Crops Research Institute, Chinese Academy of Tropical Agricultural Sciences, Zhanjiang, Guangdong, China
- 3National Key Laboratory for Tropical Crop Breeding, Sanya Research Institute, Chinese Academy of Tropical Agricultural Sciences, Sanya, China
- 4State Key Laboratory of Aridland Crop Science, Gansu Agricultural University, Lanzhou, China
- 5College of Agronomy, Gansu Agricultural University, Lanzhou, China
- 6College of Tropical Crops, Yunnan Agricultural University, Pu’er, China
Heat stress severely restricts potato tuber growth and development, yet the roles of WRKY transcription factors (TFs) in mediating heat responses remain poorly understood. Using quantitative reverse transcription PCR (qRT-PCR), we identified StWRKY75 in potato cultivars ‘Atlantic’ and ‘Desiree’ as a heat-inducible gene, showing significant upregulation under 30°C and 35°C treatments. Phylogenetic analysis classified StWRKY75 into the WRKY Group II family, with close evolutionary homology to tomato SlWRKY75, and subcellular localization confirmed its nuclear targeting. To characterize its function, we generated transgenic lines overexpressing StWRKY75 (OE) or knockdown by RNA interference (RNAi). Under heat stress conditions, OE plants demonstrated superior thermotolerance compared to non-transgenic (NT) controls, manifested by improved growth parameters (plant height, tuber weight per plant, fresh weight, dry weight, root fresh weight, and root dry weight), enhanced activities of antioxidant enzymes (ascorbate peroxidase [APX], catalase [CAT], peroxidase [POD], and superoxide dismutase [SOD]), increased proline accumulation, elevated photosynthetic efficiency, and reduced malondialdehyde (MDA) content. Conversely, RNAi lines exhibited compromised thermotolerance with reversed growth parameters and biochemical characteristics. qRT-PCR analysis revealed that StWRKY75 positively regulated key heat-stress responsive genes, including those encoding antioxidant enzymes (StAPX, StCAT, StSOD, StPOD), proline biosynthesis Pyrroline-5-carboxylate synthase (StP5CS), and heat shock proteins (StHSP20, StHSP70, StHSP90). These findings demonstrate that StWRKY75 acts as a positive regulator of potato thermotolerance by enhancing growth, antioxidant capacity, proline metabolism, photosynthesis, and heat-responsive gene expression, providing a valuable target for improving crop heat resilience in breeding programs.
1 Introduction
Potatoes (Solanum tuberosum L.), a critical component of global food systems, serve as a multifaceted solution to address food security challenges exacerbated by climate change and population growth (Devaux et al., 2020). As the fourth most widely cultivated crop worldwide, following rice, wheat, and maize, potatoes offer exceptional nutritional value, high yield potential, and adaptability across diverse agro-ecologies (Adekanmbi et al., 2024). Hailed as a “climate-smart” staple, their resilience to marginal soils and suboptimal climates positions them as a strategic choice for enhancing global food security and nutrition (Kumar et al., 2021). However, exposure to temperatures exceeding 30°C detrimentally impacts potato growth and yield, causing tuber discoloration, impaired starch accumulation, disrupted dormancy, and premature sprouting, consequences directly linked to global warming induced by climate change (Rykaczewska, 2015). As temperate crops, potatoes thrive optimally at ≤25°C; while transient exposure to 25–30°C is tolerated, productivity declines significantly under such conditions (Singh et al., 2020). Prolonged heat stress (>30°C) induces stomatal closure for water conservation, halving plant development rates (Aien et al., 2017). Extensive research identifies temperature as the primary determinant of potato productivity, with elevated heat emerging as a dominant stressor compared to other environmental variables (Bender and Sentelhas, 2020; Gonzalez-Jimenez et al., 2023; Zhu et al., 2024; Guillemette et al., 2025).
Plants have evolved sophisticated mechanisms to combat abiotic stresses, with TFs playing a central regulatory role. The WRKY TF superfamily, unique to algae and higher plants, is pivotal in mediating responses to biotic and abiotic stresses (Pandey and Somssich, 2009). Structurally, WRKY proteins are defined by an N-terminal DNA-binding domain containing a conserved heptapeptide (e.g., WRKYGQK, with variants such as WRKYGKK and WRKYGMK) and a C-terminal zinc-finger motif, primarily classified as C2H2 or C2HC types, though atypical structures like CX29HXH also exist (Eulgem et al., 2000; Phukan et al., 2016). Grouped into three categories (I, II, or III) based on the number of DNA-binding domains and zinc-finger arrangements, their modular architecture enhances regulatory functions in stress responses and plant development (Wani et al., 2021). WRKY TFs modulate gene expression through dual mechanisms: direct binding to W-box cis-elements in target promoters to activate or repress transcription, and forming protein complexes with co-factors to enhance transcriptional activity (Eulgem et al., 2000; Phukan et al., 2016). Notably, many WRKY promoters contain W-box elements, enabling autoregulation and cross-regulatory networks via homo/hetero dimerization with other WRKYs, collectively shaping plant transcriptional processes (Ciolkowski et al., 2008). Since the first WRKY gene (SPF1 in sweet potato) was identified in 1994 (Ishiguro and Nakamura, 1994), this family has been extensively studied in various plants under abiotic stresses. For example, in Arabidopsis, AtWRKY25/26/33 regulate thermotolerance (Li et al., 2011), while AtWRKY6/26/30/40/45/75 are upregulated under reactive oxygen species (ROS) treatment (Cheong et al., 2002). Transgenic rice seedlings overexpressing OsWRKY11 exhibit enhanced tolerance to both drought and heat stress (Wu et al., 2009), and OsWRKY72 mediates salt tolerance (Song et al., 2010). The tomato SlWRKY3 contributes to salt stress regulation (Hichri et al., 2017), while apple MdWRKY30 enhances resistance to salt and osmotic stress in transgenic calli through the modulation of stress-responsive genes (Dong et al., 2020). In maize, ZmWRKY106 confers heat tolerance, whereas in wheat, TaWRKY30 enhances drought resistance (Wang et al., 2018; El-Esawi et al., 2019). These studies highlight WRKY TFs as key players in maintaining plant growth under stress, often by enhancing antioxidant enzyme activities, proline accumulation, and ROS scavenging gene expression (Duan et al., 2023; Yu et al., 2023; Wang et al., 2024). The WRKY transcription factor (TF) family plays a crucial role in plant stress responses, including heat tolerance. In potato (Solanum tuberosum), 79 StWRKY genes have been identified through genome-wide analysis (Zhang et al., 2017), with sequences available in the Potato Genome Sequencing Consortium (PGSC) database (http://solanaceae.plantbiology.msu.edu/pgsc_download.shtml). However, the functional characterization of StWRKY75 in heat stress responses remains unexplored. This qRT-PCR analysis provided the evidence that StWRKY75 confers heat tolerance in potato, which is validated by the up-regulated expression of StWRKY75 genes in two cultivars of potatoes (Atlantic and Desiree) under varied heat stress treatments at various time intervals. This upregulated expression profile of the StWRKY75 gene provides a strong base for further analyzing the functional validation of this gene under high-temperature stress conditions. Furthermore, extensive evidence identifies Desiree as a potato cultivar with high sensitivity to both drought and elevated temperature (Basu and Minhas, 1991; Stark et al., 2013; Rolando et al., 2015). At the same time, Atlantic exhibits pronounced thermosensitivity, demonstrating significant reductions in both photosynthetic efficiency and tuber yield under evaluated heat stress conditions. This highlights Atlantic as a heat-sensitive cultivar, with its physiological and agronomic performance being markedly compromised under elevated temperatures (Ahn et al., 2004; Gautam et al., 2021; 2024). Given their established susceptibility to elevated temperatures, the potato cultivars Desiree and Atlantic were chosen as model genotypes in this study to assess heat stress-induced alterations in growth, physiological, and photosynthetic performance.
This study aims to elucidate the function of StWRKY75 in potato (cultivars ‘Atlantic’ and ‘Desiree’) thermotolerance by integrating multiple-sequence and phylogenetic analysis, expression profiling, subcellular localization, and transgenic validation. Specifically, we examine how StWRKY75 modulates growth-related traits, antioxidant enzyme activities, proline accumulation, MDA contents, photosynthetic efficiency, and the expression of heat-responsive genes (StAPX, StSOD, StPOD, StCAT, StP5CS, StHSP20, StHSP70, and StHSP90) to enhance stress tolerance. Our findings provide mechanistic insights into StWRKY75-mediated heat resistance, offering potential targets for improving potato resilience to climate change.
2 Materials and methods
2.1 Plant material and experimental design
The apical buds of tissue-cultured seedlings from two potato cultivars with inconsistent genetic backgrounds for heat stress, ‘Atlantic’ (heat-sensitive) and ‘Desiree’ (heat-tolerant), were transferred to MS medium and cultured for 28 days under controlled conditions (16-hour light/8-hour dark photoperiod, 2800 lux light intensity, 20°C). The seedlings were then transplanted into autoclaved substrate soil and cultivated in a growth chamber for 14 days (16-hour light/8-hour dark photoperiod, 2800 lux light intensity, 20°C, 75% relative humidity). Uniform plants were selected and transplanted into pots (26 cm × 27 cm × 18 cm) containing a soil-vermiculite mixture (1:1, v/v) and grown for five weeks under soil moisture maintained at 70–75%. To investigate the response of the StWRKY75 gene to drought, heat, and salt stress, experimental conditions included drought stress induced by 10% and 20% polyethylene glycol 6000 (PEG6000), heat stress at 30°C and 35°C, and salt stress with 75 mM and 150 mM sodium chloride (NaCl). Plants were exposed to these abiotic stresses for 0, 1, 2, 4, 8, 16, 24, and 48 hours, followed by leaf sampling for mRNA expression analysis. To assess StWRKY75 expression levels, 432 potted plants were analyzed, encompassing two potato cultivars, two drought stress treatments (10% and 20% PEG6000), two heat stress treatments (30°C and 35°C), two salt stress treatments (75 mM and 150 mM NaCl), eight time points (0–48 hours), three biological replicates, and three technical replicates (pots). For quantitative evaluation of physiological parameters, photosynthetic indices, and heat-responsive genes under heat stress, 14-day-old transgenic and NT lines of both cultivars from growth chambers were transplanted into pots (26 cm × 27 cm × 18 cm) containing a soil-vermiculite mixture (1:1, v/v), grown for five weeks under 70–75% soil moisture, and subjected to heat stress (30°C and 35°C) for 48 hours. A total of 378 seedlings were evaluated, including two cultivars, seven genotypes per cultivar (one NT line, three StWRKY75 overexpression lines, and three RNAi-mediated silencing lines), two heat treatments (30°C and 35°C), three biological replicates, and three technical replicates.
2.2 Phylogenetic and multiple-sequence alignment analysis
Multiple-sequence alignment of StWRKY75 with orthologs from 18 additional plant species was conducted using CLUSTALW (version 1.83) (Thompson et al., 1994). Conserved protein motifs were subsequently identified through the same alignment analysis. Phylogenetic reconstruction was performed using MEGA X (version 4.1) (Dereeper et al., 2010), employing the Neighbor-Joining (NJ) method with 1000 bootstrap replicates to assess nodal support.
2.3 RNA extraction and quantitative PCR analysis
Total RNA was extracted from the samples using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. First-strand cDNA was synthesized from 1 µg of total RNA using the TransScript First-Strand cDNA Synthesis Kit (TransGen Biotech, Beijing, China). The qRT-PCR was performed on a LightCycler 480 II instrument (Roche, Rotkreuz, Switzerland) in 20 µL reaction mixtures. Each reaction contained 100 ng of cDNA, 0.8 µL of gene-specific primers (see Supplementary Table S1), and 10 µL of SYBR Premix Ex Taq (2×) (Takara, Tokyo, Japan). The amplification conditions were as follows: initial denaturation at 94°C for 2 min, followed by 34 cycles of denaturation at 94°C for 30 s, primer annealing at 60°C for 34 s, and extension at 72°C for 30 s. The Stef1α gene was used as an internal control to normalize gene expression levels. Relative gene expression was quantified using the 2−ΔΔCt method (Livak and Schmittgen, 2001) based on cycle threshold (CT) values. To ensure reproducibility, three biological replicates were analyzed, each with three technical replicates. The primer sequences used in this study are provided in Supplementary Table S1.
2.4 Plasmid construction and genetic transformation
The StWRKY75 coding sequence (GenBank ID: NM_001288675.1) was PCR-amplified using specific primers (forward: 5’-CTCGACATGGAGAATTATGCAACAATAT-3’; reverse: 5’-GTCGACAAAAGGAAGTATAGATTTGCATCT-3’; Bioeditas, Shaanxi, China) and cloned into the pBI121-EGFP vector following established protocols (Li et al., 2020). RNAi lines were generated as previously described (Lu et al., 2019). The sense cDNA fragment was amplified using EcoR I/Kpn I-restriction sites primers and cloned into pHANNIBAL (pHAN-StWRKY75-R). Similarly, the antisense fragment was amplified with Hind III/Xba I primers and inserted to create pHAN-StWRKY75-RF. The pHAN-StWRKY75 construct was then sub-cloned into the pART vector at the Xho I and Xba I restriction sites, resulting in the pART-StWRKY75-RNAi construct. The construct was transformed into Agrobacterium tumefaciens LBA4404 and cultured (28°C, 48 h) for subsequent plant transformation. Transformants were verified by PCR amplification using RNAi-specific primers: 5’-TGGAGAATTATGCAACAATATTTC-3’ (forward) and 5’-CCATCTATAACCATCAAGAAT-3’ (reverse), NPT II selection marker primers: 5’-ATGACTGGGCACAACAGACAATCG-3’ (forward) and 5’-TCAGAAGAACTCGTCAAGAAGGCG-3’ (reverse). The transformed Agrobacterium tumefaciens culture was maintained in an LB medium supplemented with 50 mg/L each of spectinomycin and gentamicin at 28°C for 48 hours. Following centrifugation (5,000 × g, 10 min), bacterial cells were resuspended in MS liquid medium to achieve OD600 = 0.3. Surface-sterilized potato stem segments (2 cm) were cultured on an MS medium containing 7.4 g/L agar, 30 g/L sucrose, 0.5 mg/L 6-benzylaminopurine (6-BA), 2.0 mg/L zeatin (ZT), 0.2 mg/L gibberellic acid(GA3), 1.0 mg/L indole-3-acetic acid (IAA) (pH adjusted to 5.8). The explants were immersed in Agrobacterium suspension for 10 min, then co-cultivated in darkness (72 h, 25°C). Subsequently, samples were transferred to the selection medium (composition as above, supplemented with 300 mg/L timentin, 100 mg/L kanamycin), with media refreshed biweekly. The selection medium was replaced every two weeks to maintain antibiotic pressure. Developing shoots that exhibited stable antibiotic resistance were subsequently excised and transferred to a rooting medium consisting of MS basal salts, 7.4 g/L agar, 30 g/L sucrose, 300 mg/L timentin, and 100 mg/L kanamycin to promote adventitious root formation.
2.5 Subcellular localization of StWRKY75 in tobacco epidermal cells
The protein-coding sequence of StWRKY75 was amplified using primers (forward: 5′-ATGGAGAATTATGCAACAATAT-3′; reverse: 5′-AAAAGGAAGTATAGATTTGCATCT-3 ′) and cloned into the pCAM35s-GFP expression vector. The recombinant plasmid was introduced into Agrobacterium tumefaciens strain GV3101. Transient transformation of tobacco (Nicotiana benthamiana) epidermal cells was performed following the protocol described by Sparkes et al. (2006). GFP fluorescence was visualized after 48 hours of post-infiltration using a Leica TCS SP8 confocal laser scanning microscope (Leica Microsystems, Wetzlar, Germany).
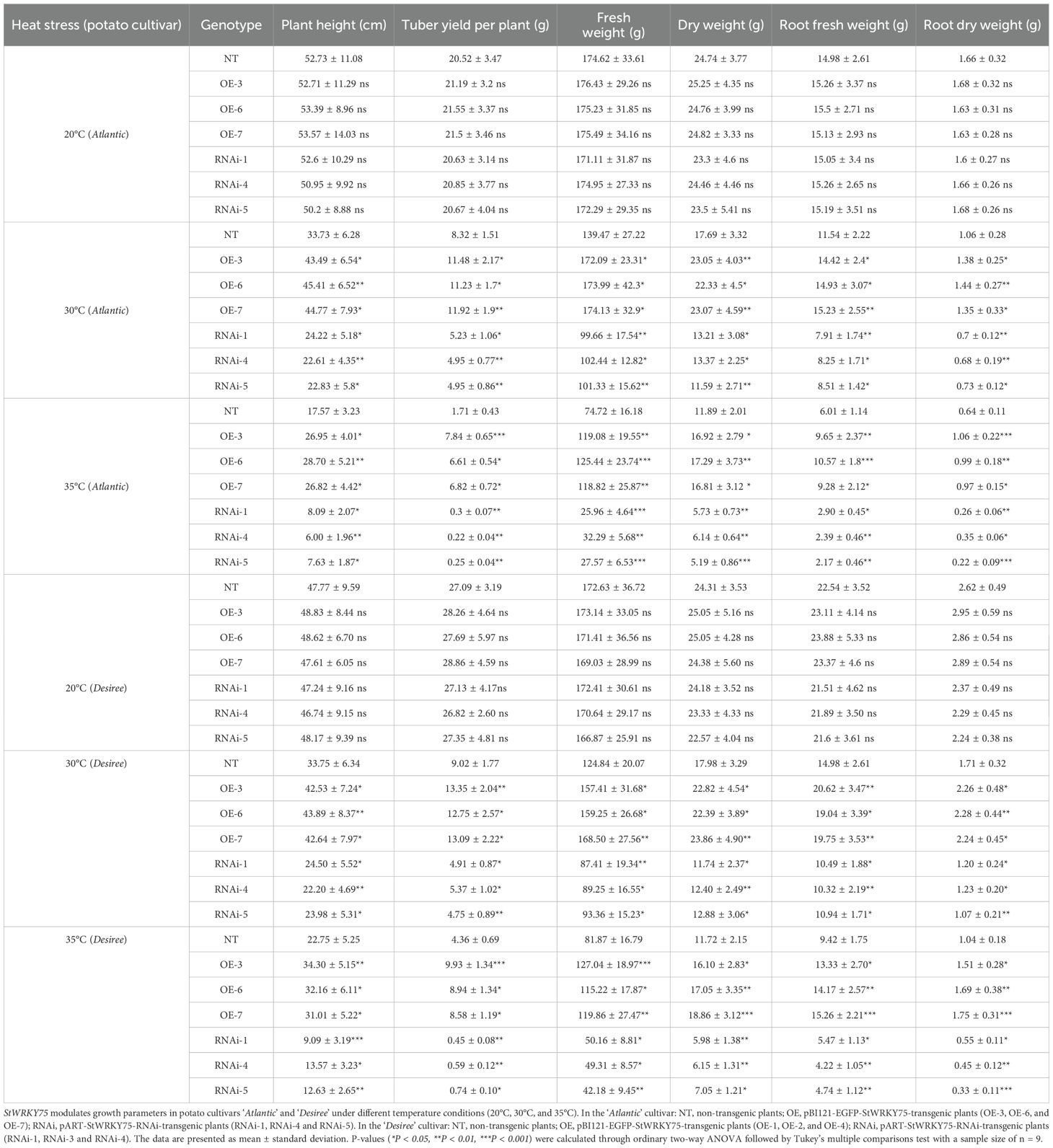
2.6 Assessment of growth-related traits
Apical buds of transgenic and NT tissue-cultured seedlings from two uniform potato cultivars, ‘Atlantic’ and ‘Desiree’, were transferred to MS medium and cultured for 28 days under controlled conditions (16-hour light/8-hour dark photoperiod, 2800 lux light intensity, 20°C). The seedlings were then transplanted into autoclaved substrate soil and cultivated in a growth chamber for 14 days (16-hour light/8-hour dark photoperiod, 2800 lux light intensity, 20°C, 70% relative humidity). Uniformly growing plants were selected and transplanted into pots (26 cm × 27 cm × 18 cm) containing a soil-vermiculite mixture (1:1, v/v) and grown for five weeks under soil moisture maintained at 70–75%. Plants with consistent growth were subjected to heat stress (30°C and 35°C) for five weeks, followed by growth parameter analysis. Plant height was measured as the distance from the soil surface to the stem apex, while tuber yield per plant and biomass (fresh and dry weights of shoots and roots) were evaluated under different heat stress conditions. Fresh weights were recorded immediately after treatment, and dry weights were determined after drying samples to constant weight in a 70°C oven. A total of 378 seedlings were assessed, encompassing two cultivars, seven genotypes per cultivar (1 NT line, 3 StWRKY75 overexpression lines, and 3 RNAi-mediated silencing lines), two heat treatments (30°C and 35°C), three biological replicates, and three technical replicates.
2.7 Assessment of physiological indicators
The key physiological parameters, including the activities of SOD (Giannopolitis and Ries, 1977), CAT (Aebi, 1984), POD (Maehly and Chance, 1954), and APX (Nakano and Asada, 1981), as well as the levels of proline (Bates et al., 1973) and MDA (Heath and Packer, 1968), were assayed using established protocols (Supplementary Data 1). The study evaluated 378 seedlings from two cultivars, each comprising seven genotypes (an NT control, three StWRKY75-overexpression lines, and three StWRKY75-RNAi lines), under two heat stress regimes (30°C and 35°C) in a completely randomized design.
2.8 Assessment of photosynthetic indicators
The photosynthetic capacity of plants was assessed by examining the third fully developed leaf from the shoot apex, with measurements conducted during mid-morning hours (9:30-11:30 AM). Gas exchange measurements, comprising net photosynthesis (Pn), transpiration (E), and stomatal conductance (Gs), were collected using Li-COR’s 6400XT portable photosynthesis monitoring equipment. Measurements were conducted under controlled environmental conditions: photosynthetic photon flux density (PPFD) of 1500 μmol·m−2·s−1, atmospheric CO2 concentration of 400 μmol·mol−1, and leaf chamber relative humidity maintained at 60-70%. The experimental design consisted of three biological replicates arranged in a completely randomized configuration.
2.9 Statistical analysis
The statistical analyses were conducted using GraphPad Prism 8.0 (GraphPad Software, California, USA) and IBM SPSS Statistics 19 (IBM Corporation, New York, USA). Before analysis, data normality was verified using Shapiro-Wilk tests, while homogeneity of variance was confirmed through Levene’s test (P>0.05). All datasets satisfied the assumptions of normality and homoscedasticity. The results are shown as mean ± standard deviation (SD). A two-way analysis of variance (two-way ANOVA) followed by Tukey’s multiple comparisons test was employed to assess differences between groups, with a significance threshold set at P < 0.05.
3 Results
3.1 Multiple-sequence alignments and phylogenetic analysis
The amino acid sequence alignment (Figure 1A) and phylogenetic tree (Figure 1B) demonstrate the evolutionary conservation of StWRKY75 among 18 other plant species. Mango (Mangifera indica) MiWRKY75, tea plant (Camellia sinensis) CsWRKY75, oil-seed camellia (Camellia lanceoleosa) ClWRKY75, Shrub althea (Hibiscus syriacus) HsWRKY75, peruvian cotton (Gossypium raimondii) GrWRKY75, silverweed (Argentina anserina) AaWRKY75, Chinese rose (Rosa chinensis) RcWRKY75, Japanese plum (Prunus mume) PmWRKY75, apple (Malus domestica) MdWRKY75, common pear (Pyrus communis) PcWRKY75, sweet potato (Ipomoea batatas) IbWRKY75, Arabidopsis (Arabidopsis thaliana) AtWRKY75, flowering tobacco (Nicotiana sylvestris) NsWRKY75, wild tobacco (Nicotiana tomentosiformis) NtWRKY75, tomato (Solanum lycopersicum) SlWRKY75, potato (Solanum tuberosum) StWRKY75, ground cherry (Physalis pubescens) PpWRKY75, wolfberry (Lycium barbarum) LbWRKY75, pepper (Capsicum annuum) CaWRKY75 and their predicted amino acid residue alignment is shown in Figure 1A. The NCBI Protein BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) was employed to retrieve the protein accession numbers of WRKY75 orthologs from diverse plant species, as listed in Supplementary Table S1. The sequence alignment analysis revealed that StWRKY75 shares significant sequence homology with orthologous proteins from diverse species, including Solanum lycopersicum (SlWRKY75), Prunus mume (PmWRKY75), Rosa chinensis (RcWRKY75), Actinidia arguta (AaWRKY75), Gossypium raimondii (GrWRKY75), and Hibiscus syriacus (HsWRKY75). All these proteins contain the conserved signature WRKYGQK at the N-terminus, followed by the C2H2 zinc-finger motif (CX5-C-X23-H-X1-H), which is characteristic of group II (Figure 1A). This conserved domain architecture confirms the evolutionary preservation of core functional foundations across these orthologs. Phylogenetic analysis revealed distinct clustering patterns among WRKY75 orthologs (Figure 1B). Notably, StWRKY75 exhibits the highest sequence homology with SlWRKY75, as they are clustered together in the phylogenetic tree. Members within each cluster exhibit conserved functional domains and significant evolutionary homology, suggesting conserved functional roles among phylogenetically related orthologs.
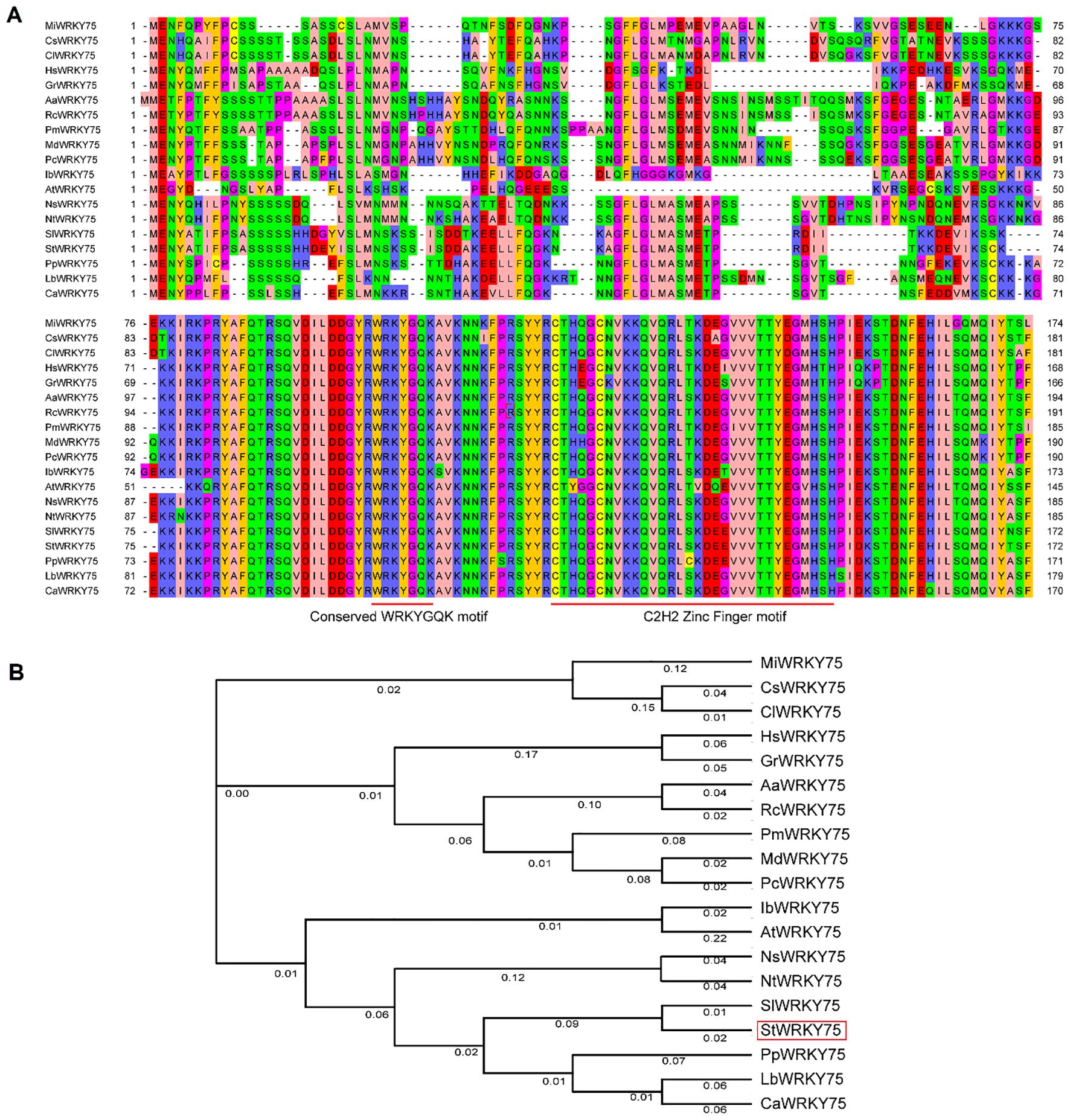
Figure 1. Multiple sequence alignment and phylogenetic relationships of StWRKY75 with other plant species. (A) Multiple sequence alignment of WRKY75 homologous proteins in potato and other plants. The different background colors represent a similar degree of amino acid sequences. The first red line indicates the WRKYGQK motif, and the second indicates the conserved C2H2 zinc-finger motif. (B) Neighbor-joining phylogenetic tree of the WRKY75 in potato and other plant species. The unrooted tree was generated using the MEGA 4.1 program with the neighbor-joining method. Bootstrap values (above 50%) from 1000 replicates are indicated at each branch.
3.2 Expression profile of StWRKY75 in potato under various abiotic stress treatments
To elucidate the transcriptional dynamics of StWRKY75 under various abiotic stresses, this study systematically analyzed the relative expression patterns in leaves of potato cultivars ‘Desiree’ and ‘Atlantic’ using qRT-PCR (Figures 2, 3). In ‘Desiree’, StWRKY75 expression showed continuous upregulation from 1 to 48 hours post-treatment (hpt) under 30°C (peaking at 48 hpt), while 35°C stress treatment induced a sustained upregulated expression pattern reaching maximum at 48 hpt (Figures 3C, D). ‘Atlantic’ displayed significant upregulation from 1–48 hpt under 30°C (peak at 2 hpt), with 35°C stress treatment triggering rapid response (peak at 4 hpt), indicating the gene’s involvement in thermotolerance regulation through rapid response to heat stress (Figures 2C, D). Under drought stress, ‘Desiree’ showed insensitivity to 10% PEG6000 (only significant downregulation at 1 hpt), whereas 20% PEG6000 treatment caused continuous suppression from 1–48 hpt (minimum at 16 hpt), suggesting its potential negative regulatory role in severe drought response (Figures 3A, B). ‘Atlantic’ exhibited significant upregulation at 4–8 hpt (peaks at 4 and 8 hpt) but downregulation at other time points (minimum at 1 hpt) under 10% PEG6000, with 20% PEG6000 inducing overall suppression (minimum at 48 hpt) (Figures 2A, B). Salt stress responses showed cultivar-specific patterns: ‘Desiree’ showed no significant changes under 75 mM NaCl but significant upregulation at 4 hpt (peak at 48 hpt) under 150 mM NaCl (Figures 3E, F); ‘Atlantic’ displayed only transient upregulation at 16 hpt under 75 mM NaCl, while 150 mM NaCl caused significant suppression from 4–16 hpt (minimum at 8 hpt) (Figures 2E, F). These findings demonstrate that StWRKY75 functions as a core regulator in heat stress response with rapid activation characteristics under both 30°C and 35°C stress treatments, while its roles in drought and salt stress may be differentially modulated by cultivar-specific genetic backgrounds and stress intensity.
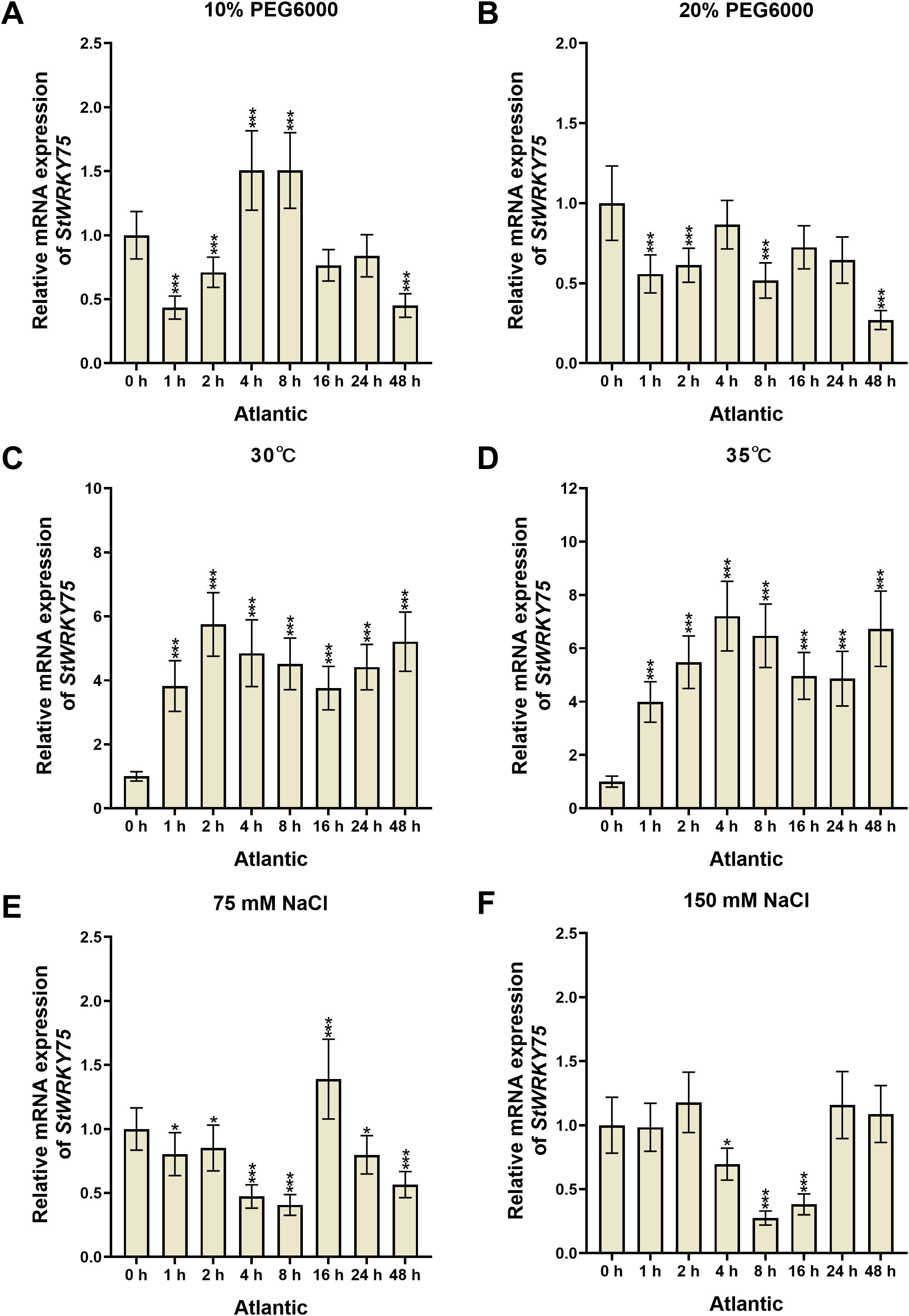
Figure 2. The expression patterns of StWRKY75 in the leaves of potato cultivar ‘Atlantic’ in response to drought (10% and 20% PEG6000), heat (30°C and 35°C), and salinity (75 mM and 150 mM NaCl), induced at the time intervals of 0, 1, 2, 4, 8, 16, 24, and 48 h. The data are presented as mean ± standard deviation. P-values (*P < 0.05, ***P < 0.001) were calculated through ordinary two-way ANOVA followed by Tukey’s multiple comparisons test with a sample size of n = 9.
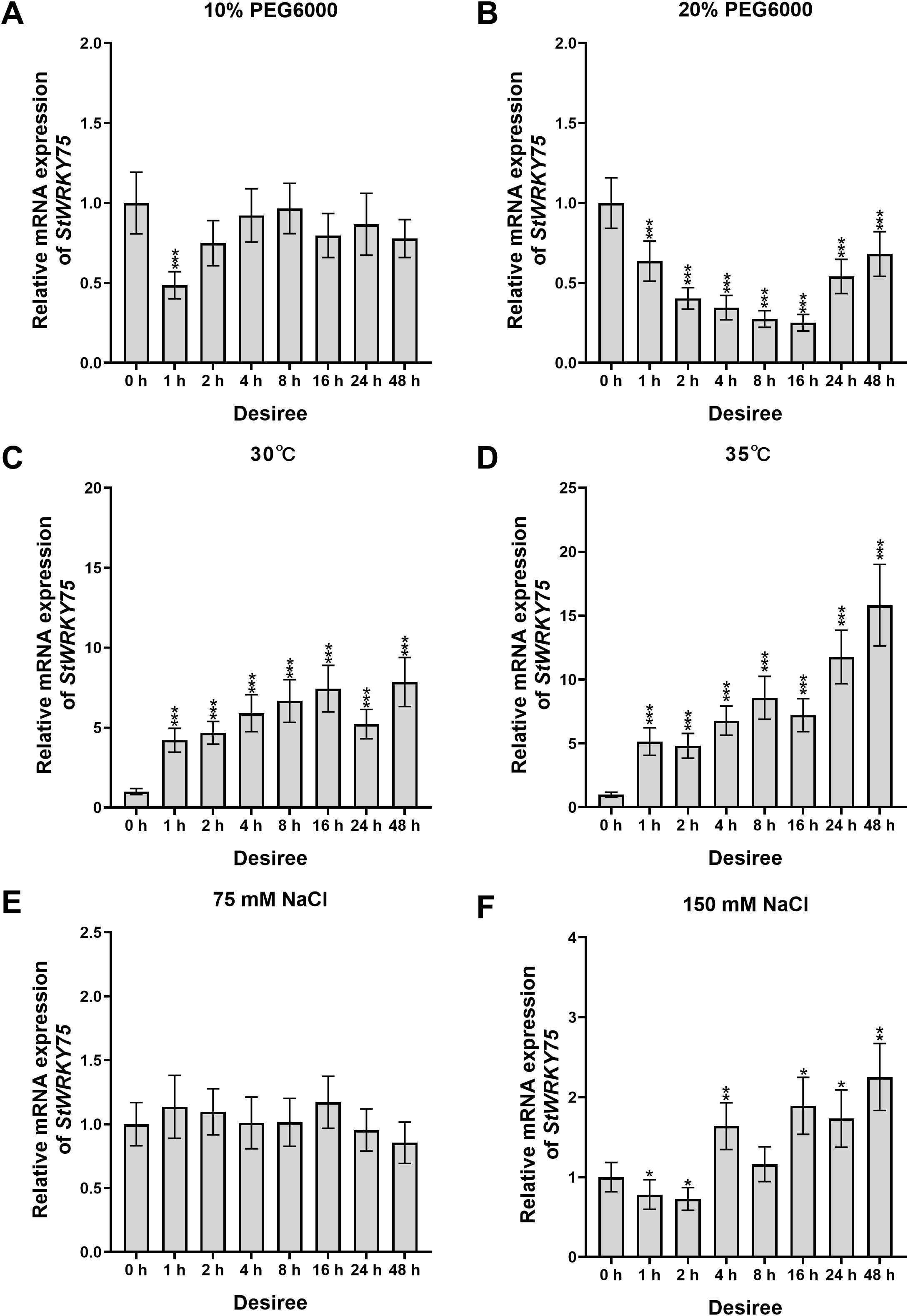
Figure 3. The expression patterns of StWRKY75 in the leaves of potato cultivar ‘Desiree’ in response to drought (10% and 20% PEG6000), heat (30°C and 35°C), and salinity (75 mM and 150 mM NaCl), induced at the time intervals of 0, 1, 2, 4, 8, 16, 24, and 48 h. The data are presented as mean ± standard deviation. P-values (*P < 0.05, **P < 0.01, ***P < 0.001) were calculated through ordinary two-way ANOVA followed by Tukey’s multiple comparisons test with a sample size of n = 9.
3.3 Subcellular localization analysis and construction of StWRKY75 transgenic potato plants
To assess the subcellular localization of StWRKY75 TF, tobacco leaves were infiltrated with Agrobacterium tumefaciens strain GV3101 carrying the recombinant plasmid pCAM35S-GFP-StWRKY75 and the empty vector control (pCAM35S-GFP). Laser confocal microscopy showed that the StWRKY75-GFP fusion protein localized exclusively to the nucleus by green protein signals (Figure 4). In contrast, the GFP vector (control) was distributed in the plasma membrane, cytoplasm, and nucleus. These results demonstrate the nuclear-specific localization of StWRKY75.
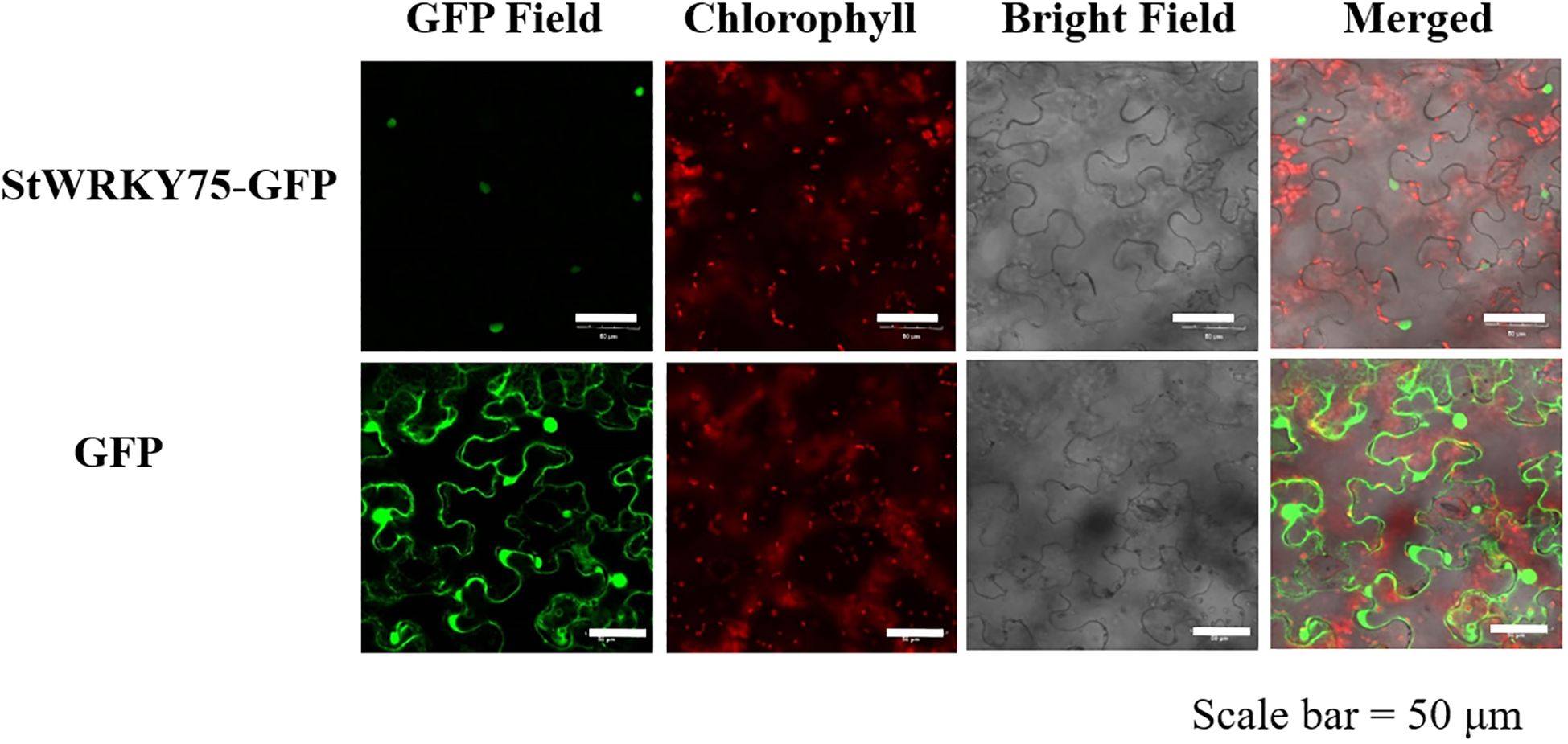
Figure 4. Subcellular localization of StWRKY75-GFP fusion protein. Confocal scanning laser microscopy analysis of tobacco transformed with pCAM35-GFP-StWRKY75. The empty vector (GFP) served as a control. Bar = 50 μm.
To functionally characterize StWRKY75 in potato, we established transgenic overexpression (OE) and RNA interference (RNAi) lines in two commercial cultivars, ‘Atlantic’ and ‘Desiree’. The qRT-PCR analysis revealed that StWRKY75 transcript levels were significantly elevated in all OE lines (OE-1 to OE-8; *** P < 0.001, Supplementary Figure S1), whereas its expression was markedly suppressed in RNAi lines (RNAi-1 to RNAi-8; ***P < 0.001, Supplementary Figure S1). Based on expression profiling, we selected three most representative lines from each group for subsequent functional studies: OE-3, OE-6, and OE-7 (showing highest overexpression) along with RNAi-1, RNAi-4, and RNAi-5 (exhibiting strongest suppression) in ‘Atlantic’(***P < 0.001, Supplementary Figures S1A, B); similarly, OE-1, OE-2, OE-4 and RNAi-1, RNAi-3, RNAi-4 were chosen from ‘Desiree’ lines (***P < 0.001, Supplementary Figures S1C, D). These selected transgenic lines were subsequently employed to investigate the functional role of StWRKY75 in thermotolerance responses.
3.4 StWRKY75 regulates the growth and biomass of potato plants under heat stress
Under optimal growth conditions (20°C), transgenic and NT control lines of potato cultivars ‘Atlantic’ and ‘Desiree’ exhibited comparable performance (P>0.05) in all evaluated growth parameters, including plant height, tuber weight per plant, fresh weight, dry weight, root fresh weight, and root dry weight (Table 1). When exposed to 30°C heat stress, all genotypes of both cultivars exhibited significant reductions in growth metrics (*P<0.05, **P<0.01). However, StWRKY75-overexpressing lines in both ‘Atlantic’ and ‘ Desiree’ demonstrated enhanced thermotolerance, with significantly greater plant height, tuber yield, and biomass accumulation compared to NT and RNAi lines (Table 1). Conversely, RNAi lines showed pronounced growth inhibition, with all measured parameters significantly lower than NT controls (*P<0.05, **P<0.01). At 35°C, heat stress, growth parameters declined more severely across all genotypes, with heat stress severely impeding potato growth. Nevertheless, OE lines in both cultivars maintained significantly superior growth performance (Table 1), while RNAi lines suffered exacerbated heat stress damage, displaying further significant reductions in biomass and tuber yield compared to NT plants (*P<0.05, **P<0.01, ***P<0.001). These findings collectively indicate that StWRKY75 overexpression confers thermotolerance by mitigating heat-induced growth suppression, while its knockdown exacerbates sensitivity, underscoring the gene’s critical role in potato heat stress adaptation.
3.5 StWRKY75 modulates antioxidant enzymes (APX, POD, CAT, SOD) activities, MDA and proline contents in potatoes under heat stress
To assess the hypothesis that StWRKY75-overexpressed potato plants of cultivars ‘Atlantic’ and ‘Desiree’ regulate antioxidant metabolism under heat stress, the activities of key antioxidant enzymes, including APX, CAT, POD, and SOD along with the oxidative stress marker MDA and the non-enzymatic osmolyte proline were investigated under varying temperature stress treatments (20°C, 30°C, and 35°C). Under control conditions (20°C), no significant differences were observed in APX, CAT, POD, or SOD activities, MDA levels, or proline content between transgenic (StWRKY75-overexpressing and RNAi) and NT plants of the potato cultivar ‘Atlantic’ (Figures 5A-F). However, when subjected to 30°C heat stress, the StWRKY75-overexpressing lines (OE-3, OE-6, OE-7) exhibited significantly enhanced activities of APX (Figure 5A), CAT (Figure 5B), POD (Figure 5C), and SOD (Figure 5D), along with increased proline accumulation (Figure 5F). In contrast, RNAi knockdown lines (RNAi-1, RNAi-4, RNAi-5) showed marked reductions in these parameters compared to NT controls. Notably, MDA content, a key indicator of oxidative damage, was significantly lower in StWRKY75-overexpressing plants (Figure 5E), whereas RNAi lines accumulated higher MDA levels, indicating greater susceptibility to oxidative stress. Under more severe heat stress (35°C), StWRKY75-overexpressing plants showed significantly greater increases in the activities of APX (Figure 5A), CAT (Figure 5B), POD (Figure 5C), and SOD (Figure 5D) activities, along with significantly elevated proline content (Figure 5E), when compared with NT control plants. Conversely, RNAi lines exhibited further suppression of these physiological indices. The StWRKY75-overexpressing lines maintained lower MDA accumulation (Figure 5F) at 35°C, demonstrating reduced oxidative damage, while RNAi lines showed exacerbated membrane lipid peroxidation due to impaired stress tolerance.
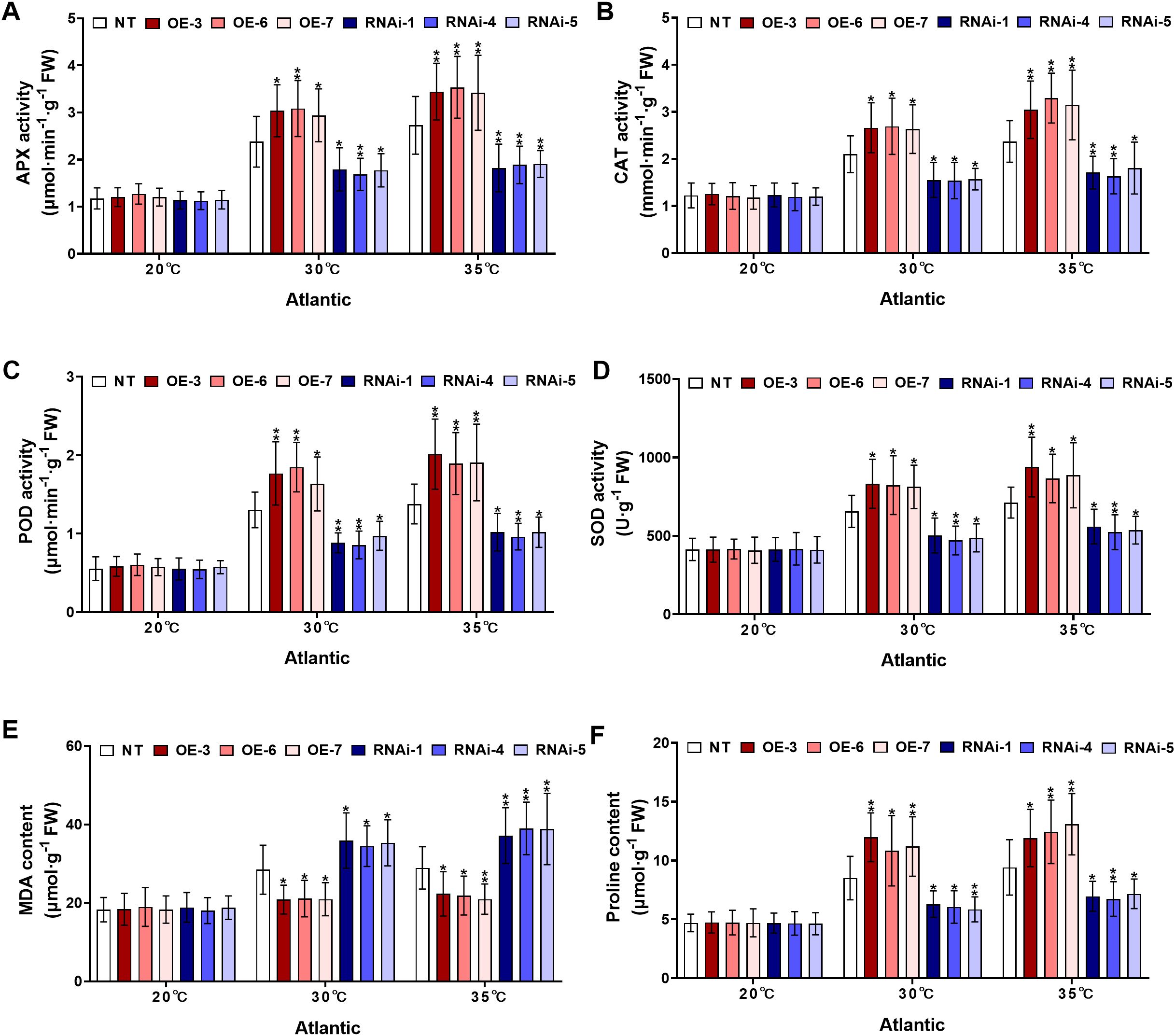
Figure 5. StWRKY75 modulates potato plants of cultivar ‘Atlantic’; (A) APX, (B) CAT, (C) POD, (D) SOD, (E) MDA, and (F) proline, after exposure to 20°C, 30°C, and 35°C of heat stress. NT, non-transgenic plants; OE, pBI121-EGFP-StWRKY75-transgenic plants (OE-3, OE-6, and OE-7); RNAi, pART-StWRKY75-RNAi-transgenic plants (RNAi-1, RNAi-4 and RNAi-5). The data are presented as mean ± standard deviation. P-values (*P < 0.05, **P < 0.01) were calculated through ordinary two-way ANOVA followed by Tukey’s multiple comparisons test with a sample size of n = 9.
Similarly, the cultivar ‘Desiree’ displayed comparable responses under high-temperature stress treatments (30°C and 35°C). Under control conditions (20°C), no significant differences were observed among all experimental potato plants for different physiological parameters (Figures 6A-F). However, when heat stress treatment was increased (30°C), the StWRKY75-overexpressing lines (OE-1, OE-2, OE-4) exhibited significantly increased activities of APX (Figure 6A), CAT (Figure 6B), POD (Figure 6C), and SOD (Figure 6D), and higher proline accumulation (Figure 6F). In contrast, RNAi-silenced lines (RNAi-1, RNAi-3, RNAi-4) showed a pronounced reduction in these physiological markers. Additionally, the MDA content was significantly lower in StWRKY75-overexpressing lines (Figure 6E), whereas RNAi lines exhibited higher MDA levels compared to NT control lines. Under severe heat stress conditions (35°C), all experimental lines exhibited exacerbated physiological damage. However, the StWRKY75-overexpressing line demonstrated significantly enhanced enzymatic activity of key antioxidant enzymes, including APX (Figure 6A), CAT (Figure 6B), POD (Figure 6C), and SOD (Figure 6D), as well as elevated proline level (Figure 6F). In contrast, StWRKY75 RNA interference (RNAi) lines exhibited a noticeable decline in these physiological parameters compared to NT plants. Likewise, the StWRKY75-overexpressing lines showed declined MDA levels (Figure 6F), and the opposite was true for StWRKY75-knockdown lines than those in NT potato lines. These findings suggest that StWRKY75 plays a critical regulatory role in enhancing thermotolerance by modulating antioxidant defense mechanisms and osmoprotectant synthesis under extreme heat stress conditions.
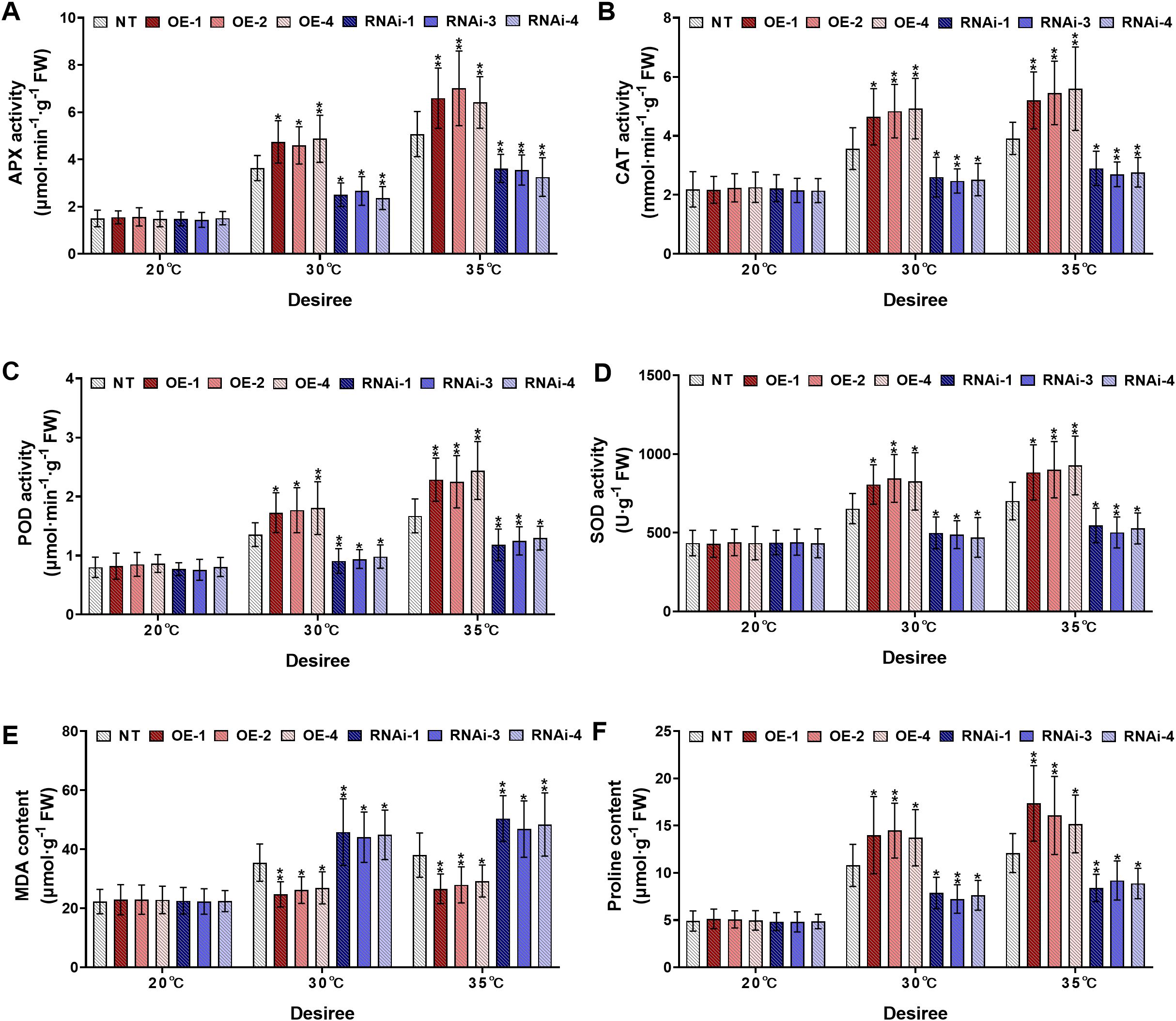
Figure 6. StWRKY75 modulates potato plants of cultivar ‘Desiree’; (A) APX, (B) CAT, (C) POD, (D) SOD, (E) MDA, and (F) proline, after exposure to 20°C, 30°C, and 35°C of heat stress. NT, non-transgenic plants; OE, pBI121-EGFP-StWRKY75-transgenic plants (OE-1, OE-2, and OE-4); RNAi, pART-StWRKY75-RNAi-transgenic plants (RNAi-1, RNAi-3 and RNAi-4). The data are presented as mean ± standard deviation. P-values (*P < 0.05, **P < 0.01) were calculated through ordinary two-way ANOVA followed by Tukey’s multiple comparisons test with a sample size of n = 9.
3.6 StWRKY75 regulates the stress-responsive genes in potatoes under heat stress
To investigate the genetic regulatory networks and biochemical mechanisms underlying heat stress adaptation in two distinct potato cultivars, ‘Atlantic’ and ‘Desiree’, this study systematically analyzed the expression profiles of key stress-responsive genes (including StAPX, StCAT, StSOD, StPOD, StP5CS, StHSP20, StHSP70, and StHSP90) across all transgenic and NT lines under different heat stress treatments (20°C, 30°C, and 35°C). The research aimed to comprehensively evaluate the differential regulation of antioxidant enzyme-related genes, proline biosynthesis-related genes, and heat shock-related genes under escalating temperature stress, thereby elucidating the genetic basis of StWRKY75-mediated thermotolerance.
Under control conditions (20°C), no significant differential expression was observed for any of the assessed stress-responsive genes (StAPX, StCAT, StSOD, StPOD, StP5CS, StHSP20, StHSP70, and StHSP90) in ‘Atlantic’ potato plants (Figures 7A-H). However, when temperature was increased to 30°C, StWRKY75-overexpressing plants (OE-3, OE-6, OE-7) showed significant upregulation in transcript levels of StAPX (Figure 7A), StCAT (Figure 7B), StSOD (Figure 7C), StPOD (Figure 7D), StP5CS (Figure 7E), StHSP20 (Figure 7F), StHSP70 (Figure 7G), and StHSP90 (Figure 7H), while StWRKY75 RNA interference lines (RNAi-1, RNAi-4, RNAi-5) exhibited marked downregulation compared to NT controls. This differential expression pattern became more pronounced under 35°C heat stress, with StWRKY75-overexpressing lines showing extremely significant upregulation of these stress-responsive genes relative to NT plants. Conversely, all target gene expressions were suppressed in StWRKY75 knockdown plants. Similarly, under standard growth conditions (20°C), the ‘Desiree’ cultivar maintained stable transcriptional levels of these key stress-responsive genes across NT and transgenic lines without significant differences (Figures 8A-H). At 30°C heat stress, however, StWRKY75-overexpressing lines (OE-1, OE-2, OE-4) demonstrated significant upregulation of antioxidant genes [StAPX (Figure 8A), StCAT (Figure 7B), StSOD (Figure 7C), StPOD (Figure 8D)], the proline biosynthesis gene StP5CS (Figure 8E), and heat shock protein genes [StHSP20 (Figure 8F), StHSP70 (Figure 8G), StHSP90 (Figure 8H)]. In contrast, these stress-responsive genes were significantly suppressed in StWRKY75 knockdown lines (RNAi-1, RNAi-3, RNAi-4) compared to NT plants. Elevated temperatures intensified these transcriptional responses, with the most prominent differential expression observed at 35°C. At this threshold, StWRKY75-overexpressing plants maintained strong upregulation of all stress-related genes (Figures 8A-H), while RNAi lines showed progressively declining expression profiles. These findings robustly demonstrate that StWRKY75 functions as a central transcriptional regulator during thermal adaptation, coordinating the upregulation of these genes (StAPX, StCAT, StSOD, StPOD, StP5CS, StHSP20, StHSP70, and StHSP90) to enhance potato thermotolerance.
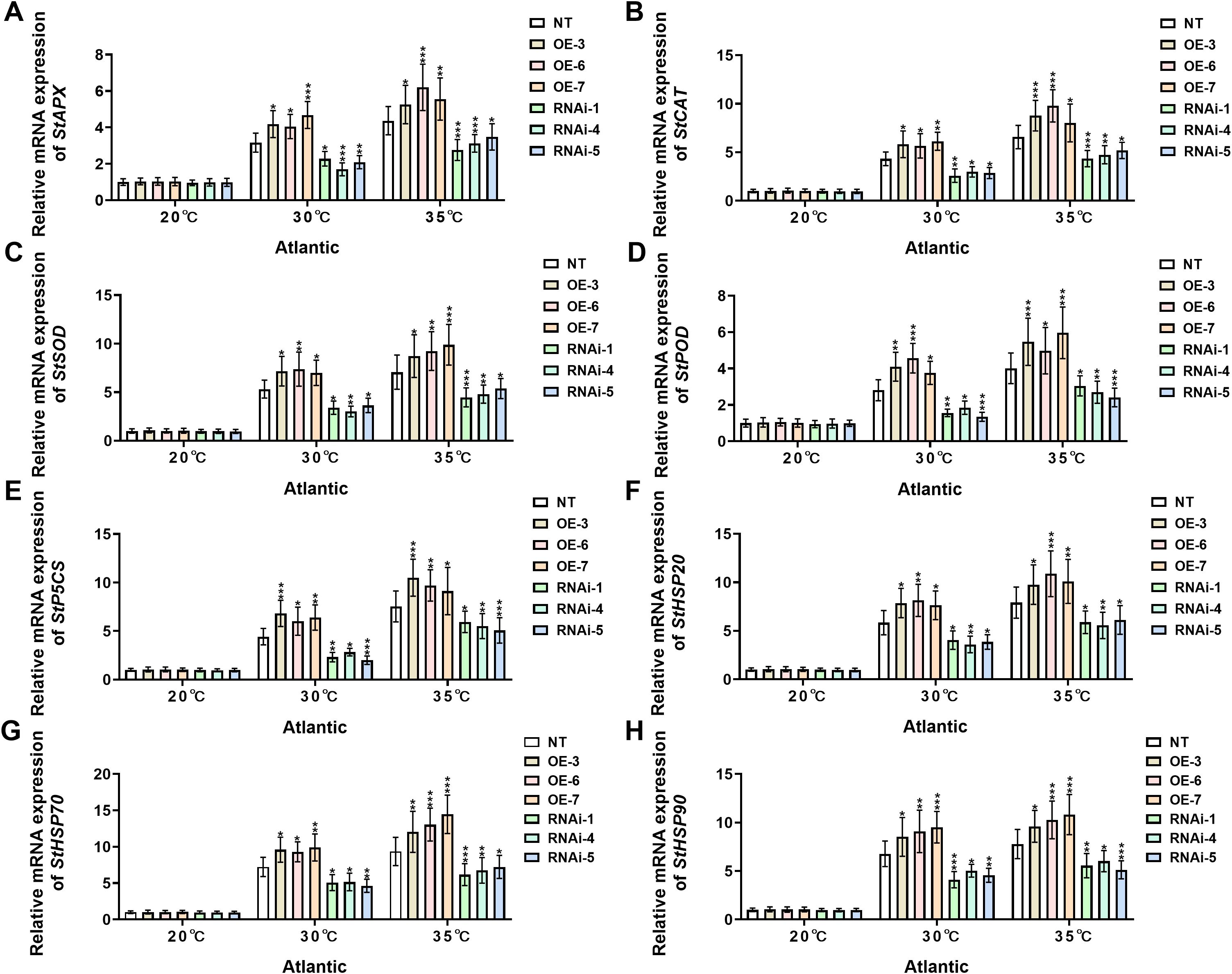
Figure 7. StWRKY75 regulates the relative mRNA expression of potato plants of cultivar ‘Atlantic’; (A) StAPX, (B) StCAT, (C) StSOD, (D) StPOD, (E) StP5CS, (F) StHSP20, (G) StHSP70, and (H) StHSP90, after exposure to 20°C, 30°C, and 35°C of heat stress. NT, non-transgenic plants; OE, pBI121-StWRKY75-transgenic plants (OE-3, OE-6, and OE-7); RNAi, pART-StWRKY75-RNAi-transgenic plants (RNAi-1, RNAi-4, and RNAi-5). The data are presented as mean ± standard deviation. P-values (*P < 0.05, **P < 0.01, ***P < 0.001) were calculated through ordinary two-way ANOVA followed by the Tukey’s multiple comparisons test with a sample size of n = 9.
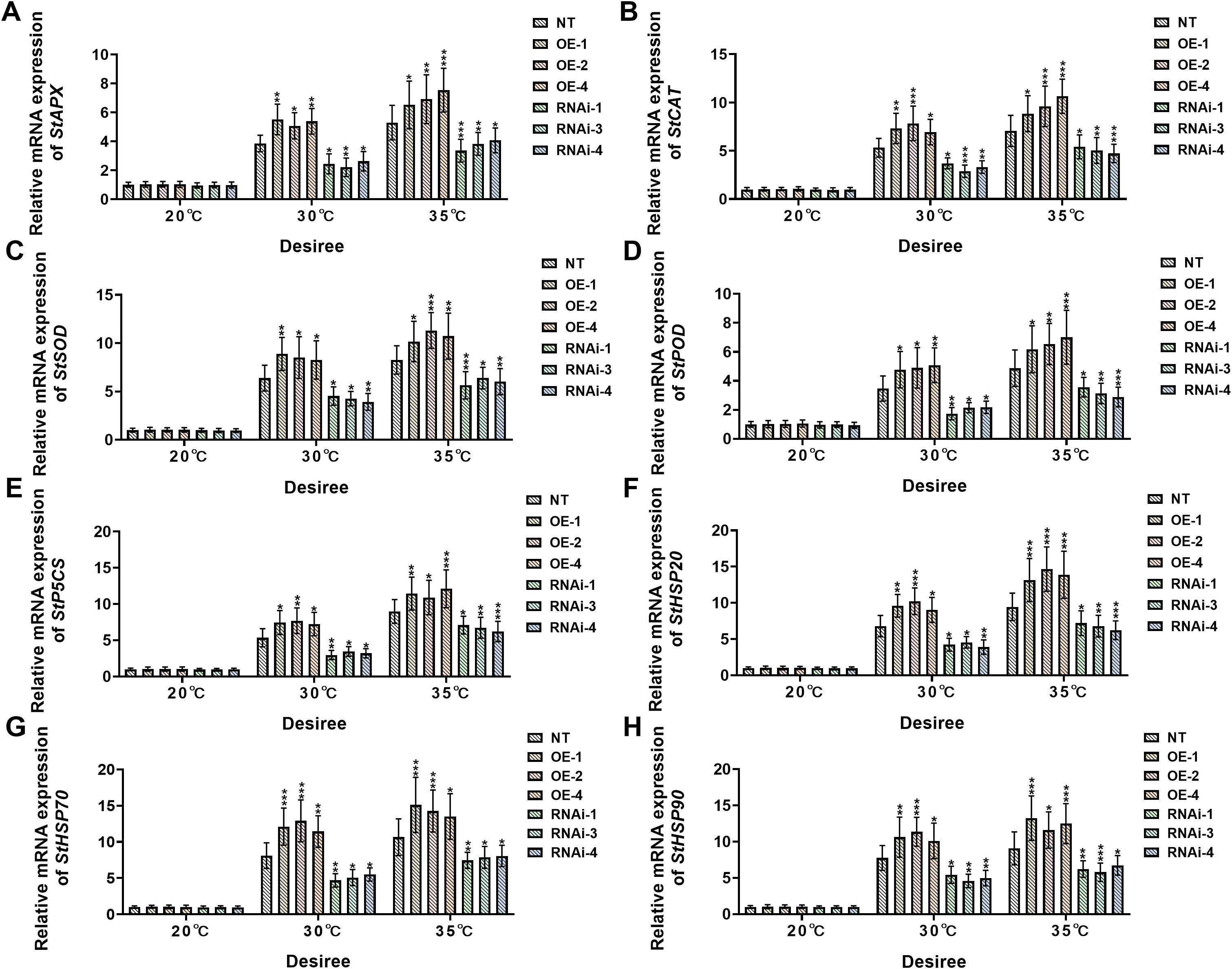
Figure 8. StWRKY75 regulates the relative mRNA expression of potato plants of cultivar ‘Desiree’; (A) StAPX, (B) StCAT, (C) StSOD, (D) StPOD, (E) StP5CS, (F) StHSP20, (G) StHSP70, and (H) StHSP90, after exposure to 20°C, 30°C, and 35°C of heat stress. NT, non-transgenic plants; OE, pBI121-StWRKY75-transgenic plants (OE-1, OE-2, and OE-4); RNAi, pART-StWRKY75-RNAi-transgenic plants (RNAi-1, RNAi-3, and RNAi-4). The data are presented as mean ± standard deviation. P-values (*P < 0.05, **P < 0.01, ***P < 0.001) were calculated through ordinary two-way ANOVA followed by the Tukey’s multiple comparisons test with a sample size of n = 9.
3.7 StWRKY75 regulates gas exchange traits of potatoes under heat stress
Under high-temperature stress regimes (30°C and 35°C), key photosynthetic parameters, including Pn, Gs, and E, were measured. Under control conditions (20°C), no significant differences were observed in gas exchange parameters between transgenic and NT plants of both ‘Atlantic’ and ‘Desiree’ potato cultivars (Figures 9A-F). However, at 30°C, StWRKY75-overexpressing lines in both cultivars exhibited significantly enhanced photosynthetic rate (Figures 9A, B), transpiration rate (Figures 9C, D), and stomatal conductance (Figures 9E, F) compared to NT plants. Conversely, StWRKY75-knockdown lines showed reduced photosynthetic performance. Under severe heat stress (35°C), growth inhibition was observed in all potato plants due to thermal damage. Nevertheless, StWRKY75-overexpressing lines maintained significantly higher photosynthetic rate (Figures 9A, B), transpiration rate (Figures 9C, D), and stomatal conductance (Figures 9E, F) compared to NT controls in both cultivars. In contrast, StWRKY75-knockdown plants displayed markedly reduced gas exchange parameters relative to NT lines, demonstrating the crucial regulatory role of StWRKY75 in maintaining photosynthetic performance under heat stress.
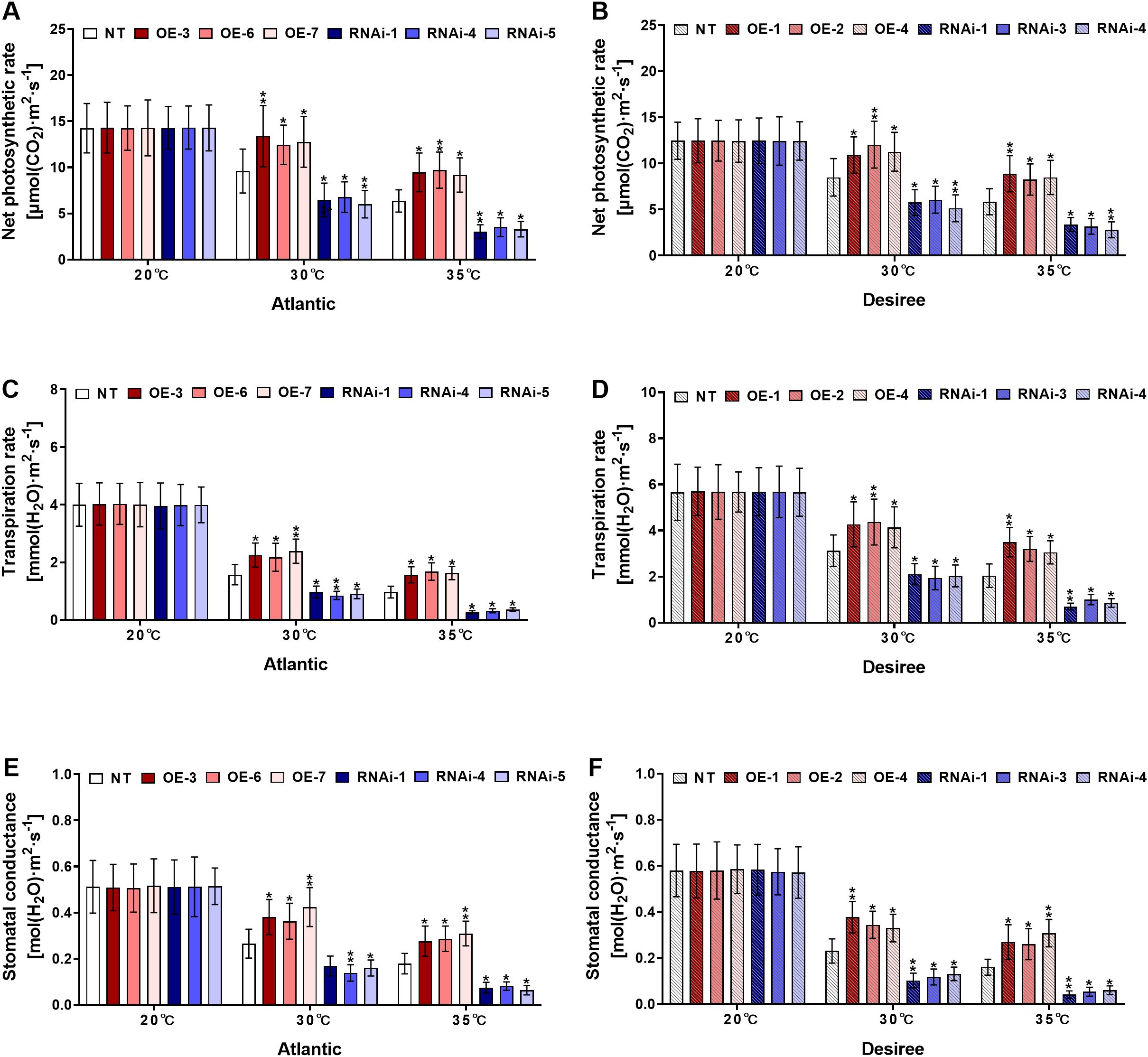
Figure 9. StWRKY75 regulates the gas exchange parameters of potato plants of cultivars, ‘Atlantic’ and ‘Desiree’; (A, B) photosynthetic rates, (C, D) transpiration rates, and (E, F) stomatal conductance, respectively, after exposure to 20°C, 30°C, and 35°C of heat stress. In the ‘Atlantic’ cultivar: NT: non-transgenic plants; OE, pBI121-EGFP-StWRKY75-transgenic plants (OE-3, OE-6, and OE-7); RNAi, pART-StWRKY75-RNAi-transgenic plants (RNAi-1, RNAi-4 and RNAi-5). In the ‘Desiree’ cultivar: NT: non-transgenic plants; OE, pBI121-EGFP-StWRKY75-transgenic plants (OE-1, OE-2, and OE-4); RNAi, pART-StWRKY75-RNAi-transgenic plants (RNAi-1, RNAi-3 and RNAi-4).The data are presented as mean ± standard deviation. P-values (*P < 0.05, **P < 0.01) were calculated through ordinary two-way ANOVA followed by the Tukey’s multiple comparisons test with a sample size of n = 9.
4 Discussion
Several WRKY TFs have been extensively studied in plants under heat stress conditions (Ding et al., 2021; Yang et al., 2022; Liu et al., 2024; Yang et al., 2025). However, there is a lack of comprehensive research on how WRKY TFs regulate heat stress responses in potatoes. The sequence alignment demonstrated that StWRKY75 harbors the highly conserved WRKYGQK domain at the N-terminus, followed by a C2H2 zinc-finger motif (CX5-C-X23-H-X1-H), a characteristic feature of Group II WRKY TFs (Figure 1A). Phylogenetic analysis classified the examined plant species into different clusters, with members within the same cluster exhibiting sequence homology and functional conservation (Figure 1B). These findings align with prior research on ZmWRKY106, which shares a 28.47% mean sequence identity with its orthologs in rice (Oryza sativa), Arabidopsis (Arabidopsis thaliana), and barley (Hordeum vulgare). Like StWRKY75, ZmWRKY106 possesses the conserved WRKYGQK domain and C2H2 zinc-finger motif, further supporting its classification within Group II. However, sequences outside these conserved regions exhibited significant divergence. Phylogenetic analysis indicated that ZmWRKY106 is most closely related to OsWRKY13 (61% bootstrap support) and HvWRKY39 (100% bootstrap support), suggesting evolutionary and functional conservation among these orthologs (Wang et al., 2018). Furthermore, through differential expression analysis under various heat stress conditions (Figures 3, 4), we confirmed that StWRKY75 was significantly induced under elevated temperatures (30°C and 35°C), suggesting its crucial functional role in heat stress response. To further understand the response of WRKY TFs to heat stress, the expression patterns of AtWRKY25, AtWRKY26, and AtWRKY33 were analyzed in Arabidopsis thaliana using qRT-PCR following heat stress treatments at 42, 45, and 48°C. The results demonstrated that AtWRKY25 and AtWRKY26 were induced at 45°C and 48°C, with expression profiles similar to those observed at 42°C. However, the fold-induction of AtWRKY26 in the wild type was consistently lower than that of AtWRKY25 at each time point following exposure to different high temperatures. In contrast, the expression of AtWRKY33 steadily decreased under heat stress, highlighting the distinct regulatory responses of these WRKY TFs to high temperatures (Li et al., 2011). Earlier investigations revealed differential expression patterns of BcWRKY22 in various Chinese cabbage tissues (roots, stems, leaves) when exposed to heat stress. The results revealed that BcWRKY22 exhibited higher expression levels in roots and stems compared to leaves, with the highest expression observed in roots under heat stress. The qRT-PCR results indicated a prompt increase in BcWRKY22 mRNA accumulation in foliar tissues 1 hour and in roots 3 hours after heat shock (HS) induction. Notably, high expression levels were maintained in both tissues even 24 hours after HS, compared to the control condition at 24°C (Wang et al., 2023). Consistent with these findings, our study demonstrated a similar research trend with different heat ingredients (30°C and 35°C), where the highest expression levels of StWRKY75 were detected in leaves of cultivars ‘Atlantic’ and ‘Desiree’ at 48 hours after heat stress treatments, as shown in Figures 2 and 3C, D.
Prior studies demonstrated that WRKY TFs mostly possess nuclear localization signals (NLS), enabling their translocation into the nucleus, and interact with DNA to regulate gene expression (Jiang et al., 2017). To further confirm the subcellular localization of WRKY proteins, a PEG-mediated transient expression system was employed in Zea mays mesophyll protoplasts using the vector p16318h-ZmWRKY106. Confocal laser scanning microscopy revealed that the p16318h-ZmWRKY106 fusion protein was predominantly localized in the nucleus after an 18-hour incubation in the dark, whereas the control vector (EGFP) exhibited fluorescence in the membrane, cytoplasm, and nucleus (Wang et al., 2018). Similarly, in this study, the subcellular localization of StWRKY75 in potato was investigated under heat stress. Green fluorescent protein (GFP) signals indicated that the StWRKY75 transcription factor was localized to the nucleus, consistent with its role as a transcriptional regulator. In contrast, the control vector (GFP) displayed fluorescence in the membrane, cytoplasm, and nucleus (Figure 4). Growth indicators, including plant height, tuber weights, shoot and root fresh and dry weights, leaf number, leaf area, root length, and total biomass, are critical parameters for evaluating the impact of abiotic stress and understanding plant adaptive mechanisms under unfavorable environmental conditions (Pandey et al., 2017). In the present study, under heat stress, StWRKY75-overexpression lines demonstrated enhanced growth across multiple indices, including plant height, tuber weight per plant, total fresh and dry weights, as well as root fresh and dry weights, relative to the NT potato lines. Conversely, RNAi-mediated gene silencing resulted in the opposite trend, as shown in Table 1. These findings align with previous research, which demonstrated that overexpression of AtWRKY30 in wheat enhances growth and biomass accumulation under heat and drought stresses. Specifically, under combined heat and drought conditions, both transformed and wild-type plants exhibited reductions in growth indicators, such as shoot fresh weight, shoot length, root fresh weight, and root length, compared to optimal growth conditions. However, AtWRKY30-overexpressing wheat plants exhibited significant improvements in growth and biomass parameters relative to wild-type plants under these stresses, highlighting the role of AtWRKY30 in enhancing stress resilience and promoting root and shoot development (El-Esawi et al., 2019).
Plants have developed sophisticated biochemical defense systems involving antioxidant enzymes (APX, SOD, CAT, POD) to combat environmental stresses. The activity levels of these enzymes, together with oxidative markers including MDA and H2O2, serve as dependable biomarkers of plant stress responses. Increased enzymatic activity of APX, SOD, CAT, and POD coupled with decreased accumulation of MDA and H2O2 demonstrates enhanced oxidative defense mechanisms and greater stress adaptation capacity (Rajput et al., 2021). Under harsh environmental conditions, plant cells exhibit marked elevation in ROS production, particularly O2-, H2O2, and HO-. The potent oxidative capacity of these molecules facilitates deleterious interactions with essential cellular constituents, resulting in the degradation of lipids, alteration of protein structures, and nucleic acid lesions that collectively compromise cell viability (García-Caparrós et al., 2021). As an important osmolyte, proline contributes significantly to ROS detoxification and improves plant resilience against multiple abiotic stressors (Hayat et al., 2012). Results from this investigation confirmed the functional role of the StWRKY75 TF in modulating oxidative stress defense mechanisms across two commercially significant potato cultivars (Solanum tuberosum L. cultivars; ‘Atlantic’ and ‘Desiree’) under differential heat stress treatments (20°C as the control and 30°C/35°C as heat stress treatments). Under control conditions, all tested plants (transgenic and NT) remained unchanged. Under varying heat stress conditions, StWRKY75-overexpressing plants exhibited enhanced activities of APX (Figures 5, 6A), CAT (Figures 5, 6B), POD (Figures 5, 6C), and SOD (Figures 5, 6D), as well as increased proline content (Figures 5, 6E), compared to non-transformed (NT) and RNA interference (RNAi) plants showed opposite outcomes. Additionally, StWRKY75-overexpressing plants displayed significantly reduced levels of MDA (Figures 5, 6F) under heat stress, indicating lower oxidative damage. In contrast, RNAi plants exhibited increased oxidative damage due to the downregulation of StWRKY75. The experimental data validate prior research, which demonstrated that overexpression of ZmWRKY106 in maize enhanced antioxidant enzyme activities (SOD, POD, and CAT) and reduced ROS levels under drought and heat stress, thereby improving abiotic tolerance (Wang et al., 2018). While the current study focused on heat stress (30°C and 35°C) rather than drought stress, the results align with earlier findings, highlighting the conserved role of WRKY TFs in enhancing antioxidant defense mechanisms under abiotic stress.
Furthermore, we analyzed the expression of stress-responsive genes, including StAPX, StSOD, StCAT, StPOD, StP5SC, StHSP20, StHSP70, and StHSP90 in two potato cultivars, ‘Atlantic’ and ‘Desiree’, StWRKY75-overexpressing and RNAi plants under varying degrees of heat stress treatments (30°C and 35°C). No differential expression of stress-related genes was observed under control conditions (20°C) (Figures 7, 8). The relative mRNA expression analysis revealed significant upregulation of StAPX (Figures 7, 8A), StCAT (Figures 7, 8C), StSOD (Figures 7, 8C), StPOD (Figures 7, 8D), StP5SC (Figures 7, 8E), StHSP20 (Figures 7, 8F), StHSP70 and (Figures 7, 8G), StHSP90 (Figures 7, 8H) in StWRKY75-overexpressing plants, while these genes were downregulated in RNAi plants after heat stress treatments. These results suggest that StWRKY75 plays a pivotal role in modulating the transcriptional cascade of stress-responsive genes, thereby enhancing heat tolerance in potato plants. Similar trends were observed in a previous study on drought stress in Arabidopsis, where overexpression of TaWRKY31 upregulated genes involved in osmotic adjustment and ROS scavenging, including AtNCED3, AtABA2, AtSnRK2.2, AtABI1, AtABF3, AtP5CS1, AtSOD(Cu/Zn), AtPOD, AtCAT, AtRD29A, and AtRD29B (Ge et al., 2024). Our study only analyzed the antioxidant-related and proline biosynthesis-related stress-responsive genes under heat stress. These data collectively underscore how WRKY TFs contribute to stress tolerance by optimizing ROS-scavenging enzymes and regulating key stress-associated genes. Heat shock proteins (HSPs) are evolutionarily conserved molecular chaperones that enhance stress adaptation by maintaining protein homeostasis under abiotic stresses, including heat, drought, and salinity (Wang et al., 2022). The current research showed an upregulation in heat shock protein-related genes, including StHSP20 (Figures 7, 8F), StHSP70 (Figures 7, 8G), and StHSP90 (Figures 7, 8H) in StWRKY75-overexpressing lines, which showed consistency with a previous report. The overexpression of PtWRKY2 significantly reduced ROS accumulation through enhanced enzymatic activity of key antioxidant enzymes related genes, including catalase-1 (CAT1), superoxide dismutase-1 (SOD1), and peroxidase-34 (POD34). Additionally, transcript levels of heat shock-associated genes (HSP70, HSP17.4) were markedly elevated in transgenic Arabidopsis lines (Liu et al., 2024). These findings demonstrate that PtWRKY2 orchestrates a coordinated upregulation of thermotolerance-related genes, enhancing both protein homeostasis and oxidative stress mitigation under heat stress conditions. Another study described that the WRKY25 knockdown slightly increased HSP20 expression, whereas WRKY33 knockdown markedly enhanced it in cotton (Ehsan et al., 2023).
Heat stress has a significant impact on photosynthesis, which is a critical physiological process in plants. The influence of high temperatures on photosynthesis parameters (Pn, Gs, and E) can vary depending on the intensity and duration of the heat, as well as the plant species and their ability to tolerate high temperatures. Additionally, heat stress negatively impacts photosynthesis by reducing the efficiency of light capture, impairing carbon fixation, increasing photorespiration, and causing oxidative damage (Zahra et al., 2023). Our investigation demonstrated that under varying degrees of heat stress conditions (20°C, 30°C, and 35°C), transgenic plants overexpressing StWRKY75 exhibited significantly enhanced photosynthetic rate (Figures 9A, B), transpiration rate (Figures 9C, D), and stomatal conductance (Figures 9E, F) compared to the NT control plants. Conversely, plants with StWRKY75 knockdown expression displayed the opposite phenotypic trends. These findings provide evidence for the regulatory role of StWRKY75 in modulating physiological responses under heat stress, thereby confirming its transcriptional regulatory function. In peanut (Arachis hypogaea), gas exchange parameters (Pn, Gs, and E) were significantly elevated in AhWRKY75 transgenic lines compared to NT control plants under salt stress conditions. Conversely, the intercellular CO2 concentration (Ci) was significantly lower in the transgenic plants relative to the controls (Zhu et al., 2021). Our current investigation demonstrated a comparable trend concerning Pn, Gs, and E; however, the key distinction lies in the application of different abiotic stress conditions. This suggests that StWRKY75 overexpression may confer enhanced photosynthetic performance and stomatal regulation across diverse stress environments, justifying further exploration into its role in plant stress tolerance mechanisms.
Although the critical role of StWRKY75 in potato heat stress response has been validated through transgenic approaches in this study, several limitations remain to be addressed in future research: Potential functional redundancy among WRKY transcription factor family members: Other homologous genes may partially compensate for StWRKY75 deficiency, possibly resulting in less pronounced phenotypic changes in RNAi lines. Future studies should employ multiplex gene knockout (CRISPR-Cas9 genome editing) or protein interaction analyses (such as yeast two-hybrid, BIFC, and Co-IP) to verify gene interaction networks. Lack of field validation: Controlled laboratory conditions (e.g., constant temperature, artificial lighting) cannot fully simulate complex field stress conditions. Subsequent multi-year and multi-location field trials under natural conditions, combined with climate data analysis, are required to evaluate the agronomic value of this gene. While StWRKY75 was identified as a central regulator of antioxidant enzymes and HSPs in thermotolerance, its direct target genes remain unverified. Additional experiments, including chromatin immunoprecipitation sequencing (ChIP-seq), yeast one-hybrid assay, and electrophoretic mobility shift assay (EMSA), should be conducted to identify DNA binding sites and further elucidate the downstream molecular mechanisms of heat tolerance. Sampling only at 48 hours after stress treatment may miss early responses (such as ROS burst within 0–48 hours) and long-term adaptation to heat stress. Additional sampling time points should be included to avoid insufficient dynamic monitoring of relevant physiological indicators.
5 Conclusion
This study demonstrates that StWRKY75, a nuclear-localized Group II WRKY transcription factor, plays a pivotal role in potato thermotolerance. Overexpression of StWRKY75 significantly enhances thermotolerance by elevating the activities of antioxidant enzymes (APX, CAT, POD, SOD), increasing proline accumulation, optimizing photosynthetic parameters (photosynthesis, transpiration, stomatal conductance), and reducing MDA accumulation. Concurrently, StWRKY75 upregulates key stress-responsive genes, including StAPX, StCAT, StPOD, StSOD, StP5CS, and heat shock protein coding genes (StHSP20, StHSP70, StHSP90). Conversely, StWRKY75-knockdown plants exhibit compromised thermotolerance with opposing physiological trends. These findings highlight StWRKY75 as a strategic target for molecular breeding to improve heat resilience in potato and related crops, offering a sustainable solution to mitigate climate change impacts on agricultural productivity.
Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Author contributions
YM: Formal analysis, Writing – review & editing, Writing – original draft, Methodology, Validation, Visualization, Investigation, Conceptualization. XZ: Visualization, Resources, Conceptualization, Writing – original draft, Formal analysis, Methodology, Validation, Supervision, Data curation, Investigation, Software, Writing – review & editing. XD: Writing – review & editing, Data curation, Formal analysis, Visualization. JL: Writing – review & editing, Visualization, Formal analysis, Methodology, Data curation. NG: Formal analysis, Visualization, Methodology, Data curation, Conceptualization, Writing – review & editing. HZh: Data curation, Writing – review & editing, Methodology, Conceptualization, Formal analysis, Visualization. HZo: Methodology, Conceptualization, Data curation, Investigation, Writing – review & editing, Visualization, Formal analysis. HJ: Data curation, Methodology, Visualization, Validation, Conceptualization, Investigation, Writing – review & editing, Software. ZC: Data curation, Visualization, Conceptualization, Writing – review & editing, Methodology. YZ: Supervision, Software, Writing – review & editing, Writing – original draft, Investigation, Funding acquisition, Conceptualization, Visualization, Project administration, Validation, Resources.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This Research Program was financially supported by the National Natural Science Foundation of China (Grant No. 32360459), the Natural Science Foundation of Guangdong Province, China (Grant No. 2024A1515010068), and the Hainan Provincial Natural Science Foundation of China (Grant No. 322MS116, 323MS095, 324MS098).
Acknowledgments
We thank Professor Si Huaijun and Professor Zhang Ning from the State Key Laboratory of Aridland Crop Science and the Life Science and Technology, Gansu Agricultural University, for their guidance and assistance in experimental techniques. Additionally, we would like to express our appreciation to Rongkai Wang (Bioediates, Shaanxi, China) for providing the plasmids pBI121-EGFP, pHANNI-BAL, and pART, as well as the construction of overexpression vectors and RNA interference expression vectors.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1617625/full#supplementary-material
References
Adekanmbi, T., Wang, X., Basheer, S., Liu, S., Yang, A., and Cheng, H. (2024). Climate change impacts on global potato yields: a review. Environ. Res. Clim. 3, 012001. doi: 10.1088/2752-5295/ad0e13
Aebi, H. (1984). “Catalase in Vitro,” in Methods in Enzymology, vol. 105 . Ed. Packer, L. (Academic Press, San Diego, California), 121–126. doi: 10.1016/s0076-6879(84)05016-3
Ahn, Y. J., Claussen, K., and Zimmerman, J. L. (2004). Genotypic differences in the heat-shock response and thermotolerance in four potato cultivars. Plant Sci. 166, 901–911. doi: 10.1016/j.plantsci.2003.11.027
Aien, A., Chaturvedi, A. K., Bahuguna, R. N., and Pal, M. (2017). Phenological sensitivity to high-temperature stress determines dry matter partitioning and yield in potato. Indian J. Plant Physiol. 22, 63–69. doi: 10.1007/s40502-016-0270-z
Basu, P. S. and Minhas, J. S. (1991). Heat tolerance and assimilate transport in different potato genotypes. J. Exp. Bot. 42, 861–866. doi: 10.1093/jxb/42.7.861
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Bender, F. D. and Sentelhas, P. C. (2020). Assessment of regional climate change impacts on Brazilian potato tuber yield. Int. J. Plant Prod. 14, 647–661. doi: 10.1007/s42106-020-00111-7
Cheong, Y. H., Chang, H. S., Gupta, R., Wang, X., Zhu, T., and Luan, S. (2002). Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129, 661–677. doi: 10.1104/pp.002857
Ciolkowski, I., Wanke, D., Birkenbihl, R. P., and Somssich, I. E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues to WRKY-domain function. Plant Mol. Boil 68, 81–92. doi: 10.1007/s11103-008-9353-1
Dereeper, A., Audic, S., Claverie, J. M., and Blanc, G. (2010). Blast-explorer helps you build datasets for phylogenetic analysis. BMC Evol. Biol. 10, 8. doi: 10.1186/1471-2148-10-8
Devaux, A., Goffart, J. P., Petsakos, A., Kromann, P., Gatto, M., Okello, J., et al. (2020). Global food security, contributions from sustainable potato agri-food systems. potato crop: Its agricult Nutr. Soc. contrib to humankind 3–35. doi: 10.1007/978-3-030-28683-5
Ding, L., Wu, Z., Teng, R., Xu, S., Cao, X., Yuan, G., et al. (2021). LlWRKY39 is involved in thermotolerance by activating LlMBF1c and interacting with LlCaM3 in lily (Lilium longiflorum). Hortic. Res. 8. doi: 10.1038/s41438-021-00473-7
Dong, Q., Zheng, W., Duan, D., Huang, D., Wang, Q., Liu, C., et al. (2020). MdWRKY30, a group IIa WRKY gene from apple, confers tolerance to salinity and osmotic stresses in transgenic apple callus and Arabidopsis seedlings. Plant Sci. 299, 110611. doi: 10.1016/j.plantsci.2020.110611
Duan, D., Yi, R., Ma, Y., Dong, Q., Mao, K., and Ma, F. (2023). Apple WRKY transcription factor MdWRKY56 positively modulates drought stress tolerance. Environ. Exp. Bot. 212, 105400. doi: 10.1016/j.envexpbot.2023.105400
Ehsan, A., Naqvi, R. Z., Azhar, M., Awan, M. J. A., Amin, I., Mansoor, S., et al. (2023). Genome-wide analysis of the WRKY gene family and negative regulation of GhWRKY25 and GhWRKY33 reveal their role in whitefly and drought stress tolerance in cotton. Genes 14, 171. doi: 10.3390/genes14010171
El-Esawi, M. A., Al-Ghamdi, A. A., Ali, H. M., and Ahmad, M. (2019). Overexpression of AtWRKY30 transcription factor enhances heat and drought stress tolerance in wheat (Triticum aestivum L.). Genes 10, 163. doi: 10.3390/genes10020163
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
García-Caparrós, P., De Filippis, L., Gul, A., Hasanuzzaman, M., Ozturk, M., Altay, V., et al. (2021). Oxidative stress and antioxidant metabolism under adverse environmental conditions: a review. Bot. Rev. 87, 421–466. doi: 10.1007/s12229-020-09231-1
Gautam, S., Scheuring, D. C., Koym, J. W., and Vales, M. I. (2024). Assessing heat tolerance in potatoes: Responses to stressful Texas field locations and controlled contrasting greenhouse conditions. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1364244
Gautam, S., Solis-Gracia, N., Teale, M. K., Mandadi, K., Silva, J. A. D., and Vales, M. I. (2021). Development of an in vitro microtuberization and temporary immersion bioreactor system to evaluate potato heat stress tolerance (Solanum tuberosum L.). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.700328
Ge, M., Tang, Y., Guan, Y., Lv, M., Zhou, C., Ma, H., et al. (2024). TaWRKY31, a novel WRKY transcription factor in wheat, regulates plant drought stress tolerance. BMC Plant Biol. 24, 27. doi: 10.1186/s12870-023-04709-7
Giannopolitis, C. N. and Ries, S. K. (1977). Superoxide dismutase: I. Occurrence in higher plants. Plant Physiol. 59, 309–314. doi: 10.1104/pp.59.2.309
Gonzalez-Jimenez, J., Andersson, B., Wiik, L., and Zhan, J. (2023). Modelling potato yield losses caused by Phytophthora infestans: Aspects of disease growth rate, infection time and temperature under climate change. Field Crops Res. 299, 108977. doi: 10.1016/j.fcr.2023.108977
Guillemette, A. M., Hernández Casanova, G., Hamilton, J. P., Pokorná, E., Dobrev, P. I., Motyka, V., et al. (2025). The physiological and molecular responses of potato tuberization to projected future elevated temperatures. Plant Physiol. 197, 664. doi: 10.1093/plphys/kiae664
Hayat, S., Hayat, Q., AlYemeni, M. N., Wani, A. S., Pichtel, J., and Ahmad, A. (2012). Role of proline under changing environments: a review. Plant Signal Behav. 7, 1456–1466. doi: 10.4161/psb.21949
Heath, R. L. and Packer, L. (1968). Photo-oxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198. doi: 10.1016/0003-9861(68)90654-1
Hichri, I., Muhovski, Y., Žižková, E., Dobrev, P. I., Gharbi, E., Franco-Zorrilla, J. M., et al. (2017). The Solanum lycopersicum WRKY3 transcription factor (SlWRKY3) is involved in salt stress tolerance in tomato. Front. Plant Sci. 8, 1343. doi: 10.3389/fpls.2017.01343
Ishiguro, S. and Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 244, 563–571. doi: 10.1007/BF00282746
Jiang, Y., Guo, L., Ma, X., Zhao, X., Jiao, B., Li, C., et al. (2017). The WRKY transcription factors PtrWRKY18 and PtrWRKY35 promote Melampsora resistance in Populus. Tree Physiol. 37, 665–675. doi: 10.1093/treephys/tpx008
Kumar, Y., Singh, R., and Kumar, A. (2021). Performance of SUBSTOR model on growth and yield of potato varieties under different planting dates in a sub-tropical environment. J. Agrometeorol 23, 213–220. doi: 10.54386/jam.v23i2.71
Li, G., Cao, C., Yang, H., Wang, J., Wei, W., Zhu, D., et al. (2020). Molecular cloning and potential role of DiSOC1s in flowering regulation in Davidia involucrata Baill. Plant Physiol. Biochem. 157, 453–459. doi: 10.1016/j.plaphy.2020.11.003
Li, S., Fu, Q., Chen, L., Huang, W., and Yu, D. (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate the induction of plant thermotolerance. Planta 233, 1237–1252. doi: 10.1007/s00425-011-1375-2
Liu, D., Cui, W., Bo, C., Wang, R., Zhu, Y., Duan, Y., et al. (2024). PtWRKY2, a WRKY transcription factor from Pinellia ternata confers heat tolerance in Arabidopsis. Sci. Rep. 14, 13807. doi: 10.1038/s41598-024-64560-0
Livak, K. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, H., Klocko, A. L., Brunner, A. M., Ma, C., Magnuson, A. C., Howe, G. T., et al. (2019). RNA Interference suppression of AGAMOUS and SEEDSTICK alters floral organ identity and impairs floral organ determinacy, ovule differentiation, and seed hair development in Populus. New Phytol. 222, 923–937. doi: 10.1111/nph.15648
Maehly, A. C. and Chance, B. (1954). “The assay of catalases and peroxidases,” in Methods of biochemical analysis, vol. 1 . Ed. Glick, D. (Interscience Publishers, New York, USA), 357–424. doi: 10.1002/9780470110171.ch14
Nakano, Y. and Asada, K. (1981). Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 22, 867–880. doi: 10.1093/oxfordjournals.pcp.a076232
Pandey, P., Irulappan, V., Bagavathiannan, M. V., and Senthil-Kumar, M. (2017). Impact of combined abiotic and biotic stresses on plant growth and avenues for crop improvement by exploiting physio-morphological traits. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00537
Pandey, S. P. and Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi: 10.1104/pp.109.138990
Phukan, U. J., Jeena, G. S., and Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00760
Rajput, V. D., Harish, Singh, R. K., Verma, K. K., Sharma, L., Quiroz-Figueroa, F. R., et al. (2021). Recent developments in the enzymatic antioxidant defense mechanism in plants with special reference to abiotic stress. Biology 10, 267. doi: 10.3390/biology10040267
Rolando, J. L., Ramírez, D. A., Yactayo, W., Monneveux, P., and Quiroz, R. (2015). Leaf greenness as a drought tolerance-related trait in potato (Solanum tuberosum L.). Environ. Exp. Bot. 110, 27–35. doi: 10.1016/j.envexpbot.2014.09.006
Rykaczewska, K. (2015). The effect of high temperature occurring in subsequent stages of plant development on potato yield and tuber physiological defects. Am. J. Potato Res. 92, 339–349. doi: 10.1007/s12230-015-9436-x
Singh, B., Kukreja, S., and Goutam, U. (2020). Impact of heat stress on potato (Solanum tuberosum L.): Present scenario and future opportunities. J. Hortic. Sci. Biotech 95, 407–424. doi: 10.1080/14620316.2019.1700173
Song, Y., Chen, L., Zhang, L., and Yu, D. (2010). Overexpression of the OsWRKY72 gene interferes with the abscisic acid signal and auxin transport pathway of. Arabidop J. Biosci. 35, 459–471. doi: 10.1007/s12038-010-0051-1
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Stark, J. C., Love, S. L., King, B. A., Marshall, J. M., Bohl, W. H., and Salaiz, T. (2013). Potato cultivar response to seasonal drought patterns. Am. J. Potato Res. 90, 207–216. doi: 10.1007/s12230-012-9285-9
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucl. Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Wang, H., Dong, Z., Chen, J., Wang, M., Ding, Y., Xue, Q., et al. (2022). Genome-wide identification and expression analysis of the Hsp20, Hsp70, and Hsp90 gene family in Dendrobium officinale. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.979801
Wang, Y., Gai, W., Yuan, L., Shang, L., Li, F., Gong, Z., et al. (2024). Heat-inducible SlWRKY3 confers thermotolerance by activating the SlGRXS1 gene cluster in tomato. Hortic. Plant J. 10, 515–531. doi: 10.1016/j.hpj.2022.12.006
Wang, H., Gao, Z., Chen, X., Li, E., Li, Y., Zhang, C., et al. (2023). BcWRKY22 activates BcCAT2 to enhance catalase (CAT) activity and reduce hydrogen peroxide (H2O2) accumulation, promoting thermotolerance in non-heading Chinese cabbage (Brassica campestris ssp. chinensis). Antioxidants 12, 1710. doi: 10.3390/antiox12091710
Wang, C. T., Ru, J. N., Liu, Y. W., Li, M., Zhao, D., Yang, J. F., et al. (2018). Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int. J. Mol. Sci. 19, 3046. doi: 10.3390/ijms19103046
Wani, S. H., Anand, S., Singh, B., Bohra, A., and Joshi, R. (2021). WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep. 40, 1071–1085. doi: 10.1007/s00299-021-02691-8
Wu, X., Shiroto, Y., Kishitani, S., Ito, Y., and Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of the HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Yang, S., Cai, W., Shen, L., Cao, J., Liu, C., Hu, J., et al. (2022). A CaCDPK29–CaWRKY27b module promotes CaWRKY40-mediated thermotolerance and immunity to Ralstonia solanacearum in pepper. New Phytol. 233, 1843–1863. doi: 10.1111/nph.17891
Yang, L., Fang, S., Liu, L., Zhao, L., Chen, W., Li, X., et al. (2025). WRKY transcription factors: Hubs for regulating plant growth and stress responses. J. Integr. Plant Biol. 67, 488–509. doi: 10.1111/jipb.13828
Yu, Y., Wu, Y., and He, L. (2023). A wheat WRKY transcription factor TaWRKY17 enhances tolerance to salt stress in transgenic Arabidopsis and wheat plants. Plant Mol. Biol. 113, 171–191. doi: 10.1007/s11103-023-01381-1
Zahra, N., Hafeez, M. B., Ghaffar, A., Kausar, A., Al Zeidi, M., Siddique, K. H., et al. (2023). Plant photosynthesis under heat stress: Effects and management. Environ. Exp. Bot. 206, 105178. doi: 10.1016/j.envexpbot.2022.105178
Zhang, C., Wang, D., Yang, C., Kong, N., Shi, Z., Zhao, P., et al. (2017). Genome-wide identification of the potato WRKY transcription factor family. PloS One 12, e0181573. doi: 10.1371/journal.pone.0181573
Zhu, H., Jiang, Y., Guo, Y., Huang, J., Zhou, M., Tang, Y., et al. (2021). A novel salt-inducible WRKY transcription factor gene, AhWRKY75, confers salt tolerance in transgenic peanut. Plant Physiol. Biochem. 160, 175–183. doi: 10.1016/j.plaphy.2021.01.014
Keywords: StWRKY75, Solanum tuberosum, heat stress, transgenic, photosynthetic efficiency
Citation: Zhu X, Duan X, Majeed Y, Luo J, Guan N, Zheng H, Zou H, Jin H, Chen Z and Zhang Y (2025) Functional analysis of StWRKY75 gene in response to heat stress in potato (Solanum tuberosum L.). Front. Plant Sci. 16:1617625. doi: 10.3389/fpls.2025.1617625
Received: 24 April 2025; Accepted: 10 June 2025;
Published: 30 June 2025.
Edited by:
Yi Wang, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Haiyang Chen, Henan Agricultural University, ChinaHaijiao Dong, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2025 Zhu, Duan, Majeed, Luo, Guan, Zheng, Zou, Jin, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasir Majeed, eWFzaXJtYWplZWQ1NDUzQGdtYWlsLmNvbQ==; Hui Jin, amluaEBjYXRhcy5jbg==; Yu Zhang, emhhbmd5dUBjYXRhcy5jbg==
 Xi Zhu
Xi Zhu Xiaoqin Duan4,5
Xiaoqin Duan4,5 Yasir Majeed
Yasir Majeed Yu Zhang
Yu Zhang