- 1Hunan Research Center of the Basic Discipline for Cell Signaling, Hunan Provincial Key Laboratory of Plant Functional Genomics and Developmental Regulation, College of Biology, Hunan University, Changsha, China
- 2Beijing Life Science Academy, Beijing, China
- 3Tobacco Chemistry Research Institute of Technology Center, China Tobacco Hunan Industrial Co., Ltd., Changsha, China
- 4Yuelushan Laboratory, Changsha, China
Strigolactones are a newly identified group of phytohormones that regulate plant growth and development and also act as communication signals in the rhizosphere. Beyond their well-known activity in stimulating parasitic weed germination, strigolactones function in regulating plant architecture, promoting symbiosis with arbuscular mycorrhizal fungi, and modulating responses to various environmental stresses. However, their low abundance, structural diversity, and instability have hindered comprehensive research and their practices. In this review, from the perspective of biological researcher, we summarize the powerful tools and strategies related to chemistry and chemical biology used in strigolactone area, covering analytical chemistry tools for isolation and structural elucidation, synthetic chemistry for structural elucidation and agricultural applications, chemical biology and biosynthetic strategies for functional characterization. Biosensors and probes used in monitoring strigolactone activity and signaling were also highlighted. Finally, we address current challenges and discuss future research perspectives, aiming to provoke more investigations on strigolactone biology and further boost their agricultural practices.
1 Introduction
The history of strigolactones (SLs) research began with the isolation of strigol from cotton root exudates, the first identified natural SL (Cook et al., 1966). Over the years, more than 35 natural SLs have been identified from various plant species, which are mostly been detected from root exudates or roots (Guercio et al., 2023; Daignan-Fornier et al., 2024). Based on their structure characters, these SLs are derived carotenoid pathways and can be divided into two groups or three types (Yoneyama et al., 2018b; Guercio et al., 2023) (Figure 1). Canonical SLs, such as Strigol and Orobanchol (Cook et al., 1966; Mori et al., 1999; Al-Babili and Bouwmeester, 2015), are characterized by a tricyclic lactone structure comprising ABC-rings linked to a butenolide group (D-ring) through an enol-ether bridge. Non-canonical SLs, on the other hand, lack the complete structure of ABC rings but retain the conserved D-ring moiety, which is crucial for their biological activities (Zwanenburg et al., 2009; Kim et al., 2014; Ueno et al., 2014; Umehara et al., 2015; Charnikhova et al., 2017; Xie et al., 2019).
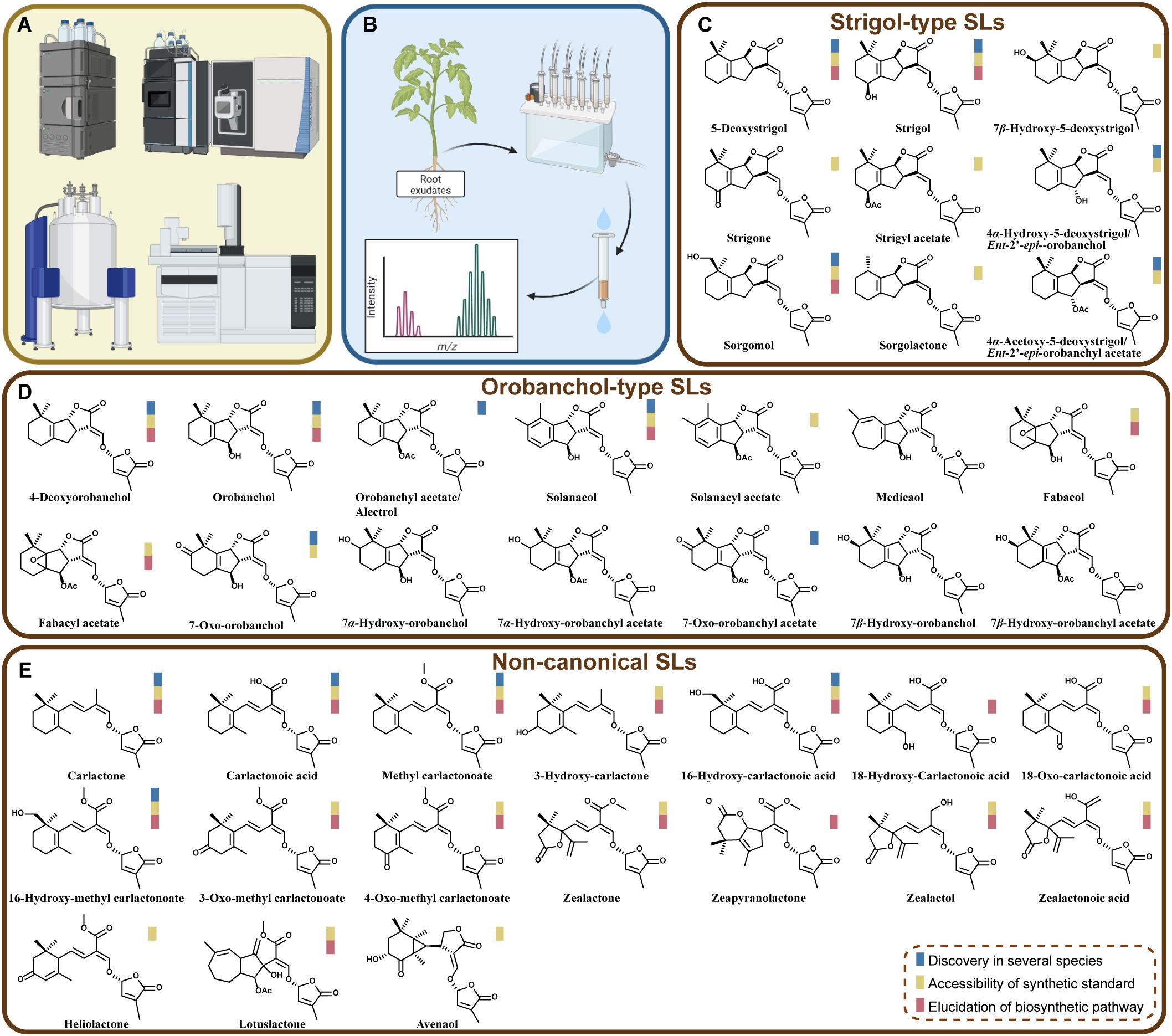
Figure 1. Analytical chemistry methods in advancing the identification of natural SLs. (A) Some main analytical chemistry instruments used in SL isolation and structural identification, including HPLC, LC-MS/MS, NMR, and GC-MS. (B) Scheme of workflow illustrating the extraction of root exudates, purification and detection. (C-E) Names and structures of natural SLs characterized from plants. On the basis of available literature, scope of their existence, the availability of synthetic standards and the progress of their biosynthesis pathways are indicated by three colored-boxes.
As mentioned, SLs were initially identified for their roles in stimulating the germination of parasitic weeds such as Striga and Orobanche (Cook et al., 1966; Parker, 2009; Westwood et al., 2010). Later in 2005, the positive biological function of SLs in promoting hyphal branching of arbuscular mycorrhizal fungi was uncovered, which further enhances nutrient uptake through symbiosis (Akiyama et al., 2005). Until 2008, SLs were recognized as key regulators of plant architecture by inhibiting shoot branching (Gomez-Roldan et al., 2008; Umehara et al., 2008). Later, more roles of SLs have been discovered, including responding to various biotic and abiotic stresses, by modulating plant growth and architecture (Brewer et al., 2013; Ruyter-Spira et al., 2013; Omoarelojie et al., 2019; Wu et al., 2022; Dun et al., 2023). For instance, in a recent study, role of SLs in safeguarding plants against abiotic stresses was uncovered, which is achieved by modulating stomatal activity, reducing transpirational water loss, enhancing nutrient uptake efficiency, and thereby strengthening overall plant resilience (Rhaman et al., 2024). Overall, these findings highlight SLs as an endogenous phytohormone and also signaling molecule in rhizosphere communications. The biosynthesis of SLs involves a core pathway starting from β-carotene and forming Carlactone (Matusova et al., 2005; Alder et al., 2012), which is catalyzed by three enzymes D27 (DWARF27), CCD7 (carotenoid cleavage dioxygenase 7), and CCD8 (carotenoid cleavage dioxygenase 8). After the production of carlactone, different structures of SLs can be biosynthesized by diversified branching pathways (Mashiguchi et al., 2020; Seto, 2023). In these steps, cytochrome P450 enzymes, methyltransferases, and other enzyme classes are involved in modifying the ABC-ring structures through a variety of reactions, including oxidations and methylations (Wu and Li, 2021; Yoda et al., 2021; Mashiguchi et al., 2022; Li et al., 2023; Kuijer et al., 2024; Li et al., 2024).
In the research lines of SLs, advances in chemical and chemical biology techniques, such as the rapid development of mass-spectrometric (MS) techniques and Nuclear Magnetic Resonance Spectroscopy (NMR), have greatly contributed to the discovery of new SLs and the novel genes/enzymes in their biosynthetic pathways (Charnikhova et al., 2017; Floková et al., 2020; Rial et al., 2020; Xie et al., 2021; Lailheugue et al., 2023; Li et al., 2023) (Figure 1). In recent years, driven by interdisciplinary approaches including analytical chemistry tools and chemical biology strategies, the isolation and detection of SLs have become more simplified and efficient, facilitating the identification of new SLs and functional characterization of related genes (Zhang et al., 2014; Wu et al., 2021; Li et al., 2023, 2024; Zhou et al., 2025) (Figure 2). The development of organic synthesis and synthetic biology has enabled the relatively large-scale production of SLs and their analogs, among which GR24 is the most widely known and used one in functional studies of SLs (Krasylenko et al., 2021; Wu et al., 2021; Pyrzanowska-Banasiak et al., 2023; Tian et al., 2024; Zhou et al., 2025). This also promote the design and application of suicidal germination inducers in combatting parasitic weeds. The emergence of biosensors and reporter systems combined with chemical probes designed to target SL receptors or biosynthetic enzymes, have opened new opportunities for dissecting the molecular mechanisms underlying SL-mediated processes (Tsuchiya et al., 2015; Samodelov et al., 2016; Chesterfield et al., 2020; Germain et al., 2022; Song et al., 2022; Wang et al., 2022a). In this review, from the perspective of biological researcher, we summarize the tools and strategies related to chemistry and chemical biology utilized in SL research, highlighting recent advancements in analytical methods and biosensor development. By providing a comprehensive overview of the latest findings, this review aims to guide future research at enhancing plant resilience against biotic and abiotic stresses, by using knowledge of phytohormones and signaling molecules.
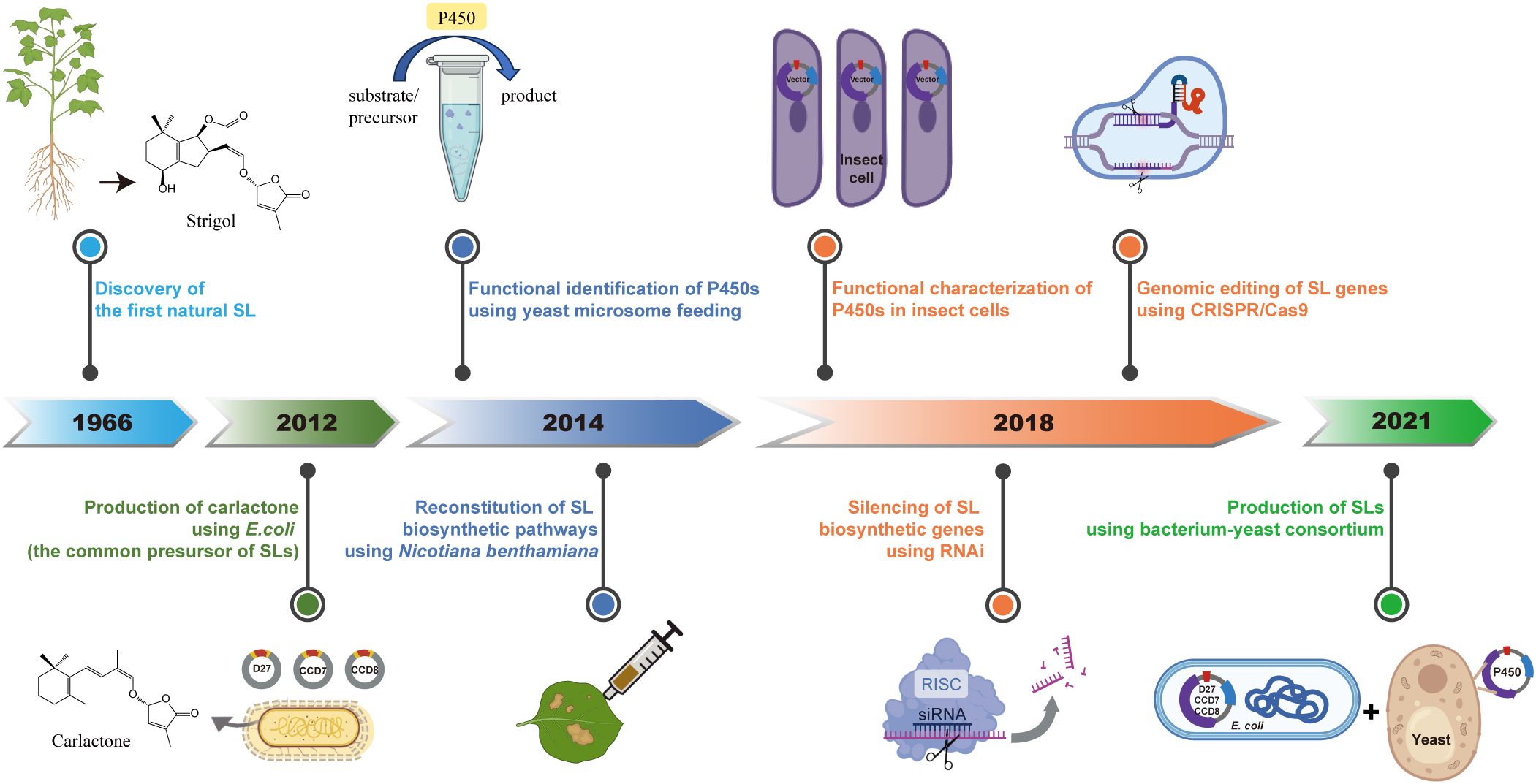
Figure 2. Milestones in SL research related to chemical biology and biosynthesis. This timeline illustrates a few key advances in SL research, from the discovery of Strigol in 1966, to modern engineering strategies. Highlights include the microbial production of Carlactone in E. coli in 2012, functional identification and reconstitution of SL biosynthetic enzymes in Nicotiana benthamiana in 2014, and the characterization of cytochrome P450s via yeast microsome feeding and insect cell systems. In 2018, RNAi-mediated silencing and CRISPR/Cas9-based genome editing enabled functional dissection of SL biosynthetic genes. More recently, a bacterium-yeast consortium has been established to produce SLs, representing a synthetic biology platform for scalable SL production.
2 Analytical chemistry tools in SL identification
High-Performance Liquid Chromatography (HPLC) has been the traditional method for SL separation, typically using C18 reversed-phase columns with mobile phases composed of water and organic solvents like methanol or acetonitrile (Halouzka et al., 2020) (Figure 1A). Besides, Ultra-High-Performance Liquid Chromatography (UHPLC) has become popular due to its higher separation efficiency and shorter analysis time (Floková et al., 2020). Coupled with MS, UHPLC-MS/MS, particularly in multiple reaction monitoring (MRM) mode, is now widely used for identification and quantification of SLs (Figure 1A)., achieving even attomolar detection limits (Rial et al., 2020) and the discovery of several new SLs (Charnikhova et al., 2017; Halouzka et al., 2020; Karniel et al., 2024; Li et al., 2024). This method could provide rather high sensitivity and specificity for SL detection. Most of natural SLs are typically analyzed in positive ion mode, with transitions of sodium adduct ions [M+H]+ or [M+Na]+ being commonly monitored (Floková et al., 2020; Halouzka et al., 2020), while a few SLs can be preferably detected in negative ion mode, such as Carlactonoic acid (Floková et al., 2020). The application of stable isotope-labeled analogs (e.g., [2H6]-5-deoxystrigol and [2H6]-2′-epi-5-deoxystrigol) as internal standards helps the correct for variations during extraction and ionization efficiency (Floková et al., 2020). It is noteworthy that there are enantiomers for natural SLs and chiral column showed great capability in separation of these isomers (Wang et al., 2022b).
NMR is another powerful tool for determining the detailed structure of SLs, especially for confirming the presence of chiral centers and functional groups (Figure 1A). It is particularly useful for structural elucidation of newly discovered SLs (Ćavar et al., 2015; Charnikhova et al., 2017; Wang and Bouwmeester, 2018; Floková et al., 2020; Li et al., 2023; Nomura et al., 2023). In addition, modern ambient techniques such as Direct Analysis in Real Time (DART) and Desorption Electrospray Ionization (DESI) offer other options for SL detection, which is faster and only require a few sample preparation steps (Halouzka et al., 2020). These techniques hold promise for rapid SL identification but require optimization to address issues like sample shrinkage and matrix effects (Halouzka et al., 2020).
Solid-phase extraction (SPE) is a widely used sample preparation technique (Poole, 2003; Khatibi et al., 2021) (Figure 1B). The broad polarity range of SLs and their low abundance in plant related samples pose significant challenges for precise detection. Nevertheless, the application of SPE helps alleviate matrix effects, crucial for improving the accuracy and sensitivity of SL quantification. Although the culture system or sample collection methods can be diverse, SPE shows attractive ability to concentrate and purify SLs from complex and relatively large volumes of biological matrices (mainly from root exudates, root tissue extracts, etc.). For instance, columns, such as C18 and HLB, have shown great performance and been applied in SL extraction in multiple plant species (López-Ráez et al., 2008; Floková et al., 2020).
3 Chemistry boosts the discovery and function analysis of SLs
The development and application of these powerful chemical tools have advanced the discovery of natural SLs from different plants (Figure 1C). According to our collected information from literature, at least 40 natural forms of SLs have been found. As mentioned, Strigol, belonging to “Strigol-type”, was the first discovered SL (Cook et al., 1966). Later on, the pictures of “Strigol-type” and “Orobanchol-type” SLs had been expanded due to the discovery of more structures. During this process, scientists had dug more into the characteristics of these natural SLs’ structures and stereochemistry (Flematti et al., 2016; Xie, 2016; Wang and Bouwmeester, 2018; Yoneyama et al., 2018b). So far, all natural SLs pose 2’ R orientation in D ring, which is essential for their biological activities, on inducing the germination of parasitic weeds or inducing the hyphal branching of symbiotic arbuscular mycorrhizal fungi (AMF) (Zwanenburg et al., 2009; Akiyama et al., 2010; Boyer et al., 2012).
It is noteworthy that in recent years (after 2012) quite several new structures from “non-canonical” group have been uncovered (Figure 1C). For instance, several maize SLs, including Zealactone, Zeapyranolactone, Zealactol, and Zealactonoic acid, were identified from a variety of maize lines (Charnikhova et al., 2017; Xie et al., 2017; Charnikhova et al., 2018; Li et al., 2023). Besides, Avenaol, Lotuslactone, and Heliolactone were discovered from root exudates of Helianthus annuus (sunflower), Lotus japonicus, and Avena strigosa, respectively (Kim et al., 2014; Ueno et al., 2014; Xie et al., 2019). It can be also noticed that several research groups have found more and more derivatives of SL precursors (i.e., Carlactone, Carlactonoic acid, and Methyl carlactonate) (Figure 1C).
In most of the cases, the natural SLs were extracted and detected from root exudates or root tissues. However, very recent, 16-Hydroxy-carlactonoic acid (16-OH-CLA) was identified to be a product by the conversion of CYP722A, by the use of a microbial consortium expression system (Wu et al., 2021; Zhou et al., 2025) (detailed description of this method is present in next section and Figure 2). This form of SL and its derivative methyl 16-Hydroxycarlactonoate (16-OH-MeCLA) were detected only in the shoot tissues of several seed plants, including Arabidopsis thaliana, poplar (Populus nigra × P. grandidentata) and pepper (Capsicum annuum), plum (Prunus mume), and Nelumbo nucifera. This SL shows bioactivity in suppressing axillary shoot branching, in a manner dependent on SL signaling (Zhou et al., 2025). Possibly more natural structures will be characterized from other plant tissues and more biological functions of these SLs could be explored in the coming future.
After the first discovery of phytohormonal function of SLs (Gomez-Roldan et al., 2008), more and more analogs or mimics of natural SLs have been designed and synthesized, which are widely used in agriculture applications. These include GR24 (the most widely applied and the most famous one) (Akiyama et al., 2005; Gomez-Roldan et al., 2008), 4-Br debranone (4BD) (Fukui et al., 2013), MPs (Methyl phenlactonoates) (Jamil et al., 2018), Nijmegen-1 (Mwakaboko and Zwanenburg, 2016; Kountche et al., 2019; Jamil et al., 2022), 2NOD (2-nitrodebranone (Li et al., 2021) and other synthetic compounds. The scope of their practices could be roughly divided into germination stimulants of parasitic weeds (Suzuki et al., 2022), crop growth/architecture regulators (Jamil et al., 2020), helper-molecules for greater stress tolerance (Bhoi et al., 2021; Qi et al., 2024). For instance, exogenous application of GR24 in several species suggested the potential of this compound in increasing drought resistance (Cao et al., 2024; Dong et al., 2024; Ge et al., 2024; Shu et al., 2024). In future, along with our better understanding of the activities based on specific groups/subsections of natural SLs and other phytohormones, more targeted and accurate designing of active forms of synthetic analogs/mimics could be achieved.
4 Chemical biology and biosynthetic strategies in SL functional characterization
Although the first SL was discovered in 1960s (Cook et al., 1966), elucidating the biosynthesis and functions had been a big challenge. With the rapid advancement of science and technology, integration of chemical biology and biosynthetic methods has provided new opportunities for tackling these issues and broaden our knowledge of SLs’ nature. Here we summarize some key discovery of this process.
In 2012, researchers elucidated the biosynthetic pathway of the SL precursor, Carlactone, by transferring three SL biosynthetic genes into Escherichia coli and supplying appropriate substrates (Alder et al., 2012). This discovery laid a foundation for SL biosynthesis research, as Carlactone is considered as the common precursor of all natural SLs. Subsequently, in 2014, through yeast microsome feeding experiments and the reconstruction of the SL synthesis pathway in Nicotiana benthamiana, the catalytic activity and function of some MAX1s (belonging to cytochrome P450 711A family) was uncovered (Abe et al., 2014; Zhang et al., 2014). This first identification of P450 involved in SL biosynthesis has inspired investigation on other members, which plant scientists are stilling working on. In 2018, utilizing RNA silencing technology, the role of tomato SlMAX1 in SL biosynthesis and plant growth was characterized (Zhang et al., 2018). Additionally, CRISPR-Cas9 technology enabled the targeted editing of rice CCD7 gene and analysis of its mutant showed the SL function in regulating plant height and tillering and further enhancing yield (Butt et al., 2018). Beyond yeast and plant expression system, the use of a baculovirus expression in insect cells has also confirmed the function of MAX1s across diverse plant species (Yoneyama et al., 2018a). These investigations provide a broader perspective on the biosynthesis and functional exploration of SLs.
The extremely low concentrations of SLs within plants have constrained both basic research and practical applications based on SLs. In 2021, a group developed a co-culture system of E. coli and yeast, establishing a microbial biosynthetic platform for the synthesis of various SLs (Wu et al., 2021). This innovative platform lays a solid foundation for the development of microbial production processes for SLs, marking a significant step toward their widespread application in agriculture and biotechnology.
5 Chemical probes and biosensors in SL activity and signaling research
Our understanding of SL signaling pathways has been significantly advanced through the development of chemical probes and biosensors. The classical probe, Yoshimulactone Green (YLG), identified ShHTLs as SL receptors in parasitic plant Striga hermonthica, with a Km value of 0.63 µM (Tsuchiya et al., 2015). Among the SLs tested in the competition hydrolysis activity assay, 5DS showed the strongest IC50 value of 0.44 µM, consistent with its higher activity in inducing Striga germination. Later, aryloxyacetyl piperazines were discovered as potential suicidal stimulants for parasitic plants at extremely low concentrations (10-8 to 10-17 M) (Wang et al., 2022a). Another group designed profluorescent SL Guillaume Clavé (GC) probes, with coumarin-based probes being highly bioactive in pea and a resorufin probe effective in moss, while YLG was less effective (Germain et al., 2022). These findings highlight the importance of SL probe specificity across different species. Another tool, genetically encoded biosensors for monitoring SL activity and signaling pathways, has been designed, among which StrigoQuant and Strigo-D2 are two notable examples (Samodelov et al., 2016; Song et al., 2022). StrigoQuant is designed to quantify SL activity and specificity. It employs a luciferase-based reporter system to detect the degradation of the SMXL6 protein, a key target in the SL signaling pathway. Among the tested SLs, 5-Deoxystrigol (5DS) was the most sensitive form, even at a low concentration of 100 fM, which is comparable to the activity observed in YLG-based assays. This provides a quantitative tool for studying SL activity in plana. Strigo-D2 is also based on SMXL6, which could monitor SL signaling patterns at cellular resolution. It was shown that different cell types respond to SLs with varying kinetics. Based on the SL receptors DAD2 from Petunia hybrida and HTL7 from Striga hermonthica, two fluorescent biosensors were constructed (Chesterfield et al., 2020). The sensitivity of both can reach nanomolar level, allowing direct detection of SLs in vitro and in vivo. Together, these biosensors provide valuable tools for studying SL signaling in various plant contexts, offering insights into the complex dynamics and specificities of SL activity.
6 Challenges and future directions
In summary, in the past decades, analytical chemistry tools such as UHPLC-MS/MS, SPE, and ambient MS techniques have significantly advanced the detection and identification of SLs (Figure 1). These methods provide the sensitivity and specificity required to study SLs in complex biological samples, paving the way for future research in plant biology and agriculture. The development of highly sensitive and specific analytical methods continues to be a focus in SL research. Combining UHPLC with high-resolution mass spectrometry (HR-MS) and optimizing sample preparation protocols will further enhance the detection and quantification of SLs and their derivatives in various plant species (Wu and Li, 2021; Zhou et al., 2025). Additionally, the application of ambient MS techniques for spatial profiling of SLs in different plant tissues is an emerging area with significant potential (Halouzka et al., 2020). Moreover, techniques for in situ detection of natural forms of SLs without complicated extraction and purification steps would be highly needed, although a few profluorescence probes and fluorescence-based biosensor have been developed for binding SL receptors (Tsuchiya et al., 2015; Samodelov et al., 2016; Chesterfield et al., 2020; Germain et al., 2022; Song et al., 2022; Wang et al., 2022a). We have to admit that this is quite challenging due to the unique characteristics of SLs, including their extremely low abundance, structural complexity, and relative instability.
As mentioned, several chemical biology has developed rapidly and boosted the discovery of SL biosynthetic pathways and also their biological functions (Figure 2). It could be noticed that the speed of discovering new enzymes/genes shows relatively slower tendency although many of the biosynthetic pathways have already been uncovered. Currently and in the coming future, with the emergence of omics and vast amount of biological big data linked with it, computational screening of putative structures and predication of candidate biosynthesis genes in phytohormone would provide us more and more accurate evidence. This could show guidance for wet-experimental designs and significantly increase the efficiency. Another trend is the combination of multiple disciplines in phytohormone research. For instance, crop populations offer us the genetic material foundation in searching new compounds; chemical tools help in standard synthesis, structural identification and mimics designing; chemical biology and synthetic biology reveal powerful ability in functional analysis and agronomic application. With all these chemistry tools, our understanding and practices based on it would be greatly enriched and expanded.
Author contributions
QZ: Writing – review & editing, Writing – original draft. CN: Visualization, Funding acquisition, Writing – original draft. LF: Writing – original draft, Visualization. MD: Writing – original draft. XL: Funding acquisition, Writing – original draft, Writing – review & editing. BK: Writing – original draft, Funding acquisition, Writing – review & editing. CL: Conceptualization, Supervision, Writing – review & editing, Funding acquisition, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This project was supported by National Key Research and Development Program of China (2023YFD1401100), Outstanding Youth Project of the Natural Science Foundation of Hunan Province (2024JJ2013), National Natural Science Foundation of China (32400258), Postgraduate Scientific Research Innovation Project of Hunan Province (CX20240386), CNTC Technology Project (110202201012(JY-12), 110202401009(JY-09) & 110202404004), and Beijing Life Science Academy Project (2023200CC0270).
Conflict of interest
Authors XL, BK were employed by China Tobacco Hunan Industrial Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abe, S., Sado, A., Tanaka, K., Kisugi, T., Asami, K., Ota, S., et al. (2014). Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. PNAS 111, 18084–18089. doi: 10.1073/pnas.1410801111
Akiyama, K., Matsuzaki, K.-i., and Hayashi, H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827. doi: 10.1038/nature03608
Akiyama, K., Ogasawara, S., Ito, S., and Hayashi, H. (2010). Structural requirements of strigolactones for hyphal branching in AM fungi. Plant Cell Physiol. 51, 1104–1117. doi: 10.1093/pcp/pcq058
Al-Babili, S. and Bouwmeester, H. J. (2015). Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 66, 161–186. doi: 10.1146/annurev-arplant-043014-114759
Alder, A., Jamil, M., Marzorati, M., Bruno, M., Vermathen, M., Bigler, P., et al. (2012). The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335, 1348–1351. doi: 10.1126/science.1218094
Bhoi, A., Yadu, B., Chandra, J., and Keshavkant, S. (2021). Contribution of strigolactone in plant physiology, hormonal interaction and abiotic stresses. Planta 254 (2), 28. doi: 10.1007/s00425-021-03678-1
Boyer, F. D., Germain, A. D., Pillot, J. P., Pouvreau, J. B., Chen, V. X., Ramos, S., et al. (2012). Structure-activity relationship studies of strigolactone-related molecules for branching inhibition in garden pea: molecule design for shoot branching. Plant Physiol. 159, 1524–1544. doi: 10.1104/pp.112.195826
Brewer, P. B., Koltai, H., and Beveridge, C. A. (2013). Diverse roles of strigolactones in plant development. Mol. Plant 6, 18–28. doi: 10.1093/mp/sss130
Butt, H., Jamil, M., Wang, J. Y., Al-Babili, S., and Mahfouz, M. (2018). Engineering plant architecture via CRISPR/Cas9-mediated alteration of strigolactone biosynthesis. BMC Plant Biol. 18 (1), 174. doi: 10.1186/s12870-018-1387-1
Cao, L., Zhang, S., Feng, L., Qiang, B., Ma, W., Cao, S., et al. (2024). Metabolic pathways regulated by strigolactones foliar spraying enhance osmoregulation and antioxidant defense in drought-prone soybean. BMC Plant Biol. 24 (1), 980. doi: 10.1186/s12870-024-05663-8
Ćavar, S., Zwanenburg, B., and Tarkowski, P. (2015). Strigolactones: occurrence, structure, and biological activity in the rhizosphere. Phytochem. Rev. 14, 691–711. doi: 10.1007/s11101-014-9370-4
Charnikhova, T. V., Gaus, K., Lumbroso, A., Sanders, M., Vincken, J. P., De Mesmaeker, A., et al. (2017). Zealactones. Novel natural strigolactones from maize. Phytochemistry 137, 123–131. doi: 10.1016/j.phytochem.2017.02.010
Charnikhova, T. V., Gaus, K., Lumbroso, A., Sanders, M., Vincken, J. P., De Mesmaeker, A., et al. (2018). Zeapyranolactone - A novel strigolactone from maize. Phytochem. Lett. 24, 172–178. doi: 10.1016/j.phytol.2018.01.003
Chesterfield, R. J., Whitfield, J. H., Pouvreau, B., Cao, D., Alexandrov, K., Beveridge, C. A., et al. (2020). Rational design of novel fluorescent enzyme biosensors for direct detection of strigolactones. ACS SYNTHETIC. Biol. 9, 2107–2118. doi: 10.1021/acssynbio.0c00192
Cook, C. E., Whichard, L. P., Turner, B., Wall, M. E., and Egley, G. H. (1966). Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154, 1189–1190. doi: 10.1126/science.154.3753.1189
Daignan-Fornier, S., Keita, A., and Boyer, F.-D. (2024). Chemistry of strigolactones, key players in plant communication. Chembiochem 25 (12), e202400133. doi: 10.1002/cbic.202400133
Dong, J., Ding, C., Chen, H., Fu, H., Pei, R., Shen, F., et al. (2024). Functions of exogenous strigolactone application and strigolactone biosynthesis genes GhMAX3/GhMAX4b in response to drought tolerance in cotton (Gossypium hirsutum L.). BMC Plant Biol. 24 (1), 1008. doi: 10.1186/s12870-024-05726-w
Dun, E. A., Brewer, P. B., Gillam, E. M. J., and Beveridge, C. A. (2023). Strigolactones and shoot branching: what is the real hormone and how does it work? Plant Cell Physiol. 64, 967–983. doi: 10.1093/pcp/pcad088
Flematti, G. R., Scaffidi, A., Waters, M. T., and Smith, S. M. (2016). Stereospecificity in strigolactone biosynthesis and perception. Planta 243, 1361–1373. doi: 10.1007/s00425-016-2523-5
Floková, K., Shimels, M., Andreo Jimenez, B., Bardaro, N., Strnad, M., Novák, O., et al. (2020). An improved strategy to analyse strigolactones in complex sample matrices using UHPLC–MS/MS. Plant Methods 16, 125. doi: 10.1186/s13007-020-00669-3
Fukui, K., Ito, S., and Asami, T. (2013). Selective mimics of strigolactone actions and their potential use for controlling damage caused by root parasitic weeds. Mol. Plant 6, 88–99. doi: 10.1093/mp/sss138
Ge, Q., Zhang, Y., Wu, J., Wei, B., Li, S., Nan, H., et al. (2024). Exogenous strigolactone alleviates post-waterlogging stress in grapevine. Plant Physiol. Biochem. 216, 109124. doi: 10.1016/j.plaphy.2024.109124
Germain, A. D., Clave, G., Schouveiler, P., Pillot, J. P., Singh, A. V., Chevalier, A., et al. (2022). Expansion of the strigolactone profluorescent probes repertory: the right probe for the right application. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.887347
Gomez-Roldan, V., Fermas, S., Brewer, P. B., Puech-Pagès, V., Dun, E. A., Pillot, J.-P., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455, 189–194. doi: 10.1038/nature07271
Guercio, A. M., Palayam, M., and Shabek, N. (2023). Strigolactones: diversity, perception, and hydrolysis. Phytochem. Rev. 22, 339–359. doi: 10.1007/s11101-023-09853-4
Halouzka, R., Zeljković, S. Ć., Klejdus, B., and Tarkowski, P. (2020). Analytical methods in strigolactone research. Plant Methods 16 (1), 76. doi: 10.1186/s13007-020-00616-2
Jamil, M., Kountche, B. A., Haider, I., Guo, X. J., Ntui, V. O., Jia, K. P., et al. (2018). Methyl phenlactonoates are efficient strigolactone analogs with simple structure. J. Exp. Bot. 69, 2319–2331. doi: 10.1093/jxb/erx438
Jamil, M., Kountche, B. A., Wang, J. Y., Haider, I., Jia, K. P., Takahashi, I., et al. (2020). A new series of carlactonoic acid based strigolactone analogs for fundamental and applied research. Front. Plant Sci. 11, 434. doi: 10.3389/fpls.2020.00434
Jamil, M., Wang, J. Y., Yonli, D., Patil, R. H., Riyazaddin, M., Gangashetty, P., et al. (2022). A new formulation for strigolactone suicidal germination agents, towards successful Striga management. Plants 11 (6), 808. doi: 10.3390/plants11060808
Karniel, U., Koch, A., Nun, N. B., Zamir, D., and Hirschberg, J. (2024). Tomato mutants reveal root and shoot strigolactone involvement in branching and broomrape resistance. Plants 13, (11), 1554. doi: 10.3390/plants13111554
Khatibi, S. A., Hamidi, S., and Siahi-Shadbad, M. R. (2021). Current trends in sample preparation by solid-phase extraction techniques for the determination of antibiotic residues in foodstuffs: a review. Crit. Rev. Food Sci. Nutr. 61, 3361–3382. doi: 10.1080/10408398.2020.1798349
Kim, H. I., Kisugi, T., Khetkam, P., Xie, X. N., Yoneyama, K., Uchida, K., et al. (2014). Avenaol, a germination stimulant for root parasitic plants from Avena strigosa. Phytochemistry 103, 85–88. doi: 10.1016/j.phytochem.2014.03.030
Kountche, B. A., Jamil, M., Yonli, D., Nikiema, M. P., Blanco-Ania, D., Asami, T., et al. (2019). Suicidal germination as a control strategy for Striga hermonthica (Benth.) in smallholder farms of sub-Saharan Africa. Plants People Planet. 1, 107–118. doi: 10.1002/ppp3.32
Krasylenko, Y., Komis, G., Hlynska, S., Vavrdová, T., Ovecka, M., Pospísil, T., et al. (2021). GR24, a synthetic strigolactone analog, and light affect the organization of cortical microtubules in Arabidopsis hypocotyl cells. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.675981
Kuijer, H. N. J., Wang, J. Y., Bougouffa, S., Abrouk, M., Jamil, M., Incitti, R., et al. (2024). Chromosome-scale pearl millet genomes reveal CLAMT1b as key determinant of strigolactone pattern and Striga susceptibility. Nat. Commun. 15 (1), 6906. doi: 10.1038/s41467-024-51189-w
Lailheugue, V., Merlin, I., Boutet, S., Perreau, F., Pouvreau, J.-B., Delgrange, S., et al. (2023). Vitislactone, a non-canonical strigolactone exudated by grapevine rootstocks in response to nitrogen starvation. Phytochemistry 215, 113837. doi: 10.1016/j.phytochem.2023.113837
Li, C., Dong, L., Durairaj, J., Guan, J. C., Yoshimura, M., Quinodoz, P., et al. (2023). Maize resistance to witchweed through changes in strigolactone biosynthesis. Science 379, 94–99. doi: 10.1126/science.abq4775
Li, C., Haider, I., Wang, J. Y., Quinodoz, P., Suarez Duran, H. G., Méndez, L. R., et al. (2024). OsCYP706C2 diverts rice strigolactone biosynthesis to a noncanonical pathway branch. Sci. Adv. 10, eadq3942. doi: 10.1126/sciadv.adq3942
Li, S. H., Li, Y. W., Chen, L. H., Zhang, C., Wang, F., Li, H. O., et al. (2021). Strigolactone mimic 2-nitrodebranone is highly active in Arabidopsis growth and development. Plant J. 107, 67–76. doi: 10.1111/tpj.15274
López-Ráez, J. A., Charnikhova, T., Gómez-Roldán, V., Matusova, R., Kohlen, W., De Vos, R., et al. (2008). Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol. 178, 863–874. doi: 10.1111/j.1469-8137.2008.02406.x
Mashiguchi, K., Seto, Y., Onozuka, Y., Suzuki, S., Takemoto, K., Wang, Y., et al. (2022). A carlactonoic acid methyltransferase that contributes to the inhibition of shoot branching in Arabidopsis. PNAS 119 (14), e2111565119. doi: 10.1073/pnas.2111565119
Mashiguchi, K., Seto, Y., and Yamaguchi, S. (2020). Strigolactone biosynthesis, transport and perception. Plant J. 105, 335–350. doi: 10.1111/tpj.15059
Matusova, R., Rani, K., Verstappen, F. W. A., Franssen, M. C. R., Beale, M. H., and Bouwmeester, H. J. (2005). The strigolactone germination stimulants of the plant-parasitic striga and orobanche spp. are derived from the carotenoid pathway. Plant Physiol. 139, 920–934. doi: 10.1104/pp.105.061382
Mori, K., Matsui, J., Yokota, T., Sakai, H., Bando, M., and Takeuchi, Y. (1999). Structure and synthesis of orobanchol, the germination stimulant for Orobanche minor. Tetrahedron. Lett. 40, 943–946. doi: 10.1016/S0040-4039(98)02495-2
Mwakaboko, A. S. and Zwanenburg, B. (2016). Strigolactone analogues with a D-ring modified at C-2. Eur. J. Org. Chem. 2016, 3495–3499. doi: 10.1002/ejoc.201600576
Nomura, T., Seto, Y., and Kyozuka, J. (2023). Unveiling the complexity of strigolactones: exploring structural diversity, biosynthesis pathways, and signaling mechanisms. J. Exp. Bot. 75, 1134–1147. doi: 10.1093/jxb/erad412
Omoarelojie, L. O., Kulkarni, M. G., Finnie, J. F., and Van Staden, J. (2019). Strigolactones and their crosstalk with other phytohormones. Ann. Bot. 124, 749–767. doi: 10.1093/aob/mcz100
Parker, C. (2009). Observations on the current status of Orobanche and Striga problems worldwide. Pest Manage. Sci. 65, 453–459. doi: 10.1002/ps.1713
Poole, C. F. (2003). New trends in solid-phase extraction. TrAC. Trends Anal. Chem. 22, 362–373. doi: 10.1016/s0165-9936(03)00605-8
Pyrzanowska-Banasiak, A., Tumer, T. B., Bukowska, B., and Krokosz, A. (2023). A multifaceted assessment of strigolactone GR24 and its derivatives: from anticancer and antidiabetic activities to antioxidant capacity and beyond. Front. Mol. Biosci. 10. doi: 10.3389/fmolb.2023.1242935
Qi, J., Mao, Y., Cui, J., Lu, X., Xu, J., Liu, Y., et al. (2024). The role of strigolactones in resistance to environmental stress in plants. Physiol. Plant 176 (4), e14419. doi: 10.1111/ppl.14419
Rhaman, M. S., Karim, M. M., Sagar, A., Asaduzzaman, M., Ye, W., and Brestic, M. (2024). Phytohormone strigolactone: involvement in guard cell signaling and abiotic stress tolerance in plants. J. Plant Growth Regul. 43, 4621–4634. doi: 10.1007/s00344-024-11421-9
Rial, C., Varela, R. M., Molinillo, J. M. G., Duran, A. G., and Macias, F. A. (2020). Quantification of strigolactones. Methods Mol. Biol. 2083, 199–208. doi: 10.1007/978-1-4939-9952-1_15
Ruyter-Spira, C., Al-Babili, S., van der Krol, S., and Bouwmeester, H. (2013). The biology of strigolactones. Trends Plant Sci. 18, 72–83. doi: 10.1016/j.tplants.2012.10.003
Samodelov, S. L., Beyer, H. M., Guo, X. J., Augustin, M., Jia, K. P., Baz, L., et al. (2016). StrigoQuant: A genetically encoded biosensor for quantifying strigolactone activity and specificity. Sci. Adv. 2 (11), e1601266. doi: 10.1126/sciadv.1601266
Seto, Y. (2023). Latest knowledge on strigolactone biosynthesis and perception. Biosci. Biotechnol. Biochem. 88, 1–7. doi: 10.1093/bbb/zbad150
Shu, H. Y., Xu, K. J., Li, X. R., Liu, J. C., Altaf, M. A., Fu, H. Z., et al. (2024). Exogenous strigolactone enhanced the drought tolerance of pepper (Capsicum chinense) by mitigating oxidative damage and altering the antioxidant mechanism. Plant Cell Rep. 43. doi: 10.1007/s00299-024-03196-w
Song, C. Z., Zhao, J., Guichard, M., Shi, D. B., Grossmann, G., Schmitt, C., et al. (2022). Strigo-D2-a bio-sensor for monitoring spatio-temporal strigolactone signaling patterns in intact plants. Plant Physiol. 188, 97–110. doi: 10.1093/plphys/kiab504
Suzuki, T., Kuruma, M., and Seto, Y. (2022). A new series of strigolactone analogs derived from cinnamic acids as germination inducers for root parasitic plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.843362
Tian, H. Y., Tang, B. X., Fan, W. W., Pan, Z. Y., Peng, J. T., Wang, Y. X., et al. (2024). The role of strigolactone analog (GR24) in endogenous hormone metabolism and hormone-related gene expression in tobacco axillary buds. Plant Cell Rep. 43 (1), 21. doi: 10.1007/s00299-023-03081-y
Tsuchiya, Y., Yoshimura, M., Sato, Y., Kuwata, K., Toh, S., Holbrook-Smith, D., et al. (2015). Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349, 864–868. doi: 10.1126/science.aab3831
Ueno, K., Furumoto, T., Umeda, S., Mizutani, M., Takikawa, H., Batchvarova, R., et al. (2014). Heliolactone, a non-sesquiterpene lactone germination stimulant for root parasitic weeds from sunflower. Phytochemistry 108, 122–128. doi: 10.1016/j.phytochem.2014.09.018
Umehara, M., Cao, M. M., Akiyama, K., Akatsu, T., Seto, Y., Hanada, A., et al. (2015). Structural Requirements of strigolactones for shoot branching inhibition in Rice and Arabidopsis. Plant Cell Physiol. 56, 1059–1072. doi: 10.1093/pcp/pcv028
Umehara, M., Hanada, A., Yoshida, S., Akiyama, K., Arite, T., Takeda-Kamiya, N., et al. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200. doi: 10.1038/nature07272
Wang, Y. and Bouwmeester, H. J. (2018). Structural diversity in the strigolactones. J. Exp. Bot. 69, 2219–2230. doi: 10.1093/jxb/ery091
Wang, Y. T., Durairaj, J., Durán, H. G. S., van Velzen, R., Flokova, K., Liao, C. Y., et al. (2022b). The tomato cytochrome P450 CYP712G1 catalyses the double oxidation of orobanchol en route to the rhizosphere signalling strigolactone, solanacol. New Phytol. 235, 1884–1899. doi: 10.1111/nph.18272
Wang, D., Pang, Z., Yu, H., Thiombiano, B., Walmsley, A., Yu, S., et al. (2022a). Probing strigolactone perception mechanisms with rationally designed small-molecule agonists stimulating germination of root parasitic weeds. Nat. Commun. 13 (1), 3987. doi: 10.1038/s41467-022-31710-9
Westwood, J. H., Yoder, J. I., Timko, M. P., and dePamphilis, C. W. (2010). The evolution of parasitism in plants. Trends Plant Sci. 15, 227–235. doi: 10.1016/j.tplants.2010.01.004
Wu, F., Gao, Y., Yang, W., Sui, N., and Zhu, J. (2022). Biological functions of strigolactones and their crosstalk with other phytohormones. Front. Plant Sci. 13, 821563. doi: 10.3389/fpls.2022.821563
Wu, S. and Li, Y. (2021). A unique sulfotransferase-involving strigolactone biosynthetic route in sorghum. Front. Plant Sci. 12, 793459. doi: 10.3389/fpls.2021.793459
Wu, S., Ma, X., Zhou, A., Valenzuela, A., Zhou, K., and Li, Y. (2021). Establishment of strigolactone-producing bacterium-yeast consortium. Sci. Adv. 7 (38), eabh4048. doi: 10.1126/sciadv.abh4048
Xie, X. (2016). Structural diversity of strigolactones and their distribution in the plant kingdom. J. Pestic. Sci. 41, 175–180. doi: 10.1584/jpestics.J16-02
Xie, X. N., Kisugi, T., Yoneyama, K., Nomura, T., Akiyama, K., Uchida, K., et al. (2017). Methyl zealactonoate, a novel germination stimulant for root parasitic weeds produced by maize. J. Pestic. Sci. 42, 58–61. doi: 10.1584/jpestics.D16-103
Xie, X. N., Mori, N., Yoneyama, K., Nomura, T., Uchida, K., Yoneyama, K., et al. (2019). Lotuslactone, a non-canonical strigolactone from Lotus japonicus. Phytochemistry 157, 200–205. doi: 10.1016/j.phytochem.2018.10.034
Xie, X., Yoneyama, K., Nomura, T., and Yoneyama, K. (2021). Evaluation and quantification of natural strigolactones from root exudates. Methods Mol. Biol. 2309, 3–12. doi: 10.1007/978-1-0716-1429-7_1
Yoda, A., Mori, N., Akiyama, K., Kikuchi, M., Xie, X. N., Miura, K., et al. (2021). Strigolactone biosynthesis catalyzed by cytochrome P450 and sulfotransferase in sorghum. New Phytol. 232, 1999–2010. doi: 10.1111/nph.17737
Yoneyama, K., Mori, N., Sato, T., Yoda, A., Xie, X., Okamoto, M., et al. (2018a). Conversion of carlactone to carlactonoic acid is a conserved function of MAX1 homologs in strigolactone biosynthesis. New Phytol. 218, 1522–1533. doi: 10.1111/nph.15055
Yoneyama, K., Xie, X. N., Yoneyama, K., Kisugi, T., Nomura, T., Nakatani, Y., et al. (2018b). Which are the major players, canonical or non-canonical strigolactones? J. Exp. Bot. 69, 2231–2239. doi: 10.1093/jxb/ery090
Zhang, Y. X., Cheng, X., Wang, Y. T., Díez-Simón, C., Flokova, K., Bimbo, A., et al. (2018). The tomato MAX1 homolog, SlMAX1, is involved in the biosynthesis of tomato strigolactones from carlactone. New Phytol. 219, 297–309. doi: 10.1111/nph.15131
Zhang, Y., van Dijk, A. D. J., Scaffidi, A., Flematti, G. R., Hofmann, M., Charnikhova, T., et al. (2014). Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat. Chem. Biol. 10, 1028–1033. doi: 10.1038/nchembio.1660
Zhou, A., Kane, A., Wu, S., Wang, K., Santiago, M., Ishiguro, Y., et al. (2025). Evolution of interorganismal strigolactone biosynthesis in seed plants. Science 387, eadp0779. doi: 10.1126/science.adp0779
Keywords: strigolactone, chemistry, chemical biology, biosynthesis, root exudate, synthetic biology
Citation: Zhou Q, Niu C, Feng L, Dong M, Li X, Kong B and Li C (2025) Chemistry and chemical biology tools contributing to the discovery and functional characterization of strigolactones. Front. Plant Sci. 16:1618437. doi: 10.3389/fpls.2025.1618437
Received: 26 April 2025; Accepted: 29 May 2025;
Published: 18 June 2025.
Edited by:
Guodong Wang, Shaanxi Normal University, ChinaReviewed by:
Mohammad Saidur Rhaman, Bangladesh Agricultural University, BangladeshCopyright © 2025 Zhou, Niu, Feng, Dong, Li, Kong and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoxu Li, bGl4eEBibHNhLmNvbS5jbg==; Bo Kong, a29uZ2JvaG56eUAxNjMuY29t; Changsheng Li, Y2hhbmdzaGVuZ2xpQGhudS5lZHUuY24=
 Qian Zhou1
Qian Zhou1 Xiaoxu Li
Xiaoxu Li Changsheng Li
Changsheng Li