- Department of Biology, Williams College, Williamstown, MA, United States
Plants synthesize a diverse array of specialized metabolites that contribute to plant development, growth, protection from biotic and abiotic stressors, and attracting pollinators and seed dispersers. Specialized metabolites are often derived from primary metabolites, such as amino acids, but also can be redirected from intermediates in primary metabolic pathways. In the L-tryptophan (Trp) biosynthetic pathway, the intermediate anthranilate is siphoned away to synthesize volatiles and specialized metabolites. Methyltransferases can produce the O-methyl ester of anthranilate, a grape aroma volatile produced in species such as grapevine, strawberry, citrus, maize, and soybean. O-Methyl anthranilate serves context-dependent roles in attracting insects and deterring herbivores. Methylation at the amine generates N-methyl anthranilate, a precursor for N-methyl anthranilate esters in citrus and antimicrobial avenacins in black oat. This Mini Review explores the regulation of anthranilate within the context of the Trp pathway and its contributions to the biosynthesis of anthranilate-containing volatiles and specialized metabolites. Also highlighted are the roles of anthranilates in plant defensive metabolism and the substrate specificity of anthranilate-using enzymes, as well as unanswered questions about the synthesis, transport, and physiological role of anthranilates.
1 Introduction
Plants synthesize a wide array of small molecules that aid in chemical defense against pathogenic microorganisms and herbivores (Maeda and Fernie, 2021). These molecules are typically secondary metabolites that are built from primary metabolites, such as amino acids, fatty acids, isoprene, nucleic acids, or intermediates in primary biosynthetic pathways (Jacobowitz and Weng, 2020). Some of the secondary metabolites produced in leaves, flowers, and roots are emitted as volatiles. These volatiles can be synthesized from five-carbon isoprene units, fatty acids, aromatic rings, or amino acids (Dudareva et al., 2013). Volatiles and secondary metabolites serve a number of roles in plants, namely in mediating interactions between plants and herbivores, pollinators, seed dispersers, and microorganisms (Schuman, 2023).
All plants synthesize anthranilate as an intermediate in the L-tryptophan (Trp) biosynthetic pathway, and anthranilate and its derivatives (i.e., anthranilates) have multiple fates in plants (Figure 1). Unlike mammals, plants have the enzymatic machinery to synthesize all 20 amino acids, and Trp is an aromatic amino acid that is essential for the synthesis of proteins, the growth hormone auxin, niacin (vitamin B3), and specialized metabolites like indole glucosinolates in the mustard family (Brassicaceae) and the monoterpene indole alkaloids vinblastine and vincristine in Madagascar periwinkle (Catharanthus roseus) (Radwanski and Last, 1995; Sønderby et al., 2010; Maeda and Dudareva, 2012; Caputi et al., 2018; Morffy and Strader, 2020). Despite the importance of Trp biosynthesis in both primary and secondary metabolism, the Trp pathway enzyme that acts on anthranilate, anthranilate phosphoribosyltransferase, was only recently biochemically characterized (Li et al., 2023).
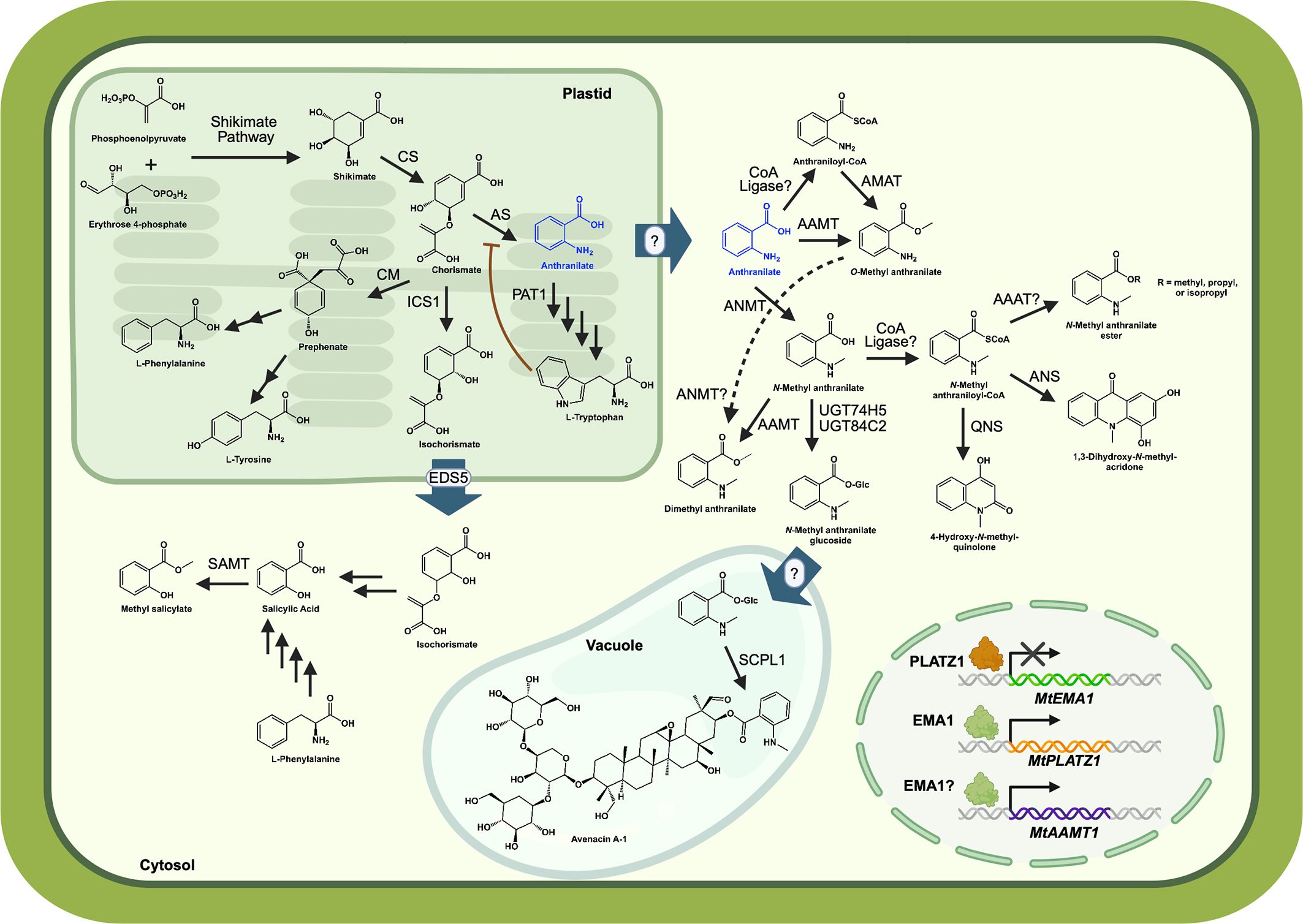
Figure 1. Anthranilate branchpoint in plants. Anthranilate (blue) is synthesized in the plastid as part of the tryptophan biosynthesis pathway, and is likely transported to the cytosol where it serves as a precursor to volatiles and specialized metabolites. Trp inhibits some anthranilate synthases (AS), denoted by the red line. The transport protein (blue arrow) is known for isochorismate, but not for anthranilate. Expression of the Medicago truncatula AAMT1 (anthranilate methyltransferase) is mediated by EMA1 (emission of methyl anthranilate 1), while PLATZ1 (plant AT-rich sequence and zinc-binding 1) represses the expression of EMA1. CS, chorismate synthase; PAT1, anthranilate phosphoribosyltransferase; CM, chorismate mutase; ICS1, isochorismate synthase 1; EDS5, enhanced disease susceptibility 5; SAMT, salicylic acid methyltransferase; ANMT, anthranilate N-methyltransferase; AMAT, anthraniloyl-CoA:methanol acyltransferase; AAAT, anthraniloyl-CoA:alcohol acyltransferase; UGT, UDP-dependent glycosyltransferase; QNS, quinolone synthase; ANS, acridone synthase; SCPL1, serine carboxypeptidase-like acyl transferase. Salicylic acid biosynthesis (Lefevere et al., 2020) and avenacin biosynthesis and localization (Orme et al., 2019) are described in more detail in recent publications. Figure was prepared using BioRender.
In at least 19 plant families, anthranilate is methylated to form a volatile ester that is responsible for grape aroma (Knudsen et al., 2006). Plant volatiles like O-methyl anthranilate (O-MeAA) are released following mechanical damage and function in mediating defense against predators. In strawberries (Frageria spp.), O-MeAA attracts Drosophila suzukii pests that preferentially oviposit on high concentrations of O-MeAA; however, O-MeAA has a concentration-dependent effect on embryo lethality (Bracker et al., 2020). In maize, an O-MeAA-containing blend of volatiles is emitted upon insect-induced tissue damage, which attracts wasps that parasitize the insect (Turlings et al., 1990; von Merey et al., 2013). In plants that don’t synthesize O-MeAA, exogenous application of the volatile on crops is effective in attracting natural enemies of insect herbivores to prevent further damage (Simpson et al., 2011). O-MeAA also functions as an effective bird repellant (Avery et al., 1995). Anthranilate methyltransferases responsible for generating O-MeAA were recently identified in Vitis spp. (grapevine), and biochemical investigations have enabled the identification of key residues that confer anthranilate activity in these enzymes (Figure 1) (Fallon et al., 2024).
Plants such as black oat (Avena strigosa) and members of the Rutaceae family, including Mexican orange blossom (Choisya ternata Kunth) and common rue (Ruta graveolens), methylate the amine of anthranilate (Figure 1) (Rohde et al., 2008; Mugford et al., 2013). In black oat, N-methyl anthranilate (N-MeAA) is incorporated into the chemical structure of avenacins, which are anti-microbial small molecules that are synthesized in root epidermal cells (Papadopoulou et al., 1999). In Rutaceae species, O-methyl esters of N-MeAA, such as dimethyl anthranilate (DiMeAA), exhibit pain-relieving activity in mice (Pinheiro et al., 2014); however, their role in mediating plant responses to biotic stress remains unclear.
This Mini-Review summarizes recent investigations of anthranilate-using enzymes in primary and secondary metabolism, as well as the substrate specificity and regulation of these enzymes. Additionally, this review highlights the need for future studies to increase our understanding of the regulation, localization, and transport of anthranilates, as well as their contributions to plant physiology.
2 Anthranilate is an intermediate in tryptophan biosynthesis
Until recently, much of what is known about Trp biosynthesis and regulation had been inferred from microbial investigations of these pathways (Maeda and Dudareva, 2012). Trp biosynthesis occurs in the plastid and emanates from the product of the shikimate pathway. The shikimate pathway starts by combining erythrose 4-phosphate (E4P) from the light-independent reactions of photosynthesis with phosphoenolpyruvate (PEP) from glycolysis. This seven-step pathway gives rise to chorismate, which serves as a precursor to the aromatic amino acids phenylalanine (Phe), tyrosine (Tyr), and Trp and to the plant hormone salicylic acid (SA) (Figure 1) (Niyogi and Fink, 1992; Westfall et al., 2014; Kroll et al., 2017; Spoel and Dong, 2024).
Anthranilate synthase (AS) catalyzes the first, and committed, step in Trp biosynthesis to convert chorismate to anthranilate and pyruvate (Figure 1) (Niyogi and Fink, 1992; Niyogi et al., 1993; Romero et al., 1995; Tozawa et al., 2001; Inaba et al., 2007). The amine that is added to chorismate has two potential sources: free ammonia (NH3) or the side chain amine of glutamine (Gln), depending on the AS subunits used to catalyze the reaction. Free ammonia is used when ASα acts as a monofunctional protein, while the ASα/β heterodimer uses Gln as the amine donor (Niyogi and Fink, 1992; Niyogi et al., 1993). AS is feedback regulated through allosteric inhibition by Trp, although ASαs that are feedback-insensitive to Trp have been identified in rice, common rue, and tobacco (Bohlmann et al., 1996; Song et al., 1998; Tozawa et al., 2001). Investigations of the source of this Trp insensitivity have identified several residues responsible for feedback inhibition in both plants and bacteria (Caligiuri and Bauerle, 1991; Bernasconi et al., 1994; Kreps et al., 1996). Transcriptional regulation of AS isozymes provides an additional layer of control. Arabidopsis contains two ASαs, and evidence suggests that ASα2 is expressed at low, constitutive levels, while ASα1 is induced during wounding and bacterial infiltration (Niyogi and Fink, 1992).
In the second step of the pathway, anthranilate phosphoribosyltransferase (PAT1) transfers a phosphoribosyl sugar onto anthranilate, forming 5-phosphoribosylanthranilate (Last and Fink, 1988; Rose et al., 1992; Radwanski and Last, 1995). The PAT1 gene was the first Trp pathway enzyme discovered in plants (Last and Fink, 1988). EMS-mutagenized Arabidopsis thaliana seedlings were grown on Trp and 5-methyl anthranilate (5-MA), which is converted to 5-methyl-Trp in plants that have a fully functional Trp pathway downstream of anthranilate. Mutant seedlings that grew on Trp and were resistant to 5-MA were named trp1-1 and were found to lack phosphoribosyl transferase activity. Because PAT1 is a single-copy gene in Arabidopsis, trp1-1 plants require Trp in the growth media. The trp1-1 mutants also exhibit blue fluorescence under UV light. The trp1 mutant plants that were supplemented with Trp appeared similar to wild type (WT) initially, but after 3-4 weeks, the plants were smaller than WT, a phenotype which could not be rescued with exogenous Trp (Last and Fink, 1988). The trp1-1 plants were bushy, had small and crinkled rosette leaves, and are virtually sterile. Another mutant allele, trp1-100, grew and developed normally without exogenous Trp, yet maintained the blue fluorescence phenotype and exhibited normal fertility (Rose et al., 1992). The source of this blue fluorescence was fortuitously discovered as a result of anthranilate β-glucoside accumulation in the trp1 mutants via glucosylation by UGT74F2, and the fluorescence phenotype was rescued in trp1 ugt74f2 double mutant plants (Quiel and Bender, 2003).
PAT1 remained biochemically uncharacterized until recently (Li et al., 2023). By comparing structural models, the putative active site residues of 82 PAT1 enzymes were compared, and the six enzymes with the most variability in the active site were characterized using steady-state kinetics. PAT1s from A. thaliana and Citrus sinensis (sweet orange) exhibited the highest catalytic efficiencies with anthranilate, followed by the PAT1s from Physcomitrium patens (spreading earth-moss), Juglans regia (English walnut), and Pistacia vera (pistachio). The PAT1 from Selaginella moellendorffii (spike moss) exhibited the lowest catalytic efficiency, with a more than 14-fold reduction relative to that of A. thaliana. The enzymes followed substrate inhibition kinetics with high concentrations of anthranilate leading to reduced enzymatic activity.
With the exception of the C. sinensis PAT1, these enzymes could also act on 3-hydroxyanthranilate (3-HAA), a Trp catabolism intermediate in mammals that has been reported in maize (Tarr and Arditti, 1982; Li et al., 2023). Comparative active site analysis between C. sinensis PAT1 and the A. thaliana PAT1, which had the highest catalytic efficiency with 3-HAA, identified Asn215 in AtPAT1 as a residue that confers 3-HAA activity (Li et al., 2023). While PAT1 is not known to have an allosteric site, Tyr inhibited the P. patens PAT1 with an IC50 value of 1180 μM, while Phe inhibited the S. moellendorfii PAT1 with an IC50 value of 1980 μM. The activity of the four other PAT1s were not modulated by aromatic amino acids.
Notably, the C. sinensis PAT1, which has a high catalytic efficiency with anthranilate, is insensitive to modulation by aromatic amino acids (Li et al., 2023). This functional data may explain how Rutaceae species balance anthranilate allocation to Trp biosynthesis with competing biosynthetic pathways for producing O-MeAA, N-MeAA esters, acridone alkaloids, and quinolones. Furthermore, enzyme-level regulation via substrate inhibition may balance flux through primary and specialized metabolism to ensure sufficient Trp is produced to promote plant growth and development.
3 Biosynthesis of anthranilate methyl esters
O-MeAA biosynthesis has only been investigated in a handful of crop plants, including soybean (Glycine max) (Lin et al., 2013), maize (Zea mays) (Köllner et al., 2010), barrel clover (Medigaco truncatula) (Pollier et al., 2019), strawberries (Frageria spp.) (Pillet et al., 2017), grapes (Vitis spp.) (Wang and De Luca, 2005; Fallon et al., 2024), and sweet orange (C. sinensis) (Huang et al., 2016). There are two biosynthetic routes for O-MeAA: one uses a one-step SAM-dependent anthranilate O-methyltransferase (AAMT) and the other uses a two-step pathway. In the two-step pathway, Coenzyme-A (CoA) is first added to the carboxylate of anthranilate by a ligase that remains to be identified (Figure 1). This anthraniloyl-CoA is then combined with methanol using an anthraniloyl-CoA:methanol acyltransferase (AMAT) to produce O-MeAA (Wang and De Luca, 2005; Yang et al., 2020).
Although O-MeAA imparts the classic grape aroma, the complete biosynthetic pathway for O-MeAA in grapes remained enigmatic until recently. While one-step AAMTs had been characterized as the mechanism for O-MeAA biosynthesis in other plants, grapes were thought to primarily rely upon the two-step pathway until the recent identification of two one-step AAMTs in Vitis (Fallon et al., 2024). In C. sinensis, a SA methyltransferase (SAMT) has activity with anthranilate, and comparisons between the newly identified grape AAMTs and the Citrus SAMT led to the identification of three residues (Gln263, Cys319, and Ala324) that confer anthranilate activity. Similar comparisons using the maize AAMT1, which is highly specific for anthranilate, identified three residues (Cys165, Tyr246, and Leu329) that increase activity with SA (Köllner et al., 2010; Fallon et al., 2024). Interestingly, one of the grape AAMTs and the strawberry AAMT have shared ancestry with jasmonate methyltransferases (Pillet et al., 2017; Fallon et al., 2024).
The regulation of AAMT expression has been shown to be induced by methyl jasmonate in maize and in the legume Medicago truncatula (Köllner et al., 2010; Pollier et al., 2019). In M. truncatula, an R2-R3 MYB transcription factor Emission of Methyl Anthranilate 1 (EMA1) was found to promote AAMT expression in hairy roots (Pollier et al., 2019). Treating seedlings with O-MeAA led to a several thousand-fold increase in EMA1 transcript levels after 24 hours. Additionally, the transcriptional repressor plant AT-rich sequence and zinc-binding 1 (PLATZ1) regulated EMA1 expression and thus volatile biosynthesis (Figure 1). Future experiments are needed to identify additional transcriptional regulators of AAMT gene expression in plants and to gain a more complete understanding of the regulatory network underlying O-MeAA biosynthesis across O-MeAA-producing species.
4 N-methyl anthranilate-containing specialized metabolites
In addition to methylation at the carboxyl group, anthranilate can also be methylated on the amine by an anthranilate N-methyltransferase (ANMT). ANMTs has been identified in black oat (A. strigosa) and common rue (R. graveolens) (Rohde et al., 2008; Mugford et al., 2013). Members of the Rutaceae family are notable for their diversity of N-MeAA metabolites, where N-MeAA serves as a precursor for acridone alkaloids and anti-malarial quinolone alkaloids (Rohde et al., 2008; Mori et al., 2013). A type III polyketide synthase, acridone synthase, condenses N-methyl anthraniloyl-CoA with three malonyl-CoA units to form 1,3-dihydroxy-N-methylacridone (Maier et al., 1993), while the condensation of N-methyl anthraniloyl-CoA with one unit of malonyl-CoA by quinoline synthase gives rise to 4-hydroxy-N-methyl-quinolone (Resmi et al., 2013). Acridone, acridine, and their derivatives are toxic to human cell lines and have broad biological activity against bacteria, viruses, and parasites and inhibit acetylcholinesterase, suggesting that they may serve a defensive role in plants (Kelly et al., 2009; Gensicka-Kowalewska et al., 2017). Quinolones are also synthesized by plants in the Rubiaceae family (Michael, 2003), and quinine from Cinchona bark has been used to treat malaria parasite (Plasmodium spp.) infections in humans (Gachelin et al., 2017).
Various N-MeAA esters including DiMeAA, propyl-N-MeAA, and isopropyl-N-MeAA, have been identified in Mexican orange blossom leaves (Choisya ternata Kunth) (Pinheiro et al., 2015). These esters are thought to be synthesized using an acyl-CoA:alchocol acyltransferase, homologous to the AMAT in grapes (Wang and De Luca, 2005); however, a CoA ligase that acts on anthranilate or N-MeAA remains to be identified in plants (Figure 1). These anthranilates exhibit dose-dependent anti-nociceptive effects, as well as anti-anxiety and anti-depressive activity in mice (Radulovic et al., 2013; Pinheiro et al., 2015). However, their role in plant metabolism and plant responses to biotic factors has yet to be investigated. DiMeAA was also identified as a quorum sensing inhibitor with antibiofilm activity against the foodborne pathogen Pseudomonas aeruginosa (Ma et al., 2021), suggesting that it may be involved in plant-microbe interactions.
Oats release avenacins, which are anti-microbial glycosylated triterpenes that are acylated at C-21 with either N-MeAA (A-1 and B-1) or benzoic acid (A-2 and B-2), from their roots to protect against pathogens in the soil (Figure 1) (Mugford et al., 2013; Owatworakit et al., 2013). The connections between the avenacins and their anti-microbial properties were made by screening oat genotypes for susceptibility to Gaeumannomyces graminis var. tritici, a fungus that causes take-all disease in oats, which unveiled that the susceptible A. longiglumis did not produce detectable levels of avenacins (Osbourn et al., 1994). Follow-up studies identified the 12-gene avenacin biosynthetic gene cluster (Li et al., 2021), which included the glycosyltransferase UGT74H5 (SAD10) that activates N-MeAA for addition onto avenacin A-1 and B-1 (Mugford et al., 2009; Owatworakit et al., 2013). N-MeAA-glucoside is imported into the vacuole, which is where it is conjugated onto avenacins via a SAD7 serine carboxypeptidase-like acyl transferase (SCPL1) (Orme et al., 2019). Recently, a nonclustered gene that encodes a glycosyltransferase, UGT84C2 (SAD4), was found that also activates N-MeAA via glucosylation for SAD7-mediated transfer onto A-1 and B-1 avenacins (Qiao et al., 2025). Aside from oats and Rutaceae species, N-methylated anthranilates have not been studied. It is likely that chemical profiling of more plants in the future will turn up additional plants that synthesize anthranilate-derived specialized metabolites.
5 Discussion
While all plants synthesize anthranilate as a Trp pathway intermediate, specialized metabolites and volatiles containing anthranilate represent an under-explored area of plant metabolism. There is still much to learn regarding how plants regulate Trp biosynthesis via anthranilate synthase and the extent to which other amino acids or plant metabolites regulate PAT1. Anthranilate is synthesized in the plastid where Trp biosynthesis occurs, but anthranilate-using enzymes in specialized metabolism, such as ANMTs, localize to the cytosol or are predicted to be cytosolic based on the absence of a signal peptide (Mugford et al., 2013; Fallon et al., 2024) (Figure 1). The plastid-to-cytosol transport mechanism for anthranilate remains to be investigated, as does the mechanisms regulating subcellular partitioning of anthranilate.
Recent advances in identifying anthranilate-using enzymes in specialized metabolism increases our understanding of the molecular basis of anthranilate recognition by enzymes such as glycosyltransferases and methyltransferases (Owatworakit et al., 2013; Fallon et al., 2024). Single amino acid changes to active site residues in the maize AAMT1 were sufficient to increase SA production 8-fold when heterologously expressed in E. coli cultures, which has enabled the identification of amino acids that confer activity with anthranilate versus SA (Fallon et al., 2024). Similarly, single mutants of the C. sinensis SAMT led to increased activity with anthranilate. Comparisons between the Ruta graveolens ANMT and a caffeate O-methyltransferase identified the molecular basis of chemoselectivity for N- versus O- methylation (Jockmann et al., 2025).
While O-MeAA mediates plant-insect interactions, there appears to be context dependence to these interactions. For example, maize induces O-MeAA biosynthesis upon insect herbivory and induces AAMT1 expression (Köllner et al., 2010), while C. sinensis plants shut off O-MeAA release when they are attacked by the psyllid-vectored bacterial pathogen Candidatus Liberibacter asiaticus (CLas) that causes citrus greening (huanglongbing) (Mann et al., 2012). Aside from the EMA1 transcriptional regulator that was identified in Medicago truncatula, additional research is needed to identify the genetic regulators of O-MeAA biosynthesis in plants in order to better understand the species-specific differences underlying O-MeAA emission. Furthermore, O-MeAA is emitted as a part of a volatile blend, and it is likely the combination of volatiles that aid in insect attraction or deterrence (von Merey et al., 2013; Zhou and Jander, 2021).
Anthranilate-containing specialized metabolites and volatiles are likely more widespread across Viridiplantae and have been undersampled in plants, especially considering how nearly all angiosperm orders have a SAMT that could conceivably also act on anthranilate (Dubs et al., 2022; Fallon et al., 2024). Except for the avenacins in A. strigosa and O-MeAA in M. truncatula, anthranilates have escaped investigation in below-ground tissues (Pollier et al., 2019; Li et al., 2021). Understanding the tissue-level and developmental patterns of the synthesis of anthranilates may aid in gaining a more comprehensive understanding in their roles in plant physiology and biotic interactions.
In summary, anthranilates are an under-investigated area of plant metabolism, and many open questions remain regarding: the identification of biosynthetic genes; the transcriptional and enzyme-level regulation of anthranilate specialized metabolic pathways; the transport of anthranilate between subcellular compartments; the diversity of plants that synthesize anthranilates; and the role of anthranilates in plant physiology.
Author contributions
CH: Conceptualization, Funding acquisition, Investigation, Project administration, Writing – original draft, Writing – review & editing. AW: Investigation, Writing – original draft, Writing – review & editing. EC: Investigation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Research in the Holland lab is supported by Williams College and the U.S. National Science Foundation (MCB-2214883 and MCB-2440307 to CH).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Avery, M. L., Decker, D. G., Humphrey, J. S., Aronov, E., Linscombe, S. D., and Way, M. (1995). Methyl anthranilate as a rice seed treatment to deter birds. J. wildlife Manage. 59 (1), 50–56. doi: 10.2307/3809115
Bernasconi, P., Walters, E. W., Woodworth, A. R., Siehl, D. L., Stone, T. E., and Subramanian, M. V. (1994). Functional expression of Arabidopsis thaliana anthranilate synthase subunit I in Escherichia coli. Plant Physiol. 106, 353–358. doi: 10.1104/pp.106.1.353
Bohlmann, J., Lins, T., Martin, W., and Eilert, U. (1996). Anthranilate synthase from Ruta graveolens. Duplicated AS alpha genes encode tryptophan-sensitive and tryptophan-insensitive isoenzymes specific to amino acid and alkaloid biosynthesis. Plant Physiol. 111, 507–514. doi: 10.1104/pp.111.2.507
Bracker, L. B., Gong, X., Schmid, C., Dawid, C., Ulrich, D., Phung, T., et al. (2020). A strawberry accession with elevated methyl anthranilate fruit concentration is naturally resistant to the pest fly Drosophila suzukii. PLoS One 15, e0234040. doi: 10.1371/journal.pone.0234040
Caligiuri, M. G. and Bauerle, R. (1991). Identification of amino acid residues involved in feedback regulation of the anthranilate synthase complex from Salmonella typhimurium. Evidence for an amino-terminal regulatory site. J. Biol. Chem. 266, 8328–8335. doi: 10.1016/S0021-9258(18)92979-0
Caputi, L., Franke, J., Farrow, S., Chung, K., Payne, R. E., Nguyen, T.-D., et al. (2018). Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle. Sci. (AAAS) 360, 1235–1239. doi: 10.1126/science.aat4100
Dubs, N. M., Davis, B. R., De Brito, V., Colebrook, K. C., Tiefel, I. J., Nakayama, M. B., et al. (2022). A collaborative classroom investigation of the evolution of SABATH methyltransferase substrate preference shifts over 120 my of flowering plant history. Mol. Biol. Evol. 39, msac007. doi: 10.1093/molbev/msac007
Dudareva, N., Klempien, A., Muhlemann, J. K., and Kaplan, I. (2013). Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 198, 16–32. doi: 10.1111/nph.12145
Fallon, M. A., Tadfie, H., Watson, A. P., Dyke, M. M., Flores, C., Cook, N., et al. (2024). Molecular basis of one-step methyl anthranilate biosynthesis in grapes, sweet orange, and maize. Plant J. 19 (5), 2363–2374. doi: 10.1111/tpj.16922
Gachelin, G., Garner, P., Ferroni, E., Tröhler, U., and Chalmers, I. (2017). Evaluating Cinchona bark and quinine for treating and preventing malaria. J. R. Soc. Med. 110, 73–82. doi: 10.1177/0141076816688411
Gensicka-Kowalewska, M., Cholewiński, G., and Dzierzbicka, K. (2017). Recent developments in the synthesis and biological activity of acridine/acridone analogues. RSC Adv. 7, 15776–15804. doi: 10.1039/C7RA01026E
Huang, R., O’donnell, A. J., Barboline, J. J., and Barkman, T. J. (2016). Convergent evolution of caffeine in plants by co-option of exapted ancestral enzymes. Proc. Natl. Acad. Sci. U.S.A. 113, 10613–10618. doi: 10.1073/pnas.1602575113
Inaba, Y., Brotherton, J. E., Ulanov, A., and Widholm, J. M. (2007). Expression of a feedback insensitive anthranilate synthase gene from tobacco increases free tryptophan in soybean plants. Plant Cell Rep. 26, 1763–1771. doi: 10.1007/s00299-007-0381-0
Jacobowitz, J. R. and Weng, J.-K. (2020). Exploring uncharted territories of plant specialized metabolism in the postgenomic era. Annu. Rev. Plant Biol. 71, 631–658. doi: 10.1146/annurev-arplant-081519-035634
Jockmann, E., Girame, H., Steinchen, W., Kind, K., Bange, G., Tittmann, K., et al. (2025). How to tell an N from an O: controlling the chemoselectivity of methyltransferases. ACS Catalysis 15, 6410–6425. doi: 10.1021/acscatal.5c00834
Kelly, J. X., Smilkstein, M. J., Brun, R., Wittlin, S., Cooper, R. A., Lane, K. D., et al. (2009). Discovery of dual function acridones as a new antimalarial chemotype. Nature 459, 270–273. doi: 10.1038/nature07937
Knudsen, J. T., Eriksson, R., Gershenzon, J., and Ståhl, B. (2006). Diversity and distribution of floral scent. botanical Rev. 72, 1–120. doi: 10.1663/0006-8101(2006)72[1:DADOFS]2.0.CO;2
Köllner, T. G., Lenk, C., Zhao, N., Seidl-Adams, I., Gershenzon, J., Chen, F., et al. (2010). Herbivore-induced SABATH methyltransferases of maize that methylate anthranilic acid using s-adenosyl-L-methionine. Plant Physiol. 153, 1795–1807. doi: 10.1104/pp.110.158360
Kreps, J. A., Ponappa, T., Dong, W., and Town, C. D. (1996). Molecular basis of alpha-methyltryptophan resistance in amt-1, a mutant of Arabidopsis thaliana with altered tryptophan metabolism. Plant Physiol. 110, 1159–1165. doi: 10.1104/pp.110.4.1159
Kroll, K., Holland, C. K., Starks, C. M., and Jez, J. M. (2017). Evolution of allosteric regulation in chorismate mutases from early plants. Biochem. J. 474, 3705–3717. doi: 10.1042/BCJ20170549
Last, R. L. and Fink, G. R. (1988). Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 240, 305–310. doi: 10.1126/science.240.4850.305
Lefevere, H., Bauters, L., and Gheysen, G. (2020). Salicylic acid biosynthesis in plants. Front. Plant Sci. 11 - 2020. doi: 10.3389/fpls.2020.00338
Li, Y., Leveau, A., Zhao, Q., Feng, Q., Lu, H., Miao, J., et al. (2021). Subtelomeric assembly of a multi-gene pathway for antimicrobial defense compounds in cereals. Nat. Commun. 12, 2563. doi: 10.1038/s41467-021-22920-8
Li, M., Tadfie, H., Darnell, C. G., and Holland, C. K. (2023). Biochemical investigation of the tryptophan biosynthetic enzyme anthranilate phosphoribosyltransferase in plants. J. Biol. Chem. 299, 105197. doi: 10.1016/j.jbc.2023.105197
Lin, J., Mazarei, M., Zhao, N., Zhu, J. J., Zhuang, X., Liu, W., et al. (2013). Overexpression of a soybean salicylic acid methyltransferase gene confers resistance to soybean cyst nematode. Plant Biotechnol. J. 11, 1135–1145. doi: 10.1111/pbi.12108
Ma, Y., Shi, Q., He, Q., and Chen, G. (2021). Metabolomic insights into the inhibition mechanism of methyl N-methylanthranilate: A novel quorum sensing inhibitor and antibiofilm agent against Pseudomonas aeruginosa. Int. J. Food Microbiol. 358, 109402. doi: 10.1016/j.ijfoodmicro.2021.109402
Maeda, H. and Dudareva, N. (2012). The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 63, 73–105. doi: 10.1146/annurev-arplant-042811-105439
Maeda, H. A. and Fernie, A. R. (2021). Evolutionary history of plant metabolism. Annu. Rev. Plant Biol. 72, 185–216. doi: 10.1146/annurev-arplant-080620-031054
Maier, W., Baumert, A., Schumann, B., Furukawa, H., and Gröger, D. (1993). Synthesis of 1,3-dihydroxy-N-methylacridone and its conversion to rutacridone by cell-free extracts of Ruta graveolens cell cultures. Phytochemistry 32, 691–698. doi: 10.1016/S0031-9422(00)95155-0
Mann, R. S., Ali, J. G., Hermann, S. L., Tiwari, S., Pelz-Stelinski, K. S., Alborn, H. T., et al. (2012). Induced release of a plant-defense volatile ‘deceptively’ attracts insect vectors to plants infected with a bacterial pathogen. PloS Pathog. 8, e1002610. doi: 10.1371/journal.ppat.1002610
Michael, J. P. (2003). Quinoline, quinazoline and acridone alkaloids. Nat. Prod Rep. 20, 476–493. doi: 10.1039/b208140g
Morffy, N. and Strader, L. C. (2020). Old Town Roads: routes of auxin biosynthesis across kingdoms. Curr. Opin. Plant Biol. 55, 21–27. doi: 10.1016/j.pbi.2020.02.002
Mori, T., Shimokawa, Y., Matsui, T., Kinjo, K., Kato, R., Noguchi, H., et al. (2013). Cloning and structure-function analyses of quinolone-and acridone-producing novel type III polyketide synthases from Citrus microcarpa. J. Biol. Chem. 288, 28845–28858. doi: 10.1074/jbc.M113.493155
Mugford, S. T., Louveau, T., Melton, R., Qi, X., Bakht, S., Hill, L., et al. (2013). Modularity of plant metabolic gene clusters: a trio of linked genes that are collectively required for acylation of triterpenes in oat. Plant Cell 25, 1078–1092. doi: 10.1105/tpc.113.110551
Mugford, S. T., Qi, X., Bakht, S., Hill, L., Wegel, E., Hughes, R. K., et al. (2009). A serine carboxypeptidase-like acyltransferase is required for synthesis of antimicrobial compounds and disease resistance in oats. Plant Cell 21, 2473–2484. doi: 10.1105/tpc.109.065870
Niyogi, K. K. and Fink, G. R. (1992). Two anthranilate synthase genes in Arabidopsis: defense-related regulation of the tryptophan pathway. Plant Cell 4, 721–733. doi: 10.1105/tpc.4.6.7212
Niyogi, K. K., Last, R. L., Fink, G. R., and Keith, B. (1993). Suppressors of trp1 fluorescence identify a new arabidopsis gene, TRP4, encoding the anthranilate synthase beta subunit. Plant Cell 5, 1011–1027. doi: 10.1105/tpc.5.9.1011
Orme, A., Louveau, T., Stephenson, M., Appelhagen, I., Melton, R., Cheema, J., et al. (2019). A noncanonical vacuolar sugar transferase required for biosynthesis of antimicrobial defense compounds in oat. Proc. Natl. Acad. Sci. (PNAS) 116, 27105–27114. doi: 10.1073/pnas.1914652116
Osbourn, A., Clarke, B., Lunness, P., Scott, P., and Daniels, M. (1994). An oat species lacking avenacin is susceptible to infection by Gaeumannomyces graminis var. tritici. Physiol. Mol. Plant Pathol. 45, 457–467. doi: 10.1016/S0885-5765(05)80042-6
Owatworakit, A., Townsend, B., Louveau, T., Jenner, H., Rejzek, M., Hughes, R. K., et al. (2013). Glycosyltransferases from oat (Avena) implicated in the acylation of avenacins. J. Biol. Chem. 288, 3696–3704. doi: 10.1074/jbc.M112.426155
Papadopoulou, K., Melton, R. E., Leggett, M., Daniels, M. J., and Osbourn, A. E. (1999). Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. (PNAS) 96, 12923–12928. doi: 10.1073/pnas.96.22.12923
Pillet, J., Chambers, A. H., Barbey, C., Bao, Z., Plotto, A., Bai, J., et al. (2017). Identification of a methyltransferase catalyzing the final step of methyl anthranilate synthesis in cultivated strawberry. BMC Plant Biol. 17, 147. doi: 10.1186/s12870-017-1088-1
Pinheiro, M. M., Miltojević, A. B., Radulović, N. S., Abdul-Wahab, I. R., Boylan, F., and Fernandes, P. D. (2015). Anti-inflammatory activity of Choisya ternata Kunth essential oil, ternanthranin, and its two synthetic analogs (methyl and propyl N-methylanthranilates). PloS One 10, e0121063. doi: 10.1371/journal.pone.0121063
Pinheiro, M. M. G., Radulović, N. S., Miltojević, A. B., Boylan, F., and Fernandes, P. D. (2014). Antinociceptive esters of N-methylanthranilic acid: Mechanism of action in heat-mediated pain. Eur. J. Pharmacol. 727, 106–114. doi: 10.1016/j.ejphar.2013.12.042
Pollier, J., De Geyter, N., Moses, T., Boachon, B., Franco-Zorrilla, J. M., Bai, Y., et al. (2019). The MYB transcription factor Emission of Methyl Anthranilate 1 stimulates emission of methyl anthranilate from Medicago truncatula hairy roots. Plant J. 99, 637–654. doi: 10.1111/tpj.14347
Qiao, X., Houghton, A., Reed, J., Steuernagel, B., Zhang, J., Owen, C., et al. (2025). Comprehensive mutant chemotyping reveals embedding of a lineage-specific biosynthetic gene cluster in wider plant metabolism. Proc. Natl. Acad. Sci. (PNAS) 122, Undefined–Undefined. doi: 10.1073/pnas.2417588122
Quiel, J. A. and Bender, J. (2003). Glucose conjugation of anthranilate by the Arabidopsis UGT74F2 glucosyltransferase is required for tryptophan mutant blue fluorescence. J. Biol. Chem. 278, 6275–6281. doi: 10.1074/jbc.M211822200
Radulovic, N. S., Miltojevic, A. B., Randjelovic, P. J., Stojanovic, N. M., and Boylan, F. (2013). Effects of methyl and isopropyl N-methylanthranilates from Choisya ternata Kunth (Rutaceae) on experimental anxiety and depression in mice. Phytother. Res. 27, 1334–1338. doi: 10.1002/ptr.4877
Radwanski, E. R. and Last, R. L. (1995). Tryptophan biosynthesis and metabolism: biochemical and molecular genetics. Plant Cell 7, 921–934. doi: 10.1105/tpc.7.7.921
Resmi, M. S., Verma, P., Gokhale, R. S., and Soniya, E. V. (2013). Identification and characterization of a type III polyketide synthase involved in quinolone alkaloid biosynthesis from Aegle marmelos correa*. J. Biol. Chem. 288, 7271–7281. doi: 10.1074/jbc.M112.429886
Rohde, B., Hans, J., Martens, S., Baumert, A., Hunziker, P., and Matern, U. (2008). Anthranilate N-methyltransferase, a branch-point enzyme of acridone biosynthesis. Plant J. 53, 541–553. doi: 10.1111/j.1365-313X.2007.03360.x
Romero, R. M., Roberts, M. F., and Phillipson, J. D. (1995). Anthranilate synthase in microorganisms and plants. Phytochemistry 39, 263–276. doi: 10.1016/0031-9422(95)00010-5
Rose, A. B., Casselman, A. L., and Last, R. L. (1992). A phosphoribosylanthranilate transferase gene is defective in blue fluorescent Arabidopsis thaliana tryptophan mutants. Plant Physiol. 100, 582–592. doi: 10.1104/pp.100.2.582
Schuman, M. C. (2023). Where, when, and why do plant volatiles mediate ecological signaling? The answer is blowing in the wind. Annu. Rev. Plant Biol. 74, 609–633. doi: 10.1146/annurev-arplant-040121-114908
Simpson, M., Gurr, G. M., Simmons, A. T., Wratten, S. D., James, D. G., Leeson, G., et al. (2011). Insect attraction to synthetic herbivore-induced plant volatile-treated field crops. Agric. For. Entomology 13, 45–57. doi: 10.1111/j.1461-9563.2010.00496.x
Sønderby, I. E., Geu-Flores, F., and Halkier, B. A. (2010). Biosynthesis of glucosinolates–gene discovery and beyond. Trends Plant Sci. 15, 283–290. doi: 10.1016/j.tplants.2010.02.005
Song, H. S., Brotherton, J. E., Gonzales, R. A., and Widholm, J. M. (1998). Tissue culture-specific expression of a naturally occurring tobacco feedback-insensitive anthranilate synthase. Plant Physiol. 117, 533–543. doi: 10.1104/pp.117.2.533
Spoel, S. H. and Dong, X. (2024). Salicylic acid in plant immunity and beyond. Plant Cell 36, 1451–1464. doi: 10.1093/plcell/koad329
Tarr, J. B. and Arditti, J. (1982). Niacin biosynthesis in seedlings of Zea mays. Plant Physiol. 69, 553–556. doi: 10.1104/pp.69.3.553
Tozawa, Y., Hasegawa, H., Terakawa, T., and Wakasa, K. (2001). Characterization of rice anthranilate synthase alpha-subunit genes OASA1 and OASA2. Tryptophan accumulation in transgenic rice expressing a feedback-insensitive mutant of OASA1. Plant Physiol. 126, 1493–1506. doi: 10.1104/pp.126.4.1493
Turlings, T. C., Tumlinson, J. H., and Lewis, W. J. (1990). Exploitation of herbivore-induced plant odors by host-seeking parasitic wasps. Science 250, 1251–1253. doi: 10.1126/science.250.4985.1251
von Merey, G. E., Veyrat, N., D’alessandro, M., and Turlings, T. C. (2013). Herbivore-induced maize leaf volatiles affect attraction and feeding behavior of Spodoptera littoralis caterpillars. Front. Plant Sci. 4, 209. doi: 10.3389/fpls.2013.00209
Wang, J. and De Luca, V. (2005). The biosynthesis and regulation of biosynthesis of Concord grape fruit esters, including ‘foxy’ methylanthranilate. Plant J. 44, 606–619. doi: 10.1111/j.1365-313X.2005.02552.x
Westfall, C. S., Xu, A., and Jez, J. M. (2014). Structural evolution of differential amino acid effector regulation in plant chorismate mutases. J. Biol. Chem. 289, 28619–28628. doi: 10.1074/jbc.M114.591123
Yang, Y., Cuenca, J., Wang, N., Liang, Z., Sun, H., Gutierrez, B., et al. (2020). A key ‘foxy’ aroma gene is regulated by homology-induced promoter indels in the iconic juice grape ‘Concord’. Hortic. Res. 7, 67. doi: 10.1038/s41438-020-0304-6
Keywords: tryptophan, anthranilate, specialized metabolism, plant defense, volatiles
Citation: Holland CK, Watson AP and Chiang E (2025) Anthranilate at the interface of tryptophan and specialized metabolite biosynthesis. Front. Plant Sci. 16:1625337. doi: 10.3389/fpls.2025.1625337
Received: 08 May 2025; Accepted: 23 June 2025;
Published: 08 July 2025.
Edited by:
Jorge El-Azaz, University of Wisconsin-Madison, United StatesReviewed by:
Jeongim Kim, University of Florida, United StatesCopyright © 2025 Holland, Watson and Chiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cynthia K. Holland, Y2toMkB3aWxsaWFtcy5lZHU=
 Cynthia K. Holland
Cynthia K. Holland Aracely P. Watson
Aracely P. Watson