- 1State Key Laboratory of Genome and Multi-omics Technologies, BGI Research, Shenzhen, China
- 2BGI Research, Wuhan, China
- 3School of Breeding and Multiplication (Sanya institute of Breeding and Multiplication), College of Tropical Agriculture and Forestry, Hainan University, Sanya, China
- 4Institute of Life Sciences, Kangwon National University, Chuncheon, Republic of Korea
Editorial on the Research Topic
Salinity and drought stress in plants: understanding physiological, biochemical and molecular responses, volume II
1 Introduction
Salinity and drought are among the most critical abiotic stressors affecting plant growth and productivity across the globe. These challenges are particularly acute in arid and semiarid regions, where climate change has intensified water scarcity and increased soil salinization (Huang and Jin; Sahu, 2024). These stresses disrupt fundamental plant functions, including photosynthesis, nutrient uptake, and cellular homeostasis. As the global population is expected to approach 10 billion by the year 2050, agricultural systems must produce approximately 70% more food to ensure food and nutritional security (Sahu and Liu, 2023; Editorial, 2024). However, the increasing severity and frequency of salinity and drought conditions threaten this target. While severe stress might entirely limit plant development, moderate stress may activate a range of adaptive responses. These include transcriptional reprogramming, osmoprotectant buildup, alterations in stomatal behavior, and the activation of antioxidant defense systems. To create crop varieties with increased resilience, it is crucial to understand these reactions at the physiological, biochemical, and molecular levels. Recent advances in spatial omics have enabled researchers to uncover stress responses with cell-type resolution, providing new insight into how plants adapt to heterogeneous stress environments (Guo et al., 2025 ; Zhong et al., 2025). Combined with CRISPR-based gene editing, high-throughput phenotyping, bioinformatics, and integrative multiomics approaches, these tools offer powerful strategies to accelerate breeding for climate-resilient crops (Purugganan and Jackson, 2021; Wang et al., 2022; Sahu et al., 2023; Wang et al.; Zhu et al.).
Volume II of “Salinity and Drought Stress in Plants: Understanding Physiological, Biochemical and Molecular Responses” builds on the foundation laid by the first Research Topic, focusing on integrative, translational research that bridges basic “omics” discoveries with applied breeding and biotechnology. Our Research Topic features an acceptance rate of approximately 44%, demonstrating the rigor of our peer review process and the high quality of the published contributions. The present editorial synthesizes 26 contributions comprising 23 original research articles and three review articles, organized into thematic categories that reflect the multidimensional nature of plant stress responses. It aims to contextualize the overall goals of the Research Topic, highlight advances in mechanistic insights, and provide a roadmap for future work in enhancing crop resilience (Figure 1).
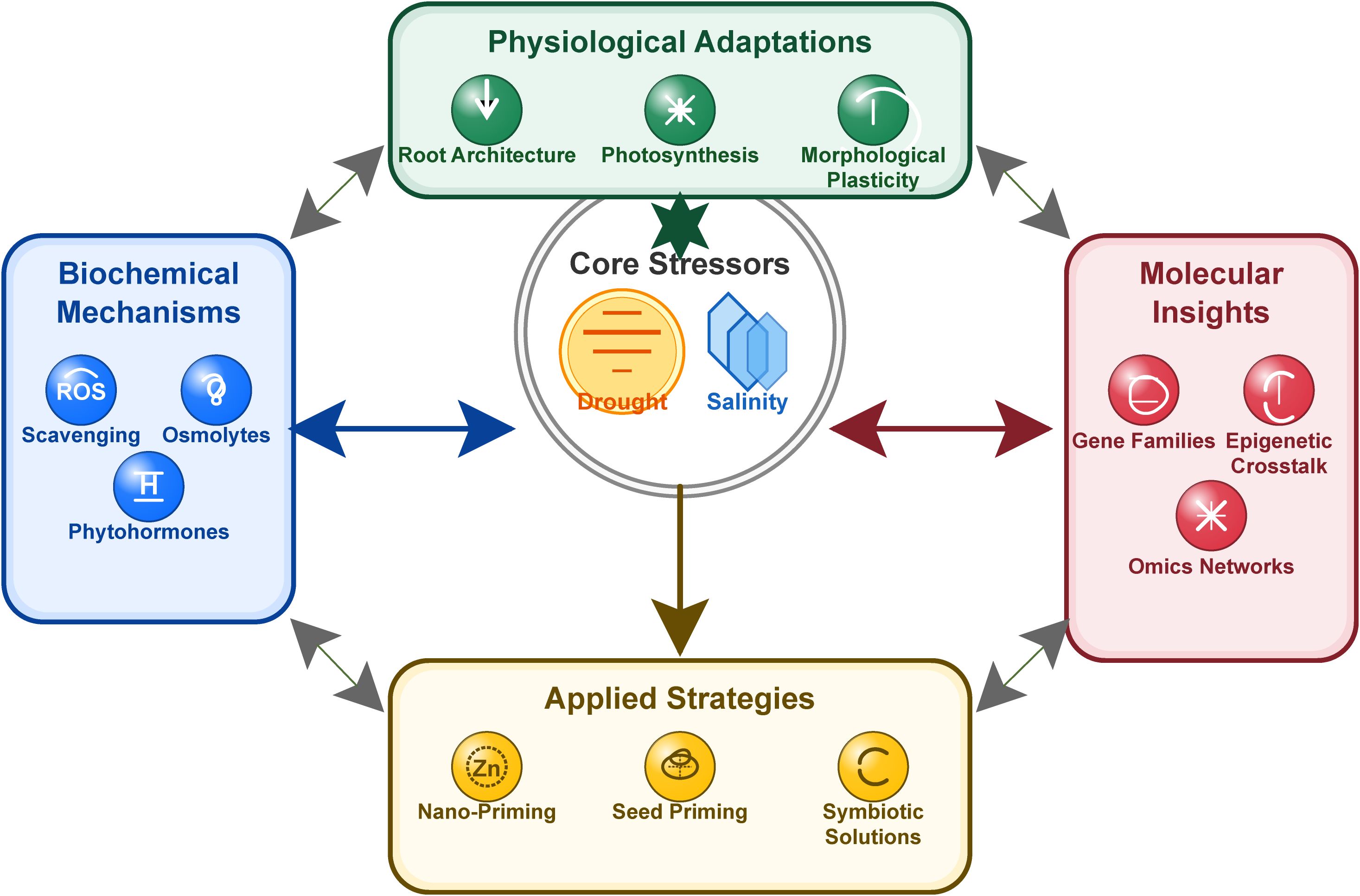
Figure 1. Interconnected mechanisms of plant stress adaptations to salinity and drought. The figure illustrates the conceptual framework of plant responses to salinity and drought stress, illustrating key physiological adjustments, biochemical pathways (e.g., osmolyte accumulation and antioxidant defense), and molecular networks (signal transduction and transcriptional regulation).
2 Physiological adaptations to drought and salinity
2.1 Root and shoot architectural traits
Morphological plasticity underpins early stress avoidance. In chickpeas, gradual drought imposed at 70 → 50 → 25 percent water- holding capacity revealed that the MCC552 and MCC696 genotypes maintained high photosynthetic water-use efficiency and developed deeper, thicker roots—traits linked to greater proline accumulation and membrane stability (Fazeli-Nasab et al.). In oats, ear organs (glumes and lemmas) sustained water status and photosynthetic activity under drought, enabling continued carbon assimilation and grain filling even when flag leaf function declined (Fang et al.).
2.2 Nocturnal Water Relations and Sap Flow
The woody shrub Cotoneaster multiflorus utilizes nocturnal sap flow and refill mechanisms to re‐establish water balance after severe drought; stomatal closure during the day, followed by nighttime replenishment, preserves cellular hydration and supports rapid recovery upon rewatering (Huang et al.).
2.3 Symbiotic and nanomaterial interventions
Beneficial microbes and nanotechnology offer novel routes for physiological enhancement. In alfalfa, inoculation with Glomus mosseae and G. etunicatum improved plant height, chlorophyll content, osmolyte levels, and antioxidant enzyme activities under salt stress, underscoring the practical potential of AM fungi in saline soils (Xu et al.). Zinc oxide nanoparticles applied via nano-priming restored biomass, water content, and ion homeostasis in Phaseolus vulgaris under salinity, with nano-priming outperforming foliar and soil applications (Gupta et al.).
3 Biochemical mechanisms and metabolite regulation
3.1 Antioxidant defense and osmotic adjustment
The overproduction of reactive oxygen species (ROS) under stress can be mitigated through enhanced antioxidant systems and osmolyte accumulation. Tobacco seedlings treated with 0.2 mg L⁻¹ strigolactone presented elevated chlorophyll content, photosynthetic efficiency, and peroxidase/catalase activity, in addition to reduced malondialdehyde (MDA) and ROS, particularly in the moisture-sensitive cultivar Y116 (Wang et al.). In rice, seedling application of uniconazole under salt stress increased peroxidase and catalase in both tolerant (HD961) and sensitive (9311) varieties, improving root growth and carbohydrate partitioning to grains and thereby increasing yield components by up to 28 percent (Du et al.).
3.2 Secondary metabolites and quality prediction
Fresh leaves of Isatis indigotica emitted distinct biophoton signals under salt and drought, which correlated with the active ingredient levels and antibacterial efficacy of the dried herb. Delayed luminescence and spontaneous photon count serve as rapid, non-destructive indicators of cultivation quality (Wang et al.).
3.3 Fulvic acid and metabolomic shifts
Fulvic acid application in oats under drought increased the leaf water content and antioxidant enzyme activity, whereas integrated transcriptome–metabolome profiling revealed that the phenylpropanoid and glutathione pathways are central to FA-mediated protection (Zhu et al.).
4 Molecular and genomic insights
4.1 Gene family characterization
Recent studies have uncovered critical gene families linked to stress responses across plant species. In Moso bamboo, researchers identified 47 DIR genes, grouping them into three categories. Among these, the PeDIR02 gene stands out— it is found in cell membranes and appears to drive rapid shoot growth while helping the plant cope with environmental stressors like drought or salinity. This gene works within a network regulated by transcription factors such as ERF, DOF, and MYB, suggesting a coordinated strategy for stress adaptation (Xuan et al.). Meanwhile, alfalfa’s nine ADF genes fall into four evolutionary subgroups. Experiments revealed that MsADF1, MsADF2/3, MsADF6, and MsADF9 are strongly induced by salt and drought, indicating that actin remodeling is involved in stress adaptation (Shi et al.). Water lilies (Nymphaea colorata), on the other hand, boast 94 class III peroxidase genes, many of which evolved through tandem duplications. These genes activate under stressors like high salt, extreme temperatures, or heavy metals, pointing to their role in detoxification and defense (Khan et al.).
4.2 Functional validation in model systems
Overexpression studies have pinpointed candidate genes for crop engineering. For example, when the AtaHMGR10 gene from Asparagus taliensis was overexpressed in Arabidopsis, the modified plants showed stronger seed germination, longer roots, and better antioxidant responses under drought, salt, and osmotic stress (Zeng et al.). Similarly, poplar trees engineered to overexpress PagSOD2a displayed higher antioxidant enzyme activity, lower oxidative damage (measured by reduced malondialdehyde levels), and improved growth in salty soils. The study also traced this trait’s regulation to upstream genes like SPL13, NGA1b, and FRS5 (Zhou et al.). In another experiment, a caffeic acid O-methyltransferase gene from Ligusticum chuanxiong boosted both lignin production and melatonin synthesis in Arabidopsis, enhancing drought tolerance through stronger antioxidant defenses (Huang et al.).
4.3 RNA-seq and population genomics
By combining RNA sequencing with population genetics, light is being shed on stress adaptation mechanisms. In flax, a merger of transcriptomics and genome-wide association studies (GWAS) pinpointed 17 salt-tolerance genes involved in pathways like phenylpropanoid biosynthesis and metal transport. Interestingly, oil flax varieties showed higher genetic diversity at these key loci compared to fiber flax, possibly explaining their greater resilience (Li et al.). For drought tolerance, transcriptome comparisons of Codonopsis pilosula seedlings highlighted cultivar-specific differences. The drought-resistant cultivar G1, for instance, activated unique genes tied to starch/sucrose metabolism, hormone signaling, and glutathione pathways—clues that could guide future breeding efforts (Wang et al.).
5 Multiomics and integrative approaches
5.1 Hexaploid triticale seedling responses
When salt-stressed triticale seedlings were analyzed using a mix of genome-wide association studies (GWAS), transcriptomics, and proteomics, researchers uncovered 81 genetic markers linked to stress tolerance and 688 genes/proteins that changed activity within 18 hours. A protein called LEA14 stood out as a fast-acting defender against stress, likely because its DNA contains regions targeted by multiple stress-related transcription factors (Wang et al.).
5.2 Dynamic salt stress regulation in maize
How do salt-tolerant and salt-sensitive maize varieties react differently to stress? A time-based study comparing the resilient SPL02 line and the sensitive Mo17 line under high salt conditions showed nearly 9,000 unique gene activity changes in each. The hardy SPL02 line relied heavily on pathways like map kinase signaling, phenylpropanoid production, and hormone regulation, while Mo17 leaned more on ABA-activated stress signaling. Five key genes, including phosphate transporters and WRKY transcription factors, were flagged as top candidates for future research (Maimaiti et al.).
6 Reviews and perspectives
6.1 Phytohormones in horticultural drought tolerance
A comprehensive review highlights the exogenous application of melatonin, salicylic acid, jasmonates, strigolactones, brassinosteroids, and γ-aminobutyric acid as modulators of molecular and physiological defense systems in horticultural crops under drought, emphasizing hormone crosstalk and integrated signaling networks. The authors stress the need to decode these complex signaling networks to improve crop resilience (Huang and Jin).
6.2 Fibrillins and redox signaling in response to multi-stress
Two studies shed light on stress adaptation strategies. One focuses on fibrillins, proteins in plant plastids that help manage lipid storage and oxidative stress. The other proposes a new model where epigenetic changes (like DNA methylation) and redox signaling work together to fine-tune plant responses to combined stressors. Both papers argue for targeting these mechanisms in breeding programs to create tougher crops (El-Sappah et al.; Shriti et al).
7 Concluding remarks
The research presented in this Research Topic showcases how blending insights from genetics, physiology, and cutting-edge technology can unlock the secrets of plant resilience. By studying everything from root and shoot structures to antioxidant systems and gene networks, scientists are uncovering how plants survive droughts and salty soils. The real challenge lies in transforming these discoveries into practical solutions. To succeed, researchers, breeders, and farmers will need to collaborate closely, harnessing tools like gene editing, precision breeding, and smarter agricultural practices. Together, these efforts could pave the way for crops that thrive in our planet’s increasingly harsh environments, helping protect food supplies and livelihoods for future generations.
Author contributions
SS: Writing – review & editing, Writing – original draft, Visualization, Conceptualization. PL: Writing – review & editing. UC: Writing – review & editing. MW: Conceptualization, Writing – review & editing, Writing – original draft.
Acknowledgments
We thank all contributing authors, reviewers, and the Frontiers in Plant Science editorial team for their support in assembling this Research Topic. SS acknowledges the support from the 10KP project and China National GeneBank.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. The Figure 1 was edited using Claude AI (https://claude.ai/)
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Editorial (2024). Feeding the future global population. Nat. Commun. 15, 222. doi: 10.1038/s41467-023-44588-y
Guo, X., Wang, Y., Zhao, C., Tan, C., Yan, W., Xiang, S., et al. (2025). An Arabidopsis single-nucleus atlas decodes leaf senescence and nutrient allocation. Cell. doi: 10.1016/j.cell.2025.03.024
Purugganan, M. D. and Jackson, S. A. (2021). Advancing crop genomics from lab to field. Nat. Genet. 53, 595–601. doi: 10.1038/s41588-021-00866-3
Sahu, S. K. (2024). Maize gets an iron boost: Biofortification breakthrough holds promise to combat iron deficiency. J. Integr. Plant Biol. 66, 635–637. doi: 10.1111/jipb.13623
Sahu, S. K. and Liu, H. (2023). A genetic solution for the global food security crisis. J. Integr. Plant Biol. 65 (6), 1359–1361. doi: 10.1111/jipb.13500
Sahu, S. K., Waseem, M., and Aslam, M. M. (2023). Bioinformatics, big data and agriculture: a challenge for the future. Front. Media SA). 14, 1271305. doi: 10.3389/fpls.2023.1271305
Wang, Y., Wang, X., Sun, S., Jin, C., Su, J., Wei, J., et al. (2022). GWAS, MWAS and mGWAS provide insights into precision agriculture based on genotype-dependent microbial effects in foxtail millet. Nat. Commun. 13, 5913. doi: 10.1038/s41467-022-33238-4
Keywords: stress response, salinity stress, drought stress, plant physiology, molecular response, plant defense
Citation: Sahu SK, Liu P, Chandrasekaran U and Waseem M (2025) Editorial: Salinity and drought stress in plants: understanding physiological, biochemical and molecular responses, volume II. Front. Plant Sci. 16:1625602. doi: 10.3389/fpls.2025.1625602
Received: 09 May 2025; Accepted: 21 May 2025;
Published: 12 June 2025.
Edited and Reviewed by:
Jai Rohila, United States Department of Agriculture, United StatesCopyright © 2025 Sahu, Liu, Chandrasekaran and Waseem. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sunil Kumar Sahu, c3VuaWxrdW1hcnNhaHVAZ2Vub21pY3MuY24=; Muhammad Waseem, MTg0MzI4QGhhaW5hbnUuZWR1LmNu
 Sunil Kumar Sahu
Sunil Kumar Sahu Pingwu Liu
Pingwu Liu Umashankar Chandrasekaran
Umashankar Chandrasekaran Muhammad Waseem
Muhammad Waseem