- State Key Laboratory of Tree Genetics and Breeding, National Key Laboratory for the Development and Utilization of Forest Food Resources, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing, China
Lipid droplets (LDs) have emerged as dynamic organelles central to plant lipid metabolism, stress adaptation, and energy homeostasis. This review synthesizes recent advances in understanding LD biogenesis and degradation in plants, highlighting conserved and divergent mechanisms relative to other eukaryotes. LD formation originates in the endoplasmic reticulum (ER), where neutral lipids synthesized by diacylglycerol acyltransferases (DGAT) and phospholipid: diacylglycerol acyltransferases (PDAT) accumulate into lens-like structures. These structures bud into the cytosol via ER machinery, including SEIPIN complexes, vesicle-associated membrane proteins, and LD-associated protein-interacting protein which regulate LD size and abundance. Degradation occurs through two major pathways: lipolysis, mainly mediated by the patatin-like lipase SUGAR-DEPENDENT1, and lipophagy, where AUTOPHAGY-RELATED proteins deliver LDs for breakdown. LDs also function as stress-responsive hubs, accumulating under abiotic stresses and during pathogen interactions, where they participate in membrane remodeling and antimicrobial defense. Extensive studies in major oilseed crops reveal that expressions of multiple genes involved in LD turnover are significantly induced under various abiotic stresses and phytohormone treatments. These genetic components operate autonomously or synergistically (e.g. DGAT and PDAT) within the TAG biosynthesis and LD metabolic pathways, effecting concurrent enhancements in stress resilience and oil production under suboptimal growth conditions. Critical knowledge gaps persist, including the interplay between lipolysis and lipophagy, the integration of energy-related signaling pathways in LD turnover, and stress-modulated post-translational control of LD proteome. Deciphering these mechanisms will advance our understanding towards LD biology.
1 Introduction
Since their identification as organelles in the 19th century, lipid droplets (LDs) have undergone various nomenclature changes. They were once referred to as lipid bodies, adiposomes, oil bodies, sphaerosomes and oleosomes, but are now commonly known as LDs (Walther and Farese, 2012). LDs are lipid-rich organelles, which possess a core of neutral lipids, predominantly triacylglycerols (TAGs) and steryl/wax esters, which is encased by a monolayer of phospholipids (PLs).
LDs are derived from the endoplasmic reticulum (ER), and the biogenesis of LDs includes the following key steps: neutral lipid synthesis at the ER; formation of a lipid lens; budding of LDs; LD growth and maturation (Mathiowetz and Olzmann, 2024). In the last decade, proteins involved in these steps have been well characterized in plants, especially model plant Arabidopsis thaliana. These proteins include TAG-synthesizing enzymes and the proteins responsible for the LDs generation, such as SEIPIN, VESICLE-ASSOCIATED MEMBRANE PROTEIN-ASSOCIATED PROTEIN 27 (VAP27) and LD-ASSOCIATED PROTEIN-INTERACTING PROTEIN (LDIP) (Barneda and Christian, 2017; Man et al., 2024). The degradation of LDs in plants is also a tightly regulated process, mainly mediated by lipolysis and lipophagy. Among the key players in lipolysis, a conserved patatin domain containing protein SUGAR-DEPENDENT1 (SDP1) stands out (Eastmond, 2006; Huang et al., 2022). In A. thaliana leaves, lipophagy occurs through microautophagy, relying on the core components of the macroautophagy pathway (Fan et al., 2019a).
The role of LDs in carbon reserve storage is fundamental to the survival and growth of plants. However, over the past decade, a paradigm shift has occurred in the perception of LDs in plant biology. Except acting as static storage organelles, LDs are now recognized as dynamic subcellular structures actively involved in multiple physiological processes. Mounting evidence has shown that LDs play a crucial role in stress adaptation. Under abiotic stress conditions such as drought, cold, and heat stress, the abundance of LDs increases in plant cells (Yang et al., 2011; Kong et al., 2013; Kim et al., 2016; Yang et al., 2024). This new understanding has highlighted the importance of lipid metabolism, lipid transport, and stress responses in plants.
Given the significance of LDs in plant physiology, this review aims to provide a comprehensive overview of the latest research advancements in the biogenesis and degradation of LDs in plants. It will also explore the importance of LDs in the stress response of plants. By integrating findings from recent studies, we hope to shed light on the complex molecular and physiological processes associated with LDs in plants, which may have implications for crop improvement, bioenergy production, and understanding plant responses to environmental changes.
2 The proteins involved in the generation of lipid droplets
LD biogenesis in plant cells shares conserved mechanisms with other eukaryotes, relying on ER-localized protein machinery to initiate LD formation and on LD surface proteins to ensure proper cytoplasmic packaging. The process begins with the synthesis of neutral lipids within the ER, where they accumulate into lens-like structures between the ER membrane leaflets (Scholz et al., 2022). Key proteins, such as SEIPIN and VAP27, facilitate the budding of nascent LDs into the cytoplasm (Guzha et al., 2023). During this step, the phospholipid monolayer of the LD becomes continuous with the outer ER membrane leaflet. Subsequently, additional proteins, including lipins and LD coat proteins, are recruited to promote LD growth. However, the mechanism underlying LD dissociation from the ER remains poorly understood (Bouchnak et al., 2023).
2.1 Enzymes for neutral lipids synthesis
In plants, the ER serves as the principal site for TAG biosynthesis, which is mainly accomplished through the glycerol-3-phosphate (G3P) pathway or the Kennedy pathway (Xu and Shanklin, 2016). Firstly, glycerol-3-phosphate acyltransferase (GPAT) catalyzes the combination of G3P and Acyl-CoA, resulting in the formation of lysophosphatidic acid (LPA). Subsequently, under the catalytic action of lysophosphatidic acid phosphatase (LDPAT), LPA combines with Acyl-CoA once more to produce phosphatidic acid (PA). Phosphatidic acid phosphatase (PAP) then dephosphorylates PA to generate diacylglycerol (DAG). Finally, DAG undergoes final acylation to form TAG through two distinct mechanisms. The Acyl-CoA-dependent pathway, catalyzed by diacylglycerol acyltransferases (DGATs), utilizes Acyl-CoA as the acyl donor (Walther and Farese, 2012). Alternatively, phospholipid: diacylglycerol acyltransferase (PDAT) drives an Acyl-CoA-independent route by transferring an acyl moiety from phosphatidylcholines (PC) to DAG, producing TAG alongside a lysophospholipid (Bates et al., 2013). Thereafter, TAGs are subsequently stored between the two leaflets of the ER. As TAG accumulates and LDs enlarge, they separate from the ER membrane and enter the cytoplasm (Figure 1, Table 1).
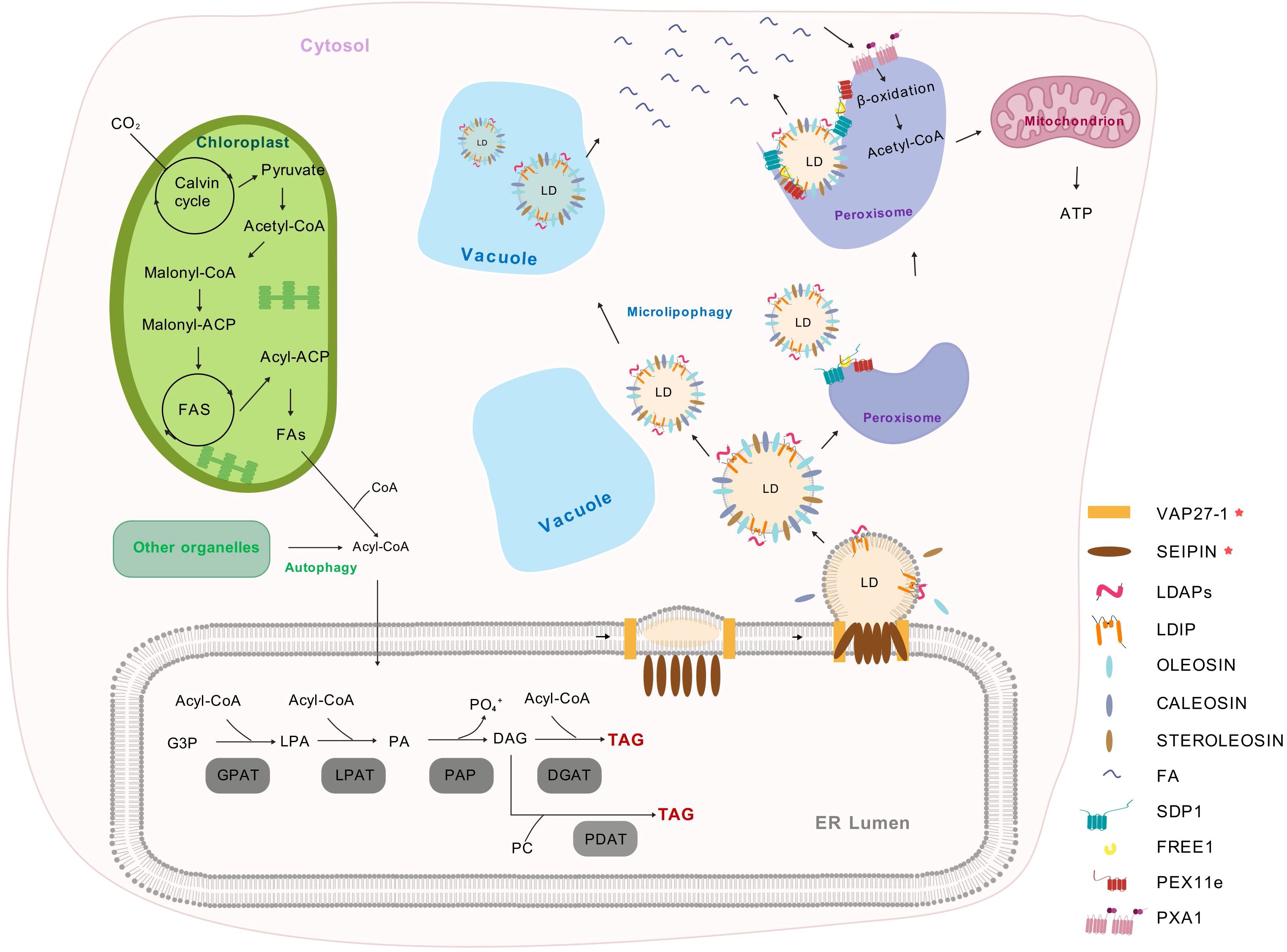
Figure 1. The biosynthesis and metabolism of plant lipid droplets (Park et al., 2013; Choi et al., 2022; Huang et al., 2022; Guzha et al., 2023). Plastids supply FAs that are transported to the cytosol and activated into Acyl-CoA. In the endoplasmic reticulum, G3P is acylated to form LPA using Acyl-CoA. LPA is further acylated by LPAT to produce PA. PAP dephosphorylates PA to DAG, which can be acylated by DGAT to form TAG. DAG can also exchange with PC, which are generated through an acyl editing cycle involving reacylation and acylation. LDs store TAG and are covered by a single layer of phospholipids and LD-associated proteins. The budding of LDs from the ER is regulated by the SEIPIN protein complex (including SEIPIN1, SEIPIN2, and SEIPIN3), which acts as an ER-localized scaffold protein to ensure proper LD formation by controlling neutral lipid synthesis and droplet size. Additionally, VAP27–1 functions as an ER-LD contact site protein, mediating phospholipid transfer to promote LD maturation and stabilize the LD formation complex. As LDs mature, they recruit proteins such as LDAP and LDIP, OLEOSIN, CALEOSIN, and STEROLEOSIN, which contribute to LD structure, stability, and function. During lipolysis, the ESCRT component FREE1 directly interacts with both PEX11e and SDP1, thereby regulating SDP1-mediated LD degradation and promoting FAs release. And these FAs are transported into peroxisome by PXA1 for β-oxidation. In contrast, lipophagy involves the selective autophagy of LDs, delivering them to vacuoles for breakdown. FAs, fatty acids; G3P, glycerol-3-phosphate; LPA, lysophosphatidic acid; LPAT, lysophosphatidic acid acyltransferase; PA, phosphatidic acid; PAP, Phosphatidic acid phosphatase; DAG, diacylglycerol; DGAT, acylated by diacylglycerol acyltransferase; PC, phosphatidylcholines; LDs, lipid droplets; TAG, triacylglycerol; ER, endoplasmic reticulum; VAP27-1, VESICLE-ASSOCIATED MEMBRANE PROTEIN-ASSOCIATED PROTEIN 27-1; LDAP, LIPID DROPLET-ASSOCIATED PROTEIN; LDIP, LDAP-INTERACTING PROTEIN; ESCRT, ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT; FREE1, FYVE DOMAIN PROTEIN REQUIRED FOR ENDOSOMAL SORTING 1; PEX11e, PEROXIN 11e; SDP1, SUGAR DEPENDENT 1; PXA1, PEROXISOMAL ABC TRANSPORTER 1.
Plants possess multiple DGAT isoform, including the ER-localized DGAT1 and DGAT2, as well as a soluble DGAT3 whose physiological role remains under investigation (Qin et al., 2023). In A. thaliana, PDAT contains two homologs, and PDAT1 is the dominant isoform in TAG biosynthesis (Fan et al., 2019b). Distinct expression patterns and functional specializations among TAG-synthesizing enzymes enable plants to adjust lipid metabolism according to developmental and environmental cues. In different plant species, such as A. thaliana, Camelina sativa and soybean, DGAT1 is the most highly expressed TAG biosynthetic enzyme (Hatanaka et al., 2022). Loss of AtDGAT1 activity in the A. thaliana dgat1–1 mutant leads to a reduction in seed oil content by at least 20% (Katavic et al., 1995; Regmi et al., 2020), while the dgat1–1 dgat2 double mutant does not display more oil reduction than dgat1–1 mutant (Zhang et al., 2009). As for PDAT genes, either oil content or FA composition is affected by the Atpdat1 mutation. The AtDGAT1 mutation causes the up-regulated expression of AtPDAT1, and the dgat1–1 pdat1–1 double mutant is lethal, indicating DGAT1 and PDAT1 have overlapping functions in A. thaliana TAG biosynthesis. The suppression of AtPDAT1 expression by RNAi interference in the dgat1–1 genetic background reduces oil accumulation by 70% to 80%, suggesting that PDAT1 rather than DGAT2 supports TAG biosynthesis when DGAT1 is lacking (Zhang et al., 2009). Furthermore, the detailed role of DGAT2, DGAT3 and PDAT2 in seed oil biosynthesis is unclear (Regmi et al., 2020).
In addition to TAGs, other forms of nonpolar lipids may also be present in LDs of some specific plant species. Wax esters (WEs), which are neutral lipids composed of a fatty alcohol esterified to a fatty acid. The WEs are synthesized through two enzymatic reactions catalyzed by fatty Acyl-CoA reductase and wax synthase. In jojoba (Simmondsia chinensis), a small shrub native to the deserts of North America, WEs can accumulate up to 60% of the seed weight (Sturtevant et al., 2020). Some algae, mosses, and pollen grains may also accumulate wax esters in LDs, though typically in smaller amounts (Guzha et al., 2023).
2.2 Lipid droplet proteins
Following their synthesis, neutral lipids — primarily TAGs — begin to accumulate between the leaflets of the ER membrane, forming small lens-like structures. These nascent lipid globules gradually expand through localized lipid synthesis and incorporation of additional neutral lipids. As these globules undergo expansion, they undergo a process of budding towards the cytosol, eventually maturing into discrete LDs (Figure 1, Table 1). This process of budding and stabilization is contingent on the recruitment of LD proteins. LD proteins are classified into two groups based on the pathways that they employ to traffic to LDs: class I LD proteins and class II LD proteins. Class I LD proteins are composed of proteins that are co-translationally inserted into the cytoplasmic face of the ER bilayer; in contrast, class II LD proteins target the LD from the cytoplasm (Mathiowetz and Olzmann, 2024).
2.2.1 OLEOSIN, CALEOSIN and STEREOLESIN
The presence of OLEOSIN proteins on the phospholipid layer of LDs plays a crucial role in LD formation and its functional regulations (Anaokar et al., 2024). The prevailing LD proteins identified in the seeds of plants are OLEOSIN, CALEOSIN, and STEREOLESIN (Guzha et al., 2023). In A. thaliana, there are a total of 16 OLEOSIN genes, which include five seed-type OLEOSIN genes. Among these, OLE1 is the most abundant OLEOSIN in A. thaliana seeds, followed by OLE2. OLEOSINs play a crucial role in preventing oil body fusion, thus maintaining the structural integrity of oil bodies. The OLEOSIN content is critical for oil body size regulation; a reduction in OLEOSIN content leads to an increase in oil body diameter due to the steric hindrance of OLEOSINs on the oil body surface inhibiting oil body fusion. Seeds of OLEOSIN single mutants (ole1 and ole2) contain larger oil bodies than those of the wild type, and seeds of an OLEOSIN double mutant (ole1 ole2) contain even larger oil bodies than those of ole1 and ole2 single mutants. This suggests that OLEOSINs are essential for normal germination and enhance plant survival during winter by inhibiting freezing stress-induced oil body fusion (Siloto et al., 2006). Recent studies have identified a low-abundance, seed-specific LD protein termed LIPID DROPLET PROTEIN OF SEEDS (LDPS), which contains an amphipathic α-helix and a proline hairpin motif that serve as LD targeting signals. A distinct domain of LDPS mediates its interaction with OLE1. ldps mutant shows smaller LDs, reduction in seed oil content, and complete absence of LD fusion during post-germinative growth. Genetic analyses using ole1 and ldps single mutants, double mutants, along with freeze-thaw experiments, demonstrated that OLE1 negatively regulates the LDPS-mediated promotion of LD expansion (Doner et al., 2025).
In comparison to OLEOSIN protein, CALEOSIN, which comprises three distinct domains, including N-terminal hydrophilic domains, C-terminal hydrophilic domains, and a central hydrophobic anchor domain, with the N-terminal domain containing a calcium-binding motif, exhibits a lower abundance (Shen et al., 2014; Liu et al., 2022). Moreover, from an evolutionary perspective, CALEOSIN protein exhibits homologous sequences in algae, fungi, and non-vascular plants, while such homology is not observed for OLEOSIN (Shen et al., 2014). The A. thaliana genome contains eight CALEOSIN genes divided into two groups: high-Mw CALEOSIN (CLO1, CLO2, CLO3 and CLO8) and low-Mw CALEOSIN (CLO4-LOL7) (Liu et al., 2022; Miklaszewska et al., 2023). Several studies have indicated that CALEOSIN proteins have overlapping functions in oil accumulation (Shen et al., 2014; Liu et al., 2022; Miklaszewska et al., 2023). The STEREOLESIN-related proteins participate in intracellular signaling during plant growth and development by being involved in the phytohormone pathways, e.g. brassinosteroids-related pathways (Shao et al., 2019).
2.2.2 Endoplasmic reticulum machinery: SEIPIN, LDIP, and VAP27
The SEIPIN complex, named after Berardinelli-Seip congenital lipodystrophy (BSCL), associates with these lipid lenses and directs the budding of nascent LDs into the cytoplasm. Most plants have multiple SEIPIN genes, in A. thaliana, three SEIPIN genes encode proteins with conserved structural features, predicted to form barrel-like complexes at the ER-LD junction (Arlt et al., 2022). Notably, AtSEIPIN2 and AtSEIPIN3 have longer N termini, with AtSEIPIN3 promoting the proliferation of very small LDs in leaves (Cai et al., 2015). The FFAT motifs present at the N termini of both SEIPIN2 and SEIPIN3 have been shown to interact with VAPs (Slee and Levine, 2019). VAPs, which are conserved across kingdoms, have been identified as the structural elements that facilitate contact sites between organelle membranes. The LD-forming complex has been demonstrated to be stabilized by VAP27–1 through a direct interaction with the N terminus of SEIPIN2 and/or SEIPIN3, a process that is deemed to be essential for LD biogenesis (Greer et al., 2020). In planta, loss of VAP27–1 results in the formation of large LDs in seeds, a phenotype similar to that observed in seipin2 seipin3 double mutants (Taurino et al., 2018). In addition, AtSEIPIN2 and AtSEIPIN3 are crucial for the modulation of the number and size of LDs by interacting with LDIP, facilitating LD biogenesis (Pyc et al., 2021).
3 Lipid droplet degradation through lipolysis by cytosolic lipases
In yeast, Drosophila, plants, and humans, stored TAGs are typically degraded by lipases, a conserved protein family with a patatin domain (Xu and Shanklin, 2016). As mentioned previously, lipolysis refers to the process of TAG degradation in LDs mediated by lipases, whereas lipophagy denotes an autophagic mechanism for LD degradation. The stored lipids are hydrolyzed by these lipases, leading to the breakdown of TAGs into components such as diacylglycerols (DAGs), monoacylglycerols (MAGs), fatty acids (FAs), and glycerol. Subsequent to this process, the hydrolytic byproducts enter diverse metabolic pathways, thereby playing pivotal roles in cellular growth, energy balance, and other physiological processes, occurring at the opportune moment (Kretzschmar et al., 2020). SDP1, the patatin-like acyl-hydrolase domain protein encoding gene, was discovered using forward genetic screening in A. thaliana (Figure 1) (Eastmond, 2006). Subsequent evidence suggests that this protein also serves as a primary enzyme for TAG hydrolysis in the leaves and roots of mature plants (Kelly et al., 2013a; Fan et al., 2014). During the early stages of seed germination in A. thaliana, SDP1 initially localizes to the surface of peroxisomes in an inactive form and subsequently extends to the surface of LDs within peroxisomes to hydrolyze TAGs (Thazar-Poulot et al., 2015). Further investigations have revealed that FYVE DOMAIN PROTEIN REQUIRED FOR ENDOSOMAL SORTING 1 (FREE1), within the ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT (ESCRT), directly interacts with both PEROXIN 11e (PEX11e) and SDP1, thereby facilitating the transport of SDP1 from peroxisomes to LDs (Huang et al., 2022). In addition to SDP1, A. thaliana possesses other patatin domain-containing lipases, such as SDP1-LIKE (SDP1L), which exhibit lipase activity and can release FAs from TAGs (Kelly et al., 2011, 2013a). Both SDP1 and SDP1L proteins are involved in the hydrolysis of TAGs during seed germination and also vegetative growth (Kelly et al., 2013a; Huang et al., 2022). In addition, the AtOBL1 gene in A. thaliana encodes for an enzyme known as OIL BODY LIPASE 1, which is associated with LDs. AtOBL1 represents the only described TAG lipase from A. thaliana that is associated with LDs, as SDP1 is regarded as a peroxisome-associated protein (Müller and Ischebeck, 2018).
Besides, biochemical analysis indicates that SDP1 and SDP1L preferentially hydrolyze TAGs over DAGs and monoacylglycerols (MAGs). The purification of oil body membranes from sdp1 sdp1L double mutant seedlings revealed a deficiency in TAG lipase activity. However, the hydrolysis of DAGs and MAGs was still observed, indicating the presence of other lipid enzymes that function in synergy with patatin-like acyl-hydrolases to complete the hydrolysis of TAG (Eastmond, 2006; Kelly et al., 2011, 2013a).
Following the liberation of FAs into the cytoplasm by SDP1, these FAs are converted into CoA esters through the action of currently unidentified Acyl-CoA synthetases (Li et al., 2016). The subsequent translocation of FAs across the peroxisomal membrane is facilitated by PXA1, an ABCD transporter belonging to the ATP-binding cassette (ABC) transporter family. Notably, PXA1 exhibits a unique intrinsic Acyl-CoA thioesterase activity (De Marcos Lousa et al., 2013). This distinctive property enables PXA1 to first bind fatty Acyl-CoAs on the cytosolic face of the peroxisomal membrane, then cleave the CoA moiety, and ultimately mediate the import of free FAs into the peroxisomal matrix for β-oxidation - a metabolic process that yields Acetyl-CoA as the end product (Kunz et al., 2009; Li et al., 2016; Fan et al., 2017). In the case of impaired β-oxidation function, pxa1 mutants exhibit delayed germination and reduced germination rate due to insufficient ATP supply required for the germination process. However, this defect can be alleviated by supplementing external carbon sources (Kunz et al., 2009). In addition, compared to the wild type, pxa1 mutant shows increased sensitivity to dark conditions and exhibits early plant death due to the compromised β-oxidation (Fan et al., 2017).
In addition, COMPARATIVE GENE IDENTIFICATION-58 (CGI-58) protein positively regulates lipid metabolism through β-oxidation-related pathway. Chapman et al. identified the homologous gene of human CGI58 in A. thaliana, referred to as CGI58-like (Yamaguchi and Osumi, 2009; James et al., 2010). In the A. thaliana mutant of this gene, plant leaves display a significantly increased TAG content of over tenfold compared to the wild type. However, unlike the sdp1 mutants, germination and growth of cgi-58 mutants do not show obvious defects (Yamaguchi and Osumi, 2009). Subsequent studies by Park et al. demonstrated that CGI-58 interacts with PXA1 to coregulate lipid homeostasis and signaling in A. thaliana (Park et al., 2013).
In the context of seedling establishment, the rapid breakdown of TAGs in planta, predominantly within four days, is particularly noteworthy. This phenomenon is further compounded by the accelerated degradation of LD-associated proteins, which may contribute to the enlargement of LDs during this critical phase. Studies have demonstrated that several LD proteins, including oleosins and steroleosins, have been observed to undergo polyubiquitination, a process associated with protein degradation (Deruyffelaere et al., 2018). This pathway, which is dependent on the removal of proteins from membranes, involves the action of the ubiquitin-proteasome system. Intriguingly, the analysis highlights the potential role of CELL DIVISION CYCLE PROTEIN 48 (CDC48) unfoldases, conserved in eukaryotes, in facilitating the unfolding and removal of membrane proteins. In planta, CDC48 has been observed to collaborate with PUX10, a scaffold protein residing at the LDs, to facilitate the degradation of ubiquitinated proteins (Kretzschmar et al., 2018; Liu et al., 2024). pux10 mutants exhibit a reduced rate of LD protein degradation and an accumulation of ubiquitinated proteins (Deruyffelaere et al., 2018). However, to date, no known degradation mechanism has been identified for LD membrane lipids.
4 Autophagic degradation of lipid droplets
Lipophagy, a selective autophagic process, first described in mammals, is a process that involves the selective uptake of LDs into the vacuole or lysosome, followed by their degradation (Figure 1, Table 1). Notably, mammalian lipophagy is a form of macroautophagy, in which autophagosomes engulf LDs, distinguishing it from microlipophagy observed in yeast. Autophagy, a self-degradative and highly conserved process, plays a crucial role in various developmental processes within cellular organisms (Zhao et al., 2020). Autophagy primarily functions through vacuolar degradation and recycling of harmful or obsolete cellular components, thereby maintaining cellular homeostasis and facilitating adaptation to environmental changes (Avin-Wittenberg et al., 2012; Couso et al., 2018; Sun et al., 2018; Bao et al., 2020). The identification of AUTOPHAGY-RELATED (ATG) genes in Saccharomyces cerevisiae revolutionized our understanding of autophagy, revealing a highly conserved eukaryotic mechanism (Marshall and Vierstra, 2018). Subsequent studies identified homologous ATG genes in plants, including A. thaliana, Oryza sativa, and Zea mays, through sequence alignment analyses (Li et al., 2015). These studies uncovered over 40 evolutionarily conserved ATG proteins that orchestrate autophagosome biogenesis and autophagy regulation across kingdoms, from yeast to mammals and plants (Marshall and Vierstra, 2018).
In A. thaliana, two independent studies, Fan et al. and Havé et al., reached the same conclusion through different approaches, thereby demonstrating the involvement of autophagy in the degradation of lipids (Fan et al., 2019a; Havé et al., 2019). Their findings suggest that, in A. thaliana leaves, basal autophagy contributes to TAG synthesis, whereas inducible autophagy under starvation contributes to LD degradation (Fan et al., 2019a). Besides, direct evidence through ultrastructural analysis has demonstrated that LDs are degraded in autophagic vacuoles (Fan et al., 2019a). In the parallel study, Havé et al. utilized protein and lipid profiling analyses on atg5 mutant, demonstrating that autophagy plays a pivotal role in the lipid metabolism of the ER and peroxisome in A. thaliana leaves (Fan et al., 2019a; Havé et al., 2019). Fan et al. investigated the role of autophagy in lipid metabolism by using mutants with auto (Havé et al., 2019). In addition to its role in A. thaliana, autophagy has been observed to contribute to the degradation of LD in other plant species. In rice, investigating osatg7 mutants has demonstrated autophagy’s crucial role during the late stages of pollen meiosis. As LDs are critical for energy supply, osatg7 mutants exhibit reduced levels of autophagy, and such deficiency leads to impaired pollen maturation (Kurusu et al., 2014).
5 Lipid droplets are involved in abiotic and biotic stress responses
5.1 Abiotic stress
It is imperative to acknowledge that plants are subject to numerous stressors throughout their life cycle, which necessitates the orchestration of adaptive responses to these environmental cues by all cellular organelles. Among these organelles, cytosolic LDs and their core set of neutral lipids and associated surface proteins play a significant yet understudied role. It has been demonstrated that environmental changes have a substantial influence on LD-related processes. For example, the abundance of LDs in A. thaliana leaves increases under drought, cold, or heat stress (Yang et al., 2011; Kong et al., 2013; Kim et al., 2016; Yang et al., 2024).
A close relationship exists between stress and TAG accumulation in plant tissues, especially the vegetative tissues (Lee et al., 2019). For instance, low-nitrogen stress and the stress hormone abscisic acid (ABA) have been observed to stimulate TAG accumulation in A. thaliana seedlings (Yang et al., 2011; Kong et al., 2013; Coulon et al., 2024). During periods of heat stress, cells undergo a process of unsaturated acyl chain replacement with saturated ones, a process that may lead to an increase in membrane fluidity (Mueller et al., 2017; Yang et al., 2024). This phenomenon suggests that LDs may absorb discarded unsaturated acyl chains from membrane lipids, resulting in the formation of triacylglycerols, thereby facilitating membrane remodeling (Yang et al., 2011; Scholz et al., 2025). Transgenic plants overexpressing LIPID DROPLET-ASSOCIATED PROTEINS (LDAPs) exhibit enhanced drought tolerance, suggesting a close relationship between stress and TAG accumulation in vegetative tissues (Zhao et al., 2023).
It has been determined that ABA signaling plays a pivotal role in the regulation of LD generation, particularly with regard to the expression of DGAT1. Tobacco transient assays have revealed a synergistic effect of ABA-insensitive 4 (ABI4) and ABI5, two important ABA-related transcription factors, in regulating DGAT1 expression under stress (Yang et al., 2011; Kong et al., 2013). Furthermore, a comprehensive transcriptome analysis has revealed that LIPID DROPLET PROTEIN (LDP) genes, including OLEOSINs and CALEOSINs, exhibited up-regulation of up to 1000-fold through the activation of ABI3. This provides compelling genetic evidence that ABI3 activates oil accumulation, most likely through up-regulating LDPs (Yang et al., 2022).
5.2 Biotic stress
Furthermore, LDs have been observed as targets by invasive organisms. Phytophthora infestans degrade LDs as energy source in guard cells to maintain stomatal opening (Yang et al., 2021). Plant RNA viruses induce endomembrane proliferation for viral replication compartments (VRCs) formation, and the host lipid metabolism is crucial for their replication. However, to date, direct links between LDs and plant virus infection have not been firmly established, and the extent of their involvement in plant defense or viral benefit remains to be elucidated (Zhang et al., 2019). In addition, during infections by pathogens such as Botrytis cinerea or Pseudomonas syringae, the leaves of plants exhibit an increased accumulation of TAGs (Sham et al., 2014; Galluzzi and Green, 2019). Besides, LDs have been proposed to function as “subcellular factories” for the production of antimicrobial compounds. For instance, two key lipid-modifying enzymes - peroxygenase (CLO3) and α-DIOXYGENASE (α-DOX) - coordinately catalyze a coupling reaction that converts α-linolenic acid into the antifungal compound 2-hydroxy-octadecanoic acid during defense responses against Colletotrichum higginsianum infection (Fernández-Santos et al., 2020). LDAP1, CLO3, and α-DOX1 are upregulated in leaves infected by Botrytis cinerea, suggesting that LD biosynthesis is induced either by the fungi or as a plant defense mechanism. The fatty acid composition of TAGs varies depending on the infecting pathogen, indicating the presence of distinct synthesis pathways. The hijacking of LDs by pathogens or their utilization by plants for defense mechanisms bears resemblance to the processes observed in animal cells (Roingeard and Melo, 2017). Meanwhile, PHYTOALEXIN DEFICIENT 3 (PAD3), a cytochrome P450 monooxygenase known to be involved in the biosynthesis of antimicrobial phytoalexins, has been observed to translocate to LDs following infection by Pseudomonas syringae (Fernández-Santos et al., 2020). This dynamic relocation of defense-related enzymes to LDs highlights the organelle’s emerging role as a critical platform for organizing plant immune responses.
6 Conclusions and prospects
This review offers a detailed examination of the processes involved in the formation and breakdown of LDs in plants, emphasizing crucial enzymes, regulatory pathways, and physiological contexts. A more profound understanding of the regulatory mechanisms governing LD-associated pathways holds considerable potential for enhancing crop yield and promoting bioenergy production. Manipulating the genes and proteins involved in LD biogenesis and turnover may lead to the development of crops with higher oil yields and improved stress resilience.
Many studies have demonstrated that LDs play a crucial role in cellular lipid homeostasis. While much attention has been paid to how LD size and number are determined in plants (Doner et al., 2025), the regulation of lipolysis and lipophagy-mediated lipid turnover, particularly during nutrient deprivation when plants rely on lipid catabolism for energy production, remains poorly understood. In mammals, the process of lipolysis is subject to stringent regulation, with the rate-limiting enzyme Adipose Triglyceride Lipase (ATGL) being subject to enhancement of up to 20-fold through its interaction with the activator CGI-58 (ABHD5) (Mathiowetz and Olzmann, 2024). Concurrently, PLIN proteins function as a regulatory mechanism, sequestering CGI-58 and thereby impeding ATGL activity (Mathiowetz and Olzmann, 2024). In contrast, plants employ SDP1 as their functional ATGL homolog, but lack both PLIN proteins and CGI-58-mediated activation of SDP1, despite the presence of a CGI-58 homolog that instead regulates PXA1 (Park et al., 2013). The current understanding of plant lipolysis regulation remains incomplete, particularly regarding whether energy-sensing pathways modulate SDP1 activity. The energy-sensing central regulators include the low-energy sensor SnRK1, the high-energy sensor TOR kinase and the sucrose-signaling metabolite T6P. These components form an intricate regulatory network where SnRK1 promotes lipolysis during energy deficit while TOR suppresses it under energy-replete conditions, with T6P fine-tuning this balance by inhibiting SnRK1 (Figueroa and Lunn, 2016; Liu and Xiong, 2022; Van Leene et al., 2022). Critical areas for future investigation include investigating the possible direct phosphorylation of SDP1 by SnRK1/TOR kinases, characterizing the functional relationship between the energy sensing module and SDP1 during lipid mobilization, and identifying potential novel components that facilitate communication between SDP1 and energy-sensing pathways. Resolution of these questions will significantly advance our understanding of the molecular mechanisms controlling LD degradation and overall plant lipid homeostasis.
Current research indicates that plants may dynamically regulate the functions of LD-associated proteins through post-translational modifications (PTMs) in response to environmental stresses. Despite extensive characterization of LD protein PTMs in animal systems (Zhang et al., 2022; Loix et al., 2024), their functional validation and molecular mechanisms remain largely unexplored in plants. Ubiquitination may regulate LD protein turnover through either proteasomal degradation or selective autophagy (e.g., lipophagy), maintaining cellular homeostasis under stress conditions. Furthermore, oxidative modifications and SUMOylation likely participate in mediating LD-organelle interactions (e.g., with peroxisomes), affecting membrane remodeling and ROS scavenging. Future investigations should integrate subcellular proteomics, PTM site-directed mutagenesis, and super-resolution imaging to systematically decipher stress-specific PTM dynamics on LD proteins and their physiological relevance. Such advances would not only elucidate the regulatory mechanisms of plant lipid metabolism under stress but may also provide novel strategies for improving crop stress tolerance.
LDs serve as critical organelles in stress response mechanisms. Numerous abiotic stressors have been shown to induce LD biogenesis (Zhao et al., 2023; Coulon et al., 2024). During senescence or stress conditions, TAG accumulation is closely linked to lipid catabolic processes. However, several key aspects remain poorly understood: the functional significance of fatty acids derived from membrane lipids like Monogalactosyldiacylglycerol (MGDG) (Fan et al., 2017); the specific roles of various lipases in stress adaptation; and the degradation mechanisms of stress-induced LDs during post-stress recovery. While LD degradation during seed germination has been well characterized (Kelly et al., 2011), the catabolic pathways of stress-induced LDs and their contributions to cellular homeostasis restoration remain elusive. Particularly, the relative importance of lipolysis versus lipophagy in TAG remobilization, the metabolic fates of neutral lipids, and the subsequent utilization of released fatty acids all require systematic investigation (Coulon et al., 2024). Elucidating these processes will not only advance our understanding of plant stress responses but also provide a theoretical framework for developing stress-resistant crops through LD manipulation.
Research has shown that the proteome of LDs in plants undergoes extensive dynamic remodeling under diverse stress conditions (Krawczyk et al., 2022). This is evidenced by the specific upregulation of stress-responsive LD-associated proteins, such as CLO3 and α-DOX1, in both wild-type plants and the tgd1–1 sdp1–4 mutant (Shimada et al., 2014). Notably, different stresses exhibit distinct regulatory effects on LD proteins: CLO3 responds to both heat stress and pathogen infection, whereas α-DOX1 is selectively activated only under pathogen infection and drought stress conditions (Scholz et al., 2025). These findings suggest that plants have evolved a stress-specific LD reprogramming mechanism, fine-tuning protein expression to adapt to different environmental threats. However, the molecular mechanisms governing LD remodeling under various stress conditions remain poorly understood and warrant further investigation.
Author contributions
YZ: Writing – original draft, Writing – review & editing. RC: Writing – review & editing, Writing – original draft. JL: Writing – original draft. YX: Writing – review & editing. LZ: Writing – review & editing. YY: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32300249 to LJ Zhou, 32200213 to YJ Ye) and Natural Science Foundation of Jiangsu Province (BK20220418 to LJ Zhou, BK20220417 to YJ Ye).
Acknowledgments
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Correction note
A correction has been made to this article. Details can be found at: 10.3389/fpls.2025.1727402.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Anaokar, S., Liang, Y., Yu, X., Cai, Y., Cai, Y., and Shanklin, J. (2024). The expression of genes encoding novel Sesame OLEOSIN variants facilitates enhanced triacylglycerol accumulation in Arabidopsis leaves and seeds. New Phytol. 243, 271–283. doi: 10.1111/nph.19548
Arlt, H., Sui, X., Folger, B., Adams, C., Chen, X., Remme, R., et al. (2022). SEIPIN forms a flexible cage at lipid droplet formation sites. Nat. Struct. Mol. Biol. 29, 194–202. doi: 10.1038/s41594-021-00718-y
Avin-Wittenberg, T., Honig, A., and Galili, G. (2012). Variations on a theme: plant autophagy in comparison to yeast and mammals. Protoplasma 249, 285–299. doi: 10.1007/s00709-011-0296-z
Aznar-Moreno, J. A., Mukherjee, T., Morley, S. A., Duressa, D., Kambhampati, S., Chu, K. L., et al. (2022). Suppression of SDP1 improves soybean seed composition by increasing oil and reducing undigestible oligosaccharides. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.863254
Bao, Y., Song, W., Wang, P., Yu, X., Li, B., Jiang, C., et al. (2020). COST1 regulates autophagy to control plant drought tolerance. Proc. Natl. Acad. Sci. U. S. A. 117, 7482–7493. doi: 10.1073/pnas.1918539117
Barneda, D. and Christian, M. (2017). Lipid droplet growth: regulation of a dynamic organelle. Curr. Opin. Cell. Biol. 47, 9–15. doi: 10.1016/j.ceb.2017.02.002
Bates, P. D., Stymne, S., and Ohlrogge, J. (2013). Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 16, 358–364. doi: 10.1016/j.pbi.2013.02.015
Bouchnak, I., Coulon, D., Salis, V., D’Andréa, S., and Bréhélin, C. (2023). Lipid droplets are versatile organelles involved in plant development and plant response to environmental changes. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1193905
Cai, Y., Goodman, J. M., Pyc, M., Mullen, R. T., Dyer, J. M., and Chapman, K. D. (2015). Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell. 27, 2616–2636. doi: 10.1105/tpc.15.00588
Cai, L., Li, X., Zhang, M., Gan, X., Yu, D., and Wang, H. (2025). Knocking out the CALEOSIN-encoding gene GmCLO1 improves soybean resistance to common cutworm. Physiol. Plant 177, e70260. doi: 10.1111/ppl.70260
Cao, Y., Zhao, L., Ying, Y., Kong, X., Hua, Y., and Chen, Y. (2015). The characterization of soybean oil body integral OLEOSIN isoforms and the effects of alkaline pH on them. Food Chem. 177, 288–294. doi: 10.1016/j.foodchem.2015.01.052
Chen, A., Hu, S., Zhu, D., Zhao, R., Huang, C., and Gao, Y. (2023). Lipid droplets proteome reveals dynamic changes of lipid droplets protein during embryonic development of Carya cathayensis nuts. Plant Sci. 334, 111753. doi: 10.1016/j.plantsci.2023.111753
Chen, J. C., Tsai, C. C., and Tzen, J. T. (1999). Cloning and secondary structure analysis of CALEOSIN, a unique calcium-binding protein in oil bodies of plant seeds. Plant Cell Physiol. 40, 1079–1086. doi: 10.1093/oxfordjournals.pcp.a029490
Choi, Y. J., Zaikova, K., Yeom, S., Kim, Y., and Lee, D. W. (2022). Biogenesis and lipase-mediated mobilization of lipid droplets in plants. Plants 11, 1243. doi: 10.3390/plants11091243
Coulon, D., Nacir, H., Bahammou, D., Jouhet, J., Bessoule, J., Fouillen, L., et al. (2024). Roles of plastoglobules and lipid droplets in leaf neutral lipid accumulation during senescence and nitrogen deprivation. J. Exp. Bot. 75, 6542–6562. doi: 10.1093/jxb/erae301
Courtois-Moreau, C. L., Pesquet, E., Sjödin, A., Muñiz, L., Bollhöner, B., Kaneda, M., et al. (2009). A unique program for cell death in xylem fibers of Populus stem. Plant J. 58, 260–274. doi: 10.1111/j.1365-313X.2008.03777.x
Couso, I., Pérez-Pérez, M. E., Martínez-Force, E., Kim, H., He, Y., Umen, J. G., et al. (2018). Autophagic flux is required for the synthesis of triacylglycerols and ribosomal protein turnover in Chlamydomonas. J. Exp. Bot. 69, 1355–1367. doi: 10.1093/jxb/erx372
De Marcos Lousa, C., van Roermund, C. W. T., Postis, V. L. G., Dietrich, D., Kerr, I. D., Wanders, R. J. A., et al. (2013). Intrinsic acyl-CoA thioesterase activity of a peroxisomal ATP binding cassette transporter is required for transport and metabolism of fatty acids. Proc. Natl. Acad. Sci. U. S. A. 110, 1279–1284. doi: 10.1073/pnas.1218034110
Demski, K., Łosiewska, A., Jasieniecka-Gazarkiewicz, K., Klińska, S., and Banaś, A. (2020). Phospholipid: Diacylglycerol Acyltransferase1 overexpression delays senescence and enhances post-heat and cold exposure fitness. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.611897
Deruyffelaere, C., Purkrtova, Z., Bouchez, I., Collet, B., Cacas, J., Chardot, T., et al. (2018). PUX10 is a CDC48A adaptor protein that regulates the extraction of ubiquitinated OLEOSINs from seed lipid droplets in Arabidopsis. Plant Cell. 30, 2116–2136. doi: 10.1105/tpc.18.00275
Doner, N. M., Clews, A. C., Esnay, N., Whitehead, P. S., Wang, Y., Romsdahl, T. B., et al. (2025). Lipid droplet protein of seeds is involved in the control of lipid droplet size in Arabidopsis seeds and seedlings. Plant Cell. 37, koaf121. doi: 10.1093/plcell/koaf121
Eastmond, P. J. (2006). SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell. 18, 665–675. doi: 10.1105/tpc.105.040543
Fan, J., Yan, C., Roston, R., Shanklin, J., and Xu, C. (2014). Arabidopsis lipins, PDAT1 acyltransferase, and SDP1 triacylglycerol lipase synergistically direct fatty acids toward β-oxidation, thereby maintaining membrane lipid homeostasis. Plant Cell. 26, 4119–4134. doi: 10.1105/tpc.114.130377
Fan, J., Yu, L., and Xu, C. (2017). A central role for triacylglycerol in membrane lipid breakdown, fatty acid β-oxidation, and plant survival under extended darkness. Plant Physiol. 174, 1517–1530. doi: 10.1104/pp.17.00653
Fan, J., Yu, L., and Xu, C. (2019a). Dual role for autophagy in lipid metabolism in Arabidopsis. Plant Cell. 31, 1598–1613. doi: 10.1105/tpc.19.00170
Fan, J., Zhou, C., Yu, L., Li, P., Shanklin, J., and Xu, C. (2019b). Diversion of carbon flux from sugars to lipids improves the growth of an Arabidopsis starchless mutant. Plants 8, 229. doi: 10.3390/plants8070229
Fernández-Santos, R., Izquierdo, Y., López, A., Muñiz, L., Martínez, M., Cascón, T., et al. (2020). Protein profiles of lipid droplets during the hypersensitive defense response of Arabidopsis against Pseudomonas infection. Plant Cell Physiol. 61, 1144–1157. doi: 10.1093/pcp/pcaa041
Figueroa, C. M. and Lunn, J. E. (2016). A tale of two sugars: trehalose 6-phosphate and sucrose. Plant Physiol. 172, 7–27. doi: 10.1104/pp.16.00417
Fox, A. R., Scochera, F., Laloux, T., Filik, K., Degand, H., Morsomme, P., et al. (2020). Plasma membrane aquaporins interact with the endoplasmic reticulum resident VAP27 proteins at ER-PM contact sites and endocytic structures. New Phytol. 228, 973–988. doi: 10.1111/nph.16743
Galluzzi, L. and Green, D. R. (2019). Autophagy-independent functions of the autophagy machinery. Cell 177, 1682–1699. doi: 10.1016/j.cell.2019.05.026
Gandla, M. L., Mähler, N., Escamez, S., Skotare, T., Obudulu, O., Möller, L., et al. (2021). Overexpression of vesicle-associated membrane protein PttVAP27–17 as a tool to improve biomass production and the overall saccharification yields in Populus trees. Biotechnol. Biofuels. 14, 43. doi: 10.1186/s13068-021-01895-0
Ghiglione, H. O., Gonzalez, F. G., Serrago, R., Maldonado, S. B., Chilcott, C., Curá, J. A., et al. (2008). Autophagy regulated by day length determines the number of fertile florets in wheat. Plant J. 55, 1010–1024. doi: 10.1111/j.1365-313X.2008.03570.x
Gidda, S. K., Park, S., Pyc, M., Yurchenko, O., Cai, Y., Wu, P., et al. (2016). Lipid droplet-associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol. 170, 2052–2071. doi: 10.1104/pp.15.01977
Greer, M. S., Cai, Y., Gidda, S. K., Esnay, N., Kretzschmar, F. K., Seay, D., et al. (2020). SEIPIN isoforms interact with the membrane-tethering protein VAP27–1 for lipid droplet formation. Plant Cell. 32, 2932–2950. doi: 10.1105/tpc.19.00771
Guzha, A., Gautam, B., Marchiafava, D., Ver Sagun, J., Garcia, T., Jarvis, B. A., et al. (2024). Targeted modulation of pennycress lipid droplet proteins impacts droplet morphology and seed oil content. Plant J. 120, 2151–2171. doi: 10.1111/tpj.17109
Guzha, A., Whitehead, P., Ischebeck, T., and Chapman, K. D. (2023). Lipid droplets: packing hydrophobic molecules within the aqueous cytoplasm. Annu. Rev. Plant Biol. 74, 195–223. doi: 10.1146/annurev-arplant-070122-021752
Hatanaka, T., Tomita, Y., Matsuoka, D., Sasayama, D., Fukayama, H., Azuma, T., et al. (2022). Different acyl-CoA: diacylglycerol acyltransferases vary widely in function, and a targeted amino acid substitution enhances oil accumulation. J. Exp. Bot. 73, 3030–3043. doi: 10.1093/jxb/erac084
Havé, M., Luo, J., Tellier, F., Balliau, T., Cueff, G., Chardon, F., et al. (2019). Proteomic and lipidomic analyses of the Arabidopsis atg5 autophagy mutant reveal major changes in endoplasmic reticulum and peroxisome metabolisms and in lipid composition. New Phytol. 223, 1461–1477. doi: 10.1111/nph.15913
Haxim, Y., Ismayil, A., Jia, Q., Wang, Y., Zheng, X., Chen, T., et al. (2017). Autophagy functions as an antiviral mechanism against geminiviruses in plants. Elife 6, e23897. doi: 10.7554/eLife.23897
Huang, S., Liu, Z., Cao, W., Li, H., Zhang, W., Cui, Y., et al. (2022). The plant ESCRT component FREE1 regulates peroxisome-mediated turnover of lipid droplets in germinating Arabidopsis seedlings. Plant Cell. 34, 4255–4273. doi: 10.1093/plcell/koac195
James, C. N., Horn, P. J., Case, C. R., Gidda, S. K., Zhang, D., Mullen, R. T., et al. (2010). Disruption of the Arabidopsis CGI-58 homologue produces chanarin-dorfman-like lipid droplet accumulation in plants. Proc. Natl. Acad. Sci. U. S. A. 107, 17833–17838. doi: 10.1073/pnas.0911359107
Jing, G., Tang, D., Yao, Y., Su, Y., Shen, Y., Bai, Y., et al. (2021). Seed specifically over-expressing DGAT2A enhances oil and linoleic acid contents in soybean seeds. Biochem. Biophys. Res. Commun. 568, 143–150. doi: 10.1016/j.bbrc.2021.06.087
Kanai, M., Yamada, T., Hayashi, M., Mano, S., and Nishimura, M. (2019). Soybean (Glycine max L.) Triacylglycerol lipase GmSDP1 regulates the quality and quantity of seed oil. Sci. Rep. 9, 8924. doi: 10.1038/s41598-019-45331-8
Katavic, V., Reed, D. W., Taylor, D. C., Giblin, E. M., Barton, D. L., Zou, J., et al. (1995). Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol. 108, 399–409. doi: 10.1104/pp.108.1.399
Kelly, A. A., Quettier, A., Shaw, E., and Eastmond, P. J. (2011). Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Plant Physiol. 157, 866–875. doi: 10.1104/pp.111.181784
Kelly, A. A., Shaw, E., Powers, S. J., Kurup, S., and Eastmond, P. J. (2013b). Suppression of the SUGAR-DEPENDENT1 triacylglycerol lipase family during seed development enhances oil yield in oilseed rape (Brassica napus L.). Plant Biotechnol. J. 11, 355–361. doi: 10.1111/pbi.12021
Kelly, A. A., van Erp, H., Quettier, A., Shaw, E., Menard, G., Kurup, S., et al. (2013a). The sugar-dependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Plant Physiol. 162, 1282–1289. doi: 10.1104/pp.113.219840
Khan, R., Gao, F., Khan, K., Shah, M. A., Ahmad, H., Fan, Z. P., et al. (2024). Evaluation of maize varieties via multivariate analysis: roles of ionome, antioxidants, and autophagy in salt tolerance. Plant Physiol. 196, 195–209. doi: 10.1093/plphys/kiae335
Kim, E. Y., Park, K. Y., Seo, Y. S., and Kim, W. T. (2016). Arabidopsis small rubber particle protein homolog SRPS play dual roles as positive factors for tissue growth and development and in drought stress responses. Plant Physiol. 170, 2494–2510. doi: 10.1104/pp.16.00165
Kim, M. J., Yang, S. W., Mao, H., Veena, S. P., Yin, J., and Chua, N. (2014). Gene silencing of Sugar-dependent 1 (JcSDP1), encoding a patatin-domain triacylglycerol lipase, enhances seed oil accumulation in Jatropha curcas. Biotechnol. Biofuels. 7, 36. doi: 10.1186/1754-6834-7-36
Kong, Y., Chen, S., Yang, Y., and An, C. (2013). ABA-insensitive (ABI) 4 and ABI5 synergistically regulate DGAT1 expression in Arabidopsis seedlings under stress. FEBS Lett. 587, 3076–3082. doi: 10.1016/j.febslet.2013.07.045
Krawczyk, H. E., Sun, S., Doner, N. M., Yan, Q., Lim, M. S. S., Scholz, P., et al. (2022). SEED LIPID DROPLET PROTEIN1, SEED LIPID DROPLET PROTEIN2, and LIPID DROPLET PLASMA MEMBRANE ADAPTOR mediate lipid droplet-plasma membrane tethering. Plant Cell. 34, 2424–2448. doi: 10.1093/plcell/koac095
Kretzschmar, F. K., Doner, N. M., Krawczyk, H. E., Scholz, P., Schmitt, K., Valerius, O., et al. (2020). Identification of low-abundance lipid droplet proteins in seeds and seedlings. Plant Physiol. 182, 1326–1345. doi: 10.1104/pp.19.01255
Kretzschmar, F. K., Mengel, L. A., Müller, A. O., Schmitt, K., Blersch, K. F., Valerius, O., et al. (2018). PUX10 is a lipid droplet-localized scaffold protein that interacts with CELL DIVISION CYCLE48 and is involved in the degradation of lipid droplet proteins. Plant Cell. 30, 2137–2160. doi: 10.1105/tpc.18.00276
Kunz, H., Scharnewski, M., Feussner, K., Feussner, I., Flügge, U., Fulda, M., et al. (2009). The ABC transporter PXA1 and peroxisomal beta-oxidation are vital for metabolism in mature leaves of Arabidopsis during extended darkness. Plant Cell. 21, 2733–2749. doi: 10.1105/tpc.108.064857
Kurusu, T., Koyano, T., Hanamata, S., Kubo, T., Noguchi, Y., Yagi, C., et al. (2014). OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10, 878–888. doi: 10.4161/auto.28279
Lee, H. G., Park, M., Park, B. Y., Kim, H. U., and Seo, P. J. (2019). The Arabidopsis MYB96 transcription factor mediates ABA-dependent triacylglycerol accumulation in vegetative tissues under drought stress conditions. Plants 8, 296. doi: 10.3390/plants8090296
Li, F., Chung, T., Pennington, J. G., Federico, M. L., Kaeppler, H. F., Kaeppler, S. M., et al. (2015). Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell. 27, 1389–1408. doi: 10.1105/tpc.15.00158
Li, N., Xu, C., Li-Beisson, Y., and Philippar, K. (2016). Fatty acid and lipid transport in plant cells. Trends Plant Sci. 21, 145–158. doi: 10.1016/j.tplants.2015.10.011
Li, Y., Zhu, K., Cui, H., Hu, Q., Wang, C., Jia, F., et al. (2025). Genome-wide association for multiple quantitative traits in forage oat germplasm based on specific length amplified fragment sequencing. Front. Plant Sci. 16. doi: 10.3389/fpls.2025.1527635
Liu, C., Hatzianestis, I. H., Pfirrmann, T., Reza, S. H., Minina, E. A., Moazzami, A., et al. (2024). Seed longevity is controlled by metacaspases. Nat. Commun. 15, 6748. doi: 10.1038/s41467-024-50848-2
Liu, Y. and Xiong, Y. (2022). Plant target of rapamycin signaling network: complexes, conservations, and specificities. J. Integr. Plant Biol. 64, 342–370. doi: 10.1111/jipb.13212
Liu, X., Yang, Z., Wang, Y., Shen, Y., Jia, Q., Zhao, C., et al. (2022). Multiple CALEOSINs have overlapping functions in oil accumulation and embryo development. J. Exp. Bot. 73, 3946–3962. doi: 10.1093/jxb/erac153
Loix, M., Zelcer, N., Bogie, J. F. J., and Hendriks, J. J. A. (2024). The ubiquitous role of ubiquitination in lipid metabolism. Trends Cell Biol. 34, 416–429. doi: 10.1016/j.tcb.2023.09.001
Luján, M.Á., Claver, A., Lorente, P., López, M. V., and Alfonso, M. (2025). Transcriptomic and proteomic analysis of oil body associated protein dynamics in the biofuel feedstock pennycress (Thlaspi arvense). Front. Plant Sci. 16. doi: 10.3389/fpls.2025.1530718
Man, Y., Zhang, Y., Chen, L., Zhou, J., Bu, Y., Zhang, X., et al. (2024). The VAMP-associated protein VAP27–1 plays a crucial role in plant resistance to ER stress by modulating ER-PM contact architecture in Arabidopsis. Plant Commun. 5, 100929. doi: 10.1016/j.xplc.2024.100929
Marshall, R. S. and Vierstra, R. D. (2018). Autophagy: the master of bulk and selective recycling. Annu. Rev. Plant Biol. 69, 173–208. doi: 10.1146/annurev-arplant-042817-040606
Mathiowetz, A. J. and Olzmann, J. A. (2024). Lipid droplets and cellular lipid flux. Nat. Cell Biol. 26, 331–345. doi: 10.1038/s41556-024-01364-4
Miklaszewska, M., Zienkiewicz, K., Klugier-Borowska, E., Rygielski, M., Feussner, I., and Zienkiewicz, A. (2023). CALEOSIN 1 interaction with AUTOPHAGY-RELATED PROTEIN 8 facilitates lipid droplet microautophagy in seedlings. Plant Physiol. 193, 2361–2380. doi: 10.1093/plphys/kiad471
Miquel, M., Trigui, G., D’Andréa, S., Kelemen, Z., Baud, S., Berger, A., et al. (2014). Specialization of OLEOSINs in oil body dynamics during seed development in Arabidopsis seeds. Plant Physiol. 164, 1866–1878. doi: 10.1104/pp.113.233262
Mueller, S. P., Unger, M., Guender, L., Fekete, A., and Mueller, M. J. (2017). Phospholipid:diacylglycerol acyltransferase-mediated triacylglyerol synthesis augments basal thermotolerance. Plant Physiol. 175, 486–497. doi: 10.1104/pp.17.00861
Müller, A. O. and Ischebeck, T. (2018). Characterization of the enzymatic activity and physiological function of the lipid droplet-associated triacylglycerol lipase AtOBL1. New Phytol. 217, 1062–1076. doi: 10.1111/nph.14902
Park, S., Gidda, S. K., James, C. N., Horn, P. J., Khuu, N., Seay, D. C., et al. (2013). The α/β hydrolase CGI-58 and peroxisomal transport protein PXA1 coregulate lipid homeostasis and signaling in Arabidopsis. Plant Cell. 25, 1726–1739. doi: 10.1105/tpc.113.111898
Pasaribu, B., Chung, T., Chen, C., Jiang, P., and Tzen, J. T. C. (2016). Identification of STEROLEOSIN in oil bodies of pine megagametophytes. Plant Physiol. Biochem. 101, 173–181. doi: 10.1016/j.plaphy.2016.02.008
Pyc, M., Cai, Y., Gidda, S. K., Yurchenko, O., Park, S., Kretzschmar, F. K., et al. (2017). Arabidopsis lipid droplet-associated protein (LDAP) - interacting protein (LDIP) influences lipid droplet size and neutral lipid homeostasis in both leaves and seeds. Plant J. 92, 1182–1201. doi: 10.1111/tpj.13754
Pyc, M., Gidda, S. K., Seay, D., Esnay, N., Kretzschmar, F. K., Cai, Y., et al. (2021). LDIP cooperates with SEIPIN and LDAP to facilitate lipid droplet biogenesis in Arabidopsis. Plant Cell. 33, 3076–3103. doi: 10.1093/plcell/koab179
Qin, Z., Wang, T., Zhao, Y., Ma, C., and Shao, Q. (2023). Molecular machinery of lipid droplet degradation and turnover in plants. Int. J. Mol. Sci. 24, 16039. doi: 10.3390/ijms242216039
Regmi, A., Shockey, J., Kotapati, H. K., and Bates, P. D. (2020). Oil-producing metabolons containing DGAT1 use separate substrate pools from those containing DGAT2 or PDAT. Plant Physiol. 184, 720–737. doi: 10.1104/pp.20.00461
Roingeard, P. and Melo, R. C. N. (2017). Lipid droplet hijacking by intracellular pathogens. Cell. Microbiol. 19, 1–8. doi: 10.1111/cmi.12688
Scholz, P., Chapman, K. D., Mullen, R. T., and Ischebeck, T. (2022). Finding new friends and revisiting old ones - how plant lipid droplets connect with other subcellular structures. New Phytol. 236, 833–838. doi: 10.1111/nph.18390
Scholz, P., Doner, N. M., Gutbrod, K., Herrfurth, C., Niemeyer, P. W., Lim, M. S. S., et al. (2025). Plasticity of the Arabidopsis leaf lipidome and proteome in response to pathogen infection and heat stress. Plant Physiol. 197, kiae274. doi: 10.1093/plphys/kiae274
Sham, A., Al-Azzawi, A., Al-Ameri, S., Al-Mahmoud, B., Awwad, F., Al-Rawashdeh, A., et al. (2014). Transcriptome analysis reveals genes commonly induced by Botrytis cinerea infection, cold, drought and oxidative stresses in Arabidopsis. PloS One 9, e113718. doi: 10.1371/journal.pone.0113718
Shao, Q., Liu, X., Su, T., Ma, C., and Wang, P. (2019). New insights into the role of seed oil body proteins in metabolism and plant development. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01568
Shen, Y., Xie, J., Liu, R., Ni, X., Wang, X., Li, Z., et al. (2014). Genomic analysis and expression investigation of CALEOSIN gene family in Arabidopsis. Biochem. Biophys. Res. Commun. 448, 365–371. doi: 10.1016/j.bbrc.2014.04.115
Shimada, T. L., Takano, Y., Shimada, T., Fujiwara, M., Fukao, Y., Mori, M., et al. (2014). Leaf oil body functions as a subcellular factory for the production of a phytoalexin in Arabidopsis. Plant Physiol. 164, 105–118. doi: 10.1104/pp.113.230185
Shomo, Z. D., Mahboub, S., Vanviratikul, H., McCormick, M., Tulyananda, T., Roston, R. L., et al. (2024). All members of the Arabidopsis DGAT and PDAT acyltransferase families operate during high and low temperatures. Plant Physiol. 195, 685–697. doi: 10.1093/plphys/kiae0
Siao, W., Wang, P., Voigt, B., Hussey, P. J., and Baluska, F. (2016). Arabidopsis SYT1 maintains stability of cortical endoplasmic reticulum networks and VAP27-1-enriched endoplasmic reticulum-plasma membrane contact sites. J. Exp. Bot. 67, 6161–6171. doi: 10.1093/jxb/erw381
Siloto, R. M. P., Findlay, K., Lopez-Villalobos, A., Yeung, E. C., Nykiforuk, C. L., and Moloney, M. M. (2006). The accumulation of OLEOSINs determines the size of seed oilbodies in Arabidopsis. Plant Cell. 18, 1961–1974. doi: 10.1105/tpc.106.041269
Slee, J. A. and Levine, T. P. (2019). Systematic prediction of FFAT motifs across eukaryote proteomes identifies nucleolar and eisosome proteins with the predicted capacity to form bridges to the endoplasmic reticulum. Contact (Thousand Oaks) 2, 1–21. doi: 10.1177/2515256419883136
Sturtevant, D., Lu, S., Zhou, Z., Shen, Y., Wang, S., Song, J., et al. (2020). The genome of jojoba (Simmondsia chinensis): A taxonomically isolated species that directs wax ester accumulation in its seeds. Sci. Adv. 6, eaay3240. doi: 10.1126/sciadv.aay3240
Sun, X., Jia, X., Huo, L., Che, R., Gong, X., Wang, P., et al. (2018). MDATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 41, 469–480. doi: 10.1111/pce.13110
Taurino, M., Costantini, S., De Domenico, S., Stefanelli, F., Ruano, G., Delgadillo, M. O., et al. (2018). SEIPIN proteins mediate lipid droplet biogenesis to promote pollen transmission and reduce seed dormancy. Plant Physiol. 176, 1531–1546. doi: 10.1104/pp.17.01430
Thazar-Poulot, N., Miquel, M., Fobis-Loisy, I., and Gaude, T. (2015). Peroxisome extensions deliver the Arabidopsis SDP1 lipase to oil bodies. Proc. Natl. Acad. Sci. U. S. A. 112, 4158–4163. doi: 10.1073/pnas.1403322112
Torabi, S., Sukumaran, A., Dhaubhadel, S., Johnson, S. E., LaFayette, P., Parrott, W. A., et al. (2021). Effects of type I diacylglycerol O-acyltransferase (DGAT1) genes on soybean (Glycine max L.) Seed composition. Sci. Rep. 11, 2556. doi: 10.1038/s41598-021-82131-5
Van Leene, J., Eeckhout, D., Gadeyne, A., Matthijs, C., Han, C., De Winne, N., et al. (2022). Mapping of the plant SnRK1 kinase signaling network reveals a key regulatory role for the class II T6P synthase-like proteins. Nat. Plants 8, 1245–1261. doi: 10.1038/s41477-022-01269-w
Veerabagu, M., Rinne, P. L. H., Skaugen, M., Paul, L. K., and van der Schoot, C. (2021). Lipid body dynamics in shoot meristems: production, enlargement, and putative organellar interactions and plasmodesmal targeting. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.674031
Walther, T. C. and Farese, R. V. J. (2012). Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81, 687–714. doi: 10.1146/annurev-biochem-061009-102430
Wang, X., Hao, T., Balamurugan, S., Yang, W., Liu, J., Dong, H., et al. (2017). A lipid droplet-associated protein involved in lipid droplet biogenesis and triacylglycerol accumulation in the oleaginous microalga phaeodactylum tricornutum. Algal Res. 26, 215–224. doi: 10.1016/j.algal.2017.07.028
Wang, S., Wang, X., Li, S., Sun, X., Xue, M., Di, D., et al. (2024). Maize lipid droplet-associated protein 2 is recruited by a virus to enhance viral multiplication and infection through regulating cellular fatty acid metabolism. Plant J. 119, 2484–2499. doi: 10.1111/tpj.16934
Xia, T., Xiao, D., Liu, D., Chai, W., Gong, Q., and Wang, N. N. (2012). Heterologous expression of ATG8c from soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis. PloS One 7, e37217. doi: 10.1371/journal.pone.0037217
Xing, X., Cao, C., Li, S., Wang, H., Xu, Z., Qi, Y., et al. (2023). α-naphthaleneacetic acid positively regulates soybean seed germination and seedling establishment by increasing antioxidant capacity, triacylglycerol mobilization and sucrose transport under drought stress. Plant Physiol. Biochem. 201, 107890. doi: 10.1016/j.plaphy.2023.107890
Xu, C. and Shanklin, J. (2016). Triacylglycerol metabolism, function, and accumulation in plant vegetative tissues. Annu. Rev. Plant Biol. 67, 179–206. doi: 10.1146/annurev-arplant-043015-111641
Xu, Y., Yan, F., Liu, Y., Wang, Y., Gao, H., Zhao, S., et al. (2021). Quantitative proteomic and lipidomics analyses of high oil content GmDGAT1–2 transgenic soybean illustrate the regulatory mechanism of lipoxygenase and OLEOSIN. Plant Cell Rep. 40, 2303–2323. doi: 10.1007/s00299-021-02768-4
Yamaguchi, T. and Osumi, T. (2009). Chanarin-dorfman syndrome: deficiency in CGI-58, a lipid droplet-bound coactivator of lipase. Biochim. Biophys. Acta 1791, 519–523. doi: 10.1016/j.bbalip.2008.10.012
Yang, Z., Liu, X., Wang, K., Li, Z., Jia, Q., Zhao, C., et al. (2022). ABA-INSENSITIVE 3 with or without FUSCA3 highly up-regulates lipid droplet proteins and activates oil accumulation. J. Exp. Bot. 73, 2077–2092. doi: 10.1093/jxb/erab524
Yang, L., Liu, H., Wang, Y., Seematti, J., Grenville-Briggs, L. J., Wang, Z., et al. (2021). Pathogen-mediated stomatal opening: a previously overlooked pathogenicity strategy in the oomycete pathogen Phytophthora infestans. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.668797
Yang, T., Niu, Q., Dai, H., Tian, X., Ma, J., Pritchard, H. W., et al. (2024). The transcription factor MYB1 activates DGAT2 transcription to promote triacylglycerol accumulation in sacha inchi (Plukenetia volubilis L.) Leaves under heat stress. Plant Physiol. Biochem. 208, 108517. doi: 10.1016/j.plaphy.2024.108517
Yang, Y., Yu, X., Song, L., and An, C. (2011). ABI4 activates DGAT1 expression in Arabidopsis seedlings during nitrogen deficiency. Plant Physiol. 156, 873–883. doi: 10.1104/pp.111.175950
Yu, L., Fan, J., and Xu, C. (2019). Peroxisomal fatty acid β-oxidation negatively impacts plant survival under salt stress. Plant Signal. Behav. 14, 1561121. doi: 10.1080/15592324.2018.1561121
Yu, X., Su, W., Zhang, H., Niu, M., Liu, X., Li, Z., et al. (2023). Genome-wide analysis of autophagy-related gene family and PagATG18a enhances salt tolerance by regulating ROS homeostasis in poplar. Int. J. Biol. Macromol. 224, 1524–1540. doi: 10.1016/j.ijbiomac.2022.10.240
Yue, W., Zhang, H., Sun, X., Su, N., Zhao, Q., Yan, Z., et al. (2022). The landscape of Autophagy-Related (ATG) Genes and functional characterization of TaVAMP727 to autophagy in wheat. Int. J. Mol. Sci. 23, 891. doi: 10.3390/ijms23020891
Zeng, X., Jiang, J., Wang, F., Liu, W., Zhang, S., Du, J., et al. (2022). Rice OsCLO5, a CALEOSIN protein, negatively regulates cold tolerance through the jasmonate signaling pathway. Plant Biol. 24, 52–61. doi: 10.1111/plb.13350
Zhang, M., Fan, J., Taylor, D. C., and Ohlrogge, J. B. (2009). DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell. 21, 3885–3901. doi: 10.1105/tpc.109.071795
Zhang, Z., He, G., Filipowicz, N. A., Randall, G., Belov, G. A., Kopek, B. G., et al. (2019). Host lipids in positive-strand RNA virus genome replication. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00286
Zhang, T., He, H., Xu, C., Fu, Q., Tao, Y., Xu, R., et al. (2021). Overexpression of type 1 and 2 diacylglycerol acyltransferase genes (JcDGAT1 and JcDGAT2) enhances oil production in the woody perennial biofuel plant Jatropha curcas. Plants 10, 699. doi: 10.3390/plants10040699
Zhang, X., Xu, W., Xu, R., Wang, Z., Zhang, X., Wang, P., et al. (2022). Plin5 bidirectionally regulates lipid metabolism in oxidative tissues. Oxid. Med. Cell. Longev. 2022, 4594956. doi: 10.1155/2022/4594956
Zhao, Y., Duan, B., Liu, Y., Wu, Y., Yu, D., Ke, L., et al. (2023). Identification and characterization of the LDAP family revealed GHLDAP2_DT enhances drought tolerance in cotton. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1167761
Keywords: lipid droplets, biogenesis, degradation, stress responses, SDP1, lipophagy
Citation: Zhao Y, Cao R, Li J, Xu Y, Zhou L and Ye Y (2025) Lipid droplets in plants: turnover and stress responses. Front. Plant Sci. 16:1625830. doi: 10.3389/fpls.2025.1625830
Received: 09 May 2025; Accepted: 06 June 2025;
Published: 27 June 2025; Corrected: 27 October 2025.
Edited by:
Jinda Wang, Fujian Agriculture and Forestry University, ChinaReviewed by:
Yuanyuan Song, Fujian Agriculture and Forestry University, ChinaAhmad Ali, Fujian Agriculture and Forestry University, China
Aomei Li, Guangxi Academy of Agricultural Science, China
Copyright © 2025 Zhao, Cao, Li, Xu, Zhou and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijuan Zhou, bGp6aG91QG5qZnUuZWR1LmNu; Yajin Ye, eWFqaW55ZUBuamZ1LmVkdS5jbg==
 Yujie Zhao
Yujie Zhao Rui Cao
Rui Cao Jincheng Li
Jincheng Li Lijuan Zhou
Lijuan Zhou Yajin Ye
Yajin Ye