- 1School of Agriculture and Aquaculture, Tra Vinh University, Vinh Long, Vietnam
- 2Functional Genomics Research Center, NTT Hi-Tech Institute, Nguyen Tat Thanh University, Ho Chi Minh City, Vietnam
- 3Center for Hi-Tech Development, Saigon Hi-Tech Park, Nguyen Tat Thanh University, Ho Chi Minh City, Vietnam
1 Introduction
Ardisia Sw. 1788 is one of 55 genera of Primulaceae and contains 739 accepted species that distribute in subtropical and tropical areas (Plants of the World Online, 2025). Ardisia species contains different phytochemical constituents such as coumarins, ardisiaquinones, and alkylphenols and was used as traditional medicine for fever, inflammation, and cancer (Kobayashi and de Mejía, 2005; de Mejía and Ramírez-Mares, 2011; Liu et al., 2022; Tian-Liang et al., 2024). Specifically, a benzoquinonoid compound was extracted from Ardisia crispa and exhibited antimetastatic and antitumor features (Kang et al., 2001). The combination of Ardisia gigantifolia leaf extract and silver nanoparticles indicated an anti-cancer activity (Le et al., 2023). Ardisia silvestris is native to Vietnam and Hainan (China) and its ethanol extract possessed the characteristics of antiphotoaging and skin-protective activities (Huang et al., 2023; Plant of the World Online, 2025). Additionally, a previous study revealed a notable anti-inflammatory characteristic of A. silvestris ethyl acetate extract (Thanh et al., 2025). Also, the antioxidant and antibacterial properties of A. silvestris leaf extract (Huynh, 2020). These previous results demonstrated the medicinal values of A. silvestris and related species in Ardisia genus. However, genomic data, including nuclear, mitochondrial, and chloroplast genomes, of A. silvestris are limited and need further investigations.
Chloroplast genome is an essential component in autotrophic plants because it encodes genes responsible for performing photosynthesis (Dobrogojski et al., 2020). The chloroplast genome had a quadripartite structure including a large single copy, a small single copy, and two inverted repeat regions, which could be altered in both autotrophic and heterotrophic plants (Daniell et al., 2016). Additionally, the genomic information of chloroplast genomes reflected the evolutionary history, which was used to explore a billion years of plant evolution (Gitzendanner et al., 2018). Previously, chloroplast genomes of Primulaceae species have been reported (Xu et al., 2020; Xie et al., 2023; Li et al., 2024). The complete chloroplast genomes of various Ardisia species such as A. crispa, A. gigantifolia, A. crenata, A. villosa, A. mamillata, A. brunnescents, A. pusilla, A. squamulosa, A. brevicaulis, and A. crenata were also published (Xie et al., 2021; Ye et al., 2024; Yuan et al., 2024). In the current study, we report the complete chloroplast genome of Ardisia silvestris, collected it Vietnam, using the Illumina sequencing flatform. The result of our study enriches the chloroplast genome data of Ardisia genus and provides initial chloroplast genomic data for further genomic studies examining phylogeny and molecular markers of A. silvestris and related taxa in Primulaceae.
2 Materials and methods
2.1 Plant sampling, DNA extraction, and next-generation sequencing
The healthy leaves of Ardisia silvestris were collected from living collection of medicinal plants at Tra Vinh University, Vinh Long Province, Vietnam (9°55’25.0”N 106°20’52.4”E). Then, the leaves were stored at −80°C in a deep freezer for further experiments. The total genomic DNA was extracted from the frozen leaves of A. silvestris using DNeasy Plant Pro Kit (Qiagen, USA) following the manufacturer’s instructions. The quality of DNA sample was checked using NanoDrop One Microvolume UV-Vis Spectrophotometer (Thermo Fisher Scientific, USA) and 1% agarose gel electrophoresis. The DNA sample selected for Nextseq550 sequencing (Illumina, USA) should have a concentration of 100 ng/µL and show a clear band on the agarose gel. The TruSeq DNA Nano kit (Illumina, USA) was used to prepare sequencing library to generate paired-end reads of 150 bp following the manufacturer’s instructions.
2.2 Assembly and annotation of chloroplast genome
The raw reads were qualified and filtered using fastp v0.24.1 to remove the adapter sequences and eliminate the reads possessing a Qscore under 20, having length shorter than 100 bp, and containing more than five N bases (Chen et al., 2018). The remaining high-quality reads were then assembled to complete chloroplast genome using NOVOPlasty v4.3.5 with the reference sequence of Ardisia fordii (NCBI accession number NC_060707) and other default settings (Dierckxsens et al., 2016). Consequently, the newly completed chloroplast genome of A. silvestris was annotated using Geseq through online interface at https://chlorobox.mpimp-golm.mpg.de/geseq.html with default settings (Tillich et al., 2017). To verify the annotation of Geseq, the annotation of protein-coding region was rechecked the start and stop codon of each gene using Geneious Prime v2024.0.1 (https://www.geneious.com/) whereas the structural formation of tRNA regions were tested using tRNAscan-SE 2.0 available at https://lowelab.ucsc.edu/tRNAscan-SE/index.html with default settings (Chan and Lowe, 2019). Additionally, the quadripartite structure of chloroplast genome, including a large single copy, a small single copy, and two inverted repeat regions, was investigated using the “Find repeat” function with the setting of minimum repeat length of 10,000 bp of Geneious Prime v0.2024.1 to locate two inverted repeat regions that flanked the large single copy and the small single copy regions. The map of chloroplast genome was illustrated using OGDRAW v1.3.1 available at https://chlorobox.mpimp-golm.mpg.de/OGDraw.html with default settings for plastid sequences (Greiner et al., 2019). The complete chloroplast genome of A. silvestris was deposited to GenBank under accession number PV608499.
3 Results
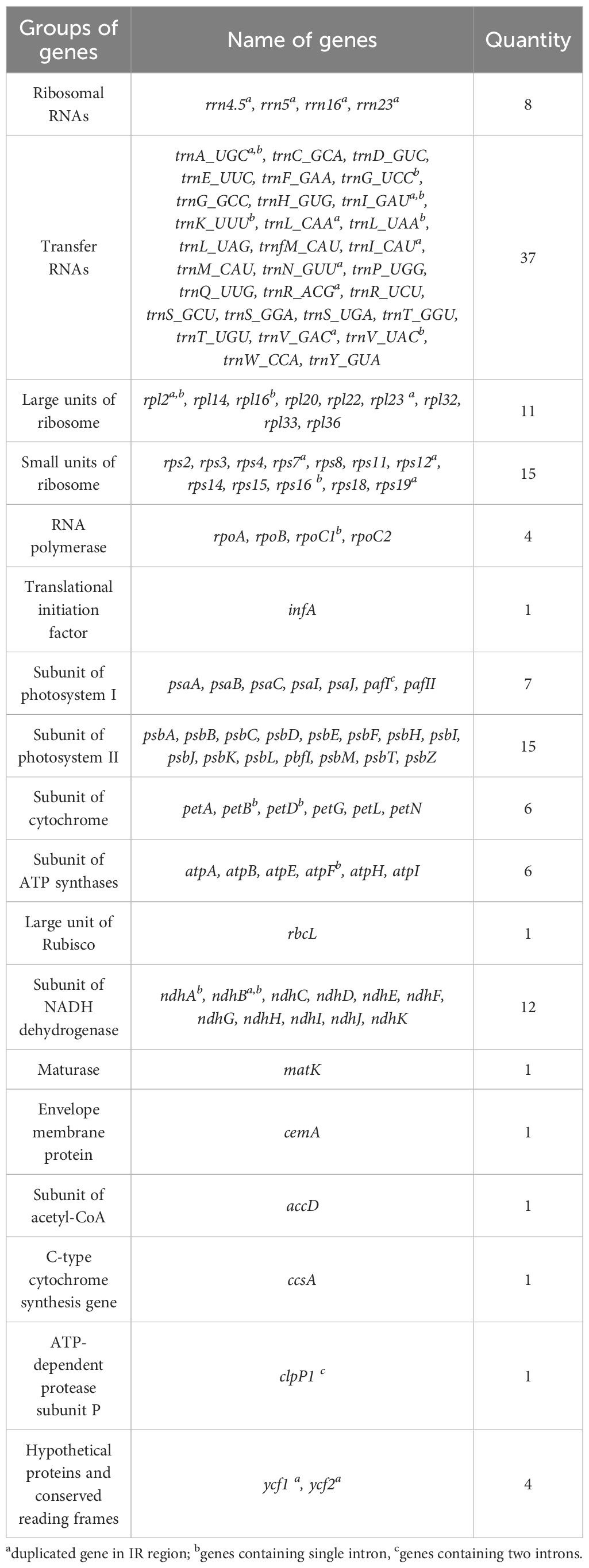
The assembly process resulted in a quadripartite chloroplast genome of A. silvestris with a mean coverage of 1642x (Figure 1). This genome was 156,640 bp in length and had 37.3% GC content. Additionally, the complete chloroplast genome of A. silvestris consisted of a large single copy (LSC) region of 85, 812 bp (35.2% GC content), a small single copy (SSC) region of 18,388 bp (30.4% GC content), and two inverted repeat (IR) regions of 26,220 bp (43.2% GC content) each. Further observation revealed that the junction between LSC and IR regions located within rps19 coding region whereas that of SSC and IR regions was in the coding region of ycf1. The complete chloroplast genome of A. silvestris encoded 79 unique protein-coding genes, 30 unique transfer RNA genes, and four unique ribosomal RNA genes (Table 1). Among 113 unique coding genes, 19 regions were duplicated in IR region including rps19, rpl2, rpl23, trnI_CAU, ycf2, trnL_CAA, ndhB, rps7, rps12, trnV_GAC, rrn16, trnI_GAU, trnA_UGC, rrn23, rrn4.5, rrn5, trnR_ACG, trnN_GUU, and ycf1. Notably, ycf1 and rps19 exhibited incomplete duplication due to expansion of IR regions. Additionally, there were nine protein genes (including rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhB, and ndhA) and six tRNAs (including trnK_UUU, trnI_GAU, trnA_UGC, trnG_UCC, trnL_UAA, and trnV_UAC) contained one intron. Meanwhile, pafI and clpP1 had two introns. The rps12 gene was trans-spliced of which the exon 2 and exon 3 located in IR regions.
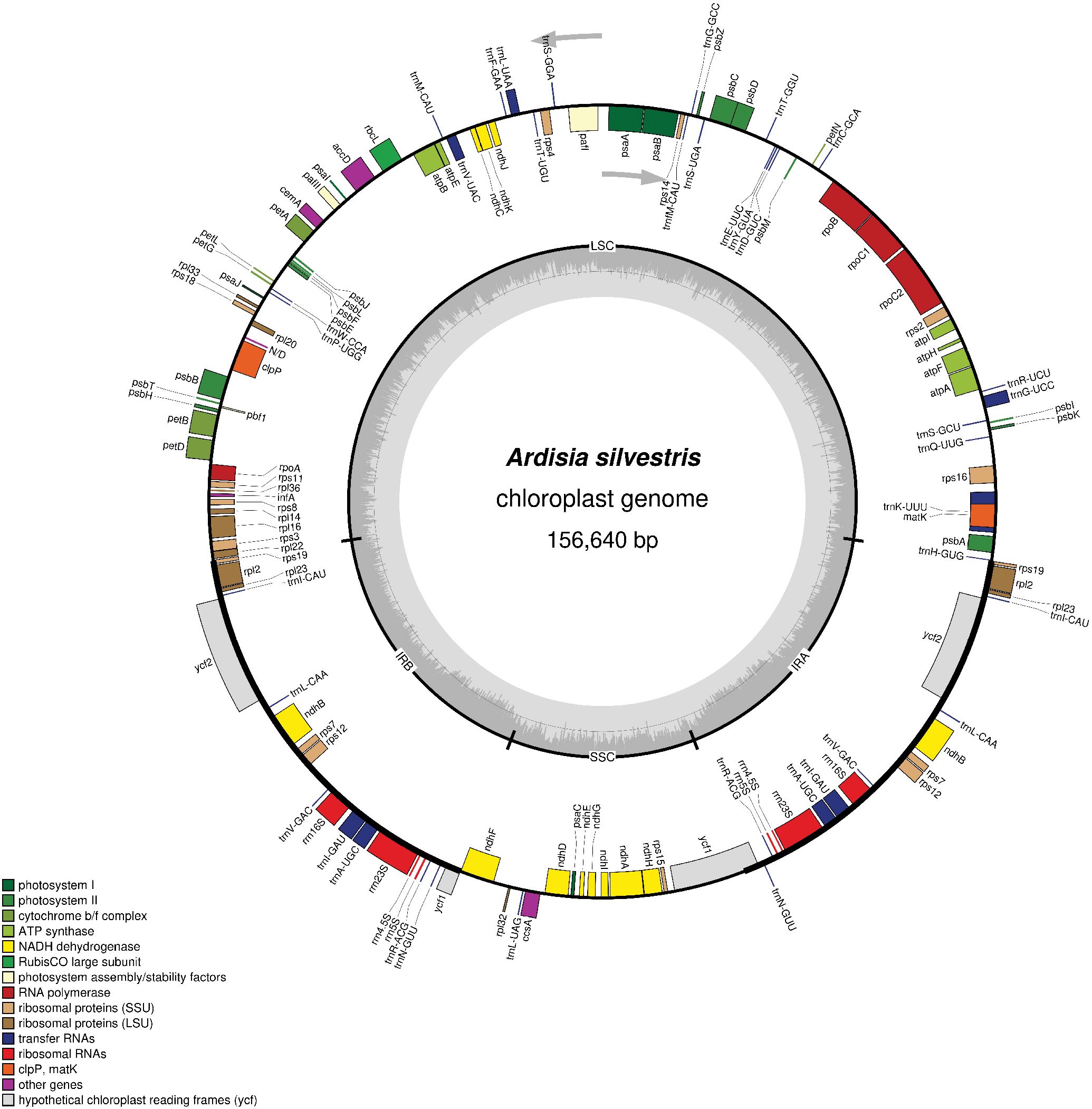
Figure 1. The chloroplast genome map of Ardisia silvestris. The arrows indicated the translation directions of inner and outer genes. The inner circle with grey color illustrates GC content. The inner circle indicates four regions of the chloroplast genome. LSC, large single copy; SSC, small single copy; IRA and IRB, inverted repeat regions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PV608499 https://www.ncbi.nlm.nih.gov/, PRJNA1261444.
Author contributions
NN: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Project administration, Writing – original draft, Writing – review & editing. HD: Conceptualization, Data curation, Formal Analysis, Methodology, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
We acknowledge Nguyen Tat Thanh University, Ho Chi Minh City, Vietnam for supporting this study. We acknowledge the support of time and facilities from Tra Vinh University (TVU) for this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Chan, P. P. and Lowe, T. M. (2019). “tRNAscan-SE: searching for tRNA genes in genomic sequences,” in Gene prediction. Methods in molecular biology. Ed. Kollmar, M. (New York, NY: Humana), 1–14. doi: 10.1007/978-1-4939-9173-0_1
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Daniell, H., Lin, C.-S., Yu, M., and Chang, W.-J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134. doi: 10.1186/s13059-016-1004-2
de Mejía, E. G. and Ramírez-Mares, M. V. (2011). Ardisia: health-promoting properties and toxicity of phytochemicals and extracts. Toxicol. Mech. Methods 21, 667–674. doi: 10.3109/15376516.2011.601355
Dierckxsens, N., Mardulyn, P., and Smits, G. (2016). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, gkw955. doi: 10.1093/nar/gkw955
Dobrogojski, J., Adamiec, M., and Luciński, R. (2020). The chloroplast genome: a review. Acta Physiol. Plant 42, 98. doi: 10.1007/s11738-020-03089-x
Gitzendanner, M. A., Soltis, P. S., Wong, G. K.-S., Ruhfel, B. R., and Soltis, D. E. (2018). Plastid phylogenomic analysis of green plants: A billion years of evolutionary history. Am. J. Bot. 105, 291–301. doi: 10.1002/ajb2.1048
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Huang, L., You, L., Aziz, N., Yu, S. H., Lee, J. S., Choung, E. S., et al. (2023). Antiphotoaging and skin-protective activities of ardisia silvestris ethanol extract in human keratinocytes. Plants 12, 1167. doi: 10.3390/plants12051167
Huynh, B. V. (2020). Phytochemical analysis of Ardisia silvestris leaf extracts and their antioxidant and antibacterial activities. J. Agric. Dev. 19, 28–35. doi: 10.52997/jad.4.04.2020
Kang, Y.-H., Kim, W. H., Park, M. K., and Han, B. H. (2001). Antimetastatic and antitumor effects of benzoquinonoid AC7–1 from Ardisia crispa. Int. J. Cancer 93, 736–740. doi: 10.1002/ijc.1384
Kobayashi, H. and de Mejía, E. (2005). The genus Ardisia: a novel source of health-promoting compounds and phytopharmaceuticals. J. Ethnopharmacol 96, 347–354. doi: 10.1016/j.jep.2004.09.037
Le, T. T. H., Ngo, T. H., Nguyen, T. H., Hoang, V. H., Nguyen, V. H., and Nguyen, P. H. (2023). Anti-cancer activity of green synthesized silver nanoparticles using Ardisia gigantifolia leaf extract against gastric cancer cells. Biochem. Biophys. Res. Commun. 661, 99–107. doi: 10.1016/j.bbrc.2023.04.037
Li, J.-T., Ju, W.-B., Li, X., Zhu, Y., Cao, T.-Y., Zhou, Y.-S., et al. (2024). The complete chloroplast genome sequence of Primula medogensis (Primulaceae) and its phylogeny. Mitochondrial DNA Part B 9, 1404–1408. doi: 10.1080/23802359.2024.2415137
Liu, B., Liu, R., Liu, Q., Ashby, C. R., Zhang, H., and Chen, Z. (2022). The ethnomedicinal and functional uses, phytochemical and pharmacology of compounds from Ardisia species: An updated review. Med. Res. Rev. 42, 1888–1929. doi: 10.1002/med.21894
Plants of the World Online (2025). Ardisia silvestris Pit. Available online at: https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:587602-1 (Accessed March 3, 2025).
Thanh, M. P., Dung, H. M., Tuan, L. A., Huong, N. T. T., Ngoc, T. T. H., Giang, L. T., et al. (2025). Anti-inflammatory effects of the extracts from Ardisia silvestris Pitard leaves in experiment. Tạp chí Nghiên cứu Y học 190, 188–195. doi: 10.52852/tcncyh.v190i5E16.3279
Tian-Liang, , Xie, Q., Shama, R., Yu, J., Xi-Gu-Ri-Gan, Q., Bao, Q., et al(2024). Ethnobotanical study of Zhuang medicinal herbs of Ardisia: variety systematization, traditional uses, phytochemistry, pharmacology, clinical application, and toxicity. J. Pharm. Pharmacol. 76, 327–353. doi: 10.1093/jpp/rgad119
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11. doi: 10.1093/nar/gkx391
Xie, C., An, W., Liu, S., Huang, Y., Yang, Z., Lin, J., et al. (2021). Comparative genomic study on the complete plastomes of four officinal Ardisia species in China. Sci. Rep. 11, 22239. doi: 10.1038/s41598-021-01561-3
Xie, Y., Yang, G., Zhang, C., Zhang, X., and Jiang, X. (2023). Comparative analysis of chloroplast genomes of endangered heterostylous species Primula wilsonii and its closely related species. Ecol. Evol. 13, e9730. doi: 10.1002/ece3.9730
Xu, W., Xia, B., and Li, X. (2020). The complete chloroplast genome sequences of five pinnate-leaved Primula species and phylogenetic analyses. Sci. Rep. 10, 20782. doi: 10.1038/s41598-020-77661-3
Ye, J., Luo, Q., Lang, Y., Ding, N., Jian, Y., Wu, Z., et al. (2024). Analysis of chloroplast genome structure and phylogeny of the traditional medicinal of Ardisia crispa (Myrsinaceae). Sci. Rep. 14, 19045. doi: 10.1038/s41598-024-66563-3
Keywords: comparative genomics, myrsinoideae, plastid genome, plastome evolution, primulaceae
Citation: Nguyen NN and Do HDK (2025) Sequencing and characterizing the complete chloroplast genome of Ardisia silvestris Pit., a potential medicinal plant in Asia. Front. Plant Sci. 16:1627578. doi: 10.3389/fpls.2025.1627578
Received: 13 May 2025; Accepted: 29 August 2025;
Published: 12 September 2025.
Edited by:
Gulmira Khassanova, S. Seifullin Kazakh AgroTechnical Research University, KazakhstanReviewed by:
Diaga Diouf, Cheikh Anta Diop University, SenegalHimanshu Sharma, Amity University, Mohali, India
Copyright © 2025 Nguyen and Do. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hoang Dang Khoa Do, ZGhka2hvYUBudHQuZWR1LnZu
†ORCID: Nhat Nam Nguyen, orcid.org/0000-0002-0505-5946
Hoang Dang Khoa Do, orcid.org/0000-0002-7970-9359
 Nhat Nam Nguyen
Nhat Nam Nguyen Hoang Dang Khoa Do
Hoang Dang Khoa Do