- Hunan Province Key Laboratory of Crop Sterile Germplasm Resource Innovation and Application, College of Life Science, Hunan Normal University, Changsha, China
Rice (Oryza sativa L.) is one of the world’s most vital staple crops, providing food for over 50% of the global population. As a salt-sensitive crop, rice is susceptible to damage from soil-soluble salt stress, which can severely reduce rice yield. Here, we aimed to elucidate the molecular mechanisms underlying salt tolerance in rice. Investigation of MADS-box genes involved in abiotic stress responses in rice led to the identification of OsMADS31. To investigate the role of OsMADS31 in salt stress tolerance, we generated its knockout mutant and overexpression lines in Nipponbare (Nip). Phenotypic analysis of T2-generation OsMADS31 knockout (osmads31) mutants revealed altered panicle morphology and significant reductions in seed-setting rate, panicle length, grain number per panicle, and 1000-grain weight. Under salt stress, both during seed germination and at the three-leaf stage, osmads31 knockout mutants exhibited markedly inhibited growth, whereas OsMADS31 overexpression (OE) lines maintained normal germination and development. At the three-leaf stage, knockout mutants showed significantly lower survival rates following salt treatment and subsequent recovery. Physiological and biochemical assays demonstrated that, compared with wild-type (WT) plants, osmads31 mutants exhibited substantially decreased catalase (CAT), peroxidase (POD), and superoxide dismutase (SOD) activities as well as reduced proline (Pro) content. Conversely, compared with WT plants, 3,3’-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) staining intensities as well as malondialdehyde (MDA) content were significantly higher in osmads31 mutants and significantly lower in OE lines. Transcriptome analysis of WT and osmads31 mutants under salt stress conditions, followed by Gene Ontology (GO) enrichment of the identified differentially expressed genes (DEGs), revealed the enrichment of genes encoding protein kinases, CATs, and transcription factors. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis identified several key pathways including carbon metabolism, amino acid biosynthesis, metabolic pathways, glycolysis/gluconeogenesis, lipid metabolism, and plant hormone signal transduction. Furthermore, weighted gene co-expression network analysis (WGCNA) of the DEGs demonstrated that OsMADS31 enhances salt tolerance by upregulating antioxidant-related genes, activating antioxidant enzymes, and reducing oxidative damage. Our results conclusively show that OsMADS31 improves salt tolerance in rice.
1 Introduction
Rice (Oryza sativa L.) is a globally vital staple crop, providing food for over half of the world’s population and playing a critical role in global food security systems (Sasaki and Burr, 2000; Wing et al., 2018). With the current global population exceeding 8 billion and projected to reach 9.7 billion by 2050, food demand will necessitate a 60% increase in crop production (Bailey-Serres et al., 2019; Hickey et al., 2019). Excessive soil salinity, an environmental stressor affecting approximately 7% of the land area globally, threatens the sustainability of crop production worldwide (Yang and Guo, 2018a). Moreover, rapid urbanization and industrialization have led to annual reductions in the size of arable land. Thus, the growing disparity between population growth and food demand, exacerbated by shrinking farmland, makes food security a critical challenge in China (Qin and Huang, 2020). China possesses abundant inland saline-alkali lands and coastal tidal flats (Esteban et al., 2016; Naveed et al., 2018). These underutilized salt-affected areas hold significant potential for future development into arable land. Among cereal crops, rice is particularly sensitive to salt stress (Hussain et al., 2018; Ma et al., 2016). Therefore, increasing the salt tolerance of crops is an urgent priority in agricultural research.
Salt stress exerts profound adverse effects on plants, disrupting physiological and biochemical processes throughout their entire life cycle from germination to senescence (Lodeyro and Carrillo, 2015; Muranaka et al., 2002; Murphy et al., 2003). Salt stress primarily induces osmotic stress and ionic stress, both of which can trigger oxidative stress in plant cells (Lodeyro and Carrillo, 2015; Zhu, 2016). To mitigate salt-induced damage, plants activate regulatory mechanisms to sustain cell growth and expansion (Fricke et al., 2004; Munns et al., 2000; Sackey et al., 2025). Salt stress rapidly induces osmotic stress in plants, since salt accumulation disrupts water and solute supply (Fricke et al., 2004; Munns et al., 2000; Qin and Huang, 2020; Zhao et al., 2020). Osmotic stress, in turn, triggers rapid stomatal closure in leaves, reducing CO2 uptake and inhibiting photosynthesis (Fricke et al., 2004). Excessive accumulation of sodium (Na+) and chloride (Cl-) ions in plant cells leads to premature leaf senescence and, in severe cases, plant death (Wegner et al., 2011; Yang and Guo, 2018a; Yang and Guo, 2018b). Elevated Na+ levels inhibit enzyme activity, disrupting metabolic processes such as the carbon cycle required for photosynthesis and other physiological pathways (Munns and Tester, 2008; Wu et al., 2018). When cytosolic Na+ concentrations exceed a critical threshold, the uptake and transport of essential elements, including potassium (K+), calcium (Ca²+), and zinc (Zn²+) is disrupted (Cheeseman, 2013; Iqbal et al., 2018; Munns et al., 2016; Razzaq et al., 2020; Seifikalhor et al., 2019; Shabala and Pottosin, 2014). Beyond osmotic and ionic stress, salt stress also induces the accumulation of reactive oxygen species (ROS) in plant cells, damaging cellular structures and macromolecules such as DNA, lipids, and enzymes (Ahanger et al., 2017; Miller et al., 2010). Under salt stress, plants must regulate ion homeostasis, osmotic balance, and oxidative stress responses to alleviate damage (Yang and Guo, 2018a; Yang and Guo, 2018b).
MADS-box genes encode transcription factors that regulate nearly all major aspects of land plant life. The MADS-box gene family participates in diverse biological processes, including vegetative development, flowering, and seed/fruit development (Saha et al., 2015), and has been extensively studied owing to its critical role in eukaryotic transcriptional regulation (de Folter and Angenent, 2006; Gramzow et al., 2010; Kaufmann et al., 2005). A defining feature of MADS-box genes is their highly conserved ~180-bp DNA sequence, which encodes the DNA-binding MADS domain (Kaufmann et al., 2005). MADS-box genes are classified as Type I or Type II, with Type II genes (termed MIKC-type MADS-box genes) predominantly governing floral organ identity (Kwantes et al., 2012; Smaczniak et al., 2012; Theissen et al., 2016). MIKC-type MADS-box transcription factors can form heterotetramers by assembling with closely related proteins. The resulting protein complexes bind to two CArG-box motifs within the same DNA loop, enhancing precise target gene recognition, and orchestrate key developmental processes in flowering plants, with distinct roles in vegetative growth and fruit development (Kappel et al., 2023). Notably, MIKC-type proteins generate intricate intra-family interaction networks, leveraging their unique ability to form multiprotein complexes comprising two or more homologous proteins that collectively function as transcriptional regulators.
MADS-box genes, as crucial regulatory factors, play pivotal roles in modulating plant developmental processes (Thompson et al., 2009; Yu et al., 2014b) and mediating hormone regulation and abiotic stress responses. In rice, genes belonging to the AGL17-like clade, including OsMADS25, OsMADS27, and OsMADS57, are nitrate-inducible (Puig et al., 2013; Yu et al., 2014a). Among these genes, OsMADS25 and OsMADS27 are critical for salt stress responses (Chen et al., 2018a; Xu et al., 2018); while OsMADS25 regulates root growth and confers salt tolerance via abscisic acid (ABA) signaling and ROS scavenging (Wu et al., 2019), OsMADS27 governs root development and osmotic stress adaptation, with its overexpression enhancing salt tolerance (Chen et al., 2018a). MADS-box transcription factor 27 (mads27) mutant, nitrate reductase-dependent nitric oxide production mediates nitrate-conferred salt tolerance in rice seedlings (Teng et al., 2025). Overexpression of the AGL17-like gene OsMADS57 has been shown to improve seed germination and root growth in both Arabidopsis and rice under salt stress, significantly boosting salinity resistance through antioxidant system activation (Wu et al., 2021). OsMADS57 also enhances cold tolerance by binding to the OsWRKY94 promoter (Chen et al., 2018b; Wu et al., 2019). In rice, OsMADS26 acts as an upstream negative regulator of stress-related genes, suppressing pathogen resistance and drought tolerance (Khong et al., 2015), whereas OsMADS87, a heat-sensitive imprinted gene associated with syncytial endosperm, participates in thermosensitivity (Chen et al., 2016). Stress sensing and signaling pathways are essential for plant survival under adverse conditions. Investigating the adaptive strategies and gene networks of plant species that lead to high-salinity tolerance may facilitate the development and transfer of salt-resistant traits to major crops (Isayenkov, 2019). However, mechanistic insights into the roles of most MADS-box genes in salt stress responses in rice remain limited.
Soil salinity, a prevalent abiotic stress factor, adversely affects rice growth and yield (Wang et al., 2018). Investigating MADS-box genes involved in abiotic stress responses in rice holds significant value, as it could help to facilitate the enhancement of stress tolerance through genetic engineering. In our previous work, we identified two salt stress-responsive jacalin-related lectin (JRL) rice genes, OsJRL45 and OsJRL40 (Gao et al., 2023a). Subsequent physiological and biochemical analyses confirmed the role of OsJRL45 and OsJRL40 in salt stress tolerance (Gao et al., 2023b, Gao et al., 2023c; Yin et al., 2024). Using a yeast two-hybrid screen, we further isolated the MADS-box transcription factor OsMADS31, which interacts with OsJRL40 and is hypothesized to participate in the salt stress response. While the MADS-box transcription factor family plays crucial roles in plant growth and development, the function of OsMADS31 in abiotic stress responses remains unclear. This study aimed to investigate whether OsMADS31 responds to salt stress. By generating OsMADS31 knockout and overexpression plants and conducting gene expression analysis, we found that OsMADS31 expression was strongly induced by NaCl, indicating that it has a critical role under salt stress conditions.
2 Materials and methods
2.1 Plant materials and growth conditions
WT rice (Oryza sativa L) cultivar Nipponbare (Nip), along with OsMADS31 knockout (osmads31) and overexpression (OE) lines generated in the Nip background, were used in this study. Seeds were germinated and cultivated hydroponically following established protocols. Plants were grown in a greenhouse under controlled conditions: 30°C day/28°C night temperature, 70% relative humidity, 14-h light/10-h dark photoperiod, and 600 μmol m-² s-¹ light intensity.
2.2 Bioinformatics analysis
2.2.1 Sequence alignment and visualization
Nucleotide sequences of the MIKC-type MADS-box genes of rice (Oryza sativa L) were retrieved from the Rice Genome Annotation Project (https://rice.uga.edu/). Subsequently, the deduced amino acid sequences of MIKC-type OsMADS-box transcription factors were aligned using MEGA 7.0.26 with the ClustalW algorithm. The aligned sequences were exported in FASTA format and visualized using ESPript 3.0 (https://espript.ibcp.fr/ESPript/ESPript/), with customized parameters for color schemes, font size (10 pt), residues per line (60), canvas dimensions, and output format (PNG). Sequence logos of conserved domains were generated using WebLogo 3 (https://weblogo.threeplusone.com/).
2.2.2 Phylogenetic analysis
Amino acid sequences of MIKC-type MADS-box proteins of rice, Arabidopsis thaliana, maize (Zea mays), and wheat (Triticum aestivum) were downloaded from the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/). After ClustalW2-based multiple sequence alignment, a maximum-likelihood (ML) phylogenetic tree with 1,000 bootstrap replicates was constructed using MEGA 7.0.26 (Kumar et al., 2018) and annotated using Evolview (https://www.evolgenius.info/).
2.2.3 Conserved domain analysis
Conserved motifs in the OsMADS31 amino acid sequence were predicted using MEME5.1.7 (https://meme-suite.org/meme/), with 12 motifs spanning the full-length protein. Domain architecture was analyzed using the NCBI Conserved Domains Database (CDD; https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi), and results were integrated with the phylogenetic tree using TBtools v2.142 for comparative visualization of evolutionary relationships and structural features (Chen et al., 2023).
2.3 Generation of mutant and transgenic lines
The CRISPR/Cas9 system was employed to generate osmads31 knockout mutants. Two 20-bp target sequences (5’-GTGATTGTGTTCTCAGGCAC-3’ and 5’-GCACCCGTTTTGAGGAGATG-3’) were selected from the OsMAS31 coding sequence using established protocols (Miao et al., 2013). The osmads31 knockout plasmids were introduced into the Nip cultivar via Agrobacterium-mediated transformation. Transgenic plants were selected for hygromycin resistance and genotyped by PCR using knockout identification primers (osmads31-F/R, Supplementary Table 1). Two independent osmads31 knockout mutant lines (KO1 and KO2) were obtained, and potential off-target effects were evaluated using CRISPR-P (http://cbi.hzau.edu.cn/cgi-bin/CRISPR) (Liu et al., 2017).
To generate OsMADS31 overexpression (OE) lines, the OsMADS31 open reading frame (ORF) was amplified from Nip cDNA using overexpression identification primers (OsMADS31OE-F/R, Supplementary Table 1) and then cloned into the pCAMBIA1390-Ubi vector. To generate pOsMADS31::GUS transgenic plants, a 2-kb fragment of the OsMADS31 promoter (upstream of ATG) was amplified from the Nip genomic DNA using GUS identification primers (OsMADS31GUS-F/R, Supplementary Table 1) and cloned into pCAMBIA1301-Ubi. All constructs were verified by sequencing (Tsingke Biotechnology) and subsequently introduced into Agrobacterium tumefaciens strain EHA105 by electroporation. The transformed Agrobacterium cells were then used to transfect Nip plants as described previously (Hiei et al., 1994).
2.4 RNA extraction and gene expression analysis
Total RNA was extracted from rice cultivar Nip plants using the TRIzol Reagent (Invitrogen, Waltham, MA, USA). To carry out the spatial expression analysis of OsMADS31, total RNA was isolated from root and shoot in Nip three-leaf stage seedlings, and different tissues of Nip heading stage including root, node, leaf, culm, anther, and young spike. The isolated total RNA was reverse-transcribed into cDNA. Subsequently, RT-qPCR was performed on the ABI PRISM 7500 system (Applied Biosystems) using Takara RR420A reagents and gene-specific primers designed with Oligo 7 (Sangon Biotech, Shanghai). OsActin served as the internal control gene.
Seedlings of the WT as well as osmads31 mutant and OsMADS31 overexpression lines were treated with 6‰ NaCl at the three-leaf-stage. RNA was extracted from the salt-treated seedlings using TRIzol Reagent, and cDNA was synthesized using HiScript II QRT SuperMix (+gDNA wiper) (Vazyme R223-01). Then, qPCR was conducted using ChamQ Universal SYBR qPCR Master Mix (Vazyme Q711-02), with OsActin serving as the reference gene. Relative gene expression levels were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001) and presented as mean ± SD. All primers used in this study are listed in Supplementary Table 2.
2.5 GUS staining assay
GUS staining was performed as previously described (Li et al., 2015). Briefly, tissues of pOsMADS31::GUS transgenic rice plants were incubated in GUS staining solution overnight at 37°C. Plant tissues were subsequently destained using 75% ethanol to remove chlorophyll. Then, the tissues were examined and photographed using the Olympus SZX7 microscope.
2.6 ROS determination
The accumulation of O2•- and H2O2 in rice plants was determined by staining with nitroblue tetrazolium chloride (NBT) and 3,3’-diaminobenzidine (DAB), respectively, as described previously (Huang et al., 2019; Jambunathan 2010). Briefly, the second leaves were collected from the KO, OE and WT plants before and after the 6‰ NaCl treatment. To perform NBT staining, 3-cm segments of leaves were prepared, vacuum-infiltrated with the NBT solution (0.1%, pH 7.8) for 30 min, and incubated at room temperature for 24 h. To perform DAB staining, leaf samples were vacuum-infiltrated with the DAB solution (1 mg/mL in 10 mM Na2HPO4 containing 0.05% Tween-20, pH 3.8) for 30 min and then incubated at 37°C with shaking for 48 h. Both the NBT- and DAB-stained samples were destained with 95% ethanol and photographed under a stereomicroscope (Nikon XAZ100).
2.7 Subcellular localization analysis
To determine the subcellular localization of OsMADS31, the coding sequence (CDS) of OsMADS31 (excluding the stop codon) was amplified from the Nip genomic DNA using gene-specific primers (Supplementary Table 1). The PCR product was cloned into the pCAMBIA1390-GFP vector. The resulting 35S::OsMADS31-GFP construct was transiently expressed in rice protoplasts via polyethylene glycol (PEG)-mediated transformation (Wang et al., 2021). GFP signal was visualized using the Zeiss LSM880 confocal microscope (Wang et al., 2022a).
2.8 Salt stress treatment of transgenic plants
After treatment with different concentrations of NaCl, the germination rate of seeds was counted on the seventh day. Transgenic and WT seeds were incubated on Murashige and Skoog (MS) medium containing varying NaCl concentrations for 12 days, and root and shoot lengths were measured subsequently.
Uniform, plump rice seeds were surface-sterilized with 0.3% sodium hypochlorite for 20 min, rinsed three times with distilled water, and placed on moist filter paper in Petri dishes at 37°C. The germinated seeds were transferred to 96-well trays containing culture solution, replaced every 4 days. After growing to the three-leaf stage (~2 weeks) under standard hydroponic conditions, seedlings were exposed to 6‰ NaCl solution for 6 days. Subsequently, the seedlings were allowed to recover for 6 days in normal solution before calculating the survival rate.
2.9 Measurement of physiological parameters
SOD, POD, and CAT activities as well as MDA and Pro contents were measured using the respective kits, as described previously (Gao et al., 2023a, Gao et al., 2023b, Gao et al., 2023c). Measurements were taken both pre-treatment and at 24 h post-salt-stress treatment. Three biological replicates were performed, and data were expressed as mean ± SD.
2.10 RNA sequencing analysis
WT and osmads31 mutant transgenic seedlings at the three-leaf stage were sampled pre-treatment and at 24 h after 6‰ NaCl exposure. Three biological replicates were performed per genotype, and the seedlings were flash-frozen in liquid nitrogen and submitted to Genedenovo Biotechnology Co., Ltd (Guangzhou, China) for RNA-seq. The obtained transcriptomic data were analyzed using established methods (Gao et al., 2023b, Gao et al., 2023c; Zhang et al., 2020). Additional data analysis was conducted using the Genedenovo Biotechnology Platform Omicsmart (https://www.omicsmart.com). The cDNA libraries were sequenced on the Illumina sequencing platform by Genedenovo Biotechnology Co., Ltd (Guangzhou, China).
2.11 Interaction network analysis
Relative expression levels of differentially expressed genes (DEGs), identified through RNA-seq, were subjected to log-transformation and normalization. Subsequently, the gene expression data were examined using correlation analysis, weighted gene co-expression network analysis (WGCNA)-based network construction, and Cytoscape visualization (Hu et al., 2016). Protein-protein interactions were predicted using STRING (https://cn.string-db.org/).
2.12 Statistical analysis
Data are presented as the mean ± SD of three or more independent replicates. The mean of replicates for each experiment was calculated using the PASW Statistics 18 software. Statistical significance was determined using one-way ANOVA and Student’s t-test at p < 0.05. Bar charts were created using GraphPad Prism 10.3.1. Heatmaps were constructed and drawn using TBtools-II v.2.142 (Chen et al., 2023).
3 Results
3.1 Conservation and amino acid sequence analysis of MIKC-type MADS domains in rice
To characterize the conserved features of MIKC-type MADS domains in rice, we performed multiple sequence alignment of their amino acid sequences. The results showed that MADS domains of the DNA binding domain are highly conserved among the 61 MIKC-type MADS-box transcription factors found in rice (Supplementary Figure 1). According to previous studies, MIKC-type MADS domains exist in two lineages (Type I and Type II) of MADS-box genes across plants, animals, and fungi. Notably, the majority of plant MADS-box genes belong to the Type II lineage and contain three additional domains compared with Type I genes: an intervening (I) domain, a keratin-like coiled-coil (K) domain, and a variable-length C-terminal (C) domain; these three domains together form the plant-specific MIKC-type structure (Nam et al., 2003). The conserved nature and distinctive characteristics of MIKC-type MADS domains provide important clues for determining the functional roles of MADS-box transcription factors in plant development and stress responses.
3.2 Analysis of conserved sequences and domains of the OsMADS31 gene in rice
The OsMADS31 CDS comprises 723 base-pairs and is predicted to encode a 240-amino acid protein. The amino acid sequence of OsMADS31 contains both the MADS and K-box domains. The MADS domain is involved in DNA binding and protein dimerization. Notably, MADS-box family proteins typically function as dimers, with their primary DNA-binding element consisting of an antiparallel coiled-coil formed by two amphipathic α-helices from each subunit (Norman et al., 1988). The MADS domain is generally associated with the K-box region. Most plant MADS-box proteins possess the typical MIKC structure, which mediates dimerization and cofactor binding, serving as the primary determinant for DNA binding. The K-box protein interaction domain, which mediates heterodimerization of MIKC-type MADS-box proteins, contains several heptad repeats with hydrophobic amino acids at the first and fourth positions. This suggests that the K-box domain forms three amphipathic α-helices, designated as K1, K2, and K3 (Yang et al., 2003). Analysis of conserved motifs among the 61 highly conserved MIKC-type MADS-box proteins identified in rice (Supplementary Figure 2) revealed that all of these 61 MADS-box proteins contain both the MADS and K-box domains, with sequence logos showing 12 conserved amino acid residue motifs. Phylogenetic analysis demonstrated that the rice gene OsMADS31 is evolutionarily closely related to its orthologs in maize (Supplementary Figure 3).
3.3 Examination of OsMADS31 gene expression and OsMADS31 protein localization
The expression of OsMADS31 was examined in the root and shoot of WT Nip three-leaf stage seedlings (Figure 1A), and different tissues (roots, nodes, leaves, stems, anthers, and young panicles) of WT Nip heading stage (Figure 1B). The results of RT-qPCR analysis showed that the expression of OsMADS31 began to increase at 4 h after the salt stress treatment at the three-leaf-stage of WT Nip and peaked at 24 h, with expression levels reaching approximately 10-fold higher than the baseline level (Figure 1C). To further examine OsMADS31 expression, a vector containing the OsMADS31 promoter-driven GUS reporter gene was constructed and transformed into Nip. GUS staining of the resulting transgenic plants demonstrated GUS activity in the coleoptile of germinating seeds as well as in whole seedlings, stems, leaves, anthers, and young panicles (Supplementary Figure 4). Quantitative analysis indicated higher expression in leaves and stem nodes, and the cross-sections of stained tissues revealed deeper staining in vascular bundles. These results suggest that OsMADS31 is expressed in multiple tissues and at different developmental stages, implying its functional importance in rice growth.
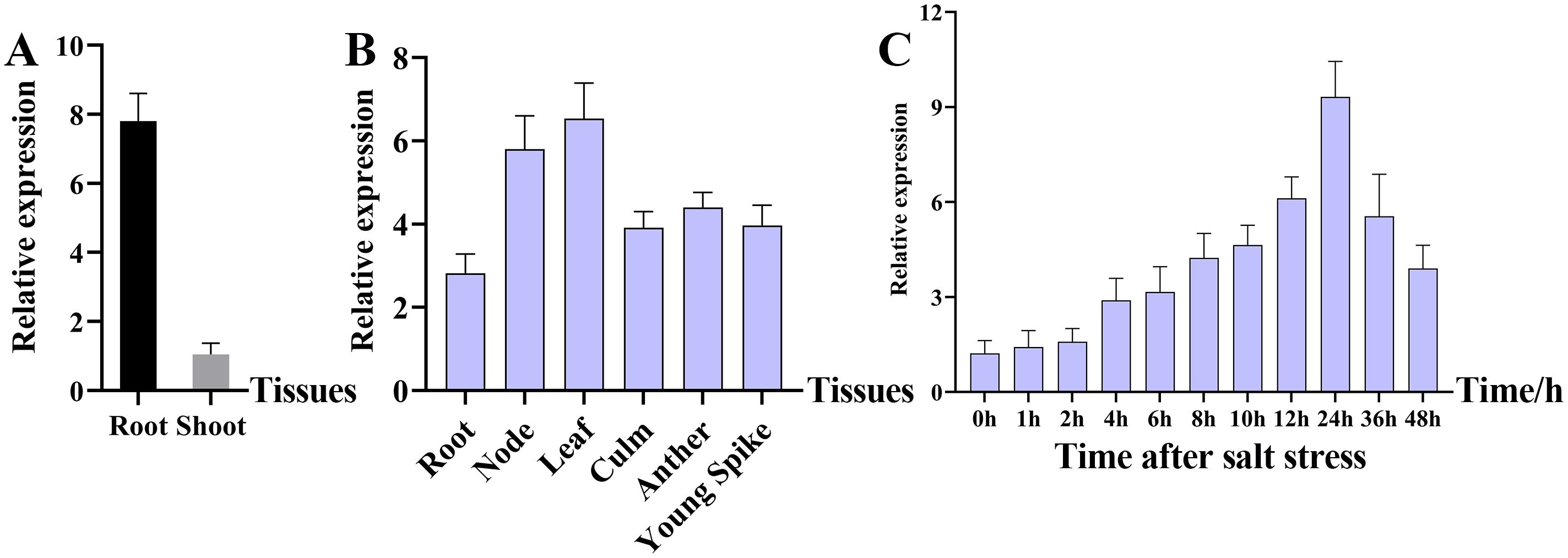
Figure 1. Characterization of the OsMADS31 gene using RT-qPCR. (A) Expression analysis of OsMADS31 at root and shoot in rice three-leaf stage seedlings by RT-qPCR, (B) expression analysis of OsMADS31 at different parts in rice heading stage by RT-qPCR, (C) expression analysis of OsMADS31 at different times under salt stress in rice three-leaf stage seedlings by RT-qPCR.
We also examined the subcellular localization pattern of OsMADS31. Online prediction tools indicated that OsMADS31 is nuclear-localized. These results were experimentally verified (Figure 2).

Figure 2. Subcellular localization analysis of OsMADS31 in transgenic rice protoplasts using confocal fluorescence microscopy. (A, B) Green fluorescence signal and nuclear localization signal (NLS) in rice protoplasts carrying the 35S::OsMADS31-GFP construct (A) and 35S::GFP construct (B). Bright, brightfield, Merge, fluorescence channel and brightfield overlay. Scale bars = 10 μm.
3.4 Analysis of osmads31 knockout mutant and OsMADS31 overexpression lines at maturity
To further investigate whether OsMADS31 is involved in salt stress tolerance, a vector for knocking-out OsMADS31 expression was constructed using the CRISPR/Cas9 technology and introduced into Nip via Agrobacterium-mediated transformation. The resultant osmads31 knockout mutants were verified by sequencing and off-target cleavage detection (Supplementary Figure 5A). Concurrently, OsMADS31 overexpression lines were generated in the Nip background. Finally, two knockout mutants (KO1 and KO2) and two overexpression lines (OE1 and OE2) were selected, based on the expression levels of OsMADS31 determined by RT-qPCR (Supplementary Figure 5B). All four genotypes were propagated to the T2 generation, and homozygous knockout mutant and transgenic plants were used for subsequent experiments.
Next, we investigated the effects of the knockout mutation of OsMADS31 on the phenotype and agronomic traits of homozygous T2-generation KO and OE plants at maturity, with the WT plants serving as controls. Knockout mutant plants exhibited significant reductions in grain length, grain width, plant height, seed-setting rate, 1000-grain weight, panicle length, and grains per panicle compared with WT plants. Conversely, overexpression lines displayed significant increases in panicle length and grain number per panicle compared with the WT (Supplementary Figure 6 & 7). These results demonstrate that the knockout mutation of OsMADS31 affects rice seed development.
3.5 Evaluation of the salt stress tolerance of osmads31 knockout mutant and OsMADS31 overexpression lines at the seed germination stage
We further investigated the salt stress tolerance of osmads31 knockout mutant and OsMADS31 overexpression plants by performing seed germination assays. Uniformly plump seeds of the WT, osmads31 knockout mutants (KO1 and KO2), and OsMADS31 overexpression lines (OE1 and OE2) were germinated on MS medium containing no NaCl (control; CK) or different NaCl concentrations (6‰, 8‰, 9‰) (Figure 3A). Under normal conditions (CK), seeds of all genotypes showed normal germination and growth, with no significant differences. At 6‰ and 8‰ NaCl concentrations, compared with the WT, KO1 and KO2 seedlings exhibited significantly reduced shoot and root lengths, whereas OE1 and OE2 seedlings showed significantly increased shoot and root lengths (Figure 3B-I). At 9‰ NaCl concentration, although KO1 and KO2 seeds could germinate, their seedlings displayed nearly no root or shoot growth; by contrast, OE1 and OE2 seedlings were maintained growth of both shoots and roots (Figure 3). These results demonstrate that OsMADS31 plays a crucial role in salt stress tolerance during seed germination.

Figure 3. Analysis of seed germination and seedling growth in osmads31 knockout mutants (KO-1 and KO-2), OsMADS31 overexpression lines (OE-1 and OE-2), and the WT under salt stress. (A) Phenotypic analysis of seed germination across the three genotypes under different NaCl concentrations. Scale bar = 5 cm. (B-I) Quantitative analysis of shoot length (B-E) and root length (F-I) of WT, KO, and OE plants under different NaCl concentrations. Data represent the mean ± SD of three independent replicates. Asterisks indicate statistically significant differences compared with the WT (*P < 0.05, **P < 0.01). ns, no significant difference.
3.6 Analysis of the salt stress tolerance of rice seedlings at the three-leaf stage
To further evaluate the salt stress tolerance of the WT, osmads31 knockout mutants, and OsMADS31 overexpression lines, seedlings at the three-leaf stage were subjected to 6‰ NaCl treatment for 6 days, followed by a 6-day recovery period, and then analyzed for phenotypic differences (Figure 4A-C). The KO mutant seedlings exhibited wilting and growth retardation under salt stress, with significantly lower survival rates than the WT after recovery; by contrast, OE plants outperformed the WT in growth and showed significantly higher survival rates (Figure 4D). These results demonstrate that OsMADS31 enhances salt stress tolerance in rice.

Figure 4. Analysis of the salt stress tolerance of the WT, osmads31 knockout mutants (KO-1 and KO-2), and OsMADS31 overexpression lines (OE-1 and OE-2) at the three-leaf stage. (A-C) Phenotypic analysis of WT, KO, and OE plants before salt stress (A), at 6 days after treatment with 6‰ NaCl (B), and at 6 days post recovery (C). (D) Survival rates of plants after recovery from salt stress treatment. (E) DAB and (F)NBT staining of leaves. Asterisks indicate statistically significant differences compared with the WT (*P < 0.05, **P < 0.01). Scale bars = 5 cm.
3.7 OsMADS31 enhances the antioxidant capacity of rice under salt stress
Next, to determine the defense response of osmads31 knockout mutants and OsMADS31 overexpression lines to salt stress, we measured the activities of antioxidant enzymes (CAT, POD, and SOD) as well as the contents of MDA and Pro in WT, KO, and OE plants at the three-leaf stage before and after treatment with 6‰ NaCl for 24 h. Compared with the WT, KO mutants exhibited significantly lower CAT, POD, SOD activities and Pro content (Figures 5A-D), but significantly higher MDA content (Figure 5E); by contrast, OE mutants showed the opposite trends. We also determined the accumulation of ROS in all three genotypes before and after the salt stress treatment by performing DAB and NBT staining of leaves, followed by destaining. The intensity of NBT and DAB staining was significantly higher in KO mutants than the WT (Figures 4E, F). Together, these results indicate that knocking-out OsMADS31 expression reduces antioxidant enzyme activities and disrupts the homeostasis of related physiological parameters under salt stress in rice.

Figure 5. Analysis of the physiological parameters of osmads31 knockout mutant (KO-1 and KO-2), OsMADS31 overexpression (OE-1 and OE-2), and WT plants at the three-leaf stage before and after treatment with 6‰ NaCl for 24 (h) (A-C) Activities of CAT (A), POD (B) and SOD enzymes (C, D) Proline quantification. (E) MDA quantification. Data represent the mean ± SD of three independent replicates. Asterisks indicate statistically significant differences compared with the WT (*P < 0.05). ns, no significant difference. CAT, catalase; POD, peroxidase; SOD, superoxide dismutase; Pro, proline; MAD, malondialdehyde; DAB, 3,3’-diaminobenzidine; NBT, nitroblue tetrazolium chloride.
3.8 Identification of genes and interaction networks regulated by OsMADS31
To further elucidate the role of OsMADS31 in salt stress response at the molecular level, we performed Gene Ontology (GO) enrichment analysis of WT and osmads31 knockout mutants before and after salt stress treatment DEGs. Comparative analysis of secondary bar plots revealed that, compared with the pre-treatment control (WT-vs-KO (CK)), salt-stressed samples (WT-vs-KO (Salt)) showed significantly fewer upregulated genes than downregulated genes. Notably, in the Biological Process category, genes involved in ‘response to stimulus’ were downregulated by more than twofold in WT-vs-KO (Salt) compared with WT-vs-KO (CK) (Supplementary Figures 8A, C). Volcano plot of differentially expressed genes after under salt stress (Supplementary Figures 8E). Comparative GO enrichment circle plots before and after the salt stress treatment demonstrated significant enrichment of genes in the Biological Process category (Supplementary Figures 8B, D). Bubble plot analysis further revealed that the WT-vs-KO (Salt) comparison was primarily enriched in GO terms such as ‘response to harmful substances’ and ‘metabolic processes’ (Supplementary Figures 9A, C). The directed acyclic graph (DAG) showed predominant enrichment of genes involved in ‘response to stimulus’ (Supplementary Figures 9B, D). Subsequent Gene Set Enrichment Analysis (GSEA) of ‘response to stimulus’-related genes after salt treatment highlighted processes related to salt stress response, while regulatory network analysis identified three key functional categories: protein kinases, catalases, and transcription factors (Supplementary Figures 9E, F). These results suggest that salt stress likely affects plant physiological responses by modulating specific gene expression patterns in rice.
Among the DEGs, 455 were upregulated and 756 were downregulated genes in KO mutants following salt treatment (Supplementary Figure 10A, B), suggesting that these DEGs may be directly associated with salt stress response. KEGG enrichment analysis identified the following top 10 pathways in WT-vs-KO (CK): ‘Biosynthesis of secondary metabolites, Cutin, suberin, and wax biosynthesis, Protein processing in the endoplasmic reticulum, Metabolic pathways, Glutathione metabolism, Carbon fixation in the calvin cycle, Photosynthesis-antenna proteins, Phenylpropanoid biosynthesis, Benzoxazinoid biosynthesis, and Betaine biosynthesis’ (Supplementary Figure 10C). Similarly, KEGG enrichment analysis of the WT-vs-KO (Salt) data showed enrichment of the following 10 pathways: ‘Phenylpropanoid biosynthesis, Biosynthesis of secondary metabolites, Plant hormone signal transduction, Pyruvate metabolism, Metabolic pathways, Alanine, aspartate and glutamate metabolism, Glycolysis/Gluconeogenesis, MAPK signaling pathway-plant, Plant-pathogen interaction, and Arginine and proline metabolism’ (Supplementary Figure 10D). KEGG hierarchy analysis of the top 20 pathways identified in WT-vs-KO (Salt) revealed carbon metabolism, amino acid biosynthesis and metabolic pathways as the three most affected pathways, with glycolysis/gluconeogenesis and lipid metabolism also potentially impacted. Subsequent GSEA revealed genes involved in plant hormone signal transduction (Supplementary Figure 10E-G). Among the genes downregulated under salt stress, several encoded transcription factors belonging to the NAC, MYB, ethylene-responsive factor (ERF), AP2/EREBP, basic helix-loop-helix (bHLH), and bZIP families (Supplementary Figure 10H). These results demonstrate that salt stress primarily affects the physiological and metabolic processes of plants by regulating genes involved in secondary metabolite biosynthesis, metabolic pathways, and plant hormone signal transduction.
To further investigate the relationship between OsMADS31 and DEGs, we performed WGCNA. WGCNA is a systematic biological method used to analyze inter-gene relationships in expression data by constructing weighted networks based on co-expression patterns for identifying gene modules associated with specific phenotypes. In this study, WGCNA was conducted using 17,695 genes with relative expression levels > 1. The results revealed 20 distinct modules, with each module containing 3 to 7,836 genes. Among these, four modules (black, brown4, magenta, and pink) showed significant associations (Supplementary Figure 11). The identification of these modules provides crucial insights for understanding the mechanism of OsMADS31-mediated salt stress tolerance in rice.
Gene regulatory network analysis enables the identification of hub genes within the regulatory network and facilitates functional prediction of unknown genes based on known gene functions. GO and KEGG enrichment analyses demonstrated that these modules were primarily involved in biosynthesis of secondary metabolites and metabolic pathways, consistent with previous KEGG enrichment findings. Although each module exhibited distinct KEGG pathways, a substantial number of genes participated in several common pathways including Biosynthesis of secondary metabolites, Plant hormone signal transduction, Metabolic pathways, and Peroxisome-related processes (Supplementary Figure 12). Co-expression network analysis was performed to identify hub (key) genes within each module and their interactions with genes in other modules (Figure 6). The black module contained 200 genes, including four hub genes (Os03g0764600, Os03g0815100, Os04g0519700, and Os01g0863300), encoding an HHO transcription factor, a stress-responsive NAC transcription factor, an auxin response factor, and a MYB family transcription factor, respectively. For example, the core gene Os03g0815100 in the Black module encodes the stress-responsive NAC transcription factor (SNAC1). Studies have shown that overexpressing SNAC1 upregulates the expression of multiple stress-related genes, thereby enhancing salt tolerance in rice (Hu et al., 2006). The brown4 module comprised 100 genes, with three hub genes (Os05g0195200, Os04g0546800, and Os01g0797600) that encode a CCCH tandem zinc finger protein, an ethylene-responsive transcription factor, and an ERF, respectively. For example, the core gene Os01g0797600 (ethylene-responsive transcription factor ERF3/OsAP37) in the Brown4 module, when its encoded protein OsAP37 is overexpressed in transgenic rice plants driven by the OsCc1 promoter, exhibited enhanced salt tolerance during the vegetative growth stage (Oh et al., 2009). The magenta module included 200 genes, with eight hub genes (Os01g0203000, Os06g0127100, Os01g0915600, Os03g0135700, Os09g0468700, Os02g0603600, Os09g0434500, and Os06g0614100), encoding a BZR1 homolog, an AP2/EREBP transcription factor, a bHLH transcription factor, an ERF, a bZIP transcription factor, and two helix-loop-helix DNA-binding proteins. For example, the core gene Os06g0127100 (AP2/EREBP transcription factor) in the Magenta module encodes dehydration-responsive element-binding proteins 1C, 1E, and 1G (DREB1C, DREB1E, DREB1G), which promote tolerance to salt stress in rice (Wang et al., 2022b). Another core gene in the Magenta module, Os09g0434500 (ethylene-responsive factor OsBIERF), belongs to a family whose members OsBIERF1, OsBIERF3, and OsBIERF4 exhibit expression regulated by salt stress (Cao et al., 2006). The pink module consisted of 150 genes, including two hub genes (Os03g0758900 and Os01g0730700), both of which encode WRKY transcription factors. These hub genes play critical regulatory roles in processes such as plant hormone signal transduction, metabolic pathways, peroxisome function, and biosynthesis of secondary derivatives, suggesting their potential involvement in salt stress response mechanisms.

Figure 6. Analysis of co-expression interaction network genes in each module. (A-D) Gene co-expression networks for the black module (four genes) (A), brown4 module (two genes) (B), magenta module (eight genes) (C), and pink module (two genes) (D).
4 Discussion
4.1 OsMADS31 is a positive regulator of plant development and stress response in rice
MADS-box proteins are a family of transcription factors characterized by a conserved MADS-box domain of approximately 58–60 amino acids at the N-terminus, which primarily functions in DNA binding (West et al., 1997; Yanofsky et al., 1990). In plants, MADS-box transcription factors are involved in nearly all critical growth and developmental processes (Alvarez-Buylla et al., 2000; Rounsley et al., 1995). Many MADS-box genes regulate plant development by interacting with other MADS-box proteins. Studies have shown that OsMADS29 in rice forms a heterodimer with its paralog OsMADS31 to cooperatively regulate seed development (Theißen and Becker, 2004; Nayar et al., 2014). In Arabidopsis, AGL24 and SVP participate in shoot development and floral transition by interacting with other proteins at different developmental stages (Michaels et al., 2003). A total of 61 MIKC-type MADS genes have been identified in rice, exhibiting highly conserved amino acid residues (Supplementary Figure 1). Conserved gene sequence and domain analyses revealed that all possess MADS and K-box structures, along with sequence signatures of 12 conserved amino acid motifs (Supplementary Figure 2). Comparative phylogenetic analysis indicated that the rice OsMADS31 gene is closely related to homologs in maize (Supplementary Figure 3). Most MIKC-type MADS genes in plants are Type II and exhibit plant-specific features (Nam et al., 2003). The conservation and specificity of the MIKC-type MADS domain provide critical insights for further functional studies in plant development and stress responses. OsMADS31 is localized to the nucleus (Figure 2). RT-qPCR and GUS staining demonstrated its widespread expression across multiple rice tissues. It responds to salt stress, with expression levels under 6‰ NaCl treatment reaching 10-fold higher than baseline after 24 hours (Figure 1, Supplementary Figure 4). Knockout of osmads31 led to significant reductions in agronomic traits, while overexpression promoted increased panicle length and grain number (Supplementary Figure 6 & 7). Analyses of the conserved specificity of the MIKC-type MADS gene domain and its expression patterns suggest that OsMADS31 acts as a positive regulator of rice development and stress responses.
4.2 OsMADS31 is a positive regulator of salt stress tolerance
The seed germination and seedling growth stages of rice are periods during which rice are most sensitive to external environmental factors (Zhang et al., 2014). Therefore, rice seed germination and seedling growth are often used as key indicators for evaluating salt stress tolerance (Yu et al., 2018). The MADS-box transcription factor AGL21 is a negative regulator of seed germination and post-germination growth and thus, plays a role in seed germination under abiotic stress. AGL21-overexpressing plants exhibit hypersensitivity to ABA, salt, and osmotic stress during seed germination and early growth, whereas agl21 mutants show reduced sensitivity (Yu et al., 2017). In a GWAS study on salt stress during rice germination, the candidate gene OsMADS31, a member of the MADS-box transcription factor family, was predicted to contribute to salt tolerance during germination despite its downregulation under salt stress conditions (Yu et al., 2018). Using mRNA in identification of unannotated salinity stress-inducible transcripts in rice (Oryza sativa L.) demonstrated that OsMADS31 expression was upregulated in roots after salt stress (Mizuno et al., 2010). The osmads31 knockout reduced salt stress tolerance, inhibiting shoot and root growth during germination, while osmads31 overexpression promoted growth (Figure 3). These results demonstrate that OsMADS31 significantly stimulates rice seed germination under salt stress. Rice is highly sensitive to salinity during early seedling growth (Nam et al., 2015), making improved salt tolerance crucial for seedling establishment and yield under stress. Under salt stress at the three-leaf stage, osmads31 knockout lines exhibited wilting and reduced survival rates, whereas osmads31overexpression enhanced survival (Figure 4). Increases in reactive oxygen species (ROS) generated by oxidative stress can damage lipids, proteins, nucleic acids, and membranes. Plants counteract salt-induced oxidative stress by deploying antioxidants such as catalase (CAT), peroxidase (POD), and superoxide dismutase (SOD) (Liu et al., 2019). In this study, osmads31 knockout lines showed significantly lower CAT, POD, and SOD activities than the wild-type (WT), which was accompanied by reduced antioxidant enzyme activity and increased oxidative damage. By contrast, osmads31 overexpression lines displayed elevated CAT, POD, and SOD levels (Figure 5), which increased antioxidant capacity and stress-responsive metabolite accumulation, facilitating ROS scavenging and mitigated membrane lipid peroxidation. These findings indicate that OsMADS31 strengthens antioxidant defenses, thereby reducing salt stress damage.
4.3 Mechanism of OsMADS31-mediated increase in salt stress tolerance in rice
Reactive oxygen species (ROS), including H2O2, O2-, and hydroxyl radicals (•OH), are chemical compounds that can cause cellular membrane damage, lipid peroxidation, and malondialdehyde (MDA) production (D’Autreaux and Toledano, 2007). The MDA level serves as a common indicator of membrane lipid peroxidation and indirectly reflects plant stress resistance (Zhang et al., 2013). Overexpression of SlMBP11 in tomato enhanced salt tolerance, as evidenced by lower relative electrolyte leakage and reduced MDA content in transgenic plants (Guo et al., 2016), suggesting that MADS-box genes may participate in antioxidant systems to confer salt tolerance. Following salt stress treatment, OsMADS31-overexpressing (OE) lines showed significantly lower MDA levels than those of wild-type (WT) and knockout lines (Figure 5E), indicating reduced oxidative damage in OE plants under salt stress. Previous studies have demonstrated that salt stress can increase antioxidant enzyme activity while inducing ROS production and accumulation, thereby regulating plant salt tolerance (Miller et al., 2010; Wahid et al., 2007; Yan et al., 2014). Under salt stress, osmads31 knockout lines exhibited more intense DAB and NBT staining than the WT, while osmads31 overexpression lines showed minimal staining (Figure 4E, F). Additionally, osmolyte accumulation is a crucial response to salt stress, with proline being the most extensively accumulated compound (Wang et al., 2015). Proline scavenges ROS, stabilizes subcellular structures, and regulates redox homeostasis (Szabados and Savoure, 2010). The osmads31 knockout lines accumulated significantly less proline than the WT under salt stress, while osmads31 overexpression lines showed the opposite trend (Figure 5D). Physiological studies on rice salt tolerance have identified proline as a key osmoprotectant (Ozturk et al., 2021) that safeguards cellular proteins and membrane structures from degradation or damage, thereby improving seedling salt tolerance (Li et al., 2018) In this study, OsMADS31-OE lines displayed markedly increased proline accumulation under salt stress at the three-leaf stage (Figure 5D), suggesting that enhanced salt tolerance in OE plants may correlate with proline biosynthesis. Transcriptome analysis revealed that salt stress alters specific gene expression patterns involving three regulatory networks: protein kinases, catalases, and transcription factors. KEGG enrichment analysis highlighted carbon metabolism and amino acid biosynthesis/metabolism as major pathways affected by salt stress, together with glycolysis/gluconeogenesis, lipid metabolism, and plant hormone signal transduction (Supplementary Figure 10). WGCNA identified differentially expressed gene modules participating in secondary metabolite biosynthesis/metabolic pathways, where hub genes played pivotal regulatory roles in plant hormone signaling, metabolic pathways, and peroxisome processes (Figure 6), indicating their involvement in salt stress responses. Together, the results provide evidence that OsMADS31 improves salt tolerance by upregulating antioxidant-related genes, activating antioxidant enzymes, and reducing oxidative damage. However, the molecular mechanisms via which it exerts these effects will require further investigation, particularly with respect to how OsMADS31-mediated stress signaling pathways enhance crop salt tolerance.
5 Conclusions
In summary, this study demonstrates a novel role of the MADS-box gene OsMADS31 in salt stress tolerance. To validate the function of OsMADS31 transcription factor in salt tolerance, we generated osmads31 knockout (KO) mutants and OsMADS31 overexpression (OE) lines in the Nipponbare background. The KO mutants showed significantly altered panicle morphology and significant reductions in seed-setting rate, panicle length, grain number per panicle, and 1000-grain weight, whereas OE lines exhibited the opposite trends. Under salt stress, KO mutants showed severely inhibited growth both during seed germination and at the three-leaf stage, whereas OE lines maintained normal germination and growth. After salt treatment and recovery at the three-leaf stage, KO mutants displayed significantly lower survival rates compared with WT plants, while OE lines grew normally. Physiological assays revealed that KO mutants exhibited significantly lower CAT, POD, and SOD activities and reduced Pro content, along with stronger DAB and NBT staining and higher MDA levels, than the WT; conversely, OE lines showed the opposite trends. Transcriptome analysis of osmads31-KO mutants under salt stress revealed significant enrichment of genes related to plant hormone signal transduction and biosynthesis pathways, suggesting their active involvement in salt stress tolerance. Based on the results of GO and KEGG enrichment analyses of DEGs, we propose that OsMADS31 plays important roles in salt stress signaling pathways.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
XY: Investigation, Writing – review & editing, Supervision, Conceptualization, Funding acquisition, Software, Methodology, Formal Analysis, Writing – original draft, Project administration, Visualization, Validation, Data curation, Resources. QG: Software, Investigation, Conceptualization, Writing – review & editing, Supervision, Data curation, Methodology. FW: Conceptualization, Investigation, Writing – review & editing. WL: Writing – review & editing. SY: Writing – review & editing. SZ: Writing – review & editing. JF: Writing – review & editing. RB: Writing – review & editing. YL: Writing – review & editing. LC: Writing – review & editing. XD: Writing – review & editing. ML: Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded by the National Key Research and Development Program of China (grant numbers 2016YFD0101107 and 2018YFD0100902), Hunan Province Key Research and Development Plan (grant number 2016JC2023), and the Natural Science Foundation of Hunan Province in China (2022JJ30377).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1628305/full#supplementary-material
References
Ahanger, M. A., Tomar, N. S., Tittal, M., Argal, S., and Agarwal., R. M. (2017). Plant growth under water/salt stress: ROS production; antioxidants and significance of added potassium under such conditions. Physiol. Mol. Biol. Plants 23, 731–744. doi: 10.1007/s12298-017-0462-7
Alvarez-Buylla, E. R., Liljegren, S. J., Pelaz, S., Gold, S. E., Burgeff, C., Ditta, G. S., et al. (2000). MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 24, 457–466. doi: 10.1046/j.1365-313x.2000.00891.x
Bailey-Serres, J., Parker, J. E., Ainsworth, E. A., Oldroyd, G. E. D., and Schroeder., J. I. (2019). Genetic strategies for improving crop yields. Nature 575, 109–118. doi: 10.1038/s41586-019-1679-0
Cao, Y., Song, F., Goodman, R. M., and Zheng., Z. (2006). Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. J. Plant Physiol. 163, 1167–1178. doi: 10.1016/j.jplph.2005.11.004
Cheeseman, J. M. (2013). The integration of activity in saline environments: problems and perspectives. Funct. Plant Biol. 40, 759–774. doi: 10.1071/FP12285
Chen, C., Begcy, K., Liu, K., Folsom, J. J., Wang, Z., Zhang, C., et al. (2016). Heat stress yields a unique MADS box transcription factor in determining seed size and thermal sensitivity. Plant Physiol. 171, 606–622. doi: 10.1104/pp.15.01992
Chen, C., Wu, Y., Li, J., Wang, X., Zeng, Z., Xu, J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Chen, H., Xu, N., Wu, Q., Yu, B., Chu, Y., Li, X., et al. (2018a). OsMADS27 regulates the root development in a NO3–Dependent manner and modulates the salt tolerance in rice (Oryza sativa L.). Plant Sci. 277, 20–32. doi: 10.1016/j.plantsci.2018.09.004
Chen, L., Zhao, Y., Xu, S., Zhang, Z., Xu, Y., Zhang, J., et al. (2018b). OsMADS57 together with OsTB1 coordinates transcription of its target OsWRKY94 and D14 to switch its organogenesis to defense for cold adaptation in rice. New Phytol. 218, 219–231. doi: 10.1111/nph.14977
D’Autreaux, B. and Toledano, M. B. (2007). ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8, 813–824. doi: 10.1038/nrm2256
de Folter, S. and Angenent, G. C. (2006). trans meets cis in MADS science. Trends Plant Sci. 11, 224–231. doi: 10.1016/j.tplants.2006.03.008
Esteban, R., Ariz, I., Cruz, C., and Moran., J. F. (2016). Review: Mechanisms of ammonium toxicity and the quest for tolerance. Plant Sci. 248, 92–101. doi: 10.1016/j.plantsci.2016.04.008
Fricke, W., Akhiyarova, G., Veselov, D., and Kudoyarova., G. (2004). Rapid and tissue-specific changes in ABA and in growth rate in response to salinity in barley leaves. J. Exp. Bot. 55, 1115–1123. doi: 10.1093/jxb/erh117
Gao, Q., Wang, H., Yin, X., Wang, F., Hu, S., Liu, W., et al. (2023a). Identification of salt tolerance related candidate genes in ‘Sea rice 86’ at the seedling and reproductive stages using QTL-seq and BSA-seq. Genes 14 (2), 458. doi: 10.3390/genes14020458
Gao, Q., Yin, X., Wang, F., Hu, S., Liu, W., Chen, L., et al. (2023b). OsJRL40, a jacalin-related lectin gene, promotes salt stress tolerance in rice. Int. J. Mol. Sci. 24 (8), 7441. doi: 10.3390/ijms24087441
Gao, Q., Yin, X., Wang, F., Zhang, C., Xiao, F., Wang, H., et al. (2023c). Jacalin-related lectin 45 (OsJRL45) isolated from ‘sea rice 86’ enhances rice salt tolerance at the seedling and reproductive stages. BMC Plant Biol. 23, 553. doi: 10.1186/s12870-023-04533-z
Gramzow, L., Ritz, M. S., and Theissen., G. (2010). On the origin of MADS-domain transcription factors. Trends Genet. 26, 149–153. doi: 10.1016/j.tig.2010.01.004
Guo, X., Chen, G., Cui, B., Gao, Q., Guo, J. E., Li, A., et al. (2016). Solanum lycopersicum agamous-like MADS-box protein AGL15-like gene, SlMBP11, confers salt stress tolerance. Mol. Breed. 36 (9), 125. doi: 10.1007/s11032-016-0544-1
Hickey, L. T., Hafeez A, N., Robinson, H., Jackson, S. A., Leal-Bertioli, S. C. M., Tester, M., et al. (2019). Breeding crops to feed 10 billion. Nat. Biotechnol. 37, 744–754. doi: 10.1038/s41587-019-0152-9
Hiei, Y., Ohta, S., Komari, T., and Kumashiro., T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. doi: 10.1046/j.1365-313x.1994.6020271.x
Hu, H., Dai, M., Yao, J., Xiao, B., Li, X., Zhang, Q., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U.S.A. 103, 12987–12992. doi: 10.1073/pnas.0604882103
Huang, Y., Jiao, Y., Xie, N., Guo, Y., Zhang, F., Xiang, Z., et al. (2019). OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 287, 110188. doi: 10.1016/j.plantsci.2019.110188
Hu, J. X., Thomas, C. E., and Brunak., S. (2016). Network biology concepts in complex disease comorbidities. Nat. Rev. Genet. 17, 615–629. doi: 10.1038/nrg.2016.87
Hussain, M., Ahmad, S., Hussain, S., Lal, R., Ul-Allah, S., and Nawaz., A. (2018). Rice in saline soils: physiology, biochemistry, genetics, and management. Adv. Agron. 148, 231–287. doi: 10.1016/bs.agron.2017.11.002
Iqbal, M. N., Rasheed, R., Ashraf, M. Y., Ashraf, M. A., and Hussain., I. (2018). Exogenously applied zinc and copper mitigate salinity effect in maize (Zea mays L.) by improving key physiological and biochemical attributes. Environ. Sci. pollut. Res. Int. 25, 23883–23896. doi: 10.1007/s11356-018-2383-6
Isayenkov, S. V. (2019). Genetic sources for the development of salt tolerance in crops. Plant Growth Regul. 89, 1–17. doi: 10.1007/s10725-019-00519-w
Jambunathan, N. (2010). Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. Methods Mol Biol. 639, 292–298. doi: 10.1007/978-1-60761-702-0_18
Kappel, S., Rumpler, F., and Theissen., G. (2023). Cracking the floral quartet code: how do multimers of MIKCC-type MADS-domain transcription factors recognize their target genes? Int. J. Mol. Sci. 24 (9), 8253. doi: 10.3390/ijms24098253
Kaufmann, K., Melzer, R., and Theissen., G. (2005). MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347, 183–198. doi: 10.1016/j.gene.2004.12.014
Khong, G. N., Pati, P. K., Richaud, F., Parizot, B., Bidzinski, P., Mai, C. D., et al. (2015). OsMADS26 negatively regulates resistance to pathogens and drought tolerance in rice. Plant Physiol. 169, 2935–2949. doi: 10.1104/pp.15.01192
Kwantes, M., Liebsch, D., and Verelst., W. (2012). How MIKC* MADS-box genes originated and evidence for their conserved function throughout the evolution of vascular plant gametophytes. Mol. Biol. Evol. 29, 293–302. doi: 10.1093/molbev/msr200
Li, J., Guo, X., Zhang, M., Wang, X., Zhao, Y., Yin, Z., et al. (2018). OsERF71 confers drought tolerance via modulating ABA signaling and proline biosynthesis. Plant Sci. 270, 131–139. doi: 10.1016/j.plantsci.2018.01.017
Li, J., Huang, Y., Tan, H., Yang, X., Tian, L., Luan, S., et al. (2015). An endoplasmic reticulum magnesium transporter is essential for pollen development in Arabidopsis. Plant Sci. 231, 212–220. doi: 10.1016/j.plantsci.2014.12.008
Liu, H., Chen, X., Song, L., Li, K., Zhang, X., Liu, S., et al. (2019). Polysaccharides from Grateloupia filicina enhance tolerance of rice seeds (Oryza sativa L.) under salt stress. Int. J. Biol. Macromol 124, 1197–1204. doi: 10.1016/j.ijbiomac.2018.11.270
Liu, H., Ding, Y., Zhou, Y., Jin, W., Xie, K., and Chen., L. L. (2017). CRISPR-P 2.0: an improved CRISPR-cas9 tool for genome editing in plants. Mol. Plant 10, 530–532. doi: 10.1016/j.molp.2017.01.003
Livak, K. J. and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-Delta Delta CT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lodeyro, A. F. and Carrillo, N. (2015). Salt stress in higher plants: mechanisms of toxicity and defensive responses. Stress Responses Plants 102, 40–48. doi: 10.1007/978-3-319-13368-3_1
Ma, X., Feng, F., Wei, H., Mei, H., Xu, K., Chen, S., et al. (2016). Genome-wide association study for plant height and grain yield in rice under contrasting moisture regimes. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01801
Miao, J., Guo, D., Zhang, J., Huang, Q., Qin, G., Zhang, X., et al. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236. doi: 10.1038/cr.2013.123
Michaels, S. D., Ditta, G., Gustafson-Brown, C., Pelaz, S., Yanofsky, M., and Amasino., R. M. (2003). AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 33, 867–874. doi: 10.1046/j.1365-313X.2003.01671.x
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., and Mittler., R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Mizuno, H., Kawahara, Y., Sakai, H., Kanamori, H., Wakimoto, H., Yamagata, H., et al. (2010). Massive parallel sequencing of mRNA in identification of unannotated salinity stress-inducible transcripts in rice (Oryza sativa L.). BMC Genomics 11, 683. doi: 10.1186/1471-2164-11-683
Munns, R., Guo, J., Passioura, J. B., and Cramer., G. R. (2000). Leaf water status controls day-time but not daily rates of leaf expansion in salt-treated barley. Funct. Plant Biol. 27, 949–957. doi: 10.1071/pp99193
Munns, R., James, R. A., Gilliham, M., Flowers, T. J., and Colmer., T. D. (2016). Tissue tolerance: an essential but elusive trait for salt-tolerant crops. Funct. Plant Biol. 43, 1103–1113. doi: 10.1071/FP16187
Munns, R. and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Muranaka, S., Shimizu, K., and Kato., M. (2002). Ionic and osmotic effects of salinity on single-leaf photosynthesis in two wheat cultivars with different drought tolerance. Photosynthetica 40, 201–207. doi: 10.1023/a:1021337522431
Murphy, L. R., Kinsey, S. T., and Durako., M. J. (2003). Physiological effects of short-term salinity changes on Ruppia maritima. Aquat. Bot. 75, 293–309. doi: 10.1016/s0304-3770(02)00206-1
Nam, M. H., Bang, E., Kwon, T. Y., Kim, Y., Kim, E. H., Cho, K., et al. (2015). Metabolite profiling of diverse rice germplasm and identification of conserved metabolic markers of rice roots in response to long-term mild salinity stress. Int. J. Mol. Sci. 16, 21959–21974. doi: 10.3390/ijms160921959
Nam, J., dePamphilis, C. W., Ma, H., and Nei., M. (2003). Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol. Biol. Evol. 20, 1435–1447. doi: 10.1093/molbev/msg152
Naveed, S. A., Zhang, F., Zhang, J., Zheng, T. Q., Meng, L. J., Pang, Y. L., et al. (2018). Identification of QTN and candidate genes for Salinity Tolerance at the Germination and Seedling Stages in Rice by Genome-Wide Association Analyses. Sci. Rep. 8, 6505. doi: 10.1038/s41598-018-24946-3
Nayar, S., Kapoor, M., and Kapoor., S. (2014). Post-translational regulation of rice MADS29 function: homodimerization or binary interactions with other seed-expressed MADS proteins modulate its translocation into the nucleus. J. Exp. Bot. 65, 5339–5350. doi: 10.1093/jxb/eru296
Norman, C., Runswick, M., Pollock, R., and Treisman., R. (1988). Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 55, 989–1003. doi: 10.1016/0092-8674(88)90244-9
Oh, S.-J., Kim, Y. S., Kwon, C.-W., Park, H. K., Jeong, J. S., and Kim, A. J.-K. (2009). Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol. 150 (3), 1368–1379. doi: 10.1104/pp.109.137554
Ozturk, M., Turkyilmaz Unal, B., Garcia-Caparros, P., Khursheed, A., Gul, A., and Hasanuzzaman., M. (2021). Osmoregulation and its actions during the drought stress in plants. Physiol. Plant 172, 1321–1335. doi: 10.1111/ppl.13297
Puig, J., Meynard, D., Khong, G. N., Pauluzzi, G., Guiderdoni, E., and Gantet., P. (2013). Analysis of the expression of the AGL17-like clade of MADS-box transcription factors in rice. Gene Expr Patterns 13, 160–170. doi: 10.1016/j.gep.2013.02.004
Qin, H. and Huang, R. F. (2020). The phytohormonal regulation of Na+/K+ and reactive oxygen species homeostasis in rice salt response. Mol. Breed. 40, 1–13. doi: 10.1007/s11032-020-1100-6
Razzaq, A., Ali, A., Safdar, L. B., Zafar, M. M., Rui, Y., Shakeel, A., et al. (2020). Salt stress induces physiochemical alterations in rice grain composition and quality. J. Food Sci. 85, 14–20. doi: 10.1111/1750-3841.14983
Rounsley, S. D., Ditta, G. S., and Yanofsky., M. F. (1995). Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 7, 1259–1269. doi: 10.1105/tpc.7.8.1259
Sackey, O. K., Feng, N., Mohammed, Y. Z., Dzou, C. F., Zheng, D., Zhao, L., et al. (2025). A comprehensive review on rice responses and tolerance to salt stress. Front. Plant Sci. 16. doi: 10.3389/fpls.2025.1561280
Saha, G., Park, J. I., Jung, H. J., Ahmed, N. U., Kayum, M. A., Chung, M. Y., et al. (2015). Genome-wide identification and characterization of MADS-box family genes related to organ development and stress resistance in Brassica rapa. BMC Genomics 16, 178. doi: 10.1186/s12864-015-1349-z
Sasaki, T. and Burr, B. (2000). International Rice Genome Sequencing Project: the effort to completely sequence the rice genome. Curr. Opin. Plant Biol. 3, 138–141. doi: 10.1016/s1369-5266(99)00047-3
Seifikalhor, M., Aliniaeifard, S., Shomali, A., Azad, N., Hassani, B., Lastochkina, O., et al. (2019). Calcium signaling and salt tolerance are diversely entwined in plants. Plant Signal Behav. 14, e1665455. doi: 10.1080/15592324.2019.1665455
Shabala, S. and Pottosin, I. (2014). Regulation of potassium transport in plants under hostile conditions: implications for abiotic and biotic stress tolerance. Physiol. Plant 151, 257–279. doi: 10.1111/ppl.12165
Smaczniak, C., Immink, R. G., Angenent, G. C., and Kaufmann., K. (2012). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139, 3081–3098. doi: 10.1242/dev.074674
Szabados, L. and Savoure, A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15, 89–97. doi: 10.1016/j.tplants.2009.11.009
Teng, Z., Zheng, Q., Peng, Y., Li, Y., Meng, S., Liu, B., et al. (2025). Nitrate reductase-dependent nitric oxide production mediates nitrate-conferred salt tolerance in rice seedlings. Plant Physiol. 197 (3), kiaf080. doi: 10.1093/plphys/kiaf080
Theißen, G. and Becker, A. (2004). Gymnosperm orthologues of class B floral homeotic genes and their impact on understanding flower origin. Crit. Rev. Plant Sci. 23, 129–148. doi: 10.1080/07352680490433240
Theissen, G., Melzer, R., and Rumpler., F. (2016). MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143, 3259–3271. doi: 10.1242/dev.134080
Thompson, B. E., Bartling, L., Whipple, C., Hall, D. H., Sakai, H., Schmidt, R., et al. (2009). bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell 21, 2578–2590. doi: 10.1105/tpc.109.067751
Wahid, A., Perveen, M., Gelani, S., and Basra., S. M. (2007). Pretreatment of seed with H2O2 improves salt tolerance of wheat seedlings by alleviation of oxidative damage and expression of stress proteins. J. Plant Physiol. 164, 283–294. doi: 10.1016/j.jplph.2006.01.005
Wang, F., Cheng, Z., Wang, J., Zhang, F., Zhang, B., Luo, S., et al. (2022a). Rice STOMATAL CYTOKINESIS DEFECTIVE2 regulates cell expansion by affecting vesicular trafficking in rice. Plant Physiol. 189, 567–584. doi: 10.1093/plphys/kiac073
Wang, X., Li, J., Li, F., Pan, Y., Cai, D., Mao, D., et al. (2021). Rice potassium transporter osHAK8 mediates K+ Uptake and translocation in response to low K+ Stress. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.730002
Wang, H., Lu, S., Guan, X., Jiang, Y., Wang, B., Hua, J., et al. (2022b). Dehydration-responsive element binding protein 1C, 1E, and 1G promote stress tolerance to chilling, heat, drought, and salt in rice. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.851731
Wang, H., Tang, X., Wang, H., and Shao., H. B. (2015). Proline accumulation and metabolism-related genes expression profiles in Kosteletzkya virginica seedlings under salt stress. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00792
Wang, J., Zhu, J., Zhang, Y., Fan, F., Li, W., Wang, F., et al. (2018). Comparative transcriptome analysis reveals molecular response to salinity stress of salt-tolerant and sensitive genotypes of indica rice at seedling stage. Sci. Rep. 8, 2085. doi: 10.1038/s41598-018-19984-w
Wegner, L. H., Stefano, G., Shabala, L., Rossi, M., Mancuso, S., and Shabala., S. (2011). Sequential depolarization of root cortical and stelar cells induced by an acute salt shock-implications for Na+ and K+ transport into xylem vessels. Plant Cell Environ. 34, 859–869. doi: 10.1111/j.1365-3040.2011.02291.x
West, A. G., Shore, P., and Sharrocks., A. D. (1997). DNA binding by MADS-box transcription factors: a molecular mechanism for differential DNA bending. Mol. Cell Biol. 17, 2876–2887. doi: 10.1128/mcb.17.5.2876
Wing, R. A., Purugganan, M. D., and Zhang., Q. (2018). The rice genome revolution: from an ancient grain to Green Super Rice. Nat. Rev. Genet. 19, 505–517. doi: 10.1038/s41576-018-0024-z
Wu, J., Yu, C., Huang, L., and Gan., Y. (2021). A rice transcription factor, OsMADS57, positively regulates high salinity tolerance in transgenic Arabidopsis thaliana and Oryza sativa plants. Physiol. Plant 173, 1120–1135. doi: 10.1111/ppl.13508
Wu, J., Yu, C., Hunag, L., Wu, M., Liu, B., Liu, Y., et al. (2019). Overexpression of MADS-box transcription factor OsMADS25 enhances salt stress tolerance in Rice and Arabidopsis. Plant Growth Regul. 90, 163–171. doi: 10.1007/s10725-019-00539-6
Wu, H., Zhang, X., Pablo, G. J., and Sergey., S. (2018). It is not all about sodium: revealing tissue specificity and signalling roles of potassium in plant responses to salt stress. Plant Soil 431, 1–17. doi: 10.1007/s11104-018-3770-y
Xu, N., Chu, Y., Chen, H., Li, X., Wu, Q., Jin, L., et al. (2018). Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA-mediated regulatory pathway and ROS scavenging. PloS Genet. 14, e1007662. doi: 10.1371/journal.pgen.1007662
Yan, H., Jia, H., Chen, X., Hao, L., An, H., and Guo., X. (2014). The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 55, 2060–2076. doi: 10.1093/pcp/pcu133
Yang, Y., Fanning, L., and Jack., T. (2003). The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 33, 47–59. doi: 10.1046/j.0960-7412.2003.01473.x
Yang, Y. and Guo, Y. (2018a). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Yang, Y. and Guo, Y. (2018b). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60 (9), 796–804. doi: 10.1111/jipb.12689
Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A., and Meyerowitz., E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. doi: 10.1038/346035a0
Yin, X., Gao, Q., Wang, F., Liu, W., Luo, Y., Zhong, S., et al. (2024). Marker-assisted selection of jacalin-related lectin genes osJRL45 and osJRL40 derived from sea rice 86 enhances salt tolerance in rice. Int. J. Mol. Sci. 25 (20), 10912. doi: 10.3390/ijms252010912
Yu, L. H., Miao, Z. Q., Qi, G. F., Wu, J., Cai, X. T., Mao, J. L., et al. (2014b). MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol. Plant 7, 1653–1669. doi: 10.1093/mp/ssu088
Yu, C., Su, S., Xu, Y., Zhao, Y., Yan, A., Huang, L., et al. (2014a). The effects of fluctuations in the nutrient supply on the expression of five members of the AGL17 clade of MADS-box genes in rice. PloS One 9, e105597. doi: 10.1371/journal.pone.0105597
Yu, L. H., Wu, J., Zhang, Z. S., Miao, Z. Q., Zhao, P. X., Wang, Z., et al. (2017). Arabidopsis MADS-box transcription factor AGL21 acts as environmental surveillance of seed germination by regulating ABI5 expression. Mol. Plant 10, 834–845. doi: 10.1016/j.molp.2017.04.004
Yu, J., Zhao, W., Tong, W., He, Q., Yoon, M. Y., Li, F. P., et al. (2018). A Genome-Wide Association Study Reveals Candidate Genes Related to Salt Tolerance in Rice (Oryza sativa) at the Germination Stage. Int. J. Mol. Sci. 19 (10), 3145. doi: 10.3390/ijms19103145
Zhang, K., Su, J., Xu, M., Zhou, Z., Zhu, X., Ma, X., et al. (2020). A common wild rice-derived BOC1 allele reduces callus browning in indica rice transformation. Nat. Commun. 11, 443. doi: 10.1038/s41467-019-14265-0
Zhang, H., Zhang, G., Lü, X., Zhou, D., and Han, X. (2014). Salt tolerance during seed germination and early seedling stages of 12 halophytes. Plant Soil 388, 229–241. doi: 10.1007/s11104-014-2322-3
Zhang, Z., Zhang, Q., Wu, J., Zheng, X., Zheng, S., Sun, X., et al. (2013). Gene knockout study reveals that cytosolic ascorbate peroxidase 2(OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PloS One 8, e57472. doi: 10.1371/journal.pone.0057472
Zhao, C., Zhang, H., Song, C., Zhu, J. K., and Shabala., S. (2020). Mechanisms of plant responses and adaptation to soil salinity. Innovation (Camb) 1, 100017. doi: 10.1016/j.xinn.2020.100017
Keywords: rice (Oryza sativa L.), MADS-box, transcription factor, OsMADS31, salt stress, antioxidant, transcriptome
Citation: Yin X, Gao Q, Wang F, Liu W, Yu S, Zhong S, Feng J, Bai R, Luo Y, Chen L, Dai X and Liang M (2025) MIKC-type MADS-box transcription factor OsMADS31 positively regulates salinity tolerance in rice. Front. Plant Sci. 16:1628305. doi: 10.3389/fpls.2025.1628305
Received: 14 May 2025; Accepted: 30 July 2025;
Published: 04 September 2025.
Edited by:
Baohua Feng, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Zhiwei Chen, Shanghai Academy of Agricultural Sciences, ChinaLong Wang, Hunan University, China
Copyright © 2025 Yin, Gao, Wang, Liu, Yu, Zhong, Feng, Bai, Luo, Chen, Dai and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojun Dai, aG5zZmR4ajAzMDlAaHVubnUuZWR1LmNu; Manzhong Liang, MTk2MmxpYW5nQDE2My5jb20=
 Xiaolin Yin
Xiaolin Yin Manzhong Liang
Manzhong Liang