- 1Translational Plant Research Laboratory, Department of Microbial Biotechnology, Bharathiar University, Coimbatore, Tamil Nadu, India
- 2Department of Bioinformatics, Alagappa University, Karaikudi, Tamil Nadu, India
- 3Centre for Research, Dhanalakshmi Srinivasan University, Tiruchirappalli, Tamil Nadu, India
- 4Department of Pharmaceutical Biology, Faculty of Pharmaceutical Sciences, UCSI University, Kuala Lumpur, Malaysia
- 5Department of Civil & Energy System Engineering, Kyonggi University, Suwon, Gyeonggi-Do, Republic of Korea
- 6Department of Microbiology, Faculty of Arts Science Commerce and Management, Karpagam Academy of Higher Education, Coimbatore, Tamil Nadu, India
- 7Department of Microbiology, Chungbuk National University Medical College, Cheongju, Republic of Korea
Millets are highly nutritious crops mostly cultivated in xeric regions of Asia and Africa. The specific agro-climatic characteristics of millets enable the rhizosphere to host diverse microorganisms that assist in the crop’s progression under harsh weather conditions. This research work intends to evaluate the plant growth-promoting (PGP) potential of the rhizospheric microbes isolated from the soil of Panicum sumatrense and Eleusine coracana from Kunnanchala and Varagampadi, residing areas of Irulas Tribal Community located in the Attapadi Hills of Kerala and Tamil Nadu, respectively. A total of 53 bacterial isolates with unique colony morphology were initially subjected to the nitrogen fixation test. Twenty-six isolates that demonstrated positive results for nitrogen fixation were characterized for phosphate solubilization, ammonia, indole acetic acid (IAA), siderophore, hydrogen cyanide (HCN), and hydrolytic enzyme production, molecular characterization, and antagonistic activity against the common plant pathogens Fusarium oxysporum and Colletotrichum gloeosporioides. The principal component analysis revealed that SA1 (Staphylococcus gallinarum) and MS6 (Kosakonia sacchari) exhibited the highest values for IAA production (0.37 ± 0.015 mg/mL) and PSI (3.36 ± 0.03), respectively. MS3 (Bacillus velezensis) demonstrated the most promising results in antagonism (91.32 ± 0.57) and ammonia production (0.11 ± 0.020 mg/mL). All the bacterial isolates exhibited a notable improvement in germination, shoot length, root length, and vigor index of Solanum lycopersicum. The most prominent results in germination studies were noted in K. sacchari (MS6), with the most pronounced effects, including a 100% germination rate, 2.58 ± 0.01 cm of shoot length, 7.61 ± 0.03 cm of root length, and a vigor index of 1019. The results of the invitro PGP traits are sufficient to support future tests on the promotion of growth invivo for these seven strains in a single or consortium.
Introduction
Millet is a small seeded cereal crop from the family Poaceae, classified under the C4 grasses, and usually cultivated in xeric and semi-xeric regions (Han et al., 2017). Major and minor millets are two categories of millets which include Eleusine coracan (finger millet), Panicum miliaceum (proso millet), Setaria italica (foxtail millet), Pennisetum glaucum (pearl millet), Echinochlo acolona (barnyard millet), Digitaria bursa (black fonio millet), Digitaria exilis (white fonio millet), Panicum miliare (little millet), Paspalum scrobiculatum (Kodo millet), Eragrostis tef (teff millet), respectively (Annor et al., 2014). Millets are known for their potential to withstand harsh, dry environments and challenging environments, including changing climates and nutrient-poor soil (Sharma and Ortiz, 2000). These characteristic features make it a wise alternative cereal crop for regions with limited rainfall, irrigation, and nutrient-poor soil. Millets typically flourish in higher temperatures and can reproduce with minimal water input, making them thermophilic and xerophilic (Saxena et al., 2018). Together with their nutritional benefits, millets offer short-term cultivation periods, reduced water requirements, and adaptability to adverse climatic conditions. When compared to major cereal crops, millets yield high on marginal land. These specific agro-climatic characteristics render millets particularly well-suited for cultivation in semi-arid regions of Asia and Africa, where other major crops frequently encounter challenges (Belton and Taylor, 2004).
The rhizosphere is an area of the soil that harbors microorganisms with a significant connection to plant physiology, involving complex organic material decomposition, resistance to stresses (both biotic and abiotic), and contributing to plant development. The resilience of millets is largely driven by the microbial community present in the rhizosphere. The term “plant growth-promoting microorganisms” (PGPM) encompasses a diverse array of organisms that reside in the rhizosphere, which includes plant growth-promoting rhizobacteria (PGPR), antagonistic microorganisms, beneficial symbionts, mycorrhizal fungi, mycoparasitic fungi, endophytic bacteria, and fungi (Meena et al., 2017). The PGPR present in the rhizosphere region boosts plant growth and development by engaging in various positive mechanisms. These include increasing the nutrient uptake, fixing the atmospheric nitrogen, suppressing the growth of phytopathogens (Wang et al., 2022; Sheteiwy et al., 2023), and improving the host plant resistance against various abiotic and biotic stressors (Selvakumar et al., 2012; Zolla et al., 2013) and results in increased plant growth and yield (Berg, 2009; Huang et al., 2014).
The principal role of PGPR in nurturing plant growth is achieved through two distinct mechanisms: direct and indirect (Kong and Liu, 2022). The direct mechanisms involve the synthesis of plant growth hormones such as cytokinin, gibberellins, and auxins, together with the solubilization of nutrients, including phosphate. The indirect mechanisms include the production of antibiosis to combat phytopathogens and ACC deaminase to resist various stressors (Barea et al., 2005). Azotobacter, Azospirillum, Acinetobacter, Azoarcus, Arthrobacter, Burkholderia, Bacillus, Beijerinckia, Enterobacter, Erwinia, Klebsiella, Gluconacetobacter, and Serratia. Some of the most extensively studied PGPR for plant growth promotion (PGP) belong to the genus Rhizobium and Pseudomonas (Murphy et al., 2003; Esitken et al., 2006). Acinetobacteria have been shown to enhance the yields of various crops, particularly those of global importance like rice, maize, and wheat (Mahmoud et al., 2024).
To address the growing food demands of an increasing population, agrochemicals use has escalated to boost crop growth and productivity. These agrochemicals are pivotal in increasing crop productivity. However, their prolonged usage may lead to soil infertility through processes such as diminishment of soil organic matter, nitrogen leaching, depletion of soil carbon, and soil compaction. To overcome the usage of hazardous chemicals in agriculture, a wise alternative is the application of biofertilizers derived from PGPR (Nader et al., 2024), while endophytes (Mahmoud et al., 2025) can also act as promising agents that enhance plant growth and support eco-friendly, sustainable agriculture. It is an environmentally friendly and cost-effective alternative that can enhance soil fertility and pave the way for sustainable agriculture. This research work intends to isolate, characterize, and identify PGPRs that possess PGP properties from the region’s millet rhizosphere.
Materials and methods
Sample collection
Rhizospheric soil samples from 2 different types of millets, at a depth of 6–10 cm, were collected: two from Kunnanchala (5 soil samples/Panicum sumatrense and 5 soil samples/Eleusine coracana), and one from Varagampadi (5 samples/P. sumatrense – wild type), a region of the Irulas tribal community located in the Attapadi Hills of Kerala and Tamil Nadu, respectively (Figure 1). The soil samples were meticulously collected in aseptic containers and immediately transported and processed in the laboratory under sterile conditions.
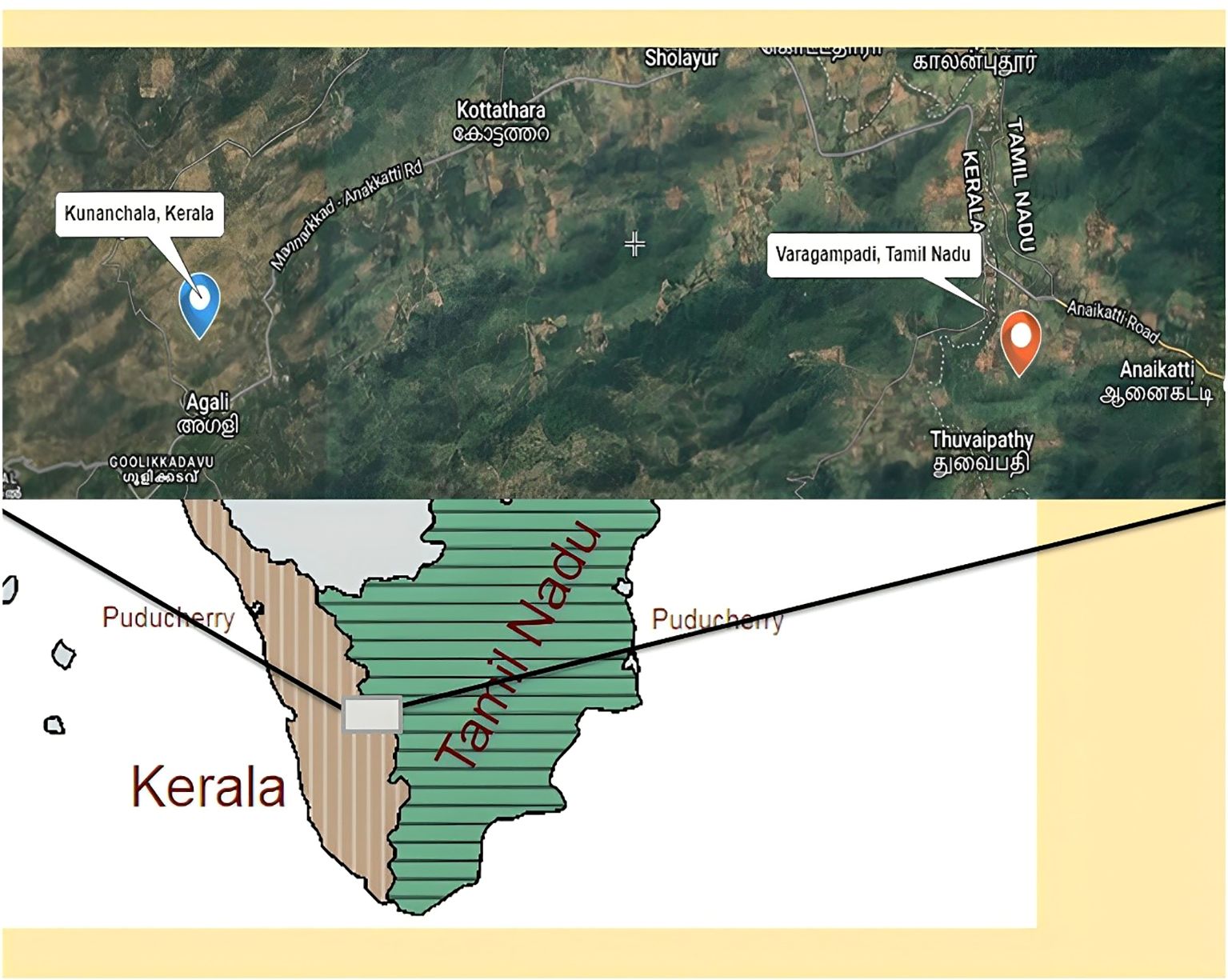
Figure 1. Rhizospheric soil sample collection area Kunnanchala (Lat 11.100603°, Long 76.609637°) and Varagampadi (Lat 11.069483°, Long 76.710132°), located in the Attapadi Hills of Kerala and Tamil Nadu, respectively.
Bacterial isolation from rhizospheric soil
The rhizospheric soil samples were obtained, transported, and shade-dried to eliminate moisture content. Thereafter, the sample was subjected to serial dilution ranging from 10-1to10-10,and each dilution was plated on the nutrient agar medium (Peptone 5 g/L, Sodium chloride 5 g/L, HM peptone B 1.5 g/L, Yeast extract 1.5 g/L, Agar 15 g/L, Final pH (at 25°C) 7.4 ± 0.2) for bacterial isolation after incubating for 2–3 days at 37°C. During this period, the colonies on the plates were compared with control plates to ensure the absence of contamination. The colonies were randomly chosen based on their distinct morphological features. Each colony was then subjected to a process of pure culture isolation. The obtained pure cultures were named with unique number codes and preserved in glycerol stocks, which were stored at a temperature of –20°C for future use.
Nitrogen fixation test
Qualitative confirmation of nitrogen fixation was achieved through the use of Jensen’s Nitrogen-free bacteria medium (JNFb) in which the inoculated pure bacterial cultures were kept under incubation at 28 ± 2°C for 4 days. The growth of bacteria on the JNFb medium indicates a positive result and qualitative confirmation for nitrogen fixation. The isolates were evaluated for their capacity to fix atmospheric nitrogen in the soil on this medium (Andy et al., 2020).
Invitro analysis of PGP activities
The isolates’ qualitative and quantitative assessment of Indole-3-acetic acid (IAA) synthesis was analyzed as per the protocol followed by Goswami et al. (2014). Based on the method followed by Cappuccino and Sherman (1992), the ammonia production was quantified by the calorimetric method. Phosphate solubilization by the isolates was analyzed by Pikovskaya medium (Pikovskaya, 1948). The halo zone formation signified phosphate solubilization (Mehta and Nautiyal, 2001) and it was quantified using the phosphate solubilization index (PSI), Goswami and Deka, 2020). Additionally, HCN and siderophore production were determined as per the protocol by Castric (1975) and Alexander and Zuberer (1991), respectively.
Hydrolytic extracellular enzyme activity
The chosen bacterial isolates were assessed for their capacity to produce catalase by adding 3% hydrogen peroxide drop by drop to a clean glass slide containing a 48-hour bacterial culture. The presence of effervescence showed a positive result for catalase activity (Kumari et al., 2018). An invitro plate assay was conducted to detect the hydrolytic extracellular enzyme production. The protease production was screened by spot-inoculating the isolated strains on a plate containing skim milk agar and incubate at 30°C for 24 to 48 hours. After incubation, the generation of a clear halo zone circle the colony indicates a positive result for the degradation of protease.
The isolates were dot-inoculated on starch agar medium and incubated for 24 to 48 hours at 30°C to determine the amylase activity. Subsequently, the plates were saturated with iodine solution, left for one minute, and discarding the excess iodine. The formation of a colorless zone surrounding the colony signifies the production of the amylase enzyme. The cellulase production was screened by inoculating the strains in carboxy methyl cellulose Congo red agar medium and incubating at 28 ± 2°C for 2–3 days. Following incubation, the formation of a halo zone indicates a positive result for the synthesis of the cellulase enzyme. For the synthesis of lipase enzyme, the isolated strains were spot-inoculated on a tributyrin agar medium and kept under incubation for 2 days at 37°C. The clear zone formed around the colony showed the ability of the isolated strains to produce lipase enzymes (Sachan et al., 2018).
In vitro screening of antifungal activity
The isolated strains were evaluated for their invitro antifungal characteristics against the fungal phytopathogens Fusarium oxysporum and Colletotrichum gloeosporioides using a dual culture technique with a potato dextrose agar (PDA) medium. The fully grown active culture of phytopathogens was prepared by cutting 5mm diameter mycelial disks and placing them at the center of fresh sterile PDA plates. The overnight-grown isolated strains were then streaked uniformly around the mycelial discs served as the control. The plates were kept under incubation at 26°C for 7 days. Following incubation, the plates were examined for the zone of inhibition against the fungal phytopathogens. All experiments were conducted in triplicate. The rate of inhibition can be calculated by the formula.
IR: inhibition rate; T: Diameter of the fungal colony in the antagonist plate, C: Diameter of the fungal colony in the control plates; (Ezrari et al., 2021).
Molecular characterization of potent PGPR
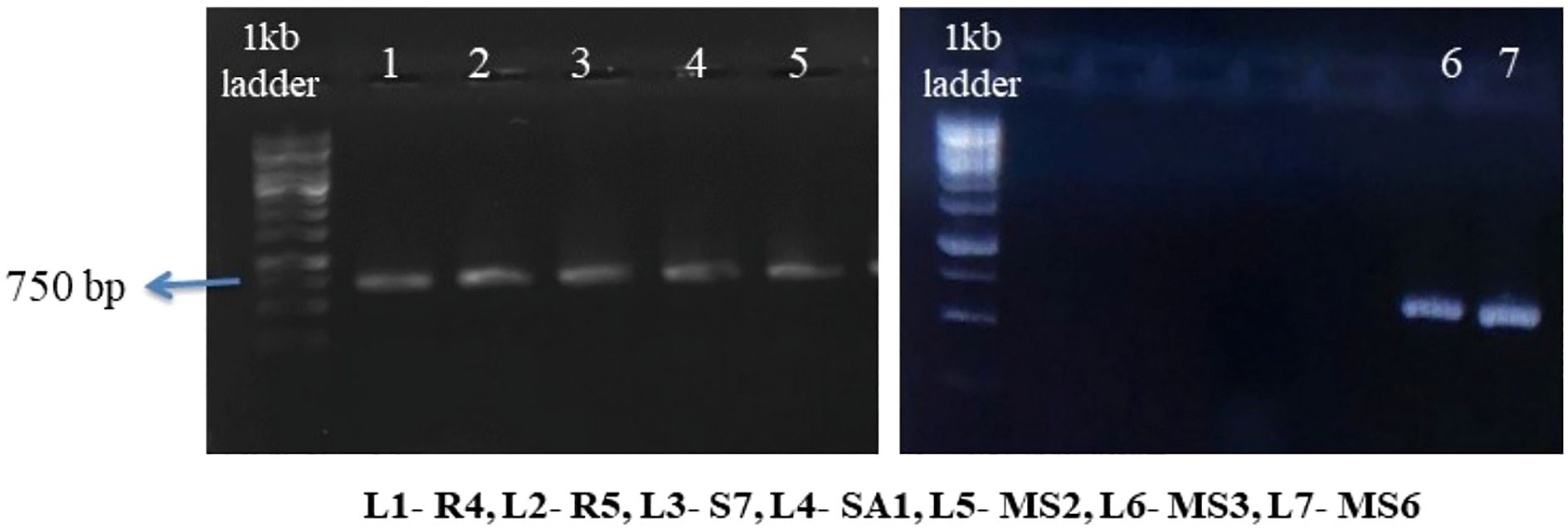
Genomic DNA was extracted from the selected strains using the phenol/chloroform method as described by Kumari et al. (2018). The overnight-grown bacterial strains were centrifuged at 10,000 rpm for 10 minutes. Following centrifugation, the supernatant was cast off and 30 μL of 10% SDS and then 3 μL of proteinase K were mixed with the pellets, which were then incubated for 1hr at 37°C. Subsequently, the mixer mentioned above was subjected to another centrifugation process, during which phenol-chloroform-isoamyl alcohol (25:24:1) was added. This step precipitated genomic DNA with ice-cold ethanol. Following the extraction of genomic DNA from the selected strains, DNA amplification was conducted using two universal primers, 16S rRNA (16SF & 16SR), via polymerase chain reaction (PCR).
Impact of bacterial isolates on seed germination
The selected seven bacterial strains were progressed for the seed germination study. The seeds of the Solanum lycopersicum PKM 1varietywere sourced from the Tamil Nadu Agricultural University, Coimbatore. Prior to the commencement of the study, the seeds were subjected to surface sterilization with 2% sodium hypochlorite for duration of 2 min. Thereafter, the seeds were rinsed 3 to 5 times with autoclaved distilled water to eliminate any residual surface disinfectant (Pandey and Gupta, 2020). The germination study was conducted as 8 treatments.
T1 - Seeds soaked in water (Control)
T2 - Seeds bioprimed with R4
T3 - Seeds bioprimed with R5
T4- Seeds bioprimed with S7
T5- Seeds bioprimed with SA1
T6- Seeds bioprimed with MS2
T7- Seeds bioprimed with MS3
T8- Seeds bioprimed with MS6
The surface-sterilized seeds were bioprimed with the seven bacterial isolates (108 CFU mL−1) separately in a sterile tube for 1 hour at a 180-rpm orbital shaker. A control set of surface-sterilized seeds was prepared by soaking them in autoclaved distilled water. Subsequently, the bioprimed and control seeds were placed on a sterile Petri plate containing wetted sterile cotton. Each plate contains 10 seeds, with three replicates per treatment. The seeds were observed for seed germination over a period of 10 days. The seed germination percentage (G%) and seed vigor index (VI) were calculated using the following formula, as outlined by Fahsi et al. (2021).
Statistical analysis
The obtained data were assessed through one-way ANOVA in SPSS Statistics version 16.0 software. The significant variance among the treatment means was calculated using Duncan’s multiple range test at a p < 0.05 significance level. The experiments were expressed as the mean of triplicate ± standard error (SE). The Principal Component Analysis (PCA) was employed to investigate the correlation between the bacterial isolates and their PGP activity. The Bar graphs were plotted using Origin 2018–64 Bit software
Results
Isolation and characterization of bacterial isolates
In the present work, 53 morphologically distinct isolates were isolated from millets (P. sumatrense and E. coracana) rhizosphere soil using the spread plate technique. categorized based on their colony morphology. All the 53 isolates were assessed for their nitrogen fixation ability, and 26 were found to be capable of fixing nitrogen when inoculated in a nitrogen-free medium.
Invitro PGP activity
Invitro assays revealed that, 14 isolates produced the phytohormone IAA, 23 isolates produced ammonia, 15 isolates were observed to solubilize phosphate, 3 isolates were identified to produce HCN, and 5 isolates were observed to produce siderophore. Based on the qualitative analysis of PGP properties, 7 of the 26 isolates (designated as R4, R5, S7, SA1, MS2, MS3, and MS6) (Figure 2) exhibited the most promising results for maximum PGP activities and were selected for further analysis of hydrolytic enzyme production, antagonistic activity, and germination assay.
The IAA production among the isolates varied from 0.05 to 0.37 mg/mL (Table 1). These results were quantified against a standard curve prepared from a precise concentration of standard IAA (Figures 3A, 4A). The highest IAA production was observed in SA1, with a concentration of 0.37 ± 0.015 mg/mL, and the lowest was in R5 (0.05 ± 0.008 mg/mL For ammonia production, the isolates R4, R5, S7, SA1, MS2, MS3, and MS6 exhibited higher production ranging from 0.06 and 0.11 mg/mL (Table 1), (Figures 3B, 4A). The highest level of ammonia was produced by MS3 with 0.11 ± 0.020 mg/mL. Upon investigating the phosphate solubilization ability of the isolates, a halo zone was observed around the bacterial colonies on Pikovskaya agar medium, with varying PSI values ranging from 2.36 to 3.80 (Table 1, Figures 3C, 4B).
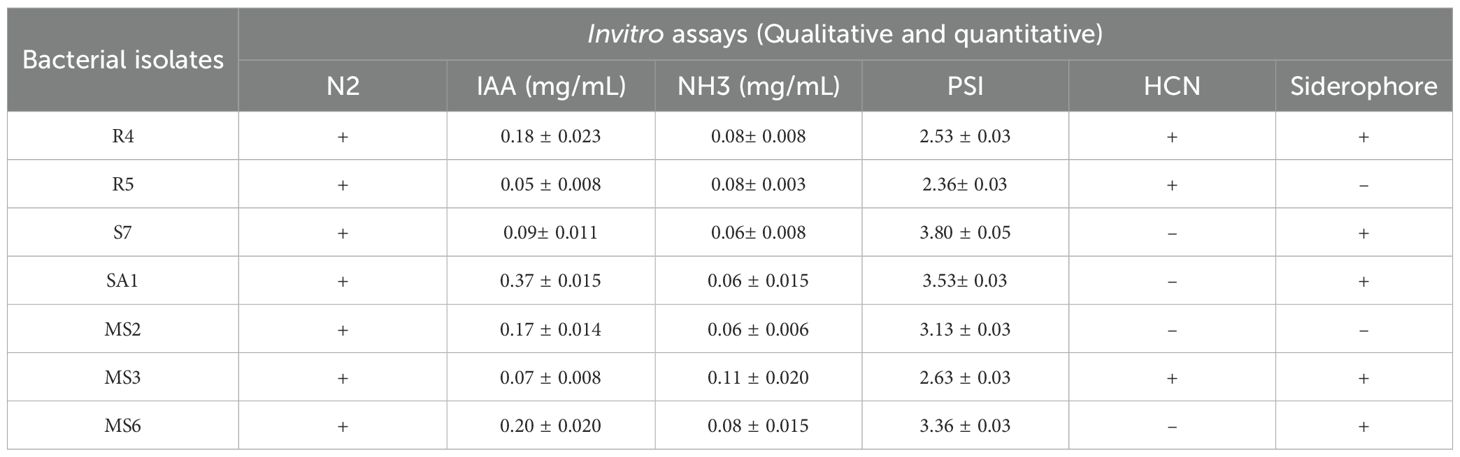
Table 1. Invitro screening of isolates for plant growth-promoting activities (qualitative and quantitative).
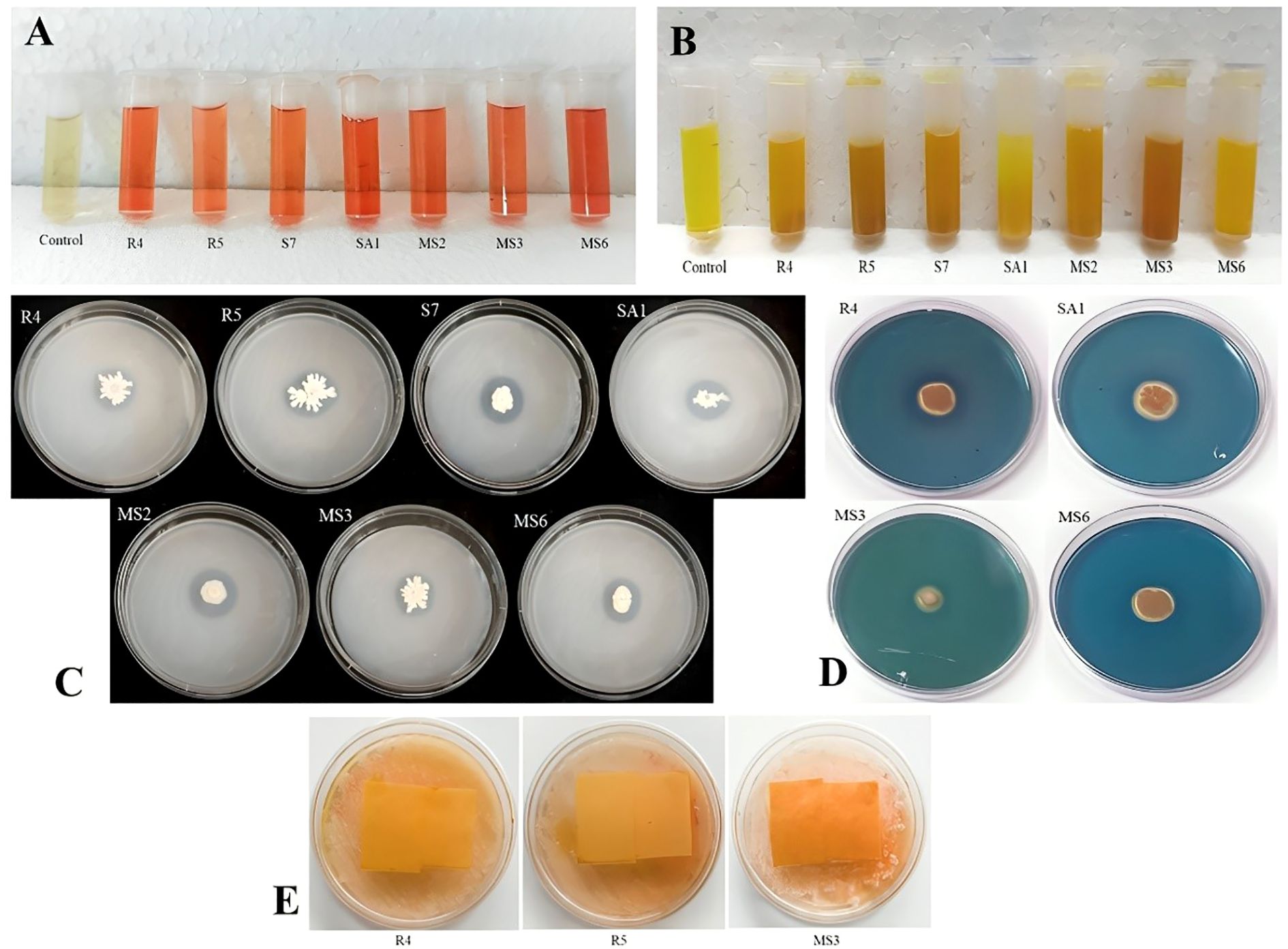
Figure 3. Invitro screening of the isolates for (A) IAA production, (B) Ammonia production, (C) phosphate solubilization activity (D) siderophore production, and (E) production of HCN.
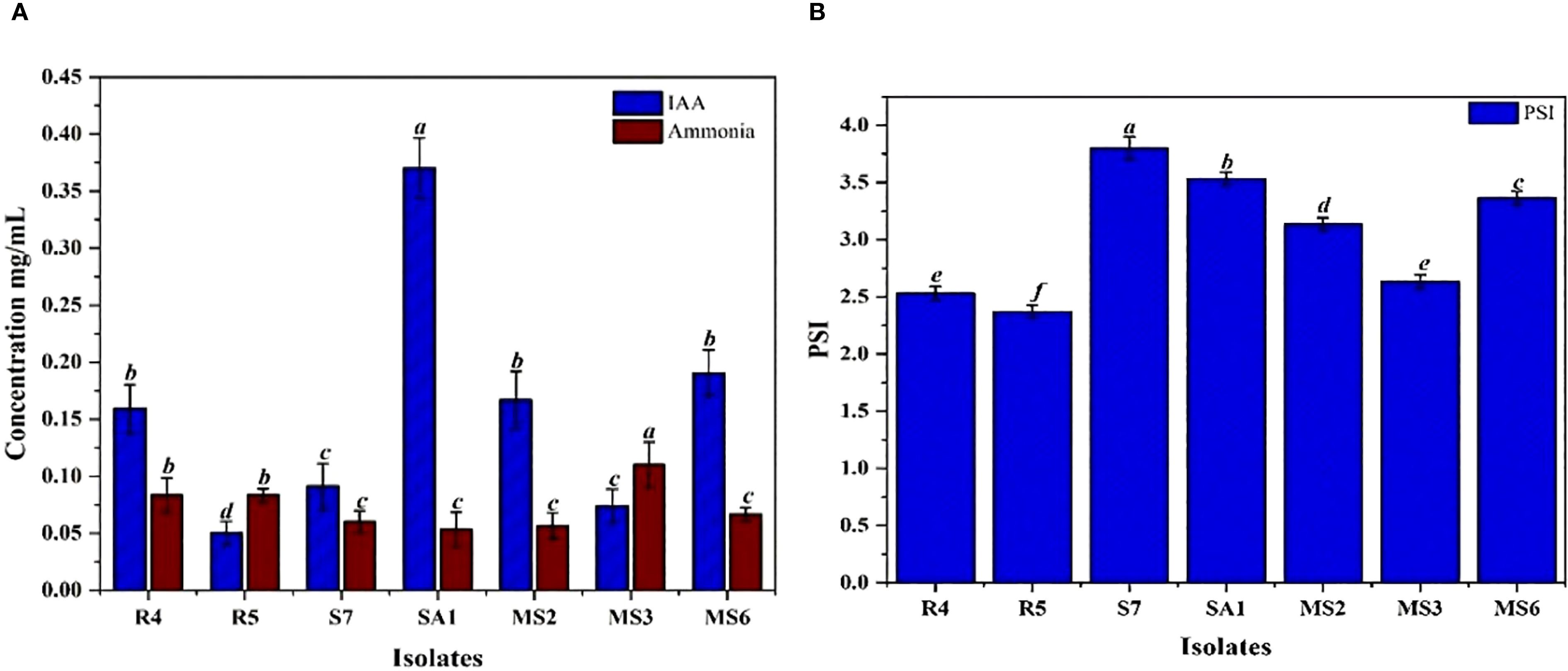
Figure 4. (A) Quantification of IAA and Ammonia production (B) phosphate solubilization index (PSI) by seven bacterial isolates, namely R4, R5, S7, SA1, MS2, MS3, and MS6. Bars with different letters show significant differences from each other (p < 0.05), and the liner bar shows the standard error.
In the present work, the isolates, namely R4, R5, and MS3, demonstrated the capacity to produce HCN and the remaining were unable to shift the color of Whatman filter paper no. 1 (Figure 3E). The synthesis of siderophore by the isolates was confirmed by the generation of an orange halo zone surrounding the bacterial colony inoculated in the chrome azurol S (CAS) agar plates. The isolates R4, S7, SA1, MS3, and MS6 were found to be capable of producing siderophore, as evidenced by the results presented in Figure 3D.
Hydrolytic enzyme activity
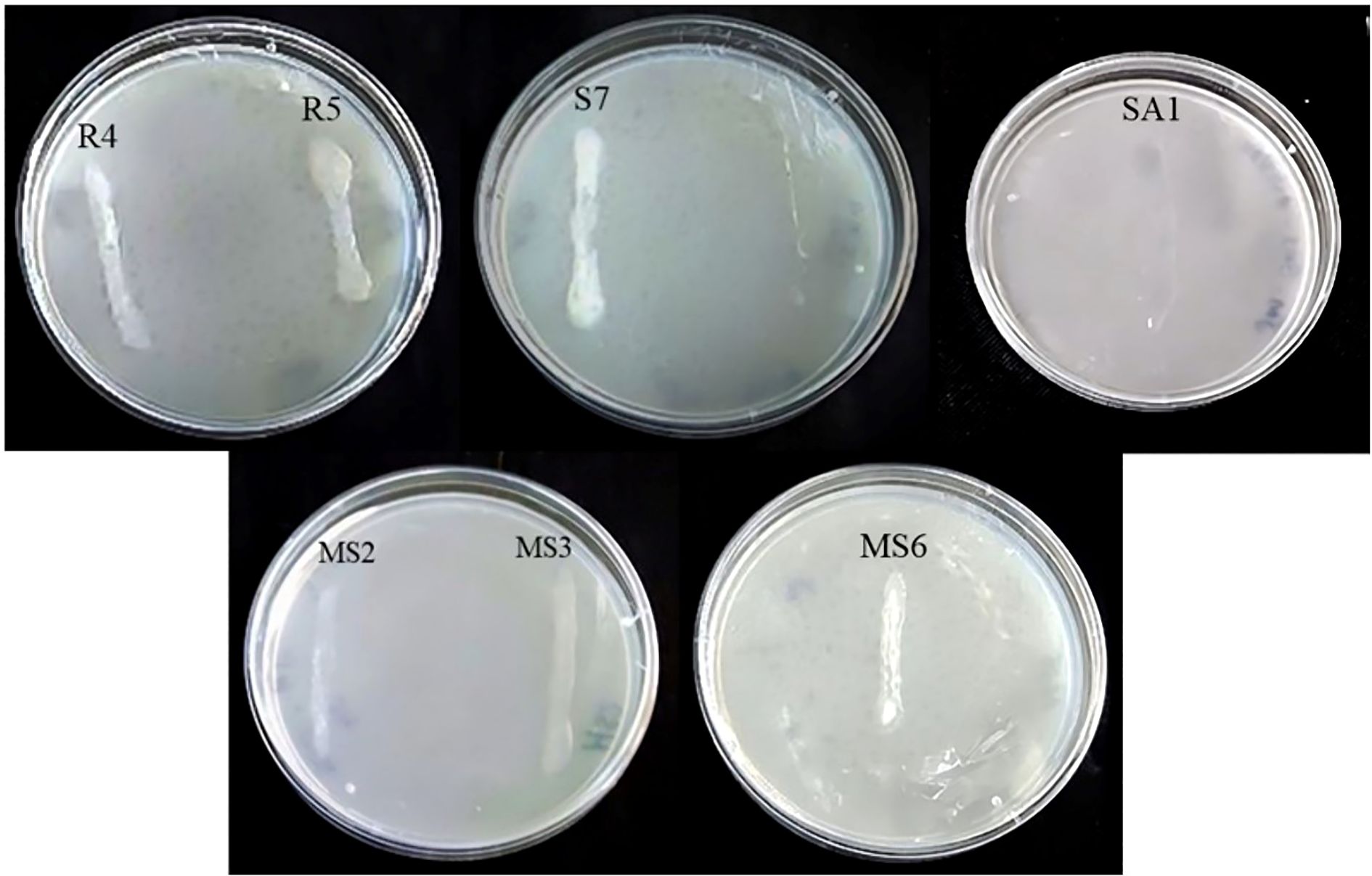
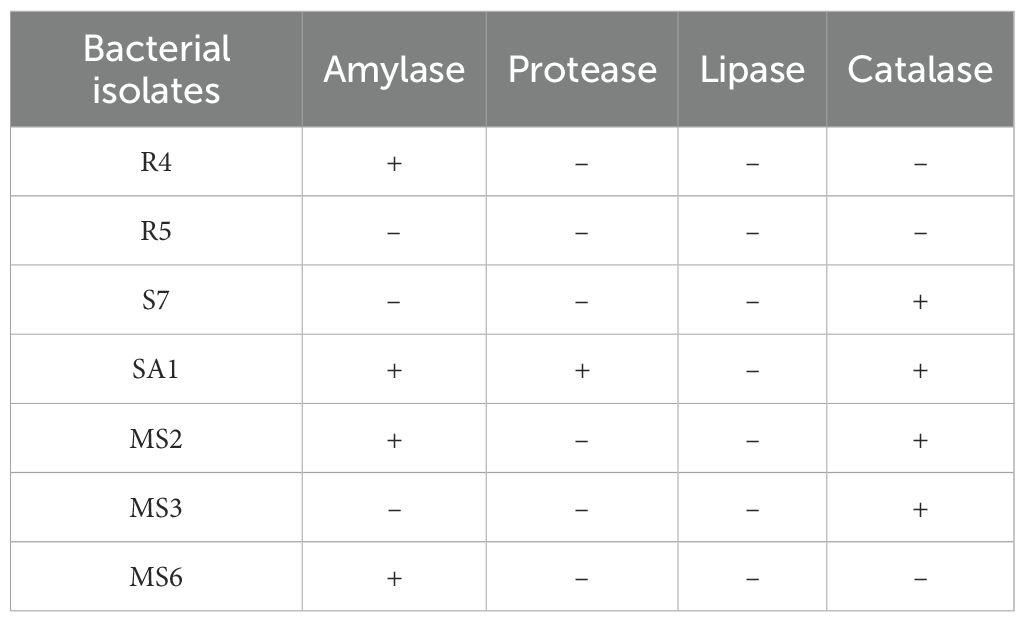
From the conducted preliminary studies, seven best isolates were selected for the enzymatic studies. In the present work, four bacterial isolates, namely R4 (E. hormaechei), SA1 (S. gallinarum), MS2 (B. amyloliquefaciens), and MS6 (K. sacchari), produced amylase (Figure 5A). While only one bacterial isolate, SA1 (S. gallinarum) exhibited positive results for protease production. The clear zone formation circling the bacterial colony on skim milk agar showed a positive result for proteolytic enzyme production (Table 2; Figure 5B). The bacterial isolates S7 (P. agglomerans), SA1 (S. gallinarum), MS2 (B. amyloliquefaciens), and MS6 (K. sacchari) exhibited positive results for catalase production (Table 2; Figure 5C). No positive results were obtained for lipase production.
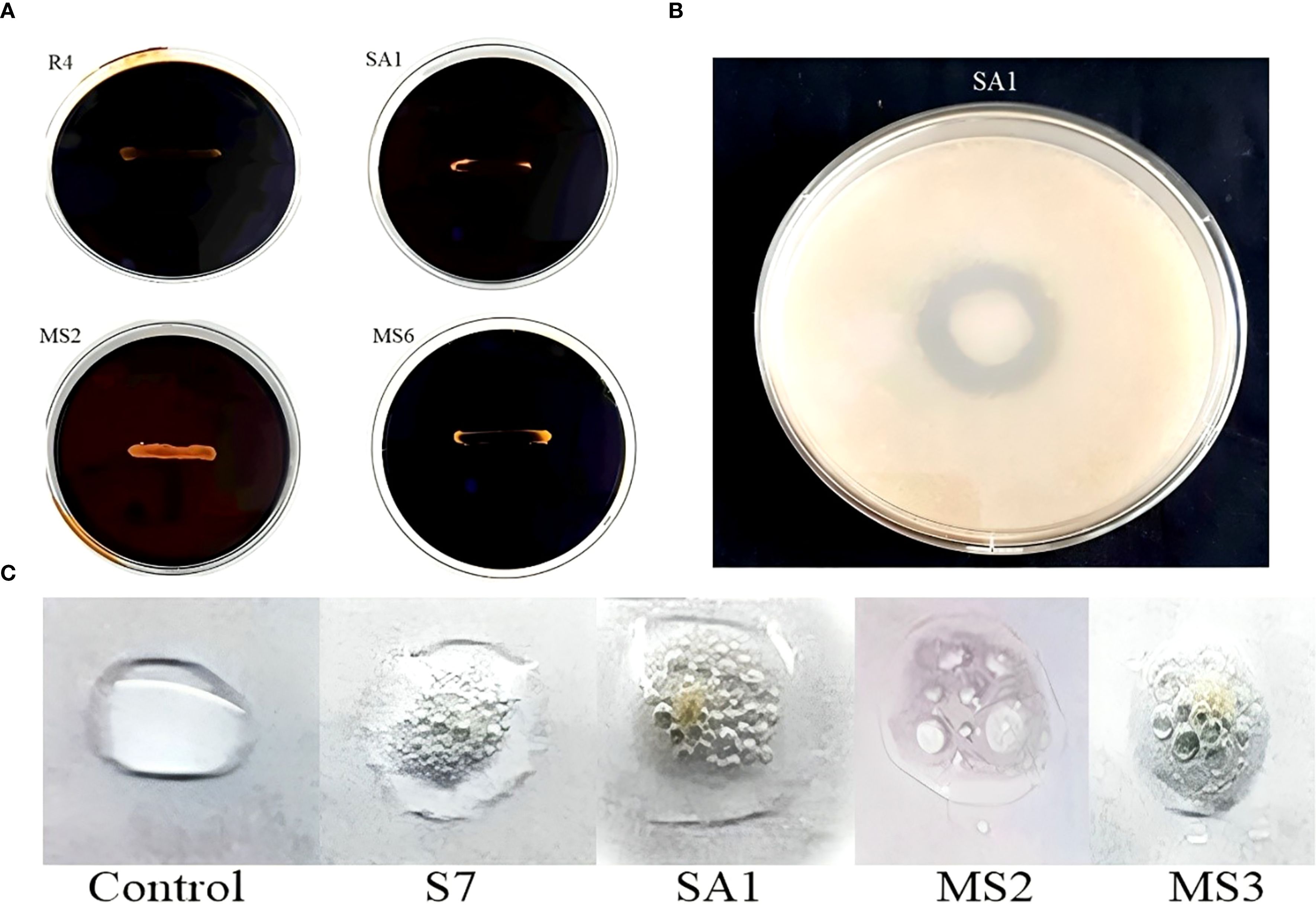
Figure 5. Enzyme activity of the isolates, confirmation of amylase (A) protease (B) and (C) catalase production.
Invitro assessment of antifungal activity
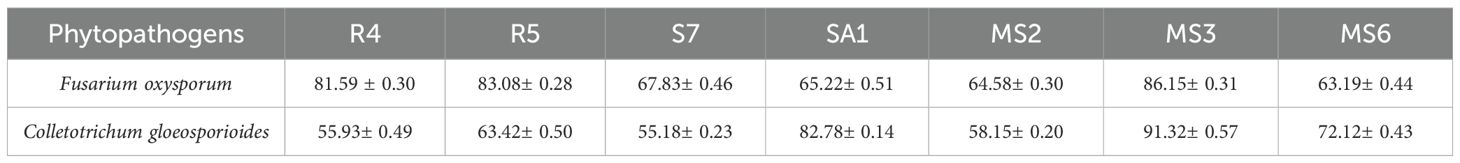
In the present study, based on the dual culture technique, all 7 bacterial isolates demonstrated the capacity to suppress the mycelial growth of the plant pathogens F.oxysporum and C.gloeosporioides, with varying degrees of efficacy. These two phytopathogens cause Fusarium wilt, Fusarium crown, and root rot (McGovern, 2015) and anthracnose (Lassois and de Bellaire, 2014) in crop plants. The antifungal activity of the bacterial strains R4, R5, S7, SA1, MS2, MS3, and MS6 exhibited a range of percent inhibition values against the pathogens, with values ranging from 63.19% to 86.15% for F. oxysporum and 55.18% to 91.32%for C. Gloeosporioides (Figures 6, 7). The highest antagonism was observed in MS3 (B. velezensis) against F. oxysporum with an inhibition rate of 86.15%, followed by R5 (B. siamensis) 83.08%, R4 (E. hormaechei) 81.59%, S7 (P. agglomerans) 67.83%, SA1 (S. gallinarum) 65.22%, MS2 (B. amyloliquefaciens) 64.58%, and MS6 (K. sacchari) 63.19%.The antagonistic activity of C.gloeosporioides was more pronounced in MS3 (B. velezensis) 91.32%, SA1 (S. gallinarum) 82.78%, MS6 (K. sacchari) 72.12%, R5 (B. siamensis) 63.42%, MS2 (B. amyloliquefaciens) 58.15%, R4 (E. hormaechei) 55.93%, and S7 (P. agglomerans) 55.18% (Table 3; Figure 8).
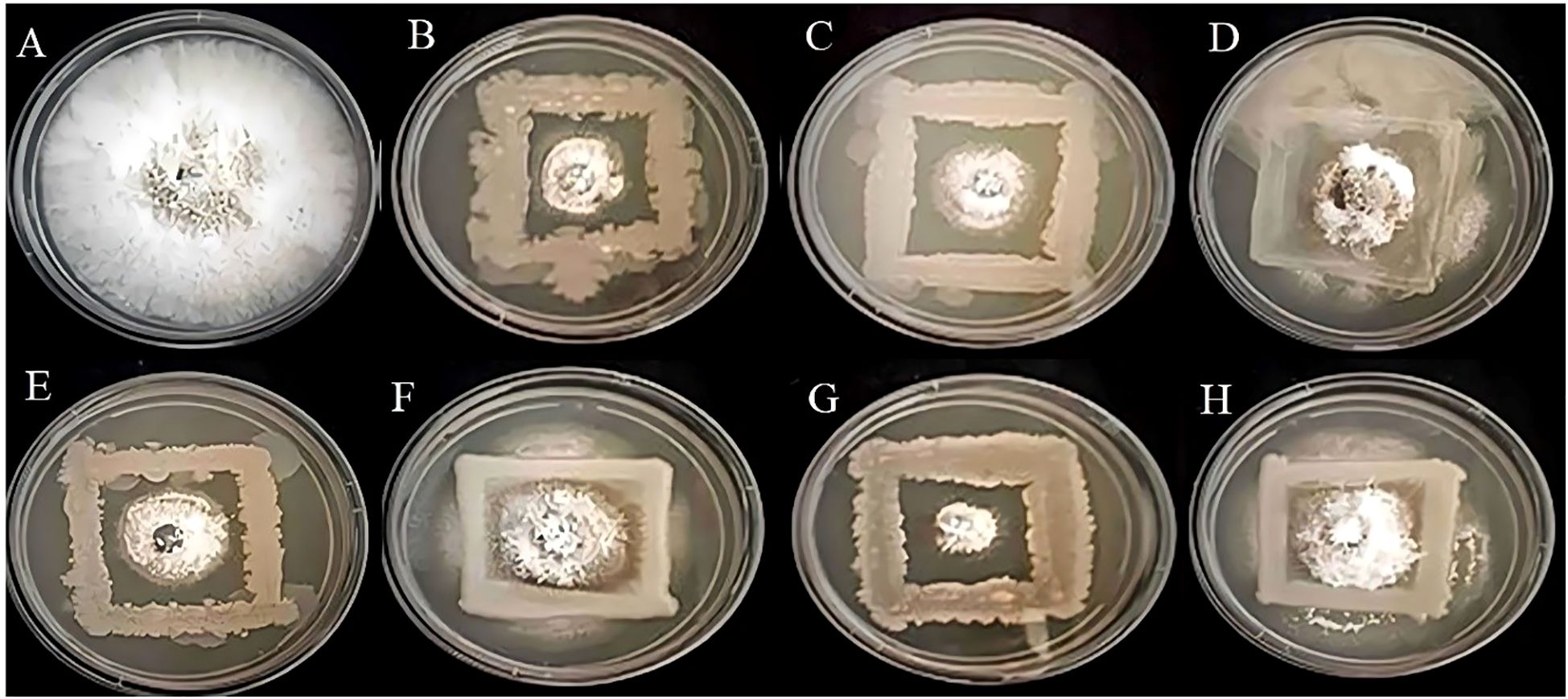
Figure 6. Antagonistic activity of Fusarium oxysporum (A) Control, (B) R4, (C) R5, (D) S7, (E) SA1, (F) MS2, (G) MS3, (H) MS6, respectively.
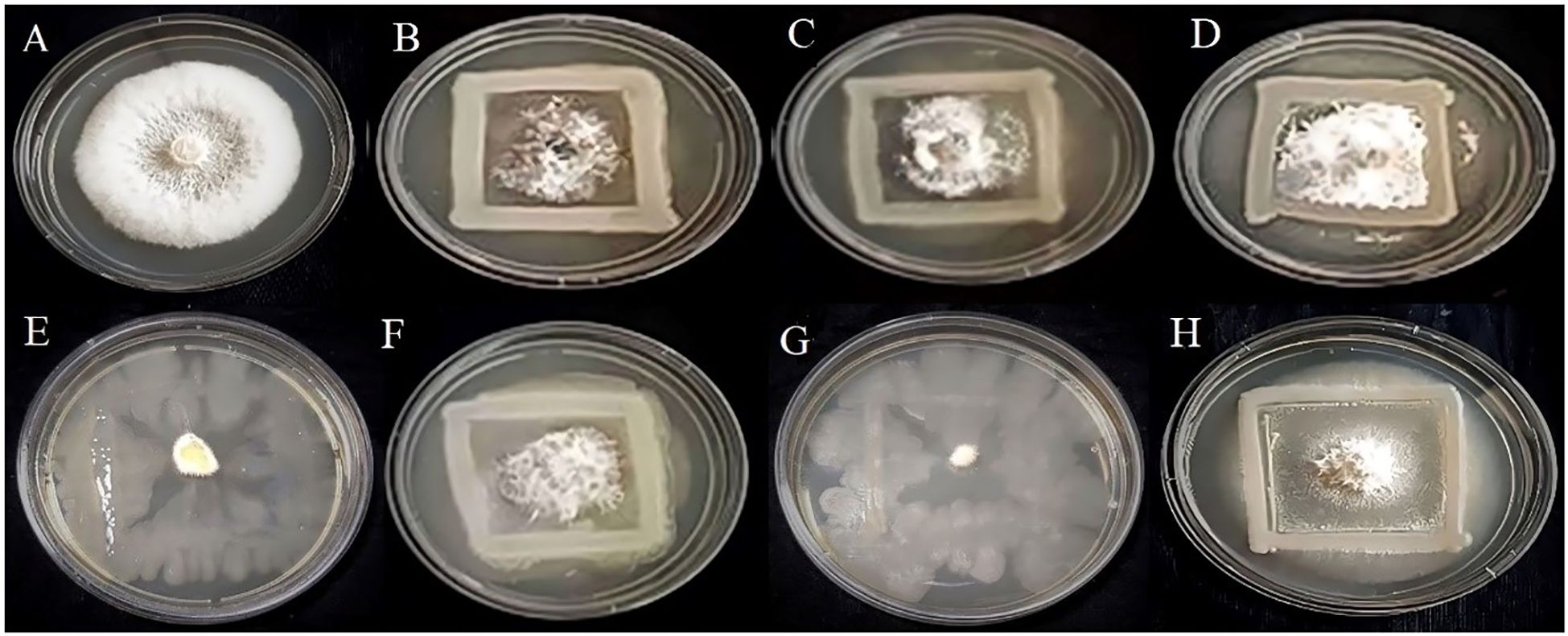
Figure 7. Antagonistic activity of Colletotrichum gloeosporioides (A) Control, (B) R4, (C) R5, (D) S7, (E) SA1, (F) MS2, (G) MS3, (H) MS6, respectively.
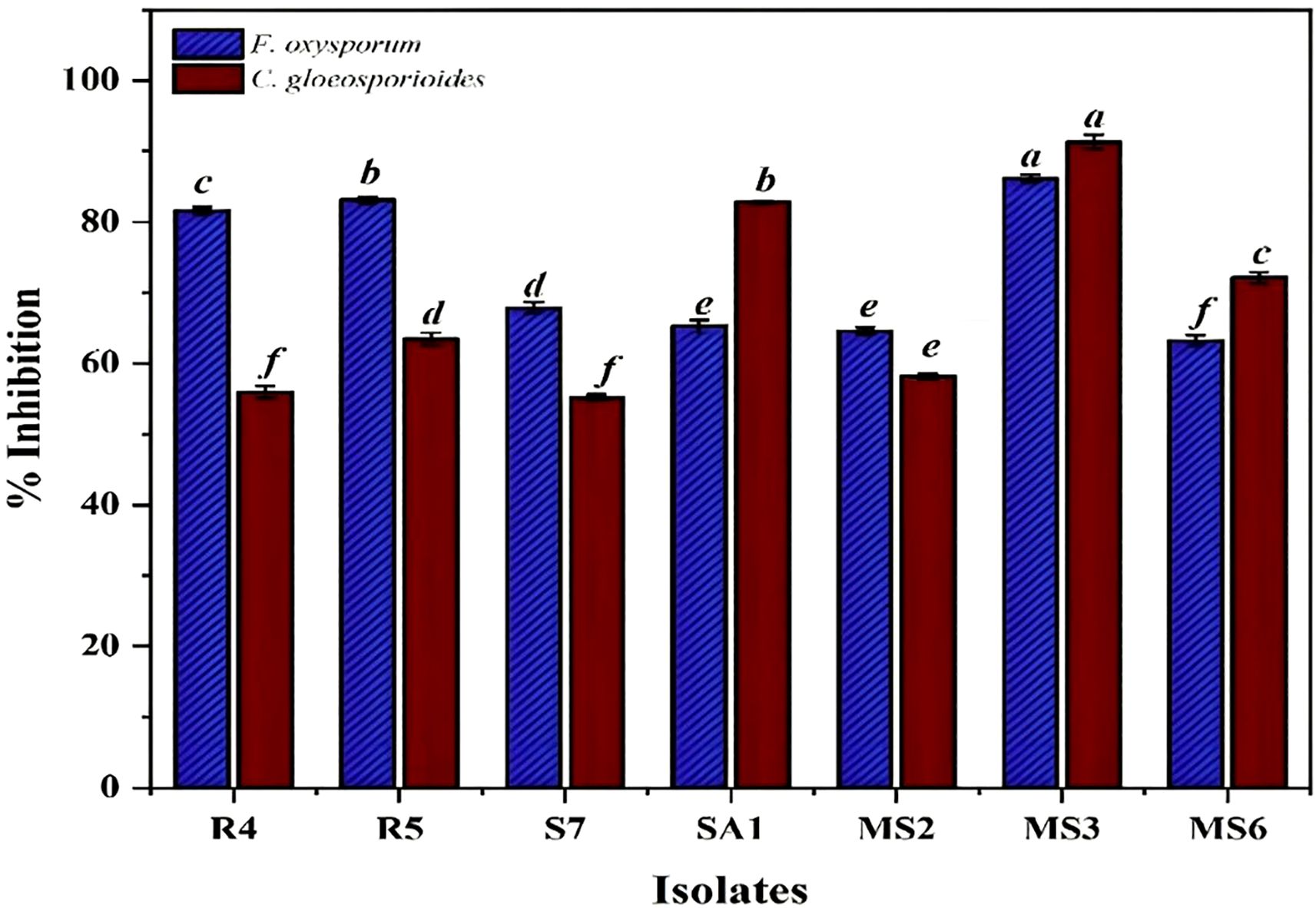
Figure 8. Invitro antagonistic activity by the seven bacterial isolates, namely R4, R5, S7, SA1, MS2, MS3, and MS6, against the phytopathogens F.oxysporum and C. gloeosporioide. Bars with different letters show significant differences from each other (p < 0.05) and the liner bar shows the standard error.
Molecular identification
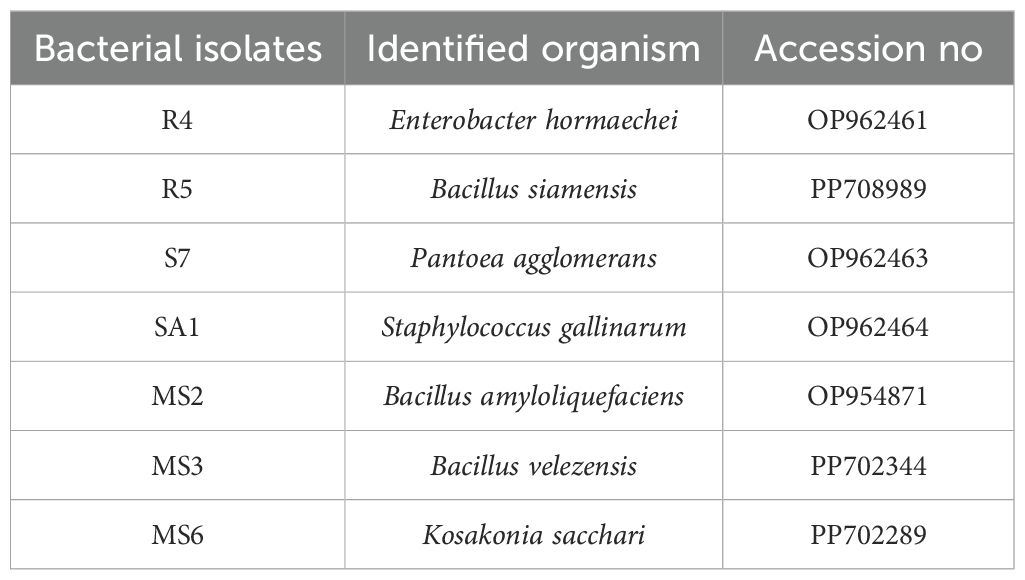
The chosen bacterial strains underwent molecular characterization through 16S rRNA sequencing (Figure 9). The resulting sequences were submitted to the NCBI GenBank BLAST (Basic Local Alignment Search Tool) Nucleotide database. Sequences were submitted to the NCBI GenBank database with the following accession numbers (Table 4): R4-Enterobacter hormaechei(OP962461), R5-Bacillus siamensis (PP708989), S7-Pantoea agglomerans (OP962463), SA1-Staphylococcus gallinarum (OP962464), MS2-Bacillus amyloliquefaciens(OP954871), MS3-Bacillus velezensis(PP702344), and MS6-K. Sacchari (PP702289).
Effect of isolates on seed germination
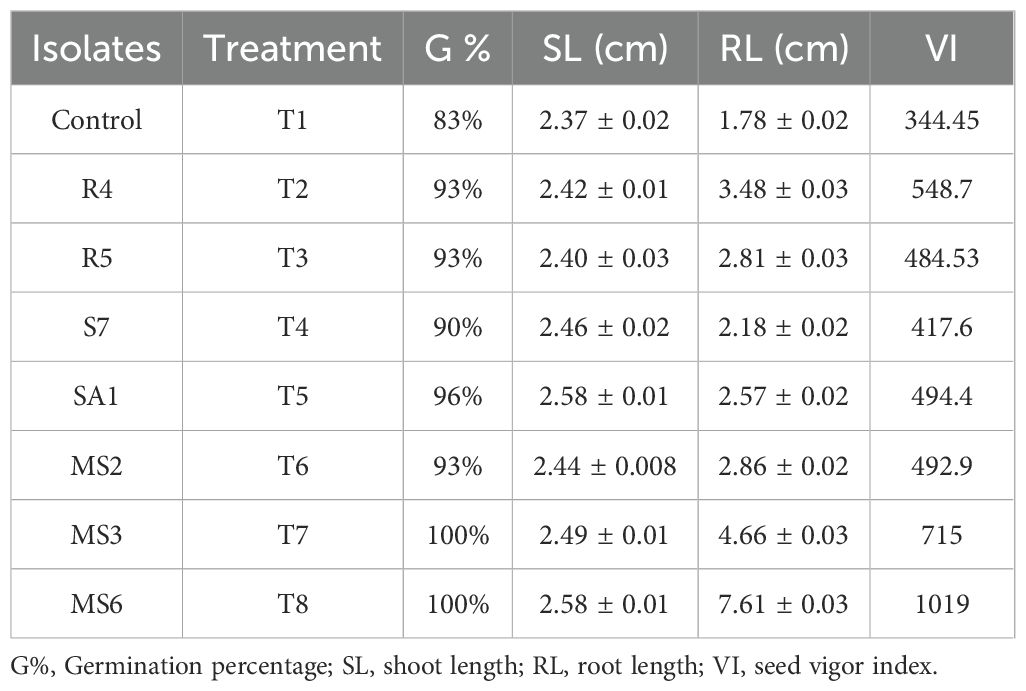
The impact of isolated bacteria on the germination of Solanum lycopersicum(tomato) seeds was noted after 10 days of sowing. The findings of the current study indicate that seeds treated with PGPB exhibited significantly enhanced seed germination, seed vigor index, root length, and shoot length of the plantlets when compared to the control (Figure 10). The highest germination percentage and seed vigor index (1019), shoot length (2.58 cm), and root length (7.61cm) were recorded in K. sacchari (MS6), which is a classified genus from the family Enterobacter (Gu et al., 2014) this Enterobacter sacchari was reclassified as K. sacchari due to its association with sugarcane as a nitrogen fixer. To date, in India, there have been only two reports published on the isolation of K. sacchari from soil samples and for its PGP activities (Giri, 2019; Shahid et al., 2021). Following this, MS3 has the seed vigor index of 715, shoot length 2.49 cm, and root length 4.66 cm and in contrast, the control exhibited a germination percentage of 83%, seed vigor index of 344.45, shoot length of 2.37 cm, and root length of 1.78 cm (Table 5; Figures 11A, B). The results of the treatment were found to be significantly greater than those of the control.
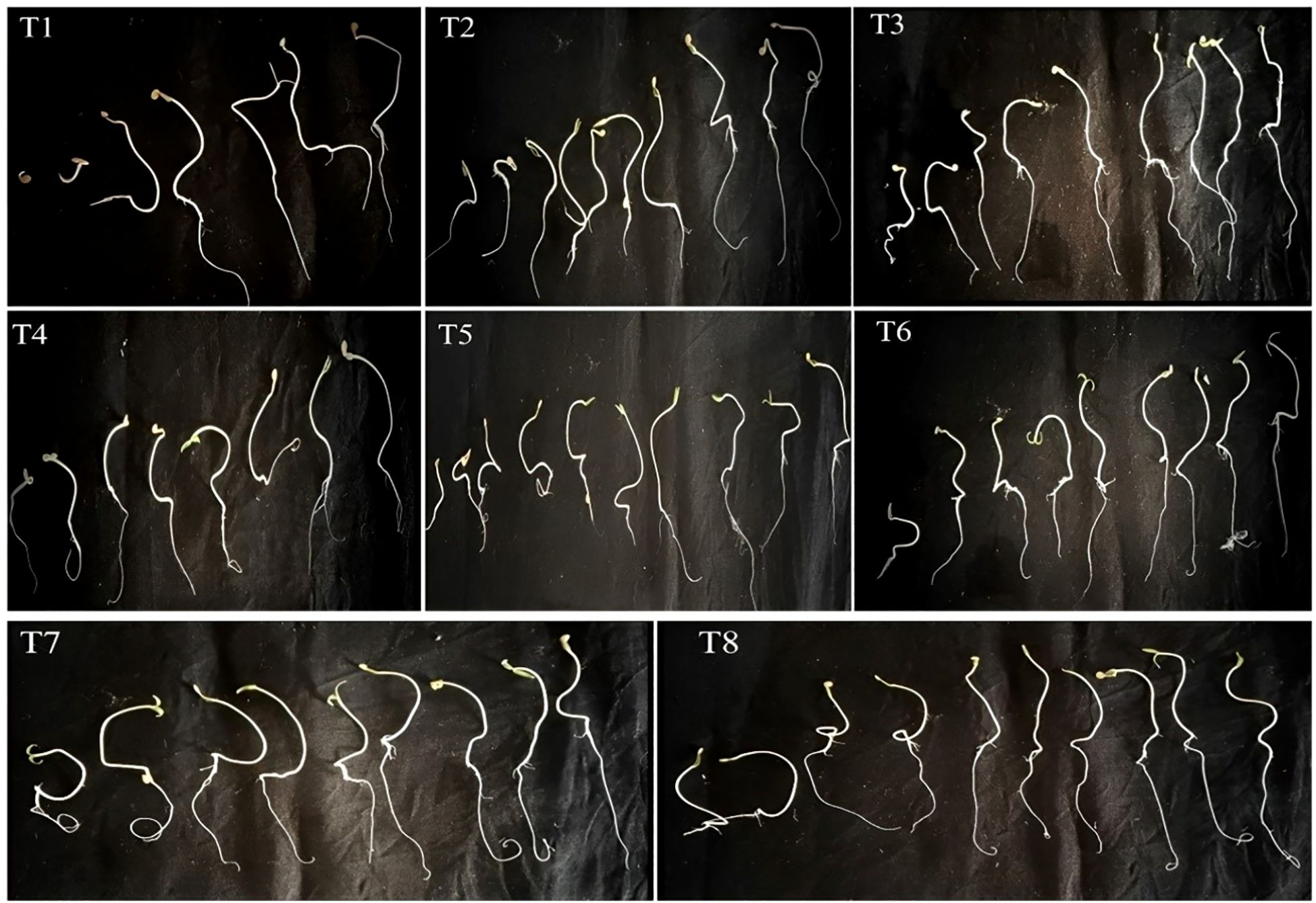
Figure 10. Ten-day-old Solanum lycopersicum treated with bacterial isolates. T1) Control, T2) Seeds treated with Enterobacter hormaechei, T3) Bacillus siamensis, T4) Pantoea agglomerans, T5) Staphylococcus gallinarum, T6) Bacillus amyloliquefaciens, T7) Bacillus velezensis, T8) Kosakonia sacchari respectively.
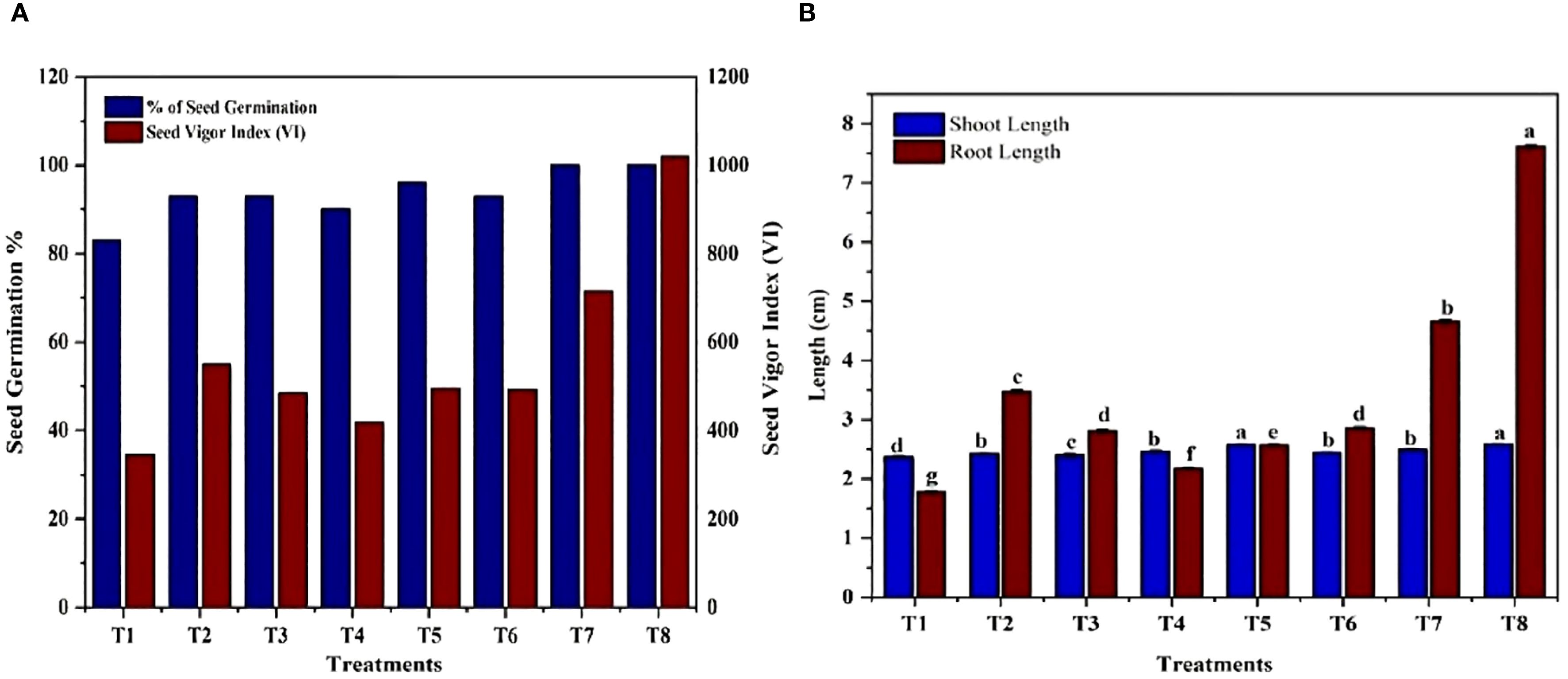
Figure 11. Impact of bacterial isolates on Solanum lycopersicum (A) seed germination percentage and seed vigor index. T1) Control, T2) Seeds treated with Enterobacter hormaechei, T3) Bacillus siamensis, T4) Pantoea agglomerans, T5) Staphylococcus gallinarum, T6) Bacillus amyloliquefaciens, T7) Bacillus velezensis, T8) Kosakonia sacchari respectively. (B) Shoot length and root length. T1) Control, T2) Seeds treated with Enterobacter hormaechei, T3) Bacillus siamensis, T4) Pantoea agglomerans, T5) Staphylococcus gallinarum, T6) Bacillus amyloliquefaciens, T7) Bacillus velezensis, T8) Kosakonia sacchari respectively.
Statistical analysis
The plant growth-promoting potential of the bacterial isolates was evaluated based on their ability to produce IAA, ammonia, antagonistic activity, and phosphate solubilization. To understand how these traits were related, Principal Component Analysis (PCA) was carried out (Figure 12). The first two principal components explained most of the variation among the isolates, with PC1 showing an eigenvalue of 59.35% and PC2 with 26.46%. Together, they accounted for 85.81% of the overall variability. PC1 was mainly influenced by IAA production and phosphate solubilization, and PC2 by ammonia production and antagonistic activity. These results suggest that different isolates promote plant growth through a distinct combination of mechanisms, with some relying more on nutrient mobilization, such as IAA production and phosphate solubilization, and the others contributing through pathogen suppression and ammonia release.
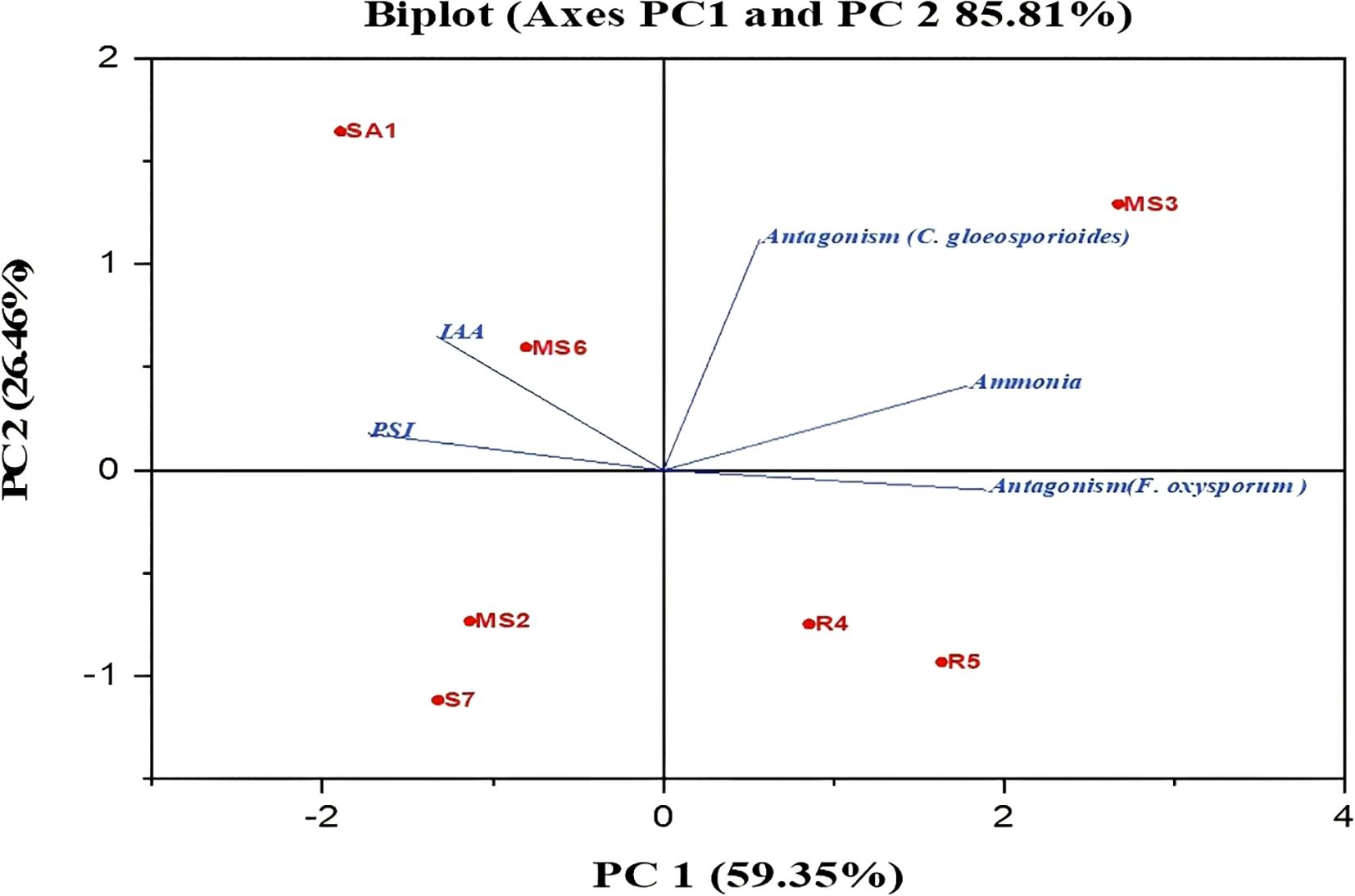
Figure 12. Principal component analysis (PCA) between the isolated bacterial stains and their PGP activity.
Discussion
In the rhizosphere, PGPRs reside on the root surface, promoting plant growth and development through both direct and indirect mechanisms (Meena et al., 2017). These mechanisms include nitrogen fixation, growth hormone production (auxins, gibberellins, cytokinins, etc.), ammonia production, inorganic phosphate solubilization, and the suppression of the growth of antagonistic microorganisms (Gerhardt et al., 2009). In the present work, 53 morphologically distinct isolates were isolated from millet rhizosphere soil using the spread plate technique. All the isolates were assessed for their nitrogen fixation ability, and 26 were found to be capable of fixing nitrogen when inoculated in a nitrogen-free medium. Nitrogen is a crucial element for the growth of the plant and yield; however, plants are unable to use nitrogen directly. The advantageous microorganisms found in the rhizosphere region are capable of converting atmospheric nitrogen into a usable form, namely ammonia, which is a major component of nucleic acids, amino acids, chlorophyll, and ATP (Andy et al., 2020). Similar results were reported by Reang et al. (2022) and Kanimozhi and Panneerselvam (2010) using the same medium, demonstrated that the isolates belonging to the genus Halomonas and Azospirillum, respectively, possess the ability to fix soil atmospheric nitrogen
It has been established that the synthesis of auxin by PGP bacteria (PGPB) represents the primary mechanism through which these bacteria can improve the growth of plants. In particular, the majority of the scientific literature focuses on IAA, the most prevalent biologically active auxin, which plays a pivotal role in plant growth by enhancing root and shoot development, elongation of roots and stems differentiation of vascular tissues, cell expansion, loosening of root cell walls, root bacterial colonization, stimulation of cell division, and defense against pathogens (Orozco-Mosqueda et al., 2023). In the present study, 14 of the 26 bacterial isolates produced IAA at varying concentrations; the highest IAA production of 0.37mg/mL was observed in SA1, and the lowest was in R5 (0.05 mg/mL). A comparable outcome was noted in the research conducted by the authors (Walpola and Arunakumara, 2016), they documented the maximum IAA production of 240 µg/mL and 332 µg/mL by Enterobacter ludwigii and Enterobacter hormaechei, respectively.
Ammonia production is a significant factor associated with the PGPB. Plants utilize the released ammonia from the PGPB as a nitrogen source, and the soil with abundant ammonia becomes alkaline, which prevents the growth of some fungi (Jha et al., 2012). In the current work, 23 bacterial isolates demonstrated positive results for ammonia production. Several reports exist on the production of ammonia in various concentrations by the genus Bacillus, particularly B. licheniformis, reported to produce nearly 4 µmol/mL (Goswami et al., 2014).
Phosphorus is a pivotal element that holds a crucial role in plant nutrition and numerous metabolic functions, including photosynthesis, respiration, energy transmission, signal transduction, and the biosynthesis of macromolecules (Anand et al., 2016). The majority of phosphorus, typically ranging from 95 to 99%, is present in an insoluble form that is not directly usable by plants. This can be achieved through the use of PGPB, which can lower the rhizosphere pH by secreting organic acids, including carboxylic acid and succinic acid. These acids bind to the phosphate group, forming calcium phosphate, thereby converting the insoluble phosphate to an available form (Sharma et al., 2013). In the present work, 15 isolates exhibited positive results with varying PSI values ranging from 2.36 to 3.80. The highest PSI was observed in P. agglomerans with a value of 3.80, and a similar result, with a 0.3 PSI difference, was reported in the research conducted by Prasad et al. (2022).
HCN is a volatile compound, a secondary metabolite, that inhibits the growth of phytopathogens (Kumar et al., 2014). Additionally, HCN production plays a pivotal role in root and shoot elongation, biomass production, and nitrogen accumulation (Marques et al., 2010). In the present work, only three isolates, namely R4 (E. hormaechei), R5 (B. siamensis), and MS3 (B. velezensis), demonstrated the capacity to produce HCN. Katiyar et al. (2017) documented the production of HCN by E. hormaechei, while Kanchiswamy et al. (2015) noted the production of HCN by the genus Bacillus.
The application of PGPB with siderophore-producing potential has been demonstrated to enhance plant growth by boosting iron intake, while simultaneously inhibiting phytopathogen growth, particularly fungi, by limiting iron availability (Kumari et al., 2018). Furthermore, the production of siderophore enables plants to take iron even in the presence of other metals like nickel and cadmium (Beneduzi et al., 2012). In the current work, 5 bacterial isolates exhibited positive results for siderophore production, namely R4 (E. hormaechei), S7 (P. agglomerans), SA1 (S. gallinarum), MS3 (B. velezensis), and MS6 (K. sacchari). The different species from the genus Enterobacter and Pseudomonas are reported by Alexander and Zuberer (1991) to produce siderophores in varying concentrations. Bhattacharyya et al. (2020) reported comparable results for the production of siderophore in varying concentrations from 99.26% to 52.31 % by various microorganisms belonging to the genus Bacillus, Arthrobacter, Micrococcus, Pseudomonas, Staphylococcus, Ochrobactrum, and Exiguobacterium. Shariati et al. (2017) reported that TonB and fepA are genes that encode for siderophore receptors in the bacteria P. agglomerans
Hydrolytic enzymes, such as protease, amylase, chitinase, lipase, and cellulase, act as defenders to protect plants from diseases by degrading the cell wall of the plant’s pathogenic microorganisms (Zerihun et al., 2019). PGPB with the potential to produce one or more of these lytic enzymes can exhibit biocontrol against plant diseases caused by harmful phytopathogenic bacteria and fungi (Rasool et al., 2021). Sritongon et al. (2023) demonstrated a comparable result, that the PGPR Bacillus subtilis produced significant production of amylase and protease. The enzyme chitinases strengthen plant defense by targeting chitin, the primary structural component of fungal cell walls, thereby inactivating pathogenic fungi without harming the host plant. In addition to enhancing resistance, these enzymes also contribute to improved plant growth and yield (Kumar et al., 2018). The genus Serratia and Halomonas are reported to produce a range of hydrolytic enzymes, cellulases, amylases, lipases, proteases, ureases, and chitinases (Ouali et al., 2024).
The antagonistic activity of the PGPB is an important feature, as it can be used as a biocontrol agent. By producing hydrolytic enzymes, siderophores, antibiotics, and inducing systemic resistance in plants, the PGPB can inhibit the growth of phytopathogens. Several studies have demonstrated the potential of PGPB isolates from the genera Bacillus and Pseudomonas, particularly B. velezensis and P. fluorescens, as effective biocontrol agents (Arkhipov et al., 2023).
The application of PGPB enhances seed germination due to the release of phytohormones, which play a crucial role in cell division (Pérez-García et al., 2023). PGPB produces phytohormones such as IAA, gibberellins, and cytokinins, which stimulate cell division and elongation during germination. Similar results for seed germination enhancement by PGPB have been reported in numerous vegetables, including Daucus carota, Allium cepa, Capsicum annuum, and Solanum lycopersicum (Lobato Ureche et al., 2021; Miljaković et al., 2022; Monalisa and Roy, 2022). The biplot PCA demonstrated a positive correlation between the IAA production and PSI. The results are comparable to those previously reported by the authors Barbaccia et al. (2022) and Sahin et al. (2004), respectively. The results demonstrated that the seven PGPBs have exhibited PGP characteristics, such as nitrogen fixation, ammonia and IAA production, phosphate solubilization, HCN production, siderophore production, hydrolytic enzyme secretion, and antagonistic activity. Further field investigations of these PGPBs will reveal their impact on PGP, the mechanism of action, and demonstrate their potential contribution to sustainable agriculture.
Conclusion
This study offers valuable insights into plant growth-promoting rhizobacteria isolated from the millet rhizosphere, highlighting their significant role in enhancing tomato seed germination. The isolates exhibited multiple beneficial PGP traits, including nitrogen fixation, IAA production, ammonia production, phosphate solubilization, HCN production, siderophore production, hydrolytic enzyme production, and antagonistic activity, reflecting their potential in supporting plant growth and development. By harnessing the native microbial diversity of the millet rhizosphere, an underexplored but rich reservoir of agriculturally beneficial microbes, this present work highlights a sustainable approach to improve crop growth and development while reducing dependence on chemical fertilizers. Further investigation of the impact of these isolates on PGP will help to uncover the molecular mechanisms underlying their plant growth-promoting effects and assess their adaptability to diverse environmental conditions. Future work will prioritize optimization of bioformulation, consortia development, and field trials to develop targeted, crop-specific microbial inoculants capable of minimizing the usage of chemical fertilizers, enhancing soil fertility, and improving sustainable agriculture practices.
Data availability statement
Sequences were submitted to the NCBI GenBank database with the following accession numbers (Table 4): R4-Enterobacter hormaechei (OP962461), R5-Bacillus siamensis (PP708989), S7-Pantoea agglomerans (OP962463), SA1-Staphylococcus gallinarum (OP962464), MS2-Bacillus amyloliquefaciens (OP954871), MS3-Bacillus velezensis (PP702344), and MS6-K. Sacchari (PP702289).
Author contributions
AS: Conceptualization, Methodology, Writing – original draft, Writing – review & editing, Data curation, Formal analysis. SB: Data curation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing, Investigation, Validation. JS: Validation, Writing – original draft, Writing – review & editing. SG: Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. RM: Conceptualization, Funding acquisition, Investigation, Supervision, Validation, Writing – original draft, Writing – review & editing. SWC: Writing – original draft, Writing – review & editing. BR: Conceptualization, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. SC: Writing – original draft, Writing – review & editing. KG: Conceptualization, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The authors are highly thankful to Bharathiar University, Coimbatore for providing University Research Fellowship-URF. We are thankful for the support provided by UCSI University for the support provided by the Center of Excellence for Research, Value Innovation and Entrepreneurship (CERVIE) and Research Excellence and Innovation Grant (REIG) with code REIG-FPS-2025/038.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alexander, D. and Zuberer, D. (1991). Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertility Soils. 12, 39–45. doi: 10.1007/BF00369386
Anand, K., Kumari, B., and Mallick, M. A. (2016). Phosphate solubilizing microbes: An effective and alternative approach as biofertilizers. Int. J. Pharm. Pharm. Sci. 8, 37–40.
Andy, A. K., Masih, S. A., and Gour, V. S. (2020). Isolation, screening and characterization of plant growth promoting rhizobacteria from rhizospheric soils of selected pulses. Biocatal Agric. Biotechnol. 27. doi: 10.1016/j.bcab.2020.101685
Annor, G. A., Marcone, M., Bertoft, E., and Seetharaman, K. (2014). Physical and molecular characterization of millet starches. Cereal Chem. 91, 286–292. doi: 10.1094/CCHEM-08-13-0155-R
Arkhipov, A., Carvalhais, L. C., and Schenk, ,. P. M. (2023). PGPR control Phytophthora capsici in tomato through induced systemic resistance, early hypersensitive response and direct antagonism in a cultivar-specific manner. Eur. J. Plant Pathol. 167, 811–832. doi: 10.1007/s10658-023-02734-8
Barbaccia, P., Gaglio, R., Dazzi, C., Miceli, C., Bella, P., Lo Papa, G., et al. (2022). Plant growth-promoting activities of bacteria isolated from an anthropogenic soil located in Agrigento Province. Microorganisms 10. doi: 10.3390/microorganisms10112167
Barea, J. M., Pozo, M. J., Azcón, R., and Azcón-Aguilar, C. (2005). Microbial co-operation in the rhizosphere. J. Exp. Bot. 56(417), 1761–1778. doi: 10.1093/jxb/eri197
Belton, P. S. and Taylor, J. R. N. (2004). Sorghum and millets: Protein sources for Africa. Trends Food Sci. Technol. 15, 94–98. doi: 10.1016/j.tifs.2003.09.002
Beneduzi, A., Ambrosini, A., and Passaglia, L. M. P. (2012). Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051. doi: 10.1590/s1415-47572012000600020
Berg, G. (2009). Plant-microbe interactions promoting plant growth and health: Perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18. doi: 10.1007/s00253-009-2092-7
Bhattacharyya, C., Banerjee, S., Acharya, U., Mitra, A., Mallick, I., Haldar, A., et al. (2020). Evaluation of plant growth promotion properties and induction of antioxidative defense mechanism by tea rhizobacteria of Darjeeling, India. Sci. Rep. 10. doi: 10.1038/s41598-020-72439-z
Cappucino, J. C. and Sherman, N., (1992). “Microbiology: A Laboratory Manual,” 3rd Edition, Benjamin/Cumming Pub. Co., New York
Castric, P. A. (1975). Hydrogen cyanide, a secondary metabolite of Pseudomonas aeruginosa. Canadian Journal of Microbiology, 21(5), 613–618.
Esitken, A., Pirlak, L., Turan, M., and Sahin, F. (2006). Effects of floral and foliar application of plant growth promoting rhizobacteria (PGPR) on yield, growth and nutrition of sweet cherry. Sci. Hortic. 110, 324–327. doi: 10.1016/j.scienta.2006.07.023
Ezrari, S., Mhidra, O., Radouane, N., Tahiri, A., Polizzi, G., Lazraq, A., et al. (2021). Potential role of rhizobacteria isolated from citrus rhizosphere for biological control of citrus dry root rot. Plants 10, 872. doi: 10.3390/plants10050872
Fahsi, N., Mahdi, I., Mesfioui, A., Biskri, L., and Allaoui, A. (2021). Plant growth-promoting rhizobacteria isolated from the jujube (Ziziphus lotus) plant enhance wheat growth, zn uptake, and heavy metal tolerance. Agric. (Switzerland) 11. doi: 10.3390/agriculture11040316
Gerhardt, K. E., Huang, X. D., Glick, B. R., and Greenberg, B. M. (2009). Phytoremediation and rhizoremediation of organic soil contaminants: Potential and challenges. Plant Sci. 176, 20–30. doi: 10.1016/j.plantsci.2008.09.014
Giri, K. (2019). The first report of indigenous free-living diazotroph kosakonia sacchari isolated from himalayan alder-based shifting cultivation system in Nagaland, India. J. Soil Sci. Plant Nutr. 19, 574–579. doi: 10.1007/s42729-019-00056-5
Goswami, M. and Deka, S. (2020). Isolation of a novel rhizobacteria having multiple plant growth promoting traits and antifungal activity against certain phytopathogens. Microbiol. Res. 240. doi: 10.1016/j.micres.2020.126516
Goswami, D., Dhandhukia, P., Patel, P., and Thakker, J. N. (2014). Screening of PGPR from saline desert of Kutch: Growth promotion in Arachis hypogea by Bacillus licheniformis A2. Microbiol. Res. 169, 66–75. doi: 10.1016/j.micres.2013.07.004
Gu, C. T., Li, C. Y., Yang, L. J., and Huo, G. C. (2014). Enterobacter xiangfangensis sp. nov., isolated from Chinese traditional sourdough, and reclassification of Enterobacter sacchari Zhu et al. 2013 as Kosakonia sacchari comb. nov. Int. J. Syst. Evol. Microbiol. 64, 2650–2656. doi: 10.1099/ijs.0.064709-0
Han, Y., Xu, L., Liu, L., Yi, M., Guo, E., Zhang, A., et al. (2017). Illumina sequencing reveals a rhizosphere bacterial community associated with foxtail millet smut disease suppression. Plant Soil 410, 411–421. doi: 10.1007/s11104-016-3042-7
Huang, X. F., Chaparro, J. M., Reardon, K. F., Zhang, R., Shen, Q., and Vivanco, J. M. (2014). Rhizosphere interactions: Root exudates, microbes, and microbial communities1. Botany 92, 267–275. doi: 10.1139/cjb-2013-0225
Jha, B., Gontia, I., and Hartmann, A. (2012). The roots of the halophyte Salicornia brachiata are a source of new halotolerant diazotrophic bacteria with plant growth-promoting potential. Plant Soil 356, 265–277. doi: 10.1007/s11104-011-0877-9
Kanchiswamy, C. N., Malnoy, M., and Maffei, M. E. (2015). Chemical diversity of microbial volatiles and their potential for plant growth and productivity. Front. Plant Sci. 6, 151. doi: 10.3389/fpls.2015.00151
Kanimozhi, K. and Panneerselvam, A. (2010). Studies on isolation and nitrogen fixation ability of Azospirillum spp. isolated from Thanjavur district. Der Chem. Sin. 1, 138–145.
Katiyar, D., Hemantaranjan, A., Singh, B., and Malakar, A. (2017). Isolation and Characterization of Plant Growth Promoting Rhizobacteria Enterobacter hormaechei and their Suppression Efficacy against Colletotrichum falcatum in Combination with Chitosan. Int. J. Plant Soil Sci. 14, 1–12. doi: 10.9734/ijpss/2017/31549
Kong, Z. and Liu, H. (2022). Modification of rhizosphere microbial communities: a possible mechanism of plant growth promoting rhizobacteria enhancing plant growth and fitness. Front. Plant Sci. 13, 920813. doi: 10.3389/fpls.2022.920813
Kumar, M., Brar, A., Yadav, M., Chawade, A., Vivekanand, V., and Pareek, N. (2018). Chitinases—potential candidates for enhanced plant resistance towards fungal pathogens. Agriculture 8, 88. doi: 10.3390/agriculture8070088
Kumar, A., Maurya, B. R., and Raghuwanshi, R. (2014). Isolation and characterization of PGPR and their effect on growth, yield and nutrient content in wheat (Triticum aestivum L.). Biocatal Agric. Biotechnol. 3, 121–128. doi: 10.1016/j.bcab.2014.08.003
Kumari, P., Meena, M., and Upadhyay, R. S. (2018). Characterization of plant growth promoting rhizobacteria (PGPR) isolated from the rhizosphere of Vigna radiata (mung bean). Biocatal Agric. Biotechnol. 16, 155–162. doi: 10.1016/j.bcab.2018.07.029
Lassois, L. and de Bellaire, L.D.L. (2014). “Crown rot disease of bananas,” in Postharvest Decay: Control Strategies, Elsevier. 103–130. doi: 10.1016/B978-0-12-411552-1.00003-X
Lobato Ureche, M. A., Pérez-Rodriguez, M. M., Ortiz, R., Monasterio, R. P., and Cohen, A. C. (2021). Rhizobacteria improve the germination and modify the phenolic compound profile of pepper (Capsicum annum L.). Rhizosphere 18. doi: 10.1016/j.rhisph.2021.100334
Mahmoud, A. M., Reyad, A. M., Khalaf, M. H., Sheteiwy, M. S., Dawood, M. F. A., El-Sawah, A. M., et al. (2024). Investigating the endophyte Actinomycota sp. JW0824 strain as a potential bioinoculant to enhance the yield, nutritive value, and chemical composition of different cultivars of anise (Pimpinella anisum L.) seeds. Biology 13, 553. doi: 10.3390/biology13080553
Mahmoud, A. M., Sheteiwy, M. S., El-Keblawy, A., Ulhassan, Z., Khalaf, M. H., Mohamed, H. S., et al. (2025). The potential biofortification role of Actinopolyspora sp. JTT-01 in enhancing the yield and tissue chemical composition of caraway plants. BMC Plant Biol. 25, 540. doi: 10.1186/s12870-025-06137-1
Marques, A. P. G. C., Pires, C., Moreira, H., Rangel, A. O. S. S., and Castro, P. M. L. (2010). Assessment of the plant growth promotion abilities of six bacterial isolates using Zea mays as indicator plant. Soil Biol. Biochem. 42, 1229–1235. doi: 10.1016/j.soilbio.2010.04.014
McGovern, R. J. (2015). Management of tomato diseases caused by Fusarium oxysporum. Crop Prot. 73, 78–92. doi: 10.1016/j.cropro.2015.02.021
Meena, M., Swapnil, P., Zehra, A., Dubey, M. K., and Upadhyay, R. S. (2017). Antagonistic assessment of Trichoderma spp. by producing volatile and non-volatile compounds against different fungal pathogens. Arch. Phytopathol. Plant Prot. 50, 629–648. doi: 10.1080/03235408.2017.1357360
Mehta, S. and Nautiyal, C. S. (2001). An efficient method for qualitative screening of phosphate-solubilizing bacteria. Curr. Microbiol. 43, 51–56. doi: 10.1007/s002840010259
Miljaković, D., Marinković, J., Tamindžić, G., Đorđević, V., Tintor, B., Milošević, D., et al. (2022). Bio-Priming of Soybean with Bradyrhizobium japonicum and Bacillus megaterium: Strategy to Improve Seed Germination and the Initial Seedling Growth. Plants 11. doi: 10.3390/plants11151927
Monalisa, S. P. and Roy, S. (2022). Research Trends in Agriculture Sciences (New Delhi: AkiNik Publications). doi: 10.22271/ed.book.1829
Murphy, J. F., Reddy, M. S., Ryu, C. M., Kloepper, J. W., and Li, R. (2003). Rhizobacteria-mediated growth promotion of tomato leads to protection against Cucumber mosaic virus. Phytopathology 93, 1301–1307. doi: 10.1094/PHYTO.2003.93.10.1301
Nader, A. A., Hauka, F. I., Afify, A. H., and El-Sawah, A. M. (2024). Drought-tolerant bacteria and arbuscular mycorrhizal fungi mitigate the detrimental effects of drought stress induced by withholding irrigation at critical growth stages of soybean (Glycine max, L.). Microorganisms 12, 1123. doi: 10.3390/microorganisms12061123
Orozco-Mosqueda, M. D. C., Santoyo, G., and Glick, B. R. (2023). Recent advances in the bacterial phytohormone modulation of plant growth. Plants 12, 606. doi: 10.3390/plants12030606
Ouali, K.O., Houali, K., Cruz, C., Melo, J., Benakli, Y., Ousmer, L., et al. (2024). Halophilic plant growth-promoting rhizobacteria as producers of antifungal metabolites under salt stress. Agronomy 14, 845. doi: 10.3390/agronomy14040845
Pandey, S. and Gupta, S. (2020). Evaluation of Pseudomonas sp. for its multifarious plant growth promoting potential and its ability to alleviate biotic and abiotic stress in tomato (Solanum lycopersicum) plants. Sci. Rep. 10. doi: 10.1038/s41598-020-77850-0
Pérez-García, L. A., Sáenz-Mata, J., Fortis-Hernández, M., Navarro-Muñoz, C. E., Palacio-Rodríguez, R., and Preciado-Rangel, P. (2023). Plant-growth-promoting rhizobacteria improve germination and bioactive compounds in cucumber seedlings. Agronomy 13, 315. doi: 10.3390/agronomy13020315
Pikovskaya, R. (1948). Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 17, 362–370.
Prasad, P., Kalam, S., Sharma, N. K., Podile, A. R., and Das, S. N. (2022). Phosphate solubilization and plant growth promotion by two Pantoea strains isolated from the flowers of Hedychium coronarium L. Front. Agron. 4. doi: 10.3389/fagro.2022.990869
Rasool, A., Imran Mir, M., Zulfajri, M., Hanafiah, M. M., Azeem Unnisa, S., and Mahboob, M. (2021). Plant growth promoting and antifungal asset of indigenous rhizobacteria secluded from saffron (Crocus sativus L.) rhizosphere. Microb. Pathog. 150. doi: 10.1016/j.micpath.2021.104734
Reang, L., Bhatt, S., Tomar, R. S., Joshi, K., Padhiyar, S., Vyas, U. M., et al. (2022). Plant growth promoting characteristics of halophilic and halotolerant bacteria isolated from coastal regions of Saurashtra Gujarat. Sci. Rep. 12, 4699. doi: 10.1038/s41598-022-08151-x
Şahin, F., Çakmakçi, R., and Kantar, F. (2004). Sugar beet and barley yields in relation to inoculation with N 2-fixing and phosphate solubilizing bacteria. Plant Soil 265, 123–129. doi: 10.1007/s11104-005-0334-8
Sachan, S., Iqbal, M. S., and Singh, A. (2018). Extracellular lipase from Pseudomonas aeruginosa JCM5962(T): Isolation, identification, and characterization. Int. Microbiol. 21, 197–205. doi: 10.1007/s10123-018-0016-z
Saxena, R., Vanga, S. K., Wang, J., Orsat, V., and Raghavan, V. (2018). Millets for food security in the context of climate change: A review. Sustainability 10. doi: 10.3390/su10072228
Selvakumar, G., Panneerselvam, P., and Ganeshamurthy, A. N. (2012). “Bacterial mediated alleviation of abiotic stress in crops,” in Bacteria in Agrobiology: Stress Management (Springer-Verlag, Berlin Heidelberg), 205–224. doi: 10.1007/978-3-642-23465-1_10
Shahid, M., Ameen, F., Maheshwari, H. S., Ahmed, B., AlNadhari, S., and Khan, M. S. (2021). Colonization of Vigna radiata by a halotolerant bacterium Kosakonia sacchari improves the ionic balance, stressor metabolites, antioxidant status and yield under NaCl stress. Appl. Soil Ecol. 158. doi: 10.1016/j.apsoil.2020.103809
Shariati, V. J., Malboobi, M. A., Tabrizi, Z., Tavakol, E., Owilia, P., and Safari, M. (2017). Comprehensive genomic analysis of a plant growth-promoting rhizobacterium Pantoea agglomerans strain P5 /631/208/212/748 /631/208/464 /38/43 /45 article. Sci. Rep. 7. doi: 10.1038/s41598-017-15820-9
Sharma, K. K. and Ortiz, R. (2000). Program for the application of genetic transformation for crop improvement in the semi-arid tropics. In Vitro Cell. Dev. Biology-Plant 36, 83–92. doi: 10.1007/s11627-000-0019-1
Sharma, S. B., Sayyed, R. Z., Trivedi, M. H., and Gobi, T. A. (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus 2, 587. doi: 10.1186/2193-1801-2-587
Sheteiwy, M. S., Ahmed, M., Kobae, Y., Basit, F., Holford, P., Yang, H., et al. (2023). The effects of microbial fertilizers application on growth, yield and some biochemical changes in the leaves and seeds of guar (Cyamopsis tetragonoloba L.). Food Res. Int. 172, 113122. doi: 10.1016/j.foodres.2023.113122
Sritongon, N., Boonlue, S., Mongkolthanaruk, W., Jogloy, S., and Riddech, N. (2023). The combination of multiple plant growth promotion and hydrolytic enzyme producing rhizobacteria and their effect on Jerusalem artichoke growth improvement. Sci. Rep. 13, 5917. doi: 10.1038/s41598-023-33099-x
Walpola, B. C. and Arunakumara, K. K. I. U. (2016). Assessment of phosphate solubilization and indole acetic acid production in plant growth promoting bacteria isolated from green house soils of Gonju-Gun, South Korea. Trop. Agric. Res. Extension 18. doi: 10.4038/tare.v18i1.5322
Wang, Y., Narayanan, M., Shi, X., Chen, X., Li, Z., Natarajan, D., et al. (2022). Plant growth-promoting bacteria in metal-contaminated soil: Current perspectives on remediation mechanisms. Front. Microbiol. 13, 966226. doi: 10.3389/fmicb.2022.966226
Zerihun, T., Birhanu, G., Genene, T., Adey, F., and Solomon, C. (2019). Isolation and biochemical characterization of Plant Growth Promoting (PGP) bacteria colonizing the rhizosphere of Tef crop during the seedling stage. J. Plant Sci. Phytopathol. 3, 013–027. doi: 10.29328/journal.jpsp.1001027
Keywords: antagonistic activity, millet, PGPR, plant growth promotion, rhizosphere
Citation: Sakthivel A, Santhanalakshmi B, Sahayarayan JJ, Sivanandhan G, Mani RR, Chang SW, Ravindran B, Chokkakula S and Kapildev G (2025) Exploring the rhizospheric bacterial community of selected millets for plant growth promotion activity in tomato. Front. Plant Sci. 16:1629184. doi: 10.3389/fpls.2025.1629184
Received: 15 May 2025; Accepted: 01 September 2025;
Published: 26 September 2025.
Edited by:
Yongjun Tan, Hunan Hybrid Rice Research Center, ChinaCopyright © 2025 Sakthivel, Santhanalakshmi, Sahayarayan, Sivanandhan, Mani, Chang, Ravindran, Chokkakula and Kapildev. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gnanajothi Kapildev, ZHJna2RldjE2QGJ1Yy5lZHUuaW4=; Ravishankar Ram Mani, UmF2aXNoYW5rYXJAdWNzaXVuaXZlcnNpdHkuZWR1Lm15; Balasubramani Ravindran, S2FsYW1yYXZpQGdtYWlsLmNvbQ==
 Anitha Sakthivel
Anitha Sakthivel Balasubramanian Santhanalakshmi
Balasubramanian Santhanalakshmi Jesudass Joseph Sahayarayan
Jesudass Joseph Sahayarayan Ganeshan Sivanandhan
Ganeshan Sivanandhan Ravishankar Ram Mani
Ravishankar Ram Mani Soon Woong Chang
Soon Woong Chang Balasubramani Ravindran
Balasubramani Ravindran Santosh Chokkakula
Santosh Chokkakula Gnanajothi Kapildev
Gnanajothi Kapildev