- 1Department of Agricultural, Forest and Food Sciences (DISAFA) – Plant Genetics, University of Turin, Turin, Italy
- 2Department of Agricultural, Environmental, Food and Forestry Science and Technology (DAGRI), University of Florence, Florence, Italy
- 3Interdepartmental Service Centre for Agricultural Chemical and Industrial Biotechnologies (CIBIACI), University of Florence, Florence, Italy
Introduction: Traditional crop varieties are vital for conserving agrobiodiversity, ensuring food security, and preserving cultural heritage. However, Italian potato (Solanum tuberosum) landraces, shaped by centuries of farmer selection and adaptation to diverse agroecological conditions, are increasingly threatened by genetic erosion and agricultural homogenization.
Methods: To support their conservation, we collected 61 accessions from 22 traditional Italian landraces, documented their traditional culinary uses, and evaluated tuber skin and pulp color. Virus presence was assessed and sanitation protocols were applied. Genetic diversity was analyzed using genotyping-by-sequencing (GBS), and comparisons were made with a global reference panel of 106 genotypes.
Results: GBS data revealed marked genetic differentiation among the Italian landraces, with three main genetic clusters showing internal structure and minimal overlap with commercial varieties. The Italian germplasm was clearly distinct from worldwide accessions, underlining its unique genetic identity.
Discussion: The observed diversity highlights the potential of traditional landraces as a valuable resource for breeding programs aimed at enhancing resilience to climate change. This integrated approach fosters both agrobiodiversity conservation and the socio-economic development of marginal areas, offering a model for safeguarding traditional crop diversity worldwide.
1 Introduction
The conservation and valorization of traditional potato varieties (Solanum tuberosum L.) are essential not only for their historical and cultural significance but also for their contributions to agricultural sustainability and biodiversity (Brush, 1995; Tobin et al., 2018). Italy, renowned for its diverse climates and rich agricultural traditions, has long served as a repository of unique plant genetic resources (Vaccino et al., 2024). The introduction of potatoes to Europe after the Columbian Exchange brought both challenges and opportunities, influencing local agricultural practices. Initial introductions of Solanum tuberosum andigena, adapted to short-day conditions and high-altitude environments in the Andes, faced limited success in most of Europe due to unfavorable photoperiods and climate conditions. It was only with the arrival of Solanum tuberosum, better suited to European latitudes, that potatoes became a staple crop, particularly in northern regions.
Over time, intensive agricultural practices and the pursuit of higher yields have led to the widespread adoption of uniform, high-yielding cultivars, often at the expense of traditional landraces (Grun, 1990; Solomon-Blackburn and Barker, 2001; Spanoghe et al., 2022). These local varieties possess adaptability to local extreme conditions and distinctive organoleptic qualities, offering valuable traits that modern cultivars lack. The loss of these local varieties has contributed to a narrowing of the genetic base, which poses significant risks to food security and crop adaptability in changing environments (Wang et al., 2019; Jo et al., 2022; Salgotra and Chauhan, 2023).
Preserving traditional potato varieties is not merely a matter of historical interest but a strategic necessity for safeguarding genetic diversity, which is essential for breeding new cultivars capable of withstanding emerging biotic threats, such as novel pathogen strains, and abiotic stressors, like drought and temperature fluctuations driven by climate change. By integrating traditional knowledge with advanced genetic and biotechnological tools, researchers can harness the unique properties of local landraces to strengthen sustainable agricultural systems (Handayani et al., 2019). The cultural and economic benefits of preserving these varieties also extend to local communities, where traditional crops are often embedded in regional identities and culinary traditions. Programs focused on recovery and valorization contribute to rural development, promote agrobiodiversity, and encourage the production and consumption of high-quality, region-specific food products. Such initiatives align with the growing trends for sustainable and locally sourced produce, highlighting traditional agriculture’s role in contemporary food systems (Johns et al., 2013).
The Italian territory, with its mountainous regions and multiple microclimates, is home to potato varieties that have developed specific adaptations, such as tolerance to marginal conditions and unique flavor profiles. Notable examples include the violet-skinned ‘Viola Calabrese,’ the renowned ‘Rossa di Cetica’ from Tuscany, and the ‘Quarantina Bianca’ from Liguria (Ghiselli et al., 2001; Manzelli et al., 2010). These local varieties have been preserved over generations by dedicated farmers who have recognized their value beyond mere productivity. Conservation efforts include in vitro culture and virus elimination techniques to ensure healthy propagation. The Department of Agricultural, Food, Environmental, and Forestry Sciences (DAGRI) at the University of Florence (Italy), has been pivotal in recovering and maintaining a collection of these landraces since the late 1990s (Angelini, 1999; Vecchio et al., 2007). Together with conservation techniques, molecular markers have been widely applied for the characterization of genetic diversity within and among these local varieties (Portis et al., 2005; Mauro et al., 2009; Carputo et al., 2013; Emanuelli et al., 2013; Vilanova et al., 2014). Indeed, compared to classical markers (i.e. SSRs), Next Generation Sequencing (NGS) technologies provide higher marker resolution, allowing higher precision in genetic and genomic analyses (Barchi et al., 2019; Saidi and Hajibarat, 2020; Toppino et al., 2020; Tripodi et al., 2021; Martina et al., 2022; Barchi et al., 2023; Martina et al., 2023, 2024). In recent years, genotype-by-sequencing (GBS) has become a key tool in potato genomics for studying genetic diversity, population structure, and trait mapping in both cultivated and wild germplasm. Applications include association studies for disease resistance traits (Sood et al., 2023) and the exploration of genome-wide variation to support conservation strategies and pre-breeding (Endelman et al., 2024; Sharma et al., 2024; Schilling et al., 2024). These studies underscore the power of GBS in revealing the complexity and richness of potato genetic resources.
We explored the genetic diversity of 61 accessions from 22 traditional potato varieties/ecotypes from Italy, aiming to collect, sanitize them from virus infection, preserve them in vitro, and finally return them to local farmers as fresh material. Genotype-by-sequencing (GBS) analyses revealed substantial genetic differentiation within Italian germplasm, and a significant level of differentiation with a set of 106 genotypes from other regions of the world. This approach safeguards biodiversity and promotes eco-friendly farming practices, supporting environmental sustainability and consumer health. Investments in traditional varieties yield socio-economic dividends, empower local communities, create jobs, uphold cultural heritage, and support high-quality, locally adapted crops sought by consumers for their unique characteristics and traceable origins. This holistic strategy preserves traditional agricultural knowledge, ensuring that future generations benefit from a resilient, diverse, and sustainable food system.
2 Materials and methods
2.1 Plant material
A total of 61 accessions, representing twenty-two traditional Italian potato varieties/ecotypes, have been collected at the Department of Agriculture, Food, Environment, and Forestry (DAGRI) at the University of Florence (Italy) since the late 1990s (Table 1, Figure 1). Tubers were characterized for pulp and skin color according to the cultivated potato descriptors (Huaman et al., 1977), with minor modifications. On the collection day, thanks to the interaction with the custodian farmers, information on the traditional culinary destination was gathered. These varieties underwent an in vitro introduction process, virus testing, and virus elimination, following the methodology previously described (Andrenelli et al., 2002, 2018; Natale et al., 2024), as detailed in the next paragraph. Additionally, six commercial cultivars (Alba, Blu di San Gallo, Desiree, Institute de Beauvais, Kennebec, and Magenta) were included in the analysis as reference standards, based on previously detected genetic identities with some local accessions using microsatellite (SSR) markers (Mandolino et al., 2015). Furthermore, sequencing data from approximately 106 genotypes from various regions worldwide were retrieved from the NCBI database (PRJNA723185 - Sood et al., 2023) for comparative analysis.
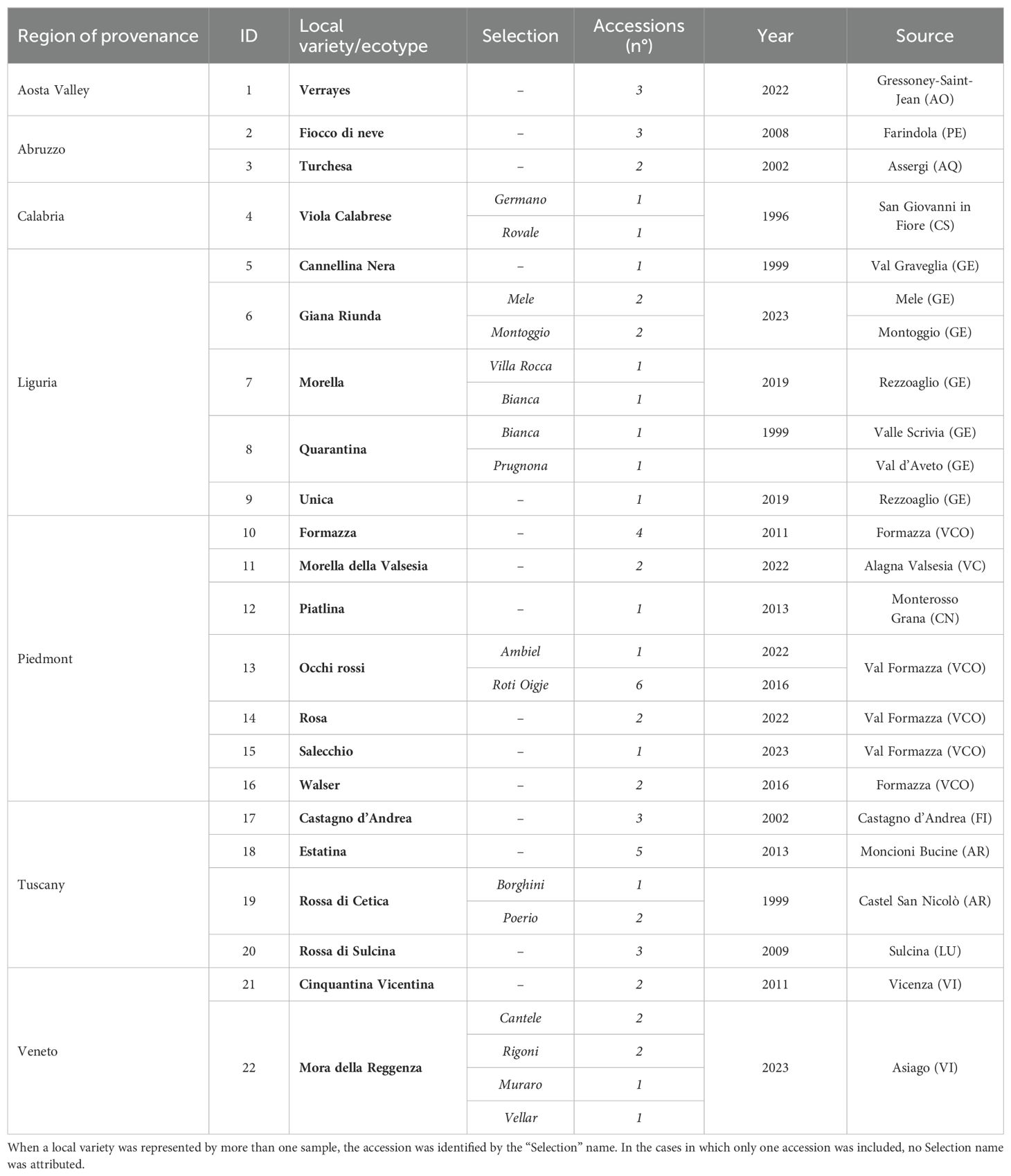
Table 1. List of Italian local potato varieties/ecotypes (in bold) and selections from the DAGRI germplasm collection, including region and locality of Q30 provenance, number of accessions, and year of introduction.
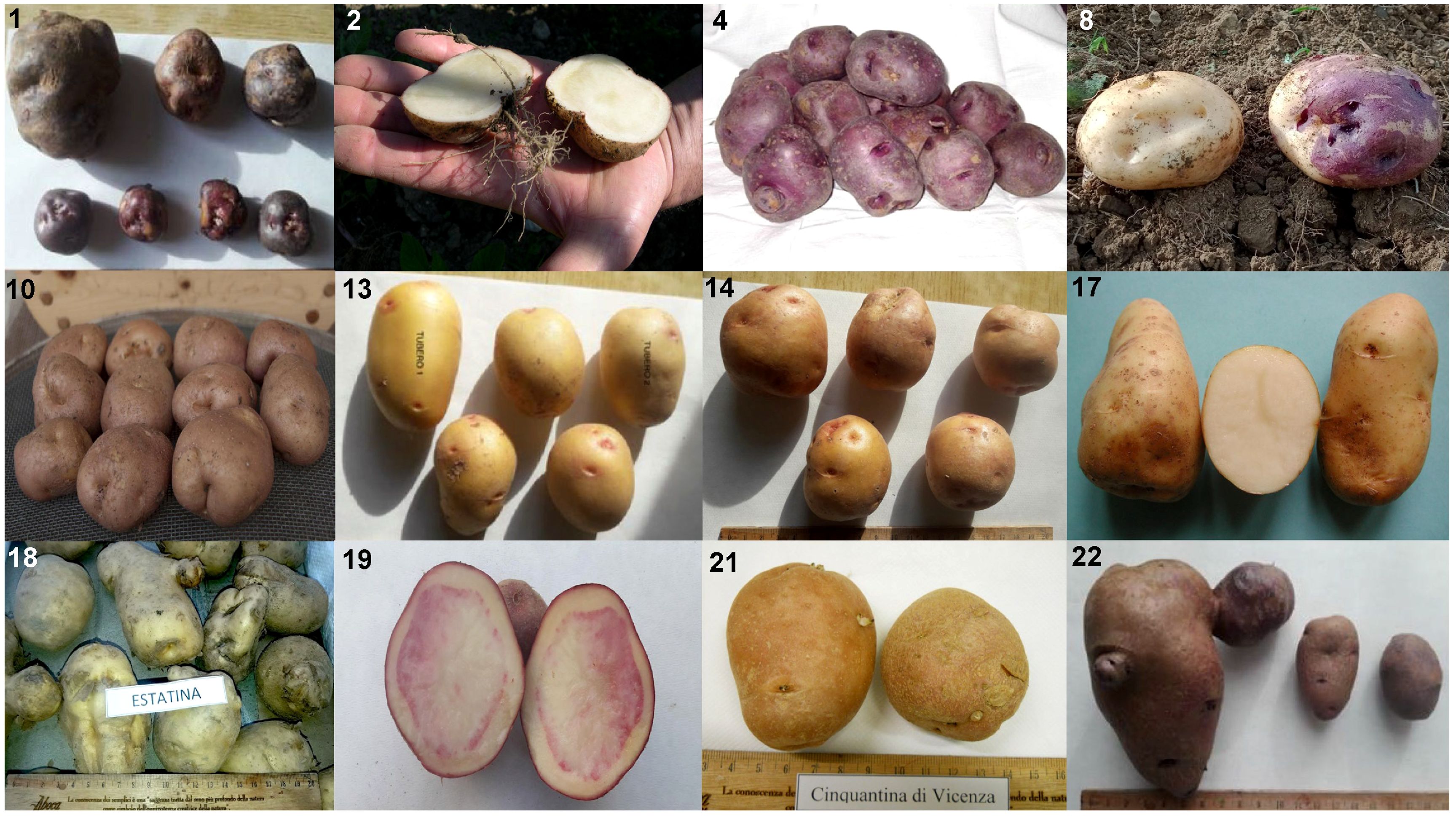
Figure 1. Example of the phenotypic variability within the analyzed collection (ID numbers refer to Table 1). From left to right, top to bottom: (1) Verrayes; (2) Fiocco di Neve; (4) Viola Calabrese; (8) Quarantina; (10) Formazza; (13) Occhi Rossi; (14) Rosa; (17) Castagno d’Andrea; (18) Estatina; (19) Rossa di Cetica; (21) Cinquantina Vicentina; (22) Mora della Reggenza.
2.2 In vitro introduction and virus eradication protocol
Collected tubers were carefully washed and allowed to sprout. Sprouts measuring 1–2 cm were harvested and disinfected by soaking in 70% ethanol for 30 seconds, followed by immersion in 2.5% sodium hypochlorite for 15 minutes. After multiple rinses with sterilized water, the sprouts were transferred to tubes containing Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) supplemented with 25 g L-1 sucrose and 2 g L-1 Phytagel for growth and propagation (Figure 1). All varieties underwent virus molecular detection testing (RT-PCR) for the main potato viruses (PVX, PVY, PVS, PLRV) according to Mortimer-Jones et al. (2009), with minor modifications. The analyses were carried out by the “Regional Phytosanitary Service” of the Tuscany region, in Italy, which exercises control and official supervision over the phytosanitary status of plants and their products during cultivation, storage, and marketing stages, to verify the possible presence of harmful organisms to plants. Those testing positive were subjected to a sanitization protocol. Two weeks-old in vitro plants were placed in a climate chamber for thermotherapy treatment, following a one-month program: 4 hours at 35°C (daylight) and 4 hours at 30°C (darkness - Griffiths et al., 1990). After this period, the apical meristem (0.1 to 0.4 mm in size) of each plant was excised and cultured on a modified MS medium containing 0.1 mg L-1 gibberellic acid (GA3) and 0.04 mg L-1 kinetin to stimulate root development and growth. The regenerated plants were subsequently tested to confirm virus eradication. The complete process is illustrated in Figure 2.
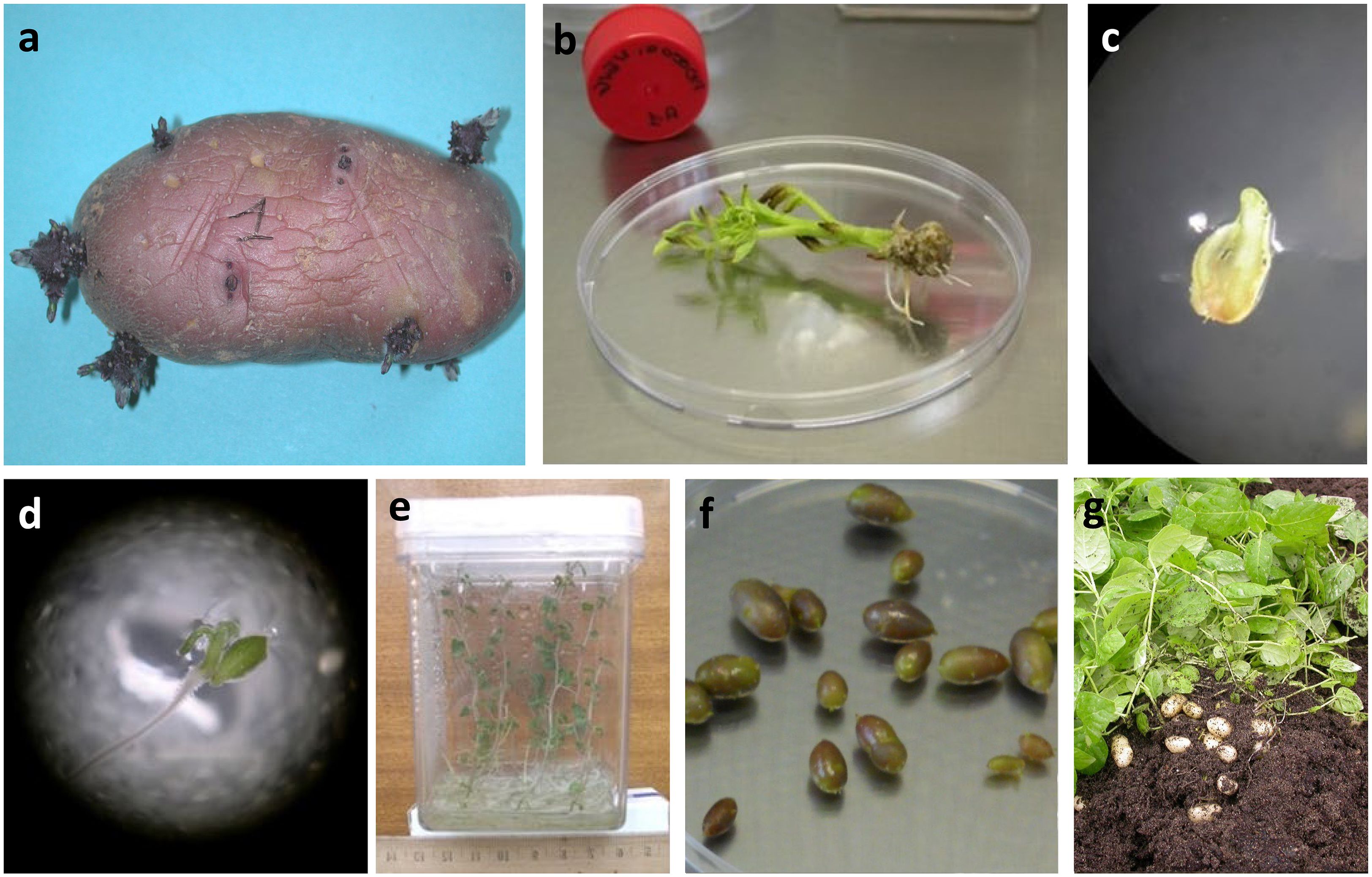
Figure 2. Main stages from in vitro introduction to seed tuber production: (a) tuber sprouting; (b) in vitro propagated plantets, obtained from tuber sprouts; (c) apical meristem growing on specific substrate; (d) apical meristem 20 days after explantation with root and leaf in initial differentiation; (e) in vitro virus free tested plantlets; (f) microtubers developed in vitro; g) vitro plant transplanted in vivo, showing minitubers production.
2.3 DNA extraction, GBS-library preparation and sequencing
Genomic DNA was isolated from in vitro plantlets using a modified CTAB method (Doyle, 1991; Li et al., 2024). The purified DNA was resuspended in 100 µL of deionized water. To remove residual RNA, DNase-free RNase was added to the sample and incubated at 35°C for one hour. DNA concentration and purity were assessed using a Qubit™ 2.0 Fluorometer and by measuring absorbance ratios at 260/280 nm and 260/230 nm with a NanoDrop™ 2000 spectrophotometer. The integrity and size distribution of the genomic DNA were evaluated by agarose gel electrophoresis. Finally, samples were digested with MseI and EcoRI restriction enzymes to assess their compatibility with subsequent library construction and sequencing.
For genotyping-by-sequencing (GBS), libraries were prepared using a dual-enzyme strategy, following a modified version of the protocol by Poland et al., 2012. Genome complexity was reduced using the restriction enzymes MseI and EcoRI, and the resulting libraries were sequenced on an Illumina NovaSeq X system at the Biotechnology Center of the University of Wisconsin-Madison.
2.4 Variants calling, filtering and phylogenic analysis
Sequencing data from both the Italian local varieties/ecotypes and the dataset from Sood et al., 2023 were quality-filtered using fastp (Chen et al., 2018) and aligned to the DM-13 Potato reference genome (v6.1; Pham et al., 2020) using the GATK pipeline (Poplin et al., 2018). After alignment, variants were filtered based on the following criteria: a minimum depth of 15 (min DP15), a minor allele frequency threshold of ≤0.5 (MAF), and a missing data rate of ≤0.05. Pruning was carried out with the SNPrelate R package (Zheng et al., 2012), using a linkage disequilibrium (LD) threshold of 0.1. The resulting set of filtered SNPs was then used for Neighbor-Joining phylogenetic analysis in MEGA11 software (Tamura et al., 2021) with the p-distance method.
3 Results and discussion
3.1 Ecotypes preservation, geographical distribution and their main characteristics
Italy provides a favorable environment for the genetic diversification of potatoes, resulting in the selection of numerous local ecotypes adapted to the specific pedoclimatic conditions of its diverse regions (Perrino and Perrino, 2024). The geographic distribution analysis of these ecotypes reveals a predominance in the northern and central regions, with some varieties also present in southern Italy (Figure 3). In the northwestern Alps, particularly in Valle d’Aosta and Piedmont, varieties such as Piatlina, Walser, Rosa, and Morella della Valsesia exhibit good adaptability to mountain environments. These varieties produce medium-small tubers, with skin colors ranging from yellow to red. In Veneto region, Mora della Reggenza and Cinquantina Vicentina are traditionally cultivated in hilly and pre-alpine environments, with tubers having either dark or yellow skin (Table 2).
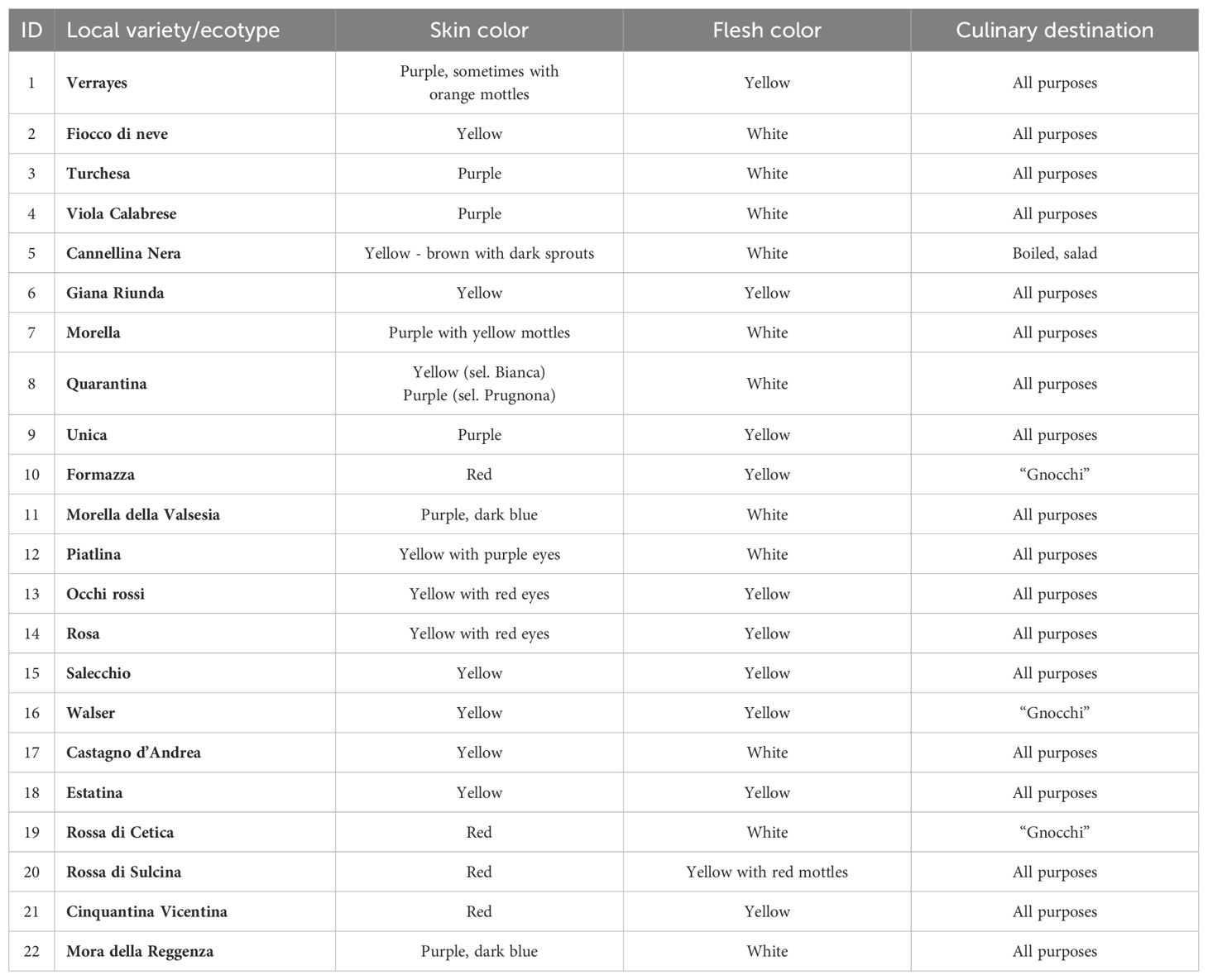
Table 2. List of Local varieties/ecotypes (in bold) along with their morphological characteristics and culinary uses.
Along the Apennine ridge, Liguria region hosts ecotypes like Quarantina and Giana Riunda, while Tuscany showcases genetic diversity with varieties such as Rossa di Cetica, Castagno d’Andrea, and Estatina, possessing flesh colors that range from white to deep yellow. In the central-southern regions, Fiocco di Neve in Abruzzo and Viola Calabrese in Calabria feature skin colors ranging from pure white to deep purple.
Italian local potato ecotypes are distinguished not only by their geographic distribution but also by a range of agronomic and morphological characteristics. Many of these ecotypes have been selected over time for their resistance to harsh environmental conditions, such as poor soil, low temperatures, and seasonal drought. Mountain varieties tend to have shorter growth cycles and smaller tubers compared to those from temperate regions, which exhibit higher productivity and a greater variety of shapes and colors (Table 2). The preservation of these ecotypes by local farmers, particularly in extreme marginal areas like the upper Valsesia valley in Piedmont, is crucial. Here, the Walser population, who migrated from Switzerland at the end of the 14th century, selected varieties like Formazza, Morella della Valsesia, and Verrayes for their adaptability to extreme pedoclimatic conditions. These varieties became integral to the seasonal diet of the Walser community, composed of small farmers and shepherds. Preserving such genetic material is vital for maintaining genetic diversity, ensuring the resilience of potato crops to environmental changes, and safeguarding Italy’s rich agricultural heritage.
3.2 In vitro introduction and virus eradication
Viral infections pose a significant challenge to the conservation and cultivation of local potato landraces, often resulting in reduced yields and, in severe cases, the loss of valuable genetic material. Aiming at assessing the phytosanitary state of the Italian landraces, the collected material was screened at molecular level, revealing a widespread presence of PVX, PLRV, PVY, and PVS, with some ecotypes exhibiting multiple infections (Table 3).
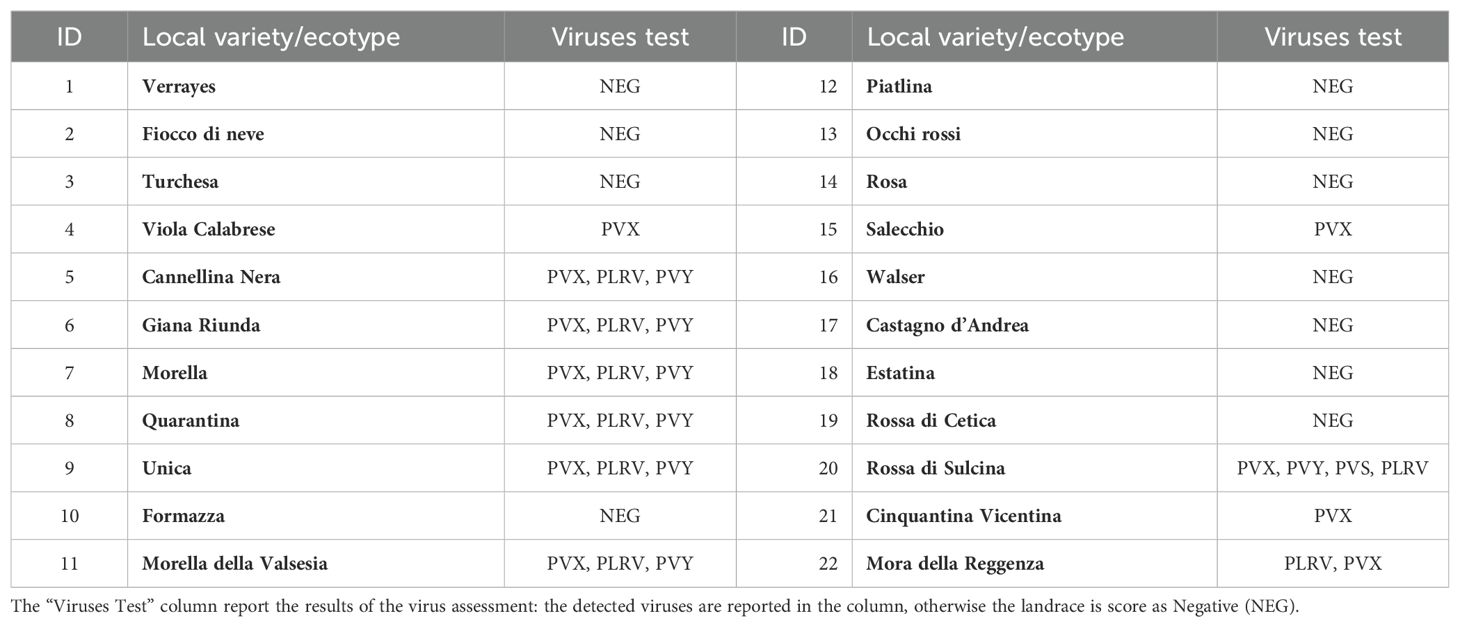
Table 3. List of Local varieties/ecotypes (in bold) with the corresponding detected virus infection.
Notably, Rossa di Cetica, Cannellina Nera, Giana Riunda, and Morella della Valsesia were among the most affected, carrying several viral strains simultaneously. This widespread viral incidence underscores the challenges associated with the long-term vegetative propagation of traditional varieties without systematic phytosanitary management. To restore the health status of infected accessions, a sanitation protocol combining thermotherapy and meristem culture was applied. Post-treatment diagnostic tests confirmed the complete elimination of viral infections in all ecotypes, demonstrating the effectiveness of this method. These findings align with previous studies on the effectiveness of meristem culture in the recovery of vegetatively propagated crops (Wang et al., 2018; Bhat and Rao, 2020; Alavijeh et al., 2025). The availability of virus-free planting material is crucial for both conservation and sustainable seed tuber production. Healthy accessions not only preserve genetic diversity but also improve agronomic performance, particularly in low-input and organic farming systems (Tegen, 2016). Moreover, the recovery and maintenance of these local varieties play a key role in safeguarding Italians potato genetic heritage, facilitating their reintroduction into traditional cultivation systems. This effort is essential for maintaining culinary traditions, regional identity, and biodiversity, while also supporting the adaptation of landraces to changing environmental conditions.
3.3 Phylogenetic analysis
Overall, approximately 17 million reads (PE150) were generated, resulting in a total of about 56.5 gigabytes of raw data. The cleaned reads were aligned to the Solanum tuberosum reference genome (v6.1; Pham et al., 2020), leading to the identification of 8.3 million SNPs. After filtering, this number was reduced to 15.872.
The resulting NJ dendrogram (Figure 4) provides a distinct molecular fingerprint for each analyzed local variety/ecotype, organizing them into three main clusters (A, B, and C), each further divided into smaller subclusters (three or four per cluster). Although selections/provenances within each variety fall within the same subcluster, they are generally distinguishable, highlighting varying levels of within-variety diversity. This diversity reflects both genetic variation and potential influences from local adaptations or microenvironmental conditions. The five commercial varieties, used as reference material, are distributed across five different subclusters (B1, B2, C1, C2, and C3), each exhibiting a unique molecular profile. This contrasts with the findings of Mandolino et al., 2015, who used 12 SSR markers for molecular characterization of a small subset of the DAGRI collection and reported perfect identity between the SSR profiles of some commercial varieties and certain local varieties included in this analysis. This discrepancy highlights the enhanced resolution of the NGS-based approach employed in our study.

Figure 4. NJ dendrogram based on 15.872 SNP markers. Selection names and Accession numbers are indicated in blue. Reference commercial varieties are indicated in red. Three different clusters have been identified (A–C), together with subclusters (in red).
Branch A includes Mora della Reggenza (Vellar’ and ‘Muraro’ selections) and Morella accessions clustered together in A1, along with Turchesa and Viola Calabrese, suggesting a shared genetic background or conserved selection practices among these ecotypes. In contrast, the Mora della Reggenza ‘Rigoni’ and ‘Cantele’ selections are separated, clustering independently in A4 and showing clear genetic differentiation from other ecotypes with similar phenological traits. This differentiation could reflect distinct historical selection pressures or genetic drift specific to its location of origin. A2 comprises a group including Unica, Salecchio, and Walser accessions, along with the three Verrayes accessions, all characterized by a single molecular profile. This uniformity across multiple accessions may indicate a stable clonal lineage, possibly conserved due to its adaptability or favorable traits. A3 contains Rosa and Occhi Rossi, which, though grouped together, remain well distinguished from each other, suggesting that these varieties, while similar, have retained distinct genetic identities.
Branch B’s sub-cluster B1 confirms the previously detected similarity between Castagno d’Andrea and the commercial variety Kennebec, consistent with the findings of Mandolino et al. (2015). However, while the SSR analysis showed complete identity between these two varieties, the higher resolution of the NGS approach used here allows us to distinguish them. This finding emphasizes the sensitivity of NGS in revealing subtle genetic differences that SSR markers may overlook. Similarly, the relationship between the Quarantina ecotype and the commercial variety Institute de Beauvais, as well as with the Fiocco di Neve accessions in B2, was also observed by Mandolino et al. (2015), who nonetheless reported complete identity between Quarantina and the French-origin commercial variety. The distinctions observed here reinforce the potential of high-resolution markers to refine our understanding of genetic relationships. Finally, Branch B includes subcluster B3, where Rossa di Cetica and Giana Riunda selections are grouped together but remain distinguishable. This indicates that, while these selections may share some genetic similarities, they have diverged enough to maintain unique profiles, showcasing how local practices can introduce variability even within closely related selections.
In Branch C, the commercial varieties Magenta and Alba cluster in C1 with Estatina accessions, each retaining distinct profiles. This demonstrates that even when commercial and local varieties cluster together, they maintain unique molecular signatures, likely reflecting the influence of their respective breeding histories. In sub-cluster C2, the commercial variety Desiree clusters with Rossa di Sulcina and Cinquantina Vicentina, each maintaining distinctive molecular profiles. This contrasts with the findings of Mandolino et al. (2015), who reported complete identity among these varieties. Within this cluster, it is interesting to note that the Rossa di Sulcina accessions show significant internal variability, with two accessions exhibiting greater genetic distance from each other than from the Cinquantina Vicentina ecotype. This internal diversity could suggest either a broader genetic base within Rossa di Sulcina or the influence of localized environmental adaptation. Finally, the commercial variety Blu di San Gallo clusters with Piatlina, Cannellina Nera, and Formazza, with the Formazza accessions well-separated from the others in the final sub-cluster (C3) of Figure 2, underlining the diversity present even within this closely grouped set.
Notably, the resolution power of the NGS technique was sufficient to distinguish only a limited number of accessions sampled from the same provenance. For instance, this was the case for two accessions of Morella della Valsesia and Giana Riunda “Mele” selection, two of the three accessions of Rossa di Sulcina, and two of the four accessions of Formazza. Similarly, it could not distinguish between different provenances within the Quarantina ecotype, even though they are characterized by different skin colors. This phenotypic difference may be due to a point mutation not captured by our SNP marker dataset. The ability of the NGS technique to detect some but not all intraspecific variations highlight both its strengths and limitations in detecting subtle genetic differences within clonally propagated crops.
3.4 Comparison of the Italian germplasm with the world-wide accessions
Building on the work of Sood et al. (2023), we examined the genetic relationships between Italian potato local varieties/ecotypes and genotypes from around the world. In their study, the neighbor-joining (NJ) method was used to categorize 222 accessions and varieties into three main clusters (A, B, and C), which were further divided into subclusters. Their results indicated that, although no strict geographic pattern was evident, Indian varieties grown in the subtropical plains tended to cluster with a subset of andigena accessions, as well as accessions from Australia, New Zealand, and Afghanistan. In contrast, the other two clusters comprised a mix of varieties from Europe, North America, and South America, suggesting some level of geographic association with notable overlaps.
In our research, we compared a collection of Italian potato varieties with approximately 106 accessions from various parts of the world, as included in Sood et al. (2023 - Supplementary Table 1). Raw sequence reads were retrieved from the NCBI database (PRJNA723185), cleaned and aligned to the reference genome (Pham et al., 2020). SNP calling and filtering were then performed, yielding a set of 10,197 high-quality SNPs for further analysis. By integrating these global accessions with our Italian dataset, we aimed to explore whether Italian varieties would exhibit similar clustering patterns or if they would stand out genetically from other regional groups.
The generated NJ dendrogram (Figure 5) reveals a marked separation between the Italian collection and the global accessions, highlighting a unique genetic signature among the Italian varieties. Unlike the findings of Sood et al. (2023), where some regional clustering patterns were observed but not strictly defined, our analysis demonstrates a clearer phylogenetic distinction. Italian varieties consistently formed a separate group, suggesting unique genetic characteristics likely shaped by local breeding practices and adaptation to regional conditions. This pronounced clustering of Italian varieties contrasts with the more intermixed clusters of international accessions, where European, North American, and South American varieties often appeared within the same groups, as reported by Sood et al. (2023).
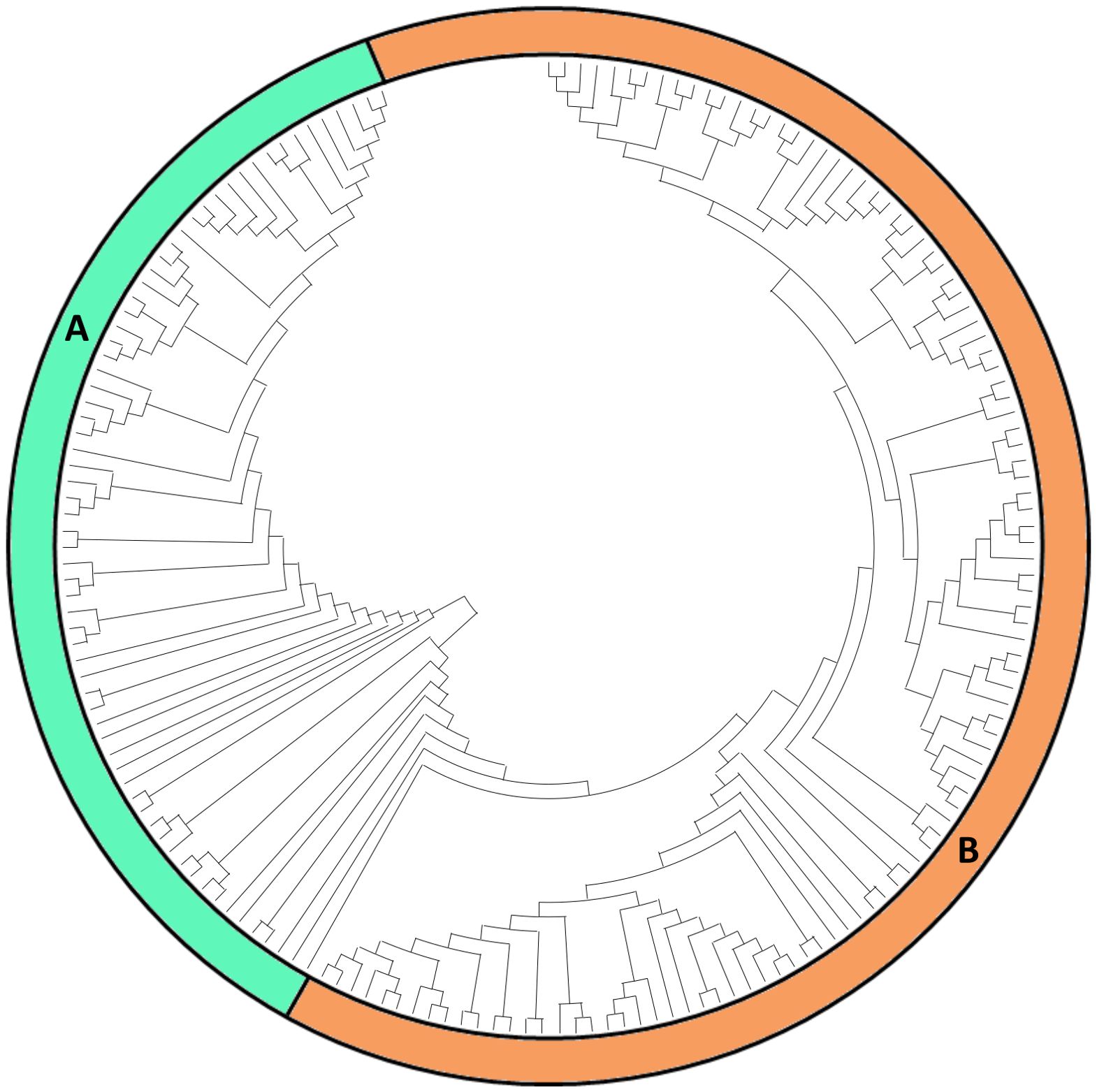
Figure 5. NJ dendrogram, constructed from 10,197 SNPs. Cluster (A) contains the accessions from Italy, while Cluster (B) contains the accessions from Sood et al., 2023.
Overall, our expanded approach, which combines Italian and international accessions, underscores a distinct phylogenetic identity for the Italian varieties. This separation may reflect historical, ecological, and selective pressures unique to Italian agricultural systems, offering valuable insights into the genetic diversity and potential breeding value of these local varieties within a global context.
4 Conclusions
The conservation and promotion of traditional potato varieties are crucial strategies for enhancing agricultural sustainability and socio-economic resilience.
In this study, we identified three major genetic clusters among 61 accessions representing 22 Italian landraces and observed a clear molecular separation from 106 international genotypes. Several landraces showed multiple viral infections, which were successfully eradicated through thermotherapy and meristem culture, enabling their recovery and safe propagation. These ecotypes, with their unique adaptations and qualities, provide invaluable genetic resources that can be harnessed to develop crops capable of facing current and future challenges, including climate change and the emergence of new pathogens. Preserving these landraces safeguards genetic diversity, ensuring a reservoir of traits that contributes to the development of resilient and productive cultivars. Moreover, the socio-economic benefits extend beyond agricultural improvements. By fostering the production and commercialization of traditional varieties, local communities can stimulate economic growth, create employment opportunities, and strengthen their cultural heritage. Initiatives that encourage collaboration among research institutions, local farmers, and policymakers, help build sustainable agricultural practices and regional networks that support the entire value chain. This, in turn, promotes rural development and ensures that high-quality, locally adapted food products reach a broader market.
The integration of traditional knowledge with modern scientific approaches is essential for maintaining biodiversity and empowering local farmers. Supporting programs that focus on the recovery, characterization, and utilization of traditional potato varieties ensure that the agricultural sector remains diverse and adaptable. These efforts not only address ecological and economic sustainability but also promote a deeper appreciation for the rich agricultural heritage and the potential of traditional crops to contribute to future food security. Investing in the conservation and valorization of traditional potato varieties is an investment in the future. It reinforces the importance of biodiversity as a foundation for sustainable development, creating a resilient agricultural system that benefits both present and future generations.
Data availability statement
Raw data supporting the conclusions of this article have been included in the present article. Sequencing data used in this study are openly available in the NCBI database (PRJNA1279639).
Author contributions
MM: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. RN: Data curation, Formal Analysis, Methodology, Resources, Writing – original draft, Writing – review & editing. EV: Formal Analysis, Software, Visualization, Writing – original draft, Writing – review & editing. AM: Investigation, Resources, Writing – original draft, Writing – review & editing. SB: Investigation, Resources, Writing – original draft, Writing – review & editing. LA: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. EP: Conceptualization, Formal Analysis, Funding acquisition, Methodology, Project administration, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was conceived as part of the “RESToRE” Project (Recovery and Valorization of Traditional Solanum tuberosum Varieties at Risk of Extinction), supported by the Piedmont Region under the PSR 2014-2024 Op. 10.2.1 framework.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1629306/full#supplementary-material
References
Alavijeh, M. K., Bayat, H., Kianpour, D., Kalantari, S., and Zarei, A. (2025). Optimization of in vitro propagation and virus eradication using meristem culture and thermotherapy in two geranium species Pelargonium X hortorum (Zonal’) and Pelargonium × domesticum (‘Regal’). BMC Plant Biol. 25, 9. doi: 10.1186/s12870-024-06027-y
Andrenelli, L., Grassi, C., Baglio, A., and Ghiselli, L. (2018). Recupero, risanamento da virus e conservazione in vitro di vecchie varietà di patata italiane (Solanum tuberosum L.). Convegno nazionale sulla Micropropagazione Pescia Pagine, 134–142.
Andrenelli, L., Palchetti, E., Ghiselli, L., and Vecchio, V. (2002). Valorizzazione di germoplasma locale e coltura in vitro per la produzione di tubero seme di patata (Solanum tuberosum L.). Italus Hortus 9, 5–6.
Angelini, M. (1999). Le patate tradizionali della Montagna Genovese. Provincia di Genova, Ufficio Attività Territoriali, via G. Maggio, 3, 16147.
Barchi, L., Acquadro, A., Alonso, D., Aprea, G., Bassolino, L., Demurtas, O., et al. (2019). Single primer enrichment technology (SPET) for high-throughput genotyping in tomato and eggplant germplasm. Front. Plant Sci. 10. Available online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01005.
Barchi, L., Aprea, G., Rabanus-Wallace, M. T., Toppino, L., Alonso, D., Portis, E., et al. (2023). Analysis of >3400 worldwide eggplant accessions reveals two independent domestication events and multiple migration-diversification routes. Plant J. 116, 1667–1680. doi: 10.1111/tpj.16455
Bhat, A. I. and Rao, G. P. (2020). “Virus elimination by meristem-tip culture,” in Characterization of Plant Viruses: Methods and Protocols. Eds. Bhat, A. I. and Rao, G. P. (Springer US, New York, NY), 465–477. doi: 10.1007/978-1-0716-0334-5_47
Brush, S. B. (1995). In situ conservation of landraces in centers of crop diversity. Crop Sci. 35, 346–354. doi: 10.2135/cropsci1995.0011183X003500020009x
Carputo, D., Alioto, D., Aversano, R., Garramone, R., Miraglia, V., Villano, C., et al. (2013). Genetic diversity among potato species as revealed by phenotypic resistances and SSR markers. Plant Genet. Resour. 11, 131–139. doi: 10.1017/S1479262112000500
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Doyle, J. (1991). “DNA protocols for plants,” in Molecular Techniques in Taxonomy. Eds. Hewitt, G. M., Johnston, A. W. B., and Young, J. P. W. (Springer, Berlin, Heidelberg), 283–293. doi: 10.1007/978-3-642-83962-7_18
Emanuelli, F., Lorenzi, S., Grzeskowiak, L., Catalano, V., Stefanini, M., Troggio, M., et al. (2013). Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol. 13, 39. doi: 10.1186/1471-2229-13-39
Endelman, J. B., Kante, M., Lindqvist-Kreuze, H., Kilian, A., Shannon, L. M., Caraza-Harter, M. V., et al. (2024). Targeted genotyping-by-sequencing of potato and data analysis with R/polyBreedR. Plant Genome 17, e20484. doi: 10.1002/tpg2.20484
Ghiselli, L., Andrenelli., L., and Vecchio, V. (2001). “La viola calabrese,” in Il Gazzettino Della Patata, 6–11.
Griffiths, H. M., Slack, S. A., and Dodds, J. H. (1990). Effect of chemical and heat therapy on virus concentrations in in vitro potato plantlets. Can. J. Bot. 68, 1515–1521. doi: 10.1139/b90-193
Handayani, T., Gilani, S. A., and Watanabe, K. N. (2019). Climatic changes and potatoes: How can we cope with the abiotic stresses? Breed Sci. 69, 545–563. doi: 10.1270/jsbbs.19070
Huaman, Z., Williams, J. T., Salhuana, W., and Vincent, L. (1977).Descriptors for the cultivated Potato. Available online at: https://www.genesys-pgr.org/descriptorlists/9e1e1aca-1003-4365-b782-cec4b09508bb (Accessed March 24, 2025).
Jo, K. R., Cho, S., Cho, J.-H., Park, H.-J., Choi, J.-G., Park, Y.-E., et al. (2022). Analysis of genetic diversity and population structure among cultivated potato clones from Korea and global breeding programs. Sci. Rep. 12, 10462. doi: 10.1038/s41598-022-12874-2
Johns, T., Powell, B., Maundu, P., and Eyzaguirre, P. B. (2013). Agricultural biodiversity as a link between traditional food systems and contemporary development, social integrity and ecological health. J. Sci. Food Agric. 93, 3433–3442. doi: 10.1002/jsfa.6351
Li, R., Cui, L., Martina, M., Bracuto, V., Meijer-Dekens, F., Wolters, A.-M. A., et al. (2024). Less is more: CRISPR/Cas9-based mutations in DND1 gene enhance tomato resistance to powdery mildew with low fitness costs. BMC Plant Biol. 24, 763. doi: 10.1186/s12870-024-05428-3
Mandolino, G., Parisi, B., Andrenelli, L., A., F., Ventisei, H., and Reid, A. (2015). Molecular fingerprinting of traditional Italian potato varieties.
Manzelli, M., Romagnoli, S., Ghiselli, L., Benedettelli, S., Palchetti, E., Andrenelli, L., et al. (2010). Typicity in potato: characterization of geographic origin. Ital. J. Agron. 5, 61–67. doi: 10.4081/ija.2010.61
Martina, M., Acquadro, A., Gulino, D., Brusco, F., Rabaglio, M., Portis, E., et al. (2022). First genetic maps development and QTL mining in Ranunculus asiaticus L. through ddRADseq. Front. Plant Sci. 13. Available online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1009206.
Martina, M., Acquadro, A., Portis, E., Barchi, L., and Lanteri, S. (2023). Diversity analyses in two ornamental and large-genome Ranunculaceae species based on a low-cost Klenow NGS-based protocol. Front. Plant Sci. 14. Available online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1187205.
Martina, M., Zayas, A., Portis, E., Di Nardo, G., Polli, M. F., Comino, C., et al. (2024). The Dark Side of the pollen: BSA-seq identified genomic regions linked to male sterility in globe artichoke. BMC Plant Biol. 24, 415. doi: 10.1186/s12870-024-05119-z
Mauro, R., Portis, E., Acquadro, A., Lombardo, S., Mauromicale, G., and Lanteri, S. (2009). Genetic diversity of globe artichoke landraces from Sicilian small-holdings: implications for evolution and domestication of the species. Conserv. Genet. 10, 431–440. doi: 10.1007/s10592-008-9621-2
Mortimer-Jones, S. M., Jones, M. G., Jones, R. A., Thomson, G., and Dwyer, G. I. (2009). A single tube, quantitative real-time RT-PCR assay that detects four potato viruses simultaneously. J. virological Methods 161, 289–296. doi: 10.1016/j.jviromet.2009.06.027
Murashige, T. and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Natale, R., Martina, M., Vergnano, E., Milani, A. M., Prina, A., Paffetti, D., et al. (2024). “Reviving forgotten treasures: a conservation effort for endangered Solanum tuberosum varieties,” in EHC2024: Genetic Resources in Horticulture: screening, propagation, use, and conservation. (In Press).
Perrino, E. V. and Perrino, P. (2024). “Conservation of plant genetic resources in Italy,” in Sustainable Utilization and Conservation of Plant Genetic Diversity. Eds. Al-Khayri, J. M., Jain, S. M., and Penna, S. (Springer Nature, Singapore), 895–971. doi: 10.1007/978-981-99-5245-8_27
Pham, G. M., Hamilton, J. P., Wood, J. C., Burke, J. T., Zhao, H., Vaillancourt, B., et al. (2020). Construction of a chromosome-scale long-read reference genome assembly for potato. GigaScience 9, giaa100. doi: 10.1093/gigascience/giaa100
Poland, J. A., Brown, P. J., Sorrells, M. E., and Jannink, J.-L. (2012). Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7, e32253. doi: 10.1371/journal.pone.0032253
Poplin, R., Ruano-Rubio, V., DePristo, M. A., Fennell, T. J., Carneiro, M. O., Auwera, G.A.V.d., et al. (2018). Scaling accurate genetic variant discovery to tens of thousands of samples. BioRxiv 201178. doi: 10.1101/201178
Portis, E., Barchi, L., Acquadro, A., Macua, J. I., and Lanteri, S. (2005). Genetic diversity assessment in cultivated cardoon by AFLP (amplified fragment length polymorphism) and microsatellite markers. Plant Breed. 124, 299–304. doi: 10.1111/j.1439-0523.2005.01098.x
Saidi, A. and Hajibarat, Z. (2020). Application of Next Generation Sequencing, GWAS, RNA seq, WGRS, for genetic improvement of potato (Solanum tuberosum L.) under drought stress. Biocatalysis Agric. Biotechnol. 29, 101801. doi: 10.1016/j.bcab.2020.101801
Salgotra, R. K. and Chauhan, B. S. (2023). Genetic diversity, conservation, and utilization of plant genetic resources. . Genes (Basel) 14, 174. doi: 10.3390/genes14010174
Schilling, F., Schumacher, C., Köhl, K., Sprenger, H., Kopka, J., Peters, R., et al. (2024). Whole-genome sequencing of tetraploid potato varieties reveals different strategies for drought tolerance. Sci. Rep. 14, 5476. doi: 10.1038/s41598-024-55669-3
Sharma, S. K., McLean, K., Hedley, P. E., Dale, F., Daniels, S., and Bryan, G. J. (2024). Genotyping-by-sequencing targets genic regions and improves resolution of genome-wide association studies in autotetraploid potato. Theor. Appl. Genet. 137, 180. doi: 10.1007/s00122-024-04651-8
Solomon-Blackburn, R. M. and Barker, H. (2001). Breeding virus resistant potatoes (Solanum tuberosum): a review of traditional and molecular approaches. Heredity 86, 17–35. doi: 10.1046/j.1365-2540.2001.00799.x
Sood, S., Bhardwaj, V., Bairwa, A., Dalamu., Dalamu Sharma, S., Sharma, A. K, et al. (2023). Genome-wide association mapping and genomic prediction for late blight and potato cyst nematode resistance in potato (Solanum tuberosum L.). Front. Plant Sci. 14. Available online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1211472.
Spanoghe, M., Marique, T., Nirsha, A., Esnault, F., and Lanterbecq, D. (2022). Genetic diversity trends in the cultivated potato: A spatiotemporal overview. Biol. (Basel) 11, 604. doi: 10.3390/biology11040604
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tegen, H. (2016). The role of plant tissue culture to supply disease free planting materials of major horticultural crops in Ethiopia. J. Biol. Agric. Healthcare. 6 (1), 122–129.
Tobin, D., Bates, R., Brennan, M., and Gill, T. (2018). Peru potato potential: Biodiversity conservation and value chain development. Renewable Agric. Food Syst. 33, 19–32. doi: 10.1017/S1742170516000284
Toppino, L., Barchi, L., Mercati, F., Acciarri, N., Perrone, D., Martina, M., et al. (2020). A new intra-specific and high-resolution genetic map of eggplant based on a RIL population, and location of QTLs related to plant anthocyanin pigmentation and seed vigour. Genes (Basel) 11, 745. doi: 10.3390/genes11070745
Tripodi, P., Rabanus-Wallace, M. T., Barchi, L., Kale, S., Esposito, S., Acquadro, A., et al. (2021). Global range expansion history of pepper (Capsicum spp.) revealed by over 10,000 genebank accessions. Proc. Natl. Acad. Sci. 118, e2104315118. doi: 10.1073/pnas.2104315118
Vaccino, P., Antonetti, M., Balconi, C., Brandolini, A., Cappellozza, S., Caputo, A. R., et al. (2024). Plant genetic resources for food and agriculture: the role and contribution of CREA (Italy) within the national program RGV-FAO. Agronomy 14, 1263. doi: 10.3390/agronomy14061263
Vecchio, V., Benedettelli, S., Palchetti, E., and Manzelli, M. (2007). Patata rossa di Cetica. Un esempio di valorizzazione dell’agro-biodiversità e del territorio.
Vilanova, S., Hurtado, M., Cardona, A., Plazas, M., Herraiz, F. J., Gramazio, P., et al. (2014). Genetic diversity and relationships in local varieties of eggplant from different cultivar groups as assessed by genomic SSR markers. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 42, 59–65. doi: 10.15835/nbha4219414
Wang, M.-R., Cui, Z.-H., Li, J.-W., Hao, X.-Y., Zhao, L., and Wang, Q.-C. (2018). In vitro thermotherapy-based methods for plant virus eradication. Plant Methods 14, 87. doi: 10.1186/s13007-018-0355-y
Wang, Y., Rashid, M. A. R., Li, X., Yao, C., Lu, L., Bai, J., et al. (2019). Collection and evaluation of genetic diversity and population structure of potato landraces and varieties in China. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00139
Keywords: biodiversity, genotyping-by-sequencing (GBS), potato, local varieties, cultural heritage
Citation: Martina M, Natale R, Vergnano E, Milani AM, Biricolti S, Andrenelli L and Portis E (2025) Conservation and valorization of Italian potato biodiversity through genotype-by-sequencing (GBS). Front. Plant Sci. 16:1629306. doi: 10.3389/fpls.2025.1629306
Received: 15 May 2025; Accepted: 23 June 2025;
Published: 21 July 2025.
Edited by:
Sunita Gupta, Sri Karan Narendra Agriculture University, IndiaReviewed by:
Barbara Pipan, Agricultural institute of Slovenia, SloveniaZagipa Sapakhova, Institute of Plant Biology and Biotechnology (IPBB), Kazakhstan
Copyright © 2025 Martina, Natale, Vergnano, Milani, Biricolti, Andrenelli and Portis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: E. Portis, ZXppby5wb3J0aXNAdW5pdG8uaXQ=
 M. Martina
M. Martina R. Natale2,3
R. Natale2,3 E. Vergnano
E. Vergnano A. M. Milani
A. M. Milani S. Biricolti
S. Biricolti E. Portis
E. Portis