- 1College of Agriculture, Anhui Science and Technology University, Chuzhou, Anhui, China
- 2International Joint Research Center of Forage Bio-Breeding in Anhui Province, Chuzhou, China
- 3The International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Hyderabad, Telangana, India
- 4Department of Biology, University of Louisiana at Lafayette, Lafayette, LA, United States
Introduction: Water is essential for plant growth, and drought is one of the most predominant constraints on crop yield. Sorghum is a well-known drought-tolerant crop model, and sorghum landraces possess novel alleles for local adaptation.
Methods: In this study, we evaluated a sorghum mini core panel of 239 landraces sampled globally for shoot and root growth under simulated drought conditions using 10% and 20% polyethylene glycol (PEG) in 2020 and 2024, and measured drought tolerance using the seedling tolerance coefficient (STC).
Results and discussion: Phenotypic analysis showed that more accessions produced more roots than longer roots when exposed to 10% PEG; however, at 20% PEG, more accessions produced longer roots than more roots, reflecting the adaptability of some accessions to drought stress. However, PEG reduced shoot growth in all accessions in both years. A genome-wide association study (GWAS) on 32 growth and 19 STC traits identified 22 loci, 19 of which were mapped to the STC traits, and 17 of these 19 were associated with STC of shoot weight. Eleven of the 22 loci were collocated with 23 previously identified mapped drought-related quantitative trait loci (QTLs); 15 of these 23 QTLs were mapped to green leaf area, total number of green leaves, or chlorophyll content. We also found 19 candidate genes for 12 of the 22 loci. Five of those genes showed either preferential or specific expression in the roots according to GeneAtlas v2. One candidate gene from a locus colocated with a previously mapped chlorophyll fluorescence QTL has been shown to increase chlorophyll fluorescence in maize in another study. The results of this study lay the foundation for further characterizing the sorghum mini core panel for novel drought-tolerant genes.
Introduction
Water is critical for plant growth and development. As with all crop plants, the growth of sorghum [Sorghum bicolor (L.) Moench] relies on an adequate water supply in the form of rainfall or irrigation well distributed throughout the growing season (Assefa et al., 2010; Eck and Musick, 1979). For example, a medium-to-late sorghum variety maturing between 110 and 130 days would require approximately 450 to 650 mm of water during the growing season, and for this reason, the average yield of dryland sorghum is approximately half that of irrigated sorghum (Assefa et al., 2010). Not surprisingly, the total water supply (available soil water at seedling emergence plus in-season precipitation) is significantly correlated with sorghum grain yield (r2 = 0.834), and for every centimeter increase of available soil water at seedling emergence and in-season precipitation, sorghum grain yield increases by 221 and 164 kg ha−1, respectively (Stone and Schlegel, 2006). This indicates that soil water at seedling emergence is slightly more important for grain yield, probably because the early stages of plant growth (germination, emergence, and seedling establishment) are potentially the most vulnerable to drought stress (Abreha et al., 2022).
Despite yield reduction by drought, sorghum is considered more drought-resistant than many other crop plants (Hadebe et al., 2017) and shows a wide range of morphological, physiological, and biochemical adaptations in response to drought stress (Liu et al., 2024). When exposed to drought, older sorghum leaves are selectively killed, while the younger leaves remain physiologically functional as a result of osmotic adjustment in the younger leaves (Blum, 2005). Sorghum plants can have higher water use efficiency because they can reduce evapotranspiration more efficiently (Tolk and Howell, 2003). This is most likely because drought-tolerant sorghums tend to produce more epicuticular wax on their leaf surface compared to sensitive ones during drought stress (Sanjari et al., 2021). Further support comes from overexpressing a sorghum WINL1, which simultaneously increases total wax/cutin content and drought tolerance in Arabidopsis (Bao et al., 2017). Another reason may be that during drought, drought-tolerant sorghum plants show more leaf rolling than the susceptible lines, reducing the effective evapotranspiration area of the uppermost leaves by approximately 75% (Matthews et al., 1990). In addition to this leaf feature, sorghum plants tend to penetrate deeper into the subsoil (40–135 cm) (Schittenhelm and Schroetter, 2014; Singh and Singh, 1995), and more roots are produced during drought (de Oliveira et al., 2022; Queiroz et al., 2019). A combination of these two factors may account for up to 90% of the total water used by sorghum (Rachidi et al., 1993). Therefore, this root feature (more and deeper roots) has been found to be a major contributor to drought tolerance in sorghum (Wright and Smith, 1983). At the physiological level, drought induces the large central vacuole to form small vesicles when the leaf water potential is at −37 bars; this maintains tonoplast integrity and allows sorghum plants to withstand drought (Giles et al., 1976). It is not surprising that drought elicits extensive genetic (Abreha et al., 2022) and proteomic responses (Li et al., 2020) in sorghum.
Because of its importance, drought tolerance has been extensively mapped in sorghum. By searching drought tolerance-related traits in the Sorghum quantitative trait locus (QTL) Atlas (Mace et al., 2019), 817 loci were identified from 19 studies published before 2019. More recently, Tsehaye et al. (2024) mapped 32 drought-related quantitative trait nucleotides (QTNs) using an association mapping panel of 216 diverse accessions and 17,637 Single Nucleotide Polymorphism (SNP) markers, four of which colocated with previously mapped drought-related QTLs. Faye et al. (2022) mapped 16 pleiotropic associations for drought responses across water stress environments using an association mapping panel of 590 predominantly West African sorghum landraces and 130,709 SNPs. Crop landraces represent local adaptations of domesticated species and contribute novel alleles for adaptation to stressful environments (Dwivedi et al., 2016). Although larger panels (Faye et al., 2022; Tao et al., 2020, 2021) have been used in a sorghum genome-wide association study (GWAS), we found that the sorghum mini core panel is effective in QTL mapping. In a previous study, we mapped and cloned a pleiotropic QTL gene for plant height, days to 50% flowering, biomass, juice yield, and juice sugar content (Upadhyaya et al., 2022) using the sorghum mini core collection of 242 global landraces (Upadhyaya et al., 2009). The mini core panel has since been used to map sorghum panicle architecture (Wang et al., 2021), sorghum plant color (Wang et al., 2024), callus induction and regeneration from mature sorghum seeds (Xu et al., 2025), and additional developmental and reproductive traits (Upadhyaya et al., 2024).
In this study, the objective was to map drought tolerance loci that are pleiotropic for more than one trait or stable across environments. Drought stress was imposed by polyethylene glycol (PEG), which is commonly used to simulate drought in sorghum (Abdel-Ghany et al., 2020; Dugas et al., 2011; Jafar et al., 2004; Pavli et al., 2013; Queiroz et al., 2019), on the sorghum mini core panel. To carry out the mapping, we evaluated seedling shoot/root length, shoot/root fresh/dry weight, germination rate with and without osmotic stress, and drought indices, which were calculated as the ratio of growth under stressed and control conditions in 2020 and 2024 and performed a GWAS on the traits as previously described (Li et al., 2018) using 6,094,317 SNP markers (Upadhyaya et al., 2022; Wang et al., 2021; Xu et al., 2025). We identified 17 QTLs for shoot fresh and dry weight and drought index, along with five QTLs for other traits. A suite of candidate genes landed on by or closest to linked SNPs was also identified.
Materials and methods
Plant materials and osmotic stress assay
A mini core panel of 239 accessions (Upadhyaya et al., 2009) was used for this study. Uniform, full, and healthy seeds, free from mechanical damage or pest or disease infection, were used. The selected seeds were surface-sterilized with a 0.1% mercuric chloride (HgCl2) solution for 15 minutes and then rinsed thoroughly three times with sterile distilled water to remove any residual HgCl2. The sterilized seeds were treated with 10% and 20% polyethylene glycol (PEG-6000) solutions in 2020 (20_10 and 20_20, respectively) and with 10% PEG in 2024 (24_10) to simulate drought stress, with distilled water as the control. The 20% PEG treatment was not repeated in 2024 because it interfered with germination and subsequent seedling growth. Each treatment consisted of 30 seeds for each of the three replicates in each accession. Both the control and treatment seeds germinated on two pieces of special blotting paper (12 × 12 cm) in a germination box and were incubated in a plant growth chamber at a constant temperature of 28°C with a 16-hour light/8-hour dark photoperiod for 10 days. On the 10th day, 10 uniformly growing seedlings were selected from each treatment. Shoot length (SL) and root length (RL) were measured using a ruler to the nearest millimeter. Fresh weights of shoots and roots (SFW and RFW, respectively) were determined using an electronic balance with a precision of 0.0001g. The shoots and roots were then dried at 75°C for 24 hours and cooled to room temperature, and their dry weights (SDW and RDW, respectively) were measured using the same balance. The germination rate (GR) was recorded for 2024.
The drought tolerance was assessed using the seedling tolerance coefficient (STC) (Qiu et al., 2007; Yu et al., 2021) and was calculated as follows:
The STC calculated for each trait was denoted as RLSTC for root length, while root length for the 10% or 20% PEG treatment conducted in 2020 was denoted as RLPEG20_10 and RLPEG20_20, respectively; the control was denoted as RL20. A list of all traits is provided in Supplementary Table S1.
Association mapping
GWAS was conducted as described (Li et al., 2018; Upadhyaya et al., 2022, 2024; Wang et al., 2021, 2024; Xu et al., 2025). In short, GWAS for the 51 traits (listed together with all Manhattan plots in Supplementary Figure S1 and Supplementary Table S1) was performed using 6,094,317 SNPs. A kinship matrix (K) was generated with EMMAX (Kang et al., 2010), and a Q matrix was calculated using STRUCTURE 2.3.4 (Pritchard et al., 2000). Both matrices were used to perform GWAS in an Mixed Linear Model (MLM) model (Yu et al., 2006). The modified Bonferroni correction was used to determine association significance thresholds. At a nominal level of α = 0.05, the threshold p-value was 8.2 × 10−9, or a −log10(p) value of 8.08. As in previous studies (Upadhyaya et al., 2022; 2024), we also included markers with p-values below 10−4 (Famoso et al., 2011; Zhao et al., 2011) to account for associations of multiple markers at a locus across more than two traits to declare an association.
QTL colocalization and identification of candidate genes
As described by Upadhyaya et al. (2024), to identify colocalizing QTLs mapped in this study based on physical location, previously mapped QTLs downloaded from the Sorghum QTL Atlas (Mace et al., 2019) and those by Faye et al. (2022) were used. The location of candidate genes was identified with the BTx623 reference sequence (whose complete genome is now available; Wei et al., 2024), S. bicolor v3.1.1 (McCormick et al., 2018) at Phytozome 13 (Goodstein et al., 2012). Genes, including linked SNP markers with p-values below 10−4, were considered candidate genes based on previous studies that showed that linked markers can land on the causal genes (Upadhyaya et al., 2022; Wang et al., 2016; Zhang et al., 2023).
Statistical analysis
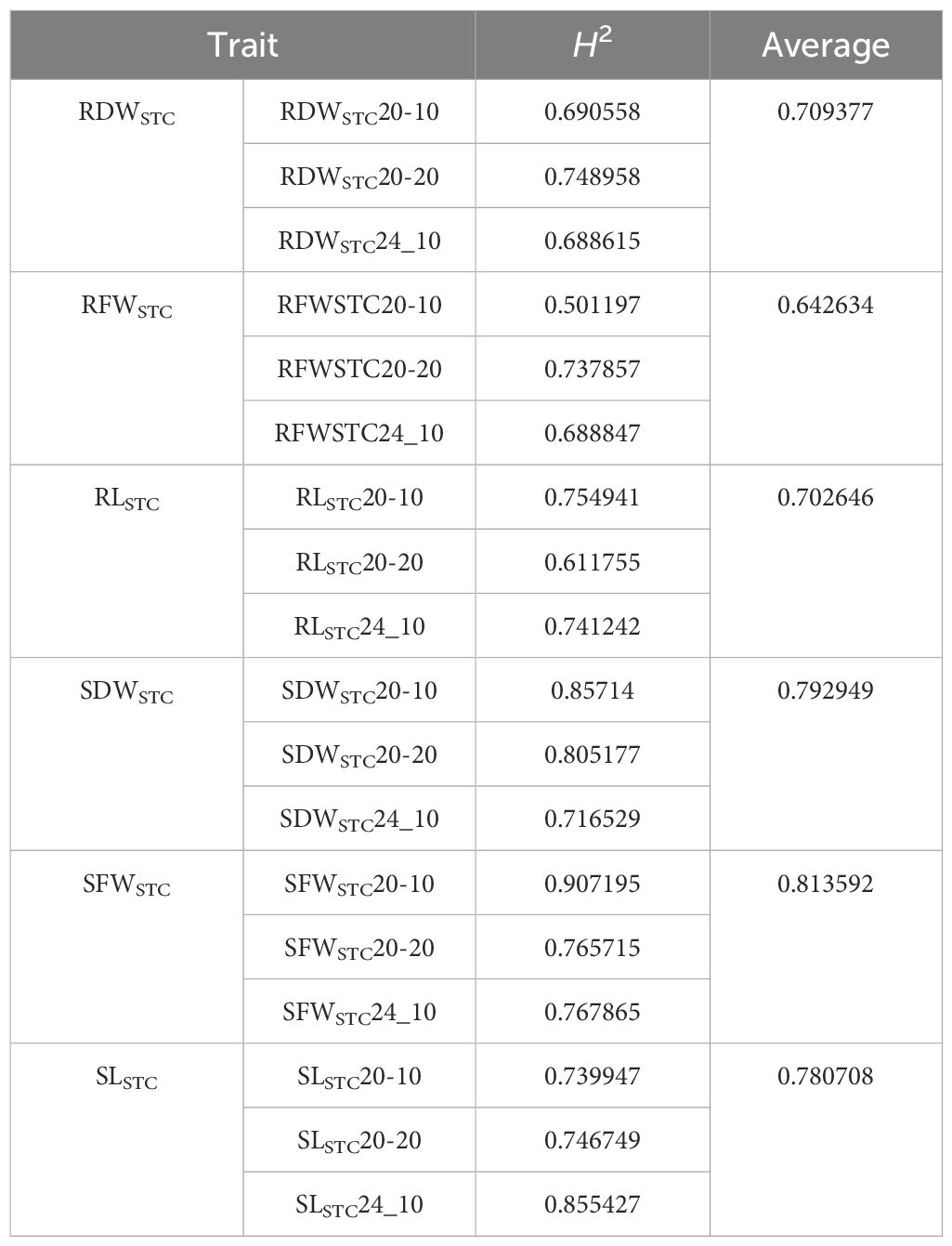
Pearson’s correlation coefficient (r) was calculated using Excel’s PEARSON function. Its significance was tested using a table of critical values. The assumptions of analysis of variance (ANOVA), i.e., data normality and variance homogeneity, were confirmed using the Kolmogorov–Smirnov test (see Supplementary Table S2). ANOVA was performed using the SPSS V29.0 statistical software with a general linear model. The variance components generated from the ANOVA were used to calculate the broad-sense heritability (H2) for the RLSTC, RDWSTC, RFWSTC, SLSTC, SDWSTC, and SFWSTC using the following formula:
where Vg, Vge, and Ve are genetic variance, the genotype × environment interaction variance, and environmental variance, respectively (Smith et al., 1998; Upadhyaya et al., 2024).
Results
Phenotypic analysis
We analyzed phenotypic variation among replicates for each accession. We found that if ranked by minimal dispersion using standard deviation (SD) for SLPEG20_10, three of the top four accessions (IS12302, IS20697, and IS2382) were all caudatum, and one (IS30466) was caudatum-bicolor (Supplementary Table S3). Pearson’s correlation between the STC traits of SFWSTC20_20 and SDWSTC20_20 was highest (r = 0.95; r = 0.77) between SFWSTC20_10 and SDWSTC20_10 and was r = 0.82 between SFWSTC24_10 and SDWSTC24_10; all significant at p < 0.001. We also observed a similar trend between RFWSTC and RDWSTC (Supplementary Table S4). This was followed by SLSTC and SFWSTC, and SLSTC and SDWSTC (both 0.84, significant at p < 0.001). We also found that SDWSTC20_20 was highly and significantly correlated with RDWSTC20_20, RFWSTC20_20, and SLSTC20_20 with r of 0.8, 0.77, and 0.84, respectively (Supplementary Table S4). Interestingly, RDWPEG20_10 was more highly correlated with SL20, RL20, SFW20, RFW20, SDW20, and RDW20 than with RDWPEG20_20 (Supplementary Table S4).
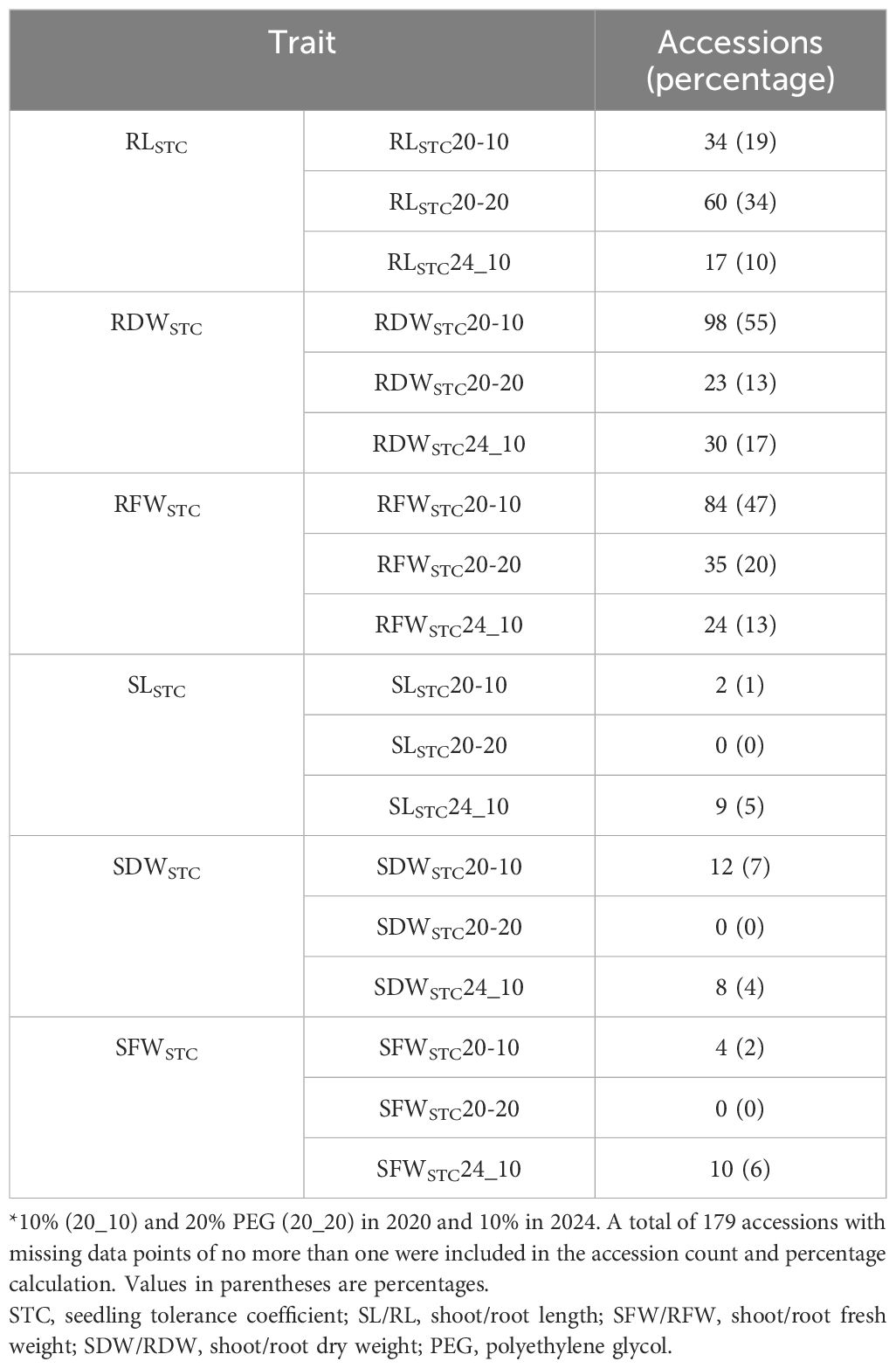
We found that 34%–55% of the mini core panel produced more root biomass and longer roots during osmotic stress (Table 1). At 10% PEG, more accessions produced more roots (47%–55%) than longer roots (19%), and this trend was also observed in the 2024 data; however, at 20% PEG, more accessions produced longer roots (34%) than more roots (13%–20%). On average, in 2020, the 10% PEG treatment reduced RL by 20%, and the 20% PEG treatment reduced RL by 72%. In 2024, when only 10% PEG was used, RL was reduced by 34% due to the treatment. When ranked by RLSTC, none of the bottom 70 accessions produced longer roots in 2020 when treatment was increased from 10% to 20% PEG (Supplementary Table S5). Furthermore, no accessions invested in shoot growth at 20% PEG, although a few random accessions (2%–7%) did show increased shoot weight at 10% PEG in both years (Table 1). This clearly demonstrates that osmotic stress greatly reduces shoot growth. Interestingly, all three root STC traits, RLSTC, RDWSTC, and RFWSTC, had slightly lower broad-sense heritability than the shoot traits (Table 2). This was most likely due to the non-genetic root response to osmotic stress, as the root was in direct contact with the stressor.
Based on RLSTC and RDWSTC, we identified one drought-tolerant (IS 30533) and one sensitive (IS 32439) accession. On average, the IS 32439 root length and dry weight were reduced by the PEG treatment by 64% and 71%, respectively, while in IS 30533, these were increased by 20% and 19%, respectively. Still, for IS 30533, shoot length and dry weight were decreased by 40% and 37%, respectively, by the treatment, while they were 65% and 60%, respectively, for IS 32439 (Supplementary Table S5).
Association mapping
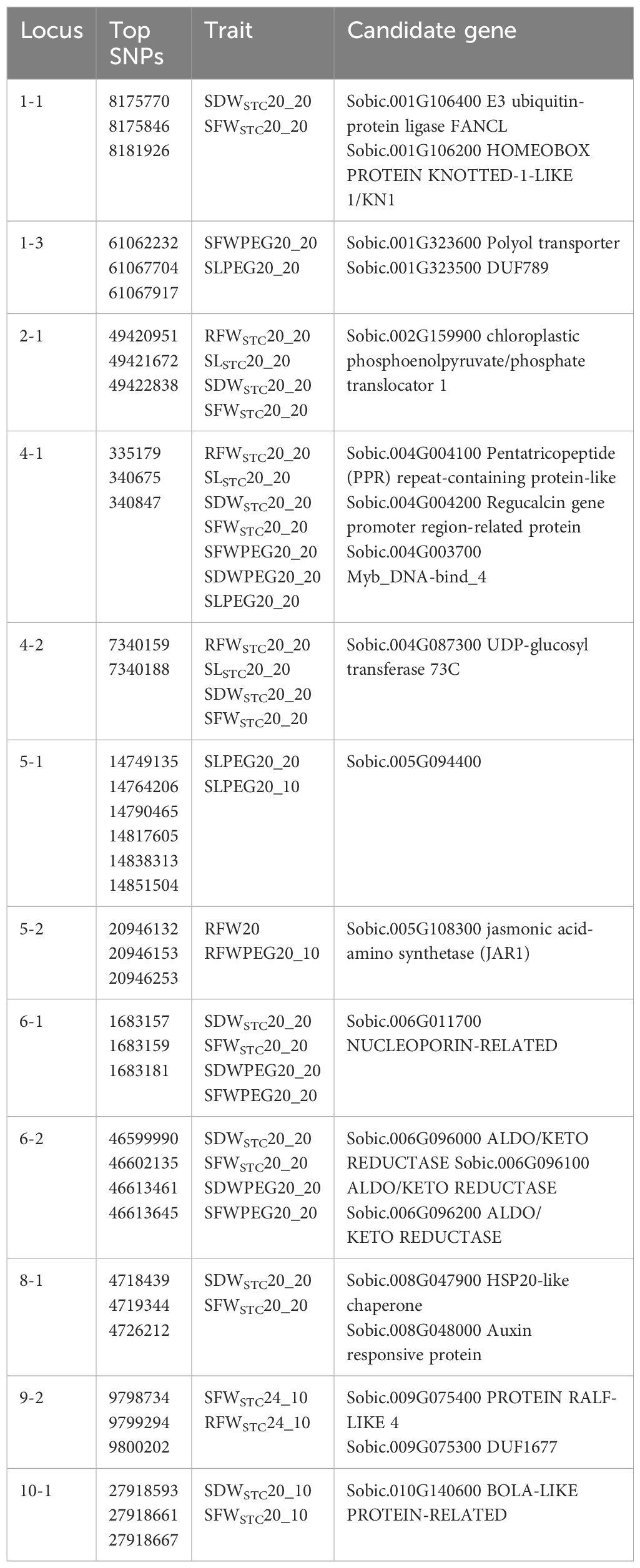
We applied the same criterion used in previous studies (Upadhyaya et al., 2022, 2024; Wang et al., 2021) for trait mapping in the sorghum mini core panel, which defines a significant association as multiple SNPs linked to a trait within the same locus with a p-value < 10−4. In addition to identifying trait-specific loci, this study also focused on pleiotropic loci—those associated with more than one trait. With this criterion, we identified 22 loci linked to 12 drought-related traits (Table 3; all 51 Manhattan plots are provided in Supplementary Figure S1). SDWSTC and SFWSTC had the highest correlation coefficient (Supplementary Table S4). Here, we report that 17 of the 22 mapped loci were pleiotropic for the two traits and that 19 of the 22 loci were mapped to the STC traits (Table 3). Seven traits (RFWSTC20_20, SLSTC20_20, SDWSTC20_20, SFWSTC20_20, SFWPEG20_20, SDWPEG20_20, and SLPEG20_20) were all mapped to the 4–1 locus. Loci 2–1 and 4-2 (RFWSTC20_20, SLSTC20_20, SDWSTC20_20, and SFWSTC20_20) and 6–1 and 6-2 (SDWSTC20_20, SFWSTC20_20, SDWPEG20_20, and SFWPEG20_20) were all pleiotropic for four traits, while all other loci were pleiotropic for two traits (Table 3).
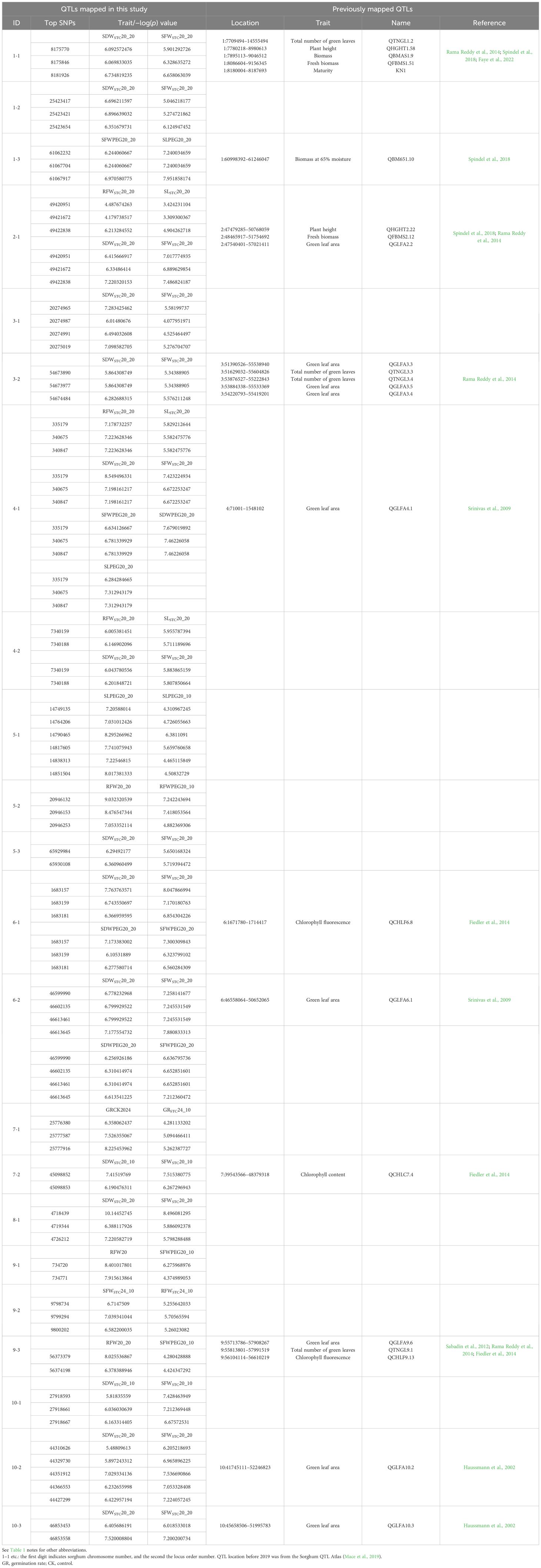
Table 3. Drought tolerance-related loci mapped in the sorghum mini core panel and their colocation with previously mapped drought-related QTLs.
Colocation with previously mapped drought-related QTLs
Eleven (1-1, 1-3, 2-1, 3-3, 4-1, 6-1, 6-2, 7-2, 9-3, 10-2, and 10-3) of the 22 loci were colocated with 23 previously mapped QTLs (Table 3). Among the 23 QTLs, 15 were associated with drought-related leaf features: nine (QGLFA2.2, QGLFA3.3, QGLFA3.5, QGLFA3.4, QGLFA4.1, QGLFA6.1, QGLFA9.6, QGLFA10.2, and QGLFA10.3) were associated with green leaf area, four (QTNGL1.2, QTNGL3.3, QTNGL3.4, and QTNGL9.1) were associated with the total number of green leaves, and two (QCHLF6.8 and QCHLF9.13) were associated with chlorophyll content (Table 3). Among the 11 loci, 1–1 and 3–2 were each colocated with five QTLs, while 2–1 and 9–3 were each colocated with three QTLs; the rest were colocated with one QTL (Table 3).
Candidate genes identified by linked SNPs
Candidate genes were identified because they had linked SNP markers that landed in coding, or 5′/3′ regions, or were closest to the linked SNPs. Using these criteria, we found 19 candidate genesmacross 12 of the 22 loci (Table 4). Five candidate genes –a transporter (Sobic.001G323600) in locus 1-3, a UDP-glucosyl transferase (Sobic.004G087300) in locus 4-2, and three aldo/keto reductases (Sobic.006G096000, Sobic.006G096100, and Sobic.006G096200) in locus 6-2 –showed consistently high expression in the roots based on data available from GeneAtlas v2 FPKM (McCormick et al., 2018). In addition, a nucleoporin gene (Sobic.006G011700) displayed shoot-specific expression (Supplementary Table S6). Haplotypes based on the three SNPs (46613461, 46613645, and 46615325) located in the promoter and coding regions of Sobic.006G096100 in locus 6-2 showed that IS 30533 had TTC while IS 34239 had CCT at these positions (Supplementary Table S7).
Discussion
In this study, we evaluated a sorghum mini core panel for shoot and root growth under simulated drought conditions imposed by 10%/and 20% PEG. The results showed that certain accessions exhibited enhanced root growth—through either increased root number or elongation—under osmotic stress. A greater number of accessions produced more roots rather than longer roots when exposed to 10% PEG. However, at 20% PEG, more accessions exhibited longer roots than increased root number, reflecting the adaptability of some accessions to drought stress. These findings are consistent with those of previous studies (Bibi et al., 2010; de Oliveira et al., 2022; Schittenhelm and Schroetter, 2014; Singh and Singh, 1995). Based on the response of root length and dry weight (RLSTC and RDWSTC), IS 30533 was identified as the most drought-tolerant, while IS 32439 was the most sensitive accession. These accessions may be of particular interest for sorghum breeding programs targeting improved drought tolerance.
Drought stress significantly and negatively impacts sorghum growth (Abreha et al., 2022), especially shoot growth (Jafar et al., 2004), which was most significantly reduced by PEG treatments as measured by shoot fresh and dry weight STC (SDWSTC and SFWSTC) (Table 1). This explains why among the 22 loci identified, 19 were mapped to the STC traits, which reflect the impact on growth, and 17 were mapped to SDWSTC and SFWSTC. Half of the mapped loci are also colocated with 23 previously mapped drought-related QTLs; 15 of these 23 QTLs were mapped to green leaf area, total number of green leaves, or chlorophyll content (Table 3). We also found 19 candidate genes for 12 of the 22 loci. Five of those genes show either preferential or specific expression in the roots (Supplementary Table S6). The relevance of some of these candidate genes to drought tolerance is explained in the following sections.
When exposed to drought stress, the immediate response must be to protect the cell. One candidate gene identified in locus 8–1 encodes Hsp20, which has been found to be induced by drought stress in sorghum (Abdel-Ghany et al., 2020; Zhang et al., 2024). As a small heat shock protein, Hsp20, may form a complex with a variety of non-native proteins to form a first line of defense against protein aggregation during stress (Haslbeck and Vierling, 2015). Drought also induces transporters for various solutes (Dong et al., 2014), and we found one polyol transporter in locus 1-3 and a chloroplastic phosphoenolpyruvate/phosphate translocator (PPT) in locus 2-1. a polyol transporter is an H+-dependent plasma membrane carrier that transports mannitol and sorbitol, which protect cells against osmotic stress (Shen et al., 1999) and are induced in grapevines by drought (Conde et al., 2015). PPT imports phosphoenolpyruvate (PEP) to the plastid from the cytosol. A loss-of-function mutant of PPT1 in Arabidopsis results in stunted roots (Staehr et al., 2014), potentially compromising the ability of roots to cope with drought. Regarding transporters, we identified in locus 6 -1 a nucleoporin that is the main transport channel between the cytoplasm and the nucleoplasm, and a maize nucleoporin, ZmNUP58, has been shown to play an important role in the stress response of maize. ZmNUP58 overexpression in maize significantly promotes both chlorophyll content and activities of antioxidant enzymes under drought conditions (Liu et al., 2022). Coincidentally, a chlorophyll fluorescence QTL (QCHLF6.8) (Fiedler et al., 2014) is also colocated in this locus (Table 3), demonstrating the effectiveness of drought QTL mapping using the mini core panel in this study. In sorghum, the stay-green trait contributes to the adaptation to post-flowering drought conditions (Abreha et al., 2022). Since drought reduces sorghum leaf chlorophyll content (Kapanigowda et al., 2013), increased chlorophyll content during drought is a sign of drought tolerance (Kassahun et al., 2010). For this reason, we found at least five QTL clusters from six studies in which stay-green loci overlap with chlorophyll content loci: one each on chromosomes 2 and 10, and three on chromosome 3 (Supplementary Table S8).
Sorghum also exhibits physiological and biochemical resistance to drought by scavenging reactive oxygen species (ROS) and changing the activity of its antioxidant enzymes (Liu et al., 2024). For ROS scavenging, a BolA protein identified in locus 10 -1 may play a negative role in ROS scavenging, as a mutation in Arabidopsis BolA causes the plant to produce longer roots and to scavenge ROS, implying an increased capacity to extract deeper soil water (Qin et al., 2015). Another example of a ROS scavenger (Yu et al., 2020) is the three aldo-keto reductases (AKR) in locus 6-2, which are mostly expressed in the roots (Supplementary Table S6). In tomatoes, the majority of AKR genes are induced by drought treatments, and silencing AKR expression reduces drought tolerance due to low proline content and high malondialdehyde content, indicating AKRs’ positive role in regulating drought tolerance in tomatoes (Guan et al., 2023). So is the E3 ubiquitin-protein ligase in locus 1-1. A rice U-box E3 ubiquitin ligase (OsPUB67) was significantly induced by drought, and its overexpression enhances the reactive oxygen species scavenging ability and stomatal closure, which improves drought tolerance (Qin et al., 2020). For antioxidant activities, we found a UDP-glucosyl transferase (UGT) in locus 4-2. Overexpressing two Arabidopsis UGTs, UGT79B2 and UGT79B3, increases drought tolerance thanks to increased anthocyanin accumulation and enhanced antioxidant activity in coping with drought (Li et al., 2017), and similar results have also been reported in rice (Dong et al., 2020).
The last candidate gene to be described is jasmonic acid-amino synthetase1 (JAR1) in locus 5-2. Jasmonic acid (JA) is of central importance in drought stress responses (Wasternack, 2014). JAR1 is involved in conjugating JA to Ile, the bioactive form of JA (Staswick and Tiryaki, 2004). JAR1 plays a major role in JA signaling (Kazan and Manners, 2008), and its expression is upregulated in the early stages of drought and decreased upon persistent drought (Chen et al., 2019). Overexpressing JAR1 reduces water loss during drought, while its mutation lowers JA–Ile content and causes hypersensitivity to drought (Mahmud et al., 2022).
In conclusion, we evaluated a sorghum mini core panel for tolerance to drought simulated by PEG. We confirmed results from previous studies that sorghum plants produced more roots than longer roots at 10% PEG, but at 20% PEG, they produced longer roots than more roots, and PEG reduced shoot growth in all accessions in both years. GWAS identified 22 loci, 19 of which were mapped to the STC traits, and 17 of the 19 were mapped to the STC of shoot weight. Eleven of the 22 loci were colocated with 15 QTLs that had been previously mapped to green leaf area, the total number of green leaves, or chlorophyll content. Of the 19 candidate genes from the 12 loci mapped, five showed either preferential or specific expression in the roots according to GeneAtlas v2. One of the candidate genes from locus 6-1, colocated with a previously mapped chlorophyll fluorescence QTL, was found to increase chlorophyll fluorescence in another study. Sorghum leaf chlorophyll content is closely associated with drought tolerance. IS 30533 was the most tolerant accession, and IS 32439 was the most sensitive accession. The results from this study will facilitate sorghum marker-assisted breeding for drought tolerance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
HM: Data curation, Writing – review & editing. KW: Data curation, Writing – review & editing. TW: Data curation, Writing – review & editing. XC: Data curation, Writing – review & editing. EH: Software, Writing – review & editing. YW: Writing – review & editing, Data curation. DH: Writing – review & editing, Data curation. YW: Software, Writing – original draft, Methodology. LW: Software, Writing – original draft, Methodology.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the National Natural Science Foundation of China (32372134), PhD Stable Talent Funding (No. NXWD202401), and the Chuzhou “Star of Innovation and Entrepreneurship” Industrial Innovation Team.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1629615/full#supplementary-material
References
Abdel-Ghany, S. E., Ullah, F., Ben-Hur, A., and Reddy, A. S. N. (2020). Transcriptome analysis of drought-resistant and drought-sensitive sorghum (Sorghum bicolor) genotypes in response to PEG-induced drought stress. Int. J. Mol. Sci. 21, 772. doi: 10.3390/ijms21030772
Abreha, K. B., Enyew, M., Carlsson, A. S., Vetukuri, R. R., Feyissa, T., Motlhaodi, T., et al. (2022). Sorghum in dryland: morphological, physiological, and molecular responses of sorghum under drought stress. Planta 255, 20. doi: 10.1007/s00425-021-03799-7
Assefa, Y., Staggenborg, S. A., and Prasad, V. P. (2010). Grain sorghum water requirement and responses to drought stress: A review. Crop Manag. 9, 1–11. doi: 10.1094/CM-2010-1109-01-RV
Bao, S. G., Shi, J. X., Luo, F., Ding, B., Hao, J. Y., Xie, X. D., et al. (2017). Overexpression of Sorghum WINL1 gene confers drought tolerance in Arabidopsis thaliana through the regulation of cuticular biosynthesis. Plant Cell Tiss. Organ Cult. 128, 347–356. doi: 10.1007/s11240-016-1114-2
Bibi, A., Sadaqat, H. A., Akram, H. M., and Mohammed, M. I. (2010). Physiological markers for screening sorghum (Sorghum bicolor) germplasm under water stress condition. Int. J. Agric. Biol. 12, 451–455.
Blum, A. (2005). Drought resistance, water-use efficiency, and yield potential—are they compatible, dissonant, or mutually exclusive? Aust. J. Agri Res. 56, 1159–1168. doi: 10.1071/AR05069
Chen, Y., Chen, Y., Shi, Z., Jin, Y., Sun, H., Xie, F., et al. (2019). Biosynthesis and signal transduction of ABA, JA, and BRs in response to drought stress of kentucky bluegrass. Int. J. Mol. Sci. 20, 1289. doi: 10.3390/ijms20061289
Conde, A., Regalado, A., Rodrigues, D., Costa, J. M., Blumwald, E., Chaves, M. M., et al. (2015). Polyols in grape berry: transport and metabolic adjustments as a physiological strategy for water-deficit stress tolerance in grapevine. J. Exp. Bot. 66, 889–906. doi: 10.1093/jxb/eru446
de Oliveira, J. P. V., Duarte, V. P., de Castro, E. M., Magalhães, P. C., and Pereira, F. J. (2022). Stomatal cavity modulates the gas exchange of Sorghum bicolor (L.) Moench. grown under different water levels. Protoplasma 259, 1081–1097. doi: 10.1007/s00709-021-01722-1
Dong, Y., Fan, G., Zhao, Z., and Deng, M. (2014). Compatible solute, transporter protein, transcription factor, and hormone-related gene expression provides an indicator of drought stress in Paulownia fortunei. Funct. Integr. Genomics 14, 479–491. doi: 10.1007/s10142-014-0373-4
Dong, N. Q., Sun, Y., Guo, T., Shi, C. L., Zhang, Y. M., Kan, Y., et al. (2020). UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 11, 2629. doi: 10.1038/s41467-020-16403-5
Dugas, D. V., Monaco, M. K., Olsen, A., Klein, R. R., Kumari, S., Ware, D., et al. (2011). Functional annotation of the transcriptome of Sorghum bicolor in response to osmotic stress and abscisic acid. BMC Genomics 12, 514. doi: 10.1186/1471-2164-12-514
Dwivedi, S. L., Ceccarelli, S., Blair, M. W., Upadhyaya, H. D., Are, A. K., and Ortiz, R. (2016). Landrace germplasm for improving yield and abiotic stress adaptation. Trends Plant Sci. 21, 31–42. doi: 10.1016/j.tplants.2015.10.012
Eck, H. V. and Musick, J. T. (1979). Plant water stress effects on irrigated grain sorghum. I. effects on yield 1. Crop Sci. 19, 589–592. doi: 10.2135/cropsci1979.0011183X001900050009x
Famoso, A. N., Zhao, K., Clark, R. T., Tung, C. W., Wright, M. H., Bustamante, C., et al. (2011). Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PloS Genet. 7, e1002221. doi: 10.1371/journal.pgen.1002221
Faye, J. M., Akata, E. A., Sine, B., Diatta, C., Cisse, N., Fonceka, D., et al. (2022). Quantitative and population genomics suggest a broad role of stay-green loci in the drought adaptation of sorghum. Plant Genome 15, e20176. doi: 10.1002/tpg2.20176
Fiedler, K., Bekele, W. A., Duensing, R., Gründig, S., Snowdon, R., Stützel, H., et al. (2014). Genetic dissection of temperature-dependent sorghum growth during juvenile development. Theor. Appl. Genet. 127, 1935–1948. doi: 10.1007/s00122-014-2350-7
Giles, K. L., Cohen, D., and Beardsell, M. F. (1976). Effects of water stress on the ultrastructure of leaf cells of sorghum bicolor. Plant Physiol. 57, 11–14. doi: 10.1104/pp.57.1.11
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Guan, X., Yu, L., and Wang, A. (2023). Genome-wide identification and characterization of aldo-keto reductase (AKR) gene family in response to abiotic stresses in solanum lycopersicum. Int. J. Mol. Sci. 24, 1272. doi: 10.3390/ijms24021272
Hadebe, S. T., Modi, A. T., and Mabhaudhi, T. (2017). Drought tolerance and water use of cereal crops: A focus on sorghum as a food security crop in sub-Saharan Africa. J. Agron. Crop Sci. 203, 177–191. doi: 10.1111/jac.12191
Haslbeck, M. and Vierling, E. (2015). A first line of stress defense: small heat shock proteins and their function in protein homeostasis. J. Mol. Biol. 427, 1537–1548. doi: 10.1016/j.jmb.2015.02.002
Haussmann, B. I., Mahalakshmi, V., Reddy, B. V., Seetharama, N., Hash, C. T., and Geiger, H. H. (2002). QTL mapping of stay-green in two sorghum recombinant inbred populations. Theor. Appl. Genet. 106, 133–142. doi: 10.1007/s00122-002-1012-3
Jafar, M. S., Nourmohammadi, G., and Maleki, A. (2004). “Effect of water deficit on seedling, plantlets and compatible solutes of forage Sorghum cv. Speedfeed,” in Proceedings of the 4th International Crop Science Congress Brisbane, Austrialia. (Brisbane, Austrialia: Australian Society of Agronomy Inc.).
Kang, H. M., Sul, J. H., Service, S. K., Zaitlen, N. A., Kong, S. Y., Freimer, N. B., et al. (2010). Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354. doi: 10.1038/ng.548
Kapanigowda, M. H., Perumal, R., Djanaguiraman, M., Aiken, R. M., Tesso, T., Prasad, P. V., et al. (2013). Genotypic variation in sorghum [Sorghum bicolor (L.) Moench] exotic germplasm collections for drought and disease tolerance. Springerplus 2, 650. doi: 10.1186/2193-1801-2-650
Kassahun, B., Bidinger, F. R., Hash, C. T., and Kuruvinashetti, M. S. (2010). Stay-green expression in early generation sorghum [Sorghum bicolor (L.) Moench] QTL introgression lines. Euphytica 172, 351–362. doi: 10.1007/s10681-009-0108-0
Kazan, K. and Manners, J. M. (2008). Jasmonate signaling: toward an integrated view. Plant Physiol. 146, 1459–1468. doi: 10.1104/pp.107.115717
Li, H., Li, Y., Ke, Q., Kwak, S. S., Zhang, S., and Deng, X. (2020). Physiological and Differential Proteomic Analyses of Imitation Drought Stress Response in Sorghum bicolor Root at the Seedling Stage. Int. J. Mol. Sci. 21, 9174. doi: 10.3390/ijms21239174
Li, P., Li, Y. J., Zhang, F. J., Zhang, G. Z., Jiang, X. Y., Yu, H. M., et al. (2017). The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 89, 85–103. doi: 10.1111/tpj.13324
Li, J., Tang, W., Zhang, Y. W., Chen, K. N., Wang, C., Liu, Y., et al. (2018). Genome-wide association studies for five forage quality-related traits in sorghum (Sorghum bicolor L.). Front. Plant Sci. 9, 1146. doi: 10.3389/fpls.2018.01146
Liu, Z., Abou-Elwafa, S. F., Xie, J., Liu, Y., Li, S., Aljabri, M., et al. (2022). A Nucleoporin NUP58 modulates responses to drought and salt stress in maize (Zea mays L.). Plant Sci. 320, 111296. doi: 10.1016/j.plantsci.2022.111296
Liu, J., Wang, X., Wu, H., Zhu, Y., Ahmad, I., Dong, G., et al. (2024). Association between reactive oxygen species, transcription factors, and candidate genes in drought-resistant sorghum. Int. J. Mol. Sci. 25, 6464. doi: 10.3390/ijms25126464
Mace, E., Innes, D., Hunt, C., Wang, X., Tao, Y., Baxter, J., et al. (2019). The Sorghum QTL Atlas: a powerful tool for trait dissection, comparative genomics and crop improvement. Theor. Appl. Genet. 132, 751–766. doi: 10.1007/s00122-018-3212-5
Mahmud, S., Ullah, C., Kortz, A., Bhattacharyya, S., Yu, P., Gershenzon, J., et al. (2022). Constitutive expression of JASMONATE RESISTANT 1 induces molecular changes that prime the plants to better withstand drought. Plant Cell Environ. 45, 2906–2922. doi: 10.1111/pce.14402
Matthews, R. B., Azam-Ali, S. N., and Peacock, J. M. (1990). Response of four sorghum lines to mid-season drought. II. Leaf characteristics. Field Crops Res. 25, 297–308. doi: 10.1016/0378-4290(90)90011-Y
McCormick, R. F., Truong, S. K., Sreedasyam, A., Jenkins, J., Shu, S., Sims, D., et al. (2018). The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 93, 338–354. doi: 10.1111/tpj.13781
Pavli, O. I., Vlachos, C. E., Kalloniati, C., Flemetakis, E., and Skaracis, G. N. (2013). Metabolite profiling reveals the effect of drought on sorghum (Sorghum bicolor L. Moench) metabolism. Plant Omics 6, 371–376.
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Qin, Q., Wang, Y., Huang, L., Du, F., Zhao, X., Li, Z., et al. (2020). A U-box E3 ubiquitin ligase OsPUB67 is positively involved in drought tolerance in rice. Plant Mol. Biol. 102, 89–107. doi: 10.1007/s11103-019-00933-8
Qin, L., Wang, M., Zuo, J., Feng, X., Liang, X., Wu, Z., et al. (2015). Cytosolic bolA plays a repressive role in the tolerance against excess iron and MV-induced oxidative stress in plants. PloS One 10, e0124887. doi: 10.1371/journal.pone.0124887
Qiu, F., Zheng, Y., Zhang, Z., and Xu, S. (2007). Mapping of QTL associated with waterlogging tolerance during the seedling stage in maize. Ann. Bot. 99, 1067–1081. doi: 10.1093/aob/mcm055
Queiroz, M. S., Oliveira, C. E., Steiner, F., Zuffo, A. M., Zoz, T., Vendruscolo, E. P., et al. (2019). Drought stresses on seed germination and early growth of maize and sorghum. J. Agric. Sci. 11, 310–318. doi: 10.5539/jas.v11n2p310
Rachidi, F., Kirkham, M. B., Stone, L. R., and Kanemasu, E. T. (1993). Soil water depletion by sunflower and sorghum under rainfed conditions. Agri Water Manag. 24, 49–62. doi: 10.1016/0378-3774(93)90061-E
Rama Reddy, N. R., Ragimasalawada, M., Sabbavarapu, M. M., Nadoor, S., and Patil, J. V. (2014). Detection and validation of stay-green QTL in post-rainy sorghum involving widely adapted cultivar, M35–1 and a popular stay-green genotype B35. BMC Genomics 15, 909. doi: 10.1186/1471-2164-15-909
Sabadin, P. K., Malosetti, M., Boer, M. P., Tardin, F. D., Santos, F. G., Guimarães, C. T., et al. (2012). Studying the genetic basis of drought tolerance in sorghum by managed stress trials and adjustments for phenological and plant height differences. Theor. Appl. Genet. 124, 1389–1402. doi: 10.1007/s00122-012-1795-9
Sanjari, S., Shobbar, Z. S., Ghanati, F., Afshari-Behbahanizadeh, S., Farajpour, M., Jokar, M., et al. (2021). Molecular, chemical, and physiological analyses of sorghum leaf wax under post-flowering drought stress. Plant Physiol. Biochem. 159, 383–391. doi: 10.1016/j.plaphy.2021.01.001
Schittenhelm, S. and Schroetter, S. (2014). Comparison of drought tolerance of maize, sweet sorghum and sorghum-Sudangrass hybrids. J. Agron. Crop Sci. 200, 46–53. doi: 10.1111/jac.12039
Shen, B., Hohmann, S., Jensen, R. G., and Bohnert, A. H. (1999). Roles of sugar alcohols in osmotic stress adaptation. Replacement of glycerol by mannitol and sorbitol in yeast. Plant Physiol. 121, 45–52. doi: 10.1104/pp.121.1.45
Singh, B. R. and Singh, D. P. (1995). Agronomic and physiological responses of sorghum, maize and pearl millet to irrigation. Field Crops Res. 42, 57–67. doi: 10.1016/0378-4290(95)00025-L
Smith, S. E., Kuehl, R. O., Ray, I. M., Hui, R., and Soleri, D. (1998). Evaluation of simple methods for estimating broad-sense heritability in stands of randomly planted genotypes. Crop Sci. 38, 1125–1129. doi: 10.2135/cropsci1998.0011183X003800050003x
Spindel, J. E., Dahlberg, J., Colgan, M., Hollingsworth, J., Sievert, J., Staggenborg, S. H., et al. (2018). Association mapping by aerial drone reveals 213 genetic associations for Sorghum bicolor biomass traits under drought. BMC Genomics 19, 679. doi: 10.1186/s12864-018-5055-5
Srinivas, G., Satish, K., Madhusudhana, R., Reddy, R. N., Mohan, S. M., and Seetharama, N. (2009). Identification of quantitative trait loci for agronomically important traits and their association with genic-microsatellite markers in sorghum. Theor. Appl. Genet. 118, 1439–1454. doi: 10.1007/s00122-009-0993-6
Staehr, P., Löttgert, T., Christmann, A., Krueger, S., Rosar, C., Rolčík, J., et al. (2014). Reticulate leaves and stunted roots are independent phenotypes pointing at opposite roles of the phosphoenolpyruvate/phosphate translocator defective in cue1 in the plastids of both organs. Front. Plant Sci. 5, 126. doi: 10.3389/fpls.2014.00126
Staswick, P. E. and Tiryaki, I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16, 2117–2127. doi: 10.1105/tpc.104.023549
Stone, L. R. and Schlegel, A. J. (2006). Yield–water supply relationships of grain sorghum and winter wheat. Agron. J. 98, 1359–1366. doi: 10.2134/agronj2006.0042
Tao, Y., Trusov, Y., Zhao, X., Wang, X., Cruickshank, A. W., Hunt, C., et al. (2021). Manipulating assimilate availability provides insight into the genes controlling grain size in sorghum. Plant J. 108, 231–243. doi: 10.1111/tpj.15437
Tao, Y., Zhao, X., Wang, X., Hathorn, A., Hunt, C., Cruickshank, A. W., et al. (2020). Large-scale GWAS in sorghum reveals common genetic control of grain size among cereals. Plant Biotechnol. J. 18, 1093–1105. doi: 10.1111/pbi.13284
Tolk, J. A. and Howell, T. A. (2003). Water use efficiencies of grain sorghum grown in three USA southern Great Plains soils. Agric. Water Manag. 59, 97–111. doi: 10.1016/S0378-3774(02)00157-9
Tsehaye, Y., Menamo, T. M., Abay, F., Tadesse, T., and Bantte, K. (2024). Multi-locus genome-wide association study for grain yield and drought tolerance indices in sorghum accessions. Plant Genome 17, e20505. doi: 10.1002/tpg2.20505
Upadhyaya, H. D., Pundir, R. P., Dwivedi, S. L., Gowda, C. L., Reddy, V. G., and Singh, S. (2009). Developing a mini core collection of sorghum for diversified utilization of germplasm. Crop Sci. 49, 1769–1780. doi: 10.2135/cropsci2009.01.0014
Upadhyaya, H. D., Wang, L., Paterson, A. H., Gowda, C. L. L., Kumar, R., Li, J., et al. (2024). Association mapping identifies stable loci containing novel genes for developmental and reproductive traits in sorghum. Genome 67, 454–463. doi: 10.1139/gen-2024-0030
Upadhyaya, H. D., Wang, L., Prakash, C. S., Liu, Y., Gao, L., Meng, R., et al. (2022). Genome-wide association mapping identifies an SNF4 ortholog that impacts biomass and sugar yield in sorghum and sugarcane. J. Exp. Bot. 73, 3584–3596. doi: 10.1093/jxb/erac110
Wang, L., Tu, W., Jin, P., Liu, Y., Du, J., Zheng, J., et al. (2024). Genome-wide association study of plant color in Sorghum bicolor. Front. Plant Sci. 15, 1320844. doi: 10.3389/fpls.2024.1320844
Wang, L., Upadhyaya, H. D., Zheng, J., Liu, Y., Singh, S. K., Gowda, C. L. L., et al. (2021). Genome-wide association mapping identifies novel panicle morphology loci and candidate genes in sorghum. Front. Plant Sci. 12, 743838. doi: 10.3389/fpls.2021.743838
Wang, X., Wang, H., Liu, S., Ferjani, A., Li, J., Yan, J., et al. (2016). Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 48, 1233–1241. doi: 10.1038/ng.3636
Wasternack, C. (2014). Action of jasmonates in plant stress responses and development–applied aspects. Biotechnol. Adv. 32, 31–39. doi: 10.1016/j.biotechadv.2013.09.009
Wei, C., Gao, L., Xiao, R., Wang, Y., Chen, B., Zou, W., et al. (2024). Complete telomere-to-telomere assemblies of two sorghum genomes to guide biological discovery. Imeta 3, e193. doi: 10.1002/imt2.193
Wright, G. C. and Smith, R. C. (1983). Differences between two grain sorghum genotypes in adaptation to drought stress. II. Root water uptake and water use. Aust. J. Agri Res. 34, 627–636. doi: 10.1071/AR9830627
Xu, J., Wang, L., Liang, Y., Shen, Q., Tu, W., Cheng, Z., et al. (2025). Association mapping and identification of candidate genes for callus induction and regeneration using sorghum mature seeds. Front. Plant Sci. 16, 1430141. doi: 10.3389/fpls.2025.1430141
Yu, J., Pressoir, G., Briggs, W. H., Vroh Bi, I., Yamasaki, M., Doebley, J. F., et al. (2006). A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38, 203–208. doi: 10.1038/ng1702
Yu, J., Sun, H., Zhang, J., Hou, Y., Zhang, T., Kang, J., et al. (2020). Analysis of aldo-keto reductase gene family and their responses to salt, drought, and abscisic acid stresses in Medicago Truncatula. Int. J. Mol. Sci. 21, 754. doi: 10.3390/ijms21030754
Yu, R., Wang, G., Yu, X., Li, L., Li, C., Song, Y., et al. (2021). Assessing alfalfa (Medicago sativa L.) tolerance to salinity at seedling stage and screening of the salinity tolerance traits. Plant Biol. (Stuttg) 23, 664–674. doi: 10.1111/plb.13271
Zhang, Q., Dai, B., Fan, M., Yang, L., Li, C., Hou, G., et al. (2024). Genome-wide profile analysis of the Hsp20 family in lettuce and identification of its response to drought stress. Front. Plant Sci. 15, 1426719. doi: 10.3389/fpls.2024.1426719
Zhang, H., Yu, F., Xie, P., Sun, S., Qiao, X., Tang, S., et al. (2023). A Gγ protein regulates alkaline sensitivity in crops. Science 379, eade8416. doi: 10.1126/science.ade8416
Keywords: sorghum, mini core, GWAS, SNPs, drought tolerance, candidate genes
Citation: Min H, Wang K, Wang T, Cheng X, Habyarimana E, Wang Y, Hu D, Wang Y-H and Wang L (2025) Association mapping and candidate gene identification for drought tolerance in sorghum. Front. Plant Sci. 16:1629615. doi: 10.3389/fpls.2025.1629615
Received: 22 May 2025; Accepted: 30 June 2025;
Published: 25 July 2025.
Edited by:
Guoquan Liu, The University of Queensland, AustraliaReviewed by:
Sofie Pearson, The University of Queensland, AustraliaYongfu Tao, Chinese Academy of Agricultural Sciences, China
Copyright © 2025 Min, Wang, Wang, Cheng, Habyarimana, Wang, Hu, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi-Hong Wang, eWlob25nLndhbmdAbG91aXNpYW5hLmVkdQ==; Lihua Wang, d2FuZ2xpaHVhZXJyQDEyNi5jb20=
 Huiting Min
Huiting Min Kang Wang1,2
Kang Wang1,2 Yi-Hong Wang
Yi-Hong Wang Lihua Wang
Lihua Wang