- Division of Marine Life Science, Faculty of Fisheries Sciences, Hokkaido University, Hakodate, Japan
Although the role of phytohormones in higher plants is well established, their role in macroalgae remains poorly understood. 1-Aminocyclopropane-1-carboxylic acid (ACC) is the immediate precursor of the plant hormone ethylene. However, recent studies suggested that ACC also acts as a signaling molecule to regulate growth and development independently of ethylene biosynthesis in land plants and red algae. This study investigated the role of ACC in the sporophytes of the red alga Pyropia yezoensis.
ACC treatment significantly inhibited the growth of P. yezoensis sporophytes, whereas ethephon, an ethylene-releasing compound, had no such effect. In addition, ACC treatment promoted the degradation of photosynthetic pigments, including chlorophyll, phycobiliprotein, and carotenoids. The investigation employed RNA sequencing to identify differentially expressed genes in response to ACC treatment in sporophytes. Notably, upregulated genes such as proteases were associated with catabolic processes. By contrast, genes related to anabolic processes such as photosynthesis, including light-harvesting complex protein and Calvin–Benson cycle enzymes, were downregulated in response to ACC treatment. ACC induced catabolic processes and repressed anabolic processes, indicating the promotion of resource reallocation in microscopic sporophytes.
Introduction
Phytohormones regulate various developmental and physiological processes in plants, including traits of agronomic importance such as plant growth, development, stress tolerance, and nutrient availability (Wilkinson et al., 2012; Fenn and Giovannoni, 2021; Jia et al., 2022). To date, knowledge on phytohormone biosynthesis and signaling pathways has allowed the use of biotechnological tools to increase the yield or improve the quality of agricultural crops (Csukasi et al., 2009). In contrast to higher plants, the lack of knowledge about the role of phytohormones in macroalgae could hinder efforts to improve algal performance and yield.
Ethylene, a gaseous phytohormone, regulates various developmental processes such as seed germination, fruit ripening, and senescence, as well as responses to biotic and abiotic stresses (Bleecker and Kende, 2000; Koyama, 2014; Binder, 2020). Ethylene biosynthesis starts with the conversion of methionine to S-adenosyl l-methionine (SAM) by SAM synthetase, and SAM is converted to the non-protein amino acid 1-aminocyclopropane-1-carboxylic acid (ACC) by the enzyme ACC synthase (Adams and Yang, 1979). ACC is then converted to ethylene by ACC oxidase (Kende, 1993). Exogenous ACC has been employed as a proxy for ethylene in plant experiments. However, an increasing number of studies indicate that ACC acts as a signaling molecule beyond its function as an ethylene precursor (Van de Poel, 2020). In land plants, ACC appears to participate in the regulation of stress responses, cell expansion, cell wall function, stomatal development, and fertilization-related events (Tsang et al., 2011; Polko and Kieber, 2019; Vanderstraeten et al., 2019; Li et al., 2020; Katayose et al., 2021).
The multicellular red algal class Bangiophyceae, which contains Porphyra and Pyropia, is among the most important marine aquaculture crops commonly used to wrap sushi and onigiri. The life cycle of Bangiophyceae generally consists of the heterogeneous alternation of macroscopic gametophytes and microscopic sporophytes. Sporophytes are classified into conchocelis, which was considered a different species before the clarification of its life cycle (Drew, 1949; Kurogi, 1953; Iwasaki, 1961). In addition to land plants, there is accumulating evidence that ACC can serve as a signaling molecule independent of its role in ethylene signaling in the gametophytes of Bangiophyceae (Uji and Mizuta, 2022a). ACC induced gametogenesis and growth suppression in the gametophytes of the monoecious species Pyropia yezoensis (formerly Neopyropia yezoensis) and the dioecious species Pyropia pseudolinearis (Uji et al., 2016; Yanagisawa et al., 2019). Exogenous ACC analogs such as ACBC also promoted sexual reproduction in the same manner as ACC, whereas ethephon, an ethylene-releasing compound, did not stimulate sexual reproduction in P. yezoensis (Uji et al., 2020; Endo et al., 2021). In RNA sequencing (RNA-seq), transcripts associated with cell division, vesicular trafficking, and extracellular matrix (ECM) were upregulated in Pyropia gametophytes treated with ACC, whereas transcripts involved in translation, plastid transcription, and photosynthesis were downregulated (Uji et al., 2016; Uji et al., 2022b). Furthermore, phospholipase D and phosphatidic acid are required for signal transduction, ultimately leading to ACC-induced sexual reproduction in P. yezoensis (Uji et al., 2022a). In addition to sexual reproduction, the activation of ACC signaling can promote heat tolerance in P. yezoensis gametophytes by activating genes associated with antioxidant defense systems (Uji and Mizuta, 2022b).
Previous observations demonstrated morphological and structural differences between gametophytic and sporophytic generations in Bangiophyceae, including differences in morphology (blade vs. filamentous), chloroplast shape (stellate vs. parietal), growth behavior (diffuse growth vs. apical growth), and pit connection structure (absent vs. present) (Pueschel and Cole, 1985). Previous studies also revealed that the gametophytes and sporophytes of Bangiophyceae differ in the composition of ECM polysaccharides (Mukai et al., 1981) and inorganic carbon use (Wang et al., 2020). However, the differences in the roles of phytohormones between gametophyte and sporophyte generations remain poorly understood. Although several studies, as described above, have investigated the role of ACC in the gametophytes of Pyropia, information on its effects in the sporophyte stage is still limited. Therefore, the differences in ACC-related responses between the gametophyte and sporophyte generations are not yet fully clarified. To address this gap, we investigated the physiological and molecular responses of P. yezoensis sporophytes to ACC.
Methods
Algal materials and chemical treatments
The filamentous sporophytes of P. yezoensis strain TU-1 initiated from free-living conchocelis (U-51 1, C-O giant) (Kuwano et al., 1996) were cultured in sterile vitamin-free Provasoli’s enriched seawater (PES (Provasoli, 1968)) at 15°C under a 14-h-light/10-h-dark photoperiod using cool-white fluorescent lamps at 40 μmol photons m−2 s−1.
For the comparative experiment on the effects of ethephon and ACC, small tufts of vegetative sporophytes (approximately 0.2 to 0.3 mm in diameter) were cultured in 55-mm tissue culture dishes with 10 mL of medium containing 0 or 50 μM ACC (Tokyo Chemical Industry, Tokyo, Japan) or 50 μM ethephon (Fujifilm Wako Pure Chemical Corporation, Osaka, Japan) at 15°C under a 14-h-light/10-h-dark photoperiod using cool-white fluorescent lamps at 40 μmol photons m−2s−1. The concentration for ACC and ethephon treatments was determined according to our previous experiments with P. yezoensis gametophytes, in which this concentration effectively induced physiological responses while avoiding severe toxic effects. The culture medium was replaced once a week, and fresh medium containing the treatment reagents was added during each medium change. Growth was determined from the volume increase of the filamentous tufts as calculated from the mean diameters at the beginning and end of the experiments. As previously reported (Lin and Stekoll, 2007), the average diameter was the mean of two measurements taken at 90° of each other, and the tuft volume was estimated using the formula for the volume of a sphere: V = (1/6)πD3 (V and D represent the volume and diameter of sporophyte tufts, respectively). The growth rate was calculated as the mean percent volume increase per day using the following formula: growth rate = [100(Vt − V0)/V0], where V0 is the initial tuft volume and Vt is the tuft volume at culture time.
Evaluation of photochemical efficiency
After 2 weeks of ACC treatment, vegetative sporophytes were used for the measurement of the maximum photochemical efficiency of photosystem II (PS II) (Fv/Fm) to evaluate the effect of ACC on photosynthesis using a portable chlorophyll fluorometer (OS1p, Opti-Science, Inc., Hudson, NH, USA). Before measurement, the samples were dark-adapted for 15 min. Measurements were taken following the manufacturer’s protocol. Data are expressed as the mean ± standard deviation (SD) of five turfs for each condition.
Quantification of pigment content
To examine the effect of ACC on the degradation of photosynthetic pigments [chlorophyll a (Chl a), phycoerythrin (PE), phycocyanin (PC), and carotenoids (Car)], sporophytes (fresh weight: ca. 0.02 g) were cultured in PES containing 0 or 50 μM ACC for 1 week at 15°C under a 14-h-light/10-h-dark photoperiod at 40 μmol photons m−2 s−1. The sporophytes were homogenized in 1 mL of 0.5 M phosphate buffer (pH 6.8) using a glass homogenizer. After centrifugation for 10 min at 12,000 ×g and 20°C, the supernatant was recovered to measure the PE and PC levels. Thereafter, 1 mL of 90% acetone was added to the homogenate, and the pellet was resuspended via vortexing. After centrifugation in the same manner, the Chl a and Car levels in the supernatant were determined. The supernatant was diluted five times with the same solvent used for extraction before light absorbance was measured using the U-1800 spectrophotometer (Hitachi, Tokyo, Japan). The PE and PC levels were calculated as described previously (Beer and Eshel, 1985), and the Chl a and Car contents were determined as previously reported (Seely et al., 1972). These experiments were repeated five times.
RNA preparation
All sporophytes from each sample were frozen in liquid nitrogen and immediately stored at −80°C until RNA extraction. Total RNA from algal samples was extracted as described previously (Drew, 1949). The quantity and integrity of the RNA samples were assessed using the NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), Qubit™ 4 fluorometer (Thermo Fisher Scientific), and Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Four libraries of complementary DNA (0 and 50 μM ACC, three replicates each) for P. yezoensis were constructed and subsequently sequenced using the Illumina NovaSeq 6000 instrument at Rhelixa Inc. (Tokyo, Japan).
RNA-seq
Low-quality reads and adapter sequences were trimmed from the obtained reads using fastp (Chen et al., 2018). After trimming, STAR (Dobin et al., 2013) was used to map the high-quality reads to an in-house gene model of P. yezoensis, which had been constructed with a reference genome (ASM982973v1) and other in-house transcriptome data using the BRAKER 2.1.6 (Brůna et al., 2021) pipeline. Next, the normalized expression of each gene was calculated using RSEM (Li and Dewey, 2011) as transcripts per million. Differentially expressed genes (DEGs) between the 0 and 50 μM ACC groups were identified by edgeR (Robinson et al., 2010) using the following criteria: false discovery rate <0.05 and |log2 fold change| >1. In addition, RNA-Seq data previously performed in our laboratory (Uji et al., 2016) were used to compare DEGs between sporophytes and gametophytes following ACC treatment.
Annotation and Gene Ontology enrichment analysis
To assess the biological significance of the DEGs, we conducted Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. GO terms were assigned to all genes using eggNOG-mapper v2 online (http://eggnog-mapper.embl.de/) (Huerta-Cepas et al., 2019; Cantalapiedra et al., 2021) with the default parameters except that the min_hit_e-value was set to 0.05. The topGO (Alexa and Rahnenfuhrer, 2023) R package was used for the GO enrichment analysis, and GO terms with P <0.05 were considered significantly enriched in the DEGs.
In addition, we employed KOBAS 3.0 software (Bu et al., 2021) to conduct KEGG annotation and enrichment analysis based on the KEGG PATHWAY Database (https://www.genome.jp/kegg/pathway.html) (Kanehisa and Goto, 2000). Pathways with corrected p-value (q-value) <0.05 were defined as significantly enriched pathways for the DEGs.
Quantitative PCR
Quantitative PCR (qPCR) analysis was performed as described previously (Uji et al., 2019). The mRNA levels were calculated using the 2−ΔΔCt method and normalized to the level of the 18S ribosomal RNA (Py18SrRNA) gene (Uji et al., 2016). The relative expression was calculated as the ratio of the mRNA level to the transcription level of samples without ACC treatment. All experiments were performed in three biological replicates. Additional File 1: Supplementary Table S1 presents the list of primers used in this study.
Statistical analysis
Data are expressed as mean ± standard deviation and analyzed using the Mann–Whitney U-test. For all analyses, P <0.05 was considered statistically significant.
Results and discussion
Effect of ACC treatment on the growth of P. yezoensis sporophytes
As a first step in clarifying the effects of ACC on P. yezoensis sporophytes, possibly independent of ethylene, we compared the effects of ACC and ethephon on growth and reproduction. When sporophytes were cultured for 40 days in a medium containing 50 μM ACC, the growth rate was 62.5%, whereas untreated (control) sporophytes exhibited a growth rate of 245.8% (Figures 1A, B). By contrast, ethephon did not significantly inhibit sporophyte growth (208.3%). The growth rate per day was 1.5% day−1 in ACC-treated sporophytes versus 6.1% day−1 in control sporophytes and 5.2% day−1 in ethephon-treated sporophytes (Figure 1C).
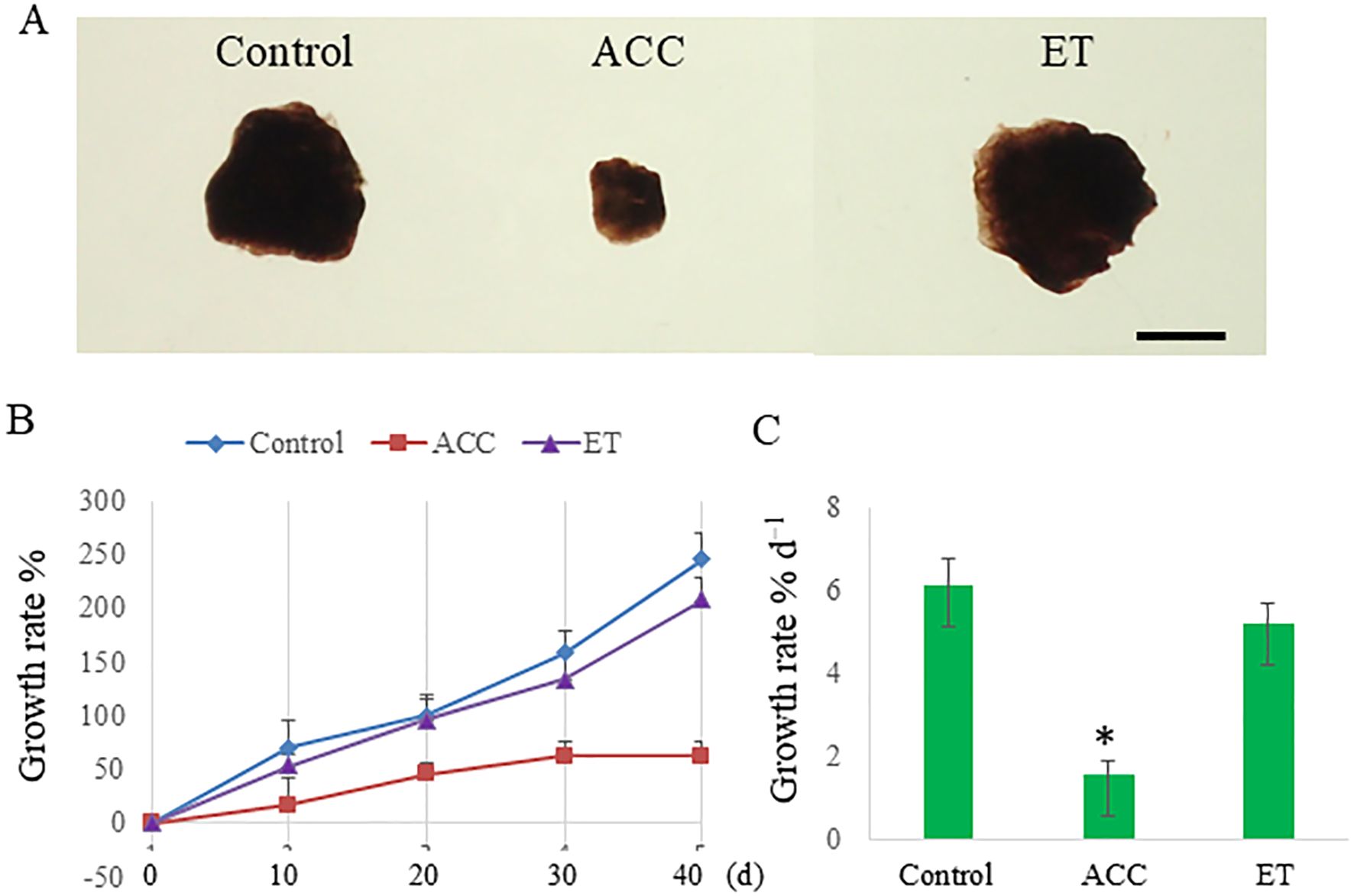
Figure 1. 1-Aminocyclopropane-1-carboxylic acid (ACC)-induced growth inhibition in Pyropia yezoensis sporophytes. (A) Representative photographs of ACC-treated P. yezoensis sporophytes. Scale bar = 5 mm. (B) Sporophytes during 40 days of culture with 0 or 50 μM ACC or 50 μM ethephon (ET). (C) Growth rate of sporophytes cultured with 0 or 50 μM ACC or 50 μM ET. Data are expressed as the mean ± standard deviation of six tufts for each condition. The asterisks indicate significant differences at P < 0.05 between the control and treatment groups.
A previous study reported increases in ACC levels during the transition from the vegetative to the reproductive stages and showed that ACC promotes the transition from vegetative conchocelis to conchosporangia in Pyropia haitanensis (Niu et al., 2024). In our study, we did not observe similar effects of ACC on the maturation of P. yezoensis sporophytes under the conditions tested (data not shown). This discrepancy may reflect species-specific differences or variations in experimental conditions, including developmental stage, ACC concentration, light, and temperature. Further studies are needed to determine whether ACC plays a broader role in the life cycle regulation of different red algal species.
Effect of ACC treatment on photosynthetic pigment content and photochemical efficiency in P. yezoensis sporophytes
Our previous study illustrated that ACC promotes the degradation of photosynthetic pigments, namely, Chl a, PE, PC, and Car, in gametophytes (Uji et al., 2016). Thus, we examined the effects of ACC on photosynthetic pigment content in sporophytes. Treatment with 50 μM ACC resulted in 17.0% and 29.4% decreases in Chl a and PE levels, respectively, compared to the control findings (Figure 2). Statistical analysis revealed significant differences in Chl a, PE, PC, and Car contents between ACC-treated and untreated sporophytes.
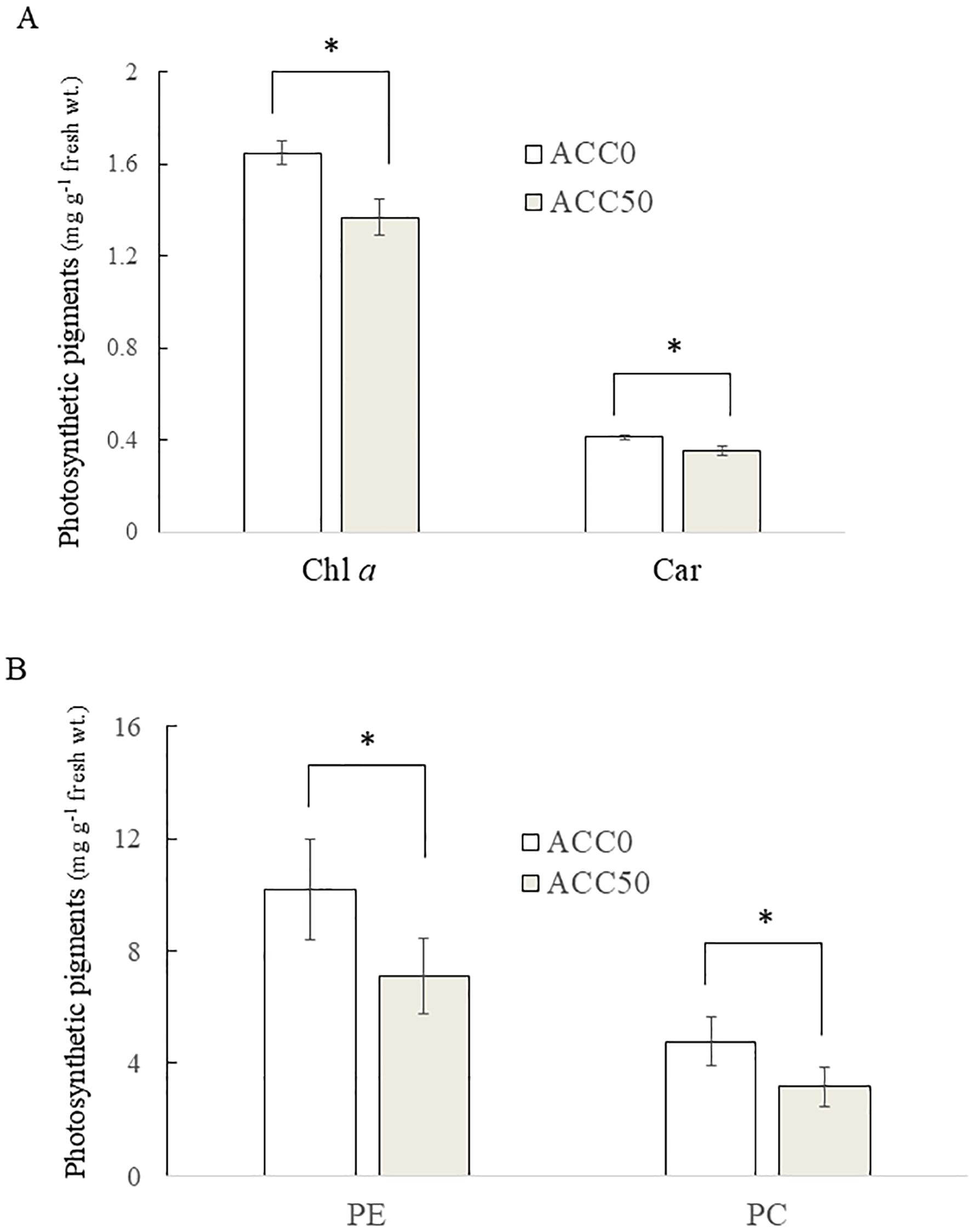
Figure 2. Effect of 1-aminocyclopropane-1-carboxylic acid (ACC) on photosynthetic pigment degradation in Pyropia yezoensis sporophytes. The contents of chlorophyll a (Chl a), carotenoids (Car) (A), phycoerythrin (PE), and phycocyanin (PC) (B) were measured using sporophytes cultured in medium containing 0 or 50 μM ACC. Data are expressed as the mean ± standard deviation of five independent experiments (mg g−1 fresh wt.). The asterisks indicate significant differences at P < 0.05 between the control and treatment groups.
Maximum quantum efficiency (Fv/Fm) using pulse amplitude modulation techniques was employed to assess the impact of ACC treatment on the photosynthetic capacity in P. yezoensis. The Fv/Fm value of sporophytes treated without ACC was 0.518, whereas that of sporophytes supplemented with ACC was 0.402 (Figure 3). Such a decline in photosynthetic efficiency is consistent with the observed reduction in photosynthetic pigment content. In higher plants, leaf senescence is typically accompanied by decreased photosynthetic activity and chlorophyll degradation as part of nutrient remobilization processes during the final stages of development (Lim et al., 2007). Our findings suggest that ACC may induce senescence-like physiological changes in the sporophytes of P. yezoensis, leading to reduced photosynthetic performance similar to the senescence processes described in terrestrial plants.
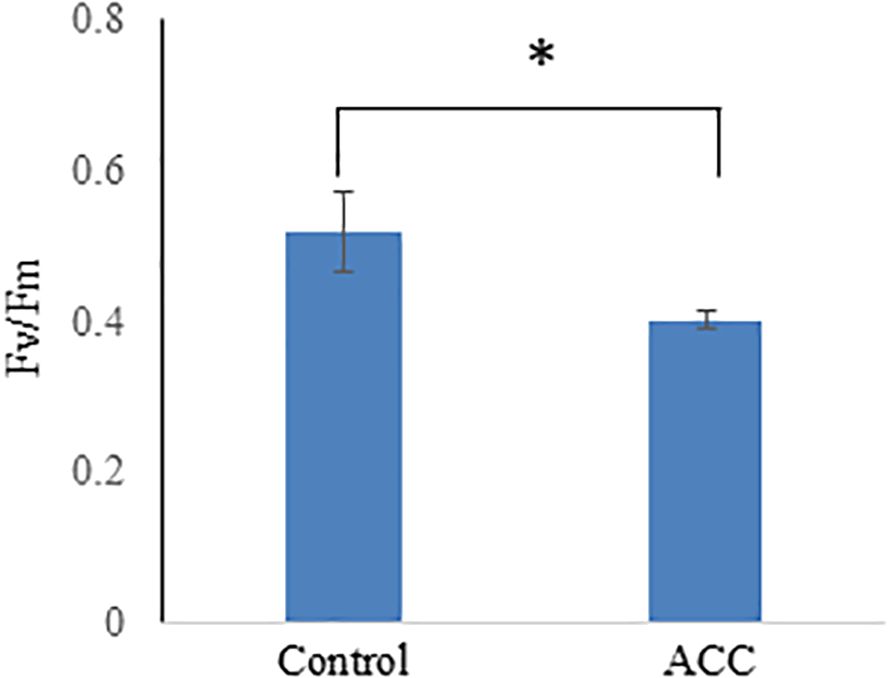
Figure 3. Effect of 1-aminocyclopropane-1-carboxylic acid (ACC) on the photosynthetic capacity in Pyropia yezoensis sporophytes. The maximum photochemical efficiency (Fv/Fm) was assessed and measured using sporophytes cultured in a medium containing 0 or 50 μM ACC. Data are expressed as the mean ± SD of five turfs for each condition. The asterisks indicate significant differences at P < 0.05 between the control and treatment groups.
Transcriptomic responses to ACC treatment
To further clarify the role of ACC in P. yezoensis sporophytes, we compared RNA-seq data between ACC-treated and untreated sporophytes. ACC-treated sporophytes were sampled at 3 days after treatment. Raw data generated by sequencing ranged from 18.0 to 21.5 million reads per sample. After filtering, 17.3–21.2 million clean reads were obtained, and the mapping rate ranged 89.0%–91.2%. A summary of the obtained RNA-seq datasets and the mapping rate of clean reads is presented in Table 1.
In total, 438 DEGs were identified between the control and ACC treatment groups, including 167 upregulated and 271 downregulated genes in response to ACC (Supplementary Tables S2, S3). Six DEGs (three upregulated and three downregulated genes) were selected for qPCR to validate the accuracy of the RNA-seq data. As illustrated in Figure 4, the expression of the selected genes was similar in the reverse transcription-qPCR and RNA-seq data, indicating that the RNA-seq results were reliable. The representative genes found to be differentially expressed in response to ACC treatment in sporophytes are presented in Tables 2, 3. The identification of these DEGs led us to perform further analyses, such as KEGG and GO enrichment, to explore their potential roles in metabolic processes and physiological changes induced by ACC, as discussed below.
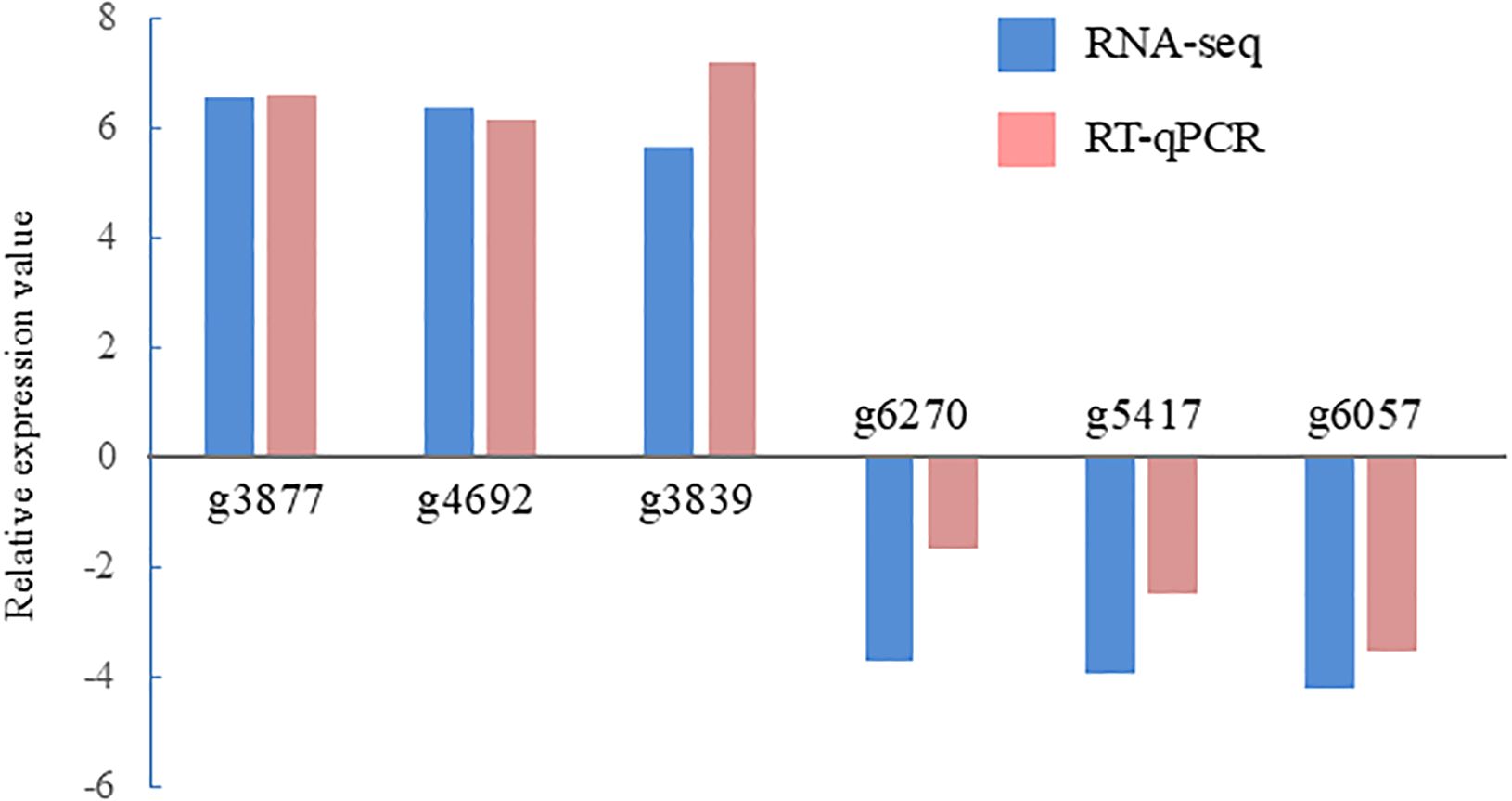
Figure 4. Validation of RNA sequencing (RNA-seq) data using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Six representative genes were selected to validate the RNA-seq data by RT-qPCR. The pink bars represent the mean log2-fold change obtained by RT-qPCR, and the blue bars represent the RNA-seq data. The results are presented as relative expression compared to that in untreated sporophytes. The primers used for RT-qPCR are listed in Supplementary Table S1.
KEGG enrichment analysis indicated that upregulated DEGs induced by ACC could be categorized into several pathways, including ubiquitin-mediated proteolysis, endocytosis, and valine, leucine, and isoleucine degradation (Figure 5). Protein degradation, which allows the recycling of nitrogen and other nutrients, is probably the most important degradation process that occurs during senescence (Roberts et al., 2012). In addition, the intracellular vesicle trafficking system transports cargo to specialized vacuolar compartments for digestive proteolysis mediated by various proteases (Wang and Schippers, 2019). Branched-chain amino acid (BCAA) degradation provides energy for plants during periods of extended darkness, early phases of germination, and late phases of senescence (Gipson et al., 2017).
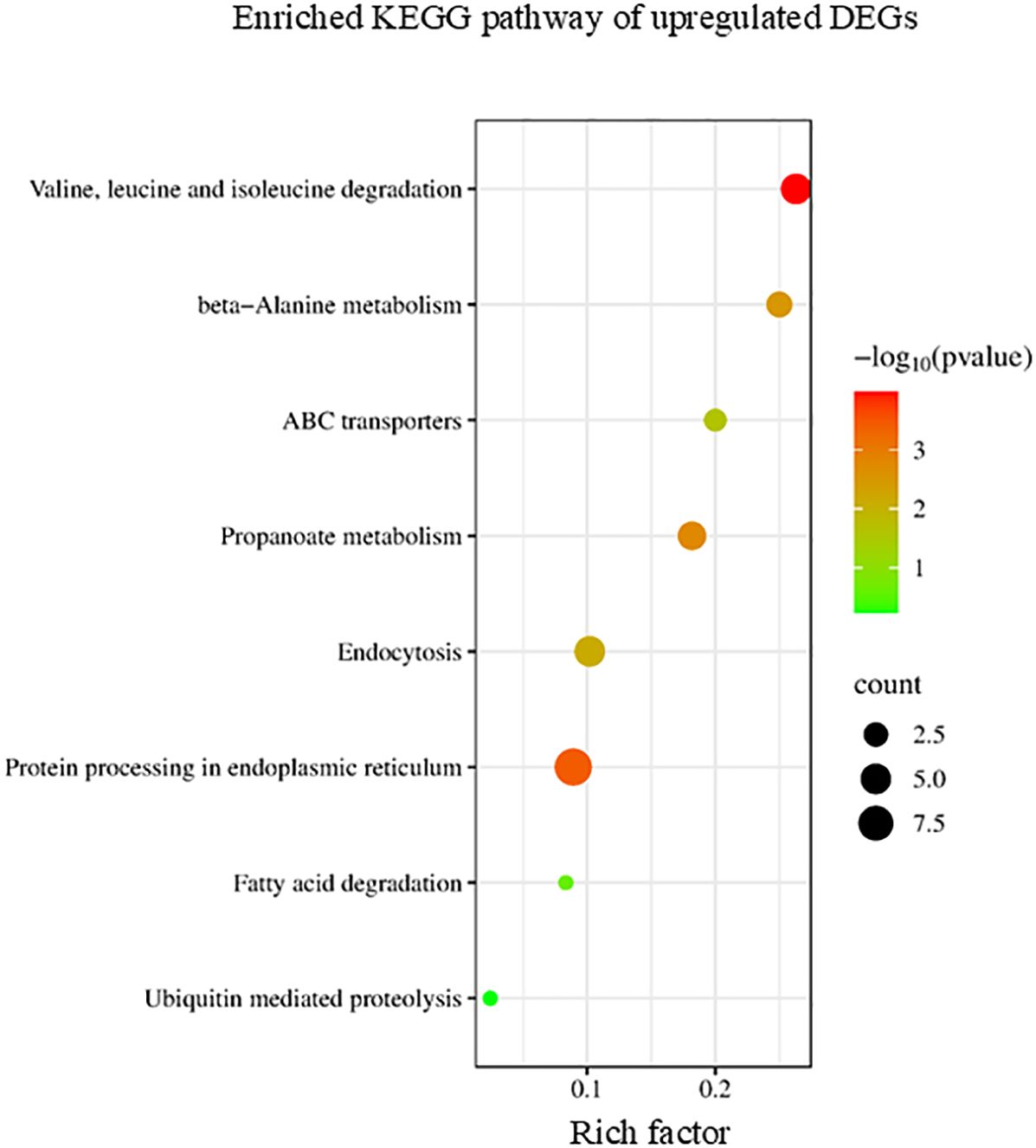
Figure 5. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the upregulated differentially expressed genes (DEGs). The degree of enrichment increased as the rich factor increased. The larger dots indicate higher numbers of differential genes enriched by the pathway.
Conversely, the KEGG enrichment analysis revealed that downregulated DEGs were categorized into pathways including photosynthesis, pentose phosphate pathway, and amino acid biosynthesis (Figure 6). In higher plants, leaf senescence is accompanied by extensive metabolic transformation from biosynthesis to degradation (Chaomurilege et al., 2023).
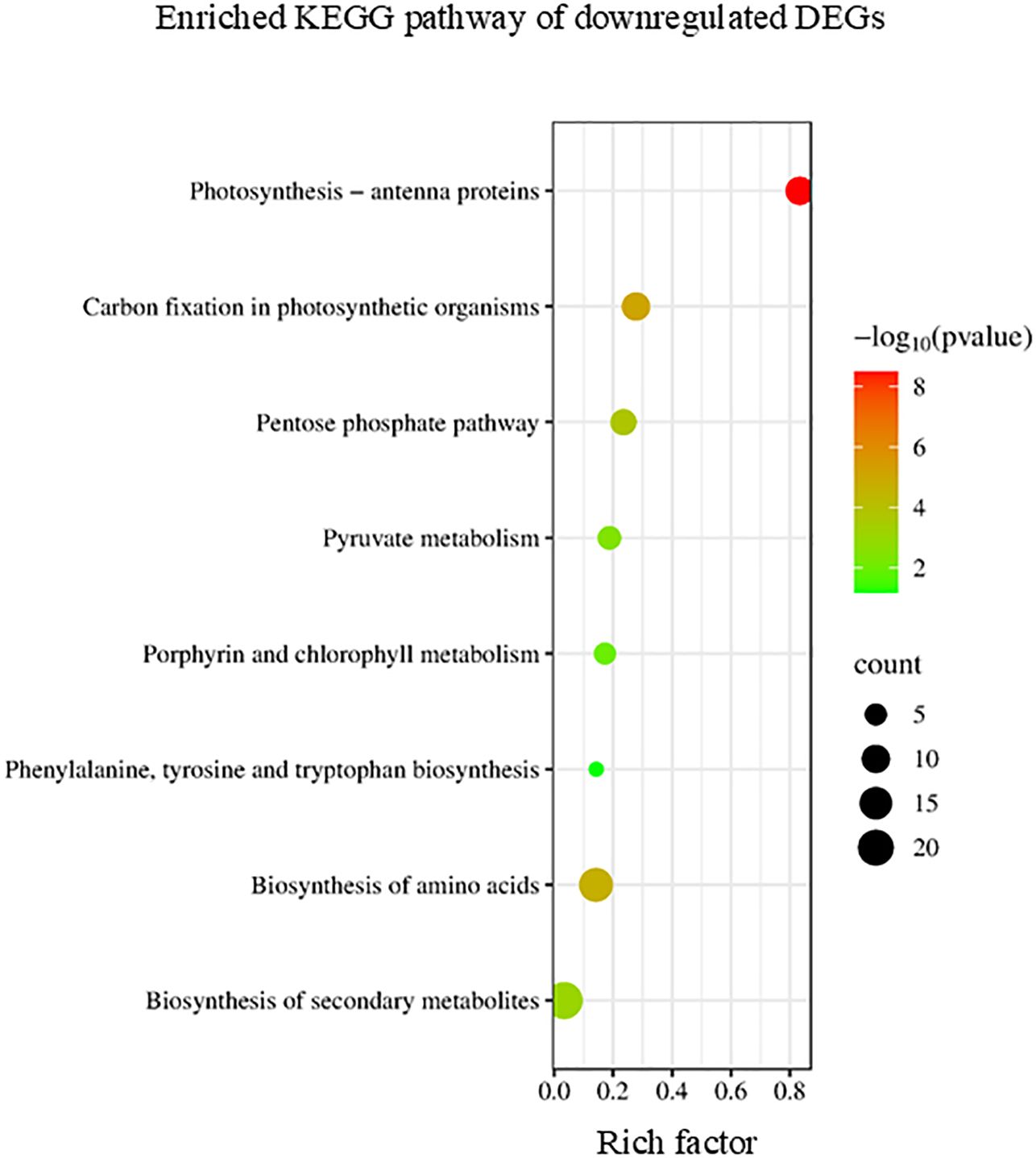
Figure 6. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the downregulated differentially expressed genes (DEGs). The degree of enrichment increased as the rich factor increased. The larger dots indicate higher numbers of differential genes enriched by the pathway.
To further elucidate the biological processes, molecular functions, and cellular components associated with DEGs, we performed GO analysis using eggNOG-mapper and topGO. In the upregulated DEGs, GO terms related to cellular catabolic processes, proteasome-related pathways, and molecular and protein transport were enriched (Figure 7), indicating that ACC induced protein catabolic processes. In the downregulated DEGs, GO terms related to photosynthesis, the photosystem, and chloroplast components were enriched (Figure 8), in line with the result that ACC induced photosynthetic pigment degradation. Consequently, KEGG enrichment analysis and GO analysis revealed that DEGs in ACC-treated sporophytes include genes associated with plant senescence.
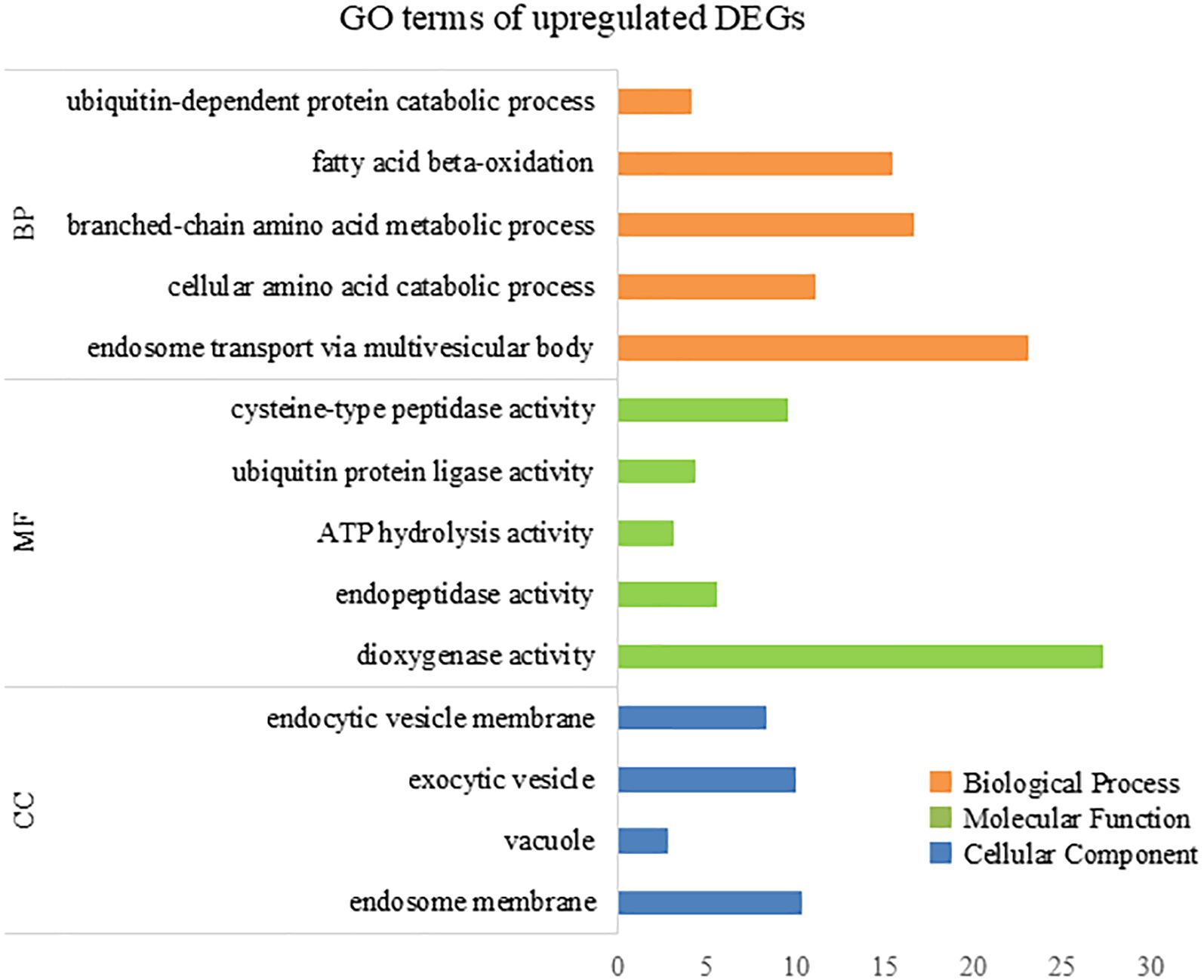
Figure 7. The numbers of enriched Gene Otology (GO) terms for the upregulated differentially expressed genes (DEGs). The GO terms are presented for three main categories: biological processes, molecular functions, and cellular components. Each GO term is listed in ascending order by p-value.
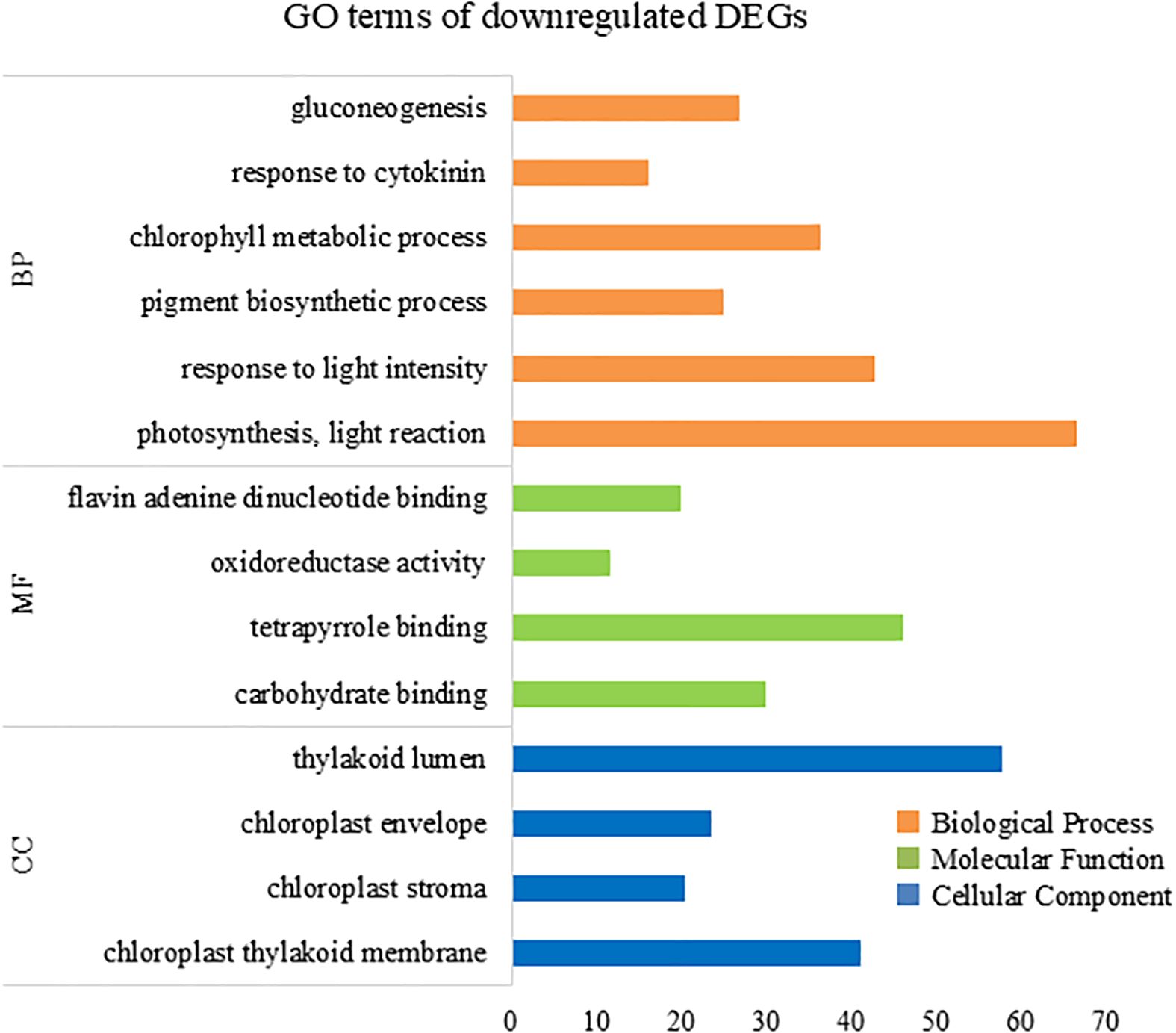
Figure 8. Numbers of enriched Gene Otology (GO) terms for the downregulated differentially expressed genes (DEGs). The GO terms are presented for three main categories: biological processes, molecular functions, and cellular components. Each GO term is listed in ascending order by p-value.
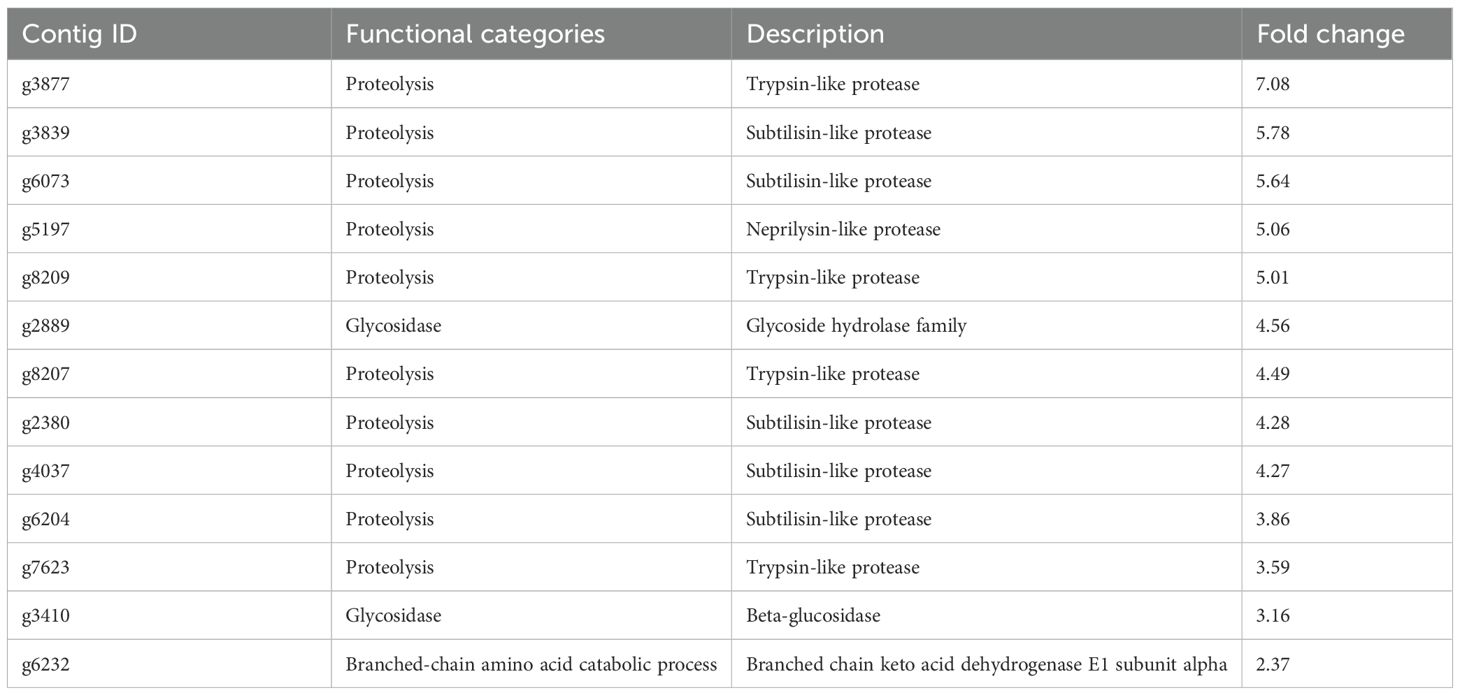
In the RNA-seq analysis, we identified 10 upregulated genes encoding proteases, including serine proteases and metalloproteases (Table 2; Additional File 1: Supplementary Figure S1). Among them, four genes encoded trypsin-like proteases, while five genes encoded subtilisin-like proteases. Subtilisin-like proteases belong to a large family found across diverse organisms and share conserved catalytic domains (Dodson and Wlodawer, 1998; Tripathi and Sowdhamini, 2006; Kim et al., 2016; Patel, 2017). Additionally, we identified one neprilysin-like metalloprotease gene (g5197) possessing typical conserved motifs, suggesting that it belongs to the M13 family of zinc metallopeptidases (Oefner et al., 2000; Turner et al., 2001; Bland et al., 2008; Yang et al., 2016). The upregulation of various proteases, including subtilisin-like and neprilysin-like proteases, in ACC-treated sporophytes suggests that ACC may enhance the protein degradation pathways. In higher plants, increased proteolytic activity during senescence facilitates nitrogen remobilization by breaking down proteins into amino acids, which can then be transported to developing tissues or storage organs (Lim et al., 2007). The observed induction of these proteases in P. yezoensis sporophytes may indicate a similar process, whereby ACC triggers catabolic pathways to recycle nitrogen and other nutrients during stress or senescence-like conditions. Further biochemical analyses will be necessary to confirm whether these transcriptomic changes translate into increased protease activity and contribute to nutrient remobilization in red algae.
In addition to proteases, the results demonstrated that the DEG (g6232) encoding the mitochondrial branched-chain alpha-ketoacid dehydrogenase complex (BCKDC) was upregulated. Among amino acids, the BCAAs leucine, isoleucine, and valine are proposed to provide their downstream catabolic products, namely, acetoacetate, acetyl-CoA, and propionyl-CoA, respectively, to the TCA cycle for energy generation (Dimou et al., 2022). In mammals, BCKDC consists of multiple copies of three proteins: keto acid dehydrogenase/carboxylase E1 (E1α and E1β), dihydrolipoyl acyltransferase E2, and dihydrolipoyl dehydrogenase E3. This complex, with a molecular weight of several megadaltons, catalyzes the second step of BCAA degradation by converting branched-chain ketoacids (intermediates in BCAA biosynthesis and catabolism) into acyl-CoA esters (Harris et al., 2001). Previous studies revealed that transcripts of BCAA catabolic genes increase during prolonged darkness, which is a well-known external stimulus capable of accelerating leaf senescence (Binder, 2010). During carbon deficiency, BCAA degradation products can serve as alternative energy sources by being fed into the TCA cycle in Arabidopsis (Angelovici et al., 2013). Thus, the induction of BCKDC from P. yezoensis suggests that ACC regulates BCAA catabolism, thereby contributing to algal fitness under energy-limited conditions.
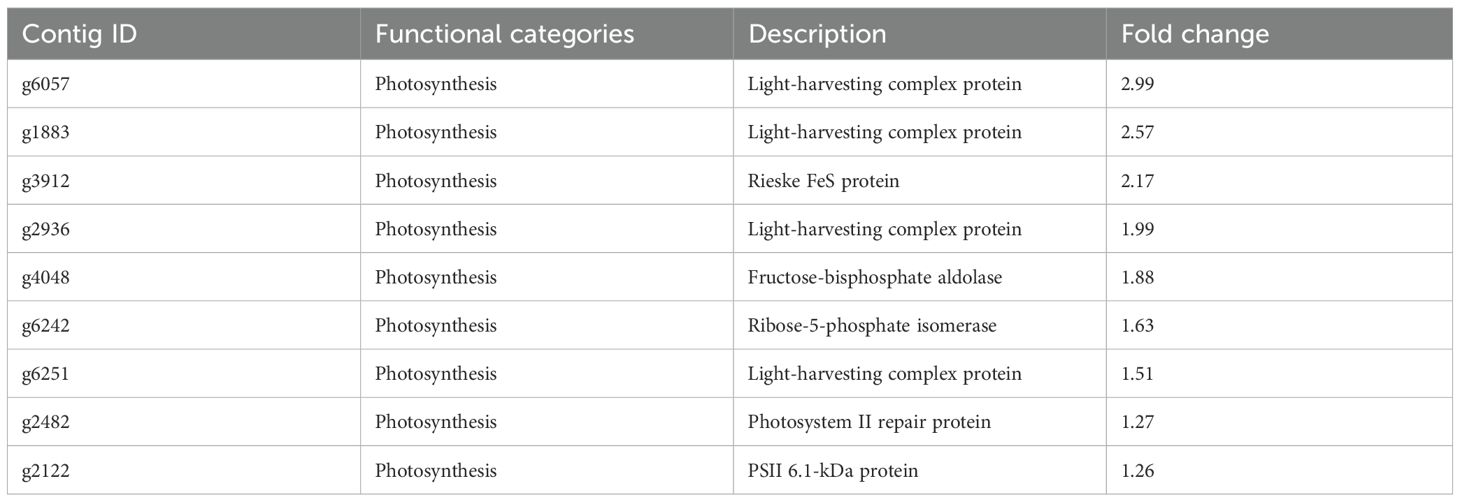
Although chloroplasts contain their own genome, a large fraction of chloroplast proteins are encoded by the nuclear genome. Chloroplasts contain up to 70% of leaf proteins, the majority of which are photosynthetic proteins, including ribulose bisphosphate, carboxylase/oxygenase, and chlorophyll-binding/light-harvesting complex proteins (Fu et al., 2022). As presented in Table 3, transcriptome analysis illustrated that nuclear genome-encoded, chloroplast-targeted proteins such as light-harvesting complex proteins (g6057, g1883, and g2936) and Rieske FeS protein (g3912) were downregulated by ACC treatment in sporophytes. In addition to the photosynthetic apparatus, genes encoding enzymes involved in the Calvin–Benson cycle, including fructose-1,6-biphosphate aldolase (g4048), were downregulated in sporophytes treated with ACC.
Consistent with the downregulation of photosynthesis-associated genes, sporophytes treated with ACC exhibited photosynthetic pigment degradation (Figure 2). In addition to a role in light harvesting, PE in red algae plays a more important role in maintaining the nitrogen pool (Mizuta et al., 2002). In higher plants, Chl degradation contributes to nitrogen remobilization during leaf senescence (Hörtensteiner, 2006). Similarly, the degradation of photosynthetic pigments, especially PE, induced by the action of ACC might be involved in nitrogen remobilization in P. yezoensis.
Senescence is the last stage of plant development, and it is accompanied by a transition from nutrient assimilation to nutrient remobilization (Roberts et al., 2012). In plants, it is known that, during senescence, many major macromolecules, including proteins, lipids, and nucleic acids, are degraded in a process triggered by the reprogramming of thousands of genes (upregulation or downregulation) in response to specific senescence-promoting factors such as the plant hormone ethylene (Kim et al., 2016). In higher plants, senescence is primarily regulated by ethylene, which acts as a key signaling molecule. ACC serves as the immediate precursor of ethylene and does not directly induce senescence processes. In contrast, our findings suggest that in P. yezoensis, ACC itself may directly trigger gene reprogramming associated with senescence-like responses, possibly independent of ethylene production.
Comparison of DEGs between sporophytes and gametophytes following ACC treatment
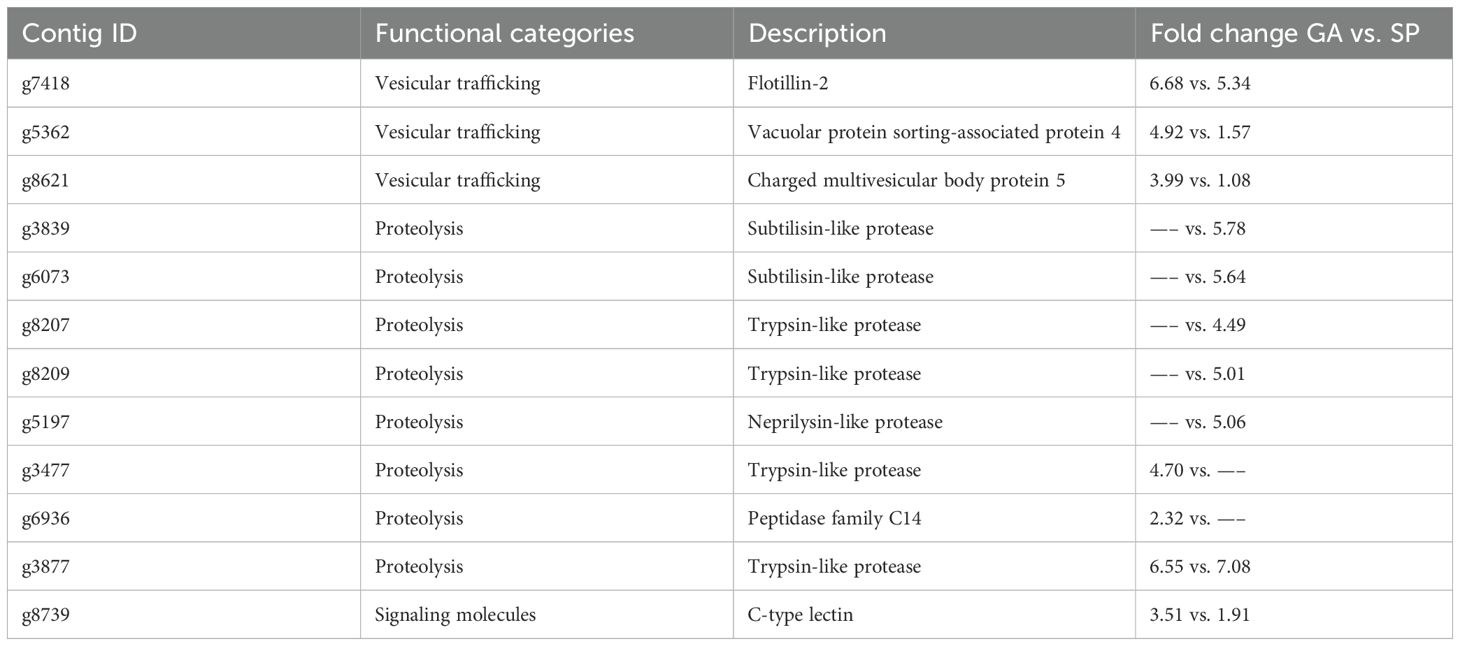
Our previous study provided the comprehensive transcriptome data of P. yezoensis gametophytes treated with ACC (Uji et al., 2016). Therefore, we compared DEGs in response to ACC exposure between gametophytic and sporophytic generations (Table 4). Endocytosis-related genes were upregulated in gametophytes and sporophytes. g7418, which encodes flotillin, was identified as a gene largely responsive to ACC in both gametophytes and sporophytes. Flotillins are membrane-associated proteins considered to function in a number of cellular contexts such as endocytosis, providing molecular scaffolding for membrane rafts that act by demarcating sites for the delivery of specific cargo (Otto and Nichols, 2011). Vacuolar protein sorting-associated protein 4 (Vps4) (g5362) and charged multivesicular body protein 5 (CHMP5) (g8621) associated with endosomal sorting complex required for transport III (ESCRT-III) (Otto and Nichols, 2011; Yang and Wang, 2022) were also upregulated in gametophytes and sporophytes treated with ACC. Nutrient starvation-induced endocytosis and degradation of various membrane proteins via the ESCRT-dependent pathway are crucial for maintaining critical amino acid levels (Alonso et al., 2016). In higher plants, the ESCRT-III subunit CHMP is required for the autophagic degradation of plastid proteins (Spitzer et al., 2015). Similarly, ACC can activate the ESCRT-dependent endosomal pathway to recycle degraded photosynthetic pigment proteins in P. yezoensis.
ACC promotes gametophyte maturation, which then releases spores and the individual disappears. By contrast, ACC suppresses the growth of sporophytes, but the individuals are maintained. The different types of proteases induced by ACC might reflect the differences in ACC responsiveness between generations. A comparison of the specific expression patterns and target substrate proteins of ACC-responsive proteases across generations will advance our understanding of the role of proteolysis in mobilizing nitrogen resources during the Pyropia life cycle.
g8739, which contains a C-type lectin domain, was upregulated in ACC-treated P. yezoensis. C-type lectins were among the first animal lectins discovered (Zelensky and Gready, 2005). In mammals, C-type lectins are secreted molecules or transmembrane proteins. Although there is little information on C-type lectins in higher plants, they are known to be involved in phytohormone signaling—for example, the Arabidopsis thaliana lectin ArathEULS3, the mRNA levels of which increase in Arabidopsis cell suspension cultures after abscisic acid (ABA) treatment, has been reported to interact with the ABA receptor PYL9, one of the most important players in ABA signaling (Dubiel et al., 2020). In addition to the C-type lectin domain, g8739 contains the von Willebrand A domain, which is a well-studied domain involved in cell adhesion ECM proteins and integrin receptors (Whittaker and Hynes, 2002). Thus, the upregulation of g8739 suggests that the gene is an important player in ACC signaling in P. yezoensis.
Taken together, these transcriptomic changes are consistent with our physiological observations that ACC treatment inhibited the growth and induced the degradation of photosynthetic pigments in P. yezoensis sporophytes. This suggests that ACC induces transcriptional changes associated with processes similar to plant senescence. However, it should be noted that these conclusions are based on transcriptomic data, and further biochemical analyses are needed to confirm whether these gene expression changes translate into functional metabolic shifts. Moreover, although ethylene is a well-known regulator of senescence in higher plants, the mechanisms by which its precursor ACC affects the sporophytes of P. yezoensis may differ due to species-specific factors and the unique biology of red algae.
Conclusions
Ethylene has long been known as a major hormone hastening leaf and flower senescence in higher plants. To our knowledge, this is the first study suggesting that ACC may have a senescence-like effect in both plants and algae, potentially independent of ethylene. Previous studies revealed that ACC promotes the transition of macroscopic gametophytes to microscopic sporophytes in P. yezoensis. In this study, ACC treatment induced catabolic processes and repressed anabolic processes in microscopic sporophytes. P. yezoensis generally grows as gametophytes during winter, when nutrients, especially nitrogen, are abundant, and as sporophytes during summer, when nutrient availability is low. Therefore, considering the effects of ACC on both gametophytes and sporophytes, it is possible that ACC contributes to reducing the metabolic activity or nutrient demand of algae under nutrient-limited conditions, potentially serving as a signal to adjust physiological processes according to environmental nutrient availability.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
TU: Investigation, Writing – original draft, Conceptualization, Writing – review & editing. SS: Writing – review & editing, Investigation. HM: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Grant-in-Aid (grant number 22K05779 to TU) from the Japan Society for the Promotion of Science.
Acknowledgments
We thank Dr. Takafumi Fujimoto and Dr. Toshiya Nishimura (Hokkaido University, Japan) for kindly providing the LightCycler 96 system.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1632530/full#supplementary-material
Abbreviations
ABA, abscisic acid; BCAA, branched-chain amino acid; BCKDC, branched-chain alpha-ketoacid dehydrogenase complex; DEG, differentially expressed genes; GO, Gene Otology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PES, Provasoli’s enriched seawater; SAM, S-adenosyl l-methionine.
References
Adams, D. O. and Yang, S. F. (1979). Ethylene biosynthesis: identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. U.S.A. 76, 170–174. doi: 10.1073/pnas.76.1.170
Alexa, A. and Rahnenfuhrer, J. (2023). topGO: Enrichment analysis for gene ontology. doi: 10.18129/B9.bioc.topGO
Alonso, Y., Adell, M., Migliano, S. M., and Teis, D. (2016). ESCRT-III and Vps4: a dynamic multipurpose tool for membrane budding and scission. FEBS J. 283, 3288–3302. doi: 10.1111/febs.13688
Angelovici, R., Lipka, A. E., Deason, N., Gonzalez-Jorge, S., Lin, H., Cepela, J., et al. (2013). Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. Plant Cell 25, 4827–4843. doi: 10.1105/tpc.113.119370
Beer, S. and Eshel, A. (1985). Determining phycoerythrin and phycocyanin concentrations in aqueous crude extracts of red algae. Aust. J. Mar. Freshwat Res. 36, 785–792. doi: 10.1071/MF9850785
Binder, S. (2010). Branched-chain amino acid metabolism in Arabidopsis thaliana. Arabidopsis Book 8, e0137. doi: 10.1199/tab.0137
Binder, B. M. (2020). Ethylene signaling in plants. J. Biol. Chem. 295, 7710–7725. doi: 10.1074/jbc.REV120.010854
Bland, N. D., Pinney, J. W., Thomas, J. E., Turner, A. J., and Isaac, R. E. (2008). Bioinformatic analysis of the neprilysin (M13) family of peptidases reveals complex evolutionary and functional relationships. BMC Evol. Biol. 8, 16. doi: 10.1186/1471-2148-8-16
Bleecker, A. B. and Kende, H. (2000). Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Brůna, T., Hoff, K. J., Lomsadze, A., Stanke, M., and Borodovsky, M. (2021). BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom Bioinform. 3, 108. doi: 10.1093/nargab/lqaa108
Bu, D., Luo, H., Huo, P., Wang, Z., Zhang, S., He, Z., et al. (2021). KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 49, 317–325. doi: 10.1093/nar/gkab447
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829. doi: 10.1093/molbev/msab293
Chaomurilege, Miyagi, A., Ishikawa, T., Yamaguchi, M., Murayama, H., and Kawai-Yamada, M. (2023). Metabolic changes associated with dark-induced leaf senescence in Arabidopsis nadk2 mutants. Plant Signal Behav. 18, 2215618. doi: 10.1080/15592324.2023.2215618
Chen, S., Chen, Y., and Gu, J. (2018). Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, 884–890. doi: 10.1093/bioinformatics/bty560
Csukasi, F., Merchante, C., and Valpuesta, V. (2009). Modification of plant hormone levels and signaling as a tool in plant biotechnology. Biotechnol. J. 4, 1293–1304. doi: 10.1002/biot.200800286
Dimou, A., Tsimihodimos, V., and Bairaktari, E. (2022). The critical role of the branched chain amino acids (BCAAs) catabolism-regulating enzymes, branched-chain aminotransferase (BCAT) and branched-chain α-keto acid dehydrogenase (BCKD), in human pathophysiology. Int. J. Mol. Sci. 23, 4022. doi: 10.3390/ijms23074022
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Dodson, G. and Wlodawer, A. (1998). Catalytic triads and their relatives. Trends Biochem. Sci. 23, 347–352. doi: 10.1016/S0968-0004(98)01254-7
Drew, K. M. (1949). Conchocelis-Phase in the life-history of Porphyra umbilicalis (L.) Kütz. Nature 164, 748–749. doi: 10.1038/164748a0
Dubiel, M., Beeckman, T., Smagghe, G., and Van Damme, E. J. M. (2020). Arabidopsis lectin EULS3 is involved in ABA signaling in roots. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00437
Endo, H., Mizuta, H., and Uji, T. (2021). α-aminoisobutyric acid mimics the effect of 1-aminocyclopropane-1-carboxylic acid to promote sexual reproduction in the marine red alga Pyropia yezoensis (Rhodophyta). J. Appl. Phycol 33, 1081–1087. doi: 10.1007/s10811-020-02326-7
Fenn, M. A. and Giovannoni, J. J. (2021). Phytohormones in fruit development and maturation. Plant J. 105, 446–458. doi: 10.1111/tpj.15112
Fu, Y., Li, X., Fan, B., Zhu, C., and Chen, Z. (2022). Chloroplasts protein quality control and turnover: A multitude of mechanisms. Int. J. Mol. Sci. 23, 7760. doi: 10.3390/ijms23147760
Gipson, A. B., Morton, K. J., Rhee, R. J., Simo, S., Clayton, J. A., Perrett, M. E., et al. (2017). Disruptions in valine degradation affect seed development and germination in Arabidopsis. Plant J. 90, 1029–1039. doi: 10.1111/tpj.13538
Harris, R. A., Kobayashi, R., Murakami, T., and Shimomura, Y. (2001). Regulation of branched-chain alpha-keto acid dehydrogenase kinase expression in rat liver. J. Nutr. 131, 841–845. doi: 10.1093/jn/131.3.841S
Hörtensteiner, S. (2006). Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 57, 55–77. doi: 10.1146/annurev.arplant.57.032905.105212
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, 309–314. doi: 10.1093/nar/gky1085
Iwasaki, H. (1961). The life cycle of Porphyra tenera in vitro. Biol. Bull. Mar. Biol. Lab. Wood’s Hole 121, 173–187. doi: 10.2307/1539469
Jia, Z. T., Giehl, R. F. H., and von Wirén, N. (2022). Nutrient-hormone relations: Driving root plasticity in plants. Mol. Plant 15, 86–103. doi: 10.1016/j.molp.2021.12.004
Kanehisa, M. and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Katayose, A., Kanda, A., Kubo, Y., Takahashi, T., and Motose, H. (2021). Distinct functions of ethylene and ACC in the basal land plant Marchantia polymorpha. Plant Cell Physiol. 62, 858–871. doi: 10.1093/pcp/pcab042
Kende, H. (1993). Ethylene biosynthesis. Annu. Rev. Plant Physiol. Mol. Biol. 44, 283–307. doi: 10.1146/annurev.pp.44.060193.001435
Kim, J., Woo, H. R., and Nam, H. G. (2016). Toward systems understanding of leaf senescence: An integrated multi-omics perspective on leaf senescence research. Mol. Plant 9, 813–825. doi: 10.1016/j.molp.2016.04.017
Koyama, T. (2014). The roles of ethylene and transcription factors in the regulation of onset of leaf senescence. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00650
Kurogi, M. (1953). Study of the life-history of Porphyra. Germination and development of the carpospores. Bull. Tohoku Reg. Fish Res. Lab. 2, 67–103.
Kuwano, K., Aruga, Y., and Saga, N. (1996). Cryopreservation of clonal gametophytic thalli of Porphyra (Rhodophyta). Plant Sci. 116, 117–124. doi: 10.1016/0168-9452(96)04380-4
Li, B. and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 323, 1471–2105. doi: 10.1186/1471-2105-12-323
Li, D. D., Flores-Sandoval, E., Ahtesham, U., Coleman, A., Clay, J. M., Bowman, J. L., et al. (2020). Ethylene-independent functions of the ethylene precursor ACC in Marchantia polymorpha. Nat. Plants. 6, 1335–1344. doi: 10.1038/s41477-020-00784-y
Lim, P. O., Kim, H. J., and Nam, H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Lin, R. L. and Stekoll, M. S. (2007). Effects of plant growth substances on the conchocelis phase of Alaskan Porphyra (Bangiales, Rhodophyta) species in conjunction with environmental variables. J. Phycol 43, 1094–1103. doi: 10.1111/j.1529-8817.2007.00388.x
Mizuta, H., Shirakura, Y., and Yasui, H. (2002). Relationship between phycoerythrin and nitrogen content in Gloiopeltis furcata and Porphyra yezoensis. . Algae 17, 89–93. doi: 10.4490/ALGAE.2002.17.2.089
Mukai, L. S., Craigie, J. S., and Brown, R. G. (1981). Chemical composition and structure of the cell walls of the conchocelis and thallus phases of Porphyra tenera (Rhodophyceae). J. Phycol 17, 192–197. doi: 10.1111/j.1529-8817.1981.tb00839.x
Niu, T., Qian, H., Chen, H., Luo, Q., Chen, J., Yang, R., et al. (2024). H2O2 drives the transition from conchocelis to conchosporangia in the red alga Pyropia haitanensis with promotion facilitated by 1-Aminocyclopropane-1-carboxylic acid. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1379428
Oefner, C., D’Arcy, A., Hennig, M., Winkler, F. K., and Dale, G. E. (2000). Structure of human neutral endopeptidase (neprilysin) complexed with phosphoramidon. J. Mol. Biol. 269, 341–349. doi: 10.1006/jmbi.1999.3492
Otto, G. P. and Nichols, B. J. (2011). The roles of flotillin microdomains–endocytosis and beyond. J. Cell Sci. 124, 3933–3940. doi: 10.1242/jcs.092015
Patel, S. (2017). A critical review on serine protease: Key immune manipulator and pathology mediator. Allergol Immunopathol. (Madr). 45, 579–591. doi: 10.1016/j.aller.2016.10.011
Polko, J. K. and Kieber, J. J. (2019). 1-aminocyclopropane 1-carboxylic acid and its emerging role as an ethylene-independent growth regulator. Front. Plant Sci. 10, 1602. doi: 10.3389/fpls.2019.01602
Provasoli, L. (1968). “Media and prospects for the cultivation of marine algae,” in Culture and collections of algae, Proc U S-Japan Conf, Hakone, September 1966. . Eds. Watanabe, A. and Hattori, A. (Tokyo: Japanese Society of Plant Physiology), 63–75.
Pueschel, C. M. and Cole, K. M. (1985). Ultrastructure of germinating carpospores of Porphyra variegata (Kjellm) Hus (Bangiales, Rhodophyta). J. Phycol 21, 146–154. doi: 10.1111/j.0022-3646.1985.00146.x
Roberts, I. N., Caputo, C., Criado, M. V., and Funk, C. (2012). Senescence-associated proteases in plants. Physiol. Plant 145, 130–139. doi: 10.1111/j.1399-3054.2012.01574.x
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Seely, G. R., Vidaver, W. E., and Duncan, M. J. (1972). Preparative and analytical extraction of pigments from brown algae with dimethyl sulfoxide. Mar. Biol. 12, 184–188. doi: 10.1007/BF00350754
Spitzer, C., Li, F., Buono, R., Roschzttardtz, H., Chung, T., Zhang, M., et al. (2015). The endosomal protein CHARGED MULTIVESICULAR BODY PROTEIN1 regulates the autophagic turnover of plastids in Arabidopsis. Plant Cell 27, 391–402. doi: 10.1105/tpc.114.135939
Tripathi, L. P. and Sowdhamini, R. (2006). Cross genome comparisons of serine proteases in Arabidopsis and rice. BMC Genomics 7, 200. doi: 10.1186/1471-2164-7-200
Tsang, D. L., Edmond, C., Harrington, J. L., and Nuehse, T. S. (2011). Cell wall integrity controls root elongation via a general 1-aminocyclopropane-1-carboxylic acid-dependent, ethylene-independent pathway. Plant Physiol. 156, 596–604. doi: 10.1104/pp.111.175372
Turner, A. J., Isaac, R. E., and Coates, D. (2001). The neprilysin (NEP) family of zinc metalloendopeptidases: genomics and function. Bioessays. 23, 261–269. doi: 10.1002/1521-1878(200103)23:3<261::AID-BIES1036>3.0.CO;2-K
Uji, T., Endo, H., and Mizuta, H. (2020). Sexual reproduction via a 1-aminocyclopropane-1-carboxylic acid-dependent pathway through redox modulation in the marine red alga Pyropia yezoensis (Rhodophyta). Front. Plant Sci. 11, 60. doi: 10.3389/fpls.2020.00060
Uji, T., Gondaira, Y., Fukuda, S., Mizuta, H., and Saga, N. (2019). Characterization and expression profiles of small heat shock proteins in the marine red alga Pyropia yezoensis. Cell Stress Chaperones 24, 223–233. doi: 10.1007/s12192-018-00959-9
Uji, T., Kandori, T., Konishi, S., and Mizuta, H. (2022a). Phospholipase D activation is required for 1-aminocyclopropane 1-carboxylic acid signaling during sexual reproduction in the marine red alga Neopyropia yezoensis (Rhodophyta). BMC Plant Biol. 22, 181. doi: 10.1186/s12870-022-03575-z
Uji, T., Matsuda, R., Takechi, K., Takano, H., Mizuta, H., and Takio, S. (2016). Ethylene regulation of sexual reproduction in the marine red alga Pyropia yezoensis (Rhodophyta). J. Appl. Phycol 28, 3501–3509. doi: 10.1007/s10811-016-0904-6
Uji, T. and Mizuta, H. (2022a). The role of plant hormones on the reproductive success of red and brown algae. Front. Plant Sci. 13, 1019334. doi: 10.3389/fpls.2022.1019334
Uji, T. and Mizuta, H. (2022b). 1-aminocyclopropane-1-carboxylic acid and its analogs alleviate heat stress damage in the marine red alga Neopyropia yezoensis (Rhodophyta). J. Appl. Phycol 34, 1527–1536. doi: 10.1007/s10811-022-02727-w
Uji, T., Ueda, S., and Mizuta, H. (2022b). Identification, characterization, and expression analysis of spondin-like and fasciclin-like genes in Neopyropia yezoensis, a marine red alga. Phycology 2, 45–59. doi: 10.3390/phycology2010003
Van de Poel, B. (2020). Ethylene’s fraternal twin steals the spotlight. Nat. Plants 6, 1309–1310. doi: 10.1038/s41477-020-00796-8
Vanderstraeten, L., Depaepe, T., Bertrand, S., and van der Straeten, D. (2019). The ethylene precursor ACC affects early vegetative development independently of ethylene signaling. Front. Plant Sci. 10, 1591. doi: 10.3389/fpls.2019.01591
Wang, H. and Schippers, J. H. M. (2019). The role and regulation of autophagy and the proteasome during aging and senescence in plants. Genes 10, 267. doi: 10.3390/genes10040267
Wang, D., Yu, X., Xu, K., Bi, G., Cao, M., Zelzion, E., et al. (2020). Pyropia yezoensis genome reveals diverse mechanisms of carbon acquisition in the intertidal environment. Nat. Commun. 11, 4028. doi: 10.1038/s41467-020-17689-1
Whittaker, C. A. and Hynes, R. O. (2002). Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13, 3369–3387. doi: 10.1091/mbc.e02-05-0259
Wilkinson, S., Kudoyarova, G. R., Veselov, D. S., Arkhipova, T. N., and Davies, W. J. (2012). Plant hormone interactions: innovative targets for crop breeding and management. J. Exp. Bot. 63, 3499–3509. doi: 10.1093/jxb/ers148
Yanagisawa, R., Sekine, N., Mizuta, H., and Uji, T. (2019). Transcriptomic analysis under ethylene precursor treatment uncovers the regulation of gene expression linked to sexual reproduction in the dioecious red alga Pyropia pseudolinearis. J. Appl. Phycol 31, 3317–3329. doi: 10.1007/s10811-019-01808-7
Yang, Y. and Wang, M. (2022). Genomic analysis of the endosomal sorting required for transport complex III pathway genes as therapeutic and prognostic biomarkers for endometrial carcinoma. Transl. Cancer Res. 11, 3108–3127. doi: 10.21037/tcr-22-660
Yang, J.-Y., Wang, P., Li, C.-Y., Dong, S., Song, X.-Y., Zhang, X.-Y., et al. (2016). Characterization of a new M13 metallopeptidase from deep-sea Shewanella sp. E525–6 and mechanistic insight into its catalysis. Front. Microbiol. 6, 1498. doi: 10.3389/fmicb.2015.01498
Keywords: Pyropia, red algae, 1-aminocyclopropane-1-carboxylic acid, ethylene, plant hormone
Citation: Uji T, Sasaki S and Mizuta H (2025) 1-Aminocyclopropane-1-carboxylic acid induces resource reallocation in Pyropia yezoensis sporophytes. Front. Plant Sci. 16:1632530. doi: 10.3389/fpls.2025.1632530
Received: 28 May 2025; Accepted: 18 July 2025;
Published: 15 August 2025.
Edited by:
Mizanur Rahman, The University of Texas Rio Grande Valley, United StatesCopyright © 2025 Uji, Sasaki and Mizuta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Toshiki Uji, dC11amlAZmlzaC5ob2t1ZGFpLmFjLmpw
 Toshiki Uji
Toshiki Uji Shun Sasaki
Shun Sasaki Hiroyuki Mizuta
Hiroyuki Mizuta