- 1Institute of Data Science and Agricultural Economics, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
- 2Agricultural Information Institute, Chinese Academy of Agricultural Sciences, Beijing, China
Gene editing technology is a revolutionary biotechnology that has shown great potential and advantages in crop breeding. Current research has proposed many technical methods and design schemes for gene editing technology in crop breeding. However, summarization and analysis are often based on the research and application of a certain technology, lacking a literature content mining perspective to summarize and analyze the application of gene editing and other technologies in crop breeding. At the same time, there is insufficient identification of future research and innovation opportunities of gene editing technology in crop breeding. This study utilized natural language processing, deep learning, and generative topographic mapping (GTM) to conduct an in-depth analysis of the literature on gene editing technology in crop breeding from the perspective of literature mining. Key technical terms in this field were identified, a literature technical map was constructed, technical blank points were identified, and innovative opportunities for blank technology combinations were analyzed. The results showed that from the literature data from 2020 to 2024, 13 technology combinations were identified. These technical contents cover the multi-technology combination strategy of molecular genetic research, the core technology of gene function research in molecular genetics of biotic and abiotic stresses, the technical means of analyzing the molecular mechanisms of stress resistance, the technical scheme of genetic improvement, etc., which provide support for revealing the potential technological innovation opportunities of gene editing technology in the field of crop breeding. This study can scientifically, objectively, and efficiently identify technological innovation opportunities from the literature. Based on the research results, future research should carry out experimental research and application exploration so as to support the application and technological innovation of gene editing technology in crop breeding.
1 Introduction
Crop breeding is an important component of agricultural biotechnology. Through selection, hybridization, and molecular biology, new varieties with excellent traits, such as high yield, disease resistance, stress resistance, and high quality, were bred to meet agricultural production and market demand (Jiang et al., 2023). With the continuous growth of the global population and the increasing impact of climate change, cultivating high-yield, high-quality, and strong stress-resistant crop varieties in a short time has become an urgent problem to be solved (Zainuddin et al., 2024). Traditional crop breeding relies on phenotypic selection, with superior offspring selected based on observed variations in crop phenotypes. This method is highly dependent on the experience of breeding experts and is time-consuming and inefficient, making it difficult to meet the rapidly changing breeding needs (Tyagi et al., 2024). With the continuous innovation and breakthroughs of breeding technology, cutting-edge biotechnologies such as transgenic technology, gene editing technology, double haploid technology, and synthetic apomixis technology have emerged, providing new opportunities for crop breeding.
As a revolutionary biotechnology, gene editing technology has shown great potential and advantages in crop breeding. By precisely modifying specific gene sequences in crop genomes, gene editing technology can achieve specific trait improvements in the precision and efficiency of breeding (Li et al., 2020; Nerkar et al., 2022; Li et al., 2024). With the deepening understanding of crop trait regulation mechanisms by scientists and the development of gene editing tools, precise gene localization, insertion, and knockout of target genes can be achieved, thereby eliminating or reducing certain adverse traits, subsequently enhancing crop disease resistance, insect resistance, and nutritional value. This technology not only shortens the breeding cycle but also reduces the research and development cost. It provides strong technical support for global food security. In addition, gene editing technology can also improve crop varieties according to specific environmental conditions, making them better at adapting to different growing environments, thereby improving the stability and sustainability of agricultural production (Gupta et al., 2024). Therefore, research on the technological innovation of gene editing technology in crop breeding will bring a revolutionary breakthrough for the sustainable development of agriculture. The precise regulation of crop genes can break through the limitations of traditional breeding and also solve global challenges such as food security, climate change, and nutritional health.
In recent years, significant progress has been made in the application research of gene editing technology in crop breeding. Dong (2024) elaborated on the application of mutations in regulatory regions via CRISPR/Cas techniques in crop breeding. Shi et al. (2023) provided an update on the application of promoter editing in crops for increased yield, enhanced tolerance to biotic and abiotic stresses, and improved quality. Yu et al. (2023) analyzed optimization methods for improving prime editing efficiency and their potential in improving crop breeding. Mukhtiar et al. (2025) highlighted CRISPR/Cas and its expanding suite of tools, such as polygenic editing, while delving into its latest innovative technologies, including tissue-specific editing, CRISPR-based gene drives, and epigenetic modifications through dCas9. Song et al. (2025) proposed to use plant prime editing (PPE) for precise plant genome modification, which overcame the limitations of traditional gene editing methods that rely on double-strand breaks and exogenous donor DNA. Liu et al. (2025) focused on three modules that drive precise DNA changes: DNA-targeting modules, effector modules, and control modules, as well as their optimization methods. At the same time, the paper also outlined innovative tools such as optogenetic systems and receptor-integrated systems that enable spatiotemporal control of genome editing expression. Li W. et al. (2025) reviewed the transformative role of large language models (LLMs) in synthetic biology (SynBio) education and research and summarized and analyzed the progress and development potential of LLMs in biomanufacturing. Zhang et al. (2025) established a vector database of over 60,000 research articles for retrieval and enhanced generation and fine-tuned the Llama3-8B model using the language data of 13,993 Arabidopsis thaliana phenotypes and 23,323 gene functions to construct a virtual expert PlantGPT for Arabidopsis phenotype gene research, providing support for functional genomics research of food crops.
The identification of technology opportunities is a process of technology monitoring and analysis based on bibliometrics and expert opinions (Liu et al., 2023). The analysis methods of technology opportunity are mainly divided into qualitative analysis and quantitative analysis. In terms of qualitative analysis methods, such as the Delphi method and technology roadmap, these methods rely on the knowledge and experience of domain experts. Experts make judgments and evaluations on technical opportunities after investigation and research. This kind of method makes full use of the collective wisdom and experience of domain experts and has been relatively mature through the comprehensive sorting, induction, and analysis of different ideas and views to identify technical opportunities. However, such methods are time-consuming, highly subjective, and easily limited by the breadth and depth of technical experts’ knowledge, resulting in the problem of result deviation. In terms of quantitative analysis methods, many experts and scholars use artificial intelligence methods such as machine learning, network analysis, and link prediction to carry out research on technology opportunity identification. Park and Yoon (2018) used the link prediction method based on citation network to construct potential technological knowledge flows (TKFs) and then extracted the converged technology opportunities by predicting the potential technological knowledge flow between different fields. Cao et al. (2023) adopted semantic analysis and dynamic network analysis methods to capture keyword semantics, mine keyword networks, and realize the potential recombination opportunities of detecting core keywords. Yang et al. (2022) identified technology opportunities for radical inventions (RIs) by measuring the value of technological novelty (VON) of each technology manifested in a patent set and the value of difficulty (VOD) of each research and development (R&D) theme contained in the patent set.
At present, there are many methods to identify technology opportunities. Among them, generative topographic mapping (GTM) is a non-linear hidden- variable model in the field of machine learning, which can realize the non-linear dimensionality reduction of high-dimensional data and is widely used in data analysis and visual analysis (Bishop et al., 1998; Kaneko, 2019). Compared with other technology recognition methods, GTM can maintain the original topological relative relationship of data in high-dimensional space as much as possible and establish a bidirectional mapping between high-dimensional and low-dimensional space. Using the GTM method to draw the technical map, the technical blank points can be visually displayed on the map. At the same time, using the reverse mapping method of GTM can realize the combination of technical words involved in obtaining the blank points in the map. Therefore, the GTM method for technology blank recognition has an excellent visualization effect and can reverse interpret the technology blank. Many scholars have carried out technology opportunity identification based on the GTM method. Zhou and Ban (2023) established a research framework for the identification and evaluation of potential technology opportunities based on GTM and system dynamics (SD) modeling from the perspective of value proposition, further improved the accuracy of technology opportunity identification, and provided theoretical and practical support for enterprises to optimize the layout of technology directions with the value orientation of technology. The research is applied in the field of new energy vehicles. Li et al. (2023) first used the GTM patent map to identify technical gaps and reverse interpret them, then clustered scientific literature to obtain scientific knowledge topics, and finally evaluated the similarity between potential technical opportunities and scientific topics through the cosine similarity value to screen out scientific and feasible technological innovation opportunities. This research is applied in the field of medical equipment. Wang et al. (2022) proposed an automated technology opportunity discovery method combining subject–action–object (SAO) and GTM. The automated method focusing on semantic information helps to understand technology opportunities and improve the accuracy of discovery. This research is applied in the field of coal bed methane extraction technology.
In summary, the current research has proposed many technical methods and design schemes for gene editing technology in crop breeding. However, it is often based on the research and application of a certain technology, lacking a literature content mining perspective to summarize and analyze the application of gene editing and other technologies in crop breeding. There is also insufficient identification of future research and innovation opportunities of gene editing technology in crop breeding. At the same time, when using a variety of data mining and analysis methods to identify and analyze technology opportunities, many scholars have put forward innovative and practical technical methods. However, there is also the possibility of omitting technical points by analyzing subject words and keywords as technical elements. In addition, the identification of technological innovation opportunities is very important for technological research and development and breakthroughs, but there is no research achievement on the identification of technology opportunities of gene editing technology in crop breeding in the public literature.
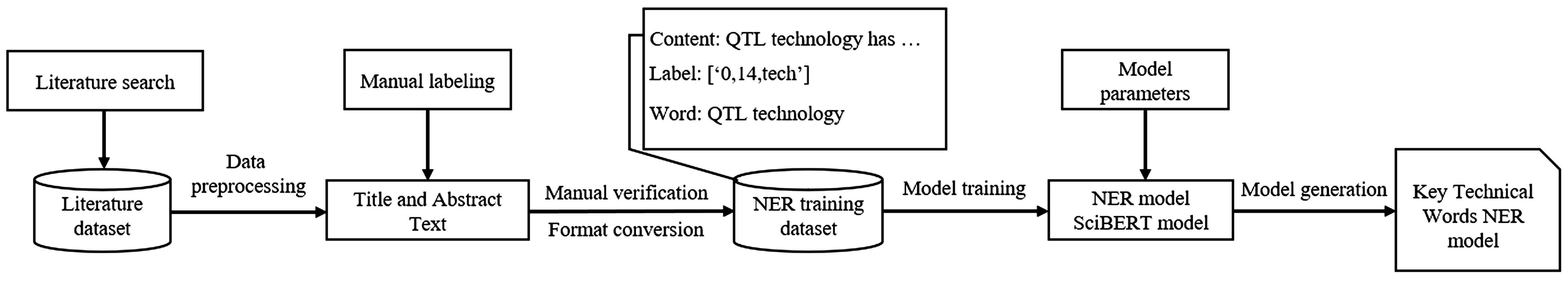
Therefore, based on the research literature dataset of gene editing technology in the field of crop breeding, this paper used spaCy, SciBERT, and GTM methods to identify potential innovation opportunities for gene editing technology and conducted an in-depth analysis of technological innovation opportunities to provide support for the future technological innovation and development of gene editing technology in crop breeding. This study used spaCy’s named entity recognition (NER) method to identify key technical words in literature titles and abstracts and chose to add the SciBERT pre-trained model to the spaCy pipeline for model training. SciBERT (Beltagy et al., 2019) is a BERT model that uses a large corpus of scientific publications, including a total of 1.14 million samples of papers in biomedicine (82%) and computer science (18%), for unsupervised pre-training, making it more suitable for natural language processing tasks on literature data. Based on the training dataset, the NER model fused with Transformer was constructed and trained.
Traditional expert judgment methods are subjective, one-sided, and easily influenced by the breadth and depth of expert knowledge. In addition, compared with the various current large language models, the advantages of using SciBERT for named entity recognition are as follows:
i. Professional advantages in semantic understanding. The big language model is an autoregressive generative model, whose training objective is to predict the probability distribution of the next word based on a given preceding text. Its training method is unidirectional, making it particularly powerful in generating coherent text. However, its understanding of context is relatively one-sided compared to SciBERT. The core idea of SciBERT is to pre- train the encoder through bidirectional context so that semantic information can be obtained from the bidirectional context, making its understanding of text more comprehensive.
ii. Professional advantages in the research field. Although the big language model has extensive knowledge, there is a problem of insufficient recognition accuracy when performing specialized terminology recognition due to the use of models trained on general corpora. SciBERT utilizes a pre- trained BERT model based on a large library of scientific publications, which provides a more accurate understanding of professional terms appearing in literature and is suitable for identifying named entities.
iii. Advantages of structured information extraction. After extracting structured information, the big language model requires additional standardization processing, which is less efficient compared to using the SpaCy+SciBERT method. The SpaCy+SciBERT method can combine multiple rules for precise extraction while providing a standardized NER pipeline to output structured entity labels for subsequent data analysis.
iv. Advantages in efficiency and cost. The fine-tuning cost of large language models is relatively high, requiring a large amount of computing power for inference, and there is also an illusion effect.
The aim of this study was to apply natural language processing technology, deep learning, and GTM methods to identify potential innovation opportunities for gene editing technology and conduct an in-depth analysis of technological innovation opportunities. By integrating multiple data mining and technology opportunity identification methods, potential technological discoveries can be provided for the development of new editing tools, precise regulation techniques, gene delivery mechanisms, off- target effects, and other directions. This has high application prospects for improving the efficiency and accuracy of genome editing technology, developing artificial intelligence (AI)-powered breeding tools, and other directions.
2 Materials and methods
2.1 Data sources
The literature data for this study were retrieved from the Web of Science Core Collection of the Science Citation Index Expanded database, and the search time was January 3, 2025. Subject keywords were used to retrieve a literature dataset of gene editing technology in crop breeding. Considering that the literature before 2020 may have deficiencies in novelty, in our study, the publication date was set from January 1, 2020, to December 31, 2024, and the literature types were limited to research papers and reviews. To ensure the quality of the literature, this study manually browsed through the titles and abstracts of the literature to determine whether the article was relevant to the selected target research topic in order to eliminate irrelevant literature. Finally, this study obtained a basic dataset of 17,234 literature, including 15,880 papers and 1,354 reviews.
2.2 Recognition of key technical words
spaCy’s NER method was used to identify key technical words in literature titles and abstracts. The spaCy is a natural language processing library written based on Python. Using spaCy’s named entity recognition method, entities with specific meanings can be extracted from unstructured text. The predefined NER tags of spaCy do not involve specific types of key technical entities, so the custom NER model was trained through spaCy to recognize key technical words of crop breeding.
2.2.1 Dataset construction
To ensure the effectiveness and accuracy of model training, a high-quality and standardized training dataset was constructed. Based on the literature title and abstract texts obtained from previous searches, 1,000 pieces of literature were randomly selected as the training dataset source. Key technical words that appeared in the dataset were annotated manually. Finally, a dataset that met the training requirements was obtained.
2.2.2 Recognition model training
spaCy allows using Transformer models (such as BERT, GPT-2, and XLNet) for natural language processing tasks. Therefore, combining the powerful context understanding capabilities of Transformer models for NER model training can produce better recognition effects. The SciBERT pre-trained model was added to the spaCy pipeline for model training. Based on the training dataset, the NER model fused with Transformer was constructed and trained. After multiple rounds of parameter tuning, a NER model that met the key technical word recognition requirements for crop breeding was finally obtained. The model construction process is shown in Figure 1.
2.3 Recognition of technical blank points
Technology map was used to identify the blank of gene editing technology. Technology map is composed of technical data information, and the original high-dimensional data space was mapped to a low-dimensional regular grid by an algorithm. The blank points displayed in the technical map represent the lack of corresponding technical combinations for the coordinate point, which are called technical blank points. By systematically analyzing and interpreting these technical blank points, potential opportunities for technological innovation can be effectively identified, providing new breakthrough directions for technological development.
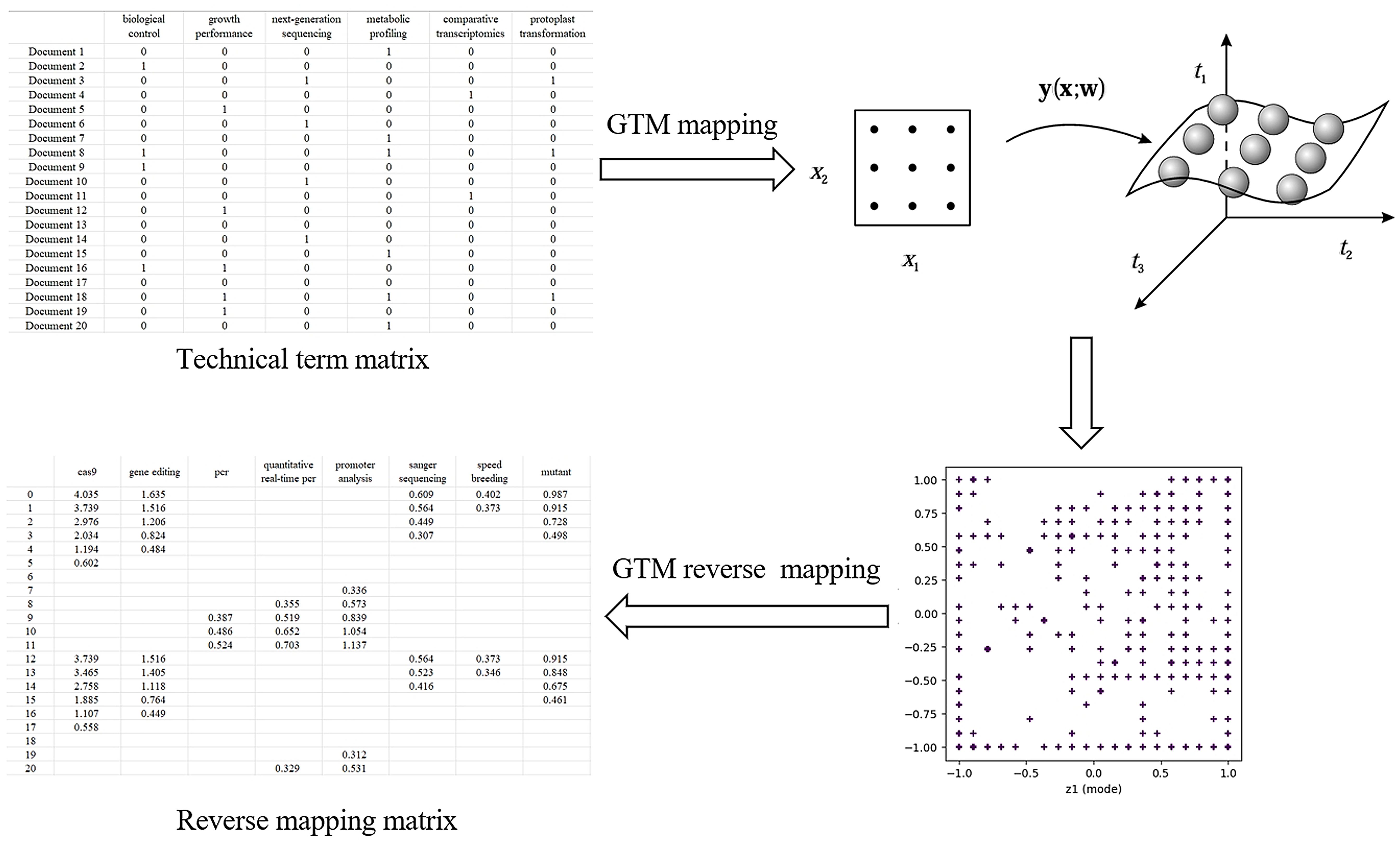
The GTM was used to draw technical maps. It helps to understand and analyze complex datasets by mapping high-dimensional data to low-dimensional latent space. The flowchart of technical blank identification is shown in Figure 2.
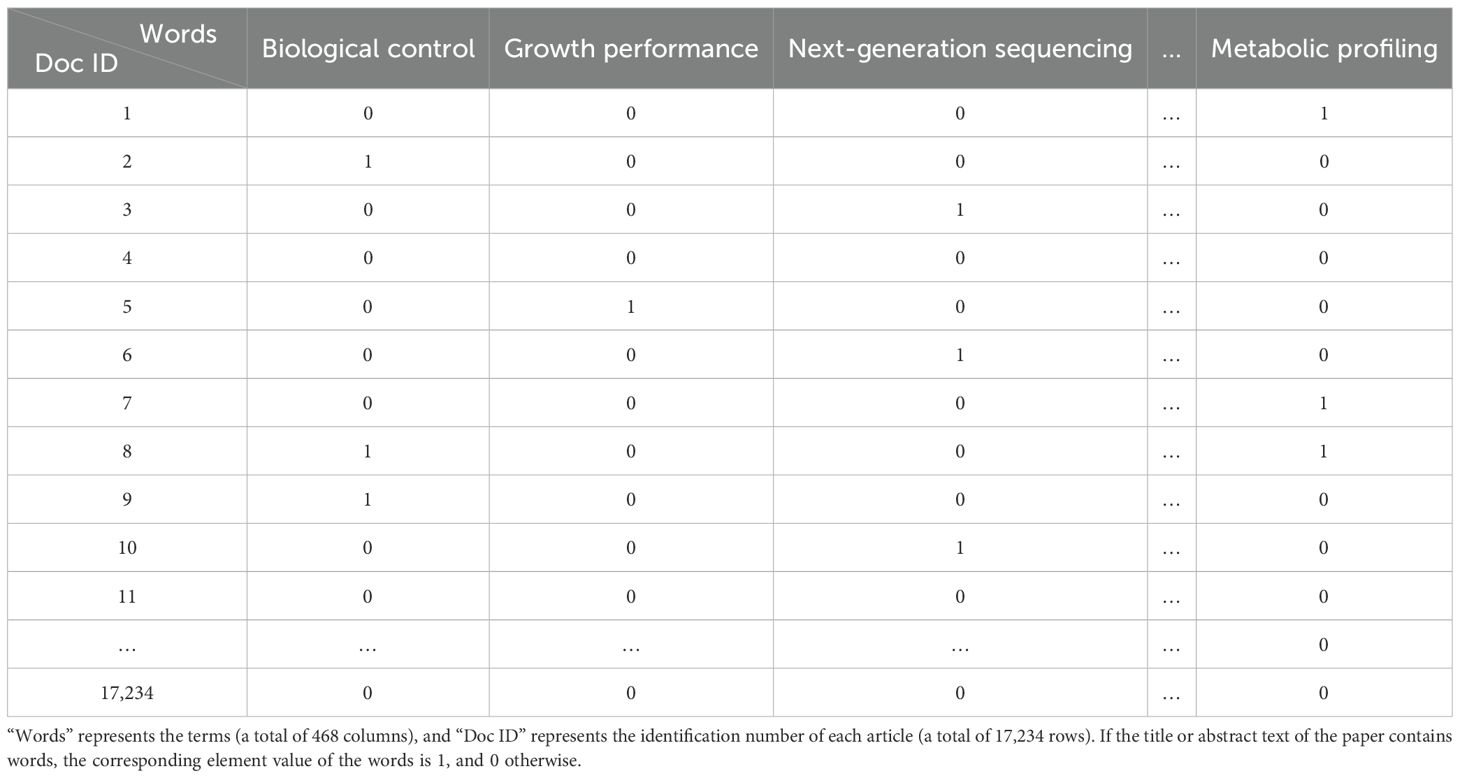
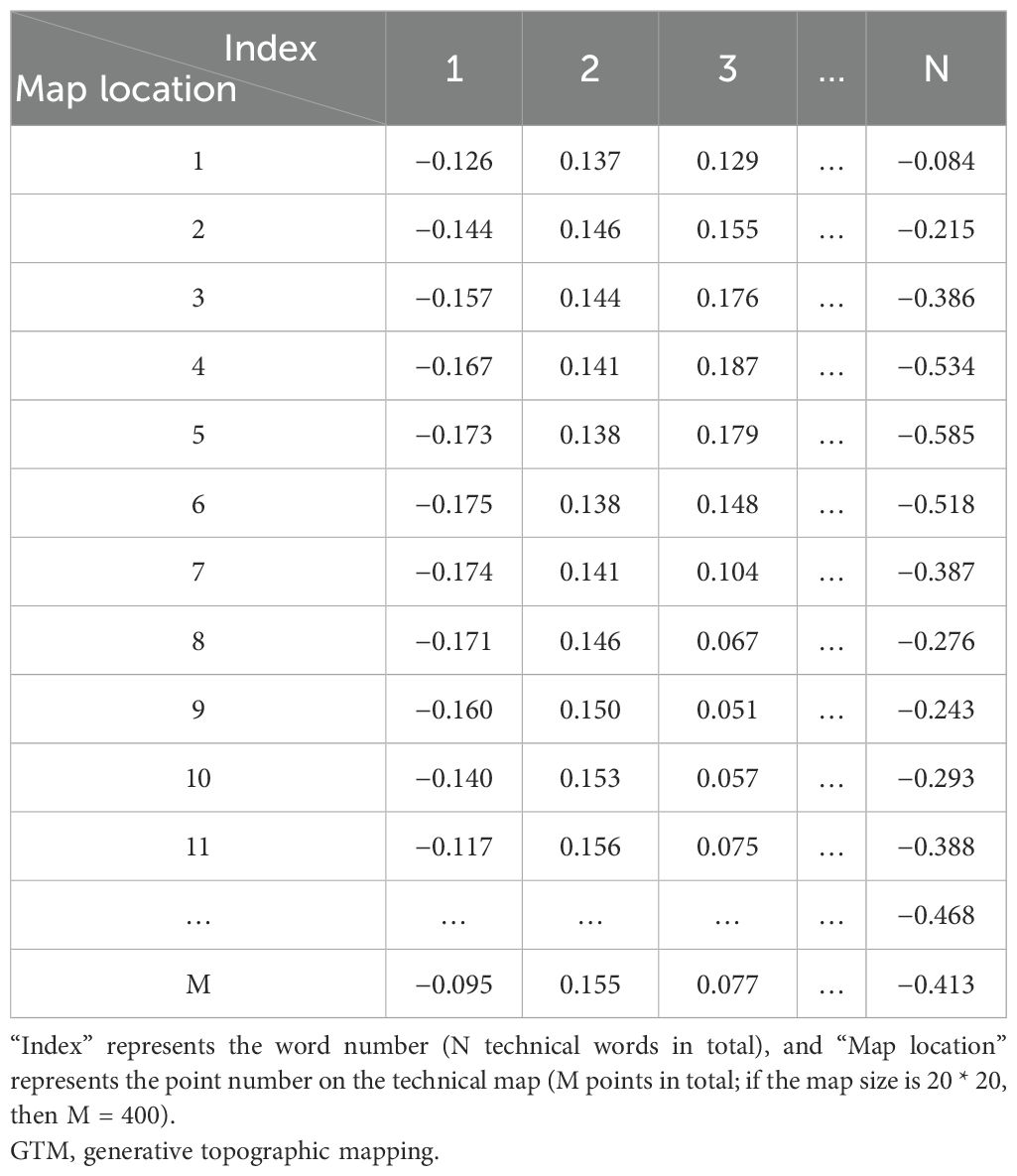
Based on the key technical word recognition model constructed in the early stage, the key technical words were recognized in the breeding literature data, and the key technical words appearing in the literature were obtained. After data deduplication and synonym merging, key technical words from the literature were selected and confirmed by domain experts. According to the literature title and abstract text, the key technical word representation vector of the binary representation was constructed. The construction method is as follows. When the title or abstract text of a paper contains the identified key technical words, the corresponding element value in the technical word vector is 1, and 0 otherwise, and finally, the technical word matrix of the paper data is formed. The technical word matrix is shown in Table 1. The constructed multi-dimensional technical feature word matrix is imported into the GTM model, and the multi-dimensional vector is reduced to two-dimensional space, in which each technical data point is mapped to a two-dimensional plane. Specific points in the map represent existing technologies in the technical field, and blank areas in the technical field are potential technology opportunities, as shown in Figure 3. On this basis, the potential technology opportunities are reversely mapped back to the multi-dimensional data blank through GTM reverse mapping, as shown in Table 2. By setting the threshold to screen technical words and combining it with the analysis of technical words by experts, the interpretation of potential technical opportunities is completed.
3 Results
3.1 Recognition results of key technical words
3.1.1 Data labeling
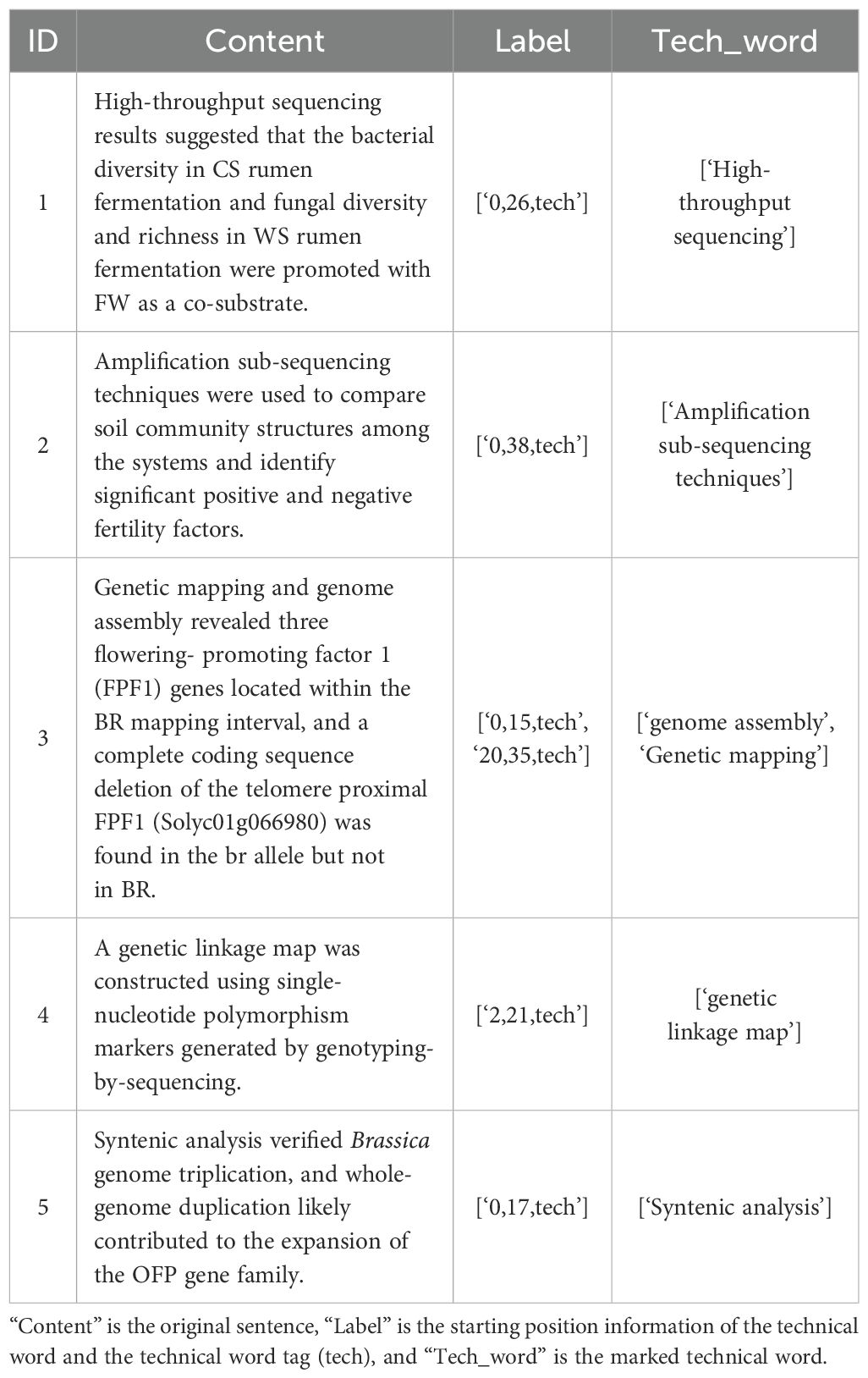
First, we specified that the marked contents are words, phrases, abbreviations of terms, etc., indicating technology. Second, we invited two experts in the field to carry out the annotation work in the way of independent annotation. Finally, we determined the final labeled entity training set by negotiating the inconsistent labeling content. In the process of annotation, we required domain experts to read and analyze the title and abstract text and mark the technical words appearing in the text. At the same time, we used natural language processing tools to extract the sentence and location information of the marked words and convert it into a format consistent with the training data. Finally, through data cleaning, manual verification, and other operations, we obtained 757 labeled data points that met the training data requirements. An example of the labeling result is shown in Table 3, where Content is the original sentence, Label is the starting position information of the technical word and the technical word tag, and Tech_word is the technical word.
3.1.2 Technical word recognition
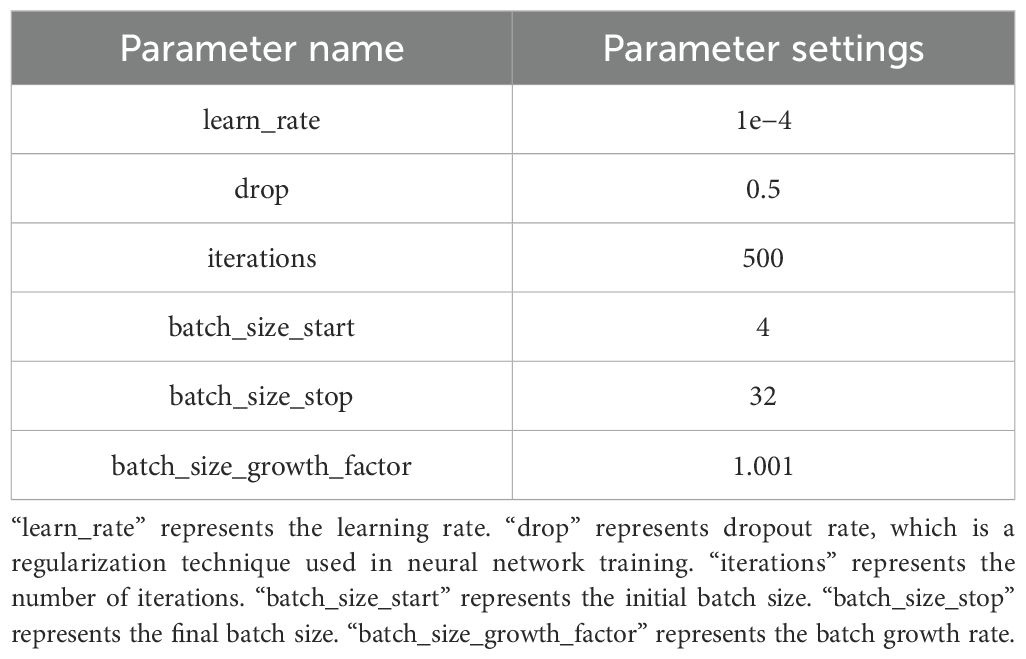
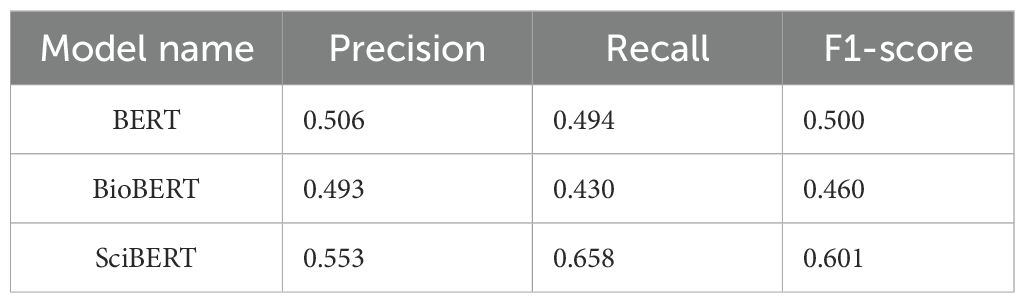
A named entity recognition model was constructed based on NER+SciBERT, and the model was trained with labeled data to generate a technical word recognition model consistent with this study. The optimization results of model training parameters are shown in Table 4. Using the key technical word recognition model, the key technical words of literature titles and abstract texts in the literature dataset in the field of crop breeding were recognized, and candidate key technical words were obtained. After data deduplication and synonym combination, 468 technical words were obtained. The BERT, BioBERT, and SciBERT models were used to compare the performance, and the precision, recall, and F1-score of the recognition task were evaluated. The experimental results are shown in Table 5. Finally, the performance of the SciBERT model for the annotation task in this study was verified.
3.2 Technology opportunity analysis
3.2.1 Technical blank identification results
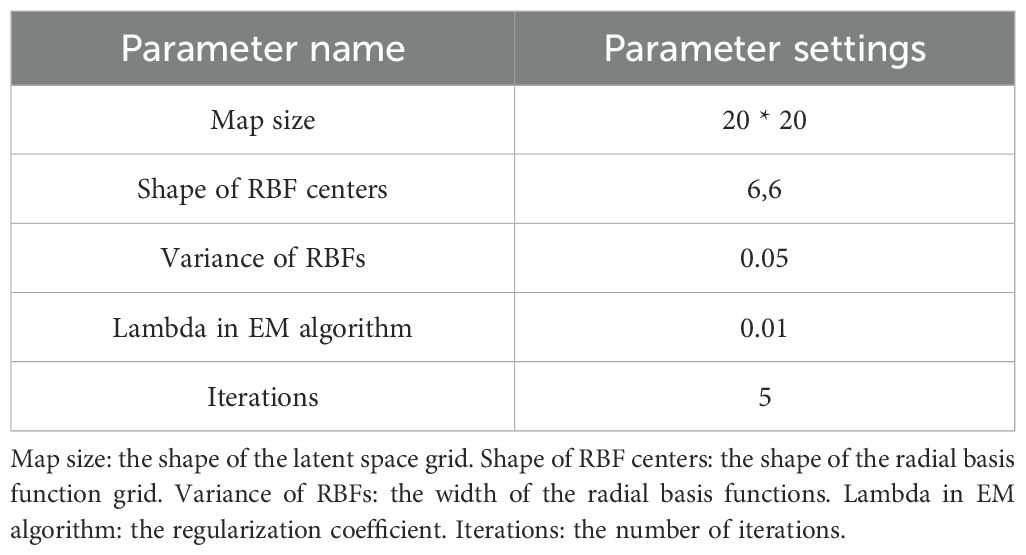
Based on the key technical words identified above, and combined with the literature information, the technical word matrix of literature data was constructed, and the multi-dimensional technical feature word matrix was input into the GTM model. The selection of GTM model parameters affected the visualization effect of the technical map. Through multiple parameter adjustments, the display effect of the technical map and the results of reverse mapping were checked, and the best GTM model parameters of this study were finally obtained. The specific parameters are shown in Table 6. Map size controls the resolution of the latent grid, affecting model complexity and detail capture. Shape of Radial Basis Function (RBF) centers determines the layout of basis functions, influencing mapping uniformity. Variance of RBFs adjusts the width of basis functions, balancing smoothness and overfitting risk. Lambda in EM algorithm regularizes model weights to trade off complexity and generalization. Iterations sets the maximum EM steps, impacting convergence and computational cost.
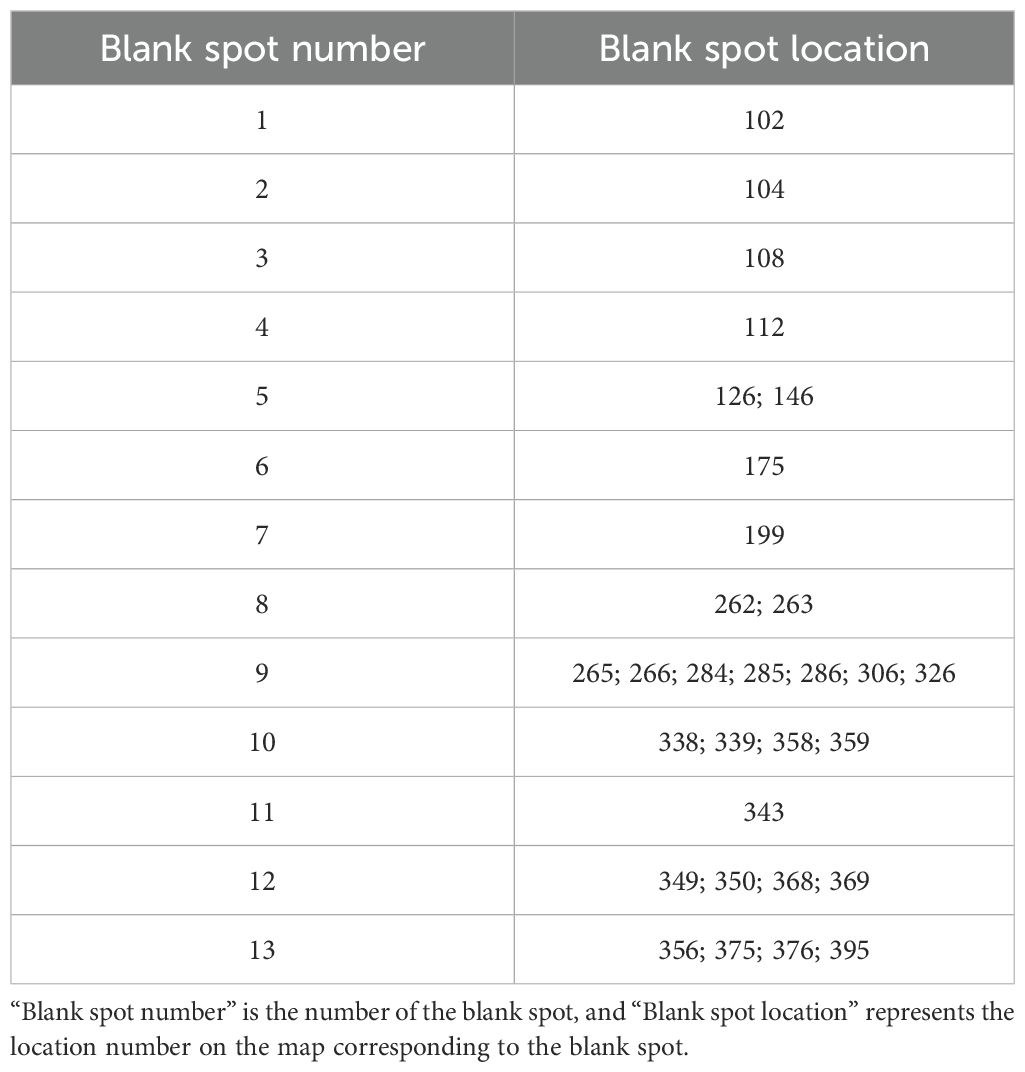
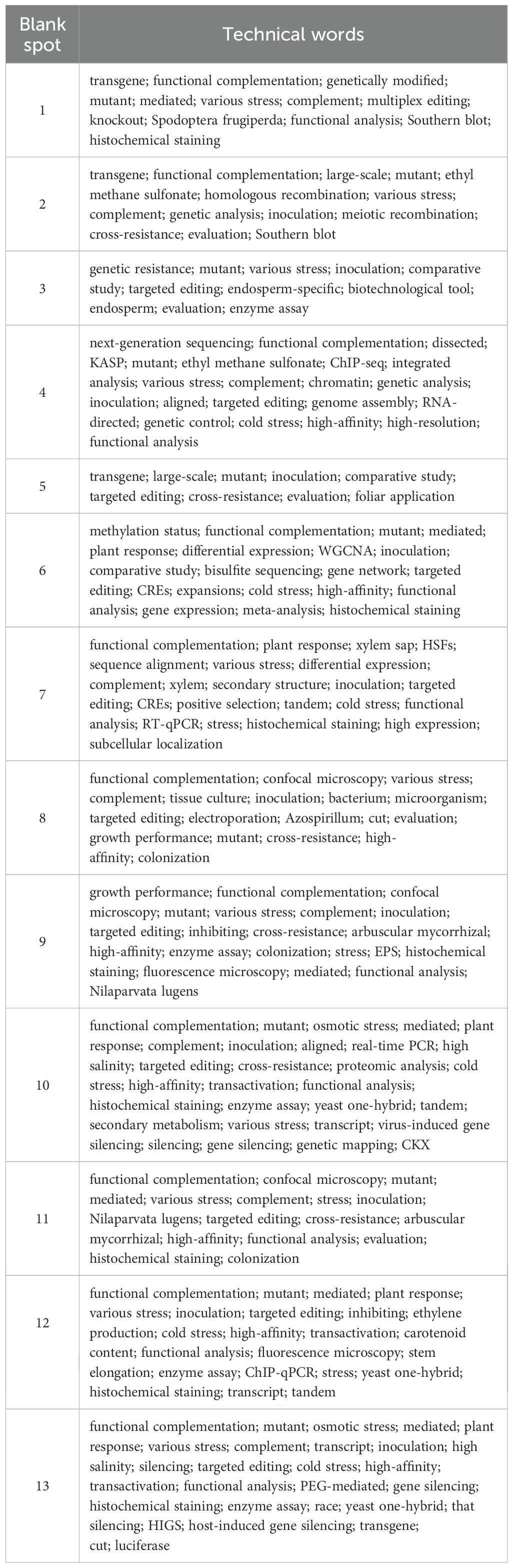
On this basis, we drew the technical map using the GTM model, as shown in Figure 4. There are 13 blank spots in the figure, and the specific positions are shown in Table 7. The blank spots are marked with Arabic numerals 1–13 in the order from left to right and from bottom to top. Blank spot location represents the location number on the map corresponding to the blank spot. For the screening of technical terms in GTM reverse mapping results, we referred to the threshold set in the experiment in the relevant literature (Zhou and Ban, 2023). At the same time, we focused on the technical words in the GTM reverse mapping results and finally determined that the threshold setting of 0.3 is more reasonable. We analyzed the blank spots, and we screened the technical words in the blank spots by setting the threshold value to 0.3. The screening results are shown in Table 8. Among them, each column represents a technical word, and each row represents the position of technical map points. Therefore, we extracted the technical combination words included in each technical blank point in combination with the blank points in Table 7. In order to facilitate intuitive viewing and analysis of technical combinations, we screened technical words, among which Table 9 shows the technical words that appeared commonly in technical combinations, and Table 10 shows the technical words that did not appear commonly in technical combinations. Domain experts can realize the interpretation and analysis of technical combinations by comprehensively analyzing technical words in the two tables.
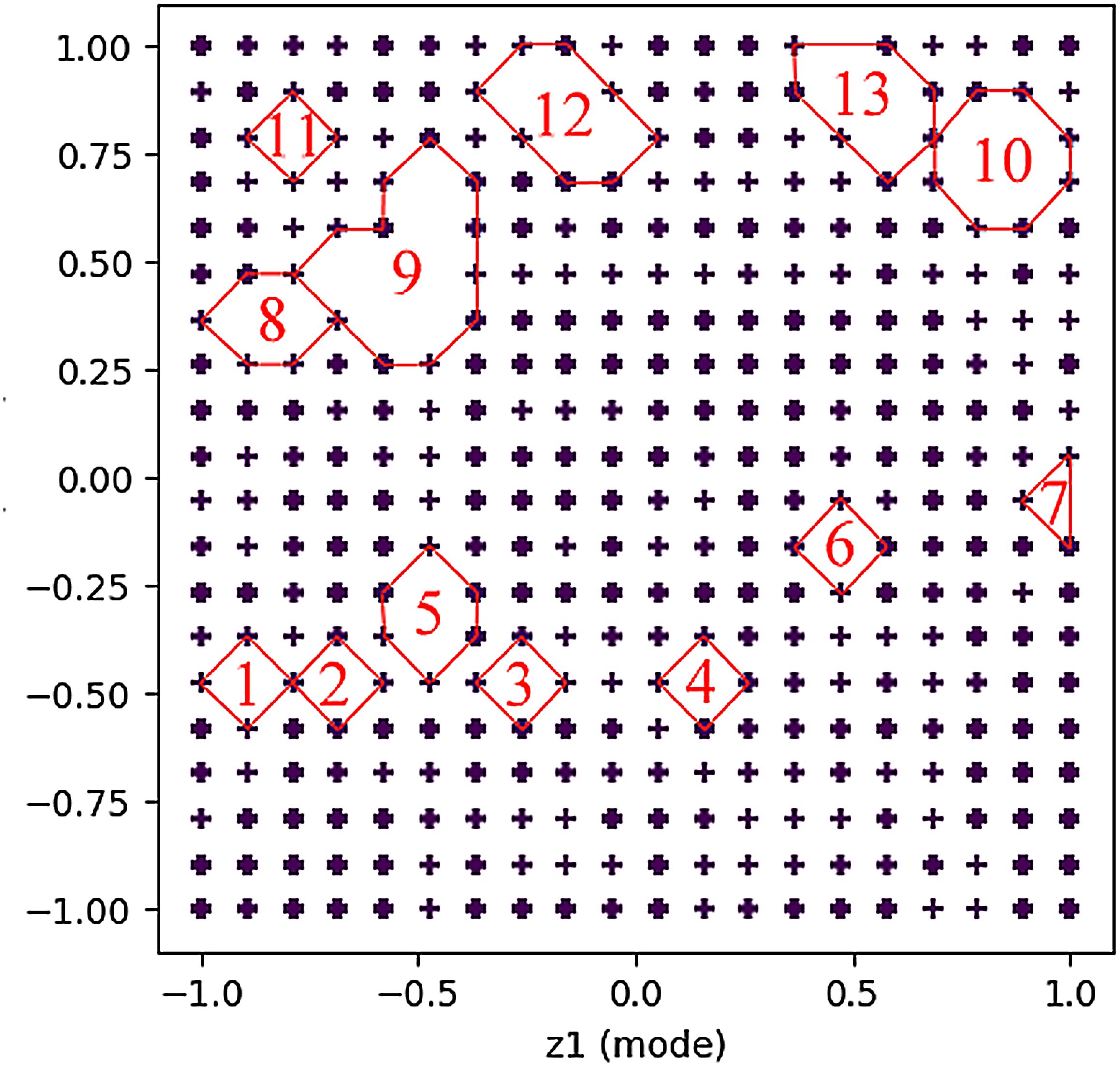
Figure 4. Technical map results generated based on GTM. A total of 13 groups of technology combinations are marked. GTM, generative topographic mapping.
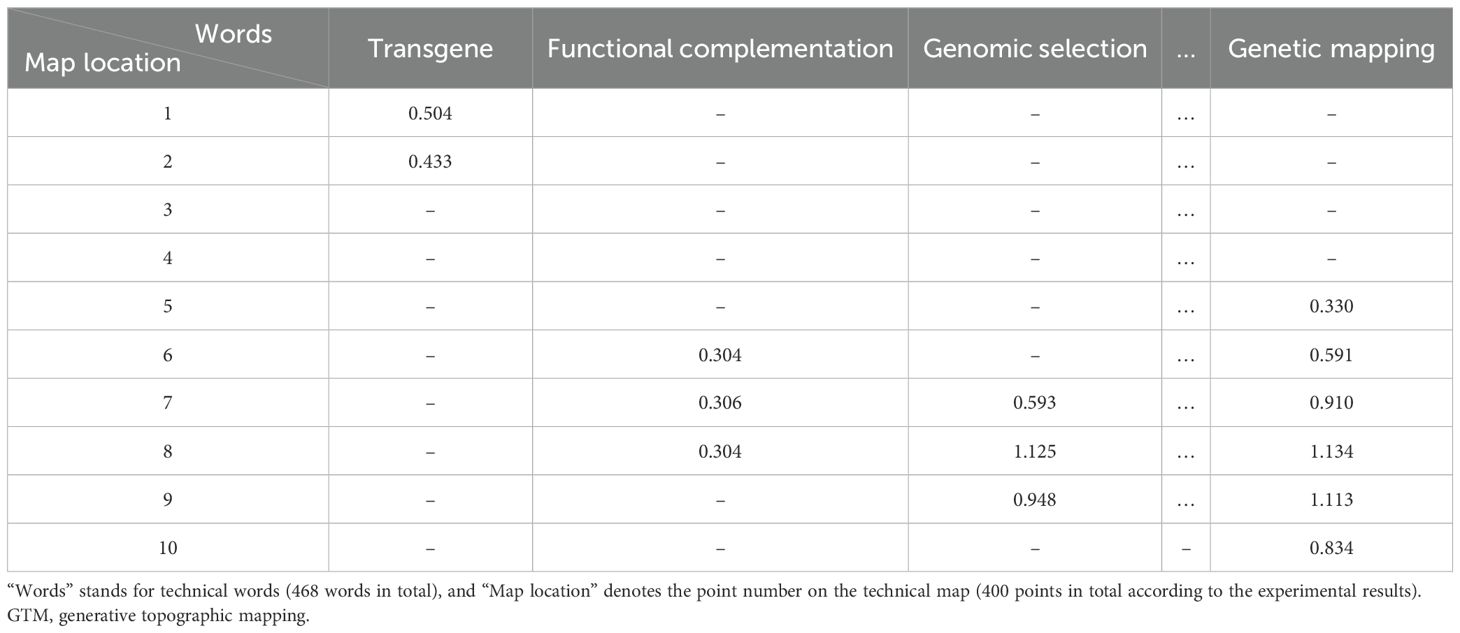
Table 8. Threshold screening based on the reverse mapping results of the GTM model (partial results of the first 10 points), and the results greater than the threshold of 0.3 are retained.
3.2.2 Technical combination analysis
3.2.2.1 Application of CRISPR/Cas9 technology in the precise regulation of agricultural biology
The first technology combination complements molecular and genetic research. By comprehensively applying technical methods such as transgene, mutant analysis, and functional complementarity analysis, the function of genes and their mechanism of action in organisms can be more comprehensively revealed. Understanding the potential uses of related technologies helps to develop new strategies for targeting key functional genes. Related research: “PEG-Delivered CRISPR-Cas9 Ribonucleoproteins System for Gene-Editing Screening of Maize Protoplasts” (Sant’Ana et al., 2020) and “CRISPR/Cas9 mediated editing of pheromone biosynthesis activating neuropeptide (PBAN) gene disrupts mating in the Fall armyworm, Spodoptera frugiperda” (Ashok et al., 2023).
3.2.2.2 Collaboration between gene editing and transformation technology promotes research on rice breeding
The second technology combination covers the core technologies of gene manipulation (transgenic and homologous recombination), mutant library construction [ethyl methane sulfonate (EMS) mutagenesis and large-scale screening], functional verification (complementary experiments and Southern blotting), and multi-dimensional evaluation (stress resistance and safety), which together constitute the core tool chain of stress resistance and disease resistance molecular genetics research. Related research: “Concurrent Disruption of Genetic Interference and Increase of Genetic Recombination Frequency in Hybrid Rice Using CRISPR/Cas9” (Liu et al., 2021) and “Agrobacterium-Mediated Genetic Transformation of Wild Oryza Species Using Immature Embryos” (Shimizu-Sato et al., 2020).
3.2.2.3 Research on the application of biotechnology in crop improvement
The third technology combination forms a research chain of targeted editing to create mutants, inoculating pathogens or applying stress, and evaluating resistance through enzyme activity and phenotype; and to compare and analyze to optimize editing strategies, providing new research strategies for analyzing molecular biology research on disease resistance and abiotic stress. Related research: “Overexpression of Nepenthesin HvNEP-1 in Barley Endosperm Reduces Fusarium Head Blight and Mycotoxin Accumulation” (Bekalu et al., 2020) and “CRISPR/Cas9-mediated editing of phytoene desaturase (PDS) gene in an important staple crop, potato” (Siddappa et al., 2023).
3.2.2.4 Technical innovation of crop physiological metabolism and quality improvement
The fourth technology combination forms a complete research chain from gene discovery [next-generation sequencing (NGS) and mutant screening] to functional verification (complementation and editing) and then to mechanism analysis [Chromatin immunoprecipitation followed by sequencing (ChIP-seq) and interaction analysis), providing a new technical method for analyzing stress- resistant molecular genetic biology research. Future research trends can focus on the combination of multiple technologies (such as single-cell sequencing combined with spatial transcriptome) and high-throughput automated analysis. Related research: “Mass screening of rice mutant populations at low CO2 for identification of lowered photorespiration and respiration rates” (Mubarak et al., 2023) and “The molecular mechanism of polyphenol oxidase 1 and the genetic improvement of wheat polyphenol oxidase activity” (Zhai et al., 2023).
3.2.2.5 Research on precision breeding strategies and tools
The fifth technology combination achieves the cultivation of new stress-resistant varieties by combining targeted editing and resistance assessment in agriculture through precise genetic manipulation, systematic phenotypic analysis, and multi-level evaluation. The combination of technologies provides experimental protocols to promote molecular mechanism analysis and genetic improvement. Related research: “Loss of Function of OsFBX267 and OsGA20ox2 in Rice Promotes Early Maturing and Semi-Dwarfism in γ-Irradiated IWP and Genome-Edited Pusa Basmati-1” (Andrew-Peter-Leon et al., 2021) and “Genome-scale targeted mutagenesis in Brassica napus using a pooled CRISPR library” (He et al., 2023). By optimizing codon, promoter, and editing conditions, the prime editors were adapted for use in plants, thereby achieving point mutations, insertions, and deletions in rice and wheat protoplasts (Lin et al., 2020).
3.2.2.6 Analysis and editing strategy of gene regulatory network for crop traits
The sixth technology combination forms a complete research chain to study the role and regulatory mechanism of key genes from epigenetics (methylation status analysis using bisulfite sequencing and differential expression using RNA-seq) to transcriptional regulation and then to functional verification. Future research can further integrate multi-omics data and spatial–temporal dynamic analysis to improve the accuracy of complex traits. Related research: “Understanding the regulatory relationship of abscisic acid and bZIP transcription factors towards amylose biosynthesis in wheat” (Kumar et al., 2021) and “CRISPR-Cas9 mediated OsMIR168a knockout reveals its pleiotropy in rice” (Zhou et al., 2021).
3.2.2.7 Research on the identification strategy of key genes for crop traits
The seventh technical combination clarifies the direct role of genes through functional verification (such as complementation and editing). Expression and localization techniques reveal spatiotemporal dynamics. Stress response analysis links gene function and phenotype. Evolutionary and structural analysis resolves the molecular basis of functional differentiation. This combination provides abundant technical means for the molecular mechanism of plant stress response and stress resistance. Related research: “The rice annexin gene OsAnn5 is involved in cold stress tolerance at the seedling stage” (Que et al., 2023) and “BnaC09.tfl1 controls determinate inflorescence trait in Brassica napus” (Zhao et al., 2024).
3.2.2.8 The complete chain of plant– microbe interaction research
The eighth technology combination forms a complete research chain from gene manipulation (targeted editing and electroporation) to functional verification (functional complementation and mutant analysis) and then to phenotypic analysis (confocal imaging and stress response evaluation). This combination provides a core technology for analyzing the molecular and genetic mechanisms of microbial interaction and resistance mechanisms. Related research: “Expression and function of the cdgD gene, encoding a CHASE-PAS-DGC-EAL domain protein, in Azospirillum brasilense” (Cruz-Pérez et al., 2021) and “Diversity and functional characterization of endophytic Methylobacterium isolated from banana cultivars of South India and its impact on early growth of tissue culture banana plantlets” (Senthilkumar et al., 2021).
3.2.2.9 Regulatory strategies for plant disease resistance
The ninth technology combination is mainly used in pathogen or pest–host interaction research, creating mutant materials through targeted editing, and revealing regulatory functions through resistance identification, enzyme activity measurement, and confocal microscopy. This combination of technologies constitutes a complete research chain from phenotypic observation to molecular mechanism analysis and provides a new strategy for the combination of multiple technologies in molecular genetic research. Related research: “Influence of Enhanced Synthesis of Exopolysaccharides in Rhizobium ruizarguesonis and Overproduction of Plant Receptor to these Compounds on Colonizing Activity of Rhizobia in Legume and Non-Legume Plants and Plant Resistance to Phytopathogenic Fungi” (Kantsurova et al., 2024) and “Functional Analysis of BcSNX3 in Regulating Resistance to Turnip Mosaic Virus (TuMV) by Autophagy in Pak-choi (Brassica campestris ssp. chinensis)” (Zhang et al., 2022).
3.2.2.10 Research on crop stress resistance and virus functional genomics
The 10th technology combination systematically reveals the technical system of the role of genes in the stress resistance pathway and promotes the analysis from single gene function to complex regulatory network through collaborative application, especially in the fields of plant stress biology and metabolic engineering. Related research: “Transcription factor TabHLH49 positively regulates dehydrin WZY2 gene expression and enhances drought stress tolerance in wheat” (Liu et al., 2020) and “Long fragment circular efficient PCR (LC-PCR): an integrated technology that modifies large plasmid constructs through site-directed gene insertion, deletion, and mutation, without the need for restriction or ligation” (Jailani et al., 2023).
3.2.2.11 Analysis of pest drug resistance mechanisms and visual detection of symbiotic fungi
The 11th technology combination combines targeted editing and resistance assessment to analyze the cross-resistance mechanism of its drug resistance genes. It can also be used for plant mycorrhizal symbioses, such as dynamic observation of mycelial colonization process by confocal microscopy and histochemical staining. Technology combination provides a new strategy for multi-technology combination in molecular genetic research. Related research: “New insights into chlorantraniliprole metabolic resistance mechanisms mediated by the striped rice borer cytochrome P450 monooxygenases: A case study of metabolic differences” (Xu et al., 2024) and “Anthocyanin pigmentation as a quantitative visual marker for arbuscular mycorrhizal fungal colonization of Medicago truncatula roots” (Kumar et al., 2022).
3.2.2.12 Research on crop development and stress response
The 12th technology combination jointly promotes research progress in gene function analysis, stress response mechanism, metabolic regulation network, and other fields through multi-level and multi-angle research and provides a complete research chain for analyzing the molecular genetic biology of disease resistance and abiotic stress. Related research: “Overexpression of SlGSNOR impairs in vitro shoot proliferation and developmental architecture in tomato but confers enhanced disease resistance” (Rasool et al., 2021) and “Excess iron accumulation affects maize endosperm development by inhibiting starch synthesis and inducing DNA damage” (Zang et al., 2024).
3.2.2.13 Research on the mechanism of crop disease resistance
The 13th technology combination plays an important role in analyzing gene function, regulatory mechanisms, and plant response to stress. Each technology has its own unique role and jointly promotes the progress of plant immunity regulation, pathogenic mechanism, crop improvement, and plant biology. Related research: “Wheat Leaf Rust Fungus Effector Protein Pt1641 Is Avirulent to TcLr1” (Chang et al., 2024) and “Host-induced gene silencing of the Verticillium dahliae thiamine transporter protein gene (VdThit) confers resistance to Verticillium wilt in cotton” (Wang et al., 2024).
3.2.3 Technology opportunity assessment
In order to effectively validate our research method, domain experts were invited to analyze 13 sets of technology combinations and rate them based on three dimensions: scientificity, feasibility, and application prospects. On the basis of the score, the comprehensive score of the evaluation index was calculated by summing the evaluation indexes. The objective evaluation of the innovation of the technology combination was ultimately achieved, and the effectiveness of this method was verified.
The technology portfolio evaluation index is described as follows: evaluate the scientificity of the technology portfolio, that is, whether it follows scientific principles and is supported by theory or empirical evidence. Evaluate the feasibility of the technology combination, that is, whether it can actually be applied or realized under the existing conditions. Evaluate the application prospects of the technology portfolio, that is, the potential value in solving practical problems in the field or promoting the development of the field.
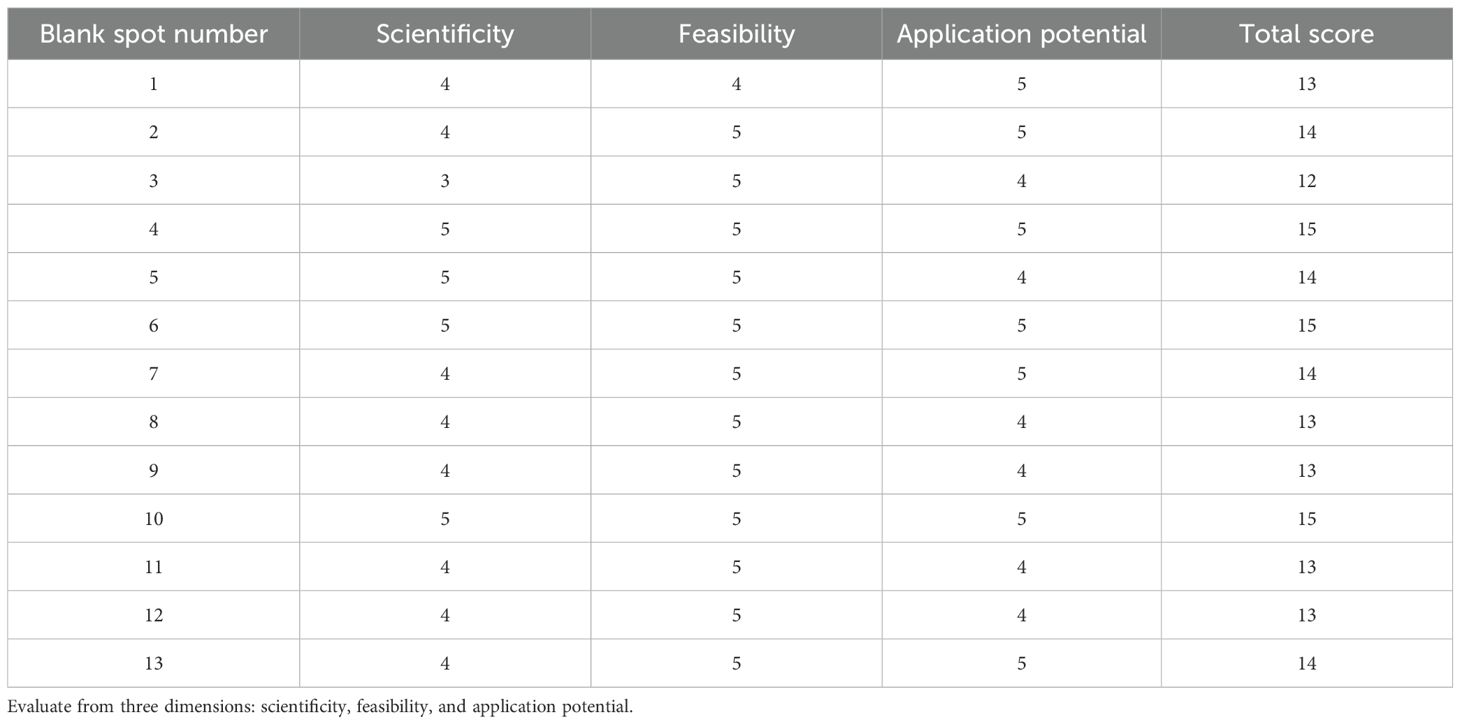
Experts in the field need to score 13 sets of technology combinations independently from 1 (poor) to 5 (excellent). The scoring results of technology combinations are shown in Table 11. The final score of the technical combination was calculated by scoring 13 groups of technical combinations by experts. From the scoring results, it can be concluded that technology combinations 4, 6, and 10 have more potential for technological innovation. Therefore, researchers can focus on and analyze the application of these technology combinations, find the experimental innovation path, and explore the innovation of technology integration.
4 Discussion
4.1 Theoretical implications
The application of gene editing technology in crop breeding is developing rapidly, but it still faces challenges in specificity, precision, delivery efficiency, and safety (Alariqi et al., 2025). Strengthening the integration of gene editing with other technologies (including high-throughput phenotypic analysis, genomic selection, and rapid breeding) is necessary to further promote its widespread application in agriculture (Atia et al., 2024). This study deepened and expanded the application research of gene editing technology in the field of crop breeding by applying new methods and technologies. Through the analysis method of literature mining, the potential research opportunities of gene editing technology in crop breeding were revealed, which provides new insights into and supplements to the existing knowledge system. At the same time, by objectively and scientifically extracting potential technology combination innovation opportunities from massive literature data, it breaks through the limitations of traditional expert experience and provides a data-driven decision-making basis for the future research direction of gene editing technology in the field of crop breeding. The method formed in this study can be extended to other technical fields, providing practical support for the innovation of related theories and technical methods.
4.2 Practical implications
Currently, relevant scholars are conducting research on core breeding technology identification, gene discovery, and utilization based on bibliometrics, Latent Dirichlet Allocation (LDA) models, network analysis, knowledge graphs, and other methods (Jia et al., 2025; Zheng, 2025; Sahu et al., 2023), providing support for planning breeding technology research and development directions. At the same time, by developing an automated tool for extracting gene–phenotype associations from literature, they explored the complex correlations between genes and phenotypes in crops, fully exploiting and utilizing data for modern molecular breeding (Gao et al., 2024). Research on innovative opportunities in the research and development direction from the perspective of literature content mining is gradually emerging. This study systematically analyzed the published literature information of gene editing technology in the field of crop breeding by integrating various text mining methods (such as natural language processing, deep learning, and generative topology mapping) and deeply mined and identified potential technological innovation opportunities, forming a set of methodological frameworks for identifying key technologies and technology combinations. This method provides scientific research institutions and enterprises with accurate decision support for R&D direction, helps them optimize scientific research layout, and reduce trial-and-error costs. At the same time, it provides decision-making reference for the improvement of the development policy system, the scientific deployment of special plans, and the investment of R&D funds. In the future, industrial data (such as patents and market reports) can be further combined to build a more comprehensive technology evaluation system to maximize the potential of gene editing technology in crop breeding.
It is worth mentioning that the literature mining provides a significant theoretical framework; additional practical validation or case studies would help to solidify the applicability and usefulness of the identified technology opportunities. Therefore, in this study, “gene editing technology in crop breeding” was selected as a case study for empirical research. The results of 13 technology combinations were based on the interpretation of domain experts and the resulting analysis. At the same time, domain experts were invited to evaluate 13 sets of technology combinations from three dimensions— scientificity, feasibility, and application prospects— to verify the effectiveness of our method.
Moreover, the three methods used in this study, namely, spaCy, SciBERT, and GTM, have cross- domain adaptability. By training specific domain data and adjusting parameters, it can be suitable for multi- domain data mining tasks. In the future, we will conduct empirical research in other fields to enhance the applicability of the methods.
4.3 Limitations and difficulties
This study quickly identified potential technology combination opportunities through literature mining methods, which avoids spending a lot of manpower to scientifically and effectively identify technology gaps. In our research, we utilized natural language processing, deep learning, and GTM to conduct an in-depth analysis of the literature on gene editing technology in crop breeding from the perspective of literature mining. The literature mining method can extract valuable research information from a large number of published works by scientists. Specific empirical research is often based on the application of one or several technologies in specific fields. Compared with specific empirical research, the method of literature content mining can provide researchers with more valuable information from a broader perspective, support their selection of specific methods and technologies, assist them in conducting specific empirical research, save their time and economic costs, and improve scientific research efficiency. It is worth noting that the results obtained from natural language processing methods still require the participation of domain experts and empirical research in specific fields to obtain more scientifically valuable results.
However, there are still some shortcomings: 1) the content differentiation of the identified technology combinations was not obvious enough, and there were multiple technical words appearing in multiple groups of technology combinations at the same time. 2) Only published academic literature was identified, and other data sources, such as invention patents and industry reports, were not involved. 3) The content analysis covered by the technical word combination and whether the technical combination can become a technological innovation opportunity for future development still needs expert judgment.
In the existing studies, researchers identify technological innovation opportunities from Science Citation Index (SCI) papers and Derwent patent data and select representative technical feature words by combining technical subject words and the Term Frequency-Inverse Document Frequency (TF-IDF) algorithm. Finally, potential technical subjects are evaluated by building a multi-dimensional feature index system and calculating the technological gap so as to realize the screening of technology opportunities (Yin et al., 2025; Li J. et al., 2025). The above research has achieved good experimental results, so it provides a reference for the in-depth research and optimization of this study. Specifically, it includes the following: 1) by obtaining keywords, calculating word TF-IDF values, and other methods, the importance of technical words is screened to obtain core technical words, thereby improving the differentiation between technical combinations; 2) expanding data sources (such as patents, policies, and reports), enriching the breadth of analysis, and improving the comprehensiveness of data coverage; and 3) by combining tools and methods such as artificial intelligence, semantic analysis, and technological innovation indicators, we can intelligently interpret and analyze the content of technology combinations, predict the possibility of technology combinations becoming opportunities for technological innovation in the future, and provide more reliable decision support for the future development of gene editing breeding.
5 Conclusions
In this study, through the comprehensive application of text mining technologies such as spaCy, Transformer, and GTM, from the perspective of literature data of gene editing technology in the field of crop breeding, the labeling of key technologies and the identification of technology combinations were carried out. The potential technological innovation opportunities of gene editing technology in the field of crop breeding were systematically mined and analyzed. The results showed that from the literature data from 2020 to 2024, 13 technology combinations were identified. Through interpretation and analysis, the content covers the multi-technology combination strategy of molecular genetic research, the core technology of gene function research in molecular genetics of biotic and abiotic stresses, the technical means of analyzing the molecular mechanisms of stress resistance, the technical scheme of genetic improvement, etc., which provides support for revealing the potential technological innovation opportunities of gene editing technology in the field of crop breeding. At the same time, the identification method of technological innovation opportunities proposed in this study can scientifically, objectively, and efficiently mine literature information and visually display the results in the form of technology map with technology combination, which provides reference significance for the identification of potential technological innovation opportunities in the field. However, there are still some shortcomings in this study: the content differentiation of identified technology combinations is not obvious enough, the sources of data mining are insufficient, and the analysis of technology combinations is not intelligent enough. In the future, we will continue to carry out in-depth research on the above issues to accelerate the application and technological innovation of gene editing technology in crop breeding.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
HZ: Data curation, Writing – original draft, Methodology, Project administration, Conceptualization, Investigation, Formal Analysis. RY: Conceptualization, Writing – review & editing, Methodology. QJ: Methodology, Writing – review & editing, Validation. SQ: Validation, Data curation, Writing – review & editing. YH: Writing – review & editing, Formal Analysis, Methodology. JZ: Project administration, Validation, Conceptualization, Writing – review & editing, Supervision. LC: Methodology, Investigation, Project administration, Conceptualization, Supervision, Formal Analysis, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was funded by the “Youth Scientific Research Fund of Beijing Academy of Agriculture and Forestry Sciences”, grant numbers QNJJ202308, and the “Science and Technology Innovation Project in Beijing Academy of Agriculture and Forestry Sciences”, grant numbers KJCX 20230208 and KJCX20230210.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alariqi, M., Ramadan, M., Yu, L., Hui, F., Hussain, A., Zhou, X., et al. (2025). Enhancing specificity, precision, accessibility, flexibility, and safety to overcome traditional CRISPR/cas editing challenges and shape future innovations. Advanced Sci. 12, e2416331. doi: 10.1002/advs.202416331
Andrew-Peter-Leon, M. T., Selvaraj, R., Kumar, K. K., Muthamilarasan, M., Yasin, J. K., and Pillai, M. A. (2021). Loss of function of osFBX267 and osGA20ox2 in rice promotes early maturing and semi-dwarfism in γ-irradiated IWP and genome-edited pusa basmati-1. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.714066
Ashok, K., Bhargava, C. N., Asokan, R., Pradeep, C., Pradhan, S. K., Kennedy, J. S., et al. (2023). CRISPR/Cas9 mediated editing of pheromone biosynthesis activating neuropeptide (PBAN) gene disrupts mating in the Fall armyworm, Spodoptera frugiperda (J. E. Smith) (Lepidoptera: Noctuidae). 3 Biotech. 13, 370. doi: 10.1007/s13205-023-03798-3
Atia, M., Jiang, W., Sedeek, K., Butt, H., and Mahfouz, M. (2024). Crop bioengineering via gene editing: reshaping the future of agriculture. Plant Cell Rep. 43, 98. doi: 10.1007/s00299-024-03183-1
Bekalu, Z. E., Krogh Madsen, C., Dionisio, G., Bæksted Holme, I., Jørgensen, L. N., S. Fomsgaard, I., et al. (2020). Overexpression of nepenthesin hvNEP-1 in barley endosperm reduces fusarium head blight and mycotoxin accumulation. Agronomy 10, 203. doi: 10.3390/agronomy10020203
Beltagy, I., Lo, K., and Cohan, A. (2019). “SciBERT: A pretrained language model for scientific text,” in Proceedings of the Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), Stroudsburg, PA, USA (Association for Computational Linguistics). doi: 10.48550/arXiv.1903.10676
Bishop, C. M., Svensén, M., and Williams, C. K. I. (1998). GTM: the generative topographic mapping. Neural Comput. 10, 215–234. doi: 10.1162/089976698300017953
Cao, X., Chen, X., Huang, L., Deng, L., Cai, Y., and Ren, H. (2023). Detecting technological recombination using semantic analysis and dynamic network analysis. Scientometrics 129, 7385–7416. doi: 10.1007/s11192-023-04812-4
Chang, J., Mapuranga, J., Li, R., Zhang, Y., Shi, J., Yan, H., et al. (2024). Wheat leaf rust fungus effector protein pt1641 is avirulent to tcLr1. Plants 13, 2255. doi: 10.3390/plants13162255
Cruz-Pérez, J. F., Lara-Oueilhe, R., Marcos-Jiménez, C., Cuatlayotl-Olarte, R., Xiqui-Vázquez, M. L., Reyes-Carmona, S. R., et al. (2021). Expression and function of the cdgD gene, encoding a CHASE–PAS-DGC-EAL domain protein, in Azospirillum brasilense. Sci. Rep. 11, 520. doi: 10.1038/s41598-020-80125-3
Dong, H. (2024). Application of genome editing techniques to regulate gene expression in crops. BMC Plant Biol. 24, 100. doi: 10.1186/s12870-024-04786-2
Gao, Y., Zhou, Q., Luo, J., Xia, C., Zhang, Y., and Yue, Z. (2024). Crop-GPA: an integrated platform of crop gene-phenotype associations. NPJ Syst. Biol. Appl. 10, 15. doi: 10.1038/s41540-024-00343-7
Gupta, D., Saini, A., van der Vyver, C., and Panda, S. K. (2024). Gene editing: paving the way for enhancing plant tolerance to abiotic stresses-mechanisms, breakthroughs, and future prospects. J. Plant Growth Regul. 43, 3986–4002. doi: 10.1007/s00344-024-11395-8
He, J., Zhang, K., Yan, S., Tang, M., Zhou, W., Yin, Y., et al. (2023). Genome-scale targeted mutagenesis inBrassica napususing a pooled CRISPR library. Genome Res. 33, 798–809. doi: 10.1101/gr.277650.123
Jailani, A. A. K., Chattopadhyay, A., Kumar, P., Singh, O. W., Mukherjee, S. K., Roy, A., et al. (2023). Accelerated long-fragment circular PCR for genetic manipulation of plant viruses in unveiling functional genomics. Viruses 15, 2332. doi: 10.3390/v15122332
Jia, Q., Zhang, H., Chuan, L., Wang, A., Qin, S., and Zhao, J. (2025). Research on core technology of crop biological breeding based on patent network analysis. World Sci-Tech R D 47, 198–214. doi: 10.16507/j.issn.1006-6055.2024.09.006
Jiang, J., Su, H., Hong, D., Yang, G., Yan, L., Xu, Y., et al. (2023). Advances and perspectives in plant biotechnology. Plant Physiol. J. 59, 1436–1462. doi: 10.13592/j.cnki.ppj.600006
Kaneko, H. (2019). Data visualization, regression, applicability domains and inverse analysis based on generative topographic mapping. Mol. Informatics Mol. Inf. 38, 1800088. doi: 10.1002/minf.201800088
Kantsurova, E. S., Bovin, A. D., Dymo, A. M., Komolkina, N. A., Shalyakina, A. A., Salnikova, E. A., et al. (2024). Influence of enhanced synthesis of exopolysaccharides in rhizobium ruizarguesonis and overproduction of plant receptor to these compounds on colonizing activity of rhizobia in legume and non-legume plants and plant resistance to phytopathogenic fungi. Curr. Microbiol. 81, 416. doi: 10.1007/s00284-024-03929-w
Kumar, A., Lin, H., Li, Q., Ruan, Y., Cousins, D., Li, F., et al. (2022). Anthocyanin pigmentation as a quantitative visual marker for arbuscular mycorrhizal fungal colonization of Medicago truncatula roots. New Phytol. 236, 1988–1998. doi: 10.1111/nph.18504
Kumar, P., Parveen, A., Sharma, H., Rahim, M. S., Mishra, A., Kumar, P., et al. (2021). Understanding the regulatory relationship of abscisic acid and bZIP transcription factors towards amylose biosynthesis in wheat. Mol. Biol. Rep. 48, 2473–2483. doi: 10.1007/s11033-021-06282-4
Li, W., Mao, Z., Xiao, Z., Liao, X., Koffas, M., Chen, Y., et al. (2025). Large language model for knowledge synthesis and AI-enhanced biomanufacturing. Trends Biotechnol. 48, 1864–1875. doi: 10.1016/j.tibtech.2025.02.008
Li, Z., Sun, Y., Chen, D., Nie., J., and Lu, X. (2023). Evaluating the technology opportunities of GTM patent map recognition based on science and technology gap. J. Intell. 42, 147–153 + 44. doi: 10.3969/j.issn.1003-2053.2025.02.008
Li, B., Sun, C., Li, J., and Gao, C. (2024). Targeted genome-modification tools and their advanced applications in crop breeding. Nat. Rev. Genet. 25, 603–622. doi: 10.1038/s41576-024-00720-2
Li, J., Xu, C., Zhang, K., and Zhou, W. (2025). Research on emerging technology identification based on deep learning and patent map from a dynamic perspective. Inf. Studies: Theory Appl. 48, 160–169. doi: 10.16353/j.cnki.1000-7490.2025.07.018
Li, S., Zheng, H., and Wang, L. (2020). Application and prospect of gene editing technology in crop breeding. Biotechnol. Bull. 36, 209–221. doi: 10.13560/j.cnki.biotech.bull.1985.2020-0328
Lin, Q., Zong, Y., Xue, C., Wang, S., Jin, S., Zhu, Z., et al. (2020). Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585. doi: 10.1038/s41587-020-0455-x
Liu, C., Cao, Y., Hua, Y., Du, G., Liu, Q., Wei, X., et al. (2021). Concurrent disruption of genetic interference and increase of genetic recombination frequency in hybrid rice using CRISPR/cas9. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.757152
Liu, Z., Feng, J., and Uden, L. (2023). Technology opportunity analysis using hierarchical semantic networks and dual link prediction. Technovation 128, 102872. doi: 10.1016/j.technovation.2023.102872
Liu, H., Yang, Y., Liu, D., Wang, X., and Zhang, L. (2020). Transcription factor TabHLH49 positively regulates dehydrin WZY2 gene expression and enhances drought stress tolerance in wheat. BMC Plant Biol. 20, 259. doi: 10.1186/s12870-020-02474-5
Liu, J., Zhang, R., Chai, N., Su, L., Zheng, Z., Liu, T., et al. (2025). Programmable genome engineering and gene modifications for plant biodesign. Plant Commun. 6, 101427. doi: 10.1016/j.xplc.2025.101427
Mubarak, A. N. M., Burgess, A. J., Pyke, K., Quick, W. P., and Murchie, E. H. (2023). Mass screening of rice mutant populations at low CO2 for identification of lowered photorespiration and respiration rates. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1125770
Mukhtiar, A., Mahmood, A., Khan, M. A., Ameen, M., Al-Khayri, J. M., and Qari, S. H. (2025). Transforming field crops with CRISPR/Cas: a new era in genome editing. Rendiconti Lincei. Sci. Fisiche e Naturali 36, 195–208. doi: 10.1007/s12210-025-01308-6
Nerkar, G., Devarumath, S., Purankar, M., Kumar, A., Valarmathi, R., Devarumath, R., et al. (2022). Advances in crop breeding through precision genome editing. Front. Genet. 13. doi: 10.3389/fgene.2022.880195
Park, I. and Yoon, B. (2018). Technological opportunity discovery for technological convergence based on the prediction of technology knowledge flow in a citation network. J. Informetrics 12, 1199–1222. doi: 10.1016/j.joi.2018.09.007
Que, Z., Lu, Q., Li, Q., and Shen, C. (2023). The rice annexin gene OsAnn5 is involved in cold stress tolerance at the seedling stage. Plant Direct 7, e539. doi: 10.1002/pld3.539
Rasool, G., Buchholz, G., Yasmin, T., Shabbir, G., Abbasi, N. A., and Malik, S. I. (2021). Overexpression of SlGSNOR impairs in vitro shoot proliferation and developmental architecture in tomato but confers enhanced disease resistance. J. Plant Physiol. 261, 153433. doi: 10.1016/j.jplph.2021.153433
Sahu, J., Chandra, T., Jaiswal, S., Iquebal, M. A., and Kumar, D. (2023). Millet research status and prospects for alleviating food insecurity through a text-mining approach. J. Agric. Sci. 161, 633–644. doi: 10.1017/s0021859623000618
Sant’Ana, R. R. A., Caprestano, C. A., Nodari, R. O., and Agapito-Tenfen, S. Z. (2020). PEG-delivered CRISPR-cas9 ribonucleoproteins system for gene-editing screening of maize protoplasts. Genes 11, 1029. doi: 10.3390/genes11091029
Senthilkumar, M., Pushpakanth, P., Arul Jose, P., Krishnamoorthy, R., and Anandham, R. (2021). Diversity and functional characterization of endophyticMethylobacteriumisolated from banana cultivars of South India and its impact on early growth of tissue culture banana plantlets. J. Appl. Microbiol. 131, 2448–2465. doi: 10.1111/jam.15112
Shi, L., Su, J., Cho, M.-J., Song, H., Dong, X., Liang, Y., et al. (2023). Promoter editing for the genetic improvement of crops. J. Exp. Bot. 74, 4349–4366. doi: 10.1093/jxb/erad175
Shimizu-Sato, S., Tsuda, K., Nosaka-Takahashi, M., Suzuki, T., Ono, S., Ta, K. N., et al. (2020). Agrobacterium-mediated genetic transformation of wild oryza species using immature embryos. Rice 13, 33. doi: 10.1186/s12284-020-00394-4
Siddappa, S., Sharma, N., Salaria, N., Thakur, K., Pathania, S., Singh, B., et al. (2023). CRISPR/Cas9-mediated editing of phytoene desaturase (PDS) gene in an important staple crop, potato. 3 Biotech. 13, 129. doi: 10.1007/s13205-023-03543-w
Song, A., Wu, S., Ma, Q., Ban, W., Liu, X., and Jin, Y. (2025). Plant prime editing: A new direction in crop breeding. Chin. J. Rice Sci. 2025, 1–24. Available online at: https://link.cnki.net/urlid/33.1146.s.20250225.1841.024.
Tyagi, A., Mir, Z. A., Almalki, M. A., Deshmukh, R., and Ali, S. (2024). Genomics-assisted breeding: A powerful breeding approach for improving plant growth and stress resilience. Agronomy 14, 1128. doi: 10.3390/agronomy14061128
Wang, J., Ding, Z., Liu, Z., and Feng, L. (2022). Technology opportunity discovery based on patent analysis: a hybrid approach of subject-action-object and generative topographic mapping. Technol. Anal. Strategic Manage. 36, 2070–2083. doi: 10.1080/09537325.2022.2126306
Wang, Q., Pan, G., Wang, X., Sun, Z., Guo, H., Su, X., et al. (2024). Host-induced gene silencing of the Verticillium dahliae thiamine transporter protein gene (VdThit) confers resistance to Verticillium wilt in cotton. J. Integr. Agric. 23, 3358–3369. doi: 10.1016/j.jia.2024.03.024
Xu, L., Zhao, J., Xu, D., Xu, G., Peng, Y., and Zhang, Y. (2024). New insights into chlorantraniliprole metabolic resistance mechanisms mediated by the striped rice borer cytochrome P450 monooxygenases: A case study of metabolic differences. Sci. Total Environ. 912, 169229. doi: 10.1016/j.scitotenv.2023.169229
Yang, W., Cao, G., Peng, Q., Zhang, J., and He, C. (2022). Effective identification of technological opportunities for radical inventions using international patent classification: application of patent data mining. Appl. Sci. 12, 6755. doi: 10.3390/app12136755
Yin, H., Li, Y., Wang, Z., Chuang, Y., Chen, X., Cai, Q., et al. (2025). Optimization study of technology opportunity identification methods from the perspective of the gap between science and technology. Stud. Sci. Sci. 43, 300–310. doi: 10.16192/j.cnki.1003-2053.20240424.001
Yu, X., Huo, G., Yu, J., Li, H., and Li, J. (2023). Prime editing: Its systematic optimization and current applications in disease treatment and agricultural breeding. Int. J. Biol. Macromolecules 253, 127025. doi: 10.1016/j.ijbiomac.2023.127025
Zainuddin, F., Ismail, M. R., Hatta, M. A. M., and Ramlee, S. I. (2024). Advancement in modern breeding and genomic approaches to accelerate rice improvement: speed breeding focus. Euphytica 220, 109. doi: 10.1007/s10681-024-03353-y
Zang, J., Yao, X., Zhang, T., Yang, B., Wang, Z., Quan, S., et al. (2024). Excess iron accumulation affects maize endosperm development by inhibiting starch synthesis and inducing DNA damage. J. Cell. Physiol. 239, e31427. doi: 10.1002/jcp.31427
Zhai, S., Liu, H., Xia, X., Li, H., Cao, X., He, Z., et al. (2023). Functional analysis of polyphenol oxidase 1 gene in common wheat. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1171839
Zhang, R., Wang, Y., Yang, W., Wen, J., Liu, W., Zhi, S., et al. (2025). PlantGPT: an arabidopsis-based intelligent agent that answers questions about plant functional genomics. Advanced Sci. e03926. doi: 10.1002/advs.202503926
Zhang, R., Zhang, C., Lyu, S., Fang, Z., Zhu, H., and Hou, X. (2022). Functional Analysis of BcSNX3 in Regulating Resistance to Turnip Mosaic Virus (TuMV) by Autophagy in Pak-choi (Brassica campestris ssp. chinensis). Agronomy 12, 1757. doi: 10.3390/agronomy12081757
Zhao, X., Zan, L., He, N., Liu, H., Xing, X., Du, D., et al. (2024). BnaC09.tfl1 controls determinate inflorescence trait in Brassica napus. Mol. Breed. 44, 68. doi: 10.1007/s11032-024-01503-7
Zheng, Q. (2025). Literature analysis of Triticum aestivum bio-breeding based on bibliometrics and machine learning. J. Zhejiang A&F Univ. 42, 210–217. doi: 10.11833/j.issn.2095-0756.20240485
Zhou, Z. and Ban, Y. (2023). Identification and evaluation of potential technical opportunities based on GTM reverse mapping and SD modeling. Inf. Studies: Theory Appl. 46, 107–114 + 95. doi: 10.16353/j.cnki.1000-7490.2023.10.014
Keywords: gene editing technology, crop breeding, literature mining, key technology, technology opportunity identification
Citation: Zhang H, Yao R, Jia Q, Qi S, He Y, Zhao J and Chuan L (2025) Innovative opportunities for gene editing technology in crop breeding: from the perspective of literature analysis. Front. Plant Sci. 16:1636024. doi: 10.3389/fpls.2025.1636024
Received: 27 May 2025; Accepted: 04 August 2025;
Published: 04 September 2025.
Edited by:
Ahmad A Omar, University of Florida, United StatesReviewed by:
Qinlong Zhu, South China Agricultural University, ChinaCheng Yuan, Yunnan Academy of Tobacco Agricultural Sciences, China
Gauri Nerkar, Indian Council of Agricultural Research, Coimbatore, India
Copyright © 2025 Zhang, Yao, Jia, Qi, He, Zhao and Chuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Limin Chuan, Y2h1YW5sbUBhZ3JpLmFjLmNu; Jingjuan Zhao, emhhb2pqQGFncmkuYWMuY24=
†These authors have contributed equally to this work and share first authorship
 Hui Zhang
Hui Zhang Ru Yao1†
Ru Yao1† Shijie Qi
Shijie Qi