Abstract
Global agriculture relies heavily on the use of synthetic nitrogen fertilizer to meet the current global food demand. Unfortunately, the average nitrogen-use efficiency (NUE) of maize (Zea mays ssp. mays) is as low as 50%. Improving the NUE of maize is essential for feeding the ever-increasing world population while also decreasing the negative environmental impacts of nitrogen fertilizer due to runoff and volatilization. Harnessing the symbiotic relationship between plants and soil microorganisms may be one method for increasing the NUE in crops such as maize. In the present study, a set of potentially beneficial bacterial species chosen based on genetic information from the host was investigated for their ability to improve NUE-related traits in maize grown under nitrogen-deficient conditions. This was carried out through non-repeated and repeated bacterial inoculations using different maize genotypes. We identified several growth-promoting bacterial isolates and observed a significant interaction between the bacterial isolates and the maize genotype, suggesting a strong interaction between the host genetics and the effects of bacterial isolates. In addition, our results showed a significant growth response to repeated inoculations with a beneficial bacterial isolate. In summary, when evaluating the plant-growth-promoting effects of a bacterial species, it is essential to consider the interaction between host plant genotype and bacterial isolate. In addition, when inoculating with bacterial isolates, multiple inoculations appear to be more effective than a single inoculation after bacterial seed priming.
1 Introduction
To meet the growing global food demand, there has been a significant increase in nitrogen (N) fertilizer use over the past century, which has directly influenced the rise in yields over the same time period (Cao et al., 2018). U.S. maize yields increased from 1930 to 1960 with an average gain of 63 kg-1 ha-1 and an average gain of 110 kg-1 ha-1 from 1960 to 2000 (Woli et al., 2018). From 1940 to 2015, N fertilizer usage also increased from 0.28 g N m-2 y-1 to 9.54 g N m-2 y-1 (Cao et al., 2018), which was a major factor responsible for increased maize yields. The increase in N fertilizer application and the genetic improvement of maize (Assefa et al., 2012) have been the key factors driving the upward trajectory of yield in maize. While N is essential for high yields, fertilization of agricultural land contributes to approximately 53 percent of the global anthropogenic emission of nitrous oxide (N2O) (Denman et al., 2007). The N2O released from N fertilization is particularly a concern for global climate change as it has a warming potential 298 times greater than CO2 (Lan et al., 2022). With the increasing application of N fertilizer, maize (Zea mays) crops only use between 25–50 percent of the applied N (Javed et al., 2022). An increase in the nitrogen-use efficiency (NUE) of global crops, such as maize, is important not only for decreasing the negative environmental and human health impacts of N fertilization but also for maintaining high crop yields.
Plants utilize N as either NH4+ or, more commonly, NO3- (Novoa and Loomis, 1981). Soil NO3- can be sensed by plant roots, allowing for the activation of NO3- transporters for uptake of NO3- (Aluko et al., 2023). Plants are equipped with various mechanisms for adapting to growth under N-deficient conditions. For example, the mechanism called stress-initiated nitrate allocation to roots (SINAR) is coordinated in part by the nitrate transporters NRT1.5 and NRT1.8 (Zhang et al., 2014). Under nitrate sufficient conditions NRT1.8 unloads nitrate into leaves and NRT1.5 loads nitrate into the xylem for long distance transport to leaves. However, under nitrate-deficient conditions, NRT1.5 gene expression is down-regulated, and NRT1.8 expression is increased in roots, leading to the unloading of nitrate from the xylem into the roots (Li et al., 2010) which increases root elongation to maximize N acquisition from the soil (Chun et al., 2005). In addition, the inducible high-affinity transport systems (IHATS) transporters are activated when NO3- concentrations are low in the soil (Crawford and Glass, 1998) to enable uptake at very low concentrations of available nitrate. Another strategy plants use to adapt to N-deficient conditions is to increase their production of indole-3-acetic acid (IAA) (Lv et al., 2021; Zhang et al., 2023), as IAA has been shown to increase root elongation, which would aid in N uptake (Zhang et al., 2023). These strategies for adaptation to N-deficient conditions and for increasing nutrient acquisition are important for maintaining yields (Abdel-Lattif et al., 2019; Raza and Farmaha, 2022).
Microorganisms in soils may also play important roles in supplying plants with N and in regulating the forms of N that are available to plants. Soils harbor a diverse variety of bacteria, archaea, fungi, and viruses, collectively making up the soil microbiome (Bardgett and van der Putten, 2014). The rhizosphere microbial communities are in direct proximity to plant roots, and are influenced by both plant uptake of nutrients and the exudation of compounds from roots, leading to an intimate host plant and microbe interaction (Trivedi et al., 2020; Chai and Schachtman, 2022b; Lopes and Schachtman, 2023b). These chemical compounds, referred to as root exudates, can influence the diversity and composition of rhizosphere microbial communities (Bertin et al., 2003). Therefore, when a plant encounters a stressful environment due to a nutrient deficiency, such as low N, it may alter its exudate composition and thereby attract a community of microbes that will aid in root growth, thereby increasing nutrient acquisition (Coskun et al., 2017).
Certain microbial species play major roles in the nitrogen cycle, such as nitrification which may benefit crop growth (Franche et al., 2009) and also lead to losses of N from soils. Nitrifying bacteria in the soil convert NH3 into NO3-, which is a form of N that plants readily utilize (Werner and Newton, 2005). Key members in the nitrification process include ammonia-oxidizing bacteria (AOB), such as Nitrosospira and Nitrosomonas, as well as nitrite-oxidizing bacteria (NOB), including the phyla Proteobacteria, Chloroflexi, Nitrospina, and Nitrospira (Clark et al., 2021). As part of the soil nitrogen cycle, bacteria also carry out denitrification, a process in which NO3- is reduced to N2O or N gas (Martienssen and Schöps, 1999) and subsequently lost to the environment. Another way in which some plant species can acquire N is from a symbiotic relationship with a bacteria that forms nodules on roots and that fix N from atmospheric N2 for use by plants (Soumare et al., 2020). There are also free-living bacterial species in the soil that fix N which may be taken up by plants and some of these include Azospirillum spp., Acetobacter diazotrophicus, Herbaspirillum seropedicae, Azoarcus spp., and Aztobacter (Steenhoudt and Vanderleyden, 2000). Because these free-living N fixing bacteria occur naturally within the soil, they can be further harnessed to provide plants with N through the design, formulation, and application of microbial inoculants.
Countries including Brazil and Argentina have adopted the use of microbial inoculants such as Azospirillum strains in their agricultural practices. Azospirillum species, such as A. brasilense and A. lipoferum, have been identified as plant-growth promoting bacteria (PGPB) in maize and wheat plants (Hungria et al., 2010). Other studies have confirmed the use of Azospirillum spp. as PGPB in maize (Galindo et al., 2019, Galindo et al., 2022; Hungria et al., 2022). In 1996 in Argentina, the first registered inoculant was developed using A. brasilense Az39, followed by the first registered inoculant in Brazil in 2010, developed with the A. brasilense Ab-V5 and Ab-V6 strains (Cassan et al., 2020). In addition to N fixation (Han and New, 1998) there have been other reports of plant-growth-promoting characteristics of Azospirillum spp. including the production of phytohormones, such as IAA (Tien et al., 1979) and gibberellins (Bottini et al., 1989). While identifying a growth-promoting function of a microbe is critical when developing a microbial inoculant, it may also be necessary to account for the interaction between the microbe and the plant genotype being inoculated (Malacrino et al., 2022). Only a few studies have demonstrated the interaction between microbial inoculants and plant genotypes (Andreote et al., 2010; French et al., 2020; Chai et al., 2021).
Past crop breeding efforts have neglected the role of the plant microbiome in crop improvement. To better integrate the microbiome with plant genetics, a previous study (Meier et al., 2022) identified a group of amplicon sequence variants (ASVs) in the rhizosphere microbiome of inbred maize plants grown under both optimal N and N-deficient conditions. These bacterial ASVs were correlated with plant traits, such as canopy coverage and agronomic traits, as well as associated with the maize genetic loci, suggesting a strong correlation between maize genetics, bacterial species in the rhizosphere, and plant performance. Based on these previous results (Meier et al., 2022) that identified ASVs associated with maize loci, we hypothesized that these bacterial ASVs may enhance maize growth in N-deficient conditions. To test this hypothesis, we used these ASVs to identify and then test 63 bacterial isolates identified from an existing culture collection. These isolates were then tested in a series of controlled environment chamber experiments to determine how their introduction assisted in the growth of maize under low N conditions. The results of this study provide insight into the potential of using culture-independent microbiome data, coupled with plant genetic information, for selecting growth-promoting isolates from a culture collection. Additionally, our results shed light on the impact of using a repeated inoculation method for applying PGPB and highlight the importance of considering the interaction between a PGPB and the plant genotype being inoculated.
2 Materials and methods
2.1 Identification of potentially beneficial isolates
2.1.1 Field-based ASV identification
Amplicon sequence variants (ASVs) of soil microbes were identified from 3,313 rhizosphere samples based on their associations with maize genes (Meier et al., 2022). ASVs that were observed across two years of sampling were identified from paired-end 16S sequencing reads collected from the samples. That analysis showed the maize genome likely underwent negative or positive selection to favor specific microbial taxa (referred to as rhizobiome traits) by removing deleterious alleles or duplicating desirable alleles. We hypothesized that plant-associated ASVs possibly contributed to increasing plant fitness because of their association with maize genes.
2.1.2 Culture collection description
The ASVs from that study were then screened against the 16S sequences from a culture collection that was developed over several years and contains plant and soil derived bacterial isolates from multiple regions of crop and grassland soils. The collection of approximately 4500 isolates was cultured from soil, rhizosphere and roots. Approximately 2700 of the isolates have had all or part of their 16S gene sequenced. That sequence was used to search databases for taxonomic assignments. In addition, 472 isolates have genome sequences that were generated by the Joint Genome Institute (Berkeley, CA) using Illumina sequencing. Those sequences and the annotations are contained in IMG/M database (https://img.jgi.doe.gov/). The agricultural fields were located near the following Nebraska cities: North Platte, Brule, Mead, North Bend, Valley. Samples from grasslands were collected in the Sandhills and that sampling was previously described (Lopes et al., 2021). Root and rhizosphere samples were collected from a range of different plant species including corn, sorghum, soybean and the grasses as described (Lopes et al., 2021).
2.1.3 Sequence alignment and matching process
To match the 724 ASVs identified from the field-based study (Meier et al., 2022) with the 16S data from the culture collection, four BLAST (Camacho et al., 2009) databases were created from the culture collection data that contained the merged, 27F, 515F, and 515R sequences. The field-based ASVs were aligned via ungapped BLAST against each database with default parameters. A custom Perl script was then used to scan all BLAST output files and collate information by ASV ID for the best alignment for individual subject IDs (i.e., microbial database sequence ID), with a requirement that the difference between the alignment length and the query (ASV) length cannot be larger than two nucleotides. Alignments to the merged sequences have preference over 27F, and 27F alignments have preference over 515F, and so forth. Of the field-identified ASVs, 629 (87%) aligned to the in-house culture collection data. The matching ASVs corresponded to 63 bacterial isolates (Supplementary Table 2) from the culture collection described above. The bacterial isolate sequences were between 100% identity to 95.205%, similar to the field-identified ASVs (Supplementary Table 1). Of these 63 isolates, 35 have draft genome sequences that may be found in the JGI IMG/M database (Supplementary Table 2). The isolates from the culture collection were then revived on nutrient-containing plates from glycerol stocks stored at -80°C and used in the experiments described below.
2.2 Maize seedling bacterial inoculation
Mo17 inbred maize was selected for the initial screening of the 63 bacterial isolates, the 14-day validation experiments, and as one of the three maize genotypes in the non-repeated and repeated inoculation experiments. The two other maize genotypes selected for the non-repeated inoculation experiment were NSL 30867 and PI 606768 and for the repeated inoculation experiment NSL 30867 and Ames 27065 were used. These maize genotypes were selected based on their root exudation of sugars (Lopes et al., 2022). The maize genotypes PI 606768 and Ames 27065 exuded relatively high concentrations of total sugars, while the maize genotype NSL 30867 exuded low concentrations of total sugars.
For all experiments, maize seeds were surface sterilized for 48 hours using chlorine gas, produced by mixing 4 mL concentrated hydrochloric acid (HCl) with 100 mL bleach in a desiccator. The chlorine gas was replaced after 24-hours. PI 606768 was sterilized for only 24 hours utilizing the same protocol, as a 48-hour sterilization decreased the germination rate of this maize genotype. Following the surface-sterilization, the seed was placed in aerated, autoclaved (20 minutes sterilization, at 121°C, 10 minutes drying) Milli-Q water to imbibe for 12–24 hours at room temperature (24°C). After imbibing, the seeds were placed in petri dishes lined with sterilized moist filter paper. The seeds were then sprayed with Captan (0.2%) to control fungal growth. The petri dishes were sealed with micropore tape and placed at 30°C in the dark for 24–48 hours until the seeds germinated.
Based on the information in the culture collection database the 63 bacterial isolates were inoculated and grown in one of the following media types depending on how they were originally isolated: yeast extract-peptone-dextrose (YPD), yeast mannitol agar (YMA), trypticase soy agar (TSA), reasoner’s 2A agar (R2A), or Ashby’s N-free medium (Stella and Suhaimi, 2010). Bacterial isolates were plated from glycerol stocks that were stored in a -80°C freezer. Germinated maize seeds were placed in a sterile petri dish when the root was about 1 cm long and approximately 8 mL of the liquid bacterial culture (OD600 of 1) was added. The petri dish was sealed with micropore tape and placed on the rotary shaker at 80 rpm for 12 hours at room temperature (24°C). This inoculation method is referred to as a seedling priming (Chai et al., 2022a). Mock-inoculated germinated seeds were treated similarly by placing them in a sterile R2A medium without any added bacteria for 12 hours and were used for the uninoculated controls grown in low and high N.
For all experiments, germinated maize seeds were inoculated using a seedling priming method with only one of the 63 bacterial isolates for each treatment. The sterility of the system ensured that only one bacterial isolate was tested in each trial for growth promoting effects on maize grown under low N conditions.
2.3 Screening of the 63 bacterial isolates and 14-day validation experiment
The screening of the 63 bacterial isolates for their plant-growth-promoting activity was carried out in eight rounds because of space limitations in the growth chamber. Initially the screening was done with n = 3 replicate Mo17 plants and then increased to n = 8 replicates per isolate tested. In each 14-day round, an uninoculated replicated set of maize plants was grown in N-deficient conditions (low N control) and N-sufficient conditions (high N control) in sterile growth bags. The germinated maize seed was inoculated with each of the 63 isolates and grown in pots (11 cm in height, 13 cm in diameter) containing 500g of calcined clay for 14 days. To ensure sterility the pots containing the calcined clay were autoclaved 3 times (25 minutes sterilization at 121°C, 10 minutes drying). A pot was then placed into a sterile growth bag (Nasco-Whirl-PAK) with an AeraSeal film placed over a hole cut in the bags to allow for gas exchange. This allowed for a sterile growth system. A volume of 450 mL of a modified (Schachtman and Munns, 1992) Hoagland’s nutrient solution with either sufficient N (14.5 mM NO3, 1 mM NH4) or with 10% NO3 (1.45 mM) and 50% NH4 (0.5 mM) was added to the calcined clay, bringing the system to 90% maximum soil water holding capacity (SWHC). The Hoagland solution was prepared with autoclaved Milli-Q water. The N-sufficient control plants (high N control) were given 450 mL full-strength Hoagland solution (15.50 mM N). Following the Hoagland solution, an inoculated maize seed was sown into the calcined clay at a 1-inch depth and then grown in a growth chamber. The planting was done in a laminar flow hood to ensure the presence of only the desired bacterial isolate. The conditions of the growth chamber were kept at 26°C during the day and 18°C during the night, with a 16-hour light period, throughout the course of all experiments. The average light intensity of the growth chamber was 350 μmol m-2 s-1. For the screening and 14-day validation experiments, no additional water or nutrients were added throughout the 14-day growth period.
Following the screening of the 63 isolates, none of the isolates imparted a statistically significant growth increase to Mo17 under low N conditions. But to test what appeared to be isolates that increased growth under low N, albeit not significantly, we chose 15 isolates to be tested again in another 14-day experiment. Plants were grown and inoculated as described for the screening experiment. However, the design of this experiment was improved using a randomized complete block design (RCBD). Due to space limitations in the growth chamber, the 15 bacterial isolates were divided into two groups for two 14-day experiments. There were 8 blocks in total, with each bacterial isolate-inoculated plant replicated per block along with two uninoculated control plants at low N and two uninoculated plants at high N. Due to some of the plants not emerging successfully, each bacterial isolate-inoculated plant had 7–8 replicates, with the low N control plants having 16 replicates in both 14-day validation experiments (n = 32 total for the low N control).
2.4 Non-repeated inoculation experiment
In the non-repeated inoculation experiment, pots (19 cm in height, 15 cm in diameter) were filled with 2/3 peat and 1/3 fine vermiculite mixture. This soil mixture was almost completely depleted of N. The pots containing the peat-vermiculite mixture were autoclaved three times (20 minutes sterilization, at 121°C, 10 minutes drying), to ensure sterility of the growth medium.
Three bacterial isolates were selected from the 15 tested in the 14-day validation experiment. One of these isolates increased the growth of Mo17 on low N significantly, while two others boosted growth, but not significantly. A 3 x 5 factorial experimental design was implemented. Three different maize genotypes were used as well as five treatments which included three bacterial isolates and the uninoculated maize in low N and in high N as controls. The treatments were arranged into 9 blocks, with each treatment containing one of the three maize genotypes and tested with three bacterial isolates and two controls in one replicate per block (n = 9). Due to poorer emergence rates, the PI 606768 low N control treatment only had 7 replicates (n = 7) and the 4589- inoculated PI 606768 treatment group only had 4 replicates (n = 4). The three maize genotypes used were Mo17, PI 606768, and NSL 30867.
Germinated seeds were treated with bacterial cultures as described for the screening experiment. Two inoculated maize seeds were sown into each pot. On the seventh day cultures for each of the three bacterial isolates was added to each pot. Cultures were resuspended in low N sterile Hoagland’s solution and adjusted to an OD600 of 0.002 and ten mls were added to each pot. The pots containing the seeds inoculated with bacterial isolates, as well as the N-deficient uninoculated plants (low N controls) were given a half-strength Hoagland nutrient solution at the time of planting, up to 85% SWHC. The high N control plants were treated with a full-strength Hoagland nutrient solution at the time of planting. Upon planting the pots were covered with plastic saucers and placed in the growth chamber. The planting process took place inside the laminar flow hood. The plants were monitored and watered to approximately 85% SWHC with low N nutrient solution twice per week throughout the 35-day growth period. At the same timepoints, full-strength Hoagland nutrient solution was given to the high N control plants.
To track the growth of the plants at 14, 21, and 28 days after planting, a phenotyping system was utilized to measure the aboveground biomass of the plants using images. The key components of this phenotyping system include a precision rotation platform (Newport Cooperation, Irvine, California), an RGB camera (Logitech BRIO 1080p Webcam) and a computer. A graphic-based LabVIEW program was used to operate the system and automatically capture multiple side views from all angles of the targeted plant within seconds. The phenotyping system captured 8 images at multiple angles over 360°. Using R (R Core Team, 2023), the aboveground biomass of each plant was accurately measured through the processing of the eight images. At 14, 21, and 28 days after planting, the plants were photographed to measure their shoot biomass. Upon the completion of the collection, the images were processed in batch from a specified directory using the “EBImage” (Pau et al., 2010), “imager” (Barthelmé and Tschumperlé, 2019), and “raster” (Hijmans, 2025) libraries in R, where each image was manually cropped and converted to a grayscale matrix based on the green channel. A binary raster grid was then generated by thresholding pixel values and applying a distance-based mask to isolate central features, with the processed binary images saved to a new directory. Edge detection was subsequently used to extract morphological traits such as shoot width, pixel-based biomass, and occupancy ratio. From the repeated inoculation experiment, a correlation coefficient between the 27-day photographed biomass measurements and the dry shoot weights of the 28-day old plants was developed in MatLab (The MathWorks Inc., 2022).
2.5 Repeated inoculation experiment
The repeated inoculation experiment used the same experimental design as the non-repeated experiment, except that the inoculation protocol was altered. The inoculation protocol consisted of seedling priming followed by 5 drenches with bacteria using the same method as described above (Figure 1). Soil drenching with a bacterial inoculant was done 14, 17, 19, 25, and 27 days after planting. The same three bacterial isolates, 111 (Arthrobacter sp.), 730 (Pseudomonas kribbensis), and 4589 (Sphingomonas sp.), were used. The three maize genotypes used in the repeated inoculation experiment were Mo17, Ames 27065, and NSL 30867. This repeated inoculation protocol was tested since the growth-promoting properties of bacteria may vary based on the concentration and recurrence of inoculation (Papin et al., 2025). The same randomized complete block design was used as described above. There were 9 blocks in the experimental design, with 1 replicate of each treatment in each block. Due to differences in germination rates, most of the treatment groups had 8 replicates (n = 8), except the NSL 20867 inoculated with isolate 730 (n = 9), NSL 30867 low N control (n = 9), Ames 27065 low N control (n = 7), and Ames 27065 inoculated with isolate 730 (n = 6). The same planting, watering, and growing conditions were used, as described above.
Figure 1
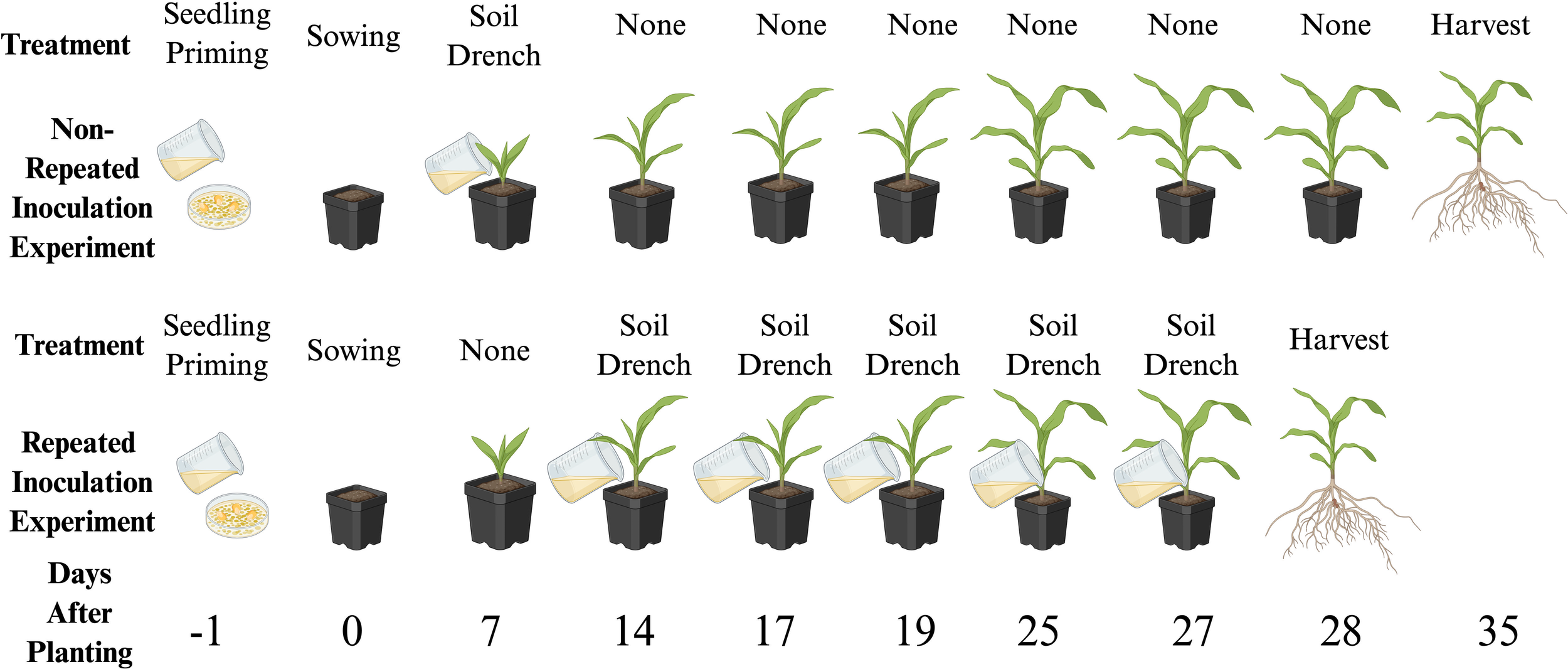
Inoculation schedule for the non-repeated and repeated inoculation experiments. In the non-repeated inoculation experiment, the maize plants were inoculated with the seedling priming technique the day before planting, given a soil drench inoculation 7 days after planting, and harvested 35 days after planting. For the repeated inoculation experiment, the maize plants were inoculated with the seedling priming technique the day before planting and then repeated soil drench inoculations were performed at 14, 17, 19, 25, and 27 days after planting and harvested 28 days after planting. Low N nutrient solution was added to both experiments twice per week. Both experiments were set up as randomized complete blocks (n = 9 blocks) and analyzed using a linear mixed model. Created in BioRender. Foster (2025). https://BioRender.com/s0lhnex.
To non-destructively measure the growth of the plants over the course of the 28-days, the same phenotyping system was utilized for phenotyping at the 14-, 21-, and 27-day timepoints. The images were analyzed using the same protocol as the non-repeated inoculation experiment.
2.6 Phylogenetic analysis and visualization
63 of the isolates were analyzed. The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed (Felsenstein, 1985). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The evolutionary distances were computed using the p-distance method (Nei and Kumar, 2000) and are in the units of the number of base differences per site. The analytical procedure encompassed 63 nucleotide sequences. The complete deletion option was applied to eliminate positions containing gaps and missing data. Evolutionary analyses were conducted in MEGA12 (Stecher et al., 2020; Kumar et al., 2024) utilizing up to 8 parallel computing threads.
2.7 Statistical analysis
The statistical analysis of the initial screening was carried out in R (R Core Team, 2023). A one-way ANOVA test was performed on the dry shoot weight of the plants in the screening assay. Inoculated plants that appeared to offer a growth effect on the dry shoot weight compared to the uninoculated low N control plants were selected for the 14-day validation experiment.
The statistical analyses of the 14-day, non-repeated and repeated growth experiments were carried out in R and SAS software (SAS Institute Inc., 2023). SAS software, Version 9.4 M8. Cary, NC: SAS Institute Inc.). A linear mixed model analysis was performed to determine if the dry shoot weight of the maize grown with each bacterial isolate was significantly higher than the dry shoot weight of the maize grown in low N conditions that lacked added bacteria. Within the linear mixed model, LS-means, also known as model-adjusted means were calculated for each treatment group. A Tukey HSD post-hoc analysis was performed to determine which specific treatment groups were significantly different from one another.
Figure 1 was created by BioRender.com. Figure 2 through 6 were prepared using the R packages “tiff” (Urbanek and Johnson, 2023), “gridExtra” (Auguie and Antonov, 2017), “readr” (Wickham et al., 2024), “ggplot2” (Wickham, 2016), and “magick” (Ooms, 2025).
Figure 2
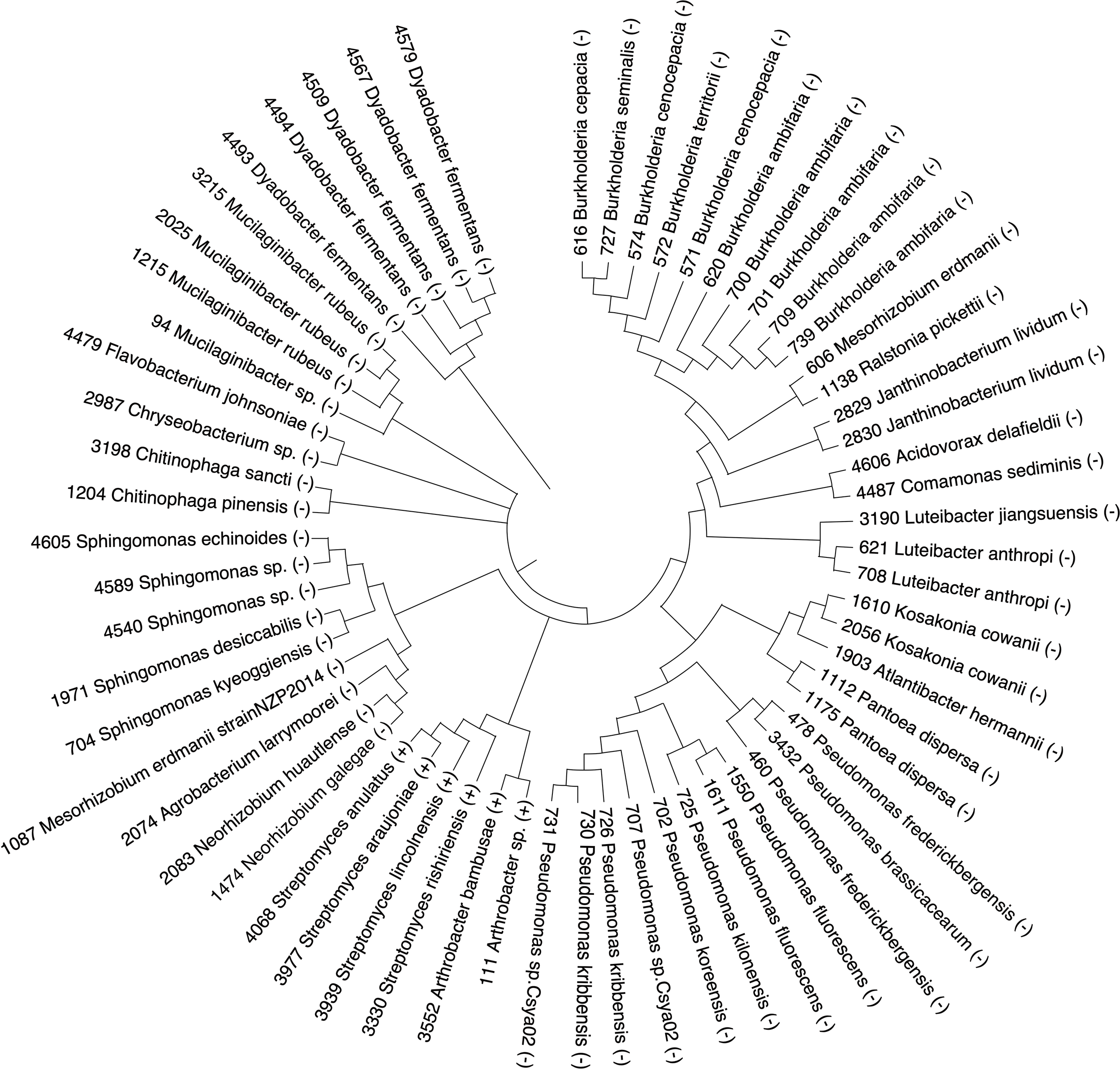
Phylogeny of the 63 bacterial taxa that were identified in a culture collection by searching with the ASVs identified in (Meier et al., 2022). Numbers that precede the genus on the phylogeny correspond to bacterial isolate numbers in our culture collection. Gram-negative bacterial isolates are indicated by the (-) at the end of the bacterial isolate name. Gram-positive bacterial isolates are indicated by the (+) at the end of the bacterial isolate name.
3 Results
3.1 Identification of 63 potentially beneficial bacterial isolates
Previous work (Meier et al., 2022) identified amplicon sequence variants (ASVs) of soil microbes from the rhizosphere soil of 240 maize inbred lines that may contribute to increasing plant fitness under low N field conditions. These ASVs were associated with maize alleles that had undergone negative or positive selection. This suggested that these host alleles may have arisen in relationship to specific microbial groups by removing harmful alleles or increasing the frequency of desirable alleles. Therefore, we used an axenic growth system to test whether microbial taxa related to these ASVs, identified under low N conditions, impart a beneficial phenotype to maize. Initially the ASVs identified from field studies (Meier et al., 2022) were used to search 16S sequences from an in-house culture collection of over 2,000 isolates collected from soil, roots, and rhizospheres of multiple plant species collected in soils from across the state of Nebraska, USA. The outcome of that search using the 724 ASV sequences identified 63 bacterial isolates that had 95 – 100% similarity to the V4 region (Supplementary Table 2). The 63 bacterial isolates were tested to determine whether they imparted a growth advantage for maize under low N conditions. These isolates included many from the genus Pseudomonas, Streptomyces, Burkholderia and Sphingomonas (Figure 2). We also identified isolates from Luteibacter, Dyadobacter, Chitinophaga, Mucilaginibacter, Pantoea, Janthinobacterium, and Kosakonia. Most of these species are gram-negative except for 7 gram-positive taxa that included Streptomyces and two isolates of Arthrobacter.
3.2 Screening of the 63 potentially beneficial bacterial isolates
The screening (n = 3–8 plant per isolate) did not identify any of the bacterial isolates that significantly increased the fresh weight or dry shoot weight of the inoculated plants compared to the uninoculated low N control plants (Figure 3).
Figure 3
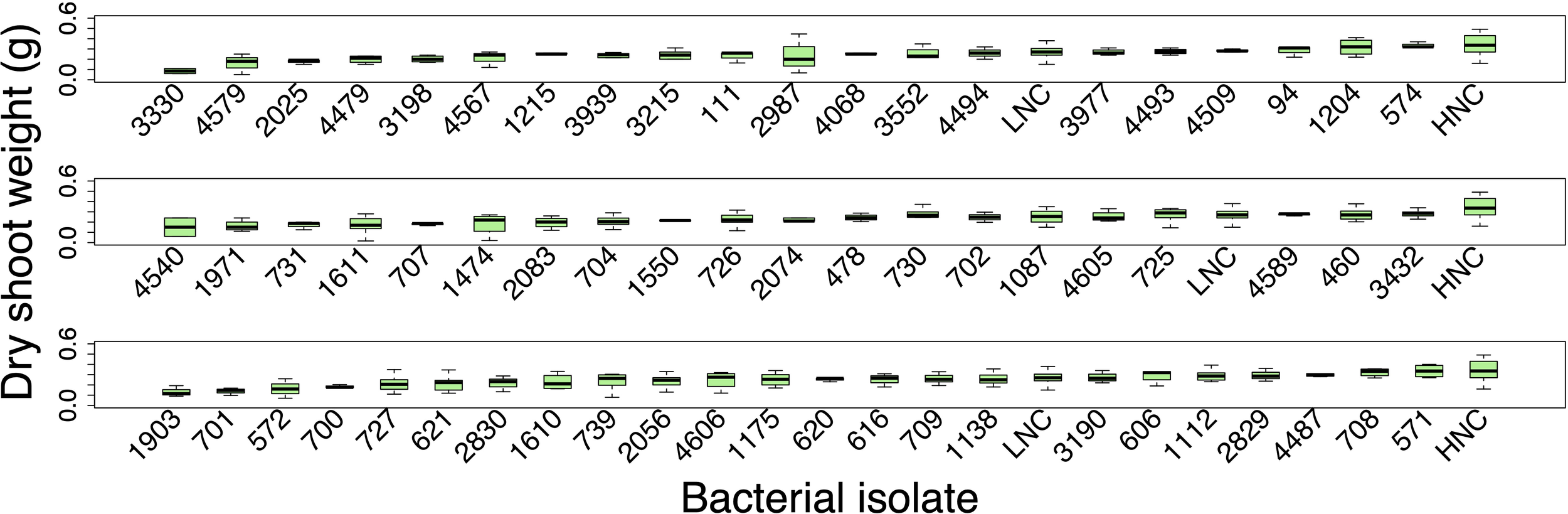
The dry shoot weight (g) results of the screening of the 63 potentially beneficial bacterial isolates. The plants were harvested at 14-days of growth when they were approximately at the V2 growth stage. Significant differences were determined using an ANOVA analysis followed by Tukey’s HSD correlation for multiple comparisons. HNC and LNC represent the high N control and low N control plants, respectively. Box plots display the median as the line within the box, as well as the lower quartile (bottom of the box) and the upper quartile (top of the box), with the whiskers showing the minimum and maximum data values). Each group is organized from lowest to highest median weight.
3.3 14-day validation experiment
Using the information from the screening, 15 bacterial isolates were selected based on increases in growth imparted to Mo17 under low N conditions (Table 1). These were then reinoculated to fresh Mo17 seeds and grown for 14 days under axenic conditions with more replicates (n = 7-8) to increase the statistical power compared to the screening. To select the 15 bacterial isolates for the 14-day experiment, the box plots of the screening results were analyzed visually to select bacterial isolates that appeared to increase the dry shoot weight.
Table 1
| Bacterial isolate ID | Isolate identity | Similarity of ASV to sequence in culture collection (%) |
|---|---|---|
| 111 | Arthrobacter sp. | 99.658 |
| 571 | Burkholderia cenocepacia | 99.315 |
| 574 | Burkholderia cenocepacia | 99.315 |
| 606 | Mesorhizobium erdmanii | 100 |
| 702 | Pseudomonas koreensis | 100 |
| 708 | Luteibacter anthropi | 99.315 |
| 726 | Pseudomonas kribbensis | 100 |
| 730 | Pseudomonas kribbensis | 100 |
| 1138 | Ralstonia picketti | 100 |
| 1204 | Chitinophaga pinensis | 96.575 |
| 2829 | Janthinobacterium lividium | 97.26 |
| 4487 | Comamonas sediminis | 97.603 |
| 4509 | Dyadobacter fermentans | 98.288 |
| 4589 | Sphingomonas sp. | 95.89 |
| 4606 | Acidovorax delafieldii | 98.63 |
List of the 15 bacterial isolates selected from the screening for further testing in the 14-day validation experiment.
The plants were grown for 14 days and harvested at approximately the (V2) stage which was when the second leaf was fully expanded. In the 14-day validation experiment, bacterial isolate 111 (Arthrobacter sp.) significantly increased the dry shoot weight of Mo17 maize compared to the uninoculated low N control (p = 0.0013) (Figure 4). Two other bacterial strains improved maize seedling growth with relatively low variation including isolate 730 (Pseudomonas kribbensis), and 4589 (Sphingomonas sp.), but the increase was not statistically significant. The remainder of the bacterial isolates had no significant growth-promoting effect on the dry shoot weight compared to the uninoculated low N control.
Figure 4
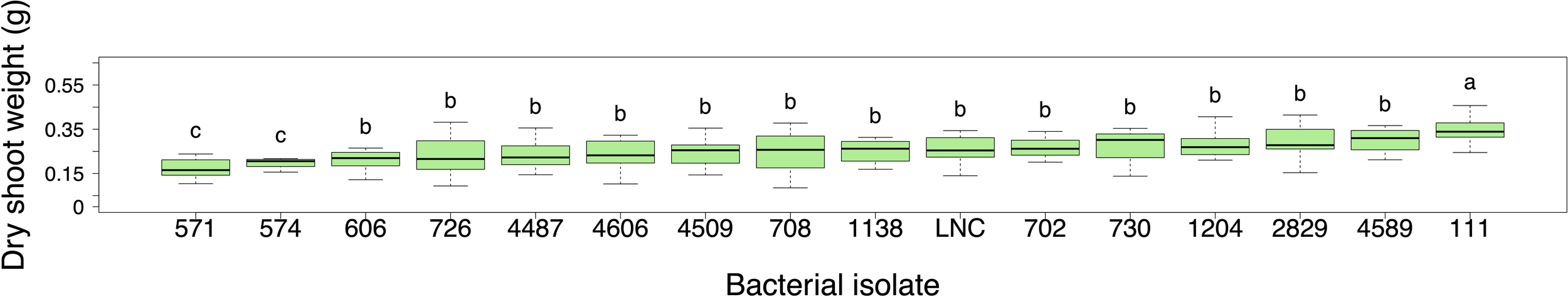
Dry shoot weight (g) of the Mo17 maize genotype inoculated with 15 selected bacterial isolates during the 14-day validation experiment. The plants were harvested at 14 days of growth, at approximately the V2 stage. A linear mixed model analysis was performed to assess the differences among treatments and the low N control. (Box plots display the median as the line within the box, as well as the lower quartile (bottom of the box) and the upper quartile (top of the box), with the whiskers showing the minimum and maximum data values. LNC represents the low N control plants. Letters above boxes represent statistically significant groupings (p < 0.05) different from the LNC). The isolates are arranged along the x-axis based on the lowest to highest median weight of the Mo17 plants. (n = 7-8, exception: n = 32 for LNC).
3.4 Non-repeated inoculation experiment
From the results of the 14-day validation experiment, three bacterial isolates were selected for a 35-day experiment to assess the effects of these potentially beneficial bacterial isolates over a longer growth period when grown in pots in ambient air. These were isolate 111 (Arthrobacter sp.), 730 (Pseudomonas kribbensis), and 4589 (Sphingomonas sp.). Bacterial isolate 111 was selected due to its significant impact on the dry shoot weight of inoculated Mo17 maize compared to the uninoculated low N control (p-value = 0.0013) (Figure 4). Bacterial isolates 730 and 4589 were selected for their apparent ability to increase the Mo17 dry shoot weight under N-deficient conditions compared to uninoculated low-N control plants. In this experiment, we also tested these three isolates using three maize genotypes: Mo17, NSL 30867, and PI 606768 to determine whether there might be a maize genotype by bacterial isolate interaction. Thirty-five days after planting when the plants were approximately at the (V3 to V4) growth stage which was when the third or fourth leaf was fully expanded, the maize plants were harvested, and their dry shoot weights were measured.
In this experiment the bacterial isolates did not have a significant effect on the dry shoot weight (p = 0.7719) (Table 2), but the maize genotype did have a significant effect on the dry shoot weight (p < 0.0001) (Table 2) under N-deficient conditions. No significant interaction between bacterial isolate and maize genotype was observed (p = 0.3132) (Table 2).
Table 2
| Effect | F-value | P-value |
|---|---|---|
| Bacterial Isolate | 0.37 | 0.7719 |
| Maize Genotype | 39.35 | < 0.0001 |
| Bacterial Isolate x Maize Genotype Maize Genotype |
1.2 | 0.3132 |
Non-repeated inoculation experiment – Linear mixed model analysis for the dry shoot weight (grams) response of each factor.
The impact of each bacterial isolate on the dry shoot weight of the inoculated plants at each specific maize genotype was further examined. However, we did not find that any of the three bacterial isolates significantly improved the growth of any of the maize genotypes grown under low N conditions (Supplementary Table 3) (Figure 5).
Figure 5
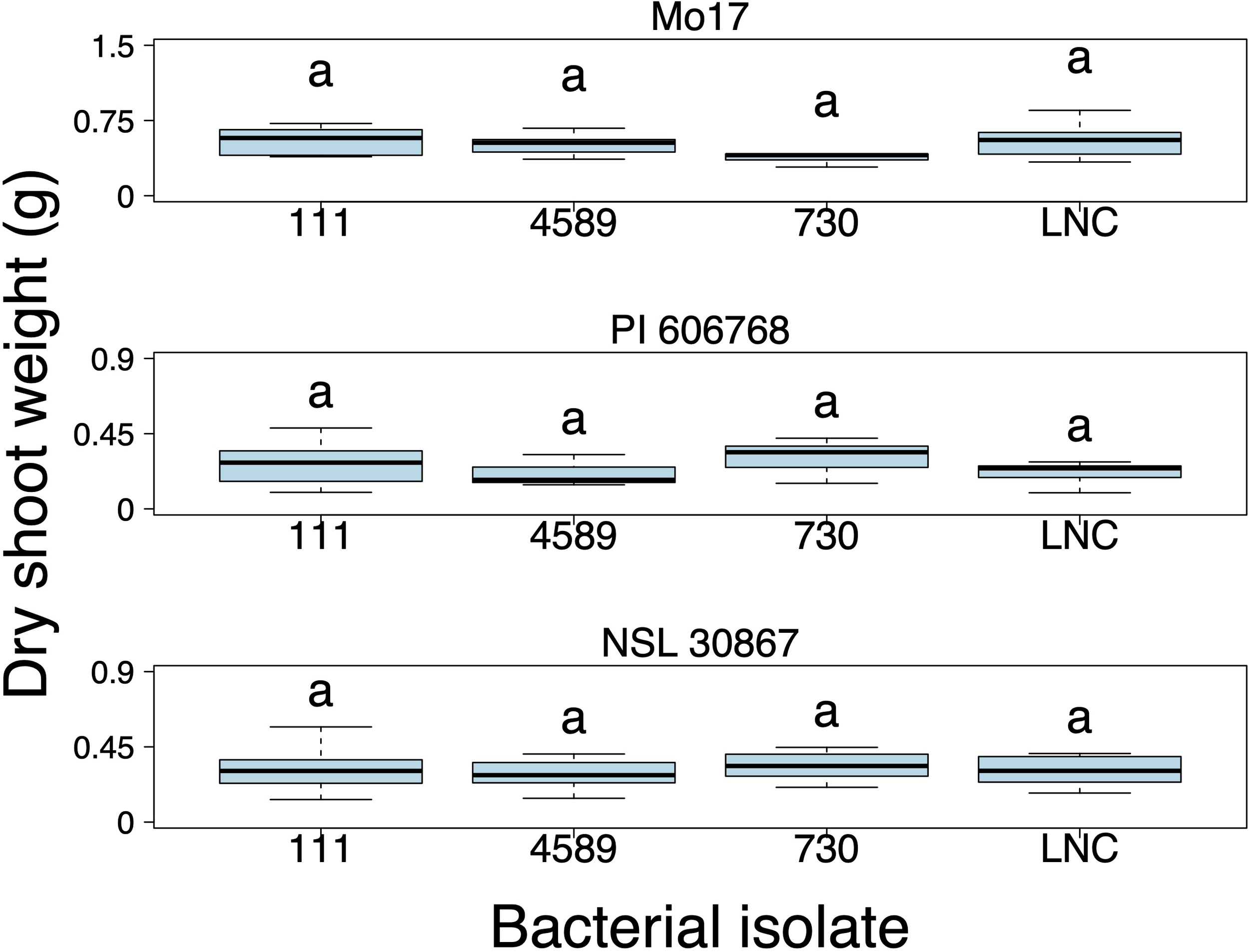
Dry shoot weight (grams) of the maize genotypes Mo17, PI 606768, and NSL 30867 inoculated with bacterial isolates during the non-repeated inoculation experiment. A linear mixed model analysis was performed to assess differences among treatments. Box plots display the median as the line within the box, as well as the lower quartile (bottom of the box) and the upper quartile (top of the box), with the whiskers showing the minimum and maximum data values. Letters above boxes represent statistically significant groupings (p < 0.05). LNC represents the low N control plants. (n = 9; Exceptions: n = 7 for LNC-PI 606768, n = 4 for 4589-PI 606768).
At the 14-, 21-, and 28 days after planting, images were collected of the plants to determine the aboveground biomass at different time points in the development of the plants. However, at none of the time points measured were there any significant differences in aboveground biomass between bacterial-inoculated plants and the uninoculated low N control plants (data not shown).
3.5 Repeated inoculation experiment
To determine whether a recurring soil drench of bacterial inoculant, as done in another study (Papin et al., 2025), may enhance the interaction between plant and bacterial isolate to reveal plant-growth-promoting abilities of the three selected bacterial isolates, a 28-day repeated inoculation experiment was carried out.
The dry shoot weight of the plants was measured 28 days after planting, when the plants were approximately at the V3-V4 stage. In this experiment the bacterial isolate alone did not have a significant effect on the dry shoot weight (p = 0.2047). (Table 3), but as in the previous experiment the maize genotype did have a significant effect on the dry shoot weight (p < 0.0001) (Table 3) under N-deficient conditions. Interestingly, there was a significant interaction effect between bacterial isolate and maize genotype (p = 0.0079) (Table 3), indicating that the dry shoot weight was dependent on the combined effect of the bacterial isolate and maize genotype. This result suggests that the bacterial isolate’s ability to act as a plant-growth-promoting species is dependent on the genetic makeup of the host plant.
Table 3
| Effect | F-value | P-value |
|---|---|---|
| Bacterial Isolate | 1.57 | 0.2047 |
| Maize Genotype | 21.22 | < 0.0001 |
| Bacterial Isolate x Maize Genotype Maize Genotype |
3.17 | 0.0079 |
Repeated inoculation experiment – linear mixed model analysis for the dry shoot weight response of each factor.
Due to the significant interaction between maize genotype and bacterial isolate (Table 3), the effect of each bacterial isolate on the dry shoot weight was analyzed for each maize genotype (Supplementary Table 4). Bacterial isolate 730 had a significantly greater effect on dry shoot weight in the Mo17 maize genotype compared to the uninoculated low N control (p-value = 0.0008) (Supplementary Table 4; Figure 6A). In contrast neither isolates 111 or 4589 had a significant positive growth impact on the Mo17 (Supplementary Table 4; Figure 6A). Conversely, in the maize genotypes Ames 27065 and NSL 30867, none of the bacterial isolates had a significant growth effect on the dry shoot weight compared to the low N uninoculated control (Supplementary Table 4; Figure 6A). Inoculation with 730 (Pseudomonas kribbensis) resulted in a significant increase in dry shoot weight of Mo17 maize plants but not Ames 27065 or NSL 30867 which highlights the strong bacterial isolate host genotype interaction.
Figure 6
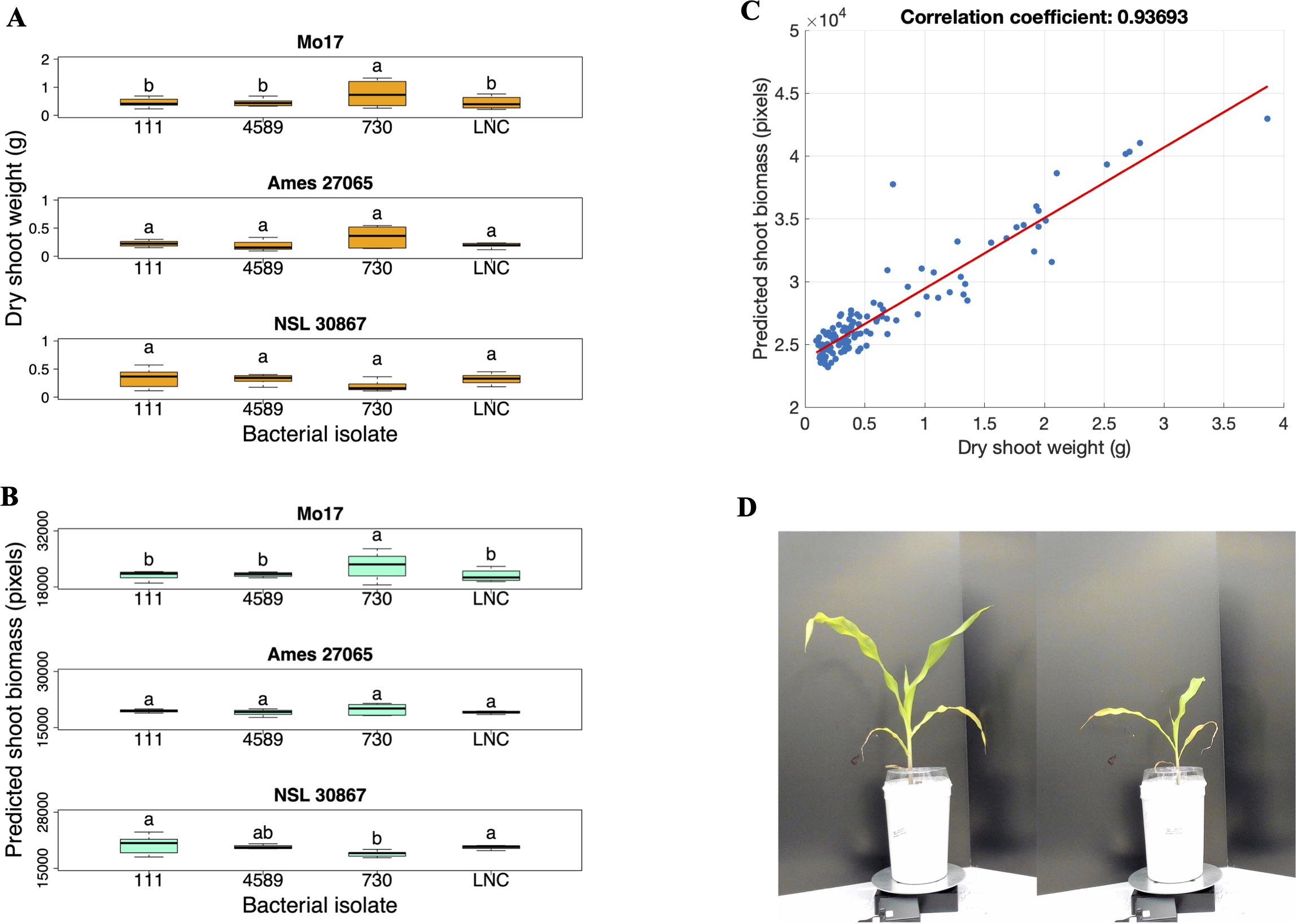
Repeated inoculation experiment. (A) Dry shoot weight (grams) and (B) Predicted biomass (pixels) of the Mo17, Ames 27065, and NSL 30867 maize genotypes inoculated with three bacterial isolates and the low N control (LNC). (C) Correlation analysis between the dry shoot weights of the 28-day old plants and the predicted shoot biomass based on image analysis at 27 days after planting (r = 0.937). (D) Two photos taken of 27-day-old, uninoculated Mo17 maize plants grown in high N (left) and low N (right) conditions in the repeated inoculation experiment. Different letters above the boxes indicate statistically significant differences at p < 0.05. LNC represents the low N control plants. Box plots display the median as the line within the box, as well as the lower quartile (bottom of the box) and the upper quartile (top of the box), with the whiskers showing the minimum and maximum data values. (n = 8; Exceptions: n = 9 for 730-NSL30867, n = 9 for LNC-NSL 30867, n = 7 for LNC-Ames 27065, and n = 6 for 730-Ames 27065.
Images were captured at 14, 21, and 27 days after planting to determine whether growth increases could be measured at earlier time points than the final destructive harvest. The aboveground biomass derived from the imaging system (day 28) was correlated to the predicted biomass computed from images (day 27). The correlation coefficient was (r = 0.937) (Figure 6B), indicating good correspondence between measured and predicted biomass. At 14 days after planting, when the plants were approximately at the V2 stage, there was no significant increase in biomass of the bacterial-inoculated plants compared to the uninoculated low-N control plants. At 21 days after planting, when the plants were approximately at the V2-V3 stage, there was a significant interaction effect between the bacterial isolate and maize genotype (p = 0.0014) (Table 4), indicating that at 21 days after planting, specific bacterial isolates influenced the growth of certain maize genotypes, highlighting a specific host-microbe relationship. The effect of each bacterial isolate was determined for each maize genotype (Supplementary Table 5). We found there was a significant increase in growth of the N-deficient Mo17 inoculated with bacterial isolate 730 (Pseudomonas kribbensis) compared to the uninoculated N-deficient Mo17 maize (p-value = 0.0006) (Figure 6C; Supplementary Table 5) at day 21. Surprisingly, we also found that NSL30867 inoculated with 730 showed a significant decrease in growth at 21 days after planting, but not at 28 days when dry shoot biomass was measured. Representative images are shown of plants grown under nitrogen-deficient and nitrogen-sufficient conditions (Figure 6D).
Table 4
| Effect | F-value | P-value |
|---|---|---|
| Bacterial Isolate | 0.79 | 0.5058 |
| Maize Genotype | 23.49 | < 0.0001 |
| Bacterial Isolate x Maize Genotype Maize Genotype |
4.05 | 0.0014 |
Repeated inoculation experiment (21-day image analysis) – linear mixed model analysis for the optically computed aboveground biomass response of each factor.
4 Discussion
4.1 Low success may be due to incorrect isolate identification
This study aimed to leverage plant genetic data and field-based 16S sequences from the maize rhizosphere to find isolates in a culture collection that enhance plant growth in low N settings. To accomplish this, we analyzed ASVs from the rhizosphere of maize cultivated in the field, which showed positive or negative links to alleles from a diverse group of maize lines (Flint-Garcia et al., 2005) in low N conditions (Meier et al., 2022). This was done to identify bacterial isolates from a laboratory culture collection containing more than 2,000 isolates. The ASVs were used to search the full or partial sequences of the 16S rRNA gene derived from each sequenced isolate in the collection. Others have identified growth-promoting bacterial species that improve maize and wheat growth, with some of these having N fixation properties (Shaharoona et al., 2008; Hungria et al., 2010; Alves et al., 2015), but there are few instances where plant genetic information has been used to identify bacterial taxa that enhance cereal plant growth. Our reasoning was that these ASVs, derived from culture-independent data associated with plant alleles that improve growth under low N conditions, would lead to the identification of unique isolates in a large culture collection that promote growth under low N conditions. From this search, we identified 63 isolates with a 95-100% match to the 16S ASV sequences from a culture-independent study. Upon further testing of the isolates on the maize genotype Mo17, only two isolates, 111 (Arthrobacter sp.) and 730 (Pseudomonas kribbensis), promoted the growth of Mo17 maize under low N conditions in a growth chamber. Bacterial isolate 730 belongs to the Pseudomonas genus and plant-growth-promoting properties of this genus have been documented, including phytohormone production, N fixation, siderophore production, and phosphate solubilization (Panpatte et al., 2016). In contrast the bacterial isolate 111 belongs to the Arthrobacter genus, and it has been reported that this genus possesses plant-growth-promoting properties, such as the ability to produce indole-3-acetic acid, solubilize phosphate, and fix N (Jiang et al., 2022). The specific plant-growth-promoting properties of 730 and 111, as well as the biological mechanisms enabling the maize genotype and bacterial inoculant interaction, remain to be further investigated. Our search revealed 63 isolates that matched the 16S ASV sequences from a culture-independent study with 95-100% accuracy. Further testing on the maize genotype Mo17 showed that only two isolates, 111 (Arthrobacter sp.) and 730 (Pseudomonas kribbensis), enhanced the growth of Mo17 maize in low N conditions within a growth chamber. The low success rate of this method in identifying growth-promoting isolates using field data may be attributed to a few factors. Initially, while the ASV sequences pinpointed culture collection isolates with up to 100% sequence identity, a key factor reducing the success rate is that numerous genes in bacterial genomes may show variation. A striking illustration of this comes from research on 19 Pseudomonas fluorescens strains collected from either the endosphere or rhizosphere of Eastern cottonwood (Populus deltoides). Despite these strains having 99% similarity in their 16S rRNA genes, notable differences were observed in the rest of the genome and in the phenotypes of the bacterial strains (Timm et al., 2015). The 16S rRNA gene is frequently utilized for phylogenetic analysis, but this approach often lacks taxonomic resolution below the genus level (Hartmann et al., 2019).
Other studies have used different approaches to identify growth-promoting bacteria. In one study, 474 bacterial isolates were obtained from the durum wheat rhizosphere and initially screened based on functional activities that may enhance plant acquisition of nutrients (Di Benedetto et al., 2019). In a first round of screening the pool of wheat rhizosphere bacteria was narrowed down to 333 and then in the second-round testing for mineralization of phosphate, production of IAA and nitrification, 16 isolates were identified as having potential plant growth promoting properties. Eventually three isolates with properties that warranted field testing were identified. Another study sampled 61 endophytic bacteria isolated from the leaves and stems of Pistachio (Pistacia vera). Through an initial screening, ten were identified as having unique functional properties and eight of the ten were identified as gram-negative. The next assessment was done based on the ability to promote root growth (Etminani and Harighi, 2018) and eight out of ten tested promoted root formation. Our study used field-derived ASVs that identified 63 isolates that were between 95 – 100% similarity to 16S sequences from a large collection of bacterial isolates. No other pre-screening tests were conducted to identify whether the bacterial isolates possess plant-growth-promoting capabilities, although testing for specific plant-growth-promoting factors may have been beneficial. The genetic approach used here was novel and identified at least two bacterial isolates that promote growth under different types of growth modalities. An improvement of this approach would have been to focus on culturing from the rhizosphere communities from which the original data was derived (Meier et al., 2022). The culture-independent microbiome-based data set with specific culturing of bacteria from the same samples would be the optimal approach that may improve the success rate in identifying growth-promoting bacteria.
4.2 Low success may be due to a lack of community
Another reason for the low success rate may be that each of the 63 bacterial isolates was inoculated individually and grown under axenic conditions without being given the opportunity to interact with other bacterial species. Cooperativity between members of a bacterial community may have been essential for observing growth-promoting effects. For example, some of the isolates can act as keystone species or assist in biofilm formation (Noirot-Gros et al., 2018), and others may recruit the beneficial bacteria to the rhizosphere (Liu et al., 2021). Within a rhizosphere community, bacterial species communicate with both the host plant (inter-kingdom communication) and with one another (intra-kingdom communication). Quorum sensing is one method of communication amongst rhizobacteria (Singh et al., 2022) that is important for biofilm formation (Hammer and Bassler, 2003; De Kievit, 2009). Auto-inducers are the signal molecules that bacterial species use in quorum sensing to communicate with other bacterial species (Schikora et al., 2016). Small, low-molecular-weight volatile organic compounds (VOCs) are also used to communicate within bacterial communities (Netzker et al., 2020) and these have been shown to promote the growth of other microbial species (Scott et al., 2019), and act directly as plant-growth promoting compounds (Park et al., 2015; Zhou et al., 2016).
Plants release exudates into the soil around their roots to influence the rhizospheric bacterial community members (Bertin et al., 2003). From there, recruited bacterial members interact with one another through cooperation or competition, influencing the plant’s overall health (Chepsergon and Moleleki, 2023). This may be important in the host-specific bacterial communities found within the rhizosphere and endosphere (Trivedi et al., 2020). Removing the dynamic relationships between bacterial species within a rhizosphere bacterial community in sterilized growth medium may also eliminate the beneficial interactions that promote the growth of the host plant. In various plant growth studies investigating the use of single bacterial isolates versus a consortium isolates, using a multi-isolate inoculum enhanced plant growth more than a single-isolate inoculum. For example, when inoculated together, Bacillus velezensis and Pseudomonas stutzeri had a beneficial growth effect on plant traits compared to plants inoculated with only one species (Sun et al., 2022). This was thought to be because the B. velezensis stimulated P. stutzeri, forming biofilms on the plant root surface, indicating cooperation between the two inoculated species. Similarly, PGPRs combined and inoculated on maize performed better than each PGPR inoculated individually (Molina-Romero et al., 2017). Moving forward, it will be necessary to simultaneously test the inoculation of multiple members of the 63 isolates or use non-sterile soils that contain native communities and would be more relevant to agroecosystems.
4.3 Repeated inoculations or very high propagule numbers are essential for growth promotion
In this study, we found that repeated inoculation of a bacterial isolate was more beneficial for enhancing plant growth with certain isolates than one or two introductions of a bacterial strain. When a novel organism is introduced to an environment, propagule pressure is a key factor in colonization success that must be considered. Propagule pressure is a concept that accounts for the frequency of introduction events as well as the number of individuals per event that are introduced (Koontz et al., 2018). This concept is often discussed in invasion biology (Stringham and Lockwood, 2021); however, propagule pressure may also be applied to help formulate the most effective bacterial inoculation strategies to maximize the plant growth-promoting effects of the bacterial species. In this study, we found that bacterial isolate 730 (Pseudomonas kribbensis) significantly increased the dry shoot weight of maize grown under N-deficient conditions under a repeated inoculation protocol. We did not find the same result in our non-repeated inoculation experiment. In the repeated inoculation experiment, six repeated inoculations were carried out, compared to the two inoculations done in the non-repeated inoculation experiment. Similar to our findings, others have shown that repeated inoculations promote plant growth in maize (Papin et al., 2025) and wheat (Chen et al., 2024), whereas one to two inoculations do not. The timing of inoculations may also be important. In one study, repeated inoculations in soil prior to maize sowing promoted the growth of aboveground biomass, while repeated inoculations carried out after maize sowing impacted the resident bacterial community but did not impact maize growth (Papin et al., 2025). In contrast, an increase in wheat growth as well as an increase in both soil bacterial diversity and functional genes related to the N cycle was reported (Chen et al., 2024) following repeated bacterial inoculations performed after sowing.
In our study, it was also noteworthy that the beneficial effects of bacterial isolate 730 (Pseudomonas kribbensis) were significant at the 21-day timepoint in the repeated inoculation experiment, but not at the 14-day timepoint. Time was not included as a variable in our statistical analysis, but it is clearly important in experimental design, as short-term experiments may be less useful in determining the growth-promoting effects of bacterial isolates. The plant growth-promoting effects of various bacteria, like Bacillus, have been reported as significant at the V6 growth stage of maize, but not at the V4 growth stage (Lin et al., 2019). In contrast to the results of the repeated inoculation experiment, bacterial isolate 111 (Arthrobacter sp.) significantly promoted the growth of Mo17 maize after just 14 days, but the growth promotion was not observed after that time point. Similarly, an Arthrobacter nicotinovorans strain isolated from the rhizosphere of Panax ginseng was found to significantly increase the shoot weight of inoculated ginseng plants after 15 days of growth (Jiang et al., 2022). Therefore, the experimental timing, multiple inoculations and the application of nondestructive phenotyping methods utilized here are important considerations when studying plant growth promotion by bacteria.
4.4 Plant genotype is essential for action of growth promotion
Plant genotype is known to influence the composition of the bacterial communities in roots, rhizosphere, and the phyllosphere (Aira et al., 2010; Bodenhausen et al., 2014; Marques et al., 2014). Two of the well-known mechanisms that shape plant bacterial interactions include the immune system (Tzipilevich et al., 2021) and root exudates (Sasse et al., 2018). It has also been shown that maize genotypes (Lopes et al., 2022; Wang et al., 2022; Lopes et al., 2023a) influence the composition of microbial communities through the release of different amounts of root exudates composed of sugars, amino acids, organic acids, and phenolic compounds (Coskun et al., 2017). Host genetics has also been demonstrated to have a strong influence on the diversity and composition of microbiomes, but less is known about the mechanisms behind the association of plant genes with microbial community structure (Peiffer et al., 2013; Wagner et al., 2016; Gehring et al., 2017). For example, when investigating the genetic variation among 302 natural accessions of Arabidopsis thaliana plants, researchers reported there were certain candidate genes, involved in important plant growth-related processes, that were also highly associated with how well the accession responded to the PGPR effects of Pseudomonas simiae WCS417r (Wintermans et al., 2016). Likewise, in another study, it was reported that the interaction between PGPB and the maize genotypes in 360 lines of tropical maize germplasm was controlled by multiple genes within the maize genotypes (Yassue et al., 2021).
Although there are interactions between members of bacterial communities and plant genotypes, there are few clear examples of this in the literature (Vargas et al., 2012; Do Amaral et al., 2016). Genotype by bacterial isolate interactions may be a critical factor limiting the widespread successful application of bacterial growth-promoting inoculants in agriculture. In a previous study, we showed genotypic differences in sorghum growth responses to a cocktail of 10 bacterial isolates (Chai et al., 2021) under low N conditions. In this current study, we found that one bacterial isolate from the genus Pseudomonas (730) only enhanced the growth of one (Mo17) and not two other maize genotypes. The other two genotypes were chosen based on total sugars and jasmonic acid in their exudates (Lopes et al., 2022). We hypothesized that because the concentrations of total sugars and jasmonic acid in the exudates impacted the rhizospheric bacterial communities in the maize plants (Lopes et al., 2022), perhaps the plant-growth-promoting effects of the bacterial isolates 111, 730, and 4589 would also be influenced by the exudate concentrations. In the repeated inoculation experiment of our study, there was a significant interaction (p = 0.0079) between the maize genotype and the bacterial isolate, indicating that maize genotypes respond differently to the plant-growth-promoting bacterial isolates tested in this study.
5 Conclusion
Our research emphasizes key factors to consider when screening soil bacteria that promote plant growth. We demonstrated that both the number and potentially the timing of inoculation significantly influence a bacterial isolate’s ability to enhance plant growth. We also confirmed the hypothesis that plant host genotype is critical to the development of positive bacterial interactions and suggest that other microbial community members are essential elements when testing inoculants under controlled conditions. Although our method of using genetically identified ASVs to select culture collection isolates did not yield a significant number of growth-promoting isolates, it is possible that culturing directly from the field-derived rhizosphere samples could have improved the success rate. In future, considering these multiple factors may significantly enhance the success rate of identifying growth-promoting bacterial isolates and may also lead to increased reliability and repeatability of biostimulants across a broader range of agroecosystems.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
LF: Visualization, Formal analysis, Writing – original draft, Data curation, Conceptualization, Writing – review & editing, Methodology, Validation. JY: Conceptualization, Writing – review & editing, Funding acquisition. J-JR: Writing – review & editing, Data curation. HM: Methodology, Software, Writing – review & editing. DS: Formal analysis, Conceptualization, Writing – original draft, Data curation, Writing – review & editing, Methodology, Funding acquisition.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work is supported by the Agriculture and Food Research Initiative (Grant Number 2022-67013-36560) from the USDA National Institute of Food and Agriculture to JY and DS.
Acknowledgments
This work was conducted as part of a master’s thesis at the University of Nebraska Lincoln (Foster, 2024). Part of the bioinformatics work was completed utilizing the Holland Computing Center of the University of Nebraska, which receives support from the UNL Office of Research and Economic Development, and the Nebraska Research Initiative. We thank Jingjie Hao for training in the laboratory on various methods. We would also like to thank Karrie Weber for her guidance and input as a graduate committee member on the development of this research project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1637156/full#supplementary-material
References
1
Abdel-Lattif H. M. Taha M. Atta M. (2019). Nitrogen use efficiency and grain protein of corn affected by low nitrogen application. Egypt. J. Agron.41, 69–78. doi: 10.21608/agro.2019.9752.1151
2
Aira M. Gómez-Brandón M. Lazcano C. Bååth E. Domínguez J. (2010). Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil. Biol. Biochem.42, 2276–2281. doi: 10.1016/j.soilbio.2010.08.029
3
Aluko O. O. Kant S. Adedire O. M. Li C. Z. Yuan G. Liu H. B. et al . (2023). Unlocking the potentials of nitrate transporters at improving plant nitrogen use efficiency. Front. Plant Sci.14. doi: 10.3389/fpls.2023.1074839
4
Alves G. C. Videira S. S. Urquiaga S. Reis V. M. (2015). Differential plant growth promotion and nitrogen fixation in two genotypes of maize by several Herbaspirillum inoculants. Plant Soil387, 307–321. doi: 10.1007/s11104-014-2295-2
5
Andreote F. D. Da Rocha U. N. Araújo W. L. Azevedo J. L. Van Overbeek L. S. (2010). Effect of bacterial inoculation, plant genotype and developmental stage on root-associated and endophytic bacterial communities in potato (Solanum tuberosum). Antonie Van Leeuwenhoek97, 389–399. doi: 10.1007/s10482-010-9421-9
6
Assefa Y. Roozeboom K. L. Staggenborg S. A. Du J. (2012). Dryland and irrigated corn yield with climate, management, and hybrid changes from 1939 through 2009. Agron. J.104, 473–482. doi: 10.2134/agronj2011.0242
7
Auguie B. Antonov A. (2017). gridExtra: miscellaneous functions for “Grid” Graphics. R package 2.3. Available online at: https://CRAN.R-project.org/package=gridExtra (Accessed August 20, 2025).
8
Bardgett R. D. van der Putten W. H. (2014). Belowground biodiversity and ecosystem functioning. Nature515, 505–511. doi: 10.1038/nature13855
9
Barthelmé S. Tschumperlé D. (2019). imager: an R package for image processinng based on Clmg. J. Open Source Software4, 1012. doi: 10.21105/joss.01012
10
Bertin C. Yang X. H. Weston L. A. (2003). The role of root exudates and allelochemicals in the rhizosphere. Plant Soil256, 67–83. doi: 10.1023/A:1026290508166
11
Bodenhausen N. Bortfeld-Miller M. Ackermann M. Vorholt J. A. (2014). A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PloS Genet.10, e1004283. doi: 10.1371/journal.pgen.1004283
12
Bottini R. Fulchieri M. Pearce D. Pharis R. P. (1989). Identification of gibberellin A(1), A(3) and Iso-A(3) in cultures of Azospirillum lipoferum. Plant Physiol.90, 45–47. doi: 10.1104/pp.90.1.45
13
Camacho C. Coulouris G. Avagyan V. Ma N. Papadopoulos J. Bealer K. et al . (2009). BLAST+: architecture and applications. BMC Bioinf.10, 421. doi: 10.1186/1471-2105-10-421
14
Cao P. Y. Lu C. Q. Yu Z. (2018). Historical nitrogen fertilizer use in agricultural ecosystems of the contiguous United States during 1850-2015: application rate, timing, and fertilizer types. ESSD10, 969–984. doi: 10.5194/essd-10-969-2018
15
Cassan F. Coniglio A. López G. Molina R. Nievas S. De Carlan C. L. et al . (2020). Everything you must know about Azospirillum and its impact on agriculture and beyond. Biol. Fert. Soils56, 461–479. doi: 10.1007/s00374-020-01463-y
16
Chai Y. N. Futrell S. Schachtman D. P. (2022a). Assessment of bacterial inoculant delivery methods for cereal crops. Front. Microbiol.13. doi: 10.3389/fmicb.2022.791110
17
Chai Y. N. Ge Y. F. Stoerger V. Schachtman D. P. (2021). High-resolution phenotyping of sorghum genotypic and phenotypic responses to low nitrogen and synthetic microbial communities. Plant Cell Environ.44, 1611–1626. doi: 10.1111/pce.14004
18
Chai Y. N. Schachtman D. P. (2022b). Root exudates impact plant performance under abiotic stress. Trends Plant Sci.27, 80–91. doi: 10.1016/j.tplants.2021.08.003
19
Chen Y. Zang H. Bai L. Lv C. Chen X. Li S. et al . (2024). Repeated inoculations improve wheat yield through modifying the rhizobacterial communities and nitrogen and phosphorus fractions. Appl. Soil Ecol.196, 105287. doi: 10.1016/j.apsoil.2024.105287
20
Chepsergon J. Moleleki L. N. (2023). Rhizosphere bacterial interactions and impact on plant health. Curr. Opin. Microbiol.73, 102297. doi: 10.1016/j.mib.2023.102297
21
Chun L. Mi G. Li J. Chen F. Zhang F. (2005). Genetic analysis of maize root characteristics in response to low nitrogen stress. Plant Soil276, 369–382. doi: 10.1007/s11104-005-5876-2
22
Clark I. M. Hughes D. J. Fu Q. L. Abadie M. Hirsch P. R. (2021). Metagenomic approaches reveal differences in genetic diversity and relative abundance of nitrifying bacteria and archaea in contrasting soils. Sci. Rep.11, 15905. doi: 10.1038/s41598-021-95100-9
23
Coskun D. Britto D. T. Shi W. Kronzucker H. J. (2017). How plant root exudates shape the nitrogen cycle. Trends Plant Sci.22, 661–673. doi: 10.1016/j.tplants.2017.05.004
24
Crawford N. M. Glass A. D. M. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci.3, 389–395. doi: 10.1016/S1360-1385(98)01311-9
25
De Kievit T. R. (2009). Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol.11, 279–288. doi: 10.1111/j.1462-2920.2008.01792.x
26
Denman K. L. Brasseur G. Chidthaisong A. Ciais P. Cox P. M. Dickinson R. E. et al . (2007). “Couplings between changes in the climate system and biogeochemistry,” in Ar4 Climate Change 2007: The Physical Science Basis. Cambridge, UK, New York, NY, USA: Cambridge University Press. 499–587.
27
Di Benedetto N. A. Campaniello D. Bevilacqua A. Cataldi M. P. Sinigaglia M. Flagella Z. et al . (2019). Isolation, screening, and characterization of plant-growth-promoting bacteria from durum wheat rhizosphere to improve N and P nutrient use efficiency. Microorganisms7, 541. doi: 10.3390/microorganisms7110541
28
Do Amaral F. P. Pankievicz V. C. S. Arisi A. C. M. De Souza E. M. Pedrosa F. Stacey G. (2016). Differential growth responses of Brachypodium distachyon genotypes to inoculation with plant growth promoting rhizobacteria. Plant Mol. Biol.90, 689–697. doi: 10.1007/s11103-016-0449-8
29
Etminani F. Harighi B. (2018). Isolation and identification of endophytic bacteria with plant growth promoting activity and biocontrol potential from wild pistachio trees. Plant Pathol. J.34, 208–217. doi: 10.5423/Ppj.Oa.07.2017.0158
30
Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution39, 783–791. doi: 10.2307/2408678
31
Flint-Garcia S. A. Thuillet A. C. Yu J. M. Pressoir G. Romero S. M. Mitchell S. E. et al . (2005). Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J.44, 1054–1064. doi: 10.1111/j.1365-313X.2005.02591.x
32
Foster L. R . (2024). Investigating the abilities of potentially beneficial bacteria for increasing nitrogen-use efficiency in maiz. Master's thesis. University of Nebraska-Lincoln.
33
Foster L . (2025). Figure 1. Inoculation schedule for the non-repeated and repeated inoculation experiments. Created in BioRender. Available online at: https://BioRender.com/s0lhnex.
34
Franche C. Lindström K. Elmerich C. (2009). Nitrogen-fixing bacteria associated with leguminous and non-leguminous plants. Plant Soil321, 35–59. doi: 10.1007/s11104-008-9833-8
35
French E. Tran T. Iyer-Pascuzzi A. S. (2020). Tomato genotype modulates selection and responses to root microbiota. Phytobiomes J.4, 314–326. doi: 10.1094/Pbiomes-02-20-0020-R
36
Galindo F. S. Rodrigues W. L. Fernandes G. C. Boleta E. H. M. Jalal A. Rosa P. et al . (2022). Enhancing agronomic efficiency and maize grain yield with Azospirillum brasilense inoculation under Brazilian savannah conditions. Eur. J. Agron.134, 126471. doi: 10.1016/j.eja.2022.126471
37
Galindo F. S. Teixeira M. C. M. Buzetti S. Pagliari P. H. Santini J. M. K. Alves C. J. et al . (2019). Maize yield response to nitrogen rates and sources associated with Azospirillum brasilense. Agron. J.111, 1985–1997. doi: 10.2134/agronj2018.07.0481
38
Gehring C. A. Sthultz C. M. Flores-Rentería L. Whipple A. V. Whitham T. G. (2017). Tree genetics defines fungal partner communities that may confer drought tolerance. Proc. Natl. Acad. Sci. U.S.A.114, 11169–11174. doi: 10.1073/pnas.1704022114
39
Hammer B. K. Bassler B. L. (2003). Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol.50, 101–114. doi: 10.1046/j.1365-2958.2003.03688.x
40
Han S. O. New P. B. (1998). Variation in nitrogen fixing ability among natural isolates of Azospirillum. Microb. Ecol.36, 193–201. doi: 10.1007/s002489900106
41
Hartmann A. Fischer D. Kinzel L. Chowdhury S. P. Hofmann A. Baldani J. I. et al . (2019). Assessment of the structural and functional diversities of plant microbiota: achievements and challenges - a review. J. Adv. Res.19, 3–13. doi: 10.1016/j.jare.2019.04.007
42
Hijmans R. J. (2025). raster: Geographic Data Analysis and Modeling. R package version 3.6-32. Available online at: https://rspatial.org/raster (Accessed August 20, 2025).
43
Hungria M. Barbosa J. Z. Rondina A. B. L. Nogueira M. A. (2022). Improving maize sustainability with partial replacement of N fertilizers by inoculation with Azospirillum brasilense. Agron. J.114, 2969–2980. doi: 10.1002/agj2.21150
44
Hungria M. Campo R. J. Souza E. M. Pedrosa F. O. (2010). Inoculation with selected strains of Azospirillum brasilense and A. lipoferum improves yields of maize and wheat in Brazil. Plant Soil331, 413–425. doi: 10.1007/s11104-009-0262-0
45
Javed T. Indu I. Singhal R. K. Shabbir R. Shah A. N. Kumar P. et al . (2022). Recent advances in agronomic and physio-molecular approaches for improving nitrogen use efficiency in crop plants. Front. Plant Sci.13. doi: 10.3389/fpls.2022.877544
46
Jiang Y. Song Y. Jiang C. Y. Li X. Liu T. T. Wang J. R. et al . (2022). Identification and characterization of Arthrobacter nicotinovorans JI39, a novel plant growth-promoting rhizobacteria strain from Panax ginseng. Front. Plant Sci.13. doi: 10.3389/fpls.2022.873621
47
Koontz M. J. Oldfather M. F. Melbourne B. A. Hufbauer R. A. (2018). Parsing propagule pressure: number, not size, of introductions drives colonization success in a novel environment. Ecol. Evol.8, 8043–8054. doi: 10.1002/ece3.4226
48
Kumar S. Stecher G. Suleski M. Sanderford M. Sharma S. Tamura K. (2024). MEGA12: molecular evolutionary genetic analysis version 12 for adaptive and green computing. Mol. Biol. Evol.41, 1–9. doi: 10.1093/molbev/msae263
49
Lan X. Y. Zhu L. Y. Yuan Q. Q. (2022). Long-term variation of greenhouse gas N2O observed by MLS during 2005-2020. Remote Sens.14, 955. doi: 10.3390/rs14040955
50
Li J. Y. Fu Y. L. Pike S. M. Bao J. Tian W. Zhang Y. et al . (2010). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem Sap and mediates cadmium tolerance. Plant Cell22, 1633–1646. doi: 10.1105/tpc.110.075242
51
Lin Y. Watts D. B. Kloepper J. W. Adesemoye A. O. Feng Y. (2019). Effect of plant growth-promoting rhizobacteria at various nitrogen rates on corn growth. Agric. Sci.10, 1542–1565. doi: 10.4236/as.2019.1012114
52
Liu Y. Gao J. Bai Z. H. Wu S. H. Li X. L. Wang N. et al . (2021). Unraveling mechanisms and impact of microbial recruitment on oilseed rape (Brassica napus L.) and the rhizosphere mediated by plant growth-promoting rhizobacteria. Microorganisms9, 161. doi: 10.3390/microorganisms9010161
53
Lopes L. D. Futrell S. L. Bergmeyer E. Hao J. J. Schachtman D. P. (2023a). Root exudate concentrations of indole-3-acetic acid (IAA) and abscisic acid (ABA) affect maize rhizobacterial communities at specific developmental stages. FEMS Microbiol. Ecol.99, 1–12. doi: 10.1093/femsec/fiad019
54
Lopes L. D. Hao J. Schachtman D. P. (2021). Alkaline soil pH affects bulk soil, rhizosphere and root endosphere microbiomes of plants growing in a Sandhills ecosystem. FEMS Microbiol. Ecol.97, fiab028. doi: 10.1093/femsec/fiab028
55
Lopes L. D. Schachtman D. P. (2023b). Rhizosphere and bulk soil bacterial community succession is influenced more by changes in soil properties than in rhizosphere metabolites across a maize growing season. Appl. Soil Ecol.189, 104960. doi: 10.1016/j.apsoil.2023.104960
56
Lopes L. D. Wang P. Futrell S. L. Schachtman D. P. (2022). Sugars and jasmonic acid concentration in root exudates affect maize rhizosphere bacterial communities. Appl. Environ. Microbiol.88, e0097122. doi: 10.1128/aem.00971-22
57
Lv X. M. Zhang Y. X. Hu L. Zhang Y. Zhang B. Xia H. Y. et al . (2021). Low-nitrogen stress stimulates lateral root initiation and nitrogen assimilation in wheat: roles of phytohormone signaling. J. Plant Growth Regul.40, 436–450. doi: 10.1007/s00344-020-10112-5
58
Malacrino A. Mosca S. Nicosia M. G. L. Agosteo G. E. Schena L. (2022). Plant genotype shapes the bacterial microbiome of fruits, leaves, and soil in olive plants. Plants11, 613. doi: 10.3390/plants11050613
59
Marques J. M. Da Silva T. F. Vollu R. E. Blank A. F. Ding G. C. Seldin L. et al . (2014). Plant age and genotype affect the bacterial community composition in the tuber rhizosphere of field-grown sweet potato plants. FEMS Microbiol. Ecol.88, 424–435. doi: 10.1111/1574-6941.12313
60
Martienssen M. Schöps R. (1999). Population dynamics of denitrifying bacteria in a model biocommunity. Water Res.33, 639–646. doi: 10.1016/S0043-1354(98)00222-X
61
Meier M. A. Xu G. Lopez-Guerrero M. G. Li G. Y. Smith C. Sigmon B. et al . (2022). Association analyses of host genetics, root-colonizing microbes, and plant phenotypes under different nitrogen conditions in maize. eLife11, e75790. doi: 10.7554/eLife.75790
62
Molina-Romero D. Baez A. Quintero-Hernández V. Castañeda-Lucio M. Fuentes-Ramírez L. E. Bustillos-Cristales M. D. et al . (2017). Compatible bacterial mixture, tolerant to desiccation, improves maize plant growth. PloS One12, e0187913. doi: 10.1371/journal.pone.0187913
63
Nei M. Kumar S. (2000). Molecular evolution and phylogenetics (New York: Oxford University Press).
64
Netzker T. Shepherdson E. M. F. Zambri M. P. Elliot M. A. (2020). Bacterial volatile compounds: functions in communication, cooperation, and competition. Annu. Rev. Microbiol.74, 409–430. doi: 10.1146/annurev-micro-011320-015542
65
Noirot-Gros M. F. Shinde S. Larsen P. E. Zerbs S. Korajczyk P. J. Kemner K. M. et al . (2018). Dynamics of aspen roots colonization by Pseudomonads reveals strain-specific and mycorrhizal-specific patterns of biofilm formation. Front. Microbiol.9. doi: 10.3389/fmicb.2018.00853
66
Novoa R. Loomis R. S. (1981). Nitrogen and plant production. Plant Soil58, 177–204. doi: 10.1007/Bf02180053
67
Ooms J. (2025). magick: Advanced Graphics and Image-Processing in R. R package version 2.8.7. Available online at: https://github.com/ropensci/magick (Accessed August 20, 2025).
68
Panpatte D. G. Jhala Y. K. Shelat H. N. Vyas R. V. (2016). “(Pseudomonas fluorescens: a promising biocontrol agent and PGPR for sustainable agriculture.),” in Microbial Inoculants in Sustainable Agricultural Productivity (New Delhi: Springer).
69
Papin M. Polrot A. Breuil M. C. Czarnes S. Dreux-Zigha A. Roux X. L. et al . (2025). Pre-sowing recurrent inoculation with Pseudomonas fluorescens promotes maize growth. Biol. Fert. Soils61, 125–140. doi: 10.1007/s00374-024-01873-2
70
Park Y. S. Dutta S. Ann M. Raaijmakers J. M. Park K. (2015). Promotion of plant growth by Pseudomonas fluorescens strain SS101 via novel volatile organic compounds. Biochem. Biophys. Res. Commun.461, 361–365. doi: 10.1016/j.bbrc.2015.04.039
71
Pau G. Fuchs F. Sklyar O. Boutros M. Huber W. (2010). EBImage-an R package for image processing with applications to cellular phenotypes. Bioinformatics26, 979–981. doi: 10.1093/bioinformatics/btq046
72
Peiffer J. A. Spor A. Koren O. Jin Z. Tringe S. G. Dangl J. L. et al . (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. U.S.A.110, 6548–6553. doi: 10.1073/pnas.1302837110
73
Raza S. Farmaha B. S. (2022). Contrasting corn yield responses to nitrogen fertilization in southeast coastal plain soils. Front. Environ. Sci.10. doi: 10.3389/fenvs.2022.955142
74
R Core Team (2023). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
75
Saitou N. Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol.4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
76
SAS Institute Inc . (2023). SAS software (Version 9.4, Maintenance Release 8). SAS Institute Inc.
77
Sasse J. Martinoia E. Northen T. (2018). Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci.23, 25–41. doi: 10.1016/j.tplants.2017.09.003
78
Schachtman D. P. Munns R. (1992). Sodium accumulation in leaves of Triticum species that differ in salt tolerance. Aust. J. Plant Physiol.19, 331–340. doi: 10.1071/Pp9920331
79
Schikora A. Schenk S. T. Hartmann A. (2016). Beneficial effects of bacteria-plant communication based on quorum sensing molecules of the N-acyl homoserine lactone group. Plant Mol. Biol.90, 605–612. doi: 10.1007/s11103-016-0457-8
80
Scott J. Sueiro-Olivares M. Ahmed W. Heddergott C. Zhao C. Thomas R. et al . (2019). Pseudomonas aeruginosa-derived volatile sulfur compounds promote distal Aspergillus fumigatus growth and a synergistic pathogen-pathogen interaction that increases pathogenicity in co-infection. Front. Microbiol.10. doi: 10.3389/fmicb.2019.02311
81
Shaharoona B. Naveed M. Arshad M. Zahir Z. A. (2008). Fertilizer-dependent efficiency of Pseudomonads for improving growth, yield, and nutrient use efficiency of wheat (Triticum aestivum L.). Appl. Microbiol. Biotechnol.79, 147–155. doi: 10.1007/s00253-008-1419-0
82
Singh K. Chandra R. Purchase D. (2022). Unraveling the secrets of rhizobacteria signaling in rhizosphere. Rhizosphere21, 100484. doi: 10.1016/j.rhisph.2022.100484
83
Soumare A. Diedhiou A. G. Thuita M. Hafidi M. Ouhdouch Y. Gopalakrishnan S. et al . (2020). Exploiting biological nitrogen fixation: a route towards a sustainable agriculture. Plants9, 1011. doi: 10.3390/plants9081011
84
Stecher G. Tamura K. Kumar S. (2020). Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol.37, 1237–1239. doi: 10.1093/molbev/msz312
85
Steenhoudt O. Vanderleyden J. (2000). Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiol. Rev.24, 487–506. doi: 10.1111/j.1574-6976.2000.tb00552.x
86
Stella M. Suhaimi M. (2010). Selection of suita ble growth medium for free-living diazotrophs isolated from compost. J. Trop. Agric. Food Sci.38, 211–219.
87
Stringham O. C. Lockwood J. L. (2021). Managing propagule pressure to prevent invasive species establishments: propagule size, number, and risk-release curve. Ecol. Appl.31, e02314. doi: 10.1002/eap.2314
88
Sun X. L. Xu Z. H. Xie J. Y. Hesselberg-Thomsen V. Tan T. M. Zheng D. Y. et al . (2022). Bacillus velezensis resident rhizosphere Pseudomonas stutzeri for plant health through metabolic interactions. ISME J.16, 774–787. doi: 10.1038/s41396-021-01125-3
89
The MathWorks Inc . (2022). MATLAB version: 23.2.0.2515942 (R2023b), Natick, Massachusetts: The MathWorks Inc. https://www.mathworks.com
90
Tien T. M. Gaskins M. H. Hubbell D. H. (1979). Plant-growth substances produced by Azospirillum brasilense and their effect on the growth of pearl millet (Pennisetum americanum L.). Appl. Environ. Microb.37, 1016–1024. doi: 10.1128/Aem.37.5.1016-1024.1979
91
Timm C. M. Campbell A. G. Utturkar S. M. Jun S. R. Parales R. E. Tan W. A. et al . (2015). Metabolic functions of Pseudomonas fluorescens strains from Populus deltoides depend on rhizosphere or endosphere isolation compartment. Front. Microbiol.6. doi: 10.3389/fmicb.2015.01118
92
Trivedi P. Leach J. E. Tringe S. G. Sa T. M. Singh B. K. (2020). Plant-microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol.18, 607–621. doi: 10.1038/s41579-020-0412-1
93
Tzipilevich E. Russ D. Dangl J. L. Benfey P. N. (2021). Plant immune system activation is necessary for efficient root colonization by auxin-secreting beneficial bacteria. Cell Host Microbe29, 1507–1520.e4. doi: 10.1016/j.chom.2021.09.005
94
Urbanek S. Johnson K. (2023). tiff: Read and Write TIFF Images. R package version 0.1-12. Available online at: https://www.rforge.net/tiff/ (Accessed August 20, 2025).
95
Vargas L. De Carvalho T. L. G. Ferreira P. C. G. Baldani V. L. D. Baldani J. I. Hemerly A. S. (2012). Early responses of rice (Oryza sativa L.) seedlings to inoculation with beneficial diazotrophic bacteria are dependent on plant and bacterial genotypes. Plant Soil356, 127–137. doi: 10.1007/s11104-012-1274-8
96
Wagner M. R. Lundberg D. S. Del Rio T. G. Tringe S. G. Dangl J. L. Mitchell-Olds T. (2016). Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun.7, 12151. doi: 10.1038/ncomms12151
97
Wang P. Lopes L. D. Lopez-Guerrero M. G. Van Dijk K. Alvarez S. Riethoven J. J. et al . (2022). Natural variation in root exudation of GABA and DIMBOA impacts the maize root endosphere and rhizosphere microbiomes. J. Exp. Bot.73, 5052–5066. doi: 10.1093/jxb/erac202
98
Werner D. Newton W . (2005). Nitrogen fixation in agriculture, forestry, ecology, and the environment. Springer, Dordrecht, Netherlands.
99
Wickham H. (2016). ggplot2: elegant graphics for data analysis (Springer-Verlag New York). Available online at: https://ggplot2.tidyverse.org (Accessed August 20, 2025).
100
Wickham H. Hester J. Bryan J. (2024). readr: Read Rectangular Text Data. R package version 2.1.5. Available online at: https://github.com/tidyverse/readrhttps://readr.tidyverse.org. (Accessed August 20, 2025).
101
Wintermans P. C. A. Bakker P. Pieterse C. M. (2016). Natural genetic variation in Arabidopsis for responsiveness to plant growth-promoting rhizobacteria. Plant Mol. Biol.90, 623–634. doi: 10.1007/s11103-016-0442-2
102
Woli K. P. Sawyer J. E. Boyer M. J. Abendroth L. J. Elmore R. W. (2018). Corn era hybrid macronutrient and dry matter accumulation in plant components. Agron. J.110, 1648–1658. doi: 10.2134/agronj2018.01.0025
103
Yassue R. M. Carvalho H. F. Gevartosky R. Sabadin F. Souza P. H. Bonatelli M. L. et al . (2021). On the genetic architecture in a public tropical maize panel of the symbiosis between corn and plant growth-promoting bacteria aiming to improve plant resilience. Mol. Breed.41, 63. doi: 10.1007/s11032-021-01257-6
104
Zhang G. B. Yi H. Y. Gong J. M. (2014). The Arabidopsis ethylene/jasmonic acid-NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell26, 3984–3998. doi: 10.1105/tpc.114.129296
105
Zhang W. J. Zhang T. Zhang J. Lei W. W. Zhao L. Wang S. et al . (2023). Low nitrogen stress promotes root nitrogen uptake and assimilation in strawberry: contribution of hormone networks. Horticulturae9, 249. doi: 10.3390/horticulturae9020249
106
Zhou J. Y. Li X. Zheng J. Y. Dai C. C. (2016). Volatiles released by endophytic Pseudomonas fluorescens promoting the growth and volatile oil accumulation in. Plant Physiol. Bioch.101, 132–140. doi: 10.1016/j.plaphy.2016.01.026
Summary
Keywords
nitrogen-use efficiency, plant-growth-promoting bacteria, rhizosphere, maize genotypes, bacterial inoculation, biostimulants
Citation
Foster LR, Yang J, Riethoven J-JM, Mukhtar H and Schachtman DP (2025) Inoculation frequency and maize genotype influence plant growth-promoting effects of soil bacteria under low nitrogen conditions. Front. Plant Sci. 16:1637156. doi: 10.3389/fpls.2025.1637156
Received
28 May 2025
Accepted
06 August 2025
Published
12 September 2025
Volume
16 - 2025
Edited by
Lénia Rodrigues, University of Évora, Portugal
Reviewed by
Fred Below, University of Illinois at Urbana-Champaign, United States
Mario Lira Junior, Federal Rural University of Pernambuco, Brazil
Updates
Copyright
© 2025 Foster, Yang, Riethoven, Mukhtar and Schachtman.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel P. Schachtman, Daniel.schachtman@unl.edu
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.