- 1Agriculture and Agri-Food Canada – Saskatoon Research and Development Center, Saskatoon, SK, Canada
- 2Global Institute for Food Security, Saskatoon, SK, Canada
Winter camelina (Camelina sativa) is a climate-resilient oilseed crop that has received attention as a feedstock crop for advanced, low-carbon-intensity biofuels. Breeding programs working on winter camelina improvement have to contend with heterogeneous germplasm, oftentimes erroneously identified as winter biotypes, and a gene pool that is much smaller than that of spring-type camelina, the latter having motivated crosses between winter and spring biotypes. For the unequivocal differentiation of winter from spring types at an early stage, breeders require a tool to track the vernalization requirement trait in segregating breeding populations as well as in putative winter cultivars, breeding lines, and accessions to be used as parental lines. Linkage mapping in a winter (‘Joelle’) × spring (‘SES0787LS’) C. sativa biparental F2 population identified two major quantitative trait loci (QTLs) for vernalization requirement on chromosomes 8 and 13. Both regions contained orthologs of Flowering Locus C (FLC), a gene known to have a significant effect on flowering time and vernalization requirement in plants. Based on the FLC gene sequences, allele-specific PCR-based markers were developed, suitable for the routine screening of C. sativa germplasm for the presence of the winter and spring alleles of all three C. sativa FLC orthologs, including a chromosome 20 locus. The analysis of the winter cultivar ‘Joelle’ and a diverse C. sativa germplasm panel uncovered greater than expected variability for the FLC alleles, with most lines possessing several different allele combinations and still undergoing genetic segregation. Contrary to previous reports, spring camelina lines can carry the spring and/or winter alleles of Csa.FLC.C20, indicating that this gene by itself only plays a subordinate role in the regulation of flowering and vernalization requirement. In winter C. sativa germplasm, combinations of Csa.FLC.C08 winter alleles with the winter alleles of one or both of Csa.FLC.C13 and Csa.FLC.C20 result in vernalization requirement, while winter Csa.FLC.C08 by itself leads to a semi-winter type. The results of this study and the tools developed herein are a first step to orchestrating the genes underlying vernalization requirement in C. sativa and developing winter camelina cultivars optimized for different winter environments.
1 Introduction
Camelina (Camelina sativa [L.] Crantz) is a short-season crucifer oilseed adapted to the temperate and continental climates of the mid-latitudes (Weiss et al., 2024). It has been shown to have potential as a low maintenance crop in North America (Blackshaw et al., 2011; Enjalbert et al., 2013; Gugel and Falk, 2006; Plessers et al., 1962; Robinson, 1987), Europe (Vollmann et al., 2007; Zanetti et al., 2017, 2021, 2024), South America (Berti et al., 2011; Solis et al., 2013), China (Gao et al., 2022; Zhang et al., 2021), and Russia (Kon’kova et al., 2021). Camelina has a number of favorable agronomic characteristics, including frost and drought tolerance (Angelini et al., 1997; Bonjean and Goffic, 1999; French et al., 2009; Hunsaker et al., 2011; Putnam et al., 1993), resistance to insect pests (Soroka et al., 2015) and diseases (reviewed in Séguin-Swartz et al., 2009), and good performance on economically marginal lands (Johnson et al., 2019; Putnam et al., 1993; Robinson, 1987). It also possesses a seed oil that has the unusual property of being both rich in unsaturated fatty acids—primarily linolenic acid (C18:3) (Budin et al., 1995; Vollmann et al., 2007; Zubr and Matthäus, 2002)—and high in antioxidants (Abramovič and Abram, 2005). Because of these characteristics, camelina has received attention for diametrically opposed applications: as a feedstock crop for advanced biofuels (Fröhlich and Rice, 2005; Shonnard et al., 2010; Wu and Leung, 2011) and as a source of healthy oil for food, feed, and nutraceutical applications (Hixson et al., 2014; Kirkhus et al., 2013; Ngo et al., 2023; Ratusz et al., 2018).
There are both spring and winter annual biotypes (Mirek, 1980; Plessers et al., 1962; Putnam et al., 1993; Zubr, 1997). Winter cultivars, which are usually seeded in the fall, possess exceptional cold hardiness and typically survive the winters in the northern USA and Canada (Gesch and Cermak, 2011; Gesch et al., 2018; Horvath et al., 2019). In the USA, winter camelina has been extensively studied for use in double- and relay-cropping systems in combination with short-season summer crops as a means to produce a biofuel feedstock crop without devoting land traditionally used for food production (Berti et al., 2015, 2017; Gesch et al., 2014; Gesch and Archer, 2013; Gesch and Johnson, 2015; Johnson et al., 2017; Sindelar et al., 2017). Winter camelina is also the only winter oilseed that can be grown on the Canadian Prairies and, together with winter cereals, allows for the establishment of true winter crop rotations. Growing winter camelina, like other crops that cover the soil over the winter months, provides environmental benefits, including the prevention of soil erosion (Lal et al., 1991) and the uptake of excess nitrogen (Staver and Brinsfield, 1998). As one of the first plants to flower in the spring, winter camelina also offers early-season feed to pollinators (Eberle et al., 2015).
To avoid flowering before the onset of winter, winter annuals have evolved to require exposure to low temperatures for several weeks, known as vernalization, to transition from vegetative to reproductive growth (Sheldon et al., 2000; Kiefer et al., 2017). In Brassicaceae, a major regulatory gene in the vernalization pathway is FLOWERING LOCUS C (FLC) (Michaels and Amasino, 1999; Swiezewski et al., 2009), a MADS-box transcription factor that under ambient temperatures suppresses the expression of the floral integrators FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) (Deng et al., 2011; Helliwell et al., 2006; Michaels and Amasino, 1999; Samach et al., 2000; Searle et al., 2006), which prevents the progression of the apical meristem from vegetative to floral development (Henderson and Dean, 2004). Vernalization brings about epigenetic silencing of FLC; in consequence, repression of FT and SOC1 is reduced, and the apical meristem transitions to produce reproductive structures (Anderson et al., 2018; Bastow et al., 2004; Sheldon et al., 2000; Schiessl et al., 2019; Takada et al., 2019). As the role of functional FLC alleles is to prevent flowering, non-functional alleles, or flc null mutants, are unresponsive to cold and result in early and vernalization-independent flowering (Michaels and Amasino, 1999). In the hexaploid genome of C. sativa, there are three orthologous copies of the FLC gene on chromosomes 8, 13, and 20 (Kagale et al., 2014). The importance of FLC for flowering time in general and vernalization requirement in camelina species has been demonstrated by quantitative trait locus (QTL) analyses, which showed that QTLs co-localized with FLC in both spring (Li et al., 2021; Lily et al., 2021) and spring × winter inter- and intra-specific mapping populations (Chaudhary et al., 2023; Kandel et al., 2024).
With a winter camelina germplasm pool that is much smaller than that of spring camelina, some breeders are resorting to winter × spring crosses to increase the genetic diversity of the winter material and to transfer traits from spring cultivars to winter breeding lines. Others have proposed winter × spring crosses to develop winter cultivars that mature earlier than current commercial varieties, which is desirable for double and relay cropping (Kandel et al., 2024). However, phenotypic differentiation between winter- and spring-type progeny is difficult, particularly when populations are grown with a vernalization period. Thus, time to flowering after vernalization is not a reliable indicator for whether a plant should be categorized as a spring, semi-winter, or winter type. Additionally, winter-type accessions deposited at gene banks are often highly variable in their expression of the winter phenotype, with many being admixtures of winter-, semi-winter-, and even spring-type plants (Chao et al., 2019). For the unequivocal differentiation of winter from spring types, breeders require a tool that affords the ability to track the vernalization requirement trait in segregating breeding populations as well as in putative winter cultivars, breeding lines, and accessions to be used as parental lines.
In this study, we report on the identification of QTLs and the development of widely applicable PCR-based markers linked to the candidate genes controlling flowering time and vernalization requirement in a winter × spring biparental population. Furthermore, we uncover an unexpected degree of genetic variation at the FLC loci in both spring- and winter-type C. sativa germplasm and propose that segregation still exists within many of the publicly available winter camelina accessions and cultivars.
2 Materials and methods
2.1 Plant materials and phenotyping for flowering behavior
2.1.1 F2 population development
Two F2 populations were developed by manual reciprocal crossing of the winter camelina cultivar ‘Joelle’ with the spring camelina cultivar ‘SES0787LS’ in 2018 and subsequent selfing of F1 plants. Seed of ‘Joelle’, a publicly available cultivar developed at Limagrain (Saint-Beauzire, France), was obtained from Dr. Russ Gesch, USDA-ARS (Morris, MN, USA). ‘SES0787LS’ was provided by Smart Earth Camelina Corporation (Saskatoon, Canada). The cross where ‘Joelle’ was used as the female and ‘SES0787LS’ was the pollen donor gave rise to F2 population 19CS1178-F2, while the F2 population 19CS1179-F2 was derived from the reciprocal cross. For crossing, closed mature buds of the female parent were manually opened, the anthers removed, and the stigma pollinated using pollen from the male parent. ‘Joelle’ plants and semi-winter-type F1 plants were placed in vernalization at 5°C for 35 days after 2 weeks in the greenhouse before being moved back to the greenhouse to induce flowering. The F2 populations were originally developed as breeding populations and are therefore composed of bulked seed derived from several F1 plants that were generated by crossing several plants of the parental genotypes.
For phenotyping, in the winter of 2020/2021, a total of 427 F2 plants (216 from 19C1178-F2 and 211 from 19CS1179-F2) plus 10 plants each of the parent lines ‘Joelle’ and ‘SES0787LS’ as well as 10 plants each of the reciprocal F1 hybrids (19CS1178 and 19CS1179) were grown in the greenhouse at 20°C/17°C day/night with a light/dark cycle of 16/8 h. Three seeds were sown in individual pots containing soilless potting mix (Stringam, 1971) amended with the controlled release fertilizer 15-9–12 Osmocote PLUS (Scotts Miracle-Gro Company, Marysville, Ohio). One week after germination, seedlings were thinned to one per pot. Plants remained in the greenhouse until the opening of the first flower, at which point days to flowering (DTF) was recorded. After 170 days, plants that had not flowered yet were assigned a DTF value of 170.
2.1.2 Diverse winter- and spring-type germplasm
The diverse camelina germplasm panel consisted of 13 winter C. sativa cultivars and accessions; 53 spring C. sativa cultivars, breeding lines, and accessions; and three Camelina microcarpa genotypes (Supplementary Table 1). The latter included one tetraploid C. microcarpa (syn. Camelina intermedia) line and one each of Type 1 and Type 2 hexaploid C. microcarpa genotypes. All lines were grown in the greenhouse without vernalization, and flowering behavior was observed. For marker analysis, DNA of leaf tissue from four plants was pooled at the six-to-eight– leaf stage.
2.2 Quantitative trait locus mapping
2.2.1 Genotyping by sequencing
Genomic DNA of F2 plants was extracted from young leaf tissue utilizing a modified sodium dodecyl sulfate method (Somers et al., 1998). A genotyping-by-sequencing (GBS) library was constructed for both the F2 mapping populations and the diverse germplasm panel following the protocol described by Fu et al. (2021), with modifications. In brief, 200 ng of DNA from each line was digested with the restriction enzymes PstI and MspI. Samples were then ligated to common adapters and size-selected via an AMPure XP (Beckman Coulter, Indianapolis, Indiana) bead cleanup. Subsequently, for each sample, 20 μL of the resulting ligation mix was added to 1× KAPA Fidelity buffer (Roche, Indianapolis, Indiana), 250 μM of KAPA dNTPs, 0.5 U KAPA HiFi HotStart DNA polymerase, and 0.5 μM of a unique NEBNext UDI (Unique Dual Index) Primer (NEB, Ipswich, Massachusetts) in a total reaction volume of 50 μL. The ensuing PCR program consisted of an initial denaturation at 98°C for 30 s, 14 cycles of denaturation at 98°C for 10 s and annealing/extension at 65°C for 75 s, and a final extension at 65°C for 5 min. All PCR samples were quantified using a Quant-iT assay (Thermo Fisher Scientific, Waltham, Massachusetts), and four samples of equal quantity were combined. The combined samples were concentrated using a Zymo DNA Clean and Concentrator-5 kit (CedarLane Labs, Burlington, Ontario) following the provided protocol. Samples were again combined, and a second AMPure XP size selection bead cleanup was completed. The final size-selected samples were again quantified with a Quant-iT PicoGreen assay, and equal amounts of each were used to create one library pool. The library was sequenced using one lane of the Illumina NovaSeq 6000 with the SE-150 sequencing protocol at the Genome Quebec Innovation Centre, Montreal, Quebec.
2.2.2 Genetic analysis of segregating populations
Sequences were de-multiplexed and trimmed of low-quality bases and adapters using Trimmomatic version 0.32 (Bolger et al., 2014) with a minimum read length of 75 bp for retention. All high-quality reads were mapped to the C. sativa reference genome (Kagale et al., 2014) using bowtie2 version 2.4.1 (Langmead and Salzberg, 2012) and SAMtools version 1.15.1 (Danecek et al., 2021), with default parameters. Subsequently, the aligned binary alignment map files were used to call single-nucleotide polymorphisms (SNPs) using the BCFtools mpileup tool (Danecek et al., 2021). The same process was followed to analyze individual ‘Joelle’ lines utilizing three separate ‘Joelle’ reference genomes [NCBI: GCA_036769185.1; DOE-JGI Phytozome Joelle v1.1 (https://phytozome.jgi.doe.gov/info/CsativaJoelle_v1_1) and the AAFC Joelle reference (unpublished)].
The genetic maps for the F2 populations were constructed using JoinMap 4.0 (Van Ooijen, 2006). Linkage groups were generated with a minimum logarithm of the odds (LOD) threshold of 5.0. The regression mapping algorithm and Kosambi mapping function were utilized to develop mapped linkage groups. QTL analysis for DTF was performed using the MQM mapping method of MapQTL 6.0 (Van Ooijen, 2009). A permutation test (10,000 permutations, 95% confidence level, mapping step size 1.0) was performed to determine the significant LOD threshold for DTF.
2.3 Marker development
Utilizing whole-genome sequences of two hexaploid and one tetraploid C. microcarpa lines and 15 spring C. sativa lines (Parkin et al., unpublished) (Supplementary Table 1), the coding region for all three FLC genes and surrounding up- and downstream sequences were isolated (Supplementary Tables 4-6). Public reference genome sequences from spring-type C. sativa lines DH55 (Kagale et al., 2014) and ‘CO46’ (GCA_036971115.1), as well as from the winter-type C. sativa cultivar ‘Joelle’ [GCA_036769185.1; DOE-JGI Phytozome Joelle v1.1 (https://phytozome.jgi.doe.gov/info/CsativaJoelle_v1_1) and AAFC Joelle reference (unpublished)], were also included in the analysis. Sequences were aligned using the EMBL-EBI online tool MUSCLE (Madeira et al., 2022). A selective Kompetitive Allele-Specific PCR (KASP) primer for the Csa.FLC.C08 allele was developed based on a SNP located 700 bp upstream of the start codon. The KASP primer that was first developed for Csa.FLC.C13 was found to be segregating in individual ‘Joelle’ plants. Because of this, several Csa.FLC.C13-specific primers were developed, and Sanger sequencing was carried out to determine the ‘Joelle’ Csa.FLC.C13 allele sequence. Subsequently, a selective KASP primer for Csa.FLC.C13 was developed based on a SNP located in the intron 637 bp before the start of exon 2. In addition, utilizing the above-mentioned whole-genome sequences and the INDEL present in exon 5 identified by Anderson et al. (2018), KASP primers were developed for Csa.FLC.C20. All primers are listed in Supplementary Table 2. Each KASP reaction contained 50 ng of genomic DNA, 4.0 μL of KASP 2× Master Mix (LGC Genomics, St. Alexandria, Minnesota), and 0.11 μL of primer assay mix in a total volume of 8.0 μL. All amplifications were performed in a CFX96 Real-Time Thermal Cycler (Bio-Rad Laboratories, Hercules, California) using the touchdown PCR protocol recommended by the manufacturer. To analyze potential heterogeneity within ‘Joelle’, 310 individual plants were tested using the FLC markers described above. Only ‘Joelle’ plants that were homozygous at all loci for the respective spring or winter alleles were kept, and self-pollination was performed using selfing bags. All diverse lines listed in Supplementary Table 1 were also examined using the FLC KASP markers.
2.4 Field experiments
In the spring of 2021, seeds of 19CS1178-F2 and 19CS1179-F2 were planted in a field trial at the AAFC Saskatoon Research Farm. Each reciprocal population was seeded in two replicates of four 20-ft. rows at a low seeding rate. Additionally, one 20-ft. row each of parent lines ‘SES0787LS’ and ‘Joelle’ was seeded in each replicate. At the rosette stage, plants of each F2 population were thinned to 125 lines per replicate. Both ‘SES0787LS’ and ‘Joelle’ were thinned to 10 plants per row. Numbered marker flags were placed next to every fifth plant to keep track of plant numbers. Leaf tissue samples were taken at the rosette stage. For each plant, the date when the first flower opened was recorded, and days to flowering calculated. After flowering, plants were cut at ground level.
3 Results
3.1 Population development and phenotyping for flowering behavior
All 427 individual F2 plants from the ‘Joelle’ × ‘SES0787LS’ (and reciprocal) cross, as well as parental lines and F1 hybrids, were evaluated for DTF under greenhouse conditions without vernalization. Plants were evaluated daily and marked as having flowered when the first open flower was detected. Plants that had not flowered after 170 days were classified as true winter types. The reciprocal crosses produced semi-winter F1 hybrids with average DTF of 59 (19CS1178) and 55 (19CS1179) days, compared to an average of 35 days for the spring-type parent, ‘SES0787LS’. There was no statistically significant difference between the DTF for the reciprocal F1 hybrids, indicating that flowering time and vernalization requirement were not influenced by maternal genetic effects.
The reciprocal populations showed a similar frequency distribution for DTF, with most of the plants exhibiting delayed flowering compared to the spring-type parent, ranging from 40 to 80 days after seeding. Both F2 populations had a mean DTF of 71 days (Figures 1A, B). Furthermore, 22% and 20% of the plants of 19CS1178-F2 and 19CS1179-F2, respectively, did not flower after 170 days and were classified as true winter types. Only two plants flowered slightly earlier than 35 days after seeding, the average DTF for the spring-type parent.
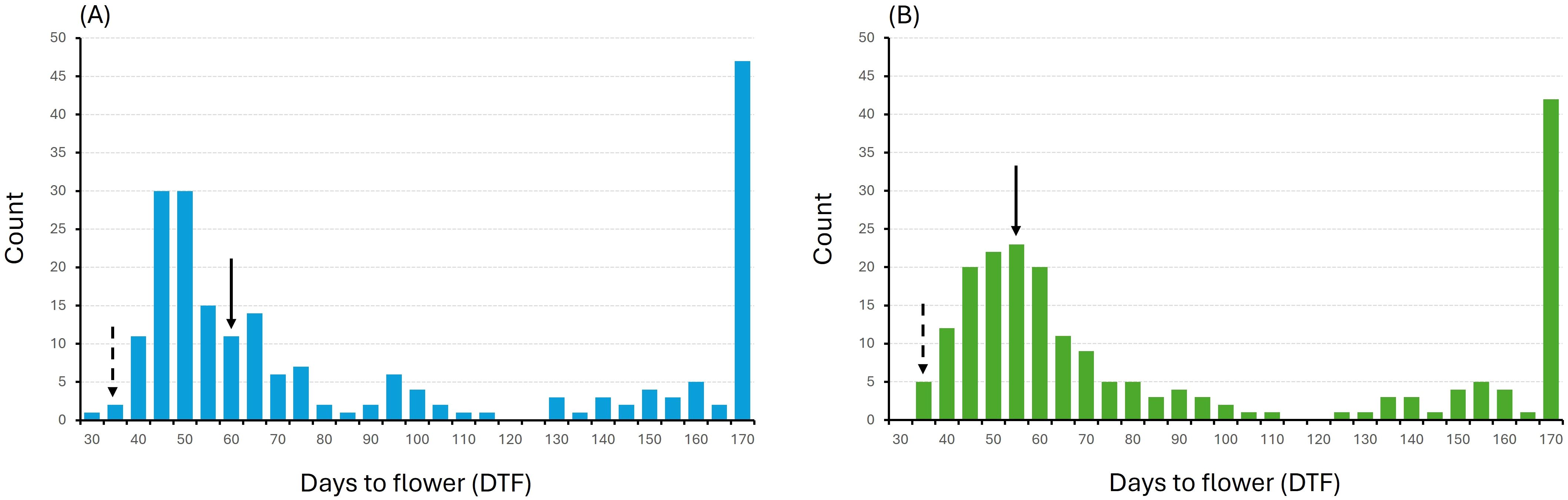
Figure 1. Frequency distribution of days to flowering (DTF) for F2 populations (A) 19CS1178-F2 (derived from ‘Joelle’ × ‘SES0787LS’) and (B) 19CS1179-F2 (derived from ‘SES0787LS’ × ‘Joelle’). Lines that did not flower after 170 days were assigned a DTF value of 170. Solid arrows point to the average DTF of F1 hybrids; dotted arrows point to average DTF of spring type parent, ‘SES0787LS’.
Segregation was detected within many of the winter lines (Figure 2) of the diverse germplasm collection; plants ranged from true-breeding winter types, where none of the plants flowered without vernalization, to spring types and various stages in between, with plants displaying different degrees of delayed flowering.

Figure 2. Camelina sativa accessions, 44 days after seeding. (A) CN113691, a mix of true winter types and plants with delayed flowering without vernalization. (B) CN113660, a mix of true winter types and spring types. (C) CN113668, a semi-winter type, with delayed flowering compared to typical spring-type plants, as shown on the left of (B).
3.2 QTL mapping
A genetic map was developed utilizing GBS data from a total of 240 randomly selected F2 plants (118 from 19CS1178-F2 and 122 from 19CS1179-F2). A total of 54,788 SNPs were detected. To ensure the accurate calling of heterozygous SNPs, extra caution was taken during SNP selection; those with more than 5% missing data or distorted segregation were removed. After filtering, a total of 1,252 SNPs were used to develop a linkage map with a total length of 1,354.4 cM (Figure 3). The average mapping interval was 1.10 cM, and the number of markers per linkage group ranged from 40 on chromosome 12 to 107 on chromosome 11 (Figure 3, Supplementary Table 3). A QTL analysis of DTF in the absence of vernalization identified two significant QTLs: the first, located on Chr13 (LOD = 12.57), explained 21.4% of the variation and peaked between the Chr13–1720684 and Chr13–4293244 SNP markers. The second QTL was located on Chr8 (LOD = 7.99), explained 14.2% of the phenotypic variance, and peaked between the Chr8–23073155 and Chr8–23387904 SNP markers. FLC gene-specific markers (described in detail below) were also mapped and used to further hone the QTL analyses. An apparent double recombination event that was ~2.0 Mb in size and contained Csa.FLC.C13 was detected. The ‘Joelle’ parental line was determined to be heterozygous in this region, and SNPs with distorted segregation were added back to the map to allow for the complete coverage of the region. The spring parental line ‘SES0787LS’ contained the winter Csa.FLC.C20 allele, and the ‘Joelle’ parental line used in the development of the F2 population was also identified to be heterozygous for the Csa.FLC.C20 marker; thus, markers in this region were treated as dominant, and those with distorted segregation were added back to the map to allow for the complete coverage of this region. A second QTL analysis with the updated map identified the same two significant QTLs, and each was associated with one of the FLC gene-specific markers. The first, located on Chr13 (LOD = 12.87), explained 21.9% of the variation and peaked between the FLC-13_KASP and Chr13–4293244 SNP markers. The second QTL was located on Chr8 (LOD = 8.07), explained 14.3% of the phenotypic variance, and peaked directly with the FLC-8_KASP marker. Two minor QTLs were also detected in both analyses: on Chr7 (LOD = 4.29), which explained 7.9% of the variation and peaked between the Chr7–20823349 and Chr7–20905187 markers, and on Chr16 (LOD = 3.75), which explained 6.9% and with the maximum peak between the Chr16–21827277 and Chr16–21959032 markers.
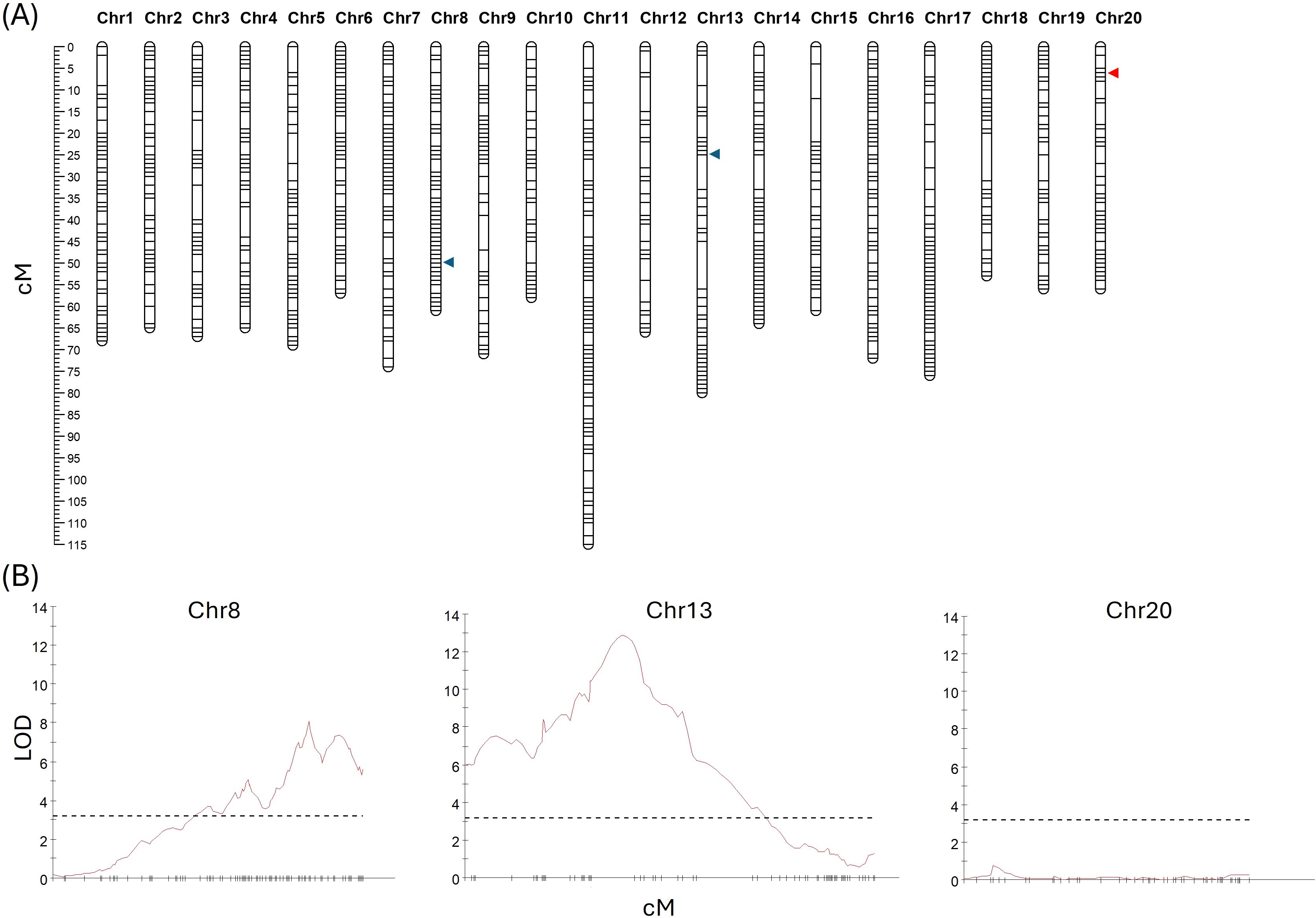
Figure 3. (A) Genetic linkage map derived from populations 19CS1178-F2 and 19CS1179-F2, with the position of quantitative trait loci (QTLs) associated with vernalization requirement (blue arrowheads) on chromosomes 8 and 13. The position of Csa.FLC.20 is marked with a red arrowhead. The distance in cM is shown on the left side. (B) QTL traces for chromosomes 8, 13, and 20. The dashed line is at 3.2 and represents the significant LOD threshold.
3.3 Marker development
FLC gene sequences were isolated from the whole-genome sequence of three C. microcarpa (winter type) and 15 spring C. sativa lines (Parkin, unpublished), as well as the DH55 reference genome (Kagale et al., 2014) and three winter-type cultivar ‘Joelle’ reference genomes [NCBI: GCA_036769185.1; DOE-JGI Phytozome Joelle v1.1; (https://phytozome.jgi.doe.gov/info/CsativaJoelle_v1_1) and AAFC ‘Joelle’ reference (unpublished)], to develop markers suitable for routine screening. The FLC.C08 sequence alignment (Supplementary Table 4) revealed an A/G SNP 700 bp upstream of the start codon. This SNP was converted to a KASP marker (Figure 4; Supplementary Table 4). The alignment of the FLC.C13 genes identified a large insert in intron 1 in all spring-type alleles (Figure 5; Supplementary Table 5). The initial KASP primer pair differentiated spring and winter genotypes, but in a sample of 10 individual ‘Joelle’ plants, segregation for winter and spring Csa.FLC.C13 alleles was detected. Two representative ‘Joelle’ plants, one with the winter and one with the spring Csa.FLC.C13 allele, were Sanger sequenced with several pairs of FLC.C13-specific primers to analyze the region from 600 bp upstream of the start codon to the end of exon 6. The results confirmed that both the spring and winter Csa.FLC.C13 alleles were present in ‘Joelle’. Aligning these sequences with those of the C. microcarpa lines resulted in the identification of 26 SNPs and 13 INDELs between the spring and winter alleles of FLC.C13. Interestingly, the hexaploid C. microcarpa line CN119205, which is a winter type, had all the SNPs and INDELs common to the spring-type lines but did not have the above-mentioned large insert in intron 1. A G/T SNP was identified 637 bp before the start of exon 2 in FLC.C13, and KASP primers were developed (Figures 4, 5; Supplementary Table 5). For FLC.C20, KASP primers were developed based on the previously identified single-base-pair INDEL in exon 5 (Anderson et al., 2018), which was confirmed with the extended sequence alignment (Figures 4, 5; Supplementary Table 6).
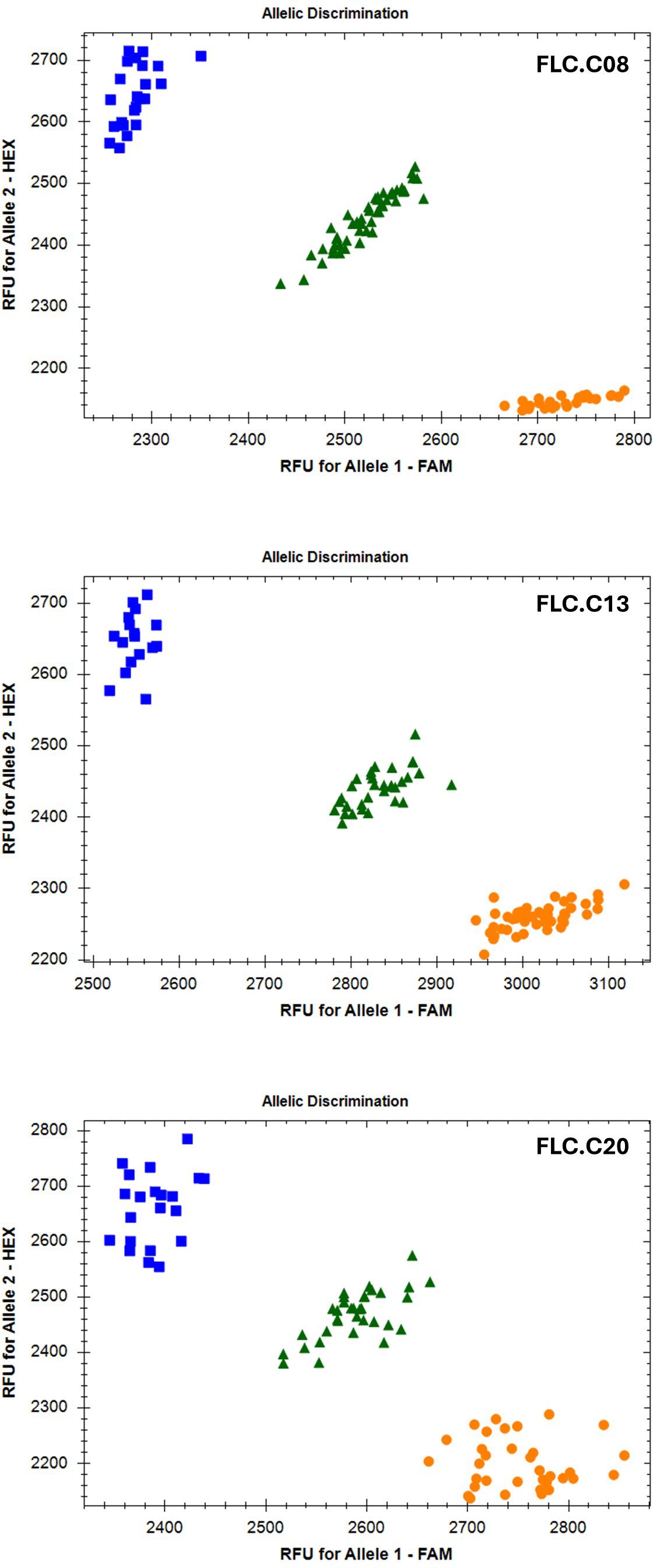
Figure 4. Bio-Rad CFX Maestro images of the Kompetitive Allele-Specific PCR (KASP) markers for the three FLC orthologs. Orange circles represent samples homozygous for the A1 allele, blue squares represent samples homozygous for the A2 allele, and green triangles represent heterozygous samples.
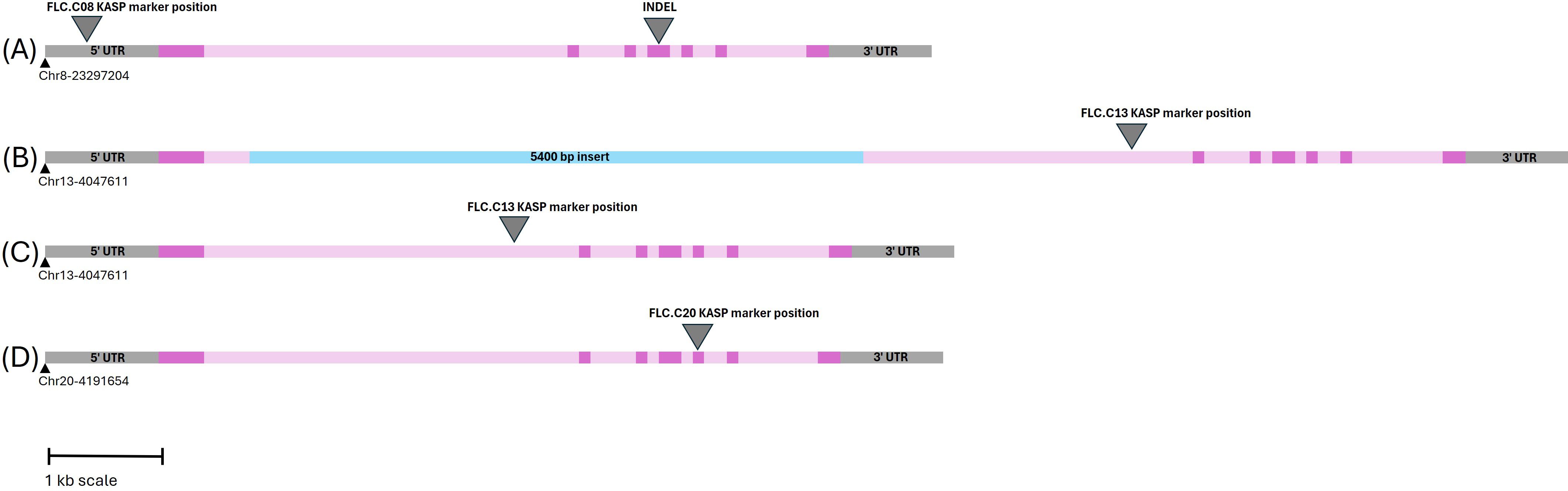
Figure 5. Schematic representation of the three Csa.FLC orthologs. Exons and introns are shown in dark and light pink, respectively. Base pair positions are based on the DH55 reference genome. (A) Csa.FLC.C08. The arrowhead on the left side points to the Kompetitive Allele-Specific PCR (KASP) marker position; the arrowhead on the right side points to the 3-bp INDEL that distinguishes the spring allele from the winter allele. (B) Spring allele of Csa.FLC.C13. The insert in intron 1 is shown in blue. (C) Winter allele of Csa.FLC.C13. The arrowhead in both panels B and C points to the KASP marker position. (D) Csa.FLC.C20. The arrowhead points to 1-bp INDEL that distinguishes the spring allele from the winter allele and serves as the KASP marker position.
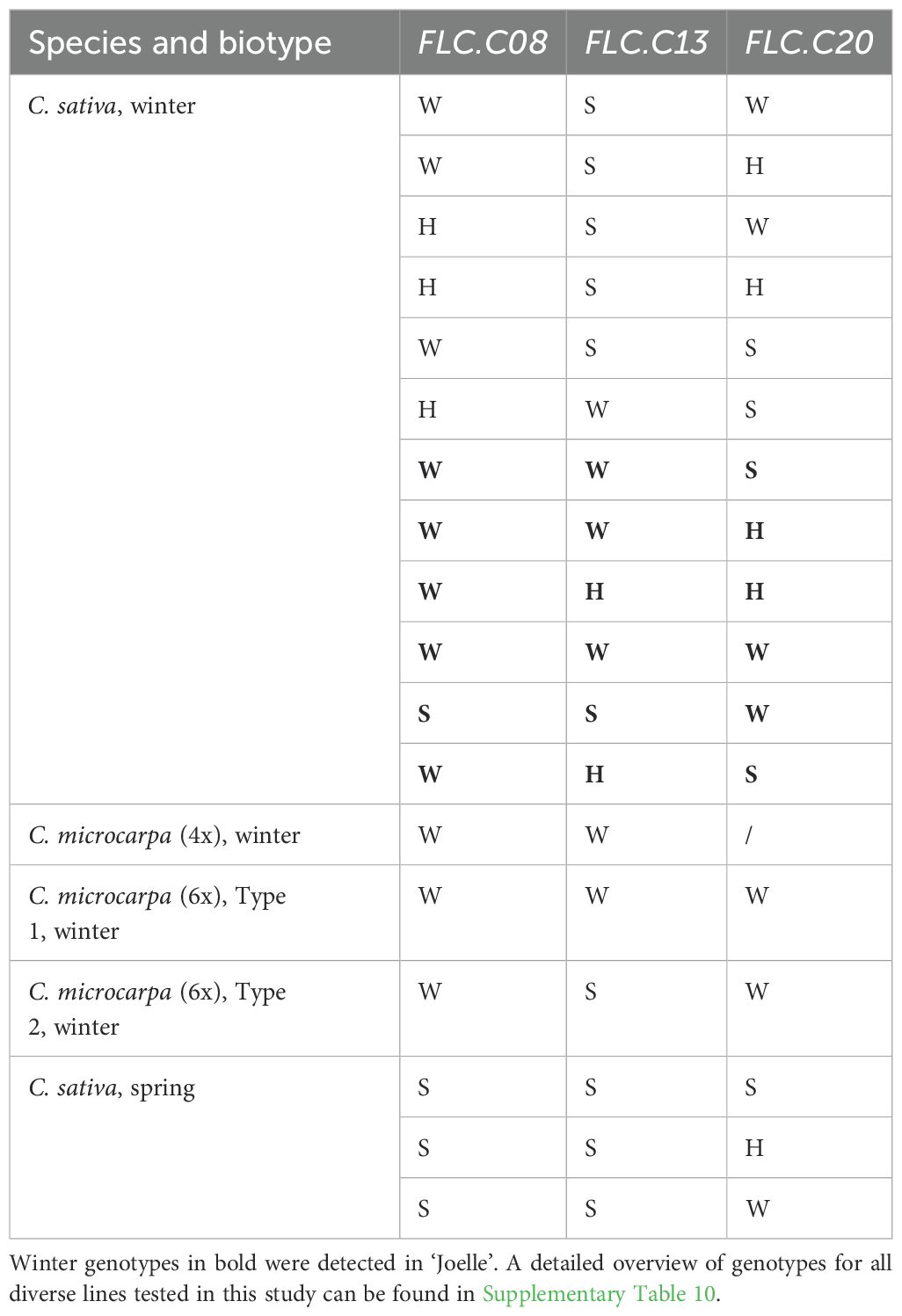
Because of the noted variation at the Csa.FLC.C13 locus, the variation of each of the FLC alleles present in ‘Joelle’ was examined by genotyping 310 individual plants (Table 1). Ten different genotypes were isolated; 130 plants had winter Csa.FLC.C08 and Csa.FLC.C20 alleles and a spring Csa.FLC.C13 allele, while 101 plants had winter Csa.FLC.C08 and Csa.FLC.C13 alleles and a spring Csa.FLC.C20 allele. Interestingly, only 30 plants, or just under 10%, were homozygous for all three winter Csa.FLC alleles. Plants that were homozygous for the Csa.FLC.C08 winter allele and the spring Csa.FLC.C13 and Csa.FLC.C20 alleles (29) were kept in the greenhouse without vernalization, and while all flowered, they were significantly delayed (>60 DTF). At least one heterozygous allele was identified in 5.8% of the samples, suggesting that some level of segregation is still occurring within this seed source.
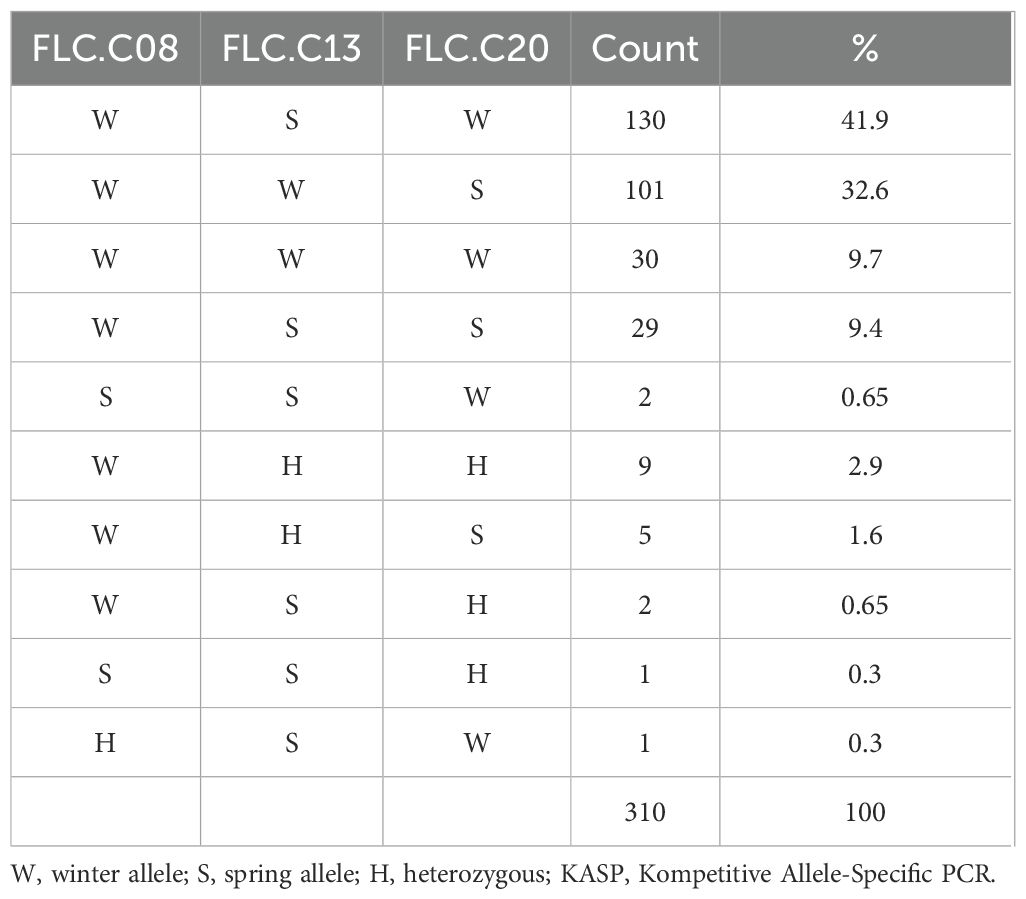
Table 1. Summary of KASP marker results for Flowering Locus C genes Csa.FLC.C08, Csa.FLC.C13, and Csa.FLC.C20 for 310 individual plants of winter camelina variety ‘Joelle’.
A GBS analysis was carried out on the 10 ‘Joelle’ lines, which were used for KASP marker optimization to determine the overall extent of variability within ‘Joelle’. All sequences were aligned to the DH55 reference genome. All SNPs with a quality score below 30 and more than 10% missing data were removed, as well as SNPs that were polymorphic only between DH55 and ‘Joelle’. After filtering, 79,740 SNPs, including 3,265 INDELs, were detected solely among the ‘Joelle’ lines (Supplementary Table 7). Two of these lines (Joelle.1 and Joelle.7) were heterozygous for both the Csa.FLC.C13 and Csa.FLC.C20 alleles. The overall percentage of heterozygous SNPs was also higher in these two lines compared to the remaining ‘Joelle’ plants, suggesting that segregation still exists within the ‘Joelle’ genome. The GBS data were also aligned to the three ‘Joelle’ reference genomes (NCBI, DOE-JGI, and AAFC). Special interest was paid to the regions of Chr13 and Chr20, which contained FLC alleles. The Chr13 double recombination event and the associated distorted segregation present in the genetic map were confirmed through the identification of a region of ~750,000 bp that was found to be segregating between individual ‘Joelle’ lines (Supplementary Table 8). The ‘Joelle’ individuals with the spring Csa.FLC.C13 allele matched the NCBI and DOE-JGI reference genomes across the entire region, while the ‘Joelle’ lines with the winter Csa.FLC.C13 allele matched the AAFC reference genome. The two ‘Joelle’ lines, which had both Csa.FLC.C13 alleles, were heterozygous for this entire region when compared to all three reference genomes (Supplementary Table 8). A similar pattern was detected in the Csa.FLC.C20 region; a region of ~1.1 Mb was identified on Chr20, which corresponds to a similar double recombination event in the genetic map (Supplementary Table 9). For Csa.FLC.C20, both the NCBI and DOE-JGI reference genomes had the winter allele, while the AAFC reference genome had the spring allele. These results agree with the Csa.FLC.C20 KASP scores, indicating that three of the ‘Joelle’ lines were heterozygous for this allele.
For markers to be useful in a breeding program, they must be polymorphic in a diverse range of breeding lines and gene bank accessions. To this end, 13 winter C. sativa, three winter-type C. microcarpa, and 53 spring C. sativa lines were assessed using the FLC-specific markers (Table 2; Supplementary Table 10). For four of the winter C. sativa accessions, two different seed batches were investigated. To ensure a representative sample, four individual plants were bulked for this analysis, except for the parental lines of the population, ‘Joelle’ and ‘SES0787LS’, for which 10 individual plants were tested. The reference genome marker scores were determined based on the SNPs in the actual sequence. All spring lines contained the spring alleles for Csa.FLC.C08 and Csa.FLC.C13, but variation was detected for the Csa.FLC.C20 INDEL, first identified by Anderson et al. (2018). Thus, 13 lines carried the winter allele, and eight were heterozygous at this locus. For the winter types, segregation was detected in many lines; only ‘Bison’, BSX, BSX-WG1, and the Type 1 hexaploid C. microcarpa accession were homozygous for either spring or winter alleles at all three FLC loci. The remainder of the lines was segregated for at least one FLC ortholog. All winter lines carried the FLC.C08 winter allele, and the FLC.C20 winter allele was present in most, except in CN113692 and CN113668. CN113692 had the FLC.C13 winter allele, as did the tetraploid and Type 1 hexaploid C. microcarpa lines, as was documented above for some of the ‘Joelle’ plants. CN113668 only carried the winter Csa.FLC.C08 gene and showed delayed flowering without vernalization.
3.4 Marker validation under field conditions
To assess the utility of the FLC markers under field conditions, 233 F2 individuals from the ‘Joelle’ × ‘SES0787LS’ cross were planted in Saskatoon, Canada, in the spring. Tissue was collected, and the lines were tested with the FLC allele-specific markers (Table 3). Days to flowering was recorded until Sept. 1; plants that had not flowered by this date were labelled as true winter types. From the population, 27 plants were classified as true winter types, and 15 lines died for unknown reasons before the flowering date could be assessed. The average DTF for the population was 51 days. Based on segregation at the three FLC loci, all 27 genotypes previously observed in the greenhouse experiment were detected, and as expected, the average DTF increased in lines with a greater number of winter FLC alleles. Plants with all three winter FLC alleles took twice as long to flower on average (mean DTF = 72) than plants with three spring FLC alleles (mean DTF = 36). Half of the plants with all three winter alleles did not flower by the end of the season and were considered true winter types.
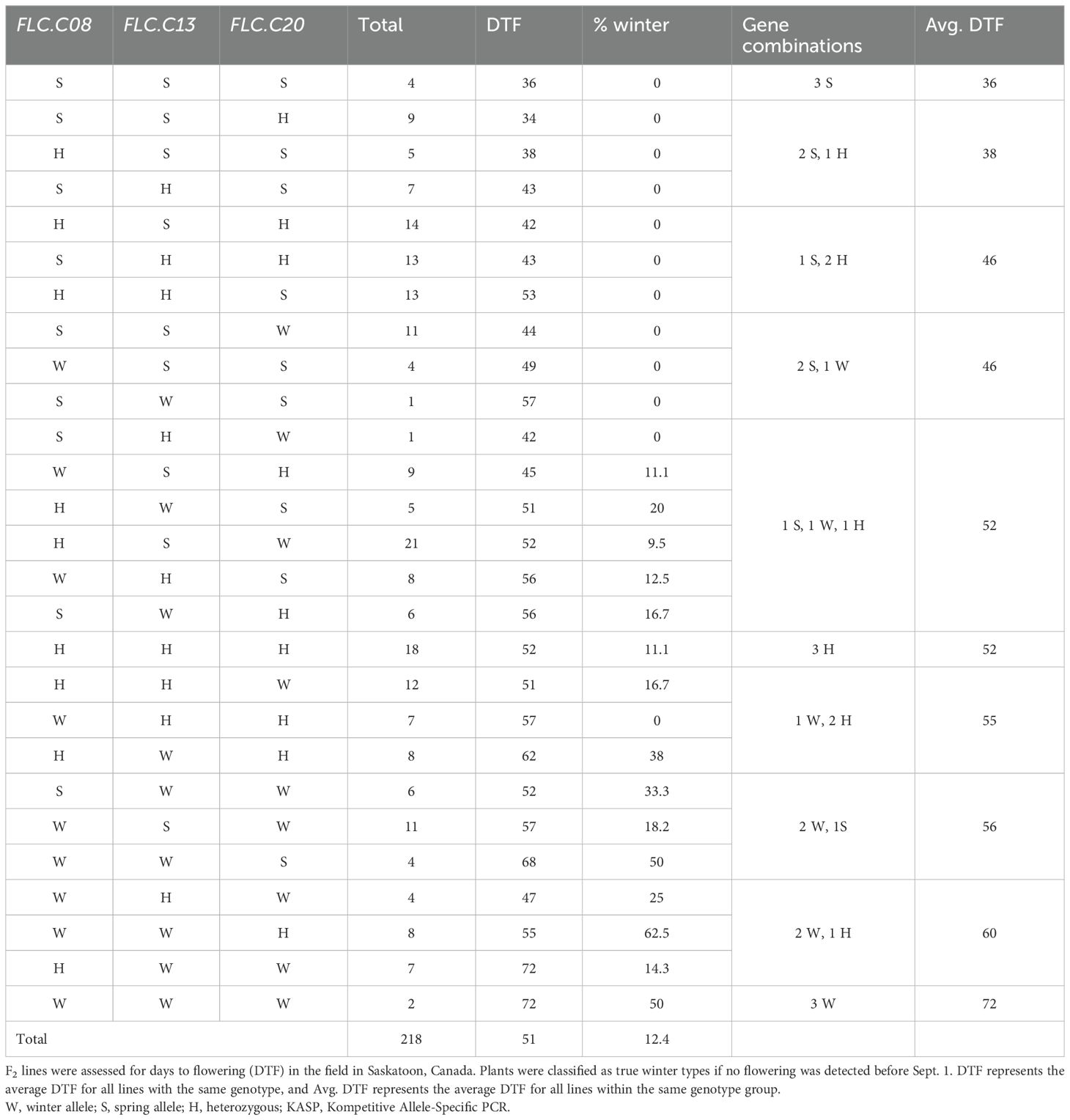
Table 3. Summary of KASP marker results for the Flowering Locus C genes Csa.FLC.C08, Csa.FLC.C13, and Csa.FLC.C20 for 218 individual F2 plants from a cross between winter camelina cultivar ‘Joelle’ with spring camelina cultivar ‘SES0787LS’.
4 Discussion
For a species’ survival, it is crucial to temporally synchronize development with the occurrence of favorable environmental conditions; accordingly, plants exhibit different life history strategies. Annuality, the completion of a plant’s life cycle in one growing season, is an adaptive evolutionary trait that emerged during the domestication of crop species from perennial ancestors, which can persist for several years (Ågren et al., 2017). Winter annuals, which require vernalization to initiate the development of reproductive tissues, have evolved in temperate climates to avoid flowering shortly before winter. Approximately a decade ago, winter annual biotypes of camelina drew the interest of plant breeders and agronomists due to their exceptional level of winter hardiness and compatibility with innovative cropping systems (Berti et al., 2015; Gesch et al., 2014). Most research on the winter camelina crop has been conducted using ‘Joelle’, a publicly available winter cultivar developed at Limagrain (France).
QTL mapping was employed to identify genomic regions underlying flowering time and gain insights into the genetic architecture of vernalization requirement in C. sativa, utilizing ‘Joelle’ as the source of winter hardiness. Two major QTLs were identified on chromosomes 8 (subgenome 1) and 13 (subgenome 2), which explained 14.3% and 21.9% of the phenotypic variation, respectively, and which both co-localized with orthologs of FLC, a well-characterized gene, which has been shown to be involved in the control of winter annual behavior in Arabidopsis thaliana (Michaels and Amasino, 1999; Swiezewski et al., 2009) and other Brassicaceae species (Schiessl et al., 2019; Takada et al., 2019), including camelina (Anderson et al., 2018; Chao et al., 2019; Chaudhary et al., 2023; Kandel et al., 2024).
Previously, one of the three C. sativa FLC orthologs, located on chromosome 20 (subgenome 3), was shown to be differentially expressed in the winter-type cultivar ‘Joelle’ when compared to the spring-type cultivar ‘CO46’, prior to and in response to vernalization. It was therefore proposed to be the main determinant for vernalization requirement in C. sativa (Anderson et al., 2018). Subsequent QTL analysis for flowering time in a population derived from a ‘Joelle’ × ‘CO46’ cross, however, did not identify a QTL on chromosome 20, but rather two major QTLs on chromosomes 8 and 13, both co-localizing with FLC genes (Kandel et al., 2024), which corroborates our own observations. Chaudhary et al. (2023) identified three major QTLs for flowering time and vernalization requirement: two on chromosomes 13 and 20 in a population derived from an interspecific cross between spring-type C. sativa and winter-type Camelina alyssum and one on chromosome 8 in a population derived from the intraspecific cross between spring-type C. sativa and semi-winter-type C. sativa ssp. pilosa. Again, all three QTL regions contained orthologs of FLC. Similarly, the mapping of QTLs for flowering time in a spring × spring population identified, among others, a QTL on chromosome 8 encompassing a FLC gene (Li et al., 2021), and a genome-wide association study (GWAS) of a spring camelina diversity panel resulted in the identification of significant SNPs located in the upstream and downstream regions of the FLC copy on chromosome 8 (Lily et al., 2021).
FLC has been proposed as candidate gene for the regulation of flowering time in previous QTL studies and GWAS in species that are members of the same botanical family as camelina, such as A. thaliana (Salomé et al., 2011; Brachi et al., 2010), Brassica napus (rapeseed and canola) (Tadege et al., 2001; Hou et al., 2012; Schiessl et al., 2019), and the latter’s progenitor species Brassica oleracea (Okazaki et al., 2007; Irwin et al., 2016) and Brassica rapa (Wu et al., 2012; Yuan et al., 2009) (reviewed in Leijten et al., 2018). A recent study on freezing tolerance in C. sativa found that QTLs for this trait also co-locate with the FLC orthologs on chromosomes 8 and 13 (Shaikh et al., 2023). This suggests that FLC may be a key regulator for multiple physiological processes in camelina, as was previously proposed for A. thaliana (Deng et al., 2011).
The co-localization of FLC with QTLs for vernalization requirement and its established role in regulating flowering time make the Csa.FLC genes ideal targets for the development of molecular markers to assist in the identification of spring- and winter-type camelina plants. For each FLC ortholog, spring and winter alleles showed distinct differences in their gene sequence, with the nucleotide sequence of spring alleles very likely rendering them non-functional or not expressed. As noted previously (Chaudhary et al., 2023), the Csa.FLC.C08 spring allele differed from the winter allele through a 3-bp deletion. This deletion causes a loss of glutamine in proximity to a binding pocket of the corresponding enzyme (unpublished data), which may impact its functionality. This hypothesis is supported by the fact that in spring types, Csa.FLC.C08 is expressed at relatively high levels independent of temperature (Chaudhary et al., 2023). The alignment of the Csa.FLC.C13 alleles identified a large insert close to the 5′ end of intron 1, which is present in all spring-type C. sativa alleles. Noteworthily, this insert is lacking from the Cmi.FLC.C13 allele of winter-type C. microcarpa CN119205, which otherwise has all of the SNPs and INDELs common to the spring-type C. sativa lines under study. This provides grounds for the hypothesis that the intron 1 insert is causal for non-functionality and/or the lack of expression of the spring allele of Csa.FLC.C13 documented by Chaudhary et al. (2023). Intron 1 of Csa.FLC.C13 may be a cis-acting gene region that, through the process of alternative splicing, contributes to the regulation of flowering time in Camelina spp., similar to instances where sequence differences in non-coding regions of FLC have been associated with differences in flowering time in Arabidopsis (Shindo et al., 2005; Li et al., 2014, 2015; Schiessl et al., 2019) and B. rapa (Yuan et al., 2009; Kitamoto et al., 2014; Wu et al., 2012) and even correlated with the divergence of annuality and perenniality in Brassica species (Kiefer et al., 2017). Although we did not identify a QTL for vernalization requirement on chromosome 20 and in association with Csa.FLC.C20, the results of previous studies (Anderson et al., 2018; Chaudhary et al., 2023) and a desire for a comprehensive analysis of FLC’s role in the genetic architecture of vernalization requirement in C. sativa led us to include Csa.FLC.C20 in subsequent investigations. Sequence analysis confirmed previous results (Anderson et al., 2018; Chao et al., 2019) that found a 1-bp frameshift mutation in exon 5 of the spring allele of Csa.FLC.C20, which results in a disrupted reading frame and consequently a non-functional enzyme. In addition to non-functionality, at ambient temperatures, Csa.FLC.C20 is expressed at a much lower level in spring types than in winter types (Anderson et al., 2018; Chaudhary et al., 2023). We used the previously described INDEL in exon 5 to develop KASP primers for Csa.FLC.C20; we chose an A/G SNP 700 bp before the start codon and a G/T SNP 673 bp before exon 2 for Csa.FLC.C08 and Csa.FLC.C13, respectively. As we were able to demonstrate, the developed co-dominant KASP markers allow for the unambiguous identification of homozygous and heterozygous Csa.FLC alleles in a high-throughput manner.
For field-grown F2 material, the marker results aligned well with observed DTF, with a greater number of winter alleles resulting in later flowering and eventually vernalization requirement. This confirms the usefulness of the markers developed herein for tracking flowering time in camelina. Overall, the results of the field trial indicated that Csa.FLC.C13 had a stronger effect on delaying flowering time than Csa.FLC.C08, which is in agreement with the results of the QTL analysis.
The F2 plants comprising the population were derived from more than one F1 plant. This certainly is unusual and generally undesired for conducting mapping studies. In addition, we inferred that the inadvertent use of genotypically different F1 plants—both homozygous for the spring Csa.FLC.C13 alleles (SS) and heterozygous (H) ones—were the cause for the distorted segregation that was observed in the Chr13 region containing Csa.FLC.C13. However, this proved to be serendipitous because it led us to deduce that the winter type parent, ‘Joelle’, was heterozygous in the region surrounding Csa.FLC.C13, which ultimately allowed us to uncover an unexpected degree of genotypic variation at all FLC loci in C. sativa. Thus, if a single F1 plant had been used to form each population, this plant would have been either homozygous for the spring Csa.FLC.C13 allele or heterozygous, with one spring and one winter allele. In the former case, no segregation would have occurred in the F2, and the QTLs on Chr13 would have been missed. In the latter case, segregation would have occurred as expected (1SS:2H:1WW). While this would have resulted in the identification of the QTLs on Chr13, it was the issue of distorted segregation that motivated us to investigate more closely the genotypic variation for FLC in ‘Joelle’ and other C. sativa germplasm.
The degree of variation we observed for the FLC alleles in ‘Joelle’, a commercial cultivar with a strong vernalization requirement and thus expected to be true breeding (homozygous) for winter alleles at all three loci, was extraordinary. Ten different allele combinations were identified, and surprisingly, only approximately 10% of the analyzed plants were indeed homozygous for the winter FLC alleles at all loci. Most plants were homozygous for a combination of the winter alleles of Csa.FLC.C08 and CsaFLC.C20 (42%) or Csa.FLC.C08 and Csa.FLC.C13 (33%), with the consistent winter locus being Csa.FLC.C08. Similarly, the three ‘Joelle’ reference genomes were homozygous for either the Csa.FLC.C13 spring and Csa.FLC.C20 winter alleles (NCBI and DOE-JGI) or the Csa.FLC.C13 winter and Csa.FLC.C20 spring alleles (AAFC reference genome), but all three had the Csa.FLC.C08 winter locus in the homozygous state. Plants with these combinations of loci needed to undergo vernalization in order to flower. Within the selected seed source, 9% of the ‘Joelle’ plants were homozygous only for the winter alleles of Csa.FLC.C08. These plants flowered without vernalization, albeit significantly delayed, indicating that one FLC winter locus leads to a semi-winter phenotype, which is in agreement with the results of Chaudhary et al. (2023). Interestingly, Csa.FLC.C13 winter alleles could only be found in combination with Csa.FLC.C08 winter alleles; 6% of ‘Joelle’ plants were heterozygous at one Csa.FLC locus at least, which corroborates our earlier findings in the ‘Joelle’ × ‘SES0787LS’ mapping population and, as does the existence of different allele combinations, also indicates that the cultivar ‘Joelle’ is still segregating. In hindsight, previous work had hinted at this phenomenon specifically for Csa.FLC.C20. Anderson et al. (2018) reported that the 1-bp deletion that is characteristic of the spring allele of this gene was present in both ‘CO46’ (spring) and ‘Joelle’ (winter), with greater frequency in the spring cultivar. This means that also in their study, ‘Joelle’ (and ‘CO46’) possessed both spring and winter Csa.FLC.C20 alleles. However, the present study is the first to draw the conclusion that the cultivar ‘Joelle’ constitutes a collection of genotypes and is still undergoing genetic segregation.
In order to validate their general utility and to determine the degree of variation at FLC in other germplasm, we tested the molecular markers developed in this study for all three FLC loci on a number of different winter C. sativa (13), winter-type C. microcarpa (3), and spring C. sativa (53) cultivars and accessions. All spring C. sativa lines were found to be homozygous for the Csa.FLC.C08 and Csa.FLC.C13 spring alleles; however, out of 53 lines, 13 were homozygous for the Csa.FLC.C20 winter allele, and eight were heterozygous at this locus. The presence of both spring and winter alleles of Csa.FLC.C20 in spring-type C. sativa germplasm clearly shows that this gene by itself only plays a subordinate role in the regulation of flowering and vernalization requirement, contrary to the proposition made by Anderson et al. (2018). Our hypothesis is corroborated by the fact that neither Kandel et al. (2024) nor the present study identified a QTL on chromosome 20. Although Chaudhary et al. (2023) did identify a chromosome 20 QTL that contained Csa.FLC.C20, it was in a population derived from an interspecific cross with C. alyssum.
The situation for winter germplasm appeared to be more complex than for spring types. Out of 13 C. sativa lines, only three were homozygous (for either spring or winter alleles) at all three FLC loci—’Bison’, BSX, and BSX-WG1—which can be traced back to one US breeding program (High Plains Crop Development). The remainder of the lines was segregated for at least one FLC ortholog. Given the strong self-fertilizing nature of C. sativa (Walsh et al., 2012), this degree of heterozygosity at the FLC loci in winter biotypes is surprising.
Only CN120025, a Type 1 hexaploid C. microcarpa genotype, had winter alleles in the homozygous state at all three loci; as shown for ‘Joelle’, this combination was remarkably rare. All other winter germplasm carried winter FLC.C08 and FLC.C20 alleles, with the exception of, not surprisingly, the tetraploid C. microcarpa accession CN119243 and C. sativa CN113692, which had FLC.C08 and FLC.C13 winter loci, as previously described for some of the ‘Joelle’ plants. Taken together, the comprehensive marker data set for ‘Joelle’ and the results for other winter C. sativa germplasm indicate that the combination of at least two winter FLC loci from different subgenomes leads to plants requiring vernalization. This may also be the case for C. microcarpa; however, additional accessions would need to be analyzed to verify this hypothesis for the wild relative. Chaudhary et al. (2023) drew similar conclusions based on their study of progeny from intra- and interspecific crosses. In the present study, combinations involving Csa.FLC.C08—Csa.FLC.C08 and Csa.FLC.C13 or CsaFLC.C08 and Csa.FLC.C20—produced a winter phenotype. This observation is consistent with work in B. napus, where different FLC composition strategies resulted in the same crop type within the Renewable Industrial Products from Rapeseed (RIPR) accession panel (Calderwood et al., 2021).
The only instance where one winter locus was found by itself was in CN113668; this accession had only the winter Csa.FLC.C08 gene. Like ‘Joelle’ plants with the same genotype, this accession flowered without vernalization, but significantly delayed, and therefore represents a semi-winter type.
Our results strongly suggest that Csa.FLC.C08 may be the most decisive ortholog for the regulation of flowering time and vernalization requirement in C. sativa. Central to this proposition is that the winter allele of this gene by itself causes a semi-winter type, and its combination with the winter alleles of one or both of the other two orthologs results in vernalization requirement. The importance of the other Csa.FLC copies is less clear, and it remains to be elucidated how the different orthologs interact with each other to bring about vernalization requirement. Both Csa.FLC.C13 and Csa.FLC.C20 act synergistically with Csa.FLC.C08 to cause vernalization requirement; however, while the results of the QTL analysis and the marker results of field-grown material suggest that Csa.FLC.C13 has the strongest effect on flowering time and vernalization requirement, the winter alleles of this gene only occur in combination with winter Csa.FLC.C08 alleles, and Csa.FLC.C20 by itself did not affect flowering time. Nevertheless, what the present study clearly shows is that all three orthologs are involved in regulating the transition to reproductive growth in C. sativa, with no indication for sub-functionalization or pseudogenization.
5 Conclusion
This study is the first to uncover an unexpected degree of variability at the FLC loci in spring- and winter-type C. sativa germplasm and to describe the development of universally applicable molecular markers that distinguish spring from winter alleles for all three orthologs. It is our hope that going forward, the developed KASP markers may serve as tools for winter camelina breeders to identify suitable parent plants to be used in crosses and to enrich segregating breeding populations for winter alleles.
An ideal winter cultivar combines a robust vernalization requirement to prevent flowering before winter with a quick resumption of growth and flowering in the spring, which is essential for early maturity. Depending on the environment—mainly defined by the length of winter—different combinations of winter FLC alleles may be required to facilitate both traits; the results of this study and the tools developed herein are a first step to designing winter camelina cultivars that are optimized for different growing regions. Near-isogenic ‘Joelle’ lines with different combinations of homozygous FLC alleles are currently being developed; future greenhouse and field experiments using these lines should yield important insights about the independent and combined effects of the different FLC copies on flowering time and vernalization requirement in winter camelina.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
VR: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Visualization, Writing – original draft. RC: Resources, Writing – review & editing. AZ: Methodology, Writing – review & editing. IP: Resources, Writing – review & editing. CE: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by funding provided by the Canadian Agricultural Partnership’s Agri-Science Program and the Sustainable Canadian Agricultural Partnership’s Agri-Science Program.
Acknowledgments
The authors gratefully acknowledge David Sarich, Lori Bobowski, Jillian Hueller, Quinn Buettner, Jordan Laturnus, and Ryan Vetter for their excellent technical support and Genome Quebec for providing the Illumina NovaSeq sequencing results. Myrtle Brkic is gratefully acknowledged for conducting protein modeling.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1639872/full#supplementary-material
Supplementary Table 1 | Camelina germplasm used in the study.
Supplementary Table 2 | Selective KASP primer sets for each of the FLC alleles. The selective primer associated with the winter phenotype is in bold.
Supplementary Table 3 | Mapping data for the F2 population derived from the ‘Joelle’ x ‘SES0787LS’ cross. Marker names are based on the SNP position in relation to the DH55 reference genome. The reference and alternative base are listed at the end with the Joelle base bolded. The regions with distorted segregation on Chr13 and Chr20 are highlighted in blue.
Supplementary Table 4 | Sequence alignment of the FLC.C08 gene. Three C. microcarpa (CN 120025, CN 119205 and CN 119243); ‘CO46’ (GCA_036971115.1) and ‘Joelle’ (GCA_036769185.1) reference sequences from NCBI; ‘Joelle’ sequences from the AAFC and the DOE-JGI phytozome reference genomes; the DH55 reference genome sequence and 15 spring type C. sativa lines were aligned using the EMBL-EBI online tool MUSCLE. All winter Camelina lines are in blue font. Exons are shown in purple font and the SNP utilized for marker development is highlighted in blue. The three-base pair INDEL that distinguishes spring- and winter alleles is highlighted in green.
Supplementary Table 5 | Sequence alignment of the FLC.C13 gene. Three C. microcarpa (CN 120025, CN 119205 and CN 119243); ‘CO46’ (GCA_036971115.1) and ‘Joelle’ (GCA_036769185.1) reference sequences from NCBI; ‘Joelle’ sequences from the AAFC and the DOE-JGI phytozome reference genomes; the DH55 reference genome sequence and 15 spring type C. sativa lines were aligned using the EMBL-EBI online tool MUSCLE. All winter Camelina lines are in blue font. Exons are shown in purple font and the SNP utilized for marker development is highlighted in blue. SNPs which distinguish the winter and spring alleles are highlighted in green.
Supplementary Table 6 | Sequence alignment of the FLC.C20 gene. Two C. microcarpa (CN 120025 and CN 119205; CN 119243 is a tetraploid C. microcarpa line that does not have a third subgenome and thus is not included); ‘CO46’ (GCA_036971115.1) and ‘Joelle’ (GCA_036769185.1) reference sequences from NCBI; ‘Joelle’ sequences from the AAFC and the DOE-JGI phytozome reference genomes; the DH55 reference genome sequence and 15 spring type C. sativa lines were aligned using the EMBL-EBI online tool MUSCLE. All winter Camelina lines are in blue font. Exons are shown in purple font and the SNP utilized for marker development is highlighted in blue (Anderson et al., 2018). A large insert only present within the ‘CO46’ reference sequence is represented by an N base highlighted in red. The complete sequence for the insert is presented at the end of the alignment and is in red font.
Supplementary Table 7 | Summary of the alignment of GBS data from 10 individual ‘Joelle’ plants with the DH55 reference genome. A total of 79740 SNPs were detected, which have a quality score over 30 and less than 20% missing data. As per standard nomenclature ‘0/0’ calls represent the reference genome SNP, ‘0/1’ heterozygous calls, ‘1/1’ represent the alternative SNPs and ‘./.’ represent a no call due to insufficient data.
Supplementary Table 8 | Comparison of GBS data from 10 individual ‘Joelle’ plants to three separate ‘Joelle’ reference genomes for the regions surrounding the Csa.FLC.C13 gene. For reference, the spring-type Csa.FLC.C13_KASP marker is present in Joelle.2, Joelle.9 & Joelle.10, the winter-type Csa.FLC.C13_KASP marker is present in Joelle.3, Joelle.4, Joelle.5, Joelle.6 & Joelle.8 and a heterozygous Csa.FLC.C13_KASP marker is present in Joelle.1 & Joelle.7. As per standard nomenclature ‘0/0’ calls represent the reference genome SNP, ‘0/1’ heterozygous calls, ‘1/1’ represent the alternative SNPs and ‘./.’ represent a no call due to insufficient data.
Supplementary Table 9 | Comparison of GBS data from 10 individual ‘Joelle’ plants to three separate ‘Joelle’ reference genomes for the regions surrounding the Csa.FLC.C20 allele. For reference, the spring-type Csa.FLC.C20_KASP marker is present in Joelle.3, Joelle.4, Joelle.6, Joelle.8, Joelle.9 & Joelle.10, the winter-type Csa.FLC.C20_KASP marker is present in Joelle.2 and a heterozygous Csa.FLC.C20_KASP marker is present in Joelle.1, Joelle.5 & Joelle.7. As per standard nomenclature ‘0/0’ calls represent the reference genome SNP, ‘0/1’ heterozygous calls, ‘1/1’ represent the alternative SNPs and ‘./.’ represent a no call due to insufficient data.
Supplementary Table 10 | KASP allele scores for a diverse panel of Camelina lines. Lines in blue font have the winter phenotype. Each line is a bulk of four plants, except for the ‘Joelle’ and ‘SES0787LS’ lines and those marked as REF (reference genomes). The REF samples are predictions based on the Csa.FLC gene sequences. Two separate bulks of several CN lines were analyzed and are labelled PRGC #1 and PGRC #2 (Plant Gene Resources of Canada). A score of “W” represents the winter FLC allele, a “S” the spring FLC allele and “H” the presence of both alleles.
References
Abramovič, H. and Abram, V. (2005). Physico-chemical properties, composition and oxidative stability of Camelina sativa oil. Food Technol. Biotechnol. 43, 63–70.
Ågren, J., Oakley, C. G., Lundemo, S., and Schemske, D. W. (2017). Adaptive divergence in flowering time among natural populations of Arabidopsis thaliana: Estimates of selection and QTL mapping. Evolution 71, 550–564. doi: 10.1111/evo.13126
Anderson, J. V., Horvath, D. P., Dogramaci, M., Dorn, K. M., Chao, W. S., Watkin, E. E., et al. (2018). Expression of FLOWERING LOCUS C and a frameshift mutation of this gene on chromosome 20 differentiate a summer and winter annual biotype of Camelina sativa. Plant Direct. 2, e00060. doi: 10.1002/pld3.60
Angelini, L. G., Moscheni, E., Colonna, G., Belloni, P., and Bonari, E. (1997). Variation in agronomic characteristics and seed oil composition of new oilseed crops in central Italy. Ind. Crop Prod. 6, 313–323. doi: 10.1016/S0926-6690(97)00022-8
Bastow, R., Mylne, J. S., Lister, C., Lippman, Z., Martienssen, R. A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. doi: 10.1038/nature02269
Berti, M., Gesch, R., Johnson, B., Ji, Y., Seames, W., and Aponte, A. (2015). Double- and relay-cropping of energy crops in the northern Great Plains, USA. Ind. Crop Prod. 75, 26–34. doi: 10.1016/j.indcrop.2015.05.012
Berti, M., Samarappuli, D., Johnson, B. L., and Gesch, R. W. (2017). Integrating winter camelina into maize and soybean cropping systems. Ind. Crop Prod. 107, 595–601. doi: 10.1016/j.indcrop.2017.06.014
Berti, M., Wilckens, R., Fischer, S., Solis, A., and Johnson, B. (2011). Seeding date influence on camelina seed yield, yield components, and oil content in Chile. Ind. Crop Prod. 34, 1358–1365. doi: 10.1016/j.indcrop.2010.12.008
Blackshaw, R., Johnson, E., Gan, Y., May, W., McAndrew, D., Barthet, V., et al. (2011). Alternative oilseed crops for biodiesel feedstock on the Canadian prairies. Can. J. Plant Sci. 91, 889–896. doi: 10.4141/cjps2011-002
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bonjean, A. and Goffic, F. L. (1999). La cameline: Camelina sativa (L.) Crantz: une opportunité pour l’agriculture et l’industrie européennes. Oléagineux Corps Gras Lipides (OCL) 6, 28–34.
Brachi, B., Faure, N., Horton, M., Flahauw, E., Vazquez, A., Nordborg, M., et al. (2010). Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PloS Genet. 6, e1000940. doi: 10.1371/journal.pgen.1000940
Budin, J. T., Breene, W. M., and Putnam, D. H. (1995). Some compositional properties of camelina (Camelina sativa L. Crantz) seeds and oils. J. Am. Oil Chem. Soc 72, 309–315. doi: 10.1007/BF02541088
Calderwood, A., Lloyd, A., Hepworth, J., Tudor, E. H., Jones, D. M., Woodhouse, S., et al (2021). Total FLC transcript dynamics from divergent paralogue expression explains flowering diversity in Brassica napus. New Phytol. 229, 3534–3548. doi: 10.1111/nph.17131
Chao, W. S., Wang, H., Horvath, D. P., and Anderson, J. V. (2019). Selection of endogenous reference genes for qRT-PCR analysis in Camelina sativa and identification of FLOWERING LOCUS C allele-specific markers to differentiate summer- and winter-biotypes. Ind. Crop Prod. 129, 495–502. doi: 10.1016/j.indcrop.2018.12.017
Chaudhary, R., Higgins, E. E., Eynck, C., Sharpe, A. G., and Parkin, I. A. P. (2023). Mapping QTL for vernalization requirement identified adaptive divergence of the candidate gene Flowering Locus C in polyploid Camelina sativa. Plant Genome 16, e20397. doi: 10.1002/tpg2.20397
Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience. 10, giab008. doi: 10.1093/gigascience/giab008
Deng, W., Ying, H., Helliwell, C. A., Taylor, J. M., Peacock, W. J., and Dennis, E. S. (2011). FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 6680–6685. doi: 10.1073/pnas.1103175108
Eberle, C. A., Thom, M. D., Nemec, K. T., Forcella, F., Lundgren, J. G., Gesch, R. W., et al. (2015). Using pennycress, camelina, and canola cash cover crops to provision pollinators. Ind. Crop Prod. 75, 20–25. doi: 10.1016/j.indcrop.2015.06.026
Enjalbert, J. N., Zheng, S., Johnson, J. J., Mullen, J. L., Byrne, P. F., and McKay, J. K. (2013). Brassicaceae germplasm diversity for agronomic and seed quality traits under drought stress. Ind. Crop Prod. 47, 176–185. doi: 10.1016/j.indcrop.2013.02.037
French, A. N., Hunsaker, D., Thorp, K., and Clarke, T. (2009). Evapotranspiration over a camelina crop at Maricopa, Arizona. Ind. Crop Prod. 29, 289–300. doi: 10.1016/j.indcrop.2008.06.001
Fröhlich, A. and Rice, B. (2005). Evaluation of Camelina sativa oil as a feedstock for biodiesel production. Ind. Crop Prod. 21, 25–31. doi: 10.1016/j.indcrop.2003.12.004
Fu, Y. B., Cober, E. R., Morrison, M. J., Marsolais, F., Peterson, G. W., and Horbach, C. (2021). Patterns of genetic variation in a soybean germplasm collection as characterized with genotyping-by-sequencing. Plants 10, 1611. doi: 10.3390/plants10081611
Gao, Y., Jiang, C., Zhang, Y., Liu, L., Wang, Y., Kim, D. S., et al. (2022). Agronomic performance of camelina genotypes selected for seed yield and quality characteristics in eastern China. Ind. Crop Prod. 184, 7. doi: 10.1016/j.indcrop.2022.115077
Gesch, R. W. and Archer, D. W. (2013). Double-cropping with winter camelina in the northern Corn Belt to produce fuel and food. Ind. Crop Prod. 44, 718–725. doi: 10.1016/j.indcrop.2012.05.023
Gesch, R. W., Archer, D. W., and Berti, M. T. (2014). Dual cropping winter camelina with soybean in the northern corn belt. Agron. J. 106, 1735–1745. doi: 10.2134/agronj14.0215
Gesch, R. W. and Cermak, S. C. (2011). Sowing date and tillage effects on fall-seeded camelina in the northern corn belt. Agron. J. 103, 980–987. doi: 10.2134/agronj2010.0485
Gesch, R. W. and Johnson, J. M. F. (2015). Water use in camelina–soybean dual cropping systems. Agron. J. 107, 1098–1104. doi: 10.2134/agronj14.0626
Gesch, R. W., Matthees, H. L., Alvarez, A. L., and Gardner, R. D. (2018). Winter camelina: Crop growth, seed yield, and quality response to cultivar and seeding rate. Crop Sci. 58, 2089–2098. doi: 10.2135/cropsci2018.01.0018
Gugel, R. K. and Falk, K. C. (2006). Agronomic and seed quality evaluation of Camelina sativa in western Canada. Can. J. Plant Sci. 86, 1047–1058. doi: 10.4141/P04-081
Helliwell, C. A., Wood, C. C., Robertson, M., James Peacock, W., and Dennis, E. S. (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46, 183–192. doi: 10.1111/j.1365-313X.2006.02686.x
Henderson, I. R. and Dean, C. (2004). Control of Arabidopsis flowering: the chill before the bloom. Development 131, 3829–3838. doi: 10.1242/dev.01294
Hixson, S. M., Parrish, C. C., and Anderson, D. M. (2014). Full substitution of fish oil with camelina (Camelina sativa) oil, with partial substitution of fish meal with camelina meal, in diets for farmed Atlantic salmon (Salmo salar) and its effect on tissue lipids and sensory quality. Lipids 49, 97–111. doi: 10.1007/s11745-013-3862-7
Horvath, D., Anderson, J. V., Chao, W. S., Zheng, P., Buchwaldt, M., Parkin, I. A., et al. (2019). Genes associated with chloroplasts and hormone-signaling, and transcription factors other than CBF s are associated with differential survival after low temperature treatments of Camelina sativa biotypes. PloS One 14, e0217692. doi: 10.1371/journal.pone.0217692
Hou, J., Long, Y., Raman, H., Zou, X., Wang, J., Dai, S., et al. (2012). A Tourist-like MITE insertion in the upstream region of the BnFLC.A10 gene is associated with vernalization requirement in rapeseed (Brassica napus L.). BMC Plant Biol. 12, 238. doi: 10.1186/1471-2229-12-238
Hunsaker, D. J., French, A. N., Clarke, T. R., and El-Shikha, D. M. (2011). Water use, crop coefficients, and irrigation management criteria for camelina production in arid regions. Irrigation Sci. 29, 27–43. doi: 10.1007/s00271-010-0213-9
Irwin, J. A., Soumpourou, E., Lister, C., Ligthart, J. D., Kennedy, S., and Dean, C. (2016). Nucleotide polymorphism affecting FLC expression underpins heading date variation in horticultural brassicas. Plant J. 87, 597–605. doi: 10.1111/tpj.13221
Johnson, J. M., Gesch, R. W., and Barbour, N. W. (2019). Spring camelina N rate: balancing agronomics and environmental risk in United States Corn Belt. Arch. Agron. Soil Sci. 65, 640–653. doi: 10.1080/03650340.2018.1519803
Johnson, G. A., Wells, M. S., Anderson, K., Gesch, R. W., Forcella, F., and Wyse, D. L. (2017). Yield tradeoffs and nitrogen between pennycress, camelina, and soybean in relay- and double-crop systems. Agron. J. 109, 2128–2135. doi: 10.2134/agronj2017.02.0065
Kagale, S., Koh, C., Nixon, J., Bollina, V., Clarke, W. E., Tuteja, R., et al. (2014). The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat. Commun. 5, 3706. doi: 10.1038/ncomms4706
Kandel, J. S., Talukder, Z. I., Shaikh, T. M., Horvath, D. P., Li, X., and Anderson, J. V. (2024). Identification of quantitative trait loci for flowering time in a Camelina biparental population developed from winter-and spring-type parents. Ind. Crop Prod. 220, 119259. doi: 10.1016/j.indcrop.2024.119259
Kiefer, C., Severing, E., Karl, R., Bergonzi, S., Koch, M., Tresch, A., et al. (2017). Divergence of annual and perennial species in the Brassicaceae and the contribution of cis-acting variation at FLC orthologues. Mol. Ecol. 26, 3437–3457. doi: 10.1111/mec.14084
Kirkhus, B., Lundon, A. R., Haugen, J. E., Vogt, G., Borge, G. I. A., and Henriksen, B. I. (2013). Effects of environmental factors on edible oil quality of organically grown Camelina sativa. J. Agric. Food Chem. 61, 3179–3185. doi: 10.1021/jf304532u
Kitamoto, N., Yui, S., Nishikawa, K., Takahata, Y., and Yokoi, S. (2014). A naturally occurring long insertion in the first intron in the Brassica rapa FLC2 gene causes delayed bolting. Euphytica. 196, 213–223. doi: 10.1007/s10681-013-1025-9
Kon’kova, N. G., Shelenga, T. V., Gridnev, G. A., Dubovskaya, A. G., and Malyshev, L. L. (2021). Stability and variability of Camelina sativa (L.) crantz economically valuable traits in various eco-geographical conditions of the Russian federation. Agronomy 11, 332. doi: 10.3390/agronomy11020332
Lal, R., Regnier, E., Eckert, D. J., Edwards, W. M., and Hammond, R. (1991). “Expectations of cover crops sustainable agriculture,” in Cover crops for clean water. Proc. Int. Conf., Jackson, TN. Ed. Hargrove, W. L. (Soil and Water Conserv. Soc. Am., Ankeny, IA), 1–11. 9–11.
Langmead, B. and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Leijten, W., Koes, R., Roobeek, I., and Frugis, G. (2018). Translating flowering time from arabidopsis thaliana to brassicaceae and asteraceae crop species. Plants. 7, 111. doi: 10.3390/plants7040111
Li, P., Filiault, D., Box, M. S., Kerdaffrec, E., Van Oosterhout, C., and Wilczek, A. M. (2014). Multiple FLC haplotypes defined by independent cis-regulatory variation underpin life history diversity in Arabidopsis thaliana. Genes Dev. 28, 1635–1640. doi: 10.1101/gad.245993.114
Li, H., Hu, X., Lovell, J. T., Grabowski, P. P., Mamidi, S., Chen, C., et al. (2021). Genetic dissection of natural variation in oilseed traits of camelina by whole-genome resequencing and QTL mapping. Plant Genome 14, e20110. doi: 10.1002/tpg2.20110
Li, P., Tao, Z., and Dean, C. (2015). Phenotypic evolution through variation in splicing of the noncoding RNA COOLAIR. Genes Dev. 29, 696–701. doi: 10.1101/gad.258814.115
Lily, Z. L., Fahlgren, N., Kutchan, T., Schachtman, D., Ge, Y., Gesch, R., et al. (2021). Discovering candidate genes related to flowering time in the spring panel of Camelina sativa. Ind. Crop Prod. 173, 114104. doi: 10.1016/j.indcrop.2021.114104
Madeira, F., Pearce, M., Tivey, A. R. N., Basutkar, P., Lee, J., Edbali, O., et al. (2022). Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 50, W276–W279. doi: 10.1093/nar/gkac240
Michaels, S. D. and Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 11, 949–956. doi: 10.1105/tpc.11.5.949
Mirek, Z. (1980). Taxonomy and nomenclature of Camelina pilosa auct. Acta Soc. Bot. Pol. 49, 553–561. doi: 10.5586/asbp.1980.050
Ngo, N. T. T., Senadheera, T. R. L., and Shahidi, F. (2023). Antioxidant Properties and Prediction of Bioactive Peptides Produced from Flixweed (sophia, Descurainis sophia L.) and Camelina (Camelina sativa (L.) Crantz) Seed Meal: Integrated In Vitro and In Silico Studies. Plants. 12, 3575. doi: 10.3390/plants12203575
Okazaki, K., Sakamoto, K., Kikuchi, R., Saito, A., Togashi, E., Kuginuki, Y., et al. (2007). Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theor. Appl. Genet. 114, 595–608. doi: 10.1007/s00122-006-0460-6
Plessers, A. G., McGregor, W. G., Carson, R. B., and Nakoneshny, W. (1962). Species trials with oilseed plants. II. Camelina. Can. J. Plant Sci. 42, 452–459. doi: 10.4141/cjps62-073
Putnam, D. H., Budin, J. T., Field, L. A., and Breene, W. M. (1993). “Camelina: a promising low input oilseed,” in New crops. Eds. Janick, J. and Simon, J. E. (Wiley, New York), 314–322.
Ratusz, K., Symoniuk, E., Wroniak, M., and Rudzińska, M. (2018). Bioactive compounds, nutritional quality and oxidative stability of cold-pressed camelina (Camelina sativa L.) oils. Appl. Sci. 8, 2606. doi: 10.3390/app8122606
Robinson, R. G. (1987). Camelina: a useful research crop and a potential oilseed crop. Available online at: https://conservancy.umn.edu/bitstream/handle/11299/141546/SB579.pdf?sequence=1 (Accessed May 15, 2025).
Salomé, P. A., Bomblies, K., Laitinen, R. A., Yant, L., Mott, R., and Weigel, D. (2011). Genetic architecture of flowering time variation in Arabidopsis thaliana. Genetics 188, 421–433. doi: 10.1534/genetics.111.126607
Samach, A., Onouchi, H., Gold, S. E., Ditta, G. S., Schwarz-Sommer, Z., Yanofsky, M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. doi: 10.1126/science.288.5471.1613
Schiessl, S. V., Quezada-Martinez, D., Tebartz, E., Snowdon, R. J., and Qian, L. (2019). The vernalisation regulator FLOWERING LOCUS C is differentially expressed in biennial and annual Brassica napus. Sci. Rep. 9, 14911. doi: 10.1038/s41598-019-51212-x
Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Kröber, et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912. doi: 10.1101/gad.373506
Séguin-Swartz, G., Eynck, C., Gugel, R. K., Strelkov, S. E., Olivier, C. Y., Li, J. L., et al. (2009). Diseases of Camelina sativa (false flax). Can. J. Plant Pathol. 31, 375–386. doi: 10.1080/07060660909507612
Shaikh, T. M., Rahman, M., Smith, T., Anderson, J. V., Chao, W. S., and Horvath, D. P. (2023). Homozygosity mapping identified loci and candidate genes responsible for freezing tolerance in Camelina sativa. Plant Genome. 16, e20318. doi: 10.1002/tpg2.20318
Sheldon, C. C., Rouse, D. T., Finnegan, E. J., Peacock, W. J., and Dennis, E. S. (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. U.S.A. 97, 3753–3758. doi: 10.1073/pnas.97.7.3753
Shindo, C., Aranzana, M. J., Lister, C., Baxter, C., Nicholls, C., Nordborg, M., et al. (2005). Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 138, 1163–1173. doi: 10.1104/pp.105.061309
Shonnard, D. R., Williams, L., and Kalnes, T. N. (2010). Camelina-derived jet fuel and diesel: Sustainable advanced biofuels. Environ. Prog. Sustain Energy. 29, 382–392. doi: 10.1002/ep.10461
Sindelar, A. J., Schmer, M. R., Gesch, R. W., Forcella, F., Eberle, C. A., Thom, M. D., et al. (2017). Winter oilseed production for biofuel in the US Corn Belt: Opportunities and limitations. GCB Bioenergy. 9, 508–524. doi: 10.1111/gcbb.12297
Solis, A., Vidal, I., Paulino, L., Johnson, B. L., and Berti, M. T. (2013). Camelina seed yield response to nitrogen, sulfur, and phosphorus fertilizer in South Central Chile. Ind. Crop Prod. 44, 132–138. doi: 10.1016/j.indcrop.2012.11.005
Somers, D. J., Friesen, K. R. D., and Rakow, G. (1998). Identification of molecular markers associated with linoleic acid desaturation in Brassica napus. Theor. Appl. Genet. 96, 897–903. doi: 10.1007/s001220050817
Soroka, J., Olivier, C., Grenkow, L., and Séguin-Swartz, G. (2015). Interactions between Camelina sativa (Brassicaceae) and insect pests of canola. Can. Entomol. 147, 193–214. doi: 10.4039/tce.2014.42
Staver, K. W. and Brinsfield, R. B. (1998). Using cereal grain winter cover crops to reduce groundwater nitrate contamination in the mid-Atlantic coastal plain. J. Soil Water Conserv. 53, 230–240. doi: 10.1080/00224561.1998.12457224
Stringam, G. R. (1971). Genetics of four hypocotyl mutants in Brassica campestris L. J. Hered. 62, 248–250. doi: 10.1093/oxfordjournals.jhered.a108161
Swiezewski, S., Liu, F., Magusin, A., and Dean, C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462, 799–802. doi: 10.1038/nature08618
Tadege, M., Sheldon, C. C., Helliwell, C. A., Stoutjesdijk, P., Dennis, E. S., and Peacock, W. J. (2001). Control of flowering time by FLC orthologues in Brassica napus. Plant J. 28, 545–553. doi: 10.1046/j.1365-313x.2001.01182.x
Takada, S., Akter, A., Itabashi, E., Nishida, N., Shea, D. J., Miyaji, N., et al. (2019). The role of FRIGIDA and FLOWERING LOCUS C genes in flowering time of Brassica rapa leafy vegetables. Sci. Rep. 9, 13843. doi: 10.1038/s41598-019-50122-2
Van Ooijen, J. W. (2006). JoinMap Version 4.0: Software for the calculation of genetic linkage maps (Wageningen, Netherlands: Plant Research International).
Van Ooijen, J. W. (2009). MapQTL 6: Software for the mapping of quantitative trait loci in experimental populations of diploid species (Wageningen, Netherlands: Kyazma BV).
Vollmann, J., Moritz, T., Kargl, C., Baumgartner, S., and Wagentristl, H. (2007). Agronomic evaluation of camelina genotypes selected for seed quality characteristics. Ind. Crop Prod. 26, 270–277. doi: 10.1016/j.indcrop.2007.03.017
Walsh, K. D., Puttick, D. M., Hills, M. J., Yang, R. C., Topinka, K. C., and Hall, L. M. (2012). First report of outcrossing rates in camelina [Camelina sativa (L.) Crantz], a potential platform for bioindustrial oils. Can. J. Plant Sci. 92, 681–685. doi: 10.4141/CJPS2011-182
Weiss, R. M., Zanetti, F., Alberghini, B., Puttick, D., Vankosky, M. A., Monti, A., et al. (2024). Bioclimatic analysis of potential worldwide production of spring-type camelina [Camelina sativa (L.) Crantz] seeded in the spring. GCB Bioenergy 16, e13126. doi: 10.1111/gcbb.13126
Wu, X. and Leung, D. Y. (2011). Optimization of biodiesel production from camelina oil using orthogonal experiment. Appl. Energy 88, 3615–3624. doi: 10.1016/j.apenergy.2011.04.041
Wu, J., Wei, K., Cheng, F., Li, S., Wang, Q., Zhao, J., et al. (2012). A naturally occurring InDel variation in BraA. FLC. b (BrFLC2) associated with flowering time variation in Brassica rapa. BMC Plant Biol. 12, 151. doi: 10.1186/1471-2229-12-151
Yuan, Y. X., Wu, J., Sun, R. F., Zhang, X. W., Xu, D. H., Bonnema, G., et al. (2009). A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J. Exp. Bot. 60, 1299–1308. doi: 10.1093/jxb/erp010
Zanetti, F., Alberghini, B., Marjanović Jeromela, A., Grahovac, N., Rajković, D., Kiprovski, B., et al. (2021). Camelina, an ancient oilseed crop actively contributing to the rural renaissance in Europe. A review. Agron. Sustain. Dev. 41, 1–18. doi: 10.1007/s13593-020-00663-y
Zanetti, F., Eynck, C., Christou, M., Krzyżaniak, M., Righini, D., Alexopoulou, E., et al. (2017). Agronomic performance and seed quality attributes of camelina (Camelina sativa L. crantz) in multi-environment trials across Europe and Canada. Ind. Crop Prod. 107, 602–608. doi: 10.1016/j.indcrop.2017.06.022
Zanetti, F., Peroni, P., Pagani, E., von Cossel, M., Greiner, B. E., Krzyżaniak, M., et al. (2024). The opportunities and potential of camelina in marginal land in Europe. Ind. Crop Prod. 211, 118224. doi: 10.1016/j.indcrop.2024.118224
Zhang, C. J., Gao, Y., Jiang, C., Liu, L., Wang, Y., Kim, D. S., et al. (2021). Camelina seed yield and quality in different growing environments in northern China. Ind. Crop Prod. 172, 71. doi: 10.1016/j.indcrop.2021.114071
Zubr, J. (1997). Oil-seed crop: Camelina sativa. Ind. Crop Prod. 6, 113–119. doi: 10.1016/S0926-6690(96)00203-8
Keywords: Camelina sativa, flowering time, vernalization, phenology, Flowering Locus C
Citation: Roslinsky V, Chaudhary R, Zatylny A, Parkin IAP and Eynck C (2025) All roads lead to Rome: QTL analysis for vernalization requirement and dissection of allelic variation uncovered unexpected diversity of FLC loci in Camelina sativa. Front. Plant Sci. 16:1639872. doi: 10.3389/fpls.2025.1639872
Received: 02 June 2025; Accepted: 07 July 2025;
Published: 25 July 2025.
Edited by:
Erik Alexandersson, Swedish University of Agricultural Sciences, SwedenReviewed by:
Anthony Richard Gendall, La Trobe University, AustraliaGurjeet Singh, Texas A&M University, United States
Copyright © 2025 Roslinsky, Chaudhary, Zatylny, Parkin and Eynck. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christina Eynck, Y2hyaXN0aW5hLmV5bmNrQGFnci5nYy5jYQ==
†ORCID: Vicky Roslinsky, orcid.org/0000-0002-2408-3328
Raju Chaudhary, orcid.org/0000-0001-5370-6945
Christina Eynck, orcid.org/000-0001-8693-2340
 Vicky Roslinsky1†
Vicky Roslinsky1† Raju Chaudhary
Raju Chaudhary Isobel A. P. Parkin
Isobel A. P. Parkin Christina Eynck
Christina Eynck