- College of Horticulture, Northwest A&F University, Xianyang, China
Introduction: Optimizing nitrogen sources and rootstock selection is crucial for sustainable watermelon production. However, the synergistic mechanisms between organic nitrogen forms and rootstocks remain poorly understood. This study investigates whether glycine, as an organic nitrogen source, modulates root-associated bacterial communities through rootstock-mediated effects to enhance watermelon growth.
Methods: Grafted watermelon plants (scion: watermelon; rootstocks: self-grafted watermelon (CK), wild watermelon (T1), bottle gourd (T2), pumpkin (T3) were cultivated under glycine (G) or ammonium nitrate (A) treatments for 25 days. Plant growth, soil enzyme activity, rhizosphere bacterial communities (16S rRNA sequencing), and root metabolomes (UPLC–MS/MS) were analyzed.
Results: Relative to ammonium nitrate, glycine to some extent increased bacterial α-diversity but there was no significant difference and altered β-diversity, whereas enhancing microbial network complexity. Rootstock genotype is the main driver of bacterial α diversity and shaped the bacterial network architecture: T1-supported networks exhibited strong associations enriched in two-component systems, whereas T3 networks reflected intensified resource competition. Rootstock identity also influenced root exudate profiles. T3 secreted high levels of amino acids and nucleotides with metabolic and defensive roles, correlating with the abundance of Edaphobacter and Actinomadura. In contrast, T1 increased Acidibacter abundance via lipid secretion. The rootstock–bacteria–metabolite interplay modulated soil enzyme activities, supported photochemical efficiency, and promoted biomass accumulation.
Discussion: These findings demonstrate the potential of glycine as a sustainable nitrogen source and identify compatible scion–rootstock combinations that enhance rhizosphere microbial dynamics and plant performance. The study provides mechanistic insights into how root exudates shape bacterial community assembly, although further work is needed to elucidate the complexity of microbe–microbe interactions.
1 Introduction
The growth of terrestrial plants is regulated by soil microbial communities, which induce roots to secrete specific compounds (Anderson et al., 2024) Rhizosphere microorganisms and root exudates serve as sensitive indicators of plant growth status. Both the aboveground and belowground compartments of terrestrial plants harbor extensive microbial populations, with the gene abundance of rhizosphere microbial communities frequently exceeding that of the host plant. Accordingly, the microbiome is described as the “second plant genome” (Mendes et al., 2013). Substantial evidence demonstrates that the rhizosphere microbiome plays a critical role in plant health (Berendsen et al., 2012).
Watermelon (Citrullus lanatus (Thumb.)), a major cucurbit horticultural crop, is valued globally for its distinctive flavor and rich nutritional content (Guo et al., 2013) and is cultivated extensively. The industrialization and specialization of modern watermelon production, combined with the challenge of excessive fertilizer application worldwide, have led to adverse effects on plant growth and yield due to persistent continuous cropping and inappropriate nitrogen management (Hakeem et al., 2011). As a nitrogen-demanding crop, watermelon is particularly susceptible to nitrogen over-application (Nawaz et al., 2018). Excessive chemical fertilizer use in protected cultivation systems frequently leads to inefficient productivity, increased production costs, and aggravated soil pollution (Pang et al., 2017). Global efforts to reduce fertilizer usage have accelerated the transition from chemical to organic fertilizers in agriculture. The application of organic fertilizers is essential for improving soil organic nitrogen content; relative to chemical fertilizers, organic fertilizers release nutrients more gradually, provide greater persistence, and are less susceptible to leaching, thereby avoiding the need for repeated excessive application. Plants can directly acquire and utilize both inorganic and organic nitrogen forms (Murphy et al., 2000). Glycine, among the most abundant free amino acids in crop soils (Wang et al., 2014), is distinguished by its minimal molecular weight and simple structure, rendering it a promising substitute for inorganic nitrogen.
Grafting is a widely used agricultural practice, particularly in watermelon production, to mitigate the adverse effects of continuous cropping, enhance disease resistance, and improve mineral uptake, thereby supporting stable yields (Edelstein et al., 2011; Rivero et al., 2003). During cultivation, auto-toxic substances may accumulate in the soil; however, root replacement via grafting can alter the rhizosphere microbiota, promoting the degradation of such substances (Wang et al., 2023). The root systems of grafted plants have the capacity to recruit and reshape functional microbial communities, thereby maintaining plant health (Poudel et al., 2019), and enhancing the stability of microbial networks, with potential reductions in pathogen populations (Ruan et al., 2020).
Nevertheless, research on organic nitrogen fertilizers has predominantly focused on non-grafted crops (Zhang et al., 2022), and limited knowledge exists regarding the effect of fully substituting inorganic nitrogen with organic nitrogen on the rhizosphere microenvironment of grafted watermelon. In particular, the variations in root exudate composition and rhizosphere microbial community diversity across different rootstocks remain poorly characterized (Hassnin et al., 2023; Ling et al., 2013). Therefore, given the potential of glycine as an organic nitrogen source and the central role of rootstock–scion interactions, we hypothesized the following:
1. Glycine application would improve soil health, plant growth, and bacterial diversity in grafted watermelon by serving as a preferential carbon and nitrogen source for beneficial rhizobacteria, thereby enhancing nutrient use efficiency and reducing environmental effect compared to inorganic fertilizers.
2. The selection of rootstock genotype would significantly modulate the root exudate profile, facilitating the recruitment of a functionally beneficial rhizosphere bacterial microbiome that enhances plant resilience and productivity under glycine nutrition.
3. The synergistic interaction between root exudates (as shaped by rootstock) and glycine-enriched rhizosphere bacterial microbiomes would be a principal driver sustaining improvements in soil health and plant performance.
This study aims to evaluate the rhizosphere microecological environment and nutrient uptake characteristics, thereby providing theoretical and technical support for practical watermelon production.
2 Materials and methods
The experiment was conducted in a sunlight greenhouse at the Horticultural Field of Northwest A&F University’s North Campus, Yangling Demonstration Zone (34°17′31.297′′ N, 108°4′26.310′′ E).
2.1 Preparation of materials and nutrient solution
The watermelon scion variety ‘8424’ was procured from Xinjiang Mingxin Kehong Co., Ltd. Rootstock varieties included ‘Warrior’ wild watermelon (Citrullus colocynthis), ‘Qinkang Shui Gua’ bottle gourd (Lagenaria siceraria), and ‘Jingxin No.2’ white-seed pumpkin (Cucurbita moschata Duchesne), sourced from Hangzhou Yihe Seedling Co., Ltd., Hunan Xuefeng Seed Industry Co., Ltd., and the Beijing Academy of Agricultural Sciences, respectively.
Seeds were sterilized and pre-germinated prior to planting (Wen et al., 2020). After germination, uniform seedlings were randomly sown into 50-cell trays. Grafting was performed 15 days post-sowing for scions and 20 days post-sowing for rootstocks using the splice-grafting method described by (Oda, 1995). Following grafting, as outlined by (Kacjan-Maršić and Osvald, 2004), grafted plants were maintained at 28°C with relative humidity above 95% for 3 days, followed by gradual ventilation and light exposure. After a 10-day graft union healing period, plants were transplanted into pots containing a substrate mixture of peat, vermiculite, and perlite at a 3:1:1 volume ratio (10 cm × 10 cm × 5 cm). Experimental treatments were carried out in a controlled climate chamber with a photosynthetic photon flux density (PPFD) of 200 μmol m⁻² s⁻¹, a 16 h light/8 h dark photoperiod, culture temperatures of 28°C (day)/18°C (night), and 60% relative humidity.
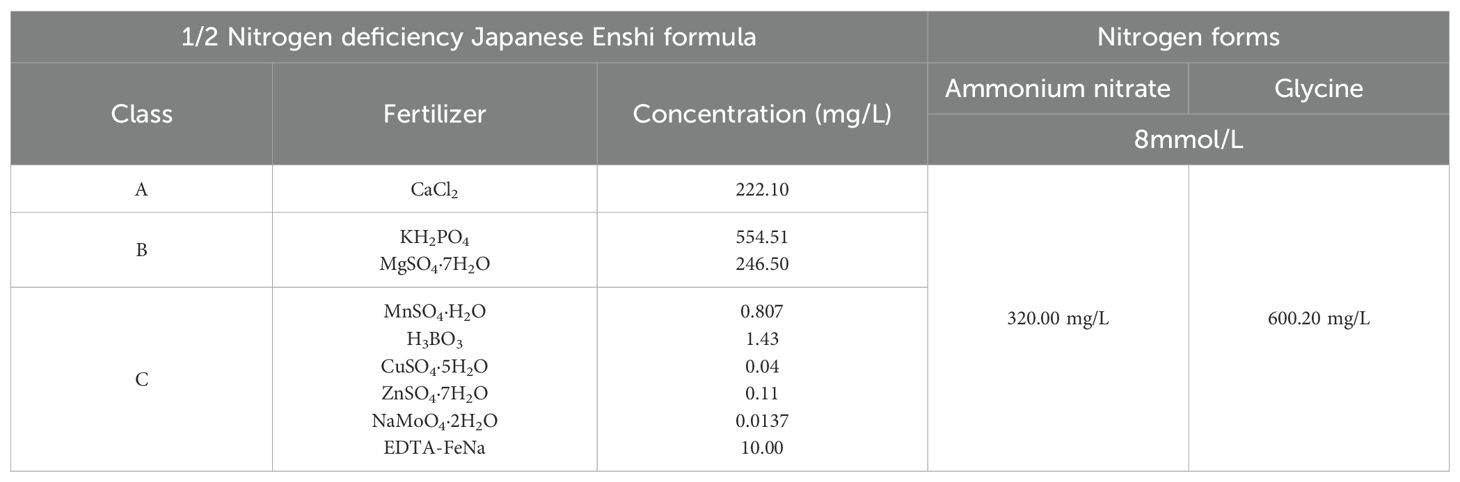
The nutrient solution was prepared based on a modified half-strength Japanese Enshi formula for horticultural crops, with all nitrogen sources omitted to create a nitrogen-deficient baseline. Organic and inorganic nitrogen sources were then supplemented, with ammonium nitrate serving as the inorganic source and glycine as the organic source. Details of the nutrient solution composition are shown in Table 1.
2.2 Experimental design and determination of physio-biochemical parameters
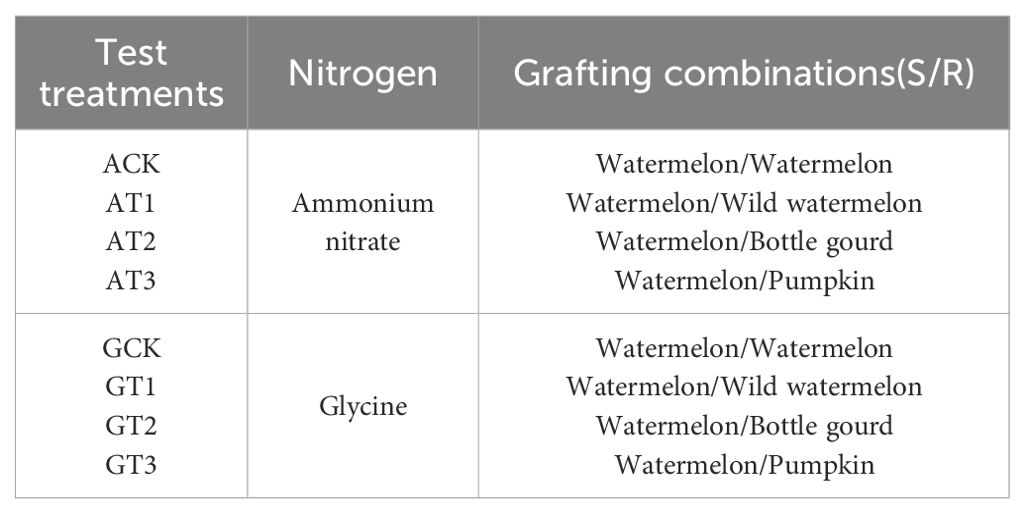
A randomized block design was employed, comprising four grafted rootstock treatments (watermelon, wild watermelon, bottle gourd, and pumpkin) and 2 nitrogen treatments (8 mmol/L ammonium nitrate and glycine), yielding 8 treatment combinations with nine plants each. Treatment details are presented in Table 2. The experiment spanned 25 days; each plant received 30 mL of nutrient solution daily and 100 mL of water every five days.
Upon completion of the cultivation period, plants and substrates were separated. Rhizosphere soil was collected by gently shaking substrate from the roots using the method described by (Wollum, 1994), immediately stored on ice, and sieved through a 2 mm mesh. Portions of the sample were air-dried for soil enzyme activity analysis.
The aboveground and belowground plant parts were separated, weighed, dried at 65°C for 48 hours, and then reweighed. All samples were collected in three replicates, each consisting of three plants (3 × 3 = 9).
A portable modulation chlorophyll fluorometer (Model PAM2500, USA) was employed to determine the fluorescence yield parameters of the leaves. For chlorophyll content determination, 0.2 g of fresh leaves (excluding main veins) was extracted with 5 mL of acetone/ethanol (2:1, v/v). Absorbance was measured at 663 nm and 645 nm (Pérez-Patricio et al., 2018), and chlorophyll content was calculated according to (Gu et al., 2016):
n the formula, A663 and A645 denote absorbance at 663 nm and 645 nm, respectively, with acetone/ethanol used as the calibration blank.
Air-dried substrate samples were used for soil enzyme activity measurement. Soil sucrase (S-SC) activity was measured using the sodium thiosulfate titration method (Gao et al., 2013); Urease activity was evaluated based on the concentration of the blue complex formed after treatment (Guo et al., 2012); Soil acid phosphatase (S-AP) activity was determined following the method of (Guan et al., 1986), utilizing toluene and disodium phenyl phosphate as assay reagents.
2.3 16S rRNA gene amplicon sequence processing
Upon completion of cultivation, plants and substrates were separated. Rhizosphere soil was collected by gently shaking the substrate from the roots using the method described by (Wollum, 1994), temporarily stored on ice, and immediately passed through a 2 mm mesh sieve. A portion of the sample was air-dried for soil enzyme activity assays, whereas another portion was stored at −80°C for microbial community analysis via 16S rRNA gene amplicon sequencing.
PCR amplification targeting the V4 region of the 16S rRNA gene was performed on extracted DNA samples using the 515F/806R primers (F: GTGCCAGCMGCCGCGGTAA; R: GGACTACHVGGGTWTCTAAT) (Walters et al., 2016). The reaction was conducted on a Bio-Rad T100 thermal cycler (Bio-Rad, USA) using the following protocol: each 30 μL reaction contained 15 μL Phusion Master Mix (2X; New England Biolabs), 0.2 μL each of forward and reverse primer (1 μM), approximately 10 ng template DNA, and ddH2O to final volume. Cycling conditions were 98°C for 10 min; 30 cycles of 98°C for 10 s, 50°C for 30 s, and 72°C for 30 s; followed by a final extension at 72°C for 5 min. PCR products were pooled in equimolar ratios according to concentration, mixed, and purified using 2% agarose gel electrophoresis. Target bands were recovered and further purified with a Qiagen Gel Extraction Kit (Qiagen, Germany) (Ahmed et al., 2021). Sequencing libraries were prepared using the TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, San Diego, CA, USA). Library quality was assessed with a Qubit® 2.0 Fluorometer (ThermoFisher Scientific, CA, USA) and Agilent Bioanalyzer 2100 (Agilent Technologies Inc., USA). Sequencing was performed on the Illumina NovaSeq 6000 platform.
For bacterial community analysis, raw 16S rRNA sequence data were quality-filtered using fastp (v0.19.6) (Chen et al., 2018), Paired-end reads were merged with FLASH v1.2.7, and clean data were further denoised in QIIME 2 using the Deblur algorithm (Knight et al., 2018) to generate high-quality amplicon sequence variants (ASVs). Taxonomic classification of ASVs was based on comparison to the SILVA reference database, with chimeric sequences removed (Haas et al., 2011). Microbial community α-diversity under different treatments was assessed by the Kruskal–Wallis test, whereas β-diversity differences between groups were evaluated using permutational multivariate analysis of variance (PERMANOVA). The Wilcoxon rank-sum test was applied to determine statistical significance of relative abundance changes at the ASV level between plant groups.
2.4 Metabolomic analysis
Separated plant root systems were thoroughly rinsed with deionized water and placed into shaded conical flasks containing 100 mL pure water for exudate collection. Following (Li et al., 2019), the collected root exudate solutions were incubated under light for 12 hours, filtered through a 0.22 μm membrane, freeze-dried (EPSILON 2–4 LSC, CHRIST, Germany), and stored at −80°C. Fresh leaves were collected at the same time.
Freeze-dried samples were reconstituted in 70% methanol containing an internal standard extraction solution at a 20× concentration ratio. The internal standard was prepared by dissolving 1 mg of standard in 1 mL of 70% methanol to yield a 1000 μg/mL stock solution, which was then diluted with 70% methanol to 250 μg/mL. After vortexing for 15 minutes and ice-water ultrasonication (KQ5200E) for 10 minutes, samples were centrifuged at 12,000 rpm and 4°C (5424R, Eppendorf) for 3 minutes. Supernatants were filtered through a 0.22 μm microporous membrane and stored in autosampler vials for subsequent UPLC–MS/MS analysis.
Sample preparation and metabolomics data processing were performed by Metware Biotechnology Co., Ltd. (Wuhan, China; https://www.metware.cn/). Instrumentation included an ultra-performance liquid chromatography (UPLC) system (ExionLCTM AD, http://sciex.com.cn/) and tandem mass spectrometry (MS/MS). Compound identification utilized the self-constructed Metware Database (MWDB) with secondary spectral information, removing isotope signals and redundant fragment ion signals from higher molecular weight compounds. Metabolite quantification was conducted using the multiple reaction monitoring (MRM) mode of triple-quadrupole mass spectrometry (Fraga et al., 2010).
LC–MS raw data were pre-processed for peak extraction, correction, metabolite identification, and annotation, followed by quality control (total ion chromatogram [TIC] overlap, coefficient of variation [CV] distribution plots, and CV filtering). High-quality data were then assessed by principal component analysis (PCA), clustering analysis, and repeatability correlation. Finally, statistical analysis (univariate and multivariate) was performed for functional prediction and interpretation of sample metabolites.
2.5 Statistical analysis
SPSS 27.0 (IBM Corp., USA) was used to perform one-way analysis of variance (ANOVA) and Duncan’s multiple range test. Adonis, namely PERMANOVA (Permutational multivariate analysis of variance), conducts multivariate analysis of variance with multiple factors. Differentially accumulated metabolites (DAMs) were identified based on two criteria: Variable Importance in Projection (VIP) score ≥ 1 and absolute value of log2(fold change) ≥ 1. Graphs were prepared using Origin 2024 (OriginLab Corp., USA).
3 Result
3.1 Effects of nitrogen source and rootstock on plant growth
The effects of the two nitrogen sources on grafted watermelon growth are illustrated in Figure 1. GCK exhibited a significant increase in both aboveground and belowground biomass compared to ACK, whereas no notable differences were observed among the other three rootstocks between the two nitrogen sources. Significant differences in biomass were found among the different grafting treatments using the same nitrogen source, with aboveground and belowground biomass increases ranging from 3.8% to 68.26% and 54.27% to 259.79%, respectively (Figures 1A, B). The T3 treatment recorded the highest aboveground biomass at 1.929 g, whereas T2 had the highest belowground biomass at 0.208 g.
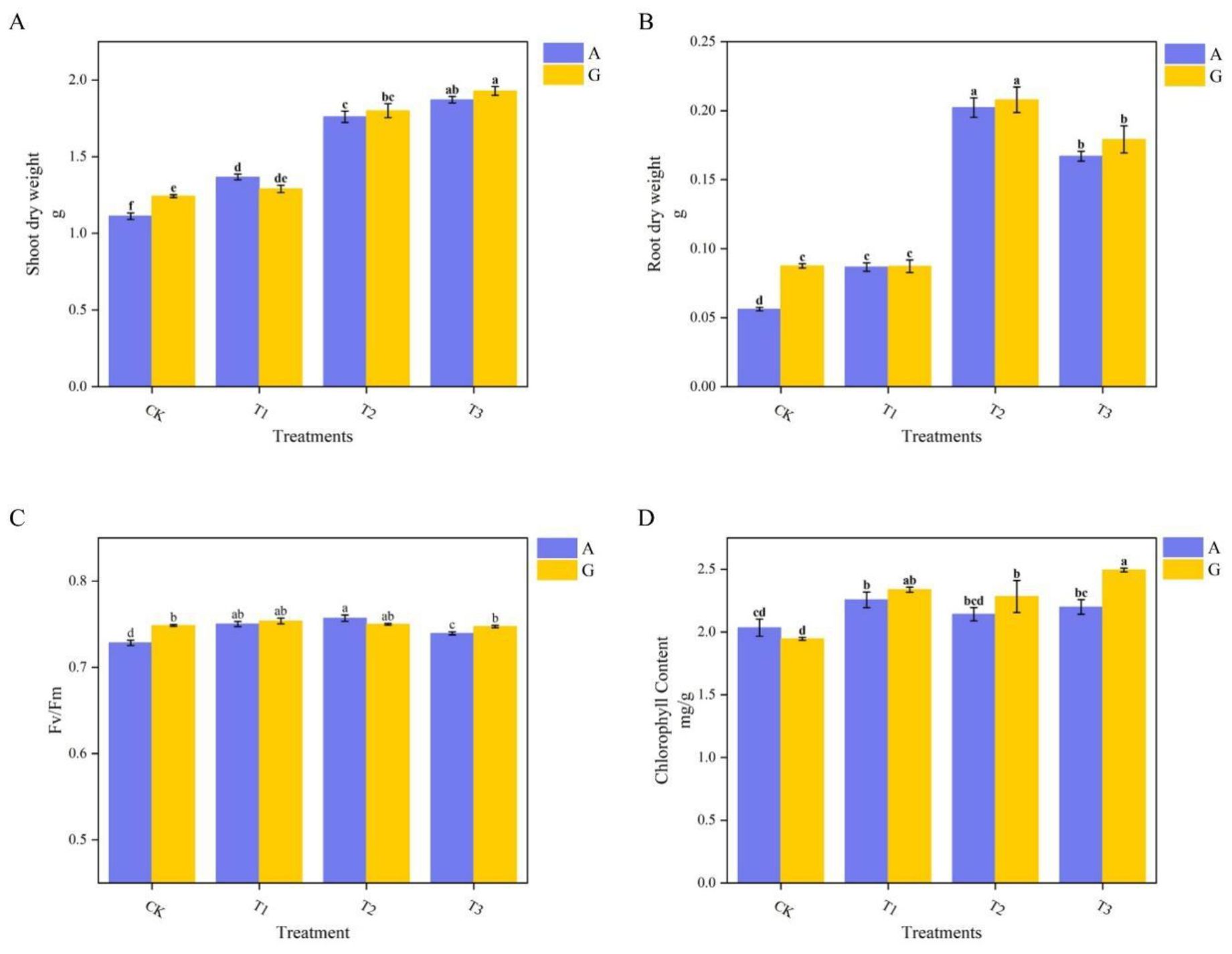
Figure 1. Growth parameters of grafted watermelon under different nitrogen treatments. (A) Aboveground dry weight; (B) Underground dry weight; (C) Chlorophyll fluorescence parameters (PSII maximum photochemical quantum yield, Fv/Fm); (D) Chlorophyll content.
Under glycine treatment, no significant differences in the maximum quantum yield of PSII photochemistry (Fv/Fm) were observed among the various grafted rootstocks, with all values approximately 0.75, which is within the normal physiological range. In contrast, under ammonium nitrate treatment, there were no differences between the T1 and T2 grafting treatments, whereas CK and T3 exhibited significant declines in Fv/Fm, with values of 0.728 and 0.739, respectively. Compared with the glycine treatment, T3 and CK displayed significant declines in Fv/Fm by 2.7% and 1.1% under ammonium nitrate (Figure 1C).
For chlorophyll content, significant changes were noted between ACK and the AT1 and AT3 treatments under the same nitrogen conditions, whereas no differences were observed among the other grafting treatments. GT3 had the highest chlorophyll content at 2.495 mg/g, whereas GCK showed the lowest at 1.947 mg/g. Across nitrogen types, no significant differences in chlorophyll content were found among the other three grafting treatments; however, GT3 exhibited chlorophyll content that was 11.8% higher than that under ammonium nitrate (Figure 1D).
According to Table 3, Adonis analysis was conducted on the above four plant growth indicators. The nitrogen source did not show significant differences, while the types of rootstocks showed extremely significant differences.

Table 3. Adonis analysis of plant growth under different nitrogen sources and rootstocks; SumsOfSqs, Sum of Squares; MeanSqs, Mean square; F.Model, F value.
3.2 Effects of nitrogen source and rootstock on soil enzyme activities
Soil urease (S-UE) activity (Figure 2A) was highest in AT3 soil, reaching 993.23 U/g, and both T3 treatments (across nitrogen sources) demonstrated higher enzyme activities than the other grafting treatments. The lowest urease activity was recorded in GCK soil (698.39 U/g). Urease activities in GCK and GT3 soils were significantly reduced by 9.04% and 7.03%, respectively, compared with ACK and AT3. No significant differences were found between T1 and T2 for either nitrogen source.
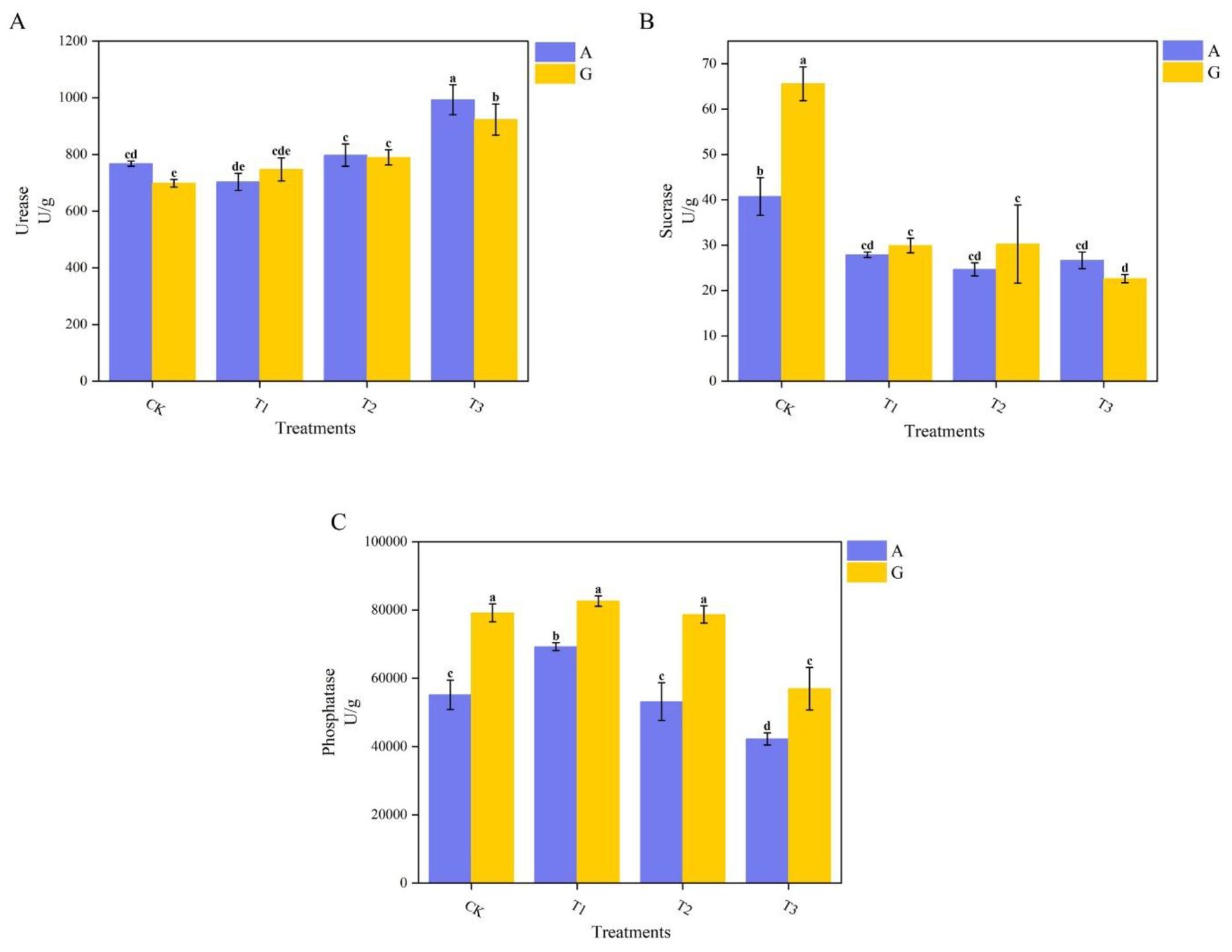
Figure 2. Enzyme activity in the rhizosphere. (A) Urease (S-UE); (B) Sucrase (S-SC); (C) Acid phosphatase (S-AP). Error bars represent standard errors of the mean. Color bars indicate nitrogen source: Ammonium Nitrate (blue), Glycine (yellow).
For S-SC and acid phosphatase (S-AP) activities (Figures 2B, C), enzyme activities were generally higher in glycine treatments compared to ammonium nitrate. Specifically, CK exhibited higher S-SC activity than the other grafting treatments, with the highest activity observed in GCK at 65.60 U/g, representing a significant increase of 61.01% over ammonium nitrate treatment. GT3 had the lowest S-SC activity. S-AP activity (Figure 2C) differed significantly (P < 0.05) among nitrogen levels, with GT1 showing the highest activity and AT3 the lowest, at 82,638.71 U/g and 42,239.21 U/g, respectively. Among the top three grafting treatments in the glycine group, no significant differences were found, nor between ACK and AT2. T3 exhibited the lowest S-AP activity across both nitrogen sources.
According to Table 4, Adonis analysis was conducted on the enzymatic activities of the above three types of soil enzymes. The nitrogen source did not show significant differences, while the type of rootstock showed extremely significant differences.

Table 4. Adonis analysis of soil enzyme activities under different nitrogen sources and rootstocks; SumsOfSqs, Sum of Squares; MeanSqs, Mean square; F.Model: F value.
3.3 Nitrogen source and rootstock effects on rhizosphere bacterial community composition
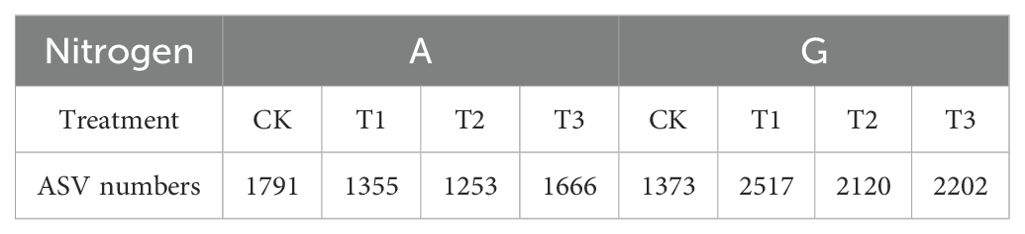
Analysis of the rhizosphere bacterial communities from all treatments yielded a total of 2,540,622 high-quality sequences, with an average of 105,859 reads per sample (range: 64,765–119,919 reads), resulting in the identification of 12,790 ASVs. Diversity indices were subsequently calculated. The rarefaction curve (Figure 3) showed that at sequencing depths between 5,000 and 40,000 reads, the curve plateaued, indicating sufficient sequencing coverage to capture the majority of sample diversity. These results suggest that the sequencing data robustly represents the ASV diversity in the soil samples, with specific ASV counts detailed in Table 5.
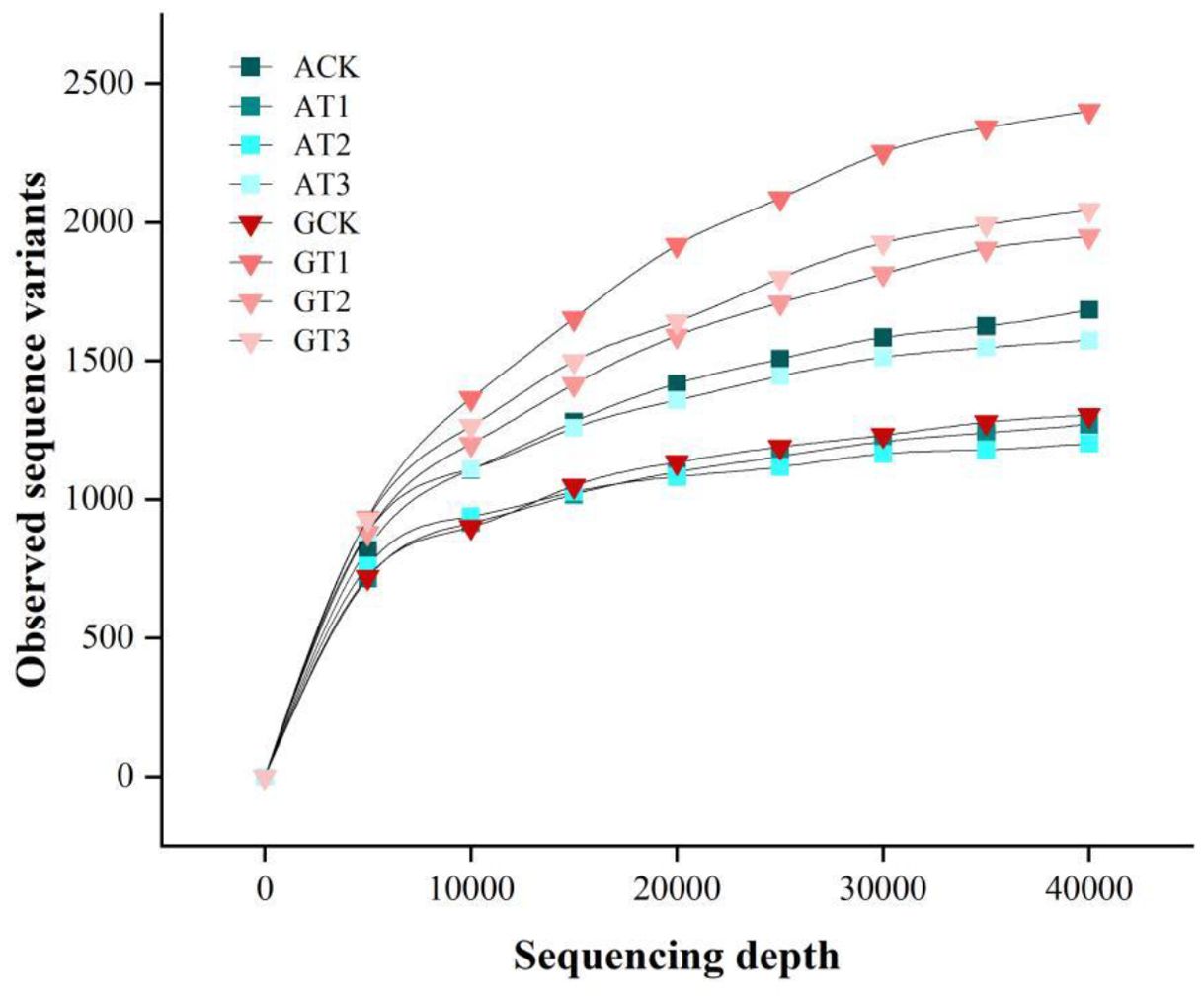
Figure 3. Rarefaction curves of ASV numbers obtained using the Deblur plugin at different non-chimeric read counts (data are randomly subsampled to demonstrate increasing sequence numbers).
Further analysis revealed the diversity and structure of the bacterial community across treatments (Figure 4). Principal coordinate analysis (PCoA) of Weighted UniFrac distances (Figure 4A; adonis, R² = 0.644, P = 0.001) explained over 50% of the total variance, demonstrating the reliability of the data and the significant effect of glycine substitution for ammonium nitrate. The Manhattan plot (Figure 4B) of G treatment samples highlighted enrichment of several phyla, including Acidobacteriota, Actinobacteria, Armatimonadota, Bdellovibrionota, Gemmatimonadota, Myxococcota, Planctomycetota, Proteobacteria, and Bacteria.
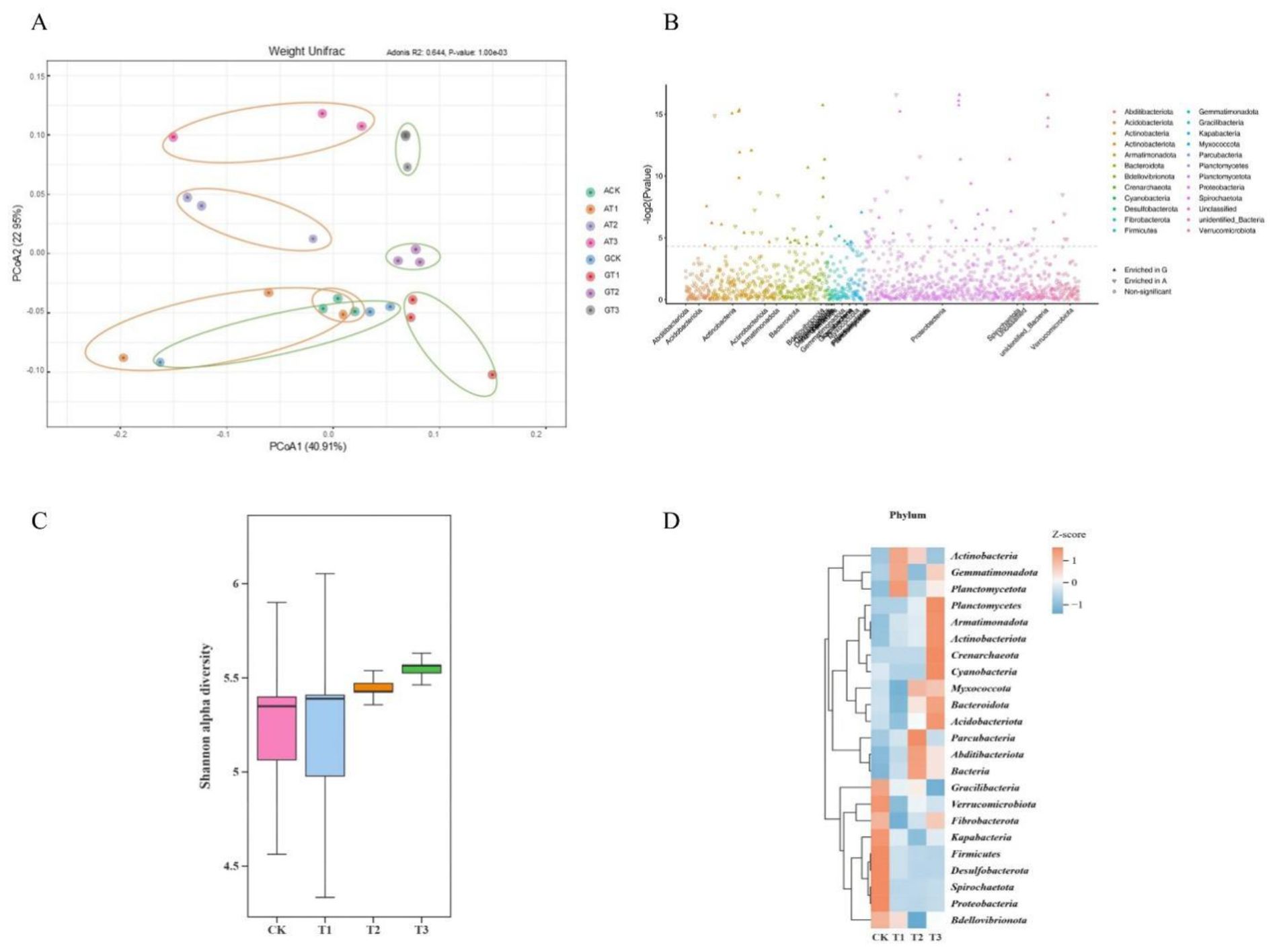
Figure 4. Bacterial community composition in rhizosphere soils under glycine versus ammonium nitrate. (A) PCoA of ASVs; (B) Manhattan plot highlighting enriched rhizobacterial phyla among differential ASVs (triangles denote differential ASVs, circles denote non-significant ASVs, P < 0.05, Wilcoxon rank-sum test); (C) Shannon index of α−diversity; (D) Heatmap of the relative abundance of differential ASVs among rootstocks under glycine application. The heatmap uses normalized z-scores for relative ASV abundance.
Within the G treatment, no significant differences in the Shannon index were observed among rootstocks (Figure 4C). However, Table 3 indicates that GCK exhibited a significantly lower ASV count than the other rootstock treatments, with GT1 exhibiting the highest count (2,517). Phylum-level heatmap analysis (Figure 4D) showed rootstock-specific enrichment: GCK (Proteobacteria, Spirochaetota, Desulfobacterota, Firmicutes, etc.), GT1 (Planctomycetota), GT2 (Parcubacteria), and GT3 (Actinobacteriota, Acidobacteriota, Armatimonadota, Crenarchaeota, Cyanobacteria, Planctomycetota, etc.).
Genus-level differential analysis was performed for each rootstock, selecting the ten most abundant genera (Figure 5A). Mitsuaria showed a relative abundance exceeding 50% in CK, whereas Acidibacter was more abundant in CK and T1. Paenarthrobacter was more prevalent in T1, T2, and T3. Edaphobaculum and Sphingomonadaceae reached their highest relative abundance in T3, with Sphingomonadaceae nearly absent in both watermelon rootstocks. LEfSe analysis identified significant differences in predicted functional gene profiles among rootstock groups, with T1 possessing the most differentially abundant genes and T2 exhibiting none (Figure 5B). Clustering analysis of the top 20 KEGG pathways (Figure 5C) showed that ABC transporters and secretion systems were enriched in CK, whereas the two-component system, glyoxylate and dicarboxylate metabolism, pyruvate metabolism, and transcription factors were enriched in T1. Oxidative phosphorylation, tRNA biogenesis, and mitochondrial biogenesis were enriched in T3. Purine metabolism was variably expressed in T1, T2, and T3.
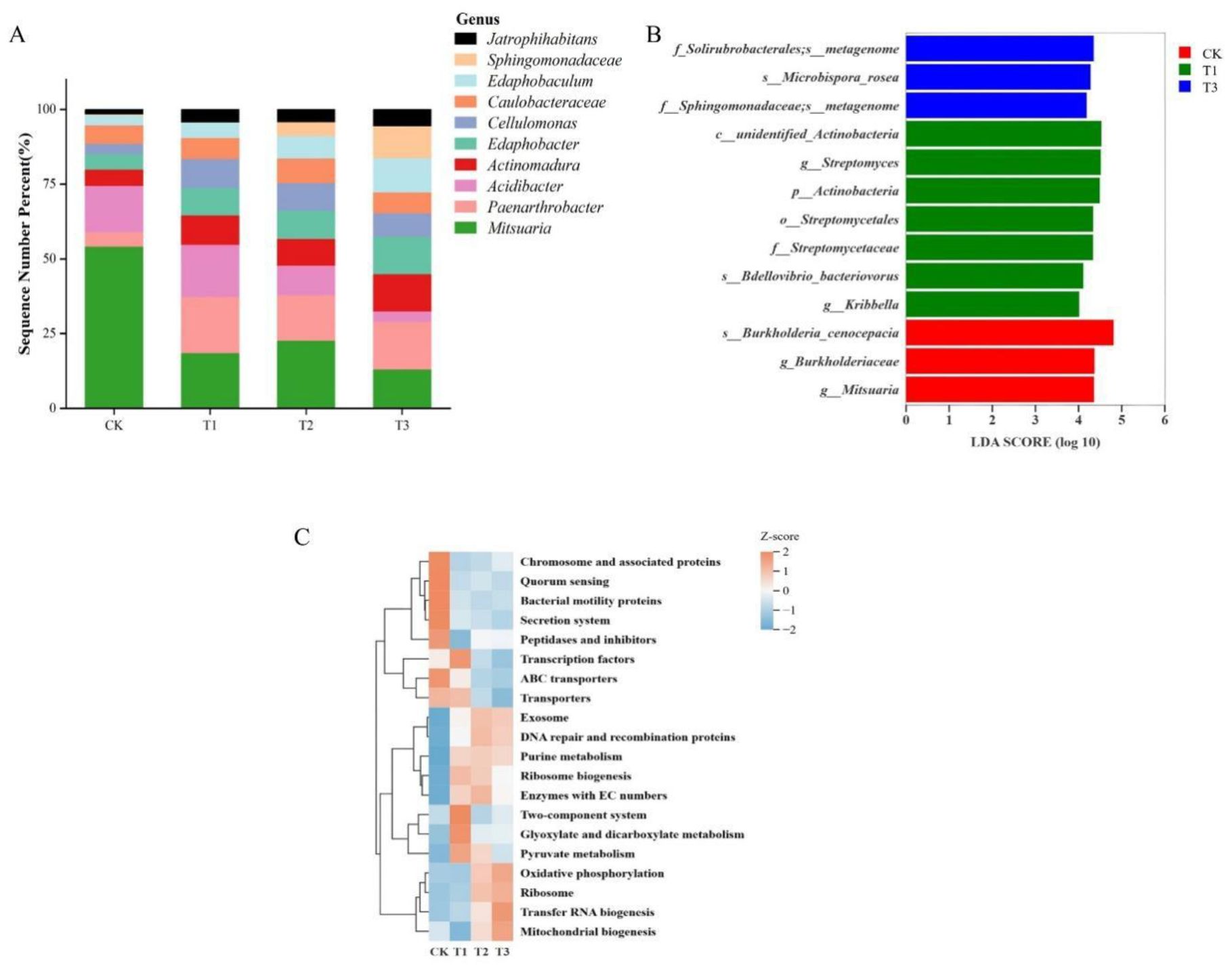
Figure 5. Differences in rhizosphere bacteria among rootstocks after glycine application. (A) Top 10 genera by relative abundance in each rootstock; (B) Differential PICRUSt functional gene predictions; (C) Heatmap of predicted functional pathways.
Network analysis, based on significant genus-level correlations (Spearman’s correlation, P < 0.05) (Figure 6), was performed to assess topological network properties and potential key associations in the rhizosphere community in response to nitrogen form (Figures 6A, B). Both node connectivity and network density were lower under ammonium nitrate than under glycine, indicating a less complex network structure in A compared to G (Table 6).
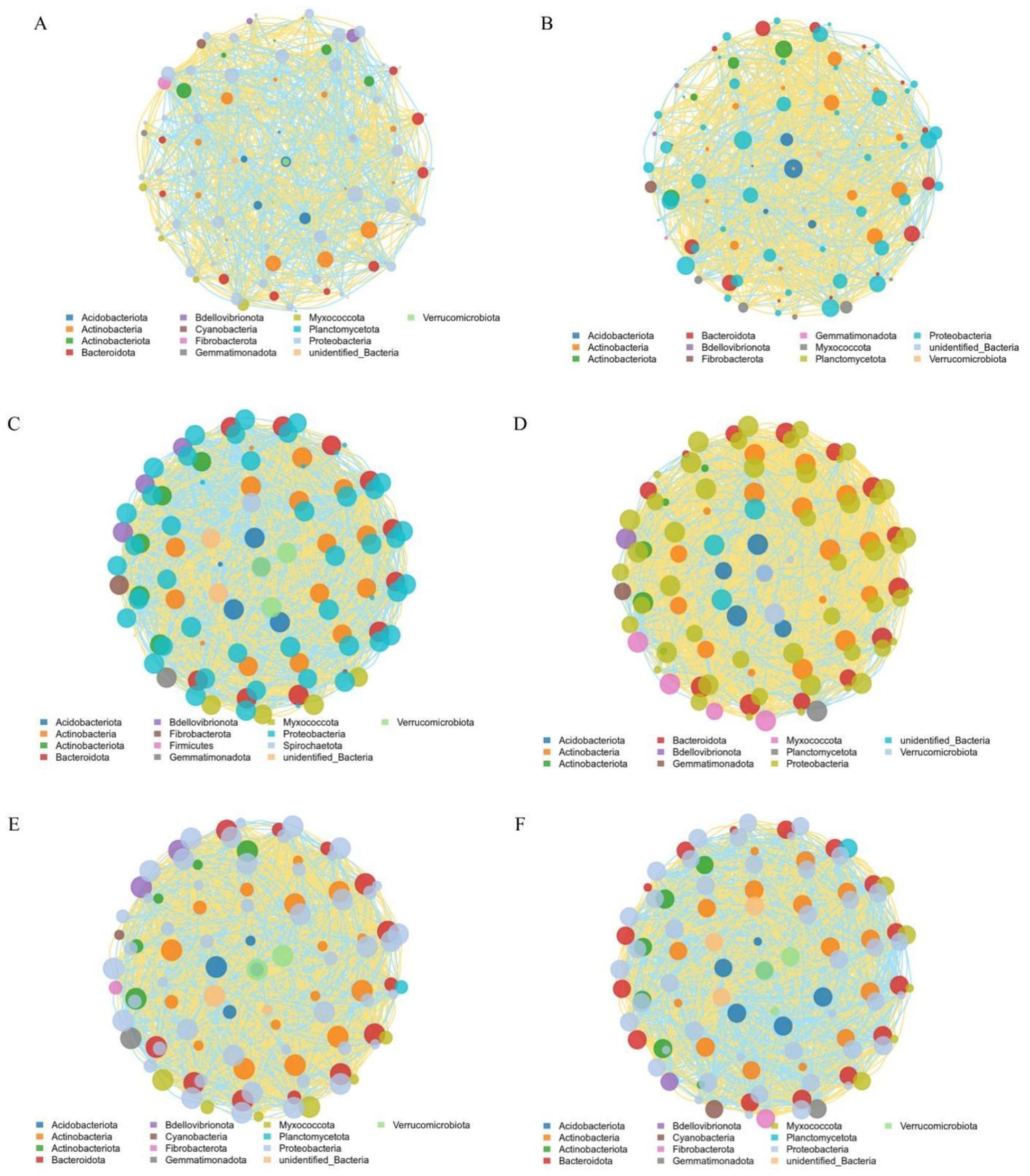
Figure 6. Co-occurrence networks of the top 100 rhizosphere bacterial genera under different nitrogen forms and rootstocks. (A) Ammonium nitrate; (B) Glycine; (C–F) Rootstock-specific networks. Nodes represent genera, colored by phylum; node size corresponds to connection degree; (C): GCK, (D): GT1, (E): GT2, (F): GT3.

Table 6. Topological characteristics of bacterial co-occurrence networks under different nitrogen sources.
Subsequently, we performed an Adonis analysis to test the effects of nitrogen source and rootstock as the two factors (Table 7). Nitrogen sources showed no significant difference, but rootstock type had a highly significant effect.

Table 7. Adonis analysis of different nitrogen sources and rootstocks; SumsOfSqs, Sum of Squares; MeanSqs, Mean square; F.Model, F value.
Within the glycine treatment (Figures 6C–F), CK displayed the highest average degree, node interconnectivity, and network density. The positive correlation ratio in CK was 50.18%, whereas T1 showed the highest ratio at 69.81%, representing a 27–40% increase over the other rootstocks. T2 and T3 exhibited highly similar patterns, with T2 displaying a slightly higher positive correlation ratio (Table 8).
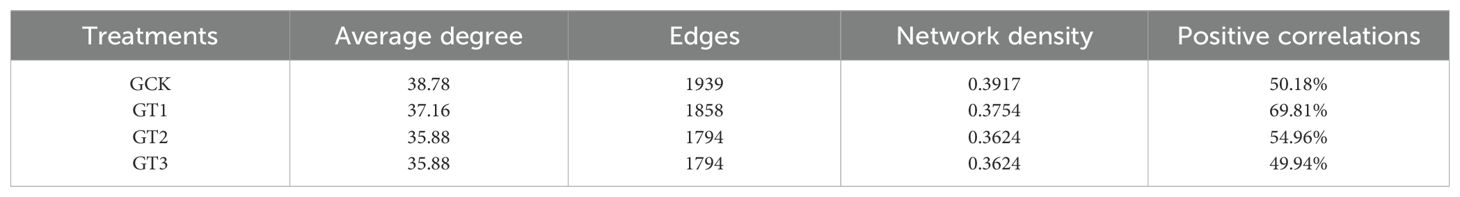
Table 8. Topological characteristics of bacterial co-occurrence networks under different rootstocks.
3.4 Metabolite profiles of roots from different grafted rootstocks
UPLC–MS/MS analysis identified 442 compounds in root exudates across all samples, with amino acids constituting the most abundant class (36.20%), followed by lipids (21.95%), organic acids (15.38%), and nucleotides (11.54%) (Figure 7A).
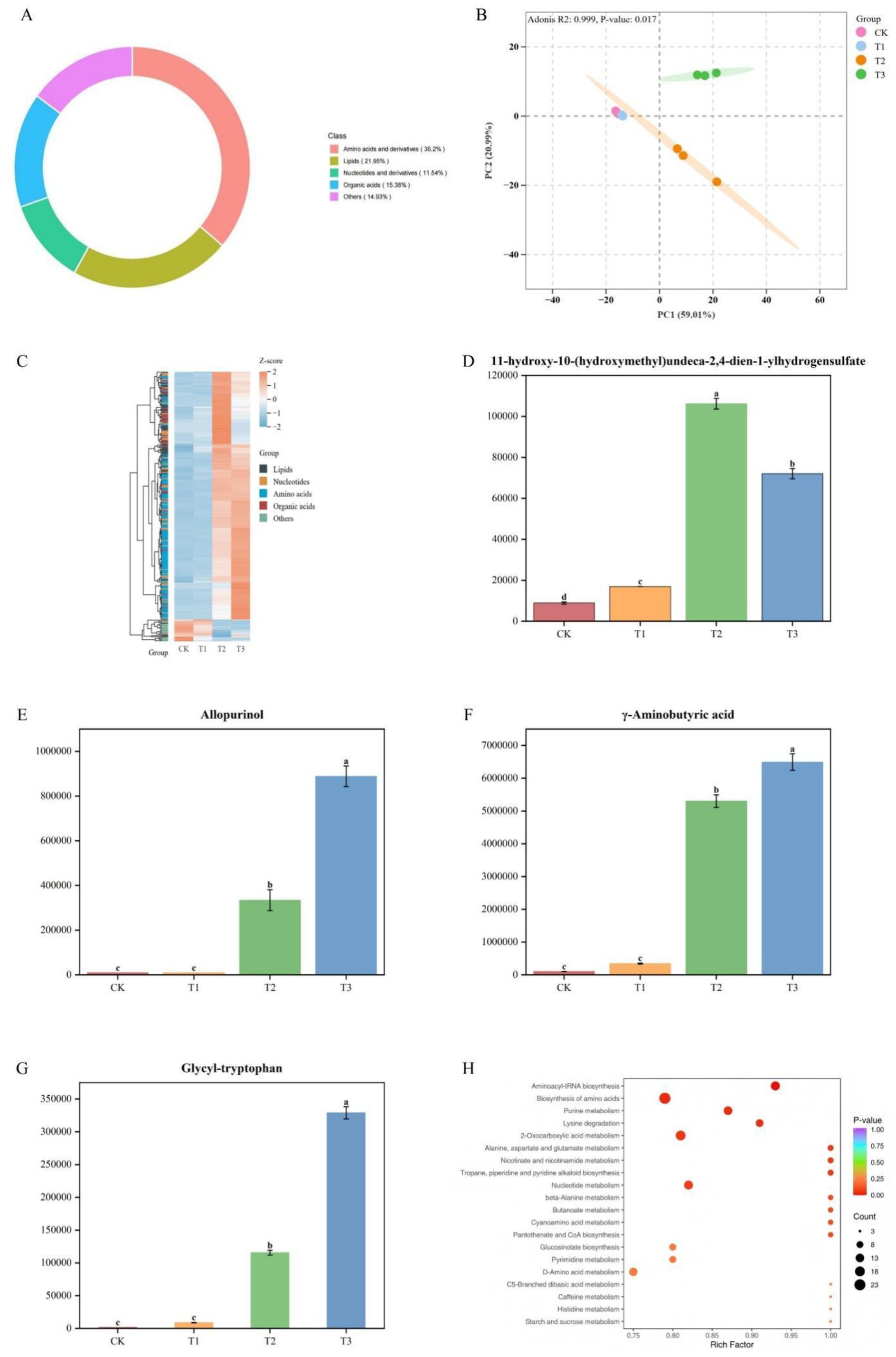
Figure 7. (A) Classification of root exudate compounds; (B) PCA of root exudates under glycine; (C) Hierarchical clustering of differential metabolites among rootstocks; (D–G) Representative metabolite concentrations in the four rootstocks; (H) Enriched metabolic pathways of root exudates. Each dot in (H) represents a metabolic pathway, with size reflecting the degree of change.
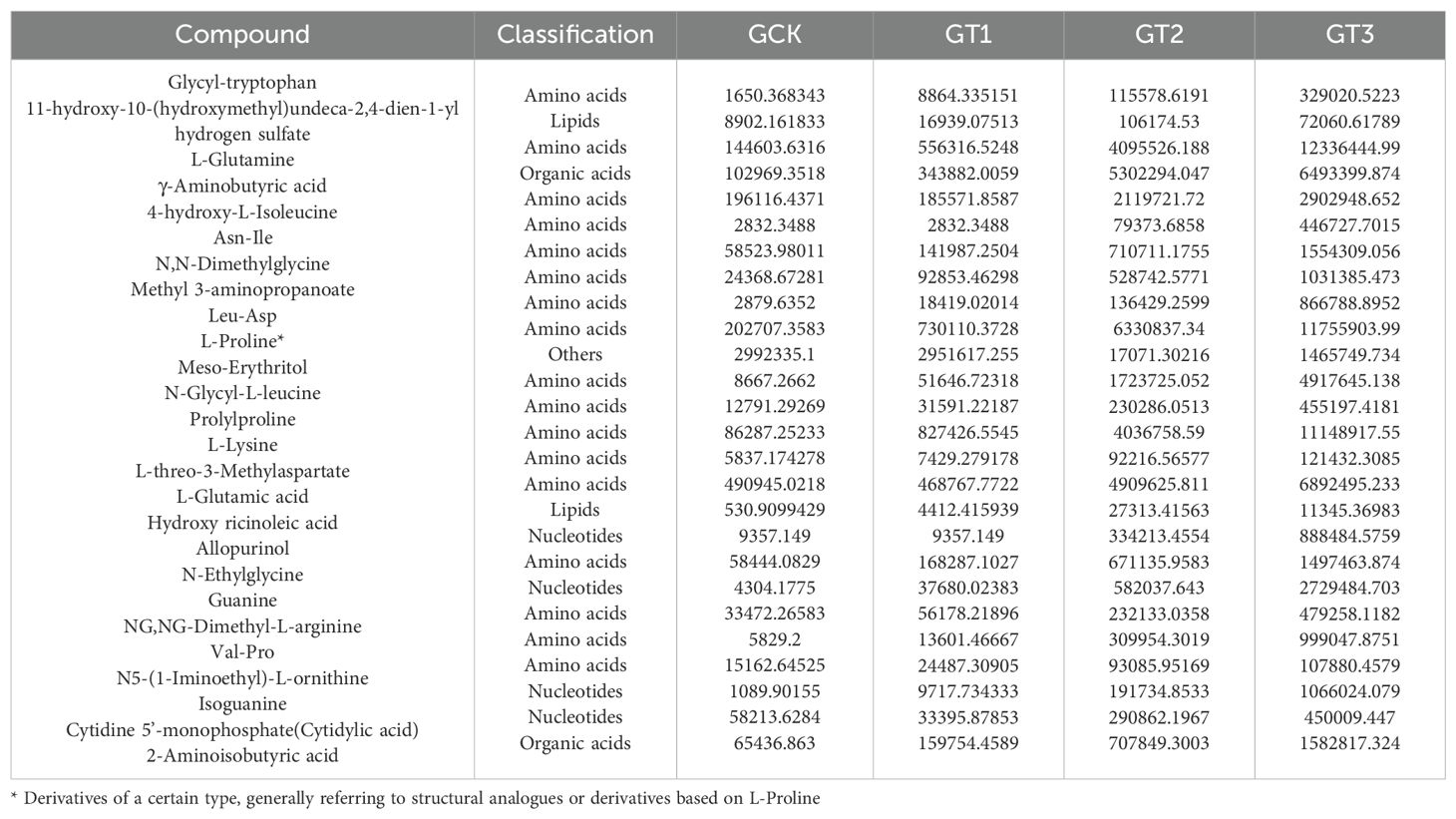
Based on differential metabolite analysis among rootstocks under glycine treatment, 261 significant metabolites were identified (VIP > 1 and P < 0.05). PCA (Figure 7B) revealed that the first two principal components accounted for 80% of the variance in root exudate composition among the four rootstocks. The results indicated minimal differences between CK and T1, whereas T2 and T3 displayed pronounced metabolic divergence from the other treatments. Hierarchical clustering (Figure 7C) confirmed this pattern, with T3 exhibiting the largest number of differential metabolites and T1 the fewest. The 26 metabolites with the smallest P-values (top 10% of 261; Table 9) were defined as highly significant. Of these, four representative metabolites—Glycyl-tryptophan, γ-aminobutyric acid, Allopurinol, and 11-hydroxy-10-(hydroxymethyl) undeca-2,4-dien-1-yl hydrogen sulfate—were selected for comparative analysis (Figures 7D–G). Glycyl-tryptophan, γ-aminobutyric acid, and Allopurinol were significantly more abundant in T3 than in other rootstocks, with no significant difference between the two watermelon rootstocks. In contrast, 11-hydroxy-10-(hydroxymethyl) undeca-2,4-dien-1-yl hydrogen sulfate was most abundant in T2, and CK had the lowest levels for all four metabolites.
Pathway enrichment analysis of root exudates under glycine treatment identified several significantly enriched metabolic pathways, including aminoacyl-tRNA biosynthesis, biosynthesis of amino acids, purine metabolism, lysine degradation, and 2-oxocarboxylic acid metabolism (Figure 7H; Wilcoxon test, p < 0.05).
3.5 Correlation between rhizosphere bacteria and root exudates
Redundancy analysis (RDA) revealed that the first two RDA axes together explained 95.34% of the variation in the association between metabolites and bacterial taxa, with RDA1 accounting for 95.13% (Figure 8A), indicating that RDA1 is the primary dimension distinguishing the association between metabolites and bacterial communities across rootstocks. Among all metabolites, lipids, nucleotides, and amino acids had substantial effects on the composition of rhizosphere bacterial genera, suggesting that these metabolite classes are key regulators of microbial communities through rootstock-mediated secretion. Acidibacter was positively correlated with lipids but negatively correlated with most other metabolites. Conversely, Edaphobaculum, Edaphobacter, Actinomadura, and several metabolite classes—including amino acids, nucleotides, and organic acids—were strongly positively correlated. Caulobacteraceae and Cellulomonas also showed positive associations. These results suggest that Edaphobacter and Actinomadura are core genera associated with amino acid and nucleotide regulation, driving the metabolic characteristics of bacteria across different rootstocks. Correlation heatmap analysis (Figure 8B) identified significant associations between 7 bacterial taxa and 22 metabolites. Acidibacter showed negative correlations with 21 metabolites (11 highly significant). Edaphobacter and Actinomadura were extremely significantly positively correlated with 20 and 16 metabolites (excluding lipids), respectively. Sphingomonadaceae, Jatrophihabitans, Edaphobaculum, and Paenarthrobacter were positively correlated with all 22 metabolites, with highly significant positive correlations for 6, 21, 18, and 20 metabolites, respectively.
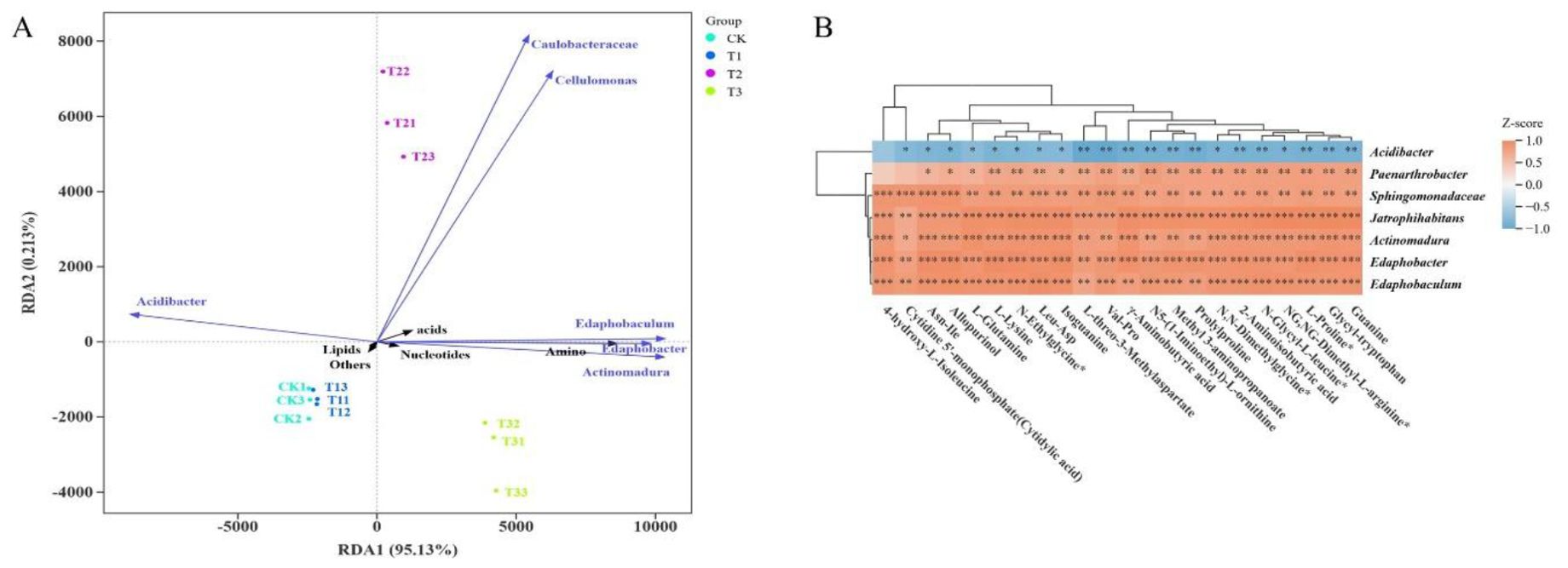
Figure 8. Correlation analysis of differential bacteria and metabolites among different rootstocks under glycine. (A) RDA; an angle less than 90° between two variables indicates a positive correlation, whereas an angle greater than 90° indicates a negative correlation. (B) Correlation heatmap of microorganisms and metabolites across four rootstocks.
Collectively, these analyses indicate that primary metabolic pathways promote the abundance of Edaphobacter and Actinomadura, and that four other genera also play critical roles in the positive regulation of metabolite dynamics. Under glycine administration, rootstocks selectively regulate metabolite distribution, which in turn shapes the structure of the rhizosphere microbial community.
4 Discussion
4.1 Glycine modulates soil enzymes and rootstock performance
Soil enzyme activity is frequently utilized as an indicator of microbial growth. S-UE plays a critical role in the nitrogen cycle (Tamaddon et al., 2020), and provides nitrogen nutrients to plants (Lee et al., 2021). The higher S-UE in treatment A compared to treatment G may be attributed to the rapid conversion of ammonium nitrogen and the subsequent release of nitrogen, which stimulates the production of urease or urease-associated microorganisms. Previous research has shown that many tested species preferentially absorb inorganic nitrogen (Zhuang et al., 2020). The application of two forms of nitrogen resulted in different microbial community structures, leading to varied effects on soil enzymatic activities. Additionally, the extensive root system and enhanced nitrogen metabolism capacity of pumpkin resulted in the highest enzyme activity observed. S-SC can convert oligosaccharides in soil into monosaccharides needed by plants and facilitate the circulation of organic nitrogen in soil (Farooq et al., 2021). Although S-SC activity in treatment G was greater than that in treatment A, the difference was not significant. The application of glycine provided not only a nitrogen source but also a carbon source, thereby altering the C/N ratio and affecting the production of sucrase. This is consistent with the findings of (Zeng et al., 2024), which demonstrated that glycine promotes sucrase activity. The higher enzyme activity in CK compared to the other grafting treatments may be attributed to compatibility issues between the scion and the rootstock, as (Wang et al., 2024) reported that heterografting can suppress the gene expression of enzymes related to sucrose metabolism. Like S-SC, the application of glycine supplied both carbon and nitrogen. Furthermore, glycine reduces the pH of the substrate, leading to lower S-AP activity in ammonium nitrate treatments. Therefore, the application of glycine and the combination of different rootstocks significantly changed the biological activity of soil enzymes, and the interaction among these enzymes was also a key factor in modifying the soil environment (Zhou et al., 2025).
Regarding biomass, bottle gourd and pumpkin demonstrated advantages, likely due to their robust root systems (Sallaku et al., 2022) or their strong adaptability, which supports better growth of watermelon (Xiong et al., 2021), ultimately increasing both aboveground and underground biomass. Overall, Fv/Fm and chlorophyll content in certain grafting treatments of treatment A significantly decreased compared to the normal levels in treatment G. The application of exogenous glycine promotes the production of endogenous compounds in plants, such as the glycine-rich protein-encoding gene GhGRPL; its upregulation enhances plant stress tolerance by thickening secondary cell walls (Yu et al., 2024) This finding is consistent with (Xu et al., 2022), which demonstrated that glycine enhances plant tolerance, helping maintain various physiological parameters at normal levels.
Based on the observed changes in S-UE, S-SC, and S-AP activities induced by glycine, suppose that glycine might can reduce the demand for nitrogen metabolism, alter the nutrient limitation pattern (Liu et al., 2022). The coordinated optimization of the photosynthetic system and the increase in plant biomass following glycine application indicate that glycine can enable efficient conversion of photosynthetic carbon assimilation to biomass by increasing light capture capacity and maintaining photochemical reaction efficiency. Although glycine has advantages as described above, the differences among rootstock types are much greater than those among nitrogen sources. Therefore, the combination of rootstock and glycine is particularly important (Tables 3, 4).
4.2 Glycine modulates rootstock-specific microbial networks
Plant-associated microbial communities play a critical role in plant health and environmental adaptation in natural ecosystems (Trivedi et al., 2021). The exogenous application of different nitrogen sources (Gallart et al., 2018) can shape diverse microbial community structures to help plants cope with environmental stresses or biotic challenges (de Vries et al., 2020; Gao et al., 2024b). IIn this study, glycine was applied as a substitute for ammonium nitrate, and rootstock replacement via grafting further modified rhizosphere microbes to enhance plant resilience. These findings support the optimization of watermelon cultivation practices and contribute to agricultural environmental sustainability. At all rarefied sampling depths (Table 5), except for the GCK treatment, glycine treatments exhibited higher ASVs, indicating that the application of glycine has the potential to promote bacterial diversity. Significant changes in β-diversity were observed in the rhizosphere (Figure 4A). Differential analysis using a Manhattan plot (Figure 4B) of bacterial communities shaped by the two nitrogen sources revealed that glycine substitution led to the enrichment of several phyla. For example, in GCK, Proteobacteria (Yang et al., 2024) exhibit rapid carbon turnover; in GT1, Planctomycetota (Klimek et al., 2024) and in GT2, Parcubacteria (Fujii et al., 2022) demonstrate graded utilization of labile and recalcitrant carbon. In GT3, Actinobacteriota and Acidobacteriota (Camargo-Montoya et al., 2025; Chen et al., 2021; Palaniyandi et al., 2013) enhance environmental adaptation and participate in biological nitrogen fixation. Previous studies (Luo et al., 2018) have demonstrated that different forms of nitrogen influence unique soil microbial compositions. Similarly, (Morais et al., 2024) found that grafting can alter a plant’s rhizosphere microbiome. Therefore, four common grafting combinations—Watermelon/Watermelon, Watermelon/Wild watermelon, Watermelon/Bottle gourd, and Watermelon/Pumpkin—were used to determine optimal scion-rootstock utilization for watermelon.
For the G treatment analysis, no significant difference was observed in the Shannon index (Figure 4C), although numerical differences were present. This may be due to the long-term response required for microbial communities to adapt to environmental changes (Davis et al., 2005), suggesting that the duration of this study may have been insufficient to capture such shifts. Cluster heatmap analysis of the four rootstocks (Figure 4D), together with differential analysis between the two nitrogen sources (Figure 4B), revealed that the significant phyla influenced by glycine were primarily present in CK, T1, and T3, as shown by the LefSe analysis (Figure 5B) of significantly different functional genes in the four rootstock microbiomes; T2 treatment exhibited no differential genes. The top ten differential microbes from the four rootstocks were analyzed. Mitsuaria was enriched by over 50% in CK, whereas the relative abundance of Paenarthrobacter was significantly lower than in other treatments. Sphingomonadaceae was enriched in the non-watermelon rootstocks T2 and T3. CK plants may recruit more Mitsuaria due to their weaker root systems, which supports nutrient absorption and helps plants withstand environmental challenges (Huang et al., 2017; Marian et al., 2018). Meanwhile, Paenarthrobacter has been identified as a degrader of organic matter, enhancing plant growth, productivity, and antioxidative capacity (Ghasemi et al., 2018; Liu et al., 2024). The recruitment of Mitsuaria in CK may have taken precedence; as shown in Figure 5B, the Mitsuaria gene in CK is significantly expressed, partially replacing the role of Paenarthrobacter and resulting in less enrichment. Sphingomonadaceae possess strong degradation capabilities, increasing plant resistance to pathogens and stress (Sultana et al., 2023; Zhou et al., 2023). Functional predictions using PICRUSt revealed significant differences in Level 3 pathways contributed by rootstock bacteria (Figure 5C).
The microbial network analysis indicated that substituting ammonium nitrate with glycine increased bacterial interactions (Figures 6A, B; Table 6). Adonis analysis (Table 7) revealed no significant difference between nitrogen sources but a highly significant difference among rootstocks (p < 0.01). Based on these results, we shifted our focus to glycine’s effects on different rootstocks. Comparisons among rootstocks showed that interactions among bacteria in wild watermelon rootstocks were stronger (Table 8) and nearly self-compatible, consistent with the findings of (Gu et al., 2024). Correlation analysis of the top 100 bacteria demonstrated that T3 treatment exhibited the highest rate of negative correlations. Studies suggest that ecological competition through negative regulation among bacteria can enhance microbial community stability (de Vries et al., 2020), benefiting plants by competing against external stresses (Hernandez et al., 2021). The high positive correlation ratio of T1 may be due to the abundance of functional genes (Figure 5B) and a well-developed two-component system (Figure 5C), resulting in enhanced environmental perception and mobilization of the microbial community, revealing a strong synergistic symbiosis induced by the rootstock. The network structure of T3 is similar, but its positive correlation ratio is not high because its energy metabolism (Figures 5B, C) is strengthened, leading to intensified resource competition and better buffering capacity under stress. CK has high ABC transporter gene expression (Figure 5C) and dominates nutrient competition.
Compared with ammonium nitrate application, although glycine treatments did not increase microbial α-diversity, it did show the specific enrichment of functional groups across different rootstocks. A more detailed analysis and functional prediction of the bacterial genera in each glycine treatment, combined with the bacterial network structure, reflect the influence of rootstocks on the energy distribution and nutrient acquisition strategies of microorganisms. These results, together with the decrease in soil enzyme S-UE activity and the increase in S-SC and S-AP activities, enhanced photosynthesis, and increased biomass. This demonstrates the adaptive division of labor of microorganisms in the soil–plant positive feedback system promoted by glycine application and rootstock optimization.
4.3 Rootstock-specific metabolites steer rhizosphere microbiome functions
Root exudates directly regulate the activity, abundance, and functional composition of rhizosphere microbial communities (Wen et al., 2020). Differential metabolite analysis under glycine and ammonium nitrate application identified significant changes in 261 of 442 metabolites. PCA of glycine-treated samples (Figure 7B) revealed minimal differences between CK and T1, both watermelon rootstocks, whereas T2 and T3 exhibited pronounced metabolic divergence, suggesting a rootstock effect on root exudate profiles. Hierarchical clustering (Figure 7C) further highlighted substantial metabolic differences between watermelon rootstocks and those of gourd and pumpkin. Notably, three of the four most differentiated metabolites were highest in pumpkin rootstock (T3; Figures 7D–G), and these metabolites serve defensive, adaptive, or buffering roles. For instance, γ-aminobutyric acid (GABA) not only mitigates stress under adverse conditions but also functions as a signaling molecule (Kaspal et al., 2021). Such organic compounds in root exudates regulate nutrient and energy flows that drive rhizosphere microbial colonization, thereby influencing the structure and dynamics of the microbial community (Iannucci et al., 2017). The results demonstrate that CK and T1 rootstocks show limited metabolic differences (Figure 7C), whereas T2 and T3 rootstocks display greater metabolic divergence, with T2’s intensity being lower. The pumpkin rootstock (T3) appears to recruit bacterial taxa such as Sphingomonadaceae through the secretion of defense-related metabolites, alleviating stress and activating GABA receptor coordination with the microbiota. This aligns with observed trends in soil enzyme activity, microbial community enrichment, network analysis, and the increase in biomass and maintenance of Fv/Fm.
RDA analysis (Figure 8A) confirmed that metabolites from CK, T1, and T3 significantly affected the bacterial community, with T3 showing the strongest metabolite–bacteria interactions. The dominant bacterial genera in these interactions were Actinomadura and Edaphobacter, which are associated with improved nutrient utilization, metabolic regulation, and stress alleviation (Hamedi and Mohammadipanah, 2015; Hemmat-Jou et al., 2018). Correlation clustering (Figure 8B) indicated that oligotrophic Acidibacter (Gao et al., 2024a) did not demonstrate a competitive advantage under these conditions, whereas other genera contributed to degradation, growth promotion, nutrient cycling, and stress adaptation (Chen et al., 2023; Gan et al., 2014; Hemmat-Jou et al., 2018; Kruczynska et al., 2023; Liu et al., 2024). Functional pathway predictions (PICRUSt; Figure 4C) and metabolite pathway analysis (Figure 7E) revealed convergence in purine metabolism, a pathway also enriched in T1 and T3. Glycine likely promotes purine metabolism by acting as a precursor for inosine monophosphate (IMP), a prerequisite for all purine nucleotides.
Both the core RDA1 axis and the correlation heatmap (Figure 8B) highlight the interactions among rootstock, microbiota, and metabolites: amino acids, nucleotides, and organic acids are enriched by Edaphobacter and Actinomadura, whereas lipids specifically promote Acidibacter. These findings indicate that rootstock genotype influences the distribution of exudate metabolites, precisely regulating the functional structure of the rhizosphere microbiome. For example, pumpkin rootstock (T3) secretes more amino acids and nucleotides, enriching Edaphobacter and Actinomadura to reinforce nitrogen cycling and stress resistance. The wild watermelon rootstock, with a highly positively correlated microbial network, enriches Edaphobacter and Actinomadura and, through lipid-enriched Acidibacter, enhances acid adaptability.
Overall, glycine to some extent improved soil enzyme activity, plant biomass, and microbial diversity, supporting the hypothesis that glycine is an effective, eco-friendly nitrogen source combined with different rootstocks for use that promotes sustainability through enrichment of beneficial bacteria and optimization of nutrient cycling. The rootstock-specific structuring of the rhizosphere microbiome via exudate-mediated recruitment and the observed correlations between specific exudate compounds and microbial abundance substantiate the central role of plant–microbe interactions in sustaining the benefits of glycine amendment.
In summary, substituting ammonium nitrate with glycine and selecting suitable scion/rootstock combinations provide new evidence for sustainable agriculture and elucidate the role of root exudates in rhizosphere microbiome assembly. Although associations between metabolites and microorganisms have been established, the complexity of microbial interactions remains to be fully elucidated, and the proposed shift in nutrient limitation patterns requires future validation through stoichiometric analysis of soil chemistry.
5 Conclusions
This study deciphered how rootstock-specific microbiome and metabolome remodeling governs glycine substitution efficacy for ammonium nitrate in grafted watermelon systems. The findings demonstrate that glycine application can moderately increase bacterial diversity. It can also enhance soil enzyme activity. Glycine improves nutrient absorption in plants and helps maintain typical plant growth. However, glycine’s effects in these areas are not significantly different from those of ammonium nitrate. Its primary advantage over ammonium nitrate is an environmental benefit. Combining specific rootstocks and scions can enhance glycine’s effectiveness. This combination also allows customization based on cultivation environment and production goals. This work advances crop domestication theory by establishing root exudate chemistry as a tunable factor for rhizosphere engineering and sustainable agricultural management.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1237532.
Author contributions
ZC: Conceptualization, Data curation, Investigation, Methodology, Supervision, Visualization, Writing – original draft, Writing – review & editing. TY: Conceptualization, Data curation, Investigation, Writing – review & editing. XB: Investigation, Writing – review & editing. YW: Investigation, Writing – review & editing. SQ: Investigation, Writing – review & editing. LT: Investigation, Writing – review & editing. HS: Investigation, Writing – review & editing. XC: Investigation, Writing – review & editing. MD: Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Key Core Technology Research Project of Agriculture in Shaanxi Province(2023NYGG008).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmed, A., Khurshid, A., Tang, X. H., Wang, J. H., Khan, T. U., and Mao, Y. X. (2021). Structural and Functional Impacts of Microbiota on Pyropia yezoensis and Surrounding Seawater in Cultivation Farms along Coastal Areas of the Yellow Sea. Microorganisms 9. doi: 10.3390/microorganisms9061291
Anderson, H. M., Cagle, G. A., Majumder, E. L. W., Silva, E., Dawson, J., Simon, P., et al. (2024). Root exudation and rhizosphere microbial assembly are influenced by novel plant trait diversity in carrot genotypes. Soil Biol. Biochem. 197. doi: 10.1016/j.soilbio.2024.109516
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. (2012). “The rhizosphere microbiome and plant health.” Trends in Plant Science 17(8), 478–486. doi: 10.1016/j.tplants.2012.04.00
Camargo-Montoya, L., Leal-Mejia, L., Arenas, N. E., Franco, D. C., Valero-Valero, N., and Vanegas, J. (2025). Metagenomic analysis reveals environmental drivers of Actinobacteriota-associated antibiotic resistance and biosynthetic genes in Colombian caribbean resource islands. J. Arid Environments 230. doi: 10.1016/j.jaridenv.2025.105431
Chen, L.-F., He, Z.-B., Zhao, W.-Z., Kong, J.-Q., and Gao, Y. (2021). Empirical evidence for microbial regulation of soil respiration in alpine forests. Ecol. Indic. 126. doi: 10.1016/j.ecolind.2021.107710
Chen, J. F., Ke, Y. C., Zhu, Y., Chen, X. L., and Xie, S. G. (2023). Deciphering of sulfonamide biodegradation mechanism in wetland sediments: from microbial community and individual populations to pathway and functional genes. Water Res. 240. doi: 10.1016/j.watres.2023.120132
Chen, S. F., Zhou, Y. Q., Chen, Y. R., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, 884–890. doi: 10.1093/bioinformatics/bty560
Davis, K. E. R., Joseph, S. J., and Janssen, P. H. (2005). Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71, 826–834. doi: 10.1128/AEM.71.2.826-834.2005
de Vries, F. T., Griffiths, R. I., Knight, C. G., Nicolitch, O., and Williams, A. (2020). Harnessing rhizosphere microbiomes for drought-resilient crop production. Science 368, 270. doi: 10.1126/science.aaz5192
Edelstein, M., Plaut, Z., and Ben-Hur, M. (2011). Sodium and chloride exclusion and retention by non-grafted and grafted melon and Cucurbita plants. J. Exp. Bot. 62, 177–184. doi: 10.1093/jxb/erq255
Farooq, T. H., Kumar, U., Mo, J., Shakoor, A., Wang, J., Rashid, M. H. U., et al. (2021). Intercropping of peanut-tea enhances soil enzymatic activity and soil nutrient status at different soil profiles in subtropical Southern China. Plants-Basel 10. doi: 10.3390/plants10050881
Fraga, C. G., Clowers, B. H., Moore, R. J., and Zink, E. M. (2010). Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography-mass spectrometry, XCMS, and chemometrics. Analytical Chem. 82, 4165–4173. doi: 10.1021/ac1003568
Fujii, N., Kuroda, K., Narihiro, T., Aoi, Y., Ozaki, N., Ohashi, A., et al. (2022). Metabolic potential of the superphylum patescibacteria reconstructed from activated sludge samples from a municipal wastewater treatment plant. Microbes Environments 37. doi: 10.1264/jsme2.ME22012
Gallart, M., Adair, K. L., Love, J., Meason, D. F., Clinton, P. W., Xue, J., et al. (2018). Host genotype and nitrogen form shape the root microbiome of pinus radiata. Microbial Ecol. 75, 419–433. doi: 10.1007/s00248-017-1055-2
Gan, H. M., Gan, H. Y., Ahmad, N. H., Aziz, N. A., Hudson, A. O., and Savka, M. A. (2014). Whole genome sequencing and analysis reveal insights into the genetic structure, diversity and evolutionary relatedness of luxI and luxR homologs in bacteria belonging to the Sphingomonadaceae family. Front. Cell. infection Microbiol. 4, 188. doi: 10.3389/fcimb.2014.00188
Gao, L., Liu, L., Lv, A.-P., Fu, L., Lian, Z.-H., Nunoura, T., et al. (2024a). Reversed oxidative TCA (roTCA) for carbon fixation by an Acidimicrobiia strain from a saline lake. Isme J. 18. doi: 10.1093/ismejo/wrae147
Gao, M., Song, W., Zhou, Q., Ma, X., and Chen, X. (2013). Interactive effect of oxytetracycline and lead on soil enzymatic activity and microbial biomass. Environ. Toxicol. Pharmacol. 36, 667–674. doi: 10.1016/j.etap.2013.07.003
Gao, Y., Yang, Q., Chen, Q., He, Y., He, W., Geng, J., et al. (2024b). Plants attacked above-ground by leaf-mining flies change below-ground microbiota to enhance plant defense. Horticulture Res. 11. doi: 10.1093/hr/uhae121
Ghasemi, Z., Ghaderian, S. M., Rodríguez-Garrido, B., Prieto-Fernández, A., and Kidd, P. S. (2018). Plant species-specificity and effects of bioinoculants and fertilization on plant performance for nickel phytomining. Plant Soil 425, 265–285. doi: 10.1007/s11104-017-3553-x
Gu, M., Jin, J., Lu, P., Yu, S., Su, H., Shang, H., et al. (2024). Regulation of root-associated microbiomes and root exudates by different tobacco species. Chem. Biol. Technol. Agric. 11. doi: 10.1186/s40538-024-00678-7
Gu, D. D., Wang, W. Z., Hu, J. D., Zhang, X. M., Wang, J. B., and Wang, B. S. (2016). Nondestructive determination of total chlorophyll content in maize using three-wavelength diffuse reflectance. J. Appl. Spectrosc. 83, 541–547. doi: 10.1007/s10812-016-0325-y
Guan, S., Zhang, D., and Zhang, Z. (1986). Soil enzyme and its research methods. Agricultural Beijing 1986, 274–297.
Guo, H., Yao, J., Cai, M. M., Qian, Y. G., Guo, Y., Richnow, H. H., et al. (2012). Effects of petroleum contamination on soil microbial numbers, metabolic activity and urease activity. Chemosphere 87, 1273–1280. doi: 10.1016/j.chemosphere.2012.01.034
Guo, S., Zhang, J., Sun, H., Salse, J., Lucas, W. J., Zhang, H., et al. (2013). The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 45, 51. doi: 10.1038/ng.2470
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hakeem, K. R., Ahmad, A., Iqbal, M., Gucel, S., and Ozturk, M. (2011). Nitrogen-efficient rice cultivars can reduce nitrate pollution. Environ. Sci. pollut. Res. 18, 1184–1193. doi: 10.1007/s11356-010-0434-8
Hamedi, J. and Mohammadipanah, F. (2015). Biotechnological application and taxonomical distribution of plant growth promoting actinobacteria. J. Ind. Microbiol. Biotechnol. 42, 157–171. doi: 10.1007/s10295-014-1537-x
Hassnin, N. M. M., Ali, A. H. H., and Kesba, H. H. (2023). Effect of Grafting on Resistance of Cucurbit Hybrids against Meloidogyne incognita Infection under Greenhouse Conditions. Pakistan J. Zoology 55, 723–733. doi: 10.17582/journal.pjz/20211226141226
Hemmat-Jou, M. H., Safari-Sinegani, A. A., Mirzaie-Asl, A., and Tahmourespour, A. (2018). Analysis of microbial communities in heavy metals-contaminated soils using the metagenomic approach. Ecotoxicology 27, 1281–1291. doi: 10.1007/s10646-018-1981-x
Hernandez, D. J., David, A. S., Menges, E. S., Searcy, C. A., and Afkhami, M. E. (2021). “Environmental stress destabilizes microbial networks.” Isme Journal 15(6), 1722–1734. doi: 10.1038/s41396-020-00882-x
Huang, X. F., Zhou, D. M., Lapsansky, E. R., Reardon, K. F., Guo, J. H., Andales, M. J., et al. (2017). Mitsuaria sp and Burkholderia sp from Arabidopsis rhizosphere enhance drought tolerance in Arabidopsis thaliana and maize (Zea mays L.). Plant Soil 419, 523–539. doi: 10.1007/s11104-017-3360-4
Iannucci, A., Fragasso, M., Beleggia, R., Nigro, F., and Papa, R. (2017). Evolution of the crop rhizosphere: impact of domestication on root exudates in tetraploid wheat (Triticum turgidum L.). Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02124
Kacjan-Maršić, N. and Osvald, J. (2004). The influence of grafting on yield of two tomato cultivars (Lycopersicon esculentum Mill.) grown in a plastic house. Acta agriculturae slovenica 83, 243–249.
Kaspal, M., Kanapaddalagamage, M. H., and Ramesh, S. A. (2021). Emerging roles of γ Aminobutyric acid (GABA) gated channels in plant stress tolerance. Plants-Basel 10. doi: 10.3390/plants10102178
Klimek, D., Herold, M., and Calusinska, M. (2024). Comparative genomic analysis of Planctomycetota potential for polysaccharide degradation identifies biotechnologically relevant microbes. BMC Genomics 25. doi: 10.1186/s12864-024-10413-z
Knight, R., Vrbanac, A., Taylor, B. C., Aksenov, A., Callewaert, C., Debelius, J., et al. (2018). Best practices for analysing microbiomes. Nat. Rev. Microbiol. 16, 410–422. doi: 10.1038/s41579-018-0029-9
Kruczynska, A., Kuzniar, A., Jacek, P., Slomczewski, A., Grzadziel, J., Marzec-Grzadziel, A., et al. (2023). Bacteroidota structure in the face of varying agricultural practices as an important indicator of soil quality - a culture independent approach. Agric. Ecosyst. Environ. 342. doi: 10.1016/j.agee.2022.108252
Lee, J. K., Park, H. J., Cha, S. J., Kwon, S. J., and Park, J. H. (2021). Effect of pyroligneous acid on soil urease, amidase, and nitrogen use efficiency by Chinese cabbage (Brassica campestris var. Pekinensis). Environ. pollut. 291. doi: 10.1016/j.envpol.2021.118132
Li, L. Y., Feng, C. X., and Zhang, Y. D. (2019). Influence of collection time on the determination of root exudates in fraxinus mandshurica by the metabolomics method. Appl. Ecol. Environ. Res. 17, 9529–9545. doi: 10.15666/aeer/1704_95299545
Ling, N., Zhang, W., Wang, D., Mao, J., Huang, Q., Guo, S., et al. (2013). Root Exudates from Grafted-Root Watermelon Showed a Certain Contribution in Inhibiting Fusarium oxysporum f. sp niveum. PLoS One 8. doi: 10.1371/journal.pone.0063383
Liu, Y., Shahbaz, M., Fang, Y., Li, B., Wei, X., Zhu, Z., et al. (2022). Stoichiometric theory shapes enzyme kinetics in paddy bulk soil but not in rhizosphere soil. Land Degradation Dev. 33, 246–256. doi: 10.1002/ldr.4141
Liu, N., Yao, Y. Y., Zhang, J., Zhang, J. G., Wu, C., Ouyang, D. J., et al. (2024). Reduction characteristic of chlorobenzene by a newly isolated Paenarthrobacter ureafaciens LY from a pharmaceutical wastewater treatment plant. Cell Biochem. Funct. 42. doi: 10.1002/cbf.3965
Luo, S., Schmid, B., De Deyn, G. B., and Yu, S. X. (2018). Soil microbes promote complementarity effects among co-existing trees through soil nitrogen partitioning. Funct. Ecol. 32, 1879–1889. doi: 10.1111/1365-2435.13109
Marian, M., Nishioka, T., Koyama, H., Suga, H., and Shimizu, M. (2018). Biocontrol potential of Ralstonia sp TCR112 and Mitsuaria sp TWR114 against tomato bacterial wilt. Appl. Soil Ecol. 128, 71–80. doi: 10.1016/j.apsoil.2018.04.005
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Morais, M. C., Torres, L. F., Kuramae, E. E., de Andrade, S. A. L., and Mazzafera, P. (2024). Plant grafting: Maximizing beneficial microbe-plant interactions. Rhizosphere 29. doi: 10.1016/j.rhisph.2023.100825
Murphy, D., Macdonald, A., Stockdale, E., Goulding, K., Fortune, S., Gaunt, J., et al. (2000). Soluble organic nitrogen in agricultural soils. Biol. Fertility Soils 30, 374–387. doi: 10.1007/s003740050018
Nawaz, M. A., Han, X. J., Chen, C., Zheng, Z. H., Shireen, F., Bie, Z. L., et al. (2018). Nitrogen Use Efficiency of Watermelon Grafted onto 10 Wild Watermelon Rootstocks under Low Nitrogen Conditions. Agronomy-Basel 8. doi: 10.3390/agronomy8110259
Oda, M. (1995). New grafting methods for fruit-bearing vegetables in Japan. Jarq-Japan Agric. Res. Q. 29, 187–194.
Palaniyandi, S. A., Yang, S. H., Zhang, L., and Suh, J.-W. (2013). Effects of actinobacteria on plant disease suppression and growth promotion. Appl. Microbiol. Biotechnol. 97, 9621–9636. doi: 10.1007/s00253-013-5206-1
Pang, G., Cai, F., Li, R., Zhao, Z., Li, R., Gu, X., et al. (2017). Trichoderma-enriched organic fertilizer can mitigate microbiome degeneration of monocropped soil to maintain better plant growth. Plant Soil 416, 181–192. doi: 10.1007/s11104-017-3178-0
Pérez-Patricio, M., Camas-Anzueto, J. L., Sanchez-Alegría, A., Aguilar-González, A., Gutiérrez-Miceli, F., Escobar-Gómez, E., et al. (2018). Optical method for estimating the chlorophyll contents in plant leaves. Sensors 18. doi: 10.3390/s18020650
Poudel, R., Jumpponen, A., Kennelly, M. M., Rivard, C. L., Gomez-Montano, L., and Garrett, K. A. (2019). Rootstocks shape the rhizobiome: rhizosphere and endosphere bacterial communities in the grafted tomato system. Appl. Environ. Microbiol. 85. doi: 10.1128/AEM.01765-18
Rivero, R. M., Ruiz, J. M., and Romero, L. (2003). Role of grafting in horticultural plants under stress conditions. J. Food Agric. Environ. 1, 70–74.
Ruan, Y., Wang, T., Guo, S., Ling, N., and Shen, Q. (2020). Plant grafting shapes complexity and co-occurrence of rhizobacterial assemblages. Microbial Ecol. 80, 643–655. doi: 10.1007/s00248-020-01532-7
Sallaku, G., Rewald, B., Sandén, H., and Balliu, A. (2022). Scions impact biomass allocation and root enzymatic activity of rootstocks in grafted melon and watermelon plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.949086
Sultana, R., Islam, S. M. N., and Sultana, T. (2023). Arsenic and other heavy metals resistant bacteria in rice ecosystem: Potential role in promoting plant growth and tolerance to heavy metal stress. Environ. Technol. Innovation 31. doi: 10.1016/j.eti.2023.103160
Tamaddon, F., Arab, D., and Ahmadi-AhmadAbadi, E. (2020). Urease immobilization on magnetic micro/nano-cellulose dialdehydes: Urease inhibitory of Biginelli product in Hantzsch reaction by urea. Carbohydr. Polymers 229. doi: 10.1016/j.carbpol.2019.115471
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., and Singh, B. K. (2021). Plant-microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 19, 72–72. (vol 18, pg 607, 2020). doi: 10.1038/s41579-020-00490-8
Walters, W., Hyde, E. R., Berg-Lyons, D., Ackermann, G., Humphrey, G., Parada, A., et al. (2016). Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. Msystems 1. doi: 10.1128/mSystems.00009-15
Wang, R. T., Yang, Y., Xu, K. X., Wang, T. J., Elsadek, M. A., Yuan, L., et al. (2024). Multi-omics analysis reveals improvement of tomato quality by grafting on goji rootstock. Food Qual. Saf. 8. doi: 10.1093/fqsafe/fyae023
Wang, X.-l., Yu, W.-j., Zhou, Q., Han, R.-f., and Huang, D.-f. (2014). Metabolic response of pakchoi leaves to amino acid nitrogen. J. Integr. Agric. 13, 778–788. doi: 10.1016/S2095-3119(13)60622-X
Wang, F., Zhan, P., Zhang, X., Xia, P., and Liang, Z. (2023). Unraveling rotational remedies: Deciphering the autotoxicity of Panax notoginseng saponins. Ind. Crops Products 206. doi: 10.1016/j.indcrop.2023.117601
Wen, T., Yuan, J., He, X., Lin, Y., Huang, Q., and Shen, Q. (2020). Enrichment of beneficial cucumber rhizosphere microbes mediated by organic acid secretion. Horticulture Res. 7, 154–154. doi: 10.1038/s41438-020-00380-3
Xiong, M., Liu, C. J., Guo, L. P., Wang, J., Wu, X. S., Li, L., et al. (2021). Compatibility evaluation and anatomical observation of melon grafted onto eight cucurbitaceae species. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.762889
Xu, W., Dou, Y., Geng, H., Fu, J., Dan, Z., Liang, T., et al. (2022). OsGRP3 enhances drought resistance by altering phenylpropanoid biosynthesis pathway in rice (Oryza sativa L.). Int. J. Mol. Sci. 23. doi: 10.3390/ijms23137045
Yang, S., Sun, J., Wang, C., Li, S., Li, Z., Luo, W., et al. (2024). Residue quality drives SOC sequestration by altering microbial taxonomic composition and ecophysiological function in desert ecosystem. Environ. Res. 250. doi: 10.1016/j.envres.2024.118518
Yu, W. T., Dai, Y. L., Chen, J. M., Liang, A. M., Wu, Y. P., Suo, Q. W., et al. (2024). Upregulation of the glycine-rich protein-encoding gene GhGRPL enhances plant tolerance to abiotic and biotic stressors by promoting secondary cell wall development. J. Integr. Agric. 23, 3311–3327. doi: 10.1016/j.jia.2024.05.025
Zeng, W. J., He, H. Z., Liu, Y., Liu, Y. Y., Luo, W. M., and Qin, W. (2024). Fertility and microbial diversity in rhizosphere soil of camellia oleifera under different intercropping systems. Pakistan J. Bot. 56, 1575–1583. doi: 10.30848/PJB2024-4(26)
Zhang, R., Liu, Q., Xu, X. T., Liao, M. A., Lin, L. J., Hu, R. P., et al. (2022). An amino acid fertilizer improves the emergent accumulator plant Nasturtium officinale R. Br. phytoremediation capability for cadmium-contaminated paddy soils. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1003743
Zhou, H. Y., Yu, Z., Zhang, S. Y., Zong, Q. H., Zhang, Y. L., Pang, Y. H., et al. (2025). Mitigating secondary salinization in grapes: long-term benefits of biochar and cow dung. Front. Plant Sci. 16. doi: 10.3389/fpls.2025.1528354
Zhou, C. R., Zhang, J. B., Miao, P. J., Dong, Q. Y., Lin, Y. X., Li, D., et al. (2023). Novel finding on how melatonin and nanoselenium alleviate 2,4-D butylate stress in wheat plants. J. Agric. Food Chem. 71, 12943–12957. doi: 10.1021/acs.jafc.3c03109
Keywords: glycine, ammonium nitrate, grafted watermelon, microbiome, bacterial metabolomics
Citation: Chen Z, Yao T, Bao X, Wang Y, Qiao S, Tan L, Shi H, Chen X and Ding M (2025) Rootstock-specific bacterial microbiome and metabolome remodeling enhances glycine substitution efficacy for ammonium nitrate in watermelon. Front. Plant Sci. 16:1640174. doi: 10.3389/fpls.2025.1640174
Received: 03 June 2025; Accepted: 12 August 2025;
Published: 05 September 2025.
Edited by:
Jie Zhou, Nanjing Agricultural University, ChinaReviewed by:
Changying Liu, Chengdu University, ChinaFoad Fatehi, Payame Noor University, Iran
Xu Zheng, Shihezi University, China
Copyright © 2025 Chen, Yao, Bao, Wang, Qiao, Tan, Shi, Chen and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Ding, ZGluZ21pbmdAbndhZnUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Zehao Chen
Zehao Chen Tian Yao†
Tian Yao† Ming Ding
Ming Ding