- 1Plant Biotechnology Resource and Outreach Center, Department of Horticulture, Michigan State University, East Lansing, MI, United States
- 2Scientific Research Center, College of Science, University of Duhok, Duhok, Kurdistan, Iraq
Manipulating the expression of flowering pathway genes holds potential for regulating tomato fruit productivity. SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) is a MADS-box gene that serves as a key integrator in the flowering pathway. In this study, two full-length SOC1 genes cloned from maize (ZmSOC1) and soybean (GmSOC1), along with a partial SOC1 gene from blueberry (VcSOC1K, containing the K-domain), were individually transformed into tomato for constitutive expression. Phenotypically, the expression of VcSOC1K and ZmSOC1, but not GmSOC1, led to early flowering. Most transgenic lines carrying any of the three constructs exhibited a significant increase in fruit number per plant, with gains of 84-161% for ZmSOC1, 72-135% for GmSOC1, and 55-96% for VcSOC1K. Notably, compared to non-transgenic controls, all three constructs enhanced fruit yield per plant to varying degrees, including ZmSOC1 by 81-169%, GmSOC1 by 60-112%, and VcSOC1K by 52-88%, primarily through enhanced branching. At the transcriptomic level, comparative analysis of GmSOC1 revealed the broader impact of the transformed genes. The increased expression of CLF and EZA1 appears to explain the unchanged flowering time of the GmSOC1 transgenic plants, while the repressed expression of DWARF genes likely contributes to enhanced branching. Additionally, numerous genes associated with biotic and abiotic stress tolerance displayed differential expression. These findings demonstrate that constitutive expression of either full-length or partial SOC1 has the potential to enhance tomato fruit production by modulating multiple pathways, at least at the transcript levels.
1 Introduction
The MADS-box gene family encodes transcription factors that are present in all eukaryotic organisms, playing essential roles in animals, plants, and fungi (Alvarez-Buylla et al., 2000; Gramzow et al., 2010; Gramzow and Theissen, 2010). In plants, these genes are crucial for a wide range of physiological and developmental processes (Colombo et al., 2008). While MADS-box proteins have been extensively studied for their roles in regulating flower development, emerging research indicates their involvement in fruit development, embryo establishment, vegetative organ development, and stress resistance. These findings highlight the significance and functional diversity of this gene superfamily in plant development (Busi et al., 2003; Ito et al., 2008; Ehlers et al., 2016).
The plant MADS-box family includes type I and II genes (Masiero et al., 2011; Gramzow and Theissen, 2013). Type I MADS box factors are crucial regulators of plant reproduction; they play key roles in the development of the female gametophyte, embryo, and endosperm development (Masiero et al., 2011). Type II genes have been extensively studied and further classified into MIKCC- and MIKC*-type two subgroups (Henschel et al., 2002). MIKCC-type genes are the most thoroughly investigated members of the plant MADS-box family and encode proteins containing four distinct domains: M, I, K, and C. The M (DNA-binding) domain is the most conserved and is shared across all MADS-box genes. The I (intervening) domain, while less conserved, facilitates the specificity of DNA-binding dimer formation. The K (keratin-like) domain is structurally conserved and mediates protein–protein interactions, while the C (C-terminal) domain, the least conserved, is involved in ternary complex formation and transcriptional activation (Henschel et al., 2002). MIKC*-type genes are believed to have originated from ancestral MIKCC-type genes through either elongation of the I region or truncation of the K box (Henschel et al., 2002; Kwantes et al., 2012; Liu et al., 2013). These genes are crucial for pollen development (Verelst et al., 2006; Liu et al., 2013). This structure-function relationship highlights the critical role of MADS-box genes in regulating diverse developmental processes, including fruit development, embryogenesis, vegetative organ formation, and stress responses.
MIKCC-type MADS-box (MIKCC) contribute floral organogenesis and flowering transition (Yanofsky et al., 1990; Weigel and Meyerowitz, 1994; Becker and Theissen, 2003). Among these, SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) serves as a conserved floral activator and integrator within the plant flowering pathway (Parenicova et al., 2003; Liu et al., 2009; Seo et al., 2009; Lee and Lee, 2010). Due to the diverse functions of the SOC1 protein in regulating plant development, its expression can be genetically engineered to improve crop yield, as evidenced in blueberry (Vaccinium corymbosum) (Song and Chen, 2018), leaf mustard (Brassica juncea cv. Varuna) (Tyagi et al., 2019), soybean (Glycine max) (Han et al., 2021), and maize (Zea mays) (Song and Han, 2021; Song et al., 2021).
Crop yield, such as in tomato, is influenced by genetic background, environmental conditions, and management practices. The potential for crop yield is genetically determined through the interactions of multiple genes and complex gene networks (Bhandari et al., 2023). Strategically, genetic improvement can be achieved by incorporating genes for biotic resistance and abiotic stress tolerance through breeding to protect yields, while modifying key genes related to growth and development can further enhance productivity (Bailey-Serres et al., 2019).
Tomato (Solanum lycopersicum L.) is an important vegetable crop and a valuable source of essential nutrients and phytochemicals, including vitamin C, lycopene, and antioxidants (Raza et al., 2022). Enhancing yield is a top breeding priority for fresh-market tomato (Bhandari et al., 2023). This can be achieved through either traditional breeding or genetic engineering (Krieger et al., 2010; Cui et al., 2022). However, since yield is a trait controlled by multiple genes, both approaches present significant challenges. Although numerous genes have been functionally characterized in tomato, only a few, particularly those involved in flowering pathways, including four tomato SOC1 genes, have shown potential for increasing yield (Krieger et al., 2010; Wang et al., 2021; Cui et al., 2022; Zahn et al., 2023). Yet the specific effects of these genes on yield remain unexplored.
Due to concerns that overexpression of the tomato SOC1 gene may lead to dosage-related effects, this study investigated the constitutive expression of two heterologous SOC1 genes, GmSOC1 from soybean (Glycine max) and ZmSOC1 from maize (Zea mays), along with a truncated derivative of blueberry (Vaccinium corymbosum) SOC1 lacking the M domain (VcSOC1K), to assess their potential for improving tomato yield.
2 Materials and methods
2.1 Constructs and plant transformation
The ZmSOC1 and GmSOC1 protein sequences were used as queries to conduct BLAST searches against the tomato proteome in Phytozome v13. The identified tomato SOC1 and SOC1-like proteins were subsequently selected for phylogenetic analysis (Figure 1).
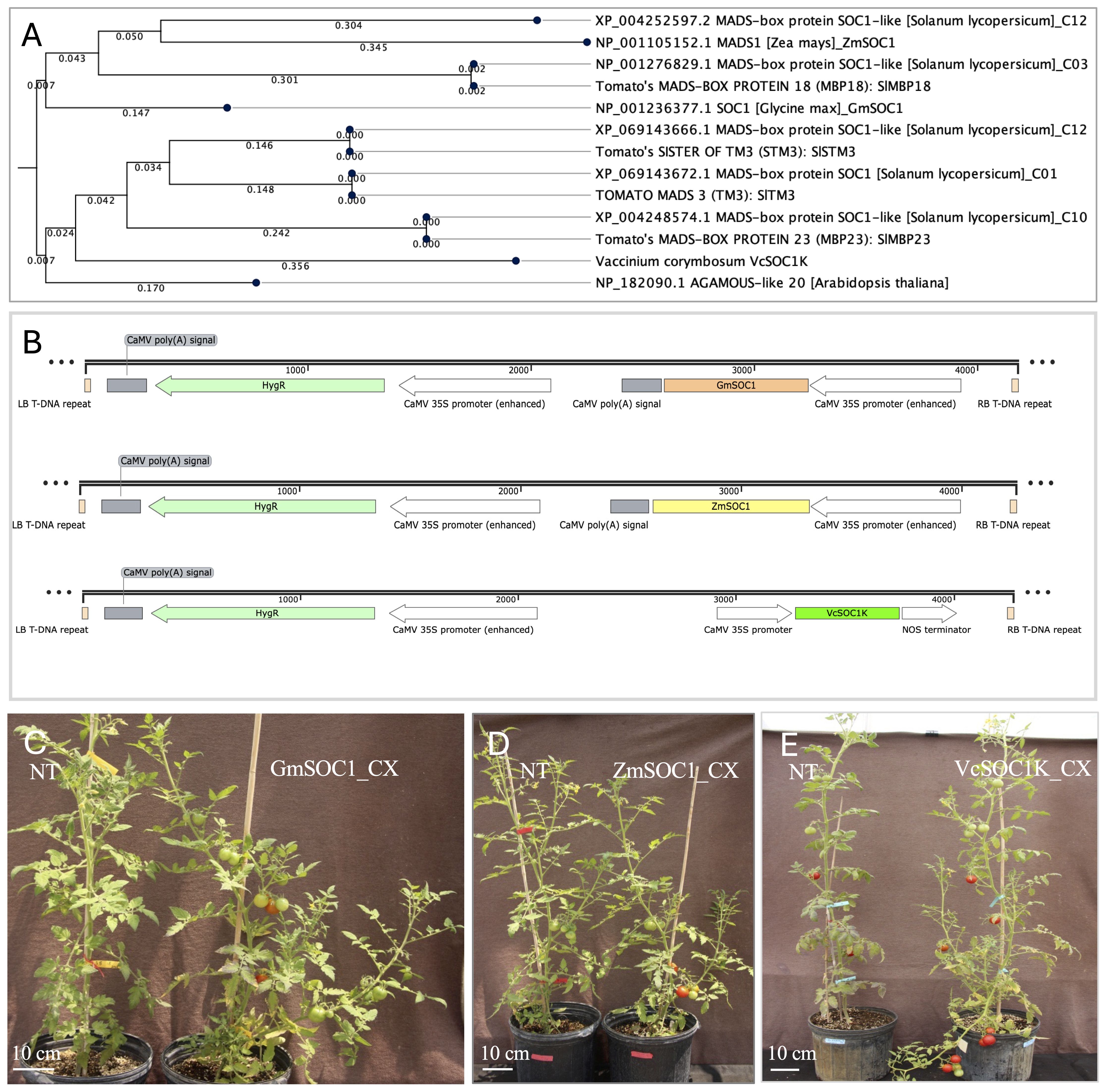
Figure 1. Genes and constructs used for generating transgenic tomato. (A) Phylogenetic analysis of five tomato SOC1 and SOC1-like proteins from four chromosomes. Protein sequences of TOMATO MADS3 (SlTM3), SISTER OF TM3 (SlSTM3), SlMBP18. and SlMBP23 are from Zahn et al., 2023. C01, C03, C10, and C12 are chromosome numbers. The analysis reveals similarities to Arabidopsis SOC1, soybean GmSOC1, maize ZmSOC1, and blueberry SOC1 K-domain (VcSOC1K), using the Neighbor Joining method with the Jukes-Cantor protein distance metric and 500 bootstrap replicates. (B) T-DNA regions of three pCAMBIA1300-derived constructs containing GmSOC1, ZmSOC1, and VcSOC1K genes. HygR: the hygromycin B resistance gene. LB and RB: the left and right borders, respectively. CaMV: Cauliflower mosaic virus. (C) Constitutive expression of GmSOC1 (GmSOC1-CX) in T0 plants and non-transgenic (NT) regenerants. (D) Constitutive expression of ZmSOC1 (ZmSOC1-CX) in T0 plants and NT regenerants. (E) Constitutive expression of VcSOC1K (VcSOC1K-CX) in T0 plants and NT regenerants.
A 696-bp ZmSOC1 (also known as ZmMADS1), identical to the sequence derived from HQ858775.1 in GenBank, was cloned using polymerase chain reaction (PCR) from the cDNA of the maize inbred line B104 (Han et al., 2021). The corresponding protein sequence matches NP_001105152.1 in GenBank (Figure 1). The fragment was inserted into a modified pCAMBIA1300 vector between a CaMV 35S promoter and a CaMV poly(A) signal at the KpnI and XbaI restriction sites (Figure 1).
Similarly, a 630-bp GmSOC1, identical to the sequence derived from NM_0011249448.2 in GenBank, was cloned from the cDNA of the soybean cultivar Thorne. The fragment was inserted into the same modified pCAMBIA1300 vector between a CaMV 35S promoter and a CaMV poly(A) signal at the KpnI and XbaI sites (Figure 1). The corresponding protein sequence is identical to NP_001236377.1 in GenBank (Figure 1).
The VcSOC1K gene was previously cloned into the T-DNA region of the binary vector pBI121, positioned between the CaMV 35S promoter and the Nos terminator for constitutive expression (Song and Chen, 2018). In this study, the VcSOC1K expression cassette was excised from pBI121 using HindIII and EcoRI digestion and subsequently ligated into the T-DNA region of a HindIII- and EcoRI-digested pCAMBIA1300 (PC1300) vector. This ensures that all three constructs share the same binary vector backbone (Figure 1).
All three constructs were confirmed by sequencing the target genes and subsequently transformed into Agrobacterium tumefaciens strain EHA105. For tomato transformation, indeterminate tomato (Lycopersicon esculentum) ‘Ailsa Craig’ was used. Seeds were sterilized in a 2.5% (v/v) sodium hypochlorite solution and germinated to produce cotyledons. Non-inoculated cotyledons were cultured on antibiotic-free regeneration medium to produce non-transgenic (NT) regenerants, which were used as controls. The regeneration medium consisted of Murashige and Skoog (MS) basal salts (Murashige and Skoog, 1962), Gamborg B5 vitamins (Gamborg et al., 1968), 2.85 μM zeatin riboside, 2.86 μM indole-3-acetic acid (IAA), 30 g/L sucrose, and 6 g/L agar. Transformation was carried out following established protocols (Danial et al., 2021). Regeenrated shoots approximately 2–3 cm in length were excised and transferred to 30 mL MS medium for rooting. For NT plants, the rooting medium lacked antibiotics, whereas for transgenic plants, it was supplemented with 10 mg/L hygromycin, 250 mg/L timentin, and 250 mg/L cefotaxime.
2.2 Transplanting, phenotyping, and genotyping of the transformants
After rooting, T0 plantlets were transplanted into 4-inch plastic pots containing water-soaked Suremix Perlite planting medium (Michigan Grower Products Inc., Galesburg, MI). The plants were covered with plastic bags and placed in a growth room maintained at 25 °C with a 16/8-hour light/dark photoperiod. Over a two-week period, the plastic bags were gradually removed to acclimate the plantlets. Once acclimated, the plants were repotted into one-gallon pots and transferred to a greenhouse.
For each construct, approximately 10–20 T0 transformants were cultivated in the greenhouse. Each transformant, derived from a separate explant, was considered an independent transgenic line. At the time of repotting into one-gallon pots, transformed and non-transformed plants of similar size, 3–5 lines per construct and non-transformants, were selected. These comparable plants were used as representatives for phenotyping of T0 plants. First-generation (T1) seeds were harvested separately from each T0 plant.
For phenotyping T1 plants, 20–30 seeds from each of three selected T0 transgenic lines per construct were germinated in soil. Ten plants from each transgenic line were transferred to one-gallon pots and grown in a greenhouse.
Phenotypic assessments for each plant included measuring the time and height of the first flower appearance, recording the time of the first mature fruit appearance, counting the total number of branches and fruits harvested, and weighing all harvested fruits. Photographs were taken to document phenotypic variations.
For genotyping of the T0 and T1 plants, genomic DNA was extracted from young leaves using the cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle, 1987). PCR was performed to detect the hygromycin phosphotransferase (hpt) transgene and the full-length sequences of GmSOC1, ZmSOC1, and VcSOC1K using the primers listed in Supplementary Table S1.
2.3 RNA sequencing and quantitative reverse transcription PCR analysis
Three groups of T1 plants, derived from three T0 transgenic lines containing GmSOC1, were selected for RNA sequencing and transcriptome analysis. Each group of T1 plants was segregated into transgenic and NT plants. For each group, two newly formed mature leaves from all transgenic plants before their flowering were pooled together to create one biological replicate for transgenic plants, while leaves from NT plants were similarly pooled to form one biological replicate for NT plants. In total, three transgenic samples and three NT samples were collected, immediately frozen in liquid nitrogen, and stored at -80°C for RNA isolation.
Total RNA from each sample was isolated using a modified CTAB method (Chang et al., 1993) and further purified with the RNeasy Mini Kit (Qiagen, Valencia, CA, United States). To eliminate any residual DNA, on-column DNase digestion was performed using the RNase-Free DNase Set (Qiagen, Valencia, CA, United States). RNA quality was assessed with the High Sensitivity RNA ScreenTape system (Agilent Technologies, Santa Clara, CA, United States). All RNA samples used for sequencing and RT-qPCR analysis had RNA integrity number equivalent scores exceeding 5.0.
The RNA samples were sequenced using the Illumina NovaSeq 6000 platform (150 bp paired-end reads) at the Research Technology Support Facility at Michigan State University (East Lansing, Michigan, United States). The quality of the sequencing reads was evaluated using the FastQC program, focusing on per-base quality scores. Each of the six biological samples yielded 19–21 million paired reads (MR), with average quality scores exceeding 35, ensuring suitability for transcriptome analysis.
A transcriptome reference was assembled from approximately 120 million paired reads (MR) obtained from all six NT and transgenic lines using Trinity v2.15.1 (Haas et al., 2013). This reference was used for differential expression analysis. Transcripts identified as differentially expressed (DETs) with a false discovery rate (FDR) below 0.05 were selected for further analysis of various pathway genes. The transcriptome reference was annotated using Trinotate/4.0.2.
Pathway genes for nine phytohormones in Arabidopsis, including auxin, cytokinin, abscisic acid (ABA), ethylene, gibberellin (GA), BRs, jasmonic acid, salicylic acid, and strigolactones, were retrieved from the RIKEN Plant Hormone Research Network. Similarly, sugar pathway genes in Arabidopsis were identified. These hormone, MADS-box, and sugar pathway genes from Arabidopsis were used as queries to perform BLAST searches against the transcriptome reference, and isoforms with e-values less than -20 were identified for transcriptome comparisons. Flowering pathway genes in Arabidopsis and cereals (Walworth et al., 2016) were used to analyze flowering-related differentially expressed transcripts (DETs) identified in this study. Gene networks of overrepresented gene ontology (GO) terms for the selected DETs were constructed using Cytoscape 3.10.3.
Six selected DEGs were further analyzed through RT-qPCR using using the SYBR Green system (LifeTechnologies, Carlsbad, CA). A tomato ACTIN gene served as the reference gene to normalize the RT-qPCR results. All primers used in the analysis are included in Supplementary Table S1. The same RNA samples used for RNA sequencing, including three biological samples and three technical replicates, were used for the RT-qPCR analysis. Fold changes were calculated using 2−ΔΔCt, where ΔΔCt = (CtTARGET – CtNOM)transgenic – (CtTARGET – CtNOM)non-transgenic.
2.4 Data analysis
Data were processed using Microsoft Office Excel. Graphing and statistical analysis were generated in R (version 4.4.0) using the ggplot, annova, and emmeans packages. Fisher’s Protected Least Significant Difference test was used to determine statistical significance at p < 0.01 and p < 0.05 levels.
3 Results
3.1 Phylogenetic analysis of tomato’s SOC1 and SOC1-like proteins
The tomato proteome in Phytozome v13 includes one SOC1 protein and four SOC1-like proteins, which have been identified as SlTM3, SlSTM3, SlMBP18, and SlMBP23 by Zahn et al (Zahn et al., 2023). These five genes are distributed across four chromosomes (Figure 1). Their protein sequences exhibit high similarity to Arabidopsis SOC1, GmSOC1, ZmSOC1, and VcSOC1K. Notably, GmSOC1 and ZmSOC1 cluster with SlMBP18 and the SlSOC1-like protein XP_004252597.2, while VcSOC1 clusters with SlTM3, SlSTM3, SlMBP23, and Arabidopsis SOC1.
In the transcriptome reference assembled from all transcripts in the leaves of the ‘Ailsa Craig’ cultivar used in this study, transcripts corresponding to SlTM3, SlSTM3, SlMBP18, and the protein identical to XP_004252597.2 were detected, while SlMBP23 was absent.
The conserved nature of SOC1 and SOC1-like genes suggests that their orthologs SlTM3, SlSTM3, SlMBP18, and SlMBP23 in tomato may play similar roles to those in other plant species. Consequently, the constitutive expression of GmSOC1, ZmSOC1, and VcSOC1K may have the potential to influence fruit production.
3.2 Phenotypic changes in T0 transgenic plants
A total of 69 hygromycin-resistant T0 transgenic lines for GmSOC1_CX, 51 for ZmSOC1_CX, and 92 for VcSOC1K_CX were obtained across the three constructs, providing sufficient material for selecting comparable lines for phenotyping and seed harvesting (Figures 1B).
The constitutive expression of GmSOC1 (GmSOC1_CX) resulted in non-significant changes, such as earlier flowering and increases in plant height and the number of flower clusters (Supplementary Figures S1A–C). Significant changes included increases in branch and fruit numbers, enhanced fruit production, larger fruit size, and earlier fruit maturation (Supplementary Figures S1D–H).
The constitutive expression of ZmSOC1 (ZmSOC1_CX) led to significant changes, including earlier flowering, earlier fruit maturation, and increased fruit number and production (Supplementary Figures S1A, E–G). Non-significant changes included decreases in plant height, numbers of flower clusters and branches, and fruit size (Supplementary Figures S1B–D, H).
The constitutive expression of VcSOC1K (VcSOC1K_CX) induced significant changes, including earlier flowering, increased numbers of flower clusters and fruits, enhanced fruit production, and reduced fruit size (Supplementary Figures S1A, C, F–H). It also caused non-significant changes, such as earlier fruit maturation and increases in plant height and branch number (Supplementary Figures S1B, D, E).
Taken together, GmSOC1_CX, ZmSOC1_CX, and VcSOC1K_CX all contributed to increased fruit numbers and production (Supplementary Figure S1).
3.3 Phenotypic changes in T1 transgenic plants
T1 transgenic plants were phenotypically compared with their corresponding plants (Figures 2A). Under greenhouse conditions, among the nine groups with constitutive expression, ZmSOC1_CX17 was the only group of T1 transgenic plants to exhibit significantly earlier flowering, while the other T1 groups showed no difference in flowering time (Figure 2).
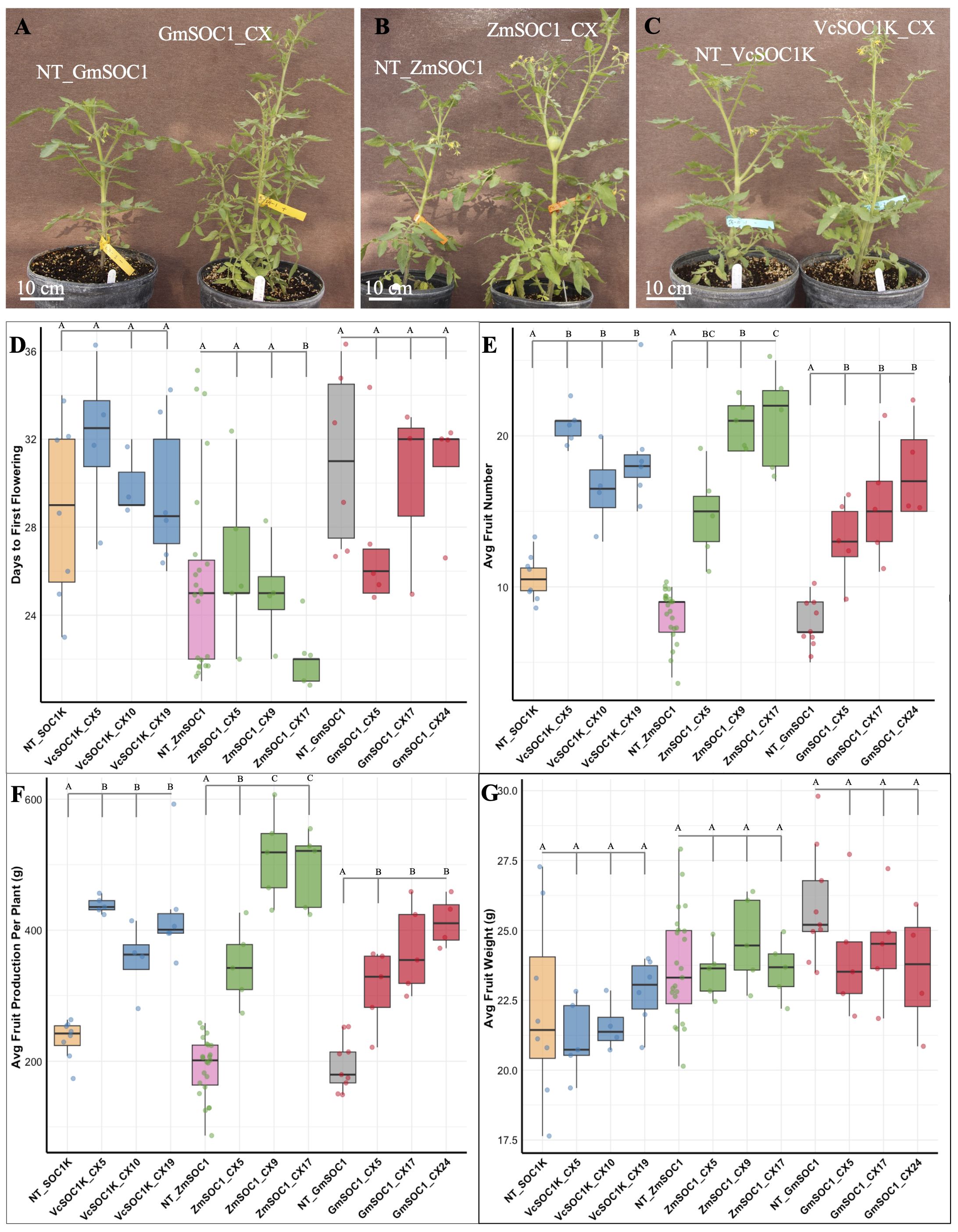
Figure 2. Phenotypic comparisons of three T1 transgenic lines for GmSOC1_CX, ZmSOC1_CX, and VcSOC1K_CX, alongside their corresponding non-transgenic (NT) seedlings (NT_GmSOC1, NT_ZmSOC1, and NT_VcSOC1K), with n > 3 for each transgenic and NT group. (A–C) Representative T1 plants. (D) Days to the appearance of the first flower after transplantation into a one-gallon pot. (E) Total number of fruits harvested. (F) Total weight of harvested fruits. (G) Average weight per fruit. The y-axis shows averages, and bars indicate standard deviations. Different letters on the bars indicate significant difference at p ≤ 0.05 by Fisher’s Protected Least Significant Difference test.
Consistent with observations in T0 plants, GmSOC1_CX, ZmSOC1_CX, and VcSOC1K_CX all resulted in increased fruit numbers and overall production, while the average fruit size showed no significant decrease (Figures 2E).
3.4 Transcriptomic analysis of T1 GmSOC1_CX plants
The T1 GmSOC1_CX plants were selected for transcriptome analysis to confirm the presence of transgenes and identify genes responsive to GmSOC1 expression in tomato. This selection builds on previous transcriptome analyses of ZmSOC1_CX in soybean and maize, as well as VcSOC1K_CX in blueberry and maize, which have been reported (Song and Chen, 2018; Han et al., 2021; Song and Han, 2021; Song et al., 2021).
The assembled transcriptome reference comprised 61,563 transcripts corresponding to 20,455 annotated genes. In GmSOC1_CX leaves, 565 differentially expressed transcripts (DETs) associated with 479 differentially expressed genes (DEGs) were identified. Among these DEGs, the two transgenes, GmSOC1 and hpt, showed fold change (FC) of 23,170 [Log2FC(transgenic/non-transgenic) = 14.5] and 7,131 [Log2FC(transgenic/non-transgenic) = 12.8], respectively, confirming the expression of GmSOC1 (Table 1). Additionally, RT-qPCR analysis of six selected DETs yielded results consistent with their RNA-seq data (Figure 3), supporting the reliability of the RNA-seq findings.
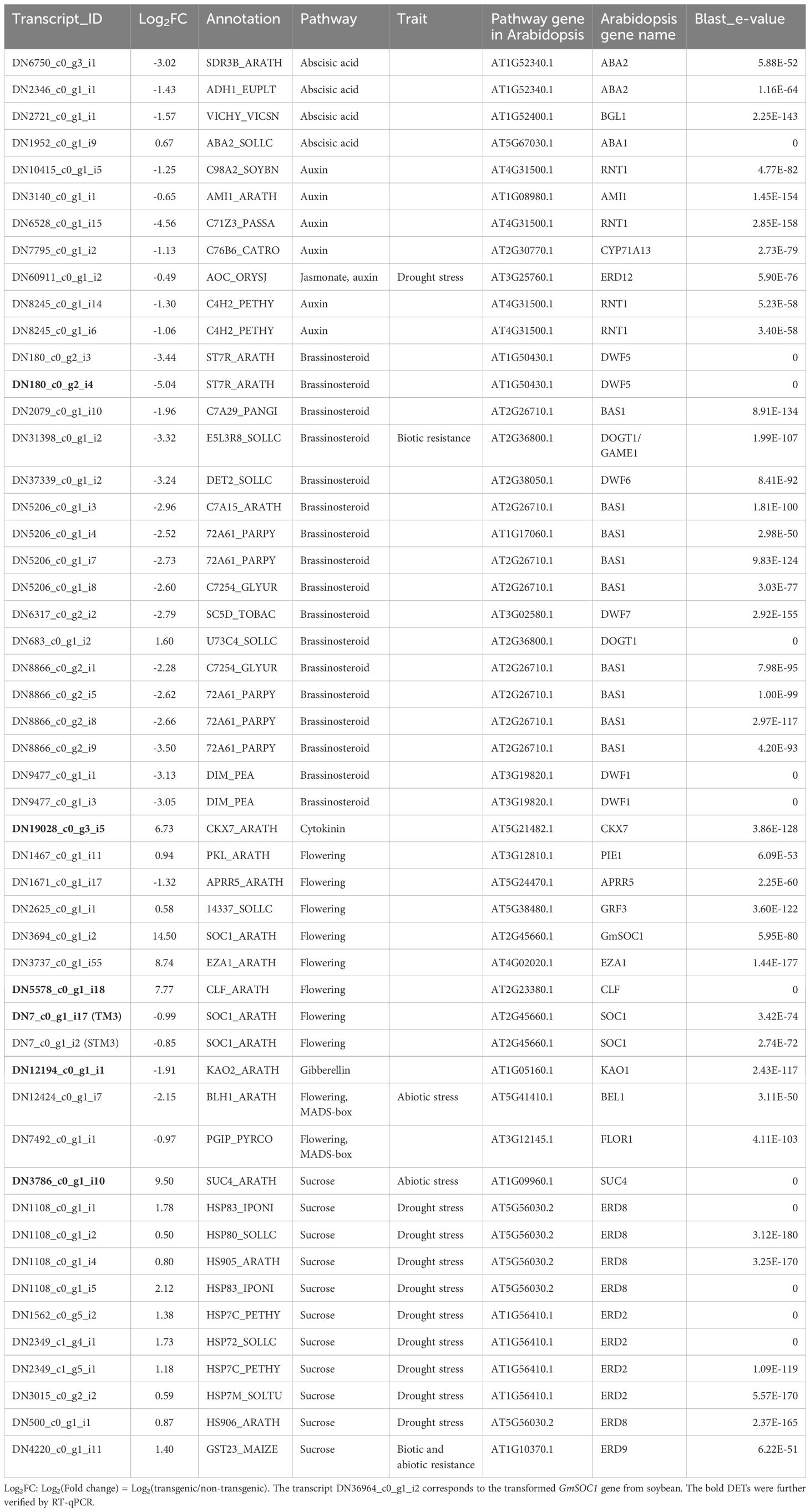
Table 1. Differentially expressed transcripts (DETs) of flowering pathway, hormone, MADS-box genes, and sucrose-related genes in leaves of GmSOC1-CX plants.
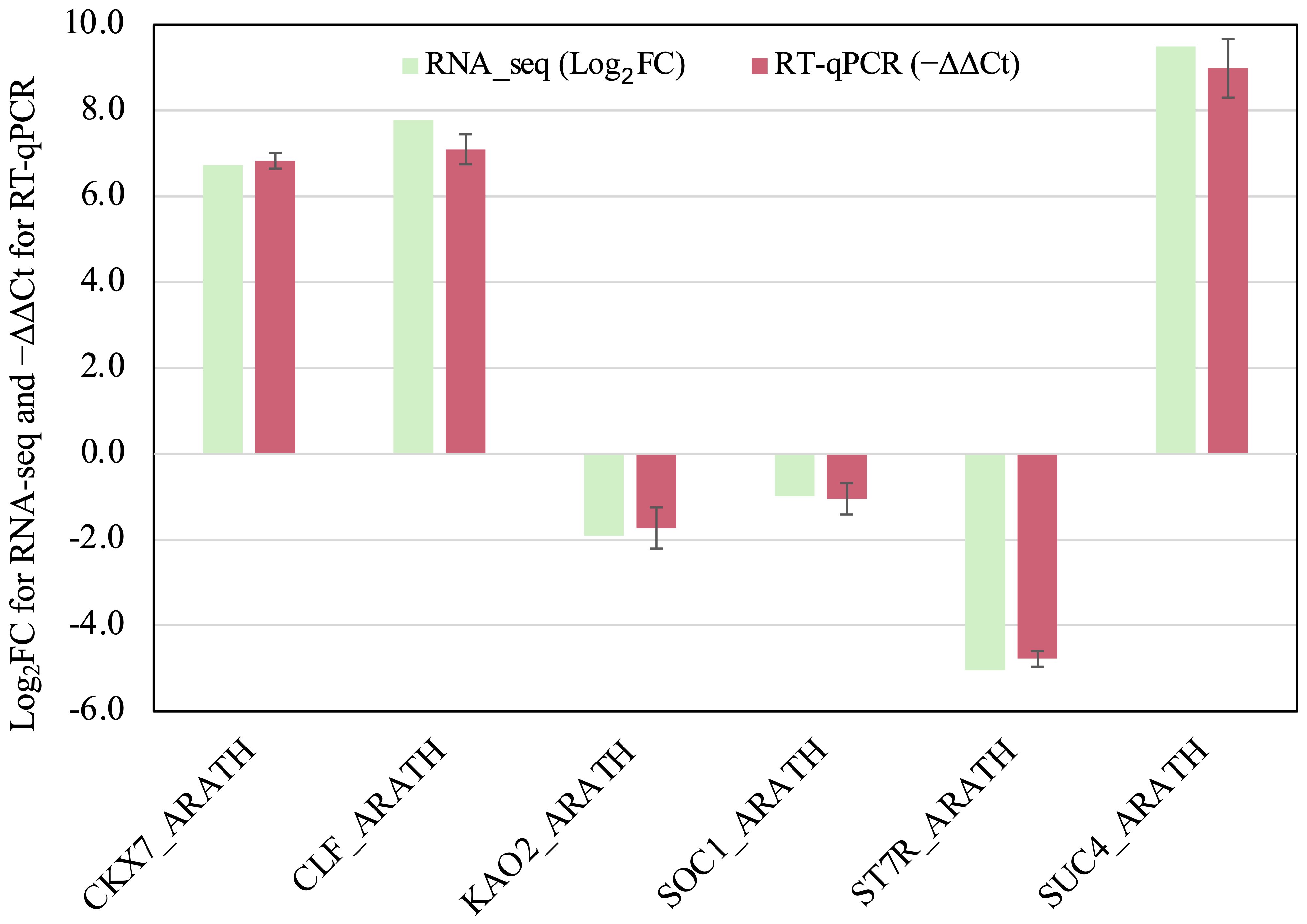
Figure 3. RT-qPCR analysis of the six selected DEGs listed in the Table 1. The −ΔΔCt values represent the average of three biological replicates and three technical replicates for each DEG. Tomato ACTIN gene SlACTIN12 (TRINITY_DN938_c0_g2_i1: ACT12_SOL) was used as the reference gene for normalization. Error bars represent standard deviation. No statistically significant differences were observed between RNA-seq and RT-qPCR data for any of the six genes analyzed (p < 0.05).
Analysis of the 479 DEGs using the GOSlim_Plants ontology file in BiNGO identified 27 overrepresented Gene Ontology (GO) terms. These included 11 terms under “Biological Process”, seven under “Molecular Function”, and nine under “Cellular Component” (Figure 4). These overrepresented GO terms suggest a broad impact of GmSOC1_CX on plant development at the transcript level, contributing to the phenotypic changes observed in GmSOC1_CX plants. For example, six overrepresented GO terms within the “Biological Process” category, namely “reproduction”, “response to abiotic stimulus”, “response to stress”, “post-embryogenic development”, “regulation of gene expression, epigenetic”, and “response to endogenous stimulus”, affect fruit production. These processes are linked through the overrepresented GO terms in the “Molecular Function” category, which regulate “transcription regulator activity” and “catalytic activity” via “binding” (Figure 4).
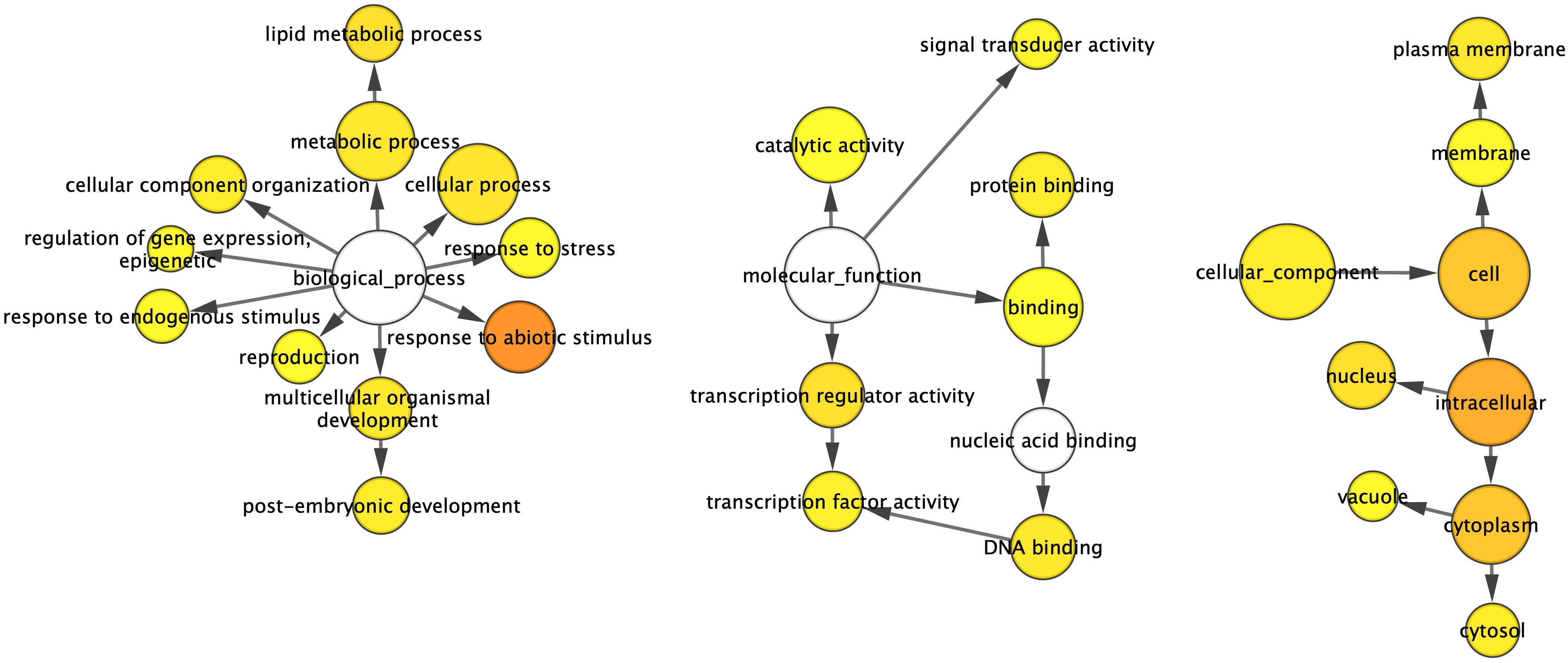
Figure 4. Gene networks of differentially expressed transcripts (DETs) identified from the comparison between the GmSOC1_CX and non-transgenic leaves. The ontology file of GOSlim_Plants in BiNGO was used to identify overrepresented GO terms (p < 0.05). Bubble size and color indicate the frequency of the GO term and the P-value, respectively.
Further analysis of the 565 DETs for key genes identified 51 key DETs associated with flowering, phytohormones, and sucrose. These were annotated to 39 DEGs and exhibited high similarity to 28 Arabidopsis genes (Table 1). These DETs were annotated to 39 DEGs and showed high similarities to 28 Arabidopsis genes (Table 1). Among the 39 DEGs, GmSOC1 was highly expressed (Log2FC(transgenic/non-transgenic) = 14.5) and suppressed the expression of two major endogenous tomato SOC1 orthologues, SlTM3 (TRINITY_DN7_c0_g1_i17) and SlSTM3 (TRINITY_DN7_c0_g1_i2). In tomato, SlTM3 and SlSTM3 promote the floral transition but serve opposing roles in inflorescence development (Zahn et al., 2023). High expression of SLSTM genes contributes to highly branched inflorescences (Wang et al., 2021).
The transcription factor BEL1-like homeodomain protein 1 (BLH1), which shares high similarity with the MADS-box gene BEL1, was downregulated. BLH family transcription factors are multifunctional, playing critical roles in plant morphogenesis, flower and fruit development, and responses to various environmental factors (Niu and Fu, 2022). Similarly, the polygalacturonase inhibitor precursor (PGIP), which exhibits a high similarity to the MADS-box gene FLOR1, known to promote flowering under long-day conditions (Torti et al., 2012), was also repressed.
The expression of two Histone-lysine N-methyltransferase genes CURLY LEAF (CLF) and ENHANCER OF ZESTE 1 POLYCOMB REPRESSIVE COMPLEX 2 SUBUNIT (EZA1), was enhanced. These genes encode catalytic subunits of the polycomb group (PcG) multiprotein complex. CLF is essential for regulating floral development by repressing the AGAMOUS homeotic gene. It achieves this by forming a nuclear complex with EZA1 and other components, targeting ABA- and glucose-responsive elements (Saleh et al., 2007; Liu et al., 2019; Shu et al., 2020; Liu et al., 2022). Notably, a loss-of-function mutant of Brassica rapa exhibits early flowering (Poza-Viejo et al., 2024), suggesting that the upregulation of CLF and EZA1 may contribute to either no significant impact or delaying flowering (Figure 5).
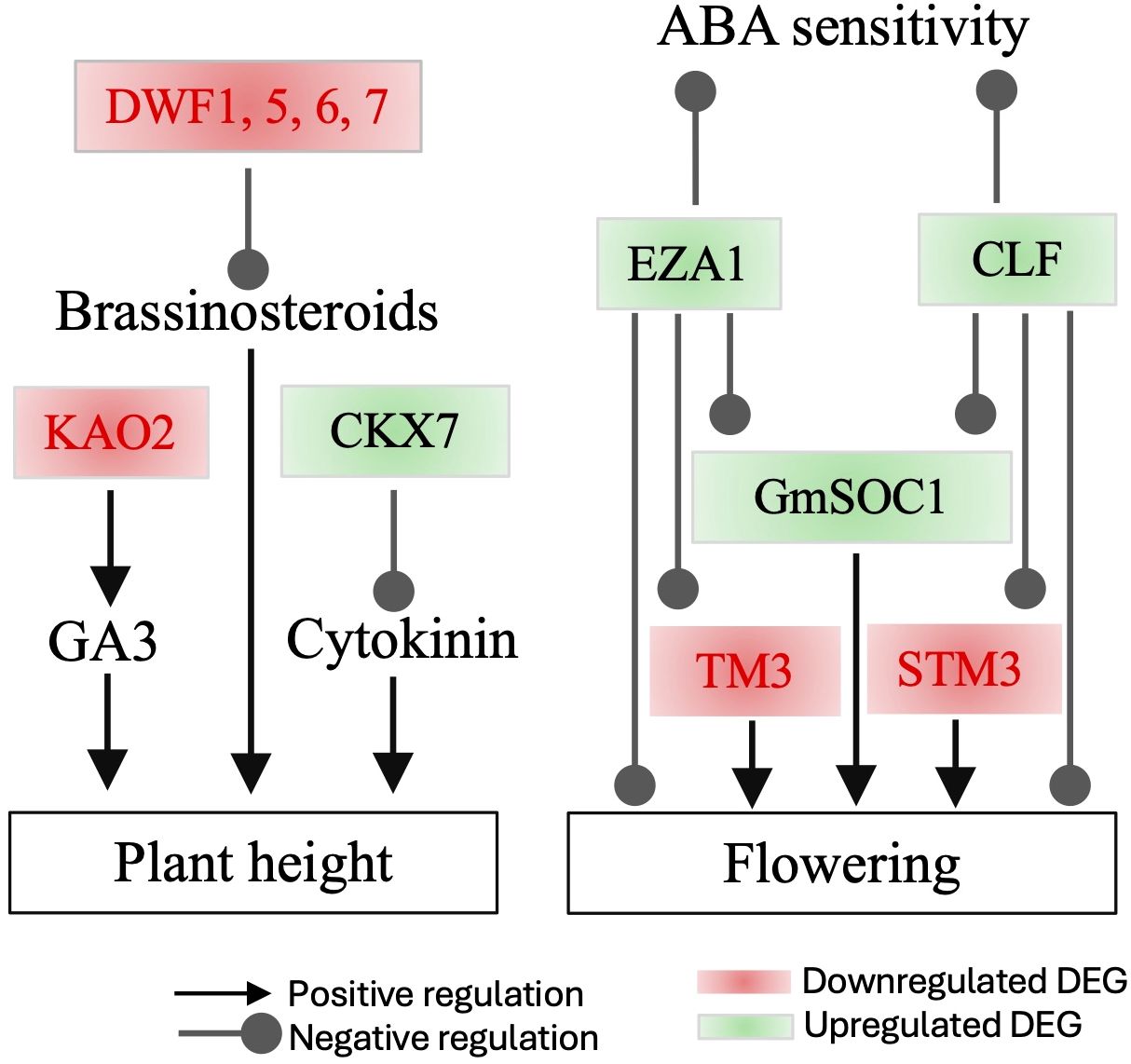
Figure 5. Interactions among major differentially expressed genes (DEGs) influencing tomato flowering and plant height in the leaves of GmSOC1_CX. The DEG data are presented in Table 1, and the proposed interactions are based on published information.
Of the genes involved in brassinosteroids (BRs) metabolism (Bajguz et al., 2020), the DEGs with repressed expressions included BAS1 and four DWARF genes (DWF1, DWF5, DWF6, and DWF7) (Table 1); these DEG can functionally lead to BRs-deficient changes, which are often associated with decreased BRs that can affect multiple agronomic traits (Bishop, 2003; Li et al., 2016; Zhan et al., 2022). The E5L3R8_SOLLC encodes tomato GLYCOALKALOID METABOLISM1 (GAME1) involved in the steroidal alkaloids (SAs) pathway (Itkin et al., 2011; Averello et al., 2025). The E5L3R8_SOLLC showed decreased expression (Table 1), suggesting a potential lower production of α-tomatine that has impact on plant defense system and fruit quality (Cárdenas et al., 2016; Sonawane et al., 2018; Averello et al., 2025).
For the cytokinin pathway genes, the expression of CKX7 was enhanced, meaning a potential of a reduced cytokinin level in the GmSOC1-CX plants (Köllmer et al., 2014). Similarly, a decreased expression of the KAO2 was found (Table 1), indicating a likely lower level of GA production (Regnault et al., 2014; Shani et al., 2024).
Among the DEGs related to sucrose, the tomato sucrose transporter SlSUT4 (SUC4_ARATH) was upregulated (Table 1). While the downregulation of SlSUT4 has been shown to promote flowering by enhancing sucrose transport to the shoot apex, its overexpression does not significantly impact flowering time or the expression of key genes in the flowering pathway (Liang et al., 2023).
Additionally, seven DEGs of HEAT SHOCK PROTEINs showed upregulation (Table 1), potentially contributing to enhanced tolerance to various abiotic stresses (Ul Haq et al., 2019). Similarly, the increased expression of the GLUTATHIONE S-TRANSFERASE (GST) 23 is associated with both biotic and abiotic resistance (Hernández Estévez and Rodríguez Hernández, 2020). In contrast, the tomato ALCOHOL DEHYDROGENASE (ADH1_EUPLT) and its homolog YFE37, annotated as SDR3B_ARATH, were both repressed. This reduction in expression may negatively impact disease resistance (Xun et al., 2022).
4 Discussion
Tomato is commonly used as a model plant to study fruit-related traits, primarily due to its suitability for genetic transformation. In this study, we demonstrate for the first time that the ectopic expression of full-length or partial SOC1 can enhance tomato fruit production per plant through a complex interaction of multiple genes and pathways. It is worth noting that ‘Ailsa Craig’ is an indeterminate tomato variety, which adds complexity to phenotyping flowering and fruiting traits in this study. When T0 plants were examined, they were of similar size at the time of re-potting. However, for T1 plants, which were grown from seed germination, plant sizes were more variable at the time of re-potting. Importantly, phenotyping was conducted prior to genotyping by PCR, which minimized potential bias in the phenotypic data and ensured its reliability.
4.1 Flowering time and the expression of Full-length SOC1 and partial SOC1
As a key integrator in the plant flowering pathway (Fornara et al., 2010; Lee and Lee, 2010; Immink et al., 2013), enhanced expression of SOC1 or its orthologues, either though overexpression or ectopic expression, can promote flowering. This phenomenon has been reported for many plant species, including tomato (Zahn et al., 2023). Among the five SOC1 and SOC1-like genes identified in tomato (Figure 1), SlTM3 and SlSTM3 play a more significant role in flowering initiation compared to SlMBP23 and SlMBP18, at least in the indeterminate cultivar Moneyberg (Zahn et al., 2023). Additionally, the high expression of SlSTM3 has been linked to a highly branched inflorescence phenotype (Wang et al., 2021).
In this study, phylogenetic analysis revealed that GmSOC1 and ZmSOC1 clustered closely with SlMBP18 and the fifth SlSOC1-like gene, while VcSOC1K showed closer similarity to SlTM3, SlSTM3, and SlMBP23 (Figure 1). Phenotypic observations demonstrated that both ZmSOC1-CX and VcSOC1-CX transgenic plants exhibited earlier flowering compared to non-transgenic controls, with statistically significant differences (P < 0.01) in the T0 generation and no significant differences (P < 0.01) in the T1 generation (Figure 2; Supplementary Figure S1A). In contrast, GmSOC1-CX plants did not show significant changes in flowering time across both T0 and T1 generations (P < 0.05). The early flowering phenotype observed in ZmSOC1-CX lines suggests that the expression of SlMBP18 and the fifth SOC1-like gene of tomato may play a role in promoting flowering. For the T1 generation, it would have been valuable to investigate additional traits such as seed germination time and seedling growth, as previous studies have shown that repressed expression of DWARF genes can delay seed germination (Li et al., 2016). Notably, while GmSOC1-CX has been reported to induce early flowering in Arabidopsis (Zhong et al., 2012), its overexpression did not significantly promote flowering in soybean. However, GmSOC1 knock-out mutants exhibited delayed flowering (Kou et al., 2022), highlighting the complex and species-specific regulatory roles of GmSOC1 in flowering time control.
Interestingly, in this study, the T0 generation of VcSOC1K-CX plants from four transgenic lines exhibited earlier flowering compared to the non-transgenic lines, whereas the T1 generation of three transgenic lines did not show this early flowering phenotype. Previous studies have reported that overexpression of VcSOC1K in blueberry and VcSOC1K-CX in tobacco promoted flowering (Song et al., 2013; Song and Chen, 2018); however, VcSOC1K-CX did not significantly induce early flowering in maize (Song and Han, 2021). This variation may be attributed to many factors, including differences in expression levels, plant species, and genotype.
At the transcript levels, GmSOC1-CX did not promote flowering, at least not significantly, is likely due to the enhanced expression of two Histone-lysine N-methyltransferase genes, CLF and EZA1 (Figure 5) (Saleh et al., 2007; Liu et al., 2019; Shu et al., 2020; Liu et al., 2022; Poza-Viejo et al., 2024).
4.2 Plant architecture and the expression of full-length SOC1 and partial SOC1
Plant architecture, including shoot and inflorescence structure, is an omnigenic trait that can significantly influence tomato productivity (Alonge et al., 2020; Gaarslev et al., 2021). Tomato SOC1 genes, such as SlTM3 and SlSTM3, serve as core regulators of inflorescence structure by interacting with other MADS-box genes (Alonge et al., 2020; Gaarslev et al., 2021; Wang et al., 2021; Zahn et al., 2023). Overexpression of SlTM3 and SlSTM3 enhances inflorescence branching (Wang et al., 2021), while their suppression reduces branching (Alonge et al., 2020; Zahn et al., 2023). In this study, transgenic lines expressing GmSOC1, ZmSOC1, and VcSOC1K exhibited no noticeable changes in inflorescence structure. For GmSOC1-CX lines, this was further supported at the transcript level by the unchanged expression of tomato FRUITFULL1 (FUL1)(Table 1), a direct activation target of SlSTM3 (Wang et al., 2021).
Phenotypically, the effect of SOC1 expression on plant height and branching has been less consistent in the literature compared to its well-established role in flowering time. In soybean, soc1 mutants have more internodes than the wild type, but the impact of GmSOC1 overexpression on plant architecture remains unclear (Kou et al., 2022). In Medicago truncatula, MtSOC1 has been shown to influence both flowering and primary stem height in both mutant and overexpression lines (Jaudal et al., 2018). Additionally, ZmSOC1-CX expression has been associated with reduced plant height in transgenic maize and soybean plants (Han et al., 2021; Song et al., 2021). In tomato, SlTM3 and SlSTM3 expression have not displayed any significant impact on plant height and branching (Zahn et al., 2023). In this study, GmSOC1-CX, ZmSOC1-CX, and VcSOC1-CX did not significantly change plant height but enhanced branching for at least the GmSOC1-CX and VcSOC1-CX plants (Figures 2A).
At the transcript level, it is noteworthy that the repressed expression of four DWARF genes could theoretically lead to reducing plant size in tomato due to less BRs production, as suggested by previous studies (Figure 5) (Bishop et al., 1999; Bishop, 2003; Montoya et al., 2005; Li et al., 2016; Zhan et al., 2022). The enhanced expression of CKX7 could result in reduced cytokinin levels, which are expected to shorten plant height, increase branching, and reduce flower number (Eckardt, 2003; Bartrina et al., 2011; Köllmer et al., 2014; Waldie and Leyser, 2018). Similarly, a decreased expression of KAO2 might reduce GA production (Regnault et al., 2014), potentially leading to delayed seed germination, stunted plant growth, and delayed flowering. In this study, although GmSOC1-CX did not exhibit phenotypic changes such as plant dwarfing, delayed flowering, or reduced flower number, it did show increased branching (Figures 1C, Figures 2A, and Supplementary Figure S1D). However, BR, cytokinin, and GA levels, as well as seed germination timing, were not investigated in this work.
4.3 Tomato fruit yield and the expression of full-length SOC1 and partial SOC1
Crop yield-defining traits vary across different crops (Bailey-Serres et al., 2019). For tomatoes, yield-defining traits include both direct factors, such as fruit number, fruit size, and fruit production efficiency per unit area, as well as related traits, including tolerance to abiotic and biotic stresses (Alonge et al., 2020; Bhandari et al., 2023). Accordingly, hormone and flowering pathway genes have become the targets for genetic improvement of yield (Krieger et al., 2010; Ariizumi et al., 2013; Wang et al., 2021; Cui et al., 2022; Kang et al., 2022). In this study, two full-length SOC1 genes from maize and soybean, along with the K-domain of the blueberry SOC1 gene, were constitutively expressed in tomato. The resulting transgenic lines exhibited an increased fruit count, leading to higher total fruit production per plant, suggesting enhanced yield potential. This rise in fruit number was associated with improved branching in the transgenic plants, likely due to decreased BRs resulting from the repression of DWARF genes. Notably, similar effects have not been reported in tomato through the overexpression of SlTM3 and SlSTM3. Interestingly, lower expression levels of GmSOC1 have been shown to enhance soybean yield (Kou et al., 2022).
In addition to its essential role in flowering, SOC1 also plays a role in other processes in Arabidopsis, such as root development and leaf senescence, both of which impact crop yield (Chen et al., 2017; Castañón-Suárez et al., 2024). However, the impact of SOC1 overexpression on root development and leaf senescence in crops remains largely unexplored. Previously, we found that constitutive expression of three SOC1 genes, either full-length or partial, has the potential to enhance yield in maize, soybean, and blueberry, primarily through the regulation of flowering pathway genes, as indicated by RNA-seq data (Song and Chen, 2018; Han et al., 2021; Song and Han, 2021; Song et al., 2021). In this study, unlike our previous findings, the observed increase in fruit production per transgenic plant is attributed to enhanced branching, likely due to the repressed expression of DWARF genes (Figure 5). Additionally, the increased expression of CLF and EZA1 in the RNA-seq data of the GmSOC1-CX lines appears to provide evidence explaining the unchanged flowering time (Figure 5).
4.4 Tomato fruit quality, biotic and abiotic tolerance, and other traits
In this study, the biotic and abiotic tolerance of the transgenic plants were not directly assessed. However, the identification of several DEGs associated with these traits suggests that the transgenes may have influenced plant resilience to biotic and abiotic stresses (Figure 4, Table 1). Additionally, fruit quality as well as the other traits may have been impacted by the DEGs.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found in the NCBI SRA repository under accession number PRJNA1280106 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1280106).
Author contributions
GD: Investigation, Writing – review & editing, Writing – original draft. JJ: Formal analysis, Writing – review & editing, Data curation. GS: Funding acquisition, Resources, Project administration, Validation, Formal analysis, Writing – original draft, Data curation, Supervision, Writing – review & editing, Conceptualization, Methodology.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
We extend our gratitude to Dr. Xue Han for her assistance in caring for the plants in the greenhouse. The work was supported partially by AgBioResearch of Michigan State University. GH study in the US were supported by Ministry of the Higer Education in the Kurdistan region in Iraq.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1640731/full#supplementary-material
Supplementary Figure 1 | Phenotypic comparisons of T0 transgenic lines (GmSOC1_CX, n = 5; ZmSOC1_CX, n = 4; and VcSOC1K_CX, n = 3) and their corresponding non-transgenic (NT) lines (NT_GmSOC1, n = 4; NT_ZmSOC1, n = 3; and NT_VcSOC1K, n = 3). (A) Days to the appearance of the first flower after potting in a one-gallon pot. (B) Plant height at the time of first flowering. (C) Number of flower clusters counted after all fruits were harvested. (D) Number of branches counted after all fruits were harvested. (E) Days to the appearance of the first mature fruit after potting in a one-gallon pot. (F) Total number of fruits harvested. (G) Total weight of harvested fruits. (H) Average weight per fruit. The y-axis shows averages, and bars indicate standard deviations.
References
Alonge, M., Wang, X., Benoit, M., Soyk, S., Pereira, L., Zhang, L., et al. (2020). Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182, 145–161.e123. doi: 10.1016/j.cell.2020.05.021
Alvarez-Buylla, E. R., Pelaz, S., Liljegren, S. J., Gold, S. E., Burgeff, C., Ditta, G. S., et al. (2000). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. U.S.A. 97, 5328–5333. doi: 10.1073/pnas.97.10.5328
Ariizumi, T., Shinozaki, Y., and Ezura, H. (2013). Genes that influence yield in tomato. Breed Sci. 63, 3–13. doi: 10.1270/jsbbs.63.3
Averello, V., Hegeman, A. D., and Chen, C. (2025). Finding the balance: Modifying the cholesterol and steroidal glycoalkaloid synthesis pathway in tomato (Solanum lycopersicum L.) for human health, fruit flavor, and plant defense. Hortic. Plant J. 11, 42–56. doi: 10.1016/j.hpj.2023.05.019
Bailey-Serres, J., Parker, J. E., Ainsworth, E. A., Oldroyd, G. E. D., and Schroeder, J. I. (2019). Genetic strategies for improving crop yields. Nature 575, 109–118. doi: 10.1038/s41586-019-1679-0
Bajguz, A., Chmur, M., and Gruszka, D. (2020). Comprehensive overview of the brassinosteroid biosynthesis pathways: substrates, products, inhibitors, and connections. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01034
Bartrina, I., Otto, E., Strnad, M., Werner, T., and Schmulling, T. (2011). Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23, 69–80. doi: 10.1105/tpc.110.079079
Becker, A. and Theissen, G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet Evol. 29, 464–489. doi: 10.1016/S1055-7903(03)00207-0
Bhandari, P., Kim, J., and Lee, T. G. (2023). Genetic architecture of fresh-market tomato yield. BMC Plant Biol. 23, 18. doi: 10.1186/s12870-022-04018-5
Bishop, G. J. (2003). Brassinosteroid mutants of crops. J. Plant Growth Regul. 22, 325–335. doi: 10.1007/s00344-003-0064-1
Bishop, G. J., Nomura, T., Yokota, T., Harrison, K., Noguchi, T., Fujioka, S., et al. (1999). The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc. Natl. Acad. Sci. 96, 1761–1766. doi: 10.1073/pnas.96.4.1761
Busi, M. V., Bustamante, C., D’Angelo, C., Hidalgo-Cuevas, M., Boggio, S. B., Valle, E. M., et al. (2003). MADS-box genes expressed during tomato seed and fruit development. Plant Mol. Biol. 52, 801–815. doi: 10.1023/a:1025001402838
Cárdenas, P. D., Sonawane, P. D., Pollier, J., Vanden Bossche, R., Dewangan, V., Weithorn, E., et al. (2016). GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nat. Commun. 7, 10654. doi: 10.1038/ncomms10654
Castañón-Suárez, C. A., Arrizubieta, M., Castelán-Muñoz, N., Sánchez-Rodríguez, D. B., Caballero-Cordero, C., Zluhan-Martínez, E., et al. (2024). The MADS-box genes SOC1 and AGL24 antagonize XAL2 functions in Arabidopsis thaliana root development. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1331269
Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116. doi: 10.1007/BF02670468
Chen, J., Zhu, X., Ren, J., Qiu, K., Li, Z., Xie, Z., et al. (2017). Suppressor of overexpression of CO 1 negatively regulates dark-induced leaf degreening and senescence by directly repressing pheophytinase and other senescence-associated genes in arabidopsis. Plant Physiol. 173, 1881–1891. doi: 10.1104/pp.16.01457
Colombo, L., Battaglia, R., and Kater, M. M. (2008). Arabidopsis ovule development and its evolutionary conservation. Trends Plant Sci. 13, 444–450. doi: 10.1016/j.tplants.2008.04.011
Cui, L., Zheng, F., Wang, J., Zhang, C., Zhang, D., Gao, S., et al. (2022). The tomato CONSTANS-LIKE protein SlCOL1 regulates fruit yield by repressing SFT gene expression. BMC Plant Biol. 22, 429. doi: 10.1186/s12870-022-03813-4
Danial, G. H., Ibrahim, D. A., and Song, G. Q. (2021). Agrobacterium-mediated transformation of two tomato cultivars (Lycopersicon esculentum Mill.) cv. Sandra and Rocky. Iraqi J. Agric. Sci. 52, 745–755. doi: 10.36103/ijas.v52i3.1366
Doyle, J. J. and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bull. 19, 5.
Eckardt, N. A. (2003). A new classic of cytokinin research: Cytokinin-deficient Arabidopsis plants provide new insights into cytokinin biology. Plant Cell 15, 2489–2492. doi: 10.1105/tpc.151110
Ehlers, K., Bhide, A. S., Tekleyohans, D. G., Wittkop, B., Snowdon, R. J., and Becker, A. (2016). The MADS box genes ABS, SHP1, and SHP2 are essential for the coordination of cell divisions in ovule and seed coat development and for endosperm formation in arabidopsis thaliana. PLoS One 11, e0165075. doi: 10.1371/journal.pone.0165075
Fornara, F., de Montaigu, A., and Coupland, G. (2010). SnapShot: control of flowering in arabidopsis. Cell 141 (3):550, 550.e1-2. doi: 10.1016/j.cell.2010.04.024
Gaarslev, N., Swinnen, G., and Soyk, S. (2021). Meristem transitions and plant architecture—learning from domestication for crop breeding. Plant Physiol. 187, 1045–1056. doi: 10.1093/plphys/kiab388
Gamborg, O. L., Miller, R. A., and Ojima, K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50, 151–158. doi: 10.1016/0014-4827(68)90403-5
Gramzow, L., Ritz, M. S., and Theißen, G. (2010). On the origin of MADS-domain transcription factors. Trends Genet. 26, 149–153. doi: 10.1016/j.tig.2010.01.004
Gramzow, L. and Theissen, G. (2010). A hitchhiker’s guide to the MADS world of plants. Genome Biol. 11, 214. doi: 10.1186/gb-2010-11-6-214
Gramzow, L. and Theissen, G. (2013). Phylogenomics of MADS-box genes in plants - two opposing life styles in one gene family. Biol. (Basel) 2, 1150–1164. doi: 10.3390/biology2031150
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. doi: 10.1038/Nprot.2013.084
Han, X., Wang, D. C., and Song, G. Q. (2021). Expression of a maize SOC1 gene enhances soybean yield potential through modulating plant growth and flowering. Sci. Rep. 11, 12758. doi: 10.1038/s41598-021-92215-x
Henschel, K., Kofuji, R., Hasebe, M., Saedler, H., Münster, T., and Theißen, G. (2002). Two ancient classes of MIKC-type MADS-box genes are present in the moss physcomitrella patens. Mol. Biol. Evol. 19, 801–814. doi: 10.1093/oxfordjournals.molbev.a004137
Hernández Estévez, I. and Rodríguez Hernández, M. (2020). Plant glutathione S-transferases: an overview. Plant Gene 23, 100233. doi: 10.1016/j.plgene.2020.100233
Immink, R. G. H., Posé, D., Ferrario, S., Ott, F., Kaufmann, K., Valentim, F. L., et al. (2013). Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators (vol 160, pg 433, 2012). Plant Physiol. 162, 2151–2151. doi: 10.1104/pp.113.900469
Itkin, M., Rogachev, I., Alkan, N., Rosenberg, T., Malitsky, S., Masini, L., et al. (2011). GLYCOALKALOID METABOLISM1 is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. Plant Cell 23, 4507–4525. doi: 10.1105/tpc.111.088732
Ito, Y., Kitagawa, M., Ihashi, N., Yabe, K., Kimbara, J., Yasuda, J., et al. (2008). DNA-binding specificity, transcriptional activation potential, and the mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 55, 212–223. doi: 10.1111/j.1365-313X.2008.03491.x
Jaudal, M., Zhang, L., Che, C., Li, G., Tang, Y., Wen, J., et al. (2018). A SOC1-like gene MtSOC1a promotes flowering and primary stem elongation in Medicago. J. Exp. Bot. 69, 4867–4880. doi: 10.1093/jxb/ery284
Kang, M. S., Kim, Y. J., Heo, J., Rajendran, S., Wang, X., Bae, J. H., et al. (2022). Newly discovered alleles of the tomato antiflorigen gene SELF PRUNING provide a range of plant compactness and yield. Int. J. Mol. Sci. 23 (13), 7149. doi: 10.3390/ijms23137149
Köllmer, I., Novák, O., Strnad, M., Schmülling, T., and Werner, T. (2014). Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J. 78, 359–371. doi: 10.1111/tpj.12477
Kou, K., Yang, H., Li, H., Fang, C., Chen, L., Yue, L., et al. (2022). A functionally divergent SOC1 homolog improves soybean yield and latitudinal adaptation. Curr. Biol. 32, 1728–1742.e1726. doi: 10.1016/j.cub.2022.02.046
Krieger, U., Lippman, Z. B., and Zamir, D. (2010). The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 42, 459–463. doi: 10.1038/ng.550
Kwantes, M., Liebsch, D., and Verelst, W. (2012). How MIKC* MADS-box genes originated and evidence for their conserved function throughout the evolution of vascular plant gametophytes. Mol. Biol. Evol. 29, 293–302. doi: 10.1093/molbev/msr200
Lee, J. and Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61, 2247–2254. doi: 10.1093/jxb/erq098
Li, X. J., Chen, X. J., Guo, X., Yin, L. L., Ahammed, G. J., Xu, C. J., et al. (2016). DWARF overexpression induces alteration in phytohormone homeostasis, development, architecture and carotenoid accumulation in tomato. Plant Biotechnol. J. 14, 1021–1033. doi: 10.1111/pbi.12474
Liang, Y., Bai, J., Xie, Z., Lian, Z., Guo, J., Zhao, F., et al. (2023). Tomato sucrose transporter SlSUT4 participates in flowering regulation by modulating gibberellin biosynthesis. Plant Physiol. 192, 1080–1098. doi: 10.1093/plphys/kiad162
Liu, Y., Bai, Y., Li, N., Li, M., Liu, W., Yun, D. J., et al. (2022). HEXOKINASE1 forms a nuclear complex with the PRC2 subunits CURLY LEAF and SWINGER to regulate glucose signaling. J. Integr. Plant Biol. 64, 1168–1180. doi: 10.1111/jipb.13261
Liu, C., Cheng, J., Zhuang, Y., Ye, L., Li, Z., Wang, Y., et al. (2019). Polycomb repressive complex 2 attenuates ABA-induced senescence in Arabidopsis. Plant J. 97, 368–377. doi: 10.1111/tpj.14125
Liu, Y., Cui, S., Wu, F., Yan, S., Lin, X., Du, X., et al. (2013). Functional conservation of MIKC*-Type MADS box genes in Arabidopsis and rice pollen maturation. Plant Cell 25, 1288–1303. doi: 10.1105/tpc.113.110049
Liu, C., Xi, W. Y., Shen, L. S., Tan, C. P., and Yu, H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16, 711–722. doi: 10.1016/j.devcel.2009.03.011
Masiero, S., Colombo, L., Grini, P. E., Schnittger, A., and Kater, M. M. (2011). The emerging importance of type I MADS box transcription factors for plant reproduction. Plant Cell 23, 865–872. doi: 10.1105/tpc.110.081737
Montoya, T., Nomura, T., Yokota, T., Farrar, K., Harrison, K., Jones, J. G. D., et al. (2005). Patterns of Dwarf expression and brassinosteroid accumulation in tomato reveal the importance of brassinosteroid synthesis during fruit development. Plant J. 42, 262–269. doi: 10.1111/j.1365-313X.2005.02376.x
Murashige, T. and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Niu, X. and Fu, D. (2022). The roles of BLH transcription factors in plant development and environmental response. Int. J. Mol. Sci. 23 (7), 3731. doi: 10.3390/ijms23073731
Parenicova, L., de Folter, S., Kieffer, M., Horner, D. S., Favalli, C., Busscher, J., et al. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15, 1538–1551. doi: 10.1105/tpc.011544
Poza-Viejo, L., Payá-Milans, M., Wilkinson, M. D., Piñeiro, M., Jarillo, J. A., and Crevillén, P. (2024). Brassica rapa CURLY LEAF is a major H3K27 methyltransferase regulating flowering time. Planta 260, 27. doi: 10.1007/s00425-024-04454-7
Raza, B., Hameed, A., and Saleem, M. Y. (2022). Fruit nutritional composition, antioxidant and biochemical profiling of diverse tomato (Solanum lycopersicum L.) genetic resource. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1035163
Regnault, T., Daviere, J. M., Heintz, D., Lange, T., and Achard, P. (2014). The gibberellin biosynthetic genes AtKAO1 and AtKAO2 have overlapping roles throughout Arabidopsis development. Plant J. 80, 462–474. doi: 10.1111/tpj.12648
Saleh, A., Al-Abdallat, A., Ndamukong, I., Alvarez-Venegas, R., and Avramova, Z. (2007). The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus. Nucleic Acids Res. 35, 6290–6296. doi: 10.1093/nar/gkm464
Seo, E., Lee, H., Jeon, J., Park, H., Kim, J., Noh, Y. S., et al. (2009). Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21, 3185–3197. doi: 10.1105/tpc.108.063883
Shani, E., Hedden, P., and Sun, T.-P. (2024). Highlights in gibberellin research: A tale of the dwarf and the slender. Plant Physiol. 195, 111–134. doi: 10.1093/plphys/kiae044
Shu, J., Chen, C., Li, C., and Cui, Y. (2020). The complexity of PRC2 catalysts CLF and SWN in plants. Biochem. Soc. Trans. 48, 2779–2789. doi: 10.1042/bst20200660
Sonawane, P. D., Heinig, U., Panda, S., Gilboa, N. S., Yona, M., Kumar, S. P., et al. (2018). Short-chain dehydrogenase/reductase governs steroidal specialized metabolites structural diversity and toxicity in the genus Solanum. Proc. Natl. Acad. Sci. 115, E5419–E5428. doi: 10.1073/pnas.1804835115
Song, G.-Q. and Chen, Q. (2018). Overexpression of the MADS-box gene K-domain increases the yield potential of blueberry. Plant Sci. 276, 10. doi: 10.1016/j.plantsci.2018.07.018
Song, G.-q. and Han, X. (2021). K-domain technology: constitutive expression of a blueberry keratin-like domain mimics expression of multiple MADS-box genes in enhancing maize grain yield. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.664983
Song, G. Q., Han, X., Ryner, J. T., Thompson, A., and Wang, K. (2021). Utilizing MIKC-type MADS-box protein SOC1 for yield potential enhancement in maize. Plant Cell Rep. 40, 1679–1693. doi: 10.1007/s00299-021-02722-4
Song, G. Q., Walworth, A., Zhao, D. Y., Hildebrandt, B., and Leasia, M. (2013). Constitutive expression of the K-domain of a Vaccinium corymbosum SOC1-like (VcSOC1-K) MADS-box gene is sufficient to promote flowering in tobacco. Plant Cell Rep. 32, 1819–1826. doi: 10.1007/S00299-013-1495-1
Torti, S., Fornara, F., Vincent, C., Andrés, F., Nordström, K., Göbel, U., et al (2012). Analysis of the arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. The Plant Cell 24, 444–462. doi: 10.1105/tpc.111.092791
Tyagi, S., Sri, T., Singh, A., Mayee, P., Shivaraj, S. M., Sharma, P., et al. (2019). SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 influences flowering time, lateral branching, oil quality, and seed yield in Brassica juncea cv. Varuna. Funct. Integr. Genomics 19, 43–60. doi: 10.1007/s10142-018-0626-8
Ul Haq, S., Khan, A., Ali, M., Khattak, A. M., Gai, W. X., Zhang, H. X., et al. (2019). Heat shock proteins: dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 20 (21), 5321. doi: 10.3390/ijms20215321
Verelst, W., Saedler, H., and Münster, T. (2006). MIKC* MADS-protein complexes bind motifs enriched in the proximal region of late pollen-specific arabidopsis promoters. Plant Physiol. 143, 447–460. doi: 10.1104/pp.106.089805
Waldie, T. and Leyser, O. (2018). Cytokinin targets auxin transport to promote shoot branching. Plant Physiol. 177, 803–818. doi: 10.1104/pp.17.01691
Walworth, A. E., Chai, B., and Song, G. Q. (2016). Transcript profile of flowering regulatory genes in vcFT-overexpressing blueberry plants. PLoS One 11, e0156993. doi: 10.1371/journal.pone.0156993
Wang, X., Liu, Z., Sun, S., Wu, J., Li, R., Wang, H., et al. (2021). SISTER OF TM3 activates FRUITFULL1 to regulate inflorescence branching in tomato. Horticulture Res. 8, 251. doi: 10.1038/s41438-021-00677-x
Weigel, D. and Meyerowitz, E. M. (1994). The ABCs of floral homeotic genes. Cell 78, 203–209. doi: 10.1016/0092-8674(94)90291-7
Xun, H., Qian, X., Wang, M., Yu, J., Zhang, X., Pang, J., et al. (2022). Overexpression of a cinnamyl alcohol dehydrogenase-coding gene, gsCAD1, from wild soybean enhances resistance to soybean mosaic virus. Int. J. Mol. Sci. 23 (23), 15206. doi: 10.3390/ijms232315206
Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A., and Meyerowitz, E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. doi: 10.1038/346035a0
Zahn, I. E., Roelofsen, C., Angenent, G. C., and Bemer, M. (2023). TM3 and STM3 promote flowering together with FUL2 and MBP20, but act antagonistically in inflorescence branching in tomato. Plants (Basel) 12 (15), 2754. doi: 10.3390/plants12152754
Zhan, H., Lu, M., Luo, Q., Tan, F., Zhao, Z., Liu, M., et al. (2022). OsCPD1 and OsCPD2 are functional brassinosteroid biosynthesis genes in rice. Plant Sci. 325, 111482. doi: 10.1016/j.plantsci.2022.111482
Keywords: brassinosteroids, flowering time, lycopersicon esculentum, MADS-box gene, plant architecture, soybean SOC1, yield enhancement
Citation: Danial GH, Jaikham J and Song G-q (2025) Constitutive expression of full-length or partial of SOC1 genes for yield enhancement in tomato. Front. Plant Sci. 16:1640731. doi: 10.3389/fpls.2025.1640731
Received: 04 June 2025; Accepted: 07 July 2025;
Published: 28 July 2025.
Edited by:
David Wm Leung, University of Canterbury, New ZealandReviewed by:
Panagiotis Madesis, University of Thessaly, GreeceVijay Sheri, Texas Tech University, United States
Copyright © 2025 Danial, Jaikham and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guo-qing Song, c29uZ2dAbXN1LmVkdQ==
 Gharbia H. Danial1,2
Gharbia H. Danial1,2 Guo-qing Song
Guo-qing Song