- Genetic Improvement for Fruits and Vegetables Laboratory, Beltsville Agricultural Research Center, U.S. Department of Agriculture-Agricultural Research Service, Beltsville, MD, United States
Foliar application of low-dose 2,4-dichlorophenoxyacetic acid (2,4-D) has been demonstrated to reduce potato common scab disease caused by phytopathogenic Streptomyces. Foliar-applied 2,4-D is translocated to the tubers but does not cause direct toxicity against the pathogen. The efficacy of 2,4-D treatment for common scab disease management is inconsistent among field trials in the literature, and the exact mode of action is unknown. Here, we identified transcriptomic responses of potato to low-dose 2,4-D treatment in the presence and absence of the pathogen and in tuber periderm and foliar tissue. Pathogen infection primarily altered transcriptomic responses in tuber periderm tissue, while foliar 2,4-D application caused larger shifts in gene expression in leaf tissue, as expected. Gene ontology (GO) terms associated with pathogen defense, stress responses, and enzymatic inhibitors were significantly enriched among differentially expressed genes in the tuber response to the pathogen. There were more differentially expressed genes and enriched GO terms in response to the pathogen when plants were treated with 2,4-D than in the non-2,4-D-treated plants, including differentially expressed genes and GO terms related to lipases, jasmonic acid signaling, and transport. Fewer differentially expressed genes were identified in tuber tissue than in leaf tissue following foliar 2,4-D treatment, but GO terms related to sucrose transport were enriched in tuber RNA samples from 2,4-D-treated, non-inoculated plants. Altered glucose and fructose, but not sucrose, levels in tuber medulla and periderm tissue, the site of common scab infection, were observed in 2,4-D-treated plants. Utilizing multiple factors, i.e., mock or 2,4-D treatments in both the presence and absence of the pathogen, in parallel transcriptional profiling experiments enabled the identification of pathways that directly respond to 2,4-D treatment in both foliar and tuber tissue and pathways with altered response in the context of pathogen infection. Identifying tools to more consistently induce these changes may enable more robust disease management than indirect foliar 2,4-D treatments.
1 Introduction
Common scab of potato is an economically costly disease of potato (Solanum tuberosum) caused by more than 10 species of Streptomyces that produce the phytotoxin thaxtomin A (Francis et al., 2010; Li et al., 2019; Thapa et al., 2019). While other virulence determinants are present in Streptomyces common scab pathogens, thaxtomin A and related toxins are the primary compounds responsible for the manifestation of the raised and pitted lesions associated with common scab of potato and other root and tuber crops (Clarke et al., 2022; Weisberg et al., 2023; Zhang et al., 2024), and the abundance of thaxtomin A-producing bacteria has been shown to be directly correlated with common scab disease severity (Shelley et al., 2023). Accordingly, much research has focused on the regulation and biosynthesis of thaxtomins by Streptomyces bacteria (Loria et al., 2008; Li et al., 2019), and comparatively little is known about the passive and active defenses responsible for host resistance to common scab. RNA sequencing has recently been used to identify transcriptomic responses to common scab infection in potato and has facilitated the identification of genes differentially regulated between susceptible and resistant cultivars (Fofana et al., 2020; Li et al., 2024), including genes putatively involved in tryptophan-induced common scab resistance (Zhao et al., 2022).
Disease management options for common scab are limited and inconsistent in their efficacy (Dees and Wanner, 2012; Braun et al., 2017). Cultural management strategies, including altered soil pH (Waterer, 2002; Marques et al., 2021), high soil moisture (Lapwood et al., 1973; Davis et al., 1976; Wilson et al., 2001; Johansen et al., 2015), and crop rotation (Larkin et al., 2011; Hunjan and Sabhikhi, 2020), have been tested, yielding inconsistent results and limited agronomic practicality. Chemical treatments such as pentachloronitrobenzene, as well as fumigants, including chloropicrin, have demonstrable efficacy for common scab disease management (Menzies, 1957; Emden and Labruyère, 1958; Rosser, 1960; Davis et al., 1976; Wilson et al., 1999; Al-Mughrabi et al., 2016) but are considered to also have substantial negative externalities, including detrimental impacts to soil health. Furthermore, several fungicides such as mancozeb and fluazinam have been reported to decrease common scab disease severity; however, field trial results are inconsistent (Mcintosh, 1973; McIntosh, 1977; Wilson et al., 1999; Al-Mughrabi et al., 2016).
Surprisingly, low-dose chemical treatments with the synthetic auxin herbicide 2,4-dichlorophenoxyacetic acid (2,4-D) and the related compound 3,5-D have been reported to attenuate disease severity for multiple cultivars of potato but also with inconsistent efficacy across trials (McIntosh et al., 1981, 1982, 1985; Waterer, 2010; Thompson et al., 2014b; Clarke et al., 2020). Treatment with 2,4-D has also been shown to enhance resistance to additional pathogenic diseases in tuber, including powdery scab (Thompson et al., 2014a; Clarke et al., 2020). Efficacy appears to be dependent on applying the foliar treatment at or shortly before tuber initiation (Thompson et al., 2013). Foliar 2,4-D treatments have been characterized for various physiological impacts on potato, including enhanced anthocyanin content in red potato skin (Fults et al., 1950; Busse and Bethke, 2020), as well as changes in sugar content (Payne and Fults, 1955). Foliar 2,4-D treatments at physiologically relevant doses associated with these phenotypic shifts have been associated with no impact, a small positive impact, or a small negative impact on total potato yield (McCubbin, 1957; Wort, 1965; Busse and Bethke, 2020; Clarke et al., 2020; Qin et al., 2021).
The mechanisms through which foliar treatment of 2,4-D leads to inhibition of common scab disease progression in potato tubers remain uncertain. Following foliar applications, 2,4-D and related compounds have been observed to translocate to and accumulate in tubers (Burrell, 1982; Tegg et al., 2008). Accumulation of 2,4-D in tubers has been associated with reduced phytotoxicity of thaxtomin A (Tegg et al., 2008, 2012), but physiologically relevant doses of 2,4-D have been shown to not directly interfere with thaxtomin A-induced tuber necrosis (Clarke et al., 2020). Furthermore, foliar 2,4-D treatments do not lead to notable changes in periderm or lenticel development, which are considered critical physiological features mediating Streptomyces infection of tubers (Tegg et al., 2008). While 2,4-D has been shown to be directly inhibitory to pathogen growth and expression of pathogenicity factors in other pathosystems (Duan et al., 2023), physiologically relevant concentrations of 2,4-D inhibit neither Streptomyces growth nor biosynthesis of thaxtomin A (McIntosh et al., 1981; Tegg et al., 2008). Therefore, 2,4-D likely suppresses common scab through an indirect mechanism. One proposed mechanism is 2,4-D-induced inhibition of thaxtomin A uptake and transport in plant cells through interference with auxin transporters. In support of this hypothesis, multiple genetic studies have demonstrated that plant genotypes with hypersensitivity to thaxtomin A are also hypersensitive in their response to auxin transport inhibitors such as 1-naphthylphthalamic acid (Tegg et al., 2013, 2016).
There are several other examples of low-dose herbicide treatments that induce disease resistance, often through unknown mechanisms (Grinstein et al., 1984; Awadalla and El-Refai, 1992; Ueno et al., 2004; Martinez et al., 2018). Because insights into the mechanisms underlying 2,4-D-induced resistance to common scab in potato are limited, in this study, we utilized a multifactor experimental approach to classify the transcriptional responses, in both potato leaf and tuber tissue, to low-dose foliar sprays of 2,4-D in the context of common scab infection.
2 Materials and methods
2.1 Plant material, Streptomyces inoculation, and 2,4-D treatment for pathogenicity assays and RNA sequencing
Twenty-four cultivar RH89-039-016 (RH89) potato plants (The Potato Genome Sequencing Consortium, 2011; tissue culture plants originally obtained from Wageningen University) were grown in 6-in. greenhouse pots, and one-half were inoculated with Streptomyces caniscabiei sp. nov. NE06-02D (“Strep”) at 2 × 106 CFU/pot as previously described (Weisberg et al., 2021) for the RNA sequencing experiment. At 4 weeks after planting (wap), six inoculated and six non-inoculated plants were sprayed until runoff with 200 mg L−1 of Weedar 64 2,4-D (Nufarm, Alsip, IL, USA) supplemented with 0.05% Tween 80 (Sigma-Aldrich, St. Louis, MO, USA). The remaining 12 plants (six inoculated and six non-inoculated) were sprayed with a mock treatment of water supplemented with 0.05% Tween 80. At 12 wap (8 weeks after 2,4-D treatments), tubers from all treatment groups were harvested for the collection of tuber periderm tissue for RNA extraction and scoring of disease severity as previously described (Clarke et al., 2020).
2.2 RNA extraction, sequencing, and analysis
For each of the four treatment conditions (“mock,” non-inoculated and mock-treated; “2,4-D,” non-inoculated and 2,4-D-treated; “Strep,” inoculated and mock-treated; “2,4-D + Strep,” inoculated and 2,4-D-treated), three biological replicates of tuber periderm tissue and three biological replicates of leaf tissue from a single experiment were sampled (except as noted in Supplementary Table S1). Each biological replicate consisted of samples pooled from two plants, with six total plants used for each treatment group. Samples were stored at −80°C. Leaf samples were taken 2 days post-2,4-D treatment, and tuber samples were taken at harvest (8 weeks post-treatment). For each sample, 100 mg of tissue was ground in liquid nitrogen using a mortar and pestle, then vortexed in 1 mL of TRI Reagent (Sigma-Aldrich). RNA extraction was conducted via RNEasy Mini Kit with on-column RNase-Free DNase Set (Qiagen, Hilden, DE) with modifications. After incubation for 30 min at room temperature (RT), samples were vortexed in 600 µL of Buffer RLT supplemented with 6 µL of β-mercaptoethanol, then incubated at RT for 5 min. Samples were shaken for 1 min with 200 µL of chloroform, then incubated at RT for 10 min, followed by centrifugation at 14,000 rpm for 30 min at 4°C; this process was repeated using 1 mL of the resulting aqueous phase. Six hundred μL of aqueous phase was gently mixed with 300 µL of 100% ethanol and transferred to RNeasy Mini spin columns, with the remaining steps carried out per manufacturer’s instructions.
The RNA sequencing pipeline was conducted by Azenta Life Sciences (South Plainfield, NJ, USA). RNA samples were quantified via Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA), and RNA integrity was verified via 4200 TapeStation (Agilent Technologies, Palo Alto, CA, USA). RNA sequencing libraries were prepared with the NEBNext Ultra II RNA Library Prep Kit for Illumina using the manufacturer’s instructions (New England Biolabs, Ipswich, MA, USA). Sequencing libraries were pooled and clustered on two lanes of a flow cell and loaded onto the Illumina HiSeq 4000 or equivalent (Illumina, San Diego, CA, USA). Samples were sequenced using a 2 × 150-bp paired-end (PE) configuration. Image analysis and base calling were conducted using HiSeq Control Software. Raw sequence data generated from Illumina HiSeq were converted into FASTQ files and de-multiplexed using Illumina’s bcl2fastq 2.17 software. One mismatch was allowed for index sequence identification.
From the resulting paired-end FASTQ files, Illumina adapter and leading 15-bp sequences were trimmed from reads via Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with default parameters and verified via FastQC reports for analysis of FASTQ file quality. For alignment of trimmed reads, genome indexing and mapping to the reference genome was performed via HISAT2 (https://daehwankimlab.github.io/hisat2/) with default options and utilized the DM 1-3–516 R44 (NCBI accession number PRJNA63145) annotated genome v6.1 (https://spuddb.uga.edu/dm_v6_1_download.shtml). Output SAM files were converted to BAM format, and mapping quality was analyzed via Samtools (http://www.htslib.org/). The High Confidence Gene Model Set annotations file for DM v6.1, in GFF3 format, was converted to GTF format via gffread (http://ccb.jhu.edu/software/stringtie/gff.shtml). For the alignment of reads to gene models, featureCounts (http://subread.sourceforge.net/) was employed using the representative, high-confidence gene model GTF and with the following options: -p (paired-end FASTQ files), –countReadpairs (count read pairs), and –transcript (assign reads at transcript-level). For pseudoalignment of trimmed reads, Salmon (https://combine-lab.github.io/salmon/) was employed to index, and assign read pairs to, the representative, high-confidence cDNA sequences for DM v.6.1.
DEG modeling and estimate dispersion analysis and principal component analysis (PCA) of assigned read pairs were performed via the “DESeq2” package (Love et al., 2014) in R Studio with R version 4.1.2. Pairwise comparison groups for DEG analysis were assigned as follows: “response to Strep,” “Strep” vs. “mock”; “response to -Strep_2,4-D,” “2,4-D + Strep” vs. “2,4-D”; “response to 2,4-D_Strep,” “2,4-D + Strep” vs. “Strep”; and “response to 2,4-D,” “2,4-D” vs. “mock” (Figure 1). Gene ontology (GO) term assignments for annotated genes for DM v6.1 were downloaded from http://spuddb.uga.edu. Additional analyses of differentially expressed genes (DEGs) were performed in R Studio with the following packages: pheatmap (DEG FPKM heat map), VennDiagram (DEG Venn diagrams), and clusterProfiler (GO term enrichment). In GO plots, “obsolete” or select, highly redundant GO terms were removed for clarity.

Figure 1. RNA-seq quality control and schema. (A) Schema for the comparison groups for RNA-seq differentially expressed gene (DEG) analysis. (B) Principal component analysis (PCA) of RNA-seq read pairs, assigned to genome features, by tuber (left) and leaf (right) samples.
2.3 Fatty acid content analysis
RH89 potato plants were grown as previously described, and at 4 wap, foliage from three plants was sprayed until runoff with each of the following treatments: 200 mg L−1 of 2,4-D, 400 mg L−1 of 2,4-D, or water (“mock”). Each treatment was supplemented with 0.05% Tween 80. At approximately 11 weeks, tubers for each sample (n = 9) were harvested for isolation of approximately 20 g of tuber skins/sample. Lipids were extracted and saponified with 0.5 N of methanolic sodium hydroxide, then methylated with 14% boron trifluoride methanol. Fatty acid methyl esters were extracted via heptane and analyzed for fatty acid content (% by sample mass) via gas chromatography-flame ionization detection (GC-FID). Sample extraction and GC-FID were performed by Eurofins Food Chemistry Testing Madison, Inc. (Madison, WI, USA).
2.4 Sucrose content analysis
RH89 potatoes were grown in 6-in. greenhouse pots and inoculated with Streptomyces as described above but utilizing strains of either S. acidiscabies ATCC49003 or S. stelliscabiei NY02-3A, separately. Streptomyces stelliscabiei is closely related to S. caniscabiei, which was used in the transcriptomic experiment, and disease from all three species is dependent on the thaxtomin A phytotoxin. 2,4-D treatments were performed as described above but included an additional 400 mg L−1 treatment group. Tubers were harvested 12 weeks after inoculation (8 weeks post-2,4-D treatment). At least two tubers were pooled from separate individual plants for the collection of each of the four biological replicate tuber periderm and tuber medulla samples from each combination of pathogen inoculum and 2,4-D treatment. Tissue samples were ground in water using Agdia (Elkhart, IN) mesh sample extraction bags. Collected samples were then assayed for sugar content using the Megazymes (Sydney, Australia) Sucrose/D-glucose/D-fructose assay kit following the manufacturer’s recommended protocols, except that samples were syringe-filtered through 0.8 µm filters instead of through filter paper.
3 Results
3.1 2,4-D treatment attenuated potato common scab disease severity in greenhouse conditions
We first sought to develop a greenhouse assay in which the efficacy of 2,4-D leaf sprays for suppressing common scab disease severity could be observed. As noted in previous field trials, low-dose 2,4-D treatments can induce minor alterations to plant foliage, including very minor leaf curling or early flowering (data not shown), and common scab infections of tubers do not present visible phenotypes in leaf tissue. Upon tuber harvesting, tuber samples are surveyed for common scab disease severity, with severe disease characterized by raised, pitted lesions and low-severity disease associated with superficial lesions. Leaf spray treatment with 200 mg L−1 of 2,4-D of 4-week-old potato plants growing in pots inoculated with pathogenic Streptomyces substantially suppressed common scab severity on tubers harvested 8 weeks following the spray treatment (Figure 2A), with tubers from 2,4-D-treated plants exhibiting significantly less severe common scab symptoms (Figure 2B). For downstream transcriptomics analyses, to avoid sampling tubers for which Streptomyces infection may not have been well established in soil, tubers with reduced disease symptoms, and not asymptomatic tubers, were selected for 2,4-D-treated, Streptomyces-inoculated plants. Accordingly, leaf samples for 2,4-D-treated, Streptomyces-inoculated plants were selected on the basis of the presence of low-severity disease symptoms observed in their corresponding tubers.

Figure 2. 2,4-D treatment significantly attenuates disease symptoms. (a) Box plots of disease scores calculated as previously described (Weisberg et al., 2021) comparing non-inoculated plants (left) to pathogen-inoculated plants (right) of both mock-treated (light gray bars) and 2,4-D-treated (dark gray bars) plants. * indicates significant difference (p < 0.05) in the Kruskal–Wallis non-parametric test. Similar results were obtained in a second independent experiment. (b) Morphology of S. caniscabiei strain NE06-02D on ISP-2 agar plates. (c) Typical disease symptoms observed on tubers in 2,4-D-treated and mock-treated plants.
3.2 RNA-seq mapping statistics and differential gene expression
To begin to survey the transcriptome in response to 2,4-D, Streptomyces (“Strep”), and combinations therein, tuber and leaf samples, each corresponding to the four treatment groups (Figure 1), were processed for RNA extraction, total RNA library preparation, and double-stranded sequencing via the Illumina platform. Following quality control trimming of Illumina adapter sequences, the resulting FASTQ files contained an average of 69,419,353 individual reads per sample (Supplementary Table S1). For alignment, HISAT2 (Kim et al., 2019) was utilized to map the reads to the doubled monoploid reference genome DM v6.1, with a mean mapping rate, including both primary and secondary mappings, of 82.46% (Supplementary Table S1). Prior to determining sets of DEGs, we separately evaluated alignment and pseudoalignment methods to assign read pairs to the reference genome. For alignment, featureCounts (Liao et al., 2014) was utilized to align read pairs to the representative gene models for DM v6.1, resulting in a mean of 25,428,076 read pairs per sample, representing an alignment rate of 72.67%, aligned to features (Supplementary Table S1). Comparatively, pseudoalignment via Salmon (Patro et al., 2017) resulted in a pseudoalignment rate of 73.55% (Supplementary Table S1). Due to the marginally higher alignment performance of Salmon, we utilized this data set for identifying DEGs. Separately, pseudoaligned read pairs from tuber (n = 12) and leaf (n = 10) samples were analyzed and normalized for differential expression via DESeq2 (Love et al., 2014) in four pairwise comparisons: “response to Strep,” “response to Strep (2,4-D),” “response to 2,4-D,” and “response to 2,4-D (Strep)” (Figure 1A). For tuber samples, PCA conveyed that PC1 and PC2 explained 56% and 30% of total variance, respectively, and that Mock and 2,4-D samples were most closely related; Strep and 2,4-D + Strep samples formed additional, separate clusters, with concordance among biological replicates within each treatment group (Figure 1B). Leaf sample PCA resulted in PC1 and PC2 explaining 57% and 19% of total variance, respectively, and illustrated clustering of 2,4-D + Strep and 2,4-D samples separate from the Mock and Strep samples (Figure 1B). Generally, large DEG counts were observed for all treatment group comparisons except for tuber “response 2,4-D” and leaf “response to Strep (2,4-D)” (Supplementary Figure S1).
3.3 Transcriptional response to Streptomyces scabiei in tuber and leaf tissue
In the tuber sample “response to Strep” comparison, we identified 800 DEGs, consisting of 772 upregulated genes and 28 downregulated genes (Figure 3A; Supplementary Table S2). Furthermore, response to Strep was also assessed via a comparison of the two groups of 2,4-D treated samples [“response to Strep (2,4-D),” i.e., response to Streptomyces infection in the context of 2,4-D treatment], yielding 1,580 DEGs, consisting of 1,135 and 445 upregulated and downregulated genes, respectively (Figure 3A; Supplementary Table S3). Overlap analysis of the DEGs from these two comparisons illustrated a specific response to Streptomyces, including 775 upregulated genes and 430 downregulated genes, in the context of foliar 2,4-D treatment, indicative of a possible enhancement to transcriptional sensitivity in tubers by foliar treatment with 2,4-D. To analyze the transcriptomic response to Streptomyces infection in tubers, we incorporated GO assignments for the DEGs identified in “response to Strep” and “response to Strep (2,4-D),” then performed GO enrichment analysis. Among upregulated DEGs in “response to Strep” were significantly enriched GO terms associated with pathogen defense, such as “response to wounding,” “cell wall modification,” “defense response to fungus,” “response to oxidative stress,” and the cellular component term “extracellular region,” with underlying DEGs corresponding to products such as pathogenesis-related (PR) proteins, chitinases, peroxidases, and jasmonate (JA) and ethylene (ET) pathway proteins (Figure 3B; Supplementary Tables S2, S4). Furthermore, several enriched GO terms were associated with fatty acid biosynthesis, with underlying genes corresponding to lipases and fatty acid desaturases. Downregulated DEGs contributed to five enriched GO terms, largely corresponding to underlying dehydration response element-binding (DREB) proteins. Analysis of upregulated DEGs in “response to Strep (2,4-D)” identified enriched GO terms largely overlapping with those from “response to Strep,” with additional terms including “defense response to bacterium,” “response to jasmonic acid,” “phospholipase,” and “acylglycerol lipase” (Figure 3B; Supplementary Table S4). Furthermore, downregulated DEGs in “response to Strep (2,4-D)” conferred significant enrichment for 13 GO terms, such as “lipid transport,” “extracellular region,” and “cell wall biogenesis.”
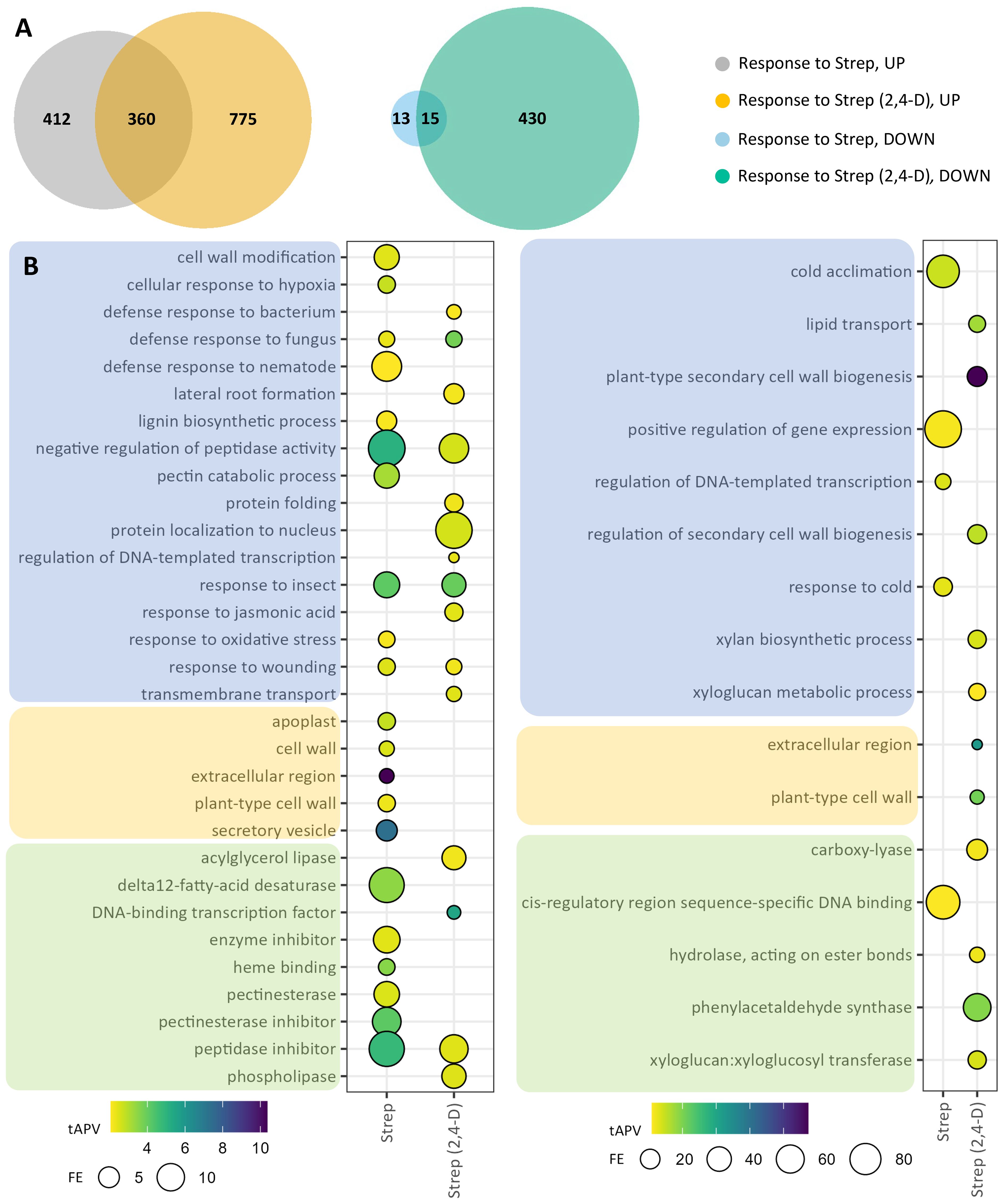
Figure 3. Differentially expressed genes (DEGs) and gene ontology (GO) enrichments for tuber samples in the response to Strep. DEGs were selected on the basis of |log2(fold change)| >2 and adjusted p-value <0.05 for the two comparison groups: “response to 2,4-D” and “response to 2,4-D (Strep).” (A) Venn diagrams for upregulated (“UP,” left) and downregulated (“DOWN,” right) DEGs. (B) GO enrichments for upregulated (left) and downregulated (right) DEGs. Enriched GO terms are grouped into “biological process” (blue), “cellular component” (orange), and “molecular function” (green) domains.
Analysis of the systemic response to Streptomyces in leaf samples yielded a smaller transcriptional response, as leaf tissue samples were collected early in the tuber development process, prior to the establishment of common scab symptoms. The foliar “response to Strep” comparison yielded 148 upregulated genes and 17 downregulated genes, and 11 upregulated DEGs and 15 downregulated DEGs were identified in “response to Strep (2,4-D)” (Supplementary Figure S2A; Supplementary Tables S5, S6). GO enrichment analysis indicated significant enrichments only from upregulated DEGs in “response to Strep,” with terms corresponding to the apoplast, extracellular matrix, and cell wall, and multiple molecular function terms associated with underlying DEGs encoding germin proteins (Supplementary Figure S2B; Supplementary Tables S5–S7).
3.4 Early, direct transcriptional responses in leaf tissue to foliar 2,4-D
To assess direct transcriptional responses to 2,4-D in leaf tissue, we sampled leaf tissue for RNA-seq 2 days after 2,4-D treatment, corresponding to 8 weeks prior to tuber harvest and preceding the development of common scab symptoms in tuber tissue. Leaf tissue was sampled within the context of Streptomyces pathogen in the soil (“response to 2,4-D (Strep)”) and without Streptomyces present in the soil (“response to 2,4-D”). In the leaf response to foliar 2,4-D alone, we identified 840 DEGs, consisting of 649 upregulated genes and 191 downregulated genes (Figure 4A; Supplementary Table S8). Of the 231 upregulated DEGs in “response to 2,4-D (Strep),” 156 overlapped with “response to 2,4-D,” as did 58 of 198 downregulated DEGs. Via GO enrichment analysis, pathways activated by 2,4-D corresponded to functions such as growth and differentiation (e.g., “cytokinin-activated signaling pathway,” “G1/S transition of mitotic cell cycle,” “meristem determinacy,” “microtubule binding”), biotic stress response (e.g., “defense response to fungus,” “innate immunity activating, cell surface,” “response to herbivore”), and carbohydrate metabolism (“carbohydrate metabolic process”) (Figure 4B; Supplementary Tables S8–S10). Accordingly, underlying genes were associated with a variety of protein classes, such as expansins, auxin efflux carriers and response proteins, cyclins, PR proteins, chitinases, peroxidases, and thioredoxins. GO enrichments derived from downregulated genes were limited to seven total terms and included “carbohydrate transport,” associated with underlying SWEET protein genes, as well as “solute:sodium symporter activity” and “positive regulation of programmed cell death” (Supplementary Figure S3; Supplementary Tables S8–S10).

Figure 4. Differentially expressed genes (DEGs) and gene ontology (GO) enrichments for leaf samples in the response to 2,4-D. DEGs were selected on the basis of |log2(fold change) >2 and adjusted p-value <0.05 for the two comparison groups: “response to 2,4-D” and “response to 2,4-D (Strep).” (A) Venn diagrams for upregulated (left) and downregulated (right) DEGs. (B) GO enrichments for upregulated DEGs. Enriched GO terms are grouped into “biological process” (blue), “cellular component” (orange), and “molecular function” (green) domains.
3.5 Foliar 2,4-D induces transcriptional changes in tubers
Because foliar 2,4-D treatment enhances resistance to common scab in tubers, we next sought to explore the transcriptional response to foliar 2,4-D treatment, alone and in the context of Streptomyces infection, in tuber tissue. 2,4-D was applied early in the tuber development process, and transcriptional response to foliar 2,4-D alone (“response to 2,4-D”) was limited, with only six upregulated DEGs identified (Figure 5A; Supplementary Table S11). Among these DEGs were two genes corresponding to Nodulin MtN3 family proteins, members of the SWEET family of sucrose transporters, and GO enrichment analysis conveyed strong (363–892-fold) enrichments for five corresponding GO terms, such as “sucrose transport,” from these two genes alone (Figure 5B; Supplementary Tables S11, S13). However, a stronger transcriptional response was observed in the “response to 2,4-D (Strep)” comparison, including 342 upregulated and 583 downregulated DEGs (Figure 5A; Supplementary Table S12). Eight significantly enriched GO terms were identified from upregulated genes, including “response to hydrogen peroxide,” associated with underlying heat shock protein and WRKY transcription factor genes, as well as “acylglycerol lipase” and “phospholipase” terms corresponding to patatin and phospholipase genes (Figure 5B; Supplementary Tables S12, S13). Downregulated genes contributed to 21 enriched GO terms, including several terms associated with growth, such as “synctium formation” and “plant-type secondary cell wall biogenesis.”
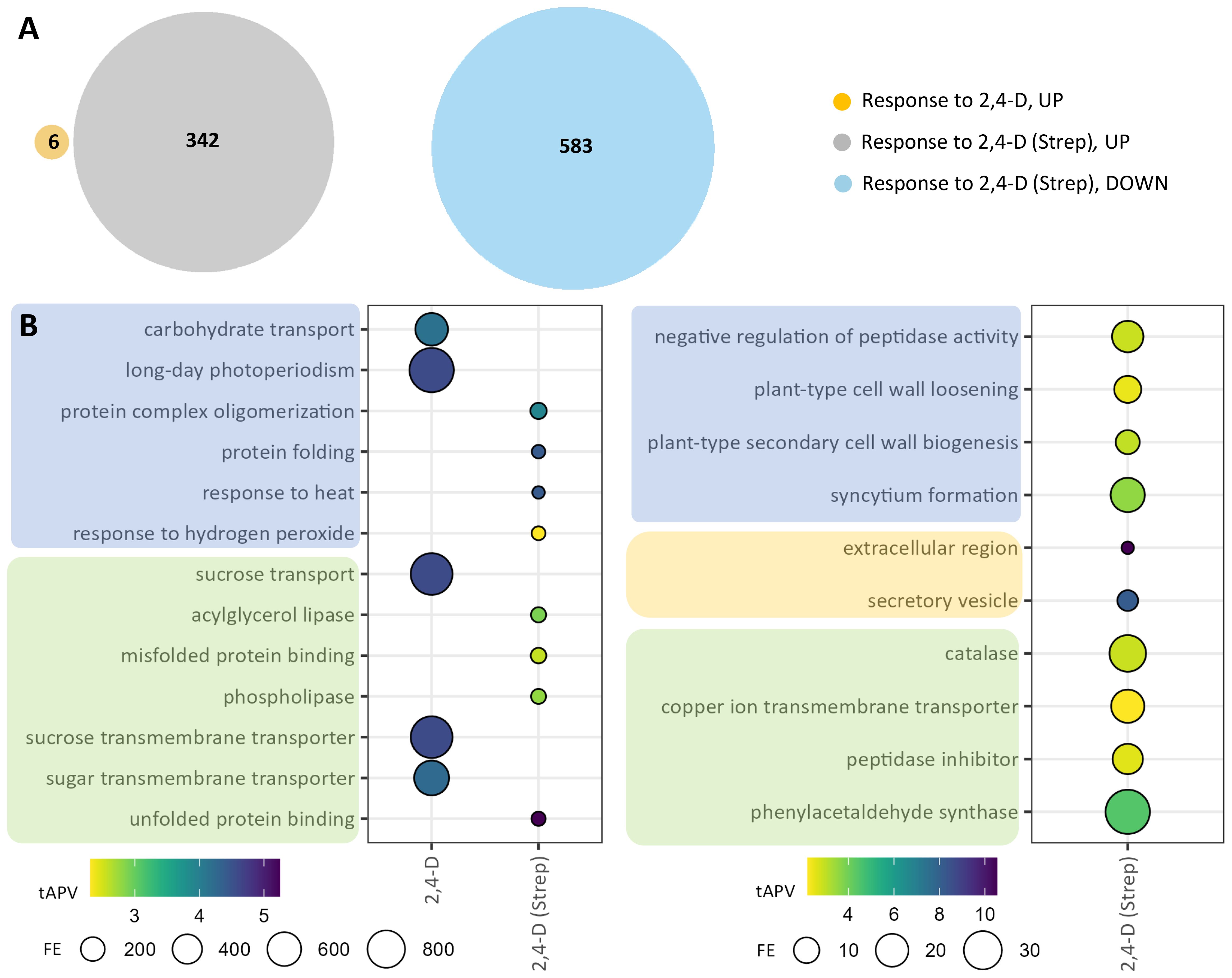
Figure 5. Differentially expressed genes (DEGs) and gene ontology (GO) enrichments for tuber samples in the response to 2,4-D. DEGs were selected on the basis of |log2(fold change)| >2 and adjusted p-value <0.05 for the two comparison groups: “response to 2,4-D” and “response to 2,4-D (Strep).” (A) Venn diagrams for upregulated (“UP,” left) and downregulated (“DOWN,” right) DEGs. (B) GO enrichments for upregulated (left) and downregulated (right) DEGs. Enriched GO terms are grouped into “biological process” (blue), “cellular component” (orange), and “molecular function” (green) domains.
3.6 Profiling predicted phenotypic changes in 2,4-D-treated tubers
Among the enriched GO terms identified in the DEGs from tuber responses to 2,4-D were the molecular function terms “acylglycerol lipase” and “phospholipase” (Figure 5B; Supplementary Table S13), and enrichment for these terms was also observed in 2,4-D-treated tubers in response to Streptomyces infection (“response to Strep (2,4-D)”; Figure 3B; Supplementary Table S4). Accordingly, the DEG sets from these comparison groups were found to include a number of genes with putative lipase, fatty acid desaturase, fatty acid hydroxylase, lipid transfer, and other functions of interest (Supplementary Tables S3, S12). To probe a potential role for tuber fatty acid metabolic profiles in the systemic response to foliar 2,4-D, we analyzed fatty acid contents in tuber tissue harvested from plants treated with 200 mg L−1 of 2,4-D, 400 mg L−1 of 2,4-D, or mock treatment (Supplementary Table S14). Esterified fatty acids from tuber skin samples were quantified via the GC-FID panel; however, most classes of fatty acids were below the detection limit, potentially indicating insufficient sensitivity of lipidomic profiling, and no significant differences between treatment groups were identified.
Upregulated DEGs in the tuber response to 2,4-D contributed to GO term enrichments associated with sucrose transport (Figure 5B; Supplementary Table S13), and among upregulated tuber DEGs in “response to Strep (2,4-D),” “response to 2,4-D,” and “response to 2,4-D (Strep)” were genes corresponding to SWEET proteins and sucrose synthases (Supplementary Tables S3, S11, S12). Therefore, we analyzed tubers for sucrose, glucose, and fructose content separately in both periderm and medulla tissue following spray treatment with 200 mg L−1 of 2,4-D, 400 mg L−1 of 2,4-D, or mock treatment. Tuber samples were collected at harvest, 12 weeks after inoculation of the potting mix with a mock inoculum, S. stelliscabiei strain NY02-3A, or S. acidiscabies strain ATCC49003 (Supplementary Figure S4). While 2,4-D treatments did lead to increased abundance of sucrose in both periderm and medulla tissue, sucrose was detected only at low levels in the periderm (the site of infection), and there was no significant dosage effect for any inoculation condition–tissue type combination based on the Kruskal–Wallis non-parametric rank-sum test. Treatment with 2,4-D significantly increased glucose abundance in the medulla and fructose abundance in the medulla and periderm tissue of mock-inoculated plants. However, these increases in glucose and fructose were not observed in pathogen-inoculated tubers.
4 Discussion
Multifactor transcriptional profiling of potato leaf and tuber tissue response to 2,4-D treatment and pathogen infection revealed multiple candidate pathways for elucidating the mechanisms through which low-dose 2,4-D treatments attenuate common scab disease severity. While previous research has focused on elucidating the impact of 2,4-D on tuber physiology, pathogen inhibition, and thaxtomin A uptake and toxicity (Tegg et al., 2008, 2012, 2013, 2016), this study sought to identify potato global transcriptomic responses to 2,4-D in the context of pathogen infection. Foliar 2,4-D treatments substantially altered transcriptional responses to the pathogen, and likewise, Streptomyces infection substantially altered transcriptional responses to 2,4-D treatments. In general, stronger transcriptional responses were seen in plants both treated with 2,4-D and infected with pathogen than either variable in isolation. Additionally, the transcriptomic response to 2,4-D treatment was only partially overlapping between the pathogen-inoculated and mock-inoculated samples, demonstrating the need for considering both experimental factors in parallel to more fully understand the complex interaction.
We observed that common scab infection caused by S. caniscabiei, which causes more raised lesions and fewer pitted lesions compared to the eponymous pathogenic species S. scabiei (Weisberg et al., 2021), induced significant transcriptional reprogramming in tuber periderm tissue, and in pathways known to be involved in pathogen defense, as previously observed following infection by the related pathogenic species S. scabiei (Fofana et al., 2020; Li et al., 2024) (Figure 3; Supplementary Tables S2–S4). In the plant response to pathogen alone (e.g., no 2,4-D treatment), the population of upregulated DEGs was enriched for GO terms corresponding to JA signaling and responses to insects, fungus, wounding, and nematodes, affirming transcriptional defense responses to pathogen infection. Such GO terms were also enriched in the plant response to the pathogen in the context of 2,4-D treatments, demonstrating the involvement of these specific, robust transcriptional responses even in tuber periderm tissue with lower disease severity resulting from the disease-suppressing effects of 2,4-D treatment.
Transcriptional response to the pathogen without 2,4-D treatment included numerous overlapping GO terms and DEGs with the transcriptional response to the pathogen with 2,4-D treatment. However, there were also many unique DEGs and GO terms in each separate analysis and a larger number of enriched GO terms and significant DEGs in response to the pathogen when the plants were previously treated with 2,4-D. This finding suggests that 2,4-D treatment elicits durable effects on the transcriptome, including cumulative effects from lower disease pressure, despite the only difference being a single 2,4-D treatment 8 weeks prior to harvest. This single treatment regimen is the current agricultural practice for low-dose foliar 2,4-D sprays to induce anthocyanin production in red potato production and is also associated with reduced levels of common scab. Examining enriched GO terms and differentially regulated DEGs that appear in the “response to Strep” comparison group but not when tubers that had been treated with 2,4-D (“response to Strep (2,4-D)”) may reveal transcriptional responses attenuated by 2,4-D, as evidenced by GO terms related to multiple classes of enzymatic inhibitors. Interestingly, several additional GO terms related to fatty acid metabolism were enriched in the tuber response to the pathogen only in the context of 2,4-D treatment, suggesting a role for fatty acids in 2,4-D-induced disease resistance. Fatty acids have been demonstrated to have critical roles in plant defenses against other pathogens, through fatty acid signaling pathways, for example (Kachroo and Kachroo, 2009; Pretorius et al., 2021). However, no differences in fatty acid content were measured in tuber periderm samples from 2,4-D-treated and mock-treated potatoes (Supplementary Table S14). Tuber periderm samples for fatty acid analysis were collected 7 weeks after 2,4-D treatment, at the time of tuber harvest and disease scoring. It is possible that fatty acid profiles and signaling activity may have been altered only in younger tubers, a developmental stage that serves as the critical window for common scab infection (Khatri et al., 2011), in the 2,4-D-treated plants.
Unsurprisingly, pathogen infection led to a comparatively smaller transcriptional response in leaf tissue in the “response to Strep” comparison group, as leaf tissue was responding systemically and before the establishment of common scab disease systems (Supplementary Figure S2; Supplementary Tables S5–S7). Multiple genes encoding Germin-like proteins, which have been linked with JA signaling and pathogen defense (Pei et al., 2019; To et al., 2022), were upregulated in foliar response to Streptomyces infection. However, 2,4-D treatment induced robust transcriptional changes in leaf tissue that were partially overlapping between the experiments with (“response to 2,4-D (Strep)”) and without (“response to 2,4-D”) pathogen infection, and the overall transcriptional response was largely represented by upregulated genes (Figures 4, Supplementary Figure S3; Supplementary Tables S8–S10). Analysis of the transcriptional responses validated the auxin and cytokinin analog functionality of 2,4-D by conferring multiple enriched GO terms related to cell growth, development, and differentiation; meristem determinacy; cytokinin signaling; and microtubule function. Upregulated DEGs and associated enriched GO terms also suggest potential immune priming by 2,4-D in leaf tissue; enhanced expression of genes corresponding to PR proteins, chitinases, thioredoxins, and peroxidases, for example, was found to be upregulated in the leaves following 2,4-D treatment. These results suggest that 2,4-D may function as an effective foliar immune priming agent, as demonstrated for multiple low-dose herbicide treatments (Martinez et al., 2018). Future work incorporating a more granular time course evaluation of potato response to 2,4-D could help to further establish the potential efficacy of 2,4-D as an immune priming agent and determine how alterations to the foliar transcriptome influence tuber development and defense against soil diseases such as common scab.
In tuber samples, while 2,4-D was shown to influence transcriptional reprogramming in response to Streptomyces infection (“response to Strep (2,4-D)”), the response to 2,4-D, with (“response to 2,4-D (Strep)”) and without (“response to 2,4-D”) the background of Streptomyces infection, elicited a less robust transcriptional response (Figure 5; Supplementary Tables S11–S13). Although 2,4-D treatments were applied 8 weeks prior to the collection of tuber periderm samples for RNA analysis, thereby possibly limiting the observable transcriptional response upon tuber harvesting, 2,4-D is thought to translocate and accumulate in tubers (Burrell, 1982; Tegg et al., 2008). The most notable transcriptional change in tuber periderm tissue induced by 2,4-D without the presence of pathogen infection was the presence of enriched GO terms associated with significant upregulation of Nodulin MtN3/SWEET genes, a gene class encoding proteins involved in the transport of sucrose and other carbohydrates and also shown to play critical roles in pathogen defense (Denancé et al., 2014; Breia et al., 2021). Interestingly, SWEET genes were largely downregulated in leaf tissue following 2,4-D treatment, conveying the possibility that 2,4-D influences the source–sink relationship for sugars in leaf and tuber tissue, respectively (Ludewig and Flügge, 2013). Some of the corresponding SWEET genes, as well as several sucrose synthases and invertases, were also found to be upregulated in the tuber response to Streptomyces infection (Supplementary Tables S2 and S3), suggesting a role for sugar transport and metabolism in the 2,4-D-induced enhancement to common scab resistance. Sucrose has been implicated in defense response and immune priming in other pathosystems and is proposed to be a mobile defense signal, elicitor of defense response, and a critical component of carbohydrate availability, which impacts pathogen growth at the plant–pathogen interface (Gómez-Ariza et al., 2007; Morkunas and Ratajczak, 2014; Li et al., 2017; Jeandet et al., 2022; Liu et al., 2022). Foliar applications of 2,4-D did lead to a moderate, but not significant, increase in sucrose content in both tuber periderm and medulla tissue for both mock-inoculated and pathogen-inoculated samples (Supplementary Figure S4). The reducing sugar hexamers glucose and fructose accumulated at significantly higher levels in tuber periderm and medulla tissue from 400 mg L−1 2,4-D-treated, mock-inoculated plants; however, this trend was not observed in tubers from pathogen-inoculated plants. Pathogen infection did not significantly increase sugar levels in the Strep-inoculated plants, in contrast to a previous work, suggesting an increase in tuber periderm reducing sugars due to common scab infection (Goto, 1981). Similar to caveats for the interpretation of the analysis of fatty acid content in tubers following 2,4-D treatment, sucrose and other carbohydrate content or signaling may be more significantly altered due to 2,4-D treatment in young tubers during the critical window of scab infection. More rigorous time-course sampling may reveal notable 2,4-D-induced phenotypic alterations, such as sugar abundance, fatty acid content, and periderm structure, in young tubers during initial Streptomyces infection.
When 2,4-D treatments were applied in the background of Streptomyces infection, substantially more DEGs and enriched GO terms were identified from tuber samples (Figure 5). Among upregulated DEGs were several genes encoding patatins, which have been implicated in disease resistance in potato and other Solanaceae vegetable crops potentially through impacts on storage and lipid metabolism (Kim et al., 2014; Bártová et al., 2019; Cheng et al., 2019). Patatins were also found to accumulate to much higher levels in a potato somaclone habituated with thaxtomin A for improved resistance to common scab (Isayenka et al., 2023). Additional phospholipase genes were also significantly upregulated, further suggesting a reprogramming of fatty acid metabolism following 2,4-D treatment. Surprisingly, the most downregulated DEG across the entire experimental platform, identified as highly downregulated in both the tuber “resp to Strep (2,4-D)” (Supplementary Table S3) and “resp to 2,4-D (Strep)” (Supplementary Table S12) analyses, was the TIR-NBS-LRR class putative R gene Soltu.DM.S001640. Why the combination of 2,4-D foliar application with Streptomyces infection led to the extreme downregulation of this putative R gene remains to be elucidated.
5 Conclusions
Multifactor analysis of potato leaf and tuber response to 2,4-D foliar application and Streptomyces infection revealed significant transcriptional reprogramming associated with reduced disease severity in 2,4-D-treated plants. Transcriptional analysis of both pathogen infection and chemical treatment facilitated the identification of differentially expressed genes and gene classes associated not only with host responses to the pathogen or 2,4-D treatment alone but also with the responses to the combined 2,4-D treatment and Streptomyces infection. Numerous gene pathways associated with canonical plant immunity and wounding response were upregulated in response to Streptomyces, affirming that multiple canonical defense pathways are being activated in response to common scab infection. Treatment with 2,4-D upregulated many genes in overlapping, canonical defense pathways, as well as in other pathways associated with immune priming. Additional phenotypic characterization of 2,4-D-treated potato is necessary to validate the molecular mechanisms responsible for 2,4-D-induced common scab disease resistance. Because 2,4-D and other auxin analog treatments also have efficacy against other potato pathogens, similar characterization of the molecular impacts of such auxin analog treatments in other potato pathosystems may reveal broad-spectrum disease resistance mechanisms.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
MF: Resources, Writing – original draft, Methodology, Validation, Investigation, Data curation, Visualization, Formal Analysis. HN: Methodology, Investigation, Writing – review & editing. JS: Supervision, Writing – review & editing. CC: Supervision, Writing – original draft, Investigation, Conceptualization, Methodology, Resources.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. Research in the Clarke Lab is funded by the U.S. Department of Agriculture, Agricultural Research Service CRIS project number 8042-21000-305. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1641317/full#supplementary-material
References
Al-Mughrabi, K. I., Vikram, A., Poirier, R., Jayasuriya, K., and Moreau, G. (2016). Management of common scab of potato in the field using biopesticides, fungicides, soil additives, or soil fumigants. Biocontrol Sci. Technol. 26, 125–135. doi: 10.1080/09583157.2015.1079809
Awadalla, O. A. and El-Refai, I. M. (1992). Herbicide-induced resistance of cotton to Verticillium wilt disease and activation of host cells to produce the phytoalexin gossypol. Can. J. Bot. 70, 1440–1444. doi: 10.1139/b92-181
Bártová, V., Bárta, J., and Jarošová, M. (2019). Antifungal and antimicrobial proteins and peptides of potato (Solanum tuberosum L.) tubers and their applications. Appl. Microbiol. Biotechnol. 103, 5533–5547. doi: 10.1007/s00253-019-09887-9
Braun, S., Gevens, A., Charkowski, A., Allen, C., and Jansky, S. (2017). Potato common scab: a review of the causal pathogens, management practices, varietal resistance screening methods, and host resistance. Am. J. Potato Res. 94, 283–296. doi: 10.1007/s12230-017-9575-3
Breia, R., Conde, A., Badim, H., Fortes, A. M., Gerós, H., and Granell, A. (2021). Plant SWEETs: from sugar transport to plant–pathogen interaction and more unexpected physiological roles. Plant Physiol. 186, 836–852. doi: 10.1093/plphys/kiab127
Burrell, M. M. (1982). The translocation of 3,5-dichlorophenoxyacetic acid in relation to its effect on potato common scab. J. Exp. Bot. 33, 215–220. doi: 10.1093/jxb/33.2.215
Busse, J. S. and Bethke, P. C. (2020). Impact of 2,4-D and potato (Solanum tuberosum L.) tuber age on anthocyanin content of skin and phellem anatomy of Red Norland. Am. J. Potato Res. 97, 102–110. doi: 10.1007/s12230-019-09760-5
Cheng, J., Song, N., and Wu, J. (2019). A patatin-like protein synergistically regulated by jasmonate and ethylene signaling pathways plays a negative role in Nicotiana attenuata resistance to Alternaria alternata. Plant Diversity 41, 7–12. doi: 10.1016/j.pld.2018.12.001
Clarke, C. R., Kramer, C. G., Kotha, R. R., and Luthria, D. L. (2022). The phytotoxin Thaxtomin A is the primary virulence determinant for scab disease of beet, carrot, and radish caused by Streptomyces scabiei. Phytopathology®. 112, 2288–2295. doi: 10.1094/PHYTO-03-22-0072-R. PHYTO-03-22-0072-R.
Clarke, C. R., Tegg, R. S., Thompson, H. K., Frederick, C., Haynes, K. G., Kramer, M., et al. (2020). Low-dose foliar treatments of the auxin analog 2,4-D reduce potato common scab and powdery scab for multiple potato cultivars and enhance root development. Crop Prot. 136, 105208. doi: 10.1016/j.cropro.2020.105208
Davis, J., McMaster, R., Callihan, R., Nissley, F., and Pavek, J. (1976). Influence of soil moisture and fungicide treatments on common scab and mineral content of potatoes. Phytopathology 66, 228–223. doi: 10.1094/Phyto-66-228
Dees, M. W. and Wanner, L. A. (2012). In search of better management of potato common scab. Potato Res. 55, 249–268. doi: 10.1007/s11540-012-9206-9
Denancé, N., Szurek, B., and Noël, L. D. (2014). Emerging functions of nodulin-like proteins in non-nodulating plant species. Plant Cell Physiol. 55, 469–474. doi: 10.1093/pcp/pct198
Duan, K., Shen, Q., Wang, Y., Xiang, P., Shi, Y., Yang, C., et al. (2023). Herbicide 2,4-dichlorophenoxyacetic acid interferes with MAP kinase signaling in Fusarium graminearum and is inhibitory to fungal growth and pathogenesis. Stress Biol. 3, 31. doi: 10.1007/s44154-023-00109-x
Emden, J. H. and Labruyère, R. E. (1958). Results of some experiments on the control of common scab of potatoes by chemical treatment of the soil. Europ. Potato J. 1, 14–24. doi: 10.1007/BF02418825
Fofana, B., Somalraju, A., Fillmore, S., Zaidi, M., Main, D., and Ghose, K. (2020). Comparative transcriptome expression analysis in susceptible and resistant potato (Solanum tuberosum) cultivars to common scab (Streptomyces scabies) revealed immune priming responses in the incompatible interaction. PloS One 15, e0235018. doi: 10.1371/journal.pone.0235018
Francis, I., Holsters, M., and Vereecke, D. (2010). The Gram-positive side of plant–microbe interactions. Environ. Microbiol. 12, 1–12. doi: 10.1111/j.1462-2920.2009.01989.x
Fults, J. L., Schaal, L. A., Landblom, N., and Payne, M. G. (1950). Stabilization and intensification of red skin color in Red McClure potatoes by use of the sodium salt of 2,4-dichloro-phenoxyacetic acid. Am. Potato J. 27, 377–395. doi: 10.1007/BF02850272
Gómez-Ariza, J., Campo, S., Rufat, M., Estopà, M., Messeguer, J., Segundo, B. S., et al. (2007). Sucrose-mediated priming of plant defense responses and broad-spectrum disease resistance by overexpression of the maize pathogenesis-related PRms protein in rice plants. MPMI 20, 832–842. doi: 10.1094/MPMI-20-7-0832
Goto, K. (1981). The relationship between common scab severity and reducing sugar contents in the peel of potato tubers. Potato Res. 24, 171–176. doi: 10.1007/BF02356237
Grinstein, A., Lisker, N., Katan, J., and Eshel, Y. (1984). Herbicide-induced resistance to plant wilt diseases. Physiol. Plant Pathol. 24, 347–356. doi: 10.1016/0048-4059(84)90008-0
Hunjan, M. S. and Sabhikhi, H. S. (2020). Designing a crop rotation strategy to manage Streptomyces scabies causing potato scab in north India. J. Phytopathol. 168, 469–477. doi: 10.1111/jph.12911
Isayenka, I., Duque-Yate, J., Goulet, M., Michaud, D., Beaulieu, C., and Beaudoin, N. (2023). Increased abundance of patatins, lipoxygenase and miraculins in a thaxtomin A-habituated potato Russet Burbank somaclone with enhanced resistance to common scab. Plant Pathol. 72, 100–111. doi: 10.1111/ppa.13650
Jeandet, P., Formela-Luboińska, M., Labudda, M., and Morkunas, I. (2022). The role of sugars in plant responses to stress and their regulatory function during development. IJMS 23, 5161. doi: 10.3390/ijms23095161
Johansen, T. J., Dees, M. W., and Hermansen, A. (2015). High soil moisture reduces common scab caused by Streptomyces turgidiscabies and Streptomyces europaeiscabiei in potato. Acta Agric. Scand. Secti. B Soil Plant Sci. 65, 193–198. doi: 10.1080/09064710.2014.988641
Kachroo, A. and Kachroo, P. (2009). Fatty acid–derived signals in plant defense. Annu. Rev. Phytopathol. 47, 153–176. doi: 10.1146/annurev-phyto-080508-081820
Khatri, B. B., Tegg, R. S., Brown, P. H., and Wilson, C. R. (2011). Temporal association of potato tuber development with susceptibility to common scab and Streptomyces scabiei-induced responses in the potato periderm. Plant Pathol. 60, 776–786. doi: 10.1111/j.1365-3059.2011.02435.x
Kim, D. S., Jeun, Y., and Hwang, B. K. (2014). The pepper patatin-like phospholipase CaPLP1 functions in plant cell death and defense signaling. Plant Mol. Biol. 84, 329–344. doi: 10.1007/s11103-013-0137-x
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Lapwood, D. H., Wellings, L. W., and Hawkins, J. H. (1973). Irrigation as a practical means to control potato common scab (Streptomyces scabies): final experiment and conclusions. Plant Pathol. 22, 35–41. doi: 10.1111/j.1365-3059.1973.tb01766.x
Larkin, R. P., Honeycutt, C. W., Griffin, T. S., Olanya, O. M., Halloran, J. M., and He, Z. (2011). Effects of different potato cropping system approaches and water management on soilborne diseases and soil microbial communities. Phytopathology 101, 58–67. doi: 10.1094/PHYTO-04-10-0100
Li, Y., Liu, J., Díaz-Cruz, G., Cheng, Z., and Bignell, D. R. D. (2019). Virulence mechanisms of plant-pathogenic Streptomyces species: an updated review. Microbiology 165, 1025–1040. doi: 10.1099/mic.0.000818
Li, Y., Wang, Y., Zhang, H., Zhang, Q., Zhai, H., Liu, Q., et al. (2017). The plasma membrane-localized sucrose transporter ibSWEET10 contributes to the resistance of sweet potato to Fusarium oxysporum. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00197
Li, C., Yuan, B., Zhang, C., Yao, Q., He, H., Wang, Q., et al. (2024). Revealing key genes and pathways in potato scab disease resistance through transcriptome analysis. Agronomy 14, 291. doi: 10.3390/agronomy14020291
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu, Y.-H., Song, Y.-H., and Ruan, Y.-L. (2022). Sugar conundrum in plant–pathogen interactions: roles of invertase and sugar transporters depend on pathosystems. J. Exp. Bot. 73, 1910–1925. doi: 10.1093/jxb/erab562
Loria, R., Bignell, D. R. D., Moll, S., Huguet-Tapia, J. C., Joshi, M. V., Johnson, E. G., et al. (2008). Thaxtomin biosynthesis: the path to plant pathogenicity in the genus Streptomyces. Antonie van Leeuwenhoek 94, 3–10. doi: 10.1007/s10482-008-9240-4
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Ludewig, F. and Flügge, U.-I. (2013). Role of metabolite transporters in source-sink carbon allocation. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00231
Marques, H. M. C., Appy, M. P., and Destéfano, S. A. L. (2021). Effect of pH soil and irrigation regimes on management of potato scab. Arq. Inst. Biol. 88, e00552020. doi: 10.1590/1808-1657000552020
Martinez, D. A., Loening, U. E., and Graham, M. C. (2018). Impacts of glyphosate-based herbicides on disease resistance and health of crops: a review. Environ. Sci. Eur. 30, 2. doi: 10.1186/s12302-018-0131-7
McCubbin, E. (1957). Proc. Florida State Hortic. Soc. 675, 93–96. Available online at: https://link.springer.com/article/10.1007/s12230-019-09760-5 (Accessed February 2025).
Mcintosh, A. H. (1973). Glasshouse tests of chemicals for control of potato common scab. Ann. Appl. Biol. 73, 189–196. doi: 10.1111/j.1744-7348.1973.tb01324.x
McIntosh, A. H. (1977). Field trials of chemicals for control of common scab by soil treatment. Potato Res. 20, 225–229. doi: 10.1007/BF02418683
McIntosh, B. A. H., Bateman, G. L., Chamberlain, K., Dawson, G. W., and Burrell, M. M. (1981). Decreased severity of potato common scab after foliar sprays of 3,5 -dichlorophenoxyacetic acid, a possible antipathogenic agent. Ann. Appl. Biol. 99, 275–281. doi: 10.1111/j.1744-7348.1981.tb04796.x
McIntosh, A. H., Burrell, M. M., and Hawkins, J. H. (1982). Field trials of foliar sprays of 3,5-dichlorophenoxyacetic acid (3,5-D) against common scab on potatoes. Potato Res. 25, 347–350. doi: 10.1007/BF02357292
McIntosh, A. H., Chamberlain, K., and Dawson, G. W. (1985). Foliar sprays against potato common scab: compounds related to 3,5-dichlorophenoxyacetic acid. Crop Prot. 4, 473–480. doi: 10.1016/0261-2194(85)90052-3
Menzies, J. D. (1957). Dosage rates and application methods with PCNB for control of potato scab and rhizoctonia. Am. Potato J. 34, 219–226. doi: 10.1007/BF02855942
Morkunas, I. and Ratajczak, L. (2014). The role of sugar signaling in plant defense responses against fungal pathogens. Acta Physiol. Plant 36, 1607–1619. doi: 10.1007/s11738-014-1559-z
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., and Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. doi: 10.1038/nmeth.4197
Payne, M. G. and Fults, J. L. (1955). The effect of maleic hydrazide and 2,4-D on reducing sugars and sucrose of Red McClure potatoes. Am. Potato J. 32, 144–149. doi: 10.1007/BF02851210
Pei, Y., Li, X., Zhu, Y., Ge, X., Sun, Y., Liu, N., et al. (2019). GhABP19, a novel germin-like protein from Gossypium hirsutum, plays an important role in the regulation of resistance to verticillium and fusarium wilt pathogens. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00583
Pretorius, C. J., Zeiss, D. R., and Dubery, I. A. (2021). The presence of oxygenated lipids in plant defense in response to biotic stress: a metabolomics appraisal. Plant Signaling Behav. 16, 1989215. doi: 10.1080/15592324.2021.1989215
Qin, R., Moparthi, S., Feldman, M., Charlton, B., and Sathuvalli, V. (2021). Effect of foliar application of 2,4-D and calcium on red-skinned potato cultivars. Agron. J. 113, 88–98. doi: 10.1002/agj2.20444
Rosser, W. R. (1960). Fungicidal control of potato common scab. Plant Pathol. 9, 61–62. doi: 10.1111/j.1365-3059.1960.tb01149.x
Shelley, B., Pandey, B., Sarwar, A., Douches, D., Qu, X., Pasche, J., et al. (2023). The role of soil abundance of TxtAB in potato common scab disease severity. Phytopathology®. 114, 1176–1185. doi: 10.1094/PHYTO-09-23-0347-R. PHYTO-09-23-0347-R.
Tegg, R. S., Corkrey, R., and Wilson, C. R. (2012). Relationship between the application of foliar chemicals to reduce common scab disease of potato and correlation with thaxtomin A toxicity. Plant Dis. 96, 97–103. doi: 10.1094/PDIS-05-11-0397
Tegg, R. S., Gill, W. M., Thompson, H. K., Davies, N. W., Ross, J. J., and Wilson, C. R. (2008). Auxin-induced resistance to common scab disease of potato linked to inhibition of thaxtomin A toxicity. Plant Dis. 92, 1321–1328. doi: 10.1094/PDIS-92-9-1321
Tegg, R. S., Shabala, S. N., Cuin, T. A., Davies, N. W., and Wilson, C. R. (2013). Enhanced resistance to the cellulose biosynthetic inhibitors, thaxtomin A and isoxaben in Arabidopsis thaliana mutants, also provides specific co-resistance to the auxin transport inhibitor, 1-NPA. BMC Plant Biol. 13, 76. doi: 10.1186/1471-2229-13-76
Tegg, R. S., Shabala, S., Cuin, T. A., and Wilson, C. R. (2016). Mechanisms of thaxtomin A-induced root toxicity revealed by a thaxtomin A sensitive Arabidopsis mutant (ucu2-2/gi-2). Plant Cell Rep. 35, 347–356. doi: 10.1007/s00299-015-1888-4
Thapa, S. P., Davis, E. W., Lyu, Q., Weisberg, A. J., Stevens, D. M., Clarke, C. R., et al. (2019). The evolution, ecology, and mechanisms of infection by gram-positive, plant-associated bacteria. Annu. Rev. Phytopathol. 57, 341–365. doi: 10.1146/annurev-phyto-082718-100124
The Potato Genome Sequencing Consortium (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. doi: 10.1038/nature10158
Thompson, H. K., Tegg, R. S., Corkrey, R., and Wilson, C. R. (2014a). Foliar treatments of 2,4-dichlorophenoxyacetic acid for control of common scab in potato have beneficial effects on powdery scab control. Sci. World J. 2014, 5. doi: 10.1155/2014/947167
Thompson, H. K., Tegg, R. S., Corkrey, R., and Wilson, C. R. (2014b). Optimal rates of 2,4-dichlophenoxyacetic acid foliar application for control of common scab in potato. Ann. Appl. Biol. 165, 293–302. doi: 10.1111/aab.12138
Thompson, H. K., Tegg, R. S., Davies, N. W., Ross, J. J., and Wilson, C. R. (2013). Determination of optimal timing of 2,4-dichlorophenoxyacetic acid foliar applications for common scab control in potato. Ann. Appl. Biol. 163, 242–256. doi: 10.1111/aab.12050
To, H. T. M., Pham, D. T., Le Thi, V. A., Nguyen, T. T., Tran, T. A., Ta, A. S., et al. (2022). The Germin-like protein OsGER4 is involved in promoting crown root development under exogenous jasmonic acid treatment in rice. Plant J. 112, 860–874. doi: 10.1111/tpj.15987
Ueno, M., Kihara, J., Honda, Y., and Arase, S. (2004). Indole-related compounds induce the resistance to rice blast fungus, Magnaporthe grisea in barley. J. Phytopathol. 152, 606–612. doi: 10.1111/j.1439-0434.2004.00903.x
Waterer, D. (2002). Impact of high soil pH on potato yields and grade losses to common scab. Can. J. Plant Sci. 82, 583–586. doi: 10.4141/P01-046
Waterer, D. (2010). Influence of growth regulators on skin colour and scab diseases of red-skinned potatoes. Can. J. Plant Sci. 90, 745–753. doi: 10.4141/CJPS10055
Weisberg, A. J., Kramer, C. G., Kotha, R. R., Luthria, D. L., Chang, J. H., and Clarke, C. R. (2021). A novel species-level group of Streptomyces exhibits variation in phytopathogenicity despite conservation of virulence loci. Mol. Plant-Microbe Interact. 34, 39–48. doi: 10.1094/MPMI-06-20-0164-R
Weisberg, A., Pearce, E. M., Kramer, C. G., Chang, J. H., and Clarke, C. R. (2023). Diverse mobile genetic elements shaped the evolution of Streptomyces virulence. Microb. Genomics. 9. doi: 10.1099/mgen.0.001127
Wilson, C. R., Pemberton, B. M., and Ransom, L. M. (2001). The effect of irrigation strategies during tuber initiation on marketable yield and development of common scab disease of potato in Russet Burbank in Tasmania. Potato Res. 44, 243–251. doi: 10.1007/BF02357902
Wilson, C. R., Ransom, L. M., and Pemberton, B. M. (1999). The relative importance of seed-borne inoculum to common scab disease of potato and the efficacy of seed tuber and soil treatments for disease control. J. Phytopathol. 147, 13–18. doi: 10.1046/j.1439-0434.1999.147001013.x
Wort, D. J. (1965). Increased tuber yield of Pontiac potatoes resulting from the foliar application 2,4-D—Mineral dusts. Am. Potato J. 42, 90–96. doi: 10.1007/BF02862448
Zhang, H., Ping, Y., Liu, X., He, X., and Du, C. (2024). Pathogenic factors of plant pathogenic streptomyces. Potato Res. 67, 621–646. doi: 10.1007/s11540-023-09660-6
Keywords: potato common scab, Streptomyces, 2,4-D, transcriptional response, disease management
Citation: Fabian ML, Nguyen HP, Stommel JR and Clarke CR (2025) Multifactor transcriptional profiling of potato during 2,4-D-induced resistance to common scab disease. Front. Plant Sci. 16:1641317. doi: 10.3389/fpls.2025.1641317
Received: 04 June 2025; Accepted: 24 July 2025;
Published: 18 August 2025.
Edited by:
Udai B. Singh, National Bureau of Agriculturally Important Microorganisms (ICAR), IndiaReviewed by:
Malkhan Singh Gurjar, Indian Agricultural Research Institute (ICAR), IndiaAbhijeet Ghatak, Bihar Agricultural University, India
Copyright © 2025 Fabian, Nguyen, Stommel and Clarke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christopher R. Clarke, Q2hyaXN0b3BoZXIuY2xhcmtlQHVzZGEuZ292
†Present address: Hien P. Nguyen, School of Medicine and Pharmacy, Duy Tan University, Danang, Vietnam
Danang Biotechnology Center (DBC), Danang Department of Science and Technology, Danang, Vietnam
 Matthew L. Fabian
Matthew L. Fabian Hien P. Nguyen†
Hien P. Nguyen† Christopher R. Clarke
Christopher R. Clarke