- Engineering Technology Research Center of National Forestry and Grassland Administration on Southwest Landscape Architecture, Yunnan Province Engineering Research Center for Functional Flower Resources and Industrialization, College of Landscape and Horticulture, Southwest Forestry University, Kunming, China
Introduction: Camellia reticulata holds cultural and horticultural significance in traditional Chinese gardens, as a regional endemic species in Yunnan Province. However, during the in vitro regeneration process of C. reticulata, there is often a phenomenon of low efficiency of adventitious bud differentiation or no adventitious bud differentiation at all.
Methods: In previous study, we observed significant morphological differences between the callus tissues of C. reticulata ‘Zipao’ and wild species. The callus of ‘Zipao’ was white and loosely textured, the wild species was green and hard. To investigate the differences between these two types of callus, we conducted transcriptome analysis and identified a differentially expressed gene GST24, which is closely related to the synthesis of glutathione (GSH). Heterologous transformation of CrGST24 into tobacco (Nicotiana tabacum) was conducted.
Results and discussion: The CrGST24 gene promoted the synthesis of endogenous auxin and cytokinin in transgenic tobacco by regulating the expression of transcription factors related to auxin and cytokinin. Exogenous IBA, 6-BA, and red-blue light treatment increased the levels of auxins and cytokinins in C. reticulata callus, thereby promoting adventitious bud differentiation. Additionally, CrGST24 interacted with CrGSHB and CrDHAR2 genes, facilitating GSH and AsA synthesis and clearing ROS.
Introduction
Camellia reticulata, also known as Yunnan camellia is among the decorative trees belonging to the Camellia genus, which are members of the Theaceae family. China is considered the origin and distribution hub of Camellia. Generally, members of the genus Camellia are classified into one of five groups. Yunnan camellia (C. reticulata) and its near relatives constitute a significant group that is primarily found in Yunnan Province. In Yunnan province, C. reticulata was regarded as an exceptional ornamental plant with high economic values. Some of them are very important flowering ornamentals that also produce oil, and have a wide variety of cultivars. In addition to its cultural and medicinal significance, C. reticulata is also a valuable source of food products. C. reticulata is mostly red-colored, and there are few accounts of color variations in the species. As a result, producers and researchers have constantly sought to breed C. reticulata of diverse colors to enhance its aesthetic value (Dai et al., 2013; Tong et al., 2015). C. reticulata is of great significance in ornamental, medicinal, and economic value. However, in breeding, its callus often has trouble differentiating into adventitious buds or may not differentiate at all.
Glutathione-S-transferase (GST) is extensively involved in various physiological and metabolic processes in plants and plays a key regulatory role when plants are exposed to stresses such as oxidation, pathogens and heavy metals. GST gene plays an important function in plant growth and development, and GST can be induced by phytohormones such as auxin, cytokinin, SA (Salicylic acid), MJ (Methyl jasmonate), and ETH (Ethylene). In tomato, the SlGST43 gene plays an extremely important role in scavenging ROS. It interacts with SlCOMT2 to regulate lignin content. Further studies indicate that SlMYB71 and SlWRKY8 interact to enhance the expression of SlGST43 (Yuan et al., 2024). In tomato, overexpression of the SlGST38 gene increased tomato resistance to viruses, and when SlGST38 was knocked out, tomato rapidly became infected with viruses, and a large accumulation of ROS resulted in tomato mortality (Méndez et al., 2023). GST genes were auxin binding proteins and influenced their function by regulating the redox state of auxin, and GST genes were involved in auxin signalling through non-catalytic effects. In addition, GST genes may regulate the concentration and gradient of auxin in plants by binding to auxin (Abel and Theologis, 1996). GST24, GST25, and GST26 genes were significantly co-expressed with the cytokinin signalling pathway, and GST24, GST25, and GST26 had important roles in cytokinin signalling (Pavlů et al., 2018). GSH acted as an antioxidant by bursted ROS and participated in the ascorbate-glutathione cycle that eliminates peroxides (Rouhier et al., 2008). GSH is a combination of glutamic acid, cysteine, and glycine. GSH is often divided into oxidised glutathione (GSSG), and reduced glutathione (GSH). Auxin induced the expression of glutathione synthetase (GS), and glutathione S-transferase (GST), thereby increased glutathione synthesis (Kong et al., 2015). Exogenous application of auxin significantly increased glutathione levels and enhanced antioxidant capacity in Arabidopsis thaliana (Zechmann et al., 2011). Ascorbic Acid (AsA), also known as Vitamin C, it had an extremely important role in antioxidant and boosting plants against biotic and abiotic stresses. In tomato seedling root systems, exogenous growth hormones and cytokinins were applied to increase AsA levels and enhance plant resistance to environmental stress (Ahanger et al., 2018). AsA affected gene expression in the auxin synthesis pathway by interacted with the auxin response factor ARF4, ARF4 promoted AsA accumulation by transcriptionally repressed MYB99, which in turn activated the expression of the AsA synthesis genes GPPS, GLDH and DHAR (Xu et al., 2023). However, research on the GST gene in C. reticulata is limited. The relationship between the GST gene and plant hormones, as well as the pathways through which the GST gene promotes the synthesis of GSH and AsA, requires further investigation.
In this study, we screened a GST gene from C. reticulata callus transcriptome data (Accession PRJNA909456 and Submission ID SUB12368224 at the NCBI BioProject database). C. reticulata callus were inoculated in medium containing different concentrations of IBA and 6-BA and placed under different light environments to analyse the expression of GST genes. The GST gene was heterologously transformed into tobacco and the function of transgenic tobacco was analysed. Finally, this experiment aimed to investigate experimental conditions that would be most conducive to the differentiation of adventitious buds from C. reticulata callus.
Materials and methods
Plant materials and growth conditions
The plant materials were C. reticulata ‘Zipao’ and wild species callus. ‘Zipao’ callus were white and loose (Supplementary Figure 1a); callus of the wild species were green and hard (Supplementary Figure 1b). Tobacco was used as the model plant for heterologous genetic transformation. Tobacco and C. reticulata callus were cultivated in a tissue culture room maintained at a constant 25°C, with a daily light exposure of 10-12 h and a humidity range of 65-75%, light intensity was 5000 LX. The red and blue light intensity required for subsequent experiments were 5000 LX.
The CrGST24 gene cloning
RNA was isolated using Vazyme Biotech RNA extraction kit (Nanjing, China), according to manufacturer’s protocol. Utilizing the NCBI online platform, specific cloning primers P1-F (AAGCAGTGGGAGTAGAGGTG) and P1-R (TTTGGCAAAGGCGAGTAGTT) were designed. The cDNA synthesized via reverse transcription was employed as the template to amplify the full-length sequence of CrGST24. The RT-PCR parameters were established as: 1 cycle of 5 min at 94°C, 35 cycles of 30 s at 94°C, 35 cycles of 30 s at 58°C, 35 cycles of 1 min at 72°C, followed by 1 cycle of 10 min at 72°C and 1 cycle of 10 min at 10°C, within a 50 μL reaction buffer. Subsequently, a gel recovery kit (Biomed, Beijing, China) was applied to purify the target bands. Eventually, the purified target band was ligated into a cloning vector and introduced into E. coli recipient cells, and then the plasmid isolated and sequenced.
Expression analysis of CrGST24 under different concentrations of IBA and 6-BA, different light environments and different light quality conditions
Three culture medium YS2, YS3, and YS4 containing different concentrations of IBA and 6-BA were prepared by increasing the concentrations of IBA and 6-BA individually or simultaneously on the basis of YS1 medium (Supplementary Table 1). Callus of C. reticulata ‘Zipao’ and wild species were inoculated on four different culture media and cultured under white light for 2 h and one week. The 2-(ΔΔCt) method was used to analyse the expression of CrGST24 gene in different types of healing tissues and under different concentrations of IBA and 6-BA treatments.
The callus of C. reticulata ‘Zipao’ and wild species were inoculated onto YS1 medium and put into light and dark environment for 8h. The 2-(ΔΔCt) method was used to analyse the expression of CrGST24 gene in different light environments.
The callus of C. reticulata ‘Zipao’ and wild species were inoculated on the YS1 medium and placed under white, red, and blue light for 8 h and three weeks. The 2-(ΔΔCt) method was used to analyse the expression of CrGST24 gene in conditions with white, red, or blue light.
Overexpression vector construction, plant transformation of CrGST24, and identification of transgenic lines
The CrGST24 gene was subjected to double digestion using the restriction enzymes BamHI and PstI. Agarose gel electrophoresis, followed by gel cutting and DNA recovery with the TAKARA Gel DNA Recovery Kit (Beijing, China), linearized the cloning vectors. The cleaved product was purified via a DNA purification kit. The target fragment was inserted into the pCAMBIA1300-35S vector. This construct was introduced into E. coli to screen positive clones and conduct sequencing. Finally, the extracted plasmids were transferred into Agrobacterium tumefaciens GV3101.
The heterologous genetic transformation of tobacco via Agrobacterium was performed according to Wang et al. (2024). Ten transgenic lines were randomly selected. RNA was extracted from the transgenic tobacco following the RNA extraction kit’s instructions, reverse transcribed into cDNA, and then verified by qPCR to screen for high-expressing positive lines.
Phenotypic analysis was conducted on high expressing transgenic tobacco lines and WT tobacco to observe any phenotypic differences between them.
In vitro regeneration growth of CrGST24 transgenic tobacco
WT tobacco and CrGST24 transgenic tobacco leaves were cut into 0.5 cm*0.5 cm leaf discs and inoculated into normal MS medium, which was observed at 5 d intervals to analyse the in vitro regeneration of transgenic tobacco leaves.
Expression analysis of auxin and cytokinin related genes in transgenic tobacco
The NCBI Primer-Blast was used to design qPCR primers for the growth hormone-related genes NtYUCCA8, NtPIN1c, NtGH3.3, NtIAA27, NtARF6, NtWOX5, and NtSAUR20; and for the cytokinin-related genes NtARR5, NtARR12, NtCRE1, NtAHP6, NtCRF6, and NtWUS qPCR primers, the NtEF1α as a reference gene (Table 1).
Expression analysis of auxin and cytokinin related genes in C.reticulata
Twelve auxin and cytokinin related genes were screened from the callus transcriptome data of C. reticulata and their expression was analyzed
Determination of auxin, cytokinin, ROS, GSH, and AsA contents in CrGST24 transgenic tobacco
Auxin and cytokinin content in transgenic tobacco and in WT were examined by ELISA enzyme-linked immunoassay. Absorbance values of auxin and cytokinin should be measured at 450 nm.
Preparation of NBT staining solution: Dissolve 50 mg NBT in 100 mL Tris buffer (pH: 7.4), and obtain NBT staining working solution after full dissolution. The prepared staining solution needs to be stored at 4°C away from light. The leaves of high expression positive transgenic tobacco and WT were cut off, and the leaves were cut into circular leaf discs with a diameter of 2 cm using a hole punch, the cut circular leaf discs were immersed in NBT staining solution, and the leaf discs were taken out after being kept away from light for 48 h. The leaf discs were immersed in 95% alcohol, and the discs were decolored with stirring while heating, and the blue areas on the leaf discs after decolorization were the areas where the ROS were accumulated. ROS, GSH, and AsA content in transgenic tobacco and in WT were examined by ELISA enzyme-linked immunoassay. Absorbance values of ROS should be measured at 450 nm; absorbance values of GSH should be measured at 412 nm; absorbance values of AsA should be measured at 265 nm.
A study on the interaction among CrGST24, CrGSHB, and CrDHAR2
Primary antibodies specific to the target protein were added and incubated, followed by incubation with secondary antibodies conjugated with fluorescent dyes to label the target protein, the nuclei were stained with DAPI. The coding sequences of CrGST24, CrGSHB, and CrDHAR2 were fused with the green fluorescent protein (GFP) to construct the corresponding fusion protein expression vectors. These vectors were then transformed into tobacco leaf cells to achieve transient expression, enabling the synthesis of the fusion proteins within the plant cells. Finally, confocal laser scanning microscopy was used to observe the distribution of fluorescent signals in the tobacco leaf cells, so as to determine the subcellular localization of the target proteins.
The full-length CDS of the CrDHAR2 (Accseeion No. PV920956 at the NCBI BioProject database) and CrGSHB (PV920957) were amplified and recombined into the pGADT7 vector to construct the prey vector. The CrGST24 (PV920955) was intercepted and inserted into the pGBKT7 vector to construct the bait vectors. The prey and bait vectors were co-transferred into the Y2H Gold yeast strain as described in the instructions (Clontech, Shanghai, China). Yeast strains carrying the indicated vectors were plated on SD/-Leu-Trp growth medium. The positive clones were diluted with 0.9% (w/v) NaCl to OD600 of 0.2, after which 10 μL of each suspension was dropped on SD/-Leu-Trp, SD/-Leu-Trp-His-Ade with X-α-Gal medium. The experiments were performed three times with similar results.
The target gene’s DNA sequence was cloned into a vector containing the luciferase (LUC) reporter gene to form a recombinant plasmid. This plasmid was then transformed into tobacco leaf cells via Agrobacterium-mediated transformation and transiently expressed there. The luciferase detection system, with the substrate added to induce luminescence, was used, and the imaging system recorded the luminescent signals.
Role of different concentrations of IBA, 6-BA, and different light qualities in the growth of C. reticulata callus
The C.reticulata ‘Zipao’ and wild species callus were placed in YS1, YS2, YS3 and YS4 callus medium, and they were placed in white light, dark, red and blue light environments cultured for 30 d.
The callus of C.reticulata ‘Zipao’ and wild species were inoculated onto YS3 medium and placed under red, blue, red-blue ratio of 2:1 and blue-red light ratio of 2:1 for one month.
Role of GSH and AsA in the induction of adventitious shoots in C. reticulata callus
The C.reticulata ‘Zipao’ and wild species callus were put into AsA1, GSH1 and A1G1 liquid medium, and cultured at 28°C and 220 R for 48 h, then inoculated into YS3 medium, and cultured for 48 h with dark place, and then put into the light environment of blue-red light ratio of 2:1. Callus inoculated into YS3 medium and cultured in blue-red light ratio of 2:1 environment without any treatment were used as control.
Method of making paraffin sections
The paraffin section method was used to observe the internal cellular structure of the callus of ‘Zipao’ and wild species C. reticulata after treatment with different light intensities, light qualities, GSH, and AsA. Making paraffin sections mainly includes: sampling, fixation, dehydration, clearing, wax infiltration, embedding, sectioning, staining (using safranin - fast green staining), dewaxing, and sealing. Microscopic observation was performed using an optical microscope (Leica DM2500), and the magnification used was 200×.
Results
Gene cloning and sequence analysis
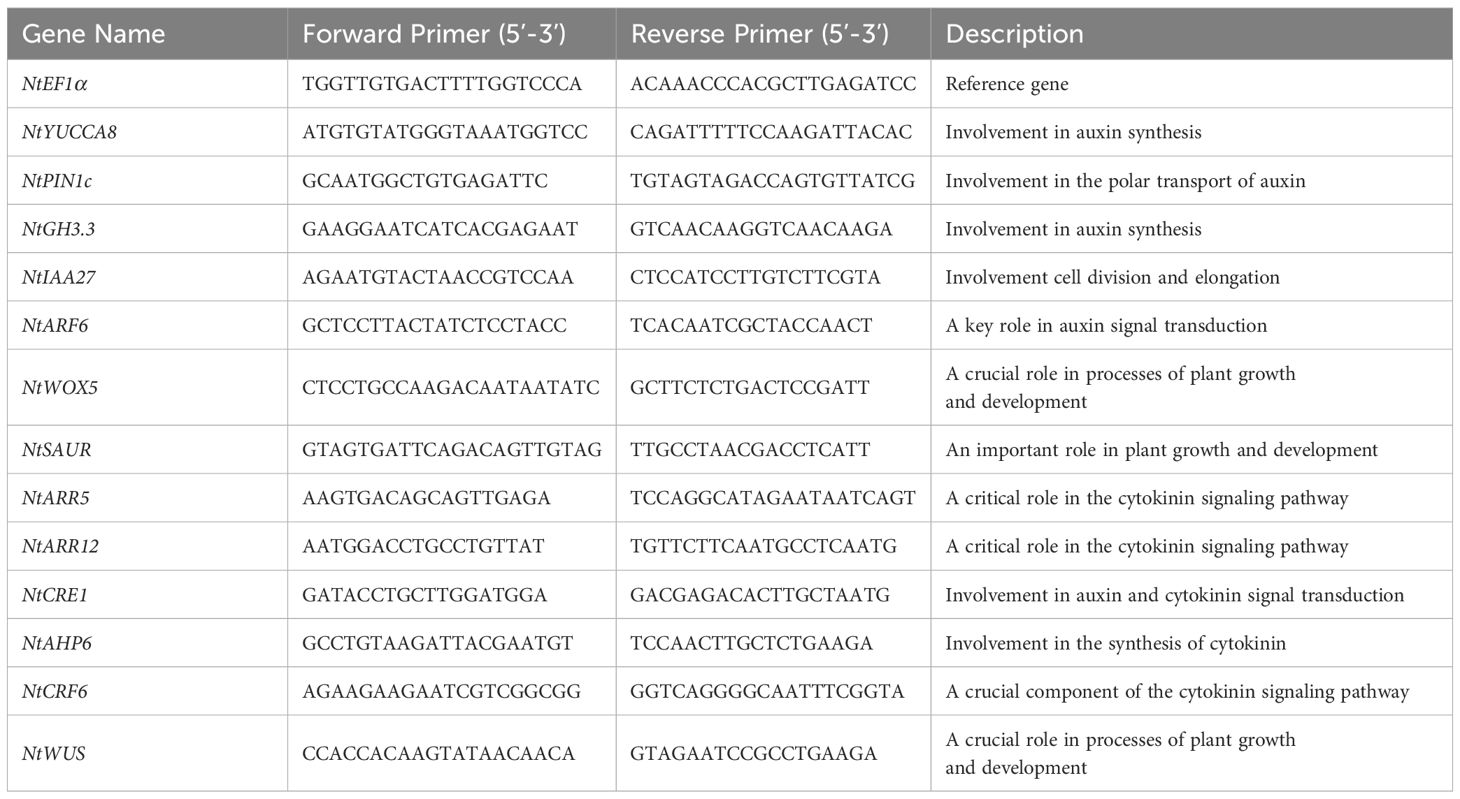
The GST24 was cloned from callus of Camellia reticulata. Multiple sequence alignment and phylogenetic analysis revealed high similarity between this gene and GST gene in Theaceae, leading to its designation as CrGST24. Based on protein homology and gene structure characteristics, the GST gene is divided into seven subfamilies: TAU, Zeta, Lambda, DHAR, Tchqd, Theta, and Phi. The CrGST24 belongs to the TAU, full length is 693 bp and contains an open reading frame encoding 213 amino acids (Figure 1a). In order to predict which proteins CrGST24 can interact with, we used the STRING database for analysis, which revealed that CrGST24 may have potential interactions with GSHB and DHAR2 (Figure 1b).
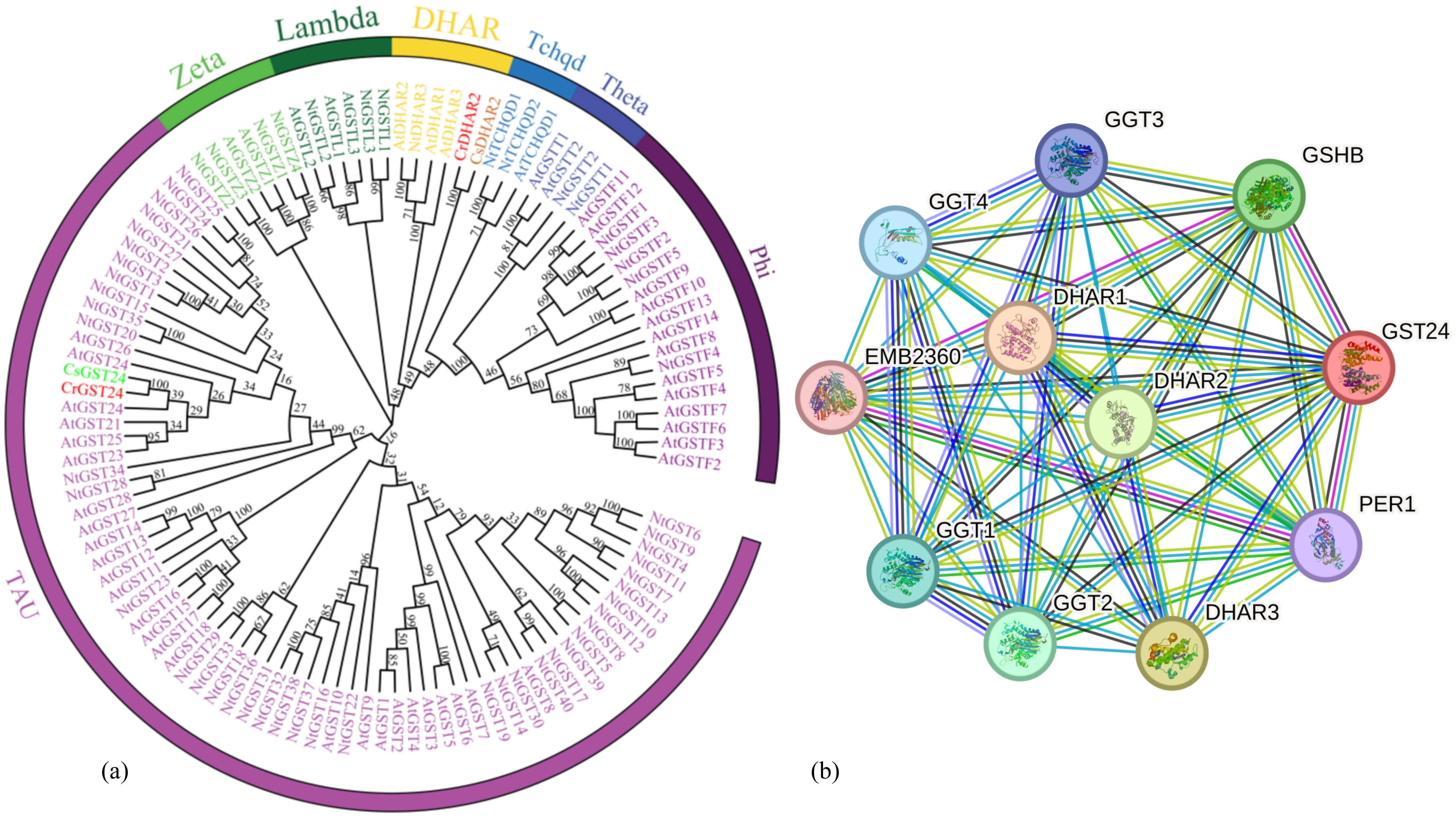
Figure 1. Bioinformatics analysis of the GST from C reticulata. Note: (a) Phylogenetic tree analysis of GST family members; (b) Protein-protein interaction network.
qPCR analysis of CrGST24 transcription factors
The callus of wild species and ‘Zipao’ of C. reticulata were placed in medium containing different concentrations of IBA and 6-BA, and treated for 2 h and a week. The expression of CrGST24 gene was significantly higher than that of the control after 2 h of treatment in medium containing different concentrations of IBA and 6-BA. The expression of CrGST24 gene in the callus of wild species was 1.55, 1.87, and 1.38 folds higher than that of the control; the expression of CrGST24 in the callus of ‘Zipao’ was 1.33, 2.03, and 1.23 folds higher than that of the control (Figure 2a). The expression of CrGST24 gene was significantly higher than that of the control after a week of treatment. The expression of CrGST24 gene in the callus of wild species was 1.30, 1.79 and, 1.33 folds higher than that of the control; the expression of CrGST24 gene in the callus of ‘Zipao’ was 1.40, 2.27, and 1.57 folds higher than that of the control (Figure 2b). The CrGST24 gene was most highly expressed in the medium of 0.1 mg/L IBA and 1.0 mg/L 6-BA.
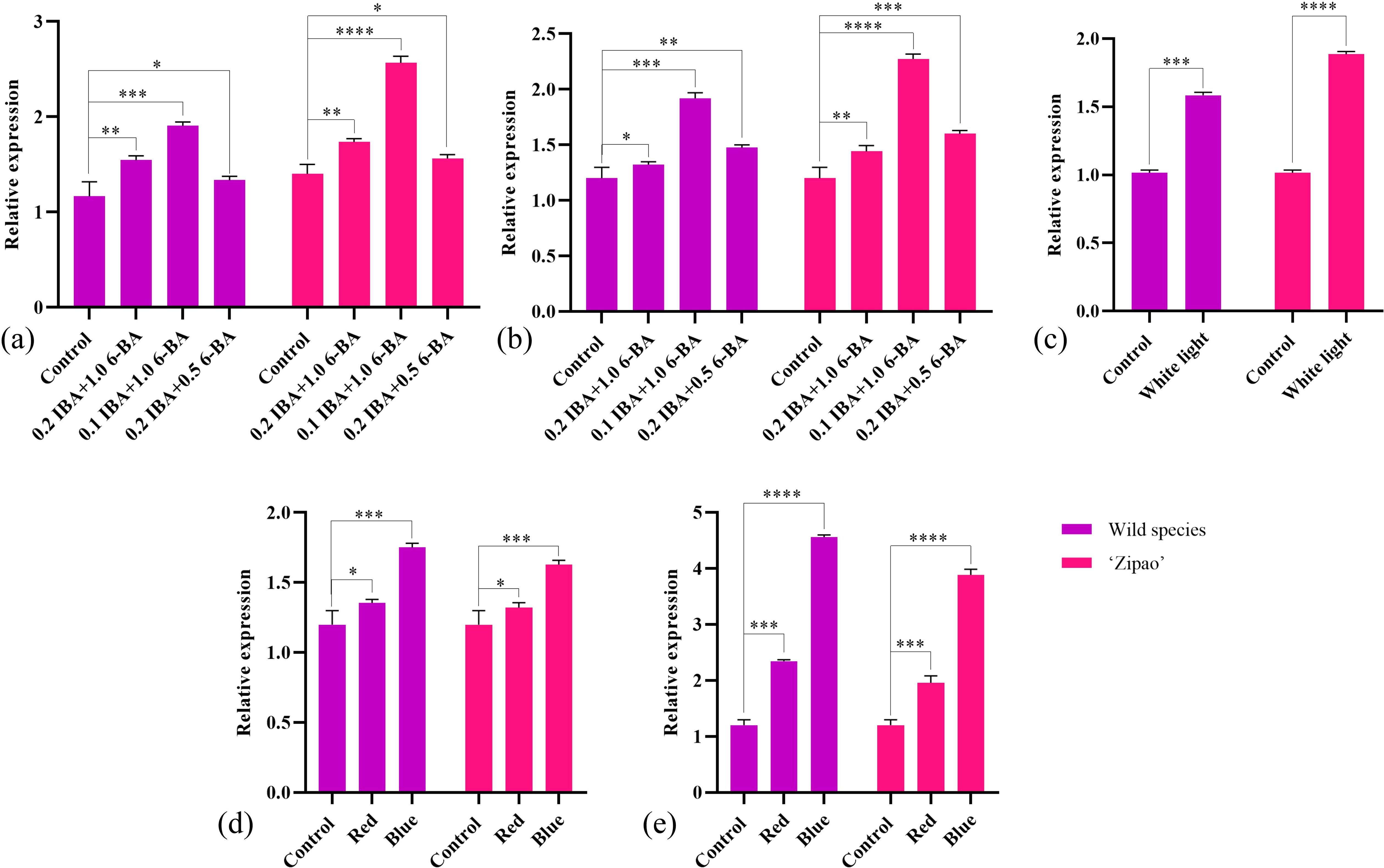
Figure 2. Expression analysis of CrGST24. (a) CrGST24 expression under different concentrations of IBA, and 6-BA (2 h); (b) CrGST24 expression under different concentrations of IBA, and 6-BA (1 week); (c) CrGST24 expression under different light conditions (8 h); (d) CrGST24 expression under different light quality (8 h); (e) CrGST24 expression under different light quality (3 d). Note: The bar chart represents the mean of three biological replicates, with error bars showed standard deviations. Asterisks indicate statistical significance based on one-way analysis of variance (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
The callus of wild species and ‘Zipao’ of C. reticulata were placed in YS1 mediun and cultured in dark and white light conditions for 8 h. The dark condition as a control. The expression of CrGST24 gene was significantly higher than that of control after 8h of treatment under different light conditions. The expression of the CrGST24 gene in the wild species was 1.53 folds higher than that of the control; the expression of the CrGST24 gene in the ‘Zipao’ was 1.76 folds higher than that of the control (Figure 2c). The CrGST24 gene responds to light induction.
The callus of wild species and ‘Zipao’ of C. reticulata were placed in YS1 medium and cultured in white, red and blue light conditions for 8 h, and 3 d. The white light as a control. The expression of CrGST24 gene was significantly higher than that of the control after 8 h of treatment in red, and blue light conditions. The expression of CrGST24 gene in the callus of wild species was 1.31 and 1.75 folds higher than that of the control; the expression of CrGST24 gene in the callus of ‘Zi Pao’ was 1.28 and 1.63 folds higher than that of the control (Figure 2d). The expression of CrGST24 gene was significantly higher than that of the control after 3 d of treatment. The expression of CrGST24 gene in the callus of wild species was 2.32 and 4.55 folds higher than that of the control; the expression of CrGST24 gene in the callus of ‘Zipao’ was 1.87 and 3.85 folds higher than that of the control (Figure 2e). The expression of the CrGST24 gene increased with treatment time, and the CrGST24 gene was responsive to both red and blue light induction.
Genetic transformation in tobacco and screening of transgenic tobacco lines
After Agrobacterium-mediated heterologous genetic transformation of tobacco, forty T1 tobacco lines were obtained. Seeds were collected and sown to generate forty T2 lines. Using DNA from the T2 transgenic tobacco as a template, primers targeting the hygromycin transferase gene were designed for PCR, screening out ten positive lines. Through qRT-PCR, three high-expression lines were identified: CrGST24-OE6, CrGST24-OE7, and CrGST24-OE22, with expression levels 1402.37, 1292.74, and 971.02 folds higher than that in WT (Figures 3a-h).
Figure 3. Genetic transformation, positive seedling screening, and phenotypic observation of transgenic tobacco. (a) Tobacco leaf discs infected with Agrobacterium tumefaciens; (b) Induction of resistant buds; (c) Expansion of resistant buds; (d) Isolation of resistant buds; (e) Formation of independent lines; (f) Expression analysis of transgenic lines; (g) Transgenic tobacco after transplanting; (h) Seeds harvested from mature transgenic tobacco for subsequent experiments; (i, m) WT tobacco plants; (j, n) CrGST24-OE6; (k, o) CrGST24-OE7; (l, p) CrGST24-OE22. The bar chart represents the mean of three biological replicates, with error bars showing standard deviations. Asterisks indicate statistical significance based on one-way analysis of variance (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Phenotype observation of the CrGST24 transgenic lines
The length and density of stem trichomes in transgenic tobacco were superior to those in WT (Figures 3i-l). Electron microscopy of freehand sections revealed that WT had approximately 35-40 trichomes per field of view (20x), with lengths ranging from 1.02 to 1.51 mm and an average length of 1.32 mm. In contrast, transgenic tobacco exhibited 75-80 trichomes, with lengths ranging from 2.11 to 3.64 mm and an average length of about 3.13 mm. This resulted in trichome lengths and densities that were 2.37 folds higher than those of the WT (Figures 3m-p).
Observations on in vitro regeneration of transgenic tobacco
The leaves of WT and transgenic tobacco were cut into uniform sizes, and then placed in normal MS medium to observe the growth. The WT leaves started to differentiate callus on the 10th day and as the treatment time increased, more callus were differentiated from the WT and by the 20th day, four callus were differentiated from the WT leaves. Leaves of CrGST24 transgenic tobacco showed signs of differentiation of adventitious roots and callus at the 5th day, with adventitious roots of about 0.1 mm in length; on the 10th day adventitious roots continued to grow and were about 0.2-0.5 mm in length; The length of the adventitious roots was about 0.4-0.7 mm on the 15th day; the length of the adventitious roots was about 0.5-0.8 mm on the 20th day. On the 35th day, both WT and transgenic tobacco plants produced adventitious buds. Compared with the WT, the adventitious buds produced by the transgenic tobacco were more obvious. On the 50th day, the adventitious buds of WT gradually grew larger, while those of transgenic tobacco had grown into independent plants (Figure 4a).
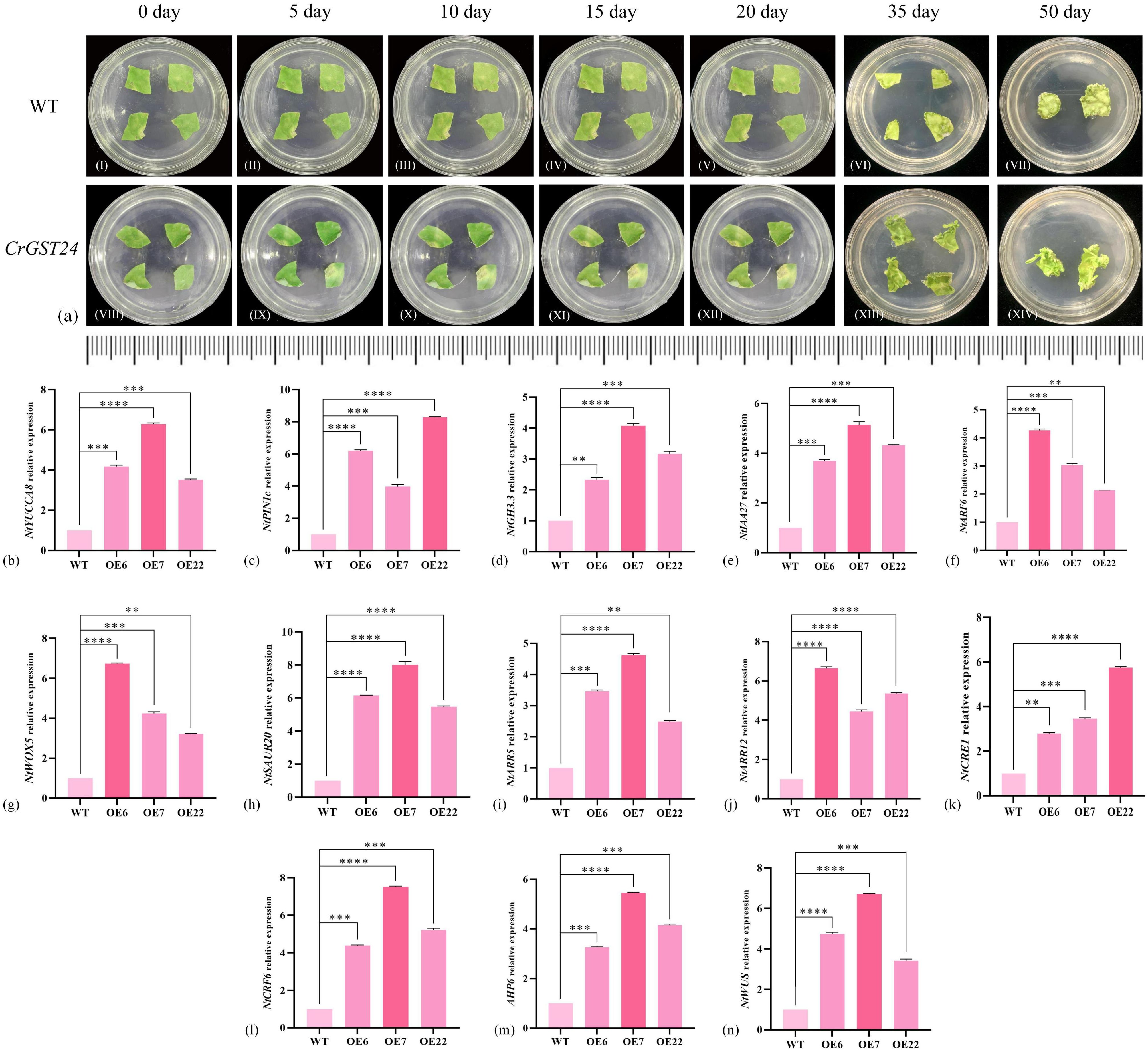
Figure 4. Observations on in vitro regeneration of CrGST24 transgenic tobacco and expression of auxin and cytokinin-related genes in CrGST24 transgenic tobacco. (a) in vitro regeneration (b-h) auxin-related genes; (i-n) cytokinin-related genes. The bar chart represents the mean of three biological replicates, with error bars showing standard deviations. Asterisks indicate statistical significance based on one-way analysis of variance (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Analysis of auxin and cytokinin-related gene expression in CrGST24 transgenic tobacco
Based on the leaf regeneration of transgenic tobacco in vitro, we found auxin and cytokinin-related genes in tobacco at the NCBI online website, and analysed the expression of these genes in transgenic tobacco by q-PCR. The expression of auxin-related genes in transgenic tobacco were all significantly increased and significantly higher than the WT. The expression of NtYUCCA8 in the three transgenic tobacco lines were 3.98, 6.07, and 3.54 folds higher than the WT (Figure 4b); NtPIN1c were 6.11, 3.87, and 7.97 folds higher than the WT (Figure 4c); NtGH3.3 were 2.32, 4.03, and 3.12 folds higher than the WT (Figure 4d); NtIAA27 were 3.68, 4.97, and 4.33 folds higher than the WT (Figure 4e); NtARF6 were 4.18, 2.89, and 1.93 folds higher than the WT (Figure 4f); NtWOX5 were 6.36, 4.02, and 3.47 folds higher than the WT (Figure 4g); NtSAUR20 were 5.87, 7.92, and 5.71 folds higher than the WT (Figure 4h).
The expression of cytokinin-related genes were significantly increased in transgenic tobacco and was significantly higher than the WT. NtARR5 were 3.24, 4.27, and 2.18 folds higher than the WT (Figure 4i); NtARR12 were 6.22, 4.12, and 5.28 folds higher than the WT (Figure 4j); NtCRE1 were 2.48, 3.67, and 5.69 folds higher than the WT (Figure 4k); NtCRF6 were 3.76, 5.71, and 3.98 folds higher than the WT (Figure 4l); NtAHP6 were 4.07, 7.39, and 5.24 folds higher than the WT (Figure 4m); NtWUS were 4.32, 6.26, and 3.77 folds higher than the WT (Figure 4n). The CrGST24 gene promoted the expression of both auxin and cytokinin-related genes.
Expression analysis of auxin and cytokinin-related genes in the callus of C. reticulata
A total of 12 auxin and cytokinin-related genes were screened in the transcriptome data of C. reticulata callus. There are seven auxin-related genes, including: CrYUCCA, CrPIN, CrGH3, CrWOX, CrARF, CrSAUR and CrIAA. The most significant differences in the expression of CrYUCCA2, CrPIN3, CrGH3.9, CrWOX13, CrARF4, CrSAUR20 and CrIAA9 were found in the callus of the wild species, which were higher than ‘Zipao’ by 1.36, 2.41, 4.17, 8.25, 11.67, 17.24 and 7.73 folds; the most significant differences in the expression of CrYUCCA10, CrPIN1, CrGH3.1, CrWOX5, CrSAUR15 and CrIAA17 were found in the callus of ‘Zipao’, which were 4.88, 2.05, 10.12, 8.85, 52.51, and 1.43 folds higher than that of the wild species (Figures 5a-g).
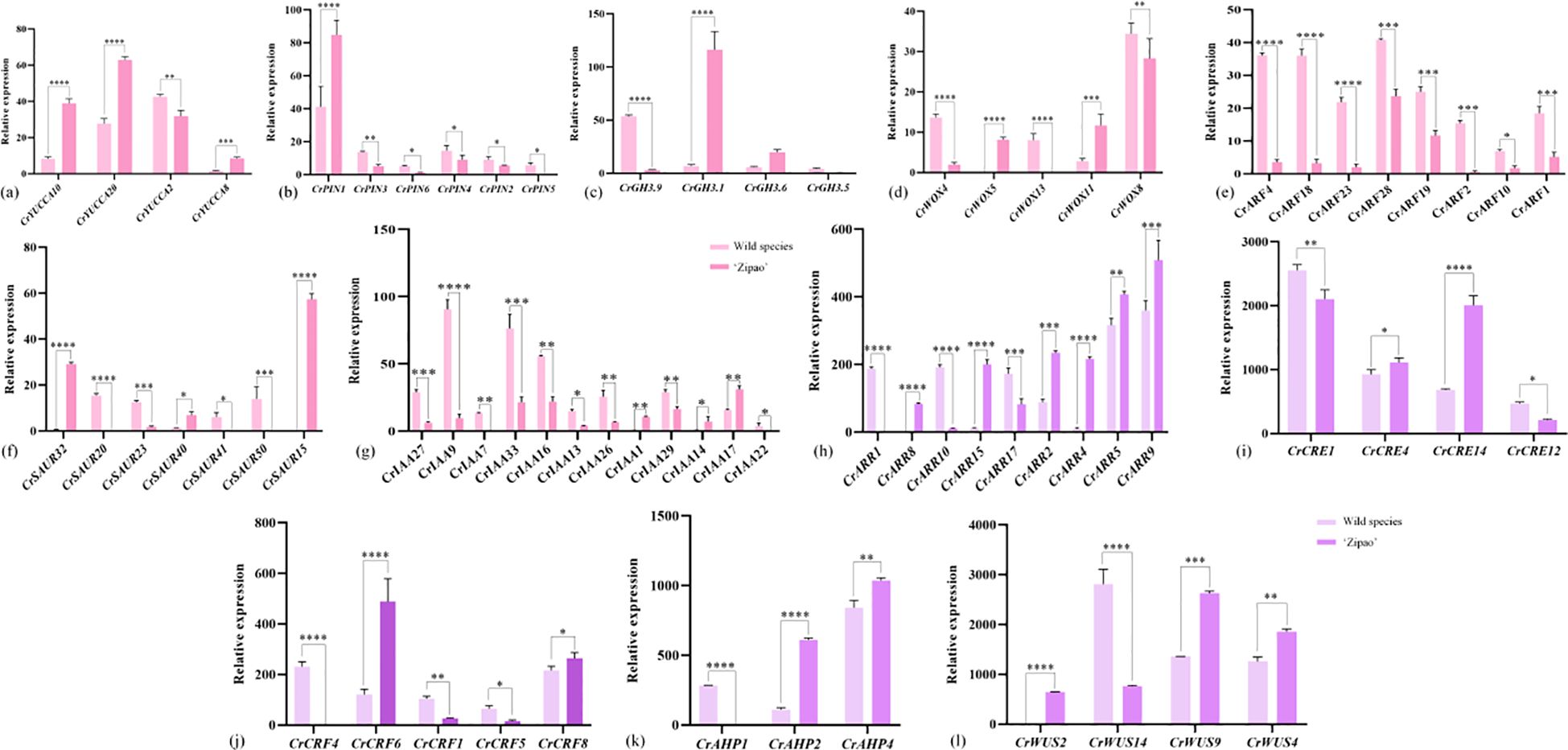
Figure 5. Expression of auxin and cytokinin-related genes in C. reticulata callus. (a-g) Auxin-related genes; (h-l) Cytokinin-related genes. The bar chart represents the mean of three biological replicates, with error bars showing standard deviations. Asterisks indicate statistical significance based on one-way analysis of variance (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
There were five cytokinin-related genes, including CrARR, CrCRE, CrCRF, CrAHP, and CrWUS. The most significant differences in the expression of CrARR1, CrCRE1, CrCRF4, CrAHP1, and CrWUS14 were found in the callus of the wild species, which were 188.95, 1.25, 205.32, 237.02, and 3.11 folds higher than that of ‘Zipao’. The most significant differences in the expression of CrARR4, CrCRE14, CrCRF6, CrAHP2, and CrWUS9 were found in ‘Zipao’ callus, which were 201.14, 2.51, 3.58, 5.67, and 2.48 folds higher than that of the wild species (Figures 5h-l).
Determination of auxin, cytokinin, ROS, GSH and AsA contents in CrGST24 transgenic tobacco
When the auxin and cytokinin contents within transgenic tobacco was examined, the auxin contents in transgenic tobacco were significantly higher than that in WT, which were 1.55, 2.09, and 1.47 folds higher than that in WT (Figure 6a); cytokinin contents in transgenic tobacco were significantly higher than that in WT, which were 1.13, 2.16, and 1.54 folds higher than that in WT (Figure 6b). The contents of auxin were significantly higher than that of cytokinin in transgenic tobacco, and the ratios of auxin and cytokinin contents in WT and transgenic tobacco were analysed, and the ratios of auxin and cytokinin contents in CrGST24 transgenic tobacco were significantly higher than those in WT, which were 1.24, 1.13, and 1.15 folds higher than those in WT (Figure 6c).
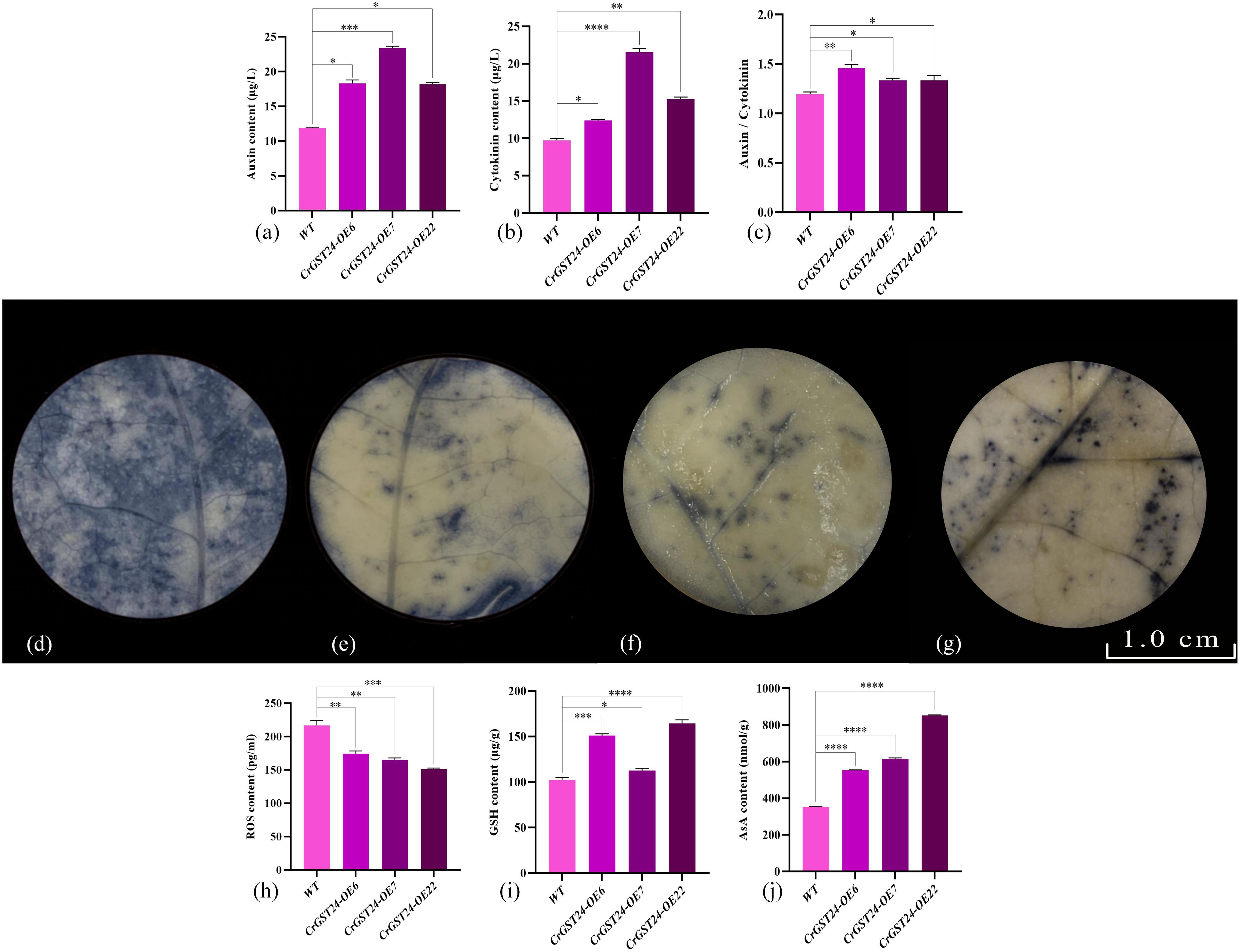
Figure 6. Expression of auxin, cytokinin, ROS, GSH, and AsA contents in CrGST24 transgenic tobacco. Note: (a) Auxin content; (b) Cytokinin content; (c) Auxin/cytokinin content; (d) WT; (e) CrGST24-OE6; (f) CrGST24-OE7; (g) CrGST24-OE22; (h) ROS content; (i) GSH content; (j) AsA content; (d-g) the blue part is the area of ROS accumulation. The bar chart represents the mean of three biological replicates, with error bars showing standard deviations. Asterisks indicate statistical significance based on one-way analysis of variance (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
NBT staining followed by decolourisation of WT and transgenic tobacco revealed that the area of ROS was significantly larger in WT than in transgenic tobacco (Figures 6d-g). The detection of ROS contents in both WT and transgenic tobacco revealed that the ROS contents of transgenic tobacco were significantly lower than that of WT, 1.15, 1.07 and 1.31 folds lower than that of WT (Figure 6h); the GSH contents of transgenic tobacco were all significantly higher than those of WT, 1.27, 1.42 and 1.35 folds higher than those of WT (Figure 6i); the AsA content of transgenic tobacco were all significant higher than those of WT, 6.09, 4.47, and 2.92 folds higher than those of WT (Figure 6j).
The interaction between CrGST24 and CrDHAR2, CrGSHB
The subcellular localization results showed that CrGST24, CrGSHB, and CrDHAR2 are all located in the nucleus (Figure 7a). We screened key genes involved in AsA and GSH synthesis by yeast two-hybrid assay, and showed that the CrGST24 gene could interact with CrGSHB and CrDHAR2 (Figures 7b, c). The LUC experiment further confirmed that CrGST24 interacted with CrGSHB and CrDHAR2 (Figures 7d, e).
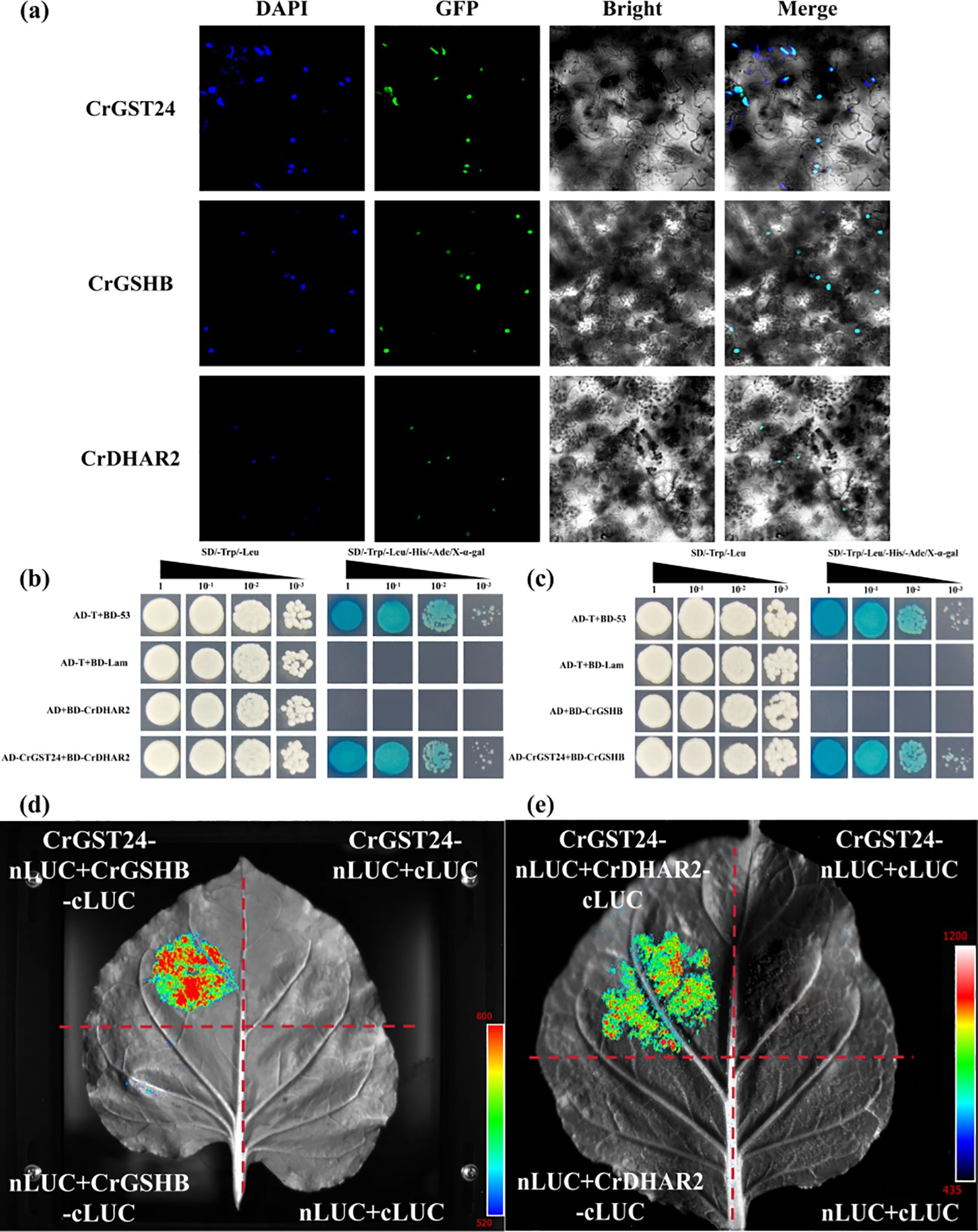
Figure 7. CrGST24 interact with CrGSHB and CrDHAR2. (a) Subcellular localization assay; (b, c) Y2H assay; (d, e) LUC assay.
Role of different concentrations of IBA, 6-BA and different light qualities in C. reticulata callus culture
The callus of ‘Zipao’ and wild species were placed on YS1, YS2, YS3 and YS4 callus medium containing different concentrations of IBA and 6-BA, and they were incubated with white light, darkness, red light, or blue light for 30 d to observe the growth of the callus. In comparison with the white loose tissue structure before treatment and other different treatments, ‘Zipao’ callus in YS3 medium and treated with red light or blue light respectively had obvious changes in colour from white to green, and the tissue structure changed from loose to denser, and the newly differentiated callus also showed obvious green colour (Figure 8a). The wild species grew better in YS3 medium and under red or blue light treatments, with denser tissue structure and subsequent newly differentiated healing tissues that were also distinctly green in colour (Figure 8b). The above results indicated that YS3 medium, as well as red or blue light were good for the C. reticulata callus growth.
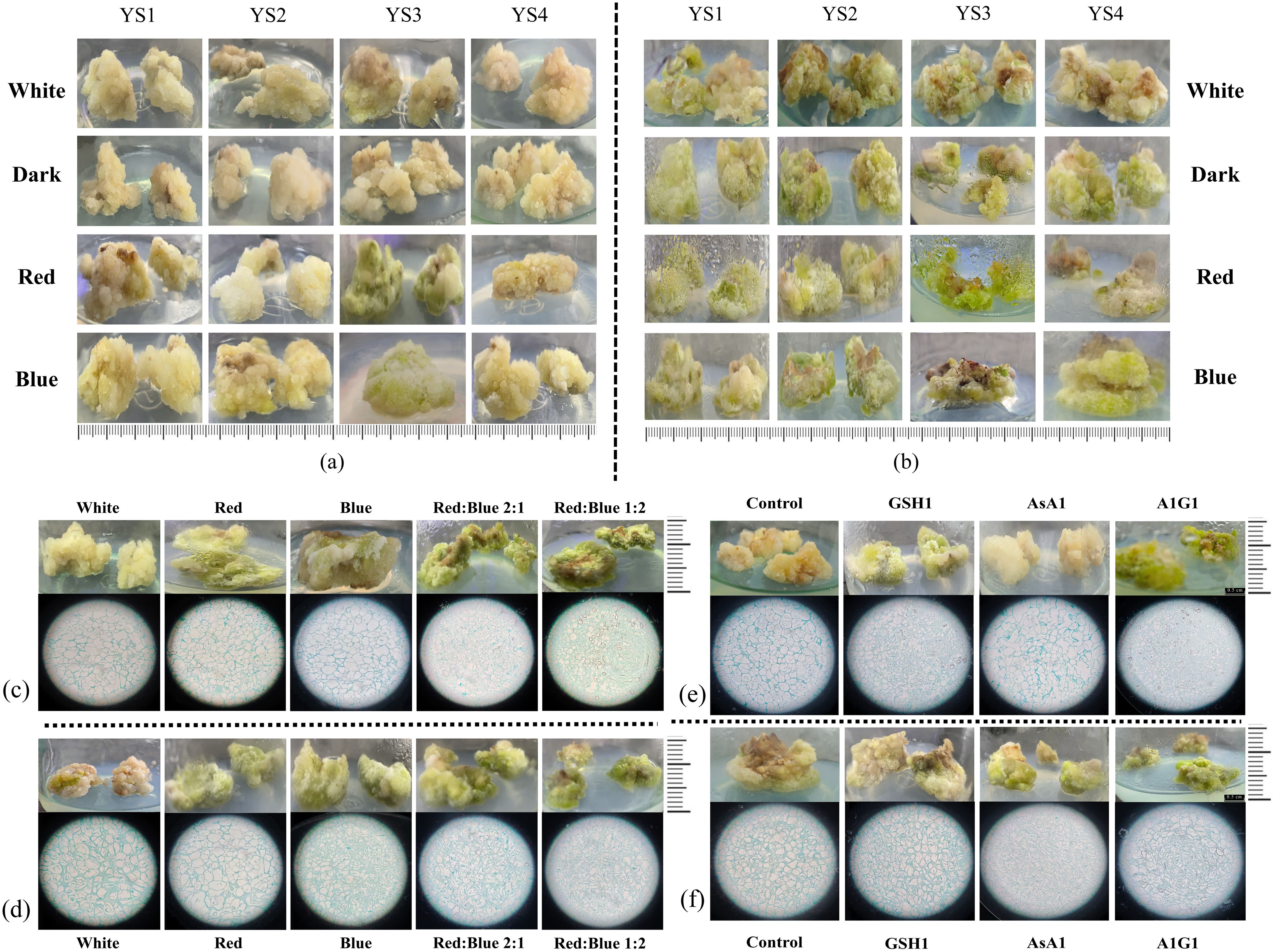
Figure 8. Growth status of callus from C. reticulata. (a, b) Callus in medium contaning different concentrations of IBA, 6-BA, and different light quality treatments; (c, d) Callus under red light, blue light, and red-blue light; (e, f) Callus under GSH and AsA treatments. (a, c, e) ‘Zipao’; (b, d, f) wild species.
Analysis of the growth of C. reticulata callus under red and blue light environments
Callus of ‘Zipao’ and wild species were inoculated on YS3 medium, or under four different light quality treatments, including red, blue, red-blue ratio of 2:1, and blue-red ratio of 2:1 for one month with white light as the control. Under the four different light quality treatments, the callus of ‘Zipao’ changed significantly from the original white loose tissue to the green firm healing tissue, and the growth condition was better under blue-red ratio of 2:1. The paraffin sections showed that the cells of callus were compact under the red or blue light treatment compared to the white light; the cells were tightly packed under red-blue ratio of 2:1 or blue-red ratio of 2:1, and compared with red-blue ratio of 2:1, the cells were much more tightly under blue-red ratio of 2:1 (Figure 8c).
There was no significant difference in the morphological characteristics of wild species callus under red or blue light compared with white light, but the morphological characteristics changed significantly after red-blue ratio of 2:1 and blue-red ratio of 2:1, with the colour changing from dark green to bright green. The paraffin sections showed that the degree of cell tightness under red or blue light was not significantly different from that after white light treatment; cells treated under red-blue ratio of 2:1 or blue-red ratio of 2:1 were tight, and the tightness of the cells was made higher under the blue-red ratio of 2:1 treatment (Figure 8d).
Role of GSH and AsA in the induction of adventitious shoots in the callus of C. reticulata “Zipao” and wild species
The wild species callus treated with GSH1 changed from light green to green and became tighter, and the cells were more tight according to the paraffin sections; the wild species callus treated with AsA1 did not change significantly; the best growth of wild species callus were observed after A1G1 treatment, with bright green and tight cells (Figure 8e).
The ‘Zipao’ callus treated with GSH1 changed from white to green and became tighter, and the cells of ‘Zipao’ callus treated with GSH1 were more tight according to the paraffin sections; there were no significant changes in the external morphology and internal structure of ‘Zipao’ callus treated with AsA1 compared to those before treatment; ‘Zipao’ callus treated with A1G1 showed the best growth, in terms of external colour the ‘Zipao’ callus treated with A1G1 were bright green in colour compared to the pre-treatment and GSH1 treatment, and in terms of cellular structure the cultivar healing cells were highly aggregated in the A1G1 treatment (Figure 8f).
Discussion
In the previous study, when the auxin was added exogenously or synthesised endogenously, it will bind to the promoter region of the downstream GST gene through a series of pathways and activate the expression of the GST gene, and when the GST gene is overexpressed, it will promote the synthesis of GSH, which will form a cycle with AsA, and thus inhibit or remove ROS (Supplementary Figure 2). The conclusions obtained in this study were consistent with previous studies that CrGST24 was able to promote the synthesis of endogenous auxin and cytokinins in transgenic tobacco, and that the content of GSH and AsA was significantly higher and the content of ROS was significantly lower in CrGST24 transgenic tobacco than in WT tobacco.
GST genes and phytohormones in plant growth
In this study, we found that CrGST24 could promote the synthesis of endogenous auxin and cytokinin. It also caused changes in the expression of auxin and cytokinin - related transcription factors such as YUCCA, ARF, GH3, WOX, WUS, CRF, AHP, CRE, and ARR. Overexpression of the YUCCA could significantly increase the auxin content in plants, thereby affecting their growth and development (Cao et al., 2019). In Arabidopsis thaliana, knocking out the YUCCA resulted in diminished adventitious bud formation capacity, whereas overexpression of the YUCCA enhanced the ability of adventitious bud differentiation (Miguel et al., 2020). In rice, overexpression of OsWOX3A led to significantly upregulated expression of PIN1. The resulting change in auxin transport direction caused by PIN1 led to local concentration gradients. This process activated the expression of YUCCA genes through feedback regulation, thereby promoting the synthesis of endogenous auxin (Sung and Paek, 2016). The WOX5 gene interacted with auxin and cytokinin signaling pathways. Specifically, by regulating the ARR genes, it affected the sensitivity to cytokinins, thereby promoting the differentiation of adventitious buds from callus (Zhai and Xu, 2021). In plant biology, the ARF transcription factor activated GH3 and SAUR genes, thereby promoted the formation of IAA conjugates and increased the levels of endogenous auxin (Dong et al., 2024). The cytokinin signal was transmitted through the AHK-AHP-ARR cascade (To and Kieber, 2008). Once activated, specific types of ARR directly bound to the promoter of CRF and upregulated their expression. Meanwhile, AHP interacted with CRF, promoting their nuclear accumulation. This formed a coordinated regulatory module between CRF and ARR, which together activated downstream cytokinin - response genes, thereby promoted cytokinin synthesis (Cutcliffe et al., 2011). Previous studies have found that the transcription factors YUCCA, PIN, WOX, SAUR, ARF, ARR, AHP, and CRF have extremely important functions in promoting the synthesis of auxin and cytokinin. In the transcriptome data of C. reticulata callus, the auxin and cytokinin-related transcription factors were also found. Combined with the results of this study, it is hypothesized that these transcription factors may bind to the promoter of the CrGST24 gene and activate its expression. Along with exogenous IBA and 6-BA, as well as red - blue light induction, this process promotes the synthesis of endogenous auxin and cytokinin.
The role of GSH and AsA in adventitious bud differentiation
GSH and AsA have extremely important roles in plant growth, especially in the scavenging of ROS (Hasanuzzaman et al., 2020). In this study we found that the CrGST24 gene promoted the synthesis of GSH and AsA in transgenic tobacco, and that the content of glutathione and ascorbic acid in CrGST24 transgenic tobacco was significantly lower than that in WT tobacco. Better growth of C. reticulata callus treated with GSH and AsA. GSH has an extremely important role in the differentiation of adventitious buds (Elaziem and Ahmed, 2020). In the induction of adventitious buds of Phoenix dactylifera, the addition of 30 mg/L GSH to the proliferation medium significantly promoted the differentiation of adventitious buds. After four generations of subculture, the number of adventitious buds significantly increased in the medium containing 30 mg/L GSH and 0.5 mg/L TDZ (Elaziem and Ahmed, 2020). GSH enhanced plant adventitious shoot differentiation by regulated intracellular iron homeostasis, participated in signal transduction pathways and improved the redox environment of the medium (Ranjana et al., 2021). Adding a certain amount of GSH to the culture medium reduced the secretion of phenolic substances, prevented browning of plant materials, and improved the induction efficiency of adventitious buds in bananas (Skogerboe and Jenderek, 2022). Glutathione is an important antioxidant in plant cells, and the dynamic balance between its reduced (GSH) and oxidised (GSSG) states (GSH/GSSG) directly affects the redox state of cells. During adventitious bud differentiation, accumulation of reactive oxygen species (ROS) inhibited cell dedifferentiation or redifferentiation. Glutathione protected DNA, proteins and membrane systems from oxidative damage by scavenging ROS, thus provided a stable microenvironment for callus formation and bud primordial differentiation (Müller et al., 2020). AsA plays an important role in cell division and growth, and AsA has an extremely important role in the growth of plant roots and adventitious shoots (Akram et al., 2017). When ascorbate peroxidase (APX1) was knocked out in oilseed rape, the differentiation rate of adventitious shoots was drastically reduced (Liang et al., 2024). Addition of AsA to the culture medium significantly increased the efficiency of inducing adventitious shoots in sorghum (Baskaran and Jayabalan, 2005). After blue light treatment of wheat seedlings, the contents of POD, SOD, GSH and AsA in wheat leaves significantly increased, while the ROS content markedly decreased. This indicated that blue light treatment could enhance the antioxidant capacity of wheat seedlings (Li et al., 2022). GST genes were closely related to GSH and AsA synthesis. In this study we found that CrGST24 promoted GSH and AsA synthesis in transgenic tobacco. The callus treated with GSH and AsA grew better. Excessive ROS could severely inhibit dedifferentiation and redifferentiation of plant callus. We hypothesised that when the GSH and AsA contents were elevated in C. reticulata callus then excessive ROS would be scavenged; exogenous addition of GSH and AsA, on the other hand, inhibits the secretion of phenolics and keeps the browning of healing tissues in culture under control. When the GSH and AsA contents in the healing tissue increased significantly, and at the same time GSH and AsA were added exogenously, a double safeguard was formed, which promoted the differentiation of adventitious shoots in the C. reticulata callus. The synthesis of GSH and AsA is a complex mechanism that requires further study.
The co-action of GST genes with other genes in plant growth
GST gene enhances plant resistance to biotic and abiotic stresses by scavenging excess ROS. In this study, overexpression of the CrGST24 gene promoted the synthesis of GSH and AsA in transgenic tobacco, and the ROS content in transgenic tobacco was significantly lower than that in WT tobacco. We found that the CrGST24 gene could interact with CrGSHB, a key gene for GSH synthesis, and CrDHAR2, a key gene for AsA synthesis, by Y2H assay. GSHB genes have an important role in plant resistance to adversity stress (Anjuman et al., 2025). Numerous studies have shown that DHAR and GSHB genes have extremely important roles in plant resistance to biotic and abiotic stresses, and in scavenging ROS. DHAR has an important role in the synthesis of AsA, DHAR maintains intracellular AsA levels in the reduced state by reducing dehydroascorbic acid (DHA) to AsA. DHAR protected cells from oxidative damage by promoted AsA synthesis and scavenged intracellular ROS (Petrova and Smith, 2015). When the DHAR gene was knocked out, the content of AsA was significantly reduced and the antioxidant capacity of the plant was reduced (Terai et al., 2020). In rice, overexpression of the DHAR gene increased rice resistance to salt stress by reducing H2O2 levels (Kim et al., 2017). GSHB is a glutathione synthetase that adds glycine to γ-glutamylcysteine to eventually produce GSH. In Brassica campestris, overexpression of the GSHB gene significantly promoted GSH synthesis, which improved antioxidant capacity and stress tolerance in Brassica campestris (Yan et al., 2013). GSHB combines γ-glutamylcysteine (γ-Glu-Cys), which is produced by catalyzing glutamate, cysteine, and γ-glutamyltransferase (GshA), with glycine to ultimately form glutathione (GSH). This process is an important mechanism for cellular antioxidation and maintenance of redox homeostasis (Copley and Dhillon, 2002). In cyanobacteria, GSH cannot be synthesized when the GSHB gene was knocked out (Okumura et al., 1997). Overexpression of the GSHB gene in Arabidopsis promoted GSH synthesis and also improved tolerance to Cd stress in Arabidopsis (Kumar and Chattopadhyay, 2018). In AsA-GSH cycle, DHAR used reduced glutathione (GSH) as a hydrogen donor to catalyzed DHA production of AsA. Under oxidative stress, the AsA synthesis pathway is limited, and DHAR compensated for the synthesis deficit by regenerated AsA. For example, under drought conditions, elevated DHAR activity promoted AsA synthesis (Hasanuzzaman et al., 2020). In transgenic tobacco, overexpression of DHAR gene increased AsA content by 2-4 folds (Wang et al., 2010). Based on the previous studies and the results of this research, we speculated that CrGST24 could utilize GSH as a cofactor to convert harmful ROS into harmless substances. CrGSHB enhanced the efficiency of the entire antioxidant system by synthesizing more GSH to provide sufficient substrates for CrGST24. When the CrGST24 gene was overexpressed, the expression of the CrGSHB gene may be promoted, thereby increased the synthesis of GSHB enzyme. More GSHB enzymes could catalyze more γ-Glu-Cys and Gly to synthesize GSH, thereby led to an increase in GSH content. When the CrGST24 gene was overexpressed, it may promote the synthesis of GSH or reduce the consumption of GSH through some mechanism, thereby providing more GSH as a substrate for CrDHAR2. Sufficient supply of GSH is conducive to CrDHAR2 catalyzing the reduction reaction of DHA more effectively, thereby increasing the content of AsA. The interaction between CrGST24 gene and CrDHAR2 gene may activate the entire AsA-GSH cycle system. Besides CrDHAR2, the activities of other related enzymes in the cycle (such as APX, GR, etc.) may also be coordinately regulated, thereby promoting the synthesis and regeneration of AsA. When the CrGST24 gene interacted with the CrGSHB and CrDHAR2 genes, it activated the entire AsA-GSH cycle system, regulated the expression of some key genes and thereby promoted the synthesis of GSH and AsA.
Conclusion
In this study, we found that the CrGST24 gene could promote the synthesis of endogenous auxin and cytokinin and influence their ratio, which was regulated by some auxin and cytokinin - related transcription factors, including CrWUS9, CrCRF4, CrAHP2, CrCRE6, CrGH3.9, CrARF4, CrPIN1, and CrWOX4. The elevation of the levels of endogenous auxin and cytokinin in the callus of C.reticulata through the treatment of exogenous IBA, 6-BA and red-blue light could increase the differentiation rate of adventitious buds, proving the above view. Moreover, the CrGST24 gene interacted with the CrGSHB and CrDHAR2 genes, thereby facilitating the synthesis of GSH and AsA and scavenging ROS. Thus, under the joint influence of the ratio of auxin and cytokinin and the scavenging ROS regulated by CrGST24, the adventitious bud differentiation was facilitated (Figure 9).
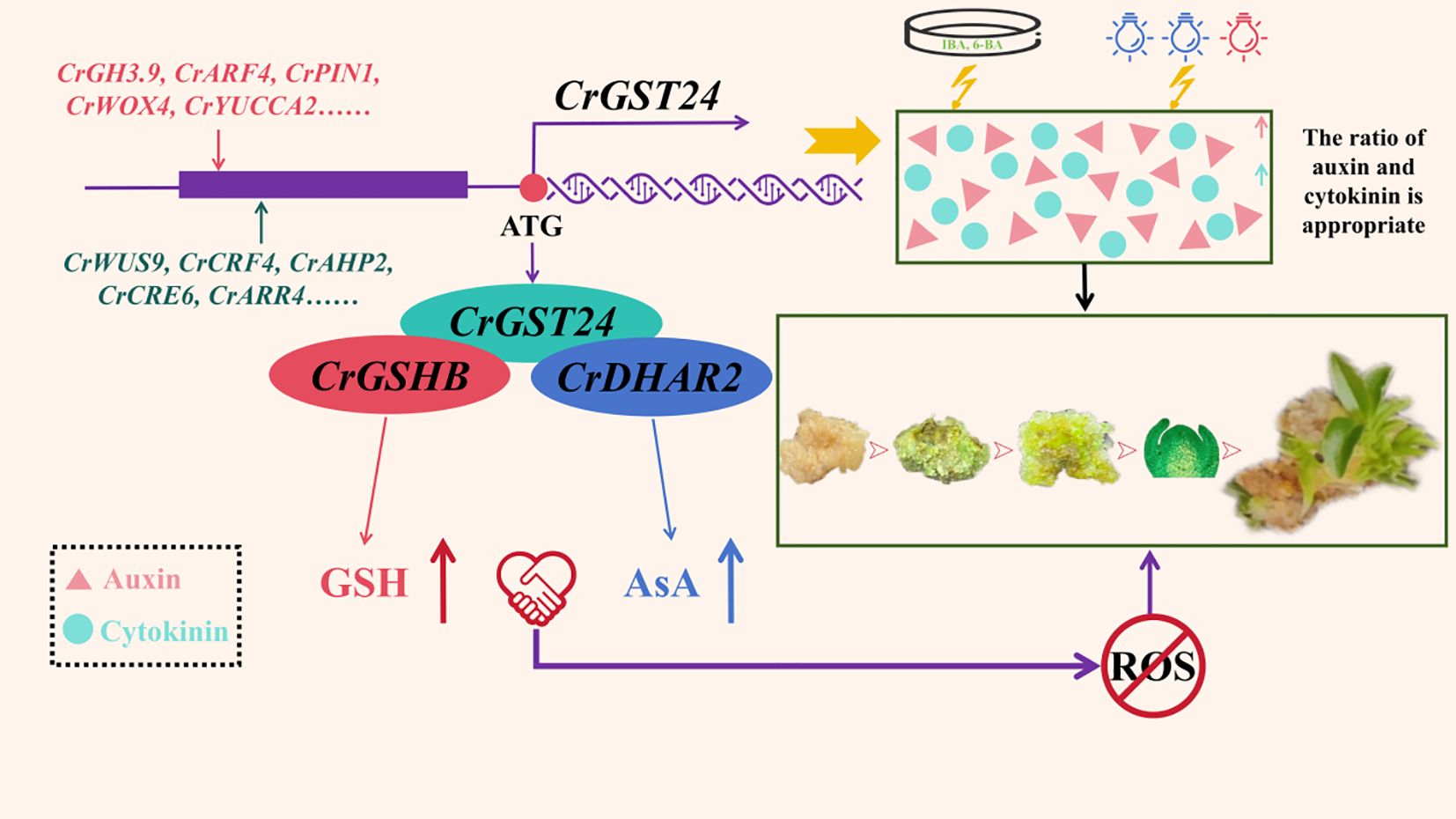
Figure 9. The mechanism of CrGST24 gene in C. reticulata promoting the formation of adventitious buds from callus differentiation.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
ZC: Conceptualization, Data curation, Formal Analysis, Methodology, Software, Writing – original draft. JW: Conceptualization, Data curation, Methodology, Software, Validation, Writing – original draft. YG: Software, Writing – original draft. XY: Software, Writing – original draft. CY: Software, Writing – original draft. TY: Software, Writing – original draft. TW: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded by Yunnan Fundamental Research Projects (202301BD070001-242) and National Key Research and Development Project (2019YFD1001005).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1641401/full#supplementary-material
Supplementary Figure 1 | Callus of Camellia reticulata used in the experiment. (a) ‘Zipao’; (b) Wild species
Supplementary Figure 2 | Schematic diagram of auxin and glutathione synthesis (Moones., 2005).
Supplementary Table 1 | The culture medium required for the research.
References
Abel, S. and Theologis, A. (1996). Early genes and auxin action. Plant Physiol. 111, 9–17. doi: 10.1104/pp.111.1.9
Ahanger, M. A., Alyemeni, M. N., Wijaya, L., Alamri, S. A., Alam, P., Ashraf, M., et al. (2018). Potential of exogenously sourced kinetin in protecting Solanum lycopersicum from NaCl-induced oxidative stress through up-regulation of the antioxidant system, ascorbate-glutathione cycle and glyoxalase system. PloS One 13, e0202175. doi: 10.1371/journal.pone.0202175
Akram, N. A., Shafiq, F., and Ashraf, M. (2017). Ascorbic Acid-A potential oxidant scavenger and its role in plant development and abiotic stress tolerance. Front. Plant Sci. 8, 613. doi: 10.3389/fpls.2017.00613
Anjuman, A., Farida, R., Amel, G., Khursheed, M., Krishna, K. Y., Sri, S. A., et al. (2025). Glutathione and biosensor technologies: Enhancing plant resilience to environmental stressors. Physiol. Mol. Plant Pathol. 136, 102570. doi: 10.1016/j.pmpp.2025.102570
Baskaran, P. and Jayabalan, N. (2005). In vitro plant regeneration and mass propagation system for Sorghum bicolor-a valuable major cereal crop. J. Agric. Technol. 1, 345–363.
Cao, X., Yang, H., Shang, C., Ma, S., Liu, L., Cheng, J., et al. (2019). The roles of auxin biosynthesis YUCCA gene family in plants. Int. J. Mol. Sci. 20, 6343. doi: 10.3390/ijms20246343
Copley, S. D. and Dhillon, J. K. (2002). Lateral gene transfer and parallel evolution in the history of glutathione biosynthesis genes. Genome Biol. 3, 0025. doi: 10.1186/gb-2002-3-5-research0025
Cutcliffe, J. W., Hellmann, E., Heyl, A., and Rashotte, A. M. (2011). CRFs form protein-protein interactions with each other and with members of the cytokinin signalling pathway in Arabidopsis via the CRF domain. J. Exp. Bot. 62, 4995–5002. doi: 10.1093/jxb/err199
Dai, S. L., Huang, H., Fu, J. X., and Hong, Y. (2013). Advances in molecular breeding of ornamental plants. Chin. Bull. Bot. 48, 589–607. doi: 10.3724/SP.J.1259.2013.00589
Dong, F. B., Sen, Q. C., Xiao, D. L., and Qi, Y. H. (2024). Advances in the study of auxin early response genes: Aux/IAA, GH3, and SAUR. Crop J. 12, 964–978. doi: 10.1016/j.cj.2024.06.011
Elaziem, A. and Ahmed, T. M. (2020). In vitro propagation for conservation of the rare date palm (Phoenix dactylifera L.) ‘Amri’ using immature inflorescence. In Vitro Cell. Dev. Biology-Plant 58, 1048–1056. doi: 10.1007/s11627-022-10296-3
Hasanuzzaman, M., Bhuyan, M. H. M. B., Parvin, K., Bhuiyan, T. F., Anee, T. I., Nahar, K., et al. (2020). Regulation of ROS metabolism in plants under environmental stress: a review of recent experimental evidence. Int. J. Mol. Sci. 21, 8695. doi: 10.3390/ijms21228695
Kim, J. J., Kim, Y. S., Park, S. I., Mok, J. E., Kim, Y. H., Park, H. M., et al. (2017). Cytosolic monodehydroascorbate reductase gene affects stress adaptation and grain yield under paddy field conditions in Oryza sativa L. japonica. Mol. Breeding: New Strategies Plant Improvement 37, 118. doi: 10.1007/s11032-017-0720-y
Kong, W., Li, Y., Zhang, M., Jin, F., and Li, J. (2015). A Novel Arabidopsis microRNA promotes IAA biosynthesis via the indole-3-acetaldoxime pathway by suppressing superroot1. Plant Cell Physiol. 56, 715–726. doi: 10.1093/pcp/pcu216
Kumar, D. and Chattopadhyay, S. (2018). Glutathione modulates the expression of heat shock proteins via the transcription factors BZIP10 and MYB21 in Arabidopsis. J. Exp. Bot. 69, 3729–3743. doi: 10.1093/jxb/ery166
Li, J., Guo, X., Zhang, S., Zhang, Y., Chen, L., Zheng, W., et al. (2022). Effects of light quality on growth, nutritional characteristics, and antioxidant properties of winter wheat seedlings (Triticum aestivum L.). Front. Plant Sci. 13, 978468. doi: 10.3389/fpls.2022.978468
Liang, X., Li, Y., Wang, L., Yi, B., Fu, T., Ma, C., et al. (2024). Knockout of stigmatic ascorbate peroxidase 1 (APX1) delays pollen rehydration and germination by mediating ROS homeostasis in Brassica napus L. Plant J. 119, 1258–1271. doi: 10.1111/tpj.16846
Méndez-López, E., Donaire, L., Gosálvez, B., Díaz-Vivancos, P., Sánchez-Pina, M. A., Tilsner, J., et al. (2023). Tomato SlGSTU38 interacts with the PepMV coat protein and promotes viral infection. New Phytol. 238, 332–348. doi: 10.1111/nph.18728
Miguel, A. U. C., Pérez-Hernández, C., Galaz-Ávalos, R. M., Brito-Argaez, L., Aguilar-Hernández, V., Loyola-Vargas, V. M., et al. (2020). YUCCA-mediated biosynthesis of the auxin IAA is required during the somatic embryogenic induction process in Coffea canephora. Int. J. Mol. Sci. 21, 4751. doi: 10.3390/ijms21134751
Müller-Schüssele, S. J., Wang, R., Gütle, D. D., Romer, J., Rodriguez-Franco, M., Scholz, M., et al. (2020). Chloroplasts require glutathione reductase to balance reactive oxygen species and maintain efficient photosynthesis[J. Plant J. 103, 1140–1154. doi: 10.1111/tpj.14791
Okumura, N., Masamoto, K., and Wada, H. (1997). The GSHB gene in the cyanobacterium Synechococcus sp. PCC 7942 encodes a functional glutathione synthetase. Microbiol. (Reading) 143, 2883–2890. doi: 10.1099/00221287-143-9-2883
Pavlů, J., Novák, J., Koukalová, V., Luklová, M., Brzobohatý, B., Černý, M., et al. (2018). Cytokinin at the crossroads of abiotic stress signalling pathways. Int. J. Mol. Sci. 19, 2450. doi: 10.3390/ijms19082450
Petrova, A. and Smith, C. M. (2015). Application of brown planthopper salivary gland extract to rice plants induces systemic host mRNA patterns associated with nutrient remobilization. PloS One 10, 0141769. doi: 10.1371/journal.pone.0141769
Ranjana, S., Soumi, G., Pinki, K., Salman, S., Chandan, R., Dibyendu, S., et al. (2021). Glutathione regulates subcellular iron homeostasis via transcriptional activation of iron responsive genes in Arabidopsis. Bio Preprint Server 5, 443283. doi: 10.1111/pce.14331
Rouhier, N., Lemaire, S. D., and Jacquot, J. P. (2008). The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 59, 143–166. doi: 10.1146/annurev.arplant.59.032607.092811
Skogerboe, D. and Jenderek, M. (2022). Improvement in micropropagation and cryopreservation of Musa. Acta Hortic. 9, 393572. doi: 10.17660/ActaHortic.2023.1359.27
Sung, H. C. and Paek, N. C. (2016). Regulatory role of the OsWOX3A transcription factor in rice root development[J. Plant Signaling Behav. 11, e1184807. doi: 10.1080/15592324.2016.1184807
Terai, Y., Ueno, H., Ogawa, T., Sawa, Y., Miyagi, A., Kawai-Yamada, M., et al. (2020). Dehydroascorbate reductases and glutathione set a threshold for high-light-induced ascorbate accumulation. Plant Physiol. 183, 112–122. doi: 10.1104/pp.19.01556
To, J. P. and Kieber, J. J. (2008). Cytokinin signaling: two-components and more[J. Trends Plant Sci. 13, 85–92. doi: 10.1016/j.tplants.2007.11.005
Tong, X., Riek, J., Guo, H., Jarvis, D., Ma, L., Long, C., et al. (2015). Impact of traditional culture on Camellia reticulata in Yunnan, China. J. Ethnobiology Ethnomedicine 11, 74. doi: 10.1186/s13002-015-0059-6
Wang, J., Gong, Y., Li, M., Bai, Y., and Wu, T. (2024). A CsWRKY48 gene from tea plants intercropped with Chinese chestnut plays an important role in resistance to biotic and abiotic stresses. Int. J. Mol. Sci. 25, 13526. doi: 10.3390/ijms252413526
Wang, Z., Xiao, Y., Chen, W., et al. (2010). Increased vitamin C content accompanied by an enhanced recycling pathway confers oxidative stress tolerance in Arabidopsis. J. Integr. Plant Biol. 52, 400–409. doi: 10.1111/j.1744-7909.2010.00921.x
Xu, X., Huang, B., Fang, X., Zhang, Q., Qi, T., Gong, M., et al. (2023). SlMYB99-mediated auxin and abscisic acid antagonistically regulate ascorbic acids biosynthesis in tomato. New Phytol. 239, 949–963. doi: 10.1111/nph.18988
Yan, H. F., Mao, P. S., and Xia, F. S. (2013). Research progress in plant antioxidant glutathione. Acta Agrestia Sin. 3, 28–35. doi: 10.11733/j.issn.1007-0435.2013.03.003
Yuan, L., Dang, J., Zhang, J., Wang, L., Zheng, H., Li, G., et al. (2024). A glutathione S-transferase regulates lignin biosynthesis and enhances salt tolerance in tomato. Plant Physiol. 196, 2989–3006. doi: 10.1093/plphys/kiae504
Zechmann, B., Koffler, B. E., and Russell, S. D. (2011). Glutathione synthesis is essential for pollen germination in vitro. BMC Plant Biol. 11, 1471–2229. doi: 10.1186/1471-2229-11-54
Keywords: Camellia reticulata, CrGST24, auxin, cytokinin, GSH, ASA, ROS
Citation: Cao Z, Wang J, Gong Y, Yin X, Yang C, Yang T and Wu T (2025) Glutathione S-transferase CrGST24 in the differentiation of adventitious buds from Camellia reticulata callus. Front. Plant Sci. 16:1641401. doi: 10.3389/fpls.2025.1641401
Received: 05 June 2025; Accepted: 24 July 2025;
Published: 15 August 2025.
Edited by:
Sergio J. Ochatt, INRA UMR1347 Agroécologie, FranceReviewed by:
Sang-Yun Han, Sungkyunkwan University, Republic of KoreaSara Yasemin, Siirt University, Türkiye
Copyright © 2025 Cao, Wang, Gong, Yin, Yang, Yang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tian Wu, d3V0aWFucG90YXRvQHN3ZnUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Zihang Cao†
Zihang Cao† Tian Wu
Tian Wu