- 1Key Laboratory of National Forestry and Grassland Administration on Plant Conservation and Utilization in Southern China, South China Botanical Garden, Chinese Academy of Sciences, Guangzhou, China
- 2South China National Botanical Garden, Guangzhou, China
- 3University of Chinese Academy of Sciences, Beijing, China
- 4College of Forestry and Landscape Architecture, South China Agricultural University, Guangzhou, China
- 5Guangxi Key Laboratory of Plant Conservation and Restoration Ecology in Karst Terrain, Guangxi Institute of Botany, CAS, Guilin, China
- 6National Gesneriaceae Germplasm Resources Bank of GXIB, Gesneriad Committee of China Wild Plant Conservation Association, Gesneriad Conservation Centre of China (GCCC), Guilin Botanical Garden, CAS, Guilin, China
The concept and definition of species are fundamental in taxonomy and evolutionary biology. Traditionally, new species have been described primarily based on the morphological species concept (MSC). However, environmental factors may drive morphological convergence among distantly related species. In addition, widespread hybridization, gene flow, and incomplete lineage sorting (ILS) among closely related taxa can further obscure phylogenetic relationships and hinder accurate species delimitation. In the current study of the genus Oreocharis Benth. (Gesneriaceae: Didymocarpoideae), we built upon a broad phylogenomic framework established in previous work. We incorporated transcriptome data of six specimens from Sichuan, China, and conducted detailed morphological comparisons and phylogenomic analyses. The integrative results reveal that these specimens represent a previously unrecognized lineage, which we formally describe here as Oreocharis yanbianensis Z. Xie & H. H. Kong, sp. nov. Our study demonstrates the importance of combining morphological and genomic data to improve the accuracy and robustness of species discovery and classification, especially in morphologically variable plant groups.
Introduction
Understanding the concept of species is crucial for the study of biodiversity, species classification, and unraveling the complexities of life on Earth (Michener et al., 2001). Over the years, scientists and taxonomists have proposed various species concepts to define and distinguish different groups of organisms (Zhou and Yang, 2011; Hong, 2016). Among the most widely recognized concepts are the Biological Species Concept (BSC, Darwin, 1859; Sokal and Crovello, 1970; Futuyma, 2013; Futuyma and Kirkpatrick, 2017), morphological species concept (MSC, Simpson, 1943), Ecological Species Concept (ESC, Valen, 1976), Phylogenetic Species Concept (PSC, Cracraft, 1983), Cohesion Species Concept (CSC, Templeton, 1989), Recognition Species Concept (RSC, Paterson, 1985; 1992), Evolutionary Species Concept (EvSC, Simpson, 1951; 1961; Wiley, 1992), and General Lineage Species Concept (GLSC, de Queiroz, 1999). Each of these concepts offers unique criteria for defining species, reflecting the diversity of life forms on our planet (Hong, 2016).
The morphological species concept (MSC), which is based on observable morphological characters, has been a favored species concept among botanists for describing new plant species (Mishler, 1985). The morphological characteristics of plants serve as valuable indicators for species identification (Wesselink and Kuiper, 2008). However, there are limitations to relying solely on morphological features in species delimitation (Wiens, 2007). Environmental influences, such as habitat conditions and selective pressures, can cause convergent evolution, leading to similar morphological traits in species that are not closely related (Losos, 2011; Stern, 2013; Hodge et al., 2025). This convergence can result in misinterpretations of species relationships, as species with similar morphological characters might be mistakenly grouped together (Bogarín et al., 2019; Karbstein et al., 2024). A prominent example of such challenges is found within the Gesneriaceae, which includes the genus Oreocharis, due to its high morphological variability and frequent misidentification.
Oreocharis Benth. is a large genus in the tribe Trichosporeae (Gesneriaceae: Didymocarpoideae) with 160 species and 15 varieties (according to the GCR, Gesneriaceae Resource Centre, https://padme.rbge.org.uk/grc/, last accessed on 15, May 2025), which is primarily found in the mountainous regions of southwest and southern China (Möller et al., 2011; Kong et al., 2022; Xie et al., 2024a). Additionally, over a dozen species have been identified in Vietnam, Myanmar, Thailand, Japan, and India (Xie et al., 2024a). The morphological complexity and diversity of Oreocharis exhibit significant variations, leading to notable disparities in classification perspectives among scholars (Wang et al., 1990; 1998; Wang et al., 2010; Möller et al., 2011; 2016; 2025; Chen et al., 2014a, 2020). The taxonomic revision of Oreocharis involved the inclusion of species from several previously segregated genera, such as Bournea Oliver (Hooker, 1893), Perantha Craib (1918), Tremacron Craib (1918), Ancylostemon Craib (1920), Isometrum Craib (1920), Dasydesmus Craib (1920), Opithandra Burtt (1956), Thamnocharis Wang (1981), Dayaoshania Wang (1983a), Schistolobos Wang (1983b), Deinocheilos Wang (1986), and Paraisometrum W.T.Wang (Weitzman et al., 1998), and the acaulescent and rosette-forming species of Briggsia Craib (1920). Additionally, some species formerly described in the genus Briggsia or Ancylostemon were incorporated into the redefined Oreocharis (Chen et al., 2012; 2014b; Möller et al., 2011; 2014). Since Möller et al. (2011) newly delineated the genus Oreocharis, a total of 61 new taxa (including 60 species and one variety) have been published (last accessed on 15, May 2025, Table 1). However, currently, many descriptions of new species within the genus Oreocharis are primarily based on morphological characteristics (Table 1). Even when molecular data is used, it is often limited to a few DNA segments, such as ITS, trnL-F, and other regions (Chen et al., 2020b; Guo et al., 2018; Hu et al., 2023; Ling et al., 2020; 2022; Lv et al., 2021).
Advancements in sequencing technologies have revolutionized the field of taxonomy, facilitating access to genome-scale molecular data (Hong et al., 2023). In the case of Oreocharis, transcriptome sequencing has been performed for 110 species (Kong et al., 2022). However, despite the potential of this molecular data, it has not been fully utilized in the process of describing new species within the genus. Taxonomists face challenges in accessing and analyzing genomic-level information, especially for those with limited bioinformatics skills. Extracting orthologous single-copy genes from transcriptome assemblies typically requires command-line tools and familiarity with a Linux environment, which poses challenges for taxonomists lacking bioinformatics training. To address this, we provide a detailed and user-friendly guide (Obtaining_single-copy_data_guide.txt on Figshare, https://doi.org/10.6084/m9.figshare.29457638.v2) on obtaining single-copy data from transcriptomes. By making this procedure accessible, taxonomists will be better equipped to incorporate molecular information effectively into species descriptions, thereby enhancing our understanding of the diversity and evolution of Oreocharis.
In this study, we aim to demonstrate the importance of integrating both morphological and molecular data for accurately describing and classifying of new species within the genus Oreocharis through six newly collected samples (XieZ3609, XieZ3631, XieZ4055, XieZ4061, XieZ4065, XieZ4071) from Sichuan, Southwest China. By combining these two complementary sources of information, we aim to provide a more robust and comprehensive framework for future taxonomic studies in the Gesneriaceae family and beyond. Our findings will not only deepen our understanding of Oreocharis but also provide a model for incorporating molecular data into plant species descriptions, with broader implications for biodiversity and evolutionary studies. As technology continues to evolve, the synergy between morphological and molecular approaches will undoubtedly play a pivotal role in deepening our understanding of plant diversity and evolution.
Materials and methods
Morphological description and comparison
Morphological data were obtained from field collections of living intact individuals in their natural habitat. Detailed data on flowers, fruits, and vegetative structures, as well as information on habitat, GPS coordinates, and elevation, were recorded. Voucher specimens and living materials were then collected for subsequent study. Morphological measurements were conducted using a vernier caliper (Yantai Greenery Tools Co., Ltd., Yantai, China) to document diagnostic characters relevant to species delimitation. This was done to meticulously document specific characters deemed pertinent for the differentiation of species. Concurrently, digital photographs—especially close-up images of floral structures — were taken to capture the key morphological traits of the new species in detail (Xie et al., 2024a). Terminology used to describe morphological features followed Harris and Harris (1994) and the Flora of China (Wang et al., 1990; 1998).
After field investigations, specimen examinations at herbaria (e.g. IBSC, KUN, PE, etc.) were also essential for obtaining additional morphological and distribution data when available specimens existed. It should be noted that species formerly assigned to the genera Bournea, Perantha, Tremacron, Ancylostemon, Isometrum, Dasydesmus, Opithandra, Thamnocharis, Dayaoshania, Schistolobos, Deinocheilos, Paraisometrum, and the acaulescent, rosette-forming species of Briggsia are of special taxonomic relevance. Although these taxa were transferred to Oreocharis by Möller et al. (2011; 2014), divergent classification systems used by different herbaria (e.g., the Hutchinson system at IBSC, KUN, and IBK; the Engler system at PE and SZ) necessitate searching for specimens under their former generic names. Voucher specimens (including type materials) were deposited at herbarium of South China Botanical Garden (IBSC), Guangzhou, China. Additional examined specimens of Oreocharis were obtained from the herbaria CDBI, IBSC, KUN, PE and SM. As a supplement, digital specimens were examined through the JSTOR Global Plants web portal (https://plants.jstor.org/) and the National Plant Specimen Resource Center (www.cvh.ac.cn/index.php).
Taxon sampling for molecular analyses
A total of 135 samples were used in this study, representing six newly collected samples, 106 previously described Oreocharis species (Supplementary Table 1 on Figshare, https://doi.org/10.6084/m9.figshare.28829327.v6), and six species from other genera of Gesneriaceae as outgroups: Aeschynanthus buxifolius Hemsl., A. moningeriae (Merr.) Chun, Anna mollifolia (W.T. Wang) W.T. Wang et K.Y. Pan, Cyrtandra hawaiensis C.B.Clarke, Didymocarpus cortusifolius (Hance) W.T. Wang and Petrocodon dealbatus Hance, selected based on previous phylogenetic analyses (Möller et al., 2011; Kong et al., 2022). During the field investigation, we meticulously collected one or more intact living plants. To protect the roots during transport, the lower portions of each plant were wrapped in moist mosses. If moss was unavailable, damp paper towels were used instead (Kong et al., 2022; Xie et al., 2024a). Prior to packing, photographs were taken and basic morphological observations and measurements were conducted. Subsequently, these living plants were sent to the greenhouse at the South China Botanical Garden, Chinese Academy of Sciences for cultivation and recovery. After recovery, fresh young leaves were harvested, cleaned of surface impurities, flash-frozen in liquid nitrogen with 50 mL EP tubes, and stored at -80°C (Kong et al., 2022; Xie et al., 2024a). All samples followed this procedure prior to transcriptome sequencing. Collection information is listed in Supplementary Table 1.
RNA extractions and illumina sequencing
Frozen leaf tissues were sent to Novogene Corporation (Tianjin, China) for RNA extraction and transcriptome sequencing. Total RNA was extracted from each individual using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA), with on-column DNase I (RNase-free, TaKaRa, Dalian, China) treatment to eliminate genomic DNA contamination. RNA integrity and concentration were assessed using agarose gel electrophoresis and a 2100 Bioanalyzer (Agilent Technologies). Purified RNA was used for cDNA library construction, followed by 150 bp paired-end sequencing on an Illumina NovaSeq 6000 platform (Kong et al., 2022). All transcriptome data are available at NCBI (Kong et al., 2022; Xie et al., 2024a), and the newly generated raw reads are publicly available under NCBI BioProject accession number PRJNA1032259, with BioSample accession numbers SAMN47951770 to SAMN47951775 (https://www.ncbi.nlm.nih.gov/sra/PRJNA1032259).
Reads filtering, 574 orthologs selection and 353 genes recognition
Quality control on the raw reads was performed to ensure the dependability of downstream analyses. In particular, fastp (Chen, 2023) was used to eliminate low-quality reads with parameter -g -q 5 -u 50 -n 15 -l 150 -overlap_diff_limit 1 -overlap_diff_percent_limit 10, and to obtain clean reads (Xie et al., 2024a). Orthologs identification was performed using GeneMiner (Xie et al., 2024b), based on a previously published dataset (https://doi.org/10.6084/m9.figshare.26927632.v1) containing 574 orthologous single-copy genes from 106 Oreocharis species, resulting in a new dataset (Dataset_1 on Figshare, https://doi.org/10.6084/m9.figshare.28829264.v2). A second dataset consisting 353 genes (Dataset_2 on Figshare, https://doi.org/10.6084/m9.figshare.28829282.v2) was generated using Easy353 with parameter -k1 31 -k2 41 -t1 5 -t2 5 -reference_number 100, based on the reference “353_ref_Gesneriaceae” downloaded using build_database.py (Zhang et al., 2022).
Phylogenetic analysis
A total of 135 individuals (including outgroups) were included in the molecular phylogenetic analysis, representing 106 described Oreocharis species, six newly collected samples and six outgroups (Supplementary Table 1). Gene alignments were performed using MAFFT 7.525 (Nakamura et al., 2018) with default parameters, and ambiguous aligned regions were trimmed using Trimal 1.4.rev15 with the automated1 setting (Capella-Gutiérrez et al., 2009). Maximum likelihood (ML) trees were inferred for each gene separately using IQ-TREE 2.0.4 (Minh et al., 2020). The resulting gene trees were then used as input for species tree inference in ASTRAL-III 5.7.8 (Zhang et al., 2018), with node support values calculated using the local posterior probability (LPP) method (Sayyari and Mirarab, 2016), based on the 574 single-copy genes dataset and 353 genes dataset, respectively.
Results
Morphology
A detailed morphological comparison between the six newly collected samples and previously described Oreocharis species revealed several shared traits such as thickly chartaceous leaf blades and yellow flowers (Figure 1), and showed remarkable resemblance to several species, including O. convexa (Craib) Mich. Möller & A. Weber, O. trichantha (B. L. Burtt & R. A. Davidson) Mich. Möller & A. Weber, O. bullata (W. T. Wang & K. Y. Pan) Mich. Möller & A. Weber, O. saxatilis (Hemsl.) Mich. Möller & A. Weber and O. concava (Craib) Mich. Möller & A. Weber (Figure 1; Table 2). Morphologically, the species most similar to XieZ4071 is O.trichantha (Figures 1-3). However, it exhibits a throat-constricted, limp, distinctly two-lipped corolla with unequal lobes; the adaxial lip is obtuse, the abaxial lip rounded, and the pistil is shorter, 3.0 ± 0.4 mm. In contrast, O. trichantha possesses throat not constricted corolla, corolla slightly 2-lipped, all lobes subequal, obovate, and pistil longer, > 10 mm (Figures 1-3 & Table 2). Further examination of herbarium specimens revealed that YLH710 was also collected from the same location, and its morphological traits were consistent with those of Xie4071.
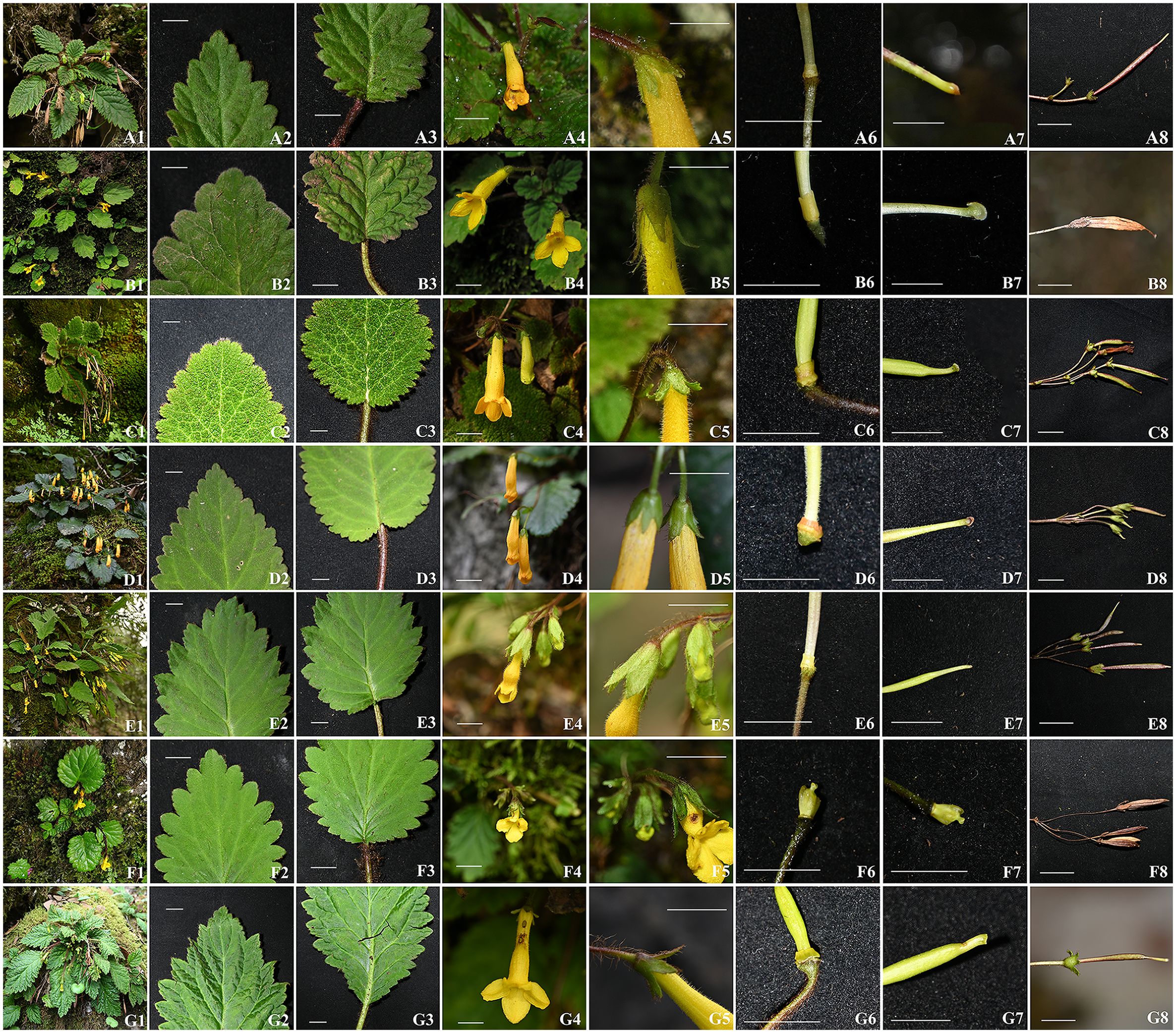
Figure 1. Morphological comparison among Oreocharis convexa (A1–A8), O. trichantha (B1–B8), O. bullata (C1–C8), O. saxatilis (D1–D8), O. concava (E1–E8), O. yanbianensis (F1–F8), and O. gamosepala (G1–G8). (A1, B1, C1, D1, E1, F1, G1) Habit; (A2, B2, C2, D2, E2, F2, G2) Leaf blade apex; (A3, B3, C3, D3, E3, F3, G3) Leaf blade base; (A4, B4, C4, D4, E4, F4, G4) Flowers; (A5, B5, C5, D5, E5, F5, G5) Calyx; (A6, B6, C6, D6, E6, F6, G6) Disc; (A7, B7, C7, D7, E7, F7, G7) Stigma; (A8, B8, C8, D8, E8, F8, G8) Fruits. All scale bars represent 1 cm. Photographs: B4–B7 by Lihua Yang, others by Zhi Xie.
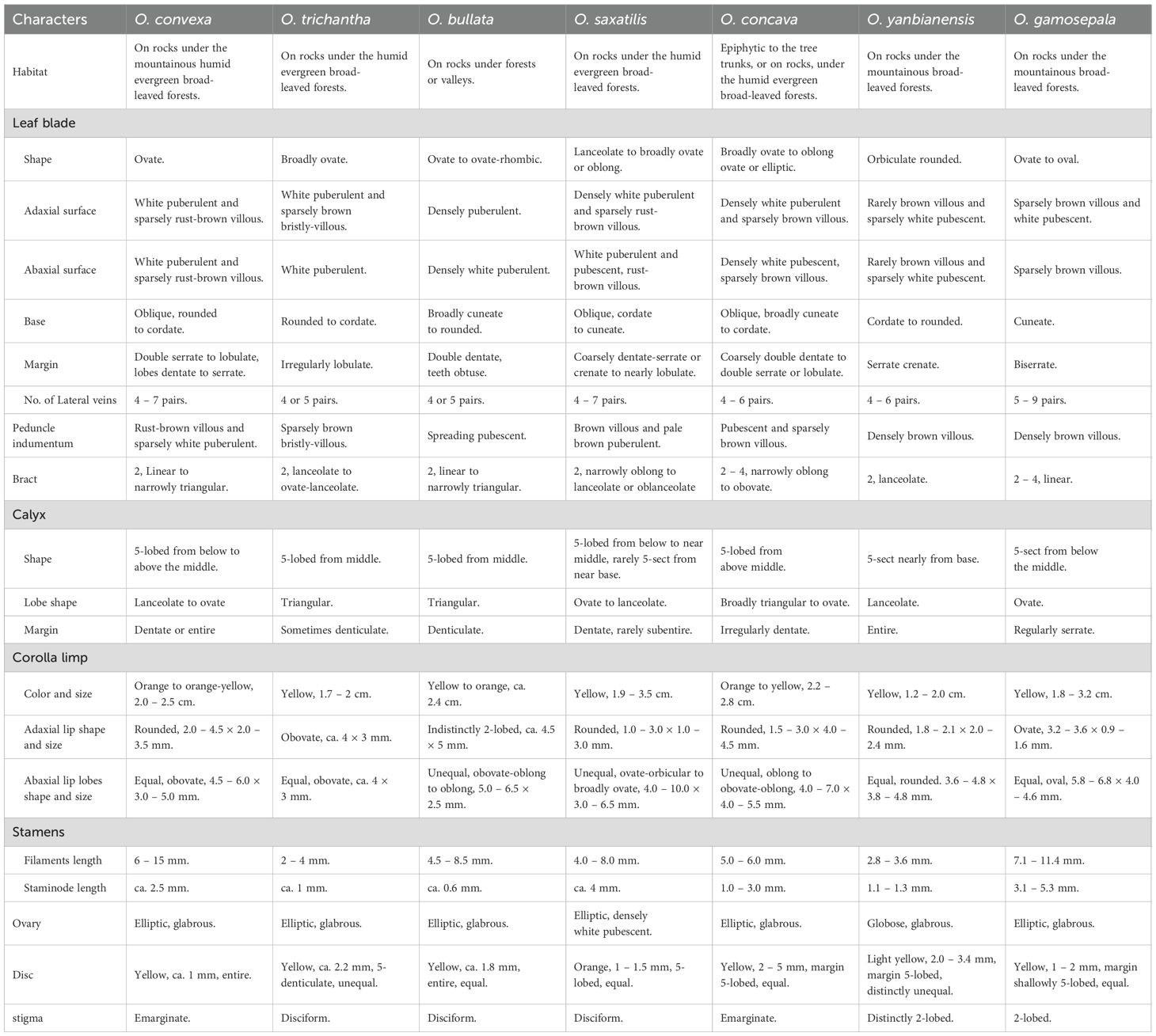
Table 2. Morphological comparison among Oreocharis convexa, O. trichantha, O. bullata, O. saxatilis, O. concava, O. yanbianensis and O. gamosepala.

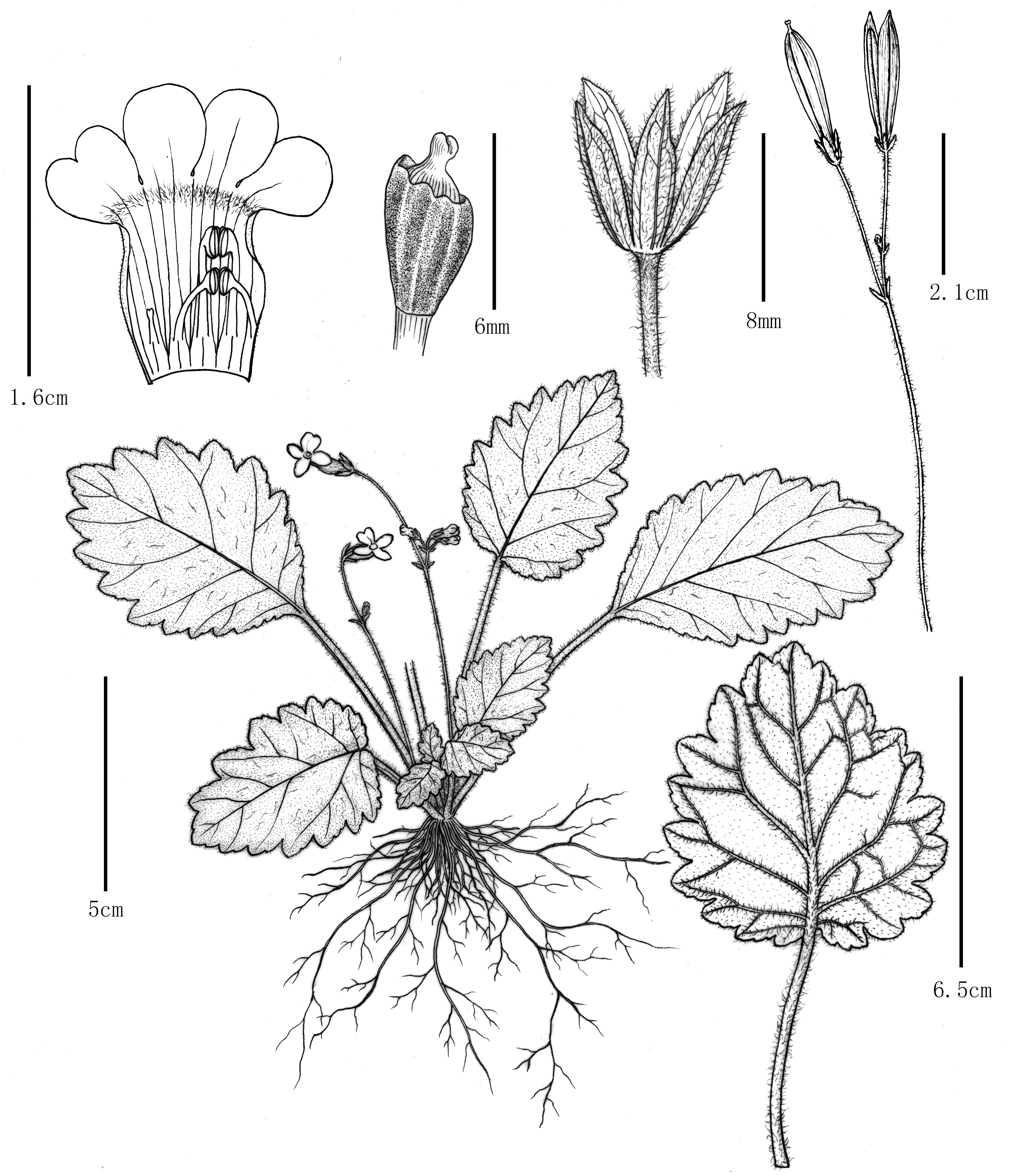
Figure 3. Oreocharis yanbianesis Z. Xie & H. H. Kong, sp. nov. (A) Habitat, the subtropical evergreen broad-leaved forests with Schima argentea E. Pritz. and Quercus spp.; (B) Population, rocks on limestone hillsides; (C) Habit; (D) individual without flower; (E) individual flowers; (F) Mature leaves: adaxially blade (left) and abaxially blade (right); (G) Cymes; (H) Bracts and pedicel; (I) Front view of flower; (J) Right side view of the flowers; (K) Calyx; (L) Opening corolla, showing stamens and staminode; (M) Disc, ovary and stigma; (N) Dehiscent capsule. Photos by Zhi Xie.
XieZ3631 resembles O. gamosepala (K. Y. Pan) Mich. Möller & A. Weber in floral morphology. Only minor differences in leaf size were observed between XieZ3631 and O. gamosepala (Figure 1 & Table 2). Comprehensive observations of wild populations have demonstrated that leaf size varies significantly both among populations and between individuals within the same population (Figure 4). We also examined the holotype of O. gamosepala (Hanyuan expedition, 0427, 11 June 1978, SM 717900003)! and paratypes (Yanyuan expedition, 368, 20 June 1978, SM 717900003! Sichuan Economic Plant expedition of Liangshan, 3508, 28 June 1959, CDBI 0130063! CDBI 0130064! KUN 0206255! PE 00030696! PE 00030697! SM 717900020! SM 717900021)!. Further field investigations and specimen examinations confirmed that multiple collections — including Qinghai-Tibetan expedition 12613 (Qinghai-Tibetan expedition, Yanyuan, 30 July 1983, PE 01173170, PE 01173171, KUN 0206235), Qinghai-Tibetan expedition, 12116 (Qinghai-Tibetan expedition, Yanyuan, 20 July 1983, PE 01173173, PE 01173172, KUN 0206237), FCY2013016 (C. Y. Feng, Yanyuan, 13 October 2013, PE 02053062), YLH712 (H. H. Kong & L. H. Yang, Yanyuan, 18 August 2018, IBSC0882883, IBSC0882884), SCYY01 (H. H. Kong, L. H. Yang & B. F. Zhou, Yanyuan, 7 September 2017, IBSC), XieZ3609 (Z. Xie & M. Zhang, Yuexi, 27 June 2023, IBSC). XieZ3641 (Z. Xie & M. Zhang, Yanyuan, 1 July 2023, IBSC), XieZ4055 (Z. Xie, Yanyuan, 25 July 2024, IBSC), XieZ4061 (Z. Xie, Muli, 25 July 2024, IBSC) and XieZ4065 (Z. Xie, Yanyuan, 26 July 2024, IBSC) are all conspecific with O. gamosepala. These specimens exhibit consistent morphological features, with only slight variation across individuals (Figure 4).

Figure 4. Morphological differences of O. gamosepala individuals between the same population (A1–A4) and different populations (B1–B4). (A1–A4) were taken from the same locality of XieZ3631, Yanyuan, Sichuan, China by Zhi Xie; (B1) was taken from Zhaotong, Yunnan, China by Youpai Zeng; (B2)was taken from the same locality of XieZ3609, Yuexi, Sichuan, China by Zhi Xie; (B3) was taken from the same locality of YLH715, Yanyuan, Sichuan, China by Lihua Yang; (B4) was taken from the same locality of XieZ4061, Muli, Sichuan, China by Zhi Xie.
YLH715 (H. H. Kong & L. H. Yang, Yanyuan, 18 August 2018, IBSC 0882896, IBSC 0882822), was previously identified as O. concava (Kong et al., 2022). However, it exhibits a cuneate leaf base, margin crenate to dentate or serrate; calyx 5-lobed, lobes equal, ovate, margin regularly dentate. These characteristics showed its similarity to O. gamosepala. In contrast, O. concava typically has oblique, broadly cuneate to cordate leaf base, margin coarsely double dentate to double serrate or lobulate; calyx 5-lobed, segments unequal, broadly triangular to ovate, margin irregularly dentate (Figure 4B3). These differences in leaf and calyx morphology cast doubt on the accuracy of the previous species identification. As flowering specimens and living plants were not observed, this identification remains provisional and should be treated with caution.
Moreover, in our prior study, YLH859 was misidentified and temporarily labeled as O. sp5. Upon revisiting the collection locality for further population observation, the sample was confirmed to match the diagnostic features O. concava. This species is characterized by broadly ovate to elliptic leaf blades, oblique to cordate leaf base; calyx 5-lobed from above middle; lobes unequal, broadly triangular to ovate; adaxial corolla lip rounded and abaxial corolla lip lobes oblong to obovate-oblong, and pistil glabrous (Figure 1; Table 2).
Furthermore, the specimens of lrh006 (R. H. Liang, Yuexi, 12 September 2006, PE 02053535, PE 02053533, PE 02053534, PE 02053521) were originally identified as O. gamosepala. However, upon examination, we found that they differ markedly from O. gamosepala in several morphological traits, including ovate leaf base, petiole sparsely white pubescent, corolla tubular, yellow to orange, adaxial lip extremely short, abaxial lip lobes ovate. These discrepancies indicate that the specimens were likely misidentified (Table 2).
Morphological comparisons led to the reidentification of XieZ3609, XieZ3631, XieZ4055, XieZ4061, and XieZ4065 as O. gamosepala, and YLH859 as O. concava. In addition, XieZ4071 and YLH710 are proposed to represent a new species, Oreocharis yanbianensis Z. Xie & H. H. Kong, sp. nov. (Figures 2, 3). All taxonomic conclusions above are based solely on morphological evidence. However, due to the widespread hybridization, gene flow, and incomplete lineage sorting (ILS) in Oreocharis, relying solely on a limited set of morphological traits may compromise identification accuracy. Therefore, integrating molecular evidence is essential to clarify the systematic position and improve the robustness of taxonomic conclusions.
Molecular analyses
A total of 114 taxa (including outgroups and two newly collected taxa) were included in the molecular phylogenetic analysis (Supplementary Table 1). Dataset_1 comprises 574 orthologous genes, which including 23.8% missing sites, and Dataset_2 consist of 353 low-copy nuclear genes, which including 0.62% missing sites.
Phylogenetic analyses based on Dataset_1 strongly supported the clustering of O. gamosepala, O. yanbianensis sp. nov., O. concava, O. saxatilis, O. bullata, O. trichantha and O. convexa (LPP = 1; Figure 5), all of which share common morphological traits such as thickly chartaceous leaf blades and tubular yellow flowers (Figure 1). Within this clade, two samples — XieZ4071 and Y710 (YLH710 labeled on specimens) — were recovered as a maximum-supported monophyletic group (LPP = 1; Figure 5); both were collected from the same locality in Yanbian county and have been identified as O. yanbianensis sp. nov. based on morphological evidence. In addition, samples XieZ3609, XieZ4065, XieZ4061, XieZ4055, and XieZ3631, which were identified as O. gamosepala, clustered together with YLH715, also with strongest support (LPP = 1; Figure 5); support value for internal nodes within this subclade were also high (LPP ≥ 0.99; Figure 5). Furthermore, O. saxatilis, O. bullata, O. trichantha, and O. convexa formed another well-supported group (LPP = 0.99; Figure 5), within which O. trichantha and O. convexa appeared as sister species, albeit with relatively low support (LPP = 0.55; Figure 5).
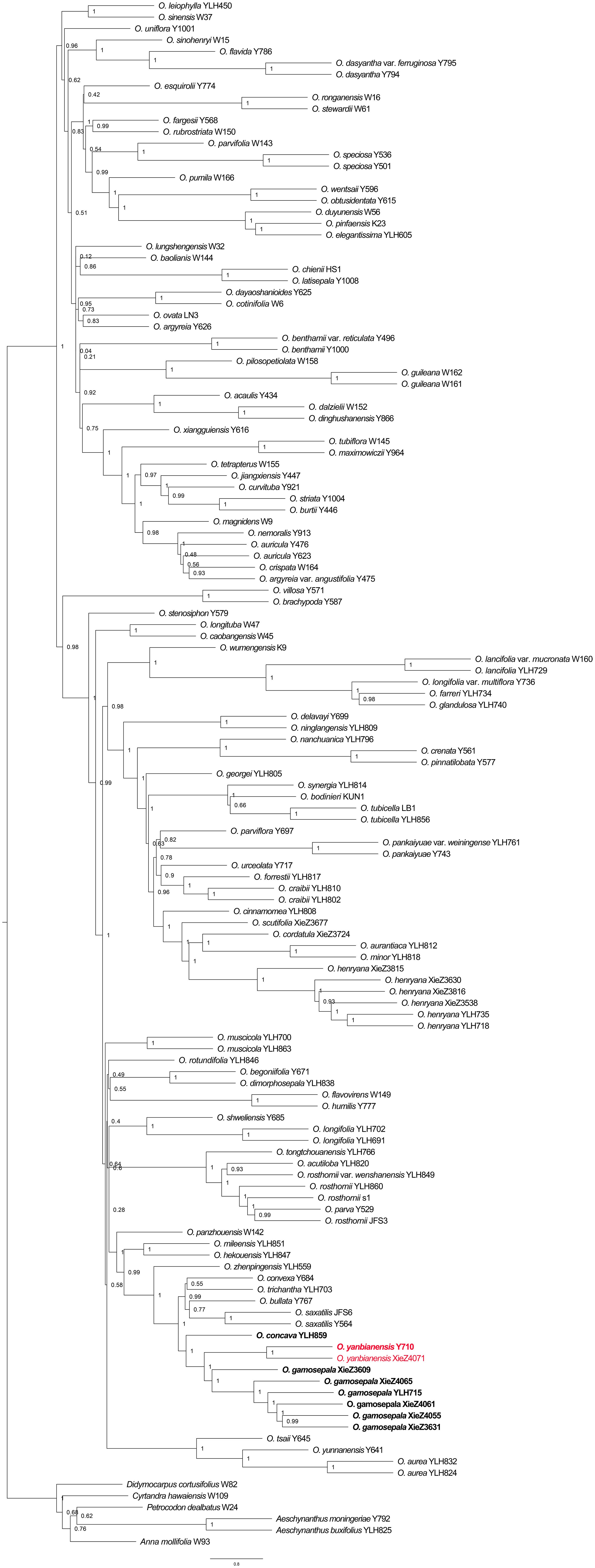
Figure 5. Phylogeny of Oreocharis based on the transcriptome dataset of 574 single-copy genes. The new species is shown in red, and the samples with species re-identification are marked in bold. The species tree was reconstructed by ASTRAL-III 5.7.8. Support for branches was evaluated with local posterior probability (LPP).
The phylogenetic analyses based on Dataset_2 also strongly supported the clustering of O. gamosepala, O. yanbianensis sp. nov., O. concava, O. saxatilis, O. bullata, O. trichantha, and O. convexa (LPP = 1; Figure 6). However, a notable difference is that several internal nodes within the O. gamosepala clade, which includes six samples exhibited lower support values (LPP ≤ 0.7; Figure 6). Additonally, O. convexa and O. concava clustered together with strong support (LPP = 1; Figure 6), with O. trichantha forming the basal lineage of this subclade. In previous studies, O. zhenpingensis J. M. Li, T. Wang & Y. G. Zhang was consistently resolved as the earliest-diverging lineage of a clade comprising several yellow-flowered species, e.g., O. concava, O. saxatilis, O. bullata, O. trichantha and O. convexa (Kong et al., 2022; Yang et al., 2022; Xie et al., 2024a). This placement was supported by both morphological traits and geographical distribution patterns (Liet al., 2017). However, in the species tree inferred from the 353 gene trees, O. zhenpingensis clustered with a strongly supported clade (LPP = 1, Figure 6) consisting of several purple-flowered species (O. lancifolia var. mucronata (K. Y. Pan) Mich. Möller & A. Weber, O. lancifolia (Franch.) Mich. Möller & A. Weber, O. longifolia var. multiflora (S. Y. Chen ex K. Y. Pan) Mich. Möller & A. Weber, O. glandulosa (Batalin) Mich. Möller & A. Weber, and O. farreri (Craib) Mich. Möller & A. Weber). This result is incongruent with previous morphological and biogeographic expectations and warrants further investigation.
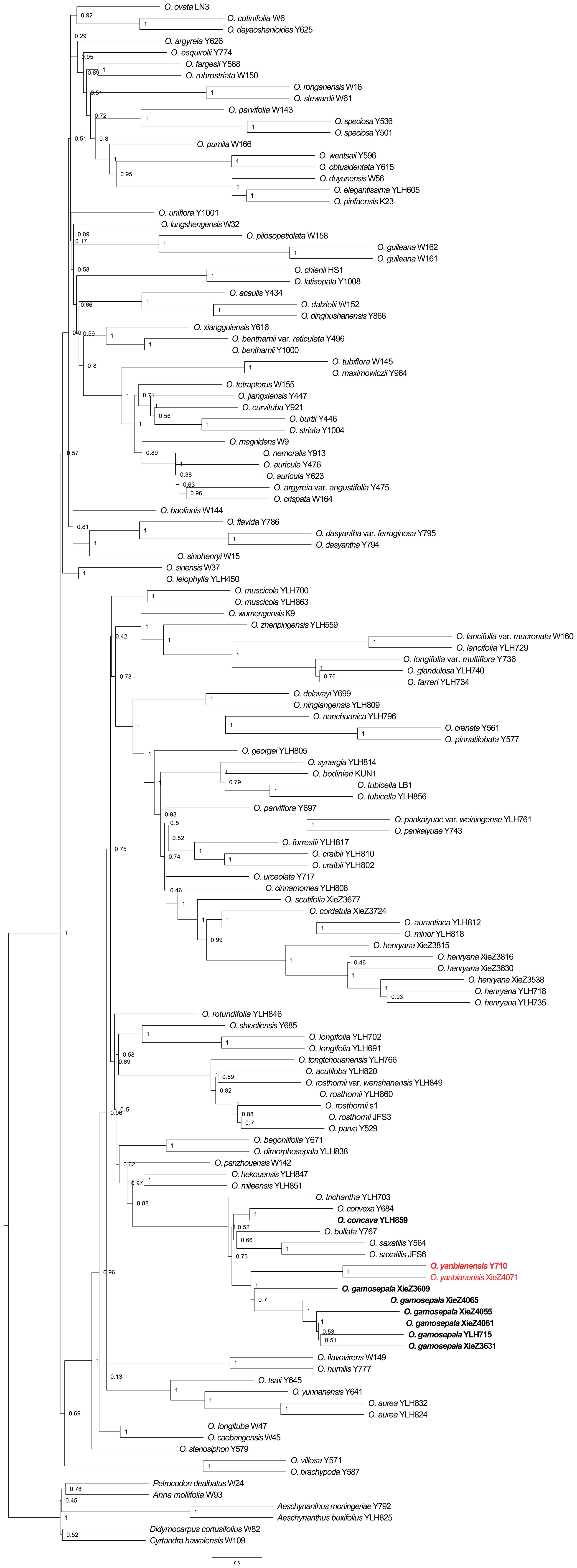
Figure 6. Phylogeny of Oreocharis based on the transcriptome dataset of 353 low-copy nuclear genes. The new species is shown in red, and the samples with species re-identification are marked in bold. The species tree was reconstructed by ASTRAL-III 5.7.8. Support for branches was evaluated with local posterior probability (LPP).
By integrating morphological and molecular evidence, we identified XieZ3609, XieZ4065, XieZ4061, XieZ4055, XieZ3631 and YLH715 as O. gamosepala (Figures 1, 4–6; Table 2), and recognized XieZ4071 and Y710 as representing a new species, O. yanbianensis Z. Xie & H. H. Kong sp. nov., which is illustrated and described below (Figures 1–3; Table 2). O. yanbianensis is morphologically most similar to O. trichantha (Figure 1; Table 2), but phylogenetically shows a closer relationship with O. gamosepala (Figures 5, 6).
Discussion
Frequent and substantial taxonomic revisions of Oreocharis have resulted in the shifting systematic positions of numerous species within this genus, accompanied by continuous changes in their scientific names (Möller et al., 2011; 2025; Fu et al., 2019c; Chen et al., 2020b). This adds a considerable layer of complexity to the studies involving this genus. Additionally, Oreocharis encompasses a wide range of plants exhibiting complex and variable morphological traits (Möller et al., 2011; Kong et al., 2022), which are often heavily influenced by environmental conditions and pollination pressures (Möller et al., 2011). Many species demonstrate significant variation in leaf shape, floral structure, and coloration. Notably, even within a single population, inter-individual trait variation can be substantial. Furthermore, striking morphological trait differences are often observed across developmental stages of the same individual (Figure 4). In contrast, convergent evolution driven by similar habitats and selective pressure can lead to analogous morphological traits in distantly related species. O. yanbianensis and O. trichantha exemplify this pattern in our present study (Figures 1, 4, 6).
For these reasons, misidentification of species is common in Oreocharis. For example, Qinghai-Tibetan expedition, 12116 (collected on 20 July 1983 from Dalin township — now Kalaba village, Baiwu township, Yanyuan county, Sichuan, China), comprises three duplicate specimens. One flowering specimen preserved in the Herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN) was identified as Ancylostemon aureus (Franch.) Burtt by Kaiyu Pan in 1986 (KUN0206237)!, while two fruiting specimens housed at the Institute of Botany, CAS (PE) and were labeled as A. rhombifolius K. Y. Pan (PE01173172! PE01173173)!, though without specified identifier and date. Interestingly, when A. rhombifolius K. Y. Pan sp. nov. was published in 1988, those specimens were not mentioned in the protologue (Pan, 1988). In 2017, we recollected this taxon (YLH715) from the same locality and initially misidentified it as Oreocharis concava, while YLH859 from Yongping, Yunnan was treated as potentially new species and labeled O. sp5. Meanwhile, specimens of YLH710 from Yanbian, Sichuan, was also misidentified as O. concava and included in previous phylogenetic analyses (Kong et al., 2022).
Since Wang et al. (2010) first constructed the phylogeny of Oreocharis, various molecular markers — such as trnL-F and ITS — have been employed in subsequent studies (Guo et al., 2018; Ling et al., 2020; 2022; Lv et al., 2021; Hu et al., 2023). In describing new species, some researchers incorporated newly sequenced samples into an existing molecular data matrix (e.g., Yang et al., 2020). However, many resulting phylogenies suffer from weak node support and potentially misidentified samples, failing to reflect the true phylogenetic relationship of this genus. This may be attributed to widespread hybridization, gene flow, and incomplete lineage sorting (ILS) in the genus (Kong et al., 2022). Furthermore, undersampling may have left many cryptic species unsampled and unsequenced, especially between poorly supported clades of Oreocharis (Kong et al., 2022).
In this study, we reconstructed the phylogeny of Oreocharis using both a 353 genes dataset and a 574 orthologous single-copy genes dataset based on transcriptome data. These analyses corrected several previous misidentifications of species. The inclusion or exclusion of a few species even altered tree topology, with some previously monophyletic groups becoming paraphyletic or polyphyletic. Y710 and YLH715 were previously classified as O. concava, demonstrating a strongly supported clade (ML = 100, Kong et al., 2022). However, in the present study, the inclusion of several O. gamosepala samples and XieZ4071 in the phylogenetic analysis led to their segregation into two distinct clades on the species tree. Y710 and XieZ4071 were clustered together with strong support (LPP = 1), indicating potential misidentification in these samples. This hypothesis was corroborated by evidence from morphological studies. We conducted multiple field expeditions over the past three years to verify these results through population-level observation and morphological comparison, combined with molecular evidence, which ultimately confirmed that YLH715 is O. gamosepala, YLH859 is O. concava, and Y710 represents an undescribed species.
Given these findings, describing new species solely based on limited morphological differences in Oreocharis is inadvisable. We advocate for comprehensive population-level studies and the integration of molecular evidence when describing new species. Such molecular evidence should not be limited to one or a few loci, but ideally include genome-scale datasets with a larger amount of information, such as the 353 genes dataset or the orthologous single-copy genes dataset. In our current phylogenetic reconstruction of Oreocharis, the 574 genes dataset appears robust, despite a few remaining topological conflicts and unresolved relationships.
Similar taxonomic issues caused by convergent evolution have been reported in other plant groups, emphasizing the broader relevance of our findings (Taha et al., 2023). Morphologically similar traits can evolve independently in unrelated taxa subjected to similar environmental pressures, such as drought, pollinator preference, or substrate type (Losos, 2011). In such cases, traditional morphology-based taxonomy alone can be misleading. A classic example lies in the striking resemblance between members of Euphorbia (Euphorbiaceae) and the cactus family Cactaceae. Although these groups are phylogenetically distant and geographically disjunct (Euphorbia in the Old World, Cactaceae in the New World), both have independently evolved succulent, spiny, columnar forms in response to arid conditions (Dorsey et al., 2013; Taha et al., 2023). These adaptations, although ecologically convergent, are not indicative of shared ancestry and have historically led to misclassification, especially in the absence of floral or molecular data (Guerrero et al., 2019).
Even within Euphorbia, recent phylogenomic work revealed at least 14 independent origins of drought-adaptive growth forms (Horn et al., 2012), underscoring the repeated emergence of misleading morphological similarity. Such findings illustrate the necessity of integrating genomic data—rather than relying on a limited number of morphological traits or DNA markers—especially in groups with high morphological plasticity or rapid ecological radiation. Consistent with this perspective, our study demonstrates how molecular evidence helped resolve misidentifications in Oreocharis. We advocate for a broader adoption of integrative taxonomy that combines morphology, molecular phylogenetics, ecology, and geography to achieve more robust and reproducible species delimitation.
Taxonomy
Oreocharis yanbianensis Z. Xie & H. H. Kong, sp. Nov. (Figures 2, 3).
Type
CHINA, Sichuan: Panzhihua (攀枝花), Yanbian (盐边), Gesala (格萨拉), alt. 2,988 m, 27°08′20″ N, 101°19′45″ E, rocks on limestone hillsides in the subtropical evergreen broad-leaved forests with Schima argentea E. Pritz. and Quercus spp., flowering, 27 July 2024, Z. Xie, XieZ 4071 (holotype, IBSC! isotype, IBK)!.
Diagnosis
This species resembles to Oreocharis trichantha in morphology, especially in leaf and flower (Figures 1F1–F8, 2, 3). However, it exhibits throat constricted corolla (vs. throat not constricted corolla), corolla limp 2-lipped, unequal, adaxial lip obtuse, abaxial lip rounded (vs. corolla slightly 2-lipped, all lobes subequal, obovate); pistil shorter, < 3 mm (vs. pistil longer, > 10 mm). On the basis of 574 single-copy genes analyses, O. yanbianensis showed closest relationships with O. gamosepala, and they formed a strongly supported sister group in phylogenetic tree.
Description
Herb perennial and stemless, rhizomatous. Leaves basal, arrangement spiral; petiole sparsely brown villous, 0.8 – 6.5 cm long; leaf blade thickly chartaceous, orbiculate rounded, 3.4 – 7.0 × 2.2 – 4.5 cm; lateral veins 4–6 on each side of midrib; adaxially green, rarely brown villous and sparsely white pubescent, midrib veins slightly depressed, lateral veins distinct; abaxially light green, rarely brown villous and sparsely white pubescent, midrib and lateral veins distinct; base cordate to rounded, apex obtuse, margin serrate crenate. Inflorescence cymose, axillary, 2–4 per plant, 2–4 flowered per cyme. Peduncle 2 branched, 2–8 cm long, densely brown villous; bracts 2, lanceolate, 3.0 – 5.0 × 0.9 – 2.2 mm. Pedicel 1.5 – 6.5 cm long, densely brown villous. Calyx 3.8 – 5.0 mm long, 5-sect nearly from base; segments equal, lanceolate, 2.6 – 4.6 × 2.0 – 2.5 mm, margin entire, apex acuminate, adaxially sparsely brown villous, abaxially glabrous. Corolla tubular, yellow, 1.2 – 2.0 cm long, outside white pubescent and inside glabrous; tube cylindric, gradually narrowing towards mouth, 8–9 mm long, throat constricted, base of tube 3.6 – 4.8 mm in diam., middle of tube 2.2 – 2.9 mm in diam.; limb 2-lipped, 4.0 – 6.2 mm long, 9.5 – 13.6 mm wide; adaxial lip 2-lobed, 1.8 – 2.1 × 2.0 – 2.4 mm, apex obtuse; abaxial lip 3-lobed, segments equal, central lobe and lateral lobes rounded, apex rounded. 3.6 – 4.8 × 3.8 – 4.8 mm. Stamens 4, filaments glabrous; adaxial 2.8 – 3.4 mm long, adnate to corolla tube 1.5 – 1.8 mm from base; abaxial 3.0 – 3.6 mm long, adnate to corolla tube 1.0 – 1.4 mm from the base; anthers coherent in 2 pairs, included, 0.8 – 1.2 mm long, elliptic, 2-loculed, dehiscing longitudinally; staminode 1, usually not present, 1.1 – 1.3 mm long, adnate to corolla tube 1.1 – 1.2 mm from base. Disc ring-like, light yellow, 2.0 – 3.4 mm high, margin 5-lobed, unequal. Pistil glabrous, 0.25 – 0.36 cm long; ovary globose, 0.20 – 0.25 cm long, 1-loculed; style short, 0.05 – 0.09 cm long; stigma 1, 2-lobed. Capsule straight, widely elliptic, glabrous, 2–3 cm × 0.5 – 0.8 cm. Seeds tiny, elliptic, 0.50 – 0.56 × 0.10 – 0.12 mm.
Distribution, habitat and phenology
This species is endemic to the Hengduan Mountains, southwest China: Sichuan (Panzhihua). On rocks of limestone hillsides in the subtropical evergreen broad-leaved forests with Schima argentea E. Pritz. and Quercus spp., alt. 2,900 – 3,000 m. Flowering is July to August, and fruiting is August to October.
Conservation status
To date, only one population of the new species was discovered in the field, located at the type locality on the limestone hillside, comprising ca. 300 mature individuals and covering an area of approximately 10,000 m2 (100 × 100 m). This habitat is well-protected by the Lvshilin scenic spot, but is still influenced by various human activities through tourism. According to the IUCN Red List Categories and Criteria, the new species is hereby assessed as “Critically Endangered (CR)” (IUCN Standards and Petitions Subcommittee, 2024).
Additional specimens examined (Paratypes)
CHINA, Sichuan: Panzhihua, Yanbian, Gesala, alt. 2,966 m, 27°8’22.39” N, 101°19’45.25” E, rocks on hillsides, flowering, 17 August 2018, H. H. Kong & L. H. Yang, YLH710 (IBSC0881928, IBSC0881929, IBSC0882485).
Etymology
The species is named after its type locality.
Vernacular name
In Chinese mandarin ‘Yán Biān Mǎ Líng Jù Tái’ (盐边马铃苣苔).
Taxonomic key to selected species of Oreocharis
1. Ovary densely white pubescent. ……………………………….…Oreocharis saxatilis
1. Ovary glabrous or pubescent. …….….………………………2
2. Ovary globose. ……………………………………………….….…… O. yanbianensis
2. Ovary elliptic. ………………………………………….……3
3. Leaf base cuneate or broadly cuneate. ……………………….4
3. Leaf base rounded to cordate. … …………………………….5
4. Abaxial corolla lip lobes equal……………………………….…….… O. gamosepala
4. Abaxial corolla lip lobes unequal. ….………………….….…6
5. Corolla puberulent, disc ca. 1 mm, entire. …………………………….…. O. convexa
5. Corolla sparsely brown villous, disc ca. 2.2 mm, 5-denticulate. ………. O. trichantha
6. Leaf ovate to ovate-rhombic.………………………………………….…… O. bullata
6. Leaf broadly ovate to oblong ovate or elliptic. …………………………… O. concava
Data availability statement
The sequences used in this study have been deposited in the National Center for Biotechnology Information (NCBI) database. The transcriptome data of all samples used in this study are openly available from NCBI: https://www.ncbi.nlm.nih.gov/sra/PRJNA649046 & https://www.ncbi.nlm.nih.gov/sra/PRJNA1032259. The list with the collected locality and GenBank accession numbers of transcriptome data of all the samples (Supplementary Table 1, https://doi.org/10.6084/m9.figshare.28829327.v6), 574 orthologous single-copy genes matrix (Dataset_1, https://doi.org/10.6084/m9.figshare.28829264.v2), 353 low-copy nuclear genes matrix (Dataset_2 on Figshare, https://doi.org/10.6084/m9.figshare.28829282.v2) and the step by step guide on obtaining single-copy data from transcriptomes (Obtaining_single-copy_data_guide.txt on Figshare, https://doi.org/10.6084/m9.figshare.29457638.v2) can be found on Figshare (https://figshare.com/).
Author contributions
ZX: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Visualization, Writing – original draft, Writing – review & editing. NP: Conceptualization, Data curation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. GX: Writing – original draft, Writing – review & editing. FW: Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing. HK: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (grant No. 32370240 and 31970231).
Acknowledgments
We are very grateful to Dr. Lihua Yang (South China Botanical Garden, Chinese Academy of Sciences) for his efforts in collecting many Oreocharis materials, and for his first discovery of the new species illustrated in this paper. He also provided five figures for this paper (Figures 1B4–B7; Figure 4B3). We thank Ms Yunxiao Liu (South China Botanical Garden, Chinese Academy of Sciences) for the excellent line drawings presented in Figure 2. We would also like to express our sincere appreciation to Miss Miao Zhang (Sichuan Normal University) for her help in the field investigation. Youpai Zeng provided Figure 4B1, special thanks. We are grateful to Mr. Junkai Huang (South China Botanical Garden, Chinese Academy of Sciences) for his insightful suggestions on the language of this article. One of the first authors, Mr. Zhi Xie wishes to expand his sincere gratitude and profound respect to Miss Wenhong Cheng for her unwavering understanding, patience, support, and love. This work was supported by the National Natural Science Foundation of China (grant No. 32370240 and 31970231).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bogarín, D., Pérez-Escobar, O. A., Karremans, A. P., Fernández, M., Kruizinga, J., Pupulin, F., et al. (2019). Phylogenetic comparative methods improve the selection of characters for generic delimitations in a hyperdiverse Neotropical orchid clade. Sci. Rep. 9, 15098. doi: 10.1038/s41598-019-51360-0
Cai, L., Chen, R. Z., Yin, Z. J., Zhang, G. X., Chen, W. H., and Shui, Y. M. (2015). Tremacron hongheense, a new species of Gesneriaceae from Southeastern Yunnan, China. Plant Diversity Resour. 37, 733–736.
Cai, L. and Dao, Z. L. (2020b). Oreocharis argentifolia (Gesneriaceae), a new species from the karst region in southeastern Yunnan, China. Nordic J. Bot. 38, e02699. doi: 10.1111/njb.02699
Cai, L., Guo, Y., Zhang, R. M., Dao, Z. L., and Wen, F. (2019). Oreocharis panzhouensis (Gesneriaceae), a new species from karst regions in Guizhou, China. Phytotaxa 393, 287–291. doi: 10.11646/phytotaxa.393.3.5
Cai, L., Huang, H., Dao, Z. L., and Wu, Z. K. (2017). Oreocharis parviflora, a new species of Gesneriaceae from northwestern Yunnan, China. Phytotaxa 329, 167–172. doi: 10.11646/phytotaxa.329.2.7
Cai, L., Huang, Z. J., Wen, F., and Dao, Z. L. (2020a). Two new species of Oreocharis (Gesneriaceae) from karst regions in Yunnan and notes on O. tetraptera and O. brachypoda from China. PhytoKeys 162, 1–12. doi: 10.3897/phytokeys.162.52174
Cai, L., Liu, F. P., Yi, X. B., and Dao, Z. L. (2020c). Oreocharis wumengensis, a new species of Gesneriaceae from northeastern Yunnan, China. PhytoKeys 157, 113–119. doi: 10.3897/phytokeys.157.33071
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chen, S. F. (2023). Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta 2, e107. doi: 10.1002/imt2.107
Chen, W. H., Chen, R. Z., Möller, M., Wen, K., and Shui, Y. M. (2016). Oreocharis ninglangensis, a showy new species of Gesneriaceae from northwestern Yunnan in China. Phytotaxa 261, 282–286. doi: 10.11646/phytotaxa.261.3.8
Chen, R. Z., Chen, W. H., Wei, Y. G., Wen, F., Yu, X. L., and Shui, Y. M. (2017b). Oreocharis crispata, a new species of Oreocharis (Gesneriaceae) from Guangxi, China. Phytotaxa 311, 195–199. doi: 10.11646/phytotaxa.311.2.8
Chen, W. H., Middleton, D. J., Nguyen, H. Q., Nguyen, H. T., Averyanov, L. V., Chen, R. Z., et al. (2017a). Two new species of Oreocharis (Gesneriaceae) from Northwest Vietnam. Gardens’ Bull. Singapore 69, 295–305. doi: 10.26492/gbs69(2).2017-08
Chen, W. H., Möller, M., Chen, R. Z., Rui, R. J., and Shui, Y. M. (2015). Oreocharis synergia, a new species of Gesneriaceae from Northwestern Yunnan, China. Phytotaxa 233, 090–093. doi: 10.11646/phytotaxa.233.1.8
Chen, W. H., Nguyen, Q. H., Chen, R. Z., Nguyen, T. H., Nguyen, S. K., Nguyen, V. T., et al. (2018). Two new species of Oreocharis (Gesneriaceae) from Fan Si Pan, the highest mountain in Vietnam. PhytoKeys 94, 95–106. doi: 10.3897/phytokeys.94.21329
Chen, W. H., Shui, Y. M., Hua, C. L., Yu, C. Y., and Wen, K. (2012). Ancylostemon dimorphosepalus (Gesneriaceae), a new species from China. Annales Botanici Fennici 49, 391–394. doi: 10.5735/085.049.0612
Chen, W. H., Shui, Y. M., and Möller, M. (2014b). Two new combinations in Oreocharis Benth. (Gesneriaceae) from China. Candollea 69, 179–182. doi: 10.15553/c2014v692a10
Chen, W. H., Shui, Y. M., Yang, J. B., Wang, H., Nishii, K., Wen, F., et al. (2014a). Taxonomic status, phylogenetic affinities and genetic diversity of a presumed extinct genus, Paraisometrum W.T. Wang (Gesneriaceae) from the karst regions of southwest China. PloS One 9, e107967. doi: 10.1371/journal.pone.0107967
Chen, W. H., Wang, H., Shui, Y. M., Möller, M., and Yu, Z. Y. (2013). Oreocharis jinpingensis (Gesneriaceae), a new species from Yunnan, China. Annales Botanici Fennici 50, 312–316. doi: 10.5735/086.050.0504
Chen, W. H., Zhang, Y. M., Guo, S. W., Zhang, Z. R., Chen, L., and Shui, Y. M. (2020b). Reassessment of Bournea Oliver (Gesneriaceae) based on molecular and palynological evidence. PhytoKeys 157, 27–41. doi: 10.3897/phytokeys.157.55254
Chen, W. H., Zhang, Y. M., He, D. M., Li, Y. L., and Shui, Y. M. (2020a). Four new species of Oreocharis (Gesneriaceae) in Yunnan province, China. PhytoKeys 157, 83–99. doi: 10.3897/phytokeys.157.32284
Craib, W. G. (1918). “Gesneracearum novitates nonnullae,” in Notes from the Royal Botanic Garden Edinburgh, vol. 10. , 211–219.
Craib, W. G. (1920). “Gesneracearum novitates,” in Notes from the Royal Botanic Garden Edinburgh, vol. 11. , 233–254.
De Queiroz, K. (1999). “The general lineage concept of species and the defining properties of the species category,” in Wilson, R. A. (ed.) Species, New interdisciplinary essays MIT Press, Cambridge. doi: 10.7551/mitpress/6396.003.0007
Do, V. T., Wei, Y. G., and Wen, F. (2017). Oreocharis caobangensis (Gesneriaceae), a new species from Cao Bang Province, northern Vietnam. Phytotaxa 302, 065–070. doi: 10.11646/phytotaxa.302.1.6
Dorsey, B. L., Haevermans, T., Aubriot, X., Morawetz, J. J., Riina, R., and Berry, P. E. (2013). Phylogenetics, morphological evolution, and classification of Euphorbia subgenus Euphorbia. Taxon 62, 291–315. doi: 10.12705/622.1
Fu, Q., Guo, Y., Huang, R., Xia, Y., and Wang, Y. Q. (2019b). Oreocharis ovatilobata (Gesneriaceae), a new species from Guizhou, China. Annales Botanici Fennici 56, 259–265. doi: 10.5735/085.056.0411
Fu, L. F., Li, S., Xin, Z. B., Wen, F., and Wei, Y. G. (2019c). The changes of the Chinese names and scientific names of gesneriaceae in China between Wang’s and Weber’s classifications for gesneriaceae. Guangxi Sci. 26, 118–131.
Fu, Q., Xia, Y., Guo, Y., Huang, R., and Wang, Y. Q. (2019a). Oreocharis odontopetala, a new species of Gesneriaceae from Guizhou, China. PhytoKeys 124, 1–9. doi: 10.3897/phytokeys.124.34609
Gong, Y. X., Ding, H. B., Yan, X. S., Wen, F., Tian, Y. H., and Tan, Y. H. (2022). Oreocharis polyneura, a new species of Gesneriaceae from southern Yunnan, China. PhytoKeys 214, 7–15. doi: 10.3897/phytokeys.214.93901
Guerrero, P. C., Majure, L. C., Cornejo-Romero, A., and Hernández-Hernández, T. (2019). Phylogenetic relationships and evolutionary trends in the cactus family. J. Hered. 110, 4–21. doi: 10.1093/jhered/esy064
Guo, Z. Y., Li, Z. Y., and Xiang, X. G. (2018). Oreocharis duyunensis (Gesneriaceae), a new species from Guizhou, China. Nordic J. Bot. 36, e01514. doi: 10.1111/njb.01514
Han, M. Q., Pan, B., Zou, L. L., and Liu, Y. (2017). Oreocharis purpurata, a new species of Gesneriaceae from Hunan, China. Phytotaxa 328, 183–188. doi: 10.11646/phytotaxa.328.2.9
Harris, J. G. and Harris, M. W. (1994). Plant identification terminology: An illustrated glossary (Spring Lake, Utah: Spring Lake Publishing), 206.
Hodge, J. R., Adams, D. S., Williams, K. L., Alencar, L. R. V., Camper, B., Larouche, O., et al. (2025). Unravelling the effects of ecology and evolutionary history in the phenotypic convergence of fishes. Syst. Biol. 2025, syaf034. doi: 10.1093/sysbio/syaf034
Hong, D. Y. (2016). Biodiversity pursuits need a scientific and operative species concept. Biodivers. Sci. 24, 979–999. doi: 10.17520/biods.2016203
Hong, K. Y., Radian, Y., Manda, T., Xu, H. B., and Luo, Y. M. (2023). The development of plant genome sequencing technology and its conservation and application in endangered gymnosperms. Plants 12, 4006. doi: 10.3390/plants12234006
Hooker, J. D. (1893). Icones Plantarum, or figures, with brief descriptive characters and remarks, of new or rare plants, selected from the author’s herbarium, Vol. 23, ser. 4, 3. Longman, Reese, Orne, Brown, Green & Longman, Kew, Bentham–Moxon Trust, London
Horn, J. W., Van Ee, B. W., Morawetz, J. J., Riina, R., Steinmann, V. W., Berry, P. E., et al. (2012). Phylogenetics and the evolution of major structural characters in the giant genus Euphorbia L. (Euphorbiaceae). Mol. Phylogenet. Evol. 63, 305–326. doi: 10.1016/j.ympev.2011.12.022
Hu, R. C., Hu, Q. M., Qin, Y., Xu, W. B., and Huang, Y. F. (2022). Oreocharis tianlinensis, a new species of Gesneriaceae from the limestone area in northwestern Guangxi, China. Taiwania 67, 479–483. doi: 10.6165/tai.2022.67.479
Hu, J., Zhang, J. Y., He, H., Yu, D. X., Jiang, H., Liu, Q., et al. (2023). Oreocharis oriolus, a new species of Gesneriaceae in a sclerophyllous oak community from Yunnan, Southwest China. Ecol. Evol. 13, e10174. doi: 10.1002/ece3.10174
Huang, X. K., Su, C. L., Yang, P., and Liu, Y. (2023). Oreocharis repenticaulis (Gesneriaceae), a new species from western Guangxi, China. Nordic J. Bot. 2023, e03636. doi: 10.1111/njb.03636
IUCN Standards and Petitions Subcommittee (2024). Guidelines for Using the IUCN Red List Categories and Criteria, version 15.1 (The Standards and Petitions Committee). Available online at: https://www.iucnredlist.org/documents/RedListGuidelines.pdf.
Karbstein, K., Kösters, L., Hodač, L., Hofmann, M., Hörandl, E., Tomasello, S., et al. (2024). Species delimitation 4.0: integrative taxonomy meets artificial intelligence. Trends Ecol. Evol. 39, 771–784. doi: 10.1016/j.tree.2023.11.002
Kong, H. H., Condamine, F. L., Yang, L. H., Harris, A. J., Feng, C., Wen, F., et al. (2022). Phylogenomic and macroevolutionary evidence for an explosive radiation of a plant genus in the Miocene. Syst. Biol. 71, 589–609. doi: 10.1093/sysbio/syab068
Le, D. K., Nguyen, T. T., Nguyen, T. P., Hoang, T. T., Wen, F., and Do, V. T. (2022). Oreocharis phuongii (Gesneriaceae), a new species from central Vietnam. PhytoKeys 193, 43–53. doi: 10.3897/phytokeys.193.77083
Li, R. F., Le, X. G., Xu, L., Maciejewski, S., Chen, B., and Wen, F. (2023). Oreocharis yangjifengensis (Gesneriaceae), a new species from Yangjifeng National Nature Reserve of Yingtan City, Jiangxi Province, China. Phytotaxa 583, 213–218. doi: 10.11646/phytotaxa.583.2.10
Li, J. M. and Li, M. Z. (2015). Oreocharis brachypodus (Gesneriaceae), a new taxon from Guizhou, China. Phytotaxa 204, 296–299. doi: 10.11646/phytotaxa.204.4.6
Li, Z. L., Ma, H. J., Ye, Z. R., Meng, D. C., Wen, F., and Hong, X. (2022). Oreocharis guangwushanensis, a new species of Gesneriaceae from Sichuan Province, China. PhytoKeys 201, 123–129. doi: 10.3897/phytokeys.201.77574
Li, X., Wang, Q. P., Yang, F., Ye, J. Y., and Wang, H. C. (2025). Oreocharis corallodiscoides (Gesneriaceae, Trichosporeae, Didymocarpinae), a new species from Yunnan, southwest China. PhytoKeys 256, 73–81. doi: 10.3897/phytokeys.256.148644
Li, J. M., Wang, T., and Zhang, Y. G. (2017). Oreocharis zhenpingensis (Gesneriaceae), a new species from Shaanxi, China. Phytotaxa 307, 292–296. doi: 10.11646/phytotaxa.307.4.7
Lin, Q. W. (2016). Beccarinda baolianis, a new species of Gesneriaceae from Fujian Province. Bull. Botanical Res. 36, 650–652.
Ling, S. J., Guan, S. P., Wen, F., Shui, Y. M., and Ren, M. X. (2020). Oreocharis jasminina (Gesneriaceae), a new species from mountain tops of Hainan Island, South China. PhytoKeys 157, 121–135. doi: 10.3897/phytokeys.157.50246
Ling, S. J., Wen, F., and Ren, M. X. (2022). Oreocharis hainanensis (Gesneriaceae), a new species from karst regions in Hainan Island, South China. Phytotaxa 538, 281–291. doi: 10.11646/phytotaxa.538.4.2
Liu, X. J. and Sun, X. G. (2021). Oreocharis wenxianensis (Gesneriaceae), a new species from Gansu Province, China. Annales Botanici Fennici 58, 181–187. doi: 10.5735/085.058.0120
Liu, Y., Xu, W. B., Huang, Y. S., Peng, C. I., and Chung, K. F. (2012). Oreocharis dayaoshanioides, a rare new species of Gesneriaceae from eastern Guangxi, China. Botanical Stud. 53, 393–399.
Losos, J. B. (2011). Convergence, adaptation, and constraint. Evolution 65, 1827–1840. doi: 10.1111/j.1558-5646.2011.01289.x
Lv, Z. Y., Yusupov, Z., Zhang, D. G., Zhang, Y. Z., Zhang, X. S., Lin, N., et al. (2021). Oreocharis xieyongii, an unusual new species of Gesneriaceae from western Hunan, China. Plant Diversity 44, 220–230. doi: 10.1016/j.pld.2021.11.008
Michener, W. K., Baerwald, T. J., Firth, P., Palmer, M. A., Rosenberger, J. L., Sandlin, E. A., et al. (2001). Defining and unraveling biocomplexity. BioScience 51, 1018–1023. doi: 10.1641/0006-3568(2001)051[1018:DAUB]2.0.CO;2
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Haeseler, A. V., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Mishler, B. D. (1985). The morphological, developmental, and phylogenetic basis of species concepts in bryophytes. Bryologist 88, 207–214. doi: 10.2307/3243030
Möller, M. (2015). Transfer of Tremacron hongheense to Oreocharis (Gesneriaceae). Phytotaxa 239, 295–296. doi: 10.11646/phytotaxa.239.3.12
Möller, M., Atkins, H. J., Bramley, G. L. C., Middleton, D. J., Baines, R., Nguyen, V. D., et al. (2018). Two new species of Oreocharis (Gesneriaceae) from Northern Vietnam. Edinburgh J. Bot. 75, 309–319. doi: 10.1017/S0960428618000148
Möller, M., Chen, W. H., Shui, Y. M., Atkins, H., and Middleton, D. J. (2014). A new genus of Gesneriaceae in China and the transfer of Briggsia species to other genera. Gardens’ Bull. Singapore 66, 195–205.
Möller, M., Middleton, D. J., Nishii, K., Wei, Y. G., and Weber, A. (2011). A new delineation for Oreocharis incorporating an additional ten genera of Chinese Gesneriaceae. Phytotaxa 23, 1–36. doi: 10.11646/phytotaxa.23.1.1
Möller, M., Middleton, D. J., Ulrich, S., and Weber, A. (2025). Bournea is not supported as separate from Oreocharis (Gesneriaceae). Edinburgh J. Bot. 82. doi: 10.24823/ejb.2025.2028
Möller, M., Wei, Y. G., Wen, F., Clark, J. L., and Weber, A. (2016). You win some you lose some: updated generic delineations and classification of Gesneriaceae-implications for the family in China. Guihaia 36, 44–60.
Nakamura, T., Yamada, K. D., Tomii, K., and Katoh, K. (2018). Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 34, 2490–2492. doi: 10.1093/bioinformatics/bty121
Nguyen, K. S., Averyanov, L. V., and Lin, C. W. (2024). Oreocharis hapii (Gesneriaceae), a new species from central Vietnam. Phytotaxa 652, 235–240. doi: 10.11646/phytotaxa.652.3.5
Pan, B., Tang, G. D., Do, T. V., Maciejewski, S., Deng, C. L., and Wen, F. (2019). Oreocharis tetrapterus (Gesneriaceae), a new species from East Guangxi, China. PhytoKeys 131, 83–89. doi: 10.3897/phytokeys.131.35434
Paterson, H. E. H. (1985). “The recognition concept of species,” in Species and Speciation. Ed. Vrba, E. (Transvaal Museum, Pretoria), 21–29.
Paterson, H. E. H. (1992). “The recognition concept of species,” in The Units of Evolution: Essays on the Nature of Species. Ed. Ereshefsky, M. (MIT Press, Cambridge), 139–158.
Qin, W. H., Ding, D. D., Li, Z. X., Gao, Y. F., Li, S., and Hong, X. (2020). Oreocharis flavovirens, a new species of Gesneriaceae from Southern Gansu Province, China. PhytoKeys 157, 101–112. doi: 10.3897/phytokeys.157.31732
Rossini, J. and Freitas, J. (2014). Oreocharis yunnanensis, a new name for the illegitimate Oreocharis glandulosa (Gesneriaceae) from China. Phytotaxa 163, 180–180. doi: 10.11646/phytotaxa.163.3.5
Sayyari, E. and Mirarab, S. (2016). Fast coalescent-based computation of local branch support from quartet frequencies. Mol. Biol. Evol. 33, 1654–1668. doi: 10.1093/molbev/msw079
Simpson, G. G. (1943). Criteria for genera, species and subspecies in zoology and paleontology. Ann. New York Acad. Sci. 44, 145–178. doi: 10.1111/j.1749-6632.1943.tb31301.x
Sokal, R. R. and Crovello, T. J. (1970). The biological species concept: a critical evaluation. Am. Nat. 104, 127–153. doi: 10.1086/282646
Stern, D. (2013). The genetic causes of convergent evolution. Nat. Rev. Genet. 14, 751–764. doi: 10.1038/nrg3483
Taha, A., Ettaqy, A., El Mderssa, M., Belaqziz, M., Fokar, M., Boukcim, H., et al. (2023). Comprehensive review of morphological adaptations and conservation strategies of cactiform succulents: A case study of Euphorbia species in arid ecosystems. Biosyst. Diversity 31, 358–367. doi: 10.15421/012342
Tan, Y. H., Li, J. W., Chen, W. H., Wen, B., and Möller, M. (2014). Additional notes on Oreocharis yunnanensis, a species of Gesneriaceae from southern Yunnan, China, including morphological and molecular data. Phytotaxa 167, 283–288. doi: 10.11646/phytotaxa.167.3.7
Tan, Y. H., Li, J. W., Pan, B., Wen, B., Yin, J. T., and Liu, Q. (2013). Oreocharis glandulosa, a new species of Gesneriaceae from southern Yunnan, China. Phytotaxa 131, 1–6. doi: 10.11646/phytotaxa.131.1.5
Tan, Y. H., Li, J. W., and Yin, J. T. (2015). Oreocharis tsaii, a new species of Gesneriaceae from southern Yunnan, China. Phytotaxa 195, 188–192. doi: 10.11646/phytotaxa.195.2.9
Templeton, A. R. (1989). “The meaning of species and speciation: A genetic perspective,” in Speciation and its Consequences. Eds. Otte, D. and Endler, J. A. (Sinauer Associates, Sunderland), 3–27.
Tran, T. P. A., Nguyen, K. S., Tan, K., and Averyanov, L. V. (2023). Oreocharis thanhii (Gesneriaceae), a new species from Northwest Vietnam. Taiwania 68, 275–280. doi: 10.6165/tai.2023.68.275
Valen, L. V. (1976). Ecological species, multispecies, and oaks. Taxon 25, 233–239. doi: 10.2307/1219444
Wang, W. T. (1981). Genus novum primitivum Gesneriacearum e Sina. Acta Phytotaxonomica Sin. 19, 485–489.
Wang, W. T. (1983b). Three new genera of Gesneriaceae from China. Botanical Res.: Contrib. Institute Botany Academia Sin. 1, 15–24.
Wang, Y. Z., Liang, R. H., Wang, B. H., Li, J. M., Qiu, Z. J., Li, Z. Y., et al. (2010). Origin and phylogenetic relationships of the Old World Gesneriaceae with actinomorphic flowers inferred from ITS and trnL-trnF sequences. Taxon 59, 1044–1052. doi: 10.1002/tax.594005
Wang, W. T., Pan, K. Y., Li, Z. Y., Weitzman, A. L., and Slog, L. E. (1998). “Gesneriaceae,” in Flora of China, vol. 18 . Eds. Wu, Z. Y. and Raven, P. H. (St. Louis, Science Press, Beijing & Missouri Botanical Garden Press) vol.18, 244–401.
Wang, W. T., Pan, K. Y., Zhang, Z. Y., Li, Z. Y., Tao, D. D., and Yin, W. C. (1990). “Gesneriaceae,” in Flora of China, vol. 69 . Eds. Wu, Z. Y., Raven, P. H., and Hong, D. Y. (Science Press, Beijing), 141–167.
Wei, J. J., Xiong, G. C., Zou, C. Y., Pan, B., and Xu, W. B. (2016). Oreocharis curvituba, a new species of Gesneriaceae from northeastern Guangxi, China. Phytotaxa 280, 190–194. doi: 10.11646/phytotaxa.280.2.9
Weitzman, A. L., Skog, L. E., Wang, W. T., Pan, K. Y., and Li, Z. Y. (1998). New taxa, new combinations, and notes on Chinese Gesneriaceae. Novon 7, 423–435. doi: 10.2307/3391777
Wesselink, M. and Kuiper, I. (2008). Species identification of botanical trace evidence using molecular markers. Forensic Sci. International: Genet. Supplement Ser. 1, 630–632. doi: 10.1016/j.fsigss.2007.10.211
Wiens, J. J. (2007). Species delimitation: new approaches for discovering diversity. Syst. Biol. 56, 875–878. doi: 10.1080/10635150701748506
Wiley, E. O. (1992). “The evolutionary species concept reconsidered,” in The Units of Evolution: Essays on the Nature of Species. Ed. Ereshefsky, M. (MIT Press, Cambridge), 79–92.
Xie, P. L., Guo, Y. L., Teng, Y., Zhou, W. B., and Yu, Y. (2024b). GeneMiner: A tool for extracting phylogenetic markers from next-generation sequencing data. Mol. Ecol. Resour. 24, e13924. doi: 10.1111/1755-0998.13924
Xie, Z., Peng, N. N., Zhang, M., Ding, G. E., Wen, F., and Kong, H. H. (2024a). Oreocharis scutifolia (Gesneriaceae), a peltate-leaved new species from the dry-hot valley of the Jinsha River Basin, Yunnan, China. Ecol. Evol. 14, e70442. doi: 10.1002/ece3.70442
Xiong, C., Chen, F., Zhang, J. H., Zhou, H. L., Zheng, C. B., and Wen, F. (2023). Oreocharis wuxiensis (Gesneriaceae), a new lithophilous species from Northeast Chongqing, China. Phytotaxa 594, 73–77. doi: 10.11646/phytotaxa.594.1.5
Yang, C. Z., Cai, D. L., and Wen, F. (2015b). Oreocharis striata (Gesneriaceae), a new Species from Fujian, China. Annales Botanici Fennici 52, 369–372. doi: 10.5735/085.052.0517
Yang, L. E., Cen, H. F., Sun, H., LoFurno, M., Maciejewski, S., Goretsky, W. J., et al. (2019). Oreocharis rubrostriata (Gesneriaceae), a new species from Guangxi, China. Kew Bull. 74, 23. doi: 10.1007/s12225-019-9810-9
Yang, L. H., Huang, J. Z., Deng, F. D., and Kang, M. (2017). Oreocharis uniflora, a new species of Gesneriaceae from Guangdong, China. Phytotaxa 295, 292–296. doi: 10.11646/phytotaxa.295.3.11
Yang, J. W., Qin, X. M., Xu, J., Li, C. R., Ren, Q. F., Yuan, M. Q., et al. (2022). Oreocharis qianyuensis, a new species of Gesneriaceae from Southwest, China based on morphological and molecular evidence. PhytoKeys 213, 119–130. doi: 10.3897/phytokeys.213.84349
Yang, L. H. and Shi, X. Z. (2021). Oreocharis reticuliflora (Gesneriaceae), a new species from southeastern Sichuan, China. Nordic J. Bot. 39, e03322. doi: 10.1111/njb.03322
Yang, L. H., Wen, F., Kong, H. H., Sun, Z. X., Su, L. Y., and Kang, M. (2020). Two new combinations in Oreocharis (Gesneriaceae) based on morphological, molecular and cytological evidence. PhytoKeys 157, 43–58. doi: 10.3897/phytokeys.157.32609
Yang, L. H., Zhou, L. X., and Kang, M. (2018). Oreocharis ovata (Gesneriaceae), a new species from Guangdong, China. Nordic J. Bot. 36, e01764. doi: 10.1111/njb.01764
Yang, L. H., Zhou, J. G., Xu, P., Chen, Z. T., Lu, Y. H., and Kang, M. (2015a). Oreocharis pilosopetiolata, a new species of Gesneriaceae from southeastern Guangdong, China. Phytotaxa 239, 287–292. doi: 10.11646/phytotaxa.239.3.10
Yi, R., Li, X. J., and Li, J. M. (2019). Oreocharis maximowiczii var. mollis (Gesneriaceae), a new variety from Fujian, China. Phytotaxa 424, 067–070. doi: 10.11646/phytotaxa.424.1.7
Zhang, C., Rabiee, M., Sayyari, E., and Mirarab, S. (2018). ASTRAL-III: Polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinf. 19, 153. doi: 10.1186/s12859-018-2129-y
Zhang, Z., Xie, P. L., Guo, Y. L., Zhou, W. B., Liu, E. Y., and Yu, Y. (2022). Easy353: A tool to get angiosperms353 genes for phylogenomic research. Mol. Biol. Evol. 39, msac261. doi: 10.1093/molbev/msac261
Zhou, G. H., Tu, R. H., Liu, A., and Yu, X. L. (2023). Oreocharis chenzhouensis, a new species of Gesneriaceae from southern Hunan, China. Phytotaxa 607, 197–204. doi: 10.11646/phytotaxa.607.3.3
Keywords: morphology, molecular analyses, Oreocharis, transcriptome data, phylogeny
Citation: Xie Z, Peng N, Xie G, Wen F and Kong H (2025) Both morphological and molecular data are crucial for describing new species — a case study on the genus Oreocharis (Gesneriaceae). Front. Plant Sci. 16:1641488. doi: 10.3389/fpls.2025.1641488
Received: 05 June 2025; Accepted: 07 August 2025;
Published: 08 September 2025.
Edited by:
Saraj Bahadur, Hainan University, ChinaReviewed by:
Jun Hu, Chinese Academy of Sciences (CAS), ChinaDeniz Aygoren Uluer, Ahi Evran University, Türkiye
Copyright © 2025 Xie, Peng, Xie, Wen and Kong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanghui Kong, a29uZ2hoQHNjYmcuYWMuY24=
†These authors have contributed equally to this work
‡ORCID: Zhi Xie, orcid.org/0000-0002-9134-3257
Nana Peng, orcid.org/0009-0000-8037-2776
Genglin Xie, orcid.org/0009-0002-1143-4923
Fang Wen, orcid.org/0000-0002-3889-8835
Hanghui Kong, orcid.org/0000-0003-2721-0449
 Zhi Xie
Zhi Xie Nana Peng1,2,3†‡
Nana Peng1,2,3†‡ Fang Wen
Fang Wen Hanghui Kong
Hanghui Kong