- 1Shandong Provincial Key Laboratory of Plant Stress, College of Life Sciences, Shandong Normal University, Jinan, China
- 2State Key Laboratory of Crop Biology, College of Agronomy, Shandong Agricultural University, Tai’an, Shandong, China
Salt stress is a major challenge to agricultural productivity and can adversely affect plant growth and development. This review examines the interaction between cell wall architecture and plant tolerance to salt stress, focusing on the mechanisms underlying growth, remodeling, and anisotropic morphogenesis. It further elucidates how the cell wall’s composition, structure, and mechanical properties affect osmotic balance, ion transport, and physiological responses to salinity in plants. Key strategies for adaptation to stress, including the synthesis of osmoprotectants and alterations in cell wall polysaccharides, are discussed to understand their role in cell integrity and expansion under salt conditions. In addition, the review emphasizes the dynamic remodeling of the cell wall, which promotes anisotropic growth patterns necessary to maintain plant structure and function under environmental stresses. Based on the current research, this review highlights potential pathways to enhance plant adaptation to salinity through targeted manipulation of cell wall properties, providing insights for future biotechnological applications to improve crop performance in a saline environment.
1 Introduction
In natural environments, plants are constantly exposed to various type of environmental stresses, both abiotic and biotic, that can significantly affect plant growth, development, and productivity (Du et al., 2024). Salt is one of the most common and harmful abiotic stress, which leading to decreased crop yield as it inhibits plant growth and limits important cellular processes (Yadav et al., 2020). Salt stress is estimated to affect over 6% of the world’s land area, approximately 800 million hectares (Mustafa et al., 2020). Salinity exhibits a huge impact on agriculture, causing substantial economic losses annually due to reduced crop yields (Chele et al., 2021). Furthermore, the irrigated agricultural land is frequently vulnerable due to salinization, which is predicted to worsen over time and potentially impact as much as 50% of irrigated land by 2050 (Singh, 2022). Salt stress typically begins with the accumulation of sodium ions (Na+) in the root zone, which creates osmotic imbalance and disrupts cellular hydration, thereby inhibiting water uptake (Atta et al., 2023; Héreil et al., 2024). The excess Na+ enters plant cells primarily through non-selective cation channels and competitive uptake mechanisms, displacing essential K+ ions and disrupting enzymatic activity and membrane integrity (Estrada et al., 2023; Zhou et al., 2024). Intracellular Na+ accumulation alters the ionic equilibrium and raises osmotic pressure, leading to metabolic hinderance, oxidative stress via reactive oxygen species (ROS), and reduced photosynthesis (Zhao et al., 2020).
Salt stress impacts various cellular structures, including the cell wall, which plays a crucial role in maintaining plant growth and development. Cell wall forms the first layer of the plant cell. The changes caused as a result of salt stress extend to the cell wall, which is a vital component of plant cells, and plays a key role in regulating growth, maintaining cell shape, and responding to environmental stresses (Cosgrove, 2022). The cell wall’s ability to adapt and remodel in response to stress is crucial to plant survival under salinity. Particularly, the accumulation of Na+ ions in the apoplast (the space outside the cell membrane) can disrupt cell wall loosening by interacting with negatively charged cell wall polymers, such as pectin and hemicelluloses. This interaction alters the pH and hinders the plant’s ability to extend its cell walls, thereby restricting growth (Dabravolski and Isayenkov, 2023).
Salt stress impacts cell wall structure not only chemically but also mechanically, disrupting anisotropic growth, which normally allows cells to expand more in one direction such as elongating hypocotyls and roots while maintaining directional stability (Colin et al., 2022). This process relies on the orientation of cellulose microfibrils, guided by cortical microtubules, which direct expansion along preferred axes. Under salt stress, there is a transient depolymerization of microtubules and removal of cell‐wall synthesis complexes (CESAs) from the plasma membrane. However, during favorable condition the microtubules and CESAs reassemble, that is facilitated by proteins like CC1/2 and SP2L, to re-establish cellulose orientation for growth recovery (Yu et al., 2022). Additionally, salt exposure often shifts growth patterns from anisotropic to radial expansion, notably in epidermal cells, a morphological change that compromises organ form. The SP2L-mediated reorientation of microtubules has been shown to induce such radial cell expansion in root transition zones, altering directional growth (Van de Peer et al., 2021; Yu et al., 2022).
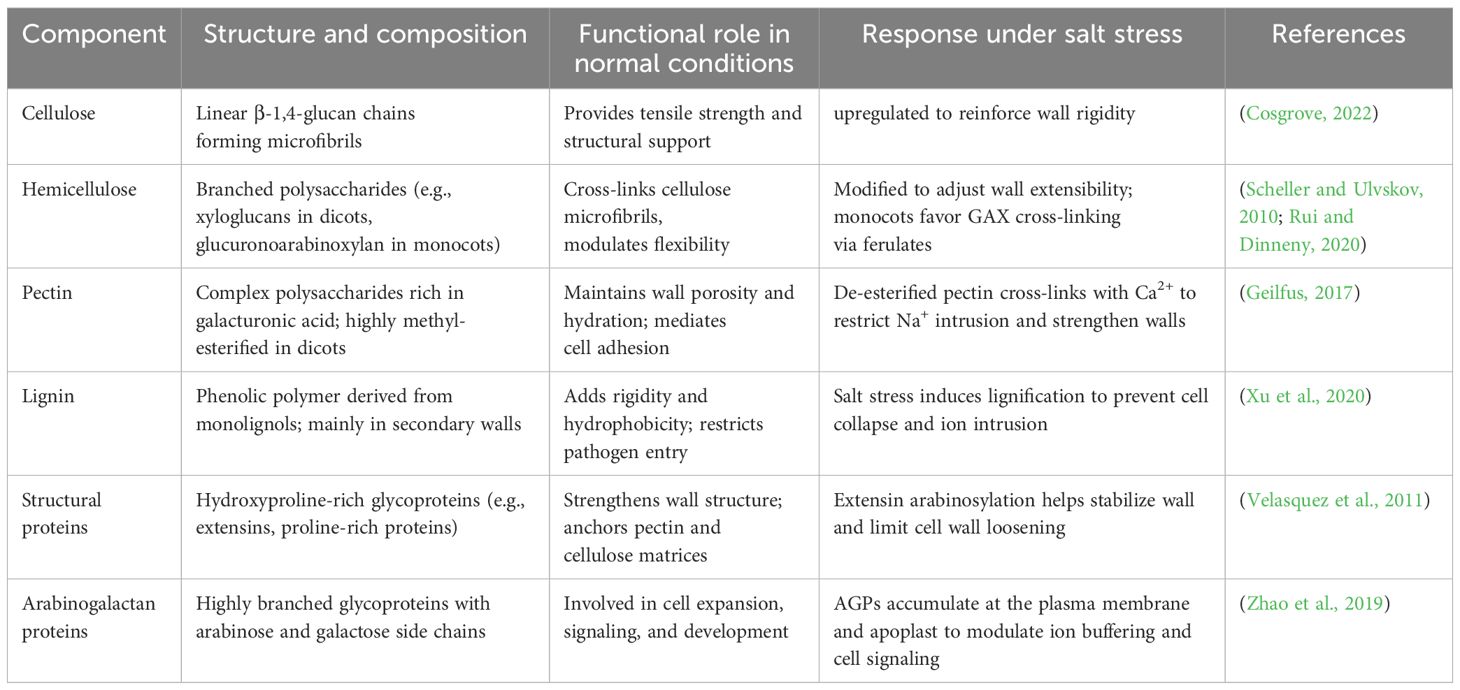
The plant cell wall is a dynamic, multifunctional structure composed of polysaccharides (cellulose, hemicellulose, pectin), structural proteins, and lignin, which collectively provide mechanical support while modulating the movement of ions, water, and signaling molecules across the cell boundary (Dokka et al., 2024). The cell wall plays a critical role in determining how plants respond to external stresses, including salinity (Dabravolski and Isayenkov, 2023). Plants have developed several defense mechanisms against the harmful consequences of salt stress, including cell wall remodeling. Through this remodeling, plants preserve cellular integrity, maintain ion homeostasis, and alter developmental patterns to avoid high salinity regions (Oliveira et al., 2020). Moreover, during salt stress, plants activate a series of biochemical pathways that modify cell wall components, such as pectin, hemicelluloses, and cellulose (Figure 1). These modifications allow the cell wall to maintain its integrity and continue supporting cell growth, even under adverse conditions (Dabravolski and Isayenkov, 2023).
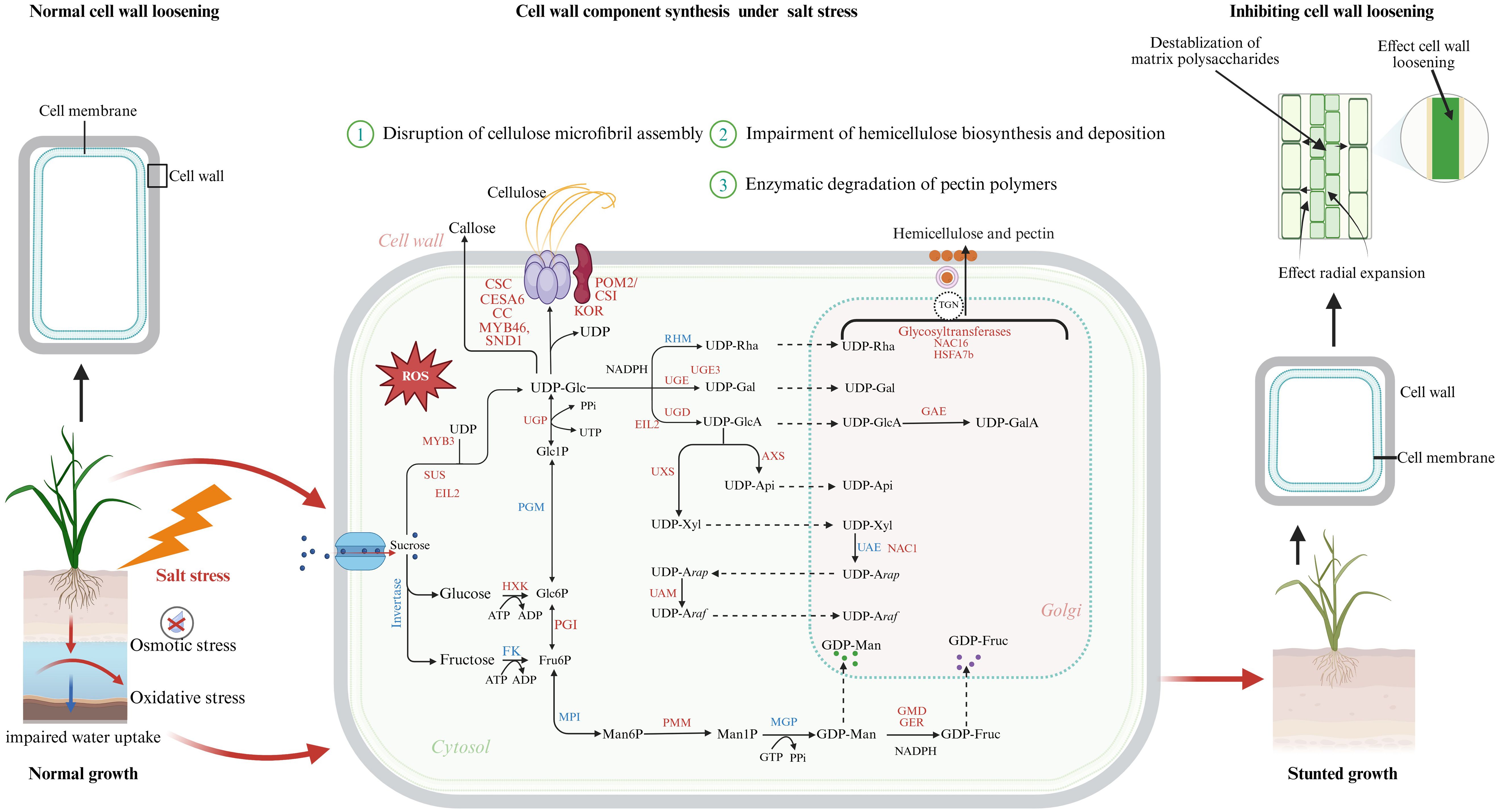
Figure 1. Overview of the effects of salt stress on plant cell wall integrity, loosening, and component biosynthesis. Salt stress in plants triggers both osmotic and oxidative stress, resulting in impaired water uptake and excessive generation of reactive oxygen species (ROS). These ROS contribute to structural damage and destabilization of matrix polysaccharides, ultimately inhibiting cell wall loosening and radial expansion. Consequently, the biosynthesis and stability of major wall components are adversely affected: cellulose becomes weakened, hemicellulose synthesis is disrupted, pectin is degraded, and lignin biosynthesis is suppressed. The intracellular sugar metabolism and biosynthetic pathways contributing to the formation of cellulose, hemicellulose, and pectin. Red-labeled genes and enzymes represent transcriptional or post-transcriptional upregulation under salt stress, including transcription factors (MYB46, SND1, NAC1, NAC16, HSFA7b, EIL2, MYB3) and key biosynthetic enzymes (UGE3, UGD, UGP, UXS, GMD, GER, GAE), which drive stress-induced remodeling of cell wall metabolism. Under normal conditions (left panel), cell wall-loosening enzymes such as XTHs and expansins facilitate expansion and growth, while salt stress (right panel) disrupts these processes, leading to reduced wall plasticity and stunted plant development. The figure integrates metabolic, transcriptional, and biophysical responses, providing a comprehensive view of how salt stress alters plant cell wall dynamics and growth.
This review focuses on the complex relationship between salt stress and plant cell wall architecture, with a focus on understanding the physiological, biochemical, and mechanical processes involved in cell wall remodeling. By examining these processes, this review aims to shed light on how plants utilize cell wall modifications to enhance their tolerance to salt stress, thus providing a foundation for developing strategies to improve crop resilience in saline environments.
2 Role of cell wall architecture in salt stress
The cell wall serves as a complex extracellular matrix surrounding most of plant cells and exhibits extreme tensile strength and extensibility. Its architecture is crucial, as it supports various aspects of plant growth and development. The cell wall frequently serves as a robust yet pliable structure, which is perpetually remodeled to direct the process of cellular expansion. The architectural composition of the plant cell wall is diverse and species-specific. The structure and composition of the plant cell wall vary between monocots and dicots. While both share key components such as cellulose, hemicelluloses, and pectin, dicot primary walls are typically rich in pectin and xyloglucans, whereas monocot (especially grasses) walls contain more glucuronoarabinoxylan and mixed-linkage glucans with lower pectin content (Cosgrove, 2022). This distinction is critical when considering cell wall remodeling under salt stress, as the biochemical pathways involved differ between plant types. While recognizing these structural distinctions between monocot and dicot cell walls, the present review aims to integrate current knowledge across both plant types to provide a comprehensive understanding of how salt stress modulates cell wall components and architecture (Table 1).
2.1 Cellulose
In terrestrial vascular plants, the cell wall is primarily composed of cellulose, a homopolymer made up of repeating glucose subunits linked by β (1-4) bonds (Ellinger and Voigt, 2014; Schneider et al., 2016). Cellulose plays a crucial role as the main load-bearing component of the cell wall, providing mechanical support. The synthesis of cellulose is an active process, with its synthesis trajectory guided by microtubules within the plant cell. During this process, a heterotrimeric rosette structure known as CesA interacts with Plant Oligosaccharide Binding Protein 2/Cellulose Synthase Interacting Protein 1 (POM2/CSI1) to facilitate cellulose production (Bringmann et al., 2012). The role of CesA in cellulose synthesis has been explored in previous studies (Joshi and Mansfield, 2007; Lampugnani et al., 2018). A mutation in the catalytic domain of CESA6 results in reduced cellulose content in plants (Huang et al., 2023). Although there is still a lack of extensive experimental evidence showing how CesA genes function under salt stress conditions in plants (Heyndrickx and Vandepoele, 2012). General Control Non-repressed Protein 5 (GCN5) plays a key role in regulating cellulose synthesis in salt-sensitive Arabidopsis thaliana by modulating the transcription of cellulose biosynthetic genes, thereby contributing to enhanced salt stress tolerance (Zheng et al., 2019). Mutants deficient in cellulose, such as cesa6, pom2/csi1, and the Companion of Cellulose Synthases (CC) mutant, display heightened sensitivity to salt stress. These mutants display a sensitive phenotype to salt stress. This increased sensitivity is primarily due to impaired cellulose biosynthesis, which compromises cell wall integrity and reduces the plant’s ability to cope with environmental stressors (Endler et al., 2015; Zhang et al., 2016a).
Salt stress inhibits cellulose synthesis, leading to altered cell wall structure and impaired plant growth (Table 2). This inhibition is partly due to the depolymerization of cortical microtubules (MTs), which disrupts the delivery and orientation of the cellulose synthase complex (CSC)—a plasma membrane-localized, multi-subunit protein complex responsible for polymerizing UDP-glucose into β-1,4-glucan chains that form cellulose microfibrils (Wang et al., 2007). The coordination between CSC and MTs is facilitated by the CC proteins, which act as molecular bridges by simultaneously binding to CSC and MTs, thereby aligning cellulose deposition with cytoskeletal dynamics. However, the precise feedback mechanism by which MTs rely on CSC—and ultimately the cell wall—for stress signal perception remains unresolved.
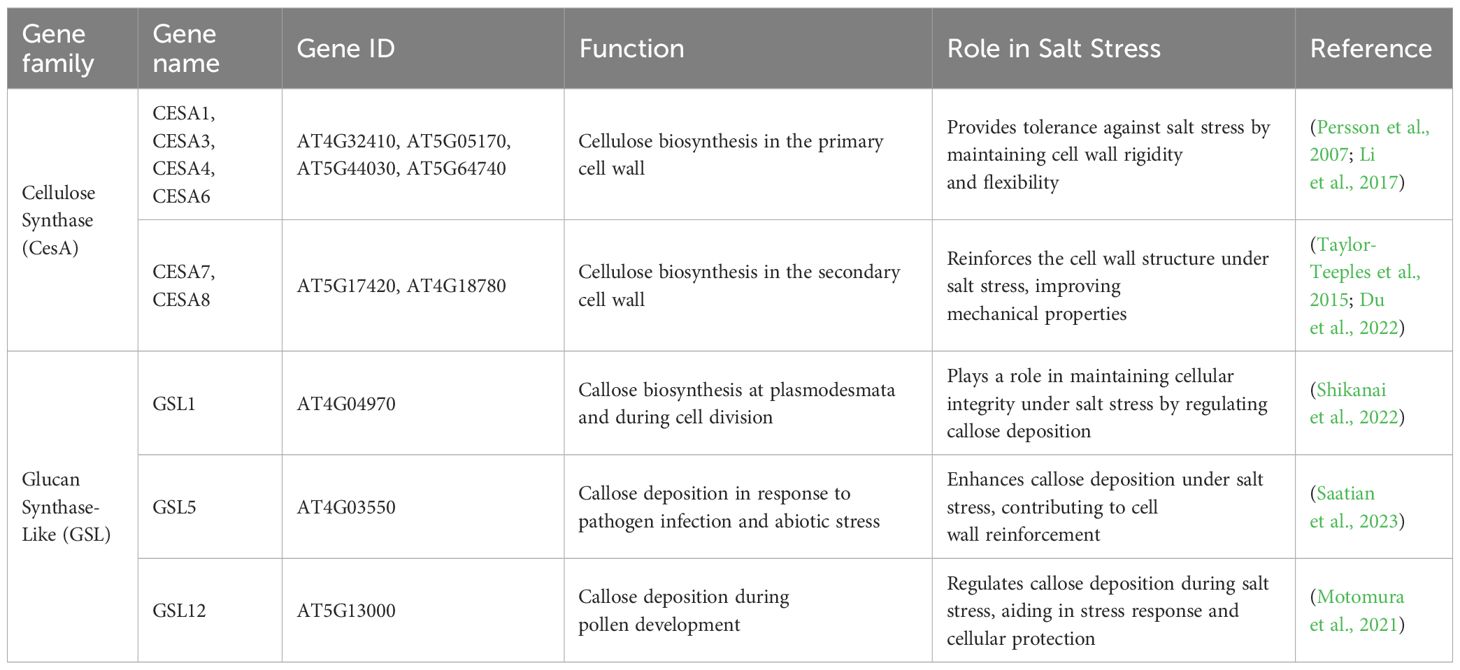
Table 2. Genes involved in cellulose biosynthesis, modification, and their roles under salt stress conditions.
Moreover, proper cellulose biosynthesis under salinity is regulated by protein glycosylation. The N-glycosylation pathway in the endoplasmic reticulum (ER) modulates salt tolerance and CSC functionality in a mature N-glycan-dependent manner. In particular, complex N-glycans are essential for the activity of KORRIGAN 1 (KOR1, also known as RADIALLY SWOLLEN2 or RSW2), a membrane-bound endo-1,4-β-glucanase that contributes to cellulose microfibril elongation (Kang et al., 2008). Additionally, transcriptomic analyses in the salt-tolerant Dendrobium officinale revealed that CELLULOSE SYNTHASE-LIKE A (CSLA) genes are upregulated under salt stress and are involved in the biosynthesis of mannan-type hemicelluloses, highlighting the broader impact of salt stress on non-cellulosic polysaccharide production (He et al., 2015).
2.2 Hemicellulose
Hemicellulose composition in the cell wall varies between species but is mainly composed of a polysaccharide backbone linked by β (1→4) linkages. Glucans with β (1→3, 1→4) links are grouped with xyloglucans, xylans, glucomannans, mannans, and other similar families (Scheller and Ulvskov, 2010). In dicotyledonous plants, xyloglucans (XyGs) typically constitute 20–25% of the primary cell wall and serve as the major hemicellulose component. In contrast, monocots possess type II primary walls in which glucuronoarabinoxylan dominate, and XyGs account for only 1–5% of total wall polysaccharides (Gigli-Bisceglia et al., 2020; Alvarez et al., 2024).
In dicots, XyGs form tightly bound complexes with cellulose, creating “hotspots” that play a central role in regulating wall mechanics. These hotspots bear localized mechanical stress during cell expansion and wall loosening, while pectins assist by modulating wall porosity and promoting cell elongation and division (Cosgrove and Jarvis, 2012). However, despite the structural significance of XyG-cellulose interactions, the precise biochemical impact of these hotspots on the mechanical properties of the cell wall remains incompletely understood and requires further investigation.
XyGs consists of a β-(1,4)-linked glucose backbone with α-(1,6)-linked xylosyl side chains. In some cases, these side chains are further modified with fucose or galactose, resulting in a complex, branched structure (Schultink et al., 2014; Pauly and Keegstra, 2016). The XyGs are a spacer polymer in the primary cell wall (Thompson, 2005; Anderson et al., 2010). Xylan is widely known as β-(1-4)-linked xylose residue decorated by glucuronic acid to form glucuroxylan. Xylan is mainly accumulated in the secondary cell wall; similarly, it is also present in some algae and monocots. The biosynthesis of XyGs takes place in the Golgi apparatus via glycan synthase and glycosyltransferases. XyGs are then exported to the plasma membrane through packing in vesicles and released into the extracellular matrix, where modification takes place via multiple extracellular enzymes, and eventually become part of the cell wall (Pauly and Keegstra, 2016).
Many studies have been conducted to better understand how hemicellulose, specifically xyloglucan, functions under salt stress (Table 3). Under salinity stress, xyloglucan is a metabolic inducer that triggers different physiological responses (Páez-Watson et al., 2020). The xyloglucan endotransglucosylase/hydrolase (XTH) enzyme plays a key role in altering cell wall morphology by cleaving and restructuring XyGs, thereby facilitating the reorganization of the cell wall matrix. The reduced end of XyG is then linked to the non-reducing end of another XyG polymer or oligomer. The gene CaXTH3 is involved in cell wall remodeling by accumulating starch content, which enables the plant mesophyll cell to retain humidity, and is actively involved in numerous cellular processes and salinity stress tolerance in transgenic Arabidopsis seedlings (Cho et al., 2006; González-Pérez et al., 2018). Additionally, the involvement of XTH19 and XTH23 in the brassinosteroid (BR) signaling elucidates how the BES1-dependent pathway affects lateral root development under salinity stress (Xu et al., 2020). Furthermore, XTH30 negatively regulates salt tolerance by promoting microtubule depolymerization and reducing crystalline cellulose deposition (Yan et al., 2019). Additionally, under salinity stress, the hemicellulose in the cell wall of Artemisia annua shows an increase in xylose content (Correa-Ferreira et al., 2019). The xyloglucan plays a significantly important role in defining the cell shape by regulating the cellulose microfibril’s loosening and tightening events during cell growth and maturation. However, little is known about how salt stress impacts the XyG structure and thus plant growth and development.
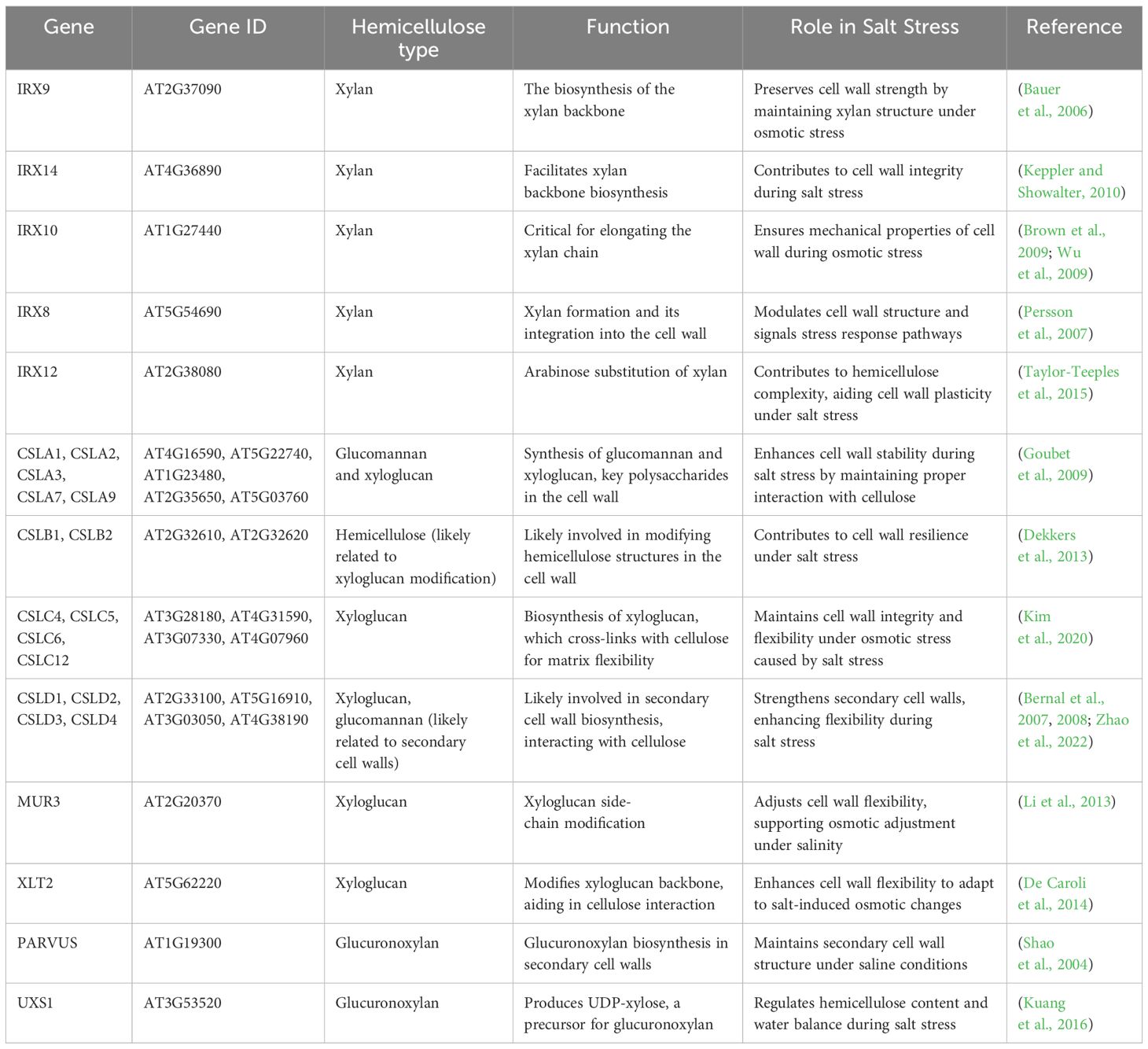
Table 3. Hemicellulose-related genes involved in cell wall synthesis and their roles under salt stress conditions.
2.3 Pectin
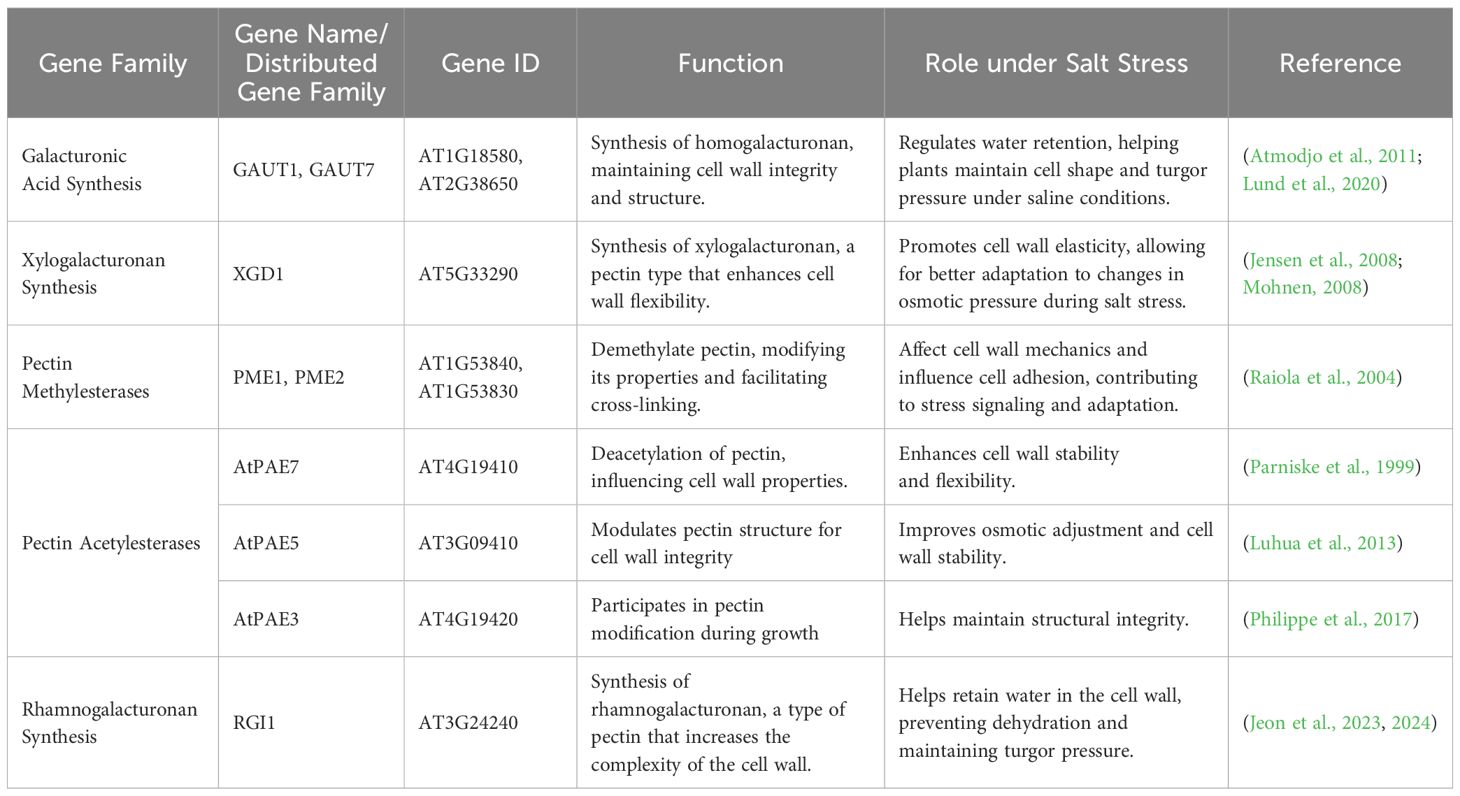
Pectin is a major component of the primary cell wall and the most diverse polysaccharide. Backbone of pectin consists of α-(1,4)-linked galacturonic acid (GalA), that are classified into three types: complex branched rhamnogalacturonan II (RG II), homogalacturonan (unbranched HG), and rhamnose-alternated (RG I), i.e., rhamnogalacturonan I α-(1,4) GalA and α-(1,2) rhamnose (Atmodjo et al., 2013; Amos et al., 2018). The pectin structure is strengthened by Ca2+-bridged HG and dimerized RG-II, which is also linked to boron ions (B3+). These interactions contribute to the strength of the primary cell wall in terrestrial plants (Liu et al., 2012; Oda and Fukuda, 2012; Kim et al., 2020). Pectin interacts with cellulose at RG-I blocks (arabinan galactan side chains) (Zykwinska et al., 2005). RG-II bonding is reinforced by the Ca2+ pectate bond. HG biosynthesis is mediated by galacturonosyl transferases (GAUTs). A detailed review of pectin biosynthesis and structure has been published by Zhang et al. (2021).
Pectin in the cell wall plays a vital role during cell proliferation and plant growth by providing flexibility (Peaucelle et al., 2012). FERONIA (FER) receptor kinases localized at the plasma membrane act as a sensor to sense pectin-associated cell wall damage during salinity stress (Feng et al., 2018; Verger and Hamant, 2018). Pectin methylesterase inhibitor (PMEI) negatively regulates salt resistance and reduces primary root growth (Jithesh et al., 2012). PMEIs maintain a high degree of pectin methyl esterification by inhibiting the activity of pectin methylesterases (PMEs). This suppression of PME activity limits the formation of calcium-mediated cross-links between homogalacturonan chains, thereby preserving cell wall plasticity. Such regulation is vital for maintaining primary root growth under saline conditions, as increased pectin methyl esterification enhances cell wall loosening and facilitates cell elongation and expansion (Peaucelle et al., 2011). The dynamic modulation of pectin structure through the PME–PMEI balance demonstrates a critical mechanism by which plants sustain root architecture and cellular integrity under abiotic stress.
Under salinity stress, the disruption of pectin structure plays a crucial role in root growth regulation (Chen et al., 2018). In developing plant cells, invasive Na+ competes with Ca2+ for binding sites on galacturonic acid residues in homogalacturonan, destabilizing the Ca2+-crosslinked structure. Similarly, the borate-crosslinked structure of GCN5 is also disrupted, weakening the integrity of the cell wall (Munarin et al., 2012). This structural compromise is sensed by FER, a cell wall integrity (CWI) sensor protein, which activates signaling pathways to mitigate the damage and restore growth (Feng et al., 2018). PMEIs, including salt tolerant Chorispora bungeana CbPMEI1, regulate PME activity, preventing excessive pectin demethylesterification that could otherwise increase the vulnerability of pectin to degradation under salt stress. Levels of pectin in the cell wall are modified by TRICHOME SPECIFIC DEFECTIVE 2 (TSD2) in rice, when exposed to salinity stress (Fang et al., 2019). Pectin methylesterase 31 (PME31), localized at the plasma membrane, positively modulates salt stress tolerance in Arabidopsis by decreasing RD29A, DREB2A, and RD29B expression, thereby reducing stress-induced ABA signaling and osmotic stress responses, ultimately contributing to cell wall stability and adaptive growth under saline conditions (Yan et al., 2018). Pectin plays a key role in regulating cell growth, and under salt stress, cellulose content increases while pectin levels decrease (Table 4). These findings suggest that salt stress enhances the rigidity of the cell wall, which inhibits root growth (An et al., 2014).
2.4 Secondary cell wall (lignin)
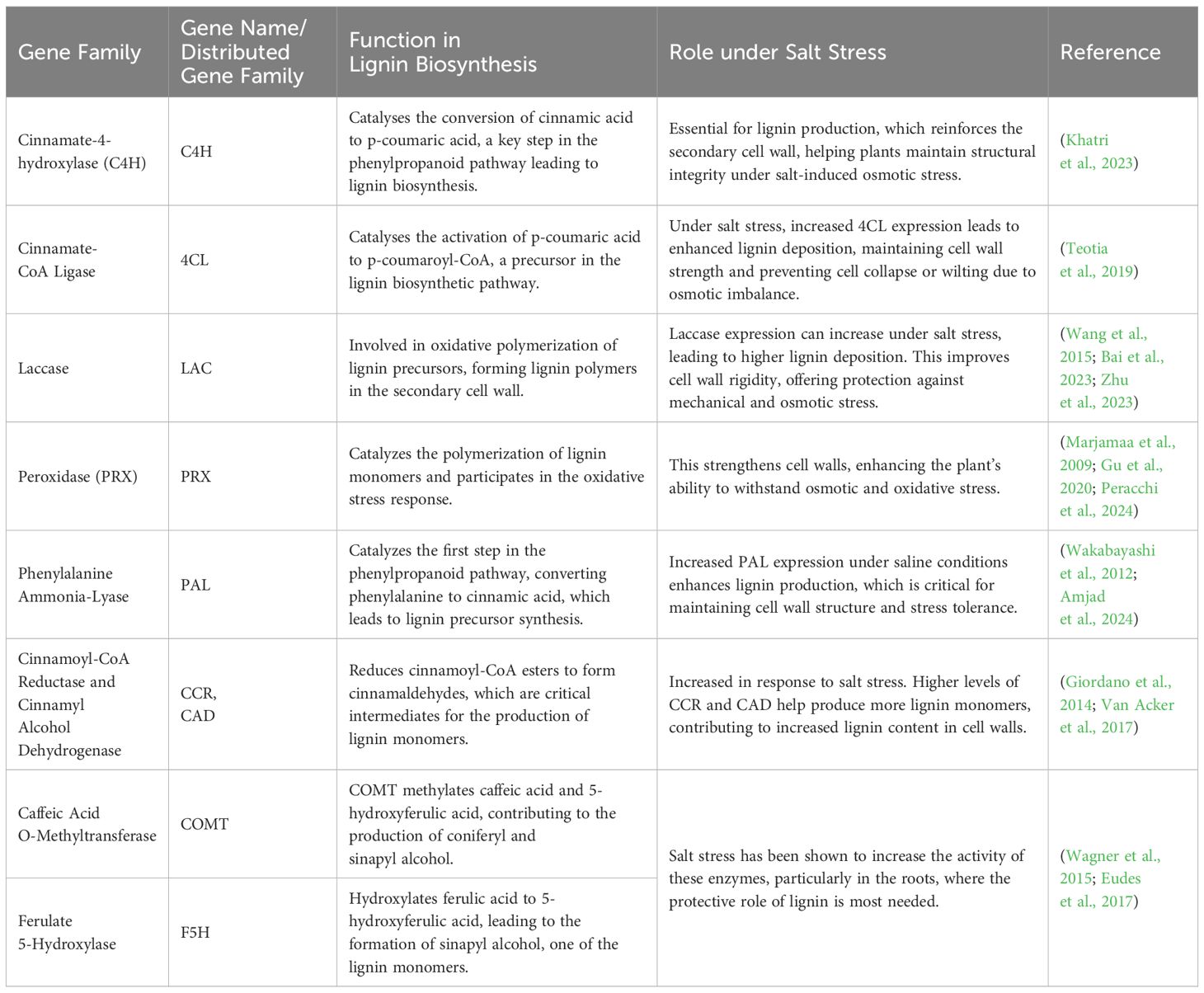
The secondary cell wall in plants mainly consists of cellulose, hemicellulose (primarily xylans and glucomannans), and lignin, along with various structural proteins and enzymes. Lignin is hydrophobic in nature, forms a substantial part of secondary cell wall biomass, and contributes to the rigidity of the plant (Scheller and Ulvskov, 2010). Both the quantity and composition of lignin varies across plant species. Lignin is structurally a complex polyphenolic polymer that integrates into cellulose and hemicellulose networks, providing mechanical strength, hydrophobicity, and rigidity to the secondary cell wall. It is primarily composed of three monolignols, i.e., coniferyl alcohol, ρ-coumaryl alcohol, and sinapyl alcohol, which are represented as guaiacyl (G), ρ-hydroxyphenyl (H), and syringyl (S), respectively, in the lignin polymer (Bonawitz and Chapple, 2010). The composition of these monolignols differs between species and cell types. The biosynthesis and acylation of lignin (monolignols) have been briefly described (Vanholme et al., 2019). Salt stress alters lignin concentration (Table 5). In response to salinity stress, maize cell walls decrease polysaccharides and arabinosyl feruloylation in arabinoxylan while increasing S-unit lignin polymers (Oliveira et al., 2020). Genotype-specific modifications in cell wall composition, including lignin and polysaccharide dynamics, play a critical role in determining plant tolerance to salt and drought stress by influencing wall plasticity and stress adaptability (Calderone et al., 2024). Salt stress increases lignin deposition in the root tracheary elements. A model was proposed in which increased lignin deposition limits ion uptake, enhances selective water transport through the symplastic pathway, reduces apoplast water flow, and restricts root growth (Sanchez-Aguayo et al., 2004). The change in lignin content and composition in response to abiotic and biotic stress has been briefly discussed by Moura et al. (2010), and recent studies suggest that selective downregulation of lignin can further improve salt stress tolerance by enhancing cell wall flexibility and facilitating better ion homeostasis in plants (Dokka et al., 2024).
Recent studies have highlighted the genetic and molecular regulation of lignin biosynthesis and its critical involvement in enhancing salt stress tolerance through diverse signaling and transcriptional networks. Mutation of histone acetyltransferase GCN5 suppresses the expression of chitinase like gene 1 (CTL1), which is essential for cellulose and lignin synthesis and salt tolerance in Arabidopsis (Zheng et al., 2019). The overexpression of Superoxide Dismutase (SOD), and Ascorbate Peroxidase (APX) in Solanum tuberosum L. induces cell wall lignification and promotes expression of different associated transcription factors during salt stress (Shafi et al., 2017). In apple, MYB46 enhances salt stress tolerance by increasing lignin accumulation and secondary cell wall synthesis and activates ABA-dependent and independent pathways that ultimately trigger various other defense responses (Chen et al., 2019). Overexpression of the sweet potato SWPA4 gene in transgenic tobacco increases lignin deposition, hydrogen peroxide (H2O2) levels, and transcription of apoplast-related genes, which promote salt tolerance. Similarly, the two cultivars of soya bean with contrasting salt tolerance showed increased cell wall cellulose, pectin, and uronic acid under salt stress (An et al., 2014). Non-methylated uronic acid in the leaf cell wall contributes to salt resistance by reducing Na+ accumulation in plant tissues (Uddin et al., 2014). The dynamic regulation of lignin content and composition under salt stress reinforces cell wall integrity, modulates water and ion transport, and enhances stress resilience, highlighting its crucial role in plant adaptation to saline environments. These structural variations shape their remodeling responses, including differences in pectin de-esterification, lignification, and reactive oxygen species (ROS) signaling (Haas and Peaucelle, 2021).
2.5 Cell wall-associated proteins
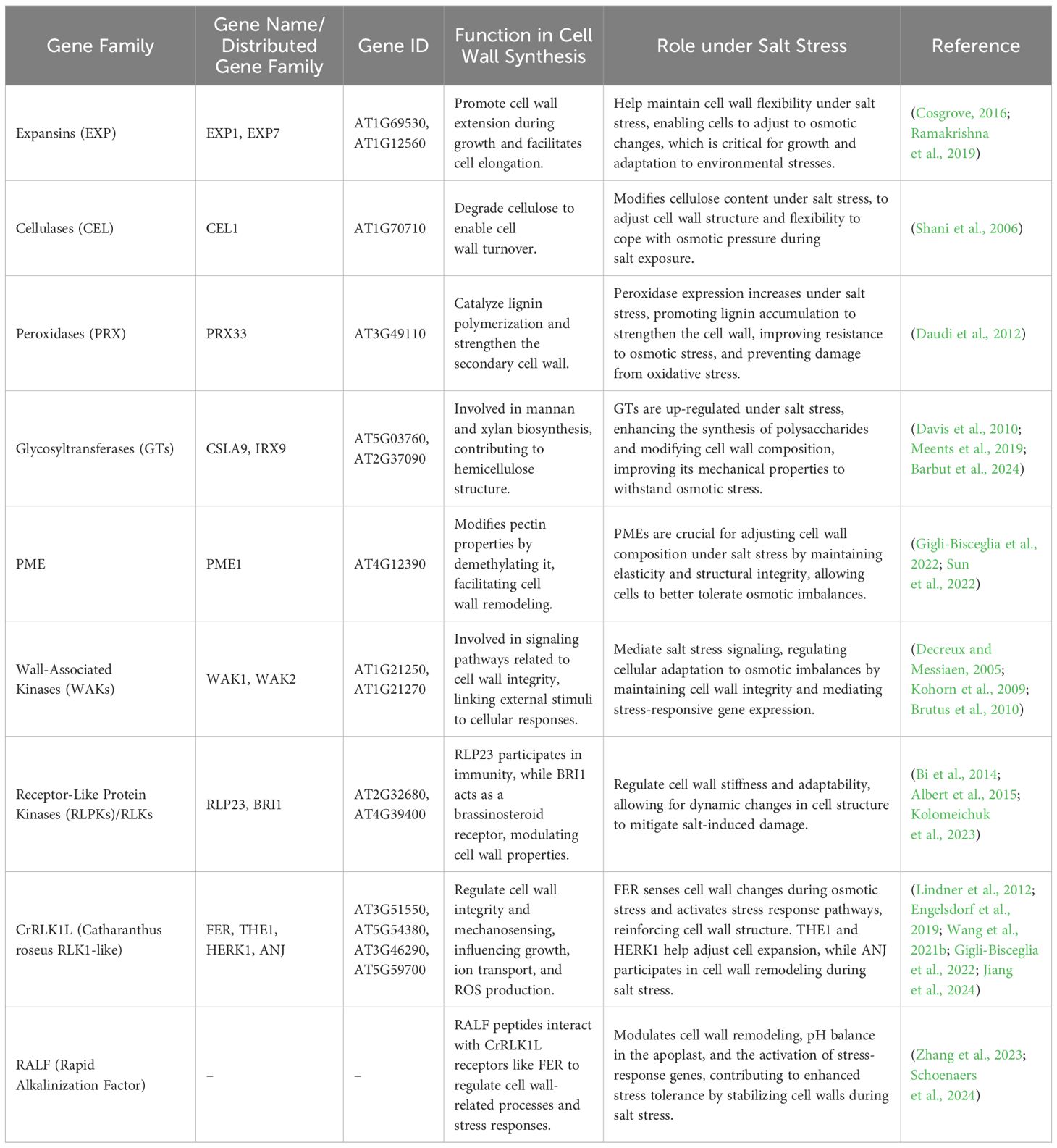
The plant cell wall is not only vital for structural integrity but also for transducing environmental signals. Mechanical measurements using atomic force microscopy and Fourier-transform infrared analysis revealed that the cell wall structure is mechanically homogeneous; nevertheless, during the growth phase, stiffness increases on average as heterogeneity grows (Radotic et al., 2012). The assembly and regulation of cell wall proteins are essential for plant function, particularly in response to salt stress (Table 6). The heterotrimeric Gβ subunit AGB1 interacts with the receptor-like kinase FER and the peptide ligand RALF1 to modulate salt stress responses, highlighting a key signaling module in cell wall-associated salinity sensing (Yu and Assmann, 2018). LRXs further contribute to signaling pathways, interacting with RALF peptides, and coordinating growth regulation under stress conditions (Zhao et al., 2018; Wang and Gou, 2020). Structural proteins such as extensins play a vital role in maintaining cell wall architecture (Lamport et al., 2011). STELLO (STL) proteins in the Golgi facilitate CSC formation and may play a role in glycosylation, although their specific substrates are unknown (Zhang et al., 2016b). The CSC protein secreted from the endomembrane to the plasma membrane is altered depending on different aspects, such as the actin cytoskeleton via polymerization and depolymerization (Zhang et al., 2019), the pH of the endomembrane system (Luo et al., 2015), and interaction with proteins such as SHOU4 and TRANVIA (Polko et al., 2018; Vellosillo et al., 2021). Cortical microtubules coordinate the delivery of CSC to the plasma membrane (Gutierrez et al., 2009). Plants tolerate salt stress by altering their protein profile, i.e., salt-tolerant cultivars often express critical proteins more efficiently than salt-sensitive plants (Dissanayake et al., 2022), which shows that the quality of proteins is more important than quantity. Under salt stress, proteins including osmotin (N. tabacum) and gramin (O. sativa) play a vital role in maintaining cellular activities, while Na+ ion toxicity is alleviated by heat shock proteins (HSPs) and late embryonic abundant (LEA) proteins (Chourey et al., 2003; Derevyanchuk et al., 2016; Kumar et al., 2022). Despite the challenges in classifying cell wall proteins due to their diverse functions, recent classification has identified various categories, including those involved in metabolism, signaling, and structure (such as extensins, cellulose synthases, and pectin-modifying enzymes). Understanding the complex interplay of these proteins in stress perception and response is crucial for developing resilient crop varieties, as ongoing research seeks to explore the precise mechanisms by which the CWPs facilitate plant adaptation to environmental challenges.
3 Cell wall integrity is compromised during salt stress
The cell wall plays a crucial role in maintaining structural stability, regulating growth, and mediating stress responses. CWI can be compromised at various levels, resulting in altered physiological and biochemical responses that differ based on species, tissue type, and stress intensity. The CWI surveillance system monitors the condition of the cell wall, which is essential for stress perception and response (Vaahtera et al., 2019; Rui and Dinneny, 2020). Several cell wall sensors involved in CWI monitoring have been identified. Plants sense salinity at multiple cellular sites, primarily the plasma membrane, cell wall, and intracellular organelles (Zhu, 2016). Salinity stress response involves a large family of plasma membrane-localized and cell wall-associated Catharanthus roseus receptor-like-kinase-1-like (CrRLK1L) proteins, of which 17 members have been found in Arabidopsis (Lindner et al., 2012). These proteins interact with cell wall components, facilitating interactions between the cell wall and interior cellular processes (Feng et al., 2018). Other proteins, such as those from the wall-associated kinases (WAK) and leucine-rich repeat (LRR) receptor kinase families, are also thought to bind cell wall components and generate intracellular responses during cell wall changes (Herger et al., 2019; Lou et al., 2020). Perturbations in CWI can affect cellular turgor pressure, leading to shifts in membrane tension and cell wall stress. This, in turn, activates mechanosensitive channels in the plasma membrane, promoting ion accumulation and initiating downstream signaling processes (Hamann et al., 2004; Hamann, 2012; Bacete and Hamann, 2020). Consequently, the composition and integrity of the primary cell wall must be tightly monitored and regulated to support cell expansion and rapidly adapt to changing environmental conditions. In addition to the cell wall, cellular organelles such as chloroplasts and the ER are involved in stress sensing. Chloroplasts not only house essential photosynthetic proteins but also play a role in detecting salt stress, with mechanisms involving the regulation of ion transport and ROS production. The ER, which manages calcium homeostasis, also processes stress-related proteins, with specific proteins like ZmSep15-like-2 shown to mitigate salt-induced oxidative stress (Zhu et al., 2019).
Salt stress alters the lipid components of the plasma membrane, affecting tension and facilitating binding of Na+ by glycosyl inositol phosphorylceramide (GIPC) sphingolipids, crucial for salt-triggered Ca2+ influx (Jiang et al., 2019). In Arabidopsis, the salt stress pathway involves calcineurin B-like protein SOS3 (CBL4), CBL-interacting protein kinase SOS2 (CIPK24), and Na+/H+ antiporter SOS1, which work together to coordinate Na+ extrusion and mitigate toxicity (Yang and Guo, 2018; Song et al., 2024). During salt and osmotic stress, vacuoles play a crucial role in regulating Na+ levels through Na+/H+ exchange, driven by proton gradients formed by vacuolar H+-ATPases. These processes help maintain cellular ion balance, while stress-induced changes like turgor reduction and membrane tension alterations trigger complex biochemical signaling cascades (Nongpiur et al., 2020; Rui and Dinneny, 2020). Proteins such as QIAN SHOU KINASE (QSK1) rapidly localize to plasmodesmata in response to osmotic stress, though their specific role in these signaling pathways remains unclear (Grison et al., 2019). Membrane tension changes are also sensed by OSCA1, which may regulate cytosolic Ca2+ levels as part of the stress response (Yuan et al., 2014). Additionally, the RAF-SnRK2 pathway is activated, mediating key processes involved in ABA and osmotic stress responses, highlighting the intricate network of mechanisms plants use to adapt to saline environments (Boudsocq et al., 2004; Takahashi et al., 2020).
3.1 ROS and the cell wall in salt stress
Reactive oxygen species (ROS) are oxygen-derived molecules that function as both essential signaling mediators and potential inducers of cellular damage under stress conditions. The major forms of ROS include superoxide anion (·O2-), hydrogen peroxide (H2O2), hydroxyl radical (·OH), and singlet oxygen (1O2). Salinity triggers osmotic, ionic, and oxidative stress in plants (van Zelm et al., 2020). Initially, osmotic stress caused by decreasing water supply reduces cellular turgor, impacting cell expansion and tissue retraction. To retain cell integrity, plants actively adjust their cell walls to reinforce structure and restore osmotic potential (Zonia and Munnik, 2007). Mutants with impaired cell wall biosynthesis, such as cesa6 and mur4, display pronounced growth inhibition under salt stress, underscoring the essential role of CWI in maintaining structural resilience and enabling adaptive responses to salinity (Zhang et al., 2016a; Zhao et al., 2020). Excessive Na+ accumulation causes ionic stress that disturbs enzymatic activity essential to metabolism and ion balance, which in turn affects the mechanical characteristics of cell walls, especially of pectin. Increased Na+ levels compete with Ca2+, disrupting pectin cross-linking, whereas salt stress activates pectin methyl esterases, affecting cell wall composition (Beauzamy et al., 2015; Gigli-Bisceglia et al., 2022). While ROS generated during salt stress can function as secondary messengers that allow peroxidases to remodel cell walls, excessive accumulation of ROS can cause oxidative damage and even plant death (Ma et al., 2012; Tenhaken, 2014). Thus, maintaining a balance of ROS levels is critical for plant survival in saline conditions. Mutants with deficient ROS regulation have revealed a complex relationship between plant ROS homeostasis, cell wall dynamics, and salt stress (Hong et al., 2020; Zhou et al., 2021). The synthesis of ROS during salt stress has both positive and negative impacts on plant growth. While high ROS can lead to oxidative damage and compromise cell integrity, low ROS can promote stress tolerance and cell wall remodeling (Liu et al., 2012). The balance between ROS generation and scavenging is crucial for maintaining CWI. Mutants with decreased ROS scavenging systems exhibit increased sensitivity to salinity (Hazman et al., 2015). Enzymes such as SOD and peroxidases help regulate oxidative stress. Furthermore, ROS-induced post-translational changes of cell wall components may influence the mechanical characteristics of the cell wall, influencing plant resilience under salt stress (Karkonen and Kuchitsu, 2015).
3.2 Microtubule dynamics under salt stress
Plant with decreased anisotropic growth under salt stress contain disrupted cellulose assembly and microtubule dynamics. Salt stress changes the dynamics of CSCs and microtubules in Arabidopsis, resulting in the removal of CesAs from the plasma membrane and microtubule depolymerization quickly after salt exposure (Komis et al., 2002; Wang et al., 2007; Gutierrez et al., 2009; Crowell et al., 2010). Moreover, the actin cytoskeleton significantly contributes to plant responses under salt stress, influencing cellulose synthase distribution and overall cell wall organization. However, after several hours of restoration of non-saline condition, microtubules reassemble and CesAs return to the membrane, indicating that plants possess adaptive mechanisms for cellulose synthesis during recovery from salt stress (Wang et al., 2007; Endler et al., 2015). The recovery of microtubule organization during salt stress depends on critical proteins like CC1 and CC2, which are essential components of CSCs. Mutants deprived of proper functioning of these proteins exhibit decreased growth, particularly in saline environments (Endler et al., 2015). Salt tolerance and CC1 function are impaired when microtubule-binding motifs are disrupted, as this prevents proper interaction between CC1 and microtubules, hindering cellular processes essential for stress adaptation (Kesten et al., 2022). While there is no direct animal tau protein homolog in plants, similarities in microtubule-binding properties suggest that plant proteins, such as CC1, may perform a similar role in regulating microtubule dynamics. Tau proteins in animals stabilize microtubules by binding to them, and it is hypothesized that salt stress might affect CC1, possibly through phosphorylation. This modification could alter the interaction between CC1 and microtubules, thereby impacting microtubule stability and cellular processes crucial for the plant’s response to salt stress (Mietelska-Porowska et al., 2014). Another key component of the CSC is KOR1, an endoglucanase found in the CSC that is internalized during salt stress and contribute to cellulose production (Nagashima et al., 2020). However, more research on KOR1 trafficking is needed, as pharmacological agents such as phenylarsine oxide and 1-butanol used to deduce its function also affect broader signaling pathways and may not specifically target KOR1 trafficking (McLoughlin et al., 2013; Gujas et al., 2017). Interestingly, phosphatidic acid (PA), associated with resistance to salt stress, functions in cellular signaling and may impact the localization of KOR1 (Zhang et al., 2012). Understanding this interaction is critical for determining plant responses to salt stress and controlling cellulose production.
Salt stress significantly disrupts cytoskeletal organization, which in turn affects CWI and plant tolerance mechanisms. Microtubule dynamics, in particular, are crucial in coordinating cellular responses to salinity. Chemical inhibitors like oryzalin (a microtubule depolymerizer) and taxol (a stabilizer) have been shown to markedly influence plant salt tolerance (Wang et al., 2007). Microtubule stability is maintained by proteins like MAP65-1, which enhances microtubule bundling, and its loss of function increases vulnerability to salt stress (Zhang et al., 2012). PA produced by phospholipase D, not only activates MAP65–1 but also interacts with MPK6, together contributing to microtubule organization and salt tolerance (Zhou et al., 2017). MPK6 further coordinates salt stress responses by linking cytoskeletal stability to broader signaling networks, including the SOS pathway (Yu et al., 2010). Additional microtubule-associated proteins, such as RIC1, facilitate microtubule reassembly after salt-induced disruption, while recently identified tropomyosin-like (TTL) proteins stabilize microtubules and interact with the CSC, directly linking cytoskeleton dynamics to cell wall biosynthesis and stress resilience (Kesten et al., 2022). Actin filaments are also critical, with salt stress-induced expression of actin depolymerizing factor 1 (ADF1) supporting filament remodeling, and loss of ADF1 leading to defective actin organization and reduced seedling survival (Wang et al., 2021a). These cytoskeletal adjustments are tightly integrated with CWI sensing, which relies on plasma membrane-localized receptors such as FER and other RLKs that detect wall perturbations and trigger Ca2+ influx during salt stress (Jose et al., 2020). The interaction of signaling modules like LRX3/4/5 with RALFs further highlights the complexity of CWI perception and its regulation under stress (Zhao et al., 2018). While these studies advance our understanding of the cytoskeleton–cell wall interface in salinity responses, the downstream intracellular signaling pathways, particularly those centered on MPK6, remain poorly understood. Elucidating these pathways will be key to developing targeted strategies for improving salt tolerance in crops.
The interplay between the cytoskeleton and CWI is crucial for plant adaptation to salt stress. Microtubule dynamics are linked to cellulose synthesis, while actin filaments also contribute to cell wall organization. Salt stress affects the organization and stability of both cytoskeletal components, leading to altered cell wall properties. For instance, ADF influences the distribution of CSC, and its expression is modulated by salt stress (Crowell et al., 2010; Wang et al., 2021a). The coordination between microtubules and actin is essential for maintaining cell shape and growth under saline conditions, highlighting the importance of cytoskeletal regulation in stress responses.
4 Cell wall remodeling under salt stress
4.1 Salt stress alters cell wall growth and extensibility
In plant cells, the cell wall and plasma membrane are kept intact through turgor pressure. The gap between them is filled with plasma membrane proteins such as Arabinogalactan proteins (AGPs), Wall-Associated Kinases (WAKs), Fasciclin-like arabinogalactan proteins (FLAs), and CesA complexes. These proteins maintain the delicate balance between cell wall extensibility and turgor pressure. This balance is critical for plant cell growth, development, and division, ensuring that plants can grow and adapt to their environments. Thus, the interaction of these components is vital to the life cycle of a plant, driving its growth and structural integrity (Wolf and Greiner, 2012; Watanabe et al., 2015).
PME plays a significant role in cell wall extensibility and remodeling. It actively contributes to the loosening of the cell wall through the action of polygalacturonase, which degrades HG. This process is ultimately essential for cell adhesion and defense responses in plants. PME modifies the ionic content, cellular adhesion, and pH of plants under stress, influencing development through a mechanism called “acid growth” (Pelloux et al., 2007). Cell expansion and cell wall synthesis include four major processes, which are outlined in the review of Wolf and Greiner (2012). ROS are released when the cell wall senses changes. As a result of internal turgor pressure, the cell wall deforms, hydrates, and relaxes, and eventually, secretes new wall material. Cell wall extension in plants is primarily regulated by several key hormones. ROS are important players in cell wall development, as discussed by Karkonen and Kuchitsu (2015) (Figure 2). In the context of the cell wall, ROS act as agents that help with loosening and signaling various biological processes. They are produced by several enzymes (NADPH oxidases, quinone reductases, amine oxidases, lipoxygenases, oxalate oxidases, class III peroxidases), while various scavenger enzymes balance ROS levels, which is crucial for plant health. Auxin, a key plant hormone, promotes cell elongation by encouraging wall stretching and modification (Majda and Robert, 2018). However, when the cell wall stops expanding, it becomes rigid. This rigidity is due to the formation of strong connections known as di- and higher oligomer bridges, which are tightened further by ferulate coupling (Fry, 2004; Harris and Trethewey, 2009). H2O2 also contributes to this process, as it can inhibit cell wall growth through certain crosslinking reactions (Cona et al., 2006).
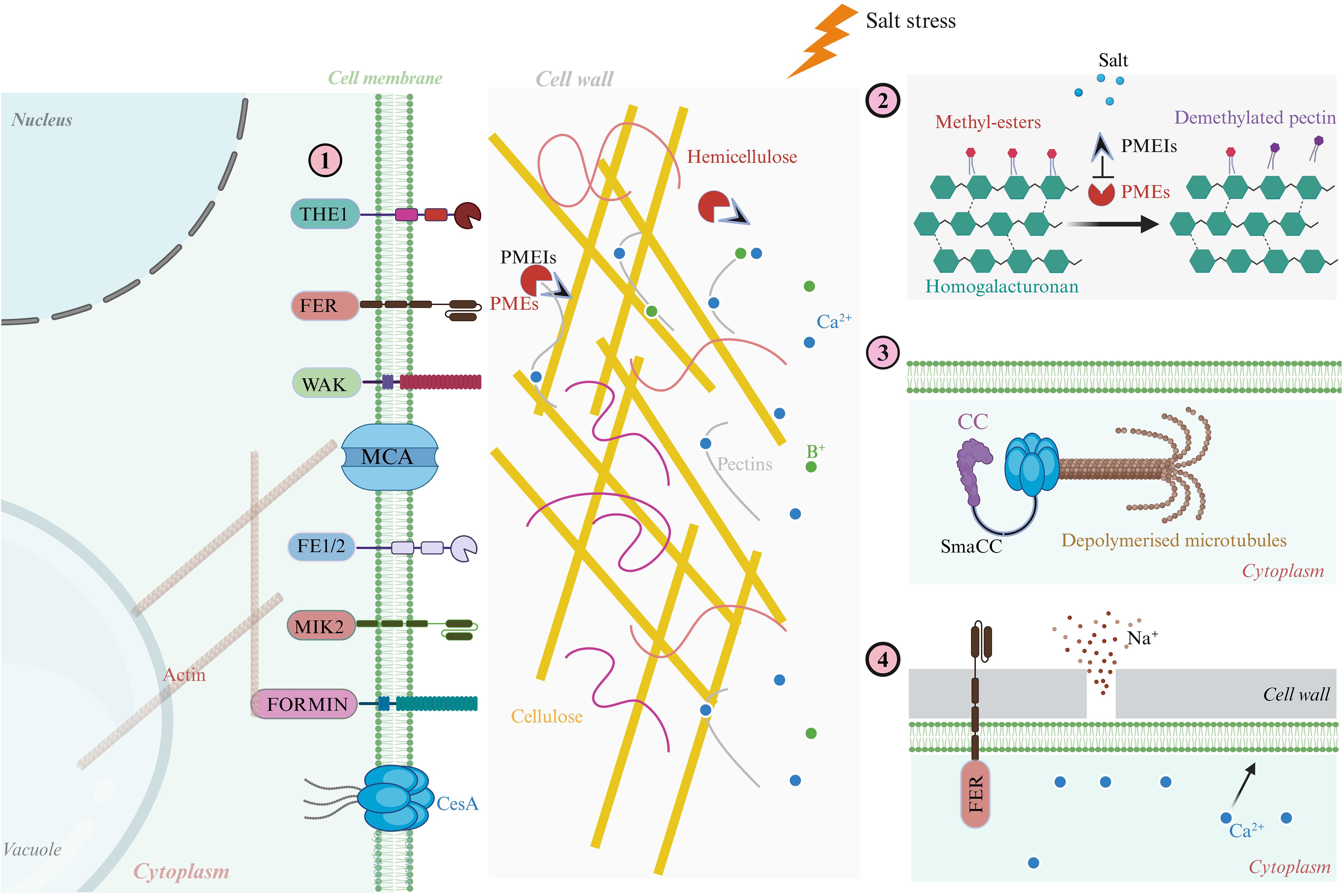
Figure 2. Cellular mechanisms of plant cell wall growth and extensibility under salt stress. This schematic illustrates how salt stress impairs cell wall extensibility by disrupting CWI, microtubule dynamics, ion balance, and calcium-pectin interactions, while also outlining mechanisms of recovery. (1) Under optimal conditions, key structural and signaling proteins such as AGPs, WAKs, FLAs protein, and CesAs, coordinate cell wall deposition and expansion, maintaining mechanical stability and enabling anisotropic growth. (2) PME modifies homogalacturonan domains, enhancing wall plasticity and promoting cell elongation. (3) Salt-induced ionic stress (notably Na+ influx) triggers cortical microtubule depolymerization, which impairs CesA trafficking and cellulose synthesis, compromising wall stiffness and directional expansion. (4) Upon stress alleviation, the receptor-like kinase FER modulates calcium–pectin cross-linking, restoring wall elasticity and supporting structural repair. Importantly, proteins such as FEI1/FEI2, MIK2, and FORMIN act as mechanosensors and regulators that bridge cytoskeletal organization with CWI maintenance, playing essential roles in signal transduction and cytoskeleton–cell wall coupling under salinity stress. These components collectively enable plants to perceive wall damage, transduce stress signals, and initiate adaptive remodeling responses crucial for survival in saline environments.
The cytoskeleton and associated proteins also play a critical role during plant cell division. Microtubule and actin filament structures are essential for this process, facilitating the synthesis of new cell walls through a specialized structure called the phragmoplast, which directs vesicles during cell division (Rasmussen et al., 2013). When plants face elevated salinity stress, they must adapt quickly to ensure proper cell expansion and resource utilization. Salt stress can lead to overaccumulation of Na+, causing K+ to efflux and increase ROS levels. This imbalance can lead to oxidative damage, disrupting turgor pressure and osmotic stability within the cells (Krasensky and Jonak, 2012; Zhang and Shi, 2013). One key player in this response is the CesA enzyme, which regulates cellulose synthesis to help manage the challenges posed by salt stress (Wang et al., 2016). Salt stress also destabilizes (depolymerizes) the microtubular structures of cells, causing CesA complexes in the membrane to become internalized (Paredez et al., 2006). A proposed model in which the CC protein interacts with microtubules and CesAs to maintain cellulose production and support plant growth during salt stress (Endler et al., 2015). BR, a class of plant hormones, also plays a vital role in this process. BR signaling may influence cell wall remodeling by regulating genes and enzymatic activities related to cell walls, particularly in grasses under stress (Rao and Dixon, 2017).
Additionally, proteins like RSA/MUR3/KAM1 help maintain the organization of actin microfilaments, enhancing salt tolerance by reducing ROS-induced damage (Li et al., 2013). During stress conditions, peroxidases utilize ROS in the apoplast to catalyze the cross-linking of phenolic compounds and glycoproteins in the cell wall, which helps to strengthen the wall and improve stress tolerance. This interaction reduces cell wall extensibility and limits cell expansion (Tenhaken, 2014). Interestingly, with prolonged salt exposure, cell wall extensibility can increase due to higher concentrations of ROS, specifically hydroxyl radicals. Enzymes such as xyloglucan-modifying enzymes and expansins in the apoplast can cleave glycosidic bonds in the cell wall, facilitating growth recovery (Tenhaken, 2014). Furthermore, the FER protein is essential for sensing salt stress and aiding cell recovery. In the absence of FER, cells may burst during recovery. It is believed that Na+ ions disrupt Ca2+ pectin cross-links in the cell wall, but FER helps balance Ca2+ levels by promoting cytosolic calcium, which is crucial for growth recovery (Feng et al., 2018). Understanding how these hormones and cellular mechanisms interact during salt stress can provide insights into enhancing plant resilience and improving agricultural practices (Mao et al., 2023).
4.2 Role of cutin and wax deposition in response to salt stress
The plant cuticle is primarily composed of cutin and waxes, forms a lipid-rich barrier over epidermal cell walls and plays a critical role in stress adaptation. Unlike the polysaccharide matrix of the inner wall, cuticular deposition is uniquely positioned to regulate non-stomatal water loss, ion exclusion, and mechanical protection traits vital under salt stress. Salt stress has been shown to induce transcriptional reprogramming of cuticle biosynthesis genes. For instance, studies in Arabidopsis thaliana and rice have demonstrated that salinity upregulates genes such as WIN1/SHN1, CER1, LACS2, and WSD1, which are involved in wax synthesis, transport, and polymerization (Liu et al., 2019; Tiika et al., 2025). Enhanced wax deposition under salinity improves leaf surface hydrophobicity and reduces passive ion influx, thereby contributing to ionic homeostasis.
Similarly, cutin biosynthesis is altered during salt exposure. The expression of genes such as GPAT6, CYP86A2, and LCR is responsive to salinity and modulates the ester-linked polyester matrix of the cuticle. Overexpression of GPAT6 in Solanum lycopersicum was found to enhance both cutin monomer levels and salt stress tolerance by maintaining water potential and delaying ion toxicity (Petit et al., 2016). In addition to structural roles, cuticle-derived signals, such as fatty acid precursors and long-chain aldehydes, activate downstream ABA-dependent pathways or interact with ROS signaling, thereby integrating surface defense with internal stress responses (Lü et al., 2009). Collectively, these findings suggest that cuticle reinforcement through wax and cutin deposition is a critical but often underappreciated component of the plant’s salt stress response. By limiting water loss and serving as a biochemical shield, the epidermal barrier enhances plant survivability in saline environments. Incorporating cuticle modifications alongside cell wall remodeling provides a comprehensive understanding of plant surface adaptations to abiotic stress.
5 Anisotropic growth under salt stress
Anisotropic growth (the directional expansion of plant cells) is required for organ formation and overall plant architecture. A plant’s ability to grow plastically depends on the interaction of its cytoskeleton, plasma membrane, and cell walls. Although the relationship between the plasma membrane and the cytoskeleton has been extensively studied, the relationship between the plasma membrane and the cell wall itself is still unclear (Feraru et al., 2011; Liu et al., 2015). Cell wall orientation and mechanical properties are key determinants of anisotropic expansion. In silico modeling shows that pectin-cellulose interactions guide cell wall alignment and support elongation, particularly in structures like hypocotyls (Bou Daher et al., 2018). The orientation of cellulose fibers has been described as similar to a ‘hoop on a barrel’, highlighting its role in determining cell elongation. Cellulose fiber orientation varies significantly inside elongating cells. In epidermal cells, for example, cellulose fibers are more perpendicular on the inner faces, providing structural resistance to expansion, while on the outer faces, they exhibit a more random orientation to facilitate outward cell elongation (Chan et al., 2010, 2011; Crowell et al., 2011).
The salt stress significantly disturbs the coordination mechanism, resulting in altered cell shape, disrupted tissue patterning, and reduced growth and yield (Figure 3). Excessive Na+ accumulation disrupts cellular homeostasis, disrupting the molecular and structural networks that control anisotropic growth. The cell wall, plasma membrane, and cytoskeleton form a dynamic interface that controls cell growth, but salt disturbs this interface on multiple levels. According to Baskin and Jensen (2013) individual cells have limited influence over their fate because they are integrated among tissues and organs. Under certain conditions, isotropic growth may result from stress-induced feedback processes. That challenges our understanding of cell wall dynamics. The cortical microtubules, which guide cellulose synthase movement, are highly sensitive to ionic imbalance (Wang et al., 2007). The dwarf mutant any1 (anisotrophy1) exhibits reduced anisotropic development in roots, shoots, and trichomes, as well as decreased cell wall crystallinity and cellulose synthase complex velocity during expansion (Fujita et al., 2013). Most plants that are exposed to salt stress cease growing [216]. Anisotropic development under these conditions is significantly affected by modifications in cell wall architecture. As demonstrated by the function of SPIRAL1 (SPR1) in preventing anisotropic growth in Arabidopsis thaliana, the architecture of cortical microtubules is sensitive to ion homeostasis and can be harmed by salt stress (Nakajima et al., 2006). According to Buschmann and Borchers (2020), a study showing that cell expansion depends on signals from both apoplastic and cytoplasmic sources, salt stress can also alter intrinsic root development directions through microtubule depolymerization.
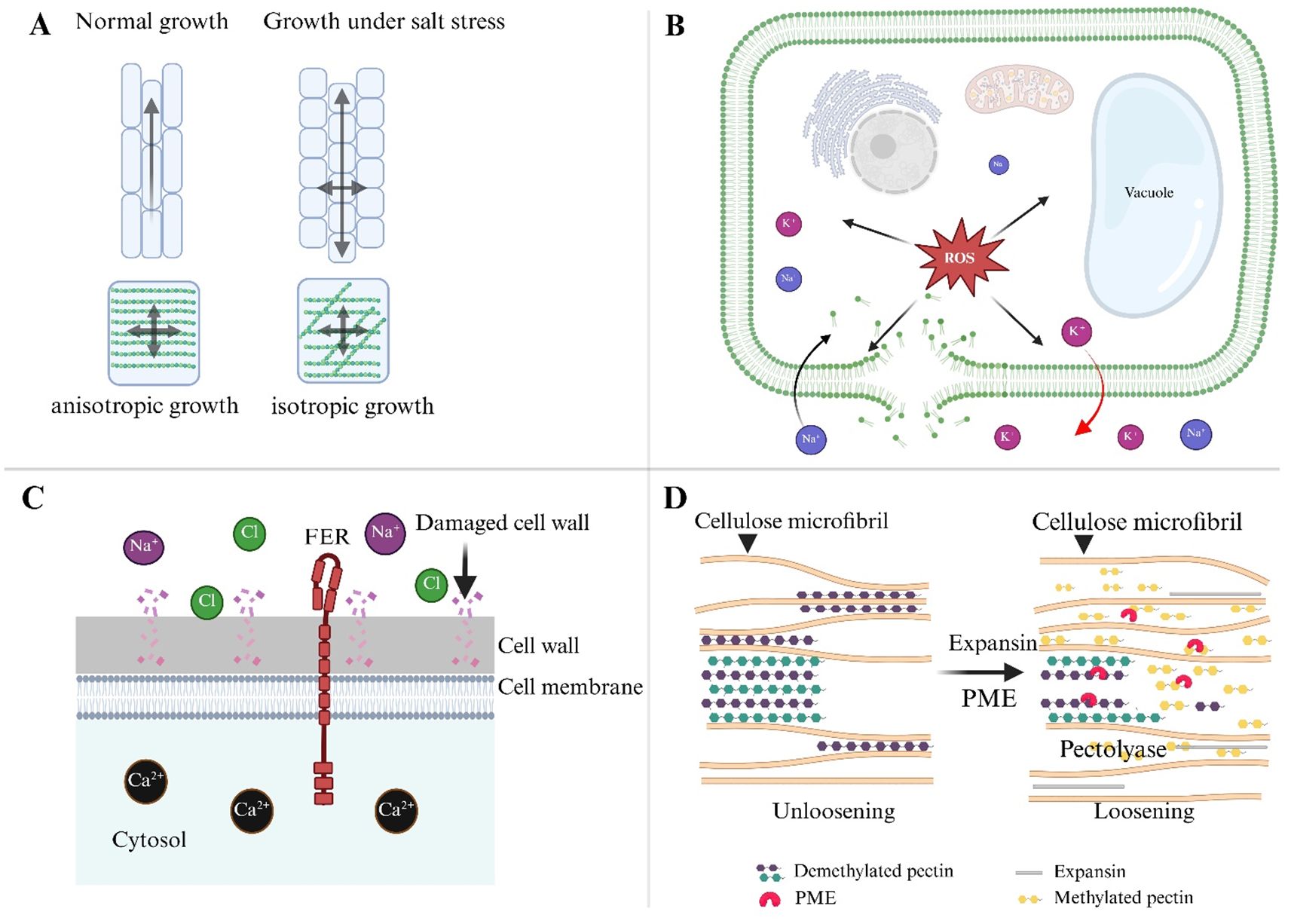
Figure 3. Mechanisms of anisotropic growth during salt stress in plants. (A) Under normal conditions, the cell wall cellulose and hemicellulose network support anisotropic growth, where cells elongate in a specific direction. In contrast, salt stress alters the orientation of cellulose fibers, resulting in isotropic growth, where cell expansion becomes less directional and more uniform across all axes. (B) Salt stress leads to an imbalance in ion homeostasis, with Na+ and K+ ions accumulating within the cell, disrupting the membrane’s integrity, and generating ROS. These ROS damage cell membranes and internal structures, inhibiting normal growth and causing cellular dysfunction. (C) The FER receptor plays a critical role in maintaining cell wall integrity during salt stress. FER interacts with extracellular signals and regulates ion homeostasis, helping to prevent excessive damage to the cell wall. By balancing Ca2+ and Cl- levels, FER prevents premature cell bursting and supports anisotropic growth under stress. (D) Expansins are key enzymes in loosening the cell wall, enabling recovery after salt stress. They interact with pectin and cellulose components to relax the wall, facilitating cell expansion. The reorganization of microtubules and cellulose fibers, coupled with expansin activity, helps restore anisotropic growth patterns, allowing plants to adapt and recover from salt-induced growth inhibition.
Furthermore, salinity alters pectin structure. Na+ binds to cell wall pectin, disrupting their charge balance and affecting wall porosity and elasticity, thus hindering expansion. The FER, a CWI sensor, helps maintain directional growth under stress. The fer mutants show abnormal root swelling and premature root initiation under salinity, indicating a failure in anisotropic expansion due to weakened wall integrity (Geng et al., 2013; Vaahtera et al., 2019). Similarly, cesa mutant, which impairs cellulose synthesis, reduces cell elongation even when microtubule orientation remains unaffected, highlighting the necessity of robust cell wall synthesis during salt stress (Fujita et al., 2013).
The actin cytoskeleton also plays a crucial role in anisotropic development under saline conditions. ARP2/3 complexes, which regulate actin meshwork formation, mediate vesicle trafficking of wall-modifying enzymes, while proteins like KINESIN-4A/FRA1 transport non-cellulosic materials along microtubules to support growth (Kong et al., 2015; Pratap Sahi et al., 2018). Disruption of these trafficking routes under salt stress further limits expansion.
On the other hand, FER seems to keep epidermal cells growing in an anisotropic form, preventing radial swelling and bursting. Importantly, certain molecular interventions can partially restore anisotropic growth under salt stress. Overexpression of RhEXPA4, a rose expansin gene, in Arabidopsis promotes root elongation and lateral root formation under saline conditions, emphasizing the role of wall-loosening proteins in maintaining directional growth (Lu et al., 2013). Nonetheless, the precise coordination of pectin de-methylesterification, xyloglucan remodeling, and PME activity during salt-induced stress remains underexplored. Finally, salt stress inhibits anisotropic growth via altering cell wall architecture, cytoskeletal dynamics, and CWI sensing. Understanding these systems is critical for developing salt-tolerant crops capable of maintaining directional growth and yield under challenging environments.
6 Conclusion and future perspectives
The plant cell wall serves as both a structural barrier and a dynamic sensor that orchestrates adaptive responses to environmental challenges such as salt stress. Salinity disrupts CWI, leading to impaired anisotropic growth by altering the deposition and organization of cellulose, hemicellulose, and pectin. These disruptions compromise wall extensibility, turgor balance, and cytoskeletal orientation, ultimately affecting plant development.
Despite advances in understanding cell wall remodeling under salt stress, the molecular mechanisms by which Na+ interferes with CWI perception and downstream signaling remain unclear. Receptor-like kinases such as FER and THE1 have emerged as key integrators of mechanical, hormonal, and immune pathways, mediating responses through ABA and JA signaling (Takahashi et al., 2020).
Future research should incorporate live-cell imaging, atomic force microscopy, and biomechanics to dissect how salt stress alters wall elasticity and polymer interactions at high resolution. Further exploration of calcium signaling, ROS dynamics, and their interactions with wall-associated kinases will deepen our mechanistic insights.
From a translational perspective, genome editing tools such as CRISPR/Cas9 and prime editing offer the potential to modify genes regulating wall biosynthesis, hormone crosstalk, and CWI sensing. Combining these with multi-omics approaches and systems biology will be critical to uncovering regulatory hubs that enable plants to maintain growth under salinity. A deeper mechanistic understanding of CWI under salt stress will ultimately facilitate the development of resilient, salt-tolerant crop varieties for sustainable agriculture.
Author contributions
FT: Data curation, Software, Investigation, Visualization, Writing – original draft, Project administration, Validation, Methodology. CM: Supervision, Writing – review & editing, Conceptualization, Investigation, Funding acquisition, Formal Analysis, Project administration. SZ: Writing – original draft, Formal Analysis, Project administration, Visualization, Data curation, Resources, Validation, Conceptualization, Methodology, Supervision, Writing – review & editing, Funding acquisition, Investigation, Software.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Agricultural Fine Seed Project of Shandong Province (grant number 2023LZGC011) and the National Natural Science Foundation of China (grant no. 32270319).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Albert, I., Bohm, H., Albert, M., Feiler, C. E., Imkampe, J., Wallmeroth, N., et al. (2015). An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1, 15140. doi: 10.1038/nplants.2015.140
PubMed Abstract | PubMed Abstract | Crossref Full Text | Google Scholar
Alvarez, V. M. Z., Fernández, P. V., and Ciancia, M. (2024). A novel substitution pattern in glucuronoarabinoxylans from woody bamboos. Carbohydr. Polymers. 323, 121356. doi: 10.1016/j.carbpol.2023.121356
Amjad, M., Wang, Y., Han, S., Haider, M. Z., Sami, A., Batool, A., et al. (2024). Genome wide identification of phenylalanine ammonia-lyase (PAL) gene family in Cucumis sativus (cucumber) against abiotic stress. BMC Genom. Data 25, 76. doi: 10.1186/s12863-024-01259-1
Amos, R. A., Pattathil, S., Yang, J. Y., Atmodjo, M. A., Urbanowicz, B. R., Moremen, K. W., et al. (2018). A two-phase model for the non-processive biosynthesis of homogalacturonan polysaccharides by the GAUT1:GAUT7 complex. J. Biol. Chem. 293, 19047–19063. doi: 10.1074/jbc.RA118.004463
An, P., Li, X., Zheng, Y., Matsuura, A., Abe, J., Eneji, A. E., et al. (2014). Effects of naCl on root growth and cell wall composition of two soya bean cultivars with contrasting salt tolerance. J. Agron. Crop Sci. 200, 212–218. doi: 10.1111/jac.12060
Anderson, C. T., Carroll, A., Akhmetova, L., and Somerville, C. (2010). Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol. 152, 787–796. doi: 10.1104/pp.109.150128
PubMed Abstract | PubMed Abstract | Crossref Full Text | Google Scholar
Atmodjo, M. A., Hao, Z., and Mohnen, D. (2013). Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 64, 747–779. doi: 10.1146/annurev-arplant-042811-105534
Atmodjo, M. A., Sakuragi, Y., Zhu, X., Burrell, A. J., Mohanty, S. S., Atwood, J. A., 3rd, et al. (2011). Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc. Natl. Acad. Sci. U.S.A. 108, 20225–20230. doi: 10.1073/pnas.1112816108
Atta, K., Mondal, S., Gorai, S., Singh, A. P., Kumari, A., Ghosh, T., et al. (2023). Impacts of salinity stress on crop plants: improving salt tolerance through genetic and molecular dissection. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1241736
Bacete, L. and Hamann, T. (2020). The role of mechanoperception in plant cell wall integrity maintenance. Plants (Basel). 9, 1–18. doi: 10.3390/plants9050574
Bai, Y., Ali, S., Liu, S., Zhou, J., and Tang, Y. (2023). Characterization of plant laccase genes and their functions. Gene 852, 147060. doi: 10.1016/j.gene.2022.147060
Barbut, F. R., Cavel, E., Donev, E. N., Gaboreanu, I., Urbancsok, J., Pandey, G., et al. (2024). Integrity of xylan backbone affects plant responses to drought. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1422701
PubMed Abstract | PubMed Abstract | Crossref Full Text | Google Scholar
Baskin, T. I. and Jensen, O. E. (2013). On the role of stress anisotropy in the growth of stems. J. Exp. Bot. 64, 4697–4707. doi: 10.1093/jxb/ert176
Bauer, S., Vasu, P., Persson, S., Mort, A. J., and Somerville, C. R. (2006). Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc. Natl. Acad. Sci. U.S.A. 103, 11417–11422. doi: 10.1073/pnas.0604632103
Beauzamy, L., Derr, J., and Boudaoud, A. (2015). Quantifying hydrostatic pressure in plant cells by using indentation with an atomic force microscope. Biophys. J. 108, 2448–2456. doi: 10.1016/j.bpj.2015.03.035
Bernal, A. J., Jensen, J. K., Harholt, J., Sorensen, S., Moller, I., Blaukopf, C., et al. (2007). Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J. 52, 791–802. doi: 10.1111/j.1365-313X.2007.03281.x
Bernal, A. J., Yoo, C. M., Mutwil, M., Jensen, J. K., Hou, G., Blaukopf, C., et al. (2008). Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol. 148, 1238–1253. doi: 10.1104/pp.108.121939
Bi, G., Liebrand, T. W., Cordewener, J. H., America, A. H., Xu, X., and Joosten, M. H. (2014). Arabidopsis thaliana receptor-like protein At RLP23 associates with the receptor-like kinase At SOBIR1. Plant Signal Behav. 9, 1–5. doi: 10.4161/psb.27937
Bonawitz, N. D. and Chapple, C. (2010). The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44, 337–363. doi: 10.1146/annurev-genet-102209-163508
Bou Daher, F., Chen, Y., Bozorg, B., Clough, J., Jonsson, H., and Braybrook, S. A. (2018). Anisotropic growth is achieved through the additive mechanical effect of material anisotropy and elastic asymmetry. Elife 7, 1–28. doi: 10.7554/eLife.38161
Boudsocq, M., Barbier-Brygoo, H., and Lauriere, C. (2004). Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279, 41758–41766. doi: 10.1074/jbc.M405259200
Bringmann, M., Li, E., Sampathkumar, A., Kocabek, T., Hauser, M. T., and Persson, S. (2012). POM-POM2/cellulose synthase interacting1 is essential for the functional association of cellulose synthase and microtubules in Arabidopsis. Plant Cell 24, 163–177. doi: 10.1105/tpc.111.093575
Brown, D. M., Zhang, Z., Stephens, E., Dupree, P., and Turner, S. R. (2009). Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J. 57, 732–746. doi: 10.1111/j.1365-313X.2008.03729.x
Brutus, A., Sicilia, F., Macone, A., Cervone, F., and De Lorenzo, G. (2010). A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U.S.A. 107, 9452–9457. doi: 10.1073/pnas.1000675107
PubMed Abstract | PubMed Abstract | Crossref Full Text | Google Scholar
Buschmann, H. and Borchers, A. (2020). Handedness in plant cell expansion: a mutant perspective on helical growth. New Phytol. 225, 53–69. doi: 10.1111/nph.16034
PubMed Abstract | PubMed Abstract | Crossref Full Text | Google Scholar
Calderone, S., Mauri, N., Manga-Robles, A., Fornalé, S., García-Mir, L., Centeno, M.-L., et al. (2024). Diverging cell wall strategies for drought adaptation in two maize inbreds with contrasting lodging resistance. Plant. Cell Environ. 47, 1747–1768. doi: 10.1111/pce.14822
Chan, J., Crowell, E., Eder, M., Calder, G., Bunnewell, S., Findlay, K., et al. (2010). The rotation of cellulose synthase trajectories is microtubule dependent and influences the texture of epidermal cell walls in Arabidopsis hypocotyls. J. Cell Sci. 123, 3490–3495. doi: 10.1242/jcs.074641
Chan, J., Eder, M., Crowell, E. F., Hampson, J., Calder, G., and Lloyd, C. (2011). Microtubules and CESA tracks at the inner epidermal wall align independently of those on the outer wall of light-grown Arabidopsis hypocotyls. J. Cell Sci. 124, 1088–1094. doi: 10.1242/jcs.086702
Chele, K. H., Tinte, M. M., Piater, L. A., Dubery, I. A., and Tugizimana, F. (2021). Soil salinity, a serious environmental issue and plant responses: A metabolomics perspective. Metabolites 11, 724. doi: 10.3390/metabo11110724
Chen, J., Chen, X., Zhang, Q., Zhang, Y., Ou, X., An, L., et al. (2018). A cold-induced pectin methyl-esterase inhibitor gene contributes negatively to freezing tolerance but positively to salt tolerance in Arabidopsis. J. Plant Physiol. 222, 67–78. doi: 10.1016/j.jplph.2018.01.003
Chen, K., Song, M., Guo, Y., Liu, L., Xue, H., Dai, H., et al. (2019). MdMYB46 could enhance salt and osmotic stress tolerance in apple by directly activating stress-responsive signals. Plant Biotechnol. J. 17, 2341–2355. doi: 10.1111/pbi.13151
Cho, S. K., Kim, J. E., Park, J. A., Eom, T. J., and Kim, W. T. (2006). Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett. 580, 3136–3144. doi: 10.1016/j.febslet.2006.04.062
Chourey, K., Ramani, S., and Apte, S. K. (2003). Accumulation of LEA proteins in salt (NaCl) stressed young seedlings of rice (Oryza sativaL.) cultivar Bura Rata and their degradation during recovery from salinity stress. J. Plant Physiol. 160, 1165–1174. doi: 10.1078/0176-1617-00909
Colin, L., Ruhnow, F., Zhu, J.-K., Zhao, C., Zhao, Y., and Persson, S. (2022). The cell biology of primary cell walls during salt stress. Plant Cell 35, 201–217. doi: 10.1093/plcell/koac292
Cona, A., Rea, G., Angelini, R., Federico, R., and Tavladoraki, P. (2006). Functions of amine oxidases in plant development and defence. Trends Plant Sci. 11, 80–88. doi: 10.1016/j.tplants.2005.12.009
Correa-Ferreira, M. L., Viudes, E. B., de Magalhaes, P. M., Paixao de Santana Filho, A., Sassaki, G. L., Pacheco, A. C., et al. (2019). Changes in the composition and structure of cell wall polysaccharides from Artemisia annua in response to salt stress. Carbohydr. Res. 483, 107753. doi: 10.1016/j.carres.2019.107753
Cosgrove, D. J. (2016). Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J. Exp. Bot. 67, 463–476. doi: 10.1093/jxb/erv511
Cosgrove, D. J. (2022). Building an extensible cell wall. Plant Physiol. 189, 1246–1277. doi: 10.1093/plphys/kiac184
Cosgrove, D. J. and Jarvis, M. C. (2012). Comparative structure and biomechanics of plant primary and secondary cell walls. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00204
Crowell, E. F., Gonneau, M., Stierhof, Y. D., Hofte, H., and Vernhettes, S. (2010). Regulated trafficking of cellulose synthases. Curr. Opin. Plant Biol. 13, 700–705. doi: 10.1016/j.pbi.2010.07.005
Crowell, E. F., Timpano, H., Desprez, T., Franssen-Verheijen, T., Emons, A. M., Hofte, H., et al. (2011). Differential regulation of cellulose orientation at the inner and outer face of epidermal cells in the Arabidopsis hypocotyl. Plant Cell 23, 2592–2605. doi: 10.1105/tpc.111.087338
Dabravolski, S. A. and Isayenkov, S. V. (2023). The regulation of plant cell wall organisation under salt stress. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1118313
Daudi, A., Cheng, Z., O’Brien, J. A., Mammarella, N., Khan, S., Ausubel, F. M., et al. (2012). The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell 24, 275–287. doi: 10.1105/tpc.111.093039
Davis, J., Brandizzi, F., Liepman, A. H., and Keegstra, K. (2010). Arabidopsis mannan synthase CSLA9 and glucan synthase CSLC4 have opposite orientations in the Golgi membrane. Plant J. 64, 1028–1037. doi: 10.1111/j.1365-313X.2010.04392.x
De Caroli, M., Lenucci, M. S., Di Sansebastiano, G. P., Tunno, M., Montefusco, A., Dalessandro, G., et al. (2014). Cellular localization and biochemical characterization of a chimeric fluorescent protein fusion of Arabidopsis cellulose synthase-like A2 inserted into Golgi membrane. ScientificWorldJournal 2014, 792420. doi: 10.1155/2014/792420
Decreux, A. and Messiaen, J. (2005). Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 46, 268–278. doi: 10.1093/pcp/pci026
Dekkers, B. J., Pearce, S., van Bolderen-Veldkamp, R. P., Marshall, A., Widera, P., Gilbert, J., et al. (2013). Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiol. 163, 205–215. doi: 10.1104/pp.113.223511
Derevyanchuk, M., Litvinovskaya, R., Khripach, V., and Kravets, V. (2016). Brassinosteroid-induced de novo protein synthesis in Zea mays under salinity and bioinformatic approach for identification of heat shock proteins. J. Plant Growth Regul. 78, 297–305. doi: 10.1007/s10725-015-0093-3
Dissanayake, B. M., Staudinger, C., Munns, R., Taylor, N. L., and Millar, A. H. (2022). Distinct salinity-induced changes in wheat metabolic machinery in different root tissue types. J. Proteomics 256, 104502. doi: 10.1016/j.jprot.2022.104502
Dokka, N., Rathinam, M., and Sreevathsa, R. (2024). Lignin lite: Boosting plant power through selective downregulation. Plant. Cell Environ. 47, 4945–4962. doi: 10.1111/pce.15060
Du, B., Haensch, R., Alfarraj, S., and Rennenberg, H. (2024). Strategies of plants to overcome abiotic and biotic stresses. Biol. Rev. Camb. Philos. Soc. 99, 1524–1536. doi: 10.1111/brv.13079
Du, J., Vandavasi, V. G., Molloy, K. R., Yang, H., Massenburg, L. N., Singh, A., et al. (2022). Evidence for plant-conserved region mediated trimeric CESAs in plant cellulose synthase complexes. Biomacromolecules 23, 3663–3677. doi: 10.1021/acs.biomac.2c00550
Ellinger, D. and Voigt, C. A. (2014). Callose biosynthesis in Arabidopsis with a focus on pathogen response: what we have learned within the last decade. Ann. Bot. 114, 1349–1358. doi: 10.1093/aob/mcu120
Endler, A., Kesten, C., Schneider, R., Zhang, Y., Ivakov, A., Froehlich, A., et al. (2015). A mechanism for sustained cellulose synthesis during salt stress. Cell 162, 1353–1364. doi: 10.1016/j.cell.2015.08.028
PubMed Abstract | PubMed Abstract | Crossref Full Text | Google Scholar
Engelsdorf, T., Kjaer, L., Gigli-Bisceglia, N., Vaahtera, L., Bauer, S., Miedes, E., et al. (2019). Functional characterization of genes mediating cell wall metabolism and responses to plant cell wall integrity impairment. BMC Plant Biol. 19, 320. doi: 10.1186/s12870-019-1934-4
Estrada, Y., Plasencia, F., Ortíz-Atienza, A., Faura, C., Flores, F. B., Lozano, R, et al. (2023). A novel function of the tomato CALCINEURIN-B LIKE 10 gene as a root-located negative regulator of salt stress. Plant Cell Environ. 46, 3433–3444. doi: 10.1111/pce.14679
Eudes, A., Dutta, T., Deng, K., Jacquet, N., Sinha, A., Benites, V. T., et al. (2017). SbCOMT (Bmr12) is involved in the biosynthesis of tricin-lignin in sorghum. PloS One 12, 1–11. doi: 10.1371/journal.pone.0178160
Fang, C., Li, K., Wu, Y., Wang, D., Zhou, J., Liu, X., et al. (2019). OsTSD2-mediated cell wall modification affects ion homeostasis and salt tolerance. Plant Cell Environ. 42, 1503–1512. doi: 10.1111/pce.13499
Feng, W., Kita, D., Peaucelle, A., Cartwright, H. N., Doan, V., Duan, Q., et al. (2018). The FERONIA Receptor Kinase Maintains Cell-Wall Integrity during Salt Stress through Ca(2+) Signaling. Curr. Biol. 28, 666–675 e665. doi: 10.1016/j.cub.2018.01.023
Feraru, E., Feraru, M. I., Kleine-Vehn, J., Martiniere, A., Mouille, G., Vanneste, S., et al. (2011). PIN polarity maintenance by the cell wall in Arabidopsis. Curr. Biol. 21, 338–343. doi: 10.1016/j.cub.2011.01.036
Fry, S. C. (2004). Oxidative coupling of tyrosine and ferulic acid residues: Intra- and extra-protoplasmic occurrence, predominance of trimers and larger products, and possible role in inter-polymeric cross-linking. Phytochem. Rev. 3, 97–111. doi: 10.1023/B:PHYT.0000047808.74647.43
Fujita, M., Himmelspach, R., Ward, J., Whittington, A., Hasenbein, N., Liu, C., et al. (2013). The anisotropy1 D604N mutation in the Arabidopsis cellulose synthase1 catalytic domain reduces cell wall crystallinity and the velocity of cellulose synthase complexes. Plant Physiol. 162, 74–85. doi: 10.1104/pp.112.211565
Geilfus, C. M. (2017). The pH of the apoplast: dynamic factor with functional impact under stress. Mol. Plant 10, 1371–1386. doi: 10.1016/j.molp.2017.09.018
Geng, Y., Wu, R., Wee, C. W., Xie, F., Wei, X., Chan, P. M., et al. (2013). A spatio-temporal understanding of growth regulation during the salt stress response in Arabidopsis. Plant Cell 25, 2132–2154. doi: 10.1105/tpc.113.112896
Gigli-Bisceglia, N., Engelsdorf, T., and Hamann, T. (2020). Plant cell wall integrity maintenance in model plants and crop species-relevant cell wall components and underlying guiding principles. Cell Mol. Life Sci. 77, 2049–2077. doi: 10.1007/s00018-019-03388-8
Gigli-Bisceglia, N., van Zelm, E., Huo, W., Lamers, J., and Testerink, C. (2022). Arabidopsis root responses to salinity depend on pectin modification and cell wall sensing. Development 149, 1–13. doi: 10.1242/dev.200363
Giordano, A., Liu, Z., Panter, S. N., Dimech, A. M., Shang, Y., Wijesinghe, H., et al. (2014). Reduced lignin content and altered lignin composition in the warm season forage grass Paspalum dilatatum by down-regulation of a Cinnamoyl CoA reductase gene. Transgenic Res. 23, 503–517. doi: 10.1007/s11248-014-9784-1
González-Pérez, L., Páez-Watson, T., Álvarez-Suarez, J. M., Obando-Rojas, M. C., Bonifaz-Arcos, E., Viteri, G., et al. (2018). Application of exogenous xyloglucan oligosaccharides affects molecular responses to salt stress in Arabidopsis thaliana seedlings. J. Soil Sci. Plant Nutr. 18, 1187–1205. doi: 10.1007/s11627-019-10048-w
Goubet, F., Barton, C. J., Mortimer, J. C., Yu, X., Zhang, Z., Miles, G. P., et al. (2009). Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J. 60, 527–538. doi: 10.1111/j.1365-313X.2009.03977.x
Grison, M. S., Kirk, P., Brault, M. L., Wu, X. N., Schulze, W. X., Benitez-Alfonso, Y., et al. (2019). Plasma membrane-associated receptor-like kinases relocalize to plasmodesmata in response to osmotic stress. Plant Physiol. 181, 142–160. doi: 10.1104/pp.19.00473
Gu, H., Wang, Y., Xie, H., Qiu, C., Zhang, S., Xiao, J., et al. (2020). Drought stress triggers proteomic changes involving lignin, flavonoids and fatty acids in tea plants. Sci. Rep. 10, 15504. doi: 10.1038/s41598-020-72596-1
Gujas, B., Cruz, T. M. D., Kastanaki, E., Vermeer, J. E. M., Munnik, T., and Rodriguez-Villalon, A. (2017). Perturbing phosphoinositide homeostasis oppositely affects vascular differentiation in Arabidopsis thaliana roots. Development 144, 3578–3589. doi: 10.1242/dev.155788
Gutierrez, R., Lindeboom, J. J., Paredez, A. R., Emons, A. M., and Ehrhardt, D. W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11, 797–806. doi: 10.1038/ncb1886
Haas, K. T. and Peaucelle, A. (2021). From monocots to dicots: the multifold aspect of cell wall expansion. J. Exp. Bot. 72, 1511–1513. doi: 10.1093/jxb/eraa573
Hamann, T. (2012). Plant cell wall integrity maintenance as an essential component of biotic stress response mechanisms. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00077
Hamann, T., Osborne, E., Youngs, H. L., Misson, J., Nussaume, L., and Somerville, C. (2004). Global expression analysis of CESA and CSL genes in Arabidopsis. Cellulose 11, 279–286. doi: 10.1023/B:CELL.0000046340.99925.57
Harris, P. J. and Trethewey, J. A. K. (2009). The distribution of ester-linked ferulic acid in the cell walls of angiosperms. Phytochem. Rev. 9, 19–33. doi: 10.1007/s11101-009-9146-4
Hazman, M., Hause, B., Eiche, E., Nick, P., and Riemann, M. (2015). Increased tolerance to salt stress in OPDA-deficient rice ALLENE OXIDE CYCLASE mutants is linked to an increased ROS-scavenging activity. J. Exp. Bot. 66, 3339–3352. doi: 10.1093/jxb/erv142
He, C., Zhang, J., Liu, X., Zeng, S., Wu, K., Yu, Z., et al. (2015). Identification of genes involved in biosynthesis of mannan polysaccharides in Dendrobium officinale by RNA-seq analysis. Plant Mol. Biol. 88, 219–231. doi: 10.1007/s11103-015-0316-z
Héreil, A., Guillaume, M., Duboscq, R., Carretero, Y., Pelpoir, E., Bitton, F., et al. (2024). Characterisation of a major QTL for sodium accumulation in tomato grown in high salinity. Plant Cell Envioron. 47, 5089–5103. doi: 10.1111/pce.15082
Herger, A., Dünser, K., Kleine-Vehn, J., and Ringli, C. (2019). Leucine-rich repeat extensin proteins and their role in cell wall sensing. Curr. Biol. 29, R851–R858. doi: 10.1016/j.cub.2019.07.039
Heyndrickx, K. S. and Vandepoele, K. (2012). Systematic identification of functional plant modules through the integration of complementary data sources. Plant Physiol. 159, 884–901. doi: 10.1104/pp.112.196725
Hong, Y., Wang, Z., Shi, H., Yao, J., Liu, X., Wang, F., et al. (2020). Reciprocal regulation between nicotinamide adenine dinucleotide metabolism and abscisic acid and stress response pathways in Arabidopsis. PloS Genet. 16, 1–21. doi: 10.1371/journal.pgen.1008892
Huang, L., Zhang, W., Li, X., Staiger, C. J., and Zhang, C. (2023). Point mutations in the catalytic domain disrupt cellulose synthase (CESA6) vesicle trafficking and protein dynamics. Plant Cell 35, 2654–2677. doi: 10.1093/plcell/koad110
Jensen, J. K., Sorensen, S. O., Harholt, J., Geshi, N., Sakuragi, Y., Moller, I., et al. (2008). Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. Plant Cell 20, 1289–1302. doi: 10.1105/tpc.107.050906
Jeon, B. W., Kang, N. Y., Park, W. Y., Oh, E., and Kim, J. (2024). Control of lateral root formation by rapamycin-induced dimerization of engineered RGI/BAK1 and by BIR3 chimera. Physiol. Plant 176, e14155. doi: 10.1111/ppl.14155
Jeon, B. W., Kim, J. S., Oh, E., Kang, N. Y., and Kim, J. (2023). ROOT MERISTEM GROWTH FACTOR1 (RGF1)-RGF1 INSENSITIVE 1 peptide-receptor pair inhibits lateral root development via the MPK6-PUCHI module in Arabidopsis. J. Exp. Bot. 74, 1475–1488. doi: 10.1093/jxb/erac495
Jiang, W., Wang, Z., Li, Y., Liu, X., Ren, Y., Li, C., et al. (2024). FERONIA regulates salt tolerance in Arabidopsis by controlling photorespiratory flux. Plant Cell 36, 4732–4751. doi: 10.1093/plcell/koae246
Jiang, Z., Zhou, X., Tao, M., Yuan, F., Liu, L., Wu, F., et al. (2019). Plant cell-surface GIPC sphingolipids sense salt to trigger Ca(2+) influx. Nature 572, 341–346. doi: 10.1038/s41586-019-1449-z
PubMed Abstract | PubMed Abstract | Crossref Full Text | Google Scholar
Jithesh, M. N., Wally, O. S. D., Manfield, I., Critchley, A. T., Hiltz, D., and Prithiviraj, B. (2012). Analysis of seaweed extract-induced transcriptome leads to identification of a negative regulator of salt tolerance in arabidopsis. Hort. Sci. 47, 704–709. doi: 10.21273/HORTSCI.47.6.704
Jose, J., Ghantasala, S., and Roy Choudhury, S. (2020). Arabidopsis transmembrane receptor-like kinases (RLKs): A bridge between extracellular signal and intracellular regulatory machinery. Int. J. Mol. Sci. 21, 1–29. doi: 10.3390/ijms21114000
Joshi, C. P. and Mansfield, S. D. (2007). The cellulose paradox–simple molecule, complex biosynthesis. Curr. Opin. Plant Biol. 10, 220–226. doi: 10.1016/j.pbi.2007.04.013
Kang, J. S., Frank, J., Kang, C. H., Kajiura, H., Vikram, M., Ueda, A., et al. (2008). Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. U.S.A. 105, 5933–5938. doi: 10.1073/pnas.0800237105
Karkonen, A. and Kuchitsu, K. (2015). Reactive oxygen species in cell wall metabolism and development in plants. Phytochemistry 112, 22–32. doi: 10.1016/j.phytochem.2014.09.016
Keppler, B. D. and Showalter, A. M. (2010). IRX14 and IRX14-LIKE, two glycosyl transferases involved in glucuronoxylan biosynthesis and drought tolerance in Arabidopsis. Mol. Plant 3, 834–841. doi: 10.1093/mp/ssq028
Kesten, C., Garcia-Moreno, A., Amorim-Silva, V., Menna, A., Castillo, A. G., Percio, F., et al. (2022). Peripheral membrane proteins modulate stress tolerance by safeguarding cellulose synthases. Sci. Adv. 8, eabq6971. doi: 10.1126/sciadv.abq6971
Khatri, P., Chen, L., Rajcan, I., and Dhaubhadel, S. (2023). Functional characterization of Cinnamate 4-hydroxylase gene family in soybean (Glycine max). PloS One 18, e0285698. doi: 10.1371/journal.pone.0285698
Kim, S. J., Chandrasekar, B., Rea, A. C., Danhof, L., Zemelis-Durfee, S., Thrower, N., et al. (2020). The synthesis of xyloglucan, an abundant plant cell wall polysaccharide, requires CSLC function. Proc. Natl. Acad. Sci. U.S.A. 117, 20316–20324. doi: 10.1073/pnas.2007245117
Kohorn, B. D., Johansen, S., Shishido, A., Todorova, T., Martinez, R., Defeo, E., et al. (2009). Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J. 60, 974–982. doi: 10.1111/j.1365-313x.2009.04016.x
Kolomeichuk, L. V., Danilova, E. D., Murgan, O. K., Sauchuk, A. L., Litvinovskaya, R. P., Khripach, V. A., et al. (2023). Endogenous brassinosteroids are involved in the formation of salt resistance in plants. Dokl. Biol. Sci. 511, 259–263. doi: 10.1134/S0012496623700485
Komis, G., Apostolakos, P., and Galatis, B. (2002). Hyperosmotic stress induces formation of tubulin macrotubules in root-tip cells of Triticum turgidum: their probable involvement in protoplast volume control. Plant Cell Physiol. 43, 911–922. doi: 10.1093/pcp/pcf114
Kong, Z., Ioki, M., Braybrook, S., Li, S., Ye, Z. H., Julie Lee, Y. R., et al. (2015). Kinesin-4 functions in vesicular transport on cortical microtubules and regulates cell wall mechanics during cell elongation in plants. Mol. Plant 8, 1011–1023. doi: 10.1016/j.molp.2015.01.004
Krasensky, J. and Jonak, C. (2012). Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 63, 1593–1608. doi: 10.1093/jxb/err460
PubMed Abstract | PubMed Abstract | Crossref Full Text | Google Scholar
Kuang, B., Zhao, X., Zhou, C., Zeng, W., Ren, J., Ebert, B., et al. (2016). Role of UDP-glucuronic acid decarboxylase in xylan biosynthesis in arabidopsis. Mol. Plant 9, 1119–1131. doi: 10.1016/j.molp.2016.04.013
Kumar, V., Roy, S., Behera, B. K., and Das, B. K. (2022). Heat shock proteins (Hsps) in cellular homeostasis: A promising tool for health management in crustacean aquaculture. Life (Basel). 12, 1–29. doi: 10.3390/life12111777
Lamport, D. T., Kieliszewski, M. J., Chen, Y., and Cannon, M. C. (2011). Role of the extensin superfamily in primary cell wall architecture. Plant Physiol. 156, 11–19. doi: 10.1104/pp.110.169011
Lampugnani, E. R., Khan, G. A., Somssich, M., and Persson, S. (2018). Building a plant cell wall at a glance. J. Cell Sci. 131, jcs207373. doi: 10.1242/jcs.207373
PubMed Abstract | PubMed Abstract | Crossref Full Text | Google Scholar
Li, W., Guan, Q., Wang, Z. Y., Wang, Y., and Zhu, J. (2013). A bi-functional xyloglucan galactosyltransferase is an indispensable salt stress tolerance determinant in Arabidopsis. Mol. Plant 6, 1344–1354. doi: 10.1093/mp/sst062
Li, S., Zhang, L., Wang, Y., Xu, F., Liu, M., Lin, P., et al. (2017). Knockdown of a cellulose synthase gene BoiCesA affects the leaf anatomy, cellulose content and salt tolerance in broccoli. Sci. Rep. 7, 41397. doi: 10.1038/srep41397
Lindner, H., Muller, L. M., Boisson-Dernier, A., and Grossniklaus, U. (2012). CrRLK1L receptor-like kinases: not just another brick in the wall. Curr. Opin. Plant Biol. 15, 659–669. doi: 10.1016/j.pbi.2012.07.003
Liu, N., Chen, J., Wang, T., Li, Q., Cui, P., Jia, C., et al. (2019). Overexpression of WAX INDUCER1/SHINE1 Gene Enhances Wax Accumulation under Osmotic Stress and Oil Synthesis in Brassica napus. Int. J. Mol. Sci. 20, 4435. doi: 10.3390/ijms20184435
Liu, Z., Persson, S., and Sanchez-Rodriguez, C. (2015). At the border: the plasma membrane-cell wall continuum. J. Exp. Bot. 66, 1553–1563. doi: 10.1093/jxb/erv019
Liu, S. G., Zhu, D. Z., Chen, G. H., Gao, X. Q., and Zhang, X. S. (2012). Disrupted actin dynamics trigger an increment in the reactive oxygen species levels in the Arabidopsis root under salt stress. Plant Cell Rep. 31, 1219–1226. doi: 10.1007/s00299-012-1242-z
Lou, H. Q., Fan, W., Jin, J. F., Xu, J. M., Chen, W. W., Yang, J. L., et al. (2020). A NAC-type transcription factor confers aluminium resistance by regulating cell wall-associated receptor kinase 1 and cell wall pectin. Plant. Cell Environ. 43, 463–478. doi: 10.1111/pce.13676
Lu, P., Kang, M., Jiang, X., Dai, F., Gao, J., and Zhang, C. (2013). RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta 237, 1547–1559. doi: 10.1007/s00425-013-1867-3
Lü, S., Song, T., Kosma, D. K., Parsons, E. P., Rowland, O., and Jenks, M. A. (2009). Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 59, 553–564. doi: 10.1111/j.1365-313X.2009.03892.x
Luhua, S., Hegie, A., Suzuki, N., Shulaev, E., Luo, X., Cenariu, D., et al. (2013). Linking genes of unknown function with abiotic stress responses by high-throughput phenotype screening. Physiol. Plant 148, 322–333. doi: 10.1111/ppl.12013
Lund, C. H., Stenbaek, A., Atmodjo, M. A., Rasmussen, R. E., Moller, I. E., Erstad, S. M., et al. (2020). Pectin synthesis and pollen tube growth in arabidopsis involves three GAUT1 golgi-anchoring proteins: GAUT5, GAUT6, and GAUT7. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.585774
Luo, Y., Scholl, S., Doering, A., Zhang, Y., Irani, N. G., Rubbo, S. D., et al. (2015). V-ATPase activity in the TGN/EE is required for exocytosis and recycling in Arabidopsis. Nat. Plants 1, 15094. doi: 10.1038/nplants.2015.94
Ma, L., Zhang, H., Sun, L., Jiao, Y., Zhang, G., Miao, C., et al. (2012). NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na(+)/K(+)homeostasis in Arabidopsis under salt stress. J. Exp. Bot. 63, 305–317. doi: 10.1093/jxb/err280
Majda, M. and Robert, S. (2018). The role of auxin in cell wall expansion. Int. J. Mol. Sci. 19, 1–21. doi: 10.3390/ijms19040951
Mao, J., Mo, Z., Yuan, G., Xiang, H., Visser, R. G. F., Bai, Y., et al. (2023). The CBL-CIPK network is involved in the physiological crosstalk between plant growth and stress adaption. Plant Cell Environ. 46, 3012–3022. doi: 10.1111/pce.14396
Marjamaa, K., Kukkola, E. M., and Fagerstedt, K. V. (2009). The role of xylem class III peroxidases in lignification. J. Exp. Bot. 60, 367–376. doi: 10.1093/jxb/ern278
McLoughlin, F., Arisz, S. A., Dekker, H. L., Kramer, G., de Koster, C. G., Haring, M. A., et al. (2013). Identification of novel candidate phosphatidic acid-binding proteins involved in the salt-stress response of Arabidopsis thaliana roots. Biochem. J. 450, 573–581. doi: 10.1042/BJ20121639
Meents, M. J., Motani, S., Mansfield, S. D., and Samuels, A. L. (2019). Organization of xylan production in the golgi during secondary cell wall biosynthesis. Plant Physiol. 181, 527–546. doi: 10.1104/pp.19.00715
Mietelska-Porowska, A., Wasik, U., Goras, M., Filipek, A., and Niewiadomska, G. (2014). Tau protein modifications and interactions: their role in function and dysfunction. Int. J. Mol. Sci. 15, 4671–4713. doi: 10.3390/ijms15034671
Mohnen, D. (2008). Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11, 266–277. doi: 10.1016/j.pbi.2008.03.006
Motomura, K., Takeuchi, H., Notaguchi, M., Tsuchi, H., Takeda, A., Kinoshita, T., et al. (2021). Persistent directional growth capability in Arabidopsis thaliana pollen tubes after nuclear elimination from the apex. Nat. Commun. 12, 2331. doi: 10.1038/s41467-021-22661-8
Moura, J. C., Bonine, C. A., de Oliveira Fernandes Viana, J., Dornelas, M. C., and Mazzafera, P. (2010). Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 52, 360–376. doi: 10.1111/j.1744-7909.2010.00892.x
Munarin, F., Tanzi, M. C., and Petrini, P. (2012). Advances in biomedical applications of pectin gels. Int. J. Biol. Macromol. 51, 681–689. doi: 10.1016/j.ijbiomac.2012.07.002
Mustafa, Y., İrem, P., Aslinur, Ç., Yasin, Ö., and Ramazan, B. (2020). “Plant responses to salt stress,” in Plant Breeding. Ed. Ibrokhim, Y. A. (IntechOpen, Rijeka), 1–18.
Nagashima, Y., Ma, Z., Liu, X., Qian, X., Zhang, X., von Schaewen, A., et al. (2020). Multiple quality control mechanisms in the ER and TGN determine subcellular dynamics and salt-stress tolerance function of KORRIGAN1. Plant Cell 32, 470–485. doi: 10.1105/tpc.19.00714
Nakajima, K., Kawamura, T., and Hashimoto, T. (2006). Role of the SPIRAL1 gene family in anisotropic growth of Arabidopsis thaliana. Plant Cell Physiol. 47, 513–522. doi: 10.1093/pcp/pcj020
Nongpiur, R. C., Singla-Pareek, S. L., and Pareek, A. (2020). The quest for osmosensors in plants. J. Exp. Bot. 71, 595–607. doi: 10.1093/jxb/erz263
Oda, Y. and Fukuda, H. (2012). Secondary cell wall patterning during xylem differentiation. Curr. Opin. Plant Biol. 15, 38–44. doi: 10.1016/j.pbi.2011.10.005
Oliveira, D. M., Mota, T. R., Salatta, F. V., Sinzker, R. C., Končitíková, R., Kopečný, D., et al. (2020). Cell wall remodeling under salt stress: Insights into changes in polysaccharides, feruloylation, lignification, and phenolic metabolism in maize. Plant Cell Environ. 43, 2172–2191. doi: 10.1111/pce.13805
Páez-Watson, T., Álvarez-Suárez, J. M., Rivas-Romero, F., Estrada, L., López, D., Pérez Pelea, L., et al. (2020). Increased salinity stress tolerance of Nicotiana tabacum L. in vitro plants with the addition of xyloglucan oligosaccharides to the culture medium. In Vitro Cell Dev. Biol. Plant 56, 325–334. doi: 10.1007/s11627-019-10048-w
Paredez, A. R., Somerville, C. R., and Ehrhardt, D. W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312, 1491–1495. doi: 10.1126/science.1126551
Parniske, M., Wulff, B. B., Bonnema, G., Thomas, C. M., Jones, D. A., and Jones, J. D. (1999). Homologues of the Cf-9 disease resistance gene (Hcr9s) are present at multiple loci on the short arm of tomato chromosome 1. Mol. Plant Microbe Interact. 12, 93–102. doi: 10.1094/MPMI.1999.12.2.93
Pauly, M. and Keegstra, K. (2016). Biosynthesis of the plant cell wall matrix polysaccharide xyloglucan. Annu. Rev. Plant Biol. 67, 235–259. doi: 10.1146/annurev-arplant-043015-112222
Peaucelle, A., Braybrook, S., and Hofte, H. (2012). Cell wall mechanics and growth control in plants: the role of pectins revisited. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00121
Peaucelle, A., Braybrook, S. A., Le Guillou, L., Bron, E., Kuhlemeier, C., and Höfte, H. (2011). Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 21, 1720–1726. doi: 10.1016/j.cub.2011.08.057
Pelloux, J., Rusterucci, C., and Mellerowicz, E. J. (2007). New insights into pectin methylesterase structure and function. Trends Plant Sci. 12, 267–277. doi: 10.1016/j.tplants.2007.04.001
Peracchi, L. M., Panahabadi, R., Barros-Rios, J., Bartley, L. E., and Sanguinet, K. A. (2024). Grass lignin: biosynthesis, biological roles, and industrial applications. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1343097
Persson, S., Paredez, A., Carroll, A., Palsdottir, H., Doblin, M., Poindexter, P., et al. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 15566–15571. doi: 10.1073/pnas.0706592104
Petit, J., Bres, C., Mauxion, J.-P., Tai, F. W. J., Martin, L. B. B., Fich, E. A., et al. (2016). The glycerol-3-phosphate acyltransferase GPAT6 from tomato plays a central role in fruit cutin biosynthesis. Plant Physiol. 171, 894–913. doi: 10.1104/pp.16.00409
Philippe, F., Pelloux, J., and Rayon, C. (2017). Plant pectin acetylesterase structure and function: new insights from bioinformatic analysis. BMC Genomics 18, 456. doi: 10.1186/s12864-017-3833-0
Polko, J. K., Barnes, W. J., Voiniciuc, C., Doctor, S., Steinwand, B., Hill, J. L., Jr., et al. (2018). SHOU4 proteins regulate trafficking of cellulose synthase complexes to the plasma membrane. Curr. Biol. 28, 3174–3182 e3176. doi: 10.1016/j.cub.2018.07.076
Pratap Sahi, V., Cifrova, P., Garcia-Gonzalez, J., Kotannal Baby, I., Mouille, G., Gineau, E., et al. (2018). Arabidopsis thaliana plants lacking the ARP2/3 complex show defects in cell wall assembly and auxin distribution. Ann. Bot. 122, 777–789. doi: 10.1093/aob/mcx178
Radotic, K., Roduit, C., Simonovic, J., Hornitschek, P., Fankhauser, C., Mutavdzic, D., et al. (2012). Atomic force microscopy stiffness tomography on living Arabidopsis thaliana cells reveals the mechanical properties of surface and deep cell-wall layers during growth. Biophys. J. 103, 386–394. doi: 10.1016/j.bpj.2012.06.046
Raiola, A., Camardella, L., Giovane, A., Mattei, B., De Lorenzo, G., Cervone, F., et al. (2004). Two Arabidopsis thaliana genes encode functional pectin methylesterase inhibitors. FEBS Lett. 557, 199–203. doi: 10.1016/s0014-5793(03)01491-1
Ramakrishna, P., Ruiz Duarte, P., Rance, G. A., Schubert, M., Vordermaier, V., Vu, L. D., et al. (2019). EXPANSIN A1-mediated radial swelling of pericycle cells positions anticlinal cell divisions during lateral root initiation. Proc. Natl. Acad. Sci. U.S.A. 116, 8597–8602. doi: 10.1073/pnas.1820882116
Rao, X. and Dixon, R. A. (2017). Brassinosteroid mediated cell wall remodeling in grasses under abiotic stress. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00806
Rasmussen, C. G., Wright, A. J., and Muller, S. (2013). The role of the cytoskeleton and associated proteins in determination of the plant cell division plane. Plant J. 75, 258–269. doi: 10.1111/tpj.12177
Rui, Y. and Dinneny, J. R. (2020). A wall with integrity: surveillance and maintenance of the plant cell wall under stress. New Phytol. 225, 1428–1439. doi: 10.1111/nph.16166
Saatian, B., Kohalmi, S. E., and Cui, Y. (2023). Localization of Arabidopsis Glucan Synthase-Like 5, 8, and 12 to plasmodesmata and the GSL8-dependent role of PDLP5 in regulating plasmodesmal permeability. Plant Signal Behav. 18, 2164670. doi: 10.1080/15592324.2022.2164670
Sanchez-Aguayo, I., Rodriguez-Galan, J. M., Garcia, R., Torreblanca, J., and Pardo, J. M. (2004). Salt stress enhances xylem development and expression of S-adenosyl-L-methionine synthase in lignifying tissues of tomato plants. Planta 220, 278–285. doi: 10.1007/s00425-004-1350-2
Scheller, H. V. and Ulvskov, P. (2010). Hemicelluloses. Annu. Rev. Plant Biol. 61, 263–289. doi: 10.1146/annurev-arplant-042809-112315
Schneider, R., Hanak, T., Persson, S., and Voigt, C. A. (2016). Cellulose and callose synthesis and organization in focus, what’s new? Curr. Opin. Plant Biol. 34, 9–16. doi: 10.1016/j.pbi.2016.07.007
Schoenaers, S., Lee, H. K., Gonneau, M., Faucher, E., Levasseur, T., Akary, E., et al. (2024). Rapid alkalinization factor 22 has a structural and signalling role in root hair cell wall assembly. Nat. Plants 10, 494–511. doi: 10.1038/s41477-024-01637-8
Schultink, A., Liu, L., Zhu, L., and Pauly, M. (2014). Structural diversity and function of xyloglucan sidechain substituents. Plants (Basel). 3, 526–542. doi: 10.3390/plants3040526
Shafi, A., Pal, A. K., Sharma, V., Kalia, S., Kumar, S., Ahuja, P. S., et al. (2017). Transgenic potato plants overexpressing SOD and APX exhibit enhanced lignification and starch biosynthesis with improved salt stress tolerance. Plant Mol. Bio Rep. 35, 504–518. doi: 10.1007/s11105-017-1041-3
Shani, Z., Dekel, M., Roiz, L., Horowitz, M., Kolosovski, N., Lapidot, S., et al. (2006). Expression of endo-1,4-β-glucanase (cel1) in Arabidopsis thaliana is associated with plant growth, xylem development and cell wall thickening. Plant Cell Rep. 25, 1067–1074. doi: 10.1007/s00299-006-0167-9
Shao, M., Zheng, H., Hu, Y., Liu, D., Jang, J.-C., Ma, H., et al. (2004). The GAOLAOZHUANGREN1 gene encodes a putative glycosyltransferase that is critical for normal development and carbohydrate metabolism. Plant Cell Physiol. 45, 1453–1460. doi: 10.1093/pcp/pch168
Shikanai, Y., Takahashi, S., Enomoto, Y., Yamagami, M., Yamaguchi, K., Shigenobu, S., et al. (2022). Arabidopsis glucan synthase-like1 (GSL1) is required for tolerance to low-calcium conditions and exhibits a function comparable to GSL10. Plant Cell Physiol. 63, 1474–1484. doi: 10.1093/pcp/pcac106
Singh, A. (2022). Soil salinity: A global threat to sustainable development. Soil Use Manage. 38, 39–67. doi: 10.1111/sum.12772
Song, R., Liao, C., Wang, L., Lu, K., Zhang, C., Wu, R., et al. (2024). SORTING NEXIN1 facilitates SALT OVERLY SENSITIVE1 protein accumulation to enhance salt tolerance in Arabidopsis. Plant Physiol. 197, kiae633. doi: 10.1093/plphys/kiae633
Sun, J., Tian, Z., Li, X., Li, S., Li, Z., Wang, J., et al. (2022). Systematic analysis of the pectin methylesterase gene family in Nicotiana tabacum and reveal their multiple roles in plant development and abiotic stresses. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.998841
Takahashi, Y., Zhang, J., Hsu, P.-K., Ceciliato, P. H., Zhang, L., Dubeaux, G., et al. (2020). MAP3Kinase-dependent SnRK2-kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat. Commun. 11, 1–12. doi: 10.1038/s41467-019-13875-y
Taylor-Teeples, M., Lin, L., de Lucas, M., Turco, G., Toal, T. W., Gaudinier, A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. doi: 10.1038/nature14099
Tenhaken, R. (2014). Cell wall remodeling under abiotic stress. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00771
Teotia, D., Gaid, M., Saini, S. S., Verma, A., Yennamalli, R. M., Khare, S. P., et al. (2019). Cinnamate-CoA ligase is involved in biosynthesis of benzoate-derived biphenyl phytoalexin in Malus × domestica ‘Golden Delicious’ cell cultures. Plant J. 100, 1176–1192. doi: 10.1111/tpj.14506
Thompson, D. S. (2005). How do cell walls regulate plant growth? J. Exp. Bot. 56, 2275–2285. doi: 10.1093/jxb/eri247
Tiika, R. J., Yang, H., Cui, G., Ma, Y., Boamah, S., Li, Y., et al. (2025). Identification and analysis of cuticular wax biosynthesis related genes in salicornia europaea under naCl treatment. Int. J. Mol. Sci. 26, 2632. doi: 10.3390/ijms26062632
Uddin, M. N., Hanstein, S., Faust, F., Eitenmuller, P. T., Pitann, B., and Schubert, S. (2014). Diferulic acids in the cell wall may contribute to the suppression of shoot growth in the first phase of salt stress in maize. Phytochemistry 102, 126–136. doi: 10.1016/j.phytochem.2014.02.014
Vaahtera, L., Schulz, J., and Hamann, T. (2019). Cell wall integrity maintenance during plant development and interaction with the environment. Nat. Plants 5, 924–932. doi: 10.1038/s41477-019-0502-0
Van Acker, R., Déjardin, A., Desmet, S., Hoengenaert, L., Vanholme, R., Morreel, K., et al. (2017). Different metabolic routes for coniferaldehyde and sinapaldehyde with CINNAMYL ALCOHOL DEHYDROGENASE1 deficiency. Plant Physiol. 175, 1018–1039. doi: 10.1104/pp.17.00834
Van de Peer, Y., Ashman, T.-L., Soltis, P. S., and Soltis, D. E. (2021). Polyploidy: an evolutionary and ecological force in stressful times. Plant Cell 33, 11–26. doi: 10.1093/plcell/koab149
Vanholme, R., De Meester, B., Ralph, J., and Boerjan, W. (2019). Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 56, 230–239. doi: 10.1016/j.copbio.2019.02.018
van Zelm, E., Zhang, Y., and Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005
Velasquez, S. M., Ricardi, M. M., Dorosz, J. G., Fernandez, P. V., Nadra, A. D., Pol-Fachin, L., et al. (2011). O-glycosylated cell wall proteins are essential in root hair growth. Science 332, 1401–1403. doi: 10.1126/science.1206657
Vellosillo, T., Dinneny, J. R., Somerville, C. R., and Ehrhardt, D. W. (2021). TRANVIA (TVA) facilitates cellulose synthase trafficking and delivery to the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 118, 1–9. doi: 10.1073/pnas.2021790118
Verger, S. and Hamant, O. (2018). Plant physiology: FERONIA defends the cell walls against corrosion. Curr. Biol. 28, R215–R217. doi: 10.1016/j.cub.2018.01.043
Wagner, A., Tobimatsu, Y., Phillips, L., Flint, H., Geddes, B., Lu, F., et al. (2015). Syringyl lignin production in conifers: Proof of concept in a Pine tracheary element system. Proc. Natl. Acad. Sci. U.S.A. 112, 6218–6223. doi: 10.1073/pnas.1411926112
Wakabayashi, K., Soga, K., and Hoson, T. (2012). Phenylalanine ammonia-lyase and cell wall peroxidase are cooperatively involved in the extensive formation of ferulate network in cell walls of developing rice shoots. J. Plant Physiol. 169, 262–267. doi: 10.1016/j.jplph.2011.10.002
Wang, J., Feng, J., Jia, W., Chang, S., Li, S., and Li, Y. (2015). Lignin engineering through laccase modification: a promising field for energy plant improvement. Biotechnol. Biofuels 8, 145. doi: 10.1186/s13068-015-0331-y
Wang, Z. and Gou, X. (2020). Receptor-like protein kinases function upstream of MAPKs in regulating plant development. Int. J. Mol. Sci. 21, 1–26. doi: 10.3390/ijms21207638
Wang, C., Li, J., and Yuan, M. (2007). Salt tolerance requires cortical microtubule reorganization in Arabidopsis. Plant Cell Physiol. 48, 1534–1547. doi: 10.1093/pcp/pcm123
Wang, T., McFarlane, H. E., and Persson, S. (2016). The impact of abiotic factors on cellulose synthesis. J. Exp. Bot. 67, 543–552. doi: 10.1093/jxb/erv488
Wang, L., Qiu, T., Yue, J., Guo, N., He, Y., Han, X., et al. (2021a). Arabidopsis ADF1 is regulated by MYB73 and is involved in response to salt stress affecting actin filament organization. Plant Cell Physiol. 62, 1387–1395. doi: 10.1093/pcp/pcab081
Wang, Z. Q., Yu, T. F., Sun, G. Z., Zheng, J. C., Chen, J., Zhou, Y. B., et al. (2021b). Genome-wide analysis of the catharanthus roseus RLK1-like in soybean and gmCrRLK1L20 responds to drought and salt stresses. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.614909
Watanabe, Y., Meents, M. J., McDonnell, L. M., Barkwill, S., Sampathkumar, A., Cartwright, H. N., et al. (2015). Visualization of cellulose synthases in Arabidopsis secondary cell walls. Science 350, 198–203. doi: 10.1126/science.aac7446
Wolf, S. and Greiner, S. (2012). Growth control by cell wall pectins. Protoplasma 249 Suppl 2, S169–S175. doi: 10.1007/s00709-011-0371-5
Wu, A. M., Rihouey, C., Seveno, M., Hornblad, E., Singh, S. K., Matsunaga, T., et al. (2009). The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J. 57, 718–731. doi: 10.1111/j.1365-313X.2008.03724.x
Xu, P., Fang, S., Chen, H., and Cai, W. (2020). The brassinosteroid-responsive xyloglucan endotransglucosylase/hydrolase 19 (XTH19) and XTH23 genes are involved in lateral root development under salt stress in Arabidopsis. Plant J. 104, 59–75. doi: 10.1111/tpj.14905
Yadav, S., Modi, P., Dave, A., Vijapura, A., Patel, D., and Patel, M. (2020). “Effect of abiotic stress on crops,” in Sustainable Crop Production. Eds. Mirza, H., Marcelo Carvalho Minhoto Teixeira, F., Masayuki, F., and Thiago Assis Rodrigues, N. (IntechOpen, Rijeka), 1–21.
Yan, J., He, H., Fang, L., and Zhang, A. (2018). Pectin methylesterase 31 positively regulates salt stress tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 496-501, 497–501. doi: 10.1016/j.bbrc.2018.01.025
Yan, J., Huang, Y., He, H., Han, T., Di, P., Sechet, J., et al. (2019). Xyloglucan endotransglucosylase-hydrolase30 negatively affects salt tolerance in Arabidopsis. J. Exp. Bot. 70, 5495–5506. doi: 10.1093/jxb/erz311
Yang, Y. and Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
PubMed Abstract | PubMed Abstract | Crossref Full Text | Google Scholar
Yu, Y. and Assmann, S. M. (2018). Inter-relationships between the heterotrimeric Gβ subunit AGB1, the receptor-like kinase FERONIA, and RALF1 in salinity response. Plant. Cell Environ. 41, 2475–2489. doi: 10.1111/pce.13370
Yu, L., Nie, J., Cao, C., Jin, Y., Yan, M., Wang, F., et al. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188, 762–773. doi: 10.1111/j.1469-8137.2010.03422.x
Yu, B., Zheng, W., Xing, L., Zhu, J.-K., Persson, S., and Zhao, Y. (2022). Root twisting drives halotropism via stress-induced microtubule reorientation. Dev. Cell 57, 2412–2425.e2416. doi: 10.1016/j.devcel.2022.09.012
Yuan, F., Yang, H., Xue, Y., Kong, D., Ye, R., Li, C., et al. (2014). OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514, 367–371. doi: 10.1038/nature13593
Zhang, W., Cai, C., and Staiger, C. J. (2019). Myosins XI are involved in exocytosis of cellulose synthase complexes. Plant Physiol. 179, 1537–1555. doi: 10.1104/pp.19.00018
Zhang, B., Gao, Y., Zhang, L., and Zhou, Y. (2021). The plant cell wall: Biosynthesis, construction, and functions. J. Integr. Plant Biol. 63, 251–272. doi: 10.1111/jipb.13055
Zhang, Q., Lin, F., Mao, T., Nie, J., Yan, M., Yuan, M., et al. (2012). Phosphatidic acid regulates microtubule organization by interacting with MAP65–1 in response to salt stress in Arabidopsis. Plant Cell 24, 4555–4576. doi: 10.1105/tpc.112.104182
Zhang, Y., Nikolovski, N., Sorieul, M., Vellosillo, T., McFarlane, H. E., Dupree, R., et al. (2016b). Golgi-localized STELLO proteins regulate the assembly and trafficking of cellulose synthase complexes in Arabidopsis. Nat. Commun. 7, 11656. doi: 10.1038/ncomms11656
Zhang, J. L. and Shi, H. (2013). Physiological and molecular mechanisms of plant salt tolerance. Photosynth. Res. 115, 1–22. doi: 10.1007/s11120-013-9813-6
Zhang, R., Shi, P. T., Zhou, M., Liu, H. Z., Xu, X. J., Liu, W. T., et al. (2023). Rapid alkalinization factor: function, regulation, and potential applications in agriculture. Stress Biol. 3, 16. doi: 10.1007/s44154-023-00093-2
Zhang, S.-S., Sun, L., Dong, X., Lu, S.-J., Tian, W., and Liu, J.-X. (2016a). Cellulose synthesis genes CESA6 and CSI1 are important for salt stress tolerance in Arabidopsis. J. Integr. Plant Biol. 58, 623–626. doi: 10.1111/jipb.12442
Zhao, H., Li, Z., Wang, Y., Wang, J., Xiao, M., Liu, H., et al. (2022). Cellulose synthase-like protein OsCSLD4 plays an important role in the response of rice to salt stress by mediating abscisic acid biosynthesis to regulate osmotic stress tolerance. Plant Biotechnol. J. 20, 468–484. doi: 10.1111/pbi.13729
Zhao, C., Zayed, O., Yu, Z., Jiang, W., Zhu, P., Hsu, C. C., et al. (2018). Leucine-rich repeat extensin proteins regulate plant salt tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, 13123–13128. doi: 10.1073/pnas.1816991115
Zhao, C., Zayed, O., Zeng, F., Liu, C., Zhang, L., Zhu, P., et al. (2019). Arabinose biosynthesis is critical for salt stress tolerance in Arabidopsis. New Phytol. 224, 274–290. doi: 10.1111/nph.15867
Zhao, C., Zhang, H., Song, C., Zhu, J. K., and Shabala, S. (2020). Mechanisms of plant responses and adaptation to soil salinity. Innovation (Camb). 1, 100017. doi: 10.1016/j.xinn.2020.100017
Zheng, M., Liu, X., Lin, J., Liu, X., Wang, Z., Xin, M., et al. (2019). Histone acetyltransferase GCN5 contributes to cell wall integrity and salt stress tolerance by altering the expression of cellulose synthesis genes. Plant J. 97, 587–602. doi: 10.1111/tpj.14144
Zhou, S., Chen, Q., Li, X., and Li, Y. (2017). MAP65–1 is required for the depolymerization and reorganization of cortical microtubules in the response to salt stress in Arabidopsis. Plant Sci. 264, 112–121. doi: 10.1016/j.plantsci.2017.09.004
Zhou, X., Lu, J., Zhang, Y., Guo, J., Lin, W., Van Norman, J. M., et al. (2021). Membrane receptor-mediated mechano-transduction maintains cell integrity during pollen tube growth within the pistil. Dev. Cell 56, 1–6. doi: 10.1016/j.devcel.2021.02.030
Zhou, H., Shi, H., Yang, Y., Feng, X., Chen, X., Xiao, F., et al. (2024). Insights into plant salt stress signaling and tolerance. J. Genet. Genomics 51, 16–34. doi: 10.1016/j.jgg.2023.08.007
Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Zhu, J., Wang, G., Li, C., Li, Q., Gao, Y., Chen, F., et al. (2019). Maize Sep15-like functions in endoplasmic reticulum and reactive oxygen species homeostasis to promote salt and osmotic stress resistance. Plant Cell Environ. 42, 1486–1502. doi: 10.1111/pce.13507
Zhu, J., Zhang, H., Huang, K., Guo, R., Zhao, J., Xie, H., et al. (2023). Comprehensive analysis of the laccase gene family in tea plant highlights its roles in development and stress responses. BMC Plant Biol. 23, 129. doi: 10.1186/s12870-023-04134-w
Zonia, L. and Munnik, T. (2007). Life under pressure: hydrostatic pressure in cell growth and function. Trends Plant Sci. 12, 90–97. doi: 10.1016/j.tplants.2007.01.006
Keywords: salt stress, cell wall composition, plant cell anisotropy, cell wall integrity, cell growth
Citation: Tariq F, Ma C and Zhao S (2025) Integrative dynamics of cell wall architecture and plant growth under salt stress. Front. Plant Sci. 16:1644412. doi: 10.3389/fpls.2025.1644412
Received: 10 June 2025; Accepted: 09 July 2025;
Published: 30 July 2025.
Edited by:
Kai-Hua Jia, Shandong Academy of Agricultural Sciences, ChinaReviewed by:
Wen-Cheng Liu, Henan University, ChinaDr. Indraneel Saha, Bose Institute, India
Jinke Chang, Northwest A&F University, China
Copyright © 2025 Tariq, Ma and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changle Ma, bWFjaGFuZ2xlQHNkbnUuZWR1LmNu; Shuangshuang Zhao, emhhb3NodWFuZ3F3MTJAMTYzLmNvbQ==
 Faheem Tariq
Faheem Tariq Changle Ma
Changle Ma Shuangshuang Zhao
Shuangshuang Zhao