- 1Indian Council of Agricultural Research (ICAR)-Directorate of Onion and Garlic Research, Pune, Maharashtra, India
- 2Mahatma Phule Krishi Vidyapeeth, Rahuri, Maharashtra, India
- 3Indian Council of Agricultural Research (ICAR)-NBPGR Regional Station, Bhowali, Uttarakhand, India
- 4Indian Council of Agricultural Research (ICAR)-IARI Regional Station, Pune, Maharashtra, India
- 5Indian Council of Agricultural Research (ICAR)-Indian Agriculture Research Institute, Hazaribagh, Jharkhand, India
Alliums, including vital crops such as onion, garlic, chives, bunching onion, and leek, are globally prized for their culinary applications and medicinal attributes. However, their genetic improvement remains constrained by large genome size, high heterozygosity, and limited characterization of genetic resources. To bridge this gap, we developed chloroplast simple sequence repeat (cp-SSR) markers, which are particularly suitable for population genetics studies because of their maternal inheritance, low recombination rates, and high variability. Leveraging the chloroplast genome of Allium fistulosum, we identified 22 cp-SSR loci, with tetranucleotides being the most prevalent, followed by di-, tri-, and pentanucleotides. Screening 96 underutilized Allium accessions using polymorphic cp-SSR markers revealed 89.2% polymorphism, indicating substantial genetic diversity. The polymorphism information content (PIC) ranged from 0.00 to 0.66 (average 0.20), confirming the utility of these markers in diversity assessments. The population structure analysis revealed three distinct genetic clusters, whereas phylogenetic analysis categorized the accessions into six major clades, mirroring their evolutionary divergence. Fixation index (FST) analysis showed high genetic differentiation (mean FST = 0.6) among accessions. These findings underscore the significance of cp-SSRs in revealing genetic structure and diversity across underutilized Allium species. This work lays a crucial foundation for integrating chloroplast markers with nuclear genomic and omics tools to drive the development of resilient, high-value cultivars suited to future agricultural challenges.
Introduction
The genus Allium, comprising approximately 750 underutilized species (Fritsch and Friesen, 2002), is the largest within the monocot group and includes numerous economically important plants. The key species include onion (Allium cepa L.), garlic (Allium sativum L.), chives (Allium schoenoprasum L.), leek (Allium porrum L.), and bunching onions (Allium fistulosum L.) (Khade et al., 2022). In addition to these well-known crops, the genus also encompasses lesser-known species, such as A. altaicum, A. ramosum, A. chinense, and A. tuberosum etc. which are of interest for their potential contributions to biodiversity research. In addition to their agricultural value, Allium species hold significant ecological importance, with some listed in the Red Book of countries such as Mongolia, Russia, and China due to concerns over their conservation status (Mandakh et al., 2020).
Underutilized Allium species exhibit significant chromosomal diversity, ranging from diploid to highly polyploid forms. This remarkable cytogenetic variability reflects the genus’ complex evolutionary history and dynamic genomic architecture, with origins tracing back to regions of Asia and Europe. Over time, these lesser-studied species have adapted to a wide array of ecological niches, resulting in a rich spectrum of phenotypic traits and specialized adaptations. Investigating their chromosomal profiles and evolutionary trajectories not only enhances our understanding of Allium genomics but also supports crop improvement efforts and biodiversity conservation. Notably, underexploited Allium populations represent untapped reservoirs of genetic diversity, offering valuable traits for sustainable breeding and long-term genetic resource management.
To date, the genetic diversity of onions and related Allium species has been examined using various molecular markers, including RAPDs (Kutty et al., 2006; dos Santos et al., 2012), ISSRs (Monteverde et al., 2015; Sudha et al., 2019; Brahimi et al., 2022, 2024), and combinations of RAPD and ISSR (Sudha et al., 2019), RAPD and PCR-RFLP (Arifin et al., 2000), and RAPD and SSR (Mohapatra et al., 2023). Additional marker systems such as RFLPs (McCallum et al., 2001), AFLPs (Van Heusden et al., 2000; Simó et al., 2014), TRAP (Kisha and Cramer, 2011; Anandhan et al., 2015), SRAP combined with ISSR (Hancı and Paşazade, 2025), ILP (Gowd et al., 2023; Jayaswall et al., 2024), and SRAP (Khade et al., 2024; Mansour et al., 2020) have also been widely applied.
More recently, SSR and SNP markers have been extensively employed to assess genetic variation in Allium fistulosum (Yamashita et al., 2010), Allium mongolicum (Hu et al., 2022), and Allium cepa (Mahajan et al., 2009; Khar et al., 2011; Khosa et al., 2014; Jayaswall et al., 2019; Gupta et al., 2020; Lyngkhoi et al., 2021; Chalbi et al., 2023; Habsatou et al., 2024), among others. Studies incorporating STS and SNPs (McCallum et al., 2012; Scholten et al., 2016; Duangjit et al., 2013; Villano et al., 2019; Mallor et al., 2014; Hanci and Gökçe, 2016; Rivera et al., 2016; Damon and Havey, 2014; Chand et al., 2018; Labate et al., 2020; Sahoo et al., 2023) further highlight the growing utility of these high-resolution markers in understanding genetic variation. Among these, SSR markers have emerged as a preferred tool due to their high polymorphism, co-dominant inheritance, reproducibility, and cross-species transferability (Son et al., 2012; Wang and Zhang, 2022). These characteristics make SSRs effective for evaluating genetic diversity and population structure in plant species.
In addition to nuclear markers, the chloroplast genome characterized by maternal inheritance offers a powerful system for elucidating evolutionary relationships, phylogeography, and population genetics within and across Allium species. As a type of SSR marker, chloroplast simple sequence repeats (cp-SSRs) are particularly advantageous due to their high mutability, conservation, variability, co-dominant inheritance, and organelle-specific transmission (Ebert and Peakall, 2009). In Allium, cp-SSR markers have successfully revealed genetic diversity, population differentiation, and evolutionary relationships among closely related taxa (Jayaswall et al., 2023). They have also been used to detect historical demographic events, such as bottlenecks and genetic drift, which are critical for understanding population dynamics (McCauley, 1995). Strikingly, cp-SSRs contributed to onion breeding programs by facilitating targeted genetic analyses and facilitating the development of conservation strategies (Sharma et al., 2020).
Cp-SSR markers offer a non-destructive and efficient means for detecting subtle genetic variations in the chloroplast genome (Powell et al., 1995). Only a few studies have reported the use of cp-SSR markers in Allium species, such as Allium cepa L., Allium sativum L., and Allium paradoxum (M. Bieb.) (Jayaswall et al., 2022). The present study aims to investigate chloroplast genetic divergence, heterozygosity, allelic diversity, population structure, and genetic relatedness across 96 underutilized Allium species using 22 cp-SSR markers. By doing so, this research seeks to provide a foundation for the strategic use of these genotypes in future breeding programs and to guide the conservation of these critical plant resources in the face of ongoing environmental challenges.
Materials and methods
Plant material and DNA extraction
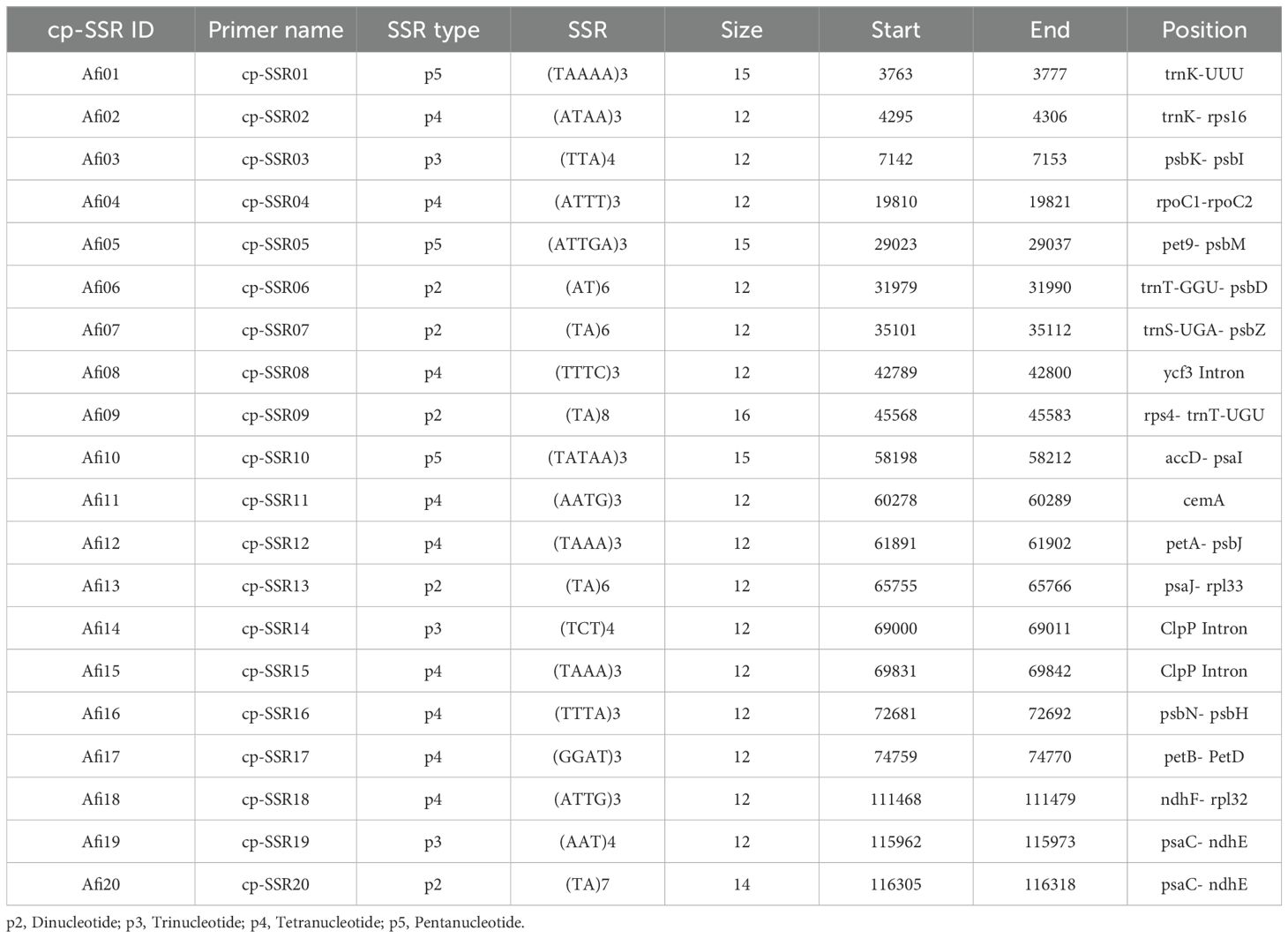
In the present study, a total of 96 underutilized Allium (A.) accessions (Table 1) were randomly collected from their primary regions of distribution across India. The collected samples were planted during the regular crop growing season at an experimental field site at the Indian Council of Agricultural Research-Directorate of Onion and Garlic Research (ICAR-DOGR) in Rajgurunagar, Pune, Maharashtra, India, which is located at geographic locations (18°52’0”N, 73°54’0”E; 645 m above sea level). Young leaf tissues from ten individuals per accession (96 underutilized Allium species) were collected randomly for genomic DNA isolation. Total genomic DNA was isolated from these samples via the CTAB method as described by Murray and Thompson (1980). Leaf tissues were homogenized in liquid nitrogen and incubated for an hour at 65°C in 1 ml of CTAB buffer, which contained 4% polyvinylpyrrolidone (PVP), 0.5% β-mercaptoethanol, 1.4 M NaCl, 100 mM Tris-HCl, 20 mM EDTA, and 2% cetyl trimethylammonium bromide. The quantity and quality of the extracted DNA were assessed by electrophoresis on a 0.8% agarose gel, using lambda HindIII marker (Thermo Fisher Scientific) used as a reference.
Table 1. Description of the samples used for the characterization of chloroplast derived simple sequence repeat markers (cp-SSR).
Chloroplast SSR marker development
The simple sequence repeats (SSR) loci within the Allium fistulosum chloroplast genome (Voucher No. PRJNA927338; NCBI Reference ID: NC_040222.1; Omelchenko et al., 2020) were identified using the MISA tool (http://misaweb.ipk-gatersleben.de/). The cp-SSR motif analyzed consisted of repeat units ranging from di- to hexanucleotides, meeting the minimum repeat thresholds set by MISA. Specifically, six motifs contained dinucleotide repeats, four contained trinucleotides, and three included tetra-, penta-, or hexanucleotide repeats. Mononucleotide repeats were excluded from further analysis. Both perfect and compound SSRs were detected via the MISA pipeline, with compound repeats defined as SSRs interrupted by non-repeat sequences of up to 100 bp. Primer pairs flanking the cp-SSR loci were designed using the BatchPrimer3 v1.0 online tool (https://probes.pw.usda.gov/batchprimer3; You et al., 2008). The primer design parameters included primer lengths of 22–27 nucleotides, amplicon sizes of 100 to 300 bp, melting temperature ranging from 48°C to 55°C, and GC content between 40% and 70%, with an optimal GC content of 50% (Table 2).
cp-SSR marker analysis
A total of 22 cp-SSR primer pairs were selected and synthesized by Eurofins Genomics (Eurofins, India). After an initial run with the newly developed primer pairs, 20 cp-SSRs exhibiting high resolution, stability, and significant polymorphism were selected for further analysis. The cp-SSR amplification was carried out in a 20 µl reaction volume, which included 2 µl of 10X reaction buffer and 50 ng of template DNA per reaction. The PCR reaction mixture consisted of 50 ng of genomic DNA (1µl), 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.2 µM of each primer (forward and reverse), and 5 U of Taq DNA polymerase. PCR amplification was performed using a Bio-Rad iCycler thermal cycler. The cycling conditions included an initial denaturation at 94°C for 4 minutes, followed by 35 cycles of denaturation at 94°C for 1 minute, annealing at the optimized temperature specific to each primer (as listed in Table 3), and extension at 72°C for 40 seconds. A final extension was carried out at 72°C for 10 minutes. The PCR products were analyzed via gel electrophoresis on a 3.2% agarose gel. Bands were visualized with a 1 kb Plus DNA ladder (Thermo Fisher Scientific) as a reference and documented using a gel documentation system.
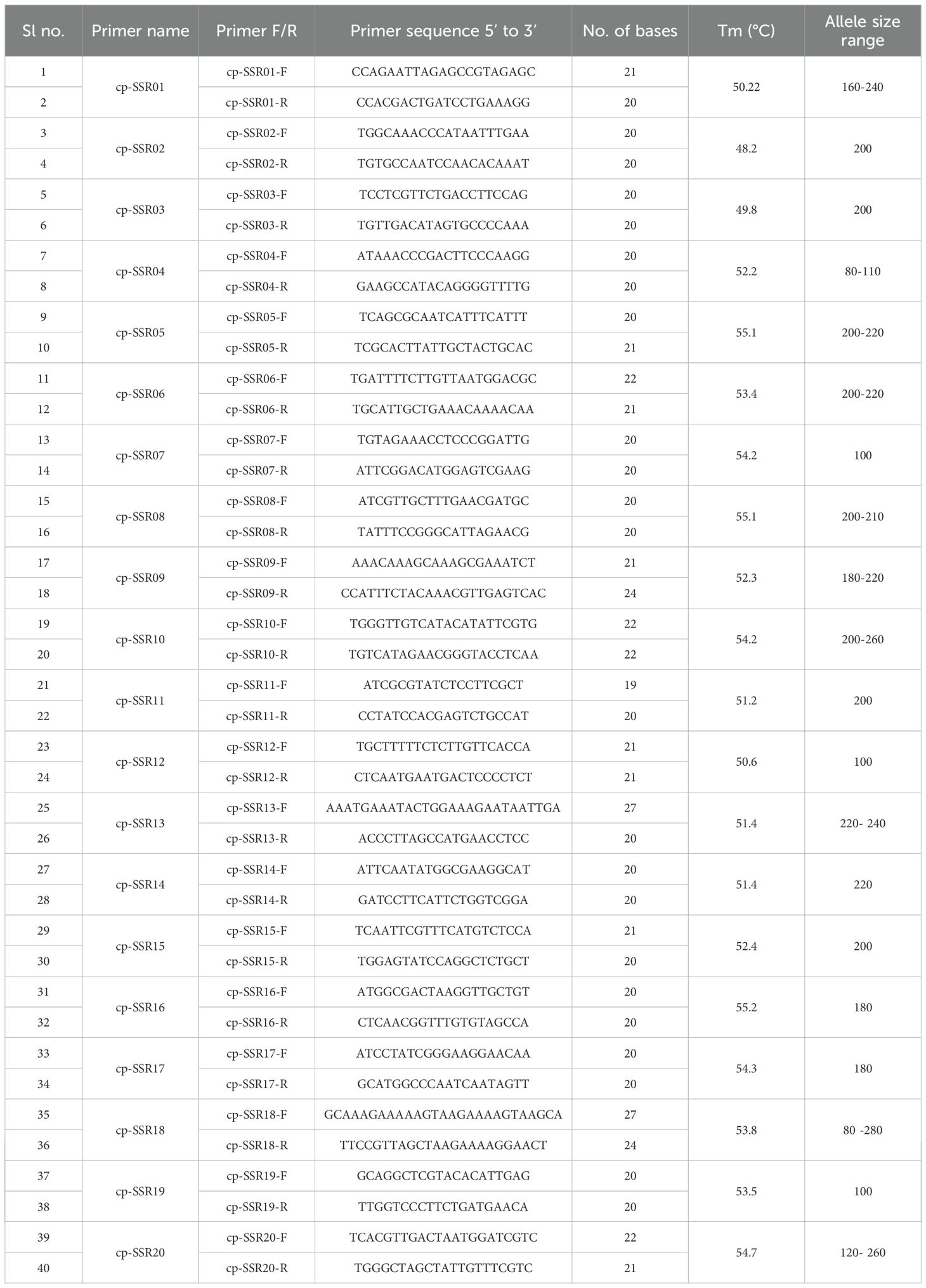
Table 3. Details of 20 chloroplast SSR markers, including sequences, annealing temperatures, and allele size.
Scoring and data analysis
To ensure the accuracy of the results, each pair of primers was used for PCR amplification and electrophoresis twice, and only the cp-SSR markers with high definition and good stability were scored. The scoring was performed on the basis of the absence (0) or presence (1) of each band for all the isolates in each primer. Genetic variation at each locus was characterized in terms of the number of alleles, and the PIC value was calculated. The binary matrix was subjected to Jaccard similarity coefficient analysis using NTSYS-pc version 2.02i (Rohlf, 1998), and the unweighted pair group method with arithmetic mean (UPGMA) clustering map based on Nei’s genetic distance was constructed using MEGA X (Kumar et al., 2018). Principal component analysis (PCA) and a cluster matrix were plotted on the basis of correlation distance and average genetic linkage via the web tool Clust-Vis (https://biit.cs.ut.ee/clustvis/) (Metsalu and Vilo, 2015). Genetic diversity parameters, including minor allele frequency (MAF), observed number of alleles (Na), observed heterozygosity (Ho), expected heterozygosity (He), and PIC, were calculated by GenAlEx 6.51 software (Peakall and Smouse, 2012). GenAlEx 6.51 was used to calculate the fixation index (FST), which measures the proportional increase in homozygosity. FST values range from 0 (no differentiation) to 1 (complete differentiation) (Wright, 1984). The population genetic structure was analyzed using Bayesian clustering methods via STRUCTURE 2.3.4 software. The number of populations (K) was tested sequentially from 1-10. Each run included a burn-in phase of 50,000 steps, followed by 200,000 Markov chain Monte Carlo (MCMC) iterations, which enhanced the reliability of clustering, as suggested by Nouri et al. (2021). The optimal K value was determined via the average lnP(K) and StructureSelector (https://lmme.ac.cn/StructureSelector/), revealing a significant peak in the ΔK values for the most suitable population grouping.
Results
Characterization of the developed cp-SSR markers
In the current study, a total of 22 cp-SSR marker pairs were identified from the Allium fistulosum chloroplast genome. Among the identified markers, tetranucleotide motifs were the most abundant (45.45%), followed by dinucleotide (27.27%), trinucleotide (13.63%), and pentanucleotide (13.63%) motifs (Table 2 and Figure 1). Notably, hexanucleotide repeats were completely absent from the chloroplast genome of A. fistulosum. The most frequently occurring motif was TA (22.73%), followed by the TAAA motif (9.09%). All other motifs were evenly distributed. Mononucleotide repeats, primarily A/T-rich, were excluded from further analysis due to their low polymorphic potential and the risk of sequencing errors caused by homopolymer runs. The average repeat lengths for the di-, tri-, tetra-, and pentanucleotide cp-SSRs were 13, 12, 12, and 15 base pairs, respectively. Among the 22 designed primer pairs, 20 (90.91%) successfully amplified clear and reproducible bands during PCR screening with underutilized Allium species (Table 3). These markers showed high polymorphism and stability indicating their potential suitability for assessing genetic diversity in Allium germplasm.
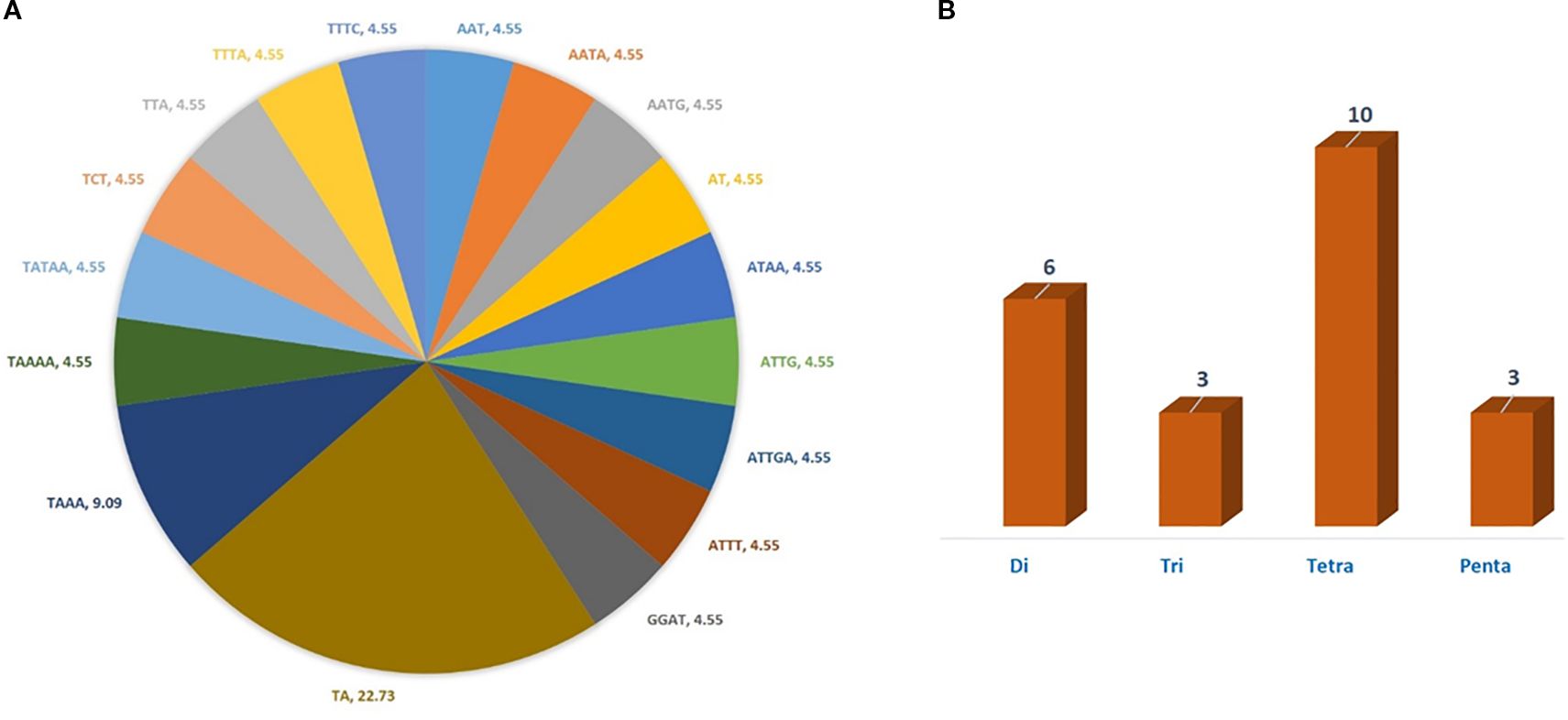
Figure 1. Type and proportion of motif repetition for cp-SSRs in the chloroplast genome of Allium fistulosum. (A) Distribution of different motif types in cp-SSRs. (B) Proportion of repeated motif types.
cp-SSR marker analysis
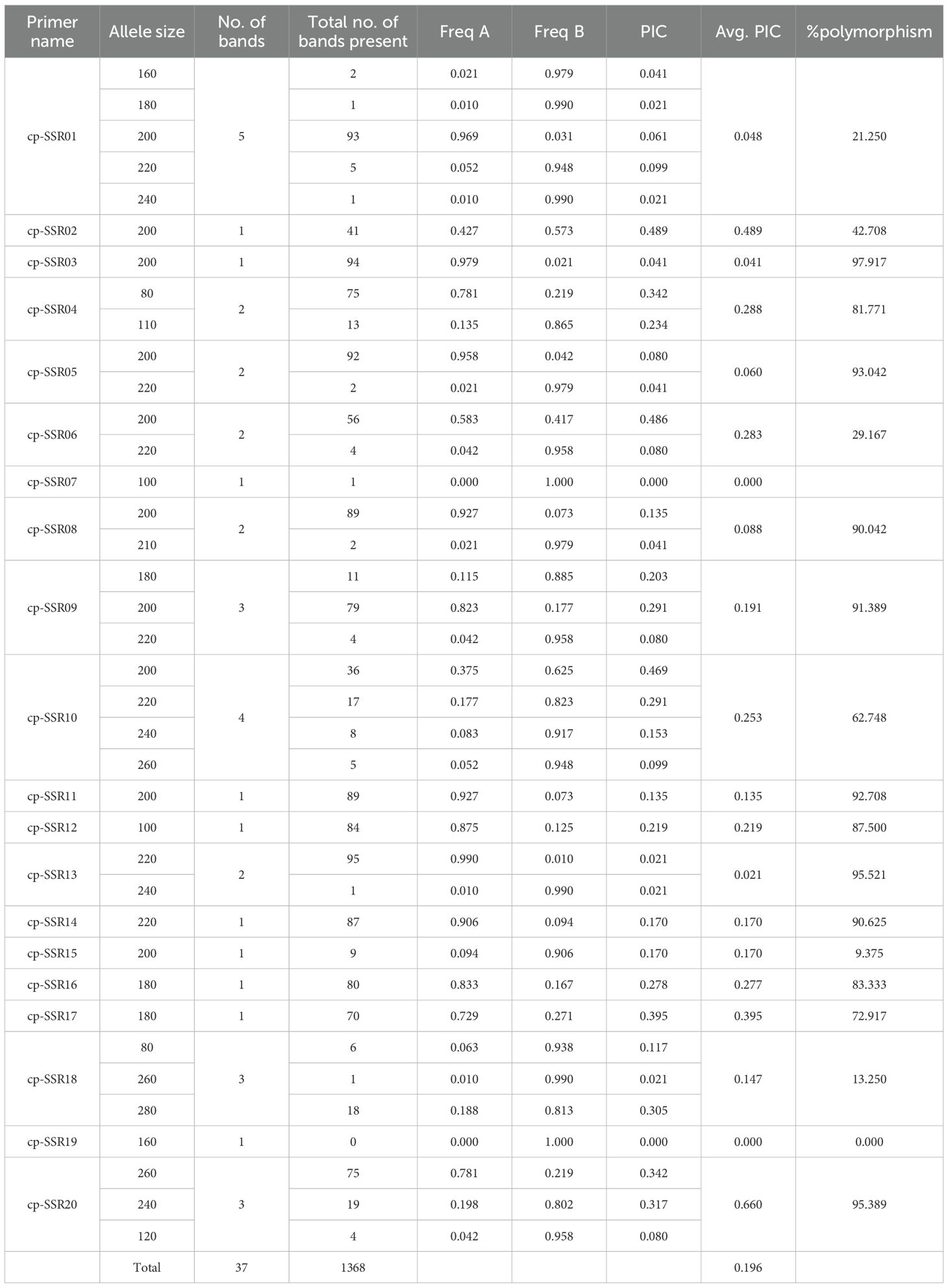
The analysis of the developed cp-SSR markers revealed a high level of polymorphism and genetic diversity among the underutilized Allium species (Supplementary Figure 1). A total of 37 allelic bands were detected using 20 polymorphic cp-SSR markers. Among these, 89.2% of the amplified alleles were polymorphic, indicating the hypervariable nature of the cp-SSR loci and the broad genetic variation in the underutilized Alliums. The observed allele sizes ranged from 80 to 280 bp (Table 3), with allele frequencies ranging from 0.00 to 0.99, demonstrating the effectiveness of these markers in capturing intra- and interspecific genetic variation. The polymorphism information content (PIC) values of the cp-SSR markers ranged from 0.00 to 0.66, with an average of 0.20 (Table 4). Notably, most of the markers exhibited more than 80% polymorphism, indicating their high utility for diversity and population genetic studies. These highly informative loci can serve as valuable molecular tools in future genetic analyses of Allium species.
Data scoring and analysis
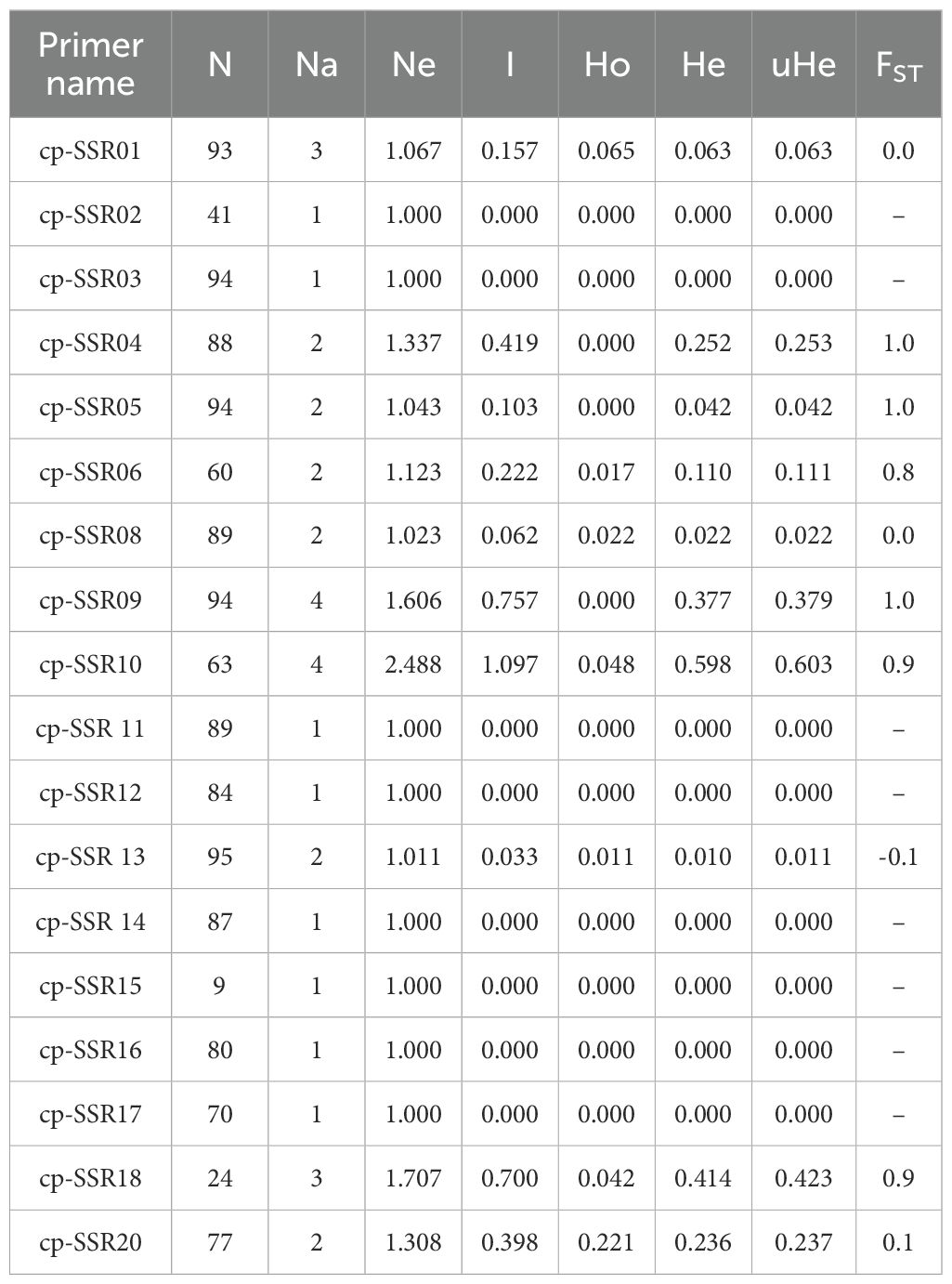
The genetic diversity analysis of 20 cp-SSR markers across 96 underutilized Allium species along and their respective accessions provided a comprehensive view of the genetic structure and variability within the genus. The key diversity indices assessed included the number of observed alleles (Na), effective number of alleles (Ne), Shannon’s information index (I), observed heterozygosity (Ho), expected heterozygosity (He), and unbiased expected heterozygosity (uHe) (Table 5). The number of alleles (Na) ranged from 1 to 4, with markers such as SSR09 and SSR10 showing the highest diversity. The effective number of alleles (Ne) ranged from 1.00 to 2.488. Shannon’s index (I) varied from 0.0 for nonpolymorphic markers to 1.1 for SSR10, indicating high intra-accession diversity for that marker.
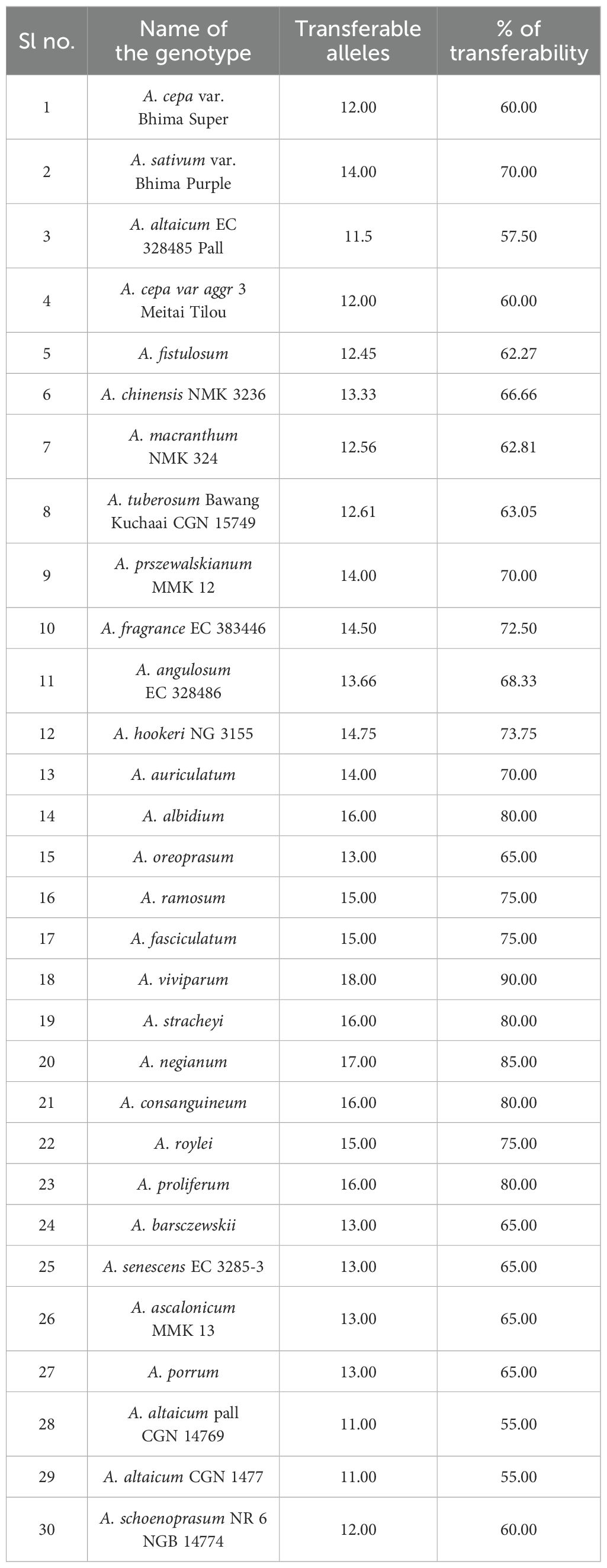
The observed heterozygosity (Ho) was predominantly 0.000 for most of the markers, suggesting low heterozygosity levels, whereas cp-SSR10 exhibited a slightly greater value of 0.048. The expected heterozygosity (He) ranged from 0.000 to 0.598, and the unbiased expected heterozygosity (uHe) ranged from 0.000 to 0.603, both of which were highest for cp-SSR10. Markers such as cp-SSR09, cp-SSR10, and cp-SSR18 exhibited high genetic diversity, whereas cp-SSR02, cp-SSR03, cp-SSR11, and cp-SSR15 were monomorphic with no diversity (Na and Ne = 1.000; I, Ho, and He = 0.000). The fixation index (FST) ranged from 0.0 to 1.0 among the accessions within the six major clades, with a mean FST value of 0.6. Cross-transferability analysis among 30 Allium genotypes (Table 6) revealed that the transferability percentage of cp-SSR alleles ranged from 55% to 90%. The highest transferability was observed in A. viviparum (90%), whereas A. altaicum Pall CGN 14769 and A. altaicum CGN 1477 presented the lowest transferability (55%). Other important species, such as A. hookeri and A. fragrance, presented intermediate transfer percentages of 73.75% and 72.5%, respectively.
Genetic relationships among underutilized and cultivated Alliums
Chloroplast microsatellite markers were utilized to assess genetic relationships among 96 underutilized Allium species through neighbor-joining (NJ) cluster analysis. The dendrogram (Figure 2) grouped the accessions into six distinct clusters (I–VI), each representing varying degrees of genetic relatedness. Cluster I was the largest, consisting of 38 accessions primarily representing Allium tuberosum and closely related taxa such as A. ramosum, A. viviparum, A. wallichi, A. consanguineum, A. albidium, and A. auriculatum. These accessions presented have high genetic similarity, likely due to shared ancestry, ecological adaptation, and geographical proximity. Cluster II included eight underutilized accessions, such as the Allium chinense MMK131, Allium chinense NMK3247, Allium chinense (RAK100), Allium chinense NMK3165, A. chinense, A. chinense NMK3236 and A. fragrans genotypes, which form a genetically cohesive group on the basis of habitat and cytoplasmic traits. Cluster III comprised 23 accessions dominated by A. fistulosum, along with related species such as A. ascalonicum, A. stracheyi, A. porrum, A. roylei etc. reflecting significant diversity and wide geographical origins. Cluster IV consisted of six cultivated Allium accessions, including landraces and hybrids (Allium cepa var. aggregatum 3, 4 and 5, Allium cepa var. Bhima Super, Allium sativum var. Bhima Purple, and Allium cepa × Allium fistulosum Beltsville Bunching, showing limited diversity due to breeding bottlenecks. Cluster V included 17 underutilized accessions, such as A. macranthum and its relatives, as well as A. porrum adapted to high-altitude environments, reflecting substantial genetic divergence. Cluster VI was the smallest, comprising four A. hookeri accessions showing a distinct genetic lineage. Notably, Clusters III and V presented the highest levels of intracluster genetic diversity, whereas Clusters I and IV were relatively homogeneous.
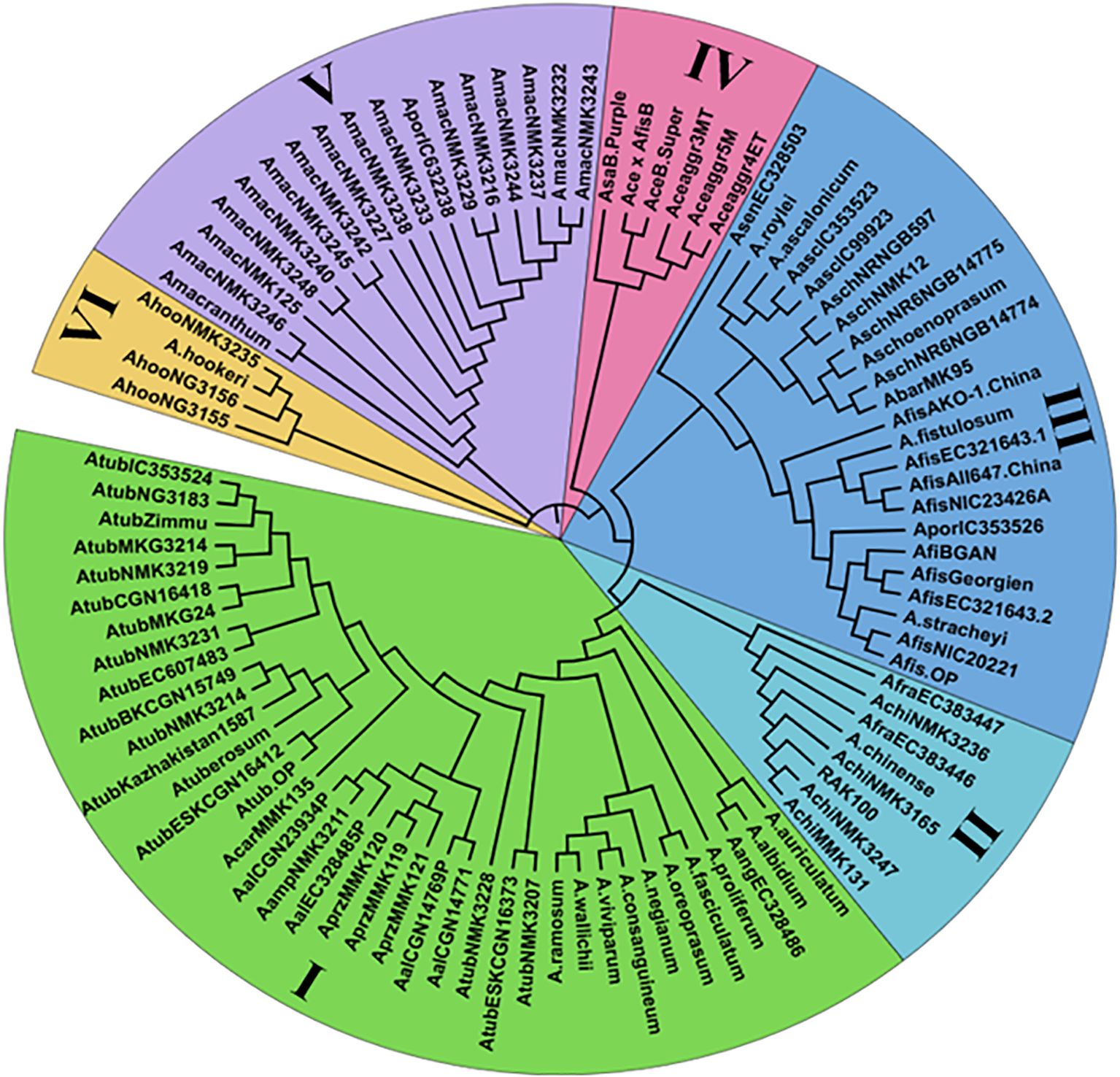
Figure 2. Dendrogram analysis of 96 underutilized Allium species based on Nei's distance of 20 novel cp-SSR markers.
Population structure analysis
The population structure was analyzed using STRUCTURE v2.3.4 software based on data from 20 cp-SSR markers. The analysis was conducted for K-values ranging from 1 to 10. As K increased, the log probability of the data [lnP(K)] also increased (Figure 3A), and the optimal number of clusters was determined using the ΔK method (Evanno et al., 2005). A clear peak at K = 3 was observed (Figure 3B), indicating the most likely number of genetic clusters. Accordingly, the accessions were grouped into three distinct sub-populations: pop1, pop2, and pop3 (Figure 3C). The mean intracluster genetic distances for these three populations were 0.2208, 0.1664, and 0.1699, respectively, whereas the average allele-frequency divergence among populations was 0.1100. The alpha mean value was 0.051, and the proportion of membership for each cluster was estimated at 0.313, 0.185, and 0.502, respectively. These results reveal a moderate level of genetic structure and highlight substantial within-population diversity among underutilized Allium species.
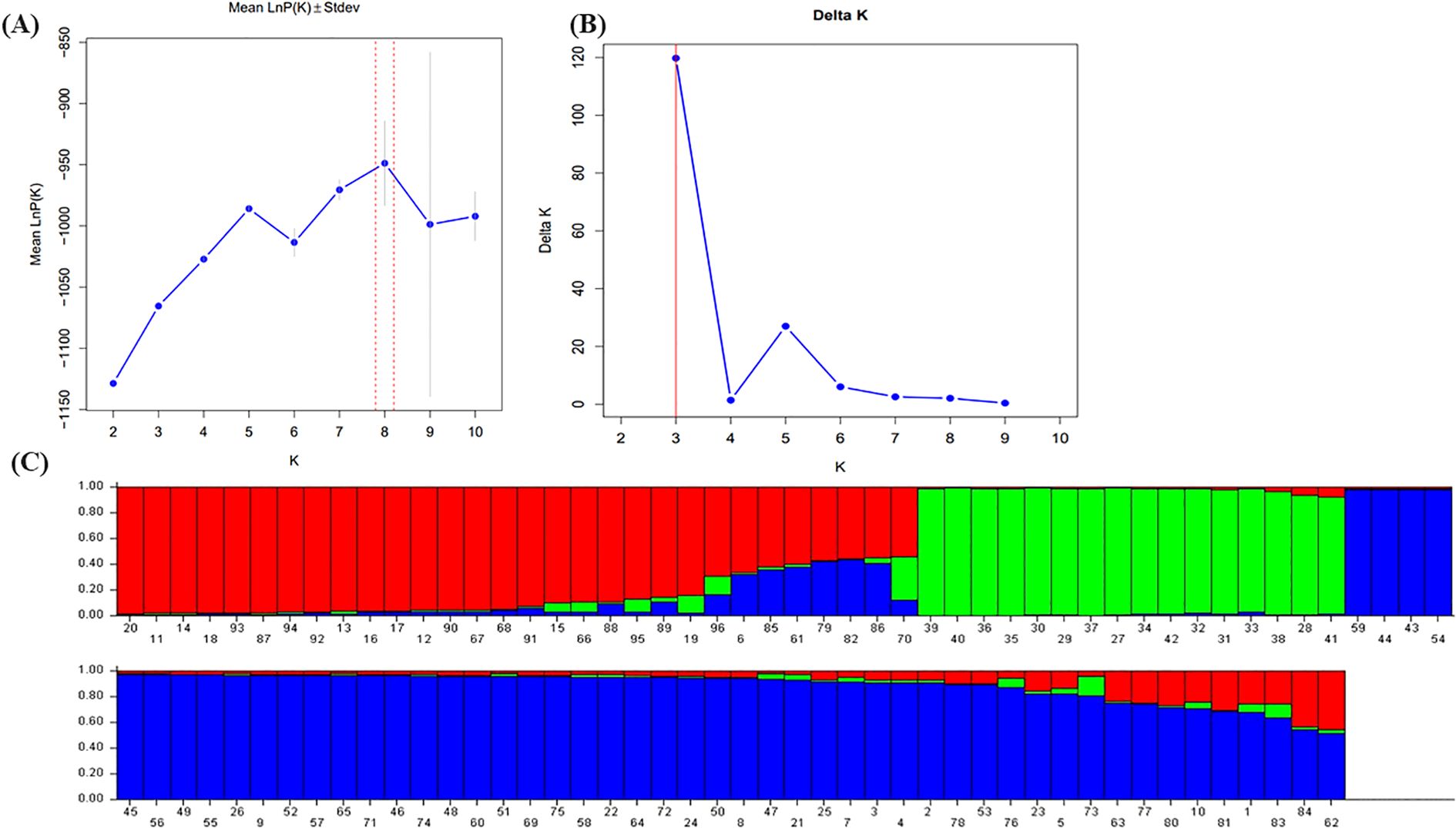
Figure 3. Population structure analysis of 96 wild Allium species based on 22 cp-SSR markers. (A) The mean value of InP (D) was used to estimate the population structure, and the range of K-values was 1-10. (B) Using the cuive of ΔK obtained by InP(K), the optimal K-value was determined to be 3. (C) The 96 underutilized Allium species studied clustered in three subgroups (subgroup I, red; subgroup II, green; and subgroup III, blue). Each histogram represents a germplasm in which different colors represent the estimated component coefficients using Q-values.
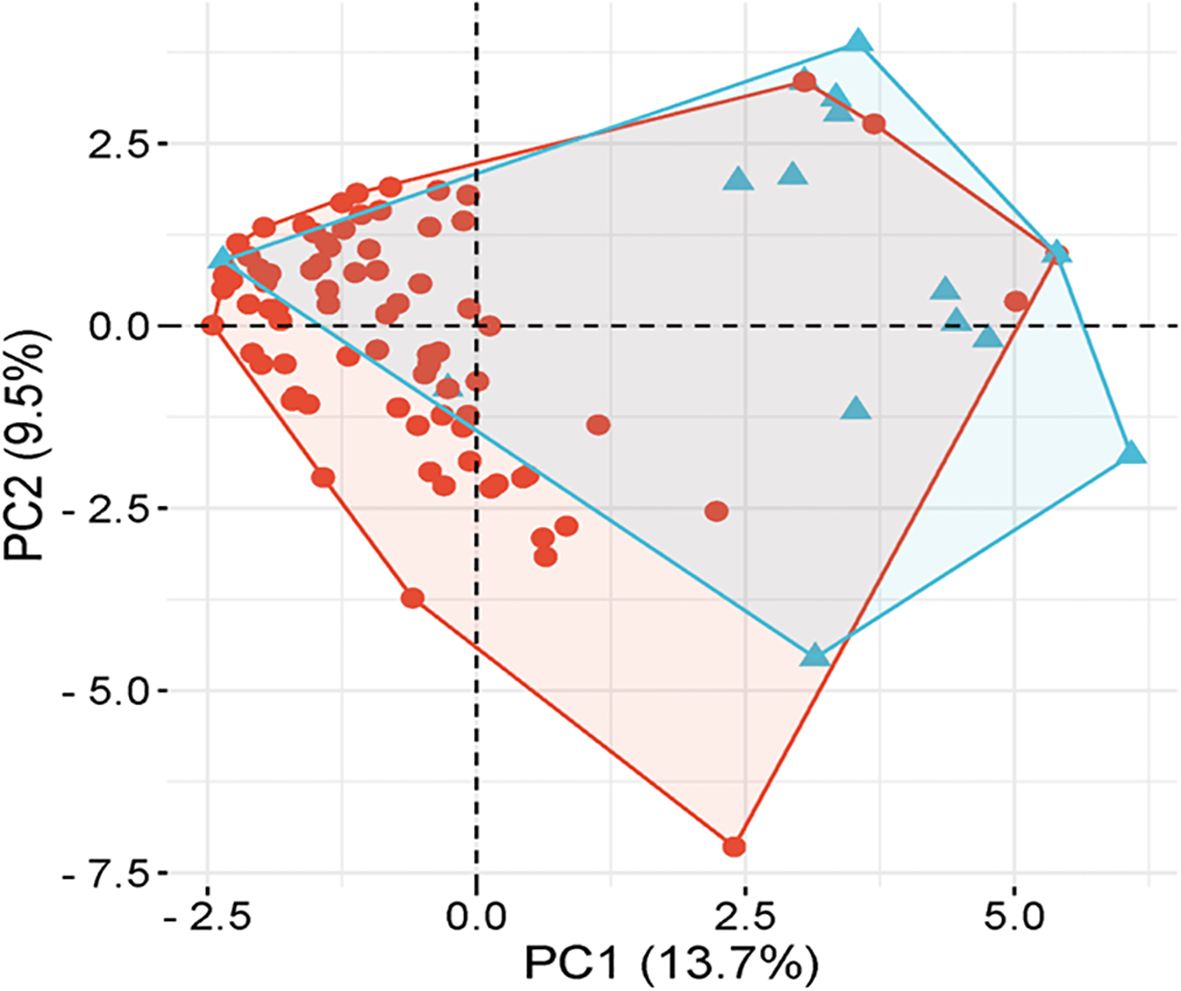
Principal component analysis and heatmap analysis
PCA analysis revealed that the first two principal components, PC1 and PC2, accounted for 13.7% and 9.5% of the total genetic variation, respectively (Figure 4). The distribution of accessions across the PCA biplot indicated the presence of two major genetic clusters. The first cluster displayed broader dispersion, primarily in the negative PC1 axis, suggesting greater genetic diversity. In contrast, the second cluster appeared more compact and tightly grouped, indicating higher genetic similarity among its members. The overlap observed between the two clusters suggested the presence of genetic admixture among certain accessions. In accordance with the PCA results, the heatmap analysis (Figure 5) presented a color-coded visualization of pairwise genetic similarity, with red shading indicating high similarity (values closer to 1.0) and gray shading denoting low similarity (values closer to -1.0). The accompanying hierarchical clustering dendrogram revealed distinct clusters of genetically similar accessions, whereas vertical patterns across the heatmap highlighted conserved genetic markers.
Discussion
Chloroplast SSR markers have proven to be highly informative in studies of plant genetic diversity, phylogenetics, and population structure because of their uniparental inheritance, low recombination rates, and conserved genomic context. The central focus of this study was the development and characterization of chloroplast simple sequence repeat (cp-SSR) markers in Allium fistulosum, with subsequent amplification testing across 96 underutilized Allium species. A total of 22 novel cp-SSR loci were identified and validated, revealing distinctive patterns in repeat motif distribution that contribute to our understanding of cp-SSR evolution and utility within the Allium genus. Our findings revealed that tetranucleotide repeats were the most abundant (45.45%), followed by dinucleotide repeats (27.27%). Notably, mononucleotide repeats were deliberately excluded due to their lower informativeness and susceptibility to sequencing errors and polymerase slippage, a strategy aligned with best practices in cp-SSR marker development (Li et al., 2020). This exclusion also avoided overrepresentation of polyA/polyT stretches, which are highly abundant but offer limited polymorphic information. Furthermore, hexanucleotide motifs were absent, consistent with previous cp-SSR studies where longer motifs are generally rare in chloroplast genomes.
Our results partly align with prior studies in Allium species. For instance, Jayaswall et al. (2022) reported the development of 22 cp-SSRs in Allium cepa and Allium sativum, and 15 cp-SSRs in Allium paradoxum. In contrast to our current findings, their work found that tri-nucleotide repeats were the most frequent motif type (50%), suggesting potential interspecific variation in repeat motif composition within the genus Allium. This difference could be attributed to genomic structural variation or differing evolutionary pressures across species. When compared to broader angiosperm studies, our cp-SSR motif composition is consistent with patterns observed in other taxa. For example, Guo et al. (2022) identified 139 cp-SSR loci across 11 tree peony plastomes, while Shukla et al. (2018) reported 21–25 cp-SSRs in various Vigna species (V. angularis, V. radiata, and V. unguiculata), and Huang et al. (2018) found 92 SSR loci across six Cupressaceae plastomes. In these studies, di- and tetranucleotide repeats also predominated, underscoring a conserved pattern of SSR distribution in chloroplast genomes across plant lineages.
Interestingly, our findings contrast with those of Feng et al. (2023) in Physalis angulata, where mononucleotide repeats were the most abundant (68.24%), followed by tetranucleotides (12.28%). The abundance of mononucleotide motifs in that study likely reflects a different analytical approach that included these repeats, which, while common, are typically avoided in marker development due to their lower polymorphic potential. Overall, the distribution of cp-SSR motif types in Allium fistulosum reflects both conserved and species-specific patterns observed across plant taxa. Our deliberate methodological choices such as excluding mononucleotide repeats support the development of highly informative, polymorphic, and stable markers, which are essential for downstream applications such as population genetics, phylogenetic reconstruction, and genetic diversity studies in Allium and related genera. This work contributes to the growing genomic toolkit for Allium research and supports future efforts in conservation and breeding of underutilized species. The high success rate of amplification (90.91%) and clear electrophoretic profiles of these cp-SSR markers demonstrate their robustness and reliability for genetic analysis. Similar success has been reported in other species, such as tree peonies, where 19 out of 21 cp-SSR markers amplified strongly (Guo et al., 2022). The cp-SSR markers developed here complement existing nuclear SSRs by capturing maternal lineage information, thereby enhancing the resolution of genetic diversity studies in Allium species. Moreover, these markers are expected to support broader applications in phylogeography, conservation genetics, and breeding. As more chloroplast genome sequences become available, the cross-transferability and expansion of cp-SSR marker sets will continue to facilitate species-specific and cross-species analyses (Huo et al., 2019; Feng et al., 2023). This study lays the groundwork for future genomic research and supports the strategic use of cp-SSR markers in Allium crop improvement and biodiversity assessment programs.
The cp-SSR marker analysis confirmed the effectiveness of the developed markers in revealing polymorphisms and genetic variation within Allium germplasm. The high rate of polymorphism (89.2%) and broad allele size range reflect the utility of these markers for studying genetic structure and relationships, especially in underutilized populations. The average PIC value (0.20) aligns with previous findings by Jayaswall et al. (2023), who reported PIC values ranging from 0.007 to 0.427 in Allium germplasm via chloroplast-derived SSR markers. Although slightly lower than in studies by Gowd et al. (2024) (PIC: 0.24–0.98; avg. 0.608), Mallor et al. (2014) (avg. 0.64), and Hanci and Gökçe (2016) (up to 0.7), the moderate PIC values in this study may be attributed to differences in genome source (chloroplast vs. nuclear SSRs), marker selection criteria, and the genetic backgrounds of the tested accessions.
Similar studies have reported varying PIC values, with Baldwin et al. (2012) and Rivera et al. (2016) reporting averages of 0.45 and 0.51, respectively. Lyngkhoi et al. (2021) reported 53 alleles using 145 SSR markers, with PIC values ranging from 0.219 to 0.715 and an average of 3.54 alleles per locus. Khar et al. (2011) reported PIC values ranging from 0.00 to 0.89 with 60 primers, detecting 54 alleles across 19 primers, with an average of 2.84 alleles per locus. These comparisons highlight the impact of population structure, genomic origin, and SSR motif type on marker informativeness. Highly polymorphic markers such as cp-SSR3 and cp-SSR14, with more than 80% polymorphism, offer strong potential for use in genetic mapping and diversity studies. Similar findings by Gowd et al. (2024), where 92 polymorphic loci were identified using 19 SSR markers across 95 Allium accessions, underscore the importance of SSRs in understanding genetic variation.
Jayaswall et al. (2023) further demonstrated the utility of chloroplast-derived SSR markers in A. cepa and A. sativum, reporting heterozygosity values ranging from 0.009 to 0.540 and PIC values ranging from 0.007 to 0.427. These markers offer a reliable platform for evaluating genetic relationships between underutilized and cultivated Allium species. The observed genetic diversity in underutilized Allium accessions holds critical value for crop improvement. Traits such as disease resistance, yield enhancement, and abiotic stress tolerance can be introgressed from underutilized relatives into cultivated backgrounds. Therefore, the conservation and characterization of underutilized Allium germplasm remains essential for the resilience and sustainability of breeding programs. Recent advances underscore the complementary role of cp-SSRs markers alongside genomic tools in exploring Allium genetic diversity and evolutionary history (Khosa et al., 2015; Jayaswall et al., 2019; Gowd et al., 2024). The integration of cp-SSR data with nuclear SSR and genome-wide SNP dataset will further enrich our understanding of the genetic makeup of Allium species and support targeted breeding and conservation strategies. The observed number of alleles (Na) and effective number of alleles (Ne) support the existence of moderate polymorphism across the cp-SSR markers used. Markers such as cp-SSR09 and cp-SSR10, which exhibited relatively higher number of alleles and diversity indices, are particularly useful in revealing genetic differences among Allium accessions. In contrast, monomorphic markers such as cp-SSR02, cp-SSR03, and cp-SSR11 are likely associated with conserved regions of the chloroplast genome, offering limited intraspecies resolution but potential value for interspecific or phylogenetic studies (Gowd et al., 2024; Karic et al., 2018).
Generally, low Ho values align with the uniparental (mostly maternal) inheritance and haploid nature of the chloroplast genome, as well as the self-pollinating behaviour of many Allium species (Lyngkhoi et al., 2021). Nevertheless, the high He and I values of cp-SSR10 demonstrate its potential for distinguishing diverse genotypes and tracking lineage relationships. These findings are consistent with previous cp-SSR studies in Allium, where allele numbers typically ranged between 2 and 5 (Lyngkhoi et al., 2021), and in other genera, such as Ziziphus (Huang et al., 2015). Compared with nuclear SSRs and EST-SSRs, cp-SSRs tend to be less polymorphic, because they are located in more conserved regions of the genome (Ricciardi et al., 2020; Baldwin et al., 2012; Khar et al., 2011). However, their high cross-species transferability and evolutionary stability make them ideal for phylogenetic studies and for characterizing maternal lineages.
The cross-transferability results indicate a broad genetic base within the genus Allium. High transferability rates in species such as A. viviparum and A. negianum suggest their close genetic relationships with other members of the genus and their potential utility in breeding programs. In contrast, lower transferability in accessions such as A. altaicum indicates possible genomic divergence or evolutionary distance. These patterns of allele sharing and divergence can be exploited for both germplasm conservation and introgression breeding strategies aimed at enhancing stress tolerance or other desirable traits. Overall, the cp-SSR markers were effective in evaluating genetic diversity, understanding evolutionary relationships, and identifying candidate accessions for conservation and breeding. The combination of highly polymorphic and conserved markers allows a dual-purpose application: detailed intraspecies diversity analysis and broader phylogenetic studies across underutilized and cultivated Allium species.
Owing to their uniparental inheritance and lack of recombination, chloroplast SSR markers are well-suited for studying genetic relationships, evolutionary history, and domestication processes in plants (Sharma et al., 2020a). In the present study, these markers effectively distinguished underutilized and cultivated Allium species into six well-defined genetic clusters. The genetic homogeneity of Cluster I highlights the close relatedness among A. tuberosum and its allies, likely due to shared ancestry and cultivation across similar ecological regions, which aligns with the findings of Jayaswall et al. (2023), who reported similar clustering patterns. Cluster II, composed of various underutilized accessions, including A. chinense and A. fragrans, reflects cytoplasmic similarity and ecological coherence, corroborating earlier findings by Gowd et al. (2024). The presence of both cultivated and underutilized species in Cluster III emphasizes its potential as a genetic bridge, with A. fistulosum and related species offering valuable traits such as disease resistance and abiotic stress tolerance. Cluster IV, comprising cultivated A. cepa genotypes, exhibited reduced diversity, a consequence of domestication and selective breeding, which is consistent with domestication bottlenecks observed by Jayaswall et al. (2023). Cluster V presented the greatest genetic divergence, harboring underutilized species adapted to niche environments such as high altitudes, in agreement with findings from Nanda et al. (2016) and Gowd et al. (2024), highlighting their value as reservoirs of unique alleles. Cluster VI, comprising A. hookeri, presented species-specific genetic uniformity, likely due to restricted geographical distribution and domestication. Despite their narrow diversity, the members of this cluster possess unique traits that are valuable for region-specific applications. The high genetic variation observed in Clusters III and V underscores the evolutionary potential of underutilized species, reaffirming the importance of integrating underutilized relatives into breeding programs to increase stress tolerance, disease resistance, and adaptability in cultivated Allium species.
Understanding population structure is critical for effective germplasm conservation, trait mapping, and breeding applications. The identification of three distinct genetic clusters among the 96 underutilized Allium accessions aligns with earlier findings by Jayaswall et al. (2023), who also reported three subpopulations using cp-SSR markers in Allium accessions. The moderate allele–frequency divergence (0.11) and varying intracluster distances observed in this study suggest both shared ancestry and independent evolutionary trajectories among the populations. The relatively high proportion of membership in pop3 (50.2%) suggests a broad and genetically diverse group, potentially encompassing accessions with mixed ancestry. In contrast, pop2, with a lower proportion (18.5%), may represent a more genetically uniform or isolated subset. The findings also correlate with those of Lyngkhoi et al. (2021), who identified two groups in 96 underutilized Allium accessions through STRUCTURE analysis and five groups via discriminant analysis, illustrating how methodology and marker type influence the resolution of population structure. Gowd et al. (2024) reported four clusters using nuclear SSRs, highlighting differences attributable to marker origin (chloroplast vs. nuclear). Similarly, Chalbi et al. (2023) reported population differentiation in Allium landraces based on accession type rather than phenotypic traits. These collective observations underscore the utility of cp-SSR markers in deciphering maternal lineage and cytoplasmic diversity, which are particularly important for breeding strategies involving cytoplasmic male sterility or other organelle-linked traits. Thus, marker-based population structure analysis not only facilitates an understanding of genetic diversity but also provides a valuable framework for selecting parental lines and managing Allium germplasm effectively.
Together, the PCA and heatmap analyses provided a nuanced understanding of the genetic diversity and structure within the underutilized Allium accessions. The separation along PC1 likely reflects deep evolutionary divergence, whereas PC2 captures more recent or subtle genetic differentiation. The broader dispersion observed in one cluster indicates high intragroup variability, potentially representing genetically diverse underutilized relatives with adaptive significance. Moreover, the tighter grouping of the second cluster suggests a subset of accessions with conserved genomic features, possibly shaped by shared ancestry or ecological adaptation. These observations are consistent with common patterns in plant population genetics, where variable levels of diversity are often observed within and among clusters (Spanoghe et al., 2020). Notably, the PCA-based clustering results corresponded well with the three subpopulations identified through STRUCTURE analysis, supporting the robustness and complementary nature of both methods. The overlapping zones in the PCA further corroborate previous studies reporting gene flow and admixture among Allium species (Xiong et al., 2022). The heatmap visualization further reinforced the PCA outcomes by graphically representing the levels of genetic similarity and divergence, with the dendrogram effectively grouping genetically close accessions. Similar integrative approaches have proven valuable in deciphering population structure and evolutionary relationships in Allium and other crop species (Anwar et al., 2017; Saina et al., 2023). Overall, these findings not only validate the genetic groupings but also emphasize the utility of multivariate and hierarchical clustering tools in germplasm characterization, aiding in the selection of genetically diverse and elite accessions for breeding and conservation programs.
Conclusion
In conclusion, this study has significantly advanced our understanding of the genetic diversity of underutilized Allium species through the development and application of novel chloroplast SSR markers. The identification of 22 cp-SSR motifs from the A. fistulosum chloroplast genome, with 20 markers exhibiting high polymorphism and stability, provides a robust toolkit for genetic analysis in Allium. The high level of polymorphism (89.2%) observed across 96 underutilized Allium species underscores the effectiveness of these markers in capturing genetic variation. The population structure analysis revealed three distinct genetic clusters, complemented by phylogenetic grouping into six major clusters, which offers valuable insights into the evolutionary relationships and genetic differentiation within the genus. These findings have important implications for Allium conservation strategies and breeding programs, highlighting the potential of underutilized germplasm as a reservoir of genetic diversity for crop improvement. This study demonstrates the utility of cp-SSR markers in revealing the complex genetic tapestry of Allium species, paving the way for precision-guided conservation efforts and the development of improved cultivars with enhanced traits such as disease resistance and stress tolerance.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
YK: Software, Methodology, Writing – review & editing, Supervision, Writing – original draft, Investigation, Conceptualization, Funding acquisition, Formal Analysis, Visualization, Data curation, Resources, Project administration, Validation. PM: Formal Analysis, Writing – original draft, Methodology, Investigation. AC: Writing – review & editing, Formal Analysis. KR: Writing – review & editing, Resources, Formal Analysis. SS: Software, Data curation, Formal Analysis, Methodology, Writing – original draft. MJ: Writing – original draft, Methodology, Software, Data curation. AP: Methodology, Writing – original draft, Software, Data curation. VH: Writing – original draft. AR: Methodology, Formal Analysis, Data curation, Writing – review & editing. SM: Writing – review & editing, Formal Analysis, Methodology, Data curation. AK: Validation, Methodology, Writing – original draft, Software, Formal Analysis. HB: Investigation, Software, Data curation, Writing – original draft, Project administration, Methodology. AG: Writing – review & editing, Methodology, Data curation. RK: Resources, Writing – review & editing, Software, Formal Analysis. KP: Methodology, Software, Data curation, Writing – review & editing. VM: Writing – review & editing, Funding acquisition, Resources, Project administration.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors thank the Indian Council of Agricultural Research- National Bureau of Plant Genetic Resources, New Delhi and its regional station, Bhowali, Uttarakhand, India for sharing underutilized species of Allium used in the research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1645145/full#supplementary-material
References
Anandhan, S., Nair, A., Kumkar, D. S., and Gopal, J. (2015). Retrotransposon based TRAP marker displays diversity among onion (Allium cepa L.) genotypes. Scientia Hortic. 190, 123–127. doi: 10.1016/j.scienta.2015.04.024
Anwar, G. M., Helmey, R. K., and Moustafa, Y. M. M. (2017). Genetic diversity among some species of the genus Allium L. using SSR and ISSR markers. Egyptian J. Genet. Cytology. 46, 221–233. doi: 10.21608/ejgc.2018.9516
Arifin, N. S., Ozaki, Y., and Okubo, H. (2000). Genetic diversity in Indonesian shallot (Allium cepa var. ascalonicum) and Allium × wakegi revealed by RAPD markers and origin of A. × wakegi identified by RFLP analyses of amplified chloroplast genes. Euphytica 111, 23–31. doi: 10.1023/A:1003732403402
Baldwin, S., Pither Joyce, M., Wright, K., Chen, L., and McCallum, J. (2012). Development of robust genomic simple sequence repeat markers for estimation of genetic diversity within and among bulb onion (Allium cepa L.) populations. Mol. Breeding. 30, 1401–1411. doi: 10.1007/s11032-012-9727-6
Brahimi, A., Hajjaj, H., and Mazouz, H. (2024). Genetic characterization of white onion genotypes (Allium cepa L.) using ISSR markers. Ecol. Genet. Genomics 33, 100287. doi: 10.1016/j.egg.2024.100287
Brahimi, A., Landschoot, S., Bekaert, B., Hajji, L., Hajjaj, H., Audenaert, K., et al. (2022). Exploring the genetic and phenotypic diversity within and between onion (Allium cepa L.) ecotypes in Morocco. J. Genet. Eng. Biotechnol. 20, 96. doi: 10.1186/s43141-022-00381-w
Chalbi, A., Chikh-Rouhou, H., Mezghani, N., Slim, A., Fayos, O., Bel-Kadhi, M. S., et al. (2023). Genetic diversity analysis of onion (Allium cepa L.) from the arid region of Tunisia using phenotypic traits and SSR markers. Horticulturae 9, 1098. doi: 10.3390/horticulturae9101098
Chand, S. K., Nanda, S., and Joshi, R. K. (2018). Genetics and molecular mapping of a novel purple blotch-resistant gene ApR1 in onion (Allium cepa L.) using STS and SSR markers. Mol. Breed. 38, 109. doi: 10.1007/s11032-018-0864-4
Damon, S. J. and Havey, M. J. (2014). Quantitative trait loci controlling amounts and types of epicuticular waxes in onion. J. Am. Soc. Hortic. Sci. 139, 597–602. doi: 10.21273/JASHS.139.5.597
dos Santos, M. D., Ragassi, C. F., Fonseca, M. E., Buzar, A. G., Oliveira, V. R., de Melo, P. C., et al. (2012). Genetic diversity of tropical-adapted onion germplasm assessed by RAPD markers. Horticultura Bras. 30, 112–118.
Duangjit, J., Bohanec, B., Chan, A. P., Town, C. D., and Havey, M. J. (2013). Transcriptome sequencing to produce SNP-based genetic maps of onion. TAG. Theor. Appl. Genet. 126, 2093–2101. doi: 10.1007/s00122-013-2121-x
Ebert, D. and Peakall, R. O. D. (2009). Chloroplast simple sequence repeats (cp-SSRs): technical resources and recommendations for expanding cp-SSR discovery and applications to a wide array of plant species. Mol. Ecol. Resources. 9, 673–690. doi: 10.1111/j.1755-0998.2008.02319.x
Evanno, G., Regnaut, R., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365294x.2005.02553.x
Feng, S., Jiao, K., Zhang, Z., Yang, S., Gao, Y., Jin, Y., et al. (2023). Development of chloroplast microsatellite markers and evaluation of genetic diversity and population structure of cutleaf groundcherry (Physalis angulata L.) in China. Plants 12, 1755. doi: 10.3390/plants12091755
Fritsch, R. M. and Friesen, N. (2002). “Evolution, domestication and taxonomy,” in Allium crop science: recent advances (CABI, Wallingford UK), 5–30. doi: 10.1079/9780851995106.0005
Gowd, T. Y. M., Deo, C., Manjunathagowda, D. C., Mahajan, V., Dutta, R., Bhutia, N. D., et al. (2024). Deciphering genetic diversity phylogeny and assembly of Allium species through micro satellite markers on nuclear DNA. Heliyon 10, e31650. doi: 10.1016/j.heliyon.2024.e31650
Gowd, T. Y. M., Manjunathagowda, D. C., Mahajan, V., Dutta, R., Bhutia, N. D., Singh, B., et al. (2023). Deployment of intron length polymorphic (ILP) markers in dissipating diversity of Allium species. South Afr. J. Bot. 160, 157–165.
Guo, Q., Guo, L., Li, Y., Yang, H., Hu, X., Song, C., et al. (2022). Development and characterization of microsatellite markers based on the chloroplast genome of tree peony. Genes 13, 1543. doi: 10.3390/genes13091543
Gupta, A. J., Anandhan, S., Kad, S. K., and Singh, M. (2020). Confirmation of hybridity in DOGR hybrids of onion (Allium cepa L.) using SSR markers. Vegetable Sci. 47, 183–188. doi: 10.61180/vegsci.2020.v47.i2.03
Habsatou, B., Broum, P., Bellanger, L., and Crouzillat, D. (2024). Genetic diversity of red onion using microsatellite (SSRs) markers. Scholars Acad. J. Biosci. 8, 278–284. doi: 10.36347/sajb.2024.v12i08.007
Hanci, F. and Gökçe, A. F. (2016). Molecular characterization of Turkish onion germplasm using SSR markers. Czech J. Genet. Plant Breeding. 52, 71–76. doi: 10.17221/162/2015-CJGPB
Hancı, F. and Paşazade, E. (2025). A comparison of efficiency parameters of SRAP and ISSR markers in revealing variation in Allium germplasm. Horticulturae 11, 294. doi: 10.3390/horticulturae11030294
Hu, X., Hu, J., Zhang, Y., Jiang, S., and Yu, Q. (2022). Genetic structure and demographic history of Allium mongolicum based on SSR markers. Plant Systematics Evol. 308, 12. doi: 10.1007/s00606-021-01802-y
Huang, L. S., Sun, Y. Q., Jin, Y., Gao, Q., Hu, X. G., Gao, F. L., et al. (2018). Development of high transferability cp-SSR markers for individual identification and genetic investigation in Cupressaceae species. Ecol. Evol. 8, 4967–4977. doi: 10.1002/ece3.4053
Huang, J., Yang, X., Zhang, C., Yin, X., Liu, S., and Li, X. (2015). Development of chloroplast microsatellite markers and analysis of chloroplast diversity in Chinese jujube (Ziziphus jujuba Mill.) and wild jujube (Ziziphus acidojujuba Mill.). PloS One 10, e0134519. doi: 10.1371/journal.pone.0134519
Huo, Y., Gao, L., Liu, B., Yang, Y., Kong, S., Sun, Y., et al. (2019). Complete chloroplast genome sequences of four Allium species: comparative and phylogenetic analyses. Sci. Rep. 9, 12250. doi: 10.1038/s41598-019-48708-x
Jayaswall, K., Bhandawat, A., Sharma, H., Yadav, V. K., Mahajan, V., and Singh, M. (2019). Characterization of Allium germplasms for conservation and sustainable management using SSR markers. Indian J. Tradit Knowl. 18, 193–199.
Jayaswall, K., Sagar, R., Jayaswal, D., Kumar, A., Singh, S. P., Seth, R., et al. (2024). Development of Allium cepa potential intron polymorphism markers for molecular breeding of Alliums. South Afr. J. Bot. 164, 209–220. doi: 10.1016/j.sajb.2023.11.050
Jayaswall, K., Sharma, H., Bhandawat, A., Sagar, R., Jayaswal, D., Kumar, A., et al. (2022). Chloroplast derived SSRs reveals genetic relationships in domesticated alliums and wild relatives. Genet. Resour Crop Evol. 69, 363–372. doi: 10.1007/s10722-021-01235-z
Jayaswall, K., Sharma, H., Jayaswal, D., Sagar, R., Bhandawat, A., Kumar, A., et al. (2023). Development of chloroplast derived SSR markers for genus Allium and their characterization in the allies for genetic improvement of Alliums. South Afr. J. Botany. 162, 304–313. doi: 10.1016/j.sajb.2023.09.021
Karic, L., Golzardi, M., and Glamoclija, S. (2018). Genetic diversity assessment of Allium cepa L. cultivars from Bosnia and Herzegovina using SSR makers. Genet. Mol. Res. 17, gmr16039870. doi: 10.4238/gmr16039870
Khade, Y. P., Salunkhe, S. R., Manjunathagowda, D. C., Sinhasane, S. R., Gowd, T. Y. M., Mahajan, V., et al. (2022). Molecular characterization of short-day onion genotypes by intron length polymorphic (ILP) markers. Genet. Resour Crop Evol. 69, 2077–2086. doi: 10.1007/s10722-022-01398-3
Khade, Y. P., Sinhasane, S. R., Mainkar, P., Rai, K. M., Salunkhe, S., Singh, P. R., et al. (2024). Exploring sequence-related amplified polymorphism (SRAP) markers for assessing genetic diversity in onion (Allium cepa L.) genotypes and their wild relatives. Genet. Resour. Crop Evol. 72, 399–415. doi: 10.1007/s10722-024-01978-5
Khar, A., Lawande, K. E., and Negi, K. S. (2011). Microsatellite marker based analysis of genetic diversity in short day tropical Indian onion and cross amplification in related Allium spp. Genet. Resour. Crop evolution. 58, 741–752. doi: 10.1007/s10722-010-9616-y
Khosa, J. S., Dhatt, A. S., Negi, K. S., and Khar, A. (2014). Utility of simple sequence repeat (SSR) markers to realize worth of germplasm in genus Allium. Indian J. Plant Genet. Resour. 27, 238–245. doi: 10.5958/0976-1926.2014.00020.5
Khosa, J. S., McCallum, J., Dhatt, A. S., and Macknight, R. C. (2015). Enhancing onion breeding using molecular tools. Plant Breeding. 135, 9–20. doi: 10.1111/pbr.12330
Kisha, T. J. and Cramer, C. S. (2011). Determining redundancy of short-day onion accessions in a germplasm collection using microsatellite and targeted region amplified polymorphic markers. J. Am. Soc. Hortic. Sci. 136, 129–134. doi: 10.21273/JASHS.136.2.129
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. evolution. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kutty, M., Veere Gowda, R., and Anand, L. (2006). Analysis of genetic diversity among Indian short-day onion (Allium cepa L.) cultivars using RAPD markers. J. Hortic. Sci. Biotechnol. 81, 774–778. doi: 10.1080/14620316.2006.11512136
Labate, J. A., Glaubitz, J. C., and Havey, M. J. (2020). Genotyping by sequencing for SNP marker development in onion. Genome 63, 607–613. doi: 10.1139/gen-2020-0011
Li, C., Zheng, Y., and Huang, P. (2020). Molecular markers from the chloroplast genome of rose provide a complementary tool for variety discrimination and profiling. Sci. Rep. 10, 12188. doi: 10.1038/s41598-020-68092-1
Lyngkhoi, F., Saini, N., Gaikwad, A. B., Thirunavukkarasu, N., Verma, P., Silvar, C., et al. (2021). Genetic diversity and population structure in onion (Allium cepa L.) accessions based on morphological and molecular approaches. Physiol. Mol. Biol. Plants. 27, 2517–2532. doi: 10.1007/s12298-021-01101-3
Mahajan, V., Jakse, J., Havey, M. J., and Lawande, K. E. (2009). Genetic fingerprinting of onion cultivars using SSR markers. Indian J. Horticulture 66, 62–68.
Mallor, C., Arnedo-Andres, M. S., and Garces-Claver, A. (2014). Assessing the genetic diversity of Spanish Allium cepa landraces for onion breeding using microsatellite markers. Scientia Horticulturae. 170, 24–31. doi: 10.1016/j.scienta.2014.02.040
Mandakh, U., Battseren, M., Ganbat, D., Ayanga, T., Adiya, Z., Borjigidai, A., et al. (2020). Folk nomenclature of plants in Cistanche deserticola-associated community in South Gobi, Mongolia. Plant Diversity. 42, 434–442. doi: 10.1016/j.pld.2020.09.008
Mansour, E. M., Elballa, M. M., El Hussein, A. A., Abdalla, A. W. H., Gadir, I. K. A., Abbo, A. S., et al. (2020). Assessment of genetic variability among onion (Allium cepa L.) cultivars using RAPD and SRAP molecular markers. Univ. Khartoum J. Agric. Sci. 28, 1–24. doi: 10.53332/uofkjas.v28i.177
McCallum, J., Baldwin, S., Shigyo, M., Deng, Y., van Heusden, S., PitherJoyce, M., et al. (2012). Allium Map-A comparative genomics resource for cultivated Allium vegetables. BMC Genomics 13, 168. doi: 10.1186/1471-2164-13-168
McCallum, J., Leite, D., PitherJoyce, M., and Havey, M. J. (2001). Expressed sequence markers for genetic analysis of bulb onion (Allium cepa L.). Theor. Appl. Genet. 103, 979–991. doi: 10.1007/s001220100630
McCauley, D. E. (1995). The use of chloroplast DNA polymorphism in studies of gene flow in plants. Trends Ecol. evolution. 10, 198–202. doi: 10.1016/S0169-5347(00)89052-7
Metsalu, T. and Vilo, J. (2015). ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 43, W566–W570. doi: 10.1093/nar/gkv468
Mohapatra, P. P., Seleiman, M. F., Mandal, R., Pramanik, K., Maity, T. K., Tarafdar, J., et al. (2023). Efficiency of RAPD and SSR markers in assessing genetic diversity in summer onion (Allium cepa L.) genotypes. Notulae Botanicae Horti Agrobotanici ClujNapoca 51, 13369. doi: 10.15835/nbha51313369
Monteverde, E., Galvan, G. A., and Speranza, P. (2015). Genetic diversification of local onion populations under different production systems in Uruguay. Plant Genet. Resour. 13, 238–246. doi: 10.1017/S1479262114000963
Murray, M. G. and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Nanda, S., Chand, S. K., Mandal, P., Tripathy, P., and Joshi, R. K. (2016). Identification of novel source of resistance and differential response of Allium genotypes to purple blotch pathogen, Alternaria porri (Ellis) Ciferri. Plant Pathol. J. 32, 519. doi: 10.5423/PPJ.OA.02.2016.0034
Nouri, N., Devineni, N., Were, V., and Khanbilvardi, R. (2021). Explaining the trends and variability in the United States tornado records using climate teleconnections and shifts in observational practices. Sci. Rep. 11, 1741. doi: 10.1038/s41598-021-81143-5
Omelchenko, D. O., Krinitsina, A. A., Belenikin, M. S., Konorov, E. A., Kuptsov, S. V., Logacheva, M. D., et al. (2020). Complete plastome sequencing of Allium paradoxum reveals unusual rearrangements and the loss of the ndh genes as compared to Allium ursinum and other onions. Gene 726, 144154. doi: 10.5423/PPJ.OA.02.2016.0034
Peakall, R. and Smouse, P. E. (2012). GenAlEx 6.5: Genetic Analysis in Excel. Population genetic software for teaching and research—An update. Bioinf. Appl. Note. 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Powell, W., Morgante, M., Andre, C., McNicol, J. W., Machray, G. C., Doyle, J. J., et al. (1995). Hypervariable microsatellites provide a general source of polymorphic DNA markers for the chloroplast genome. Curr. Biol. 5, 1023–1029. doi: 10.1016/S0960-9822(95)00206-5
Ricciardi, L., Mazzeo, R., Marcotrigiano, A. R., Rainaldi, G., Iovieno, P., Zonno, V., et al. (2020). Assessment of genetic diversity of the “Acquaviva red onion” (Allium cepa L.) apulian landrace. Plants 9, 260. doi: 10.3390/plants9020260
Rivera, A., Mallor, C., Garcés-Claver, A., García-Ulloa, A., Pomar, F., and Silvar, C. (2016). Assessing the genetic diversity in onion (Allium cepa L.) landraces from northwest Spain and comparison with the European variability. New Z. J. Crop Hortic. Science. 44, 103–120. doi: 10.1080/01140671.2016.1150308
Rohlf, F. J. (1998). “NTSYSpc,” in Numerical taxonomy and multivariate analysis system. Version 2.0 (User Guide: Exeter Software, Setauket, New York), 31.
Sahoo, J., Mahanty, B., Mishra, R., and Joshi, R. K. (2023). Development of SNP markers linked to purple blotch resistance for marker-assisted selection in onion (Allium cepa L.) breeding. 3 Biotech. 13, 137. doi: 10.1007/s13205-023-03562-7
Saina, J. K., Li, Z. Z., Ngarega, B. K., Gituru, R. W., Chen, J. M., and Liao, Y. Y. (2023). Exploring the genetic diversity and population structure of Ailanthus altissima using chloroplast and nuclear microsatellite DNA markers across its native range. Front. Plant Science. 14. doi: 10.3389/fpls.2023.1197137
Scholten, O. E., van Aauwen, M. P. W., Shahin, A., Hendrickx, P. M., Keizer, L. C. P., Burger, K., et al. (2016). SNP-markers in Allium species to facilitate introgression breeding in onion. BMC Plant Biol. 16, 187. doi: 10.1186/s12870-016-0879-0
Sharma, H., Bhandawat, A., and Rawat, S. (2020a). Cross-transferability of SSR markers developed in Rhododendron species of Himalaya. Mol. Biol. Rep. 47, 6399–6406. doi: 10.1007/s11033-020-05606-0
Sharma, H., Hyvonen, J., and Poczai, P. (2020). Development of chloroplast microsatellite markers for giant ragweed (Ambrosia trifida). Appl. Plant Sci. 8, e11313. doi: 10.1002/aps3.11313
Shukla, N., Kuntal, H., Shanker, A., and Sharma, S. N. (2018). Mining and analysis of simple sequence repeats in the chloroplast genomes of genus Vigna. Biotechnol. Res. Innovation. 2, 9–18. doi: 10.1016/j.biori.2018.08.001
Simó, J., Pascual, L., Canizares, J., and Casanas, F. (2014). Spanish onion landraces (Allium cepa L.) as sources of germplasm for breeding calcots: a morphological and molecular survey. Euphytica 195, 287–300. doi: 10.1007/s10681-013-0995-y
Son, J. H., Park, K. C., Lee, S. I., Kim, J. H., and Kim, N. S. (2012). Species relationships among Allium species by ISSR analysis. Horticulture Environment Biotechnol. 53, 256–262. doi: 10.1007/s13580-012-0130-3
Spanoghe, M. C., Marique, T., Rivière, J., Moulin, M., Dekuijper, C., Nirsha, A., et al. (2020). Genetic patterns recognition in crop species using self-organizing map: The example of the highly heterozygous autotetraploid potato (Solanum tuberosum L.). Genet. Resour. Crop Evolution. 67, 947–966. doi: 10.1007/s10722-020-00894-8
Sudha, G. S., Ramesh, P., Sekhar, A. C., Krishna, T. S., Bramhachari, P. V., and Riazunnisa, K. (2019). Genetic diversity analysis of selected onion (Allium cepa L.) germplasm using specific RAPD and ISSR polymorphism markers. Biocatalysis Agric. Biotechnol. 17, 110–118. doi: 10.1016/j.bcab.2018.11.007
Van Heusden, A. W., Van Ooijen, J. W., Vrielinkvan Ginkel, R., Verbeek, W. H. J., Wietsma, W. A., and Kik, C. (2000). A genetic map of an interspecific cross in Allium based on amplified fragment length polymorphism (AFLPTM) markers. Theor. Appl. Genet. 100, 118–126. doi: 10.1007/s001220050017
Villano, C., Esposito, S., Carucci, F., Iorizzo, M., Frusciante, L., Carputo, D., et al. (2019). High-throughput genotyping in onion reveals structure of genetic diversity and informative SNPs useful for molecular breeding. Mol. Breed. 39, 1–11. doi: 10.1007/s11032-018-0912-0
Wang, Y. and Zhang, Y. (2022). Advances in molecular breeding of vegetable crops. Horticulturae 8, 821. doi: 10.3390/horticulturae8090821
Wright, S. (1984). Evolution and the genetics of populations, volume 4: variability within and among natural populations (Chicago: University of Chicago Press).
Xiong, Y., Xiong, Y., Shu, X., Yu, Q., Lei, X., Li, D., et al. (2022). Molecular phylogeography and intraspecific divergences in siberian wildrye (Elymus sibiricus L.) wild populations in China, inferred from chloroplast DNA sequence and cp-SSR markers. Front. Plant Science. 13. doi: 10.3389/fpls.2022.862759
Yamashita, K., Tsukazaki, H., Kojima, A., Ohara, T., and Wako, T. (2010). Inheritance mode of male sterility in bunching onion (Allium fistulosum L.) accessions. Euphytica 173, 357–367. doi: 10.1007/s10681-009-0101-7
Keywords: cp-SSR, Allium fistulosum, underutilized species, population structure, cross transferability
Citation: Khade YP, Mainkar P, Chandanshive A, Rai KM, Sinhasane SR, Jadhav M, Patil A, Hembade VL, Radhakrishna A, More SJ, Khar A, Bhandari HR, Gupta AJ, Kale RB, Prakash K and Mahajan V (2025) Harnessing chloroplast SSRs to decipher genetic diversity in underutilized Allium species. Front. Plant Sci. 16:1645145. doi: 10.3389/fpls.2025.1645145
Received: 12 June 2025; Accepted: 18 August 2025;
Published: 15 September 2025.
Edited by:
Mohan Lal, North East Institute of Science and Technology (CSIR), IndiaReviewed by:
Shivani Rohilla, Forest Research Institute, IndiaEbubekir Paşazade, Orta Karadeniz Geçit Kuşağı Tarımsal Araştırma Enstitüsü Müdürlüğü, Türkiye
Copyright © 2025 Khade, Mainkar, Chandanshive, Rai, Sinhasane, Jadhav, Patil, Hembade, Radhakrishna, More, Khar, Bhandari, Gupta, Kale, Prakash and Mahajan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yogesh P Khade, eW9nZXNoLmlhcmlAZ21haWwuY29t; Hem Raj Bhandari, aGVtcGJnQGdtYWlsLmNvbQ==; Vijay Mahajan, dmlqYm1haGFAeWFob28uY29t
†These authors have contributed equally to this work
 Yogesh P. Khade
Yogesh P. Khade Pawan Mainkar
Pawan Mainkar Aniket Chandanshive
Aniket Chandanshive Krishna Madhav Rai3
Krishna Madhav Rai3 Amol Patil
Amol Patil Vivekanand L. Hembade
Vivekanand L. Hembade Auji Radhakrishna
Auji Radhakrishna Sanket J. More
Sanket J. More Anil Khar
Anil Khar Hem Raj Bhandari
Hem Raj Bhandari Rajiv B. Kale
Rajiv B. Kale