- 1College of Life Science and Technology, Inner Mongolia Normal University, Hohhot, China
- 2Key Laboratory of Biodiversity Conservation and Sustainable Utilization in Mongolian Plateau for College and University of Inner Mongolia Autonomous Region, Hohhot, China
- 3College of Computer Science and Technology, Inner Mongolia Normal University, Hohhot, China
- 4Department of Botany, National Museum of Natural History, Smithsonian Institution, Washington, DC, United States
- 5College of Architecture and Environment, Sichuan University, Chengdu, China
The genus Oxytropis DC. comprises about 310 species distributed in Asia, Europe, and North America. Previous studies based on evidences from morphology or a few molecular markers are helpful for understanding the classification and systematic evolution of Oxytropis. However, a scarcity of chloroplast genomic resources for Oxytropis has hindered the understanding of the genus’s systematic classification and chloroplast genome evolution. Here comparative genomic analyses were conducted on chloroplast genomes of 24 Oxytropis species. Chloroplast genomes of Oxytropis species showed the triad structure due to the loss of one copy of the IR, with the size range from 121854 bp to 125271 bp. The Oxytropis cp genomes encoded a total of 110 genes, including 76 protein-coding genes (PCGs), 30 transfer RNA (tRNA) genes, and four ribosomal RNA (rRNA) genes. It was found that the atpF intron, one clpP intron, one rps12 intron, rpl22 gene, rps16 gene, and infA gene were lost in the Oxytropis cp genomes. Seven regions (5’-rps12-clpP, clpP intron, psbM-petN, rpl23-trnI-CAU, ndhJ-trnF-GAA,trnQ-UUG-accD, trnL-UAA-trnT-UGU) were chosen as potential molecular markers, which will contribute to species identification, population genetics and phylogenetic studies of Oxytropis. The phylogenetic relationships among Oxytropis species provided some implications for the classification of Oxytropis. Congruent with studies based on the morphological evidence, the close relationships between O. neimonggolica and O. diversifolia, as well as O. filiformis and O. coerulea were revealed. The results supported the treatment of O. daqingshanica as a separate species and refuted the inclusion of O. daqingshanica in O. ochrantha as conspecific taxa. In addition, it was suggested that O. chiliophylla should be considered as a separate species rather than its inclusion in O. microphylla. The 16 positively selected genes (rps3, rps4, rps7, rps11, rps12, rpl2, rpl20, rpl32, rpoC2, psbC, rbcL, atpF, clpP, accD, ycf1, ycf2) are related to important biological processes for instance self-replication, photosynthesis and metabolite biosynthesis, which may contribute to the adaptation of Oxytropis to its habitats. This study will lay a solid foundation for further studies on species identification, taxonomy, and systematic evolution of Oxytropis.
1 Introduction
As the third largest flowering plant family after Asteraceae Bercht. & J.Presl and Orchidaceae Juss., the Fabaceae Lindl. comprises about 751 genera and 19,500 species worldwide (LPWG (The Legume Phylogeny Working Group), 2013). Combined with a series of diagnostic characteristics, phylogenetic analyses based on matK sequences supported the classification system with six monophyletic subfamilies within the Fabaceae, with Papilionoideae DC. (503 genera, ca. 14,000 species) and Caesalpinioideae DC. (148 genera, ca. 4400 species) as the largest two subfamilies, followed by Detarioideae Burmeist. (84 genera, ca. 760 species), Cercidoideae LPWG (12 genera, ca. 335 species), Dialioideae LPWG (17 genera, ca. 85 species), and Duparquetioideae LPWG (one genus, one species) (LPWG (The Legume Phylogeny Working Group), 2017). Phylogenetic analyses based on plastomes and nuclear genes strongly support the classification system of six Fabaceae subfamilies (Zhang et al., 2020; Zhao et al., 2021), which has currently gained widespread acceptance and consensus among scholars. Oxytropis DC. belongs to the Astragalean clade under the inverted-repeat-lacking clade (IRLC) of the subfamily Papillonoideae (LPWG (The Legume Phylogeny Working Group), 2017; Zhao et al., 2021; Duan et al., 2024). The genus Oxytropis has about 310 species distributed in Asia, Europe, and North America, with a concentrated distribution in Central Asia (Zhang, 1998; Zhu et al., 2010). Oxytropis is an important component of the flora in the alpine and arid regions of the Northern Hemisphere temperate zones, and is one of the common groups in alpine, desert, and semi desert regions (Li and Ni, 1985). The Oxytropis plants have certain feeding, medicinal, and ornamental value (Kholina et al., 2021a; Sandanov et al., 2023; Wang B. et al., 2024). Due to the extremely similar morphology between Astragalus and Oxytropis, the Oxytropis species were included in Astragalus defined by Linnaeus (1753). De Candolle (1802) first separated Oxytropis from Astragalus based on the characteristics of keel petal shape and pod septum shape. Delimitation of the subgenera and sections were conducted by taxonomist since the establishment of Oxytropis, and although there is a certain consensus, different perspectives also exist (e.g., Bunge, 1874; Vasil’chenko et al., 1948; Pavlov, 1961; Zhang, 1998; Zhu et al., 2010). Micromorphological evidence has been applied to the classification of Oxytropis and some insights have been gained (Karaman et al., 2009; Ceter et al., 2013; Erkul et al., 2015; Zhao et al., 2022, 2023). With the development of sequencing technology, molecular markers have been used to address the questions on systematic evolution of Oxytropis, however, most studies involved a few molecular markers and referred species sampling with limited geographical ranges due to the focus on regional treatments (e.g., Archambault and Strömvik, 2012; Tekpinar et al., 2016a, 2016b; Kholina et al., 2016, 2021a, 2022). In addition, the cp genome sequences of the Oxytropis that can be used for study on systematic evolution are still scarce (Su et al., 2019; Liu et al., 2021; Bei et al., 2022; Tavares et al., 2022). Some progress has been achieved in the phylogenetic study of Oxytropis, but there is still a long way to uncover the systematic evolutionary questions for this complex taxonomic group with a large number of species, wide distribution, diverse morphology, and a relatively recent diversification history (Shavvon et al., 2017). The lack of effective molecular markers has to some extent hindered the phylogenetic study of Oxytropis, thus employing highly variable molecular markers coupled with increased taxon sampling promise advances in the issues on its taxonomy and evolution. The adaptation of Oxytropis species to special habitats makes it an excellent model for studying adaptive evolution, which is still an open issue for Oxytropis.
Chloroplast (cp) is a vital organelle in green plants, having a crucial role in photosynthesis and a myriad of metabolic activities (Neuhaus and Emes, 2000; Daniell et al., 2016; Wang J. et al., 2024). In angiosperms, the cp genomes are mostly a quadripartite structure: a large single-copy (LSC) region and a small single-copy region (SSC) separated by two inverted repeats (IRs) (Wicke et al., 2011), however, losses of the IR exist in a few angiosperm families, such as Geraniaceae (Guisinger et al., 2011; Ruhlman et al., 2017), Cactaceae (Sanderson et al., 2015), Arecaceae (Barrett et al., 2016), Fabaceae (Choi et al., 2019), Lophopyxidaceae and Putranjivaceae (Jin et al., 2020), and Passifloraceae (Cauz-Santos et al., 2020). Chloroplast genome has been widely used in studies on taxonomy, phylogeny and evolution of angiosperm (e.g., Kan et al., 2024; Li et al., 2024a; Yan et al., 2024; Wang et al., 2025; Yan et al., 2025), due to its own advantages such as uniparental inheritance, small size, lack of recombination, and moderate nucleotide substitution rate (Palmer, 1985; Wicke et al., 2011; Mower and Vickrey, 2018).
Through comparative genomics analyses of the cp genomes of 24 species of Oxytropis, this study aims to (1) explore the basic characteristics of Oxytropis cp genomes, (2) screen the hotspot regions as potential molecular markers of Oxytropis, (3) provide preliminary insights into the current classification of some Oxytropis species, and (4) understand the adaptation of Oxytropis species to the environment at the molecular level. Our study will lay a solid foundation for future studies on cp genome evolution, species identification, genetic diversity, and systematic evolution of Oxytropis.
2 Materials and methods
2.1 Plant material, DNA extraction and sequencing
Materials for the 19 Oxytropis species in the present study were collected during field trips, with the collected Oxytropis plants pressed into herbarium specimens, and fresh and tender leaves dried in silica gel without affecting identification. The collected specimens were identified by referring to relevant reference books (e.g., Li and Ni, 1985; Zhang, 1998; Zhu et al., 2010; Zhao et al., 2019), and all the voucher specimens were preserved in the herbarium of the Inner Mongolia Normal University (NMTC) (Table 1). Total genomic DNA was isolated from the silica-dried leaves according to the protocol of Doyle and Doyle (1987). The extracted DNA was fragmented by sonication and then used for construction of short-insert library (insert size, 300 bp) by NEBNext® Ultra™ II DNA Library Prep Kit for Illumina®. Finally, the pooled libraries were sequenced by the Illumina NovoSeq platform in Novogene (Beijing, China).
2.2 Chloroplast genome assembly and annotation
Trimomatic v. 0.33 (Bolger et al., 2014) was used to remove adapters in the obtained raw sequencing data. The filtered raw reads for each species were then used to assemble the cp whole genome sequence by NOVOPlasty v. 4.3.1 (Dierckxsens et al., 2017), with the cp genome sequence of O. bicolor (GenBank accession no. MN255323) (Su et al., 2019) as the reference and its rbcL sequence as the seed. The cp genome of O. myriophylla obtained from our previous study (Niu et al., 2024) was used as the reference to conduct annotations of the 19 Oxytropis cp genomes in the present study. The brief procedure for annotating the cp genome of O. myriophylla was as follows: following the annotation method of Zhang et al. (2022), GeSeq (Tillich et al., 2017) and CPGAVAS2 (Shi et al., 2019) were used to annotate the cp genome of O. myriophylla, with the cp genomes of O. bicolor (MN255323), O. arctobia (MT409175) and O. spelendens (MT409174) designated as custom reference genomes. The annotation results obtained from GeSeq and CPGAVAS2 were imported into Geneious Prime (Kearse et al., 2012) to check the intron/exon boundaries and the start and stop codon positions. If necessary, manual corrections were performed to obtain the elaborated annotated cp genome of O. myriophylla. The brief workflow for annotations of the 19 Oxytropis cp genomes was as follows: in Geneious Prime, MAFFT (Katoh and Standley, 2013) alignment was performed between the cp genome sequence of O. myriophylla with the complete annotation information and the cp genome sequence of other Oxytropis species. Based on the alignment results, transferring annotations function in Geneious Prime was used for annotation, and the annotation results were manually checked and proofread to finally generate the complete annotated cp genome of other Oxytropis species. The cp genome sequences of Oxytropis species with annotation information in gb format were imported into OrganellarGenomeDRAW (Greiner et al., 2019) to draw their cp genome circular maps. Moreover, annotation of other cp genomes obtained from GenBank were checked before being used for analysis.
2.3 Comparative chloroplast genome analyses
Comparative analysis was conducted on the basic characteristics of cp genome lengths, GC contents, and gene quantities in 27 cp genomes of 24 Oxytropis species using Geneious Prime (Table 1). The 27 Oxytropis cp genomes were aligned in MAUVE ver. 2.4.0 under the progressiveMauve algorithm (Darling et al., 2004, 2010). Due to cp genomes of O. falcata and O. arctobia with inversion, the two cp genome sequences were not used in molecular marker identification. The coding and noncoding regions in 25 cp genomes of 22 Oxytropis species were extracted by Geneious Prime, and all the homologous sequences were aligned one by one in MAFFT v. 7.490 (Katoh and Standley, 2013). The final aligned homologous sequences in fasta format were imported into DnaSP v. 6.12.03 (Rozas et al., 2017) and their nucleotide variability (Pi) values were calculated, and finally candidate molecular markers were screened based on the Pi values and sequence lengths.
2.4 Phylogenetic analyses
To reconstruct the phylogenetic relationships among Oxytropis species under the phylogenetic background of the Astragaglean clade, a total of 46 cp genome sequences from 43 species under the IRLC of the subfamily Papilionoideae were selected for phylogenetic analysis based on Zhao et al. (2021) (Supplementary Table S1). Considering the phenomena of gene/intron loss and inversion in the cp genomes of the IRLC of Papilionoideae (Jansen et al., 2008), only protein coding genes (PCGs) were selected for phylogenetic tree construction. Seventy-six PCGs were extracted from the cp genomes by Geneious Prime and each PCG was aligned separately using MAFFT v. 7.490 (Katoh and Standley, 2013). Alignments of genes that were not common to all species and genes with significant length differences (accD, clpP, psbL, ycf1, and ycf2) were removed. Finally, alignments of the remaining 71 PCGs were concatenated to form the phylogenetic dataset. Bayesian inference (BI) and the maximum likelihood (ML) methods were employed to construct the phylogenetic trees. GTR+I+G was recommended as best-fit model by PartitionFinder2 (Lanfear et al., 2017), and MrBayes v. 3.2.7a (Ronquist et al., 2012) was then used to construct BI tree based on the method of Zhang et al. (2022). The ML tree was constructed by RAxML v. 8.2.12 (Stamatakis, 2014) following the method of Li et al. (2024b). Finally, these two phylogenetic trees were visualized using FigTree v. 1.4.4 (Rambaut, 2018), and Alhagi sparsifolia, Caragana arborescens, Chesneya acaulis, Corethrodendron multijugum, and Tibetia liangshanensis were designated as the outgroup to root the trees.
2.5 Adaptive evolution analyses
Based on the results of phylogenetic analyses, 31 cp genomes involving 24 species of Oxytropis and its four related taxa (Carmichaelia australis, Lessertia frutescens, Phyllolobium chinense, and Sphaerophysa salsula) were selected for selection pressure analyses. The CodeML program in the PAML software package (Yang, 2007) is currently the most widely used bioinformatics tool for selection pressure analyses. EasyCodeML (Gao et al., 2019) can offer a user-friendly graphical interface for executing CodeML. Site models from CodeML were performed in EasyCodeML with the purpose of detecting positive selection sites of PCGs in Oxytropis cp genomes. Seventy-six PCGs shared by Oxytropis and its related taxa were firstly extracted from the cp genomes using Geneious Prime. MAFFT (Katoh and Standley, 2013) was then employed to perform multiple alignment of each PCG according to its codons and stop codons were manually deleted in the final alignment matrix. The final alignment of each PCG was concatenated into a supermatrix, which was exported into fasta format as an input file for EasyCodeML. The ML tree constructed based on the supermatrix by RAxML v. 8.2.12 (Stamatakis, 2014) was as an input tree in EasyCodeML (Supplementary Figure S1). The likelihood ratio test (LRT) was used to detect positive selection sites with four comparison models: M0 vs. M3, M1a vs. M2a, M7 vs. M8, and M8a vs. M8. With LRT threshold p<0.05, Bayesian empirical Bayes (BEB) (Yang et al., 2005) or Naïve empirical Bayes (NEB) (Nielsen and Yang, 1998) analysis was adopted to detect positive selection sites with posterior probabilities ≥0.95.
3 Results and discussion
3.1 Features of Oxytropis chloroplast genome
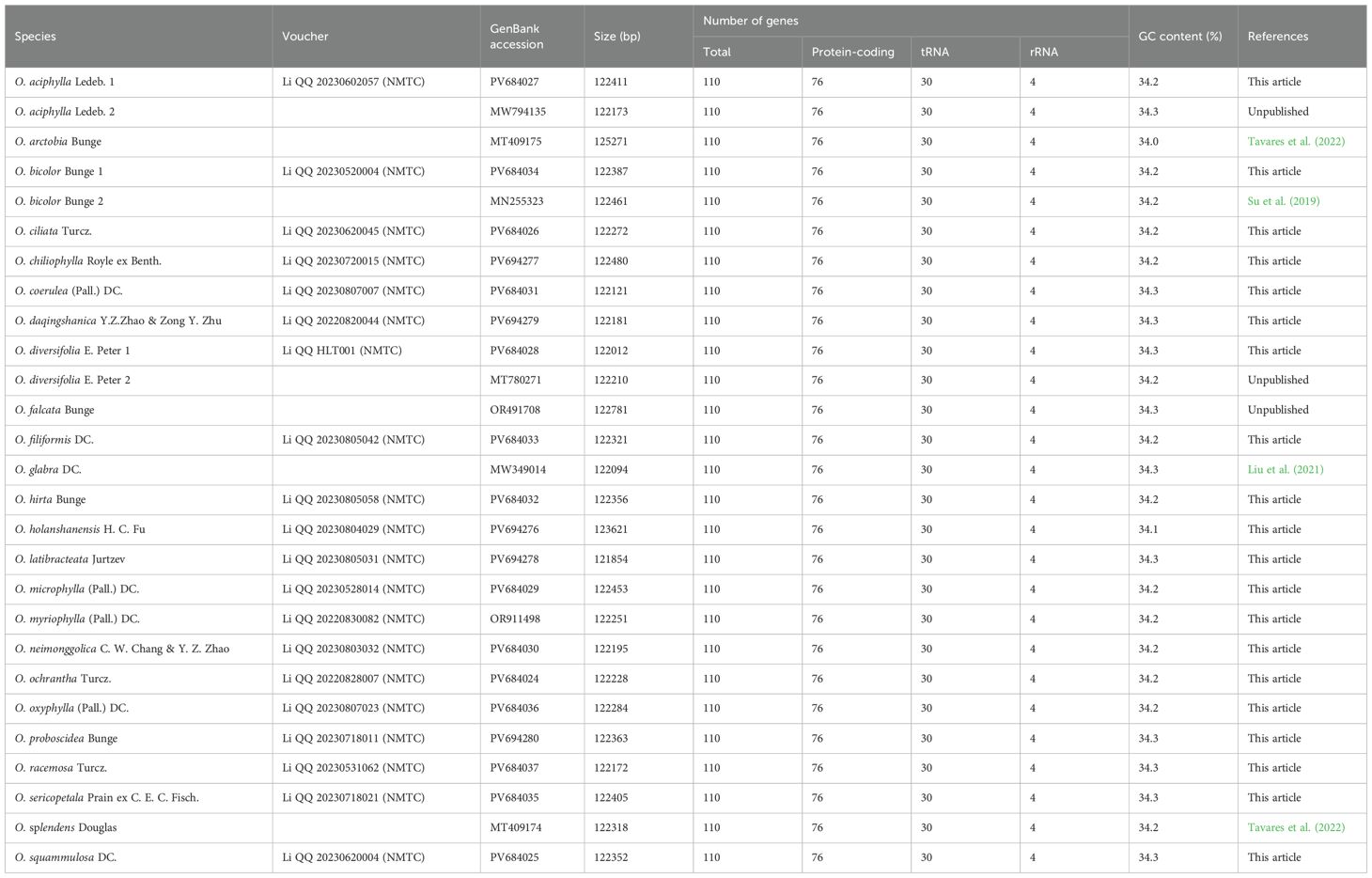
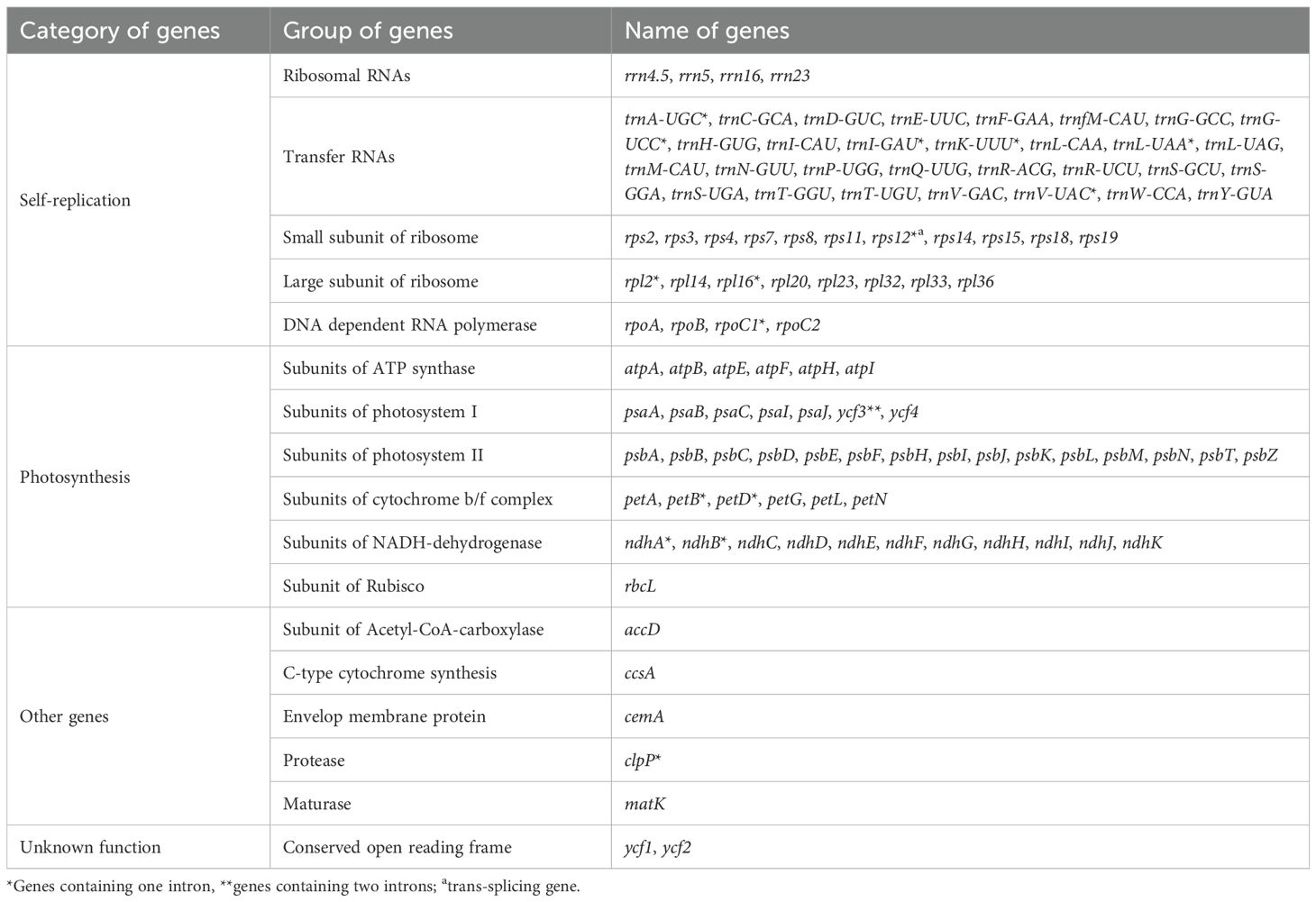
The size range of cp genomes of 24 Oxytropis species was from 121854 bp (O. latibracteata) to 125271 bp (O. Arctobia) (Table 1; Figure 1). Compared with the cp genomes of some Papilionoideae taxa such as Cyamopsis (Kaila et al., 2017), Ormosia (Liu et al., 2019), and Campylotropis (Feng et al., 2022) with typical quadripartite structure, the cp genomes of Oxytropis species showed the triad structure due to the loss of approximately 25 kb IR. The cp genomes of the IRLC groups in Papillonoideae have lost one IR copy and exhibit the triad structure (Wojciechowski et al., 2004; Jansen et al., 2008). The GC content in the cp genomes of 24 Oxytropis species (34.0%–34.3%) was roughly equivalent to that in cp genomes of other IRLC taxa such as Glycyrrhiza, Astragalus, and Galega (Duan et al., 2020; Su et al., 2021; Feng et al., 2023). The cp genomes of Oxytropis species encoded a total of 110 genes, including 76 protein-coding genes (PCGs), 30 transfer RNA (tRNA) genes, and four ribosomal RNA (rRNA) genes (Tables 1, 2). The 110 genes can be classified into four categories according to their functions: 57 genes related to self-replication, 46 associated with photosynthesis, five for other genes, and two genes with unknown function. Moreover, 15 genes contained one intron (ndhB, clpP, ndhA, rpl16, petB, rpoC1, rpl2, petD, rps12, trnI-GAU, trnG-UCC, trnL-UAA, trnK-UUU, trnA-UGC, trnV-UAC), while gene ycf3 possessed two introns. Gene rps12 had trans-splicing in the Oxytropis cp genome, like in most other angiosperms.
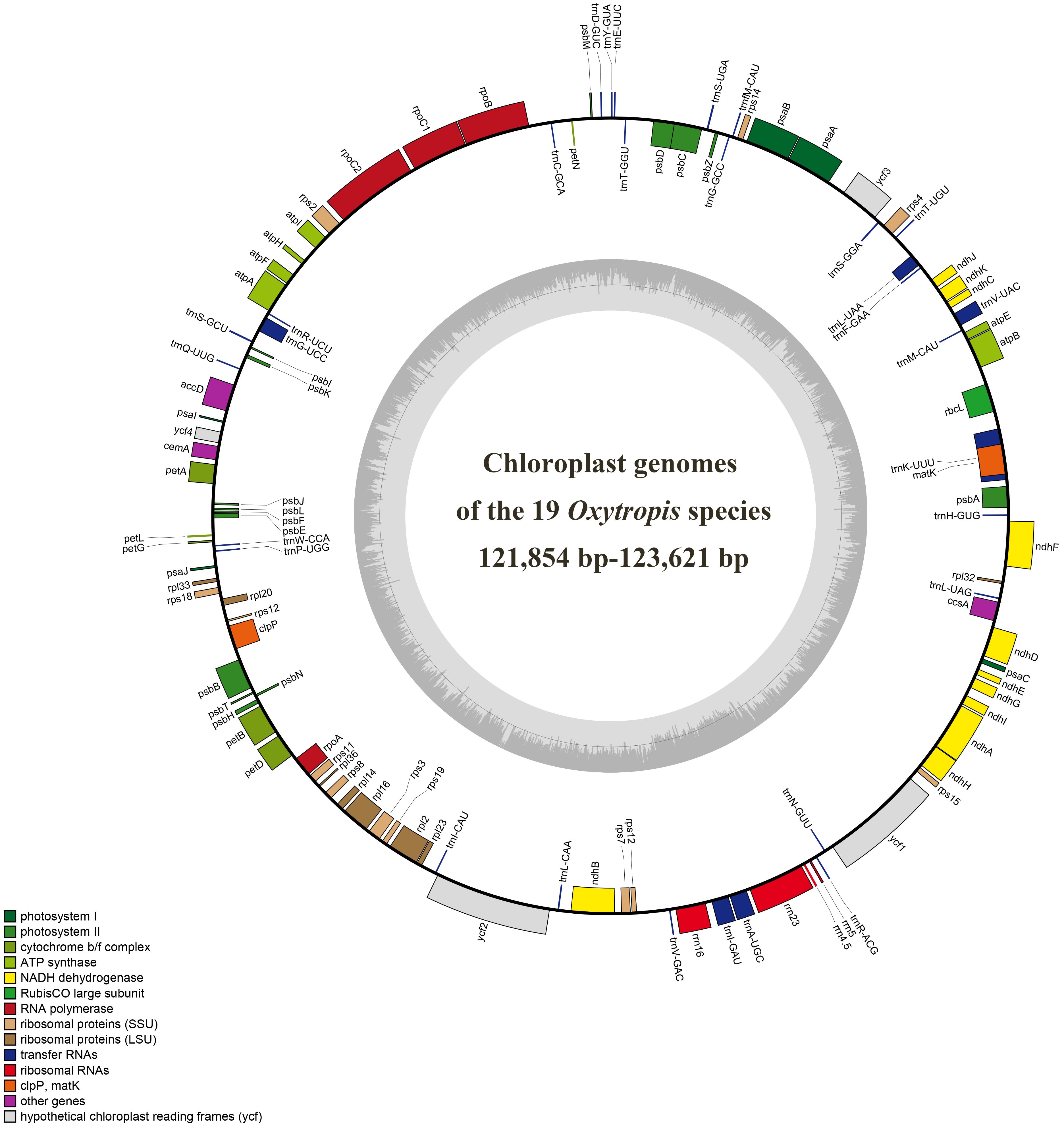
Figure 1. Chloroplast genome map of the 19 Oxytropis species generated in this study. Genes outside the circle are transcribed counterclockwise, and those inside are transcribed clockwise.
There are phenomena such as intron loss, gene loss, and inversion in the cp genome evolution of Papilionoideae (Jansen et al., 2008). Our study found the absence of the atpF intron, clpP intron, and rps12 intron in the cp genomes of Oxytropis and its closely related species. The atpF intron is lost in the cp genomes of Oxytropis, Lessertia, and Phaerophysa species, while it is present in Phyllopium, Carmichaelia, and Astragalus species. The absence or presence of the atpF intron in the cp genome could be used as a potential molecular marker for distinguishing the morphologically highly similar genera Oxytropis and Astragalus. Loss of one clpP intron was detected in the cp genomes of Oxytropis and its closely related taxa, including Astragalus, Carmichaelia, Lessertia, Phyllolobium, and Sphaerophysa species. The absence of clpP intron has also occurred in other IRLC groups of Papillonoideae, for example, one clpP intron was lost in Alhagi, Caragana, and Vicia species; and two clpP introns were lost in Tibetia, Corethrodendron, and Glycyrrhiza species (Jansen et al., 2008; Li et al., 2020; Lee et al., 2021), while the two clpP introns are present in other angiosperms genera such as Uncaria (Dai et al., 2023), Alisma (Lan et al., 2024), and Argentina (Li et al., 2024b). The rps12-3′-end intron was lost in the cp genomes of Oxytropis and its closely related taxa, and the loss of rps12 intron is a common phenomenon in the IRLC group (Jansen et al., 2008; Lee et al., 2021). Most angiosperm cp genomes contain genes rpl22, rps16, and infA, all of which are lost in the Oxytropis cp genomes. The gene rpl22 is absent in the cp genomes of all legumes (Jansen et al., 2008). We detected that genes rps16 and infA are also not present in the cp genomes of related taxa of the Oxytropis, including Alhagi, Astragalus, Caragana, Carmichaelia, Chesneya, Corethrodendron, Lessertia, Phyllolobium, Sphaerophysa, and Tibetia species. Analysis by Mauve showed that among the 27 cp genomes of Oxytropis, cp genomes of O. falcata and O. arctobia had inversion, while the remaining 25 cp genomes have the same gene order with no obvious reorganization (Supplementary Figure S2). The gene order rearrangement did not affect the sequences of any of the involving genes in cp genomes of O. falcata and O. arctobia.
3.2 Divergence hotspots
DnaSP v. 6.12.03 (Rozas et al., 2017) was employed to calculate the Pi values of a total of 253 regions of Oxytropis cp genome with the aim to screen the highly divergent regions. The Pi values ranged from 0%-4.244%, with a mean value of 0.436%, showing Oxytropis cp genomes with a high level of similarity (Supplementary Table S2; Figure 2). As a whole, 42 regions with Pi =0, 132 regions with 0%<Pi ≤ 0.5%, 50 regions with 0.5%<Pi ≤ 1%, 20 regions with 1%<Pi ≤ 1.5% (petG-trnW-CCA, atpF, trnT-GGU-trnE-UUC, rps8-rpl14, atpA-trnR-UCU, trnF-GAA-trnL-UAA, trnQ-UUG-accD, rpl33-rps18, trnV-GAC-rrn16, rps11-rpl36, rps4-trnS-GGA, atpH-atpF, trnL-UAA-trnT-UGU, rpoA-rps11, trnR-ACG-trnN-GUU, psbK-trnQ-UUG, psaB-rps14, ycf1-rps15, trnH-GUG-psbA, trnP-UGG-psaJ), and nine regions with Pi>1.5% (5’-rps12-clpP, trnR-UCU-trnG-UCC, clpP intron, psbM-petN, trnfM-CAU-trnG-GCC, trnI-CAU-ycf2, ndhI-ndhG, rpl23-trnI-CAU, ndhJ-trnF-GAA). Among the 29 regions with Pi>1%, 27 regions (excluding clpP intron and atpF) were located in the intergenic region, indicating that the non-coding regions exhibited higher variation compared to the coding regions, and regions located in the intergenic spacers (IGS) with greater potential for development of molecular markers.
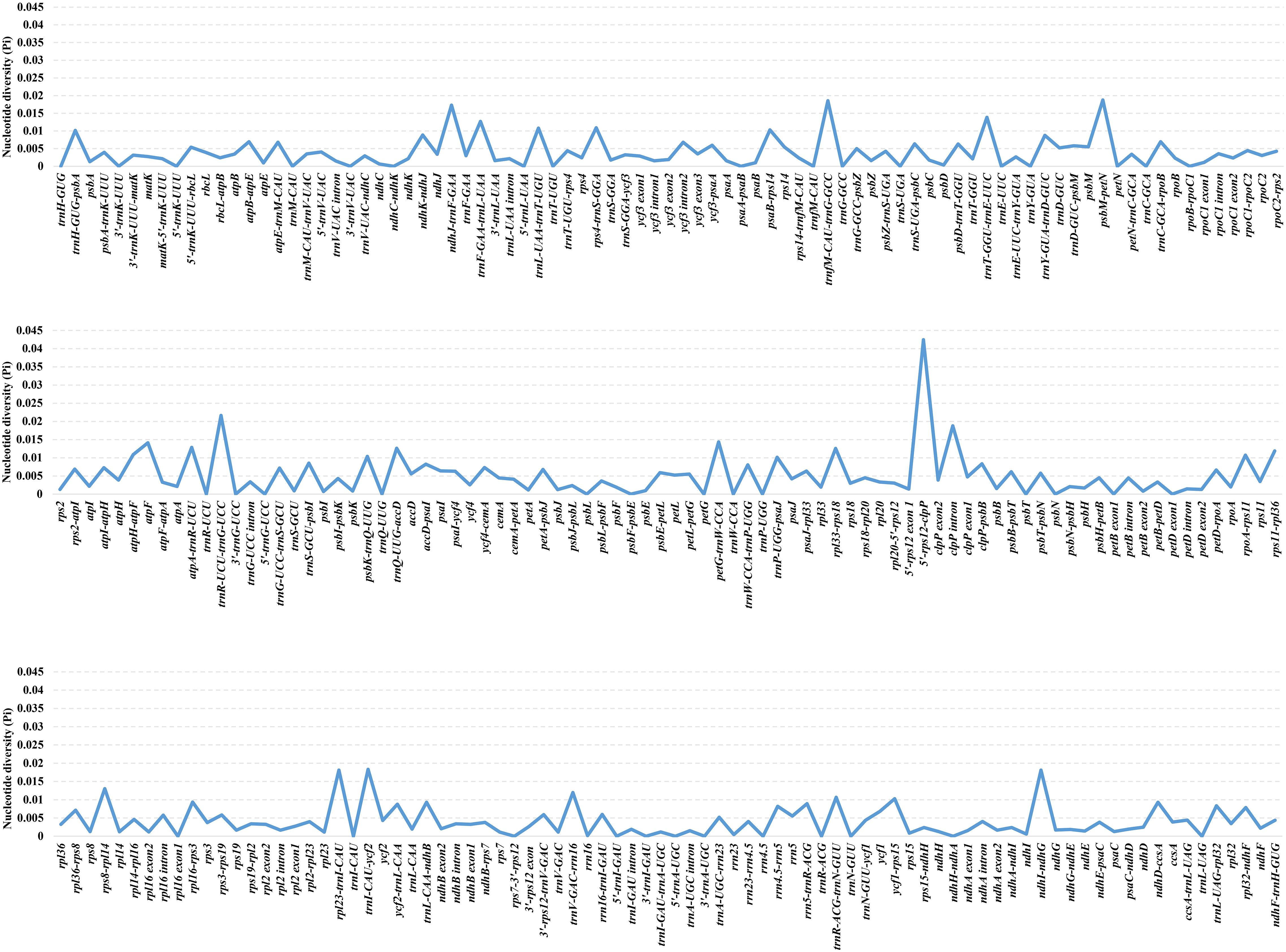
Figure 2. The nucleotide diversity (Pi) values of shared regions in 25 Oxytropis chloroplast genomes.
In order to screen molecular markers with potential for development, 19 regions with Pi >1% and alignment lengths >300bp were identified as candidate molecular markers for Oxytropis, namely 5’-rps12-clpP, clpP intron, psbM-petN, ndhI-ndhG, rpl23-trnI-CAU, ndhJ-trnF-GAA, atpF, trnT-GGU-trnE-UUC, rps8-rpl14, trnQ-UUG-accD, rpl33-rps18, rps11-rpl36, atpH-atpF, trnL-UAA-trnT-UGU, trnR-ACG-trnN-GUU, psbK-trnQ-UUG, ycf1-rps15, trnH-GUG-psbA, and trnP-UGG-psaJ. Among these 19 markers, taking into account both Pi value and sequence alignment length, seven regions (5’-rps12-clpP, clpP intron, psbM-petN, rpl23-trnI-CAU, ndhJ-trnF-GAA,trnQ-UUG-accD, trnL-UAA-trnT-UGU) were selected as potential molecular markers for Oxytropis. The cp molecular markers used in previous phylogenetic studies of Oxytropis (Kulshreshtha et al., 2004; Wojciechowski et al., 2004; Artyukova et al., 2011; Tekpinar et al., 2016a, 2016b; Kholina et al., 2016, 2018a, 2018b, 2020, 2021a, 2021b, 2021c, 2022; Chen et al., 2020; Kozyrenko et al., 2020; Sandanov et al., 2023) included matK, rpoC1, rpoC2, trnL intron, trnV intron, trnL-trnF, trnH-psbA, petG-trnP, and trnS-trnG. All other markers except trnH-psbA were not among the developed candidate molecular markers for Oxytropis, which suggested the significance of developing molecular markers for specific taxonomic groups. Overall, our newly screened potential molecular markers will contribute to species identification, population genetics and phylogenetic studies of Oxytropis.
3.3 Phylogenetic analyses
Overall, compared to previous phylogenetic studies of Oxytropis using several molecular markers obtained with Sanger sequencing (e.g., Kholina et al., 2016; Tekpinar et al., 2016b; Shavvon et al., 2017), relatively high phylogenetic resolution was obtained in our study by utilizing the plastid genome data. Phylogenetic trees inferred from BI and ML analyses were almost identical in topology, and the difference mainly lied in the relative position of O. squammulosa versus O. filiformis and O. coerulea (Figure 3; Supplementary Figures S3, S4). Both the BI and ML trees showed that the outgroup species were robustly separated from the Astragaglean clade (PP = 1.00, ML BS = 100%). Within the Astragaglean clade, there are three major clades, namely Oxytropis, Astragalus, and Coluteoid clades. Phylogenetic tree showed that Oxytropis species were well clustered together (PP = 1.00, ML BS = 100%), which corroborated the previous studies that Oxytropis was monophyletic (e.g., Archambault and Strömvik, 2012; Tekpinar et al., 2016a; Kholina et al., 2016; Shavvon et al., 2017). Consistent with studies of Su et al. (2021); Tian et al. (2021) and Moghaddam et al. (2023) based on cp genome, our result indicated that Oxytropis was sister to Coluteoid clade and Oxytropis+Coluteoid clade had a sister relationship with Astragalus. However, studies of Moghaddam et al. (2016) using ITS, matK and rpl32-trnL data and Zhao et al. (2021) based on low-copy nuclear genes revealed that Oxytropis+Astragalus clade was sisters to Coluteoid clade. The nuclear-cytoplasmic conflict on the phylogenetic position of Oxytropis may reflect the complex evolutionary history of this genus. Although the current taxon sampling was still limited, the systematic relationships among Oxytropis species showed in our study still provided some insights into the classification of Oxytropis. In the phylogenetic trees, O. neimonggolica was clustered together with O. diversifolia (PP = 1.00, ML BS = 100%), which supported their close relationship based on morphological study (Zhu et al., 2010; Zhao et al., 2019). Oxytropis ochrantha and O. myriophylla were clustered together and was sister to O. daqingshanica, which supported the treatment of O. daqingshanica as a separate species (Zhao et al., 2019) and disapproved the inclusion of O. daqingshanica in O. ochrantha as conspecific taxa (Zhu et al., 2010). Oxytropis filiformis was clustered with O. coerulea (PP = 1.00, ML BS = 99%), suggesting their close affinity, which was congruent with studies based on the morphological evidences (Zhu et al., 2010; Zhao et al., 2019). Oxytropis microphylla and O. ciliata grouped together and O. chiliophylla was distantly related to these two species, which suggested that O. chiliophylla should be considered as a separate species rather than including it in O. microphylla. Further taxonomic treatments in Oxytropis should be conducted by combining evidence from morphology, anatomy, ecology and palynology.
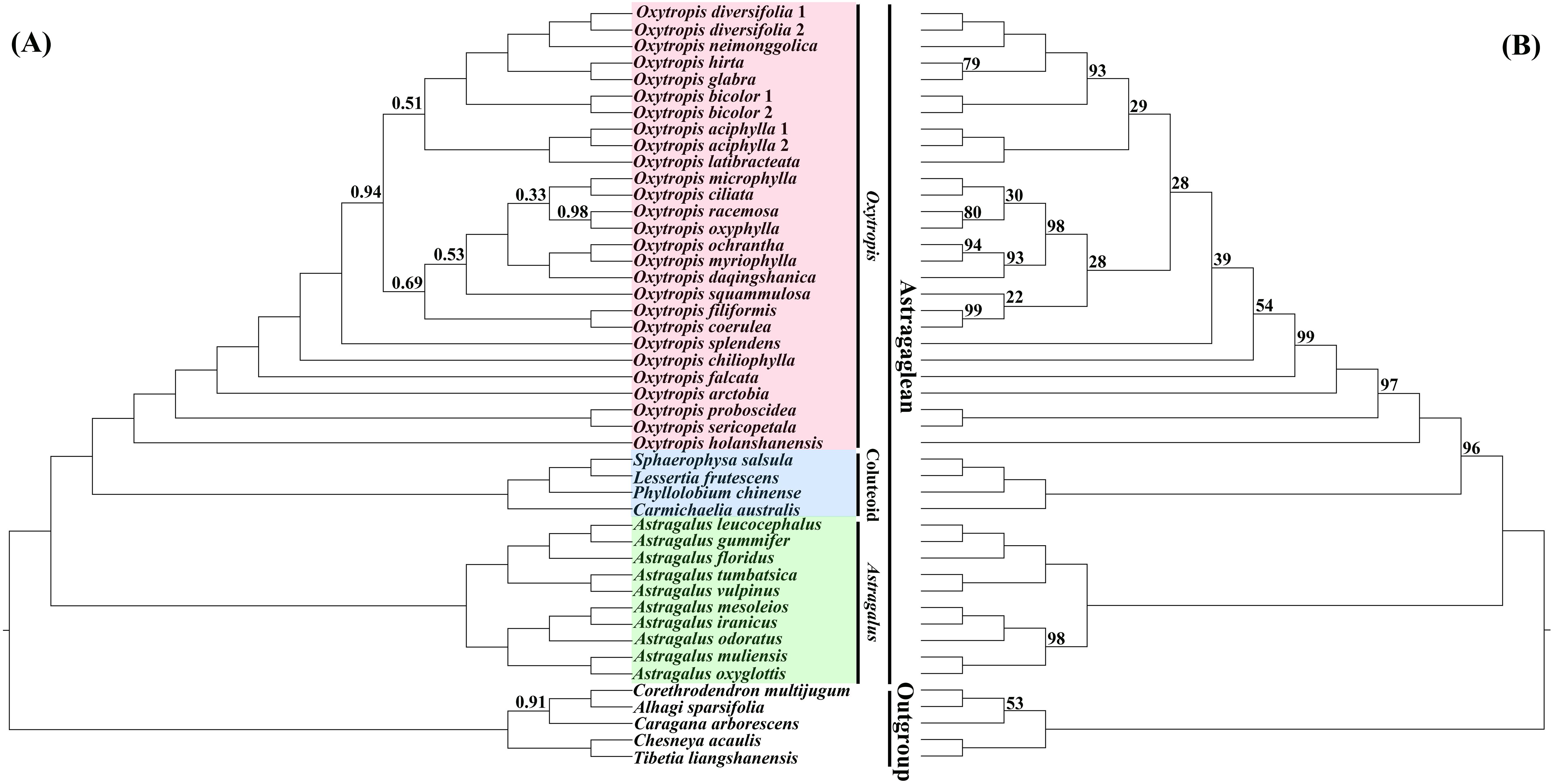
Figure 3. Phylogenetic trees of Oxytropis and its related taxa based on the dataset of 71 concatenated protein-coding genes (PCGs) of the chloroplast genomes. (A) Bayesian inference (BI) tree, (B) maximum likelihood (ML) tree. Values along branches indicate Bayesian posterior probabilities (only PP < 1.00 are shown) and ML bootstrap percentages (only values < 100% are shown), respectively.
3.4 Adaptive evolution
The p-values of LRTs for compared models M0 vs. M3, M1a vs. M2a, M7 vs. M8, and M8a vs. M8 is below threshold 0.05, suggesting adaptation signatures within Oxytropis cp genomes (Supplementary Tables S3, S4; Table 3). According to the manual of PAML (Yang, 2007), M0 vs. M3 was not suggested as a test of positive selection but as a test of variable ω among sites. In addition, M1a vs. M2a seems to be more stringent compared with M7 vs. M8 which has been confirmed in our results. Therefore, we relied on result under model M8 to discuss positive selection sites in Oxytropis cp genomes. Sixteen genes with positive selection sites were detected according to BEB analysis under model M8. The number of positive selection sites in these genes ranged from 1 to 47: nine genes (rps4, psbC, rpl20, rps12, rps11, rps3, rpl2, rps7, rpl32) with one site, two genes (rpoC2, atpF) having two sites, clpP possessing three sites, rbcL with five sites, ycf2 containing eight sites, accD with 12 sites, and ycf1 harboring the largest number of sites. According to their functional category, nine genes (rps3, rps4, rps7, rps11, rps12, rpl2, rpl20, rpl32, rpoC2) were associated with self-replication, three genes (psbC, rbcL, atpF) were responsible for photosynthesis, genes clpP and accD belonged to other genes, and genes ycf1 and ycf2 are functionally unknown.
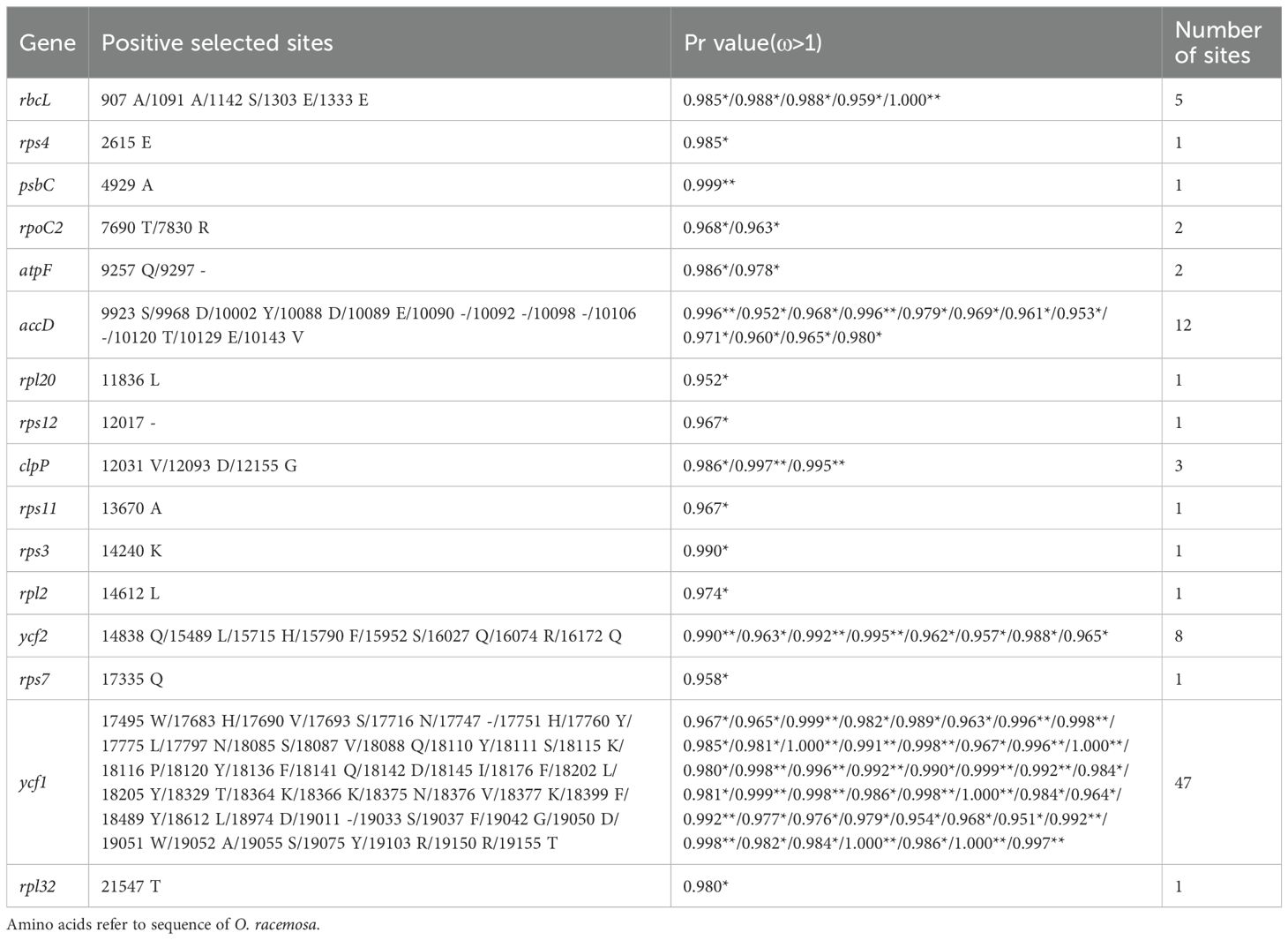
Table 3. Positively selected sites (*: P>95%; **: P>99%) detected in the Oxytropis chloroplast genomes in comparisons of M7 vs. M8 and M8a vs. M8 under Bayes empirical Bayes (BEB) analysis.
The adaptive evolution of these 16 genes may help Oxytropis species adapt to their habitats. Among them, rps3, rps4, rps7, rps11, rps12, rpl2, rpl20, and rpl32 encoded ribosomal subunit proteins. Chloroplast ribosomal proteins are essential for cp ribosome assembly, which plays an important role in plant survival, acclimation and adaptation (Schmid et al., 2024). DNA dependent RNA polymerase subunit beta’’ encoded by rpoC2, is one of the components of the core of plastid-encoded polymerase (PEP) which acts as the major transcription machinery of mature chloroplasts (Zhelyazkova et al., 2012; Kindgren and Strand, 2015). The 43-kDa chlorophyll a-binding protein (CP43) encoded by psbC, together with CP47 encoded by psbB, binds chlorophyll, as an inner light-harvesting complex of photosystem II (PSII) (Landi and Guidi, 2022). The large subunit of Rubisco was encoded by rbcL (Wicke et al., 2011). Rubisco mediates the fixation of inorganic carbon from CO2 into organic compounds during photosynthesis (Wilson and Hayer-Hartl, 2018). In most lineages of terrestrial land plants, rbcL is under positive selection (Kapralov and Filatov, 2007). ATP synthase CF0 B subunit encoded by atpF is one of the important constituents of chloroplast ATP synthase, which using the proton gradient produces ATP that is indispensable for photosynthesis and plant growth (Hahn et al., 2018; Yamamoto et al., 2023). Gene clpP in chloroplast is essential for plant development, with an indispensable function for cell viability (Shikanai et al., 2001; Kuroda and Maliga, 2003). The chloroplast gene clpP together with a nuclear multi gene family encodes the Clp protease that degrades damaged proteins during environmental stresses (Clarke, 1999; Adam and Clarke, 2002). The key enzyme acetyl-CoA carboxylase (ACCase) regulates de novo synthesis of fatty acids in plants (Rawsthorne, 2002). The accD gene encodes one of the four subunits of ACCase, which is essential for cell viability, leaf development, and seed development (Madoka et al., 2002; Kode et al., 2005; Caroca et al., 2021). Products encoded by essential genes ycf1 and ycf2 of higher plants are essential for plant cell survival (Drescher et al., 2000).
Previous work suggested that Oxytropis arctobia accD gene was under positive selection, which might be related to its adaptation to the cold environment in the Arctic (Tavares et al., 2022). Positively selected genes in Oxytropis detected in our study were also found under positive selection in some other Fabaceae genera. For example, rpl2, rpoC2 and accD were under positive selection in Pueraria (Zhou et al., 2023), and so were rps11, clpP, accD and ycf1 in Astragalus (Moghaddam et al., 2023), rps4, rpl32, accD and ycf2 in Pterocarpus (Hong et al., 2020), rps4, rps7, rpl32 and clpP in Vicia (Li et al., 2020), rps7, rpl20, atpF, ycf1 and ycf2 in Caragana (Cui et al., 2024), and rps3, rps12, rpoC2, psbC, rbcL, clpP, accD, ycf1 and ycf2 in Dalbergia (Li et al., 2022). Oxytropis species spread in temperate and cold regions of the Northern Hemisphere in Asia, Europe, and North America, usually thriving in harsh environments such as the Arctic areas and alpine ecosystems (Zhu et al., 2010; Archambault and Strömvik, 2012; Kholina et al., 2016; Tavares et al., 2022). Oxytropis species grow in various habitats such as mountains, steppes, prairies, meadows, deserts, semi deserts, forest-steppes, and forests (Zhu et al., 2010; Sandanov et al., 2023; Welsh, 2023). The origin of Oxytropis was dated to about 5.6 million years ago, with 95% highest posterior density intervals ranging from 3.61 to 8.07 Ma, which coincides with climate modifications around the Miocene-Pliocene boundary (Shavvon et al., 2017). It was inferred that Oxytropis experienced a recent rapid radiation based on its recent age estimates, short interior branch on gene tree, little genetic differences, and diverse morphology and ecological habitats (Shavvon et al., 2017). The 16 positively selected genes in the Oxytropis cp genome are related to important biological processes for instance self-replication, photosynthesis and metabolite biosynthesis, which may contribute to the adaptation of Oxytropis to diverse habitats, especially under extreme arid and cold conditions. The adaptation of Oxytropis to diverse habitats may have to some extent promoted the rapid diversification of Oxytropis in its relatively recent evolutionary history.
4 Conclusion
In this study, comparative analysis of cp genomes of 24 Oxytropis species revealed that their cp genomes exhibited a triad structure, and the cp genome size, GC content, and gene content were conserved. Seven highly divergent regions (5’-rps12-clpP, clpP intron, psbM-petN, rpl23-trnI-CAU, ndhJ-trnF-GAA,trnQ-UUG-accD, trnL-UAA-trnT-UGU) identified in this study may be potentially utilized as high-resolution DNA barcodes, which will facilitate species identification and phylogenetic and phylogeographic studies of Oxytropis. Phylogenetic analysis based on the cp genome sequences supported the monophyly of Oxytropis and provided some new insights into the classification of Oxytropis. The results indicated that the cp genome can be utilized as an informative molecular marker for enhancing our understandings of evolutionary diversification in Oxytropis. Sixteen protein-coding genes (rps3, rps4, rps7, rps11, rps12, rpl2, rpl20, rpl32, rpoC2, psbC, rbcL, atpF, clpP, accD, ycf1, ycf2) showed evidence for positive selection, which may contribute to the adaptation of Oxytropis to its diverse habitats. Overall, our study improved the understanding of cp genome features, phylogenetic relationships, and adaptive evolution in Oxytropis. Employing single-copy nuclear genes coupled with more detailed taxon sampling will facilitate future work on the phylogeny, biogeography, and adaptive evolution of Oxytropis.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank of NCBI (https://www.ncbi.nlm.nih.gov/genbank/), PV684024-PV684037, and PV694276-PV694280.
Author contributions
Q-QL: Conceptualization, Formal Analysis, Investigation, Project administration, Writing – original draft. YN: Formal Analysis, Investigation, Writing – original draft. Z-PZ: Formal Analysis, Investigation, Writing – original draft. JW: Conceptualization, Writing – review & editing. C-YL: Conceptualization, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by the Fundamental Research Funds for Inner Mongolia Normal University (No. 2022JBBJ012), the National Natural Science Foundation of China (No. 32260053), and the Natural Science Foundation of Inner Mongolia, China (No. 2022LHQN03004).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1645582/full#supplementary-material
References
Adam, Z. and Clarke, A. K. (2002). Cutting edge of chloroplast proteolysis. Trends Plant Sci. 7, 451–456. doi: 10.1016/s1360-1385(02)02326-9
Archambault, A. and Strömvik, M. V. (2012). Evolutionary relationships in Oxytropis species, as estimated from the nuclear ribosomal internal transcribed spacer (ITS) sequences point to multiple expansions into the Arctic. Botany 90, 770–779. doi: 10.1139/B2012-023
Artyukova, E. V., Kozyrenko, M. M., Kholina, A. B., and Zhuravlev, Y. N. (2011). High chloroplast haplotype diversity in the endemic legume Oxytropis chankaensis may result from independent polyploidization events. Genetica 139, 221–232. doi: 10.1007/s10709-010-9539-8
Barrett, C. F., Baker, W. J., Comer, J. R., Conran, J. G., Lahmeyer, S. C., Leebens-Mack, J. H., et al. (2016). Plastid genomes reveal support for deep phylogenetic relationships and extensive rate variation among palms and other commelinid monocots. New Phytol. 209, 855–870. doi: 10.1111/nph.13617
Bei, Z., Zhang, L., and Tian, X. (2022). Characterization of the complete chloroplast genome of Oxytropis aciphylla Ledeb. (Leguminosae). Mitochondrial DNA Part B 7, 1756–1757. doi: 10.1080/23802359.2022.2124822
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bunge, A. (1874). Species generis Oxytropis. Mémoires de l’Académie impériale des sciences de St.-Pétersbourg. 7e série. 22, 1–166.
Caroca, R., Howell, K. A., Malinova, I., Burgos, A., Tiller, N., Pellizzer, T., et al. (2021). Knockdown of the plastid-encoded acetyl-CoA carboxylase gene uncovers functions in metabolism and development. Plant Physiol. 185, 1091–1110. doi: 10.1093/plphys/kiaa106
Cauz-Santos, L. A., da Costa, Z. P., Callot, C., Cauet, S., Zucchi, M. I., Bergès, H., et al. (2020). A repertory of rearrangements and the ross of an inverted repeat region in Passiflora chloroplast genomes. Genome Biol. Evol. 12, 1841–1857. doi: 10.1093/gbe/evaa155
Ceter, T., Erkul, S. K., Aytaç, Z., and Başer, B. (2013). Pollen morphology of the genus Oxytropis DC. in Turkey. Bangladesh J. Bot. 42, 167–174. doi: 10.3329/bjb.v42i1.15908
Chen, J. T., Zhang, D. G., Lv, Z. Y., Huang, X. H., Liu, P. J., Yang, J. N., et al. (2020). Oxytropis shennongjiaensis (Fabaceae), a new species from Hubei, Central China. PhytoKeys 149, 117–128. doi: 10.3897/phytokeys.149.49533
Choi, I. S., Jansen, R., and Ruhlman, T. (2019). Lost and found: return of the inverted repeat in the legume clade defined by its absence. Genome Biol. Evol. 11, 1321–1333. doi: 10.1093/gbe/evz076
Clarke, A. K. (1999). ATP-dependent Clp proteases in photosynthetic organisms—a cut above the rest! Ann. Bot. 83, 593–599. doi: 10.1006/anbo.1999.0878
Cui, X., Liu, K., Li, E., Zhang, Z., and Dong, W. (2024). Chloroplast genomes evolution and phylogenetic relationships of Caragana species. Int. J. Mol. Sci. 25, 6786. doi: 10.3390/ijms25126786
Dai, J., Liu, Q., Xu, X., Tan, Z., Lin, Y., Gao, X., et al. (2023). Comparative and phylogenetic analysis of the complete chloroplast genomes of Uncaria (Rubiaceae) species. Front. Plant Sci. 14, 1271689. doi: 10.3389/fpls.2023.1271689
Daniell, H., Lin, C. S., Yu, M., and Chang, W. J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134. doi: 10.1186/s13059-016-1004-2
Darling, A. C. E., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Darling, A. E., Mau, B., and Perna, N. T. (2010). ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS One 5, e11147. doi: 10.1371/journal.pone.0011147
De Candolle, A. P. (1802). Astragalogia nempe Astragali, Biserrulae et Oxytropidis, nec non Phacae, Colutae et Lessertiae historia iconibus illustrata(Paris: Joann. Bapt. Garnery), 324. doi: 10.5962/bhl.title.517
Dierckxsens, N., Mardulyn, P., and Smits, G. (2017). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18. doi: 10.1093/nar/gkw955
Doyle, J. J. and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Drescher, A., Ruf, S., Calsa, T., Jr., Carrer, H., and Bock, R. (2000). The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 22, 97–104. doi: 10.1046/j.1365-313x.2000.00722.x
Duan, L., Harris, A. J., Su, C., Zhang, Z. R., Arslan, E., Ertuğrul, K., et al. (2020). Chloroplast phylogenomics reveals the intercontinental biogeographic history of the liquorice genus (Leguminosae: Glycyrrhiza). Front. Plant Sci. 11, 793. doi: 10.3389/fpls.2020.00793
Duan, L., Su, C., Wen, J., Ji, Y.-W., Jiang, Y., Zhang, T., et al. (2024). New insights into the phylogenetic relationships of tribe Astragaleae (Fabaceae subfamily Papilionoideae) and Astragalus—the largest genus of angiosperm. Biol. Div. 1, 136–146. doi: 10.1002/bod2.12021
Erkul, S. K., Celep, F., and Aytaç, Z. (2015). Seed morphology and its systematic implications for genus Oxytropis DC. (Fabaceae). Plant Biosyst. 149, 875–883. doi: 10.1080/11263504.2014.969353
Feng, J., Xiong, Y., Su, X., Liu, T., Xiong, Y., Zhao, J., et al. (2023). Analysis of complete chloroplast genome: structure, phylogenetic relationships of Galega orientalis and evolutionary inference of Galegeae. Genes (Basel) 14, 176. doi: 10.3390/genes14010176
Feng, Y., Gao, X. F., Zhang, J. Y., Jiang, L. S., Li, X., Deng, H. N., et al. (2022). Complete chloroplast genomes provide insights into evolution and phylogeny of Campylotropis (Fabaceae). Front. Plant Sci. 13, 895543. doi: 10.3389/fpls.2022.895543
Gao, F., Chen, C., Arab, D. A., Du, Z., He, Y., and Ho, S. Y. W. (2019). EasyCodeML: a visual tool for analysis of selection using CodeML. Ecol. Evol. 9, 3891–3898. doi: 10.1002/ece3.5015
Greiner, S., Lehwark, P., and Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Guisinger, M. M., Kuehl, J. V., Boore, J. L., and Jansen, R. K. (2011). Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol. Biol. Evol. 28, 583–600. doi: 10.1093/molbev/msq229
Hahn, A., Vonck, J., Mills, D. J., Meier, T., and Kühlbrandt, W. (2018). Structure, mechanism, and regulation of the chloroplast ATP synthase. Science 360, eaat4318. doi: 10.1126/science.aat4318
Hong, Z., Wu, Z., Zhao, K., Yang, Z., Zhang, N., Guo, J., et al. (2020). Comparative analyses of five complete chloroplast genomes from the genus Pterocarpus (Fabacaeae). Int. J. Mol. Sci. 21, 3758. doi: 10.3390/ijms21113758
Jansen, R. K., Wojciechowski, M. F., Sanniyasi, E., Lee, S. B., and Daniell, H. (2008). Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol. Phylogenet. Evol. 48, 1204–1217. doi: 10.1016/j.ympev.2008.06.013
Jin, D. M., Wicke, S., Gan, L., Yang, J. B., Jin, J. J., and Yi, T. S. (2020). The loss of the inverted repeat in the Putranjivoid clade of Malpighiales. Front. Plant Sci. 11, 942. doi: 10.3389/fpls.2020.00942
Kaila, T., Chaduvla, P. K., Rawal, H. C., Saxena, S., Tyagi, A., Mithra, S. V. A., et al. (2017). Chloroplast genome sequence of clusterbean (Cyamopsis tetragonoloba L.): genome structure and comparative analysis. Genes (Basel) 8, 212. doi: 10.3390/genes8090212
Kan, J., Nie, L., Wang, M., Tiwari, R., Tembrock, L. R., and Wang, J. (2024). The Mendelian pea pan-plastome: insights into genomic structure, evolutionary history, and genetic diversity of an essential food crop. Genomics Commun. 1, e004. doi: 10.48130/gcomm-0024-0004
Kapralov, M. V. and Filatov, D. A. (2007). Widespread positive selection in the photosynthetic Rubisco enzyme. BMC Evol. Biol. 7, 73. doi: 10.1186/1471-2148-7-73
Karaman, S., Suludere, Z., Pinar, M., and Aytac, Z. (2009). The leaflets micromorphology of the genus Oxytropis DC. (Leguminosae) in Turkey by SEM. Bot. Res. J. 2, 7–23.
Katoh, K. and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kholina, A. B., Kozyrenko, M. M., Artyukova, E. V., Koldaeva, M. N., Sandanov, D. V., and Selyutina, I. (2021c). Phylogenetic relationships of the species of Asian Russia of the subgenera Phacoxytropis and Tragacanthoxytropis genus Oxytropis based on the polymorphism of markers of the chloroplast and nuclear genomes. Russ. J. Genet. 57, 1042–1056. doi: 10.1134/S1022795421090052
Kholina, A. B., Kozyrenko, M. M., Artyukova, E. V., and Pozdnyakova, T. E. (2021a). Variability of chloroplast DNA in Oxytropis section Polyadena (Fabaceae) from Asian Russia: population analysis and phylogenetic relationships. Biol. Bull. 48, 16–25. doi: 10.1134/S1062359021010076
Kholina, A. B., Kozyrenko, M. M., Artyukova, E. V., and Sandanov, D. V. (2018b). Modern state of populations of endemic Oxytropis species from Baikal Siberia and their phylogenetic relationships based on chloroplast DNA markers. Russ. J. Genet. 54, 805–815. doi: 10.1134/S1022795418070050
Kholina, A. B., Kozyrenko, M. M., Artyukova, E. V., Sandanov, D. V., and Andrianova, E. A. (2016). Phylogenetic relationships of the species of Oxytropis DC. subg. Oxytropis and Phacoxytropis (Fabaceae) from Asian Russia inferred from the nucleotide sequence analysis of the intergenic spacers of the chloroplast genome. Russ. J. Genet. 52, 780–793. doi: 10.1134/S1022795416060065
Kholina, A., Kozyrenko, M., Artyukova, E., Sandanov, D., and Selyutina, I. (2021b). Genetic diversity of Oxytropis section Xerobia (Fabaceae) in one of the centres of speciation. Genetica 149, 89–101. doi: 10.1007/s10709-021-00115-9
Kholina, A., Kozyrenko, M., Artyukova, E., Sandanov, D., Selyutina, I., and Chimitov, D. (2018a). Plastid DNA variation of the endemic species Oxytropis glandulosa Turcz. (Fabaceae). Turk. J. Bot. 42, 38–50. doi: 10.3906/bot-1706-11
Kholina, A. B., Kozyrenko, M. M., Artyukova, E. V., Yakubov, V. V., Khoreva, M. G., Andrianova, E. A., et al. (2020). Phylogenetic relationships of Oxytropis section Arctobia of Northeast Asia according to sequencing of the intergenic spacers of chloroplast and ITS of nuclear genomes. Russ. J. Genet. 56, 1424–1434. doi: 10.1134/S1022795420120091
Kholina, A., Kozyrenko, M., Artyukova, E., Yakubov, V., Khoreva, M., Andrianova, E., et al. (2022). The species of Oxytropis DC. of section Gloeocephala Bunge (Fabaceae) from Northeast Asia: genetic diversity and relationships based on sequencing of the intergenic spacers of cpDNA and ITS nrDNA. Genetica 150, 117–128. doi: 10.1007/s10709-022-00152-y
Kindgren, P. and Strand, A. (2015). Chloroplast transcription, untangling the Gordian Knot. New Phytol. 206, 889–891. doi: 10.1111/nph.13388
Kode, V., Mudd, E. A., Iamtham, S., and Day, A. (2005). The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 44, 237–244. doi: 10.1111/j.1365-313X.2005.02533.x
Kozyrenko, M. M., Kholina, A. B., Artyukova, E. V., Koldaeva, M. N., Yakubov, V. V., and Prokopenko, S. V. (2020). Molecular phylogenetic analysis of the endemic Far Eastern closely related Oxytropis species of section Orobia (Fabaceae). Russ. J. Genet. 56, 429–440. doi: 10.1134/S1022795420040043
Kulshreshtha, S., Creamer, R., and Sterling, T. M. (2004). Phylogenetic relationships among New Mexico Astragalus mollissimus varieties and Oxytropis species by restriction fragment analysis. Weed Sci. 52, 984–988. doi: 10.1614/WS-03-143R1
Kuroda, H. and Maliga, P. (2003). The plastid clpP1 protease gene is essential for plant development. Nature 425, 86–89. doi: 10.1038/nature01909
Lan, Z. Q., Zheng, W., Talavera, A., Nie, Z. L., Liu, J., Johnson, G., et al. (2024). Comparative and phylogenetic analyses of plastid genomes of the medicinally important genus Alisma (Alismataceae). Front. Plant Sci. 15, 1415253. doi: 10.3389/fpls.2024.1415253
Landi, M. and Guidi, L. (2022). Effects of abiotic stress on photosystem II proteins. Photosynthetica 61, 148–156. doi: 10.32615/ps.2022.043
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., and Calcott, B. (2017). PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. doi: 10.1093/molbev/msw260
Lee, C., Choi, I. S., Cardoso, D., de Lima, H. C., de Queiroz, L. P., Wojciechowski, M. F., et al. (2021). The chicken or the egg? Plastome evolution and an independent loss of the inverted repeat in papilionoid legumes. Plant J. 107, 861–875. doi: 10.1111/tpj.15351
Li, Q. Q., Khasbagan, Zhang, Z. P., Wen, J., and Yu, Y. (2024a). Plastid phylogenomics of the tribe Potentilleae (Rosaceae). Mol. Phylogenet. Evol. 190, 107961. doi: 10.1016/j.ympev.2023.107961
Li, C., Liu, Y., Lin, F., Zheng, Y., and Huang, P. (2022). Characterization of the complete chloroplast genome sequences of six Dalbergia species and its comparative analysis in the subfamily of Papilionoideae (Fabaceae). PeerJ 10, e13570. doi: 10.7717/peerj.13570
Li, P. C. and Ni, C. C. (1985). “Oxytropis DC,” in Flora Xizangica, vol. 2. Comprehensive Scientific Expedition Team of Tibetan Plateau, Chinese Academy of Sciences (Science Press, Beijing), 846–871.
Li, Q. Q., Zhang, Z. P., Aogan, and Wen, J. (2024b). Comparative chloroplast genomes of Argentina species: genome evolution and phylogenomic implications. Front. Plant Sci. 15, 1349358. doi: 10.3389/fpls.2024.1349358
Li, C., Zhao, Y., Xu, Z., Yang, G., Peng, J., and Peng, X. (2020). Initial characterization of the chloroplast genome of Vicia sepium, an important wild resource plant, and related inferences about its evolution. Front. Genet. 11, 73. doi: 10.3389/fgene.2020.00073
Liu, H., Su, Z., Yu, S., Liu, J., Yin, X., Zhang, G., et al. (2019). Genome comparison reveals mutation hotspots in the chloroplast genome and phylogenetic relationships of Ormosia species. BioMed. Res. Int. 2019, 7265030. doi: 10.1155/2019/7265030
Liu, S., Wei, Y. L., Si, W., Qu, W. R., Yang, T. G., Wu, Z. H., et al. (2021). Complete chloroplast genome sequence of Oxytropis glabra (Leguminosae). Mitochondrial DNA Part B 6, 2478–2479. doi: 10.1080/23802359.2021.1914228
LPWG (The Legume Phylogeny Working Group) (2013). Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species-rich clades. Taxon 62, 217–248. doi: 10.12705/622.8
LPWG (The Legume Phylogeny Working Group) (2017). A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon 66, 44–77. doi: 10.12705/661.3
Madoka, Y., Tomizawa, K. I., Mizoi, J., Nishida, I., Nagano, Y., and Sasaki, Y. (2002). Chloroplast transformation with modified accD operon increases acetyl-CoA carboxylase and causes extension of leaf longevity and increase in seed yield in tobacco. Plant Cell Physiol. 43, 1518–1525. doi: 10.1093/pcp/pcf172
Moghaddam, M., Kazempour Osaloo, S., Hosseiny, H., and Azimi, F. (2016). Phylogeny and divergence times of the Coluteoid clade with special reference to Colutea (Fabaceae) inferred from nrDNA ITS and two cpDNAs, matK and rpl32-trnL(UAG) sequences data. Plant Biosyst. 151, 1082–1093. doi: 10.1080/11263504.2016.1244120
Moghaddam, M., Wojciechowski., M. F., and Kazempour-Osaloo, S. (2023). Characterization and comparative analysis of the complete plastid genomes of four Astragalus species. PloS One 18, e0286083. doi: 10.1371/journal.pone.0286083
Mower, J. P. and Vickrey, T. L. (2018). Structural diversity among plastid genomes of land plants. Adv. Bot. Res. 85, 263–292. doi: 10.1016/bs.abr.2017.11.013
Neuhaus, H. E. and Emes, M. J. (2000). Nonphotosynthetic metabolism in plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 111–140. doi: 10.1146/annurev.arplant.51.1.111
Nielsen, R. and Yang, Z. (1998). Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148, 929–936. doi: 10.1093/genetics/148.3.929
Niu, Y., Wang, X. M., Zhang, Z. P., and Li, Q. Q. (2024). The complete chloroplast genome of Oxytropis myriophylla (Fabaceae). J. Inner Mongolia Norm. Univ. (Nat. Sci. Edn.) 53, 284–288 + 297. doi: 10.3969/j.issn.1001-8735.2024.03.009
Palmer, J. D. (1985). Comparative organization of chloroplast genomes. Annu. Rev. Genet. 19, 325–354. doi: 10.1146/annurev.ge.19.120185.001545
Rambaut, A. (2018). FigTree version 1.4.4. Available online at: https://github.com/rambaut/figtree/releases (Accessed November 03, 2023).
Rawsthorne, S. (2002). Carbon flux and fatty acid synthesis in plants. Prog. Lipid Res. 41, 182–196. doi: 10.1016/S0163-7827(01)00023-6
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Ruhlman, T. A., Zhang, J., Blazier, J. C., Sabir, J. S. M., and Jansen, R. K. (2017). Recombination-dependent replication and gene conversion homogenize repeat sequences and diversify plastid genome structure. Am. J. Bot. 104, 559–572. doi: 10.3732/ajb.1600453
Sandanov, D. V., Kholina, A. B., Kozyrenko, M. M., Artyukova, E. V., and Wang, Z. (2023). Genetic diversity of Oxytropis species from the center of the genus origin: insight from molecular studies. Diversity 15, 244. doi: 10.3390/d15020244
Sanderson, M. J., Copetti, D., Burquez, A., Bustamante, E., Charboneau, J. L. M., Eguiarte, L. E., et al. (2015). Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): loss of the ndh gene suite and inverted repeat. Am. J. Bot. 102, 1115–1127. doi: 10.3732/ajb.1500184
Schmid, L. M., Manavski, N., Chi, W., and Meurer, J. (2024). Chloroplast ribosome biogenesis factors. Plant Cell Physiol. 65, 516–536. doi: 10.1093/pcp/pcad082
Shavvon, R. S., Osaloo, S. K., Maassoumi, A. A., Moharrek, F., Erkul, S. K., Lemmon, A. R., et al. (2017). Increasing phylogenetic support for explosively radiating taxa: the promise of high-throughput sequencing for Oxytropis (Fabaceae). J. Syst. Evol. 55, 385–404. doi: 10.1111/jse.12269
Shi, L., Chen, H., Jiang, M., Wang, L., Wu, X., Huang, L., et al. (2019). CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47, W65–W73. doi: 10.1093/nar/gkz345
Shikanai, T., Shimizu, K., Ueda, K., Nishimura, Y., Kuroiwa, T., and Hashimoto, T. (2001). The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 42, 264–273. doi: 10.1093/pcp/pce031
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Su, C., Duan, L., Liu, P., Liu, J., Chang, Z., and Wen, J. (2021). Chloroplast phylogenomics and character evolution of eastern Asian Astragalus (Leguminosae): tackling the phylogenetic structure of the largest genus of flowering plants in Asia. Mol. Phylogenet. Evol. 156, 107025. doi: 10.1016/j.ympev.2020.107025
Su, C., Liu, P. L., Chang, Z. ,. Y., and Wen, J. (2019). The complete chloroplast genome sequence of Oxytropis bicolor Bunge (Fabaceae). Mitochondrial DNA Part B 4, 3762–3763. doi: 10.1080/23802359.2019.1682479
Tavares, J. C. C., Achakkagari, S. R., Archambault, A., and Strömvik, M. V. (2022). The plastome of Arctic Oxytropis arctobia (Fabaceae) is significantly different from that of O. splendens and other related species. Genome 65, 301–313. doi: 10.1139/gen-2021-0059
Tekpinar, A. D., Erkul, S. K., Aytaç, Z., and Kaya, Z. (2016a). Phylogenetic relationships between Oxytropis DC. and Astragalus L. species native to an Old World diversity center inferred from nuclear ribosomal ITS and plastid matK gene sequences. Turk. J. Biol. 40, 250–263. doi: 10.3906/biy-1502-5
Tekpinar, A., Erkul, S. K., Aytaç, Z., and Kaya, Z. (2016b). Phylogenetic relationships among native Oxytropis species in Turkey using the trnL intron, trnL-F IGS, and trnV intron cpDNA regions. Turk. J. Biol. 40, 472–479. doi: 10.3906/bot-1506-45
Tian, C., Li, X., Wu, Z., Li, Z., Hou, X., and Li, F. Y. (2021). Characterization and comparative analysis of complete chloroplast genomes of three species from the genus Astragalus (Leguminosae). Front. Genet. 12, 705482. doi: 10.3389/fgene.2021.705482
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11. doi: 10.1093/nar/gkx391
Vasil’chenko, I. T., Fedchenko, B. A., and Shishkin, B. K. (1948). “Oxytropis DC,” in Flora of the USSR, vol. 13 . Eds. Shishkin, B. K. and Bobrov, E. G. (Izdatel’stvo Akademii Nauk SSSR, Moskva-Leningrad), 1–229.
Wang, J., Kan, S., Liao, X., Zhou, J., Tembrock, L. R., Daniell, H., et al. (2024). Plant organellar genomes: much done, much more to do. Trends Plant Sci. 29, 754–769. doi: 10.1016/j.tplants.2023.12.014
Wang, Y., Li, E., Sun, J., Zhang, Z., and Dong, W. (2025). Phylogenetic diversity and interspecies competition shaped species diversity in adaptive radiated Ligustrum (Oleaceae). J. Syst. Evol. 63, 229–244. doi: 10.1111/jse.13117
Wang, B., Tian, Z., Lang, S., Kong, Q., Liu, X., Chen, Y., et al. (2024). The genus Oxytropis DC: application, phytochemistry, pharmacology, and toxicity. J. Pharm. Pharmacol. 76, 1079–1114. doi: 10.1093/jpp/rgae048
Welsh, S. L. (2023). “Oxytropis DC,” in Flora of North America North of Mexico, vol. 11. Flora of North America Editorial Committee (Oxford University Press, New York and Oxford), 557–583.
Wicke, S., Schneeweiss, G. M., depamphilis, C. W., Müller, K. F., and Quandt, D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297. doi: 10.1007/s11103-011-9762-4
Wilson, R. H. and Hayer-Hartl, M. (2018). Complex chaperone dependence of Rubisco biogenesis. Biochemistry 57, 3210–3216. doi: 10.1021/acs.biochem.8b00132
Wojciechowski, M. F., Lavin, M., and Sanderson, M. J. (2004). A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am. J. Bot. 91, 1846–1862. doi: 10.3732/ajb.91.11.1846
Yamamoto, H., Cheuk, A., Shearman, J., Nixon, P. J., Meier, T., and Shikanai, T. (2023). Impact of engineering the ATP synthase rotor ring on photosynthesis in tobacco chloroplasts. Plant Physiol. 192, 1221–1233. doi: 10.1093/plphys/kiad043
Yan, X. L., Kan, S. L., Wang, M. X., Li, Y. Y., Tembrock, L. R., He, W. C., et al. (2024). Genetic diversity and evolution of the plastome in allotetraploid cotton (Gossypium spp.). J. Syst. Evol. 62, 1118–1136. doi: 10.1111/jse.13070
Yan, H. F., Liu, T. J., Yuan, X., Xu, Y., Zhang, S. Y., Hao, G., et al. (2025). Revisiting the phylogeny of Primulaceae s.l. using whole plastid genomes: highlighting phylogenetic conflicts and their implications. J. Syst. Evol. 63, 788–802. doi: 10.1111/jse.13154
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yang, Z., Wong, W. S. W., and Nielsen, R. (2005). Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118. doi: 10.1093/molbev/msi097
Zhang, Z. W. (1998). “Oxytropis DC,” in Flora Reipublicae Polularis Sinicae, vol. 42. Editorial Committee of Flora Reipublicae Popularis Sinicae, Chinese Academy of Sciences (Science Press, Beijing), 1–146.
Zhang, R., Wang, Y. H., Jin, J. J., Stull, G. W., Bruneau, A., Cardoso, D., et al. (2020). Exploration of plastid phylogenomic conflict yields new insights into the deep relationships of Leguminosae. Syst. Biol. 69, 613–622. doi: 10.1093/sysbio/syaa013
Zhang, G. J., Zhang, Z. P., and Li, Q. Q. (2022). Comparative analysis of chloroplast genomes of Sanguisorba species and insights into phylogenetic implications and molecular dating. Nord. J. Bot. 2022, e03719. doi: 10.1111/njb.03719
Zhao, X., Hou, Q., Du, M., Zhang, H., Jia, L., Zhang, Z., et al. (2022). Micromorphological leaf epidermal traits as potential taxonomic markers for infrageneric classification of Oxytropis (Fabaceae). PhytoKeys 201, 51–76. doi: 10.3897/phytokeys.201.85154
Zhao, X., Liu, Y., Li, J., Zhang, H., Jia, L., Hou, Q., et al. (2023). Numerical analyses of seed morphology and its taxonomic significance in the genus Oxytropis DC. (Fabaceae) from northwestern China. PhytoKeys 222, 49–67. doi: 10.3897/phytokeys.222.96990
Zhao, Y., Zhang, R., Jiang, K. W., Qi, J., Hu, Y., Guo, J., et al. (2021). Nuclear phylotranscriptomics and phylogenomics support numerous polyploidization events and hypotheses for the evolution of rhizobial nitrogen-fixing symbiosis in Fabaceae. Mol. Plant 14, 748–773. doi: 10.1016/j.molp.2021.02.006
Zhao, Y. Z., Zhao, L. Q., and Cao, R. (Eds.) (2019). Flora Intramongolica. 3rd ed Vol. 3 (Huhhot: Typis Intramongolicae Popularis), 33–67.
Zhelyazkova, P., Sharma, C. M., Forstner, K. U., Liere, K., Vogel, J., and Borner, T. (2012). The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24, 123–136. doi: 10.1105/tpc.111.089441
Zhou, Y., Shang, X. H., Xiao, L., Wu, Z. D., Cao, S., and Yan, H. B. (2023). Comparative plastomes of Pueraria montana var. lobata (Leguminosae: Phaseoleae) and closely related taxa: insights into phylogenomic implications and evolutionary divergence. BMC Genomics 24, 299. doi: 10.1186/s12864-023-09356-8
Keywords: Oxytropis, adaptive evolution, chloroplast genome, comparative analyses, phylogeny
Citation: Li Q-Q, Niu Y, Zhang Z-P, Wen J and Liao C-Y (2025) Comparative chloroplast genome analyses of Oxytropis DC. species: new insights into genome evolution and phylogenomic implications. Front. Plant Sci. 16:1645582. doi: 10.3389/fpls.2025.1645582
Received: 12 June 2025; Accepted: 08 August 2025;
Published: 28 August 2025.
Edited by:
Xiaohua Jin, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Zhiqiang Wu, Chinese Academy of Agricultural Sciences, ChinaEnze Li, Beijing Forestry University, China
Liang-Liang Yue, Southwest Forestry University, China
Copyright © 2025 Li, Niu, Zhang, Wen and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qin-Qin Li, bGlxcUBpbW51LmVkdS5jbg==; Chen-Yang Liao, Y2hlbnlhbmdsaWFvQHNjdS5lZHUuY24=
†These authors have contributed equally to this work
 Qin-Qin Li
Qin-Qin Li Yan Niu1,2†
Yan Niu1,2† Jun Wen
Jun Wen